SUMMARY:
Environmental pathogens move from ecological niches to mammalian hosts, requiring adaptation to dramatically different environments. Microbes that disseminate farther, including the fungal meningitis pathogen Cryptococcus neoformans, require additional adaptation to diverse tissues. We demonstrate that formation of a small C. neoformans morphotype – called “seed” cells due to their colonizing ability – is critical for extrapulmonary organ entry. Seed cells exhibit changes in fungal cell size and surface expression that result in enhanced macrophage update. Seed cell formation is triggered by environmental factors, including C. neoformans’ environmental niche, pigeon guano with phosphate playing central roles. Seed cells show enhanced expression of phosphate acquisition genes and mutants unable to acquire phosphate fail to adopt the seed cell morphotype. Additionally, phosphate can be released by tissue damage, potentially establishing a feed-forward loop of seed cell formation and dissemination. Thus, C. neoformans’ size variation represent inducible morphotypes that change host interactions to facilitate microbe spread.
eTOC Blurb
Environmental microbes that disseminate during infection must adjust to disparate environments. Denham et al. identifies an inducible morphotype that facilitates organ entry by the pathogenic fungus Cryptococcus neoformans. Alterations in phosphate concentration induce this small morphotype, facilitating fungal exit from the lung and dissemination to other organs.
Graphical Abstract
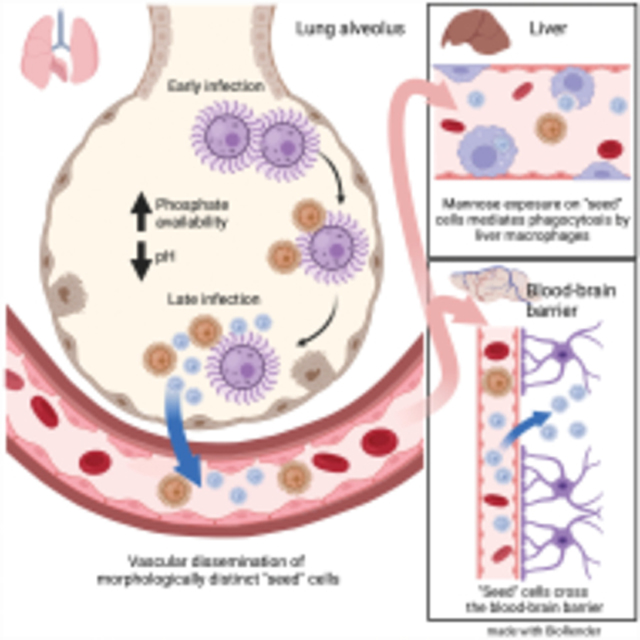
INTRODUCTION:
Environmental pathogens must adapt to a wide variety of conditions as they transition from their ecological niches to host infection. This is particularly challenging for pathogens that cause disseminated disease, such as the primary cause of fungal meningitis, Cryptococcus neoformans (Rajasingham et al., 2017). As C. neoformans escapes from the lungs following inhalation (Maziarz and Perfect, 2016), it must evade the immune system (Bojarczuk et al., 2016; Bulmer and Sans, 1967; Chun et al., 2011; Luberto et al., 2003; Okagaki et al., 2010; Stano et al., 2009; Walsh et al., 2019; Zaragoza et al., 2010), survive in the bloodstream and/or host cells (Botts and Hull, 2010; Charlier et al., 2009; Diamond and Bennett, 1973; Feldmesser et al., 2000; Gaylord et al., 2020; Walsh et al., 2019), and finally enter and grow in the central nervous system (CNS) (Chang et al., 2004; Chen et al., 2003; Chen et al., 2021; Huang et al., 2011; Liu et al., 2013; Liu et al., 2014; Maruvada et al., 2012; Olszewski et al., 2004; Santiago-Tirado et al., 2017; Vu et al., 2013). The resulting fungal meningoencephalitis drives cryptococcosis patient mortality, killing ~180,000 people each year worldwide (Rajasingham et al., 2017). In its environmental niche, C. neoformans is a ubiquitous saprophytic fungus associated with trees and bird guano (Emmons, 1955; Lazera et al., 1996). Human exposure thought to be as high as 70% by age five (Goldman et al., 2001). Preventing infection is therefore challenging for the patients most at risk: those with CD4+ T cell deficiency (Rajasingham et al., 2017).
Many microbes employ phenotypic switching and/or heterogeneity to survive in fluctuating host environments (Altamirano et al., 2020; Weigel and Dersch, 2018). These strategies include the antigen switching employed by bacteria (Haas and Meyer, 1986; Swanson et al., 1986) and parasites (Roberts et al., 1992; Smith et al., 1995), which allows avoidance of adaptive immune responses. Fungi do not exhibit antigen switch but adjust exposure levels of immunogenic cell surface entities such as β-glucan (Ballou et al., 2016).
Phenotypic heterogeneity is a mechanism of bet-hedging against environmental stressors (Altamirano et al., 2020), including varied host responses (Davis, 2018). For example, microbial cells at the foci perimeters are be more open to immune cell attack; while the core may be hypoxic and nutrient-depleted (Davis et al., 2015; Kowalski et al., 2019). Variation within populations provide phenotypic robustness and can influence which cells in a localized population are more likely to spread to new host sites, such as during cancer metastasis (San Juan et al., 2019).
Fungi undergo dramatic phenotypic changes that influence pathogenesis, such as the yeast / hyphal switches of dimorphic fungi (Garfoot et al., 2016; Guimarães et al., 2011; Kanetsuna and Carbonell, 1971; Mukaremera et al., 2017; Viriyakosol et al., 2013; Whiston et al., 2012), spherule development in Coccidioides species (spp.) (Viriyakosol et al., 2013; Whiston et al., 2012), and conidia sporulation in Aspergillus spp. (Aimanianda et al., 2009; Hohl et al., 2005). The differences between yeast and filamentous forms are superficially obvious due to their extreme morphological differences, but we are only beginning to understand more subtle variations within morphological states that have profound influences on infectivity (Pande et al., 2013; Tao et al., 2014). Cryptococcus neoformans grows primarily as a budding yeast with an inducible, immunomodulatory capsule (Casadevall et al., 2019; O’Meara and Alspaugh, 2012); a filamentous form is generally associated with mating and not virulence (Wang et al., 2012). Here we use the seemingly subtle differences, such as capsule and cell body size, between of C. neoformans yeast cells probe the role of phenotypic heterogeneity in intra-host dissemination.
An important yeast form morphotype is the polyploid (≥4C) “titan” cell, with cell body diameters of 10–100 μm (Dambuza et al., 2018; Hommel et al., 2018; Okagaki and Nielsen, 2012; Okagaki et al., 2011; Trevijano-Contador et al., 2018; Zaragoza et al., 2010; Zhou and Ballou, 2018). Titans are more resistant to phagocytosis and oxidative stress and skew immunity toward a non-protective Th2 response (Crabtree et al., 2012; Okagaki and Nielsen, 2012; Okagaki et al., 2010).
However, titans are a minority of cryptococcal cells within the lungs (Zhou and Ballou, 2018) and are rarely observed outside of the lungs. At extrapulmonary sites, including the brain, overall cell and capsule sizes are smaller (Charlier et al., 2005; Denham et al., 2018; Rivera et al., 1998; Xie et al., 2012). While the C. neoformans populations in extrapulmonary organs become heterogeneous over time (Lee et al., 1996), the C. neoformans population that appears early in the murine brain during dissemination is striking in its homogeneity and small median size (Denham et al., 2018; Fernandes et al., 2018; Fernandes and Carter, 2020; Fernandes et al., 2016; Zaragoza, 2011). We therefore hypothesize that smaller morphotypes are more likely to disseminate and enter extrapulmonary organs.
RESULTS:
Small cell formation correlates with extrapulmonary dissemination
Cryptococcus spp. isolates vary in pathogenesis and hetergeneity (Fernandes et al., 2018). C. gattii is less likely than C. neoformans to disseminate and produces larger cells in the lungs (Ngamskulrungroj et al., 2012; Okubo et al., 2013). To systematically assess morphological shifts, we inoculated ~8-week-old female B6 mice with a panel of common Cryptococcus spp. reference strains (2.5×104 cells / mouse). At 3 and 17 days post-inoculation (dpi), we homogenized lungs and measured fungal cell body and capsule size (Figures 1A,B). Size varied widely. While KN99 and Bt63 cells (both C. neoformans) shifted smaller between 3 and 17 dpi, R265 (C. deuterogattii) and WM276 (C. gattii) continued to increase in size—largely due to increased capsule thickness. Smaller median size at 17 dpi correlated with increased extrapulmonary burden (Figures 1C,D). These data suggest that among Cryptococcus isolates, the emergence of pulmonary small cells (<10 μm) is a strong predictor of extrapulmonary dissemination.
Figure 1: Morphological transitions during pulmonary infections correlate with fungal dissemination.
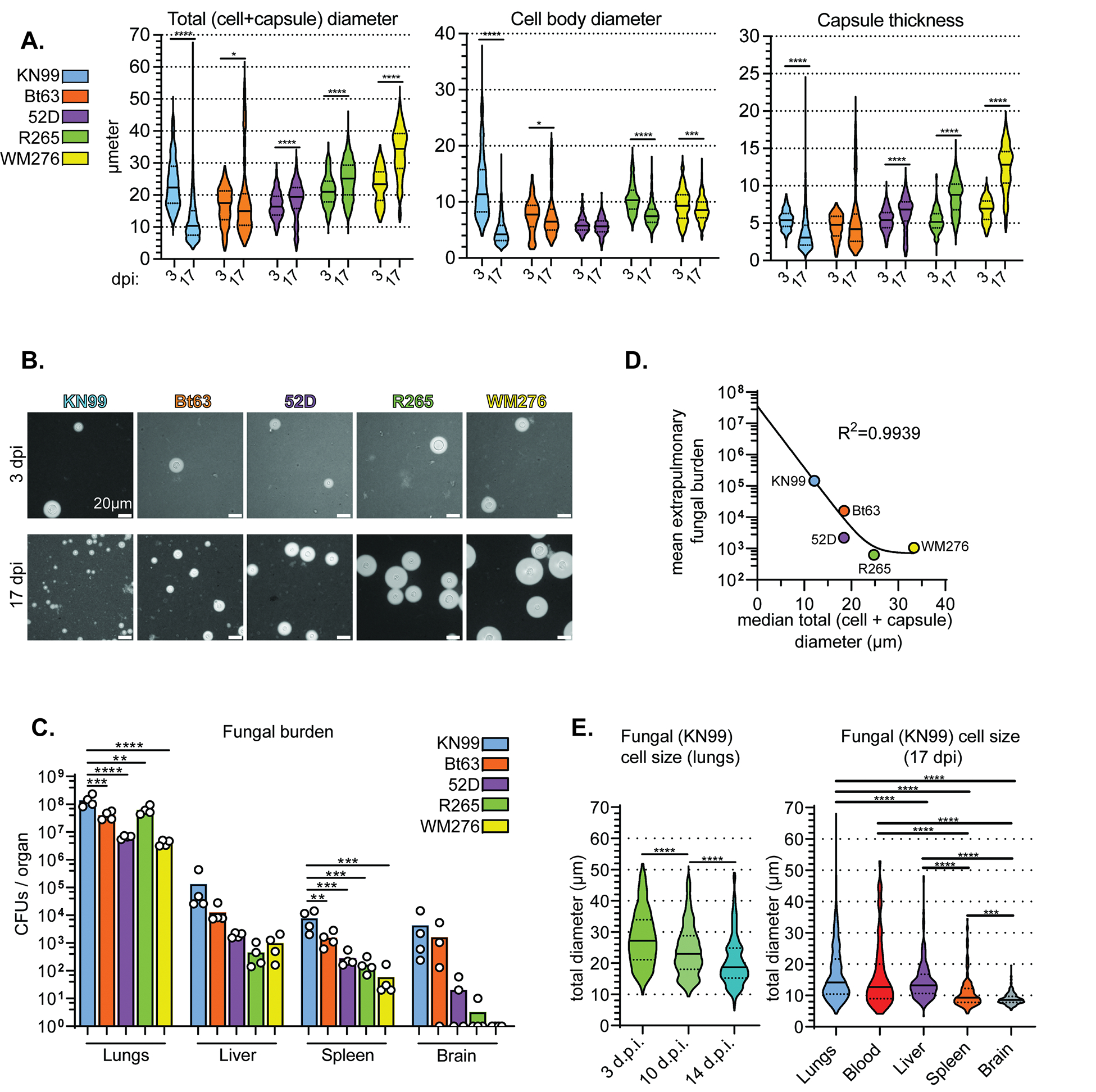
(A) Total (cell + capsule) diameter, cell body diameter, and capsule thickness measurements at 3 and 17 days post-intranasal inoculation (dpi) of mice (Mann-Whitney U test; N=4 mice per time point, 57–65 fungal cells measured per mouse at 3 dpi and 117–266 fungal cells per mouse at 17 dpi). (B) Representative India ink images of fungal cells in 1A. (C) Fungal burden in mice at 17 dpi (One-way ANOVA and uncorrected Fisher’s LSD). The bar graph represents the mean. (D) Nonlinear regression of mean extrapulmonary (liver + spleen + brain) fungal burden and median total diameter. (E) Total diameter in KN99-infected lungs at 3, 10, and 14 dpi and total diameter in KN99-infected organs at 17 dpi (Mann-Whitney U-test; N=4 mice per timepoint; fungal cells measured per mouse in the lungs: 275, blood: 38, liver: 72, spleen: 43, brain: 72). For all violin plots, solid lines in the violin plots indicate the median and dotted lines mark quartiles. In addition to the p-value cutoff, violin plot comparisons are only marked as significant if the 95% confidence interval does not overlap. For all panels in all figures: ns: not significant *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
We also measured cell and capsule size of fungi from the blood, liver, spleen, and brains of mice inoculated with KN99 (Figure 1E). The cell size range in the blood and liver – rare fungal cells measured 50 μm across – was surprisingly broad considering that murine microcapillaries are as small as 3.5 μm. This could reflect growth within the organ, particularly in the early colonized extrapulmonary organs (i.e. the liver). Median total diameter in the blood at 17 dpi was 12.7 μm, smaller than median total diameter in the 17 dpi lungs (14.2 μm). Fungal cell size in the spleen and brain was far more restricted. The vast majority of splenic fungi were <15 μm total diameter; brain fungi rarely exceeded 10 μm. Fungal cells usually enter the brain at 10 dpi (Denham et al., 2018) and we measured at 17 dpi, so this could represent growth within the organ. However, the number of pulmonary ~10 μm total diameter fungal cells substantially increases at 10 dpi (Figure 1E), the same time fungi appear in the brain. Together, these data suggest that small cell formation in the lungs predicts extrapulmonary dissemination.
Cells isolated from the lungs display size-dependent differences in their cell surfaces
When we compared the labor-intensive microscopy-based size measurements with a flow cytometry approach, we still observed a fungal size shift over the course of infection (Figure S1). We thus isolated KN99-mCherry cells from infected lung tissue (14 dpi) via fluorescence-activated cell sorting (FACS) using forward scatter area (FSC-A) as a stand-in for size. We sorted FSC-A cross-sections corresponding to the smallest (small ex vivo: median total diameter of 7.6 μm), intermediate (mid ex vivo: median total diameter of 14.0 μm), and largest (large ex vivo: median total diameter of 21.8 μm) ~20% of the population (Figure 2A). We gated for single cells using forward scatter height vs area (FSC-H vs FSC-A); > 95% of ex vivo cell populations were single cells. This was to prevent the irregular shape of a budding yeast cell from confounding the analysis. We further confirmed that we isolated distinct populations based on cell and capsule size (Figure 2B,C). Fungal cells from each gate were equivalently viable (Figure 2D).
Figure 2: Small ex vivo cells disseminate to extrapulmonary organs at a higher rate than larger cells.
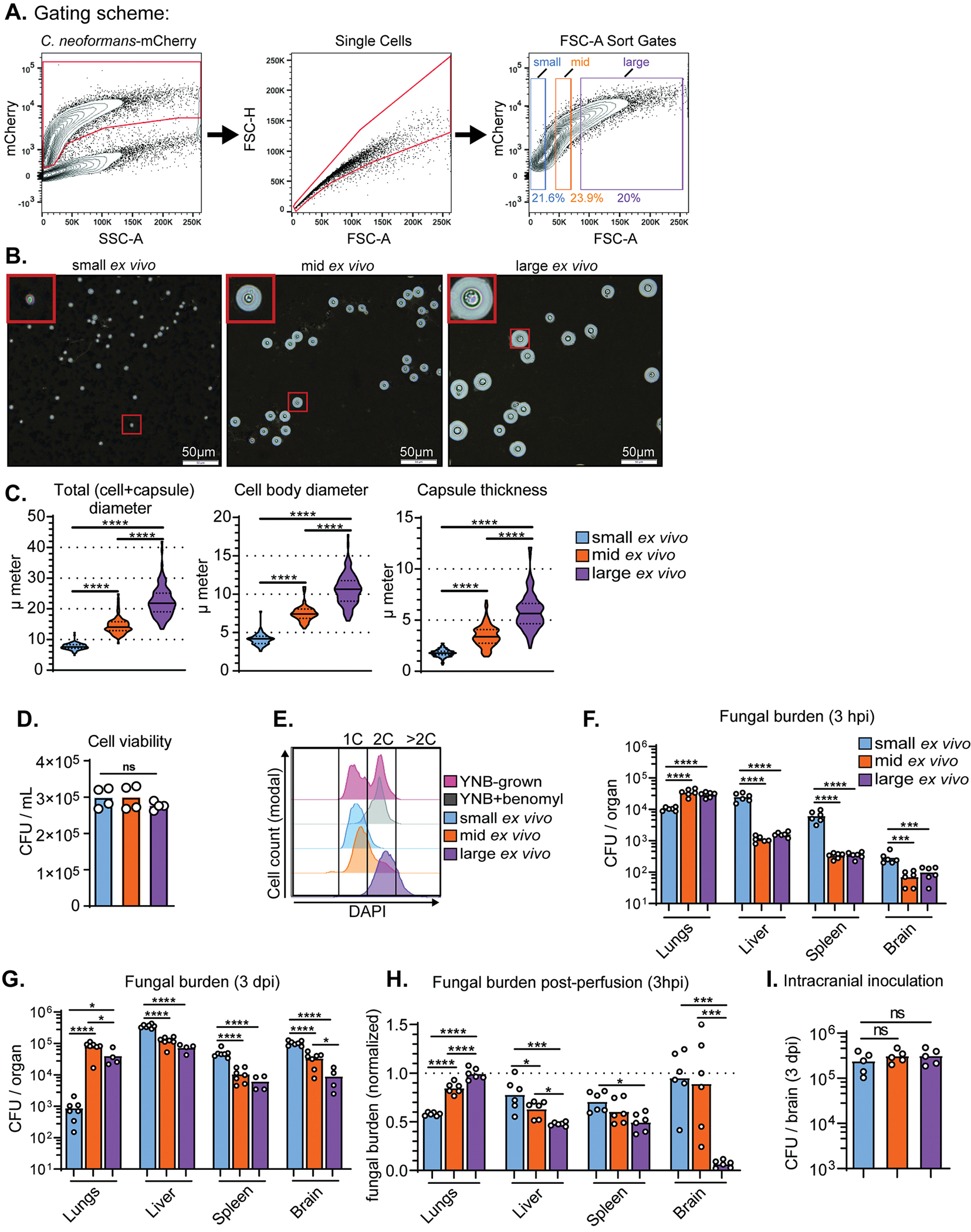
(A) FACS gating scheme for sorting KN99-mCherry cells from infected mouse lungs into populations of small, mid, and large cells using forward scatter-area (FSC-A) to approximate size. (B) Representative India ink images of cells sorted from small, mid, and large FSC-A gates. (C) Violin plots of measurements of cells from mouse lungs. At least 100 cells were measured per mouse and data from three mice is shown on the graph. Solid lines in the violin plots indicate the median and dotted lines mark quartiles. P-values shown were calculated using a Mann-Whitney U-test. (D) Cell viability post-sort quantified by colony-forming unit (CFU) count (N=4). (E) Representative flow plot of DNA-content measured with DAPI staining. (F) Fungal burden, measured by colony forming unit counts (CFU), at 3 hours post-inoculation (hpi) and (G) 3 days post-inoculation (dpi) in mice intravenously-inoculated with small, mid, or large ex vivo cells (One-way ANOVA and uncorrected Fisher’s LSD). (H) Fungal burden in mice inoculated with ex vivo populations and perfused at 3 hpi. Fungal burden was normalized to inoculated, non-perfused control mice (Mann-Whitney U-test). (I) Fungal burden at 3 dpi in mice intracranially inoculated with sorted ex vivo fungal cells (One-way ANOVA and uncorrected Fisher’s LSD).
We measured DNA ploidy with 4′,6-diamidino-2-phenylindole (DAPI) staining (Figure 2E). We used haploid cells cultured in yeast nitrogen base (YNB) medium +/− benomyl as a control, since benomyl halts mitosis and traps cells at 2C (Jacobs et al., 1988). Cells with >2C content are likely titan cells. Only the large population contained an appreciable number of titan cells (mean: 21.4% >2C) (Figure S2). Intermediate cells averaged 74.2% 1C and 22.8% 2C, while small cells averaged 89.2% 1C and 10.0% 2C.
Small ex vivo cells enter extrapulmonary organs at a higher rate than larger ex vivo cells
Most infection models—including intravenous dissemination models—inoculate with C. neoformans yeast cells collected from nutrient-rich laboratory media (Mukaremera et al., 2019; Ngamskulrungroj et al., 2012; Sabiiti et al., 2012), which are phenotypically homogenous. Here we accounted for lung infection-generated heterogeneity by isolating cells directly from infected lung tissue. We intravenously inoculated naïve mice with small, intermediate, and large ex vivo cells (105 cells / mouse). Blood flow from the tail vein runs directly to the heart, then to the lungs for reoxygenation. The lungs harbor the first major bed of microcapillaries fungi encounter following tail vein inoculation. Therefore, sorting fungal cells from infected lung tissue and inoculating via tail vein roughly replicates fungi entering the bloodstream from the lungs. As we hypothesized, a greater number of small ex vivo cells reached the liver, spleen, and brain by 3 hours post-inoculation (hpi) than intermediate and large ex vivo cells (Figure 2F). In contrast, the majority of intermediate and large cells remained in the lungs at 3 hpi. We observed the same trends at 3 dpi; small ex vivo cells are thus well-capable of proliferating in extrapulmonary tissues (Figure 2G).
Although brain fungal burden was equivalent in mice inoculated with intermediate and large ex vivo cells at 3 hpi, there were more intermediate cells in the brain by 3 dpi. We asked whether large cells were deficient in blood-brain barrier crossing or proliferation in the brain. We intracardially perfused mice at 3 hpi to remove circulating fungal cells and normalized the fungal burden in perfused mice to nonperfused mice. While perfusion did not significantly affect brain fungal burden in mice inoculated with small or intermediate cells, perfusion drove nearly all of the large cells from the brain (Figure 2H). All ex vivo cells grew equally well in the brain following intracranial inoculation (Figure 2I). Access to and entry into extrapulmonary organs are therefore both dissemination bottlenecks and small cells have advantages at both stages.
Small ex vivo cells have altered cell surface architecture that result in preferential uptake by macrophages.
Morphological changes in C. neoformans are accompanied by changes in immune recognition and cell surface architecture. We screened the binding rate (% total fungal cells bound) of a panel of soluble receptors to small, intermediate, and large ex vivo cells (Figure 3A, Figure S3A–D). As in Figure 2A, we gated for single cells to prevent the irregular shape of yeast buds from confounding analysis. Large cells were more likely to be bound by complement protein C3 than intermediate or small cells (p < 0.05). In contrast, lung surfactant protein D (SPD) preferentially bound small and intermediate cells (p < 0.01). SPD binding to C. neoformans is deleterious for the host, potentially because SPD agglutinates fungal cells and disrupts destruction by macrophages (Geunes-Boyer et al., 2012). We saw low-level, uniformly distributed binding of IgA and IgM class antibodies and little binding by mannose-binding lectins.
Figure 3: Mannose recognition by macrophages contributes to organ uptake by small ex vivo cells.
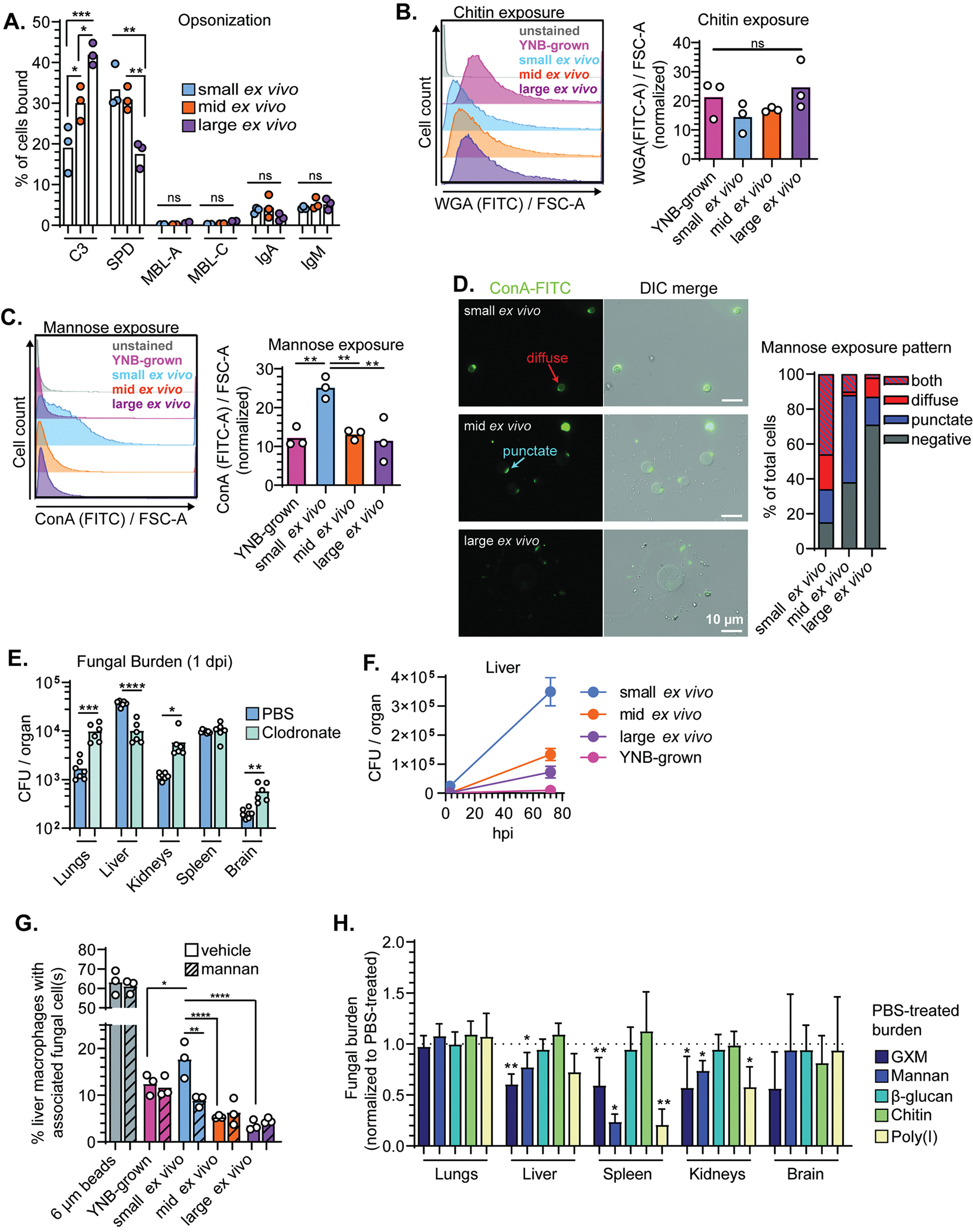
(A) Percentage of total cells bound by complement (C3), surfactant protein D (SPD), mannose binding lectins A and C (MBL-A; MBL-C), IgA, and IgM. (B) Representative flow plots and quantification of wheat-germ agglutinin (WGA) stained cells (chitin exposure). (C) Representative flow plots and quantification of concanavalin A (ConA) stained cells (mannose exposure). (D) Representative images and characterization of mannose exposure pattern after staining with ConA (N=50 fungal cells). All comparisons in Figure 2 were analyzed using one-way ANOVA and uncorrected Fisher’s LSD. All bar graphs display the mean. (E) Fungal burden at 1 dpi in mice pretreated with either clodronate or vehicle control (PBS) liposomes and then inoculated with small ex vivo cells and (Unpaired t test). (F) Fungal burden in livers from mice (N=4–6) inoculated with ex vivo fungal cells or YNB-grown fungi at 3 and 72 hpi. Points indicate the mean and error bars the standard deviation. (G) Percentage of primary cultured murine liver macrophages with physically associated plastic beads or fungal cells 4 hpi. Dashed lines indicate 10-minute pretreatment with exogenous mannan (One-way ANOVA and uncorrected Fisher’s LSD; N=2263–3197 total macrophages counted across 3 independent replicates). (H) Fungal burden in mice intravenously treated with indicated ligand 2–3 minutes prior to intravenous inoculation with small ex vivo fungal cells. Mice were perfused 10 minutes post-inoculation and fungal burden was normalized to vehicle (PBS) treated controls (Mann Whitney U-test; N=6 mice per group). Bar graphs display the mean and error bars the standard deviation.
Host factor binding data suggested that cell surface architecture differs between sorted populations. We used two lectins to measure microbial feature exposure in different layers of the cell wall. Wheat germ agglutinin (WGA) recognizes chitin in the inner cell wall and concanavalin A (conA) binds mannose on mannoproteins in the outer cell wall and inner capsule. Both dyes measure exposure rather than total quantity; the bulky lectin might not be able to access targets deep within the cell wall, so the absence of signal is not the absence of chitin or mannose but lack of exposure. There were no difference in chitin exposure as measured by flow cytometry (Figure 3B). However, mannose was significantly more exposed on small ex vivo cells compared with intermediate and large cells and YNB-grown cells, which lack obvious capsules (Figure 3C).
We visualized mannose exposure patterns microscopically and characterized staining as 1) negative, 2) punctate, 3) diffuse, or 4) both punctate and diffuse (Figure 3D). Mannose was more likely to be exposed both at specific puncta and diffusely across the surface of small ex vivo cells. The puncta are likely bud scars (Panepinto et al., 2007). The diffuse mannose exposure on small cells may be due thin capsules. Altogether, these results suggest that size heterogeneity correlates with cell surface heterogeneity.
As small ex vivo cells most efficiently entered extrapulmonary organs, we next examined organ entry mechanisms. The liver plays a major role in removing microbes and microbial components from the blood (Macpherson et al., 2016) due to microbial-associated molecular patterns (MAMPs) recognition (Kubes and Jenne, 2018). Intravital imaging revealed that liver macrophages remove C. neoformans cells from the bloodstream (Sun et al., 2019). However, the inoculum for those studies consisted of in vitro-cultured C. neoformans cells. Since we found that small ex vivo cells differ from intermediate and large ex vivo cells and cells cultured in YNB, we tested whether host cell recognition of fungi varies with subpopulation.
Platelets and phagocytes are critical for the removal of pathogens from the blood by sequestration in organs. To determine which cell types are important for small cell organ entry, we systematically depleted cell types and measured organ distribution of ex vivo populations following tail vein injection. First, we depleted liver, spleen, and bone marrow macrophages with intravenous injection of clodronate liposomes 48 hours prior to inoculation (Figures S3E,F). Macrophage depletion reduced small ex vivo cell fungal burden in the liver, suggesting that liver-resident macrophages are important for liver entry (Figure 3E). These results corroborate Sun et al.’s findings (Sun et al., 2019). However, small ex vivo cells grow rapidly in the liver compared with in vitro-grown cells or other ex vivo populations (Figure 3F), suggesting that attempts by the host to contain C. neoformans cells by sequestering them backfires. There is also an increase in fungal burden in downstream organs such as the kidneys and brain, presumably because more fungi remain in the blood.
We then tested whether platelets or neutrophils impact organ entry by small ex vivo cells. Prior work found that platelets do not mediate the removal of in vitro-grown C. neoformans cells from the blood to organs (Sun et al., 2019). When we depleted platelets, we also did not see any effect on fungal organ entry (Figures S4A–C). Neutrophil depletion greatly increased fungal burden in the lungs and brain when in vitro-grown fungal cells were injected intravenously (Sun et al., 2016; Zhang et al., 2016). However, neutrophil-depletion only slightly increased lung fungal burden following small ex vivo cell inoculation and did not affect fungal burden in other organs (Figures S4D–F).
Macrophage recognition of small ex vivo cells is partially mediated by mannose exposure.
Since exposed mannose was the major cell surface difference between small and other ex vivo populations, we hypothesized that mannose mediates macrophage recognition (Figure 3C). We isolated liver macrophages from healthy ~8-week-old B6 mice (Li et al., 2014) and co-cultured them with 6 μm plastic beads, YNB-grown cells, and sorted ex vivo populations (2:1 multiplicity of infection). Half received pretreatment with exogenous mannan. After 4 hours we washed away free fungal cells, leaving only “macrophage-associated” fungi: those bound or internalized by macrophages.
Approximately 60% of macrophages associated with 6 μm plastic beads, indicating that the macrophages maintained phagocytic ability (Figure 3G). 18% of macrophages associated with small ex vivo cells, more than twice the frequency of intermediate and large ex vivo cells. Pretreatment with exogenous mannan reduced macrophage association with small cells by ~40% but did not affect association with intermediate and large cells, YNB-grown cells, or beads.
We then tested whether mannose exposure on small ex vivo cells affects organ entry in vivo by intravenously injecting either 400 μg of exogenous mannan or vehicle (PBS) 2–3 minutes prior to intravenous inoculation with 105 small ex vivo cells. 10 minutes later, we perfused mice to remove bloodstream fungal cells and plated organs for colony forming units (CFUs). If mannose recognition facilitated fungal entry into an organ, we expected reduced fungal burden in mice that received exogenous mannan compared to the vehicle control. We repeated this experiment with different fungal cell wall and capsule components (GXM, β-glucan, chitin) and the scavenger recepter ligand polyinosinic acid (poly(I)) (Pearson et al., 1993).
Mannan reduced fungal uptake to the liver and kidneys by ~20% and in the spleen by ~75% (Figures 3H and S3G) but was not the only fungal ligand that influenced organ entry: Poly(I) reduced organ uptake as much as mannan. The purified capsule polysaccharide GXM reduced entry into the liver, spleen, kidneys by ~40%. β-glucan and chitin did not appreciably affect fungal organ entry, possibly because they are shielded by the capsule. Fungal burden in the brain was too low – <100 CFU per mouse - to detect changes. These data demonstrate that small ex vivo cells’ organ entry involves multiple cell surface ligands and that tissue-specific dependencies mediate organ entry.
Enhanced small ex vivo cell dissemination is not solely dependent on size.
We next assessed whether small ex vivo cells disseminate more efficiently due to their size alone. We inoculated mice via the tail vein with either large ex vivo cells, small ex vivo cells or size-matched inert polystyrene beads. Large cells disseminated similarly to size-matched 25 μm beads: most resided in the lungs at 3 hpi (Figure 4A). More 6 μm beads traveled past the lungs to reach the liver and spleen compared to 25 μm beads. However, small ex vivo cells reached the liver and spleen at an even higher rate, indicating that a fungal-specific factor, in addition to size, influences dissemination and extrapulmonary organ entry.
Figure 4: Small cells generated in vitro replicate the dissemination profile of small ex vivo cells.
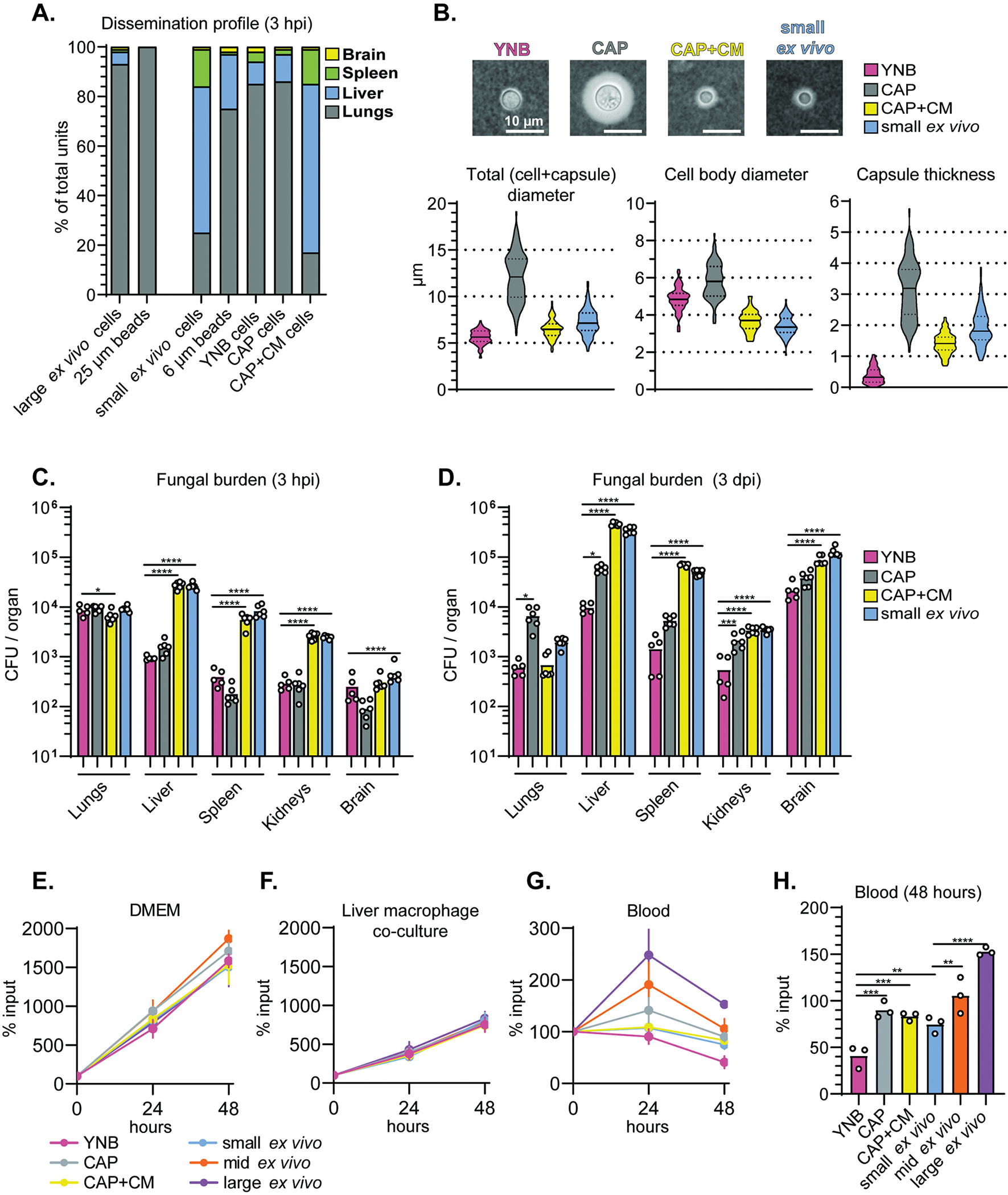
(A) Percentage of total plastic beads or live cells in the indicated organ 3 hpi (intravenous inoculation). (B) Representative India ink images and quantification of cell and capsule size (N=100 fungal cells). CAP: capsule inducing medium. CAP+CM: CAP + 10% conditioned medium. (C) Fungal burden measured by CFU in mice at 3 hpi and (D) 3 days post-inoculation (dpi). (E) Fungal cell survival in DMEM and (F) liver macrophage co-culture (multiplicity of infection of 1:1) (% of input). (G) Fungal cell survival in mouse blood measured as a % of input over 48 hours and (H) at 48 hours growth. All comparisons in Figure 4 were analyzed using one-way ANOVA and uncorrected Fisher’s LSD. All bar graphs display the mean.
We then identified in vitro conditions that reproduced the small ex vivo cell morphotype and extrapulmonary organ entry profile. We previously observed that cells grown in capsule-inducing medium (CAP; 10% Sabouraud’s dextrose broth buffered to pH 7.4) shifted toward smaller cell and capsule size if supplemented with conditioned medium (CM) (Denham et al., 2018). We collected CM by filtering the supernatant from cultures grown 24 hours in YNB. CM consists of unspent nutrients and secreted fungal factors but not fungal cells. YNB growth suppresses capsule formation, resulting in uniform cells <10 μm in diameter.
We compared the dissemination profile of small ex vivo cells to three in vitro populations: (1) YNB-grown cells (2) CAP-grown cells, and (3) CAP-grown cells supplemented with CM (CAP+CM). To control for size, we sorted in vitro-grown populations the same way as ex vivo cells. YNB- and CAP+CM-grown cells were sorted using the “small” FSC-A gate (5.6 μm and 6.5 μm median total diameters, respectively; few YNB or CAP+CM cells fell in the other FSC-A gates) and CAP-grown cells were sorted using the “mid” FSC-A gate (12.1 μm median total diameter; few CAP-grown cells fell into the other FSC-A gates) (Figure 4B).
Morphology and culture conditions heavily impacted organ distribution. Only CAP+CM cells replicated the dissemination profile of small ex vivo cells (Figure 4C,D). Despite being roughly the same size as small ex vivo cells, YNB-grown cells displayed similar organ distributions to CAP-grown cells at 3 hpi, and lower fungal burden in extrapulmonary organs than all other populations at 3 dpi. Therefore, both absolute fungal cell size and cell morphology influence the outcome of vascular dissemination.
Host-adapted cells survive better in the blood than cells grown in nutrient-replete medium.
The caveat to using fungal burden as a readout for dissemination and organ entry is that we cannot easily distinguish between the ability of cells to reach certain organs and their ability to survive in those organs. YNB-grown cells may not be as primed for survival in the bloodstream as host tissue-grown or CAP-grown cells. To control for blood survival differences, we examined the survival of in vitro-grown and ex vivo-sorted cells in whole mouse blood or liver macrophage co-culture (multiplicity of infection of 1:1). DMEM (Figure 4E) served as a control. Liver macrophages suppressed in vitro and ex vivo cell growth equivalently (Figure 4F). YNB-grown cells exhibited a decrease in fungal burden in blood (Figure 4G,H). Large and intermediate ex vivo populations fared the best. These results support the idea that size and physiology are critical for bloodstream dissemination: small ex vivo cells potentially combine the stress resistance required for increased bloodstream survival with the smaller cell size to facilitate widespread dissemination via microcapillaries.
Expression of phosphate acquisition genes differs between small cells and larger C. neoformans cells.
Since CAP+CM-grown cells and small ex vivo cells share organ entry profiles, we hypothesized that they resemble each other transcriptionally. We performed RNA-seq of cells grown in YNB, CAP, and CAP+CM medium and small, intermediate and large ex vivo cells.
The YNB-grown transcriptome is the outlier (Figure 5A,B). There were 1085 upregulated and 1297 downregulated genes shared only among the ex vivo populations relative to YNB-grown cells. Core sets of 699 and 291 genes were up- and down-regulated respectively among the ex vivo populations and CAP/CAP+CM populations relative to YNB-grown cells. Among the ex vivo populations, small ex vivo cells were the most transcriptionally distinct (Figure 5A). 928 genes were differentially expressed in small vs intermediate ex vivo cells, and 2,365 genes were differentially expressed in small vs large ex vivo cells. Only 93 genes were differentially expressed in intermediate vs large ex vivo cells (Figure 5A).
Figure 5: Expression of phosphate acquisition genes correlate with cell and capsule size changes.
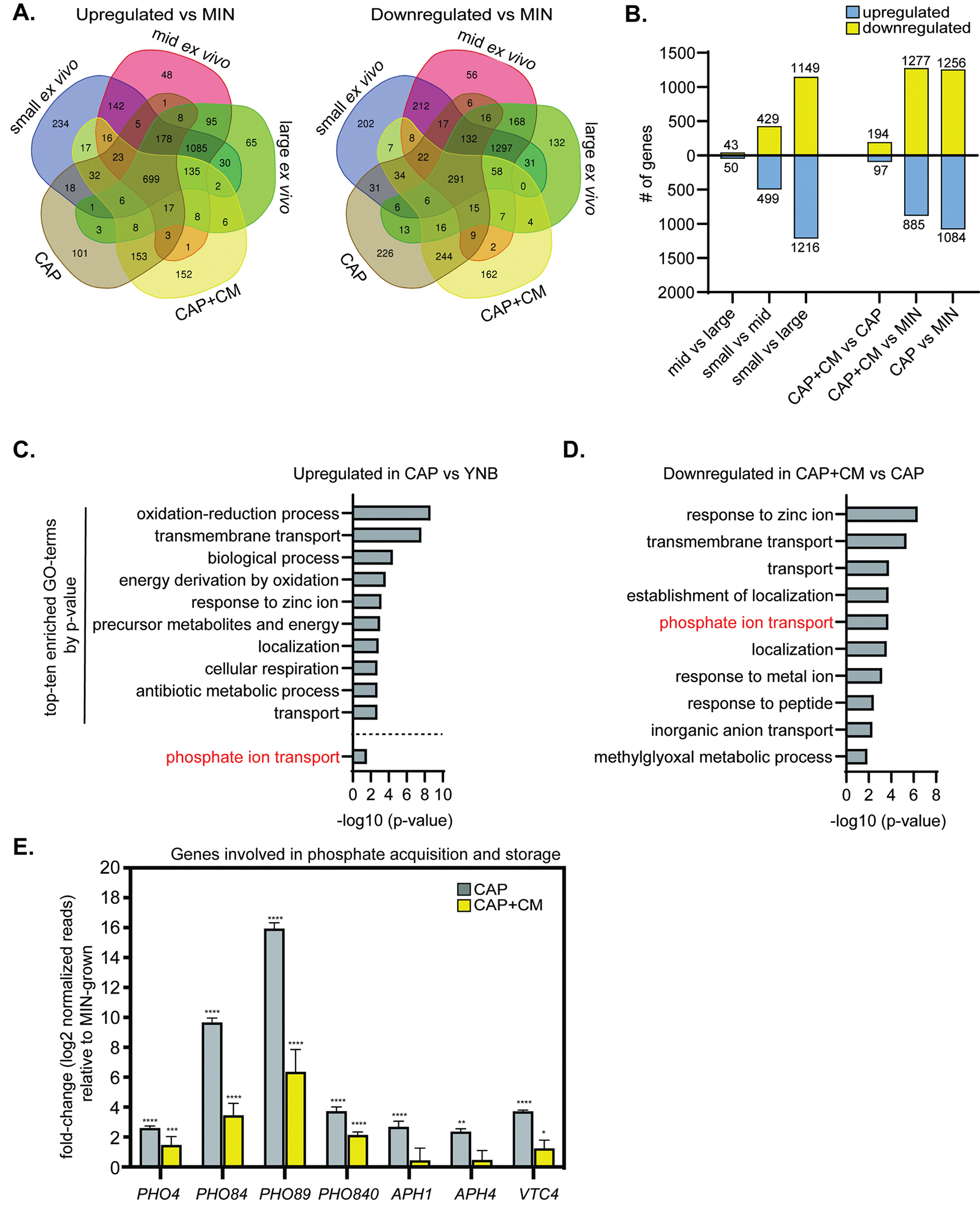
(A) Venn diagram of genes upregulated or downregulated in different conditions relative to YNB-grown cells. (B) Comparisons of the total number of differentially regulated genes among in vitro-grown populations. (C) Top-ten most significantly enriched gene ontology (GO) terms representing genes upregulated in CAP vs YNB-grown cells (−log10 (p-value)). Phosphate ion transport was #46. (D) Top-ten most significantly enriched GO-terms representing genes downregulated in CAP+CM vs CAP-grown cells (−log10 (p-value)). (E) Select phosphate acquisition and storage RNA-seq gene expression relative to YNB-grown cells (adjusted p values). All bar graphs display the mean and error bars the standard deviation.
Gene ontology term (GO-term) enrichment analysis revealed that phosphate ion transport genes were induced in CAP-grown cells vs YNB-grown cells (Figure 5C), and suppressed in CAP+CM vs CAP-grown cells (Figure 5D). Genes involved in phosphate acquisition and storage showed the same trends (Figure 5E). In phosphate-starved C. neoformans cells, the phosphate acquisition transcription factor, Pho4, translocates to the nucleus. There it activates expression of genes involved in phosphate acquisition and storage, including PHO4. These include phosphate transporters: PHO84, PHO89, and PHO840; phosphatases: APH1 (secreted/vacuolar acid phosphatase) and APH4 (predicted intracellular acid phosphatase); and VTC4 (vacuolar transport chaperone involved in processing polyphosphate). Pho4-dependent genes were upregulated in CAP vs YNB-grown cells and then suppressed in CAP+CM, although many phosphate acquisition genes remained more highly expressed in CAP+CM-grown cells than YNB-grown cells.
Phosphate drives C. neoformans populations toward smaller morphotypes
We hypothesized that phosphate is a critical signal for small cell induction in CAP+CM media, but components of YNB and YNB-based conditioned medium are also candidates. We screened every component of YNB individually (including phosphate) to identify components sufficient to induce small cell formation CAP medium-grown cells. We also tested two soluble fungal-derived factors that accumulate during growth in YNB medium: exo-GXM (Denham et al., 2018) and the quorum sensing-like peptide Qsp1 (Homer et al., 2016; Lee et al., 2007).
We followed our standard in vitro small cell induction protocol. After 24 hours grown in YNB, we sub-cultured 105 cells/mL into CAP medium for another 24 hours. We further subcultured 1:1 into fresh CAP medium + 10% (final volume) CM, a YNB component, a fungal-derived factor, or water (vehicle control). 24 hours later, we measured cell size (FSC-A) by flow cytometry.
Only phosphate was sufficient to stimulate a shift toward smaller cell size (Figure S5A), and did so in a concentration-dependent manner (Figures 6A,B) at concentrations below that of phosphate in blood (Beis and Newsholme, 1975). Phosphate-supplemented cells decreased both total (cell+capsule) diameter (Figure 6C) as well as cell (Figure S5B) and capsule (Figure S5C) size individually. Qsp1 did not induce smaller cells (Figures S5A,B). We previously reported that exo-GXM may trigger the formation of smaller cells (Denham et al., 2018). However, we were unable to reproduce those results (Figure S5A), possibly due to differences in exo-GXM preparation, but more likely because we previously solubilized exo-GXM in phosphate buffered saline.
Figure 6: Phosphate is sufficient to drive C. neoformans towards smaller morphotypes.
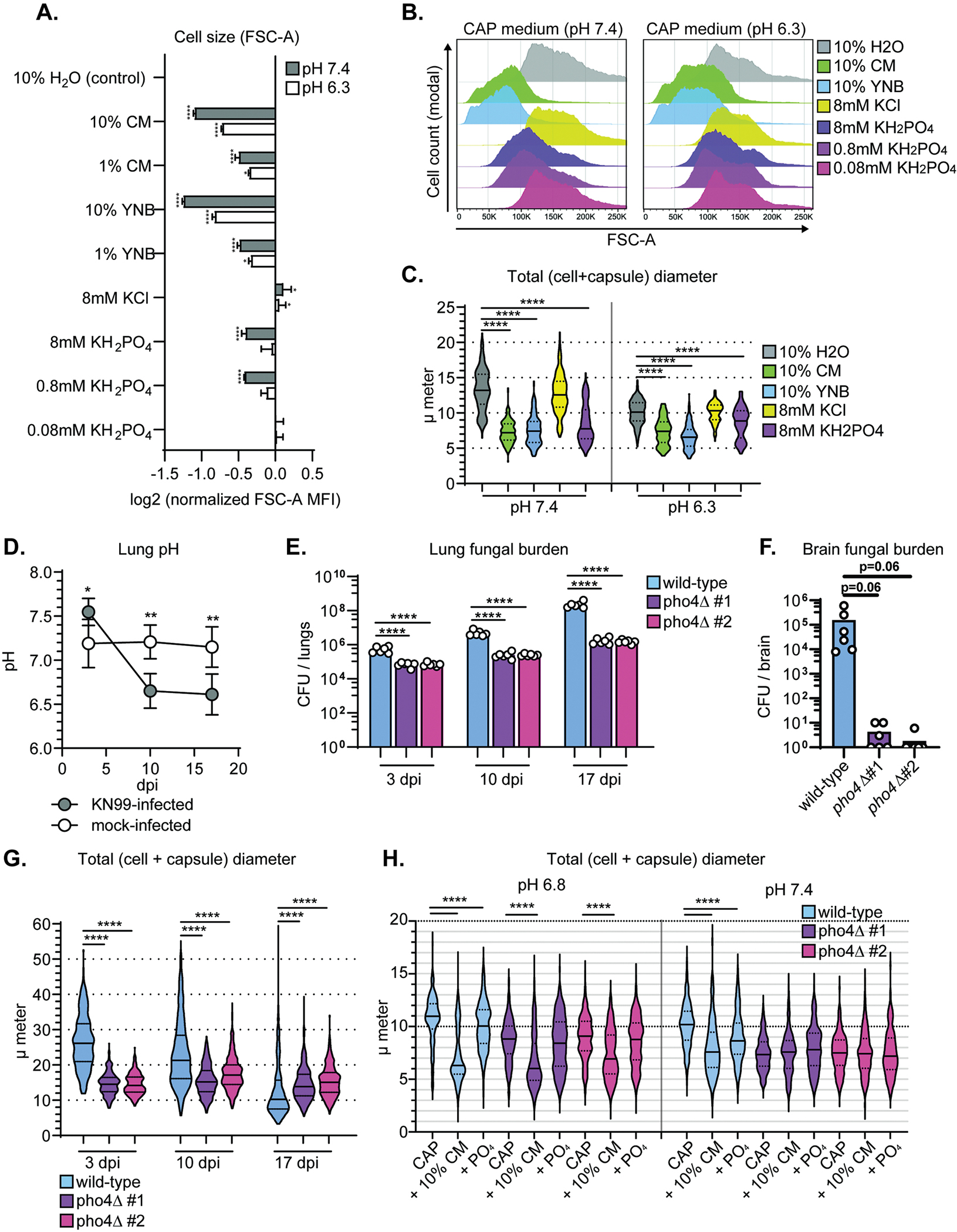
(A) Cell size (FSC-A) measured using flow cytometry after growth in CAP-medium pH 7.4 and supplementation with the indicated factor. FSC-A median fluorescence intensity (MFI) was normalized to control (10% H2O)-treated cells (one-way ANOVA and uncorrected Fisher’s LSD). Bar graphs display the mean. (B) Representative flow cytometry plots for conditions in Figure 6A. (C) Total (cell + capsule) diameter measurements representing conditions in Figure 6A (Mann-Whitney U-test; N=200 fungal cells). Solid lines in the violin plots indicate the median and dotted lines mark quartiles. (D) pH in C. neoformans strain KN99-infected and mock-infected mouse lungs at 3, 10, and 17 dpi (N=4 mice; one-way ANOVA and uncorrected Fisher’s LSD; error bars represent standard deviation). (E) Lung fungal burden in mice at 3, 10, and 17 dpi (One-way ANOVA and uncorrected Fisher’s LSD). The bar graph represents the mean. (F) Brain fungal burden in mice at 17 dpi (One-way ANOVA and uncorrected Fisher’s LSD). The bar graph represents the mean. (G) Total diameter measurements at 3, 10, and 17 dpi (Mann-Whitney U test; N=6 mice per timepoint; 50, 100, and 150 wild-type cells measured per mouse at days 3, 10, and 17, respectively; 50, 50, and 100 pho4Δ mutant cells measured per mouse at days 3, 10, and 17, respectively) (H) Total diameter measurements of wild-type and pho4Δ mutant cells after growth in CAP-medium at labeled pH and with supplementation with the indicated factor (10% CM or 800 μM PO4) (Mann-Whitney test; N = 3 biological replicates per experiment and >100 cells measured per replicate. Solid lines in the violin plots indicate the median and dotted lines mark quartiles.
pH is important for phosphate acquisition because phosphate transporters Pho84 and Pho840 are phosphate/H+ symporters, requiring a proton gradient to efficiently transport phosphate into the cell (Kretschmer et al., 2014). In alkaline pH, C. neoformans inefficiently imports phosphate and upregulates phosphate acquisition machinery (Kretschmer et al., 2014; Lev et al., 2017). C. neoformans releases metabolites that acidify the local microenvironment and increase nutrient uptake (Himmelreich et al., 2003; Wright et al., 2002); cryptococcomas in the brain acidify to levels as low as pH 5.5 (Himmelreich et al., 2003; Wright et al., 2002).
We measured lung pH in mice infected intranasally with C. neoformans. pH decreased from ~7.4 to as low as ~6.3 between 3 and 17 dpi (Figure 6D). We measured a modest, but significant decrease in total diameter by light microscopy when pH 6.3 CAP-grown cells were supplemented with phosphate (Figure 6C), which was mostly due to a decrease in capsule thickness (Figure S5). Phosphate did not induce significant shifts in FSC-A-measured cell size CAP medium pH 6.3, potentially because phosphate availability at pH 6.3 is not as limiting as pH 7.4 (Figures 6A and 6B). When phosphate availability suddenly increases due to higher extracellular concentrations or lower pH, C. neoformans cells shift toward smaller morphotypes.
Limiting phosphate acquisition genetically suppresses morphogenesis in vivo
We hypothesized that limiting phosphate acquisition in vivo would alter morphogenesis, perhaps delaying the appearance of small morphotypes. To test this hypothesis, we intranasally inoculated mice with wild-type KN99, or either of two independently constructed KN99:pho4Δ strains. pho4Δ cells fail to adequately upregulate genes involved in phosphate acquisition in phosphate-limiting conditions and become hypersensitive to alkaline conditions. pho4Δ cells grow poorly in the lungs and are attenuated for CNS dissemination due to poor survival in alkaline blood (Lev et al., 2017). We observed a similar pattern (Figure 6E,F). We also found less cell and capsule size variation in pho4Δ cells than wild-type cells (Figures 6G, S5D–F). Total median wild-type diameter was far larger than the pho4Δ cells at day 3 (wild-type: 26 μm; pho4Δ #1: 15 μm; pho4Δ #2: 14 μm) and smaller than the pho4Δ cells at day 17 (wild-type: 10 μm; pho4Δ #1: 14 μm; pho4Δ #2: 15 μm). pho4Δ cells therefore appear to be deficient in titan cell formation in vivo (Figure 6G). However, this result potentially obscures whether pho4Δ mutant cells are able to form small cells.
Titan cell formation is not required for small cell induction in vitro. We therefore induced small cells in vitro with either conditioned medium or inorganic phosphate. At pH 7.4, pho4Δ cells exhibit constitutively small cell bodies and capsules (wild-type: 10.2 μm, pho4Δ #1: 7.3 μm; pho4Δ #2: 7.5 μm) (Figure 6H). At pH 6.8, we found that pho4Δ cells’ total diameters were only slightly smaller than wild-type cells at pH 6.8 (wild-type: 10.6 μm, pho4Δ #1: 9 μm; pho4Δ #2: 9 μm). At pH 6.8, both wild-type and pho4Δ cells decrease in total diameter in response to conditioned medium but not phosphate, while pH 7.4-grown pho4Δ did not exhibit a cell size shift in response to either conditioned medium or phosphate (Figure 6H). Together, these data demonstrate that PHO4 is necessary for phosphate-induced small cell formation at pH 7.4 and that this phosphate-induced pathway could well be the only molecular trigger of seed cell formation at pH 7.4. That pho4Δ cells’ ability to form small cells at pH 6.8 could indicate that two pathways exist to drive small cell formation.
Finally, we measured phosphate levels within C. neoformans cells during growth under capsule-inducing conditions (10% Sabouraud’s pH 7.4 or pH 6.8) and following in vitro small cell induction. Small cells contained a higher amount of phosphate on a per cell basis, despite their smaller cell volume (Figure S5G). Small cell formation therefore involves increased acquisition of a limited nutrient and is triggered by multiple common signals, strongly supporting the idea that it is a major pathway that can be activated under a variety of conditions.
Components of C. neoformans’s environmental niche induce small cells
Phosphate is available from a number of sources that C. neoformans could encounter in the environment or during infection. These include nucleotide pools within fungal and mammalian cells (Beis and Newsholme, 1975; Harris et al., 1958) and phosphate in the blood at millimolar concentrations (Beis and Newsholme, 1975). Moreover, the infection process itself could increase the amount of extracellular phosphate. Neutrophil extracellular traps (NETs) include extruded neutrophil DNA (Brinkmann et al., 2004). Cell lysis releases intracellular nucleoside mono-, di-, or triphosphates and free ATP acts as a damage, inflammation, and apoptotic signal (Dosch et al., 2018; Elliott et al., 2009; Gault et al., 2014; Grygorczyk et al., 2021). Extracellular ATP is released by macrophages in response to bacterial infection (Ren et al., 2014). Outside the host, C. neoformans is associated with pigeon guano (Emmons, 1955), which is rich in phosphate (Otero et al., 2018).
Using our in vitro small cell induction protocol, we tested which phosphate sources trigger small cell formation. E. coli genomic DNA and sheared salmon sperm DNA (ssDNA) both induced modest significant shift in the median total diameter, from 9.4 microns (at pH 7.4) to 8.0 microns for E. coli genomic DNA and 8.3 microns for ssDNA (Figure 7A). Of the nucleoside mono-, di-, and triphosphates, nucleoside triphosphates (NTPs) induced the greatest reduction in median population diameter at both pH 6.8 (8.4 to 7.1 μm) and pH 7.4 (9.4 to 6.7 μm) (Figure 7B,C). This appears to be synergistic, as the median diameter shift induced by all four NTPs together (at 200 μM total concentration, or 50 μM each) was greater than any individual NTP at 200 μM concentration (Figure S6A,B). Among the nucleotide sugars, UDP-GlcNAc, a precursor for chitin, induced the largest shift in the median population diameter at both pH 6.8 and pH 7.4 (Fig 7D) (8.9 to 7.4 μm at pH 7.4; 9.3 to 7.0 μm at pH 6.8).
Figure 7: Diverse phosphate sources, including pigeon guano, induce small cells.
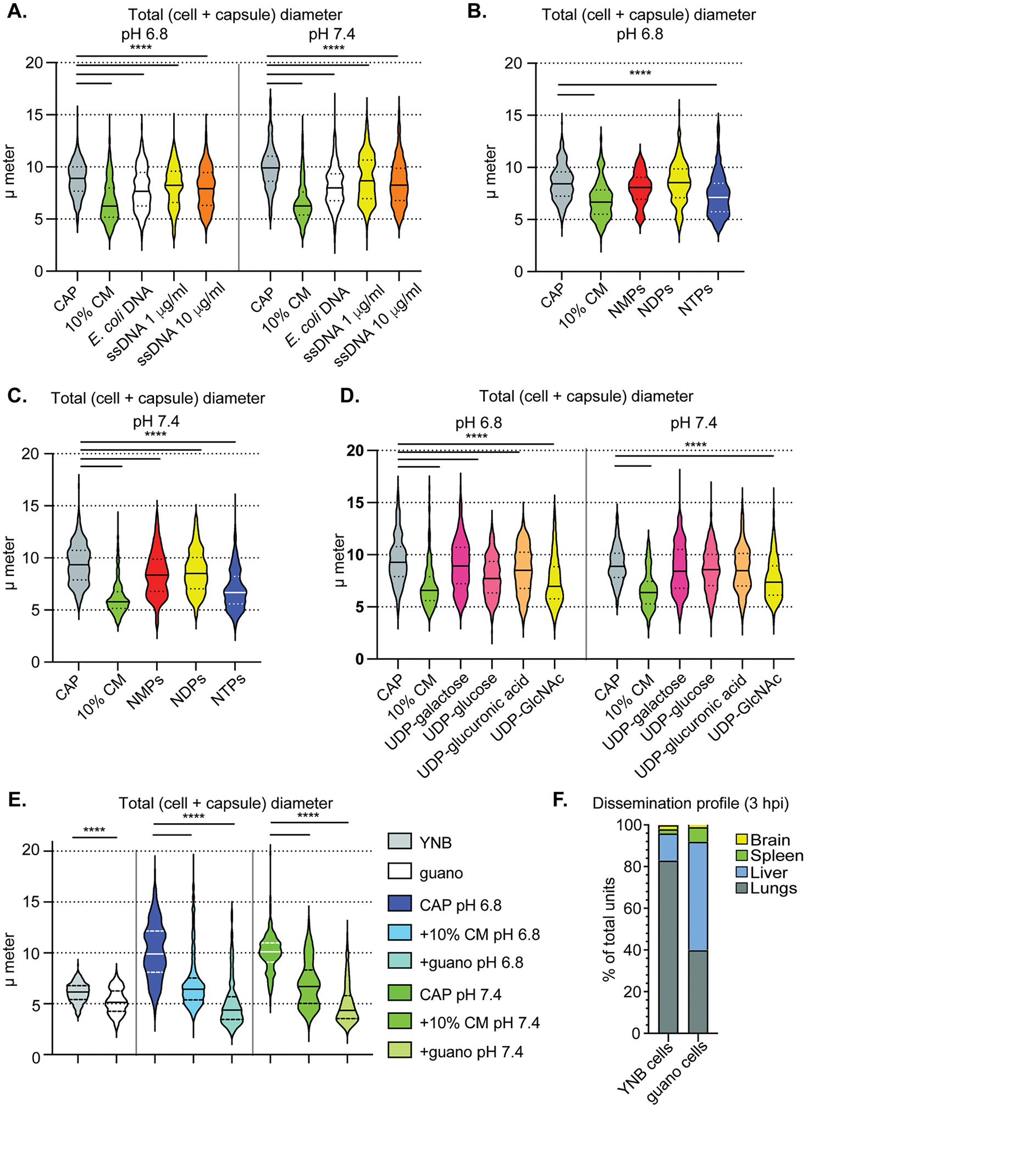
(A) Total diameter measurements of cells after growth in CAP-medium at pH 6.8 (left) or 7.4 (right) and supplementation with different DNA sources. E. coli DNA concentration was 1 μg/ml and ssDNA at either 1 μg/ml (yellow) or 10 μg/ml (orange). (B) Total diameter measurements of CAP-grown cells after exposure to nucleotide mono-, di-, or triphosphate pools at 200 μM total concentration (50 μM each of adenine, cysteine, guanine, and uridine mono-, di-, or triphosphates) following growth at pH 6.8 or (C) pH 7.4. (D) Total diameter measurements of CAP-grown cells after exposure to UDP sugars at pH 6.8 (left) or pH 7.4 (right). (E) Total diameter measurements of CAP-grown cells after exposure to pigeon guano extract medium. (F) Organ distribution following tail vein infection of C. neoformans cells grown in either YNB or pigeon guano extract medium. Panels A-E: P-values calculated using Mann-Whitney test; N = 3 biological replicates per experiment and >100 cells measured per replicate. The solid line represents the population median and the dotted lines the 25th and 75th percentiles. Solid lines in the violin plots indicate the median and dotted lines mark quartiles.
However, the largest shift in median total diameter was induced by pigeon guano (Figure 7E, Figure S6C,D; 10.1 to 4.3 μm at pH 7.4, 9.9 to 4.4 μm at pH 6.8), which surpassed even conditioned medium as an inducing agent. This shift was smaller but still significant in pho4Δ cells (Figure S6E), suggesting that guano contains a phosphate-independent inducing factor. Since volume is a function of the radius cubed (V = 4/3 × π × r3) and radius = ½ × diameter, the volume of capsule-induced cells is 11-fold greater than the volume of a guano-induced small cell. Even relatively modest reductions in diameter, such as that induced by UDP-GlcNAc at pH 7.4, represents an almost 2-fold reduction in volume compared to CAP-grown cells.
In addition to inducing a substantial change in size and volume, growth in pigeon guano medium changes organ entry abilities of C. neoformans cells. We found that growing cells in 10% guano medium, even without inducing small cell formation, resulted in more entry into extra pulmonary organs than YNB-grown cells (Figure 7F). Together, these data demonstrate that C. neoformans’s environmental niche can make C. neoformans cells more prone to extrapulmonary organ entry and demonstrates the importance of considering environmental niches when studying these opportunistic pathogens.
DISCUSSION:
Here we demonstrate the formation of an inducible morphotype that readily enters and survives in extrapulmonary organs. Since these cells are transcriptionally distinct (Figure 5) and formed in response to extracellular signals (Figure 5, 6, 7), we argue that they represent a separate morphotype rather than part of a continuum of variously sized cells. Given their importance for dissemination and extrapulmonary organ invasion and proliferation, we suggest the name “seed cells.”
While seed cells are not the only smaller-sized morphotype in C. neoformans, they are distinct from other morphotypes. Microcells, at approximately 1 micron in diameter, are smaller than seed cells (Feldmesser et al., 2001). Drop cells, identified by high non-cytoplasmic levels of oxidative stress response stain CMFDA, are similar in shape and size. However, drop cells are metabolically inactive (Alanio et al., 2015), while cell growth is required for seed cell induction: if we do not add fresh growth medium when inducing small cells in vitro, the median population diameter does not shift (Figure 6B). Seed cells and titan daughter cells (“titanides”) (Zhou et al., 2020) might be the same, independently described morphotype. We cannot completely eliminate the possibility in vivo, but a key difference in vitro is that we can obtain small cells in vitro without inducing titan cells beforehand (Figure 6). In other ways, titanides are similar to seed cells, particularly their hypothesized role in dissemination.
This work and others’ underscore the critical importance of phenotypic heterogeneity in the dissemination process. Heterogeneity in the form of antigen or phase switching is common in bacteria (van der Woude and Bäumler, 2004) and parasite infections (Deitsch et al., 1999; Ward et al., 1999) but not fungi. Instead, fungi shift their surface antigens in response to environmental changes (Ballou et al., 2016; Hommel et al., 2018; López-Fuentes et al., 2018; Trevijano-Contador et al., 2018) or through morphological changes (Alanio et al., 2015; Botts et al., 2009; Feldmesser et al., 2001; Fernandes et al., 2018; Okagaki et al., 2010).
Increased mannose exposure on the C. neoformans cell surface is important for seed cells’ interactions with the host immune system. Prior data concerning mannose recognition and mannose-binding lectin during cryptococcosis is contradictory (Eisen et al., 2008; Fang et al., 2015). Mannose receptor-deficient mice are sensitive to C. neoformans infection (Dan et al., 2008). Mannose recognition via the mannose receptor was dispensable for murine macrophage cryptococcal phagocytosis in vitro but primary blood monocyte-derived human macrophages showed reduced phagocytosis when pre- treated with exogenous mannose. (Lim et al., 2018). Increased mannose exposure may result in a balance between host and microbe: in places such as the liver, increased mannose-dependent uptake (Figure 3G) is countered by robust seed cell growth within the organ, with the attempted fungal sequestration backfiring on the host (Figure 3F). Increased seed cell phagocytosis might suggest a Trojan horse model of dissemination, with fungal cells spreading while contained within a host cell. “Naked” C. neoformans cells have been observed entering organs within two hours of intravenous infection (Shi et al., 2010) and arrest in capillaries within one minute of inoculation (Gibson et al., 2022; Shi et al., 2010), then are internalized by brain endothelial cells (Chen et al., 2021). Depletion of liver-resident macrophages by clodronate (Figure 3D) decreases liver CFU. This indicates only that liver-resident macrophages are important for liver entry. The Trojan horse might therefore be more important for organ barrier crossing than transport to organs. We think that our data currently supports either Trojan horse or “naked” crossing models.
Our data also indicate that growth conditions dramatically influence organ entry ability. Cells from more stressful, host or host-like environments were better primed for dissemination. Capsule formation was not sufficient to facilitate organ entry (Figure 4A) but allowed slightly increased blood survival (Figure 4E); YNB-grown cells neither entered organs nor survived blood growth well. We thus propose that efficient organ entry requires smaller cell and capsule morphologies (Figure 4A), stress resistance to prime fungi for survival in the blood and tissue (Alanio et al., 2015; Ngamskulrungroj et al., 2012), and cell surface changes. We see all these changes in seed cells.
Phosphate is essential for cell homeostasis and growth and its acquisition is tightly controlled in fungi (Kohler et al., 2020; Lev and Djordjevic, 2018). Cryptococcus relies on a proton gradient to import phosphate (Lev and Djordjevic, 2018) and therefore survives poorly in the alkaline (pH 7.4) bloodstream due to the inability to upregulate phosphate acquisition machinery (Lev et al., 2017). Our results suggest that increased phosphate availability also impacts earlier dissemination by mediating pulmonary seed cells emergence (Figure 7) and inducing seed cells at phosphate-limiting pHs (Figure 6). In vivo phosphate concentrations seem to correlate with observed C. neoformans morphotypes. Phosphate is low (≤50 μM) in bronchoalveolar lavage fluid of uninfected mice (Saito et al., 2015), which corresponds with titan cell-forming conditions early in infection. Lung pH decreased by an order of magnitude over the course of infection (Figure 6D), which could also enhance phosphate acquisition (Lev et al., 2017) or repress capsule growth (Farhi et al., 1970; O’Meara and Alspaugh, 2012) while still permitting seed cell formation.
Fungal cell ploidy is another source of population variation that can correlate with morphology and drives adaptation, in-host survival, and pathogenicity in multiple fungal species (Gerstein et al., 2015; Okagaki and Nielsen, 2012; Selmecki et al., 2015). Of our ex vivo populations, only large cells contain potential titan (>2C) cells (Figure S2). Intermediate cells display the DNA content profile of actively growing haploid yeast cells (Todd et al., 2018). We therefore think that comparisons between seed and intermediate ex vivo cells are independent of ploidy.
While C. neoformans cell size morphotypes have been extensively observed in mice, their presence during human infection remains controversial. Autopsies of human patients occur at the end of infection, when titan cells are rare in the mouse model (Figure 1) and so they might be uncommon in humans. Titan-like cells have been observed in the lungs (Cruickshank et al., 1973) and formation of smaller cells correlates with higher intracranial pressure and thus dissemination (Fernandes et al., 2018). Xie et al. (Xie et al., 2012) even found titan-like cells in the lungs and smaller cells in the brain in humans, similar to our mouse data. Since titan cell formation is not required for seed cell formation in vitro, so even the absence of titan cells does not exclude a role for seed cells in cryptococcosis.
There are some notable limitations to our study. For example, we initially define subpopulations based on sorted cross-sections of fungal cells within the lungs rather than biological criteria. Small ex vivo “seed” cells correspond with the size of disseminated cells in the brain, but intermediate and large cells are based on distinguishing but arbitrary size cutoffs. Further studies of fungal subpopulations within the lungs and analysis of their dissemination would enhance our understanding of systemic cryptococcosis. Furthermore, C. neoformans faces multiple potential bottlenecks during dissemination, including pulmonary escape. C. neoformans can access the bloodstream by crossing lung epithelial cells or via the Trojan horse mechanism within macrophages (Denham and Brown, 2018). We focused on extrapulmonary organ entry rather than examining lung escape by seed cells directly. However, seed cells are more likely to be phagocytosed, which may lead to increased Trojan horse dissemination or might only impact organ entry.
Phenotypic heterogeneity is an important microbial property that facilitates severe, disseminated infections (Zaragoza, 2011; Zhou and Ballou, 2018). Here we demonstrate that an individual morphotype, the seed cell, is prone to disseminate and shows increased extrapulmonary organ entry. Seed cells’ dissemination ability is consistent with observations in humans (Fernandes et al., 2018) in that phenotypic heterogeneity strongly influences clinical outcome. Small C. neoformans cells are also found in the brains of human patients with cryptococcal meningitis (Xie et al., 2012). Overall, seed cells represent a inducible morphotype formed in response to phosphate and other signals. Infection-induced phosphate availability could establish a feed-forward loop that results in self-propagation of C. neoformans dissemination. Moreover, conditions in C. neoformans’s environmental niche can induce seed cell formation and increase organ entry, thus increasing disease-causing potential for even an “accidental” pathogen and suggesting a broader role for seed cells in the C. neoformans lifecycle.
STAR METHODS:
RESOURCE AVAILABILITY:
Lead contact
Requests for resources or information should be directed to the lead contact, Jessica Brown (jessica.brown@path.utah.edu).
Materials availability
All materials generated for this paper are available without restriction. Please request from the lead contact.
Data and code availability
RNA-seq data have been deposited at NCBI GEO database (Accession number: GSE152784) and are publicly available.
There is not original code associated with this paper.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL and SUBJECT DETAIL
Mouse infection models and fungal burden analysis
For intranasal inoculations, ~8-week-old female C57BL/6NJ mice (Jackson Laboratory; RRID:IMSR_JAX:005304) were anesthetized with ketamine/dexmedetomidine hydrochloride (Dexdomitor) delivered intraperitoneally. Animals for all experiments were ordered from Jackson labs and randomly assigned to experimental groups. They were then suspended by their front incisors on a horizontal strand of thread. Unless otherwise indicated, mice were inoculated intranasally with 2.5×104 Cryptococcus cells in 50 μl of 1XPBS using a micropipette. The inoculum was placed dropwise onto a nasal flare before being inhaled by mice. Ten minutes later, mice were intraperitoneally administered the reversal agent atipamezole (Antisedan).
For intravenous inoculations, ~8-week-old female C57BL/6NJ mice (Jackson Laboratory) were warmed under a heat lamp before being placed in a restraint. Mice were inoculated via the tail vein with 105 C. neoformans cells or beads in 200 μl of 1XPBS using 28-gauge × 12.7 mm syringes. In order to competitively inhibit host interactions with fungal components in vivo, we administered 400 μg of GXM (see “GXM isolation), mannan (Sigma-Aldrich catalog no. M7504), β-glucan (Millipore Sigma catalog no. 1048288), the chitin monomer N-acetyl glucosamine (Vector Laboratories S-9002), or polyinosinic acid (Poly(I)) (Sigma-Aldrich catalog no. 26936–41-4) in 200 μl of 1XPBS 2–3 minutes prior to inoculation with fungal cells.
For intracranial inoculations, ~6-week-old female C57BL/6NJ mice (Jackson Laboratories) were anesthetized with ketamine/dexmedetomidine hydrochloride as described above. They were inoculated intracranially with 103 C. neoformans cells in 30 μl of 1XPBS via a 26-gauge 1/2-inch needle. Following inoculation, mice were intraperitoneally administered the reversal agent atipamezole (Antisedan; ~0.0125 mg/g).
Nonperfused mice were euthanized by CO2 asphyxiation and cervical dislocation. Mice that were intracardially perfused at time of death were anesthetized with isoflurane and perfused in a nonrecirculating fashion before cervical dislocation. Fungal burden was assessed by excising organs and homogenizing them in 5 mL 1XPBS, washing the probe in between samples: 30 seconds 10% bleach, 45 seconds 70% EtOH, and 10 seconds sterile H2O. Ten-fold serial dilutions of organ homogenate were plated on Sabouraud’s agar containing 10 mg/ml gentamicin and 100 mg/ml carbenicillin and stored at 30°C for 3 days before counting CFUs.
All animal procedures were approved by the University of Utah Institutional Animal Care and Use Committee.
Fungal strains and growth media
The following strains were used in this study: KN99α (Brown lab stock which was also the background for mutant strains), Bt63 (Brown lab stock, NCBI:txid1295841), 52D (ATCC 24067), R265 (ATCC MYA4093), WM276 (ATCC MYA-4071). C. neoformans mCherry+ contains a version of H2B tagged at the C-terminus with mCherry and was a gift from Alexander Idnurm (Verma, 2014). All strains are listed in the Key Resources Table.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| α-complement C3: mAb 11H9 | ThermoFisher | Cat# MA1-40046 |
| Mouse anti-Rat IgG2a, FITC | ThermoFisher | Cat# 11-4817-82 |
| α-surfactant protein D, polyclonal | Abcam | Cat# ab203309 |
| Donkey anti-Rabbit IgG, AlexaFluor 405 | Abcam | Cat# ab175651 |
| α-mannose binding lectin A, mAb 8G6 | Hycult | Cat# HM1035 |
| Mouse anti-Rat IgG2a, FITC | ThermoFisher | Cat# 11-4817-82 |
| α-mannose binding lectin C, mAb 14D12 | Abcam | Cat# ab106046 |
| α-IgM mAb II/41, FITC | BD Biosciences | Cat# 553437 |
| α-IgA mAb 11-44-2, FITC | SouthernBiotech | Cat# 1165-02 |
| anti-Ly6G clone 1A8 | BioXCell | Cat# BP0075-1 |
| anti-CD45-efluor450 | eBiosciences | Cat# 48-0451-82 |
| anti-CD11b-APC | eBiosciences | Cat# 17-0112-82 |
| anti-Ly6G-FITC | eBiosciences | Cat# 11-5931-82 |
| Mouse anti-GPIbα antibody | Emfret | Cat# R300 |
| Fluorescein-conjugated concanvalin A | Vector Laboratories | Cat# FL-1001 |
| Fluorescein-conjugated wheat germ agglutinin | Vector Laboratories | Cat# FL-1021 |
| Bacterial and virus strains | ||
| C. neoformans KN99α pho4::NAT #1 | This manuscript | |
| C. neoformans KN99α pho4::NAT #2 | This manuscript | |
| C. neoformans KN99α H2B-mCherry NAT+ | Gift from Alexander Idnurm | Verma and Idnurm, 2014 |
| C. neoformans KN99α | Fungal Genetics Stock Center | Cat# 10369 |
| C. neoformans Bt63a | Heitman lab | NCBI:txid1295841 |
| C. neoformans 52D | ATCC | Cat# 24067 |
| C. deuterogattii R265 | ATCC | Cat# MYA4093 |
| C. gattii WM276 | ATCC | Cat# MYA-4071 |
| Chemicals, peptides, and recombinant proteins | ||
| fluorescein-conjugated concanavalin A | Vector Laboratories | Cat# FL-1001 |
| fluorescein-conjugated wheat-germ agglutinin | Vector Laboratories | Cat# FL-1021 |
| Clodronate liposomes | Liposoma BV | Cat# C-010 |
| Control liposomes | Liposoma BV | Cat# P-010 |
| TRIzol | ThermoFisher Scientific | Cat# 15596026 |
| Recombinant Qsp1 | Peptide 2.0 | |
| Scrambled Qsp1 | Peptide 2.0 | |
| Control liposomes | Liposoma BV | Cat# P-010 |
| Pigeon guano | Michael Shapiro, University of Utah | |
| Green fluorescent polystyrene microspheres, 6 μm | Polysciences, Inc. | Cat# 17156-2 |
| Green fluorescent polystyrene microspheres, 25 μm | Polysciences, Inc. | Cat# 18241-2 |
| India ink | Higgins | Cat# 44201 |
| UDP-glucose | Sigma Aldrich | Cat# 670120 |
| UDP-galactose | Sigma Aldrich | Cat# 670111-M |
| UDP-glucuronic acid | Sigma Aldrich | Cat# U5625 |
| UDP-GlcNAc | Sigma Aldrich | Cat# 670107 |
| Sabouraud’s dextrose agar | Difco | Cat# 238230 |
| Yeast Nitrogen Base without amino acids | Difco | Cat# 291940 |
| 10% fetal calf serum | GenClone | Cat# 25-550 |
| RPMI 1640 | GenClone | Cat# 25-506 |
| Dulbecco’s Modified Eagle Medium | GenClone | Cat# 25-500 |
| DAPI | Sigma Aldrich | Cat# 28718-90-3 |
| Percoll | GE Healthcare | Cat# 17-0891-01 |
| benomyl | Agilent | Cat# PST-1245 |
| Direct Yellow 96 | AK Scientific | Cat# 61725-08-4 |
| Accutase | Corning | Cat# 25-058-CI |
| Trypan blue | HyClone | Cat# SV30084.01 |
| Accutase | Corning | Cat# 25-058-CI |
| mannan | Sigma Aldrich | Cat# M7504 |
| β-glucan | Millipore Sigma | Cat# 1048288 |
| N-acetyl glucosamine | Vector Laboratories | Cat# S-9002 |
| Polyinosinic acid (Poly(I)) | Sigma Aldrich | Cat# 26936-41-1 |
| Dexdomitor | Zoetis, Inc | Cat# 122692-5 |
| Zetamine | VetOne | Cat# 13985-584-10 |
| Antisedan | Zoetis, Inc | Cat# 87219-02296-2 |
| Critical commercial assays | ||
| Phosphate colorimetric kit | Sigma Aldrich | Cat# MAK030-1KT |
| NEBNext Ultra II Directional RNA Library Prep with rRNA Depletion Kit | New England BioLabs | Cat# E7760 |
| Qiagen RNeasy kit | Qiagen | Cat# 74104 |
| 0.5 mm zirconium beads | Biospec Products | Cat# 11079105z |
| 1 mm zirconium beads | Biospec Products | Cat# 11079110z |
| Deposited data | ||
| RNA-seq dataset | NCBI-GEO | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE152784 |
| Experimental models: Organisms/strains | ||
| Mice: C57Bl/6NJ | Jackson labs | RRID:IMSR_JAX:005304 |
| Oligonucleotides | ||
| pho4KO F: 5’ AAAACGGCTGAAGGCTCGTTCT3’ | This manuscript | |
| pho4KO R: CTTCTGCAAGGTGAAGTTCACG | This manuscript | |
| pho4KO ORF.F: 5’CCATCTCAGATACCAACTCGCC’ | This manuscript | |
| pho4KO ORF.R 5’CAATTTGCTGAGAGCCATAGGC3’: | This manuscript | |
| pho4KO 5’verF: 5’AGAGCTATATGGTATGACGAAC3’ | This manuscript | |
| pho4KO 5’verR: 5’TGTGCTGATCATCCGATGCCAC3 | This manuscript | |
| pho4KO 3’verF: 5’TGTGGAGGATGGTGGGGAATAG3 | This manuscript | |
| pho4KO 3’verR: 5’GGACGTGAGCCAATAAGTTCCT3 | This manuscript | |
| Software and algorithms | ||
| GraphPad Prism 9 | Dotmatics | |
| FlowJo | BD Life Sciences | |
| STAR version 2.7.2c | Dobin et al. 2013 | |
| Venn diagram tool | Van de Peer group, Ghent University | Bioinformatics.psb.ugent.be/webtools/Venn/ |
The following growth media were used in this study: YNB medium (yeast nitrogen base without amino acids [Difco catalog no. 291940], 2% glucose), CAP medium (10% Sabouraud’s dextrose [Difco catalog no. 238230], buffered with 50 mM HEPES (pH 7.4) or MES (pH 6.3)), Titan cell medium (5% Sabouraud’s dextrose, 10% fetal calf serum (GenClone catalog no. 25–550), 15 μM sodium azide, 50 mM HEPES (pH 7.4)).
Liver macrophage isolation and infection
Liver macrophages were isolated from 8- to 10-week-old female C57BL/6NJ mice (Jackson Laboratory) according to established procedures (Li et al., 2014). We euthanized mice, then perfused the liver with 10 ml 1x PBS. We removed and minced the liver, then incubated in RPMI 1640 + 0.1% type IV collagenase for 30 minutes at 37°C. Liver pieces were then run through a serological pipet for ~10 minutes to disperse tissue, then filtered through a 70 μm cell strainer (Corning) to separate single cells. Tissue remaining on the filter was mashed against the filter with the flat end of a sterile 5 ml syringe plunger and washed with 10 ml RPMI. Samples were then subjected to differential centrifugation to purify liver macrophages using the following steps: 1) Centrifuged at 300 × g for 10 minutes at 4°C, resuspended pellet in 10 mls cold RPMI. 2) Centrifuged at 300 × g for 5 minutes at 4°C, resuspended pellet in 10 mls cold RPMI. 3) Resuspended cell pellet in 10 mls cold RPMI, then centrifuged at 50 × g for 3 minutes at 4°C. 4) Transferred aqueous phase, then centrifuged 300 × g for 5 minutes at 4°C. These cell pellets contained primarily macrophages but also endothelial and other cells. To isolate liver macrophages, we selected for adherence to plastic by resuspending in complete DMEM (Dulbecco’s Modified Eagle’s Medium, GenClone catalog no. 25–500) supplemented with 10% fetal bovine serum (FBS, (GenClone catalog no. 25–550) and 100 U/mL Penicillin/Streptomycin and seeded into a T-25 flask at a density of 8–10 × 106 cells/flask. Macrophages were allowed to settle and adhere for 4 hours in a mammalian tissue culture incubator (37 °C, 5% CO2). Nonadherent cells were then removed from the dish by gently washing 3 times with 1XPBS (without divalent cations), leaving adherent macrophages. The media was replaced with 5 mL complete DMEM, and the cells were rested for 4 days. The media was replaced on the day after initial seeding, and then as needed.
Fungal infection and association with liver macrophages was assessed using the following procedures. The media was aspirated from rested liver macrophages and the cells were washed once with 1XPBS. Macrophages were lifted from the plate by treating them with 1mL Accutase (Corning catalog no. 25–058-CI) for ~5 minutes, tapping the plate to dislodge cells. 1 mL complete DMEM was added to the Accutase-treated cells, which were then centrifuged at 300×g for 5 minutes. Liver macrophages were resuspended cells in 0.5–1 mL complete DMEM and live cells were counted by mixing trypan blue (HyClone catalog no. SV30084.01) with cell suspension at a 1:1 ratio and counting clear cells on a hemocytometer. 40,000 live cells/well were seeded into 96-well plates in complete DMEM. The liver macrophages were allowed to settle and adhere overnight.
The next day, the inoculum was prepared by resuspending C. neoformans cells in 500 μl of 0.01% Direct Yellow 96 (AK Scientific 61725–08-4), vortexing briefly to mix, and incubating for 5 minutes on the benchtop. C. neoformans cells stained with Direct Yellow 96 were pelleted, washed once with 1 mL 1XPBS, and resuspended at a concentration of 8×106 cells/mL in serum-free DMEM. The supernatant was aspirated from the macrophages seeded in the 96-well plates and was replaced with 200 μl of serum-free DMEM. 10 μl of the inoculum (8×104 cells) was added to each well, resulting in a multiplicity of infection of 1:1. Macrophages and C. neoformans cells were co-cultured for 4 hours in a mammalian tissue culture incubator (37 °C, 5% CO2). The medium was then aspirated from each well, and the cells were washed three times with 100 μl of 1XPBS to remove non-macrophage associated fungal cells. The remaining cells were fixed in 100 μl of 4% paraformaldehyde for 10 min. The paraformaldehyde was then removed and the cells were washed twice with 100 μl of 1XPBS and stained with100 μl of DAPI (Sigma Aldrich catalog no. 28718–90-3) working solution (600 nM DAPI; 0.1% Triton X-100; in 1XPBS) for 5 minutes. The DAPI working solution was then removed and cells were washed twice with 100 μl of 1XPBS. Finally, 100 μl of 1XPBS was added to the wells to prevent desiccation. Cells were imaged on a Nikon widefield microscope, and the number of macrophages with associated C. neoformans cell(s) were counted (green-fluorescent due to Direct Yellow 96). Six images were taken per well in the same relative positions and every imaged macrophage was scored as having a physically associated fungal cell(s) or not, resulting in 2263–3197 macrophages counted per experimental condition across three independent replicates.
In order to asses liver macrophage killing of C. neoformans cells, liver macrophages were seeded in 96-well plates and infected with C. neoformans cells as described above. The plates containing C. neoformans and macrophages were incubated for 24 or 48 hours. After the desired time, the supernatant was collected from each well and transferred to a microcentrifuge tube. 200 μl of sterile distilled water was then added to each well in order to lyse the macrophages for 40 minutes at 37°C. The contents of each well were then mixed to collect every C. neoformans cell and combined with the respective supernatant now in a microcentrifuge tube. The wells were rinsed with 200 μl sterile PBS and added to the respective microcentrifuge tube. Serial dilutions of each of the well contents were plated on YPAD agar and incubated at 30°C for 2–3 days before counting CFUs. CFUs quantified at each timepoint were normalized to the CFUs in the inoculum in order to calculate the “% of input”. Each macrophage killing experiment was repeated on three different days with each experimental condition being performed in duplicate wells.
Cell-type specific depletion in mice
Hepatic and splenic macrophages were depleted from mice using clodronate liposomes. Mice were intravenously administered 200 μl of clodronate liposomes (Liposoma BV catalog no. C-010) or PBS control liposomes (Liposoma BV catalog no. P-010) 48 hours prior to inoculation with C. neoformans cells. Macrophage depletion efficiency in the liver and spleen was assessed on the day of inoculation via flow cytometry. Macrophages were identified as CD45+ (anti-CD45-efluor450, eBiosciences catalog no. 48–0451-82) and F4/80+ (anti-F4/80-APC, eBiosciences catalog no. 17–4801-80).
Neutrophils were depleted from mice using anti-Ly6G antibodies. Mice were intraperitoneally administered 200 ug of anti-Ly6G (clone 1A8 BioXcell catalog no. BP0075–1) antibodies or PBS control at 24 and 2 hours prior to inoculation with C. neoformans cells. Neutrophil depletion efficiency in the blood was assessed on the day of inoculation via flow cytometry. Neutrophils were identified as CD45+ (anti-CD45-efluor450, eBiosciences catalog no. 48–0451-82), CD11b+ (anti-CD11b-APC, eBiosciences catalog no. 17–0112-82), and Ly6G+ (anti-Ly6G-FITC, eBiosciences catalog no. 11–5931-82).
Platelets were depleted from mice using anti-GPIbα antibodies. Mice were intravenously administered 80 μl of anti-GPIbα antibodies (Emfret catalog no. R300) or PBS control 24 hours prior to inoculation with C. neoformans cells. Platelet depletion efficiency was determined by Hemavet 950FS (Drew Scientific Group) analysis of blood collected by cheek bleed 10 minutes prior to anti-GPIbα administration and on the day of inoculation.
METHODS DETAILS
In vitro small cell induction assay
To screen for factors that facilitated shifts in cell size, we picked single colonies from C. neoformans cells streaked on YPAD agar and cultured them overnight (12–18 hours) in YNB medium at 30 °C. We then sub-cultured those cells into YNB medium, CAP medium, or titan cell medium for 24 hours at an initial cell density of 105 cells/mL. YNB- and CAP-grown cells were cultured at 37 °C, while titan cell medium-grown cells were cultured at 37 °C and 5% CO2. At this stage, conditioned medium (CM) was collected from the 24-hour YNB cultures by pelleting the fungal cells and filtering (0.22 μm pore size) the supernatant. 450 μl of the CAP and titan cell media cultures were sub-cultured into 450 μl fresh media and 50 μl (10% of the final volume) of one of the following supplements solubilized in H2O: 10% H2O (vehicle control), CM, 1% CM, 50 μg GXM, 50 μM Qsp1 peptide (Peptide 2.0: NFGAPGGAYPW), 50 μM Qsp1 scrambled peptide (Peptide 2.0: AWAGYFPGPNG), 10% 1x PBS (phosphate-buffered saline), 10% YNB medium; the following YNB medium supplements were added to a final concentration that equaled their concentrations in YNB medium: 2% glucose, 4.531 nM folic acid, 8.186 nM biotin, 250.6 nM copper sulfate pentahydrate, 531.4 nM riboflavin, 602.4 nM potassium iodide, 839.5 nM calcium pantothenate, 971.3 nM Sodium molybdate dihydrate, 1.186 μM thiamine hydrochloride, 1.233 μM ferric chloride hexahydrate, 1.458 μM p-aminobenzoic acid, 1.945 μM pyridoxine hydrochloride, 2.477 μM zinc sulfate heptahydrate, 2.649 μM manganese sulfate monohydrate, 3.249 μM niacin, 8.087 μM boric acid, 11.10 μM inositol, 901.1 μM calcium chloride dihydrate, 1.711 mM sodium chloride, 4.154 mM magnesium sulfate heptahydrate, 7.348 mM potassium phosphate monobasic, and 37.8mM ammonium sulfate. The supplemented cultures were incubated for another 24 hours at 37 °C, following which 400 μl was aliquoted to estimate cell size by flow cytometry and another 400 μl was aliquoted to measure cell and capsule size (see “fungal cell size measurements”) in India ink (Higgins catalog no. 44201).
Pigeon guano medium
Pigeon guano was a gift from Michael Shapiro’s lab (University of Utah Department of Biology). Following collection, it was lyophilized, then stored at room temperature until use. Lyophilized guano was ground in a coffee grinder until a powder. Guano medium consisted of a 20% w/v solution in water, which was boiled for 10 minutes, then filtered through first Whatman 3 mm filter paper, then a 0.5 μm polystyrene filter, and finally a 0.2 μm polystyrene filter. Resultant medium was stored at room temperature for up to four weeks.
Phosphate source small cell induction assay
We grew fresh colonies of KN99 cells overnight in YNB at 37°C, then subcultured at 105 cells/ml in CAP-medium 10% Sabouraud’s buffered to either pH 7.4 with 50 mM HEPES or pH 6.8 with 50 mM Tris HCl). Cultures were grown 24 hours at 37°C, then diluted 1:1 with either 20% (w/v) pigeon guano medium or fresh 10% Sabouraud’s of the specified pH containing the phosphate source of interest (DNA; nucleotide mono-, di-, or triphosphates; or UDP sugars).
Fungal cell size measurements
C. neoformans cells harvested from laboratory growth medium or infected mouse lungs were fixed in 2% paraformaldehyde for 20 minutes. To visualize capsule, 4 μl of India ink is mixed with 4 μl of cell suspension on a microscope slide. At least 10 successive images were taken starting at one edge of the coverslip and moving across to the opposite side, as smaller cells tended to drift to the edge of the coverslips. Total diameter was measured from one edge of the capsule to the other. Cell body diameter was measured from one edge of the cell wall to the other. Capsule thickness was calculated as follows: capsule thickness = (total diameter – cell body diameter) / 2. The total number of cells counted for a given experiment is indicated in the figure legends.
FACS isolation of fungal cells
Mice were intranasally inoculated with C. neoformans KN99-mCherry (see “Mouse infection models”). At 14 dpi, lungs from 3 mice were excised and placed in 15 mL of 1XPBS. The three lungs were homogenized together using a mechanical tissue homogenizer. The homogenate was filtered first through a 70 μM cell strainer and then through a 40 μM cell strainer to prevent downstream clogging of the flow cytometer. Cells were centrifuged at 2195×g for 10 min. The supernatant was discarded and the pellet was resuspended in ~1mL 1XPBS. The suspension was filtered once more through a 70 μM cell strainer and was resuspended at ~1×107 cells/mL in 1XPBS for FACS (BD FACS Aria). The FACS gating scheme is represented in Figure 2. To control for the irregular shape of budding cells, we gated for single cells (routinely >95% of the population) after sorting for mCherry+ cells. Gating strategy is shown in Figure 2A. Data was analyzed using FlowJo (BD Life Sciences).
Isolation of beads from mouse organs
Mice were intravenously administered (see “Mouse infection models”) 105 green fluorescent polystyrene beads (Polysciences, Inc. catalog no. 17156–2 and 18241–2), and 3 hours later the indicated organs were harvested. Organs were homogenized with a mechanical tissue homogenizer in 5 mL tissue lysis buffer (100 mM Tris-Cl pH 8.0, 200mM NaCl, 5mM EDTA, 0.2% (w/v) SDS), washing the probe in between samples: 15 seconds H2O, 15 seconds 70% EtOH, and 15 seconds fresh H2O.
Beads from each organ were concentrated via differential centrifugation. The homogenates were poured into a fresh 50 mL conical. The homogenization tubes were washed with 30 mL of H2O and combined with the respective homogenized sample. The samples were filtered through cell strainers (70 μM pore size), after which another 10 mL of H2O was passed through each strainer. The samples were centrifuged at 4480 × g for 40 minutes. The supernatant was aspirated to the 5 mL mark and the samples were transferred to fresh 15 mL tubes containing 2 mL of 100% Percoll (GE Healthcare catalog no. 17–0891-01). Water was added to the 10 mL mark. The tubes were vortexed to mix and allowed to settle for 5 minutes before centrifugation at 1120 × g for 45 minutes. The supernatant was aspirated to the 2 mL mark. 10 mL of H2O was added to the samples before centrifuging at 4480 × g for 20 minutes. The supernatant was aspirated and resuspended in 200–1000 μl 1XPBS for flow cytometry. Entire samples were analyzed to determine the number of beads per organ.
Fungal cell staining and analysis
Between 105 and 106 C. neoformans cells harvested from laboratory growth medium or infected mouse lungs were pelleted and fixed in 2% paraformaldehyde for 20 minutes. Fixed cells were pelleted and washed twice with 1XPBS. Cells were then resuspended in 1XPBS and stained with the desired antibodies/lectins/fluorescent dye.
To estimate DNA content, cells were stained for 10 minutes (room temperature) in 0.3 μg/mL 1XPBS+0.1% triton X-100. Mid-log phase fungal cells grown in YNB medium +/− 80 μg/mL benomyl (Agilent catalog no. PST-1245) for 24 hours were used to set 1C and 2C gates. Untreated mid-log phage fungal cells display two DAPI peaks (1C ad 2C), while benomyl prevents cell division and traps cells at 2C.
To estimate fungal microbial associated molecular pattern exposure, cells were stained at room temperature with 5 μg/mL fluorescein-conjugated wheat-germ agglutinin (WGA, Vector Laboratories catalog no. FL-1021) for 15 minutes to detect exposed chitin or 50 μg/mL fluorescein-conjugated concanavalin A (ConA, Vector Laboratories catalog no. FL-1001) for 5 minutes to detect exposed mannose.
To estimate the percentage of cells bound by soluble host factors, we used the following antibodies: α-complement C3 (primary: mAb 11H9, ThermoFisher catalog no. MA1–40046; secondary: Mouse anti-Rat IgG2a, FITC, Thermofisher catalog no. 11–4817-82), α-surfactant protein D (primary: polyclonal, Abcam catalog no. ab203309; secondary: Donkey anti-Rabbit IgG, AlexaFluor 405, Abcam catalog no. ab175651), α-mannose binding lectin A (primary: mAb 8G6, Hycult catalog no. HM1035; secondary: Mouse anti-Rat IgG2a, FITC, Thermofisher catalog no. 11–4817-82), α-mannose binding lectin C (primary: mAb 14D12, Abcam catalog no. ab106046; secondary: Mouse anti-Rat IgG2a, FITC, Thermofisher catalog no. 11–4817-82), α-IgM (mAb II/41, FITC, BD Biosciences catalog no. 553437), α-IgA (mAb 11–44-2, FITC, SouthernBiotech catalog no. 1165–02). Cells were stained with primary antibodies (1:50 dilution) for 30 minutes on ice. When a secondary antibody was required, primary antibody labeled cells were pelleted, washed twice with 1XPBS and stained with the secondary antibody (1:50 dilution) for 20 minutes on ice.
After staining, cells were pelleted and washed twice with 1mL 1XPBS before being resuspended in 200–400 μl of 1XPBS for flow cytometry. C. neoformans cells were gated for single cells following mCherry+ gating to remove budded cells, which might confound the analysis.
GXM isolation
GXM was isolated as described previously (Wozniak and Levitz, 2009) with slight modifications. C. neoformans (KN99) cells were grown in 100 mL for 3 days at 37°C in YNB medium (YNB+2% glucose). Cells were centrifuged for 15 minutes at 12,000×g. The supernatant was filtered (0.45 μM pore size) and 50 mL of supernatant was transferred to a 250 mL bottle. ~150 mL of 95% EtOH was added to the samples in 4–5 aliquots, mixing after each addition. Samples were stored at 4°C overnight to precipitate GXM. Samples were centrifuged for 30 minutes at 15,000×g, 4°C. The pellets were resuspended in 0.2M NaCl to a concentration of about 10 mg/mL (~50 mL per 250 mL bottle). Samples were transferred to a clean beaker stirred until samples were completely resuspended. 3 mg hexadecyltrimethylammonium bromide (CTAB) per 1mg precipitate was slowly added with stirring and low heat. Once solubilized, the samples were removed from heat and 250 mL of 0.05% CTAB was slowly added with stirring. The samples were moved to fresh 250 mL bottles and centrifuged at for 2 hours at 11,000×g, 4°C. The supernatant was discarded and the pellet was washed in 150 mL of 10% EtOH. The samples were centrifuge for 20 minutes at 15,000×g, room temperature. The supernatant was discarded and the pellets were resuspended in 50 mL of 1M NaCl. The samples were transferred to a clean beaker and stirred until completely resuspended. Approximately 2 volumes of 95% EtOH was added dropwise with stirring to precipitate the GXM while leaving the CTAB in solution. The samples were centrifuged in fresh 250 mL bottles for 20 minutes at 11,000×g, room temperature. The supernatant was discarded and the precipitate was dissolved in 2M NaCl. The resulting solution was placed in a snake-skin dialysis cassette with a 3,500 molecular weight cutoff (Thermo Fisher Scientific catalog no. 68035) and dialyzed overnight against 1M NaCl. The samples were subsequently dialyzed against distilled H2O for 14 days at 4°C. The distilled H2O was changed every 1–2 hours for the first 2 days, and then 4–5 times a day for the next 6 days. For the last 2–6 days, the water was changed 2–3 times a day. The dialyzed samples were lyophilized and stored at −80 °C.
Fungal survival in blood
We collected whole blood from 8- to 10-week-old female C57BL/6NJ mice (Jackson Laboratory) to assess C. neoformans survival in blood. Mice were anesthetized with isoflurane, and whole blood was collected by cardiac puncture using 28-gauge × 12.7 mm syringes pre-coated with 0.5M EDTA. Blood was immediately transferred to tubes containing heparin to achieve a final concentration of 30 units of heparin per mL of blood. 100 μl of blood was added to individual wells of a 96-well plate. 100 C. neoformans cells in 5 ul of 1XPBS were used to inoculate each well. The plates were incubated in a mammalian tissue culture incubator (37 °C, 5% CO2) without shaking. At the indicated timepoint, the well contents were transferred to fresh microcentrifuge tubes. The wells were washed with 100 ul of 0.05M EDTA, which was then added to the respective microcentrifuge tube. The entire contents of the microcentrifuge tubes were plated on YPAD agar and incubated at 30°C for 2–3 days before counting CFUs. CFUs quantified at each timepoint were normalized to the CFUs in the inoculum in order to calculate the “% of input”. Each experiment was repeated on three different days with each experimental condition being performed in duplicate wells.
RNA isolation, sequencing, and analysis
RNA sequencing data is publicly available at NCBI GEO (Accession number: GSE152784). C. neoformans cells were harvested from laboratory media and from infected mouse lungs for RNA isolation and sequencing. Three replicates were sequenced for each sample with replicates being defined as follows. Approximately 107 cells were harvested from the indicated in vitro growth condition on three separate days. At least 106 ex vivo cells were isolated per sort gate each day (see “FACS isolation of fungal cells”). Ex vivo cells were isolated from the pooled lung homogenate of three mice. Ex vivo sample populations were isolated on two consecutive days and pooled for RNA isolation and sequencing, with the end result being that each sequenced ex vivo replicate represents fungal cells isolated from pools of six mice.
RNA was isolated using phenol-chloroform extraction, followed by Qiagen RNeasy column-based cleanup. Fungal cells were pelleted, flash frozen in liquid nitrogen, and lyophilized until completely dry. Lyophilized cells were resuspended in 750 μl of TRIzol (ThermoFisher Scientific catalog no. 15596026) inside screw-cap microcentrifuge tubes. Approximately 50 μl of 1-mm and 150 μl of 0.5mm zirconium beads (BioSpec Products catalog no. 11079110z and 11079105z) were added to each sample and placed in a Biospec Products mini bead beater to lyse (12 × 2 min beating pulses, using sample blocks stored at −20 °C to prevent overheating). 150 μl of chloroform was added to the lysed samples and incubated for 2–3min at room temperature. Samples were centrifuged for 15 minutes at 12,000 × g, 4°C. The aqueous phase was transferred to a fresh tube and combined with 5–10 μg RNase-free glycogen as a carrier. 0.375 mL of isopropanol was added to each sample before incubating at least ~20 minutes on ice. The samples were centrifuged for 15 minutes at 12,000 × g, 4°C. The supernatant was discarded, and the isopropanol was removed by pipetting. The samples were then briefly air-dried before resuspension in 350 μl of RLT buffer (Qiagen RNeasy) and 350 μl 70% EtOH. At this junction, samples were run through Qiagen’s RNAeasy kit (Qiagen catalog no. 74104) with on-column DNase treatment.
cDNA libraries were generated with the NEBNext Ultra II Directional RNA Library Prep with rRNA Depletion Kit (New England BioLabs catalog no. E7760) by the High Throughput Genomics Core at the University of Utah. Sample libraries were sequenced on the Illumina NovaSeq 6000. One set of samples (primarily the ex vivo small, medium, and large cells) were stored by the High Throughput Genomics Core at −80°C for six months prior to library construction and showed more degradation products than replicates stored at −80°C for one month. We therefore limited our analysis and interpretation of the data from these samples.
The Cryptococcus neoformans var. grubii H99 genome and gene feature files were downloaded from Ensembl Fungi release 46 and the reference database was created using STAR version 2.7.2c with splice junctions optimized for 150 base pair reads (Dobin et al., 2013). Optical duplicates were removed from the paired end FASTQ files using clumpify v38.34 and reads were trimmed of adapters using cutadapt 1.16 (Martin, 2011). The trimmed reads were aligned to the reference database using STAR in two pass mode to output a BAM file sorted by coordinates. Mapped reads were assigned to annotated genes in the gene feature file using featureCounts version 1.6.3 (Liao et al., 2014). The output files from cutadapt, FastQC, Picard CollectRnaSeqMetrics, STAR and featureCounts were summarized using MultiQC to check for any sample outliers (Ewels et al., 2016). Differentially expressed genes were identified using a 5% false discovery rate with DESeq2 version 1.24.0 (Love et al., 2014). Differentially expressed genes were run through FungiDB Gene Ontology (GO) Enrichment analysis, selecting for “biological process” with a p-value cutoff of 0.05 (Basenko et al., 2018; Stajich et al., 2012). Venn Diagrams were made with software from the Van de Peer group at Ghent University (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Lung pH measurements
To measure lung tissue pH, mouse lungs were excised and immediately pierced with a micro pH electrode (Orion (ThermoFisher Scientific) catalog no. 9863BN). The electrode was completely embedded in the lung tissue before taking pH readings. Two readings were taken per mouse. One reading was taken from the left lobe of the lungs and a second reading was taken from one of the right lobes. These two readings were averaged to estimate lung pH for a given mouse.
Construction of the KN99:pho4Δ strains
The KN99:pho4Δ (CNAG_06751Δ) strains were constructed via homologous recombination-based replacement of the pho4 genomic sequence with a nourseothicin resistance marker. The KN99:pho4Δ #1 strain was obtained from the KN99 gene deletion library (Fungal Genetics Stock Center, 2015 Madhani plates). The KN99:pho4Δ #2 strain was generated by PCR amplification of the nourseothicin resistance marker and ~1,000 base pair flanking regions from the KN99:pho4Δ #1 strain (primers used to amplify the knock-out construct: Forward: 5’ AAAACGGCTGAAGGCTCGTTCT3’, Reverse: CTTCTGCAAGGTGAAGTTCACG). The resulting pho4 knock-out cassette was used to transform wild-type KN99 cells via biolistic transformation as described previously (Davidson et al., 2000). Putative transformants were PCR-verified by confirming the absence of the pho4 open reading frame (Primers to verify the absence of the open reading frame: Forward: 5’CCATCTCAGATACCAACTCGCC’, Reverse: 5’CAATTTGCTGAGAGCCATAGGC3’) and the integration of the knock-out construct at the correct site (Primers to verify the correct 5’ integration site: Forward: 5’AGAGCTATATGGTATGACGAAC3’, Reverse: 5’TGTGCTGATCATCCGATGCCAC3’ and the correct 3’ integration site: Forward 5’TGTGGAGGATGGTGGGGAATAG3’, Reverse: 5’GGACGTGAGCCAATAAGTTCCT3’.
Phosphate extraction and measurement
To determine the amount of phosphate within cells, we grew cells in CAP medium and performed a standard small cell induction assay. We harvested 3 × 107 total cells by centrifugation, washed briefly in water to remove any excess growth medium, then froze in liquid nitrogen. Samples were then lyophilized overnight, then resuspended in 300 μl MilliQ water, vortexed for 30 seconds on/30 seconds off (on ice) for 30 minutes, then tested for phosphate concentration using the Phosphate Colorimetric Kit (Sigma-Aldrich, catalog no. MAK030–1KT) according to the manufacturer’s instructions.
QUANTIFICATION and STATISTICAL ANALYSIS
All statistical analyses, with the exception of RNA sequencing analysis, were performed with GraphPad Prism 8 software. Hypothesis-driven experimental comparisons were analyzed by unpaired t-test or one-way ANOVA and uncorrected Fisher’s LSD if the data held a Gaussian distribution and a Mann-Whitney U-test if the data were not Gaussian. For experiments with large sample sizes (e.g. cell size measurements), statistical significance is shown only if the 95% confidence intervals of the median compared samples does not overlap. In vitro experiments were replicated three times unless otherwise stated. In vivo (murine infection) experiments were replicated twice unless otherwise stated. Sample sizes are stated in the figure legend.
Supplementary Material
Highlights:
Cryptococcus neoformans exhibits an inducible morphotype within the lungs.
This “seed cell” is critical for extrapulmonary organ invasion and dissemination.
Alterations in pH and phosphate concentration support seed cell formation in the lungs.
ACKNOWLEDGEMENTS:
This work was supported by NIH research grant R01AI130248 to J.C.S.B. S.T.D. was supported by NIH grant T32AI055434. Flow cytometry work was supported by the University of Utah Flow Cytometry Facility through the NCI Award Number 5P30CA042014–24 and by the NIH National Center for Research Resources Award Number 1S10RR026802–01. RNA sequencing and analysis was performed by the High-Throughput Genomics and Bioinformatics Analysis Facility at the Huntsman Cancer Institute at the University of Utah. pho4Δ #1 clone was constructed in the Madhani laboratory with support from NIH funding (R01AI100272). We thank Dr. Kyla Ost for her valuable input regarding opsonin-binding and Dr. Matthew Rondina and Dr. Alicia Eustes for their assistance with in vivo platelet depletion.
Footnotes
DECLARATION OF INTERESTS:
The authors have no competing interests to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES:
- Aimanianda V, Bayry J, Bozza S, Kniemeyer O, Perruccio K, Elluru SR, Clavaud C, Paris S, Brakhage AA, Kaveri SV, et al. (2009). Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 460, 1117–1121. 10.1038/nature08264. [DOI] [PubMed] [Google Scholar]
- Alanio A, Vernel-Pauillac F, Sturny-Leclère A, and Dromer F (2015). Cryptococcus neoformans host adaptation: toward biological evidence of dormancy. mBio 6. 10.1128/mBio.02580-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altamirano S, Jackson KM, and Nielsen K (2020). The interplay of phenotype and genotype in Cryptococcus neoformans disease. Biosci Rep 40, BSR20190337. 10.1042/BSR20190337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballou ER, Avelar GM, Childers DS, Mackie J, Bain JM, Wagener J, Kastora SL, Panea MD, Hardison SE, Walker LA, et al. (2016). Lactate signalling regulates fungal β-glucan masking and immune evasion. Nat Microbiol 2, 16238. 10.1038/nmicrobiol.2016.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basenko EY, Pulman JA, Shanmugasundram A, Harb OS, Crouch K, Starns D, Warrenfeltz S, Aurrecoechea C, Stoeckert CJ Jr., Kissinger JC, et al. (2018). FungiDB: An Integrated Bioinformatic Resource for Fungi and Oomycetes. J Fungi (Basel) 4, 39. 10.3390/jof4010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beis I, and Newsholme EA (1975). The contents of adenine nucleotides, phosphagens and some glycolytic intermediates in resting muscles from vertebrates and invertebrates. Biochem J 152, 23–32. 10.1042/bj1520023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojarczuk A, Miller KA, Hotham R, Lewis A, Ogryzko NV, Kamuyango AA, Frost H, Gibson RH, Stillman E, May RC, et al. (2016). Cryptococcus neoformans intracellular proliferation and capsule size determines early macrophage control of infection. Sci Rep 6, 21489. 10.1038/srep21489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botts MR, Giles SS, Gates MA, Kozel TR, and Hull CM (2009). Isolation and Characterization of Cryptococcus neoformans Spores Reveal a Critical Role for Capsule Biosynthesis Genes in Spore Biogenesis. Eukaryot Cell 8, 595–605. 10.1128/ec.00352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botts MR, and Hull CM (2010). Dueling in the lung: how Cryptococcus spores race the host for survival. Curr Opin Microbiol 13, 437–442. S1369–5274(10)00064–0 [pii] 10.1016/j.mib.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, and Zychlinsky A (2004). Neutrophil extracellular traps kill bacteria. Science 303, 1532–1535. 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- Bulmer GS, and Sans MD (1967). Cryptococcus neoformans. II. Phagocytosis by human leukocytes. J Bacteriol 94, 1480–1483. 10.1128/jb.94.5.1480-1483.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, Coelho C, Cordero RJB, Dragotakes Q, Jung E, Vij R, and Wear MP (2019). The capsule of Cryptococcus neoformans. Virulence 10, 822–831. 10.1080/21505594.2018.1431087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Stins MF, McCaffery MJ, Miller GF, Pare DR, Dam T, Paul-Satyaseela M, Kim KS, and Kwon-Chung KJ (2004). Cryptococcal yeast cells invade the central nervous system via transcellular penetration of the blood-brain barrier. Infect Immun 72, 4985–4995. 10.1128/iai.72.9.4985-4995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlier C, Chrétien F, Baudrimont M, Mordelet E, Lortholary O, and Dromer F (2005). Capsule structure changes associated with Cryptococcus neoformans crossing of the blood-brain barrier. Am J Pathol 166, 421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlier C, Nielsen K, Daou S, Brigitte M, Chretien F, and Dromer F (2009). Evidence of a role for monocytes in dissemination and brain invasion by Cryptococcus neoformans. Infect Immun 77, 120–127. 10.1128/iai.01065-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SHM, Stins MF, Huang SH, Chen YH, Kwon-Chung KJ, Chang Y, Kim KS, Suzuki K, and Jong AY (2003). Cryptococcus neoformans induces alterations in the cytoskeleton of human brain microvascular endothelial cells. J Med Microbiol 52, 961–970. 10.1099/jmm.0.05230-0. [DOI] [PubMed] [Google Scholar]
- Chen Y, Li C, Sun D, Strickland AB, Liu G, and Shi M (2021). Quantitative analysis reveals internalisation of Cryptococcus neoformans by brain endothelial cells in vivo. Cell Microbiol 23, e13330. 10.1111/cmi.13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun CD, Brown JCS, and Madhani HD (2011). A Major Role for Capsule-Independent Phagocytosis-Inhibitory Mechanisms in Mammalian Infection by Cryptococcus neoformans. Cell Host Microbe 9, 243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree JN, Okagaki LH, Wiesner DL, Strain AK, Nielsen JN, and Nielsen K (2012). Titan cell production enhances the virulence of Cryptococcus neoformans. Infect Immun 80, 3776–3785. 10.1128/IAI.00507-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshank JG, Cavill R, and Jelbert M (1973). Cryptococcus neoformans of unusual morphology. Applied Microbiology 25, 309–312. 10.1128/am.25.2.309-312.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambuza IM, Drake T, Chapuis A, Zhou X, Correia J, Taylor-Smith L, LeGrave N, Rasmussen T, Fisher MC, Bicanic T, et al. (2018). The Cryptococcus neoformans Titan cell is an inducible and regulated morphotype underlying pathogenesis. PLoS pathogens 14, e1006978–e1006978. 10.1371/journal.ppat.1006978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan JM, Kelly RM, Lee CK, and Levitz SM (2008). Role of the mannose receptor in a murine model of Cryptococcus neoformans infection. Infect Immun 76, 2362–2367. IAI.00095–08 [pii] 10.1128/IAI.00095-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RC, Cruz MC, Sia RA, Allen B, Alspaugh JA, and Heitman J (2000). Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fungal Genet Biol 29, 38–48. 10.1006/fgbi.1999.1180S1087-1845(99)91180-5 [pii]. [DOI] [PubMed] [Google Scholar]
- Davis KM (2018). All Yersinia Are Not Created Equal: Phenotypic Adaptation to Distinct Niches Within Mammalian Tissues. Front Cell Infect Microbiol 8, 261. 10.3389/fcimb.2018.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KM, Mohammadi S, and Isberg RR (2015). Community behavior and spatial regulation within a bacterial microcolony in deep tissue sites serves to protect against host attack. Cell Host Microbe 17, 21–31. 10.1016/j.chom.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitsch KW, del Pinal A, and Wellems TE (1999). Intra-cluster recombination and var transcription switches in the antigenic variation of Plasmodium falciparum. Mol Biochem Parasitol 101, 107–116. 10.1016/s0166-6851(99)00062-6. [DOI] [PubMed] [Google Scholar]
- Denham ST, and Brown JCS (2018). Mechanisms of Pulmonary Escape and Dissemination by Cryptococcus neoformans. J Fungi (Basel) 4, 25. 10.3390/jof4010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denham ST, Verma S, Reynolds RC, Worne CL, Daugherty JM, Lane TE, and Brown JCS (2018). Regulated release of cryptococcal polysaccharide drives virulence and suppresses immune infiltration into the central nervous system. Infect Immun 86. 10.1128/iai.00662-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond RD, and Bennett JE (1973). Growth of Cryptococcus neoformans Within Human Macrophages In Vitro. Infect Immun 7, 231–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosch M, Gerber J, Jebbawi F, and Beldi G (2018). Mechanisms of ATP Release by Inflammatory Cells. Int J Mol Sci 19. 10.3390/ijms19041222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen DP, Dean MM, O’Sullivan MV, Heatley S, and Minchinton RM (2008). Mannose-binding lectin deficiency does not appear to predispose to cryptococcosis in non-immunocompromised patients. Med Mycol 46, 371–375. 10.1080/13693780701874515. [DOI] [PubMed] [Google Scholar]
- Elliott MR, Chekeni FB, Trampont PC, Lazarowski ER, Kadl A, Walk SF, Park D, Woodson RI, Ostankovich M, Sharma P, et al. (2009). Nucleotides released by apoptotic cells act as a find-me signal to promote phagocytic clearance. Nature 461, 282–286. 10.1038/nature08296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons CW (1955). Saprophytic sources of Cryptococcus neoformans associated with the pigeon (Columba livia). Am J Hyg 62, 227–232. 10.1093/oxfordjournals.aje.a119775. [DOI] [PubMed] [Google Scholar]
- Ewels P, Magnusson M, Lundin S, and Kaller M (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W, Fa Z, and Liao W (2015). Epidemiology of Cryptococcus and cryptococcosis in China. Fungal Genet Biol 78, 7–15. 10.1016/j.fgb.2014.10.017. [DOI] [PubMed] [Google Scholar]
- Farhi F, Bulmer GS, and Tacker JR (1970). Cryptococcus neoformans IV. The Not-So-Encapsulated Yeast. Infect Immun 1, 526–531. 10.1128/IAI.1.6.526-531.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmesser M, Kress Y, and Casadevall A (2001). Dynamic changes in the morphology of Cryptococcus neoformans during murine pulmonary infection. Microbiology 147, 2355–2365. doi: 10.1099/00221287-147-8-2355. [DOI] [PubMed] [Google Scholar]
- Feldmesser M, Kress Y, Novikoff P, and Casadevall A (2000). Cryptococcus neoformans Is a facultative intracellular pathogen in murine pulmonary infection. Infect Immun 68, 4225–4237. 10.1128/iai.68.7.4225-4237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes KE, Brockway A, Haverkamp M, Cuomo CA, van Ogtrop F, Perfect JR, and Carter DA (2018). Phenotypic Variability Correlates with Clinical Outcome in Cryptococcus Isolates Obtained from Botswanan HIV/AIDS Patients. mBio 9. 10.1128/mBio.02016-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes KE, and Carter DA (2020). Cellular plasticity of pathogenic fungi during infection. PLoS Pathog 16, e1008571. 10.1371/journal.ppat.1008571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes KE, Dwyer C, Campbell LT, and Carter DA (2016). Species in the Cryptococcus gattii Complex Differ in Capsule and Cell Size following Growth under Capsule-Inducing Conditions. mSphere 1. 10.1128/mSphere.00350-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfoot AL, Shen Q, Wüthrich M, Klein BS, and Rappleye CA (2016). The Eng1 β-Glucanase Enhances Histoplasma Virulence by Reducing β-Glucan Exposure. mBio 7, e01388–01315. 10.1128/mBio.01388-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gault WJ, Enyedi B, and Niethammer P (2014). Osmotic surveillance mediates rapid wound closure through nucleotide release. J Cell Biol 207, 767–782. 10.1083/jcb.201408049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaylord EA, Choy HL, and Doering TL (2020). Dangerous Liaisons: Interactions of Cryptococcus neoformans with Host Phagocytes. Pathogens 9. 10.3390/pathogens9110891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein AC, Fu MS, Mukaremera L, Li Z, Ormerod KL, Fraser JA, Berman J, and Nielsen K (2015). Polyploid titan cells produce haploid and aneuploid progeny to promote stress adaptation. mBio 6, e01340. 10.1128/mBio.01340-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geunes-Boyer S, Beers MF, Perfect JR, Heitman J, and Wright JR (2012). Surfactant protein D facilitates Cryptococcus neoformans infection. Infect Immun 80, 2444–2453. 10.1128/IAI.05613-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson JF, Bojarczuk A, Evans RJ, Kamuyango AA, Hotham R, Lagendijk AK, Hogan BM, Ingham PW, Renshaw SA, and Johnston SA (2022). Blood vessel occlusion by Cryptococcus neoformans is a mechanism for haemorrhagic dissemination of infection. PLoS Pathog 18, e1010389. 10.1371/journal.ppat.1010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman DL, Khine H, Abadi J, Lindenberg DJ, Pirofski L, Niang R, and Casadevall A (2001). Serologic evidence for Cryptococcus neoformans infection in early childhood. Pediatrics 107, E66. 10.1542/peds.107.5.e66. [DOI] [PubMed] [Google Scholar]
- Grygorczyk R, Boudreault F, Ponomarchuk O, Tan JJ, Furuya K, Goldgewicht J, Kenfack FD, and Yu F (2021). Lytic Release of Cellular ATP: Physiological Relevance and Therapeutic Applications. Life (Basel) 11. 10.3390/life11070700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães AJ, de Cerqueira MD, and Nosanchuk JD (2011). Surface architecture of histoplasma capsulatum. Front Microbiol 2, 225. 10.3389/fmicb.2011.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas R, and Meyer TF (1986). The repertoire of silent pilus genes in Neisseria gonorrhoeae: evidence for gene conversion. Cell 44, 107–115. 10.1016/0092-8674(86)90489-7. [DOI] [PubMed] [Google Scholar]
- Harris G, Davies JW, and Parsons R (1958). Nucleotide pool of brewers’ yeast during growth and fermentation. Nature 182, 1565–1567. 10.1038/1821565a0. [DOI] [PubMed] [Google Scholar]
- Himmelreich U, Allen C, Dowd S, Malik R, Shehan BP, Mountford C, and Sorrell TC (2003). Identification of metabolites of importance in the pathogenesis of pulmonary cryptococcoma using nuclear magnetic resonance spectroscopy. Microbes and Infection 5, 285–290. 10.1016/S1286-4579(03)00028-5. [DOI] [PubMed] [Google Scholar]
- Hohl TM, Van Epps HL, Rivera A, Morgan LA, Chen PL, Feldmesser M, and Pamer EG (2005). Aspergillus fumigatus triggers inflammatory responses by stage-specific beta-glucan display. PLoS Pathog 1, e30. 10.1371/journal.ppat.0010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer CM, Summers DK, Goranov AI, Clarke SC, Wiesner DL, Diedrich JK, Moresco JJ, Toffaletti D, Upadhya R, Caradonna I, et al. (2016). Intracellular Action of a Secreted Peptide Required for Fungal Virulence. Cell Host Microbe 19, 849–864. 10.1016/j.chom.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hommel B, Mukaremera L, Cordero RJB, Coelho C, Desjardins CA, Sturny-Leclère A, Janbon G, Perfect JR, Fraser JA, Casadevall A, et al. (2018). Titan cells formation in Cryptococcus neoformans is finely tuned by environmental conditions and modulated by positive and negative genetic regulators. PLoS Pathog 14, e1006982. 10.1371/journal.ppat.1006982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SH, Long M, Wu CH, Kwon-Chung KJ, Chang YC, Chi F, Lee S, and Jong A (2011). Invasion of Cryptococcus neoformans into human brain microvascular endothelial cells is mediated through the lipid rafts-endocytic pathway via the dual specificity tyrosine phosphorylation-regulated kinase 3 (DYRK3). J Biol Chem 286, 34761–34769. 10.1074/jbc.M111.219378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs CW, Adams AE, Szaniszlo PJ, and Pringle JR (1988). Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J Cell Biol 107, 1409–1426. 10.1083/jcb.107.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanetsuna F, and Carbonell LM (1971). Cell wall composition of the yeastlike and mycelial forms of Blastomyces dermatitidis. J Bacteriol 106, 946–948. 10.1128/jb.106.3.946-948.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler JR, Acosta-Zaldivar M, and Qi W (2020). Phosphate in Virulence of Candida albicans and Candida glabrata. J Fungi (Basel) 6, 40. 10.3390/jof6020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski CH, Kerkaert JD, Liu KW, Bond MC, Hartmann R, Nadell CD, Stajich JE, and Cramer RA (2019). Fungal biofilm morphology impacts hypoxia fitness and disease progression. Nat Microbiol 4, 2430–2441. 10.1038/s41564-019-0558-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer M, Reiner E, Hu G, Tam N, Oliveira DL, Caza M, Yeon JH, Kim J, Kastrup CJ, Jung WH, and Kronstad JW (2014). Defects in phosphate acquisition and storage influence virulence of Cryptococcus neoformans. Infect Immun 82, 2697–2712. 10.1128/iai.01607-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubes P, and Jenne C (2018). Immune Responses in the Liver. Annu Rev Immunol 36, 247–277. 10.1146/annurev-immunol-051116-052415. [DOI] [PubMed] [Google Scholar]
- Lazera MS, Pires FD, Camillo-Coura L, Nishikawa MM, Bezerra CC, Trilles L, and Wanke B (1996). Natural habitat of Cryptococcus neoformans var. neoformans in decaying wood forming hollows in living trees. J Med Vet Mycol 34, 127–131. 10.1080/02681219680000191. [DOI] [PubMed] [Google Scholar]
- Lee H, Chang YC, Nardone G, and Kwon-Chung KJ (2007). TUP1 disruption in Cryptococcus neoformans uncovers a peptide-mediated density-dependent growth phenomenon that mimics quorum sensing. Mol Microbiol 64, 591–601. 10.1111/j.1365-2958.2007.05666.x. [DOI] [PubMed] [Google Scholar]
- Lee SC, Dickson DW, and Casadevall A (1996). Pathology of cryptococcal meningoencephalitis: analysis of 27 patients with pathogenetic implications. Hum Pathol 27, 839–847. 10.1016/s0046-8177(96)90459-1. [DOI] [PubMed] [Google Scholar]
- Lev S, and Djordjevic JT (2018). Why is a functional PHO pathway required by fungal pathogens to disseminate within a phosphate-rich host: A paradox explained by alkaline pH-simulated nutrient deprivation and expanded PHO pathway function. PLoS Pathog 14, e1007021. 10.1371/journal.ppat.1007021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev S, Kaufman-Francis K, Desmarini D, Juillard PG, Li C, Stifter SA, Feng CG, Sorrell TC, Grau GE, Bahn YS, and Djordjevic JT (2017). Pho4 Is Essential for Dissemination of Cryptococcus neoformans to the Host Brain by Promoting Phosphate Uptake and Growth at Alkaline pH. mSphere 2. 10.1128/mSphere.00381-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PZ, Li JZ, Li M, Gong JP, and He K (2014). An efficient method to isolate and culture mouse Kupffer cells. Immunol Lett 158, 52–56. 10.1016/j.imlet.2013.12.002. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Lim J, Coates CJ, Seoane PI, Garelnabi M, Taylor-Smith LM, Monteith P, Macleod CL, Escaron CJ, Brown GD, Hall RA, and May RC (2018). Characterizing the Mechanisms of Nonopsonic Uptake of Cryptococci by Macrophages. J Immunol 200, 3539–3546. 10.4049/jimmunol.1700790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TB, Kim JC, Wang Y, Toffaletti DL, Eugenin E, Perfect JR, Kim KJ, and Xue C (2013). Brain inositol is a novel stimulator for promoting Cryptococcus penetration of the blood-brain barrier. PLoS Pathog 9, e1003247. 10.1371/journal.ppat.1003247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TB, Subbian S, Pan W, Eugenin E, Xie J, and Xue C (2014). Cryptococcus inositol utilization modulates the host protective immune response during brain infection. Cell Communication and Signaling 12, 51. 10.1186/s12964-014-0051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Fuentes E, Gutiérrez-Escobedo G, Timmermans B, Van Dijck P, De Las Peñas A, and Castaño I (2018). Candida glabrata’s Genome Plasticity Confers a Unique Pattern of Expressed Cell Wall Proteins. J Fungi (Basel) 4. 10.3390/jof4020067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luberto C, Martinez-Marino B, Taraskiewicz D, Bolanos B, Chitano P, Toffaletti DL, Cox GM, Perfect JR, Hannun YA, Balish E, and Poeta MD (2003). Identification of App1 as a regulator of phagocytosis and virulence of Cryptococcus neoformans. J Clin Invest. 112, 1080–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson AJ, Heikenwalder M, and Ganal-Vonarburg SC (2016). The Liver at the Nexus of Host-Microbial Interactions. Cell Host Microbe 20, 561–571. 10.1016/j.chom.2016.10.016. [DOI] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet 17, 10–12. [Google Scholar]
- Maruvada R, Zhu L, Pearce D, Zheng Y, Perfect J, Kwon-Chung KJ, and Kim KS (2012). Cryptococcus neoformans phospholipase B1 activates host cell Rac1 for traversal across the blood-brain barrier. Cell Microbiol 14, 1544–1553. 10.1111/j.1462-5822.2012.01819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maziarz EK, and Perfect JR (2016). Cryptococcosis. Infect Dis Clin North Am 30, 179–206. 10.1016/j.idc.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaremera L, Lee KK, Mora-Montes HM, and Gow NAR (2017). Candida albicans Yeast, Pseudohyphal, and Hyphal Morphogenesis Differentially Affects Immune Recognition. Front Immunol 8, 629. 10.3389/fimmu.2017.00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaremera L, McDonald TR, Nielsen JN, Molenaar CJ, Akampurira A, Schutz C, Taseera K, Muzoora C, Meintjes G, Meya DB, et al. (2019). The Mouse Inhalation Model of Cryptococcus neoformans Infection Recapitulates Strain Virulence in Humans and Shows that Closely Related Strains Can Possess Differential Virulence. Infect Immun 87. 10.1128/iai.00046-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngamskulrungroj P, Chang Y, Sionov E, and Kwon-Chung KJ (2012). The primary target organ of Cryptococcus gattii is different from that of Cryptococcus neoformans in a murine model. mBio 3. 10.1128/mBio.00103-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara TR, and Alspaugh JA (2012). The Cryptococcus neoformans capsule: a sword and a shield. Clin Microbiol Rev 25, 387–408. 10.1128/CMR.00001-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okagaki LH, and Nielsen K (2012). Titan cells confer protection from phagocytosis in Cryptococcus neoformans infections. Eukaryot Cell 11, 820–826. 10.1128/EC.00121-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okagaki LH, Strain AK, Nielsen JN, Charlier C, Baltes NJ, Chrétien F, Heitman J, Dromer F, and Nielsen K (2010). Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog 6, e1000953–e1000953. 10.1371/journal.ppat.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okagaki LH, Wang Y, Ballou ER, O’Meara TR, Bahn Y-S, Alspaugh JA, Xue C, and Nielsen K (2011). Cryptococcal titan cell formation is regulated by G-protein signaling in response to multiple stimuli. Eukaryot Cell 10, 1306–1316. 10.1128/EC.05179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo Y, Wakayama M, Ohno H, Yamamoto S, Tochigi N, Tanabe K, Kaneko Y, Yamagoe S, Umeyama T, Shinozaki M, et al. (2013). Histopathological study of murine pulmonary cryptococcosis induced by Cryptococcus gattii and Cryptococcus neoformans. Jpn J Infect Dis 66, 216–221. 10.7883/yoken.66.216. [DOI] [PubMed] [Google Scholar]
- Olszewski MA, Noverr MC, Chen GH, Toews GB, Cox GM, Perfect JR, and Huffnagle GB (2004). Urease expression by Cryptococcus neoformans promotes microvascular sequestration, thereby enhancing central nervous system invasion. Am J Pathol 164, 1761–1771. 10.1016/s0002-9440(10)63734-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero XL, De La Peña-Lastra S, Pérez-Alberti A, Ferreira TO, and Huerta-Diaz MA (2018). Seabird colonies as important global drivers in the nitrogen and phosphorus cycles. Nat Commun 9, 246. 10.1038/s41467-017-02446-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande K, Chen C, and Noble SM (2013). Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat Genet 45, 1088–1091. 10.1038/ng.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panepinto JC, Komperda KW, Hacham M, Shin S, Liu X, and Williamson PR (2007). Binding of serum mannan binding lectin to a cell integrity-defective Cryptococcus neoformans ccr4Delta mutant. Infect Immun 75, 4769–4779. 10.1128/IAI.00536-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson AM, Rich A, and Krieger M (1993). Polynucleotide binding to macrophage scavenger receptors depends on the formation of base-quartet-stabilized four-stranded helices. J Biol Chem 268, 3546–3554. [PubMed] [Google Scholar]
- Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP, Chiller TM, Denning DW, Loyse A, and Boulware DR (2017). Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis. 10.1016/S1473-3099(17)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Teng Y, Tan B, Zhang X, Jiang W, Liu M, Jiang W, Du B, and Qian M (2014). Toll-like receptor-triggered calcium mobilization protects mice against bacterial infection through extracellular ATP release. Infect Immun 82, 5076–5085. 10.1128/iai.02546-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera J, Feldmesser M, Cammer M, and Casadevall A (1998). Organ-Dependent Variation of Capsule Thickness in Cryptococcus neoformans during Experimental Murine Infection. Infect Immun 66, 5027–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DJ, Craig AG, Berendt AR, Pinches R, Nash G, Marsh K, and Newbold CI (1992). Rapid switching to multiple antigenic and adhesive phenotypes in malaria. Nature 357, 689–692. 10.1038/357689a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabiiti W, May RC, and Pursall ER (2012). Experimental models of cryptococcosis. Int J Microbiol 2012, 626745. 10.1155/2012/626745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A, Nikolaidis NM, Amlal H, Uehara Y, Gardner JC, LaSance K, Pitstick LB, Bridges JP, Wikenheiser-Brokamp KA, McGraw DW, et al. (2015). Modeling pulmonary alveolar microlithiasis by epithelial deletion of the Npt2b sodium phosphate cotransporter reveals putative biomarkers and strategies for treatment. Sci Transl Med 7, 313ra181. 10.1126/scitranslmed.aac8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Juan BP, Garcia-Leon MJ, Rangel L, Goetz JG, and Chaffer CL (2019). The Complexities of Metastasis. Cancers (Basel) 11, 1575. 10.3390/cancers11101575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Tirado FH, Onken MD, Cooper JA, Klein RS, and Doering TL (2017). Trojan Horse Transit Contributes to Blood-Brain Barrier Crossing of a Eukaryotic Pathogen. mBio 8. 10.1128/mBio.02183-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki AM, Maruvka YE, Richmond PA, Guillet M, Shoresh N, Sorenson AL, De S, Kishony R, Michor F, Dowell R, and Pellman D (2015). Polyploidy can drive rapid adaptation in yeast. Nature 519, 349–352. 10.1038/nature14187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Li SS, Zheng C, Jones GJ, Kim KS, Zhou H, Kubes P, and Mody CH (2010). Real-time imaging of trapping and urease-dependent transmigration of Cryptococcus neoformans in mouse brain. J Clin Invest 120, 1683–1693. 10.1172/jci41963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JD, Chitnis CE, Craig AG, Roberts DJ, Hudson-Taylor DE, Peterson DS, Pinches R, Newbold CI, and Miller LH (1995). Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell 82, 101–110. 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich JE, Harris T, Brunk BP, Brestelli J, Fischer S, Harb OS, Kissinger JC, Li W, Nayak V, Pinney DF, et al. (2012). FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res 40, D675–681. 10.1093/nar/gkr918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stano P, Williams V, Villani M, Cymbalyuk ES, Qureshi A, Huang Y, Morace G, Luberto C, Tomlinson S, and Del Poeta M (2009). App1: an antiphagocytic protein that binds to complement receptors 3 and 2. J Immunol 182, 84–91. 10.4049/jimmunol.182.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Sun P, Li H, Zhang M, Liu G, Strickland AB, Chen Y, Fu Y, Xu J, Yosri M, et al. (2019). Fungal dissemination is limited by liver macrophage filtration of the blood. Nat Commun 10, 4566. 10.1038/s41467-019-12381-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Zhang M, Liu G, Wu H, Li C, Zhou H, Zhang X, and Shi M (2016). Intravascular clearance of disseminating Cryptococcus neoformans in the brain can be improved by enhancing neutrophil recruitment in mice. Eur J Immunol 46, 1704–1714. 10.1002/eji.201546239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson J, Bergström S, Robbins K, Barrera O, Corwin D, and Koomey JM (1986). Gene conversion involving the pilin structural gene correlates with pilus+ in equilibrium with pilus- changes in Neisseria gonorrhoeae. Cell 47, 267–276. 10.1016/0092-8674(86)90449-6. [DOI] [PubMed] [Google Scholar]
- Tao L, Du H, Guan G, Dai Y, Nobile CJ, Liang W, Cao C, Zhang Q, Zhong J, and Huang G (2014). Discovery of a “white-gray-opaque” tristable phenotypic switching system in candida albicans: roles of non-genetic diversity in host adaptation. PLoS Biol 12, e1001830. 10.1371/journal.pbio.1001830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd RT, Braverman AL, and Selmecki A (2018). Flow Cytometry Analysis of Fungal Ploidy. Curr Protoc Microbiol 50, e58–e58. 10.1002/cpmc.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevijano-Contador N, de Oliveira HC, García-Rodas R, Rossi SA, Llorente I, Zaballos Á, Janbon G, Ariño J, and Zaragoza Ó (2018). Cryptococcus neoformans can form titan-like cells in vitro in response to multiple signals. PLoS Pathog 14, e1007007. 10.1371/journal.ppat.1007007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Woude MW, and Bäumler AJ (2004). Phase and antigenic variation in bacteria. Clin Microbiol Rev 17, 581–611, table of contents. 10.1128/cmr.17.3.581-611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S (2014). Disconnecting a role for DNA repair in the pathogenicity of the human pathogen Cryptococcus neoformans. PhD (University of Missouri; - Kansas City: ). [Google Scholar]
- Viriyakosol S, Singhania A, Fierer J, Goldberg J, Kirkland TN, and Woelk CH (2013). Gene expression in human fungal pathogen Coccidioides immitis changes as arthroconidia differentiate into spherules and mature. BMC Microbiol 13, 121. 10.1186/1471-2180-13-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu K, Eigenheer RA, Phinney BS, and Gelli A (2013). Cryptococcus neoformans promotes its transmigration into the central nervous system by inducing molecular and cellular changes in brain endothelial cells. Infect Immun 81, 3139–3147. 10.1128/iai.00554-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh NM, Botts MR, McDermott AJ, Ortiz SC, Wüthrich M, Klein B, and Hull CM (2019). Infectious particle identity determines dissemination and disease outcome for the inhaled human fungal pathogen Cryptococcus. PLoS Pathog 15, e1007777. 10.1371/journal.ppat.1007777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhai B, and Lin X (2012). The link between morphotype transition and virulence in Cryptococcus neoformans. PLoS Pathog 8, e1002765. 10.1371/journal.ppat.1002765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward CP, Clottey GT, Dorris M, Ji DD, and Arnot DE (1999). Analysis of Plasmodium falciparum PfEMP-1/var genes suggests that recombination rearranges constrained sequences. Mol Biochem Parasitol 102, 167–177. 10.1016/s0166-6851(99)00106-1. [DOI] [PubMed] [Google Scholar]
- Weigel WA, and Dersch P (2018). Phenotypic heterogeneity: a bacterial virulence strategy. Microbes Infect 20, 570–577. 10.1016/j.micinf.2018.01.008. [DOI] [PubMed] [Google Scholar]
- Whiston E, Zhang Wise H, Sharpton TJ, Jui G, Cole GT, and Taylor JW (2012). Comparative transcriptomics of the saprobic and parasitic growth phases in Coccidioides spp. PLoS One 7, e41034. 10.1371/journal.pone.0041034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak KL, and Levitz SM (2009). Isolation and purification of antigenic components of Cryptococcus. Methods Mol Biol 470, 71–83. 10.1007/978-1-59745-204-5_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright L, Bubb W, Davidson J, Santangelo R, Krockenberger M, Himmelreich U, and Sorrell T (2002). Metabolites released by Cryptococcus neoformans var. neoformans and var. gattii differentially affect human neutrophil function. Microbes Infect 4, 1427–1438. 10.1016/s1286-4579(02)00024-2. [DOI] [PubMed] [Google Scholar]
- Xie S, Sao R, Braun A, and Bottone EJ (2012). Difference in Cryptococcus neoformans cellular and capsule size in sequential pulmonary and meningeal infection: a postmortem study. Diagnostic Microbiology and Infectious Disease 73, 49–52. 10.1016/j.diagmicrobio.2012.01.008. [DOI] [PubMed] [Google Scholar]
- Zaragoza O (2011). Multiple Disguises for the Same Party: The Concepts of Morphogenesis and Phenotypic Variations in Cryptococcus neoformans. Front Microbiol 2, 181. 10.3389/fmicb.2011.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, García-Rodas R, Nosanchuk JD, Cuenca-Estrella M, Rodríguez-Tudela JL, and Casadevall A (2010). Fungal cell gigantism during mammalian infection. PLoS Pathog 6, e1000945–e1000945. 10.1371/journal.ppat.1000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Sun D, Liu G, Wu H, Zhou H, and Shi M (2016). Real-time in vivo imaging reveals the ability of neutrophils to remove Cryptococcus neoformans directly from the brain vasculature. J Leukoc Biol 99, 467–473. 10.1189/jlb.4AB0715-281R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, and Ballou ER (2018). The Cryptococcus neoformans Titan Cell: From In Vivo Phenomenon to In Vitro Model. Curr Clin Microbiol Rep 5, 252–260. 10.1007/s40588-018-0107-9. [DOI] [Google Scholar]
- Zhou X, Zafar H, Sephton-Clark P, Mohamed SH, Chapuis A, Makarova M, MacCallum DM, Drummond RA, Dambuza IM, and Ballou ER (2020). Host environmental conditions induce small fungal cell size and alter population heterogeneity in Cryptococcus neoformans. bioRxiv, 2020.2001.2003.894709. 10.1101/2020.01.03.894709. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited at NCBI GEO database (Accession number: GSE152784) and are publicly available.
There is not original code associated with this paper.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


