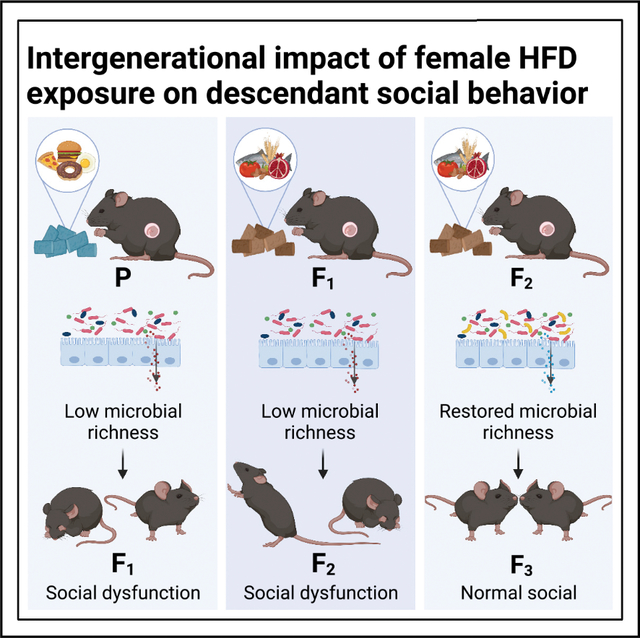
SUMMARY
Dysbiosis of the maternal gut microbiome during pregnancy is associated with adverse neurodevelopmental outcomes. We previously showed that maternal high-fat diet (MHFD) in mice induces gut dysbiosis, social dysfunction, and underlying synaptic plasticity deficits in male offspring (F1). Here, we reason that, if HFD-mediated changes in maternal gut microbiota drive offspring social deficits, then MHFD-induced dysbiosis in F1 female MHFD offspring would likewise impair F2 social behavior. Metataxonomic sequencing reveals reduced microbial richness among female F1 MHFD offspring. Despite recovery of microbial richness among MHFD-descendant F2 mice, they display social dysfunction. Post-weaning Limosilactobacillus reuteri treatment increases the abundance of short-chain fatty acid-producing taxa and rescues MHFD-descendant F2 social deficits. L. reuteri exerts a sexually dimorphic impact on gut microbiota configuration, increasing discriminant taxa between female cohorts. Collectively, these results show multigenerational impacts of HFD-induced dysbiosis in the maternal lineage and highlight the potential of maternal microbiome-targeted interventions for neurodevelopmental disorders.
In brief
Di Gesù, Matz et al. find that maternal high-fat diet results in intergenerational (F1, F2), but not transgenerational (F3), descendant social dysfunction. Metataxonomic sequencing and fecal microbiota transplant data implicate dysbiosis of the maternal gut microbiome in intergenerational social deficits, which can be restored by post-weaning Limosilactobacillus reuteri-6475 supplementation.
Graphical Abstract
INTRODUCTION
Gut microbiota are emerging as significant contributors to healthy brain function, as well as neurodysfunction and neurological disease (Buffington et al., 2021; Hertz-Picciotto et al., 2018; Lopez-Aranda et al., 2021; Morais et al., 2021; Nagpal and Cryan, 2021; Parenti et al., 2020; Rosenberg et al., 2007). Early-life neurodevelopment is particularly vulnerable to the adverse effects of disruption of the gut microbial community, or “dysbiosis” (Champagne-Jorgensen et al., 2020; Coley and Hsiao, 2021; Oliphant et al., 2021), as demonstrated by recent findings that dysbiosis of the maternal gut microbiome during pregnancy caused by infection or antibiotic-mediated microbial depletion alters fetal neurodevelopment, contributing to abnormal brain structure and function underlying maladaptive, autism-like behaviors in offspring (Di Gesu et al., 2021; Kim et al., 2017, 2022; Vuong et al., 2020). Notably, gut microbiome composition is driven primarily by diet (Bisanz et al., 2019; Carmody et al., 2015; David et al., 2014; Gacesa et al., 2022), and Western diet (Kopp, 2019), with high refined carbohydrate and fat but low fermentable fiber content, is increasingly implicated in the growing burden of non-communicable chronic disorders, including neuropsychiatric diseases (Blaser, 2017; Sonnenburg and Sonnenburg, 2019). Western diet is likewise implicated in the growing prevalence of metabolic disorders and obesity (Franks and McCarthy, 2016; Klurfeld and Kritchevsky, 1986; Kopp, 2019), and epidemiological studies identify maternal obesity as a significant risk factor for neurodevelopmental disorders, including autism spectrum disorder (ASD) (Kong et al., 2018; Li et al., 2016a, 2016b; Panjwani et al., 2019). Whether Western diet-induced dysbiosis of the maternal gut microbiome and the corresponding disruption of its remodeling during pregnancy (Gohir et al., 2015, 2019) are causally related to behavioral phenotypes and the underlying neuropathology in offspring remains unclear, as does the potential for diet-induced dysbiosis in a single generation to affect neurodevelopment and behavior across multiple descendant generations.
Working with both human populations and preclinical animal models, we and others have shown that high-fat, low-fiber diets drive functional, disease-associated changes in mammalian gut microbial ecology—generally characterized by an overall decrease in microbial (alpha) diversity and accompanying loss of beneficial functionalities afforded to the host by a complex microbial ecosystem (Afroz et al., 2021; Carmody et al., 2015; Di Gesu et al., 2021; Turnbaugh et al., 2008). Yet, the precise mechanisms mediating the relationship between westernized maternal diet and offspring neurobehavioral outcomes remains unestablished. Intriguingly, a recent study including 213 mother-child dyads found that alpha diversity of the prenatal maternal gut microbiome was predictive of child behavioral health outcomes at age 2 years (Dawson et al., 2021). Specifically, the abundance of short-chain fatty acid (SCFA)-producing bacterial taxa from families Lachnospiraceae and Ruminococcaceae in the maternal prenatal microbiome positively correlated with normative behavior in children and healthy maternal diet. SCFAs are essential microbially-derived metabolites that are synthesized through degradation of dietary plant fiber (i.e., microbiota-accessible carbohydrates [MACs]). SCFAs modulate activity of the host nervous and immune systems (Erny et al., 2021) and regulate histone deacetylase activity (Dupraz et al., 2021), while antibiotic- and diet-driven reduction in SCFA production is associated with disease pathology (reviewed in Dalile et al., 2019; O’Riordan et al., 2022). Notably, low-fiber diet-induced loss of microbial richness has been shown to compound over generations. Successive generations of low MAC diet consumption led to the extinction of community members that could not be restored by reintroduction of dietary MACs alone, instead requiring active restoration of negatively selected taxa (Sonnenburg et al., 2016). Western diet-induced dysbiosis of the maternal gut microbiome during pregnancy and lactation in a single generation could thereby drive a sustained negative impact on neurodevelopment and behavior across multiple descendant generations.
Existing epidemiological studies on the multigenerational effects of maternal nutritional status during pregnancy primarily focus on populations affected by famine. These studies have identified specific epigenetic modifications that confer heightened transgenerational risk for neuropsychiatric disorders, including schizophrenia (Hoek et al., 1998; St Clair et al., 2005). Multigenerational clinical studies of maternal overnutrition mostly focus on descendant metabolic metrics (Campisano et al., 2019; Chang et al., 2019; Cirillo et al., 2021; Dudley et al., 2011; Friedman, 2018), while preclinical studies of parental (P) overnutrition largely follow the paternal lineage, investigating the role of germ-line reprogramming (Rodgers et al., 2015; Sarker et al., 2018), leaving complexities presented by the mother-offspring dyad relatively unexplored. However, given that the pathology underlying neurodevelopmental disorders begins in utero (Courchesne et al., 2020; de la Torre-Ubieta et al., 2016; Usui et al., 2021; Willsey et al., 2013), it is critical to elucidate the complex mechanisms by which maternal diet impacts fetal neurodevelopment (reviewed in Di Gesu et al., 2021). Working in a preclinical mouse model for maternal diet-induced obesity (Buffington et al., 2016), we recently demonstrated that maternal high-fat diet (MHFD) induced functional changes in gut microbiome composition as well as autism-like social deficits and underlying changes in synaptic plasticity in dopaminergic neurons within the ventral tegmental area—a key locus in the social reward circuit (Bariselli et al., 2018; Gunaydin et al., 2014; Hung et al., 2017) —of male offspring. Yet, the impact of MHFD on female offspring gut microbiome composition and, relatedly, the influence of ancestral HFD-induced dysbiosis in the maternal lineage on descendant brain function and behavior beyond the first generation (F1) remain completely unexplored.
Given that MHFD-induced dysbiosis of the gut microbiome is vertically transmitted to F1 offspring (Buffington et al., 2016; Chu et al., 2016; Ma et al., 2014), we hypothesized that social deficits would be observed in F2 offspring, even in the absence of direct exposure to HFD. To address this hypothesis, we took an inter-disciplinary approach combining 16S ribosomal RNA (rRNA) gene amplicon (metataxonomic) sequencing, behavioral neuroscience, fecal microbiota transplants (FMTs) in gnotobiotic recipients, and anaerobic microbiology. As predicted, we observed dysbiosis of the gut microbiome in F1 MHFD-descendant females and social dysfunction in their offspring (F2), despite recovery of F2 gut microbial richness. FMTs from MHFD-descendant F2 donors were sufficient to rescue baseline social deficits in gnotobiotic mice, and we observed normal social behavior in F3 MHFD descendants. Consistent with our finding in MHFD F1 male offspring, F2 social deficits were rescued by treatment with the probiotic Limosilactobacillus reuteri-6475. Surprisingly, we observed a sexually dimorphic impact of L. reuteri on host microbiota configuration, reflected by an increased number of discriminant taxa between F2 female cohorts, revealing a relative malleability of the female gut microbiome in response to probiotic intervention, which may inform future therapeutic development.
RESULTS
MHFD-descendant F2 mice display social dysfunction despite no direct exposure to MHFD
We and others previously reported detrimental effects of MHFD exposure during gestation and lactation on social behavior in first-generation (F1) offspring and identified underlying deficits in the mesocorticolimbic dopaminergic social reward circuit of MHFD F1 males (Bordeleau et al., 2021; Buffington et al., 2016; Kang et al., 2014). Here, we sought to determine whether social deficits observed in the F1 generation were recapitulated in F2, even in the absence of direct exposure to MHFD. To generate F2 (Figure 1A), we fed C57Bl/6N females a regular diet (RD) or HFD for 6 weeks prior to mating to produce the F1 generation. Total weight and lean mass of P generation females did not differ between diet cohorts at time of mating (Figures S1A and S1B), but fat mass was significantly increased in the HFD-fed group as early as 1 week on diet (Figure S1C). Shifts in gut community structure were apparent within 1 week on HFD and sustained at 4 weeks (Figure S1D), while alpha diversity metrics, including observed operational taxonomic units (OTUs) (Figure S1E) and the Shannon index (Figure S1F), and the ratio of the two most abundant phyla occupying the mammalian gut microbiome, Firmicutes:Bacteroidota (Figure S1G), differed significantly between RD- versus HFD-fed females by 4 weeks on diet. We maintained the P generation females on their assigned prepregnancy diet throughout pregnancy and lactation, sires had minimal exposure to HFD prior to F1 conception, and all F1 mice were weaned onto RD chow, regardless of maternal diet. We collected stool samples from MRD and MHFD F1 females to characterize their gut microbial ecology using 16S rRNA gene amplicon (metataxonomic) sequencing, then bred MRD control (N = 5 pairs) and MHFD (N = 4 pairs) F1 offspring to produce MRD- and MHFD-descendant F2 generation mice, respectively (Figure 1A). We caged the F2 mice according to sex and maternal lineage at weaning, then assessed social and anxiety-like behavior in 7- to 10-week-old male and female MRD and MHFD F2 descendants. We collected stool samples from the respective F2 cohorts at the time of behavior (Figure 1B).
Figure 1. MHFD-descendant F2 offspring display social dysfunction.
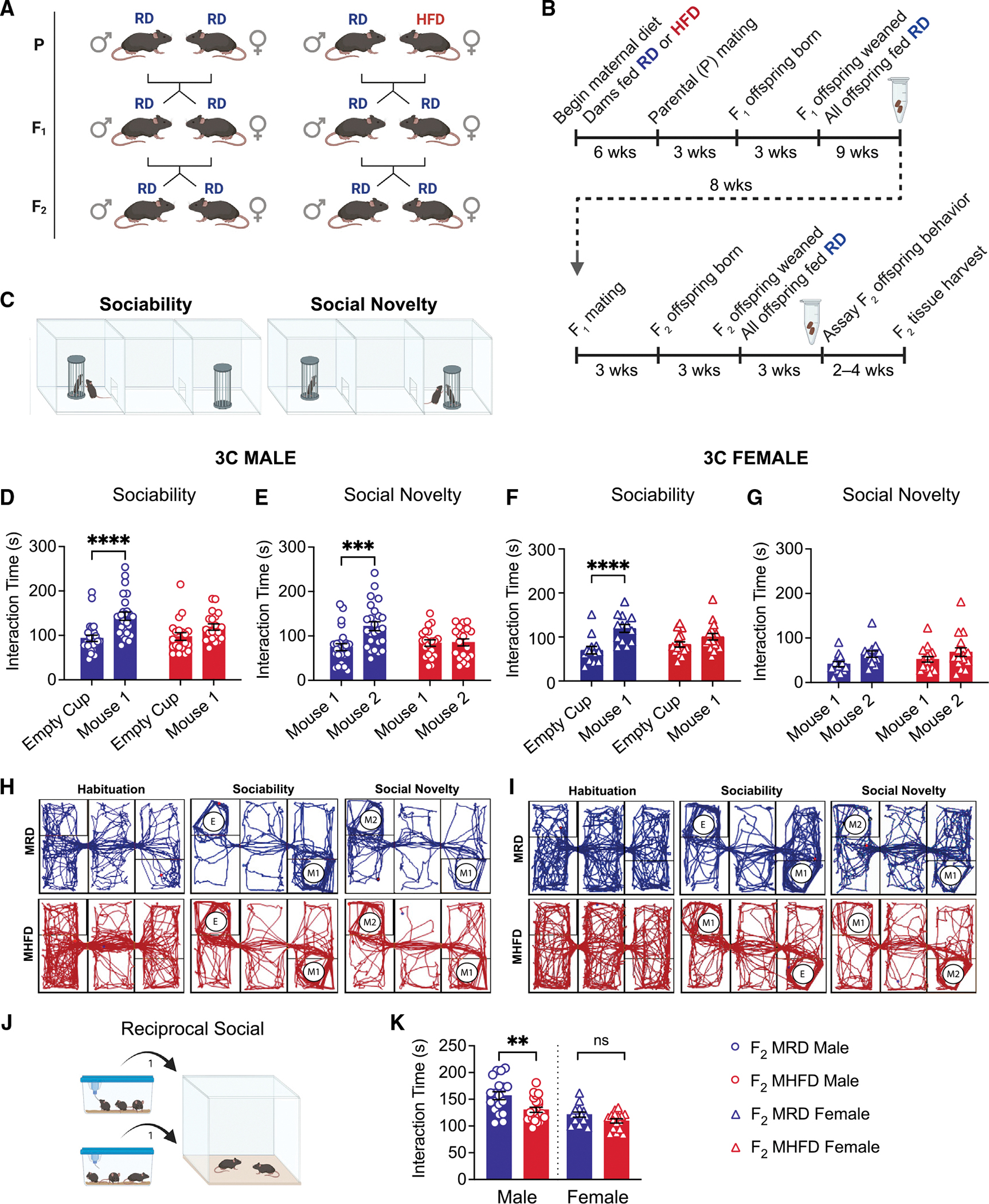
(A) Schematic of breeding scheme to produce MRD- and MHFD-descendant F2 generations with HFD exposure indicated.
(B) Timeline of mating and behavior. Eppendorf tubes represent time of stool sample collection.
(C) Schematic of Crawley’s 3 chamber (3C) test for sociability and preference for social novelty.
(D and E) (D) MHFD F2 male offspring show impaired sociability (MRD: t(86) = 4.517, p < 0.0001; MHFD: t(86) = 1.735, p = 0.1651) and (E) preference for social novelty (MRD: t(43) = 4.208, p = 0.0003; MHFD: t(43) = 0.1159, p = 0.9916) compared with MRD F2 males.
(F) MRD F2 females show a strong preference in 3C sociability (MRD: t(28) = 5.406, p < 0.0001), with MHFD F2 female offspring showing mild deficits in sociability (MHFD: t(28) = 2.181, p = 0.0741).
(G–I) (G) Neither MRD nor MHFD F2 female offspring showed a statistically significant preference for social novelty (MRD: t(28) = 1.934, p = 0.1224; MHFD: t(28) = 1.613, p = 0.222). Representative MRD-descendant (blue) or MHFD-descendant (red) F2 offspring 3C track plots for (H) males and (I) females.
(J) Reciprocal social interaction schematic.
(K) Comparison of reciprocal social interaction times revealed decreased interaction among MHFD-descendant F2 male, but not female, stranger pairs (male MHFD versus MRD: t(39) = 3.103, p = 0.0036; female MHFD versus MRD: t(28) = 1.918, p = 0.0653). Bar graphs show mean ± SEM with individual data points representing biological replicates. (D–G) N = 13–25 subjects per group; (K) N = 14–22 pairs per group.
We assessed MRD- versus MHFD-descendant F2 cohort social behavior using Crawley’s three chamber (3C) test for sociability and preference for social novelty (Figure 1C). We analyzed male and female behavior separately to allow for identification of sexually dimorphic phenotypes, given the disproportionate prevalence of ASD among human males (Constantino and Charman, 2012) and the need to deepen our understanding of how biological sex contributes to ASD risk. Consistent with our hypothesis, while MRD-descendant F2 males showed strong sociability and preference for social novelty, MHFD-descendant F2 males were significantly impaired (Figures 1D, 1E, 1H, S2A, S2B, S2E, and S2F). Conversely, while MHFD-descendant F2 females displayed a mild reduction in sociability compared with their F2 MRD counterparts (Figures 1F, 1I, S2C, and S2G), neither MRD nor MHFD F2 females showed a statistically significant preference for interaction with a novel over a familiar mouse in the social novelty phase (Figures 1G, 1I, S2D, and S2H).
We next measured reciprocal social interaction between sex-matched stranger pairs (Figure 1J). Consistent with our 3C results, MHFD-descendant F2 male pairs spent significantly less time interacting than MRD-descendant F2 male pairs, while no difference in interaction time was observed between pairs of MRD- versus MHFD-descendant F2 females (Figure 1K). MHFD female F2 offspring did show a reduction in contact duration, while no differences in number of contacts were observed for either sex (Figures S2I and S2J). Taken together, these data identify a strong social dysfunction phenotype among MHFD-descendent F2 males, despite no direct exposure to MHFD. In contrast, we detected only a mild social dysfunction phenotype in F2 females and carefully consider this finding in the discussion and the limitations of the study.
While social dysfunction is a core behavioral symptom of ASD, a proportion of individuals with ASD have comorbid anxiety disorders and/or attention-deficit/hyperactivity disorder (ADHD) (Avni et al., 2018). We thus used the open-field test to assess hyperactivity and anxiety-like behavior in the F2 generation (Figure S3A). Neither cohort demonstrated changes in locomotor, as measured by distance traveled (Figure S3B) and speed (Figure S3C), or anxiety-like behavior, interpreted from time spent in the center of the open-field arena (Figure S3D), regardless of sex. Females displayed increased distance and speed compared with males, independent of diet. These results are consistent with data obtained during the habituation phase of the 3C task (Figure S3E), which showed no differences in locomotor activity driven by either sex or diet (Figures S3F and S3G). Thus, the social deficits in MHFD-descendant F2 males and females (Figures 1 and S3) are not due to hyperactivity or increased anxiety-like behavior.
MHFD-descendant F1 and F2 mice harbor a distinct gut microbiome composition, with recovery of richness in the F2 generation
Maternal gut microbiota play a critical role in modulating fetal brain development in ways that impact the function of behaviorally relevant neural circuits in adolescent and adult offspring (Di Gesu et al., 2021; Kim et al., 2017, 2022; Vuong et al., 2020). We previously showed that alterations in alpha and beta diversity of the HFD-fed adult female mouse microbiome are reflected in long-term gut dysbiosis in their male MHFD offspring (F1) (Buffington et al., 2016). Notably, however, we did not sequence the MHFD-descendant F1 female gut microbiome in prior work. Given that gut microbiota are vertically transmitted from mammalian mothers to their offspring (Ferretti et al., 2018), we hypothesized that (1) MHFD would similarly affect the gut microbiome composition of MHFD-descendant F1 females and, consequently, (2) changes observed in MHFD F1 females would be likewise reflected in the MHFD F2 generation. We thus performed 16S rRNA gene amplicon (metataxonomic) sequencing to characterize the microbial gut composition and community structure of MRD- versus MHFD-descendant F1 mothers and their offspring (F2).
We first assessed beta diversity—the variation of microbial communities between samples—of averaged rarefied 16S rRNA gene sequences detected in F1 and F2 fecal samples. Principal-coordinate analysis (PCoA) using unweighted UniFrac, which incorporates phylogenetic distances but does not account for the abundance of operational taxonomic units (OTUs), revealed statistically significant clusters (Figure 2A). Specifically, fecal microbial communities of F1 MHFD females clustered together and were distinct from both F1 and F2 MRD cohorts. F2 MHFD offspring communities likewise clustered according to cohort, but the MHFD-descendant F2 gut microbiota shifted away from that of the F1 MHFD cohort toward the F2 MRD cohort, demonstrated by partially overlapping 95% confidence intervals between the two groups. As expected, F1 and F2 MRD communities show low degrees of variation, appearing more similar to each other than the MHFD groups. Distinct clustering was not observed by weighted UniFrac, which accounts for the relative taxa abundance, but did reach statistical significance (Figure S4A).
Figure 2. F2 MHFD descendants harbor a distinct gut microbiome from F2 MRD descendants but show a partial recovery of richness.
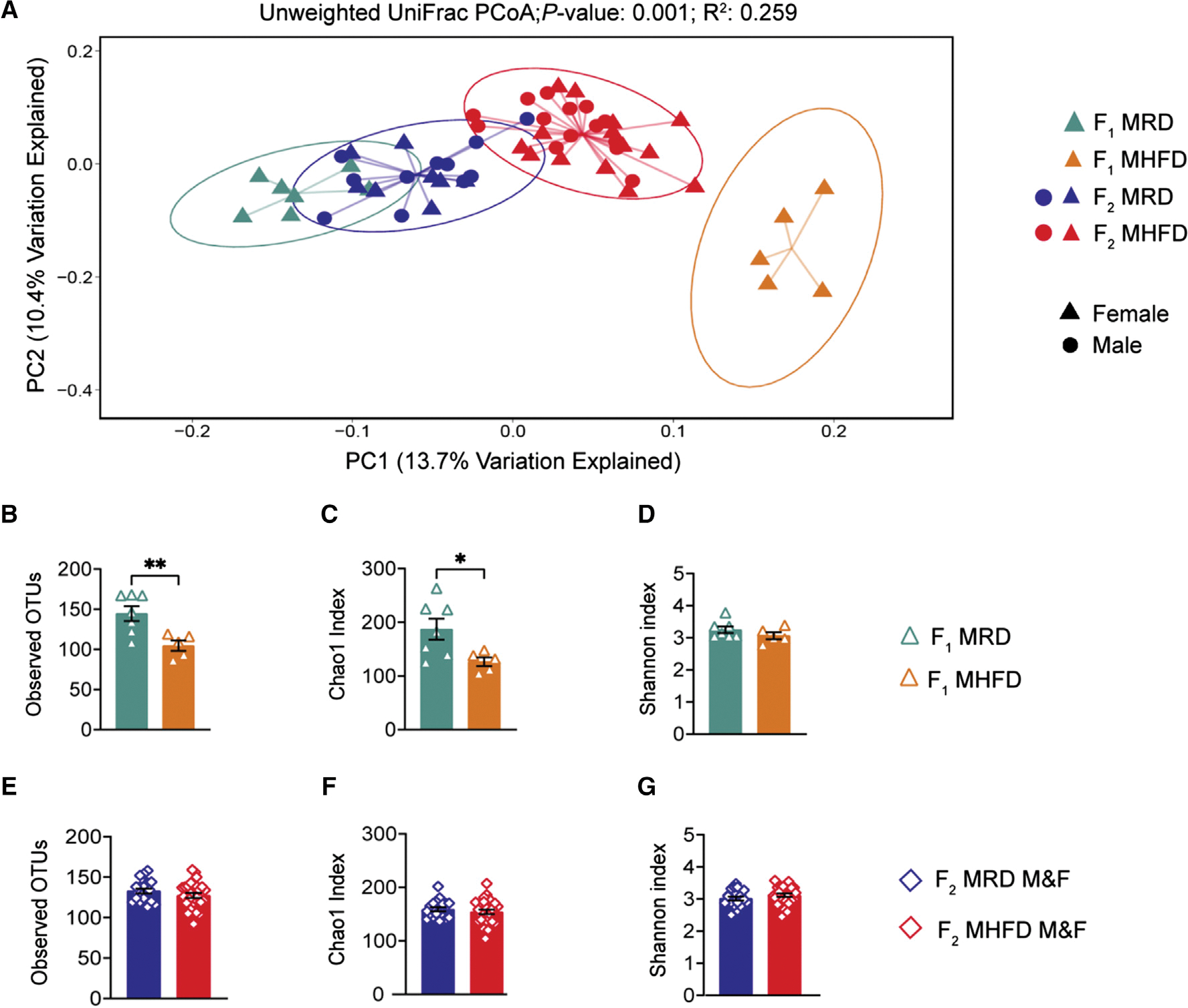
(A) Principal-coordinate analysis (PCoA) of unweighted UniFrac distances from the averaged rarefied 16S rRNA gene amplicon sequencing dataset (8,310 reads/sample; n = 1,000 rarefactions) revealed statistically significant clusters based on diet and generation (p = 0.001, R2 = 0.259).
(B–D) Alpha diversity metrics of F1 females used to generate F2 offspring revealed a statistically significant loss of microbial diversity in MHFD compared with MRD dams, as measured by observed OTUs (MHFD F1 versus MRD F1: t(10) = 3.272, p = 0.0084) and Chao1 index (MHFD F1 versus MRD F1: t(10) = 2.494, p = 0.0317). The Shannon diversity index did not differ between cohorts (MHFD F1 versus MRD F1: t(10) = 1.204, p = 0.2564).
(E–G) No significant differences in alpha diversity between F2 groups were observed, as measured by observed OTUs (MHFD F2 versus MRD F2: t(45) = 1.192, p = 0.2395), Chao1 index (MHFD F2 versus MRD F2: t(45) = 0.8097, p = 0.4224), and Shannon index (MHFD F2 versus MRD F2: t(45) = 1.274, p = 0.2093). Bar graphs show mean ± SEM with individual data points representing biological replicates. (A–G) N = 5–15 subjects per group.
Next, we assessed within-sample diversity (alpha diversity) by comparing observed OTU counts, Chao1 index, and Shannon index in F1 females (Figures 2B–2D). Consistent with our observation in the P generation HFD-fed dams, the average number of OTUs identified per sample, which reflects the richness of each microbial community, was significantly reduced in MHFD-descendant F1 females(Figure 2B). The Chao1 index, a richness estimator based on low-abundance OTUs, was likewise decreased in MHFD-descendant compared with MRD-descendant F1 females (Figure 2C), while the Shannon index, which accounts for both abundance and evenness, did not indicate a statistically significant difference between cohorts (Figure 2D). Surprisingly, when we analyzed the MRD versus MHFD F2 generation 16S sequencing dataset, however, no statistically significant differences were identified across any alpha diversity metric (Figures 2E–2G), revealing a recovery of microbial richness in the F2 generation. Further assessment of beta diversity by linear discriminant analysis (LDA) effect size (LEfSe) identified 19 differentially abundant genus-level taxa between maternal diets in the F1 generation (Figure 3A). Likewise, comparison of F2 offspring cohorts revealed statistically significant differences in the abundance of 20 taxa (Figure 3B), including enrichment in genera Faecalibacterium and Parabacteroides, with diminished abundance of genera Alistipes and family Prevotellaceae and Erysipelotrichaceae, between the MHFD- versus MRD-descendant F2 generation. Taken together, these results demonstrate a comparable effect of MHFD on the male and female F1 offspring gut microbiome that is only partially recapitulated in F2, and a potential role for low abundance species in driving phenotypic differences observed between F2 descendant groups.
Figure 3. LEfSe analysis indicates differentially abundant genus-level taxa between diet cohorts in F1 and F2 generations.
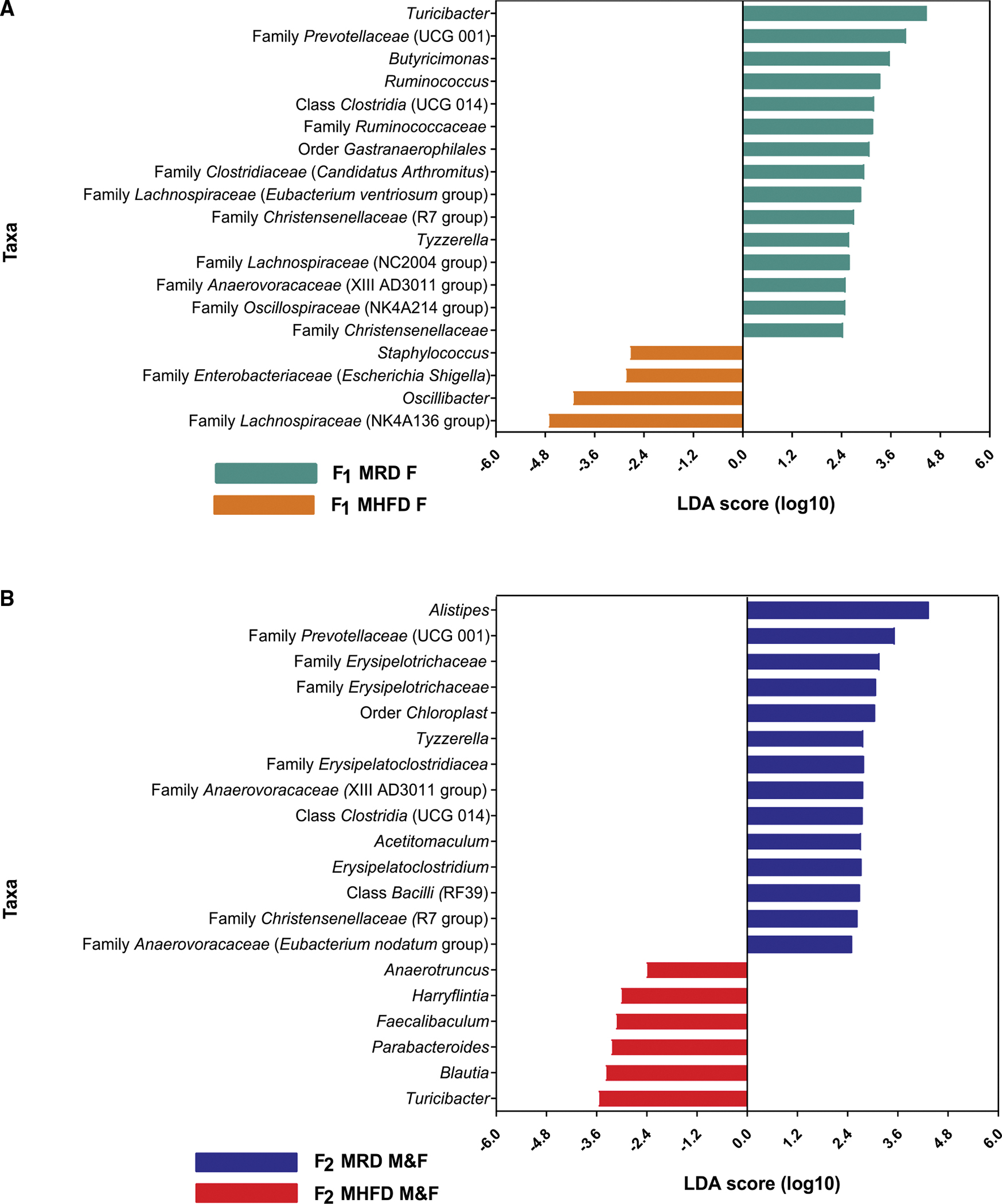
(A) Histogram of the LDA scores (log10) computed for genus-level taxa with differential abundance in MHFD and MRD F1 dams.
(B) Histogram of the LDA scores (log10) computed for genus-level taxa with differential abundance in MHFD and MRD F2 offspring. LDA, linear discriminant analysis; LEfSe, LDA effect size. (A and B) N = 5–15 subjects per group.
FMT from either MRD- or MHFD-descendant F2 donors rescues social dysfunction in formerly germ-free recipients
Gut microbiota metabolize dietary nutrients into bioactive metabolites that modulate host physiology, including the function of neural circuits underlying behavior (Buffington et al., 2021; Dodd et al., 2017; Williams et al., 2014). To establish the relationship between the functional composition of the F2 MHFD-descendant microbiome and their social deficits, we performed FMT from either MRD- or MHFD-descendent F2 donors into germ-free (GF) recipient mice. We chose to perform FMTs into GF recipients because they are the current gold standard for establishing the causality of microbiota-led effects on host behavioral outcomes (Nagpal and Cryan, 2021). Furthermore, we previously found that FMT from only MRD, but not MHFD, F1 male donors was sufficient to rescue social deficits in GF-recipient males (Buffington et al., 2016). Here, we transplanted juvenile male GF mice with anaerobically prepared fecal donor slurry prepared from stool isolated from MRD- or MHFD-descendant F2 males via oral gavage. We performed this study exclusively with male donors and recipients, given the robustness of the male social dysfunction phenotype. FMT recipients were housed by donor cohort in sterile caging and provided with sterile food and water for 5 weeks before behavioral analysis (Figure 4A). Consistent with previous reports (Buffington et al., 2016; Desbonnet et al., 2014), we observed impaired social behavior in age-matched, non-colonized GF control mice in the 3C assay (Figures 4B–4D and S5A–S5D). Remarkably, in contrast with our F1 results, FMT from either MRD- or MHFD-descendant F2 donors rescued GF social deficits in the 3C task, as indicated by a statistically significant social preference in both sociability and social novelty (Figures 4B–4D) and corroborated by automated time in chamber data (Figures S5A and S5B). Furthermore, GF hyperactivity in the 3C habituation phase and the open-field task were rescued in each recipient cohort (Figures S5E–S5H). Open-field time in center data was comparable between GF control and the two FMT-colonized cohorts (Figure S5I).
Figure 4. Fecal microbiota transplants from both F2 MRD and MHFD descendants rescue germ-free social deficits.
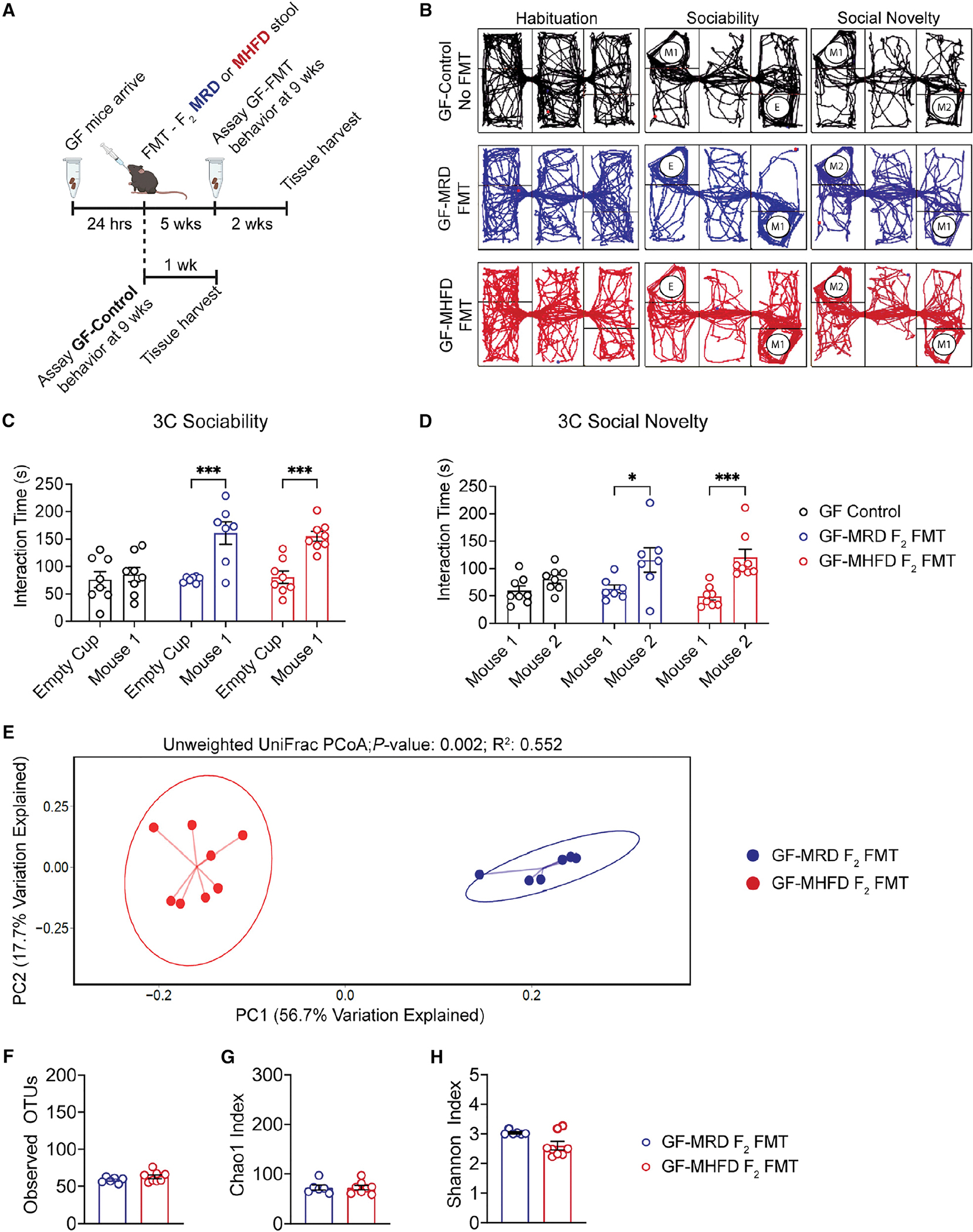
(A) Timeline of germ-free (GF) fecal microbiota transplant (FMT), stool collection for 16S rRNA gene sequencing, and behavioral analysis.
(B–D) (B) Representative 3C track plots for GF-Control (black), GF MRD-FMT (blue), or GF MHFD-FMT (red) males. 3C interaction times for (C) sociability (GF-Control: t(20) = 0.5364, p = 0.9349; MRD-FMT: t(20) = 4.688, p = 0.0004; MHFD-FMT: t(20) = 4.429, p = 0.0008) and (D) preference for social novelty (GF-Control: t(20) = 1.355, p = 0.4696; MRD-FMT: t(20) = 3.155, p = 0.0149; MHFD-FMT: t(20) = 4.558, p = 0.0006) showing social dysfunction in GF males is rescued by FMT from either MRD- or MHFD-descendant F2 donors. Bar graphs show mean ± SEM with individual data points.
(E) PCoA of unweighted UniFrac distances from the averaged rarefied 16S rRNA gene amplicon sequencing dataset (3,370 reads/sample; n = 1,000 rarefactions) revealed statistically significant clusters based on diet (p = 0.002, R2 = 0.552).
(F–H) Alpha diversity metrics did not differ between GF MRD-FMT or GF MHFD-FMT males, as measured by observed OTUs (GF MRD-FMT versus GF MHFD-FMT: t(12) = 1.391, p = 0.1895), Chao1 index (GF MRD-FMT versus GF MHFD-FMT: Mann-Whitney U = 22, p = 0.8518), and Shannon diversity index (GF MRD-FMT versus GF MHFD-FMT: Mann-Whitney U = 12, p = 0.1419). Bar graphs show mean ± SEM with individual data points representing biological replicates.
(C–H) N = 7–8 subjects per group.
While the assessment of beta diversity revealed statistically significant clusters between fecal microbial communities from GF mice colonized with either MRD- or MHFD-descendant F2 offspring microbiota (Figures 4E and S4B), no differences were observed in alpha diversity metrics between groups (Figures 4G and 4H), consistent with the results of alpha and beta diversity analysis in the F2 generation offspring 16S sequencing dataset (Figures 2A and 2E–2G). These data indicate that the adolescent state of the male MHFD F2 gut microbiome, although distinct from that of the MRD lineage, is not the primary cause of F2 male social deficits. Importantly, however, these data do not rule out the therapeutic potential of targeting the adolescent gut microbiome to resolve social deficits in MHFD descendants.
Post-weaning L. reuteri treatment rescues MHFD-descendant F2 social deficits
In our previous work, metagenomic whole-genome shotgun sequencing revealed a marked decrease in the abundance of the commensal species L. reuteri (Buffington et al., 2016) in the male F1 MHFD gut. Interestingly, L. reuteri had been shown to increase both peripheral and central oxytocin (Poutahidis et al., 2013), a hormone that regulates social behavior through modulating the mesolimbic dopaminergic reward system (Buffington et al., 2016; Hung et al., 2017). Precision treatment with a strain of L. reuteri isolated from human breastmilk, L. reuteri (ATCC PTA-6475), rescued social behavior in MHFD F1 males, as well as in multiple mouse models for ASD, including idiopathic and genetic models (Buffington et al., 2021; Sgritta et al., 2019). Here, we sought to determine whether L. reuteri could likewise rescue MHFD-descendant F2 social deficits.
Monogamous pairs of F1 MRD- or MHFD-descendant mice were bred to generate F2 offspring which, upon weaning, were administered L. reuteri in drinking water daily through the remainder of the experiment (Figure 5A). Consistent with our results in the MHFD-descendant F1 males, L. reuteri rescued MHFD-descendant F2 male social deficits in reciprocal social interaction (Figures 5B, S6I, and S6J) and the 3C task (Figures 5C, 5D, 5G, S6A, S6B, S6E, and S6F). Remarkably, we observed an unexpected and dramatic increase in social behavior among L. reuteri-treated MHFD-descendant F2 females, including a significant increase in reciprocal social interaction (Figures 5B, S6I, and S6J) and instatement of preference for social novelty in the 3C task (Figures 5F, 5H, S6C, and S6H), metrics which we found to be indistinguishable between untreated MRD- versus MHFD-descendant F2 females (Figures 1G, 1I, 1K, S2D, and S2H). 3C sociability was also rescued in the L. reuteri-treated MHFD-descendant F2 females (Figures 5E, 5H, S6C, and S6G). L. reuteri treatment did not impact locomotor (Figures S6K–S6N) or anxiety-like behavior (Figure S6O) in F2 descendants of either sex or lineage. Together, these results demonstrate that post-weaning L. reuteri treatment is sufficient to rescue social dysfunction in MHFD-descendant F2 males and stimulate social function in MHFD-descendant F2 females, thus providing further support for early adolescence as a viable therapeutic window for microbiota-targeted treatments for disorders of social dysfunction.
Figure 5. L. reuteri rescues social deficits in male F2 MHFD-descendant offspring and exerts differential effects on F2 MRD versus MHFD offspring gut microbiota composition.
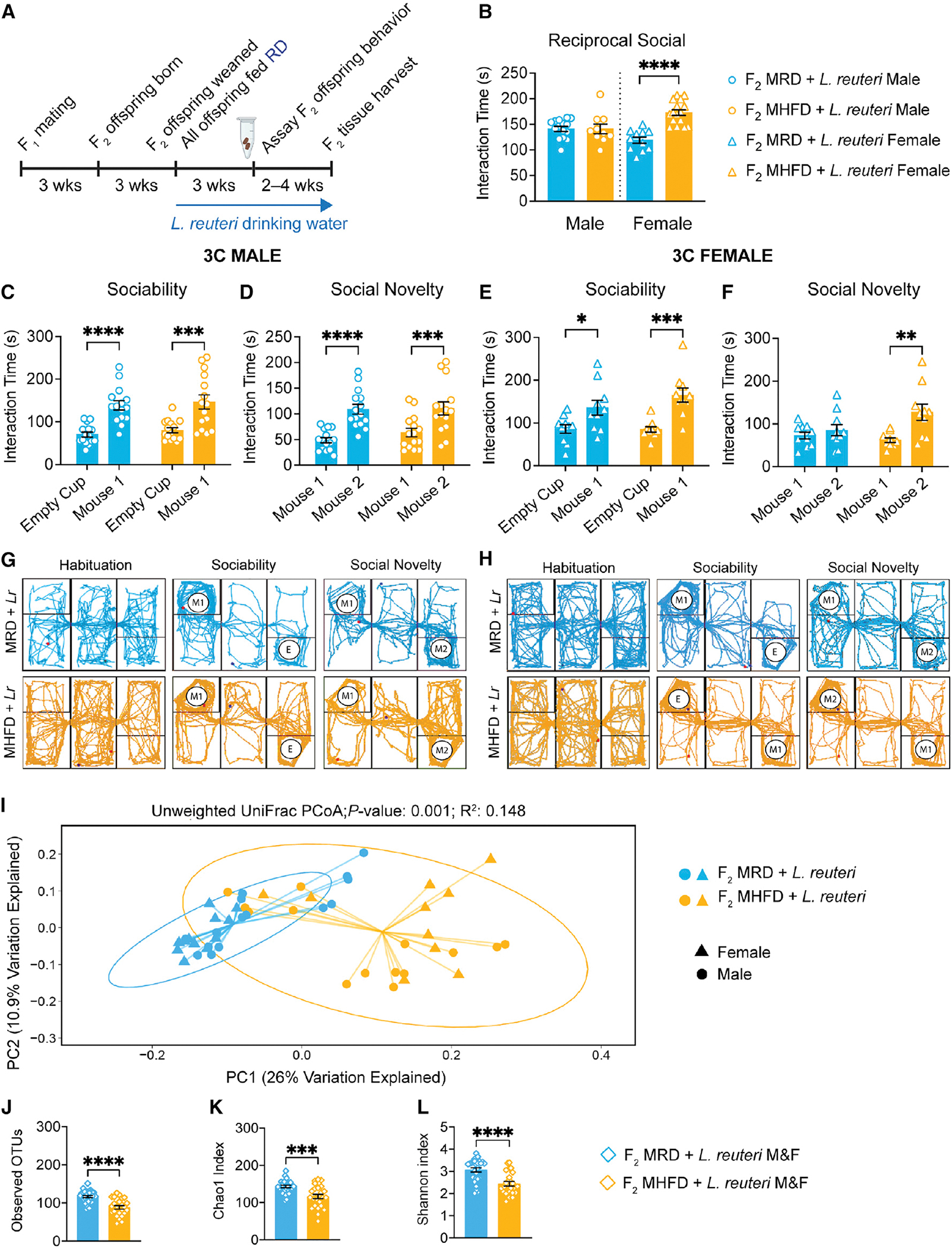
(A) Experimental schematic. Three monogamous MRD lineage or MHFD lineage F1 breeder cages were used to establish MRD- and MHFD-descendant F2 generations, respectively. Upon weaning at 3 weeks old, all mice were given drinking water containing 108 CFU/mL L. reuteri daily through behavior assessment.
(B) Analysis of the reciprocal social interaction times between pairs revealed no differences between males (male MHFD + L reuteri versus MRD + L. reuteri: t(22) = 0.01855, p = 0.9854), with increased interaction times between F2 MHFD + L. reuteri females compared with F2 MRD + L. reuteri females (female MHFD + L. reuteri versus MRD + L. reuteri: t(25) = 6.340, p < 0.0001).
(C and D) (C) F2 male MHFD + L. reuteri offspring show typical preference during sociability (MRD: t(56) = 4.515, p < 0.0001; MHFD: t(56) = 4.373, p = 0.0001) and (D) social novelty (MRD: t(28) = 5.472, p < 0.0001; MHFD: t(28) = 4.28, p = 0.0004).
(E–H) (E) Likewise, MHFD + L. reuteri F2 female offspring show a statistically significant preference during sociability (MRD: t(36) = 2.633, p = 0.0246; MHFD: t(36) = 4.253, p = 0.0003) and (F) preference for social novelty (MRD: t(18) = 0.7226, p = 0.7228; MHFD: t(18) = 3.763, p = 0.0028). Representative MRD + L. reuteri (light blue) or MHFD + L. reuteri (light orange) track plots for (G) males and (H) females, respectively. Bar graphs show mean ± SEM with individual data points.
(I) PCoA of unweighted UniFrac distances from the averaged rarefied 16S rRNA gene amplicon sequencing dataset (5,508 reads/sample; n = 1,000 rarefactions) revealed statistically significant clusters based on diet (p = 0.001, R2 = 0.148).
(J–L) L. reuteri administration differentially affects MRD and MHFD F2 gut microbiome alpha diversity, as measured by observed OTUs (MHFD F2 + L. reuteri versus MRD F2 + L. reuteri: t(50) = 5.285, p < 0.0001), Chao1 index (MHFD F2 + L. reuteri versus MRD F2 + L. reuteri: t(52) = 4.143, p = 0.0001), and Shannon index (MHFD F2 + L. reuteri versus MRD F2 + L. reuteri: t(50) = 4.436, p < 0.0001). Bar graphs show mean ± SEM with individual data points representing biological replicates. B, N = 10–16 pairs per group; (C–F) N = 10–15 subjects per group; (I–L) N = 24–28 subjects per group.
L. reuteri exerts differential impacts on the community ecology of the MRD- versus MHFD-descendant F2 gut microbiota
We next assessed whether L. reuteri administration, which restores sociability and preference for social novelty in MHFD F2 mice, also alters microbiota composition in the F2 generation. L. reuteri-6475 is a moderate producer of the antimicrobial metabolite reuterin, which is generated through anaerobic fermentation of glycerol. Reuterin has been shown to reduce the abundance of a subset of species in the gut, including opportunistic pathogens, such as enterohemorrhagic Escherichia coli, that cause hemolytic-uremic syndrome (Eaton et al., 2011), and to prevent necrotizing enterocolitis in premature infants (Shelby et al., 2021). Remarkably, we observed a differential impact of L. reuteri treatment on both alpha and beta diversity in MRD versus MHFD F2 descendants (Figures 5 and 6). Alpha diversity metrics observed OTU counts, Chao1 index, and Shannon index (Figures 5J–5L) each showed significant differences between MRD- versus MHFD-descendant F2 cohorts. The distinct effect of L. reuteri supplementation on gut microbial ecology between groups was also shown by changes in the relative abundance of 36 taxa identified by LEfSe (Figure 6A). Notably, we observed a significant increase in the proportion of SCFA-producing taxa, such as Limosilactobacillus, Ligilactobacillus, and Ruminococcus genera among L. reuteri-treated F2 MHFD descendants, compared with their L. reuteri-treated F2 MRD-descendant counterparts. PCoA of unweighted and weighted UniFrac dissimilarities revealed statistically significant clusters of fecal microbial communities between maternal lineage cohorts (Figures 5I and S4C); however, visually these clusters overlapped. We thus stratified the microbiome sequencing data by sex and found a sexually dimorphic impact of L. reuteri on host microbiome composition, with 38 discriminant taxa identified between MRD- versus MHFD-descendant F2 females and only 13 between male cohorts (Figure S7). Taken together, these data identify a substantial impact of L. reuteri treatment on the overall composition of the host gut microbiome, thus challenging the idea that the effects of L. reuteri on host brain and behavior are independent of its impact on the native microbial community and, moreover, revealing a relative instability of the MHFD-descendant gut microbiome, a state that could paradoxically heighten both disease susceptibility as well as therapeutic efficacy.
Figure 6. LEfSe analysis indicates differentially abundant genus-level taxa between diet lineage in L. reuteri-treated F2 groups.
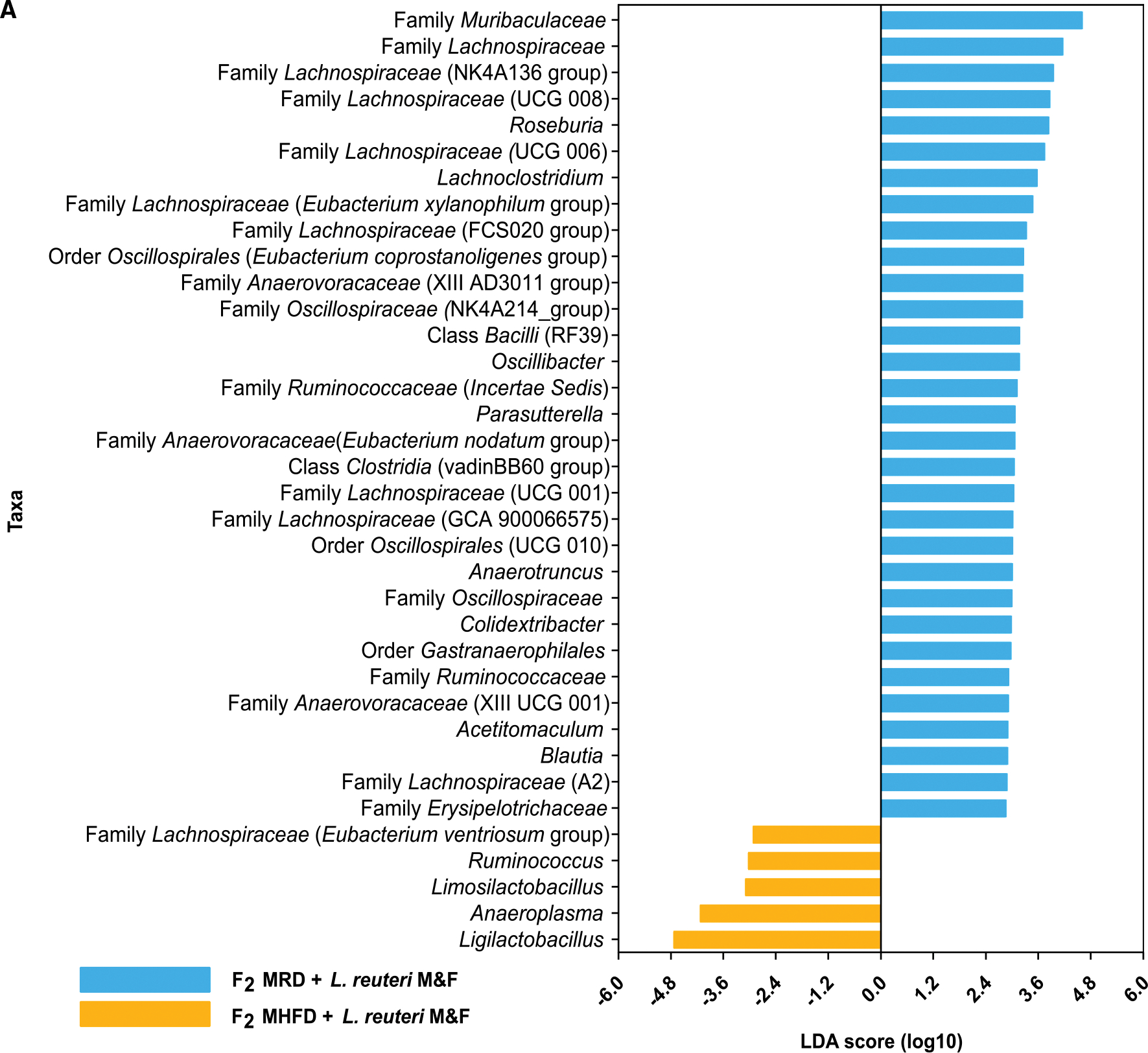
Histogram of the LDA scores (log10) computed for genus-level taxa with differential abundance in MHFD and MRD F2 offspring treated with L. reuteri. LDA, linear discriminant analysis; LEfSe, LDA effect size. N = 24–28 subjects per group.
F3 generation MHFD descendants display normal social behavior
Having found that MHFD-descendant F2 gut microbiota are sufficient to rescue baseline social deficits in GF FMT recipients, despite clear social dysfunction in the F2 males themselves, we hypothesized that HFD-associated dysbiosis of the maternal gut microbiome is causal in descendant social deficits, consistent with early-life pathology underlying neurodevelopmental disorders (de la Torre-Ubieta et al., 2016). Accordingly, we reasoned that MHFD-descendant F3 males would display neurotypical social behavior given the partial restoration, including the increase in microbial richness, of the MHFD-descendant F2 (F3 maternal) gut microbiome.
F3 subjects were generated by monogamous breeding of MRD- and MHFD-descendant F2 mice and weaned into litter-specific cages separated by sex and maternal lineage at 3 weeks of age (Figure 7A). Consistent with our hypothesis, we observed comparable reciprocal social interaction between maternal diet lineages (Figures 7B, S8I, and S8J) and statistically significant social preferences in MHFD-descendant F3 mice in the 3C task (Figures 7C–7H and S8A–S8H). We did not observe any differences in locomotor (Figures S8K–S8N) or anxiety-like behavior (Figure S8O) between the F3 cohorts of either sex. Collectively, these data demonstrate that the effects of HFD-induced dysbiosis in the maternal lineage can be resolved within two generations upon restoration of a healthy diet.
Figure 7. Neurotypical social behavior is observed in the MHFD-descendant F3 generation.
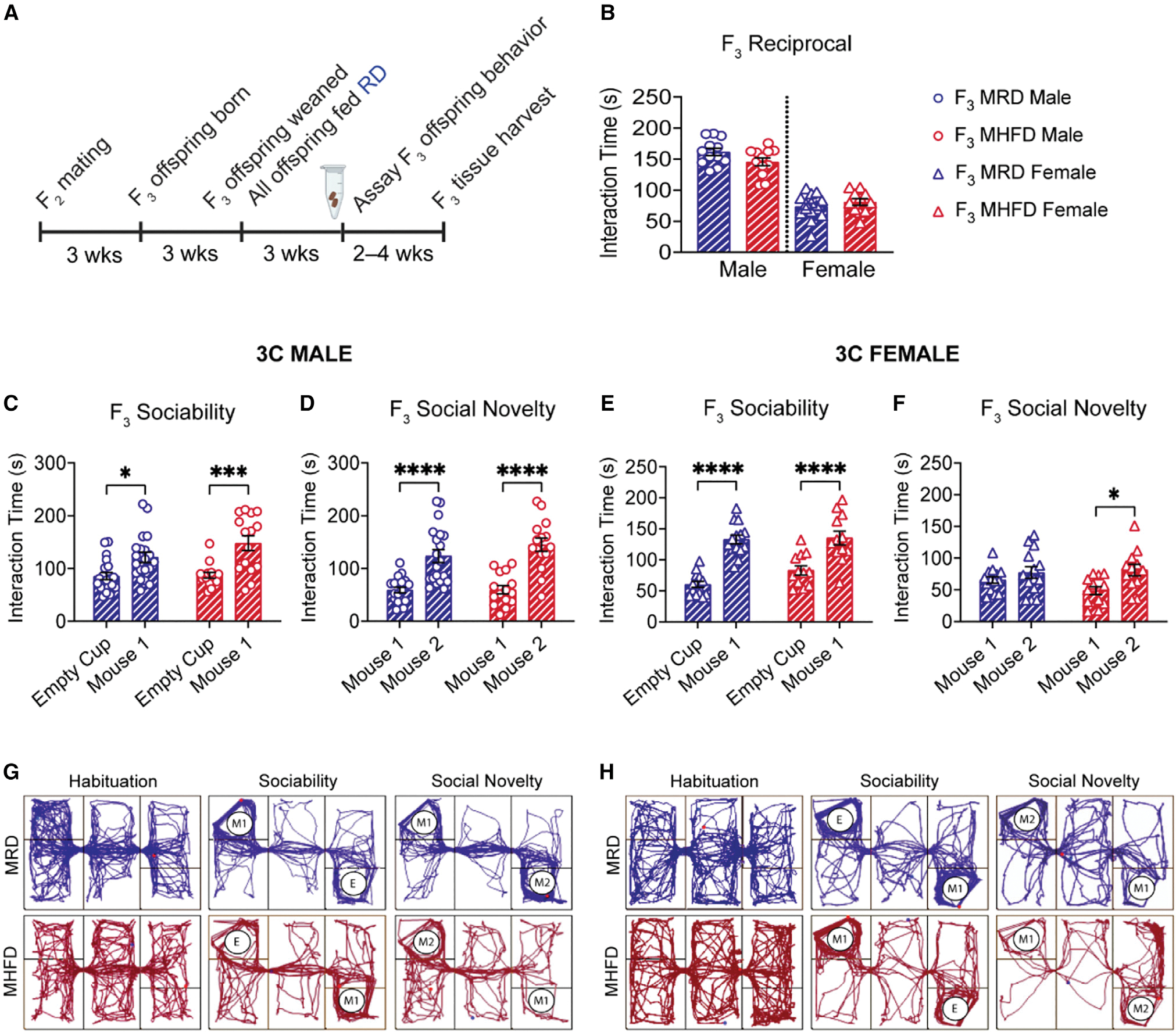
(A) Breeding schematic. MRD control and MHFD-descendant F2 mice were bred to produce the MRD- and MHFD-descendant F3 generations, respectively.
(B–H) (B) Comparison of reciprocal social interaction times revealed no differences between male (F3 MRD versus MHFD: t(22) = 1.824, p = 0.0817) or female pairs (F3 MRD versus MHFD: t(24) = 0.9925, p = 0.3309). Both MRD- and MHFD-descendant F3 males demonstrate normal sociability (C) (MRD: t(33) = 2.934, p = 0.0120; MHFD: t(33) = 4.322, p = 0.0003) and preference for social novelty (D) (MRD: t(33) = 5.493, p < 0.0001; MHFD: t(33) = 6.271, p < 0.0001) in the 3C task. F3 MRD and MHFD female offspring show a statistically significant preference for mouse 1 during the sociability phase of the 3C task (E) (MRD: t(48) = 6.979, p < 0.0001; MHFD: t(48) = 4.663, p < 0.0001), while only MHFD-descendant F3 females show a preference for social novelty (F) (MRD: t(48) = 1.722, p = 0.4632; MHFD: t(48) = 2.879, p = 0.0118). Representative MRD and MHFD track plots for (G) males and (H) females, respectively. Bar graphs show mean ± SEM with individual data points representing biological replicates. (B) N = 11–15 pairs per group; (C–F) N = 12–20 subjects per group.
DISCUSSION
Traditionally, neurodevelopmental disorders and the underlying pathology were exclusively attributed to variation in the human genome; however, it is becoming increasingly evident that dysbiosis of the gut microbiome can likewise contribute to host brain and behavioral dysfunction. Microbiome-directed therapies are emerging as an innovative avenue for ameliorating maladaptive behaviors associated with neurodevelopmental disorders (Stewart Campbell et al., 2022). Here, we provide evidence in an environmental mouse model for social dysfunction that diet-induced dysbiosis of the maternal gut microbiome could causally contribute to behavioral deficits in descendants.
We report significant social deficits in the MHFD-descendant F2 generation (Figures 1D–1I, 1K, and S2), with increased severity in the male sex, recapitulating the disproportionate prevalence of ASD in human males (Christensen et al., 2018; Ferri et al., 2018). Consistent with previous findings in male MHFD offspring (F1) and other mouse models for social dysfunction (Buffington et al., 2016, 2021; Sgritta et al., 2019), L. reuteri supplementation was sufficient to normalize MHFD-descendant F2 social behavior in both 3C and reciprocal social interaction (Figures 5B–5H and S6A–S6J). Conversely, while we observed impaired sociability in MHFD-descendant F2 females in the 3C task (Figures 1F and 1I), neither MRD- nor MHFD-descendant F2 female offspring demonstrated preference for social novelty (Figures 1G and 1I), and we observed no differences in reciprocal social interaction between groups (Figure 1K). Unexpectedly, L. reuteri administration led to a highly significant increase in both reciprocal social interaction (Figure 5B) and preference for social novelty (Figure 5F) among MHFD-descendant F2 females, metrics that did not differ between the non-treated F2 cohorts (Figures 1G, 1I, and 1K). We anticipate that probiotic species other than L. reuteri that exert similar effects on the microbial community ecology of the host and, relatedly, host physiology, would exert similar behavioral effects. The remarkable effect of L. reuteri on female social behavior was accompanied by a near-tripling of discriminant taxa between the MRD- versus MHFD-descendant F2 females relative to their male counterparts (Figure S7). Given that the males and females within the respective cohorts originate from common litters, the disproportionate impact on the female gut microbiome provides a rich opportunity for future investigation into the effects of host sex on microbiome composition and malleability. The sexually dimorphic impact of grandmaternal HFD on descendant social behavior, consistent with the milder behavioral phenotypes in human females with ASD compared with males (Baron-Cohen et al., 2005; Werling and Geschwind, 2013), may reveal redundancy in female social circuitry to ensure maternal care for young. Taken together, these data add to a growing body of evidence that host responses to probiotics are sex specific (Karunasena et al., 2014), which has important therapeutic implications.
Consistent with the finding that loss of microbial richness driven by a low MAC diet is largely recoverable within a single generation (Sonnenburg et al., 2016), we likewise observed recovery of microbial richness in MHFD-descendant F2 males and females (Figures 2E and 2F). Nonetheless, changes in the abundance of specific taxa were observed in the MHFD-descendant F2 generation (Figures 2A and 3), revealing enduring “scars” on microbiome configuration due to HFD exposure in the maternal lineage that could have functional consequences for the host. Finally, the absence of social deficits in the MHFD-descendant F3 generation (Figures 7 and S8) suggests that the diet-mediated recovery of gut microbial richness in the MHFD-descendant F2 females is sufficient to normalize fetal neurodevelopment and behavioral outcomes in their descendants.
FMT from F2 donors to GF recipients (Figures 4 and S5) provided insight into the relative contribution of dysbiosis of the offspring versus maternal gut microbiome to offspring social deficits. MHFD-descendant F2 male-derived fecal microbiota was sufficient to rescue baseline social deficits in formerly GF recipients (Figures 4 and S5), suggesting that it is not the adolescent state of the F2 gut microbiome driving their social dysfunction, but instead dysbiosis of the maternal (F1) gut microbiome. Multi-omic analyses to determine the molecular and cellular mechanisms by which HFD-induced scars on the microbiome in the maternal lineage affect brain development, function, and behavior will provide powerful insight into how diet-induced prepregnancy obesity predisposes offspring to neurodevelopmental disorders.
The differential impact of L. reuteri on gut community structure of MRD- versus MHFD-descendant F2 offspring (Figures 5, 6, and S4C) reveals a relative instability of the MHFD-descendant F2 gut microbiota when challenged by probiotic exposure. Surprisingly, we found Limosilactobacillus to be more abundant in the gut of treated MHFD versus MRD F2 descendants, despite equivalent dosing (Figure 6). HFD-induced dysbiosis in the maternal lineage (Figures 2E–2G) may thus allow for L. reuteri to exert a greater impact on the native community. Conversely, in the absence of an environmental stressor (i.e., diet-induced dysbiosis in the maternal lineage), the effect of L. reuteri may be limited due to ecological stability. Interestingly, L. reuteri promoted the expansion of other SCFA-producing taxa, including Ruminococcus and Ligilactobacillus, which we identified as discriminant taxa preferentially increased among the MHFD versus MRD lineage F2 animals treated with L. reuteri (Figures 5 and S7). In this light, we hypothesize that the L. reuteri-mediated and MHFD lineage-specific increase in biosynthetic potential for SCFAs might be key to the resulting behavioral rescue in treated F2 MHFD descendants, given that SCFA receptors are broadly expressed throughout the immune and nervous systems, SCFA supplementation relieves heightened responses to social stress (van de Wouw et al., 2018), and gut microbiota-derived SCFAs regulate the expression of the cytokine IL-17a, which is associated with social behavior (Dupraz et al., 2021; Reed et al., 2020; Shin Yim et al., 2017). Moreover, given the exaggerated effect of L. reuteri on the female gut microbiome and that maternal prenatal abundance of SCFA-producing bacterial taxa was recently found to be positively correlated with child behavioral outcomes in a human cohort (Dawson et al., 2021), we anticipate that targeted maternal probiotic treatment during pregnancy could have a protective role in offspring neurodevelopment. Together, these results identify an intergenerational impact of grandmaternal HFD on the stability of descendant microbial communities in response to environmental pressures, with implications for host disease susceptibility as well as therapeutic targeting of the gut microbiome.
Given that we observed (1) significant changes in female MHFD offspring F1 gut microbiome composition, (2) MHFD-descendant F2 male social dysfunction (Figures 1 and S2), despite partial recovery of gut microbiome diversity (Figures 2, 3, and S4), (3) resolution of baseline social deficits upon transplant of GF mice with MHFD F2 fecal content (Figures 4 and S5), and (4) normal social behavior in the MHFD-descendant F3 generation (Figures 7 and S8), we propose a model in which HFD-induced dysbiosis of the maternal gut microbiome and its vertical transmission to the MHFD-descendant F1 females are respectively causal in MHFD-descendant F1 and F2 social dysfunction. Longitudinal metagenomic sequencing and functional profiling of the maternal gut microbiome during pregnancy and lactation will improve our understanding of the impact of diet on remodeling of the maternal gut microbiome and metabolome during the antenatal period and the precise mechanisms underlying its influence on offspring neurodevelopment.
Here, we report dysbiosis of the gut microbiome and behavioral dysfunction across multiple descendant generations in response to a single generation of HFD consumption. Translationally, our findings emphasize the need to consider family history of diet-associated metabolic dysfunction and obesity in the maternal lineage as potential risk factors for neurodevelopmental disorders, independent of the mother’s own diet, lifestyle, and medical history, and highlight the potential for the implementation of policies providing universal access to nutritious food, particularly for expecting mothers, to yield multigenerational returns. Finally, they identify the maternal gut microbiome and its remodeling during pregnancy as promising therapeutic targets for improving long-term neurobehavioral outcomes in descendants.
Limitations of the study
Here, we show that HFD-induced dysbiosis in the maternal lineage contributes to social dysfunction across multiple generations of descendants and that probiotic strains can exert a sexually dimorphic impact on host gut microbial ecology. Additional experiments will be required to fully elucidate the mechanisms underlying these phenomena. Integrated multi-omic studies, including metagenomic, metabolomic, transcriptomic, and epigenomic approaches interrogating changes in both the mothers and their offspring, in each respective generation, will provide key mechanistic insight. We provide evidence that L. reuteri modifies the configuration of the host gut microbiome in a sex-dependent fashion. It will be interesting to pursue the therapeutic implications of this finding. For instance, could distinct probiotic, prebiotic, or synbiotic formulations provide improved care for male versus female patients, not only in the context of neurodevelopment and behavior, but also in other disease contexts, such as metabolic disorders or irritable bowel disease? Relatedly, given the heightened remodeling of the female gut microbiome in response to L. reuteri and our finding that dysbiosis of the maternal gut microbiome modulates behavioral outcomes in descendants, the prenatal period is an obvious target for probiotic-mediated intervention aimed at reducing risk for adverse neurodevelopmental outcomes in offspring. Preclinical investigation into critical intervention periods will be of both scientific and translational significance. Cross-fostering experiments, to tease apart the impacts of gestational versus lactational exposure, will be key to identifying optimal windows for intervention. Our metataxonomic sequencing data identify multiple candidate species that may exert similar effects on host neurodevelopment and behavior as L. reuteri, and their therapeutic utility should be explored. Finally, careful assessment of female-specific behaviors, such as alloparenting (Carcea et al., 2021), may reveal complexity in female social behavior that is undetectable in the 3C task and reciprocal social interaction, tasks historically optimized to assess male social behaviors in genetic mouse models for ASD (Silverman et al., 2010). In this vein, how maternal environmental exposures, including HFD, affect maternal behavior, and female-specific behavior, in general, is a neglected topic of research that merits investigation, particularly in the context of the intergenerational effects of maternal environmental stressors on descendant neurobehavioral health outcomes.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Shelly Buffington (shbuffin@utmb.edu).
Materials availability
This study did not generate new unique reagents. Information on reagents used in this study is available in the key resources table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Bacterial and virus strains | ||
|
| ||
| Limosilactobacillus reuteri PTA-6475 | ATCC | Strain MM4-1A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| De Man, Rogosa, and Sharpe (MRS) Media | MilliporeSigma | Cat# MHA00MRS2 |
| De Man, Rogosa, and Sharpe (MRS) Agar | MilliporeSigma | Cat# 110660 |
| 5% H2, 5% CO2, 90% N2 anaerobic gas mixture | Linde Gas & Equipment, Praxair Distribution | Cat# BI NICDHYC1-K |
|
| ||
| Critical commercial assays | ||
|
| ||
| MagAttract PowerSoil DNA Kit | Qiagen | Cat #27100-4-EP |
|
| ||
| Deposited data | ||
|
| ||
| 16S ribosomal RNA (rRNA) gene amplicon sequencing data | NCBI SRA | BioProject PRJNA878523 |
| Original unprocessed data | This paper; Mendeley | https://doi.org/10.17632/z6wz8kpk3n.1 |
| Galaxy Computation Tool | Huttenhower Lab | http://huttenhower.sph.harvard.edu/galaxy |
| Agile Toolkit for Incisive Microbial Analyses (ATIMA2) Platform | Petrosino Lab | https://atima.research.bcm.edu |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| C57Bl/6N | Taconic Biosciences | Cat# B6-M; Cat# B6-F |
| Germ-free mice (C57Bl/6N background) | UTMB Germ Free Mouse Facility | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| ANY-maze Video Tracking Software | Stoelting | https://www.stoeltingco.com/anymaze/video-tracking/software.html |
| Prism 9.0 | Graphpad | https://www.graphpad.com/scientific-software/prism |
| Adobe Illustrator CS6 | Adobe | https://www.adobe.com |
| Miseq Software | Illumina | https://support.illumina.com/sequencing/sequencing_software/miseq_reporter/downloads.html |
| R | The R Foundation | https://www.r-project.org/about.html |
| R Studio | R Studio | https://rstudio.com/ |
|
| ||
| Other | ||
|
| ||
| Ugo Basile Sociability Cage, Mouse, w/Gray Enclosures | Stoelting | Cat# 60450 |
| Small Mouse ‘Stranger’ Enclosures (wire cages), set/2, Gray | Stoelting | Cat# 60451 |
| Open Field, 40cm, Gray | Stoelting | Cat# 60101 |
| Reciprocal Social Interaction Box, Clear Acrylic | The Container Store | Cat# 10071077 |
| Regular diet chow | Lab Diets | Cat# 5001 |
| High-fat diet chow | Research Diets | Cat# D12492 |
| Bed-O’cobs 1/8 mouse home cage bedding | The Andersons Plant Nutrient | Cat# 8B |
Data and code availability
The accession number for the 16S rRNA gene sequencing data reported in this paper is NCBI Sequence Read Archive: BioProject PRJNA878523. The original, unprocessed data reported in this manuscript can be accessed via Mendeley Data: https://doi.org/10.17632/z6wz8kpk3n.1.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse husbandry and maternal diet
WT C57BL/6N mice were obtained from Taconic Laboratories (B6), maintained at animal biosafety level-1 under specific pathogen free (SPF) conditions, and kept on a 12-hour light/dark cycle in polysulfone cages with access to sterile food and water ad libitum. Six-week-old females were placed on either a regular diet (RD) consisting of 13.4% kcal from fat, 30% kcal from protein, and 57% kcal from carbohydrates (Lab Diets, #5001) or HFD consisting of 60% kcal from fat, 20% kcal from protein, and 20% kcal from carbohydrates (Research Diets, #D12492). After six weeks on diet, females were randomly assigned to monogamous pairs with WT C57BL/6N adult males to produce subject offspring. Resulting offspring (F1) were weaned at 3 weeks of age and all placed on RD, regardless of maternal diet (RD or HFD). Resulting F1 maternal regular diet (MRD) pairs and MHFD offspring pairs were monogamously bred to generate F2 offspring (Figures 1A and 1B). F2 offspring were weaned at 3 weeks of age and fed RD. Behavioral tests were performed on male and female F2 offspring at 7–8 weeks of age, with handling for three days prior in conjunction with stool collection. Behaviorally naïve MRD-descendant F2 pairs and MHFD-descendant F2 pairs were monogamously bred to generate the F3 generation (Figures 1A and 1B). F3 descendants were weaned at 3 weeks of age and fed RD. Behavioral tests were performed on male and female F3 offspring at 7–8 weeks of age, with handling for three days prior in conjunction with stool collection. C57Bl/6N germ-free mice were acquired from the Baylor College of Medicine Gnotobiotic Core Facility. Animal care and experimental procedures were approved by The University of Texas Medical Branch Institutional Animal Care and Use Committee in accordance with all guidelines set forth by the U.S. National Institutes of Health.
METHOD DETAILS
Behavior
Behavioral assays were performed as previously described (Buffington et al., 2016), as detailed in brief below. Animals were acclimated to the behavioral suite for 30–60 minutes prior to initiation of all experiments. Apparatuses were spot cleaned with 70% ethanol after each animal and thoroughly cleaned at the end of the day. ANY-maze automated video tracking system software version 6.33 (Stoelting) was used for video recording and automated data acquisition.
Reciprocal social interaction
Mice were placed in a neutral, 30 cm3 Plexiglass arena containing a shallow layer of corncob bedding with a previously unencountered age- and sex-matched conspecific matched for maternal diet. Time spent engaged in social interaction (close following, touching, nose-to-nose sniffing, nose-to-anus sniffing, and/or crawling over/under each other) over a ten-minute period was recorded by a blinded human observer and analyzed via ANY-maze.
Crawley’s three-chamber (3C) test for sociability and preference for social novelty
Crawley’s 3C test for sociability and preference for social novelty was performed as described (Silverman et al., 2010). Briefly, animals were habituated for 10 minutes in a 60 × 40 × 23 cm arena divided into three inter-connected chambers. Sociability was measured during a second 10-minute interval in which the test subject could interact either with an empty wire cup (Empty) or a wire cup containing an age- and sex-matched stranger conspecific (Mouse 1). Stranger mice were acclimated to restraint in the wire cup during three training periods, over a period of two days prior to the test. Preference for social novelty was assayed by introducing a second stranger mouse (Mouse 2) into the previously empty wire cup. The location of the Empty Cup/Mouse 2 versus Mouse 1 were counterbalanced within cohorts. Interactions were again recorded by ANY-maze software and an independent observer.
Open field test
Animal test subjects were gently lowered into an open arena (40 × 40 × 20 cm) and allowed to explore freely for 10 minutes. ANY-maze automatically measured distance traveled, speed, and position in the arena, as well as time spent in the center of the arena (defined as the interior 20 × 20 cm).
16S ribosomal RNA (rRNA) gene amplicon sequencing
Stool samples were aseptically collected in sterile 2mL Eppendorf tubes, immediately placed on dry ice, and stored at −80°C until further processing. Bacterial DNA was extracted and sequenced by the Alkek Center for Metagenomics and Microbiome Research at Baylor College of Medicine using protocols adapted from those developed for the NIH-Human Microbiome Project (Human Microbiome Project Consortium, 2012), as described previously (Buffington et al., 2016). Briefly, bacterial genomic DNA was extracted using MagAttract PowerSoil DNA Kit (Qiagen) followed by PCR amplification of the 16S rDNA V4 region. The primers used for amplification include MiSeq adapters and single-end barcodes allowing for pooling and direct sequencing of PCR products. Sequencing was performed on the Illumina MiSeq platform using the 2 × 250 bp paired-end protocol yielding overlapping paired-end reads. The 16S rRNA gene read pairs were demultiplexed and merged using USEARCH v7.0.1090 (Edgar, 2010), allowing zero mismatches and a minimum overlap of 50 bases. Merged reads were trimmed at first base with Q5. A quality filter was applied to the resulting merged reads and reads containing above 0.05 expected errors were discarded. 16S rRNA gene sequences were clustered into Operational Taxonomic Units (OTUs) at a similarity cutoff value of 97% using the UPARSE algorithm (Edgar, 2013). OTUs were mapped to an optimized version of the SILVA Database (Quast et al., 2013) containing only the 16S v4 region to determine taxonomies. Abundances were recovered by mapping the demultiplexed reads to the UPARSE OTUs. A custom script constructed a rarefied OTU table from the output files generated in the previous two steps for downstream analyses of alpha-diversity, beta-diversity, and phylogenetic trends using the Agile Toolkit for Incisive Microbial Analyses (ATIMA2) platform (atima.research.bcm.edu).
Limosilactobacillus (L.) reuteri culture and treatment
L. reuteri PTA-6475™ was purchased from ATCC (MM4–1A). Cultures were grown anaerobically in De Man, Rogosa, and Sharpe (MRS) broth (MilliporeSigma) for 48 hours in a 5% CO2, 5% H2, remainder N2 environment. Stocks were aliquoted and frozen at −80°C then moved to −20°C storage the week of use. Stock viability was confirmed and colony forming units (CFU) quantified by serial plating on MRS agar and growing anaerobically at 37°C for 48 hours. For treatment of F2 offspring, 200 μL L. reuteri was added to 200 mL sterile drinking water at a concentration of 1 × 108 CFU/mL at time of weaning. Treated water was refreshed daily, Monday – Friday, for at least four weeks prior to behavior, and was continued throughout behavioral assessment until time of tissue collection.
Fecal microbiota transplants
Two-to-three fresh fecal samples were collected from each donor mouse (5 MRD- and 4 MHFD-descendant F2 male subjects), pooled together according to diet, and homogenized in sterile, deoxygenated PBS (1 mL PBS per stool) in an anaerobic chamber. The resulting slurry was vortexed, rested on ice for 15 minutes, and then spun at 1200 rpm for 5 minutes. Supernatants were transferred to sterile 50mL conical tubes within the anaerobic chamber and thoroughly sealed prior to exit from the chamber for transit to the recipients. 16 four-week-old C57BL/6N germ-free (GF) recipient mice were immediately colonized (8 per group) by a single 200 μL gavage. Mice were housed for 4 weeks before handling, stool collection, and behavior. Fecal samples were snap frozen on dry ice and stored at −80°C until prepared for sequencing. 8 six-week-old GF control (GF-C) mice, not subjected to FMT, were used to establish a baseline for behavioral and molecular analysis.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses
Power analyses to establish group size were performed in GPower 3.1. Data were analyzed and visualized using GraphPad Prism version 8.4.3 for Mac OS X, GraphPad Software, San Diego, California USA, www.graphpad.com. Data are presented as individual data points throughout the figures. Bar graphs include background columns indicating group mean. Error bars represent mean ± SEM, unless otherwise indicated, with each point representing an individual mouse (biological replicate) or pair of mice, in the reciprocal social interaction task. Sample sizes are indicated in the figure legends. Litter numbers are as follows: For MRD/MHFD F2 male and female offspring, 6–7 and 4–6 litters per group were used, respectively (Figures 1, 2, 3, S2, S3 and S4). For GF male mice, 2 litters per group were used (Figures 4 and S5). For MRD/MHFD + L. reuteri F2 male and female offspring, 3–4 and 3 litters per group were used, respectively (Figures 5, 6, S6 and S7). For MRD/MHFD F3 male and female offspring, 5 and 3 litters per group were used, respectively (Figures 7 and S8). Statistics are reported in figure legends as t(degrees of freedom) = t-statistic or q-statistic, p = p-value. p < 0.05 was considered significant, where *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. For data without repeated measures (3C locomotor data and open field test data), data were visualized as column data but analyzed using a two-tailed nested t-test to account for cage effects. No significant differences between sex-matched cages within the same cohort were identified. For Crawley’s 3C assay data, to retain the precision of estimated variability while still accounting for litter effects, a mixed-effects model with repeated measures was applied, with the Šídák correction for multiple comparisons (Jimenez and Zylka, 2021; Lazic and Essioux, 2013). Random (litter) effects with a variance of zero were removed from the model and analyzed by repeated measures two-way ANOVA. For reciprocal social experiments, an unpaired, two-tailed t-test was performed unless otherwise indicated. In cases where data were not normally distributed, the non-parametric Mann-Whitney test was used. Locomotor data was analyzed by one-way ANOVA with Tukey’s adjustment for multiple comparisons. Social Preference Indexes were calculated as in (Rein et al., 2020) and analyzed using a nested t-test. Principal coordinate analysis (PCoA) plots were constructed based on calculated UniFrac distances and the Monte Carlo permutation test was used to estimate p values. Beta diversity was assessed based on both unweighted and weighted UniFrac distances, visualized by principal coordinate analysis (PCoA), and statistically analyzed using permutational multivariate analysis of variance (PERMANOVA). Differentially abundant bacterial taxa were determined by Linear Discriminant Analysis (LDA) Effect Size (LEfSe) (Segata et al., 2011) using the Galaxy computational tool (http://huttenhower.sph.harvard.edu/galaxy) (Afgan et al., 2018) and applying the all-against-all strategy with a minimum logarithmic LDA score (i.e., biomarker effect size) of 2.0.
Summary of outliers and excluded data
A limited number of animals were removed from analysis due to statistical identification as outliers and/or failure to meet pre-established criteria. Outliers were identified by Grubbs’ test (alpha = 0.05, http://graphpad.com/quickcalcs/grubbs1/) and were removed from that specific dataset (e.g., OF, RS, or 3C); therefore, sample sizes may vary slightly by test. For 3C, Grubbs’ test was run on the difference between interaction times (M1 − E, and M2 − M1). Additionally, mice that failed to interact for a minimum of 10 seconds with either social or non-social object in the 3C task were removed from the entire 3C dataset.
Supplementary Material
Highlights.
MHFD reduces microbial richness in female offspring (F1)
Social deficits persist in MHFD-descendant F2, despite recovery of microbial richness
Post-weaning L. reuteri treatment rescues MHFD-descendant F2 social deficits
L. reuteri exerts a sexually dimorphic impact on host microbiota configuration
ACKNOWLEDGMENTS
We would like to acknowledge the UTMB Animal Resource Center and GermFree Mouse Facility (supported by the Sealy Center for Microbiome Research) and Dr. Maki Wakamiya, as well as the Baylor College of Medicine Gnotobiotic Core Facility (supported in part by PHS grant P30DK056338) and Dr. Stephanie W. Fowler for providing excellent care for the mice involved in this study, Dr. Heidi Spratt for statistical consultation, and The Alkek Center for Metagenomics and Microbiome Research at Baylor College of Medicine for RNA extraction and 16S rRNA gene amplicon sequencing. Body composition analyses were performed in the UTMB Center for Addiction Research (CAR) Rodent In Vivo Assessment (RIVA) Core. Figures were created using Adobe Illustrator and schematics using BioRender (BioRender.com; publication and licensing rights agreement QR23HLBKNP). Research reported in this publication was supported with funding from the National Institutes of Health (R01HD109095), the Brain & Behavior Research Foundation (NARSAD Young Investigator Grant 28298), the Scott-Gentle Foundation, the UTMB Institute for Human Infections and Immunity, and the Gulf Coast Center for Precision Environmental Health, via Center Core Grant from the National Institute for Environmental Health Sciences of the National Institutes of Health (NIH) under award no. P30ES030285 (the content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH), to S.A.B. Salary support for L.M.M. during a portion of the project was provided by NIH 1T32AG067952–01 (UTMB Mitchell Center for Neurodegenerative Diseases).
Footnotes
DECLARATION OF INTERESTS
S.A.B. is an inventor on a patent granted to Baylor College of Medicine related to the use of Limosilactobacillus reuteri for treating disorders characterized by social dysfunction (US Patent No. 11135252). The authors declare no other competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111461.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
REFERENCES
- Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Grüning BA, et al. (2018). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afroz KF, Reyes N, Young K, Parikh K, Misra V, and Alviñ a K (2021). Altered gut microbiome and autism like behavior are associated with parental high salt diet in male mice. Sci. Rep. 11, 8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avni E, Ben-Itzchak E, and Zachor DA (2018). The presence of comorbid ADHD and anxiety symptoms in autism spectrum disorder: clinical presentation and predictors. Front. Psychiatry 9, 717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bariselli S, Hörnberg H, Pré vost-Solié C, Musardo S, Hatstatt-Burklé L, Scheiffele P, and Bellone C (2018). Role of VTA dopamine neurons and neuroligin 3 in sociability traits related to nonfamiliar conspecific interaction. Nat. Commun. 9, 3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron-Cohen S, Knickmeyer RC, and Belmonte MK (2005). Sex differences in the brain: implications for explaining autism. Science 310, 819–823. [DOI] [PubMed] [Google Scholar]
- Bisanz JE, Upadhyay V, Turnbaugh JA, Ly K, and Turnbaugh PJ (2019). Meta-analysis reveals reproducible gut microbiome alterations in response to a high-fat diet. Cell Host Microbe 26, 265–272.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser MJ (2017). The theory of disappearing microbiota and the epidemics of chronic diseases. Nat. Rev. Immunol. 17, 461–463. [DOI] [PubMed] [Google Scholar]
- Bordeleau M, Fernández de Cossío L, Lacabanne C, Savage JC, Vernoux N, Chakravarty M, and Tremblay MÉ (2021). Maternal high-fat diet modifies myelin organization, microglial interactions, and results in social memory and sensorimotor gating deficits in adolescent mouse offspring. Brain Behav. Immun. Health 15, 100281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffington SA, Di Prisco GV, Auchtung TA, Ajami NJ, Petrosino JF, and Costa-Mattioli M (2016). Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell 165, 1762–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffington SA, Dooling SW, Sgritta M, Noecker C, Murillo OD, Felice DF, Turnbaugh PJ, and Costa-Mattioli M (2021). Dissecting the contribution of host genetics and the microbiome in complex behaviors. Cell 184, 1740–1756.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisano S, La Colla A, Echarte SM, and Chisari AN (2019). Interplay between early-life malnutrition, epigenetic modulation of the immune function and liver diseases. Nutr. Res. Rev. 32, 128–145. [DOI] [PubMed] [Google Scholar]
- Carcea I, Caraballo NL, Marlin BJ, Ooyama R, Riceberg JS, Mendoza Navarro JM, Opendak M, Diaz VE, Schuster L, Alvarado Torres MI, et al. (2021). Oxytocin neurons enable social transmission of maternal behaviour. Nature 596, 553–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody RN, Gerber GK, Luevano JM Jr., Gatti DM, Somes L, Svenson KL, and Turnbaugh PJ (2015). Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17, 72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne-Jorgensen K, Mian MF, Kay S, Hanani H, Ziv O, McVey Neufeld KA, Koren O, and Bienenstock J (2020). Prenatal low-dose penicillin results in long-term sex-specific changes to murine behaviour, immune regulation, and gut microbiota. Brain Behav. Immun 84, 154–163. [DOI] [PubMed] [Google Scholar]
- Chang E, Hafner H, Varghese M, Griffin C, Clemente J, Islam M, Carlson Z, Zhu A, Hak L, Abrishami S, et al. (2019). Programming effects of maternal and gestational obesity on offspring metabolism and metabolic inflammation. Sci. Rep. 9, 16027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen DL, Braun KVN, Baio J, Bilder D, Charles J, Constantino JN, Daniels J, Durkin MS, Fitzgerald RT, Kurzius-Spencer M, et al. (2018). Prevalence and characteristics of autism spectrum disorder among children aged 8 Years - autism and developmental disabilities monitoring network, 11 sites, United States, 2012. MMWR. Surveill. Summ. 65, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu DM, Antony KM, Ma J, Prince AL, Showalter L, Moller M, and Aa-gaard KM (2016). The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Med. 8, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo PM, La Merrill MA, Krigbaum NY, and Cohn BA (2021). Grandmaternal perinatal serum DDT in relation to granddaughter early menarche and adult obesity: three generations in the child health and development studies cohort. Cancer Epidemiol. Biomarkers Prev. 30, 1480–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coley EJL, and Hsiao EY (2021). Malnutrition and the microbiome as modifiers of early neurodevelopment. Trends Neurosci. 44, 753–764. [DOI] [PubMed] [Google Scholar]
- Constantino JN, and Charman T (2012). Gender bias, female resilience, and the sex ratio in autism. J. Am. Acad. Child Adolesc. Psychiatry 51, 756–758. [DOI] [PubMed] [Google Scholar]
- Courchesne E, Gazestani VH, and Lewis NE (2020). Prenatal origins of ASD: the when, what, and how of ASD development. Trends Neurosci. 43, 326–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalile B, Van Oudenhove L, Vervliet B, and Verbeke K (2019). The role of short-chain fatty acids in microbiota-gut-brain communication. Nat. Rev. Gastroenterol. Hepatol. 16, 461–478. [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson SL, O’Hely M, Jacka FN, Ponsonby AL, Symeonides C, Loughman A, Collier F, Moreno-Betancur M, Sly P, Burgner D, et al. (2021). Maternal prenatal gut microbiota composition predicts child behaviour. EBioMedicine 68, 103400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre-Ubieta L, Won H, Stein JL, and Geschwind DH (2016). Advancing the understanding of autism disease mechanisms through genetics. Nat. Med. 22, 345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbonnet L, Clarke G, Shanahan F, Dinan TG, and Cryan JF (2014). Microbiota is essential for social development in the mouse. Mol. Psychiatry 19, 146–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Gesu CM, Matz LM, and Buffington SA (2021). Diet-induced dysbiosis of the maternal gut microbiome in early life programming of neurodevelopmental disorders. Neurosci. Res. 168, 3–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, Le A, Cowan TM, Nolan GP, Fischbach MA, et al. (2017). A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley KJ, Sloboda DM, Connor KL, Beltrand J, and Vickers MH (2011). Offspring of mothers fed a high fat diet display hepatic cell cycle inhibition and associated changes in gene expression and DNA methylation. PLoS One 6, e21662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupraz L, Magniez A, Rolhion N, Richard ML, Da Costa G, Touch S, Mayeur C, Planchais J, Agus A, Danne C, et al. (2021). Gut microbiota-derived short-chain fatty acids regulate IL-17 production by mouse and human intestinal gammadelta T cells. Cell Rep. 36, 109332. [DOI] [PubMed] [Google Scholar]
- Eaton KA, Honkala A, Auchtung TA, and Britton RA (2011). Probiotic Lactobacillus reuteri ameliorates disease due to enterohemorrhagic Escherichia coli in germfree mice. Infect. Immun. 79, 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. [DOI] [PubMed] [Google Scholar]
- Edgar RC (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. [DOI] [PubMed] [Google Scholar]
- Erny D, Dokalis N, Mezö C, Castoldi A, Mossad O, Staszewski O, Frosch M, Villa M, Fuchs V, Mayer A, et al. (2021). Microbiota-derived acetate enables the metabolic fitness of the brain innate immune system during health and disease. Cell Metab. 33, 2260–2276.e7. [DOI] [PubMed] [Google Scholar]
- Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, Armanini F, Truong DT, Manara S, Zolfo M, et al. (2018). Mother-to-Infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 24, 133–145.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri SL, Abel T, and Brodkin ES (2018). Sex differences in autism spectrum disorder: a review. Curr. Psychiatry Rep. 20, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks PW, and McCarthy MI (2016). Exposing the exposures responsible for type 2 diabetes and obesity. Science 354, 69–73. [DOI] [PubMed] [Google Scholar]
- Friedman JE (2018). Developmental programming of obesity and diabetes in mouse, monkey, and man in 2018: where are we headed? Diabetes 67, 2137–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gacesa R, Kurilshikov A, Vich Vila A, Sinha T, Klaassen MAY, Bolte LA, Andreu-Sánchez S, Chen L, Collij V, Hu S, et al. (2022). Environmental factors shaping the gut microbiome in a Dutch population. Nature 604, 732–739. [DOI] [PubMed] [Google Scholar]
- Gohir W, Kennedy KM, Wallace JG, Saoi M, Bellissimo CJ, Britz-McKibbin P, Petrik JJ, Surette MG, and Sloboda DM (2019). High-fat diet intake modulates maternal intestinal adaptations to pregnancy and results in placental hypoxia, as well as altered fetal gut barrier proteins and immune markers. J. Physiol. 597, 3029–3051. [DOI] [PubMed] [Google Scholar]
- Gohir W, Whelan FJ, Surette MG, Moore C, Schertzer JD, and Sloboda DM (2015). Pregnancy-related changes in the maternal gut microbiota are dependent upon the mother’s periconceptional diet. Gut Microb. 6, 310–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaydin LA, Grosenick L, Finkelstein JC, Kauvar IV, Fenno LE, Adhikari A, Lammel S, Mirzabekov JJ, Airan RD, Zalocusky KA, et al. (2014). Natural neural projection dynamics underlying social behavior. Cell 157, 1535–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz-Picciotto I, Schmidt RJ, and Krakowiak P (2018). Understanding environmental contributions to autism: causal concepts and the state of science. Autism Res. 11, 554–586. [DOI] [PubMed] [Google Scholar]
- Hoek HW, Brown AS, and Susser E (1998). The Dutch famine and schizophrenia spectrum disorders. Soc. Psychiatry Psychiatr. Epidemiol. 33, 373–379. [DOI] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium (2012). A framework for human microbiome research. Nature 486, 215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung LW, Neuner S, Polepalli JS, Beier KT, Wright M, Walsh JJ, Lewis EM, Luo L, Deisseroth K, Dö len G, et al. (2017). Gating of social reward by oxytocin in the ventral tegmental area. Science 357, 1406–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez JA, and Zylka MJ (2021). Controlling litter effects to enhance rigor and reproducibility with rodent models of neurodevelopmental disorders. J. Neurodev. Disord. 13, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SS, Kurti A, Fair DA, and Fryer JD (2014). Dietary intervention rescues maternal obesity induced behavior deficits and neuroinflammation in offspring. J. Neuroinflammation 11, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunasena E, McMahon KW, Chang D, and Brashears MM (2014). Host responses to the pathogen Mycobacterium avium subsp. paratuberculosis and beneficial microbes exhibit host sex specificity. Appl. Environ. Microbiol. 80, 4481–4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Paik D, Ramirez RN, Biggs DG, Park Y, Kwon HK, Choi GB, and Huh JR (2022). Maternal gut bacteria drive intestinal inflammation in offspring with neurodevelopmental disorders by altering the chromatin landscape of CD4(+) T cells. Immunity 55, 145–158.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim H, Yim YS, Ha S, Atarashi K, Tan TG, Longman RS, Honda K, Littman DR, Choi GB, et al. (2017). Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature 549, 528–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klurfeld DM, and Kritchevsky D (1986). The Western diet: an examination of its relationship with chronic disease. J. Am. Coll. Nutr. 5, 477–485. [DOI] [PubMed] [Google Scholar]
- Kong L, Norstedt G, Schalling M, Gissler M, and Lavebratt C (2018). The risk of offspring psychiatric disorders in the setting of maternal obesity and diabetes. Pediatrics 142. e20180776. [DOI] [PubMed] [Google Scholar]
- Kopp W (2019). How western diet and lifestyle drive the pandemic of obesity and civilization diseases. Diabetes Metab. Syndr. Obes. 12, 2221–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazic SE, and Essioux L (2013). Improving basic and translational science by accounting for litter-to-litter variation in animal models. BMC Neurosci. 14, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Fallin MD, Riley A, Landa R, Walker SO, Silverstein M, Caruso D, Pearson C, Kiang S, Dahm JL, et al. (2016a). The association of maternal obesity and diabetes with autism and other developmental disabilities. Pediatrics 137. e20152206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YM, Ou JJ, Liu L, Zhang D, Zhao JP, and Tang SY (2016b). Association between maternal obesity and autism spectrum disorder in offspring: a meta-analysis. J. Autism Dev. Disord. 46, 95–102. [DOI] [PubMed] [Google Scholar]
- Lopez-Aranda MF, Chattopadhyay I, Boxx GM, Fraley ER, Silva TK, Zhou M, Phan M, Herrera I, Taloma S, Mandanas R, et al. (2021). Postnatal immune activation causes social deficits in a mouse model of tuberous sclerosis: role of microglia and clinical implications. Sci. Adv. 7, eabf2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Prince AL, Bader D, Hu M, Ganu R, Baquero K, Blundell P, Alan Harris R, Frias AE, Grove KL, et al. (2014). High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. Nat. Commun. 5, 3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais LH, Schreiber HL 4th, and Mazmanian SK (2021). The gut microbiota-brain axis in behaviour and brain disorders. Nat. Rev. Microbiol. 19, 241–255. [DOI] [PubMed] [Google Scholar]
- Nagpal J, and Cryan JF (2021). Microbiota-brain interactions: moving toward mechanisms in model organisms. Neuron 109, 3930–3953. [DOI] [PubMed] [Google Scholar]
- O’Riordan KJ, Collins MK, Moloney GM, Knox EG, Aburto MR, Fülling C, Morley SJ, Clarke G, Schellekens H, and Cryan JF (2022). Short chain fatty acids: microbial metabolites for gut-brain axis signalling. Mol. Cell. Endocrinol. 546, 111572. [DOI] [PubMed] [Google Scholar]
- Oliphant K, Ali M, D’Souza M, Hughes PD, Sulakhe D, Wang AZ, Xie B, Yeasin R, Msall ME, Andrews B, et al. (2021). Bacteroidota and Lachnospiraceae integration into the gut microbiome at key time points in early life are linked to infant neurodevelopment. Gut Microb. 13, 1997560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panjwani AA, Ji Y, Fahey JW, Palmer A, Wang G, Hong X, Zuckerman B, and Wang X (2019). Maternal obesity/diabetes, plasma branched-chain amino acids, and autism spectrum disorder risk in urban low-income children: evidence of sex difference. Autism Res. 12, 1562–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenti I, Rabaneda LG, Schoen H, and Novarino G (2020). Neurodevelopmental disorders: from genetics to functional pathways. Trends Neurosci. 43, 608–621. [DOI] [PubMed] [Google Scholar]
- Poutahidis T, Kleinewietfeld M, Smillie C, Levkovich T, Perrotta A, Bhela S, Varian BJ, Ibrahim YM, Lakritz JR, Kearney SM, et al. (2013). Microbial reprogramming inhibits Western diet-associated obesity. PLoS One 8, e68596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, and Glöckner FO (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed MD, Yim YS, Wimmer RD, Kim H, Ryu C, Welch GM, Andina M, King HO, Waisman A, Halassa MM, et al. (2020). IL-17a promotes sociability in mouse models of neurodevelopmental disorders. Nature 577, 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein B, Ma K, and Yan Z (2020). A standardized social preference protocol for measuring social deficits in mouse models of autism. Nat. Protoc. 15, 3464–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers AB, Morgan CP, Leu NA, and Bale TL (2015). Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proc. Natl. Acad. Sci. USA 112, 13699–13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E, Koren O, Reshef L, Efrony R, and Zilber-Rosenberg I (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362. [DOI] [PubMed] [Google Scholar]
- Sarker G, Berrens R, von Arx J, Pelczar P, Reik W, Wolfrum C, and Peleg-Raibstein D (2018). Transgenerational transmission of hedonic behaviors and metabolic phenotypes induced by maternal overnutrition. Transl. Psychiatry 8, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, and Huttenhower C (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgritta M, Dooling SW, Buffington SA, Momin EN, Francis MB, Britton RA, and Costa-Mattioli M (2019). Mechanisms underlying microbialmediated changes in social behavior in mouse models of autism spectrum disorder. Neuron 101, 246–259.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby RD, Mar P, Janzow GE, Mashburn-Warren L, Tengberg N, Navarro JB, Allen JM, Wickham J, Wang Y, Bailey MT, et al. (2021). Antibacterial and anti-inflammatory effects of Lactobacillus reuteri in its biofilm state contribute to its beneficial effects in a rat model of experimental necrotizing enterocolitis. J. Pediatr. Surg. 57, 1382–1390. [DOI] [PubMed] [Google Scholar]
- Shin Yim Y, Park A, Berrios J, Lafourcade M, Pascual LM, Soares N, Yeon Kim J, Kim S, Kim H, Waisman A, et al. (2017). Reversing behavioural abnormalities in mice exposed to maternal inflammation. Nature 549, 482–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Yang M, Lord C, and Crawley JN (2010). Behavioural phenotyping assays for mouse models of autism. Nat. Rev. Neurosci. 11, 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, and Sonnenburg JL (2016). Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg ED, and Sonnenburg JL (2019). The ancestral and industrialized gut microbiota and implications for human health. Nat. Rev. Microbiol. 17, 383–390. [DOI] [PubMed] [Google Scholar]
- St Clair D, Xu M, Wang P, Yu Y, Fang Y, Zhang F, Zheng X, Gu N, Feng G, Sham P, et al. (2005). Rates of adult schizophrenia following prenatal exposure to the Chinese famine of 1959–1961. JAMA 294, 557–562. [DOI] [PubMed] [Google Scholar]
- Stewart Campbell A, Needham BD, Meyer CR, Tan J, Conrad M, Preston GM, Bolognani F, Rao SG, Heussler H, Griffith R, et al. (2022). Safety and target engagement of an oral small-molecule sequestrant in adolescents with autism spectrum disorder: an open-label phase 1b/2a trial. Nat. Med. 28, 528–534. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Bäckhed F, Fulton L, and Gordon JI (2008). Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3, 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui N, Berto S, Konishi A, Kondo M, Konopka G, Matsuzaki H, and Shimada S (2021). Zbtb16 regulates social cognitive behaviors and neocortical development. Transl. Psychiatry 11, 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wouw M, Boehme M, Lyte JM, Wiley N, Strain C, O’Sullivan O, Clarke G, Stanton C, Dinan TG, and Cryan JF (2018). Short-chain fatty acids: microbial metabolites that alleviate stress-induced brain-gut axis alterations. J. Physiol. 596, 4923–4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong HE, Pronovost GN, Williams DW, Coley EJL, Siegler EL, Qiu A, Kazantsev M, Wilson CJ, Rendon T, and Hsiao EY (2020). The maternal microbiome modulates fetal neurodevelopment in mice. Nature 586, 281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werling DM, and Geschwind DH (2013). Sex differences in autism spectrum disorders. Curr. Opin. Neurol. 26, 146–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BB, Van Benschoten AH, Cimermancic P, Donia MS, Zimmermann M, Taketani M, Ishihara A, Kashyap PC, Fraser JS, and Fischbach MA (2014). Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe 16, 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, Reilly SK, Lin L, Fertuzinhos S, Miller JA, et al. (2013). Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 155, 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the 16S rRNA gene sequencing data reported in this paper is NCBI Sequence Read Archive: BioProject PRJNA878523. The original, unprocessed data reported in this manuscript can be accessed via Mendeley Data: https://doi.org/10.17632/z6wz8kpk3n.1.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


