Abstract
Pseudomonas aeruginosa is an opportunistic bacterial pathogen that often encounters hypoxic/anoxic environments within the host, which increases its tolerance to many conventional antibiotics. Towards identifying novel treatments, we explored the therapeutic potential of chlorate, a pro-drug that kills hypoxic/anoxic, antibiotic-tolerant P. aeruginosa populations. While chlorate itself is relatively nontoxic, it is enzymatically reduced to the toxic oxidizing agent, chlorite, by hypoxically-induced nitrate reductase. To better assess chlorate’s therapeutic potential, we investigated mechanisms of chlorate toxicity and resistance in P. aeruginosa. We used transposon mutagenesis to identify genes that alter P. aeruginosa fitness during chlorate treatment, finding that methionine sulfoxide reductases (Msr), which repair oxidized methionine residues, support survival during chlorate stress. Chlorate treatment leads to proteome-wide methionine oxidation, which is exacerbated in a ΔmsrAΔmsrB strain. In response to chlorate, P. aeruginosa upregulates proteins involved in a wide range of functions, including metabolism, DNA replication/repair, protein repair, transcription, and translation, and these newly synthesized proteins are particularly vulnerable to methionine oxidation. The addition of exogenous methionine partially rescues P. aeruginosa survival during chlorate treatment, suggesting that widespread methionine oxidation contributes to death. Finally, we found that mutations that decrease nitrate reductase activity are a common mechanism of chlorate resistance.
Graphical Abstract
Chlorate is a pro-drug that kills pathogens in low-oxygen environments where conventional antibiotics are less effective. This study reveals that chlorate reduction to chlorite (catalyzed by Nar) kills cells by causing proteome-wide methionine oxidation (MetSO), which can be partially repaired by methionine sulfoxide reductase (Msr) enzymes. Mutations that decrease Nar activity are a primary mechanism of chlorate resistance.

Introduction
It is widely known that many conventional antibiotics are ineffective at killing slow- or non-growing bacterial cells (1). One key physiological constraint that dictates the growth rate of many pathogens is oxygen availability. Many pathogens grow slower, and thus display increased antibiotic tolerance, under hypoxic/anoxic conditions (2–4). Slow growth and antibiotic tolerance are also defining features of biofilms, where bacteria grow as dense multicellular aggregates with oxygen-limited interior populations (5, 6).
The link between hypoxic/anoxic environments and antibiotic failure has devastating consequences for treating infections. For example, the mucus that coats the airways of cystic fibrosis (CF) patients is largely hypoxic/anoxic (7) and supports biofilm growth of opportunistic pathogens, such as Pseudomonas aeruginosa (8, 9). Chronic lung infections caused by bacteria like P. aeruginosa are the primary cause of mortality in CF patients and persist for decades despite aggressive antibiotic treatment regimens (10), likely in part because some fraction of the bacterial population enters an antibiotic-tolerant state. Chronic wounds, which affect ~2% of the US population (~6.5M people) (11), also contain large regions of hypoxic/anoxic tissue (12) and support biofilm growth (8). Importantly, chronic wound infections must be resolved before healing can proceed (13), and our inability to eradicate chronic wound infections has massive repercussions, frequently leading to lower-extremity amputations in diabetic patients (14). It is estimated that such amputations occur every 30s worldwide, and these amputations are associated with a 48-74% 5-year mortality rate across various studies (15, 16).
There is a clear need to develop alternative therapeutics that are effective at killing pathogens that inhabit hypoxic/anoxic host environments. One approach is to identify drugs that target processes that are specific to bacteria, such as anaerobic nitrate respiration. Nitrate is a potent terminal electron acceptor that sustains energy conservation, and the respiratory nitrate reductase (Nar) is conserved and widespread across facultative anaerobes (17). There is strong evidence that nitrate respiration is a relevant metabolism used by opportunistic pathogens in vivo. First, appreciable nitrate concentrations (up to ~0.4 mM) can be found in host environments, including the inflamed gut, CF sputum, and chronic wounds (7, 18–21). Further, P. aeruginosa nar transcripts have been detected in the sputa, and anti-Nar antibodies have been detected in the sera of CF patients (22, 23). The downstream products of nitrate respiration (i.e. denitrification products such as nitrous oxide) have also been detected in CF sputum samples (24). Finally, work in a mouse colitis model has directly shown that respiration of host-derived nitrate supports the expansion of enteric pathogens via Nar activity (21, 25).
The nitrate analog, chlorate, can effectively kill Nar-containing pathogens. While the physiological function of Nar is to reduce nitrate to nitrite, it has been known since the 1960s that Nar can also reduce chlorate to the toxic oxidizing agent, chlorite (26, 27). Previously, we showed that chlorate kills hypoxic/anoxic, tobramycin-tolerant biofilm populations of P. aeruginosa (28). This finding supports chlorate’s potential to serve as an effective pro-drug for combatting antibiotic tolerance in chronic infections. In addition to our work, a number of studies from the early 2000s showed that feeding chlorate to a variety of different livestock led to a decrease in fecal enteric counts (reviewed in (29)). Those studies also demonstrated that chlorate could be ingested at levels that were safe for animals but toxic to bacteria. Together, these findings motivate further exploration of chlorate’s therapeutic use for resolving infections found in hypoxic/anoxic host environments.
In this study, we sought to determine the mechanism(s) by which chlorate kills bacteria and how bacteria might develop resistance to chlorate; such knowledge is important to evaluate chlorate’s potential as a pro-drug. We used an unbiased genetic screen (transposon insertion sequencing) to identify mutations that either increase or decrease P. aeruginosa fitness in the presence of chlorate. Our findings suggest that proteome-wide methionine oxidation plays a key role in chlorate-mediated death, and that the primary mechanism of chlorate resistance requires mutations that inactivate or reduce Nar activity.
Results
Chlorate Tn-Seq experimental design
To identify mechanisms of chlorate toxicity and resistance, we took an unbiased genetic approach known as transposon insertion sequencing (Tn-Seq). In Tn-Seq, a transposon mutant library is exposed to a condition of interest, and changes in the relative abundance of each mutant is quantified using next-generation sequencing (30). Transposon mutants that increase or decrease in relative abundance confer a fitness advantage or disadvantage, respectively, under the condition of interest.
We grew triplicate aliquots of a P. aeruginosa transposon library of ~150,000 unique mutants (31) under anoxic conditions (LB with 40 mM KNO3). Overnight cultures were pelleted and washed to remove residual nitrate, split in half, and exposed to either no (control) or 1 mM chlorate for 30 minutes. The chlorate-treated library cultures consumed 12 ± 1% (mean ± SEM) of the provided chlorate, which led to a 0.7 ± 0.1 log reduction (mean ± SEM) in viable cell counts compared to untreated samples. After an aerobic outgrowth step, cultures were prepared for sequencing.
One caveat of this experiment is that a large fraction of the sequencing reads (53 ± 6% reads, mean ± SEM) mapped to the Escherichia coli vector that was used to generate the P. aeruginosa transposon library, rather than to the P. aeruginosa genome. This suggests that residual E. coli was present in the library. However, all library cultures were plated for viable counts following chlorate treatment and aerobic outgrowth, and E. coli colonies were not detected (via colony morphology) on LB plates. This suggests that abundance of E. coli was low relative to P. aeruginosa during the experiment, but that perhaps vector reads were amplified during the sequencing preparation steps. Regardless of this caveat, there are many indicators that our Tn-seq results are biologically relevant, including the identification of positive control genes previously shown to confer a fitness advantage (ΔnarGHJI) or disadvantage (ΔlasR) during chlorate exposure (28), and validation tests performed in this study that support the hypotheses generated from our Tn-Seq screen.
Modulating Nar activity is a primary mechanism of tuning chlorate sensitivity
Our Tn-Seq screen identified 131 or 55 genes that, when mutated, increased or decreased fitness by at least 2-fold during chlorate treatment, respectively (Table S1). Broadly, these genes could be grouped into several functional categories: denitrification, transcriptional regulation, other energy conservation pathways, and protein degradation and repair (Fig. 1A). Prior to this experiment, it was known that Nar activity is central to chlorate toxicity, so we expected that mutations that decrease Nar activity would likely increase chlorate resistance and mutations that increase Nar activity would likely increase chlorate sensitivity. Thus, it was unsurprising to find that the disruption of genes encoding Nar (narGHJI) confer a fitness advantage, consistent with our previous observation that nar mutants are completely resistant to chlorate (28).
Figure 1:
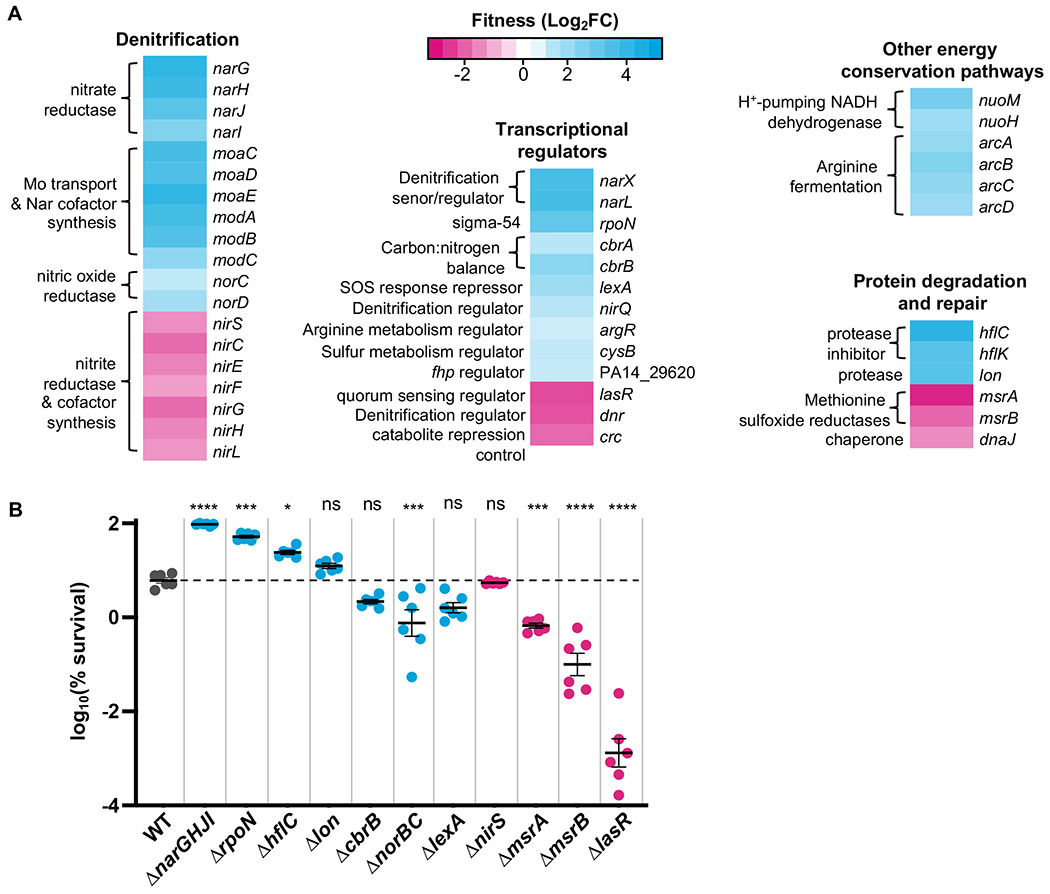
Chlorate Tn-seq results. A. Representative Tn-seq results showing genes that, when mutated, increase (cyan) or decrease (magenta) fitness during chlorate exposure (fitness is determined as log2(fold change CPM chlorate-treated vs. untreated). The complete data set can be viewed in Table S1. B. Deletion mutant strains were generated for select Tn-seq hits and exposed to chlorate to determine percent survival. Results for WT are colored in gray, and the dashed horizontal line indicates the mean percent survival for WT. Mutants predicted by the Tn-seq screen to increase or decrease in percent survival relative to WT are colored cyan and magenta, respectively. Shown are six biological replicates (three replicates per two independent experiments) with the mean ± standard error of the mean (SEM). The difference between WT and mutant log10(% survival) was determined by one-way ANOVA; ns = not significant, * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
In addition to nar mutations, our Tn-Seq screen identified many other genes whose activities affect nar/Nar expression/activity. For example, within the denitrification category, we found that disruption of genes involved in molybdenum transport (modABC) and synthesis of Nar’s molybdenum-containing cofactor (moaCDE), both of which are required for anaerobic growth on nitrate (32, 33), provides a fitness advantage during chlorate treatment. Additionally, our screen identified mutations in downstream denitrification genes (nir, nor) that affect fitness, likely because the activity of other denitrification enzymes affects nar expression. For instance, a nitric oxide reductase (nor) mutant accumulates toxic nitric oxide, which in turn downregulates nar expression (34), and nor mutants are unable to grow via anaerobic nitrate respiration (33). Within the transcriptional regulators category, there are several genes known to affect nar/Nar expression/activity or the expression of other denitrification genes (nir, nor), including narXL, dnr, nirQ (35), lasR (28, 36, 37), and cysB, whose mutation leads to increased levels of the Nar activity-repressing Pseudomonas quinolone signal (38, 39).
In addition to the Nar enzyme, chlorate reduction also requires a source of electrons which originate from carbon catabolism, enter the membrane via NADH dehydrogenase, and are ultimately carried to Nar via quinol. Thus, it is reasonable to predict that mutations disrupting electron flow to Nar would increase fitness during chlorate treatment, while mutations increasing electron flow to Nar would decrease fitness. Consistent with this prediction, our screen identified NADH dehydrogenase mutants (nuoH, nuoM) as having increased fitness, and likewise this enzyme has been shown to be required for growth via anaerobic nitrate respiration in P. aeruginosa (33, 40). Genes involved in regulating catabolism, such as those that encode the carbon catabolite CbrAB/Crc system, were also identified in our screen. Although the mechanistic connection between the CbrAB/Crc system and Nar remains unclear, it was previously shown that ΔcbrB and Δcrc strains have altered expression of many denitrification genes (41), which could affect chlorate sensitivity. Future studies could explore whether carbon catabolite mutants, which have altered catabolic rates (42), also have altered electron flux towards Nar; changes in Nar activity (nitrite generation) are known to affect the expression of downstream denitrification genes (43).
Experimental validation of Tn-Seq Results
We validated our Tn-Seq findings by generating unmarked deletions in a number of genes identified through this screen. The resulting eleven mutant strains were exposed to chlorate and their percent survival was compared to WT (Fig. 1B). Of the mutant strains predicted to be more fit during chlorate exposure (Fig. 1B, cyan), ΔnarGHJI, ΔrpoN and ΔhflC strains showed the greatest increase in chlorate resistance. Of the mutant strains predicted to be less fit during chlorate exposure (Fig. 1B, magenta), ΔmsrA, ΔmsrB, and ΔlasR strains showed the greatest increase in chlorate sensitivity. We and others previously showed that strains with inactivating mutations in lasR, which encodes a quorum sensing regulator, have higher rates of Nar activity (28, 36), which likely explains its increased chlorate sensitivity.
Our Tn-seq approach was successful in identifying a novel set of genes for exploring mechanisms of chlorate toxicity and resistance, however, not all tested mutants behaved as predicted by our Tn-Seq results (Fig. 1B). One reason for this discrepancy may be that different conditions were used for each experiment. For Tn-seq, dense stocks of the transposon library (which was constructed under aerobic conditions) were thawed, grown anaerobically overnight, washed, and treated with chlorate. For our validation experiments, mutants were grown aerobically overnight (because some deletion mutants do not grow well anaerobically (e.g. ΔnarGHJI, ΔnorBC)) and then moved to the anaerobic chamber for chlorate treatment. There are many physiological differences between aerobically- and anaerobically-grown cells, including differences in nar expression (35), which might account for some of the discrepancies between these experiments. Additionally, although the transposon includes an outward-facing promoter to reduce polar effects, we cannot rule out the possibility that transposon insertion disrupted the expression of neighboring genes, such that unmarked deletion strains might not recapitulate transposon mutant findings.
Chlorate exposure leads to increased levels of methionine oxidation across the proteome
While our Tn-seq results support our understanding that chlorate sensitivity is primarily mediated through Nar activity, we opted to explore other processes to identify novel factors related to chlorate toxicity and resistance. The category of protein repair and degradation (Fig. 1A) particularly intrigued us because we did not see a direct connection to Nar activity. Notably, methionine sulfoxide reductase mutants (ΔmsrA, ΔmsrB) were more sensitive to chlorate than WT (Fig. 1B), so we chose to focus on these mutants to deepen our understanding of how chlorate treatment affects the cell.
Methionine sulfoxide reductases are responsible for repairing oxidized methionine residues by catalyzing the reduction of methionine sulfoxide (MetSO) to methionine (44). Each enzyme (MsrA, MsrB) reduces a specific stereoisomer of MetSO, although msrA and msrB have different expression profiles and confer different levels of protection to specific oxidants in P. aeruginosa (45). We hypothesized that Nar-generated chlorite oxidizes methionine side chains to MetSO, and that widespread methionine oxidation of the proteome contributes to cell death. To test this hypothesis, we measured MetSO levels across the proteome of untreated and chlorate treated cultures. We treated anaerobically-grown P. aeruginosa cultures (WT, ΔmsrAΔmsrB) with or without 1 mM chlorate for 8 hours before collecting cell pellets for proteomic analysis. After cell lysis, we considered the possibility that high levels of spurious, chlorate-independent methionine oxidation might occur during proteome sample preparation, which would skew our results. Accordingly, we used a method developed by Bettinger et. al (46) where lysates were treated with 18O-H2O2 to oxidize remaining methionine residues to MetS18O, thereby blocking chlorate-independent methionine oxidation and differentially labeling these residues (i.e. chlorate-dependent oxidation is MetS16O, while chlorate-independent oxidation is MetS18O). However, we found that even with the addition of the oxidizing agent H2O2, methionine residues were not fully oxidized in our lysates (Table S2), suggesting that high levels of chlorate-independent oxidation do not occur during sample preparation. Thus, we calculated the fraction oxidation for each methionine residue as the quantity of MetS16O divided by the sum of all unoxidized and oxidized methionine residues (Met + MetS18O + MetS16O).
Using these lysates, we identified 3,197 proteins from the P. aeruginosa proteome (5,886 possible proteins) by the direct quantitative observation of 25,760 unique peptide sequences. Among these peptides, 2,951 contained at least one methionine residue, of which 2,611 (88.5%) showed some level of oxidation. Ultimately, 1,861 unique methionine residues were identified as showing some level of oxidation (i.e. different peptide forms can account for the same methionine residue) (Table S3). We found that chlorate-treated cultures had substantially higher levels of methionine oxidation compared to untreated cultures (Fig. 2A, Table S2). On average, 10% of detected methionine residues were oxidized in the proteomes of untreated WT cultures, whereas 42% of detected residues were oxidized in chlorate-treated cultures. An even more dramatic effect was observed when performing this experiment with a ΔmsrAΔmsrB strain. Approximately 7% of detected methionine residues were oxidized in untreated ΔmsrAΔmsrB cultures, whereas 59% were oxidized in chlorate-treated cultures. The increased oxidation in chlorate-treated ΔmsrAΔmsrB compared to WT cultures is consistent with a role for these enzymes in repairing oxidized methionine residues during chlorate treatment.
Figure 2:

Chlorate-treated cells have increased levels of proteome-wide methionine oxidation. A. Percentage of methionine residues that are oxidized (methionine sulfoxide, MetSO) in proteomes of three biological replicates of WT and ΔmsrAΔmsrB cells treated with or without 1 mM chlorate for 8 hours. Differences between % MetSO were determined by one-way ANOVA; ns = not significant, **** = p < 0.0001. B. Hierarchical clustering diagram of 361 methionine residues identified across all four treatment conditions. Methionine residues coloration indicates extent of oxidation under each condition, which shows that some methionine residues appear to be oxidation targets during chlorate treatment. C. Identity of highly oxidized proteins from WT and/or ΔmsrAΔmsrB chlorate-treated samples organized by KEGG category. Highly oxidized proteins were defined as having at least one highly oxidized methionine, which was defined as a residue showing at least 20% oxidation in chlorate-treated samples and the level of oxidation was 1.5-fold higher in chlorate-treated compared to untreated samples. The complete data set can be found in Table S4.
To learn more about chlorate-mediated methionine oxidation, we focused on a subset of 361 methionine residues that were detected in all four conditions (WT −/+ chlorate, ΔmsrAΔmsrB −/+ chlorate). Focusing on this subset, we observed distinct methionine oxidation trends (Fig. 2B). Some methionine residues were highly oxidized (~100% oxidation) across all four conditions, whereas other methionine residues appeared to be targets of oxidation specifically during chlorate treatment. For instance, some methionine residues showed ~0% oxidation in untreated samples but ~100% oxidation in chlorate-treated samples. Additionally, there appeared to be a core set of methionine residues that were highly oxidized in both WT and ΔmsrAΔmsrB chlorate-treated samples, as well as highly oxidized residues that were unique to each strain during chlorate treatment (particularly in ΔmsrAΔmsrB, where more residues were highly oxidized in response to chlorate). To identify the proteins that are oxidation-prone during chlorate treatment, we defined highly oxidized proteins as those with at least one highly oxidized methionine residue; a methionine residue was considered highly oxidized when ≥ 20% of the residue was oxidized in chlorate-treated samples (75% of methionine residues showed < 20% oxidation in untreated WT cultures) and the level of methionine oxidation was ≥ 1.5-fold higher in chlorate-treated samples compared to untreated samples. Using these criteria, we identified 144 proteins that were highly oxidized in chlorate-treated WT and/or ΔmsrAΔmsrB samples (Table S4). When this analysis was performed in reverse, only 18 and 14 proteins were found to be more oxidized in untreated compared to chlorate-treated WT and ΔmsrAΔmsrB samples, respectively.
Using the KEGG Database, we assigned highly oxidized proteins to functional categories (Fig. 2C, Table S4). We found that many highly oxidized proteins were also expected to be highly abundant (47–49). For instance, many highly oxidized proteins are involved in transcription and translation: RNA polymerase (RpoB, RpoC), transcription elongation and termination factors (NusG, Rho), 5 ribosomal proteins, translation initiation and elongation factors (IF-2, EF-Tu, Ef-Ts, EF-G), and 4 aminoacyl-tRNA biosynthesis proteins. Additionally, highly oxidized proteins included those that might be synthesized in response to chlorate treatment, such as proteases (HslU, HslV, PepN), chaperones (ClpX, ClpAB, DnaK), and cysteine/methionine metabolism proteins (MetE, PA14_05220, PA14_05230) (Fig. 2C, Table S4). Interestingly, when we explored which KEGG categories were induced in response to chlorate, we found that each KEGG category in Figure 2C (excluding “Other”) showed increased protein abundance in response to chlorate stress in WT and/or ΔmsrAΔmsrB cells, suggesting that in addition to predicted categories (e.g. protein repair), cells upregulate a variety of functions in response to chlorate (Fig. S1). Accordingly, we explored whether these newly synthesized proteins were targets of methionine oxidation. We found that highly oxidized methionine residues were more abundant in chlorate-treated compared to untreated samples (t test, p < 0.01) (Fig. 3, closed circles). This pattern was observed in both WT and ΔmsrAΔmsrB cultures, but again, a larger fraction of methionine residues were oxidized in the mutant. Our data suggests that newly synthesized, nascent proteins may be particularly vulnerable to methionine oxidation compared to pre-existing proteins.
Figure 3:
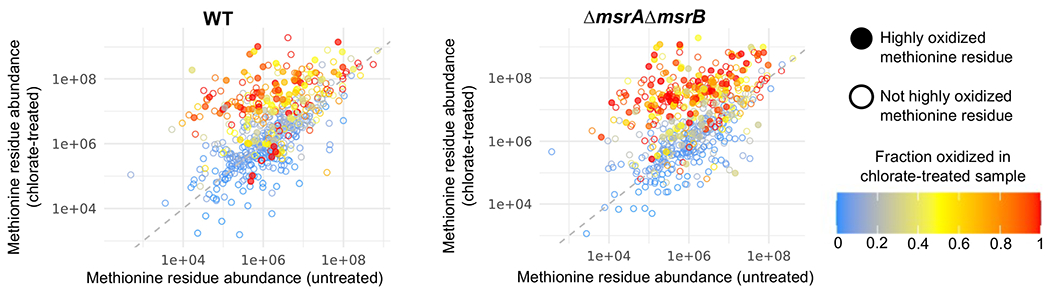
Proteins synthesized in response to chlorate treatment become methionine oxidation targets. Abundance of 361 methionine residues (those found across all four conditions) in untreated vs. chlorate-treated samples of WT (left panel) or ΔmsrAΔmsrB (right panel) cultures. Methionine residue coloration indicates the extent of residue oxidation in chlorate-treated conditions. Closed circles represent highly oxidized methionine residues (defined as fraction oxidation ≥ 0.2 in chlorate treated samples and fraction oxidation ≥ 1.5-fold higher in chlorate-treated compared to untreated samples); open circles represent methionine residues defined as not highly oxidized. Highly oxidized methionine residues (closed circles) are more abundant in chlorate-treated samples compared to untreated samples (t test, p < 0.01).
Of the 144 highly oxidized proteins, 76 were found to be highly oxidized in both WT and ΔmsrAΔmsrB chlorate-treated samples, while 12 and 56 were unique to WT and msrAΔmsrB chlorate-treated samples, respectively (Table S4). Although these strains did not show complete overlap in highly oxidized proteins, all highly oxidized proteins fell into the same functional categories, independent of the strain. In other words, it appears that the same types of proteins are oxidation targets in both strains, although higher levels of methionine oxidation and cell death occur in chlorate-treated ΔmsrAΔmsrB cultures compared to WT.
Limited periplasmic protein oxidation may occur during chlorate treatment
While chlorite is the product of chlorate reduction by Nar, chlorite’s interaction with other cellular components might generate other forms of reactive chlorine species. For example, chlorite is known to react with cysteine to generate hypochlorous acid (HOCl) (50). Unlike chlorite, HOCl is expected to remain uncharged at cytosolic pH, theoretically allowing it to diffuse across the membrane and react with periplasmic proteins; we therefore searched our data for signatures of oxidative stress in the periplasm. In addition to cytoplasmic Msr enzymes, P. aeruginosa also encodes YedYZ, where YedY is a periplasmic Msr and YedZ is an inner membrane-bound complex that supplies YedY with electrons from the quinone pool (51, 52). Our Tn-seq data showed that yedY and yedZ mutants had small fitness defects during chlorate treatment, with abundances decreasing by 1.3- and 1.2-fold, respectively (p ≤ 0.05). We next turned to our proteomic data set (Table S3) to further explore whether periplasmic protein oxidation occurs during chlorate treatment. Of 179 P. aeruginosa periplasmic proteins (protein localization predicted via www.pseudomonas.com), our approach identified 17 unique methionine residues (representing 16 periplasmic proteins) in at least one untreated/chlorate-treated pairing (i.e. WT +/− chlorate and/or ΔmsrAΔmsrB +/− chlorate). Of the 17 methionine residues, 2 were highly oxidized in chlorate-treated ΔmsrAΔmsrB cultures: M208 of the hypothetical protein encoded by PA14_07250, and M168 of AotJ which is involved in arginine/ornithine transport (53). We could not determine whether these methionine residues were highly oxidized in chlorate-treated WT cells because they were not identified in both untreated and chlorate-treated WT samples. Taken together, our data suggest that chlorate treatment might damage periplasmic targets, however we cannot exclude the possibility that these residues were oxidized in the cytoplasm prior to protein translocation to the periplasm.
Exogenous methionine partially rescues chlorate toxicity
Having found that chlorate treatment is correlated with increased levels of MetSO, we sought to determine whether widespread proteome oxidation causally contributes to cell death. To test this, we incubated cells with chlorate in the presence of exogenous methionine, which would act as a toxin sponge, protecting the proteome by absorbing some of the oxidative damage. This has been shown to be an effective strategy in other systems, where bacteria have evolved mechanisms of absorbing oxidative damage by expressing methionine-rich proteins (51, 54). We found that methionine addition increased chlorate survival in WT cultures, but the effect was mild, with 50 mM methionine addition producing a 5-fold increase in percent survival compared to no addition (Fig. 4). However, methionine addition had a much stronger protective effect on mutants with increased chlorate sensitivity. For instance, 50 mM methionine addition increased percent survival by 10-fold and >1000-fold in chlorate-treated ΔmsrAΔmsrB and ΔlasR cultures, respectively (Fig. 4). When this experiment was performed with leucine supplementation rather than methionine, there was no rescue effect in WT, ΔmsrAΔmsrB, or ΔlasR (Fig. S2A). However, the supplementation of cysteine, another sulfur-containing amino acid, also partially rescued all strains from chlorate killing, although cysteine itself was toxic at high concentrations (Fig. S2B). Finally, we also showed that amino acid addition in the absence of chlorate treatment is not toxic to P. aeruginosa (with the exception of high cysteine concentrations, Fig. S2C). The observation that adding sulfur-containing compounds (methionine, cysteine) increases survival during chlorate treatment supports the model that Nar-generated chlorite oxidizes sulfur compounds (e.g. causing proteome-wide methionine oxidation), which contributes to cell death.
Figure 4:

Exogenous methionine addition increases survival during chlorate exposure in WT cultures. This rescuing effect is even more pronounced in strains with increased sensitivity to chlorate (ΔmsrAΔmsrB, ΔlasR). Shown are three biological replicates with the mean ± SEM. Differences between chlorate-only and chlorate plus methionine log10(% survival) were determined by one-way ANOVA; ns = not significant, *** = p < 0.001, **** = p < 0.0001.
Chlorate resistance is correlated with decreased rates of chlorate reduction
In addition to identifying mechanisms of chlorate toxicity, we also wanted to learn about mechanisms of chlorate resistance beyond mutation of the nitrate reductase itself. In addition to ΔnarGHJI, we identified ΔrpoN and ΔhflC as strains with the greatest increase in chlorate resistance compared to WT (Fig. 1B). RpoN is an alternative sigma factor (σ54) that regulates genes involved in a wide range of functions, including nitrogen metabolism, motility (55), quorum sensing (56, 57), and secretion systems (58). HflC works with HflK to inhibit activity of the inner membrane-associated protease, FtsH (59, 60). FtsH has been shown to support tolerance of another reactive chlorine species (hypochlorous acid) and regulates a variety of functions, including quorum sensing molecules known to inhibit Nar activity (61).
Our prior work showed that nar mutants are fully resistant to chlorate because they do not reduce chlorate. Thus, we decided to test whether ΔrpoN and ΔhflC resistance was also linked to lower rates of chlorate reduction. When strains were grown aerobically and then shifted to an anoxic environment for chlorate treatment, we observed a trend where resistant mutants had increased survival and reduced less chlorate compared to WT, although differences between WT and ΔhflC chlorate consumption did not reach statistical significance (Fig. 5A). Interestingly, we obtained surprising results when the same experiment was performed using anaerobically-grown (rather than aerobically-grown) cells (Fig. 5B). Under these conditions, ΔrpoN was more sensitive to chlorate than WT, and likewise had reduced more chlorate than WT. Meanwhile, ΔhflC remained more resistant and reduced less chlorate compared to WT. While we do not yet understand the differing phenotypes of the ΔrpoN strain, given that RpoN is a global regulator (55, 62), we expect that its deletion will lead to pleiotropic effects with conditionally-sensitive phenotypes. Important here is the observed trend: cells that reduce more chlorate are more sensitive, whereas cells that reduce less chlorate are more resistant. Although other factors might also support chlorate resistance in ΔrpoN and ΔhflC strains (e.g. increased rates of repair of chlorite-damaged targets) in addition to changes in chlorate reduction rates, these results suggests that reduced Nar activity is a common mechanism of chlorate resistance.
Figure 5:
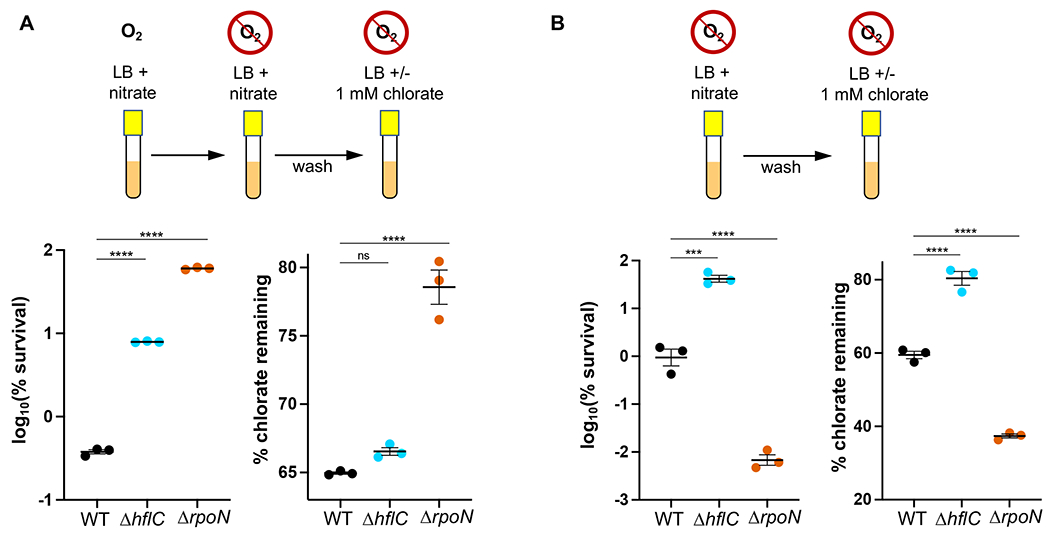
Chlorate resistance is correlated with lower rates of chlorate reduction. A. When grown aerobically, shifted to anoxia, and treated with chlorate, both ΔhflC and ΔrpoN cultures show increased survival and decreased chlorate reduction compared to WT. B. When grown anaerobically and treated with chlorate, ΔhflC has increased survival and decreased chlorate reduction, while ΔrpoN has decreased survival and increased chlorate reduction compared to WT. Shown are three biological replicates with the mean ± SEM. Differences between WT and mutant log10(% survival) or % chlorate remaining were determined by one-way ANOVA; ns = not significant, *** = p < 0.001, **** = p < 0.0001.
For completeness, we also note that our ΔrpoN strain has a small colony phenotype, and sheds large colony suppressor mutants (Fig. S3). Because these suppressor mutants accounted for a small percentage of the population in these experiments (~ 0.05%), it seems unlikely that they contributed to our experimental results. The nuances of ΔrpoN physiology are interesting and worthy of follow up given that P. aeruginosa rpoN mutants are frequently isolated from chronic infection sites (63, 64).
Discussion
The aim of this study was to better understand mechanisms of toxicity and resistance for the novel pro-drug, chlorate, which kills antibiotic-tolerant populations of P. aeruginosa biofilms (28). Our chlorate Tn-seq results confirmed that Nar, the enzyme that reduces chlorate thereby generating toxic chlorite, is the primary mediator of chlorate sensitivity. Many of the genes identified and validated in our screen, such as those related to denitrification, transcriptional regulation, or catabolism/energy-conservation, likely modulate Nar (and thus chlorite production) by affecting its catalytic activity, regulation, or access to reductant. Beyond factors that control Nar, our results implicate methionine oxidation as playing an important role in chlorate-mediated death.
Methionine sulfoxide reductase genes (msrA, msrB) were some of the strongest hits in our chlorate Tn-seq screen and deletion mutant strains were more sensitive to chlorate than WT. We found that chlorate-treated cultures showed extensive proteome-wide methionine oxidation, and this was exacerbated in a ΔmsrAΔmsrB strain. Further, exogenous methionine partially rescued P. aeruginosa survival during chlorate treatment. These findings mirror how bacteria are known to respond to other reactive chlorine species, such as chlorite and hypochlorous acid (HOCl)/hypochlorite (HOCl pKa = 7.5) (65). For instance, (per)chlorate-reducing bacteria generate periplasmic chlorite as a byproduct of their mode of growth. Ultimately, this chlorite is enzymatically dismutated to oxygen and chloride (66), although some hypochlorite is inadvertently released during this process (67). Accordingly, (per)chlorate-reducing bacteria must contend with both chlorite and hypochlorite stress. In response, Azospira suillum expresses a methionine-rich peptide (MrpX) to sacrificially absorb the oxidative damage generated by these molecules. A. suillum also synthesizes a periplasmic methionine sulfoxide reductase under these conditions to repair oxidized MrpX (51). Additionally, bacteria can encounter HOCl in host environments, where innate immune cells generate this oxidizing agent as means of controlling bacterial populations (68). HOCl induces msr expression in many bacteria (65, 69, 70), and msr mutant strains of Escherichia coli and Helicobacter pylori are more sensitive to HOCl (54, 71). In H. pylori, catalase was shown to be important for surviving HOCl treatment not because of its catalytic activity, but because it is a methionine-rich protein. Similar to our findings, exogenous addition of enzymatically-dead, methionine-rich catalase was able to quench HOCl-imposed oxidative stress in H. pylori (54).
The parallels between bacterial responses to chlorate and HOCl treatment are intriguing and lead us to consider whether Nar-generated chlorite is further reduced to HOCl within the cell. We do not have direct evidence to support this idea, although reactions between cysteine and chlorite are expected to form HOCl, as well as chlorine dioxide (50), both of which readily oxidize methionine residues (65, 72). In addition to its reactivity with cysteine, chlorite was shown to directly oxidize protein methionine residues in vitro (51). Thus, it seems plausible that chlorite-mediated methionine oxidation might also generate HOCl (as in the case of cysteine oxidation), but to our knowledge, this has not been experimentally demonstrated. If HOCl is generated during chlorate treatment, one important distinction from classical HOCl studies is that the oxidant is generated intracellularly rather than supplied extracellularly. It is thought that exogenously-supplied HOCl is most toxic once it reacts with components of the inner membrane (65), which should be more easily accomplished when reactive chlorine species are produced within the cytoplasm by an inner membrane-bound protein.
Although generated in the cytoplasm, there is interest in whether chlorite (or other reactive chlorine species) can escape Nar+ pathogen cells to potentially damage nearby host cells. In prior work, we did not detect extracellular chlorite in chlorate-treated P. aeruginosa cultures (28) suggesting that chlorite is contained within the cell or converted to a different chemical species. In line with the above considerations, if generated, HOCl would more readily traverse the inner membrane to cause periplasmic protein oxidation compared to negatively charged chlorite. However, we did not observe extensive oxidation of periplasmic proteins during chlorate treatment; we identified only two highly oxidized methionine residues from periplasmic proteins, and we cannot rule out that these residues were oxidized in the cytoplasm prior to the protein’s translocation to the periplasm. Future studies should address these questions more directly: which reactive chlorine species arise during chlorate treatment, and to what extent do they escape the cytosol?
We found that proteins upregulated in response to chlorate were targets of methionine oxidation. Although we do not know why, it seems plausible that nascent peptides, such as those newly synthesized during an oxidative stress event, might present more exposed methionine residues, whose hydrophobic side chain might otherwise be buried within a folded protein structure. In line with this thinking, others have observed a cysteine residue that was only accessible for oxidation once the protein was partially unfolded (73). We also showed that proteins involved in a wide range of functions (metabolism of various substrates, DNA replication/repair, protein repair, transcription, and translation) were induced in response to chlorate, suggesting that cells respond to chlorate stress in a variety of different ways. Importantly, while we chose to explore the role of methionine oxidation in chlorate-mediated death, we acknowledge that other factors (e.g. DNA damage, lipid oxidation) likely also contribute to death during chlorate treatment. Indeed, there is some evidence to support further exploration of such hypotheses, including our Tn-Seq finding that a lexA (SOS response repressor) transposon mutant is more resistant to chlorate than WT (although this finding was not recapitulated in a deletion mutant strain, Fig. 1B), and our finding that proteins involved in DNA repair and lipid metabolism increase in abundance and are highly oxidized during chlorate treatment. Future studies are needed to define the role that these and other processes might play in chlorate-mediated death.
Finally, our Tn-seq approach also identified mutations that lead to increased chlorate resistance. Here again our results reinforce the connection between chlorate reduction (i.e. Nar activity) and sensitivity. How Nar activity is altered in chlorate-resistant strains (ΔhflC, ΔrpoN) remains unclear, and we cannot rule out that other factors might also contribute to increased resistance in these strains. Yet, we find these results to be encouraging: considering the evidence that nitrate respiration is an important metabolism that supports P. aeruginosa growth and survival during infection (22–24, 74), becoming resistant to chlorate could hinder pathogen fitness in such environments. A priority for future work will be to explore whether and/or how chlorate resistance evolves in pathogens in the context of the host environment.
Methods
Bacterial strain, growth conditions, and percent survival
Strains used in this study include P. aeruginosa UCBPP- PA14 (wild type (WT)) and isogenic ΔnarGHJI (28), ΔlasR (31), Δlon (75), ΔnirS and ΔnorBC (76) strains, as well as a variety of other mutants constructed in the WT background for this study (strain construction described below). All experiments used Luria-Bertani medium (LB; Difco) as a growth medium, supplemented with KNO3 as specified. Aerobic liquid cultures were incubated at 37°C with shaking at 250 rpm. Anaerobic work was conducted in an anaerobic glove box with a 95% N2–5% H2 atmosphere, and anaerobic cultures were incubated at 37°C without shaking. For relevant experiments, percent survival was calculated for each strain/condition by dividing CFU-per milliliter values from each replicate culture containing chlorate by the average CFU-per milliliter value from triplicate control cultures lacking chlorate and multiplying by 100. The conditions used for chlorate treatment (anoxia without nitrate) are non-growing conditions (28, Figure 4D), such that any decrease in percent survival results from death rather than growth inhibition relative to the untreated sample.
Transposon library chlorate exposure
The transposon library used in this work was generated previously and contains ~150,000 unique transposon mutants (averaging 1 insertion per ~40 bases) (31). Three aliquots (three replicates) of the transposon library were thawed on ice, and each diluted into 20 mL LB with 40 mM KNO3 (final OD500 = 0.05). Cultures were grown anaerobically for 13.5 hours, and the following work was also conducted in the anaerobic glove box until otherwise indicated. Each culture was split in half (control sample, chlorate treated sample), pelleted, and washed with LB twice to remove the KNO3. Cells were pelleted again and resuspended either in LB (control; final OD500 = 2) or LB with 1 mM NaClO3 (treatment; final OD500 = 2). Cells were incubated +/− NaClO3 for 30 minutes, after which cultures were removed from the anaerobic glove box, pelleted, and washed with LB three times to remove NaClO3. Resuspension were diluted into 25 mL of LB at OD500 = 0.02 for control samples and OD500 = 0.04 for chlorate treated samples (assumed ~50% cell death with chlorate treatment). Cultures were grown aerobically for 4.4 ± 0.3 and 5.4 ± 0.7 doublings for control and chlorate treated samples, respectively (mean ± SEM). After aerobic outgrowth, cultures were pelleted and stored at −80°C. The chlorate exposure conditions for this experiment (30 minute exposure to 1 mM NaClO3) were chosen to minimize killing in chlorate-treated library samples; we reasoned that significant death would make it harder to distinguish between the relative importance of different functions during early-stage chlorate stress, thus shorter exposure effectively improved the sensitivity of our measurement.
Transposon library sequencing and data analysis
Genomic DNA was extracted from pelleted samples and prepared for Illumina sequencing (DNA shearing, end-repair, polyC tailing, enrichment of transposon-genome junctions and addition of adapter for Illumina sequencing by PCR) as described previously (31). DNA was sequenced using 100 bp single-end reads on the Illumina HiSeq 2500 platform at the Millard and Muriel Jacobs Genetics and Genomics Laboratory at Caltech. Sequence derived from the transposon (TATAAGAGTCAG) was trimmed from each read using a custom perl script, and reads were mapped to the P. aeruginosa genome UCBPP-PA14 genome using Bowtie 2 (version 2.3.5.1, “bowtie2 -x <bt2.idx> -U <input.fastq> -S <output.sam>”) (77). The output sam files were converted to bam files using samtools (version 1.9, “samtools view -bSh <input.sam> > <output.bam>”) (78). featureCounts, from the subread package, was used to determine the total number of insertions per gene (version 2.0.0, “featureCounts -f –read2pos 5 -F SAF -a <input.saf.txt> -o <output.txt> <input.bam>”) (79). Statistical significance of differences in read counts per gene was assessed using the voom/limma packages (80) developed for differential expression analysis, and implemented though the Degust website (http://degust.erc.monash.edu) (81). Genes with ≤ 3 reads across 5 or more samples (of 6 total samples) were removed from the data. To be considered significant, a gene/mutant must have a fitness change ≥ 2 between chlorate-treated and untreated samples, and p < 0.05 (Table S1).
Mutant strain construction
UCBPP-PA14 markerless gene deletions were constructed by PCR-amplifying 0.5-1 kb fragments immediately upstream and downstream of the target locus (primers used in this study are listed in Table S5). Fragments were Gibson cloned (NEB E2611) (82) into HindIII- and SacI-digested pMQ30 (83), and the resulting plasmids were transformed into E. coli DH10B cells using LB supplemented with 20 μg ml−1 gentamicin for selection. These plasmids were then moved to UCBPP-PA14 via triparental conjugation (with E. coli +pRK2013), and merodiploids were selected by plating on VBMM medium supplemented with 50 μg ml−1 gentamicin (84). Streak-purified merodiploid strains were resuspended in PBS and plated on LB supplemented with 10% sucrose. Plates were incubated at room temperature and sucrose resistant/gentamicin sensitive colonies were PCR-screened to confirm that the genomic region had been deleted.
Chlorate sensitivity assays
To confirm Tn-Seq results, a select number of mutants were constructed based on Tn-Seq findings (described above), and cultures of each mutant strain were grown aerobically in LB with 40 mM KNO3. Overnight cultures were moved to the anaerobic glove box and incubated at 37°C for 2 hours to adapt to anoxia. Within the anaerobic glove box, cultures were then pelleted, washed twice with LB to remove residual KNO3, and resuspended in LB without (control) or with 10 mM NaClO3 to final OD500 = 2. Cultures were incubated at 37°C for 8 hours, after which they were removed from the anaerobic glove box to determine viable cell counts for calculating percent survival. For these experiments, cultures were grown aerobically and shifted to anoxia because some mutant strains (ΔnarGHJI, ΔnorBC) are unable to grow anaerobically. A higher chlorate concentration was used (10 mM) because aerobically-grown, anoxia-adapted cells are less sensitive to chlorate and we aimed to achieve 1-2 log killing in WT cultures so that resistant or sensitive mutants could be easily distinguished from WT.
Similar experiments were performed with chlorate resistant mutant strains (including ΔhlfC and ΔrpoN) to test chlorate reduction rates. Strains were grown aerobically in LB with 40 mM KNO3, and overnight cultures were moved to the anaerobic glove box and incubated at 37°C for 1 hour to adapt to anoxia. Cultures were then pelleted, washed twice with LB to remove residual KNO3, and resuspended in LB without (control) or with 1 mM NaClO3 to final OD500 = 2. Cultures were incubated at 37°C for 48 hours, after which they were removed from the anaerobic glove box to determine viable cell counts, and to collect supernatants to quantify remaining chlorate concentration (described below). In additional experiments, resistant mutant strains were grown anaerobically in LB with 40 mM KNO3 and these overnight cultures were pelleted, washed twice with LB to remove residual KNO3, and resuspended in LB without (control) or with 1 mM NaClO3 to final OD500 = 2. Cultures were incubated for 24 hours after which they were removed from the anaerobic glove box to determine viable cell counts, and to collect supernatants to quantify remaining chlorate concentration (described below). Longer chlorate exposure times (24-48 hours) were chosen for follow-up experiments with resistant mutants to test their ability to demonstrate resistance over longer timescales and harsher treatment conditions. Between these two approaches (oxic-anoxic adapted vs. anoxic only), we chose a longer incubation time for oxic-anoxic adapted conditions because these cells are less sensitive to chlorate than anoxically-grown cells, and we aimed to achieve roughly equal killing in chlorate-treated WT cultures under both conditions.
Chlorate quantification via ion chromatography
Culture samples were pelleted and the supernatant was diluted 1:10 in dH2O in a 0.5 mL vial (Thermo Fisher Scientific catalog numbers 038010 and 038011). Chlorate concentrations were determined using a Dionex ICS 2000 ion chromatography as described previously (28) at the Resnick Water and Environment Laboratory at Caltech.
MetSO proteome quantification
Pellet Collection:
WT and ΔmsrAΔmsrB strains were grown anaerobically in LB + 40 mM KNO3, and overnight cultures were pelleted and resuspended in LB without (control) or with 1 mM NaClO3 to final OD500 = 2. Cultures were incubated for 8 hours and then removed from the anaerobic glove box. Cultures were pelleted, washed once with PBS, and then stored at −80°C until sample preparation. The chlorate exposure conditions (8 hour exposure to 1 mM NaClO3) were chosen to achieve ~50% killing in WT cultures to indicate that cells were experiencing chlorate stress, while a sufficient number of intact cells remained for proteomic analysis.
Sample preparation:
These methods were adapted from Bettinger et al. (46). Cell pellets were thawed and lysed on ice with 200 μL lysis buffer (5% SDS/50mM triethylammonium bicarbonate) containing 1x Halt protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific #78447), followed by tip sonication at 20% power for cycles of 30 second on and 10 second off for a total of 5 minutes using a Fisher Scientific 550 Sonic Dismembrator. The samples were then clarified by centrifugation at 16,000xg for 10 minutes at 4°C and protein concentration of the supernatants measured by BCA assay (Thermo Fisher Scientific #23227). To inactivate catalase activity, 50 μL of each sample was heated at 95°C for 30 minutes. To induce labeled methionine oxidation, 20 μg of protein from each sample was volumed to 10 μL with lysis buffer (final concentration of 0.5 mg/mL) and treated with 10 μL 2.5% 18O-H2O2 (Sigma #609978; Cambridge Isotope Laboratories #OLM-7640-PK; final concentration of 1.25%) for 3 hours at room temperature. To prepare proteomes for LC-MS analysis, 18O-H2O2 treated samples were incubated with 2mM dithiothreitol (Sigma #D5545) for 20 minutes at room temperature, followed by incubation with 10mM iodoacetamide (Sigma #I1149) for 20 minutes at room temperature in the dark. Samples were then loaded onto a micro-size S-trap column (ProtiFi, LLC) as per manufacturer instructions. Briefly, samples were acidified by adding 27.5% phosphoric acid at 1/10 the sample volume, then 100 mM triethylammonium bicarbonate (TEAB) in 90% methanol was added at 6x the sample volume, and samples were loaded onto the column by centrifugation at 4000g for 30 seconds, followed by reapplying the flow-through to the S-trap two additional times. Columns were then washed 4x with 100 mM TEAB in 90% methanol by centrifugation at 4,000xg for 30 second, followed by a final centrifugation at 4,000xg for 1 minute. 2 μg trypsin (Thermo Fisher Scientific #20233) in 20 μL 50mM TEAB was then added (1:10 trypsin:protein) to each column and samples were incubated at 37°C overnight in a humidified incubator. Peptides were eluted from the column by 40 μL 50mM TEAB, followed by 40 μL 0.2% formic acid (Sigma #5.33002), and finally 40 μL 50% acetonitrile (ACN) (Sigma-Aldrich #1.00029). The 120 μL eluate of each sample was then lyophilized, then resuspended in 20 μL 0.2% formic acid by vortexing and sonication. Peptides were quantified using the Pierce quantitative colorimetric peptide assay kit (Thermo Fisher Scientific #23275) and portions of each sample were diluted to 100 ng/μL in 2% acetonitrile/0.2% formic acid.
Mass spectrometry:
LC-MS analysis of digested peptides was performed on an EASY-nLC 1200 (Thermo Fisher Scientific, San Jose, CA) coupled to a Q Exactive HF Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) equipped with a Nanospray Flex ion source. 500 ng peptides of each sample were directly loaded onto an Aurora 25cm x 75μm ID, 1.6μm C18 column (Ion Opticks) heated to 50°C. The peptides were separated with a 2hr gradient at a flow rate of 350 nL/min as follows: 2–6% Solvent B (7.5 min), 6-25% B (82.5 min), 25-40% B (30 min), 40-98% B (1 min), and held at 98% B (12 min). Solvent A consisted of 97.8 % H2O, 2% ACN, and 0.2% formic acid and solvent B consisted of 19.8% H2O, 80 % ACN, and 0.2% formic acid. The Q Exactive HF was operated in data dependent mode with Tune (version 2.8 SP1 build 2806) instrument control software. Spray voltage was set to 1.6 kV, S-lens RF level at 50, and heated capillary at 275°C. Full scan resolution was set to 60,000 at m/z 200. Automatic gain control target was 3 × 106 with a maximum injection time of 15 ms. Mass range was set to 375–1500 m/z and charge state inclusion set to select precursors of charge state 2-5 for DDA analysis. For data dependent ms2 scans the loop count was 12, AGC target was set at 1 × 105, intensity threshold was kept at 1 × 105, and dynamic exclusion set to exclude precursors after one time for 45 seconds. Isolation width was set at 1.2 m/z and a fixed first mass of 100 was used. Normalized collision energy was set at 28. Peptide match was set to off, and isotope exclusion was on. Data acquisition was controlled by Xcalibur (4.0.27.42), with ms1 data acquisition in profile mode and ms2 data acquisition in centroid mode.
MetSO Peptide Analysis:
LC-MS data was reduced and quantified using the MaxQuant (85) bioinformatics platform to achieve a 0.05 FDR at the PSM and protein inference level. Data was searched utilizing the UniProt curated proteome for Pseudomonas aeruginosa (UCBPP-PA14), consisting of 5,886 proteins (downloaded 2022.02.09), along with 158 known contaminants. The enzyme selected was trypsin. Modifications were selected as fixed Carbamidomethyl (C), and dynamic Oxidation (M), Oxidation O18 (M), and Acetyl (Protein N-term). Match tolerances were set to 20 ppm for precursors and 0.5 Da for fragments. Quantitative assessment of oxidation was performed using the method and python code proposed by Bettinger et al.(46). The R statistical compute language (86) was used to normalize quantitative values between data sets and compute protein expression differences using the limma package (87) as the difference in log2 mean values between binary classes and tested for significance using an empirical Bayes method with multiple hypothesis correction adjusted by the Benjamini-Hochberg method (88). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (89) partner repository with the dataset identifier PXD033396.
Methionine rescue experiments
WT and mutant cultures were grown anaerobically in LB with 40 mM KNO3, after which overnight cultures were pelleted and washed twice with LB to remove residual KNO3, then resuspended in LB without (control) or with 1 mM NaClO3 to final OD500 = 2. Cultures were also supplemented with one amino acid (methionine, leucine, or cysteine) at concentrations ranging from 0 – 50 mM. Cultures were incubated for 48 hours before being removed from the anaerobic glove box to determine viable cell counts for calculating percent survival. A longer chlorate exposure time (48 hours) was chosen for this experiment to achieve high levels of killing; this allowed us to test the rescue effect of amino acid addition across several orders of magnitude viability.
Supplementary Material
Supporting Information: Figures S1-S3
Table S1. Chlorate Tn-seq results
Table S2. Proteome labeling efficiency and average methionine oxidation by replicate
Table S3. Quantification of methionine oxidation by residue
Table S4. Highly oxidized proteins
Table S5. Primers used in this study
Acknowledgements
Grants to D.K.N. from the NIH (1R21AI146987-02) and the Doren Family Foundation supported this research. M.A.S. was supported by a postdoctoral fellowship from the Cystic Fibrosis Foundation (SPERO19F0). We thank Megan Bergkessel (University of Dundee) for help with Tn-seq analyses, including providing custom scripts, and Nathan Dalleska and the Resnick Water and Environment Laboratory (Caltech) for help with metabolite analyses. We also thank John Bettinger for providing the python code used to analyze methionine oxidation levels via the 18O-H2O2 labeling method that he and colleagues developed (Bettinger et al.) The Proteome Exploration Laboratory was supported by NIH OD010788, NIH OD020013, the Betty and Gordon Moore Foundation through grant GBMF775 and the Beckman Institute at Caltech.
Data Availability Statement
The proteomic data that supports the findings of this study are openly available in PRIDE at http://www.ebi.ac.uk/pride, accession number PXD033396.
References
- 1.Brauner A, Fridman O, Gefen O, Balaban NQ, Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nature reviews. Microbiology 14, 320–330 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Gutierrez A et al. , Understanding and sensitizing density-dependent persistence to quinolone antibiotics. Mol Cell 68, 1147–1154 e1143 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Hamad MA et al. , Adaptation and antibiotic tolerance of anaerobic Burkholderia pseudomallei. Antimicrobial agents and chemotherapy 55, 3313–3323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narten M, Rosin N, Schobert M, Tielen P, Susceptibility of Pseudomonas aeruginosa urinary tract isolates and influence of urinary tract conditions on antibiotic tolerance. Curr Microbiol 64, 7–16 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Borriello G et al. , Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrobial agents and chemotherapy 48, 2659–2664 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pabst B, Pitts B, Lauchnor E, Stewart PS, Gel-entrapped Staphylococcus aureus bacteria as models of biofilm infection exhibit growth in dense aggregates, oxygen limitation, antibiotic tolerance, and heterogeneous gene expression. Antimicrobial agents and chemotherapy 60, 6294–6301 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowley ES, Kopf SH, LaRiviere A, Ziebis W, Newman DK, Pediatric cystic fibrosis sputum can be chemically dynamic, anoxic, and extremely reduced due to hydrogen sulfide formation. mBio 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bjarnsholt T et al. , The in vivo biofilm. Trends Microbiol 21, 466–474 (2013). [DOI] [PubMed] [Google Scholar]
- 9.DePas WH et al. , Exposing the three-dimensional biogeography and metabolic states of pathogens in cystic fibrosis sputum via hydrogel embedding, clearing, and rRNA labeling. mBio 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyczak JB, Cannon CL, Pier GB, Lung infections associated with cystic fibrosis. Clinical microbiology reviews 15, 194–222 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sen CK et al. , Human skin wounds: a major and snowballing threat to public health and the economy. Wound repair and regeneration : official publication of the Wound Healing Society [and] the European Tissue Repair Society 17, 763–771 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.James GA et al. , Microsensor and transcriptomic signatures of oxygen depletion in biofilms associated with chronic wounds. Wound repair and regeneration : official publication of the Wound Healing Society [and] the European Tissue Repair Society 24, 373–383 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han G, Ceilley R, Chronic wound healing: A review of current management and treatments. Advances in therapy 34, 599–610 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martins-Mendes D et al. , The independent contribution of diabetic foot ulcer on lower extremity amputation and mortality risk. J Diabetes Complications 28, 632–638 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robbins JM et al. , Mortality rates and diabetic foot ulcers: is it time to communicate mortality risk to patients with diabetic foot ulceration? Journal of the American Podiatric Medical Association 98, 489–493 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Bharara M, Mills JL, Suresh K, Rilo HL, Armstrong DG, Diabetes and landmine-related amputations: a call to arms to save limbs. Int Wound J 6, 2–3 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cook GM, Greening C, Hards K, Berney M, Energetics of pathogenic bacteria and opportunities for drug development. Advances in Microbial Physiology, Vol 65: Advances in Bacterial Pathogen Biology 65, 1–62 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Palmer KL, Aye LM, Whiteley M, Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol 189, 8079–8087 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Line L et al. , Physiological levels of nitrate support anoxic growth by denitrification of Pseudomonas aeruginosa at growth rates reported in cystic fibrosis lungs and sputum. Front Microbiol 5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernatchez SF et al. , Nitric oxide levels in wound fluid may reflect the healing trajectory. Wound repair and regeneration : official publication of the Wound Healing Society [and] the European Tissue Repair Society 21, 410–417 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Winter SE et al. , Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–711 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beckmann C et al. , Use of phage display to identify potential Pseudomonas aeruginosa gene products relevant to early cystic fibrosis airway infections. Infect Immun 73, 444–452 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quinn RA et al. , Biogeochemical forces shape the composition and physiology of polymicrobial communities in the cystic fibrosis lung. mBio 5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolpen M et al. , Nitrous oxide production in sputum from cystic fibrosis patients with chronic Pseudomonas aeruginosa lung infection. Plos One 9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez CA et al. , Phage-mediated acquisition of a type III secreted effector protein boosts growth of Salmonella by nitrate respiration. mBio 3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hochstein LI, Tomlinson GA, The enzymes associated with denitrification. Annual review of microbiology 42, 231–261 (1988). [DOI] [PubMed] [Google Scholar]
- 27.Puig J, Azoulay E, Pichinoty F, Gendre J, Genetic mapping of the chl C gene of the nitrate reductase A system in Escherichia coli K12. Biochemical and biophysical research communications 35, 659–662 (1969). [DOI] [PubMed] [Google Scholar]
- 28.Spero MA, Newman DK, Chlorate specifically targets oxidant-starved, antibiotic-tolerant populations of Pseudomonas aeruginosa biofilms. mBio 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith DJ, Oliver CE, Taylor JB, Anderson RC, Invited review: Efficacy, metabolism, and toxic responses to chlorate salts in food and laboratory animals. Journal of animal science 90, 4098–4117 (2012). [DOI] [PubMed] [Google Scholar]
- 30.van Opijnen T, Camilli A, Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nature reviews. Microbiology 11, 435–442 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basta DW, Bergkessel M, Newman DK, Identification of fitness determinants during energy-limited growth arrest in Pseudomonas aeruginosa. mBio 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pederick VG et al. , Acquisition and role of molybdate in Pseudomonas aeruginosa. Appl Environ Microbiol 80, 6843–6852 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filiatrault MJ, Picardo KF, Ngai H, Passador L, Iglewski BH, Identification of Pseudomonas aeruginosa genes involved in virulence and anaerobic growth. Infect Immun 74, 4237–4245 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoon SS et al. , Two-pronged survival strategy for the major cystic fibrosis pathogen, Pseudomonas aeruginosa, lacking the capacity to degrade nitric oxide during anaerobic respiration. Embo J 26, 3662–3672 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schreiber K et al. , The anaerobic regulatory network required for Pseudomonas aeruginosa nitrate respiration. J Bacteriol 189, 4310–4314 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffman LR et al. , Nutrient availability as a mechanism for selection of antibiotic tolerant Pseudomonas aeruginosa within the CF airway. PLoS pathogens 6, e1000712 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toyofuku M et al. , Quorum sensing regulates denitrification in Pseudomonas aeruginosa PAO1. J Bacteriol 189, 4969–4972 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farrow JM 3rd, Hudson LL, Wells G, Coleman JP, Pesci EC, CysB negatively affects the transcription of pqsR and Pseudomonas quinolone signal production in Pseudomonas aeruginosa. J Bacteriol 197, 1988–2002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toyofuku M et al. , Influence of the Pseudomonas quinolone signal on denitrification in Pseudomonas aeruginosa. J Bacteriol 190, 7947–7956 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torres A et al. , NADH Dehydrogenases in Pseudomonas aeruginosa growth and virulence. Front Microbiol 10, 75 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sonnleitner E et al. , Novel targets of the CbrAB/Crc carbon catabolite control system revealed by transcript abundance in Pseudomonas aeruginosa. Plos One 7, e44637 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolff JA, MacGregor CH, Eisenberg RC, Phibbs PV Jr., Isolation and characterization of catabolite repression control mutants of Pseudomonas aeruginosa PAO. J Bacteriol 173, 4700–4706 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arai H, Igarashi Y, Kodama T, Nitrite activates the transcription of the Pseudomonas aeruginosa nitrite reductase and cytochrome c-551 operon under anaerobic conditions. Febs Lett 288, 227–228 (1991). [DOI] [PubMed] [Google Scholar]
- 44.Ezraty B, Aussel L, Barras F, Methionine sulfoxide reductases in prokaryotes. Biochimica et biophysica acta 1703, 221–229 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Romsang A, Atichartpongkul S, Trinachartvanit W, Vattanaviboon P, Mongkolsuk S, Gene expression and physiological role of Pseudomonas aeruginosa methionine sulfoxide reductases during oxidative stress. J Bacteriol 195, 3299–3308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bettinger JQ, Welle KA, Hryhorenko JR, Ghaemmaghami S, Quantitative analysis of in vivo methionine oxidation of the human proteome. J Proteome Res 19, 624–633 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mori M et al. , From coarse to fine: the absolute Escherichia coli proteome under diverse growth conditions. Mol Syst Biol 17, e9536 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidt A et al. , The quantitative and condition-dependent Escherichia coli proteome. Nat Biotechnol 34, 104–110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arike L et al. , Comparison and applications of label-free absolute proteome quantification methods on Escherichia coli. J Proteomics 75, 5437–5448 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Darkwa J, Olojo R, Chikwana E, Simoyi RH, Antioxidant chemistry: Oxidation of L-cysteine and its metabolites by chlorite and chlorine dioxide. J Phys Chem A 108, 5576–5587 (2004). [Google Scholar]
- 51.Melnyk RA et al. , Novel mechanism for scavenging of hypochlorite involving a periplasmic methionine-rich peptide and methionine sulfoxide reductase. mBio 6, e00233–00215 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gennaris A et al. , Repairing oxidized proteins in the bacterial envelope using respiratory chain electrons. Nature 528, 409–412 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishijyo T, Park SM, Lu CD, Itoh Y, Abdelal AT, Molecular characterization and regulation of an operon encoding a system for transport of arginine and ornithine and the ArgR regulatory protein in Pseudomonas aeruginosa. J Bacteriol 180, 5559–5566 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benoit SL, Maier RJ, Helicobacter catalase devoid of catalytic activity protects the bacterium against oxidative stress. J Biol Chem 291, 23366–23373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Totten PA, Lara JC, Lory S, The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J Bacteriol 172, 389–396 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heurlier K, Denervaud V, Pessi G, Reimmann C, Haas D, Negative control of quorum sensing by RpoN (sigma54) in Pseudomonas aeruginosa PAO1. J Bacteriol 185, 2227–2235 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thompson LS, Webb JS, Rice SA, Kjelleberg S, The alternative sigma factor RpoN regulates the quorum sensing gene rhll in Pseudomonas aeruginosa. Fems Microbiol Lett 220, 187–195 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Sana TG, Soscia C, Tonglet CM, Garvis S, Bleves S, Divergent control of two type VI secretion systems by RpoN in Pseudomonas aeruginosa. Plos One 8, e76030 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bandyopadhyay K, Parua PK, Datta AB, Parrack P, Escherichia coli HflK and HflC can individually inhibit the HflB (FtsH)-mediated proteolysis of lambdaCII in vitro. Arch Biochem Biophys 501, 239–243 (2010). [DOI] [PubMed] [Google Scholar]
- 60.van Bloois E et al. , Detection of cross-links between FtsH, YidC, HflK/C suggests a linked role for these proteins in quality control upon insertion of bacterial inner membrane proteins. Febs Lett 582, 1419–1424 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Kamal SM et al. , Two FtsH proteases contribute to fitness and adaptation of Pseudomonas aeruginosa clone C strains. Front Microbiol 10, 1372 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Damron FH et al. , Analysis of the Pseudomonas aeruginosa regulon controlled by the sensor kinase KinB and sigma factor RpoN. J Bacteriol 194, 1317–1330 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith EE et al. , Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A 103, 8487–8492 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vanderwoude J et al. , The evolution of virulence in Pseudomonas aeruginosa during chronic wound infection. Proc Biol Sci 287, 20202272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gray MJ, Wholey WY, Jakob U, Bacterial responses to reactive chlorine species. Annual review of microbiology 67, 141–160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Youngblut MD, Wang O, Barnum TP, Coates JD, (Per)chlorate in biology on Earth and beyond. Annual review of microbiology 70, 435–457 (2016). [DOI] [PubMed] [Google Scholar]
- 67.Hofbauer S et al. , Transiently produced hypochlorite is responsible for the irreversible inhibition of chlorite dismutase. Biochemistry-Us 53, 3145–3157 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Winterbourn CC, Kettle AJ, Redox reactions and microbial killing in the neutrophil phagosome. Antioxid Redox Signal 18, 642–660 (2013). [DOI] [PubMed] [Google Scholar]
- 69.Groitl B, Dahl JU, Schroeder JW, Jakob U, Pseudomonas aeruginosa defense systems against microbicidal oxidants. Molecular microbiology 106, 335–350 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Basu Thakur P et al. , Complex responses to hydrogen peroxide and hypochlorous acid by the probiotic bacterium Lactobacillus reuteri. mSystems 4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rosen H et al. , Methionine oxidation contributes to bacterial killing by the myeloperoxidase system of neutrophils. Proc Natl Acad Sci U S A 106, 18686–18691 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Loginova IV, Rubtsova SA, Kuchin AV, Oxidation by chlorine dioxide of methionine and cysteine derivatives to sulfoxides. Chem Nat Compd 44, 752–754 (2008). [Google Scholar]
- 73.Winter J, Linke K, Jatzek A, Jakob U, Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol Cell 17, 381–392 (2005). [DOI] [PubMed] [Google Scholar]
- 74.Palmer KL, Brown SA, Whiteley M, Membrane-bound nitrate reductase is required for anaerobic growth in cystic fibrosis sputum. J Bacteriol 189, 4449–4455 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Basta DW, Angeles-Albores D, Spero MA, Ciemniecki JA, Newman DK, Heat-shock proteases promote survival of Pseudomonas aeruginosa during growth arrest. Proc Natl Acad Sci U S A 117, 4358–4367 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilbert SA, Newman DK, The double-edged role of nitric oxide drives predictable microbial community organization according to the microenvironment. bioRxiv 2021.12.09.472001 (2021). [Google Scholar]
- 77.Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2. Nature methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li H et al. , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liao Y, Smyth GK, Shi W, featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 80.Ritchie ME et al. , limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Powell DR (2019) Degust v4.1.1. (Zenodo; ). [Google Scholar]
- 82.Gibson DG, Enzymatic assembly of overlapping DNA fragments. Methods in enzymology 498, 349–361 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O’Toole GA, Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol 72, 5027–5036 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Choi KH, Schweizer HP, mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nature protocols 1, 153–161 (2006). [DOI] [PubMed] [Google Scholar]
- 85.Tyanova S, Temu T, Cox J, The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nature protocols 11, 2301–2319 (2016). [DOI] [PubMed] [Google Scholar]
- 86.R_Core_Team (2021) R: A language and environment for statistical computer. (R Foundation for Statistical Computing; ), pp https://www.R-project.org. [Google Scholar]
- 87.Smyth GK, “limma: Linear Models for Microarray Data” in Bioinformatics and computational biology solutions using R and Bioconductor, Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, Eds. (Springer, New York, 2005), pp. 397–420. [Google Scholar]
- 88.Benjamini Y, Hochberg Y, Controlling the false discovery rate - a practical and powerful approach to multiple testing. J Roy Stat Soc B Met 57, 289–300 (1995). [Google Scholar]
- 89.Perez-Riverol Y et al. , The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res 50, D543–D552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information: Figures S1-S3
Table S1. Chlorate Tn-seq results
Table S2. Proteome labeling efficiency and average methionine oxidation by replicate
Table S3. Quantification of methionine oxidation by residue
Table S4. Highly oxidized proteins
Table S5. Primers used in this study
Data Availability Statement
The proteomic data that supports the findings of this study are openly available in PRIDE at http://www.ebi.ac.uk/pride, accession number PXD033396.


