Abstract
Nucleic acid amplification was performed for five loci in the cag pathogenicity island (PAI) of Helicobacter pylori (comprising cagA, the cagA promoter region, cagE, cagT, and the left end of cagII [LEC]), and gastric inflammation in patients was evaluated. Of 204 H. pylori isolates from Japanese patients (53 with peptic ulcer, 55 with gastric cancer, and 96 with chronic gastritis), 197 (96.6%) were positive for all five loci. Two isolates (1%) were negative for all five loci, and five isolates (2.4%) were positive for only cagA and LEC. These latter seven isolates were all from patients with mild chronic gastritis. Neutrophil infiltration in gastric mucosa was significantly milder in patients infected with partially or totally deleted-PAI strains than in those with intact-PAI strains. The cagE gene was a more accurate marker of an intact cag PAI than the cagA gene, and cagE seemed to be more useful in discriminating between H. pylori strains causing different rates of disease progression.
Helicobacter pylori is a gram-negative, spiral-shaped, microaerophilic bacterium that infects human gastric mucosa and is recognized as a major cause of chronic active gastritis, peptic ulcer disease, gastric adenocarcinoma, and gastric mucosa-associated lymphoid tissue lymphoma 10, 16, 17, 18, 20, 23, 33. Although the pathogenesis of H. pylori infection is not well understood, there are several putative virulence factors that may contribute to mucosal damage by H. pylori infection.
The cytotoxin-associated-gene (cag) pathogenicity island (PAI) is an approximately 40-kb cluster of genes in the H. pylori chromosome 4, 29 and is divided into two regions, cagI and cagII. There are at least 14 and 16 open reading frames (ORFs) in cagI and cagII, respectively. Some of the ORFs in the cag PAI are believed to encode proteins which have similarities to other bacterial secretion systems, such as the Bordetella pertussis toxin secretion system 4.
The cag PAI is considered to be one of the major virulence factors of H. pylori 4. Extensive studies of the cagA gene, located in the most downstream portion of the cag PAI, have indicated that the CagA protein is associated with peptic ulcer disease, gastric cancer, and mucosa-associated lymphoid tissue lymphoma in the stomach 3, 5, 11, 13, 14, 22, 24, 25, 27, 30, 32. Blaser et al. 3 revealed that CagA antibodies were more frequently detected in H. pylori-infected patients with gastric cancer than in those without gastric cancer (odds ratio, 1.9). Furthermore, Parsonnet et al. 24 showed that subjects infected with H. pylori who had CagA antibodies were more likely to develop gastric cancer as compared with uninfected subjects (odds ratio, 5.8), while H. pylori-infected subjects without CagA antibodies were at only slightly and not significantly increased risk for cancer (odds ratio, 2.2). Thus, the cagA gene is conventionally used as a marker of pathogenic strains. However, several studies suggest that the cagA gene cannot be used as a suitable marker for cag PAI-associated virulence for the following reasons: (i) although cag PAI-intact H. pylori strains are shown to induce interleukin-8 secretion from gastric epithelial cells 1, 4, 6, 7, 15, 26, an inactivation of some cag PAI genes such as cagE but not cagA causes a marked reduction in the ability of H. pylori to induce interleukin-8 induction 1, 4, 8, 21, 31; (ii) we have previously shown that some Japanese strains obtained from patients with nonulcer dyspepsia lack most of the cag PAI genes, including the promoter region of the cagA gene, despite the presence of the cagA gene itself, indicating that the presence of the cagA gene does not always signify the presence of an intact cag PAI and an ability to produce CagA protein 15. Although recent studies have revealed that the CagA protein is translocated into the host cells and tyrosine phosphorylated, the precise role of the CagA protein in H. pylori pathogenesis is still unknown 2, 19, 28. These findings may suggest that a gene other than cagA can be used as a marker for cag PAI-associated virulence.
By using our recombinant CagA protein and antibodies, we previously showed a high prevalence of CagA-producing H. pylori strains in Japan 13, 14. Furthermore, by using Southern blot hybridization with DNA probes obtained by cloning 15 different ORFs in cag PAI, we clarified the details of DNA structure in the entire cag PAI 15 and found that Japanese strains deficient in the cagA gene lacked most of the cag PAI genes, including the cagE gene and the promoter region of the cagA gene. However, it is troublesome to check all cag PAI genes by Southern blotting.
Thus, in the present study, we attempted to establish a simple and practical method for determining the structure of cag PAI and consequently discriminating between Japanese H. pylori strains causing different rates of disease progression.
A total of 204 H. pylori isolates was obtained from H. pylori-infected adults who had undergone upper gastrointestinal endoscopy at Tokyo University Hospital. The patients consisted of 145 men and 59 women with a mean age of 58.5 years (ranging from 22 to 85 years). Patient endoscopic findings were as follows: gastric cancer in 55 patients, gastric ulcer in 22, duodenal ulcer in 17, both gastric and duodenal ulcers in 14, and chronic gastritis in 96.
Gastric biopsy specimens were cultured on Columbia agar with 5% (vol/vol) horse blood and Dent antibiotic supplement (Oxoid, Basingstoke, United Kingdom) at 37°C for 5 days under microaerobic conditions (CampyPak System; BBL, Cockeysville, Md.). Organisms were identified as H. pylori by colony morphology and gram staining, as well as positive activity for urease, catalase, and oxidase. The isolates were stored at −80°C in brucella broth with 5% (vol/vol) fetal bovine serum containing 16% (vol/vol) glycerol. DNA was prepared as described previously 15.
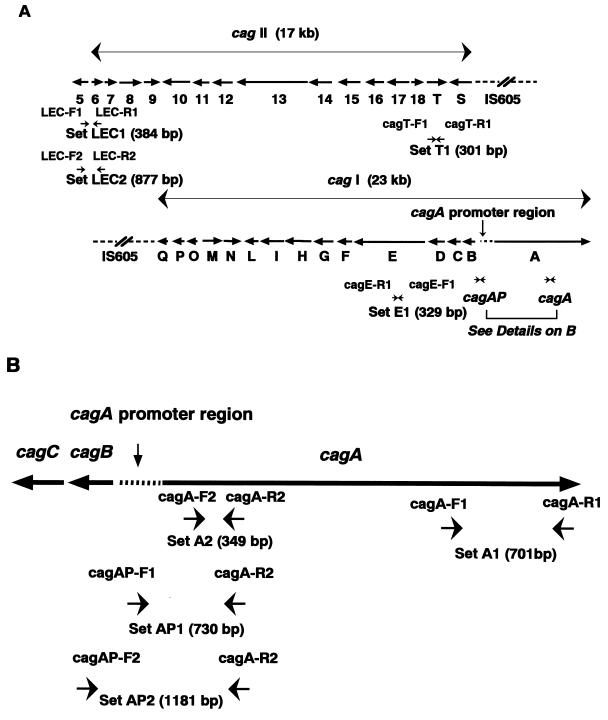
Five different loci allowing for structure screening of cag PAI were selected on the basis of our previous Southern blot analysis 15. The previous study revealed that when the ORFs of cag PAI were deleted, the deletions started from the region between cagA and the cagA promoter region through cagQ (cagI) and continued from cagS through cag-13 or cag-8 (cagII) 15. Thus, cagA, the cagA promoter region, and cagE were selected to represent cagI, and cagT and the left end of cagII (LEC) were selected to represent cagII. Therefore, overall five loci were selected.
Pairs of oligonucleotide primers were used to detect the presence of the cag PAI genes cagA, the cagA promoter region, cagE, cagT, and the LEC, containing both inside and outside genes of cag PAI, and these primer pairs were designed on the basis of published sequences reported by Censini et al. (GenBank accession number, U60176), Akopyants et al. (GenBank accession number, AC000108), Tomb et al. (GenBank accession number, AE000511), and ourselves (GenBank accession number, AF001357) (Table 1; Fig. 1). As shown in Fig. 1A and B, two sets of primers were used to detect the cagA gene (sets A1 and A2), the cagA promoter region (sets AP1 and AP2), and the LEC (sets LEC1 and LEC2). To detect cagE and cagT, one set of primers was used for each gene, set E1 and set T1, respectively. H. pylori strains ATCC 43526 and 43579, which have been determined to have the entire cag PAI 15, were used as positive controls for each PCR. Since eight cagA gene-negative strains from Western countries, including Tx30a, kindly provided by J. C. Atherton (Nottingham University, United Kingdom), were determined to lack the entire cag PAI 15, these were used as negative controls. The genomic DNAs from other bacterial species—Escherichia coli, Pseudomonas aeruginosa, Serratia marcescens, Haemophilus influenzae, Streptococcus pneumoniae, Campylobacter fetus, Campylobacter jejuni, Klebsiella pneumoniae, Citrobacter freundii, and Enterobacter aerogenes—were tested using each primer set to assess the specificity of each PCR.
TABLE 1.
Sequences and locations of oligonucleotide primers
| Primer | Primer sequence | Locationa |
|---|---|---|
| cagA-F1 | 5′-AACAGGACAAGTAGCTAGCC-3′ | 2,700–2,719* |
| cagA-F2 | 5′-GATAACAGGCAAGCTTTTGA-3′ | 157–176* |
| cagA-R1 | 5′-TATTAATGCGTGTGTGGCTG-3′ | 3,400–3,381* |
| cagA-R2 | 5′-CTGCAAAAGATTGTTTGGCAGA-3′ | 505–484* |
| cagE-F1 | 5′-GCGATTGTTATTGTGCTTGTAG-3′ | 16,891–16,870† |
| cagE-R1 | 5′-GAAGTGGTTAAAAAATCAATGCCCC-3′ | 16,563–16,587† |
| cagT-F1 | 5′-CCATGTTTATACGCCTGTGT-3′ | 442–461† |
| cagT-R1 | 5′-CATCACCACACCCTTTTGAT-3′ | 723–742† |
| cagAP-F1 | 5′-GTGGGTAAAAATGTGAATCG-3′ | 18,738–18,757† |
| cagAP-F2 | 5′-CTACTTGTCCCAACCATTTT-3′ | 18,495–18,514† |
| LEC-F1 | 5′-ACATTTTGGCTAAATAAACGCTG-3′ | 3,920–3,942‡ |
| LEC-F2 | 5′-ATAGCGTTTTGTGCATAGAA-3′ | 3,856–3,875‡ |
| LEC-R1 | 5′-TCTCCATGTTGCCATTATGCT-3′ | 4,303–4,283‡ |
| LEC-R2 | 5′-ATCTTTAGTCTCTTTAGCTT-3′ | 4,732–4,713‡ |
FIG. 1.
Structure of cag pathogenicity island and locations of PCR primers. (A) Locations of PCR primers for cagE, cagT, and the LEC. Two sets of primers were used to detect the cagA gene: primer set A1 (cagA-F1 and cagA-R1) and set A2 (cagA-F2 and cagA-R2). For detection of the promoter region of cagA, primer sets AP1 (cagAP-F1 and cagA-R2) and AP2 (cagAP-F2 and cagA-R2) were used, and for the left end of cagII, primer sets LEC1 (LEC-F1 and LEC-R1) and LEC2 (LEC-F2 and LEC-R2) were used. The expected lengths of PCR products using each primer set are shown. The sequence and location of each primer are shown in Table 1. (B) Locations of PCR primers for the cagA gene and its promoter region. The expected lengths of PCR products using each primer set are also shown.
For histological analysis, biopsy specimens from corpus and antrum were embedded in paraffin, stained with hematoxylin and eosin, and examined by two pathologists blinded to the patient's clinical diagnosis or characteristics of the H. pylori strain. The presence of chronic active gastritis was determined by scoring the following parameters on the basis of the updated Sydney System 9: density of inflammatory infiltration (0 to 3) and density of neutrophil infiltration (0 to 3). For each parameter, 0 is none, 1 is mild, 2 is moderate, and 3 is severe.
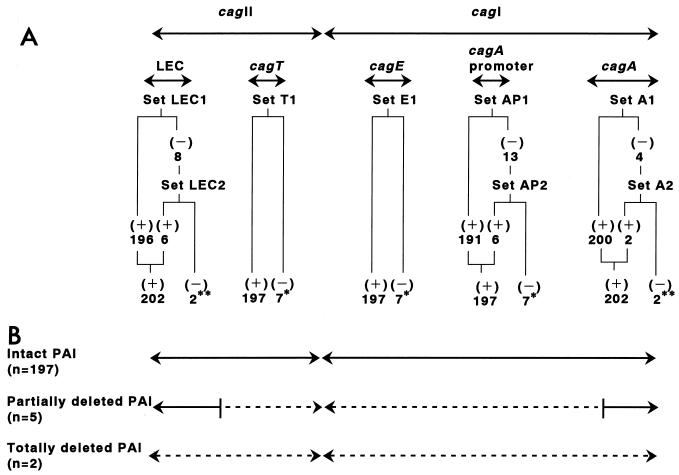
PCR amplification specificity for cagA, the cagA promoter region, cagE, cagT, and the LEC was assessed by testing H. pylori strains ATCC 43526 and 43579 and eight cagA gene-negative strains from Western countries, as well as 10 other bacterial species. Only H. pylori strains ATCC 43526 and 43579 were positive for PCR amplification of all five loci. Eight cagA gene-negative Western strains and the other bacterial species tested were all negative for PCR of all five loci. Thus, the specificity of PCR for each PAI locus was 100%.
To assess the sensitivity of PCR for each locus, excluding the cagA promoter region, PCR was performed with 30 H. pylori isolates from Japanese patients whose cag PAI gene status was determined by Southern blot analysis in our previous study 15. PCR results for cagA (primer sets A1 and A2), cagE (primer set E1), cagT (primer set E1), and the LEC (primer sets LEC1 and LEC2) were completely consistent with those of previous Southern blot analyses 15. Thus, if at least two sets of primers were used, the specificity of PCR for each PAI locus was 100%.
As shown in Fig. 2 and Table 2, 202 out of 204 (99.0%) isolates were positive for cagA and LEC, and 197 out of 204 (96.6%) isolates were positive for the cagA promoter region, cagE, and cagT. Since two cagA-negative strains were also negative for all other genes tested and the remaining five out of seven cagA promoter-negative strains were negative for cagE and cagT, the cag PAI genes present in Japanese H. pylori isolates were divided into three types; intact-PAI, partially deleted-PAI, and totally deleted-PAI genes (Fig. 2).
FIG. 2.
Results of cag PAI gene PCR in 204 Japanese H. pylori isolates. (A) Positivity of cag PAI gene PCR by each primer set: ∗, seven cagA promoter PCR-negative isolates also negative for both cagE and cagT PCR; ∗∗, two isolates negative for LEC PCR and also negative for cagA PCR. (B) Types of cag PAI structures determined in this study.
TABLE 2.
Relationship between presence of cag PAI genes and clinical diagnosis
| Diagnosis (total patients) | Patients with cag type infection:
|
||
|---|---|---|---|
| Intact PAI | Partially deleted PAI | Totally deleted PAI | |
| Peptic ulcer disease (53) | 53 | 0 | 0 |
| Gastric cancer (55) | 55 | 0 | 0 |
| Chronic gastritis (96) | 89 | 5 | 2 |
| Total (204) | 197 | 5 | 2 |
Recently, Jenk et al. reported that the presence of the entire cag PAI is highly related to duodenal ulcers but that the clinical outcome of H. pylori infection is not reliably predicted by analyzing several genes of the cag PAI, including cagA, cagE, and cagT 12. In their study, the presence of cagE was completely consistent with that of cagA but not cagT. In contrast, our study revealed consistency in the presence of cagE with cagT but not cagA, indicating that the strain diversity may exist in relation to cag PAI genes among Western countries and Japan.
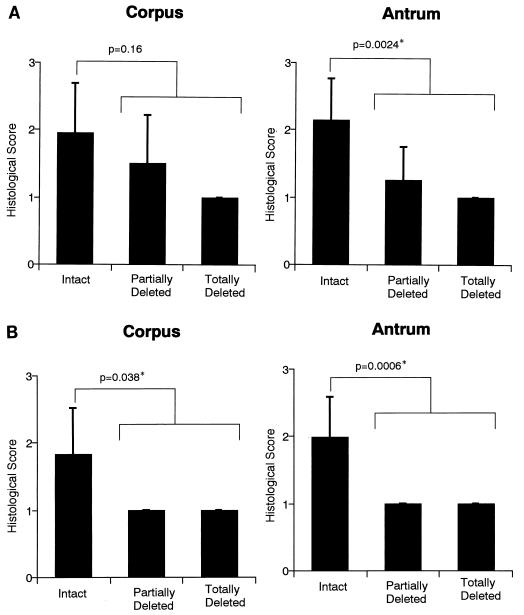
Strains with partially or totally deleted cag PAIs, which lack both cagE and cagT, were more frequently found in more patients with chronic gastritis only (7 out of 96 patients [7.3%]) than with peptic ulcer disease (0 out of 53; P = 0.042) or with gastric cancer (0 out of 55; P = 0.039) (Table 2). Furthermore, by assessing inflammation activity in the gastric mucosa of 64 patients (59 infected with intact-PAI-type strains, 4 with partially deleted-PAI-type strains, and 1 with a totally deleted-PAI-type strain), we found no significant differences in inflammatory infiltration of corpus between patients with intact type strains and those with partially or totally deleted type strains. However, inflammatory infiltration in antrum (Fig. 3A) and neutrophil infiltration in corpus and antrum (Fig. 3B) were significantly milder in patients with partially or totally deleted type strains than in those with intact type strains. These findings suggest that the strains with partially or totally deleted PAI may have weaker ability to cause disease progression than those with intact PAI.
FIG. 3.
Relationship between gastric inflammation and the presence of cag PAI genes of infected H. pylori strains. A total of 64 patients (59 infected with strains with intact type, 4 with partially deleted type, and 1 with totally deleted type) were assessed for (A) inflammatory infiltration and (B) neutrophil infiltration in corpus and antral mucosa. Each valuable was scored on a four-point histological scale (0, none; 1, mild; 2, moderate; and 3, severe). ∗, statistically significant by Mann-Whitney U test.
In the present study, the partially or totally deleted type strains in Japan lacked cagE, cagT, and the cagA gene promoter region, regardless of the presence of the cagA gene itself. Therefore, they could be discriminated from intact type strains by detection of cagE, cagT, or the cagA gene promoter region but not by detection of the cagA gene itself. Although the cagA gene is conventionally used as a marker for virulence, especially with the PCR amplification method, our results indicate that not the cagA gene itself but the promoter region of the cagA gene could be a better marker. However, due to the diversity of the cagA gene promoter region sequences, designing specific primers to detect this region may be difficult. Since the cagE gene is located near the cagA gene promoter region and retained consistently within this region, it seems valid to choose the cagE gene as a substitute for the cagA gene promoter region. Since the primer sets designed for cagE PCR in this study were extremely specific and sensitive, at least for Japanese strains, we conclude that cagE PCR can be used as a practical method for screening the status of the cag PAI structure, which may be related to disease progression, for a large number of samples in order to test clinical significance or to conduct an epidemiological survey.
In conclusion, the results of the present study indicate that cagE is more accurate, as a marker of an intact cag PAI, than the cagA gene and that it seems to be more useful in discriminating between H. pylori strains with different rates of disease progression in Japan. Detection of the cagE gene by PCR amplification with specific primers can be used as a simple and practical method for their discrimination.
REFERENCES
- 1.Akopyants N S, Clifton S W, Kersulyte D, Crabtree J E, Youree B E, Reece C A, Bukanov N O, Drazek E S, Roe B A, Berg D E. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol Microbiol. 1998;28:37–53. doi: 10.1046/j.1365-2958.1998.00770.x. [DOI] [PubMed] [Google Scholar]
- 2.Asahi M, Azuma T, Ito S, Suto H, Nagai Y, Tsubokawa M, Tohyama Y, Maeda S, Omata M, Suzuki T, Sasakawa C. Helicobacter pylori CagA protein can be tyrosine phosphorylated in gastric epithelial cells. J Exp Med. 2000;191:593–602. doi: 10.1084/jem.191.4.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaser M J, Perez-Perez G I, Kleanthous H, Cover T L, Peek R M, Chyou P H, Stemmermann G N, Nomura A. Infection with Helicobacter pylori strains possessing cagA associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55:2111–2115. [PubMed] [Google Scholar]
- 4.Censini S, Lange C, Xiang Z, Crabtree J E, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type-specific and disease associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N, Rappuoli R. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA. 1993;90:5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crabtree J E, Farmery S M, Lindley I J, Figura N, Peichl P, Tompkins D S. CagA/cytotoxic strains of Helicobacter pylori and interleukin-8 in gastric epithelial cell lines. J Clin Pathol. 1994;47:945–950. doi: 10.1136/jcp.47.10.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crabtree J E, Covacci A, Farmery S M, Xiang Z, Tompkins D S, Perry S, Lindley I J, Rappuoli R. Helicobacter pylori induced interleukin-8 expression in gastric epithelial cells is associated with CagA positive phenotype. J Clin Pathol. 1995;48:41–45. doi: 10.1136/jcp.48.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crabtree J E, Xiang Z, Lindley I J, Tompkins D S, Rappuoli R, Covacci A. Induction of interleukin-8 secretion from gastric epithelial cells by a cagA negative isogenic mutant of Helicobacter pylori. J Clin Pathol. 1995;48:967–969. doi: 10.1136/jcp.48.10.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixon M F, Genta R M, Yardley J H, Correa P. Classification and grading of gastritis: the updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol. 1996;20:1161–1181. doi: 10.1097/00000478-199610000-00001. [DOI] [PubMed] [Google Scholar]
- 10.Graham D Y, Lew G M, Klein P D, Evans D G, Evans D J, Saeed Z A, Malaty H M. Effect of treatment of Helicobacter pylori infection on the long-term recurrence of gastric or duodenal ulcer. Ann Intern Med. 1992;116:705–708. doi: 10.7326/0003-4819-116-9-705. [DOI] [PubMed] [Google Scholar]
- 11.Graham D Y, Genta R M, Graham D P, Crabtree J E. Serum CagA antibodies in asymptomatic subjects and patients with peptic ulcer: lack of correlation of IgG antibody in patients with peptic ulcer or asymptomatic Helicobacter pylori gastritis. J Clin Pathol. 1996;49:829–832. doi: 10.1136/jcp.49.10.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenks P J, Mégraud F, Labigne A. Clinical outcome after infection with Helicobacter pylori does not appear to be reliably predicted by the presence of any of the genes of the cag pathogenicity island. Gut. 1998;43:752–758. doi: 10.1136/gut.43.6.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maeda S, Kanai F, Ogura K, Yoshida H, Ikenoue T, Takahashi M, Kawabe T, Shiratori Y, Omata M. High seropositivity of anti-CagA antibody in Helicobacter pylori-infected patients irrelevant to peptic ulcers and normal mucosa in Japan. Dig Dis Sci. 1997;42:1841–1847. doi: 10.1023/a:1018846723379. [DOI] [PubMed] [Google Scholar]
- 14.Maeda S, Ogura K, Yoshida H, Kanai F, Ikenoue T, Kato N, Shiratori Y, Omata M. Major virulence factors, VacA and CagA, are commonly positive in Helicobacter pylori isolates in Japan. Gut. 1998;42:338–343. doi: 10.1136/gut.42.3.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maeda S, Yoshida H, Ikenoue T, Ogura K, Kanai F, Kato N, Shiratori Y, Omata M. Structure of cag pathogenicity island in Japanese Helicobacter pylori isolates. Gut. 1998;44:336–341. doi: 10.1136/gut.44.3.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marshall B J, Warren J R. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;i:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 17.Miehlke S, Kibler K, Kim J G, Figura N, Small S M, Graham D Y, Go M F. Allelic variation in cagA gene of Helicobacter pylori obtained from Korea compared to the United States. Am J Gastroenterol. 1996;91:1322–1325. [PubMed] [Google Scholar]
- 18.Nomura A, Stemmermann G N, Chyou P H, Kato I, Perez-Perez G I, Blaser M J. Helicobacter pylori infection and gastric carcinoma in a population of Japanese-Americans in Hawaii. N Engl J Med. 1991;325:1132–1136. doi: 10.1056/NEJM199110173251604. [DOI] [PubMed] [Google Scholar]
- 19.Odenbreit S, Puls J, Sedlmaier B, Gerland E, Fischer W, Haas R. Translocation of Helicobacter pylori CagA protein into gastric epithelial cells by type IV secretion. Science. 2000;287:1497–1500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 20.Ogura K, Kanai F, Maeda S, Yoshida H, Ogura M, Lan K H, Hirota K, Kawabe T, Shiratori Y, Omata M. High prevalence of cytotoxin-positive Helicobacter pylori in patients irrelevant to the presence of peptic ulcers in Japan. Gut. 1997;41:463–468. doi: 10.1136/gut.41.4.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogura K, Takahashi M, Maeda S, Ikenoue T, Kanai F, Yoshida H, Shiratori Y, Mori K, Mafune K, Omata M. Interleukin-8 production in primary cultures of human gastric epithelial cells induced by Helicobacter pylori. Dig Dis Sci. 1998;43:2738–2743. doi: 10.1023/a:1026671815512. [DOI] [PubMed] [Google Scholar]
- 22.Pan Z J, van der Hulst R W M, Feller M, Xiao S D, Tytgat G N J, Dankert J, van der Ende A. Equally high prevalences of infection with cagA-positive Helicobacter pylori in Chinese patients with peptic ulcer disease and those with chronic gastritis-associated dyspepsia. J Clin Microbiol. 1997;35:1344–1347. doi: 10.1128/jcm.35.6.1344-1347.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parsonnet J, Friedman G D, Vandersteen D P, Chang Y, Vogelman J H, Orentreich N, Richard K S. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 24.Parsonnet J, Friedman G D, Orentreich N, Vogelman H. Risk for gastric cancer in people with CagA positive or CagA negative Helicobacter pylori infection. Gut. 1997;40:297–301. doi: 10.1136/gut.40.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peek R M, Miller G G, Tham K T, Perez-Perez G I, Zhao X M, Atherton J C, Blaser M J. Heightened inflammatory response and cytokine expression in vivo cagA+Helicobacter pylori strains. Lab Invest. 1995;71:760–770. [PubMed] [Google Scholar]
- 26.Sharma S A, Tummuru M K R, Miller G G, Blaser M J. Interleukin-8 response of gastric epithelial cell lines to Helicobacter pylori stimulation in vitro. Infect Immun. 1995;63:1681–1687. doi: 10.1128/iai.63.5.1681-1687.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimoyama T, Fukuda S, Tanaka M, Minami T, Saito Y, Munakata A. High prevalence of the CagA-positive Helicobacter pylori strains in Japanese asymptomatic patients and gastric cancer patients. Scand J Gastroenterol. 1997;32:465–468. doi: 10.3109/00365529709025082. [DOI] [PubMed] [Google Scholar]
- 28.Stein M, Rappuoli R, Covacci A. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc Natl Acad Sci USA. 2000;97:1263–1268. doi: 10.1073/pnas.97.3.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenny K, Fitzegerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;338:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 30.Tummuru M K R, Cover T L, Blaser M J. Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori: evidence of linkage to cytotoxin production. Infect Immun. 1993;61:1799–1809. doi: 10.1128/iai.61.5.1799-1809.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tummuru M K R, Sharma S A, Blaser M J. Helicobacter pylori picB, a homologue of the Bordetella pertussis toxin secretion protein, is required for induction of IL-8 in gastric epithelial cells. Mol Microbiol. 1995;18:867–876. doi: 10.1111/j.1365-2958.1995.18050867.x. [DOI] [PubMed] [Google Scholar]
- 32.Weel J E L, van der Hulst R W M, Gerrits Y, Roorda P, Feller M, Dankert J, Tytgat G N J, van der Ende A. The interrelationship between cytotoxin-associated gene A, vacuolating cytotoxin, and Helicobacter pylori-related diseases. J Infect Dis. 1996;173:1171–1175. doi: 10.1093/infdis/173.5.1171. [DOI] [PubMed] [Google Scholar]
- 33.Wotherspoon A C, Doglioni C, Diss T C, Pan L, Moschini A, de Boni M, Isaacson P G. Regression of primary low-grade B-cell gastric lymphoma of mucosa-associated lymphoid tissue type after eradication of Helicobacter pylori. Lancet. 1993;342:575–577. doi: 10.1016/0140-6736(93)91409-f. [DOI] [PubMed] [Google Scholar]