ABSTRACT
Epigenetic clocks quantify regular changes in DNA methylation that occur with age, or in relation to biomarkers of ageing, and are strong predictors of morbidity and mortality. Here, we assess whether measures of fetal nutrition and growth that predict adult chronic disease also predict accelerated biological ageing in young adulthood using a suite of commonly used epigenetic clocks. Data come from the Cebu Longitudinal Health and Nutrition Survey (CLHNS), a long-running cohort followed since birth in metropolitan Cebu, Philippines. Past work has shown that birth weight (BW) and the mother’s arm fat during pregnancy (a measure of pregnancy energy status) relate inversely to health outcomes in the CLHNS but primarily in males. Genome-wide DNA methylation was assessed in whole blood using the Infinium EPIC array. Participants included males (n=895) and females (n=803) measured in 2005 (20.8–22.5 years). Clocks included the Hannum and Horvath clocks trained on chronological age, the DNAmPhenoAge and DNAmGrimAge clocks trained on clinical biomarkers, the Dunedin pace of ageing (DunedinPACE) clock trained on longitudinal changes in ageing biomarkers, and the DNAmTL clock trained on leukocyte telomere length. In males, lower BW predicted advanced biological ageing using the Hannum, DNAmPhenoAge, DunedinPoAm, and DNAmTL clocks. In contrast, BW did not predict any clock in female participants. Participants’ mothers’ pregnancy arm fat only predicted DNAmTL in males. These findings suggest that epigenetic clocks are a useful tool for gauging long-term outcomes predicted by fetal growth, and add to existing evidence in the CLHNS for sex differences in these relationships.
KEYWORDS: DOHaD, ageing, pregnancy, epigenetic clocks, senescence
Introduction
In humans, being born small, as a result of fetal growth restriction or preterm birth increases risk for adverse adult health outcomes including hypertension, stroke, diabetes, and cardiovascular disease mortality [1–3]. Lower birth weight also appears to accelerate degenerative, senescent decline, and increase all-cause mortality, a phenomenon often referred to as ‘biological aging’ [4]. Animal model experiments have shown that restricting fetal access to nutrients during gestation can replicate many of these outcomes, heightening interest in the role of prenatal nutrition and growth as influences on lifelong health [5,6].
The recent development of ‘epigenetic clocks’ has opened up new opportunities to explore the long-term impacts of early environments on the pace of ageing in humans [7]. DNA methylation, or the attachment of a methyl group to DNA, is important for cellular differentiation and gene regulatory control. DNA methylation at a subset of loci changes predictably with age, allowing prediction of chronological age [8,9]. However, deviations between an individual’s age predicted using epigenetic clocks and their true chronological age may provide insights into biological ageing. An individual that looks epigenetically older than their chronological age, defined as epigenetic age acceleration (EAA), is at higher risk for a range of chronic diseases and all-cause mortality [8,9]. More recently, epigenetic clocks (e.g., DNAmGrimAge, DNAmPhenoAge) have been trained on suites of clinical measures linked to chronic and metabolic disease risk that are strongly tied to mortality [10,11], or on leukocyte telomere length, a measure of replicative senescence (DNAmTL) [12]. Finally, a new clock has been trained using the longitudinal pace of change in a suite of age-related biomarkers to track the rate of ageing through time (DunedinPACE) [13].
Because epigenetic clocks predict outcomes like life expectancy, they provide a potentially powerful tool to link early life experiences, such as prenatal nutrition and growth, to measures that are in turn predictive of late life health and biological ageing. However, to date, few studies have explored relationships between markers of prenatal nutrition and growth and adult epigenetic clock measures. Some [14], but not all [15], studies have found evidence that epigenetic age acceleration relates positively to developmental or maturational tempo during adolescence, indicating that it likely serves as a developmental marker prior to the transition to adulthood. Using data from the longitudinal ALSPAC Cohort study in the UK, Simpkin [16] found that birth weight was positively related to EAA using the Horvath clock measured at age 7, but inversely related to EAA at age 17 years, supporting the idea that the biological meaning of epigenetic age acceleration shifts during late adolescence or with the transition to adulthood. In one of the few studies to link birth weight to adult epigenetic age acceleration, Madden [17] found that lower birth weights predicted age acceleration in adults using the DNAmGrimAge clock, along with predicting shorter telomeres using a DNA methylation clock trained on telomere length [11] but was unrelated to several others (the Horvath, Hannum, and DNAmPhenoAge clocks). These studies provide mixed support for the utility of epigenetic clocks as biomarkers of intermediate pathways linking early life experiences to long-term health outcomes and longevity.
Here, we assess whether measures reflecting the nutritional and growth environment in utero predict several measures of EAA in early adulthood. Data come from the Cebu Longitudinal Health and Nutrition Survey (CLHNS), which has followed a large birth cohort living in metropolitan Cebu, Philippines, since prior to their own births in 1983–1984 [18]. Follow-up interviews and blood samples were obtained in 2005, when participants were 20–22 years of age, allowing assessment of the role of early life nutrition and growth as predictors of EAA measured in early adulthood. After measuring genome-wide DNA methylation using the Infinium Methylation EPIC array, we selected epigenetic clocks trained on biological and health outcomes that reflect multiple dimensions of biological ageing. For comparability with prior work, we use linear regression to evaluate the links between birth weight, adjusted for gestational age, and age acceleration in five clocks that have previously been used to gauge the long-term impacts of prenatal nutrition and birth size [17]: Horvath’s clock, Hannum’s clock, DNAmPhenoAge, DNAmGrimAge, and a clock trained on telomere length (DNAmTL). We also include a recently described clock that gauges the longitudinal pace of ageing (the Dunedin pace of ageing or DunedinPACE clock). Finally, we consider relationships between epigenetic clock measures and the mother’s energy status during pregnancy (arm fat area), which has previously been shown to predict multiple cardiometabolic risk factors among offspring in the CLHNS [19,20].
Methods
Study population
Data come from the CLHNS, a longitudinal survey of 3,080 singletons whose mothers were recruited during pregnancy between 1983 and 1984 in metropolitan Cebu, Philippines [18,21]. In 2005, participants were visited in-home for anthropometric and questionnaire assessments, along with a collection of fasting venous blood samples for biomarker and genetic analyses. This research was conducted under conditions of written informed consent, with the approval of the Institutional Review Boards of the University of North Carolina at Chapel Hill (1983 and 2005 surveys and sample collection) and Northwestern University (Evanston, Illinois; DNA methylation analysis).
Pregnancy and birth measures. Birth weight was measured by birth attendants or trained interviewers at birth. Gestational age at birth was estimated from the date of the mother’s last menstrual period (LMP) recorded at the baseline survey. Trained nurses performed Ballard clinical assessments to determine if pregnancy complications occurred or if the infant’s birth weight was <2.5 kg.
Other covariates: In 1983, the participants’ mothers, and in 2005, the participants themselves, answered questions about assets, education, income, and educational attainment. Interviewers measured height, weight, triceps skinfold thickness, and mid-upper arm circumference during pregnancy using standard techniques (Lohman et al. 1998). Because we have previously reported evidence that EAA is altered during pregnancy [22], we adjusted for reproductive status at the time of blood draw. Women were asked about their current pregnancy status; we used subsequent reproductive histories in 2007 and 2009 to confirm 2005 pregnancy status; in the event that women were pregnant but were not yet aware (or did not report that they were pregnant) they were recoded as pregnant. We adjusted for each individual’s adult smoking (defined as a dichotomous variable, indicating currently smoking 5 or more sticks daily), education (highest grade completed), household income (pesos), and the body mass index (calculated as weight in kg/height in m2). Among women who were pregnant during the 2005 interview (when DNAm was measured), we used non-pregnant BMIs obtained during the next available survey (2007 or 2009). CLHNS women on average gain weight but maintain a relatively stable within-population rank for BMI as they age (e.g., r > 0.8 for 2005 BMI vs. 2009 BMI). Following our prior work with the sample [23], for each female we calculated within-population BMI z-scores for each survey year and used the most recently measured, non-pregnant estimate in models (raw BMI data from 2005 were used in male models). Full regression models with all covariates are reported in the supplemental materials.
DNA methylation data. Blood samples were obtained during the 2005 survey for DNA extraction, which was used for DNA methylation analysis. Briefly, genomic DNA underwent bisulphite conversion (EZ DNA Methylation kit, Zymo Research), converting CpG methylation information into sequence information by converting unmethylated cytosines into uracils while leaving methylated cytosines intact. Subsequently, genome-wide DNA methylation was assessed with the Infinium Methylation EPIC array following manufacturer’s instructions (Illumina, Inc.). Samples were randomized on runs by baseline neighbourhood (capturing dimensions of socioeconomic status and urban-periurban-rural variation) to minimize any batch effects. Quality control involved first confirming participant sex and replicate status. This was followed by quantile normalization using lumi on all probes including SNP-associated and XY multiple binding probes [24]. To maximize the number of sites available for the epigenetic age calculator, probes with detection p-values above 0.01 were coded as NA for poor performing samples only, and were otherwise retained. A subset of 30,084 probes from the EPIC array (including rows with Illumina IDs only if not found on the EPIC array) were then constructed and uploaded with accompanying age, sex, and tissue information to the online epigenetic clock calculator (http://labs.genetics.ucla.edu/horvath/dnamage/) using the ‘Advanced analysis’ option. The Belsky PACE clock was calculated using the DunedinPACE R package (https://github.com/danbelsky/DunedinPACE). Immune cell composition was imputed from the DNAm data using previously described methods [25–27].
Data Availability Statement. The analysis plan and dataset used for these analyses are publicly available through the Center for Open Science OSF portal at the following address: https://osf.io/nu6mf/
Sample selection
Of the 3080 original liveborn singletons in the birth cohort, 1888 were interviewed in the 2005 survey (at ages 20–22), and of these 1698 provided blood samples and had valid DNAm data and no other missing variables. Compared to the singleton liveborns who were either lost to follow-up or who were otherwise not included in the final analysis sample (n = 1382), the mother’s baseline height (150.8 vs. 150.7 cm; p < 0.69), 1983 household income (280 vs. 288 pesos; p < 0.677) and baseline education levels in 1983 (7.45 vs. 7.70 years completed, p < 0.072) were similar. The analysis sample had birth weights that were slightly heavier on average than those lost to follow-up (3011 vs. 2962 grams; p < 0.0021) but lived in neighbourhoods with lower baseline urbanicity scores (29.6 vs. 31.8; p < 0.00001).
Statistical analysis
All statistical analyses were conducted using Stata 16.0 (College Station, TX). We first report unadjusted means and standard deviations (or % for count variables) for the sample, before running a series of regression models linking prenatal nutrition measures to epigenetic clocks. In the CLHNS sample, there is precedent for sex differences in the short- and long-term health correlates of early environments, with males generally showing evidence for stronger long-term health effects of markers like birth weight [19,20,28,29]. In the current data, there were significant or borderline significant sex * birth weight interactions in the models predicting DNAmPhenoAge (interaction p < 0.025) and Hannum’s clock upweighted for cell composition (EEAA; interaction p < 0.092), with interactions in a similar direction but not significant for several other outcomes. To account for differences in relationship by sex, and also to allow adjustment for current pregnancy status among females, separate linear regressions were run for males and females linking 1) birth weight (adjusted for gestational age at delivery), 2) maternal arm fat area during pregnancy, and 3) both birth weight and maternal arm fat considered simultaneously, to each clock, adjusting for 2005 survey data on current household income, highest grade completed, smoking status, BMI, and (in females) current pregnancy status.
Results
As expected, there were significant sex differences in anthropometrics (Table 1). Compared to females, males showed more advanced epigenetic measures of ageing for Horvath, Hannum, DNAmGrimAge and DNAmTL clocks, with females showing higher DNAmPhenoAge and DunedinPACE (for reference, means and standard deviations for the epigenetic clock acceleration values are presented in Supplemental Table 1).
Table 1.
Sample Characteristics.
| Male (n = 895) | SE | Female (n = 803) | SE | p-value | |
|---|---|---|---|---|---|
| Height (cm) | 163.09 | 5.84 | 151.17 | 5.42 | 0.00001 |
| Weight (kg) | 56.16 | 9.45 | 46.82 | 8.20 | 0.00001 |
| BMI (kg/m2)a | 21.07 | 3.09 | 20.42 | 3.25 | 0.00001 |
| Pregnant (%) | - | - | 12.0% | - | - |
| Years of schooling | 9.92 | 3.19 | 11.13 | 2.77 | 0.00001 |
| Household incomes (pesos) | 572 | 885 | 643 | 1621 | 0.256 |
| Smoking (daily 5+ sticks) | 2.5% | 4.0% | 0.073 | ||
| Birth weight (g) | 3031 | 431 | 2988 | 416 | 0.035 |
| Gest. Age at birth (weeks) | 38.7 | 2.1 | 38.9 | 2.1 | 0.027 |
| Mother’s preg. arm fat area (cm2) | 14.8 | 5.5 | 14.7 | 5.4 | 0.751 |
| Age (years) | 21.67 | 0.34 | 21.68 | 0.36 | 0.443 |
| Epigenetic clocks | |||||
| Horvath (years) | 35.04 | 3.83 | 33.65 | 3.92 | 0.00001 |
| Hannum (years) | 37.24 | 3.48 | 36.61 | 3.41 | 0.0002 |
| DNAmPhenoAge (years) | 22.97 | 4.00 | 23.76 | 4.35 | 0.0001 |
| DNAmGrimAge (years) | 43.86 | 2.56 | 42.40 | 2.35 | 0.00001 |
| DunedinPACE (bio year/chronol. year |
1.07 | 0.10 | 1.11 | 0.12 | 0.00001 |
| DNAmTL (kilobase) | 7.39 | 0.14 | 7.47 | 0.14 | 0.00001 |
aDescriptive statistics for BMI for females is limited to the 707 women who were not pregnant during the 2005 blood draw, and the T-test evaluating male-female differences in BMI was also calculated among this subset. Please see methods for discussion of how non-pregnant BMIs used in regression models were calculated.
We next ran a series of regression models linking the participants’ birth weights (adjusted for gestational age at delivery) and their mother’s energy status while pregnant with them (maternal arm fat area, MAFA), to adult epigenetic clock measures. In males (Table 2 and Figure 1; full models are reported in supplemental Tables 2–9), birth weight was significantly inversely related to Hannum EAA, DNAmPhenoAge, DunedinPACE, and positively related to DNAmTL, all consistent with evidence for more rapid biological ageing among individuals who were lighter at birth. DNAmGrimAge was inversely related to birth but not significantly (p < 0.117 in BW only model) and was borderline inversely related to birth weight (p < 0.089) in the model also adjusting for mother’s pregnancy arm fat area. The mother’s arm fat area is related to DNAmTL in a curvilinear relationship, with shorter telomeres (consistent with more rapid biological ageing) among both lower and higher birth weight individuals.
Table 2.
Linear regression models linking male epigenetic clocks at 20–22 years of age to birth weight (adjusted for gestational age) and their mother’s arm fat area (MAFA) during pregnancya.
| Model 1 | Model 2 | Full model | |||
|---|---|---|---|---|---|
| Horvath | |||||
| Birth weight | 0.012 (0.306) | 0.029 (0.308) | |||
| MAFA | −0.011 (0.024) | −0.012 (0.024) | |||
| Model adj. R2 | 0.000 | 0.000 | 0.000 | ||
| Hannum | |||||
| Birth weight | −0.660 (0.277)** | −0.620 (0.279)** | |||
| MAFA | −0.033 (0.021) | −0.027 (0.022) | |||
| Model adj. R2 | 0.019 | 0.015 | 0.019 | ||
| DNAmTL | |||||
| Birth weight | 0.025 (0.011)** | 0.025 (0.011)** | |||
| MAFA | 0.012 (0.003)**** −0.0003 |
0.012 (0.003)**** −0.0003 |
|||
| MAFA[2] | (0.00008)**** | (0.00008)**** | |||
| Model adj. R2 | 0.048 | 0.057 | 0.061 | ||
| DNAmPhenoAge | |||||
| Birth weight | −0.907 (0.318)*** | −0.902 (0.321)*** | |||
| MAFA | −0.011 (0.025) | −0.003 (0.025) | |||
| Model adj. R2 | 0.018 | 0.009 | 0.017 | ||
| DNAmGrimAge | |||||
| Birth weight | −0.314 (0.200) | −0.343 (0.202)* | |||
| MAFA | 0.017 (0.015) | 0.02 (0.016) | |||
| Model adj. R2 | 0.057 | 0.056 | 0.058 | ||
| DunedinPACE | |||||
| Birth weight | −0.019 (0.008)** | −0.019 (0.008)** | |||
| MAFA | 0.0004 (0.0006) | 0.0005 (0.0006) | |||
| Model adj. R2 | 0.031 | 0.026 | 0.031 | ||
aregression coefficients (SE). All models adjust for adult BMI, highest grade completed, household income, and daily smoking (see Supplemental Tables 2–9 for full models). *p<0.1 **p<0.05 ***p<0.01 ****p<0.0002. Because DNAmTL related to MAFA in a curvilinear fashion in males, a quadratic term (MAFA [2]) was included in models predicting DNAmTL (p < 0.0007 for joint F test for MAFA and MAFA [2] in Models 2 and 3)
Figure 1.
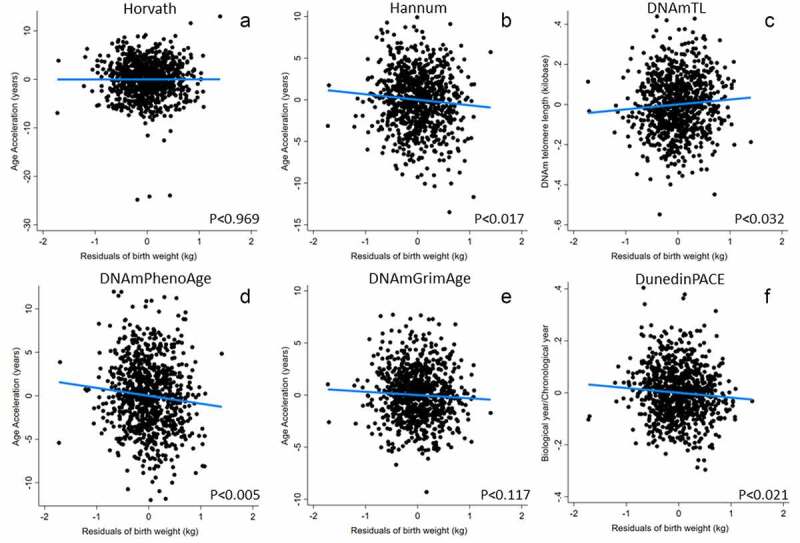
Epigenetic clocks predicted by birth weight in males. Birth weight was adjusted for gestational age before inclusion in models. Models adjust for adult BMI, highest grade completed, household income, and daily smoking (from Table 2, Model 1). Note that, unlike all other measures, higher values of DNAmTL (telomere length) imply slower biological ageing.
Hannum’s clock (which was significantly and inversely related to birth weight) was trained using blood leukocytes. Because differences in immune cell composition may underlie some of the variations in the Hannum epigenetic age prediction, we investigated whether statistically controlling for this variation using bioinformatically imputed immune cell composition affected the link between birth weight and the Hannum clock. Consistent with the effect of immune cell composition, Hannum intrinsic epigenetic age acceleration (IEAA-Hannum, derived from the residuals of a linear regression of the Hannum epigenetic age estimate on chronological age and measures of blood cell count), was no longer related to birth weight. In contrast, Hannum extrinsic epigenetic age acceleration (Hannum-EEAA, a measure that incorporates age-associated changes in immune cell composition as well as age to the clock estimate and better reflects immune ageing) was more strongly related than the base Hannum clock. To clarify these findings, we further ran post hoc models assessing relationships between birth weight and DNAm-imputed cell composition (Supplemental Table 10). In males, birth weight was positively related to the proportion of CD8T, CD4T, and borderline positively related to B-Cells, was inversely related to granulocytes, and unrelated to monocytes and natural killer cells. In females, immune cell composition measures were strongly related to current pregnancy status, but were unrelated to birth weight.
In females, no clock was significantly related to either birth weight adjusted for gestational age or the mother’s pregnancy energy status (Table 3 and Figure 2). In addition, in models evaluating gestational age at delivery as an independent predictor of adult epigenetic clocks, gestational age only predicted the DNAmTL clock and in females only (Supplemental Tables 11–12).
Table 3.
Linear regression models linking female epigenetic clocks at 20–22 years of age to birth weight (adjusted for gestational age) and their mother’s arm fat area (MAFA) during pregnancya.
| Model 1 | Model 2 | Full model | |||
|---|---|---|---|---|---|
| Horvath | |||||
| Birth weight | 0.183 (0.350) | 0.230 (0.357) | |||
| MAFA | −0.015 (0.026) | −0.019 (0.027) | |||
| Model adj. R2 | 0.000 | 0.000 | 0.000 | ||
| Hannum | |||||
| Birth weight | −0.043 (0.297) | 0.24 (0.303) | |||
| MAFA | −0.026 (0.022) | −0.026 (0.023) | |||
| Model adj. R2 | 0.038 | 0.039 | 0.038 | ||
| DNAmTL | |||||
| Birth weight | 0.002 (0.012) | 0.002 (0.012) | |||
| MAFA | −0.0002 (0.0009) | −0.0002 (0.0009) | |||
| Model adj. R2 | 0.060 | 0.060 | 0.059 | ||
| DNAmPhenoAge | |||||
| Birth weight | 0.265 (0.376) | 0.368 (0.374) | |||
| MAFA | −0.036 (0.028) | −0.041 (0.028) | |||
| Model adj. R2 | 0.092 | 0.093 | 0.093 | ||
| DNAmGrimAge | |||||
| Birth weight | −0.283 (0.193) | −0.233 (0.196) | |||
| MAFA | −0.023 (0.015) | −0.020 (0.015) | |||
| Model adj. R2 | 0.154 | 0.155 | 0.155 | ||
| DunedinPACE | |||||
| Birth weight | −0.012 (0.01) | −0.012 (0.01) | |||
| MAFA | 0.0002 (0.0007) | 0.0003 (0.0007) | |||
| Model adj. R2 | 0.194 | 0.192 | 0.193 | ||
aregression coefficients (SE). All models adjust for adult BMI, highest grade completed, household income, current pregnancy status, and daily smoking (see Supplemental Tables 2–9 for full models).
Figure 2.
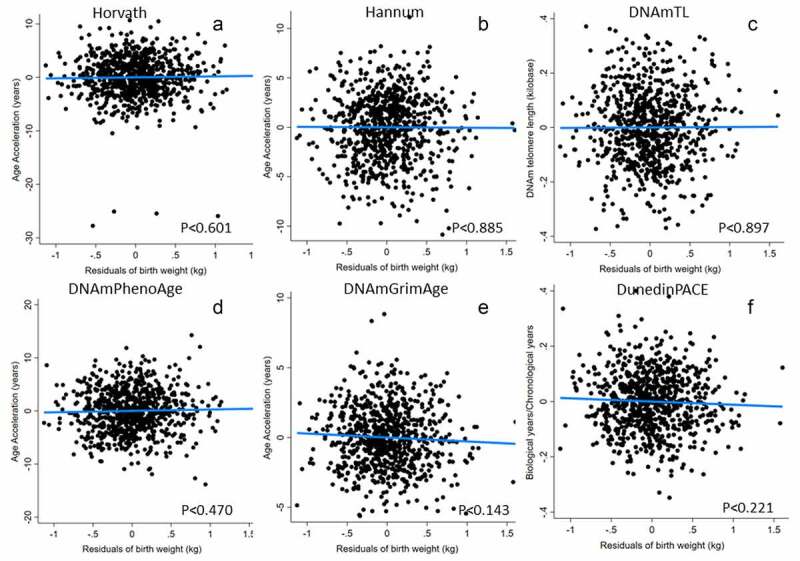
Epigenetic clocks predicted by birth weight in females. Birth weight was adjusted for gestational before inclusion in models. Models adjust for adult BMI, highest grade completed, household income, current pregnancy status and daily smoking (from Table 3, Model 1). Note that, unlike all other measures, higher values of DNAmTL (telomere length) imply slower biological ageing.
Discussion
In the CLHNS sample, measures of prenatal nutrition and growth predict male markers of accelerated biological ageing using epigenetic clocks that index several dimensions of ageing and long-term health. Among men, birth weight had a significant inverse linear relationship with Hannum EAA and DNAmPhenoAge, and the DunedinPACE clocks, a weaker inverse relationship with the DNAmGrimAge clock and was also positively related to DNAmTL. These five relationships are consistent with accelerated biological ageing or heightened long-term disease risk in relation to lower birth weight. Only the Horvath clock was clearly unrelated to either birth weight or the mother’s arm fat during pregnancy in males. In contrast, neither birth weight or the mother’s arm fat during pregnancy related to any epigenetic clock measure in females. These findings point to potential long-term implications of fetal growth and birth size among the CLHNS cohort that are limited, at this young adult age, to males.
Our finding of sex differences in relationships between birth weight and adult epigenetic clock measures of biological ageing, with relationships present in males only, is consistent with previous findings from the CLHNS linking birth weight and other prenatal measures to a range of biological and health outcomes in adolescence and adulthood. Prior CLHNS work has found that birth weight is inversely related to outcomes such as blood pressure [19] and lipid levels [20] among males but not females. Similarly, circadian cortisol rhythms were flatter, indicating a more adverse hormone profile, among men born premature but not women born premature [30]. We have also reported that pregnancy metabolic and endocrine characteristics in the current generation of mothers (generation 2) tend to have stronger relationships with birth outcomes among male compared to female offspring (generation 3): compared to female offspring, maternal fasting glucose was a stronger, positive, and significant predictor of birth weight in male offspring [29], while maternal cortisol levels were stronger negative predictors of birth outcomes in male offspring [28]. Taken together, this past work points to potential sex differences in sensitivity to the prenatal environment, with gestational factors having more consistent short- and long-term impacts on male development and health. Our current findings using epigenetic clocks add to this evidence and show that males born lighter tend to have measures consistent with accelerated age and mortality risk using multiple clocks that were trained on different sets of ageing and health-related outcomes.
The pattern of relationships observed in the Cebu cohort provides insights into potential pathways linking early environments with biological ageing markers measured in males. Hannum’s clock was significantly inversely related to birth weight, and this relationship was weakened after adjustment for immune cell composition (IEAAHannum) and strengthened using a version of Hannum’s clock that incorporates age-related changes in immune cell composition (EEAA). Because Hannum’s clock was developed based upon DNA collected from blood samples (immune cells), these relationships point to changes in immune cell composition in relation to birth weight as the primary driver of relationships with Hannum’s clock. In post hoc analyses, we found that birth weight is a significant predictor of multiple immune parameters in males but unrelated to immune measures in females. Specifically, as birth weights increased, the proportion of CD4, CD8, and B Cells cells increased, while the proportion of neutrophils decreased. This finding builds on our prior work on a smaller subset of the CLHNS sample [31], and supports a shift away from innate and towards adaptive immunity in relation to favourable prenatal nutrition and growth [32]. The relationship with DNAmTL that we documented is consistent with prior work linking lower birth weights to shorter qPCR-measured telomere length in the study population [33], and also point to evidence of more rapid ageing of immune cells as reflected in shorter telomeres (a proxy for cell replication), among males who were born lighter. An effect specific to immune cell composition is consistent with our observation of a lack of relationship between birth weight and Horvath’s pan-tissue clock, which is less subject to differences between cells and tissues (Horvath 2013). In addition to relationships with immune cell composition and replicative senescence, higher birth weights predicted reduced biological ageing based upon the DNAmPhenoAge and DNAmGrim clocks, which are trained on suites of biochemical and clinical markers that predict morbidity and mortality, and the DunedinPACE clock, which is trained on the longitudinal pace of change in multiple clinical ageing markers. Taken together, our findings suggest that being born at lower birth weight predicts multiple measures of accelerated biological ageing among males in the CLHNS, including changes in immune cell composition and replicative potential, along with a suite of clinical ageing biomarkers that are strong predictors of mortality. Because there were few relationships with maternal arm fat area during pregnancy, it appears that birth size, perhaps as a reflection of fetal growth rate, is of primary importance in the relationships with long-term male health.
These findings add to a small but growing list of studies exploring links between birth weight and adult epigenetic clock measures. In general, the studies published thus far point to accelerated epigenetic ageing in relation to lower birth weight, with some evidence for sex differences in this effect. Using the Horvath clock, Lieshout [34] found that men born extremely low birth weight had an accelerated epigenetic age in adulthood compared to normal birth weight men, with no differences in birth weight status among women. In a large cohort from Scotland, Madden and colleagues [17] found that epigenetic age was accelerated in relation to lower birth weights using the DNAmGrimAge and DNAmTL clocks but was unrelated to the Horvath, Hannum, and DNAmPhenoAge clocks. This paper pooled male and female samples, and it was not reported whether they tested for possible interactions by sex. Similarly, an analysis of the ALSPAC cohort found evidence that epigenetic age was accelerated in relation to lower birth weights at 17 years of age, but there was no indication that the authors tested for heterogeneity by sex [16].
The causes of the sex differences in birth weight-adult epigenetic clock acceleration noted here are not known, but several factors could be important. Given that epigenetic age appears to serve as an index of maturational tempo during development, it is possible that accelerated maturation triggered by lower birth weights, which has been shown in some populations [35], including the females at Cebu [36], could contribute to accelerated epigenetic age measured in adulthood. The age at which epigenetic measures were obtained (20–22 years) was early enough in adulthood that a male bias in speeding up of maturity or rapid catch-up growth after small birth size might in theory contribute to the sex differences that we see. Counter to this hypothesis, we previously found that men who were born heavier tended to mature earlier at Cebu [37].
A sex difference in developmental sensitivity has been widely described in the anthropological and zoology literatures [38,39] and could contribute to the findings noted here. In the context of other DOHaD-inspired work, it has been noted that males tend to already be larger at birth, and there is evidence that male birth size is more sensitive to maternal nutritional supplementation during pregnancy [40]. It has been proposed that greater requirements for nutrition, to sustain dimorphic growth, could leave males more susceptible to comparable levels of shortfall [41,42]. To date, the studies that report testing for sex differences in relationships between birth weight and adult epigenetic age acceleration have found evidence for such differences, and it will be important for future studies of these relationships to explore this possibility systematically across a wider set of populations and age ranges.
Strengths of this study include not only the availability of prospectively collected, longitudinal data, but a relatively large sample size. With a combined sample of nearly 1700, this study represents among the largest analyses of the relationship between prenatal growth and nutrition measures and adult epigenetic clock measures. This large sample size also meant that we were powered to detect even relatively modest relationships as a result. The relationships that we report are generally weak, and it is not clear how biologically important they are in comparison to other factors known to influence the pace of biological ageing. The relationships present in males were consistent in direction and effect across nearly all of the measures that we investigated, including measures that are dependent on mitotic division (telomere length), immune cell ageing (Hannum), and composite measures of physiological and metabolic dysregulation (DNAmPhenoAge, DNAmGrim, and DunedinPACE). This consistency in findings reinforces the likelihood that we are detecting real biological relationships that influence the pace of ageing in males. Another limitation of this study stems from the relatively young age at which epigenetic age measures were obtained; future work that assesses the cohort at a later age will be necessary to clarify whether the relationships that we document persist into later life, including the noted sex differences, and if so whether they are amplified in interaction with ongoing exposures across the lifecycle or diminished in strength as individuals age.
Overall, we find evidence that slower fetal growth rate, as reflected in birth weight adjusted for gestational age at delivery, predicts markers consistent with accelerated biological ageing in young adult men, but not women. The direction of these relationships and the sex differences in how they manifest are consistent with past work predicting other chronic disease markers in the study population, and share similarities with the few other studies that explore birth weight-adult epigenetic age associations in other populations. These findings demonstrate epigenetic clocks as a potentially useful integrative set of tools to explore multiple downstream health and ageing effects of prenatal nutrition and growth, and point to likely sex differences in how these effects manifest.
Acknowledgments
We thank the researchers at the USC-Office of Population Studies Foundation, Inc., University of San Carlos, Cebu City, Philippines, for their role in the study design and data collection, and the study participants, who generously provided their time. Two anonymous authors provided feedback that improved the final manuscript. Funding: NIH R01AG061006; HL085144; HD054501; P2CHD050924.
Funding Statement
NIH Office of Extramural Research, National Institutes of Health R01AG061006; HL085144; HD054501; P2CHD050924
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Barker D. Mothers, Babies, and Disease in Later Life. London: BMJ Publishing; 1994. [Google Scholar]
- [2].Gluckman PD, Hanson MA. Developmental Origins of Health and Disease. Cambridge: Cambridge University Press; 2006. [Google Scholar]
- [3].Hoffman DJ, Powell TL, Barrett ES, et al. Developmental origins of metabolic diseases. Physiol Rev. 2021;101(3):739–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jylhava J, Pedersen NL, Hagg S. Biological Age Predictors. EBioMedicine. 2017;21:29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Langley-Evans SC, Langley-Evans AJ, Marchand MC. Nutritional programming of blood pressure and renal morphology. Arch Physiol Biochem. 2003;111:8–16. [DOI] [PubMed] [Google Scholar]
- [6].Reynolds CM, Vickers MH. Utility of Small Animal Models of Developmental Programming. Methods Mol Biol. 2018;1735:145–163. [DOI] [PubMed] [Google Scholar]
- [7].Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev. 2018;19(6):371–384. [DOI] [PubMed] [Google Scholar]
- [8].Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Levine ME, Lu AT, Quach A, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY). 2018;10(4):573–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lu AT, Quach A, Wilson JG, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY). 2019;11(2):303–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lu AT, Seeboth A, Tsai PC, et al. DNA methylation-based estimator of telomere length. Aging (Albany NY). 2019;11(16):5895–5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Belsky DW, Caspi A, Corcoran DL, et al. DunedinPACE, a DNA methylation biomarker of the pace of aging. Elife. 2022;11: e73420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Binder AM, Corvalan C, Mericq V, et al. Faster ticking rate of the epigenetic clock is associated with faster pubertal development in girls. Epigenetics. 2018;13(1):85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Simpkin AJ, Howe LD, Tilling K, et al. The epigenetic clock and physical development during childhood and adolescence: longitudinal analysis from a UK birth cohort. Int J Epidemiol. 2017;46(2):549–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Simpkin AJ, Hemani G, Suderman M, et al. Prenatal and early life influences on epigenetic age in children: a study of mother-offspring pairs from two cohort studies. Hum Mol Genet. 2016;25(1):191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Madden RA, McCartney DL, Walker RM, et al. Birth weight associations with DNA methylation differences in an adult population. Epigenetics. 2021;16(7):783–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kuzawa CW, Adair L, Bechayda SA, et al. Evolutionary life history theory as an organising framework for cohort studies: insights from the Cebu Longitudinal Health and Nutrition Survey. Ann Hum Biol. 2020;47(2):94–105. [DOI] [PubMed] [Google Scholar]
- [19].Adair LS, Kuzawa CW, Borja J. Maternal energy stores and diet composition during pregnancy program adolescent blood pressure. Circulation. 2001;104(9):1034–1039. [DOI] [PubMed] [Google Scholar]
- [20].Kuzawa CW, Adair LS. Lipid profiles in adolescent Filipinos: relation to birth weight and maternal energy status during pregnancy. Am J Clin Nutr. 2003;77(4):960–966. [DOI] [PubMed] [Google Scholar]
- [21].Adair LS, Popkin BM, Akin JS, et al. Cohort profile: the Cebu longitudinal health and nutrition survey. Int J Epidemiol. 2011;40(3):619–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ryan CP, Hayes MG, Lee NR, et al. Reproduction predicts shorter telomeres and epigenetic age acceleration among young adult women. Sci Rep. 2018;8(1):11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kuzawa CW, Fried RL, Borja JB, et al. Maternal pregnancy C-reactive protein predicts offspring birth size and body composition in metropolitan Cebu, Philippines. J Dev Orig Health Dis. 2017;8(6):674–681. [DOI] [PubMed] [Google Scholar]
- [24].Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. [DOI] [PubMed] [Google Scholar]
- [25].Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13(1):86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Horvath S, Levine AJ. HIV-1 Infection Accelerates Age According to the Epigenetic Clock. J Infect Dis. 2015;212(10):1563–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Thayer ZM, Feranil AB, Kuzawa CW. Maternal cortisol disproportionately impacts fetal growth in male offspring: evidence from the Philippines. Am J Hum Biol. 2012;24(1):1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Fried RL, Mayol NL, McDade TW, et al. Maternal metabolic adaptations to pregnancy among young women in Cebu, Philippines. Am J Hum Biol. 2017;29(5). DOI: 10.1002/ajhb.23011 [DOI] [PubMed] [Google Scholar]
- [30].Lee J, Fried R, Thayer Z, et al. Preterm delivery as a predictor of diurnal cortisol profiles in adulthood: evidence from Cebu, Philippines. Am J Hum Biol. 2014;26(5):598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].McDade TW, Jones MJ, Miller G, et al. Birth weight and postnatal microbial exposures predict the distribution of peripheral blood leukocyte subsets in young adults in the Philippines. J Dev Orig Health Dis. 2018;9(2):198–207. [DOI] [PubMed] [Google Scholar]
- [32].McDade TW, Georgiev AV, Kuzawa CW. Trade-offs between acquired and innate immune defenses in humans. Evol Med Public Health. 2016;2016(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Masterson EE, Hayes MG, Kuzawa CW, et al. Early life growth and adult telomere length in a Filipino cohort study. Am J Hum Biol. 2019;31(6):e23299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Van Lieshout RJ, McGowan PO, de Vega WC, et al. Extremely Low Birth Weight and Accelerated Biological Aging. Pediatrics. 2021;147. [DOI] [PubMed] [Google Scholar]
- [35].Ibanez L, Ferrer A, Marcos MV, et al. Early puberty: rapid progression and reduced final height in girls with low birth weight. Pediatrics. 2000;106(5):E72. [DOI] [PubMed] [Google Scholar]
- [36].Adair LS. Size at birth predicts age at menarche. Pediatrics. 2001;107(4):E59. [DOI] [PubMed] [Google Scholar]
- [37].Kuzawa CW, McDade TW, Adair LS, et al. Rapid weight gain after birth predicts life history and reproductive strategy in Filipino males. Proceedings of the National Academy of Sciences of the United States of America 2010; 107:16800–16805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Stinson S. Sex differences in environmental sensitivity during growth and development. American Journal of Physical Anthropology. 1985;28(S6):123–147. [Google Scholar]
- [39].Clutton-Brock TH, Guinness FE, Albon SD. Red deer: behavior and ecology of two sexes. Chicago: University of Chicago Press; 1982. [Google Scholar]
- [40].Adair LS, Pollitt E. Outcome of maternal nutritional supplementation: a comprehensive review of the Bacon Chow study. Am J Clin Nutr. 1985;41(5):948–978. [DOI] [PubMed] [Google Scholar]
- [41].Kuzawa CW. Developmental origins of life history: growth, productivity, and reproduction. Am J Hum Biol. 2007;19(5):654–661. [DOI] [PubMed] [Google Scholar]
- [42].Kuzawa CW. Fetal origins of developmental plasticity: are fetal cues reliable predictors of future nutritional environments? Am J Hum Biol. 2005;17(1):5–21. [DOI] [PubMed] [Google Scholar]


