ABSTRACT
HIV-exposed uninfected infants (HEU) have abnormal immunologic functions and increased infectious morbidity in the first 6 months of life, which gradually decreases thereafter. The mechanisms underlying HEU immune dysfunctions are unknown. We hypothesized that unique characteristics of the HEU gut microbiota associated with maternal HIV status may underlie the HEU immunologic dysfunctions. We characterized the infant gut, maternal gut, and breast milk microbiomes of mother-infant pairs, including 123 with HEU and 117 with HIV-uninfected infants (HUU), from South Africa. Pan-bacterial 16S rRNA gene sequencing was performed on (i) infant stool at 6, 28, and 62 weeks; (ii) maternal stool at delivery and 62 weeks; and (iii) breast milk at 6 weeks. Infant gut alpha and beta diversities were similar between groups. Microbial composition significantly differed, including 12 genera, 5 families and 1 phylum at 6 weeks; 12 genera and 2 families at 28 weeks; and 2 genera and 2 families at 62 weeks of life. Maternal gut microbiomes significantly differed in beta diversity and microbial composition, and breast milk microbiomes differed in microbial composition only. Infant gut microbiotas extensively overlapped with maternal gut and minimally with breast milk microbiotas. Nevertheless, exclusively breastfed HEU and HUU had less divergent microbiomes than nonexclusively breastfed infants. Feeding pattern and maternal gut microbiome imprint the HEU gut microbiome. Compared to HUU, the HEU gut microbiome prominently differs in early infancy, including increased abundance of taxa previously observed to be present in excess in adults with HIV. The HEU and HUU gut microbiome compositions converge over time, mirroring the kinetics of HEU infectious morbidity risk.
KEYWORDS: HIV, HIV-exposed uninfected infants, gut microbiome, breast milk microbiome, pregnant women with HIV, human immunodeficiency virus
INTRODUCTION
In utero-HIV-exposed uninfected infants (HEU) have a higher incidence of hospitalization and 2- to 4-fold higher mortality rate than HIV-unexposed uninfected infants (HUU) in low-and-middle-income countries (1–15). In the United States, Canada, and other areas with improved health care, the risk of death is similar in HEU and HUU, but recent studies have reported more frequent hospitalizations due to infections in HEU (16–20). The infections with high morbidity and mortality in HEU are caused by common childhood pathogens, including respiratory syncytial virus (RSV) and Streptococcus pneumoniae (14, 21–25). Insufficient passive protection due to low transplacental transport of maternal antibodies in women with HIV might account for the increased morbidity of infections in HEU during early infancy (22, 26). However, we showed that concentrations at birth of antibodies against respiratory pathogens, including RSV and S. pneumoniae, were similar in HEU who developed medically attended lower respiratory tract infections in the first 6 months of life and those who did not, suggesting that the immune responses of HEU to infections are insufficient to control these pathogens (27). Multiple phenotypic and functional immunologic characteristics differentiate HEU and HUU in the first year of life, particularly in cell-mediated immunity (19, 20, 28–30). These dysfunctions may explain the increased vulnerability of HEU infants to infections. However, the mechanisms underlying immunologic dysfunctions in HEU are not currently understood.
The gut microbiome has an important role in the education of the infant immune system. The human gut harbors 12% to 20% of the total lymphocytes, and most importantly, it is a site of innate and adaptive T cell maturation second only to the thymus (31, 32). Myeloid and lymphoid immune cells acquire specific characteristics through transcriptional and epigenetic reprogramming and modulation of transcription factor binding directed by the gut microbiota (33–35). Gut-imprinted myeloid and lymphoid cells exit the gut to populate intestinal lymph nodes, the peripheral blood, and other tissues (36–41). The composition of the infant gut microbiome undergoes sequential changes after birth, influenced primarily by the delivery mode, maternal microbiome, and maternal and infant diet (42–50). Other factors, including gestational age at birth, antibiotic therapy, genetic background of the host, and geographic location, also contribute to the composition and structure of the microbiome (33, 49–58). Although subject to many changes in the first 1 to 2 years of life, the infant gut microbiome evolves in a scripted fashion until it acquires adult characteristics (59, 60).
Pioneering work by Bender et al. showed differences in gut microbial alpha diversity of 1- to 4-month-old HEU and HUU examined cross-sectionally at a single time point (61). The authors did not find sufficient differences in either breast milk or skin microbiotas of mothers with and without HIV to explain HEU gut dysbiosis and did not analyze the maternal gut microbiome. Subsequently, D’Souza et al. compared the structure of the gut microbiome at 6, 16, and 24 weeks of age in HEU receiving co-trimoxazole prophylaxis or not and showed that co-trimoxazole administration may also contribute to the dysbiosis observed in HEU (62). Two other cross-sectional studies that compared HEU and HUU gut microbiomes at approximately 2 years of life found similar alpha or beta diversities in the two groups (63, 64).
Breast milk, oral, vaginal, and maternal gut microbiotas contribute to the formation of the infant microbiome in the beginning of life, with the gut microbiota making the largest contribution (65). In addition, strictly anaerobic bacteria are shared by the breast milk and maternal gut, which led to the proposal of a gut-breast axis whereby dendritic cells transport bacteria from the gut to the mammary gland (66, 67). Due to pH and other ecologic requirements, the maternal vaginal microbiota may only transiently colonize the infant gut (65). Previous studies have shown differences in the composition of the gut microbiotas of people with and without HIV (68, 69). After early controversy whether the differences originated from HIV infection or sexual practices of males who have sex with males, it was established that HIV infection by itself was associated with specific microbiome characteristics, including in pregnant and nonpregnant women (68). These conclusions were consistent with the bidirectional relationship between microbiota and host immune system, whereby the host immune responses to commensals play an important role in shaping the microbiome (52).
The overarching hypothesis of this study is that gut dysbiosis in HEU contributes to immunologic dysfunctions in early life. Moreover, the composition of the gut microbiome can be modified by adjusting the diet and other determining factors. Here, we report on the evolution of the gut microbiome in HEU in comparison with that in HUU during the first 62 weeks of life and the relationship of the maternal gut, breast milk, and infant gut microbiotas in a longitudinal cohort of 240 mother-infant dyads.
RESULTS
Characteristics of the study population.
This study enrolled 240 mother-infant pairs, including 123 mothers with HIV and 117 without HIV, between June and December 2017. Notable differences between the mothers with and without HIV were higher chronological age and parity and lower body mass index (BMI) in mothers with HIV (Table 1). Mothers with HIV had medians of 347 CD4+ T cells/μL of blood and <20 HIV RNA copies/mL of plasma. All but one of the mothers with HIV reported antiretroviral compliance during pregnancy. There were no differences in alcohol or tobacco use or level of education between the two groups.
TABLE 1.
Participant characteristics at deliverya
| Characteristic | Value for: |
P | |
|---|---|---|---|
| HIV or exposed group | Non-HIV or unexposed group | ||
| Mothers | With HIV (n = 123) | Without HIV (n = 117) | |
| Age (yrs) (median [Q1, Q3]) | 30.0 [26.0, 34.5] | 25.0 [22.0, 30.0] | <0.01 |
| Previous pregnancies | |||
| Median [Q1, Q3] | 2.00 [1.00, 3.00] | 1.00 [0, 2.00] | <0.01 |
| No. (%) missing | 2 (1.6) | 5 (4.3) | |
| BMI at 62 wks | |||
| Median [Q1, Q3] | 23.9 [20.5, 28.1] | 27.1 [21.7, 31.1] | 0.02 |
| No. (%) missing | 51 (41.5) | 59 (50.4) | |
| No. (%) with no smoking during pregnancy | 114 (92.7) | 113 (96.6) | 0.18 |
| No. (%) with no alcohol during pregnancy | 112 (91.1) | 109 (93.2) | 0.55 |
| CD4+ cells/μL | |||
| Median [Q1, Q3] | 347 [227, 499] | NA | |
| No. (%) missing | 7 (5.7) | NA | |
| Log HIV RNA copies/mL | |||
| Median [Q1, Q3] | 1.00 [0, 2.06]b | NA | |
| No. (%) missing | 10 (8.1) | NA | |
| No. (%) compliant with ART | 122 (99.2) | NA | |
| Infants | HEU (n = 123) | HUU (n = 117) | |
| No. (%) female | 65 (52.8) | 56 (47.9) | 0.44 |
| No. (%) with vaginal delivery | 116 (94.3) | 116 (99.1) | 0.07 |
| Gestational age (wks) | |||
| Mean (SD) | 39.1 (2.22) | 39.3 (1.52) | 0.48 |
| No. (%) missing | 4 (3.3) | 1 (0.9) | |
| Birth wt (g) [mean (SD)] | 3,070 (423) | 3,250 (437) | <0.01 |
ART, antiretroviral treatment; NA, not evaluated.
A target that was not detected was assigned a value of 0, and those with values of <20 were assigned a value of 10 copies/mL.
At birth, HEU and HUU had similar gestational ages by study design, with an average of 39 weeks. The sex distribution was also similar (Table 1). HEU had significantly lower weight at birth, with a mean of 3,070 g compared with 3,250 g in HUU. None of the infants were small or large for gestational age. Seven HEU and one HUU were delivered by emergency cesarean section for obstetrical indications identified after initiation of labor.
Feeding pattern and antibiotic usage were recorded at each visit (Table 2). At 6 weeks of life, >80% of infants were exclusively breastfed in each group and 15% HEU and 10% HUU, respectively, were exclusively formula fed. At 28 weeks, 61% of HEU and 50% of HUU were still exclusively breastfed, whereas exclusive formula feeding was reported in 22% of HEU and 11% of HUU. At 62 weeks, all infants received solid foods. Co-trimoxazole was used in 5%, 90%, and 87% of the HEU at 6, 28, and 62 weeks.
TABLE 2.
Changes in breastfeeding and co-trimoxazole use over time in infants with gut microbiome information
| Parameter | No. (%) of infants at: |
|||||
|---|---|---|---|---|---|---|
| 6 wks |
28 wks |
62 wks |
||||
| HEU (n = 98) | HUU (n = 88) | HEU (n = 86) | HUU (n = 80) | HEU (n = 74) | HUU (n = 74) | |
| Feeding method | ||||||
| Exclusive breast feeding | 83 (84.7) | 72 (81.8) | 52 (60.5) | 40 (50.0) | 0 (0) | 0 (0) |
| Exclusive formula feeding | 13 (13.3) | 4 (4.5) | 19 (22.1) | 9 (11.3) | 0 (0) | 0 (0) |
| Mixeda | 2 (2.0) | 12 (13.6) | 15 (17.4) | 31 (38.8) | 74 (100) | 74 (100) |
| Co-trimoxazole | ||||||
| No | 93 (94.9) | 88 (100) | 8 (9.3) | 79 (98.8) | 10 (13.5) | 74 (100) |
| Yes | 5 (5.1) | 0 (0) | 77 (89.5) | 0 (0) | 64 (86.5) | 0 (0) |
| Missing | 0 (0) | 0 (0) | 1 (1.2) | 1 (1.3) | 0 (0) | 0 (0) |
Any combination of breast milk, formula, and/or solids.
Infant gut microbiota.
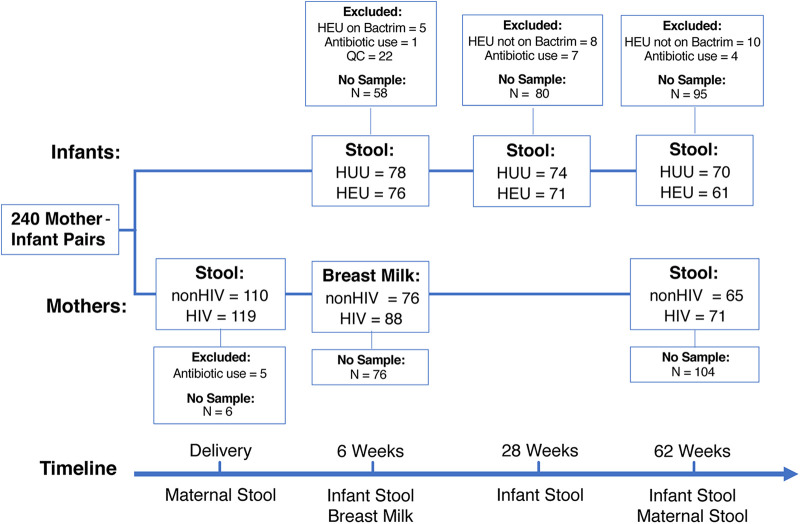
Figure 1 shows the number of samples collected for the microbiome analyses and the reasons for exclusions. Among infants, 61 to 78 rectal samples/group of HEU or HUU were included in the final analysis.
FIG 1.
Consort diagram. “QC” indicates samples that were excluded because they did not pass sequence quality control (details are provided in Materials and Methods).
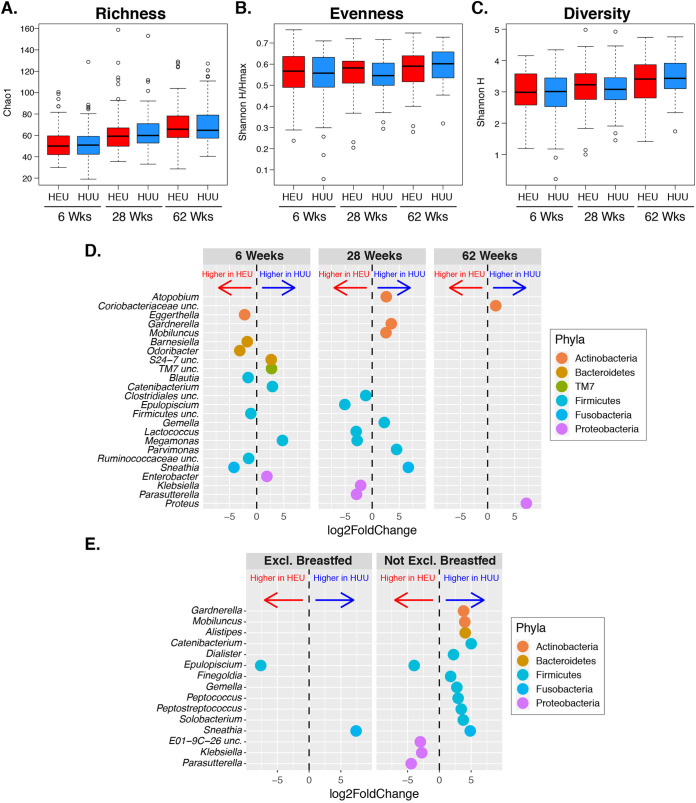
No significant differences were found in either alpha diversity (richness, evenness, and Shannon diversity) or beta diversity between HEU and HUU at any time point (Fig. 2). An evaluation of the relationship between diversity of the HEU gut microbiome at 6 weeks and the maternal CD4+ T cell counts or viral loads at delivery did not reveal significant associations (data not shown).
FIG 2.
Infant gut microbiome. (A to C) Alpha diversity analysis of community richness (Chao1), evenness (Shannon H/Hmax), and diversity (Shannon H). There were no significant differences between HEU and HUU. (D) Genera with significantly different relative abundances between HEU and HUU at the indicated time points (FDR P < 0.05). (E) Genera with significantly different relative abundances between HEU and HUU exclusively (Excl.) breastfed at 28 weeks of life and not exclusively breastfed.
At 6 weeks, 12 out of the 224 genus-level taxa significantly differed between HEU and HUU, including higher abundance in HEU of Eggerthella sp, Barnesiella sp, Odoribacter sp, Blautia sp, Sneathia sp, one Firmicutes uncl, and one Ruminococcaceae uncl, and lower abundance in HEU of Catenibacterium sp, Megamonas sp, Enterobacter sp, TM7, and Bacteroidetes uncl (Fig. 2). In addition, compared with HUU, the HEU microbiome contained higher overall abundance of Firmicutes uncl, Carnobacteriaceae and Enterococcaceae families; lower abundance of the Coriobacteriaceae, Bacteroidetes and S24-7 families; and lower abundance of the phylum Proteobacteria (Table S1).
At 28 weeks, 12 taxa significantly differed between HEU and HUU. However, the divergent taxa were not conserved between 6 and 28 weeks (Fig. 2). Compared with HUU, HEU had higher abundance of unclassified Clostridiales, Epulopiscium sp., Lactococcus sp., Megamonas sp., Klebsiella sp., and Parasutterella sp. and lower abundance of Atopobium sp., Gardnerella sp., Mobiluncus sp., Gemella sp., Parvimonas sp., and Sneathia sp. At the family level, the Leptotrichiaceae and family XI incertae sedis (Firmicutes) were significantly less abundant in HEU than HUU (see Table S1 in the supplemental material). There were no differences in abundance at the phylum level. Considering the difference between HEU and HUU in the proportions of exclusively breastfed infants at 28 weeks of life, we performed separate comparisons in exclusively breastfed and nonexclusively breastfed HEU and HUU (Fig. 2). Regardless of breastfeeding status, Epulopiscium sp. was significantly more abundant and Sneathia sp. was less abundant in HEU than HUU. All other significantly different taxa were present only among nonexclusively breastfed infants. Exclusively breastfed HEU also had a lower abundance of the family Leptotrichiaceae than HUU.
(A) Differences in infant gut microbiome; (B) differences in maternal gut microbiome; (C) differences in breast milk microbiome. Download Table S1, DOCX file, 0.05 MB (49.7KB, docx) .
Copyright © 2022 Jackson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
At 62 weeks of life, 2 taxa differed significantly between the HEU and HUU gut microbiomes, Proteus sp. and unclassified Coriobacteriaceae, both with lower abundance in HEU. At the family level, the Lachnospiraceae and unclassified Clostridiales were more abundant in HEU. There were no differences at the phylum level (Table S1).
The longitudinal analysis of the infants’ microbiome changes over time showed that richness increased significantly from 6 to 28 weeks in HEU (P = 0.02) and HUU (P = 0.002) (Fig. S1), but after adjustment for breastfeeding status at 28 weeks, the changes lost statistical significance. In contrast, from 28 to 62 weeks, evenness and Shannon diversity increased significantly in HUU (P values of 0.005 and 0.003, respectively) but not in HEU (Fig. S1). After adjustment for exclusive-breastfeeding status at 28 weeks, there were no statistically significant differences in either group of infants. In addition to the decrease in the number of significantly different taxa from 12 at 6 weeks and 28 weeks to 2 at 62 weeks, we also observed a decrease in the variability of the HEU versus HUU fold changes obtained from the DESeq analyses from both 6 weeks and 28 weeks relative to 62 weeks. The variance of the log2 fold change estimates for all 224 filtered taxa was significantly greater at 6 weeks (P value = 0.01) and at 28 weeks (P value = 0.02) than at 62 weeks, further indicating more similarity in the gut microbiome between HEU and HUU over time.
Longitudinal changes in alpha diversity in HEU and HUU gut microbiomes. Richness increased significantly from 6 to 28 weeks in HEU (FDR = 0.02) and HUU (FDR = 0.002); evenness and Shannon diversity increased significantly only in HUU (FDR P values of 0.005 and 0.003, respectively). Download FIG S1, PDF file, 0.1 MB (83.3KB, pdf) .
Copyright © 2022 Jackson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Maternal gut microbiome.
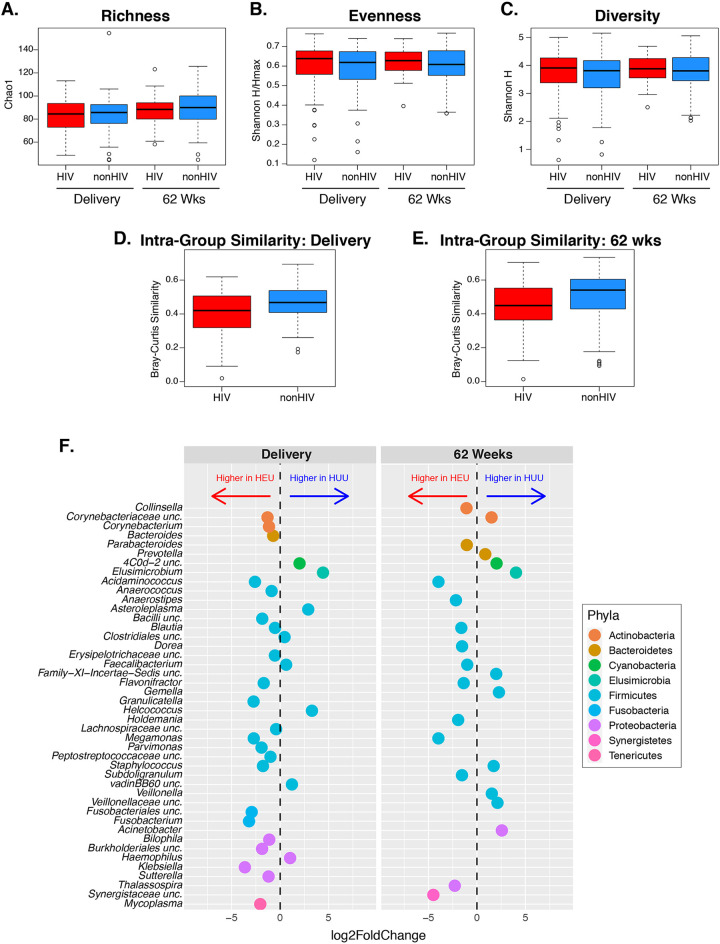
To determine the extent to which the maternal gut microbiome was responsible for differences in the HEU and HUU gut microbiomes, we analyzed the maternal gut microbiota at delivery and 62 weeks postpartum. Alpha diversity of the maternal gut microbiome was similar in mothers with and without HIV at both time points (Fig. 3A to C). However, we found a significant difference in beta diversity between mothers with and without HIV at delivery and 62 weeks postpartum with regard to the microbial composition in the two groups (P values of 0.02 and <0.01, respectively) (Fig. S2). To further evaluate the community composition, we assessed interindividual similarity within groups and found that mothers with HIV were less similar to each other when compared to mothers without HIV at either time point, with this difference being statistically significant at delivery (P = 0.03) (Fig. 3D) but not at 62 weeks (P = 0.06) (Fig. 3E). Finally, the intraindividual similarity for each mother between delivery and postpartum was not different between groups.
FIG 3.
Maternal gut microbiome. (A to C) Alpha diversity analysis of community richness (Chao1), evenness (Shannon H/Hmax), and diversity (Shannon H). There were no significant differences between mothers with and without HIV. (D and E) Beta diversity showing significantly less similarity (higher diversity) among mothers with HIV than among mothers without HIV at delivery (FDR P = 0.03) but not postpartum (FDR P = 0.06). (F) Genera with significantly different relative abundances between mothers with and without HIV at the indicated time points (FDR P < 0.05).
Maternal gut microbiome composition. Bray-Curtis analysis showed significant differences at delivery (FDR = 0.02) and at 62 weeks postpartum (FDR < 0.01). Download FIG S2, PDF file, 0.4 MB (388.1KB, pdf) .
Copyright © 2022 Jackson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
At delivery, there were 30 taxa significantly different between mothers with and without HIV, 22 of which were significantly more abundant in mothers with HIV (Fig. 3F). Sixteen families differed between mothers with and without HIV, including a higher abundance of Enterobacteriaceae and Carnobacteriaceae in mothers with HIV, which mirrored the differences between HEU and HUU at 6 weeks (Table S1). Three phyla, Cyanobacteria, Elusimicrobia, and Synergistetes, differed between mothers with and without HIV (Table S1).
At 62 weeks postpartum, 23 taxa significantly differed in abundance between mothers with and without HIV, including 13 with higher abundance in mothers with HIV (Fig. 3F). Consistent differences in microbiome composition between delivery and postpartum included higher abundances of Blautia sp., Megamonas sp., Acidaminococcus sp., and Flavonifractor sp. and lower abundance of Elusimicrobium sp. in mothers with HIV. Six families and one phylum differed between mothers with and without HIV, all with higher abundance in mothers without HIV (Table S1).
To determine if maternal characteristics, including those associated with HIV infection, played a role in the structure of the maternal gut microbiota and contributed to the differences between mothers with and without HIV, we analyzed the effects of diet, BMI, and age in both groups and of CD4+ T cell counts and plasma HIV load at delivery in mothers with HIV. With respect to diet, the analysis of daily consumption of fruit, red meat, any meat, milk, and yogurt showed that mothers with HIV had significantly higher fruit consumption than mothers without HIV at 62 weeks postpartum (averages of 4.66 and 3.92 servings per week) and no other differences (Table S2). We found a significant effect of red meat consumption on overall maternal gut composition (residual permutational multivariate analysis of variance [PERMANOVA] test P value < 0.01), but no significant effect of fruit or of any other food group. BMI, which was lower in mothers with HIV (Table 1), also had a significant effect on the overall composition of the maternal gut microbiome at 62 weeks postpartum (residual PERMANOVA test P value < 0.01). Although significantly different between mothers with and without HIV, maternal age and parity did not affect the overall composition of maternal gut microbiota. CD4+ T cell counts negatively correlated with the intraindividual gut microbiome similarity from delivery to 62 weeks postpartum (P = 0.05), but not with the interindividual similarity at delivery (Fig. S3). Plasma HIV load did not have an appreciable effect on the gut microbiome.
Relationship between the gut microbiome beta diversity and HIV disease characteristics in mothers with HIV. The data show the relationship of intraindividual similarity between delivery and 62 weeks with CD4 count (A) and log HIV load (B) and the relationship between interindividual similarity at delivery and CD4 count (C) and log HIV load (D). Download FIG S3, PDF file, 0.1 MB (94.1KB, pdf) .
Copyright © 2022 Jackson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Maternal diet was measured as the number of servings per day and then calculated as relative percentage per day. Both counts of the number of servings per day and the relative percentage based on all servings are presented. P values are shown for both counts of servings and relative percentage, although only counts were used to determine statistical differences between groups (Wilcoxon test). As these 10 comparisons were planned before the study, they are presented as raw P values without adjustment for multiple comparisons. Only fruit consumption at 62 weeks postpartum differed between groups (P values = 0.04). Download Table S2, DOCX file, 0.02 MB (19.2KB, docx) .
Copyright © 2022 Jackson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Maternal breast milk.
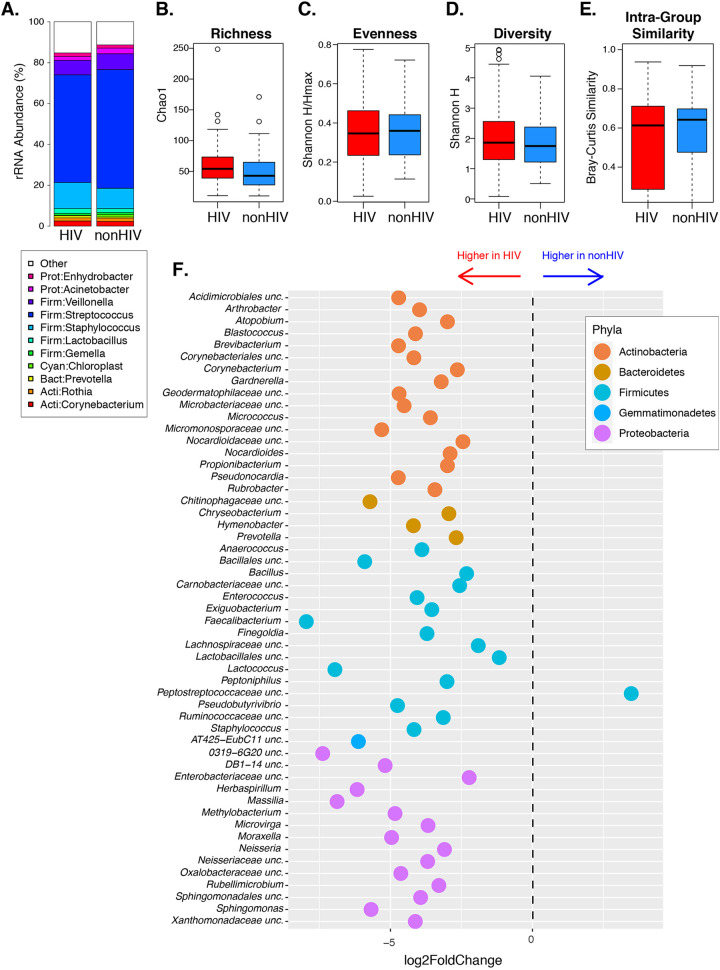
Analysis of breast milk-associated bacteria revealed that 50% of the relative abundance across all samples was attributed to Streptococcus or Staphylococcus (mean relative abundance of 55.22% and 11.41%, respectively) (Fig. 4A). The alpha and beta diversity measurements of the breast milk microbiotas were similar in mothers with and without HIV, and there was no difference in interindividual similarity between groups (Fig. 4B to E). Fifty-three taxa were differentially abundant between groups, including 52 with higher abundance in mothers with HIV. Notably, the differentially abundant taxa included the following genera that were also higher in the gut microbiotas of mothers with HIV than without HIV at delivery: Staphylococcus, Anaerococcus, and Corynebacterium. At the family level, 54 taxa were more abundant in mothers with HIV and none were less abundant compared to mothers without HIV (Table S1). Among the 54 families with differential abundance in maternal breast milk, Carnobacteriaceae and Enterococcaceae were also more abundant in HEU than HUU gut microbiomes at 6 weeks of life, but the Coriobacteriaceae were less abundant in HEU than HUU. There were no differences at the phylum level.
FIG 4.
Breast milk microbiome at 6 weeks postpartum. (A) Average relative abundance of bacterial breast milk taxa, stratified by HIV status. (B to D) Alpha diversity analysis of community richness (Chao1), evenness (Shannon H/Hmax), and diversity (Shannon H) showing no significant differences between mothers with and without HIV. (E) Beta diversity showing no differences between mothers with and without HIV. (F) Genera with significantly different abundances in mothers with and without HIV (FDR P < 0.05).
We performed additional analyses of maternal characteristics that might affect the breast milk microbiome in mothers with and without HIV. Similar to the gut microbiome, red meat consumption was significantly associated with the breast milk microbiome composition. There was no effect of BMI or parity, but there was a significant effect of age on the overall maternal breast milk composition. The analysis of the relationship between the maternal stool and breast milk microbiota composition (intraindividual similarity) did not reveal statistically significant differences between mothers with and without HIV (Fig. 5). There were no statistically significant associations of CD4+ T cell count or plasma HIV load at delivery with the breast milk microbiome composition at 6 weeks postpartum.
FIG 5.
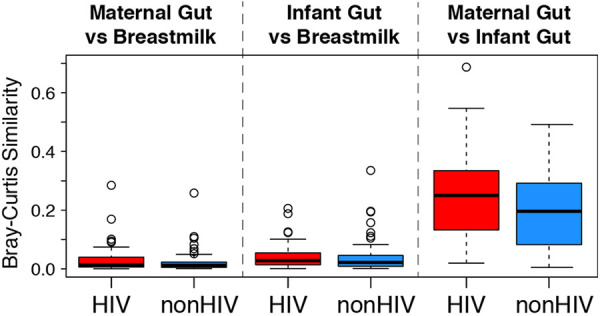
Similarities between breast milk and gut microbiomes, stratified by HIV status. Data were derived from paired analysis of Bray-Curtis similarity scores. The composition of infant gut and breast milk microbiomes at 6 weeks of life had equally little similarity in HEU and HUU mother-infant dyads, which was comparable to the compositions of maternal gut at delivery and breast milk at 6 weeks postpartum. There was equally high similarity between infant and maternal gut microbiomes at 6 weeks of life and delivery, respectively, in HEU and HUU mother-infant dyads.
Relationship of infant gut microbiota with maternal gut and breast milk microbiota.
At 6 weeks, gut microbiotas in breastfed HEU and HUU were more similar to their respective maternal gut microbiotas sampled at delivery than to maternal breast milk microbiotas (Fig. 5). Moreover, the similarities of the maternal breast milk with both maternal and infant gut microbiotas had low but equal magnitudes.
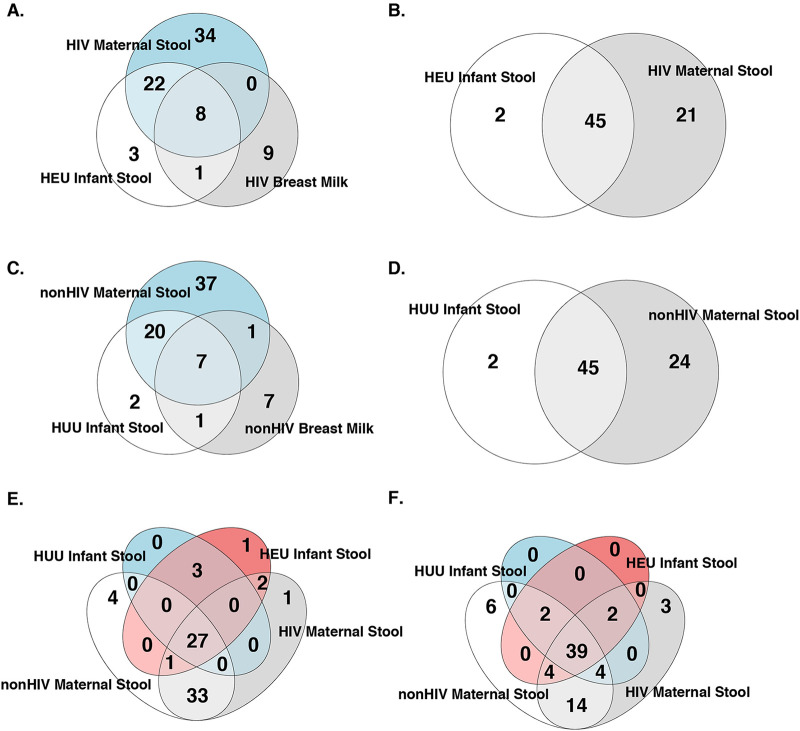
At 6 weeks, among 86 core taxa defined by 51% prevalence within a group and ≥0.01% relative abundance, 30 genera were shared between HEU and maternal gut microbiomes, including 8 genera that were also present in the breast milk microbiome. A single genus, Actinomyces, was present in HEU gut and breast milk microbiomes and was absent in the maternal gut (Fig. 6A). A similar analysis in HUU revealed 27 genera that were shared between infant and maternal gut microbiomes, including 7 also present in breast milk; the genus that was exclusively shared between HEU gut and breast milk microbiomes, Actinomyces, was also exclusively shared between infant gut and breast milk microbiomes in HUU, and a single genus, Haemophilus, was exclusively shared between maternal gut and breast milk microbiomes (Fig. 6C). Twenty-seven core taxa were shared by the gut microbiomes of HEU and HUU infants and their mothers. Two infant gut taxa (Peptostreptococcus and Fusobacterium) were exclusively shared by HEU and their mothers, but none were exclusively shared by HUU and their mothers (Fig. 6E). In addition, compared with HUU and their mothers, HEU and their mothers at delivery had higher abundances of Blautia sp. in the gut microbiome at 6 weeks and Klebsiella sp. and Megamonas sp. at 28 weeks. Genera with overlapping differential abundance in breast milk and infant gut microbiome were represented by Lactococcus sp. at 28 weeks.
FIG 6.
Venn diagrams of core bacterial taxa in infant gut, maternal gut, and breast milk microbiomes. The numerals indicate the numbers of taxa in the shared compartments. (A) HEU gut and breast milk microbiomes at 6 weeks postpartum and maternal gut microbiome at delivery. (B) HEU infant and maternal gut microbiomes at 62 weeks postpartum. (C) HUU gut and breast milk microbiomes at 6 weeks postpartum and maternal gut microbiome at delivery. (D) HUU infant and maternal gut microbiomes at 62 weeks postpartum. (E) HEU and HUU gut microbiomes at 6 weeks of life and maternal gut microbiomes at delivery. (F) HEU, HUU, and maternal gut microbiomes at 62 weeks of life.
At 62 weeks, 45 taxa were shared between HEU and HUU maternal-infant dyads (Fig. 6B and D). Thirty-nine taxa were shared by the gut microbiomes of HEU with those of HUU and with their mothers (Fig. 6F). No taxa were exclusively shared between HEU or HUU mother-infant gut microbiomes at 62 weeks postpartum.
DISCUSSION
In this study, which to our knowledge represents the largest analysis of the gut microbiome in HEU infants, we found differences in the relative abundances of multiple individual taxa between HEU and HUU infant gut microbiota, but not in alpha or beta diversity. Our results contrast with a previous report, which showed lower alpha diversity in HEU than HUU gut microbiotas in the first 4 months of life (61). Multiple differences between the two studies may explain the divergent findings, the most notable of which are the following: (i) our study had approximately 5-fold more participants than the previous study (240 versus 48); (ii) our study had a prospective design with inclusion criteria that ensured similar gestational ages at birth and modes of delivery, and homogeneous feeding habits, eliminating many of the confounders that the previous study had to contend with; and (iii) the studies were conducted in different geographic locations (Haiti versus South Africa) (61). We found only two other studies in the literature that compared the gut microbiomes in HEU and HUU. Although each study enrolled only 9 to 25 children/group at around 2 years of age, they found similar alpha and beta diversity in the two groups, supporting our results (63, 64).
As expected, both HEU and HUU groups exhibited significant increases in alpha diversity between 6 and 28 weeks of life. Rapid increases in the diversity of bacterial communities is a well-recognized characteristic of the infant gut microbiome (49, 56, 70). However, in contrast to the HUU gut microbiome, which continued to show increases in evenness and Shannon diversity between 28 and 62 weeks, no significant changes were noted in HEU. This observation was unexpected considering that a higher proportion of HEU than HUU transitioned from exclusive breastfeeding at 28 weeks of life to mixed feeding at 62 weeks. However, all HEU received co-trimoxazole prophylaxis between 28 and 62 weeks, which may have hampered the diversification of the gut microbial communities. A previous study that compared HEU with and without co-trimoxazole prophylaxis showed an increase in richness and prevalence of antibiotic resistance genes associated with the use of co-trimoxazole prophylaxis between 6 and 24 weeks of life, but no information was obtained after 24 weeks (62). Thus, the results of our study could be viewed as complementary to those of the previous study.
Differences in the abundances of specific bacterial taxa between HEU and HUU may be relevant to the immune system development and infectious vulnerability of HEU. For example, Eggerthella lenta, belonging to a genus that was higher in HEU than HUU at 6 weeks of life, has been associated with Th17 cell stimulation, increased risk of rheumatoid arthritis and ulcerative colitis, and enhanced microbial translocation (71, 72). A recent study showed evidence of higher microbial translocation in HEU than HUU in early life, which might be related to the microbiome and, possibly, to the excess of Eggerthella sp. in the HEU microbiome in early life (73). Excess abundances of Carnobacteriaceae and Lachnospiraceae, which were higher in HEU than HUU at 6 and 62 weeks, respectively, were also described in the gut microbiome of adults with HIV versus those without HIV (74). Notably, members of the family Lachnospiraceae are producers of butyrate, a short-chain fatty acids that promotes regulatory-T-cell (Treg) differentiation, as are other members of the Clostridiales, which were higher in HEU than HUU at 6 weeks of life (34, 75). Increased proportions of Treg may translate into a lower ability to clear infections. Additional research in HEU is needed to better understand relationships between specific gut microbes and the immune responses and infectious disease morbidity in this population.
It is important to note that the abundances of pathobionts, such as Klebsiella sp. and Enterobacter sp., did not differ appreciably between HEU and HUU infants either before or during co-trimoxazole prophylaxis. These findings suggest that gut pathobionts may not contribute to the increased infectious morbidity and mortality observed in HEU.
We found extensive overlap between infant and maternal gut microbiota both in HEU and HUU. However, HEU mother-infant dyads had more taxa in common than HUU dyads at 6 weeks of life, including two taxa, Peptostreptococcus sp. and Fusobacterium sp., that were exclusively shared by HEU mother-infant pairs. Notably, in this highly treated maternal population with HIV, the maternal CD4+ T cell count and HIV plasma RNA at delivery did not influence the maternal or HEU gut microbiota composition. No unique taxa were found in HUU mother-infant dyads that were not present in HEU mother-infant dyads. We found several bacterial taxa with higher abundance in HEU than in HUU maternal-infant dyads, including Blautia, Klebsiella, and the family Lachnospiraceae, while Gemella spp. were less abundant. Other taxa, such as Clostridiales, Megamonas, and Parvimonas, manifested divergent relationships between HEU and HUU compared to mothers with and without HIV. The mechanism underlying the higher similarity between gut microbiota of HEU mother-infant dyads at 6 weeks compared with HUU dyads as well as the biologic relevance of the taxa shared by one group but not the other need to be further investigated, as they may offer clues to HEU-associated immunopathogenesis.
There were more significant differences in the gut microbiome structure between mothers with and without HIV than between HEU and HUU. Significant differences in mothers included higher beta diversity at delivery and 62 weeks postpartum and higher dissimilarity among mothers with HIV than among mothers without HIV. Importantly, although considerable dissimilarity in gut microbiotas were evident between mothers with and without HIV both at delivery and postpartum, the dissimilarity between HEU and HUU gut microbiotas decreased over time. Notably, the excessive morbidity and mortality of infections in HEU also decreases over time, establishing a temporal association with the maturation of the gut microbiome. It will be important to determine in future studies the characteristics of the HEU gut microbiome that are most closely associated with improved protection against infection.
Breast milk plays an important role in shaping the infant gut microbiome. However, at 6 weeks postpartum, we found very little overlap between breast milk and infant gut microbial composition in exclusively breastfed HEU or HUU. In both HEU and HUU, only one genus, Actinomyces, was shared between breast milk and infant gut microbiomes but was not also present in the maternal gut microbiome. Actinomyces are common commensals found on mucosae and skin and may have been skin contaminants of breast milk. All other 7 or 8 shared taxa were also present in the maternal gut. These findings suggest that the effect of the breast milk on the infant gut microbiota is mediated by its biochemical and immunologic properties, which favor the development of specific microbial taxa in the infant gut rather than by seeding the infant gut with breast milk bacteria. Although it was proposed that the oligosaccharide composition differed in mothers with and without HIV, exclusive breastfeeding was a unifying factor for the gut microbiota in HEU and HUU, suggesting that differences in oligosaccharide composition may not impact the gut microbiome of HEU versus HUU (76).
Although the primary objective of this study was to compare the HEU and HUU gut microbiomes in the first year of life, the investigation of the maternal gut and milk microbiomes as they related to the infant gut microbiomes revealed notable differences between mothers with and without HIV that are worth emphasizing. The gut microbiomes of mothers with HIV were less similar to one another among members of the group than within groups of mothers without HIV. We hypothesized that mothers with HIV as a group had more diverse immunologic conditions than mothers without HIV, which reduced the constraints normally operating on the gastrointestinal microbiome and thereby expanded person-to-person variation. Since the relationship between the gut microbiome and the host immune system is bidirectional, we thought that immunologic and microbiome dissimilarities within this group might be correlated. We did not find an association between maternal CD4+ T cell counts and microbiomes in mothers with HIV. Although this finding does not support our hypothesis, it does not completely refute it either, because the CD4+ T cell composition in the intestinal mucosa and/or secondary lymphoid organs may be the key determinant of the gut microbiome, and these parameters cannot be assessed by studying exclusively the peripheral blood. In addition, we did not collect full maternal medical histories, which may have contained important explanatory information, such as occurrence of gastrointestinal infections, nadir CD4+ T cell counts in the peripheral blood, or signs of inflammation. The breast milk microbiotas differed between mothers with and without HIV, including an excessive number of taxa with higher abundance in mothers with HIV than in mothers without HIV. Although we did not find any associations of the breast milk microbiome and HIV disease characteristics during the study, differences in immune responses to commensal bacteria between women with HIV and women without HIV might have contributed to the differential abundance of specific taxa. Collectively, these findings indicate that additional studies are needed to fully define the parameters that affect the gut and breast milk microbiomes in mothers with and without HIV.
A limitation of this study was the loss to follow-up among HEU and HUU mother-infant dyads. The use of co-trimoxazole in HEU was an unavoidable confounder. Strengths included the large number of participants enrolled in the study and the excellent quality of the samples that were collected.
We conclude that the gut microbiomes of HEU and HUU are heavily influenced by the maternal gut microbiome, whereas breast milk-associated microbes do not play an apparent, differentiating role. The microbiotas of HEU and HUU converged over time, mirroring the decrease in the excess of infectious morbidity and mortality in HEU. Thus, the immunologic effects of taxa that differentiate the HEU from HUU gut microbiomes in early life deserve further investigation, because to the extent to which they are related to excess infectious morbidity in HEU, they may uncover potential strategies to decrease the vulnerability of HEU to infectious complications in early life.
MATERIALS AND METHODS
Study design.
Women with and without HIV were recruited during labor at Chris Hani Baragwanath Hospital in Johannesburg, South Africa. Inclusion criteria for all women were singleton term gestation, planned vaginal delivery, and intent to breastfeed. Women with HIV had to have been prescribed antiretrovirals but not co-trimoxazole during pregnancy. After written informed consent was given, maternal and infant metadata were collected from medical records and by interviewing the study participants at study visits 6, 28, and 62 weeks postpartum. Maternal blood was obtained at delivery for CD4+ T cells and HIV plasma RNA measurements at the local laboratory. Infant rectal swabs were obtained at 6, 28, and 62 weeks of life, and maternal rectal swabs were collected at delivery and 62 weeks postpartum for microbiome analysis. Breast milk collected at 6 weeks postpartum from lactating mothers was also submitted to microbiome analysis.
Microbiome analyses.
DNA was extracted from rectal swabs and breast milk using the QIAamp Powerfecal DNA isolation kit (Qiagen INC, Hilden, Germany). Bacterial profiles were determined by broad-range PCR amplification and sequence analysis of 16S rRNA genes following our previously described methods (77, 78). In brief, amplicons were generated using barcoded primers that target the V3V4 variable region of the 16S rRNA gene: primers 338F (5′-ACTCCTACGGGAGGCAGCAG) and 806R (5′-GGACTACHVGGGTWTCTAAT). PCR products were normalized using a SequalPrep kit (Invitrogen, Carlsbad, CA), pooled, lyophilized, purified, and concentrated using a DNA Clean & Concentrator kit (Zymo, Irvine, CA). Pooled amplicons were quantified using a Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA). The pool was diluted to 4 nM and denatured with 0.2 N NaOH at room temperature. The denatured DNA was diluted to 15 pM and spiked with 25% Illumina PhiX control DNA prior to loading the sequencer. Illumina paired-end sequencing was performed on the MiSeq platform using a 600-cycle version 3 reagent kit.
Paired-end reads were aligned to human reference genome hg19 with bowtie2, and matching sequences were discarded (79, 80). Demultiplexed paired reads were assembled using Phrap (81, 82) and pairs that did not assemble were discarded. Assembled sequences were trimmed over a moving window of 5 nucleotides until average quality met or exceeded 20. Trimmed sequences with more than one ambiguity or shorter than 350 nucleotides were discarded. Potential chimeras identified with Uchime (usearch6.0.203_i86linux32) (83) using the Schloss (84) Silva reference sequences were removed from subsequent analyses. Assembled sequences were aligned and classified with SINA (1.3.0-r23838) (85) using the 418,497 bacterial sequences in Silva 115NR99 (86) as a reference configured to yield the Silva taxonomy. Taxonomic annotations were based on the default lowest common ancestor parameters used by Silva. Closed-reference operational taxonomic units (OTU) were produced by binning sequences with identical taxonomic assignments. For the gut microbiome, this process generated 89,921,190 sequences for 795 samples (median, 106,528 sequences/sample; interquartile range [IQR], 83,402 to 126,470), and the median Good’s coverage score was ≥99.8%. For the breast milk microbiome, this process generated 11,308,330 sequences for 164 samples (median, 76,650 sequences/sample; IQR, 42,540 to 94,492), and the median Good’s coverage score was ≥99.5%. The software package Explicet (v2.10.5; www.explicet.org) (87) was used to calculate rarefied alpha diversity indices.
Statistics.
Microbiome analyses were conducted largely within the framework of the microbiome package (version 1.12.0) (88) in R (version 4.0.2) (89), which utilizes both phyloseq (version 1.34.0) (90) and vegan (version 2.5-7) (91). Prior to analysis, the OTU were filtered based on prevalence (at least 5% of libraries are represented) and relative abundance (at least one library must have a relative abundance of 0.01%). Analyses for differences in abundances were conducted using DESeq2 (92), and all significantly different OTU were determined based on a false-discovery-rate (FDR)-adjusted level of 0.05. To determine how the variability of the estimates from DESeq2 changed over time, we followed a multistep procedure: (i) the log2 fold change estimates were extracted from each time point for all taxa; (ii) Bartlett’s test was used to test for homogeneity of variances; (iii) an F test was conducted in a pairwise fashion between the three time points if the result of step 2 found a difference in variances. For assessing differences in alpha diversity among two-group comparisons, either Welch’s t test or a Wilcoxon test was used as appropriate based on the distribution of the data. Longitudinal alpha diversity measurements were assessed by first calculating the difference in alpha diversity between time points and then fitting a linear model with group as a covariate to determine how each group changed over time. To adjust for breastfeeding, a linear model without a group variable was fit, and then the residuals from that model were used in a regression as the outcome with group as a covariate. A variety of methods were used for the assessment of beta diversity. Overall PERMANOVA were conducted using the Bray-Curtis dissimilarity index and the adonis function (1,000 permutations) within the vegan package (91), followed by pairwise FDR tests if appropriate. To measure both intra- and interindividual similarity, the divergence function within the microbiome package (88) was used with the methodology for beta diversity analyses as described previously (93), with the exception of reporting the similarity instead of dissimilarity by subtracting the calculated divergence from 1.
To assess the global effect of covariates such as diet and BMI on the microbiome, a three-step process was used. First, a logistic regression with group (HIV versus non-HIV) as the outcome and the variable of interest was fitted individually for each variable (red meat, fruit, BMI, etc.), and the residuals were extracted. Next, a matrix of residuals from the DESeq2 analyses for each OTU was extracted. Finally, a PERMANOVA with the adonis function was used with the matrix of the residuals from step 2 on the left side of the equation and the residuals from the model in step 1 on the right side of the equation. The final PERMANOVA yields a P value for the global effect of that variable on the microbiome, although further interpretation of this result is not possible from this output alone. Correlations between HIV data and beta diversity used the Spearman rank correlation test to account for the nonparametric nature of the data. Shared core taxa were determined using the eulerr package (version 6.1.1) (94) with the method described on the microbiome package github (88). A prevalence of 51% and a detection limit of 0.0001 were used to match the OTU filtering conducted during the processing of the data. Code for all analyses is available upon request.
Study approval.
The study was approved by the Human Research Ethics Committee at the University of the Witwatersrand (approval number M171185) and the Colorado Multiple Institutions Review Board. Written informed consent was received prior to participation in the study.
Data availability.
All sequences and corresponding metadata were deposited in the NCBI Sequence Read Archive under BioProject accession number PRJNA816484.
ACKNOWLEDGMENT
This study was funded by the National Institute of Allergy and Infectious Diseases, U01AI131360-01 (AW).
Contributor Information
Adriana Weinberg, Email: Adriana.Weinberg@ucdenver.edu.
Ilhem Messaoudi, University of Kentucky.
REFERENCES
- 1.Epalza C, Goetghebuer T, Hainaut M, Prayez F, Barlow P, Dediste A, Marchant A, Levy J. 2010. High incidence of invasive group B streptococcal infections in HIV-exposed uninfected infants. Pediatrics 126:e631–e638. doi: 10.1542/peds.2010-0183. [DOI] [PubMed] [Google Scholar]
- 2.Izadnegahdar R, Fox MP, Jeena P, Qazi SA, Thea DM. 2014. Revisiting pneumonia and exposure status in infants born to HIV-infected mothers. Pediatr Infect Dis J 33:70–72. doi: 10.1097/INF.0b013e31829f0ade. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koyanagi A, Humphrey JH, Ntozini R, Nathoo K, Moulton LH, Iliff P, Mutasa K, Ruff A, Ward B. 2011. Morbidity among human immunodeficiency virus-exposed but uninfected, human immunodeficiency virus-infected, and human immunodeficiency virus-unexposed infants in Zimbabwe before availability of highly active antiretroviral therapy. Pediatr Infect Dis J 30:45–51. doi: 10.1097/INF.0b013e3181ecbf7e. [DOI] [PubMed] [Google Scholar]
- 4.Kuhn L, Kasonde P, Sinkala M, Kankasa C, Semrau K, Scott N, Tsai WY, Vermund SH, Aldrovandi GM, Thea DM. 2005. Does severity of HIV disease in HIV-infected mothers affect mortality and morbidity among their uninfected infants? Clin Infect Dis 41:1654–1661. doi: 10.1086/498029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McNally LM, Jeena PM, Gajee K, Thula SA, Sturm AW, Cassol S, Tomkins AM, Coovadia HM, Goldblatt D. 2007. Effect of age, polymicrobial disease, and maternal HIV status on treatment response and cause of severe pneumonia in South African children: a prospective descriptive study. Lancet 369:1440–1451. doi: 10.1016/S0140-6736(07)60670-9. [DOI] [PubMed] [Google Scholar]
- 6.Mussi-Pinhata MM, Freimanis L, Yamamoto AY, Korelitz J, Pinto JA, Cruz ML, Losso MH, Read JS, National Institute of Child Health and Human Development International Site Development Initiative Perinatal Study Group . 2007. Infectious disease morbidity among young HIV-1-exposed but uninfected infants in Latin American and Caribbean countries: the National Institute of Child Health and Human Development International Site Development Initiative Perinatal Study. Pediatrics 119:e694-704–e704. doi: 10.1542/peds.2006-1856. [DOI] [PubMed] [Google Scholar]
- 7.Mussi-Pinhata MM, Motta F, Freimanis-Hance L, de Souza R, Szyld E, Succi RC, Christie CD, Rolon MJ, Ceriotto M, Read JS. 2010. Lower respiratory tract infections among human immunodeficiency virus-exposed, uninfected infants. Int J Infect Dis 14(Suppl 3):e176–E182. doi: 10.1016/j.ijid.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shapiro RL, Lockman S. 2010. Mortality among HIV-exposed infants: the first and final frontier. Clin Infect Dis 50:445–447. doi: 10.1086/649887. [DOI] [PubMed] [Google Scholar]
- 9.Slogrove A, Reikie B, Naidoo S, De Beer C, Ho K, Cotton M, Bettinger J, Speert D, Esser M, Kollmann T. 2012. HIV-exposed uninfected infants are at increased risk for severe infections in the first year of life. J Trop Pediatr 58:505–508. doi: 10.1093/tropej/fms019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slogrove AL, Cotton MF, Esser MM. 2010. Severe infections in HIV-exposed uninfected infants: clinical evidence of immunodeficiency. J Trop Pediatr 56:75–81. doi: 10.1093/tropej/fmp057. [DOI] [PubMed] [Google Scholar]
- 11.Kourtis AP, Wiener J, Kayira D, Chasela C, Ellington SR, Hyde L, Hosseinipour M, van der Horst C, Jamieson DJ. 2013. Health outcomes of HIV-exposed uninfected African infants. AIDS 27:749–759. doi: 10.1097/QAD.0b013e32835ca29f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taron-Brocard C, Le Chenadec J, Faye A, Dollfus C, Goetghebuer T, Gajdos V, Labaune JM, Perilhou A, Mandelbrot L, Blanche S, Warszawski J, France REcherche Nord&Sud Sida-HIV Hepatites–Enquete Perinatale Francaise–CP1/CO11 Study Group . 2014. Increased risk of serious bacterial infections due to maternal immunosuppression in HIV-exposed uninfected infants in a European country. Clin Infect Dis 59:1332–1445. doi: 10.1093/cid/ciu586. [DOI] [PubMed] [Google Scholar]
- 13.Kelly MS, Wirth KE, Steenhoff A, Cunningham C, Arscott-Mills T, Boiditswe S, Patel M, Shah S, Finalle R, Makone I, Feemster K. 2014. Treatment failures and excess mortality among HIV-exposed, uninfected children with pneumonia. J Pediatric Infect Dis Soc 4:e117–e126. doi: 10.1093/jpids/piu092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Mollendorf C, von Gottberg A, Tempia S, Meiring S, de Gouveia L, Quan V, Lengana S, Avenant T, du Plessis N, Eley B, Finlayson H, Reubenson G, Moshe M, O’Brien KL, Klugman KP, Whitney CG, Cohen C, Group for Enteric, Respiratory and Meningeal Disease Surveillance in South A . 2015. Increased risk for and mortality from invasive pneumococcal disease in HIV-exposed but uninfected infants aged <1 year in South Africa, 2009-2013. Clin Infect Dis 60:1346–1356. doi: 10.1093/cid/civ059. [DOI] [PubMed] [Google Scholar]
- 15.Evans C, Chasekwa B, Ntozini R, Majo FD, Mutasa K, Tavengwa N, Mutasa B, Mbuya MNN, Smith LE, Stoltzfus RJ, Moulton LH, Humphrey JH, Prendergast AJ, Sanitation Hygiene Infant Nutrition Efficacy (SHINE) Trial Team . 2021. Mortality, human immunodeficiency virus (HIV) transmission, and growth in children exposed to HIV in rural Zimbabwe. Clin Infect Dis 72:586–594. doi: 10.1093/cid/ciaa076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Labuda SM, Huo Y, Kacanek D, Patel K, Huybrechts K, Jao J, Smith C, Hernandez-Diaz S, Scott G, Burchett S, Kakkar F, Chadwick EG, Van Dyke RB, Pediatric HIV/AIDS Cohort Study . 2019. Rates of hospitalization and infection-related hospitalization among HIV-exposed uninfected children compared to HIV-unexposed uninfected children in the United States, 2007-2016. Clin Infect Dis 71:332–339. doi: 10.1093/cid/ciz820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson K, Kalk E, Madlala HP, Nyemba DC, Kassanjee R, Jacob N, Slogrove A, Smith M, Eley BS, Cotton MF, Muloiwa R, Spittal G, Kroon M, Boulle A, Myer L, Davies MA. 2021. Increased infectious-cause hospitalization among infants who are HIV-exposed uninfected compared with HIV-unexposed. AIDS 35:2327–2339. doi: 10.1097/QAD.0000000000003039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li SNJ, Albert A, Piske M, Janssen PA, Alimenti A, Jesson J, Côté HCF, Sauvé L. 2022. Higher hospitalization rates in children born HIV-exposed uninfected in British Columbia, Canada, between 1990 and 2012. Pediatr Infect Dis J 41:124–130. doi: 10.1097/INF.0000000000003365. [DOI] [PubMed] [Google Scholar]
- 19.Jalbert E, Williamson KM, Kroehl ME, Johnson MJ, Cutland C, Madhi SA, Nunes MC, Weinberg A. 2019. HIV-exposed uninfected infants have increased regulatory T cells that correlate with decreased T cell function. Front Immunol 10:595. doi: 10.3389/fimmu.2019.00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith C, Jalbert E, de Almeida V, Canniff J, Lenz LL, Mussi-Pinhata MM, Cohen RA, Yu Q, Amaral FR, Pinto J, Alarcon JO, Siberry G, Weinberg A. 2017. Altered natural killer cell runction in HIV-exposed uninfected Infants. Front Immunol 8:470. doi: 10.3389/fimmu.2017.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen C, Moyes J, Tempia S, Groome M, Walaza S, Pretorius M, Naby F, Mekgoe O, Kahn K, von Gottberg A, Wolter N, Cohen AL, von Mollendorf C, Venter M, Madhi SA. 2016. Epidemiology of acute lower respiratory tract infection in HIV-exposed uninfected infants. Pediatrics 137 doi:10.1542/peds.2015-3272. doi: 10.1542/peds.2015-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weinberg A, Mussi-Pinhata M, Yu Q, et al. 2016. Severe respiratory infections in HIV-exposed uninfected infants: serologic analysis, abstr ••••. CROI 2016, Feb 22–25, 2016, Boston, MA.
- 23.Cutland CL, Schrag SJ, Thigpen MC, Velaphi SC, Wadula J, Adrian PV, Kuwanda L, Groome MJ, Buchmann E, Madhi SA. 2015. Increased risk for group B Streptococcus sepsis in young infants exposed to HIV, Soweto, South Africa, 2004-2008. Emerg Infect Dis 21:638–645. doi: 10.3201/eid2104.141562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moyes J, Cohen C, Pretorius M, Groome M, von Gottberg A, Wolter N, Walaza S, Haffejee S, Chhagan M, Naby F, Cohen AL, Tempia S, Kahn K, Dawood H, Venter M, Madhi SA, South African Severe Acute Respiratory Illness Surveillance Group . 2013. Epidemiology of respiratory syncytial virus-associated acute lower respiratory tract infection hospitalizations among HIV-infected and HIV-uninfected South African children, 2010-2011. J Infect Dis 208(Suppl 3):S217–S226. doi: 10.1093/infdis/jit479. [DOI] [PubMed] [Google Scholar]
- 25.Weinberg A, Mussi-Pinhata MM, Yu Q, Cohen RA, Almeida VC, Amaral F, Pinto J, Teixeira ML, Succi RC, Freimanis L, Read JS, Siberry G, Nisdi Perinatal LCP . 2017. Excess respiratory viral infections and low antibody responses among HIV-exposed, uninfected infants. AIDS 31:669–679. doi: 10.1097/QAD.0000000000001393. [DOI] [PubMed] [Google Scholar]
- 26.Dangor Z, Kwatra G, Izu A, Adrian P, van Niekerk N, Cutland CL, Adam Y, Velaphi S, Lala SG, Madhi SA. 2015. HIV-1 is associated with lower group B Streptococcus capsular and surface-protein IgG antibody levels and reduced transplacental antibody transfer in pregnant women. J Infect Dis 212:453–462. doi: 10.1093/infdis/jiv064. [DOI] [PubMed] [Google Scholar]
- 27.Weinberg A, Mussi-Pinhata MM, Yu Q, Cohen RA, Almeida VC, Amaral FR, Freimanis L, Harris DR, Smith C, Siberry G. 2018. Factors associated with lower respiratory tract infections in HIV-exposed uninfected infants. AIDS Res Hum Retroviruses 34:527–535. doi: 10.1089/AID.2017.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abu-Raya B, Kollmann TR, Marchant A, MacGillivray DM. 2016. The Immune System of HIV-Exposed Uninfected Infants. Front Immunol 7:383. doi: 10.3389/fimmu.2016.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kidzeru EB, Hesseling AC, Passmore JA, Myer L, Gamieldien H, Tchakoute CT, Gray CM, Sodora DL, Jaspan HB. 2014. In-utero exposure to maternal HIV infection alters T-cell immune responses to vaccination in HIV-uninfected infants. AIDS 28:1421–1430. doi: 10.1097/QAD.0000000000000292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith C, Moraka NO, Ibrahim M, Moyo S, Mayondi G, Kammerer B, Leidner J, Gaseitsiwe S, Li S, Shapiro R, Lockman S, Weinberg A. 2020. Human immunodeficiency virus exposure but not early cytomegalovirus infection is associated with increased hospitalization and decreased memory T-cell responses to tetanus vaccine. J Infect Dis 221:1167–1175. doi: 10.1093/infdis/jiz590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ganusov VV, De Boer RJ. 2007. Do most lymphocytes in humans really reside in the gut? Trends Immunol 28:514–518. doi: 10.1016/j.it.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 32.Ma H, Tao W, Zhu S. 2019. T lymphocytes in the intestinal mucosa: defense and tolerance. Cell Mol Immunol 16:216–224. doi: 10.1038/s41423-019-0208-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levy M, Thaiss CA, Elinav E. 2015. Metagenomic cross-talk: the regulatory interplay between immunogenomics and the microbiome. Genome Med 7:120. doi: 10.1186/s13073-015-0249-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skelly AN, Sato Y, Kearney S, Honda K. 2019. Mining the microbiota for microbial and metabolite-based immunotherapies. Nat Rev Immunol 19:305–323. doi: 10.1038/s41577-019-0144-5. [DOI] [PubMed] [Google Scholar]
- 35.Correa-Oliveira R, Fachi JL, Vieira A, Sato FT, Vinolo MA. 2016. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunol 5:e73. doi: 10.1038/cti.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Constantinides MG, Link VM, Tamoutounour S, Wong AC, Perez-Chaparro PJ, Han SJ, Chen YE, Li K, Farhat S, Weckel A, Krishnamurthy SR, Vujkovic-Cvijin I, Linehan JL, Bouladoux N, Merrill ED, Roy S, Cua DJ, Adams EJ, Bhandoola A, Scharschmidt TC, Aube J, Fischbach MA, Belkaid Y. 2019. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science 366:eaax6624. doi: 10.1126/science.aax6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Legoux F, Bellet D, Daviaud C, El Morr Y, Darbois A, Niort K, Procopio E, Salou M, Gilet J, Ryffel B, Balvay A, Foussier A, Sarkis M, El Marjou A, Schmidt F, Rabot S, Lantz O. 2019. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science 366:494–499. doi: 10.1126/science.aaw2719. [DOI] [PubMed] [Google Scholar]
- 38.Ravens S, Fichtner AS, Willers M, Torkornoo D, Pirr S, Schoning J, Deseke M, Sandrock I, Bubke A, Wilharm A, Dodoo D, Egyir B, Flanagan KL, Steinbruck L, Dickinson P, Ghazal P, Adu B, Viemann D, Prinz I. 2020. Microbial exposure drives polyclonal expansion of innate gammadelta T cells immediately after birth. Proc Natl Acad Sci USA 117:18649–18660. doi: 10.1073/pnas.1922588117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barbi J, Pardoll D, Pan F. 2014. Treg functional stability and its responsiveness to the microenvironment. Immunol Rev 259:115–139. doi: 10.1111/imr.12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanoue T, Atarashi K, Honda K. 2016. Development and maintenance of intestinal regulatory T cells. Nat Rev Immunol 16:295–309. doi: 10.1038/nri.2016.36. [DOI] [PubMed] [Google Scholar]
- 41.Torow N, Yu K, Hassani K, Freitag J, Schulz O, Basic M, Brennecke A, Sparwasser T, Wagner N, Bleich A, Lochner M, Weiss S, Forster R, Pabst O, Hornef MW. 2015. Active suppression of intestinal CD4+TCRαβ+ T-lymphocyte maturation during the postnatal period. Nat Commun 6:7725. doi: 10.1038/ncomms8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brumbaugh DE, Arruda J, Robbins K, Ir D, Santorico SA, Robertson CE, Frank DN. 2016. Mode of delivery determines neonatal pharyngeal bacterial composition and early intestinal colonization. J Pediatr Gastroenterol Nutr 63:320–328. doi: 10.1097/MPG.0000000000001124. [DOI] [PubMed] [Google Scholar]
- 43.Krebs NF, Sherlock LG, Westcott J, Culbertson D, Hambidge KM, Feazel LM, Robertson CE, Frank DN. 2013. Effects of different complementary feeding regimens on iron status and enteric microbiota in breastfed infants. J Pediatr 163:416–423. doi: 10.1016/j.jpeds.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lemas DJ, Young BE, Baker PR, II, Tomczik AC, Soderborg TK, Hernandez TL, de la Houssaye BA, Robertson CE, Rudolph MC, Ir D, Patinkin ZW, Krebs NF, Santorico SA, Weir T, Barbour LA, Frank DN, Friedman JE. 2016. Alterations in human milk leptin and insulin are associated with early changes in the infant intestinal microbiome. Am J Clin Nutr 103:1291–1300. doi: 10.3945/ajcn.115.126375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soderborg TK, Carpenter CM, Janssen RC, Weir TL, Robertson CE, Ir D, Young BE, Krebs NF, Hernandez TL, Barbour LA, Frank DN, Kroehl M, Friedman JE. 2020. Gestational diabetes is uniquely associated with altered early seeding of the infant gut microbiota. Front Endocrinol (Lausanne) 11:603021. doi: 10.3389/fendo.2020.603021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soderborg TK, Clark SE, Mulligan CE, Janssen RC, Babcock L, Ir D, Young B, Krebs N, Lemas DJ, Johnson LK, Weir T, Lenz LL, Frank DN, Hernandez TL, Kuhn KA, D’Alessandro A, Barbour LA, El Kasmi KC, Friedman JE. 2018. The gut microbiota in infants of obese mothers increases inflammation and susceptibility to NAFLD. Nat Commun 9:4462. doi: 10.1038/s41467-018-06929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang M, Frank DN, Tshefu A, Lokangaka A, Goudar SS, Dhaded SM, Somannavar MS, Hendricks AE, Ir D, Robertson CE, Kemp JF, Lander RL, Westcott JE, Hambidge KM, Krebs NF. 2019. Different gut microbial profiles in sub-Saharan African and South Asian women of childbearing age are primarily associated with dietary intakes. Front Microbiol 10:1848. doi: 10.3389/fmicb.2019.01848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang M, Matz KL, Berman LM, Davis KN, Melanson EL, Frank DN, Hendricks AE, Krebs NF. 2021. Effects of complementary feeding with different protein-rich foods on infant growth and gut health: study protocol. Front Pediatr 9:793215. doi: 10.3389/fped.2021.793215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, D Lieber A, Wu F, Perez-Perez GI, Chen Y, Schweizer W, Zheng X, Contreras M, Dominguez-Bello MG, Blaser MJ. 2016. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med 8:343ra82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laforest-Lapointe I, Arrieta MC. 2017. Patterns of early-life gut microbial colonization during human immune development: an ecological perspective. Front Immunol 8:788. doi: 10.3389/fimmu.2017.00788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, Al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Tremaroli V, Yin Y, Bergman S, Xu X, Madsen L, Kristiansen K, Dahlgren J, Wang J. 2015. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17:852. doi: 10.1016/j.chom.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 52.Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. 2017. Dysbiosis and the immune system. Nat Rev Immunol 17:219–232. doi: 10.1038/nri.2017.7. [DOI] [PubMed] [Google Scholar]
- 53.Mao K, Baptista AP, Tamoutounour S, Zhuang L, Bouladoux N, Martins AJ, Huang Y, Gerner MY, Belkaid Y, Germain RN. 2018. Innate and adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. Nature 554:255–259. doi: 10.1038/nature25437. [DOI] [PubMed] [Google Scholar]
- 54.Nash MJ, Frank DN, Friedman JE. 2017. Early microbes modify immune system development and metabolic homeostasis—the “restaurant” hypothesis revisited. Front Endocrinol (Lausanne) 8:349. doi: 10.3389/fendo.2017.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neu J. 2016. The microbiome during pregnancy and early postnatal life. Semin Fetal Neonatal Med 21:373–379. doi: 10.1016/j.siny.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 56.Palmer C, Bik EM, Digiulio DB, Relman DA, Brown PO. 2007. Development of the human infant intestinal microbiota. PLoS Biol 5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reyman M, van Houten MA, van Baarle D, Bosch A, Man WH, Chu M, Arp K, Watson RL, Sanders EAM, Fuentes S, Bogaert D. 2019. Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat Commun 10:4997. doi: 10.1038/s41467-019-13014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zimmermann P, Curtis N. 2018. Factors influencing the intestinal microbiome during the first year of life. Pediatr Infect Dis J 37:e315–e335. doi: 10.1097/INF.0000000000002103. [DOI] [PubMed] [Google Scholar]
- 59.Robertson RC, Manges AR, Finlay BB, Prendergast AJ. 2019. The human microbiome and child growth—first 1000 days and beyond. Trends Microbiol 27:131–147. doi: 10.1016/j.tim.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 60.Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, Benezra A, DeStefano J, Meier MF, Muegge BD, Barratt MJ, VanArendonk LG, Zhang Q, Province MA, Petri WA, Jr, Ahmed T, Gordon JI. 2014. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 510:417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bender JM, Li F, Martelly S, Byrt E, Rouzier V, Leo M, Tobin N, Pannaraj PS, Adisetiyo H, Rollie A, Santiskulvong C, Wang S, Autran C, Bode L, Fitzgerald D, Kuhn L, Aldrovandi GM. 2016. Maternal HIV infection influences the microbiome of HIV-uninfected infants. Sci Transl Med 8:349ra100. doi: 10.1126/scitranslmed.aaf5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.D’Souza AW, Moodley-Govender E, Berla B, Kelkar T, Wang B, Sun X, Daniels B, Coutsoudis A, Trehan I, Dantas G. 2020. Cotrimoxazole prophylaxis increases resistance gene prevalence and alpha diversity but decreases beta diversity in the gut microbiome of human immunodeficiency virus-exposed, uninfected infants. Clin Infect Dis 71:2858–2868. doi: 10.1093/cid/ciz1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amenyogbe N, Dimitriu P, Cho P, Ruck C, Fortuno ES, 3rd, Cai B, Alimenti A, Côté HCF, Maan EJ, Slogrove AL, Esser M, Marchant A, Goetghebuer T, Shannon CP, Tebbutt SJ, Kollmann TR, Mohn WW, Smolen KK. 2020. Innate immune responses and gut microbiomes distinguish HIV-exposed from HIV-unexposed children in a population-specific manner. J Immunol 205:2618–2628. doi: 10.4049/jimmunol.2000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Machiavelli A, Duarte RTD, Pires M, Zárate-Bladés CR, Pinto AR. 2019. The impact of in utero HIV exposure on gut microbiota, inflammation, and microbial translocation. Gut Microbes 10:599–614. doi: 10.1080/19490976.2018.1560768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, Armanini F, Truong DT, Manara S, Zolfo M, Beghini F, Bertorelli R, De Sanctis V, Bariletti I, Canto R, Clementi R, Cologna M, Crifo T, Cusumano G, Gottardi S, Innamorati C, Mase C, Postai D, Savoi D, Duranti S, Lugli GA, Mancabelli L, Turroni F, Ferrario C, Milani C, Mangifesta M, Anzalone R, Viappiani A, Yassour M, Vlamakis H, Xavier R, Collado CM, Koren O, Tateo S, Soffiati M, Pedrotti A, Ventura M, Huttenhower C, Bork P, Segata N. 2018. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 24:133–145.E5. doi: 10.1016/j.chom.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lyons KE, Ryan CA, Dempsey EM, Ross RP, Stanton C. 2020. Breast milk, a source of beneficial microbes and associated benefits for infant health. Nutrients 12:1039. doi: 10.3390/nu12041039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ruiz L, Garcia-Carral C, Rodriguez JM. 2019. Unfolding the human milk microbiome landscape in the omics era. Front Microbiol 10:1378. doi: 10.3389/fmicb.2019.01378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Z, Usyk M, Sollecito CC, Qiu Y, Williams-Nguyen J, Hua S, Gradissimo A, Wang T, Xue X, Kurland IJ, Ley K, Landay AL, Anastos K, Knight R, Kaplan RC, Burk RD, Qi Q. 2020. Altered gut microbiota and host metabolite profiles in women with human immunodeficiency virus. Clin Infect Dis 71:2345–2353. doi: 10.1093/cid/ciz1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dillon SM, Lee EJ, Kotter CV, Austin GL, Dong Z, Hecht DK, Gianella S, Siewe B, Smith DM, Landay AL, Robertson CE, Frank DN, Wilson CC. 2014. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol 7:983–994. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, Al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Tremaroli V, Yin Y, Bergman S, Xu X, Madsen L, Kristiansen K, Dahlgren J, Wang J, Jun W. 2015. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 71.Balakrishnan B, Luckey D, Taneja V. 2019. Autoimmunity-associated gut commensals modulate gut permeability and immunity in humanized mice. Mil Med 184:529–536. doi: 10.1093/milmed/usy309. [DOI] [PubMed] [Google Scholar]
- 72.Alexander M, Ang QY, Nayak RR, Bustion AE, Sandy M, Zhang B, Upadhyay V, Pollard KS, Lynch SV, Turnbaugh PJ. 2022. Human gut bacterial metabolism drives Th17 activation and colitis. Cell Host Microbe 30:17–30.E9. doi: 10.1016/j.chom.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dzanibe S, Lennard K, Kiravu A, Seabrook MSS, Alinde B, Holmes SP, Blish CA, Jaspan HB, Gray CM. 2021. Stereotypic expansion of T regulatory and Th17 cells during infancy is disrupted by HIV exposure and gut epithelial damage. J Immunol 208:27–37. doi: 10.4049/jimmunol.2100503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.San-Juan-Vergara H, Zurek E, Ajami NJ, Mogollon C, Peña M, Portnoy I, Vélez JI, Cadena-Cruz C, Diaz-Olmos Y, Hurtado-Gómez L, Sanchez-Sit S, Hernández D, Urruchurtu I, Di-Ruggiero P, Guardo-García E, Torres N, Vidal-Orjuela O, Viasus D, Petrosino JF, Cervantes-Acosta G. 2018. A Lachnospiraceae-dominated bacterial signature in the fecal microbiota of HIV-infected individuals from Colombia, South America. Sci Rep 8:4479. doi: 10.1038/s41598-018-22629-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vacca M, Celano G, Calabrese FM, Portincasa P, Gobbetti M, De Angelis M. 2020. The controversial role of human gut Lachnospiraceae. Microorganisms 8:573. doi: 10.3390/microorganisms8040573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weinberg A, Mussi-Pinhata MM, Yu Q, Cohen RA, Almeida VC, Amaral F, Pinto J, Teixeira ML, Succi RC, Freimanis L, Read JS, Siberry G, NISDI Perinatal, LILAC, CIRAI Protocols. 2017. Excess respiratory viral infections and low antibody responses among HIV-exposed, uninfected infants. AIDS. 31:669–679. doi: 10.1097/QAD.0000000000001393. [DOI] [PubMed] [Google Scholar]
- 77.Hara N, Alkanani AK, Ir D, Robertson CE, Wagner BD, Frank DN, Zipris D. 2012. Prevention of virus-induced type 1 diabetes with antibiotic therapy. J Immunol 189:3805–3814. doi: 10.4049/jimmunol.1201257. [DOI] [PubMed] [Google Scholar]
- 78.Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, von Bergen M, McCoy KD, Macpherson AJ, Danska JS. 2013. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 79.Illumina. iGenomes: ready-to-use reference sequences and annotations. http://support.illumina.com/sequencing/sequencing_software/igenome.ilmn. Accessed 21 March, 2022.
- 80.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ewing B, Green P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194. doi: 10.1101/gr.8.3.186. [DOI] [PubMed] [Google Scholar]
- 82.Ewing B, Hillier L, Wendl MC, Green P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 83.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schloss PD, Westcott SL. 2011. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl Environ Microbiol 77:3219–3226. doi: 10.1128/AEM.02810-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pruesse E, Peplies J, Glockner FO. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Robertson CE, Harris JK, Wagner BD, Granger D, Browne K, Tatem B, Feazel LM, Park K, Pace NR, Frank DN. 2013. Explicet: graphical user interface software for metadata-driven management, analysis and visualization of microbiome data. Bioinformatics 29:3100–3101. doi: 10.1093/bioinformatics/btt526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shetty L. 2012. microbiome R package. http://microbiome.github.io.
- 89.R Core Team. 2020. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. [Google Scholar]
- 90.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oksanen J, Blanchet G, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MH, Szoecs E, Wagner H. 2019. Vegan: Community Ecology Package. R package version 2.5–7. http://vegan.r-forge.r-project.org.
- 92.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lahti L, Shetty L. 2019. Beta diversity and microbiome divergence. https://microbiome.github.io/tutorials/Betadiversity.html.
- 94.Larsson J. 2021. eulerr: area-proportional Euler and Venn diagrams with ellipses. https://CRAN.R-project.org/package=eulerr.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Differences in infant gut microbiome; (B) differences in maternal gut microbiome; (C) differences in breast milk microbiome. Download Table S1, DOCX file, 0.05 MB (49.7KB, docx) .
Copyright © 2022 Jackson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Longitudinal changes in alpha diversity in HEU and HUU gut microbiomes. Richness increased significantly from 6 to 28 weeks in HEU (FDR = 0.02) and HUU (FDR = 0.002); evenness and Shannon diversity increased significantly only in HUU (FDR P values of 0.005 and 0.003, respectively). Download FIG S1, PDF file, 0.1 MB (83.3KB, pdf) .
Copyright © 2022 Jackson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Maternal gut microbiome composition. Bray-Curtis analysis showed significant differences at delivery (FDR = 0.02) and at 62 weeks postpartum (FDR < 0.01). Download FIG S2, PDF file, 0.4 MB (388.1KB, pdf) .
Copyright © 2022 Jackson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relationship between the gut microbiome beta diversity and HIV disease characteristics in mothers with HIV. The data show the relationship of intraindividual similarity between delivery and 62 weeks with CD4 count (A) and log HIV load (B) and the relationship between interindividual similarity at delivery and CD4 count (C) and log HIV load (D). Download FIG S3, PDF file, 0.1 MB (94.1KB, pdf) .
Copyright © 2022 Jackson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Maternal diet was measured as the number of servings per day and then calculated as relative percentage per day. Both counts of the number of servings per day and the relative percentage based on all servings are presented. P values are shown for both counts of servings and relative percentage, although only counts were used to determine statistical differences between groups (Wilcoxon test). As these 10 comparisons were planned before the study, they are presented as raw P values without adjustment for multiple comparisons. Only fruit consumption at 62 weeks postpartum differed between groups (P values = 0.04). Download Table S2, DOCX file, 0.02 MB (19.2KB, docx) .
Copyright © 2022 Jackson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All sequences and corresponding metadata were deposited in the NCBI Sequence Read Archive under BioProject accession number PRJNA816484.