Summary
Eukaryotes express three nuclear RNA polymerases (Pols I, II, and III) that are essential for cell survival. Despite extensive investigation of the three Pols, significant knowledge gaps regarding their biochemical properties remain because each Pol has been evaluated independently under disparate experimental conditions and methodologies. To advance our understanding of the Pols, we employed identical in vitro transcription assays for direct comparison of their elongation rates, elongation complex (EC) stabilities, and fidelities. Pol I is the fastest, most likely to misincorporate, forms the least stable EC, and is most sensitive to alterations in reaction buffers. Pol II is the slowest of the Pols, forms the most stable EC, and negligibly misincorporated an incorrect nucleotide. The enzymatic properties of Pol III were intermediate between Pols I and II in all assays examined. These results reveal unique enzymatic characteristics of the Pols that provide new insights into their evolutionary divergence.
Subject areas: Biochemistry, Molecular genetics, Evolutionary mechanisms
Graphical abstract

Highlights
-
•
Transient-state kinetic analyses reveal the mechanism of AMP incorporation by Pol III
-
•
In vitro biochemical assays uncover unique properties of Pols I, II, and III
Biochemistry; Molecular genetics; Evolutionary mechanisms
Introduction
In contrast to Bacteria and Archaea that express a single RNA polymerase, eukaryotes express multiple, nuclear, DNA-dependent RNA polymerases (Pols) with specialized transcriptional responsibilities (Werner, 2007; Werner and Grohmann, 2011; Kwapisz et al., 2008). Despite the discovery of the Pols over five decades ago (Roeder and Rutter, 1969), few comparative studies have been performed between the Pols (Lisica et al., 2016; Fan et al., 2005). Therefore, we lack a full understanding of their divergent enzymatic properties. Understanding the distinct biochemical properties of the Pols is critical to our understanding of eukaryotic gene expression.
Pol I is a 14-subunit enzyme (Engel et al., 2013; Fernández-Tornero et al., 2013) responsible for synthesis of the 35 S precursor ribosomal RNA (rRNA) from the ribosomal DNA (rDNA) in the nucleolus (Petes, 1979). The rDNA is organized in ∼200 tandem repeats in yeast, each 9.1 kb in length (Houseley and Tollervey, 2011; Nomura et al., 2000). In actively growing cells, approximately half of the repeats are undergoing Pol I transcription at any given time. These active repeats are thought to be free of ordered nucleosomes (Conconi et al., 1989; Merz et al., 2008; Rattner et al., 1982; Scheer, 1978). Pol I is regulated by a Pol I-specific factor, Rrn3, during transcription initiation in a growth dependent manner (Milkereit and Tschochner, 1998). In rapidly growing yeast cells, ribosomes can make up a staggering 40% of the total cells’ volume (Warner, 1999). Sixty percent of total transcription is committed to rRNA synthesis (Warner, 1999). As a result, Pol I transcription is among the most energetically demanding activities of eukaryotic cells (Warner, 1999; Laferté et al., 2006).
Pol II synthesizes messenger RNA (mRNA) and most regulatory non-coding RNAs including long non-coding RNA, micro-RNA, small nuclear RNA, and small nucleolar RNA. The length of Pol II-synthesized transcripts in yeast vary substantially, ranging from 20 to over 4000 nt (Hurowitz and Brown, 2003; Lee et al., 2004). Unlike the largely nucleosome-free rDNA transcribed by Pol I, Pol II-transcribed loci are bound by nucleosomes (Studitsky et al., 2004; Kulaeva et al., 2013). Pol II and Pol II gene elements recruit an extensive number of transcription-associated proteins and transcription factors (TFs) to finely control gene expression (Petesch and Lis, 2012; Hahn, 2004; Schier and Taatjes, 2020; Burton, 2014). Despite being responsible for the majority of eukaryotic genes, the relative activity of Pol II is dwarfed by the activities of Pols I and III that make up over 80% of the transcriptional activity in a growing yeast cell (Paule and White, 2000; Warner, 1999).
Finally, Pol III synthesizes small, non-coding RNAs, including transfer RNA (tRNA), 5 S rRNA, U6 spliceosomal RNA, and 7SL1 RNA of the signal recognition particle (Dieci et al., 2007, 2013). Pol III transcribes more genes than Pol I, but fewer than Pol II, there are approximately 300 Pol III-transcribed genes in yeast (Harismendy et al., 2003). Although Pol III transcribes a diverse subset of genes, the length of its RNA products is consistently short, approximately 100 nt (Turowski et al., 2016). Half of Pol III transcribed genes are nucleosome-free, whereas the other half are nucleosome-bound (Shukla and Bhargava, 2018). Pol III transcription is primarily controlled through three distinct promoters (Type 1, 2, and 3) (Schramm and Hernandez, 2002; Geiduschek and Kassavetis, 2001) that recruit different combinations of Pol III TFs (Acker et al., 2013; Huang and Maraia, 2001; Arimbasseri et al., 2014) and a negative regulator, Maf1 (Boguta et al., 1997). Like Pol I, Pol III’s activity is highly specialized for synthesis of translation machinery; therefore, it is tightly linked to cell proliferation (Płonka et al., 2019).
Over the course of evolution, we hypothesize that eukaryotic Pols have evolved distinct biochemical properties that specialize them for their unique cellular roles. To define and compare the enzymatic properties of the Pols, we have examined the mechanisms of transcription using transient-state kinetic techniques under defined experimental conditions. This approach fills a void in the gene expression field because it allows for direct comparisons between the Pols. The data revealed that Pol I is the fastest of the Pols, but highly sensitive to the DNA environment and reaction conditions. Although Pol II was the slowest Pol, it formed the most stable EC, and was the least error prone. Pol III shares biochemical properties similar to Pols I and II. Pol III was second to either Pol I or II regarding single-nucleotide addition speed, EC stability, fidelity, and sensitivity to varying reaction conditions. These data reveal that Pols I, II, and III possess distinct biochemical properties and enhance our understanding of their evolutionary divergence.
Results
Model-independent analysis of Pol III single-nucleotide addition
To elucidate the mechanism of Pol III nucleotide addition, we utilized a rapid mixing approach previously utilized for the investigation of Pols I and II (Appling et al., 2015; Jacobs et al., 2021). Pol III was purified from Saccharomyces cerevisiae (yeast) and incubated with a pre-annealed hybrid composed of a 9-mer RNA and a complementary 64 nt DNA template strand (DNAt), in buffer A (Appling et al., 2015). Elongation complexes (ECs) were formed with the addition of the complementary DNA non-template strand, DNAnt (Figure S1). The 9-mers were 3′ radiolabeled with α-32P-CTP and Mg2+, generating 10-mers and the reaction was halted before collecting time points. The halted radiolabeled ECs were loaded into one syringe of a mixing instrument, the chemical-quenched flow. An NTP mix containing ATP and Mg2+ was loaded opposite of the EC syringe. ECs and NTPs were rapidly mixed together and allowed to incubate for fixed amounts of time, (0.01–200) s. Reactions were halted with 1 M HCl and subsequently loaded into sequencing gels to separate the RNAs by length. Gels were exposed to phosphor screens, imaged, and analyzed.
In the image of the gel, we resolve 10-mers, generating during the labeling step, and 11-mers, synthesized with the addition of AMP in the chemical-quenched flow (Figure 1A). Based on our DNAt, we expected incorporation of a single AMP (Figure S1 and STAR methods). Surprisingly, in our last three time points, (60, 100, 200) s, we observe bands that correspond to 12-mers. This observation was a consequence of either trace GTP contamination in ATP stocks or misincorporation of AMP. We are unable to differentiate between the two possibilities. Because the amount of 12-mer was less than 5% of the product formed, we summed the 12-mer signal with the 11-mer signal. The logic being that any 12-mers were previously 11-mers. Therefore, there is no loss of information in the time course for the 11-mer. Two dinucleotide species were also observed (Figure 1A). The GC and CA dimers are present because of Pol III’s intrinsic nuclease activity, conferred by the C11 subunit. Unlike the CA dimer signal that increases over time, the GC dimer signal is constant over time course because it was formed during the labeling step.
| (Equation 1) |
| (Equation 2) |
Figure 1.
Pol III AMP addition time courses
(A) Representative gel of Pol III AMP addition at 50 μM ATP.
(B) Plot of the fraction of 11-mer + 12-mer over time at 50 μM ATP, fit to Equation 1.
(C) Plot of the fraction of CA dimer over time at 50 μM ATP, fit to Equation 2.
(D) 11-mer + 12-mer time courses collected as a function of [ATP] and fit to Equation 1.
(E) CA dimer time courses collected as a function [ATP] and fit according to Equation 2.
(F) kobs,addition plotted as a function of [ATP]. Data were fit to Equation 3.
(G–J) Observed rate constants, kobs,decay,1, kobs,decay,2, kobs,cleavage,1, and kobs,cleavage,2, plotted as a function of [ATP]. Data were fit using weighted NLLS to a constant. All time courses were collected in triplicate and the average is plotted with error bars corresponding to the standard deviation about the mean.
To determine the observed rate constants governing Pol III AMP addition, we plotted the fraction of 11-mer + 12-mer over time (Figure 1B). We observed a rise, (∼0.01 – ∼0.1) s, and biphasic decay, (∼0.2 – ∼6) s and (∼6 – ∼200) s. Therefore, the data were fit to a sum of three exponentials (Equation 1). The rise in the fraction of 11-mer + 12-mer was governed by kobs,addition, whereas its decay was described by two rate constants, kobs,decay,1 and kobs,decay,2. To analyze Pol III’s cleavage activity alone, we fit the fraction of CA dimer to a sum of two exponentials (Equation 2), revealing a fast and slow rate constant, kobs,cleavage,fast and kobs,cleavage,slow, respectively (Figure 1C).
Figure 1C indicates that the concentration of the CA dinucleotide is still increasing at 200 s. We hypothesize that the slow kobs,cleavage,2 rate constant accounts for this phenomenon. This interpretation is supported by an extended time course, (110–2000) s, that reveals a slow increase in the fraction of CA dimer and slow decay of 11-mer + 12-mer that continues beyond 2000 s (Figure S2). These data reveal that Pol III can cleave the RNA via at least two distinct mechanisms and these cleavage events are governed by rate constants that vary more than 10-fold. In this experimental design, we cause Pol III molecules to pause after AMP incorporation because we do not provide the next cognate nucleotide. It is possible that under extended pausing in vitro, Pol III undergoes a conformational change that is less favorable for cleavage and this results in the slow cleavage rate constant, kobs,cleavage,2.
| (Equation 3) |
To determine the elementary rate constants for the nucleotide addition cycle catalyzed by Pol III, we perturbed the reaction by varying [ATP] (10–2000) μM. Time courses were collected in triplicate and fit to Equation 1 or 2 for 11-mer + 12-mer (Figure 1D) and CA dimer (Figure 1E) fractions, respectively. A secondary plot of kobs,addition as a function of [ATP] was generated and fit to Equation 3 (Figure 1F). Our analyses produced a kobs,max = (60 ± 10) s−1 and K1/2 = (200 ± 100) μM (Figure 1F). Secondary plots of kobs,decay,1, and kobs,decay,2 were generated and fit using weighted non-linear least squares (NLLS) to a constant (Figures 1G and 1H). Both 11-mer + 12-mer decay rate constants were revealed to be constants with a mean value of kobs,decay,1 = (1.1 ± 0.03) s−1 and kobs,decay,2 = (0.003 ± 0.0002) s−1 (Figures 1G and 1H). Similarly, the calculated CA dimer rate constants were constant over [ATP], kobs,cleavage,fast = (1.2 ± 0.3) s−1 and kobs,cleavage,slow = (0.04 ± 0.006) s−1 (Figures 1I and 1J). The first rate constant describing the decay of the 11-mer + 12-mer is within error of the fast cleavage rate constant, kobs,cleavage,fast. This observation supports the conclusion that kobs,decay,1 and kobs,cleavage,fast describe Pol III’s cleavage activity. The slower cleavage rate constants kobs,decay,2 and kobs,cleavage,slow are not within error. We are unable to determine the identity of either of these steps with this model-independent analysis alone.
Model-dependent analysis of Pol III single-nucleotide addition
To determine the elementary rate constants governing individual steps in nucleotide addition, we performed model-dependent analyses, globally fitting the time courses as a function of [ATP] to a minimal reaction scheme. We found that Scheme 1 best describes the [ATP] dependent time courses (Figure 2A). First, Pol III ECs with a radiolabeled 10-mer, EC10, bind ATP reversibly. Once ATP is bound, phosphodiester bond formation occurs, yielding an 11-mer. EC11 species existed in two forms, (EC11)1 immediately undergoes RNA cleavage whereas the other requires a potential conformation change before RNA cleavage, (EC11)2. It is important to note that although Scheme 1 defines the rate constant k3 as the bond formation step, we cannot rule out the possibility that this step is a slow conformational change followed by bond formation, which is fast and not detected. We utilized our custom MATLAB tool box, MENOTR (Ingram et al., 2021b), that combines genetic algorithm and NLLS to globally fit the [ATP] dependent time courses using Scheme 1. The 11-mer + 12-mer and CA dimer time courses were plotted with their corresponding best fit lines (Figures 2B and 2C). Parameter values k1-k7 are reported (Table 1).
Figure 2.
Global fit of Pol III AMP time courses to Scheme 1
(A) Minimal kinetic model describing Pol III AMP addition.
(B) Global fit of 11-mer + 12-mer time courses to Scheme 1.
(C) Global fit of CA dimer time courses to Scheme 1. Time courses were collected in triplicate and the error corresponds to the standard deviation about the mean.
Table 1.
Scheme 1 fitted parameter values and bounds
| Kinetic Parameter | Fitted value | Lower bound | Upper bound |
|---|---|---|---|
| k1 | 1 x 108M−1s−1 | ||
| k2 | 2.3 x 103s−1 | 1.4 x 103s−1 | 3.8 x 103s−1 |
| k3 | 54 s−1 | 48 s−1 | 62 s−1 |
| k4 | 14 s−1 | 10 s−1 | 19 s−1 |
| k5 | 1.4 s−1 | 1.3 s−1 | 1.5 s−1 |
| k6 | 0.45 s−1 | 0.40 s−1 | 0.51 s−1 |
| k7 | 0.005 s−1 | 0.003 s−1 | 0.007 s−1 |
Pol III AMP addition time courses were collected in triplicate with varying [ATP], (10–2000) μM. Data were fit simultaneously using Scheme 1 in MENOTR (Ingram et al., 2021b). ATP binding, described by k1, was assumed to be diffusion controlled and constrained to 1 × 108M−1s−1. A range of k1 values (1 × 105 - 1 × 108) M−1 s−1 were evaluated and the results support diffusion controlled binding (Table S7). Upper and lower bounds were determined using a parameter grid searching strategy in MENOTR with a 68% confidence interval as previously described (Appling et al., 2015).
The model-independent and-dependent analyses are consistent. The kobs,max = (60 ± 10) s−1, determined through model-independent analysis, is consistent with k3 = ∼54 s−1, determined by model-dependent analysis (Table 1). Similarly, the observed rate constants describing Pol III cleavage, kobs,decay,1 ∼1.1 s−1 and kobs,cleavage,fast ∼1.2 s−1, are consistent with k5 = ∼1.4 s−1. Of interest, the second 11-mer + 12-mer decay observed rate constant, kobs,decay,2 ∼0.003 s−1 is in agreement with k7 = ∼0.005 s−1. This suggests that kobs,decay,1 corresponds to the population of EC11 that immediately undergoes RNA cleavage, whereas kobs,decay,2 describes RNA cleavage that occurs after one or more conformational changes.
Based on previous research on Pols I (Scull et al., 2019) and II (Malagon et al., 2006; Dangkulwanich et al., 2013), we hypothesized that the rate-limiting step of Pol III AMP addition was phosphodiester bond formation. To test this, we compared time courses collected with ATP and a slowly hydrolysable analog, Sp-ATP-α-S. If kobs,addition corresponds to phosphodiester bond formation, we expect at least a 10-fold decrease in the observed rate constant (Erie et al., 1993; Mizrahi et al., 1985; Patel et al., 1991). Sp-ATP-α-S time courses were collected in triplicate, a representative gel is shown in Figure S3A. We fit ATP and Sp-ATP-α-S time courses to Equation 1 and Equation 2 for 11-mer + 12-mer and CA dimer, respectively (Figures S3B and S3C). There was a ∼40-fold decrease in kobs,addition in the presence of Sp-ATP-α-S compared to ATP (Table S1). These data suggest that the rate-limiting step for nucleotide addition, kobs,addition and k3, corresponds to phosphodiester bond formation.
Pol III incorporated AMP faster than Pol II, slower than Pol I
| (Equation 4) |
| (Equation 5) |
With Pol III time courses collected, we compared Pol III AMP addition at 1 mM ATP to previously published Pol I and II results (Jacobs et al., 2021). For Pol I single-nucleotide addition, we observed a rise in the intensity of 11-mer over the first half of the time course, (0.005–0.1) s, and a decrease in the second half, (0.2–10) s (Figure 3A, top). Like Pol III, Pol I possesses intrinsic cleavage activity because of its A12 subunit, therefore GC and CA dimers were resolved at the bottom of the gel. We fit Pol I time courses to a sum of two exponentials (Appling et al., 2015; Jacobs et al., 2021) to calculate two observed rate constants, kobs,addition describing the appearance of the 11-mer, and kobs,decay describing the disappearance of the 11-mer (Equation 4). The rise of the fraction CA dimer over time is described by a single exponential (Equation 5).
| (Equation 6) |
Figure 3.
AMP addition catalyzed by Pols I, II, and III at 1 mM ATP
(A) Representative gel of Pol I AMP addition at 1 mM ATP, top. Plot of the fraction of 11-mer over time, fit to Equation 4, bottom.
(B) Representative gel of Pol II AMP at 1 mM ATP, top. Plot of the fraction 11-mer over time, fit to Equation 6, bottom.
(C) Representative gel of Pol III AMP addition at 1 mM ATP, top. Plot of the fraction of 11-mer + 12-mer over time, fit to Equation 1, bottom.
(D) Plot of the fraction of CA dimer synthesized by Pol I over time, fit to Equation 5.
(E) Plot of the fraction of CA dimer synthesized by Pol III over time, fit to Equation 2. All time courses were collected in triplicate and the average is plotted with error bars corresponding to the standard deviation about the mean.
For Pol II, we observed a rise and plateau of the fraction of 11-mer over time (Figure 3B). Because TFIIS is not provided in the reaction, Pol II is unable to synthesize dimers. The data display two rise phases, thus, the data were fit to a sum of two exponentials as described previously (Jacobs et al., 2021) (Equation 6). An ongoing study reveals that kobs,addition,1 describes bond formation, whereas kobs,addition,2 corresponds to a conformational change (unpublished).
It is important to note that we failed to observe synthesis of 12-mers in Pol I and II experiments due to the shorter time courses (0.005–10) s (Figures 3A and 3B). Pol III required a longer time course to describe the nucleotide addition reaction (0.01–200) s (Figures 1 and 3C). Extended Pol I and II time courses revealed the presence of 12-mers (Figure S4). These data suggest that all Pols either misincorporate because of the high concentration of ATP and the lack of the next cognate nucleotide or incorporate cognate G because of low concentrations of GTP contamination, at extended time points (>60 s) and at a high concentration of ATP.
At 1 mM ATP, Pol I AMP incorporation was described by kobs,addition = (221 ± 81) s−1 (Jacobs et al., 2021). Pol II time courses revealed two addition rate constants, kobs,addition,1 = (38 ± 8.8) s−1 and kobs,addition,2 (4.0 ± 0.6) s−1 (Jacobs et al., 2021). In this study, we determined Pol III kobs,addition = (91.2 ± 5) s−1 at 1 mM ATP. Pol II nucleotide addition was significantly slower than both Pols I and III, whereas Pol I was the fastest. These findings reveal extensive divergence in the kinetics of nucleotide addition by the Pols.
Pol III cleaves RNA faster than Pol I
Because Pol II lacks intrinsic cleavage activity, we compared the nuclease activity of Pols I and III conferred by bona fide subunits A12 and C11, respectively. The nuclease activity of Pol I was described by a single observed rate constant, kobs,cleavage = (0.2 ± 0.01) s−1 (Figure 3D and Equation 5). Although the appearance of the CA dimer by Pol III was described by two observed rate constants, kobs,cleavage,fast = (1.5 ± 0.1) s−1 and kobs,cleavage,slow = (0.02 ± 0.02) s−1 (Figure 3E and Equation 2). The amplitude terms for Pol III fast and slow cleavage, 0.56 ± 0.02 and 0.16 ± 0.02, respectively, suggest that ∼78% of Pol III ECs can cleave RNA nearly an order of magnitude faster than Pol I.
Pols I, II, and III are differentially affected by reaction conditions
To investigate the impact of reaction buffer conditions, we tested Pols I, II, and III AMP addition in six different buffers selected from the Pol literature, buffers A – F (Table S2). Time courses for each Pol were collected in triplicate, and the mean and standard deviation of the fraction of 11-mer at each time point were plotted (Figure S5). The time courses were fit using Equations 4, 6 and 1 for Pols I, II, and III, respectively.
To compare the observed rate constants governing nucleotide addition of Pols I, II, and III, we compared kobs,addition for Pol I (Equation 4), kobs,addition,1 for Pol II (Equation 6), and kobs,addition for Pol III (Equation 1). We omitted Pol II’s kobs,addition,2 rate constant from the comparison because it likely corresponds to a conformation change (unpublished).
We found that Pol I kobs,addition rate constants were highly variable across buffers A – F. Pol I was the fastest in buffer A, ∼220 s−1, and the slowest in buffers B and D, ∼31 s−1 (Figure S5A and Table S3). Pol II 11-mer observed rate constants were more consistent than Pol I values, ranging from ∼39 s−1 in buffer A to 10 s−1 in buffer D (Figure S5B and Table S4). The kobs,addition rate constants describing Pol III’s 11-mer + 12-mer formation across buffers A – F were similarly consistent, with the fastest observed rate constant revealed in buffer C, ∼140 s−1, and the slowest in buffer B, ∼56 s−1 (Figure S5C and Table S5).
In addition, we compared the observed rate constants describing Pol I and III cleavage, kobs,clevage for Pol I (Equation 5) and kobs,cleavage,fast for Pol III (Equation 2), across buffers A – F. We found that Pol III cleavage exceeded Pol I in all buffers except for buffer E, but they are nearly within error of each other in buffer E (Figures S5D and S5E, Table S3 and Table S5).
These data suggest that each Pol was influenced by alterations in reaction conditions. Pol I displayed the fastest observed rate constant across buffers A – F, but its activity was pH sensitive. Pol II was consistently the slowest of the Pols and was the least impacted by buffer variation. Finally, Pol III observed rate constants were altered more than Pol II’s but less than Pol I’s. These experiments demonstrate that these enzymes not only possess divergent AMP addition rates, but they also have evolved different tolerances to reaction conditions. This observation may reveal vulnerabilities inherent to one enzyme compared to the others, and these vulnerabilities may be therapeutically useful.
Mean observed rate constants describing Pol I and III multi-nucleotide addition are similar
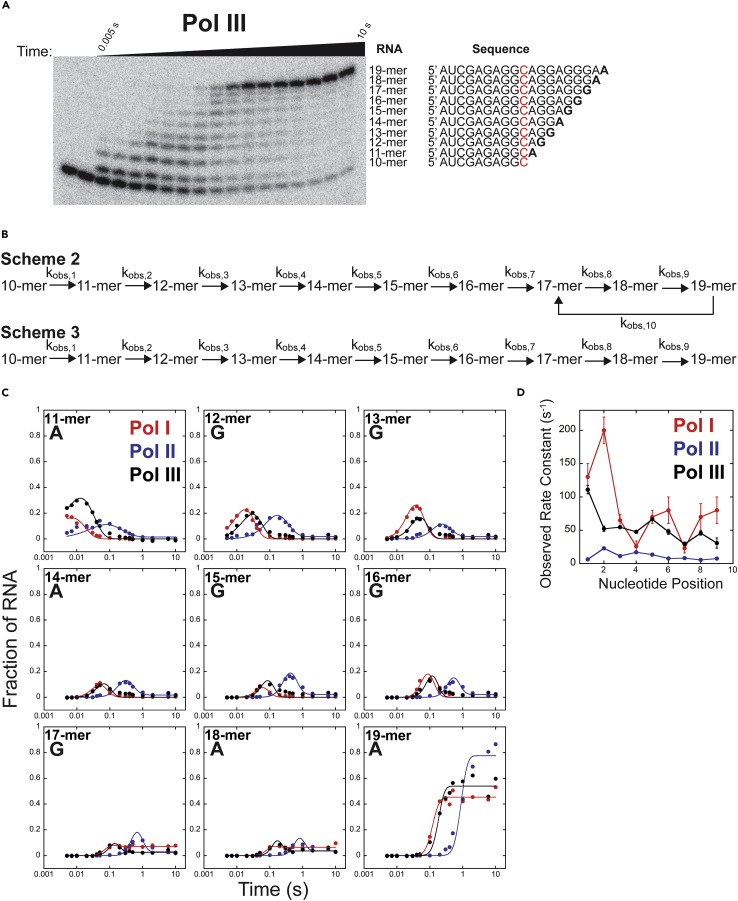
We aimed to determine if our AMP addition results are indicative of average elongation speed over multiple nucleotide addition events. For these experiments, we mixed Pol I, II, and III ECs with saturating concentrations of ATP and GTP in buffer A, as previously detailed (Ingram et al., 2021a). Providing ATP and GTP allowed the Pols to incorporate nine successive nucleotides, generating a 19-mer from the starting 10-mer (Figure 4A). Time courses were collected in triplicate for Pols I, II, and III at 1 mM ATP and 1 mM GTP.
Figure 4.
Pol I, II, and III multi-nucleotide addition time courses at 1 mM ATP and 1 mM GTP
(A) Representative gel of Pol III AMP and GMP multi-nucleotide addition at 1 mM ATP and 1 mM GTP.
(B) Reaction schemes describing Pol I, II, and III multi-nucleotide addition. Scheme 2 describes Pol I and III multi-nucleotide addition and Scheme 3 describes Pol II multi-nucleotide addition.
(C) Pols I, II, and III representative data sets describing the appearance and disappearance of each RNA species over time, fit to their respective schemes.
(D) Plot of Pols I, II, and III average kobs values for each nucleotide position. Each point represents the mean of three independent reactions with error bars corresponding to the standard deviation about the mean.
To analyze the appearance of each RNA, we formulated two kinetic models, Schemes 2 and 3 (Figure 4B). Resultant observed rate constants are first order and independent of NTP because of the high NTP concentration utilized. Scheme 2, previously developed to describe Pol I (Jacobs et al., 2021; Ingram et al., 2021a), was used to fit data from both Pols I and III because they require kobs,10 to describe their nuclease activity. Pol II time courses were fit using a simpler model, Scheme 3 (Jacobs et al., 2021). Parameter values were optimized using MENOTR (Ingram et al., 2021b) and the best fit lines are shown, describing the appearance and disappearance of each RNA (Figure 4C). Experimental time courses describing the fraction of each RNA were fit globally. The mean and standard deviation of each observed rate constant were calculated for each Pol (Table S6). It is important to highlight that Scheme 2 and 3 are distinct from Scheme 1 (Figure 2A). Scheme 1 models the nucleotide concentration dependence of a single-nucleotide addition. The model uses elementary rate constants accounting for the binding and any additional rate-limiting steps in the single incorporation. In contrast, Scheme 2 and 3 model the extension of 10-mer to 19-mer with a unimolecular observed rate constant between each incorporation. In reality, each cycle of incorporation must consist of NTP binding, bond formation, pyrophosphate release, conformational changes, and translocation. However, the multi-nucleotide addition experiments performed at one fixed nucleotide concentration are only sensitive to the slowest step in each repeating cycle (Lucius and Lohman, 2004; Miller et al., 2013) of nucleotide addition, and that value is reported in the kobs,n parameter.
To compare Pol I, II, and III multi-nucleotide addition, we plotted the observed rate constant over nucleotide positions (position 1, 11-mer, to position 9, 19-mer) (Figure 4A). The weighted average kobs values for Pols I, II, and III were (53 ± 4) s−1, (8.6 ± 0.1) s−1, and (56.8 ± 0.8) s−1, respectively. We found that Pol II was the slowest of the Pols, consistent with AMP addition results (Figures 3 and S5). Of interest, the mean observed rate constant describing Pol I and Pol III multi-nucleotide addition were within error. Although this observation is inconsistent with our AMP addition data, those experiments do not take into consideration translocation, the influence of DNA sequence on nucleotide addition kinetics, or the incorporation of GMP.
Pol I cleavage, kobs,10 = (12 ± 3) s−1, was larger than Pol III’s value, kobs,10 = (2 ± 0.6) s−1 (Table S6). Pol I cleavage of the 19-mer to a 17-mer is evident in the 17-mer plot. The fraction of 17-mer peaks at 0.07 and plateaus, it does not return to 0, unlike 11-mer – 16-mer plots (Figure 4C). In addition, we found that Pol I observed rate constants were highly heterogeneous over the nucleotide positions, ranging from (200–23) s−1 (Figure 4C and Table S6). The observed rate constants for Pols II and III fluctuated less. This suggests that Pol I nucleotide addition and transcript cleavage is more sensitive to the flanking DNA sequence, identity of the encoded NTP, or the length/structure of the nascent RNA than Pols II and III. The features of the template that affect Pol I elongation kinetics are under further investigation.
Pol III elongation complex stability is higher than Pol I, lower than Pol II
We revealed substantial differences between the nucleotide addition kinetics of the Pols (Figures 3, 4, and S5). The relative simplicity of these in vitro assays empowers us to measure other properties inherent to the Pols. Using an RNase protection assay (Appling et al., 2018), we measured the stability of Pol I, II, and III ECs. It is critical that Pols are stably bound to the template DNA, allowing for processive transcription elongation, without being bound too tightly because that can cause difficulty in EC disengagement or termination. Because Pols I, II, and III transcribe DNA that vary in gene length and topological properties, we hypothesized that their EC stabilities would vary.
To test this hypothesis, we mixed radiolabeled Pol I, II, or III ECs with 10 μM RNase A and 750 mM KCl at t = 0 in buffer A, as previously described (Appling et al., 2018; Scull et al., 2020; Jacobs et al., 2021). If an EC collapses, the 10-mer will be cleaved by RNase A into a 7-mer and an unlabeled, undetectable 3-mer (Figure 5A). In contrast, if the EC is intact, it will protect the 10-mer from RNase A. Therefore, by quantifying the amount of 10-mer and 7-mer over a time course, we can monitor Pol I, II, and III EC collapse.
Figure 5.
EC stability experiments at 750 mM KCl for Pols I, II, and III
(A) Pol I, II, and III ECs are mixed with 10 μM RNase A and 750 mM KCl at t = 0. Representative EC stability gels of Pols I (8 s–40 min), II (8 s–72 h), and III (8 s–24 h) resolve 10-mers (intact ECs) and 7-mers (collapsed ECs). The fraction of EC collapse is determined by calculating the fraction of 7-mer over total RNA.
(B) Plot of the fraction of EC collapse over time for Pols I, II, and III. Three independent reactions were collected in triplicate and the mean is plotted with error bars reflecting the standard deviation about the mean.
We observed that complete disassembly of Pol I ECs occurred rapidly, by 40 min, whereas Pol III ECs remained intact for 9 h (Figure 5B). Remarkably, we found complete collapse of Pol II ECs occurred only after three days (Figure 5B). These extensive differences between the Pols may suggest that the chromatin environment navigated by the Pols in vivo has exerted selective pressure on their EC stabilities during evolution. Pol I transcribes nucleosome-free DNA whereas Pol II genes and half of Pol III genes are nucleosome-bound. Pol II molecules must endure extended pausing during productive rounds of transcription (Core and Adelman, 2019). Perhaps these template properties explain why Pol II forms the most stable EC whereas Pol I ECs are highly unstable.
Pol I more readily incorporates incorrect nucleotides than Pols II or III
A critical feature of Pol transcription is nucleotide misincorporation. We define misincorporation as the incorporation of an incorrect or non-cognate nucleotide. To evaluate this property, we formed ECs in buffer A with a 9-mer and incubated them with the next cognate nucleotide, α-32P-CTP, or, in a separate reaction, a non-cognate nucleotide, α-32P-ATP (Scull et al., 2019; Jacobs et al., 2021). Incubation with α-32P-CTP for 10 min served as a control for the number of active ECs. Time points were collected over (1–60) min in the presence of α-32P-ATP.
It is important to highlight a key difference between this assay and single-nucleotide addition experiments (Figure 3). AMP addition experiments were executed at a high ATP concentration (1 mM), whereas we provided very low concentrations of α-32P-ATP and α-32P-CTP, ∼5 nM, for misincorporation experiments. By providing an extremely sub-saturating concentration of nucleotides, it likely renders any trace amounts of contaminating NTPs undetectable.
As expected, Pols I and III synthesized 10-mers upon incubation with α-32P-CTP (Figures 6A and 6C). A fraction of those 10-mers, ∼0.1, were cleaved and resolved as GC dimers. In the presence of only a non-cognate nucleotide, α-32P-ATP, we resolved 10-mers and GA dimers, produced from cleavage of the incorrect 10-mer. Pol II only synthesized 10-mers for both α-32P-CTP and α-32P-ATP time points (Figure 6B).
Figure 6.
Time courses of AMP misincorporation by Pols I, II, and III
(A–C) Representative gels of Pol I, II, or Pol III ECs provided with α-32P-CTP for 10 min or α-32P-ATP for (1–60) min.
(D) Plot of the fraction of incorrect 10-mers synthesized with α-32P-ATP by Pols I, II, and III.
(E) Plot of the fraction of GA dimer produced by Pols I and III. Time courses were collected in triplicate and the mean is plotted with error corresponding to the standard deviation about the mean.
In Figure 6D, we plotted the fraction of incorrect 10-mers, synthesized with α-32P-ATP, produced by Pols I, II, and III over time. At the first time point (1 min), we see the highest fraction of incorrect 10-mers for Pols I and III, ∼0.012 and ∼0.007, respectively. The fraction of incorrect 10-mer gradually decreased over the time course, until ultimately, negligible 10-mer signal remains, ∼0.008 and ∼0.003, for Pols I and III, respectively, at 60 min. As incorrect 10-mer signal decreased, the fraction of GA dimer increased (Figure 6E). This suggests that >90% misincorporation events were followed by cleavage for Pols I and III. We observed the opposite trend for Pol II. Pol II gradually misincorporated α-32P-ATP, generating the incorrect 10-mer over the time course, until it plateaued at ∼0.006.
Because the vast majority of incorrect 10-mers were rapidly cleaved by Pols I and III, it is likely that cleavage would allow for the resolution of incorrect additions in vivo when all NTPs are available. The final transcriptional fidelity of the Pols in vivo is similar, which is consistent with this theory (Gout et al., 2017). Ultimately, we determined that Pols I, II, and III have similar fidelity, but different mechanisms to achieve it.
Discussion
Pols I, II, and III are enzymatically distinct
We defined a minimal kinetic model that describes AMP addition catalyzed by Pol III (Figures 1 and 2). On comparison to Pols I and II, we found that Pol I incorporates AMP significantly faster than Pols II and III (Figure 3). The gap in speed between Pol I and III was closed in the context of multi-nucleotide addition compared to single-nucleotide addition (Figure 4). Outside of nucleotide addition kinetics, we revealed that Pol II forms the most stable EC (Figure 5) and is the least error prone Pol (Figure 6). Our results support the notion that each Pol possesses unique biochemical features that suit them for their specialized transcriptional roles.
Comparison of Pol I, II, and III nucleotide addition mechanisms
Our group has investigated the nucleotide addition mechanism of Pols I (Appling et al., 2015), II (unpublished), and III (Figures 1 and 2) under identical experimental conditions, allowing for direct comparisons. Pol I AMP incorporation is described by kobs,max = (270 ± 30) s−1 and K1/2 = (170 ± 20) μM (Appling et al., 2015). Interestingly, Pol II exhibits two partially rate-limiting steps, kobs,fast, kobs,fast,max = (50 ± 7) s−1 and K1/2 = (110 ± 30) μM, and kobs,slow, kobs,slow,max = (4.9 ± 0.4) s−1 and K1/2 = (57 ± 7) μM (unpublished). Similar to Figure S3, we previously utilized Sp-ATP-α-S to determine that kobs,fast likely corresponds to bond formation (unpublished). In this study, we determined that Pol III AMP incorporation is described by kobs,max = (60 ± 10) s−1 and K1/2 = (200 ± 100) μM (Figure 1F).
We hypothesize that the high degree of sequence conservation around the active site of Pols I, II, and III (Carter and Drouin, 2009) is responsible for their similar ATP binding affinity, ∼150 μM. In contrast, there are small differences in and around the mobile domains that may account for their significant differences in nucleotide addition kinetics (Tan et al., 2008; Kaplan, 2013). One critical mobile element, the trigger loop, alternates between a closed and open conformation, coordinating incoming NTPs and allowing pyrophosphate release and translocation, respectively (Cheung and Cramer, 2012; Kaplan and Kornberg, 2008). Changes within the trigger loop (Viktorovskaya et al., 2013) have been shown to modulate the dynamics of the NTP coordination in the active site, bond formation, and translocation (Larson et al., 2012). Point mutations within the trigger loop support this theory. The Kashlev group found that a mutation at the base of Pol II’s trigger loop, rpb1-E1103G, increased nucleotide addition rate (Kireeva et al., 2008). The homologous mutation in Pol I’s trigger loop, rpa190-E1124G, resulted in decreased elongation rate (Scull et al., 2019; Viktorovskaya et al., 2013). The homologous mutation in Pol III, rpc160-E1070G, has not been characterized yet in vitro. These data suggest that minor differences within or around the trigger loops of the Pols may contribute to their substantial divergence in nucleotide addition kinetics.
Pol I and III in vitro transcription elongation rates are consistent in vivo
We found that Pol I was significantly faster than Pols II and III at AMP addition (Figure 3). Because it is impossible to measure single-nucleotide addition in vivo, we compared our multi-nucleotide addition results (Figure 4) to processive elongation measurements made in vivo to evaluate whether our biochemical findings are consistent with the living system.
Pol I in vivo elongation rate estimations have been made using electron microscopy (EM) of Miller chromatin spreads. An estimated elongation rate of 60 nt s−1 (French et al., 2003) was calculated by determining how fast Pol I molecules must transcribe the rDNA to synthesize rRNA for 2000 ribosomes per minute (Warner, 1999). Similarly, Pol III elongation rate estimations were made using EM analysis of Miller chromatin spreads, yielding an estimated rate of 61 nt s−1 (French et al., 2008). The average elongation rate of Pol II in vivo was determined to be 33 nt s−1 by monitoring the “last wave” of Pol II molecules to dissociate from an 8 kb gene, under a GAL1 promoter, following glucose addition (Mason and Struhl, 2005).
Pol I and III average multi-nucleotide addition observed rate constants, ∼53 s−1 and ∼57 s−1 respectively (Figure 4), are consistent with in vivo estimates. For Pol II, our calculated values are much lower than in vivo calculations, ∼8 s−1 (Figure 4) vs. 33 nt s−1 (Mason and Struhl, 2005), but the trend is identical—Pol II is the slowest of the Pols. How do we reconcile that difference? It is likely if TFIIS were provided in vitro, there would be an increase in Pol II multi-nucleotide addition rate constants (Sheridan et al., 2019). One group found that in absence of TFIIS, Pol II elongation rate was reduced by 50% (Noe Gonzalez et al., 2021). Many other factors also interact with Pol II and likely enhance nucleotide addition in vivo, including Spt4/5, PAF1, and FACT (Vos et al., 2020; Saunders et al., 2003; Crickard et al., 2017). Therefore, it is probable that the rate of Pol II elongation in vivo is increased by its association with Pol II-specific TFs. The mechanisms of action of Pol II-associated TFs will be interesting to reveal in future investigations.
Pol I is pH sensitive
We found that although Pol I displayed the fastest AMP addition observed rate constant across buffers A – F (Figue S5 and Table S3), it was the most sensitive Pol to varying reaction conditions. Pol I nucleotide addition kinetics was particularly sensitive to pH changes. Buffer A and D consist of identical components with the exception of pH, buffer A (7.9) and buffer D (7.5) (Table S2). This sole difference resulted in a seven-fold change in Pol I kobs,addition values (Table S3). Interestingly, Pol III single-nucleotide addition was faster than Pol I in three buffer conditions, buffers B – D (Table S5). The common factor across buffers B – D is a lower pH, ranging from 7.5–7.6. Surprisingly, Pol II nucleotide addition in buffers B and C were within error of Pol I’s (Tables S3 and S4). This suggests that the catalytic activity of Pol I is highly sensitive to pH alterations. Can fluctuation in pH regulate rRNA synthesis in vivo?
We know that cellular stress induces chemical and morphological changes in the nucleolus (Boulon et al., 2010). As a result of cellular stress, ribosome biogenesis is downregulated (Grummt, 2013). The downregulation of Pol I transcription of the rDNA is effective in saving cellular energy and resources to restore homeostasis. Although there is no direct evidence that pH fluctuations in vivo directly contribute to the downregulation of ribosome biogenesis, this connection has been established for other nucleolar proteins. Lee and colleagues showed that the activity and location of ubiquitin ligases are regulated by physiological fluctuations in cellular pH (Mekhail et al., 2004, 2005). Therefore, it is plausible that pH is an unexpected regulator of Pol I transcription under stress.
DNA sequence affects the catalytic activity of Pols I, II, and III in absence of trans-acting factors
It is well established that DNA-protein interactions are critical regulators in gene expression. Specific DNA sequences (e.g. promoters, enhancers, silencers) are bound by TFs that regulate transcriptional processes such as Pol initiation complex assembly, chromatin remodeling, and recruitment of coactivators and corepressors (Kadonaga, 2004). Here, we observe that nucleotide addition kinetics vary widely because of DNA sequence, independently of trans-acting factors (Figure 4). Over nine nucleotide incorporation positions, rate constants varied 7.7-fold for Pol I, 2.1-fold for Pol II, and 2.7-fold for Pol III (Figure 4D and Table S6).
It would not be surprising if Pols I, II, and III vary in their sensitivity to certain DNA sequences. Over the course of evolution, selective pressures have maintained certain stretches of DNA sequences at many genomic loci, each unique to each Pol. For example, the rDNA of higher eukaryotes, metazoans, is more GC-rich than yeast (Escobar et al., 2011). From native-elongating sequencing studies, we know that Pol I occupancy decreases upon an increase in GC content (Scull et al., 2020). This observation can be interpreted to indicate that GC-rich DNA is transcribed faster than AT-rich DNA. It is possible that these stretches of fast elongation in GC-rich regions, and slow elongation in AT-rich regions, are critical features of Pol I transcription elongation that facilitate the formation of RNA secondary structures and co-transcriptional rRNA processing.
We have also known since the mid-1980s that Pol II transcription elongation is a key target for regulating gene expression (Eick and Bornkamm, 1986; Bentley and Groudine, 1986). AT-rich DNA is enriched within introns, whereas GC-rich DNA is enriched within coding gene regions (Fenouil et al., 2012; Close et al., 2012). It is theorized that Pol II slows down and pauses more during AT-rich regions which may be critical for the formation of secondary RNA structures, maintenance of permissible euchromatin architecture, rapid gene expression under stress conditions, and coordination with splicing machinery (Nechaev and Adelman, 2008; Close et al., 2012; Zamft et al., 2012; Adelman and Lis, 2012).
Similar to prokaryotic transcription termination, Pol III can terminate transcription in a factor-independent manner. When Pol III ECs encounter T-tracts in the DNAnt, it destabilizes the EC resulting in Pol III disengagement (Adelman and Lis, 2012; Arimbasseri et al., 2013). Therefore, Pol III clearly has altered response to DNA sequences compared to Pols I and II. How DNA sequence affects Pol I, II, and III nucleotide addition in vitro is a topic of ongoing investigation.
Potential consequences of misincorporation frequencies in vivo
We found that Pol II is the least error prone of the Pols (Figure 6). This is interesting because the Pols have similar transcriptional error rates in vivo, 4.3 × 10−6 per bp for Pol I, 3.9 × 10−6 per bp for Pol II, and 1.7 × 10−5 per bp for Pol III (Gout et al., 2017). It is important to emphasize that our results do not contradict these in vivo measurements. Rather, our data suggests that Pol II is the least likely Pol to misincorporate. Because we only provided the non-cognate nucleotide, our assay is only sensitive to proofreading (evidenced by cleavage RNA products) not incorporation of the correct nucleotide following misincorporation because the cognate nucleotide was not provided in those reactions. This suggests that Pols I and III must invest more energy in reconciling those errors to achieve a final transcriptional error rate similar to Pol II. This suggestion is reasonable given that unlike Pols I and III that possess bona fide proofreading subunits, Pol II must recruit a trans-acting factor, TFIIS. Perhaps the slow rate of transcription elongation by Pol II allows for accurate selection of incoming NTPs, resulting in higher transcription fidelity. In support of this hypothesis, mutations in RPB1 that speed up Pol II elongation also increase misincorporation (Irvin et al., 2014).
Uncovering the unique features of the Pols contribute to modern anti-cancer strategies
Our investigation of the Pols is critical to both basic and translational science. Fundamentally, it is of paramount importance to understand the molecular mechanisms that govern these essential enzymes. Our work is also relevant to ongoing efforts to selectively inhibit Pol I for cancer chemotherapy (Ferreira et al., 2020; Laham-Karam et al., 2020; Peltonen et al., 2010, 2014; Haddach et al., 2012). To selectively target Pol I, without interfering with the activities of Pols II and III, we must identify and exploit the unique features of Pol I. One example where this strategy has been successful is BMH-21, a small molecule DNA intercalator (Peltonen et al., 2014; Wei et al., 2018; Jacobs et al., 2022). Although the mechanism of action of BMH-21 is well understood (Jacobs et al., 2022), we do not know the degree of specificity to Pol I. Preliminary data show that Pol I is uniquely sensitive to BMH-21 compared to Pols II and III. We hypothesize that Pol I’s heightened vulnerability to BMH-21 is because of its rapid elongation rate and low EC stability elucidated in this study. These findings illustrate how basic, biochemical characterization can support and empower efforts to develop and deploy new therapeutic interventions.
Limitations of the study
The rigorous in vitro transcription system deployed in this study revealed critical biochemical differences in the Pols. By nature, biochemical and biophysical approaches are performed under conditions optimized in vitro and these conditions are not identical to the in vivo environment. Currently, there are no in vivo methods that would detect the properties evaluated in this study. Nevertheless, biochemical characterization, especially comparative analyses like those presented here, provide deep insight into fundamental functions of key enzymes like the eukaryotic RNA polymerases. If the goal is to understand how these enzymes are controlled, it is critically important to understand their core functions and limitations.
Future studies will examine the effects of template sequence, temperature, ionic content of the buffer, and longer elongation events on Pol function. Because of the malleability of our in vitro system, these modifications are straightforward and will continue to drive our understanding of the mechanisms of eukaryotic gene expression.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Experimental models: Organisms/strains | ||
| Pol I: Saccharomyces cerevisiae: CB010 Mata, pep4∷HIS3/prb1∷LEU2, prc1∷HISG, can1, ade2, trp1, ura3, his3, leu2–3, 112, RPA135 C-terminal TEV 3HA 10his | (Jacobs et al., 2021) | DAS1084 |
| Pol II: Saccharomyces cerevisiae: CB010 Mata, pep4∷HIS3/prb1∷LEU2, prc1∷HISG, can1, ade2, trp1, ura3, his3, leu2–3, 112, RPB2 C-terminal TEV 3HA 10his | (Jacobs et al., 2021) | DAS1085 |
| Pol III: Saccharomyces cerevisiae: CB010 Mata, pep4∷HIS3/prb1∷LEU2, prc1∷HISG, can1, ade2, trp1, ura3, his3, leu2–3, 112, RPC2 C-terminal TEV 3HA 10his | This paper | DAS1086 |
| Oligonucleotides | ||
|
DNAt5′ ACCAGCAGGCCGATTGGGATGGG TATTCCCTCCTGCCTCTCGATGGCTGTAAG TATCCTATAGG |
Integrated DNA Technologies (IDT) | DAS1719 |
|
DNAnt5′ CCTATAGGATACTTACAGCC ATCGAGAGGCAGGAGGGAATACCCA TCCCAATCGGCCTGCTGGT |
Integrated DNA Technologies (IDT) | DAS1718 |
| RNA 5′ AUCGAGAGG | Integrated DNA Technologies (IDT) | DAS1551 |
| Software and algorithms | ||
| ImageQuant Tl (IQTL) 8.2 | Fisher Scientific | RRID:SCR_014246; https://www.fishersci.com/shop/products/iqtl-imagequant-tl-8-2/501963627 |
| KaleidaGraph v4.5.4 | Synergy Software | RRID:SCR_014980; https://www.synergy.com |
| MENOTR written in MATLAB (MathWorks, Inc., Natick MA) | (Ingram et al., 2021b) | https://github.com/ZachIngram/2021-MENOTR |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, David Schneider (dschneid@uab.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Pol I, II, and III purification
Saccharomyces cerevisiae (yeast) Pols I, II, and III were purified using an identical strategy. The second largest subunit of the Pols were C-terminally tagged with: TEV cleavage site, three HA repeats, and 10 histidine residues. Yeast was grown at the Bioexpression and Fermentation Facility, Department of Biochemistry and Molecular Biology at University of Georgia.
Purification buffers
Breakage buffer: 400 mM (NH4)2SO4, 50 mM Tris-SO4 pH 7.8, 10 mM MgCl2, 10 μM ZnCl2, 10% glycerol.
KCl low imidazole buffer: 100 mM KCl, 50 mM Tris-SO4 pH 7.6, 5 mM MgCl2, 10 mM imidazole, 20% glycerol.
KCl high imidazole buffer: 100 mM KCl, 50 mM Tris-SO4 pH 7.6, 5 mM MgCl2, 250 mM imidazole, 20% glycerol.
Low KCl buffer: 100 mM KCl, 20 mM HEPES pH 7.8, 1 mM MgCl2, 10 μM ZnCl2, 20% glycerol.
High KCl buffer: 2 M KCl, 20 mM HEPES pH 7.8, 1 mM MgCl2, 10 μM ZnCl2, 20% glycerol.
MonoQ A buffer: 200 mM KOAc, 20 mM HEPES pH 7.8, 1 mM MgCl2, 10 μM ZnCl2, 10% glycerol.
MonoQ B buffer: 2 M KOAc, 20 mM HEPES pH 7.8, 1 mM MgCl2, 10 μM ZnCl2, 10% glycerol.
Purification strategy
Each Pol was purified from approximately 150 g of wet cell mass. Cells were washed and lysed in breakage buffer. Cells were ultracentrifuged and the supernatant was loaded on five Ni columns (GE Healthcare). Columns were washed with KCl low imidazole buffer and eluted with KCl high imidazole buffer onto a heparin column (GE Healthcare). Ni columns were removed, and the heparin column was washed with low KCl buffer. Sample was eluted from the heparin column with high KCl buffer. To change into MonoQ A buffer, the eluate was run over desalting columns. Sample was manually loaded onto a MonoQ column (GE Healthcare). Fractions were collected with a 0 to 100% MonoQ B buffer gradient, 0.25 mL/min over 100 min.
Identity of Pol fractions
We confirmed the presence of each Pol I, II, or III subunit with mass spectrometry (Figures S6A–S6C). Each Pol preparation only contained subunits specific to that Pol. Complete Pol I, II, and III mass spectrometry results are displayed in Tables S8–S10. Purity of Pol I, II, and III preparations were examined with SDS PAGE (Figure S6D) and Western Blots.
Method details
Single-nucleotide addition assay
Buffer A
Buffer A is composed of 40 mM KCl, 20 mM Tris-OAc pH 7.9, 10 mM Mg(OAc)2, 2 mM DTT, and 0.2 mg/mL BSA. Pols I, II, and III demonstrated efficient nucleotide addition in buffer A, so we executed all experiments in buffer A with the exception of Figure S5.
EC formation
Purified Pol I, II, or III (∼16 nM) was incubated with a pre-annealed RNA (162.75 nM) and DNAt (54.26 nM) hybrid in buffer A for 10 min at 4°C followed by 20 min 25°C. The DNAnt (162.75 nM) was added and allowed to incubate at 25°C for 40 min. Each oligonucleotide was ordered from Integrated DNA Technologies:
DNAnt 5’ –CCTATAGGATACTTACAGCCATCGAGAGGCAGGAGGGAATACCCATCCCAATCGGCCTGCTGGT
RNA 5’ – AUCGAGAGG
DNAt 5’ –
ACCAGCAGGCCGATTGGGATGGGTATTCCCTCCTGCCTCTCGATGGCTGTAAGTATCCTATAGG
EC labeling
Formed ECs are labeled with 5 nM α-32P-CTP and 100 μM Mg(OAc)2 for 10 min at 25°C. The labeling reaction is stopped with 1 mM EDTA.
Quenched-flow time courses
Radiolabeled ECs (580 μL) was loaded into one syringe of a rapid mixing instrument, opposite of the NTP mixing syringe. The NTP mix (580 μL) contains: ATP (20–4000 μM), 18 mM Mg(OAc)2, and 1 mM EDTA in buffer A. The EC and NTP substrates were mixed in a 1:1 ratio within 2 ms and allowed to incubate for a fixed amount of time, (0.01–200) s for Pol III and (0.005–10) s for Pols I and II. Because mixing is 1:1, final concentrations are two-fold lower than described above. Each time point reaction was stopped, or quenched, with 1 M HCl. Equalizing HCl was added to each reaction and neutralized. An aliquot of each reaction was mixed with loading dye, boiled for 5 min, and loaded onto a 28% sequencing gel. Each gel was exposed to a phosphorimager screen for 12–24 h. Screens were scanned and the images were quantified using ImageQuant. The fraction of each RNA over time was fit using MENOTR (Ingram et al., 2021b).
Single-nucleotide addition in buffers A – F
Single-nucleotide addition assays were executed in triplicate in each buffer condition, A – F (Table S2), across Pols I, II, and III as previously described above. Buffer A contains 40 mM KCl, 20 mM Tris-OAc pH 7.9, 10 mM Mg(OAc)2, 2 mM DTT, and 0.2 mg/mL BSA (Appling et al., 2015). Buffer B contains 80 mM KOAc, 50 mM HEPES pH 7.5, 5 mM MgSO4, 10 mM DTT, and 10% glycerol (Fazal et al., 2015). Buffer C contains 60 mM (NH4)2SO4, 20 mM HEPES pH 7.6, 8 mM MgSO4, 10 μM ZnCl2, and 10% glycerol (Lisica et al., 2016). Buffer D contains 40 mM KCl, 20 mM Tris-OAc pH 7.5, 10 mM Mg(OAc)2, 2 mM DTT, and 0.2 mg/mL BSA. Buffer E contains 100 mM NaCl, 40 mM HEPES pH 7.8, 42 mM MgCl2, 3 mM BME, 0.1 mg/mL BSA, and 5% glycerol (Arimbasseri and Maraia, 2015). Buffer F contains 100 mM NaCl, 40 mM Tris-HCl pH 8, 7 mM MgCl2, 3 mM BME, 0.1 mg/mL BSA, and 7.5% glycerol (Kassavetis et al., 1999).
Multi-nucleotide addition assay
Multi-nucleotide addition assays were performed as detailed above with ATP and GTP provided in the NTP syringe in buffer A. Radiolabeled ECs were formed, with a 10-mer starting RNA, and mixed with 1 mM ATP, 1 mM GTP, and 9 mM Mg(OAc)2 (final concentrations) in the chemical-quenched flow. Reactions were run on sequencing gels to resolve the RNAs, 10-mer – 19-mer, by size. Model-dependent analysis of Pol I, II, and III multi-nucleotide addition time courses was performed with MENOTR (Ingram et al., 2021b).
Elongation complex stability assay
Radiolabeled Pol I, II, or III ECs were mixed with 10 μM RNase A (Thermo Scientific EN0531) and 750 mM KCl at t = 0 in buffer A. Time points were collected continuously, transferring an aliquot of the reaction mix to a tube with loading dye. EC disassembly was monitored by resolving 10-mers and 7-mers on sequencing gels.
Misincorporation assay
Pol I, II, or III ECs were incubated with a non-cognate nucleotide, α-32P-ATP (5 nM), and 100 μM Mg(OAc)2 for (1–60) min in buffer A. For our control, we incubated the Pols with the cognate nucleotide, α-32P-CTP (5 nM), to gage the amount of active ECs for 10 min. To account for differences in the emissions of the radioactive nucleotides, α-32P-CTP and α-32P-ATP, we performed a spot test of different dilutions of the nucleotides and normalized our signal to their ratio. We compared the amount of misincorporated nucleotides to the amount of correct incorporations to calculate the fraction of incorrect 10-mer.
Quantification and statistical analysis
Model-dependent analyses
MENOTR, Multi-start Evolutionary Nonlinear OpTimizeR, was utilized for all model-dependent fitting (Ingram et al., 2021b). MENOTR is a parameter optimization MATLAB toolbox that leverages a hybrid genetic/NLLS algorithm. The strengths of both algorithms are utilized to achieve an optimized model that would otherwise be difficult to ascertain. MENTOR is available on GitHub: https://github.com/ZachIngram/2021-MENOTR.
Acknowledgments
We thank the present and past members of the Schneider and Lucius labs for their critical evaluation of this work. This study was supported by National Institutes of Health grants R35-GM140710 (to D.A.S.), T32-GM109780 (to R.Q.J.), T32-GM008111 (to R.Q.J.), and by NSF grant MCB1817749 (to A.L.L. and D.A.S.).
Author contributions
R.Q.J., Z.M.I., A.L.L., and D.A.S. conceived and designed experiments; R. Q. J. performed experiments; R. Q. J. and Z. M. I. analyzed results; and R.Q.J., Z.M.I., A.L.L., and D.A.S. wrote and revised the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: November 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105306.
Contributor Information
Aaron L. Lucius, Email: allucius@uab.edu.
David A. Schneider, Email: dschneid@uab.edu.
Supplemental information
Data and code availability
-
•
Data: All data reported in the article will be shared by the lead contact on request.
-
•
Code: We used our custom-built software package written in MATLAB, MENOTR (Ingram et al., 2021b), to perform model-dependent analyses. MENTOR is available on GitHub: https://github.com/ZachIngram/2021-MENOTR.
-
•
All other items: Any additional information required to reanalyze the data reported in this article is available from the lead contact on request.
References
- Acker J., Conesa C., Lefebvre O. Yeast RNA polymerase III transcription factors and effectors. Biochim. Biophys. Acta. 2013;1829:283–295. doi: 10.1016/j.bbagrm.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Adelman K., Lis J.T. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appling F.D., Lucius A.L., Schneider D.A. Transient-state kinetic analysis of the RNA polymerase I nucleotide incorporation mechanism. Biophys. J. 2015;109:2382–2393. doi: 10.1016/j.bpj.2015.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appling F.D., Scull C.E., Lucius A.L., Schneider D.A. The A12.2 subunit is an intrinsic destabilizer of the RNA polymerase I elongation complex. Biophys. J. 2018;114:2507–2515. doi: 10.1016/j.bpj.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimbasseri A.G., Maraia R.J. Mechanism of transcription termination by RNA polymerase III utilizes a non-template strand sequence-specific signal element. Mol. Cell. 2015;58:1124–1132. doi: 10.1016/j.molcel.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimbasseri A.G., Rijal K., Maraia R.J. Transcription termination by the eukaryotic RNA polymerase III. Biochim. Biophys. Acta. 2013;1829:318–330. doi: 10.1016/j.bbagrm.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimbasseri A.G., Rijal K., Maraia R.J. Comparative overview of RNA polymerase II and III transcription cycles, with focus on RNA polymerase III termination and reinitiation. Transcription. 2014;5:e27639. doi: 10.4161/trns.27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D.L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986;321:702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Boguta M., Czerska K., Zoładek T. Mutation in a new gene MAF1 affects tRNA suppressor efficiency in Saccharomyces cerevisiae. Gene. 1997;185:291–296. doi: 10.1016/s0378-1119(96)00669-5. [DOI] [PubMed] [Google Scholar]
- Boulon S., Westman B.J., Hutten S., Boisvert F.M., Lamond A.I. The nucleolus under stress. Mol. Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton Z.F. The Old and New Testaments of gene regulation. Evolution of multi-subunit RNA polymerases and co-evolution of eukaryote complexity with the RNAP II CTD. Transcription. 2014;5:e28674. doi: 10.4161/trns.28674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter R., Drouin G. Structural differentiation of the three eukaryotic RNA polymerases. Genomics. 2009;94:388–396. doi: 10.1016/j.ygeno.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Cheung A.C.M., Cramer P. A movie of RNA polymerase II transcription. Cell. 2012;149:1431–1437. doi: 10.1016/j.cell.2012.06.006. [DOI] [PubMed] [Google Scholar]
- Close P., East P., Dirac-Svejstrup A.B., Hartmann H., Heron M., Maslen S., Chariot A., Söding J., Skehel M., Svejstrup J.Q. DBIRD complex integrates alternative mRNA splicing with RNA polymerase II transcript elongation. Nature. 2012;484:386–389. doi: 10.1038/nature10925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conconi A., Widmer R.M., Koller T., Sogo J.M. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989;57:753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- Core L., Adelman K. Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev. 2019;33:960–982. doi: 10.1101/gad.325142.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crickard J.B., Lee J., Lee T.H., Reese J.C. The elongation factor Spt4/5 regulates RNA polymerase II transcription through the nucleosome. Nucleic Acids Res. 2017;45:6362–6374. doi: 10.1093/nar/gkx220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangkulwanich M., Ishibashi T., Liu S., Kireeva M.L., Lubkowska L., Kashlev M., Bustamante C.J. Complete dissection of transcription elongation reveals slow translocation of RNA polymerase II in a linear ratchet mechanism. Elife. 2013;2:e00971. doi: 10.7554/eLife.00971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G., Conti A., Pagano A., Carnevali D. Identification of RNA polymerase III-transcribed genes in eukaryotic genomes. Biochim. Biophys. Acta. 2013;1829:296–305. doi: 10.1016/j.bbagrm.2012.09.010. [DOI] [PubMed] [Google Scholar]
- Dieci G., Fiorino G., Castelnuovo M., Teichmann M., Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Eick D., Bornkamm G.W. Transcriptional arrest within the first exon is a fast control mechanism in c-myc gene expression. Nucleic Acids Res. 1986;14:8331–8346. doi: 10.1093/nar/14.21.8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel C., Sainsbury S., Cheung A.C., Kostrewa D., Cramer P. RNA polymerase I structure and transcription regulation. Nature. 2013;502:650–655. doi: 10.1038/nature12712. [DOI] [PubMed] [Google Scholar]
- Erie D.A., Hajiseyedjavadi O., Young M.C., Von Hippel P.H. Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science. 1993;262:867–873. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- Escobar J.S., Glémin S., Galtier N. GC-biased gene conversion impacts ribosomal DNA evolution in vertebrates, angiosperms, and other eukaryotes. Mol. Biol. Evol. 2011;28:2561–2575. doi: 10.1093/molbev/msr079. [DOI] [PubMed] [Google Scholar]
- Fan X., Shi H., Lis J.T. Distinct transcriptional responses of RNA polymerases I, II and III to aptamers that bind TBP. Nucleic Acids Res. 2005;33:838–845. doi: 10.1093/nar/gki212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazal F.M., Meng C.A., Murakami K., Kornberg R.D., Block S.M. Real-time observation of the initiation of RNA polymerase II transcription. Nature. 2015;525:274–277. doi: 10.1038/nature14882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenouil R., Cauchy P., Koch F., Descostes N., Cabeza J.Z., Innocenti C., Ferrier P., Spicuglia S., Gut M., Gut I., Andrau J.C. CpG islands and GC content dictate nucleosome depletion in a transcription-independent manner at mammalian promoters. Genome Res. 2012;22:2399–2408. doi: 10.1101/gr.138776.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Tornero C., Moreno-Morcillo M., Rashid U.J., Taylor N.M.I., Ruiz F.M., Gruene T., Legrand P., Steuerwald U., Müller C.W. Crystal structure of the 14-subunit RNA polymerase I. Nature. 2013;502:644–649. doi: 10.1038/nature12636. [DOI] [PubMed] [Google Scholar]
- Ferreira R., Schneekloth J.S., Jr., Panov K.I., Hannan K.M., Hannan R.D. Targeting the RNA polymerase I transcription for cancer therapy comes of age. Cells. 2020;9:266. doi: 10.3390/cells9020266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French S.L., Osheim Y.N., Cioci F., Nomura M., Beyer A.L. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell Biol. 2003;23:1558–1568. doi: 10.1128/MCB.23.5.1558-1568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French S.L., Osheim Y.N., Schneider D.A., Sikes M.L., Fernandez C.F., Copela L.A., Misra V.A., Nomura M., Wolin S.L., Beyer A.L. Visual analysis of the yeast 5S rRNA gene transcriptome: regulation and role of La protein. Mol. Cell Biol. 2008;28:4576–4587. doi: 10.1128/MCB.00127-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek E.P., Kassavetis G.A. The RNA polymerase III transcription apparatus. J. Mol. Biol. 2001;310:1–26. doi: 10.1006/jmbi.2001.4732. [DOI] [PubMed] [Google Scholar]
- Gout J.-F., Li W., Fritsch C., Li A., Haroon S., Singh L., Hua D., Fazelinia H., Smith Z., Seeholzer S., et al. The landscape of transcription errors in eukaryotic cells. Sci. Adv. 2017;3:e1701484. doi: 10.1126/sciadv.1701484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I. The nucleolus—guardian of cellular homeostasis and genome integrity. Chromosoma. 2013;122:487–497. doi: 10.1007/s00412-013-0430-0. [DOI] [PubMed] [Google Scholar]
- Haddach M., Schwaebe M.K., Michaux J., Nagasawa J., O'brien S.E., Whitten J.P., Pierre F., Kerdoncuff P., Darjania L., Stansfield R., et al. Discovery of CX-5461, the first direct and selective inhibitor of RNA polymerase I, for cancer therapeutics. ACS Med. Chem. Lett. 2012;3:602–606. doi: 10.1021/ml300110s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harismendy O., Gendrel C.G., Soularue P., Gidrol X., Sentenac A., Werner M., Lefebvre O. Genome-wide location of yeast RNA polymerase III transcription machinery. EMBO J. 2003;22:4738–4747. doi: 10.1093/emboj/cdg466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J., Tollervey D. Repeat expansion in the budding yeast ribosomal DNA can occur independently of the canonical homologous recombination machinery. Nucleic Acids Res. 2011;39:8778–8791. doi: 10.1093/nar/gkr589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Maraia R.J. Comparison of the RNA polymerase III transcription machinery in Schizosaccharomyces pombe, Saccharomyces cerevisiae and human. Nucleic Acids Res. 2001;29:2675–2690. doi: 10.1093/nar/29.13.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurowitz E.H., Brown P.O. Genome-wide analysis of mRNA lengths in Saccharomyces cerevisiae. Genome Biol. 2003;5:R2. doi: 10.1186/gb-2003-5-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram Z.M., Schneider D.A., Lucius A.L. Transient-state kinetic analysis of multi-nucleotide addition catalyzed by RNA polymerase I. Biophys. J. 2021;120:4378–4390. doi: 10.1016/j.bpj.2021.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram Z.M., Scull N.W., Schneider D.S., Lucius A.L. Multi-start Evolutionary Nonlinear OpTimizeR (MENOTR): a hybrid parameter optimization toolbox. Biophys. Chem. 2021;279:106682. doi: 10.1016/j.bpc.2021.106682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvin J.D., Kireeva M.L., Gotte D.R., Shafer B.K., Huang I., Kashlev M., Strathern J.N. A genetic assay for transcription errors reveals multilayer control of RNA polymerase II fidelity. PLoS Genet. 2014;10:e1004532. doi: 10.1371/journal.pgen.1004532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs R.Q., Huffines A.K., Laiho M., Schneider D.A. The small-molecule BMH-21 directly inhibits transcription elongation and DNA occupancy of RNA polymerase I in vivo and in vitro. J. Biol. Chem. 2022;298:101450. doi: 10.1016/j.jbc.2021.101450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs R.Q., Ingram Z.M., Lucius A.L., Schneider D.A. Defining the divergent enzymatic properties of RNA polymerases I and II. J. Biol. Chem. 2021;296:100051. doi: 10.1074/jbc.RA120.015904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga J.T. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell. 2004;116:247–257. doi: 10.1016/s0092-8674(03)01078-x. [DOI] [PubMed] [Google Scholar]
- Kaplan C.D., Kornberg R.D. A bridge to transcription by RNA polymerase. J. Biol. 2008;7:39. doi: 10.1186/jbiol99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan C.D. Basic mechanisms of RNA polymerase II activity and alteration of gene expression in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2013;1829:39–54. doi: 10.1016/j.bbagrm.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G.A., Letts G.A., Geiduschek E.P. A minimal RNA polymerase III transcription system. EMBO J. 1999;18:5042–5051. doi: 10.1093/emboj/18.18.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva M.L., Nedialkov Y.A., Cremona G.H., Purtov Y.A., Lubkowska L., Malagon F., Burton Z.F., Strathern J.N., Kashlev M. Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol. Cell. 2008;30:557–566. doi: 10.1016/j.molcel.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulaeva O.I., Hsieh F.-K., Chang H.-W., Luse D.S., Studitsky V.M. Mechanism of transcription through a nucleosome by RNA polymerase II. Biochim. Biophys. Acta. 2013;1829:76–83. doi: 10.1016/j.bbagrm.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwapisz M., Beckouët F., Thuriaux P. Early evolution of eukaryotic DNA-dependent RNA polymerases. Trends Genet. 2008;24:211–215. doi: 10.1016/j.tig.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Laferté A., Favry E., Sentenac A., Riva M., Carles C., Chédin S. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 2006;20:2030–2040. doi: 10.1101/gad.386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laham-Karam N., Pinto G.P., Poso A., Kokkonen P. Transcription and translation inhibitors in cancer treatment. Front. Chem. 2020;8:276. doi: 10.3389/fchem.2020.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson M.H., Zhou J., Kaplan C.D., Palangat M., Kornberg R.D., Landick R., Block S.M. Trigger loop dynamics mediate the balance between the transcriptional fidelity and speed of RNA polymerase II. Proc. Natl. Acad. Sci. USA. 2012;109:6555–6560. doi: 10.1073/pnas.1200939109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Kim M., Han J., Yeom K.-H., Lee S., Baek S.H., Kim V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisica A., Engel C., Jahnel M., Roldán É., Galburt E.A., Cramer P., Grill S.W. Mechanisms of backtrack recovery by RNA polymerases I and II. Proc. Natl. Acad. Sci. USA. 2016;113:2946–2951. doi: 10.1073/pnas.1517011113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucius A.L., Lohman T.M. Effects of temperature and ATP on the kinetic mechanism and kinetic step-size for E.coli RecBCD helicase-catalyzed DNA unwinding. J. Mol. Biol. 2004;339:751–771. doi: 10.1016/j.jmb.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Malagon F., Kireeva M.L., Shafer B.K., Lubkowska L., Kashlev M., Strathern J.N. Mutations in the Saccharomyces cerevisiae RPB1 gene conferring hypersensitivity to 6-azauracil. Genetics. 2006;172:2201–2209. doi: 10.1534/genetics.105.052415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason P.B., Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Mekhail K., Khacho M., Carrigan A., Hache R.R.J., Gunaratnam L., Lee S. Regulation of ubiquitin ligase dynamics by the nucleolus. J. Cell Biol. 2005;170:733–744. doi: 10.1083/jcb.200506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhail K., Gunaratnam L., Bonicalzi M.E., Lee S. HIF activation by pH-dependent nucleolar sequestration of VHL. Nat. Cell Biol. 2004;6:642–647. doi: 10.1038/ncb1144. [DOI] [PubMed] [Google Scholar]
- Merz K., Hondele M., Goetze H., Gmelch K., Stoeckl U., Griesenbeck J. Actively transcribed rRNA genes in S. cerevisiae are organized in a specialized chromatin associated with the high-mobility group protein Hmo1 and are largely devoid of histone molecules. Genes Dev. 2008;22:1190–1204. doi: 10.1101/gad.466908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit P., Tschochner H. A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during transcription. EMBO J. 1998;17:3692–3703. doi: 10.1093/emboj/17.13.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.M., Lin J., Li T., Lucius A.L. E. coli ClpA catalyzed polypeptide translocation is allosterically controlled by the protease ClpP. J. Mol. Biol. 2013;425:2795–2812. doi: 10.1016/j.jmb.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrahi V., Henrie R.N., Marlier J.F., Johnson K.A., Benkovic S.J. Rate-limiting steps in the DNA polymerase I reaction pathway. Biochemistry. 1985;24:4010–4018. doi: 10.1021/bi00336a031. [DOI] [PubMed] [Google Scholar]
- Nechaev S., Adelman K. Promoter-proximal Pol II: when stalling speeds things up. Cell Cycle. 2008;7:1539–1544. doi: 10.4161/cc.7.11.6006. [DOI] [PubMed] [Google Scholar]
- Noe Gonzalez M., Blears D., Svejstrup J.Q. Causes and consequences of RNA polymerase II stalling during transcript elongation. Nat. Rev. Mol. Cell Biol. 2021;22:3–21. doi: 10.1038/s41580-020-00308-8. [DOI] [PubMed] [Google Scholar]
- Nomura M., Nogi Y., Oakes M. Madame Curie Bioscience Database; 2000-2013. Transcription of rDNA in the Yeast Saccharomyces cerevisiae. [Google Scholar]
- Patel S.S., Wong I., Johnson K.A. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991;30:511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- Paule M.R., White R.J. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltonen K., Colis L., Liu H., Jäämaa S., Moore H.M., Enbäck J., Laakkonen P., Vaahtokari A., Jones R.J., af Hällström T.M., Laiho M. Identification of novel p53 pathway activating small-molecule compounds reveals unexpected similarities with known therapeutic agents. PLoS One. 2010;5:e12996. doi: 10.1371/journal.pone.0012996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltonen K., Colis L., Liu H., Trivedi R., Moubarek M.S., Moore H.M., Bai B., Rudek M.A., Bieberich C.J., Laiho M. A targeting modality for destruction of RNA polymerase I that possesses anticancer activity. Cancer Cell. 2014;25:77–90. doi: 10.1016/j.ccr.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes T.D. Yeast ribosomal DNA genes are located on chromosome XII. Proc. Natl. Acad. Sci. USA. 1979;76:410–414. doi: 10.1073/pnas.76.1.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petesch S.J., Lis J.T. Overcoming the nucleosome barrier during transcript elongation. Trends Genet. 2012;28:285–294. doi: 10.1016/j.tig.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Płonka M., Wawrzycka D., Wysocki R., Boguta M., Cieśla M. Coupling of RNA polymerase III assembly to cell cycle progression in Saccharomyces cerevisiae. Cell Cycle. 2019;18:500–510. doi: 10.1080/15384101.2019.1578134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner J.B., Saunders C., Davie J.R., Hamkalo B.A. Ultrastructural organization of yeast chromatin. J. Cell Biol. 1982;93:217–222. doi: 10.1083/jcb.93.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder R.G., Rutter W.J. Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature. 1969;224:234–237. doi: 10.1038/224234a0. [DOI] [PubMed] [Google Scholar]
- Saunders A., Werner J., Andrulis E.D., Nakayama T., Hirose S., Reinberg D., Lis J.T. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science. 2003;301:1094–1096. doi: 10.1126/science.1085712. [DOI] [PubMed] [Google Scholar]
- Scheer U. Changes of nucleosome frequency in nucleolar and non-nucleolar chromatin as a function of transcription: an electron microscopic study. Cell. 1978;13:535–549. doi: 10.1016/0092-8674(78)90327-6. [DOI] [PubMed] [Google Scholar]
- Schier A.C., Taatjes D.J. Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev. 2020;34:465–488. doi: 10.1101/gad.335679.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm L., Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- Scull C.E., Clarke A.M., Lucius A.L., Schneider D.A. Downstream sequence-dependent RNA cleavage and pausing by RNA polymerase I. J. Biol. Chem. 2020;295:1288–1299. doi: 10.1074/jbc.RA119.011354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scull C.E., Ingram Z.M., Lucius A.L., Schneider D.A. A novel assay for RNA polymerase I transcription elongation sheds light on the evolutionary divergence of eukaryotic RNA polymerases. Biochemistry. 2019;58:2116–2124. doi: 10.1021/acs.biochem.8b01256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan R.M., Fong N., D'alessandro A., Bentley D.L. Widespread backtracking by RNA Pol II is a major effector of gene activation, 5' pause release, termination, and transcription elongation rate. Mol. Cell. 2019;73:107–118.e4. doi: 10.1016/j.molcel.2018.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A., Bhargava P. Regulation of tRNA gene transcription by the chromatin structure and nucleosome dynamics. Biochim. Biophys. Acta. Gene Regul. Mech. 2018;1861:295–309. doi: 10.1016/j.bbagrm.2017.11.008. [DOI] [PubMed] [Google Scholar]
- Studitsky V.M., Walter W., Kireeva M., Kashlev M., Felsenfeld G. Chromatin remodeling by RNA polymerases. Trends Biochem. Sci. 2004;29:127–135. doi: 10.1016/j.tibs.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Tan L., Wiesler S., Trzaska D., Carney H.C., Weinzierl R.O.J. Bridge helix and trigger loop perturbations generate superactive RNA polymerases. J. Biol. 2008;7:40. doi: 10.1186/jbiol98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turowski T.W., Leśniewska E., Delan-Forino C., Sayou C., Boguta M., Tollervey D. Global analysis of transcriptionally engaged yeast RNA polymerase III reveals extended tRNA transcripts. Genome Res. 2016;26:933–944. doi: 10.1101/gr.205492.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viktorovskaya O.V., Engel K.L., French S.L., Cui P., Vandeventer P.J., Pavlovic E.M., Beyer A.L., Kaplan C.D., Schneider D.A. Divergent contributions of conserved active site residues to transcription by eukaryotic RNA polymerases I and II. Cell Rep. 2013;4:974–984. doi: 10.1016/j.celrep.2013.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos S.M., Farnung L., Linden A., Urlaub H., Cramer P. Structure of complete Pol II–DSIF–PAF–SPT6 transcription complex reveals RTF1 allosteric activation. Nat. Struct. Mol. Biol. 2020;27:668–677. doi: 10.1038/s41594-020-0437-1. [DOI] [PubMed] [Google Scholar]
- Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Wei T., Najmi S.M., Liu H., Peltonen K., Kucerova A., Schneider D.A., Laiho M. Small-molecule targeting of RNA polymerase I activates a conserved transcription elongation checkpoint. Cell Rep. 2018;23:404–414. doi: 10.1016/j.celrep.2018.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner F., Grohmann D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat. Rev. Microbiol. 2011;9:85–98. doi: 10.1038/nrmicro2507. [DOI] [PubMed] [Google Scholar]
- Werner F. Structure and function of archaeal RNA polymerases. Mol. Microbiol. 2007;65:1395–1404. doi: 10.1111/j.1365-2958.2007.05876.x. [DOI] [PubMed] [Google Scholar]
- Zamft B., Bintu L., Ishibashi T., Bustamante C. Nascent RNA structure modulates the transcriptional dynamics of RNA polymerases. Proc. Natl. Acad. Sci. USA. 2012;109:8948–8953. doi: 10.1073/pnas.1205063109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data: All data reported in the article will be shared by the lead contact on request.
-
•
Code: We used our custom-built software package written in MATLAB, MENOTR (Ingram et al., 2021b), to perform model-dependent analyses. MENTOR is available on GitHub: https://github.com/ZachIngram/2021-MENOTR.
-
•
All other items: Any additional information required to reanalyze the data reported in this article is available from the lead contact on request.