Abstract
The gastrointestinal tract is involved in coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The gut microbiota has important roles in viral entry receptor angiotensin-converting enzyme 2 (ACE2) expression, immune homeostasis, and crosstalk between the gut and lungs, the ‘gut–lung axis’. Emerging preclinical and clinical studies indicate that the gut microbiota might contribute to COVID-19 pathogenesis and disease outcomes; SARS-CoV-2 infection was associated with altered intestinal microbiota and correlated with inflammatory and immune responses. Here, we discuss the cutting-edge evidence on the interactions between SARS-CoV-2 infection and the gut microbiota, key microbial changes in relation to COVID-19 severity and host immune dysregulations with the possible underlying mechanisms, and the conceivable consequences of the pandemic on the human microbiome and post-pandemic health. Finally, potential modulatory strategies of the gut microbiota are discussed. These insights could shed light on the development of microbiota-based interventions for COVID-19.
Subject terms: SARS-CoV-2, Gastroenterology, SARS-CoV-2
The gastrointestinal tract is involved in COVID-19, gastrointestinal symptoms can occur and marked changes in the gut microbiota have been observed. This Perspective highlights interactions between the gut microbiota and SARS-CoV-2 infection and increasing interest in the gut–lung axis.
Introduction
With the coronavirus disease 2019 (COVID-19) pandemic now in its third year, it seems possible that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) will be here for the long term. However, there remain many unknowns, including the emergence of viral mutants, duration of natural and post-vaccination immunity, and reasons for persistent antigen-specific memory responses in COVID-19 (refs.1,2). Although SARS-CoV-2 primarily infects the respiratory tract, several lines of evidence point towards substantial involvement of the gastrointestinal tract, including the ability of SARS-CoV-2 to infect and replicate in intestinal enterocytes3, increased expression of the viral entry receptor (angiotensin-converting enzyme 2 (ACE2) receptor) and of several membrane-bound serine proteases (such as transmembrane protease serine 2 (TMPRSS2) and TMPRSS4) in intestinal epithelial cells4, as well as the presence of SARS-CoV-2 genomic and sub-genomic RNA in mucosal and faecal samples of individuals with SARS-CoV-2 infection5–8. In a longitudinal study of 113 patients with COVID-19, SARS-CoV-2 RNA was found in the faecal samples of nearly 50% of individuals during acute infection and faecal RNA shedding persisted for 7 months in 3.8% of patients7. Moreover, a wide range of gastrointestinal symptoms, including diarrhoea, nausea, vomiting, loss of appetite and abdominal pain, have been reported in patients with COVID-19, with a pooled prevalence of 17.6% according to a meta-analysis of 60 studies involving a total of 4,243 patients9. Another meta-analysis of 35 studies involving 6,686 patients suggested that COVID-19 with gastrointestinal involvement had a worse disease course10 and, surprisingly, that gastrointestinal symptoms could even precede respiratory symptoms11,12.
Emerging studies have begun to bridge the gap between the gut microbiome and the pathophysiology of COVID-19. In fact, the gastrointestinal tract is regarded as the largest immune organ in the body and its resident microbiota can regulate host immunity, defend against pathogens and support nutrient metabolism13. The composition of gut microbiota is determined and could be dynamically altered by numerous factors, including genes, diet, lifestyle, disease and ageing14,15. Gut dysbiosis with a reduction in microbial diversity is commonly linked to immune-mediated inflammatory and autoimmune diseases16,17. The gut microorganisms could also regulate local and systemic inflammatory activity17, and studies have demonstrated that respiratory infections are associated with both compositional and functional alterations of the gut microbiota18 through vital crosstalk between gut microorganisms and the pulmonary system, otherwise known as the ‘gut–lung axis’19. The bi-directional interactions between the gut microbiota and the lungs have been linked with host immune responses to SARS-CoV-2 infection20. In particular, COVID-19 has been shown to be associated with microbiome alterations and gut barrier dysfunction in human studies, which could increase the translocation of bacterial products and toxins into the circulatory system and exacerbate the systemic inflammatory response20,21. Disruption of the gut microbiota could negatively influence the recruitment of immune cells to the lung, which could then increase the susceptibility of developing respiratory tract infections22–24. Furthermore, gut microorganisms could lead to a decreased expression of ACE2, a key regulator of innate immunity and microbial ecology, thereby influencing viral invasion and replication25,26. Overall, state-of-the-art microbiome analyses have advanced our understanding towards the critical but clinically underappreciated roles that gut microorganisms play in SARS-CoV-2 pathogenesis and COVID-19 severity and prognosis.
In this Perspective, we summarize the latest data on gut microbiota alterations in patients with COVID-19 globally and discuss evidence of the mechanical roles of these microorganisms in SARS-CoV-2 infectivity and disease outcomes. When available, we review clinical studies, in vitro and in vivo studies, systematic reviews, and high-quality narrative reviews on COVID-19 and the gut microbiome published between 31 December 2019 and 28 August 2022 (Supplementary Box 1). We also highlight knowledge gaps and share our viewpoints on the potential clinical applications of modulating the gut microbiota in the prevention and treatment of acute COVID-19 and post-acute COVID-19 conditions.
COVID-19 and gut microbiota
Acute COVID-19
Human studies
As of August 2022, a total of 46 human observational studies have reported alterations in gut microbiota composition in patients with COVID-19 at the time of diagnosis and during the disease course compared with individuals without COVID-19 (Supplementary Table 1). Amongst these studies, which originated from different populations in China, USA, Japan, Bangladesh, United Arab Emirates, Portugal, Germany and Hungary, the faecal microbiome of patients with COVID-19 showed decreased bacterial diversity27–30 and reduced abundance of short-chain fatty acid (SCFA)-producing bacteria from the Lachnospiraceae, Ruminococcaceae and Eubacteriaceae families as well as increased opportunistic pathogens from Enterobacteriaceae families27–29,31–37 compared with the faecal microbiome of healthy individuals. Specifically, the abundance of Faecalibacterium27,28,31,33–35,37, Eubacterium28,34,37,38, Coprococcus27,35,36,38, Ruminococcus27,28,35, Lachnospira27,35,38 and Roseburia27,35 was decreased, whereas that of Enterococcus32,34–36, Rothia28,32,35,36 and Lactobacillus27,35,36 was increased. Intriguingly, a number of studies with a modest sample size of typically ~30 patients have reported that patients with COVID-19 had a distinct gut microbiome composition compared with patients with community-acquired viral pneumonia39 and influenza virus infection28,35. In a study consisting of 62 patients with COVID-19, 33 patients with seasonal flu and 40 healthy individuals as controls, patients with COVID-19 had significantly decreased (P = 0.0124) bacterial diversity with enrichment of opportunistic pathogens, such as Streptococcus, Veillonella, Fusobacterium and Escherichia, as well as a higher faecal concentration of the pro-inflammatory cytokine IL-18 compared with patients with seasonal flu35. Another study of 30 patients with COVID-19 found a significantly higher relative abundance of phyla Actinobacteria (now known as Actinomycetota) and Firmicutes (now known as Bacillota) compared with 24 patients with H1N1 influenza infection28. Hospitalized patients with COVID-19 (n = 15) also showed an increased abundance of Enterococcus faecium and Clostridium ramosum compared with those hospitalized with other types of viral pneumonia (n = 7)39. Although the sample size of these studies remains small, these findings highlight that SARS-CoV-2 might have specific effects on the gut microbiota.
Although consistent gut microbial changes have been reported in patients with COVID-19, these studies had some limitations. First, most studies were cross-sectional and interindividual demographic differences could have greatly influenced the gut microbiome. Second, clinical information and symptoms at the time of sample collection were not always well presented and, in most studies, faecal samples before infection were not collected. Third, it remains unclear whether diet and medications, such as antibiotics and proton pump inhibitors, could influence these microbial changes specifically in the setting of COVID-19. Most studies have focused on adult populations and the effect of SARS-CoV-2 on the gut microbiome in children is comparatively less evident33,37. Lastly, the effect of variants of concern, such as Omicron, on the gut microbiome is still largely unknown as most published studies so far have focused on earlier variants.
Studies in children
The gut microbiota in early childhood is less mature and more vulnerable to environmental exposures compared with that in adults40; hence, microbiome alterations during early life, whereby children’s immune, metabolic and cognition systems are under development, have been associated with subsequent risks of several childhood illnesses, including infections, asthma, allergy and diabetes41–44. Preliminary data showed that an altered gut microbiome in nine children with COVID-19 aged between 7 and 139 months was dominated by the genus Pseudomonas, and this change persisted for weeks after hospital discharge45. In 12 asymptomatic young children aged 0–24 months with SARS-CoV-2 infection, depletion of Bifidobacterium bifidum and Akkermansia muciniphila, which are linked to protection against inflammation, were observed in SARS-CoV-2-positive faecal samples46. Multisystem inflammatory syndrome in children (MIS-C) associated with COVID-19 has been increasingly recognized47. Interestingly, substantial changes in the composition of the gut microbiota were found in children with MIS-C (n = 25) and COVID-19 (n = 20), including a reduction of Faecalibacterium prausnitzii48. Nevertheless, whether the development of MIS-C might, in part, be attributed to COVID-19-related microbial dysbiosis remains unclear.
Beyond bacteria
Beyond bacteria, the gastrointestinal tract is also home to a large number of fungi, viruses and archaea, which regulate host homeostasis, pathophysiological processes, immune regulation and assembly of the co-residing gut bacterial microbiome49,50. Two observational studies reported that SARS-CoV-2 infection was linked to altered composition of fungal microbiota51,52. Both patients with COVID-19 (n = 67) and patients with H1N1 infection (n = 35) were found to have an increased fungal load and enrichment of Aspergillus and Penicillium in their faecal samples compared with 48 healthy individuals as controls51. Patients with COVID-19 also harboured highly heterogeneous mycobiome configurations and a higher abundance of opportunistic fungal pathogens, including Candida albicans, Candida auris and Aspergillus flavus52. Fungal infection caused by Candida spp. has been reported as a major complication in severe COVID-19, with the prevalence ranging from 0.7% to 23.5%53,54. This co-infection was associated with a high mortality rate due to a longer intensive care unit stay, catheterization and broad-spectrum antibiotic use53. Additionally, two studies showed the distinct composition of gut viral community in patients with SARS-CoV-2 infection compared with that of healthy individuals55,56. Metagenomic profiling of RNA and DNA viruses of faecal samples from 100 patients with COVID-19 and 78 individuals without COVID-19 found that faecal viruses in COVID-19 were characterized by the depletion of multiple bacteriophage lineages (DNA viruses)55. Interestingly, the abundance of four gut viruses (Myxococcus phage, Bacteroides phage, Murmansk poxvirus and Sphaerotilus phage) inversely correlated with both severity of COVID-19 and host age55. This observation could partly explain why older individuals (aged above 65 years) were more susceptible to severe and worse COVID-19 outcomes; however, this aspect needs to be further explored in animal studies. In a mouse model, SARS-CoV-2 infection can cause a shift in the composition of gut viruses with altered expression of immune-related or infection-related genes in gut epithelial cells, including host–virus or host–bacteria crosstalk genes IL15, C3 and MUC2 (ref.56). Mechanistic studies will be necessary to explore the contribution of specific microbial communities (such as bacteria, fungi and viruses) to SARS-CoV-2 pathogenesis and clinical outcomes.
Severity of COVID-19
In SARS-CoV-2 infection, increased severity of the disease is attributed not only to the virus itself but also to an aggressive immune response marked by a cytokine storm leading to systemic inflammation and tissue damage57 (Fig. 1). It is believed that an ‘abnormal’ community of gut microorganisms is associated with hyperactive inflammatory responses in severe COVID-19 as well as with autoinflammatory and autoimmune disorders that occur during or after the infection58. It is assumed that, during the infection, commensal bacteria are replaced by opportunistic pathogens39,58,59. It remains unclear, however, whether it is the absence of beneficial bacteria or an active role played by dysbiotic bacteria that triggers and leads to sustained and excessive inflammation during severe COVID-19. It has been hypothesized that damage to the gut epithelium caused by SARS-CoV-2 infection leads to translocation of opportunistic pathogens and endotoxins in the gut, which could further exacerbate the systemic inflammatory response and predispose the individual to secondary infections, acute respiratory distress syndrome, multi-organ failure and even death20,21. Dysbiosis could also negatively influence the balance and recruitment of immune cells, such as mucosal-associated T cells, in the lungs through the gut–lung axis, which contributes to the development of respiratory tract infections22–24.
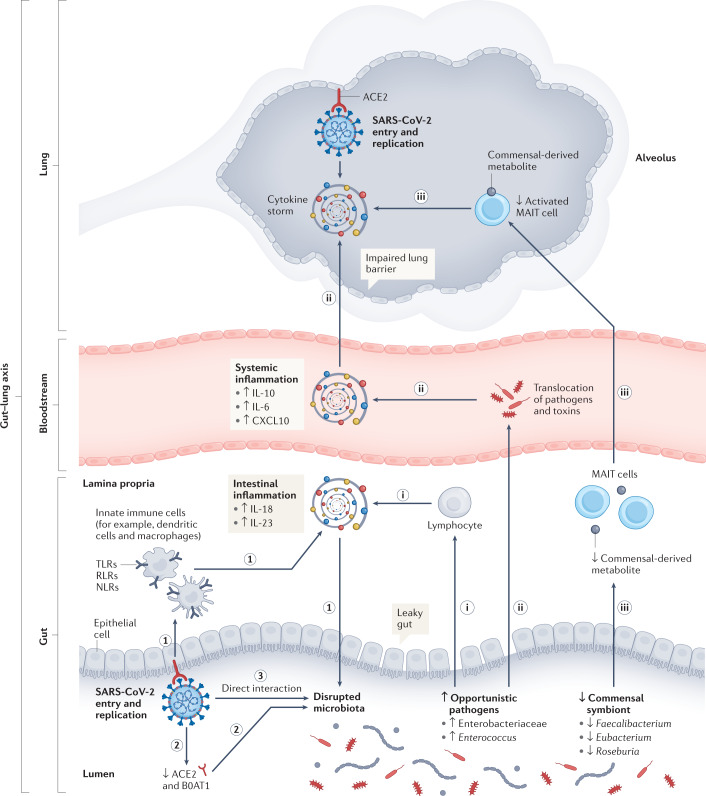
Fig. 1. Possible roles of the gut microbiota in dysfunctional immune responses and COVID-19 severity.
We propose potential mechanisms by which the gut microbiota can contribute to dysfunctional immune responses and coronavirus disease 2019 (COVID-19) severity. The first part hypothesizes that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection could lead to gut dysbiosis by several possible mechanisms: (1) invasion of SARS-CoV-2 can activate pattern-recognition receptors (Toll-like receptors (TLRs), RLRs, NLRs), which are recognized by innate immune cells, resulting in the release of various pro-inflammatory cytokines109. These activated immune responses could impair gut permeability, disrupt gut microbiota equilibrium and result in an increased abundance of opportunistic pathogens (for example, Enterobacteriaceae and Enterococcus) and decreased abundance of commensal symbionts (for example, Faecalibacterium, Eubacterium and Roseburia). (2) SARS-CoV-2 infection was found to downregulate the expression of angiotensin-converting enzyme 2 (ACE2) and B0AT1 (a molecular ACE2 chaperone) on the luminal surfaces of intestinal epithelial cells, which may facilitate the growth of the pathogens25,113,114. (3) An in vitro study found that SARS-CoV-2 might directly infect bacteria115. In the second part, we propose that specific intrinsic ‘microbiome signatures’ at the point of SARS-CoV-2 infection could influence the severity of infection and host immune response by several putative mechanisms: (i) increased opportunistic pathogens might be further recognized by innate lymphocytes and intensify gut pro-inflammatory responses185. (ii) Opportunistic pathogens and toxins could translocate into the circulatory system, causing bacteraemia and exacerbating systematic inflammation and disease severity20,21. (iii) Depleted commensal symbionts could negatively influence the recruitment of immune cells, such as activated mucosal-associated invariant T (MAIT) cells, to affect susceptibility and severity of respiratory tract infections22–24. CXCL10, C-X-C motif ligand 10.
In patients with COVID-19, studies (including a preprint) have shown a higher level of several plasma markers of gut permeability, including fatty acid-binding protein 2 (FABP2), peptidoglycan, lipopolysaccharide-binding protein (LBP) and faecal calprotectin, compared with healthy controls, indicating the disruption of gut barrier integrity during SARS-CoV-2 infection60,61. Interestingly, in a cohort of 60 patients with COVID-19, elevated levels of zonulin, a protein that modulates tight junctions of the gastrointestinal tract, were found to be associated with higher mortality, more severe illness and increased levels of markers of systemic inflammation (IL-6), highlighting the relationship between impaired intestinal barrier function and COVID-19 severity62. Furthermore, opportunistic pathogens from the Enterobacteriaceae family (such as Escherichia coli and Klebsiella pneumoniae) and the Enterococcus genus (such as Enterococcus faecalis) were found to be over-represented in the gastrointestinal tract of patients with COVID-19 who have critical illness27,34. Increased permeability and expansion of opportunistic pathogenic microorganisms in the gut might contribute to a high risk of bloodstream infections in COVID-19 (ref.63). Co-infections have been identified in 3–25% of patients in various studies64–67, including Klebsiella spp., Enterococcus spp. and E. coli infections, which generally accounted for up to 50% of death in patients with critical illness68.
In addition, several human studies reported that the abundance of butyrate-producing genera Faecalibacterium33,37 and Roseburia33,38,69 negatively correlated with disease severity. In a prospective study of 117 patients with COVID-19 from Germany, the abundance of Faecalibacterium and Roseburia negatively correlated with disease severity33. It remains unclear whether these microbial signals precede the infection or represent a consequence of the disease as only a few studies had included asymptomatic individuals. The relative abundance of a specific bacterial species, Spirochaetes, was higher in asymptomatic individuals with SARS-CoV-2 infection than in healthy individuals70. A study from Hong Kong of 100 patients with COVID-19 and 78 individuals without COVID-19 as controls showed that changes in the composition of the gut microbiota correlated with severity of COVID-19 and altered levels of blood inflammatory markers37. Depletion of bacterial species with immunomodulatory potential, including F. prausnitzii and Eubacterium rectale, was linked to increased serum levels of pro-inflammatory mediators, plasma TNF, IL-10, C-X-C motif ligand 10 (CXCL10) and C-X-C motif ligand 2 (CXCL2)37. These data suggest that some microorganisms, such as F. prausnitzii and E. rectale, which are commonly abundant in healthy individuals, might have a role in limiting or preventing inflammation37. Intriguingly, altered gut microbiota composition persisted for at least 30 days after SARS-CoV-2 infection had been cleared37, and the changes seen, such as depletion of F. prausnitzii, were similar to those reported in other chronic inflammatory conditions58. Others have reported that the abundance of Eubacterium ramulus showed a negative correlation with IL-6, and E. faecalis had a negative correlation with IL-4 and plasma CD8+ T cells in patients with COVID-19 (ref.71). These cytokines and chemokines are associated with an interferon-driven T helper 1 (TH1) response72, indicating that the gut microbiota might modulate the systemic TH1 response in COVID-19. Moreover, faecal levels of IL-18, which contributed to the breakdown of barrier integrity, showed a positive correlation with the abundance of Peptostreptococcus, Citrobacter and Fusobacterium, highlighting a possible role of the gut microbiota in affecting gut permeability and inflammation during SARS-CoV-2 infection35,73. Further studies to dissect the relationship between cytokine release and shifts in gut microbiota will facilitate our understanding of whether these microbial changes directly contribute to cytokine storms in patients with COVID-19 or represent a consequence of critical disease.
The oral cavity has been shown to be an important site for SARS-CoV-2 infection and human saliva could be a potential route of SARS-CoV-2 transmission74. Accumulating data have reported associations between the oral microbiome and COVID-19 severity, host immune response, and SARS-CoV-2 viral load36,75–78. Similar to the gastrointestinal tract, the oral microbiome of patients with COVID-19 was characterized by reduced diversity of bacteria36,75. Interestingly, some bacterial genera and species, such as Rothia mucilaginosa36, Granulicatella36, Veillonella28,36 and Campylobacter36,79, were found to be significantly (all P < 0.05) enriched in both oral and faecal samples of patients with COVID-19. The oral microbiome of patients with COVID-19 also showed elevations in xenobiotic biodegradation and amino acid metabolism78. Importantly, a number of bacteria of the genus Veillonella that were enriched in the oral cavity of 31 patients with COVID-19 were reported to be over-represented in the bronchoalveolar lavage fluid (BALF) of a patient with COVID-19 (ref.78). These data suggest that the oral cavity might serve as a possible reservoir for microorganisms to induce co-infections in the lungs and as a potential source through which translocated species contribute to gut dysbiosis in COVID-19.
Post-acute COVID-19 syndrome
Features
Following acute infection with SARS-CoV-2, a systematic review of 45 studies including 9,751 participants with COVID-19 concluded that 73% of patients develop persisting debilitating symptoms, a condition termed post-acute COVID-19 syndrome (PACS), commonly known as ‘long COVID’80. PACS can affect multiple organs, including persistent respiratory symptoms (such as coughing or shortness of breath), cardiovascular symptoms (such as chest pain or palpitations), gastrointestinal symptoms (such as loss of appetite or diarrhoea), neuropsychiatric symptoms (such as anxiety or insomnia), musculoskeletal symptoms (such as joint pain or muscle weakness) and dermatological symptoms (such as skin rash or hair loss)81–83 (Fig. 2). Patients with PACS had a higher risk of prolonged health impairments 6 months after the acute infection, and PACS is associated with reduced health-related quality of life and physical functions81,84. Although the exact cause of PACS is not clear, it has been hypothesized that several mechanisms, either alone or in combination, could contribute to this syndrome, including the persistence of residual viral structures, perturbations of inflammatory and immune responses, and tissue or organ damage85. Some of these potential mechanisms resemble that of post-infectious irritable bowel syndrome (PI-IBS), a comparable condition whereby long-term gastrointestinal symptoms persist following acute gastroenteritis86,87. It was reported that between 1.02% (15 of 1,475) and 7.14% (20 of 280) of patients with COVID-19 developed symptoms of IBS after acute SARS-CoV-2 infection86,87. We hypothesize that these symptoms might be part of an emerging phenomenon known as ‘post-COVID IBS’88,89. Interestingly, gastrointestinal sequelae were reported to be greater in patients with acute gastrointestinal symptoms during the acute phase of SARS-CoV-2 infection than in patients without acute gastrointestinal symptoms88. The roles of the gut microbiota and host immune response that underlie the physiological manifestations of PI-IBS could also be a factor and warrant further investigations. Vaccination against SARS-CoV-2 only conferred limited protection against PACS when comparing 6-month risks of incident post-acute sequelae in 16,035 patients with COVID-19 with prior vaccination and 48,536 patients with COVID-19 without prior vaccination90. Thus, reliance on vaccination as a sole alleviation approach might only partially reduce the long-term health consequences of SARS-CoV-2 infection. As the human gut microbiota has a crucial role in the maturation and modulation of the immune system91, an aberrant immune response to SARS-CoV-2 infection induced by resident microorganisms could also affect the convalescent phase (Fig. 2).
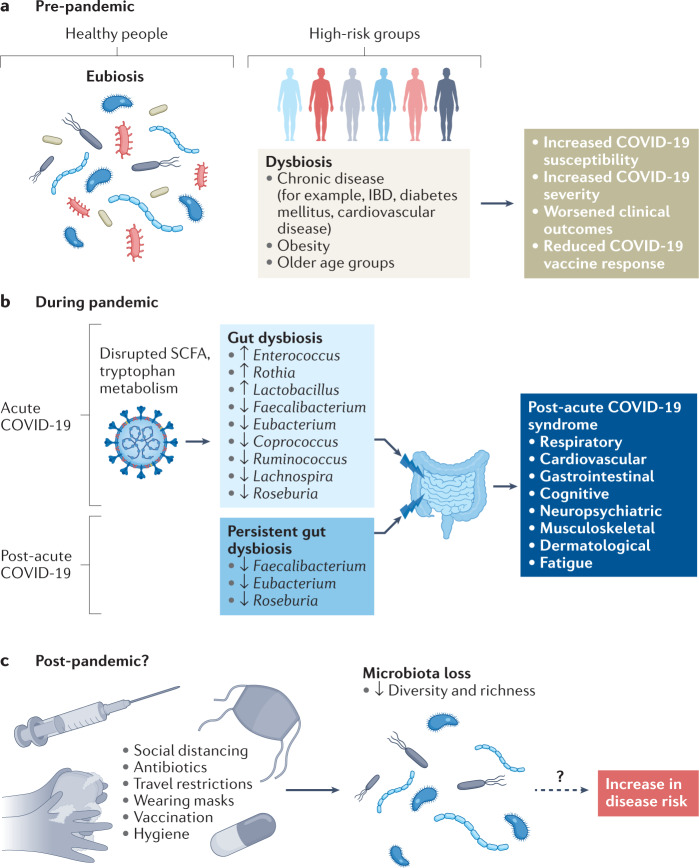
Fig. 2. Proposed model of gut microbiome changes pre-pandemic, during pandemic, and post-pandemic and how COVID-19 measures influence microbiota diversity during an individual’s lifetime.
a, Prior to the coronavirus disease 2019 (COVID-19) pandemic, the gut microbiota in a healthy individual was characterized by ‘eubiosis’, a balanced gut ecosystem with rich microbial diversity, whilst certain individuals, including older age groups and those with chronic diseases such as inflammatory bowel disease (IBD), diabetes mellitus, cardiovascular disease and obesity, had an altered gut ecosystem with reduced microbial diversity and altered gut microbial composition104,105. ‘Dysbiosis’ could contribute to increased susceptibility to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, increased COVID-19 severity, worsened clinical outcomes and/or decreased COVID-19 vaccine response. b, During the pandemic, acute COVID-19 infection was associated with consistent gut microbiota composition changes and impaired short-chain fatty acid (SCFA) biosynthesis20,120,124 and disrupted tryptophan metabolism127–130. Dysbiosis seen in the initial infection was also associated with post-acute COVID-19 syndrome, including chronic respiratory symptoms (for example, coughing or shortness of breath), cardiovascular symptoms (for example, chest pain or palpitation), gastrointestinal symptoms (for example, loss of appetite or diarrhoea), neuropsychiatric symptoms (for example, anxiety or insomnia), musculoskeletal symptoms (for example, joint pain or muscle weakness), and dermatological symptoms (for example, skin rash or hair loss)81–83. In the post-acute COVID-19 phase, the gut microbiota remained persistently disrupted, characterized by persistent depletion of SCFA-producing bacteria Faecalibacterium, Eubacterium and Roseburia83,93. Alterations of gut microbiota composition in the post-acute stage were also associated with multi-organ post-acute COVID-19 syndrome83. c, Beyond the pandemic, existing pandemic control practices with strict implementation of social distancing, extensive hygiene measures, regular vaccination and restricted travel could negatively affect microbiome diversity in infants and have substantial effects on early-life bacterial colonization in the gut, with unknown consequences for disease risk. The key microbial changes consistently reported in multiple human studies are presented.
Gut microbiome analysis
Several groups have reported differences in the gut microbiome in individuals who developed PACS compared with those who made a complete recovery from COVID-19 (refs.83,92–95). Microbial differences were detected at the time of diagnosis but were exaggerated after 6 months83,92. In particular, microbiota richness was not restored to normal conditions after discharge in patients with PACS. Persistent symptoms can be associated with the presence of a small quantity of residual virus in immunopriviledged tissues, especially regions of the body such as the gut that are not directly protected by antibodies96,97. A striking observation was that dysbiosis persisted for months after the clearance of the virus. Compared with healthy individuals as controls, those recovered from COVID-19 had reduced bacterial diversity and richness at 3 months, accompanied by a lower abundance of beneficial commensals and a higher abundance of opportunistic pathogens83,98. The relative abundance of members of Bifidobacterium and Ruminococcus remained significantly (P < 0.001) depleted in patients with COVID-19 compared with controls at 6-month follow-up83.
An observational study showed that respiratory dysfunction after discharge in 85 patients with COVID-19 was associated with dysbiosis based on 16S ribosomal RNA sequencing of faecal samples as well as raised plasma LBP levels93. Respiratory dysfunction assessed by diffusing capacity of the lungs for carbon monoxide (DLCO) after 3 months of discharge from hospital was linked to increased abundance of the genera Flavonifractor and Veillonella (potentially linked to fibrosis) and reduced abundance of several members of the Lachnospiraceae family (Lachnospiraceae FCS020 group, Eubacterium ventriosum group, Fusicatenibacter, Lachnospiraceae ND3007 group) and Ruminococcaceae family (Ruminococcus, Subdoligranulum); the latter microbial communities were butyrate producers. These data point to the potential involvement of gut–lung crosstalk in relation to long-term pulmonary dysfunction and long COVID symptoms.
In a prospective study that tracked longitudinal dynamics of gut microbiota of 106 hospitalized patients with COVID-19 in Hong Kong, approximately three-quarters of patients developed PACS (commonly fatigue, poor memory and anxiety) 6 months after the infection83. Shotgun metagenomic analysis of faecal samples showed significantly lower microbial diversity and reduced types of bacteria compared with individuals without PACS and healthy individuals as controls83. Patients with PACS also had an increased abundance of Bacteroides vulgatus and Ruminococcus gnavus and a reduced abundance of F. prausnitzii. Interestingly, B. vulgatus was also shown to be sixfold elevated in the faecal samples of patients (n = 11) with PI-IBS compared with healthy individuals (n = 11)86,99; this finding suggests that B. vulgatus is potentially linked to the pathogenesis of both PACS and PI-IBS. Moreover, having respiratory symptoms at 6 months post-SARS-CoV-2 infection was associated with increased levels of opportunistic pathogenic species such as Streptococcus vestibularis and Streptococcus anginosus, whereas fatigue and neuropsychiatric symptoms were associated with nosocomial pathogens such as Clostridium innocuum and Actinomyces naeslundii83. Butyrate-producing bacteria were substantially decreased in patients with hair loss and certain bacteria, such as Bifidobacterium pseudocatenulatum and F. prausnitzii, had the largest inverse correlations with PACS development83. Bacterial species, including B. longum and Blautia wexlerae, at admission were negatively associated with the development of PACS at 6 months, implying a potential protective role of these species in the convalescent phase83. By contrast, Atopobium parvulum, Actinomyces johnsonii and Actinomyces sp. S6 Spd3 were enriched in patients with PACS. These findings suggest that a person’s gut microbiome configuration at the time of infection might affect their susceptibility to long-term complications of COVID-19. Nonetheless, these changes could represent reactive changes to PACS, and future research needs to include prospective longitudinal studies of non-hospitalized patients followed from the time of infection until symptom development to delineate the exact contribution of dysbiosis to PACS symptoms.
Mechanistic links
ACE2 and the gut microbiota
ACE2, an entry receptor for SARS-CoV-2, is highly expressed in small bowel enterocytes100. ACE2 can regulate microbial ecology, innate immunity and dietary amino acid homeostasis by maintaining the balance of the renin–angiotensin system and amino acid transporter B0AT1 (also known as SLC6A19)–ACE2 complex25,26. Several animal studies have explored the effect of gut microbiota on intestinal ACE2 expression. Intestinal and lung ACE2 expression levels were markedly higher in germ-free mice than in conventional mice101. Gnotobiotic mice colonized with different microbiota showed variability in intestinal ACE2 expression, which could partially be attributable to differences in types of microbiome-encoded proteases and peptidases101. A study published in 2021 identified transcriptional factors regulating ACE2 expression in the gut102, including GATA4, which was known to be regulated by the gut microbiota70. These data suggest that the gut microbiota might play a part in regulating ACE2 expression.
Moreover, specific bacterial species, such as Bacteroides dorei and Bifidobacterium longum, were shown to inhibit colonic ACE2 expression in mouse models21,103. In addition, four Bacteroides species associated with downregulation of ACE2 expression in the mouse colon103 showed significant (P < 0.05) inverse correlations with faecal SARS-CoV-2 load39. High-risk groups succumbing to COVID-19 were often those with comorbid conditions, such as diabetes, cardiovascular disease and obesity, which were also associated with microbiome abnormalities, characterized by reduced bacterial diversity104,105. This finding suggests that gut microbiota prior to infection might contribute to host susceptibility to SARS-CoV-2 and ACE expression106,107 (Fig. 2). In addition, other SARS-CoV-2 entry factors, such as host glycosaminoglycan heparan sulfate necessary for virus cellular binding, can be modified by bacteria such as Bacteroides108.
SARS-CoV-2 infection and dysbiosis
To date, the majority of clinical studies have shown associations between SARS-CoV-2 infection and an altered gut microbiome, although it remains unclear whether changes were a cause or effect of the infection. We hypothesize several putative mechanisms by which SARS-CoV-2 can result in gut dysbiosis. Invasion of SARS-CoV-2 into the lungs could cause tissue damage and activate strong pro-inflammatory pathways characterized by upregulation of NF-κB and TNF pathways to induce the cytokine storm57,109,110. Alternatively, gut infection could cause direct impairment of intestinal structure and breakdown of the intestinal epithelial barrier and promote intestinal inflammation111. Activated systematic and intestinal inflammation might contribute to changes in gut microbiota112. SARS-CoV-2 infection could downregulate the expression of ACE2 and B0AT1 (a molecular ACE2 chaperone) on the luminal surfaces of intestinal epithelial cells, which might then facilitate pathogen growth25,113,114. An in vitro study observed that SARS-CoV-2 might function like bacteriophages and directly infect bacteria115, revealing another possible mechanism of SARS-CoV-2 affecting the gut microbiota.
Four animal studies have provided evidence that SARS-CoV-2 infection might play a part in driving alterations in gut microbiota ecology56,116–118. When a non-human primate model of rhesus macaques and cynomolgus macaques was challenged with SARS-CoV-2, the gut microbiome changed gradually from day 0 until day 13 post-infection117. An increase in abundance of Acinetobacter and genera of the Ruminococcaceae family was associated with the presence of SARS-CoV-2 in the upper respiratory tract, whereas a decrease in SCFA levels and changes in the levels of tryptophan and several bile acid metabolites were observed in infected animals117. In addition, hamster models were able to recapitulate some of the hallmark features of severe COVID-19 seen in humans. SARS-CoV-2 infection resulted in an over-representation of deleterious bacterial taxa, such as Desulfovibrionaceae and Enterobacteriaceae, a lower abundance of SCFA-producing bacteria, and faecal SCFA, in line with findings seen in humans118. However, the infectious virus has not been detected in the gut of hamsters suggesting that alterations of gut microbiota could be, in part, secondary to systematic inflammation caused by SARS-CoV-2 infection in the lungs118. Supplementation of a combination of sodium acetate, propionate and butyrate in the hamster model challenged by SARS-CoV-2 failed to interfere with SARS-CoV-2 replication in the lungs or ameliorate gut inflammation at the gene expression level118. In a study using human colon biopsy samples obtained from healthy individuals, the addition of SCFAs (including acetate, propionate and butyrate) had minimal effects on the expression of antiviral and anti-inflammatory-related genes or viral load in intestinal cells119. Whether changes in the gut microbiota can directly affect the clinical manifestations of COVID-19 remains largely unknown although strong associations between COVID-19 severity and specific bacteria and metabolites have been consistently observed in human studies.
Microbial metabolites in COVID-19
Emerging human studies determining the functional capacity of gut microbiota and the faecal metabolomic and proteomic profile in patients with COVID-19 have provided deeper insights into understanding microbiota–host interactions and their consequences on disease outcomes. SARS-CoV-2 infection was associated with alterations in carbohydrate, lipid and amino acid metabolism of the gut microbiota38,45,120–123. Several studies have shown impaired SCFA biosynthesis in faecal samples of patients with COVID-19 (refs.20,120,124). In a metagenomic analysis of 66 antibiotics-naive patients with COVID-19 and 70 individuals without COVID-19 (ref.124), patients with SARS-CoV-2 infection showed a reduced capacity of their gut microbiota for SCFA biosynthesis, which negatively correlated with disease severity and elevated plasma concentrations of the pro-inflammatory cytokine IL-10 and the chemokine CXCL10 (ref.124). Consistently, 19 patients with COVID-19-related severe and/or critical illness showed reduced faecal concentrations of SCFAs, including acetate, propionate, butyrate, valeric acid and caproic acid, through measurement of faecal metabolites124. SCFAs can activate anti-inflammatory responses of immune cells, inhibit inflammatory signalling pathways125 and maintain the integrity of the gut barrier to prevent translocation of gut endotoxins and bacteria into the circulation, thereby alleviating local and systemic inflammatory responses126. Given the importance of SCFAs in regulating host immune response, the deficiency of SCFA biosynthesis in COVID-19 could be associated with disease pathogenesis and severity. However, whether SCFA depletion is a cause or consequence of COVID-19 infection remains to be elucidated.
Several studies measuring plasma metabolites of patients with COVID-19 have shown disrupted tryptophan metabolism and augmented activation of the kynurenine pathway, which is involved in tryptophan metabolism, when compared with healthy individuals as controls127–130. Tryptophan metabolism is linked to autoimmunity, viral infections and gut health through regulation of the ratio of regulatory T cells to TH17 cells and B cell activity131. Increased metabolites of the kynurenine pathway transported into the brain could trigger symptoms such as fatigue, poor memory and depression in both human and animal studies130,132, which are common symptoms of ‘long COVID’130. Importantly, tryptophan metabolites are key mediators of the host–microbiota interface. The gut microbiota can directly use tryptophan as a substrate and influence host tryptophan absorption and metabolism to regulate host physiological and immune responses according to evidence from both human and animal studies133. Endogenous host tryptophan metabolites can profoundly influence gut microbiota composition and functions such as Akkermansia and Lactobacillus134,135. Taken together, these data suggest tryptophan metabolism as a possible mechanism by which gut microbiota is involved in COVID-19.
A reduced concentration of sphingolipids in sera136 and faeces121 and altered gut microbial sphingolipid metabolism120 were reported in patients with COVID-19. Sphingolipids are components of biomembranes, mediating signal transduction and immune activation137. Sphingolipids produced by Bacteroides can increase exogenous sphingolipids138 and thus enhance differentiation of regulatory T cells as observed in in vitro or in vivo studies139, which could inhibit replication of coronaviruses. This observation supports the hypothesis that gut microbiota-derived sphingolipids might regulate host defence against SARS-CoV-2 infection.
Additionally, increased faecal sucrose and depleted faecal glucose levels were reported in 56 patients with COVID-19 compared with 47 healthy individuals as controls122. Abnormal levels of sucrose and glucose might be associated with impaired sucrase-isomaltase activity140,141. This change could be associated with common intestinal symptoms, such as diarrhoea, vomiting, flatulence and abdominal pain, in COVID-19 (ref.140). Flatulence is typically caused by the fermentation of unabsorbed carbohydrates in the intestine by bacteria142. Increased levels of sucrose have been associated with enriched levels of Actinomyces and Streptococcus parasanguinis122, implying that dysbiosis in COVID-19 might disrupt intestinal fermentation and contribute to gastrointestinal symptoms.
Vaccine immune response and dysbiosis
As of the end of August 2022, over 12.4 billion doses of SARS-CoV-2 vaccines have been administered (156 vaccines per 100 people) worldwide according to the World Health Organization. Different types of vaccines developed to elicit effective immune responses to protect from COVID-19 or to reduce disease severity if infected have changed the course of the pandemic143,144. Yet, there is a great variation in vaccine response among different individuals in different populations. A growing number of animal and clinical studies have shown the critical role of gut microbiota composition and function in modulating responses to vaccines such as trivalent influenza vaccine and oral rotavirus vaccine145. The gut microbiota can influence vaccine immunogenicity, including the activation of antigen-presenting cells through pattern-recognition receptors by bacteria-derived molecules, such as flagellin and peptidoglycan, that act as potent natural adjuvants to enhance immune response146. In particular, bacterial flagellin was shown to be a critical contributor to the antibody response to the flu vaccine through Toll-like receptor 5 (TLR5) sensing147. In germ-free and antibiotic-treated mice, NOD2-mediated sensing of bacterial peptidoglycan enhanced immune responses to intranasal cholera toxin vaccine148. The gut microbiota could also modulate vaccine response via bacteria-derived metabolites such as SCFAs, which upregulate gene expression for antibody response and promote plasma B cell differentiation according to in vivo evidence149.
A prospective study published in 2022 characterized the gut microbiome associated with immune responses against an inactivated vaccine (CoronaVac, Sinovac) or an mRNA vaccine (BNT162b2, BioNTech, Comirnaty) and associated adverse events in 138 individuals who had received a COVID-19 vaccine150. The relative abundance of Bifidobacterium adolescentis in high responders of the CoronaVac vaccine was significantly (P = 0.023) higher than that of low responders prior to vaccination, with enrichment of carbohydrate pathways related to B. adolescentis150. One month following the second dose of CoronaVac vaccination, B. adolescentis remained more abundant in high responders150. Furthermore, the neutralizing antibody response was positively correlated with the abundance of butyrate-producing bacteria with flagella and fimbriae, including Roseburia faecis150. Interestingly, the abundance of Prevotella copri and two Megamonas species before COVID-19 vaccination were associated with fewer adverse events among both groups of vaccinees, implying that these bacteria might play anti-inflammatory roles in immune response150. A prospective study in 43 patients with inflammatory bowel disease also showed associations between gut dysbiosis and serological response against BNT162b2 or ChAdOx1 nCoV-19 (Vaxzevria, AstraZeneca) vaccines151. Bilophila abundance and levels of the faecal metabolite trimethylamine were positively associated with vaccine response, whereas Streptococcus abundance and levels of metabolites, including phenylalanine, phenylacetate and succinate, showed a negative association151. These findings indicate the importance of SCFAs, pattern-recognition receptor-mediated sensing and disease-associated gut dysbiosis in modulating COVID-19 vaccine immunogenicity. A study also showed a slight shift in the gut microbiota after the first dose of vaccine, although it is unclear if microbial changes were caused by vaccination or due to COVID-19-related measures as a control group who did not receive vaccines was not studied150. Others have shown that an mRNA vaccine led to enriched oral microbial diversity, decreased abundance of Bacteroides and changes in the gut microbiome during the course of SinoVac vaccine in 40 healthy individuals152.
To date, one of the main challenges regarding COVID-19 vaccination is the waning of protection over time, even for booster doses153,154. Those aged 60 years or above, those who are immunocompromised, or those with comorbidities, such as obesity, are highly susceptible to the rapid waning of vaccine immunogenecity154,155. It has been hypothesized that gut dysbiosis could be one of the mechanisms by which these risk factors cause weakened defences against SARS-CoV-2 infection following vaccination. In young children aged 3–5 years, COVID-19 vaccine (CoronaVac, Sinovac) effectiveness has been shown to be suboptimal156. Thus, these vulnerable individuals or those with contraindications to COVID-19 vaccines might potentially benefit from microbiota-based interventions in complementing the effectiveness of COVID-19 vaccines. Clinical trials on the efficacy of microbiota-based interventions in improving COVID-19 vaccine immunogenicity, reducing vaccine-associated adverse events and prolonging vaccine effectiveness over time are warranted.
Consequences on the human microbiome
Current pandemic control measures have been proposed to have large, uneven and potentially long-term detrimental effects on the human microbiome globally with strict implementation of social distancing, extensive hygiene measures, restricted travel and other measures that influence overall microbial loss157. A small study comparing pre-pandemic gut microbiota of 23 healthy individuals from Buenos Aires, Argentina, and its variation during social isolation found a significant (P < 0.05) decrease in phylogenetic diversity and in the phylum Verrucomicrobiota, which contributes to intestinal health and glucose homeostasis, during isolation158. The COVID-19 pandemic has been associated with a markedly lower Actinobacteria (Actinomycetota) richness, higher Bacteroidetes (now known as Bacteroidota) richness, and an altered abundance of antibiotic-resistance genes in faecal samples collected from 32 healthy individuals before and during the first wave of the pandemic159.
Birth and early infancy are critical windows for microbiome establishment and development, and microbial colonization in early life can influence subsequent risks of common illnesses, including eczema, asthma, autoimmunity, allergy and diabetes41–44. It has been hypothesized that the pandemic could have disrupted the gut bacterial composition of infants and created other long-term complications in their later life due to increased hygiene, restricted social exposure and decreased travel, which are essential factors affecting early-life bacterial colonization in the gut160. Preliminary analyses of faecal microbiome (n = 307) collected from infants born before and during the pandemic in a neonatal intensive care unit in the USA showed marked differences in microbiome diversity, highlighting that the COVID-19 pandemic might cause a loss of microbial diversity and richness in infants161. Studies have demonstrated that the pandemic increased the risk of childhood diseases, including obesity, myopia and mental health disorders with prolonged home confinement160,162–165. More work is needed to investigate and confirm potential links between the pandemic, microbiota loss and future disease risk. Although our knowledge of COVID-19 and the microbiome is incomplete, future infection control measures that result in microbial loss need to be balanced with strategies that promote microbial diversity to ensure health benefits for future generations (Fig. 2).
Microbiota-based interventions
Clinical trials using dietary modification, probiotics, prebiotics and faecal microbiota transplantation (FMT) are under way to determine their effectiveness in the treatment of acute COVID-19 and in enhancing the effectiveness of the SARS-CoV-2 vaccine. Supplementary Table 2 summarizes clinical trials involving modulation of the gut microbiome for COVID-19 management.
Antibiotic stewardship
In the early pandemic, antibiotics were commonly used. A meta-analysis estimated that three in four patients with COVID-19 globally had been prescribed antibiotics, and such proportion is markedly above the estimated incidence of bacterial co-infections in COVID-19, which stands at only 8.6%166. Clinical outcomes in 45 patients with moderate COVID-19 showed no difference between those who had and had not received antibiotics, suggesting that antibiotics were not beneficial in improving clinical outcomes of COVID-19 (ref.37). Marked gut dysbiosis has been reported in patients with COVID-19 who have received antibiotics during hospitalization37, and antibiotic-induced gut dysbiosis impaired immune responses to seasonal influenza vaccines in humans167. A longitudinal study of 200 patients with COVID-19 showed that less intake of antibiotics in the year prior to COVID-19 was associated with milder disease severity and more rapid clearance of SARS-CoV-2 (ref.168). Thus, antimicrobial stewardship is critical to preventing antibiotic-induced dysbiosis, severe COVID-19 and the risk of antimicrobial resistance in patients with COVID-19.
Dietary modification
Dietary modification has been proposed as a means to modulate the gut microbiota and improve the clinical outcomes of COVID-19 (Fig. 3). Several population studies have provided epidemiological evidence that specific dietary habits were associated with COVID-19-related mortality and disease outcomes. In a large prospective survey of 592,571 participants in the UK and USA, a diet rich in plant food was linked to reduced risk and severity of COVID-19 (ref.169). A retrospective study of 509 patients with COVID-19 found statistically significant associations between a vegetarian diet and milder COVID-19 severity in people aged >65 years, whilst non-vegetarian individuals were found to be more prone to developing critically severe COVID-19 (ref.170). Higher intake of prebiotic-rich food and less intake of sugar in the year prior to COVID-19 was also associated with milder disease severity and faster clearance of SARS-CoV-2 in a cohort of 200 people168. A multicentre study of 7,337 individuals showed that, in patients with COVID-19 with type 2 diabetes mellitus, clinical outcomes could be improved by well-controlled blood glucose, highlighting the potential of low glycaemic index dietary modification with increased intake of vegetables and fruit in improving outcomes among hospitalized patients171. Dietary patterns across European countries were found to be associated with ACE2 activity and death rate172, suggesting the possible mechanistic link between diet and COVID-19 mortality. A case report revealed improvement in severe post-acute gastrointestinal symptoms in a patient with COVID-19 with high-fibre intervention containing brans, inulin and Fibersol-2, which was associated with gut bacteria compositional shifts with increased SCFA-producing bacteria, including Oscillibacter sp., Anaerofustis sp., Blautia spp. and Eubacterium hallii173. Understanding the effect of diet on microbial changes in response to coronavirus will pave novel dietary interventions during the COVID-19 pandemic174.
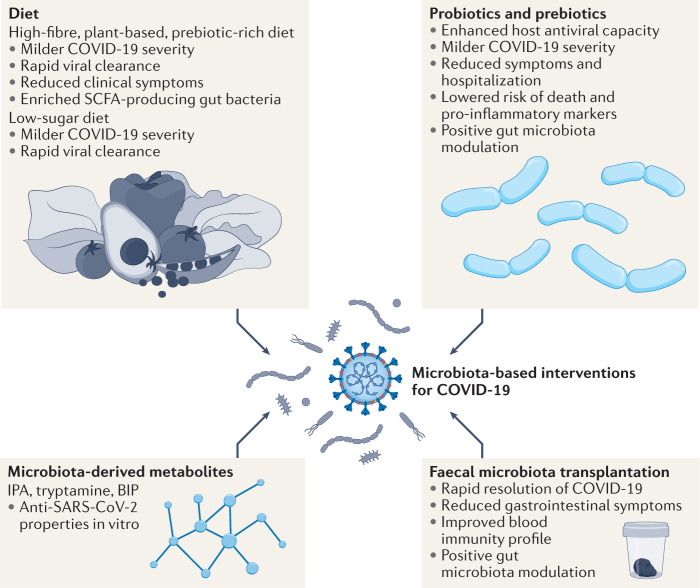
Fig. 3. Microbiota-based interventions in COVID-19.
Multiple studies have suggested the role of diet, probiotics, and microbiota-derived metabolites and faecal microbiota transplantation in enhancing antiviral capacity and improving clinical outcomes of coronavirus disease 2019 (COVID-19), such as severity and symptoms, potentially through the modulation of gut microbiota. Nevertheless, many of these preliminary conclusions were drawn based on association and retrospective analyses. Further animal and clinical studies are warranted to elucidate the mechanistic links underlying the therapeutic effects of these microbiota-based interventions. BIP, 2,5-bis(3-indolylmethyl)pyrazine; IPA, N6-(Δ2-isopentenyl)adenosine; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SCFA, short-chain fatty acid.
Probiotics and prebiotics
Several small to medium retrospective studies and clinical trials with sample sizes ranging from 55 to 411 have demonstrated that administration of oral probiotics and prebiotics induced antiviral activity and had positive effects on gut microbial composition and diversity and was associated with improved clinical outcomes in patients with COVID-19 (refs.175–180). Two retrospective studies including 200 and 411 hospitalized patients with COVID-19 have shown that use of the probiotic species Bifidobacterium, Enterococcus, Lactobacillus and Streptococcus was associated with more rapid clinical improvement by 3 days, decreased hospitalization length and reduced risk of death in patients with severe pneumonia175,176. Results of a randomized controlled trial involving 200 patients with COVID-19 showed that a 14-day probiotic supplementation of Bacillus coagulans, Bacillus subtilis and Bacillus clausii combined with systemic enzymes with immunomodulatory potential (serratiopeptidase, bromelain, amylase, lysozyme, peptidase, catalase, papain, glucoamylase and lactoferrin) improved physical and mental fatigue in individuals recovered from COVID-19 (ref.177). Another randomized controlled trial of 300 participants also found that supplementation of a probiotic formula consisting of Lactiplantibacillus plantarum and Pediococcus acidilactici in patients with COVID-19 was associated with a reduction in nasopharyngeal viral load, lung infiltrates, shortened duration of clinical symptoms by 2–7 days and improved immune response in the intervention group compared with the control group receiving placebo178. Mechanisms of probiotics were related to improved immune response and positive compositional shifts of the gut microbiota with an increased similarity to the microbial composition of the gut microbiota in healthy individuals as well as a decrease in COVID-19-associated bacteria as shown in pilot studies179,180. In an open-label study of 55 hospitalized patients with COVID-19, a higher proportion of individuals (88% versus 63.3%) who received 4 weeks of a synbiotic formula consisting of Bifidobacterium strains and prebiotics (SIM01) achieved resolution of clinical symptoms, increased IgG antibodies against SARS-CoV-2, and reduced blood pro-inflammatory markers such as IL-6, CCL2, M-CSF, TNF and IL-1RA, than individuals in the standard treatment arm180. There was also an increase in the abundance of commensal bacteria, such as Bifidobacterium, Eubacterium and Faecalibacterium species, and a decrease of opportunistic pathogens, such as E. coli and Bacteroides species, in the gut microbiota of individuals who received SIM01 (ref.180). A longitudinal cohort study of 200 individuals showed that regular intake of probiotic yoghurt in the year prior to COVID-19 was associated with milder disease severity168.
As of August 2022, 31 clinical studies have been registered on ClinicalTrials.gov and Supplementary Table 2 summarizes all current clinical trials evaluating the efficacy of probiotics or synbiotics in treating and preventing COVID-19. The majority of clinical trials focused on the use of probiotic species in the Lactobacillus, Bifidobacterium and Enterococcus genera for COVID-19 treatment or prevention. Live biotherapeutic products, regarded as biological medicinal products that contain live microorganisms for human therapeutic use (such as a Lactococcus lactis engineered to secrete IL-10)181, have not been studied for COVID-19 but represent a promising avenue because of a multifactorial mode of action. An in vitro study using bacterial extracts obtained from human samples has identified three novel anti-SARS-CoV-2 metabolites, N6-(Δ2-isopentenyl)adenosine, tryptamine and 2,5-bis(3-indolylmethyl)pyrazine182, which are structurally or functionally comparable to synthetic COVID-19 drugs, including remdesivir, fluvoxamine and favipiravir. Further in vivo investigations and clinical trials are needed to determine the safety and efficacy of live biotherapeutic products and microbiota-derived metabolites for COVID-19.
Faecal microbiota transplantation
FMT aims to restore microbial dysbiosis via the transfer of a healthy microbiome to an individual with a disease. Two case reports have shown the use of FMT in COVID-19 (refs.183,184). Improved gastrointestinal symptoms were reported in 5 of 11 individuals with COVID-19 (ref.183), and favourable improvement was observed in blood immunity markers and gut microbiota composition with increased abundance of Bifidobacterium and Faecalibacterium. In two patients with COVID-19 and concurrent recurrent Clostridioides difficile infection, FMT treatment seemed safe and COVID-19-related respiratory symptoms rapidly resolved within 1 month after FMT184. Nonetheless, many of these preliminary conclusions regarding the use of FMT and probiotics were drawn based on association and retrospective analyses. Further animal and clinical studies are warranted to elucidate the mechanistic links underlying the therapeutic effects of these microbiota-based interventions.
Conclusions
Rapidly expanding knowledge of the importance of the gut microbiota in COVID-19 represents a paradigm shift in understanding the contribution of specific microorganisms to host susceptibility and immune response to SARS-CoV-2 infection. Observational studies have provided important insights into the role of dysbiosis in acute and post-acute COVID-19 conditions and their relationship with disease severity and vaccine immunogenicity. Preliminary clinical studies have revealed potential modulatory effects of probiotic bacteria, mainly Bifidobacterium and Lactobacillus species, against SARS-CoV-2 infection. Although there are limited large-scale clinical trials, we believe this field is growing with tremendous opportunities, and future research and directions have been illustrated in Box 1. Collectively, these data have provided an unprecedented opportunity to drive microbiota discoveries towards clinical applications during the different waves of the pandemic. With more breakthroughs in metagenomics, metabolomics, viral immunology and novel microbiome therapeutics, these microbiota discoveries will hopefully take us into a new paradigm to combat COVID-19 in the near future.
Box 1 Future research and directions.
Prevention of COVID-19
To determine microbial risk factors for susceptibility to SARS-CoV-2 infection
To understand the interplay between keystone species with SARS-CoV-2 viral entry factors, including ACE2 and heparan sulfate
To understand the role of the gut microbiota as a potential reservoir for recurrent or breakthrough SARS-CoV-2 infection and continuous immune cell stimulation
To develop dietary and microbiota-based interventions to reduce host susceptibility to SARS-CoV-2 infections
To evaluate the efficacy of microbiota-based interventions in improving vaccine immunogenicity, prolonging vaccine effectiveness and reducing vaccine-associated adverse events
Acute COVID-19
To explore mechanisms of host–microorganism interactions in response to SARS-CoV-2 infection and severity
To determine the effects of SARS-CoV-2 infection on other microbial components such as fungi, viruses and archaea
To understand how the gut virome and mycobiome calibrate host immunity and regulate severity of SARS-CoV-2 infection
To identify keystone bacteria and mechanisms specific to COVID-19 in different populations
To explore the effect of different variants of concern (delta, omicron) on the gut microbiota
To develop novel oral live biotherapeutics in improving COVID-19 outcomes in hospitalized patients
Post-acute COVID-19
To elucidate how gut microbiota dysbiosis contributes to PACS and persistent symptoms affecting different organs
To define microbial enterotypes for predicting the risk of developing post-acute complications
To evaluate the effects of residual SARS-CoV-2 viral particles on gut microbiota and PACS
To develop microbial therapeutic approaches for the prevention and treatment of PACS
Beyond COVID-19 pandemic
To track changes in the gut microbiome during COVID-19 and its effect on long-term health
To understand the effects of hygienic measures during the pandemic on microbiome acquisition, microbiome loss and reinoculation
To study the effect of SARS-CoV-2 infection on the newborns’ gut microbiome and their long-term health consequences
ACE2, angiotensin-converting enzyme 2; COVID-19, coronavirus disease 2019; PACS; post-acute COVID-19 syndrome; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Supplementary information
Acknowledgements
The authors thank Whitney Tang from Microbiota I-Center (MagIC) for drawing the initial drafts of the figures. The authors are partially supported by InnoHK, The Government of Hong Kong, Special Administrative Region of the People’s Republic of China. R.I.L. received additional support from the Hong Kong PhD Fellowship Scheme (HKPFS) and CUHK Vice-Chancellor HKPFS Scholarship.
Author contributions
F.Z. and R.I.L. researched data for the article. S.C.N., F.Z., R.I.L. and Q.L. contributed substantially to discussion of the content. F.Z., R.I.L., Q.L. and Q.S. wrote the article. S.C.N., F.Z., R.I.L. and F.K.L.C. reviewed and/or edited the manuscript before submission.
Peer review
Peer review information
Nature Reviews Gastroenterology & Hepatology thanks Gianluca Ianiro, Lanjuan Li and Benjamin Mullish for their contribution to the peer review of this work.
Competing interests
S.C.N. and F.K.L.C. are the scientific co-founders and sit on the board of Directors of GenieBiome. S.C.N. has served as an advisory board member for Pfizer, Ferring, Janssen and Abbvie, and as a speaker for Ferring, Tillotts, Menarini, Janssen, Abbvie and Takeda. She has received research grants from Olympus, Ferring and Abbvie. F.K.L.C. has served as an adviser and lecture speaker for Eisai, AstraZeneca, Pfizer, Takeda Pharmaceutical, and Takeda (China) Holdings. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Fen Zhang, Raphaela I. Lau.
Change history
1/12/2023
A Correction to this paper has been published: 10.1038/s41575-023-00742-x
Supplementary information
The online version contains supplementary material available at 10.1038/s41575-022-00698-4.
References
- 1.Tregoning JS, Flight KE, Higham SL, Wang Z, Pierce BF. Progress of the COVID-19 vaccine effort: viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat. Rev. Immunol. 2021;21:626–636. doi: 10.1038/s41577-021-00592-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Files JK, et al. Duration of post-COVID-19 symptoms is associated with sustained SARS-CoV-2-specific immune responses. JCI Insight. 2021;6:e151544. doi: 10.1172/jci.insight.151544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamers MM, et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zang R, et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5:eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuo T, et al. Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut. 2020;70:276–284. doi: 10.1136/gutjnl-2020-322294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao F, et al. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg. Infect. Dis. 2020;26:1920. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Natarajan A, et al. Gastrointestinal symptoms and fecal shedding of SARS-CoV-2 RNA suggest prolonged gastrointestinal infection. Med. 2022;3:371–387. doi: 10.1016/j.medj.2022.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zollner A, et al. Post-acute COVID-19 is characterized by gut viral antigen persistence in inflammatory bowel diseases. Gastroenterology. 2022;163:495–506. doi: 10.1053/j.gastro.2022.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung KS, et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020;159:81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao R, et al. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: a systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2020;5:667–678. doi: 10.1016/S2468-1253(20)30126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song Y, et al. SARS-CoV-2 induced diarrhoea as onset symptom in patient with COVID-19. Gut. 2020;69:1143–1144. doi: 10.1136/gutjnl-2020-320891. [DOI] [PubMed] [Google Scholar]
- 12.Wang D, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert JA, et al. Current understanding of the human microbiome. Nat. Med. 2018;24:392–400. doi: 10.1038/nm.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothschild D, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 15.Kurilshikov A, et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 2021;53:156–165. doi: 10.1038/s41588-020-00763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolter M, et al. Leveraging diet to engineer the gut microbiome. Nat. Rev. Gastroenterol. Hepatol. 2021;18:885–902. doi: 10.1038/s41575-021-00512-7. [DOI] [PubMed] [Google Scholar]
- 17.Ruff WE, Greiling TM, Kriegel MA. Host–microbiota interactions in immune-mediated diseases. Nat. Rev. Microbiol. 2020;18:521–538. doi: 10.1038/s41579-020-0367-2. [DOI] [PubMed] [Google Scholar]
- 18.Groves HT, Higham SL, Moffatt MF, Cox MJ, Tregoning JS. Respiratory viral infection alters the gut microbiota by inducing inappetence. mBio. 2020;11:e03236-–19. doi: 10.1128/mBio.03236-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang D, et al. The cross-talk between gut microbiota and lungs in common lung diseases. Front. Microbiol. 2020;11:301. doi: 10.3389/fmicb.2020.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Z, et al. Gut microbiome alterations and gut barrier dysfunction are associated with host immune homeostasis in COVID-19 patients. BMC Med. 2022;20:24. doi: 10.1186/s12916-021-02212-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vatanen T, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165:842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anand S, Mande SS. Diet, microbiota and gut-lung connection. Front. Microbiol. 2018;9:2147. doi: 10.3389/fmicb.2018.02147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parrot T, et al. MAIT cell activation and dynamics associated with COVID-19 disease severity. Sci. Immunol. 2020;5:eabe1670. doi: 10.1126/sciimmunol.abe1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Legoux F, Salou M, Lantz O. MAIT cell development and functions: the microbial connection. Immunity. 2020;53:710–723. doi: 10.1016/j.immuni.2020.09.009. [DOI] [PubMed] [Google Scholar]
- 25.Hashimoto T, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Viana SD, Nunes S, Reis F. ACE2 imbalance as a key player for the poor outcomes in COVID-19 patients with age-related comorbidities–role of gut microbiota dysbiosis. Ageing Res. Rev. 2020;62:101123. doi: 10.1016/j.arr.2020.101123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaibani P, et al. The gut microbiota of critically ill patients with COVID-19. Front. Cell. Infect. Microbiol. 2021;11:670424. doi: 10.3389/fcimb.2021.670424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu S, et al. Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 influenza. Clin. Infect. Dis. 2020;71:2669–2678. doi: 10.1093/cid/ciaa709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren Z, et al. Alterations in the human oral and gut microbiomes and lipidomics in COVID-19. Gut. 2021;70:1253–1265. doi: 10.1136/gutjnl-2020-323826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu R, et al. Temporal association between human upper respiratory and gut bacterial microbiomes during the course of COVID-19 in adults. Commun. Biol. 2021;4:240. doi: 10.1038/s42003-021-01796-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizutani T, et al. Correlation analysis between gut microbiota alterations and the cytokine response in patients with coronavirus disease during hospitalization. Microbiol. Spectr. 2022;10:e0168921. doi: 10.1128/spectrum.01689-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rafiqul Islam S, et al. Dysbiosis of oral and gut microbiomes in SARS-CoV-2 infected patients in Bangladesh: elucidating the role of opportunistic gut microbes. Front. Med. 2022;9:163. doi: 10.3389/fmed.2022.821777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reinold J, et al. A pro-inflammatory gut microbiome characterizes SARS-CoV-2 infected patients and a reduction in the connectivity of an anti-inflammatory bacterial network associates with severe COVID-19. Front. Cell. Infect. Microbiol. 2021;11:1154. doi: 10.3389/fcimb.2021.747816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang L, et al. Clinical significance of the correlation between changes in the major intestinal bacteria species and COVID-19 severity. Engineering. 2020;6:1178–1184. doi: 10.1016/j.eng.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tao W, et al. Analysis of the intestinal microbiota in COVID-19 patients and its correlation with the inflammatory factor IL-18. Med. Microecol. 2020;5:100023. doi: 10.1016/j.medmic.2020.100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu Y, et al. Altered oral and gut microbiota and its association with SARS-CoV-2 viral load in COVID-19 patients during hospitalization. NPJ Biofilms Microbiomes. 2021;7:61. doi: 10.1038/s41522-021-00232-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeoh YK, et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698–706. doi: 10.1136/gutjnl-2020-323020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li S, et al. Microbiome profiling using shotgun metagenomic sequencing identified unique microorganisms in COVID-19 patients with altered gut microbiota. Front. Microbiol. 2021;12:712081. doi: 10.3389/fmicb.2021.712081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuo T, et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020;159:944–955.e8. doi: 10.1053/j.gastro.2020.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolte EE, Moorshead D, Aagaard KM. Maternal and early life exposures and their potential to influence development of the microbiome. Genome Med. 2022;14:4. doi: 10.1186/s13073-021-01005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Steenhuijsen Piters WA, Binkowska J, Bogaert D. Early life microbiota and respiratory tract infections. Cell Host Microbe. 2020;28:223–232. doi: 10.1016/j.chom.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 42.Renz H, Skevaki C. Early life microbial exposures and allergy risks: opportunities for prevention. Nat. Rev. Immunol. 2021;21:177–191. doi: 10.1038/s41577-020-00420-y. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X-S, et al. Maternal cecal microbiota transfer rescues early-life antibiotic-induced enhancement of type 1 diabetes in mice. Cell Host Microbe. 2021;29:1249–1265.e9. doi: 10.1016/j.chom.2021.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarkar A, Yoo JY, Valeria Ozorio Dutra S, Morgan KH, Groer M. The association between early-life gut microbiota and long-term health and diseases. J. Clin. Med. 2021;10:459. doi: 10.3390/jcm10030459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu R, et al. Progressive deterioration of the upper respiratory tract and the gut microbiomes in children during the early infection stages of COVID-19. J. Genet. Genomics. 2021;48:803–814. doi: 10.1016/j.jgg.2021.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nashed L, et al. Gut microbiota changes are detected in asymptomatic very young children with SARS-CoV-2 infection. Gut. 2022 doi: 10.1136/gutjnl-2021-326599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang L, et al. COVID-19 and multisystem inflammatory syndrome in children and adolescents. Lancet Infect. Dis. 2020;20:e276–e288. doi: 10.1016/S1473-3099(20)30651-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suskun C, et al. Intestinal microbiota composition of children with infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and multisystem inflammatory syndrome (MIS-C) Eur. J. Pediatr. 2022;181:3175–3191. doi: 10.1007/s00431-022-04494-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nash AK, et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome. 2017;5:153. doi: 10.1186/s40168-017-0373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lv L, et al. Gut mycobiota alterations in patients with COVID-19 and H1N1 infections and their associations with clinical features. Commun. Biol. 2021;4:480. doi: 10.1038/s42003-021-02036-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zuo T, et al. Alterations in fecal fungal microbiome of patients with COVID-19 during time of hospitalization until discharge. Gastroenterology. 2020;159:1302–1310.e5. doi: 10.1053/j.gastro.2020.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roudbary M, et al. Overview on the prevalence of fungal infections, immune response, and microbiome role in COVID-19 patients. J. Fungi. 2021;7:720. doi: 10.3390/jof7090720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arastehfar A, et al. COVID-19-associated candidiasis (CAC): an underestimated complication in the absence of immunological predispositions? J. Fungi. 2020;6:211. doi: 10.3390/jof6040211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zuo T, et al. Temporal landscape of human gut RNA and DNA virome in SARS-CoV-2 infection and severity. Microbiome. 2021;9:91. doi: 10.1186/s40168-021-01008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao J, et al. Integrated gut virome and bacteriome dynamics in COVID-19 patients. Gut Microbes. 2021;13:1887722. doi: 10.1080/19490976.2021.1887722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matheson NJ, Lehner PJ. How does SARS-CoV-2 cause COVID-19? Science. 2020;369:510–511. doi: 10.1126/science.abc6156. [DOI] [PubMed] [Google Scholar]
- 58.Katz-Agranov N, Zandman-Goddard G. Autoimmunity and COVID-19–the microbiotal connection. Autoimmun. Rev. 2021;20:102865. doi: 10.1016/j.autrev.2021.102865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vignesh R, et al. Could perturbation of gut microbiota possibly exacerbate the severity of COVID-19 via cytokine storm? Front. Immunol. 2021;11:607734. doi: 10.3389/fimmu.2020.607734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prasad R, et al. Plasma microbiome in COVID-19 subjects: an indicator of gut barrier defects and dysbiosis. bioRxiv. 2021 doi: 10.1101/2021.04.06.438634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Effenberger M, et al. Faecal calprotectin indicates intestinal inflammation in COVID-19. Gut. 2020;69:1543–1544. doi: 10.1136/gutjnl-2020-321388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Giron LB, et al. Plasma markers of disrupted gut permeability in severe COVID-19 patients. Front. Immunol. 2021;12:686240. doi: 10.3389/fimmu.2021.686240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chhibber-Goel J, Gopinathan S, Sharma A. Interplay between severities of COVID-19 and the gut microbiome: implications of bacterial co-infections? Gut Pathog. 2021;13:14. doi: 10.1186/s13099-021-00407-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Langford BJ, et al. Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin. Microbiol. Infect. 2020;26:1622–1629. doi: 10.1016/j.cmi.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saade A, et al. Infectious events in patients with severe COVID-19: results of a cohort of patients with high prevalence of underlying immune defect. Ann. Intensive Care. 2021;11:83. doi: 10.1186/s13613-021-00873-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garcia-Vidal C, et al. Incidence of co-infections and superinfections in hospitalized patients with COVID-19: a retrospective cohort study. Clin. Microbiol. Infect. 2021;27:83–88. doi: 10.1016/j.cmi.2020.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou F, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moreira-Rosário A, et al. Gut microbiota diversity and C-reactive protein are predictors of disease severity in COVID-19 patients. Front. Microbiol. 2021;12:1820. doi: 10.3389/fmicb.2021.705020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu Y, et al. Distinct metagenomic signatures in the SARS-CoV-2 infection. Front. Cell. Infect. Microbiol. 2021;11:1019. doi: 10.3389/fcimb.2021.706970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou Y, et al. Gut microbiota dysbiosis correlates with abnormal immune response in moderate COVID-19 patients with fever. J. Inflamm. Res. 2021;14:2619. doi: 10.2147/JIR.S311518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brodin P. Immune determinants of COVID-19 disease presentation and severity. Nat. Med. 2021;27:28–33. doi: 10.1038/s41591-020-01202-8. [DOI] [PubMed] [Google Scholar]
- 73.Nowarski R, et al. Epithelial IL-18 equilibrium controls barrier function in colitis. Cell. 2015;163:1444–1456. doi: 10.1016/j.cell.2015.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang N, et al. SARS-CoV-2 infection of the oral cavity and saliva. Nat. Med. 2021;27:892–903. doi: 10.1038/s41591-021-01296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Soffritti I, et al. Oral microbiome dysbiosis is associated with symptoms severity and local immune/inflammatory response in COVID-19 patients: a cross-sectional study. Front. Microbiol. 2021;12:687513. doi: 10.3389/fmicb.2021.687513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Castilhos J, et al. Severe dysbiosis and specific Haemophilus and Neisseria signatures as hallmarks of the oropharyngeal microbiome in critically ill COVID-19 patients. Clin. Infect. Dis. 2021;75:e1063–e1071. doi: 10.1093/cid/ciab902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miller EH, et al. Oral microbiome alterations and SARS-CoV-2 saliva viral load in patients with COVID-19. Microbiol. Spectr. 2021;9:e00055–21. doi: 10.1128/Spectrum.00055-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ma S, et al. Metagenomic analysis reveals oropharyngeal microbiota alterations in patients with COVID-19. Signal. Transduct. Target. Ther. 2021;6:191. doi: 10.1038/s41392-021-00614-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Newsome RC, et al. The gut microbiome of COVID-19 recovered patients returns to uninfected status in a minority-dominated United States cohort. Gut Microbes. 2021;13:1926840. doi: 10.1080/19490976.2021.1926840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nasserie T, Hittle M, Goodman SN. Assessment of the frequency and variety of persistent symptoms among patients with COVID-19: a systematic review. JAMA Netw. Open. 2021;4:e2111417. doi: 10.1001/jamanetworkopen.2021.11417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang C, et al. 6-month consequences of COVID-19 in patients discharged from hospital: a cohort study. Lancet. 2021;397:220–232. doi: 10.1016/S0140-6736(20)32656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nalbandian A, et al. Post-acute COVID-19 syndrome. Nat. Med. 2021;27:601–615. doi: 10.1038/s41591-021-01283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu Q, et al. Gut microbiota dynamics in a prospective cohort of patients with post-acute COVID-19 syndrome. Gut. 2022;71:544–552. doi: 10.1136/gutjnl-2021-325989. [DOI] [PubMed] [Google Scholar]
- 84.Evans RA, et al. Physical, cognitive, and mental health impacts of COVID-19 after hospitalisation (PHOSP-COVID): a UK multicentre, prospective cohort study. Lancet Respir. Med. 2021;9:1275–1287. doi: 10.1016/S2213-2600(21)00383-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Choutka J, Jansari V, Hornig M, Iwasaki A. Unexplained post-acute infection syndromes. Nat. Med. 2022;28:911–923. doi: 10.1038/s41591-022-01810-6. [DOI] [PubMed] [Google Scholar]
- 86.Lee YY, Annamalai C, Rao SS. Post-infectious irritable bowel syndrome. Curr. Gastroenterol. Rep. 2017;19:56. doi: 10.1007/s11894-017-0595-4. [DOI] [PubMed] [Google Scholar]
- 87.Holtmann GJ, Ford AC, Talley NJ. Pathophysiology of irritable bowel syndrome. Lancet Gastroenterol. Hepatol. 2016;1:133–146. doi: 10.1016/S2468-1253(16)30023-1. [DOI] [PubMed] [Google Scholar]
- 88.Austhof E, et al. Persisting gastrointestinal symptoms and post-infectious irritable bowel syndrome following SARS-CoV-2 infection: results from the Arizona CoVHORT. Epidemiol. Infect. 2022;150:e136. doi: 10.1017/S0950268822001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ghoshal UC, et al. Post‐infection functional gastrointestinal disorders following coronavirus disease‐19: a case–control study. J. Gastroenterol. Hepatol. 2022;37:489–498. doi: 10.1111/jgh.15717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Al-Aly Z, Bowe B, Xie Y. Long COVID after breakthrough SARS-CoV-2 infection. Nat. Med. 2022;28:1461–1467. doi: 10.1038/s41591-022-01840-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hu J, et al. Probiotics, prebiotics and dietary approaches during COVID-19 pandemic. Trends Food Sci. Technol. 2021;108:187–196. doi: 10.1016/j.tifs.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen Y, et al. Six-month follow-up of gut microbiota richness in patients with COVID-19. Gut. 2022;71:222–225. doi: 10.1136/gutjnl-2021-324090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vestad B, et al. Respiratory dysfunction three months after severe COVID‐19 is associated with gut microbiota alterations. J. Intern. Med. 2022;291:801–812. doi: 10.1111/joim.13458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Su Q, Lau RI, Liu Q, Chan FKL, Ng SC. Post-acute COVID-19 syndrome and gut dysbiosis linger beyond 1 year after SARS-CoV-2 clearance. Gut. 2022 doi: 10.1136/gutjnl-2022-328319. [DOI] [PubMed] [Google Scholar]
- 95.Cui GY, et al. Characterization of oral and gut microbiome and plasma metabolomics in COVID-19 patients after 1-year follow-up. Mil. Med. Res. 2022;9:32. doi: 10.1186/s40779-022-00387-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cervia C, et al. Immunoglobulin signature predicts risk of post-acute COVID-19 syndrome. Nat. Commun. 2022;13:446. doi: 10.1038/s41467-021-27797-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Marx V. Scientists set out to connect the dots on long COVID. Nat. Methods. 2021;18:449–453. doi: 10.1038/s41592-021-01145-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhou Y, Zhang J, Zhang D, Ma W-L, Wang X. Linking the gut microbiota to persistent symptoms in survivors of COVID-19 after discharge. J. Microbiol. 2021;59:941–948. doi: 10.1007/s12275-021-1206-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jalanka-Tuovinen J, et al. Faecal microbiota composition and host–microbe cross-talk following gastroenteritis and in postinfectious irritable bowel syndrome. Gut. 2014;63:1737–1745. doi: 10.1136/gutjnl-2013-305994. [DOI] [PubMed] [Google Scholar]
- 100.Zhang H, et al. Specific ACE2 expression in small intestinal enterocytes may cause gastrointestinal symptoms and injury after 2019-nCoV infection. Int. J. Infect. Dis. 2020;96:19–24. doi: 10.1016/j.ijid.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Koester ST, Li N, Lachance DM, Morella NM, Dey N. Variability in digestive and respiratory tract Ace2 expression is associated with the microbiome. PLoS ONE. 2021;16:e0248730. doi: 10.1371/journal.pone.0248730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hirayama M, et al. Intestinal Collinsella may mitigate infection and exacerbation of COVID-19 by producing ursodeoxycholate. PLoS ONE. 2021;16:e0260451. doi: 10.1371/journal.pone.0260451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Geva-Zatorsky N, et al. Mining the human gut microbiota for immunomodulatory organisms. Cell. 2017;168:928–943.e11. doi: 10.1016/j.cell.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vijay A, Valdes AM. Role of the gut microbiome in chronic diseases: a narrative review. Eur. J. Clin. Nutr. 2021;76:489–501. doi: 10.1038/s41430-021-00991-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 105.O’Toole PW, Jeffery IB. Gut microbiota and aging. Science. 2015;350:1214–1215. doi: 10.1126/science.aac8469. [DOI] [PubMed] [Google Scholar]
- 106.Li Y, et al. Systematic profiling of ACE2 expression in diverse physiological and pathological conditions for COVID‐19/SARS‐CoV‐2. J. Cell. Mol. Med. 2020;24:9478–9482. doi: 10.1111/jcmm.15607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gracia-Ramos AE. Is the ACE2 overexpression a risk factor for COVID-19 infection? Arch. Med. Res. 2020;51:345–346. doi: 10.1016/j.arcmed.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Martino C, et al. Bacterial modification of the host glycosaminoglycan heparan sulfate modulates SARS-CoV-2 infectivity. Preprint at. bioRxiv. 2020 doi: 10.1101/2020.08.17.238444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Merad M, Martin JC. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kircheis R, et al. NF-κB pathway as a potential target for treatment of critical stage COVID-19 patients. Front. Immunol. 2020;11:3446. doi: 10.3389/fimmu.2020.598444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guo Y, et al. SARS-CoV-2 induced intestinal responses with a biomimetic human gut-on-chip. Sci. Bull. 2021;66:783–793. doi: 10.1016/j.scib.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cerf-Bensussan N, Gaboriau-Routhiau V. The immune system and the gut microbiota: friends or foes? Nat. Rev. Immunol. 2010;10:735–744. doi: 10.1038/nri2850. [DOI] [PubMed] [Google Scholar]
- 113.Yan R, et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lecarpentier Y, Vallée A. The key role of the level of ACE2 gene expression in SARS-CoV-2 infection. Aging. 2021;13:14552. doi: 10.18632/aging.203181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Brogna C, et al. Could SARS-CoV-2 have bacteriophage behavior or induce the activity of other bacteriophages? Vaccines. 2022;10:708. doi: 10.3390/vaccines10050708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Seibert B, et al. Mild and severe SARS-CoV-2 infection induces respiratory and intestinal microbiome changes in the K18-hACE2 transgenic mouse model. Microbiol. Spectr. 2021;9:e00536–21. doi: 10.1128/Spectrum.00536-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sokol H, et al. SARS-CoV-2 infection in nonhuman primates alters the composition and functional activity of the gut microbiota. Gut Microbes. 2021;13:1893113. doi: 10.1080/19490976.2021.1893113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sencio V, et al. Alteration of the gut microbiota following SARS-CoV-2 infection correlates with disease severity in hamsters. Gut Microbes. 2022;14:2018900. doi: 10.1080/19490976.2021.2018900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pascoal LB, et al. Microbiota-derived short-chain fatty acids do not interfere with SARS-CoV-2 infection of human colonic samples. Gut Microbes. 2021;13:1874740. doi: 10.1080/19490976.2021.1874740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhou T, et al. SARS‐CoV‐2 triggered oxidative stress and abnormal energy metabolism in gut microbiota. MedComm. 2022;3:41–56. doi: 10.1002/mco2.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.He F, et al. Fecal multi-omics analysis reveals diverse molecular alterations of gut ecosystem in COVID-19 patients. Anal. Chim. Acta. 2021;1180:338881. doi: 10.1016/j.aca.2021.338881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lv L, et al. The faecal metabolome in COVID-19 patients is altered and associated with clinical features and gut microbes. Anal. Chim. Acta. 2021;1152:338267. doi: 10.1016/j.aca.2021.338267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Al Bataineh MT, et al. Gut microbiota interplay with COVID-19 reveals links to host lipid metabolism among Middle Eastern populations. Front. Microbiol. 2021;12:761067. doi: 10.3389/fmicb.2021.761067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang F, et al. Prolonged impairment of short-chain fatty acid and L-isoleucine biosynthesis in gut microbiome in patients with COVID-19. Gastroenterology. 2022;162:548–561.e4. doi: 10.1053/j.gastro.2021.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Geirnaert A, et al. Butyrate-producing bacteria supplemented in vitro to Crohn’s disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Sci. Rep. 2017;7:11450. doi: 10.1038/s41598-017-11734-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yao Y, et al. The role of short-chain fatty acids in immunity, inflammation and metabolism. Crit. Rev. Food Sci. Nutr. 2020;62:1–12. doi: 10.1080/10408398.2020.1854675. [DOI] [PubMed] [Google Scholar]
- 127.Lionetto L, et al. Increased kynurenine-to-tryptophan ratio in the serum of patients infected with SARS-CoV2: an observational cohort study. Biochim. Biophys. Acta Mol. Basis Dis. 2021;1867:166042. doi: 10.1016/j.bbadis.2020.166042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Barberis E, et al. Large-scale plasma analysis revealed new mechanisms and molecules associated with the host response to SARS-CoV-2. Int. J. Mol. Sci. 2020;21:8623. doi: 10.3390/ijms21228623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Robertson J, et al. Serum neopterin levels in relation to mild and severe COVID-19. BMC Infect. Dis. 2020;20:942. doi: 10.1186/s12879-020-05671-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Eroğlu İ, Eroğlu BÇ, Güven GS. Altered tryptophan absorption and metabolism could underlie long-term symptoms in survivors of coronavirus disease 2019 (COVID-19) Nutrition. 2021;90:111308. doi: 10.1016/j.nut.2021.111308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dagenais-Lussier X, et al. Latest developments in tryptophan metabolism: understanding its role in B cell immunity. Cytokine Growth Factor Rev. 2021;59:111–117. doi: 10.1016/j.cytogfr.2021.02.003. [DOI] [PubMed] [Google Scholar]
- 132.Cervenka I, Agudelo LZ, Ruas JL. Kynurenines: tryptophan’s metabolites in exercise, inflammation, and mental health. Science. 2017;357:eaaf9794. doi: 10.1126/science.aaf9794. [DOI] [PubMed] [Google Scholar]
- 133.Agus A, Planchais J, Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. 2018;23:716–724. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 134.Gao J, et al. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front. Cell. Infect. Microbiol. 2018;8:13. doi: 10.3389/fcimb.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sonner JK, et al. Dietary tryptophan links encephalogenicity of autoreactive T cells with gut microbial ecology. Nat. Commun. 2019;10:4877. doi: 10.1038/s41467-019-12776-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shen B, et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182:59–72.e15. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rohrhofer J, Zwirzitz B, Selberherr E, Untersmayr E. The impact of dietary sphingolipids on intestinal microbiota and gastrointestinal immune homeostasis. Front. Immunol. 2021;12:635704. doi: 10.3389/fimmu.2021.635704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Brown EM, et al. Bacteroides-derived sphingolipids are critical for maintaining intestinal homeostasis and syMbiosis. Cell Host Microbe. 2019;25:668–680.e7. doi: 10.1016/j.chom.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kue CS, et al. C6-ceramide in combination with transforming growth factor-β enhances Treg cell differentiation and stable FoxP3 expression in vitro and in vivo. Immunobiology. 2013;218:952–959. doi: 10.1016/j.imbio.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 140.Gericke B, Amiri M, Naim HY. The multiple roles of sucrase-isomaltase in the intestinal physiology. Mol. Cell. Pediatr. 2016;3:2. doi: 10.1186/s40348-016-0033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Deb C, et al. Sucrase-isomaltase gene variants in patients with abnormal sucrase activity and functional gastrointestinal disorders. J. Pediatr. Gastroenterol. Nutr. 2021;72:29–35. doi: 10.1097/MPG.0000000000002852. [DOI] [PubMed] [Google Scholar]
- 142.Kalantar-Zadeh K, Berean KJ, Burgell RE, Muir JG, Gibson PR. Intestinal gases: influence on gut disorders and the role of dietary manipulations. Nat. Rev. Gastroenterol. Hepatol. 2019;16:733–747. doi: 10.1038/s41575-019-0193-z. [DOI] [PubMed] [Google Scholar]
- 143.Nasreen S, et al. Effectiveness of COVID-19 vaccines against symptomatic SARS-CoV-2 infection and severe outcomes with variants of concern in Ontario. Nat. Microbiol. 2022;7:379–385. doi: 10.1038/s41564-021-01053-0. [DOI] [PubMed] [Google Scholar]
- 144.Andrews N, et al. Effectiveness of COVID-19 booster vaccines against covid-19 related symptoms, hospitalisation and death in England. Nat. Med. 2022;28:831–837. doi: 10.1038/s41591-022-01699-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lynn DJ, Benson SC, Lynn MA, Pulendran B. Modulation of immune responses to vaccination by the microbiota: implications and potential mechanisms. Nat. Rev. Immunol. 2021;22:33–46. doi: 10.1038/s41577-021-00554-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pulendran B, S Arunachalam P, O’Hagan DT. Emerging concepts in the science of vaccine adjuvants. Nat. Rev. Drug Discov. 2021;20:454–475. doi: 10.1038/s41573-021-00163-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Oh JZ, et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity. 2014;41:478–492. doi: 10.1016/j.immuni.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kim D, et al. Nod2-mediated recognition of the microbiota is critical for mucosal adjuvant activity of cholera toxin. Nat. Med. 2016;22:524–530. doi: 10.1038/nm.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kim M, Qie Y, Park J, Kim CH. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe. 2016;20:202–214. doi: 10.1016/j.chom.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ng SC, et al. Gut microbiota composition is associated with SARS-CoV-2 vaccine immunogenicity and adverse events. Gut. 2022;71:1106–1116. doi: 10.1136/gutjnl-2021-326563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Alexander JL, et al. The gut microbiota and metabolome is associated with diminished COVID-19 vaccine-induced antibody responses in immunosuppressed inflammatory bowel disease patients. Lancet Gastroenterol. Hepatol. 2022;7:342–352. doi: 10.1016/S2468-1253(22)00005-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Uehara O, et al. Alterations in the oral microbiome of individuals with a healthy oral environment following COVID-19 vaccination. BMC Oral Health. 2022;22:50. doi: 10.1186/s12903-022-02093-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Levine-Tiefenbrun M, et al. Waning of SARS-CoV-2 booster viral-load reduction effectiveness. Nat. Commun. 2022;13:1237. doi: 10.1038/s41467-022-28936-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ward H, et al. Population antibody responses following COVID-19 vaccination in 212,102 individuals. Nat. Commun. 2022;13:907. doi: 10.1038/s41467-022-28527-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pérez-Alós L, et al. Modeling of waning immunity after SARS-CoV-2 vaccination and influencing factors. Nat. Commun. 2022;13:1614. doi: 10.1038/s41467-022-29225-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Araos R, et al. Effectiveness of CoronaVac in children 3 to 5 years during the omicron SARS-CoV-2 outbreak. Nat. Med. 2022;28:1377–1380. doi: 10.1038/s41591-022-01874-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Finlay BB, et al. The hygiene hypothesis, the COVID pandemic, and consequences for the human microbiome. Proc. Natl Acad. Sci. USA. 2021;118:e2010217118. doi: 10.1073/pnas.2010217118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Aguilera P, et al. A two-time point analysis of gut microbiota in the general population of buenos aires and its variation due to preventive and compulsory social isolation during the COVID-19 pandemic. Front. Microbiol. 2022;13:803121. doi: 10.3389/fmicb.2022.803121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Peng Y, et al. Gut microbiome and resistome changes during the first wave of the COVID-19 pandemic in comparison with pre-pandemic travel-related changes. J. Travel Med. 2021;28:taab067. doi: 10.1093/jtm/taab067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Romano-Keeler J, Zhang J, Sun J. COVID-19 and the neonatal microbiome: will the pandemic cost infants their microbes? Gut Microbes. 2021;13:1912562. doi: 10.1080/19490976.2021.1912562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Xia JS, Oliphant K, Claud E. The pandemic effects on the microbiome of infants in the neonatal intensive care unit (NICU) FASEB J. 2022 doi: 10.1096/fasebj.2022.36.S1.0R562. [DOI] [Google Scholar]
- 162.Rundle AG, Park Y, Herbstman JB, Kinsey EW, Wang YC. COVID-19 related school closings and risk of weight gain among children. Obesity. 2020;28:1008. doi: 10.1002/oby.22813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liu JJ, Bao Y, Huang X, Shi J, Lu L. Mental health considerations for children quarantined because of COVID-19. Lancet Child. Adolesc. Health. 2020;4:347–349. doi: 10.1016/S2352-4642(20)30096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wang J, et al. Progression of myopia in school-aged children after COVID-19 home confinement. JAMA Ophthal. 2021;139:293–300. doi: 10.1001/jamaophthalmol.2020.6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bauer KW, et al. A safety net unraveling: Feeding young children during COVID-19. Am. J. Public Health. 2021;111:116–120. doi: 10.2105/AJPH.2020.305980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Langford BJ, et al. Antibiotic prescribing in patients with COVID-19: rapid review and meta-analysis. Clin. Microbiol. Infect. 2021;27:520–531. doi: 10.1016/j.cmi.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hagan T, et al. Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell. 2019;178:1313–1328.e13. doi: 10.1016/j.cell.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hegazy M, et al. Beyond probiotic legend: ESSAP gut microbiota health score to delineate SARS-COV-2 infection severity. Br. J. Nutr. 2021;127:1180–1189. doi: 10.1017/S0007114521001926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Merino J, et al. Diet quality and risk and severity of COVID-19: a prospective cohort study. Gut. 2021;70:2096–2104. doi: 10.1136/gutjnl-2021-325353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hou Y-C, Su W-L, Chao Y-C. COVID-19 illness severity in the elderly in relation to vegetarian and non-vegetarian diets: a single-center experience. Front. Nutr. 2022;9:837458. doi: 10.3389/fnut.2022.837458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhu L, et al. Association of blood glucose control and outcomes in patients with COVID-19 and pre-existing type 2 diabetes. Cell Metab. 2020;31:1068–1077.e3. doi: 10.1016/j.cmet.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bousquet J, et al. Is diet partly responsible for differences in COVID‐19 death rates between and within countries? Clin. Transl. Allergy. 2020;10:16. doi: 10.1186/s13601-020-00323-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang Y, Wu G, Zhao L, Wang W. Nutritional modulation of gut microbiota alleviates severe gastrointestinal symptoms in a patient with post-acute COVID-19 syndrome. mBio. 2022;13:e03801–21. doi: 10.1128/mbio.03801-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Schupack DA, Mars RA, Voelker DH, Abeykoon JP, Kashyap PC. The promise of the gut microbiome as part of individualized treatment strategies. Nat. Rev. Gastroenterol. Hepatol. 2021;19:7–25. doi: 10.1038/s41575-021-00499-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhang L, et al. Probiotics use is associated with improved clinical outcomes among hospitalized patients with COVID-19. Ther. Adv. Gastroenterol. 2021;14:17562848211035670. doi: 10.1177/17562848211035670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ceccarelli G, et al. Oral bacteriotherapy in patients with COVID-19: a retrospective cohort study. Front. Nutr. 2021;7:341. doi: 10.3389/fnut.2020.613928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Rathi A, Jadhav SB, Shah N. A randomized controlled trial of the efficacy of systemic enzymes and probiotics in the resolution of post-COVID fatigue. Medicines. 2021;8:47. doi: 10.3390/medicines8090047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Gutiérrez-Castrellón P, et al. Probiotic improves symptomatic and viral clearance in Covid19 outpatients: a randomized, quadruple-blinded, placebo-controlled trial. Gut Microbes. 2022;14:2018899. doi: 10.1080/19490976.2021.2018899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wu C, et al. The volatile and heterogeneous gut microbiota shifts of COVID‐19 patients over the course of a probiotics‐assisted therapy. Clin. Transl. Med. 2021;11:e643. doi: 10.1002/ctm2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zhang L, et al. Gut microbiota‐derived synbiotic formula (SIM01) as a novel adjuvant therapy for COVID‐19: An open‐label pilot study. J. Gastroenterol. Hepatol. 2022;37:823–831. doi: 10.1111/jgh.15796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Heavey MK, Durmusoglu D, Crook N, Anselmo AC. Discovery and delivery strategies for engineered live biotherapeutic products. Trends Biotechnol. 2021;40:254–369. doi: 10.1016/j.tibtech.2021.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Piscotta FJ, et al. Metabolites with SARS-CoV-2 inhibitory activity identified from human microbiome commensals. mSphere. 2021;6:e00711–21. doi: 10.1128/mSphere.00711-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Liu F, et al. Gastrointestinal disturbance and effect of fecal microbiota transplantation in discharged COVID-19 patients. J. Med. Case Rep. 2021;15:60. doi: 10.1186/s13256-020-02583-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Biliński J, et al. Rapid resolution of COVID-19 after faecal microbiota transplantation. Gut. 2022;71:230–232. doi: 10.1136/gutjnl-2021-325010. [DOI] [PubMed] [Google Scholar]
- 185.Constantinides MG. Interactions between the microbiota and innate and innate‐like lymphocytes. J. Leukoc. Biol. 2018;103:409–419. doi: 10.1002/JLB.3RI0917-378R. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.