ABSTRACT
Throughout the course of evolution, bacteria have developed signal transduction tools such as two-component systems (TCSs) to meet their demands to thrive even under the most challenging environmental conditions. One TCS called MtrAB is commonly found in Actinobacteria and is implicated in cell wall metabolism, osmoprotection, cell proliferation, antigen secretion, and biosynthesis of secondary metabolites. However, precisely how the MtrAB TCS regulates the bacterial responses to external environments remains unclear. Here, we report that the MtrAB TCS regulates the cell envelope response of alkali-tolerant bacterium Dietzia sp. strain DQ12-45-1b to extreme alkaline stimuli. We found that under alkaline conditions, an mtrAB mutant exhibited both reduced growth and abnormal morphology compared to the wild-type strain. Electrophoretic mobility shift assay analysis showed that MtrA binds the promoter of the mraZ gene critical for cell wall homeostasis, suggesting that MtrA directly controls transcription of this regulator. In conclusion, our findings demonstrate that MtrAB TCS is involved in controlling the bacterial response to alkaline stimuli by regulating the expression of the cell wall homeostasis regulator MraZ in Dietzia sp. DQ12-45-1b, providing novel details critical for a mechanistic understanding of how cell wall homeostasis is controlled.
IMPORTANCE Microorganisms can be found in most extreme environments, and they have to adapt to a wide range of environmental stresses. The two-component systems (TCSs) found in bacteria detect environmental stimuli and regulate physiological pathways for survival. The MtrAB TCS conserved in Corynebacterineae is critical for maintaining the metabolism of the cell wall components that protects bacteria from diverse environmental stresses. However, how the MtrAB TCS regulates cell wall homeostasis and adaptation under stress conditions is unclear. Here, we report that the MtrAB TCS in Dietzia sp. DQ12-45-1b plays a critical role in alkaline resistance by modulating the cell wall homeostasis through the MtrAB-MraZ pathway. Thus, our work provides a novel regulatory pathway used by bacteria for adaptation and survival under extreme alkaline stresses.
KEYWORDS: Dietzia sp. DQ12-45-1b, two-component system, MtrAB, alkaline environment, MraZ, cell wall homeostasis, Dietzia
INTRODUCTION
Bacteria comprise approximately 15% of the entire biomass on Earth and can be found even in the most extreme environments where higher plants and animals fail to thrive (1). The cell envelope acts as a barrier to protect bacteria from external stresses, as well as from attacks. To maintain the integrity of the cell envelope in these extreme environments, bacteria utilize a wide variety of signal transduction regulatory systems, such as two-component systems (TCSs) (2). These TCSs are able to detect multiple environmental stimuli including pH, temperature, osmotic pressure, oxidative stress, and different ions (3–7). Prototypical TCSs typically consist of a histidine kinase sensor (HK) and a response regulator (RR). In general, the HK senses a stimulus, which leads to ATP hydrolysis and phosphorylation of a conserved histidine residue of the HK. Subsequently, the phosphorylated HK transfers the phosphoryl group to a conserved aspartic acid residue of the cognate RR, ultimately resulting in an activated form of the RR that in turn controls activation of genes further downstream (2, 8, 9).
The MtrAB comprises a highly conserved TCS that is specifically expressed in Actinobacteria. The MtrAB consists of MtrA, which functions as RR, and MtrB, which functions as HK (10, 11). The MtrAB TCS plays multiple roles in various cellular processes, including cell cycle control, cell wall homeostasis, and environmental stress resistance. For example, in Mycobacterium, the MtrAB TCS is essential and is involved in regulation of the cell cycle progression (i.e., replication and cell division), cell proliferation, cell wall homeostasis, and biofilm formation (12–14). Previous studies reported that the MtrA regulon comprises key players involved in cell division and cell wall metabolisms, such as ftsI, dacB1, ripA, fbpBC, and rpfABC (15–17). Two studies showed that MtrA interacts with the promoter of dnaA involved in DNA replication (18, 19). In contrast, the MtrAB TCS of Corynebacterium glutamicum is involved in osmoregulation and cell wall metabolism. The MtrAB TCS regulates cell wall metabolism via repression of mepA and nlpC and is involved in osmoregulation by directly activating betP and proP. (20, 21). Furthermore, in Streptomyces species, the MtrAB TCS plays an important role in the developmental life cycle (i.e., aerial mycelium formation and spore maturation). The phosphorylated MtrA regulates aerial mycelium formation by activating chp, rdl, and ramCSAB genes, as well as the bldK operon at the early phase. In addition, MtrA represses bldD to activate BldN, whose targets include chp and rdl genes, to regulate aerial mycelium formation. MtrA also affects Streptomyces differentiation, namely, by regulating whiI directly and whiH indirectly, both of which are essential for the activation of whiE (22). These observations strongly suggest that MtrAB TCS both directly and indirectly affects cell wall homeostasis, osmoprotection, cell cycle progression, and the developmental life cycle.
Peptidoglycan biosynthesis and metabolism represents an important adaptation to stress. For example, Exiguobacterium sibiricum increases the expression of peptidoglycan biosynthesis genes (murADEI) at −2.5°C, suggesting that the thickening of the cell wall may protect the cell against disruption by ice formation and/or osmotic pressure, on account of the subzero temperatures (23). In comparison, in Listeria monocytogenes, genes involved in peptidoglycan biosynthesis (murABCE) are downregulated, which is presumably related to the reduced cellular growth rate when experiencing cold stress (4°C) (24). In Mycobacteria, the PknB-CwlM-MurA regulatory pathway functions to swiftly repress peptidoglycan synthesis during nutrient limitation. This regulatory pathway possibly contributes to antibiotic tolerance during growth and under changing environmental conditions (25). Peptidoglycan modification is also involved in bacterial response to environmental pH perturbation. Increased expression of peptidoglycan biosynthesis has been reported for Escherichia coli under acid stress. This result possibly attempts to maintain cell wall structural integrity (26). In natural environments, bacteria usually encounter pH changes in the alkaline range, rather than acidic range. Several Listeria monocytogenes genes involved in cell division (i.e., ftsE and iap) are repressed, and the cell shape becomes filamentous or an elongated chain forms under alkaline stress. These changes in either cell division or peptidoglycan metabolism may represent an adaptive response resulting in resistance to alkaline growth conditions (27, 28). However, the effects on regulatory pathways responsible for changed peptidoglycan metabolism or cell division under alkaline pH stress remain to be elucidated.
The actinobacterial genus Dietzia is widely distributed in terrestrial and aquatic habitats and in clinical materials, even in the most extreme environments, such as deep seas and cold deserts (29–31). Most Dietzia species can grow under high-alkaline (pH 10 to 12), high-saline (7 to 16% [wt/vol] NaCl), and low-temperature (4°C) conditions. Moreover, Dietzia species have great potential for applications in a wide range of industries. They are thought to be good candidates for bioremediation in alkaline, salty, and frigid environments (32). In addition, Dietzia species were reported to be used as potential probiotics to inhibit Mycobacterium subsp. paratuberculosis growth in cultures in vitro (33). Dietzia sp. strain DQ12-45-1b is one of the most studied Dietzia strains; it was isolated from oil production water (34). It can degrade a wide range of aliphatic hydrocarbons and aromatic compounds and can grow under high-alkaline and high-saline conditions (34, 35). To understand its different adaptive mechanisms in alkaline environments, we set out to investigate the role of MtrAB TCS in cell wall homeostasis under the alkaline stress in Dietzia sp. DQ12-45-1b. We show that the MtrAB TCS controls the cellular shape and cell division in response to the alkaline stress, namely, by directly regulating the expression of the regulator MraZ, which contributes to cell wall metabolism and cell division.
RESULTS
MtrAB is essential for maintaining growth under alkaline conditions.
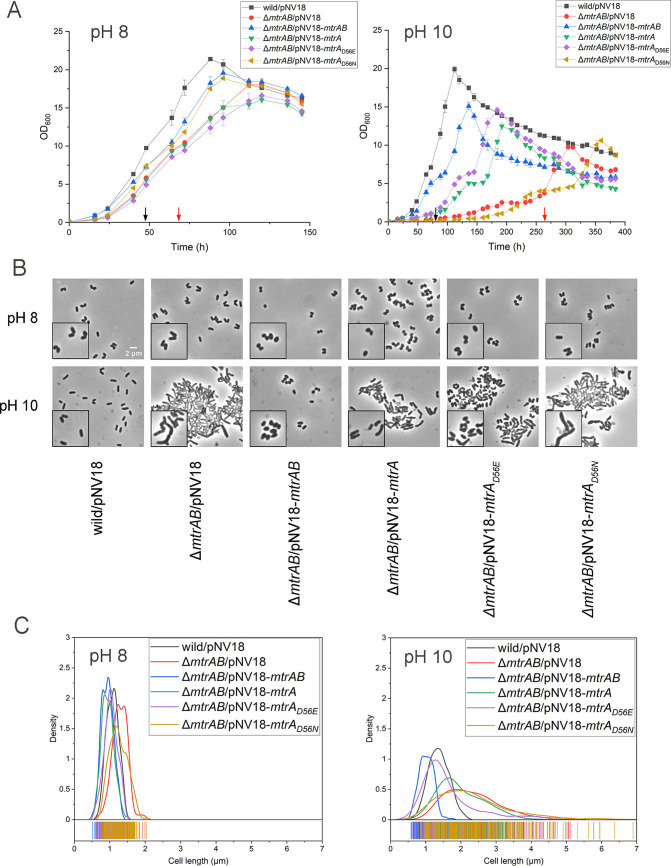
Dietzia sp. DQ12-45-1b thrives under diverse environmental conditions, including extreme stress such as highly alkaline pH (34). Dietzia sp. DQ12-45-1b harbors 14 complete pairs of TCSs, as well as a gene encoding one orphan response regulator. We found that inactivation of the genes encoding MtrAB TCS homologs GJR88_01482/01483 significantly suppressed the growth rate at pH 10 (Fig. 1A). MtrA homolog GJR88_01482 in Dietzia sp. DQ12-45-1b (denoted as MtrADz) exhibits 81% amino acid sequence identity to MtrA of Mycobacterium tuberculosis, and GJR88_01483 (denoted as MtrBDz) exhibits 56% identity to MtrB of M. tuberculosis. The complement of mtrAB genes recovered the growth of the ΔmtrAB mutant at pH 10 (Fig. 1A), suggesting that the TCS MtrAB plays a critical role in establishing resistance to alkaline conditions in Dietzia sp. DQ12-45-1b.
FIG 1.
Characterization of ΔmtrAB mutants overexpressing wild-type, phosphorylation-defective, or phosphorylation-mimicking MtrA strains at different pHs. (A) Growth curves of the Dietzia sp. DQ12-45-1b wild-type strain carrying pNV18 or the ΔmtrAB mutant carrying pNV18 (empty plasmid), pNV18-mtrAB, pNV18-mtrA, pNV18-mtrAD56E, or pNV18-mtrAD56N, respectively, at pH 8 and pH 10. Sampling points for RNA-seq analyses of wild-type and ΔmtrAB mutant strains are indicated by black and red arrows, respectively. (B) Representative micrographs of the above-mentioned strains examined by bright-field microscopy. (C) Distribution curves of cell lengths at pH 8 and pH 10 of ~200 cells/sample from panel B were measured using Digimizer software.
To investigate whether phosphorylation of MtrA is required for alkaline resistance in Dietzia sp. DQ12-45-1b, we compared the amino acid sequences of other potential MtrA and MtrB sequences from different genera. Our analysis showed that MtrA and MtrB from Dietzia sp. DQ12-45-1b contained highly conserved amino acid residues with other MtrAB proteins, including the potential phosphorylation sites at Asp56 of MtrA and His322 of MtrB (36, 37) (see Fig. S1 and S2 in the supplemental material). Next, we replaced the conserved Asp56 with either asparagine (MtrAD56N) to disrupt phosphorylation of MtrA or glutamate (MtrAD56E) to mimic phosphorylation (36). We found that the ΔmtrAB mutant strain overexpressing mtrAD56E partially restored the delayed growth at pH 10. In addition, overexpressing phosphorylation-defective mtrAD56N was similar to the ΔmtrAB mutant strain (Fig. 1A). These results suggested that phosphorylation of MtrA is required to establish resistance to alkaline stress in Dietzia sp. DQ12-45-1b. Importantly, overexpressing mtrA also restored the growth of the ΔmtrAB mutant strain at pH 10, suggesting that there is potential branched regulation of MtrA by other kinases under in vivo conditions.
MtrAB is essential for maintaining regular cell morphology under alkaline conditions.
We found that under alkaline conditions, our ΔmtrAB mutant strain was irregularly shaped and elongated (Fig. 1B and C). The cell lengths of both wild-type strain and the ΔmtrAB mutant were similar at pH 8. However, at pH 10, the length of ΔmtrAB cells (2.79 ± 0.99 μm) was significantly (P < 0.001) greater than that of wild-type cells (1.34 ± 0.23 μm) (Fig. 1B and C). We also noticed that the cells of ΔmtrAB mutant were irregularly shaped, with a small percentage exhibiting branched phenotypes (Fig. 1B; see also Fig. S3). At pH 10, the complement of mtrAB recovered cell length similar to that observed for the wild-type strain. Together, these results suggested that the MtrAB TCS is essential for maintaining normal cell morphology under alkaline stress conditions.
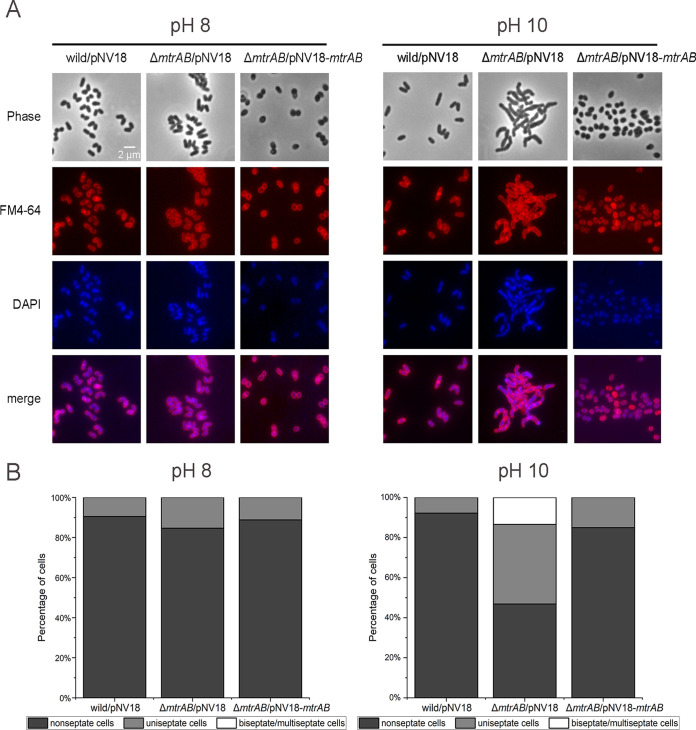
Dividing cells are known to possess one or more septa and DNA foci detectable by fluorescent labeling. Correspondingly, nonseptate cells (nondividing cells) are cells with a detectable gap between the membranes (38). To investigate whether elongation of ΔmtrAB cells was due to incomplete cell division, we used the fluorescent dyes FM4-64 and DAPI (4′,6′-diamidino-2-phenylindole) that detect the septa and DNA foci within dividing cells. Our results showed that nondividing cells dominated in all strains (i.e., the wild-type, ΔmtrAB, and complementary strains) at pH 8, which accounted for more than 80% of the total cells (Fig. 2). While this ratio significantly decreased in cells of ΔmtrAB at pH 10, it remained unchanged in cells of both wild-type and mtrAB complementary strains. Moreover, we detected multiple septa only in ΔmtrAB cells at pH 10, which account for ~13% of the total cells (Fig. 2B), indicating that the ΔmtrAB strain displayed cell division defects at pH 10. Furthermore, the complement of MtrAD56N failed to recover the cell morphology of ΔmtrAB cells at pH 10. However, MtrA and MtrAD56E partially recovered the cell shape of ΔmtrAB cells under alkaline conditions, with the phenotypes of the latter strain being closer to that of the wild-type strain (Fig. 1B and C). Together, these results indicated that the MtrAB TCS plays an important role in normal cell division under alkaline stress conditions.
FIG 2.
MtrAB TCS is required for septation in Dietzia sp. DQ12-45-1b at pH 10. (A) Fluorescence microscopy of septation in the Dietzia sp. DQ12-45-1b wild-type strain, the ΔmtrAB mutant, and complementary strain cells in the exponential growth phase at pH 8 and pH 10. The cell membrane and DNA were marked using FM4-64 (red) and DAPI (blue), respectively. (B) Stacked columns of the percentages of cells containing a different number of septa from A were analyzed at pH 8 and pH 10 (~200 cells/sample).
MtrAB regulates peptidoglycan metabolism in Dietzia sp. DQ12-45-1b.
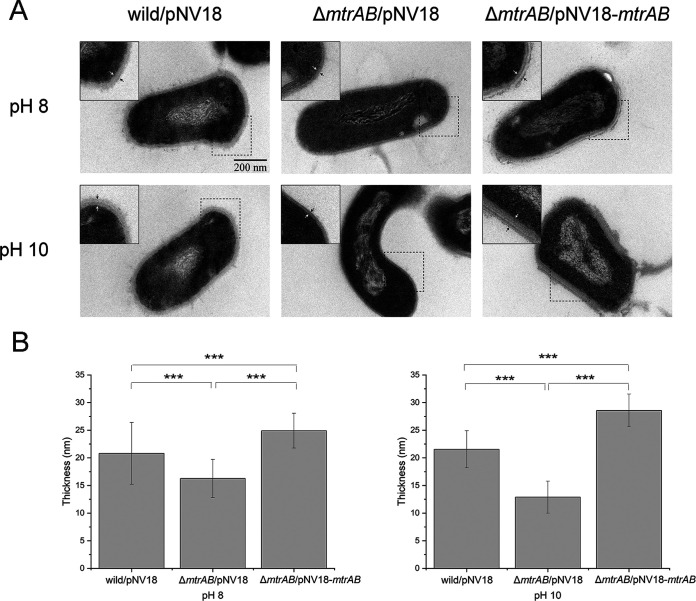
Since the regulation of peptidoglycan synthesis plays an important role in bacterial growth and morphology (39), we speculated that MtrAB controls the growth and cell division under alkaline stress by regulating peptidoglycan metabolism. To probe the ultrastructure of the cell envelopes of strains under alkaline conditions, we used thin-section transmission electron microscopy. As shown in Fig. 3, deletion of mtrAB significantly (P < 0.001) decreased the thickness of the cell envelope at both pH 8 and pH 10. Although high-pH conditions failed to affect the thickness of the cell envelope in the wild-type strain, they significantly reduced the thickness of the cell envelope in the ΔmtrAB mutant (Fig. 3). The change in the thickness of the cell envelope might result from a change in peptidoglycan metabolism, implying that MtrAB is involved in the regulation of peptidoglycan metabolism under alkaline stress conditions.
FIG 3.
Ultrastructure of the cell envelope of Dietzia cells. (A) Representative thin-section transmission electron microscopy of the Dietzia sp. DQ12-45-1b wild-type strain, the ΔmtrAB mutant, and complementary strain cells under different pH conditions. The distance between two arrows represents the thickness of the cell envelope. Insets show small portions of cell cross sections for each image. (B) Histogram of cell envelope thickness measurements obtained by analyzing ~10 cells/sample in transmission electron microscopic cross sections from panel A. Every cross-section selected multiple regions for measurement using Digimizer (unpaired t test for panel B; ***, P < 0.001).
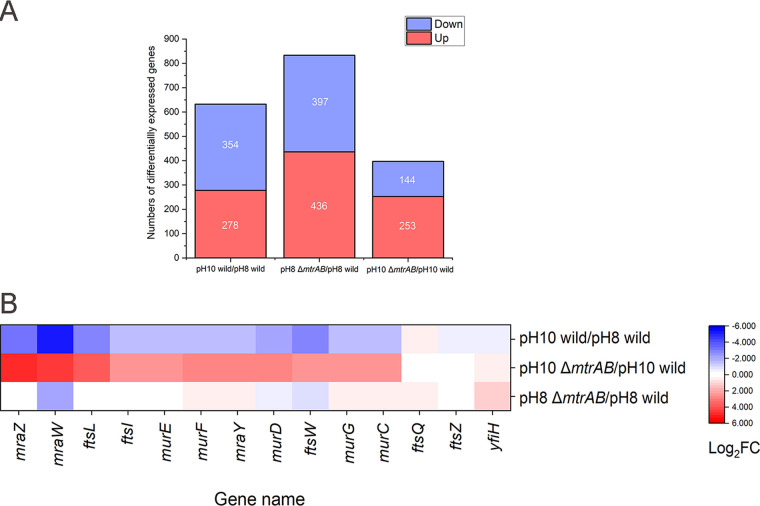
To confirm the roles of mtrAB on the regulation of peptidoglycan metabolism under alkaline stress, we compared the transcriptional profiles of the wild-type and ΔmtrAB strains at different pH conditions. We found 278 upregulated genes and 354 downregulated genes of the wild-type strain at pH 10 compared to pH 8 conditions. In addition, we identified 144 genes with at least a 2-fold decrease and 253 genes with at least a 2-fold increase in the ΔmtrAB mutant compared to the wild-type strains under the alkaline stress (Fig. 4A). Based on the scatterplots, we confirmed that the parallel samples have a good linear correlation, suggesting that the data are reliable (see Fig. S4). To seek out the pathways altered by deletion of MtrAB under the alkaline stress, we analyzed transcriptomic data by Gene Ontology (GO) enrichment analysis (see Fig. S5). We found that a large number of enriched GO terms were related to cell division, cell wall biosynthesis, and metabolic process. Upon further analysis, we observed significantly downregulated expression of the genes involved in peptidoglycan biosynthesis (murCDEFGJ, mraY, and ftsWI), hydrolysis (ripA, cwlO, mepA), cell division (ftsEXKL, sepF, and sepIVA), and related regulators (mraZ, envC, and whmD), especially the division cell wall (dcw) gene cluster in the wild-type strain at pH 10 compared to pH 8 (Fig. 4B; see also Table S1). These results implied that the regulation of peptidoglycan synthesis, hydrolysis, and cell division is important for alkaline resistance in the wild-type strain. The ΔmtrAB mutant exhibited significantly higher expression of the above-mentioned genes compared to wild-type strain at pH 10 (Fig. 4B; see also Table S1). The results suggested that MtrAB represses the genes involved in peptidoglycan homeostasis and cell division, especially the dcw gene cluster under alkaline growth conditions. Next, we overexpressed these genes involved in peptidoglycan homeostasis, cell division, and related regulators in the Dietzia sp. DQ12-45-1b wild-type strain. However, these overexpressed strains failed to cause abnormal cell morphology except the mraZ-overexpressing strain (see Fig. S6 and S7), indicating that at pH 10, a number of genes participating in peptidoglycan synthesis, hydrolysis, and cell division simultaneously act on the phenotypes of ΔmtrAB mutant. Together, these results suggested that MtrA represses specific genes involved in peptidoglycan homeostasis and cell division at pH 10, presumably to ensure that Dietzia sp. DQ12-45-1b maintains normal cell morphology.
FIG 4.
Transcriptomic analysis of the dcw cluster wild-type and ΔmtrAB mutant strains at pH 8 and pH 10. (A) Stacked columns of the number of DEGs (fold change ≥ 2, FDR ≤ 0.05). (B) Heatmap of the dcw cluster induced and repressed in wild-type and ΔmtrAB mutant strains under different pH conditions. Red represents upregulated genes, and blue represents downregulated genes.
MtrAB controls cell wall homeostasis at pH 10 by directly repressing mraZ gene expression.
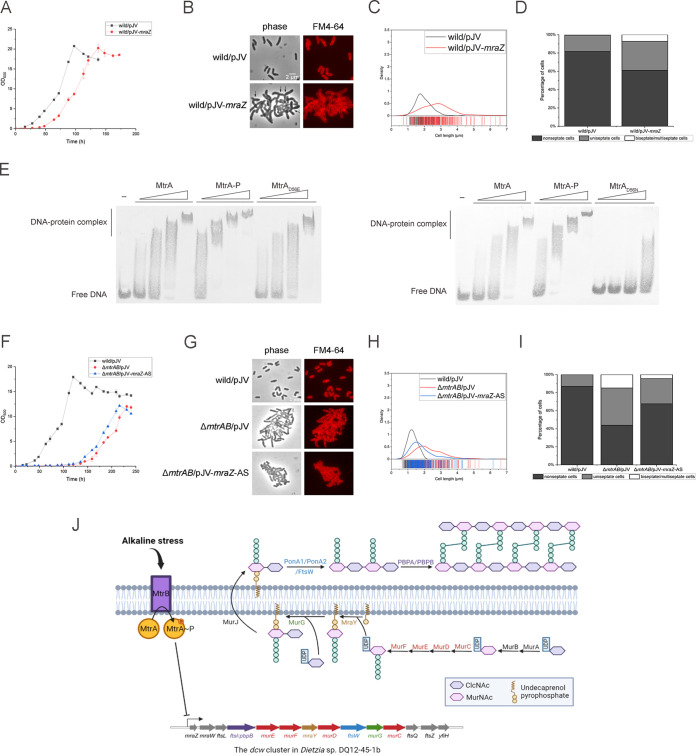
Since mraZ is the head gene of the dcw cluster that potentially regulates downstream genes (Fig. 4B) (40), we wondered whether MtrAB regulates the expression of the mraZ gene in response to alkaline stress in Dietzia sp. DQ12-45-1b. We first confirmed the function of MraZ by overexpressing this gene in the Dietzia sp. DQ12-45-1b wild-type strain. We found that at pH 10, the strain overexpressing the mraZ gene exhibited a significantly delayed growth and an elongated and partially branched cell shape (Fig. 5A to C). We also noticed that overexpression of the mraZ gene resulted in the formation of multiseptate cells. These cells comprised approximately 7% of the total population during the early exponential phase at pH 10 (Fig. 5B and D). These results clearly demonstrated that overexpression of MraZ results in morphological deformation together with delayed growth. Since the expression of mraZ was induced in the ΔmtrAB mutant at pH 10, these results suggested that the function of the MtrAB TCS is to repress the expression of MraZ to maintain cell wall homeostasis under alkaline stress.
FIG 5.
Characterization of wild-type strains overexpressing mraZ, ΔmtrAB strains silencing mraZ at pH 10 stress, and binding of MtrA to the mraZ promoter analyzed by EMSA. (A and F) Growth curves of the Dietzia sp. DQ12-45-1b wild-type strain containing the mraZ overexpression plasmid and the ΔmtrAB strain containing antisense mRNA of mraZ plasmid from the acetamide-inducible promoter at pH 10. (B and G) Fluorescence microscopy of septation in wild type/pJV-mraZ (wild/pJV-mraZ) and ΔmtrAB/pJV-mraZ-AS under pH 10 stress. The branched phenotype is marked with arrows. (C, D, H, and I) Distribution curves and stacked columns of the percentage of cells containing a different number of septa from panels B and G were analyzed (~200 cells/sample). (E) EMSA analysis of MtrA, MtrA-P, MtrAD56E, and MtrAD56N binding to the mraZ promoter. MtrA, MtrAD56E, MtrAD56N, and MtrA~P at 0.5, 1, 2.5, and 5 μM were incubated for 30 min with 10 pM DIG-labeled Pmraz. (J) Proposed mechanism’s diagram illustrating the relationship between MtrAB TCS and the dcw cluster in Dietzia sp. DQ12-45-1b.
To investigate whether MtrA protein regulates mraZ directly, we prepared MtrA protein, phosphorylation-defective MtrAD56N, and phosphorylation-mimic MtrAD56E, phosphorylated the MtrA with EnvZ histidine kinase in vitro, and carried out an electrophoretic mobility shift assay (EMSA) with a digoxigenin (DIG)-labeled 100-bp DNA fragment upstream of the mraZ gene. We found that when the protein concentrations were up to 5 μM for MtrA and MtrAD56E and 2.5 μM for phosphorylated MtrA, a slower shift band was observed, demonstrating that both the MtrA and the phosphorylated MtrA directly bind to the promoter region of the mraZ gene (Fig. 5E). On the other hand, we found that MtrAD56N fails to bind to the promoter of mraZ. Previous studies reported that the replacement of the phosphorylation site of the response regulator with asparagine does not significantly affect protein conformation (36, 41), so the functional change of D56N mutant may result from the disruption of the phosphorylation site. Moreover, MtrA and MtrA-P cannot bind to the negative-control DNA (see Fig. S8). Compared to phosphorylation-defective MtrAD56N, MtrA can bind to the probe. One possible reason is that MtrA is slightly phosphorylated by other kinase sensors in Escherichia coli before being purified because of HK-RR branched regulation (42). Together, these results strongly suggested that phosphorylation is required for the MtrA-DNA binding activity, at least under our in vitro conditions.
We next disrupted the expression of the mraZ gene in the ΔmtrAB mutant to assess whether repression of MraZ is necessary for the regulation of MtrAB in responding to alkaline stress. We failed to delete the mraZ gene in the ΔmtrAB mutant. We speculated that the mraZ gene is essential in the ΔmtrAB mutant and then silenced the mraZ gene in the ΔmtrAB mutant under the control of the acetamide-inducible promoter through antisense RNA technology (43, 44). We found that under pH 10 conditions, the cell morphology of ΔmtrAB containing antisense mRNA of mraZ was partially recovered when grown with 0.5% acetamide, but the growth was similar to that observed with the ΔmtrAB mutant strain (Fig. 5F to I). This result suggested that MraZ participates in both peptidoglycan biosynthesis and cell division of MtrAB under alkaline stress. Together, since MtrAB directly regulates the mraZ gene, we believe that MtrAB controls cell wall homeostasis under alkaline stress by directly repressing the mraZ gene (Fig. 5J).
DISCUSSION
In this study, we demonstrated that the MtrAB TCS regulates the cell wall homeostasis in Dietzia sp. DQ12-45-1b responding to environmental alkaline stress, namely, in a MraZ-dependent manner. This is the first report showing that the MtrAB TCS of Dietzia bacteria plays a critical role in peptidoglycan biosynthesis and cell division under environmental alkaline stress conditions.
The MtrAB TCS maintains the cell wall homeostasis which may be related to the universal responses to diverse environmental stresses. For example, in Mycobacterium tuberculosis, MtrB is required to withstand acid stress (pH 5.5) (14). In Corynebacterium glutamicum MtrAB is involved in osmoregulation, as well as cell wall metabolism (20). Here, we also demonstrated that MtrAB is essential for maintaining growth under alkaline condition in Dietzia sp. DQ12-45-1b. In addition, based on the transcriptomic data, we found that the expression levels of mtrA and mtrB increased ~1.5-fold (<2-fold) in pH 10 compared to pH 8 in the wild-type strain (see Table S1). Moreover, in Dietzia sp. DQ12-45-1b, deletion of mtrAB also resulted in growth defect, increased cell length, and branched cell shape in response to various cell envelope stresses, such as oxidative, sodium dodecyl sulfate (SDS), hyperosmotic, and cell-envelope-targeting antibiotic stresses (data not shown). These findings suggest that MtrAB may play a critical role in response to the diverse environmental stresses. Interestingly, the MtrAB deficiency also resulted in abnormal cell morphology in the above-mentioned bacteria. It has been reported previously that ΔmtrA and ΔmtrB mutants of Mycobacterium smegmatis both exhibit filamentous with branches and buds (15, 45). In addition, a ΔmtrAB mutant form of Corynebacterium glutamicum exhibited increased cell length and irregular septum formation (20). In addition, the absence of mtrB in Mycobacterium tuberculosis resulted in a chain-like phenotype and increased bulbous regions (13, 37). In summary, deletion of MtrAB TCS results in elongated cells with irregular septa in Corynebacterineae, indicating that the universal pathways that MtrAB TCS regulates may play a critical role in cell morphology. These findings are also important for the future research of the signals recognized by MtrB. MtrAB TCS is involved in response to diverse cell envelope stresses and regulates cell wall homeostasis, which suggests that cell wall fragments are sensed by MtrB and are likely to be universally present in Corynebacterineae.
Based on our transcriptomic analysis, we found that at pH 10 condition, the expression levels of genes involved in peptidoglycan biosynthesis (murCDEFGJ, mraY, and ftsWI), hydrolysis (ripA, cwlO, and mepA), cell division (ftsEXKL, sepF, and sepIVA), and related regulators (mraZ, envC, and whmD) significantly changed in ΔmtrAB mutant compared to the wild-type strain, a process that was accompanied by the appearance of irregular cell morphology, as well as delayed growth, in our ΔmtrAB mutant. It is well known that cooperation between peptidoglycan synthetases and hydrolases is critical for cell elongation and division (46, 47). In addition, the enzymes involved in peptidoglycan synthesis and hydrolysis are coregulated to ensure the balance between two processes (25, 48, 49). It also has been reported that imbalance between peptidoglycan synthesis and hydrolysis results in cell lysis (50). Therefore, abnormal transcriptional changes of genes involved in peptidoglycan synthesis and hydrolysis might lead to Dietzia sp. DQ12-45-1b ΔmtrAB cell lysis, thereby reducing the rate of growth. Moreover, the transcriptomic analysis showed that the expression levels of previously known sodium/proton antiporter genes such as mrpACDEFG related to alkaline resistance did not significantly change in the ΔmtrAB mutant (see Table S1), which suggested that MtrAB regulated a novel pathway for alkaline resistance (35). Although the mechanisms need to be further determined, the abnormal cell morphology of mtrAB-deficient strains in Corynebacterineae suggested that the cell wall homeostasis maintained by MtrAB may be related to the responses to alkaline stress, even to other diverse cell envelope stress conditions.
The MtrA-MraZ regulatory pathway may be universal but not exclusive to Corynebacterineae. Based on our findings, we conclude that MtrAB TCS in our Dietzia sp. DQ12-45-1b strain is directly involved in peptidoglycan biosynthesis, as well as cell division, that depends on the regulator MraZ during responses to alkaline stress. Since mraZ has been reported to belong to putative MtrA targets using a global CHIP-seq method in Mycobacterium tuberculosis (15), the MtrA-MraZ regulatory pathway may represent one universal pathway used by Corynebacterineae to achieve normal cell morphology. However, our results also suggest that the MtrA-MraZ regulatory pathway is not an exclusive pathway in response to alkaline stress. We showed that the phenotypes of overexpression of mraZ were similar to the ΔmtrAB mutant, but silencing of mraZ in the ΔmtrAB mutant was partially offset in response to alkaline stress. Together, these results suggest that additional proteins in addition to MraZ levels are critical for the phenotype of our ΔmtrAB mutant strain under alkaline conditions. Transcriptomic analysis showed that other genes belonging to the MtrA regulon, such as mepA and whmD, were also upregulated in ΔmtrAB mutant strain under alkaline conditions (15, 20). Previous studies reported that overexpression of the hydrolase gene mepA in Escherichia coli resulted in an elongated cell morphology, presumably because of interference with the localization or activity of certain PG synthetases (20, 51). Overexpression of the regulator gene whmD in the Mycobacterium smegmatis wild-type strain resulted in mislocated septa, presumably because WhmD levels are important for the efficiency or correct location of septum formation, perhaps by affecting FtsZ localization or assembly (52, 53). However, in contrast to previous reports, overexpression of either MepA or WhmD in Dietzia sp. DQ12-45-1b failed to change cell morphology (see Fig. S7). These results suggest that MtrAB regulates redundant pathways to maintain the cell wall homeostasis and normal cell morphology to respond to the environmental stresses.
In conclusion, the relationship of MtrAB TCS and MraZ in this study should greatly assist future investigations into the transcriptional regulators of MraZ. We established here the direct relationship between MtrAB TCS and MraZ involved in cell wall synthesis and cell division. The dcw cluster, including the mraZ gene, is well conserved in most bacteria and thus is important for investigating the regulatory pathways of the mraZ gene in more detail.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used are listed in Table S2 in the supplemental material.
All Dietzia strains were cultured in GPY medium (glucose [10 g/L], tryptone [10 g/L], and yeast extract [5 g/L]) with the appropriate antibiotics (kanamycin [50 μg/mL] and streptomycin [30 μg/mL]) at 30°C and shaken at 150 rpm. For pH stress experiments, GPY medium was added with a mixture of 0.2 M Na2HPO4·12H2O and 0.2 M NaH2PO4·2H2O to pH 8 and with a mixture of 0.2 M Na2CO3 and 0.2 M NaHCO3 to pH 10. To eliminate the effects of different sodium concentrations, we added 0.05 M NaCl into pH 10 GPY medium. All Escherichia coli strains were cultured in Luria-Bertani (LB) medium (NaCl [10 g/L], tryptone [10 g/L], and yeast extract [5 g/L]) with the appropriate antibiotics (kanamycin [50 μg/mL]) at 37°C with shaking at 150 rpm. Since the differences in cell length had little effect on the optical density measurement, we monitored the growth by measuring optical density instead of CFU. We measured the optical density at 600 nm (OD600) by using a UV spectrophotometer (Shimadzu, Japan). All growth curves experiments were performed in three biological replicates. Moreover, we added 0.5% acetamide to cultures of the strains harboring pJV-related plasmids to exclude the potential effects of acetamide.
Construction of the ΔmtrAB mutant and complementary strains.
The preparation method of electrocompetent cells of Dietzia sp. DQ12-45-1b was described previously (35). Briefly, Dietzia strains were cultured at OD600 of 0.05 in the presence of the appropriate antibiotics and 0.5% glycine overnight at 30°C with 150-rpm shaking. We added 0.2% Tween 80, penicillin (0.5 μg/mL), and isoniazid (0.8 mg/mL) to the cultures when they had grown to an OD600 of 0.4 to 0.6. In addition to these substances, 0.2% acetamide was added to the culture of the Dietzia sp. DQ12-45-1b wild type/pJV53 (wild/pJV53) strain. After 4 h, the cells were harvested and washed twice with 10% glycerin by centrifugation at 1,500 × g for 10 min at 4°C. The electrocompetent cells were resuspended in 10% glycerin and stored at −80°C for further experiments.
The ΔmtrAB mutant strain was obtained by the double homologous recombination method (54). In brief, the upstream and downstream homologous fragments of mtrAB (~500 bp) were PCR amplified from the genomic DNA of Dietzia sp. DQ12-45-1b using the primer pairs mtrABLF/mtrABLR and mtrABRF/mtrABRR (see Table S3). Next, two homologous fragments were fused to a streptomycin cassette by PCR using the primers fusionF and fusionR (see Table S3). The fused DNA fragment containing the upstream and downstream homologous regions of mtrAB flanking the streptomycin cassette was then transformed into Dietzia sp. DQ12-45-1b wild/pJV53 competent cells by electroporation (2,000 V/mm, 12 ms) (55, 56). Using this method, mtrAB was successfully replaced by the streptomycin cassette to obtain the ΔmtrAB mutant strain.
The target genes mtrA and mtrAB were amplified from genomic DNA of Dietzia sp. DQ12-45-1b using the primer pairs mtrAF/mtrAR and mtrAF/mtrABR with HindIII restriction sites on both sides. Next, the DNA fragment of the vector pNV18 was amplified with the HindIII restriction sites using the primers pNVF and pNVR from the pNV18-DsRed plasmid to delete the gene encoding DsRed (see Table S3). The HindIII-digested DNA fragments of mtrA and mtrAB were then ligated into the HindIII-digested fragment from pNV18-DsRed. The recombinant plasmids pNV18-mtrA and pNV18-mtrAB were transformed into ΔmtrAB mutant competent cells by electroporation to produce the complementary strains.
Construction of acetamide-inducible mraZ overexpression vector.
The complete mraZ gene fragment was cloned from genomic DNA of Dietzia sp. DQ12-45-1b by PCR using the primers mraZF and mraZR. Also, the DNA fragment of the vector pJV was amplified using the primers pJVF and pJVR from the pJV53 plasmid (see Table S3). Next, the mraZ PCR fragment was ligated into pJV, which contains an acetamide-inducible promoter, using the homologous recombination method with a Hieff Clone Plus One Step cloning kit (Yeasen). Dietzia sp. DQ12-45-1b was electroporated with the recombinant overexpression plasmid pJV-mraZ.
Construction of mraZ antisense expression vector.
Portion of the mraZ gene was amplified from genomic DNA of Dietzia sp. DQ12-45-1b by PCR using the primers mraZASF and mraZASR. Next, the plasmid vector pJV53 was linearized by PCR using the primers pJVF and pJVR (see Table S3). The purified mraZ PCR fragment was ligated into pJV in an antisense orientation using the same homologous recombination method by Hieff Clone Plus One Step cloning kit (Yeasen). The recombinant plasmid pJV-mraZ-AS was obtained, whose expression can be induced by acetamide. The pJV-mraZ-AS was electroporated into Dietzia sp. DQ12-45-1b ΔmtrAB mutant competent cells.
Construction of overexpression vectors carrying genes involved in peptidoglycan homeostasis and cell division.
All genes involved in peptidoglycan homeostasis and cell division were cloned from genomic DNA of Dietzia sp. DQ12-45-1b by PCR using the primers shown in Table S3. The plasmid vector pNV18 was linearized by PCR using the primers pNVF2 and pNVR2 (see Table S3). Next, cell wall-related genes were ligated into pNV18, which contains a p45 promoter, using the Hieff Clone Plus One Step cloning kit (Yeasen) homologous recombination method. All recombinant overexpression vectors were electroporated into Dietzia sp. DQ12-45-1b wild-type competent cells.
Construction of mtrA phosphorylation-defective and phosphorylation-mimicking vectors.
The methods which cause mtrA to lose and mimic phosphorylation activities by point mutant were already verified (36). The primers mtrAD56NF/mtrAD56NR and pNVD56NF/pNVD56NR were used to amplify by PCR from pNV18-mtrA plasmid to replace the aspartic acid at codon 56 with asparagine to create plasmid pNV18-mtrAD56N. Next, the primer pairs mtrAD56EF/mtrAD56NR and pNVD56NF/pNVD56ER were used to amplify from pNV18-mtrA to replace the aspartic acid at codon 56 with glutamate to create plasmid pNV18-mtrAD56E (see Table S3). To construct the vectors pNV18-mtrAD56N and pNV18-mtrAD56E, homologous recombination was performed using a Hieff Clone Plus One Step cloning kit. Two recombinant plasmids were electroporated into Dietzia sp. DQ12-45-1b ΔmtrAB mutant competent cells.
Construction of EnvZdel, MtrA, MtrAD56E, and MtrAD56N expression vectors.
To purified EnvZdel, the region of envZ without transmembrane region (envZdel) was PCR amplified from E. coli K-12 substrain MG1655 with the primers envZF and envZR. The plasmid vector pET-28a was linearized by PCR using the primers pETF and pETR (see Table S3). The purified envZdel PCR fragment was ligated into pET-28a using the same homologous recombination method by the Hieff Clone Plus One Step cloning kit. For purification of the MtrA protein, the complete mtrA gene was cloned from Dietzia sp. DQ12-45-1b using the primers mtrAF and mtrAR incorporating NdeI and HindIII restriction sites, respectively. Next, the mtrAD56E and mtrAD56N fragments were cloned from pNV18-mtrAD56E and pNV18-mtrAD56N plasmids using the same primers, mtrAF and mtrAR. Simultaneously, the vector pET-28a was linearized using the primers pETF2 and pETR2 (see Table S3). The amplified mtrA, mtrAD56E, and mtrAD56N DNA fragments were ligated into the NdeI- and HindIII-digested pET-28a fragment. Then, the recombinant plasmids pET-28a-envZdel, pET-28a-mtrA, pET-28a-mtrAD56E, and pET-28a-mtrAD56N were transformed into the E. coli expression strain BL21(DE3) (TransGen).
Transmission electron microscopy.
Cells that grew to the midlog phase were collected by centrifugation at 5,000 rpm for 5 min at 4°C after being washed three times with phosphate-buffered saline (PBS; pH 7.4) and then fixed with fresh 2.5% glutaraldehyde overnight at 4°C. The cells were then rinsed three times and concentrated with ddH2O. The cells were placed on a carbon-filmed, 300-mesh copper grid (Electron Microscopy China) for 2 min. Next, the grids were negatively stained using 2% (wt/vol) uranyl acetate in water for 1 min. The excess sample was removed using Whatman filter paper. Grids were examined using a transmission electron microscope (Hitachi HT7700, Japan) operated at 80 kV.
Bright-field and fluorescence microscopy.
For staining procedures, cells were washed three times with PBS buffer (pH 7.4) and collected by centrifugation at 5,000 rpm for 5 min at 4°C. Membrane dye FM4-64 (Thermo Fisher) and DNA dye DAPI (Beyotime) were added into bacterial suspension at 37°C for 10 min in the dark. Bright-field microscopy images were acquired using a microscope with 100× oil immersion lens. Cell length and cell wall thickness quantitative measurements were made using Digimizer software (MedCalc, Ltd., Ostend, Belgium) (57). An unpaired t test was used for statistical analysis. The branched cells were not included in the cell length measurement analysis.
Expression and purification of EnvZdel, MtrA, MtrAD56E, and MtrAD56N.
For EnvZdel, MtrA, MtrAD56E, and MtrAD56N protein overproduction, E. coli BL21(DE3) cells transformed with pET-28a vector derivatives expressing EnvZdel, MtrA, MtrAD56E, and MtrAD56N were grown overnight in Luria-Bertani (LB) medium with kanamycin at 37°C with shaking at 150 rpm. Cultures were diluted 1:100 and grown in fresh medium until the OD600 reached to 0.4 to 0.6. Next, 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added to the cultures for 16 h at 20°C to induce protein production. Cells were harvested by centrifugation at 8,000 × g for 10 min at 4°C. After the pellet was resuspended in 20 mL of lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 5% glycerin [pH 8.0]), the cells were disrupted via sonication, followed by centrifugation at 12,000 × g for 30 min at 4°C. The purification of EnvZdel, MtrA, MtrAD56E, and MtrAD56N proteins used a gravity-flow column prepacked with 1 mL of nickel-nitrilotriacetate (Ni-NTA) agarose (Qiagen). After being washed with 10 mL of wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, 5% glycerin [pH 8.0]), the proteins were eluted by applying 500 μL of elution buffer three times (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, 5% glycerin [pH 8.0]). The proteins were then ultrafiltered using ultrafiltration buffer (50 mM Tris-HCl [pH 7.5], 300 mM NaCl, 1 mM dithiothreitol [DTT], 10% glycerol) and stored at −80°C for further experiments.
Phosphorylation of MtrA by EnvZdel kinase.
The MtrA was phosphorylated by EnvZ, as previously described (36). In brief, purified His-EnvZdel was diluted with ultrafiltration buffer. The reaction premixture (total volume, 100 μL) for the autophosphorylation assay was composed of 50 mM Tris-HCl (pH 7.5), 50 mM KCl, 20 mM MgCl2, 1 mM DTT, and 30 μM diluted His-EnvZdel. The reaction was initiated by adding 10 μL of 100 mM ATP at a final concentration of 10 mM at room temperature. After a 1-h reaction, a phosphorylated form of His-EnvZdel was obtained. The reaction mixture described above was then mixed with 120 μM diluted His-MtrA, followed by incubation at room temperature for 1 h. Lastly, the phosphorylated form of His-MtrA was prepared for EMSA.
Electrophoretic mobility shift assay.
DNA fragment was amplified using the primers mraZDIGF and mraZDIGR containing DIG label (see Table S3). Then, a 10 pM concentration of purified DNA fragment was incubated with 0 to 5 μM MtrA, MtrAD56E, MtrAD56N, or MtrA-P in binding buffer [5 mM Tris-HCl (pH 7.6), 50 mM KCl, 0.5 mM EDTA, 5 mM (NH4)2SO4, 0.5 mM DTT, 0.1% Tween 20, 5 mM MgCl2, 5 mM CaCl2] at room temperature for 30 min. The reaction was stopped by adding 10× loading buffer (50 mM Tris-HCl [pH 6.8], 30% glycerol, 0.12% bromophenol blue). The reaction mixtures were electrophoresed on a 5% nondenaturing polyacrylamide gel for 3 h at 40 V. Next, the DNA and proteins were transferred to nylon membranes using semidry transfer method for 10 min at 10 V. The subsequent immunological detection was using a DIG High Prime DNA Labeling and Detection Starter Kit I (Roche, Mannheim, Germany) according to the manufacturer’s instructions.
RNA isolation and sequencing.
The cultures were mixed immediately with 10% (vol/vol) STOP solution (90% [vol/vol] ethanol, 10% [vol/vol] phenol) for 10 min on the ice when grown to the midexponential phase. Next, the pH of the cultures was adjusted to pH 6 using HCl. The cultures were then pelleted by centrifugation at 8,000 × g for 20 min at 4°C. Subsequently, cells were ground in liquid nitrogen and resuspended with RNAiso Plus (TaKaRa), followed by the addition of chloroform. After centrifugation (12,000 × g, 15 min, 4°C), the upper layer of the phenol-chloroform was collected, and the same volume of isopropanol was added. The RNA was obtained by centrifugation (12,000 × g, 10 min, 4°C), followed by the addition of 75% ethanol to precipitate RNA again. The genomic DNA was removed with recombinant DNase I (TaKaRa) in combination with the recombinant RNase inhibitor (TaKaRa). The quality and integrity of total RNA was assessed with an Agilent 4200 bioanalyzer. Sequencing was performed using the Illumina NovaSeq6000 platform (Magigene, Ltd., Guangzhou, China). Three biological replicates per condition were used in RNA-seq experiments.
Transcriptomic data analysis.
The differentially expressed genes (DEGs) were defined by a threshold of fold change ≥2 and a false discovery rate (FDR) of ≤0.05. Heatmaps and stacked columns were produced using Origin software. The Gene Ontology (GO) annotation of DEGs was obtained using Blast2GO software. Enrichment analysis of DEGs in GO pathways (Q ≤ 0.05) was performed using OmicShare tools.
Data availability.
The transcriptomic raw sequence has been deposited to National Microbiology Data Center under accession number NMDC10018150.
ACKNOWLEDGMENTS
We thank T. Juelich (University of Chinese Academy of Sciences, Beijing, China) for linguistic assistance during preparation of the manuscript. We thank the entire Wu lab for critical discussion of our manuscript.
This study was supported by grants from the National Key R&D Program of China (2018YFA0902100 and 2021YFA0910300) and the National Natural Science Foundation of China (32130004, 91951204, 32161133023, and 32170113).
Footnotes
Supplemental material is available online only.
Contributor Information
Yong Nie, Email: nieyong@pku.edu.cn.
Xiao-Lei Wu, Email: xiaolei_wu@pku.edu.cn.
Kathryn T. Elliott, College of New Jersey
REFERENCES
- 1.Bar-On YM, Phillips R, Milo R. 2018. The biomass distribution on Earth. Proc Natl Acad Sci USA 115:6506–6511. doi: 10.1073/pnas.1711842115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y, Rose J, Huang S, Hu Y, Wu Q, Wang D, Li C, Liu M, Zhou P, Jiang L. 2017. A pH-gated conformational switch regulates the phosphatase activity of bifunctional HisKA-family histidine kinases. Nat Commun 8:1–10. doi: 10.1038/s41467-017-02310-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakraborty S, Li M, Chatterjee C, Sivaraman J, Leung KY, Mok Y-K. 2010. Temperature and Mg2+ sensing by a novel PhoP-PhoQ two-component system for regulation of virulence in Edwardsiella tarda. J Biol Chem 285:38876–38888. doi: 10.1074/jbc.M110.179150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tipton KA, Rather PN. 2017. An ompR-envZ two-component system ortholog regulates phase variation, osmotic tolerance, motility, and virulence in Acinetobacter baumannii strain AB5075. J Bacteriol 199:e00705-16. doi: 10.1128/JB.00705-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCluskey J, Hinds J, Husain S, Witney A, Mitchell T. 2004. A two-component system that controls the expression of pneumococcal surface antigen A (PsaA) and regulates virulence and resistance to oxidative stress in Streptococcus pneumoniae. Mol Microbiol 51:1661–1675. doi: 10.1111/j.1365-2958.2003.03917.x. [DOI] [PubMed] [Google Scholar]
- 7.Glover RT, Kriakov J, Garforth SJ, Baughn AD, Jacobs WR. 2007. The two-component regulatory system senX3-regX3 regulates phosphate-dependent gene expression in Mycobacterium smegmatis. J Bacteriol 189:5495–5503. doi: 10.1128/JB.00190-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao R, Stock AM. 2009. Biological insights from structures of two-component proteins. Annu Rev Microbiol 63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoch JA. 2000. Two-component and phosphorelay signal transduction. Curr Opin Microbiol 3:165–170. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 10.Hoskisson PA, Hutchings MI. 2006. MtrAB–LpqB: a conserved three-component system in actinobacteria? Trends Microbiol 14:444–449. doi: 10.1016/j.tim.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Friedland N, Mack TR, Yu M, Hung L-W, Terwilliger TC, Waldo GS, Stock AM. 2007. Domain orientation in the inactive response regulator Mycobacterium tuberculosis MtrA provides a barrier to activation. Biochemistry 46:6733–6743. doi: 10.1021/bi602546q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen HT, Wolff KA, Cartabuke RH, Ogwang S, Nguyen L. 2010. A lipoprotein modulates activity of the MtrAB two-component system to provide intrinsic multidrug resistance, cytokinetic control and cell wall homeostasis in Mycobacterium. Mol Microbiol 76:348–364. doi: 10.1111/j.1365-2958.2010.07110.x. [DOI] [PubMed] [Google Scholar]
- 13.Plocinska R, Martinez L, Gorla P, Pandeeti E, Sarva K, Blaszczyk E, Dziadek J, Madiraju MV, Rajagopalan M. 2014. Mycobacterium tuberculosis MtrB sensor kinase interactions with FtsI and Wag31 proteins reveal a role for MtrB distinct from that regulating MtrA activities. J Bacteriol 196:4120–4129. doi: 10.1128/JB.01795-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banerjee SK, Lata S, Sharma AK, Bagchi S, Kumar M, Sahu SK, Sarkar D, Gupta P, Jana K, Gupta UD, Singh R, Saha S, Basu J, Kundu M. 2019. The sensor kinase MtrB of Mycobacterium tuberculosis regulates hypoxic survival and establishment of infection. J Biol Chem 294:19862–19876. doi: 10.1074/jbc.RA119.009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorla P, Plocinska R, Sarva K, Satsangi AT, Pandeeti E, Donnelly R, Dziadek J, Rajagopalan M, Madiraju MV. 2018. MtrA response regulator controls cell division and cell wall metabolism and affects susceptibility of mycobacteria to the first line antituberculosis drugs. Front Microbiol 9:2839. doi: 10.3389/fmicb.2018.02839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chatterjee A, Sharma AK, Mahatha AC, Banerjee SK, Kumar M, Saha S, Basu J, Kundu M. 2018. Global mapping of MtrA-binding sites links MtrA to regulation of its targets in Mycobacterium tuberculosis. Microbiology (Reading) 164:99–110. doi: 10.1099/mic.0.000585. [DOI] [PubMed] [Google Scholar]
- 17.Sharma AK, Chatterjee A, Gupta S, Banerjee R, Mandal S, Mukhopadhyay J, Basu J, Kundu M. 2015. MtrA, an essential response regulator of the MtrAB two-component system, regulates the transcription of resuscitation-promoting factor B of Mycobacterium tuberculosis. Microbiology (Reading) 161:1271–1281. doi: 10.1099/mic.0.000087. [DOI] [PubMed] [Google Scholar]
- 18.Fol M, Chauhan A, Nair NK, Maloney E, Moomey M, Jagannath C, Madiraju MV, Rajagopalan M. 2006. Modulation of Mycobacterium tuberculosis proliferation by MtrA, an essential two-component response regulator. Mol Microbiol 60:643–657. doi: 10.1111/j.1365-2958.2006.05137.x. [DOI] [PubMed] [Google Scholar]
- 19.Purushotham G, Sarva KB, Blaszczyk E, Rajagopalan M, Madiraju MV. 2015. Mycobacterium tuberculosis oriC sequestration by MtrA response regulator. Mol Microbiol 98:586–604. doi: 10.1111/mmi.13144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Möker N, Brocker M, Schaffer S, Krämer R, Morbach S, Bott M. 2004. Deletion of the genes encoding the MtrA–MtrB two-component system of Corynebacterium glutamicum has a strong influence on cell morphology, antibiotics susceptibility and expression of genes involved in osmoprotection. Mol Microbiol 54:420–438. doi: 10.1111/j.1365-2958.2004.04249.x. [DOI] [PubMed] [Google Scholar]
- 21.Brocker M, Mack C, Bott M. 2011. Target genes, consensus binding site, and role of phosphorylation for the response regulator MtrA of Corynebacterium glutamicum. J Bacteriol 193:1237–1249. doi: 10.1128/JB.01032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang P, Wu L, Zhu Y, Liu M, Wang Y, Cao G, Chen X-L, Tao M, Pang X. 2017. Deletion of MtrA inhibits cellular development of Streptomyces coelicolor and alters expression of developmental regulatory genes. Front Microbiol 8:2013. doi: 10.3389/fmicb.2017.02013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodrigues DF, Ivanova N, He Z, Huebner M, Zhou J, Tiedje JM. 2008. Architecture of thermal adaptation in an Exiguobacterium sibiricum strain isolated from 3-million-year-old permafrost: a genome and transcriptome approach. BMC Genomics 9:547–517. doi: 10.1186/1471-2164-9-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hingston P, Chen J, Allen K, Truelstrup Hansen L, Wang S. 2017. Strand specific RNA-sequencing and membrane lipid profiling reveals growth phase-dependent cold stress response mechanisms in Listeria monocytogenes. PLoS One 12:e0180123. doi: 10.1371/journal.pone.0180123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boutte CC, Baer CE, Papavinasasundaram K, Liu W, Chase MR, Meniche X, Fortune SM, Sassetti CM, Ioerger TR, Rubin EJ. 2016. A cytoplasmic peptidoglycan amidase homologue controls mycobacterial cell wall synthesis. Elife 5:e14590. doi: 10.7554/eLife.14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shayanfar S, Broumand A, Pillai S. 2018. Acid stress induces differential accumulation of metabolites in Escherichia coli O26: H11. J Appl Microbiol 125:1911–1919. doi: 10.1111/jam.14081. [DOI] [PubMed] [Google Scholar]
- 27.Giotis ES, Muthaiyan A, Blair IS, Wilkinson BJ, McDowell DA. 2008. Genomic and proteomic analysis of the alkali-tolerance response (AlTR) in Listeria monocytogenes 10403S. BMC Microbiol 8:102–111. doi: 10.1186/1471-2180-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giotis ES, Blair IS, McDowell DA. 2007. Morphological changes in Listeria monocytogenes subjected to sublethal alkaline stress. Int J Food Microbiol 120:250–258. doi: 10.1016/j.ijfoodmicro.2007.08.036. [DOI] [PubMed] [Google Scholar]
- 29.Koerner RJ, Goodfellow M, Jones AL. 2009. The genus Dietzia: a new home for some known and emerging opportunist pathogens. FEMS Immunol Med Microbiol 55:296–305. doi: 10.1111/j.1574-695X.2008.00513.x. [DOI] [PubMed] [Google Scholar]
- 30.Wang W, Cai B, Shao Z. 2014. Oil degradation and biosurfactant production by the deep sea bacterium Dietzia maris As-13-3. Front Microbiol 5:711. doi: 10.3389/fmicb.2014.00711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayilraj S, Suresh K, Kroppenstedt R, Saini H. 2006. Dietzia kunjamensis sp. nov., isolated from the Indian Himalayas. Int J Syst Evol Microbiol 56:1667–1671. doi: 10.1099/ijs.0.64212-0. [DOI] [PubMed] [Google Scholar]
- 32.Fang H, Xu JB, Nie Y, Wu XL. 2021. Pan-genomic analysis reveals that the evolution of Dietzia species depends on their living habitats. Environ Microbiol 23:861–877. doi: 10.1111/1462-2920.15176. [DOI] [PubMed] [Google Scholar]
- 33.Click RE, Van Kampen CL. 2010. Assessment of Dietzia subsp. C79793-74 for treatment of cattle with evidence of paratuberculosis. Virulence 1:145–155. doi: 10.4161/viru.1.3.10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X-B, Chi C-Q, Nie Y, Tang Y-Q, Tan Y, Wu G, Wu X-L. 2011. Degradation of petroleum hydrocarbons (C6–C40) and crude oil by a novel Dietzia strain. Bioresour Technol 102:7755–7761. doi: 10.1016/j.biortech.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 35.Fang H, Qin X-Y, Zhang K-D, Nie Y, Wu X-L. 2018. Role of the group 2 Mrp sodium/proton antiporter in rapid response to high alkaline shock in the alkaline-and salt-tolerant Dietzia sp. DQ12-45-1b. Appl Microbiol Biotechnol 102:3765–3777. doi: 10.1007/s00253-018-8846-3. [DOI] [PubMed] [Google Scholar]
- 36.Al Zayer M, Stankowska D, Dziedzic R, Sarva K, Madiraju MV, Rajagopalan M. 2011. Mycobacterium tuberculosis mtrA merodiploid strains with point mutations in the signal-receiving domain of MtrA exhibit growth defects in nutrient broth. Plasmid 65:210–218. doi: 10.1016/j.plasmid.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plocinska R, Purushotham G, Sarva K, Vadrevu IS, Pandeeti EV, Arora N, Plocinski P, Madiraju MV, Rajagopalan M. 2012. Septal localization of the Mycobacterium tuberculosis MtrB sensor kinase promotes MtrA regulon expression. J Biol Chem 287:23887–23899. doi: 10.1074/jbc.M112.346544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemmer KC, Alberge F, Myers KS, Dohnalkova AC, Schaub RE, Lenz JD, Imam S, Dillard JP, Noguera DR, Donohue TJ. 2020. The NtrYX two-component system regulates the bacterial cell envelope. mBio 11:e00957-20. doi: 10.1128/mBio.00957-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Typas A, Banzhaf M, Gross CA, Vollmer W. 2011. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eraso JM, Markillie LM, Mitchell HD, Taylor RC, Orr G, Margolin W. 2014. The highly conserved MraZ protein is a transcriptional regulator in Escherichia coli. J Bacteriol 196:2053–2066. doi: 10.1128/JB.01370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reyrat J-M, David M, Batut J, Boistard P. 1994. FixL of Rhizobium meliloti enhances the transcriptional activity of a mutant FixJD54N protein by phosphorylation of an alternate residue. J Bacteriol 176:1969–1976. doi: 10.1128/jb.176.7.1969-1976.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laub MT, Goulian M. 2007. Specificity in two-component signal transduction pathways. Annu Rev Genet 41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 43.Deol P, Vohra R, Saini AK, Singh A, Chandra H, Chopra P, Das TK, Tyagi AK, Singh Y. 2005. Role of Mycobacterium tuberculosis Ser/Thr kinase PknF: implications in glucose transport and cell division. J Bacteriol 187:3415–3420. doi: 10.1128/JB.187.10.3415-3420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chandolia A, Rathor N, Sharma M, Saini NK, Sinha R, Malhotra P, Brahmachari V, Bose M. 2014. Functional analysis of mce4A gene of Mycobacterium tuberculosis H37Rv using antisense approach. Microbiol Res 169:780–787. doi: 10.1016/j.micres.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 45.Sarva K, Satsangi AT, Plocinska R, Madiraju M, Rajagopalan M. 2019. Two-component kinase TrcS complements Mycobacterium smegmatis mtrB kinase mutant. Tuberculosis 116:S107–S113. doi: 10.1016/j.tube.2019.04.017. [DOI] [PubMed] [Google Scholar]
- 46.Rohs PD, Bernhardt TG. 2021. Growth and division of the peptidoglycan matrix. Annu Rev Microbiol 75:315–336. doi: 10.1146/annurev-micro-020518-120056. [DOI] [PubMed] [Google Scholar]
- 47.Egan AJ, Errington J, Vollmer W. 2020. Regulation of peptidoglycan synthesis and remodelling. Nat Rev Microbiol 18:446–460. doi: 10.1038/s41579-020-0366-3. [DOI] [PubMed] [Google Scholar]
- 48.Antignac A, Sieradzki K, Tomasz A. 2007. Perturbation of cell wall synthesis suppresses autolysis in Staphylococcus aureus: evidence for coregulation of cell wall synthetic and hydrolytic enzymes. J Bacteriol 189:7573–7580. doi: 10.1128/JB.01048-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hett EC, Chao MC, Rubin EJ. 2010. Interaction and modulation of two antagonistic cell wall enzymes of mycobacteria. PLoS Pathog 6:e1001020. doi: 10.1371/journal.ppat.1001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen S, Wu Y, Niu H, Sun J, Han X, Zhang L. 2021. Imbalance between peptidoglycan synthases and hydrolases regulated lysis of Lactobacillus bulgaricus in batch culture. Arch Microbiol 203:4571–4578. doi: 10.1007/s00203-021-02433-0. [DOI] [PubMed] [Google Scholar]
- 51.Lange R, Hengge-Aronis R. 1994. The nlpD gene is located in an operon with rpoS on the Escherichia coli chromosome and encodes a novel lipoprotein with a potential function in ceil wall formation. Mol Microbiol 13:733–743. doi: 10.1111/j.1365-2958.1994.tb00466.x. [DOI] [PubMed] [Google Scholar]
- 52.Gomez JE, Bishai WR. 2000. whmD is an essential mycobacterial gene required for proper septation and cell division. Proc Natl Acad Sci USA 97:8554–8559. doi: 10.1073/pnas.140225297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhattacharya D, Kumar A, Panda D. 2017. WhmD promotes the assembly of Mycobacterium smegmatis FtsZ: a possible role of WhmD in bacterial cell division. Int J Biol Macromol 95:582–591. doi: 10.1016/j.ijbiomac.2016.11.056. [DOI] [PubMed] [Google Scholar]
- 54.Van Kessel JC, Hatfull GF. 2007. Recombineering in Mycobacterium tuberculosis. Nat Methods 4:147–152. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 55.Lu S, Nie Y, Tang Y-Q, Xiong G, Wu X-L. 2014. A critical combination of operating parameters can significantly increase the electrotransformation efficiency of a Gram-positive Dietzia strain. J Microbiol Methods 103:144–151. doi: 10.1016/j.mimet.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 56.Liang J-L, JiangYang J-H, Nie Y, Wu X-L. 2016. Regulation of the alkane hydroxylase CYP153 gene in a Gram-positive alkane-degrading bacterium, Dietzia sp. strain DQ12-45-1b. Appl Environ Microbiol 82:608–619. doi: 10.1128/AEM.02811-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alqaisi MR, Mohsin RM, Ahmed TS. 2020. Efficiency of image analysis as a direct method in spore dimensions measurement. Plant Cell Biotechnol Mol Biol 21:57–64. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.02311-22-s0001.pdf, PDF file, 2.9 MB (3MB, pdf)
Data Availability Statement
The transcriptomic raw sequence has been deposited to National Microbiology Data Center under accession number NMDC10018150.