Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has posed a serious threat to global public health. A novel vaccine made from messenger RNA (mRNA) has been developed and approved for use at an unprecedented pace. However, an increased risk of myocarditis has been reported after BNT162b2 mRNA vaccination due to unknown causes. In this study, we used single-cell RNA sequencing and single-cell T cell receptor sequencing analyses of peripheral blood mononuclear cells (PBMCs) to describe, for the first time, changes in the peripheral immune landscape of a patient who underwent myocarditis after BNT162b2 vaccination. The greatest changes were observed in the transcriptomic profile of monocytes in terms of the number of differentially expressed genes. When compared to the transcriptome of PBMCs from vaccinated individuals without complications, increased expression levels of IL7R were detected in multiple cell clusters. Overall, results from this study can help advance research into the pathogenesis of BNT162b2-induced myocarditis.
Keywords: BNT162b2, myocarditis, peripheral blood mononucleear cells, severe acute respiratory syndrome coronavirus 2, single-cell RNA sequencing
INTRODUCTION
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic is ongoing, with a total of 241 million patients and approximately 5 million deaths worldwide. Fortunately, messenger RNA (mRNA)-based vaccines, which were developed less than a year after the start of the pandemic, confer 95% protection against SARS-CoV-2 infection (Arunachalam et al., 2021; Polack et al., 2020; Shah and Woo, 2021). Currently, more than six billion doses of vaccines, including the Pfizer-BioNTech BNT162b2 mRNA vaccine, have been administered. The BNT162b2 vaccine has recently been linked to an increased risk of myocarditis (Barda et al., 2021; Li et al., 2022). However, little is known about its mechanism of action.
Myocarditis is caused by inflammation of the cardiac muscle, which may result in decreased cardiac output and irregular heart rhythm. According to the Vaccine Adverse Event Reporting System, myocarditis after BNT162b2 mRNA vaccination usually occurs within a week of vaccination, particularly in male adolescents and young adults. In this study, we report the changes in the peripheral immune landscape via single-cell RNA sequencing (RNA-seq) analysis in a patient with myocarditis after BNT12b2 vaccination. A cardiac biopsy sample from the patient showed infiltration of immune cells, highlighting the importance of understanding the role of peripheral blood mononuclear cells (PBMCs) in the pathogenesis of vaccination-induced myocarditis. We also describe a wide range of transcriptomic changes at the time of acute myocarditis among different cell clusters and compare the results with PBMCs of vaccinated individuals without complications, offering new perspectives on vaccination-induced myocarditis and possibly helping in overcoming the pandemic.
MATERIALS AND METHODS
Ethics statement
The study was conducted in accordance with the Declaration of Helsinki (2013) and approved by the Institutional Review Board (IRB) of Uijeongbu St. Mary’s Hospital (IRB approval No. UC19TIDE0142). Written informed consent was obtained from the participant.
Isolation of PBMCs
Blood in an EDTA tube was mixed with the same amount of phosphate-buffered saline (PBS). The mixture of blood and PBS was then transferred to a Leucosep tube. The mixture was then centrifuged at 1,000 × g for 15 min at room temperature. The supernatant was collected in a 50 ml conical tube. The cells were washed by centrifugation at 400 × g for 10 min at room temperature. After removing the supernatant, cells were washed twice. The cells were counted and resuspended in stock solution (1:9 = DMSO:fetal bovine serum). After 24 h in a cell container in a –80°C deep freezer, the stock was stored in a liquid nitrogen tank.
Chromium Next GEM Single Cell 5′v2 (Dual Index)
To obtain information on cell preparation, we used the LUNA-FL™ automated fluorescence cell counter (Logos Biosystems, Korea) to consult the 10X Genomics single-cell protocols cell preparation guide and the guidelines for optimal sample preparation flowchart (documents CG00053 and CG000126, respectively).
Libraries were prepared using the chromium controller according to the 10X Chromium Next GEM Single Cell V(D)J User Guide (CG000331). Briefly, cell suspensions were diluted in nuclease-free water to achieve a targeted cell count of 10,000. The cell suspension was mixed with the master mix and loaded with Single Cell 5′ Gel Beads and Partitioning Oil into a Next GEM Chip K. RNA transcripts from single cells were uniquely barcoded and reverse-transcribed within droplets. The cDNA molecules were pooled and enriched using polymerase chain reaction (PCR). For the V(D)J-enriched library, the enriched cDNA pool was first amplified using T Cell Mix1 primer or B Cell Mix1 and then amplified using T Cell Mix2 or B Cell Mix2 primers. For the 5′ gene expression library, the cDNA pool undergoes an end repair process, the addition of a single ‘A’ base, and ligation of the adapters. The products were then purified and enriched by PCR to create a 5′ gene expression library. The purified libraries were quantified using qPCR according to the qPCR Quantification Protocol Guide (Kapa Biosystems, USA) and qualified using an Agilent Technologies 4200 TapeStation (Agilent Technologies, USA). The libraries were then sequenced using the HiSeq platform (Illumina, USA) according to the read length provided in the user guide.
Single cell RNA-seq and VDJ analysis
The fastq files from single-cell 5′ profiling were further analyzed using the 10X Genomics CellRanger software (v.6.1.1). The 5′ transcriptome profiling and VDJ analysis were performed using the Cellranger multi command. The latest version of the human reference gene (GRCh38) was downloaded for analyzing gene expression. The expected cell number was 10,000. The latest human reference (GRCh38-vdj) was used as the VDJ reference. The R package “Seurat” was used to establish the object, detect cell clusters based on gene expression, and identify subpopulation-enriched markers (Stuart et al., 2019). Cells detected with fewer than 25 and more than 6,000 features and those with mitochondrial genes higher than 20% were excluded from the analysis. A scale factor of 10,000 was used for data normalization. Variable genes were computed using the “vst” method with 2000 features. After scale and PCA (principal component anlaysis) calculations, the Harmony package was used to remove batch effects and integrate samples with 1 to 50 dimensions. Cell clustering was performed at a resolution of 0.8 and the results were visualized using uniform manifold approximation and projection (UMAP). A total of 26 clusters were identified, and cell type was manually defined using the marker gene list. For differential genes, the FindMarkers function was utilized with the following options: only.pos = FALSE, min.pct = 0.15, and logfc.threshold = 0.15. The AverageExpression function was used to measure the expression in each cluster and sample. Post-analysis, including pathway analysis, was conducted using the differential gene list and expression dataset. The RunALRA function was used for data restoration for pathway analysis. The gene sets shown in the figure are as follows: T-cell receptor complex (GO:0042101), transcription factor AP-1 complex (GO:0035976), adaptive immune response (GO:0002250), IL-2 signaling pathway (WP:WP49), response to stimulus (GO:0050896), I-kappaB/NF-kappaB complex (GO:0033256), and signaling by TGF-beta Receptor Complex (REAC:R-HSA-170834). VDJ profiling was extracted from CellRanger multi-output and added to the R object using a custom script.
Data availability
The data relevant to the manuscript have been uploaded to the Sequence Read Archive (SRA) (SRR18209603, SRR18209602, SRR18209601, and SRR18209600).
RESULTS
Clinical information of the patient with myocarditis after BNT12b2 vaccination
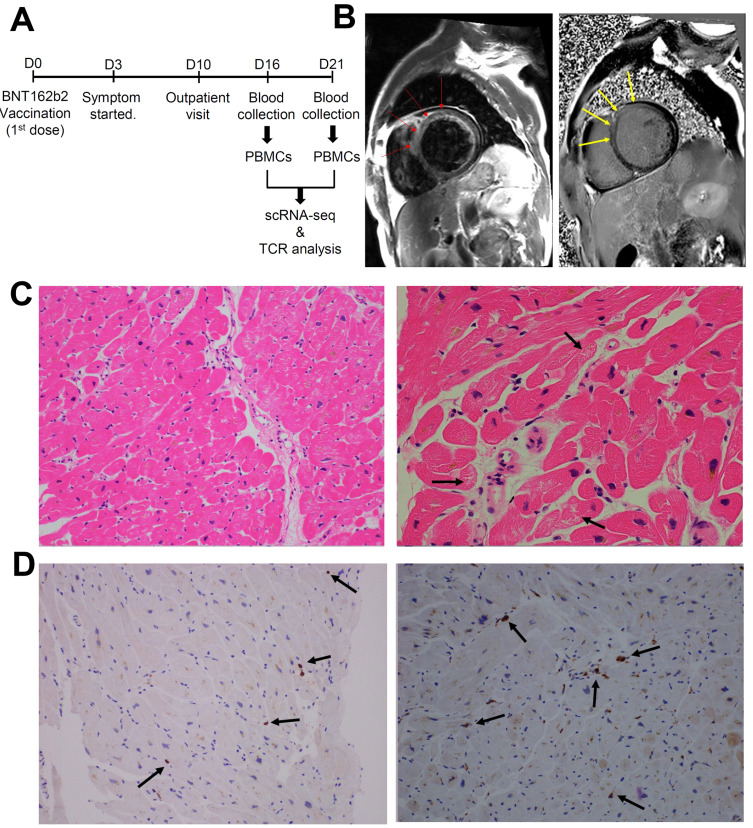
A 59-year-old male visited the hospital with shortness of breath. He received his first dose of BNT162b2 vaccine three days prior to the onset of symptoms (Fig. 1A). After 5 days of prescribing furosemide 40 mg once a day, transthoracic echocardiography showed a dilated LV cavity (left ventricular internal diameter end diastole = 62.0 mm), a left ventricular ejection fraction of 24%, and global hypokinesia of the LV wall. Laboratory findings also revealed elevated creatine kinase-myocardial band (CK-MB), NT-proBNP (pro-brain natriuretic peptide), creatine phosphokinase, and C-reactive protein levels (Table 1). Although the patient showed slight elevation of CK-MB level on day 20 due to chronic and mild heart failure, CK-MB level decreased from 13.2 ng/ml on day 16 to 3.86 ng/ml on day 18, validating the time points for blood collection. Cardiac magnetic resonance imaging showed myocardial edema and abnormal subepicardial and mid-wall hyperenhancement (Fig. 1B). Endomyocardial biopsy from the right ventricular septum showed a few microfoci of myofiber eosinophilic changes, disrupted myocytes, and inflammatory infiltrates (Fig. 1C). The biopsy sample was immunopositive for CD3, a T-cell marker, and for CD68, a pan-macrophage marker. (Fig. 1D). In addition, an alternative etiology of myocarditis beyond the temporary association with COVID-19 vaccination was not observed.
Fig. 1. Clinical manifestation of the patient with myocarditis after BNT162b2 vaccination.
(A) Timeline for the collection of samples. (B) Cardiac magnetic resonance imaging findings. T2-weighted black blood image (left) and delayed enhancement image (right) show diffuse increased signal intensity (as compared with skeletal muscles) due to edema and abnormal subepicardial and mid-wall hyperenhancement, particularly in the anterior and antero-septal portions of the left ventricle (red and yellow arrows). (C) Endomyocardial biopsy showed active multifocal lympho-histocytic myocarditis with a few microfoci of myofiber eosinophilic changes and disrupted myocytes (marked with black arrows; H&E stain, ×200). The inflammatory infiltrates show mild extent and focal distribution of the lymphocytic type without fibrosis (left and right). (D) Left: CD3 immunostaining highlights that inflammatory infiltrates are mainly composed of T lymphocytes (in brown, black arrows) (×200). Right: CD68 immunostaining highlights that inflammatory infiltrates are mainly composed of macrophages (in brown, black arrows) (×200).
Table 1.
Clinical information of the patient with myocarditis after BNT162b2 vaccination
| Day | Test | Result | Reference | |
|---|---|---|---|---|
| 10 | NT-proBNP (pg/ml) | 1,113.00 | 5-125 | |
| CK-MB (ng/ml) | 22.70 | 0.3-4.94 | ||
| Troponin-T (ng/ml) | 0.019 | <0.099 | ||
| C-reactive protein (mg/dl) | 0.18 | <0.5 | ||
| ECG | PVC | |||
| T-wave inversion at V5-6 | ||||
| ST-segment elevation at V1-4 | ||||
| 15 | Transthoracic echocardiography | Diastolic left ventricular internal diameter (mm) | 62 | |
| Left ventricular ejection fraction (%) | 24 | |||
| 16 | Coronary angiography | Normal | ||
| Endocardial biopsy | A few microfoci of myofiber eosinophilic changes | |||
| Disrupted myocytes | ||||
| Inflammatory infiltrates showing mild extent and focal distribution | ||||
| Lymphocytic type without fibrosis | ||||
| CK-MB (ng/ml) | 13.2 | 0.3-4.94 | ||
| 17 | Cardiac magnetic resonance imaging | Myocardial edema on T2-weighted study | ||
| Late gadolinium enhancement at basal to mid antero-septal wall of LV | ||||
| CK-MB (ng/ml) | 6.69 | 0.3-4.94 | ||
| 18 | CK-MB (ng/ml) | 3.86 | 0.3-4.94 | |
| 20 | CK-MB (ng/ml) | 10.60 | 0.3-4.94 | |
| ECG | T-wave inversion at V5-6 | |||
| ST-segment elevation at V1-4 | ||||
| 24 | C-reactive protein (mg/dl) | 1.02 | <0.5 | |
Immune cell profiling of the patient with myocarditis after BNT162b2 vaccination
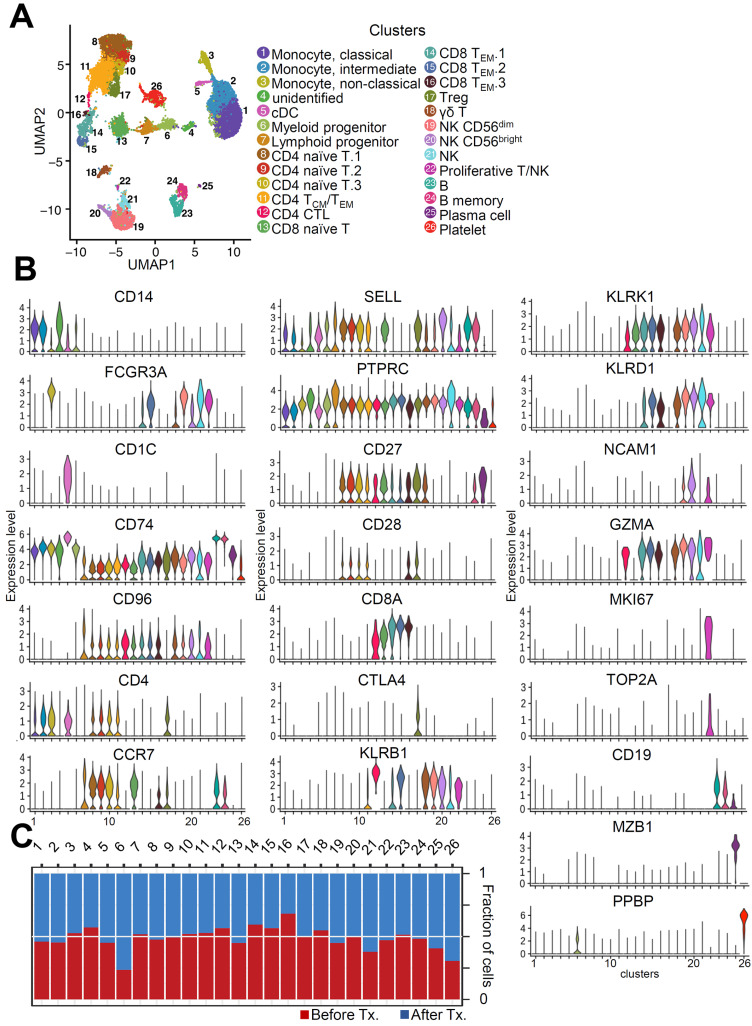
To determine the immune cell profile, we conducted single-cell RNA sequencing analysis (scRNA-seq), including VDJ analysis, on PBMCs from a patient with myocarditis as a side effect of BNT162b2 vaccination. To investigate the changes in transcriptomic dynamics at the single-cell level, we collected blood at two time points: before treatment (Tx.) and after treatment (Fig. 1A). We obtained transcriptome datasets from 22,585 PBMCs collected at two different time points after quality control (Fig. 2A). To uncover the changes at the time of myocarditis (day 16), we performed unsupervised clustering and obtained 26 cell clusters after batch correction using Harmony (Korsunsky et al., 2019; Fig. 2A). Classical dendritic cells (cDC) are located next to monocytes and CD4 naïve T cells, and CD8 effector memory T (TEM) cells are divided into subpopulations. To validate each cluster, we evaluated the expression levels of immune cell markers of each cell cluster (Fig. 2B). Next, we analyzed the fraction of cells in each cluster under different conditions. While cluster 6 and 26 were largely enriched in “After Tx.,” cluster 16 was markedly enriched in “Before Tx.” (Fig. 2C). However, the fractions of most peripheral immune cells were similar between the two groups, implying that immune composition would not be a major factor in the development of acute myocarditis with the BNT162b2 vaccine.
Fig. 2. Single-cell transcriptional characteristics in the patient with myocarditis after BNT162b2 vaccination.
(A) UMAP representation of cell types annotated by transcriptional profiling. cDC, classical dendritic cells; TCM, central memory T cells; TEM, effector memory T cells; NK, natural killer cells. (B) The violin plots were drawn with immune cell markers. (C) The fraction of cells in each cluster was counted.
Cell clusters show difference in transcriptomic profile change at the time of myocarditis
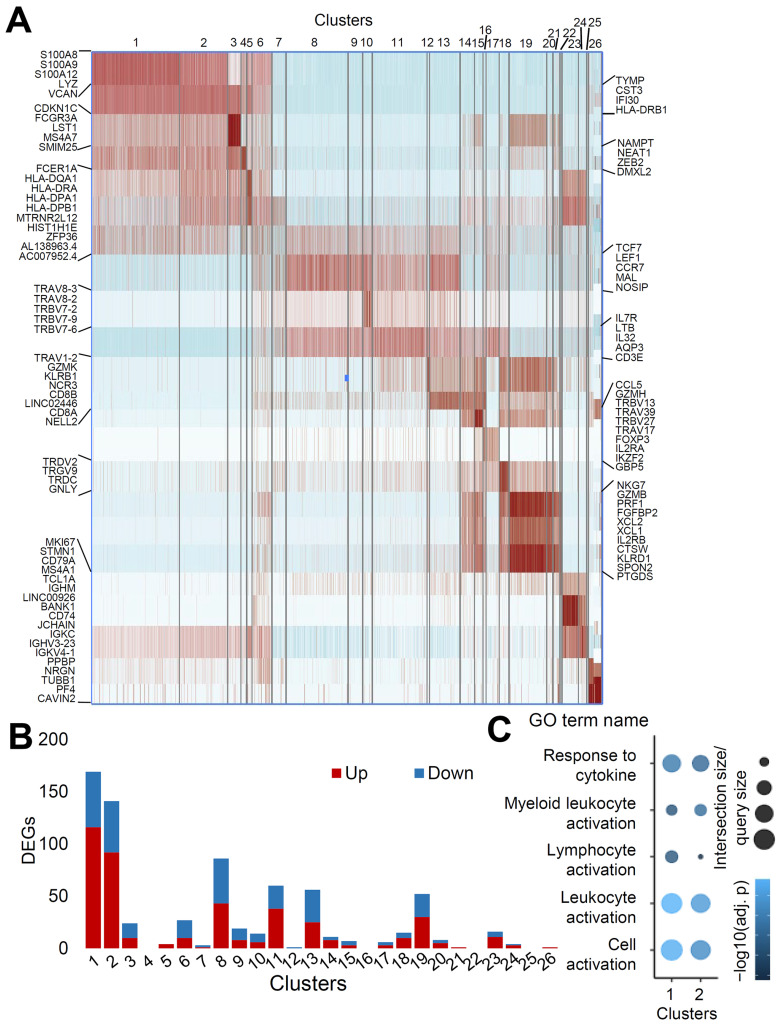
Well-annotated cell types showed transcriptomic profiles that differed from each other in the heatmap, with genes upregulated in each cluster (Fig. 3A). Most upregulated genes included marker genes for each immune cell type, validating the clustering results. Moreover, subpopulations of CD4 naïve T cells and CD8 TEM cells, which are located close to each other on the UMAP representation, showed substantial differences in T cell receptor (TCR)-related genes (Egorov et al., 2018). We then compared the transcriptional responses among clusters with the number of differentially expressed genes (DEGs) with an adjusted p-value of less than 0.05 (Fig. 3B). Classical and intermediate monocytes showed the greatest degree of transcriptomic changes in terms of the number of DEGs (Fig. 3B).
Fig. 3. Degree of changes in transcriptomic profile differs according to the cell clusters.
(A) Heatmap with the gene markers from each cluster. (B) The number of DEGs satisfying the condition (minimum percentage of expressing cells > 15%, logFC > 0.15, adjusted P < 0.05) was counted. (C) Pathway analysis with DEGs of raw P value less than 0.01 in cluster 1 and 2.
Next, we performed pathway analysis on DEGs with a raw p-value of less than 0.01 in clusters 1 (classical monocyte) and 2 (intermediate monocyte). Pathway analysis of up and downregulated DEGs showed changes in pathways related to response to cytokines, myeloid leukocyte activation, lymphocyte activation, leukocyte activation, and cell activation, implying that monocytes have the potential to play an important role in the pathogenesis of myocarditis in BNT162b2-vaccinated individuals (Fig. 3C).
Characterization of clonal VDJ rearrangement in the condition of myocarditis after BNT162b2 vaccination
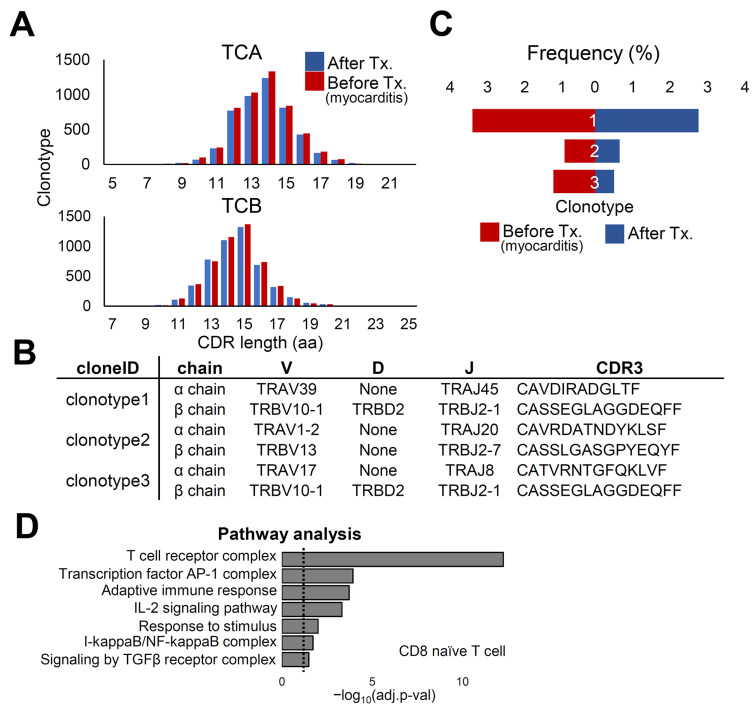
T cell immunity is an important mechanism for clearing viral infections, including SARS-CoV-2 (Le Bert et al., 2020; Luo et al., 2021). In addition, identification of disease-relevant TCR would lead to the identification of effective biomarkers that enable the exclusion of individuals who are highly likely to progress to myocarditis from vaccination (Mitchell and Michels, 2020). Therefore, we performed single-cell TCR sequencing analysis to profile TCR repertoire diversity. TCR, both α and β, is formed by the rearrangement of variable (V), diversity (D), junctional (J), and constant (C) regions. Recombination of these regions results in high diversity in the complementarity-determining region 3 (CDR3), which is responsible for peptide recognition. The distribution of CDR3 length in T cell clones showed minimal changes between the two time points (Fig. 4A). Using single-cell TCR analysis, we introduced the three most abundant CDR3 sequences with VDJ combinations in PBMCs from patients with myocarditis after vaccination (Fig. 4B). Although clonotype 1 was the most abundant in both conditions, clonotype 3 was the second most abundant at the time of myocarditis, while clonotype 2 was the second most frequent after treatment (Fig. 4C). Although TCR analysis revealed insignificant changes in the clonal expansion of T cells, there were statistically significant differences in several gene pathways after data restoration (Linderman et al., 2022), including the adaptive immune response pathway (Fig. 4D).
Fig. 4. Analysis of TCR repertoire in the patient with myocarditis at two time points.
(A) Distribution of CDR3 length of T cell clones after the treatment (blue) and before the treatment (myocarditis; red). (B) Single cell TCR analysis was performed. The three clonotypes with the highest frequency are described with sequences. (C) The frequency of clonotypes is shown depending on the condition. (D) Pathway analysis was performed after data restoration in cluster 13 CD8 naïve T cell. DEGs with adjusted P value smaller than 0.1 were used. Dotted line shows where adjusted P value is 0.05. TCA, T cell receptor A; TCB, T cell receptor B; AP-1, activator protein 1; IL-2, interleukin-2; TGFβ, transforming growth factor-β.
IL7R increased in the patient with myocarditis after vaccination
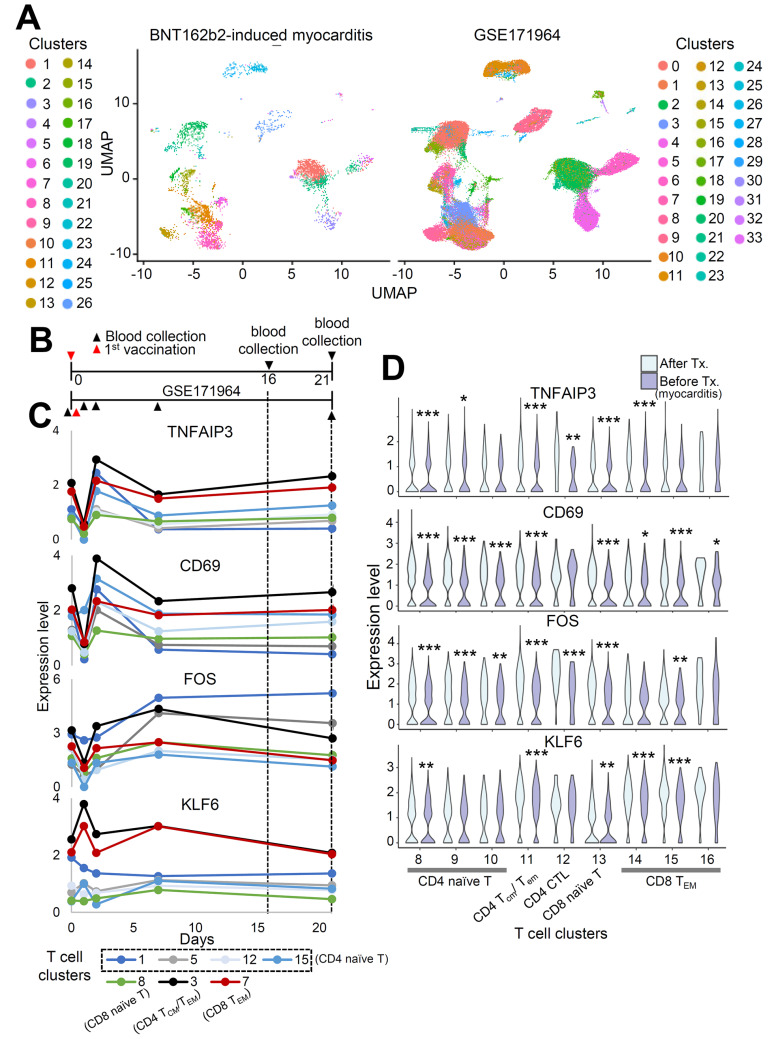
To validate the clustering results and investigate the findings specific to patients with myocarditis after vaccination, we combined and analyzed single-cell RNA sequencing data from this study with publicly available data (GSE171964), analyzing PBMC samples from vaccinated individuals (Arunachalam et al., 2021) after batch correction using Harmony (Korsunsky et al., 2019; Fig. 5A). Cell clusters overlapped between the datasets, validating the results of this study (Fig. 5A). GSE171964 includes PBMCs collected at different time points. As our study analyzed PBMCs 16 and 21 days after the 1st vaccination, PBMCs at days 16 and 21 were considered to be the appropriate experimental controls (Fig. 5B).
Fig. 5. Single-cell RNA seq data of BNT162b2-induced myocarditis case and vaccinated individuals are integrated.
(A) UMAP representation of PBMCs from this study combined with PBMCs from GSE171964. Different colors denote different clusters. (B) Timeline indicating different time points of sample collection for two datasets. (C) The expression levels of TNFAIP3, CD69, FOS, and KLF6 are marked according to the time after vaccination in T cell subpopulations (GSE171964). Time points when PBMCs from this study were collected were marked with dashed line. (D) Violin plot shows the expression levels of TNFAIP3, CD69, FOS, and KLF6 in T cell subpopulations. *P < 0.05, **P < 0.01, ***P < 0.001; Wilcox test.
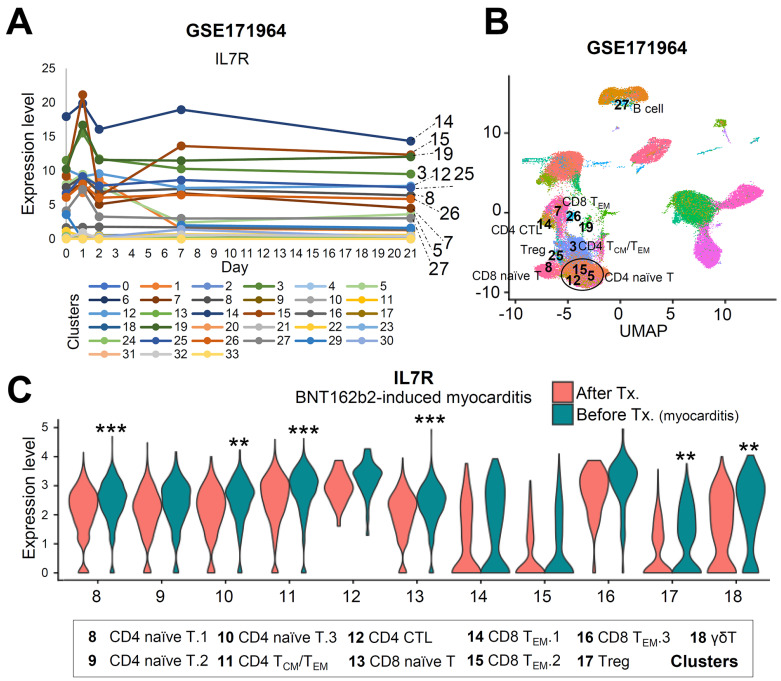
Contributing to the previous report that SARS-CoV-2 mRNA vaccination induces an adaptive immune response coordinated by early CD4 T cells facilitating B cells and CD8 effector T cells (Sureshchandra et al., 2021) and that gene signatures in adaptive immune response pathways changed between two time points (Fig. 4D), we investigated changes in the T cell transcriptome after vaccination via GSE171964 and at the time of myocarditis in an attempt to find a gene that is differentially expressed only at the time of myocarditis. The T-cell subpopulation was annotated after integration with our data (Supplementary Fig. S1). The expression levels of key genes, TNFAIP3, CD69 (T cell activation markers), FOS, and KLF6 (memory development markers), in the T cell subpopulation were not upregulated in either vaccinated individuals or patients with myocarditis (Figs. 5C and 5D). However, IL7R was upregulated only in BNT162b2-induced myocarditis cases (Supplementary Fig. S2A). Interleukin-7 is important for T cell survival, homeostasis, and activation (Colpitts et al., 2009; Lawson et al., 2015). IL7R, which was upregulated across multiple cell subtypes at the time of acute myocarditis, was high in the T cell subpopulation (Figs. 6A and 6B, Supplementary Fig. S2B) and was not increased in terms of the expression level in GSE171964, implying the possibility that the upregulation of IL7R is related to myocarditis after vaccination (Fig. 6C, Supplementary Fig. S2C).
Fig. 6. The mRNA expression level of IL7R is increased at the time of acute myocarditis.
(A) T Single-cell RNA sequencing data of PBMCs of vaccinated individuals was analyzed with automatic clustering. The expression level of IL7R of each cluster is shown with different colors denoting different clusters. (B) Cluster information marked in 6A are shown on the UMAP representation. (C) The expression level of IL7R in BNT162b2-induced myocarditis case is shown in T cell clusters. **P < 0.01, ***P < 0.001; Wilcox test.
In conclusion, we present, for the first time, a transcriptomic landscape of PBMCs at the single-cell level from a patient with myocarditis after BNT162b2 vaccination. Cardiac biopsy at the time of acute myocarditis revealed immunopositivity for CD3 and CD68. PBMCs were collected before and after treatment to determine characteristics specific to the time of acute myocarditis. A high number of DEGs were identified in classical and intermediate monocytes, with changes in pathways related to immune cell activation. Although there were several limitations in the current study, including the number of samples, our data will help investigate peripheral immune cells contributing to the severe side effects of BNT162b2 vaccination.
DISCUSSION
Although the mRNA-based vaccine, BNT162b2, has shown good safety profile in multiple studies, BNT162b2 vaccination has a positive correlation with an increased risk of myocarditis (Barda et al., 2021). However, the related mechanism is not yet known. Understanding the mechanism would contribute to identifying the risk factors and preventing the occurrence of the side effects of BNT162b2. This is the first study to provide single-cell RNA sequencing data for PBMCs from the patient with myocarditis after vaccination. Recently, it has been reported that cancer immunotherapy significantly increases myocarditis (Makunts et al., 2021). Although myocarditis is a “local” inflammation of myocardium, “systemic” activation of immune response, such as activation via cancer immunotherapy, can contribute to myocarditis occurrence, implying the possibility that myocarditis is a local event easily affected by the systemic immune landscape. Therefore, it is important to examine changes in peripheral immune cells, which can contribute to the pathogenesis of vaccine-induced myocarditis.
Inflammatory responses are carried out by immune cells, and their activity is largely dependent on various inflammatory mediators (Bhusal et al., 2020). Particularly, cytokines, including numerous interleukins, are important modulators of the immune response (Turner et al., 2014). Interleukin-7 is a highly important inflammatory cytokine in patients with stable and unstable angina (Damas et al., 2003). Plasma levels of interleukin-7 are elevated not only in patients with unstable angina pectoris (Damas et al., 2003) but also in patients in the acute and convalescent stages of SARS-CoV-2 infection (Wang et al., 2021). Our study showed elevation of IL7R in several subpopulations at the time of myocarditis, which was not observed in the peripheral immune cells of vaccinated individuals at the corresponding time points. Considering that IL7R is elevated shortly after vaccination (Arunachalam et al., 2021) and that interleukin-7-driven inflammation plays a role in several cardiac diseases, we need to pay more attention to the effect of hyperactivation of IL7R on the development of myocarditis after vaccination.
However, this study had some limitations. Owing to the scarcity of cases, the number of samples was not sufficient. Further evaluation of PBMCs from other patients, including further analysis of the role of IL7R, is necessary to confirm the characteristics of the peripheral immune landscape specific for myocarditis after vaccination. However, this study is a meaningful first step towards understanding the major side effects of BNT162b2.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This study was supported by Korea Health Technology R&D Project (HR18C0012) from the Ministry of Health & Welfare to S.F. and the National Research Foundation of Korea (NRF) grant funded by the Korean Government (MSIT) (NRF-2018R1A5A2025079). B.K.Y. was supported by the Seok-San Biomedical Science Scholarship, Yonsei University College of Medicine.
Footnotes
AUTHOR CONTRIBUTIONS
B.K.Y. was involved in the execution of the study and was primarily responsible for writing the manuscript. T.G.O. performed single-cell and VDJ analyses. S.B. was involved in study design. K.J.S. and S.H.K. gathered clinical information of the patients. J.Y.L. and Y.K. provided technical support for this project. J.-w.K. was involved in the conceptualization of the study. H.-S.A. and S.F. were primarily involved in data collection, hypothesis development, and manuscript writing and editing.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Arunachalam P.S., Scott M.K.D., Hagan T., Li C., Feng Y., Wimmers F., Grigoryan L., Trisal M., Edara V.V., Lai L., et al. Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature. 2021;596:410–416. doi: 10.1038/s41586-021-03791-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barda N., Dagan N., Ben-Shlomo Y., Kepten E., Waxman J., Ohana R., Hernan M.A., Lipsitch M., Kohane I., Netzer D., et al. Safety of the BNT162b2 mRNA Covid-19 vaccine in a nationwide setting. N. Engl. J. Med. 2021;385:1078–1090. doi: 10.1056/NEJMoa2110475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhusal R.P., Foster S.R., Stone M.J. Structural basis of chemokine and receptor interactions: key regulators of leukocyte recruitment in inflammatory responses. Protein Sci. 2020;29:420–432. doi: 10.1002/pro.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colpitts S.L., Dalton N.M., Scott P. IL-7 receptor expression provides the potential for long-term survival of both CD62Lhigh central memory T cells and Th1 effector cells during Leishmania major infection. J. Immunol. 2009;182:5702–5711. doi: 10.4049/jimmunol.0803450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damas J.K., Waehre T., Yndestad A., Otterdal K., Hognestad A., Solum N.O., Gullestad L., Froland S.S., Aukrust P. Interleukin-7-mediated inflammation in unstable angina: possible role of chemokines and platelets. Circulation. 2003;107:2670–2676. doi: 10.1161/01.CIR.0000070542.18001.87. [DOI] [PubMed] [Google Scholar]
- Egorov E.S., Kasatskaya S.A., Zubov V.N., Izraelson M., Nakonechnaya T.O., Staroverov D.B., Angius A., Cucca F., Mamedov I.Z., Rosati E., et al. The changing landscape of naive T cell receptor repertoire with human aging. Front. Immunol. 2018;9:1618. doi: 10.3389/fimmu.2018.01618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P.R., Raychaudhuri S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson B.R., Gonzalez-Quintial R., Eleftheriadis T., Farrar M.A., Miller S.D., Sauer K., McGavern D.B., Kono D.H., Baccala R., Theofilopoulos A.N. Interleukin-7 is required for CD4(+) T cell activation and autoimmune neuroinflammation. Clin. Immunol. 2015;161:260–269. doi: 10.1016/j.clim.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- Li C., Chen Y., Zhao Y., Lung D.C., Ye Z., Song W., Liu F.F., Cai J.P., Wong W.M., Yip C.C., et al. Intravenous injection of COVID-19 mRNA vaccine can induce acute myopericarditis in mouse model. Clin. Infect. Dis. 2022;74:1933–1950. doi: 10.1093/cid/ciab707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linderman G.C., Zhao J., Roulis M., Bielecki P., Flavell R.A., Nadler B., Kluger Y. Zero-preserving imputation of single-cell RNA-seq data. Nat. Commun. 2022;13:192. doi: 10.1038/s41467-021-27729-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L., Liang W., Pang J., Xu G., Chen Y., Guo X., Wang X., Zhao Y., Lai Y., Liu Y., et al. Dynamics of TCR repertoire and T cell function in COVID-19 convalescent individuals. Cell Discov. 2021;7:89. doi: 10.1038/s41421-021-00321-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makunts T., Saunders I.M., Cohen I.V., Li M., Moumedjian T., Issa M.A., Burkhart K., Lee P., Patel S.P., Abagyan R. Myocarditis occurrence with cancer immunotherapy across indications in clinical trial and post-marketing data. Sci. Rep. 2021;11:17324. doi: 10.1038/s41598-021-96467-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A.M., Michels A.W. T cell receptor sequencing in autoimmunity. J. Life Sci. (Westlake Village) 2020;2:38–58. doi: 10.36069/JoLS/20201203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Perez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M., Woo H.G. Molecular perspectives of SARS-CoV-2: pathology, immune evasion, and therapeutic interventions. Mol. Cells. 2021;44:408–421. doi: 10.14348/molcells.2021.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., 3rd, Stoeckius M., 3rd, Smibert P., 3rd, Satija R., 3rd Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureshchandra S., Lewis S.A., Doratt B.M., Jankeel A., Coimbra Ibraim I., Messaoudi I. Single-cell profiling of T and B cell repertoires following SARS-CoV-2 mRNA vaccine. JCI Insight. 2021;6:e153201. doi: 10.1172/jci.insight.153201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M.D., Nedjai B., Hurst T., Pennington D.J. Cytokines and chemokines: at the crossroads of cell signalling and inflammatory disease. Biochim. Biophys. Acta. 2014;1843:2563–2582. doi: 10.1016/j.bbamcr.2014.05.014. [DOI] [PubMed] [Google Scholar]
- Wang G.L., Gao H.X., Wang Y.L., Wei X., Liu Y.Z., Lu J.H., Li L., Wang H.B., Zhao L., Rong Y.X., et al. Serum IP-10 and IL-7 levels are associated with disease severity of coronavirus disease 2019. Cytokine. 2021;142:155500. doi: 10.1016/j.cyto.2021.155500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data relevant to the manuscript have been uploaded to the Sequence Read Archive (SRA) (SRR18209603, SRR18209602, SRR18209601, and SRR18209600).