Abstract
Plant hormones are signaling compounds regulating critical aspects of growth, development, and environmental stress responses. Abiotic stresses, such as drought, salinity, heat, cold and flooding have profound effects on plants. Adaptations to such stresses require sophisticated sensing, signaling and response mechanisms for stress tolerance. Here we review recent advances in hormonal control of abiotic stress responses in plants and highlight points of hormonal crosstalk during abiotic stress signaling. Specific topics are addressed, including osmotic stress sensory and signaling mechanisms, hormonal control of gene regulation and plant development during stress, hormone-regulated submergence tolerance and stomatal movements. We further explore how innovative imaging approaches are providing insights into single cell and tissue hormone dynamics. Recent advances open new opportunities for agricultural applications.
Introduction
Plants are a major source for food, fuel and fiber, and important contributors to the ecological diversity and sustainability of our planet. To optimize their growth and productivity under changing environmental conditions, that are intensified due to climate change, plants have developed sophisticated mechanisms to sense and respond to external stresses1,2. Among them, abiotic stresses appear in various forms, associated either with weather conditions, i.e. rainfall, temperature and irradiation from the sun, or to the quality of the soil in which plants grow, i.e. the content of water, nutrients and soil contaminants1. In particular, changes in water availability and temperature, e.g. heat stress, have been associated with climate change2.
To develop concepts and approaches for protecting plants from the negative effects of abiotic stresses, and to secure the future demands for plant products, we need to understand the mechanisms of plant stress responses at the molecular level. Plant hormones, i.e. abscisic acid (ABA), auxin, brassinosteroid (BR), cytokinin (CK), ethylene (ET), gibberellic acid (GA), jasmonic acid (JA), salicylic acid (SA) and strigolactone (SL) are well known plant growth regulators that mediate adaptations to environmental conditions. For an overview on their functions and respective signaling mechanisms, please refer to a recent review3 and references therein. The diverse roles of phytohormones in abiotic stress responses are reviewed here, and many of these roles are listed in Table 1.
Table 1 |.
Examples of roles of phytohormones in abiotic stress responses.
| Phytohormone | Ref. |
|---|---|
| Abscisic acid (ABA) | |
| Drought stress in roots induces ABA biosynthesis in shoots via hydraulic signals, and CLE25 peptide-mediated induction of NCED3 expression. | 26,27,32 |
| Osmotic- and salt stress induce the activation of SnRK2-type protein kinases, and SnRK2 activation is mediated by Subgroup B Raf-like kinases. | 34,36,39,42,44 |
| Root hydrotropism requires ABA signaling in the cortex of the elongation zone. | 101 |
| Salt stress inhibits lateral root formation, which depends on ABA synthesis and endodermal ABA signaling. ABA inhibits lateral root formation through interfering with auxin signaling. | 104,116 |
| Salt stress, K+ and SO42− deficiency induce endodermal suberization in an ABA-dependent manner. | 207 |
| Cold stress responses are modulated via SnRK2.6/OST1 phosphorylation of the transcription factor ICE1. | 208 |
| Heat stress tolerance is reduced in mutants deficient in ABA signaling or biosynthesis. | 209 |
| Auxin (IAA) | |
| Salt stress induces halotropism, the preferential growth away from areas of high salinity, which is mediated by auxin redistribution to induce root bending. | 105,106 |
| Hydropatterning, the preferential formation of lateral roots near water, is initiated by auxin signaling, and depends on the auxin response factor ARF7. | 114,115 |
| Drought stress induces the expression of IAA5 and IAA19, two transcriptional repressors of auxin responses. Additionally, iaa mutants have reduced survival during osmotic stress. | 76 |
| Heat stress induces auxin biosynthesis via PIF4, and the stabilization of auxin co-receptors. Auxin signaling via ARFs mediates high temperature-dependent hypocotyl elongation. | 210–212 |
| Brassinosteroid (BR) | |
| Drought stress responses interfere with BR signaling via BR and ABA crosstalk at the level of BES1 and RD26 mediated transcriptional regulation. | 71 |
| Cold acclimation and freezing tolerance involve BR signaling through its effect on COR and CBF gene expression. | 213,214 |
| During Thermomorphogenesis, PIF4 induces BR biosynthesis, whereas the BR activated transcription factor BZR1 functions in a feedforward loop downstream of auxin and PIF4 to further induce PIF4 expression. | 215,216 |
| Cytokinin (CK) | |
| Drought- and salt-stress induce the reduction of CK content and signaling, leading to an increased ABA sensitivity, likely via interaction of SnRK2s with type-A and type-B ARRs. | 74,181,217 |
| The osmotic stress-dependent hydrotropic response depends on the asymmetric distribution of CK signaling in the root tip, which is enhanced at the lower water potential side. | 202 |
| Ethylene (ET) | |
| Salt stress induces the production of ET and ET signaling. ET signaling promotes salt tolerance. | 121 |
| Flooding or submergence adaptation depends on ET production and the function of group VII Ethylene Response Factors (ERFs) in Arabidopsis and other related ERF genes in rice (Sub1A and SNORKEL), likely inducing GA biosynthesis. | 128,129,135 |
| Metal deficiency reduces endodermal suberization in an ET-dependent manner. | 207 |
| Gibberellic acid (GA) | |
| Under drought stress conditions GA signaling interferes with ABA signaling via DELLA protein interactions with the ABA-regulated TF ABF2. | 218 |
| Salt stress reduces the levels of bioactive GAs, likely via ABA signaling. della-quadruple mutants are hypersensitive to salt stress. | 121 |
| Cold stress responses are mediated via DELLA accumulation and interactions with GRF-type TFs. | 219 |
| Heat stress induces GA biosynthesis and the degradation of DELLAs in a COP1-dependent manner to regulate hypocotyl elongation. | 220 |
| Water submergence triggers GA production to induce internode elongation in rice. | 135–137 |
| Jasmonic acid (JA) | |
| Cold stress induces the production of JA. JAZ degradation in response to cold releases ICE1 and ICE2 from JAZ-mediated repression. | 221 |
| Heat stress promotes the accumulation of the JA receptor COI1 to enhance downstream JA responses. | 185 |
| Strigolactone (SL) | |
| Drought- and salt stress responses are positively modulated by SL via ABA-dependent and ABA-independent pathways. | 179,222 |
Abbreviations:
CLE25, CLAVATA3/ENHANCER OF SHOOT REGENERATION-RELATED 25
NCED3, NINE-CIS-EPOXYCAROTENOID DIOXIGENASE 3
SnRK2, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE
ARF, AUXIN RESPONSE FACTOR
ICE1, INDUCER OF CBF EXPRESSION 1
IAA5 and IAA9, INDOLE-3-ACETIC ACID INDUCIBLE 5 and 9
PIF4, PHYTOCHROME-INTERACTING FACTOR4
BRL3, BRI1-LIKE 3
BES1, BRI1-EMS-SUPPRESSOR 1
RD26, RESPONSIVE TO DESSICATION 26
COR, COLD REGULATED
CBF, C-REPEAT BINDING FACTOR
ARR, ARABIDOPSIS RESPONSE REGULATOR
ERF, ETHYLENE RESPONSE FACTOR
DELLA, plant-specific GRAS family proteins functioning as repressors of the GA signaling pathway
ABF, ABRE-BINDING FACTOR
GRF, GROWTH REGULATORY FACTOR
COP1, CONSTITUTIVE PHOTOMORPHOGENETIC 1
JAZ, JASMONATE-ZIM-DOMAIN PROTEIN
COI1, CORONATINE-INSENSITIVE 1
Here, we review how plant hormones and other signaling compounds mediate plant responses to abiotic stresses, including drought, osmotic stress and flooding. We further summarize the current view on how osmotic stress is sensed by plants, and how this leads to the activation of SnRK2-type protein kinases and interactions with plant hormone signaling modules. Then we elaborate on phytohormone-dependent gene regulatory mechanisms that mediate abiotic stress responses of plants. We also highlight how hormones regulate seed germination, how ABA and auxin coordinate root growth under stress, the effects of stress-dependent hormone responses on flowering time, the ethylene- and gibberellic acid-mediated regulation of plant responses to flooding, and the ABA and abiotic stress sensing mechanisms regulating stomatal aperture. Abiotic stresses further cause bud dormancy, leaf senescence, and organ abscission, which is reviewed elsewhere4. Finally, we review how biosensor-based hormone imaging techniques are contributing to the elucidation of hormone dynamics under abiotic stress.
Osmotic stress sensing and signaling
Water uptake from the soil and water movements within plants are driven by water potential gradients. Hypoosmotic stress, such as flooding, leads to cell swelling, whereas hyperosmotic stress, such as drought and salinity, leads to plant wilting. Plants have evolved osmotic stress adaptive mechanisms, including the regulation of cellular osmoticum concentrations, stomatal movements, and plant development through ABA-dependent and ABA-independent pathways1,5. Here, we summarize the current understanding of osmotic sensory and signaling mechanisms in plants.
Osmotic- and salt stress sensing.
Plants can sense the alteration of turgor, the mild change of solute concentrations in cells, and the mechanical effects on cellular structures caused by osmotic stress. Calcium signaling is suggested to play a key role in osmosensing because cytosolic-free calcium concentrations ([Ca2+]cyt) in plants rapidly and transiently increase within seconds of exposure to osmotic shock6,7. The roles of mechanosensitive ion channels in osmotic stress sensing have been investigated (FIG. 1a). MECHANOSENSITIVE CHANNELS OF SMALL CONDUCTANCE-LIKE (MSLs) are non-selective ion channels activated by membrane tension for osmoregulation during hypo-osmolality in organelles, hydration and germination in pollen, touch responses in roots, and cell swelling8–10. MID1-COMPLEMENTING ACTIVITY (MCA)-type Ca2+-permeable channels are activated by membrane tension and are suggested to mediate hypo-osmotic shock and touch sensing in roots11. They also function in mediating cold tolerance and cold-induced [Ca2+]cyt increases12. Another mechanosensitive ion channel PIEZO1 (PZO1) is required for mechanotransduction at root tips13. Interestingly, plant PIEZOs are localized in the vacuolar membrane to regulate [Ca2+]cyt oscillations and tip growth in Physcomitrium patens caulonemal cells and mediate vacuole tubulation in the tips of Arabidopsis pollen tubes14, suggesting a potential function under hypo-osmotic stress.
Figure 1 |. Osmotic stress and salinity sensing and signaling in plants.
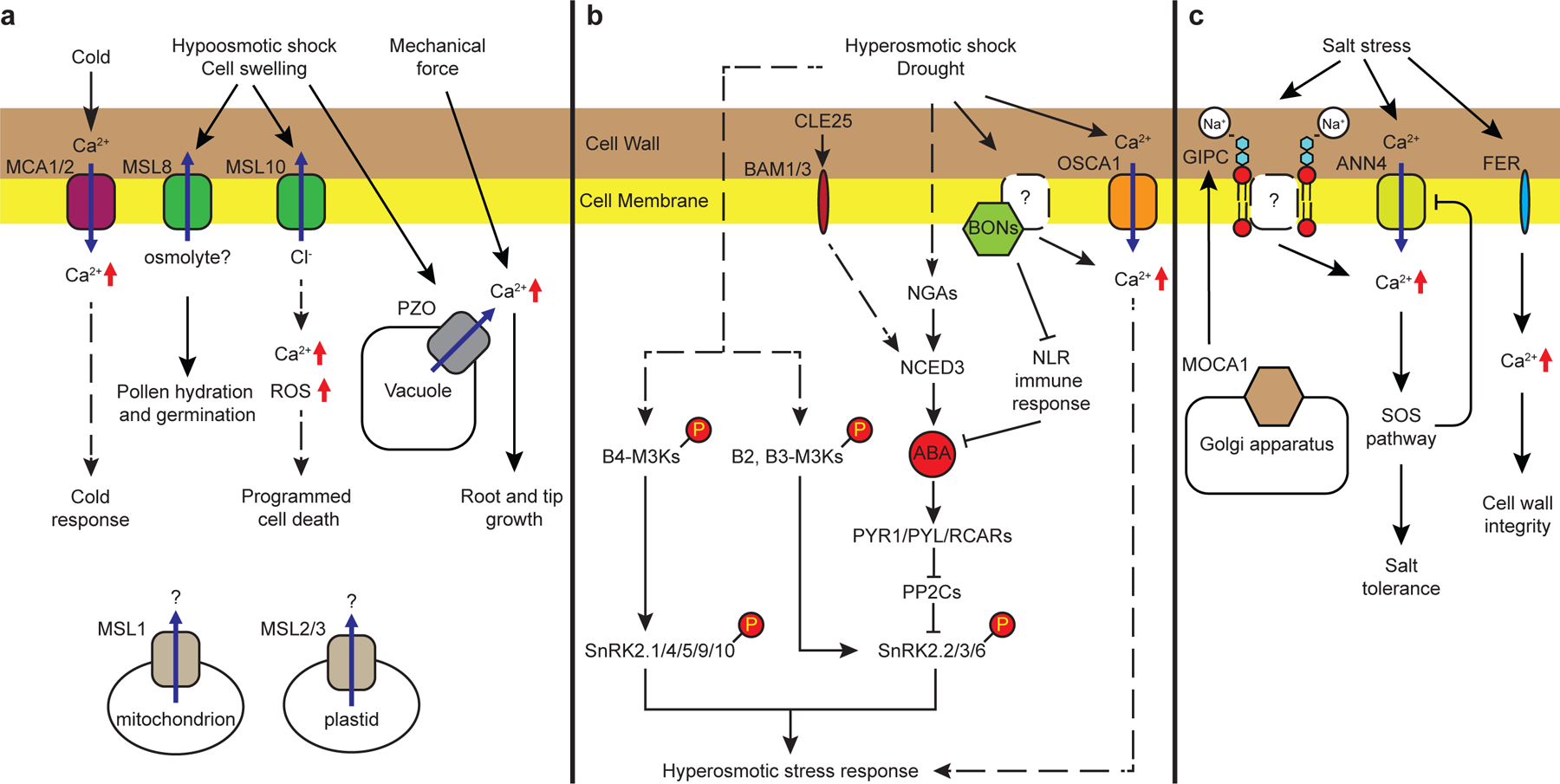
a | Mechanosensitive channels have been proposed to be involved in sensing the alterations of membrane tension caused by hypoosmotic stress and other abiotic stresses. MSL8 prevents bursting of pollen during hydration and germination8. MSL10 potentiates hypoosmotic/cell swelling-induced [Ca2+]cyt transient increases, ROS production, and programmed cell death10. MSL1 and MSL2/3 control mitochondrial and plastidial osmotic pressure223. MCA1 and MCA2 function in hypoosmotic and cold-induced [Ca2+]cyt transients and regulate cold acclimation responses12. Tonoplast-localized PIEZOs (PZO) are required for [Ca2+]cyt oscillations in tip-growing cells14, and mechanical-induced [Ca2+]cyt increases in the root tip to regulate root penetration into denser barriers13. b | Hyperosmotic stress-induced [Ca2+]cyt increases have been reported to function in early hyperosmotic-stress signaling. OSCA1 is an osmotic/mechanical-sensitive channel required for hyperosmotic-induced [Ca2+]cyt increases15. Ca2+-responsive phospholipid-binding BONZAI (BON) proteins regulate hyperosmotic-induced [Ca2+]cyt increases and suppress NLR immune signaling to trigger a hyperosmotic stress response18. Drought induces ABA biosynthesis NCED3 gene expression, leading to ABA accumulation. Root-derived CLE25 peptides activate NCED3 gene expression in the shoot in response to dehydration likely through receptor-like kinases BAM1 and BAM332. NGATHA (NGA) transcription factors are responsible for the drought-induced transcriptional activation of NCED330. Hyperosmotic stress activates Raf-like M3Ks via phosphorylation through an unknown osmotic stress sensor-mediated signal transduction mechanism. Members of the B2 and B3 subgroups of Raf-like M3Ks mediate both the rapid osmotic stress-induced and slower, post-ABA synthesis, activation of SnRK2.2/2.3/2.6, whereas the B4 subgroup of Raf-like M3Ks only activate osmotic stress-responsive SnRK2.1/2.4/2.5/2.9/2.1020,42–44. c | A Salt-induced [Ca2+]cyt increase triggers tolerance responses through the salt overly sensitive (SOS) pathway. Glycosyl inositol phosphorylceramide sphingolipids (GIPCs) synthesized by Inositol Phosphorylceramide Glucuronosyltransferase (MOCA1/IPUT1) are involved in Na+ sensing24. The Annexin 4 (ANN4)-medicated [Ca2+]cyt increase is feedback inhibited by the SOS pathway for fine-tuning salt tolerance25. FERONIA (FER) is required for maintaining cell wall integrity under salt stress22.
A potential osmosensor REDUCED HYPEROSMOLALITY-INDUCED [Ca2+]cyt INCREASE 1 (OSCA1) is involved in hyperosmotic stress-induced [Ca2+]cyt increases for osmotic stress tolerance15 (FIG. 1b). OSCA1 was initially characterized as a hyperosmolality-activated Ca2+-permeable cation channel15. Later studies reported that OSCA family proteins function as stretch-activated channels16,17. Ca2+-responsive phospholipid-binding BONZAI (BON) proteins were recently reported to mediate hyperosmotic stress tolerance by positively regulating osmotic stress-induced [Ca2+]cyt increases, ABA accumulation, and gene expression18. These membrane-associated Ca2+-responsive BON proteins may be involved in osmotic sensing and signaling by regulating the initial [Ca2+]cyt elevation together with plasma membrane Ca2+ transporters19 (FIG. 1b). Defects in plant growth and ABA accumulation under osmotic stress in bon mutants can be restored by crossing bon mutants with snc1–11 and pad4 mutant alleles that are impaired in nucleotide-binding domain and leucine-rich repeat (NLR) immune signaling18. Therefore, BONs may confer osmotic stress responses by suppressing NLR immune signaling. Although several osmotic stress-linked mechanisms have been characterized, further research is needed to dissect their differential functions. Notably, the activation of SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2 (SnRK2) kinases in response to hyperosmotic shock is not impaired in osca septuple and bon1 bon2 bon3 triple mutants18,20 indicating the need to identify additional osmotic stress sensing and signaling mechanisms.
Plants sense salinity and induce rapid [Ca2+]cyt transients6,21 to trigger salt tolerance responses through the Salt Overly Sensitive (SOS) pathway1 (FIG. 1c). Under salt stress, the receptor-like kinase FERONIA may sense cell wall defects caused by salinity, and elicits cell-specific [Ca2+]cyt transients for maintaining cell wall integrity22. FERONIA potentially also interferes with ABA signaling via interaction with the PROTEIN PHOSPHATASE TYPE 2C (PP2C) ABA INSENSITIVE 2 (ABI2)23. In Arabidopsis, the current hypothesis is that plasma membrane glycosyl inositol phosphorylceramide sphingolipids (GIPCs) function in salt sensing24. In this model, GIPC lipid formation is catalyzed by the protein MOCA1/IPUT1 (MONOCATION-INDUCED [Ca2+]i INCREASES 1/Inositol Phosphorylceramide Glucuronosyltransferase 1), and binding of Na+ ions to GIPC lipids activates Ca2+ influx channels24. Disruption of the ANNEXIN gene AtANN4, a putative Ca2+ permeable channel component, impairs salt stress-induced [Ca2+]cyt transients by ~40%, while Ca2+-activated SOS2-SCaBP8/CBL10 complexes negatively regulate AtANN4 by phosphorylation to fine-tune salt tolerance responses25.
Osmotic stress-induced ABA biosynthesis.
Endogenous ABA concentrations increase approximately 2.5 to 6 hours after exposure to water deficiency26–29. Stress-induced de novo ABA synthesis depends on the induction of the NCED3 gene that encodes a 9-cis-epoxycatoteinoid dioxygenase catalyzing the rate-limiting step for ABA biosynthesis30. Posttranslational processing of NCED3 in the chloroplast has also been reported to regulate ABA accumulation31. In response to water deficiency in roots, a hydraulic signal contributes to a rapid root-to-shoot water deficiency signal to trigger ABA biosynthesis in Arabidopsis leaves and stomatal closure26. In addition, a small peptide CLAVATA3/EMBRYO-SURROUNDING REGION-RELATED 25 (CLE25) is induced in the root vasculature during drought stress and moves to aerial tissues to induce NCED3 gene expression likely through BARELY ANY MERISTEM (BAM1 and BAM3) receptor-like kinases32 (FIG. 1b). At the transcriptional level, an Arabidopsis NAC transcription factor ATAF1 was suggested to regulate NCED3 expression to enhance ABA accumulation33. Moreover, the NGATHA (NGA) protein family, including four members in Arabidopsis, were identified as transcriptional activators regulating NCED3 expression through direct binding to a cis-acting element (CACTTG) in the 5’ UTR of the NCED3 promoter30.
An emerging role of Raf-like MAPKKKs.
An ABA-independent rapid osmotic stress signal transduction pathway and an ABA-dependent pathway converge at the level of SnRK2-type protein kinase activation. The Arabidopsis genome encodes ten SnRK2 genes. Except for SnRK2.9, the other SnRK2s are activated by osmotic stress, whereas only SnRK2.2, SnRK2.3, and SnRK2.6/OST1 are clearly activated by ABA34–37. ABA-dependent SnRK2 activation through the PYRABACTIN RESISTANCE 1 (PYR1)/PYR1-LIKE (PYL)/REGULATORY COMPONENT OF ABA RECEPTOR (RCAR) and PP2C ABA sensing module has been well described38. Since SnRK2 activation by osmotic stress is not impaired in ABA-insensitive dominant negative PP2C Arabidopsis mutants34,39, osmotic stress employs other signaling mechanisms to activate SnRK2 kinases. This ABA-independent pathway is still largely unknown including the identity of the contributing osmotic stress sensors.
SnRK2s have an auto-phosphorylation activity which enhances the kinase activity itself40. Previous in vitro studies identified a key phosphorylation site at Ser-175 within the activation-loop of the SnRK2.6/OST1 kinase domain40. In vivo analyses revealed that both ABA and osmotic stress induce phosphorylation at Ser-171 and Ser-175 residues41.
Recent studies identified Raf-like MAP kinase kinase kinases (Raf-like M3Ks) to be required for phosphorylation-dependent SnRK2 activation via osmotic stress and ABA signaling 20,42–44 (FIG. 1b). SnRK2.6/OST1 was found to be impaired in auto-activation after dephosphorylation by PP2Cs42. The re-activation of SnRK2.6/OST1 requires the initial trans-phosphorylation at Ser-171 or Ser-175 by members of the Arabidopsis B2 and B3 subgroup of Raf-like M3Ks42,44. Moreover, the raf-like m3kδ1/δ6/δ7 triple knock-out mutant exhibited not only a reduced SnRK2 kinase activation by ABA but also impairment in SnRK2 activation by osmotic stress42. Important functions of B4 subgroup Raf-like M3Ks in osmotic stress-, but not in ABA-induced rapid SnRK2 activation, were identified20,43 (FIG. 1b).
Roles for Raf-like M3Ks in ABA responses were initially identified in genetic ABA/stress response screens in Arabidopsis and in moss45,46. In the moss Physcomitrium patens, the ABA and abiotic stress-responsive Raf-like kinase (ARK/ANR), a single ancestral gene similar to the Arabidopsis B3 subgroup, has a role in osmotic stress- and ABA signaling46–48. A recent study reported that this ARK/ANR kinase is activated by ABA47. The Arabidopsis B3 Raf-like M3K subgroup contains another well-studied gene, CONSTITUTIVE TRIPLE RESPONSE 1 (CTR1), functioning as a negative regulator of ethylene signaling. Ethylene deactivates CTR1 through ethylene receptors, a family of histidine kinases including ETHYLENE RESPONSE 1 (ETR1), which directly binds to CTR149. Interestingly, Physcomitrium patens ARK/ANR (also known as PpCTR1L) mediates not only ABA signaling in moss, but also ethylene responses50, which might suggest a role of Raf-like M3Ks as a signaling “hub”. How osmotic stress sensors are linked to Raf-like M3Ks, SnRK2 activation and downstream components remains to be determined.
Gene regulation under abiotic stress
Changes in gene expression mediate many of the effects of phytohormones. Early genomic technologies revealed that abiotic stress-linked ABA increases change the mRNA levels of thousands of genes (e.g. REF.51). This, together with the discovery that many classic ABA-insensitive mutations were mapped to transcriptional regulators suggested a prominent role for gene regulation in abiotic stress resistance52.
ABA-mediated transcriptional regulation and hormone crosstalk.
Early studies discovered a conserved cis-acting regulatory element known as the ABA-RESPONSIVE ELEMENT (ABRE) in the promoters of drought-induced genes53. ABREs are recognized by bZIP-type transcription factors (TFs), including a family of four ABRE-binding proteins/ABRE-binding factors (AREBs/ABFs)54, and the closely related ABA-INSENSITIVE 5 (ABI5)52,55,56 (FIG. 2). During ABA signaling, ABA-dependent SnRK2-type protein kinases directly phosphorylate and activate AREBs/ABFs and ABI557–59. In Arabidopsis, the four partially redundant AREBs/ABFs are responsible for most of the transcriptional responses to ABA during vegetative growth60, whereas ABI5 is more important during seed germination61 (see below). Many of the AREB/ABF targets are other TF genes, implying that a multilevel transcription factor hierarchy controls ABA-dependent transcriptome remodeling62–65. A seminal study using ChIP-Seq to profile the genome-wide binding sites of 21 TFs during the ABA response revealed that many binding events are dynamic, and that multiple TFs can target the same gene64. Crucially, the ABA-induced binding of some TFs was positively correlated with the presence of adjacent ABRE sites, suggesting that some TFs may act cooperatively with AREBs/ABFs. Indeed, several NAC family transcription factors are required for ABA-dependent transcription events including ANAC096, which interacts with ABF2 to activate RD29A transcription62,66,67.
Figure 2 |. Hormonal crosstalk through transcriptional regulation.
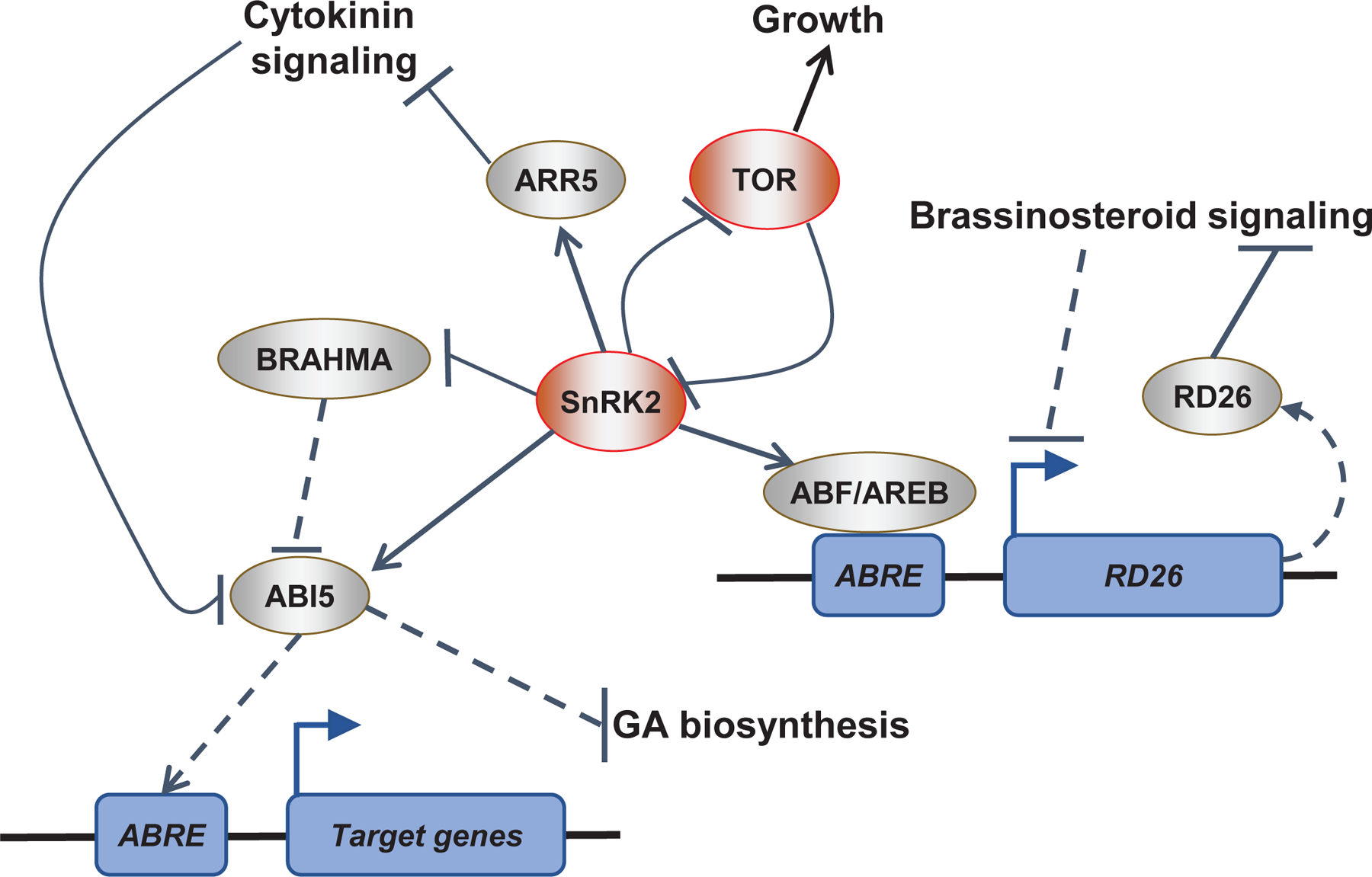
Plant hormones control abiotic stress response by altering transcriptional programs. The ABA signaling system intersects with many other hormone pathways during transcription. This figure summarizes the relationships between ABA signaling and key transcriptional regulators during hormonal signaling. During ABA-responses, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2 (SnRK2) protein kinases phosphorylate ABA-RESPONSIVE ELEMENT (ABRE)-binding proteins/ABRE-binding factors (AREBs/ABFs) and ABA-INSENSITIVE 5 (ABI5) transcription factors. AREBs/ABFs and ABI5 activate target genes with ABREs in their promotors to drive ABA responses. For instance, during drought stress AREBs/ABFs activate transcription of RESPONSE TO DESSICATION 26 (RD26), a transcription factor that can repress brassinosteroid signaling. Additionally, in dormant seeds, ABI5 target genes repress gibberellic acid (GA) biosynthesis and thereby block germination. Cytokinin signaling can repress ABA responses possibly by triggering the degradation of ABI5. ABA-activated SnRK2-type protein kinases promote transcriptional ABA responses by phosphorylating type-A ARABIDOPSIS RESPONSE REGULATOR5 (ARR5), a negative regulator of the cytokinin pathway. Finally, in unstressed conditions the protein kinase TARGET OF RAPAMYCIN (TOR) promotes plant growth by inhibiting SnRK2-type protein kinase-mediated ABA responses through phosphorylation of ABA receptors. Conversely, during stress SnRK2-type protein kinases phosphorylate and inhibit the TOR regulatory protein RAPTOR1B leading to growth repression. Note that not all mechanisms shown here are necessarily present at the same time or in the same cell/tissue.
In the absence of abiotic stress, repression of ABA signaling promotes optimal growth. For instance, under non-stress conditions mRNA levels of ABI5 are low, and ABI5 transcription is increased upon exposure to ABA or osmotic stress55. This repression of ABI5 requires the SWI2/SNF2 chromatin remodeling ATPase BRAHMA68. In the absence of ABA, BRAHMA inhibits the transcription of ABI5 by promoting nucleosome occupancy at the transcription start site of the ABI5 gene. Interestingly, phosphorylation of BRAHMA by SnRK2.2/2.3/2.6 appears to inhibit its action and in turn, this inhibitory phosphate is removed by group A PP2Cs69 (FIG. 2).
Different hormone pathways interact to control numerous aspects of plant life in the absence of abiotic stress and we direct readers to a review on this topic for a concentrated discussion70. Here we focus on how transcriptional regulation enables hormone crosstalk during drought stress responses. For instance, the ABA-induced TF RESPONSE TO DESSICATION 26 (RD26) represses a subset of brassinosteroid-induced genes62,71. Furthermore, brassinosteroid-activated TFs repress the expression of RD26, suggesting that antagonistic crosstalk between ABA and brassinosteroid signaling contributes to drought stress responses72 (FIG. 2). Intriguingly, overexpression of the vascular-enriched brassinosteroid receptor BRL3 causes constitutive expression of some drought-induced genes, including RD26, and promotes drought resistance73. The cytokinin and ABA pathways also converge on the level of transcriptional control. SnRK2-mediated phosphorylation of the type-A ARABIDOPSIS RESPONSE REGULATOR ARR5, a negative regulator of cytokinin signaling, promotes its protein stability thereby downregulating cytokinin responses during drought stress74. Oppositely, cytokinin can trigger the degradation of ABI5 to promote seed germination75 (FIG. 2). Furthermore, dehydration induces the expression of IAA5 and IAA19, two transcriptional repressors of the auxin signaling pathway, indicating that auxin responses are repressed during drought stress76.
Post-transcriptional abiotic stress responses.
Post-transcriptional processes expand gene regulatory possibilities beyond transcriptional control, and recent research has uncovered the contributions of such mechanisms in shaping ABA responses. The discovery of a mutant in the mRNA cap-binding protein abh1 exhibiting ABA-hypersensitivity established an early link between mRNA processing and ABA signaling77. Alternative mRNA splicing, a process producing multiple distinct mRNA isoforms from a single gene, is modulated by abiotic stress and regulates ABA responses78–82. HAB1 for instance, a group A PP2C gene, encodes multiple splice isoforms, of which HAB1.2 retains an intron leading to a non-functional protein and ABA hypersensitivity79,80.
Recently mRNA decay has emerged as an additional mechanism contributing to abiotic stress responses. Degradation of mRNA molecules is mediated by mRNA decapping, the removal of the 5’ methyl-guanosine cap83. During osmotic stress, ABA-independent subclass I SnRK2-type protein kinases phosphorylate the decapping activator VARICOSE (VCS) leading to the destabilization of some transcripts84. The 5’ end of mRNAs can be alternatively modified by the addition of a nicotinamide adenine dinucleotide (NAD+). In plants, the NAD+ cap occurs on many transcripts and is thought to down-regulate gene expression by promoting the degradation of marked mRNAs85,86. A recent study demonstrated that the NAD+-capped transcriptome undergoes extensive changes in response to ABA and that many ABA-induced transcripts lose their NAD+ caps following ABA treatment possibly promoting their stability87.
Growth regulation under abiotic stress
Phytohormones regulate many aspects of plant growth and development. Recent discoveries have begun to illuminate how they control different strategies of plant growth and development in response to abiotic stress.
TOR interaction with abiotic stress responses.
The protein kinase TARGET OF RAPAMYCIN (TOR) is a central developmental and metabolic regulator in plants88. A reciprocal regulation between ABA and TOR pathways has been proposed to coordinate plant growth and abiotic stress responses89. TOR phosphorylates PYL/RCAR ABA receptors to inhibit ABA signaling and promote growth under non-stress conditions, whereas ABA-activated SnRK2 kinases phosphorylate the REGULATORY-ASSOCIATED PROTEIN OF TOR 1B (RAPTOR1B) to inhibit TOR kinase activity and repress growth in response to abiotic stress conditions (FIG. 2).
Gibberellic acid, ABA and the decision to germinate.
The regulation of seed germination promotes seedling survival by coordinating embryo development and emergence with environmental conditions. The balance of two competing hormone signaling pathways, gibberellic acid and ABA, dominates the decision to germinate52. During seed maturation a network of transcription factors including the ABA-regulated TFs ABI3, ABI4, and ABI5 induce genes required for seed desiccation and ABA biosynthesis and repress GA biosynthesis genes. Environmental signals, such as cold and light, that trigger seeds to break dormancy do so by flipping the balance towards GA52. This antagonistic relationship between ABA and GA arises at multiple points in their respective pathways (FIG. 3a).
Figure 3 |. Hormonal control of growth and development during abiotic stress.
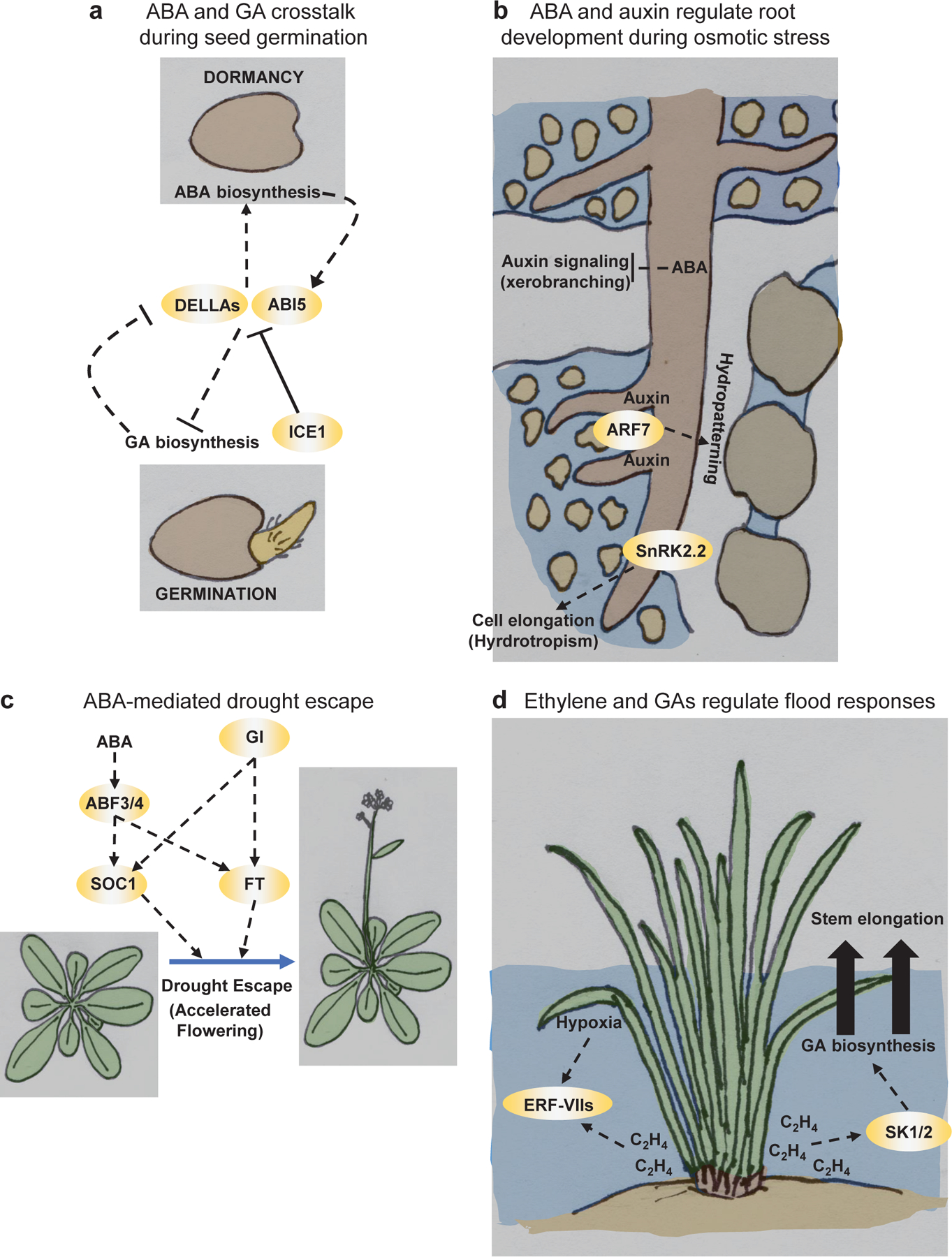
a | ABA and GA signaling pathways antagonistically control germination. In dormant seeds, DELLA proteins and ABI5 promote ABA signaling by stimulating expression of ABA biosynthesis genes and the ABI5 gene and inhibit GA responses by repressing GA biosynthesis. INDUCER OF CBF EXPRESSION1 (ICE1) antagonizes DELLA and ABI5 activity to promote germination. During germination, GA levels increase, and GA triggers the destruction of DELLA proteins leading to decreased ABA signaling. b | Water is unevenly distributed in soil and large air pockets form between soil particles. Primary roots display hydrotropism or biased growth towards areas of higher water. This process depends on SnRK2.2 protein kinase activity in cortex cells of the elongation zone. When roots enter air spaces lateral root formation is repressed (xerobranching), a process that depends on the ABA inhibition of auxin signaling. Roots growing in areas where water is asymmetrically distributed display a growth program known as hydropatterning, where lateral roots preferentially form on the water contacting side. In hydropatterning, the auxin response factor ARF7 stimulates preferential lateral root initiation. c | During prolonged drought, plants will accelerate flowering to reproduce in a process called drought escape. Under drought stress, the ABA-activated transcription factors ABF3/4 and the floral regulator GIGANTEA (GI) stimulate expression of the flowering inducers SUPRESSOR OF OVEREXPRESSION OF CO1 (SOC1) and FLOWERING LOCUS T (FT) to advance flowering. d | Submerged plant tissues experience hypoxia and elevated ethylene gas (C2H4). These cues activate transcription factors known as group VII ETHYLENE RESPONSE FACTORs (ERF-VIIs). ERF-VIIs initiate a conserved hypoxia-induced transcriptional program. In deepwater rice varieties, elevated ethylene activates the ERFs SNORKEL1 and 2 (SK1/2) which induce GA biosynthesis. GA signaling promotes a flood escape strategy where stems elongate to emerge into the air.
DELLA proteins are members of the plant-specific GRAS (GIBBERELLIN-INSENSITIVE, REPRESSOR of ga1–3, SCARECROW) family of transcriptional regulators that lack DNA-binding activity and function by interacting with other TFs90. DELLAs inhibit GA responses, and GA signaling inactivates DELLAs in part by triggering their proteasomal degradation90,91. DELLAs interact with ABI3 and ABI5, and together these protein complexes stimulate the transcription of SOMNUS, a key dormancy promoting factor that activates ABA biosynthesis genes and represses GA biosynthesis genes92. Interestingly, the action of the DELLA-ABI5 complex is inhibited by the bHLH TF INDUCER OF CBF EXPRESSION1 (ICE1)93. Binding of ICE1 blocks the DNA binding activity of ABI5, and this interaction is stimulated by GA treatment, possibly due to the degradation of DELLAs providing a possible mechanism through which prior exposure to cold temperatures may promote germination.
The control of ABI5 expression appears to be a major regulatory point for multiple environmental signals during germination. The light signaling component ELONGATED HYPCOTYL 5 (HY5) directly activates ABI5 transcription in response to light94. The DELLA protein RGL2 further promotes ABA signaling by enhancing the transcription of ABI5. GA production during germination initiation could then reduce ABI5 expression through RGL2 degradation95. High salinity inhibits seed germination, and two different transcription factors, AGL21 and RSM1, were reported recently to enhance ABI5 expression during exposure to NaCl96,97.
Auxin, ABA and root growth under stress.
Root development is shaped by environmental conditions and this topic has been the subject of multiple excellent reviews (e.g. REF.98). Here we focus on the mechanisms by which auxin and ABA control the architecture of the root system in response to water and salinity stresses (FIG. 3b).
While high concentrations of exogenous ABA inhibit root growth, lower (nM range) concentrations stimulate primary root growth99. Water distribution in soil is uneven, and plants partly address this situation through hydrotropism, the directional growth of roots towards water. Hydrotropism is impaired in ABA deficient mutants and ABA accumulates in root tissues during water stress suggesting an important role for ABA signaling100. For hydrotropism the SnRK2.2 kinase is required specifically in cortical cells of the root elongation zone where it promotes the cell elongation necessary for differential growth101 (FIG. 3b). Low concentrations of ABA (100 nM) stimulate primary root growth by abating PP2C-mediated inhibition of apoplastic H+ efflux through the AUTOINHIBITED H+-ATPase 2 (AHA2)102. This mechanism provides a contribution to the hydrotropic response102. Two recent studies have implicated brassinosteroid signaling in the hydrotropic response as well, although the mechanism is currently unclear73,103. By contrast, in high saline environments, lateral roots enter a prolonged growth arrest which requires endodermal ABA signaling104. Roots also exhibit preferential growth away from areas of high-salinity – a phenomenon called halotropism105. Salt treatment induces internalization of the auxin transporter PIN-FORMED 2 (PIN2) and when roots encounter a longitudinal salinity gradient, auxin accumulates on the side of the root furthest away from the salt source which then leads to root bending105,106. Interestingly, hydrotropism does not appear to act through auxin redistribution suggesting that halotropism is a distinct process107,108.
ABA also regulates root tissue patterning during water stress. Endodermal ABA signaling stimulates xylem differentiation by inducing the expression of the microRNAs miR165/166, two key regulators of vascular development109,110. ABA functions within xylem cells as well, where it activates expression of several VASCULAR-RELATED NAC DOMAIN (VND) TFs which promote xylem differentiation111.
Research on two related water-dependent root-branching strategies, hydropatterning and xerobranching, has uncovered requirements for auxin and ABA signaling98. Lateral roots initiate from pericycle cells within the primary root and their initiation is timed by an auxin-regulated transcriptional network112,113. The position of these initiation events was shown to respond to water availability in a process termed hydropatterning114. In hydropatterning, differences in water content across the circumference of primary roots lead to preferential lateral root initiation where water is available. Hydropatterning was correlated with auxin biosynthesis and signaling on the side of the root in contact with water114. A recent study demonstrated that hydropatterning requires the auxin response factor ARF7115 (FIG. 3b). Removal of seedlings from agar plates and their exposure to air triggered the post-translational modification of ARF7 with a SUMO protein and SUMOylated-ARF7 had reduced DNA-binding activity. Roots can encounter large air spaces in soil, and in these regions lateral root formation is repressed. This repression of branching along the entire root circumference has been termed xerobranching. A recent study implicated ABA signaling in the xerobranching response116. The roots of barley plants were found to accumulate ABA following a transient water deficit. Short-term ABA treatment of aeroponically grown maize and barley roots led to a zone of lateral root repression showing that ABA can mimic a xerobranching response. Furthermore, ABA treatment disrupted auxin signaling in roots suggesting a possible mechanism for lateral root repression116 (FIG. 3b).
Gibberellin, ABA and ethylene regulate flowering during abiotic stress.
A core genetic network regulates flowering time in plants, and this network receives inputs from endogenous, environmental, and seasonal cues117. Here we explore how hormone signaling intersects with core flowering regulators to mediate the effects of abiotic stress on flowering time.
During periods of prolonged drought many species will accelerate the flowering transition to reproduce before death and this response is known as drought escape118. The exact role of ABA during the flowering transition is currently unclear, and puzzlingly snrk2.2/2.3/2.6 triple mutants are early flowering119 while ABA-deficient mutants and areb1/areb2/abf3/abf1 quadruple mutants are late flowering60,118. Here we focus on the case of drought-accelerated flowering, where emerging evidence suggests a positive role for ABA signaling. Under long-day conditions flowering time is delayed in ABA biosynthesis mutants and advanced in an ABA hypersensitive pp2c triple mutant118. Crucially, drought stress magnifies this delay, suggesting that ABA is required to promote drought escape118. This positive role of ABA in drought-induced flowering requires the core photoperiod dependent flowering regulator GIGANTEA (GI)118,120. Additionally, the drought escape response is abolished in abf3/abf4 transcription factor double mutants and ABF3/ABF4 can directly induce the transcription of SUPPRESSOR OF CONSTANS1 (SOC1), another key flowering gene65 (FIG. 3c).
In contrast to drought, salt stress causes an ethylene dependent delay in flowering time in Arabidopsis121,122. Salt stress leads to ethylene accumulation by inducing the expression of ethylene biosynthesis genes121. Although the underlying mechanism is unclear, ethylene interferes with GA signaling leading to the accumulation of DELLA proteins. DELLAs can then delay flowering by inhibiting the flowering stimulating TF CONSTANS123. In addition, salt stress also represses flowering by inducing the degradation of GIGANTEA124.
Ethylene and gibberellin control flooding responses.
Floods are a major cause for crop loss in agriculture and a clear environmental challenge for some plants in natural ecosystems125. Plants possess an array of developmental and physiological strategies to adapt to flooding with different strategies exhibited in different species. Here we discuss advances related to hormone signaling during submergence and provide focused coverage of recent work on the hormonal control of a flood-escape strategy in rice. We further direct readers to recent reviews on the broader topic of flooding responses126,127.
The submergence of plant tissues impedes cellular access to O2 and CO2 which can severely disrupt metabolism. Additionally, restricted gas diffusion underwater leads to an accumulation of ethylene within flooded plant tissues125. Prolonged flooding can cause hypoxia which activates a conserved gene expression program that supports plant survival in limiting O2. In Arabidopsis, the transcription of these hypoxia-responsive genes requires five transcription factors known as group VII ETHYLENE RESPONSE FACTORs (ERF-VIIs)128. Additionally, Submergence1, a major QTL associated with improved flood tolerance in rice, contains a cluster of three related ERF genes129. Both hypoxia and high concentrations of ethylene enhance the protein stability of ERF-VIIs leading to target gene transcription130–133 (FIG. 3d). ERF-VII TFs bind to conserved cis-regulatory elements and the chromatin accessibility at these regulatory elements increases in response to flooding128,134. Interestingly, accessible ERF-VII binding sites were more prevalent in the flood-adapted rice genome than in the dryland-adapted tomato Solanum pennellii134.
Some flood adapted species display an escape strategy where underwater shoots and leaves elongate to emerge into the air125. Research on a flooding-tolerant rice variety known as deepwater-rice has begun to reveal how ethylene and gibberellin signaling control this underwater growth response. GA stimulates stem elongation by promoting internode growth, and this relationship has been exploited during plant domestication. In rice, ethylene accumulates in submerged stem and leaf tissues and this elevated ethylene concentration induces the expression of two ERFs known as SNORKEL1 and SNORKEL2, two major QTLs associated with deepwater internode elongation135. SNORKEL1 and SNORKEL2 may stimulate stem elongation by inducing GA biosynthesis (FIG. 3d). More recently, an additional locus associated with internode elongation was mapped to SEMIDWARF1 (SD1), a key GA biosynthesis gene136. In contrast to the more common semidwarf rice variety which carries a null allele of SD1, deepwater-rice plants induce SD1 expression in submerged tissues. The resulting increased GA levels act together with an additional locus called ACCELERATOR OF INTERNODE ELONGATION1 (ACE1) to promote cell division in the intercalary meristem137.
Interestingly, a recent study reported that compacted soil leads to ethylene accumulation in roots which subsequently inhibits further growth, possibly allowing plants to avoid regions with poor soil aeration138. This suggests that elevated ethylene concentration may be a common and early cue for air deficiency stress that plants use to adapt their growth.
Regulation of stomatal movements
Stomatal pores formed by guard cells in the leaf epidermis allow the uptake of CO2 for photosynthesis in exchange for water. To optimize plant water-use-efficiency, guard cells sense and respond to several abiotic factors, including light, CO2, and drought. ABA is a central regulator of guard cell physiology (FIG. 4a), and here we discuss the crosstalk with abiotic factors and other hormones.
Figure 4 |. Guard cell signal transduction and stomatal responses to environmental stimuli.
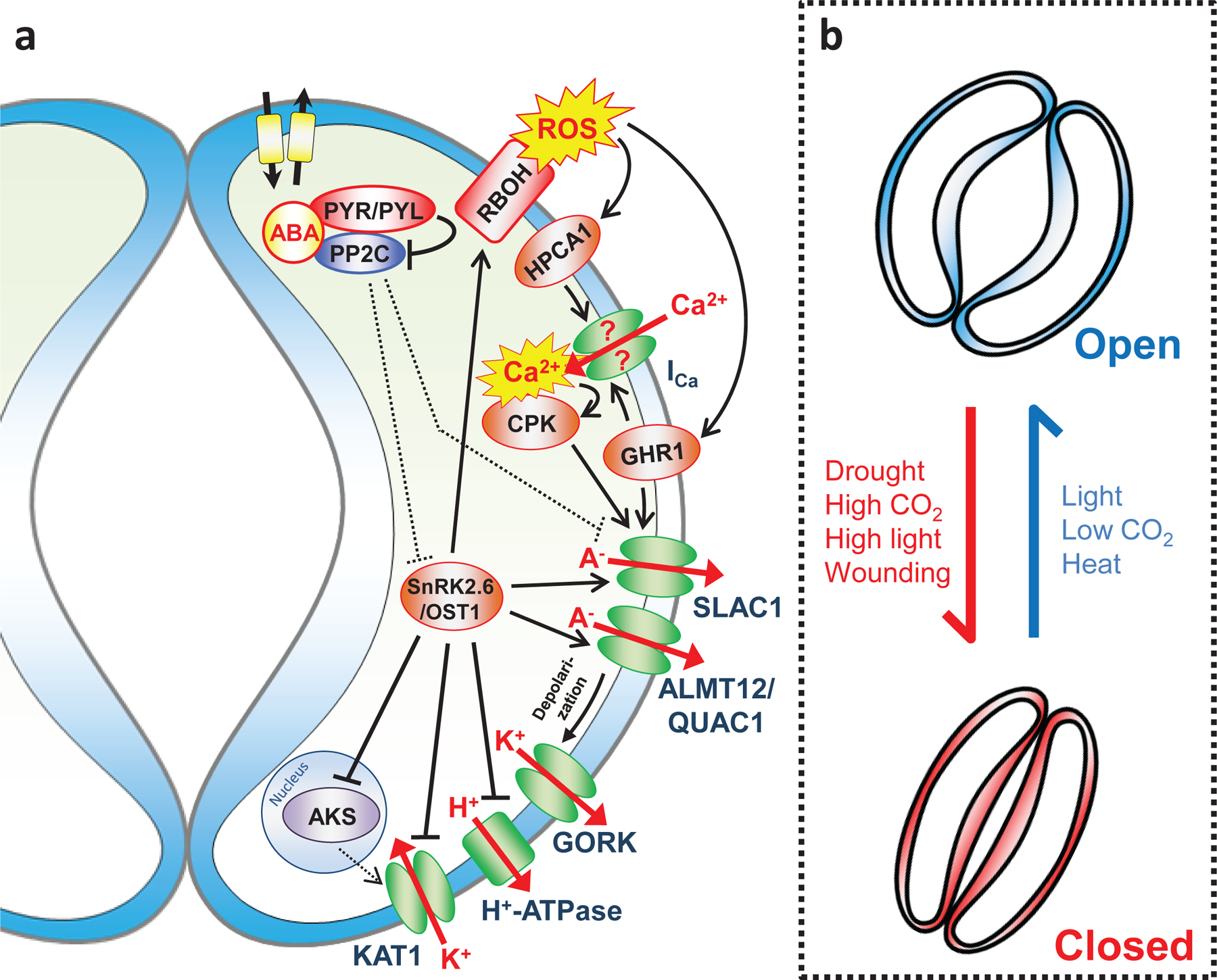
a | Schematic model of abscisic acid (ABA) signal transduction in guard cells. ABA transporters mediate ABA import or export from guard cells. In the presence of ABA, the key regulator SNF1-RELATED PROTEIN KINASE 2.6/OPEN STOMATA 1 (SnRK2.6/OST1) phosphorylates and activates SLOW ANION CHANNEL-ASSOCIATED 1 (SLAC1), ALUMINUM-ACTIVATED MALATE TRANSPORTER 12/QUICK-ACTIVATING ANION CHANNEL 1 (ALMT12/QUAC1), and RESPIRATORY BURST OXIDASE HOMOLOGs (RBOHs). Activation of the S-type anion channel SLAC1 and the R-type anion channel ALMT12/QUAC1 leads to long-term plasma membrane depolarization, which causes K+ efflux through the voltage-dependent K+out channel GUARD CELL OUTWARD RECTIFYING K+ CHANNEL (GORK). Activated RBOH NADPH oxidases produce ROS that mediate HYDROGEN-PEROXIDE-INDUCED Ca2+ INCREASES 1 (HPCA1) sensor-dependent activation of Ca2+-permeable ICa channels, resulting in the elevation of the cytosolic Ca2+ concentration ([Ca2+]cyt). Elevated [Ca2+]cyt activates Ca2+-sensor proteins including Ca2+-DEPENDENT PROTEIN KINASEs (CPKs) that phosphorylate and activate SLAC1. The (pseudo-)kinase GUARD CELL HYDROGEN PEROXIDE-RESISTANT 1 (GHR1) mediates the activation of ICa and SLAC1 channels through an unknown mechanism, possibly as scaffolding protein. SnRK2.6/OST1 inhibits the K+in channel K+ CHANNEL IN ARABIDOPSIS THALIANA 1 (KAT1), that mediates K+ uptake, by direct phosphorylation. In addition, SnRK2.6/OST1 causes a long-term decrease of KAT1 expression by inhibition of ABA-RESPONSIVE KINASE SUBSTRATE (AKS) transcription factors. ABA also inhibits H+-ATPase activity through SnRK2.6/OST1, but the detailed mechanism is unknown. Dashed lines indicate steps that are inhibited in the presence of ABA. Note that only guard cell ABA signaling regulating ion transport across the plasma membrane is depicted in this figure. b | In addition to drought stress, several other environmental stimuli can be perceived by guard cells and affect stomatal aperture though sophisticated signaling crosstalk and integration.
Stomatal response to drought.
Drought stress has been reported to trigger ABA synthesis in vascular tissues and guard cells139,140. ABA signaling in guard cells regulates plasma membrane ion channels to trigger long-term efflux of anions and K+, resulting in the reduction of guard cell turgor and stomatal closure. Anion release from guard cells and subsequent plasma membrane depolarization is mediated by slow-type (S-type) and rapid-type (R-type) anion channels141. A major S-type anion channel in Arabidopsis guard cells is encoded by SLOW ANION CHANNEL-ASSOCIATED 1 (SLAC1)142,143. ALUMINUM-ACTIVATED MALATE TRANSPORTER 12/QUICK-ACTIVATING ANION CHANNEL 1 (ALMT12/QUAC1) contributes to 40 percent of R-type anion currents144. The anion channel-triggered depolarization in turn induces K+ efflux through the voltage-dependent outward-rectifying K+ (K+out) channel145 GUARD CELL OUTWARD RECTIFYING K+ CHANNEL (GORK)146. The protein kinase SnRK2.6/OST1 is a major positive regulator of ABA signaling in guard cells35. SnRK2.6/OST1 phosphorylates and activates both, SLAC1147,148 and ALMT12/QUAC1149. Group A PP2Cs, as negative ABA signaling regulators, directly dephosphorylate and inactivate not only SnRK2.6/OST1150,151 but also SnRK2.6/OST1 substrates, such as SLAC1152. Ion transport at the vacuolar membrane is also required for ABA-induced stomatal closure and detailed information has been reviewed153.
Cytosolic Ca2+ fine-tunes ABA-mediated stomatal closure by regulating Ca2+-sensor proteins, such as Ca2+-dependent protein kinases (CPKs), that contribute to the activation of SLAC1152,154. ABA can induce elevations in [Ca2+]cyt in guard cells, and one of the underlying mechanisms is plasma membrane Ca2+ influx through hyperpolarization-activated Ca2+-permeable cation (ICa) channels155,156. ABA-activation of ICa channels involves several steps. First, SnRK2.6/OST1 triggers extracellular reactive oxygen species (ROS) production which includes SnRK2 regulation of NADPH oxidases157,158. ROS mediates ICa channel activation in the plasma membrane via the hydrogen peroxide (H2O2) sensor kinase HYDROGEN-PEROXIDE-INDUCED Ca2+ INCREASES 1 (HPCA1)159 and the receptor-like (pseudo-)kinase GUARD CELL HYDROGEN PEROXIDE-RESISTANT1 (GHR1)160. GHR1 also contributes to SLAC1 activation161. In addition to Ca2+ and ROS, other small molecules such as hydrogen sulfide162,163 and gamma-aminobutyric acid164 have been recently shown to modulate guard cell ABA signaling.
Abiotic signal integration in guard cells.
Guard cells can perceive and integrate several environmental stimuli (FIG. 4b). Amongst them, light and CO2 are major abiotic stimuli that regulate stomatal aperture. Blue and red light induce stomatal opening mechanisms to maximize photosynthesis. Light-induced stomatal opening is mediated by H+-ATPase activation and subsequent K+ uptake through voltage-dependent inward-rectifying (K+in ) channels at the guard cell plasma membrane145,165,166. ABA suppresses light-induced stomatal opening via inhibition of H+-ATPases and K+in channels. Group D PP2Cs (PP2C.Ds) and their negative regulators, the SMALL AUXIN-UP RNAs (SAURs) also contribute to the regulation of H+-ATPases in Arabidopsis guard cells167,168. To which extent auxin is involved in this mechanism remains to be elucidated. Rapid downregulation of K+in channels is mediated by SnRK2.6/OST1-dependent phosphorylation of the K+in channel KAT1169 and also by [Ca2+]cyt elevation170. On a slower time-scale, the expression of K+in channel-encoding genes, including KAT1, is inhibited via SnRK2-dependent inactivation of ABA-RESPONSIVE KINASE SUBSTRATE (AKS) transcription factors171.
High CO2 induces stomatal closure, whereas low CO2 induces stomatal opening. In response to ABA, stomata close and do not easily re-open in the short term. In contrast, high CO2-mediated stomatal closure is rapidly reversible (e.g. REF.172). The mechanisms by which high CO2 mediates stomatal closure and activates SLAC1 differ from those of ABA172–174. In contrast to ABA signaling, elevated CO2 does not rapidly activate SnRK2-type protein kinases172,175. Raf-like kinases, such as CONVERGENCE OF BLUE LIGHT AND CO2 1 (CBC1), CBC2165 and HIGH LEAF TEMPERATURE 1 (HT1)176,177, inhibit S-type anion channel activation via an unknown mechanism. CBCs function at a convergence point between blue light and low CO2-induced stomatal opening signaling pathways165.
Signaling crosstalk between ABA and other hormones contributes to guard cell abiotic stress responses. The F-box protein MORE AXILLARY GROWTH 2 (MAX2) is a central regulator of both strigolactone and karrikin signaling178. MAX2-dependent signaling induces the upregulation of ABA-signaling genes, such as SnRK2.6/OST1, thereby enhancing ABA-induced stomatal closure and drought tolerance179,180. The type-B ARRs ARR1, ARR10, and ARR12, acting as positive transcriptional regulators of cytokinin signaling, negatively regulate stomatal closure and drought tolerance181. ABA and water deficit suppress cytokinin signaling via downregulation of type-B ARRs, as a proposed adaptive mechanism to survive drought181. It was reported that excess high light stress triggers local and whole-plant systemic stomatal closure, which is likely mediated by the NADPH oxidase RBOHD in coordination with ABA, salicylic acid, and jasmonate signaling182. Darkness and high CO2 however, do not induce stomatal closure in systemic leaves of Arabidopsis183. Under heat stress, plants open stomata to cool down the leaves by transpiration. Jasmonate has been suggested to fine-tune stomatal apertures during a combination of heat and other stresses such as high light and wounding184,185. Brassinosteroids can positively mediate stomatal opening186. In the brassinosteroid-biosynthesis mutant dwarf5 (dwf5), KAT1 expression is downregulated via an AKS-independent pathway and the light-driven activation of H+-ATPases remains intact, suggesting that brassinosteroid regulation of stomatal opening is independent of ABA signaling186.
Monitoring hormone responses in plants
To understand phytohormone signaling processes during abiotic stress, it is important to determine under which stress conditions, and in which cell-types or tissues and time frames phytohormone responses appear. Furthermore, it is relevant to determine at which level (i.e. biosynthesis, transport, perception, transduction, and transcriptional response) abiotic stresses interfere with a certain hormone signaling pathway. Genetically encoded phytohormone indicators (GEPHIs) are biosensors that allow the in vivo monitoring of cellular hormone responses at high spatiotemporal resolution and at various levels. Their functional principles, advantages over other methodologies and their limitations have been extensively discussed3,187,188 and are summarized in BOX 1. Here we review their contribution to abiotic stress response analyses in plants.
Box 1 |. Genetically encoded phytohormone indicators.
Genetically encoded phytohormone indicators (GEPHIs) enable the in vivo analysis of hormone concentration changes and subsequent downstream signaling processes at tissue- and cellular resolution. Several GEPHIs have been developed and employed in plants, and more comprehensive information can be found elsewhere3,187,188.
FRET-based GEPHIs.
To directly monitor hormone concentration changes, Förster Resonance Energy Transfer (FRET)-based indicators have been developed for ABA (ABACUS and ABAleon)28,189,224, auxin (AuxSen)192 and gibberellins (GPS1)190. These indicators consist of a sensory domain that changes its structure in a hormone-bound configuration, thereby affecting the distance, orientation, and the fluorescence emission ratio of a fluorescent protein (FP) FRET pair upon excitation of the FRET donor FP. ABAleon-type ABA indicators have recently received two updates. Dual-reporting indicators, consisting of ABAleonSD1–3L21 fused via the self-cleaving P2A peptide linker to the red-fluorescing Ca2+ indicator R-GECO1 or the pH indicator PA-17, allow the simultaneous monitoring of ABA together with Ca2+ or pH224. Whereas, ABAleon2.1_Tao3s, that harbor a nanobody recognition domain and a secretion signal, can be recruited by a subcellular targeted nanobody to either side of the endoplasmic reticulum (ER) membrane194. For the analysis of signaling processes downstream of ABA perception, the FRET-based SnRK2 Activity Sensor (SNACS) has been developed, providing an approach to investigate the activation of SnRK2-type protein kinases in response to abiotic stresses and SnRK2 interaction with other hormone signaling pathways175.
Degradation-based hormone reporters.
Several plant hormones induce downstream responses by regulating the ubiquitination and proteasomal degradation of transcriptional repressors3. Degradation-based GEPHIs, monitor their protein levels using FP fusions. More sophisticated reporters employ only a hormone-dependent degradation domain (degron-motif) fused to a FP to report an increased hormone concentration and signaling strength via a decrease in FP stability. For achieving a ratiometric readout, reporters for auxin, such as R2D2225 and qDII226 co-express a non-degradable FP as a reference.
Synthetic Hormone-activated Cas9-based Repressors (HACRs).
HACRs consist of a deactivated dCas9 fused to a hormone-dependent degradation domain and a fragment of the repressor TOPLESS. The dCas9 component associates with a guide RNA (gRNA) and recruits the HACR to a target promoter. Hormone-dependent degradation of the HACR then leads to a de-repression of the target (reporter) gene227.
A reporter for ethylene-dependent translational regulation.
The ethylene signaling pathway involves the EIN2 C-terminal domain (CEND) association with 3’ UTRs of EIN3-binding F-box protein (EBF) mRNAs to repress their translation228,229. Based on this mechanism, a translational reporter has been designed consisting of a FP coding sequence followed by three tandems of Ethylene Responsive RNA elements containing Poly-Uridylates (EPUs). Upon induction of EIN2, the translation of this 6x EPU reporter is inhibited, and transgenic plants expressing this construct are insensitive towards ethylene228.
Hormone-activated transcriptional reporters.
Several marker genes for plant hormone signaling have been identified and their promoters have been employed to drive the expression of reporter genes. In addition, the identification of cis-elements that are targeted by hormone-specific transcriptional regulators, lead to the development of synthetic promoters (SPs). SPs contain multiple repeats of cis-element-containing promoter fragments upstream of a minimal 35S promoter to drive reporter gene expression. They have been developed for almost any plant hormone3,187,188.
GEPHIs that enable the direct detection of phytohormone concentration changes have been initially developed for ABA28,189, followed by reporters for GA, SL and auxin190–192. However, only the FRET-based ABA indicators ABACUS1–2µ and ABAleon have thus far been employed in research related to abiotic stress28,189. Analyses in Arabidopsis using ABAleon2.1 under non stress conditions revealed the existence of an ABA gradient in roots and comparably higher basal ABA concentrations in guard cells, in the root-hypocotyl junction, and the root-tip28,193. Experiments using ABAleon2.1_Tao3s, that were targeted to either side of the ER membrane, indicated that in tobacco protoplasts, ABA levels might be higher in the ER compared to the cytosol194. How ABA gradients are maintained, and to what extent ABA biosynthesis and transport pathways contribute to distinct ABA concentration patterns could be further analyzed using ABA biosensors, similar to research previously conducted on gibberellin gradients in Arabidopsis roots195. There is increasing evidence that water deficit in Arabidopsis roots first induces the biosynthesis of ABA in leaves, via long-distance signals, before ABA accumulation is detected in roots26,27,32. Consistent with these findings, ABA indicator analyses could not detect rapid osmotic- or salt stress-induced ABA elevations in roots under imposed experimental conditions. Instead, ABA elevations were observed only several hours after exposure to stress28,189. Further analyses in Arabidopsis also revealed that sulfate and cysteine trigger ABA level increases in guard cells196, whereas CO2 elevation did not cause a rapid ABA concentration increase172,175. More detailed analyses are required to determine the spatiotemporal parameters of ABA elevation in response to water deficit and the intercellular transport routes of ABA. In the future it will also be interesting to investigate whether recently developed indicators for auxin192, GA190 and SL191 can detect respective hormone dynamics in response to abiotic stress.
Complementary to direct ABA indicators, the FRET-based SNACS reporter monitors downstream SnRK2-type protein kinase activity175. In Arabidopsis guard cells, SNACS responded to ABA, but not to elevated CO2 concentrations, or treatments with methyl jasmonate. These results were consistent with a lack of ABA accumulation under the same experimental conditions, providing evidence for the hypothesis that basal ABA signaling rather than SnRK2 activation contributes to elevated CO2 and methyl jasmonate responses in guard cells172,175.
Early on, promoter fragments of marker genes, or synthetic hormone-responsive promoters, were used to drive reporter gene expression as a readout for the detection of phytohormone signaling patterns3,187,188 (BOX 1). Although several transcriptional reporters were employed for the analyses of abiotic stress responses, most of the research related to abiotic stress focused on ABA. In this context, ABA signaling reporters were employed for the analyses of drought-, osmotic-, salt-, cold and high CO2 responses26,172,193,197, contributing to the hypothesis that in response to water shortage, ABA is largely synthesized in shoots rather than in roots of Arabidopsis26. Furthermore, the proRD29A-based ABA signaling reporter was employed as a readout in genetic screens197, contributing to the identification of ABA synthesis genes in Arabidopsis198. Also synthetic hormone-responsive promoter (SP) reporters were recently utilized for the reconstitution of ABA signaling in yeast199 and for the analysis of ABA-mediated transcriptional regulation in Arabidopsis200. The latter 6xABRE SPs reported basal ABA-independent activity in the root quiescent center, and ABA-, salt- and osmotic stress-dependent increases in other root tissues200, albeit with an apparent relatively low dynamic range compared to the proRAB18:GFP reporter193.
Reporters for other phytohormones also contributed to important observations on the roles of auxin, cytokinin and gibberellin in osmotic stress201, the contribution of cytokinin signaling to the hydrotropic response202, and the involvement of gibberellin signaling in the salt stress response121. The utilization of hormone reporters in species other than Arabidopsis is beginning to emerge (e.g. Kirschner et al.203) and will likely aid in determining differences and similarities in hormone signaling between different taxa.
Conclusions
Due to climate change, abiotic stresses, such as drought, salt, heat, and flooding are becoming increasingly challenging for plants1,2. Climate change and abiotic stresses can also intensify plant diseases204. Such alarming conditions demand innovative approaches. Recent advances in plant biology are providing crucial new insights into how plants sense and respond to abiotic stresses. While translating such findings into field applications remains challenging205, the advanced understanding of individual hormone-regulated abiotic stress responses, reviewed here, has the potential to provide key insights for developing more resilient crops through both, engineering and mining of traits from more resistant wild crop relatives (e.g. REF.125). The elucidation of mechanisms, genes and pathways that control these traits, can provide road maps for applications and translational research into enhancing or protecting yields in response to abiotic stressors. Many of the advances we discuss in this review were made in the model system Arabidopsis thaliana. Therefore, research will be needed to determine whether similar or divergent mechanisms are used in crops. Moreover, it has become clear that specific cell types have specific hormone signaling pathways and outputs, and therefore alteration of cell- or tissue-targeted traits will require investigation of hormone signaling mechanisms in those cell types. New tools, including hormone reporters, protein complex identifications, single cell sequencing and other approaches will enable the dissection of abiotic stress-linked cell-type specific and species-specific signal transduction mechanisms. Genetic approaches, including genomics-accelerated breeding and CRISPR-Cas9 gene editing provide new opportunities to accelerate the development of abiotic stress resilient traits. Furthermore, the genomics revolution combined with automated phenotyping are enhancing our ability to understand or predict which of these genes and mechanisms could be primarily used by resilient wild relatives. This can lead to targeted breeding of improved traits into crops. Moreover, enhancing yields of climate change resilient wild varieties through knowledge-guided de-novo domestication of crops206 provides an important new avenue for incorporating beneficial hormone signaling traits. Continued advances at understanding the interplay of plant hormones in diverse responses to abiotic stress will be important for developing abiotic stress resilient crops.
Acknowledgements
We apologize to those authors whose research we have not cited, due to limitations on the number of references. Research in the authors’ laboratories was supported by grants from the National Institutes of Health (GM060396-ES010337) and the National Science Foundation (MCB-1900567) (to J.I.S.), and from the Japan Society for the Promotion of Science (18K05557 and 18KK0425) (to S.M.).
Glossary
- Abiotic stress
Environmental stresses that are associated to the non-living environment such as weather conditions or the quality of the soil in which plants grow.
- Phytohormones
Phytohormones are plant-derived compounds that function as plant growth regulators either locally or over long distances and at low (sub µM) concentrations.
- Osmotic stress
Osmotic stress derives from differences in the water potential between plant cells and their environment. Hypoosmotic stress leads to cell swelling, whereas hyperosmotic stress leads to plant wilting.
- Mechanosensitive ion channels
Ion channels that respond to mechanical forces, e.g. induced by membrane tension.
- CLE25
A small peptide that is induced in the root vasculature during drought stress and moves to aerial tissues to induce NCED3 gene expression.
- SnRK2s
A family of protein kinases, including the ABA-activated SnRK2.2/2.3/2.6, that are activated in response to hyperosmotic stress.
- B2, B3 and B4 subgroup Raf-like M3Ks
Members of these subgroups of Raf-like M3Ks are involved in the osmotic stress- and ABA-dependent activation of SnRK2-type protein kinases.
- Seed dormancy
Inhibition of seed germination during unfavorable environmental conditions requires the downregulation of gibberellic acid signal transduction.
- Hydrotropism
The directional growth of roots towards regions of the soil environment with higher water content.
- Halotropism
The directional growth of roots away from regions of high salinity.
- Xerobranching
A water-responsive root developmental program, where the formation of lateral roots is repressed in regions of the soil environment that lacks water.
- Hydropatterning
A water-responsive root developmental program active when water is asymmetrically available around the circumference of the root. Lateral roots preferentially form on the water contacting side.
- Drought escape
An adaptive response to prolonged drought stress where plants accelerate the transition to flowering in order to reproduce.
- Deepwater rice
Specific varieties of rice (Oryza sativa) can survive periods of prolonged flooding by activating a developmental program, that depends on ethylene and gibberellin, to promote stem elongation into the air.
- Stomata
Small pores in the leaf epidermis that are formed by guard cells to allow the uptake of CO2 for photosynthesis in exchange for water loss.
- Depolarization and hyperpolarization
Changes in the cell membrane potential, making it more positive or more negative, respectively.
- GEPHI
Genetically encoded phytohormone indicators that allow the in vivo monitoring of hormone levels and downstream hormone signaling responses.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Zhu JK Abiotic Stress Signaling and Responses in Plants. Cell 167, 313–324 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamann E et al. Review: Plant eco-evolutionary responses to climate change: Emerging directions. Plant Sci 304, 110737 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Waadt R Phytohormone signaling mechanisms and genetic methods for their modulation and detection. Curr. Opin. Plant Biol 57, 31–40 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Zhao Y et al. Control of plant water use by ABA induction of senescence and dormancy: An overlooked lesson from evolution. Plant Cell Physiol 58, 1319–1327 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Yoshida T, Mogami J & Yamaguchi-Shinozaki K ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol 21, 133–139 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Knight H, Trewavas AJ & Knight MR Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J 12, 1067–1078 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Stephan AB, Kunz HH, Yang E & Schroeder JI Rapid hyperosmotic-induced Ca2+ responses in Arabidopsis thaliana exhibit sensory potentiation and involvement of plastidial KEA transporters. Proc. Natl. Acad. Sci. U. S. A 113, E5242–E5249 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamilton ES et al. Mechanosensitive channel MSL8 regulates osmotic forces during pollen hydration and germination. Science 350, 438–441 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee CP et al. MSL1 is a mechanosensitive ion channel that dissipates mitochondrial membrane potential and maintains redox homeostasis in mitochondria during abiotic stress. Plant J 88, 809–825 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basu D & Haswell ES The Mechanosensitive Ion Channel MSL10 Potentiates Responses to Cell Swelling in Arabidopsis Seedlings. Curr. Biol 30, 2716–2728.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Yoshimura K, Iida K & Iida H MCAs in Arabidopsis are Ca2+-permeable mechanosensitive channels inherently sensitive to membrane tension. Nat. Commun 12, 1–9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mori K et al. Ca2+-permeable mechanosensitive channels MCA1 and MCA2 mediate cold-induced cytosolic Ca2+ increase and cold tolerance in Arabidopsis. Sci. Rep 8, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mousavi SAR et al. PIEZO ion channel is required for root mechanotransduction in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A 118, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radin I et al. Plant PIEZO homologs modulate vacuole morphology during tip growth. Science 373, 586–590 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Yuan F et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 514, 367–371 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Jojoa-Cruz S et al. Cryo-EM structure of the mechanically activated ion channel OSCA1.2. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang M et al. Structure of the mechanosensitive OSCA channels. Nat. Struct. Mol. Biol 25, 850–858 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Chen K et al. BONZAI Proteins Control Global Osmotic Stress Responses in Plants. Curr. Biol 30, 4815–4825.e4 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Yang DL et al. Calcium pumps and interacting BON1 protein modulate calcium signature, stomatal closure, and plant immunity. Plant Physiol 175, 424–437 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Z et al. A RAF-SnRK2 kinase cascade mediates early osmotic stress signaling in higher plants. Nat. Commun 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi WG, Toyota M, Kim SH, Hilleary R & Gilroy S Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl. Acad. Sci. U. S. A 111, 6497–6502 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng W et al. The FERONIA Receptor Kinase Maintains Cell-Wall Integrity during Salt Stress through Ca2+ Signaling. Curr. Biol 28, 666–675.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J et al. FERONIA interacts with ABI2-type phosphatases to facilitate signaling cross-talk between abscisic acid and RALF peptide in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A 113, E5519–E5527 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang Z et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 572, 341–346 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Ma L et al. The SOS2-SCaBP8 Complex Generates and Fine-Tunes an AtANN4-Dependent Calcium Signature under Salt Stress. Dev. Cell 48, 697–709.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Christmann A, Weiler EW, Steudle E & Grill E A hydraulic signal in root-to-shoot signalling of water shortage. Plant J 52, 167–174 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Ikegami K, Okamoto M, Seo M & Koshiba T Activation of abscisic acid biosynthesis in the leaves of Arabidopsis thaliana in response to water deficit. J. Plant Res 122, 235–243 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Waadt R et al. FRET-based reporters for the direct visualization of abscisic acid concentration changes and distribution in Arabidopsis. Elife 2014, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urano K et al. Analysis of plant hormone profiles in response to moderate dehydration stress. Plant J 90, 17–36 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Sato H et al. Arabidopsis thaliana NGATHA1 transcription factor induces ABA biosynthesis by activating NCED3 gene during dehydration stress. Proc. Natl. Acad. Sci. U. S. A 115, E11178–E11187 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalladan R et al. Natural variation in 9-cis-epoxycartenoid dioxygenase 3 and ABA accumulation. Plant Physiol 179, 1620–1631 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi F et al. A small peptide modulates stomatal control via abscisic acid in long-distance signaling. Nature 556, 235–238 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Jensen MK et al. ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana. FEBS Open Bio 3, 321–327 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida R et al. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem 281, 5310–5318 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Mustilli AC, Merlot S, Vavasseur A, Fenzi F & Giraudat J Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14, 3089–3099 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujii H, Verslues PE & Zhu JK Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc. Natl. Acad. Sci. U. S. A 108, 1717–1722 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boudsocq M, Barbier-Brygoo H & Laurière C Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem 279, 41758–41766 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Cutler SR, Rodriguez PL, Finkelstein RR & Abrams SR Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol 61, 651–679 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Boudsocq M, Droillard MJ, Barbier-Brygoo H & Laurière C Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol. Biol 63, 491–503 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Belin C et al. Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol 141, 1316–1327 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vlad F et al. Phospho-site mapping, genetic and in planta activation studies reveal key aspects of the different phosphorylation mechanisms involved in activation of SnRK2s. Plant J 63, 778–790 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Takahashi Y et al. MAP3Kinase-dependent SnRK2-kinase activation is required for abscisic acid signal transduction and rapid osmotic stress response. Nat. Commun 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soma F, Takahashi F, Suzuki T, Shinozaki K & Yamaguchi-Shinozaki K Plant Raf-like kinases regulate the mRNA population upstream of ABA-unresponsive SnRK2 kinases under drought stress. Nat. Commun 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin Z et al. Initiation and amplification of SnRK2 activation in abscisic acid signaling. Nat. Commun 12, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hauser F et al. A genomic-scale artificial MicroRNA library as a tool to investigate the functionally redundant gene space in Arabidopsis. Plant Cell 25, 2848–2863 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saruhashi M et al. Plant Raf-like kinase integrates abscisic acid and hyperosmotic stress signaling upstream of SNF1-related protein kinase2. Proc. Natl. Acad. Sci. U. S. A 112, E6388–E6396 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Islam M et al. Activation of SnRK2 by Raf-like kinase ARK represents a primary mechanism of ABA and abiotic stress responses. Plant Physiol 185, 533–546 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stevenson SR et al. Genetic analysis of Physcomitrella patens identifies ABSCISIC ACID NON-RESPONSIVE, a regulator of ABA responses unique to basal land plants and required for desiccation tolerance. Plant Cell 28, 1310–1327 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Binder BM Ethylene signaling in plants. J. Biol. Chem 295, 7710–7725 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yasumura Y et al. An ancestral role for CONSTITUTIVE TRIPLE RESPONSE1 proteins in both ethylene and abscisic acid signaling. Plant Physiol 169, 283–298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goda H et al. The AtGenExpress hormone and chemical treatment data set: Experimental design, data evaluation, model data analysis and data access. Plant J 55, 526–542 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Finkelstein R Abscisic Acid Synthesis and Response. in The Arabidopsis Book vol. 11 e0166 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marcotte WR, Russell SH & Quatrano RS Abscisic acid-responsive sequences from the em gene of wheat. Plant Cell 1, 969–976 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fujita Y, Yoshida T & Yamaguchi-Shinozaki K Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiologia Plantarum vol. 147 15–27 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Brocard IM, Lynch TJ & Finkelstein RR Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol 129, 1533–1543 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carles C et al. Regulation of Arabidopsis thaliana Em genes: Role of ABI5. Plant J 30, 373–383 (2002). [DOI] [PubMed] [Google Scholar]
- 57.Furihata T et al. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. U. S. A 103, 1988–1993 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fujii H, Verslues PE & Zhu JK Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19, 485–494 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sirichandra C et al. The Arabidopsis ABA-activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14–3-3 binding site involved in its turnover. PLoS One 5, 1–13 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshida T et al. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant, Cell Environ 38, 35–49 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Finkelstein R, Gampala SSL, Lynch TJ, Thomas TL & Rock CD Redundant and distinct functions of the ABA response loci ABA-insensitive(ABI)5 and ABRE-binding factor (ABF)3. Plant Mol. Biol 59, 253–267 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Fujita M et al. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39, 863–876 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Kim JS et al. An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol 52, 2136–2146 (2011). [DOI] [PubMed] [Google Scholar]
- 64.Song L et al. A transcription factor hierarchy defines an environmental stress response network. Science 354, aag1550–aag1550 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hwang K, Susila H, Nasim Z, Jung JY & Ahn JH Arabidopsis ABF3 and ABF4 Transcription Factors Act with the NF-YC Complex to Regulate SOC1 Expression and Mediate Drought-Accelerated Flowering. Mol. Plant 12, 489–505 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Xu ZY et al. The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 25, 4708–4724 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takasaki H et al. SNAC-As, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. Plant J 84, 1114–1123 (2015). [DOI] [PubMed] [Google Scholar]
- 68.Han SK et al. The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell 24, 4892–4906 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peirats-Llobet M et al. A Direct Link between Abscisic Acid Sensing and the Chromatin-Remodeling ATPase BRAHMA via Core ABA Signaling Pathway Components. Mol. Plant 9, 136–147 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Vanstraelen M & Benkov E Hormonal interactions in the regulation of plant development. Annu. Rev. Cell Dev. Biol 28, 463–487 (2012). [DOI] [PubMed] [Google Scholar]
- 71.Ye H et al. RD26 mediates crosstalk between drought and brassinosteroid signalling pathways. Nat. Commun 8, 1–13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun Y et al. Integration of Brassinosteroid Signal Transduction with the Transcription Network for Plant Growth Regulation in Arabidopsis. Dev. Cell 19, 765–777 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fàbregas N et al. Overexpression of the vascular brassinosteroid receptor BRL3 confers drought resistance without penalizing plant growth. Nat. Commun 9, 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang X et al. The Antagonistic Action of Abscisic Acid and Cytokinin Signaling Mediates Drought Stress Response in Arabidopsis. Mol. Plant 11, 970–982 (2018). [DOI] [PubMed] [Google Scholar]
- 75.Guan C et al. Cytokinin antagonizes abscisic acid-mediated inhibition of cotyledon greening by promoting the degradation of ABSCISIC ACID INSENSITIVE5 protein in Arabidopsis. Plant Physiol 164, 1515–1526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shani E et al. Plant Stress Tolerance Requires Auxin-Sensitive Aux/IAA Transcriptional Repressors. Curr. Biol 27, 437–444 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hugouvieux V, Kwak JM & Schroeder JI An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106, 477–487 (2001). [DOI] [PubMed] [Google Scholar]
- 78.Cui P, Zhang S, Ding F, Ali S & Xiong L Dynamic regulation of genome-wide pre-mRNA splicing and stress tolerance by the Sm-like protein LSm5 in Arabidopsis. Genome Biol 15, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang Z et al. ABA signalling is fine-tuned by antagonistic HAB1 variants. Nat. Commun 6, (2015). [DOI] [PubMed] [Google Scholar]
- 80.Zhan X et al. An Arabidopsis PWI and RRM motif-containing protein is critical for pre-mRNA splicing and ABA responses. Nat. Commun 6, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Carrasco-López C et al. Environment-dependent regulation of spliceosome activity by the LSM2–8 complex in Arabidopsis. Nucleic Acids Res 45, 7416–7431 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ma Y et al. Arabidopsis exoribonuclease USB1 interacts with the PPR-domain protein SOAR1 to negatively regulate abscisic acid signaling. J. Exp. Bot 71, 5837–5851 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schoenberg DR & Maquat LE Regulation of cytoplasmic mRNA decay. Nature Reviews Genetics 13, 246–259 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Soma F et al. ABA-unresponsive SnRK2 protein kinases regulate mRNA decay under osmotic stress in plants. Nat. Plants 3, (2017). [DOI] [PubMed] [Google Scholar]
- 85.Wang Y et al. NAD+-capped RNAs are widespread in the Arabidopsis transcriptome and can probably be translated. Proc. Natl. Acad. Sci. U. S. A 116, 12094–12102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang H et al. NAD tagSeq reveals that NAD+-capped RNAs are mostly produced from a large number of protein-coding genes in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A 116, 12072–12077 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yu X et al. Messenger RNA 5′ NAD+ Capping Is a Dynamic Regulatory Epitranscriptome Mark That Is Required for Proper Response to Abscisic Acid in Arabidopsis. Dev. Cell 56, 125–140.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xiong Y & Sheen J The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiol 164, 499–512 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang P et al. Reciprocal Regulation of the TOR Kinase and ABA Receptor Balances Plant Growth and Stress Response. Mol. Cell 69, 100–112.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun TP The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol 21, (2011). [DOI] [PubMed] [Google Scholar]
- 91.Davière JM & Achard P A Pivotal Role of DELLAs in Regulating Multiple Hormone Signals. Mol. Plant 9, 10–20 (2016). [DOI] [PubMed] [Google Scholar]
- 92.Lim S et al. ABA-insensitive3, ABA-insensitive5, and DELLAs interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 25, 4863–4878 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hu Y et al. The transcription factor INDUCER OF CBF EXPRESSION1 interacts with ABSCISIC ACID INSENSITIVE5 and DELLA proteins to fine-tune abscisic acid signaling during seed germination in Arabidopsis. Plant Cell 31, 1520–1538 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen H et al. Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc. Natl. Acad. Sci. U. S. A 105, 4495–4500 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu X et al. The NF-YC-RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat. Commun 7, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yu LH et al. Arabidopsis MADS-Box Transcription Factor AGL21 Acts as Environmental Surveillance of Seed Germination by Regulating ABI5 Expression. Mol. Plant 10, 834–845 (2017). [DOI] [PubMed] [Google Scholar]
- 97.Yang B et al. RSM1, an Arabidopsis MYB protein, interacts with HY5/HYH to modulate seed germination and seedling development in response to abscisic acid and salinity. PLoS Genet 14, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dinneny JR Developmental responses to water and salinity in root systems. Annu. Rev. Cell Dev. Biol 35, 239–257 (2019). [DOI] [PubMed] [Google Scholar]
- 99.Zhang H et al. ABA promotes quiescence of the quiescent centre and suppresses stem cell differentiation in the Arabidopsis primary root meristem. Plant J 64, 764–774 (2010). [DOI] [PubMed] [Google Scholar]
- 100.Takahashi N, Goto N, Okada K & Takahashi H Hydrotropism in abscisic acid, wavy, and gravitropic mutants of Arabidopsis thaliana. Planta 216, 203–211 (2002). [DOI] [PubMed] [Google Scholar]
- 101.Dietrich D et al. Root hydrotropism is controlled via a cortex-specific growth mechanism. Nat. Plants 3, (2017). [DOI] [PubMed] [Google Scholar]
- 102.Miao R et al. Low ABA concentration promotes root growth and hydrotropism through relief of ABA INSENSITIVE 1-mediated inhibition of plasma membrane H+-ATPase 2. Sci. Adv 7, 4113–4130 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Miao R et al. Comparative analysis of Arabidopsis ecotypes reveals a role for brassinosteroids in root hydrotropism. Plant Physiol 176, 2720–2736 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Duan L et al. Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 25, 324–341 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Galvan-Ampudia CS et al. Halotropism is a response of plant roots to avoid a saline environment. Curr. Biol 23, 2044–2050 (2013). [DOI] [PubMed] [Google Scholar]
- 106.Korver RA et al. Halotropism requires phospholipase Dζ1-mediated modulation of cellular polarity of auxin transport carriers. Plant Cell Environ 43, 143–158 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kaneyasu T et al. Auxin response, but not its polar transport, plays a role in hydrotropism of Arabidopsis roots. J. Exp. Bot 58, 1143–1150 (2007). [DOI] [PubMed] [Google Scholar]
- 108.Shkolnik D, Krieger G, Nuriel R & Fromm H Hydrotropism: Root Bending Does Not Require Auxin Redistribution. Mol. Plant 9, 757–759 (2016). [DOI] [PubMed] [Google Scholar]
- 109.Ramachandran P, Wang G, Augstein F, De Vries J & Carlsbecker A Continuous root xylem formation and vascular acclimation to water deficit involves endodermal ABA signalling via miR165. Dev 145, (2018). [DOI] [PubMed] [Google Scholar]
- 110.Bloch D, Puli MR, Mosquna A & Yalovsky S Abiotic stress modulates root patterning via ABA-regulated microRNA expression in the endodermis initials. Dev 146, (2019). [DOI] [PubMed] [Google Scholar]
- 111.Ramachandran P et al. Abscisic acid signaling activates distinct VND transcription factors to promote xylem differentiation in Arabidopsis. Curr. Biol 31, 3153–3161.e5 (2021). [DOI] [PubMed] [Google Scholar]
- 112.De Smet I et al. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134, 681–690 (2007). [DOI] [PubMed] [Google Scholar]
- 113.Moreno-Risueno MA et al. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329, 1306–1311 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bao Y et al. Plant roots use a patterning mechanism to position lateral root branches toward available water. Proc. Natl. Acad. Sci. U. S. A 111, 9319–9324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Orosa-Puente B et al. Root branching toward water involves posttranslational modification of transcription factor ARF7. Science 362, 1407–1410 (2018). [DOI] [PubMed] [Google Scholar]
- 116.Orman-Ligeza B et al. The Xerobranching Response Represses Lateral Root Formation When Roots Are Not in Contact with Water. Curr. Biol 28, 3165–3173.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 117.Andrés F & Coupland G The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet 13, 627–639 (2012). [DOI] [PubMed] [Google Scholar]
- 118.Riboni M, Galbiati M, Tonelli C & Conti L GIGANTEA enables drought escape response via abscisic acid-dependent activation of the florigens and SUPPRESSOR of OVEREXPRESSION of CONSTANS. Plant Physiol 162, 1706–1719 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang P et al. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. U. S. A 110, 11205–11210 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Riboni M, Test AR, Galbiati M, Tonelli C & Conti L ABA-dependent control of GIGANTEA signalling enables drought escape via up-regulation of FLOWERING LOCUS T in Arabidopsis thaliana. J. Exp. Bot 67, 6309–6322 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Achard P et al. Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94 (2006). [DOI] [PubMed] [Google Scholar]
- 122.Achard P et al. The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc. Natl. Acad. Sci. U. S. A 104, 6484–6489 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang H et al. The DELLA-CONSTANS transcription factor cascade integrates gibberellic acid and photoperiod signaling to regulate flowering. Plant Physiol 172, 479–488 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kim WY et al. Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat. Commun 4, (2013). [DOI] [PubMed] [Google Scholar]
- 125.Voesenek LACJ & Bailey-Serres J Flood adaptive traits and processes: An overview. New Phytol 206, 57–73 (2015). [DOI] [PubMed] [Google Scholar]
- 126.Hartman S, Sasidharan R & Voesenek LACJ The role of ethylene in metabolic acclimations to low oxygen. New Phytol 229, 64–70 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lee TA & Bailey-Serres J Conserved and nuanced hierarchy of gene regulatory response to hypoxia. New Phytol 229, 71–78 (2021). [DOI] [PubMed] [Google Scholar]
- 128.Gasch P et al. Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in Arabidopsis. Plant Cell 28, 160–180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Xu K et al. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442, 705–708 (2006). [DOI] [PubMed] [Google Scholar]
- 130.Gibbs DJ et al. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479, 415–418 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Licausi F et al. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479, 419–422 (2011). [DOI] [PubMed] [Google Scholar]
- 132.Hartman S et al. Ethylene-mediated nitric oxide depletion pre-adapts plants to hypoxia stress. Nat. Commun 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lin CC et al. Regulatory cascade involving transcriptional and N-end rule pathways in rice under submergence. Proc. Natl. Acad. Sci. U. S. A 116, 3300–3309 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Reynoso MA et al. Evolutionary flexibility in flooding response circuitry in angiosperms. Science 365, 1291–1295 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hattori Y et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460, 1026–1030 (2009). [DOI] [PubMed] [Google Scholar]
- 136.Kuroha T et al. Ethylene-gibberellin signaling underlies adaptation of rice to periodic flooding. Science 361, 181–186 (2018). [DOI] [PubMed] [Google Scholar]
- 137.Nagai K et al. Antagonistic regulation of the gibberellic acid response during stem growth in rice. Nature 584, 109–114 (2020). [DOI] [PubMed] [Google Scholar]
- 138.Pandey BK et al. Plant roots sense soil compaction through restricted ethylene diffusion. Science 371, 276–280 (2021). [DOI] [PubMed] [Google Scholar]
- 139.Endo A et al. Drought induction of Arabidopsis 9-cis-epoxycarotenoid dioxygenase occurs in vascular parenchyma cells. Plant Physiol 147, 1984–1993 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bauer H et al. The stomatal response to reduced relative humidity requires guard cell-autonomous ABA synthesis. Curr. Biol 23, 53–57 (2013). [DOI] [PubMed] [Google Scholar]
- 141.Schroeder JI & Keller BU Two types of anion channel currents in guard cells with distinct voltage regulation. Proc. Natl. Acad. Sci. U. S. A 89, 5025–5029 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Negi J et al. CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 452, 483–486 (2008). [DOI] [PubMed] [Google Scholar]
- 143.Vahisalu T et al. SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452, 487–491 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Meyer S et al. AtALMT12 represents an R-type anion channel required for stomatal movement in Arabidopsis guard cells. Plant J 63, 1054–1062 (2010). [DOI] [PubMed] [Google Scholar]
- 145.Ward JM, M̈aser P & Schroeder JI Plant ion channels: Gene families, physiology, and functional genomics analyses. Annu. Rev. Physiol 71, 59–82 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hosy E et al. The Arabidopsis outward K+ channel GORK is involved in regulation of stomatal movements and plant transpiration. Proc. Natl. Acad. Sci. U. S. A 100, 5549–5554 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Geiger D et al. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. U. S. A 106, 21425–21430 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lee SC, Lan W, Buchanan BB & Luan S A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc. Natl. Acad. Sci. U. S. A 106, 21419–21424 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Imes D et al. Open stomata 1 (OST1) kinase controls R-type anion channel QUAC1 in Arabidopsis guard cells. Plant J 74, 372–382 (2013). [DOI] [PubMed] [Google Scholar]
- 150.Vlad F et al. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21, 3170–3184 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Umezawa T et al. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A 106, 17588–17593 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Brandt B et al. Calcium specificity signaling mechanisms in abscisic acid signal transduction in Arabidopsis guard cells. Elife 4, 1–25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Eisenach C & De Angeli A Ion transport at the vacuole during stomatal movements. Plant Physiol 174, 520–530 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Geiger D et al. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl. Acad. Sci. U. S. A 107, 8023–8028 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hamilton DWA, Hills A, Köhler B & Blatt MR Ca2+ channels at the plasma membrane of stomatal guard cells are activated by hyperpolarization and abscisic acid. Proc. Natl. Acad. Sci. U. S. A 97, 4967–4972 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pei ZM et al. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406, 731–734 (2000). [DOI] [PubMed] [Google Scholar]
- 157.Sirichandra C et al. Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett 583, 2982–2986 (2009). [DOI] [PubMed] [Google Scholar]
- 158.Han JP et al. Fine-tuning of RBOHF activity is achieved by differential phosphorylation and Ca2+ binding. New Phytol 221, 1935–1949 (2019). [DOI] [PubMed] [Google Scholar]
- 159.Wu F et al. Hydrogen peroxide sensor HPCA1 is an LRR receptor kinase in Arabidopsis. Nature 578, 577–581 (2020). [DOI] [PubMed] [Google Scholar]
- 160.Hua D et al. A plasma membrane receptor kinase, GHR1, mediates abscisic acid- and hydrogen peroxide-regulated stomatal movement in Arabidopsis. Plant Cell 24, 2546–2561 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sierla M et al. The receptor-like pseudokinase GHR1 is required for stomatal closure. Plant Cell 30, 2813–2837 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wang L, Wan R, Shi Y & Xue S Hydrogen Sulfide Activates S-Type Anion Channel via OST1 and Ca2+ Modules. Mol. Plant 9, 489–491 (2016). [DOI] [PubMed] [Google Scholar]
- 163.Pantaleno R, Scuffi D & García-Mata C Hydrogen sulphide as a guard cell network regulator. New Phytol 230, 451–456 (2021). [DOI] [PubMed] [Google Scholar]
- 164.Xu B et al. GABA signalling modulates stomatal opening to enhance plant water use efficiency and drought resilience. Nat. Commun 12, 1952 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hiyama A et al. Blue light and CO2 signals converge to regulate light-induced stomatal opening. Nat. Commun 8, 1–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ando E & Kinoshita T Red light-induced phosphorylation of plasma membrane H+-ATPase in stomatal guard cells. Plant Physiol 178, 838–849 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Inoue SI & Kinoshita T Blue light regulation of stomatal opening and the plasma membrane H+-ATPase. Plant Physiol 174, 531–538 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wong JH et al. SAUR proteins and PP2C.D phosphatases regulate H+-ATPases and K+ channels to control stomatal movements. Plant Physiol 185, 256–273 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sato A et al. Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem. J 424, 439–448 (2009). [DOI] [PubMed] [Google Scholar]
- 170.Siegel RS et al. Calcium elevation-dependent and attenuated resting calcium-dependent abscisic acid induction of stomatal closure and abscisic acid-induced enhancement of calcium sensitivities of S-type anion and inward-rectifying K+ channels in Arabidopsis guard cells. Plant J 59, 207–220 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Takahashi Y et al. BHLH transcription factors that facilitate K+ uptake during stomatal opening are repressed by abscisic acid through phosphorylation. Sci. Signal 6, ra48–ra48 (2013). [DOI] [PubMed] [Google Scholar]
- 172.Hsu PK et al. Abscisic acid-independent stomatal CO2 signal transduction pathway and convergence of CO2 and ABA signaling downstream of OST1 kinase. Proc. Natl. Acad. Sci. U. S. A 115, E9971–E9980 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yamamoto Y et al. The transmembrane region of guard cell SLAC1 channels perceives CO2 signals via an ABA-independent pathway in Arabidopsis. Plant Cell 28, 557–567 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhang J et al. Identification of SLAC1 anion channel residues required for CO2/bicarbonate sensing and regulation of stomatal movements. Proc. Natl. Acad. Sci. U. S. A 115, 11129–11137 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhang L et al. FRET kinase sensor development reveals SnRK2/OST1 activation by ABA but not by MeJA and high CO2 during stomatal closure. Elife 9, 1–74 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hashimoto-Sugimoto M et al. Dominant and recessive mutations in the Raf-like kinase HT1 gene completely disrupt stomatal responses to CO2 in Arabidopsis. J. Exp. Bot 67, 3251–3261 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hõrak H et al. A dominant mutation in the ht1 kinase uncovers roles of MAP kinases and GHR1 in CO2-induced stomatal closure. Plant Cell 28, 2493–2509 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Waters MT, Gutjahr C, Bennett T & Nelson DC Strigolactone Signaling and Evolution. Annu. Rev. Plant Biol 68, 291–322 (2017). [DOI] [PubMed] [Google Scholar]
- 179.Ha C. Van et al. Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc. Natl. Acad. Sci. U. S. A 111, 851–856 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Li W et al. Comparative functional analyses of DWARF14 and KARRIKIN INSENSITIVE 2 in drought adaptation of Arabidopsis thaliana. Plant J 103, 111–127 (2020). [DOI] [PubMed] [Google Scholar]
- 181.Nguyen KH et al. Arabidopsis type B cytokinin response regulators ARR1, ARR10, and ARR12 negatively regulate plant responses to drought. Proc. Natl. Acad. Sci. U. S. A 113, 3090–3095 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Devireddy AR, Zandalinas SI, Gómez-Cadenas A, Blumwald E & Mittler R Coordinating the overall stomatal response of plants: Rapid leaf-to-leaf communication during light stress. Sci. Signal 11, (2018). [DOI] [PubMed] [Google Scholar]
- 183.Ehonen S, Hölttä T & Kangasjärvi J Systemic signaling in the regulation of stomatal conductance. Plant Physiol 182, 1829–1832 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zandalinas SI et al. Systemic signaling during abiotic stress combination in plants. Proc. Natl. Acad. Sci. U. S. A 117, 13810–13820 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Havko NE et al. Insect herbivory antagonizes leaf cooling responses to elevated temperature in tomato. Proc. Natl. Acad. Sci. U. S. A 117, 2211–2217 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Inoue SI et al. Brassinosteroid involvement in Arabidopsis thaliana stomatal opening. Plant Cell Physiol 58, 1048–1058 (2017). [DOI] [PubMed] [Google Scholar]
- 187.Isoda R et al. Sensors for the quantification, localization and analysis of the dynamics of plant hormones. Plant J 105, 542–557 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhao C, Yaschenko A, Alonso JM & Stepanova AN Leveraging synthetic biology approaches in plant hormone research. Curr. Opin. Plant Biol 60, 101998 (2021). [DOI] [PubMed] [Google Scholar]
- 189.Jones AM et al. Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. Elife 3, e01741 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Rizza A, Walia A, Lanquar V, Frommer WB & Jones AM In vivo gibberellin gradients visualized in rapidly elongating tissues. Nat. Plants 3, 803–813 (2017). [DOI] [PubMed] [Google Scholar]
- 191.Chesterfield RJ et al. Rational Design of Novel Fluorescent Enzyme Biosensors for Direct Detection of Strigolactones. ACS Synth. Biol 9, 2107–2118 (2020). [DOI] [PubMed] [Google Scholar]
- 192.Herud-Sikimić O et al. A biosensor for the direct visualization of auxin. Nature 592, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Waadt R, Hsu PK & Schroeder JI Abscisic acid and other plant hormones: Methods to visualize distribution and signaling. BioEssays 37, 1338–1349 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhou Y, Wang Y, Li J & Liang J In vivo FRET-FLIM reveals ER-specific increases in the ABA level upon environmental stresses. Plant Physiol 186, 1545–1561 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Rizza A et al. Differential biosynthesis and cellular permeability explain longitudinal gibberellin gradients in growing roots. Proc. Natl. Acad. Sci. U. S. A 118, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Batool S et al. Sulfate is incorporated into cysteine to trigger ABA production and stomatal closure. Plant Cell 30, 2973–2987 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ishitani M, Xiong L, Stevenson B & Zhu JK Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: Interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell 9, 1935–1949 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Xiong L, Lee H, Ishitani M & Zhu JK Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J. Biol. Chem 277, 8588–8596 (2002). [DOI] [PubMed] [Google Scholar]
- 199.Ruschhaupt M et al. Rebuilding core abscisic acid signaling pathways of Arabidopsis in yeast. EMBO J 38, 1–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wu R et al. The 6xABRE synthetic promoter enables the spatiotemporal analysis of ABA-mediated transcriptional regulation. Plant Physiol 177, 1650–1665 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Rowe JH, Topping JF, Liu J & Lindsey K Abscisic acid regulates root growth under osmotic stress conditions via an interacting hormonal network with cytokinin, ethylene and auxin. New Phytol 211, 225–239 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Chang J et al. Asymmetric distribution of cytokinins determines root hydrotropism in Arabidopsis thaliana. Cell Res 29, 984–993 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Kirschner GK, Stahl Y, Imani J, von Korff M & Simon R Fluorescent reporter lines for auxin and cytokinin signalling in barley (Hordeum vulgare). PLoS One 13, e0196086 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.IPCC Secretariat. Plant health and climate change. FAO on behalf of the Secretariat of the International Plant Protection Convention #2 (2021) doi: 10.1201/b14056-16. [DOI] [Google Scholar]
- 205.Nuccio ML, Paul M, Bate NJ, Cohn J & Cutler SR Where are the drought tolerant crops? An assessment of more than two decades of plant biotechnology effort in crop improvement. Plant Sci 273, 110–119 (2018). [DOI] [PubMed] [Google Scholar]
- 206.Zsögön A et al. De novo domestication of wild tomato using genome editing. Nat. Biotechnol 36, 1211–1216 (2018). [DOI] [PubMed] [Google Scholar]
- 207.Barberon M et al. Adaptation of Root Function by Nutrient-Induced Plasticity of Endodermal Differentiation. Cell 164, 447–459 (2016). [DOI] [PubMed] [Google Scholar]
- 208.Ding Y et al. OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell 32, 278–289 (2015). [DOI] [PubMed] [Google Scholar]
- 209.Larkindale J, Hall JD, Knight MR & Vierling E Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol 138, 882–897 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wang R et al. HSP90 regulates temperature-dependent seedling growth in Arabidopsis by stabilizing the auxin co-receptor F-box protein TIR1. Nat. Commun 7, 1–11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Reed JW et al. Three auxin response factors promote hypocotyl elongation. Plant Physiol 178, 864–875 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Sun J, Qi L, Li Y, Chu J & Li C PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet 8, e1002594 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Eremina M et al. Brassinosteroids participate in the control of basal and acquired freezing tolerance of plants. Proc. Natl. Acad. Sci. U. S. A 113, E5982–E5991 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Li H et al. BZR1 Positively Regulates Freezing Tolerance via CBF-Dependent and CBF-Independent Pathways in Arabidopsis. Mol. Plant 10, 545–559 (2017). [DOI] [PubMed] [Google Scholar]
- 215.Ibañez C et al. Brassinosteroids Dominate Hormonal Regulation of Plant Thermomorphogenesis via BZR1. Curr. Biol 28, 303–310.e3 (2018). [DOI] [PubMed] [Google Scholar]
- 216.Martínez C et al. PIF4‐induced BR synthesis is critical to diurnal and thermomorphogenic growth. EMBO J 37, e99552 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Nishiyama R et al. Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 23, 2169–2183 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wang Z et al. GAI functions in the plant response to dehydration stress in Arabidopsis thaliana. Int. J. Mol. Sci 21, 819 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Lantzouni O, Alkofer A, Falter-Braun P & Schwechheimer C Growth-regulating factors interact with DELLAs and regulate growth in cold stress. Plant Cell 32, 1018–1034 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Blanco-Touriñán N et al. COP1 destabilizes DELLA proteins in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A 117, 13792–13799 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Hu Y, Jiang L, Wang F & Yu D Jasmonate regulates the INDUCER OF CBF expression-C-repeat binding factor/DRE binding factor1 Cascade and freezing tolerance in Arabidopsis. Plant Cell 25, 2907–2924 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Yang T et al. The suppressor of MAX2 1 (SMAX1)-like SMXL6, SMXL7 and SMXL8 act as negative regulators in response to drought stress in Arabidopsis. Plant Cell Physiol 61, 1477–1492 (2020). [DOI] [PubMed] [Google Scholar]
- 223.Lee JS, Wilson ME, Richardson RA & Haswell ES Genetic and physical interactions between the organellar mechanosensitive ion channel homologs MSL1, MSL2, and MSL3 reveal a role for inter-organellar communication in plant development. Plant Direct 3, e00124 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Waadt R et al. Dual-reporting transcriptionally linked genetically encoded fluorescent indicators resolve the spatiotemporal coordination of cytosolic abscisic acid and second messenger dynamics in Arabidopsis. Plant Cell 32, 2582–2601 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Liao CY et al. Reporters for sensitive and quantitative measurement of auxin response. Nat. Methods 12, 207–210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Galvan-Ampudia CS et al. Temporal integration of auxin information for the regulation of patterning. Elife 9, 1–65 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Khakhar A, Leydon AR, Lemmex AC, Klavins E & Nemhauser JL Synthetic hormone-responsive transcription factors can monitor and reprogram plant development. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Li W et al. EIN2-Directed Translational Regulation of Ethylene Signaling in Arabidopsis. Cell 163, 670–683 (2015). [DOI] [PubMed] [Google Scholar]
- 229.Merchante C et al. Gene-Specific Translation Regulation Mediated by the Hormone-Signaling Molecule EIN2. Cell 163, 684–697 (2015). [DOI] [PubMed] [Google Scholar]


