Abstract
Cereal crops including maize, rice, wheat, sorghum, barley, millet, oats and rye are the major calorie sources in our daily life and also important bioenergy sources of the world. The rapidly advancing and state-of-the-art genome-editing tools such as zinc finger nucleases, TAL effector nucleases, and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated systems (CRISPR-Cas9-, CRISPR-Cas12a- and CRISPR/Cas-derived base editors) have accelerated the functional genomics and have promising potential for precision breeding of grass crops. With the availability of annotated genomes of the major cereal crops, application of these established genome-editing toolkits to grass plants holds promise to increase the nutritional value and productivity. Furthermore, these easy-to-use and robust genome-editing toolkits have advanced the reverse genetics for discovery of novel gene functions in crop plants. In this review, we document some of important progress in development and utilization of genome-editing tool sets in grass plants. We also highlight present and future uses of genome-editing toolkits that can sustain and improve the quality of cereal grain for food consumption.
Keywords: Genome editing, Cereal crops, ZFNs, TALENs, CRISPR/Cas9, CRISPR/Cas12a, Base editor
Introduction
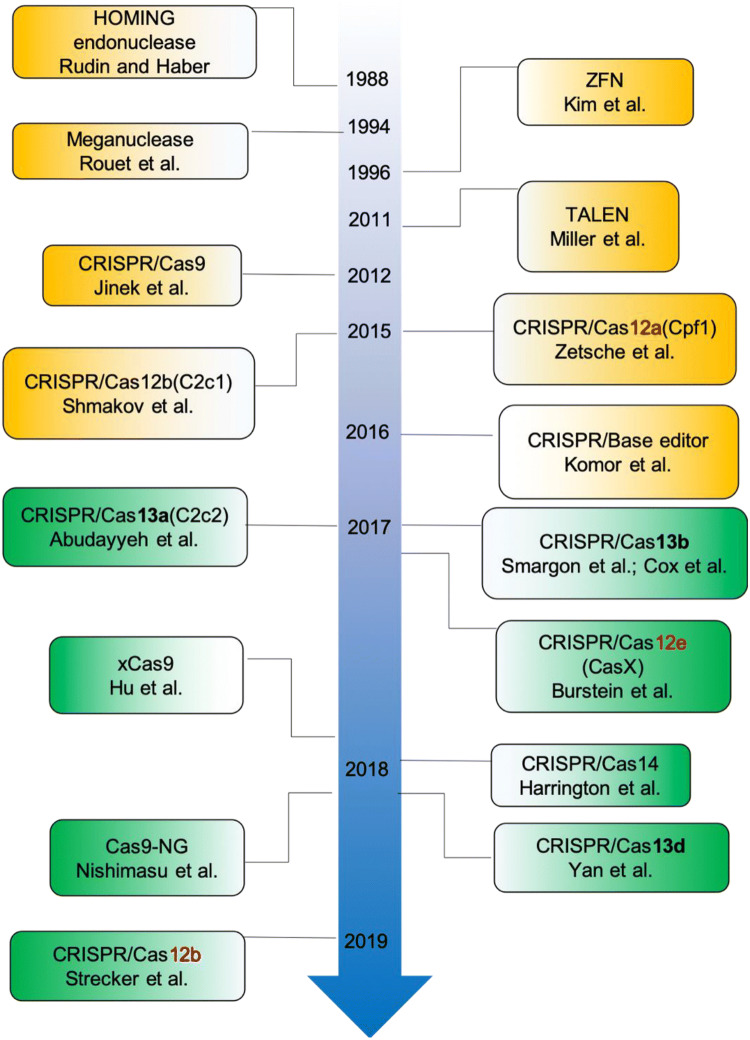
The basis of genome-editing technologies is to use programmable endonucleases to introduce site-specific double-stranded DNA breaks (DSBs) in vivo and exploit the intrinsic DSB repair mechanisms for desired DNA alterations within the genomes of interest. Several types of engineered nucleases (e.g., ZFNs, zinc finger nucleases; TALENs, transcription activator-like effector nucleases; CRISPR/Cas, clustered regularly interspaced short palindromic repeats and associated proteins) have been applied to introduce DSBs in eukaryotic cells. Subsequently, non-homologous end joining (NHEJ) and homology directed repair (HDR), as two major cellular DNA repair mechanisms, seal the DNA breakages. Random insertions and deletions (INDELs) occurring at the DNA cleavage sites are the outcomes of the error-prone NHEJ repair pathway. HDR, on the other hand, can lead to the desired and precise gene replacement if the homologous exogenous template/donor DNA with desired changes is present. Prevalent genome-editing technologies make use of NHEJ repair mechanism to create random mutations at the site-specific genomic loci that eventually lead to frameshift changes in the targeted gene, so-called targeted mutagenesis for gene inactivation. Several breakthroughs in genome editing by developing and applying customized endonucleases in eukaryotes have been made in the last two decades, particularly in the past 8 years (Fig. 1). This review will recapitulate the development and utilization of some of the readily developed genome-editing tools in grass plants (Table 1) and intend to have a comprehensive overview of the fast-paced and new frontiers in agricultural advancement.
Fig. 1.
Milestone and timeline of targeted mutagenesis methods
Table 1.
Examples of published genome editing methods in grass plants
| Grain crops | Meganuclease | ZFNs | TALENs | CRISPR/Cas9 | CRISPR/Cas12a | xCas9/Cas9-NG | Base editors (ABE/CBE) |
|---|---|---|---|---|---|---|---|
| Barley | ✓ (Wendt et al. 2013) | ✓ (Lawrenson et al. 2015) | |||||
| Brachypodium | ✓ (Shan et al. 2013a) | ||||||
| Maize | ✓ (Gao et al. 2010) | ✓ (Shukla et al. 2009) | ✓ (Liang et al. 2014) | ✓ (Lee et al. 2019) | ✓ (Zong et al. 2017) | ||
| Rice | ✓ (Jung et al. 2018) | ✓ (Li et al. 2012) | ✓ (Jiang et al. 2013) | ✓ (Xu et al. 2017) | ✓ (Wang et al. 2019) | ✓ (Zong et al. 2017) | |
| Sorghum | ✓ (Jiang et al. 2013) | ||||||
| Wheat | ✓ (Wang et al. 2014) | ✓ (Wang et al. 2014) | ✓ (Zong et al. 2017) |
Early discovery of ZFNs for genome editing
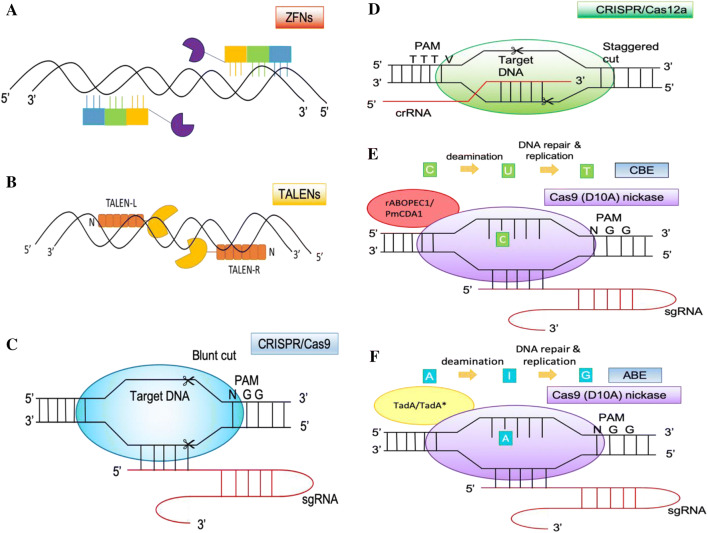
Zinc finger nuclease is the early and prominent tool for genome editing. ZFN comprises of a DNA binding domain of tandem zinc finger repeats fused with a DNA cleavage domain from the type II restriction endonuclease, FokI, as illustrated in Fig. 2A. Each zinc finger can bind to a triplet of DNA sequences (Carroll 2011). An array of three–six individual zinc finger repeats can recognize nine to eighteen base pairs of preselected DNA target (Carroll 2011). The zinc finger repeats can be custom-engineered and assembled to recognize the intended DNA sequences.
Fig. 2.
Currently established programmable nucleases including ZFNs, TALENs, CRISPR/Cas9-, Cas12a- and CRISPR-derived base editors
In general, three–six zinc fingers are selected from a premade library and assembled together as DNA-binding domain to recognize specific DNA sequences (Davies et al. 2017). Furthermore, a pair of assembled zinc finger DNA-binding domains is fused with FokI nuclease domains to target the preselected DNA sequences. Next, FokI nuclease domains dimerize to make the double-stranded DNA breaks at the target sites precisely.
ZFNs have been shown to be an efficient tool for targeted mutagenesis in maize and rice (Jung et al. 2018; Shukla et al. 2009) (Table 2). For example, two paralogues of ZmIPK1 gene that were implied in phytate biosynthesis pathway in maize seeds were chosen for targeted mutagenesis (Shukla et al. 2009). In rice, SSIVa locus, participating in starch biosynthesis process, was edited using zinc finger nucleases, and the mutagenized plants displayed the low starch, no grain filling and dwarf phenotype (Jung et al. 2018). Unfortunately, ZFNs have some disadvantages in recognizing specific DNA sequence accurately and generating off target DSBs, in addition to technical challenge to engineering. One improvement such as engineering heterodimer FokI nuclease domains was made to minimize off-target effects and increase the specificity of ZFNs (Miller et al. 2007). Unlike the CRISPR/Cas (see below) that requires PAM sequence at the target sites, ZFNs do not need this requirement, which has become one advantage of this technology.
Table 2.
Grass plants with established genome editing and application
| Speciesa | Targeted mutagenesis toolkits | Target gene(s) | Traits/phenotypes | Delivery methods | Editing efficiency | Reference |
|---|---|---|---|---|---|---|
| Rice | ZFNs | SSIVa | Low starch and dwarf | Agrobacterium | Jung et al. (2018) | |
| TALEN | OsSWEET11 &14 | Resistance to bacterial blight | Agrobacterium | T1: 48–63% | Li et al. (2012) | |
| Cas9/sgRNA |
OsBADH2 OsMPK2 OsPDS |
Albino and dwarf phenotype | Biolistic | 7.1%; 9.4% | Shan et al. (2013b) | |
| Cas9/sgRNA | OsSWEET11 &14 | Resistance to bacterial blight | Rice protoplasts | Jiang et al. (2013) | ||
| Cas9/sgRNA |
OsCAO1 OsLAZY1 |
Pale green leaf phenotype; tiller-spreading phenotype | Agrobacterium | T1: 83.3–91.6% | Miao et al. (2013) | |
| Cas9/sgRNA |
OsROC5 OsSPP OsYSA |
YSA target showing albino leaf phenotype | Agrobacterium |
T1: 26% 84% 5% |
Feng et al. (2013) | |
| Cas9/sgRNA | OsMPK5 | Rice protoplasts | 3–8% | Xie and Yang (2013) | ||
| Cas9/sgRNA | OsMYB1 | Agrobacterium | 50% | Mao et al. (2013) | ||
| Cas9/sgRNA |
OsSWEET1a OsSWEET1b OsSWEET11 OsSWEET13 |
Agrobacterium | 87–100% | Zhou et al. (2014) | ||
| Cas9/sgRNA |
OsDMC1A OsDMC1B OsCDK |
Agrobacterium | Mikami et al. (2016) | |||
| Cas9/sgRNA |
OsNAL1 OsLPA1 OsLG1 OsGL1-1 |
Narrow leave, increased tiller angle, loss of specialized organs (laminar joint, auricle and ligule) and disrupted formation of leaf cuticular wax | Agrobacterium | 2.1–23.4% | Hu et al. (2016) | |
| Cas9/sgRNA |
OsDL OsCYP72A33 OsCYP72A32 OsVIP1 OsVIP1-like |
Drooping leaves | Agrobacterium | Kaya et al. (2016) | ||
| Cas12a | OsPDS, OsBEL | Albino phenotype | Agrobacterium | T0: 13.6–21.4%; 20–41.2% | Xu et al. (2017) | |
| Cas12a | OsNAL1, OsLG1 | Agrobacterium | 5–12.1% | Hu et al. (2017) | ||
| Cas12a |
OsPDS OsDEP1 OsROC5 |
Albino phenotype; curly leaves | Agrobacterium | 93.8–100% | Tang et al. (2017) | |
| xCas9 | MONOCULM1 (MOC1), DWARF14 (D14) and PHYTOENE DESATURASE (PDS) | Agrobacterium | 2.08–29.17% | Wang et al. (2019) | ||
| xCas9 |
OsGS3 OsDEP1 |
Agrobacterium | 6.7–21.1% | Zhong et al. (2019) | ||
| Cas9-NG |
OsDEP1 OsPDS |
Agrobacterium | 3.5–56.3% | Zhong et al. (2019) | ||
| Brachypodium | TALENs |
BdABA1, BdCKX2, BdSMC6, BdSPL, BdSBP, BdCOI1, BdRHT, BdHTA1 |
Agrobacterium | Shan et al. (2013a) | ||
| Maize | I-CREI homing endonuclease | LIGULELESS1 (LG1) | Agrobacterium | 3% | Gao et al. (2010) | |
| ZFNs | IPK1 | Plasmid DNA delivery | Shukla et al. (2009) | |||
| TALENs | Gl2 | Agrobacterium | 10% | Char et al. (2015) | ||
|
TALEN & Cas9/sgRNA |
ZmPDS, ZmIPK1A, ZmIPK, ZmMRP4 | Enzyme in the phytic acid biosynthetic pathway | Agrobacterium | 13.3–39.1% in plants | Liang et al. (2014) | |
| Cas9/sgRNA | ZmHKT1 | Agrobacterium | 100% | Xing et al. (2014) | ||
| Cas9/sgRNA |
LIG, ALS2, MS26, MS45 |
ALS2: resistance to chlorsulfuron, MS45: male sterile | Biolistic | 2.4–9.7% | Svitashev et al. (2016) | |
| Cas9/sgRNA | Zmzb7 | Albino, leave chlorosis | Agrobacterium | 19–31%. | Feng et al. (2016) | |
| Cas9/sgRNA | phytoene synthase gene (PSY1) | Albino phenotype | Agrobacterium | 65.80%–86.87% | Zhu et al. (2016) | |
| Cas9/sgRNA |
ZmAgo18A, ZmAgo18B A1, A4 |
Agrobacterium | 70% | Char et al. (2017) | ||
| Cas9/sgRNA | LIGULELESS1 (LG1) | Smaller leaf angles | Agrobacterium | 51.5%–91.2% | Li et al. (2017a) | |
| Cas9/sgRNA | ARGOS8 | Improved grain yield under drought conditions | Biolistic | Shi et al. (2017) | ||
|
Cas9/sgRNA & Cas12a |
Gl2 | Glossy leave | Agrobacterium |
90–100% in the Cas9 0–60% in Cas12a |
Lee et al. (2019) | |
| Cas9/sgRNA | MS8 | Male sterile | Agrobacterium | Chen et al. (2018) | ||
| Cas9/sgRNA | Zmzb7 | Albino, chlorosis | Agrobacterium | 66% | Feng et al. (2018) | |
| Wheat | TALENs | TaMLO | Resistance to powdery mildew | Biolistic | 3.4–6.0% | Wang et al. (2014) |
| Cas9/sgRNA | TaMLO | Resistance to powdery mildew | Biolistic | 5.6% | Wang et al. (2014) | |
| Cas9/sgRNA |
TaGASR7 TaGW2 TaDEP1 TaNAC2 TaPIN1 TaLOX2 |
TaGASR7: control grain length and weight; TaDEP1: dwarf phenotype |
TECCDNA IVTs |
1–9.5% | Zhang et al. (2016) | |
| Cas9/sgRNA | α-Gliadin | Low-gluten | Biolistic | 62.3–75.1% | Sanchez-Leon et al. (2018) | |
| Cas9/sgRNA | phytoene desaturase (PDS) | Chimeric photobleaching phenotype | Agrobacterium | 11–17% | Howells et al. (2018) | |
| Cas9/sgRNA | TaGW2, TaLpx-1, and TaMLO | TaGW2: increase in seed size and grain weight | Particle bombardment | Wang et al. (2018) | ||
| Cas9/sgRNA | TaGASR7 | Particle bombardment | 5.2% | Hamada et al. (2018) | ||
| Cas9/sgRNA | TaCKX2-1, TaGLW7, TaGW2, and TaGW8, | Increased grain number per spikelet | Agrobacterium | 10% | Zhang et al. (2019b) | |
| Sorghum | Cas9/sgRNA | Reporter gene encoding for red fluorescence | Agrobacterium | Jiang et al. (2013) | ||
| Cas9/sgRNA | SbCENH3 | Potential lethality | Agrobacterium | Che et al. (2018) | ||
| Cas9/sgRNA | klC | Increases in digestibility and protein quality | Agrobacterium | 96% | Li et al. (2018a) | |
| Cas9/sgRNA | cinnamyl alcohol dehydrogenase(CAD) & phytoene desaturase(PDS) | Biolistic | Liu et al. (2019) | |||
| Barley | TALENs | HvPAPhy_a | Agrobacterium | Wendt et al. (2013) | ||
| Cas9/sgRNA | HvPM19 | Agrobacterium | 10%–23% | Lawrenson et al. (2015) | ||
| Switchgrass | Cas9/sgRNA | tb1a, tb1b, PGM | tb1a: tb1b: increased tiller production | Agrobacterium |
95.5% 11% 13.7% |
Liu et al. (2018) |
aOnly selected publications were listed
Use of TALENs for targeted mutagenesis
TALENs are another genetic tool for targeted mutagenesis just after ZFNs. TAL effectors (TALEs) originate from the plant pathogens of Xanthomonas and Ralstonia. They are a group of virulence factors from the Gram-negative bacterial pathogens entering into host plant cells via a type III secretion system (Boch and Bonas 2010). After translocated into the host cells, TALEs recognize and transcriptionally activate host target genes to condition disease susceptibility or trigger host resistance response based on the nature of target genes in plants (Boch and Bonas 2010). TALEs have a conserved central repetitive DNA binding domain consisting of multiple repeats of 33–35 amino acids (Boch and Bonas 2010). Two variable amino acids at positions twelve and thirteen of each repeat are known as repeat variable di-residues (RVDs). Four predominant RVDs such as NI, NG, NN and HD recognize the nucleotides adenine (A), thiamine (T), guanine (G), and cytosine (C), respectively (Boch et al. 2009; Moscou and Bogdanove 2009). At the N-terminus of TALE, it contains a type III translocation signal. At the C-terminus, TALE has acidic transcription activation domain (AD) and nuclear localization signals (NLS) (Zhu et al. 1998).
TALEs have been harnessed into biotechnology for accurate and site-specific mutagenesis in a plethora of grass plants (Table 2). Similar to ZFN, TALEN also is fusion protein derived from the programmable TALE DNA-binding domain and the nuclease domain of FokI. The DNA-binding domain is formed with tandem repeats of 34 amino acids; each repeat binds to a single base of the targeted DNA according to the TALE DNA recognition code. TALENs function as dimer and are apart by 16-bp to 20-bp of spacer (Christian et al. 2010; Li et al. 2011). DNA target site for TALENs is more specific due to the lengthy DNA sequences (32–48 bp) bound by the paired TALENs. Dimerized FokI nuclease domains induce a double-stranded DNA break within the spacer region after recognizing two subsites of the targeted region as simplified in Fig. 2B (Bogdanove and Voytas 2011). A variety of TALEN modular assembly methods including Golden Gate modular assembly and PCR-based methods have been well set up to engineer TALENs over the years (Akmammedov et al. 2016; Li et al. 2014; Zhang et al. 2013).
TALENs earned the Nature’s “Method of the Year” laurel in 2011 and were listed as the “breakthrough of 2012″ in Science. It holds a great promise to alleviate the devastating plant diseases including rice bacterial blight caused by Xanthomonas oryzae pv. oryzae (Xoo) (Li et al. 2012). The disruption of TAL effector (AvrXa7 and PthXo3) binding sites in the promoter region of OsSWEET14 has made the rice plants become strongly resistant to the blight disease (Li et al. 2012). In addition, TALENs together with CRISPR systems were utilized to edit the key enzymes (ZmPDS, ZmIPK1A, ZmIPK and ZmMRP4) engaged in phytic acid biosynthetic pathway in maize protoplasts (Liang et al. 2014). TALENs were also demonstrated to be an efficient targeted mutagenesis tool in Hi-II maize, targeting the endogenous Glossy2 gene with about 10% editing efficiency (Char et al. 2015). TALENs were also deployed to edit three homeologous alleles of MILDEW-RESISTANCE LOCUS (MLO) genes in hexaploid bread wheat that conferred resistance to the powdery mildew (Wang et al. 2014). Furthermore, TALENs were employed for gene modification in Brachypodium, including BdABA1 (Bradi5g11750), BdCKX2 (Bradi2g06030), BdSMC6 (Bradi4g08527), BdSPL (Bradi2g03740), BdSBP (Bradi4g33770), BdCOI1 (Bradi2g23730), BdRHT (Bradi1g11090), and BdHTA1 (Bradi1g25390) (Shan et al. 2013b). Last but not least, TALENs were tested to induce mutations in the specific genomic locus in barley (Wendt et al. 2013). In summary, TALEN technology has become a great tool in solving the problematic and pathogenic challenges that reduce the agricultural production. Also, the TALEN technology can advance the functional analysis of different genes in crops plants that feed the world. However, it is worthy to note that the process of TALEN vector construction involving several tedious steps that are time-consuming, cumbersome and labor-intensive has become a hurdle for many users.
Use of engineered CRISPR/Cas9 for genome editing
The function of CRISPR/Cas9 system as an acquired immunity to ward off intruding virus and foreign DNA materials is an important mechanism to protect archaea and bacteria (Barrangou 2013). The main components of CRISPR/Cas9 system for genome editing include the DNA cutting enzyme, Cas9, and the single chimeric guide RNA (sgRNA) derived from the combination of trans-activating crRNA (tracrRNA) and CRISPR RNA (crRNA) (Jinek et al. 2012) (Fig. 2C). Cas9 protein from S. pyogenes is the widely used nuclease for genome editing (Tsai and Joung 2016). Cas9 proteins from Streptococcus thermophilus, Staphylococcus aureus and Neisseria meningitidis have also been applied in the field of genome editing (Penewit et al. 2018; Muller et al. 2016). This class II type II Cas protein comprises of NHN domain and RuvC domain. NHN domain cleaves the complementary strand; whereas RuvC domain cleaves the non-complementary strand of the gRNA guide region (Nishimasu et al. 2014). Cas9 scans the target genome and searches first for the 3–8-nt-long protospacer-adjacent motif (PAM) sequence (5′-NGG-3′ for SpCas9; 5′-NNGRRT-3′ for SaCas9; 5′-NNNNGATT-3′ for NmCas9; 5′-NNAGAAW-3′ or 5′-NGGNG-3′ for StCas9), then uses the gRNA guide sequence (20-nt) for target recognition and generates site-specific DSBs at the target sequence three nucleotides upstream of the PAM (Jinek et al. 2012; Muller et al. 2016; Penewit et al. 2018). NHEJ DNA repair mechanism seals the broken DNA with the results of INDELs at the site of DSBs.
Easy assembly of CRISPR/Cas9 constructs and high efficiency of genome editing have made this system surpass ZFNs and TALENs just in a few years. Moreover, several creative and distinct CRISPR designs using CRISPR/Cas9 technology have made this system an even more appealing tool to many users. For instances, CRISPR/Cas9 system was shown to be effective in creating large chromosomal deletion in rice plants (Zhou et al. 2014). Also, multiplex genome editing using CRISPR/Cas9 is developed to target a gene family (OsMAPK) simultaneously using an endogenous tRNA processing system (Xie et al. 2015), and a single transcript unit CRISPR/Cas9 (STU CRISPR/Cas9) expression system is driven by a single Pol II promoter to express the two components of the reagents (Tang et al. 2019). These convenient tool kits in plants have the users make their desired CRISPR reagents much easier.
CRISPR-associated protein 9 (CRISPR/Cas9) has been extensively harnessed for site-specific mutagenesis in a plethora of grass plant species including rice, sorghum, maize, wheat, switch grass, and many other important grass plants (Jiang et al. 2013; Liang et al. 2014; Shan et al. 2014; Liu et al. 2018; Wang et al. 2014; Svitashev et al. 2015) (Table 2). Results from whole genome sequencing (WGS) in Japonica rice reveals that off-target mutations created by Cas9 is relatively low and carefully design intended sgRNA can reduce off-targeting effects (Tang et al. 2018). Tissue culture involving Agrobacterium transformation, on the other hand, generated high level of spontaneous mutations in different plants species (Tang et al. 2018). Similarly, off target effects of Cas9 in maize were evaluated using CIRCLE-seq and off-target mutations were barely found in edited lines when examined (Lee et al. 2019). Efforts have also been made to reduce the off-target effect of CRISPR systems. For example, altered versions of Cas9 [SpCas9-HF1 and eSpCas9 (1.1)] could reduce cleavage activity of Cas9 on substrate having mismatched and greatly reduced the off-target effects resulted from the CRISPR/Cas9 system (Chen et al. 2017). Design and use of shortened guide sequences of gRNA (e.g., 17–18 nt) could significantly reduce the off-target mutation without compromising on-target efficiency (Fu et al. 2014).
Moreover, some derivatives from CRISPR systems have been repurposed and adapted in the areas of epigenetic engineering (e.g., methylation, demethylation, acetylation, deacetylation), transcriptional regulation (activation and repression) and cell imaging in mammalian cells, but have not yet been fully materialized in plants (Kwon et al. 2017; Hilton et al. 2015; Gilbert et al. 2013; Chen et al. 2013; Knight et al. 2018; Vojta et al. 2016). In plants, some initial and pioneer work in development of transcriptional activation using VP64 and transcriptional repression using SRDX were established and applied in the monocots and dicots (Lowder et al. 2015). Followed by that, an improved version of CRISPR Act2.0 and mTALE-Act also has been made and used in multiplex genes activation (Lowder et al. 2018). Besides that, a newly developed dCas9-TAD that was named as dcas9-TV was shown to have stronger activation activity in plant and mammalian cells (Li et al. 2017c). In addition, live cell imaging in plants using CRISPR-Cas9 system to track the movements of telomere has been reported (Dreissig et al. 2017). These elegant methods made the sgRNAs, when fused with the dead Cas9 or nickase Cas9 system, a mini gadget with multi-functions and multi-tasks in basic scientific research. Evidently, CRISPR/Cas9 technology was nominated as the “breakthrough of the year” in 2015 by Science magazine. It has become much versatile in basic biology.
CRISPR/Cas12a, an alternative system, for genome editing
Furthermore, newly emerged Cas12a (formerly named as Cpf1) is a promising nuclease that has an efficient genome-editing frequency that only contains a single RuvC domain (Zetsche et al. 2015). Cas12a recognizes TTTV PAM (V could be A, C or G nucleotide) that expands the targetable regions in the grass plant genomes and provides broader choices for users when choosing the target sites within the gene of interest (Table 2). Various forms of Cas12a system include FnCas12a (Francisella novicida U112), AsCas12a (Acidaminococcus sp. BV3L6), LbCas12a (Lachnospiraceae bacterium ND2006), and many other Cas12a nucleases were demonstrated to function for genome editing in mammalian cells and has been successfully applied in rice and maize (Zetsche et al. 2015; Xu et al. 2017; Wang et al. 2017; Tang et al. 2017; Lee et al. 2019).
This system is extremely useful when targeting T-rich genomic region with a PAM sequence of 5′-TTTV-3′ (Zetsche et al. 2015). One distinct feature of Cas12a system is that the trans-activating crRNA (tracrRNA) is not part of guiding RNA, only the CRISPR RNA (crRNA) is needed for Cas12a (Zetsche et al. 2015)(Fig. 2D). Another difference of the CRISPR/Cas12a system is Cas12a nuclease creates staggered cut instead of blunt end cut of DNA target by CRISPR/Cas9 system (Zetsche et al. 2015). Staggered cut will be created distally from the PAM sequence, at the eighteenth nucleotide on the non-targeted strand and the twenty-third nucleotide on the targeted strand at the DNA cleavage site. These five-nucleotide overhangs at the 5′ ends are believed to increase efficiency of HDR mediated gene replacement in eukaryotic systems (Moreno-Mateos et al. 2017; Begemann et al. 2017a). Another advantage of the Cas12a system is less off-target effects as reported (Kim et al. 2016). This was further confirmed using whole genome sequencing in Cas12a edited rice with the results of no off-targeting mutations (Tang et al. 2018). Hence, unwanted off-target effects could be reduced using Cas12a nuclease. Also, this class II type V endonuclease itself can process pre-crRNA, so this system can be easily utilized as a multiplexing approach (Zetsche et al. 2017). Users may be optimistic and more favorable to use Cas12a system in engineering their crop plants of interest.
Another feature of Cas12a is the nuclease is thermosensitive. The activity of Cas12a decreases significantly below the temperature of 28 °C as first demonstrated in zebrafish embryos, while Cas12a has high cleavage activity at 37 °C (Moreno-Mateos et al. 2017). Similarly, Cas12a was also shown to be temperature sensitive in plant systems. AsCas12a was proved to have increased activity at high-temperature regime (above 28 °C) in rice; the editing efficiency could reach up to 93% in T0 plants (Malzahn et al. 2019). In maize and Arabidopsis, they were found to have the frequency of 100% edited mutants at 28 °C in T1 generation using LbCas12a (Malzahn et al. 2019). Therefore, using different temperature to modulate Cas12a activity will be an excellent approach in altering the genome-editing activity of Cas12a temporally and conditionally. It is known that different grass species prefer to have their favorable growing conditions at various temperatures. Moreover, transcriptional repression using CRISPR-Cas12a system has also been manifested in Arabidopsis that result in transcriptional reduction of miRNA gene specifically the miR195b (Tang et al. 2017).
Base editors induce genomic mutations without double-stranded breaks and in the absence of template DNA
Cytosine base editor (CBE) and adenine base editor (ABE) are two novel breakthrough technologies for targeted base editing. CBE and ABE enable single nucleotide conversion without DSBs or requiring donor template (Kang et al. 2018). The concept for CBE is to utilize mutant Cas9 (enzymatically dead Cas9 or Cas9 nickase in a later version) fused with cytosine deaminase that catalyzes cytosine deamination (Fig. 2E). Cytosine deaminase converses cytidine (C) to thymine (T) within certain ranges or windows of nucleotides that are located upstream of the PAM sequence (Komor et al. 2016). Cytosine deaminase has the ability to catalyze the conversion of cytosine into uracil (Komor et al. 2016). DNA repair mechanism then incorporates a T into the DNA during DNA replication (Fig. 2E). On the other hand, adenine base editor (ABE) utilizes the adenosine deaminase to convert A–G (Komor et al. 2016) (Fig. 2F). Adenosine is deaminated into inosine, G will be incorporated during DNA repair and replication processes as illustrated in Fig. 2F.
Different base editors have been well established in the past few years. The first generation of Base Editor was named as BE1, using the rat cytidine deaminase, APOBEC1, to convert the C–T within a window of approximately five nucleotides that are located 13-bp upstream from the PAM sequences (Komor et al. 2016). The subsequent generation of base editor was called BE2. It was improved with the fusion of uracil glycosylase inhibitor (UGI) that could inhibit the base excision repair mechanism and avoid unintended deletion (Komor et al. 2016). In the third generation of base editor (BE3), the dcas9 was substituted by Cas9 D10A nickase, leading to the efficiency of base editing with two- to sixfold increase relative to BE2 (Komor et al. 2016). Besides using defective Cas9 from S. pyogenes, Cas9 homolog from S. aureus (Sa) with the NNGRRT PAM also was used in base editing (SaBE3) (Kim et al. 2017). The conversion frequencies range from 50 to 75% (Kim et al. 2017). Lastly, the latest version of BE4 was appended with two UGIs at the C-terminus. Not surprisingly, BE4 is 1.5-fold more efficient than BE3 (Komor et al. 2017). To reduce INDEL frequency in base editing, GAM protein from bacteriophage Mu was utilized to fuse at the N-terminal regions of SpBE3, SaBE3, SpBE4, and SaBE4 (Komor et al. 2017). At the current stage, SaBE4-GAM combined all the desired improvements with reduced INDELs frequencies, increasing product purity and base editing efficiency (Komor et al. 2017).
On the other hand, to introduce conversion from A to G, the TadA: TadA* heterodimer was deployed to act as the adenosine deaminase to turn the base of A–G and T–C (Gaudelli et al. 2017). The current versions of adenine base editor include version ABE 0.1 to version ABE 7.10. Each of them has the slightly varied activity window upstream from the PAM sequences and editing efficiencies. Version ABE7.10 has the highest editing efficiency with an average of 53% and has the activity window of 4-bp to 7-bp upstream from the PAM sequence (Gaudelli et al. 2017). Base editing technology was first proved to work in human cells; it was then developed, modified and applied in rice plants (Ren et al. 2018; Li et al. 2017b). Recently, rBE14 with the transfer RNA adenosine deaminase was developed to facilitate the conversion of A into G and T into C in rice with the editing efficiency of 17.64% (Yan et al. 2018a). The human AID (hAID) has been rice codon optimized to increase the base editing efficiency of high GC content; it is also applicable to AC-, TC- and CC-rich region as well (Ren et al. 2018). Cas9 fusion with activation-induced cytidine deaminase (Target-AID) was demonstrated to induce point mutation in herbicide resistance gene (Shimatani et al. 2017). Furthermore, fusion protein of Cas9 nickase together with plant codons optimized APOBEC3A (A3A-PBE) that can expand the base editing window to 17-nt have been shown to convert C into T in wheat, rice and potato (Zong et al. 2018).
More lately, the performance of base editor efficiency was optimized by adopting bipartite nuclear localization signals (bpNLS)-Anc689 APOBEC-32 aa Linker and bpNLS-adenine deaminase of ABE7.10-32 aa Linker, respectively, in rice, improving the efficiency of C to T replacement up to 72.4% (Wang et al. 2019). Using base editor toolkit is a good strategy to introduce point mutations in different plant species including wheat and maize (Zong et al. 2017). Most recently, transgene-free herbicide resistance wheat was generated by base editing the acetolactate synthase (ALS) and acetyl-coenzyme A carboxylase genes (Zhang et al. 2019a). Apart from this, expanding the CRISPR/Cas9 base editing to CRISPR/Cas12a base editing would increase the scope of PAM sequences when choosing the target regions of the gene of interest (Li et al. 2018b).
Nonetheless, some drawbacks of base editors include low editing efficiency, and high off-target effects by base editors, especially CBE (cytosine base editor) (Jin et al. 2019), calling for improvement in specificity in future. In particular, the activity window of the base editors is the key point to determine whether the conversions of the specific and intended nucleotides perform accurately in the purpose of targeted mutagenesis.
Engineered Cas nuclease variants with altered PAM requirement
Modified versions of Cas9 or Cas12a nucleases can expand and tolerate a variety of PAM sequences. The naturally occurring S. pyogene Cas9 and L. bacterium ND2006 Cas12a can only recognize NGG and TTTV PAM (N could be A, C and G) sequences, respectively, which restricts choice of target sites when used for targeting the region of interest. Engineered Cas9 and Cas12a variants can provide more options of PAMs when choosing the intended target sites. For instance, NAG, NG, NGA and NGCG PAM sequence were shown to function for Cas9 variants (Hu et al. 2016). On the other hand, AsCas12a variants with TYTC and TATV PAMs and LbCas12a variants recognizing TATG PAM broaden the range of targeted sites substantially (Gao et al. 2017; Zhong et al. 2018). More recently, an engineered version of Cas12a (enAsCas12a) was demonstrated to have an improved genome-editing activity in human cells (Kleinstiver et al. 2019). This Cas12a variant was shown to work in specific gene targeting, epigenetics and base editing (Kleinstiver et al. 2019) and thus can be potentially applied in grass plants.
xCas9 and Cas9-NG are the most recently developed Cas9 variants that can increase flexibility of PAMs while maintaining relatively high efficiency in genome editing (Hu et al. 2018; Nishimasu et al. 2018). Broad range of PAMs including NG, GAA, and GAT were displayed to work in mammalian cells. Very recently, xCas9 was proved to function in rice with low efficiency (Wang et al. 2019). Further efforts in optimization including conditions of genome editing are needed to have better performance in this system (Wang et al. 2019). Engineered version of SpCas9 named as SpCas9-NGv1 was demonstrated to efficiently edit the target genes using the NG PAMs in rice and Arabidopsis (Endo et al. 2019). SpCas9-NGv1 nickase can also be fused with cytidine deaminase to convert C into T in rice acetolactate synthase (ALS) and drooping leaf (DL) genes. In another study, SpCas9-NGv1 can be used in the adenine base editor in rice (Negishi et al. 2019). Similar reports showed that xCas9 and Cas9-NG can be utilized to expand the scope of base editing in rice (Ren et al. 2019; Hua et al. 2019; Zhong et al. 2019). Indeed, this is the next generation of genome-editing tool that expands the range of genome editing in plants since it can tolerate more variety and different PAMs.
High-throughput genome editing and low-cost genotyping/mutant screening
One of the ultimate goals in genome editing is to develop high-throughput and robotic platforms for vector construction and plant mutant screening. This novel and robust technology requires labor-intensive works to deliver the CRISPR constructs into the living organisms and select for different types of mutations. Therefore, robotic automation is ideal and necessary to reduce technical error caused by humankind. Owing to the high mutation frequencies created using CRISPR/Cas9 and other alternative nucleases, large-scale mutations by multiplexing can be a readily obtained nowadays.
On the other hand, a variety of simple and rapid mutant screening methods have been developed to identify the mutants induced by CRISPR system. Traditionally, T7 endonuclease 1 (T7E1) assay, polyacrylamide gel electrophoresis (PAGE)-based analysis, restriction enzyme (RE)-based assay, high resolution melting (HRM) analysis, Sanger sequencing, next-generation sequencing (NGS) were used to detect mutations. More recently, single-strand conformational polymorphism (SSCP), annealing at critical temperature PCR (ACT-PCR), PCR followed by RNP digestion method and Mutation Sites-Based Specific Primers Polymerase Chain Reaction (MSBSP-PCR) were demonstrated in genotyping the genome-edited mutants (Zheng et al. 2016; Hua et al. 2017; Guo et al. 2018; Liang et al. 2018). Each of them has different characteristics. For example, SSCP is a great genotyping method for identifying small indels including 1-bp indels that is commonly resulted from CRISPR/Cas9 editing system (Zheng et al. 2016). ACT-PCR on the other hand utilizes the traditional annealing at critical temperature PCR to screen the CRISPR-induced mutants (Hua et al. 2017). PCR/RNP mutation detection methods can be used in polyploidy plants (Liang et al. 2018). MSBSP-PCR is also a simple and easy method to detect the biallelic mutants/homozygous mutants induced by CRISPR/Cas9 (Guo et al. 2018).
Users can make use of all the established genome-editing and mutant screening platforms to advance the exploration of functional genomics. Genome-wide targeted mutagenesis methods using CRISPR/Cas9 have been extensively explored in rice breeding program (Lu et al. 2017). Characterization of numerous genes that engaged in different biological processes will be a low-hanging fruit in near future.
Transgene-free delivery methods
Agrobacterium tumefaciens-mediated DNA delivery, biolistic particle bombardment and viral vector delivery are the three common ways to introduce the CRISPR/Cas9 reagents into the plant cells. However, developing transgene-free CRISPR-edited crops is a crucial topic and critical issue particularly in European countries. To mitigate this concern, ribonucleoprotein (RNP) complex is a good choice to introduce Cas9 and sgRNA into plants cells such as protoplasts through somatic embryogenesis for DNA-free genome editing. The Cas9 and sgRNA reagents can easily be degraded in a short time after delivered into plant cells without leaving any footprints in plant genome. This technique has been successfully applied in a few grass species including rice and wheat (Liang et al. 2017; Woo et al. 2015). On the other hand, the use of mRNA in vitro transcription has also been utilized to deliver CRISPR reagents into the plant tissue (Zhang et al. 2016, 2019b). This non-transgenic approach can potentially escape the regulation set-up by the USDA APHIS. Collectively, this transgene-free genome-editing method is more favorable to the public consumers to commercialize and disseminate the improved-quality crop plants. Nevertheless, the labor-intensive and time-consuming tissue culture works to regenerate plants from protoplasts are a bottleneck and required to invest more efforts.
Regulatory status for genome-editing crops
TALENs and the CRISPR-derived systems are powerful and robust tools for precise genome editing. They can be deployed for improving specific and important agronomical traits. However, the regulatory status of genome-edited crops is a hot topic to debate among scientific community and the regulators. In maize, CRISPR/Cas9-mediated waxy gene (Wx1)-edited corn with starch enrichment developed by Dupont Pioneer has been exempted from GMO regulations (Firko 2016). Another example is the TALEN-based SWEET gene-edited Xoo resistance rice that also received a similar ruling from the USDA (Firko 2015). In short, TALENs and CRISPR technologies hold a great promise to edit desirable traits and these novel plant-breeding approaches provide an additional way to improve the economically important crops. Thus, regulatory status of genome-editing crops is essential for dissemination of these breakthrough technologies and their derived plant products throughout the world.
Future perspectives
Plant genome-editing database (PGED)
Recently, one publication from Boyce Thompson Institute for Plant Research called for the submission of the CRISPR-edited mutants to the online resource (Zheng et al. 2019). This platform or repository provides researchers a pool of the genome-editing mutants’ information including the gene IDs, transformation information, mutant phenotypes, types of mutations, gRNA sequences, seeds availability and other detailed information in the online database (Zheng et al. 2019). Indeed, it will benefit the plant science community to share different mutants for the purpose of individual research. This idea is quite similar with the Arabidopsis Information Resource (TAIR) with the differences of having a molecular biology database in other plant species and different targeted mutagenesis methods. This is a good idea and may be extremely useful for the researchers to access the data, information of the mutant plants, and plant materials (e.g., mutant seeds) created through CRISPR technology.
Cas12a-mediated base editors in plants
Cas12a base editor was shown to work in human cells (Li et al. 2018b); this work can be extended into plants system by converting C–T in TTT-rich genomic region. It is known that CRISPR/Cas9-derived base editor has the limitation of G-rich targeted region, CRISPR/Cas12a base editor can overcome such constrains of Cas9 base editor and further elaborates the scope of gene editing within the T-rich genomic region in crop plants.
Combination of different nucleases
FokI endonuclease was shown to work with guide RNA-based Cas9 to have much higher specificity and high efficiency of genome editing in human cells (Tsai et al. 2014). This was carried out to reduce the undesired off-target mutations in line with the highly accurate genome editing (Tsai et al. 2014). On the other hand, combining nuclease Cas9 protein and Cas12a protein for genome engineering will be another possible experimental approach in grass plants. This will provide users with broader choice of the PAM sequence selection when designing the desired guide RNA and crRNA. One recent study has reported the fusion guide RNA (fgRNA) that combines nuclease Cas9 and Cas12a work in human cells and yet to be demonstrated in plants (Kweon et al. 2017).
Base editing of RNA
Most recently, RNA base editors just started burgeoning using the dCas13b (~ 1100aa) and ADAR (adenosine deaminase acting on RNA) enzymes to catalyze the hydrolysis of adenosine to inosine using RNA as templates in mammalian cells (Cox et al. 2017; Chaudhary 2018). This system was named as RNA Editing for Programmable A to I Replacement (REPAIR) with no restricted PAM at the editing site (Cox et al. 2017). This newly developed system is able to correct two human disease-related mutations including X-linked nephrogenic diabetes insipidus and Fanconi Anemia with 35% and 23% of the editing efficiency (Cox et al. 2017). Indeed, RNA base editor provides a new approach in the field of genome engineering and can be extrapolated in the plants system in near future.
Concluding remarks
A suite of genome-editing tool sets including meganucleases, ZFNs, TALENs, CRISPR-Cas9-, -Cas12a-, -Cas12e-, -Cas13a-, -Cas13b-, -Cas13d-, -Cas14- and CRISPR/Cas-derived base editors have been well developed and overwhelmed the scientific journals of high impact factors in the last decade (Silva et al. 2011; Bedell et al. 2012; Doudna and Charpentier 2014; Urnov et al. 2010; Smargon et al. 2017; Harrington et al. 2018; Abudayyeh et al. 2017; Yan et al. 2018b; Gaudelli et al. 2017; Zetsche et al. 2015; Begemann et al. 2017b). All of these tools have their distinguishable pros and cons. Therefore, further improvements and optimization are needed to make the custom-designed constructs suitable for different grass plants. In short, genome-editing technologies are robust and formidable genetic and biotech tools to study functional genomics in grass plants (Table 2). Furthermore, crop improvements can be achieved through gene/trait discovery and ingression with readily genome-editing platforms in grass crops. With the genome-editing tools becoming popular and widely used, the fundamental biology and application in crop plants will advance in a pace that is unimaginable ten years ago.
Acknowledgements
The authors gratefully acknowledge grant support from the National Science Foundation (1936492 to B.Y.) and a subaward to MU from Heinrich Heine University of Dusseldorf, which was funded by the Bill & Melinda Gates Foundation [OPP1155704] (B.Y.).
References
- Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, Lander ES, Voytas DF, Ting AY, Zhang F. RNA targeting with CRISPR-Cas13. Nature. 2017;550(7675):280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akmammedov A, Katsuyama T, Paro R. A rapid TALEN assembly protocol. Methods Mol Biol. 2016;1480:269–281. doi: 10.1007/978-1-4939-6380-5_23. [DOI] [PubMed] [Google Scholar]
- Barrangou R. CRISPR-Cas systems and RNA-guided interference. Wiley Interdiscip Rev RNA. 2013;4(3):267–278. doi: 10.1002/wrna.1159. [DOI] [PubMed] [Google Scholar]
- Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, 2nd, Tan W, Penheiter SG, Ma AC, Leung AY, Fahrenkrug SC, Carlson DF, Voytas DF, Clark KJ, Essner JJ, Ekker SC. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491(7422):114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begemann MB, Gray BN, January E, Gordon GC, He Y, Liu H, Wu X, Brutnell TP, Mockler TC, Oufattole M. Precise insertion and guided editing of higher plant genomes using Cpf1 CRISPR nucleases. Sci Rep. 2017;7(1):11606. doi: 10.1038/s41598-017-11760-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begemann MB, Gray BN, January E, Singer A, Kesler DC, He Y, Liu H, Guo H, Jordan A, Brutnell TP, Mockler TC, Oufattole M. Characterization and validation of a novel group of type V, class 2 nucleases for in vivo genome editing. BioRxiv:192799. 2017 doi: 10.1101/192799. [DOI] [Google Scholar]
- Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol. 2010;48:419–436. doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326(5959):1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333(6051):1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- Burstein D, Harrington LB, Strutt SC, Probst AJ, Anantharaman K, Thomas BC, Doudna JA, Banfield JF. New CRISPR-Cas systems from uncultivated microbes. Nature. 2017;542(7640):237–241. doi: 10.1038/nature21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188(4):773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Char SN, Unger-Wallace E, Frame B, Briggs SA, Main M, Spalding MH, Vollbrecht E, Wang K, Yang B. Heritable site-specific mutagenesis using TALENs in maize. Plant Biotechnol J. 2015;13(7):1002–1010. doi: 10.1111/pbi.12344. [DOI] [PubMed] [Google Scholar]
- Char SN, Neelakandan AK, Nahampun H, Frame B, Main M, Spalding MH, Becraft PW, Meyers BC, Walbot V, Wang K, Yang B. An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotechnol J. 2017;15(2):257–268. doi: 10.1111/pbi.12611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary K. RNA base editing: programmable protein machine. Curr Sci. 2018;114(9):1808–1809. doi: 10.18520/cs/v114/i09/1808-1809. [DOI] [Google Scholar]
- Che P, Anand A, Wu E, Sander JD, Simon MK, Zhu W, Sigmund AL, Zastrow-Hayes G, Miller M, Liu D, Lawit SJ, Zhao ZY, Albertsen MC, Jones TJ. Developing a flexible, high-efficiency Agrobacterium-mediated sorghum transformation system with broad application. Plant Biotechnol J. 2018;16(7):1388–1395. doi: 10.1111/pbi.12879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, Huang B. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155(7):1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, Sternberg SH, Joung JK, Yildiz A, Doudna JA. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550(7676):407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Xu Q, Liu Y, Zhang J, Ren D, Wang G, Liu Y. Generation of transgene-free maize male sterile lines using the CRISPR/Cas9 system. Front Plant Sci. 2018;9:1180. doi: 10.3389/fpls.2018.01180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186(2):757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, Zhang F. RNA editing with CRISPR-Cas13. Science. 2017;358(6366):1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JP, Kumar S, Sastry-Dent L. Use of zinc-finger nucleases for crop improvement. Prog Mol Biol Transl Sci. 2017;149:47–63. doi: 10.1016/bs.pmbts.2017.03.006. [DOI] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Dreissig S, Schiml S, Schindele P, Weiss O, Rutten T, Schubert V, Gladilin E, Mette MF, Puchta H, Houben A. Live-cell CRISPR imaging in plants reveals dynamic telomere movements. Plant J. 2017;91(4):565–573. doi: 10.1111/tpj.13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Mikami M, Endo A, Kaya H, Itoh T, Nishimasu H, Nureki O, Toki S. Genome editing in plants by engineered CRISPR-Cas9 recognizing NG PAM. Nat Plants. 2019;5(1):14–17. doi: 10.1038/s41477-018-0321-8. [DOI] [PubMed] [Google Scholar]
- Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P, Cao F, Zhu S, Zhang F, Mao Y, Zhu JK. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013;23(10):1229–1232. doi: 10.1038/cr.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C, Yuan J, Wang R, Liu Y, Birchler JA, Han F. Efficient targeted genome modification in maize using CRISPR/Cas9 system. J Genet Genom. 2016;43(1):37–43. doi: 10.1016/j.jgg.2015.10.002. [DOI] [PubMed] [Google Scholar]
- Feng C, Su H, Bai H, Wang R, Liu Y, Guo X, Liu C, Zhang J, Yuan J, Birchler JA, Han F. High-efficiency genome editing using a dmc1 promoter-controlled CRISPR/Cas9 system in maize. Plant Biotechnol J. 2018;16(11):1848–1857. doi: 10.1111/pbi.12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firko MJ (2015) Inquiry regarding APHIS position on null-segregant TALEN-mutagenized rice lines as non-regulated articles. Received by Bing Yang. https://www.aphis.usda.gov/biotechnology/downloads/reg_loi/aphis_resp_isu_ting_rice.pdf
- Firko MJ (2016) Confirmation of regulatory status of waxy corn developed by CRISPR-Cas technology. Received by Daria H. Schmidt. https://www.aphis.usda.gov/biotechnology/downloads/reg_loi/15-352-01_air_response_signed.pdf
- Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Smith J, Yang M, Jones S, Djukanovic V, Nicholson MG, West A, Bidney D, Falco SC, Jantz D, Lyznik LA. Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J. 2010;61(1):176–187. doi: 10.1111/j.1365-313X.2009.04041.x. [DOI] [PubMed] [Google Scholar]
- Gao L, Cox DBT, Yan WX, Manteiga JC, Schneider MW, Yamano T, Nishimasu H, Nureki O, Crosetto N, Zhang F. Engineered Cpf1 variants with altered PAM specificities. Nat Biotechnol. 2017;35(8):789–792. doi: 10.1038/nbt.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. 2017;551(7681):464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154(2):442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Li K, Jin L, Xu R, Miao K, Yang F, Qi C, Zhang L, Botella JR, Wang R, Miao Y. A simple and cost-effective method for screening of CRISPR/Cas9-induced homozygous/biallelic mutants. Plant Methods. 2018;14:40. doi: 10.1186/s13007-018-0305-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H, Liu Y, Nagira Y, Miki R, Taoka N, Imai R. Biolistic-delivery-based transient CRISPR/Cas9 expression enables in planta genome editing in wheat. Sci Rep. 2018;8(1):14422. doi: 10.1038/s41598-018-32714-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington LB, Burstein D, Chen JS, Paez-Espino D, Ma E, Witte IP, Cofsky JC, Kyrpides NC, Banfield JF, Doudna JA. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362(6416):839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33(5):510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howells RM, Craze M, Bowden S, Wallington EJ. Efficient generation of stable, heritable gene edits in wheat using CRISPR/Cas9. BMC Plant Biol. 2018;18(1):215. doi: 10.1186/s12870-018-1433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Wang C, Fu Y, Liu Q, Jiao X, Wang K. Expanding the range of CRISPR/Cas9 genome editing in rice. Mol Plant. 2016;9(6):943–945. doi: 10.1016/j.molp.2016.03.003. [DOI] [PubMed] [Google Scholar]
- Hu X, Wang C, Liu Q, Fu Y, Wang K. Targeted mutagenesis in rice using CRISPR-Cpf1 system. J Genet Genom. 2017;44(1):71–73. doi: 10.1016/j.jgg.2016.12.001. [DOI] [PubMed] [Google Scholar]
- Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, Zeina CM, Gao X, Rees HA, Lin Z, Liu DR. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556(7699):57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Wang C, Huang J, Wang K. A simple and efficient method for CRISPR/Cas9-induced mutant screening. J Genet Genom. 2017;44(4):207–213. doi: 10.1016/j.jgg.2017.03.005. [DOI] [PubMed] [Google Scholar]
- Hua K, Tao X, Han P, Wang R, Zhu JK. Genome engineering in rice using Cas9 variants that recognize NG PAM sequences. Mol Plant. 2019;12(7):1003–1014. doi: 10.1016/j.molp.2019.03.009. [DOI] [PubMed] [Google Scholar]
- Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41(20):e188. doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Zong Y, Gao Q, Zhu Z, Wang Y, Qin P, Liang C, Wang D, Qiu JL, Zhang F, Gao C. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019;364(6437):292–295. doi: 10.1126/science.aaw7166. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y-J, Nogoy FM, Lee S-K, Cho Y-G, Kang K-K. Application of ZFN for site directed mutagenesis of rice SSIVa gene. Biotechnol Bioprocess Eng. 2018;23(1):108–115. doi: 10.1007/s12257-017-0420-9. [DOI] [Google Scholar]
- Kang BC, Yun JY, Kim ST, Shin Y, Ryu J, Choi M, Woo JW, Kim JS. Precision genome engineering through adenine base editing in plants. Nat Plants. 2018;4(7):427–431. doi: 10.1038/s41477-018-0178-x. [DOI] [PubMed] [Google Scholar]
- Kaya H, Mikami M, Endo A, Endo M, Toki S. Highly specific targeted mutagenesis in plants using Staphylococcus aureus Cas9. Sci Rep. 2016;6:26871. doi: 10.1038/srep26871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93(3):1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim J, Hur JK, Been KW, Yoon SH, Kim JS. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol. 2016;34(8):863–868. doi: 10.1038/nbt.3609. [DOI] [PubMed] [Google Scholar]
- Kim YB, Komor AC, Levy JM, Packer MS, Zhao KT, Liu DR. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol. 2017;35(4):371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Sousa AA, Walton RT, Tak YE, Hsu JY, Clement K, Welch MM, Horng JE, Malagon-Lopez J, Scarfo I, Maus MV, Pinello L, Aryee MJ, Joung JK. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat Biotechnol. 2019;37(3):276–282. doi: 10.1038/s41587-018-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SC, Tjian R, Doudna JA. Genomes in focus: development and applications of CRISPR-Cas9 imaging technologies. Angew Chem Int Ed Engl. 2018;57(16):4329–4337. doi: 10.1002/anie.201709201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533(7603):420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Zhao KT, Packer MS, Gaudelli NM, Waterbury AL, Koblan LW, Kim YB, Badran AH, Liu DR. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T: a base editors with higher efficiency and product purity. Sci Adv. 2017;3(8):eaao4774. doi: 10.1126/sciadv.aao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweon J, Jang AH, Kim DE, Yang JW, Yoon M, Rim Shin H, Kim JS, Kim Y. Fusion guide RNAs for orthogonal gene manipulation with Cas9 and Cpf1. Nat Commun. 2017;8(1):1723. doi: 10.1038/s41467-017-01650-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon DY, Zhao YT, Lamonica JM, Zhou Z. Locus-specific histone deacetylation using a synthetic CRISPR-Cas9-based HDAC. Nat Commun. 2017;8:15315. doi: 10.1038/ncomms15315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrenson T, Shorinola O, Stacey N, Li C, Ostergaard L, Patron N, Uauy C, Harwood W. Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 2015;16:258. doi: 10.1186/s13059-015-0826-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Zhang Y, Kleinstiver BP, Guo JA, Aryee MJ, Miller J, Malzahn A, Zarecor S, Lawrence-Dill CJ, Joung JK, Qi Y, Wang K. Activities and specificities of CRISPR/Cas9 and Cas12a nucleases for targeted mutagenesis in maize. Plant Biotechnol J. 2019;17(2):362–372. doi: 10.1111/pbi.12982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, Yang B. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011;39(1):359–372. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Liu B, Spalding MH, Weeks DP, Yang B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol. 2012;30(5):390–392. doi: 10.1038/nbt.2199. [DOI] [PubMed] [Google Scholar]
- Li T, Liu B, Chen CY, Yang B. TALEN utilization in rice genome modifications. Methods. 2014;69(1):9–16. doi: 10.1016/j.ymeth.2014.03.019. [DOI] [PubMed] [Google Scholar]
- Li C, Liu C, Qi X, Wu Y, Fei X, Mao L, Cheng B, Li X, Xie C. RNA-guided Cas9 as an in vivo desired-target mutator in maize. Plant Biotechnol J. 2017;15(12):1566–1576. doi: 10.1111/pbi.12739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Sun Y, Du J, Zhao Y, Xia L. Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol Plant. 2017;10(3):526–529. doi: 10.1016/j.molp.2016.12.001. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang D, Xiong X, Yan B, Xie W, Sheen J, Li JF. A potent Cas9-derived gene activator for plant and mammalian cells. Nat Plants. 2017;3(12):930–936. doi: 10.1038/s41477-017-0046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Jia S, Yobi A, Ge Z, Sato SJ, Zhang C, Angelovici R, Clemente TE, Holding DR. Editing of an alpha-kafirin gene family increases, digestibility and protein quality in sorghum. Plant Physiol. 2018;177(4):1425–1438. doi: 10.1104/pp.18.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang Y, Liu Y, Yang B, Wang X, Wei J, Lu Z, Zhang Y, Wu J, Huang X, Yang L, Chen J. Base editing with a Cpf1-cytidine deaminase fusion. Nat Biotechnol. 2018;36(4):324–327. doi: 10.1038/nbt.4102. [DOI] [PubMed] [Google Scholar]
- Liang Z, Zhang K, Chen K, Gao C. Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J Genet Genom. 2014;41(2):63–68. doi: 10.1016/j.jgg.2013.12.001. [DOI] [PubMed] [Google Scholar]
- Liang Z, Chen K, Li T, Zhang Y, Wang Y, Zhao Q, Liu J, Zhang H, Liu C, Ran Y, Gao C. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun. 2017;8:14261. doi: 10.1038/ncomms14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Chen K, Yan Y, Zhang Y, Gao C. Genotyping genome-edited mutations in plants using CRISPR ribonucleoprotein complexes. Plant Biotechnol J. 2018;16(12):2053–2062. doi: 10.1111/pbi.12938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Merrick P, Zhang Z, Ji C, Yang B, Fei SZ. Targeted mutagenesis in tetraploid switchgrass (Panicum virgatum L.) using CRISPR/Cas9. Plant Biotechnol J. 2018;16(2):381–393. doi: 10.1111/pbi.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Li J, Godwin ID. Genome editing by CRISPR/Cas9 in sorghum through biolistic bombardment. Methods Mol Biol. 2019;1931:169–183. doi: 10.1007/978-1-4939-9039-9_12. [DOI] [PubMed] [Google Scholar]
- Lowder LG, Zhang D, Baltes NJ, Paul JW, 3rd, Tang X, Zheng X, Voytas DF, Hsieh TF, Zhang Y, Qi Y. A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 2015;169(2):971–985. doi: 10.1104/pp.15.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder LG, Zhou J, Zhang Y, Malzahn A, Zhong Z, Hsieh TF, Voytas DF, Zhang Y, Qi Y. Robust transcriptional activation in plants using multiplexed CRISPR-Act2.0 and mTALE-act systems. Mol Plant. 2018;11(2):245–256. doi: 10.1016/j.molp.2017.11.010. [DOI] [PubMed] [Google Scholar]
- Lu Y, Ye X, Guo R, Huang J, Wang W, Tang J, Tan L, Zhu JK, Chu C, Qian Y. Genome-wide targeted mutagenesis in rice using the CRISPR/Cas9 system. Mol Plant. 2017;10(9):1242–1245. doi: 10.1016/j.molp.2017.06.007. [DOI] [PubMed] [Google Scholar]
- Malzahn AA, Tang X, Lee K, Ren Q, Sretenovic S, Zhang Y, Chen H, Kang M, Bao Y, Zheng X, Deng K, Zhang T, Salcedo V, Wang K, Zhang Y, Qi Y. Application of CRISPR-Cas12a temperature sensitivity for improved genome editing in rice, maize, and Arabidopsis. BMC Biol. 2019;17(1):9. doi: 10.1186/s12915-019-0629-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Zhang H, Xu N, Zhang B, Gou F, Zhu JK. Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol Plant. 2013;6(6):2008–2011. doi: 10.1093/mp/sst121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J, Guo D, Zhang J, Huang Q, Qin G, Zhang X, Wan J, Gu H, Qu LJ. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013;23(10):1233–1236. doi: 10.1038/cr.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami M, Toki S, Endo M. Precision targeted mutagenesis via Cas9 paired nickases in rice. Plant Cell Physiol. 2016;57(5):1058–1068. doi: 10.1093/pcp/pcw049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, Gregory PD, Pabo CO, Rebar EJ. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25(7):778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29(2):143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Moreno-Mateos MA, Fernandez JP, Rouet R, Vejnar CE, Lane MA, Mis E, Khokha MK, Doudna JA, Giraldez AJ. CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Nat Commun. 2017;8(1):2024. doi: 10.1038/s41467-017-01836-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326(5959):1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Muller M, Lee CM, Gasiunas G, Davis TH, Cradick TJ, Siksnys V, Bao G, Cathomen T, Mussolino C. Streptococcus thermophilus CRISPR-Cas9 systems enable specific editing of the human genome. Mol Ther. 2016;24(3):636–644. doi: 10.1038/mt.2015.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi K, Kaya H, Abe K, Hara N, Saika H, Toki S. An adenine base editor with expanded targeting scope using SpCas9-NGv1 in rice. Plant Biotechnol J. 2019 doi: 10.1111/pbi.13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156(5):935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Shi X, Ishiguro S, Gao L, Hirano S, Okazaki S, Noda T, Abudayyeh OO, Gootenberg JS, Mori H, Oura S, Holmes B, Tanaka M, Seki M, Hirano H, Aburatani H, Ishitani R, Ikawa M, Yachie N, Zhang F, Nureki O. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 2018;361(6408):1259–1262. doi: 10.1126/science.aas9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penewit K, Holmes EA, McLean K, Ren M, Waalkes A, Salipante SJ. Efficient and scalable precision genome editing in Staphylococcus aureus through conditional recombineering and CRISPR/Cas9-mediated counterselection. MBio. 2018 doi: 10.1128/mbio.00067-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Yan F, Kuang Y, Li N, Zhang D, Zhou X, Lin H, Zhou H. Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9-guided hyperactive hAID mutant. Mol Plant. 2018;11(4):623–626. doi: 10.1016/j.molp.2018.01.005. [DOI] [PubMed] [Google Scholar]
- Ren B, Liu L, Li S, Kuang Y, Wang J, Zhang D, Zhou X, Lin H, Zhou H. Cas9-NG greatly expands the targeting scope of the genome-editing toolkit by recognizing NG and other atypical PAMs in rice. Mol Plant. 2019;12(7):1015–1026. doi: 10.1016/j.molp.2019.03.010. [DOI] [PubMed] [Google Scholar]
- Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. 1994;14(12):8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin N, Haber JE. Efficient repair of HO-induced chromosomal breaks in Saccharomyces cerevisiae by recombination between flanking homologous sequences. Mol Cell Biol. 1988;8(9):3918–3928. doi: 10.1128/mcb.8.9.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Leon S, Gil-Humanes J, Ozuna CV, Gimenez MJ, Sousa C, Voytas DF, Barro F. Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol J. 2018;16(4):902–910. doi: 10.1111/pbi.12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Chen K, Liang Z, Li J, Zhang Y, Zhang K, Liu J, Voytas DF, Zheng X, Zhang Y, Gao C. Rapid and efficient gene modification in rice and Brachypodium using TALENs. Mol Plant. 2013;6(4):1365–1368. doi: 10.1093/mp/sss162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu JL, Gao C. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol. 2013;31(8):686–688. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Li J, Gao C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc. 2014;9(10):2395–2410. doi: 10.1038/nprot.2014.157. [DOI] [PubMed] [Google Scholar]
- Shi J, Gao H, Wang H, Lafitte HR, Archibald RL, Yang M, Hakimi SM, Mo H, Habben JE. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol J. 2017;15(2):207–216. doi: 10.1111/pbi.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H, Teramura H, Yamamoto T, Komatsu H, Miura K, Ezura H, Nishida K, Ariizumi T, Kondo A. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol. 2017;35(5):441–443. doi: 10.1038/nbt.3833. [DOI] [PubMed] [Google Scholar]
- Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, Zhang F, Koonin EV. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60(3):385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng X, Choi VM, Rock JM, Wu YY, Katibah GE, Zhifang G, McCaskill D, Simpson MA, Blakeslee B, Greenwalt SA, Butler HJ, Hinkley SJ, Zhang L, Rebar EJ, Gregory PD, Urnov FD. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature. 2009;459(7245):437–441. doi: 10.1038/nature07992. [DOI] [PubMed] [Google Scholar]
- Silva G, Poirot L, Galetto R, Smith J, Montoya G, Duchateau P, Paques F. Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr Gene Ther. 2011;11(1):11–27. doi: 10.2174/156652311794520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smargon AA, Cox DBT, Pyzocha NK, Zheng K, Slaymaker IM, Gootenberg JS, Abudayyeh OA, Essletzbichler P, Shmakov S, Makarova KS, Koonin EV, Zhang F. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol Cell. 2017;65(4):618–630.e617. doi: 10.1016/j.molcel.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strecker J, Jones S, Koopal B, Schmid-Burgk J, Zetsche B, Gao L, Makarova KS, Koonin EV, Zhang F. Engineering of CRISPR-Cas12b for human genome editing. Nat Commun. 2019;10(1):212. doi: 10.1038/s41467-018-08224-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitashev S, Young JK, Schwartz C, Gao H, Falco SC, Cigan AM. Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 2015;169(2):931–945. doi: 10.1104/pp.15.00793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitashev S, Schwartz C, Lenderts B, Young JK, Mark Cigan A. Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat Commun. 2016;7:13274. doi: 10.1038/ncomms13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Lowder LG, Zhang T, Malzahn AA, Zheng X, Voytas DF, Zhong Z, Chen Y, Ren Q, Li Q, Kirkland ER, Zhang Y, Qi Y. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants. 2017;3:17103. doi: 10.1038/nplants.2017.103. [DOI] [PubMed] [Google Scholar]
- Tang X, Liu G, Zhou J, Ren Q, You Q, Tian L, Xin X, Zhong Z, Liu B, Zheng X, Zhang D, Malzahn A, Gong Z, Qi Y, Zhang T, Zhang Y. A large-scale whole-genome sequencing analysis reveals highly specific genome editing by both Cas9 and Cpf1 (Cas12a) nucleases in rice. Genome Biol. 2018;19(1):84. doi: 10.1186/s13059-018-1458-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Ren Q, Yang L, Bao Y, Zhong Z, He Y, Liu S, Qi C, Liu B, Wang Y, Sretenovic S, Zhang Y, Zheng X, Zhang T, Qi Y, Zhang Y. Single transcript unit CRISPR 2.0 systems for robust Cas9 and Cas12a mediated plant genome editing. Plant Biotechnol J. 2019;17(7):1431–1445. doi: 10.1111/pbi.13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Joung JK. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat Rev Genet. 2016;17(5):300–312. doi: 10.1038/nrg.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32(6):569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- Vojta A, Dobrinic P, Tadic V, Bockor L, Korac P, Julg B, Klasic M, Zoldos V. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016;44(12):5615–5628. doi: 10.1093/nar/gkw159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol. 2014;32(9):947–951. doi: 10.1038/nbt.2969. [DOI] [PubMed] [Google Scholar]
- Wang M, Mao Y, Lu Y, Tao X, Zhu JK. Multiplex gene editing in rice using the CRISPR-Cpf1 system. Mol Plant. 2017;10(7):1011–1013. doi: 10.1016/j.molp.2017.03.001. [DOI] [PubMed] [Google Scholar]
- Wang W, Pan Q, He F, Akhunova A, Chao S, Trick H, Akhunov E. Transgenerational CRISPR-Cas9 activity facilitates multiplex gene editing in allopolyploid wheat. Crispr j. 2018;1(1):65–74. doi: 10.1089/crispr.2017.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Meng X, Hu X, Sun T, Li J, Wang K, Yu H. xCas9 expands the scope of genome editing with reduced efficiency in rice. Plant Biotechnol J. 2019;17(4):709–711. doi: 10.1111/pbi.13053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt T, Holm PB, Starker CG, Christian M, Voytas DF, Brinch-Pedersen H, Holme IB. TAL effector nucleases induce mutations at a pre-selected location in the genome of primary barley transformants. Plant Mol Biol. 2013;83(3):279–285. doi: 10.1007/s11103-013-0078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo JW, Kim J, Kwon SI, Corvalan C, Cho SW, Kim H, Kim SG, Kim ST, Choe S, Kim JS. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol. 2015;33(11):1162–1164. doi: 10.1038/nbt.3389. [DOI] [PubMed] [Google Scholar]
- Xie K, Yang Y. RNA-guided genome editing in plants using a CRISPR-Cas system. Mol Plant. 2013;6(6):1975–1983. doi: 10.1093/mp/sst119. [DOI] [PubMed] [Google Scholar]
- Xie K, Minkenberg B, Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci USA. 2015;112(11):3570–3575. doi: 10.1073/pnas.1420294112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014;14:327. doi: 10.1186/s12870-014-0327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Qin R, Li H, Li D, Li L, Wei P, Yang J. Generation of targeted mutant rice using a CRISPR-Cpf1 system. Plant Biotechnol J. 2017;15(6):713–717. doi: 10.1111/pbi.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F, Kuang Y, Ren B, Wang J, Zhang D, Lin H, Yang B, Zhou X, Zhou H. Highly efficient A.T to G.C base editing by Cas9n-guided tRNA adenosine deaminase in rice. Mol Plant. 2018;11(4):631–634. doi: 10.1016/j.molp.2018.02.008. [DOI] [PubMed] [Google Scholar]
- Yan WX, Chong S, Zhang H, Makarova KS, Koonin EV, Cheng DR, Scott DA. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol Cell. 2018;70(2):327–339.e325. doi: 10.1016/j.molcel.2018.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Heidenreich M, Mohanraju P, Fedorova I, Kneppers J, DeGennaro EM, Winblad N, Choudhury SR, Abudayyeh OO, Gootenberg JS, Wu WY, Scott DA, Severinov K, van der Oost J, Zhang F. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol. 2017;35(1):31–34. doi: 10.1038/nbt.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang F, Li X, Baller JA, Qi Y, Starker CG, Bogdanove AJ, Voytas DF. Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol. 2013;161(1):20–27. doi: 10.1104/pp.112.205179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liang Z, Zong Y, Wang Y, Liu J, Chen K, Qiu JL, Gao C. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun. 2016;7:12617. doi: 10.1038/ncomms12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Liu J, Chai Z, Chen S, Bai Y, Zong Y, Chen K, Li J, Jiang L, Gao C. Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nat Plants. 2019;5(5):480–485. doi: 10.1038/s41477-019-0405-0. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Hua L, Gupta A, Tricoli D, Edwards KJ, Yang B, Li W. Development of an Agrobacterium-delivered CRISPR/Cas9 system for wheat genome editing. Plant Biotechnol J. 2019 doi: 10.1111/pbi.13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Yang S, Zhang D, Zhong Z, Tang X, Deng K, Zhou J, Qi Y, Zhang Y. Effective screen of CRISPR/Cas9-induced mutants in rice by single-strand conformation polymorphism. Plant Cell Rep. 2016;35(7):1545–1554. doi: 10.1007/s00299-016-1967-1. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Zhang N, Martin GB, Fei Z. Plant genome editing database (PGED): a call for submission of information about genome-edited plant mutants. Mol Plant. 2019;12(2):127–129. doi: 10.1016/j.molp.2019.01.001. [DOI] [PubMed] [Google Scholar]
- Zhong Z, Zhang Y, You Q, Tang X, Ren Q, Liu S, Yang L, Wang Y, Liu X, Liu B, Zhang T, Zheng X, Le Y, Zhang Y, Qi Y. Plant genome editing using FnCpf1 and LbCpf1 nucleases at redefined and altered PAM sites. Mol Plant. 2018;11(7):999–1002. doi: 10.1016/j.molp.2018.03.008. [DOI] [PubMed] [Google Scholar]
- Zhong Z, Sretenovic S, Ren Q, Yang L, Bao Y, Qi C, Yuan M, He Y, Liu S, Liu X, Wang J, Huang L, Wang Y, Baby D, Wang D, Zhang T, Qi Y, Zhang Y. Improving plant genome editing with high-fidelity xCas9 and non-canonical PAM-targeting Cas9-NG. Mol Plant. 2019;12(7):1027–1036. doi: 10.1016/j.molp.2019.03.011. [DOI] [PubMed] [Google Scholar]
- Zhou H, Liu B, Weeks DP, Spalding MH, Yang B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014;42(17):10903–10914. doi: 10.1093/nar/gku806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Yang B, Chittoor JM, Johnson LB, White FF. AvrXa10 contains an acidic transcriptional activation domain in the functionally conserved C terminus. Mol Plant Microbe Interact. 1998;11(8):824–832. doi: 10.1094/mpmi.1998.11.8.824. [DOI] [PubMed] [Google Scholar]
- Zhu J, Song N, Sun S, Yang W, Zhao H, Song W, Lai J. Efficiency and inheritance of targeted mutagenesis in maize using CRISPR-Cas9. J Genet Genom. 2016;43(1):25–36. doi: 10.1016/j.jgg.2015.10.006. [DOI] [PubMed] [Google Scholar]
- Zong Y, Wang Y, Li C, Zhang R, Chen K, Ran Y, Qiu JL, Wang D, Gao C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol. 2017;35(5):438–440. doi: 10.1038/nbt.3811. [DOI] [PubMed] [Google Scholar]
- Zong Y, Song Q, Li C, Jin S, Zhang D, Wang Y, Qiu JL, Gao C. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat Biotechnol. 2018 doi: 10.1038/nbt.4261. [DOI] [PubMed] [Google Scholar]