Abstract
The global incidence of nonalcoholic fatty liver disease (NAFLD) is mounting incessantly, and it is emerging as the most frequent cause of chronic and end stage liver disorders. It is the starting point for a range of conditions from simple steatosis to more progressive nonalcoholic steatohepatitis (NASH) and associated hepatocellular carcinoma (HCC). Dysregulation of insulin secretion and dyslipidemia due to obesity and other lifestyle variables are the primary contributors to establishment of NAFLD. Onset and progression of NAFLD is orchestrated by an interplay of metabolic environment with genetic and epigenetic factors. An incompletely understood mechanism of NAFLD progression has greatly hampered the progress in identification of novel prognostic and therapeutic strategies. Emerging evidence suggests altered DNA methylation pattern as a key determinant of NAFLD pathogenesis. Environmental and lifestyle factors can manipulate DNA methylation patterns in a reversible manner, which manifests as changes in gene expression. In this review we attempt to highlight the importance of DNA methylation in establishment and progression of NAFLD. Development of novel diagnostic, prognostic and therapeutic strategies centered around DNA methylation signatures and modifiers has also been explored.
Keywords: Nonalcoholic fatty liver disease (NAFLD), Nonalcoholic steatohepatitis (NASH), Fibrosis, Epigenetics, DNA methylation, CpG islands, 5-methyl cytosine, Ten-eleven translocation (TET) enzymes, DNA methyl Transferase (DNMT), Diet
Nonalcoholic fatty liver disease (NAFLD); Nonalcoholic steatohepatitis (NASH); Fibrosis; Epigenetics; DNA methylation; CpG islands; 5-methyl cytosine; Ten-eleven translocation (TET) enzymes; DNA methyl transferase (DNMT); Diet.
1. Introduction
Nonalcoholic fatty liver disease (NAFLD) is increasing at an alarming rate and has become a major health concern worldwide. NAFLD is the leading cause of liver fibrosis, cirrhosis and hepatocellular carcinoma (HCC) [1]. NAFLD is defined as a clinical condition with the evidence of hepatic steatosis in more than 5% of hepatocytes in individuals with a history of very little or no alcohol consumption [2]. Histologically NAFLD is broadly categorized into nonalcoholic fatty liver (NAFL) and nonalcoholic steatohepatitis (NASH) based on its pathophysiology. NAFL exhibits a more benign course, having hepatic steatosis with no evidence of hepatocellular injury, whereas NASH is the more aggressive form displaying steatosis, hepatocyte inflammation and ballooning with or without fibrosis which might progress to cirrhosis, liver failure and mortality [3]. Metabolic derangements in NAFLD along with its heterogeneous pathogenesis have led to modification in its nomenclature by several experts to metabolic fatty liver disease (MAFLD) [4]. The diagnosis of MAFLD in patient population can be sub-grouped into obese-MAFLD, lean-MAFLD and diabetic-MAFLD having variable severity, prognosis, and diagnostic parameters. This sub-grouping of patients may help in defining the heterogeneity of NAFLD. Dietary habits, physical activity, genetic, epigenetic, and immunological factors contribute to NAFLD heterogeneity [5]. Identification of these additional factors that could modulate the clinical outcomes of NAFLD may help in understanding the disease heterogeneity and approaches to personalized medicine.
Globally, the occurrence of NAFLD is estimated to be 15–30% of the total population, however in the obese population the prevalence of NAFLD is 50–90% [1, 6]. The prevalence of NAFLD is increasing even in pediatric population with White and Asian children exhibiting a higher prevalence than African American children [7]. A recent mathematical model has projected the prevalence of NASH to rise by 56% by the year 2030 in various countries [8]. According to epidemiological data, 3–15% of obese patients having NASH develop cirrhosis and 4–27% of obese-NASH-cirrhosis patients progress to HCC. HCC has also been observed in patients suffering from NASH without cirrhosis [9]. It has been projected that NAFLD is poised to become the prime trigger for development of end stage liver diseases and HCC over the next decade [10].
Early diagnosis of NAFLD is imperative for this silently progressive disease which is mostly asymptomatic till its transition to NASH or other degenerative liver disorders. NAFLD diagnosis has traditionally been confirmed by liver biopsy which is sparingly used being invasive, expensive and not well accepted by the patients [11]. Various diagnostic tests and biomarkers based on combination of several indicators of steatosis exist that can be employed in clinical practice for early diagnosis and have been extensively reviewed [12]. Noninvasive or minimally invasive diagnostic methods include Visceral adiposity index (VAI), Fatty liver index (FLI), NAFLD liver fat score (NLFS), Triglyceride and Glucose Index (TyG), and Hepatic steatosis index (HSI). Fatty liver index which comprises body mass index (BMI), waist circumference, serum triglyceride, and gamma-glutamyltransferase levels is considered as the most practical, economical and precise in diagnosing NAFLD [12]. Imaging techniques like magnetic resonance imaging-Proton density fat fraction (MRI-PDFF), elastography, and controlled attenuation parameter (CAP) are emerging as definitive diagnostic criterion for assessing liver fat content [13].
Genetic predisposition as well as epigenetic modifications are of paramount significance in the development and pathogenesis of various diseases including NAFLD [14]. DNA methylation is a key epigenetic event that plays a pivotal role in regulation of transcription, embryonic development, genomic imprinting, genomic stability as well as chromatin structure [15, 16]. Abnormalities in the DNA methylation system has been implicated in the pathogenesis of various human malignancies [16, 17]. Aberrant DNA methylation patterns affect cellular homoeostasis with hypermethylation and hypomethylation of global genomic DNA resulting in altered gene expression [18, 19]. DNA methylation patterns appear to be reversible and have been demonstrated to be modulated by various environmental and lifestyle stimuli in a number of studies [20]. Interplay of genetic and epigenetic factors, with obesogenic exposures, sedentary lifestyle and high calorie/carbohydrate diet seem to be operational in fatty liver diseases [21]. In this review we highlight recent studies on aberrant DNA methylation profiles found in NAFLD and NASH patients and their alteration by lifestyle determinants. Vast clinical spectrum and diverse outcomes associated with NAFLD warrant an in-depth study of its pathogenesis and epigenetic aspects for better management strategies and development of novel therapeutic approaches.
For this review, DNA methylation studies dealing with the establishment and progression of NAFLD to end stage liver disorders were searched in the PUBMED and NLM database during July 2021–May 2022. The keywords used were DNA methylation and NAFLD; DNA methylation and NASH; DNA methylation and HCC; Epigenetics and NAFLD; Epigenetics and NASH; NAFLD and Sex; NAFLD diagnosis and management; NAFLD Progression; NAFLD/NASH and Epidemiology; Epigenetics and Liver disease; DNA methylation and obesity; DNA methylation patterns and epigenetic memory; Exercise and DNA methylation; NAFLD/NASH and DNMT; TET enzymes and NAFLD/NASH; Homocysteine and NAFLD/NASH; Dietary methyl deficiency and liver carcinogenesis; Nutrition epigenetic and liver diseases. A wide range of reports were found in the database from which initially abstracts were screened after which relevant studies, review articles were selected and consulted for this review. Both animal and human studies were included in the review. Major findings of these studies have been discussed in the following sections.
2. Pathogenesis and risk factors of NAFLD
NAFLD is a multifactorial disease exhibiting complex and incompletely understood pathophysiology. The progression of NAFLD was initially explained using a “two hit hypothesis”, where the “first hit” is lipid accumulation in hepatocytes enhancing the vulnerability of the liver to “second hit” including a variety of damaging factors such as oxidative stress, genetic polymorphisms, epigenetic modifications, apoptosis, inflammatory pathways, dysregulated adipokine secretion, and stimulation of hepatic stellate cells [22]. With time, it has evolved into “parallel multiple hit theory” of NAFLD progression [23]. Insulin resistance acts as the “first hit” resulting in augmented uptake and synthesis of free fatty acids leading to ectopic fat deposition and steatosis in hepatocytes. Varied parallel processes contribute to the development of steatosis and liver inflammation including adipokine deregulation, endoplasmic reticulum stress, oxidative stress, mitochondrial dysfunction, inflammatory cascades, unfolded protein response, lipotoxicity, altered gut microbiota, chemokines, cytokines and innate immunity [24].
Dyslipidemia, insulin resistance (IR), metabolic syndrome, obesity and type-2 diabetes mellitus (T2DM) are noteworthy risk factors for NAFLD [25]. IR is the most familiar irregularity associated with NAFLD and a useful predictor of its severity and progression [26, 27]. IR leads to NAFLD either through increased de novo lipogenesis (DNL) or by increased free fatty acid (FFA) transport to the liver from adipose tissue [26]. Also, hyperinsulinemia secondary to IR leads to increased expression and activity of the sterol response element binding protein 1c (SREBP-1c), a transcription factor, which stimulates the expression of all enzymes essential for DNL [26, 28]. Therefore, the gravity of IR is strongly linked to liver steatosis and the prevalence of NAFLD among individuals having hyperglycemia and T2DM [29, 30]). Obesity is known to cause excessive levels of circulating FFA, augmenting the risk of fatty liver and development of IR. Notable association of NAFLD with overall fat mass, waist circumference and abdominal adiposity has been well documented [31]. The endocrine function of adipose tissue specifically visceral fat has been implicated in producing multiple pro-inflammatory cytokines mediating the clinical manifestations of NAFLD and its progression [32]. Most observational studies suggest a bi-directional correlation of NAFLD with metabolic syndrome [33]. NAFLD seems to display sexual dimorphism with higher prevalence in males, although in females this protection seems to wane after menopause [34, 35, 36]. NAFLD has been found to be widespread in non-obese asymptomatic population as well, further confounding its diagnosis and management [37].
Hypernutrition and high fat diet (HFD) can greatly increase the circulating levels of FFAs which get deposited in adipose tissue and liver leading to low grade chronic inflammation and IR [38]. The contributors of fats that lead to hepatic steatosis include lipolysis from peripheral adipose tissue adding to the unesterified pool, DNL and dietary fatty acids. According to a study by Donnelly et al, most of the fat (nearly 60%) arises from the non-esterified fatty acids pool in the blood (the source of this being adipose tissue); a quarter from de novo lipogenesis; and only a small fraction (approximately 15%) from the diet [39].
NAFLD represents a heterogeneous patient population where some individuals exhibit steatosis alone, while others develop hepatocyte damage and advanced fibrosis. This heterogeneous pathophysiology of NAFLD could be explained by genetic and epigenetic variants, sex, and comorbidities such as obesity, T2DM and many others that critically influence disease progression, prognosis, and differential response to treatments. Genome wide association studies reveal a connection between specific genetic variants and NAFLD onset and severity. Some of the key genetic variants associated with the NAFLD/NASH risk are TM6SF2, PNPLA3, MBOAT7, GCKR, APOB, lysophospholipase-like protein 1 (LYPLAL1), neurocan (NCAN), HSD17B1 and protein phosphatase-1 regulatory subunit-3B (PPP1R3B) [40, 41, 42]. These genetic variants do not account for all NAFLD individuals. Polygenic risk scores (PRS) is being explored as a helpful tool to recognize patients with high risk of developing NAFLD and its associated complications [43, 44]. The association of genetic variants with NAFLD development can be used to predict the disease trajectories which might aid diagnosis and better prognostication [45]. A recent study by Bianco et al., using a PRS system involving MBOAT7, TM6SF2, GCKR and PNPLA3 might enhance HCC risk gradation in NAFLD [44]. Genetic risk scores using genetic variants PNPLA3, TM6SF2, and HSD17B13 was also shown to increase the risk of cirrhosis by 12 folds in patients with NAFLD [46]. Variants of PNPLA3 have been shown to correspond with NAFLD prevalence across ethnicities [47]. Mutations in PNPLA3, which encodes for triacylglycerol lipase correlated with the presence of NAFLD and NASH [48].
Different genetic variants of NAFLD were found to be associated with specific comorbidities. SREBF-2, TM6SF2, and transcription factor-7-like 2 (TCFL2) are predominantly present in individuals having increased risk of T2DM whereas adiponectin gene (ADIPOQ) and Src homology (SH2B1) are associated with obesity [49, 50, 51]. These genes are also linked with the dysregulated glucose and lipid homeostasis, IR, NAFLD, obesity and T2DM indicating greatly intertwined, shared mechanisms of disease pathogenesis [52, 53]. Therefore, these genetic variants are in agreement with the view that metabolic dysfunction is the prime driver of NAFLD. Interestingly some of the genetic variants associated with increased risk of NAFLD were associated with decreased risk of certain other metabolic diseases, for instance, GCRK P446L variant was reported to reduce the risk of T2DM [54]. The proposed mechanism for this observation was induction of DNL leading to decreased triglycerides levels, concomitant with improved hepatic glucose metabolism [55]. Similarly, PNPLA3I148M, and TM6SF2 variants were found to have inverse relationship with metabolic disorders such as T2DM and cardiovascular diseases [54, 56]. Thus, the genetics of fatty liver disease with metabolic variables is complex and warrants further studies. Pathogenesis of NAFLD might be cumulative of both metabolic as well as a genetic components which are fundamentally different [57].
Apart from the variants of a gene, its epigenetic modifications under different environmental conditions results in altered cell physiology. Epigenetics deals with reversible alterations in expression of gene and associated phenotype that are not due to the alteration of DNA sequence. Association of epigenetic dysregulation with the onset and progression of NAFLD is being extensively investigated [58]. Available literature has shown the association of circulating PPARγ methylation level with fibrosis in NAFLD patients and deacetylation of SIRT1was observed to improve hepatic steatosis [58, 59]. Therefore, targeting the epigenetic factors may be one of the mechanisms for drug action to attenuate the onset of NAFLD, for example resveratrol and glucagon like peptide-1 receptor agonists (GLP-1RAs), as activators for SIRT1 alleviate the diet induced fatty liver in animal models [60, 61, 62]. Besides, vorinostat, an inhibitor of Histone deacetylase (HDAC) that inhibits the lipid synthesis in liver cells in vitro, can be a candidate drug in the treatment of NAFLD [63]. Epigenetic modifiers are being investigated as potential drug targets for NAFLD. Over the past two decades epigenetic alterations leading to NAFLD have been investigated and are being recognized as major determinants in its pathogenesis [15, 64]. Recently Lonardo et al., have deliberated on the approaches targeting the genetic background, epigenetic modifiers and clinical features to achieve the goal of precision medicine in NAFLD patients [65]. Therefore, genetic markers and their epigenetic modification with clinical information would help in precise NAFLD classification and its management.
3. DNA methylation in NAFLD
Epigenetics is an indispensable biological process dealing with alteration in expression of gene or modification of cellular pathways by adaptive mechanism that are not associated with alteration in DNA sequence [66]. It encompasses many factors such as DNA methylation, acetylation, phosphorylation, copy number alterations, miRNA and chromatin modifications. Changes in gene expression due to aberrant epigenetic alterations leads to the development of various diseases [67]. DNA methylation is one of the prime epigenetic mechanisms that can regulate gene expression. During this modification methyl group is added to the fifth carbon of cytosine resulting in 5-methylcytosine (5mC) at CpG islands of DNA [68, 69]. CpG islands found in the regulatory sites of gene are mostly unmethylated whereas, those present in non-regulatory regions are typically methylated [70, 71, 72, 73]. It has been found that approximately 1% of DNA bases are methylated cytosines representing 70–80% of CpG dinucleotides [69].
DNA methylation is critically involved in gene expression with 5mC as an established and inherited epigenetic feature which typically suppresses transcription. Mechanism operating in the inhibition of gene expression by cytosine methylation is either the prevention of the interaction of transcription factors or through recruitment of DNA binding domains having affinity for 5mC for example, 5-methyl-CpG binding domain 1-4, methyl CpG binding protein 2 and Kaiso initiate the formation of compact structure by interacting with histone-modifying enzymes, thereby affecting the cell/tissue specific gene expression [74, 75, 76]. Also, cytosine methylation can affect the chromatin structure and participate in X chromosome inactivation, genome imprinting as well as cellular differentiation [77].
The process of DNA methylation is mainly carried out by a set of enzymes collectively called DNA methyl transferases (DNMTs). During their catalytic reactions they transfer the methyl group to 5mC from S-adenosyl L-methionine (SAM) to cytosine [78]. Among the different DNMTs present in humans, DNMT1, DNMT3A and DNMT3B are implicated in methyltransferase activities [73]. DNMT1 copies CpG methylation patterns and is vital for carrying forward such methylated patterns to the newly synthesized daughter strand during DNA replication. DNMT3A and DNMT3B are de novo methyltransferases responsible for installing DNA methylation patterns during gametogenesis and early embryogenesis [79]. For the stable maintenance of methylation status DNMT3A and DNMT3B conjoin with DNMT1.
Another base modification in DNA namely, 5-hydroxymethylation of cytosine (55 hmC) has added new layers to the understanding of epigenetic mechanisms in development of diseases. It influences both activation as well as repression of gene transcription by influencing DNA demethylation [80]. 55 hmC is enriched in expressed genes and enhancer sequences in DNA. 55 hmC exhibits tissue specific expression and depending on the cellular conditions it undergoes epigenetic modification in accordance with the environmental and metabolic state. 55 hmC is abundant in human hepatic genome especially in genes actively involved in energy metabolism [81]. Formation of 55 hmC in DNA occurs through the oxidation of 5mC by a group of enzymes known as Ten-Eleven translocation (TET) enzymes. These iron-dependent enzymes have a dioxygenase activity. They act as 5mC hydroxylases using alpha-ketoglutarate as their co-substrate. TETs have the ability to reverse locus-specific DNA methylation. These 5mC oxidation products then mediate the transformation of 5mC to unmodified Cs [82]. TETs act as regulators of epigenetic processes as generation of 55 hmC is critical for DNA demethylation [83]. Three TET enzymes have been identified in mammals so far, namely, TET1, TET2 and TET3 [82].
The mechanisms operating in methylation and demethylation of DNA, influenced by myriads of known and unknown factors clearly point toward a very complex and dynamic landscape of DNA methylation. DNA methylation is implicated in numerous cellular processes such as somatic, germline and embryonic development, repression and expression of genes, transposition and pathogenesis of diverse disorders. With the rapid pace of technical advancement and newer genetic manipulation tools, the future of mammalian DNA methylation and demethylation research holds great promise in deciphering novel mechanisms and therapeutic tools for various human diseases [84].
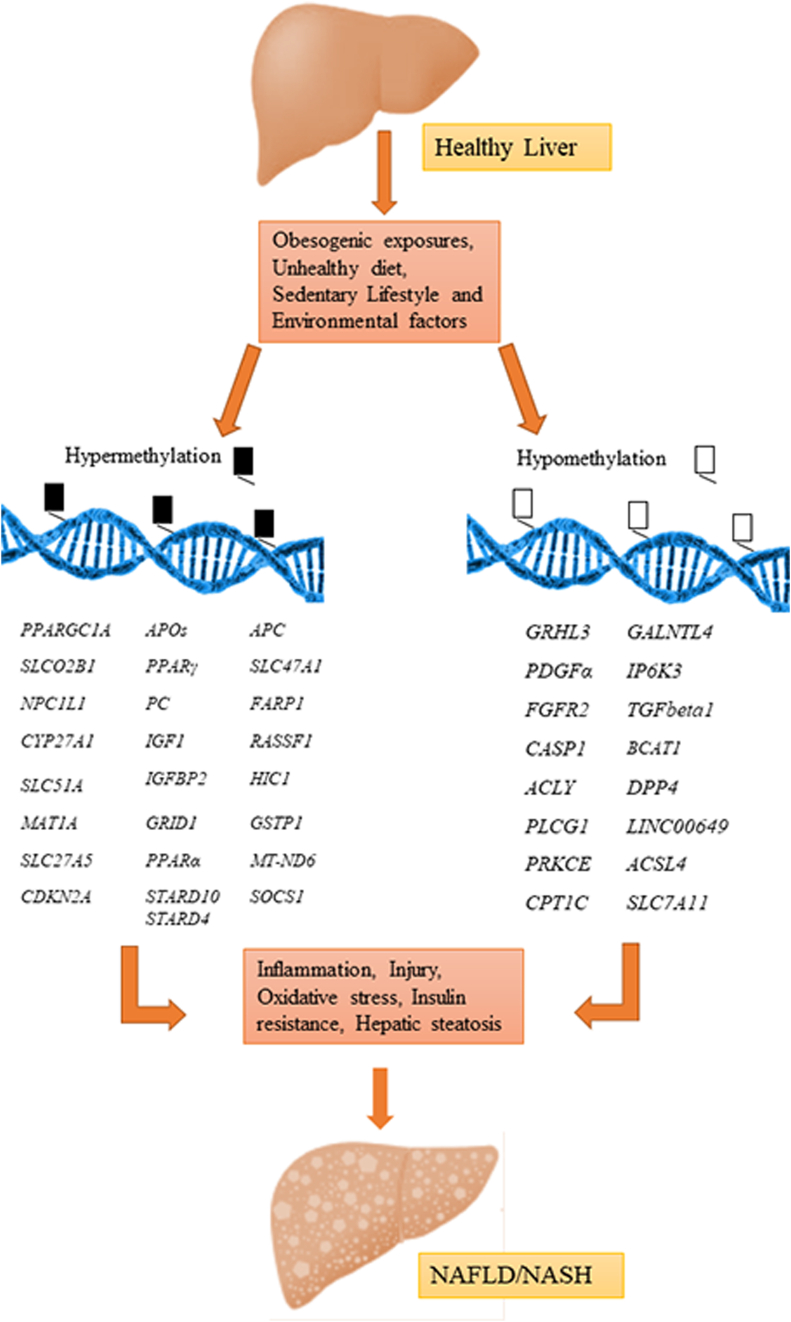
Altered methylation patterns of hepatic DNA for various genes has been depicted in Figure 1 [85]. Understanding the aberrations in DNA methylation system which might account for some principal features of NAFLD pathobiology will greatly augment our knowledge of the DNA methylation landscape in disease process. This has been dealt in the following sections.
Figure 1.
Aberrant DNA methylation in NAFLD: An illustration of the genes modified by epigenetic regulation due to their hypo-and hypermethylation in context of development and progression of NAFLD/NASH. Transforming growth factor beta1 (TGFβ1), platelet derived growth factor-α (PDGFα), peroxisome proliferator activated receptor-α (PPARα), peroxisome proliferator activated receptor-δ (PPARδ), mitochondrial encoded gene NADH dehydrogenase-6 (MT-ND6), PPARγ coactivator 1a (PPARGC1A), hepatic dipeptidyl peptidase 4 (DPP4), pyruvate carboxylase (PC), ATP citrate lyase (ACLY), phospholipase C gamma 1 (PLCG1), insulin like growth factor 1 (IGF1), Insulin like growth factor binding protein-2 (IGFBP2), protein kinase C epsilon (PRKCE), polypeptide N acetylgalactosamninyltransferase like 4 (GALNTL4), glutamate ionotropic receptor delta type subunit 1 (GRID1) and inositol hexakisphosphate kinase 3 (IP6K3), Fibroblast growth factor 2 (FGFR2), caspase 1 (CASP1), methionine adenosyl methyltransferase (MAT1A), Acyl-CoA synthetase long chain family member 4 (ACSL4), carnitine palmitoyltransferase 1 C (CPT1C), ARH/RhoGEF, pleckstrin domain protein1 (FARP1) Apolipoprotein (APOs), Niemann-Pick C1-Like 1 (NPCC1L1), STAR related lipid transfer domain 10 (STARD10), STAR related lipid transfer domain 4 (STARD4), Grainy head like transcription factor-3 (GRHL3), Cyclin dependent kinase inhibitor 2A (CDKN2A), Cytochrome P450 family 27 subfamily A member 1 (CYP27A1), Solute Carrier family 51 subunit alpha (SLC51A), Solute Carrier family 27 member 5 (SLC27A5), Solute carrier organic anion transporter family member 2B1 (SLCOB1), Suppressor of cytokine signaling 1 (SOCS1), Ras association domain family member 1 (RASSF1), Hypermethylated in cancer (HIC1), Glutathione S-transferase pi1 (GSTP1), Solute Carrier family 47 member 1 (SLC47A1), Solute Carrier family 7 member 11 (SLC7A11), Branched chain amino acid transaminase-1 (BCAT-1), Long intergenic nonprotein coding RNA 649 (LINC000649), Adenomatous polyposis coli (APC).
3.1. Establishment and progression of NAFLD to NASH
Liver is the central organ participating in various metabolic pathways of carbohydrates, lipids, and proteins metabolism and is also involved in detoxification reactions [32]. Liver fat content is regulated by a dedicated set of enzymes and proteins involved in transport of lipids, lipogenesis and their oxidation inside the liver as well as their secretion from liver to extra hepatic tissues. Dysregulation in any of these processes can manifest in liver steatosis and NAFLD development [86]. As mentioned earlier, NAFLD can be broadly categorized into NAFL exhibiting simple benign form of steatosis and NASH representing the more advanced form resulting in progressive fibrosis, cirrhosis and mortality [3]. Epigenetic mechanisms have been implicated in these changes in the liver physiology. The well-studied epigenetic modulation being DNA methylation and demethylation. Methyl donor for these methylation reactions comes from different pathways and methionine metabolism is prime among them, where SAM is sequentially demethylated to S-adenosylhomocysteine and homocysteine [87]. Elevated homocysteine levels have been observed in NAFLD patients compared to healthy controls suggesting an impaired methionine metabolism leading to decreased concentration of methyl donors such as methionine, choline and betaine in liver [88]. This could be a manifestation of hepatic injury, inflammation, oxidative stress and unfolded protein response owing to hepatic fat accumulation [88]. Elevated hepatic homocysteine levels with methionine deficiency, resulting from impaired homocysteine remethylation and aberrant methyltransferase reactions has been observed in diet induced NAFLD in mice. Further, decreased expression of DNMT3A gene was noticed in these mice although global pattern of methylation of DNA was unaltered [89]. Lai et al., observed significantly lower levels of global DNA methylation in liver biopsies along with increased serum homocysteine levels in NAFLD patients compared to overweight controls. Furthermore, lower hepatic global DNA methylation levels and hyper-homocystenemia was associated with increased hepatic inflammation grade, disease progression and histological severity of NAFLD [90]. This clearly points towards elevated levels of homocysteine to be an effect of aberrant DNA methylation rather than being a cause of metabolic abnormalities.
In a study of liver mitochondrial DNA methylation by Pirola et al., significantly higher methylation of mitochondrial gene NADH dehydrogenase-6 (MT-ND6) and lower mRNA levels was reported in NASH patients in comparison to simple steatosis patients. This points towards an association of hepatic methylation and transcriptional downregulation of MT-ND-6 with the severity of NAFLD [91]. It was also observed that epigenetic alterations of mtDNA were potentially reversible using intervention programs like physical activity as status of hepatic MT-ND6 methylation in NASH group inversely correlated with regular physical activity [91]. Another observational study by Pirola et al., suggested that 5-hmC level was associated with development of NAFLD via dysregulation of the expression of PPARγ coactivator 1a (PGC-1α) [92]. Also, while exploring the levels of PPARGC1A, an association was observed with a reduction in nonnuclear 5-hmC in NAFLD patients whereas liver PPARGC1A mRNA levels were negatively associated with global 5-hmC levels. This suggested that its methylation status modulates its transcriptional activity [92]. PPARGC1A encoded PGC-1α acts as a transcriptional coactivator regulating genes involved in cellular respiration, energy homeostasis, mitochondrial biogenesis, adaptive thermogenesis, adipocyte development, and lipid and glucose metabolism. Environmental signals have a great impact on its activity and synchronize the genes involved in metabolism by interacting with certain transcription factors and chromatin remodelers [93]. TET variants namely, p. Ile1123Met missense (TET1-rs3998860) and p. Ile1762Val substitution (TET2-rs2454206) were appreciably correlated with serum content of CK-18 fragment (cell death biomarker) and liver PPARGC1A methylation as well as transcript level of the same. These genetic variants of TET demonstrate the involvement of epigenetic modification in regulation of hepatocyte apoptosis and TET induced regulation of transcription of PPARGC1A in response to cellular metabolic milieu in NAFLD patients [92]. In another study, insulin resistance and liver transcriptional activity of PPARGC1A displayed a close interaction in NAFLD where liver PPARGC1A promoter methylation directly correlated with plasma fasting insulin levels and inversely correlated with liver mitochondrial DNA methylation level [94].
The advent of Genome wide DNA methylation profile analysis has facilitated screening large sample sizes from various human tissues [95]. Ahrens et al., using genome wide DNA methylation analysis from liver samples of 45 morbidly obese patients in different stages of NAFLD, extracted 9 NAFLD associated genes exhibiting variation in methylation. These genes are primarily involved in intermediary metabolism and insulin signaling including pyruvate carboxylase (PC), insulin like growth factor-1 (IGF1), ATP citrate lyase (ACLY), glutamateionotropic receptor delta type subunit-1 (GRID1), phospholipase C gamma-1 (PLCG1), Insulin like growth factor binding protein-2 (IGFBP2), inositol hexakisphosphate kinase-3 (IP6K3), polypeptide N-acetylgalactosamninyltransferase like-4 (GALNTL4), and protein kinase C epsilon (PRKCE) [96]. In a study of gene cohorts, methylation drifts in forty one genes involved in lipid metabolism and fourteen genes in vitamin D and energy balance were reported in NAFLD including novel genes namely, members of APO family (lipid transport), NPC1L1, STARD10, STARD4 (cholesterol transport) and GRHL3 (energy balance) [97]. In a related study using systematic analysis of drug metabolism and bile acid homeostasis genes, it was shown that 64 genes were differentially methylated in NAFLD. Among those, altered methylation was significantly correlated with transcription of 26 genes including CYP27A1, OSTα, SLC 27A5, SLC02B1, SLC47A1 and several UGT and CYP genes [98]. In another study, levels of hepatic dipeptidyl peptidase 4 (DPP4), a hepatokine secreted by hepatocytes, was also found to be elevated in NAFLD and NASH patients [99]. In a diet induced obesity model of young mice higher DPP4 expression and decreased methylation of 4 intronic CpG sites of DPP4 gene was observed which amplified the glucose induced transcription of DPP4. Also, a relationship of increased hepatic DPP4 expression levels and corresponding reduction in DNA methylation was observed in human liver biopsies to different hepatosteatosis and NASH stages which suggested a very complex intertwined regulation of fuel metabolism through epigenetic modulation [100].
Assessment of DNA methylation of peripheral blood mononuclear cells (PBMC) is emerging as a potential biomarker for the diagnosis of NAFLD. Using genome-wide DNA methylation investigation of PBMC DNA in NAFLD patients, promoter hypomethylation of SEC14 like lipid binding 3 (SEC14L3) and PRKCE were reported [101]. Another study published a panel of 863 CpG sites showed varied levels of methylation, with global hypomethylation predominating in plasma circulating cell free DNA (ccDNA) of NAFLD patients. Hypomethylation of Acyl-CoA synthetase long chain family member 4 (ACSL4) and carnitinepalmitoyltransferase 1 C (CPT1C) were linked to a heightened risk of NAFLD and might serve as biomarkers for NAFLD. Further, leukocytichypomethylatedACSL4 might be a useful biomarker for pathological traits of NAFLD [102]. In an exploration by Wu et al., using epigenome wide association studies (EWAS) six differentially methylated CpG sites in ACSL4, CRLS1, CTP1A, SIGIRR, SSBP1 and ZNF622 were identified which could serve as serum biomarkers to distinguish between those with NASH and simple steatosis [103]. Evaluation of peripheral blood DNA identified 22 CpG sites with differential DNA methylation which correlated well with hepatic fat accumulation. The risk of NAFLD and T2DM was also found to be correlated with decreased methylation of a CpG island in the long intergenic nonprotein coding RNA (LINC00649) [104]. Nano et al., using EWAS identified eight genes associated with markers of liver function, among which altered DNA methylation at SLC7A11 was linked with reduced incidence of hepatic steatosis, through favorable association with a panel of genes involved in lipid metabolism [105].
3.2. Sexual dimorphism in NAFLD and related liver disorders
The liver accounts for highest level of sexual dimorphism, with male and female livers varying in the expression of over 72% of the liver genes [106]. As mentioned earlier, sexual dimorphism exists in susceptibility, progression and outcomes of NAFLD and other hepatic diseases [36, 107, 108, 109]. Differential gene expression based on sex might partly account for this difference in susceptibility and progression of NAFLD. Wegermann et al., identified genetic loci belonging to energy metabolism affecting NAFLD related fibrosis to be modified by sex and menopause [110]. The role of estrogen, androgen, growth hormone and their receptors in influencing the pathophysiology and sex-dependent incidence of NAFLD has been extensively reviewed by Torre [111]. One of the mechanisms by which growth hormone mediates its sexual differentiating actions is by modulating DNA methylation in a sex specific manner [112]. In a study on rats by Strakovsky, et al., sex specific variations were observed in liver gene expression and epigenome at birth. In this report developmental bisphenol A (BPA) exposure aggravated high fat (HF) diet induced adult hepatic steatosis [113]. It was demonstrated that Cpt1a mRNA was downregulated in males but not in female offspring at birth upon prenatal BPA exposure and was connected to hepatic steatosis susceptibility in adulthood. This was potentially through aberrant methylation and histone modifications of Cpt1a, which encodes a crucial regulatory enzyme for β-oxidation in mitochondria. Besides Cpt1a, several other differentially methylated regions were observed in males but not in female rats [113]. Zhou et al., demonstrated sex linked protective capability of modest doses of aspirin on NAFLD in female but not in male mouse having increased NAFLD risk [114]. One of the mechanisms of observed Aspirin effects was the inhibition of Wnt signaling via hypo-methylation of CpG islands upstream of start codon of APC leading to its higher expression. Due to its interactions with β-catenin and Axin, the APC protein is known to block Wnt signaling thus forming APC/Axin/β-catenin destruction complex thereby displaying its anti-proliferative effect. This effect was not observed in male mice, suggesting that NAFLD development and its epigenetic regulation varies with sex [114]. Since most studies have used only male animals, it’s difficult to assess the extent of the sex-based differences in epigenetic alterations, warranting further investigation.
3.3. Progression from NASH to fibrosis and cirrhosis
Heightened inflammation in NASH can give rise to progressive fibrosis which can result in cirrhosis. 152 differentially methylated CpG islands for transcription factors and developmental pathways were observed between NASH subjects and controls. Subjects with NASH also exhibited epigenetic age progression correlating with hepatic collagen content and extent of fibrosis [115]. Hence, DNA methyl signature is being suggested as a measure of chronological age as well as biological age [116].
Regulation of specialized enzymes including DNMTs and TETs, that catalyze DNA methylation system in a series of chemical reactions have the potential to reprogram genome-wide methylation patterns [117]. Any changes in their activity or expression can greatly influence global as well as gene specific methylation of DNA. In a study by Page et al., global alterations in 5mC, 55 hmC and reduced expression of TETs with overall increased levels of DNMT3A, DNMT3B and DNMT1 were observed in animal and human fibrotic livers as well as in activated hepatic stellate cells. It was observed that a directed knock down of DNMT3A expression led to decreased pro-fibrotic phenotype in activated hepatic stellate cells [118]. In a targeted DNA methylation study of CpGs of five genes implicated in modulating fibrogenesis, hypomethylation of pro-fibrogenic genes including transforming growth factor beta1 (TGFβ1) and platelet derived growth factor-α (PDGFα); while hypermethylation of anti-fibrogenic genes including peroxisome proliferator activated receptor-α, (PPARα) and peroxisome proliferator activated receptor-δ (PPARδ) was reported in advanced NAFLD patients compared to mild NAFLD patients. Thus at specific CpGs, methylation status of DNA might serve to predict severity and progression of NAFLD to liver fibrosis [119]. Decreased methylation of CpG26 in Parvin beta variant1 (PARVB variant1) and increased methylation of CpG99 in patatin like phospho lipase domain containing protein 3 (PNPLA3) was observed in livers of patients with severe NAFLD (stages 3 and 4) as compared with those having mild NAFLD (stages 1 and 2) and were shown to be correlated with severity of NAFLD [120]. In a study by Murphy et al., 69,247 differentially methylated CpG sites (76% hypomethylated, 24% hypermethylated) were reported in advanced NAFLD patients as compared to mild patients. It was found that 7% of these differentially methylated CpG sites correlated with transcript levels establishing a relationship between methylome and transcriptome of NAFLD patients. Various genes were found to be hypomethylated and over expressed including Fibroblast growth factor 2 (FGFR2), involved in epithelial mesenchymal transition and caspase 1 (CASP1) known to be implicated in inflammation and liver fibrosis. Also, genes involved in 1-carbon metabolism including methionine adenosylmethyltransferase (MAT1A) which is involved in synthesis of SAM, showed reduced expression due to increased methylation in advanced NAFLD compared to mild disease (based on histological severity of fibrosis) [121]. In a study comparing NAFLD patients with or without severe fibrosis using EWAS aberrant methylation of seven fibrosis related CpGs was reported. Cell type proportions of liver cells was well correlated with the development of fibrosis where natural killer cells were augmented whereas epithelial cell number dropped with increasing severity of fibrosis [122]. Hardy et al., reported promoter hypermethylation of PPARγ using plasma circulating cell free DNA (ccDNA) from NAFLD patients exhibiting severe form of fibrosis in comparison to NAFLD subjects having mild fibrosis [58].
Overexpression of branched chain amino-acid transaminase 1 (BCAT1) which converts alpha-ketoglutarate to glutamate plays vital role in citric acid cycleand oxidative phosphorylation has been linked to the occurrence and severity of NAFLD and NASH [123]. BCAT1was found to be over expressed and hypomethylated in NAFLD patients experiencing adverse clinical outcomes [124]. Thus, it is evident that DNA methylation patterns could be used for detecting and predicting fibrosis in NAFLD patients.
3.4. The hepatocellular carcinoma (HCC) risk
The liver exhibits a unique characteristic of regeneration which concurrently makes it vulnerable to tumorogenesis. It has been observed that enduring hepatic disorders, damage or fatty infiltration frequently leads to liver cancer [125]. NAFLD along with NASH are well recognized hepatic pathologies which significantly increase the risk for development of HCC [1, 126, 127]. Distinctive modulations in gene expression due to anomalous DNA methylation patterns of some genes might promote the progression of NAFLD/NASH associated HCC. . In a study by Dreval et al, investigating liver gene expression along with DNA methylation changes in mice using Stelic Animal Model (STAM) of NASH derived hepatocarcinogensis, it was found that aberrantly expressed genes had greater accrual of DNA methylation abnormalities with majority of atypically methylated genes found in fully developed HCC. One of these genes, tubulin beta 2B class IIB (Tubb2b), was found hypomethylated and overexpressed as liver carcinogenesis progressed [128]. This gene is a member of a family that encodes several microtubule cytoskeleton α- and β-tubulin proteins playing essential roles in cell differentiation, motility and shape which are abnormally expressed in human cancers including HCC [129, 130]. Another study by Margai et al., using relevant mice models (diet induced NAFLD mice, STAM of NASH derived HCC as well as choline and folate deficient diet mice models exhibiting NAFLD to HCC progression that resemble carcinogenesis in humans) characterized progressive increase in glycine N-methyltransferase (Gnmt) promoter methylation and corresponding reduction of Gnmt expression [131]. One consequence of GNMT decrease was found to be an increase in methylation along with an increase in SAM levels. GNMT is an abundant SAM dependent hepatic methyltransferase having folate binding and tumor suppressing features that transfers methyl group to glycine from SAM forming sarcosine and SAH [132, 133]. Decreased GNMT level was also observed in human HCC tissues as well as liver cancer cell lines [131]. In GNMT knockout mice, it was previously demonstrated that aberrant methylations result in steatosis and hepatocarcinogenic pathways in mice [134]. The authors suggested that assessing Gnmt methylation could be used for NAFLD stratification.
In a meta-analysis, decreased mRNA levels of PNPLA3 while increased expression of PARVB was correlated with NAFLD succession to HCC [135]. PNPLA3 protein is a lipid droplet associated protein having triglyceride hydrolase activity. Altered expression and hypermethylation of PNPLA3 is associated with live fat accumulation, inflammation, fibrosis, cirrhosis and HCC development [120, 136]. In another study it was shown that the liver biopsies of NAFLD patients with increased hepatocyte ballooning exhibited accrued oxidative DNA damage that could lead to promoter hypermethylation of tumor suppressor genes including HIC1, GSTP1, SOCS1, RASSF1, CDKN2A and APC, which could increase the risk of hepatocarcinogenesisin NAFLD subjects [137]. In a recent study, by performing differentially methylated region (DMR) network analysis, Kurokawa et al., identified two NAFLD specific DMR networks in NAFLD, viral hepatitis and HCC. The authors demonstrated that the methylation status of DMR-2 altered in a cooperative and/or synergistic manner during HCC establishment. Aberrant methylation levels of fatty acid binding protein-1 (FABP1), serum/glucocorticoid regulated kinase-2 (SGK2) and hepatocyte nuclear factor 4 alpha (HNF4A) were found from fibrosis to cancer making these as attractive therapeutic targets in NAFLD and associated HCC [138]. Using genome-wide DNA methylation analysis as well as mRNA expression levels in tissue samples of HCCs derived from precancerous background of NASH, Tian el al. found that deviant DNA methylation patterns resulted in altered gene expression in NASH associated HCC. It was observed that DNA hypomethylation as well as mRNA overexpression of tumor related genes includingDCAF4L2, CKLF, UBE2C, TUBA1B, TRIM4 and PRC1 occurred in NASH related HCC which were frequently linked to the necroinflammatory status of NASH and with poor tumor differentiation [139].
4. Lifestyle variables as DNA methylations modifiers
Over the past few decades, changes in lifestyle patterns have led to global rise in cluster of metabolic diseases including obesity, IR, T2DM and NAFLD. Dietary pattern and macronutrient composition impact theattenuation/development of NAFLD. Hypercaloric and HFD have been linked to the development of NAFLD by increasing the intrahepatic fat content [140, 141]. Several studies have shown that saturated fat consumption promotes hepatic fat accumulation [142]. Saturated fats are mostly found in processed food, high-fat dairy products red meat, cakes, cookies etc. [143]. Decreased consumption of carbohydrates having a high glycemic index, fructose content, sweeteners and sugar sweetened beverages have been reported to ameliorate NAFLD symptoms [144, 145]. Studies exploring the role of dietary proteins in relation to NAFLD have given conflicting results. Some reports have demonstrated positive association with NAFLD, whereas others have shown an inverse relationship, these inconsistencies may be due to type of protein consumed [146, 147]. High intake of meat, particularly red meat and processed meats is well recognized to be associated with cardiovascular diseases, insulin resistance and T2DM [148]. Proteins from animal sources seem to promote NAFLD, whereas those from plant sources have the opposite effect [149]. Excessive intake of calories, sugars, saturated fats, fructose, animal protein, salt as well as cholesterol increase the incidence and progression of NAFLD and NASH [150]. Hence, dietary habits characterize the overall amalgamation of foods that might lead to synergistic health effects. One of the most well studied dietetic pattern is the conventional Mediterranean diet, characterized by low intake of red meat, processed food, sugar and refined carbohydrate. Mediterranean diet has been consistently shown to protect against not only cardiovascular diseases and T2DM but also NAFLD [151, 152].
“Nutriepigenomics” a fast-evolving field that involves the study of nutrient-genome interactions via epigenetic alterations is rapidly gaining prominence in various complex human disorders [153, 154]. Food intake influences epigenome remodeling considerably all throughout life. Maternal dietary habits appear to be significant during pregnancy as well as postpartum, that leads to epigenetic alterations affecting the susceptibility to metabolic diseases by reprogramming the metabolic milieu [154]. In a study by Wankhade et al., it was concluded that maternal HFD negatively influences littermates DNA methylation and gut microbiota to augment the development of NAFLD and its advanced sequelae [155]. Wang et al., showed that high glucose (HG) induced lipid accumulation in hepatocytes in cell culture and increased levels of nuclear 25-hydroxycholestrol (25HC), an oxysterol, which specially activated the DNMT1. This significantly increased methylation of over 2000 genes belonging to 57 pathways of fuel metabolism such as PI3K, cAMP mediated insulin secretion, diabetes and NAFLD signaling [156]. HG diet induced cholesterol as well as triglyceride biosynthesis and increased 25HC levels. The HG diet also leads to excess accumulation of acetyl-CoA which is utilized for cholesterol as well as oxysterols (25HC) biosynthesis. 25HC has been shown to be a ligand of liver-X-receptors which are important in lipid metabolism. This increases the expression of vital enzymes of lipid biosynthesis like, ACC and FAS and increases the concentration of intracellular lipids through SREBP-1C cascade. Oxysterols have adverse effects on liver and are associated with the pathogenesis of various disorders such as metabolic syndrome and NAFLD, thus exacerbating the role of HG in liver damage [157].
Dietary deficiencies of methyl donors like choline, Vitamin B12, folic acid and betaine are linked with altered methylation of DNA that favors the progression of NAFLD and liver carcinogenesis. These methyl donors are required for SAM synthesis thus diet can greatly influence DNA methylation [158]. A methyl deficient diet induces liver injury in mice analogous to human NASH. In an investigation using mouse gene expression model and CpG microarrays, it was found that CpG islands of 164 genes were demethylated in mouse livers, displaying an inverse relationship with expression of specific genes following a diet devoid of methyl donors. These 164 genes are involved in glucose and lipid metabolism, DNA repair, apoptosis, fibrosis and liver tissue remodeling [159]. In a study by Cordeno et al., newborn rats whose mothers were on an obesogenic diet had greater intrahepatic fat content. Hyperhomocysteinemia induced by maternal high fat-sucrose diet was prevented by maternal methyl donor supplementation. DNA methylation metabolism in liver was also affected by maternal dietary pattern due to perturbation in one-carbon metabolism [160]. Cordeno et al., also demonstrated in mice that methyl donor supplementation could revert HFD induced hepatic triglyceride buildup and altered DNA methylation pattern of fatty acid synthase (FASN) promoter [161]. Mice on choline deficient diet leading to disturbed one-carbon metabolism exhibited reduced PPARα expression and hypermethylation of PPARα promoter [162]. It was shown that betaine supplementation in HFD fed mice resulted in global methylation and decreased promoter methylation of microsomal triglyceride transfer protein (MTTP) gene, promoting liver triglyceride export and reducing hepatic steatosis [163]. In another report it was shown that SAM supplementation attenuated triglyceride accumulation in alcoholic micropigs with fatty liver disease [164]. McCurdy et al., showed that HFD feeding in non-primate mothers makes the developing fetus highly vulnerable to excess lipids triggered lipotoxicity in fetal livers increasing the risk of pediatric NAFLD [165]. It was shown by Wesolowski et al., that western style diet of mothers induces oxidative stress and altered intracellular transport while, shifting obese mothers to a healthy diet improved liver metabolites and lipotoxicity in primate models [166].
NAFLD is speculated to be reversed by modulating dietary patterns. In study by Kim et al., HFD fed mice were subsequently fed with a normal chow diet resulting in reversal of diet-induced NAFLD with altered DNA methylation in important hepatic genes through Genome wide DNA methylation analysis, although lipid regulating APOA4 gene remained hypomethylated [167]. In another study it was shown that maternal obesity during pregnancy and lactation in dams animal model, programs the development of NAFLD phenotype in offspring [168]. Offspring of HFD fed mothers in rats exhibit CpG hypomethylation and increased expression of hepatic cell cycle inhibitor CDKN1A suggestive of hepatic dysfunction [169]. In a related study by Chen et al., maternal high fat/cholesterol diet exacerbated the establishment of NAFLD in male offspring by low hepatic ApoB gene expression and hypermethylation of CpG dinucleotides in the promoter region of ApoB gene compared to male offspring born to chow fed mice. Increased methylation of ApoB gene that is essential for VLDL assembly results in low expression of Apo-B in livers contributing to altered lipid profile and lipid transport from liver [170]. Deregulated Nuclear factor erythroid related factor2 (NRF2) signaling cascades play an important role in liver diseases. In HFD fed mice resveratrol led to hepatic hypomethylation of Nrf2 promoter which correlated with reduction in triglyceride levels [171]. Thus, consumption of polyphenols and anti-oxidants like resveratrol with potent anti-oxidant effects could be beneficial in NAFLD. Lyall et al., showed that diet induced hepatic steatosis in mice was linked to reversible 5-hmC changes in functionally essential genes, hence 5-hmC profile could be a potential marker for NAFLD state [172]. In mice fed on lipogenic, methyl donor deficient diet caused liver injury similar to human NASH. In this model, deviations in the expression of DNMT1 and de novo DNMT3a proteins were observed linking epigenetic alterations to hepatic steatotic pathogenesis [173].
A sedentary lifestyle is an important risk factor for NAFLD, obesity and T2DM [174]. Exercise training noticeably impacts DNA methylation in a tissue and locus specific manner [175]. A recent study by Wu et al., low-carbohydrate diet (LCD), and exercise plus low carbohydrate diet (ELCD) intervention, in both humans and mice NAFLD models showed GAB2 promoter hypermethylation thereby repressing mRNA expression. GAB2 plays an important role in differentiation, proliferation and cell migration by recruitment of factors like Ras, p85 and Shp2. This study concluded ELCD intervention as an effective approach for the management of NASH through epigenetic modulation by environmental factors [176]. In an investigation assessing the role of low-carbohydrate/Mediterranean and low-fat diets with or without physical activity in 120 human participants, differential DNA methylation patterns and increased intrahepatic fat accumulation was observed (measured by MRI) after 18 months of lifestyle intervention. At the baseline, specific CpG methylation of AC074286.1, calcium release activated channel regulator2A (CRACR2A), alpha-2-macroglobulin psuedogene1 (A2MP1), ARH/RhoGEF and pleckstrin domain protein1 (FARP1) loci were inversely correlated with intrahepatic fat where FARP1 methylation achieved similar predictive value for NAFLD as obtained by MRI based-diagnosis. After 18 months of dietary intervention an altered pattern of DNA methylation in A2MP1 was observed whereas CpGs of AC074286.1, CRACR2A and FARP1 were altered in different physical activity groups [177]. Thus, this study demonstrates a putative role of epigenetic remodeling subsequent to lifestyle interventions. Zhou et al., demonstrated that fast-food diet in mice elevated liver epigenome susceptibility to metabolic disorders concomitant with genome wide differential DNA methylation. Physical exercise was observed confer protection against this fast food diet induced reprogramming of the epigenome and DNA hypermethylation [178]. Role of physical exercise in liver health emphasizing the role of epigenetics has been extensively reviewed by Stevanovic et al. [179]. Ahrens et al., in an intraindividual comparison of liver biopsies pre and post bariatric surgery observed NAFLD-linked methylation signatures to be distinct and partially reversible, providing an example of treatment induced epigenetic organ remodeling [96].
5. Conclusion
In the present review we have summarized the recent studies on the effects of DNA methylation patterns in NAFLD pathogenesis. Altered CpG methylation patterns known till now may be a snapshot of a highly active state that may be more dynamic than earlier hypothesized, encompassing an amalgamation of genetic, epigenetic, inflammatory, environmental and nutritional effects. There may be more multilayered alterations in the global and gene specific methylation patterns in NAFLD. Numerous studies in human and animal models suggest altered DNA methylation patterns at global and locus specific levels leading to hepatic lipid accumulation, steatosis, inflammation, and injury responsible for establishment and progression of NAFLD. Altered DNA hyper and hypo methylation patterns are correlated with altered expression of specific genes. The changes that lead to alterations in DNA methylation and expression of enzymes regulating these are beginning to get unraveled. DNA methylation status seems to be modifiable particularly with methyl donor supplementation in diet. Lifestyle variations like exercise, losing weight and switching from high fat/sugar diet to low carbohydrate diet has the potential to reverse the altered liver methylome in NAFLD. Further studies are required to decipher the associated mechanisms and to comprehend the complex epigenetic alterations in NAFLD establishment and progression. Thus, DNA methylation holds promise in understanding the interplay of genetic, lifestyle and metabolic factors in NAFLD development and progression. The augmented understanding would help in developing new diagnostic and therapeutic strategies for the timely management and risk stratification of NAFLD and NASH.
Declarations
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data included in article/supp. material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Footnotes
This article is a part of the “Epigenetic Mechanisms” Special issue.
References
- 1.Powell E.E., Wong V.W., Rinella M. Non-alcoholic fatty liver disease. Lancet. 2021;397(10290):2212–2224. doi: 10.1016/S0140-6736(20)32511-3. [DOI] [PubMed] [Google Scholar]
- 2.Chalasani N., et al. The diagnosis and management of non-alcoholic fatty liver disease: practice guideline by the American association for the study of liver diseases, American college of gastroenterology, and the American gastroenterological association. Hepatology. 2012;55(6):2005–2023. doi: 10.1002/hep.25762. [DOI] [PubMed] [Google Scholar]
- 3.Singh S., et al. Fibrosis progression in nonalcoholic fatty liver vs nonalcoholic steatohepatitis: a systematic review and meta-analysis of paired-biopsy studies. Clin. Gastroenterol. Hepatol. 2015;13(4):643–654. doi: 10.1016/j.cgh.2014.04.014. e1-9; quiz e39-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eslam M., Sanyal A.J., George J. MAFLD: a consensus-driven proposed nomenclature for metabolic associated fatty liver disease. Gastroenterology. 2020;158(7):1999–2014. doi: 10.1053/j.gastro.2019.11.312. [DOI] [PubMed] [Google Scholar]
- 5.Lonardo A., et al. Effect of cofactors on NAFLD/NASH and MAFLD. A paradigm illustrating the pathomechanics of organ dysfunction. Metab Target Organ Damage. 2022;2(3) doi: 10.20517/mtod.2022.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Divella R., et al. Obesity, nonalcoholic fatty liver disease and adipocytokines network in promotion of cancer. Int. J. Biol. Sci. 2019;15(3):610–616. doi: 10.7150/ijbs.29599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y.Y., Yeh M.M. Non-alcoholic fatty liver disease: a review with clinical and pathological correlation. J. Formos. Med. Assoc. 2021;120(1 Pt 1):68–77. doi: 10.1016/j.jfma.2020.07.006. [DOI] [PubMed] [Google Scholar]
- 8.Estes C., et al. Modeling NAFLD disease burden in China, France, Germany, Italy, Japan, Spain, United Kingdom, and United States for the period 2016-2030. J. Hepatol. 2018;69(4):896–904. doi: 10.1016/j.jhep.2018.05.036. [DOI] [PubMed] [Google Scholar]
- 9.Dhamija E., Paul S.B., Kedia S. Non-alcoholic fatty liver disease associated with hepatocellular carcinoma: an increasing concern. Indian J. Med. Res. 2019;149(1):9–17. doi: 10.4103/ijmr.IJMR_1456_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perumpail R.B., et al. Pathogenesis of hepatocarcinogenesis in non-cirrhotic nonalcoholic fatty liver disease: potential mechanistic pathways. World J. Hepatol. 2015;7(22):2384–2388. doi: 10.4254/wjh.v7.i22.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drescher H.K., Weiskirchen S., Weiskirchen R. Current status in testing for nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH) Cells. 2019;8(8) doi: 10.3390/cells8080845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National Guideline Centre (UK). Non-Alcoholic Fatty Liver Disease: Assessment And Management 2016: London. [PubMed]
- 13.Piazzolla V.A., Mangia A. Noninvasive diagnosis of NAFLD and NASH. Cells. 2020;9(4) doi: 10.3390/cells9041005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovalic A.J., et al. Genetic and epigenetic culprits in the pathogenesis of nonalcoholic fatty liver disease. J Clin Exp Hepatol. 2018;8(4):390–402. doi: 10.1016/j.jceh.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Del Campo J.A., et al. Genetic and epigenetic regulation in nonalcoholic fatty liver disease (NAFLD) Int. J. Mol. Sci. 2018;19(3) doi: 10.3390/ijms19030911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robertson K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005;6(8):597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 17.Nishiyama A., Nakanishi M. Navigating the DNA methylation landscape of cancer. Trends Genet. 2021;37(11):1012–1027. doi: 10.1016/j.tig.2021.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Li Y.Y. Genetic and epigenetic variants influencing the development of nonalcoholic fatty liver disease. World J. Gastroenterol. 2012;18(45):6546–6551. doi: 10.3748/wjg.v18.i45.6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sui B.D., et al. Epigenetic regulation of mesenchymal stem cell homeostasis. Trends Cell Biol. 2020;30(2):97–116. doi: 10.1016/j.tcb.2019.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Alegria-Torres J.A., Baccarelli A., Bollati V. Epigenetics and lifestyle. Epigenomics. 2011;3(3):267–277. doi: 10.2217/epi.11.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Younossi Z., et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2018;15(1):11–20. doi: 10.1038/nrgastro.2017.109. [DOI] [PubMed] [Google Scholar]
- 22.Day C.P., James O.F. Steatohepatitis: a tale of two "hits"? Gastroenterology. 1998;114(4):842–845. doi: 10.1016/s0016-5085(98)70599-2. [DOI] [PubMed] [Google Scholar]
- 23.Tilg H., Moschen A.R. Evolution of inflammation in nonalcoholic fatty liver disease: the multiple parallel hits hypothesis. Hepatology. 2010;52(5):1836–1846. doi: 10.1002/hep.24001. [DOI] [PubMed] [Google Scholar]
- 24.Bessone F., Razori M.V., Roma M.G. Molecular pathways of nonalcoholic fatty liver disease development and progression. Cell. Mol. Life Sci. 2019;76(1):99–128. doi: 10.1007/s00018-018-2947-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loomba R., Sanyal A.J. The global NAFLD epidemic. Nat. Rev. Gastroenterol. Hepatol. 2013;10(11):686–690. doi: 10.1038/nrgastro.2013.171. [DOI] [PubMed] [Google Scholar]
- 26.Sakurai Y., et al. Role of insulin resistance in MAFLD. Int. J. Mol. Sci. 2021;22(8) doi: 10.3390/ijms22084156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Atabek M.E., Selver Eklioglu B., Akyurek N. Which metabolic syndrome criteria best predict non-alcoholic fatty liver disease in children? Eat. Weight Disord. 2014;19(4):495–501. doi: 10.1007/s40519-014-0129-0. [DOI] [PubMed] [Google Scholar]
- 28.Tanase D.M., et al. The intricate relationship between type 2 diabetes mellitus (T2DM), insulin resistance (IR), and nonalcoholic fatty liver disease (NAFLD) J. Diabetes Res. 2020;2020 doi: 10.1155/2020/3920196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holterman A., et al. Nonalcoholic fatty liver disease and bariatric surgery in adolescents. Semin. Pediatr. Surg. 2014;23(1):49–57. doi: 10.1053/j.sempedsurg.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 30.Silveira L.S., et al. Intra-abdominal fat is related to metabolic syndrome and non-alcoholic fat liver disease in obese youth. BMC Pediatr. 2013;13:115. doi: 10.1186/1471-2431-13-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mencin A.A., Lavine J.E. Advances in pediatric nonalcoholic fatty liver disease. Pediatr. Clin. 2011;58(6):1375–1392. doi: 10.1016/j.pcl.2011.09.005. x. [DOI] [PubMed] [Google Scholar]
- 32.Vachher M., et al. Contribution of organokines in the development of NAFLD/NASH associated hepatocellular carcinoma. J. Cell. Biochem. 2022 doi: 10.1002/jcb.30252. In press. [DOI] [PubMed] [Google Scholar]
- 33.Lonardo A., et al. History of nonalcoholic fatty liver disease. Int. J. Mol. Sci. 2020;21(16) doi: 10.3390/ijms21165888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J.D., et al. Patient sex, reproductive status, and synthetic hormone use associate with histologic severity of nonalcoholic steatohepatitis. Clin. Gastroenterol. Hepatol. 2017;15(1):127–131 e2. doi: 10.1016/j.cgh.2016.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ballestri S., et al. NAFLD as a sexual dimorphic disease: role of gender and reproductive status in the development and progression of nonalcoholic fatty liver disease and inherent cardiovascular risk. Adv. Ther. 2017;34(6):1291–1326. doi: 10.1007/s12325-017-0556-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lonardo A., et al. Sex differences in nonalcoholic fatty liver disease: state of the art and identification of research gaps. Hepatology. 2019;70(4):1457–1469. doi: 10.1002/hep.30626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye Q., et al. Global prevalence, incidence, and outcomes of non-obese or lean non-alcoholic fatty liver disease: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2020;5(8):739–752. doi: 10.1016/S2468-1253(20)30077-7. [DOI] [PubMed] [Google Scholar]
- 38.Gupta D., Krueger C.B., Lastra G. Over-nutrition, obesity and insulin resistance in the development of beta-cell dysfunction. Curr. Diabetes Rev. 2012;8(2):76–83. doi: 10.2174/157339912799424564. [DOI] [PubMed] [Google Scholar]
- 39.Donnelly K.L., et al. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J. Clin. Invest. 2005;115(5):1343–1351. doi: 10.1172/JCI23621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trepo E., Valenti L. Update on NAFLD genetics: from new variants to the clinic. J. Hepatol. 2020;72(6):1196–1209. doi: 10.1016/j.jhep.2020.02.020. [DOI] [PubMed] [Google Scholar]
- 41.Krawczyk M., Liebe R., Lammert F. Toward genetic prediction of nonalcoholic fatty liver disease trajectories: PNPLA3 and beyond. Gastroenterology. 2020;158(7):1865–1880 e1. doi: 10.1053/j.gastro.2020.01.053. [DOI] [PubMed] [Google Scholar]
- 42.Anstee Q.M., Day C.P. The genetics of NAFLD. Nat. Rev. Gastroenterol. Hepatol. 2013;10(11):645–655. doi: 10.1038/nrgastro.2013.182. [DOI] [PubMed] [Google Scholar]
- 43.Yki-Jarvinen H., Luukkonen P.K. Heterogeneity of non-alcoholic fatty liver disease. Liver Int. 2015;35(12):2498–2500. doi: 10.1111/liv.12970. [DOI] [PubMed] [Google Scholar]
- 44.Bianco C., et al. Non-invasive stratification of hepatocellular carcinoma risk in non-alcoholic fatty liver using polygenic risk scores. J. Hepatol. 2021;74(4):775–782. doi: 10.1016/j.jhep.2020.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romeo S., Sanyal A., Valenti L. Leveraging human genetics to identify potential new treatments for fatty liver disease. Cell Metabol. 2020;31(1):35–45. doi: 10.1016/j.cmet.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 46.Gellert-Kristensen H., et al. Combined effect of PNPLA3, TM6SF2, and HSD17B13 variants on risk of cirrhosis and hepatocellular carcinoma in the general population. Hepatology. 2020;72(3):845–856. doi: 10.1002/hep.31238. [DOI] [PubMed] [Google Scholar]
- 47.Gorden D.L., et al. Biomarkers of NAFLD progression: a lipidomics approach to an epidemic. J. Lipid Res. 2015;56(3):722–736. doi: 10.1194/jlr.P056002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smagris E., et al. Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology. 2015;61(1):108–118. doi: 10.1002/hep.27242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahajan A., et al. Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes. Nat. Genet. 2018;50(4):559–571. doi: 10.1038/s41588-018-0084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lyssenko V., et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J. Clin. Invest. 2007;117(8):2155–2163. doi: 10.1172/JCI30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perez-Diaz-Del-Campo N., et al. Association of the SH2B1 rs7359397 gene polymorphism with steatosis severity in subjects with obesity and non-alcoholic fatty liver disease. Nutrients. 2020;12(5) doi: 10.3390/nu12051260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Musso G., et al. Transcription factor 7-like 2 polymorphism modulates glucose and lipid homeostasis, adipokine profile, and hepatocyte apoptosis in NASH. Hepatology. 2009;49(2):426–435. doi: 10.1002/hep.22659. [DOI] [PubMed] [Google Scholar]
- 53.Ramya K., et al. Genetic association of ADIPOQ gene variants with type 2 diabetes, obesity and serum adiponectin levels in south Indian population. Gene. 2013;532(2):253–262. doi: 10.1016/j.gene.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 54.Vaxillaire M., et al. The common P446L polymorphism in GCKR inversely modulates fasting glucose and triglyceride levels and reduces type 2 diabetes risk in the DESIR prospective general French population. Diabetes. 2008;57(8):2253–2257. doi: 10.2337/db07-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Speliotes E.K., et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7(3):e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Franko A., et al. Dissociation of fatty liver and insulin resistance in I148M PNPLA3 carriers: differences in diacylglycerol (DAG) FA18:1 lipid species as a possible explanation. Nutrients. 2018;10(9) doi: 10.3390/nu10091314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luukkonen P.K., et al. Distinct contributions of metabolic dysfunction and genetic risk factors in the pathogenesis of non-alcoholic fatty liver disease. J. Hepatol. 2021;76(3):526–535. doi: 10.1016/j.jhep.2021.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hardy T., et al. Plasma DNA methylation: a potential biomarker for stratification of liver fibrosis in non-alcoholic fatty liver disease. Gut. 2017;66(7):1321–1328. doi: 10.1136/gutjnl-2016-311526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cao Y., et al. Hepatic menin recruits SIRT1 to control liver steatosis through histone deacetylation. J. Hepatol. 2013;59(6):1299–1306. doi: 10.1016/j.jhep.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 60.Sodum N., et al. Epigenetics in NAFLD/NASH: targets and therapy. Pharmacol. Res. 2021;167 doi: 10.1016/j.phrs.2021.105484. [DOI] [PubMed] [Google Scholar]
- 61.Ahn J., et al. Dietary resveratrol alters lipid metabolism-related gene expression of mice on an atherogenic diet. J. Hepatol. 2008;49(6):1019–1028. doi: 10.1016/j.jhep.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 62.Xu F., et al. SIRT1 mediates the effect of GLP-1 receptor agonist exenatide on ameliorating hepatic steatosis. Diabetes. 2014;63(11):3637–3646. doi: 10.2337/db14-0263. [DOI] [PubMed] [Google Scholar]
- 63.Cheng Z., et al. Gene expression profile-based drug screen identifies SAHA as a novel treatment for NAFLD. Mol Omics. 2019;15(1):50–58. doi: 10.1039/c8mo00214b. [DOI] [PubMed] [Google Scholar]
- 64.Rodriguez-Sanabria J.S., et al. An update in epigenetics in metabolic-associated fatty liver disease. Front. Med. 2021;8 doi: 10.3389/fmed.2021.770504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lonardo A., Arab J.P., Arrese M. Perspectives on precision medicine approaches to NAFLD diagnosis and management. Adv. Ther. 2021;38(5):2130–2158. doi: 10.1007/s12325-021-01690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Portela A., Esteller M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010;28(10):1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 67.Moosavi A., Motevalizadeh Ardekani A. Role of epigenetics in biology and human diseases. Iran. Biomed. J. 2016;20(5):246–258. doi: 10.22045/ibj.2016.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Illingworth R.S., Bird A.P. CpG islands--'a rough guide'. FEBS Lett. 2009;583(11):1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 69.Reik W., Dean W. DNA methylation and mammalian epigenetics. Electrophoresis. 2001;22(14):2838–2843. doi: 10.1002/1522-2683(200108)22:14<2838::AID-ELPS2838>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 70.Kim J.K., Samaranayake M., Pradhan S. Epigenetic mechanisms in mammals. Cell. Mol. Life Sci. 2009;66(4):596–612. doi: 10.1007/s00018-008-8432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Auclair G., Weber M. Mechanisms of DNA methylation and demethylation in mammals. Biochimie. 2012;94(11):2202–2211. doi: 10.1016/j.biochi.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 72.Choi S.W., Friso S. Epigenetics: a new bridge between nutrition and health. Adv. Nutr. 2010;1(1):8–16. doi: 10.3945/an.110.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Klose R.J., Bird A.P. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31(2):89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 74.Samblas M., Milagro F.I., Martinez A. DNA methylation markers in obesity, metabolic syndrome, and weight loss. Epigenetics. 2019;14(5):421–444. doi: 10.1080/15592294.2019.1595297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 76.Bogdanovic O., Veenstra G.J. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma. 2009;118(5):549–565. doi: 10.1007/s00412-009-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gopalakrishnan S., Van Emburgh B.O., Robertson K.D. DNA methylation in development and human disease. Mutat. Res. 2008;647(1-2):30–38. doi: 10.1016/j.mrfmmm.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018;19(2):81–92. doi: 10.1038/nrg.2017.80. [DOI] [PubMed] [Google Scholar]
- 79.Ren W., Gao L., Song J. Structural basis of DNMT1 and DNMT3A-mediated DNA methylation. Genes. 2018;9(12) doi: 10.3390/genes9120620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Laird A., et al. 5-hydroxymethylcytosine profiling as an indicator of cellular state. Epigenomics. 2013;5(6):655–669. doi: 10.2217/epi.13.69. [DOI] [PubMed] [Google Scholar]
- 81.Thomson J.P., et al. Dynamic changes in 5-hydroxymethylation signatures underpin early and late events in drug exposed liver. Nucleic Acids Res. 2013;41(11):5639–5654. doi: 10.1093/nar/gkt232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rasmussen K.D., Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016;30(7):733–750. doi: 10.1101/gad.276568.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Delatte B., Deplus R., Fuks F. Playing TETris with DNA modifications. EMBO J. 2014;33(11):1198–1211. doi: 10.15252/embj.201488290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Greenberg M.V.C., Bourc'his D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019;20(10):590–607. doi: 10.1038/s41580-019-0159-6. [DOI] [PubMed] [Google Scholar]
- 85.Hyun J., Jung Y. DNA methylation in nonalcoholic fatty liver disease. Int. J. Mol. Sci. 2020;21(21) doi: 10.3390/ijms21218138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Naik A., et al. Molecular interactions between NAFLD and xenobiotic metabolism. Front. Genet. 2013;4:2. doi: 10.3389/fgene.2013.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mato J.M., Lu S.C. Role of S-adenosyl-L-methionine in liver health and injury. Hepatology. 2007;45(5):1306–1312. doi: 10.1002/hep.21650. [DOI] [PubMed] [Google Scholar]
- 88.de Carvalho S.C., et al. Plasmatic higher levels of homocysteine in non-alcoholic fatty liver disease (NAFLD) Nutr. J. 2013;12:37. doi: 10.1186/1475-2891-12-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pacana T., et al. Dysregulated hepatic methionine metabolism drives homocysteine elevation in diet-induced nonalcoholic fatty liver disease. PLoS One. 2015;10(8):e0136822. doi: 10.1371/journal.pone.0136822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lai Z., et al. Association of hepatic global DNA methylation and serum one-carbon metabolites with histological severity in patients with NAFLD. Obesity. 2020;28(1):197–205. doi: 10.1002/oby.22667. [DOI] [PubMed] [Google Scholar]
- 91.Pirola C.J., et al. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut. 2013;62(9):1356–1363. doi: 10.1136/gutjnl-2012-302962. [DOI] [PubMed] [Google Scholar]
- 92.Pirola C.J., et al. Epigenetic modifications in the biology of nonalcoholic fatty liver disease: the role of DNA hydroxymethylation and TET proteins. Medicine (Baltim.) 2015;94(36):e1480. doi: 10.1097/MD.0000000000001480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu C., Lin J.D. PGC-1 coactivators in the control of energy metabolism. Acta Biochim. Biophys. Sin. 2011;43(4):248–257. doi: 10.1093/abbs/gmr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sookoian S., et al. Epigenetic regulation of insulin resistance in nonalcoholic fatty liver disease: impact of liver methylation of the peroxisome proliferator-activated receptor gamma coactivator 1alpha promoter. Hepatology. 2010;52(6):1992–2000. doi: 10.1002/hep.23927. [DOI] [PubMed] [Google Scholar]
- 95.Bibikova M., et al. Genome-wide DNA methylation profiling using Infinium(R) assay. Epigenomics. 2009;1(1):177–200. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- 96.Ahrens M., et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metabol. 2013;18(2):296–302. doi: 10.1016/j.cmet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 97.Mwinyi J., et al. NAFLD is associated with methylation shifts with relevance for the expression of genes involved in lipoprotein particle composition. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2017;1862(3):314–323. doi: 10.1016/j.bbalip.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 98.Schioth H.B., et al. A targeted analysis reveals relevant shifts in the methylation and transcription of genes responsible for bile acid homeostasis and drug metabolism in non-alcoholic fatty liver disease. BMC Genom. 2016;17:462. doi: 10.1186/s12864-016-2814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Baumeier C., et al. Elevated hepatic DPP4 activity promotes insulin resistance and non-alcoholic fatty liver disease. Mol. Metabol. 2017;6(10):1254–1263. doi: 10.1016/j.molmet.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Baumeier C., et al. Hepatic DPP4 DNA methylation associates with fatty liver. Diabetes. 2017;66(1):25–35. doi: 10.2337/db15-1716. [DOI] [PubMed] [Google Scholar]
- 101.Pan X., et al. Genome-wide DNA methylation profiling in nonalcoholic fatty liver reveals predictive aberrant methylation in PRKCE and SEC14L3 promoters. Dig. Liver Dis. 2022;54(4):521–528. doi: 10.1016/j.dld.2021.05.013. [DOI] [PubMed] [Google Scholar]
- 102.Zhang R.N., et al. Genome-wide analysis of DNA methylation in human peripheral leukocytes identifies potential biomarkers of nonalcoholic fatty liver disease. Int. J. Mol. Med. 2018;42(1):443–452. doi: 10.3892/ijmm.2018.3583. [DOI] [PubMed] [Google Scholar]
- 103.Wu J., et al. Altered DNA methylation sites in peripheral blood leukocytes from patients with simple steatosis and nonalcoholic steatohepatitis (NASH) Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. 2018;24:6946–6967. doi: 10.12659/MSM.909747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ma J., et al. A peripheral blood DNA methylation signature of hepatic fat reveals a potential causal pathway for nonalcoholic fatty liver disease. Diabetes. 2019;68(5):1073–1083. doi: 10.2337/DB18-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nano J., et al. Epigenome-wide association study identifies methylation sites associated with liver enzymes and hepatic steatosis. Gastroenterology. 2017;153(4):1096–1106 e2. doi: 10.1053/j.gastro.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 106.Yang X., et al. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16(8):995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lonardo A., Suzuki A. Sexual dimorphism of NAFLD in adults. Focus on clinical aspects and implications for practice and translational research. J. Clin. Med. 2020;9(5) doi: 10.3390/jcm9051278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Balakrishnan M., et al. Women have a lower risk of nonalcoholic fatty liver disease but a higher risk of progression vs men: a systematic review and meta-analysis. Clin. Gastroenterol. Hepatol. 2021;19(1):61–71 e15. doi: 10.1016/j.cgh.2020.04.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Phan H., et al. The association of sex steroid hormone concentrations with non-alcoholic fatty liver disease and liver enzymes in US men. Liver Int. 2021;41(2):300–310. doi: 10.1111/liv.14652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wegermann K., et al. Sex and menopause modify the effect of single nucleotide polymorphism genotypes on fibrosis in NAFLD. Hepatol Commun. 2021;5(4):598–607. doi: 10.1002/hep4.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Della Torre S. Beyond the X Factor: relevance of sex hormones in NAFLD pathophysiology. Cells. 2021;10(9) doi: 10.3390/cells10092502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ramirez M.C., et al. Expression and methylation status of female-predominant GH-dependent liver genes are modified by neonatal androgenization in female mice. Mol. Cell. Endocrinol. 2014;382(2):825–834. doi: 10.1016/j.mce.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 113.Strakovsky R.S., et al. Developmental bisphenol A (BPA) exposure leads to sex-specific modification of hepatic gene expression and epigenome at birth that may exacerbate high-fat diet-induced hepatic steatosis. Toxicol. Appl. Pharmacol. 2015;284(2):101–112. doi: 10.1016/j.taap.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhou Y., et al. Sex-associated preventive effects of low-dose aspirin on obesity and non-alcoholic fatty liver disease in mouse offspring with over-nutrition in utero. Lab. Invest. 2019;99(2):244–259. doi: 10.1038/s41374-018-0144-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Loomba R., et al. DNA methylation signatures reflect aging in patients with nonalcoholic steatohepatitis. JCI Insight. 2018;3(2) doi: 10.1172/jci.insight.96685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Horvath S., Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 2018;19(6):371–384. doi: 10.1038/s41576-018-0004-3. [DOI] [PubMed] [Google Scholar]
- 117.Reik W., Dean W., Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293(5532):1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 118.Page A., et al. Hepatic stellate cell transdifferentiation involves genome-wide remodeling of the DNA methylation landscape. J. Hepatol. 2016;64(3):661–673. doi: 10.1016/j.jhep.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zeybel M., et al. Differential DNA methylation of genes involved in fibrosis progression in non-alcoholic fatty liver disease and alcoholic liver disease. Clin. Epigenet. 2015;7:25. doi: 10.1186/s13148-015-0056-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kitamoto T., et al. Targeted-bisulfite sequence analysis of the methylation of CpG islands in genes encoding PNPLA3, SAMM50, and PARVB of patients with non-alcoholic fatty liver disease. J. Hepatol. 2015;63(2):494–502. doi: 10.1016/j.jhep.2015.02.049. [DOI] [PubMed] [Google Scholar]
- 121.Murphy S.K., et al. Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology. 2013;145(5):1076–1087. doi: 10.1053/j.gastro.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Johnson N.D., et al. Differential DNA methylation and changing cell-type proportions as fibrotic stage progresses in NAFLD. Clin. Epigenet. 2021;13(1):152. doi: 10.1186/s13148-021-01129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mardinoglu A., et al. Genome-scale metabolic modelling of hepatocytes reveals serine deficiency in patients with non-alcoholic fatty liver disease. Nat. Commun. 2014;5:3083. doi: 10.1038/ncomms4083. [DOI] [PubMed] [Google Scholar]
- 124.Wegermann K., et al. Branched chain amino acid transaminase 1 (BCAT1) is overexpressed and hypomethylated in patients with non-alcoholic fatty liver disease who experience adverse clinical events: a pilot study. PLoS One. 2018;13(9):e0204308. doi: 10.1371/journal.pone.0204308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Karin M., Dhar D. Liver carcinogenesis: from naughty chemicals to soothing fat and the surprising role of NRF2. Carcinogenesis. 2016;37(6):541–546. doi: 10.1093/carcin/bgw060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lazarus J.V., et al. Advancing the global public health agenda for NAFLD: a consensus statement. Nat. Rev. Gastroenterol. Hepatol. 2022;19(1):60–78. doi: 10.1038/s41575-021-00523-4. [DOI] [PubMed] [Google Scholar]
- 127.Huang D.Q., El-Serag H.B., Loomba R. Global epidemiology of NAFLD-related HCC: trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2021;18(4):223–238. doi: 10.1038/s41575-020-00381-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dreval K., et al. Gene expression and DNA methylation alterations during non-alcoholic steatohepatitis-associated liver carcinogenesis. Front. Genet. 2019;10:486. doi: 10.3389/fgene.2019.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gadadhar S., et al. The tubulin code at a glance. J. Cell Sci. 2017;130(8):1347–1353. doi: 10.1242/jcs.199471. [DOI] [PubMed] [Google Scholar]
- 130.Sun Q., et al. Expression profiling reveals dysregulation of cellular cytoskeletal genes in HBx-induced hepatocarcinogenesis. Cancer Biol. Ther. 2007;6(5):668–674. doi: 10.4161/cbt.6.5.3955. [DOI] [PubMed] [Google Scholar]
- 131.Borowa-Mazgaj B., et al. Gene expression and DNA methylation alterations in the Glycine N-methyltransferase gene in diet-induced nonalcoholic fatty liver disease-associated carcinogenesis. Toxicol. Sci. 2019;170(2):273–282. doi: 10.1093/toxsci/kfz110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Simile M.M., et al. Alterations of methionine metabolism in hepatocarcinogenesis: the emergent role of glycine N-methyltransferase in liver injury. Ann. Gastroenterol. 2018;31(5):552–560. doi: 10.20524/aog.2018.0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen M., et al. Tumor suppressor gene glycine N-methyltransferase and its potential in liver disorders and hepatocellular carcinoma. Toxicol. Appl. Pharmacol. 2019;378 doi: 10.1016/j.taap.2019.114607. [DOI] [PubMed] [Google Scholar]
- 134.Martinez-Chantar M.L., et al. Loss of the glycine N-methyltransferase gene leads to steatosis and hepatocellular carcinoma in mice. Hepatology. 2008;47(4):1191–1199. doi: 10.1002/hep.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ryaboshapkina M., Hammar M. Human hepatic gene expression signature of non-alcoholic fatty liver disease progression, a meta-analysis. Sci. Rep. 2017;7(1) doi: 10.1038/s41598-017-10930-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dong X.C. PNPLA3-A potential therapeutic target for personalized treatment of chronic liver disease. Front. Med. 2019;6:304. doi: 10.3389/fmed.2019.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nishida N., et al. Unique features associated with hepatic oxidative DNA damage and DNA methylation in non-alcoholic fatty liver disease. J. Gastroenterol. Hepatol. 2016;31(9):1646–1653. doi: 10.1111/jgh.13318. [DOI] [PubMed] [Google Scholar]
- 138.Kurokawa S., et al. Two differentially methylated region networks in nonalcoholic fatty liver disease, viral hepatitis, and hepatocellular carcinoma. BMC Gastroenterol. 2022;22(1):278. doi: 10.1186/s12876-022-02360-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tian Y., et al. Aberrant DNA methylation results in altered gene expression in non-alcoholic steatohepatitis-related hepatocellular carcinomas. J. Cancer Res. Clin. Oncol. 2020;146(10):2461–2477. doi: 10.1007/s00432-020-03298-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hernandez E.A., et al. Acute dietary fat intake initiates alterations in energy metabolism and insulin resistance. J. Clin. Invest. 2017;127(2):695–708. doi: 10.1172/JCI89444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bortolotti M., et al. High protein intake reduces intrahepatocellular lipid deposition in humans. Am. J. Clin. Nutr. 2009;90(4):1002–1010. doi: 10.3945/ajcn.2008.27296. [DOI] [PubMed] [Google Scholar]
- 142.Rosqvist F., et al. Overeating saturated fat promotes fatty liver and ceramides compared with polyunsaturated fat: a randomized trial. J. Clin. Endocrinol. Metab. 2019;104(12):6207–6219. doi: 10.1210/jc.2019-00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Luukkonen P.K., et al. Saturated fat is more metabolically harmful for the human liver than unsaturated fat or simple sugars. Diabetes Care. 2018;41(8):1732–1739. doi: 10.2337/dc18-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Howard B.V., Wylie-Rosett J. Sugar and cardiovascular disease: a statement for healthcare professionals from the committee on nutrition of the council on nutrition, physical activity, and metabolism of the American heart association. Circulation. 2002;106(4):523–527. doi: 10.1161/01.cir.0000019552.77778.04. [DOI] [PubMed] [Google Scholar]
- 145.Jensen T., et al. Fructose and sugar: a major mediator of non-alcoholic fatty liver disease. J. Hepatol. 2018;68(5):1063–1075. doi: 10.1016/j.jhep.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Markova M., et al. Isocaloric diets high in animal or plant protein reduce liver fat and inflammation in individuals with type 2 diabetes. Gastroenterology. 2017;152(3):571–585 e8. doi: 10.1053/j.gastro.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 147.Lang S., et al. High protein intake is associated with histological disease activity in patients with NAFLD. Hepatol Commun. 2020;4(5):681–695. doi: 10.1002/hep4.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Turner K.M., Keogh J.B., Clifton P.M. Red meat, dairy, and insulin sensitivity: a randomized crossover intervention study. Am. J. Clin. Nutr. 2015;101(6):1173–1179. doi: 10.3945/ajcn.114.104976. [DOI] [PubMed] [Google Scholar]
- 149.Zelber-Sagi S., et al. High red and processed meat consumption is associated with non-alcoholic fatty liver disease and insulin resistance. J. Hepatol. 2018;68(6):1239–1246. doi: 10.1016/j.jhep.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 150.Berna G., Romero-Gomez M. The role of nutrition in non-alcoholic fatty liver disease: pathophysiology and management. Liver Int. 2020;40(Suppl 1):102–108. doi: 10.1111/liv.14360. [DOI] [PubMed] [Google Scholar]
- 151.Ryan M.C., et al. The Mediterranean diet improves hepatic steatosis and insulin sensitivity in individuals with non-alcoholic fatty liver disease. J. Hepatol. 2013;59(1):138–143. doi: 10.1016/j.jhep.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 152.Bozzetto L., et al. Liver fat is reduced by an isoenergetic MUFA diet in a controlled randomized study in type 2 diabetic patients. Diabetes Care. 2012;35(7):1429–1435. doi: 10.2337/dc12-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zaiou M., et al. Dietary patterns influence target gene expression through emerging epigenetic mechanisms in nonalcoholic fatty liver disease. Biomedicines. 2021;9(9) doi: 10.3390/biomedicines9091256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Remely M., et al. Nutriepigenomics: the role of nutrition in epigenetic control of human diseases. Curr. Opin. Clin. Nutr. Metab. Care. 2015;18(4):328–333. doi: 10.1097/MCO.0000000000000180. [DOI] [PubMed] [Google Scholar]
- 155.Wankhade U.D., et al. Enhanced offspring predisposition to steatohepatitis with maternal high-fat diet is associated with epigenetic and microbiome alterations. PLoS One. 2017;12(4):e0175675. doi: 10.1371/journal.pone.0175675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang Y., et al. High glucose induces lipid accumulation via 25-hydroxycholesterol DNA-CpG methylation. iScience. 2020;23(5) doi: 10.1016/j.isci.2020.101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ma Y., et al. 25-Hydroxycholesterol-3-sulfate regulates macrophage lipid metabolism via the LXR/SREBP-1 signaling pathway. Am. J. Physiol. Endocrinol. Metab. 2008;295(6):E1369–E1379. doi: 10.1152/ajpendo.90555.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Starlard-Davenport A., et al. Dietary methyl deficiency, microRNA expression and susceptibility to liver carcinogenesis. J. Nutrigenetics Nutrigenomics. 2010;3(4-6):259–266. doi: 10.1159/000324362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tryndyak V.P., et al. Coupling global methylation and gene expression profiles reveal key pathophysiological events in liver injury induced by a methyl-deficient diet. Mol. Nutr. Food Res. 2011;55(3):411–418. doi: 10.1002/mnfr.201000300. [DOI] [PubMed] [Google Scholar]
- 160.Cordero P., et al. Maternal methyl donors supplementation during lactation prevents the hyperhomocysteinemia induced by a high-fat-sucrose intake by dams. Int. J. Mol. Sci. 2013;14(12):24422–24437. doi: 10.3390/ijms141224422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Cordero P., et al. Dietary supplementation with methyl donors reduces fatty liver and modifies the fatty acid synthase DNA methylation profile in rats fed an obesogenic diet. Genes Nutr. 2013;8(1):105–113. doi: 10.1007/s12263-012-0300-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mikael L.G., et al. Disturbed one-carbon metabolism causing adverse reproductive outcomes in mice is associated with altered expression of apolipoprotein AI and inflammatory mediators PPARalpha, interferon-gamma, and interleukin-10. J. Nutr. 2012;142(3):411–418. doi: 10.3945/jn.111.151753. [DOI] [PubMed] [Google Scholar]
- 163.Wang L.J., et al. Betaine attenuates hepatic steatosis by reducing methylation of the MTTP promoter and elevating genomic methylation in mice fed a high-fat diet. J. Nutr. Biochem. 2014;25(3):329–336. doi: 10.1016/j.jnutbio.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 164.Zivkovic A.M., et al. Quantitative lipid metabolomic changes in alcoholic micropigs with fatty liver disease. Alcohol Clin. Exp. Res. 2009;33(4):751–758. doi: 10.1111/j.1530-0277.2008.00892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.McCurdy C.E., et al. Maternal high-fat diet triggers lipotoxicity in the fetal livers of nonhuman primates. J. Clin. Invest. 2009;119(2):323–335. doi: 10.1172/JCI32661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wesolowski S.R., et al. Switching obese mothers to a healthy diet improves fetal hypoxemia, hepatic metabolites, and lipotoxicity in non-human primates. Mol. Metabol. 2018;18:25–41. doi: 10.1016/j.molmet.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kim H., et al. Persistent changes in liver methylation and microbiome composition following reversal of diet-induced non-alcoholic-fatty liver disease. Cell. Mol. Life Sci. 2019;76(21):4341–4354. doi: 10.1007/s00018-019-03114-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Oben J.A., et al. Maternal obesity during pregnancy and lactation programs the development of offspring non-alcoholic fatty liver disease in mice. J. Hepatol. 2010;52(6):913–920. doi: 10.1016/j.jhep.2009.12.042. [DOI] [PubMed] [Google Scholar]
- 169.Dudley K.J., et al. Offspring of mothers fed a high fat diet display hepatic cell cycle inhibition and associated changes in gene expression and DNA methylation. PLoS One. 2011;6(7) doi: 10.1371/journal.pone.0021662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chen H.C., et al. The nonalcoholic fatty liver disease-like phenotype and lowered serum VLDL are associated with decreased expression and DNA hypermethylation of hepatic ApoB in male offspring of ApoE deficient mothers fed a with Western diet. J. Nutr. Biochem. 2020;77 doi: 10.1016/j.jnutbio.2019.108319. [DOI] [PubMed] [Google Scholar]
- 171.Hosseini H., et al. Resveratrol alleviates non-alcoholic fatty liver disease through epigenetic modification of the Nrf2 signaling pathway. Int. J. Biochem. Cell Biol. 2020;119 doi: 10.1016/j.biocel.2019.105667. [DOI] [PubMed] [Google Scholar]
- 172.Lyall M.J., et al. Non-alcoholic fatty liver disease (NAFLD) is associated with dynamic changes in DNA hydroxymethylation. Epigenetics. 2020;15(1-2):61–71. doi: 10.1080/15592294.2019.1649527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Pogribny I.P., et al. Hepatic epigenetic phenotype predetermines individual susceptibility to hepatic steatosis in mice fed a lipogenic methyl-deficient diet. J. Hepatol. 2009;51(1):176–186. doi: 10.1016/j.jhep.2009.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Dongiovanni P., Valenti L. A nutrigenomic approach to non-alcoholic fatty liver disease. Int. J. Mol. Sci. 2017;18(7) doi: 10.3390/ijms18071534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Voisin S., et al. Exercise training and DNA methylation in humans. Acta Physiol. 2015;213(1):39–59. doi: 10.1111/apha.12414. [DOI] [PubMed] [Google Scholar]
- 176.Wu N., et al. Effect of exercise and diet intervention in NAFLD and NASH via GAB2 methylation. Cell Biosci. 2021;11(1):189. doi: 10.1186/s13578-021-00701-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yaskolka Meir A., et al. Effects of lifestyle interventions on epigenetic signatures of liver fat: central randomized controlled trial. Liver Int. 2021;41(9):2101–2111. doi: 10.1111/liv.14916. [DOI] [PubMed] [Google Scholar]
- 178.Zhou D., et al. High fat diet and exercise lead to a disrupted and pathogenic DNA methylome in mouse liver. Epigenetics. 2017;12(1):55–69. doi: 10.1080/15592294.2016.1261239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Stevanovic J., et al. Physical exercise and liver "fitness": role of mitochondrial function and epigenetics-related mechanisms in non-alcoholic fatty liver disease. Mol. Metabol. 2020;32:1–14. doi: 10.1016/j.molmet.2019.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supp. material/referenced in article.