ABSTRACT
The type III secretion system (T3SS) is a syringe-like virulence factor that delivers bacterial proteins directly into the cytoplasm of host cells. An essential component of the system is the translocon, which creates a pore in the host cell membrane through which proteins are injected. In Pseudomonas aeruginosa, the translocation pore is formed by proteins PopB and PopD and attaches to the T3SS needle via the needle tip protein PcrV. The structure and stoichiometry of the multimeric pore are unknown. We took a genetic approach to map contact points within the system by taking advantage of the fact that the translocator proteins of P. aeruginosa and the related Aeromonas hydrophila T3SS are incompatible and cannot be freely exchanged. We created chimeric versions of P. aeruginosa PopB and A. hydrophila AopB to intentionally disrupt and restore protein-protein interactions. We identified a chimeric B-translocator that specifically disrupts an interaction with the needle tip protein. This disruption did not affect membrane insertion of the B-translocator but did prevent formation of the translocation pore, arguing that the needle tip protein drives the formation of the translocation pore.
KEYWORDS: T3SS, translocon, pore formation
INTRODUCTION
The syringe-like type III secretion system (T3SS) is used by Pseudomonas aeruginosa to inject host cells with effector proteins—ExoS, ExoT, ExoU, and ExoY—that weaken epithelial barriers and hinder immune clearance by neutrophils and macrophages (1). This system is a prime target for antivirulence drug development (2–4). Particularly attractive as a target, the translocon creates a pore in the host cell membrane and is partially accessible from the extracellular space. However, the translocon is poorly understood (5–7). Complex processes, such as the needle tip-mediated insertion of translocator proteins into the host cell membrane or triggering of effector secretion, cannot be recapitulated in vitro. The structure of the translocation pore remains elusive. The intact translocation pore has yet to be purified. The Salmonella translocon was visualized in the context of an infected cell by cryo-electron tomography, but the structure lacked molecular resolution (8). A further complication, in the case of P. aeruginosa, is that the copy number of the assembled system is low, hampering direct in situ imaging studies (9, 10).
The formation of the translocation pore has been studied in vitro using purified proteins (11–14). In the case of P. aeruginosa, two transmembrane proteins, PopB and PopD, oligomerize to form a pore in the host cell membrane (Fig. 1A). PopB and PopD can also form pores in lipid bilayers (15–17), under the right conditions, and these pores require both translocator proteins, just as both PopB and PopD are required for translocation in the context of host cells. The stoichiometry of translocator proteins in in vitro-formed pores was estimated using photobleaching, and the results suggest that these pores consist of 8 copies of PopB and 8 copies of PopD (18). Genetic studies have also shed light on the composition and function of the translocon. The needle tip protein is required for translocator insertion in every T3SS examined to date (19–24). Indeed, the P. aeruginosa needle tip protein PcrV connects the translocation pore to the T3SS needle and is needed for the insertion of PopB and PopD into the host cell membrane. Recent data suggest that the insertion of PopD is further facilitated by PopB (15). The needle tip protein plays a role in sensing host cell contact, so that effector proteins are unleashed only after the translocon is fully assembled (25–27). Triggering effector secretion requires a conformational change in the translocation pore that, in the case of Pseudomonas, is transmitted via the C-terminus of PopD to the collar domain of PcrV (25). Conformational changes likely also trigger effector secretion in other species. This has been best characterized in Shigella, where intermediate filament binding and actin polymerization mediate translocon stability and the opening of the translocation pore through distinct conformational changes in the translocon (28, 29). These systems differ in the nature of the host factors required to trigger effector secretion, however, since interfering with actin polymerization does not inhibit effector translocation in Pseudomonas (25, 30). The nature of the host cell process that triggers effector secretion in the P. aeruginosa system remains enigmatic.
FIG 1.
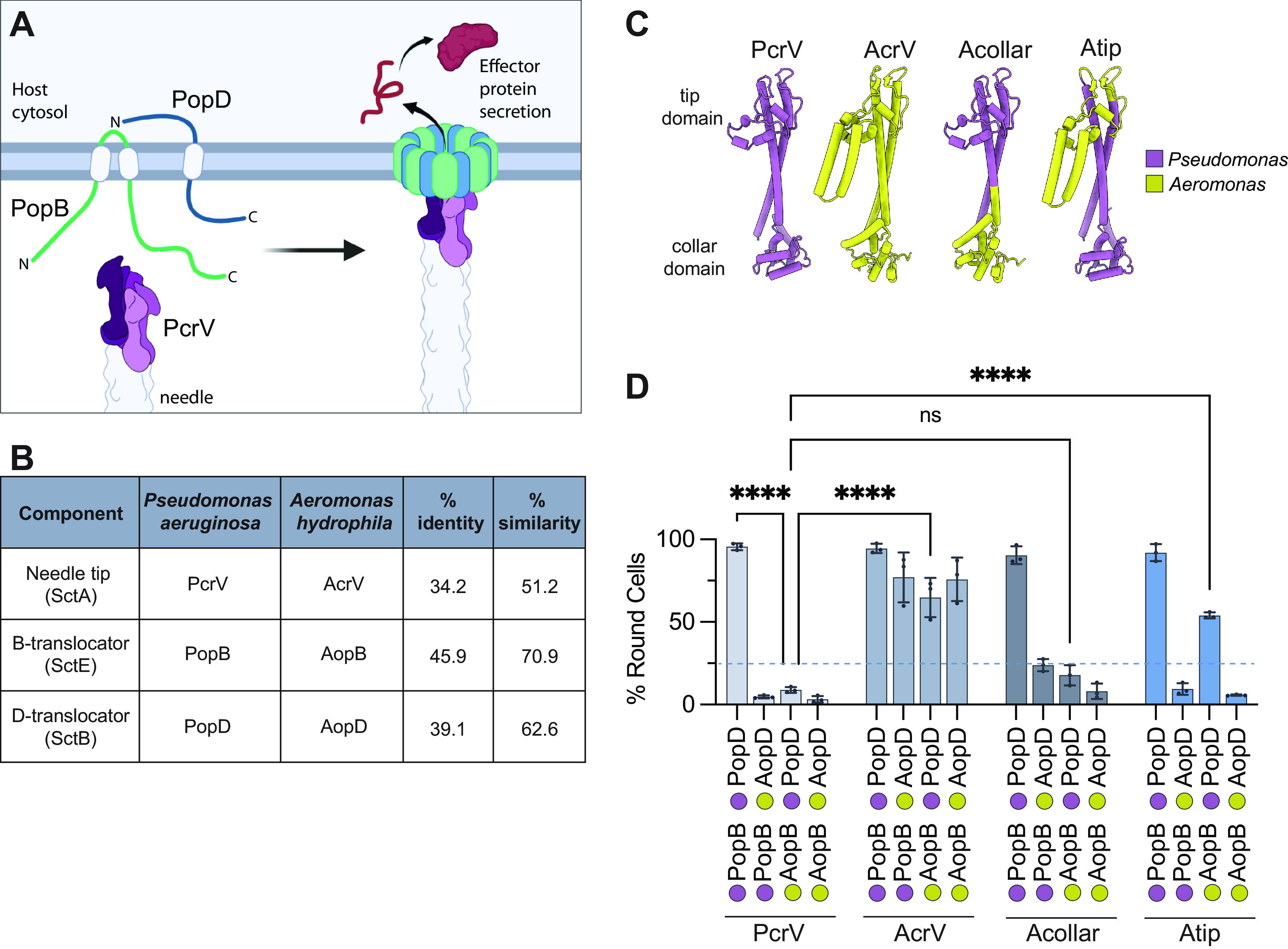
Translocator proteins from P. aeruginosa and A. hydrophila are not interchangeable. (A) Model of component parts of the translocon (left) and assembled translocation pore (right). PopB is inserted in the membrane with N- and C-termini toward the extracellular space (54). PopD has its C terminus in the extracellular space (25). The pentameric arrangement of PcrV is based on alignment to the Salmonella needle tip structure (55). (B) Amino acid sequence identity and similarity for P. aeruginosa (accession number AE004091) and A. hydrophila AH-3 (accession number AY528667) translocators. Sequences were aligned with ClustalW. (C) AlphaFold2 models of the needle tip protein from P. aeruginosa (PcrV), A. hydrophila strain AH-3 (AcrV), and two chimeric-fusion V proteins. The hybrid Acollar protein has an AcrV substitution in the N-terminal globular domain [residues 1 to F143 from AcrV are fused to PcrV(Q124–end)]. The hybrid Atip contains the bulkier tip domain of AcrV substituted for the tip domain of PcrV (residues 1 to A158 from PcrV, A179 to S310 from AcrV, and V244 to the end from PcrV). Monomer models were created from protein sequences using AlphaFold2 implemented in Google Colab Notebooks (33, 56) and visualized with UCSF ChimeraX (57). The position of the collar domain is uncertain and varies across models. The tip domain has a high degree of confidence and is static between models (55). Pseudomonas segments are shown in purple, and Aeromonas segments in yellow. (D) All strains lack the pore-forming translocator operon (ΔpcrH-popBD) and produce the indicated needle tip protein: PcrV (strain RP3624), AcrV (strain RP7360), Acollar (strain RP6166), or Atip (strain RP6425). These background strains were complemented with plasmids carrying the indicated translocator proteins along with both export chaperone homologs, pcrH and acrH (Table S1). A549 epithelial cells were infected, and cellular rounding (indicating delivery of ExoS) was monitored by microscopy after 2 h. In our analysis, we consider a translocator system nonfunctional if it leads to less than 25% cell rounding at 2 h (dotted line). Statistical significance was calculated by two-way analysis of variance (ANOVA) with the Tukey multiple-comparison test. n = 3 biological replicates. Error bars show standard deviations. ****, P < 0.0001; ns, not statistically significant (P > 0.05). Statistical data for all comparisons are listed in Table S3. Schematic figures were created using BioRender.com.
List of bacterial strains and plasmids used in the study. Download Table S1, PDF file, 0.05 MB (52.9KB, pdf) .
Copyright © 2022 Kundracik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
All pairwise comparisons for translocator exchange cytotoxicity testing presented in Fig. 1D. Download Table S3, XLSX file, 0.01 MB (15.1KB, xlsx) .
Copyright © 2022 Kundracik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In this study, we took a genetic approach to map interactions between translocon proteins. We exploited the incompatibilities between the homologous T3SS systems of P. aeruginosa and Aeromonas hydrophila to map an interaction between the needle tip protein and two small regions of PopB. Functional assays of our genetic hybrids revealed that the PopB-PcrV interaction is crucial for the formation of the translocation pore, but not for insertion of the translocators into the host membrane. We therefore propose for the first time that the needle tip protein is actively involved in assembling the translocation pore.
RESULTS
P. aeruginosa and A. hydrophila translocator proteins are incompatible.
P. aeruginosa translocator proteins are not interchangeable with their Yersinia homologs (21, 25, 31). Previously, we used this observation to genetically map the protein-protein contacts that were disrupted by combining Pseudomonas and Yersinia translocators. This led to the discovery of several new protein-protein interactions, two of which—an interaction between PopD and PcrV and a PopD homodimer interaction—are important for sensing host cell contact. Here, we extend this analysis by examining the incompatibility between P. aeruginosa and A. hydrophila translocator homologs (Fig. 1B and Fig. S1 and S2 in the supplemental material). Compared to PcrV, AcrV has a 49-residue insertion in the hypervariable portion of the tip domain (32). This segment is predicted by AlphaFold2 to fold into an alpha-helical hairpin (Fig. 1C) (33). The rest of the tip domain and central helices are predicted to fold similarly in PcrV and AcrV.
Protein sequence alignment for PcrV-AcrV. Related to Fig. 1 and 2. PcrV (GenBank accession no. AE004091) and AcrV (GenBank accession no. AY528667) were aligned using ClustalW. The amino acid sequences are 36% identical (50% similar). The construct Atip is indicated by purple (segments from PcrV) and yellow (segments from AcrV). Download FIG S1, TIF file, 0.9 MB (970.9KB, tif) .
Copyright © 2022 Kundracik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Protein sequence alignment for PopB-AopB. Related to Fig. 1 and 2. PopB (GenBank accession no. AE004091) and AopB (GenBank accession no. AY528667) were aligned using ClustalW. The amino acid sequences are 46% identical (71% similar). The construct Bmix is indicated by green (segments from PopB) and yellow (segments from AopB). Boxes indicate predicted transmembrane domains from the data with UniProt accession numbers Q9I324 (PopB) and Q699Q8 (AopB). Download FIG S2, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2022 Kundracik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To determine whether translocator proteins from A. hydrophila can substitute for their respective P. aeruginosa homologs, we constructed P. aeruginosa strains carrying various combinations of the translocator proteins and needle tips, including PcrV-AcrV chimeras designated “Atip” and “Acollar.” These strains were assayed for their ability to intoxicate epithelial cells. Compatible sets of proteins induce cell rounding by mediating the delivery of ExoS, which induces actin depolymerization. Incompatible systems induce cell rounding more slowly or not at all (Fig. S3).
Additional controls and kinetics for cytotoxicity assay. Related to Fig. 1. (A) The cytotoxicity experiment was performed as described in the legend to Fig. 1 using background strains RP3624 (PcrV) and RP6425 (Atip). The wild-type strain is RP2318. The ΔpcrV strain is RP3223. The ΔpopB and ΔpopD strains have the background strain RP3624 complemented with a plasmid carrying either popB or popD along with both export chaperone homologs, pcrH and acrH. n = 3 biological replicates. Error bars show standard deviations. (B and C) Kinetics of cell rounding in response to P. aeruginosa infection. The cytotoxicity experiment was performed as described in the legend to Fig. 1 but was stopped at various time points up to 3 h. HBD denotes pcrH-popB-popD. n = 4 biological replicates. Error bars show standard deviations. The data set is split into two graphs for clarity. Download FIG S3, TIF file, 0.6 MB (651.9KB, tif) .
Copyright © 2022 Kundracik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The native Pseudomonas translocators (PopB and PopD) are compatible with both PcrV and AcrV, as well as both hybrid needle tip proteins (Fig. 1D). In contrast, Aeromonas AopB and AopD each display specific incompatibilities. AopD is only compatible with AcrV. That is, the two systems containing both AopD and AcrV induce ~75% cell rounding within 2 h, whereas the strain producing AopD and PcrV is nonfunctional. Interestingly, AopD function is not fully restored by either hybrid needle tip, arguing that AopD disrupts interactions with both the collar and tip domains of PcrV. AopB is similarly incompatible with PcrV but functional when produced in the context of its cognate needle tip protein, AcrV. Notably, AopB is incompatible with PcrV and Acollar but significantly rescued by AcrV or Atip. This finding argues for an interaction between the B-translocator and the tip domain of the needle tip protein. We decided to pursue this interaction further, since no function has been assigned to the tip domain of the needle tip protein for any T3SS.
The tip domain of the needle tip protein interacts with PopB.
We next sought to narrow down the specific region of AopB that clashes with the PcrV tip domain. We used an iterative approach to create a panel of PopB-AopB chimeric proteins, which were assayed in the context of the wild-type PcrV and the chimeric needle tip protein Atip (Fig. 2A and B and Fig. S4 and S5). Notably, this approach allowed us to exclude proteins that are likely misfolded, degraded, or no longer secreted, since these chimeras cannot be complemented through the addition of the permissive Atip needle tip protein.
FIG 2.
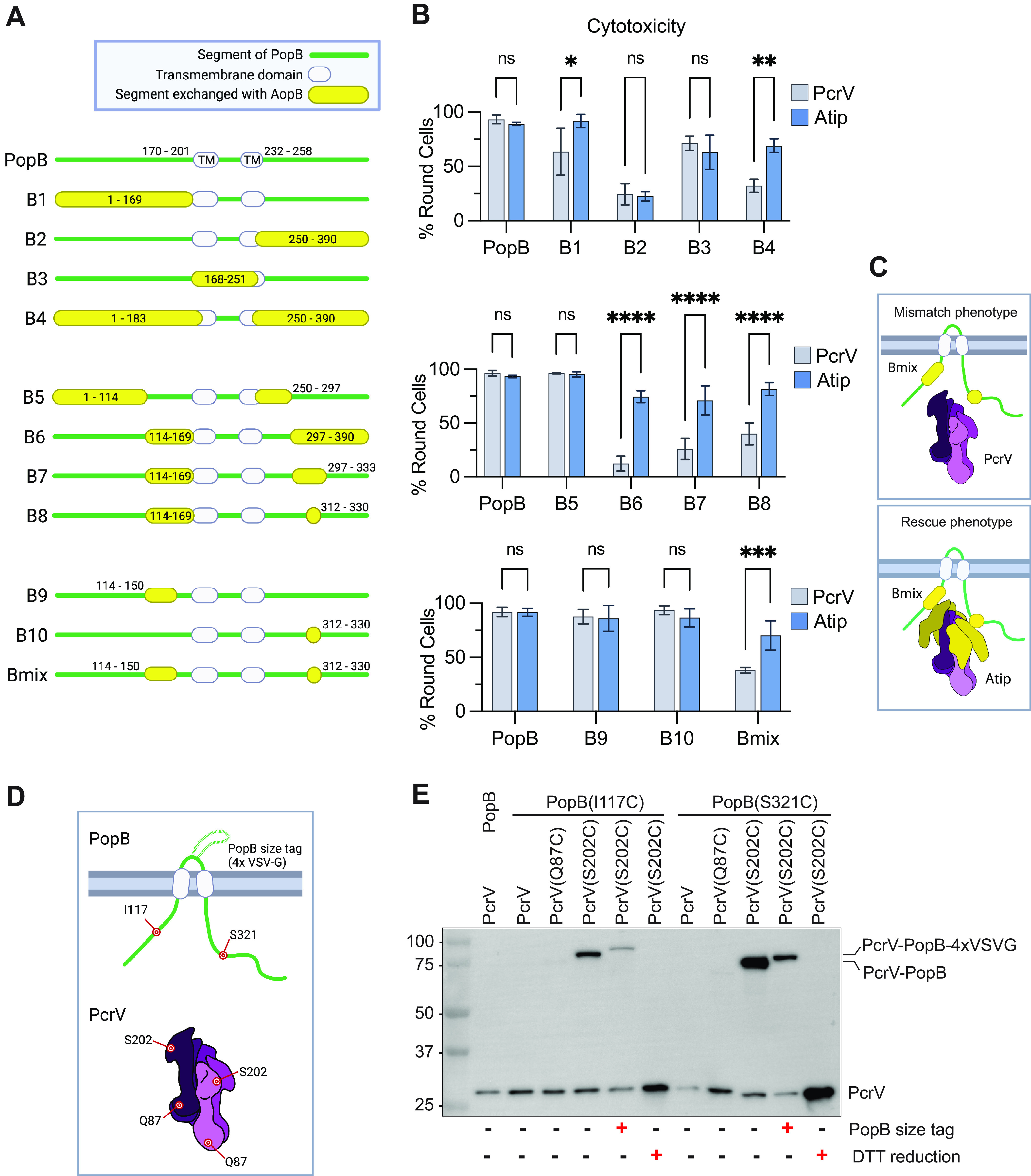
The AopB-Atip interaction is mapped to two small regions of the B-translocator. (A) Schematic diagram of PopB-AopB chimeras. Residues are numbered according to PopB amino acid sequence. PopB transmembrane domains are predicted to be residues 170 to 201 (TM1) and residues 232 to 258 (TM2) (58). Green lines indicate PopB. Yellow bubbles indicate AopB. Transmembrane domains are indicated by white bubbles. (B) Background strains PAO1F ΔexsE ΔexoT exoY ΔpcrH-popBD with wild-type pcrV (strain RP3624) or Atip (strain RP6425) were complemented with acrH, pcrH, and popD and the indicated popB-aopB chimera. A549 cells were infected for 2 h, and the proportion of round cells assessed. n = 3 biological replicates. Error bars show standard deviations. Statistical significance was calculated using two-way ANOVA with the Sidak multiple-comparison test. Significance thresholds: *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ****, P < 0.0001; ns, not statistically significant (P > 0.05). (C) Schematic diagram of Bmix chimera with wild-type PcrV or chimeric Atip, illustrating the tip-dependent phenotype. The Bmix/PcrV combination has reduced cytotoxicity, presumably due to incompatibilities introduced by the homologous substitutions (mismatch phenotype). The Bmix/Atip combination performs significantly better in the cytotoxicity assay (rescue phenotype). (D) Cysteine mutations were introduced into PopB and PcrV in the locations shown schematically. In subsequent experiments, PcrV proteins having a single cysteine mutation (Q87C or S202C) were paired with PopB mutants having a single cysteine mutation (I117C or S321C). A variant of PopB includes a 5.3-kDa size tag consisting of four copies of the vesicular stomatitis virus glycoprotein (4×VSV-G) inserted after residue 225 (25). (E) Background strains PAO1F ΔexsE exoS(G/A−) ΔexoT exoY ΔpcrH-popBD with wild-type pcrV (strain RP3670), pcrV(Q87C) (strain RP12661), or pcrV(S202C) (strain RP12598) were complemented with acrH, pcrH, and popD and the indicated version of popB. A549 cells were infected in the presence of copper for 2 h. After infection, off-target disulfide bond formation was blocked by the addition of iodoacetamide. Membrane extract samples were prepared in SDS sample buffer lacking or containing dithiothreitol (DTT) as indicated. The blot is representative of three independent experiments. Molecular-weight size markers (in kilodaltons) are indicated. “exoS(G/A−)” denotes the mutant exoS(R146K/E379D/E381D), which is enzymatically inactive (30). Schematic figures were created using BioRender.com.
Refining the region of interest for the PopB-AopB interaction. Related to Fig. 2. (A) Schematic diagrams of PopB-AopB chimeras. This set of chimeras was designed to refine the boundaries of the PopB-PcrV interaction, focusing on the N-terminal substitution. Created with BioRender.com. (B) Cytotoxicity data for PopB-AopB chimeras. The experiment was performed as described in the legend to Fig. 1 using background strains RP3624 (PcrV) and RP6425 (Atip). n = 3 biological replicates. Error bars show standard deviations. Statistical differences were analyzed with two-way ANOVA and the Sidak multiple-comparison test: *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ****, P < 0.0001; ns, not statistically significant (P > 0.05). Download FIG S4, TIF file, 0.7 MB (749.4KB, tif) .
Copyright © 2022 Kundracik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Evidence for a PopB-PopB interaction. Related to Fig. 2. (A) Schematic diagrams of PopB-AopB chimeras. (B) Cytotoxicity data for PopB-AopB chimeras. The experiment was performed as described in the legend to Fig. 1 using background strains RP3624 (PcrV) and RP6425 (Atip). n = 3 biological replicates. Error bars show standard deviations. Statistical differences were analyzed with two-way ANOVA and the Sidak multiple-comparison test: *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ***, P < 0.0001; ns, not statistically significant (P > 0.05). (C) Schematic diagram showing two possible PopB-PopB interactions. In the first scenario (left), two adjacent monomers of PopB interact with each other. In the second scenario (right), PopB folds on itself and has an intramolecular interaction. Created with BioRender.com. Download FIG S5, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2022 Kundracik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The initial junctions correspond approximately to the two transmembrane domains in PopB (residues 170 to 201 and 232 to 258). Constructs B1 and B2, which replace the N-terminal and C-terminal extracellular portions of PopB with the corresponding regions of AopB, resulted in a weak tip-dependent phenotype and loss of function, respectively (Fig. 2B). We therefore decided to replace either the central portion of PopB (creating construct B3) or both the N- and C-terminal domains (construct B4). Construct B4 retained the tip-dependent phenotype of AopB. We therefore focused on further narrowing down the portions of the extracellular regions of AopB that clash with the wild-type PcrV needle tip. We subdivided the N- and C-terminal extracellular domains and replaced those regions with the homologous portions of AopB (constructs B5 and B6). While B5 was functional in the context of PcrV, construct B6 still required the hybrid Atip needle tip for function. Further reduction of the C-terminal substitution (constructs B7 and B8) and of the N-terminal substitution (Fig. S4) maintained the tip-dependent phenotype.
Our final chimeric construct, Bmix, had residues 114 to 150 and 312 to 330 of PopB replaced with the homologous segments of AopB (Fig. 2C). While neither substitution on its own displayed a significant defect in T3SS-mediated cytotoxicity, the combination of both substitutions resulted in Atip-dependent cell rounding, arguing that both segments are required for a contact with the tip domain. To verify this interaction, we assessed disulfide bond formation between cysteine mutant PopB(I117C) (PopB bearing a mutation of Ile to Cys at position 117) or PopB(S321C) paired with a tip domain cysteine mutant, PcrV(S202C) or a collar domain cysteine mutant, PcrV(Q87C) (Fig. 2D and E). Both PopB cysteine mutants were able to form a disulfide bond with PcrV(S202C) but not PcrV(Q87C), confirming that these regions of PopB interact with the needle tip protein tip domain. These data, together with the genetic rescue by the Atip needle tip, also argue that the loss of function of the Bmix protein in the context of wild-type PcrV is due to the loss of the interaction with the needle tip, rather than the result of misfolding of the chimeric Bmix protein.
In the course of mapping the region of incompatibility between AopB and PcrV, we noticed that many of the constructs that began with PopB and ended with AopB were inactive (Fig. S5). The converse fusions retained activity, suggesting that the choice of fusion joint was not to be blamed for the defect. Indeed, function was restored in sandwich fusions in which the PopB N- and C-termini were paired together (e.g., construct B4), suggesting that the mismatch interrupted a critical interaction between the N- and C-termini of PopB. Whether this interaction was intra- or intermolecular (i.e., needed for PopB dimer formation) was unclear. Additionally, we examined whether the AcrV hypervariable domain was responsible for the tip dependence of AopB. Deletion of the two 25-amino-acid helices unique to AcrV did not result in incompatibility with Bmix (Fig. S6), arguing that the incompatibility stemmed from a different, more conserved portion of the PcrV tip.
The nonhomologous segment of AcrV is not required for rescue of Bmix. Related to Fig. 2. (A) Residues 220 to 269 of AcrV, which have no homologous segment in PcrV, were deleted from the Atip construct to create the PcrV-AcrV chimera “Atip-trim.” Models of PcrV, Atip, and Atip-trim were created as described in the legend to Fig. 1. (B) The experiment was performed as described in the legend to Fig. 1, using background strains RP3624 (PcrV), RP6425 (Atip), and RP12517 (Atip-trim). Statistical significance was calculated by two-way ANOVA with the Tukey multiple-comparison test. Data bars are a summary of the results from three biological replicates with error bars showing standard deviations. ns, not significant (P > 0.05). Download FIG S6, TIF file, 0.9 MB (955KB, tif) .
Copyright © 2022 Kundracik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The PopB-tip domain interaction is not required for membrane insertion of PopB.
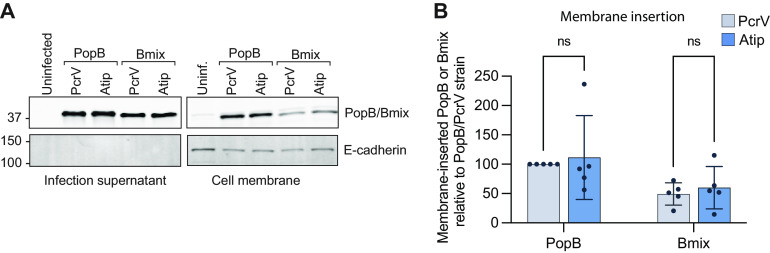
We next sought to understand which stages of translocon assembly were affected by the PopB/PcrV mismatch. Since the needle tip was required for translocator insertion, we hypothesized that the tip domain interaction was critical for membrane insertion of PopB and PopD. We infected A549 epithelial cells with P. aeruginosa strains carrying the chimeric Bmix protein and either wild-type PcrV or chimeric Atip and assessed the amount of B-translocator in the host membrane by Western blotting. There was no significant difference in the amounts of membrane-inserted Bmix in the context of wild-type PcrV or chimeric Atip, indicating that the tip domain interaction was not required for membrane insertion of PopB (Fig. 3).
FIG 3.
The Bmix-PcrV mismatch does not hinder membrane insertion of PopB. A549 cells were infected for 2 h with P. aeruginosa cells producing PopD and the indicated needle tip/B-translocator combination (background strains RP3670 and RP11222). The control strain lacks pcrH, popB, and popD. After infection, cells were washed with 1 M KCl to remove nonspecifically adhered protein, and membrane-inserted translocator proteins were extracted using Triton X-100 (see Materials and Methods). (A) Translocator proteins, as well as E-cadherin (fractionation/loading control), were detected by Western blotting in membrane extract and supernatant samples. The blot is representative of five independent experiments. The positions of molecular-weight size markers (in kilodaltons) are indicated. (B) Quantification of 5 biological replicates. The amount of PopB/Bmix is normalized to that of E-cadherin or EGFR. Error bars show standard deviations. Statistical significance was assessed with two-way ANOVA using the Sidak multiple-comparison test. ns, not statistically significant (i.e., P > 0.05).
Pore formation is hindered by disruption of the interaction between PopB and the tip domain of PcrV.
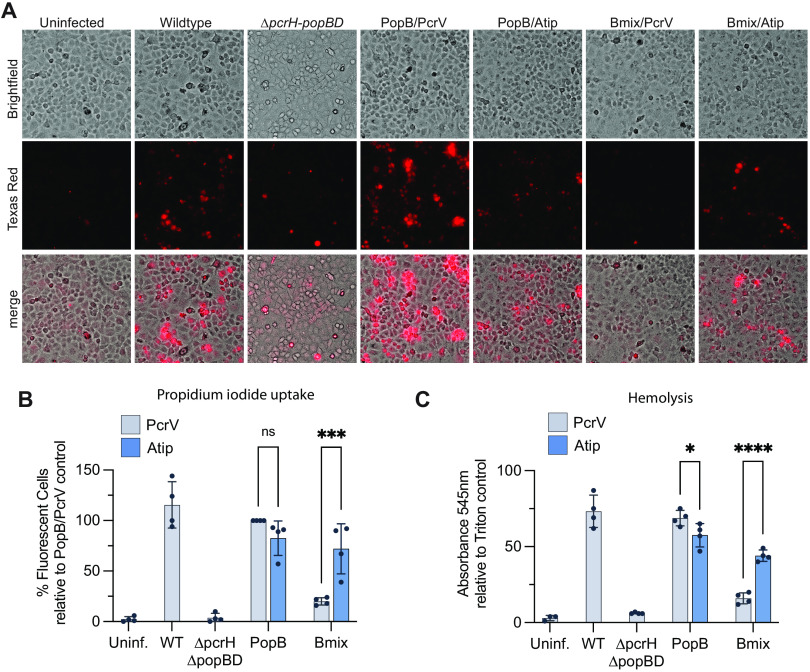
To investigate whether disruption of the PopB-PcrV interaction would affect pore formation, A549 cells were infected in the presence of propidium iodide (PI), which is small enough (668 Daltons) to enter cells through translocation pores (34–36). There was no significant difference in PI uptake between strains producing wild-type PopB and either PcrV or Atip, reiterating our finding from the cytotoxicity assay that PopB can be paired with either version of the needle tip protein (Fig. 4A and B). On the other hand, the Bmix/PcrV strain averaged less than 25% the amount of PI uptake as the PopB/PcrV strain, but this defect was significantly corrected in the Bmix/Atip strain. We employed a pore formation assay in sheep red blood cells to corroborate these findings (Fig. 4C). In the hemolysis assay, Bmix/Atip performed significantly better than Bmix/PcrV. We also observed a small but statistically significant defect for PopB/Atip compared to PopB/PcrV, consistent with the importance of this interaction for pore formation. Together, these findings indicate that a mismatch between PopB and the tip domain of PcrV impedes the formation of the translocation pore.
FIG 4.
Pore formation is disrupted by mismatch between PopB and PcrV. (A, B) A549 cells were infected in the presence of propidium iodide with RP11946 (PAO1 ΔexsE ΔexoSTY pcrV+ ΔpcrH-popBD) or RP11948 [PAO1 ΔexsE ΔexoSTY pcrV::acrV(tip) ΔpcrH-popBD] producing PopD and the indicated translocators. RP2317 (PAO1 ΔexsE ΔexoSTY pcrV+) was the wild-type control. After 2 h, cells were washed and imaged (bright field and fluorescence with Texas red filter). (A) Representative micrographs from one biological replicate are shown. (B) Four biological replicates are quantified. Fluorescent cells were scored manually, and the results of each experiment were normalized to the results for the PopB/PcrV strain. Error bars show standard deviations. Statistical differences were analyzed with two-way ANOVA and the Sidak multiple-comparison test: *, P < 0.05;ns, not statistically significant (P > 0.05). (C) Sheep erythrocytes were pretreated with papain, which cleaves surface glycoproteins and exposes cryptic binding sites for P. aeruginosa hemagglutinin (53). The erythrocytes were infected for 1 h with strain RP3670 [PAO1 ΔexsE exoS(G/A−) pcrV+ ΔpcrH-popBD] or strain RP11222 [PAO1 ΔexsE exoS(G/A−) pcrV::Atip ΔpcrH-popBD] producing PopD and the indicated translocators. The erythrocyte suspension was then mixed, unbroken erythrocytes pelleted, and supernatant UV-Vis absorbance measured at 545 nm. The results of each experiment were normalized to the results for a Triton X-100-lysed control. n = 4 biological replicates. Statistical differences were analyzed with two-way ANOVA and the Sidak multiple-comparison test. Significance thresholds: *, P < 0.05; ****, P < 0.0001; ns, not statistically significant (P > 0.05).
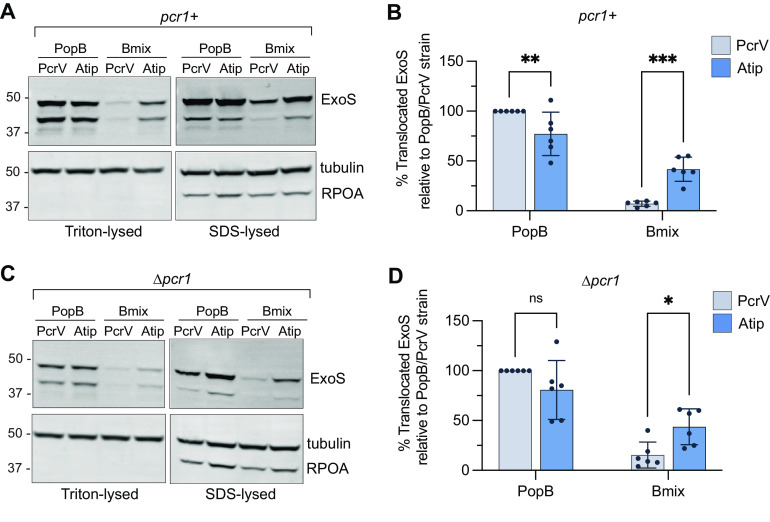
Defective translocation in the mismatched Bmix-PcrV strain was not overcome by bypassing secretion regulation.
A hallmark of all T3SS is that secretion of effector proteins is triggered by host cell contact (37, 38). The nature of the trigger is unknown but, in P. aeruginosa, involves a conformational change in the translocon that alters a contact between PopD and the PcrV collar domain (25). We set out to test whether the mismatch between PcrV and Bmix interrupts this triggering mechanism. In P. aeruginosa, the PopN-Pcr1 complex prevents effector secretion before host cell contact (39). Deletion of pcr1 results in constitutive effector secretion, regardless of host cell contact (25). To test whether the PopB-PcrV interaction was important for secretion regulation, we measured the translocation of the effector ExoS in strains lacking the secretion regulator Pcr1. If the PopB-PcrV interaction was primarily required for sensing host cell contact, deletion of pcr1 should restore translocation in the Bmix/PcrV strain, since a pcr1 mutant no longer required host cell contact to initiate effector export. Consistent with the cytotoxicity assays, translocation of ExoS was impaired in the Bmix/PcrV strain but significantly restored in the Bmix/Atip strain (Fig. 5). However, deletion of pcr1 was not sufficient to restore ExoS translocation in the mismatched Bmix/PcrV strain. Secretion into the extracellular milieu was not affected (Fig. S7). Taken together, these results indicated that the translocation defect of the mismatched Bmix/PcrV strain was primarily due to the defect in pore formation. An additional defect in host cell sensing cannot be ruled out since inhibition of pore assembly could be masking this step.
FIG 5.
Deletion of the secretion regulator Pcr1 does not restore translocation for the translocator-mismatched (Bmix-PcrV) strain. A549 cells were infected for 2 h with strain RP3670 [PAO1 ΔexsE exoS(G/A−) pcrV+ ΔpcrH-popBD], strain RP11222 [PAO1 ΔexsE exoS(G/A−) pcrV::Atip ΔpcrH-popBD], strain RP6370 [PAO1 ΔexsE exoS(G/A−) pcrV+ ΔpcrH-popBD Δpcr1], or strain RP11226 [PAO1 ΔexsE exoS(G/A−) pcrV::Atip ΔpcrH-popBD Δpcr1] complemented with popD and the indicated B-translocator, along with both translocator chaperones, pcrH and acrH. The amount of translocated ExoS was assessed by lysing the host cells with either Triton X-100, which lyses the eukaryotic cell membrane but does not lyse the bacteria, or lysing with SDS, which lyses both the eukaryotic cell and peripherally attached bacteria. ExoS was also monitored in supernatant samples and in bacteria collected from the supernatant after infection (shown in Fig. S7). The presence of ExoS, tubulin (host cell cytoplasmic content), and RNA polymerase subunit α (RPOA, bacterial cytoplasmic content) was assessed by Western blotting. (A and C) Western blots of pcr1+ strains (A) and Δpcr1 strains (C). The amount of ExoS translocated into A549 cells is shown by the Triton-lysed samples. The lower-molecular-weight band is attributable to intracellular cleavage of ExoS (59). The positions of molecular-weight size markers (in kilodaltons) are indicated. SDS-lysed samples show the total amount of ExoS both translocated into host cells and remaining in attached bacteria. (B and D) Quantification of ExoS translocation in Triton-lysed samples for pcr1+ strains (B) and Δpcr1 strains (D). The amount of translocated ExoS is normalized to the amount of tubulin in the same strain and to the amount of ExoS in the strain producing PopB/PcrV. n = 6 biological replicates. Error bars show standard deviations. Statistical differences were analyzed with two-way ANOVA and the Sidak multiple-comparison test. Significance thresholds: *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ns, not statistically significant (P > 0.05).
Supernatant samples for the translocation assay. Related to Fig. 5. The experiment was performed as described in the legend to Fig. 5 using background strains RP3670 (PcrV), RP11222 (Atip), RP6370 (PcrV Δpcr1), and RP11226 (Atip Δpcr1). (A) Schematic diagram of experimental design. (B) Supernatant samples show the amounts of ExoS secreted before host cell contact. Deletion of the secretion regulator pcr1 causes premature secretion of ExoS. (C) Bacterial samples show the amounts of ExoS contained in the bacteria cytosol at the end of the infection. Download FIG S7, TIF file, 1.9 MB (1.9MB, tif) .
Copyright © 2022 Kundracik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Mapping interactions among translocator proteins has been challenging. Biochemical assays have been used to demonstrate interactions by using purified pore-forming translocator proteins (18, 40). Recently, a combination of accessibility of cysteine mutants to chemical modification and disulfide bond formation between adjacent residues was used to map interactions in the Shigella flexneri translocation pore assembled in the plasma membrane of infected cells (28, 41, 42). Here, we used a genetic approach to identify several functionally important interactions that were disrupted by pairing P. aeruginosa and A. hydrophila translocator proteins (Fig. 1). First, our data indicated that AopD failed to interact productively with the P. aeruginosa needle tip protein PcrV. AopD, just like YopD, harbors a phenylalanine residue at a position corresponding to alanine 292 of PopD, which we had shown previously disrupts a critical contact with the collar domain of PcrV (25). However, unlike the YopD-PcrV incompatibility, producing AopD in the context of a hybrid needle tip in which the collar domain of PcrV was replaced with the corresponding region of AcrV (Acollar) only partially restored function. Since AopD fully supported translocation in the context of AcrV, this result argued that AopD made two contacts with the needle tip: one with the collar domain, and a second interaction with the tip domain of the needle tip protein. Second, while mapping the incompatibility between AopB and PcrV, we discovered an internal N- and C-termini of PopB that was also disrupted in PopB-AopB hybrids (Fig. S5). Function could be restored by having both the N-terminus and C-terminus of the hybrid protein derive from PopB. The interaction that was disrupted in the PopB-AopB hybrids could be intramolecular, i.e., required for the folding of individual PopB monomers, or intermolecular, i.e., required for PopB oligomerization. Further research will be needed to distinguish between these possibilities. Finally, we found that the tip domain of the needle tip protein interacted with two regions of the B-translocator (Fig. 2). The needle tip-dependent translocation defect was only evident when both regions of PopB (residues 114 to 150 and 312 to 330) were replaced with the corresponding regions of AopB (resulting in the construct Bmix), suggesting that the two regions both interacted with the needle tip and that a partial substitution was not divergent enough to block the contact with the P. aeruginosa needle tip protein PcrV. As with the internal PopB interaction, the two interacting regions we identified here could be involved in an intermolecular interaction that allows a PopB monomer to interact with the needle tip. However, PopB can dimerize (25), and so the two regions could also be part of an epitope that is formed by two adjacent PopB monomers, which is then recognized by the needle tip. A better structural understanding of the translocon will be necessary to resolve this question.
Antibodies directed against the tip of PcrV are protective in vivo and have been used in clinical trials to treat Pseudomonas infections (43–45). Since this portion of the needle tip had not been implicated in a particular translocon function, we decided to delve more deeply into the interaction between PopB and the tip domain of PcrV. Using a construct that specifically disrupts this interaction (Bmix), as well as a hybrid needle tip that restores translocation in the context of Bmix (Atip), we assessed several aspects of translocon function to pinpoint the role of this interaction in effector translocation. First, we assessed the ability of PcrV to insert PopB or Bmix into the host cell membrane. While we recovered less Bmix than PopB in these experiments, the amount was not affected by the needle tip, demonstrating that the Bmix-PcrV incompatibility did not affect Bmix insertion (Fig. 3). We next assessed the ability of P. aeruginosa to form pores in membranes by detecting translocon-dependent uptake of propidium iodide. Unlike in the translocon insertion experiments, we observed a defect in pore formation by the Bmix/PcrV strain, which was rescued in the Bmix/Atip strain (Fig. 4). Taken together, these data argue that the interaction identified here is required for the assembly of the translocation pore. We also assayed downstream steps in translocation by detecting translocated ExoS protein in lysates of infected epithelial cells. While we could recapitulate the tip-dependent translocation phenotype of bacteria producing the Bmix translocator protein, this defect could not be restored by removing a negative regulator of effector secretion, Pcr1, from the system (Fig. 5). Restoration of effector injection by removing Pcr1 would have indicated that the Bmix-PcrV mismatch interfered with host cell sensing. The defect in pore assembly appears to be the primary block in translocon function incurred by the Bmix-PcrV mismatch. However, an additional role of this interaction in the docking of the tip to the translocation pore or in host cell sensing cannot be ruled out, since a block in an earlier step of the translocation process could mask a defect in these later steps.
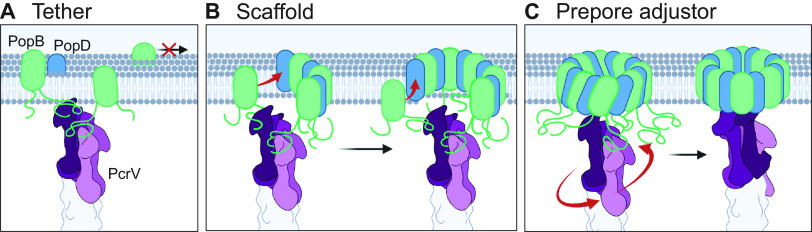
Our genetic analysis of translocon function has uncovered a novel step in the translocon assembly process: needle tip-guided formation of the translocation pore. We envision three different models for this activity (Fig. 6). First, the needle tip could simply serve as a tether for inserted translocator proteins, increasing their local concentration and thereby driving pore formation. Second, the needle tip could serve as a scaffold to actively fold and assemble inserted translocator proteins into the nascent pore. Third, the translocator proteins could insert and assemble into a prepore, which is opened through the interaction with the needle tip. The latter is reminiscent of an assembly pathway that was suggested for the Shigella T3SS, where it was proposed that IpaB may insert into the plasma membrane as a prepore, based on the observation that an ipaC null mutant still retains residual ability to lyse red blood cells (11, 46). Which model is correct, or whether multiple aspects of these models (e.g., tethering and chaperoned folding/assembly) play a role in the assembly of the translocation pore awaits further research. It is also not clear whether PopD similarly relies on the needle tip for assembly into the translocation pore. Intriguingly, our genetic analysis indicates that AopD also requires the tip domain of AcrV for its function. While we have not fully mapped this interaction yet, it will be interesting to see if this interaction is also specifically required for pore formation. Notably, purified PopB and PopD form oligomeric complexes on their own in vitro, as well as heterooligomeric complexes if administered simultaneously. However, PopD complexes cannot be disrupted through the subsequent addition of PopB (18), arguing that this homomeric complex would likely have to be avoided during assembly on the cell surface. Perhaps needle tip-guided pore assembly avoids this dead end? Our results also have implications for interpreting in vitro models of translocation pore formation. In such studies, PopB and PopD are used to form pores in lipid vesicles, but in the absence of the needle tip. If the contribution of the needle tip is to increase the local concentration of translocator proteins, then this function could likely be overcome by increasing the concentration of translocator proteins in those in vitro reactions. However, if the needle tip serves as a scaffold that helps organize the pore, then this raises the question of how faithfully translocation pores assembled in vitro recapitulate pores formed by infecting bacteria in host cells. Clearly, assembling this key interface between the host cell and the bacterium is more complex than initially anticipated.
FIG 6.
Models of PcrV-assisted translocation pore formation. (A) Tether model: PcrV prevents lateral diffusion of PopB and PopD in the membrane, encouraging oligomerization. (B) Scaffold model: the pore proteins could oligomerize around the PcrV needle tip as they are secreted. (C) Prepore model: PopB and PopD could form a prepore structure which is converted by PcrV to its final form. Schematic figures were created using BioRender.com.
MATERIALS AND METHODS
Bacterial strains, cells, and growth conditions.
All bacterial strains used in the study are listed in Table S1. E. coli strains were grown in LB medium (10 g/L tryptone, 5 g/L yeast extract, 5 g/L NaCl). P. aeruginosa strains were grown in high-salt LB (LB with 11.7 g/L NaCl, 5 mM MgCl2, and 0.5 mM CaCl2). Plasmids were retained with 15 μg/mL gentamicin. The expression of plasmid-borne genes under the control of the lacUV5 promoter was induced with 100 μM IPTG (isopropyl β-d-1-thiogalactopyranoside).
A549 cells (ATCC CCL-185) were grown in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) (RP10 medium) at 37°C with 5% CO2. The cells were maintained with penicillin/streptomycin. Before infection, A549 cells were rinsed once with Dulbecco’s phosphate-buffered saline (DPBS), and the medium was exchanged with RP10 without antibiotics [RP10(−)].
Plasmid and strain construction.
The plasmids used in this study are listed in Table S1. The parent strain was PAO1 (47). Purified DNA from Aeromonas hydrophila strain AH-3 (48) was used as the template for acrH, aopB, aopD, and acrV. Plasmids were constructed using standard molecular biology techniques. Wild-type and chimeric translocator genes were cloned by splicing by overlap extension (SOE)-PCR using the primers listed in Table S2. PCR products were digested and ligated into plasmid pEXG2 (allelic exchange vector), pPSV37 (a plasmid that can replicate in P. aeruginosa with the lacUV5 promoter and lacIq), or pPGEH (a plasmid that can replicate in P. aeruginosa with the T3SS pcrG promoter). Mutations in the P. aeruginosa chromosome were introduced by allelic exchange as described previously (49). pPG-popD was generated by amplifying three fragments from pP37-popD (50) using primers dblatoOri/pGtoRepA, dblatoGent/dlacItoGent, and dlacItoTerm/pGtoPopD that were then combined by Gibson assembly (51), thereby removing the lacI gene and remnants of the bla open reading frame (ORF) in pPSV37, as well as replacing the lacUV5 promoter with the promoter upstream from pcrG and introducing a BbvCI site between the gentamicin resistance gene and the replication origin. pPGEH was generated by amplifying the vector backbone of pPG-popD using primers pGpolyFor and pGpolyRev and recircularizing the resultant PCR product using Gibson assembly.
Primers used for cloning in this study. Download Table S2, XLSX file, 0.02 MB (22.4KB, xlsx) .
Copyright © 2022 Kundracik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Cytotoxicity.
Cytotoxicity assays were performed as described previously (25). Briefly, A549 cells were seeded in a 24-well plate at a density of 7.5e4 cells/well. The next day, A549 cells were rinsed twice with DPBS and the medium was replaced with RP10(−) supplemented with 100 μM IPTG. A549 cells were infected with mid-log-phase bacteria at a multiplicity of infection (MOI) of 25. The infection was stopped by fixation with formaldehyde after 2 h, except for the kinetics experiments, which were stopped at various time points up to 3 h. Round versus flat (“healthy”) cells were counted manually by low-power phase-contrast microscopy.
Disulfide cross-linking.
Disulfide cross-linking assays were as described previously (25), with some modifications. Pseudomonas strains producing cysteine mutants of PopB and/or PcrV were used to infect A549 epithelial cells for 2 h in the presence of 25 μM copper-(1,10)-phenanthroline. After infection, the cells were washed with 10 mM iodoacetamide for 10 min to alkylate free cysteine residues. The cells were rinsed with PBS-MC (phosphate buffered saline supplemented with 5 mM MgCl2 and 0.5 mM CaCl2) and then harvested into 1 mL PBS-MC with 2 mM phenylmethylsulfonyl fluoride (PMSF) and Roche cOmplete mini protease inhibitor. The cells were pelleted (5 min at 2,300 × g). The cell membranes were solubilized with 0.1% Triton X-100 in the presence of 2 mM PMSF for 15 min at room temperature. The cellular debris was pelleted (3 min at 9,300 × g), and the supernatant was mixed with 4× SDS sample buffer with or without dithiothreitol (DTT) as indicated. The samples were heated for 10 min at 80°C and analyzed by Western blotting.
A549 membrane insertion.
A549 cells were infected with strain PAO1 harboring the indicated combinations of translocator proteins. After infection, supernatant samples were cleared of bacteria by centrifugation. The A549 cells were rinsed in high-salt buffer (PBS-MC with 1 M KCl) to remove peripherally associated proteins from the membrane. To release intracellular proteins, the A549 cells were treated for 30 min with streptolysin O (Millipore) preactivated with 10 mM DTT. The cells were harvested by scraping and treated with Triton X-100. Cellular debris was removed by centrifugation, and the solubilized membrane was analyzed by Western blotting. Blots were probed for PopB, and the membrane protein E-cadherin or epidermal growth factor receptor (EGFR) was used as a loading control.
Propidium iodide uptake.
The day prior to infection, a 24-well plate was seeded with 9e5 A549 cells/well. The day of infection, the medium was changed to RPMI 1640 (without phenol red) supplemented with 6 μg/mL propidium iodide (Biotium). Cells were infected at an MOI of 100 for 2 h and then rinsed twice with PBS-MC. The unfixed cells were imaged with a Cytation 5 imager (BioTek) using a 20× objective for bright-field and Texas red fluorescence imaging. Red cells were manually counted.
Hemolysis.
Following the previously described protocol (52), sheep erythrocytes in sodium citrate buffer (Quad Five) were washed several times with PBS until the supernatant was clear and then treated with 0.1% papain and 0.01% cysteine for 30 min to promote adhesion by P. aeruginosa (53). Erythrocytes were washed again and resuspended in RPMI 1640 (without phenol red) to a concentration of 5e8 cells/mL. P. aeruginosa strains at mid-log phase were pelleted and resuspended in PBS-MC, the optical density at 600 nm (OD600) was measured, and bacteria were resuspended to a concentration of 2.5e9 cells/mL. Bacteria and red blood cells were mixed thoroughly 1:1 (MOI of 5). Bacteria and erythrocytes were centrifuged (2,000 × g for 5 min). After 2 h at 37°C, the infection mixtures were resuspended and unbroken cells were removed by centrifugation. The absorbance at 545 nm was read with a BioTek Synergy HT plate reader. Hemolysis was calculated as follows: % hemolysis = (Asample − APBS blank)/(ATriton − APBS blank).
Translocation.
ExoS regulates its own secretion through a feedback loop (30, 34). To remove feedback regulation from the equation, we used strains producing a version of ExoS in which the enzymatic activities have been inactivated by point mutations (30). The day before the experiment, A549 cells were seeded in 10-cm2 tissue culture-treated dishes at 1.5e6 cells/dish. A549 epithelial cells were infected with mid-log-phase P. aeruginosa cells for 2 h at an MOI of 25, washed three times with PBS-MC, and rinsed with 1 mL of 250 μg/mL proteinase K in PBS-MC. The protease solution was removed promptly, and the cells were incubated at room temperature for 15 min to digest extracellular protein. Protease-treated cells were resuspended in 1 mL of PBS-MC with 2 mM PMSF, pelleted (3 min at 8,000 × g), and resuspended in 95 μL of PBS-MC with 0.1% Triton X-100. After incubation on ice for 15 min, 45 μL of the cell suspension was removed and mixed with 15 μL of 4× SDS sample buffer (SDS sample). The remaining cells were pelleted, and 45 μL of supernatant was removed and combined with 15 μL of 4× SDS sample buffer (Triton sample). Samples were heated for 10 min at 95°C, separated by SDS-PAGE, and analyzed by Western blotting. Membranes were probed with antibodies directed against ExoS, tubulin, and RNA polymerase α (RpoA).
Western blotting.
Samples were separated on 10% SDS-PAGE gels (Bio-Rad) alongside a molecular-weight marker (Bio-Rad precision plus protein dual color standard). Samples were transferred to Immobilon-FL PVDF (Millipore) for fluorescence imaging or Immobilon-PSQ PVDF (Millipore) for chemiluminescence imaging. For fluorescence detection, blots were incubated with LI-COR anti-mouse secondary antibody for detection at 700 nm or anti-rabbit secondary antibody for detection at 800 nm, both at 1:10,000 concentration, and imaged with a LI-COR Odyssey system. Densitometry analysis was performed with LI-COR Odyssey software and ImageJ. For chemiluminescence detection, blots were incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit or goat anti-mouse secondary antibodies. Detection was performed with chemiluminescent substrate (Advansta Sirius) with image detection on a GE ImageQuant LAS 4000 imager.
Statistical analysis.
Statistical analysis was performed in GraphPad Prism using at least three biological replicates performed on independent days using independent bacterial cultures. Statistical tests are listed in figure legends.
ACKNOWLEDGMENTS
This work was supported by NIH grants number R21 AI107131, R21 AI168028, and 5R01EY022052. E.K. was supported in part by NIH grant number T32 GM007250.
Cells were stored in the Visual Sciences Research Center tissue culture core supported by NEI grant number P30 EY011373. We thank Juan Tomas (Universitat de Barcelona, Department de Microbiologia) for strain Aeromonas hydrophila AH-3. Anti-EGFR antibody was a gift from Cathleen Carlin (Case Western Reserve University). We are grateful to George Dubyak (Case Western Reserve University) for the use of the BioTek Cytation 5 plate reader.
Author contributions were as follows. E. Kundracik: investigation, data curation, formal analysis, writing - original draft, writing - review & editing. J. Trichka: investigation. J. E. Díaz Aponte: investigation. A. Roistacher: resources. A. Rietsch: conceptualization, investigation, data curation, writing - original draft, writing - review & editing, supervision, funding acquisition.
Contributor Information
Arne Rietsch, Email: arne.rietsch@case.edu.
Joanna B. Goldberg, Emory University School of Medicine
REFERENCES
- 1.Berube BJ, Rangel SM, Hauser AR. 2016. Pseudomonas aeruginosa: breaking down barriers. Curr Genet 62:109–113. doi: 10.1007/s00294-015-0522-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lyons BJE, Strynadka NCJ. 2019. On the road to structure-based development of anti-virulence therapeutics targeting the type III secretion system injectisome. Medchemcomm 10:1273–1289. doi: 10.1039/c9md00146h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sawa T, Ito E, Nguyen VH, Haight M. 2014. Anti-PcrV antibody strategies against virulent Pseudomonas aeruginosa. Hum Vaccin Immunother 10:2843–2852. doi: 10.4161/21645515.2014.971641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dickey SW, Cheung GYC, Otto M. 2017. Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat Rev Drug Discov 16:457–471. doi: 10.1038/nrd.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Umrekar TR, Cohen E, Drobnič T, Gonzalez-Rodriguez N, Beeby M. 2021. CryoEM of bacterial secretion systems: a primer for microbiologists. Mol Microbiol 115:366–382. doi: 10.1111/mmi.14637. [DOI] [PubMed] [Google Scholar]
- 6.Dey S, Chakravarty A, Guha Biswas P, De Guzman RN. 2019. The type III secretion system needle, tip, and translocon. Protein Sci 28:1582–1593. doi: 10.1002/pro.3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenkins J, Worrall LJ, Strynadka NCJ. 2022. Recent structural advances towards understanding of the bacterial type III secretion injectisome. Trends Biochem Sci 47:795–809. doi: 10.1016/j.tibs.2022.04.013. [DOI] [PubMed] [Google Scholar]
- 8.Park D, Lara-Tejero M, Waxham MN, Li W, Hu B, Galán JE, Liu J. 2018. Visualization of the type III secretion mediated Salmonella-host cell interface using cryo-electron tomography. Elife 7:e39514. doi: 10.7554/eLife.39514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lombardi C, Tolchard J, Bouillot S, Signor L, Gebus C, Liebl D, Fenel D, Teulon J-M, Brock J, Habenstein B, Pellequer J-L, Faudry E, Loquet A, Attrée I, Dessen A, Job V. 2019. Structural and functional characterization of the type three secretion system (T3SS) needle of Pseudomonas aeruginosa. Front Microbiol 10:573. doi: 10.3389/fmicb.2019.00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rietsch A, Mekalanos JJ. 2006. Metabolic regulation of type III secretion gene expression in Pseudomonas aeruginosa. Mol Microbiol 59:807–820. doi: 10.1111/j.1365-2958.2005.04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickenson NE, Choudhari SP, Adam PR, Kramer RM, Joshi SB, Middaugh CR, Picking WL, Picking WD. 2013. Oligomeric states of the Shigella translocator protein IpaB provide structural insights into formation of the type III secretion translocon. Protein Sci 22:614–627. doi: 10.1002/pro.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hume PJ, McGhie EJ, Hayward RD, Koronakis V. 2003. The purified Shigella IpaB and Salmonella SipB translocators share biochemical properties and membrane topology. Mol Microbiol 49:425–439. doi: 10.1046/j.1365-2958.2003.03559.x. [DOI] [PubMed] [Google Scholar]
- 13.Hayward RD, Cain RJ, McGhie EJ, Phillips N, Garner MJ, Koronakis V. 2005. Cholesterol binding by the bacterial type III translocon is essential for virulence effector delivery into mammalian cells. Mol Microbiol 56:590–603. doi: 10.1111/j.1365-2958.2005.04568.x. [DOI] [PubMed] [Google Scholar]
- 14.Kueltzo LA, Osiecki J, Barker J, Picking WL, Ersoy B, Picking WD, Middaugh CR. 2003. Structure-function analysis of invasion plasmid antigen C (IpaC) from Shigella flexneri. J Biol Chem 278:2792–2798. doi: 10.1074/jbc.M208383200. [DOI] [PubMed] [Google Scholar]
- 15.Tang Y, Romano FB, Breña M, Heuck AP. 2018. The Pseudomonas aeruginosa type III secretion translocator PopB assists the insertion of the PopD translocator into host cell membranes. J Biol Chem 293:8982–8993. doi: 10.1074/jbc.RA118.002766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wager B, Faudry E, Wills T, Attree I, Delcour AH. 2013. Current fluctuation analysis of the PopB and PopD translocon components of the Pseudomonas aeruginosa type III secretion system. Biophys J 104:1445–1455. doi: 10.1016/j.bpj.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faudry E, Vernier G, Neumann E, Forge V, Attree I. 2006. Synergistic pore formation by type III toxin translocators of Pseudomonas aeruginosa. Biochemistry 45:8117–8123. doi: 10.1021/bi060452+. [DOI] [PubMed] [Google Scholar]
- 18.Romano FB, Tang Y, Rossi KC, Monopoli KR, Ross JL, Heuck AP. 2016. Type 3 secretion translocators spontaneously assemble a hexadecameric transmembrane complex. J Biol Chem 291:6304–6315. doi: 10.1074/jbc.M115.681031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goure J, Pastor A, Faudry E, Chabert J, Dessen A, Attree I. 2004. The V antigen of Pseudomonas aeruginosa is required for assembly of the functional PopB/PopD translocation pore in host cell membranes. Infect Immun 72:4741–4750. doi: 10.1128/IAI.72.8.4741-4750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Picking WL, Nishioka H, Hearn PD, Baxter MA, Harrington AT, Blocker A, Picking WD. 2005. IpaD of Shigella flexneri is independently required for regulation of Ipa protein secretion and efficient insertion of IpaB and IpaC into host membranes. Infect Immun 73:1432–1440. doi: 10.1128/IAI.73.3.1432-1440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broz P, Mueller CA, Müller SA, Philippsen A, Sorg I, Engel A, Cornelis GR. 2007. Function and molecular architecture of the Yersinia injectisome tip complex. Mol Microbiol 65:1311–1320. doi: 10.1111/j.1365-2958.2007.05871.x. [DOI] [PubMed] [Google Scholar]
- 22.Mueller CA, Broz P, Cornelis GR. 2008. The type III secretion system tip complex and translocon. Mol Microbiol 68:1085–1095. doi: 10.1111/j.1365-2958.2008.06237.x. [DOI] [PubMed] [Google Scholar]
- 23.Matteï P-J, Faudry E, Job V, Izoré T, Attree I, Dessen A. 2011. Membrane targeting and pore formation by the type III secretion system translocon: membrane targeting and pore formation by the T3SS. FEBS J 278:414–426. doi: 10.1111/j.1742-4658.2010.07974.x. [DOI] [PubMed] [Google Scholar]
- 24.Ekestubbe S, Bröms JE, Edgren T, Fällman M, Francis MS, Forsberg Å. 2016. The amino-terminal part of the needle-tip translocator LcrV of Yersinia pseudotuberculosis is required for early targeting of YopH and in vivo virulence. Front Cell Infect Microbiol 6:175. doi: 10.3389/fcimb.2016.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armentrout EI, Rietsch A. 2016. The type III secretion translocation pore senses host cell contact. PLoS Pathog 12:e1005530. doi: 10.1371/journal.ppat.1005530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veenendaal AKJ, Hodgkinson JL, Schwarzer L, Stabat D, Zenk SF, Blocker AJ. 2007. The type III secretion system needle tip complex mediates host cell sensing and translocon insertion. Mol Microbiol 63:1719–1730. doi: 10.1111/j.1365-2958.2007.05620.x. [DOI] [PubMed] [Google Scholar]
- 27.Roehrich AD, Martinez-Argudo I, Johnson S, Blocker AJ, Veenendaal AKJ. 2010. The extreme C terminus of Shigella flexneri IpaB is required for regulation of type III secretion, needle tip composition, and binding. Infect Immun 78:1682–1691. doi: 10.1128/IAI.00645-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russo BC, Duncan-Lowey JK, Chen P, Goldberg MB. 2021. The type 3 secretion system requires actin polymerization to open translocon pores. PLoS Pathog 17:e1009932. doi: 10.1371/journal.ppat.1009932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russo BC, Duncan JK, Wiscovitch AL, Hachey AC, Goldberg MB. 2019. Activation of Shigella flexneri type 3 secretion requires a host-induced conformational change to the translocon pore. PLoS Pathog 15:e1007928. doi: 10.1371/journal.ppat.1007928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cisz M, Lee P-C, Rietsch A. 2008. ExoS controls the cell contact-mediated switch to effector secretion in Pseudomonas aeruginosa. J Bacteriol 190:2726–2738. doi: 10.1128/JB.01553-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frithz-Lindsten E, Holmström A, Jacobsson L, Soltani M, Olsson J, Rosqvist R, Forsberg A. 1998. Functional conservation of the effector protein translocators PopB/YopB and PopD/YopD of Pseudomonas aeruginosa and Yersinia pseudotuberculosis. Mol Microbiol 29:1155–1165. doi: 10.1046/j.1365-2958.1998.00994.x. [DOI] [PubMed] [Google Scholar]
- 32.Heuck AP, Brovedan MA. 13 June 2022. Evolutionary conservation, variability, and adaptation of type III secretion systems. J Membr Biol doi: 10.1007/s00232-022-00247-9. [DOI] [PubMed] [Google Scholar]
- 33.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Armentrout EI, Kundracik EC, Rietsch A. 2021. Cell-type-specific hypertranslocation of effectors by the Pseudomonas aeruginosa type III secretion system. Mol Microbiol 115:305–319. doi: 10.1111/mmi.14617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dacheux D, Goure J, Chabert J, Usson Y, Attree I. 2001. Pore-forming activity of type III system-secreted proteins leads to oncosis of Pseudomonas aeruginosa-infected macrophages. Mol Microbiol 40:76–85. doi: 10.1046/j.1365-2958.2001.02368.x. [DOI] [PubMed] [Google Scholar]
- 36.Schoehn G, Di Guilmi AM, Lemaire D, Attree I, Weissenhorn W, Dessen A. 2003. Oligomerization of type III secretion proteins PopB and PopD precedes pore formation in Pseudomonas. EMBO J 22:4957–4967. doi: 10.1093/emboj/cdg499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosqvist R, Magnusson KE, Wolf-Watz H. 1994. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J 13:964–972. doi: 10.1002/j.1460-2075.1994.tb06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hauser AR. 2009. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat Rev Microbiol 7:654–665. doi: 10.1038/nrmicro2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee P-C, Zmina SE, Stopford CM, Toska J, Rietsch A. 2014. Control of type III secretion activity and substrate specificity by the cytoplasmic regulator PcrG. Proc Natl Acad Sci USA 111:E2027–E2036. doi: 10.1073/pnas.1402658111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis R, Marquart ME, Lucius D, Picking WD. 1998. Protein-protein interactions in the assembly of Shigella flexneri invasion plasmid antigens IpaB and IpaC into protein complexes. Biochim Biophys Acta 1429:45–56. doi: 10.1016/s0167-4838(98)00213-1. [DOI] [PubMed] [Google Scholar]
- 41.Russo BC, Duncan JK, Goldberg MB. 2019. Topological analysis of the type 3 secretion system translocon pore protein IpaC following its native delivery to the plasma membrane during infection. mBio 10:e00877-19. doi: 10.1128/mBio.00877-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen P, Russo BC, Duncan-Lowey JK, Bitar N, Egger KT, Goldberg MB. 2021. Topology and contribution to the pore channel lining of plasma membrane-embedded Shigella flexneri type 3 secretion translocase IpaB. mBio 12:e03021-21. doi: 10.1128/mBio.03021-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tabor DE, Oganesyan V, Keller AE, Yu L, McLaughlin RE, Song E, Warrener P, Rosenthal K, Esser M, Qi Y, Ruzin A, Stover CK, DiGiandomenico A. 2018. Pseudomonas aeruginosa PcrV and Psl, the molecular targets of bispecific antibody MEDI3902, are conserved among diverse global clinical isolates. J Infect Dis 218:1983–1994. doi: 10.1093/infdis/jiy438. [DOI] [PubMed] [Google Scholar]
- 44.François B, Luyt C-E, Dugard A, Wolff M, Diehl J-L, Jaber S, Forel J-M, Garot D, Kipnis E, Mebazaa A, Misset B, Andremont A, Ploy M-C, Jacobs A, Yarranton G, Pearce T, Fagon J-Y, Chastre J. 2012. Safety and pharmacokinetics of an anti-PcrV PEGylated monoclonal antibody fragment in mechanically ventilated patients colonized with Pseudomonas aeruginosa: a randomized, double-blind, placebo-controlled trial. Crit Care Med 40:2320–2326. doi: 10.1097/CCM.0b013e31825334f6. [DOI] [PubMed] [Google Scholar]
- 45.Jain R, Beckett VV, Konstan MW, Accurso FJ, Burns JL, Mayer-Hamblett N, Milla C, VanDevanter DR, Chmiel JF, KB001-A Study Group . 2018. KB001-A, a novel anti-inflammatory, found to be safe and well-tolerated in cystic fibrosis patients infected with Pseudomonas aeruginosa. J Cyst Fibros 17:484–491. doi: 10.1016/j.jcf.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 46.Barta ML, Tachiyama S, Muthuramalingam M, Arizmendi O, Villanueva CE, Ramyar KX, Geisbrecht BV, Lovell S, Battaile KP, Picking WL, Picking WD. 2018. Using disruptive insertional mutagenesis to identify the in situ structure-function landscape of the Shigella translocator protein IpaB. Protein Sci 27:1392–1406. doi: 10.1002/pro.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bleves S, Soscia C, Nogueira-Orlandi P, Lazdunski A, Filloux A. 2005. Quorum sensing negatively controls type III secretion regulon expression in Pseudomonas aeruginosa PAO1. J Bacteriol 187:3898–3902. doi: 10.1128/JB.187.11.3898-3902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vilches S, Urgell C, Merino S, Chacón MR, Soler L, Castro-Escarpulli G, Figueras MJ, Tomás JM. 2004. Complete type III secretion system of a mesophilic Aeromonas hydrophila strain. Appl Environ Microbiol 70:6914–6919. doi: 10.1128/AEM.70.11.6914-6919.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rietsch A, Vallet-Gely I, Dove SL, Mekalanos JJ. 2005. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc Natl Acad Sci USA 102:8006–8011. doi: 10.1073/pnas.0503005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomalka AG, Stopford CM, Lee P-C, Rietsch A. 2012. A translocator-specific export signal establishes the translocator-effector secretion hierarchy that is important for type III secretion system function. Mol Microbiol 86:1464–1481. doi: 10.1111/mmi.12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 52.Costa TRD, Francis MK, Farag SI, Edgren T, Francis MS. 2019. Measurement of Yersinia translocon pore formation in erythrocytes. Methods Mol Biol 2010:211–229. doi: 10.1007/978-1-4939-9541-7_15. [DOI] [PubMed] [Google Scholar]
- 53.Gilboa-Garber N. 1972. Purification and properties of hemagglutinin from Pseudomonas aeruginosa and its reaction with human blood cells. Biochim Biophys Acta 273:165–173. doi: 10.1016/0304-4165(72)90204-8. [DOI] [PubMed] [Google Scholar]
- 54.Discola KF, Förster A, Boulay F, Simorre J-P, Attree I, Dessen A, Job V. 2014. Membrane and chaperone recognition by the major translocator protein PopB of the type III secretion system of Pseudomonas aeruginosa. J Biol Chem 289:3591–3601. doi: 10.1074/jbc.M113.517920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo EZ, Galán JE. 2021. Cryo-EM structure of the needle filament tip complex of the Salmonella type III secretion injectisome. Proc Natl Acad Sci USA 118:e2114552118. doi: 10.1073/pnas.2114552118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. 2022. ColabFold: making protein folding accessible to all. Nat Methods 19:679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, Ferrin TE. 2021. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci 30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.UniProt Consortium. 2021. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res 49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pederson KJ, Pal S, Vallis AJ, Frank DW, Barbieri JT. 2000. Intracellular localization and processing of Pseudomonas aeruginosa ExoS in eukaryotic cells. Mol Microbiol 37:287–299. doi: 10.1046/j.1365-2958.2000.01990.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of bacterial strains and plasmids used in the study. Download Table S1, PDF file, 0.05 MB (52.9KB, pdf) .
Copyright © 2022 Kundracik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
All pairwise comparisons for translocator exchange cytotoxicity testing presented in Fig. 1D. Download Table S3, XLSX file, 0.01 MB (15.1KB, xlsx) .
Copyright © 2022 Kundracik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Protein sequence alignment for PcrV-AcrV. Related to Fig. 1 and 2. PcrV (GenBank accession no. AE004091) and AcrV (GenBank accession no. AY528667) were aligned using ClustalW. The amino acid sequences are 36% identical (50% similar). The construct Atip is indicated by purple (segments from PcrV) and yellow (segments from AcrV). Download FIG S1, TIF file, 0.9 MB (970.9KB, tif) .
Copyright © 2022 Kundracik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Protein sequence alignment for PopB-AopB. Related to Fig. 1 and 2. PopB (GenBank accession no. AE004091) and AopB (GenBank accession no. AY528667) were aligned using ClustalW. The amino acid sequences are 46% identical (71% similar). The construct Bmix is indicated by green (segments from PopB) and yellow (segments from AopB). Boxes indicate predicted transmembrane domains from the data with UniProt accession numbers Q9I324 (PopB) and Q699Q8 (AopB). Download FIG S2, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2022 Kundracik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Additional controls and kinetics for cytotoxicity assay. Related to Fig. 1. (A) The cytotoxicity experiment was performed as described in the legend to Fig. 1 using background strains RP3624 (PcrV) and RP6425 (Atip). The wild-type strain is RP2318. The ΔpcrV strain is RP3223. The ΔpopB and ΔpopD strains have the background strain RP3624 complemented with a plasmid carrying either popB or popD along with both export chaperone homologs, pcrH and acrH. n = 3 biological replicates. Error bars show standard deviations. (B and C) Kinetics of cell rounding in response to P. aeruginosa infection. The cytotoxicity experiment was performed as described in the legend to Fig. 1 but was stopped at various time points up to 3 h. HBD denotes pcrH-popB-popD. n = 4 biological replicates. Error bars show standard deviations. The data set is split into two graphs for clarity. Download FIG S3, TIF file, 0.6 MB (651.9KB, tif) .
Copyright © 2022 Kundracik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Refining the region of interest for the PopB-AopB interaction. Related to Fig. 2. (A) Schematic diagrams of PopB-AopB chimeras. This set of chimeras was designed to refine the boundaries of the PopB-PcrV interaction, focusing on the N-terminal substitution. Created with BioRender.com. (B) Cytotoxicity data for PopB-AopB chimeras. The experiment was performed as described in the legend to Fig. 1 using background strains RP3624 (PcrV) and RP6425 (Atip). n = 3 biological replicates. Error bars show standard deviations. Statistical differences were analyzed with two-way ANOVA and the Sidak multiple-comparison test: *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ****, P < 0.0001; ns, not statistically significant (P > 0.05). Download FIG S4, TIF file, 0.7 MB (749.4KB, tif) .
Copyright © 2022 Kundracik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Evidence for a PopB-PopB interaction. Related to Fig. 2. (A) Schematic diagrams of PopB-AopB chimeras. (B) Cytotoxicity data for PopB-AopB chimeras. The experiment was performed as described in the legend to Fig. 1 using background strains RP3624 (PcrV) and RP6425 (Atip). n = 3 biological replicates. Error bars show standard deviations. Statistical differences were analyzed with two-way ANOVA and the Sidak multiple-comparison test: *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ***, P < 0.0001; ns, not statistically significant (P > 0.05). (C) Schematic diagram showing two possible PopB-PopB interactions. In the first scenario (left), two adjacent monomers of PopB interact with each other. In the second scenario (right), PopB folds on itself and has an intramolecular interaction. Created with BioRender.com. Download FIG S5, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2022 Kundracik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The nonhomologous segment of AcrV is not required for rescue of Bmix. Related to Fig. 2. (A) Residues 220 to 269 of AcrV, which have no homologous segment in PcrV, were deleted from the Atip construct to create the PcrV-AcrV chimera “Atip-trim.” Models of PcrV, Atip, and Atip-trim were created as described in the legend to Fig. 1. (B) The experiment was performed as described in the legend to Fig. 1, using background strains RP3624 (PcrV), RP6425 (Atip), and RP12517 (Atip-trim). Statistical significance was calculated by two-way ANOVA with the Tukey multiple-comparison test. Data bars are a summary of the results from three biological replicates with error bars showing standard deviations. ns, not significant (P > 0.05). Download FIG S6, TIF file, 0.9 MB (955KB, tif) .
Copyright © 2022 Kundracik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supernatant samples for the translocation assay. Related to Fig. 5. The experiment was performed as described in the legend to Fig. 5 using background strains RP3670 (PcrV), RP11222 (Atip), RP6370 (PcrV Δpcr1), and RP11226 (Atip Δpcr1). (A) Schematic diagram of experimental design. (B) Supernatant samples show the amounts of ExoS secreted before host cell contact. Deletion of the secretion regulator pcr1 causes premature secretion of ExoS. (C) Bacterial samples show the amounts of ExoS contained in the bacteria cytosol at the end of the infection. Download FIG S7, TIF file, 1.9 MB (1.9MB, tif) .
Copyright © 2022 Kundracik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used for cloning in this study. Download Table S2, XLSX file, 0.02 MB (22.4KB, xlsx) .
Copyright © 2022 Kundracik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.