Abstract
Background and Aims
To determine prevalence and clinical utility of pathogenic germline variants (PGV) in gastric and esophageal cancer patients using universal genetic testing approach.
Methods
We undertook a prospective study of germline sequencing using an > 80 gene next-generation sequencing platform among patients with gastric and esophageal cancers receiving care at Mayo Clinic Cancer Center between April 1, 2018, and March 31, 2020. Patients were not selected based on cancer stage, family history of cancer, ethnicity, or age. Family cascade testing was offered at no cost.
Results
A total of 96 patients were evaluated. Median age was 66 years, 80.2% were male, 89.6% were white. Nearly 39% of the cohort had esophageal cancer, 35.4% gastric cancer and 26% gastroesophageal junction cancers. Approximately half (52%) of the patients had metastatic disease. Pathogenic germline variants (PGV) were detected in 15.6% (n = 15) patients. The prevalence of PGV was 10.8% in esophageal cancer, 17.6% in gastric cancer and 20% in gastroesophageal cancer. Eighty percent of patients with a positive result would not have been detected by screening with standard guidelines for genetic testing. Most PGV detected included genes with high and moderate penetrance related to DNA damage response including BRCA1, BRCA2, PALB2 and ATM.
Conclusions
Universal multi-gene panel testing in gastric and esophageal cancers was associated with detection of heritable mutations in 15% of patients. The majority of PGV would not be detected with current screening guidelines and are related to DNA damage response.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10620-022-07387-x.
Keywords: Gastric cancer, Esophageal cancer, Germline testing, BRCA
Introduction
Gastric and esophageal cancers jointly cause significant morbidity and mortality. Worldwide it was estimated that there were more than 1 million new cases of gastric cancer in 2020 [1]. In the same period, it is estimated that there were 768,793 deaths related to the disease, ranking third in cancer-related deaths in the world [1]. However, in the USA and Western Europe, the incidence of gastric cancer has decreased in the last decade [2]. Gastric cancer is often related to environmental factors including Helicobacter Pylori infection, smoking and diet [3–5] and it is estimated that only 5% are related to inherited cancer predisposition syndromes [2]. National Comprehensive Cancer Network (NCCN) guidelines recommend that genetic risk assessment and counseling should be based on age of diagnosis and number of relatives affected with gastric cancer or a family history of juvenile polyps or Lynch Syndrome [2]. One of the most common hereditary syndromes associated with gastric cancer is hereditary diffuse gastric cancer, where 30–50% of the families carry germline aberrations in the tumor suppressor gene CDH1 [6].
The decrease in incidence of gastric cancers in the USA and Western Europe are related to changes in food conservation and preservatives, diet, hygiene, and eradication of Helicobacter Pylori [2, 7, 8]. Interestingly, the incidence of esophageal and gastroesophageal junction (GEJ) cancers in the USA is on the rise [2]. Globally, esophageal and GEJ tumors have high incidence in middle- and low-income countries, with an estimated incidence of 604,100 new cases in 2020 [1]. Smoking and alcohol are the most common risk factors associated with squamous esophageal carcinoma, with high incidence in Eastern Europe and Asia [9, 10], whereas, esophageal and GEJ adenocarcinoma are more common in North America and Western Europe, generally related to obesity and gastroesophageal reflux disease possibly associated with Barrett’s esophagus [11–13]. Several familial high-risk syndromes may be related to the development of esophageal cancers; however, no specific guidelines or recommendations for hereditary cancer risk assessment for these cancers are available [2].
Little is known about germline genetic testing in unselected patients with gastric cancer and esophageal cancer. Expanded germline testing in these patients could improve the current understanding of the disease and help establish specific guidelines. In this article, we report the findings and clinical characteristics of a multi-center prospective cohort of gastric and esophageal cancer patients who underwent germline testing with next-generation sequencing using a > 80 gene panel [14].
Methods
Patient Selection: Inclusion and Exclusion Criteria
From April 1, 2018 through March 31, 2020, a total of 2,984 adult (18 years of age or older) patients with a new or active diagnosis of cancer were recruited from medical oncology, radiation oncology, dermatology and surgical oncology clinics at any of the four Mayo Clinic destination Cancer Centers in Phoenix, AZ; Jacksonville, FL or Rochester, MN, and a community oncology practice (Eau Claire, WI) as part of the Interrogating Cancer Etiology Using Proactive Genetic Testing (INTERCEPT) program [14]. Patients undergoing surveillance post-curative cancer or with hematological malignances were excluded. Research coordinators of each site recruited patients using central lists of daily oncology clinic visits. Germline sequencing using a next-generation sequencing (NGS) panel of 83 genes (84 genes as of July 2019) was offered at no cost for all participants and had disclosure of results. This panel included all cancer-predisposing genes identified in the American College of Medical Genetics and Genomics guidelines. Patients in this study were not selected based on clinical characteristics, including family or personal history of cancer, cancer type, multifocal tumor, stage of disease, ethnicity or age at diagnosis. Patients with a previously established molecular diagnosis of a cancer genetic syndrome were excluded from the INTERCEPT study; however, none of the patients in this cohort had a prior genetic diagnosis. From this cohort, 96 patients with a diagnosis of gastric, esophageal, and GEJ cancers were analyzed in this study. Detailed methodology is provided in the following manuscript [14].
Prior to undergoing genetic testing, all patients viewed a standard pretest education video and were offered additional pretest genetic counseling if they desired. All test results were reviewed by a certified genetic counselor or physicians with expertise in cancer genetics. Individuals with pathogenic or likely pathogenic variants were invited for genetic counseling. Family cascade testing at no additional cost was offered.
Clinical, demographic, family history, treatment types and clinical outcome information were collected on all patients in this study either from medical records or self-administered electronic questionnaires for family pedigree information.
This study was approved by the Mayo Clinic Institutional Review Board (IRB 18–000,326). All patients provided written informed consent. Data were de-identified except to investigators of the study.
Sequencing, Variant Calling and Result Reporting
All patients underwent next-generation sequencing (NGS) germline genetic testing with a multi-gene cancer panel of 83 genes (84 genes as of July 2019) on the Invitae Multicancer panel. Full gene sequencing, deletion/duplication analysis, copy number variant detection and variant interpretation were performed at Invitae (San Francisco, CA), as previously described [14]. All patients had their variant findings source verified and confirmed by independent review of the test results by a medical geneticist. Pathogenic germline variants (PGV) were classified as highly (relative risk [RR] > 4), intermediate (RR 2–4) or low (RR < 2) penetrant, recessive or of uncertain clinical actionability based on disease risks and prior modeling. Investigators and treating clinicians were not blinded to the genetic testing results which were returned in real time to inform clinical management.
Statistical Analysis
Descriptive statistics for demographic, clinical and treatment-related characteristics of the cohort were examined. Survival time was calculated as the number of months from the date of cancer diagnosis to the date of death or last follow-up. Due to the small group sizes and clinical differences among these cancer sites, formal statistical tests were not performed. Rates of detection of clinically actionable findings using 2018 and 2020 NCCN guidelines were calculated. Rates of uptake of family variant testing (FVT) and mutation rates of tested family members were examined.
Results
Cohort Characteristics
Demographic distribution of the cohort is shown in Table 1, stratified by primary tumor location. Overall, 80.2% of included patients are male, 89.6% white, the median age at diagnosis was 66 years. More than half (55.2%) were smokers, 49% had hypertension, 20.8% had a history of diabetes and 15.6% had a body mass index higher than 30 (Supplementary table 1).
Table 1.
Cohort characteristics
| Esophageal (N = 37) | Gastric (N = 34) | GEJ (N = 25) | Total (N = 96) | |
|---|---|---|---|---|
| Region | ||||
| Midwest | 7 (18.9%) | 7 (20.6%) | 5 (20.0%) | 19 (19.8%) |
| Southeast | 8 (21.6%) | 12 (35.3%) | 7 (28.0%) | 27 (28.1%) |
| Southwest | 22 (59.5%) | 15 (44.1%) | 13 (52.0%) | 50 (52.1%) |
| Gender | ||||
| Male | 33 (89.2%) | 23 (67.6%) | 21 (84.0%) | 77 (80.2%) |
| Female | 4 (10.8%) | 11 (32.4%) | 4 (16.0%) | 19 (19.8%) |
| Age | ||||
| Mean (SD) | 68.3 (8.1) | 62.0 (14.0) | 62.4 (8.4) | 64.5 (10.9) |
| Median | 70.0 | 63.5 | 63.0 | 66.0 |
| Range | 49.0–80.0 | 18.0–80.0 | 34.0–75.0 | 18.0–80.0 |
| Race (dichotomized) | ||||
| White | 35 (94.6%) | 28 (82.4%) | 23 (92.0%) | 86 (89.6%) |
| Non-white | 2 (5.4%) | 6 (17.6%) | 2 (8.0%) | 10 (10.4%) |
| Ethnicity (dichotomized) | ||||
| Hispanic/Latino | 1 (2.7%) | 2 (5.9%) | 1 (4.0%) | 4 (4.2%) |
| Non-Hispanic | 36 (97.3%) | 32 (94.1%) | 24 (96.0%) | 92 (95.8%) |
| Germline Result | ||||
| Positive | 4 (10.8%) | 6 (17.6%) | 5 (20.0%) | 15 (15.6%) |
| Negative | 13 (35.1%) | 9 (26.5%) | 5 (20.0%) | 27 (28.1%) |
| VUS | 20 (54.1%) | 19 (55.9%) | 15 (60.0%) | 54 (56.2%) |
| Histological type | ||||
| Adenocarcinoma | 33 (89.2%) | 31 (91.2%) | 25 (100.0%) | 89 (92.7%) |
| Squamous cell carcinoma | 3 (8.1%) | 1 (2.9%) | 0 (0.0%) | 4 (4.2%) |
| Other | 1 (2.7%) | 2 (5.9%) | 0 (0.0%) | 3 (3.1%) |
| Barrett’s esophagus | ||||
| Yes | 20 (54.1%) | 3 (8.8%) | 9 (36.0%) | 32 (33.3%) |
| No | 17 (45.9%) | 31 (91.2%) | 16 (64.0%) | 64 (66.7%) |
| Helicobacter Pylori | ||||
| Yes | 2 (5.4%) | 7 (20.6%) | 1 (4.0%) | 10 (10.4%) |
| No | 35 (94.6%) | 27 (79.4%) | 24 (96.0%) | 86 (89.6%) |
| Result (dichotomized) | ||||
| Positive | 4 (10.8%) | 6 (17.6%) | 5 (20.0%) | 15 (15.6%) |
| VUS/Negative | 33 (89.2%) | 28 (82.4%) | 20 (80.0%) | 81 (84.4%) |
| Staging AJCC 8th edition at diagnosis (Clinical stage) | ||||
| 1 | 5 (13.5%) | 4 (11.8%) | 1 (4.0%) | 10 (10.4%) |
| 2 | 5 (13.5%) | 7 (20.6%) | 2 (8.0%) | 14 (14.6%) |
| 3 | 6 (16.2%) | 7 (20.6%) | 9 (36.0%) | 22 (22.9%) |
| 4 | 21 (56.8%) | 16 (47.1%) | 13 (52.0%) | 50 (52.1%) |
| Staging AJCC 8th edition (Early vs. Late) | ||||
| Early Stage (0–2) | 10 (27.0%) | 11 (32.4%) | 3 (12.0%) | 24 (25.0%) |
| Late Stage (3–4) | 27 (73.0%) | 23 (67.6%) | 22 (88.0%) | 72 (75.0%) |
| Deceased | ||||
| Yes | 13 (35.1%) | 13 (38.2%) | 11 (44.0%) | 37 (38.5%) |
| No | 24 (64.9%) | 21 (61.8%) | 14 (56.0%) | 59 (61.5%) |
| Overall Survival (months) | ||||
| Mean (SD) | 23.3 (16.6) | 22.7 (21.9) | 17.2 (13.8) | 21.5 (18.0) |
| Median | 18.8 | 17.0 | 15.8 | 17.3 |
| Range | 4.4–74.5 | 1.7–99.4 | 2.5–52.6 | 1.7–99.4 |
Percentages calculated by column
Legend: GEJ: gastroesophageal junction tumors, AJCC: American Joint Committee on Cancer, VUS: variant of uncertain significance
Primary tumor location included esophagus (n = 37, 38.5%), gastric (n = 34,35.4%) and gastroesophageal junction (n = 25, 26%). The proportion of patients with early stages (I and II) was 25%, and late-stage disease (III and IV) was 75%. Complete pedigree was obtained for 44 (45.8%) patients. Family history of cancer in a first-degree relative was reported in 33 patients (34.4%).
The median follow-up time across all patients in this cohort was 17.3 months. In that time, 38.5% of patients expired. The median survival time for patients with esophageal cancer was 18.8 months (range: 4.4–74.5), for gastric cancer was 17 months (range: 1.7–99.4) and for gastroesophageal junction tumors was 15.8 months (range: 2.5–52.6).
Variants Detection
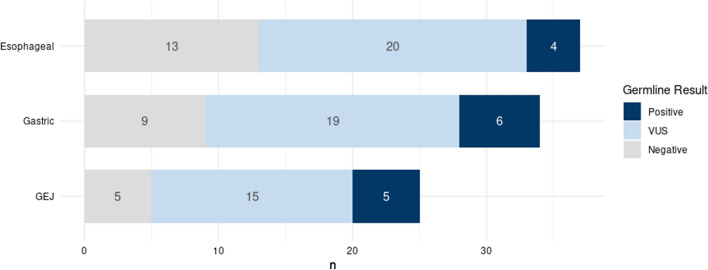
Of the 96 patients undergoing germline analysis, 15 patients (15.6%) carried pathogenic/likely pathogenic variants (PGV) conferring cancer predisposition (Fig. 1), with 11 (73.3%) of the PGV in high and moderate penetrance genes (Table 2). The most common variants were found in DNA damage repair (DDR) genes including BRCA1, BRCA2 (26.7%) and ATM (13.3%). No PGVs were detected in the Lynch syndrome associated genes (Fig. 2).
Fig. 1.
Prevalence of pathogenic germline variants
Table 2.
Distribution of pathogenic germline variants by penetrance status
| Pathogenic variant | Total (n = 15) | |
|---|---|---|
| High Penetrance | BRCA1 | 2 (13.3%) |
| BRCA2 | 2 (13.3%) | |
| CDH1 | 1 (6.7%) | |
| PALB2 | 1 (6.7%) | |
| SDHA | 1 (6.7%) | |
| Moderate Penetrance | ATM | 2 (13.3%) |
| HOXB13 | 1 (6.7%) | |
| MITF | 1 (6.7%) | |
| Low Penetrance | MUTYH (monoallelic) | 1 (6.7%) |
| Recessive Alleles | FH | 2 (13.3%) |
| RECQL4 | 1 (6.7%) |
Fig. 2.
Pathogenic germline variants stratified by primary tumor location
The PGV prevalence in esophageal cancer was 10.8%, gastric cancer was 17.6% and gastroesophageal junction tumors was 20%. Median age of patients with a PGV was 70 years (mean age 64.5 years old, 93% were ≥ 50 y/o), and only four of fifteen patients (26.7%) with a PGV had a family history of cancer in first-degree relative (Supplementary table 2). Of the 15 patients found to have a PGV, 80% (12) had PGVs that are qualifiers for potential clinically actionable management or treatment changes (Supplementary table 3). These can be categorized into potential eligibility for clinical treatment trials (47%) or published guideline management recommendations (80%). All systemic treatments are reviewed in Supplementary Table 4.
Applying the 2020 National Comprehensive Cancer Network (NCCN), National Society of Genetic Counselors (NSCG) or American College of Medical Genetics and Genomics (ACMG) genetic testing referral criteria, 80% (12 of 15) carriers of a PGV would not have been detected. Only two of the PGV carriers met guidelines based on family history regardless of personal history (Table 3). Of the seven patients potentially eligible for clinical trials, five (71%) would be missed by strict adherence to testing criteria (Supplementary table 5).
Table 3.
Screening guidelines met by patients with pathogenic germline variant
| 2018 (N = 15) | 2020 (N = 15) | |
|---|---|---|
| Did they meet NCCN/NSGC/ACMG testing guidelines? | ||
| Yes | 3 (20.0%) | 3 (20.0%) |
| No | 12 (80.0%) | 12 (80.0%) |
| Of those that met a screening guideline, which guidelines were met? | ||
| NCCN | ||
| Yes | 1 (6.7%) | 2 (13.3%) |
| No | 14 (93.3%) | 13 (86.7%) |
| NSGC/ACMG | ||
| Yes | 2 (13.3%) | 2 (13.3%) |
| No | 13 (86.7%) | 13 (86.7%) |
| Did they meet guidelines based on family history regardless of personal history? | ||
| Yes | 2 (13.3%) | 2 (13.3%) |
| No | 9 (60.0%) | 10 (66.7%) |
| Not available | 4 (26.7%) | 3 (20.0%) |
No cost family variant testing (FVT) was offered to all blood relatives of affected participants. Only two patients (13.3%) with PGV had family members undergo FVT within a 3-month window of their test result.
Discussion
Universal germline genetic testing in unselected patients with gastric, gastroesophageal junction and esophageal cancers was able to identify PGV in 15.6% of patients with the majority in high and moderate penetrance genes, including those in DNA damage repair pathways where precision targeted therapies exist. The incidence of PGVs was higher in GEJ tumors when compared to gastric and esophageal cancers. The population frequency of PGVs in BRCA1, BRCA2, CDH1, PALB2, SDHA, ATM, HOXB13, MITF, and FH is estimated to be less than 1%, and for MUTYH heterozygotes to be 1–2% [15–17]. Accordingly, the frequencies of PGVs in this cohort in high penetrance genes BRCA1, BRCA2, CDH1, PALB2 and SDHA, moderate penetrance genes ATM, HOXB13 and MITF, and low penetrance/recessive genes MUTYH (monoallelic) and FH all appear to be overrepresented in these patients with cancer compared to the general population.
Importantly, 80% of the PGV would not have been detected using current (2020) guidelines for genetic testing from national societies or professional organizations. For example, NCCN guidelines for considering germline genetic testing of individuals with gastric cancer require patients meet a complicated combination of criteria for eligibility [2]. This includes any one of the following: being diagnosed with diffuse gastric cancer (DGC) before 50 years of age without a family history, having a personal or family history of DGC and lobular breast cancer (one diagnosed < 70 years of age), two gastric cancer cases in a family (one confirmed DGC regardless of age), two cases of lobular breast cancer in family members < 50 years of age, DGC in any age in individuals of Maori ethnicity (or with personal or family history of cleft lip/cleft palate), or bilateral lobular breast cancer < 70 years of age.
For patients with GEJ or esophageal cancers, the NCCN does not have any guidelines recommending germline genetic testing, indicating that, “Although early age of onset, multiple family members with the same or related cancer, and individuals with multiple primary cancers are all signs of hereditary cancer, specific referral guidelines for esophageal and [GEJ] cancers risk assessment are not possible at this time.” Accordingly, none of the patients with GEJ or esophageal cancers would be recommended for germline genetic testing because the NCCN does not have guidelines that apply to these patients [2].
The incidence of gastric cancer worldwide is relatively low in countries such as the USA and highest in East Asian countries such as Japan. However, the rate of germline mutations in CDH1 in patients with gastric cancer varies inversely with the incidences of gastric cancer, with high CDH1 mutation detection rates in the USA and low detection rates in Japan [18]. Prior to identification of CDH1 germline mutations in Japanese patients, nearly all cases of gastric cancer were attributed to past exposure to H. pylori or some other carcinogen such as food and smoking, in the setting of a widely implemented screening program for gastric cancer. Identification of CDH1 PGVs in Japanese gastric cancer patients, though low in frequency, led to the recommendation to screen for CDH1 PGVs, as part of medical management in gastric cancer patients in Japan [19]. However, due in part to low rates of detection for CDH1 PGVs in countries such as Japan, genetic testing is often under-utilized. The higher rate of PGVs in CDH1 and other cancer risk genes in US patients, as reported here, may provide motivation to offer germline genetic testing to all patients with gastric, GEJ and esophageal cancers.
Although the majority of gastric cancer cases are felt to be sporadic, up to 20% of gastric cancer patients may also have relatives with gastric cancer suggesting a familial risk. From those patients, only 3–5% are found to have an inherited germline variant detected [20, 21]. Multiple cancer predisposition syndromes may be associated with the development of gastric cancers such as Li Fraumeni syndrome, Lynch syndrome and CDH1-associated hereditary diffuse gastric cancer [20, 22, 23]. Most patients with families that meet clinical criteria for hereditary diffuse gastric cancer will harbor CDH1 germline mutations [24]. However, for CDH1-mutation negative patients, application of targeted sequencing has identified pathogenic variants in other related genes including CTNNA1, BRCA1, BRCA2, STK11, SDHB, PRSS1, ATM, MSR1, RAD51 and PALB2 [24–30]. DDR genes are of particular interest considering possible targeted treatment strategies in this group of patients with homologous recombination deficient tumors, including platinum-based therapies and poly-ADP-ribose polymerase (PARP) inhibitors such as olaparib. In the current manuscript, application of a universal multi-gene panel testing found 17.6% of gastric cancer patients harbored a PGV. Interestingly along with a PGV in the gene CDH1, PGV in DDR genes including BRCA1 and BRCA2 were identified. Exploiting DDR genes have already been the subject of a randomized trial in ATM-negative metastatic gastric cancer [31]. However, the combination paclitaxel plus olaparib did not improve overall survival [32]. Nowadays other combinations with olaparib are being developed, including with anti-programmed death-ligand 1 (PD-L1) durvalumab and anti-vascular endothelial growth factor receptor-2 (VEGFR2) ramucirumab [31–33].
Among patients with esophageal and GEJ cancers, PGV were identified in 10.8 and 20% with the majority being in DDR-related genes including ATM, BRCA2 and PALB2. Some esophageal cancers have been found to be related to hereditary cancer predisposition syndromes such as tylosis, familial Barrett’s esophagus, Bloom syndrome and Fanconi anemia, with the latter related to BRCA2 and PALB2 [34–37]. The association between BRCA and esophageal cancer (both squamous cell carcinoma and adenocarcinoma) was also observed by other groups [38–40]. However, currently, referral to a cancer genetics professional is recommended only for patients fulfilling criteria for a known high-risk syndrome. Considering current guidelines including NCCN, NSCG or ACMG in the year 2020, only 20% of patients with a PGV would have been detected. This limits the opportunities for future cancer prevention in the proband and their relatives. Several tumor agnostic trials are already underway or being developed for patients that harbor a deleterious mutation in homologous recombination repair (HRR) genes. As an example, a phase II study evaluating the efficacy of rucaparib (PARP inhibitor) for advanced solid tumors that have a deleterious mutation (germline or somatic) in BRCA1, BRCA2, PALB2, RAD51C, RAD51D, BARD1, BRIP1, FANCA, NBN, RAD51 or RAD51B (NCT04171700). With this new horizon of options for patients with advanced and recurrent disease, this study in unselected gastric and esophageal cancer patients highlights the usefulness of universal germline testing. Despite major advances in the systemic treatment of metastatic gastric and esophageal cancers with the incorporation of immunotherapy in the first line of systemic treatment, median overall survival remains poor, with most patients with advanced disease dying within 2 years of diagnosis [41–43].
Some limitations of the current study include the demographic characteristics of patients seen at the multiple Mayo Clinic sites participating, as they may not reflect populations in other areas of the USA or other countries. Further, no integrative somatic tumor analysis was performed. The combination of universal germline and somatic testing may detect additional HRR deficient tumors. Due to a limited number of cases with positive PGV in each of the three different primary tumor locations, an analysis and comparison of outcomes and response to treatments was not permissible. Larger cohorts are necessary to address important questions related to treatment outcomes of gastric, esophageal, and GEJ cancer patients with PGVs.
In this prospective, multi-site study in unselected patients with gastric, esophageal, and GEJ cancers, implementation of universal germline testing identified that 1 in 6 patients carry a PGV. Incorporation of universal germline testing in these patients could improve development of protocols for cancer risk assessment, personalized medicine, and family counseling.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgments
We would like to thank the following persons for their assistance with this project—Sydney Welp, Jessie Fox, Plush Guiterrez, Sara Hernandez, Sharon Levy, Eric Nelson, Anne Bofferding, Arta Palaj, Lorelei Bandel, Megan Mulcahy and David Upjohn.
Author contributions
Drs. Samadder and Kunze had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analyses. Study concept and design (NJS, KAS, PLSUJ, TBS); acquisition, analysis and interpretation of data (NJS, PLSUJ, DRJ, LB, EDE, RLN, KLK); drafting of the manuscript (PLSUJ, NJS); critical revision of the manuscript for important intellectual content (NJS, DRJ, LB, KLK, MAG, EDE, RLN, MB, DA, MBS, TBS); statistical analysis (KLK, MAG); obtained funding (NJS and KAS).
Funding
Support for this project was provided by Mayo Transform the Practice Grant, Mayo Clinic Center for Individualized Medicine, Desert Mountain Members’ CARE Foundation, David and Twila Woods Foundation and a Faculty Career Development Award from the Gerstner Foundation (NJS). Role of the Funding Source: The study was funded by Mayo Transform the Practice Grant, Mayo Clinic Center for Individualized Medicine, Desert Mountain Members’ CARE Foundation, David and Twila Woods Foundation. The funding sources did not play a role in the design, conduct or reporting of the study or in the decision to submit the manuscript for publication.
Declarations
Conflict of interest
NJS is a consultant for Jansen Research and Development and Cancer Prevention Pharmaceuticals. EDE and RLN are employees and stockholders of Invitae. RLN is also a consultant for Pfizer and a consultant and stockholder in Genome Medical and Maze Therapeutics. No other authors have a conflict of interest to disclose.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CACancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Gastric cancer. NCCN guidelines version 2.2021. In: https://www.nccn.org/professionals/physician_gls/pdf/gastric.pdf. Accessed 19 March 2021.
- 3.Zhao JK, Wu M, Kim CH, et al. Jiangsu Four Cancers Study: A large case-control study of lung, liver, stomach and esophageal cancers in Jiangsu Province. China. Eur J Cancer Prev. 2017;26:357–364. doi: 10.1097/CEJ.0000000000000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yusefi A, Lankarani KB, Bastani P, Radinmanesh M, Kavosi Z. Risk factors for gastric cancer: a systematic review. Asian Pac J Cancer Prev. 2018;19:591–603. doi: 10.22034/APJCP.2018.19.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He Z, Zhao TT, Xu HM, et al. Association between alcohol consumption and the risk of gastric cancer: a meta-analysis of prospective cohort studies. Oncotarget. 2017;8:84459–84472. doi: 10.18632/oncotarget.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts ME, Ranola JMO, Marshall ML, et al. Comparison of CDH1 penetrance estimates in clinically ascertained families vs families ascertained for multiple gastric cancers. JAMA Oncol. 2019;5:1325–1331. doi: 10.1001/jamaoncol.2019.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sitarz R, Skierucha M, Mielko J, Offerhaus GJA, Maciejewski R, Polkowski WP. Gastric cancer: epidemiology, prevention, classification, and treatment. Cancer Manag Res. 2018;10:239–248. doi: 10.2147/CMAR.S149619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.National Cancer Institute. Surveillance, Epidemiology, and End Results (SEER) Program. Cancer Stat Facts: Stomach Cancer. 2020. In: https://seer.cancer.gov/statfacts/html/stomach.html. Accessed 19 March 2021.
- 9.Freedman ND, Abnet CC, Leitzmann MF, et al. A prospective study of tobacco, alcohol, and the risk of esophageal and gastric cancer subtypes. Am J Epidemiol. 2017;165:1424–1433. doi: 10.1093/aje/kwm051. [DOI] [PubMed] [Google Scholar]
- 10.Engel LS, Chow WH, Vaughan TL, et al. Population attributable risks of esophageal and gastric cancers. J Natl Cancer Inst. 2003;95:1404–1413. doi: 10.1093/jnci/djg047. [DOI] [PubMed] [Google Scholar]
- 11.Turati F, Tramacere I, La Vecchia C, Negri E. A meta-analysis of body mass index and esophageal and gastric cardia adenocarcinoma. Ann Oncol. 2013;24:609–617. doi: 10.1093/annonc/mds244. [DOI] [PubMed] [Google Scholar]
- 12.Cossentino MJ,Wong RK. Barrett's esophagus and risk of esophageal adenocarcinoma. Sem Gastroint Dis. Vol. 14. No. 3. 2003. [PubMed]
- 13.Lagergren J, Bergstrom R, Lindgren A, Nyren O. Symptomatic gastroesophageal reflux as a risk factor for esophageal adenocarcinoma. N Engl J Med. 1999;340:825–831. doi: 10.1056/NEJM199903183401101. [DOI] [PubMed] [Google Scholar]
- 14.Samadder NJ, Riegert-Johnson D, Boardman L, et al. Comparison of universal genetic testing vs guideline-directed targeted testing for patients with hereditary cancer syndrome. JAMA Oncol. 2021;7:230–237. doi: 10.1001/jamaoncol.2020.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrucelli N, Daly MB, Pal T. BRCA1-and BRCA2-associated hereditary breast and ovarian cancer. GeneReviews®[Internet] 2016.
- 16.Nielsen M, Morreau H, Vasen HFA, Hes FJ. MUTYH-associated polyposis (MAP) Crit Rev Oncol Hematol. 2011;79(1):1–16. doi: 10.1016/j.critrevonc.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 17.Haverfield EV, Esplin ED, Aguilar SJ, et al. Physician-directed genetic screening to evaluate personal risk for medically actionable disorders: a large multi-center cohort study. BMC Med. 2021;19:199. doi: 10.1186/s12916-021-01999-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugimoto S, Komatsu H, Morohoshi Y, Kanai T. Recognition of and recent issues in hereditary diffuse gastric cancer. J Gastroenterol. 2015;50:831–843. doi: 10.1007/s00535-015-1093-9. [DOI] [PubMed] [Google Scholar]
- 19.Yamada H, Shinmura K, Ito H, et al. Germline alterations in the CDH1 gene in familial gastric cancer in the Japanese population. Cancer Sci. 2011;102:1782–1788. doi: 10.1111/j.1349-7006.2011.02038.x. [DOI] [PubMed] [Google Scholar]
- 20.van der Post RS, Vogelaar IP, Carneiro F, et al. Hereditary diffuse gastric cancer: updated clinical guidelines with an emphasis on germline CDH1 mutation carriers. J Med Gen. 2015;52:361–374. doi: 10.1136/jmedgenet-2015-103094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Post RS, Oliveira C, Guilford P, Carneiro F. Hereditary gastric cancer: What’s new? Update 2013–2018. Fam Cancer. 2019;18:363–367. doi: 10.1007/s10689-019-00127-7. [DOI] [PubMed] [Google Scholar]
- 22.Masciari S, Dewanwala A, Stoffel EM, et al. Gastric cancer in individuals with Li-Fraumeni syndrome. Genet Med. 2011;13:651–657. doi: 10.1097/GIM.0b013e31821628b6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capelle LG, Van Grieken NC, Lingsma HF, et al. Risk and epidemiological time trends of gastric cancer in Lynch syndrome carriers in the Netherlands. Gastroenterology. 2010;138:487–492. doi: 10.1053/j.gastro.2009.10.051. [DOI] [PubMed] [Google Scholar]
- 24.Hansford S, Kaurah P, Li-Chang H, et al. Hereditary diffuse gastric cancer syndrome: CDH1 mutations and beyond. JAMA Oncol. 2015;1:23–32. doi: 10.1001/jamaoncol.2014.168. [DOI] [PubMed] [Google Scholar]
- 25.Sahasrabudhe R, Lott P, Bohorquez M, et al. Germline mutations in PALB2, BRCA1, and RAD51C, which regulate DNA recombination repair, in patients with gastric cancer. Gastroenterol. 2017;152:983–986. doi: 10.1053/j.gastro.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jakubowska A, Nej K, Huzarski T, Scott RJ, Lubinski J. BRCA2 gene mutations in families with aggregations of breast and stomach cancers. Br J Cancer. 2002;87:888–891. doi: 10.1038/sj.bjc.6600562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedenson B. BRCA1 and BRCA2 pathways and the risk of cancers other than breast or ovarian. Med Gen Med. 2005;7:60. [PMC free article] [PubMed] [Google Scholar]
- 28.Fewings E, Larianov A, Redman J, et al. Germline pathogenic variants in PALB2 and other cancer-predisposing genes in families with hereditary diffuse gastric cancer without CDH1 mutation: a whole-exome sequencing study. Lancet Gastroenterol Hepatol. 2018;3:489–498. doi: 10.1016/S2468-1253(18)30079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ichikawa H, Wakai T, Nagahashi M, et al. Pathogenic germline BRCA1/2 mutations and familial predisposition to gastric cancer. JCO Prec Oncol. 2018 doi: 10.1200/PO.18.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ku GY, Kemel Y, Maron SB, et al. Prevalence of Germline Alterations on Targeted Tumor-Normal Sequencing of Esophagogastric Cancer. JAMA Net Open. 2021;4(7):e2114753–e2114753. doi: 10.1001/jamanetworkopen.2021.14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bang YJ, Xu RH, Chin K, et al. Olaparib in combination with paclitaxel in patients with advanced gastric cancer who have progressed following first-line therapy (GOLD): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2017;18:1637–1651. doi: 10.1016/S1470-2045(17)30682-4. [DOI] [PubMed] [Google Scholar]
- 32.Bang YJ, Kaufman B, Geva R et al. An open-label, phase II basket study of olaparib and durvalumab (MEDIOLA): Results in patients with relapsed gastric cancer. J Clin Oncol 2019; 37: abstr 140–140.
- 33.Cecchini M, LoRusso P, Shyr Y et al. NCI 10066: A phase 1/2 study of olaparib in combination with ramucirumab in metastatic gastric and gastroesophageal junction (GEJ) adenocarcinoma. J Clin Oncol 2018; 36: abstr TPS4137-TPS4137.
- 34.Varela AB, Rodríguez MMB, Boullosa PE, Silva JG. Tylosis A with squamous cell carcinoma of the oesophagus in a Spanish family. Eur J Gastroenterol Hepatol. 2011;23:286–288. doi: 10.1097/MEG.0b013e328344042d. [DOI] [PubMed] [Google Scholar]
- 35.Verbeek RE, Spittuler LF, Peute A, et al. Familial clustering of Barrett's esophagus and esophageal adenocarcinoma in a European cohort. Clin Gastroenterol Hepatol. 2014;12:1656–1663. doi: 10.1016/j.cgh.2014.01.028. [DOI] [PubMed] [Google Scholar]
- 36.Cunniff C, Bassetti JA, Ellis NA. Bloom's syndrome: clinical spectrum, molecular pathogenesis, and cancer predisposition. Mol Syndromol. 2017;8:4–23. doi: 10.1159/000452082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D'Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nature Rev Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 38.Ko JMY, Ning L, Zhao XK, et al. BRCA2 loss-of-function germline mutations are associated with esophageal squamous cell carcinoma risk in Chinese. Int J Cancer. 2020;146:1042–1051. doi: 10.1002/ijc.32619. [DOI] [PubMed] [Google Scholar]
- 39.Akbari MR, Malekzadeh R, Nasrollahzadeh D, et al. Germline BRCA2 mutations and the risk of esophageal squamous cell carcinoma. Oncogene. 2008;27:1290–1296. doi: 10.1038/sj.onc.1210739. [DOI] [PubMed] [Google Scholar]
- 40.Secrier M, Li X, Silva N, et al. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat genet. 2016;48:1131–1141. doi: 10.1038/ng.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moehler M, Shitara K, Garrido M, et al. LBA6_PR Nivolumab (nivo) plus chemotherapy (chemo) versus chemo as first-line (1L) treatment for advanced gastric cancer/gastroesophageal junction cancer (GC/GEJC)/esophageal adenocarcinoma (EAC): First results of the CheckMate 649 study. Ann Oncol. 2020;31:S1191. doi: 10.1016/j.annonc.2020.08.2296. [DOI] [Google Scholar]
- 42.Kato K, Sun J, Shah MA, et al. Pembrolizumab plus chemotherapy versus chemotherapy as first-line therapy in patients with advanced esophageal cancer: The phase 3 KEYNOTE-590 study. Ann Oncol. 2020;31:S1142. doi: 10.1016/j.annonc.2020.08.2298. [DOI] [Google Scholar]
- 43.de Aguiar VG, Segatelli V, Macedo ALV, et al. Signet ring cell component, not the Lauren subtype, predicts poor survival: an analysis of 198 cases of gastric cancer. Future Oncol. 2019;15:401–408. doi: 10.2217/fon-2018-0354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.