ABSTRACT
The IL-15 superagonist N-803 has been shown to enhance the function of CD8 T cells and NK cells. We previously found that in a subset of vaccinated, ART-naive, SIV+ rhesus macaques, N-803 treatment led to a rapid but transient decline in plasma viremia that positively correlated with an increase in the frequency of CD8 T cells. Here, we tested the hypothesis that prophylactic vaccination was required for the N-803 mediated suppression of SIV plasma viremia. We either vaccinated rhesus macaques with a DNA prime/Ad5 boost regimen using vectors expressing SIVmac239 gag with or without a plasmid expressing IL-12 or left them unvaccinated. The animals were then intravenously infected with SIVmac239M. 6 months after infection, the animals were treated with N-803. We found no differences in the control of plasma viremia during N-803 treatment between vaccinated and unvaccinated macaques. Interestingly, when we divided the SIV+ animals based on their plasma viral load set-points prior to the N-803 treatment, N-803 increased the frequency of SIV-specific T cells expressing ki-67+ and granzyme B+ in animals with low plasma viremia (<104 copies/mL; SIV controllers) compared to animals with high plasma viremia (>104 copies/mL; SIV noncontrollers). In addition, Gag-specific CD8 T cells from the SIV+ controllers had a greater increase in CD8+CD107a+ T cells in ex vivo functional assays than did the SIV+ noncontrollers. Overall, our results indicate that N-803 is most effective in SIV+ animals with a preexisting immunological ability to control SIV replication.
IMPORTANCE N-803 is a drug that boosts the immune cells involved in combating HIV/SIV infection. Here, we found that in SIV+ rhesus macaques that were not on antiretroviral therapy, N-803 increased the proliferation and potential capacity for killing of the SIV-specific immune cells to a greater degree in animals that spontaneously controlled SIV than in animals that did not control SIV. Understanding the mechanism of how N-803 might function differently in individuals that control HIV/SIV (for example, individuals on antiretroviral therapy or spontaneous controllers) compared to settings where HIV/SIV are not controlled, could impact the efficacy of N-803 utilization in the field of HIV cure.
KEYWORDS: HIV, IL-15 superagonist, N-803, SIV, infection, macaque, vaccine
INTRODUCTION
While nearly all individuals mount a CD8 T cell-mediated immune response to acute human immunodeficiency virus/simian immunodeficiency virus (HIV/SIV) infection, few individuals spontaneously control HIV/SIV replication (1, 2). Several studies comparing the immune responses of elite controllers to those of progressors have implied that polyfunctional CD8 T cells, consisting of both cytolytic and noncytolytic responses, are responsible, at least in part, for the control of HIV/SIV infections (3–5).
Prophylactic vaccine studies have been performed to elicit polyfunctional CD8 T cells that mimic those present in elite controllers (6, 7). Despite this, most of these strategies fail, and the CD8 T cells lose their potential to kill infected target cells and to suppress virus replication (8–10).
Recently, many investigators have turned to the use of immunotherapeutic agents which could, in combination with vaccines, improve the abilities of CD8 T cells to target and destroy infected cells during a HIV/SIV infection (11). One such class of immunotherapeutic agents that can boost the CD8 T cell response to HIV/SIV is the class of interleukin-15 (IL-15) agonists (12, 13). IL-15 is a cytokine that is normally produced by antigen-presenting cells during viral infections and promotes the development and growth of several innate and adaptive immune cells, in particular, NK and CD8 T cells (14–16).
One member of the class of IL-15 agonists that has gained enthusiasm in immunotherapeutic studies and clinical trials recently is N-803. N-803 is a soluble IL-15 superagonist in which a constitutively active IL-15 molecule containing a single amino acid mutation (N72D) is bound to the sushi domain of IL-15Rα and is fused to the Fc region of IgG1 (17, 18). N-803 is in use in clinical trials in cancer patients, as it improves the abilities of CD8 T cells and NK cells to target and destroy tumor cells (19).
There is a growing body of evidence suggesting that N-803 may be a promising immunotherapeutic agent in HIV+ individuals. Previous in vivo studies of SIV+ macaques (20–22) indicated that N-803 treatment increased CD8 T cell and NK cell frequencies. N-803 also increased the frequencies of these cells in the lymph nodes, which can likely be attributed to an increased expression of lymph node homing markers, such as CXCR5 (21). These promising features of N-803 have been a part of the rationale to test it in a phase II clinical trial in HIV+ individuals in Thailand under the name of Anktiva (https://immunitybio.com/immunitybio-announces-launch-of-phase-2-trial-of-il-15-superagonist-anktiva-with-antiretroviral-therapy-to-inhibit-hiv-reservoirs/), as well as in clinical trials in the United States (23) (https://actgnetwork.org/studies/a5386-n-803-with-or-without-bnabs-for-hiv-1-control-in-participants-living-with-hiv-1-on-suppressive-art/).
While N-803 has the potential to boost the frequency and cytolytic function of CD8 T cells in vivo (20, 22), the subsequent impact on HIV/SIV replication is less clear. We previously treated four ART-naive SIV+ rhesus macaques with N-803, and they all exhibited transient control of SIV plasma viremia within 7 days of the N-803 treatment (20). The suppression of SIV replication in these four animals, in the absence of ART, did not completely extend to other SIV+ animal studies (21). This could be partly attributed to the latency reversing activity of N-803 (24, 25). Understanding the conditions under which N-803 can successfully boost immune responses to control actively replicating HIV/SIV or reduce the viral reservoir is necessary for the improvement of the clinical relevance of this agent.
Here, we begin to define the features of SIV+ macaques that are associated with the N-803-mediated suppression of SIV replication. Previous associations include a lower chronic viral load set point (≤104 SIV gag copies/mL plasma) prior to N-803 treatment, host MHC genetics associated with spontaneous SIV control, and prior vaccination (20, 21). In this study, we tested the hypothesis that preexisting vaccine-elicited CD8 T cells were required for the N-803-mediated suppression of SIV replication. We used rhesus macaques (RM) expressing the Mamu-A*001 MHC class I allele, which is not associated with the natural control of SIV infections (26–28). Unfortunately, treatment with N-803 failed to reduce plasma SIV viremia in both vaccinated and unvaccinated macaques.
We then rearranged the animal groups to determine whether the spontaneous initial control of SIV replication was associated with the improved immunological responsiveness of polyfunctional virus-specific CD8+ T cells to N-803. We characterized the functions of peripheral SIV-specific CD8 T cells prior to and during the first 7 days after the N-803 treatment. We found that the proliferative and cytolytic capacities of the SIV-specific cells from the SIV controllers were more robust compared to those from the SIV noncontrollers after the N-803 treatment. Our results imply that N-803 efficacy is likely maximized in a host who has not succumbed to the severe immunological dysfunction associated with HIV/SIV pathogenesis.
RESULTS
Treatment with N-803 did not suppress chronic SIV replication in vaccinated or unvaccinated rhesus macaques.
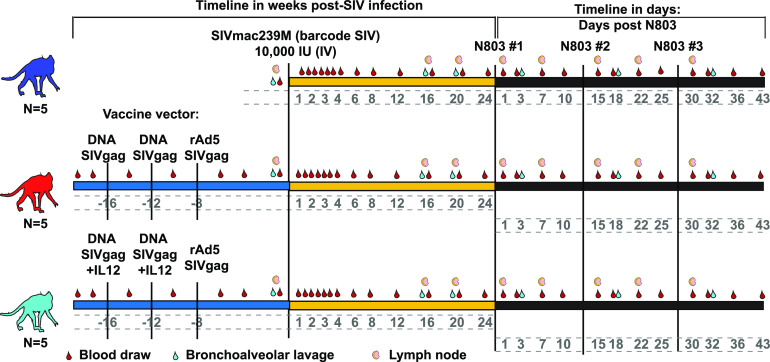
Our initial goal was to test the hypothesis that prophylactic vaccination elicits CD8 T cells that are recalled during N-803 treatment and suppress plasma viremia. We designed a study with 15 rhesus macaques that expressed the Mamu-A1*001 MHC class I allele but did not express the Mamu-B*008 or-B*017 alleles that are associated with SIV viral control (27, 29). 10 animals were vaccinated with a DNA prime/rAd5 boost regimen to generate a high frequency of SIV-specific CD8 T cells (Fig. 1, red and light blue). We coadministered a plasmid expressing rhesus macaque IL-12 along with the DNA vaccine in five of the animals (Fig. 1, light blue). All vaccine vectors expressed the SIVmac239 gag gene. Five animals remained unvaccinated (Fig. 1, dark blue). This specific vaccine strategy was chosen to elicit CD8 T cells without preventing infection with SIV (30, 31). All 15 of the animals were then infected intravenously with 10,000 infectious units (IU) of the molecularly barcoded SIVmac239M strain (Fig. 1) (32). The population of the barcodes present in circulating viruses was examined from 7 days up to 168 days (~5 to 6 months) postinfection between the vaccinated and unvaccinated macaques, and no apparent differences were observed between the 3 cohorts (data not shown).
FIG 1.
Outline for Mamu-A*001+ macaque vaccine study. Mamu-A*001+ rhesus macaques were either left unvaccinated (blue) or vaccinated with a heterologous prime/boost regimen with (light blue) or without (red) an IL-12 DNA vector adjuvant, as indicated. Animals were infected with SIVmac239M for ~6 months and then treated with 3 doses of 0.1 mg/kg N-803, delivered subcutaneously, separated by 2 weeks per dose. Samples were collected as indicated.
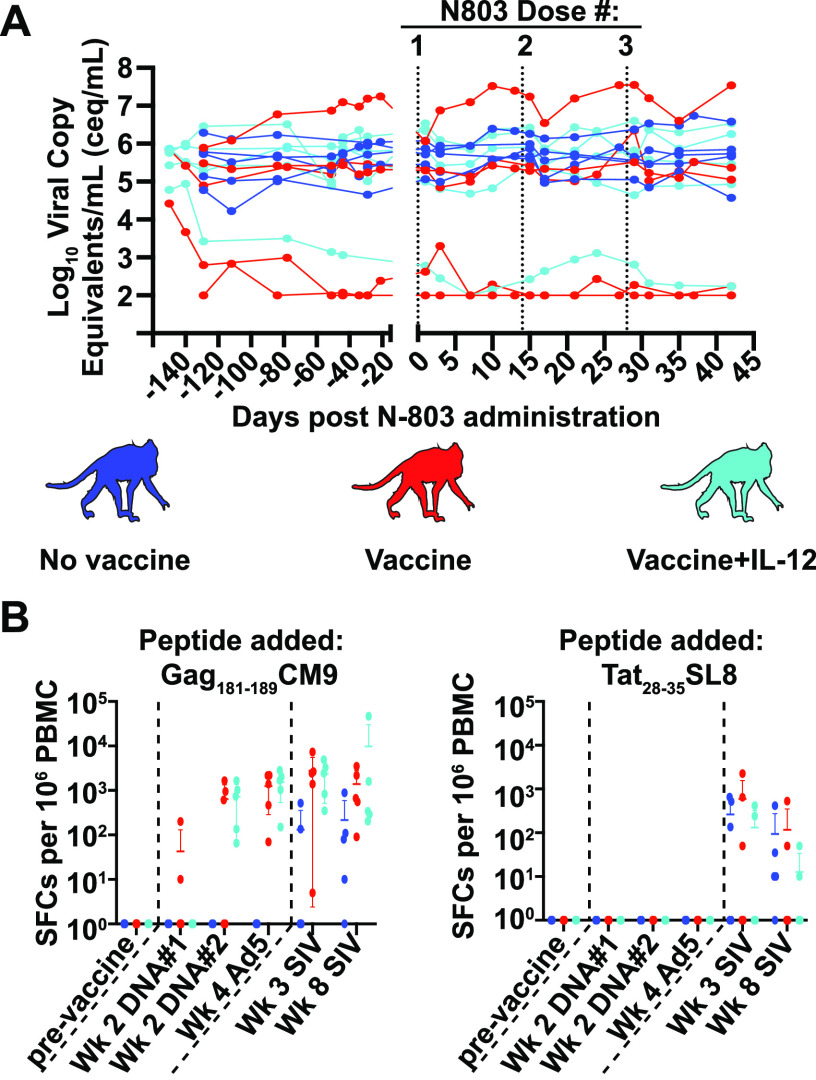
As expected, the vaccine did not protect from SIV infection. However, the viral load set point in 3 of the 15 animals was below 104 copies/mL (Fig. 2A). These three animals were all vaccinated. In contrast, chronic plasma viremia in the other 12 animals was between 105 and 107 copies/mL (Fig. 2A). Approximately 6 months after infection, all 15 animals received 3 doses of 0.1 mg/kg N-803, separated by 14 days each (Fig. 1). N-803 treatment did not affect the plasma viral loads for any of the animals (Fig. 2A). We also examined the frequencies and absolute counts of SIV specific and bulk CD8 T cells present in the peripheral blood and lymph nodes. In concordance with previous findings (21, 22), there was a decrease in the absolute counts of SIV-specific and bulk CD8 T cells 1 day after N-803 treatment, followed by 2 to 10-fold increases by day 7 in the peripheral blood. These changes in CD8 T cell counts were transient, and they were not different between the vaccinated and unvaccinated macaques (Fig. S1A). Webb and colleagues also previously observed moderate increases in the frequency of CD8 T cells in the lymph nodes following an N-803 treatment (21, 22). In the present study, while there was a trend toward increased frequencies of CD8 T cells in the lymph nodes by day 7 after the N-803 treatment, these changes were not statistically significant and did not differ between the vaccinated and unvaccinated macaques (Fig. S1B).
FIG 2.
Viral loads for vaccinated and unvaccinated macaques are unchanged and did not differ during the N-803 treatment. (A) Plasma was isolated from whole blood samples from the unvaccinated (blue) and vaccinated (red and light blue) animals from the indicated time points post-N-803 administration, and the Log10 virus copy equivalents/mL (ceq/mL) were determined as described in the methods. Vertical dashed lines indicate a time point at which N-803 was delivered. (B) IFN-γ ELISpots were performed on frozen PBMCs as indicated in the methods. Briefly, PBMC from the indicated time points post-vaccination or post-SIV infection were thawed and then incubated overnight with 1 μM of either the indicated peptides or media alone as a negative control. The plates were developed according to the manufacturer's instructions. A positive response was defined as the greater of the number of spot-forming colonies (SFCs) per 106 PBMCs that were 2 standard deviations above the average value for the negative-control, or 50 SFCs/106 PBMC.
We performed IFN-γ-ELISpot assays using peripheral blood mononuclear cells (PBMCs) collected pre- and post-vaccination, as well as pre- and post-SIV infection (Fig. 2B), to confirm that the vaccine did elicit Gag-specific T cells. The majority of the vaccinated animals had a positive IFN-γ ELISpot response when stimulated with the Gag181-189CM9 peptide by 2 weeks after the second DNA vaccination, and all of the animals had detectable Gag181-189CM9-specific T cells at 4 weeks after vaccination with the rAd5 particles (Fig. 2B, left graph). The unvaccinated macaques only produced Gag181-189CM9-specific T cells after the SIV infection (Fig. 2B, left panel). No animals had detectable T cells specific for Tat28-35SL8 until after the SIV infection (Fig. 2B, right panel).
Reevaluation of N-803 efficacy based on prior immune mediated viral control.
Prior vaccination did not predict whether the macaques would respond to the N-803 treatment, suggesting that this was not the sole factor responsible for the N-803-mediated virus suppression that we observed in our previous study (20). All of the animals in our previous study spontaneously controlled SIV earlier during infection, a result typically associated with cytotoxic T cell function (33). Therefore, we decided to restructure the current study to test the alternative hypothesis that prior spontaneous SIV control predicts whether N-803 treatment improves CD8 T cell function with the potential to control SIV replication. Along the same lines, we expected that the CD8 T cells from the SIV noncontrollers would not exhibit improved function after the N-803 treatment.
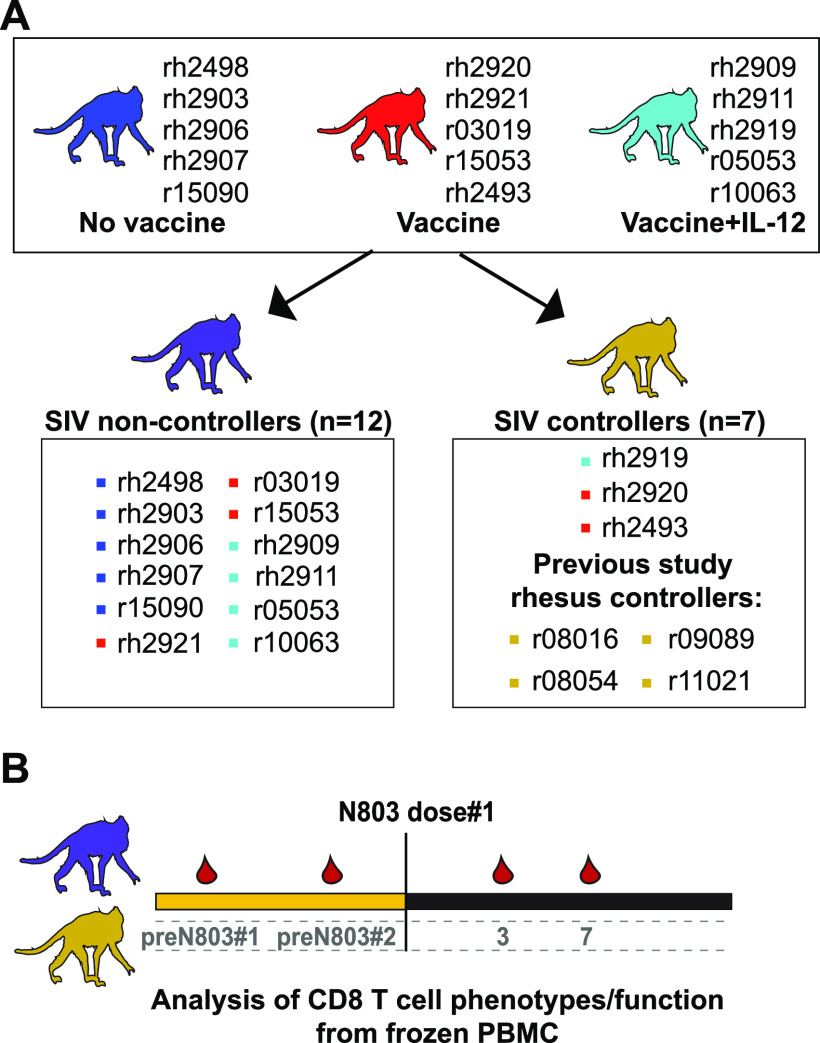
To test this alternative hypothesis, we rearranged our animal groups into SIV controllers and noncontrollers based on the viral load set point established prior to treatment with N-803. We included the 12 SIV noncontrollers from this study (Fig. 3A, purple) whose viral load setpoint was above 104 copies/mL. There were only 3 animals with a viral load set point lower than 104 copies/mL in the current study, so we included samples from the 4 animals who responded to N-803 in the 2018 study to increase the size of the controller group (20) (Fig. 3A, gold). These animals had been infected with non-barcoded SIVmac239 for 1.5 to 3 years, and they had viral loads of approximately 103 to 104 copies/mL at the time of N-803 treatment (Table 1) (20). We only included PBMC collected during the first 7 days after the first dose of N-803 treatment from the 2018 study (Fig. 3B) because the N-803 regimens were different for these two studies after this time point (described in Table 1).
FIG 3.
Rearrangement of animals into SIV controllers and SIV noncontrollers. (A) Animals from the original vaccine study (blue, red, and light blue) were grouped according to viral loads. SIV noncontrollers (n = 12, purple monkey) are shown in the box on the left and had viral loads above 104 ceq/mL (Fig. 2). SIV controllers (n = 7, gold monkey) are shown in the box on the right and had viral loads at or below 104 ceq/mL (20) (Fig. 2). (B) Timeline of samples used for studies comparing CD8 T cells from the SIV controllers and noncontrollers. Frozen PBMC collected from the time points indicated from the animal groups described in (A) are shown on the timeline and were used for the downstream analysis.
TABLE 1.
Animals included in the studya
| Animal ID | Sex | Age at time of N-803 (yrs) | Infecting SIV strain | Length of SIV infection | N-803 dosing regimen | Samples were used for the indicated figures |
|---|---|---|---|---|---|---|
| rh2903 | M | 5 | SIVmac239M | 6 mo | 0.1 mg/kg SubQ every 2 wks | 1 to 8; S1; S4 |
| rh2906 | F | 4 | SIVmac239M | 6 mo | 1 to 8; S1; S4 | |
| rh2907 | F | 4 | SIVmac239M | 6 mo | 1 to 8; S1; S4 | |
| rh2909 | M | 5.5 | SIVmac239M | 6 mo | 1 to 8; S1; S4 | |
| rh2911 | M | 5.4 | SIVmac239M | 6 mo | 1 to 8; S1; S4 | |
| rh2919 | F | 4.6 | SIVmac239M | 6 mo | 1 to 8; S1; S4 | |
| rh2920 | M | 6.5 | SIVmac239M | 6 mo | 1 to 8; S1; S4 | |
| rh2921 | F | 4.5 | SIVmac239M | 6 mo | 1 to 8; S1; S4 | |
| r05053 | F | 15 | SIVmac239M | 6 mo | 1 to 8; S1; S4 | |
| r15053 | M | 3.5 | SIVmac239M | 6 mo | 1 to 8; S1; S4 | |
| r15090 | F | 3.3 | SIVmac239M | 6 mo | 1 to 8; S1; S4 | |
| r03019 | F | 16 | SIVmac239M | 6 mo | 1 to 8; S1; S4 | |
| r10063 | M | 10 | SIVmac239M | 6 mo | 1 to 8; S1; S4 | |
| rh2493 | M | 11.9 | SIVmac239M | 6 mo | 1 to 8; S1 to S4 | |
| rh2498 | M | 11.9 | SIVmac239M | 6 mo | 1 to 8; S1; S3 to S4 | |
| r08016 | M | 8 | SIVmac239 | 3 yr | 0.1 mg/kg SubQ weekly | 3 to 8; S4 |
| r08054 | M | 6 | SIVmac239 | 3 yr | 3 to 8; S4 | |
| r09089 | M | 7 | SIVmac239 | 3 yr | 3 to 8; S2, S4 | |
| r11021 | M | 5 | SIVmac239 | 1.5 yr | 3 to 8; S4 |
S1 to S4, Supplementary figures 1 to 4; yr, year; mo, month. Length of SIV infection indicates the length of the SIV infection prior to the N-803 treatment.
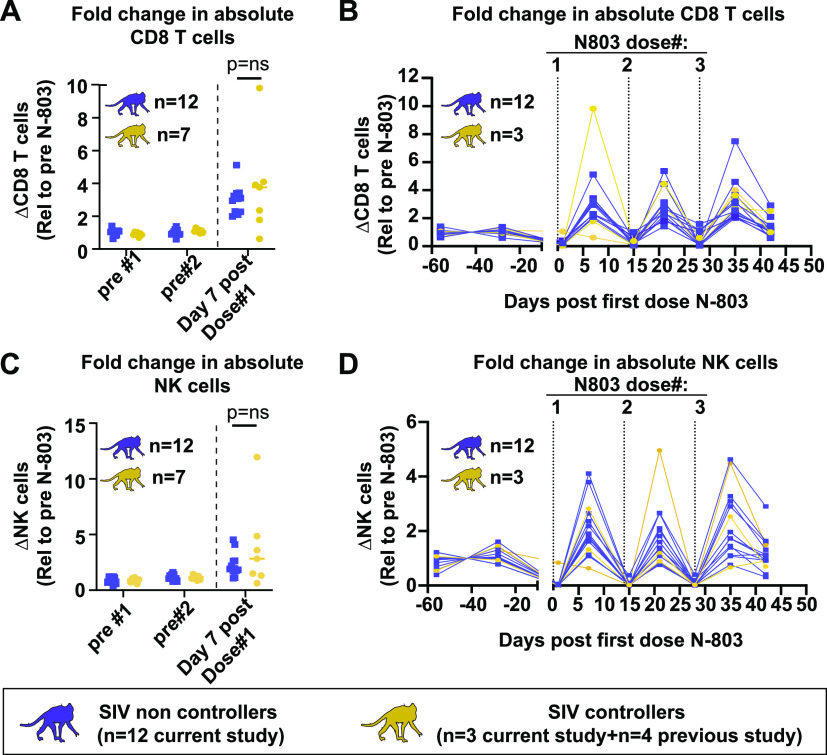
SIV controller status does not impact N-803 mediated increases in bulk CD8 T cells or NK cells.
We measured the increases in absolute CD8 T cells (Fig. 4A) and NK cells (Fig. 4C) in the peripheral blood from the SIV controllers and noncontrollers. There was no difference in the N-803 mediated increase in either cell type between the controllers (gold) and the noncontrollers (purple) 7 days after the first dose of N-803. We found similar results when we extended this analysis to the same cell populations for all three doses of N-803 used in the animal study described here (Fig. 4B and D).
FIG 4.
No differences were observed in the increases of bulk CD8 T cells or NK cells between the SIV controllers and noncontrollers during the N-803 treatment. (A–D). PBMC from SIV noncontrollers (purple) or SIV controllers (gold) collected at the indicated time points post-N-803 treatment were stained with the panel originally described in Table 3 of (20). The frequency of CD8 T cells (defined as CD3+CD8+ cells; panels A and B) and NK cells (defined as CD3−CD8+NKG2A+ cells; panels C and D) were determined. Complete white blood cell counts (CBC) were used to quantify the absolute number of each cell population. Then, the data were normalized to the average value for the pretreatment controls for each population and are displayed as the fold changes in absolute cell counts, relative to the pretreatment average. For panels A and C, Mann-Whitney tests were performed to determine statistical significance. P = ns, not significant.
SIV controller status does not impact N-803 mediated expansion of SIV-specific cells.
We wanted to determine if the virus-specific CD8 T cells from the SIV controllers were uniquely more responsive to treatment with N-803, compared to their noncontroller counterparts. To characterize the SIV-specific cells in greater detail, we utilized the Mamu-A*001 tetramers Gag181-189CM9 and Tat28-35SL8. We also included the Mamu-B*008-tetramer Nef137-146RL10 for the animals who expressed Mamu-B*008 but not Mamu-A*001 (r08016, r09089, and r08084 from Fig. 3). See Fig. S2 for the gating schematics.
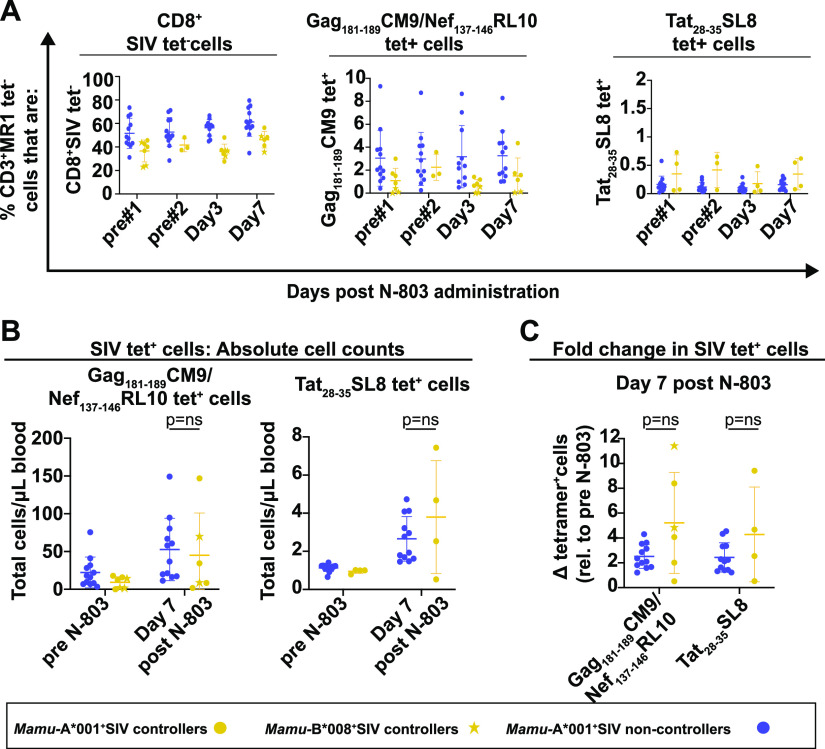
Within each group, we examined the frequencies of the CD3+ cells that were CD8+SIV tetramer negative (CD8+SIVtet−), Gag181-189CM9/Nef137-146RL10 tetramer positive, or Tat28-35SL8 tetramer positive (Fig. 5A). At each time point, there were somewhat higher frequencies of CD3+CD8+SIV tetramer negative T cells in the SIV noncontrollers compared to the SIV controllers, and their frequencies were unaffected by the N-803 treatment (Fig. 5A, left panel). When we examined the SIV tetramer+ cells, there were no statistically significant differences between the two groups in the frequencies of any tetramer+ cells before or after the N-803 treatment (Fig. 5A, middle and right panels). We also calculated the absolute numbers of Gag181-189CM9, Nef137-146RL10, and Tat28-35SL8+ CD8 T cells pre-and post-N-803 treatment (Fig. 5B). While N-803 did increase the absolute number of these antigen-specific T cells in the peripheral blood 7 days after its administration, the fold change in antigen-specific T cells was similar for both the SIV controllers and the noncontrollers (Fig. 5C).
FIG 5.
Frequencies of SIV-specific cells or memory populations between SIV controllers and noncontrollers do not differ, and they are not altered during N-803 treatment. (A) Frozen PBMC from the SIV noncontrollers (purple) and SIV controllers (gold) that were collected from the indicated time points post-N-803 were thawed and then stained with the panel described in Table 3. Flow cytometric analysis was performed to determine the frequencies of the CD3+MR1 tetramer-cells that were: CD8+SIV tet– (left panel), CD8+Gag181-189CM9 (gold/purple circles), CD8+Nef137-146RL10 (gold stars) tet+ (middle panel), or CD8+Tat28-35SL8 tet+ (right panel) for each time point. (B and C) Frozen PBMC from the SIV noncontrollers (purple) or SIV controllers (gold) were collected from the indicated time points as described above. Complete white blood cell counts (CBC) were used to quantify the absolute number of each cell population (B). Then, the data were normalized to the average value for the pretreatment controls for each population and are displayed as the fold changes in absolute cell counts, relative to the pretreatment average (C). Mann-Whitney tests were performed to determine statistical significance. P = ns, not significant.
The frequencies of vaccine-elicited T cells expressing granzyme B and ki-67 are increased more in SIV controllers compared to noncontrollers.
We hypothesized that the vaccine-elicited CD8 T cells from SIV controllers may exhibit improved function after N-803 treatment, compared to those of noncontrollers. This includes increased proliferation and cytolytic potential. The proliferation of cells can be measured by examining the frequency expressing ki-67, which is an intracellular marker that aids in cell division (34). N-803 is known to increase the frequency of ki-67+ CD8 T cells from HIV-naive humans in vitro and SIV+ macaques in vivo (20, 21). granzyme B and perforin are molecules involved in the degranulation and destruction of infected target cells (35, 36). N-803 and other IL-15 agonists increase the expression of granzyme B and perforin in the NK cells and CD8 T cells of healthy individuals (18, 37, 38). We do not know how chronic immune activation, a feature common among SIV noncontrollers (39), reduces the expansion of CD8 T cells producing ki-67, granzyme B, or perforin upon N-803 treatment, compared to the SIV controllers.
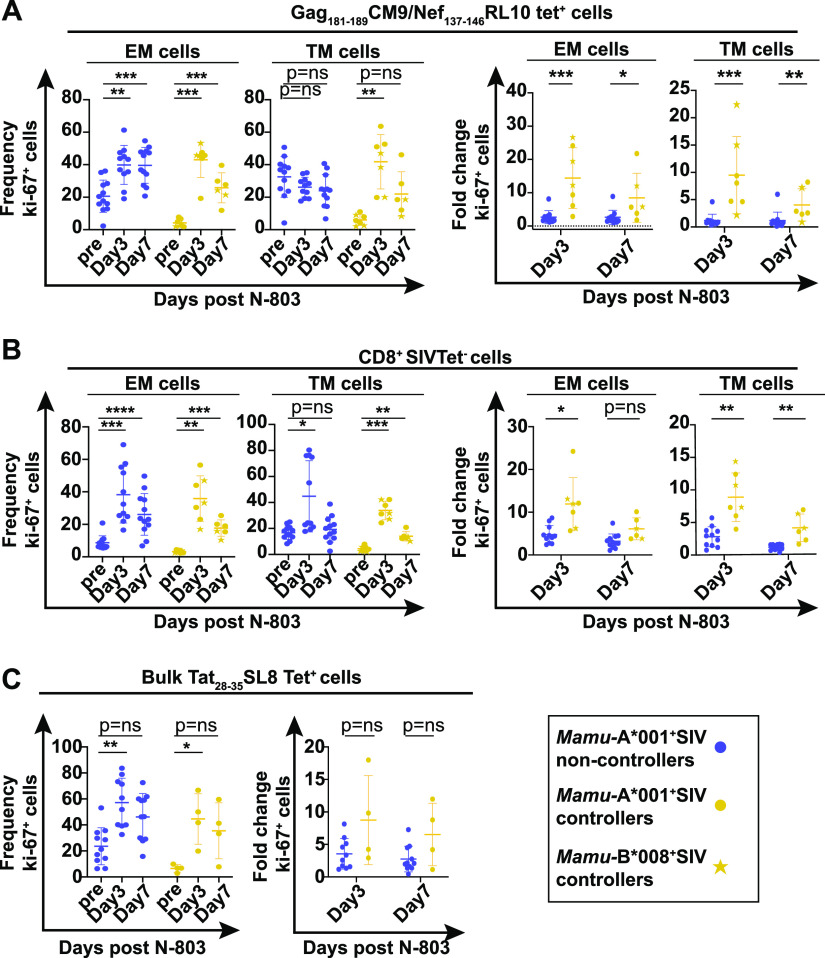
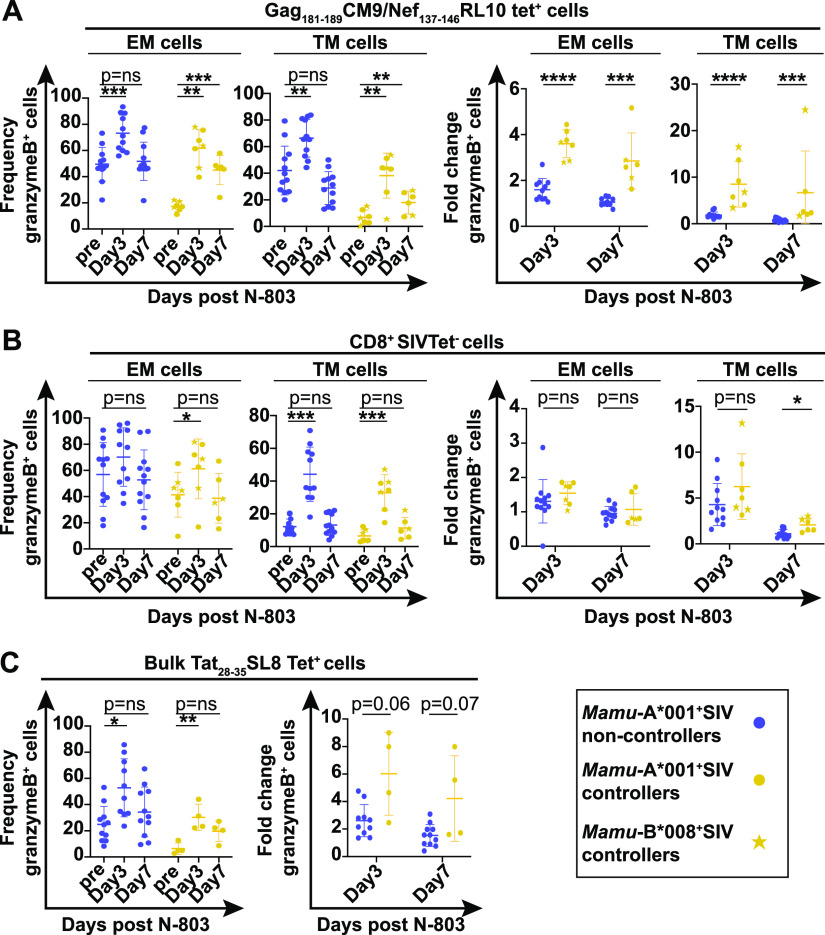
We measured the frequencies of CD8+SIV tetramer+ and CD8+tetramer-negative cells expressing granzyme B and ki-67 before and after receiving N-803 (gating is shown in Fig. S3). We found the frequency of Gag181-189CM9 tetramer+ EM and TM cells producing ki-67 or granzyme B was increased most notably on day 3 after N-803 treatment (Fig. 6A and 7A, left two panels). The fold increases in Gag181-189CM9 tetramer+ cells expressing these markers was most apparent in the SIV controllers, compared to the SIV noncontrollers (Fig. 6A and 7A, right two panels). This was attributed to the fact that the baseline frequencies of the Gag181-189CM9 tetramer+ EM and TM cells expressing ki-67 or granzyme B were much higher in the SIV noncontrollers than in the controllers (Fig. 6 and 7A, left panels; Fig. S3). This is likely a result of ongoing antigenic stimulation by circulating virus. Central memory cells were too rare to characterize (data not shown).
FIG 6.
N-803 treatment increases the frequency of SIV-specific cells expressing the proliferation marker ki-67 to a greater extent in SIV controllers compared to SIV noncontrollers. Frozen PBMC from SIV noncontrollers (purple) and SIV controllers (gold circles and stars) that were collected from the indicated time points post-N-803 treatment were thawed, and flow cytometry was performed using the panel described in Table 2. The frequencies of ki-67+ cells (left panels) and the fold changes in ki-67 relative to the pre-N-803 treatment controls (right panels) were examined at the indicated time points for effector memory (CD28−CD95+CCR7–; EM), and transitional memory (CD28+CD95+CCR7–; TM), cells for the following parent populations of cells: (A) CD8+Gag181-189CM9 (gold/purple circles) or CD8+Nef137-146RL10 (gold stars) tet+ cells or (B) CD8+SIV tet–cells. (C) CD8+Tat28-35SL8 tet+ cells were analyzed in bulk for the frequencies (left panel) and fold changes (right panel) of the ki-67+ cells. For the graphs of the ki-67 frequencies within a cohort, repeated measures analysis of variance (ANOVA) nonparametric tests were performed with Dunnett’s multiple comparisons for individuals across multiple time points. For graphs comparing the fold changes in the percent ki67+ cells between controllers and noncontrollers for a given time point, Mann-Whitney tests were performed to determine statistical significance: P = ns, not significant; *, P ≤ 0.05; **, P ≤ 0.005; ***, P ≤ 0.0005.
FIG 7.
N-803 treatment increases the frequency of SIV-specific cells expressing the proliferation marker granzyme B to a greater extent in SIV controllers compared to SIV noncontrollers. Frozen PBMC from SIV noncontrollers (purple) and SIV controllers (gold circles and stars) that were collected from the indicated time points post N-803 were thawed, and flow cytometry was performed using the panel described in Table 2. The frequencies of granzyme B+ cells (left panels) or the fold changes in granzyme B+ cells relative to the pre-N-803 controls (right panels) were examined at the indicated time points for effector memory (CD28–CD95+CCR7–; EM), and transitional memory (CD28+CD95+CCR7-; TM), cells for the following parent populations of cells: (A) CD8+Gag181-189CM9 (gold/purple circles) or CD8+Nef137-146RL10 (gold stars) tet+ cells or (B) CD8+SIV tet–cells. (C) CD8+Tat28-35SL8 tet+ cells were analyzed in bulk for the frequency (left panel) and fold change (right panel) of granzyme B+ cells. For the graphs of the granzyme B frequencies within a cohort, repeated measures ANOVA nonparametric tests were performed with Dunnett’s multiple comparisons for individuals across multiple time points. For graphs comparing the fold changes in the percent granzyme B+ cells between controllers and noncontrollers for a given time point, Mann-Whitney tests were performed to determine statistical significance: P = ns, not significant; *, P ≤ 0.05; **, P ≤ 0.005; ***, P ≤ 0.0005.
In contrast to the Gag181-189CM9 tetramer+ cells, the changes in the frequencies of the CD8+ SIV tetramer negative EM and TM cells expressing ki-67 or granzyme B after the N-803 treatment were similar in all of the animals (Fig. 6B and 7B, left two panels). This resulted in fewer differences between the SIV noncontrollers and the SIV controllers. The population of Tat28-35SL8 tetramer+ cells was analyzed in bulk, as there were too few cells to examine individual memory populations. The frequencies of Tat28-35SL8 tetramer+ cells expressing ki-67 and granzyme B increased similarly after the N-803 treatment across animals (Fig. 6C and 7C).
N-803 treatment of SIV controllers increases the frequency of virus-specific CD8 T cells expressing CD107a.
We performed intracellular cytokine staining (ICS) assays using PBMC collected pre- and post-N-803 treatment. We compared the frequency of antigen-specific CD8 T cells producing TNF-α, IFN-γ, and CD107a between the SIV controllers and the noncontrollers. A representative gating schematic for CD107a, TNF-α, and IFN-γ is shown in Fig. S4 for an SIV controller (gold, top panels) and for an SIV noncontroller (purple, bottom panels).
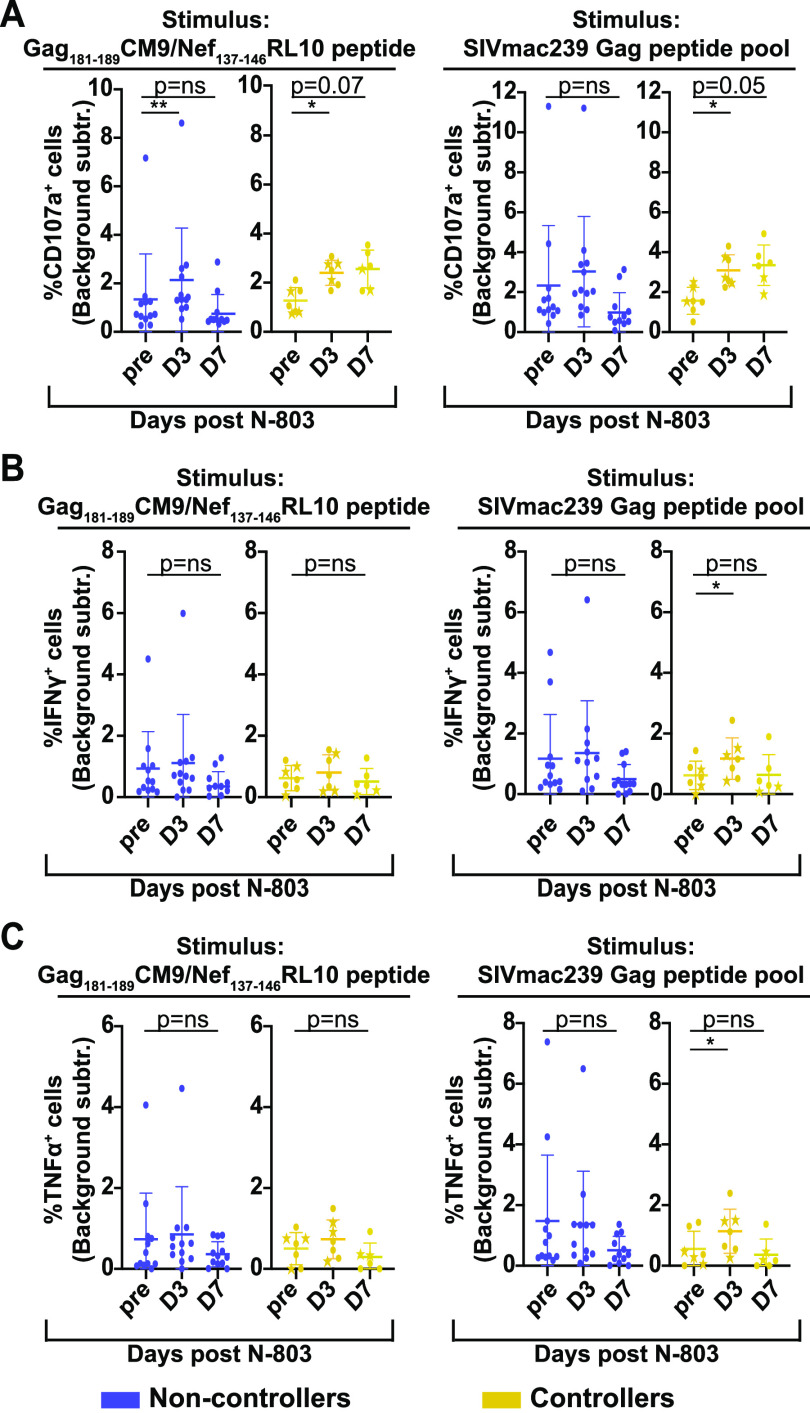
We found that all animals had an increased frequency of CD8+CD107a+, IFN-γ+, and TNF-+ cells in response to Gag or Nef peptides or a Gag peptide pool, relative to unstimulated controls (Fig. S4B). We normalized the data from stimulated to unstimulated controls collected from matched time points (Fig. 8A to C).
FIG 8.
N-803 treatment in SIV controllers, but not in SIV noncontrollers, leads to an improvement in CD107a production in functional assays. Frozen PBMC from SIV noncontrollers (purple) and SIV controllers (gold) that were collected from the pre-N-803 treatment (pre), day 3 post-N-803 treatment (D3), and day 7 post-N-803 treatment (D7) time points were thawed and incubated overnight with either media alone, 0.5 μg/mL Gag181-189CM9, Nef137-146RL10 peptides, or 0.5 μg/mL of the SIVmac239 Gag peptide pool. The next day, flow cytometry was performed using the panel described in Table 4. The data were normalized by background subtraction. The frequency of cells producing CD107a (A), IFN-γ (B), or TNF-α (C) after background subtraction in response to the indicated antigen are shown. For all statistical analyses, repeated measures ANOVA nonparametric tests were performed with Dunnett’s multiple comparisons. For individuals for whom samples from time points were missing, mixed-effects ANOVA tests were performed using the Geisser-Greenhouse correction. *, P ≤ 0.05; **, P ≤ 0.005.
Similar to the frequencies of CD8 T cells expressing ki-67 or granzyme B, the SIV noncontrollers (purple) had a higher background of CD8 T cells expressing CD107a compared to the controllers (Fig. S4B). After eliminating the background signal, we found that the N-803 treatment had little impact on the frequency of CD107a+ cells from the SIV noncontrollers, relative to the pre-N-803 time points (Fig. 8A, purple). Similarly, after the elimination of background signal, there were no statistically significant differences in the frequencies of IFN-γ+ or TNF-α+ cells after N-803 treatment for the SIV noncontrollers, relative to the pre-N-803 time points (Fig. 8B and C, purple).
In contrast, cells collected from SIV controllers at day 3 post-N-803 treatment had a statistically significant increase in the frequency of antigen-specific CD107a+ CD8 T cells. The frequency of CD107a+ antigen-specific cells collected from day 7 post-N-803 remained elevated but did not reach statistical significance (Fig. 8A, right panel, gold). We found minor but statistically significant increases in IFN-γ+ (Fig. 8B, right panel, gold) and TNF-α+ (Fig. 8C, right panel, gold) CD8 T cells with Gag peptide pool stimulation from the cells collected from the day 3 post-N-803 treatment time point, compared to the pre-N-803 time point.
DISCUSSION
We previously found that the administration of the IL-15 superagonist N-803 to SIV+ ART-naive vaccinated macaques led to the rapid, but transient, control of plasma viremia (20). Here, we tested the hypothesis that vaccination, prior to SIV infection, was necessary for the N-803 treatment to induce suppression of plasma viremia. Unfortunately, we found that prior vaccination alone did not confer N-803-mediated control of plasma viremia during N-803 treatments in macaques (Fig. 2). When divided into groups based on viral load, we found that the N-803 treatment led to larger increases in the frequencies of ki-67 and granzyme B+ Gag181-189CM9 tetramer+ cells of SIV controllers compared to those of SIV noncontrollers (Fig. 6 and 7). Additionally, in the SIV controllers but not in the noncontrollers, the CD8 T cells collected after the N-803 treatment were more likely to produce CD107a after Gag peptide stimulation. These findings suggest that Gag-specific CD8 T cells from SIV controllers may have the potential to exhibit improved proliferation and cytotoxicity with the N-803 treatment, compared to those of SIV noncontrollers.
Our study could have important implications for the clinical use of N-803 to boost CD8 T cell function in HIV+ individuals. Those with uncontrolled plasma viremia have dysregulated HIV/SIV specific CD8 T cells (8, 39). We observed that the SIV-specific CD8 T cells of noncontrollers had a chronically activated phenotype, with higher frequencies of cells expressing granzyme B and ki-67 before the N-803 treatment (Fig. 6 and 7; Fig. S3). N-803 did not lead to a substantial increase of CD8 T cells with this activated phenotype during treatment (Fig. 6 and 7). Chronic immune activation leads to a type of immune “exhaustion” that leaves HIV/SIV specific cells unable to combat infection (39–42). It is possible that the SIV-specific CD8 T cells from the SIV noncontrollers exhibit some phenotypes associated with immune exhaustion, which could lead to the inhibition of signaling pathways downstream of the IL-15 receptor activation. For example, STAT-5 is activated as a part of the IL-15 receptor signaling pathway (43). Chronic HIV/SIV infection can lead to an increase in the frequency of PD-1 expression on immune cells (44, 45), which may reduce STAT-5 activation (46, 47) and thus inhibit the IL-15 receptor signaling pathway.
Alternatively, chronic viral infection and/or immune exhaustion in our macaque model could have direct effects on the surface expression of the IL-15 receptor. The IL-15R is one member of the γC receptor cytokine family, which also includes the IL-7R and IL-2R (43). HIV infection in humans can lead to the downregulation of IL-7 receptor expression (CD127) on CD8 and CD4 T cells (48, 49). Studies in LCMV-infected mice have shown that chronic LCMV infection and a subsequent activation/exhaustion of immune cells can lead to the downregulation of the IL-15/IL-2 β chain receptor, CD122 (50). We previously found that multiple doses of N-803 led to the downregulation of CD132 (common γ chain receptor) and CD122 on CD8 T cells and NK cells (20). We did not have enough frozen PBMC from the SIV controllers to measure the expression of CD132 and CD122 across all of the animals in the present study. Future studies could determine whether the chronic immune activation of antigen-specific CD8 T cells of SIV noncontrollers leads to the inhibition of IL-15 receptor expression or the dysfunction of IL-15 signaling pathways after the N-803 treatment, compared to those of SIV controllers. Furthermore, while antiretroviral treatment (ART) may not fully restore the function and phenotype of HIV/SIV-specific CD8 T cells (51), future studies could determine whether ART can restore the defects associated with IL-15 signaling that are ascribed to the chronic immune activation of antigen-specific CD8 T cells in SIV noncontrollers.
One other factor that may be driving chronic immune activation in the SIV noncontrollers is the type I interferon (IFN-I). There is a growing body of evidence that IFN-I can augment immune activation during chronic viral infections, including HIV/SIV infection (52, 53). In addition to the dysregulation of CD8 and CD4 T cells in the periphery, IFN-I signaling during chronic SIV infection can also impair the development of B cells and the antibody response in the lymph node follicles (54). These defects could further drive virus proliferation and subsequent immune cell activation and/or exhaustion (54). If chronic IFN-I signaling is responsible for the phenotypes we observed in our study here (Fig. 6 and 8), future studies could investigate the blocking of IFN-I signaling prior to or during N-803 treatment to reverse chronic immune cell activation and improve T cell function. There have been success in LCMV-infected mice showing that blocking IFN-I signaling could result in faster viral clearance and reduced viral loads (55). Some recent studies have shown that blocking IFN-I signaling during chronic SIV infection could improve T cell function and reduce SIV reservoir size (56, 57). Overall, exploring mechanisms by which we could reverse chronic immune activation and increase responsiveness to N-803 could be valuable for individuals who do not naturally control HIV/SIV infection.
While vaccination did not always lead to the control of SIV infection in our study, the vaccine-elicited Gag181-189CM9 tetramer+ cells from the SIV controllers exhibited greater increases in the frequencies of cells expressing ki-67 and granzyme B during the N-803 treatment, compared to those of the noncontrollers (Fig. 6 and 7A). Furthermore, the Gag-specific CD8 T cells in the ICS assays from the SIV controllers displayed a greater ability to produce CD107a, compared to the Gag-specific CD8 T cells from the SIV noncontrollers (Fig. 8). These improvements to the proliferative and cytotoxic capabilities suggests that vaccination, combined with N-803 immunotherapy, may impact the SIV reservoir. N-803 was found in two separate studies to be unable to reduce the SIV reservoir, but both of these studies were in unvaccinated macaques (22). However, we observed differences in the functions of CD8 T cells in the peripheral blood in the SIV controllers during the N-803 treatment, compared to those of the noncontrollers (Fig. 6 and 8). While our vaccination approach was not successful, our data could indicate that a different, more robust vaccination approach plus N-803 treatment could improve the cytotoxic function of CD8 T cells in the lymph nodes and tissues. Since the lymph nodes are critical sites of SIV replication, it will be interesting to focus on the dynamics of N-803-boosted, vaccine-elicited CD8 T cells from SIV controllers and noncontrollers in future, larger animal studies.
Overall, our study begins to explore the host conditions under which N-803 may be most effective at improving the cytotoxic function of CD8 T cells. We found that prophylactic vaccination alone did not condition animals to respond to N-803. However, animals predisposed to viral control had a population of CD8 T cells that were more responsive to N-803, compared to animals that never controlled virus replication. Specifically, it appears as though N-803 increases the function of the SIV-specific CD8 T cells from the SIV controllers to a greater degree than those from the SIV noncontrollers. Our findings support that N-803 could have potential as an immunotherapeutic agent for HIV/SIV+ individuals, particularly in the setting of HIV/SIV control, such as during ART treatment.
MATERIALS AND METHODS
Animals and reagents.
(i) Animals. All 15 of the newly infected Indian rhesus macaques involved in this study were genotyped for the MHC class I allele Mamu-A1*001 using methods described previously (58). All of the animals involved in this study were cared for and housed at the Wisconsin National Primate Resource Center (WNPRC), following practices that were approved by the University of Wisconsin Graduate School Institutional Animals Care and Use Committee (IACUC; protocol number G005507). All procedures, such as the administration of vaccines, biopsies, blood draw collections, and the administration of N-803, were performed as written in the IACUC protocol, under anesthesia to minimize suffering.
Frozen samples were also analyzed from rhesus macaques included in a previously published study (20) that was approved under protocol number G005507. Table 1 shows a list of the animal ID, sex, infecting SIV strain, length of SIV infection prior to N-803 treatment, N-803 dosing regimen, and the figures in which samples from each animal were included.
(ii) DNA plasmid vector. The endotoxin-free DNA plasmid vaccine vector (hCMV/R-SIVmac239 gag) utilized in this study consisted of the SIVmac239 gag gene produced under the control of a CMV promoter (hCMV/R), and it was constructed as previously described (59). This plasmid vector was kindly provided by John Mascola, Robert Seder, and Wing-Pui Kong at the NIH Vaccine Research Center.
(iii) Rhesus IL-12 DNA plasmid vector. The endotoxin-free rhesus IL-12 DNA plasmid vector (plasmid AG157) was kindly provided by George Pavlakis and Barbara Felber. This plasmid vector consisted of the two subunits of the rhesus IL-12 gene expressed from dual promoters in the plasmid, and it was constructed as previously described (60).
(iv) rAd5 SIVmac239gag vaccine vector. All vaccinated animals received 1010 particles of a replication-incompetent rAD5 vaccine vector expressing SIVmac239 gag (ViraQuest; North Liberty, IA).
(v) N-803 reagent. N-803 was provided by ImmunityBio (San Diego, CA), and it was produced using previously described methods (18).
Clinical procedures and viral loads.
(i) Vaccination of rhesus macaques. The vaccine regimen consisted of a heterologous DNA prime/Ad5 boost strategy. Briefly, all vaccinated macaques received 2 doses of 2 mg of a DNA vaccine vector expressing the SIVmac239 gag gene, with or without 0.2 mg of a rhesus macaque IL-12 plasmid expression vector, delivered by electroporation (described below). Each dose was separated by 4 weeks. 4 weeks after the second DNA vaccination, the animals were given a boost vaccine consisting of 1010 particles of an rAD5 vaccine vector expressing SIVmac239 gag, delivered intramuscularly.
(ii) Delivery of DNA vectors by electroporation. As described above, the animals were given two doses of the DNA vaccine vector, delivered approximately 4 weeks apart, following previously described protocols (31). Briefly, 2 mg of the hCMV/R-SIVmac239 gag plasmid, with or without 0.2 mg of the rhesus macaque IL-12 plasmid (AG157), were prepared in 500 μL of endotoxin-free PBS. The plasmids were delivered intramuscularly via electroporation. The electroporation device was provided by Ichor Medical Systems (San Diego, CA). The electroporation method has been utilized in previous studies (61) and consisted of an electrical impulse of 40 ms over a 400 ms duration at 250 V/cm.
(iii) SIVmac239M infection. All 15 animals in this study were infected intravenously with 10,000 infectious units (IU) of a barcoded SIVmac239, termed SIVmac239M. In the case of the vaccinated macaques, the SIV infection occurred eight weeks after the rAd5 vaccination. The SIVmac239M virus used in this study was kindly provided by Brandon Keele and was constructed as previously described (32).
(iv) N-803 administration. The N-803 dose and route of administration used in this study were previously determined to be safe and efficacious in macaques (18, 20). In the present study, all macaques received N-803 approximately 6 months after the SIV infection. They received three doses of 0.1 mg/kg N-803, administered subcutaneously. Each dose was separated by 2 weeks.
(v) Viral loads. Plasma viral loads were quantified as previously described (20, 62). Briefly, viral RNA (vRNA) was isolated from plasma samples using the Maxwell Viral Total Nucleic Acid Purification Kit (Promega, Madison WI). Then, vRNA was reverse transcribed using the TaqMan Fast Virus 1-Step qRT-PCR Kit (Invitrogen) and quantified on a LightCycler 480 or LC96 instrument (Roche, Indianapolis, IN).
IFN-γ ELISpot assays.
To validate that vaccination elicited Gag-specific immune responses, IFN-γ ELISPOT assays were used as previously described (63, 64). Briefly, frozen PBMCs from the indicated time points pre- and post-vaccination were thawed, and 1×105 cells were then added to each well of a monkey IFN-γ ELISPOT plate (Mabtech, Sweden), along with 1 μM of either Gag181-189CM9 or Tat28-35SL8 peptides in RPMI media supplemented with 10% fetal bovine serum (FBS). As negative and positive controls, cells were incubated with either RPMI media with 10% FBS alone or with media supplemented with 10 μg/mL concanavalin A, respectively. The plates were incubated overnight at 37°C and 5% CO2, and they were developed the following day, according to the manufacturer’s instructions. The plates were read using an AID robotic ELISPOT reader (AID, Strassberg, Germany). A positive response was defined as the greater of the number of spot-forming colonies (SFCs) per 106 PBMCs that were 2 standard deviations above the average value for the negative-control or 50 SFCs/106 PBMC.
Flow cytometric analysis.
(i) Tetrameric reagents. The Mamu-B*008 Nef137-146RL10 and Mamu-A1*001 Tat28-35SL8 biotinylated monomers were produced by the NIH Tetramer Core Facility at Emory University (Atlanta, GA). The Mamu-A1*001 Gag181-189CM9 biotinylated monomer was purchased from MBL International (Woburn, MA). The Mamu-B*008 Nef137-146RL10 and Mamu-A*001 Gag181-189CM9 monomers were then tetramerized with streptavidin-PE (0.5 mg/mL, BD Biosciences) at a 4:1 molar ratio of monomer: streptavidin. Briefly, 1/10 volumes of streptavidin-PE were added to the monomer every 10 min and incubated in the dark at 4°C until the 4:1 molar ratio was achieved. The Mamu-A*001 Tat28-35SL8 monomer was tetramerized to streptavidin-BV605 (0.1 mg/mL, BD Biosciences) or streptavidin-APC (0.25 mg/mL, Agilent Technologies) at 8:1 monomer:streptavidin molar ratios for both fluorochromes. The tetramerization protocol was identical to that described above.
(ii) In vitro characterization of NK cells. Using freshly isolated PBMC isolated from whole blood samples, NK cells were characterized using the antibodies, staining panel, and methods described in Table 2 of our previous manuscript (20). Flow cytometric analysis was performed on a BD Symphony A3 (Becton, Dickinson, Franklin Lakes, NJ), and the data were analyzed using the FlowJo software package for Macintosh (version 10.7.1).
(iii) In vitro characterization of SIV-specific CD8 T cell phenotype and function. For all characterizations of CD8 T cell phenotypes and functional analyses performed in this study, frozen PBMC isolated from whole blood samples were used. The panels used are indicated in Tables 2 to 4.
TABLE 2.
Cytolytic granule/proliferation panel
| Antibody | Clone | Tetramer/surface/intracellular |
|---|---|---|
| Gag181-189CM9-PE or Nef137-146RL10-PE | —b | Tetramer |
| Tat28-35SL8-BV605 | — | Tetramer |
| MR1 5OPRU-BV421 | — | Tetramera |
| CD3-AF700 | SP34-2 | Surface |
| CD4-BUV737 | SK3 | Surface |
| CD8-BUV395 | RPA-T8 | Surface |
| CD28-BV510 | CD28.2 | Surface |
| CD95-PE Cy7 | DX2 | Surface |
| CCR7-BV711 | G043H7 | Surface |
| LIVE/DEAD Near IR ARD | — | — |
| ki-67-AF488 | B56 | Intracellular |
| GranzymeB-PE CF594 | GB11 | Intracellular |
| Perforin-AF647 | PF344 | Intracellular |
The MR1 5OPRU tetramer staining was used to characterize MAIT cells, which were excluded from the manuscript.
—, indictaed “not applicable” (N/A).
TABLE 3.
Chemokine/trafficking panel
| Antibody | Clone | Tetramer/surface/intracellular |
|---|---|---|
| Gag181-189CM9-PE | —b | Tetramer |
| Tat28-35SL8-BV605 | — | Tetramer |
| MR1 5OPRU-APC | — | Tetramera |
| CD3-AF700 | SP34-2 | Surface |
| CD4-BUV737 | SK3 | Surface |
| CD8-BUV395 | RPA-T8 | Surface |
| CXCR3-BV650 | G025H7 | Surface |
| CCR6-PE Cy7 | 11A9 | Surface |
| CXCR5-PerCP Efluor710 | MU5UBEE | Surface |
| CD122-BV421 | Mik-β3 | Surface |
| CD132-BB515 | AG184 | Surface |
| LIVE/DEAD Near IR ARD | — | — |
The MR1 5OPRU tetramer staining was used to characterize MAIT cells, which were excluded from the manuscript.
—, indicated “not applicable” (N/A).
TABLE 4.
Antibodies used for ICS assay
| Antibody | Clone | Tetramer/surface/intracellular |
|---|---|---|
| CD3-V500 | SP34-2 | Surface |
| CD4-BUV737 | SK3 | Surface |
| CD8-BUV395 | RPA-T8 | Surface |
| CD107a-BV605 | H4A3 | Surface |
| TNF-α-AF700 | Mab11 | Intracellular |
| IFN-γ-FITC | 4S.B3 | Intracellular |
| LIVE/DEAD Near IR ARD | —a | — |
—, indicated “not applicable” (N/A).
For all panels, tetramer staining was performed prior to additional surface and intracellular staining. Tetramer stains were performed at room temperature in the dark for 45 min in RPMI media supplemented with 10% FBS and 50 nM dasatinib (Thermo Fisher Scientific, Waltham, MA). After 45 min, the cells were washed in a solution of FACS buffer (2% FBS in a 1× PBS solution) containing 50 nM dasatinib, and surface stains were performed using the antibodies indicated in Tables 2 to 4 for 20 min at room temperature in the dark. Cells were fixed in a 2% paraformaldehyde solution. Following a 20-minute incubation, samples were either run on a BD Symphony A3 or permeabilized and stained for 20 min at room temperature in medium B (Thermo Fisher Scientific, Waltham, MA) for intracellular markers. Flow cytometric analysis was performed as described above.
(iv) Intracellular cytokine staining (ICS) assay. Intracellular cytokine staining assays were performed as previously described, with slight modifications for optimal responses (20, 63). Briefly, frozen PBMC were thawed and rested for approximately 4 to 6 h in RPMI media containing 15% FBS. Then, Gag181-189CM9 or Tat28-35SL8 peptides, or the SIVmac239 Gag peptide pool, were added to the cells at a final concentration of 0.5 μg/mL. The cells and peptides were incubated at 37°C for 90 min, and then 1 μg/mL Brefeldin A (BioLegend, San Diego, CA) and 2 μM monensin (BioLegend, San Diego, CA) were added along with CD107a-BV605 and incubated for 16 h (overnight) at 37°C and 5% CO2. The following day, the cells were stained according to the methods described above with the surface and intracellular antibodies provided in Table 4. Flow cytometric analysis was performed as described above.
Statistical analysis.
For statistical analyses performed with the same individuals across time, repeated measures analysis of variance (ANOVA) nonparametric tests with Dunnett’s multiple comparisons were performed. For individuals with missing samples for a subset of time points, mixed-effects ANOVA tests were performed, using the Geisser-Greenhouse correction. For statistical analyses comparing two groups of animals from the same time point, Mann-Whitney tests were used.
ACKNOWLEDGMENTS
We thank the NIH Tetramer Core Facility (contract number 75N93020D00005) for providing the Mamu-B*008 Nef137-146RL10 and Mamu-A*001 Tat28-35SL8 biotinylated monomers. We thank John Mascola, Robert Seder, and Wing-Pui Kong for the hCMV/R-SIVmac239 gag DNA vaccine vector. We also thank George Pavlakis and Barbara Felber for the rhesus IL-12 plasmid vector. We thank Brandon Keele for the use of the SIVmac239M barcoded virus and Athena Golfinos for sequencing and analyzing the SIVmac239M barcode.
We thank the staff at the Wisconsin National Primate Resource Center (WNPRC) for their excellent veterinary care of the animals involved in this study.
This study was supported by funding supplied through the National Institutes of Health (NIH R01 grant number AI108415). This research was conducted at a facility constructed with support from the Research Facilities Improvement Program grant numbers RR15459-01 and RR020141-01. The Wisconsin National Primate Research Center is also supported by grants P51RR000167 and P51OD011106.
J.T.S. is an employee of ImmunityBio.
Footnotes
Supplemental material is available online only.
Contributor Information
Shelby L. O’Connor, Email: slfeinberg@wisc.edu.
Guido Silvestri, Emory University.
REFERENCES
- 1.Gebara NY, El Kamari V, Rizk N. 2019. HIV-1 elite controllers: an immunovirological review and clinical perspectives. J Virus Erad 5:163–166. 10.1016/S2055-6640(20)30046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deeks SG, Walker BD. 2007. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity 27:406–416. 10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Owen RE, Heitman JW, Hirschkorn DF, Lanteri MC, Biswas HH, Martin JN, Krone MR, Deeks SG, Norris PJ, NIAID CFHIVVI . 2010. HIV+ elite controllers have low HIV-specific T-cell activation yet maintain strong, polyfunctional T-cell responses. Aids 24:1095–1105. 10.1097/QAD.0b013e3283377a1e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen S, Deleage C, Darko S, Ransier A, Truong DP, Agarwal D, Japp AS, Wu VH, Kuri-Cervantes L, Abdel-Mohsen M, Del Rio Estrada PM, Ablanedo-Terrazas Y, Gostick E, Hoxie JA, Zhang NR, Naji A, Reyes-Terán G, Estes JD, Price DA, Douek DC, Deeks SG, Buggert M, Betts MR. 2019. Elite control of HIV is associated with distinct functional and transcriptional signatures in lymphoid tissue CD8+ T cells. Sci Transl Med 11. 10.1126/scitranslmed.aax4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hersperger AR, Migueles SA, Betts MR, Connors M. 2011. Qualitative features of the HIV-specific CD8+ T-cell response associated with immunologic control. Curr Opin HIV AIDS 6:169–173. 10.1097/COH.0b013e3283454c39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Panagioti E, Klenerman P, Lee LN, van der Burg SH, Arens R. 2018. Features of effective T cell-inducing vaccines against chronic viral infections. Front Immunol 9:276. 10.3389/fimmu.2018.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korber B, Fischer W. 2020. T cell-based strategies for HIV-1 vaccines. Hum Vaccin Immunother 16:713–722. 10.1080/21645515.2019.1666957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, Mackey EW, Miller JD, Leslie AJ, DePierres C, Mncube Z, Duraiswamy J, Zhu B, Eichbaum Q, Altfeld M, Wherry EJ, Coovadia HM, Goulder PJ, Klenerman P, Ahmed R, Freeman GJ, Walker BD. 2006. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature 443:350–354. 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 9.Zhang JY, Zhang Z, Wang X, Fu JL, Yao J, Jiao Y, Chen L, Zhang H, Wei J, Jin L, Shi M, Gao GF, Wu H, Wang FS. 2007. PD-1 up-regulation is correlated with HIV-specific memory CD8+ T-cell exhaustion in typical progressors but not in long-term nonprogressors. Blood 109:4671–4678. 10.1182/blood-2006-09-044826. [DOI] [PubMed] [Google Scholar]
- 10.Collins DR, Gaiha GD, Walker BD. 2020. CD8+ T cells in HIV control, cure and prevention. Nat Rev Immunol 20:471–482. 10.1038/s41577-020-0274-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seddiki N, Picard F, Dupaty L, Lévy Y, Godot V. 2020. The potential of immune modulation in therapeutic HIV-1 vaccination. Vaccines (Basel) 8:419. 10.3390/vaccines8030419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Younes SA, Freeman ML, Mudd JC, Shive CL, Reynaldi A, Panigrahi S, Estes JD, Deleage C, Lucero C, Anderson J, Schacker TW, Davenport MP, McCune JM, Hunt PW, Lee SA, Serrano-Villar S, Debernardo RL, Jacobson JM, Canaday DH, Sekaly RP, Rodriguez B, Sieg SF, Lederman MM. 2016. IL-15 promotes activation and expansion of CD8+ T cells in HIV-1 infection. J Clin Invest 126:2745–2756. 10.1172/JCI85996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mastroianni CM, d'Ettorre G, Forcina G, Vullo V. 2004. Teaching tired T cells to fight HIV: time to test IL-15 for immunotherapy. Trends Immunol 25:121–125. 10.1016/j.it.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 14.Lu J, Giuntoli RL, Omiya R, Kobayashi H, Kennedy R, Celis E. 2002. Interleukin 15 promotes antigen-independent in vitro expansion and long-term survival of antitumor cytotoxic T lymphocytes. Clin Cancer Res 8:3877–3884. [PubMed] [Google Scholar]
- 15.Weng NP, Liu K, Catalfamo M, Li Y, Henkart PA. 2002. IL-15 is a growth factor and an activator of CD8 memory T cells. Ann N Y Acad Sci 975:46–56. 10.1111/j.1749-6632.2002.tb05940.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhang M, Wen B, Anton OM, Yao Z, Dubois S, Ju W, Sato N, DiLillo DJ, Bamford RN, Ravetch JV, Waldmann TA. 2018. IL-15 enhanced antibody-dependent cellular cytotoxicity mediated by NK cells and macrophages. Proc Natl Acad Sci USA 115:E10915–E10924. 10.1073/pnas.1811615115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han KP, Zhu X, Liu B, Jeng E, Kong L, Yovandich JL, Vyas VV, Marcus WD, Chavaillaz PA, Romero CA, Rhode PR, Wong HC. 2011. IL-15:IL-15 receptor alpha superagonist complex: high-level co-expression in recombinant mammalian cells, purification and characterization. Cytokine 56:804–810. 10.1016/j.cyto.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhode PR, Egan JO, Xu W, Hong H, Webb GM, Chen X, Liu B, Zhu X, Wen J, You L, Kong L, Edwards AC, Han K, Shi S, Alter S, Sacha JB, Jeng EK, Cai W, Wong HC. 2016. Comparison of the superagonist complex, ALT-803, to IL15 as cancer immunotherapeutics in animal models. Cancer Immunol Res 4:49–60. 10.1158/2326-6066.CIR-15-0093-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong HC, Jeng EK, Rhode PR. 2013. The IL-15-based superagonist ALT-803 promotes the antigen-independent conversion of memory CD8+ T cells into innate-like effector cells with antitumor activity. Oncoimmunology 2:e26442. 10.4161/onci.26442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellis-Connell AL, Balgeman AJ, Zarbock KR, Barry G, Weiler A, Egan JO, Jeng EK, Friedrich T, Miller JS, Haase AT, Schacker TW, Wong HC, Rakasz E, O'Connor SL. 2018. ALT-803 transiently reduces simian immunodeficiency virus replication in the absence of antiretroviral treatment. J Virol 92 10.1128/JVI.01748-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Webb GM, Li S, Mwakalundwa G, Folkvord JM, Greene JM, Reed JS, Stanton JJ, Legasse AW, Hobbs T, Martin LD, Park BS, Whitney JB, Jeng EK, Wong HC, Nixon DF, Jones RB, Connick E, Skinner PJ, Sacha JB. 2018. The human IL-15 superagonist ALT-803 directs SIV-specific CD8+ T cells into B-cell follicles. Blood Adv 2:76–84. 10.1182/bloodadvances.2017012971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Webb GM, Molden J, Busman-Sahay K, Abdulhaqq S, Wu HL, Weber WC, Bateman KB, Reed JS, Northrup M, Maier N, Tanaka S, Gao L, Davey B, Carpenter BL, Axthelm MK, Stanton JJ, Smedley J, Greene JM, Safrit JT, Estes JD, Skinner PJ, Sacha JB. 2020. The human IL-15 superagonist N-803 promotes migration of virus-specific CD8+ T and NK cells to B cell follicles but does not reverse latency in ART-suppressed, SHIV-infected macaques. PLoS Pathog 16:e1008339. 10.1371/journal.ppat.1008339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller JS, Davis ZB, Helgeson E, Reilly C, Thorkelson A, Anderson J, Lima NS, Jorstad S, Hart GT, Lee JH, Safrit JT, Wong H, Cooley S, Gharu L, Chung H, Soon-Shiong P, Dobrowolski C, Fletcher CV, Karn J, Douek DC, Schacker TW. 2022. Safety and virologic impact of the IL-15 superagonist N-803 in people living with HIV: a phase 1 trial. Nat Med 28:392–400. 10.1038/s41591-021-01651-9. [DOI] [PubMed] [Google Scholar]
- 24.Jones RB, Mueller S, O'Connor R, Rimpel K, Sloan DD, Karel D, Wong HC, Jeng EK, Thomas AS, Whitney JB, Lim S-Y, Kovacs C, Benko E, Karandish S, Huang S-H, Buzon MJ, Lichterfeld M, Irrinki A, Murry JP, Tsai A, Yu H, Geleziunas R, Trocha A, Ostrowski MA, Irvine DJ, Walker BD. 2016. A subset of latency-reversing agents expose HIV-infected resting CD4+ T-cells to recognition by cytotoxic T-lymphocytes. PLoS Pathog 12:e1005545. 10.1371/journal.ppat.1005545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McBrien JB, Wong AKH, White E, Carnathan DG, Lee JH, Safrit JT, Vanderford TH, Paiardini M, Chahroudi A, Silvestri G. 2020. Combination of CD8β depletion and interleukin-15 superagonist N-803 induces virus reactivation in simian-human immunodeficiency virus-infected, long-term ART-treated rhesus macaques. J Virol 94. 10.1128/JVI.00755-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang ZQ, Fu TM, Casimiro DR, Davies ME, Liang X, Schleif WA, Handt L, Tussey L, Chen M, Tang A, Wilson KA, Trigona WL, Freed DC, Tan CY, Horton M, Emini EA, Shiver JW. 2002. Mamu-A*01 allele-mediated attenuation of disease progression in simian-human immunodeficiency virus infection. J Virol 76:12845–12854. 10.1128/jvi.76.24.12845-12854.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loffredo JT, Maxwell J, Qi Y, Glidden CE, Borchardt GJ, Soma T, Bean AT, Beal DR, Wilson NA, Rehrauer WM, Lifson JD, Carrington M, Watkins DI. 2007. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J Virol 81:8827–8832. 10.1128/JVI.00895-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loffredo JT, Bean AT, Beal DR, León EJ, May GE, Piaskowski SM, Furlott JR, Reed J, Musani SK, Rakasz EG, Friedrich TC, Wilson NA, Allison DB, Watkins DI. 2008. Patterns of CD8+ immunodominance may influence the ability of Mamu-B*08-positive macaques to naturally control simian immunodeficiency virus SIVmac239 replication. J Virol 82:1723–1738. 10.1128/JVI.02084-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yant LJ, Friedrich TC, Johnson RC, May GE, Maness NJ, Enz AM, Lifson JD, O'Connor DH, Carrington M, Watkins DI. 2006. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol 80:5074–5077. 10.1128/JVI.80.10.5074-5077.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casimiro DR, Wang F, Schleif WA, Liang X, Zhang Z-Q, Tobery TW, Davies M-E, McDermott AB, O'Connor DH, Fridman A, Bagchi A, Tussey LG, Bett AJ, Finnefrock AC, Fu T-m, Tang A, Wilson KA, Chen M, Perry HC, Heidecker GJ, Freed DC, Carella A, Punt KS, Sykes KJ, Huang L, Ausensi VI, Bachinsky M, Sadasivan-Nair U, Watkins DI, Emini EA, Shiver JW. 2005. Attenuation of simian immunodeficiency virus SIVmac239 infection by prophylactic immunization with dna and recombinant adenoviral vaccine vectors expressing Gag. J Virol 79:15547–15555. 10.1128/JVI.79.24.15547-15555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winstone N, Wilson AJ, Morrow G, Boggiano C, Chiuchiolo MJ, Lopez M, Kemelman M, Ginsberg AA, Mullen K, Coleman JW, Wu CD, Narpala S, Ouellette I, Dean HJ, Lin F, Sardesai NY, Cassamasa H, McBride D, Felber BK, Pavlakis GN, Schultz A, Hudgens MG, King CR, Zamb TJ, Parks CL, McDermott AB. 2011. Enhanced control of pathogenic Simian immunodeficiency virus SIVmac239 replication in macaques immunized with an interleukin-12 plasmid and a DNA prime-viral vector boost vaccine regimen. J Virol 85:9578–9587. 10.1128/JVI.05060-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fennessey CM, Pinkevych M, Immonen TT, Reynaldi A, Venturi V, Nadella P, Reid C, Newman L, Lipkey L, Oswald K, Bosche WJ, Trivett MT, Ohlen C, Ott DE, Estes JD, Del Prete GQ, Lifson JD, Davenport MP, Keele BF. 2017. Genetically-barcoded SIV facilitates enumeration of rebound variants and estimation of reactivation rates in nonhuman primates following interruption of suppressive antiretroviral therapy. PLoS Pathog 13:e1006359. 10.1371/journal.ppat.1006359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McBrien JB, Kumar NA, Silvestri G. 2018. Mechanisms of CD8+ T cell-mediated suppression of HIV/SIV replication. Eur J Immunol 48:898–914. 10.1002/eji.201747172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun X, Kaufman PD. 2018. Ki-67: more than a proliferation marker. Chromosoma 127:175–186. 10.1007/s00412-018-0659-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voskoboinik I, Whisstock JC, Trapani JA. 2015. Perforin and granzymes: function, dysfunction and human pathology. Nat Rev Immunol 15:388–400. 10.1038/nri3839. [DOI] [PubMed] [Google Scholar]
- 36.Chowdhury D, Lieberman J. 2008. Death by a thousand cuts: granzyme pathways of programmed cell death. Annu Rev Immunol 26:389–420. 10.1146/annurev.immunol.26.021607.090404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosario M, Liu B, Kong L, Collins LI, Schneider SE, Chen X, Han K, Jeng EK, Rhode PR, Leong JW, Schappe T, Jewell BA, Keppel CR, Shah K, Hess B, Romee R, Piwnica-Worms DR, Cashen AF, Bartlett NL, Wong HC, Fehniger TA. 2016. The IL-15-based ALT-803 complex enhances FcγRIIIa-triggered NK cell responses and in vivo clearance of B cell lymphomas. Clin Cancer Res 22:596–608. 10.1158/1078-0432.CCR-15-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Picker LJ, Reed-Inderbitzin EF, Hagen SI, Edgar JB, Hansen SG, Legasse A, Planer S, Piatak M, Lifson JD, Maino VC, Axthelm MK, Villinger F. 2006. IL-15 induces CD4 effector memory T cell production and tissue emigration in nonhuman primates. J Clin Invest 116:1514–1524. 10.1172/JCI27564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sodora DL, Silvestri G. 2008. Immune activation and AIDS pathogenesis. AIDS 22:439–446. 10.1097/QAD.0b013e3282f2dbe7. [DOI] [PubMed] [Google Scholar]
- 40.Wherry EJ, Kurachi M. 2015. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 15:486–499. 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wherry EJ. 2011. T cell exhaustion. Nat Immunol 12:492–499. 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 42.Migueles SA, Osborne CM, Royce C, Compton AA, Joshi RP, Weeks KA, Rood JE, Berkley AM, Sacha JB, Cogliano-Shutta NA, Lloyd M, Roby G, Kwan R, McLaughlin M, Stallings S, Rehm C, O'Shea MA, Mican J, Packard BZ, Komoriya A, Palmer S, Wiegand AP, Maldarelli F, Coffin JM, Mellors JW, Hallahan CW, Follman DA, Connors M. 2008. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity 29:1009–1021. 10.1016/j.immuni.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fehniger TA, Caligiuri MA. 2001. Interleukin 15: biology and relevance to human disease. Blood 97:14–32. 10.1182/blood.v97.1.14. [DOI] [PubMed] [Google Scholar]
- 44.Petrovas C, Casazza JP, Brenchley JM, Price DA, Gostick E, Adams WC, Precopio ML, Schacker T, Roederer M, Douek DC, Koup RA. 2006. PD-1 is a regulator of virus-specific CD8+ T cell survival in HIV infection. J Exp Med 203:2281–2292. 10.1084/jem.20061496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petrovas C, Price DA, Mattapallil J, Ambrozak DR, Geldmacher C, Cecchinato V, Vaccari M, Tryniszewska E, Gostick E, Roederer M, Douek DC, Morgan SH, Davis SJ, Franchini G, Koup RA. 2007. SIV-specific CD8+ T cells express high levels of PD1 and cytokines but have impaired proliferative capacity in acute and chronic SIVmac251 infection. Blood 110:928–936. 10.1182/blood-2007-01-069112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor S, Huang Y, Mallett G, Stathopoulou C, Felizardo TC, Sun MA, Martin EL, Zhu N, Woodward EL, Elias MS, Scott J, Reynolds NJ, Paul WE, Fowler DH, Amarnath S. 2017. PD-1 regulates KLRG1+ group 2 innate lymphoid cells. J Exp Med 214:1663–1678. 10.1084/jem.20161653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Franceschini D, Paroli M, Francavilla V, Videtta M, Morrone S, Labbadia G, Cerino A, Mondelli MU, Barnaba V. 2009. PD-L1 negatively regulates CD4+CD25+Foxp3+ Tregs by limiting STAT-5 phosphorylation in patients chronically infected with HCV. J Clin Invest 119:551–564. 10.1172/JCI36604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paiardini M, Cervasi B, Albrecht H, Muthukumar A, Dunham R, Gordon S, Radziewicz H, Piedimonte G, Magnani M, Montroni M, Kaech SM, Weintrob A, Altman JD, Sodora DL, Feinberg MB, Silvestri G. 2005. Loss of CD127 expression defines an expansion of effector CD8+ T cells in HIV-infected individuals. J Immunol 174:2900–2909. 10.4049/jimmunol.174.5.2900. [DOI] [PubMed] [Google Scholar]
- 49.Sirskyj D, Thèze J, Kumar A, Kryworuchko M. 2008. Disruption of the gamma c cytokine network in T cells during HIV infection. Cytokine 43:1–14. 10.1016/j.cyto.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 50.Shin H, Blackburn SD, Blattman JN, Wherry EJ. 2007. Viral antigen and extensive division maintain virus-specific CD8 T cells during chronic infection. J Exp Med 204:941–949. 10.1084/jem.20061937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Migueles SA, Weeks KA, Nou E, Berkley AM, Rood JE, Osborne CM, Hallahan CW, Cogliano-Shutta NA, Metcalf JA, McLaughlin M, Kwan R, Mican JM, Davey RT, Connors M. 2009. Defective human immunodeficiency virus-specific CD8+ T-cell polyfunctionality, proliferation, and cytotoxicity are not restored by antiretroviral therapy. J Virol 83:11876–11889. 10.1128/JVI.01153-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hardy GA, Sieg S, Rodriguez B, Anthony D, Asaad R, Jiang W, Mudd J, Schacker T, Funderburg NT, Pilch-Cooper HA, Debernardo R, Rabin RL, Lederman MM, Harding CV. 2013. Interferon-α is the primary plasma type-I IFN in HIV-1 infection and correlates with immune activation and disease markers. PLoS One 8:e56527. 10.1371/journal.pone.0056527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Le Saout C, Hasley RB, Imamichi H, Tcheung L, Hu Z, Luckey MA, Park JH, Durum SK, Smith M, Rupert AW, Sneller MC, Lane HC, Catalfamo M. 2014. Chronic exposure to type-I IFN under lymphopenic conditions alters CD4 T cell homeostasis. PLoS Pathog 10:e1003976. 10.1371/journal.ppat.1003976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wood MP, Jones CI, Lippy A, Oliver BG, Walund B, Fancher KA, Fisher BS, Wright PJ, Fuller JT, Murapa P, Habib J, Mavigner M, Chahroudi A, Sather DN, Fuller DH, Sodora DL. 2021. Rapid progression is associated with lymphoid follicle dysfunction in SIV-infected infant rhesus macaques. PLoS Pathog 17:e1009575. 10.1371/journal.ppat.1009575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teijaro JR, Ng C, Lee AM, Sullivan BM, Sheehan KC, Welch M, Schreiber RD, de la Torre JC, Oldstone MB. 2013. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science 340:207–211. 10.1126/science.1235214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swainson LA, Sharma AA, Ghneim K, Ribeiro SP, Wilkinson P, Dunham RM, Albright RG, Wong S, Estes JD, Piatak M, Deeks SG, Hunt PW, Sekaly RP, McCune JM. 2022. IFN-α blockade during ART-treated SIV infection lowers tissue vDNA, rescues immune function, and improves overall health. JCI Insight 7. 10.1172/jci.insight.153046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harper J, Huot N, Micci L, Tharp G, King C, Rascle P, Shenvi N, Wang H, Galardi C, Upadhyay AA, Villinger F, Lifson J, Silvestri G, Easley K, Jacquelin B, Bosinger S, Müller-Trutwin M, Paiardini M. 2021. IL-21 and IFNα therapy rescues terminally differentiated NK cells and limits SIV reservoir in ART-treated macaques. Nat Commun 12:2866. 10.1038/s41467-021-23189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shortreed CG, Wiseman RW, Karl JA, Bussan HE, Baker DA, Prall TM, Haj AK, Moreno GK, Penedo MCT, O'Connor DH. 2020. Characterization of 100 extended major histocompatibility complex haplotypes in Indonesian cynomolgus macaques. Immunogenetics 72:225–239. 10.1007/s00251-020-01159-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barouch DH, Yang ZY, Kong WP, Korioth-Schmitz B, Sumida SM, Truitt DM, Kishko MG, Arthur JC, Miura A, Mascola JR, Letvin NL, Nabel GJ. 2005. A human T-cell leukemia virus type 1 regulatory element enhances the immunogenicity of human immunodeficiency virus type 1 DNA vaccines in mice and nonhuman primates. J Virol 79:8828–8834. 10.1128/JVI.79.14.8828-8834.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jalah R, Rosati M, Ganneru B, Pilkington GR, Valentin A, Kulkarni V, Bergamaschi C, Chowdhury B, Zhang GM, Beach RK, Alicea C, Broderick KE, Sardesai NY, Pavlakis GN, Felber BK. 2013. The p40 subunit of interleukin (IL)-12 promotes stabilization and export of the p35 subunit: implications for improved IL-12 cytokine production. J Biol Chem 288:6763–6776. 10.1074/jbc.M112.436675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keane-Myers AM, Bell M, Hannaman D, Albrecht M. 2014. DNA electroporation of multi-agent vaccines conferring protection against select agent challenge: triGrid delivery system. Methods Mol Biol 1121:325–336. 10.1007/978-1-4614-9632-8_29. [DOI] [PubMed] [Google Scholar]
- 62.Ellis AL, Balgeman AJ, Larson EC, Rodgers MA, Ameel C, Baranowski T, Kannal N, Maiello P, Juno JA, Scanga CA, O'Connor SL. 2020. MAIT cells are functionally impaired in a Mauritian cynomolgus macaque model of SIV and Mtb co-infection. PLoS Pathog 16:e1008585. 10.1371/journal.ppat.1008585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ellis A, Balgeman A, Rodgers M, Updike C, Tomko J, Maiello P, Scanga CA, O'Connor SL. 2017. Characterization of T cells specific for CFP-10 and ESAT-6 in Mycobacterium tuberculosis-infected Mauritian cynomolgus macaques. Infect Immun 85. 10.1128/IAI.01009-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O'Connor SL, Lhost JJ, Becker EA, Detmer AM, Johnson RC, Macnair CE, Wiseman RW, Karl JA, Greene JM, Burwitz BJ, Bimber BN, Lank SM, Tuscher JJ, Mee ET, Rose NJ, Desrosiers RC, Hughes AL, Friedrich TC, Carrington M, O'Connor DH. 2010. MHC heterozygote advantage in simian immunodeficiency virus-infected Mauritian cynomolgus macaques. Sci Transl Med 2:22ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4. Download jvi.01185-22-s0001.pdf, PDF file, 1.5 MB (1.5MB, pdf)