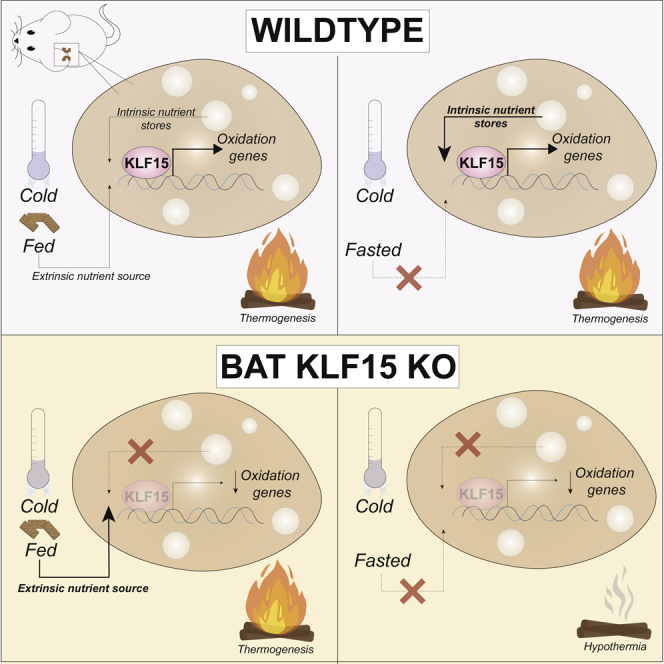
Summary
Brown adipose tissue (BAT) is a specialized metabolic organ responsible for non-shivering thermogenesis. Recently, its activity has been shown to be critical in systemic metabolic health through its utilization and consumption of macronutrients. In the face of energetically demanding states, metabolic flexibility and systemic coordination of nutrient partitioning is requisite for health and survival. In this study, we elucidate BAT’s differential transcriptional adaptations in response to multiple nutrient challenges and demonstrate its context-dependent prioritization of lipid, glucose, and amino acid metabolism. We show that the transcription factor Krüppel-like factor 15 (KLF15) plays a critical role in BAT metabolic flexibility. BAT-specific loss of KLF15 results in widespread changes in circulating metabolites and severely compromised thermogenesis in response to high energy demands, indicative of impaired nutrient utilization and metabolic flexibility. Together, our data demonstrate KLF15 in BAT plays an indispensable role in partitioning resources to maintain homeostasis and ensure survival.
Subject areas: Human metabolism, Molecular biology, Cell biology
Graphical abstract
Highlights
-
•
KLF15 is a critical regulator of BAT thermogenesis and metabolism
-
•
BAT transcriptionally prioritizes thermogenesis over fasting
-
•
BAT metabolic flexibility is significantly impaired with KLF15 deficiency
-
•
BAT KLF15 knockout alters circulating lipids and amino acids
Human metabolism; Molecular biology; Cell biology.
Introduction
Brown adipose tissue’s (BAT) ability to generate heat via non-shivering thermogenesis allows endothermic organisms to maintain a narrow body temperature range, a phenomenon crucial to individual survival and the evolutionary success of mammals at large (Kajimura and Saito, 2014; Nedergaard and Cannon, 1985). Both circulating and BAT-intrinsic lipids, glucose, and branched chain amino acids (BCAA) serve as critical thermogenic fuel sources, and BAT utilization of these macronutrients is important for systemic metabolic health (Bartelt et al., 2011; Cannon and Nedergaard, 2004; Haemmerle et al., 2006; Simcox et al., 2017; Stanford et al., 2013; Yoneshiro et al., 2019). In humans, increased BAT activity is associated with improved insulin sensitivity and decreased circulating lipids, whereas, BAT dysfunction or absence is linked to the development of obesity, insulin resistance, and dyslipidemia (Bartelt et al., 2011; Chondronikola et al., 2016; Dong et al., 2018; Saito et al., 2009, 2020; Vijgen et al., 2011). As such, concerted efforts are now underway to understand BAT physiology in hopes of shedding light on BAT nutrient utilization and its systemic impact on metabolic health. Over the last few decades, the transcription factor Krüppel-like factor 15 (KLF15) has emerged as a critical regulator of systemic homeostasis in the face of varying metabolic demands (Hsieh et al., 2019). In this study, we find that proper substrate switching is requisite for non-shivering thermogenesis in response to chronic cold exposure and fasting. Loss of BAT KLF15 and the subsequent lack of BAT metabolic flexibility results in widespread changes in circulating metabolites and severely compromised thermogenesis in response to high energy demands. Collectively, our findings shed light on BAT nutrient utilization in various energetic states and add to our growing understanding of KLF15 as an orchestrator of systemic metabolism.
Results
Characterization of BAT transcriptome in response to cold and fasting
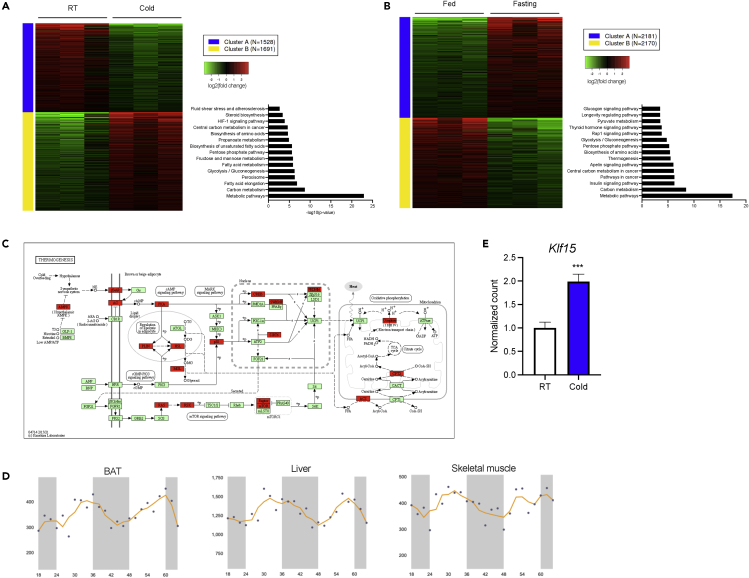
Maintaining body temperature in cold exposure requires systemic coordination of nutrient mobilization and selective utilization to ensure survival. To gain a comprehensive understanding of the transcriptional adaptations that must occur for successful non-shivering thermogenesis in cold, we performed RNA-sequencing (RNA-seq) studies on BAT isolated from control mice at room temperature (RT, fed ad libitum) and after a chronic cold challenge (10 days at 4°C, fed ad libitum). RNA-seq analysis of BAT comparing RT versus cold revealed 3219 differentially expressed genes (DEGs). As expected from previous physiological data demonstrating BAT energy utilization in response to cold, K-means clustering followed by KEGG pathway analysis of upregulated genes revealed enrichment of pathways related to the metabolism of all three macronutrients, with a preponderance of pathways related to lipid metabolism (Figure 1A).
Figure 1.
Characterization of BAT transcriptome in response to cold and fasting
(A and B) K-means clustering of DEGs followed by KEGG pathway analysis in (A) RT vs Cold (n = 3) and (B) RT (Fed) vs Fasting (n = 3) in K15 F/F control mice.
(C) KEGG pathway for thermogenesis highlighting downregulated genes in K15 F/F Fasting condition in red and green.
(D) Klf15 expression data mined from CircaDB showing similar rhythmic oscillations in BAT, liver, and skeletal muscle.
(E) Relative BAT Klf15 expression in room temperature (RT) vs cold (n = 3–4). Data represent mean ± SEM. Comparisons between groups were performed using an unpaired, 2-tailed Student’s t test. ∗p < 0.05, ∗∗p < 0.01.
This upregulation of lipid metabolism pathways is reminiscent of BAT adaptations in states of fasting where survival again is dependent on appropriate and selective nutrient mobilization and utilization (Nabatame et al., 2021). We performed RNA-seq on BAT harvested from control mice (RT, fed ad libitum) and on BAT harvested after an overnight fast (12 h, no food) and identified 4351 DEGs. Pathway analysis of downregulated genes in fasting revealed significant changes in metabolic pathways, most notably for glucose metabolism (Figure 1B). Notably, thermogenesis pathways were among the topmost enriched pathways as well, supporting observations that body temperature decreases with fasting (Figure 1C) (Chatamra et al., 1984). The RNA-seq data are concordant with past studies that have shown decreased BAT Pdk4 transcription, PDK4 activity, and glucose oxidation in fasting with a concurrent increase in lipid metabolism gene expression (Nabatame et al., 2021). These findings also suggest that fasting induces a nutrient switch in BAT like that seen in liver and muscle, where nutrient consumption is downregulated to preserve circulating glucose for glucose obligate organs, such as the brain.
To allow for time-of-day changes in nutrient availability, BAT activity and core body temperature exhibits circadian rhythmicity (Adlanmerini et al., 2019; Chappuis et al., 2013; Machado et al., 2018). Using publicly available circadian expression datasets (CircaDB), we noted that Klf15 oscillates rhythmically in BAT in a pattern similar to those observed in liver and in skeletal muscle (Figure 1D) (“CIRCA: Circadian gene expression profiles.,” n.d.; Su et al., 2004). Klf15 expression increases throughout the light phase and decreases throughout the dark phase, suggesting that KLF15 function in BAT is more analogous to that in these highly oxidative tissues than to nutrient storing tissues such as white adipose tissue (WAT) (Figure 1D) (Fan et al., 2021; Haldar et al., 2012). Concordant with the pattern observed during the light phase (i.e., when food intake and activity are decreased in mice), Klf15 expression acutely increases in BAT in response to fasting (Marmol et al., 2020; Nabatame et al., 2021). Additionally, mice that are chronically cold challenged at 4°C show significant upregulation of BAT Klf15 expression as well (Figure 1E). These responses to energetically demanding states suggest that KLF15 plays an important role in BAT metabolism.
K15-BKO mice demonstrate disturbances in lipid and amino acid metabolic pathways
To explore the role of KLF15 in BAT metabolism, we generated a BAT-specific KLF15 knockout (K15-BKO) mouse by mating the K15 F/F line with the Ucp1-cre line (The Jackson Laboratory) (Figure S1A). Of note, expression of Ucp1, which encodes the key protein responsible for BAT non-shivering thermogenesis, was unchanged between K15 F/F controls and K15-BKO BAT (Figure S1B). Measurement of core body temperature using implanted temperature telemetry devices showed appropriate oscillation of temperature throughout the 12 h day/night cycle with no significant differences between K15 F/F and K15-BKO mice (Figure S1C). Furthermore, K15-BKO mice were developmentally normal and demonstrated similar weight gain, insulin sensitivity, and glucose tolerance compared to K15 F/F at 8 weeks of age (Figures S1D–S1F).
Interestingly, despite these findings, K15-BKO mice demonstrated a significant deviation from the transcriptional adaptions seen in response to cold and to fasting in K15 F/F BAT. We performed parallel RNA-seq studies on BAT isolated from K15-BKO mice at RT, after chronic cold challenge, and after an overnight fast. While between 40% and 55% of DEGs are shared between the two genotypes under each condition, a large portion of expected up- and downregulated genes (between 25% and 30% depending on stimulus) are lost in the absence of KLF15 (Figure S2).
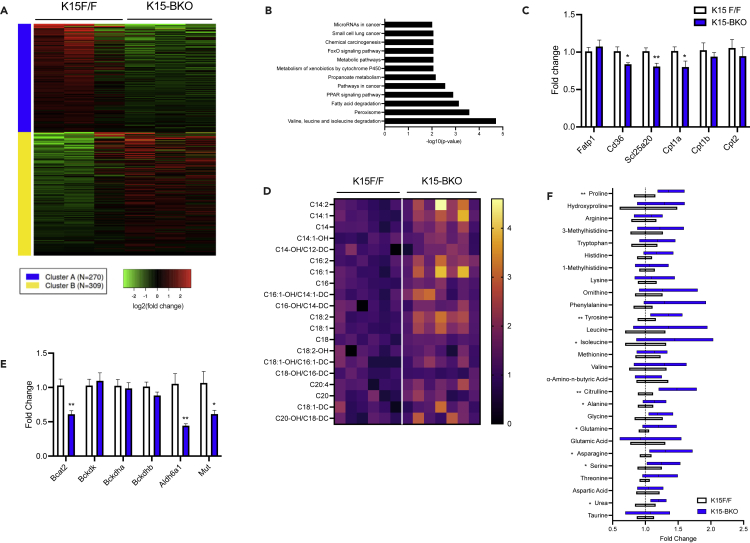
RNA-seq analysis comparing K15 F/F and K15-BKO BAT at RT identified 579 DEGs (Figure 2A). Pathway analysis of downregulated genes in K15-BKO BAT revealed a strong signature for metabolic pathways, fatty acid (FA) degradation, and PPAR signaling pathway, suggesting a fundamental defect in energy utilization pathways in K15-BKO BAT (Figure 2B). Indeed, a number of genes involved in lipid uptake into the cell and transport of long-chain acylcarnitines (LCACs) into the mitochondria (e.g. Cd36, Slc25a20, and Cpt1a) were downregulated in K15-BKO BAT (Figure 2C). These impairments in lipid uptake and LCAC mitochondrial transport were reflected in plasma acylcarnitine analysis using tandem mass spectrometry: K15-BKO mice demonstrated a significant increase in many circulating acylcarnitine species, particularly LCACs, which are a major fuel source for BAT thermogenesis (Figure 2D) (Simcox et al., 2017). In addition to lipids, recent studies have demonstrated that BCAA metabolism plays a significant role in BAT thermogenesis capabilities and to systemic BCAA clearance (Yoneshiro et al., 2019). We noted that BCAA metabolic pathways (valine, leucine, and isoleucine degradation) were also significantly enriched from K15 F/F vs K15-BKO RT RNA-seq studies (Figure 2B). In K15-BKO BAT, confirmatory qPCR analysis showed significant downregulation of genes involved in BCAA catabolism, including Bcat2, Aldh6a1, and Mut (Figure 2E). Of note, several plasma amino acids in K15-BKO mice were increased at baseline, suggesting either a block in amino acid utilization or increased proteolysis (Figure 2F).
Figure 2.
K15-BKO mice demonstrate disturbances in lipid and amino acid metabolic pathways
(A) Heatmap and k-means clustering of DEGs comparing K15 F/F and K15-BKO BAT at RT (n = 3).
(B) KEGG pathway analysis of downregulated genes in K15-BKO.
(C) qPCR of select lipid metabolism genes in BAT (n = 6–7).
(D) Heatmap depicting circulating acylcarnitine levels K15 F/F and K15-BKO at RT, normalized to K15 F/F RT (n = 7).
(E) qPCR of BCAA catabolism genes in BAT (n = 6–7).
(F) Circulating amino acid levels at RT, normalized to K15 F/F (n = 6). Data represent mean ± SEM. Comparisons between groups were performed using an unpaired, 2-tailed Student’s t test. ∗p < 0.05, ∗∗p < 0.01.
KLF15 is critical in the regulation of BAT response to cold
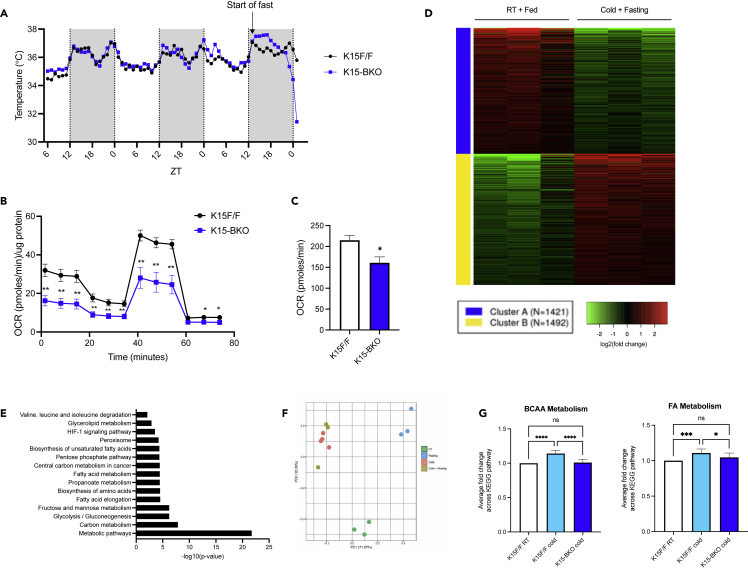
To assess BAT non-shivering thermogenesis capability, K15 F/F and K15-BKO mice were exposed to a chronic cold challenge for 10 days at 4°C. K15-BKO mice were surprisingly able to maintain body temperature comparable to K15 F/F (Figure S3A). We noted that past studies showed that substrates derived from food consumption are able to compensate for a lack of tissue-intrinsic fuels for BAT thermogenesis during cold exposure (Shin et al., 2017, 2018). Additionally, compared with the fed state, food deprivation induces a greater drop in body temperature throughout the circadian cycle (Tokizawa et al., 2009). Given that K15-BKO mice had increased food intake throughout the day (Figure S3B), we subjected mice to chronic cold challenge for 10 days followed by an overnight fast to explore BAT activity while excluding the potential effects of feeding. Strikingly, K15-BKO mice were incapable of maintaining body temperature in the absence of exogenous fuel sources and quickly succumbed to severe hypothermia within 12 h of fasting (Figure 3A). To gain further functional insights on how KLF15 affects BAT fuel consumption, we performed Seahorse assays in both cultured brown adipocytes and brown adipose tissue isolated from K15 F/F and K15-BKO mice. Loss of KLF15 resulted in significantly decreased oxygen consumption rates in both primary cells and whole tissue, reflecting an impaired ability to oxidize macronutrients (Figures 3B and 3C). Beiging or browning of white adipose tissue has been shown to have a profound impact on thermoregulation; however, subcutaneous white adipose tissue isolated from K15 F/F and K15-BKO did not reveal any significant differences in expression of browning genes, Klf15, or lipid metabolism genes (Figures S3C–S3E). These findings suggest that the observed phenotype is driven by the absence of BAT KLF15 specifically, and these animals appear to be unable to mount appropriate nutrient switches to maintain adequate thermogenesis in the absence of exogenous fuel sources.
Figure 3.
KLF15 is critical in the regulation of BAT response to cold
(A) Continuous core body temperature measurement using implantable temperature telemetry devices in K15 F/F vs K15-BKO during chronic cold challenge followed by fasting (n = 6).
(B) Seahorse Mito Stress assay depicting the oxygen consumption rate (OCR) from primary brown adipocytes isolated from K15 F/F and K15-BKO mice (n = 6). Brown adipocyte oxygen consumption rate was normalized to protein concentration per well.
(C) Basal oxygen consumption rate from BAT from K15 F/F and K15-BKO at RT (n = 6).
(D) K-means clustering of DEGs from RNA-seq of K15 F/F RT (fed) vs C + F (n = 3).
(E) KEGG pathway analysis of upregulated DEGs from RNA-seq of K15 F/F RT (fed) vs C + F.
(F) PCA of top 10,000 highest expressed genes in K15 F/F showing clustering of cold and C + F conditions together.
(G) Average fpkm of all genes within KEGG pathway gene set for BCAA metabolism and fatty acid metabolism for K15 F/F at RT, K15 F/F in cold, and K15-BKO in cold. Comparisons between groups were performed using unpaired, 2-tailed Student’s t test for panels (B) and (C) and two-way ANOVA for (G). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
We again turned to the K15 F/F control animal to investigate what transcriptional changes must occur in the context of cold exposure and fasting (C + F) to maintain body temperature. RNA-seq analysis resulted in 2913 DEGs, with significant upregulation of genes related to FA and BCAA metabolism (Figures 3D and 3E). Interestingly, principal component analysis revealed that the transcriptional response to C + F is much more like that of cold challenge alone than to fasting, further supported by a comparison of K15 F/F RNA-seq data between cold and C + F which only returned 302 DEGs (Figure 3F). Thus, when an animal is confronted with both cold and fasting stimuli, a state that is akin to field conditions, BAT thermogenic transcriptional programs predominate over those of fasting. This prioritization of macronutrient uptake and catabolism in BAT is necessary for stable body temperature and survival. In response to cold, K15-BKO mice demonstrate a significant decrease in overall expression of genes involved in both BCAA and FA metabolism pathways compared to K15 F/F mice. In fact, expression levels are similar to those seen in K15 F/F at RT, indicating that in the absence of KLF15, mice are unable to mount the appropriate metabolic transcriptional response to cold (Figure 3G).
BAT-intrinsic and extrinsic fuel sources are severely altered in K15-BKO mice
Recent work has uncovered elegant systems of BAT cross-organ signaling, substrate production, and BAT nutrient consumption for thermogenesis. For example, cold stimulates WAT lipolysis, and released FAs go on to activate transcriptional programs in the liver that lead to acylcarnitine production. These acylcarnitines then serve as important oxidative fuel for BAT thermogenesis (Simcox et al., 2017; Yoneshiro et al., 2019). We observed that in K15-BKO mice, there is significant downregulation of numerous BCAA and lipid metabolism genes in response to C + F in BAT (Figures S4A and S4B). This, paired with increases in circulating LCACs and amino acids downstream of these genes, suggests a disconnect between peripheral substrate production and BAT consumption (Figures S4C and S4D).
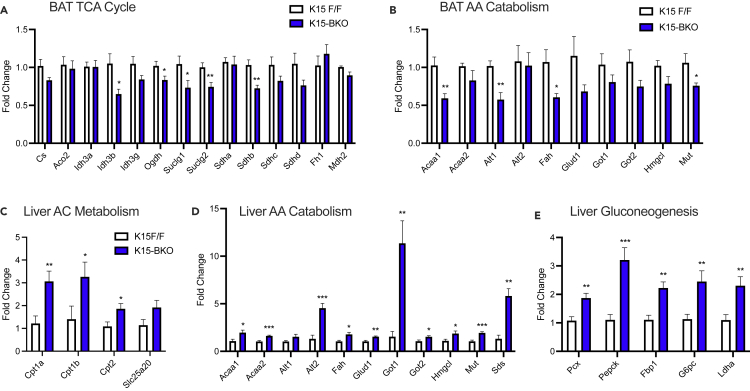
Both BAT-intrinsic nutrients (e.g., lipids through lipolysis) and those taken up from the circulation enter the TCA cycle and are necessary for non-shivering thermogenesis. Interestingly, we observed that numerous TCA cycle genes (e.g. Idh3b, Ogdh, Suclg1, Suclg2, and Sdhb) were significantly downregulated in K15-BKO compared to K15 F/F (Figure 4A). Additionally, the expression of several amino acid catabolism genes that participate in TCA cycle anaplerosis was also decreased in K15-BKO BAT. These include Mut and Acaa1, which encode for enzymes involved in BCAA catabolism and can generate succinyl-CoA and acetyl CoA, respectively; and Alt1 and Fah, which encode enzymes that can generate pyruvate and fumarate and acetoacetate, respectively (Figures 4 and S4). These data indicated that loss of BAT KLF15 results in impaired ability to utilize various macronutrients including amino acids, posing a challenge for the animal in nutrient scarce conditions.
Figure 4.
BAT-intrinsic and extrinsic fuel sources are severely altered in K15-BKO mice
(A) qPCR of genes showing fold change in K15-BKO relative to K15 F/F for (A) TCA cycle genes in BAT (n = 7–8).
(B) AA catabolism genes in BAT (n = 6–8).
(C) Acylcarnitine (AC) genes in liver (n = 6–8).
(D) AA catabolism genes in liver (n = 6–7).
(E) Gluconeogenesis genes in liver (n = 7). Data represent mean ± SEM. Comparisons between groups were performed using an unpaired, 2-tailed Student’s t test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Under the C + F condition, we observed substantial shifts in K15-BKO systemic metabolism in response to deficient BAT function. In the liver, K15-BKO animals had significant elevations in genes related to acylcarnitine metabolism (e.g. Cpt1a, Cpt1b, and Cpt2) and in numerous amino acid catabolic genes (e.g. Acaa1, Acaa2, Alt2, Fah, Glud1, Got1, Got2, Hmgcl, Mut, and Sds), likely as compensation for insufficient BAT function (Figures 4C and 4D). Interestingly, the most dramatic increases in gene expression occurred with respect to glucogenic amino acid catabolic genes (Alt2, Sds, and Got1). Subsequent qPCR analyses revealed upregulation of both Pcx and Pepck, in addition to other key gluconeogenesis genes (Fbp1 and G6pc) in K15-BKO liver (Figure 4E). Ldha expression, encoding lactate dehydrogenase, was also upregulated, suggesting that there may be increased utilization of lactate for glucose generation in K15-BKO mice. These data together suggest that K15-BKO mice require increased hepatic gluconeogenesis to maintain blood glucose levels with cold exposure and fasting. Thus, in the context of impaired BAT lipid and BCAA metabolism, there appears to be an increased reliance on the liver to provide substrates for systemic metabolic homeostasis.
Discussion
Brown adipose tissue is an instrumental mediator of systemic metabolic homeostasis through its partitioning and utilization of nutrients in response to various energetic demands. In the current study, we demonstrate the widespread transcriptional changes that take place in BAT to enable metabolic flexibility and survival when an organism is faced with challenges such as fasting and cold. Additionally, we establish BAT KLF15 as a critical driver of proper transcriptional and metabolic adaptations to cold exposure in the absence of exogenous fuel sources.
The transcription factor KLF15 has been shown to be a major regulator of metabolism and metabolic adaptions in a variety of tissues. For example, fasting induces Klf15 expression in liver and in skeletal muscle, where it acts to promote amino acid degradation for gluconeogenesis (Fan et al., 2021; Gray et al., 2007). In contrast, Klf15 expression in WAT decreases in response to fasting and acts to promote lipid storage and other anabolic functions in the fed state (Matoba et al., 2017). However, comparably little is known about KLF15 activity and function in BAT, which shares qualities of both skeletal muscle and WAT. Like muscle, BAT is mitochondria-laden, highly oxidative, and thermogenic. Yet, it can also robustly partake in FA synthesis, storage, and release much like WAT (Festuccia et al., 2011; Yu et al., 2002). Here, we found that concordant with past studies, BAT Klf15 expression is upregulated in fasting conditions like that in liver and muscle (Marmol et al., 2020; Nabatame et al., 2021). Furthermore, KLF15 exhibits a similar circadian rhythmicity across BAT, liver, and muscle, which corresponds with times of relative nutrient abundance and deficit, suggesting a similar function in energy mobilization and utilization.
While the peripheral response to cold and fasting are similar in many regards (i.e., mobilization of fats and glucose from WAT and liver, breakdown of amino acids in skeletal muscle), we find here that BAT transcriptional programs in response to cold and to fasting are drastically different. Cold challenge induced expression of transcriptional programs involved in the metabolism of all three macronutrients, especially that of lipid metabolism, while fasting suppressed transcription of genes involved in glucose metabolism, presumably to preserve circulating glucose for glucose-obligate organs in the face of nutrient scarcity. Interestingly, in fasting, we observed that genes involved in thermogenesis were also downregulated, which prompted the question of what happens when animals are exposed to both fasting and cold, a condition that is frequently endured in the wild. Our RNA-seq studies revealed that the combination of cold and fasting induced a transcriptional profile that is more like that of cold alone than that of fasting alone, suggesting that there may be biological prioritization to address adequate thermogenesis.
Our study utilized an in vivo BAT-specific KLF15 knockout model for the first time to demonstrate the effects of BAT KLF15 deficiency on whole organism physiology. In concordance with past cell-based studies, we found that lack of KLF15 in BAT led to widespread downregulation of BAT lipid and BCAA metabolism as well as systemic impacts such as increases in liver acylcarnitine metabolism and gluconeogenesis, likely as a compensatory mechanism for insufficient BAT function (Nabatame et al., 2021; Simcox et al., 2017). Furthermore, we see clear evidence of decreased macronutrient oxidation as lack of BAT KLF15 resulted in substantially decreased oxygen consumption rates in both primary cultures and whole tissue. These aberrations ultimately resulted in a striking hypothermic phenotype in response to resource-limited conditions as K15-BKO mice are unable to properly utilize the required nutrients for thermogenesis. A recent study by Marmol et al. (2020) provides an important and interesting complement to our current knockout studies. They found that BAT-specific loss of activin receptor ALK7 results in increased BAT expression of KLF15 with subsequent increases in BAT lipid and amino acid catabolism in conditions of nutrient stress (Marmol et al., 2020). Taken together, our findings and those of Marmol et al. (2020), establish a role for KLF15 in the regulation of macronutrient metabolism in the context of BAT adaptation in response to varying energetic demands.
In summary, our study demonstrates that 1) the predominant metabolic transcriptional programs in BAT change vastly depending on stimulus; 2) BAT prioritizes thermogenesis when confronted with competing stimuli; and 3) KLF15 is a critical regulator of BAT nutrient metabolism and thermogenesis. Without KLF15, BAT is unable to drive the activation and balance of transcriptional programs needed for proper function in response to energetically challenging states. Our work sheds light on the selectivity and metabolic flexibility of BAT fuel utilization and provides a more complete understanding of KLF15 as an orchestrator of systemic metabolism.
Limitations of the study
In the present study, we showed that KLF15 deficiency in brown adipose tissue and brown adipocytes results in markedly decreased oxygen consumption, serving as a measure for altered BAT metabolism. In vivo assessment of BAT metabolism in the absence of KLF15 under cold, fasting, and cold + fasting would be ideal. However, current techniques are limited by availability of metabolic cages capable of maintaining adequate cold challenge temperatures as well as conceptual issues of using radiolabeled-glucose positron emission tomography scans in assessing metabolic alterations in the setting of cold exposure and fasting. Additionally, while we observed an increase in measured circulating amino acids and long-chain fatty acids, we have not yet directly established where these metabolites come from (e.g. build up in the BAT, increased intestinal absorption, or increased production from other peripheral organs). Future studies using radiolabeled substrates in the setting of cold, fasting, and cold + fasting would shed light on this question to further clarify the interplay of different organs in response to energetic stressors and provide additional insights with regard to metabolic flux.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Collagenase, Type 1 | Worthington Biochemical Corporation | Cat#LS004196 |
| Bovine Serum Albumin | Thermo Fisher Scientific | Cat#BP1600-1 |
| Insulin solution | Sigma-Aldrich | Cat#I9278 |
| Rosiglitazone | Sigma-Aldrich | Cat#557366 |
| 3,3′,5-Triiodo-L-thyronine (T3) | Sigma-Aldrich | Cat#T2877 |
| HEPES | Sigma-Aldrich | Cat#H4034 |
| D-(+)-Glucose | Sigma-Aldrich | Cat#G8270 |
| D-(+)-Glucose solution | Sigma-Aldrich | Cat#G8769 |
| Sodium pyruvate solution | Sigma-Aldrich | Cat#S8636 |
| L-Glutamine solution | Sigma-Aldrich | Cat#G7513 |
| Critical commercial assays | ||
| Aurum Total RNA Fatty and Fibrous Tissue Kit | Bio-Rad | Cat#7326830 |
| iScript Reverse Transcription Supermix | Bio-Rad | Cat#1708841 |
| Seahorse XFp Cell Mito Stress Test Kit | Agilent | Cat#103010-100 |
| Seahorse XFp FluxPak | Agilent | Cat#103022-100 |
| Seahorse XFe24 Islet Capture FluxPak | Agilent | Cat#103518-100 |
| Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#23227 |
| Deposited data | ||
| RNA sequencing data | NCBI: Gene Expression Omnibus | NCBI GEO: GSE178720 |
| CircaDB: Circadian gene expression profiles data base | Publicly available | http://circadb.hogeneschlab.org/mouse |
| Experimental models: Cell lines | ||
| Primary mouse brown adipocytes | mouse brown adipose tissue | |
| Experimental models: Organisms/strains | ||
| Mouse: K15F/F:Klf15tm2Jain | Jain laboratory | |
| Mouse: K15-BKO: Klf15tm2Jain x B6.FVB-Tg(Ucp1-cre)1Evdr/J | This manuscript | |
| Mouse: Ucp1-cre: B6.FVB-Tg(Ucp1-cre)1Evdr/J | Jackson Laboratories | |
| Oligonucleotides | ||
| See Table S1 | ||
| Software and algorithms | ||
| Prism 9 | GraphPad Prism | |
| TopHat2 | Johns Hopkins University Center for Computational Biology | |
| Cufflinks | Cole Trapnell’s lab - University of Washington | |
| Shinyheatmap | https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0176334 | |
| iDEP.96 | https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-018-2486-6 | |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Mukesh Jain (Mukesh_jain@brown.edu).
Materials availability
The K15-BKO (Klf15tm2Jain x B6.FVB-Tg(Ucp1-cre)1Evdr/J) mouse model is available from our laboratory with a completed Material Transfer Agreement.
Experimental model and subject details
Mice
The brown adipose specific KLF15 knockout (K15-BKO) mouse was created by mating the KLF15flox/flox (K15 F/F) line with the B6.FVB-Tg(Ucp1-cre)1Evdr/J line (Ucp1-cre) from Jackson Laboratories. The K15 F/F (Klf15tm2Jain) mouse line was generated by Ozgene by inserting LoxP sites flanking exon 2 of the Klf15 gene and maintained at our facilities as an inbred strain (Lu et al., 2013). All K15-BKO mice have been backcrossed to KLF15flox/flox for >6 generations. Age matched, male, non-littermate K15 F/F mice were used as controls for all experiments utilizing K15-BKO mice. Adult male mice approximately 8–12 weeks in age were utilized for all experiments. Mice are kept in a temperature-controlled facility at 22°C, on a daily 12 h light/dark schedule, fed with tap water and standard chow ad libitum. Animals were weighed weekly for body weight assessment. Food intake was monitored by food hopper weight throughout 5 days of measurements. Where indicated, mice were subjected to 4°C cold challenge for 10 days, kept on a daily 12 h light/dark schedule, fed with tap water ad libitum and standard chow. For fasting experiments, mice were fasted for 12 h overnight with free access to water. For cold exposure experiments and temperature telemetry experiments, mice were singly housed. All experiments involving animals were conducted under protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Case Western Reserve University.
Primary murine cell culture
Brown preadipocytes were isolated from the interscapular, axillary, and cervical BAT depots of K15 F/F and K15-BKO male mice. Tissues were minced and then digested in collagenase solution (0.2% collagenase I (Worthington), 2% BSA in DMEM high glucose, pyruvate (Gibco)). The minced tissue-collagenase solution was incubated at 37οC in a shaking water bath (100 rpm) for 30 min and vortexed every 5 min. The cells were then filtered through a 40uM filter (Falcon), following addition of 100% filtered FBS. Cells were spun down and resuspended in growth media (DMEM high glucose, pyruvate, 15% FBS, 1%PSG). Cells were grown to confluency for use in subsequent mitochondrial assays. For mitochondrial assays, cells were switched to differentiation media (growth medium containing 50nM insulin, 5nM T3, and 1 uM rosiglitazone (Sigma)). Cells were kept in a carbon dioxide incubator (Thermo) with 10% CO2 at 37°C.
Method details
Intraperitoneal glucose and insulin tolerance tests (IPGTT and IPITT)
Intraperitoneal injection of glucose (2 g/kg body weight) or insulin (0.5 U/kg body weight) was performed following 5 h fast. Blood glucose was measured at baseline, 15, 30, 60, 90, and 120 min by tail bleed by glucometer reading.
RNA isolation and quantitative real-time PCR
Tissue samples were disrupted in PureZOL in a Tissue-lyzer (Qiagen) using stainless steel beads (30Hz for total 2 min). Total RNA was isolated using Bio-Rad Aurum Total RNA Fatty and Fibrous Tissue Kit using manufacturer’s instructions and transcribed to cDNA using iScript (Bio-Rad). Quantitative real-time PCR was performed using Taqman method and appropriate probes from Roche Universal Probe Library System. Gene expression was normalized to cyclophilin B and compared using ΔΔCt method. All primers were efficiency tested and validated. Primer sequences used in this study can be found in Table S1.
RNA-sequencing
Brown adipose tissue (BAT) was isolated from K15 F/F and K15-BKO mice under room temperature, cold, fasting, and cold + fasting conditions. RNA isolation was performed, as described above. cDNA sequencing libraries were prepared by Macrogen NGS using the TruSeq Stranded Total RNA with Ribo-Zero Gold Human/Mouse/Rat kit and sequenced on an Illumina platform. Reads from each sample were aligned to the mouse genome (UCSC mm10) using TopHat2 (Kim et al., 2013). Gene expression was quantitated as fragments per kilobase of transcript per million mapped reads (FPKM) using Cufflinks (Trapnell et al., 2010). Differentially expressed genes (DE-Gs) were identified with an adjusted p value cutoff q < 0.05. DE-Gs were input into shinyheatmap to generate heatmap clustering (Khomtchouk et al., 2017). Significantly enriched KEGG were generated by iDEP (Ge et al., 2018). The RNA sequencing data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and available through GSE178720.
Cell-based mitochondrial assay
Upon confluence, the brown preadipocytes were passaged into a Seahorse XFp cell culture miniplate (1500 cells/well) and kept in growth media for 24 h. Media was then switched to differentiation media (growth medium containing 50nM insulin, 5nM T3, and 1 uM rosiglitazone (Sigma)) for 4–6 days, with daily media changes. Brown adipocyte mitochondrial respiration rate was evaluated with the Seahorse XFp Cell Mito Stress Kit utilizing the Agilent Seahorse XFp Analyzer (Agilent). On the day of the assay, cells were washed once with Seahorse assay media (Seahorse XF base medium minimal DMEM (Agilent) supplemented with 25mM glucose, 1.0mM pyruvate, 3.97mM glutamine (Sigma), pH adjusted to 7.4) prior to the assay. Oxygen consumption rate was assessed with 3 measurement cycles (each comprised of 3 min mixing and 3 min measuring) for the basal oxygen consumption rate and following the respective injections of oligomycin, FCCP, and rotenone and antimycin A. The final per well drugs concentrations used were 5uM oligomycin, 1uM FCCP, and 1uM rotenone + antimycin A (Agilent). After the assay, cells were rinsed with PBS and collected into RIPA buffer for protein quantification. Samples were kept on ice, vortexed every 10 min for 30 min, and then centrifuged at 4°C for 10 min at 15,000g. The supernatant was collected, and protein was quantified with Pierce BCA assay kit (Thermo). All cell-based assays were run in duplicate and then averaged.
Brown adipose tissue-based mitochondrial assay
A single interscapular BAT depot was harvested from K15 F/F and K15-BKO mice. A 1.2mm Uni-Core biopsy puncher (Qiagen) was used to generate 5 equally sized tissue pieces per animal. Each tissue punch was placed in one well of an XF24 islet capture microplate and covered with an islet screen (Agilent). Then, 450uL of wash media (DMEM, high glucose, pyruvate (Gibco) supplemented with 25X10−3M glucose and 25X10−3M HEPES) was added to each well. Tissues pieces were washed once with wash media and then once with assay media (Seahorse XF base medium minimal DMEM supplemented (Agilent) with 25mM glucose, 1.0mM pyruvate, 3.97mM glutamine (Sigma), pH adjusted to 7.4). The plate was then incubated at 37°C for 45 min with 450uL of assay media per well. Basal oxygen consumption rate was measured with the Seahorse XFe24 Analyzer and corresponding XFe24 sensor cartridges (Agilent). Basal oxygen consumption was assessed with 10 measurement cycles, each comprised of 3 min of mixing, 2 min of waiting, and 3 min of measuring. The average of the 5 technical replicates was taken and each measurement was averaged to determine the basal oxygen consumption rate.
Acylcarnitine measurements
Whole blood was collected from K15 F/F and K15-BKO mice, spun down at 2000rpm for 20 min, and plasma was collected. Plasma acylcarnitine species were analyzed by tandem mass spectrometry using sample preparation methods described previously (An et al., 2004; Ferrara et al., 2008). The data were acquired using a Waters TQD mass spectrometer equipped with Acquity UPLC system and controlled by MassLynx 4.1 operating system (Waters, Milford, MA).
Amino acids measurements
Whole blood was collected from K15 F/F and K15-BKO mice in heparin and spun down at 2000rpm for 20 min. Plasma was collected, frozen, and sent to Vanderbilt MMPC for analysis, which was performed by HPLC using Biochrom 30 amino acid analyzer.
Temperature telemetry
Mice were anesthetized using isoflurane. A midline incision was made to open the abdomen, and temperature telemetry transmitters (DSI, TA-F10) were implanted into the abdominal cavity of animals. Incision was then closed with 7-0 suture, and mice were kept on heat for recovery and given analgesia for 72 h. Mice were then singly caged, and continuous temperature monitoring was started 2-week post-surgery once animals fully recovered.
Quantification and statistical analysis
Assays were performed independently and data plotted showing mean +/− SE. Statistical comparisons were made by either using 2-way ANOVA or student’s t-test, as indicated in the figure legends, with GraphPad Prism 9. The significance of difference was set at p values <0.05. In all figures, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. RNA-seq data analyzed using methods detailed above. Additional statistical details of experiments, including sample sizes, tests used, significance levels, and technical replicates can be found within figure legends and in the method details.
Acknowledgments
This work was supported by NIH grants R35HL135789 (M.K.J.), R01HL086548 (M.K.J.), R01DK111468/RES511821 (M.K.J. and C.B.N.), T32GM007250 (L.F., D.R.S., and A.F.L.), T32HL134622 (L.F. and A.F.L.), F30HL139014 (DRS). This work was also supported by American Heart Association Established Investigator Award (M.K.J.) and AHA-Allen Frontiers Award (M.K.J.) and Leducq Foundation Transatlantic Network of Excellence Award (M.K.J.).
Author contributions
L.F., A.F.L., D.R.S., and M.K.J. designed the studies. L.F., A.F.L., D.R.S., K.S.K., Y.L., O.I., T.D., and C.E.B. performed the experiments. L.F., A.F.L., K.S.K., E.R.C., and V.V. analyzed the data. D.R.S. and M.J.K. provided critical research guidance. L.F. and A.F.L. prepared the figures and drafted the manuscript. D.R.S. and M.K.J. edited the manuscript. M.K.J. approved the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: November 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105292.
Supplemental information
Data and code availability
-
•
RNA sequencing data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Adlanmerini M., Carpenter B.J., Remsberg J.R., Aubert Y., Peed L.C., Richter H.J., Lazar M.A. Circadian lipid synthesis in brown fat maintains murine body temperature during chronic cold. Proc. Natl. Acad. Sci. USA. 2019;116:18691–18699. doi: 10.1073/pnas.1909883116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J., Muoio D.M., Shiota M., Fujimoto Y., Cline G.W., Shulman G.I., Koves T.R., Stevens R., Millington D., Newgard C.B. Hepatic expression of malonyl-CoA decarboxylase reverses muscle, liver and whole-animal insulin resistance. Nat. Med. 2004;10:268–274. doi: 10.1038/nm995. [DOI] [PubMed] [Google Scholar]
- Bartelt A., Bruns O.T., Reimer R., Hohenberg H., Ittrich H., Peldschus K., Kaul M.G., Tromsdorf U.I., Weller H., Waurisch C., et al. Brown adipose tissue activity controls triglyceride clearance. Nat. Med. 2011;17:200–205. doi: 10.1038/nm.2297. [DOI] [PubMed] [Google Scholar]
- Cannon B., Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- Chappuis S., Ripperger J.A., Schnell A., Rando G., Jud C., Wahli W., Albrecht U. Role of the circadian clock gene Per2 in adaptation to cold temperature. Mol. Metabol. 2013;2:184–193. doi: 10.1016/j.molmet.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatamra K., Daniel P.M., Lam D.K.C. The effects of fasting on core temperature, blood glucose and body and organ weights in rats. Q. J. Exp. Physiol. 1984;69:541–545. doi: 10.1113/expphysiol.1984.sp002840. [DOI] [PubMed] [Google Scholar]
- Chondronikola M., Volpi E., Børsheim E., Porter C., Saraf M.K., Annamalai P., Yfanti C., Chao T., Wong D., Shinoda K., et al. Brown adipose tissue activation is linked to distinct systemic effects on lipid metabolism in humans. Cell Metabol. 2016;23:1200–1206. doi: 10.1016/j.cmet.2016.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CIRCA Circadian gene expression profiles. http://circadb.hogeneschlab.org/mouse
- Dong M., Lin J., Lim W., Jin W., Lee H.J. Role of brown adipose tissue in metabolic syndrome, aging, and cancer cachexia. Front. Med. 2018;12:130–138. doi: 10.1007/s11684-017-0555-2. [DOI] [PubMed] [Google Scholar]
- Fan L., Sweet D.R., Prosdocimo D.A., Vinayachandran V., Chan E.R., Zhang R., Ilkayeva O., Lu Y., Keerthy K.S., Booth C.E., et al. Muscle Krüppel-like factor 15 regulates lipid flux and systemic metabolic homeostasis. J. Clin. Invest. 2021;131 doi: 10.1172/JCI139496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara C.T., Wang P., Neto E.C., Stevens R.D., Bain J.R., Wenner B.R., Ilkayeva O.R., Keller M.P., Blasiole D.A., Kendziorski C., et al. Genetic networks of liver metabolism revealed by integration of metabolic and transcriptional profiling. PLoS Genet. 2008;4:e1000034. doi: 10.1371/journal.pgen.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festuccia W.T., Blanchard P.-G., Deshaies Y. Control of Brown adipose tissue glucose and lipid metabolism by PPARγ. Front. Endocrinol. 2011;2 doi: 10.3389/fendo.2011.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S.X., Son E.W., Yao R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinf. 2018;19:534. doi: 10.1186/s12859-018-2486-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S., Wang B., Orihuela Y., Hong E.-G., Fisch S., Haldar S., Cline G.W., Kim J.K., Peroni O.D., Kahn B.B., Jain M.K. Regulation of gluconeogenesis by krüppel-like factor 15. Cell Metabol. 2007;5:305–312. doi: 10.1016/j.cmet.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haemmerle G., Lass A., Zimmermann R., Gorkiewicz G., Meyer C., Rozman J., Heldmaier G., Maier R., Theussl C., Eder S., et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 2006;312:734–737. doi: 10.1126/science.1123965. [DOI] [PubMed] [Google Scholar]
- Haldar S.M., Jeyaraj D., Anand P., Zhu H., Lu Y., Prosdocimo D.A., Eapen B., Kawanami D., Okutsu M., Brotto L., et al. Kruppel-like factor 15 regulates skeletal muscle lipid flux and exercise adaptation. Proc. Natl. Acad. Sci. USA. 2012;109:6739–6744. doi: 10.1073/pnas.1121060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh P.N., Fan L., Sweet D.R., Jain M.K. The krüppel-like factors and control of energy homeostasis. Endocr. Rev. 2019;40:137–152. doi: 10.1210/er.2018-00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S., Saito M. A new era in brown adipose tissue biology: molecular control of brown fat development and energy homeostasis. Annu. Rev. Physiol. 2014;76:225–249. doi: 10.1146/annurev-physiol-021113-170252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khomtchouk B.B., Hennessy J.R., Wahlestedt C. shinyheatmap: ultra fast low memory heatmap web interface for big data genomics. PLoS One. 2017;12:e0176334. doi: 10.1371/journal.pone.0176334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Zhang L., Liao X., Sangwung P., Prosdocimo D.A., Zhou G., Votruba A.R., Brian L., Han Y.J., Gao H., et al. Kruppel-like factor 15 is critical for vascular inflammation. J. Clin. Invest. 2013;123:4232–4241. doi: 10.1172/JCI68552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado F.S.M., Zhang Z., Su Y., de Goede P., Jansen R., Foppen E., Coimbra C.C., Kalsbeek A. Time-of-Day effects on metabolic and clock-related adjustments to cold. Front. Endocrinol. 2018;9 doi: 10.3389/fendo.2018.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmol P., Krapacher F., Ibáñez C.F. Control of brown adipose tissue adaptation to nutrient stress by the activin receptor ALK7. Elife. 2020;9:e54721. doi: 10.7554/eLife.54721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba K., Lu Y., Zhang R., Chen E.R., Sangwung P., Wang B., Prosdocimo D.A., Jain M.K. Adipose KLF15 controls lipid handling to adapt to nutrient availability. Cell Rep. 2017;21:3129–3140. doi: 10.1016/j.celrep.2017.11.032. [DOI] [PubMed] [Google Scholar]
- Nabatame Y., Hosooka T., Aoki C., Hosokawa Y., Imamori M., Tamori Y., Okamatsu-Ogura Y., Yoneshiro T., Kajimura S., Saito M., Ogawa W. Kruppel-like factor 15 regulates fuel switching between glucose and fatty acids in brown adipocytes. J. Diabetes Invest. 2021;12:1144–1151. doi: 10.1111/jdi.13511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedergaard J., Cannon B. [3H]GDP binding and thermogenin amount in brown adipose tissue mitochondria from cold-exposed rats. Am. J. Physiol. Cell Physiol. 1985;248:C365–C371. doi: 10.1152/ajpcell.1985.248.3.C365. [DOI] [PubMed] [Google Scholar]
- Saito M., Matsushita M., Yoneshiro T., Okamatsu-Ogura Y. Brown adipose tissue, diet-induced thermogenesis, and thermogenic food ingredients: from mice to men. Front. Endocrinol. 2020;11 doi: 10.3389/fendo.2020.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M., Okamatsu-Ogura Y., Matsushita M., Watanabe K., Yoneshiro T., Nio-Kobayashi J., Iwanaga T., Miyagawa M., Kameya T., Nakada K., et al. High incidence of metabolically active Brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes. 2009;58:1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H., Ma Y., Chanturiya T., Cao Q., Wang Y., Kadegowda A.K.G., Jackson R., Rumore D., Xue B., Shi H., et al. Lipolysis in Brown adipocytes is not essential for cold-induced thermogenesis in mice. Cell Metabol. 2017;26:764–777.e5. doi: 10.1016/j.cmet.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H., Shi H., Xue B., Yu L. What activates thermogenesis when lipid droplet lipolysis is absent in brown adipocytes? Adipocyte. 2018;7:143–147. doi: 10.1080/21623945.2018.1453769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simcox J., Geoghegan G., Maschek J.A., Bensard C.L., Pasquali M., Miao R., Lee S., Jiang L., Huck I., Kershaw E.E., et al. Global analysis of plasma lipids identifies liver-derived acylcarnitines as a fuel source for brown fat thermogenesis. Cell Metabol. 2017;26:509–522.e6. doi: 10.1016/j.cmet.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford K.I., Middelbeek R.J.W., Townsend K.L., An D., Nygaard E.B., Hitchcox K.M., Markan K.R., Nakano K., Hirshman M.F., Tseng Y.-H., Goodyear L.J. Brown adipose tissue regulates glucose homeostasis and insulin sensitivity. J. Clin. Invest. 2013;123:215–223. doi: 10.1172/JCI62308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokizawa K., Uchida Y., Nagashima K. Thermoregulation in the cold changes depending on the time of day and feeding condition: physiological and anatomical analyses of involved circadian mechanisms. Neuroscience. 2009;164:1377–1386. doi: 10.1016/j.neuroscience.2009.08.040. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen G.H.E.J., Bouvy N.D., Teule G.J.J., Brans B., Schrauwen P., Lichtenbelt W.D. van M. Brown adipose tissue in morbidly obese subjects. PLoS One. 2011;6:e17247. doi: 10.1371/journal.pone.0017247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneshiro T., Wang Q., Tajima K., Matsushita M., Maki H., Igarashi K., Dai Z., White P.J., McGarrah R.W., Ilkayeva O.R., et al. BCAA catabolism in brown fat controls energy homeostasis through SLC25A44. Nature. 2019;572:614–619. doi: 10.1038/s41586-019-1503-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.X., Lewin D.A., Forrest W., Adams S.H. Cold elicits the simultaneous induction of fatty acid synthesis and β-oxidation in murine brown adipose tissue: prediction from differential gene expression and confirmation in vivo. Faseb. J. 2002;16:155–168. doi: 10.1096/fj.01-0568com. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
RNA sequencing data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.