Summary
A model was previously proposed that DA neurons provide SHH ligand to striatal interneurons, which in turn support the survival of DA neurons through the release of trophic factors such as Glial cell-derived neurotrophic factor (GDNF). However, some key clinical observations do not support this proposed model, and a recent independent study shows that striatal cholinergic neuron survival does not rely on intact DA neuron projections. To resolve this discrepancy, we generated several independent mouse lines to examine the exact role of DA neuron-derived Shh signaling in the maintenance of the basal ganglia circuit and to identify the Shh-producing cells in the adult brain. Our data suggest that the deletion of Shh in DA neurons does not affect DA neuron survival or locomotive function in cKO mice during aging, nor does it affect the long-term survival of cholinergic or FS PV + interneurons in the striatum (STR).
Subject areas: Natural sciences, biological sciences, neuroscience, cellular neuroscience
Graphical abstract
Highlights
-
•
Shh-expressing neurons are identified in multiple brain regions
-
•
Dopaminergic neurons are not Shh-expressing neurons in the adult mouse brain
-
•
Dopaminergic Shh deletion does not affect striatal PV or cholinergic neuron survival
Natural sciences; Biological sciences; Neuroscience; Cellular neuroscience.
Introduction
Basal ganglia neuronal circuits are critical for movement control, integrating cortical glutaminergic neuron input, nigrostriatal dopaminergic (DA) neuron input, and local cholinergic and fast-spiking (FS) GABA neuronal (PV+) input. It has been proposed that these different projections and neuronal types can have non-autonomous neurotrophic regulation on each other (Ortega-de San Luis et al., 2018) and that DA neuron-derived Sonic hedgehog (SHH) ligand might play an essential role in the homeostasis of these circuits (Gonzalez-Reyes et al., 2012). The phylogenetically conserved Shh pathway is critical for the development of DA neurons during early embryonic stages (Ye et al., 1998; Hynes et al., 1995) and promotes the survival of DA neurons in adulthood under stress (Miao et al., 1997; Tsuboi and Shults, 2002). Shh signaling has also been shown to be required for the survival of striatal cholinergic interneurons (Ortega-de San Luis et al., 2018). Therefore, Shh has been suggested to play an important role in maintaining the homeostasis of the basal ganglia, and DA neurons have been proposed as a critical source of ligand for striatal interneurons (Gonzalez-Reyes et al., 2012).
The SHH ligand is translated as a ∼45kDa precursor and undergoes autocatalytic processing to produce a ∼20kDa N-terminal signaling domain (referred to as SHH-N) and a ∼25kDa C-terminal domain with no known signaling role. SHH can signal in both an autocrine (Komada et al., 2008) and paracrine fashion. Binding of the SHH ligand to the Patched-1 (PTCH1) receptor releases the inhibition of the Smoothened (SMO) receptor which subsequently leads to the activation of glioma-associated oncogene (GLI) transcription factors (Briscoe and Therond, 2013). Previous work has shown that Shh and Fibroblast growth factor (Fgf) signaling is critical in defining the early progenitors that give rise to DA neurons (Ye et al., 1998). The exact role of Shh signaling in the later differentiation and maturation of DA neurons is less defined. Although several studies have indicated that Shh signaling regulates the expansion of DA progenitors and the subsequent generation of mature DA neurons, this prevailing view has been based primarily on in vitro culture results, and the exact in vivo function of Shh signaling in the patterning and neurogenesis of the ventral midbrain (VMB) remains unclear. Additionally, owing to the lethal phenotype of Shh knockout embryos at or before birth, the exact role of Shh signaling in the maintenance and survival of DA neurons in adult brain has not been examined until recently. Using a Smoothened (Smo) (the receptor for Shh signaling) conditional knockout (cKO) mouse line under the control of the Dopamine transporter (Slc6a3, commonly referred to as DAT) promoter with knockout induced at E15.5, we (Zhou et al., 2016) and others (Gonzalez-Reyes et al., 2012) have shown that autonomous Shh signaling in DA neurons is not critical for their maturation or survival.
One previous study reported that the SHH ligand produced by DA neurons is instead required to maintain the survival of striatal cholinergic and FS PV + neurons, which in turn are required for the survival of adult DA neurons by providing GDNF trophic support (Gonzalez-Reyes et al., 2012). However, several key findings in the Gonzalez-Reyes et al. study have not been replicated (Ortega-de San Luis et al., 2018). In addition, the proposed mechanism of regulation of the DA-Shh/cholinergic/FS PV + neuronal circuit is contradicted by several lines of evidence in the literature. For example, there is a lack of degenerative phenotype in striatal cholinergic neurons in patients with Parkinson disease (PD) (Girasole and Nelson, 2015) or 6-OHDA lesion models (Girasole and Nelson, 2015; DeBoer et al., 1993). To examine the exact role of DA neuron-derived Shh signaling in the maintenance of the basal ganglia circuit and to identify the Shh-producing cells in adult brain, we generated DA- (DATcre) and catecholaminergic- (THcre) specific Shh cKO mice (termed DATcre-shh cKO or THcre-shh cKO). These mice allowed us to investigate the reliance of DA neurons, striatal cholinergic neurons, or FS PV + neurons on DA neuron-derived SHH ligand. We also used fate mapping in neonatal and adult mice to identify the SHH-producing cells at the neonatal and adult stages, as previous studies have only mapped Shh-producing cells up to E12.5 during mouse embryo development (Blaess et al., 2011; Ihrie et al., 2011). Our data suggest that DA neurons are not a critical source for striatal or subventricular zone (SVZ) Shh signaling, and that DA neuron-specific deletion of Shh does not affect striatal Shh RNA levels, nor does it affect the long-term survival of cholinergic or FS PV + interneurons in the striatum (STR). We also report that the deletion of Shh in DA neurons does not affect DA neuron survival or locomotive function in cKO mice during aging. Instead, cortical and striatal neurons might be the source of SHH ligand for striatal interneurons in the adult brain.
Results
Dopaminergic-specific gene recombination mediated by the DAT-IRES-cre mouse line
To examine the role of DA neuron-derived SHH ligand, we generated DA neuron-specific Shh cKO mice. To achieve this, we crossed DATIREScre mice (JAX, 006,660, referred to as DATcre in this study) containing cre recombinase knocked into the 3′ end of the endogenous DAT allele following an internal ribosomal entry site (IRES) sequence with Shh fl/fl mice (JAX, 004,293) containing alleles of endogenous Shh flanked by loxP sites on exon 2. This Shh fl/fl mouse line (Lewis et al., 2001) was first described in 2001 and has been widely used to study the role of Shh in a variety of cell types in vivo (Zhao et al., 2014; Castillo-Azofeifa et al., 2017; Wagstaff et al., 2021).
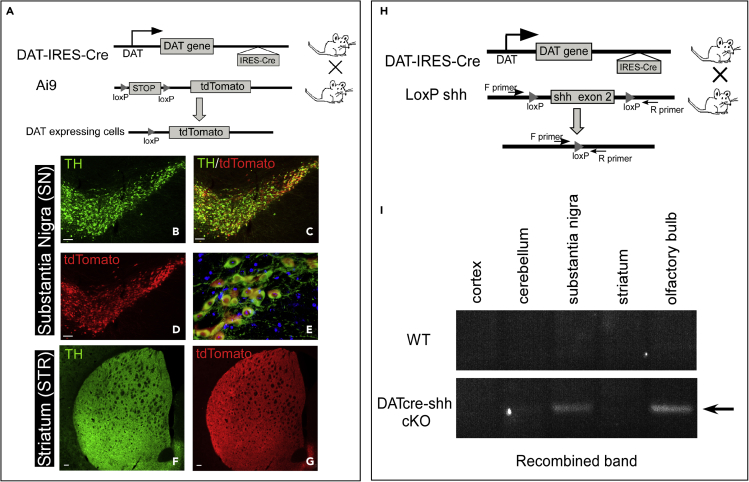
To validate the DA neuron-specific cKO of Shh in the DATcre mouse line, we first generated the DATcre-Ai9-tdTomato mouse line in which the tdTomato reporter gene is positioned 3′ to a floxed Stop cassette. In Figure 1, we show that tdTomato expression is detected specifically in Tyrosine hydroxylase (TH+) DA neurons in the substantia nigra (SN) (>90% of TH + neurons are tdTomato+) (Figures 1B–1E). In addition, we also observe tdTomato + axonal projections in the striatum (STR) that colocalize with TH immunostaining (Figures 1F and 1G) (refer to Table 1 for antibody information). We only observe tdTomato + neurons in the SN, ventral tegmental area (VTA), and olfactory bulb (OB) which is consistent with the known DAT expression pattern in the adult brain. As we could not validate several commercially available SHH antibodies by immunostaining in wild-type (WT) or Shh constitutive KO embryonic tissues, in addition to reporter gene expression, we also validated the recombination of the floxed Shh allele by PCR using a set of primers flanking the floxed exon 2 (Figure 1H). Our data (Figure 1I) show that the recombined band (delta band) is detected only in the OB and SN of DATcre-shh cKO mice but not in other brain regions. As a control, WT mice do not show the recombined band in any brain regions examined, confirming the DA neuron-specific deletion of Shh in our DATcre-shh cKO mice.
Figure 1.
DA neuron-specific gene recombination mediated by the DATcre mouse model
(A) Expression of the TdTomato reporter gene in DA neurons.
(B–E) mediated by DAT promoter-driven IRES-cre in the DATcre-TdTomato reporter line.
(F and G) tdTomato expression is also observed in striatal DA neuron axons colocalizing with TH expression. Scale bar = 100um.
(H) Generation of DA neuron-specific Shh conditional knockout mice (DATcre-shh cKO mice) using the same DAT promoter-driven cre line but crossed with Shh floxed mice.
(I) deletion of Shh exon 2 confirmed by the detection of recombined delta band using primers flanking outside of the two loxP sites. Note that the recombined delta band is only present in DATcre-shh cKO mouse tissue in the SN and OB as expected and is absent in WT tissue. Representative results from at least 3 mice from each phenotype (at the age of 3 months). Scale bar = 100μm.
Table 1.
Primer/probe sets used in qRT-PCR
| Primer/probe set: | ||
| Hmbs: | Forward Primer: | 5′-TCCCTGAAGGATGTGCCTAC-3′ |
| Reverse Primer: | 5′-ACAAGGGTTTTCCCGTTTG-3′ | |
| Probe: | Universal Probe Library: Probe 79 - Roche | |
| CHAT: | Forward Primer: | 5'- GGTTCGGTGCGTAACAGC-3′ |
| Reverse Primer: | 5'- GCGATTCTTAATCCAGAGTAGCA-3′ | |
| Probe: | Universal Probe Library: Probe 108 - Roche | |
| VCHAT | Forward Primer: | 5'- AGAGCCCTACCCTGATCTCTG-3′ |
| Reverse Primer: | 5'- CAAGTAGGCGCTGGCATTAG-3′ | |
| Probe: | Universal Probe Library: Probe 70 - Roche | |
| shh: | Forward Primer: | 5′-ACCCCGACATCATATTTAAGGA-3′ |
| Reverse Primer: | 5′-TTAACTTGTCTTTGCACCTCTGA-3′ | |
| Probe: | Universal Probe Library: Probe 32 - Roche | |
| Gli | Forward Primer: | 5′-CTGACTGTGCCCGAGAGTG-3′ |
| Reverse Primer: | 5′-CGCTGCTGCAAGAGGACT-3′ | |
| Probe: | Universal Probe Library: Probe 84 - Roche | |
| Drd1a: | Forward Primer: | 5′-TCTGGTTTACCTGATCCCTCA-3′ |
| Reverse Primer: | 5′-GCCTCCTCCCTCTTCAGGT-3′ | |
| Probe: | Universal Probe Library: Probe 82 - Roche | |
| Drd2: | Forward Primer: | 5′-CTCTTTGGACTCAACAACACAGA-3′ |
| Reverse Primer: | 5′-AAGGGCACGTAGAACGAGAC-3′ | |
| Probe: | Universal Probe Library: Probe 25 - Roche | |
| GDNF: | Forward Primer: | 5′-TCCAACTGGGGGTCTACG-3′ |
| Reverse Primer: | 5′-GACATCCCATAACTTCATCTTAGAGTC-3′ | |
| Probe: | Universal Probe Library: Probe 70 - Roche | |
| PGK1 (gDNA): | Forward Primer: | 5′-TACCTGCTGGCTGGATGG-3′ |
| Reverse Primer: | 5′-CACAGCCTCGGCATATTTCT-3′ | |
| Probe: | Universal Probe Library: Probe 108 - Roche | |
| Floxed Shh intron 1 (gDNA): | Forward Primer: | 5′-GCTGCCTAGACCAAGAATGT-3′ |
| Reverse Primer: | 5′-TGAGGAAACCCATAGCAACTC-3′ | |
| Probe: | FAM/CTTGTCCTT/ZEN/GACCCACTGCCTACC/3IABkFQ- | |
| Unfloxed Shh exon 3 (gDNA): | Forward Primer: | 5′-ATTGGCACCTGGCTGTT-3′ |
| Reverse Primer: | 5′-GCTTTCCCATCAGTTCCTTATTTC-3′ | |
| Probe: | FAM/CGAGACCAT/ZEN/GCATCCCTTGGGAAT/3IABkFQ | |
Dopaminergic neuron shh deletion does not affect dopaminergic neuron survival during aging
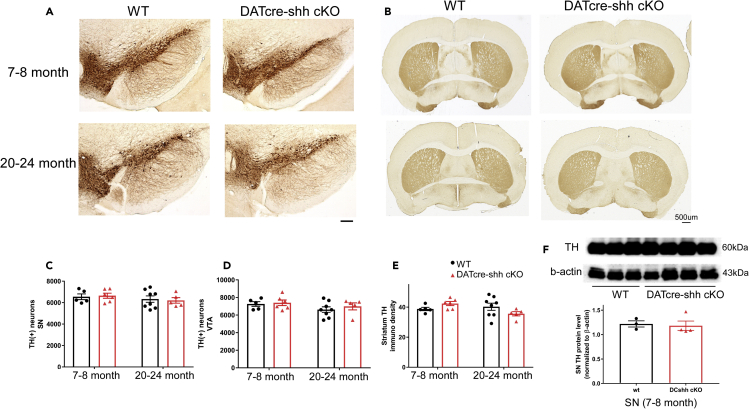
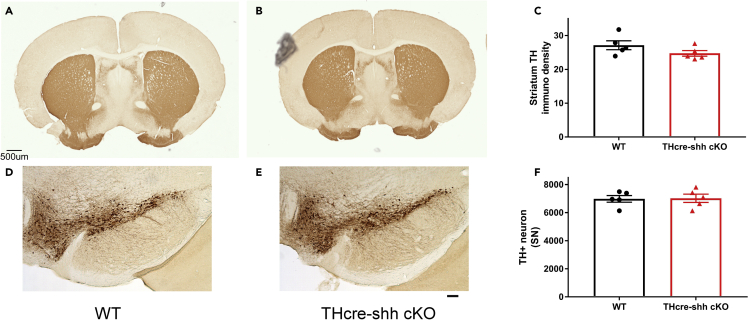
It has been reported previously that DA neuron-specific deletion of Shh leads to the degeneration of DA neurons during the process of aging starting as early as 4 months of age (Gonzalez-Reyes et al., 2012). We carried out unbiased stereological counts of TH + neurons in both the SN and VTA in young and aged control (DATcre-shhwt/wt) or DATcre-shh cKO (DATcre-shhfl/fl) mice in which cre expression is initiated by the DAT promoter at ∼ E15.5. We did not detect any loss of DA neurons in the SN of young adult (7–8 months old) or aged (20–24 month old) DATcre-shh cKO mice (Representative SN immunostaining images in Figure 2A, quantification in Figures 2C and 2D). Consistent with the lack of DA neuron degeneration in the SN, we did not observe any differences in the immunoreactivity of TH fiber density in the STR of young adult or aged DATcre-shh cKO mice compared to control (DATcre-shh wt/wt) mice (Figure 2B representative images and 2E with quantification). Similarly, we did not observe any differences in TH protein levels in the SN of 7–8-month-old control or DATcre-shh cKO mice using Western blot. As our results contradict those of the Gonzalez-Reyes et al. study (Gonzalez-Reyes et al., 2012) and to further ensure the rigor of our study, we generated an additional conditional Shh knockout line (THcre-shh cKO) in which cre is driven by the Th promoter via knock in of an IRES-cre cassette into the endogenous Th gene. Using this line, we induced the expression of cre in all catecholaminergic neurons starting at E11.5. We quantified the number of (TH+) DA neurons in the SN using unbiased stereology in 7–8 month old control or THcre-shh cKO mice. Consistent with results from our DATcre-shh cKO mouse line, we do not observe any difference in the total number of DA neurons in the SN (Figure 3 for representative images and quantification). Similarly, we did not observe any differences in (TH+) fiber density in the STR of THcre-shh cKO mice compared to control mice (Figure 3). The previous study (Gonzalez-Reyes et al., 2012) used a different DATcre line (which is a heterozygous knockout for DAT gene) and a different Shh loxP mouse line (that has not been validated by independent groups) which might account for the different phenotype in their DATcre-shh cKO line compared to our DATcre or THcre-shh cKO lines. Although the Shh loxP line used in our experiments has been published in many studies, to further increase the rigor of our research we wanted to confirm our Shh loxP line is able to mediate cre-induced gene deletion of Shh-exon2 and leads to functional loss of Shh. Therefore, we generated the CMV-CRE-shh KO mouse line in which ubiquitous cre expression is driven by the human cytomegalovirus (CMV) immediate-early enhancer and promoter. Embryonic and ubiquitous expression of cre in the CMV-CRE line indeed leads to deficits in embryonic development at E10.5 which phenocopies global Shh KO embryos (n > 3 WT and KO embryos compared from two litters), validating our Shh loxP line (Figure S1). Therefore, we conclude that DA neuron-specific Shh signaling is not required for the survival of DA neurons during aging when it is deleted at early embryonic stage (E11.5 through Th promoter-driven cre) or at a later stage (E15.5 through DAT promoter-driven cre expression).
Figure 2.
DA neuron-specific deletion of Shh does not affect DA neuron survival in the SN or DA neuron axonal projection in the STR in adult or aged mice
Representative images from WT or DATcre-shh cKO mice at 7–8 months or 20–24 months of age in the SN (A) or STR (B) immunostained for Tyrosine hydroxylase (TH). Quantification using unbiased stereological method of (TH+) DA neurons in the SN (C), VTA (D), and striatal (TH+) fiber density (E) in WT and DATcre-shh cKO mice.
(F) Expression of TH protein detected by Western blot from SN in WT or DATCre-shh cKO at 7–8 months of age does not show any difference in TH protein levels. Each data point represents an individual mouse. Mean + SEM. No statistical significance for any of the groups using either two-Way ANOVA(for multiple groups in C, D, E) or Student’s t test (for comparison between two groups in F). Scale bar = 100μm.
Figure 3.
Deletion of Shh in catecholaminergic neurons mediated by Th-driven IRES-cre does not affect DA neuron survival in 7–8-month-old mice
Representative images from WT or THcre-shh cKO mice at 7–8 months of age in the STR (A and B) and SN (D and E) immunostained for TH. Quantification of striatal TH fiber density (C) or Unbiased stereological counts of DA neurons in the SN (F). Each data point represents an individual mouse. Mean + SEM. No significant difference between groups (Student’s t test). Scale bar = 50μm.
Dopaminergic neuron-specific Sonic hedgehog ligand is not required for the survival of striatal cholinergic (CHAT+) or fast-spiking (PV+) interneuron survival in adult brains
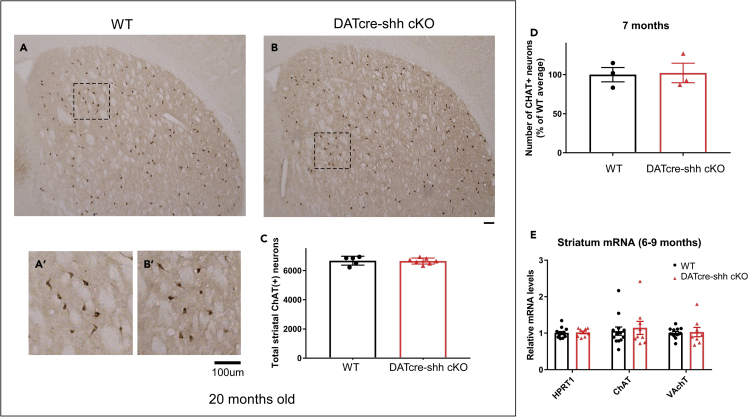
It has also been reported previously that DA neuron-derived SHH ligand could be important to maintain striatal interneuron survival via trophic factor regulation (Gonzalez-Reyes et al., 2012). However, later studies have reported that striatal FS PV + neurons do not rely on Shh signaling for survival and that cholinergic neurons do rely on Shh signaling for survival, though this Shh is not provided by DA neurons (Ortega-de San Luis et al., 2018). To further investigate this proposed reliance of cholinergic or PV + interneurons on DA neuron-derived SHH ligand, we examined whether DA neuron-specific deletion of Shh affects cholinergic interneurons in the STR in our mouse model. We report that choline acetyltransferase (ChAT) positive interneuron number does not differ between control or DATcre-shh cKO mice in aged (20months old, Figures 4A–4C) or young (7–8 months, Figure 4D) mice. Consistent with this finding, mRNA levels of Chat and Vesicular acetylcholine transporter (Vchat) do not differ in the STR of control or cKO mice (Figure 4E), suggesting that striatal cholinergic neurons do not rely on DA neuron-derived SHH ligand. These data are in agreement with the Ortega-de San Luis et al. study (Ortega-de San Luis et al., 2018). As PV + interneurons are also an important component of the striatal circuits, we also examined the total number of PV + interneurons in the STR of control or DATcre-shh cKO mice using unbiased stereology. Our results show no difference in PV + interneuron number in the STR in control or DATcre-shh cKO mice, suggesting that DA neuron-derived SHH ligand is not critical for PV + interneuron development or maintenance (Figure 5).
Figure 4.
Deletion of Shh in DA neurons mediated by DAT promoter-driven IRES-cre does not affect cholinergic neuron number in the STR in aged mice (20 months)
Representative images from WT or DATcre-shh cKO mice at 20 months of age in the STR (A and B) immunostained for CHAT (A′-B′ higher magnification images of the small box in A or B). Unbiased stereological quantification of (CHAT+) neurons in striatum of WT or DATCre-shh cKO mice (C). Similar results are also observed in young adult WT or DATcre-shh cKO mice at 7 months of age (D). mRNA expression of cholinergic neuron marker genes is not different between WT or DATcre-shh cKO mice at 6–9 months of age (E). For each gene, expression is normalized to housekeeping gene HMBS expression. Each data point represents an individual mouse. Mean + SEM. No significant difference between groups (Student’s t test). (n = 13 for control and n = 9 for DATcre-shh cKO for Panel E).
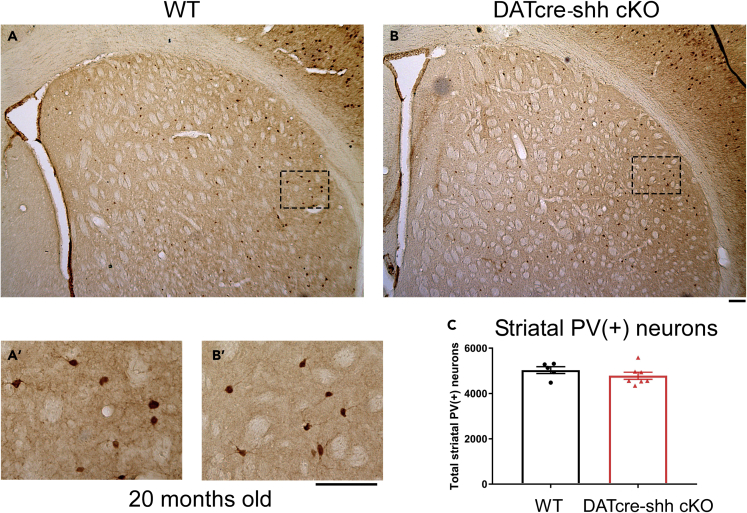
Figure 5.
Deletion of Shh in DA neurons (in DATCre-shh cKO mice) does not affect PV + neuron number in the STR in aged mice (20 months)
Representative images from WT or DATcre-shh cKO mice at 20 months of age in the STR (A and B) immunostained for PV (Parvalbumin) (A′-B′ higher magnification images of the small box in A or B). Unbiased stereological quantification of PV + neurons in the STR of WT or DATcre-shh cKO mice (C). Each data point represents an individual mouse. Mean + SEM. No significant difference between groups (Student’s t test). Scale bar = 100μm.
Dopaminergic neurons are not the critical source of Sonic hedgehog ligand for the maintenance of subventricular zone neural stem cells
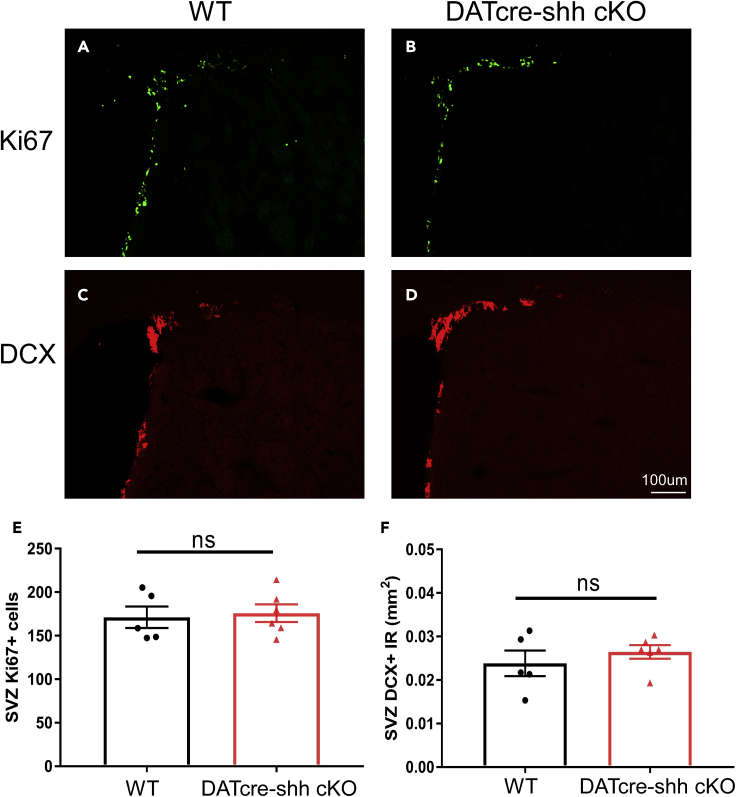
It has been reported by others and by our group that Shh signaling in postnatal neural stem cells (NSCs) is critical for maintaining adult neurogenesis (Lai et al., 2003; Petrova et al., 2013; Wang et al., 2022). We have recently reported that the deletion of the Shh receptor Smoothened (SMO) in Glial fibrillary acidic protein (GFAP) positive NSCs at the perinatal stage leads to premature aging-related decline of adult neurogenesis (Wang et al., 2022). DA neuron projections have been described at the SVZ region where the adult neural stem cells reside and have been suggested to regulate SVZ neurogenesis through the release of factors such as dopamine (Garcia-Garrote et al., 2021; Hoglinger et al., 2004). Therefore, we decided to examine whether DA neuron projections from the midbrain to the STR/SVZ region could be a critical source of SHH ligand for maintaining Shh signaling in NSCs. In adult control and DATcre-shh cKO mice, we examined both the number of proliferating cells (using an antibody against Marker of Proliferation Ki67 (Ki67)) and the number of newly born neuroblasts (using an antibody against Doublecortin (DCX)) in the SVZ. Our data show no difference in Ki67 + proliferating cells (Figures 6A, 6B, and 6E) or DCX + cells (Figures 6C, 6D, and 6F) in control or DATcre-shh cKO mice. This suggests that DA neurons are not a critical source for the Shh signaling that is critical for NSCs to maintain adult neurogenesis.
Figure 6.
DA neurons are not a critical source of SHH ligand for SVZ NSCs
We observe no difference in the number of proliferating cells (A and B) or neuroblasts (C and D) in 3-month-old WT or DATcre-shh cKO mice at the SVZ, suggesting that DA neurons are not an active source of SHH ligand for SVZ NSCs. Mean + SEM.
(E and F) Quantification from multiple sections averaged in each animal. Each data point represents the average from an individual mouse. Mean + SEM. No significant difference between groups (Student’s t test). Scale bar = 100um.
Shh-expressing cells in the neonatal and adult mouse brain are non-Dopaminergic neurons (NeuN + but TH- neurons) in various brain regions
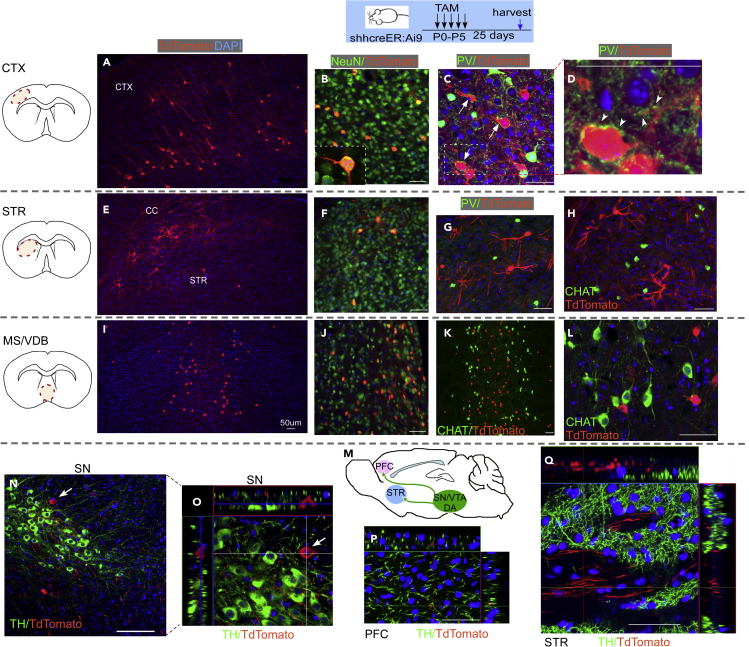
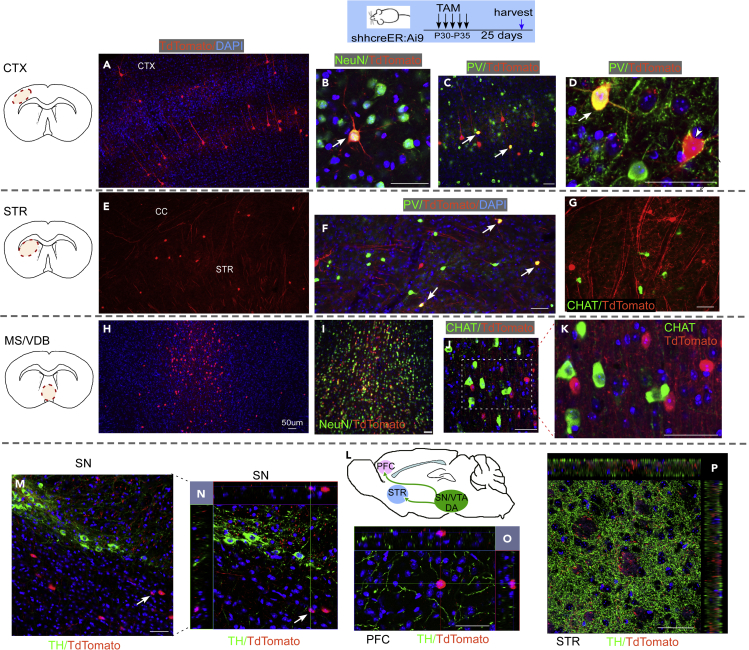
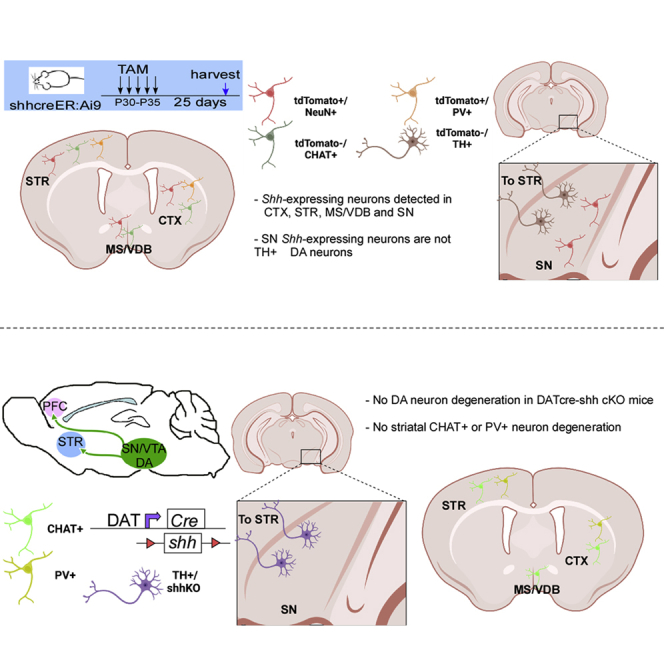
As we did not notice any difference in DA neuron, striatal cholinergic neuron, or PV + interneuron survival or SVZ neurogenesis in DATcre-shh cKO mice, we wanted to identify the Shh-expressing cells in the neonatal and adult mouse brain and investigate whether postnatal DA neurons express Shh. As we could not validate the specificity of several SHH antibodies, we used a genetic inducible fate mapping (GIFM) strategy by crossing ShhcreER mice (in which the creER gene is inserted into the endogenous loci of the Shh gene) with the Ai9 tdTomato reporter line. By Tamoxifen (TAM) administration (180 mg/kg X 5 daily gavage administration), we induced tdTomato expression in Shh-expressing cells in neonates or adult mice. We harvested the brains at either 1 or 25 days after TAM administration and examined the brain for tdTomato positive cells and expression of other cellular markers. tdTomato expression pattern was similar among brains that were harvested immediately (day 1) or later (25 days) after TAM treatment, with brains harvested at day 25 after TAM showing higher tdTomato fluorescent intensity. Therefore, in this study, we present data of tdTomato + cells at 25 days after TAM administration. In ShhcreER-Ai9 tdTomato mice that were treated with TAM from the P0-P5 neonatal stage, we observe TdTomato + cells at day 30 depicting Shh-expressing cells from P0-P5 in cortical neurons (Figures 7A–7D), striatal neurons (Figures 7E–7H), and medial septum (MS)/vertical diagonal band of Broca (VDB) neurons (Figures 7I–7L). tdTomato + neurons are also present in the SN (visualized via RBFOX3 staining, commonly referred to as NeuN, data not shown); however, they are not colocalized to (TH+) DA neurons (Figures 7N and 7O). Consistent with this data, tdTomato + neuronal terminals in the prefrontal cortex (PFC, Figure 7P) or STR (Figure 7Q) do not colocalize with (TH+) neuron fibers. Similarly, in ShhcreER-Ai9 tdTomato mice that were treated with TAM from P30-P35, we observed a similar pattern of neurons that are tdTomato + at multiple brain regions (cortex, STR, and MS/VDB, Figure 8). Some of the tdTomato + neurons are also PV+ (in cortex Figure 8D), but most are not ChAT+ (in MS Figures 8H–8K). Again, although tdTomato + cells are detected in the SN and are colocalized with the neuronal marker NeuN (data not shown), they are not TH+ (Figures 8M and 8N), suggesting DA neurons are not the Shh-expressing neurons in the adult mouse brain. Similarly, TH + neuronal terminals in the PFC and STR do not colocalize with tdTomato + fibers in one month TAM-labeled Shh-expressing cells (Figures 8O and 8P). Therefore, we conclude by genetic tracing method that midbrain DA neurons are not Shh-expressing either at the early neonatal stage (P0-P5) or at the young adult stage (P30-P35).
Figure 7.
Shh-expressing cells in postnatal mouse brain labeled by Shh-CreER-tdTomato line treated with TAM at P0-P5 age
tdTomato + cells at 25 days post TAM depicting Shh-expressing cells at P0-P5 stage are observed in (A–D) cortical neurons that are NeuN+ and receive PV + projections, (E–H) striatal neurons, and (I–L) MS/VDB neurons that are NeuN + but CHAT or PV negative. tdTomato + neurons are also present in the SN (NeuN+, not shown) but are not colocalized with TH + DA neurons (N and O). Consistent with these findings, tdTomato + neuronal terminals in the PFC (P) or STR (Q) do not colocalize with TH + fibers. Scale bar = 50μm).
Figure 8.
Shh-expressing cells in the young adult mouse brain labeled by ShhCreER-tdTomato line treated with TAM at P30-P35 age
tdTomato + cells at 25 days post TAM depicting Shh-expressing cells at P30-P35 stage are observed in (A–D) cortical PV + neurons and (E–G) striatal PV + neurons, but not CHAT + neurons. tdTomato + cells are also observed in (H–K) MS/VDB neurons but mostly not in CHAT + cholinergic neurons. tdTomato + neurons are also present in the SN (NeuN+, image not shown) but are not colocalized to TH + DA neurons (M and N). Consistent with this data, tdTomato + neuronal terminals in the PFC (O) and STR (P) do not colocalize with TH + fibers. Scale bar = 50um).
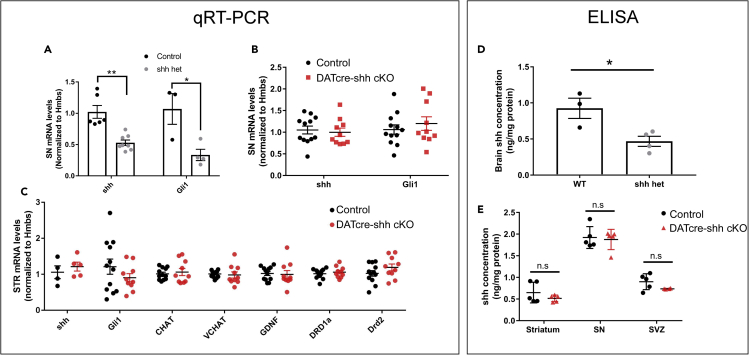
Shh transcripts and Sonic hedgehog peptide levels are not altered in the substantia nigra or striatum in DATcre-shh cKO mouse brains
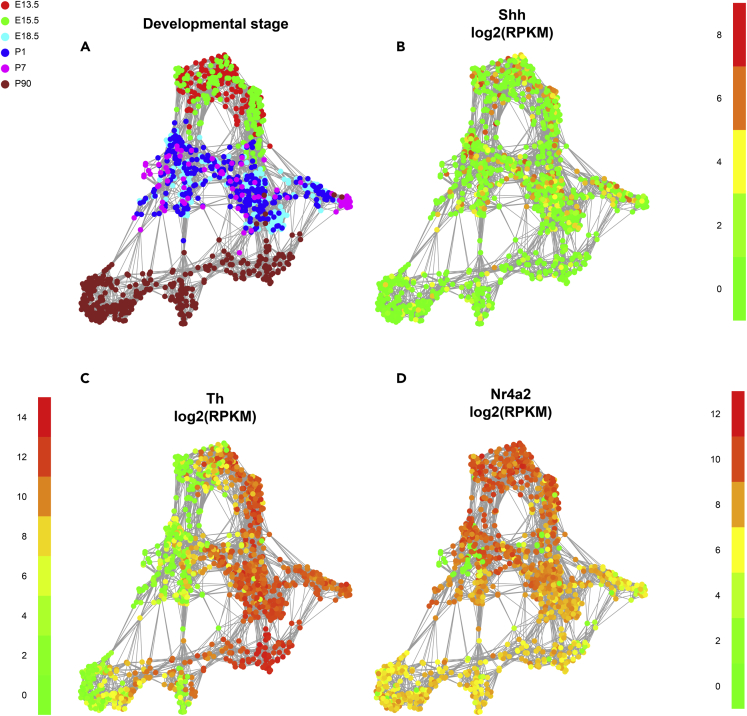
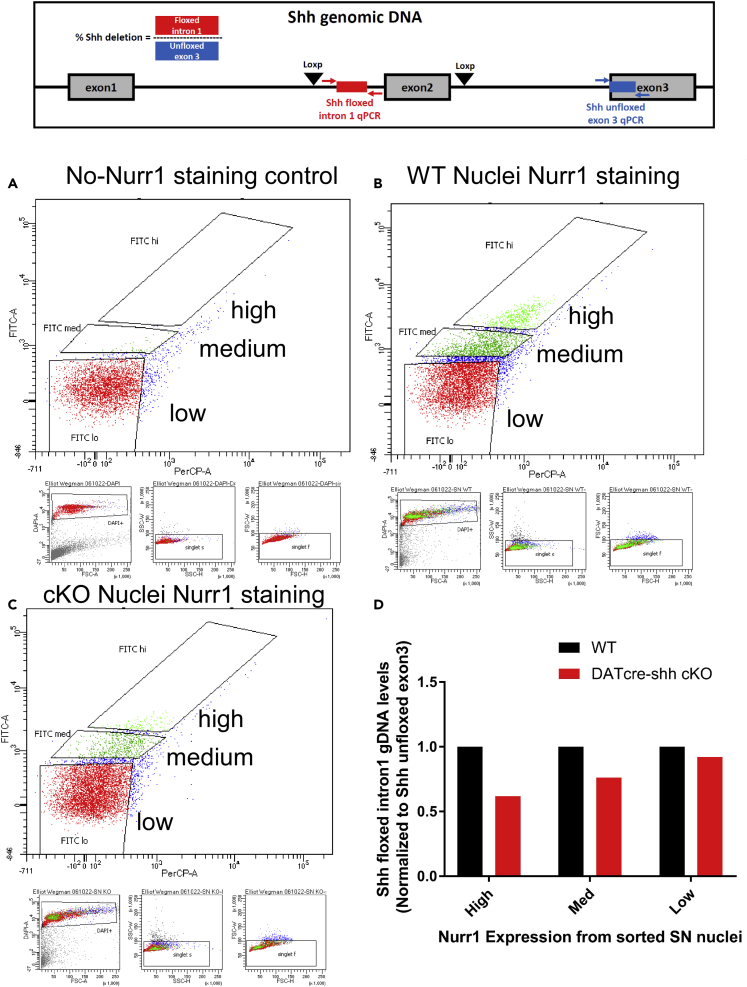
To further and more rigorously examine whether DA neurons contribute to Shh levels in the adult brain in the SN and STR, we measured the mRNA levels of Shh and its target gene Gli1 in the SN where DA neurons reside. Our data show that total levels of Shh mRNA in the SN are not altered in DATcre-shh cKO mice, suggesting that DA neurons are not the major contributors of Shh mRNA in the SN (Figure 9A). Consistent with our previous report, we did not observe any difference in Gli1 mRNA levels in the SN in DATcre-shh cKO mice, suggesting that the deletion of Shh in DA neurons does not affect Shh signaling in the SN (Figure 9A). It has been reported previously in a different DATcre-shh cKO mouse line that GDNF levels significantly decrease in the STR of DATcre-shh cKO mice, which is speculated to cause the degeneration of DA neurons. To examine this in our mouse model, we looked at mRNA levels of Gdnf in the STR. We found no difference in Gdnf transcripts in the STR of control and DATcre-shh cKO mice (Figure 9B). Consistent with our immunohistochemistry data, there is no difference in Chat or Vchat mRNA levels (Figure 9B), supporting a lack of degeneration of cholinergic neurons in the STR. Additionally, we did not detect any difference in Shh or Gli1 levels in the STR of control and DATcre-shh cKO mice (Figure 9B). To further examine SHH protein levels in brain lysate, we used a SHH ELISA kit to measure SHH levels from brain lysates harvested from the STR, SN, and SVZ regions in control or DATcre-shh cKO mice. We first validated the specificity and sensitivity of the ELISA kit using WT or Shh heterozygous knockout mice. Total brain homogenates from the Shh heterozygous knockout mice show a 50% reduction of SHH protein level compared to WT littermate controls measured by this ELISA kit (Figure 9D). Using this kit, we did not detect any difference in SHH protein levels in the STR, SN, or SVZ regions of WT and DATcre-shh cKO mice (Figure 9E). This data further supports our conclusion that DA neurons are not a major or critical source of SHH ligand in the adult brain, and that the deletion of Shh in DA neurons does not affect Shh levels or signaling in vivo. To further confirm that Shh is not expressed or is expressed at very low levels in DA neurons in the adult brain, we analyzed single-cell RNA-Seq data generated from previous studies with a focus on midbrain DA neurons (Tiklová et al., 2019; Kamath et al., 2022). Here we present data from the Tiklová et al. study showing that during early development, DA neurons do express Shh transcript; however, in adult DA neurons this expression is largely downregulated (Figures 10A and 10B) while other DA neuron-specific marker genes (such as TH or Nurr1, Figures 10C and 10D) sustained high expression levels into adulthood. Another study using single-nuclear RNA-seq to characterize DA neurons in adult rats also shows little to no Shh expression in adult DA neurons (Kamath et al., 2022). In addition to our analysis of publicly available RNA-seq data, we also utilized a recently developed protocol to enrich DA nuclei from the SN by Nurr1+ fluorescence-activated nuclear sorting (FANS) (Kamath et al., 2022). Using this protocol, we sorted nuclei from two WT and three DATcre-shh cKO mice based on Nurr1-expression level (Figures 11A–11C). This protocol has been reported to result in an abundance of ∼25% DA neuron nuclei in the total population of sorted Nurr1+ nuclei (Kamath et al., 2022). Interestingly, our results show that nuclei isolated from the SN of the cKO mice expressing high or medium levels of Nurr1 protein have a 30% decrease in intron 1 of Shh gDNA (which is within the floxed region) compared to WT mice as determined by quantitative real-time PCR (Figure 11D). This 30% decrease in Shh floxed intron 1 abundance is in line with our expectation that these populations of Nurr1+ nuclei should contain ∼25% DA neuron nuclei. As a control, we show that Nurr1-negative nuclei (non-DA neurons) have similar levels of Shh floxed intron 1 gDNA in WT or cKO mice (Figure 11D, ‘low’ population). These results provide further validation that the floxed Shh gDNA sequence is deleted in DA neurons in our DATcre-shh cKO mice.
Figure 9.
Deletion of Shh in DA neurons does not affect total Shh mRNA or protein levels in the SN
(A) The specificity of Shh and Gli1 qPCR primer/probe sets are validated in Shh global heterozygous KO mice. Using these validated primer/probe sets, we did not detect any changes in the levels of Shh or Gli1 mRNA levels in DATcre-shh cKO mice in the SN or STR, suggesting that Shh is not expressed in DAT + DA neurons (B). Consistent with these results, there is no difference in striatal mRNA expression levels of Gli1, Chat, Vchat, Gdnf, Drd1a or Drd2 between WT or DATcre-shh cKO mice (C). Similarly, to validate the specificity of the SHH ELISA kit, we measured SHH levels in brain lysate in WT or heterozygous global Shh KO mice. The results from this assay show a 50% decrease in SHH protein expression in brain lysates in Shh heterozygous knockout mice (D). However, using the same ELISA kit, we did not detect any changes in SHH levels in WT or DATcre-shh cKO mice in the STR, SN or SVZ regions (E), suggesting that Shh is not actively expressed in DA neurons under physiological conditions in the adult mouse brain. Each data point represents an individual mouse (3–6 month old). Mean + SEM. ∗<0.05, Student’s t test.
Figure 10.
DA neuron expression of Shh at different developmental stages. scRNA-seq data of DA neurons from different age groups of mice show that adult SN DA neurons (magenta group > P90 in panel (A)) have no or very low levels of Shh expression (B), while they show abundant expression of other DA neuron markers such as Th (C) and Nr4a2 (D)
Note that the Y axis scales for Th and Nr4a2 are larger than the scale for Shh expression. Plot was generated using scRNA-seq data obtained from Tiklová et al., a previous study (Tiklová et al., 2019) with permission from Dr. Thomas Perlmann.
Figure 11.
Validation of DA neuron Shh deletion
DA neuron nuclei were enriched by Nurr1 immunoreactivity fluorescence-activated nuclear sorting (FANS) from the SN of two WT and three DATcre-shh cKO mice. Nuclei were sorted into three categories: high expression (1.5–2.5k nuclei), medium expression (17k-18k nuclei), and low expression (150–200k nuclei) of Nurr1. Depicted are the gates for the high, medium, and low expression populations in (A) a control sample not stained for Nurr1, (B) the pooled WT sample, and (C) the pooled DATcre-shh cKO sample. Genomic DNA from the sorted nuclei was harvested and qPCR performed for the floxed Shh intron 1 and unfloxed exon 3 to confirm the deletion of the floxed site in DATcre-shh cKO DA nuclei. Shh floxed intron 1 gDNA levels were normalized to unfloxed exon 3 gDNA levels in each sample. The data show a ∼30% decrease in floxed Shh intron 1 gDNA harvested from Nurr1+ nuclei in DATcre-shh KO mice compared to WT mice (D) while levels of Shh floxed intron 1 do not change in Nurr1-negative nuclei. These results are in line with a previous study that used this method to enrich for DA nuclei and observed ∼25% abundance of DA neuron nuclei in the sorted Nurr1+ population (Kamath et al., 2022).
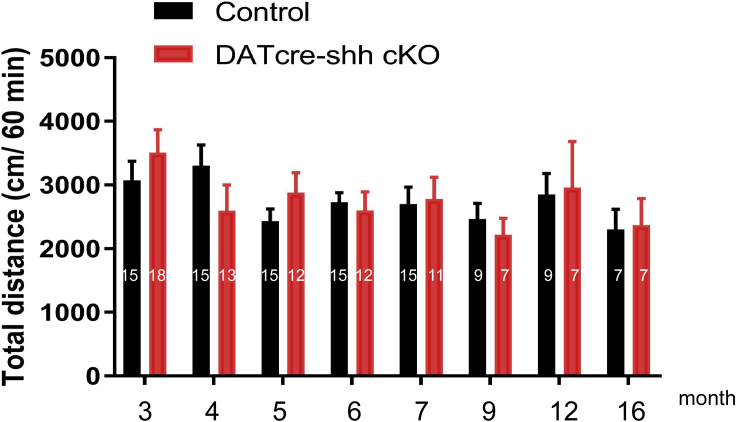
DA-specific deletion of shh gene does not affect locomotor function in mice
Lastly, we wanted to evaluate whether the DATcre-shh cKO mice exhibit changes in general locomotive function. We measured spontaneous locomotion in an automated open field chamber for 1 h and did not observe any difference between control and DATcre-shh cKO mice at any of the time points tested (3–16 months of age, Figure 12). Therefore, our behavioral data also supports a lack of motor deficits in the DATcre-shh cKO mice, which is in conflict with the previous report (Gonzalez-Reyes et al., 2012).
Figure 12.
DA neuron-specific Shh deletion does not affect total distance traveled in the locomotion chamber over a period of 60 min
Measurements were taken from mice that range between 3 and 16 months of age. Mean + SEM. Number on bar shows n number for each group/time point. There is no significant difference between genotype or time points. two-way ANOVA.
Discussion
Early in the development of DA neurons, Shh is known to induce the commitment of floor plate progenitors in the midbrain to a DA neuron fate. However, the role of autonomous Shh signaling in DA neurons during differentiation, and especially during later maturation stages, has recently come into question. Our previous study has demonstrated that abolishing Shh signaling in DA neurons during late gestation stages (around E15.5) in DAT + neurons does not affect their maturation and survival into adulthood or during aging (Zhou et al., 2016). In this current study, we further investigate whether DA neurons are a critical source of SHH ligand through their nigrostriatal projections and whether striatal interneurons (cholinergic or FS PV + GABA neurons) rely on DA neuron-derived SHH ligand for survival. Previous studies investigating this issue have generated conflicting results. One study (Gonzalez-Reyes et al., 2012) has reported that DA neuron-derived SHH is critical for striatal cholinergic and PV + neuron survival, and DA neuron-specific deletion of Shh leads to the degeneration of cholinergic neurons and PV + FS interneurons in the STR, which in turn leads to the degeneration of midbrain DA neurons via loss of the trophic factor GDNF. In contrast to these findings, a recent independent study using PV-specific deletion of Smo (Shh receptor) showed that striatal PV + neurons do not require Shh signaling for their survival (Ortega-de San Luis et al., 2018). The latter study further demonstrated that striatal cholinergic neurons require Shh signaling for their survival by cholinergic-specific Smo deletion; however, cholinergic interneurons do not rely on nigrostriatal DA neuron projections for survival, suggesting that DA neurons might not be the source of SHH ligand for striatal cholinergic neurons. To further investigate whether striatal interneurons rely on DA neuron-derived SHH ligand, we independently generated a DAT-IRES-cre Shh cKO mouse line. Our data support the finding of the Ortega-de San Luis et al. study, suggesting that DA neurons are not the main source of Shh signaling in the STR and that DA neuron-derived SHH ligand is not critical for cholinergic or FS interneuron survival in adulthood or through the aging process.
Our data show that DA neuron-specific deletion of Shh does not affect DA neuron survival in the VTA and SN in adult mice or aged mice. Combined with our previous report that autonomous Shh signaling is not required for DA neuron survival, we conclude that neither Shh expression nor Smo expression is critical for DA neuron survival in the adult brain. The discrepancy between our results and the previous study (Gonzalez-Reyes et al., 2012) could stem from the two different DATcre-shh cKO mouse models used. The previous study used a DATcre mouse line that knocked cre into the endogenous DAT loci; however, the insertion of the cre expression cassette is at the 5′ UTR of the endogenous DAT gene. Therefore, the DATcre-shh cKO mice in their study is a compound Shh knockout (homozygous for Shh) and DAT heterozygous knockout mice (owing to the disruption of DAT by the CRE cassette). It has been reported that DAT heterozygous knockout mice (Giros et al., 1996; Spielewoy et al., 2000, 2001; Hall et al., 2003) show certain behavioral and biochemical changes compared to wild-type mice. Combination of the DAT heterozygous knockout and the deletion of Shh might have generated some unexpected confound in combination with the Shh deletion. In our study, the DAT-IREScre mouse line has the insertion of the cre expression cassette in the 3′ UTR of the DAT gene following an IRES sequence. Heterozygous DAT-IRES-cre mice have been previously reported to not induce a significant decrease in the DAT protein level in vivo (Backman et al., 2006). Therefore, in our study, there is no or minimal decrease in DAT expression in DA neurons in our control (DATcreShh w/w) or DATcre-shh cKO (DATIREScreShh fl/fl) mice. An additional difference between our model and that of the previous study is the Shh floxed mice. We used a Shh floxed mouse line that has been widely used in many studies investigating the function of Shh in a variety of cell types (Zhao et al., 2014; Castillo-Azofeifa et al., 2017; Wagstaff et al., 2021). The Shh floxed mouse line used in the previous study is a new mouse model that has not been validated by any independent group. In this model, an IRES LacZ cassette is inserted into the Shh loci. Whether the insertion of the IRES-lacZ cassette altered the regulation of the endogenous Shh loci is unknown.
Although cholinergic interneurons only comprise about 1–2% of all striatal neurons, they extend extensive axonal arborization throughout the entire STR (Contant et al., 1996). Through a “volume transmission” mechanism, cholinergic interneurons, although sparse, play a critical modulatory role in the basal ganglia circuit and regulate movement and reward-related learning (Mallet et al., 2019). Previously, a reciprocal trophic reliance between SN DA neurons and striatal cholinergic neurons has been proposed: DA neurons provide SHH ligand for the survival of striatal cholinergic neurons, and the latter, in turn, provide GDNF for the survival of SN DA neurons (Gonzalez-Reyes et al., 2012). However, several lines of evidence in human patients and animal studies conflict with the previously proposed role of DA neuron-derived SHH ligand in maintaining cholinergic neuron survival. There have been no reports that cholinergic tone is diminished in patients with PD when at least 60–80% of SN DA neuron projections are lost, which will likely affect secreted ligands from DA neurons. Instead, anticholinergics are still widely used for the symptomatic treatment of PD, either as a monotherapy or combined with other treatments (Katzenschlager et al., 2003). Additionally, nigrostriatal denervation leads to increased acetylcholine (Ach) release, (Ding et al., 2006) and in 6-OHDA lesioned animals, cholinergic tone is also increased (Girasole and Nelson, 2015; DeBoer et al., 1993). Our data is in line with these previous findings that DA neuron projections are not a critical source for striatal interneurons. In DA neuron-specific Shh cKO mice, cholinergic neuron number does not decrease compared to WT mice in both young and old mice. Consistent with this lack of cholinergic degeneration, Chat and Vchat mRNA levels are unchanged in DATshh cKO mice compared to WT mice. Similarly, FS GABA interneurons (PV+) do not degenerate in our DATshh cKO mice. To further validate that the widely used Shh loxP mouse line results in Shh loss of function in the presence of cre activity, we generated a ubiquitous Shh KO mouse line by crossing the constitutive CMVcre line with the Shh floxed line (CMVcre-shh KO). As expected, the CMVcre-shh KO mice phenocopies the global Shh KO mouse during embryonic development, presenting deficits in neuronal tube formation. This validates cre-mediated loss of Shh function in vivo in this Shh floxed mouse line.
Another well-known effect of Shh signaling is the maintenance of adult neural stem cells. We and others have reported that abolishing Shh signaling in the mGFAPcre mouse line results in decreased adult neurogenesis in both the SVZ and SGZ, and our recent study shows that Shh KO further accelerates the age-related decline in adult neurogenesis and contributes to increased anxiety in mGFAP-smo cKO mice (Wang et al., 2022). As SN DA neuron projections have been previously reported to regulate SVZ NSCs at least partially through the release of dopamine (Garcia-Garrote et al., 2021; Hoglinger et al., 2004), we examined whether midbrain DA neuron projections could potentially provide SHH ligand to the SVZ NSCs in order to maintain their homeostasis. However, our data show that at 3 months of age, there is no difference in either the population of proliferating cells (Ki67+) or newly differentiated neuroblasts (DCX+) in the SVZ, suggesting midbrain DA neuron projections are not a critical source for maintaining Shh signaling in SVZ NSCs. One other potential source of SHH ligand for SVZ NSCs could be the cerebral spinal fluid (CSF). It has been reported previously that the transventricular delivery of SHH is essential for cerebellar ventricular zone development (Huang et al., 2010). Our data show local Shh-expressing neurons in the STR, some of which are in close proximity to the SVZ zone. These local Shh-expressing neurons could also be a potential source to maintain SVZ NSCs Shh signaling.
Our data suggest that neither striatal interneurons nor SVZ NSCs require DA neurons as a source for SHH ligand; therefore, we speculate that DA neurons are not a major source of Shh in the adult brain. A recent study has reported that striatal cholinergic neurons are dependent on Shh signaling for survival. As DA neuron Shh deletion does not affect this process, we further investigated what cell types are expressing Shh in the adult brain and whether TH + DA neurons are among them. We employed a genetic inducible fate mapping strategy (GIFM) that has recently been used to fate map the development of DA neuron progenitors (Blaess et al., 2011). In that study, Shh-expressing cells during development were labeled genetically by Shh promoter-driven cre-mediated reporter expression which allows fate mapping of Shh-producing cells and tracing of their final development into DA neurons in the ventral mesencephalon (VM). It was reported that during early development, Shh-producing cells from E7.5 up to E11.5 develop into mature DA neurons examined at P21-P30 (Blaess et al., 2011). However, at later embryonic stages such as E12.5, Shh-expressing neurons that mature into TH + cells (analyzed at E18.5) are dramatically decreased, suggesting that when VM progenitors commit to the DA fate, they might downregulate Shh expression. However, GIFM has only been used in one previous study in the adult mouse brain using the more stringent R26YFP mouse line (Ihrie et al., 2011). In addition, GIFM in the neonatal mouse brain has not been used to examine whether TH + DA neurons express Shh in the neonatal brain. Using GIFM, we investigated postnatal Shh-expressing cells at the neonatal stage (P0-P5) or in the young adult brain (P30-P35). We chose the Ai9-tdTomato as a reporter because of its robust expression in all cell types and strong fluorescence to minimize the underestimation of Shh-expressing cells. Our data shows no expression of Shh in TH + cells of perinatal or young adult mice (visualized by Shh-cre driven tdTomato expression). Sparse tdTomato + cells are present and colocalize with NeuN + neurons in the ventral midbrain, however, we did not find TH and tdTomato double-positive cells, supporting our conclusion that DA neurons do not express Shh in vivo in the neonatal or adult brain. These results agree with a previous study using a ShhcreER-R26YFP mouse line in the adult brain to examine Shh-expressing cells (YFP+) at only 4 days after TAM administration. They show that Shh-expressing cells (YFP+) are not DA neurons (TH+) (Ihrie et al., 2011). Our study uses a stronger fluorescent reporter that is more sensitive to cre activity (Stifter and Greter, 2020), and we examined both immediate (5 days after TAM) or delayed (30 days after TAM treatment) timepoints to allow for stronger expression of the fluorescent reporter. Still, our study also shows similar results supporting the hypothesis that Shh-expressing cells in the neonatal and adult mouse brain (tdTomato+) are not TH + DA neurons. Consistent with these findings, tdTomato + axon terminals do not colocalize with TH + axonal fibers in the PFC or STR. Interestingly, we identified NeuN+ and tdTomato + local neurons in the cortex and STR, supporting the hypothesis that cortical-striatal projections or local striatal interneurons could be the source for SHH ligand to support cholinergic neuron survival. Future studies using neuronal type specific- and cortical or striatal specific- Shh conditional knockout mouse models are warranted to further characterize the potential source of striatal SHH ligand from these two brain regions.
To further increase the rigor of our study, we also measured Shh mRNA levels in the SN and SHH protein levels in SN, STR, and SVZ (ELISA). Our data show that while the ELISA kit successfully detected a 50% decrease in SHH protein levels in global Shh heterozygous knockout mice, we did not detect any difference in SHH protein levels in the SN, STR, or SVZ between WT and DATcre-shh cKO mice. This is consistent with our qRT-PCR results showing no difference in the total level of Shh mRNA in the SN between WT and DATcre-shh cKO mice. These results are further supported by our data showing decreased abundance of floxed Shh intron 1 in gDNA extracted from sorted Nurr1+ DA neuron-enriched nuclei of DATcre-shh cKO mice, and publicly available scRNA-seq data showing little to no expression of Shh in mature rodent DA neurons (Kamath et al., 2022; Tiklová et al., 2019).
In summary, based on our multiple analyses, we conclude that mature DA neurons are not Shh-expressing neurons. We also conclude that autonomous Shh expression is not required for DA neuron survival, nor is DA neuron-derived SHH ligand critical for striatal cholinergic or FS GABA PV + neuron survival. These findings are consistent with previous reports of the phenotype of increased cholinergic tone in patients with PD and mouse models of PD. We also report that GDNF levels are not changed in DATcre-shh cKO mice, consistent with a lack of degeneration of midbrain DA neurons in DATcre-shh cKO mice. We propose that instead, cortical or local striatal neurons might be an important source of SHH ligand for maintaining cholinergic neuronal survival and feedback regulation on DA neuron function.
Limitations of the study
Owing to the low expression of Shh mRNA in DA neurons (demonstrated by multiple previous scRNA Seq studies referenced here), we were unable to unequivocally confirm the deletion of the floxed exon 2 specifically in DA neurons at a single cell resolution in vivo in our genetic model using RNA in-situ hybridization methods such as RNAScope. Similarly, because of the small size of the floxed genomic DNA region (exon 2), detection of the deleted floxed region in DA neurons by co-labeling fluorescent DNA in-situ hybridization and immunohistochemistry to mark DA neurons is currently unfeasible. These caveats are the main limitations of this study. However, the minimal levels of Shh mRNA in DA neurons supports our hypothesis and corroborate the lack of phenotype in DA-specific Shh cKO mice.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit Anti-Tyrosine Hydroxylase antibody | Millipore | Cat#AB152; RRID: AB_390204 |
| Mouse Anti-NeuN antibody | Millipore | Cat# MAB377; RRID:AB_2298772 |
| Goat Anti-Choline Acetyltransferase antibody | Millipore | Cat# AB144P;RRID:AB_2079751 |
| Rabbit Doublecortin Antibody | Cell Signaling | Cat# 4604; RRID:AB_561007 |
| Ki-67 Monoclonal Antibody (SolA15) | Thermo Fisher Scientific | Cat# 14-5698-82; RRID:AB_10854564 |
| Rabbit anti-Parvalbumin antibody | Swant | Cat# PV27; RRID:AB_2631173 |
| Goat Nurr1 Polyclonal antibody | R & D Systems | Car#AF2156; RRID:AB_2153894 |
| VECTASTAIN ABC-Peroxidase Kit | Vector Laboratories | Cat# PK-4001; RRID:AB_2336810 |
| Mouse Nurr1 monoclonal antibody FITC conjugated | Santa Cruz Biotechnology | Cat# SC-376984 FITC |
| Chemicals, peptides, and recombinant proteins | ||
| Tamoxifen | Sigma | Cat# T5648 |
| 3, 3′-Diaminobenzidine | Electron Microscopy Science | Cat#13080 |
| Critical commercial assays | ||
| Mouse ShhN ELISA Kit (For Lysates) | RayBio | Cat#: ELM-ShhN-CL |
| Deposited data | ||
| scRNAseq Data | Tiklová et al., 2019 | GEO: GSE116138 |
| Experimental models: Organisms/strains | ||
| Mouse B6.SJL-Slc6a3tm1.1(cre)Bkmn/J | The Jackson Laboratory | IMSR Cat# JAX:006660 RRID: IMSR_JAX:006660 |
| Mouse: B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J | The Jackson Laboratory | IMSR Cat# JAX:007909, RRID:IMSR_JAX:007909 |
| Mouse B6;129-Shhtm2Amc/J | The Jackson Laboratory | IMSR Cat# JAX:004293, RRID: MMRRC_004293-UCD |
| Mouse C57BL/6J | The Jackson Laboratoory | IMSR Cat# JAX: 000664, RRID:ISMR_JAX: 000664 |
| Oligonucleotides | ||
| For qRT-PCR sequence please see Table 1 | This paper | N/A |
| For genotyping sequence please see mouse lines in methods | The Jackson Laboratory | N/A |
| Software and algorithms | ||
| Stereo Investigator | MBF Bioscience | RRID:SCR_017667 |
| ImageJ | NIH | RRID:SCR_003070 |
| Sigma Plot software | Systat Software | RRID:SCR_003210 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yu Luo (luoy2@ucmail.uc.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Animals
All animal protocols were conducted under National Institutes Health (NIH) Guidelines using the NIH handbook Animals in Research and were approved by the Institutional Animal Care and Use Committee of University of Cincinnati. The mice were housed in the animal facility of University of Cincinnati on a 14-h light/10-h dark diurnal cycle. Food was provided ad libitum. Both sexes were used for all experiments and we did not observe sex-related differences. For confirmation of DATcre-Shh recombination by PCR and TH/tdTomato immunohistochemistry, 3mo mice were used. For unbiased stereological counts of DA neurons, TH Western blot and TH staining in the STR, 7-8mo and 20mo mice were used. For TH staining in the STR following deletion of Shh in catecholaminergic neurons, 7-8mo mice were used. For unbiased stereological counts of PV+ neurons following Shh deletion, 20mo mice were used. For counts of proliferating NSCs following Shh deletion, 3mo mice were used. For qRT-PCR and ELISA assays, 3-6mo mice were used. For validation of our Shh floxed mouse model by using a global CMV promoter, embryos at E10.5 and E13.5 were harvested. Specific information for the age of mice used for each experiment is also available in the figure legends. Mouse line DATcre-Ai9-tdTomato were generate by crossing DATIREScre knock-in mice (JAX stock number 006660), with a tdTomato reporter line (Ai9) (JAX stock number: 007909).DA neuron-specific Shh knockout mice (DATcre-shh cKO, Figure 1H) were generated by crossing floxedShhc (JAX stock number: 004,293 but backcrossed with C57BL/6J [JAX stock number: 000664] for more than 10 generations in house), with DATIREScre knock-in mice to obtain DATcre(+/−)Shh loxP/loxP (DATcre-Shh cKO mice) or DATcre (+/−)Shh wt/wt (WT control mice). THcre-shh cKO mice were generated by crossing TH-IRESCre knock-in mice (Lindeberg et al., 2004) with the the floxedShhc mouse line to generate the THcre(+/−)Shh loxP/loxP (THcre-Shh cKO mice) or THcre (+/−)Shh wt/wt (WT control mice). The CMVcre-shh KO mice were generated by crossing the CMVcre (JAX stock number: 006054) with the floxedShhc mouse line to generate the CMVcre(+)ShhloxP/loxP (CMVcre-shh KO mice) or CMVcre (+/−)Shh wt/wt (WT control mice). TheShhcreER mouse line (JAX stock number: 005623) with the tdTomato reporter line (Ai9) to generateShhcreER (+/−) Ai9 (+/−) mice. The DAT-IRES-Cre gene was genotyped using primers (5′ TGG CTG TTG GTG TAA AGT GG -3′; 5′ GGA CAG GGA CAT GGT TGA CT -3′; 5′ CCA AAA GAC GGC AAT ATG GT -3′). tdTomato were genotyped using primers: (5′ AAG GGA GCT GCA GTG GAG TA -3′; 5′ CCG AAA ATC TGT GGG AAG TC -3′; 5' CTG TTC CTG TAC GGC ATG G -3′; 5' GGC ATT AAA GCA GCG TAT CC -3′). Shh floxed alleles were genotyped by the primer set (5′-GGA CAC CAT TCT ATG CAG GG -3′; 5′ TCT CTG CCA GGC TTG TCT GG -3′). TH-IRES-cre allele was genotyped using primers (5′ CTT TCC TTC CTT TAT TGA GAT -3′; 5′-CAC CCT GAC CCA AGC ACT-3′). CMVcre allele was genotyped using (5′ GGT TAG CAC CGC AGG TGT AG -3′; 5′ CTA ATC GCC ATC TTC CAG CAG -3′).ShhcreER allele was genotyped using (5′ CTA GGC CAC AGA ATT GAA AGA TCT -3′; 5′ GTA GGT GGA AAT TCT AGC ATC ATC C -3′; 5′ ATA CCG GAG ATC ATG CAA GC -3′; 5′ AGG TGG ACC TGA TCA TGG AG -3′).Shh delta band detection forShh recombined gene (5′GCC AGA AGT CCT GTC TCA CC -3′; 5′ CAT GGT CCT TGG TGG TCT CT -3′).
Tamoxifen treatment in vivo
ShhCreERT2-R26R-Ai9 tdTomato mice (P0 or 1 month old) were given tamoxifen dissolved in 10% EtOH/90% sunflower oil by direct stomach injection (Pitulescu et al., 2010) (for P0-P5 neonatal pups) or gavage feeding (for 1 month old mice) at a dose of 180 mg/kg daily for 5 consecutive days. This dosing regimen was previously demonstrated to provide maximal recombination with minimal mortality and successfully monitored the activated recombined alleles in our previous studies (Lagace et al., 2007; Jin et al., 2015; Li et al., 2010). We harvested at two time points after TAM treatment: immediately after the 5 day TAM treatment or 25 days after the last TAM treatment day for Shh-expressing cell fate mapping.
Method details
Immunohistochemistry
Mice were perfused transcardially with cold phosphate buffer followed by a solution of 4% paraformaldehyde (PFA, pH 7.2) in 0.1 M phosphate buffer (pH 7.2). Brains were removed from the skull, post-fixed in 4% PFA overnight at 4°C, and sequentially transferred to 20 and 30% sucrose in 0.1 M phosphate buffer (pH 7.2) solutions overnight. Brains were frozen on dry ice and sectioned on a cryostat to obtain coronal sections of 30 μm in thickness. The sections were then incubated with blocking buffer (4% bovine serum albumin and 0.3% Triton x-100 in 0.1M PB) for one hour at room temperature. The primary antibodies were prepared in the blocking buffer and the sections were incubated in the solution overnight: Rabbit anti-Tyrosine Hydroxylase (1:1000, Millipore), rabbit anti-Double-Cortin (1:1000, Cell Signaling), rat anti-Ki67 (1:200, Invitrogen), goat anti-Choline Acetyltransferase (1:1000, Millipore), mouse anti-NeuN (1:500, Millipore Sigma) and rabbit anti-parvalbumin (1:1000, Swant). After incubation with primary antibody solution for 18–48 hours, the sections were washed in 0.1 M phosphate buffer (pH 7.2), and incubated for 4 hours at room temperature in diluted secondary antibody prepared with blocking solution (secondary antibody conjugated with Alexa 488 or Alexa 555, 1:1000; Life Technologies, Carlsbad, CA, USA). The slides were then washed with phosphate buffer (pH 7.2) and cover-slipped. Images were acquired using a Zeiss LSM 710 Live DUO Leica or Stellaris 8 confocal microscope. Omission of primary or secondary antibodies resulted in no staining and served as negative controls. To quantify total number of DCX+ and Ki67+, SVZ region was imaged at 100 × magnification for each animal. 5 sections containing SVZ per animal were imaged from all animals for quantification of DCX and Ki67 positive cells. Counts were averaged for each mouse and used as a single data point for figures and statistical analysis. TH, CHAT and PV positive neurons were quantified using unbiased stereology method with DAB immunostaining as described below.
Immunohistochemistry with DAB
Mice were perfused transcardially with cold phosphate buffer followed by a solution of 4% paraformaldehyde (PFA, pH 7.2) in 0.1 M phosphate buffer (pH 7.2). Brains were removed from the skull, post-fixed in 4% PFA overnight at 4°C, and sequentially transferred to 20 and 30% sucrose in 0.1 M phosphate buffer (pH 7.2) solutions overnight. Brains were frozen on dry ice and sectioned on a cryostat to obtain coronal sections of 30 μm in thickness. The sections were then incubated with H2O2 in phosphate buffer for 30 minutes, followed by an incubation in blocking buffer (4% bovine serum albumin and 0.3% Triton x-100 in 0.1M PB) for one hour. The primary antibodies were prepared in the blocking buffer and the sections were incubated in the solution overnight: Rabbit anti-Tyrosine Hydroxylase (1:1000, Millipore), goat anti-Choline Acetyltransferase (1:1000, Millipore) or rabbit anti-parvalbumin (1:1000, Swant). After incubation with primary antibody solution for 24 hours, the sections were washed and incubated for 1 hour at room temperature in diluted secondary antibody prepared with blocking solution. The sections were rinsed in 0.1M PB and incubated in biotinylated antibody anti-rabbit or anti-goat (1 : 200; Vector Laboratories, Burlingame CA, USA) for 1 hour, followed by incubation for 1 hour with avidin-biotin-horseradish peroxidase complex. Staining was developed with 2,3′ diaminobenzidine tetrahydrochloride (DAB 0.5 mg/mL in 50 mM Tris-HCl buffer 7.4). All staining were carried out in one experiment to avoid differences caused by batch effect. Brain sections from each mouse were developed for the exact same duration in the DAB reaction. TH+ fiber density in the striatum was quantified from at least 4 sections that are consistent among all animals using the same method as described in our previous publication (Filichia et al., 2016).
Unbiased stereological analysis
Unbiased stereological counts of TH-positive (TH+) neurons within the SN pas compacta were performed using stereological principles and analyzed with StereoInvestigator software (Microbrightfield, Williston, VT, USA), as previously described(Filichia et al., 2016). Optical fractionator sampling was carried out on a Leica DM5000B microscope (Leica Microsystems, Bannockburn, IL, USA) equipped with a motorized stage and Lucivid attachment (40× objective). Midbrain DA groups were outlined on the basis of TH immunolabelling, with reference to a coronal atlas of the mouse brain (Franklin and Paxinos, 1997). 4 complete sets of brain sections were collected for each brain at 40μm thickness. One set of 8–10 brain sections that expand the whole SN or striatum were analyzed for each mouse. For each tissue section analyzed, section thickness was assessed in each sampling site and guard zones of 2.5 μm were used at the top and bottom of each section. Pilot studies were used to determine suitable counting frame and sampling grid dimensions prior to counting. The following stereologic parameters were used in the final study: grid size, (X) 220 μm, (Y) 166 μm; counting frame, (X) 68.2 μm, (Y) 75 μm, depth was 20 μm. Gundersen coefficients of error for m = 1 were all less than 0.10. Stereologic estimations were performed with the same parameters in the SN and pas compacta and VTA of WT or DATcre-Shh KO mice. For CHAT+ or PV+ neurons, striatum was outline based on anatomical landmarks, the following stereologic parameters were used in the final study: grid size, (X) 570 μm, (Y) 570 μm; counting frame, (X) 240 μm, (Y) 240 μm, depth was 20 μm. Gundersen coefficients of error for m = 1 were all less than 0.10.
Western-blot analysis
Brain tissue of substantia nigra and striatum were harvested and lysed in the following lysis buffer (10 mM HEPES-NaOH, pH 7.5, 150 mM NaCl, 1 mM EGTA, 1% Triton X-100, 1 : 300 protease inhibitor cocktail, 1 : 300 phosphatase inhibitor cocktail). After 20 min of incubation on ice, homogenates were spun at 12 000 g for 20 min at 4°C. The supernatants correspond to the total cell lysates. Protein concentrations were determined by Bradford assay. 30 μg of proteins were resuspended in Laemmli buffer, loaded on sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes. Membranes were probed with the indicated antibodies followed by visualization by enhanced chemiluminescence, and then were quantitated using NIH ImageJ software (NIH, Bethesda, MD, USA). The antibodies used in this study was Rabbit anti-Tyrosine Hydroxylase (1:1000, Millipore) and Mouse anti-beta-actin (1:10000, Sigma Aldrich).
qRT-PCR
RNA expression levels were quantified in each animal groups by qRT-PCR as described in our previous publication (Sheng et al., 2016). Tissues and RNA were extracted form SN or STR using the same method described previously(Luo et al., 2010). Total RNA (1 ug) was treated with RQ-1 Rnase-free Dnase I and reverse transcribed into cDNA using random hexamers by Superscript III reverse transcriptase (Life Sciences). cDNA levels for Hmbs (hydroxymethylbilane synthase) and various target genes were determined, using specific primer/probe sets by quantitative RT-PCR using a Roche Light Cycler II 480. Relative expression level was calculated using the delta Ct method compared to Hmbs as a reference gene. Primers and carboxyfluorescein (FAM) labeled probes used in the quantitative RT-PCR for each gene are listed in Table 1.
ELISA
SHH levels in tissue samples were determined using a RayBiotech ELISA kit (catalog #: ELM-ShhN-CL). Mouse brains were harvested and quickly frozen. Tissue punchouts were collected from different brain regions (SN, STR and SVZ) using cryostat sectioning and Leica brain punch tissue kit. The punched tissues were sonicated with 50 μL of lysis buffer included in the kit with added protease inhibitors and then centrifuged. Supernatants were collected and total protein concentration was determined by BCA assay (Pierce). Tissue lysates were then analyzed for SHH protein levels per the manufacturer’s instructions. SHH concentrations were calculated based on standard curves produced by recombinant SHH protein standards provided in the kit and were reported in pg/mL. Based on the total protein concentration of each sample, the final SHH concentration in each lysate was reported as ng/mg total protein. Duplicates from the tissue lysates were analyzed for each sample and the average was presented as a single data point for each mouse in the figure and for statistical analysis.
scRNA-seq data analysis
scRNA-seq data from Tiklova et al. was analyzed for Shh expression in DA neurons at different developmental stages. Data from this previous study [22] was reanalyzed using 2D shiny platform (available publicly at https://rshiny.nbis.se/shiny-server-apps/shiny-apps-scrnaseq/perlmannlab_mouseDA/) with permission from Dr. Thomas Perlmann.
FANS enrichment of DA neuron nuclei
The SN from two WT and three DATcre-shh cKO mice were microdissected and dissociated in 0.5mL detergent lysis buffer (low sucrose buffer, 10% Triton X-100) by dounce homogenization as previously described (Matson et al., 2018). A Wheaton 1mL homogenizer was used with ∼10 dounces of the loose pestle followed by ∼10 dounces of the tight pestle until no large tissue chunks remained. A 40mm strainer was wetted using 1mL of low sucrose buffer (0.32M sucrose, 10mM HEPES, 5mM CaCl2, 3mM MgAc, 0.1mM EDTA, 1mM DTT, Water fill to 25mL). Samples were filtered through the strainer using 1mL of low sucrose buffer. An additional 1mL of low sucrose buffer was used to wash the strainer. The dissociated nuclei were pooled into two samples: the two WT SN into one WT sample, and the three cKO SN into one cKO sample. Samples were centrifuged at 3,200xg for 10 min at 4C. Following centrifugation, the supernatant was decanted and the samples were resuspended in low sucrose buffer. Samples were then sonicated on the lowest setting for 15s on ice. Using a serological pipette, 12.5mL of density sucrose buffer (1M sucrose, 10mM HEPES, 3mM MgAc, 1mM DTT, Water fill to 25mL) was layered underneath each sample. The samples were then centrifuged at 3200xg for 20 min at 4C. Following centrifugation, the supernatant was decanted, and the samples were resuspended in 0.6mL of resuspension buffer (0.4 mg/mL BSA in 1xPBS). We then performed FANS using an antibody against Mouse anti Nurr1 (1:50, 1hr incubation at 4C, Santa Cruz) to enrich for DA neuron nuclei as previously described [23]. Nuclei were sorted into three separate populations: high, medium, and low expression of Nurr1. An unstained sample was used to set up the gate parameters. For the WT sample, 148,412 nuclei were sorted into the low expression group, 17,808 nuclei into the medium expression group, and 2,602 into the high expression group. For the cKO sample, 202,403 nuclei were sorted into the low expression group, 16,839 nuclei into the medium expression group, and 1,673 nuclei into the high expression group. Nuclei were sorted directly into lysis buffer and gDNA was isolated from each sample (Zymo Research Quick DNA MicroPrep). Abundance of floxed Shh intron 1 gDNA was determined by real time-qPCR. gDNA abundance was analyzed using the delta-delta-CT method. Floxed Shh intron 1 abundance was normalized to unfloxed Shh exon 3 levels in each gDNA sample. WT normalized Shh floxed intron 1 levels were normalized to 1 for each group (low, medium, or high Nurr1 expression), and normalized cKO Shh floxed intron 1 levels were compared to WT levels of the corresponding group.
Locomotor function
Mice motor activities were assessed using automated open field Accuscan activity monitors (Columbus, OH, USA) in different age groups of mice that are littermates of control or DATcre-shh cKO mice as previously described (Shen et al., 2008). Different cohorts of littermates were analyzed as they age and data were pooled together. N number for each time points were listed in the figure. No mice were excluded. There are 16 horizontal and 8 vertical infrared sensors (interval 2.5 cm) in each chamber. Each mouse was put into a 42 × 42 × 31 cm Plexiglas open box for 1 hour with food and water supply. To avoid observer bias, this locomotor test was automatically monitored by the computer and software. Locomotor activity was calculated by automated Fusion software (Accuscan, Columbus, OH, USA). Total distance traveled (cm, the distance traveled by the animals) was reported for the 1-hour time period.
Quantification and statistical analysis
All studies were analyzed using SigmaStat software. Results are expressed by mean±SEM of the indicated number of experiments. Specific information for each experiment can be found in the figure legends. In general average or stereological results for each animal was used a single data point in statistical analysis. Statistical analysis was performed using the Student’s t test, and one- or two-way analysis of variance (ANOVA), as appropriate, with Tukey post hoc tests or Bonferroni post hoc tests for repeated behavioral measurements. A p value equal to or less than 0.05 was considered significant.
Acknowledgments
The TH-IRES-Cre mice were obtained originally from Dr. Ted Ebendel and we would like to thank Dr. Ebendel for sharing this mouse line. We would like to thank Dr. Thomas Perlmann for permission to generate Figure 10 from their previously published scRNAseq data (Tiklová et al., 2019). We would also like to thank Dr. Chet Closson from UC live microscopy core for assistance with confocal microscope images. Dr. Yu Luo is supported by NIH grants (R01NS091213 and R01NS107365). Research reported in this publication was also supported by the Office Of The Director, National Institutes Of Health of the National Institutes of Health under Award Number S10OD030402. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
YL conceptualized the study. FT, EW, and YL designed the experiments. FT performed most of the unbiased stereological counts and behavioral tests. EW performed the scRNAseq data analysis, RT-PCR gene analysis, and FACS-enriched qRT-PCR. TL and NF performed Western blot and ELISA experiments. All authors recorded and analyzed data with help from YL. FT, EW, and YL drafted the article. All authors read, edited, and approved the final version of the article.
Declaration of interests
The authors declare no competing interests.
Published: November 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105278.
Supplemental information
Data and code availability
-
•
No new Single-cell RNA-seq data was generated in this study. Plot in Figure 10 was generated using scRNA-seq data obtained from Tiklová et al., (2019), a previous study with permission from Dr. Thomas Perlmann (GEO: GSE116138). Microscopy data and behavioral test data reported in this paper will be shared by the lead contact upon request.
-
•
No original code was generated in this study.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Backman C.M., Malik N., Zhang Y., Shan L., Grinberg A., Hoffer B.J., Westphal H., Tomac A.C. Characterization of a mouse strain expressing Cre recombinase from the 3' untranslated region of the dopamine transporter locus. Genesis. 2006;44:383–390. doi: 10.1002/dvg.20228. [DOI] [PubMed] [Google Scholar]
- Blaess S., Bodea G.O., Kabanova A., Chanet S., Mugniery E., Derouiche A., Stephen D., Joyner A.L. Temporal-spatial changes in Sonic Hedgehog expression and signaling reveal different potentials of ventral mesencephalic progenitors to populate distinct ventral midbrain nuclei. Neural Dev. 2011;6:29. doi: 10.1186/1749-8104-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J., Therond P.P. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- Castillo-Azofeifa D., Losacco J.T., Salcedo E., Golden E.J., Finger T.E., Barlow L.A. Sonic hedgehog from both nerves and epithelium is a key trophic factor for taste bud maintenance. Development. 2017;144:3054–3065. doi: 10.1242/dev.150342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contant C., Umbriaco D., Garcia S., Watkins K.C., Descarries L. Ultrastructural characterization of the acetylcholine innervation in adult rat neostriatum. Neuroscience. 1996;71:937–947. doi: 10.1016/0306-4522(95)00507-2. [DOI] [PubMed] [Google Scholar]
- Deboer P., Abercrombie E.D., Heeringa M., Westerink B.H. Differential effect of systemic administration of bromocriptine and L-dopa on the release of acetylcholine from striatum of intact and 6-OHDA-treated rats. Brain Res. 1993;608:198–203. doi: 10.1016/0006-8993(93)91459-6. [DOI] [PubMed] [Google Scholar]
- Ding J., Guzman J.N., Tkatch T., Chen S., Goldberg J.A., Ebert P.J., Levitt P., Wilson C.J., Hamm H.E., Surmeier D.J. RGS4-dependent attenuation of M4 autoreceptor function in striatal cholinergic interneurons following dopamine depletion. Nat. Neurosci. 2006;9:832–842. doi: 10.1038/nn1700. [DOI] [PubMed] [Google Scholar]
- Filichia E., Hoffer B., Qi X., Luo Y. Inhibition of Drp1 mitochondrial translocation provides neural protection in dopaminergic system in a Parkinson's disease model induced by MPTP. Sci. Rep. 2016;6:32656. doi: 10.1038/srep32656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin K.B.J., Paxinos G. The Mouse Brain in Stereotaxic Coordinates. Academic Press; 1997. San Diego. [Google Scholar]
- Garcia-Garrote M., Parga J.A., Labandeira P.J., Labandeira-Garcia J.L., Rodriguez-Pallares J. Dopamine regulates adult neurogenesis in the ventricular-subventricular zone via dopamine D3 angiotensin type 2 receptor interactions. Stem Cell. 2021;39:1778–1794. doi: 10.1002/stem.3457. [DOI] [PubMed] [Google Scholar]
- Girasole A.E., Nelson A.B. Probing striatal microcircuitry to understand the functional role of cholinergic interneurons. Mov. Disord. 2015;30:1306–1318. doi: 10.1002/mds.26340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giros B., Jaber M., Jones S.R., Wightman R.M., Caron M.G. Hyperlocomotion and indifference to cocaine and amphetamine in mice lacking the dopamine transporter. Nature. 1996;379:606–612. doi: 10.1038/379606a0. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reyes L.E., Verbitsky M., Blesa J., Jackson-Lewis V., Paredes D., Tillack K., Phani S., Kramer E.R., Przedborski S., Kottmann A.H. Sonic hedgehog maintains cellular and neurochemical homeostasis in the adult nigrostriatal circuit. Neuron. 2012;75:306–319. doi: 10.1016/j.neuron.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall F.S., Sora I., Uhl G.R. Sex-dependent modulation of ethanol consumption in vesicular monoamine transporter 2 (VMAT2) and dopamine transporter (DAT) knockout mice. Neuropsychopharmacology. 2003;28:620–628. doi: 10.1038/sj.npp.1300070. [DOI] [PubMed] [Google Scholar]
- Hoglinger G.U., Rizk P., Muriel M.P., Duyckaerts C., Oertel W.H., Caille I., Hirsch E.C. Dopamine depletion impairs precursor cell proliferation in Parkinson disease. Nat. Neurosci. 2004;7:726–735. doi: 10.1038/nn1265. [DOI] [PubMed] [Google Scholar]
- Huang X., Liu J., Ketova T., Fleming J.T., Grover V.K., Cooper M.K., Litingtung Y., Chiang C. Transventricular delivery of Sonic hedgehog is essential to cerebellar ventricular zone development. Proc. Natl. Acad. Sci. USA. 2010;107:8422–8427. doi: 10.1073/pnas.0911838107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes M., Porter J.A., Chiang C., Chang D., Tessier-Lavigne M., Beachy P.A., Rosenthal A. Induction of midbrain dopaminergic neurons by Sonic hedgehog. Neuron. 1995;15:35–44. doi: 10.1016/0896-6273(95)90062-4. [DOI] [PubMed] [Google Scholar]
- Ihrie R.A., Shah J.K., Harwell C.C., Levine J.H., Guinto C.D., Lezameta M., Kriegstein A.R., Alvarez-Buylla A. Persistent sonic hedgehog signaling in adult brain determines neural stem cell positional identity. Neuron. 2011;71:250–262. doi: 10.1016/j.neuron.2011.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Raviv N., Barnett A., Bambakidis N.C., Filichia E., LUO Y. The shh signaling pathway is upregulated in multiple cell types in cortical ischemia and influences the outcome of stroke in an animal model. PLoS One. 2015;10:e0124657. doi: 10.1371/journal.pone.0124657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath T., Abdulraouf A., Burris S.J., Langlieb J., Gazestani V., Nadaf N.M., Balderrama K., Vanderburg C., Macosko E.Z. Single-cell genomic profiling of human dopamine neurons identifies a population that selectively degenerates in Parkinson's disease. Nat. Neurosci. 2022;25:588–595. doi: 10.1038/s41593-022-01061-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzenschlager R., Sampaio C., Costa J., Lees A. Anticholinergics for symptomatic management of Parkinson's disease. Cochrane Database Syst. Rev. 2003:CD003735. doi: 10.1002/14651858.CD003735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komada M., Saitsu H., Kinboshi M., Miura T., Shiota K., Ishibashi M. Hedgehog signaling is involved in development of the neocortex. Development. 2008;135:2717–2727. doi: 10.1242/dev.015891. [DOI] [PubMed] [Google Scholar]
- Lagace D.C., Whitman M.C., Noonan M.A., Ables J.L., Decarolis N.A., Arguello A.A., Donovan M.H., Fischer S.J., Farnbauch L.A., Beech R.D., et al. Dynamic contribution of nestin-expressing stem cells to adult neurogenesis. J. Neurosci. 2007;27:12623–12629. doi: 10.1523/JNEUROSCI.3812-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai K., Kaspar B.K., Gage F.H., Schaffer D.V. Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat. Neurosci. 2003;6:21–27. doi: 10.1038/nn983. [DOI] [PubMed] [Google Scholar]
- Lewis P.M., Dunn M.P., McMahon J.A., Logan M., Martin J.F., St-Jacques B., McMahon A.P. Cholesterol modification of sonic hedgehog is required for long-range signaling activity and effective modulation of signaling by Ptc1. Cell. 2001;105:599–612. doi: 10.1016/s0092-8674(01)00369-5. [DOI] [PubMed] [Google Scholar]
- Li L., Harms K.M., Ventura P.B., Lagace D.C., Eisch A.J., Cunningham L.A. Focal cerebral ischemia induces a multilineage cytogenic response from adult subventricular zone that is predominantly gliogenic. Glia. 2010;58:1610–1619. doi: 10.1002/glia.21033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeberg J., Usoskin D., Bengtsson H., Gustafsson A., Kylberg A., Söderström S., Ebendal T. Transgenic expression of Cre recombinase from the tyrosine hydroxylase locus. Genesis. 2004;40:67–73. doi: 10.1002/gene.20065. [DOI] [PubMed] [Google Scholar]
- Luo Y., Wang Y., Kuang S.Y., Chiang Y.H., Hoffer B. Decreased level of Nurr1 in heterozygous young adult mice leads to exacerbated acute and long-term toxicity after repeated methamphetamine exposure. PLoS One. 2010;5:e15193. doi: 10.1371/journal.pone.0015193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet N., Leblois A., Maurice N., Beurrier C. Striatal cholinergic interneurons: how to elucidate their function in Health and disease. Front. Pharmacol. 2019;10:1488. doi: 10.3389/fphar.2019.01488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson K.J.E., Sathyamurthy A., Johnson K.R., Kelly M.C., Kelley M.W., Levine A.J. Isolation of adult spinal cord nuclei for massively parallel single-nucleus RNA sequencing. J. Vis. Exp. 2018:58413. doi: 10.3791/58413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao N., Wang M., Ott J.A., D'alessandro J.S., Woolf T.M., Bumcrot D.A., Mahanthappa N.K., Pang K. Sonic hedgehog promotes the survival of specific CNS neuron populations and protects these cells from toxic insult in vitro. J. Neurosci. 1997;17:5891–5899. doi: 10.1523/JNEUROSCI.17-15-05891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-de San Luis C., Sanchez-Garcia M.A., Nieto-Gonzalez J.L., García-Junco-Clemente P., Montero-Sanchez A., Fernandez-Chacon R., Pascual A. Substantia nigra dopaminergic neurons and striatal interneurons are engaged in three parallel but interdependent postnatal neurotrophic circuits. Aging Cell. 2018;17:e12821. doi: 10.1111/acel.12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova R., Garcia A.D.R., Joyner A.L. Titration of GLI3 repressor activity by sonic hedgehog signaling is critical for maintaining multiple adult neural stem cell and astrocyte functions. J. Neurosci. 2013;33:17490–17505. doi: 10.1523/JNEUROSCI.2042-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitulescu M.E., Schmidt I., Benedito R., Adams R.H. Inducible gene targeting in the neonatal vasculature and analysis of retinal angiogenesis in mice. Nat. Protoc. 2010;5:1518–1534. doi: 10.1038/nprot.2010.113. [DOI] [PubMed] [Google Scholar]
- Shen H., Luo Y., Kuo C.C., Wang Y. BMP7 reduces synergistic injury induced by methamphetamine and ischemia in mouse brain. Neurosci. Lett. 2008;442:15–18. doi: 10.1016/j.neulet.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y., Filichia E., Shick E., Preston K.L., Phillips K.A., Cooperman L., Lin Z., Tesar P., Hoffer B., Luo Y. Using iPSC-derived human DA neurons from opioid-dependent subjects to study dopamine dynamics. Brain Behav. 2016;6:e00491. doi: 10.1002/brb3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielewoy C., Biala G., Roubert C., Hamon M., Betancur C., Giros B. Hypolocomotor effects of acute and daily d-amphetamine in mice lacking the dopamine transporter. Psychopharmacology. 2001;159:2–9. doi: 10.1007/s002130100901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielewoy C., Roubert C., Hamon M., Nosten-Bertrand M., Betancur C., Giros B. Behavioural disturbances associated with hyperdopaminergia in dopamine-transporter knockout mice. Behav. Pharmacol. 2000;11:279–290. doi: 10.1097/00008877-200006000-00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stifter S.A., Greter M. STOP floxing around: specificity and leakiness of inducible Cre/loxP systems. Eur. J. Immunol. 2020;50:338–341. doi: 10.1002/eji.202048546. [DOI] [PubMed] [Google Scholar]
- Tiklová K., Björklund Å.K., Lahti L., Fiorenzano A., Nolbrant S., Gillberg L., Volakakis N., Yokota C., Hilscher M.M., Hauling T., et al. Single-cell RNA sequencing reveals midbrain dopamine neuron diversity emerging during mouse brain development. Nat. Commun. 2019;10:581. doi: 10.1038/s41467-019-08453-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi K., Shults C.W. Intrastriatal injection of sonic hedgehog reduces behavioral impairment in a rat model of Parkinson's disease. Exp. Neurol. 2002;173:95–104. doi: 10.1006/exnr.2001.7825. [DOI] [PubMed] [Google Scholar]
- Wagstaff L.J., Gomez-Sanchez J.A., Fazal S.V., Otto G.W., Kilpatrick A.M., Michael K., Wong L.Y., Ma K.H., Turmaine M., Svaren J., et al. Failures of nerve regeneration caused by aging or chronic denervation are rescued by restoring Schwann cell c-Jun. Elife. 2021;10:e62232. doi: 10.7554/eLife.62232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Ware K., Bedolla A., Allgire E., Turcato F.C., Weed M., Sah R., Luo Y. Disruption of sonic hedgehog signaling accelerates age-related neurogenesis decline and abolishes stroke-induced neurogenesis and leads to increased anxiety behavior in stroke mice. Transl. Stroke Res. 2022;13:830–844. doi: 10.1007/s12975-022-00994-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W., Shimamura K., Rubenstein J.L., Hynes M.A., Rosenthal A. FGF and Shh signals control dopaminergic and serotonergic cell fate in the anterior neural plate. Cell. 1998;93:755–766. doi: 10.1016/s0092-8674(00)81437-3. [DOI] [PubMed] [Google Scholar]
- Zhao H., Feng J., Seidel K., Shi S., Klein O., Sharpe P., Chai Y. Secretion of shh by a neurovascular bundle niche supports mesenchymal stem cell homeostasis in the adult mouse incisor. Cell Stem Cell. 2014;14:160–173. doi: 10.1016/j.stem.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Pace J., Filichia E., Lv T., Davis B., Hoffer B., Selman W., Luo Y. Effect of the sonic hedgehog receptor smoothened on the survival and function of dopaminergic neurons. Exp. Neurol. 2016;283:235–245. doi: 10.1016/j.expneurol.2016.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
No new Single-cell RNA-seq data was generated in this study. Plot in Figure 10 was generated using scRNA-seq data obtained from Tiklová et al., (2019), a previous study with permission from Dr. Thomas Perlmann (GEO: GSE116138). Microscopy data and behavioral test data reported in this paper will be shared by the lead contact upon request.
-
•
No original code was generated in this study.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.