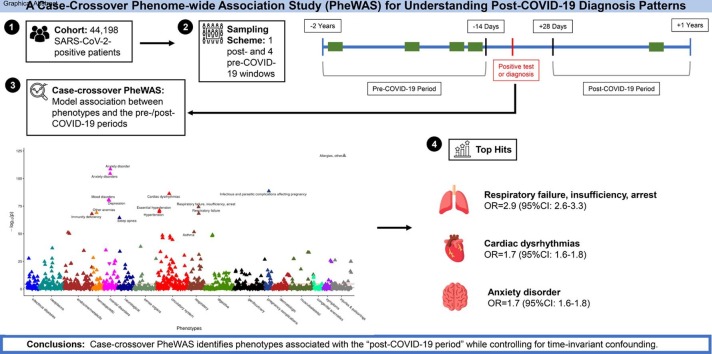
Graphical abstract
Keywords: Electronic Health Records, Flu positive control, Healthcare utilization, Case-crossover, Multiple testing, Phenome-wide association study, Post-COVID-19, Test-negative controls, Within-subject confounding, Vaccination
Abstract
Background
Post COVID-19 condition (PCC) is known to affect a large proportion of COVID-19 survivors. Robust study design and methods are needed to understand post-COVID-19 diagnosis patterns in all survivors, not just those clinically diagnosed with PCC.
Methods
We applied a case-crossover Phenome-Wide Association Study (PheWAS) in a retrospective cohort of COVID-19 survivors, comparing the occurrences of 1,671 diagnosis-based phenotype codes (PheCodes) pre- and post-COVID-19 infection periods in the same individual using a conditional logistic regression. We studied how this pattern varied by COVID-19 severity and vaccination status, and we compared to test negative and test negative but flu positive controls.
Results
In 44,198 SARS-CoV-2-positive patients, we found enrichment in respiratory, circulatory, and mental health disorders post-COVID-19-infection. Top hits included anxiety disorder (p = 2.8e-109, OR = 1.7 [95 % CI: 1.6–1.8]), cardiac dysrhythmias (p = 4.9e-87, OR = 1.7 [95 % CI: 1.6–1.8]), and respiratory failure, insufficiency, arrest (p = 5.2e-75, OR = 2.9 [95 % CI: 2.6–3.3]). In severe patients, we found stronger associations with respiratory and circulatory disorders compared to mild/moderate patients. Fully vaccinated patients had mental health and chronic circulatory diseases rise to the top of the association list, similar to the mild/moderate cohort. Both control groups (test negative, test negative and flu positive) showed a different pattern of hits to SARS-CoV-2 positives.
Conclusions
Patients experience myriad symptoms more than 28 days after SARS-CoV-2 infection, but especially respiratory, circulatory, and mental health disorders. Our case-crossover PheWAS approach controls for within-person confounders that are time-invariant. Comparison to test negatives and test negative but flu positive patients with a similar design helped identify enrichment specific to COVID-19. This design may be applied other emerging diseases with long-lasting effects other than a SARS-CoV-2 infection. Given the potential for bias from observational data, these results should be considered exploratory. As we look into the future, we must be aware of COVID-19 survivors’ healthcare needs.
1. Introduction
Though most patients with Coronavirus Disease 2019 (COVID-19) recover, [1] many survivors report symptoms long after disease onset, a condition commonly referred to as “long COVID” or “post COVID-19 condition” (hereinafter abbreviated as PCC). [2], [3], [4] While initially the names and definitions of PCC were highly heterogeneous, the consensus clinical case definition [3] proposed by the WHO in October 2021 represented a significant step towards reaching global consistency. A recent meta-analysis estimated that 43 % (95 % CI: 39 %–46 %) of COVID-19 survivors experience at least one lingering condition post-COVID-19. [5] This, paired with estimates for global COVID-19 reported case counts, [6] the estimated prevalence of PCC among initially asymptomatic cases, [7] and the fraction of unreported COVID-19 infections, [8], [9] forms the basis that hundreds of millions of people may have or have had post-COVID-19-related health complications.
Female sex, older age, severe COVID-19, and comorbidities such as asthma are claimed to be associated with PCC. [5] Common symptoms include fatigue, brain fog/memory issues, headache, heart conditions, respiratory conditions, sleep disorders, and mental health conditions, [4] but PCC symptomatology still remains heterogeneous. Recent research has shown that COVID-19 may increase risk for cardiovascular events, kidney-related outcomes, and diabetes sometimes long after infection [10], [11], [12] and that PCC can persist for months after infection. [13], [14] Regardless of a formal diagnosis, several surveys indicated that post-COVID-19-related disabilities have affected a large proportion of the population. [15], [16], [17]
However, there are also skepticisms and contradictions in the literature. One recent study suggested that not every new or persistent symptom post-infection can be attributed to a confirmed COVID-19 diagnosis. [18] Another important question is whether vaccination or later SARS-CoV-2 variants reduces PCC development. To date, results have been inconsistent, with some studies finding vaccination to confer a protective effect, but others finding the contrary. [19], [20], [21], [22]
While a proper population-based survivorship cohort with adequate follow-up time is the ideal study design to understand post-COVID-19 clinical outcomes, electronic health records (EHRs) offer snapshots of patients’ health status and thus allow comparisons of the medical phenome of COVID-19-positive patients before and after COVID-19 diagnosis. EHRs are easily accessible and enabled many studies on post COVID-19 complications. [10], [11], [12], [14], [23], [24] Phenome-Wide Association Studies (PheWAS) are an increasingly common EHR-based method to agnostically find associations between hundreds of phenotypes and some other health-related factor. [25] Recently, PheWAS have been used to understand the genetic and phenotypic risk factors for COVID-19 outcomes. [26], [27], [28], [29] Such studies can be error-prone due to lack of a suitable control group or confounding due to differences in other patient characteristics determining who is getting tested and diagnosed for COVID-19 as well as who is seeking post-COVID-19 care. Researchers may consider matching, weighting or regression adjustment as potential remedies to this problem, but these methods are only able to adjust for a limited set of measured confounders. [30], [31]
The case-crossover design is an elegant design-based solution which reduces potential confounding by using events observed for the same person during suitably defined case and control periods. [32], [33] This design can be thought of as a matched case-control design that controls for both observed and unobserved person-level confounders that are invariant over the case and control windows. Case-crossover designs have been used to study early COVID-19 detection and post-COVID-19-vaccination cerebral venous thrombosis. [34], [35] One particular study used claims data to estimate the association between patient diagnoses and the time period after COVID-19 infection, [36] and another used EHR data to conduct a post-COVID-19 PheWAS. [37]
In October 2021 a new diagnosis code specifically for PCC was introduced, [38] thus facilitating the clear identification of PCC patients, but in this study we took an agnostic look across hundreds of diagnoses to understand which ones are more commonly seen post-COVID-19 using a case-crossover design with more than two years of follow-up data. We conduct analyses stratified by COVID-19 severity and vaccination status. We compare these results to the results of the same analysis applied to test negative controls and a test negative flu positive cohort to discern unique contributions of COVID-19. Using this approach, we aim to improve our understanding of post-COVID-19 diagnosis patterns and consequently to advance healthcare and societal support for all COVID-19 survivors.
2. Methods
2.1. COVID-19-positive cohort
Data were extracted retrospectively from EHRs for patients in the Michigan Medicine (MM) health system. Ethical review and approval were waived for this study due to its qualification for a federal exemption as secondary research for which consent is not required. Determination for exemption was made by the University of Michigan Medical School Institutional Review Board (study ID: HUM00180294). Individual-level data included de-identified information regarding reverse transcription polymerase chain reaction (RT-PCR) testing for SARS-CoV-2, patient demographics, diagnoses, vaccinations, hospitalizations, ICU admission, and death. We included all adult individuals with either 1) positive RT-PCR test result or 2) diagnosis of COVID-19 infection based on International Classification of Disease (ICD)-10-CM codes U07.1 or U07.2 between March 10, 2020, to August 1, 2022. We defined the date of the first positive test or diagnosis as the index test date for each patient. For patients with multiple positive tests, we considered their first positive test as the index test date. Patients with missing test dates were excluded from this analysis.
2.2. Test negative controls
We also measured test negative controls – patients tested, but who never received a positive RT-PCR result nor a COVID-19 diagnosis. We matched negative to positive patients at a 4:1 ratio on age, gender, and Charlson Comorbidity Index. [39] The index test date for negative patients who were tested multiple times was defined as the date of their first COVID-19 test to ensure sufficient follow-up post-test. A sub-cohort of test negative patients who were diagnosed with other forms of the flu (defined using PheCode 481; PheCode system described below) during the same period were also measured, where the date of flu diagnosis (if multiple, one was randomly chosen) served as their index date for choosing the case-control windows.
2.3. Study design
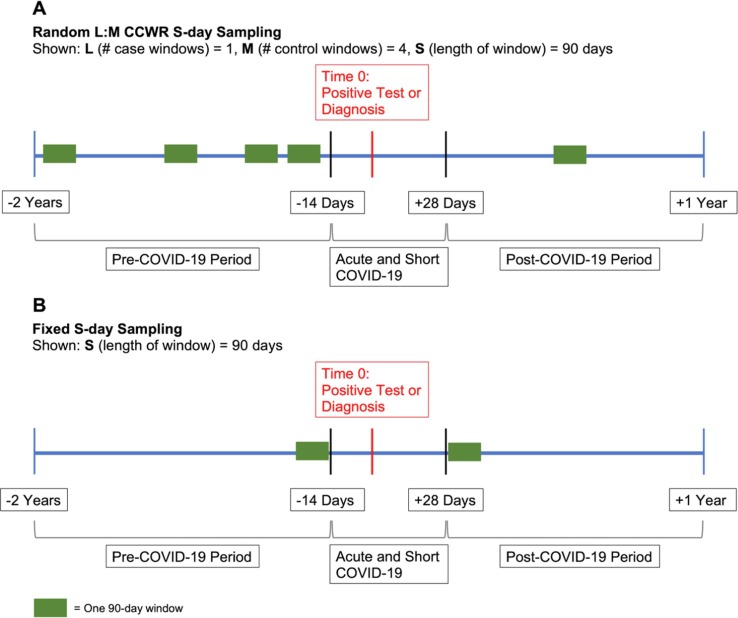
We used a case-crossover design where each COVID-19-positive case served as its own control. We defined three time periods relative to the index test date (time zero): “pre-COVID-19 period” (−2 years to −14 days), “acute and short COVID-19 period” (−14 days to + 28 days), and “post-COVID-19 period” (+ 28 days to + 1 year; Fig. 1 ). Thus, the “post-COVID-19 period” did not include the acute phase of COVID-19. We included 14 days prior to the index test date in the “acute and short COVID-19 period” to account for individuals who may have had COVID-19 and related symptoms before testing positive. Patients were included in the study if they had at least one EHR encounter with a diagnosis in both the “pre-” and “post-COVID-19 period.”
Fig. 1.
Sampling Schematic for Case-Crossover Design. Panel A depicts the random L:M CCWR (Case:Control Window Ratio) sampling design used in our primary analysis, wherein we randomly sampled L = 1 case window and M = 4 control windows (by randomly choosing a window start date), each with S = 90 days in length. A patient’s index test date is denoted by the red line. The “Acute and Short COVID-19 period” is from −14 days to + 28 days, the “post-COVID-19 period” is from + 28 days to + 1 year, and the “pre-COVID-19 period” is from −14 days to −2 years from the index test date. In this instance, one 90-day case window is randomly selected from the “post-COVID-19 period,” and four 90-day control windows are selected from the “pre-COVID-19 period.” Panel B depicts the fixed scheme where two windows of S = 90 days length are selected from each of the periods with fixed start dates. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We implemented two sampling schemes to be used in the case-crossover design-based PheWAS. Primarily, we used a random L:M case:control window ratio (CCWR) design in which we randomly sampled (without replacement) up to L case windows (“cases”) and up to M control windows (“controls”), each S days in length, from each study participant’s “post-COVID-19 period” and “pre-COVID-19 period”, respectively (termed random L:M CCWR S-day analysis; Fig. 1 A). Windows of length S days were selected by randomly choosing window start dates. We also used a fixed window design where we selected 1 case and 1 control window (of length S days) from a fixed start date, defined as the date most proximal to the index test (termed fixed S-day analysis; Fig. 1 B).
2.4. Demographic and clinical variables
Age, gender, race, and Body Mass Index (BMI) were reported from patients’ EHRs. Patients aged >= 90 years were coded as being exactly 90 years old for confidentiality reasons. A patient was considered a MM primary care patient if they received primary care at MM in the last two years. We also computed the Charlson Comorbidity Index using pre-existing conditions 14 days prior to the index date.
2.5. COVID-19 severity
COVID-19-related hospital and ICU admission were defined for COVID-19 positive patients as having each respective outcome within 30 days following the index test date. [40] COVID-19-related death was defined as death within 60 days following the index test date. These outcomes describe 30-day all-cause-hospitalization and 60-day all-cause-mortality following a COVID positive test. We define the composite outcome as “severe COVID-19” if a COVID-19 patient experienced a COVID-19-related hospitalization, ICU admission, or death as defined above. A patient is considered “mild/moderate COVID-19” otherwise. See eFigure 1 for details.
2.6. COVID-19 vaccination
The date on which a person was considered fully vaccinated was after either 1) two doses of Moderna, Pfizer-BioNTech, or Astrazeneca or 2) one dose of Johnson and Johnson - Janssen vaccine, and 21 or more days had elapsed after their last dose. [41] Patients were considered unvaccinated if they had exactly zero or an unknown number of doses at index test date. Partially vaccinated patients were not included in the stratified analysis but were included in the overall analysis. We note that MM’s vaccine eligibility criteria changed over time and mirrored the CDC’s recommendations. Thus, most patients diagnosed before 2021 were unvaccinated at their index test date. eFigure 2 details how vaccination status was determined.
2.7. Diagnosis code mapping
ICD diagnosis codes were extracted for each patient and mapped to their corresponding PheCodes according to the PheWAS catalog ICD maps. [42] Standard PheCode exclusions were applied, and one observed PheCode during a corresponding time window was considered the presence of a diagnosis. The totality of observed PheCodes for an individual was termed their “phenome.” We grouped PheCodes into symptom groups as defined in the PheWAS catalog. [43]
2.8. Descriptive analysis of diagnosis patterns
We tabulated presence of any new PheCodes (and PCC-related PheCodes as defined in eTable 1) [4] as well as the number of new PheCodes received during the “post-COVID-19 period.” A PheCode was considered new if it was present in the “post-COVID-19 period” but not present during the “pre-COVID-19 period.” Additionally, we counted visits per month and follow-up time (in weeks) during both the “pre-“ and “post-COVID-19 periods”. A visit was defined as any unique day on which at least one diagnosis was recorded, and follow-up time was computed by taking the difference between the date most proximal to the index test date for a period (−14 days for “pre-COVID-19 period”, + 28 days for “post-COVID-19 period”) and the most distal date on which they received a diagnosis in their “pre-“ and “post-COVID-19 periods” (up to −2 years for “pre-COVID-19 period”, up to + 1 year for “post-COVID-19 period”).
2.9. Statistical analysis for PheWAS
We used a PheWAS approach with a case-crossover design. To account for the within-subject matched analysis, conditional logistic regression was used to model the association between case and control windows and patients’ phenomes. Let us consider a 1:M case-crossover design with N patients analyzing K PheCodes. Let index patients, index case and control windows of a patient, and index Phecodes. Patient i’s case window () is matched to multiple randomly selected control windows (). For each PheCode , we fit the following model:
where is an indicator for whether PheCode is present in window of patient and denotes the case/control window for patient i. The conditional logistic regression conditions on the matched design or the fact that is a case window and are control windows for the same individual , such that the patient-specific intercept is eliminated and the conditional likelihood only retains , the coefficient of PheCode shared by all patients. The resulting conditional likelihood for PheCode k takes the following form:
For a model to be run, we specified that at least 10 subjects (5 for cohorts with < 5,000 subjects) in the analytic dataset should have a given PheCode in their case (control) periods. We used Manhattan plots to visualize the p-values corresponding to the null hypotheses and the directions of the association.
For each sampling scheme, a PheWAS was run on the entire COVID-19-positive cohort (termed “overall” cohort) and several subgroups – severe, mild/moderate, fully vaccinated, and unvaccinated patients. Random 1:4 CCWR 90-day sampling was used in the primary analysis. We chose 90 days as it aligns with the WHO’s PCC case definition as well as recent research. [3], [5] Sensitivity analyses regarding the length of the window and the case:control ratio included fixed 90-day, fixed 30-day, random 1:2 CCWR 180-day, random 2:4 CCWR 90-day. We also conducted a random 1:4 CCWR 90-day analysis on test negative and test negative but flu positive controls, and random 1:4 CCWR 90-day analysis stratified by year of infection (2020, 2021, 2022). For test negative controls, we performed PheWAS on controls matched to the overall cohort and to the severe cohort. We formally compared cohorts by testing for a difference in effect sizes (eMethods 1).
All analyses were performed in R (version 4.1.2), [44] and the PheWAS package was used. [45] Summary statistics are reported as median (interquartile range [IQR]) for continuous variables or n (%) for categorical variables. Odds Ratios (ORs) with Wald-type 95 % Confidence Intervals (CIs) and p-values are reported from each conditional logistic regression model. Phenome-wide significance (“hits”) was determined by the Holm-Bonferroni method. [46] We reported on both the Bonferroni and Holm-Bonferroni hits in PheWAS plots.
3. Results
3.1. Cohort description
Between March 10, 2020 and August 1, 2022, 353,648 patients were tested or diagnosed for COVID-19 at MM. Of these, 44,198 COVID-19-positive patients were included in our study, to which 160,399 test negative controls were matched. In addition, 1,328 test negative patients with an index flu infection during the same period were also included as a second set of controls (see eFigure 1 for a flow diagram defining the analytic cohort). Median (IQR) age was 48 (31–63) and 61 % of the cohort was female (Table 1 ). Of the positive patients, 2,569 (5.8 %) patients experienced severe COVID-19, and 41,629 (94.2 %) had mild/moderate COVID-19. 16,468 (37 %) patients were fully vaccinated and 25,736 (58 %) were unvaccinated at their index test date.
Table 1.
Cohort Summary. Summary statistics for the cohort are presented as median (IQR) for continuous variables and n (%) for categorical variables. The table is stratified by vaccination status at index test date. Missing values are reported for each variable.
| Variable | Overall (n = 44,198) | Fully Vaccinated (n = 16,468)a | Unvaccinated (n = 25,736)a, b |
|---|---|---|---|
| Age | 48 (31, 63) | 51 (34, 65) | 45 (29, 61) |
| Gender | |||
| Female | 26,880 (61 %) | 10,148 (62 %) | 15,544 (60 %) |
| Male | 17,316 (39 %) | 6,320 (38 %) | 10,191 (40 %) |
| (Missing) | 2 (<0.1 %) | 0 (0 %) | 1 (<0.1 %) |
| Race | |||
| African American | 4,926 (11 %) | 1,429 (8.7 %) | 3,262 (13 %) |
| Asian | 1,574 (3.6 %) | 790 (4.8 %) | 706 (2.7 %) |
| Caucasian | 34,579 (78 %) | 13,132 (80 %) | 19,927 (77 %) |
| Other | 1,919 (4.3 %) | 631 (3.8 %) | 1,206 (4.7 %) |
| (Missing) | 1,200 (2.7 %) | 486 (3.0 %) | 635 (2.5 %) |
| BMI | 28 (24, 33) | 28 (24, 33) | 28 (24, 34) |
| (Missing) | 2,624 (5.9 %) | 677 (4.1 %) | 1,835 (7.1 %) |
| Charlson Comorbidity Index | 1.00 (0.00, 3.00) | 1.00 (0.00, 4.00) | 1.00 (0.00, 3.00) |
| (Missing) | 1,114 (2.5 %) | 535 (3.2 %) | 498 (1.9 %) |
| Primary Care Patientc | 23,871 (54 %) | 9,940 (60 %) | 12,928 (50 %) |
| COVID-19 Severityd | |||
| Mild/Moderate | 41,629 (94.2 %) | 15,784 (95.8 %) | 23,989 (93.2 %) |
| Severe | 2,569 (5.8 %) | 684 (4.2 %) | 1,747 (6.8 %) |
1,994 partially vaccinated patients not represented.
Includes those with unknown vaccination status.
Received primary care at MM in last 2 years.
Severe if experienced COVID-19-related hospitalization, ICU admission or death; mild/moderate otherwise.
3.2. Descriptive diagnosis patterns
Both COVID-19-positive (Table 2 ) and COVID-19-negative patients (eTable 2) received a similar number and rate of diagnoses in the “post period”, and we saw a similar trend even when looking only at PCC-related diagnoses (eTable 1). The flu positive cohort had an increased number and rate of diagnoses in the “post period” (eTable 3). Increasing COVID-19 severity led to increased numbers and rates of diagnosis (i.e., 90 % of severe vs 79 % of mild/moderate with 1 + new diagnosis). Positives and negatives (including the flu positive cohort) both most commonly received circulatory, mental, and digestive disorders in the “post period” (eTables 4–6).
Table 2.
Summary of Diagnosis Patterns. This table includes six outcomes: follow-up time in weeks, visits per month, individuals with at least one new diagnosis in the “post-COVID-19 period,” individuals with at least one new PCC-related diagnosis in the “post-COVID-19 period,” the number of new diagnoses per month in the “post-COVID-19 period,” and the number of new PCC-related diagnoses per month in the “post-COVID-19 period.” Each outcome is stratified by both COVID-19 severity, “pre-“/”post-COVID-19 period,” and vaccination status. Statistics are presented as median (IQR) for continuous variables and n (%) for categorical variables, and sample sizes for cohorts are provided.
|
Overall (n = 44,198)a |
Fully Vaccinated (n = 16,468)a |
Unvaccinated (n = 25,736)a, b |
|||||
|---|---|---|---|---|---|---|---|
| Outcome | Cohort | Pre-COVID-19 | Post-COVID-19 | Pre-COVID-19 | Post-COVID-19 | Pre-COVID-19 | Post-COVID-19 |
| Follow-up Time (Weeks) | Overall (n = 44,198) | 90.86 (59.04, 99.71) | 25.14 (13.29, 41) | 94.29 (67.43, 100.29) | 17.71 (7.43, 25.29) | 88 (54.29, 99.14) | 34.71 (19.29, 44.86) |
| Mild/Moderate (n = 41,629) | 90.43 (58.86, 99.57) | 25 (13.29, 40.57) | 94.07 (67.29, 100.29) | 17.57 (7.43, 25.14) | 87.29 (53.86, 98.86) | 34.57 (19.29, 44.71) | |
| Severe (n = 2,569) | 96.43 (66.86, 101) | 29.71 (14.14, 45.29) | 98.07 (70.82, 101.29) | 19.14 (6.82, 27.57) | 95.71 (64.71, 100.86) | 38.14 (19.29, 46.43) | |
| Hospitalized, No ICU (n = 1,900) | 96.14 (66.68, 101) | 29.36 (15.71, 45) | 97.86 (69.04, 101.14) | 20.29 (7.71, 27.57) | 95.29 (64.71, 100.86) | 38.14 (20.29, 46.29) | |
| Hospitalized and ICU (n = 588) | 97.71 (70.32, 101.14) | 35.71 (15.11, 46.43) | 98.71 (81.79, 101.29) | 18.86 (6.43, 30.21) | 97.43 (68.29, 101.14) | 41.57 (21.43, 47) | |
| Deceased (n = 136) | 96.64 (64.64, 101.43) | 2.29 (0.86, 4.46) | 99.64 (69.96, 101.82) | 1.79 (0.75, 3.11) | 96.43 (60.89, 101.07) | 2.43 (0.86, 5.57) | |
| Visits Per Month | Overall (n = 44,198) | 0.64 (0.25, 1.44) | 0.54 (0.18, 1.26) | 0.93 (0.42, 1.91) | 0.45 (0.18, 0.99) | 0.51 (0.21, 1.15) | 0.54 (0.18, 1.35) |
| Mild/Moderate (n = 41,629) | 0.59 (0.25, 1.36) | 0.45 (0.18, 1.17) | 0.89 (0.38, 1.83) | 0.45 (0.18, 0.99) | 0.47 (0.17, 1.06) | 0.54 (0.18, 1.26) | |
| Severe (n = 2,569) | 1.44 (0.59, 3.06) | 1.44 (0.54, 3.25) | 2.17 (0.98, 3.95) | 1.35 (0.45, 2.64) | 1.27 (0.51, 2.55) | 1.53 (0.54, 3.43) | |
| Hospitalized, No ICU (n = 1,900) | 1.44 (0.59, 2.93) | 1.26 (0.45, 2.8) | 2.08 (0.98, 3.66) | 1.13 (0.45, 2.37) | 1.23 (0.51, 2.42) | 1.35 (0.45, 2.89) | |
| Hospitalized and ICU (n = 588) | 1.61 (0.64, 3.65) | 2.75 (0.99, 5.33) | 2.85 (0.91, 5.10) | 2.53 (0.9, 4.69) | 1.4 (0.55, 3.23) | 2.89 (0.99, 5.6) | |
| Deceased (n = 136) | 1.36 (0.51, 3.53) | 1.9 (0.95, 4.99) | 2.61 (1.2, 3.95) | 2.85 (0.95, 12.83) | 1.13 (0.47, 3.45) | 0.95 (0.95, 4.51) | |
| 1 + New Diagnosisc | Overall (n = 44,198) | 34,257 (79 %) | 11,917 (75 %) | 20,809 (82 %) | |||
| Mild/Moderate (n = 41,629) | 31,950 (79 %) | 11,324 (74 %) | 19,222 (82 %) | ||||
| Severe (n = 2,569) | 2,307 (90 %) | 593 (87 %) | 1,587 (91 %) | ||||
| Hospitalized, No ICU (n = 1,900) | 1,682 (89 %) | 454 (86 %) | 1,127 (90 %) | ||||
| Hospitalized and ICU (n = 588) | 567 (97 %) | 126 (97 %) | 421 (97 %) | ||||
| Deceased (n = 136) | 101 (76 %) | 24 (73 %) | 70 (76 %) | ||||
| 1 + New PCC-Related Diagnosisc | Overall (n = 44,198) | 16,205 (59 %) | 5,469 (51 %) | 9,930 (64 %) | |||
| Mild/Moderate (n = 41,629) | 14,784 (58 %) | 5,128 (51 %) | 8,930 (63 %) | ||||
| Severe (n = 2,569) | 1,421 (71 %) | 341 (63 %) | 1,000 (74 %) | ||||
| Hospitalized, No ICU (n = 1,900) | 1,005 (68 %) | 251 (59 %) | 692 (71 %) | ||||
| Hospitalized and ICU (n = 588) | 403 (80 %) | 86 (77 %) | 303 (81 %) | ||||
| Deceased (n = 136) | 29 (49 %) | 9 (43 %) | 16 (48 %) | ||||
| New Diagnoses Per Month | Overall (n = 44,198) | 0.36 (0.09, 0.90) | 0.27 (0, 0.72) | 0.36 (0.09, 0.90) | |||
| Mild/Moderate (n = 41,629) | 0.27 (0.09, 0.81) | 0.27 (0, 0.72) | 0.36 (0.09, 0.90) | ||||
| Severe (n = 2,569) | 0.99 (0.27, 2.26) | 0.9 (0.27, 2.08) | 1.08 (0.36, 2.44) | ||||
| Hospitalized, No ICU (n = 1,900) | 0.81 (0.27, 1.90) | 0.63 (0.18, 1.62) | 0.9 (0.27, 1.90) | ||||
| Hospitalized and ICU (n = 588) | 2.17 (0.90, 4.06) | 2.17 (0.90, 3.81) | 2.17 (0.95, 4.15) | ||||
| Deceased (n = 136) | 1.9 (0.95, 13.31) | 6.65 (0, 19.01) | 0.95 (0.95, 10.69) | ||||
| New PCC-Related Diagnoses Per Month | Overall (n = 44,198) | 0.09 (0, 0.18) | 0.09 (0, 0.18) | 0.09 (0, 0.18) | |||
| Mild/Moderate (n = 41,629) | 0.09 (0, 0.18) | 0.09 (0, 0.18) | 0.09 (0, 0.18) | ||||
| Severe (n = 2,569) | 0.09 (0, 0.27) | 0.09 (0, 0.18) | 0.09 (0, 0.27) | ||||
| Hospitalized, No ICU (n = 1,900) | 0.09 (0, 0.27) | 0.09 (0, 0.18) | 0.09 (0, 0.27) | ||||
| Hospitalized and ICU (n = 588) | 0.18 (0.09, 0.36) | 0.18 (0.09, 0.36) | 0.18 (0.09, 0.36) | ||||
| Deceased (n = 136) | 0 (0, 1.90) | 0 (0, 1.90) | 0 (0, 0.95) | ||||
Median (IQR) or Frequency (%).
Includes those with unknown vaccination status.
In the ∼ 11 month-long “post-COVID-19 period”.
3.3. Overall Case-Crossover PheWAS analysis
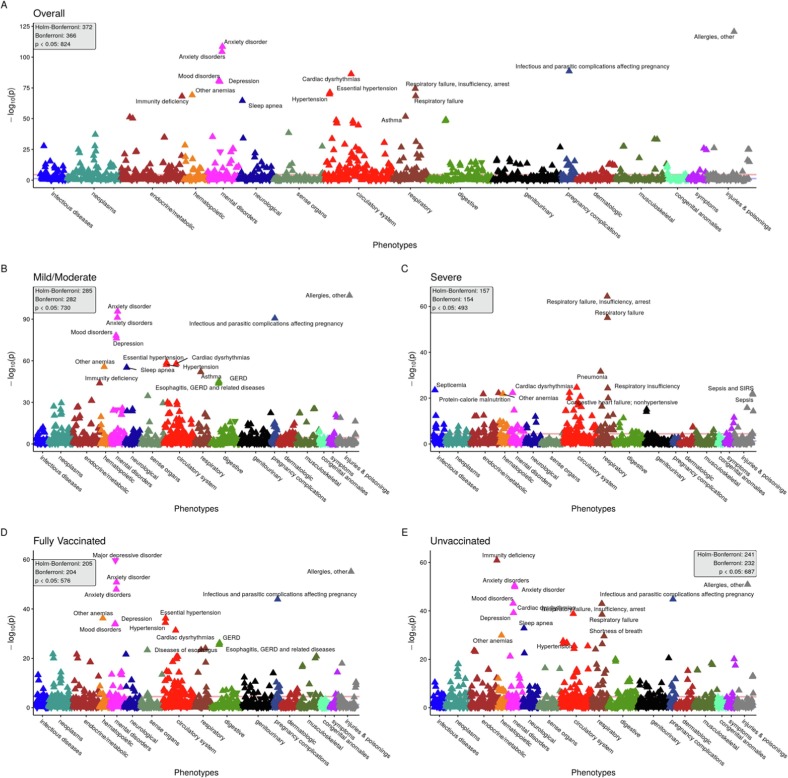
1,671 PheCodes were evaluated in the primary analysis for the overall cohort (Fig. 2 A), and a total of 372 PheCodes reached phenome-wide significance according to Holm-Bonferroni multiple testing rule. We saw the highest proportion of phenome-wide significant hits in circulatory (73 hits/total of 171 circulatory codes; 43 %), mental disorders (24/76; 32 %), and respiratory (27/85; 32 %; Table 3 ). The top hits in each of these groups were anxiety disorder (p = 2.8e-109, OR = 1.7 [95 % CI: 1.6–1.8]), cardiac dysrhythmias (p = 4.9e-87, OR = 1.7 [95 % CI: 1.6–1.8]), and respiratory failure, insufficiency, arrest (p = 5.2e-75, OR = 2.9 [95 % CI: 2.6–3.3]).
Fig. 2.
Random 1:4 CCWR 90-day analysis Manhattan plots. Panel of PheWAS Manhattan plots showing overall (panel A) and stratified by COVID-19 severity (panels B and C) and vaccination status (panels D and E). PheCodes (grouped by category) are on the x-axis and the -log10(p-value) is on the y-axis. The Bonferroni-adjusted p-value threshold line (in red) is shown, and the nominal p-value threshold (0.05) is also shown in blue. For each panel, the number of hits at the Bonferroni, Holm-Bonferroni and nominal p-value threshold are provided. Some of the top hits for each plot are annotated. For each hit, an upward pointing triangle represents a positive association (OR greater than 1), and a downward facing triangle represents a negative association (OR < 1). Note: The following two PheCodes were removed from plots for better visualization due to their extreme p-values: “Other infectious and parasitic diseases” (p = 1.2e-119 in overall cohort) and “Other headache syndromes” (p = 1.9e-139 in overall cohort). The former is a PheCode connected to COVID-19 infection and sequelae, [47] so its low p-value is unsurprising. The extreme association seen for “Other headache syndromes” is somewhat more surprising because it had a negative association with the “post-COVID-19 period”, perhaps relating to patients being less willing to visit the doctor for a “mild” symptom like headache during a pandemic. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 3.
PheWAS Hits by Symptom Group. The first and second columns give PheCode symptom groups as defined by the PheWAS catalog and the total number of PheCodes in each group. The other columns give the number of phenome-wide significant hits and the proportion of hits to the total number of PheCodes in each symptom group for each cohort in the primary analysis including the two control cohorts.
|
Phenome-Wide Significant Hitsa, b |
||||||||
|---|---|---|---|---|---|---|---|---|
| Symptom Group | Total PheCodes in Groupc | Overall (n = 44,198) | Mild/Moderate (n = 41,629) | Severe (n = 2,569) | Fully Vaccinated (n = 16,468) | Unvaccinated (n = 25,736) | Negative (n = 160,399) | Flu (n = 1,328) |
| circulatory system | 171 | 73 (43 %) | 58 (34 %) | 36 (21 %) | 51 (30 %) | 45 (26 %) | 121 (71 %) | 1 (1 %) |
| congenital anomalies | 56 | 5 (9 %) | 3 (5 %) | – | – | – | 7 (12 %) | – |
| dermatologic | 95 | 10 (11 %) | 15 (16 %) | 2 (2 %) | 6 (6 %) | 7 (7 %) | 35 (37 %) | – |
| digestive | 162 | 26 (16 %) | 20 (12 %) | 9 (6 %) | 12 (7 %) | 21 (13 %) | 88 (54 %) | – |
| endocrine/metabolic | 169 | 43 (25 %) | 28 (17 %) | 26 (15 %) | 17 (10 %) | 31 (18 %) | 97 (57 %) | – |
| genitourinary | 173 | 27 (16 %) | 21 (12 %) | 3 (2 %) | 15 (9 %) | 16 (9 %) | 71 (41 %) | – |
| hematopoietic | 62 | 13 (21 %) | 7 (11 %) | 9 (15 %) | 5 (8 %) | 10 (16 %) | 31 (50 %) | – |
| infectious diseases | 69 | 16 (23 %) | 8 (12 %) | 8 (12 %) | 10 (14 %) | 7 (10 %) | 33 (48 %) | – |
| injuries & poisonings | 122 | 13 (11 %) | 6 (5 %) | 7 (6 %) | 7 (6 %) | 6 (5 %) | 45 (37 %) | – |
| mental disorders | 76 | 24 (32 %) | 22 (29 %) | 15 (20 %) | 17 (22 %) | 18 (24 %) | 52 (68 %) | – |
| musculoskeletal | 132 | 11 (8 %) | 12 (9 %) | 4 (3 %) | 9 (7 %) | 9 (7 %) | 53 (40 %) | – |
| neoplasms | 141 | 39 (28 %) | 32 (23 %) | 7 (5 %) | 22 (16 %) | 23 (16 %) | 72 (51 %) | – |
| neurological | 85 | 18 (21 %) | 14 (16 %) | 5 (6 %) | 9 (11 %) | 14 (16 %) | 46 (54 %) | – |
| pregnancy complications | 46 | 13 (28 %) | 12 (26 %) | – | 9 (20 %) | 9 (20 %) | 19 (41 %) | – |
| respiratory | 85 | 27 (32 %) | 13 (15 %) | 21 (25 %) | 8 (9 %) | 18 (21 %) | 52 (61 %) | – |
| sense organs | 127 | 8 (6 %) | 9 (7 %) | 1 (1 %) | 5 (4 %) | 4 (3 %) | 24 (19 %) | – |
| symptoms | 46 | 6 (13 %) | 5 (11 %) | 4 (9 %) | 3 (7 %) | 3 (7 %) | 26 (57 %) | – |
n (% of total PheCodes in group).
According to the Holm-Bonferroni method.
Not every available PheCode was evaluated in each PheWAS due to case/control thresholds.
3.4. Stratified analyses
3.4.1. By COVID-19 severity status
Top groups for the mild/moderate cohort (Fig. 2 B) were circulatory system (58/171; 34 %), mental disorders (22/76; 29 %), and pregnancy complications (12/46; 26 %, Table 3). Essential hypertension (p = 2.6e-59, OR = 1.5 [95 % CI: 1.4–1.5]), anxiety disorder (p = 2.7e-96, OR = 1.6 [95 % CI: 1.6–1.7]), and infectious and parasitic complications affecting pregnancy (p = 2.4e-91, OR = 9.8 [95 % CI: 7.8–12.2]) were top hits in these groups. For the severe cohort, we saw a different pattern of hits (Fig. 2 C), with respiratory conditions being a top category (21/85; 25 %). Other top groups include circulatory system (36/171; 21 %) and mental disorders (15/76; 20 %), and the top hit from these groups were respiratory failure, insufficiency, arrest (p = 4.2e-65, OR = 6.3 [95 % CI: 5.1–7.7]), cardiac dysrhythmias (p = 2.4e-25, OR = 2.3 [95 % CI: 1.9–2.6]), and neurological disorders (p = 4.6e-23, OR = 2.8 [95 % CI: 2.3–3.4]).
3.4.2. By vaccination status
Among those fully vaccinated at index test date (Fig. 2 D), we saw circulatory system (51/171; 30 %), mental disorders (17/76; 22 %), and pregnancy complications (9/46; 20 %, Table 3). Essential hypertension (p = 6.3e-37, OR = 1.6 [95 % CI: 1.5–1.7]), major depressive disorder (p = 2.3e-60, OR = 0.4 [95 % CI: 0.3–0.4]), and infectious and parasitic complications affecting pregnancy (p = 1.3e-44, OR = 12.8 [95 % CI: 8.9–18.2]) were top hits in these groups. The unvaccinated cohort (Fig. 2 E) was largely similar to the overall cohort with circulatory (45/171; 26 %), mental disorders (18/76; 24 %), and respiratory (18/85; 21 %) being the top groups. Top hits in these groups were cardiac dysrhythmias (p = 1.5e-39, OR = 1.6 [95 % CI: 1.5–1.7]), anxiety disorders (p = 3.2e-51, OR = 1.6 [95 % CI: 1.5–1.7]), and respiratory failure, insufficiency, arrest (p = 1.1e-43, OR = 2.9 [95 % CI: 2.5–3.3]).
3.4.3. Summary of comparison between severity and vaccination subgroups
A large proportion of circulatory hits was common across all cohorts. The most striking observation is the strength of association for respiratory conditions in the severe cohort. Comparing the top 20 hits from each subgroup revealed septicemia and protein-calorie malnutrition were unique to the severe cohort in addition to several severe respiratory disorders; shortness of breath was unique to those unvaccinated (eFigure 3). Bearing in mind that p-value magnitudes are directly influenced by sample sizes (which are dissimilar across cohorts), we note that the p-value ranks/patterns of the mild/moderate, fully vaccinated, and unvaccinated subgroups appeared similar to the overall cohort, but the unvaccinated group was largely driving the strongest associations, and the top enriched categories in the unvaccinated were identical to the overall cohort as well.
3.5. Comparison with test negative controls
Circulatory (121/171; 71 %), mental disorders (52/76; 68 %), and respiratory (52/85; 61 %) were the top groups in the PheWAS analysis for the test negative cohort (Table 3 , eFigure 4). Top hits in these groups were cardiac dysrhythmias (p = 3.3e-254, OR = 1.7 [95 % CI: 1.6–1.7]), anxiety disorders (p = 9.8e-221, OR = 1.5 [95 % CI: 1.4–1.5]), and respiratory failure, insufficiency, arrest (p = 2.5e-129, OR = 2.4 [95 % CI: 2.3–2.6]).
The top symptom groups in negatives were similar to that seen in the overall and unvaccinated cohort. Viral pneumonia, disturbances of the sensation of smell and taste, and chronic fatigue syndrome were hits in the positive but not negative cohort (eFigure 5).
3.6. Comparison with test negative flu positive controls
Ischemic heart disease (p = 1.6e-5, OR = 2.5 [95 % CI: 1.7–3.9]), a circulatory disease (Table 3), was the sole phenome-wide significant hit in the flu positive cohort (eFigure 6).
Depression and sleep apnea were in the top 20 phenotypes for the COVID-19-positive but not the flu positive cohort, while ischemic heart disease, calculus of the kidney and gout were seen in the flu positive cohort (eFigure 7).
Details regarding odds ratios and p-values for the test negative PheWASs as well as other PheWASs from the primary analysis are in eTable 7.
3.7. Sensitivity analyses
We also conducted several sensitivity analyses to evaluate the effect our design and analytic choices made on the primary analysis. Increasing the number of cases and controls used resulted in higher power (more phenome-wide significant hits; eFigures 8–9). Using the fixed sampling scheme resulted in lower power and a different pattern of hits, although respiratory and circulatory conditions still gave a strong signal (eFigure 10–11). Those diagnosed in 2021 and beyond closely resembled the fully vaccinated cohort, as severe respiratory illnesses waned, and common chronic diseases became more pronounced over time (eFigure 12).
3.8. Formal comparison of effect sizes
3.8.1. By severity and vaccination status
The severe cohort had larger effect sizes than the mild/moderate cohort for the vast majority of PheCodes (eFigure 13A). Groups that tended to exhibit very large differences include respiratory (OR:6.2 vs 2.0 for respiratory failure, insufficiency, arrest; p = 9.6e-19) and circulatory system (OR:7.4 vs 2.3 for acute pulmonary heart disease; p = 2.2e-7). When looking at vaccination status (eFigure 13B), those unvaccinated were more likely to be diagnosed with shortness of breath (OR:1.7 vs 1.2; p = 2.4e-6) and immunity deficiency (OR:3.7 vs 1.7; p = 4.0e-14) in the “post-COVID-19 period.”.
eFigures 13C, 13D, and 14 give the results of an effect size comparison between COVID-19 positives and negatives, COVID-19 positives and the test negative flu positive cohort, and the COVID-19-positive severe cohort and test negatives matched to the severe cohort, respectively. Briefly, respiratory and mental disorders generally have larger effect sizes in the COVID-19-positive cohort, and endocrine/metabolic and circulatory disorders have similar effect sizes between COVID-19 positives and negatives. eTable 8 gives full details of the effect size comparisons.
4. Discussion
4.1. Strengths and principal findings
In this study, we present a case-crossover PheWAS approach to characterize changes in diagnosis patterns after a COVID-19 infection. Our results show that the “post-COVID-19 period,” defined as + 28 days to + 1 year from a positive COVID-19 test or diagnosis, is associated with a wide variety of diagnoses across many organ systems. Despite our analysis being an agnostic screen, results are remarkably congruent with existing PCC literature in that we found respiratory, circulatory, and mental health disorders to be highly enriched post-COVID-19-infection in COVID-19 positives, but also in negatives. Patients with severe COVID-19 were more likely to receive a wide variety of diagnoses, but particularly respiratory and circulatory diagnoses, in the “post-COVID-19 period,” compared to those with mild/moderate COVID-19. Fully vaccinated patients were more likely than those unvaccinated to be diagnosed with chronic conditions like hypertension in the “post-COVID-19 period.” This MM cohort has been extensively studied in the past, [26], [40], [48], [49], [50] but the current study provides the longest follow-up time (over 2 years) to date and includes a “post-COVID-19 period.”
Our approach offers an advantage over traditional case-control PheWAS methods in that it controls for time-invariant confounding. Our results generally concur with those reported in a similar post-COVID-19 PheWAS without a case-crossover design, [37] but mental health conditions appear more prominently in our results. Future research may use and refine this approach to continue studying post-COVID-19 manifestations, but this pre/post design could be applied to any event, not just a SARS-CoV-2 infection. This method could prove useful in elucidating long-lasting sequelae for future emerging infectious diseases, especially in the early stages where such consequences are poorly understood, and data warehouses are being used to tease out post-infection patterns in an agnostic way. A case-crossover design may also be applied to other EHR-enabled association studies such as LabWAS and DrugWAS.
4.2. Contextualization of results
Healthcare utilization metrics (Table 2 , eTable 2–3) were very similar between COVID-19 positives, negatives, and the test negative flu positive cohort. However, SARS-CoV-2 positives were receiving different categories of diagnoses than both the control cohorts. We observed that post-flu manifestations were distinct from post-COVID-19 manifestations during the same time period, but this comparison was severely limited by sample size. We observed much stronger effect sizes for many respiratory and mental diagnoses in COVID-19 positives compared to negatives. Further, as results for the overall cohort are the composition of distinct association patterns of the subgroups therein, we note that strong respiratory signals we observed appear to have been driven by those with severe COVID-19. Severe patients also had stronger effect sizes for respiratory conditions than their matched controls. The common hits between COVID-19 positives and negatives, including many endocrine/metabolic and circulatory hits, may be a result of our design’s inability to control for time-varying factors, such as pandemic-driven changes in health-related behavior and the effects of aging. These findings highlight the need for strict diagnostic criteria for PCC such that coincidental diagnoses are not attributed to the COVID-19 infection. However, the current lack of understanding about the causal mechanisms of PCC hampers such a clear differentiation.
We found fully vaccinated patients with breakthrough infections had similar association patterns to the mild/moderate cohort, likely due to significant overlap between these groups. Many phenotypes with large effect sizes for fully vaccinated individuals (hypertension, anxiety disorder) were chronic disorders common across all included patients (eTables 4–6) and may be more related to willingness to see a physician and healthcare access over time rather than COVID-19 disease. It is worth noting that the COVID-19 virus itself was also different over time. During 2020, the Alpha variant was dominant, while in 2021 and 2022 (when vaccines were widely available in the US) the Delta and Omicron variants were dominant. Temporal variation in symptomatology may be because different variants attack different parts of the body. [51]
It is interesting to note that allergies were strongly associated with the “post-COVID-19 period” in all cohorts including COVID-19-negative patients. Some new evidence suggests PCC responds to treatment with antihistamines. [52] Our finding that mental health disorders were highly enriched in the “post-COVID-19 period” in positives and negatives is consistent with the notion that the COVID-19 pandemic introduced new mental health challenges, partly due to social changes and partly due to how COVID-19 affects the brain. [53], [54] The negative cohort showed a pronounced effect for cancer-related diagnoses, perhaps pointing to the reality that cancer treatment was delayed for many, especially high-risk patients, during the pandemic. [55] Some research proposes a link between influenza infection and ischemic heart disease, the top hit in the influenza cohort. [56]
4.3. Limitations
This study is limited by the implicit assumption in a case-crossover design that there exists no within-person time-varying confounders. However, many aspects of human behavior changed during the COVID-19 pandemic. For example, health-seeking behavior decreased after the pandemic started due to fear of the virus, government restrictions, and lack of healthcare resources. [57] The presence of this specific type of time-varying confounding, especially for those diagnosed early in the pandemic, could bias our results against seeing an effect because this confounding would result in a relative reduction in diagnoses during the “post-COVID-19 period”. This effect may be less pronounced for those diagnosed in the later stages of the pandemic. Our analysis stratified by year also gives us confidence that this method is picking up a true signal. The fact that our fixed 30-day results are similar to the fixed 90-day results might suggest time trends play a relatively small role in this analysis. Some alternative solutions could be to add time-varying covariates to the models (i.e. prevalence of cases during the period), confidence interval calibration, [58] and a case-time-control design which can account for time-varying confounding. [59]
We focused on individuals tested for COVID-19, but there exists a well-documented testing bias which could make our cohort non-representative, especially considering that testing at the beginning of the pandemic was restricted to symptomatic or at-risk individuals. [60] Additionally, some cases in our cohort presented for COVID-19 symptoms (“for COVID-19”), but others presented for something else and just happened to have COVID-19 (“with incidental COVID-19”), which may help explain the strong effect sizes we observed for pregnancy complications and congenital anomalies. We treated unknown vaccination status as being unvaccinated, but some patients may have received vaccination outside of the MM system from which the vaccination data came. [40] By requiring included patients to have encounters both pre- and post-COVID-19, we may have selected MM primary care patients or patients with more complex health history than the general population of those tested for COVID-19, hampering generalizability. We hoped to alleviate some of this concern by matching positives and negatives on Charlson Comorbidity Index. Test negative controls are a useful, but imperfect method of control given the potential baseline differences between COVID-19 positives and negatives. The flu positive cohort represents a more suitable control group, but we were unfortunately underpowered to detect associations using this group. EHRs are also prone to selection and classification bias. [61]
Our analysis involved choosing the values for several design parameters including the CCWR, the minimum case/control count, and the window size. It is difficult to know whether the parameters we chose were “correct,” but sensitivity analyses show our matching scheme is robust to the CCWR and window size. We chose to censor diagnosis records at −2 and + 1 years from the index test date, but it is possible that even if an individual has a healthcare visit during the follow-up, the diagnosis codes received during the visit do not comprehensively reflect their health state. We chose not to censor the small number of patients with multiple COVID-19 infections, which potentially added noise to our results. Further, diagnosis codes may be poor reflections of the course of disease. Finally, some spurious associations potentially appeared in our results due to biases we discussed, despite applying the Holm-Bonferroni correction.
For the above reasons, this analysis should be considered exploratory, and no causal conclusions can be deduced. We propose that future investigations can further explore the validity and applicability of this approach and replicate our findings under a similar design in other analytical cohorts.
5. Conclusions
We present a case-crossover PheWAS framework as a plausible agnostic screen that can be used to identify phenotypes associated with the “post-COVID-19 period” while controlling for time-invariant confounders. We discussed several potential sources of bias in our analyses. Consequently, the results should be considered exploratory. Future investigations may to refine and improve this approach to address such biases and replicate our findings. Epidemiologic studies that translate data into actionable clinical knowledge are crucial to advancing the field of biomedical informatics. Future research should investigate the mechanisms by which COVID-19 sequelae can occur and the myriad factors that might put a patient at risk of new post-COVID-19 symptoms.
Funding
The research presented here was funded by the National Science Foundation (https://www.nsf.gov/) under grant DMS 1712933 (BM), the National Institutes of Health (https://www.nih.gov) under grant 5R01HG008773-05 (BM) and 5P30CA046592-30 (BM), and the Michigan Collaborative Addiction Resources & Education System (https://micaresed.org) under grant 1UG3CA267907-01 (BM). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
CRediT authorship contribution statement
Spencer R. Haupert: Methodology, Writing – original draft, Writing – review & editing. Xu Shi: Conceptualization, Methodology, Supervision, Writing – original draft, Writing – review & editing. Chen Chen: Writing – original draft, Writing – review & editing. Lars G. Fritsche: Conceptualization, Methodology, Supervision, Writing – original draft, Writing – review & editing. Bhramar Mukherjee: Conceptualization, Methodology, Supervision, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Icons used in the graphical abstract were designed by Freepik, GOWI, and mynamepong downloaded from flaticon.com.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jbi.2022.104237.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Pei S., Yamana T.K., Kandula S., Galanti M., Shaman J. Burden and characteristics of COVID-19 in the United States during 2020. Nature. 2021;598(7880):338–341. doi: 10.1038/s41586-021-03914-4. [DOI] [PubMed] [Google Scholar]
- 2.Nalbandian A., Sehgal K., Gupta A., Madhavan M.V., McGroder C., Stevens J.S., Cook J.R., Nordvig A.S., Shalev D., Sehrawat T.S., Ahluwalia N., Bikdeli B., Dietz D., Der-Nigoghossian C., Liyanage-Don N., Rosner G.F., Bernstein E.J., Mohan S., Beckley A.A., Seres D.S., Choueiri T.K., Uriel N., Ausiello J.C., Accili D., Freedberg D.E., Baldwin M., Schwartz A., Brodie D., Garcia C.K., Elkind M.S.V., Connors J.M., Bilezikian J.P., Landry D.W., Wan E.Y. Post-acute COVID-19 syndrome. Nat. Med. 2021;27(4):601–615. doi: 10.1038/s41591-021-01283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coronavirus disease (COVID-19): Post COVID-19 condition. World Health Organization. Accessed March 14, 2022. http://www.who.int/news-room/questions-and-answers/item/coronavirus-disease-(covid-19)-post-covid-19-condition.
- 4.CDC. COVID-19 and Your Health. Centers for Disease Control and Prevention. Published February 11, 2020. Accessed March 16, 2022. https://www.cdc.gov/coronavirus/2019-ncov/long-term-effects/index.html.
- 5.Chen C, Haupert SR, Zimmermann L, Shi X, Fritsche LG, Mukherjee B. Global Prevalence of Post-Coronavirus Disease 2019 (COVID-19) Condition or Long COVID: A Meta-Analysis and Systematic Review. J Infect Dis. Published online April 16, 2022:jiac136. doi:10.1093/infdis/jiac136. [DOI] [PMC free article] [PubMed]
- 6.WHO Coronavirus (COVID-19) Dashboard. Accessed February 21, 2022. https://covid19.who.int.
- 7.Huang Y, Pinto MD, Borelli JL, et al. COVID Symptoms, Symptom Clusters, and Predictors for Becoming a Long-Hauler: Looking for Clarity in the Haze of the Pandemic. Published online March 5, 2021:2021.03.03.21252086. doi:10.1101/2021.03.03.21252086. [DOI] [PMC free article] [PubMed]
- 8.Estimated COVID-19 Burden. Centers for Disease Control and Prevention. Accessed February 21, 2022. https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/burden.html.
- 9.Aronna MS, Guglielmi R, Moschen LM. Estimate of the rate of unreported COVID-19 cases during the first outbreak in Rio de Janeiro. Published online October 9, 2021:2021.10.08.21264741. doi:10.1101/2021.10.08.21264741. [DOI] [PMC free article] [PubMed]
- 10.Xie Y., Xu E., Bowe B., Al-Aly Z. Long-term cardiovascular outcomes of COVID-19. Nat. Med. 2022;28(3):583–590. doi: 10.1038/s41591-022-01689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowe B., Xie Y., Xu E., Al-Aly Z. Kidney Outcomes in Long COVID. J. Am. Soc. Nephrol. 2021;32(11):2851–2862. doi: 10.1681/ASN.2021060734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie Y., Al-Aly Z. Risks and burdens of incident diabetes in long COVID: a cohort study. Lancet Diabetes Endocrinol. 2022;10(5):311–321. doi: 10.1016/S2213-8587(22)00044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans RA, Leavy OC, Richardson M, et al. Clinical characteristics with inflammation profiling of Long-COVID and association with one-year recovery following hospitalisation in the UK: a prospective observational study. Published online December 20, 2021:2021.12.13.21267471. doi:10.1101/2021.12.13.21267471. [DOI] [PMC free article] [PubMed]
- 14.Xie Y., Bowe B., Al-Aly Z. Burdens of post-acute sequelae of COVID-19 by severity of acute infection, demographics and health status. Nat. Commun. 2021;12(1):6571. doi: 10.1038/s41467-021-26513-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.After COVID-19 illness, Michiganders experienced increased disabilities. University of Michigan News. Published February 10, 2022. Accessed March 14, 2022. https://news.umich.edu/after-covid-19-illness-michiganders-experienced-increased-disabilities/.
- 16.Bach K. Is ‘long Covid’ worsening the labor shortage? Brookings. Published January 11, 2022. Accessed March 14, 2022. https://www.brookings.edu/research/is-long-covid-worsening-the-labor-shortage/.
- 17.Davis H.E., Assaf G.S., McCorkell L., Wei H., Low R.J., Re’em Y., Redfield S., Austin J.P., Akrami A. Characterizing long COVID in an international cohort: 7 months of symptoms and their impact. eClinicalMedicine. 2021;38:101019. doi: 10.1016/j.eclinm.2021.101019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matta J., Wiernik E., Robineau O., Carrat F., Touvier M., Severi G., de Lamballerie X., Blanché H., Deleuze J.-F., Gouraud C., Hoertel N., Ranque B., Goldberg M., Zins M., Lemogne C., Kab S., Renuy A., Le-Got S., Ribet C., Wiernik E., Goldberg M., Zins M., Artaud F., Gerbouin-Rérolle P., Enguix M., Laplanche C., Gomes-Rima R., Hoang L., Correia E., Barry A.A., Senina N., Severi G., Allegre J., Szabo de Edelenyi F., Druesne-Pecollo N., Esseddik Y., Hercberg S., Touvier M., Charles M.-A., Ancel P.-Y., Benhammou V., Ritmi A., Marchand L., Zaros C., Lordmi E., Candea A., de Visme S., Simeon T., Thierry X., Geay B., Dufourg M.-N., Milcent K., Rahib D., Lydie N., Lusivika-Nzinga C., Pannetier G., Lapidus N., Goderel I., Dorival C., Nicol J., Carrat F., Lai C., Belhadji L., Esperou H., Couffin-Cadiergues S., Gagliolo J.-M., Blanché H., Sébaoun J.-M., Beaudoin J.-C., Gressin L., Morel V., Ouili O., Deleuze J.-F., Ninove L., Priet S., Saba Villarroel P.M., Fourié T., Mohamed Ali S., Amroun A., Seston M., Ayhan N., Pastorino B., de Lamballerie X. Association of Self-reported COVID-19 Infection and SARS-CoV-2 Serology Test Results With Persistent Physical Symptoms Among French Adults During the COVID-19 Pandemic. JAMA Intern Med. 2022;182(1):19–25. doi: 10.1001/jamainternmed.2021.6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taquet M, Dercon Q, Harrison PJ. Six-month sequelae of post-vaccination SARS-CoV-2 infection: a retrospective cohort study of 10,024 breakthrough infections. Published online November 8, 2021:2021.10.26.21265508. doi:10.1101/2021.10.26.21265508. [DOI] [PMC free article] [PubMed]
- 20.Antonelli M., Penfold R.S., Merino J., Sudre C.H., Molteni E., Berry S., Canas L.S., Graham M.S., Klaser K., Modat M., Murray B., Kerfoot E., Chen L., Deng J., Österdahl M.F., Cheetham N.J., Drew D.A., Nguyen L.H., Pujol J.C., Hu C., Selvachandran S., Polidori L., May A., Wolf J., Chan A.T., Hammers A., Duncan E.L., Spector T.D., Ourselin S., Steves C.J. Risk factors and disease profile of post-vaccination SARS-CoV-2 infection in UK users of the COVID Symptom Study app: a prospective, community-based, nested, case-control study. Lancet Infect. Dis. 2022;22(1):43–55. doi: 10.1016/S1473-3099(21)00460-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.P. Kuodi, Y. Gorelik, H. Zayyad, et al., Association between vaccination status and reported incidence of post-acute COVID-19 symptoms in Israel: a cross-sectional study of patients tested between March 2020 and November 2021. Published online January 17, 2022:2022.01.05.22268800. 10.1101/2022.01.05.22268800. [DOI]
- 22.The Effectiveness of Vaccination Against Long COVID. UK Health Security Agency. https://ukhsa.koha-ptfs.co.uk/cgi-bin/koha/opac-retrieve-file.pl?id=fe4f10cd3cd509fe045ad4f72ae0dfff.
- 23.Taquet M., Dercon Q., Luciano S., Geddes J.R., Husain M., Harrison P.J., Kretzschmar M.E.E. Incidence, co-occurrence, and evolution of long-COVID features: A 6-month retrospective cohort study of 273,618 survivors of COVID-19. PLoS Med. 2021;18(9):e1003773. doi: 10.1371/journal.pmed.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jovanoski N., Chen X., Becker U., Zalocusky K., Chawla D., Tsai L., Borm M., Neighbors M., Yau V. Severity of COVID-19 and adverse long-term outcomes: a retrospective cohort study based on a US electronic health record database. BMJ Open. 2021;11(12):e056284. doi: 10.1136/bmjopen-2021-056284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Denny J.C., Ritchie M.D., Basford M.A., Pulley J.M., Bastarache L., Brown-Gentry K., Wang D., Masys D.R., Roden D.M., Crawford D.C. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene–disease associations. Bioinformatics. 2010;26(9):1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salvatore M., Gu T., Mack J.A., Prabhu Sankar S., Patil S., Valley T.S., Singh K., Nallamothu B.K., Kheterpal S., Lisabeth L., Fritsche L.G., Mukherjee B. A Phenome-Wide Association Study (PheWAS) of COVID-19 Outcomes by Race Using the Electronic Health Records Data in Michigan Medicine. J Clin Med. 2021;10(7):1351. doi: 10.3390/jcm10071351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verma A., Tsao N.L., Thomann L.O., Ho Y.-L., Iyengar S.K., Luoh S.-W., Carr R., Crawford D.C., Efird J.T., Huffman J.E., Hung A., Ivey K.L., Levin M.G., Lynch J., Natarajan P., Pyarajan S., Bick A.G., Costa L., Genovese G., Hauger R., Madduri R., Pathak G.A., Polimanti R., Voight B., Vujkovic M., Zekavat S.M., Zhao H., Ritchie M.D., Chang K.-M., Cho K., Casas J.P., Tsao P.S., Gaziano J.M., O’Donnell C., Damrauer S.M., Liao K.P., Barsh G.S. A Phenome-Wide Association Study of genes associated with COVID-19 severity reveals shared genetics with complex diseases in the Million Veteran Program. PLoS Genet. 2022;18(4):e1010113. doi: 10.1371/journal.pgen.1010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song R.J., Ho Y.-L., Schubert P., Park Y., Posner D., Lord E.M., Costa L., Gerlovin H., Kurgansky K.E., Anglin-Foote T., DuVall S., Huffman J.E., Pyarajan S., Beckham J.C., Chang K.-M., Liao K.P., Djousse L., Gagnon D.R., Whitbourne S.B., Ramoni R., Muralidhar S., Tsao P.S., O’Donnell C.J., Gaziano J.M., Casas J.P., Cho K., Marconi V.C. Phenome-wide association of 1809 phenotypes and COVID-19 disease progression in the Veterans Health Administration Million Veteran Program. PLoS ONE. 2021;16(5):e0251651. doi: 10.1371/journal.pone.0251651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Regan J.A., Abdulrahim J.W., Bihlmeyer N.A., Haynes C., Kwee L.C., Patel M.R., Shah S.H. Phenome-Wide Association Study of Severe COVID-19 Genetic Risk Variants. J Am Heart Assoc. 2022;11(5):e024004. doi: 10.1161/JAHA.121.024004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith P.G., Day N.E. The Design of Case-Control Studies: The Influence of Confounding and Interaction Effects. Int. J. Epidemiol. 1984;13(3):356–365. doi: 10.1093/ije/13.3.356. [DOI] [PubMed] [Google Scholar]
- 31.Schulz K.F., Grimes D.A. Case-control studies: research in reverse. The Lancet. 2002;359(9304):431–434. doi: 10.1016/S0140-6736(02)07605-5. [DOI] [PubMed] [Google Scholar]
- 32.Lumley T., Levy D. Bias in the case – crossover design: implications for studies of air pollution. Environmetrics. 2000;11(6):689–704. doi: 10.1002/1099-095X(200011/12)11:6<689::AID-ENV439>3.0.CO;2-N. [DOI] [Google Scholar]
- 33.Maclure M. The case-crossover design: a method for studying transient effects on the risk of acute events. Am. J. Epidemiol. 1991;133(2):144–153. doi: 10.1093/oxfordjournals.aje.a115853. [DOI] [PubMed] [Google Scholar]
- 34.McKeigue P.M., Burgul R., Bishop J., Robertson C., McMenamin J., O’Leary M., McAllister D.A., Colhoun H.M. Association of cerebral venous thrombosis with recent COVID-19 vaccination: case-crossover study using ascertainment through neuroimaging in Scotland. BMC Infect. Dis. 2021;21(1) doi: 10.1186/s12879-021-06960-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brakenhoff T.B., Franks B., Goodale B.M., van de Wijgert J., Montes S., Veen D., Fredslund E.K., Rispens T., Risch L., Dowling A.V., Folarin A.A., Bruijning P., Dobson R., Heikamp T., Klaver P., Cronin M., Grobbee D.E., Denaxas S., Reitsma J.B., Simon C., Kuchta A., Stolk P., Downward G., van Lier R., Kjellberg J., Risch M., Grossmann K., Conen D., Aeschbacher S. A prospective, randomized, single-blinded, crossover trial to investigate the effect of a wearable device in addition to a daily symptom diary for the remote early detection of SARS-CoV-2 infections (COVID-RED): a structured summary of a study protocol for a randomized controlled trial. Trials. 2021;22(1) doi: 10.1186/s13063-021-05241-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murk W., Gierada M., Fralick M., Weckstein A., Klesh R., Rassen J.A. Diagnosis-wide analysis of COVID-19 complications: an exposure-crossover study. CMAJ. 2021;193(1):E10–E18. doi: 10.1503/cmaj.201686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kerchberger VE, Peterson JF, Wei WQ. Scanning the medical phenome to identify new diagnoses after recovery from COVID-19 in a US cohort. J Am Med Inf Assoc. Published online August 2022:ocac159. doi:10.1093/jamia/ocac159. [DOI] [PMC free article] [PubMed]
- 38.Emergency use ICD codes for COVID-19 disease outbreak. World Health Organization. Accessed March 14, 2022. http://www.who.int/standards/classifications/classification-of-diseases/emergency-use-icd-codes-for-covid-19-disease-outbreak.
- 39.Charlson M.E., Pompei P., Ales K.L., MacKenzie C.R. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40(5):373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 40.Roberts EK, Gu T, Mukherjee B, Fritsche LG. Estimating COVID-19 Vaccination Effectiveness Using Electronic Health Records of an Academic Medical Center in Michigan. Published online January 31, 2022:2022.01.29.22269971. doi:10.1101/2022.01.29.22269971. [DOI] [PMC free article] [PubMed]
- 41.CDC. COVID-19 Vaccination. Centers for Disease Control and Prevention. Published May 6, 2022. Accessed May 9, 2022. https://www.cdc.gov/coronavirus/2019-ncov/vaccines/stay-up-to-date.html.
- 42.Wu P., Gifford A., Meng X., Li X., Campbell H., Varley T., Zhao J., Carroll R., Bastarache L., Denny J.C., Theodoratou E., Wei W.-Q. Mapping ICD-10 and ICD-10-CM Codes to Phecodes: Workflow Development and Initial Evaluation. JMIR Med Inform. 2019;7(4):e14325. doi: 10.2196/14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.PheWAS - Phenome Wide Association Studies. Accessed June 28, 2022. https://phewascatalog.org/phecodes.
- 44.R Core Team. R: The R Project for Statistical Computing. Published online 2021. Accessed May 9, 2022. https://www.r-project.org/.
- 45.Carroll R.J., Bastarache L., Denny J.C. R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics. 2014;30(16):2375–2376. doi: 10.1093/bioinformatics/btu197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holm S. A Simple Sequentially Rejective Multiple Test Procedure. Scand. J. Stat. 1979;6(2):65–70. [Google Scholar]
- 47.CDC. Healthcare Workers. Centers for Disease Control and Prevention. Published February 11, 2020. Accessed May 9, 2022. https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-care/post-covid-public-health-recs.html.
- 48.Gu T., Mack J.A., Salvatore M., Prabhu Sankar S., Valley T.S., Singh K., Nallamothu B.K., Kheterpal S., Lisabeth L., Fritsche L.G., Mukherjee B. Characteristics Associated With Racial/Ethnic Disparities in COVID-19 Outcomes in an Academic Health Care System. JAMA Netw Open. 2020;3(10):e2025197. doi: 10.1001/jamanetworkopen.2020.25197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu Y., Gu T., Valley T.S., Mukherjee B., Fritsche L.G. Changes in COVID-19-related outcomes, potential risk factors and disparities over time. Epidemiol. Infect. 2021;149 doi: 10.1017/S0950268821001898. [DOI] [Google Scholar]
- 50.Shen C., Risk M., Schiopu E., Hayek S.S., Xie T., Holevinski L., Akin C., Freed G., Zhao L. Efficacy of COVID-19 vaccines in patients taking immunosuppressants. Ann. Rheum. Dis. 2022;81(6):875–880. doi: 10.1136/annrheumdis-2021-222045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao H., Lu L.u., Peng Z., Chen L.-L., Meng X., Zhang C., Ip J.D., Chan W.-M., Chu A.-H., Chan K.-H., Jin D.-Y., Chen H., Yuen K.-Y., To K.-W. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg Microbes Infect. 2022;11(1):277–283. doi: 10.1080/22221751.2021.2023329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Glynne P., Tahmasebi N., Gant V., Gupta R. Long COVID following mild SARS-CoV-2 infection: characteristic T cell alterations and response to antihistamines. J. Invest. Med. 2022;70(1):61–67. doi: 10.1136/jim-2021-002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pfefferbaum B., North C.S. Mental Health and the Covid-19 Pandemic. N. Engl. J. Med. 2020;383(6):510–512. doi: 10.1056/NEJMp2008017. [DOI] [PubMed] [Google Scholar]
- 54.Douaud G., Lee S., Alfaro-Almagro F., Arthofer C., Wang C., McCarthy P., Lange F., Andersson J.L.R., Griffanti L., Duff E., Jbabdi S., Taschler B., Keating P., Winkler A.M., Collins R., Matthews P.M., Allen N., Miller K.L., Nichols T.E., Smith S.M. SARS-CoV-2 is associated with changes in brain structure in UK Biobank. Nature. 2022;604(7907):697–707. doi: 10.1038/s41586-022-04569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Al-Quteimat O.M., Amer A.M. The Impact of the COVID-19 Pandemic on Cancer Patients. Am. J. Clin. Oncol. 2020;43(6):452–455. doi: 10.1097/COC.0000000000000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bainton DAVID, Jones G.R., Hole DAVID. Influenza and Ischaemic Heart Disease-a Possible Trigger for Acute Myocardial Infarction? Int. J. Epidemiol. 1978;7(3):231–239. doi: 10.1093/ije/7.3.231. [DOI] [PubMed] [Google Scholar]
- 57.Reduced Access to Care - Research and Development Survey - COVID-19. Centers for Disease Control and Prevention. Published August 6, 2021. Accessed February 24, 2022. https://www.cdc.gov/nchs/covid19/rands/reduced-access-to-care.htm.
- 58.Schuemie M.J., Hripcsak G., Ryan P.B., Madigan D., Suchard M.A. Empirical confidence interval calibration for population-level effect estimation studies in observational healthcare data. Proc Natl Acad Sci U S A. 2018;115(11):2571–2577. doi: 10.1073/pnas.1708282114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schneeweiss S., Stürmer T., Maclure M. Case-crossover and case-time-control designs as alternatives in pharmacoepidemiologic research. Pharmacoepidemiol. Drug Saf. 1997;6(Suppl 3):S51–S59. doi: 10.1002/(SICI)1099-1557(199710)6:3+<S51::AID-PDS301>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 60.Griffith G.J., Morris T.T., Tudball M.J., Herbert A., Mancano G., Pike L., Sharp G.C., Sterne J., Palmer T.M., Davey Smith G., Tilling K., Zuccolo L., Davies N.M., Hemani G. Collider bias undermines our understanding of COVID-19 disease risk and severity. Nat. Commun. 2020;11(1) doi: 10.1038/s41467-020-19478-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beesley L.J., Mukherjee B. Statistical inference for association studies using electronic health records: handling both selection bias and outcome misclassification. Biometrics. 2022;78(1):214–226. doi: 10.1111/biom.13400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.