Abstract
Ventricular arrhythmias are the primary cause of sudden cardiac death and one of the leading causes of mortality worldwide. Whole-heart computational modeling offers a unique approach for studying ventricular arrhythmias, offering vast potential for developing both a mechanistic understanding of ventricular arrhythmias and clinical applications for treatment. In this review, the fundamentals of whole-heart ventricular modeling and current methods of personalizing models using clinical data are presented. From this foundation, the authors summarize recent advances in whole-heart ventricular arrhythmia modeling. Efforts in gaining mechanistic insights into ventricular arrhythmias are discussed, in addition to other applications of models such as the assessment of novel therapeutics. The review emphasizes the unique benefits of computational modeling that allow for insights that are not obtainable by contemporary experimental or clinical means. Additionally, the clinical impact of modeling is explored, demonstrating how patient care is influenced by the information gained from ventricular arrhythmia models. The authors conclude with future perspectives about the direction of whole-heart ventricular arrhythmia modeling, outlining how advances in neural network methodologies hold the potential to reduce computational expense and permit for efficient whole-heart modeling.
INTRODUCTION
Ventricular arrhythmias (VA), consisting of ventricular tachycardia (VT) and ventricular fibrillation (VF), are life-threatening electrical rhythm disorders that are the primary cause of sudden cardiac death (SCD).1 Understanding the mechanisms underlying VAs is important both for the treatment and management of VAs and necessary for the development of novel therapeutics. However, it remains challenging to translate the empirical outcomes to clinical approaches because insights derived from experimental conditions may not always be applicable to in vivo conditions in the human heart. Clinical studies, on the other hand, are limited in the mechanistic insights that can be gleaned due to the lack of precision on controlling for experimental conditions.
Computational heart modeling is an emergent technology that is well-poised both to shed light on mechanisms underlying VAs and to aid clinical decision making in the management of VAs. Biophysically detailed ventricular models offer unique insights into the physiology of ventricular arrhythmias which can then be used to inform clinicians in treating VAs in various ways. These models have shown promising results in determining VA risk, ablation targets, disease mechanisms, and exploring novel therapies. Furthermore, computational technologies have great potential for being integrated into contemporary clinical workflows.
In this article, we review recent studies of VA that use computational whole-heart models to elucidate VA mechanisms and advancements in patient-specific clinical applications, including VA treatment and risk stratification. The general workflow for ventricular whole-heart modeling is shown in Fig. 1. We first discuss the fundamentals of personalized, whole-heart modeling, highlighting the underlying modeling principles and the contemporary approaches to creating patient-specific models. We then proceed to summarize studies that provide mechanistic insights into VA from the perspectives of arrhythmia initiation, and scar-related and functional-type re-entries. We then discuss recent studies using computational ventricular models for investigating novel therapeutics in VAs and clinical applications of VA modeling including arrhythmia risk stratification and catheter-ablation planning of VT.
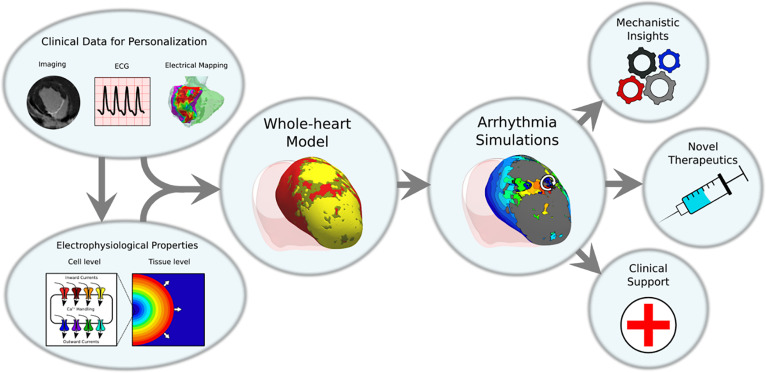
FIG. 1.
Overview of whole-heart ventricular arrhythmia modeling. Cell-level and tissue-level properties of ventricular myocyte electrophysiology are incorporated into whole-organ level ventricular heart models. These ventricular heart models can also incorporate personalized information from clinical data modalities such as medical imaging, electrocardiogram (ECG), and invasive electrical mapping. These models are then used to run simulations of ventricular arrhythmias (VA) which can be used to gain insights into fundamental biophysical mechanisms, to predict results in novel therapeutic modalities, and to aid in clinical decision making in the management of VAs.
FUNDAMENTAL CONCEPTS OF WHOLE-HEART VENTRICULAR ARRHYTHMIA MODELING
Whole-heart ventricular models integrate information from multiple scales ranging from cell-level ionic properties to whole-organ tissue distributions. Here we review the fundamental concepts of constructing whole-heart ventricular models, including methods of model personalization using clinical data.
Biophysically detailed models of cellular electrophysiology
Biophysically detailed models of ventricular myocyte electrophysiology typically follow Hodgkin–Huxley type formulations.2 Briefly, the membrane dynamics are modeled as an RC circuit where the resistances (more often represented as conductances) represent ion flux through membrane channels, pumps, and transporters, and the capacitor represents the cell membrane phospholipid bilayer. From this representation, a system of ordinary differential equations can be derived to describe the change in membrane voltage over time. Among the most common human ventricular myocyte models used in whole-heart models are the Ten Tusscher–Panfilov3 and the O'Hara–Rudy4 that were calibrated to human experimental data.
Bidomain and monodomain equations
The bidomain equations are the most explicit mathematical description of electrical wave propagation through cardiac tissues and consider both intracellular and extracellular current.5 Changes in the intracellular potential (φi) and extracellular potential (φe) are coupled via membrane dynamics that involves ion channels, pumps, and other transporters. The mathematical equations consist of a system of partial differential equations with respect to space and time [Eqs. (1)–(3)],
| (1) |
| (2) |
| (3) |
In the equations, Vm represents the transmembrane voltage and the difference between φi and φe. σi and σe are the intracellular and extracellular conductivity tensors, respectively. β is the ratio between the membrane surface area to the volume. Is represents an external stimulus applied to the intracellular space. Cm is the membrane capacitance per unit area, Iion is the transmembrane ionic current density, and η represents the gating variables that govern the kinetics of the different ionic currents,
| (4) |
The monodomain equations are a simplification of the bidomain equations and are derived by assuming a proportionality between σi and σe. With this assumption, the bidomain system above [Eqs. (1)–(3)] is simplified into a single equation [Eq. (4)]. In the above formulation, σm is the effective bulk conductivity that relates σi and σe. In electrophysiological simulation, monodomain equations are often used in place of the bidomain system due to enormous savings in computational costs.6
The Purkinje system, the component of the cardiac conduction system responsible for fast synchronous activation of the ventricular myocardium, can also be incorporated into whole-heart models. It can be represented as a one-dimensional branching cable system with an increased conduction velocity that couples with the ventricular myocardium at Purkinje-myocardial junctions along the endocardial surface.7 Due to the inherent difficulties in producing anatomically correct or patient-specific Purkinje trees, most ventricular whole-heart models do not include Purkinje fibers.
Using medical imaging to personalize ventricular models
Computational heart models can be personalized by combining information from medical imaging modalities. The biophysics of different imaging modalities allows for the characterization of different pathological tissue types which can then be incorporated into heart models to study VAs in the context of the diseased heart.8 Two major imaging modalities that can be used for model personalization include magnetic resonance imaging (MRI) and computed tomography (CT).
MRI can be combined with contrast agents such as gadolinium to improve characterization of diseased tissue types. Such sequences, called late gadolinium-enhanced MRIs (LGE-MRIs), are used in routine clinical workflows and are considered the gold standard in cardiac imaging.8 With LGE-MRI, scar tissue arising from infarction can be detected as regions of hyper-enhancement.9–12 The spatial pattern of this scar distribution can then be readily incorporated into computational heart models and can be assigned various electrophysiological properties based upon available experimental evidence. One inherent limitation of LGE-MRI is image artifacts caused by implanted cardioverter defibrillators (ICDs), which are often present in patients at risk of VAs. Protocols such as the wideband sequence are being optimized to minimize artifact burden and have shown success in substrate characterization.13 LGE has also been used in the assessment of various nonischemic substrates.14,15 Unlike the post-infarct substrate, which is localized to one myocardial region, nonischemic cardiomyopathy tends to involve diffuse fibrosis, and hence scar tissue can be more difficult to identify. More reproducible, quantitative T1 mapping which does not require contrast injection has emerged to help characterize the myocardial substrate.16
CT is another imaging modality commonly acquired in clinical workflows. Compared to LGE MRI, conventional CT has limitations in differentiating the scar from healthy myocardium due to the limited inherent soft-tissue contrast. Similar to LGE-MRI, contrast agents are also used in conjunction with CT to improve delineation of myocardial structures. Previous studies showed that successful scar quantification could be achieved by delayed enhanced CT protocols, but such images have poor signal-to-noise ratios and may be difficult to obtain clinically.17 Despite the limited contrast within the myocardium, contrast-enhanced (CE) CT offers a sharp contrast between blood and the myocardium. This clear distinction between blood and cardiac tissue allows for accurate assessment of wall thickness using CE-CT.18–21 Regions of wall thinning have been shown to correlate with regions of scar and electrophysiological abnormalities.20,21 In addition to wall thinning, infiltrating adiposity, an arrhythmogenic substrate involved in certain myocardial diseases, can be quantified on CT.22–25 Infiltrating fat on CT appears as darker, hypoattenuated regions within the myocardium. Similar to LGE-MRI, CE-CT can also be affected by lead artifacts that can preclude visualization of parts of the ventricular myocardium. However, the image quality of CE-CT tends to be more consistent than LGE-MRI, and there are methods for reducing artifact burden.26
Using electrical measurements to personalize ventricular models
Personalization of electrophysiology requires electrical measurements that can be obtained either noninvasively or invasively. Electrocardiograms (ECGs) noninvasively measure cardiac electrical activity and are frequently obtained clinically. This electrical information reflects the heart's conduction and repolarization properties which can be used to calibrate model parameters to capture the patient-specific electrophysiology.27–31 One specific technique called electrocardiographic imaging (ECGI) reconstructs electrical information on epicardial surface using electrical measurements obtained at the body surface.32,33 This information is then used to noninvasively assess cardiac conduction patterns to localize arrhythmias. These activation maps can then be used to tune computational models to better represent the patient-specific electrophysiology.34,35 However, such techniques are still in their infancy and require further development in terms of accuracy, computational costs, and generalizability to larger patient cohorts. Invasive electrical information can be obtained in the form of intracardiac electrograms measured intraprocedurally. Electrograms are measured locally at locations throughout the endocardial or epicardial surface, providing a glimpse at cardiac conduction patterns throughout the heart. This information can be used to adjust parameters of computational models to better reflect patient-specific electrophysiology.36–39 However, such invasive measurements are not usually present pre-procedurally; it is also non-trivial to determine which model parameters need to be changed to reflect the electrical measurements, and overfitting a model reduces its generalizability.
Nonetheless, even without patient-specific electrical data, whole-heart ventricular models with non-calibrated electrophysiological properties can still yield useful predictions that are consistent with clinical data.40–42 One such sensitivity analysis reported that post-infarct VT predictions were largely robust to a range of different conduction velocities and action potential durations.43 Further sensitivity analyses and approaches to incorporate uncertainty30 into the electrophysiological parameterization of these whole-heart models should be investigated.
MECHANISTIC INSIGHTS INTO VENTRICULAR ARRHYTHMIAS USING BIOPHYSICALLY DETAILED VENTRICULAR MODELS
Biophysically detailed computational models can provide mechanistic insights into arrhythmia pathophysiology that are not easily assessed experimentally. Parameters can be changed and the resultant effects quantified to establish mechanistic underpinnings of disease processes. Here we discuss the use of whole-heart modeling in several key subjects pertaining to VA.
Ventricular arrhythmia initiation
Computational modeling of ventricular electrophysiology has been used to improve understanding about the mechanisms of arrhythmia initiation. Re-entry, one of the dominant mechanisms of arrhythmias, necessitates conduction slowing and unidirectional conduction block to be present.44 In the context of VA pathophysiology, VA initiation depends on the interaction between correctly timed ectopic beats with the local anatomical/functional heterogeneities present in the diseased myocardium.
Failing hearts often undergo significant remodeling processes that predispose patients to VAs. Remodeled substrates can often give rise to early afterdepolarizations that create ectopic beats and can initiate VAs. A recent combined computational and clinical study sought to understand mechanisms between a clinically recognized phenomenon called low-amplitude action potential voltage alternans and ventricular arrhythmias.45 Figure 2(a) shows a representative graphic of their study. Using multiscale computational models of failing human ventricles, the authors were able to define a mechanism linking abnormal calcium handling in heart failure to low-amplitude voltage alternans which resulted in initiation of ventricular fibrillation. The models demonstrated how increasing pacing frequency created a pro-arrhythmogenic substrate, which combined with a correctly timed premature stimulus would initiate a re-entrant arrhythmia. Other studies have also looked at the role of calcium in arrhythmia initiation. One such study investigated the conditions in which calcium-mediated ectopy could cause VT arrhythmogenesis in infarcted substrates.46 The authors utilized both 2D and 3D ventricular models incorporating scar and infarct border zone tissue types. Infarct border zone tissue types were modeled as having abnormal electrophysiological properties consistent with experimental evidence. Increasing fibrosis was shown to be correlated with the probability of ectopic activity likely due to alterations in the local electrotonic conditions.
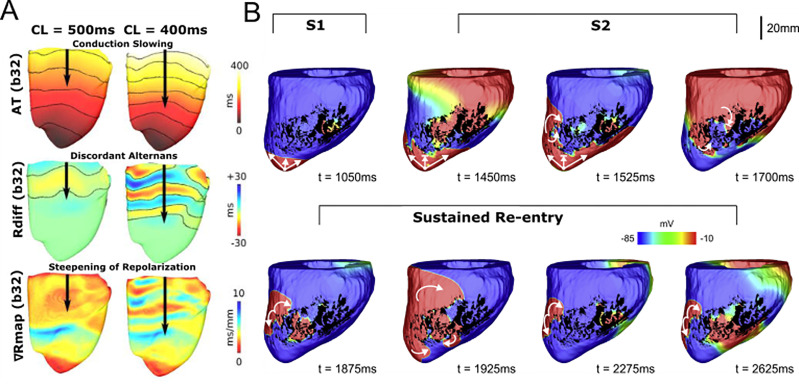
FIG. 2.
Insights into mechanisms of ventricular arrhythmia initiation. (a) Investigating mechanisms of arrhythmia initiation. Decreasing pacing stimulus cycle length results in conduction slowing, increased alternans, and a steeper repolarization gradient, all of which creates an arrhythmogenic substrate for VA initiation. Reproduced with permission from Bayer et al., Heart Rhythm 13, 1922 (2016). Copyright 2016 Elsevier.45 (b) Determining electrophysiological factors sufficient for VA initiation. In whole-heart swine models, VA could be initiated by infarct border zone model with fibrosis and decreased sodium conductance. Using an S1 and S2 pacing protocol, sustained re-entry was achieved. Reproduced with permission from Campos et al., Biophys. J. 117, 2361 (2019). Copyright 2019 Author(s), licensed under a Creative Commons Attribution (CC BY) license.47
Other pro-arrhythmic ionic mechanisms have also been examined. A recent study examined the effects of decreasing sodium conductance and fibrosis density on re-entry formation.47 Using both 2D and 3D ventricular models, they ascertained that decreases in sodium conductance could interact synergistically with fibrosis to establish conduction block and promote re-entry. Figure 2(b) shows an example of re-entry being initiated in an animal-specific whole-heart model. Using an S1–S2 stimulus protocol, sustained re-entry was induced due to conduction block that was mediated only by decreased sodium conductance and non-conducting fibrosis in the border zone [Fig. 2(b)].
The Purkinje system may also have an arrhythmogenic role in certain VAs. Due to the electrotonic interactions present at Purkinje-myocardial junctions, the Purkinje system can facilitate re-entry. One such study investigated how ectopic beats originating from various parts of the Purkinje system could initiate VAs.48 The authors conclude that ectopic beats originating in the distal rather than proximal branches of the Purkinje tree were more likely to induce re-entry. In a separate study, simulations were used to investigate the role of the Purkinje system during post-shock VAs.49 Here, the Purkinje system tended to be proarrhythmic by stabilizing re-entry and providing alternate pathways for wave propagation through the Purkinje fibers.
Elucidating the relationship between scar remodeling and the VT circuit
Image-based computational heart models have emerged as a unique way to examine the remodeled scar distribution as it relates to VT. Simulations provide insights beyond simple structural analyses and allow for evaluation of the electrophysiological and functional aspects of the VT circuit.
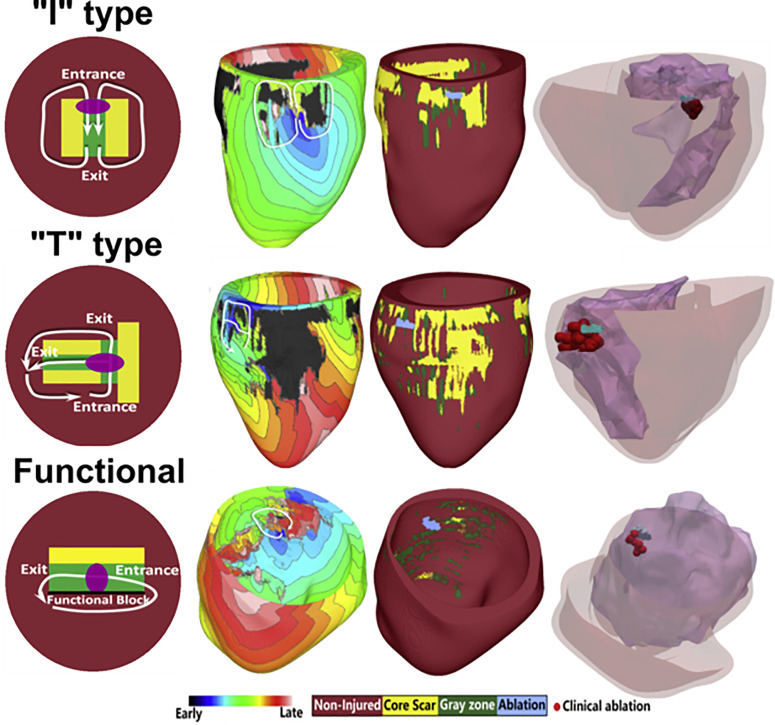
In a study examining animal-specific swine models, the authors investigated the structural characteristics of the scar surrounding the VT circuit.50 Heart models were reconstructed from high resolution ex vivo MRIs, and the VT conduction pathway was characterized by quantifying the distance from the surrounding scar. From this analysis, the authors identified a distribution of channel widths that the VT circuit critical isthmus was most likely to be found in. A recent study involving patient data examined the VT circuits induced in six LGE-CMR-based digital heart models and their corresponding ablation lesions.51 In this study, the authors characterized the various types of conducting channel phenotypes and identified three distinct classes: I-type, T-type, and functional-type channels. Figure 3 illustrates these three types of channels and the corresponding whole-heart simulation results. Each type of conducting channel was consistent with VT morphologies that have been reported in the experimental and clinical literature. Furthermore, this study also validated the simulation results by analyzing how clinical ablation lesions corresponded with virtual-heart VT circuits (Fig. 3). These results highlight how computational modeling can be used to develop greater mechanistic insights into how the scar distribution gives rise to arrhythmias.
FIG. 3.
Relationship between scar remodeling and ventricular tachycardia circuits. Three types of conducting channels arising from scar remodeling were characterized and identified in patient-specific virtual-heart models. I-type channels involve non-conducting scar surrounding a central isthmus gray zone, resulting in a figure-of-eight VT morphology. T-type channels involve a more complex structure with multiple exit and entrance sites. Finally, the functional-type channel involves a combination of functional and anatomical block. Model predictions were validated by comparing virtual-heart ablation targets with clinical ablation lesions. These results suggest that if the type of conducting channel can be identified, corresponding optimal ablation strategies can be applied to effectively terminate VT. Reproduced with permission from Deng et al., Biophys. J. 117, 2361 (2019). Copyright 2019 Cell Press.51
The infarct border zone also plays a critical role in VT arrhythmogenesis. On MRI, the infarct border zone is identified as gray zone, tissues with an intermediate signal intensity between scar and non-injured myocardium. In patient-specific models with scar and gray zone distributions, it was demonstrated how arrhythmia activity primarily concentrated in regions of the gray zone and was largely dependent on the morphology and size of these remodeled tissue regions.52 Variations in the structural heterogeneity of the gray zone did not seem to have a major impact on arrhythmogenicity. A separate study similarly found that the gray zone geometry had a large impact on arrhythmia dynamics, and additionally increased amounts of gray zone in fibrotic regions tend to destabilize the VT circuit.53 Another study also investigated the effects of different fibrosis distribution on arrhythmia vulnerability and identified several geometric configurations that seem to be more arrhythmogenic.54 Finally, yet another study examined what characteristics of the infarct border zone were significant for arrhythmogenesis using simplistic 3D toy heart models.55 They applied arrhythmia induction protocols in two toy infarct distributions: transmural and subendocardial. From the transmural infarct geometry, they determined that the extent of scar and repolarization properties of the border zone were heavily important for the initiation of VT. From the subendocardial infarct distribution, they concluded that the re-entry propagation followed the predominant fiber directions and the location of the premature stimulus. Collectively, these studies implicate that the geometric structure of gray zone plays a vital role in determining arrhythmogenesis.
New work has also explored the scar distribution in non-ischemic cardiomyopathy. A novel methodology of modeling interstitial fibrosis, a major hallmark of fibrosis development in non-ischemic cardiomyopathy, was recently developed.56 LGE-MRI-based computational heart models were reconstructed, and the image intensity information on LGE-MRI is used to derive the patient-specific fibrotic regions. Mesh elements within the fibrotic regions are randomly disconnected as a function of the intensity values to create a heterogeneous tissue distribution, resembling the non-ischemic substrate. All simulations with successful arrhythmia induction resulted from micro-re-entry within the interstitial fibrosis region, offering possible insight into the structure of non-ischemic VT circuits.
Ventricular arrhythmias with functional re-entrant patterns
Functional re-entries arise as a result of electrical heterogeneities and do not require the presence of anatomical obstacles to propagate. Here, we discuss how whole-heart computational models have been used to understand various VAs with functional re-entrant patterns.
Myocardial ischemia resulting from an acute infarct affects the membrane dynamics via alterations, among others, in potassium concentrations. The resultant electrophysiological changes can give rise to significant repolarization heterogeneities and form an arrhythmogenic substrate that allows for functional re-entries to arise. The arrhythmogenic properties of this functional substrate following acute ischemia were examined in whole-heart models.57 The authors first determined that sodium channel availability was an important factor in regulating the arrhythmogenicity of the ischemic zone. Decreased sodium channel availability increases arrhythmic risk and the probability of focal ectopic beats from the right ventricle and ventricular base. Second, in a separate study, the same authors modeled both subendocardial and transmural ischemic distributions.58 The authors identified two distinct mechanisms in both ischemic region distributions: macro-re-entry around the region for transmural ischemic and micro-re-entry in the ischemic region border zone for subendocardial ischemia.
Ventricular fibrillation (VF) is another type of VA that is often believed to involve functional-type re-entrant patterns. However, due to its complexity and lethality, it is difficult to characterize VF dynamics experimentally or clinically. Multiple-wavelet and/or mother-rotor have been hypothesized to be the main mechanisms of VF. These dynamics are in turn governed by the shape of the action potential duration (APD) restitution (APDR) and conduction velocity (CV) restitution (CVR) curves. In one such computational study, a nonionic “rule-based” (Wei–Harumi whole-heart model) whole-heart model was used to examine the effects of varying APDR and CVR curves on VF organization and conversion.59 The results show how having a flattened APDR tends to cause multiple-wavelet VF to organize into VT whereas VT degenerates into VF due to spatial heterogeneity of APDR. This study shows how the synergy between APDR and CVR contributes to the transition between multiple-wavelet and mother-rotor mechanisms in VF. A separate study demonstrated how in a 3D heart model with the patient-specific scar distribution represented functional re-entry rotors preferentially anchored to regions with fibrosis, emphasizing the importance of fibrosis in sustaining re-entries.60
Polymorphic VTs represent another type of VAs. Unlike monomorphic VTs, polymorphic VTs tend to have more functional-type re-entrant patterns and do not typically possess a fixed rotor. Torsades de pointes (TdP) is an example of a clinically recognized polymorphic VT that arises from increased repolarization dispersion. Computational models can reveal deeper insights into the mechanisms of how TdP manifests.61 In this study, the authors induced repolarization heterogeneities in the whole-heart models and examined the resultant arrhythmia dynamics. From these experiments, they identified two potential mechanisms of TdP genesis: initiation via multiple ectopic foci or early afterdepolarizations, inducing block and subsequent re-entry. In a combined experimental and computational study, it was demonstrated how phase singularities of the TdP re-entrant wave initiated in areas of regional repolarization gradients and anchored to areas with the greatest difference in local repolarization properties.62 Collectively, these studies demonstrate the advantages of computational modeling over experimental or clinical approaches in elucidating complex arrhythmia mechanisms.
INSIGHTS INTO NOVEL THERAPIES
Novel emergent cardiac therapies hold significant promise but are difficult to assess in vivo and in vitro. Carefully designed, numerical studies built upon basic biophysical principles can offer predictions beyond what is currently capable of being assessed experimentally. These predictions can help direct further research and offer a roadmap for future experimental designs as technologies become available.
Assessing arrhythmogenicity of cell-based regenerative therapies
Cell-based cardiac regenerative therapies, a promising treatment to reverse cardiac remodeling in the post-infarct heart, have been found to be arrhythmogenic. However, why these newly engrafted cells can be arrhythmogenic is poorly understood. In a novel study to better elucidate these mechanisms, the authors devised a computational multi-scale whole-heart modeling framework to simulate the consequences of different cell-based therapy modalities.63 Figure 4 highlights how patch engraftment can be simulated in whole-heart models, and the subsequent arrhythmogenicity can be evaluated. Several unprecedented arrhythmogenic mechanisms of stem cell engraftment were revealed by the simulation results. First, for pluripotent stem cell-derived cardiomyocytes (PSC-CMs) injection, parameters such as injection location, cell dosage, and engraftment spatial distribution are decisive in the occurrence of ectopic propagations. Finally, In PSC-CM cell sheet transplantation, computational models using various parameter settings showed that the engraftment location and its impact on substrate heterogeneity primarily determines VT inducibility (Fig. 4). A recent study assessed the arrhythmogenic effects of stem cell-derived cardiomyocyte engraftment in models with patient-specific fibrotic distributions.64 They determined that arrhythmias arising from engraftment were likely to be from re-entrant, not focal, mechanisms, and that the location of the patch engraftment relative to the patient-specific fibrotic distribution was important in determining arrhythmogenicity.
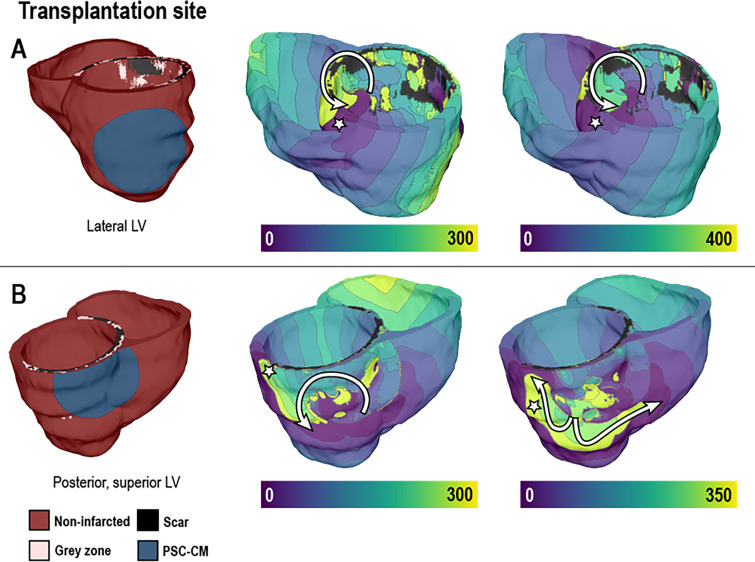
FIG. 4.
Examining arrhythmogenic mechanisms in post-infarct hearts with pluripotent stem cell-derived cardiomyocyte (PSC-CM) transplantation. Colors indicate the sequence of activation (ms). Pacing to induce arrhythmia was delivered from sites marked with the white star. Transplantation of stem-cell-derived cardiomyocytes resulted in re-entrant arrhythmias (pathway traced with the white arrow), suggesting that the location of this cell therapy could be arrhythmogenic. Reproduced with permission from Yu et al., Sci. Rep. 9, 9238 (2019). Copyright 2019 Author(s), licensed under a Creative Commons Attribution (CC BY) license.63
Determining feasibility of optogenetics for arrhythmia treatment
Optogenetics-based defibrillation has been proposed as a novel potential alternative to ICD therapy in ventricular fibrillation (VF) due to its noninvasive and less distressing therapeutic delivery.65 However, the feasibility of using light stimuli to pace or intervene in arrhythmic activity on human-scale clinical applications and the ideal opsin properties for terminating VF in humans remains unclear.66 To address these unanswered questions, a recent computational simulation study modeled optogenetic therapy in the context of the whole human ventricle.67 Four parameters (opsin variants, optrode grid densities, light pulse duration, and light pulse timings) were manipulated to construct 96 different configurations of ventricular simulations. The therapeutic efficacy depended on the extent of the propagating wavefront, which was equivalently quantified as the volume of tissue excited by the light source. In these numerical experiments, red light successfully terminated VF, while blue light was not able to do so in any combination. Opsin red light sensitivity primarily determined the successfulness of VF termination, LED array density and longer pulse duration being the subsidiary factors of defibrillation efficacy.
CLINICAL APPLICATIONS
Computational heart models can be used for personalized and noninvasive arrhythmia prognosis for post-infarct patients. The main steps in personalized virtual heart model construction are the acquisition of images, segmentation and labeling of diseased tissues from imaging, construction of a 3D heart geometry, incorporation of fiber orientation, and assignment of electrophysiological property in each region.41–43,68,69 Pacing protocols adapted from clinical procedures are then applied to induce VT in the virtual heart. The outcome of these virtual electrophysiological studies is then used to establish a patient's arrhythmic risk or to determine ablation targets.
Assessment of patient arrhythmic risk
Accurate scar segmentation relies on cardiac MR image acquisition protocol as well as the image post-processing techniques. This variability was studied by building personalized computational heart models using imaging protocol-based variations and examining the corresponding differences in the image-based virtual heart model outcomes (n = 25).69 The sensitivity and specificity of virtual models were over 66% for the clinical outcomes based on VT inducibility regardless of the imaging sequence. Multi-contrast late enhancement (MCLE), a quantitative T1 mapping technique, had higher specificity (>80%) and sensitivity (>80%) for the clinical outcomes. This result suggests that quantitative imaging protocols, which are sensitive to the native tissue contrast, might overcome the current issues with LGE-MRI, including manual scar segmentation and thresholding errors.
In a retrospective computational study consisting of 41 post-infarct patients with reduced ejection fraction, the authors demonstrated that virtual heart inducibility was more predictive for re-entrant arrhythmia than the multiple standard clinical metrics.42 Patients with inducible virtual hearts were more likely to correspond to those who suffered arrhythmic outcomes than patients with non-inducible virtual hearts. This study highlighted how the virtual-heart approach could be used to noninvasively determine a patient's arrhythmia risk. In a separate proof-of-concept study, the authors investigated VT risk in a small cohort of myocardial infarction (MI) patients (n = 4) with preserved ejection fraction.70 Even though the patients in this cohort were not candidates for ICD placement, one patient had a VT history, and the personalized simulation results matched the clinical result, indicating the generalizability of using virtual electrophysiological studies to assess VA risk.
Virtual-heart arrhythmia risk stratification has also been adapted for non-infarct related arrhythmias. In a pediatric cohort, virtual-heart technology was used to investigate the VT induction propensity for patients with acute myocarditis (n = 12).71 Models successfully determined the VT inducibility for all patients and outperformed the clinical metrics. However, LGE MRI cannot distinguish acute (edema) and chronic states (scar or fibrosis) of myocardial injury from myocarditis. This study showed that modeling both conditions with altered conductivity and action potential duration yields highly predictive personalized heart models. In a separate non-ischemic disease process, patients with tetralogy of Fallot (rTOF) who underwent surgical intervention in their childhood are at higher risk of VT after the procedure due to fibrotic remodeling. A recent study applied virtual-heart technology to arrhythmia risk assessment in rTOF patients.72 In this cohort, the authors examined VT inducibility for seven patients who were deemed to be at low risk according to the clinical guidelines (prolonged QRS duration). Virtual pacing in both ventricles resulted in the re-entrant VTs for patients with clinically detected VT (n = 2), while clinically VT-negative patient models were not inducible. These studies illustrate the vast generalizability of using virtual-heart technology for risk stratification.
More recently, machine learning (ML) techniques have been combined with image-based virtual heart simulations to understand disease mechanisms and improve VT risk stratification. One study integrated medical images with ECG data to group hypertrophic cardiomyopathy (HCM) patients into different phenotypes.73 The aim of the study was to understand the mechanisms underlying abnormal ECG patterns and evaluate the risk associated with each phenotype. An unsupervised clustering algorithm assigned patients into four groups based on their ECG characteristics. Various hypotheses were tested to virtually reproduce the phenotypic ECG characteristics with the LGE MRI image-based simulations. The authors found two distinct mechanisms underlying ECG abnormalities in HCM, associated with ionic remodeling and abnormal conduction, respectively, and the subgroup with ionic remodeling expression had the highest SCD risk score. These findings led to a better HCM patient stratification and benefit the clinical therapy selection. Although not directly pertaining to VTs, a new study developed a novel ML based-approach for arrhythmia risk stratification by combining simulation and raw-image (LGE-MRI) based features.74 An ML classifier had high specificity and sensitivity (>80%) for arrhythmia recurrence risk with features from simulations with a minimal contribution from raw image-based features. The methodology from this study could be readily adopted for VT risk stratification in the future.
Guiding VT ablation therapy
Catheter ablation is a major adjunct in the contemporary management of VTs. This minimally invasive procedure involves the use of catheters that are maneuvered and placed into the cardiac chambers. Radiofrequency energy is then delivered to specific diseased areas of the myocardium to terminate the source of the VA. Identification of these specific locations is difficult and requires careful characterization of the arrhythmogenic substrate through a laborious process called electroanatomic mapping (EAM). Ablation lesions are then delivered at sites of abnormal electrical signals according to the EAM. This procedure is time-consuming and does not guarantee VT termination. Consequently, ablation targets can be inaccurate and may lead to VA recurrence. Patient-specific computational heart modeling can aid in improving ablation precision by proposing ablation targets, providing noninvasive localization of abnormal electrical signals, and/or identifying VT exit sites.
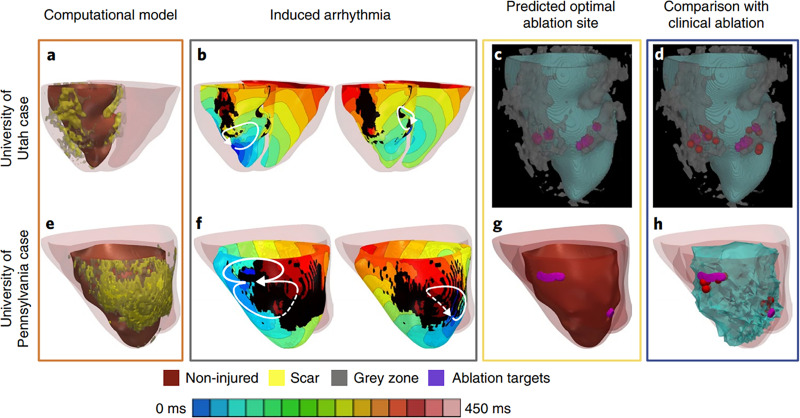
Virtual-heart technology has seen great success in identifying optimal ablation targets to terminate VT. A retrospective feasibility study with 13 patients who underwent ablation showed that ablation targets from image-based simulations were consistent with the clinical targets.75 These results highlight how in silico mapping strategies can achieve similar results noninvasively and provide more mechanistic insights into the disease. The authors concluded that in addition to the slow conducting border zone, the infarct core rim was also part of the VT re-entry circuit. Recently, this work was extended into the first prospective study that used cardiac electrophysiological whole-heart modeling to affect patient care.41 The authors successfully pinpointed ablation targets with virtual hearts in the first virtual-heart prospective study with five patients, along with 21 retrospective human and animal studies. Figure 5 depicts the simulation results and clinical outcomes of two of the prospective patients who underwent VT ablation. Although these models were by design not calibrated to the patient's electrophysiology, the predicted ablation lesions still successfully terminated VT in these prospective patients. Further sensitivity analyses demonstrated that these ablation targets were largely robust to various changes in electrophysiological properties.43 This landmark work highlights the vast potential for virtual-heart technology to impact the clinical management of VAs. In addition, the virtual heart approach can also be used to assess the efficacy of emerging technologies that may not be ready for clinical use. A recent study evaluated the VT termination success of an augmented reality-based catheter navigation system using a virtual heart approach.76 Using virtual-heart modeling, this study demonstrated how the augmented reality system could improve ablation targeting.
FIG. 5.
Virtual-heart technology for guiding ventricular tachycardia ablation. Patient-specific computational models reconstructed from clinical images. Electrophysiological properties are assigned to both non-diseased and diseased tissue. From these models, ventricular tachycardia is simulated and corresponding ablation targets that terminate re-entry are determined. These ablation targets can then be incorporated into electroanatomic mapping systems where they can be used to guide VT ablation therapy. Reproduced with permission from Prakosa et al., Nat. Biomed. Eng. 2, 732 (2018). Copyright 2018 Author(s), licensed under a Creative Commons Attribution (CC BY) license.41
Subsequent studies have also demonstrated the potential for patient-specific computational heart models to delineate the VT circuit and help determine ablation targets. In a study consisting of seven MRI-based patient-specific heart models, the authors gained mechanistic insights into the VT circuit physiology.77 They estimated the re-entry circuits with simulations and validated these estimations with electroanatomical mapping data. VT inducibility was accurately simulated for all patients, and VT entry and exit sites were associated with the heterogeneous distribution of action potential duration restitution and conductivity. A separate study extensively personalized a single 3D ventricular model to accurately reproduce the patient-specific VT circuit.78 Using clinical data, they manually fine-tuned their model parameters until the simulated VT morphology was consistent with the clinically induced VT morphology. They investigated the border zone electrical properties with varying fibrosis constituents (from 10% to 30%). A combination of the border zone with 30% replacement fibrosis, heterogeneity in action potential durations, and reduced conduction velocity resulted in the most realistic representation of the patient-specific VT. This study highlights how computational heart models can help elucidate the VT circuit which in turn could aid pre-procedural ablation planning. Finally, a recent study compared automated ECG-based localization algorithm with image-based virtual-heart predictions of VT circuits in four post-infarct patients.79 Overall, the authors found reasonable spatial concordance between VT exit sites predicted by the ECG-based algorithm and the image-based virtual heart approach, demonstrating a synergistic nature between the two methodologies. This study highlights the utility of using virtual-heart modeling to delineate the VT circuit and hence determine ablation targets.
Patient-specific, computational models can also provide noninvasive characterization of the electrical substrate to aid in pre-procedural ablation planning. Pace mapping is a clinical electrophysiological technique used during substrate mapping to localize the VT exit site, which in some cases can represent good targets for ablation. A new study offered a framework for how simulations of pace mapping in whole-heart models could be compared with clinically recorded electrograms to aid in pre-procedural planning.80 The noninvasive methodology that they outlined involved first simulating VT in human whole-heart models that included porcine infarct geometries. Then, pseudo-ECGs were then computed from these VT morphologies as well as pseudo electrograms from an implantable cardioverter defibrillator (ICD). They showed that the simulated pace mapping could theoretically be used to identify the VT exit sites and slow conducting isthmuses, potentially offering a valuable noninvasive tool to aid pre-procedural ablation planning. Efforts have also been taken to reproduce intracardiac electrograms that would be recorded during substrate mapping. In one such study, patient-specific heart models successfully reproduced the abnormal patterns in intracardiac electrograms which could represent targets for ablation.81 The goal of this study was to explore the biophysical mechanisms that lead to the fractioned border zone electrograms. For each electrogram, several statistics were computed to summarize each signal. These statistics were then compared between normal and abnormal cases of both the clinical and simulated electrograms. The difference between normal and abnormal pseudo-electrograms showed resemblance to the clinical counterparts. Similar to previous studies, this simulation technique could potentially be used to identify ablation targets noninvasively. The authors further advanced this methodology by combining this biophysical modeling approach with ML.82 The authors sought to develop an ML classifier that could accurately distinguish between normal and abnormal electrogram signals. The classifier, trained with image-based and simulation-based features, is able to accurately identify abnormal intracardiac electrograms. Although not all regions with abnormal electrograms need to be ablated, this study represents a stark advancement in computational heart modeling approaches.
In addition to LGE-CMR, CT has also been used in the pre-procedural assessment of arrhythmogenic substrate. A recent study outlined a different approach for virtual-heart reconstruction than previous studies, using CT images and assuming regions of wall thinning to be scar.83 The scar was modeled with reduced conduction speed as a function of myocardial wall thickness. This study included five patients with chronic infarct and thinning myocardial wall. This computational workflow was designed to be computationally efficient and integrated into clinical workflows as a noninvasive, intra-operative mapping tool for ablation therapy. Infiltrating adipose tissue on CT has been identified as substrates based on intensity values on CT and combined with virtual heart technology to predict VT ablation targets.40 Rapid pacing was then used to induce VTs. Each VT was analyzed, and corresponding ablation targets were determined to terminate each VT pathway. The authors further validated this approach in a retrospective study consisting of 29 post-infarct patients who underwent VT ablation. Overall, the predicted ablation targets by virtual-heart were concordant with the clinical ablation targets and required overall less ablation volumes. Moreover, since CT is more accessible across a broad range of clinical centers, such technologies could be readily deployed prospectively to improve VT ablation strategies.
Previous computational studies have evaluated heterogeneity in action potential duration as a VT susceptibility metric. A recent study examined the interaction between activation and repolarization wavefronts into in silico mapping experiments.84 A novel technique called re-entry vulnerability index (RVI) pinpointed the slow conducting and abnormal repolarized sites without needing the induced VT. The authors evaluated the RVI algorithm's performance at different electrophysiological measurement conditions and showed the potential use of RVI to target re-entrant circuits in the clinical setting. The same authors later simulated substrate mapping using a porcine heart model and attempted to identify the ablation targets using RVI and endocardial electrogram features without inducing VT.85 Activation time (AT) gradients combined with voltage cutoffs successfully identified the VT exit sites, while RVI based maps determined the region near the VT exit sites.
FUTURE PERSPECTIVES
In this review, we have summarized recent achievements and advancements of whole-heart computational modeling in uncovering mechanisms of VA, predicting results of novel therapeutics not currently attainable by experimental means, and improving clinical VA management. These applications highlight the transformative nature of computational whole-heart modeling in VA and its role in precision cardiovascular medicine. As high-performance computing and machine learning become increasingly sophisticated, newer advancements will likely develop in whole-heart modeling. Machine-learning based approaches are becoming more and more commonplace in the field of cardiac electrophysiology.86 Such tools are well-designed to tackle several of the deficiencies in cardiac modeling. For instance, physics-based deep neural network methodologies are being developed to bypass the computationally expensive nature of executing bidomain and monodomain simulations and may eventually be extended to whole-heart modeling.87 Having such tools would allow for greater flexibility in assessing a multitude of model parameters which would allow for fine-tuned, personalized simulations of the patient's electrophysiology to inform clinical providers in real-time. With the advent of such improvements, computational whole-heart modeling is well-poised to become an integral part in both mechanistic understanding and clinical care of VAs.
DATA AVAILABILITY
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- 1. Narayan S. M., Wang P. J., and Daubert J. P., “ New concepts in sudden cardiac arrest to address an intractable epidemic: JACC state-of-the-art review,” J. Am. Coll. Cardiol. 73(1), 70–88 (2019). 10.1016/j.jacc.2018.09.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hodgkin A. L. and Huxley A. F., “ A quantitative description of membrane current and its application to conduction and excitation in nerve,” J. Physiol. 117(4), 500–544 (1952). 10.1113/jphysiol.1952.sp004764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. ten Tusscher K. H. W. J. and Panfilov A. V., “ Alternans and spiral breakup in a human ventricular tissue model,” Am. J. Physiol.—Heart Circ. Physiol. 291(3), H1088 (2006). 10.1152/ajpheart.00109.2006 [DOI] [PubMed] [Google Scholar]
- 4. O'Hara T., Virág L., Varró A., and Rudy Y., “ Simulation of the undiseased human cardiac ventricular action potential: Model formulation and experimental validation,” PLoS Comput. Biol. 7(5), e1002061 (2011). 10.1371/journal.pcbi.1002061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vigmond E. J., Aguel F., and Trayanova N. A., “ Computational techniques for solving the bidomain equations in three dimensions,” IEEE Trans. Biomed. Eng. 49(11), 1260–1269 (2002). 10.1109/TBME.2002.804597 [DOI] [PubMed] [Google Scholar]
- 6. Bishop M. J. and Plank G., “ Representing cardiac bidomain bath-loading effects by an augmented monodomain approach: Application to complex ventricular models,” IEEE Trans. Biomed. Eng. 58(4), 1066–1075 (2011). 10.1109/TBME.2010.2096425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vigmond E. J. and Clements C., “ Construction of a computer model to investigate sawtooth effects in the Purkinje system,” IEEE Trans. Biomed. Eng. 54(3), 389–399 (2007). 10.1109/TBME.2006.888817 [DOI] [PubMed] [Google Scholar]
- 8. Mahida S., Sacher F., Dubois R. et al. , “ Cardiac imaging in patients with ventricular tachycardia,” Circulation 136(25), 2491–2507 (2017). 10.1161/CIRCULATIONAHA.117.029349 [DOI] [PubMed] [Google Scholar]
- 9. Schmidt A., Azevedo C. F. et al. , “ Infarct tissue heterogeneity by magnetic resonance imaging identifies enhanced cardiac arrhythmia susceptibility in patients with left ventricular dysfunction,” Circulation 115, 2006 (2007). 10.1161/CIRCULATIONAHA.106.653568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Okada D. R., Miller J., Chrispin J. et al. , “ Substrate spatial complexity analysis for the prediction of ventricular arrhythmias in patients with ischemic cardiomyopathy,” Circ.: Arrhythmia Electrophysiol. 13(4), 281–290 (2020). 10.1161/CIRCEP.119.007975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Disertori M., Rigoni M., Pace N. et al. , “ Myocardial fibrosis assessment by LGE is a powerful predictor of ventricular tachyarrhythmias in ischemic and nonischemic LV dysfunction: A meta-analysis,” JACC: Cardiovasc. Imaging 9, 1046 (2016). 10.1016/j.jcmg.2016.01.033 [DOI] [PubMed] [Google Scholar]
- 12. Zegard A., Okafor O., de Bono J. et al. , “ Myocardial fibrosis as a predictor of sudden death in patients with coronary artery disease,” J. Am. Coll. Cardiol. 77(1), 29–41 (2021). 10.1016/j.jacc.2020.10.046 [DOI] [PubMed] [Google Scholar]
- 13. Roca-Luque I., van Breukelen A., Alarcon F. et al. , “ Ventricular scar channel entrances identified by new wideband cardiac magnetic resonance sequence to guide ventricular tachycardia ablation in patients with cardiac defibrillators,” EP Europace 22(4), 598–606 (2020). 10.1093/europace/euaa021 [DOI] [PubMed] [Google Scholar]
- 14. Wu K. C., Weiss R. G., Thiemann D. R. et al. , “ Late gadolinium enhancement by cardiovascular magnetic resonance heralds an adverse prognosis in nonischemic cardiomyopathy,” J. Am. Coll. Cardiol. 51(25), 2414–2421 (2008). 10.1016/j.jacc.2008.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Becker M. A. J., Cornel J. H., van de Ven P. M., van Rossum A. C., Allaart C. P., and Germans T., “ The prognostic value of late gadolinium-enhanced cardiac magnetic resonance imaging in nonischemic dilated cardiomyopathy: A review and meta-analysis,” JACC: Cardiovasc. Imaging 11(9), 1274–1284 (2018). 10.1016/j.jcmg.2018.03.006 [DOI] [PubMed] [Google Scholar]
- 16. Muser D., Nucifora G., Castro S. A. et al. , “ Myocardial substrate characterization by CMR T1 mapping in patients with NICM and no LGE undergoing catheter ablation of VT,” JACC: Clin. Electrophysiol. 7, 831 (2021). 10.1016/j.jacep.2020.10.002 [DOI] [PubMed] [Google Scholar]
- 17. Chang H. J., George R. T., Schuleri K. H. et al. , “ Prospective electrocardiogram-gated delayed enhanced multidetector computed tomography accurately quantifies infarct size and reduces radiation exposure,” JACC: Cardiovasc. Imaging 2(4), 412–420 (2009). 10.1016/j.jcmg.2008.12.019 [DOI] [PubMed] [Google Scholar]
- 18. Yamashita S., Sacher F., Hooks D. A. et al. , “ Myocardial wall thinning predicts transmural substrate in patients with scar-related ventricular tachycardia,” Heart Rhythm 14(2), 155–163 (2017). 10.1016/j.hrthm.2016.11.012 [DOI] [PubMed] [Google Scholar]
- 19. Komatsu Y., Cochet H., Jadidi A. et al. , “ Regional myocardial wall thinning at multidetector computed tomography correlates to arrhythmogenic substrate in postinfarction ventricular tachycardia: Assessment of structural and electrical substrate,” Circ.: Arrhythmia Electrophysiol. 6(2), 342–350 (2013). 10.1161/CIRCEP.112.000191 [DOI] [PubMed] [Google Scholar]
- 20. Takigawa M., Duchateau J., Sacher F. et al. , “ Are wall thickness channels defined by computed tomography predictive of isthmuses of postinfarction ventricular tachycardia?,” Heart Rhythm 16, 1661 (2019). 10.1016/j.hrthm.2019.06.012 [DOI] [PubMed] [Google Scholar]
- 21. Takigawa M., Martin R., Cheniti G. et al. , “ Detailed comparison between the wall thickness and voltages in chronic myocardial infarction,” J. Cardiovasc. Electrophysiol. 30(2), 195–204 (2019). 10.1111/jce.13767 [DOI] [PubMed] [Google Scholar]
- 22. Raney A. R., Saremi F., Kenchaiah S. et al. , “ Multidetector computed tomography shows intramyocardial fat deposition,” J. Cardiovasc. Comput. Tomogr. 2(3), 152–163 (2008). 10.1016/j.jcct.2008.01.004 [DOI] [PubMed] [Google Scholar]
- 23. Aliyari Ghasabeh M., te Riele A. S. J. M., James C. A. et al. , “ Epicardial fat distribution assessed with cardiac CT in arrhythmogenic right ventricular dysplasia/cardiomyopathy,” Radiology 289(3), 641–648 (2018). 10.1148/radiol.2018180224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sasaki T., Calkins H., Miller C. F. et al. , “ New insight into scar-related ventricular tachycardia circuits in ischemic cardiomyopathy: Fat deposition after myocardial infarction on computed tomography—A pilot study,” Heart Rhythm 12(7), 1508–1518 (2015). 10.1016/j.hrthm.2015.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cheniti G., Sridi S., Sacher F. et al. , “ Post-myocardial infarction scar with fat deposition shows specific electrophysiological properties and worse outcome after ventricular tachycardia ablation,” J. Am. Heart Assoc. 8(15), e012482 (2019). 10.1161/JAHA.119.012482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kalisz K., Buethe J., Saboo S. S., Abbara S., Halliburton S., and Rajiah P., “ Artifacts at cardiac CT: Physics and solutions,” RadioGraphics 36(7), 2064–2083 (2016). 10.1148/rg.2016160079 [DOI] [PubMed] [Google Scholar]
- 27. Villongco C. T., Krummen D. E., Stark P., Omens J. H., and McCulloch A. D., “ Patient-specific modeling of ventricular activation pattern using surface ECG-derived vectorcardiogram in bundle branch block,” Prog. Biophys. Mol. Biol. 115(2-3), 305–313 (2014). 10.1016/j.pbiomolbio.2014.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pezzuto S., Prinzen F. W., Potse M. et al. , “ Reconstruction of three-dimensional biventricular activation based on the 12-lead electrocardiogram via patient-specific modelling,” EP Europace 23(4), 640–647 (2021). 10.1093/europace/euaa330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gillette K., Gsell M. A. F., Prassl A. J. et al. , “ A framework for the generation of digital twins of cardiac electrophysiology from clinical 12-leads ECGs,” Med. Image Anal. 71, 102080 (2021). 10.1016/j.media.2021.102080 [DOI] [PubMed] [Google Scholar]
- 30. Ushenin K., Kalinin V., Gitinova S., Sopov O., and Solovyova O., “ Parameter variations in personalized electrophysiological models of human heart ventricles,” PLoS One 16(4), e0249062 (2021). 10.1371/journal.pone.0249062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grandits T., Effland A., Pock T., Krause R., Plank G., and Pezzuto S., “ GEASI: Geodesic-based earliest activation sites identification in cardiac models,” Int. J. Numer. Methods Biomed. Eng. 37, e3505 (2021). 10.1002/CNM.3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rudy Y., “ Noninvasive mapping of repolarization with electrocardiographic imaging,” J. Am. Heart Assoc. 10(9), e021396 (2021). 10.1161/JAHA.121.021396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rudy Y., “ Noninvasive ECG imaging (ECGI): Mapping the arrhythmic substrate of the human heart,” Int. J. Cardiol. 237, 13–14 (2017). 10.1016/j.ijcard.2017.02.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Giffard-Roisin S., Delingette H., Jackson T. et al. , “ Transfer learning from simulations on a reference anatomy for ECGI in personalized cardiac resynchronization therapy,” IEEE Trans. Biomed. Eng. 66(2), 343–353 (2019). 10.1109/TBME.2018.2839713 [DOI] [PubMed] [Google Scholar]
- 35. Giffard-Roisin S., Jackson T., Fovargue L. et al. , “ Noninvasive personalization of a cardiac electrophysiology model from body surface potential mapping,” IEEE Trans. Biomed. Eng. 64(9), 2206–2218 (2017). 10.1109/TBME.2016.2629849 [DOI] [PubMed] [Google Scholar]
- 36. Sánchez C., D'Ambrosio G., Maffessanti F. et al. , “ Sensitivity analysis of ventricular activation and electrocardiogram in tailored models of heart-failure patients,” Med. Biol. Eng. Comput. 56(3), 491–504 (2018). 10.1007/s11517-017-1696-9 [DOI] [PubMed] [Google Scholar]
- 37. Potse M., Krause D., Kroon W. et al. , “ Patient-specific modelling of cardiac electrophysiology in heart-failure patients,” Europace 16(Suppl 4), iv56–iv61 (2014). 10.1093/europace/euu257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lubrecht J. M., Grandits T., Gharaviri A. et al. , “ Automatic reconstruction of the left atrium activation from sparse intracardiac contact recordings by inverse estimate of fibre structure and anisotropic conduction in a patient-specific model,” Europace 23(Supplement_1), I63–I70 (2021). 10.1093/europace/euaa392 [DOI] [PubMed] [Google Scholar]
- 39. Aronis K. N., Prakosa A., Bergamaschi T. et al. , “ Characterization of the electrophysiologic remodeling of patients with ischemic cardiomyopathy by clinical measurements and computer simulations coupled with machine learning,” Front. Physiol. 12, 1079 (2021). 10.3389/FPHYS.2021.684149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sung E., Prakosa A., Aronis K. N. et al. , “ Personalized digital-heart technology for ventricular tachycardia ablation targeting in hearts with infiltrating adiposity,” Circ.: Arrhythmia Electrophysiol. 13, e008912 (2020). 10.1161/CIRCEP.120.008912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Prakosa A., Arevalo H. J., Deng D. et al. , “ Personalized virtual-heart technology for guiding the ablation of infarct-related ventricular tachycardia,” Nat. Biomed. Eng. 2(10), 732–740 (2018). 10.1038/s41551-018-0282-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Arevalo H. J., Vadakkumpadan F., Guallar E. et al. , “ Arrhythmia risk stratification of patients after myocardial infarction using personalized heart models,” Nat. Commun. 7, 11437 (2016). 10.1038/ncomms11437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Deng D., Prakosa A., Shade J., Nikolov P., and Trayanova N. A., “ Sensitivity of ablation targets prediction to electrophysiological parameter variability in image-based computational models of ventricular tachycardia in post-infarction patients,” Front. Physiol. 10, 628 (2019). 10.3389/fphys.2019.00628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. de Bakker J. M. T., van Capelle F. J. L., Janse M. J. et al. , “ Reentry as a cause of ventricular tachycardia in patients with chronic ischemic heart disease: Electrophysiology and anatomic correlation,” Circulation 77(3), 589–606 (1988). 10.1161/01.CIR.77.3.589 [DOI] [PubMed] [Google Scholar]
- 45. Bayer J. D., Lalani G. G., Vigmond E. J., Narayan S. M., and Trayanova N. A., “ Mechanisms linking electrical alternans and clinical ventricular arrhythmia in human heart failure,” Heart Rhythm 13(9), 1922–1931 (2016). 10.1016/j.hrthm.2016.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Campos F. O., Shiferaw Y., Weber dos Santos R., Plank G., and Bishop M. J., “ Microscopic isthmuses and fibrosis within the border zone of infarcted hearts promote calcium-mediated ectopy and conduction block,” Front. Phys. 6, 57 (2018). 10.3389/fphy.2018.00057 [DOI] [Google Scholar]
- 47. Campos F. O., Whitaker J., Neji R. et al. , “ Factors Promoting conduction slowing as substrates for block and reentry in infarcted hearts,” Biophys. J. 117(12), 2361–2374 (2019). 10.1016/j.bpj.2019.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Deo M., Boyle P. M., Kim A. M., and Vigmond E. J., “ Arrhythmogenesis by single ectopic beats originating in the Purkinje system,” Am. J. Physiol.: Heart Circ. Physiol. 299(4), 1002–1011 (2010) 10.1152/AJPHEART.01237.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Deo M., Boyle P., Plank G., and Vigmond E., “ Arrhythmogenic mechanisms of the Purkinje system during electric shocks: A modeling study,” Heart Rhythm 6(12), 1782–1789 (2009). 10.1016/j.hrthm.2009.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pashakhanloo F., Herzka D. A., Halperin H., McVeigh E. R., and Trayanova N. A., “ Role of 3-dimensional architecture of scar and surviving tissue in ventricular tachycardia: Insights from high-resolution ex vivo porcine models,” Circ.: Arrhythmia Electrophysiol. 11(6), e006131 (2018). 10.1161/CIRCEP.117.006131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Deng D., Prakosa A., Shade J., Nikolov P., and Trayanova N. A., “ Characterizing conduction channels in postinfarction patients using a personalized virtual heart,” Biophys. J. 117, 2287 (2019). 10.1016/j.bpj.2019.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Arevalo H., Plank G., Helm P., Halperin H., and Trayanova N., “ Tachycardia in post-infarction hearts: Insights from 3D image-based ventricular models,” PLoS One 8, e68872 (2013). 10.1371/journal.pone.0068872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ringenberg J., Deo M., Filgueiras-Rama D. et al. , “ Effects of fibrosis morphology on reentrant ventricular tachycardia inducibility and simulation fidelity in patient-derived models,” Clin. Med. Insights: Cardiol. 8(Suppl 1), 1–13 (2014). 10.4137/CMC.S15712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liang C., Wang K., Li Q., Bai J., and Zhang H., “ Influence of the distribution of fibrosis within an area of myocardial infarction on wave propagation in ventricular tissue,” Sci. Rep. 9(1), 14151 (2019). 10.1038/s41598-019-50478-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Colli-Franzone P., Gionti V., Pavarino L. F., Scacchi S., and Storti C., “ Role of infarct scar dimensions, border zone repolarization properties and anisotropy in the origin and maintenance of cardiac reentry,” Math. Biosci. 315, 108228 (2019). 10.1016/j.mbs.2019.108228 [DOI] [PubMed] [Google Scholar]
- 56. Balaban G., Costa C. M., Porter B. et al. , “ 3D electrophysiological modeling of interstitial fibrosis networks and their role in ventricular arrhythmias in non-ischemic cardiomyopathy,” IEEE Trans. Biomed. Eng. 67(11), 3125–3133 (2020). 10.1109/TBME.2020.2976924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Martinez-Navarro H., Zhou X., Bueno-Orovio A., and Rodriguez B., “ Electrophysiological and anatomical factors determine arrhythmic risk in acute myocardial ischaemia and its modulation by sodium current availability,” Interface Focus 11(1), 20190124 (2021). 10.1098/rsfs.2019.0124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Martinez-Navarro H., Mincholé A., Bueno-Orovio A., and Rodriguez B., “ High arrhythmic risk in antero-septal acute myocardial ischemia is explained by increased transmural reentry occurrence,” Sci. Rep. 9(1), 16803 (2019). 10.1038/s41598-019-53221-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zheng Y., Wei D., Zhu X., Chen W., Fukuda K., and Shimokawa H., “ Ventricular fibrillation mechanisms and cardiac restitutions: An investigation by simulation study on whole-heart model,” Comput. Biol. Med. 63, 261–268 (2015). 10.1016/j.compbiomed.2014.06.014 [DOI] [PubMed] [Google Scholar]
- 60. Vandersickel N., Watanabe M., Tao Q., Fostier J., Zeppenfeld K., and Panfilov A. v, “ Dynamical anchoring of distant arrhythmia sources by fibrotic regions via restructuring of the activation pattern,” PLoS Comput. Biol. 14(12), e1006637 (2018). 10.1371/journal.pcbi.1006637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vandersickel N., de Boer T. P., Vos M. A., and Panfilov A. v, “ Perpetuation of torsade de pointes in heterogeneous hearts: Competing foci or re-entry?,” J. Physiol. 594(23), 6865–6878 (2016). 10.1113/JP271728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rivaud M. R., Bayer J. D., Cluitmans M. et al. , “ Critical repolarization gradients determine the induction of reentry-based torsades de pointes arrhythmia in models of long QT syndrome,” Heart Rhythm 18(2), 278–287 (2021). 10.1016/j.hrthm.2020.09.020 [DOI] [PubMed] [Google Scholar]
- 63. Yu J. K., Franceschi W., Huang Q., Pashakhanloo F., Boyle P. M., and Trayanova N. A., “ A comprehensive, multiscale framework for evaluation of arrhythmias arising from cell therapy in the whole post-myocardial infarcted heart,” Sci. Rep. 9(1), 9238 (2019). 10.1038/s41598-019-45684-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yu J. K., Liang J. A., Franceschi W. H. et al. , “ Assessment of arrhythmia mechanism and burden of the infarcted ventricles following remuscularization with pluripotent stem cell-derived cardiomyocyte patches using patient-derived models,” Cardiovasc. Res. (published online) (2021). 10.1093/cvr/cvab140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Boyle P. M., Karathanos T. V., and Trayanova N. A., “ Cardiac Optogenetics 2018,” JACC: Clin. Electrophysiol. 4(2), 155–167 (2018). 10.1016/j.jacep.2017.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Boyle P. M., Karathanos T. V., Entcheva E., and Trayanova N. A., “ Computational modeling of cardiac optogenetics: Methodology overview & review of findings from simulations,” Comput. Biol. Med. 65, 200–208 (2015). 10.1016/j.compbiomed.2015.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Karathanos T. V., Bayer J. D., Wang D., Boyle P. M., and Trayanova N. A., “ Opsin spectral sensitivity determines the effectiveness of optogenetic termination of ventricular fibrillation in the human heart: A simulation study,” J. Physiol. 594(23), 6879–6891 (2016). 10.1113/JP271739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ukwatta E., Arevalo H., Rajchl M. et al. , “ Image-based reconstruction of three-dimensional myocardial infarct geometry for patient-specific modeling of cardiac electrophysiology,” Med. Phys. 42(8), 4579–4590 (2015). 10.1118/1.4926428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ukwatta E., Nikolov P., Zabihollahy F., Trayanova N. A., and Wright G. A., “ Virtual electrophysiological study as a tool for evaluating efficacy of MRI techniques in predicting adverse arrhythmic events in ischemic patients,” Phys. Med. Biol. 63(22), 225008 (2018). 10.1088/1361-6560/aae8b2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Deng D., Arevalo H. J., Prakosa A., Callans D. J., and Trayanova N. A., “ A feasibility study of arrhythmia risk prediction in patients with myocardial infarction and preserved ejection fraction,” Europace 18(suppl_4), iv60–iv66 (2016). 10.1093/europace/euw351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cartoski M. J., Nikolov P. P., Prakosa A., Boyle P. M., Spevak P. J., and Trayanova N. A., “ Computational identification of ventricular arrhythmia risk in pediatric myocarditis,” Pediatr. Cardiol. 40(4), 857–864 (2019). 10.1007/s00246-019-02082-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shade J. K., Cartoski M. J., Nikolov P. et al. , “ Ventricular arrhythmia risk prediction in repaired tetralogy of Fallot using personalized computational cardiac models,” Heart Rhythm 17(3), 408–414 (2020). 10.1016/j.hrthm.2019.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lyon A., Mincholé A., Bueno-Orovio A., and Rodriguez B., “ Improving the clinical understanding of hypertrophic cardiomyopathy by combining patient data, machine learning and computer simulations: A case study,” Morphologie 103(343), 169–179 (2019). 10.1016/j.morpho.2019.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Shade J. K., Ali R. L., Basile D. et al. , “ Preprocedure application of machine learning and mechanistic simulations predicts likelihood of paroxysmal atrial fibrillation recurrence following pulmonary vein isolation,” Circ.: Arrhythmia Electrophysiol. 13(7), 617–627 (2020). 10.1161/CIRCEP.119.008213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ashikaga H., Arevalo H., Vadakkumpadan F. et al. , “ Feasibility of image-based simulation to estimate ablation target in human ventricular arrhythmia,” Heart Rhythm 10(8), 1109–1116 (2013). 10.1016/j.hrthm.2013.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Prakosa A., Southworth M. K., Avari Silva J. N., Silva J. R., and Trayanova N. A., “ Impact of augmented-reality improvement in ablation catheter navigation as assessed by virtual-heart simulations of ventricular tachycardia ablation,” Comput. Biol. Med. 133, 104366 (2021). 10.1016/j.compbiomed.2021.104366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chen Z., Cabrera-Lozoya R., Relan J. et al. , “ Biophysical modeling predicts ventricular tachycardia inducibility and circuit morphology: A combined clinical validation and computer modeling approach,” J. Cardiovasc. Electrophysiol. 27(7), 851–860 (2016). 10.1111/jce.12991 [DOI] [PubMed] [Google Scholar]
- 78. Lopez-Perez A., Sebastian R., Izquierdo M., Ruiz R., Bishop M., and Ferrero J. M., “ Personalized cardiac computational models: From clinical data to simulation of infarct-related ventricular tachycardia,” Front. Physiol. 10, 580 (2019). 10.3389/fphys.2019.00580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhou S., Sung E., Prakosa A. et al. , “ Feasibility study shows concordance between image‐based virtual‐heart ablation targets and predicted ECG‐based arrhythmia exit‐sites,” Pacing Clin. Electrophysiol. 44(3), 432–441 (2021). 10.1111/pace.14181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Monaci S., Strocchi M., Rodero C. et al. , “ In-silico pace-mapping using a detailed whole torso model and implanted electronic device electrograms for more efficient ablation planning,” Comput. Biol. Med. 125, 104005 (2020). 10.1016/j.compbiomed.2020.104005 [DOI] [PubMed] [Google Scholar]
- 81. Cabrera-Lozoya R., Berte B., Cochet H., Jaïs P., Ayache N., and Sermesant M., “ Image-based biophysical simulation of intracardiac abnormal ventricular electrograms,” IEEE Trans. Biomed. Eng. 64(7), 1446–1454 (2017). 10.1109/TBME.2016.2562918 [DOI] [PubMed] [Google Scholar]
- 82. Lozoya R. C., Berte B., Cochet H., Jaïs P., Ayache N., and Sermesant M., “ Model-based feature augmentation for cardiac ablation target learning from images,” IEEE Trans. Biomed. Eng. 66(1), 30–40 (2019). 10.1109/TBME.2018.2818300 [DOI] [PubMed] [Google Scholar]
- 83. Cedilnik N., Duchateau J., Dubois R. et al. , “ Fast personalized electrophysiological models from computed tomography images for ventricular tachycardia ablation planning,” EP Europace 20(suppl_3), iii94–iii101 (2018). 10.1093/europace/euy228 [DOI] [PubMed] [Google Scholar]
- 84. Campos F. O., Orini M., Taggart P. et al. , “ Characterizing the clinical implementation of a novel activation-repolarization metric to identify targets for catheter ablation of ventricular tachycardias using computational models,” Comput. Biol. Med. 108, 263–275 (2019). 10.1016/j.compbiomed.2019.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Campos F. O., Orini M., Arnold R. et al. , “ Assessing the ability of substrate mapping techniques to guide ventricular tachycardia ablation using computational modelling,” Comput. Biol. Med. 130, 104214 (2021). 10.1016/j.compbiomed.2021.104214 [DOI] [PubMed] [Google Scholar]
- 86. Trayanova N. A., Popescu D. M., and Shade J. K., “ Machine learning in arrhythmia and electrophysiology,” Circ. Res. 128, 544–566 (2021). 10.1161/CIRCRESAHA.120.317872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fresca S., Manzoni A., Dedè L., and Quarteroni A., “ Deep learning-based reduced order models in cardiac electrophysiology,” PLoS One 15, e0239416 (2020). 10.1371/journal.pone.0239416 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.