Opinion statement
Melanoma is caused by a variety of somatic mutations, and among these mutations, BRAF mutation occurs most frequently and has routinely been evaluated as a critical diagnostic biomarker in clinical practice. The introduction of targeted agents for BRAF-mutant melanoma has significantly improved overall survival in a large proportion of patients. However, there is BRAF inhibitor resistance in most patients, and its mechanisms are complicated and need further clarification. Additionally, treatment approaches to overcome resistance have evolved rapidly, shifting from monotherapy to multimodality treatment, which has dramatically improved patient outcomes in clinical trials and practice. This review highlights the mechanisms of BRAF inhibitor resistance in melanoma and discusses the current state of its therapeutic approaches that can be further explored in clinical practice.
Keywords: Melanoma, BRAF mutation, BRAF inhibitor, Targeted therapy, Resistance mechanism, Combination therapy
Introduction
Melanoma is a tumor caused by malignant transformation of melanocytes and a large variety of somatic mutations. Initiated by receptor tyrosine kinase (RTK) and RAS activation, the RAF-MEK-ERK axis is involved in regulating the main physiological processes, such as proliferation, cell cycle, and apoptosis [1]. Activation of BRAF, accounting for 41–55%, is the most common genetic alteration in the occurrence of melanoma [2]. Among BRAF mutations, over 90% are at codon 600, and among these, over 90% are single nucleotide mutations resulting in substitution of glutamic acid for valine (BRAFV600E: GTG>GAG). The second most common mutation is BRAFV600K (GTG>AAG), which represents 5–6%, followed by BRAFV600R (GTG>AGG), BRAFV600'E2' (GTG>GAA), and BRAFV600D (GTG>GAT) [3].
Many BRAF inhibitors (BRAFi) have been developed and approved by the US Food and Drug Administration (FDA), including vemurafenib, dabrafenib, and encorafenib. However, resistance to BRAFi develops quickly, with a median progression-free survival (PFS) of nine months [4]. Thanks to the use of MEK inhibitors (MEKi) including trametinib, binimetinib, and cobimetinib with BRAFi, the metastasis-free survival (MFS) and PFSs for combined therapies increased [5]. Unfortunately, combined agents also develop acquired resistance at a median of 9–11 months [6], which remains a challenge. Therefore, in this review, we will provide deep insight into the BRAF gene to further describe resistance mechanisms and effective strategies for overcoming resistance and to explore potential treatments with the aim of improving outcomes in patients with BRAF-mutations.
Mechanisms of BRAF inhibitor resistance
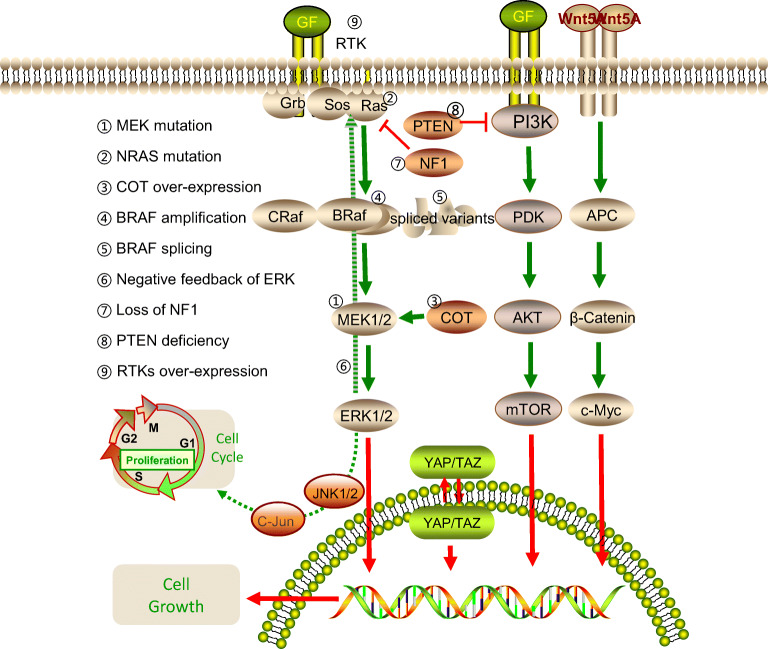
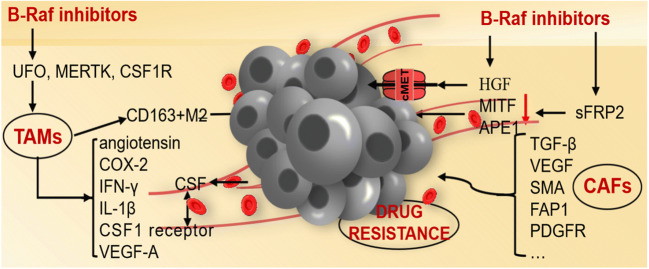
Multiple mechanisms have been identified to result in resistance which include genomic (Figure 1) or epigenetic abnormalities and the tumor microenvironment (Figure 2).
Fig. 1.
The genetic mechanisms of BRAF inhibitor resistance.
Fig. 2.
The role of tumor microenvironment in BRAF inhibitor resistance.
Genetic and epigenetic changes serve critical functions
Reactivation of the BRAF/MEK transduction pathway
The recovery of MAPK signaling is the most common mechanism of BRAFi resistance. Deregulation at each point may contribute to resistance [7].
RTKs
The hyperactivation of RTKs could promote resistance by activating parallel pathways or directly activating RAS, involving receptors such as PDGFRb, EGFR, MET, KIT, and IGF-1R [8, 9].
RAS gene
Constitutive activation of mutated RAS increases BRAF dimerization and subsequent reactivation of MAPK [10]. For example, mutation of NRAS could induce the dimerization of BRAF and CRAF [11].
BRAF gene
Overexpression of mutated BRAF, favoring dimerization, results in BRAFi inefficiency and reactivation of the ERK pathway [12]. Moreover, the splicing variant of BRAFV600E, like p61BRAFV600E, can form dimers independently of RAS, making BRAFi ineffective as they only block monomers [13]. Additionally, activation of ARAF and CRAF induces BRAFi resistance, as all RAF isoforms can do the same. Biochemically after treatment with vemurafenib, USP28 expression increased which connects with FBW7 to regulate the stability of RAF and forms a complex to target BRAF for degradation [14].
MEK
Mutations in MEK1/MEK2 make BRAFi ineffective regardless of its inhibition as signals below BRAF in the MAPK pathway can still be transduced. The type of MEK1/2 mutations presented varying states of resistance. MEK1 point mutations in C121S, E203K, Q56P, K57E, and MEK2 point mutations, including E207K and Q60P, are associated with BRAFi resistance [15, 16], activating downstream ERK, jumping over BRAF stimulation and leading to resistance. Additionally, overexpression of COT can reactivate MEK, developing resistance to BRAFi [17].
ERK
A resetting event of ERK1/2 pathway in adaptive response to BRAFi was proven. Theoretically, the existence of BRAFi is associated with low expression of active RAS, but with the reduction of SPRY2, DUSP, and SPRY expression tested by microarray analysis after vemurafenib, RAS activation reoccurs [18]. Additionally, BRAFi inhibits tumor growth by inhibiting ERK, which in turn inhibits the negative feedback inhibition of ERK on RAS, partially restoring RAS activity and leading to the formation of BRAF dimers induced by RAS [10]. Additionally, the loss of NF1 can result in RAS resistance to negative feedback [19], and NF1 inactivation in melanoma harboring BRAF mutation results in a selective advantage by reversing oncogene-mediated suppression of RAS, which is driven by ERK-induced negative feedback [20].
Upregulation of bypass activation
The PI3K-AKT-mTOR pathway
Despite continuous targeted inhibition, bypass tracks could eventually lead to the abnormal activation of the downstream pathway. Notably, the more potent target inhibitors are used, the more frequently bypass tracks are likely to be developed [21]. There are four major oncogenic signaling pathways that drive cell growth and proliferation: the PI3K, YAP/TAZ, STAT/JAK, and WNT5A/β-catenin pathways.
The PI3K/AKT/mTOR pathway, which provides antiapoptotic signals and increases proliferation, interacts with MAPK pathways at multiple points. Adaptive PI3K/AKT activity may occur when ERK signaling is blocked, which compensates for BRAFi. The PI3K/AKT pathway is activated by growth factors binding RTKs, so when BRAF is blocked, tumor cells can overexpress RTK, leading to permanent PI3K/AKT signaling [22]. Additionally, PDGFRβ, IGFR1 and EGFR overexpression were reported to cause this pathway reactivation in BRAFi-resistant melanoma [23]. Additionally, mutations in the PI3K/AKT genes induce AKT phosphorylation, which increases anti-apoptotic signaling and the expression of key proliferation genes, independent of BRAF [24]. It is necessary for signal transduction to switch from PIP2 to PIP3, which is promoted by PI3K and negatively regulated by PTEN in PI3K signaling [25], so the activation of AKT after PTEN deficiency is also necessary for intrinsic resistance to BRAFi [26]. These changes make it possible for melanoma cells to proliferate independently of BRAF and are significantly involved in adaptive resistance to BRAFi.
YAP/TAZ pathway
Regulated by the homologous proteins Yes-associated protein (YAP) and the transcriptional coactivator with PDZ-binding motif (TAZ), the Hippo pathway is involved in malignant transformation and the growth and metastasis of cancer stem cells. In melanoma cells resistant to BRAFi, the occurrence of nuclear translocation of YAP and TAZ increased and the expression of cell cycle molecules was then promoted [27, 28]. Additionally, the knockdown of YAP or TAZ was reported to suppress the viability of melanoma cells resistant to BRAFi [29]. Therefore, the activation of the YAP/TAZ pathway renders resistance to BRAFi [28], which was proven to be correlated with continuous ERK1/2 activity [27].
JNK/c-Jun pathway
The JNK/c-Jun pathway is recognized as an important regulator of cell proliferation, metabolism, and death, and JNKs also belong to the MAPK pathway [30]. C-Jun in melanoma is thought to function downstream of ERK by promoting the transcription of cyclin D1, a positive regulator of the G1-S cell cycle transition. Single-cell analysis suggests that p-c-Jun upregulation contributes to resistance to vemurafenib in some populations by decoupling the inhibition of proliferation from the induction of apoptosis. Additionally, exposing cells to vemurafenib and JNK inhibitors such as JNK-IN-8 results in synergistic cell killing [30, 31].
WNT5A/β-catenin pathway
Numerous studies now identify aberrations in β-catenin-independent WNT pathways in melanoma, most notably activated by WNT5A [32]. WNT5A protein and transcript levels were dramatically increased in BRAFi-resistant cells. In vitro studies demonstrated that a loss of WNT5A reduced the viability of cells in the presence of BRAFi. WNT5A-dependent signaling promotes the resistance of melanoma cells to BRAFi via its receptors RYK and FZD7 and the activation of PI3K/AKT signaling has also been reported [33].
Epigenetic mechanisms
Epigenetic alterations refer to heritable changes that modulate gene transcription without causing changes in DNA sequences [34]. Epigenetic mechanisms regarding BRAFi resistance include DNA methylation, noncoding RNAs, histone-modifying enzymes, and histone modifications.
DNA methylation
Hypomethylation, defined as the absence of methyl groups from cytosines, can open tightly packed chromatin, leading to genomic instability. In contrast, hypermethylation can lead to transcriptional repression [35]. Drug resistance corresponding to transcriptomic and methylomic alterations was identified [36] and transcriptomic analysis revealed differential mRNA expression in genes connected with differential methylation at CpG clusters and short DNA sequences that are mainly CG-rich and remain unmethylated [37], suggesting key connections between drug resistance and epigenetic regulation of DNA methylation.
Mutations in DNA methyltransferases (DNMTs), such as DNMT3B, considered a de novo DNA methyltransferase for depositing and maintaining methyl marks [38], have a role in tumor progression. Low global DNA methylation levels appeared in drug-tolerant melanoma cells following targeted treatment as DNMT3A, DNMT3B, and DNMT1 were differentially expressed [39].
Histone-modifying enzymes (HMEs) and posttranslational modifications (PTMs)
The N-terminal tails of histone proteins forming a histone octamer can be covalently and reversibly changed with various kinds of PTMs. These dynamic histone PTMs and specific histone proteins can promote either transcriptional activation or repression of targeted genes by remodeling their chromatin structures [36].
The expression of histone demethylases like KDM6A, KDM6B, KDM1B, JARID1A, and JARID1B is elevated in melanoma with a drug-tolerant state, which is accompanied by increased levels of H3K9me3 and lower levels of H3K4me3 and H3K27me3, indicating selected gene silencing and epigenetic activation. SETDB1 and SETDB2, histone methyltransferases, are also upregulated after treatment with BRAFi and MEKi and their knockdown restored drug sensitivity [39]. In addition, the histone deacetylase SIRT6 was downregulated in BRAFi-resistant melanoma cells, leading to upregulation of the IGF-1 receptor (IGF-1R) and subsequent AKT pathway activation [40].
Noncoding RNAs
Previous studies have identified miRNAs and lncRNAs as effectors of resistance to BRAFi (Table 1).
Table 1.
Non-coding RNAs associated with BRAFi resistance in melanomas
| Types of ncRNA | Name | Expression after resistance | Mechanisms in BRAFi resistance | Ref |
|---|---|---|---|---|
| miRNA | miR-514a | ↑ | Decreased expression of the tumor suppressor NF1 | [41] |
| miR-125a | ↑ | Inhibiting pro-apoptotic parts of intrinsic apoptosis pathway | [42] | |
| miR-211-5p | ↑ | Increased expression of MITF regulating TRPM1 gene resulting in activation of the survival pathway | [43] | |
| miR-34a, miR-100, and miR-125b | ↑ | Involvement in the control of cell proliferation and apoptosis | [44] | |
| miR-204-5p and miR-211-5p | ↑ | Stimulation in Ras and MAPK upregulation | [45] | |
| miRNA-204 and miRNA-211 | ↑ | Reducing expression of NUAK1/ARK5 proteins | [46] | |
| miR-1246 | ↑ | G2/M arrest and autophagy | [47] | |
| miR-7 | ↓ | Increased expressions of EGFR, IGF-1R, and CRAF and further activation of MAPK and PI3K/AKT pathway | [48] | |
| miR-579-3p | ↓ | Negative correlation with BRAF in resistant cells | [49] | |
| miR-200c | ↓ | Deactivation of the PI3K/AKT and MAPK signaling cascades, and acquisition of epithelial-mesenchymal transition-like phenotypes | [50] | |
| lncRNA | EMICERI | ↑ | Regulating MOB3B whose over-expression downregulates LATS1 to activate the Hippo signaling pathway | [51] |
| MIRAT | ↑ | Binding to IQGAP1 and facilitating signaling through the MAPK pathway | [52] | |
| TUG1 | ↑ | Acting as an oncogene sponging miR-129-5p and inducing cell growth and invasion | [46] | |
| TSLNC8 | ↑ | Binding with the catalytic sub-unit of PP1α to regulate its distribution, and re-activating the MAPK signaling | [53] | |
| SAMMSON | ↑ | Interacting with p32 and regulating the metabolism of mitochondria and CARF-p53 signaling pathway | [54] [55] | |
| RMEL3 | Unknown | A positive regulator of PI3K and MAPK signaling in melanoma | [56] | |
| IGF2AS, MEG3, and Zeb2NAT | ↑ | Might serve as prognostic markers of response to vemurafenib treatment in melanoma patients | [57] |
In detail, it contains information regarding the expression levels after BRAFi resistance (↑or↓), the mechanisms in BRAFi resistance the references in which they are described.
Influences of the tumor microenvironment
Resistance to BRAFi in melanoma is known to develop not only as a result of genomic or epigenetic abnormalities, but the role of tumor microenvironment is also important. Recent research has identified the role of intratumoral fibroblasts and macrophages in the development of resistance.
Cancer-associated fibroblasts (CAFs)
CAFs, differing from normal fibroblasts by upregulated expression of vimentin, fibroblast activation protein-1 (FAP1), and α-smooth-muscle actin (SMA), as well as PDGFR and TGFβ signaling [58], have been reported to allow therapeutic escape from BRAFi.
In the neighborhood of CAFs, melanoma cells present an aggressive and dedifferentiated mesenchymal phenotype. After treatment with BRAFi, melanoma cells maintain high levels of active mTOR signaling, facilitating protein synthesis, cell growth, and utilization of nutrients from the microenvironment [59]. They also respond to growth factors and cytokines secreted by CAFs, including TGF-β and VEGF, which promote cell survival and growth [58]. Vemurafenib directly activates fibroblasts to secrete hepatocyte growth factor (HGF), which activates both the MAPK/ERK and PI3K/AKT signaling pathways and downregulates the expression of proapoptotic genes [60]. It is interesting to discover that aging fibroblasts related to aged melanoma are more invasive and were shown to secrete frizzled related protein 2 (sFRP2), inhibiting β-catenin and downregulate expression of MITF and apurinic endonuclease (APE1), rendering cells resistant to BRAFi [61].
CAFs also secrete extracellular matrix (ECM) components. For example, ECM-induced integrin signaling promotes resistance to BRAFi. In regard to interaction, BRAFi-resistant melanoma cells release TGF-β, promoting CAF differentiation, which in turn increases the expression of ECM molecules and further develops to BRAFi resistance [62].
Tumor-associated macrophages (TAMs)
TAMs, which are activated M2 macrophages, express various anti-inflammatory factors and an immune-suppressive microenvironment was built. A high number of intratumoral CD163+ macrophages correlate with BRAFi resistance. The transition from macrophages to CD163+ M2 macrophages is induced by exosome-derived growth factors and interleukins released by melanoma cells, T-regulatory cells, and other macrophages. In the paracrine mode, macrophages that express the colony-stimulating factor 1 receptor (CSF1R) respond to CSF secreted by melanoma cells, which also stimulates resistance in autocrine manners [58]. In turn, TAMs secrete the melanoma-stimulating molecules angiotensin, COX-2, IFN-γ, and IL-1β, supporting melanoma growth and metastasis [63].
Moreover, BRAFi itself also stimulates TAMs. For instance, it induces the production of VEGF-A, which stimulates not only angiogenesis in the tumor but also macrophage survival and tumor immune escape [64]. Additionally, TAMs secrete TNFα, which promotes MITF expression and inhibits BRAF protein to block apoptosis in melanoma [65]. For vemurafenib, TAMs protected melanoma cells from BRAFi-induced apoptosis but did not disturb the G2/M phase [66].
Current treatment strategies to overcome BRAFi resistance
One current strategy to prevent or delay resistance is to develop combination therapies. A complementary strategy is to develop novel MAPKi to tackle resistance mechanisms and offer new options for future therapeutic development.
Development of combination therapies
Current clinical trials of combination therapies to overcome BRAF inhibitor resistance in patients with melanoma are listed in Table 2.
Table 2.
Current clinical trials of combination therapies to overcome the BRAF inhibitors resistance in patients with melanoma
| TrialID | Public title | Year | Recruitment Status | Phase | Intervention | Drug target |
|---|---|---|---|---|---|---|
| NCT04903119 | Nilotinib Plus Dabrafenib/Trametinib in Metastatic Melanoma | 2021 | Recruiting | 1 | Trametinib, Dabrafenib, Nilotinib | MEK, BRAF, KIT, PDGFR |
| NCT04557956 | Testing the Addition of the Anti-cancer Drug, Tazemetostat, to the Usual Treatment (Dabrafenib and Trametinib) for Metastatic Melanoma That Has Progressed on the Usual Treatment | 2020 | Recruiting | 1/2 | Trametinib, Dabrafenib, Tazemetostat | MEK, BRAF, EZH2 |
| NCT04527549 | Testing Dabrafenib and Trametinib With or Without Hydroxychloroquine in Stage IIIC or IV BRAF V600E/K Melanoma | 2020 | Recruiting | 2 | Trametinib, Dabrafenib, Hydroxychloroquine | MEK, BRAF, Autophagy |
| NCT04375527 | Binimetinib and Nivolumab for the Treatment of Locally Advanced Unresectable or Metastatic BRAF V600 Wildtype Melanoma | 2020 | Recruiting | 2 | Binimetinib, Nivolumab | MEK, PD-1 |
| NCT03972046 | Neoadjuvant Use of Talimogene Laherparepvec and BRAF/MEK Inhibitor for Advanced Nodal BRAF Mutant Melanoma | 2019 | Active, not recruiting | 2 | Trametinib, Dabrafenib, Talimogene laherparepvec (T-Vec) | MEK, BRAF, GM-CSF |
| NCT04201457 | A Trial of Dabrafenib, Trametinib and Hydroxychloroquine for Patients With Recurrent LGG or HGG With a BRAF Aberration | 2019 | Recruiting | 1/2 | Trametinib, Dabrafenib, Hydroxychloroquine | MEK, BRAF, Autophagy |
| NCT03580382 | Study of CDX-3379, a Human Monoclonal Antibody Targeting ERBB3, in Combination With the MEK Inhibitor, Trametinib, in Patients With Advanced Stage NRAS Mutant and BRAF/NRAS Wildtype (WT) Melanoma | 2018 | Active, not recruiting | 1/2 | Trametinib, CDX-3379 | MEK, ERBB3 |
| NCT03543969 | Adaptive BRAF-MEK Inhibitor Therapy for Advanced BRAF Mutant Melanoma | 2018 | Recruiting | 1 | Cobimetinib, Vemurafenib | MEK, BRAF |
| NCT03455764 | MCS110 With BRAF/MEK Inhibition in Patients With Melanoma | 2018 | Active, not recruiting | 1/2 | Trametinib, Dabrafenib, MCS110 | MEK, BRAF, CSF-1 |
| NCT03668431 | Dabrafenib + Trametinib + PDR001 In Colorectal Cancer | 2018 | Recruiting | 2 | Trametinib, Dabrafenib, PDR001 | MEK, BRAF, PD-1 |
| NCT03693170 | Encorafenib, Binimetinib and Cetuximab in Subjects With Previously Untreated BRAF-mutant ColoRectal Cancer ANCHOR-CRC | 2018 | Active, not recruiting | 2 | Binimetinib, encorafenib, Cetuximab | MEK, BRAF, EGFR |
| NCT03101254 | LY3022855 With BRAF/MEK Inhibition in Patients With Melanoma | 2017 | Active, not recruiting | 1/2 | Cobimetinib, Vemurafenib, LY3022855 | MEK, BRAF, CSF-1 |
| NCT03026517 | Clinical Trial of Phenformin in Combination With BRAF Inhibitor + MEK Inhibitor for Patients With BRAF-mutated Melanoma | 2017 | Recruiting | 1 | Trametinib, Dabrafenib, Phenformin | MEK, BRAF, Activation of AMPK |
| NCT03272464 | INCB039110 in Combination With Dabrafenib and Trametinib in Patients With BRAFmutant Melanoma and Other Solid Tumors. | 2017 | Active, not recruiting | 1 | Trametinib, Dabrafenib, INCB039110 | MEK, BRAF, JAK1 |
| NCT02967692 | A Study of the Anti-PD1 Antibody PDR001, in Combination With Dabrafenib and Trametinib in Advanced Melanoma | 2016 | Active, not recruiting | 3 | Trametinib, Dabrafenib, Spartalizumab | MEK, BRAF, PD-1 |
| NCT02858921 | Neoadjuvant Dabrafenib, Trametinib and/or Pembrolizumab in BRAF Mutant Resectable Stage III Melanoma Neo Trio | 2016 | Active, not recruiting | 2 | Trametinib, Dabrafenib, Pembrolizumab | MEK, BRAF, PD-1 |
| NCT02382549 | A Clinical Trial to Evaluate a Melanoma Helper Peptide Vaccine Plus Dabrafenib and Trametinib | 2015 | Recruiting | 1 | Trametinib, Dabrafenib, 6MHP | MEK, BRAF, Induce T cell and Ab responses |
Combination of BRAF and MEK inhibitors
Combining BRAFi with MEKi delayed the emergence of resistance to a single BRAFi and significantly inhibited melanoma growth [67], which also reduced the occurrence of proliferative skin lesions and secondary malignancies [68].
Findings from clinical trials confirmed that combination treatment (dabrafenib/trametinib and vemurafenib/cobimetinib) had higher response rates (approximately 65–70%), improved progression-free survival (median, approximately 12 months), and improved overall survival (median, approximately 24 months) with less cutaneous toxicity, and such combinations are now the standard targeted therapy for patients with BRAF-mutant melanoma.
Combination of PI3K/AKT inhibitors and MAPK inhibitors
The sensitivity of BRAF-mutant melanoma to BRAFi or MEKi can be enhanced through combined inhibition of the PI3K/AKT pathway, which has synergistic effects in inducing apoptosis [69]. In a study using uveal melanomas harboring activated Q209 L/P mutations, a combination of MEKi GSK1120212 and pan-PI3K inhibitor GSK2126458 induced significant cell apoptosis in comparison with inhibition of either pathway alone [70].
Aside from boosting the efficacy of MAPK inhibitors, addition of PI3K/AKT inhibitors may overcome their resistance. A study showed that the phosphorylation of AKT and MAPK downstream target S6 did not decrease in resistant melanoma, which means that an increased AKT pathway may contribute to resistance. In support of this, the combination of BRAFi and MEKi with an AKT inhibitor reversed the resistance of melanoma cells to vemurafenib [71]. Similarly, the addition of the PI3K inhibitor GSK2118136 to either dabrafenib or trametinib also delayed resistance [67].
As BRAF-mutant melanoma resistant to treatment with BRAFi or MEKi showed suppression of mTOR activity [72], targeting downstream effector mTOR was also included to disrupt aberrant PI3K/AKT signaling. Strangely, the inhibition of BRAF, PI3K, and mTOR suppressed melanoma cell growth but was ineffective at inducing cell death, while a combination of PI3K and mTOR inhibitors with a MEKi induced apoptosis [9].
Despite promising preclinical data, the clinical combination of BRAFi or MEKi with PI3K/AKT inhibitors was disappointing. A phase 1 trial (NCT01476137) testing the safety of trametinib in combination with the AKT inhibitor afuresertib showed only a partial response (PR). Other clinical trials are currently undergoing, such as a combination of the PI3K inhibitor copanlisib with a MEKi in melanoma (NCT01392521). However, due to their early-phase nature, it is not possible to determine the efficacy of these combinations until initiation of phase 2 and 3 trials with larger patient cohorts [73].
Combination of BRAF and/or MEK inhibitors with immunotherapy
Immune checkpoint inhibitors (ICIs), involving inhibitors targeting cytotoxic T lymphocyte antigen 4 (CTLA-4) and programmed death 1 (PD-1) receptors (i.e., ipilimumab, nivolumab, and pembrolizumab), have lower response rates but more durable responses than BRAFi. Preclinical and translational data indicate that combining BRAFi and MEKi with ICI could exceed the limitations of each class and potentially lead to longer-lasting responses [74••].
Combined treatment with vemurafenib and adoptive lymphocyte transfer therapy in vivo showed better antitumor effects than either treatment alone [75]. Mechanistically, treatment with a BRAF inhibitor alone or in combination with a MEK inhibitor is associated with increased CD8 + T-cell infiltration, increased T-cell cytotoxicity, and decreased expression of immunosuppressive molecules such as interleukins-6 and interleukins-8, which further supports the potential of combining BRAF/MEK inhibitors with immunotherapy [76].
PD-L1, an immunosuppressive ligand of PD-1 receptors, is also associated with PI3K-mediated resistance to the BRAFi vemurafenib [77], and its upregulation in tumor cells that have acquired resistance to BRAFi has been reported [78]. Hence, concurrent inhibition of the MAPK pathway and immune checkpoint signaling may prove a potent antitumor combination therapy, which means that the combination of trametinib with immune checkpoint inhibitors targeting PD-1, PD-L1, or CTLA-4 was more effective than use of each inhibitor alone.
Clinical trials combining MAPK dual-targeted inhibitors with immune checkpoint inhibitors for melanoma are currently underway (NCT01940809, NCT02130466, and NCT02858921) [79, 80]. Among the results of Keynote-022, IMspire 150, and COMBI-i, COMBI-i achieved the highest response rate of 78%, but there was no significant difference in the objective response rate (ORR) between the triple-drug group and dual-target group in all three studies, indicating that the dual-target ORR reached the ideal effective rate. In addition, according to the COMBI-i study, triple-drug combination may be more applicable to patients with a larger tumor burden [LDH ≥ 2 ULN, number of metastases ≥ 3, sum of lesion diameters ≥ 66 mm], and higher tumor mutation load (TMB) ≥ 10 mt/M. However, complementarily, with the comparison of these three clinical studies, the incidence of adverse reactions of the triplet regimens was relatively high, with an incidence of adverse reactions of grade 3 and above of more than 70%, and the IMspire150 study even reached nearly 80%. Therefore, we can conclude that targeted combined immunization can be better applied and promoted if the adverse reaction rates can be effectively controlled.
Combination with synergistic small molecules
Several synergistic small molecules have emerged to restore the efficacy of vemurafenib sensitivity. HSP90 inhibition may be a highly effective strategy for managing vemurafenib resistance [81]. Fluvastatin, the BET-bromodomain inhibitor JQ1, and the natural product neferine have been shown to reduce vemurafenib resistance in melanoma [82]. CHMFL-BMX-078, the BMX inhibitor, significantly decreased the phosphorylation of AKT, and had high selectivity for vemurafenib-resistant melanoma cells. Combination treatment with vemurafenib and CHMFL-BMX-078 inhibited AKT pathway and ERK signaling pathway, which explains their enhanced inhibitory effects. Furthermore, the aptamer LL4A selectively inhibits vemurafenib-resistant melanoma by binding to the CD63 protein [83]. However, there are currently no viable methods for entirely reversing vemurafenib resistance, and the development of novel combinations is still urgently needed.
Strategies for blocking MAPK signaling
Inhibition of mutant RAF signaling
Two different strategies are currently under investigation for the inhibition of mutant RAF and mitigation of paradoxical activation — pan-RAF inhibitors and paradox breakers.
Type II pan-RAF inhibitors target both active RAF dimers and monomers and therefore inhibit ERK signaling including AZ628, CCT241161, and TAK-580 [84]. However, their uses in vivo have been more limited because of their lack of selectivity for mutant BRAF, in contrast with vemurafenib and dabrafenib, which could lead to greater toxicity and disruption of WT RAF signaling [85].
A third generation of RAF inhibitors, known as “paradox breakers,” inhibits BRAF without promoting dimerization, thereby preventing paradoxical upregulation of ERK signaling [86]. Paradox breakers have the potential to be effective against V600E mutation splice variants and upstream RAS mutations and are currently under development [87]. PLX8394, a BRAF-specific dimer breaker, selectively disrupts BRAF homo and BRAF-CRAF heterodimers, which will be effective for treatment of tumors with class 1 or 2 BRAF mutants, BRAF fusions and RAS-independent, BRAF dimer-dependent resistance to current BRAFi [88]. Unfortunately, this drug has been shown to paradoxically activate RAF signaling, and BRAF fusions could drive acquired resistance to PLX8394 noted in some preclinical trials [89].
Vertical blockade of MAPK signaling
Adequate adaptive barriers can be ensured in vertical blockade of MAPK signaling through polytherapy to suppress BRAFi resistance. Interestingly, preclinical modeling suggests that intermittent BRAF inhibitor therapy may delay resistance [90]. However, S1320, a phase 2 clinical trial evaluating whether intermittent dosing of dabrafenib and trametinib improves PFS in patients with metastatic and unresectable BRAFV600 melanoma, demonstrates that continuous dosing yields superior PFS compared to intermittent dosing in patients with BRAFV600E and BRAFV600K melanoma [91]. A similar vertical blockade strategy was adopted in a phase II clinical trial of metastatic colon cancer where EGFR, BRAF, and MEK were coinhibited. The results indicated not only a superior ORR but also PFS and OS, as described above [92, 93]. Moreover, recent studies have shown that SHP2 inhibitors are effective in bypassing the paradoxical activation of MAPK signaling upon BRAF inhibition in RAS-mutant cancers [94]. These studies highlight the importance of molecular classification and profiling to better predict therapy regimens.
Potential and future direction
In view of the complex drug resistance mechanism, individualized and precise management should be an inevitable choice in clinical practice. However, at the same time, as researchers, there are still many issues that need to be explored. The recently resolved BRAF-MEK1-14-3-3 complexes uncovered molecular details of how BRAF is regulated by phosphorylation and protein-protein interactions [95], which may provide information for the assembly of complexes targeting MAPK signaling pathways. These interactions may serve as good candidates for combination approaches to provide synergistic effects and reduce possible side effects. Moreover, with a handful of FDA-approved RAF/MEK inhibitors widely used in the clinic, the impact of inhibiting RAF and/or MEK on somatic tissue and tumor-infiltrating immune cells has recently been brought into the spotlight. Another challenge is to understand the dimerization mechanism which will enable further improvement of the current inhibitors in designing tailored drugs. Deeper investigation into these issues will lead to the development of new strategies to achieve therapeutic goals.
New innovative therapies are also needed to address drug resistance. Proteolysis targeting chimeras (PROTACs), a novel strategy to knock down proteins of interest [96], have potential advantages over conventional small molecule inhibitors [97]. To address the limitations of BRAFi therapies, a study [98••] using vemurafenib-based PROTACs achieved low nanomolar degradation of all classes of BRAF mutants but WT RAF and outperformed vemurafenib in inhibiting cancer cell growth. Additionally, the modular design of PROTACs makes it ideal for providing tailored drugs and personalized medicine. On the other hand, the discovery of the key mechanisms regulating TME, it may have a beneficial antitumor effect. At the same time, attention should be paid to the relationship between changes in tumor cell intrinsic signaling pathways and T-cell rejection or infiltration. The development of new targeted therapy based on TME can also be a new therapeutic strategy.
Acknowledgements
Jingqin Zhong would like to extend his sincere gratitude to his supervisor, Yong Chen, for his instructive advice and useful suggestions on Jingqin Zhong's thesis. He is deeply grateful of his help in the completion of this thesis. High tribute shall be paid to Wei Sun, whose profound knowledge of English triggers Jingqin Zhong's love for research. He is also deeply indebted to all the other contributors. Finally, Jingqin Zhong is indebted to his parents for their continuous support and encouragement.
Declarations
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Conflict of Interest
This work was supported by Shanghai Science and Technology Development Funds (19411951700), the Lingang Laboratory (Grant No. LG-QS-202205-11), and the national natural science foundation of China (81802636).
JZ wrote and edited this manuscript and created tables and figure. JZ, ZZ, WS, WY, CW, WL, XL, and YC reviewed and revised the manuscript. WS and YC provided direction and guidance throughout the preparation of the manuscript. All authors read and approved the final manuscript.
Footnotes
This article is part of the Topical Collection on Skin Cancer
The original online version of this article was revised to update the sequence of author list, conflict of interest text and addition of Acknowledgment section.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
12/2/2022
A Correction to this paper has been published: 10.1007/s11864-022-01038-z
Contributor Information
Wei Sun, Email: wsun14@fudan.edu.cn.
Yong Chen, Email: chenyong@fudan.edu.cn.
References and Recommended Reading
Papers of particular interest, published recently, have been highlighted as: •• Of major importance
- 1.Guo YJ, Pan WW, Liu SB, Shen ZF, Xu Y, Hu LL. ERK/MAPK signalling pathway and tumorigenesis. Exp Ther Med. 2020;19(3):1997–2007. doi: 10.3892/etm.2020.8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siroy AE, Boland GM, Milton DR, Roszik J, Frankian S, Malke J, Haydu L, Prieto VG, Tetzlaff M, Ivan D, Wang WL, Torres-Cabala C, Curry J, Roy-Chowdhuri S, Broaddus R, Rashid A, Stewart J, Gershenwald JE, Amaria RN, Patel SP, Papadopoulos NE, Bedikian A, Hwu WJ, Hwu P, Diab A, Woodman SE, Aldape KD, Luthra R, Patel KP, Shaw KR, Mills GB, Mendelsohn J, Meric-Bernstam F, Kim KB, Routbort MJ, Lazar AJ, Davies MA. Beyond BRAFV600: clinical mutation panel testing by next-generation sequencing in advanced melanoma. J Investig Dermatol. 2015;135(2):508–515. doi: 10.1038/jid.2014.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Institute, W.T.S . Catalogue of somatic mutations in cancer (COSMIC) 2017. [Google Scholar]
- 4.Long G, et al. Dabrafenib plus trametinib versus dabrafenib monotherapy in patients with metastatic BRAF V600E/K-mutant melanoma: long-term survival and safety analysis of a phase 3 study. Ann Oncol. 2017;28(7):1631–1639. doi: 10.1093/annonc/mdx176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ascierto PA, Dummer R, Gogas HJ, Flaherty KT, Arance A, Mandala M, Liszkay G, Garbe C, Schadendorf D, Krajsova I, Gutzmer R, de Groot JWB, Loquai C, Gollerkeri A, Pickard MD, Robert C. Update on tolerability and overall survival in COLUMBUS: landmark analysis of a randomised phase 3 trial of encorafenib plus binimetinib vs vemurafenib or encorafenib in patients with BRAF V600–mutant melanoma. Eur J Cancer. 2020;126:33–44. doi: 10.1016/j.ejca.2019.11.016. [DOI] [PubMed] [Google Scholar]
- 6.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, Hamid O, Schuchter L, Cebon J, Ibrahim N, Kudchadkar R, Burris HA, III, Falchook G, Algazi A, Lewis K, Long GV, Puzanov I, Lebowitz P, Singh A, Little S, Sun P, Allred A, Ouellet D, Kim KB, Patel K, Weber J. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367(18):1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanchez JN, Wang T, Cohen MS. BRAF and MEK inhibitors: use and resistance in BRAF-mutated cancers. Drugs. 2018;78(5):549–566. doi: 10.1007/s40265-018-0884-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gopal YV, et al. Basal and treatment-induced activation of AKT mediates resistance to cell death by AZD6244 (ARRY-142886) in Braf-mutant human cutaneous melanoma cells. Cancer Res. 2010;70(21):8736–8747. doi: 10.1158/0008-5472.CAN-10-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi H, Kong X, Ribas A, Lo RS. Combinatorial treatments that overcome PDGFRβ-driven resistance of melanoma cells to V600EB-RAF inhibition. Cancer Res. 2011;71(15):5067–5074. doi: 10.1158/0008-5472.CAN-11-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartnik E, Fiedorowicz M, Czarnecka AM. Mechanisms of melanoma resistance to treatment with BRAF and MEK inhibitors. Nowotwory. Journal of Oncology. 2019;69(3-4):133-141.
- 11.Atefi M, Titz B, Tsoi J, Avramis E, le A, Ng C, Lomova A, Lassen A, Friedman M, Chmielowski B, Ribas A, Graeber TG. CRAF R391W is a melanoma driver oncogene. Sci Rep. 2016;6(1):1–11. doi: 10.1038/srep27454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson DB, Menzies AM, Zimmer L, Eroglu Z, Ye F, Zhao S, Rizos H, Sucker A, Scolyer RA, Gutzmer R, Gogas H, Kefford RF, Thompson JF, Becker JC, Berking C, Egberts F, Loquai C, Goldinger SM, Pupo GM, Hugo W, Kong X, Garraway LA, Sosman JA, Ribas A, Lo RS, Long GV, Schadendorf D. Acquired BRAF inhibitor resistance: a multicenter meta-analysis of the spectrum and frequencies, clinical behaviour, and phenotypic associations of resistance mechanisms. Eur J Cancer. 2015;51(18):2792–2799. doi: 10.1016/j.ejca.2015.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vido MJ, et al. BRAF splice variant resistance to RAF inhibitor requires enhanced MEK association. Cell Rep. 2018;25(6):1501–1510. e3. doi: 10.1016/j.celrep.2018.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saei A, Palafox M, Benoukraf T, Kumari N, Jaynes PW, Iyengar PV, Muñoz-Couselo E, Nuciforo P, Cortés J, Nötzel C, Kumarakulasinghe NB, Richard JLC, Bin Adam Isa ZF, Pang B, Guzman M, Siqin Z, Yang H, Tam WL, Serra V, Eichhorn PJA. Loss of USP28-mediated BRAF degradation drives resistance to RAF cancer therapies. J Exp Med. 2018;215(7):1913–1928. doi: 10.1084/jem.20171960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, Johnson NL, Granger DA, Jordan NV, Darr DB, Usary J, Kuan PF, Smalley DM, Major B, He X, Hoadley KA, Zhou B, Sharpless NE, Perou CM, Kim WY, Gomez SM, Chen X, Jin J, Frye SV, Earp HS, Graves LM, Johnson GL. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149(2):307–321. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu P-K, Park J-I. MEK1/2 inhibitors: molecular activity and resistance mechanisms. in Seminars in oncology. Elsevier; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma V, Young L, Cavadas M, Owen K, Reproducibility Project: Cancer Biology Registered Report: COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Elife. 2016;5:e11414. doi: 10.7554/eLife.11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kugel CH, 3rd, Aplin AE. Adaptive resistance to RAF inhibitors in melanoma. Pigment Cell Melanoma Res. 2014;27(6):1032–1038. doi: 10.1111/pcmr.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, Solit DB, Rosen N. V600EBRAF is associated with disabled feedback inhibition of RAF–MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci. 2009;106(11):4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiuru M, Busam KJ. The NF1 gene in tumor syndromes and melanoma. Lab Investig. 2017;97(2):146–157. doi: 10.1038/labinvest.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Redaelli S, Ceccon M, Zappa M, Sharma GG, Mastini C, Mauri M, Nigoghossian M, Massimino L, Cordani N, Farina F, Piazza R, Gambacorti-Passerini C, Mologni L. Lorlatinib treatment elicits multiple on-and off-target mechanisms of resistance in ALK-driven cancer. Cancer Res. 2018;78(24):6866–6880. doi: 10.1158/0008-5472.CAN-18-1867. [DOI] [PubMed] [Google Scholar]
- 22.Caporali S, Alvino E, Lacal PM, Levati L, Giurato G, Memoli D, Caprini E, Cappellini GCA, D’atri S. Targeting the PI3K/AKT/mTOR pathway overcomes the stimulating effect of dabrafenib on the invasive behavior of melanoma cells with acquired resistance to the BRAF inhibitor. Int J Oncol. 2016;49(3):1164–1174. doi: 10.3892/ijo.2016.3594. [DOI] [PubMed] [Google Scholar]
- 23.Chan XY, et al. Role played by signalling pathways in overcoming BRAF inhibitor resistance in melanoma. Int J Mol Sci. 2017;18(7):1527. doi: 10.3390/ijms18071527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Czarnecka AM, Bartnik E, Fiedorowicz M, Rutkowski P. Targeted therapy in melanoma and mechanisms of resistance. Int J Mol Sci. 2020;21(13):4576. doi: 10.3390/ijms21134576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Juric D, Castel P, Griffith M, Griffith OL, Won HH, Ellis H, Ebbesen SH, Ainscough BJ, Ramu A, Iyer G, Shah RH, Huynh T, Mino-Kenudson M, Sgroi D, Isakoff S, Thabet A, Elamine L, Solit DB, Lowe SW, Quadt C, Peters M, Derti A, Schegel R, Huang A, Mardis ER, Berger MF, Baselga J, Scaltriti M. Convergent loss of PTEN leads to clinical resistance to a PI (3) Kα inhibitor. Nature. 2015;518(7538):240–244. doi: 10.1038/nature13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nathanson KL, Martin AM, Wubbenhorst B, Greshock J, Letrero R, D'Andrea K, O'Day S, Infante JR, Falchook GS, Arkenau HT, Millward M, Brown MP, Pavlick A, Davies MA, Ma B, Gagnon R, Curtis M, Lebowitz PF, Kefford R, Long GV. Tumor genetic analyses of patients with metastatic melanoma treated with the BRAF inhibitor dabrafenib (GSK2118436) Clin Cancer Res. 2013;19(17):4868–4878. doi: 10.1158/1078-0432.CCR-13-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fisher ML, Grun D, Adhikary G, Xu W, Eckert RL. Inhibition of YAP function overcomes BRAF inhibitor resistance in melanoma cancer stem cells. Oncotarget. 2017;8(66):110257–110272. doi: 10.18632/oncotarget.22628. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 28.Kim MH, Kim J, Hong H, Lee SH, Lee JK, Jung E, Kim J. Actin remodeling confers BRAF inhibitor resistance to melanoma cells through YAP/TAZ activation. EMBO J. 2016;35(5):462–478. doi: 10.15252/embj.201592081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim H, Frederick DT, Levesque MP, Cooper ZA, Feng Y, Krepler C, Brill L, Samuels Y, Hayward NK, Perlina A, Piris A, Zhang T, Halaban R, Herlyn MM, Brown KM, Wargo JA, Dummer R, Flaherty KT, Ronai Z’A. Downregulation of the ubiquitin ligase RNF125 underlies resistance of melanoma cells to BRAF inhibitors via JAK1 deregulation. Cell Rep. 2015;11(9):1458–1473. doi: 10.1016/j.celrep.2015.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karin M, Gallagher E. From JNK to pay dirt: jun kinases, their biochemistry, physiology and clinical importance. IUBMB Life. 2005;57(4-5):283–295. doi: 10.1080/15216540500097111. [DOI] [PubMed] [Google Scholar]
- 31.Muthusamy V, Piva TJ. The UV response of the skin: a review of the MAPK, NFκB and TNFα signal transduction pathways. Arch Dermatol Res. 2010;302(1):5–17. doi: 10.1007/s00403-009-0994-y. [DOI] [PubMed] [Google Scholar]
- 32.Weeraratna AT. A Wnt-er wonderland—the complexity of Wnt signaling in melanoma. Cancer Metastasis Rev. 2005;24(2):237–250. doi: 10.1007/s10555-005-1574-z. [DOI] [PubMed] [Google Scholar]
- 33.Anastas JN, Kulikauskas RM, Tamir T, Rizos H, Long GV, von Euw EM, Yang PT, Chen HW, Haydu L, Toroni RA, Lucero OM, Chien AJ, Moon RT. WNT5A enhances resistance of melanoma cells to targeted BRAF inhibitors. J Clin Invest. 2014;124(7):2877–2890. doi: 10.1172/JCI70156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alipour S, Nouri M, Sakhinia E, Samadi N, Roshanravan N, Ghavami A, Khabbazi A. Epigenetic alterations in chronic disease focusing on Behçet’s disease. Biomed Pharmacother. 2017;91:526–533. doi: 10.1016/j.biopha.2017.04.106. [DOI] [PubMed] [Google Scholar]
- 35.Plass C, Pfister SM, Lindroth AM, Bogatyrova O, Claus R, Lichter P. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat Rev Genet. 2013;14(11):765–780. doi: 10.1038/nrg3554. [DOI] [PubMed] [Google Scholar]
- 36.Hugo W, Shi H, Sun L, Piva M, Song C, Kong X, Moriceau G, Hong A, Dahlman KB, Johnson DB, Sosman JA, Ribas A, Lo RS. Non-genomic and immune evolution of melanoma acquiring MAPKi resistance. Cell. 2015;162(6):1271–1285. doi: 10.1016/j.cell.2015.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Girault I, Tozlu S, Lidereau R, Bièche I. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin Cancer Res. 2003;9(12):4415–4422. [PubMed] [Google Scholar]
- 39.Al Emran A, et al. Distinct histone modifications denote early stress-induced drug tolerance in cancer. Oncotarget. 2018;9(9):8206–8222. doi: 10.18632/oncotarget.23654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strub T, Ghiraldini FG, Carcamo S, Li M, Wroblewska A, Singh R, Goldberg MS, Hasson D, Wang Z, Gallagher SJ, Hersey P, Ma’ayan A, Long GV, Scolyer RA, Brown B, Zheng B, Bernstein E. SIRT6 haploinsufficiency induces BRAF V600E melanoma cell resistance to MAPK inhibitors via IGF signalling. Nat Commun. 2018;9(1):1–13. doi: 10.1038/s41467-018-05966-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stark MS, et al. miR-514a regulates the tumour suppressor NF1 and modulates BRAFi sensitivity in melanoma. Oncotarget. 2015;6(19):17753. doi: 10.18632/oncotarget.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koetz-Ploch L, Hanniford D, Dolgalev I, Sokolova E, Zhong J, Díaz-Martínez M, Bernstein E, Darvishian F, Flaherty KT, Chapman PB, Tawbi H, Hernando E. Micro RNA-125a promotes resistance to BRAF inhibitors through suppression of the intrinsic apoptotic pathway. Pigment Cell Melanoma Res. 2017;30(3):328–338. doi: 10.1111/pcmr.12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lunavat TR, Cheng L, Einarsdottir BO, Olofsson Bagge R, Veppil Muralidharan S, Sharples RA, Lässer C, Gho YS, Hill AF, Nilsson JA, Lötvall J. BRAFV600 inhibition alters the microRNA cargo in the vesicular secretome of malignant melanoma cells. Proc Natl Acad Sci. 2017;114(29):E5930–E5939. doi: 10.1073/pnas.1705206114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vergani E, di Guardo L, Dugo M, Rigoletto S, Tragni G, Ruggeri R, Perrone F, Tamborini E, Gloghini A, Arienti F, Vergani B, Deho P, de Cecco L, Vallacchi V, Frati P, Shahaj E, Villa A, Santinami M, de Braud F, Rivoltini L, Rodolfo M. Overcoming melanoma resistance to vemurafenib by targeting CCL2-induced miR-34a, miR-100 and miR-125b. Oncotarget. 2016;7(4):4428–4441. doi: 10.18632/oncotarget.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Díaz-Martínez M, Benito-Jardón L, Alonso L, Koetz-Ploch L, Hernando E, Teixidó J. miR-204-5p and miR-211-5p contribute to BRAF inhibitor resistance in melanoma. Cancer Res. 2018;78(4):1017–1030. doi: 10.1158/0008-5472.CAN-17-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Long J, Menggen Q, Wuren Q, Shi Q, Pi X. Long noncoding RNA taurine-upregulated gene1 (TUG1) promotes tumor growth and metastasis through TUG1/Mir-129-5p/astrocyte-elevated gene-1 (AEG-1) axis in malignant melanoma. Medl Sci Monit Int Med J Exp Clin Res. 2018;24:1547–1559. doi: 10.12659/MSM.906616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim J-H, Ahn J-H, Lee M. Upregulation of microRNA-1246 is associated with BRAF inhibitor resistance in melanoma cells with mutant BRAF. Cancer Res Treat Off J Korean Cancer Assoc. 2017;49(4):947–959. doi: 10.4143/crt.2016.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun X, et al. miR-7 reverses the resistance to BRAFi in melanoma by targeting EGFR/IGF-1R/CRAF and inhibiting the MAPK and PI3K/AKT signaling pathways. Oncotarget. 2016;7(33):53558. doi: 10.18632/oncotarget.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fattore L, Mancini R, Acunzo M, Romano G, Laganà A, Pisanu ME, Malpicci D, Madonna G, Mallardo D, Capone M, Fulciniti F, Mazzucchelli L, Botti G, Croce CM, Ascierto PA, Ciliberto G. miR-579-3p controls melanoma progression and resistance to target therapy. Proc Natl Acad Sci. 2016;113(34):E5005–E5013. doi: 10.1073/pnas.1607753113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu S, Tetzlaff MT, Wang T, Yang R, Xie L, Zhang G, Krepler C, Xiao M, Beqiri M, Xu W, Karakousis G, Schuchter L, Amaravadi RK, Xu W, Wei Z, Herlyn M, Yao Y, Zhang L, Wang Y, Zhang L, Xu X. miR-200c/Bmi1 axis and epithelial–mesenchymal transition contribute to acquired resistance to BRAF inhibitor treatment. Pigment Cell Melanoma Res. 2015;28(4):431–441. doi: 10.1111/pcmr.12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Joung J, Engreitz JM, Konermann S, Abudayyeh OO, Verdine VK, Aguet F, Gootenberg JS, Sanjana NE, Wright JB, Fulco CP, Tseng YY, Yoon CH, Boehm JS, Lander ES, Zhang F. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature. 2017;548(7667):343–346. doi: 10.1038/nature23451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanlorenzo M, Vujic I, Esteve-Puig R, Lai K, Vujic M, Lin K, Posch C, Dimon M, Moy A, Zekhtser M, Johnston K, Gho D, Ho W, Gajjala A, Oses Prieto J, Burlingame A, Daud A, Rappersberger K, Ortiz-Urda S. The lincRNA MIRAT binds to IQGAP1 and modulates the MAPK pathway in NRAS mutant melanoma. Sci Rep. 2018;8(1):1–9. doi: 10.1038/s41598-018-27643-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han Y, Fang J, Xiao Z, Deng J, Zhang M, Gu L. Downregulation of lncRNA TSLNC8 promotes melanoma resistance to BRAF inhibitor PLX4720 through binding with PP1α to re-activate MAPK signaling. J Cancer Res Clin Oncol. 2021;147(3):767–777. doi: 10.1007/s00432-020-03484-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leucci E, Vendramin R, Spinazzi M, Laurette P, Fiers M, Wouters J, Radaelli E, Eyckerman S, Leonelli C, Vanderheyden K, Rogiers A, Hermans E, Baatsen P, Aerts S, Amant F, van Aelst S, van den Oord J, de Strooper B, Davidson I, Lafontaine DLJ, Gevaert K, Vandesompele J, Mestdagh P, Marine JC. Melanoma addiction to the long non-coding RNA SAMMSON. Nature. 2016;531(7595):518–522. doi: 10.1038/nature17161. [DOI] [PubMed] [Google Scholar]
- 55.Han S, Yan Y, Ren Y, Hu Y, Wang Y, Chen L, Zhi Z, Zheng Y, Shao Y, Liu J. lncRNA SAMMSON mediates adaptive resistance to RAF inhibition in BRAF-mutant melanoma cells. Cancer Res. 2021;81(11):2918–2929. doi: 10.1158/0008-5472.CAN-20-3145. [DOI] [PubMed] [Google Scholar]
- 56.Cardoso C, Serafim RB, Kawakami A, Gonçalves Pereira C, Roszik J, Valente V, Vazquez VL, Fisher DE, Espreafico EM. The lncRNA RMEL3 protects immortalized cells from serum withdrawal-induced growth arrest and promotes melanoma cell proliferation and tumor growth. Pigment Cell Melanoma Res. 2019;32(2):303–314. doi: 10.1111/pcmr.12751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kolenda T, Rutkowski P, Michalak M, Kozak K, Guglas K, Ryś M, Galus Ł, Woźniak S, Ługowska I, Gos A, Teresiak A, Mackiewicz A, Lamperska K, Mackiewicz J. Plasma lncRNA expression profile as a prognostic tool in BRAF-mutant metastatic melanoma patients treated with BRAF inhibitor. Oncotarget. 2019;10(39):3879–3893. doi: 10.18632/oncotarget.26989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Almeida FV, Douglass SM, Fane ME, Weeraratna AT. Bad company: microenvironmentally mediated resistance to targeted therapy in melanoma. Pigment Cell Melanoma Res. 2019;32(2):237–247. doi: 10.1111/pcmr.12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karbowniczek M, Spittle CS, Morrison T, Wu H, Henske EP. mTOR is activated in the majority of malignant melanomas. J Investig Dermatol. 2008;128(4):980–987. doi: 10.1038/sj.jid.5701074. [DOI] [PubMed] [Google Scholar]
- 60.Porta C, Paglino C, Mosca A. Targeting PI3K/Akt/mTOR signaling in cancer. Front Oncol. 2014;4:64. doi: 10.3389/fonc.2014.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaur A, Webster MR, Marchbank K, Behera R, Ndoye A, Kugel CH, Dang VM, Appleton J, O’Connell MP, Cheng P, Valiga AA, Morissette R, McDonnell NB, Ferrucci L, Kossenkov AV, Meeth K, Tang HY, Yin X, Wood WH, Lehrmann E, Becker KG, Flaherty KT, Frederick DT, Wargo JA, Cooper ZA, Tetzlaff MT, Hudgens C, Aird KM, Zhang R, Xu X, Liu Q, Bartlett E, Karakousis G, Eroglu Z, Lo RS, Chan M, Menzies AM, Long GV, Johnson DB, Sosman J, Schilling B, Schadendorf D, Speicher DW, Bosenberg M, Ribas A, Weeraratna AT. sFRP2 in the aged microenvironment drives melanoma metastasis and therapy resistance. Nature. 2016;532(7598):250–254. doi: 10.1038/nature17392. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 62.Fedorenko IV, Wargo JA, Flaherty KT, Messina JL, Smalley KSM. BRAF inhibition generates a host–tumor niche that mediates therapeutic escape. J Investig Dermatol. 2015;135(12):3115–3124. doi: 10.1038/jid.2015.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang H, Yang L, Wang D, Zhang Q, Zhang L. Pro-tumor activities of macrophages in the progression of melanoma. Hum Vaccines Immunother. 2017;13(7):1556–1562. doi: 10.1080/21645515.2017.1312043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Atzori MG, Ceci C, Ruffini F, Trapani M, Barbaccia ML, Tentori L, D'Atri S, Lacal PM, Graziani G. Role of VEGFR-1 in melanoma acquired resistance to the BRAF inhibitor vemurafenib. J Cell Mol Med. 2020;24(1):465–475. doi: 10.1111/jcmm.14755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ruffell B, Coussens LM. Macrophages and therapeutic resistance in cancer. Cancer Cell. 2015;27(4):462–472. doi: 10.1016/j.ccell.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang T, Xiao M, Ge Y, Krepler C, Belser E, Lopez-Coral A, Xu X, Zhang G, Azuma R, Liu Q, Liu R, Li L, Amaravadi RK, Xu W, Karakousis G, Gangadhar TC, Schuchter LM, Lieu M, Khare S, Halloran MB, Herlyn M, Kaufman RE. BRAF inhibition stimulates melanoma-associated macrophages to drive tumor growth. Clin Cancer Res. 2015;21(7):1652–1664. doi: 10.1158/1078-0432.CCR-14-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Greger JG, Eastman SD, Zhang V, Bleam MR, Hughes AM, Smitheman KN, Dickerson SH, Laquerre SG, Liu L, Gilmer TM. Combinations of BRAF, MEK, and PI3K/mTOR inhibitors overcome acquired resistance to the BRAF inhibitor GSK2118436 dabrafenib, mediated by NRAS or MEK mutations. Mol Cancer Ther. 2012;11(4):909–920. doi: 10.1158/1535-7163.MCT-11-0989. [DOI] [PubMed] [Google Scholar]
- 68.Larkin J, Ascierto PA, Dréno B, Atkinson V, Liszkay G, Maio M, Mandalà M, Demidov L, Stroyakovskiy D, Thomas L, de la Cruz-Merino L, Dutriaux C, Garbe C, Sovak MA, Chang I, Choong N, Hack SP, McArthur GA, Ribas A. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med. 2014;371(20):1867–1876. doi: 10.1056/NEJMoa1408868. [DOI] [PubMed] [Google Scholar]
- 69.Sánchez-Hernández I, Baquero P, Calleros L, Chiloeches A. Dual inhibition of V600EBRAF and the PI3K/AKT/mTOR pathway cooperates to induce apoptosis in melanoma cells through a MEK-independent mechanism. Cancer Lett. 2012;314(2):244–255. doi: 10.1016/j.canlet.2011.09.037. [DOI] [PubMed] [Google Scholar]
- 70.Khalili JS, Yu X, Wang J, Hayes BC, Davies MA, Lizee G, Esmaeli B, Woodman SE. Combination small molecule MEK and PI3K inhibition enhances uveal melanoma cell death in a mutant GNAQ-and GNA11-dependent manner. Clin Cancer Res. 2012;18(16):4345–4355. doi: 10.1158/1078-0432.CCR-11-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Atefi M, von Euw E, Attar N, Ng C, Chu C, Guo D, Nazarian R, Chmielowski B, Glaspy JA, Comin-Anduix B, Mischel PS, Lo RS, Ribas A. Reversing melanoma cross-resistance to BRAF and MEK inhibitors by co-targeting the AKT/mTOR pathway. PLoS ONE. 2011;6(12):e28973. doi: 10.1371/journal.pone.0028973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Corcoran RB, et al. TORC1 suppression predicts responsiveness to RAF and MEK inhibition in BRAF-mutant melanoma. Sci Transl Med. 2013;5(196):196ra98. doi: 10.1126/scitranslmed.3005753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tolcher AW, Patnaik A, Papadopoulos KP, Rasco DW, Becerra CR, Allred AJ, Orford K, Aktan G, Ferron-Brady G, Ibrahim N, Gauvin J, Motwani M, Cornfeld M. Phase I study of the MEK inhibitor trametinib in combination with the AKT inhibitor afuresertib in patients with solid tumors and multiple myeloma. Cancer Chemother Pharmacol. 2015;75(1):183–189. doi: 10.1007/s00280-014-2615-5. [DOI] [PubMed] [Google Scholar]
- 74.Bai X, Flaherty K. Targeted and immunotherapies in BRAF mutant melanoma: where we stand and what to expect. Br J Dermatol. 2021;185(2):253–262. doi: 10.1111/bjd.19394. [DOI] [PubMed] [Google Scholar]
- 75.Koya RC, Mok S, Otte N, Blacketor KJ, Comin-Anduix B, Tumeh PC, Minasyan A, Graham NA, Graeber TG, Chodon T, Ribas A. BRAF inhibitor vemurafenib improves the antitumor activity of adoptive cell immunotherapy. Cancer Res. 2012;72(16):3928–3937. doi: 10.1158/0008-5472.CAN-11-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frederick DT, Piris A, Cogdill AP, Cooper ZA, Lezcano C, Ferrone CR, Mitra D, Boni A, Newton LP, Liu C, Peng W, Sullivan RJ, Lawrence DP, Hodi FS, Overwijk WW, Lizée G, Murphy GF, Hwu P, Flaherty KT, Fisher DE, Wargo JA. BRAF inhibition is associated with enhanced melanoma antigen expression and a more favorable tumor microenvironment in patients with metastatic melanoma. Clin Cancer Res. 2013;19(5):1225–1231. doi: 10.1158/1078-0432.CCR-12-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Atefi M, Avramis E, Lassen A, Wong DJL, Robert L, Foulad D, Cerniglia M, Titz B, Chodon T, Graeber TG, Comin-Anduix B, Ribas A. Effects of MAPK and PI3K pathways on PD-L1 expression in melanoma. Clin Cancer Res. 2014;20(13):3446–3457. doi: 10.1158/1078-0432.CCR-13-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu L, Mayes PA, Eastman S, Shi H, Yadavilli S, Zhang T, Yang J, Seestaller-Wehr L, Zhang SY, Hopson C, Tsvetkov L, Jing J, Zhang S, Smothers J, Hoos A. The BRAF and MEK inhibitors dabrafenib and trametinib: effects on immune function and in combination with immunomodulatory antibodies targeting PD-1, PD-L1, and CTLA-4. Clin Cancer Res. 2015;21(7):1639–1651. doi: 10.1158/1078-0432.CCR-14-2339. [DOI] [PubMed] [Google Scholar]
- 79.Puzanov I. Combining targeted and immunotherapy: BRAF inhibitor dabrafenib (D)±the MEK inhibitor trametinib (T) in combination with ipilimumab (Ipi) for V600E/K mutation-positive unresectable or metastatic melanoma (MM) J Transl Med. 2015;13(1):1–2. [Google Scholar]
- 80.Ribas A, Hodi FS, Callahan M, Konto C, Wolchok J. Hepatotoxicity with combination of vemurafenib and ipilimumab. N Engl J Med. 2013;368(14):1365–1366. doi: 10.1056/NEJMc1302338. [DOI] [PubMed] [Google Scholar]
- 81.Paraiso KH, et al. The HSP90 inhibitor XL888 overcomes BRAF inhibitor resistance mediated through diverse mechanisms. Clin Cancer Res. 2012;18(9):2502–2514. doi: 10.1158/1078-0432.CCR-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wei H, Guan YD, Zhang LX, Liu S, Lu AP, Cheng Y, Cao DS. A combinatorial target screening strategy for deorphaning macromolecular targets of natural product. Eur J Med Chem. 2020;204:112644. doi: 10.1016/j.ejmech.2020.112644. [DOI] [PubMed] [Google Scholar]
- 83.Li H, Liu J, Xiao X, Sun S, Zhang H, Zhang Y, Zhou W, Zhang B, Roy M, Liu H, Ye M, Wang Z, Liu-Smith F, Liu J. A novel aptamer LL4A specifically targets vemurafenib-resistant melanoma through binding to the CD63 protein. Mol Ther Nucleic Acids. 2019;18:727–738. doi: 10.1016/j.omtn.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cook FA, Cook SJ. Inhibition of RAF dimers: it takes two to tango. Biochem Soc Trans. 2021;49(1):237–251. doi: 10.1042/BST20200485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Noeparast A, Giron P, de Brakeleer S, Eggermont C, de Ridder U, Teugels E, de Grève J. Type II RAF inhibitor causes superior ERK pathway suppression compared to type I RAF inhibitor in cells expressing different BRAF mutant types recurrently found in lung cancer. Oncotarget. 2018;9(22):16110–16123. doi: 10.18632/oncotarget.24576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jin T, Lavoie H, Sahmi M, David M, Hilt C, Hammell A, Therrien M. RAF inhibitors promote RAS-RAF interaction by allosterically disrupting RAF autoinhibition. Nat Commun. 2017;8(1):1–16. doi: 10.1038/s41467-017-01274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Basile KJ, le K, Hartsough EJ, Aplin AE. Inhibition of mutant BRAF splice variant signaling by next-generation, selective RAF inhibitors. Pigment Cell Melanoma Res. 2014;27(3):479–484. doi: 10.1111/pcmr.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yao Z, Gao Y, Su W, Yaeger R, Tao J, Na N, Zhang Y, Zhang C, Rymar A, Tao A, Timaul NM, Mcgriskin R, Outmezguine NA, Zhao HY, Chang Q, Qeriqi B, Barbacid M, de Stanchina E, Hyman DM, Bollag G, Rosen N. RAF inhibitor PLX8394 selectively disrupts BRAF dimers and RAS-independent BRAF-mutant-driven signaling. Nat Med. 2019;25(2):284–291. doi: 10.1038/s41591-018-0274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Botton T, et al. Genetic heterogeneity of BRAF fusion kinases in melanoma affects drug responses. Cell Rep. 2019;29(3):573–588. e7. doi: 10.1016/j.celrep.2019.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moriceau G, Hugo W, Hong A, Shi H, Kong X, Yu CC, Koya RC, Samatar AA, Khanlou N, Braun J, Ruchalski K, Seifert H, Larkin J, Dahlman KB, Johnson DB, Algazi A, Sosman JA, Ribas A, Lo RS. Tunable-combinatorial mechanisms of acquired resistance limit the efficacy of BRAF/MEK cotargeting but result in melanoma drug addiction. Cancer Cell. 2015;27(2):240–256. doi: 10.1016/j.ccell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Algazi AP, Othus M, Daud AI, Lo RS, Mehnert JM, Truong TG, Conry R, Kendra K, Doolittle GC, Clark JI, Messino MJ, Moore DF, Jr, Lao C, Faller BA, Govindarajan R, Harker-Murray A, Dreisbach L, Moon J, Grossmann KF, Ribas A. Continuous versus intermittent BRAF and MEK inhibition in patients with BRAF-mutated melanoma: a randomized phase 2 trial. Nat Med. 2020;26(10):1564–1568. doi: 10.1038/s41591-020-1060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Corcoran RB, André T, Atreya CE, Schellens JHM, Yoshino T, Bendell JC, Hollebecque A, McRee AJ, Siena S, Middleton G, Muro K, Gordon MS, Tabernero J, Yaeger R, O'Dwyer PJ, Humblet Y, de Vos F, Jung AS, Brase JC, Jaeger S, Bettinger S, Mookerjee B, Rangwala F, van Cutsem E. Combined BRAF, EGFR, and MEK inhibition in patients with BRAFV600E-mutant colorectal cancer. Cancer Discov. 2018;8(4):428–443. doi: 10.1158/2159-8290.CD-17-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ebi H. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition. Ann Oncol. 2012;23:xi41. doi: 10.1016/S0923-7534(20)32033-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nichols RJ, Haderk F, Stahlhut C, Schulze CJ, Hemmati G, Wildes D, Tzitzilonis C, Mordec K, Marquez A, Romero J, Hsieh T, Zaman A, Olivas V, McCoach C, Blakely CM, Wang Z, Kiss G, Koltun ES, Gill AL, Singh M, Goldsmith MA, Smith JAM, Bivona TG. RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1-and RAS-driven cancers. Nat Cell Biol. 2018;20(9):1064–1073. doi: 10.1038/s41556-018-0169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liau NP, et al. Negative regulation of RAF kinase activity by ATP is overcome by 14-3-3-induced dimerization. Nat Struct Mol Biol. 2020;27(2):134–141. doi: 10.1038/s41594-019-0365-0. [DOI] [PubMed] [Google Scholar]
- 96.Martín-Acosta P, Xiao X. PROTACs to address the challenges facing small molecule inhibitors. Eur J Med Chem. 2020;210:112993. doi: 10.1016/j.ejmech.2020.112993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang C, Zhang Y, Xing D, Zhang R. PROTACs technology for targeting non-oncoproteins: advances and perspectives. Bioorg Chem. 2021:105109. [DOI] [PubMed]
- 98.Alabi S, et al. Mutant-selective degradation by BRAF-targeting PROTACs. Nat Commun. 2021;12(1):1–11. doi: 10.1038/s41467-021-21159-7. [DOI] [PMC free article] [PubMed] [Google Scholar]