Abstract
Gene editing technologies such as CRISPR/Cas9 have been used to improve many agricultural traits, from disease resistance to grain quality. Now, emerging research has used CRISPR/Cas9 and other gene editing technologies to target plant reproduction, including major areas such as flowering time and seed dormancy. Traits related to these areas have important implications for agriculture, as manipulation of flowering time has multiple applications, including tailoring crops for regional adaptation and improving yield. Moreover, understanding seed dormancy will enable approaches to improve germination upon planting and prevent pre-harvest sprouting. Here, we summarize trends and recent advances in using gene editing to gain a better understanding of plant reproduction and apply the resulting information for crop improvement.
Keywords: Genome editing, CRISPR/Cas9, Flowering time, Florigen, Seed dormancy
Introduction
Supplementing traditional breeding and selection with new genome manipulation technologies, such as plant transformation and (more recently) targeted genome editing, could substantially accelerate crop improvement (Borisjuk et al. 2019; Chen et al. 2019). Genome editing using specific targeted nucleases is a relatively young, burgeoning technology that is rapidly becoming an integral part of research and development in many areas of life science. Boosted by the advent of CRISPR/Cas9 nuclease systems based on target recognition by RNA:DNA complementarity, gene editing has had a huge impact on plant biology in less than 10 years, as it was readily adopted to introduce specific genetic changes in plant genomes in experiments that helped resolve difficult scientific questions and improved important traits in major agricultural crops. The rapid adoption of this technology for basic and applied research on the world’s most important crops is well illustrated by the number of NCBI-registered publications yielded by searches using the crop’s name “AND CRISPR” as a query; as of September 2020, this query yielded: 526 hits for rice (Oryza sativa), 127 for wheat (Triticum aestivum), 156 for maize (Zea mays), and 376 for Arabidopsis thaliana.
A variety of comprehensive, recent reviews have focused on advances in gene editing technologies (Razzaq et al. 2019; Bilichak et al. 2020; Gürel et al. 2020; Hahn et al. 2020; He and Zhao 2020; Hsieh-Feng and Yang 2020; Li and Xia 2020), and the application of these techniques to select crops such as rice (Oryza sativa, Biswal et al. 2019), bread wheat (Triticum aestivum; Borisjuk et al. 2019; Kumar et al. 2019; Hensel 2020), maize (Zea mays; Agarwal et al. 2018,), soybean (Glycine max; Bao et al. 2020), sorghum (Sorghum bicolor; Char and Yang 2020), and the improvement of certain traits, such as abiotic stress tolerance (Abdelrahman et al. 2018), disease resistance (Zaidi et al. 2016; Borrelli et al. 2018; Bisht et al. 2019), and grain quality (Fiaz et al. 2019).
However, the use of gene editing technologies in the field of plant reproduction such as flowering time and seed dormancy, which determine crop yield and sustainability in various environments, has not been highlighted. Flowering/bolting time is critical for plant reproduction and a key contributor to crop productivity, seed size, and grain nutritional quality (Gaudinier and Blackman 2020). For example, early bolting can limit vegetative growth and severely decrease yields, but late flowering can inhibit seed production. Flowering time is one of the most important agronomic traits determining grain yield and regional adaptation, as plants adapted to specific day lengths may not flower at different latitudes. Therefore, modulating the seasonal timing of reproduction is a major goal of scientists and breeders focused on developing novel plant varieties that are adapted to local environments and the changing climate (Jung and Müller 2009).
Mature seeds generally undergo a period of dormancy, followed by germination and production of the next generation of offspring. The strength of dormancy, defined as the seed’s resistance to germination, is an important agronomic trait, as a high level of dormancy will lead to non-uniform, variable germination after seed sowing in the field. However, certain combinations of environmental and genetic factors may eliminate or reduce seed dormancy, resulting in seeds that germinate on the spike, a phenomenon called pre-harvest sprouting (PHS). PHS presents a considerable problem for agriculture, particularly in regions where the rainy season overlaps with the harvest season. Therefore, finding the optimal balance of seed dormancy to prevent PHS but allow uniform germination in the field is pivotal for crop productivity.
Here, inspired by recent advances in the area and by reports presented at the 6th International Symposium on Plant Reproductive Development in the Summer of 2019 in Shanghai, we try to fill in the gaps and reflect on the current state of research involving the use of gene editing to modulate flowering time and seed dormancy, with a focus on major crop species.
Flowering time
Flowering time genetic network
The induction of flowering in most plants depends on seasonal cues, such as day length (photoperiod) and temperature, with plants integrating environmental signals to define flowering time. The long-day crops, such as wheat, barley (Hordeum vulgare), pea (Pisum sativum), and lentils (Lens culinaris), which mostly originated in the Fertile Crescent (Nakamichi 2014), flower in response to lengthening days in spring when the light period extends to a certain critical length. To flower, long-day plants usually need a period of low temperature in winter (vernalization) when they remain in a relatively cold-tolerant vegetative state. The short-day crops, such as rice, maize, sugarcane (Saccharum sp.), sorghum, and soybean, originate from areas closer to the equator and initiate flowering based on long dark periods.
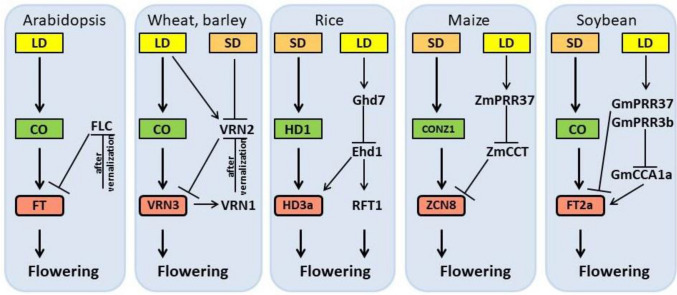
The molecular basis of the photoperiodic regulation of flowering time has been primarily studied in the long-day model plant Arabidopsis. In Arabidopsis, the switch from vegetative to reproductive development is implemented by a complex network of approximately 180 genes (Fornara et al. 2010), with some accelerating flowering and others repressing flowering. In Arabidopsis, the age, circadian clock, gibberellin biosynthesis and signaling, ambient temperature, vernalization, autonomous and photoperiod pathways converge on the floral integrator genes FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1). The molecular principles and pathways revealed in Arabidopsis (Fornara et al. 2010) have helped us understand flowering in major crops, and rapid progress in genomics has uncovered numerous genes involved in flowering time in rice (Kojima et al. 2002), maize (Meng et al. 2011), wheat, barley (Yan et al. 2006), sorghum (Wolabu et al. 2016), and tomato (Solanum lycopersicum, Lifschitz et al. 2006). Our current knowledge of the genetic and molecular/physiological mechanisms of flowering and flowering time in dicots and monocots is represented in Fig. 1 and summarized in detail in several reviews (Brambilla et al. 2017a; Li and Xu 2017; Leijten et al 2018).
Fig. 1.
Simplified flowering regulatory networks in the model plant Arabidopsis and major crops wheat, barley, rice, maize, and soybean. Arrows indicate gene activation and flat-ended lines indicate gene repression. Green boxes represent homologs of Arabidopsis CONSTANS (CO) and red boxes represent homologs of Arabidopsis FLOWERING LOCUS T (FT). Yellow and orange boxes represent long-day (LD) and short-day (SD) condition, respectively
The manipulation of flowering genes began with the activating targeting of two inflorescence meristem identity genes, APETALA1 (AP1) and LEAFY (LFY), and the florigen gene FT (Kardailsky et al. 1999; Kobayashi et al. 1999). FT, which is also referred to as florigen, acts as an integrator of pathways controlling flowering (Turck et al. 2008; Andrés and Coupland 2012). Arabidopsis FT was among the first targets of RNA-guided gene editing in plants when Hyun et al. used the CRISPR/Cas system to generate a heritable ft null allele in Arabidopsis, which had a late flowering phenotype, similar to other ft null alleles (Hyun et al. 2015). That same year, Ma et al. (2015) developed a multiplex CRISPR/Cas9-based gene editing system targeting 11 of the 13 FT-like genes in rice to study their functions. This system induced frame-shift mutations in the majority of targeted genes, resulting in premature leaf senescence phenotypes; however, the flowering phenotypes remain unclear (Ma et al. 2015).
Modulating flowering time in the major crops: maize, soybean, and rice.
Maize (Zea mays) was first domesticated in Mexico as a tropical species requiring a daylength of less than 13 h to flower, but was adapted to flower in the long-day environments of the USA, Canada, and Chile over the course of domestication (Hung et al. 2012). While the natural genetic variability of flowering time in maize allows wide adaption to diverse geographic zones, a detailed understanding of the basic principles might allow us to control flowering and thus maximize crop yield, especially in response to current challenges around climate change (Parent et al. 2018).
Flowering time control in maize is highly polygenic compared to the relatively simple regulation of flowering time in wheat and barley, which were domesticated in the Fertile Crescent (Cockram et al. 2007). Although many loci affecting natural variation in flowering time have been detected in maize (Buckler et al. 2009), the CCT (CO, CO-like and TIMING OF CAB1) domain gene ZmCCT is the best-characterized locus related to maize flowering time (Hung et al. 2012). ZmCCT is a homolog of Ghd7, a key regulator of the photoperiod response in rice. Ghd7 is expressed at maximum levels only in certain lines grown under long-day conditions (Xue et al. 2008). ZmCCT9 exhibits distinct diurnal expression and negatively regulates the expression of the FT ortholog ZCN8, thereby resulting in late flowering under long days (Huang et al. 2018); consistent with this, knockout of ZmCCT9 by CRISPR/Cas9 caused early flowering under long days (Huang et al. 2018).
Soybean, an important legume crop, is a typical short-day dicotyledon that flowers when the daylength is shorter than a certain threshold. This natural sensitivity to photoperiod limits its cultivation range. Therefore, generating daylength-insensitive soybean varieties is crucial for increasing the cultivation area of this crop to include lower and/or higher latitudes (Sedivy et al. 2017). The FT homologs GmFT2a and GmFT5a play similar, important roles in flowering, as overexpression of these two genes in soybean induced early flowering under long-day conditions (Nan et al. 2014). The CRISPR/Cas9-mediated T2 soybean mutant ft2a exhibits late flowering under both long-day and short-day conditions (Cai et al. 2018). Detailed analysis of ft2a, ft5a, and ft2a ft5a mutants under short- and long-day conditions (Cai et al. 2020b) revealed that GmFT2a has a greater effect than GmFT5a under short-day conditions, whereas GmFT5a has a greater effect under long-day conditions. GmFT5a is also essential for the adaptation of soybean to high-latitude regions. The ft2a ft5a double mutants showed a flowering time shift of approximately 31 days under short-day conditions, and produced significantly more pods and seeds per plant than the wild type, pointing to the huge yield potential of these mutants in the tropics.
In a follow-up study, the authors targeted GmFT2a and GmFT4 by base editing mediated by the nickase Cas9n (D10A) fused with rat cytosine deaminase and uracil glycosylase inhibitor as the base editor (Cai et al. 2020a). The C-to-G transition in GmFT2a in the ft2a-C7G-BE plants did not generate a frame-shift mutation, but it led to an amino acid change from proline to alanine. The base-edited mutants showed late flowering, but the flowering time shift was milder than that of the knockout mutants, demonstrating the ability to fine-tune flowering time using various CRISPR/Cas9-based tools.
In addition to directly targeting FT homologs, other studies in soybean have targeted regulators of GmFT expression. For example, soybean E1 encodes a B3 domain transcription factor that suppresses GmFT; E1 truncated by CRISPR/Cas failed to inhibit GmFT2a/5a causing early flowering (Han et al. 2019). Other studies have targeted Pseudo-Response Regulator (PRR) proteins, which play conserved roles in photoperiod responses in dicots and monocots (Wang et al. 2020). Two recent studies (Wang et al. 2020; Li et al. 2020) explored the roles of PRR proteins in regulating flowering time in soybean. The GmPRR proteins contain a C-terminal CCT domain and an N-terminal response regulator receiver domain. CRISPR/Cas9-induced mutations in GmPRR37 (Wang et al. 2020) and GmPRR3b (Li et al. 2020) promoted early flowering in soybean, whereas overexpressing GmPRR37 or GmPRR3b significantly delayed flowering. GmPRR37 downregulates the expression of the flowering-promoting genes GmFT2a and GmFT5a and upregulates the expression of the flowering inhibitor gene GmFT1a under long-day conditions, whereas GmPRR3b directly represses the expression of the flowering enhancer GmCCA1a. In both cases, the inhibitory effects of these proteins on flowering required the CCT domain, which was truncated in the gene-edited mutants due to added stop codons. Together, these studies shed light on the pathway linking the central circadian clock to flowering time regulation in soybean.
Rice, another crop domesticated in the tropics, also has a complex genetic network regulating flowering (Hori et al. 2016; Kong et al. 2016). Because the key components of flowering regulation in rice were identified and characterized prior to the widespread adoption of gene editing (Shrestha et al. 2014), newer gene editing studies have focused on some of the recently identified novel flowering control factors. For example, Brambilla et al. (2017b) used CRISPR/Cas9 to confirm the role of two bZIP transcription factors, Hd3a BINDING REPRESSOR FACTOR 1 (HBF1) and HBF2, in downregulating the rice flowering-promoting genes Early heading date 1(Ehd), Heading date 3a (Hd3a), and Rice Flowering Locus 1(RFT1) (Fig. 1). The double hbf1 hbf2 loss-of-function mutants flowered earlier than the wild type, and this phenotype was stronger when plants were grown under long-day compared to short-day conditions. Moreover, using electrophoretic mobility shift assays (EMSAs), the authors demonstrated that the HBFs most likely act by binding to the Abscisic Acid Responsive Elements (ABREs) in the Ehd1 promoter.
A recent study by Wu et al. (2020) targeted the Ehd1 gene itself with the aim to adapt Japonica rice, traditionally cultivated in the mid-latitude area to the lower latitude of Southern China. The mid-latitude Japonica varieties, which increase popularity over Indica because of their superior grain quality, commonly display early flowering when growing under short-day photoperiod and high temperature in low latitudes, resulting in low grain yield because of shortened vegetative growth period (Wei et al. 2016). The generated frame-shift Ehd1 mutants by CRISPR/Cas9 editing in four japonica varieties Nipponbare, Longdao16, Longdao24, and Xiushui134 demonstrated significantly longer vegetative growth periods compared with the wild-type plants, when planting under low-latitude conditions. The in-frame mutants exhibited intermediate-long vegetative growth periods. The field trials showed that both the in-frame and frame-shift mutant lines had significantly improved yield compared with wild-type plants, demonstrating the potential of proposed gene editing approach for adapting elite Japonica varieties for production in low latitude (Wu et al. 2020).
Co-expression analysis of photoperiodic flowering gene networks predicted that the Golden2 (G2)-like transcription factor OsPHL3 regulates flowering time in rice (Zeng et al. 2018). This role of OsPHL3 was confirmed by overexpression and CRISPR/Cas9-mediated knockdown. Rice lines overexpressing OsPHL3 showed delayed flowering, whereas knocking out OsPHL3 promoted flowering regardless of genetic background or photoperiod. These findings indicate that in addition to their diverse roles in regulating numerous processes, G2-like transcription factors also play critical roles as negative regulators of flowering time in rice.
Studies of genes related to other agronomic traits have also revealed unexpected effects on flowering. For example, editing of the yield-related gene GS3 by CRISPR/Cas9 in rice caused not only an increase in seed size (Li et al. 2016) but also an unexpected early flowering phenotype (Meng et al. 2018). Further research of the obtained plants can provide a better understanding of the genetic pathways linking flowering and yield.
Manipulating flowering genes in other crops
Following the major crops, CRISPR/Cas systems have been established to modify the expression of flowering genes in various minor crops such as sorghum, apple, pear (Pyrus sp.), rapeseed (Brassica napus), and tomato.
Genome editing technologies allow rapid functional analysis of homologs of flowering time regulators identified in Arabidopsis by facilitating the generation of loss-of-function mutations in just about any species for which a system for delivery editing reagents has been established. For example, a newly established Agrobacterium-mediated CRISPR/Cas9 delivery system was used to target a candidate FT gene in sorghum, resulting in a frame-shift mutation. The mutant exhibited a 10-day delay in flowering time, confirming that this gene functions in the regulation of flowering time (Char et al. 2020). Another example involved examination of homologs of the phosphatidyl-ethanolamine binding (PEPB)-like protein TERMINAL FLOWER 1 (TFL1), which prevents the expression of LFY and AP1. Overexpressing TFL1 led to late flowering in Arabidopsis (Ratcliffe et al. 1998). RNA interference of MdTFL1 led to precocious flowering in apple (Malus x domestica) (Kotoda et al. 2006). An efficient CRISPR/Cas delivery system has been developed for apple and European pear (Pyrus communis L.) (Charrier et al. 2019). Using this system, the authors successfully knocked out TFL1 expression in both species, which resulted in extreme phenotypes including the compete loss of vegetative growth and continuous flowering after only a few months of regeneration in vitro. Early flowering was observed in 93% of the apple lines targeting MdTFL1.1 and 9% of the pear lines targeting PcTFL1.1, where the majority of edited alleles harbored deletions of one or more bases.
To modulate flowering time in the oilseed crop Brassica napus, Jiang et al. (2018a) took advantage of information about the role of chromatin methylation in regulating flowering in Arabidopsis, i.e., that the methylation of histone H3 lysine is involved in activating FLOWERING LOCUS C (FLC) (He et al. 2004; Zhou et al. 2020). The Arabidopsis methyltransferase SET DOMAIN GROUP8 (SDG8) controls flowering time by directly altering the H3K36 m2/3 levels at the FLC locus, and the authors targeted the SDG8 homologs BnaSDG8.A and BnaSDG8.C. CRISPR/Cas9-mediated knockdown of these genes led to a drastic reduction in the number of days to flowering (from 120 days in wild type to 60 days) due to reduced H3K36 m2/3 levels in chromatin at the BnaFLC loci. Their results demonstrate that BnaSDG8.A/C directly participate in regulating flowering time by epigenetically modifying the chromatin at BnaFLCs (Jiang et al. 2018a). The approach of targeting factors involved in epigenetic regulation of gene expression could be used to control the floral transition via epigenetic chromatin modification; such approaches could be directly used to breed early flowering varieties of Brassica species and perhaps other crops.
Lippman’s group at Cold Spring Harbor Laboratory (Soyk et al. 2017) used CRISPR/Cas9 to engineer mutations in tomato SELF-PRUNING 5G (SP5G), which is a paralog of SINGLE-FLOWER TRUSS (SFT), a major inducer of flowering in tomato. SP5G differs from SFT by several amino acids within a domain determining florigenic activity; these differences converted SP5G into a flowering repressor or anti-florigen (Cao et al. 2016; Lifschitz et al. 2014). Mutations in SP5G resulted in the elimination of daylength sensitivity and the creation of an early yielding tomato variety. The authors speculate that targeting SP5G homologs in other crops could allow daylength sensitivity to be customized in a single step to expand the geographical cultivation range of elite varieties.
Vernalization in temperate cereals
The agricultural success of the temperate cereals wheat and barley relies on their adaptation to a wide range of environments. This adaptation is in part due to allelic diversity in the VRN vernalization genes, which regulate plant growth habits. VRN1 is an MADS-box transcription factor homologous to Arabidopsis AP1 that promotes flowering and VRN2 is a ZCCT domain-containing protein that acts as a floral repressor and shares sequence similarity with rice Ghd7.
Differences in VRN1, VRN2, and VRN3 underlie the separation of wheat and barley cultivars into winter and spring varieties (Distelfeld et al. 2009). The winter varieties are planted in the fall and require long exposure to low temperatures to induce flowering in the spring (vernalization requirement), whereas the spring varieties do not require vernalization and are planted in the spring. Both types have advantages and disadvantages for cultivation under certain conditions. For example, in China, winter wheat is cultivated in 5 out of 10 agricultural climatic zones, spring wheat is grown in 3 zones, and a mixture of spring and winter varieties is cultivated in 2 zones (Zhang et al. 2012).
According to the current model (Distelfeld et al. 2009; Brambilla et al. 2017a), the expression of VRN1 is induced by low temperatures and VRN1 directly binds to the VRN2 promoter, reducing its expression during vernalization. Suppressing VRN2 levels and/or the sufficient expression of VRN1 is required to induce the expression of VRN3 in temperate cereals; this gene is a homolog of Arabidopsis and rice FT. VRN1 expression is regulated by the binding of transcriptional repressors to two cis-elements in its promoter (a VRN-box and CArG-box) (Kane et al. 2007; Distelfeld et al. 2009) and by the interaction of its first intron with RNA-binding proteins (Xiao et al, 2014).
These repressor-binding sites are logical targets for gene editing aimed at converting winter wheat into a crop able to flower and produce seeds without vernalization. Zong et al. (2018) recently explored this notion using Cas9 nickase fused with a cytidine deaminase (A3A-PBE) for C-to-T base conversion of the VRN- and CArG-boxes in the TaVRN1-A1 promoter. Deep sequencing of amplicons from wheat protoplasts transfected with the A3A-PBE vectors identified mutations in these cis-elements with efficiencies ranging from 1.2–27.7%. These initial results pave the way for producing valuable mutants for further analyzing both the regulation of VRN1 expression and the process of vernalization in cereals as a whole.
Pre-harvest sprouting and seed dormancy
Molecular control of seed dormancy and PHS
PHS, dormancy of seeds, and efficient seed germination rely on intertwined pathways regulated by genetic and environmental factors. The genetic control of seed dormancy has not yet been completely elucidated, but the major structural and regulatory genes that form the broad genetic networks have been described (Reviewed in: Nonogaki and Nonogaki 2017; Tuan et al. 2018; Nakamura 2018; Vetch et al. 2019). The hormonal balance in seeds, particularly gibberellic acid (GA) and abscisic acid (ABA), also affects seed dormancy. The genetic control of GA/ABA sensitivity involves Viviparous-1 (Vp1), the ortholog of ABA INSENSITIVE 3 (ABI3), and MOTHER OF FT AND TFL1 (MFT), which encode proteins from the PEPB superfamily. Overexpressing TaMFT in wheat resulted in significantly longer dormancy and the absence of PHS (Nakamura et al. 2011). By contrast, RNA interference-mediated knockdown of TaMFT led to rapid seed germination and increased PHS, confirming the role of this gene in seed dormancy (Liu et al. 2013). Vp1 and MFT are positive regulators of ABA sensitivity but negative regulators of GA sensitivity (Jiang et al. 2018b) (see Fig. 2).
Fig. 2.
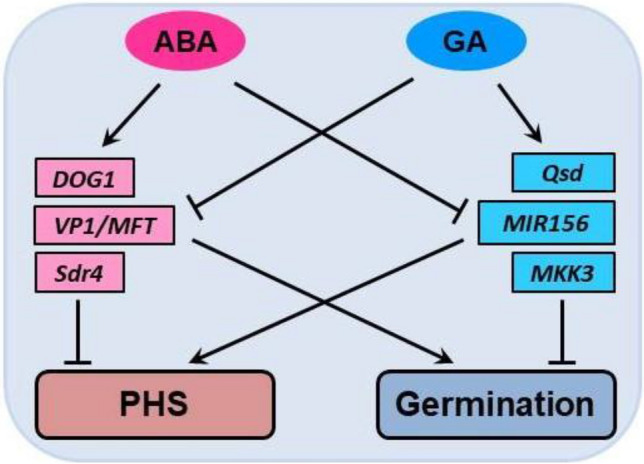
Simplified networks regulating pre-harvest sprouting (PHS) and germination. Phytohormones abscisic acid (ABA) and gibberellic acid (GA) play antagonistic role in regulation of PHS and germination. Arrows indicate 264 activation and flat-ended lines indicate repression
DELAY OF GERMINATION 1 (DOG1) plays a central role in controlling seed maturation and dormancy via the ABA-dependent inhibition of HYPERSENSITIVE GERMINATION 1 (a negative regulator of ABA responses in germinating seeds) (Vetch et al. 2019). MFT and DOG1 are members of GA- and ABA-regulated networks that negatively modulate seed germination and could therefore be effectively used against PHS (Reviewed in: Tuan et al. 2018; Nonogaki 2019; Vetch et al. 2019). Additionally, DOG1 is a major component of ABA signaling in seeds in a highly complex regulatory network involving the mitochondrial alternative respiration pathway and NADH dehydrogenase (Nonogaki 2019).
Seed dormancy 4 (Sdr 4) encodes a zinc finger protein that controls the expression of seed dormancy-related genes. OsSdr4 is a member of a regulatory network in rice together with OsDOG1 and OsVp1 (Sugimoto et al. 2010). Transformation of a wheat cultivar that did not show PHS with an allele of Mitogen Activated Kinase 3 (TaMKK3) from a PHS-susceptible cultivar strongly increased susceptibility to PHS. Genetic transformation of wheat genotypes with different TaMKK3 alleles resulted in changes in seed dormancy (Torada et al. 2016), indicating that this protein plays a vital role in PHS, possibly via protein phosphorylation and signal transduction (Danquah et al. 2015).
In general, red wheats are more PHS resistant than white wheats, an effect thought to be related to the presence of the red pigment precursor catechin, which inhibits germination (Himi et al. 2002). TaMyb10, encoding an R2R3-type transcription factor involved in flavonoid biosynthesis, is a promising candidate gene for altering grain color and avoiding PHS (Kato et al. 2017).
The alanine aminotransferase gene Qsd also deserves a special attention in efforts to limit PHS. Barley HvQsd strongly affects seed dormancy (Sato et al. 2016). In wheat, certain TaQsd1-B allelic variants produce significantly longer seed dormancy periods compared to others; for example, the TaQsd1-B allele in cv. Chinese Spring produces longer dormancy (Onishi et al. 2017).
Gene editing related to PHS and seed dormancy
Loss-of-function mutations of TaQsd1 were produced in all three homoeoalleles of bread wheat cv. Fielder via Agrobacterium-mediated CRISPR/Cas9 gene editing (Abe et al. 2019). After conventional crossing, triple heterozygotes (AaBbDd) were selected, and a triple homozygous mutant was developed, confirmed, and propagated. Sequence analysis revealed a single-nucleotide insertion in the target site in each of three homoeologous chromosomes causing shifts in the reading frames in TaQsd1-A, -B, and -D, thereby leading to the production of defective polypeptides. The triple-recessive homozygous CRISPR/Cas9 mutant of TaQsd1 showed no PHS phenotype and longer seed dormancy than the wild type. This study also demonstrated that novel breeding materials in bread wheat could be produced via genome editing using peptide-mediated delivery of gRNA/Cas9 protein complexes, thereby producing new, transgene-free, “non-GM” (non-Genetically Modified) cultivars (Abe et al. 2019). Indeed, the production of non-GM plants that lack transgenes might be one practical application of genome-editing technology in crop breeding.
Longer dormancy is an important trait for preventing PHS in rice; however, this might also negatively influence seed germination. The targeted mutagenesis of OsVP1 in rice via CRISPR/Cas9 gene editing successfully produced mutant lines with shorter dormancy and improved germination (Jung et al. 2019). The expression vector OsU3::vp1-sgRNA/pBOsC designed based on the OsVP1 sequence was delivered to callus tissue from seeds of the japonica rice cv. Dongjin using Agrobacterium-mediated transformation. Eighteen T0 mutants were produced with small insertion/deletions and substitutions in the target fragment of OsTP1, and four transgene-free homozygous knockout T1 lines were selected. Seeds of the four T1 gene-edited OsVP1 mutant lines showed significantly improved germination, with shorter seed dormancy compared to the wild type. These results are promising for the production of novel non-GM rice cultivars (Jung et al., 2019).
MIR156 from rice was identified as a microRNA that targets IDEAL PLANT ARCHITECTURE 1 (IPA1); downregulating MIR156 significantly improved grain yield. Mutations in five recently identified MIR156 subfamily members, MIR156a–MIR156c, MIR156k, and MIR156l, strongly suppressed PHS and led to longer dormancy (Miao et al. 2019). These results were achieved via CRISPR/Cas9 gene editing. Eleven MIR156 genes were targeted with six CRISPR/Cas9 vectors in rice cultivars Nipponbare and XS134 via Agrobacterium-mediated transformation. Five independent knockout mutant MIR156 lines were obtained in each background. The mutants showed significantly increased IPA1 expression, and their progenies showed strong improvements in PHS (Miao et al. 2019).
These successful examples of gene editing to eliminate PHS and prolong dormancy could be used as a guideline for further investigation of target genes involved in controlling PHS and seed dormancy.
Conclusions and perspectives
In this review, we describe the recent progress made in manipulating genes related to flowering time, seed dormancy, and PHS by gene editing technologies in major crops. The targeted genes and affected crop species are summarized in the Table 1.
Table 1.
List of genes targeted to manipulate flowering time, seed germination and dormancy
| Plant species | Targeted gene | Gene product/function | System for editing | Types of editing events | Phenotype manifestation | References |
|---|---|---|---|---|---|---|
| Arabidopsis thaliana | AtFT | PEPB /florigen, flowering induction | Embryo specific Cas9 expression | Small indels | Late flowering | Hyun et al. (2015) |
| A. thaliana | AtAP1 | MADS-box transcription factor/inflorescence meristem identity gene | Germ-line-specific Cas9 system | Small indels | – | Mao et al. (2016) |
| A. thaliana | AtTFL1 | PEPB-like protein/prevents the expression of inflorescence meristem identity genes | Dual-CRISPR/Cas9 | Mostly long deletions between targets, inversion mutation in target (2.2%) | – | Zhang et al. (2017) |
| Brassica napus | BnaSDG8.A and BnaSDG8.C | Histone 3 lysine 36 (H3K36) methyltransferase SDG8/activation of central repressor of FLOWERING LOCUS C (FLC) | CRISPR/Cas9 | Small indels | Early flowering | Jiang et al. (2018a, b) |
| Glycine max | GmFT2a | Homolog of AtFT/florigen, flowering induction | CRISPR/Cas9 | Mostly small indels | Late flowering under both LD and SD | Cai et al. (2018) |
| G. max | GmFT2a and GmFT4 | Homolog of AtFT, PEBP, putative kinase inhibitor/pathway integrator activates floral organ identity genes | Cas9-APOBEC1-UGI | Point mutations: C-to-G or C-to-T substitutions | Late flowering under both LD and SD | Cai et al. (2020a) |
| G. max | GmE1 | B3 domain transcription factor/regulation of photoperiodic flowering | CRISPR/Cas9 | Long deletions | Early flowering | Han et al. (2019) |
| G. max | GmPRR37 | Pseudo‐response regulator protein/regulation of photoperiodic flowering and circadian clock | CRISPR/Cas9 | Small indels | Early flowering under LD | Wang et al. (2020) |
| G. max | GmPRR3b | Pseudo‐response regulator protein/ regulation of photoperiodic flowering and circadian clock | CRISPR/Cas9 | Short deletions | Early flowering | Li et al. (2020) |
| Solanum lycopersicum | SlSP5G | Homolog of AtTFL1, PEPB-like protein/inhibition of inflorescence meristem identity genes | Dual-CRISPR/Cas9 | Small and long deletions | Rapid flowering under LD | Soyk et al. (2017) |
| Malus x domestica | MdTFL1.1 | Homolog of AtTFL1, PEPB-like protein/repressor of inflorescence meristem identity genes LFY and AP1 | Dual-CRISPR/Cas9 | Small indels located in the target sequence | Flowering short after in vitro regenerating | Charrier et al. (2019) |
| Oryza sativa | OsFTL1-11 | Homolog of AtFT, PEBP, putative kinase inhibitor/pathway integrator activates floral organ identity genes | Multiplex-CRISPR/Cas | Small indels | Premature leaf senescence | Ma et al. (2015) |
| O. sativa | OsMADS15 | Homolog of AtAP1, MADS-box transcription factor/inflorescence meristem identity gene CRISPR/Cas9 | CRISPR/Cas9 | Short deletions | Abnormalities in spikelet development | Song et al. (2017) |
| O. sativa | OsEhd1 | B-type response regulator/regulation of Hd3a and RFT1 expression | CRISPR/Cas9 | Mostly short deletions | Prolonged vegetative growth, late flowering, higher yield | Wu et al. (2020) |
| O. sativa | OsHBF1 and OsHBF2 | bZIP transcription factor Hd3a BINDING REPRESSOR FACTOR1 and 2/repress flowering | CRISPR/Cas9 | Small indels | Early flowering of double hbf1 hbf2 mutants | Brambilla et al. (2017b) |
| O. sativa | OsGS3 | Gamma subunit of G protein/grain size | CRISPR/Cas9 | – | Early flowering | Meng et al. (2018) |
| O. sativa | OsPHL3 | G2-like MYB-CC transcription factor/ regulation of chloroplast development and photosynthesis | CRISPR/Cas9 | Small indels | Early flowering under LD and SD conditions | Zeng et al. (2018) |
| O. sativa | OsVP1 | Viviparous-1 transcription factor/regulation of gibberellic acid and abscisic acid signaling | CRISPR/Cas9 | Small indels | Speeding-up of germination and reduction of seed dormancy | Jung et al. (2019) |
| O. sativa | OsMIR156 | miR156/suppress gibberellic acid signaling | Multiplex-CRISPR/Cas9 | Small indels | Enhanced seed dormancy and suppression of PHS | Miao et al. (2019) |
| Triticum aestivum | TaVRN1-A1 | Homolog of AtAP1, MADS-box AP-like transcription factor/inflorescence meristem identity gene | Cas9-APOBEC3A (RNP, transient test on protoplasts) | C-to-T substitutions | – | Zong et al. 2018 |
| T. aestivum | TaQsd1 | Alanine amino transferase/quantitative trait locus on seed dormancy 1 | CRISPR/Cas9 | Small indels | Changed germination rates | Abe et al. (2019) |
| Zea mays | ZmCCT9 | CCT domain-containing gene/ photoperiod response | Dual-CRISPR/Cas9 | Long deletions | Early flowering under LD | Huang et al. (2018) |
‘–’ not tested or data not provided
Flowering time. Flowering time is an important agronomic trait that helps to determine the geographic adaptation and productivity of crops. Since 1991, when the classic ABC model for floral organ identity was introduced (Coen and Meyerowitz, 1991) and numerous flowering time mutants were characterized in Arabidopsis (Koornneef et al. 1991), our understanding of the molecular mechanisms controlling the transition from the vegetative to reproductive state and flower development has progressed and expanded from a few model plant species to many agricultural crops (Blümel et al. 2015). Arabidopsis FT homologs in many crops have been established as major players in the floral transition and integrators of several flowering pathways. For example, during domestication and natural selection, maize became adapted to a short-day photoperiod by alterations in FT-related gene expression though a transposon integration in the ZmFT promoter. By editing FT homologs and other genetic determinants of flowering using the CRISPR/Cas system, flowering time was successfully modulated in a number of major crops such as maize, rice, and soybean, as well as other crops such as rapeseed, apple, pear, tomato, and sorghum. Based on these successes, we expect that flowering time could be adjusted in many other cultivated crops and likely in some valuable species that are currently not in widespread agricultural use.
Despite the lack of data on the effects of editing genes related to winter/spring growth habits in the temperate cereals wheat and barley, looking at progress in our understanding of the molecular mechanisms of vernalization requirements, we predict that we will soon see these crops engineered to convert from winter to spring growth and vice versa.
Pre-harvest sprouting and seed dormancy. PHS, seed germination on spikes prior to harvesting, is a serious problem that decreases yield and grain quality in wet Asian monsoon areas and is occurring in Europe with increasing frequency. Our understanding of PHS directly relies on our knowledge of seed dormancy, a process controlled by multiple gene networks and hormones. While our understanding of these networks is far from complete, recent studies have revealed a few key genes controlling seed dormancy. CRISPR/Cas targeting of these genes produced promising results, allowing this trait to be manipulated in wheat (Abe et al. 2019) and rice (Jung et al. 2019). We hope to see further progress in this area, which should help limit the losses caused by PHD and facilitate the optimization of seed germination.
Even though gene editing relies on numerous techniques used to generate transgenic plants (GM Technologies), in most cases, guided nucleases (such as RNA-guided CRISPR/ Cas9) produce small deletions/insertions or nucleotide transitions similar to those found in naturally occurring populations or produced by the conventional chemical mutagenesis (Voytas and Gao 2014). In classic GM plants, the new trait is associated with the introduced DNA sequence. By contrast, in gene-edited plants, once any transgenes encoding the genome-editing reagents have been segregated out (or in lines produced by emerging transgene-free editing methods), there is no way to distinguish between a naturally occurring mutation and a gene edit. Therefore, genome-edited plants are much more readily accepted by safety regulators in many countries (Dobrovidova 2019; Friedrichs et al 2019). Thus, we hope that the public acceptance of genome editing and its introduction into modern breeding programs will promote the rapid, precise improvement of major staple crops, minor horticultural crops, and emerging, not-yet-domesticated crops.
Acknowledgements
We thank the staff and students of Huaiyin Normal University, Huai’an (China), Flinders University of South Australia, SA (Australia), and S. Seifullin Kazakh AgroTechnical University, Astana (Kazakhstan) for their support in this study. We also thank Carly Schramm (Flinders University of South Australia, Adelaide, Australia) for critical comments on the manuscript.
Funding
This research was supported by a personal grant to NB from Huaiyin Normal University, Huai’an (China). The Ministry of Education and Science (Kazakhstan) also provided financial support for this research through Research Program BR05236500 (SJ).
Compliance with ethical standards
Conflicts of interest
The authors declare that the research was conducted in the absence of any potential conflict of interest.
References
- Abdelrahman M, Al-Sadi AM, Pour-Aboughadareh A, Burritt DJ, Tran L-SP. Genome editing using CRISPR/Cas9-targeted mutagenesis: An opportunity for yield improvements of crop plants grown under environmental stresses. Plant Physiol Biochem. 2018;131:31–36. doi: 10.1016/j.plaphy.2018.03.012. [DOI] [PubMed] [Google Scholar]
- Abe F, Haque E, Hisano H, Tanaka T, Kamiya Y, Mikami M, Kawaura K, Endo M, Onishi K, Hayashi T, Sato K. Genome-edited triple-recessive mutation alters seed dormancy in wheat. Cell Rep. 2019;28:1362–1369.e4. doi: 10.1016/j.celrep.2019.06.090. [DOI] [PubMed] [Google Scholar]
- Agarwal A, Yadava P, Kumar K, Singh I, Kaul T, Pattanayak A, Agrawal PK. Insights into maize genome editing via CRISPR/Cas9. Physiol Mol Biol Plants. 2018;24:175–183. doi: 10.1007/s12298-017-0502-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012;13:627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- Bao A, Tran L-S, Cao D. CRISPR/Cas9-based gene editing in soybean. In: Jain M, Garg R, editors. Legume Genomics. New York, NY: Humana; 2020. pp. 349–364. [Google Scholar]
- Bilichak A, Gaudet D, Laurie J. Emerging genome engineering tools in crop research and breeding. Methods Mol Biol. 2020;2072:165–181. doi: 10.1007/978-1-4939-9865-4_14. [DOI] [PubMed] [Google Scholar]
- Bisht DS, Bhatia V, Bhattacharya R. Improving plant-resistance to insect-pests and pathogens: The new opportunities through targeted genome editing. Semin Cell Dev Biol. 2019;96:65–76. doi: 10.1016/j.semcdb.2019.04.008. [DOI] [PubMed] [Google Scholar]
- Biswal AK, Mangrauthia SK, Reddy MR, Yugandhar P. CRISPR mediated genome engineering to develop climate smart rice: Challenges and opportunities. Semin Cell Dev Biol. 2019;96:100–106. doi: 10.1016/j.semcdb.2019.04.005. [DOI] [PubMed] [Google Scholar]
- Blümel M, Dally N, Jung C. Flowering time regulation in crops—what did we learn from Arabidopsis? Curr Opin Biotechnol. 2015;32:121–129. doi: 10.1016/j.copbio.2014.11.023. [DOI] [PubMed] [Google Scholar]
- Borisjuk N, Kishchenko O, Eliby S, Schramm C, Anderson P, Jatayev S, Kurishbayev A, Shavrukov Y. Genetic modification for wheat improvement: From transgenesis to genome editing. Biomed Res Int. 2019;2019:1–18. doi: 10.1155/2019/6216304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrelli VMG, Brambilla V, Rogowsky P, Marocco A, Lanubile A. The enhancement of plant disease resistance using CRISPR/Cas9 technology. Front Plant Sci. 2018;9:1245. doi: 10.3389/fpls.2018.01245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambilla V, Gomez-Ariza J, Cerise M, Fornara F. The importance of being on time: regulatory networks controlling photoperiodic flowering in cereals. Front Plant Sci. 2017;8:665. doi: 10.3389/fpls.2017.00665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambilla V, Martignago D, Goretti D, Cerise M, Somssich M, de Rosa M, Galbiati F, Shrestha R, Lazzaro F, Simon R, Fornara F. Antagonistic transcription factor complexes modulate the floral transition in rice. Plant Cell. 2017;29:2801. doi: 10.1105/tpc.17.00645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C, Ersoz E, Flint-Garcia S, Garcia A, Glaubitz JC, Goodman MM, Harjes C, Guill K, Kroon DE, Larsson S, Lepak NK, Li H, Mitchell SE, Pressoir G, Peiffer JA, Rosas MO, Rocheford TR, Romay MC, Romero S, Salvo S, Villeda HS, Sofia da Silva H, Sun Q, Tian F, Upadyayula N, Ware D, Yates H, Yu J, Zhang Z, Kresovich S, McMullen MD. The genetic architecture of maize flowering time. Science. 2009;325:714. doi: 10.1126/science.1174276. [DOI] [PubMed] [Google Scholar]
- Cai Y, Chen L, Liu X, Guo C, Sun S, Wu C, Jiang B, Han T, Hou W. CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotechnol J. 2018;16:176–185. doi: 10.1111/pbi.12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Chen L, Zhang Y, Yuan S, Su Q, Sun S, Wu C, Yao W, Han T, Hou W. Target base editing in soybean using a modified CRISPR/Cas9 system. Plant Biotechnol J. 2020 doi: 10.1111/pbi.13386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Wang L, Chen L, Wu T, Liu L, Sun S, Wu C, Yao W, Jiang B, Yuan S, Han T, Hou W. Mutagenesis of GmFT2a and GmFT5a mediated by CRISPR/Cas9 contributes for expanding the regional adaptability of soybean. Plant Biotechnol J. 2020;18:298–309. doi: 10.1111/pbi.13199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao K, Cui L, Zhou X, Ye L, Zou Z, Deng S. Four tomato FLOWERING LOCUS T-like proteins act antagonistically to regulate floral initiation. Front Plant Sci. 2016;6:1213. doi: 10.3389/fpls.2015.01213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Char SN, Yang B. Genome editing in grass plants. aBIOTECH. 2020;1:41–57. doi: 10.1007/s42994-019-00005-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Char SN, Wei J, Mu Q, Li X, Zhang ZJ, Yu J, Yang B. An Agrobacterium-delivered CRISPR/Cas9 system for targeted mutagenesis in sorghum. Plant Biotechnol J. 2020;18:319–321. doi: 10.1111/pbi.13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier A, Vergne E, Dousset N, Richer A, Petiteau A, Chevreau E. Efficient targeted mutagenesis in apple and first-time edition of pear using the CRISPR-Cas9 system. Front Plant Sci. 2019;10:40. doi: 10.3389/fpls.2019.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Wang Y, Zhang R, Zhang H, Gao C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol. 2019;70:667–697. doi: 10.1146/annurev-arplant-050718-100049. [DOI] [PubMed] [Google Scholar]
- Cockram J, Jones H, Leigh FJ, O’Sullivan D, Powell W, Laurie DA, Greenland AJ. Control of flowering time in temperate cereals: genes, domestication, and sustainable productivity. J Exp Bot. 2007;58:1231–1244. doi: 10.1093/jxb/erm042. [DOI] [PubMed] [Google Scholar]
- Coen E, Meyerowitz E. The war of the whorls: genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- Danquah A, de Zélicourt A, Boudsocq M, Neubauer J, Freidit Frey N, Leonhardt N, Pateyron S, Gwinner F, Tamby J-P, Ortiz-Masia D, Marcote MJ, Hirt H, Colcombet J. Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J. 2015;82:232–244. doi: 10.1111/tpj.12808. [DOI] [PubMed] [Google Scholar]
- Distelfeld A, Li C, Dubcovsky J. Regulation of flowering in temperate cereals. Curr Opin Plant Biol. 2009;12:178–184. doi: 10.1016/j.pbi.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Dobrovidova O. Russia joins in global gene-editing bonanza. Nature. 2019;569:319–320. doi: 10.1038/d41586-019-01519-6. [DOI] [PubMed] [Google Scholar]
- Fiaz S, Ahmad S, Noor MA, Wang X, Younas A, Riaz A, Riaz A, Ali F. Applications of the CRISPR/Cas9 system for rice grain quality improvement: perspectives and opportunities. Int J Mol Sci. 2019;20:888. doi: 10.3390/ijms20040888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, de Montaigu A, Coupland G. SnapShot: Control of flowering in Arabidopsis. Cell. 2010;141:550–550.e2. doi: 10.1016/j.cell.2010.04.024. [DOI] [PubMed] [Google Scholar]
- Friedrichs S, Takasu Y, Kearns P, Dagallier B, Oshima R, Schofield J, Moreddu C. An overview of regulatory approaches to genome editing in agriculture. Biotec Res Innov. 2019;3:208–220. doi: 10.1016/j.biori.2019.07.001. [DOI] [Google Scholar]
- Gaudinier A, Blackman BK. Evolutionary processes from the perspective of flowering time diversity. New Phytol. 2020;225:1883–1898. doi: 10.1111/nph.16205. [DOI] [PubMed] [Google Scholar]
- Gürel F, Zhang Y, Sretenovic S, Qi Y. CRISPR-Cas nucleases and base editors for plant genome editing. aBIOTECH. 2020;1:74–87. doi: 10.1007/s42994-019-00010-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn F, Korolev A, Sanjurjo Loures L, Nekrasov V. A modular cloning toolkit for genome editing in plants. BMC Plant Biol. 2020;20:179. doi: 10.1186/s12870-020-02388-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Guo B, Guo Y, Zhang B, Wang X, Qiu L-J. Creation of early flowering germplasm of soybean by CRISPR/Cas9 technology. Front Plant Sci. 2019;10:1446. doi: 10.3389/fpls.2019.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Zhao Y. Technological breakthroughs in generating transgene-free and genetically stable CRISPR-edited plants. aBIOTECH. 2020;1:88–96. doi: 10.1007/s42994-019-00013-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Doyle M, Amasino R. PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 2004;18:2774–2784. doi: 10.1101/gad.1244504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel G. Genetic transformation of Triticeae cereals – Summary of almost three-decade’s development. Biotechnol Adv. 2020;40:107484. doi: 10.1016/j.biotechadv.2019.107484. [DOI] [PubMed] [Google Scholar]
- Himi E, Mares D, Yanagisawa A, Noda K. Effect of grain colour gene (R) on grain dormancy and sensitivity of the embryo to abscisic acid (ABA) in wheat. J Exp Bot. 2002;53:1569–1574. doi: 10.1093/jxb/erf005. [DOI] [PubMed] [Google Scholar]
- Hori K, Matsubara K, Yano M. Genetic control of flowering time in rice: integration of Mendelian genetics and genomics. Theor Appl Genet. 2016;129:2241–2252. doi: 10.1007/s00122-016-2773-4. [DOI] [PubMed] [Google Scholar]
- Hsieh-Feng V, Yang Y. Efficient expression of multiple guide RNAs for CRISPR/Cas genome editing. aBIOTECH. 2020;1:123–134. doi: 10.1007/s42994-019-00014-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Sun H, Xu D, Chen Q, Liang Y, Wang X, Xu G, Tian J, Wang C, Li D, Wu L, Yang X, Jin W, Doebley JF, Tian F. ZmCCT9 enhances maize adaptation to higher latitudes. Proc Natl Acad Sci U S A. 2018;115:E334–E341. doi: 10.1073/pnas.1718058115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung H-Y, Shannon L, Tian F, Bradbury P, Chen C, Flint-Garcia S, McMullen M, Buckler E, Doebley J, Holland J. ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc Natl Acad Sci USA. 2012;109:E1913–E1921. doi: 10.1073/pnas.1203189109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y, Kim J, Cho SW, Choi Y, Kim J-S, Coupland G. Site-directed mutagenesis in Arabidopsis thaliana using dividing tissue-targeted RGEN of the CRISPR/Cas system to generate heritable null alleles. Planta. 2015;241:271–284. doi: 10.1007/s00425-014-2180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Li D, Jin L, Ruan Y, Shen W-H, Liu C. Histone lysine methyltransferases BnaSDG8.A and BnaSDG8.C are involved in the floral transition in Brassica napus. Plant J. 2018;95:672–685. doi: 10.1111/tpj.13978. [DOI] [PubMed] [Google Scholar]
- Jiang H, Zhao LX, Chen XJ, Cao JJ, Wu ZY, Liu K, Zhang C, Wei WX, Xie HY, Li L, Gan YG, Lu J, Chang C, Zhang HP, Xia XC, Xiao S-H, Ma CX. A novel 33-bp insertion in the promoter of TaMFT-3A is associated with pre-harvest sprouting resistance in common wheat. Mol Breed. 2018;38:69. doi: 10.1007/s11032-018-0830-1. [DOI] [Google Scholar]
- Jung C, Müller A. Flowering time control and applications in plant breeding. Trends Plant Sci. 2009;14:563–573. doi: 10.1016/j.tplants.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Jung Y, Lee H, Bae S, Kim J, Kim D, Kim H, Nam K, Nogoy FM, Yongbo D, Kang K. Acquisition of seed dormancy breaking in rice (Oryza sativa L.) via CRISPR/Cas9-targeted mutagenesis of OsVP1 gene. Plant Biotechnol Rep. 2019;13:511–520. doi: 10.1007/s11816-019-00580-x. [DOI] [Google Scholar]
- Kane NA, Agharbaoui Z, Diallo AO, Adam H, Tominaga Y, Ouellet F, Sarhan F. TaVRT2 represses transcription of the wheat vernalization gene TaVRN1. Plant J. 2007;51:670–680. doi: 10.1111/j.1365-313X.2007.03172.x. [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. Activation tagging of the floral inducer FT. Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- Kato K, Maruyama-Funatsuki W, Yanaka M, Ban Y, Takata K. Improving preharvest sprouting resistance in durum wheat with bread wheat genes. Breed Sci. 2017;67:466–471. doi: 10.1270/jsbbs.17042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M. Hd3a, a rice ortholog of the Arabidopsis FT Gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002;43:1096–1105. doi: 10.1093/pcp/pcf156. [DOI] [PubMed] [Google Scholar]
- Kong D, Chen S, Zhou L, Gao H, Luo L, Liu Z (2016) Research progress of photoperiod regulated genes on flowering time in rice. Hereditas (Beijing) 38:532–542. https://doi.org/10.16288/j.yczz.15-478 [DOI] [PubMed]
- Koornneef M, Hanhart C, Veen J. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- Kotoda N, Iwanami H, Takahashi S, Abe K. Antisense Expression of MdTFL1, a TFL1-like gene, reduces the juvenile phase in apple. J Am Soc Hort Sci. 2006;131:74–81. doi: 10.21273/JASHS.131.1.74. [DOI] [Google Scholar]
- Kumar R, Kaur A, Pandey A, Mamrutha HM, Singh GP. CRISPR-based genome editing in wheat: a comprehensive review and future prospects. Mol Biol Rep. 2019;46:3557–3569. doi: 10.1007/s11033-019-04761-3. [DOI] [PubMed] [Google Scholar]
- Leijten W, Koes R, Roobeek I, Frugis G. Translating flowering time from Arabidopsis thaliana to Brassicaceae and Asteraceae Crop Species. Plants. 2018;7:111. doi: 10.3390/plants7040111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xu M. CCT family genes in cereal crops: A current overview. Crop J. 2017;5:449–458. doi: 10.1016/j.cj.2017.07.001. [DOI] [Google Scholar]
- Li S, Xia L. Precise gene replacement in plants through CRISPR/Cas genome editing technology: current status and future perspectives. aBIOTECH. 2020;1:58–73. doi: 10.1007/s42994-019-00009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Li X, Zhou Z, Wu P, Fang M, Pan X, Lin Q, Luo W, Wu G, Li H. Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front Plant Sci. 2016;7:377. doi: 10.3389/fpls.2016.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Li Y, Li Y, Lu H, Hong H, Tian Y, Li H, Zhao T, Zhou X, Liu J, Zhou X, Jackson SA, Liu B, Qiu L. A domestication-associated gene GmPRR3b regulates the circadian clock and flowering time in soybean. Mol Plant. 2020;13:745–759. doi: 10.1016/j.molp.2020.01.014. [DOI] [PubMed] [Google Scholar]
- Lifschitz E, Eviatar T, Rozman A, Shalit A, Goldshmidt A, Amsellem Z, Alvarez JP, Eshed Y. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc Natl Acad Sci USA. 2006;103:6398–6403. doi: 10.1073/pnas.0601620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz E, Ayre BG, Eshed Y. Florigen and anti-florigen—a systemic mechanism for coordinating growth and termination in flowering plants. Front Plant Sci. 2014;5:465. doi: 10.3389/fpls.2014.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Sehgal S, Li J, Lin M, Trick H, Yu J, Gill B. Cloning and characterization of a critical regulator for pre-harvest sprouting in wheat. Genetics. 2013;195:263–273. doi: 10.1534/genetics.113.152330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu Y-G. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant. 2015;8:1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Mao Y, Zhang Z, Feng Z, Wei P, Zhang H, Botella JR, Zhu J-K. Development of germ-line-specific CRISPR-Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol J. 2016;14:519–532. doi: 10.1111/pbi.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Muszynski MG, Danilevskaya ON. The FT-like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell. 2011;23:942–960. doi: 10.1105/tpc.110.081406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng S, Xu P, Zhang Y, Wang H, Cao L, Cheng S (2018) CRISPR/Cas9-mediated editing of GS3 to improve flowering time in japonica Rice. Chin J Rice Sci 32(2): 119–127. 10.16819/j.1001-7216.2018.7112 (In Chinese with English abstract)
- Miao C, Wang Z, Zhang L, Yao J, Hua K, Liu X, Shi H, Zhu J-K. The grain yield modulator miR156 regulates seed dormancy through the gibberellin pathway in rice. Nat Commun. 2019;10:3822. doi: 10.1038/s41467-019-11830-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N. Adaptation to the local environment by modifications of the photoperiod response in crops. Plant Cell Physiol. 2014;56:594–604. doi: 10.1093/pcp/pcu181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Abe F, Kawahigashi H, Nakazono K, Tagiri A, Matsumoto T, Utsugi S, Ogawa T, Handa H, Ishida H, Mori M, Kawaura K, Ogihara Y, Miura H. A wheat homolog of MOTHER of FT and TFL1 acts in the regulation of germination. Plant Cell. 2011;23:3215–3229. doi: 10.1105/tpc.111.088492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S. Grain dormancy genes responsible for preventing pre-harvest sprouting in barley and wheat. Breed Sci. 2018;68:295–304. doi: 10.1270/jsbbs.17138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan H, Cao D, Zhang D, Li Y, Lu S, Tang L, Yuan X, Liu B, Kong F. GmFT2a and GmFT5a redundantly and differentially regulate flowering through interaction with and upregulation of the bZIP transcription factor GmFDL19 in soybean. PLoS ONE. 2014;9:e97669. doi: 10.1371/journal.pone.0097669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonogaki H. The long-standing paradox of seed dormancy unfolded? Trends Plant Sci. 2019;24:989–998. doi: 10.1016/j.tplants.2019.06.011. [DOI] [PubMed] [Google Scholar]
- Nonogaki M, Nonogaki H. Prevention of preharvest sprouting through hormone engineering and germination recovery by chemical biology. Front Plant Sci. 2017;8:90. doi: 10.3389/fpls.2017.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi K, Yamane M, Yamaji N, Tokui M, Kanamori H, Wu J, Komatsuda T, Sato K. Sequence differences in the seed dormancy gene Qsd1 among various wheat genomes. BMC Genomics. 2017;18:497. doi: 10.1186/s12864-017-3880-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent B, Leclere M, Lacube S, Semenov MA, Welcker C, Martre P, Tardieu F. Maize yields over Europe may increase in spite of climate change, with an appropriate use of the genetic variability of flowering time. Proceedings of the National Academy of Sciences. 2018;115(42):10642–10647. doi: 10.1073/pnas.1720716115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe O, Amaya I, Vincent C, Rothstein S, Carpenter R, Coen E, Bradley D. A common mechanism controls the life cycle and architecture of plants. Development (Cambridge, England) 1998;125:1609–1615. doi: 10.1242/dev.125.9.1609. [DOI] [PubMed] [Google Scholar]
- Razzaq A, Saleem F, Kanwal M, Mustafa G, Yousaf S, Imran Arshad HM, Hameed MK, Khan MS, Joyia FA. Modern trends in plant genome editing: an inclusive review of the CRISPR/Cas9 toolbox. Int J Mol Sci. 2019;20:4045. doi: 10.3390/ijms20164045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Yamane M, Yamaji N, Kanamori H, Tagiri A, Schwerdt J, Fincher G, Matsumoto T, Takeda K, Komatsuda T. Alanine aminotransferase controls seed dormancy in barley. Nat Commun. 2016;7:11625. doi: 10.1038/ncomms11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedivy EJ, Wu F, Hanzawa Y. Soybean domestication: the origin, genetic architecture and molecular bases. New Phytol. 2017;214:539–553. doi: 10.1111/nph.14418. [DOI] [PubMed] [Google Scholar]
- Shrestha R, Gómez-Ariza J, Brambilla V, Fornara F. Molecular control of seasonal flowering in rice, Arabidopsis and temperate cereals. Ann Bot. 2014;114:1445–1458. doi: 10.1093/aob/mcu032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song F, Li Q, Lu D, Wang L, Yuan Z (2017) Genome editing of rice OsMADS15 gene by using CRISPR-Cas9 system. Plant Physiol J 53 (6): 969–978. 10.13592/j.cnki.ppj.2017.0131(In Chinese with English abstract)
- Soyk S, Müller NA, Park SJ, Schmalenbach I, Jiang K, Hayama R, Zhang L, Van Eck J, Jiménez-Gómez JM, Lippman ZB. Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat Genet. 2017;49:162–168. doi: 10.1038/ng.3733. [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Takeuchi Y, Ebana K, Miyao A, Hirochika H, Hara N, Ishiyama K, Kobayashi M, Ban Y, Hattori T, Yano M. Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc Natl Acad Sci USA. 2010;107:5792–5797. doi: 10.1073/pnas.0911965107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torada A, Koike M, Ogawa T, Takenouchi Y, Tadamura K, Wu J, Matsumoto T, Kawaura K, Ogihara Y. A causal gene for seed dormancy on wheat chromosome 4A encodes a MAP kinase kinase. Curr Biol. 2016;26:782–787. doi: 10.1016/j.cub.2016.01.063. [DOI] [PubMed] [Google Scholar]
- Tuan P, Kumar R, Rehal P, Toora P, Ayele B. Molecular Mechanisms Underlying Abscisic Acid/Gibberellin Balance in the Control of Seed Dormancy and Germination in Cereals. Front Plant Sci. 2018;9:668. doi: 10.3389/fpls.2018.00668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- Vetch J, Stougaard R, Martin J, Giroux M. Review: Revealing the genetic mechanisms of pre-harvest sprouting in hexaploid wheat (Triticum aestivum L.) Plant Sci. 2019;281:180–185. doi: 10.1016/j.plantsci.2019.01.004. [DOI] [PubMed] [Google Scholar]
- Voytas DF, Gao C. Precision genome engineering and agriculture: opportunities and regulatory challenges. PLoS Biol. 2014;12:e1001877. doi: 10.1371/journal.pbio.1001877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Sun S, Wu T, Liu L, Sun X, Cai Y, Li J, Jia H, Yuan S, Chen L, Jiang B, Wu C, Hou W, Han T. Natural variation and CRISPR/Cas9-mediated mutation in GmPRR37 affect photoperiodic flowering and contribute to regional adaptation of soybean. Plant Biotechnol J. 2020 doi: 10.1111/pbi.13346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F-J, Tsai Y-C, Wu H-P, Huang L-T, Chen Y-C, Chen Y-F, Wu C-C, Tseng Y-T, Hsing YC. Both Hd1 and Ehd1 are important for artificial selection of flowering time in cultivated rice. Plant Sci. 2016;242:187–194. doi: 10.1016/j.plantsci.2015.09.005. [DOI] [PubMed] [Google Scholar]
- Wolabu TW, Zhang F, Niu L, Kalve S, Bhatnagar-Mathur P, Muszynski MG, Tadege M. Three FLOWERING LOCUS T -like genes function as potential florigens and mediate photoperiod response in sorghum. New Phytol. 2016;210:946–959. doi: 10.1111/nph.13834. [DOI] [PubMed] [Google Scholar]
- Wu M, Liu H, Lin Y, Chen J, Fu Y, Luo J, Zhang Z, Liang K, Chen S, Wang F. In-frame and frame-shift editing of the Ehd1 gene to develop japonica rice with prolonged basic vegetative growth periods. Front Plant Sci. 2020;11:307. doi: 10.3389/fpls.2020.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Xu S, Li C, Xu Y, Xing L, Niu Y, Huan Q, Tang Y, Zhao C, Wagner D, Gao C, Chong K. O-GlcNAc-mediated interaction between VER2 and TaGRP2 elicits TaVRN1 mRNA accumulation during vernalization in winter wheat. Nat Commun. 2014;5:4572. doi: 10.1038/ncomms5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, Zhang Q. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA. 2006;103:19581–19586. doi: 10.1073/pnas.0607142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SS-A, Tashkandi M, Mansoor S, Mahfouz MM. Engineering plant immunity: using CRISPR/Cas9 to generate virus resistance. Front Plant Sci. 2016;7:1673. doi: 10.3389/fpls.2016.01673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Liu X, Zhou Z, Li D, Zhao X, Zhu L, Yingfeng L, Hu S. Identification of a G2-like transcription factor, OsPHL3, functions as a negative regulator of flowering in rice by co-expression and reverse genetic analysis. BMC Plant Biol. 2018;18:157. doi: 10.1186/s12870-018-1382-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wang Y, Wu S, Yang J, Liu H, Zhou Y. A single nucleotide polymorphism at the Vrn-D1 promoter region in common wheat is associated with vernalization response. Theor Appl Genet. 2012;125:1697–1704. doi: 10.1007/s00122-012-1946-z. [DOI] [PubMed] [Google Scholar]
- Zhang C, Liu C, Weng J, Cheng B, Liu F, Li X, Xie C. Creation of targeted inversion mutations in plants using an RNA-guided endonuclease. Crop J. 2017;5:83–88. doi: 10.1016/j.cj.2016.08.001. [DOI] [Google Scholar]
- Zhou H, Liu Y, Liang Y, Zhou D, Li S, Lin S, Dong H, Huang L. The function of histone lysine methylation related SET domain group proteins in plants. Protein Sci. 2020;29:1120–1137. doi: 10.1002/pro.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Y, Song Q, Li C, Jin S, Zhang D, Wang Y, Qiu J-L, Gao C. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat Biotechnol. 2018 doi: 10.1038/nbt.4261. [DOI] [PubMed] [Google Scholar]