Abstract
The emergence of large-scale phenotypic, genetic, and other multi-model biochemical data has offered unprecedented opportunities for drug discovery including drug repurposing. Various knowledge graph-based methods have been developed to integrate and analyze complex and heterogeneous data sources to find new therapeutic applications for existing drugs. However, existing methods have limitations in modeling and capturing context-sensitive inter-relationships among tens of thousands of biomedical entities. In this paper, we developed KG-Predict: a knowledge graph computational framework for drug repurposing. We first integrated multiple types of entities and relations from various genotypic and phenotypic databases to construct a knowledge graph termed GP-KG. GP-KG was composed of 1,246,726 associations between 61,146 entities. KG-Predict then aggregated the heterogeneous topological and semantic information from GP-KG to learn low-dimensional representations of entities and relations, and further utilized these representations to infer new drug–disease interactions. In cross-validation experiments, KG-Predict achieved high performances [AUROC (the area under receiver operating characteristic) = 0.981, AUPR (the area under precision–recall) = 0.409 and MRR (the mean reciprocal rank) = 0.261], outperforming other state-of-art graph embedding methods. We applied KG-Predict in identifying novel repositioned candidate drugs for Alzheimer’s disease (AD) and showed that KG-Predict prioritized both FDA-approved and active clinical trial anti-AD drugs among the top (AUROC = 0.868 and AUPR = 0.364).
Keywords: Drug repurposing, Knowledge graph, Computational prediction framework, Alzheimer’s disease
1. Introduction
Traditional drug discovery often takes 10–17 years and costs ~$2 billion to bring a drug to market, with a high attrition rate of 90% [1-3]. On the other hand, drug repurposing, which is the process of finding new indications for existing drugs, has demonstrated advantages over de novo drug design, including faster development time and reduced risk [4-6]. In the past few years, drug repurposing research has benefited greatly from the explosion of large-scale biomedical databases. Numerous computational approaches have been developed to systematically analyze various biomedical data to infer new indications for a given drug or find new treatments for a given disease [7-12].
Various network-based approaches have been developed for drug repurposing in the past decades. Several traditional studies identified new indications of an existing drug through constructing drug or disease similarity networks [13-16]. A key idea behind these methods is that similar drugs may treat similar diseases and vice versa. However, it is hard to provide a universal definition of similarity [17] and the similarity scores only reflect the strength of connections among entities while ignoring how two entities are connected [18]. Several computational strategies have been introduced to extract functional information from heterogeneous data sources to strengthen the prediction of new drug indications. For example, Dai et al. [19] built a three-layer network by integrating drug–gene interactions, disease–gene interactions and gene–gene interactions and proposed a matrix factorization model to predict novel drug indications. RWHNDR proposed by Luo et al. [20] constructed a heterogeneous network by combining multiple sub-networks including drug network, disease network, target network, drug–disease network, drug–target network and target–disease network. They applied a random walk model to capture the global information of the heterogeneous network to predict candidate pharmacological treatments for diseases. Despite the impressive performances of these models, these methods were limited to capture the semantics within different types of relationships between biomedical entities. It is proposed that knowledge graph (KG) approaches would be of value in identifying the semantic connections between multiple databases [21-23]. Knowledge graph embedding, which aims to embed the entities and relations of a knowledge graph into low-dimensional vector spaces while maximally preserving its topological properties and leverages these representations for link prediction, is an emerging computational approach for biomedical discovery [24-26], including drug repurposing [27]. Moon et al. [28] constructed a heterogeneous knowledge graph including a variety of drug-related information and utilized TransE [29] model to infer drug–disease–target or drug–target–side effect relationships. Mohamed et al. [30] extracted drugs, target proteins, action pathways and targeted diseases from drug target knowledge bases to build a knowledge graph and extended the DistMult [31] and ComplEx [32] to propose a computational approach for predicting new associations between drugs and their targets. Other studies [33,34] constructed biomedical knowledge graphs and applied several knowledge graph completion algorithms (i.e., TransE, DistMult, ConvE [35], RotateE [36] to predict drug candidates for COVID-19 or Parkinson’s Disease. The key to any knowledge graph-based drug repurposing approach is the types, quantity, and quality of semantic relationships captured in the underlying knowledge graphs.
While knowledge-graph methods have been successfully used in drug repurposing, they are limited by the fact that the underlying knowledge graphs mainly captured genetic and genomic information of diseases and drugs such as drug targets, pathways, and disease genes . In addition, these methods also have limitations in capturing heterogeneous topology information and feature interactions to predict new interactions. In recent years, phenotypic information has become available as a new source of drug repositioning [37]. This type of information can be integrated with other biomedical data sources to enrich associations between drugs, genes, and diseases; and potentially boost the accuracy of drug repositioning [38,39].
In this paper, we addressed aforementioned limitations and proposed KG-Predict, a knowledge graph computational framework for inferring new drug indications. We first constructed GP-KG by connecting various genotypic and phenotypic databases. GP-KG composed 9 kinds of phenotypic and genotypic relations and 7 kinds of entities including drugs, genes, diseases, and phenotypic annotations. The embedding module of KG-Predict was trained to learn low-dimensional representations of entities and relations in GP-KG. The predicting module of KG-Predict captured rich heterogeneous feature interactions between entity and relation embeddings to infer new drug–disease interactions. We compared KG-Predict with several state-of-the-art models. We also used Alzheimer’s disease (AD) as a case study and evaluated predicted drugs by clinical trials.
2. Materials and methods
2.1. Knowledge graph construction
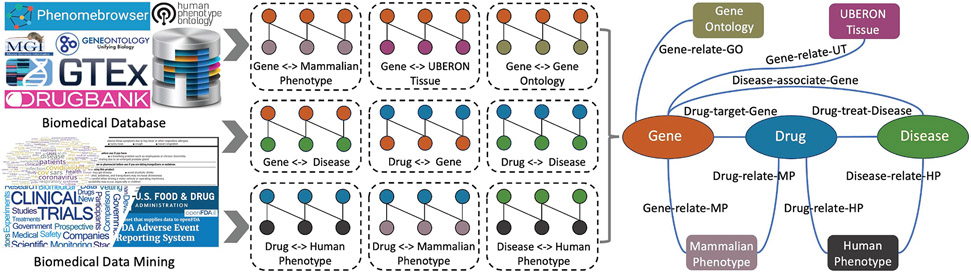
GP-KG consisted of nine types of semantic relationships among seven biomedical entity types. It was built from publicly available phenome-level databases, genome-level databases, and text-mined knowledge bases. Each of these relationships (e.g., disease–gene, drug–gene, drug–disease, and gene–phenotype interactions) and databases (e.g., Drugbank, TreatKB, MGI, HPO, GAO) has widely been used for drug repurposing [11,22,27,38,39]. In this study, we integrated these heterogeneous semantic relationships into one unified knowledge graph GP-KG. Table 1 listed statistics of extracted entities and relations.
Table 1.
Statistics of entities and interactions in GP-KG.
| Knowledge type | Data source | Interaction type | Interaction number |
Node type | Node number |
|---|---|---|---|---|---|
| Phenome-level knowledge | Phenomebrowser Database | Drug-relate-MP | 36,422 | Drug | 1,228 |
| Mammalian Phenotype | 1,363 | ||||
| Drug-relate-HP | 175,713 | Drug | 1,429 | ||
| Human Phenotype Ontology | 3,003 | ||||
| Mouse Genome Informatics (MGI) Database | Gene-relate-MP | 187,304 | Gene | 12,219 | |
| Mammalian Phenotype | 9,916 | ||||
| Gene Ontology Annotation (GOA) Database | Gene-relate-GO | 204,862 | Gene | 16,283 | |
| Gene Ontology | 15,924 | ||||
| Genotype-Tissue Expression (GTEx) database | Gene-relate-UT | 539,845 | Gene | 16,579 | |
| Tissue Uberon | 51 | ||||
| Human Phenotype Ontology (HPO) database | Disease-relate-HP | 87,154 | Disease | 7,172 | |
| Human Phenotype Ontology | 6,784 | ||||
| Genome-level knowledge | DrugBank database | Drug–target–Gene | 5,280 | Drug | 985 |
| Gene | 1,365 | ||||
| Mouse Genome Informatics (MGI) Database | Disease-associate-Gene | 7,382 | Disease | 4,350 | |
| Gene | 3,363 | ||||
| Text-mined knowledge | TreatKB | Drug–treat–Disease | 2,764 | Drug | 639 |
| Disease | 371 |
2.1.1. Phenome-level databases
Gene–phenotype interactions were derived from abnormal phenotypes observed in mouse models, the normal functions of gene products and anatomical location of gene expressions. Specifically,
Mouse Genome Informatics (MGI) database for Gene-relate-MP interactions. MGI [40] is the online database and bioinformatics resource, which provides access to data on the genetics, genomics, and biology of the laboratory mouse to facilitate the study of human health and disease. File “MGI_GenePheno.rpt” in the MGI database provided the connection between genotypes and mammalian phenotype (MP) annotations. We mapped each mouse gene to its human ortholog using mouse–human orthologs with phenotype annotations to obtain Gene-relate-MP interactions.
Gene Ontology Annotation (GOA) Database for Gene-relate-GOA interactions. The GOA database provides annotations to the UniProt Knowledgebase using the standardized vocabulary of the Gene Ontology [41]. We downloaded file “goa_human.gaf.gz” from the GOA database to extract Gene-relate-GO interactions. Some of pairs inferred from electronic annotation or with no available biological data were removed.
Genotype-Tissue Expression (GTEx) database for Gene-relate-UT interactions. GTEx [42] Program provides a data resource and tissue bank that includes the relationship between genetic variants and gene expression in multiple human tissues and across individuals. We chose a cutoff of 4.0 transcripts per million as a threshold [39] to extract genes expressed in each tissue and mapped each tissue to the Uberon Anatomy Ontology to obtain human Gene-relate-UT interactions.
Disease-relate-HP interactions were collected from Human Phenotype Ontology (HPO) database. HPO [43] database provides a standardized vocabulary of phenotypic abnormalities that have been seen in human disease. Phenotype annotations of human diseases with the human phenotype (HP) ontology were obtained from file “phenotype.hpoa” in the HPO database and these annotations were used to construct Disease-relate-HP interactions.
We collected Drug-relate-HP and Drug-relate-MP interactions from file “sider_drug_phenotype.txt” in the Phenomebrowser database. Phenomebrowser [44] is a platform that aggregates phenotype connections with biomedical concepts. This platform provided drug-phenotype datasets which include mammalian phenotype (MP) annotations [45] and human phenotype (HP) ontology [43] associating drugs.
2.1.2. Genome-level databases
We collected Drug–target–Gene interactions from the DrugBank database. DrugBank [46] is a comprehensive online database that provides information on drugs and drug targets. It has been widely used in many bioinformatics and cheminformatics tasks. In our study, we extracted FDA-approved drugs and their target genes from file “drugbank_all_target_polypeptide_ids.csv” in Drug-Bank.
We extracted Disease-associate-Gene interactions from the file “MGI_DO.rpt” in MGI database. We excluded disease–gene pairs where the organism of the gene were not human.
2.1.3. Text-mined knowledge bases
In our previous studies [47,48], we developed natural language processing techniques to extract Drug–treat–Disease interactions from records of patients in FAERS, FDA drug labels, MEDLINE abstracts and clinical trial studies. From the above databases, we collected a drug set including 1430 drugs and a disease set including 7784 diseases. We extracted drug–disease pairs from TreatKB in which drugs and diseases appeared in the drug set and disease set.
2.1.4. Data integrating and processing
We mapped and integrated data from the above resources using standard biomedical terminologies. Drug names were mapped to their active ingredients using PubChem identifiers. Diseases that were represented using their OMIM (online mendelian inheritance in man) [49] identifiers in MGI and HPO databases were mapped using UMLS (unified medical language system) CUIs (Concept Unique Identifiers) [50, 51]. For example, Alzheimer’s disease was denoted by its OMIM identifier (104300) in MGI and HPO databases. In GP-KG, we converted AD’s OMIM identifier (104300) into its CUI (C0002395) using mapping file “MRCONSO.RRF” downloaded from UMLS.
Fig. 1 showed the structure of the knowledge graph. All interaction information extracted from different data sources were merged into a KG, in which each type of biomedical concepts (i.e., drugs, genes, diseases, and phenotype annotations) was considered as a node type and each type of interactions (i.e., Drug–target–Gene, Drug–treat–Disease, and Drug-relate-MP) was considered as an edge type. Table 1 showed statistics of entities and interactions in GP-KG. The knowledge graph contained 61,146 nodes, 1,246,726 edges, 7 node types, and 9 semantic relationships. Among 9 kinds of relationships, two of them were derived from genome-level knowledge databases, e.g., “Drug–target-Gene”, “Disease-associate-Gene”. One was from text-mined knowledge bases, e.g., “Drug-treat-Disease”. Other six relations were derived from phenome-level knowledge databases.
Fig. 1.
Flow chart of the GP-KG construction: (a) extracted raw interactions from biomedical databases and text-mined knowledge bases, (b) mapped entities into standard identifiers and merged raw interactions into a knowledge graph.
2.2. KG-Predict Model
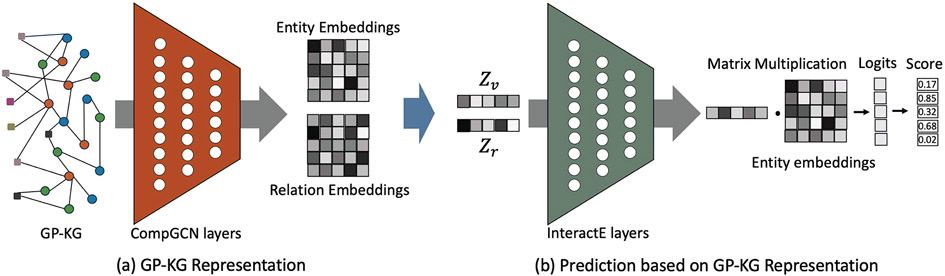
Fig. 2 provided an overview of KG-Predict Model. KG-Predict included a embedding module and a predicting module. The embedding module took GP-KG as input and learned low-dimensional embeddings of entities and relations. For a drug node, the embedding module aggregated information from the drug identifier, topological structure of the drug’s neighborhoods (such as genes, diseases and phenotype annotations) and semantic relations between the drug and its neighbors to learn the drug’s embedding. Once learned, the predicting module concatenated embeddings of entities and relations and used three operations to extract topologic and semantic information to make link predictions. For each triple (eg., Drug–Treat–Disease), the predicting module could be represented as a ranking function which generated higher scores for true triples and lower scores for false triples.
Fig. 2.
The pipeline of the KG-Predict consists of: (a) embedding: using a stack of CompGCN layers to capture the heterogeneous topologic structures and semantic features to learn embeddings of entities and relations, (b) prediction or ranking: using InteractE to rank candidate entities (drugs in this study) for given input entity (a specific disease in this study) and relation embeddings “Drug–treat–Disease” in this study).
2.2.1. GP-KG representation
The embedding module of KG-Predict is used to transform entities (e.g., drugs, diseases), relations (e.g., Drug–treat–disease), and their features into low-dimensional vector representations whilst maximally preserving properties like graph structure and information. These entity and relation representations are used to predict unseen interactions in the knowledge graph. More specifically, we first define the knowledge graph as,
| (1) |
where V and E denote the set of entities and relations, respectively. T ⊆ V × E × V denotes the set of triplets, X represents features of nodes, R denotes the set of relations, S denotes the initial relation features. KG-Predict takes G as input and learns embeddings of entities and relations by aggregating multi-relational information in the knowledge graph.
We extend composition-based multi-relational graph convolutional networks (CompGCN) [52] to learn representations of entities and relations. The pseudo code of the GP-KG embedding algorithm is illustrated in Algorithm 1. The procedure has three steps:
Input: the feature vector Xυ of node υ (e.g., drugs, diseases) and the feature vector Sr of edge r (e.g., Drug–treat–disease) is used as the initial input representation vector of entity υ and relation r.
CompGCN layer: our model stacks several convolutional layers. These layers update the representation vectors of entities by aggregating heterogeneous information through their neighboring entities and neighboring semantic relations. Specifically, let represents the feature vector of the entity υ in the nth layer, KG-Predict in nth layer aggregates the embeddings of υ’s neighboring entities and relations using a aggregator function AGGREGAREn(.). For a drug node, AGGREGAREn(.) generates the aggregated feature vector for the drug by aggregating information from its neighboring entities (e.g., genes, diseases, human phenotype ontology, mammalian phenotype annotations) and relations (e.g., Drug–target–Gene, Drug–treat–Disease, Drug-relate-HP, Drug-relate-MP). It then uses a concat function CONCAT(.) to sum up these corresponding embeddings and υ’s embedding in previous layer to obtain the embedding of the entity υ in the nth layer. Similarly, let denote the representation of a relation r after n − 1 layers. KG-Predict utilizes a learnable transformation matrix which projects all the relations to the same embedding space as entities to update relation r’s embedding in the nth layer. It is an end-to-end learning process, the aggregator function AGGREGAREn(.) aggregates each entity’s information from its neighboring entities and relations, and the concat function CONCAT(.) sums up all neighboring information and maps the entity into a low dimensional vector. Both functions are achieved via neural networks.
Output: the embeddings of entity υ and relation r are generated after N iterations. ZV and ZR are denoted as the set of entity embeddings and the set of relation embeddings, respectively.
| Algorithm 1: GP-KG Embedding Algorithm | |
|---|---|
|
|
2.2.2. Prediction based on GP-KG representation
KG-Predict utilizes the InteractE model [53] to capture heterogeneous feature interactions preserved in entity and relation embeddings to infer new drug–disease interactions. Denoted υ is a drug entity and r is the relation of ‘Drug–treat–Disease’, The predicting modular of KG-Predict is input with node embedding Zυ and relation embedding Zr and predicts another suitable disease entity u that can composes a correct triplet, that is predicting disease entity u given subject entity and relation (υ, r). The goal is achieved through defining a score function ϕ(υ, r, u) and score a correct triplet higher than incorrect triplets.
The pseudo code of the interaction prediction algorithm is illustrated in Algorithm 2. The procedure has three steps:
Input: the embeddings of entities and relations obtained from the embedding module are as input of predicting module. For example, to infer whether a triple (Memantine, Drug–treat–Disease, Alzheimer’s Disease) is true, the embedding vector Zυ of drug υ (e.g., Memantine), the embedding vector Zr of relation r (e.g., Drug–treat–Disease) and the embedding vector Zu of disease u (e.g., Alzheimer’s Disease) are input to our predicting module.
InteractE layer: the layer uses neural networks to capture variety of heterogeneous interactions preserved in the entity and relation embeddings, which is beneficial to prediction performance. Specifically, let w ∈ Rk×k be a convolutional kernel of size k, three operations (feature permutation, checkered feature reshaping, and circular convolution) are used in the predicting module. The first operation is to generate t-random permutations of both Zυ and Zr, denoted by . KG-Predict conducts the reshaping operation and defines , φ(.)is the reshaping function which is used to capture maximum heterogeneous interactions between entity and relation features. KG-Predict stacks the reshaped matrices into a 3D tensor, that is then processed with depth-wise circular convolution. Our model flattens and concatenates the output of each circular convolution into a vector ζ(υ, r).
Output: we design a score function ϕ(υ, r, u) to get the score of triple (υ, u, r) and utilize a sigmoid function p(υ, r, u) to calculate the probability of the triple (υ, r, u). Dυ,r was denoted as probability distribution of entity υ connecting to other entities with relation r. The output scores are then passed to the loss function to calculate training loss. We use the standard cross entropy loss as the loss function.
| Algorithm 2: Interaction Prediction Algorithm | |
|---|---|
|
|
2.3. Evaluation and comparison
We conducted cross-validation to evaluate the model performance for predicting drug–disease interactions. All drug–disease interactions were randomly shuffled five times, and split into training (60%), validation (20%) and test (20%) set with the same ratios. Thus, five training, validation and test sets were created for the cross-validation experiments. We used each training set to build the model, used each corresponding validation set to optimize the parameter setting of the model, and used the test set to verify the model performance. In order to provide a comprehensive comparison, we also constructed the similarity-based heterogeneous networks (S-HNs) that included six types of relations: drug–drug, drug–gene, drug–disease, disease–disease, gene–disease, gene–gene. The S-HNs shared the same drug–gene, drug–disease, and gene-disease interactions with GP-KG. Other Drug–drug, gene–gene and disease–disease similarity networks were from DTINet [54], PPIN [55] and TargetPredict [38]. The S-HNs included 22,784 nodes of 3 types and 624,855 relationships of 6 types.
We also added negative samples for cross-validation experiment. We generate negative counterparts for each positive triple in each test set by enumerating the complement set of positive examples. We treated drug–disease pairs that are not in GP-KG as negatives. For each known triple (υs, r, υo) in the test set where υs was a drug entity, r was the relation of ‘TREAT’ and υo was a disease entity, we created a set of negative samples (υs, r, υn) by replacing the subject υo to subject υn. υn was another disease entity. These generated negative samples have not been in the original knowledge graph. We enumerated all disease entities for each triple in the test set to construct the negative dataset.
Several state-of-the-art computational methods for drug–disease prediction were used to compared with our model. DeepWalk model [56] was a traditional graph embedding method which used random walks (RWs) to learn embeddings for nodes in graphs and input these embeddings to a logistic regression function to predict new relations. Matrix factorization model [56] aimed to factorize a data matrix into lower dimensional matrices and built a Logistic regression predictor for link prediction. TransE [29] was a translation-based method for link prediction, which used the relation for translating the head entity to a tail entity. DistMult [31] interpreted interaction prediction as a task of tensor decomposition. ConvE [35] was a deep learning model which used 2D convolutions over embeddings to predict new interactions in knowledge graphs. RotatE [36] mapped the entities and relations to the complex vector space and took predications as rotations from subjects to objects in complex space. Several widely used evaluation metrics were adopted to measure performances of these models including Hits@N and MRR (mean reciprocal rank). Hits@N was the hit percentage of true triples in a test set being ranked by a model within the top N positions, it evaluated the ability for “early recognition” of true predictions. MRR was the average inverse rank for true triples. A higher MRR value indicated a better model. In addition, we also calculated AUROC (Area Under Receiver Operating Characteristic curve) and AUPR (Area Under Precision-Recall Curve) to compare performances of these models. These metrics afforded comprehensive assessments of a model and has been recognized in bioinformatics applications.
Hyperparameters for KG-Predict were tuned using the grid search on the validation set. We tuned the learning rate η ∈ {0.0001, 0.001, 0.01}, embedding dimensions κ ∈ {100, 200, 400 }, Number of GCN Layer ℓ ∈ {1, 2, 3 }, batch size β ∈ {64, 128, 256 }, and dropout σ ∈ {0.1, 0.2, 0.3 }. For baseline methods, we downloaded their implementations from the original authors’ websites and used their default model parameters to train these baseline models.
3. Results
3.1. Experimental setup
KG-Predict was built on PyTorch and used Adam optimizers to generate gradients and update embeddings and parameters. ReLU was used as activation function f. We used the validation sets to perform a grid search to learn the model’s best hyperparameters. The optimal values were selected based on MRR results. We first checked how the number of GCN layers affected the performance. We found that deeper GCN layers did not improve the performance while substantially increased the computational cost. Thus ℓ was set to 1. We also investigated the influence of embedding dimensions κ. We found that increasing κ boosted the performance marginally but too large κ substantially increased both memory and computation costs. Therefor we chose the middle value 200 as embedding dimension κ setting. Other optimal hyperparameters η and β were set to 0.001 and 128. To avoid over-fitting, the dropout setting after each convolution layer was 0.1 in the embedding module and 0.3 in the predicting module. This procedure was performed iteratively for 500 iterations. These settings were used throughout all results reported in this paper.
3.2. Overall performance evaluation with cross-validation
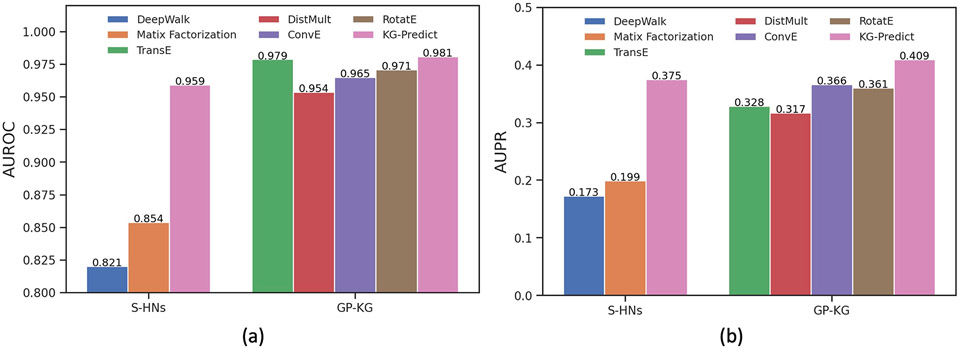
We evaluated the performances of KG-Predict and six state-of-art models using cross-validation. Statistics of the average score of Hits@N and MRR for KG-Predict and baseline models were given in Table 2. We first compared KG-Predict with two traditional methods using S-HM. KG-Predict outperformed DeepWalk and matrix factorization approaches by a large margin. Compared with four knowledge graph embedding techniques on GP-KG, KG-Predict achieved the best performance in terms of Hits@N and MRR. For example, RotatE achieved a 2.4% improvement in terms of Hits@10 value compared with DistMult. ConvE achieved a competitive performance with RotatE. Our method outperformed ConvE by 4.5% (MRR), 4.8% (Hits@1), 3.4% (Hits@3) and 4.8% (Hits@10). It also improved over RotatE with 4.9% on MRR and 4.4% on Hits@10. This validated KG-Predict that captured rich heterogeneous interactions can improve the predicting performance. In addition, KG-Predict using GP-KG achieved better performances on drug–disease prediction than that using S-HNs. It showed that incorporating phenotypic data can boost predicting accuracy.
Table 2.
Overall predictive performance of drug–disease associations.
| Data | Method | Hits@1 | Hits@3 | Hits@10 | MRR |
|---|---|---|---|---|---|
| S-HNs | DeepWalk | 0.048 | 0.086 | 0.137 | 0.081 |
| Matrix factorization | 0.057 | 0.102 | 0.194 | 0.095 | |
| KG-Predict | 0.147 | 0.255 | 0.411 | 0.234 | |
| GP-KG | TransE | 0.116 | 0.226 | 0.399 | 0.209 |
| DistMult | 0.103 | 0.207 | 0.379 | 0.191 | |
| ConvE | 0.126 | 0.232 | 0.399 | 0.216 | |
| RotatE | 0.119 | 0.231 | 0.403 | 0.212 | |
| KG-Predict | 0.174 | 0.266 | 0.447 | 0.261 |
We then used the AUROC and AUPR to evaluate how these known positive drug–disease pairs ranked among all the possible pairs. Fig. 3 showed the average AUROC and AUPR of each model using cross-validation. KG-Predict got an excellent performance on AUROC and AUPR compared with DeepWalk and matrix factorization methods using S-HNs. We further compared KG-Predict with four knowledge graph embedding models using GP-KG. KG-Predict performed the best in discriminating positive and negative pairs in drug repurposing tasks, outperforming that of TransE, DistMult, ConvE, and RotatE. Comparing the AUROC and AUPR of KG-Predict trained by the GP-KG and S-HNs, KG-Predict (GP-KG) achieved a significantly higher overall AUROC of 0.981 and AUPR of 0.409 compared with 0.959 and 0.375 obtained by the KG-Predict (S-HNs).
Fig. 3.
Performance of KG-Predict as compared with baseline models: (a) The bar graph of AUROC, (b) The bar graph of AUPR. KG-Predict achieved the best performance on AUROC and AUPR underlying both S-HNs and GP-KG.
3.3. Visualization of entity embeddings with GP-KG representation
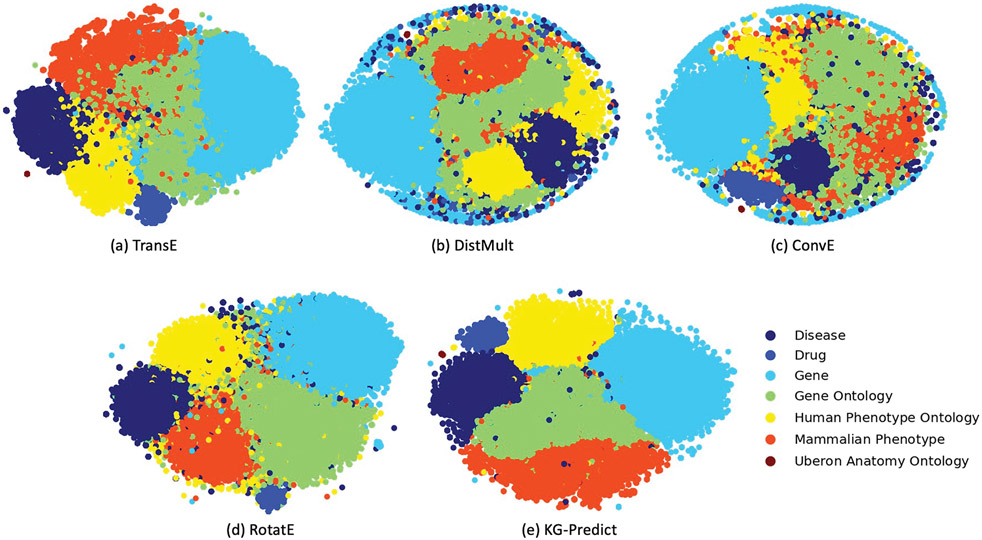
In the following, we evaluated the quality of entity representations produced by each method to qualitatively interpret models’ learning abilities. We learned entity embeddings into 200-dimensional vector spaces for each model and input them into t-SNE [57] to reduce the dimensionality to 2 and visualized nodes in a 2-D space. This method can reveal the local and global features encoded in the embedding vectors and thus can be used to visualize clusters within the knowledge graph. Fig. 4 showed 2-D entity embeddings produced by KG-Predict and four state-of-art models, with colors corresponding to different entity groups. Compared to TransE, DistMult, ConvE, and RotatE, the GP-KG had visibly distinct clusters with clear group separation, demonstrating that KG-Predict was capable of learning and preserving the high-level structural information embedded in the GP-KG and had strong ability to distinguish the differences between multiple type of entities.
Fig. 4.
Visualization of entity embeddings using the t-SNE package. Each dot represents an entity, with its color indicating node type. From this graph, we can clearly see that KG-Predict, but not the baseline models, can distinguish embeddings of different type of nodes, demonstrating that KG-Predict is able to capture high-level structural information from GP-KG.
4. Case study: drug repurposing for Alzheimer’s disease
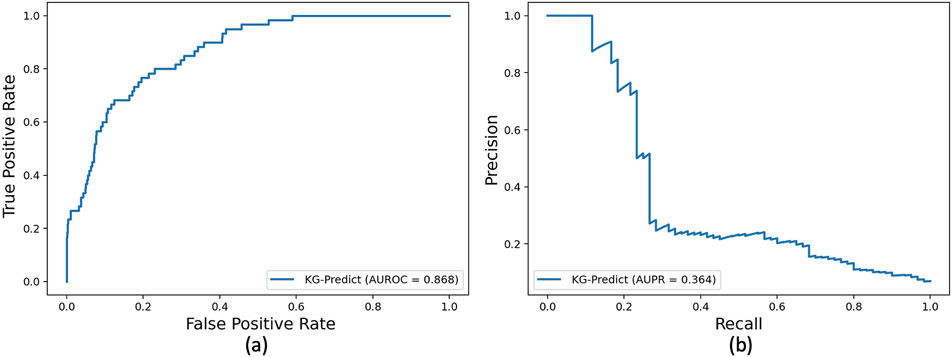
To further validate the efficacies of our model, we conducted a case study to infer novel drug repurposing for Alzheimer’s disease (AD). For objective performance evaluation, we collected clinically reported drugs that have been tested for treating AD from the ClinicalTrials.gov database as an external validation set and validated the top ranked drugs against evidence from clinical trials [58,59]. The external validation set carried 60 drugs. We treated these 60 drugs as positive samples. Other drugs in GP-KG were considered to be negative samples. Fig. 5 gave the AUROC and AUPR curves for this task. KG-Predict achieved an AUROC value of 0.868 and an AUPR value of 0.364.
Fig. 5.
Performance of KG-Predict in drug repurposing for Alzheimer’s disease evaluated using FDA-approved drugs: (a) AUROC, (b) AUPR.
Table 3 showed the top 10 highest-scoring novel drug repurposing candidates for AD with the canonical name of the drug, original indication, and the reported evidence. Among the top 10 drugs, memantine has been approved for Alzheimer’s disease. Eight of the top-ranked drugs have been tested in clinical trials for treating AD. These results further confirmed the validity of KG-Predict for drug repositioning.
Table 3.
Top 10-ranked repositioned drug candidates for AD.
| Drug | Indication | Evidence (Status) | Studying period |
|---|---|---|---|
| Risperidone | Schizophrenia, Bipolar Mania | NCT00034762 (Completed) | 56 days |
| Acitretin | Psoriasis | NCT01078168 (Completed) | 28 days |
| Memantine | Alzheimer’s Disease | FDA_approved | |
| Aripiprazole | Schizophrenia, Bipolar disorder, Depressive disorder | NCT01438060 (Completed) | 70 days–980 days |
| Ibuprofen | Pain, Fever, Inflammation | NCT04570644 (Completed) | 60 days |
| Minocycline | wide variety of infections | NCT01463384 (Completed) | 180 days |
| Thalidomide | Multiple Myeloma, Erythema Nodosum Leprosum | NCT01094340 (Unknown) | 168 days |
| Fluoxetine | Depressive Disorder, Obsessive compulsive disorder | ||
| Citalopram | Depression | NCT00898807 (Completed) | 63 days |
| Dasatinib | Myeloid leukemia, Lymphoblastic leukemia | NCT04063124 (Recruiting) | 84 days |
Note: NCT**: AD drugs from clinical trials. FDA-approved AD drug is highlighted in bold.
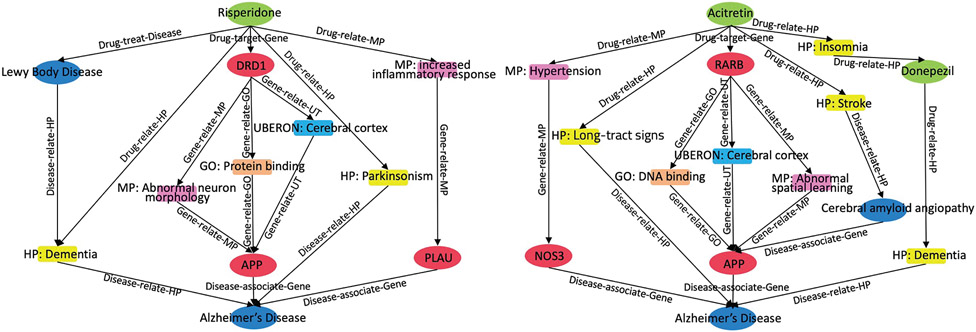
To better understand how our model made novel link predictions, we selected top predicted drugs to analyze their mechanism in original GP-KG. Our method hypothesized that risperidone and acitretin were potential drugs with top-2 ranked for AD. Risperidone was used to treat schizophrenia and acitretin was used to treat psoriasis. In Fig. 6, several reasoning paths were provided to support these hypotheses. For instance, side effects of two drugs were directly mapped to phenotypes of AD. It suggested that they may share similar underlying pathways [9]. Recent research studies indicated that more than 50% of individuals with AD had Lewy body diseases [60]. Drugs related to Lewy bodies may also work on AD due to the interdependence between the two diseases. Several observational evidences also showed that antihypertensive treatments can reduce the risk of AD [61]. In addition, APP and DRD1 have been reported to be involved in AD pathology [62]. This indicated that drugs that targets to DRD1 may potentially treat AD. This case study illustrated that KG-Predict can capture heterogeneous network information and phenotypic features from the GP-KG to infer new drug indications.
Fig. 6.
Visualization of supportive paths between the top-2 predicted drugs and the Alzheimer’s disease.
5. Discussion and conclusion
In this paper, we proposed KG-Predict, a knowledge graph computational framework, which embedded a knowledge graph into low-dimensional vector spaces and inferred novel drug indications based on latent vectors. We first constructed a knowledge graph named GP-KG that contains over 1 million interconnections between 61,146 entities including drugs, genes, diseases, and phenotypic annotations. KG-Predict captured multiple types of genotypic and phenotypic features embedded in GP-KG, and significantly improved the predicting accuracy of unseen interactions. Extensive experiments demonstrated that our model achieved competitive performance on the task of drug re-purposing. In addition, we externally validated the potential ability of our method in drug repositioning for Alzheimer’s disease.
KG-Predict has several potential limitations under the current deep learning framework. First, it is hard to build gold-standard unknown pairs as negative samples due to the inherent lack of negative drug–disease pairs in the public databases and literature. Although we made efforts to assemble drug–disease interactions from publicly available databases, it may incorrectly classify a drug–disease association as negative when the association is not yet known. We will further design natural language technique-based relationship mining methods to extract potential drug–disease pairs to decrease false-negative rates. In addition, we currently used the PubChem identifiers of drugs, Entrez gene identifiers, and phenotypic annotations in constructing the knowledge graph and have not considered non-topological domain information, e.g., chemical structure of drugs and gene expressions. In the future, we will expand GP-KG by merging additional domain-specific information embedded using contrastive learning to further improve predictive performances. Finally, we used KG-Predict to predict repurposed drugs for diseases in this study. However, KG-Predict is highly flexible in predicting other semantic relationships among biomedical entities, including disease–disease, drug–drug, drug–gene, disease–gene among others. For example, KG-Predict can be used to understand how COVID-19 and Alzheimer’s disease are semantically connected based on both genetic and phenotypic relevance, which can complement existing epidemiologic studies of COVID-19-disease relationships, including our recent patient electronic health record-based studies of Alzheimer disease and COVID-19 [63,64]. Predicting mechanistic links between disease pairs such as COVID-19 and Alzheimer disease offers new drug repurposing opportunities by targeting these critical links.
Funding
This work has been supported by NIH National Institute of Aging, USA R01 AG057557, R01 AG061388, R56 AG062272, National Institute on Alcohol Abuse and Alcoholism, USA (grant no. R01AA029831), National Eye Institute, USA (EY029297), National Institute on Drug Abuse, USA (UG1DA049435, CTN-0114), American Cancer Society Research Scholar, USA Grant RSG-16-049-01-MPC, The Clinical and Translational Science Collaborative (CTSC) of Cleveland, USA (UL1TR002548-01).
Abbreviations and terms
We listed all terms and their definitions in Table 4.
Table 4.
Definitions of abbreviations and terms.
| Term | Definition |
|---|---|
| KG-Predict | The knowledge graph computational framework |
| GP-KG | The knowledge graph integrating various genotypic and phenotypic databases |
| S-HNs | The similarity-based heterogeneous networks |
| MP | Mammalian phenotype |
| HPO | Human phenotype ontology |
| MGI | Mouse genome informatics |
| GTEx | Genotype-tissue expression |
| GOA | Gene ontology annotation |
| UAO | Uberon anatomy ontology |
| CompGCN | Composition-based multi-relational graph convolutional network |
| MRR | Mean reciprocal rank |
| AUROC | Area under receiver operating characteristic curve |
| AUPR | Area under precision–recall curve |
| AD | Alzheimer’s Disease |
Footnotes
CRediT authorship contribution statement
Zhenxiang Gao: Conceived the study, Designed and performed the experiment, Writing – original draft. Pingjian Ding: Provided inputs on method design, Evaluation, Case studies, Manuscript. Rong Xu: Conceived the study, Edited the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Availability and implementation
The code, data and supplementary material are available at http://nlp.case.edu/public/data/GPKG-Predict/
References
- [1].Zeng X, Tu X, Liu Y, Fu X, Su Y, Toward better drug discovery with knowledge graph, Curr. Opin. Struct 72 (2022) 114–126. [DOI] [PubMed] [Google Scholar]
- [2].Saberian N Peyvandipour A, Donato M Ansari S Draghici S A new computational drug repurposing method using established disease–drug pair knowledge, Bioinform 35 (2019) 3672–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kola I, Landis J, Can the pharmaceutical industry reduce attrition rates? Nat. Rev. Drug Discov 3 (2004) 711–716. [DOI] [PubMed] [Google Scholar]
- [4].Choudhury C Murugan NA, Priyakumar UD, Structure-based drug repurposing: Traditional and advanced AI/ML-aided methods, Drug Discov. (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hua Y, Dai X, Xu Y, Xing G, Liu H, Lu T, Zhang Y, Drug repositioning: Progress and challenges in drug discovery for various diseases, Eur. J. Med. Chem 234 (2022) 114239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Correia AS, Gärtner F, Vale N, Drug combination and repurposing for cancer therapy: The example of breast cancer, Heliyon 7 (2021) e05948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pan X, Lin X, Cao D, Zeng X, Yu PS, He L, Cheng F, Deep learning for drug repurposing: Methods, databases, and applications, Wiley Interdiscip. Rev. Comput. Mol. Sci e1597 (2022) 1–21. [Google Scholar]
- [8].Luo H, Li M, Yang M, Wu FX, Li Y, Wang J, Biomedical data and computational models for drug repositioning: a comprehensive review, Brief. Bioinformatics 22 (2021) 1604–1619. [DOI] [PubMed] [Google Scholar]
- [9].Jarada TN, Rokne JG, Alhajj R, A review of computational drug repositioning: Strategies, approaches, opportunities, challenges, and directions, J. Cheminform 12 (2020) 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Pushpakom S, Iorio F, Eyers PA, Escott KJ, Hopper S, Drug repurposing: Progress, challenges and recommendations, Nat. Rev. Drug Discov 18 (2019) 41–58. [DOI] [PubMed] [Google Scholar]
- [11].Bagherian M, Sabeti E, Wang K, Sartor MA, Nikolovska-Coleska Z, Najarian K, Machine learning approaches and databases for prediction of drug–target interaction: A survey paper, Brief. Bioinformatics 22 (2021) 247–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hamed AA, Leszczynska A, Schreiber M, MolecRank: A specificity-based network analysis algorithm, in: International Conference on Advanced Machine Learning Technologies and Applications, AMLTA, 2019, pp. 159–168. [Google Scholar]
- [13].Dudley JT, Deshpande T, Butte AJ, Exploiting drug–Disease relationships for computational drug repositioning, Brief. Bioinformatics 12 (2011) 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Li J, Lu Z, A new method for computational drug repositioning using drug pairwise similarity, in: IEEE Int. Conf. Bioinformatics Biomed., 2012, pp. 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cheng F, Liu C, Jiang J, Lu W, Li W, Liu G, Tang Y, Prediction of drug-target interactions and drug repositioning via network-based inference, PLoS Comput. Biol 8 (2012) e1002503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhang W, Yue X, Lin W, Wu W, Liu R, Huang F, Liu F, Predicting drug-disease associations by using similarity constrained matrix factorization, BMC Bioinform. 19 (2018) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ye Q, Hsieh CY, Yang Z, Kang Y, Chen J, Cao D, A unified drug–Target interaction prediction framework based on knowledge graph and recommendation system, Nature Commun. 12 (2021) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhou M, Chen Y, Xu R, A drug-side effect context-sensitive network approach for drug target prediction, Bioinform 35 (2019) 2100–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dai W, Liu X, Gao Y, Chen L, Song J, Matrix factorization-based prediction of novel drug indications by integrating genomic space, Comput. Math. Methods Med 1 (2015) 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Luo H, Wang J, Li M, Luo J, Ni P, Computational drug repositioning with random walk on a heterogeneous network, IEEE/ACM Trans. Comput. Biol. Bioinform 16 (2018) 1890–1900. [DOI] [PubMed] [Google Scholar]
- [21].Li Z, Zhong Q, Yang J, Duan Y, Wang W, Wu C, He K, Deepkg: an end-to-end deep learning-based workflow for biomedical knowledge graph extraction, optimization and applications, Bioinform 38 (2022) 1477–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bonner S, Barrett IP, Ye C, Swiers R, Engkvist O, Bender A, A review of biomedical datasets relating to drug discovery: A knowledge graph perspective, 2021, arXiv. [DOI] [PubMed] [Google Scholar]
- [23].Nicholson DN, Greene CS, Constructing knowledge graphs and their biomedical applications, Comput. Struct. Biotechnol. J 18 (2020) 1414–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Su X, You ZH, Huang DS, Wang L, Wong L, Ji B, Zhao B, Biomedical knowledge graph embedding with capsule network for multi-label drug-drug interaction prediction, IEEE Trans. Knowl. Data Eng 14 (2022) 1–13. [Google Scholar]
- [25].Rossi A, Barbosa D, Firmani D, Matinata A, Merialdo P, Knowledge graph embedding for link prediction: A comparative analysis, ACM Trans. Knowl. Discov. Data 15 (2021) 1–49. [Google Scholar]
- [26].Mohamed SK, Nounu A, Nováček V, Biological applications of knowledge graph embedding models, Brief. Bioinformatics 22 (2021) 1679–1693. [DOI] [PubMed] [Google Scholar]
- [27].Zheng S, Rao J, Song Y, Zhang J, Xiao X, Fang EF, Niu Z, PharmKG: A dedicated knowledge graph benchmark for bomedical data mining, Brief. Bioinformatics 22 (2021) bbaa344. [DOI] [PubMed] [Google Scholar]
- [28].Moon C, Jin C, Dong X, Abrar S, Zheng W, Chirkova RY, Tropsha A, Learning drug-disease-target embedding (DDTE) from knowledge graphs to inform drug repurposing hypotheses, J. Biomed. Inform 119 (2021) 103838. [DOI] [PubMed] [Google Scholar]
- [29].Bordes A, Usunier N, Garcia-Duran A, Weston J, Yakhnenko O, Translating embeddings for modeling multi-relational data, in: Adv. Neural Inf. Process. Syst, 2013, p. 26. [Google Scholar]
- [30].Mohamed SK, Nováček V, Nounu A, Discovering protein drug targets using knowledge graph embeddings, Bioinform 36 (2020) 603–610. [DOI] [PubMed] [Google Scholar]
- [31].Yang B, Yih WT, He X, Gao J, Deng L, Embedding entities and relations for learning and inference in knowledge bases, in: Int. Conf. Learn. Represent, 2015, p. 6575. [Google Scholar]
- [32].Trouillon T, Welbl J, Riedel S, Gaussier É, Bouchard G, Complex embeddings for simple link prediction, in: Int. Conf. Mach. Learn., 2016, pp. 2071–2080. [Google Scholar]
- [33].Zhang R, Hristovski D, Schutte D, Kastrin A, Fiszman M, Kilicoglu H, Drug repurposing for COVID-19 via knowledge graph completion, J. Biomed. Inform 115 (2021) 103696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhang X, Che C, Drug repurposing for Parkinson’s disease by integrating knowledge graph completion model and knowledge fusion of medical literature, Future Internet 13 (2021) 14. [Google Scholar]
- [35].Dettmers T, Minervini P, Stenetorp P, Riedel S, Convolutional 2D knowledge graph embeddings, in: Int. Thirty-Second AAAI Conf. Artif. Intell., 2018, pp. 1811–1818. [Google Scholar]
- [36].Sun Z, Deng ZH, Nie JY, Tang J, Rotate: Knowledge graph embedding by relational rotation in complex space, in: Int. Conf. Learn. Represent., 2019. [Google Scholar]
- [37].Park K, A review of computational drug repurposing, Transl. Clin 27 (2019) 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhou M, Zheng C, Xu R, Combining phenome-driven drug-target interaction prediction with patients’ electronic health records-based clinical corroboration toward drug discovery, Bioinform 36 (2020) 436–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chen J, Althagafi A, Hoehndorf R, Predicting candidate genes from phenotypes, functions and anatomical site of expression, Bioinform 37 (2021) 853–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Eppig JT, Smith CL, Blake JA, Ringwald M, Kadin JA, Richardson JE, Bult CJ, Mouse genome informatics (MGI): Resources for mining mouse genetic, genomic, and biological data in support of primary and translational research, Syst. Genet (2017) 47–73.. [DOI] [PubMed] [Google Scholar]
- [41].Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Sherlock G, Gene ontology: Tool for the unification of biology, Nat. Genet 25 (2000) 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].GTEx Consortium, Ardlie KG, Deluca DS, Segrè AV, Sullivan TJ, Young TR, Dermitzakis ET, The genotype-tissue expression (GTEx) pilot analysis: Multitissue gene regulation in humans, Science 348 (2015) 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Robinson PN, Mundlos S, The human phenotype ontology, Clin. Genet 77 (2010) 525–534. [DOI] [PubMed] [Google Scholar]
- [44].OntoSIML, Phenomebrowser,, 2021, http://phenomebrowser.net/archive/sider_{d}rug_{p}henotype.txt.
- [45].Smith CL, Eppig JT, The mammalian phenotype ontology: enabling robust annotation and comparative analysis, Wiley Interdiscip. Rev. Syst. Biol. Med 1 (2009) 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Hassanali M, DrugBank: A knowledgebase for drugs, drug actions and drug targets, Nucleic Acids Res. 36 (2008) D901–D906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Xu R, Wang Q, Automatic signal extraction, prioritizing and filtering approaches in detecting post-marketing cardiovascular events associated with targeted cancer drugs from the FDA adverse event reporting system (FAERS), J. Biomed. Inform 47 (2014) 171–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Xu R, Wang Q, Large-scale extraction of accurate drug-disease treatment pairs from biomedical literature for drug repurposing, BMC Bioinform. 14 (2013) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA, Online mendelian inheritance in man (OMIM), a knowledgebase of human genes and genetic disorders, Nucleic Acids Res. 33 (2005) D514–D517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Campbell KE, Oliver DE, Spackman KA, Shortliffe EH, Representing thoughts, words, and things in the UMLS, J. Am. Med. Inform. Assoc 5 (1998) 421–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lindberg DA, Humphreys BL, McCray AT, The unified medical language system, Yearb. Med. Inform 2 (1993) 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Vashishth S, Sanyal S, Nitin V, Talukdar P, Composition-based multi-relational graph convolutional networks, in: Int. Conf. Learn. Represent., 2019. [Google Scholar]
- [53].Vashishth S, Sanyal S, Nitin V, Agrawal N, Talukdar P, Interacte: Improving convolution-based knowledge graph embeddings by increasing feature interactions, in: Int. AAAI Conf. Artif. Intell., 2020, pp. 3009–3016. [Google Scholar]
- [54].Luo Y, Zhao X, Zhou J, Yang J, Zhang Y, Kuang W, Zeng J, A network integration approach for drug-target interaction prediction and computational drug repositioning from heterogeneous information, Nature Commun. 8 (2017) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Von Mering C, STRING v10: Protein–protein interaction networks, integrated over the tree of life, Nucleic Acids Res. 43 (2015) D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Yue X, Wang Z, Huang J, Parthasarathy S, Moosavinasab S, Huang Y, Graph embedding on biomedical networks: Methods, applications and evaluations, Bioinform 36 (2020) 1241–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Van der Maaten L, Hinton G, Visualizing data using t-SNE, J. Mach. Learn. Res 9 (2008) 2579–2605. [Google Scholar]
- [58].Zarin DA, Tse T, Williams RJ, Califf RM, Ide NC, The ClinicalTrials. gov results database—Update and key issues, N. Engl. J. Med 364 (2011) 852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Gates LE, Hamed AA, The anatomy of the SARS-CoV-2 biomedical literature: Introducing the covidx network algorithm for drug repurposing recommendation, J. Med. Internet Res 22 (2020) e21169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Bayram E, Shan G, Cummings JL, Associations between comorbid TDP-43, Lewy body pathology, and neuropsychiatric symptoms in Alzheimer’s disease, J. Alzheimer’s Dis 69 (2019) 953–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Barthold D, Joyce G, Wharton W, Kehoe P, Zissimopoulos J, The association of multiple anti-hypertensive medication classes with alzheimer’s disease incidence across sex, race, and ethnicity, PLoS One 13 (2018) e0206705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Song C, Zhang Y, Huang W, Shi J, Huang Q, Jiang M, Circular RNA Cwc27 contributes to Alzheimer’s disease pathogenesis by repressing Pur-α activity, Cell Death Differ. 29 (2021) 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wang Q, Davis PB, Gurney ME, Xu R, COVID-19 and dementia: Analyses of risk, disparity, and outcomes from electronic health records in the US, Alzheimer’s Dementia 17 (2021) 1297–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Wang L, Davis PB, Xu R, COVID-19 breakthrough infections and hospitalizations among vaccinated patients with dementia in the United States between december 2020 and august 2021, Alzheimer’s Dementia (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]