Abstract
N6-methyladenosine (m6A) is a reversible epigenetic modification of mRNA and other RNAs that plays a significant role in regulating gene expression and biological processes. However, m6A abundance, dynamics, and transcriptional regulatory mechanisms remain unexplored in the context of soybean resistance to Meloidogyne incognita. In this study, we performed a comparative analysis of transcriptome-wide m6A and metabolome profiles of soybean root tissues with and without M. incognita infection. Global m6A hypermethylation was widely induced in response to M. incognita infection and was enriched around the 3′ end of coding sequences and in 3′ UTR regions. There were 2069 significantly modified m6A sites, 594 differentially expressed genes, and 103 differentially accumulated metabolites between infected and uninfected roots, including coumestrol, psoralidin, and 2-hydroxyethylphosphonate. Among 101 m6A-modified DEGs, 34 genes were hypomethylated and upregulated, and 39 genes were hypermethylated and downregulated, indicating a highly negative correlation between m6A methylation and gene transcript abundance. A number of these m6A-modified DEGs, including WRKY70, ERF60, POD47 and LRR receptor-like serine/threonine-protein kinases, were involved in plant defense responses. Our study provides new insights into the critical role of m6A modification in early soybean responses to M. incognita.
Supplementary Information
The online version contains supplementary material available at 10.1007/s42994-022-00077-2.
Keywords: m6A methylation, Soybean, Meloidogyne incognita, m6A-seq, RNA-seq, Metabolome
Introduction
N6-methyladenosine (m6A) RNA methylation is an abundant internal modification of messenger RNAs (mRNAs) and long noncoding RNAs (lncRNAs) in eukaryotes (Vanyushin et al. 1970). Since the first discovery of m6A methylation in mammalian mRNA in 1975 (Wei et al. 1975), m6A has been found in various eukaryotes, including yeasts (Clancy et al. 2002), flies (Levis and Penman 1978), fish (Zhao et al. 2017), mammals (Dominissini et al. 2012; Meyer et al. 2012), and plants (Luo et al. 2014). More than 80% of all eukaryotic RNA methylation modifications have been reported to be m6A modifications (Kierzek and Kierzek 2003), and this modification is usually clustered in the stop codon and 3′ UTR, although it is also found in the start codon, 5′ UTR, and coding regions (Ke et al. 2015; Meyer et al. 2012). m6A plays a vital role in developmental regulation (Bodi et al. 2010; Li et al. 2014) and stress responses (Li et al. 2014; Ok et al. 2005; Scutenaire et al. 2018; Wei et al. 2018) by affecting multiple aspects of RNA function such as mRNA translation, splicing, stability, and degradation and miRNA biogenesis (Pontier et al. 2019; Visvanathan and Somasundaram 2018; Wang et al. 2014, 2015). In mammals, m6A is reported to regulate embryo development, fertility, neuronal functions, hematopoietic stem cell and progenitor cell development, adipogenesis, and the circadian clock (Du et al. 2016; Fukusumi et al. 2008; Fustin et al. 2013; Zheng et al. 2013). In plants, m6A also plays major roles in the regulation of photosynthesis, floral transition, fruit ripening, and seed vitality (Duan et al. 2017; Fray and Simpson 2015; Shen et al. 2019; Vespa et al. 2004; Zhang et al. 2019; Zhou et al. 2019).
m6A is a dynamic and reversible modification. Three distinct groups of proteins are involved in m6A modification: m6A methyltransferases (writers), demethylases (erasers), and m6A binding proteins (readers) (Shao et al. 2021). m6A is methylated by the binding of the writer to a highly conserved consensus sequence, RRACH (R = G or A; H: U > A > C). In plants, the writer is a large methyltransferase complex containing MTA70-like proteins (MTA, MTB), FKBP12 INTERACTING PROTEIN 37 KD (FIP37), HAKAI, and other components (Vespa et al. 2004; Shen et al. 2016; Růžička et al. 2017). The removal of m6A is catalyzed by erasers, which include the ALKBH family proteins ALKBH9B, ALKBH10B, and SLALKBH2 (Martínez-Pérez et al. 2021; Duan et al. 2017; Tang et al. 2021; Zhou et al. 2019). The m6A modification is recognized by readers, which mediates pre-mRNA splicing and other specific functions. They incorporate YTH-domain family proteins such as EVOLUTIONARILY CONSERVED C-TERMINAL REGIONS (ECT1–11) and a variant of the 30-kDa subunit of cleavage and polyadenylation specificity factor (CPSF30) (Wei et al. 2018; Pontier et al. 2019; Song et al. 2021; Hou et al. 2021).
An increasing number of studies have revealed that m6A methylation participates in responses to various abiotic stresses. Anderson et al. reported that m6A modification could promote the mRNA stability of key transcripts under salt stress in Arabidopsis (Anderson et al. 2018). Arabidopsis ECT2 improved heat stress tolerance by relocalizing stress granules in the cytoplasm (Scutenaire et al. 2018). Overexpression of the m6A methyltransferase ClMTB enhances drought tolerance in tobacco by mitigating oxidative stress and photosynthetic inhibition (He et al. 2021b). In addition, evidence indicates that m6A methylation also plays important roles in regulating plant immunity to biotic invasion (Mondo et al. 2017). For example, the Arabidopsis demethylase atALKBH9B can remove m6A from single-stranded RNA and reduce Alfalfa mosaic virus (AMV) infection (Martinez-Perez et al. 2017). In watermelons, the m6A level was more significantly reduced in a resistant variety than a susceptible variety after inoculation with Cucumber green mottle mosaic virus (CGMMV) (He et al. 2017). In rice, m6A modification was mainly associated with genes that were not actively expressed in virus-infected plants, and there was an increased level of m6A methylation during rice interaction with Rice black-streaked dwarf virus (RBSDV) and Rice stripe virus (RSV) (Zhang et al. 2021a). Guo et al. reported that apples showed enhanced powdery mildew resistance after overexpression of MhYTP2, which may act by regulating MdMLO19 mRNA stability and the translation of antioxidant genes (Guo et al. 2022). Overall, these studies suggest that m6A modification influences plant stress responses, but the specific functions of m6A in plant–pathogen interactions remain largely unknown.
Root-knot nematodes (RKNs, Meloidogyne spp.) are sedentary endoparasites with a wide host range that cause over USD 75 billion in economic losses annually. When infective J2 larvae penetrate the plant root, they migrate towards the vasculature and finally initiate permanent feeding structures known as giant cells, which are important for the development and reproduction of the nematodes (Favery et al. 2016; Hewezi 2020). The plant defense system is activated during RKN parasitism, and immune responses towards the nematodes are coordinated by various signaling pathways that involve reactive oxygen species (ROS), plant hormones, extracellular receptor-like proteins, and kinases (Goverse and Smant 2014). However, m6A abundance, dynamics, and topology during the RKN–host plant interaction remain unexplored.
In this study, we surveyed the changes in soybean m6A in response to M. incognita infection by whole-genome m6A sequencing (m6A-seq), RNA sequencing (RNA-seq), and metabolomics approaches. We combined parallel RNA-seq and metabolome analyses to analyze the putative functions of differentially expressed m6A-modified genes (DMGs) and metabolites derived from the M. incognita-infected soybean roots. We found that there was a highly negative correlation between m6A methylation and the expression levels of some genes involved in the regulation of plant immune responses, including genes encoding transcription factors (TFs), oxidoreductase activity-related proteins, and kinases. Our study provides new insights for understanding changes in mRNA modification in response to biotic stresses in soybean.
Results
Transcriptome-wide mapping of m6A methylation in soybean roots after M. incognita infection
To create a transcriptome-wide m6A methylation modification map in soybean, three replicate samples of control and M. incognita-infected soybean roots were collected and sequenced (Fig. 1). After removing adaptors and low-quality data, 45.46–49.12 million high-quality m6A-IP-seq reads per replicate were aligned to the soybean reference genome, with a mapping rate of over 90%. Furthermore, 39.24–42.1 million reads were uniquely aligned to the genome, and 84.42–97.17 million were mapped to splice reads (Table S1).
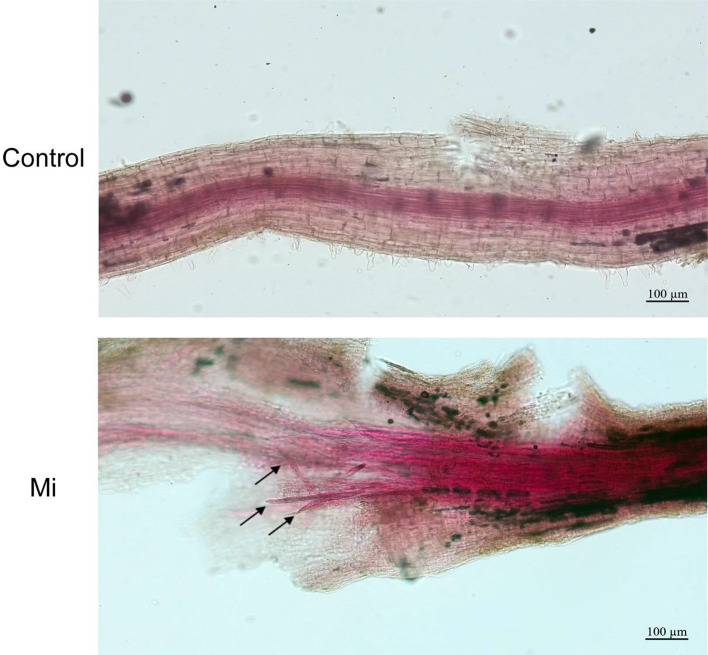
Fig. 1.
Sodium hypochlorite-acid fuchsin-stained soybean roots with and without M. incognita infection. The upper roots were uninfected (Control), and the lower roots were inoculated with M. incognita for 3 days (Mi)
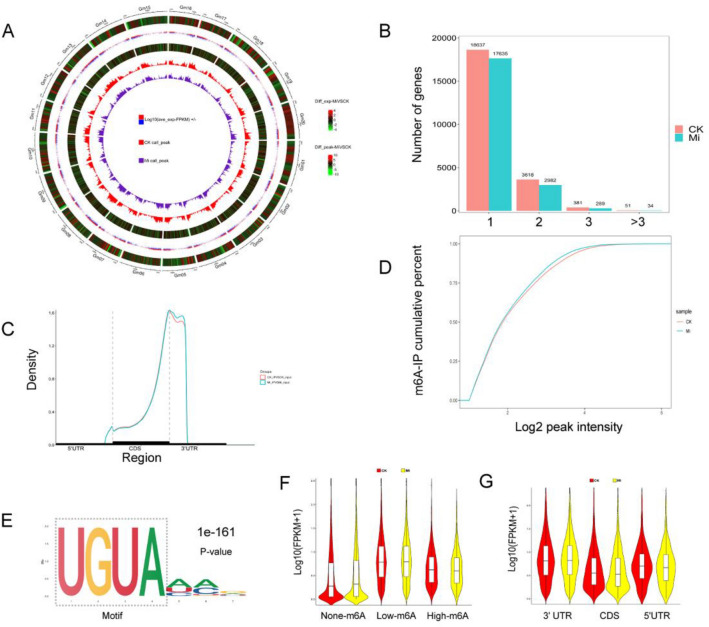
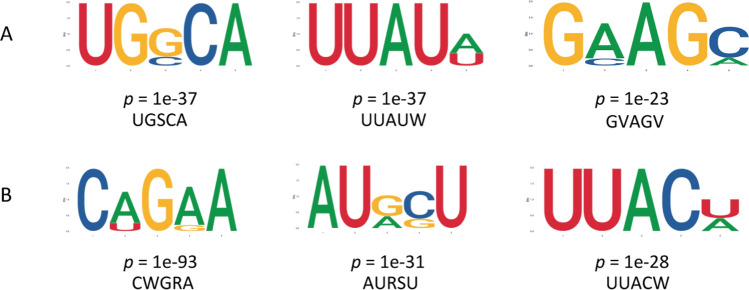
At the genome level, 42 170 m6A peak calls in 20 940 genes were identified from the M. incognita-infected roots, and 42 816 peaks in 22 687 genes were identified from control roots, with an average of 1.95 m6A sites per gene. Peak calls were enriched at the transcription start sites (TSS) and transcription end sites (TES) (Fig. 2A). Among the methylated transcripts, 83.14% contained one m6A site, 15.13% contained two sites, and 1.73% contained three or more sites (Fig. 2B). Read distribution analysis showed that m6A peaks were highly enriched in the 3′ terminal region, including the 3′ end of the coding sequence (CDS) and the 3′ UTR regions (Fig. 2C), consistent with earlier reports from rice and barley (Zhang et al. 2021a; Su et al. 2022). Global m6A hypermethylation was slightly induced by M. incognita infection compared with the control (Fig. 2D). To identify whether there were conserved motifs within m6A peaks in the control and infected soybean roots, the MEME2 and HOMER suites were used to perform de novo searches for enriched motifs. The most significant consensus motif of m6A peaks in soybean was 5′-UGUAHYY-3′ (A = m6A, H = A/C/U, and Y = A/G/U/C) (Fig. 1E). The UGSCA (S = G/C, 52.23%), UUAUW (W = A/U, 53.88%), and GVAGV (V = A/C, 54.26%) motifs were also enriched in control samples, and the CWGRA (R = A/G, 58.15%), AURSU (68.12%), and UUACW (45.78%) motifs were also enriched in infected samples (Fig. 3). To explore the correlation between gene expression and m6A modification, the expression levels of genes with or without m6A modifications were compared. From Fig. 2F, we can see that genes with m6A modification had higher expression levels than those without this modification in both infected and control roots. We also examined the correlation between expression level and m6A binding sites in the m6A-modified genes and found that genes with m6A modifications in the 3′-UTR regions had significantly higher expression than genes that were modified in other regions (Fig. 2G).
Fig. 2.
Overview of the m6A methylome and transcriptome of soybean roots under M. incognita infection (Mi) and control (CK) conditions. (A) Circos plot of m6A peak density and expression of genes on soybean chromosomes. The six rings from the outside to the inside show (1) the lengths of the soybean chromosomes, (2) a heatmap of differentially expressed genes between Mi and CK, (3) the average gene expression (FPKM) in Mi and CK, (4) a heatmap of differentially modified m6A peaks between Mi and CK, (5) a bar graph of CK peak enrichment, and (6) a bar graph of Mi peak enrichment. (B) Numbers of m6A-methylated genes with different numbers of m6A peaks under Mi and CK conditions. (C) Density of m6A peaks under Mi and CK conditions in three non-overlapping regions: 5′ UTR, CDS, and 3′ UTR. UTR, untranslated region; CDS, coding sequence. (D) Cumulative distribution of Log2(peak intensity) of m6A modification under Mi and CK conditions. (E) The predominant consensus motif for m6A methylation sites. (F) All m6A-modified genes were classified into three categories based on the number of m6A sites in each gene: non-m6A genes (m6A sites = 0), low-m6A genes (m6A sites < 3), and high-m6A genes (m6A sites ≥ 3). (G) Genes were classified into three categories (3′ UTR, CDS, and 5′ UTR) based on the location of the m6A peak in the gene
Fig. 3.
Identification of specific m6A methylation sites and sequence motifs in control and M. incognita-infected soybean roots identified using the DREME and MEME suites. (A) The significantly enriched consensus motifs in control samples. (B) The significantly enriched consensus motifs in M. incognita-infected samples
Differentially methylated genes (DMGs) in response to M. incognita are involved in multiple signaling pathways and cellular processes
Two thousand sixty-nine genes showed differentially modified m6A methylation peaks (DMPs, log2|FC|≥ 1, P ≤ 0.05) between M. incognita-infected and control soybean roots. Among these DMGs, 1007 were hypermethylated and 1062 were hypomethylated under M. incognita infection.
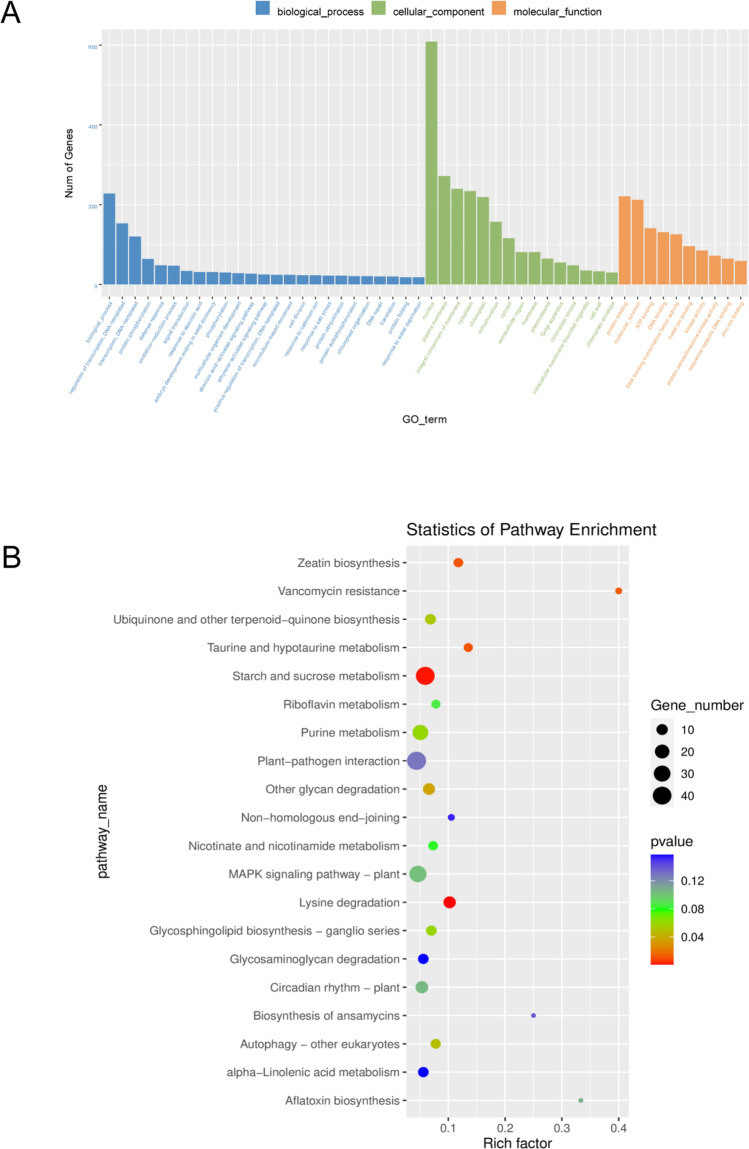
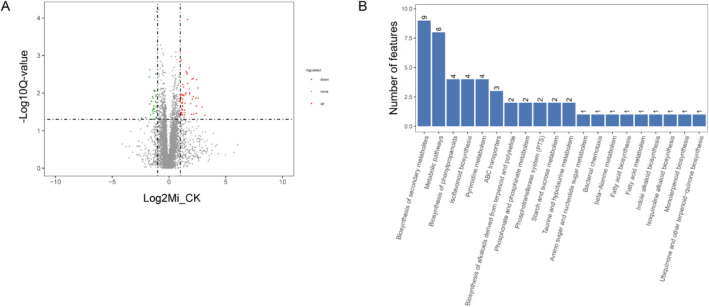
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed to explore the potential functional roles of DMGs in response to M. incognita infection. GO analysis showed that DMGs were involved in multiple biological processes such as regulation of transcription, protein phosphorylation, defense response, oxidation reduction process, signal transduction, and so forth (Fig. 4C). Notably, 85 genes were involved in kinase activity (GO:0016301), 2 in cytokinin transport (GO:0010184), and 4 in regulation of the jasmonic acid-mediated signaling pathway (GO:2000022). Moreover, 25 genes were enriched in the ethylene-activated signaling pathway (GO:0009873) (Supplementary data 1). KEGG analysis showed that DMGs were enriched mainly in eight pathways, and the top four were lysine degradation (KO00310), starch and sucrose metabolism (KO00500), zeatin biosynthesis (KO00908), and taurine and hypotaurine metabolism (KO00430). Seven genes were enriched in autophagy (KO04136), and 9 genes were enriched in the ubiquinone and other terpenoid-quinone biosynthesis pathway (KO00130) (Fig. 4D, Supplementary data 2).
Fig. 4.
m6A-methylated transcripts were differentially expressed in soybean roots after M. incognita infection. (A) GO enrichment of differentially methylated genes (DMGs) under M. incognita-infected (Mi) and control (CK) conditions. (B) KEGG enrichment of DMGs under M. incognita-infected (Mi) and control (CK) conditions
Differentially expressed genes (DEGs) and differentially accumulated metabolites (DAMs) related to M. incognita infection
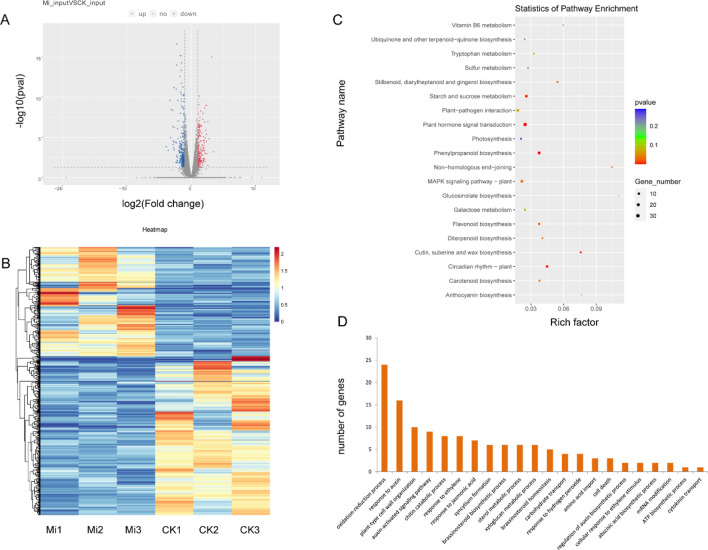
RNA-seq analysis was performed to quantify changes in gene expression levels under M. incognita infection. A total of 594 DEGs (log2|FC|≥ 1, P ≤ 0.05) were identified between M. incognita-infected and controlled soybean roots, 352 downregulated and 242 upregulated (Fig. 5A, B). Among these DEGs, 44.61% contained m6A peaks. KEGG analysis revealed that the DEGs were significantly enriched in 11 pathways, including phenylpropanoid biosynthesis, plant hormone signal transduction, circadian rhythm, MAPK signaling pathway, carotenoid biosynthesis, plant–pathogen interaction, and so forth (Fig. 5C, Supplementary data 3). GO analysis showed that multiple DEGs were annotated with the terms chitin catabolic process, plant-type cell wall organization, syncytium formation, and jasmonic acid metabolic process (Fig. 5D, Supplementary data 4).
Fig. 5.
Differentially expressed genes (DEGs) in soybean roots under M. incognita-infected (Mi) and control (CK) conditions. (A) Volcano plot of up- (red) and downregulated (blue) genes between Mi and CK. Genes with |Log2(fold change)|≥ 1 and P ≤ 0.05 were considered to be differentially expressed. (B) Heatmap of RNA-seq data from Mi and CK samples. Rows, individual mRNA transcripts; columns, individual Mi and CK samples. Orange and blue represent upregulation and downregulation of mRNA levels in Mi and control samples, respectively. (C) KEGG enrichment of DEGs. (D) GO analysis of DEGs assigned to biological processes
The metabolite compositions of control and M. incognita-infected soybean roots were analyzed by LC–MS/MS, and 3757 and 2217 total compounds were identified in positive and negative ion modes, respectively. We performed DAM analysis and identified 69 metabolites that were upregulated under M. incognita infection. These included sulfoacetate, d-(-)-fructose, malonic acid, 4-(beta-d-glucosyloxy) benzoate, secologanin, coumestrol, psoralidin, 2-hydroxyethylphosphonate, diethyl phosphate, and orthophosphate, which were mainly assigned to the pathways taurine and hypotaurine metabolism, amino sugar and nucleotide sugar metabolism, fatty acid biosynthesis, ubiquinone and other terpenoid-quinone biosynthesis, indole alkaloid biosynthesis, isoflavonoid biosynthesis, and oxidative phosphorylation. We also identified 34 metabolites that were downregulated under M. incognita infection. These included d-( +)-maltose, α,α-trehalose, thymidine 5′-monophosphate, 3′-UMP, actinidine, and 5-l-glutamyl-taurine, which were mainly assigned to the pathways starch and sucrose metabolism, pyrimidine metabolism, and taurine and hypotaurine metabolism (Fig. 6, Supplementary data 5).
Fig. 6.
Analysis of differentially accumulated metabolites (DAMs) in soybean roots under M. incognita-infected (Mi) and control (CK) conditions. (A) Volcano plot of up- (red) and downregulated (green) metabolites between Mi and CK. Metabolites with |Log2(fold change)|≥ 1 and P < 0.05 were defined as differentially abundant. (B) The top 20 KEGG pathways to which the DAMs were assigned
Conjoint analysis of m6A-modified gene expression in response to M. incognita infection
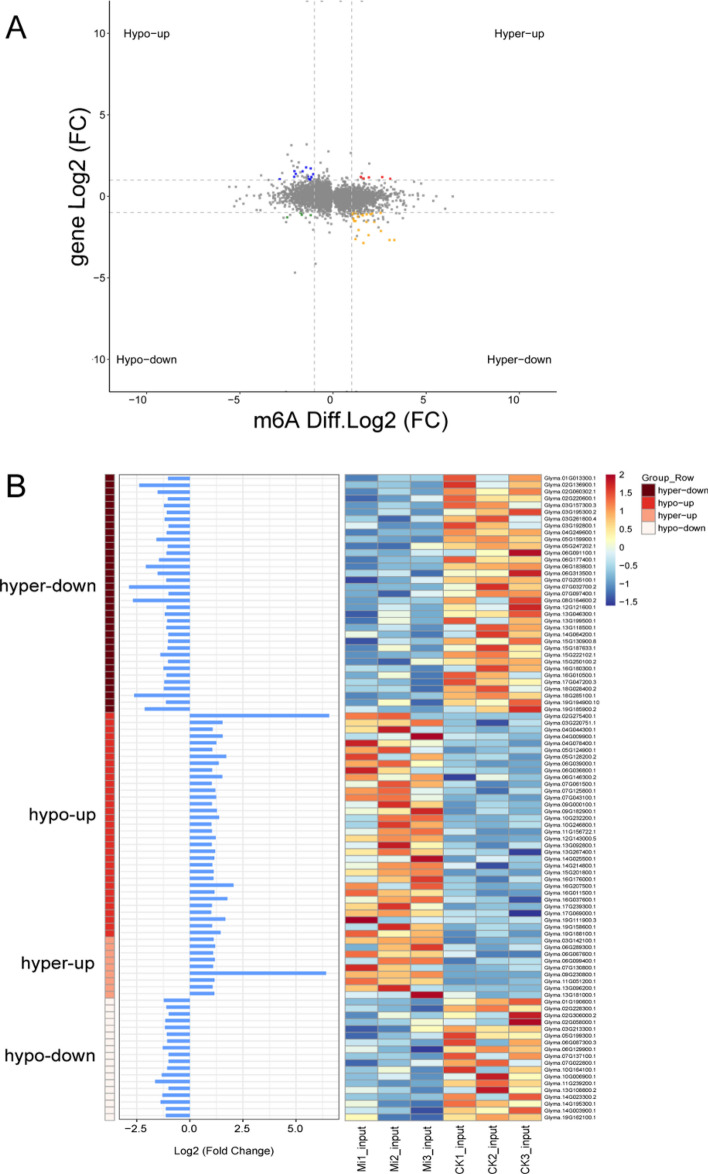
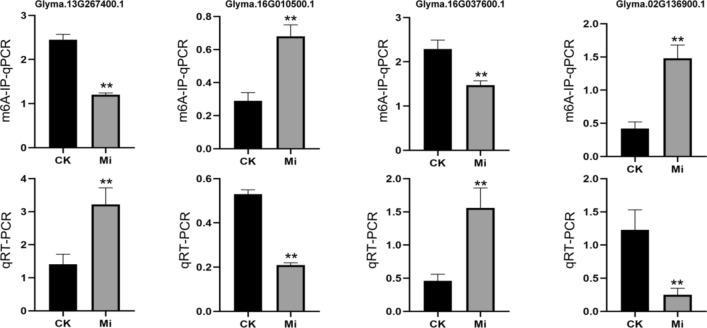
To explore whether m6A modifications were involved in regulating gene expression under M. incognita infection, a conjoint analysis of DMGs and DEGs was performed, and 101 DEGs with DMPs (i.e., DEPs) were identified. Among them, 34 genes were hypomethylated and upregulated (hypo-up) (33.66%), 9 were hypermethylated and upregulated (hyper-up) (8.9%), 19 were hypomethylated and downregulated (hypo-down) (18.81%), and 39 were hypermethylated and downregulated (hyper-down) (38.61%). These results revealed that m6A methylation peaks specific to control or M. incognita-infected roots were strongly correlated with their corresponding mRNA expression levels (P ≤ 0.05) (Fig. 7). Functional annotation analyses indicated that these DEPs were mainly categorized into four signaling pathways: regulation of transcription, oxidoreductase activity, defense response signaling pathway, and ubiquitin–proteasome pathway. Six transcription factors (TFs) were identified among these genes, including WRKY transcription factor 70 (WRKY 70, Glyma.13G267400.1), heat stress transcription factor A-7a (HSF A-7a, Glyma.03G157300.3), MYB transcription factor 114 (MYB114, Glyma.03G261800.4), MYB transcription factor 124 (MYB124, Glyma.06G036800.1), zinc finger protein (ZFP, Glyma.16G010500.1), and ethylene-responsive transcription factor 60 (ERF60, Glyma.11G239200.1). The m6A methylation levels and transcript levels of these TFs were negatively correlated, with the exception of ERF60. Five of the genes that were related to oxidoreductase activity, including berberine bridge enzyme-like 28 (BBE-like 28, Glyma.05G124900.1), plant cysteine oxidase 2 (PCO2, Glyma.16G037600.1), and peroxidase 47 (POD 47, Glyma.16G207500.1) showed reduced m6A modification and increased transcript levels. By contrast, putative respiratory burst oxidase homolog protein (Glyma.07G130800.1), 9-cis-epoxycarotenoid dioxygenase (NCED1, Glyma.15G250100.2), and TPR repeat-containing thioredoxin (TDX, Glyma.05G247202.1) showed increased m6A modification and reduced transcript levels. Five of the DEPs were involved in the defense response signaling pathway, including two LRR receptor-like serine/threonine-protein kinases (Glyma.05G128200.2, hypo-up; Glyma.15G130900.8, hyper-down), one G-type lectin S-receptor-like serine/threonine-protein kinase (Glyma.12G143000.5, hypo-up), and two cytochrome P450 genes (CYP94A1, Glyma.19G162100.1, hypo-up; CYP93A3, Glyma.03G142100.1, hyper-up). Finally, four of these genes participated in the ubiquitin–proteasome pathway, including ubiquitin carboxyl-terminal hydrolase (Glyma.02G220600.1, hyper-down), U-box domain-containing protein 18 (Glyma.06G183800.1, hyper-down), NADH-ubiquinone oxidoreductase (Glyma.15G187633.1, hyper-down) and putative RING finger protein P32A8.03c (Glyma.15G201800.1, hypo-up) (Fig. 8, Table S2). Moreover, we verified the m6A methylation and transcript levels of Glyma.13G267400.1, Glyma.16G010500.1, Glyma.16G037600 and Glyma.02G136900.1 by m6A-IP-qPCR and qRT-PCR (Fig. 9). The expression patterns of these genes were consistent with the results of m6A-IP-seq and RNA-seq.
Fig. 7.
Conjoint analysis of m6A-seq and RNA-seq data. (A) A four-image map of differentially expressed genes (DEGs) and differential peaks that significantly changed in both m6A and mRNA levels in M. incognita-infected and control samples (fold change ≥ 1, P < 0.05). (B) Bar chart and heat map of the hyper-down, hypo-up, hyper-up, and hypo-down genes shown in A. Left, m6A-seq; right, RNA-seq
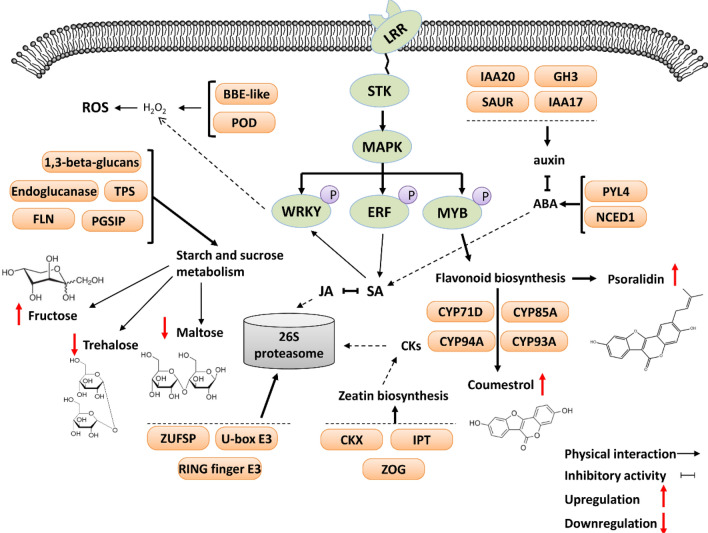
Fig. 8.
Model illustrating the proposed regulatory networks based on transcriptome-wide m6A and metabolome profiles of soybean roots under M. incognita infection
Fig. 9.
Validation of m6A peaks of four randomly selected genes in M. incognita-infected (Mi) and uninfected (CK) soybean roots using m6A-immunoprecipitation (IP)-qPCR (m6A-IP-qPCR) and RT-PCR (qRT-PCR). Each bar represents the mean plus SD of three biological replicates, with four technical replicates for each independent experiment, and the mean values significantly different from the control are demoted by ∗∗ as determined by independent samples t-test (P < 0.01)
Discussion
Topology and features of m6A modification in soybean
Using m6A-seq and transcriptome analysis, we first revealed an increase in overall m6A modification levels of soybean under M. incognita infection. The m6A distribution on the gene body was highly selective, and almost all soybean m6A peaks were located around the 3′ end of the coding sequence (CDS) and throughout the 3′ UTR region, similar to results in Arabidopsis thaliana, rice, maize, barley, and tomato (Su et al. 2022; Wan et al. 2015; Zhang et al. 2021a; Zhou et al. 2019) but different from cotton, whose m6A methylation is highly enriched in the stop codon and CDS region. In general, the expression level of genes was associated with m6A modification in the 3′ UTR, implying that m6A methylation in the 3′ UTR may play a regulatory role in transcriptional processes in soybean.
In previous work, RRACH (R = A/G, H = A/C/U) was the most significantly enriched motif in plant m6A peaks, and Kane and Beemon demonstrated that when the highly conserved GAC was mutated to GAU, m6A modifications no longer occurred in Rous sarcoma virus mRNA (Kane and Beemon 1987). URUAY is another plant-specific consensus motif exhibited in m6A peaks and was identified from Arabidopsis, rice, watermelon, and tomato (He et al. 2021a, b; Wan et al. 2015; Zhang et al. 2021a; Zhou et al. 2019). Previous studies indicated that there may be higher enrichment E-values for the URUAY motif than the RRACH motif in dicot plants (Luo et al. 2014; He et al. 2021a, b). Here, we identified the consensus motif UGUAHYY (Y = A/G/U/C) that was significantly enriched in control and M. incognita-infected soybean samples, consistent with results in Arabidopsis, watermelon, and tomato. We also found that UGSCA (S = G/C) and UUAUW (W = A/U) motifs were enriched in control samples, and CWGRA, AURSU, and UUACW motifs were enriched in M. incognita-infected samples. These results reveal potential consensus motifs around m6A peaks in infected and uninfected roots, and the functions of these consensus motifs deserve further exploration.
Potential roles of m6A modification in early responses to M. incognita infection
The m6A modification is the most common and conserved type of RNA methylation; it is involved in mRNA processing (Visvanathan and Somasundaram 2018; Pontier et al. 2019), embryo development (Tzafrir et al. 2003), and stress responses (Dominissini et al. 2012; Jia et al. 2013; Ok et al. 2005). However, the precise roles played by m6A modification in these processes are obscure. Imam et al. revealed that m6A modification influenced the viral life cycle and viral replication in mammalian host–virus interactions (Imam et al. 2020). Li et al. found that m6A modification was strongly associated with the occurrence and development of liver hepatocellular carcinoma, and some RNA methylation-related genes play an important role in tumorigenesis and metastasis (Li et al. 2021). The m6A modification process has been reported to regulate the expression levels of genes involved in key pathways in Arabidopsis, tobacco, rice, and watermelon upon virus infection (He et al. 2021a, b; Li et al. 2018; Martinez-Perez et al. 2021; Zhang et al. 2021a).
In the current study, 2069 differentially methylated genes (DMGs) were identified in M. incognita-infected and control soybean roots. These DMGs were associated with multiple signaling pathways involved in host defense responses. Interestingly, KEGG analysis showed that zeatin biosynthesis was the most significantly enriched phytohormone signaling pathway. Zeatin is a type of cytokinin (CK) derivative. CKs are a class of phytohormones that promote plant cell division and differentiation and participate in the regulation of plant growth, physiological activities, and metabolic processes (Kieber and Schaller, 2018; Schafer et al. 2015). Recently, many studies have reported changes in CK contents and CK signaling during plant–pathogen interactions. In Arabidopsis, elevated levels of cytokinin enhanced defense responses to Pseudomonas syringae pv. tomato DC3000 by cross talk with salicylic acid (SA) signaling (Choi et al. 2010). The overexpression of cytokinin oxidase genes (AtCKX3 and ZmCKX1) in Lotus japonicas roots disturbed the formation of giant cells during M. incognita infection, and there were fewer female nematodes in the transgenic roots than in control roots (Absmanner et al. 2013). Trdá et al. reported that invasion of the fungus Leptosphaeria maculans can alter the activity of O-glucosyltransferase and N-glucosyltransferases in infected Brassica napus tissues, leading to the transformation of the CK isopentyladenine (iP) into iP-9-riboside-5′-monophosphate (iPRMP) or of cis-zeatin into cis-zeatin O-glucoside. This modification of CK profiles contributed to the infection process (Trdá et al. 2017). Here, we found that some genes related to CK processes were modified by m6A, although the specific roles played by these genes during M. incognita infection, especially their interactions in phytohormone networks, remain to be fully characterized. Two other enriched pathways, autophagy (KO04136) and ubiquinone and other terpenoid-quinone biosynthesis (KO00130), are also important for plant fitness and immunity, and there have been reports that pathogens can hijack these pathways to facilitate survival or replication (Choi et al. 2018; Kud et al. 2019).
Multiple omics approaches integration will speed up the identification efficient of candidate genes for deeply research (Cao et al. 2022; Han et al. 2022; Lu et al. 2022). To further explore the relationship between m6A methylation levels and transcript levels in response to M. incognita infection, we identified 101 highly hypermethylated or hypomethylated DEGs, most of which have been reported to participate in the regulation of plant immune defense responses. For example, WRKY and MYB TFs can act as activators or repressors in plant hormone signal transduction, playing important roles in pathogen-associated molecular pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) (Dubos et al. 2010; Jiang et al. 2017; Rushton et al. 2010; Wani et al. 2021). MYB transcription factors have been proposed to regulate the biosynthesis of flavonoids (Anwar et al. 2019; Ma and Constabel 2019; Wang et al. 2018, 2020), which are one of the most important secondary metabolites that contribute to plant development and responses to biotic and abiotic stimuli (Petrussa et al. 2013; Silva et al. 2018; Zakaryan et al. 2017). Their transcriptional regulatory network is complex, and the MYBs may be involved in different steps of the flavonoid biosynthetic pathway. In Arabidopsis, Myb11, Myb12, and Myb111 can activate four flavonol biosynthesis genes and play an important role in regulating the early steps of the flavonoid pathway (Xu et al. 2015). Moreover, seedlings of the triple mutant myb11 myb12 myb111 do not form flavonols (Stracke et al. 2007). By contrast, some MYBs are negative regulators of the flavonoid biosynthetic pathway. For example, Arabidopsis MYB4 represses flavonoid biosynthesis by inhibiting the transcriptional activity of Arogenate dehydratase 6 (ADT6) (Wang et al. 2020). There have also been reports on MYB repressors in other species, such as AmMYB308 in Antirrhinum majus (Tamagnone et al. 1998), FaMYB1 in strawberry (Aharoni et al. 2001; Salvatierra et al. 2013), and PtrMYB182 and PtrMYB57 in poplar (Yoshida et al. 2015; Wan et al. 2017). In this study, we found that two flavonoid metabolites, coumestrol and psoralidin, showed significantly higher accumulation in M. incognita-infected soybean roots. We speculate that the induction of flavonoid synthesis may have been associated with m6A methylation of MYB TFs; these changes in m6A methylation levels then influenced MYB TF expression, thereby regulating the flavonoid biosynthesis pathway. In addition, two genes with essential roles in reactive oxygen species (ROS) homeostasis, BBE-like 28 and POD 47, were hypomethylated and upregulated in M. incognita-infected soybean roots. Basal defense responses to biotic stress can be activated by the production of ROS, and ROS accumulation can be detected around the M. incognita feeding site in the early stage of infection. The changes in expression levels of these genes may have altered root ROS concentrations, thereby activating resistance responses towards M. incognita (Fig. 8).
In conclusion, this study uncovered important features of the m6A methylome during the interaction between soybean and M. incognita. Combining parallel m6A-seq, RNA-seq, and metabolome analysis, we found that m6A-methylated genes were involved in multiple signaling pathways and cellular processes, such as zeatin biosynthesis and autophagy. The hypomethylation or hypermethylation of some transcription factors and oxidoreductases was consistent with the accumulation of flavonoid metabolites and ROS in root tissues, which play important roles in M. incognita resistance. It would be interesting to further characterize the mechanism of m6A methylation in soybean in response to M. incognita, as the results could be used to exploit epitranscriptomic RNA modifications for crop improvement.
Materials and methods
Plant materials and M. incognita inoculation
Meloidogyne incognita nematodes were propagated on cucumber plants (Cucumis sativus L., Zhongnong 6). Meloidogyne incognita susceptible Williams 82 soybean plants were planted in a growth chamber under controlled conditions (16-h light/8-h dark) at 26 °C. For M. incognita inoculation, egg masses were collected from the cucumber roots and hatched in water. Soybean seedlings with two true leaves were inoculated with 1000 J2 larvae per plant, and uninfected seedlings served as the controls. At 3 days post-inoculation (during the large-scale invasion of roots by larvae) (Martínez-Medina et al. 2017), nine replicate root samples were collected from infected and uninfected soybean plants; each replicate consisted of pooled root material from three individual plants. The pooled samples were immediately frozen in liquid nitrogen and stored at − 80 °C for use in m6A-IP-seq, RNA-seq, and metabolome analysis.
RNA extraction and fragmentation
Three biological replicates per treatment were used for the m6A-IP-seq and RNA-seq experiments. Total RNA was isolated from the frozen root samples using the TRIzol reagent (Invitrogen, USA). The concentration and quality of the total RNA were determined on a NanoDrop ND-1000 spectrophotometer (NanoDrop, USA). Poly(A) RNA was enriched by two rounds of purification using Dynabeads Oligo (dT)25–61,005 (Thermo Fisher, USA). Finally, the poly(A) RNA was chemically fragmented into small pieces using a Magnesium RNA Fragmentation Module (NEB, USA).
m6A-IP-seq library construction and sequencing
Fragmented RNA was incubated with affinity purified anti-m6A polyclonal antibody (Synaptic Systems, Germany) in IP buffer for 2 h at 4 °C. The IP RNA was reverse transcribed to cDNA with SuperScript II Reverse Transcriptase (Invitrogen, USA). cDNA library construction was performed as described previously (Zhang et al. 2021b), and m6A-seq was performed on the Illumina NovaSeq 6000 platform at Hangzhou LC-Bio Technology Co., Ltd.
m6A sequencing and analysis
The raw reads were trimmed to remove adapters, low-quality bases, and undetermined bases using the default parameters of fastp software (https://github.com/OpenGene/fastp), and the sequence quality of the IP and Input samples was assessed with FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc) and RseQC (http://rseqc.sourceforge.net/). The reads were aligned to the reference genome of Glycine max. The exomePeak2 R package (https://bioconductor.org/packages/release/bioc/html/exomePeak2.html) was used to identify m6A peaks, and the peaks were annotated with the R package ANNOVAR (http://www.openbioinformatics.org/annovar/). The discovery of m6A-enriched motifs was performed using MEME (http://meme-suite.org) and HOMER (http://homer.ucsd.edu/homer/motif). StringTie (https://ccb.jhu.edu/software/stringtie) was used to calculate FPKM (total exon fragments/million mapped reads × exon length in kb) for all genes or peaks from the input libraries. Differentially expressed genes or peaks were identified using the criteria of absolute log2(fold change) ≥ 1 and P-value < 0.05 with the edgeR R package (https://bioconductor.org/packages/edgeR). We used the Blast2GO website and the KEGG Automated Annotation Server (KAAS) to perform GO functional enrichment analysis and KEGG enrichment analysis with default parameters (Conesa et al. 2005; Mao et al. 2005).
Metabolome analysis
Six biological replicates were used for metabolome analysis. Sample processing, extraction, and ultra-performance liquid chromatography–tandem mass spectrometry (LC–MS/MS) were performed by LC-Bio Technology Co., Ltd. (Hangzhou). The frozen samples were ground in liquid nitrogen, then resuspended in prechilled 80% methanol and 0.1% formic acid. After incubation on ice for 5 min, the samples were centrifuged at 15 000 rpm and 4 °C for 10 min. The supernatants were filtered through 0.22-μm microfilters and used for LC–MS/MS analysis. The LC–MS/MS analyses were performed using a Q Exactive instrument (Thermo Fisher Scientific, USA) in positive/negative polarity mode, and data were processed using Compound Discoverer 3.1.0 (Thermo Fisher Scientific, USA) for peak alignment and selection, gap filling, and metabolite identification. Partial least squares-discriminant analysis (PLS-DA) was performed using the metaX R package (Wen et al. 2017). Metabolites with a ratio ≥ 2 or ≤ 1/2, P-value < 0.05, and VIP > 1 were considered to be differentially abundant.
Validation of m6A sites using m6A-IP-qPCR and qRT-PCR
Total RNA was extracted from infected and uninfected soybean roots. mRNA purification, fragmentation, and m6A-IP experiments were performed as previously described (Zhang et al. 2021a, b; He et al. 2021a). The immunoprecipitated mRNA (IP mRNA) and pre-immunoprecipitated mRNA (input mRNA) were reverse transcribed to cDNA using the FastQuant RT Kit with gDNase (Tiangen, China). The cDNA was used as a template for m6A-IP-qPCR and qRT-PCR. The soybean ubiquitin gene (Glyma.20G141600) served as an internal control, and the primers are listed in Table S3. Gene expression changes were calculated using the 2−ΔΔCt method (Livak and Schmittgen 2001).
Raw sequencing data have been deposited at the China National GeneBank database CNGBdb-EBB under project accession number CNP0002790 (https://db.cngb.org/ebb/bio_resources/; submission ID sub029557).
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This study was financially supported by the National Natural Science Foundation of China (31901859, 31901858) and the Syngenta-NEAU union foundation.
Author contributions
XH: Conceptualization, Data curation, Writing—Original draft preparation; QS: Data curation, Visualization, Investigation, Writing—Original draft preparation; ZH: Data curation, Visualization, Investigation; WS: Reviewing and Supervision; QC: Supervision; ZQ: Conceptualization, Methodology, Supervision, Writing, Reviewing and Editing.
Data availability
All data generated or analysed during this study are included in this published article (and its supplementary information files).
Declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Compliance with ethical requirements
This article does not contain any studies with human or animal subjects.
Footnotes
Xue Han, Qianqian Shi and Ziyi He have contributed equally to this work.
References
- Absmanner B, Stadler R, Hammes UZ. Phloem development in nematode-induced feeding sites: the implications of auxin and cytokinin. Front Plant Sci. 2013;4:241. doi: 10.3389/fpls.2013.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni A, De Vos CH, Wein M, Sun Z, Greco R, Kroon A, et al. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 2001;28(3):319–332. doi: 10.1046/j.1365-313X.2001.01154.x. [DOI] [PubMed] [Google Scholar]
- Anderson SJ, Kramer MC, Gosai SJ, Yu X, Vandivier LE, Nelson A, et al. N6-Methyladenosine inhibits local ribonucleolytic cleavage to stabilize mRNAs in Arabidopsis. Cell Rep. 2018;25(5):1146–1157. doi: 10.1016/j.celrep.2018.10.020. [DOI] [PubMed] [Google Scholar]
- Anwar M, Yu W, Yao H, Zhou P, Allan AC, Zeng L. NtMYB3, an R2R3-MYB from narcissus, regulates flavonoid biosynthesis. Int J Mol Sci. 2019;20:21. doi: 10.3390/ijms20215456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodi Z, Button JD, Grierson D, Fray RG. Yeast targets for mRNA methylation. Nucleic Acids Res. 2010;38(16):5327–5335. doi: 10.1093/nar/gkq266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao P, Zhao Y, Wu F, Xin D, Liu C, Wu X, et al. Multi-Omics techniques for soybean molecular breeding. Int J Mol Sci. 2022;23(9):4994. doi: 10.3390/ijms23094994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Huh SU, Kojima M, Sakakibara H, Paek KH, Hwang I. The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev Cell. 2010;19(2):284–295. doi: 10.1016/j.devcel.2010.07.011. [DOI] [PubMed] [Google Scholar]
- Choi Y, Bowman JW, Jung JU. Autophagy during viral infection-a double-edged sword. Nat Rev Microbiol. 2018;16(6):341–354. doi: 10.1038/s41579-018-0003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy MJ, Shambaugh ME, Timpte CS, Bokar JA. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 2002;30(20):4509–4518. doi: 10.1093/nar/gkf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626. doi: 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan HC, Wei LH, Zhang C, Wang Y, Chen L, Lu Z, et al. ALKBH10B is an RNA N6-methyladenosine demethylase affecting Arabidopsis floral transition. Plant Cell. 2017;29(12):2995–3011. doi: 10.1105/tpc.16.00912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15(10):573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Favery B, Quentin M, Jaubert-Possamai S, Abad P. Gall-forming root—knot nematodes hijack key plant cellular functions to induce multinucleate and hypertrophied feeding cells. J Insect Physiol. 2016;84:60–69. doi: 10.1016/j.jinsphys.2015.07.013. [DOI] [PubMed] [Google Scholar]
- Fray RG, Simpson GG. The Arabidopsis epitranscriptome. Curr Opin Plant Biol. 2015;27:17–21. doi: 10.1016/j.pbi.2015.05.015. [DOI] [PubMed] [Google Scholar]
- Fukusumi Y, Naruse C, Asano M. Wtap is required for differentiation of endoderm and mesoderm in the mouse embryo. Dev Dyn. 2008;237(3):618–629. doi: 10.1002/dvdy.21444. [DOI] [PubMed] [Google Scholar]
- Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, et al. RNA- methylation- dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155(4):793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- Goverse A, Smant G. The activation and suppression of plant innate immunity by parasitic nematodes. Annu Rev Phytopathol. 2014;52:243–265. doi: 10.1146/annurev-phyto-102313-050118. [DOI] [PubMed] [Google Scholar]
- Guo T, Liu C, Meng F, Hu L, Fu X, Yang Z, et al. The m6A reader MhYTP2 regulates MdMLO19 mRNA stability and antioxidant genes translation efficiency conferring powdery mildew resistance in apple. Plant Biotechnol J. 2022;20(3):511–525. doi: 10.1111/pbi.13733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Li J, Zhao Y, Zhang Z, Jiang H, Wang J, et al. Integrated transcriptomic and proteomic characterization of a chromosome segment substitution line reveals a new regulatory network controlling the seed storage profile of soybean. Food Energy Secur. 2022;11:e381. doi: 10.1002/fes3.381. [DOI] [Google Scholar]
- He S, Wang H, Liu R, He M, Che T, Jin L, et al. mRNA N6-methyladenosine methylation of postnatal liver development in pig. PLoS ONE. 2017;12(3):e173421. doi: 10.1371/journal.pone.0173421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Li L, Yao Y, Li Y, Zhang H, Fan M. Transcriptome-wide N6-methyladenosine (m6A) methylation in watermelon under CGMMV infection. BMC Plant Biol. 2021;21(1):516. doi: 10.1186/s12870-021-03289-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Li Y, Yao Y, Zhang H, Wang Y, Gao J, et al. Overexpression of watermelon m6A methyltransferase ClMTB enhances drought tolerance in tobacco by mitigating oxidative stress and photosynthesis inhibition and modulating stress-responsive gene expression. Plant Physiol Biochem. 2021;168:340–352. doi: 10.1016/j.plaphy.2021.10.007. [DOI] [PubMed] [Google Scholar]
- Hewezi T. Epigenetic mechanisms in nematode-plant interactions. Annu Rev Phytopathol. 2020;58:119–138. doi: 10.1146/annurev-phyto-010820-012805. [DOI] [PubMed] [Google Scholar]
- Hou Y, Sun J, Wu B, Gao Y, Nie H, Nie Z, et al. CPSF30-L-mediated recognition of mRNA m6A modification controls alternative polyadenylation of nitrate signaling-related gene transcripts in Arabidopsis. Mol Plant. 2021;14(4):688–699. doi: 10.1016/j.molp.2021.01.013. [DOI] [PubMed] [Google Scholar]
- Imam H, Kim GW, Siddiqui A. Epitranscriptomic (N6-methyladenosine) modification of viral RNA and Virus-Host interactions. Front Cell Infect Microbiol. 2020;10:584283. doi: 10.3389/fcimb.2020.584283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, He C. Reversible RNA adenosine methylation in biological regulation. Trends Genet. 2013;29(2):108–115. doi: 10.1016/j.tig.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Ma S, Ye N, Jiang M, Cao J, Zhang J. WRKY transcription factors in plant responses to stresses. J Integr Plant Biol. 2017;59(2):86–101. doi: 10.1111/jipb.12513. [DOI] [PubMed] [Google Scholar]
- Kane SE, Beemon K. Inhibition of methylation at two internal N6-methyladenosine sites caused by GAC to GAU mutations. J Biol Chem. 1987;262(7):3422–3427. doi: 10.1016/S0021-9258(18)61520-0. [DOI] [PubMed] [Google Scholar]
- Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, et al. A majority of m6A residues are in the last exons, allowing the potential for 3’ UTR regulation. Genes Dev. 2015;29(19):2037–2053. doi: 10.1101/gad.269415.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Schaller GE. Cytokinin signaling in plant development. Development. 2018;145:4. doi: 10.1242/dev.149344. [DOI] [PubMed] [Google Scholar]
- Kierzek E, Kierzek R. The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res. 2003;31(15):4472–4480. doi: 10.1093/nar/gkg633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kud J, Wang W, Gross R, Fan Y, Huang L, Yuan Y, et al. The potato cyst nematode effector RHA1B is a ubiquitin ligase and uses two distinct mechanisms to suppress plant immune signaling. PLoS Pathog. 2019;15(4):e1007720. doi: 10.1371/journal.ppat.1007720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R, Penman S. 5'-terminal structures of poly(A)+ cytoplasmic messenger RNA and of poly(A)+ and poly(A)- heterogeneous nuclear RNA of cells of the dipteran Drosophila melanogaster. J Mol Biol. 1978;120(4):487–515. doi: 10.1016/0022-2836(78)90350-9. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang X, Li C, Hu S, Yu J, Song S. Transcriptome-wide N6-methyladenosine profiling of rice callus and leaf reveals the presence of tissue-specific competitors involved in selective mRNA modification. RNA Biol. 2014;11(9):1180–1188. doi: 10.4161/rna.36281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Shi J, Yu L, Zhao X, Ran L, Hu D, et al. N6-methyl-adenosine level in Nicotiana tabacum is associated with tobacco mosaic virus. Virol J. 2018;15(1):87. doi: 10.1186/s12985-018-0997-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Qi D, Zhu B, Ye X. Analysis of m6A RNA methylation-related genes in liver hepatocellular carcinoma and their correlation with survival. Int J Mol Sci. 2021;22:3. doi: 10.3390/ijms22031474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu S, Fang C, Abe J, Kong F, Liu B. Current overview on the genetic basis of key genes involved in soybean domestication. aBIOTECH. 2022;3:126–139. doi: 10.1007/s42994-022-00074-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo GZ, MacQueen A, Zheng G, Duan H, Dore LC, Lu Z, et al. Unique features of the m6A methylome in Arabidopsis thaliana. Nat Commun. 2014;5:5630. doi: 10.1038/ncomms6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Constabel CP. MYB repressors as regulators of phenylpropanoid metabolism in plants. Trends Plant Sci. 2019;24(3):275–289. doi: 10.1016/j.tplants.2018.12.003. [DOI] [PubMed] [Google Scholar]
- Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21(19):3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- Martínez-Medina A, Fernandez I, Lok GB, Pozo MJ, Pieterse CM, Van Wees SC. Shifting from priming of salicylic acid- to jasmonic acid-regulated defences by Trichoderma protects tomato against the root knot nematode Meloidogyne incognita. New Phytol. 2017;213(3):1363–1377. doi: 10.1111/nph.14251. [DOI] [PubMed] [Google Scholar]
- Martinez-Perez M, Aparicio F, Lopez-Gresa MP, Belles JM, Sanchez-Navarro JA, Pallas V. Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc Natl Acad Sci U S A. 2017;114(40):10755–10760. doi: 10.1073/pnas.1703139114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez M, Gomez-Mena C, Alvarado-Marchena L, Nadi R, Micol JL, Pallas V, et al. The m6A RNA demethylase ALKBH9B plays a critical role for vascular movement of alfalfa mosaic virus in Arabidopsis. Front Microbiol. 2021;12:745576. doi: 10.3389/fmicb.2021.745576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149(7):1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondo SJ, Dannebaum RO, Kuo RC, Louie KB, Bewick AJ, LaButti K, et al. Widespread adenine N6-methylation of active genes in fungi. Nat Genet. 2017;49(6):964–968. doi: 10.1038/ng.3859. [DOI] [PubMed] [Google Scholar]
- Ok SH, Jeong HJ, Bae JM, Shin JS, Luan S, Kim KN. Novel CIPK1-associated proteins in Arabidopsis contain an evolutionarily conserved C-terminal region that mediates nuclear localization. Plant Physiol. 2005;139(1):138–150. doi: 10.1104/pp.105.065649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrussa E, Braidot E, Zancani M, Peresson C, Bertolini A, Patui S, et al. Plant flavonoids-biosynthesis, transport and involvement in stress responses. Int J Mol Sci. 2013;14(7):14950–14973. doi: 10.3390/ijms140714950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier D, Picart C, El BM, Roudier F, Xu T, Lahmy S, et al. The m6A pathway protects the transcriptome integrity by restricting RNA chimera formation in plants. Life Sci Alliance. 2019;2:3. doi: 10.26508/lsa.201900393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010;15(5):247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Růžička K, Zhang M, Campilho A, Bodi Z, Kashif M, Saleh M, et al. Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 2017;215(1):157–172. doi: 10.1111/nph.14586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatierra A, Pimentel P, Moya-León MA, Herrera R. Increased accumulation of anthocyanins in Fragaria chiloensis fruits by transient suppression of FcMYB1 gene. Phytochemistry. 2013;90:25–36. doi: 10.1016/j.phytochem.2013.02.016. [DOI] [PubMed] [Google Scholar]
- Schafer M, Brutting C, Meza-Canales ID, Grosskinsky DK, Vankova R, Baldwin IT, et al. The role of cis-zeatin-type cytokinins in plant growth regulation and mediating responses to environmental interactions. J Exp Bot. 2015;66(16):4873–4884. doi: 10.1093/jxb/erv214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scutenaire J, Deragon JM, Jean V, Benhamed M, Raynaud C, Favory JJ, et al. The YTH domain protein ECT2 is an m6A reader required for normal trichome branching in Arabidopsis. Plant Cell. 2018;30(5):986–1005. doi: 10.1105/tpc.17.00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Wong CE, Shen L, Yu H. N6-methyladenosine modification underlies messenger RNA metabolism and plant development. Curr Opin Plant Biol. 2021;63:102047. doi: 10.1016/j.pbi.2021.102047. [DOI] [PubMed] [Google Scholar]
- Shen L, Liang Z, Gu X, Chen Y, Teo ZW, Hou X, et al. N(6)-methyladenosine RNA modification regulates shoot stem cell fate in arabidopsis. Dev Cell. 2016;38(2):186–200. doi: 10.1016/j.devcel.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Liang Z, Wong CE, Yu H. Messenger RNA modifications in plants. Trends Plant Sci. 2019;24(4):328–341. doi: 10.1016/j.tplants.2019.01.005. [DOI] [PubMed] [Google Scholar]
- Silva EC, Abhayawardhana PL, Lygin AV, Robertson CL, Liu M, Liu Z, et al. Coumestrol confers partial resistance in soybean plants against cercospora leaf blight. Phytopathology. 2018;108(8):935–947. doi: 10.1094/PHYTO-05-17-0189-R. [DOI] [PubMed] [Google Scholar]
- Song P, Yang J, Wang C, Lu Q, Shi L, Tayier S, et al. Arabidopsis N6-methyladenosine reader CPSF30-L recognizes FUE signals to control polyadenylation site choice in liquid-like nuclear bodies. Mol Plant. 2021;14(4):571–587. doi: 10.1016/j.molp.2021.01.014. [DOI] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, et al. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007;50(4):660–677. doi: 10.1111/j.1365-313X.2007.03078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T, Fu L, Kuang L, Chen D, Zhang G, Shen Q, et al. Transcriptome-wide m6A methylation profile reveals regulatory networks in roots of barley under cadmium stress. J Hazard Mater. 2022;423(Pt A):127140. doi: 10.1016/j.jhazmat.2021.127140. [DOI] [PubMed] [Google Scholar]
- Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K, et al. The AmMYB308 and AmMYB330 transcription factors from antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell. 1998;10(2):135–154. doi: 10.1105/tpc.10.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Yang J, Duan H, Jia G. ALKBH10B, an mRNA m6A demethylase, modulates aba response during seed germination in Arabidopsis. Front Plant Sci. 2021;12:712713. doi: 10.3389/fpls.2021.712713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trda L, Baresova M, Sasek V, Novakova M, Zahajska L, Dobrev PI, et al. Cytokinin metabolism of pathogenic fungus Leptosphaeria maculans involves isopentenyltransferase, adenosine kinase and cytokinin oxidase/dehydrogenase. Front Microbiol. 2017;8:1374. doi: 10.3389/fmicb.2017.01374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Dickerman A, Brazhnik O, Nguyen Q, McElver J, Frye C, et al. The Arabidopsis seedgenes project. Nucleic Acids Res. 2003;31(1):90–93. doi: 10.1093/nar/gkg028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanyushin BF, Tkacheva SG, Belozersky AN. Rare bases in animal DNA. Nature. 1970;225(5236):948–949. doi: 10.1038/225948a0. [DOI] [PubMed] [Google Scholar]
- Vespa L, Vachon G, Berger F, Perazza D, Faure JD, Herzog M. The immunophilin-interacting protein AtFIP37 from Arabidopsis is essential for plant development and is involved in trichome endoreduplication. Plant Physiol. 2004;134(4):1283–1292. doi: 10.1104/pp.103.028050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan A, Somasundaram K. mRNA traffic control reviewed: N6-Methyladenosine (m6A) takes the driver's seat. BioEssays. 2018;40:1. doi: 10.1002/bies.201700093. [DOI] [PubMed] [Google Scholar]
- Wan Y, Tang K, Zhang D, Xie S, Zhu X, Wang Z, et al. Transcriptome-wide high-throughput deep m6A-seq reveals unique differential m6A methylation patterns between three organs in Arabidopsis thaliana. Genome Biol. 2015;16:272. doi: 10.1186/s13059-015-0839-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan S, Li C, Ma X, Luo K. PtrMYB57 contributes to the negative regulation of anthocyanin and proanthocyanidin biosynthesis in poplar. Plant Cell Rep. 2017;36(8):1263–1276. doi: 10.1007/s00299-017-2151-y. [DOI] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N6-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161(6):1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WL, Wang YX, Li H, Liu ZW, Cui X, Zhuang J. Two MYB transcription factors (CsMYB2 and CsMYB26) are involved in flavonoid biosynthesis in tea plant [Camellia sinensis (L.) O. Kuntze] BMC Plant Biol. 2018;18(1):288. doi: 10.1186/s12870-018-1502-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XC, Wu J, Guan ML, Zhao CH, Geng P, Zhao Q. Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. Plant J. 2020;101(3):637–652. doi: 10.1111/tpj.14570. [DOI] [PubMed] [Google Scholar]
- Wani SH, Anand S, Singh B, Bohra A, Joshi R. WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 2021;40(7):1071–1085. doi: 10.1007/s00299-021-02691-8. [DOI] [PubMed] [Google Scholar]
- Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5' terminus of HeLa cell messenger RNA. Cell. 1975;4(4):379–386. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- Wei LH, Song P, Wang Y, Lu Z, Tang Q, Yu Q, et al. The m6A reader ECT2 controls trichome morphology by affecting mRNA stability in Arabidopsis. Plant Cell. 2018;30(5):968–985. doi: 10.1105/tpc.17.00934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B, Mei Z, Zeng C, Liu S. MetaX: A flexible and comprehensive software for processing metabolomics data. BMC Bioinformatics. 2017;18(1):183. doi: 10.1186/s12859-017-1579-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Dubos C, Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015;20(3):176–185. doi: 10.1016/j.tplants.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Ma D, Constabel CP. The MYB182 protein down-regulates proanthocyanidin and anthocyanin biosynthesis in poplar by repressing both structural and regulatory flavonoid genes. Plant Physiol. 2015;167(3):693–710. doi: 10.1104/pp.114.253674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakaryan H, Arabyan E, Oo A, Zandi K. Flavonoids: Promising natural compounds against viral infections. Arch Virol. 2017;162(9):2539–2551. doi: 10.1007/s00705-017-3417-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Zhang YC, Liao JY, Yu Y, Zhou YF, Feng YZ, et al. The subunit of RNA N6-methyladenosine methyltransferase OsFIP regulates early degeneration of microspores in rice. PLoS Genet. 2019;15(5):e1008120. doi: 10.1371/journal.pgen.1008120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Zhuang X, Dong Z, Xu K, Chen X, Liu F, et al. The dynamics of N6-methyladenine RNA modification in interactions between rice and plant viruses. Genome Biol. 2021;22(1):189. doi: 10.1186/s13059-021-02410-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TY, Wang ZQ, Hu HC, Chen ZQ, Liu P, Gao SQ, et al. Transcriptome-Wide N6-Methyladenosine (m6A) profiling of susceptible and resistant wheat varieties reveals the involvement of variety-specific m6A modification involved in virus-host interaction pathways. Front Microbiol. 2021;12:656302. doi: 10.3389/fmicb.2021.656302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao BS, Wang X, Beadell AV, Lu Z, Shi H, Kuuspalu A, et al. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature. 2017;542(7642):475–478. doi: 10.1038/nature21355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Tian S, Qin G. RNA methylomes reveal the m6A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening. Genome Biol. 2019;20(1):156. doi: 10.1186/s13059-019-1771-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its supplementary information files).