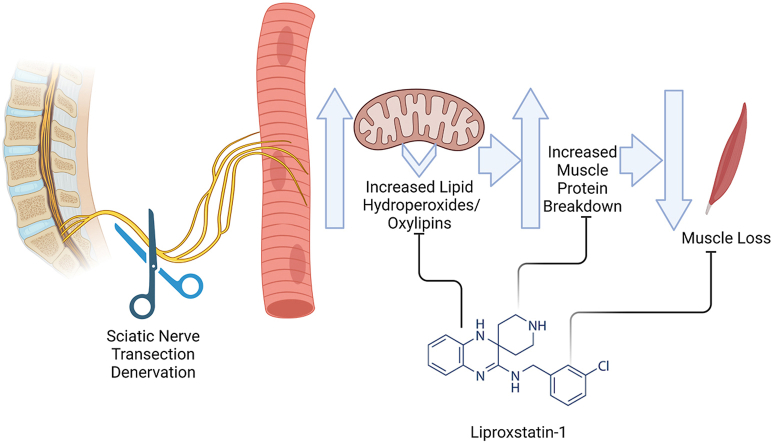
Abstract
Loss of innervation is a key driver of age associated muscle atrophy and weakness (sarcopenia). Our laboratory has previously shown that denervation induced atrophy is associated with the generation of mitochondrial hydroperoxides and lipid mediators produced downstream of cPLA2 and 12/15 lipoxygenase (12/15-LOX). To define the pathological impact of lipid hydroperoxides generated in denervation-induced atrophy in vivo, we treated mice with liproxstatin-1, a lipid hydroperoxide scavenger. We treated adult male mice with 5 mg/kg liproxstain-1 or vehicle one day prior to sciatic nerve transection and daily for 7 days post-denervation before tissue analysis. Liproxstatin-1 treatment protected gastrocnemius mass and fiber cross sectional area (∼40% less atrophy post-denervation in treated versus untreated mice). Mitochondrial hydroperoxide generation was reduced 80% in vitro and by over 65% in vivo by liproxstatin-1 treatment in denervated permeabilized muscle fibers and decreased the content of 4-HNE by ∼25% post-denervation. Lipidomic analysis revealed detectable levels of 25 oxylipins in denervated gastrocnemius muscle and significantly increased levels for eight oxylipins that are generated by metabolism of fatty acids through 12/15-LOX. Liproxstatin-1 treatment reduced the level of three of the eight denervation-induced oxylipins, specifically 15-HEPE, 13-HOTrE and 17-HDOHE. Denervation elevated protein degradation rates in muscle and treatment with liproxstatin-1 reduced rates of protein breakdown in denervated muscle. In contrast, protein synthesis rates were unchanged by denervation. Targeted proteomics revealed a number of proteins with altered expression after denervation but no effect of liproxstain-1. Transcriptomic analysis revealed 203 differentially expressed genes in denervated muscle from vehicle or liproxstatin-1 treated mice, including ER stress, nitric oxide signaling, Gαi signaling, glucocorticoid receptor signaling, and other pathways. Overall, these data suggest lipid hydroperoxides and oxylipins are key drivers of increased protein breakdown and muscle loss associated with denervation induced atrophy and a potential target for sarcopenia intervention.
Keywords: Skeletal, Mitochondria, ROS, Neuromuscular junction, Lipoxygenase
Graphical abstract
Highlights
-
•
Liproxstatin-1 ameliorates denervation-induced muscle atrophy.
-
•
Liproxstatin-1 reduces the basal hydroperoxides and oxylipins in denervated muscle.
-
•
Liproxstatin-1 reduces muscle protein breakdown rates in denervated muscle.
1. Introduction
Sarcopenia is characterized by progressive loss of skeletal muscle mass and strength with an increased risk of adverse outcomes such as physical disability, poor quality of life and even death [[1], [2], [3], [4], [5], [6]]. In an effort to reveal underlying mechanisms of sarcopenia and potential effective interventions, our laboratory and others have clearly shown that loss of motor neurons and reduced motor neuron innervation at neuromuscular junctions (NMJs) are key drivers of sarcopenia [[7], [8], [9], [10], [11], [12], [13]]. A key finding from our previous studies is that significant levels of hydroperoxides are generated from both isolated mitochondria and permeabilized muscle fiber bundles in skeletal muscle that has lost innervation [14,15], and we found that the production of hydroperoxides is strongly correlated to the extent of muscle loss in different models of neurogenic atrophy [16]. In an effort to determine if reducing hydroperoxides protects against denervation-induced muscle loss, we performed a sciatic nerve transection in mice overexpressing mitochondrial targeted H2O2 scavengers. To our surprise overexpression of mitochondrial hydrogen peroxide scavengers did not reduce hydroperoxides associated with denervation or protect against denervation-induced muscle loss [16]. In contrast, inhibition of cytosolic phospholipase A2 (cPLA2) reduced hydroperoxide generation, and protected against denervation-induced muscle loss [16,17]. These data led us to explore whether oxygenated lipids, as opposed to electron transport chain generated reactive oxygen species (ROS), are the primary species of hydroperoxides generated in denervated muscle [16,17].
Oxylipins are generated by oxygenation of polyunsaturated fatty acids such as arachidonic acid, eicosapentaenoic acid (EPA), docosahexaenoic acid (DHA), linoleic acid (LA), and other fatty acids [18] initially formed by chemical oxidation events or enzymatically. Oxylipins can be mono-oxygenated or di-oxygenated (lipid hydroperoxides), and oxylipins are a key component of lipid signaling [19]. In a previous study designed to explore whether oxygenated lipids contribute to denervation-induced atrophy, our laboratory showed that reducing enzymatic generation of oxylipins by genetic deletion of 12/15-lipoxygenase (12/15-LOX) protected against denervation-induced muscle loss [20]. However, genetic deletion of 5-lipoxygenase did not protect against denervation-induced muscle atrophy [20]. Lipid hydroperoxides are well known for their ability to promote oxidative damage and pathologic conditions. For example, lipid peroxidation leads to the production of toxic signaling molecules such as 4-hydroxynonenal (4-HNE) [21,22], which has been shown to trigger skeletal muscle pathology [[21], [22], [23]]. Lipid hydroperoxides can be further converted by an array of secondary reactions to form several classes of mono-oxygenated oxylipins [18]. Overexpression of the phospholipid hydroperoxide glutathione peroxidase (Gpx4), an enzyme that reduces lipid hydroperoxides within membranes, ameliorated denervation-induced muscle loss [17]. Together, these data support that oxygenated lipids may contribute to denervation-induced muscle loss.
The underlying mechanisms of lipid hydroperoxide-induced muscle degeneration and the potential for oxylipins as a viable intervention target for age related muscle loss are not defined. Researchers discovered liproxstatin-1 from screening of small molecules that could prevent cell death in response to deletion of glutathione peroxidase 4 (GPx4, an enzyme which neutralizing lipid hydroperoxides in membranes) in kidney [24]. Liproxstatin-1 decreased sporadic lipid peroxidation and enzymatic insertion of polyunsaturated fatty acids into membranes by ACSL4 (Acyl-CoA synthetase 4) [25,26]. Liproxstatin-1 acts as a radical-trapping antioxidant and is ∼10 fold more reactive towards lipid hydroperoxides than other forms of reactive oxygen species [26]. We have previously shown that Liproxstatin-1 can inhibit the Amplex Red signal in vitro in response to both hydrogen peroxide and 15-(S) HpETE [16]. Therefore, treatment with liproxstatin-1 has the potential to prevent the accumulation of lipid hydroperoxides and prevent muscle lipid peroxide damage in muscle. The goal of the current study is to test if neutralizing lipid hydroperoxides via treatment with liproxstatin-1 can inhibit denervation-induced muscle loss. We hypothesize that treatment with liproxstatin-1 will ameliorate denervation-induced muscle atrophy. Our data suggest that lipid hydroperoxides and oxylipins generation in skeletal muscle are key potential contributors to denervation-induced protein degradation and muscle atrophy.
2. Materials and methods
Animals. We conducted all animal experiments in accordance with the guidelines for the care and use of laboratory animals for the Oklahoma Medical Research Foundation (OMRF). The Institutional Animal Care and Use Committees at the OMRF approved the study. All experiments used mice on a C57BL/6J genetic background currently housed in the mouse colony of Dr. Van Remmen at the OMRF.
Sciatic nerve transection surgeries and drug treatment. We performed Sciatic nerve transection and sham surgeries on 6–8-month-old male C57BL/6J mice as previously described [27,28]. We euthanized mice seven days after the surgery, and we collected tissues. We treated mice with liproxstatin-1 (5 mg/kg BW) or vehicle (10% DMSO and 90% corn oil) following sciatic nerve transection surgery. We dissolved Liproxstatin-1 (Selleck Chemicals LLC, Houston, TX 77014) in DMSO (10 mg/ml) and further diluted in corn oil to 1 mg/ml. We gave mice intraperitoneal (ip) injections of vehicle control or 5 mg/kg liproxstatin-1 once daily beginning with the first injection one day prior to surgery and continuing until euthanasia seven days after surgery. After sacrifice, we compared gastrocnemius muscle from the following groups: sham vehicle, sham liproxstatin-1, denervated vehicle, and denervated liproxstatin-1.
Lipid extraction and LC/MS/MS analysis. We homogenized tissue samples with ceramic beads in 1 ml antioxidant buffer containing 100 μM diethylenetriaminepentaacetic acid (DTPA) and 100 μM M butylated hydroxytoluene (BHT) in phosphate buffered saline using a Bead Ruptor Elite for 30 s at 6 m/s, under cooled nitrogen gas (4 °C). We spiked samples with 12-HETE-d8 (2.5 ng), 15-HETE-d8 (2.5 ng), 13-HODE-d4 (2.3 ng), standards (Cayman Chemical) and 10 ng of PE 15:0–18:1-d7 (Avanti) prior to homogenization. Lipids were extracted by adding a 2.5 ml solvent mixture (1 M acetic acid/isopropanol/hexane; 2:20:30, v/v/v) to 1 ml homogenate in a glass extraction vial and vortexed for 60 sec. 2.5 ml hexane was then added to samples and after vortexing for 60 s, tubes were centrifuged (500 g for 5 min at 4 °C) to recover lipids in the upper hexane layer (aqueous phase), which was transferred to a clean tube. We re-extracted aqueous samples as described above by addition of 2.5 ml hexane and combined upper layers. We performed the Bligh and Dyer technique for lipid extraction from the lower aqueous layer. Specifically, we added 3.75 ml of a 2:1 ratio of methanol: chloroform followed by vortexing for 60 s. We added subsequent additions of 1.25 ml chloroform and 1.25 ml water followed with a vortexing step for 60 s, and we recovered the lower layer following centrifugation as described above and combined the lower layer with the upper layers from the first stage of extraction. We dried solvent under vacuum and we reconstituted lipid extract in 100 μl HPLC grade methanol. For oxylipin analysis, we separated lipids by liquid chromatography (LC) using a gradient of 30–100% B over 20 min (A: Water: Mob B 95:5 + 0.1% glacial acetic acid, B: Acetonitrile: Methanol – 80:15 + 0.1% glacial acetic acid) on an Eclipse Plus C18 Column (Agilent), and analyzed on a Sciex QTRAP® 6500 LC-MS/MS system. Source conditions: TEM 475 °C, IS -4500, GS1 60, GS2 60, CUR 35. Lipids were detected in negative ion mode using MRM monitoring with the following parent to daughter ion transitions for lipids that are reported in this study: 12-HETE [M − H]- 319.2/179.1, 15-HETE [M − H]- 319.2/219.1, 13-HODE [M − H]- 295.2/195.1, 15-HETrE [M − H]- 321.2/221.1, 15-HEPE [M − H]- 317.2/219.1, 14-HDOHE [M − H]- 343.2/205.1, 17-HDOHE [M − H]- 343.2/201.1, 13-HOTrE [M − H]- 293.2/195.1. For full details of this scheduled MRM method which detects >100 lipids see: [29]. We monitored deuterated internal standards using precursor to product ions transitions of 12-HETE-d8 [M − H]- 327.2/184.1, 15-HETE-d8 [M − H]- 327.2/226.1 and 13-HODE-d4 [M − H]- 299.2/198.1. We integrated chromatographic peaks using Multiquant 3.0.2 software (Sciex). The criteria for assigning a peak was signal: noise of at least 5:1 and with at least 7 points across a peak. We calculated the ratio of analyte peak areas to internal standard and quantified lipids using a standard curve made up and run at the same time as the samples. We then standardized each oxylipin per mg of tissue. For oxPL measurements, lipids were separated by liquid chromatography (LC) using a gradient of 50–100% B over 10 min, followed by 30 min at 100% B (A: methanol: acetonitrile: water, 1 mM ammonium acetate, 60:20:20. B: methanol, 1 mM ammonium acetate), with a flow rate of 0.2 ml/min on a Luna C18 column (15 cm × 2 mm, 3 μm) (Phenomenex), and analyzed on a Sciex QTRAP® 6500 LC-MS/MS system. Source conditions, negative ion mode: TEM 500 °C, IS -4500, GS1 40, GS2 30, CUR 35. Lipids were detecting using MRM monitoring with the following parent to daughter ion transitions: PE 18:0a_20:4(O) and PC 16:0a_20:4(O) [M − H]- 782.6/319.2, PE 18:0p_20:4(O) [M − H]- 766.6/319.2, PE 18:1p_20:4(O) [M − H]- 764.6/319.2, PE 16:0p_20:4(O) [M − H]- 738.6/319.2 and PC 18:0a_20:4(O) [M − H]- 810.7/319.2. We monitored PE 15:0–18:1-d7 as internal standard using precursor to production ion transition of: [M − H]- 709.5/288.2. We integrated chromatographic peaks using Analyst 1.7 software (Sciex). The criteria for assigning a peak was signal: noise of at least 5:1 and with at least 7 points across a peak. We integrated these MRM transitions which gave rise to multiple peaks indicating the presence of isomers as a group and expressed them as fold-change between sample types, for analyte: internal standard ratios.
Gastrocnemius cross sectional area. We sectioned gastrocnemius muscle after sacrifice, mounted in optimum cutting temperature compound (OCT), and flash frozen in liquid nitrogen-cooled isopentane. We cut sections (10 μm), mounted, and stained the sections with laminin. Briefly, we blocked sections with 10% normal goat serum and 2% bovine serum albumin in Phosphate Buffered Saline (PBS) for 1 h. We then stained sections overnight with laminin antibody (Sigma L9393) diluted in PBS with 1% BSA. The next day we washed the sections in PBS 3 times for 5 min followed by incubation with a FITC conjugated secondary antibody. We washed sections in PBS 3 times for 5 min. We visualized and captured images with Zeiss Axiovert 200 M microscope, Zeiss Axiocam MRC camera, and Zeiss AxioVision software V4.8.2.0 (Carl Zeiss AG, Oberkochen, Germany) at 10 × magnification. We then measured the cross-sectional area of the muscle fibers using an ImageJ automated analysis.
Muscle fiber permeabilization, oxygen consumption, and hydroperoxide production rate measurement. Muscle fiber bundles were permeabilized with saponin for the measurement of oxygen consumption and hydroperoxide production using Amplex UltraRed and the Oroboros Oxygraph-2k (O2k, OROBOROS Instruments, Innsbruck, Austria) with fluorometer as previously described [27,30,31]. We added H2O2, 12-HpETE, 15-HpETE, 12-HETE, and 15-HETE into the O2K chamber to test if the oxylipins react with the Amplex Red probe (Fig. 1A). In Fig. 1B, we removed denervated gastrocnemius muscle from each animal and separated into 4 permeabilized fiber bundles per animal. Fiber bundles were either untreated or treated with 3 μM liproxstatin-1 in vitro. We added the treatments directly to the Eppendorf tubes during the preparation and in the O2K chamber. In Fig. 1C, sham and denervated fiber bundles from mice treated in vivo with either vehicle or liproxstatin-1 were permeabilized and washed. We measured hydroperoxide generation and oxygen consumption in the O2K respirometer. Briefly, OCR and peroxide production were measured in permeabilized fiber bundles in buffer Z media containing 10 μM Amplex UltraRed (Molecular Probes, Eugene, OR), 1 U/ml horseradish peroxidase, superoxide dismutase and blebbistatin (25 μM) at 37 °C. Rates of respiration and peroxide production were determined using the following sequential additions of substrates and inhibitors: glutamate (10 mM), malate (2 mM), pyruvate (5 mM), ADP (5 mM), succinate (10 mM), rotenone (1 μM), Antimycin A (1 μM), and TMPD (0.5 mM) immediately followed by ascorbate (5 mM, ascorbate is added to ensure TMPD is reduced, so TMPD can continue to donate electrons). We normalized respiration measurements to Antimycin A to account for non-mitochondrial oxygen consumption. We normalized data for both OCR and rates of hydroperoxide generation by milligrams of muscle bundle wet weights weighed on Acculab AL-104 scale.
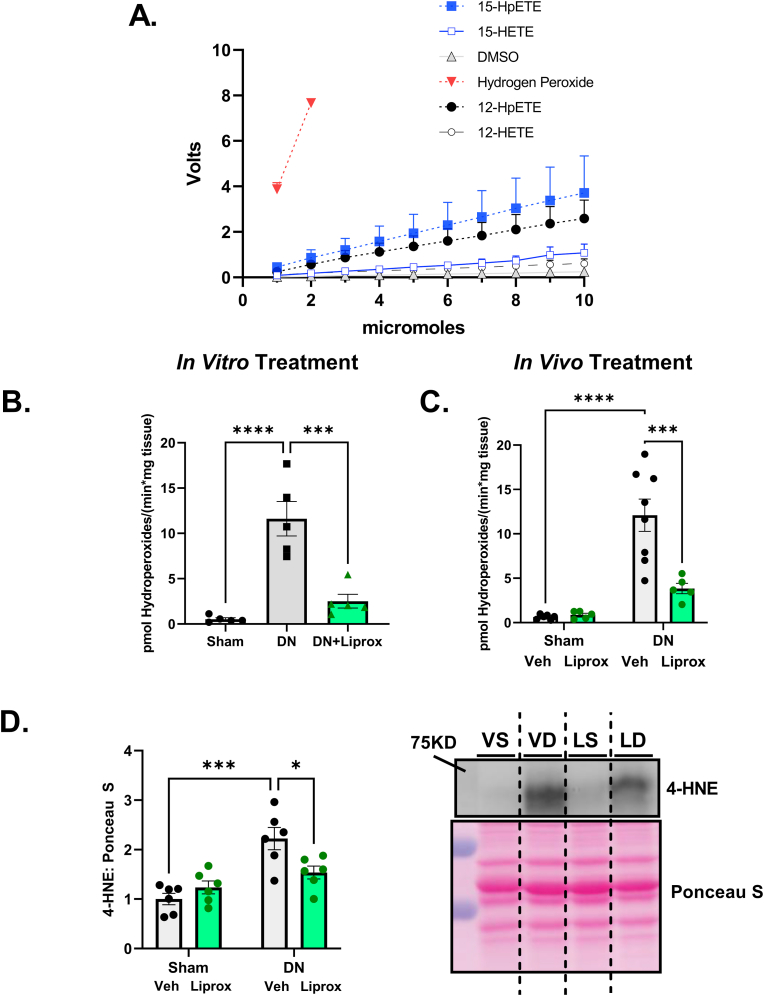
Fig. 1.
Liproxstatin-1 reduces hydroperoxide generation in denervated muscle. (A) Lipid Hydroperoxides and mono-oxygenated oxylipins react with amplex red. (B) Hydroperoxide generation in fiber bundles from sham and denervated muscle with liproxstatin-1 treatment in vitro. (C) Hydroperoxide generation in fiber bundles from mice treated daily with either Veh or Liprox. (D) 4-HNE content in gastrocnemius muscle. (E) Representative 4-HNE image. An n = 5–6 per group was used. DN, denervated; Liprox, Liproxstatin-1. Asterisk denotes post hoc differences at an alpha set at P < 0.05.
Targeted quantitative mass spectrometry for proteins. We used targeted quantitative mass spectrometry to measure protein abundance as previously described [[32], [33], [34]]. Briefly, we homogenized gastrocnemius samples in RIPA buffer containing 10 mM Tris‐Cl (pH 8.0), 1 mM EDTA, 1% Triton X‐100 (v/v), 0.1% sodium deoxycholate (w/v), 0.1% sodium dodecyl sulfate (w/v), 140 mM NaCl, and 1 mM phenylmethylsulfonyl fluoride, with protease inhibitor cocktail (Calbiochem Set III, EDTA‐free; EMD Millipore; Billerica, MA, USA), and we used Bradford assay to determine protein concentration. For targeted proteomic analysis at the Oklahoma Medical Research Foundation core facility, we used 60 μG protein, as previously described [32]. We added 200 μl of 1% SDS and 20 μl of internal standard to each sample. We then mixed each sample and heated for 15 min in a water bath at 70° Celsius. We added acetone to precipitate in freezer overnight. In preparation of the gel, we removed samples from the freezer and spun down for 10 min at 10 000×g in the microfuge. We poured off the supernatant and dried in the speed vac before we reconstituted in DSB at 1 μg/μL. We mixed the samples and heated once again before loading the gel. After running the gel, we fixed and stained the gel with GelCode blue. We cut sections, divided into smaller pieces and washed/destained. We reduced the proteins with DTT and alkylated with iodoacetamide. We then washed with ethanol and bicarb and then digested with 1 μg of trypsin overnight at room temperature. We extracted the peptides produced from the gel, the extract evaporated to dryness, and reconstituted in 1% acetic acid for analysis. We analzyed the digest samples using a Q-Exactive orbitrap system (ThermoScintific). We injected 5 μl aliquots at 1.5 μL/min with 0.1% formic acid. We eluted the column at 150 nL/min with a linear gradient of CH3CN in water with 0.1% formic acid (2% CH3CN to 65% CH3CN in 60 min). The orbitrap mass spectrometer acquired full scan mass spectra with a m/z resolution of 280 000. Ion source settings included a spray voltage of 1.5 kV, ion transfer tube temperature of 300 °C, and positive ions mode. We analyzed data using Skyline. Skyline is an application for targeted proteomics method creation and quantitative data analysis.
Measurement of protein turnover. We determined protein synthesis rates according to our previously described methods [[35], [36], [37], [38]]. Mice received a bolus i.p. injection (equivalent to approximately 5% body water) of 99% deuterium oxide (D2O) immediately after surgery. We supplemented drinking water thereafter with 8% D2O in drinking water until euthanasia. We homogenized skeletal muscle tissue 1:20 in isolation buffer (100 mM KCl, 40 mM Tris HCl, 10 mM Tris base, 5 mM MgCl2, 1 mM EDTA, 1 mM Adenosine triphosphate, pH = 7.5) with phosphatase and protease inhibitors (HALT, Thermo Fisher Scientific) using a bead homogenizer (Next Advance Inc., Averill Park, NY, USA). After homogenization, we isolated subcellular fractions via differential centrifugation as previously described [[36], [37], [38]]. The pentafluorobenzyl‐N,N‐di (pentafluorobenzyl) derivative of alanine was analyzed by an Agilent 7890A GC coupled to an Agilent 5975C MS as previously described [[36], [37], [38]]. We analyzed distilled plasma on a Liquid Water Isotope Analyzer (LWIA‐45‐EP, Los Gatos Research, Inc., San Jose, CA, USA). We calculated the newly synthesized fraction (f) of proteins from the enrichment of alanine bound in muscle proteins over the entire labelling period, divided by the true precursor enrichment (p), using plasma D2O enrichment with mass isotopomer distribution analysis adjustment [39]. The period of D2O measurement was a period of muscle loss, which violates the steady state assumptions of isotopic labelling. To account for this non‐steady state condition, we calculated protein synthesis (Ksyn) and protein breakdown (Kdeg) based on our previously published work [[40], [41], [42]].
RNA Isolation and RNA Sequencing. We extracted total RNA from gastrocnemius using TRI reagent solution (Invitrogen, Carlsbad, CA, United States) as previously described [31,43]. RNA sequencing was performed at the OMRF Clinical Genomics Core and analyzed by the Geroinformarics Core in the Oklahoma Nathan Shock Center of Excellence in the Biology of Aging as previously described [44].
Differential Gene Expression Analysis. We quantified gene expression by the number of reads mapped to the sense-strand exons and converted to reads per kilobase per million (RPKM). We flagged genes as detectable with an empirical minimum RPKM of 0.1 and we computed fold changes as the ratio between the arithmetic mean RPKM values of the groups. We reported genes with a fold change ≥1.5 in either direction or with a p < 0.05 as significantly differentially expressed genes (DEGs).
Ingenuity Pathway Analysis. Using ingenuity upstream analysis (IPA, QIAGEN Redwood City, CA, USA, www.qiagen.com/ingenuity we compared the genes that changed the most between vehicle-treated and Liproxstatin-1-treated denervated muscle. We reported the most significant analysis results based on p < 0.0001 and activation z-score > 2.
Western blot analysis. We performed western blots as previously described [30,43,45]. Briefly, we homogenized gastrocnemius muscle in RIPA buffer. Protein concentrations were determined using a Bradford assay. We resolved 20–40 μg of protein by sodium dodecyl sulfate‐polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane and blocked in 5% weight by volume bovine serum albumin in Tris‐buffered saline with 0.2% Tween 20. We probed membranes overnight for antibodies specific to 4-HNE. Imaging was performed with G:BOX imaging system (Syngene) and quantified using Alpha View: Protein Simple analysis software.
Statistical analysis. For animal experiments, independent factors were Surgery (Sham or Denervated) and Treatment (Vehicle or Liproxstatin-1). A Two-Way ANOVA was employed for each dependent variable for animal experiments. Where we found significant F‐ratios, we used Tukey Kramer post hoc test to determine differences among means. For all experiments, the comparison‐wise error rate, α, was set at 0.05 for all statistical tests. We analyzed data and compiled figures using GraphPad Prism (La Jolla, CA, USA) and data expressed as mean ± SEM.
3. Results
Liproxstatin-1 reduces hydroperoxide generation in denervated muscle. We have previously shown that Amplex Red reacts with lipid hydroperoxides [16] and that only a portion of the Amplex Red derived signal following denervation is inhibited by the hydrogen peroxide scavenger catalase, even at very high levels of catalase. In Fig. 1A, we expanded on our prior findings by showing the Amplex Red signal in vitro responded to the lipid hydroperoxides 12-HpETE and 15-HpETE, as well as the mono-oxygenated oxylipins 12-HETE and 15-HETE. Although the response is less robust than hydrogen peroxide, the lipid hydroperoxide/oxylipin species show a clear reaction with Amplex Red in agreement with our previous report [16].
Here we asked whether liproxstatin-1 treatment in vivo and in vitro could inhibit the Amplex Red signal in permeabilized denervated muscle fiber bundles. As shown in Fig. 1B, treatment of permeabilized fiber bundles from denervated muscle with 3 μM liproxstatin-1 inhibited the Amplex Red signal associated with denervation by approximately 80%. To test whether liproxstatin-1 treatment in vivo could reduce hydroperoxides after denervation, we treated mice with daily injections of either vehicle or liproxstatin-1 in vivo beginning one day prior to and once daily for 7 days after sciatic nerve transection surgery. Denervation elevated the Amplex Red signal several folds at 7 days post denervation in permeabilized fiber bundles in denervated versus sham muscle from vehicle treated mice. Liproxstatin-1 lowered the hydroperoxide signal nearly 70% in fibers from denervated mice (Fig. 1C). Consistent with the reduced Amplex Red signal in muscle from liproxstatin-1 treated mice, denervation elevated 4-hydroxyneoneal (4-HNE) content, a marker of lipid peroxidation, and liproxstatin-1 reduced 4-HNE content (Fig. 1D/1E).
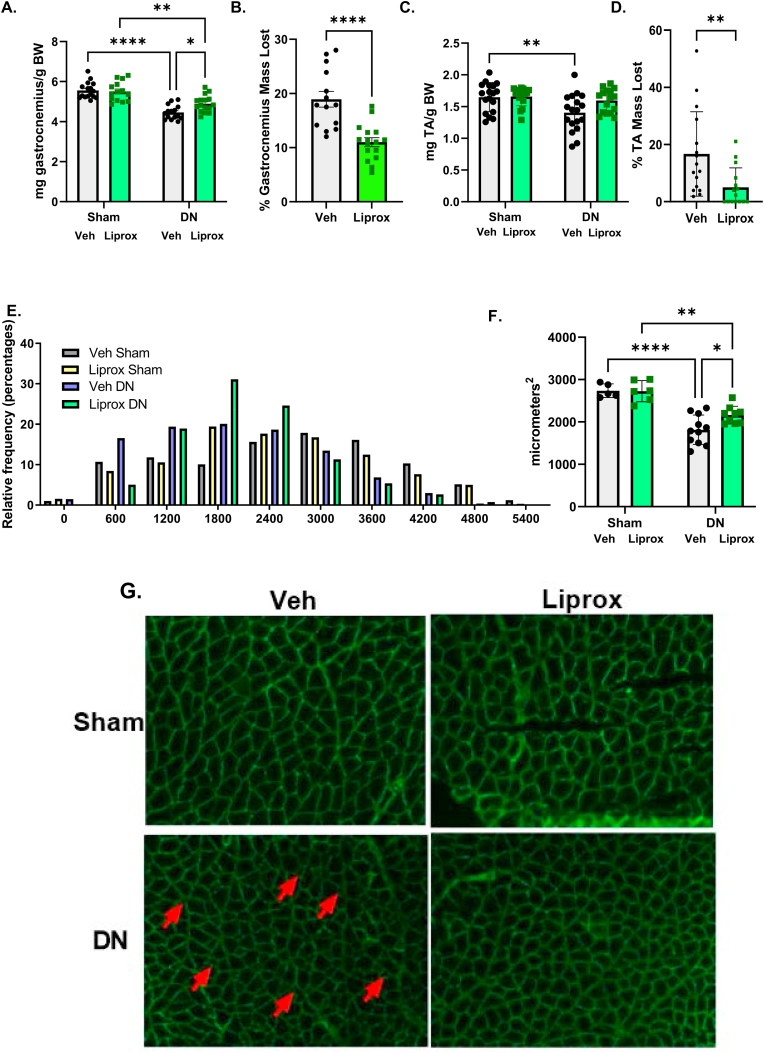
Liproxstatin-1 treatment ameliorates denervation-induced muscle loss. We measured gastrocnemius and tibialis anterior mass and fiber cross sectional area in gastrocnemius muscle in response to daily treatment with liproxstatin-1 beginning one day before surgery and continuing for 7 days post-surgery. The vehicle treated mice lost close to 20% gastrocnemius muscle mass in response to denervation. In contrast, liproxtatin-1 treatment reduced the percentage of muscle mass loss in the denervated gastrocnemius muscle by almost 50% (Fig. 2A and B). The tibialis anterior muscle showed a similar response to denervation and liproxstatin-1 treatment (Fig. 2C and D). There were also fewer small sized fibers after denervation in liproxstatin-1 treatment mice, while there was no change in the fiber size distribution with the sham surgery (Fig. 2E). The mean cross-sectional area was higher in liproxstatin-1 treated denervated muscle (Fig. 2F). Fig. 2G displays the representative images. The arrows are highlighting small muscle fibers in denervated muscle from vehicle.
Fig. 2.
Liproxstatin-1 treatment ameliorates denervation-induced muscle loss. (A) Gastrocnemius muscle mass. (B) Percent gastrocnemius mass loss in denervated muscle from contralateral sham control. (C) Tibialis anterior muscle mass. (D) Percent Tibialis anterior mass loss in denervated muscle from contralateral sham control. (E) Histogram showing the distribution of fiber sizes in denervated muscle from mice treated with either vehicle or liproxstatin-1. (F) Mean cross sectional area of gastrocnemius muscle. (G) Representative images for cross sectional area analysis. DN, denervated; Veh, Vehicle; Liprox, Liproxstatin-1. Veh, grey bar; Liprox, green bar. Asterisk denotes post hoc differences at an alpha set at P < 0.05. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
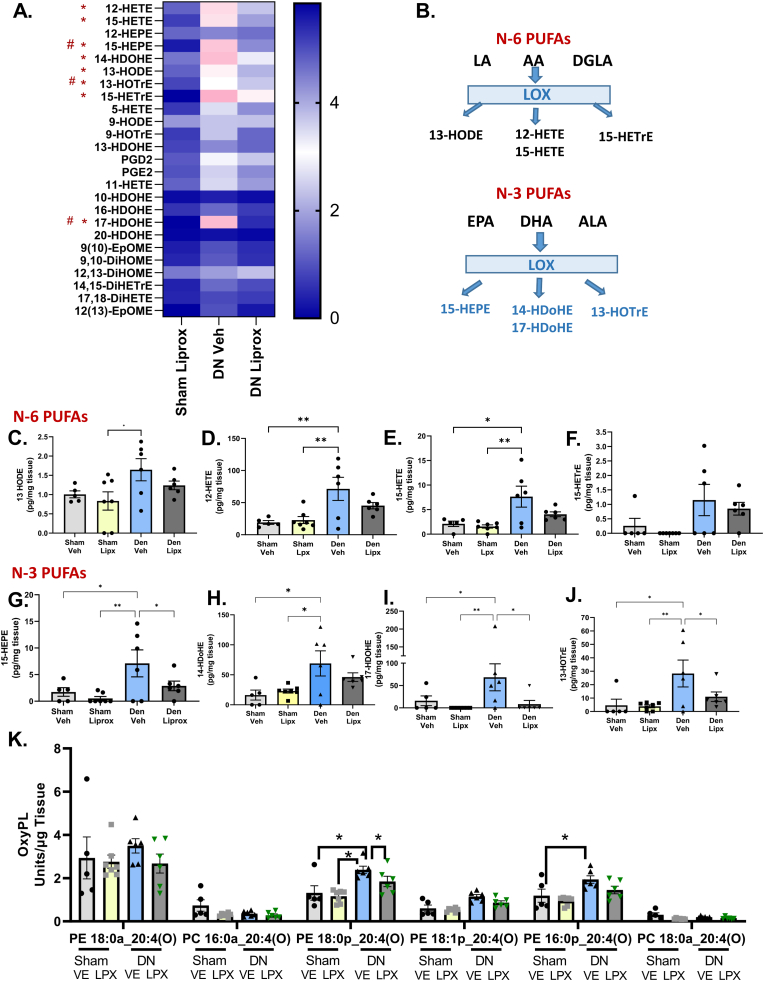
Denervation elevated oxylipin mediators in muscle. To better understand the potential role of oxylipins in skeletal muscle in response to loss of innervation, we used lipidomic analysis. Fig. 3 shows the content of oxylipins in sham and denervated gastrocnemius muscle from mice treated with vehicle or liproxstatin-1. Of the 25 oxylipin species detected in gastrocnemius muscle after sciatic nerve transection, denervation increased 8 in vehicle treated mice (Fig. 3A). Denervation elevated the oxylipins 13-HODE, 12-HETE, 15-HETE, 15-HETrE, 15-HEPE, 14-HDOHE, 17-HDOHE, and 13-HOTrE (Fig. 3 C-J). Denervation did not elevate oxylipins in liproxstatin-1 treated mice (Fig. 3 C-J). Interestingly, denervation elevated oxylipins generated via 12/15-LOX or platelet type 12-LOX and originate from both n3 and n6 polyunsaturated fatty acids, and an enzymatic origin of oxylipins likely since the most abundant were the 12-HETE and 14-HDOHE (Fig. 3B). Treatment with liproxstatin-1 reduced the content of three of the eight elevated oxylipins in denervated muscle, specifically 13-HOTrE, 15-HEPE, and 17-HDOHE (Fig. 3 G/I/J).
Fig. 3.
Oxylipin content is elevated in denervated muscle and treatment with liproxstatin-1 reduces oxylipin content. (A) Heat map showing changes in oxylipin content relative to Sham Veh. * indicates a statistical difference between vehicle sham and vehicle denervated. # indicates a statistical difference between vehicle denervated and liproxstatin-1 denervated. (B) Schematic showing where the oxylipins are generated from. (C–J) Statistically different oxylipins individually graphed out. (K) Graph showing the different species of oxidized phospholipids. N of 5–7 per group was used. DN, denervated; Veh, Vehicle; Liprox, Liproxstatin-1. Asterisk denotes post hoc differences at an alpha set at P < 0.05.
Next, we analyzed muscle for the presence of oxidized phospholipids (oxPL). First, we performed a precursor LC/MS/MS run, scanning for the product ion at m/z 319.2, which represents mono-oxygenated forms of arachidonate, including HETEs, esterified to phospholipids (Supplementary Fig. 1). The LC/MS/MS analysis showed the presence of ions, at m/z 782.6, 766.6 and 738.6, with weaker ions at m/z 810.7 and 764.6. These are consistent with phosphatidylethanolamines (PE) and phosphatidylcholines (PC) previously identified as originating from 12/15-LOX in monocytes and macrophages, or 12-LOX in platelets [46,47]. The precursor scan signal was weak indicating low abundance of the lipids (Supplementary Fig. 1). Next, we monitored the precursor to product ion pairs, in multiple reaction monitoring (MRM) mode. For each MRM transition, there were several similar sized peaks seen, indicating that the lipids are comprised of several positional isomers (Supplementary Fig. 2). To confirm this, we acquired enhanced product ion spectra (EPI) for the most abundant series, detected at m/z 782, which showed the presence of expected ions for PE 18:0_20:4(O) including: 18:0 at Sn1 (m/z 283) and 20:4(O) at Sn2 (m/z 319) in all peaks. Furthermore, we detected fragment ions that originate from 5-HETE (m/z 115, 14.1 min), 12-HETE (m/z 179, 13.4 min), 11-HETE (m/z 167, 13.2 min) and 15-HETE (m/z 219, 13.0 min) in product ion spectra, however these were very low abundance (Supplementary Fig. 3). We also detected the 8-HETE ion at m/z 155, but the signal was extremely low (Supplementary Figure 3). The remaining precursor ions at m/z 738, 764, 766 and 810 were less abundant than m/z 782, and so EPI spectra could not fully assign structure, but based on their relative retention times, they are putatively assigned as PE and PCs containing mixed HETE isomers: PE 18:0p_20:4(O) (m/z 766), PE 18:1p_20:4(O) (m/z 764), PE 16:0p_20:4(O) (m/z 738), PC 18:0a_20:4(O) (m/z 810).
To compare the impact of denervation, we monitored the lipids by the HETE product ion (m/z 319), since this method was more sensitive. Thus, we integrated each molecular species as a group of peaks (e.g., not distinguishing positional isomers for HETEs), and expressed their levels as ratio of analyte: internal standard (area). Although there was variation across samples, the data showed that denervation elevated the levels of four PE families, and liproxsatin-1 treatment suppressed the PE families (Fig. 3K).
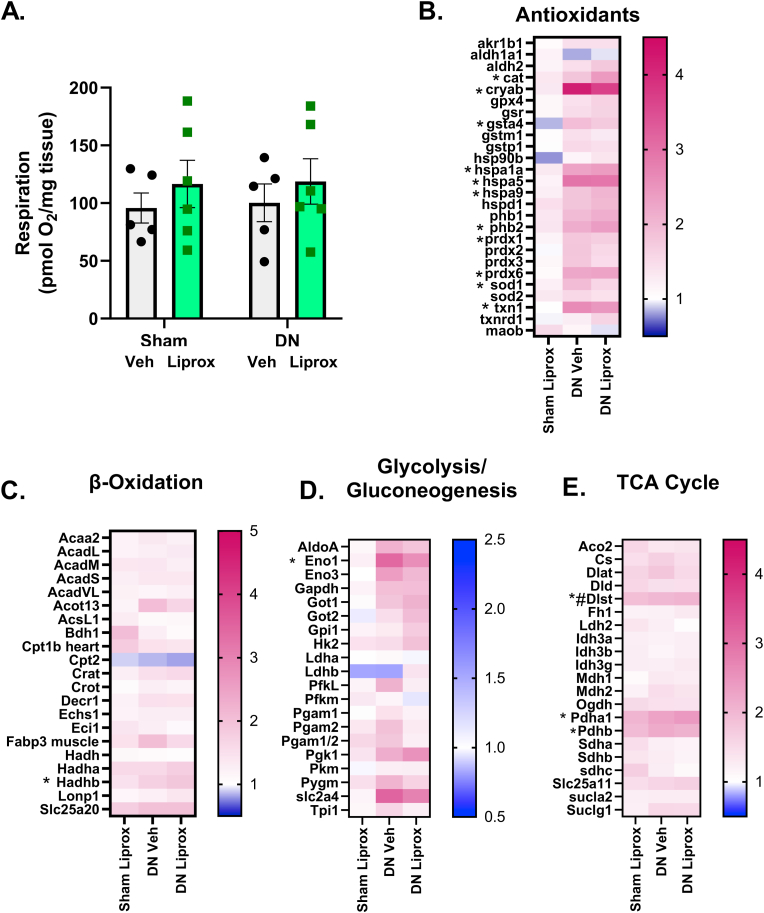
Denervation does not alter maximally stimulated mitochondrial respiration or proteins involved in antioxidant response. Because we previously showed elevated lipid hydroperoxides in isolated mitochondria from muscle post-denervation [16], we asked whether denervation and/or intervention with liproxstatin-1 alters mitochondrial function, mitochondrial protein expression or expression of key metabolic pathways. Mitochondrial respiration measured in permeabilized fiber bundles was not different between groups (Fig. 4A). Using selected reaction monitoring mass spectrometry analysis to measure targeted expression of several panels of related proteins [48,49], we found an increase in the protein expression of 11 out of the 27 proteins measured in an antioxidant panel in response to denervation (Fig. 4B). However, liproxstatin-1 treatment did not alter the expression of antioxidant proteins (Fig. 4B). Denervation and liproxstatin-1 minimally affected proteins involved in β oxidation, glycolysis/gluconeogenesis, and the TCA cycle (Fig. 4 C-E).
Fig. 4.
Denervation does not alter mitochondrial respiration. (A) Maximally stimulated respiration from complex I and II. (B) Heat map for antioxidant proteins. (C) Heat map for beta oxidation proteins. (D) Heat map for glycolysis and gluconeogenesis proteins. (E) Heat map for Krebs cycle proteins. N = 5–6 per group. DN, denervated; Veh, Vehicle; Liprox, Liproxstatin-1. Veh, grey bar; Liprox, green bar. Asterisk denotes post hoc differences at an alpha set at P < 0.05. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
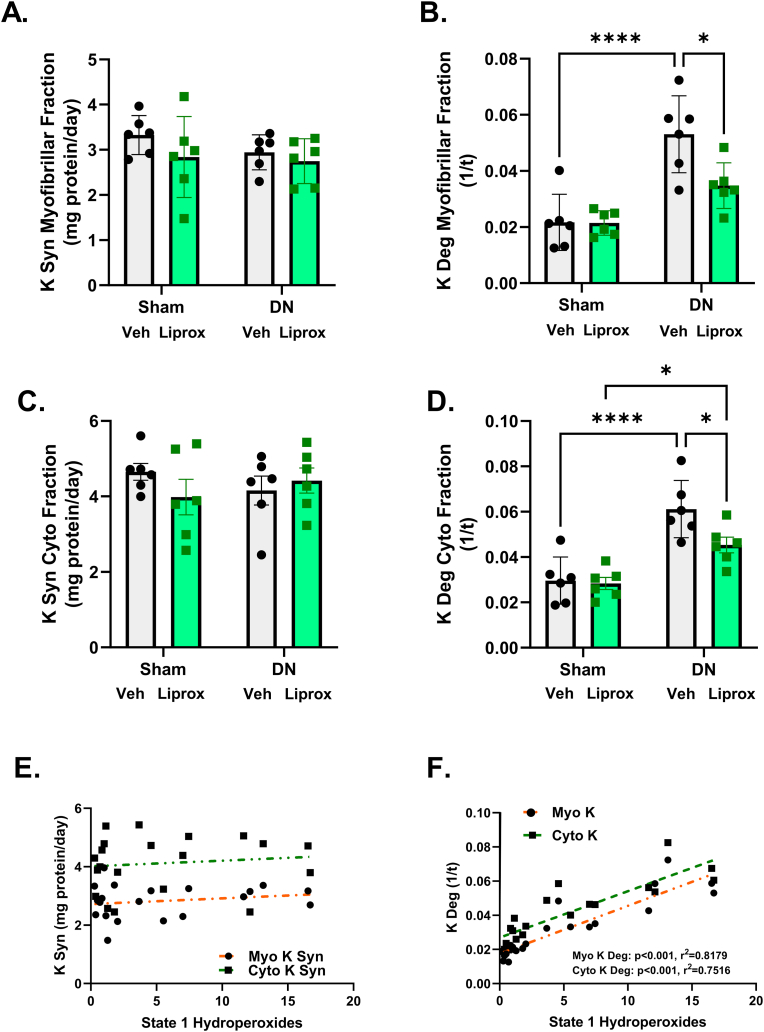
Liproxstatin-1 inhibits a denervation-induced increase in protein breakdown rates. Because treatment with liproxstatin-1 protects against denervation-induced muscle loss, we asked whether our liproxstatin-1 intervention changed rates of protein synthesis or breakdown in subcellular fractions from denervated muscle. Using in vivo labeling with D2O, we found that myofibrillar protein synthesis rates were not different among groups (Fig. 5A); however, denervation elevated myofibrillar protein breakdown by close to 2-fold and liproxstatin-1 treatment blunted myofibrillar protein breakdown more than 30% (Fig. 5B). Similarly, cytosolic protein synthesis rates were not different between groups (Fig. 5C); however, denervation elevated cytosolic protein breakdown by 2-fold (Fig. 5D) and lowered approximately 30% by liproxstatin-1 treatment (Fig. 5D). Interestingly, rates of protein breakdown but not protein synthesis were correlated with basal hydroperoxide generation (Fig. 5E and F).
Fig. 5.
Liproxstatin-1 ameliorates denervation-induced increase in protein breakdown rates in subcellular fractions. (A) Rate of myofibrillar protein synthesis. (B) Rate of myofibrillar protein breakdown. (C) Rate of cytosolic protein synthesis. (D) Rate of cytosolic protein breakdown. (E) Correlation between rates of protein synthesis and hydroperoxide generation in myofibrillar and cytosolic fractions. (F) Correlation between rates of protein breakdown and hydroperoxide generation in myofibrillar and cytosolic fractions. N = 6 per group. DN, denervated; Veh, Vehicle; Liprox, Liproxstatin-1. Veh, grey bar; Liprox, green bar. Asterisk denotes post hoc differences at an alpha set at P < 0.05. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
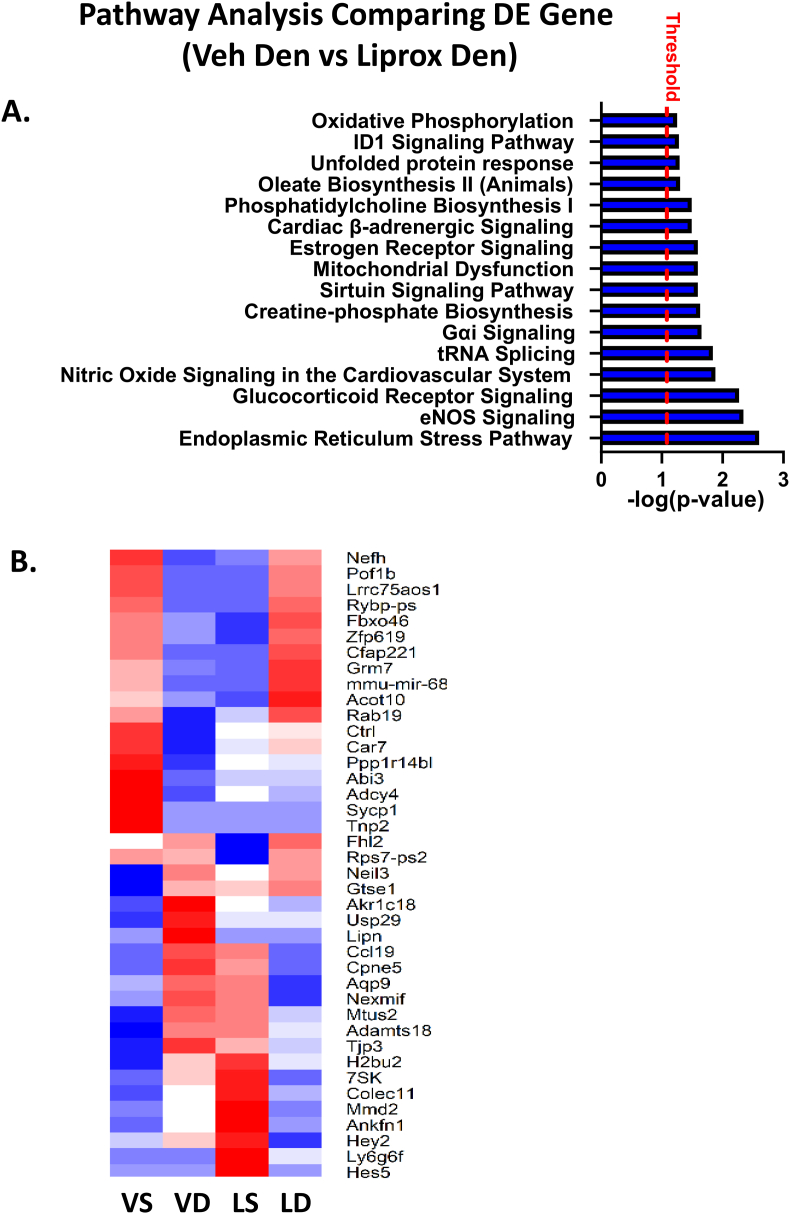
Gene expression analysis in gastrocnemius muscle from vehicle and liproxstatin-1 treated mice in response to denervation. To further explore the changes in muscle which may contribute the protective effect of liproxstatin-1 in muscle, we performed transcriptomic analysis in gastrocnemius muscle tissue. We found a total of 203 differentially expressed genes in a comparison of denervated muscle from vehicle treated and liproxstatin-1 treated mice. Fig. 6A shows pathway analysis comparing differentially expressed pathways comparing denervated muscle from vehicle and liproxstatin-1 treated mice. Pathways that were significantly altered in denervated muscle from liproxstatin-1 treated mice when compared to the vehicle included ER stress, unfolded protein response, protein ubiquitination pathway, eNOS signaling, and mitochondrial dysfunction (Fig. 6A). Fig. 6B shows a heat map of 40 differentially expressed genes expressed in opposite directions comparing vehicle versus liproxstatin-1 treated muscle following denervation.
Fig. 6.
RNA sequencing comparing differentially expressed genes in denervated muscle from mice treated with either vehicle or liproxstatin-1. (A) Pathway analysis comparison between DN Veh and DN Liprox. (B) Heat map showing differentially expressed genes between DN Veh and DN Liprox. N = 3 per group. Alpha for differentially expressed genes was set at p < 0.05.
4. Discussion
The loss of innervation is a potential target for age and disease-related muscle loss. The goal of this study was to determine whether liproxstatin-1, a lipid hydroperoxide scavenger, can modulate muscle atrophy phenotypes associated with denervation by reducing levels of mitochondrial lipid hydroperoxide and oxylipins. Our results showed that daily treatment with the lipid hydroperoxide scavenger liproxstatin-1 in vivo was effective at reducing the generation of mitochondrial hydroperoxides, inhibited muscle protein degradation and blunted the muscle atrophy that occurred in response to denervation. In addition, we showed that liproxstatin-1 treatment changes the oxylipin profile in denervated muscle.
In a prior study, we showed that the fluorogenic probe Amplex Red probe interacts with lipid hydroperoxides in addition to hydrogen peroxide [16]. Here we extended these results to show that Amplex Red can also react with mono-oxygenated oxylipins such as 12-HETE and 15-HETE. Since Amplex Red is a peroxidase substrate, it is not clear how mono-oxygenated oxylipins react with Amplex Red. The signal generated by the Amplex Red probe is a combination of a reaction with the electron transport chain generated reactive oxygen species, lipid hydroperoxides, and mono-oxygenated oxylipins. Because the addition of catalase and the genetic overexpression of hydrogen peroxide scavengers in muscle does not reduce the basal Amplex Red signal in denervated muscle [16], we hypothesized that the State 1 Amplex Red signal in denervated muscle was primarily composed of oxylipins and lipid hydroperoxides.
Our prior studies showed that denervation elevated lipid hydroperoxides and oxylipins [16,17]; therefore, we asked whether liproxstatin-1 could reduce lipid hydroperoxides and oxylipins in denervated mice in vivo. In vivo treatment with liproxstatin-1 reduced the Amplex Red signal in permeabilized fiber bundles by ∼70%. Our prior studies showed that overexpression of hydrogen peroxide scavengers in muscle and in vitro addition of exogenous catalase to permeabilized fiber bundles did not reduce the Amplex Red signal in permeabilized fiber bundles [16], supporting the fact that the signal measured by the Amplex Red in response to denervation is not primarily H2O2 generated by the mitochondrial electron transport chain. To further support these findings, we showed that treatment with liproxstatin-1 reduces 4-HNE content in denervated muscle. Together these data suggest that oxylipins and lipid hydroperoxides compose most of the State 1 hydroperoxide Amplex Red signal in denervated muscle, and that treatment with liproxstatin-1 can reduce oxylipins and lipid hydroperoxides in denervated muscle.
Prior work has shown that denervation induces muscle mitochondrial release of H2O2 as well as oxidized phospholipids [50]. To detect the impact of H2O2 specifically, our laboratory used mouse models targeting overexpression of hydrogen peroxide scavengers (catalase and peroxiredoxin 3) confined to muscle mitochondria. Elevated muscle levels of these mitochondrial H2O2 scavengers did not protect muscle mass in a 7-day sciatic nerve transection model, which may be because oxidized lipid and oxylipin levels were not impacted by these interventions. In contrast, elevated levels of Prdx3 and mitochondrial targeted catalase were protective in mitigating muscle loss in the CuZnSOD knockout mouse model (Sod1KO) of accelerated sarcopenia and frailty [51,52]. In the case of overexpression of muscle mitochondrial targeted catalase, the neuromuscular junction (NMJ) disruption seen in the Sod1KO mice was completely prevented preserving muscle mass and contractile function in the Sod1KO mice [51]. However, overexpression of PRDX3 did not protect the NMJ; however, there still was a protection in muscle mass and force [52]. There is evidence that supports that PRDX3 may directly reduce lipid peroxidation in liver tissue (reduced F2 isoprostanes and 4-HNE adducts) [53]. Therefore, it is possible that PRDX3 can reduce lipid hydroperoxides/oxylipins in muscle. We still do not know if PRDX3 has a direct effect on oxidized lipids or an indirect effect of lipid peroxidation by reducing overall levels of oxidation via scavenging H2O2. Together these data suggest that reducing H2O2 preserves the NMJ thus preventing denervation induced atrophy and increased generation of oxidized lipids. However, after denervation occurs, reducing oxidized lipids may be a better strategy to preserve muscle mass than using H2O2 scavengers [16].
The effect of oxylipins and lipid hydroperoxides on muscle atrophy is currently understudied. Elevated lipid hydroperoxides can lead to membrane damage, DNA mutations, and the production of toxic signaling molecules such as 4-hydroxynonenal that can trigger muscle pathology [[21], [22], [23],25,54]. The presence of allylic hydrogens in the polyunsaturated fatty acid (PUFA) components of phospholipids that make up cellular membranes leave phospholipid membranes sensitive to oxidation [55]. Lipid peroxidation in membranes can lead to loss of PUFAs, decreased membrane fluidity, and altered membrane permeability or other membrane functions [56,57]. Lipid hydroperoxides generated in membranes can also cause damage to other cell membranes through induction of chain reaction lipid peroxidation and lead to further generation of toxic lipid hydroperoxides metabolites including 4-HNE [[58], [59], [60]]. In the mitochondria, membrane damage can lead to loss of calcium activation of caspases and increased apoptosis [61,62]. Lipid hydroperoxides from membrane bilayers can be relatively stable allowing them to move within the cell to the mitochondria or to the nucleus to cause DNA damage by lipid hydroperoxide-derived radicals [63,64].
In the current study, we showed that daily treatment with liproxstatin-1 protects against denervation-induced muscle loss. Smaller fiber cross sectional area, reduced fiber number, or a combination of reduced cross-sectional area and reduced fiber number can lead to reduced muscle mass. We showed that treatment with liproxstatin-1 reduced atrophy in denervated muscle. We noted that there was only a partial protection against denervation-induced muscle mass loss in mice treated with liproxstatin-1. In fact, there are likely multiple cellular mechanisms that trigger muscle loss associated with denervation. For example, a recent study identified Coactivator-associated arginine methyltransferase 1 (CARM1) as a key driver of muscle wasting associated with neurological deficits [65]. Skeletal muscle atrophy occurs by an imbalance in protein turnover favoring protein degradation over protein synthesis. We did not find any change in protein synthesis rates in response to denervation in either myofibrillar or cytosolic protein fractions. However, denervation significantly elevated protein breakdown rates and that were inhibited by treatment with liproxstatin-1. These data indicate that liproxstatin-1 treatment reduces atrophy in denervated muscle by reducing the increase in muscle protein breakdown associated with denervation. We need further studies to uncover the mechanisms involved.
Oxylipins are oxygenated products of PUFAs that have signaling roles [19]. We showed for the first time that denervation elevated specific oxylipins in muscle. Our data also showed that denervation elevated three oxylipins, which treatment with liproxstatin-1 reduced. It is likely that both enzymatic generation of oxylipins and autoxidation are responsible for the increase in oxylipin content. Prior reports show that liproxstatin-1 does not inhibit 12/15-Lipoxygenase and thus should not alter enzymatic generation of lipid hydroperoxides through this pathway [26]; therefore, the reduction in oxylipin content in muscle from liproxstatin-1 treated mice was likely due to scavenging by liproxstatin-1. From our understanding, researchers have not assessed if liproxstatin-1 induced inhibition of mouse 12/15-LOX or platelet type 12-LOX. Our laboratory previously showed partial protection from denervation-induced atrophy in 12/15-LOX knockout mice, supporting 12/15-LOX as a source of deleterious lipid hydroperoxides in denervated muscle [16,20]. In support of this, the lipidomic dataset in the current study shows that all eight oxylipins elevated during denervation are products of 12/15-LOX. We hypothesize that some of the oxylipins elevated during denervation can help trigger muscle atrophy programing. In fact, 15-HETE, an oxylipin that was elevated in denervation and reduced with treatment of liproxstatin-1, has been shown to trigger muscle loss via the ubiquitin proteasome system in vitro [66]. It should be noted that some oxylipins may elicit positive benefits to skeletal muscle. In fact, researchers have shown that linoleic acid treatment may protect against oxidative stress dependent mechanisms of denervation-induced muscle wasting [67]. Our data shows that the substrate for many of the oxylipins elevated in denervated muscle is predominately arachidonic acid, so the substrate may be important in determining if oxylipins generated are harmful or beneficial. We need more research to understand how specific oxylipins can trigger muscle pathology.
As we detected multiple positional isomers in the lipidomic analysis for oxPL, these lipids won't have originated from direct LOX catalysis, but more likely via secondary peroxidation of phospholipids that occurred downstream of the enzyme. It is well known that LOX enzymes are not 100% efficient, and lipid radicals that form during turnover can leak off [68], and then these are available to mediate secondary peroxidation, e.g. of phospholipids. To assess if secondary peroxidation could contribute to the oxPL profile, we next tested the impact of denervation and liproxstatin-1 on oxPL levels in muscle. Our data showed that denervation tended to elevate four oxidized PE families, with this suppressed by liproxstatin-1 treatment. We hypothesize that enzymatic activity drove the overall lipid profile; however, secondary oxidation occurred when enzymes were not adequately controlled. The antioxidant activity of liproxstatin-1 may help control secondary lipid peroxidation.
In our prior studies we reported that mitochondria are a major source of lipid hydroperoxide/oxylipin generation in denervated muscle [16]. Therefore, we wanted to determine if denervation or liproxstatin-1 treatment altered mitochondrial respiration and mitochondrial proteins. Neither denervation nor liproxstatin-1 intervention altered maximally stimulated mitochondrial respiration; however, there may be changes in mitochondrial respiration at reduced ADP concentrations. However, prior literature has shown that denervation does not result in increased muscle hydroperoxides at low ADP concentrations [69]. Denervation increased antioxidant protein content, while liproxstatin-1 had no effect on protein content for antioxidant proteins, or proteins involved in beta oxidation, gluconeogenesis/glycolysis, or the krebs cycle. It is likely that treatment with liproxstatin-1 has very little effect on the translation of metabolic proteins based on our targeted proteomics measurements. Since neither denervation nor liproxstatin-1 had altered mitochondrial respiration, and there was a minimal effect on protein content of metabolic proteins, we shifted our focus to analyzing the transcriptome in denervated muscle from mice treated with liproxstatin-1.
To begin to define the molecular mechanisms in which treatment with liproxstatin-1 protects against denervation-induced muscle loss, we performed transcriptomic analysis. Ingenuity Pathway Analysis showed that liproxstatin-1 treatment differentially expressed pathways associated with ER stress, unfolded protein response, protein ubiquitination pathway, eNOS signaling, and mitochondrial dysfunction in denervated muscle. Prior literature has shown that ER stress, unfolded protein response pathways, and protein ubiquitination pathways contribute to denervation-induced muscle loss [70,71] and next steps will include determining whether liproxstain-1 alters denervation-induced atrophy through these pathways. This is the first report of differentially expressed genes in denervated muscle with and without liproxstatin-1 treatment, and we need to conduct more experiments to determine if liproxsatin-1 alters protein breakdown signaling in muscle. It is possible that liproxstatin-1 prevents muscle damage, which would then reduce the breakdown response associated with muscle damage.
5. Limitations
With the current approaches, we cannot distinguish whether changes in muscle were due directly to a reduction in oxylipins or other indirect antioxidant effect of liproxstatin-1. However, we view this study as the first step in understanding the role of oxylipins in muscle atrophy and our next steps will define the effect of specific oxylipins on key pathways important in maintenance of muscle mass. We also cannot distinguish between oxidized lipids generated enzymatically or via auto-oxidation, and future studies should try to distinguish between this. Prior studies have shown that 12/15-LOX is a major source of the generation of oxidized lipids that may contribute to muscle atrophy [16,20]. Our data shows that liproxstatin-1 decreases oxylipin profile regardless of substrate. Substrates such as linoleic acid actual protect against muscle wasting [67]; therefore, targeting oxylipins generated specifically from arachidonic acid may be a more beneficial therapeutic strategy. Finally, treatment with liproxstatin-1 does not completely rescue skeletal muscle mass loss associated with denervation, which indicates that other factors besides oxidized lipids contribute to denervation-induced muscle atrophy [65].
6. Conclusions
In summary, we have shown that reducing lipid hydroperoxides/oxylipins via treatment with liproxstatin-1 minimizes denervation-induced atrophy. Further, we have shown that liproxstatin-1 treatment reduces oxylipin content in denervated muscle. Liproxstatin-1 treatment prevented the increase in muscle protein breakdown associated with denervation. Genes associated with ER stress, unfolded protein response, and the ubiquitin proteasome system may contribute to the protective mechanisms of liproxstatin-1 treatment. Surprisingly, denervation or liproxstatin-1 treatment did not affect maximally stimulated mitochondrial respiration; however, we need to perform ADP titration assays in future studies to fully understand if denervation or liproxstatin-1 treatment impacts mitochondrial respiration. Future studies should examine underlying mechanisms of lipid hydroperoxide pathology in denervated muscle.
Declaration of competing interest
All authors declare that there are no conflicts of interest.
Acknowledgements
Support for this work has been provided by National Institute on Aging P01AG051442. Dr. Van Remmen is the recipient of a VA Senior Research Career Scientist award (1 IK6 BX005234). Contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. The authors would like to thank the members of the Van Remmen Laboratory for their contributions to the experiments presented here. Dr. Jacob L. Brown's postdoctoral training was supported by NIA T32 AG052363. Dr. Jacob L. Brown is currently supported by a VA Career Development Award (1 IK2 BX005620-01A1). Sêr Cymru Project Sepsis grant funded by Welsh Government/European Union (European Research Development Fund) supported Dr. Victoria J.Tyrrell.
We would also like to extend our gratitude to the numerous other faculties, staff and other researchers at the Oklahoma Medical Research Foundation and OUHSC for helpful discussions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102518.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Delmonico M.J., Harris T.B., Lee J.S., Visser M., Nevitt M., Kritchevsky S.B., Tylavsky F.A., Newman A.B. Alternative definitions of sarcopenia, lower extremity performance, and functional impairment with aging in older men and women. J. Am. Geriatr. Soc. 2007;55(5):769–774. doi: 10.1111/j.1532-5415.2007.01140.x. [DOI] [PubMed] [Google Scholar]
- 2.Goodpaster B.H., Park S.W., Harris T.B., Kritchevsky S.B., Nevitt M., Schwartz A.V., Simonsick E.M., Tylavsky F.A., Visser M., Newman A.B. The loss of skeletal muscle strength, mass, and quality in older adults: the health, aging and body composition study. J. Gerontol. A Biol. Sci. Med. Sci. 2006;61(10):1059–1064. doi: 10.1093/gerona/61.10.1059. [DOI] [PubMed] [Google Scholar]
- 3.Baumgartner R.N., Koehler K.M., Gallagher D., Romero L., Heymsfield S.B., Ross R.R., Garry P.J., Lindeman R.D. Epidemiology of sarcopenia among the elderly in New Mexico. Am. J. Epidemiol. 1998;147(8):755–763. doi: 10.1093/oxfordjournals.aje.a009520. [DOI] [PubMed] [Google Scholar]
- 4.Lorenzo C. Body composition and physical function in older adults. Obesity. 2009;17(2):211–212. doi: 10.1038/oby.2008.506. [DOI] [PubMed] [Google Scholar]
- 5.Schneider S.M., Al-Jaouni R., Filippi J., Wiroth J.B., Zeanandin G., Arab K., Hébuterne X. Sarcopenia is prevalent in patients with Crohn's disease in clinical remission. Inflamm. Bowel Dis. 2008;14(11):1562–1568. doi: 10.1002/ibd.20504. [DOI] [PubMed] [Google Scholar]
- 6.Janssen I., Shepard D.S., Katzmarzyk P.T., Roubenoff R. The healthcare costs of sarcopenia in the United States. J. Am. Geriatr. Soc. 2004;52(1):80–85. doi: 10.1111/j.1532-5415.2004.52014.x. [DOI] [PubMed] [Google Scholar]
- 7.Jang Y.C., Van Remmen H. Age-associated alterations of the neuromuscular junction. Exp. Gerontol. 2011;46(2–3):193–198. doi: 10.1016/j.exger.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell M.J., McComas A.J., Petito F. Physiological changes in ageing muscles. J. Neurol. Neurosurg. Psychiatr. 1973;36(2):174–182. doi: 10.1136/jnnp.36.2.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowan S.L., Rygiel K., Purves-Smith F.M., Solbak N.M., Turnbull D.M., Hepple R.T. Denervation causes fiber atrophy and myosin heavy chain co-expression in senescent skeletal muscle. PLoS One. 2012;7(1) doi: 10.1371/journal.pone.0029082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deepa S.S., Van Remmen H., Brooks S.V., Faulkner J.A., Larkin L., McArdle A., Jackson M.J., Vasilaki A., Richardson A. Accelerated sarcopenia in Cu/Zn superoxide dismutase knockout mice. Free Radic. Biol. Med. 2019;132:19–23. doi: 10.1016/j.freeradbiomed.2018.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deschenes M.R., Roby M.A., Eason M.K., Harris M.B. Remodeling of the neuromuscular junction precedes sarcopenia related alterations in myofibers. Exp. Gerontol. 2010;45(5):389–393. doi: 10.1016/j.exger.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spendiff S., Vuda M., Gouspillou G., Aare S., Perez A., Morais J.A., Jagoe R.T., Filion M.E., Glicksman R., Kapchinsky S., MacMillan N.J., Pion C.H., Aubertin-Leheudre M., Hettwer S., Correa J.A., Taivassalo T., Hepple R.T. Denervation drives mitochondrial dysfunction in skeletal muscle of octogenarians. J Physiol. 2016;594(24):7361–7379. doi: 10.1113/JP272487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsson L., Ansved T. Effects of ageing on the motor unit. Prog. Neurobiol. 1995;45(5):397–458. doi: 10.1016/0301-0082(95)98601-z. [DOI] [PubMed] [Google Scholar]
- 14.Sataranatarajan K., Pharaoh G., Brown J.L., Ranjit R., Piekarz K.M., Street K., Wren J.D., Georgescu C., Kinter C., Kinter M., Freeman W.M., Richardson A., Van Remmen H. Molecular changes in transcription and metabolic pathways underlying muscle atrophy in the CuZnSOD null mouse model of sarcopenia. Geroscience. 2020;42(4):1101–1118. doi: 10.1007/s11357-020-00189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muller F.L., Song W., Jang Y.C., Liu Y., Sabia M., Richardson A., Van Remmen H. Denervation-induced skeletal muscle atrophy is associated with increased mitochondrial ROS production. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007;293(3):R1159–R1168. doi: 10.1152/ajpregu.00767.2006. [DOI] [PubMed] [Google Scholar]
- 16.Pharaoh G., Brown J.L., Sataranatarajan K., Kneis P., Bian J., Ranjit R., Hadad N., Georgescu C., Rabinovitch P., Ran Q., Wren J.D., Freeman W., Kinter M., Richardson A., Van Remmen H. Targeting cPLA(2) derived lipid hydroperoxides as a potential intervention for sarcopenia. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-70792-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhattacharya A., Muller F.L., Liu Y., Sabia M., Liang H., Song W., Jang Y.C., Ran Q., Van Remmen H. Denervation induces cytosolic phospholipase A2-mediated fatty acid hydroperoxide generation by muscle mitochondria. J. Biol. Chem. 2009;284(1):46–55. doi: 10.1074/jbc.M806311200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shearer G.C., Walker R.E. An overview of the biologic effects of omega-6 oxylipins in humans. Prostaglandins Leukot. Essent. Fatty Acids. 2018;137:26–38. doi: 10.1016/j.plefa.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 19.Gabbs M., Leng S., Devassy J.G., Monirujjaman M., Aukema H.M. Advances in our understanding of oxylipins derived from dietary PUFAs. Adv. Nutr. 2015;6(5):513–540. doi: 10.3945/an.114.007732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhattacharya A., Hamilton R., Jernigan A., Zhang Y., Sabia M., Rahman M.M., Li Y., Wei R., Chaudhuri A., Van Remmen H. Genetic ablation of 12/15-lipoxygenase but not 5-lipoxygenase protects against denervation-induced muscle atrophy. Free Radic. Biol. Med. 2014;67:30–40. doi: 10.1016/j.freeradbiomed.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Schwarzer E., Arese P., Skorokhod O.A. Role of the lipoperoxidation product 4-hydroxynonenal in the pathogenesis of severe malaria anemia and malaria immunodepression. Oxid. Med. Cell. Longev. 2015;2015 doi: 10.1155/2015/638416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zablocka-Slowinska K., Placzkowska S., Skorska K., Prescha A., Pawelczyk K., Porebska I., Kosacka M., Grajeta H. Oxidative stress in lung cancer patients is associated with altered serum markers of lipid metabolism. PLoS One. 2019;14(4) doi: 10.1371/journal.pone.0215246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dalle-Donne I., Rossi R., Giustarini D., Milzani A., Colombo R. Protein carbonyl groups as biomarkers of oxidative stress. Clin. Chim. Acta. 2003;329(1–2):23–38. doi: 10.1016/s0009-8981(03)00003-2. [DOI] [PubMed] [Google Scholar]
- 24.Friedmann Angeli J.P., Schneider M., Proneth B., Tyurina Y.Y., Tyurin V.A., Hammond V.J., Herbach N., Aichler M., Walch A., Eggenhofer E., Basavarajappa D., Rådmark O., Kobayashi S., Seibt T., Beck H., Neff F., Esposito I., Wanke R., Förster H., Yefremova O., Heinrichmeyer M., Bornkamm G.W., Geissler E.K., Thomas S.B., Stockwell B.R., O'Donnell V.B., Kagan V.E., Schick J.A., Conrad M. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 2014;16(12):1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao J.Y., Dixon S.J. Mechanisms of ferroptosis. Cell. Mol. Life Sci. : CM. 2016;73(11–12):2195–2209. doi: 10.1007/s00018-016-2194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zilka O., Shah R., Li B., Friedmann Angeli J.P., Griesser M., Conrad M., Pratt D.A. On the mechanism of cytoprotection by ferrostatin-1 and liproxstatin-1 and the role of lipid peroxidation in ferroptotic cell death. ACS Cent. Sci. 2017;3(3):232–243. doi: 10.1021/acscentsci.7b00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pharaoh G., Brown J.L., Sataranatarajan K., Kneis P., Bian J., Ranjit R., Hadad N., Georgescu C., Rabinovitch P., Ran Q., Wren J.D., Freeman W., Kinter M., Richardson A., Van Remmen H. Targeting cPLA(2) derived lipid hydroperoxides as a potential intervention for sarcopenia. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-70792-7. 13968-13968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhattacharya A., Hamilton R., Jernigan A., Zhang Y., Sabia M., Rahman M.M., Li Y., Wei R., Chaudhuri A., Van Remmen H. Genetic ablation of 12/15-lipoxygenase but not 5-lipoxygenase protects against denervation-induced muscle atrophy. Free Radic. Biol. Med. 2014;67:30–40. doi: 10.1016/j.freeradbiomed.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Misheva M., Kotzamanis K., Davies L.C., Tyrrell V.J., Rodrigues P.R.S., Benavides G.A., Hinz C., Murphy R.C., Kennedy P., Taylor P.R., Rosas M., Jones S.A., McLaren J.E., Deshpande S., Andrews R., Schebb N.H., Czubala M.A., Gurney M., Aldrovandi M., Meckelmann S.W., Ghazal P., Darley-Usmar V., White D.A., O'Donnell V.B. Oxylipin metabolism is controlled by mitochondrial β-oxidation during bacterial inflammation. Nat. Commun. 2022;13(1):139. doi: 10.1038/s41467-021-27766-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown J.L., Lawrence M.M., Ahn B., Kneis P., Piekarz K.M., Qaisar R., Ranjit R., Bian J., Pharaoh G., Brown C., Peelor F.F., 3rd, Kinter M.T., Miller B.F., Richardson A., Van Remmen H. Cancer cachexia in a mouse model of oxidative stress. J. Cachexia Sarcopenia Muscle. 2020;11(6):1688–1704. doi: 10.1002/jcsm.12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown J.L., Rosa-Caldwell M.E., Lee D.E., Blackwell T.A., Brown L.A., Perry R.A., Haynie W.S., Hardee J.P., Carson J.A., Wiggs M.P., Washington T.A., Greene N.P. Mitochondrial degeneration precedes the development of muscle atrophy in progression of cancer cachexia in tumour-bearing mice. J. Cachexia Sarcopenia Muscle. 2017;8(6):926–938. doi: 10.1002/jcsm.12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown J.L., Lawrence M.M., Ahn B., Kneis P., Piekarz K.M., Qaisar R., Ranjit R., Bian J., Pharaoh G., Brown C., Peelor F.F., 3rd, Kinter M.T., Miller B.F., Richardson A., Van Remmen H. J Cachexia Sarcopenia Muscle; 2020. Cancer Cachexia in a Mouse Model of Oxidative Stress. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahn B., Pharaoh G., Premkumar P., Huseman K., Ranjit R., Kinter M., Szweda L., Kiss T., Fulop G., Tarantini S., Csiszar A., Ungvari Z., Van Remmen H. Nrf2 deficiency exacerbates age-related contractile dysfunction and loss of skeletal muscle mass. Redox Biol. 2018;17:47–58. doi: 10.1016/j.redox.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pharaoh G., Sataranatarajan K., Street K., Hill S., Gregston J., Ahn B., Kinter C., Kinter M., Van Remmen H. Metabolic and stress response changes precede disease onset in the spinal cord of mutant SOD1 ALS mice. Front. Neurosci. 2019;13:487. doi: 10.3389/fnins.2019.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown J.L., Lawrence M.M., Ahn B., Kneis P., Piekarz K.M., Qaisar R., Ranjit R., Bian J., Pharaoh G., Brown C., Peelor F.F., 3rd, Kinter M.T., Miller B.F., Richardson A., Van Remmen H. Cancer cachexia in a mouse model of oxidative stress. J.Cachexia Sarcopenia Muscle. 2020;11(6):1688–1704. doi: 10.1002/jcsm.12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drake J.C., Bruns D.R., Peelor F.F., 3rd, Biela L.M., Miller R.A., Hamilton K.L., Miller B.F. Long-lived crowded-litter mice have an age-dependent increase in protein synthesis to DNA synthesis ratio and mTORC1 substrate phosphorylation. Am. J. Physiol. Endocrinol. Metab. 2014;307(9):E813–E821. doi: 10.1152/ajpendo.00256.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drake J.C., Peelor F.F., 3rd, Biela L.M., Watkins M.K., Miller R.A., Hamilton K.L., Miller B.F. Assessment of mitochondrial biogenesis and mTORC1 signaling during chronic rapamycin feeding in male and female mice. J. Gerontol. A Biol. Sci. Med. Sci. 2013;68(12):1493–1501. doi: 10.1093/gerona/glt047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drake J.C., Bruns D.R., Peelor F.F., 3rd, Biela L.M., Miller R.A., Miller B.F., Hamilton K.L. Long-lived Snell dwarf mice display increased proteostatic mechanisms that are not dependent on decreased mTORC1 activity. Aging Cell. 2015;14(3):474–482. doi: 10.1111/acel.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Busch R., Kim Y.K., Neese R.A., Schade-Serin V., Collins M., Awada M., Gardner J.L., Beysen C., Marino M.E., Misell L.M., Hellerstein M.K. Measurement of protein turnover rates by heavy water labeling of nonessential amino acids. Biochim. Biophys. Acta. 2006;1760(5):730–744. doi: 10.1016/j.bbagen.2005.12.023. [DOI] [PubMed] [Google Scholar]
- 40.Miller B.F., Wolff C.A., Peelor F.F., 3rd, Shipman P.D., Hamilton K.L. Modeling the contribution of individual proteins to mixed skeletal muscle protein synthetic rates over increasing periods of label incorporation. J. Appl. Physiol. 2015;118(6):655–661. doi: 10.1152/japplphysiol.00987.2014. 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller B.F., Hamilton K.L., Majeed Z.R., Abshire S.M., Confides A.L., Hayek A.M., Hunt E.R., Shipman P., Peelor F.F., 3rd, Butterfield T.A., Dupont-Versteegden E.E. Enhanced skeletal muscle regrowth and remodelling in massaged and contralateral non-massaged hindlimb. J Physiol. 2018;596(1):83–103. doi: 10.1113/JP275089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lawrence M.M., Van Pelt D.W., Confides A.L., Hunt E.R., Hettinger Z.R., Laurin J.L., Reid J.J., Peelor F.F., 3rd, Butterfield T.A., Dupont-Versteegden E.E., Miller B.F. Massage as a mechanotherapy promotes skeletal muscle protein and ribosomal turnover but does not mitigate muscle atrophy during disuse in adult rats. Acta Physiol. 2020;229(3) doi: 10.1111/apha.13460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown J.L., Lee D.E., Rosa-Caldwell M.E., Brown L.A., Perry R.A., Haynie W.S., Huseman K., Sataranatarajan K., Van Remmen H., Washington T.A., Wiggs M.P., Greene N.P. Protein imbalance in the development of skeletal muscle wasting in tumour-bearing mice. J. Cachexia Sarcopenia Muscle. 2018;9(5):987–1002. doi: 10.1002/jcsm.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qaisar R., Pharaoh G., Bhaskaran S., Xu H., Ranjit R., Bian J., Ahn B., Georgescu C., Wren J.D., Van Remmen H. Restoration of sarcoplasmic reticulum Ca(2+) ATPase (SERCA) activity prevents age-related muscle atrophy and weakness in mice. Int. J. Mol. Sci. 2020;22(1):37. doi: 10.3390/ijms22010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown J.L., Rosa-Caldwell M.E., Lee D.E., Brown L.A., Perry R.A., Shimkus K.L., Blackwell T.A., Fluckey J.D., Carson J.A., Dridi S., Washington T.A., Greene N.P. PGC-1α4 gene expression is suppressed by the IL-6-MEK-ERK 1/2 MAPK signalling axis and altered by resistance exercise, obesity and muscle injury. Acta Physiol. 2017;220(2):275–288. doi: 10.1111/apha.12826. [DOI] [PubMed] [Google Scholar]
- 46.Thomas C.P., Morgan L.T., Maskrey B.H., Murphy R.C., Kühn H., Hazen S.L., Goodall A.H., Hamali H.A., Collins P.W., O'Donnell V.B. Phospholipid-esterified eicosanoids are generated in agonist-activated human platelets and enhance tissue factor-dependent thrombin generation. J. Biol. Chem. 2010;285(10):6891–6903. doi: 10.1074/jbc.M109.078428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maskrey B.H., Bermúdez-Fajardo A., Morgan A.H., Stewart-Jones E., Dioszeghy V., Taylor G.W., Baker P.R., Coles B., Coffey M.J., Kühn H., O'Donnell V.B. Activated platelets and monocytes generate four hydroxyphosphatidylethanolamines via lipoxygenase. J. Biol. Chem. 2007;282(28):20151–20163. doi: 10.1074/jbc.M611776200. [DOI] [PubMed] [Google Scholar]
- 48.Kinter C.S., Lundie J.M., Patel H., Rindler P.M., Szweda L.I., Kinter M. A quantitative proteomic profile of the Nrf2-mediated antioxidant response of macrophages to oxidized LDL determined by multiplexed selected reaction monitoring. PLoS One. 2012;7(11) doi: 10.1371/journal.pone.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., Kern R., Tabb D.L., Liebler D.C., MacCoss M.J. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Staunton C.A., Owen E.D., Pollock N., Vasilaki A., Barrett-Jolley R., McArdle A., Jackson M.J. HyPer2 imaging reveals temporal and heterogeneous hydrogen peroxide changes in denervated and aged skeletal muscle fibers in vivo. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-51035-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu H., Ranjit R., Richardson A., Van Remmen H. Muscle mitochondrial catalase expression prevents neuromuscular junction disruption, atrophy, and weakness in a mouse model of accelerated sarcopenia. J. Cachexia Sarcopenia Muscle. 2021;12(6):1582–1596. doi: 10.1002/jcsm.12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahn B., Ranjit R., Kneis P., Xu H., Piekarz K.M., Freeman W.M., Kinter M., Richardson A., Ran Q., Brooks S.V., Van Remmen H. Scavenging mitochondrial hydrogen peroxide by peroxiredoxin 3 overexpression attenuates contractile dysfunction and muscle atrophy in a murine model of accelerated sarcopenia. Aging Cell. 2022;21(3) doi: 10.1111/acel.13569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen L., Na R., Gu M., Salmon A.B., Liu Y., Liang H., Qi W., Van Remmen H., Richardson A., Ran Q. Reduction of mitochondrial H2O2 by overexpressing peroxiredoxin 3 improves glucose tolerance in mice. Aging Cell. 2008;7(6):866–878. doi: 10.1111/j.1474-9726.2008.00432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gentile F., Arcaro A., Pizzimenti S., Daga M., Cetrangolo G.P., Dianzani C., Lepore A., Graf M., Ames P.R.J., Barrera G. DNA damage by lipid peroxidation products: implications in cancer, inflammation and autoimmunity. AIMS Genet. 2017;4(2):103–137. doi: 10.3934/genet.2017.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leitinger N. Oxidized phospholipids as modulators of inflammation in atherosclerosis. Curr. Opin. Lipidol. 2003;14(5):421–430. doi: 10.1097/00041433-200310000-00002. [DOI] [PubMed] [Google Scholar]
- 56.Catalá A., Díaz M. Editorial: impact of lipid peroxidation on the physiology and pathophysiology of cell membranes. Front. Physiol. 2016;7:423. doi: 10.3389/fphys.2016.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gaschler M.M., Stockwell B.R. Lipid peroxidation in cell death. Biochem. Biophys. Res. Commun. 2017;482(3):419–425. doi: 10.1016/j.bbrc.2016.10.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Esterbauer H., Schaur R.J., Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic. Biol. Med. 1991;11(1):81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 59.Spickett C.M. The lipid peroxidation product 4-hydroxy-2-nonenal: advances in chemistry and analysis. Redox Biol. 2013;1(1):145–152. doi: 10.1016/j.redox.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schneider C., Tallman K.A., Porter N.A., Brash A.R. Two distinct pathways of formation of 4-hydroxynonenal. Mechanisms of nonenzymatic transformation of the 9- and 13-hydroperoxides of linoleic acid to 4-hydroxyalkenals. J. Biol. Chem. 2001;276(24):20831–20838. doi: 10.1074/jbc.M101821200. [DOI] [PubMed] [Google Scholar]
- 61.Kroemer G., Galluzzi L., Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 2007;87(1):99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- 62.Duchen M.R. Mitochondria and calcium: from cell signalling to cell death. J Physiol. 2000;529 Pt 1(Pt 1):57–68. doi: 10.1111/j.1469-7793.2000.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Negre-Salvayre A., Coatrieux C., Ingueneau C., Salvayre R. Advanced lipid peroxidation end products in oxidative damage to proteins. Potential role in diseases and therapeutic prospects for the inhibitors. Br. J. Pharmacol. 2008;153(1):6–20. doi: 10.1038/sj.bjp.0707395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ayala A., Muñoz M.F., Argüelles S. Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell. Longev. 2014;2014 doi: 10.1155/2014/360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stouth D.W., vanLieshout T.L., Ng S.Y., Webb E.K., Manta A., Moll Z., Ljubicic V. CARM1 regulates AMPK signaling in skeletal muscle. iScience. 2020;23(11) doi: 10.1016/j.isci.2020.101755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whitehouse A.S., Khal J., Tisdale M.J. Induction of protein catabolism in myotubes by 15(S)-hydroxyeicosatetraenoic acid through increased expression of the ubiquitin-proteasome pathway. Br. J. Cancer. 2003;89(4):737–745. doi: 10.1038/sj.bjc.6601184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee M.H., Lee J.H., Kim W.J., Kim S.H., Kim S.Y., Kim H.S., Kim T.J. Linoleic acid attenuates denervation-induced skeletal muscle atrophy in mice through regulation of reactive oxygen species-dependent signaling. Int. J. Mol. Sci. 2022;23(9) doi: 10.3390/ijms23094778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ivanov I., Saam J., Kuhn H., Holzhütter H.G. Dual role of oxygen during lipoxygenase reactions. FEBS J. 2005;272(10):2523–2535. doi: 10.1111/j.1742-4658.2005.04673.x. [DOI] [PubMed] [Google Scholar]
- 69.Pharaoh G., Brown J., Ranjit R., Ungvari Z., Van Remmen H. Reduced adenosine diphosphate sensitivity in skeletal muscle mitochondria increases reactive oxygen species production in mouse models of aging and oxidative stress but not denervation. JCSM Rapid Communications. 2021;4(1):75–89. doi: 10.1002/rco2.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bohnert K.R., McMillan J.D., Kumar A. Emerging roles of ER stress and unfolded protein response pathways in skeletal muscle health and disease. J. Cell. Physiol. 2018;233(1):67–78. doi: 10.1002/jcp.25852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baumann C.W., Liu H.M., Thompson L.V. Denervation-induced activation of the ubiquitin-proteasome system reduces skeletal muscle quantity not quality. PLoS One. 2016;11(8) doi: 10.1371/journal.pone.0160839. e0160839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.