Multimodal single-cell profiling reveals clonal architecture and genetic drivers in the progression of MDS to AML.
Abstract
Clonal evolution in myelodysplastic syndrome (MDS) can result in clinical progression and secondary acute myeloid leukemia (sAML). To dissect changes in clonal architecture associated with this progression, we performed single-cell genotyping of paired MDS and sAML samples from 18 patients. Analysis of single-cell genotypes revealed patient-specific clonal evolution and enabled the assessment of single-cell mutational cooccurrence. We discovered that changes in clonal architecture proceed via distinct patterns, classified as static or dynamic, with dynamic clonal architectures having a more proliferative phenotype by blast count fold change. Proteogenomic analysis of a subset of patients confirmed that pathogenic mutations were primarily confined to primitive and mature myeloid cells, though we also identify rare but present mutations in lymphocyte subsets. Single-cell transcriptomic analysis of paired sample sets further identified gene sets and signaling pathways involved in two cases of progression. Together, these data define serial changes in the MDS clonal landscape with clinical and therapeutic implications.
Significance:
Precise clonal trajectories in MDS progression are made possible by single-cell genomic sequencing. Here we use this technology to uncover the patterns of clonal architecture and clonal evolution that drive the transformation to secondary AML. We further define the phenotypic and transcriptional changes of disease progression at the single-cell level.
See related article by Menssen et al., p. 330 (31).
See related commentary by Romine and van Galen, p. 270.
This article is highlighted in the In This Issue feature, p. 265
INTRODUCTION
For patients with myelodysplastic syndromes (MDS), the transformation to secondary acute myeloid leukemia (sAML) brings poor therapeutic response and a significantly shortened lifespan (1–3). Mutated hematopoietic cells exist within a complex ecosystem, where both intrinsic and extrinsic forces influence the process of clonal evolution (4, 5). This clonal evolution is fundamental to disease progression and therapeutic resistance in MDS and sAML (6–11). Foundational studies using bulk next-generation sequencing have leveraged mutational frequencies and statistical modeling to impute the clonal identity and extract patterns of clonal evolution (10, 12, 13). Whole-genome sequencing data first demonstrated that approximately 85% of bone marrow cells in seven MDS patients were clonal and that at least one new pathogenic mutation accompanied progression to sAML (8). In each of the cases reported, clonal evolution occurred through the linear acquisition and expansion of new mutant subclones. A subsequent study investigated 11 patients with samples at MDS and sAML by whole-exome sequencing combined with a larger cohort of 44 MDS-to-sAML samples analyzed via targeted sequencing (12). This analysis demonstrated that clonal evolution could occur through either the linear evolution of subclones or branching evolution, with clonal competition from parallel subclones (12). A recent review summarized these and other published studies comparing the genetics of paired MDS and sAML samples and confirmed that linear and branching patterns occur (14). Furthermore, they revealed that signaling and transcription factor mutations are more abundant at sAML, whereas almost all mutations present in MDS samples persist in sAML. Ultimately, however, bulk DNA sequencing (DNA-seq) is not able to directly confirm cell-specific mutational identities.
Novel approaches using single-cell DNA-seq (scDNA-seq) have enabled precise clonal assignments (13). This technology provides a key advantage by defining mutational presence within single cells of mutationally diverse samples. Single-cell sequencing has already provided insights regarding the heterogeneity of the MDS stem cell pool and the presence of clonal competition within this pool (13). High-throughput scDNA-seq has also recently been leveraged by a number of researchers to investigate clonal evolution in clonal hematopoiesis, as well as myeloid and lymphoid malignancies (15–22). Results from these studies have revealed mutational associations, genotype/phenotype relationships, and the increasing clonal complexity of myeloid malignancies throughout the course of therapy (15, 16, 22). Furthermore, a recent study analyzing a large cohort of AML data has found that clonal heterogeneity, mutational frequency, evolution pattern, and mutational order correlate with clinical outcomes (23). Thus, understanding clonal architecture and change over time could lead to translational impact in myeloid malignancies.
Given these advances in single-cell genotyping and potential clinical impact, we hypothesized that scDNA-seq would better characterize clonal shifts upon MDS progression. Here, we use single-cell genomics to analyze clonal, phenotypic, and transcriptional changes in paired samples of MDS and sAML.
RESULTS
Single-Cell DNA Sequencing of Paired Samples Resolves Mutational Characteristics of MDS and sAML
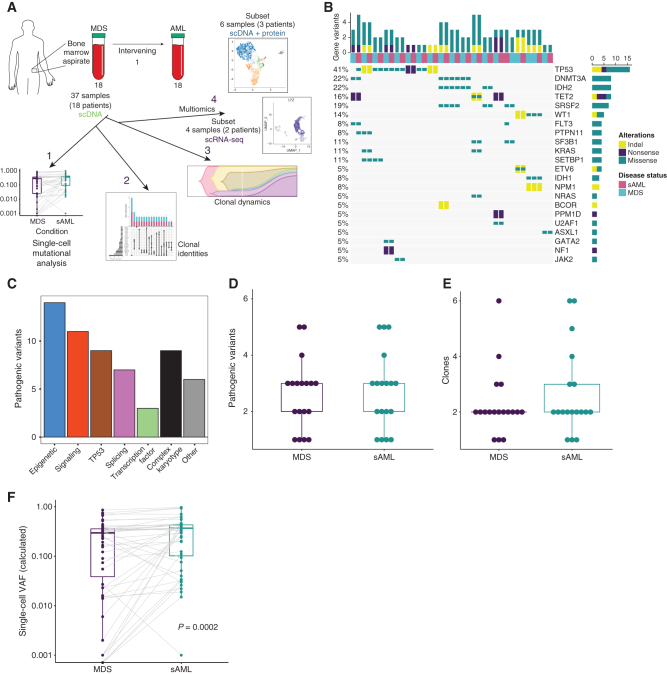
Using an scDNA-seq amplicon panel composed of 45 commonly mutated genes, we analyzed 37 paired longitudinal bone marrow samples from 18 patients (Fig. 1A; Supplementary Fig. S1A). The mutational landscape for all samples is depicted in Fig. 1B, whereas patient characteristics for the cohort are shown in Supplementary Table S1. A total of 136,710 individual cells were genotyped, averaging 3,695 cells per sample (range, 209–9,792 cells/sample) and 169,352 reads per cell (range, 12,016–1,834,736; Supplementary Fig. S1B and S1C; Supplementary Table S2). Single-cell variant allele frequencies (VAF) were overall well correlated with results from bulk-sequencing (R = 0.75, P ≤ 0.0001, Pearson correlation; Supplementary Fig. S1D). Variants in ASXL1 were genotyped at a low frequency by scDNA-seq, which others have observed (18). The calculated median allele dropout (ADO) rate was between 1.9% and 15.8% per sample, with a median of 6.80% overall, consistent with previously published ADO rates for this platform (Supplementary Table S3; refs. 17, 24, 25). We identified 50 pathogenic or likely pathogenic recurrent mutations (44 unique mutations) within 50 clones (Fig. 1B and C; Supplementary Fig. S1C; Supplementary Table S4). As expected, the number of mutations correlated with the number of clones per sample (Supplementary Fig. S1E). We further characterized gene functional categories in our cohort. We found mutations in genes from six functional categories, similar to previously published categories, but with some minor differences in gene composition from these studies (Supplementary Table S5; refs. 26–29). Using functional categories for mutations in myeloid malignancy, 14 of the mutations were epigenetic (which included mutations in DNMT3a, IDH1/2, TET2, ASXL1, and BCOR), 11 were signaling (which included FLT3, JAK2, NF1, NRAS, KRAS, and PTPN11), 9 were in TP53, 7 were splicing (which included U2AF1, SRSF2, and SF3B1), 3 were in transcription factors (which included GATA2 and ETV6), and 6 mutations in genes outside of these categories (which included SETBP1, WT1, and NPM1; Fig. 1C; refs. 14, 26). The number of mutations and clones did not differ between MDS and sAML (Fig. 1D and E). TP53 (18%, 9/50 mutations) and DNMT3A (8%, 4/50) were the most frequently mutated genes in the cohort. All DNMT3A variants were the R882H missense variant, whereas variants in TP53 differed in each instance. Missense mutations accounted for most variants detected by single-cell sequencing (37 total, 31 unique; Fig. 1B). The mean scVAF for identified pathogenic mutations at MDS increased at sAML (24.2% vs. 33.1%, P = 0.0002 by the paired Wilcoxon rank-sum test; Fig. 1F). Within the patient cohort, all but one patient was treated with a DNA methyltransferase inhibitor (DNMTi; 17/18), either decitabine or azacitidine for MDS, but therapy differed thereafter and at sAML (Supplementary Table S1).
Figure 1.
Longitudinal analysis using scDNA-seq of patients progressing from MDS to sAML. A, Illustration depicting the sample workflow. B, Oncoprint of 37 patient samples generated using scDNA-seq data. Each column is a unique sample in the cohort and disease status per legend at right. Mutated genes are listed on the right of the Oncoprint, and each type of alteration is color-coded (indels/yellow, nonsense/purple, missense/teal). The percentage of samples mutated for each gene is listed on the left, and the number of genes with variants per sample is along the top. C, Bar chart depicting the number of mutations in each category in the cohort. D, Box plot indicating the number of pathogenic variants identified at each disease state (for all box plots in this figure, centerline represents the median, box represents the interquartile range (IQR), whiskers, 1.5 × IQR). E, Box plot depicting the number of clones identified at each disease state. F, Box plots representing the calculated VAFs for pathogenic variants at MDS and their respective increase or decrease at sAML. The overall difference in VAFs was calculated using the Wilcoxon rank-sum test (P = 0.0002).
Single-Cell Mutational Identity Defines MDS Clonal Architecture
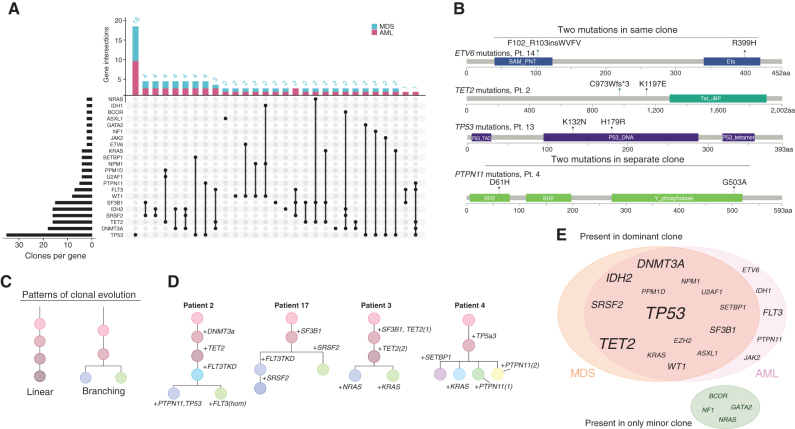
scDNA-seq clarified the cooccurrence of pathogenic variants within individual cells. The two most common intraclonal mutational cooccurrences were IDH2/DNMT3A (three individuals) and IDH2/SRSF2 (three individuals; Fig. 2A; Supplementary Fig. S2A). Other mutational cooccurrences were notable, including both cases of subclonal SETBP1 mutations (patients 4 and 12) coexisting with TP53 mutations. Four patients had two pathogenic mutations in the same gene. Of these, three cooccurred in the same subclone (ETV6, TET2, and TP53), and one was found in two separate subclones (PTPN11; Fig. 2B). Such subclonal competition was common in cases with RAS pathway mutations (classified as PTPN11, KRAS, NRAS, and NF1). Two cases of MDS/sAML harbored at least two distinct subclones with different RAS pathway mutations (patients 3 and 4; Fig. 2C). Competing parallel subclones with RAS pathway mutations were also observed in an independent cohort in six additional RAS family mutants, as well as in previous scDNA investigations of AML (15, 30, 31). The presence of signaling mutations was enriched in samples with branching clonal evolution in our cohort (6/6 cases branching cases with signaling mutations, 0/12 without, Fisher exact test, P = 1e−5; Fig. 2C and D). We further referenced publicly available genomic data from a data set of MDS samples representing 4,231 samples/patients using the cBioPortal to analyze RAS pathway mutation cooccurrence (19, 32, 33). Here, we confirmed statistically significant cooccurrences of two RAS pathway mutational pairs, including NRAS and KRAS (q < 0.001), NRAS and PTPN11 (q < 0.001), and KRAS and PTPN11 (q < 0.038). NRAS and NF1 were mutually exclusive (q = 0.003; data set was queried on KRAS, NRAS, PTPN11, and NF1 mutations; Table 1).
Figure 2.
Single-cell mutational identities define clonal evolution. A, UpSet plot depicting comutational occurrences in MDS and sAML clones. The number of times a gene was involved in a clone is listed on the left side of the plot for the entire cohort. The number of times each gene combination was detected in a clone is listed at the top of the plot. B, Illustration of multiple mutations within the same gene. This phenomenon was observed in four patients, three instances occurring within the same clone and one occurring in different clones. Specific gene residues are listed in each instance. C, Patterns of clonal evolution depicted as phylogenies. D, Phylogenies for samples with a branching evolution and signaling mutations. E, Venn diagram of variants in the dominant clone. Mutations in the genes listed in the green circle occurred only in minor clones at either MDS or sAML. Font size is representative of the number of times mutations were observed in each gene.
Table 1.
Cooccurrence of RAS pathway mutations in MDS
| Gene A | Gene B | Neither | A not B | B not A | Both | Log2 Odds ratio | P value | q value | Tendency |
|---|---|---|---|---|---|---|---|---|---|
| NRAS | KRAS | 3,600 | 447 | 139 | 45 | 1.383 | <0.001 | <0.001 | Cooccurrence |
| NRAS | PTPN11 | 3,566 | 448 | 173 | 44 | 1.018 | <0.001 | <0.001 | Cooccurrence |
| KRAS | PTPN11 | 3,846 | 168 | 201 | 16 | 0.866 | 0.025 | 0.038 | Cooccurrence |
| NRAS | NF1 | 3,062 | 442 | 94 | 3 | −2.177 | 0.001 | 0.003 | Mutual exclusivity |
Overall, the mutational number and heterogeneity did not differ between MDS and sAML. The mean number of mutations per clone was similar between disease states (2.15 and 2.25, respectively; two-tailed Student t test, P = 0.62; Supplementary Fig. S2B), as was the Shannon diversity index of subclones (Supplementary Fig. S2C). We further analyzed mutations in dominant, or most abundant, clones in each case. DNMT3A and TET2 mutations were present in the founding, or earliest detectable clone if founding clone was not identified, except for two cases in which a TET2 mutation occurred following a previous DNMT3A or TET2 mutation (patients 2 and 3; Supplementary Fig. S3A–S3C). When present, DNMT3A and TET2 mutations were always found in the dominant clone in sAML (5/5 cases). TP53 variants appeared in both the dominant and founding clones in 87.5% (7/8) of cases with at least one TP53 mutation. Similarly, variants in TP53 were the most common to occur as the sole variant in a clone, with 18 TP53-only clones detected in the cohort (Fig. 2A). Variants in the genes PTPN11, IDH1, and FLT3 occurred in the dominant clone only upon progression to sAML, though they were found in a small number of cells in the corresponding MDS samples (Fig. 2E; Supplementary Fig. S4).
Progression of MDS Occurs via Distinct Patterns of Clonal Evolution and Architecture Change
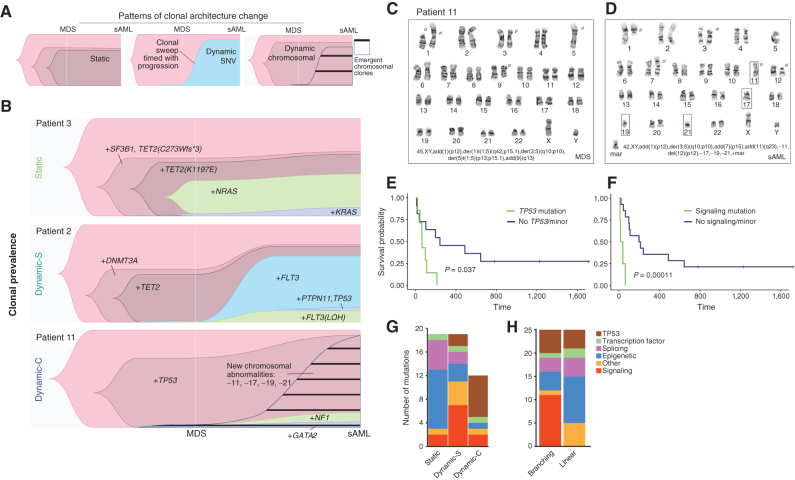
To test the hypothesis that changes in the mutational landscape accompany disease progression, we analyzed the clonal phylogenies and changes from MDS to sAML in each patient. In this analysis, we distinguished between the pattern of clonal evolution, which can be inferred from scDNA-seq data from one sample and articulates the sample's phylogenetic history, and the pattern of clonal architecture change, which is defined by comparison of clonal frequencies between at least two samples (Figs. 2C and 3A). For each case, we analyzed both patterns of clonal evolution and clonal architecture change. We found that most patients (67%, 12/18) displayed linear clonal evolution (defined based on scDNA data), in which only one subclone arises from any previous clone or subclone. The remaining six cases demonstrated branching clonal evolution (Supplementary Fig. S3). With respect to clonal architecture change, we identified three distinct patterns: static and dynamic, which were further classified based on the type of clonal change (SNV or chromosomal; Fig. 3A). Six of 18 patients (33.3%) exhibited a Static clonal architecture, which displayed minimal clonal change (<10% change for any minor clone) and without the emergence of a new dominant clone (e.g., patient 3; Fig. 3B). The second pattern consisted of an emergent dominant clone in sAML (5/18, 27.8%), indicative of a subclonal sweep coinciding with progression (e.g., patient 2; Fig. 3B; ref. 5). Here, we define an emergent dominant subclone as a clone that was small or undetectable in the MDS sample. Given that these variants were detected by scDNA-seq, we call this pattern Dynamic SNV (Dynamic-S). In all Static and all but two Dynamic-S cases, karyotypes were normal or had fewer than three abnormalities (Supplementary Table S1). The third group (38.9%, 7/18) displayed minimal changes in single-cell variant-defined clonal architecture, like the Static group; however, this group was characterized by karyotypes which increased in complexity at sAML, representing a dynamic-chromosomal landscape (e.g., patient 11; Fig. 3B). Thus, we termed this group Dynamic-Chromosomal (Dynamic-C). Though performed on a small number of single cells, clinical karyotypes can identify the subclonal architecture of large genetic changes. Dynamic-C cases demonstrated clonal evolution marked by increasing loss or gain of chromosomal material, including new deletions of partial and whole chromosomes, duplications, and translocations. Rather than small driver mutations (detected via amplicon sequencing) defining the clonal structure, as in the Dynamic-S group, transformation in this pattern coincided with a deteriorating chromosomal landscape, with numerous gains or losses of chromosomal material (karyotype shown for patient 11; Fig. 3C and D; list of changes for all Dynamic-C patients in Supplementary Table S6).
Figure 3.
scDNA-seq characterizes clonal trajectories during disease progression. A, Representative Timescape plots of the three types of clonal progression observed in the cohort. B, Clonal prevalence over time of three patients, each with a distinct pattern of clonal progression from MDS to sAML: Static, Dynamic SNV, and Dynamic Chromosomal, respectively. The leftmost clone in each plot is the parent clone from which all cells were derived (no mutations detected). Karyotypes at (C) MDS and (D) sAML for patient 11 demonstrating dynamic-chromosomal clonal progression. E, Kaplan–Meier curve of sAML survival when the signaling mutation was present in the dominant clone. F, Kaplan–Meier curve for TP53 samples when present in the dominant clone (for both Kaplan–Meier curve, log-rank test P value is shown). G and H, Mutation type distribution per clonal architecture and clonal evolution pattern.
We further investigated the existence of these patterns in MDS progression using data from an independent cohort (31). Among 12 cases that were analyzed with whole-genome sequencing for which a clonal analysis was done, we identified one case of static evolution (UPN 280498), whereas the remaining 11 represented a dynamic change in clonal architecture (6 Dynamic-S and 5 Dynamic-C cases; ref. 31).
Clinical Correlates of Clonal Evolution
There was a significantly higher blast fold change (log2 scale) in the combined Dynamic group (Supplementary Fig. S4A). Blast fold change did not correlate with time to progression, and time between samples was similar among the groups (Supplementary Fig. S4B). A comparison of blast fold change within each group demonstrated a significantly greater blast increase in Dynamic-S compared with Static cases (Supplementary Fig. S4C and S4D). The Dynamic-C group had significantly fewer pathogenic mutations and clones (by scDNA-seq) compared with patients with Static or Dynamic-S clonal evolution (Supplementary Fig. S4E and S4F). The cohort had three patients with treatment-related MDS, 2 of 7 patients in the Dynamic-C group and one patient in the Dynamic-S group. There was no statistically significant difference in survival after sAML among the groups, though median survival for all patients was dismal at only 106 days (Supplementary Fig. S4G). Though survival did not vary based on the clonal architecture change, we hypothesized that the mutational identity of the sAML-dominant clone could correlate with the outcome. In a univariate analysis, the presence of a signaling mutation or the presence of a TP53 mutation in the dominant sAML subclone was found to correlate with sAML survival (Fig. 3E and F). To understand the multivariate effect of these mutations, we calculated a Cox proportional hazards model for death based on age over 60, sex, TP53 status, and IPSS-R. In this model, signaling (HR = 559; 95% CI, 15.6–2 × 104) and TP53 (HR = 15.7; 95% CI, 2–118) mutations in the dominant sAML subclone were both associated with increased risk of death (P = 0.0005 and P = 0.007, respectively; Supplementary Table S7; Supplementary Fig. S5A–S5E; residuals of model). An IPSS-R group of intermediate (I) was associated with decreased risk of death in the model (HR 0.003; 95% CI, 0.00003–0.25 and HR 0.03; 95% CI, 0.003–0.47, respectively; Supplementary Table S7).
Characteristic Genomic Changes Accompany Dynamic Clonal Architecture
We next characterized the genomic alterations of each architectural change or clonal evolution pattern (Fig. 3G and H; Supplementary Fig. S3). The Dynamic-C group was enriched for TP53 mutations (6/7 within the group vs. 2/11 in other groups, Fisher exact test, P = 0.009; Fig. 3G). As the disease progressed, the allele burden of TP53 mutations or deletions increased in all cases (Supplementary Table S6; Supplementary Fig. S3C). Increased TP53 allelic state at the sample level has recently been associated with worse outcomes in MDS; thus, it is not surprising that single-cell TP53 allelic state would increase at progression (34, 35). By contrast, signaling mutations often accompanied Dynamic-S architectural change (36). Here, we observed an enrichment of signaling mutations in the dominant, or largest, subclone of Dynamic-S sAML samples compared with all other samples (3/5 vs. 0/13, Fisher exact test, P = 0.01; Fig. 3G), which included two FLT3 tyrosine kinase domain (TKD) mutations (patients 2 and 17) and a PTPN11 mutation (patient 4; Supplementary Fig. S3B). We also observed a similar enrichment of signaling mutations in cases with branching clonal evolution (Fig. 3H).
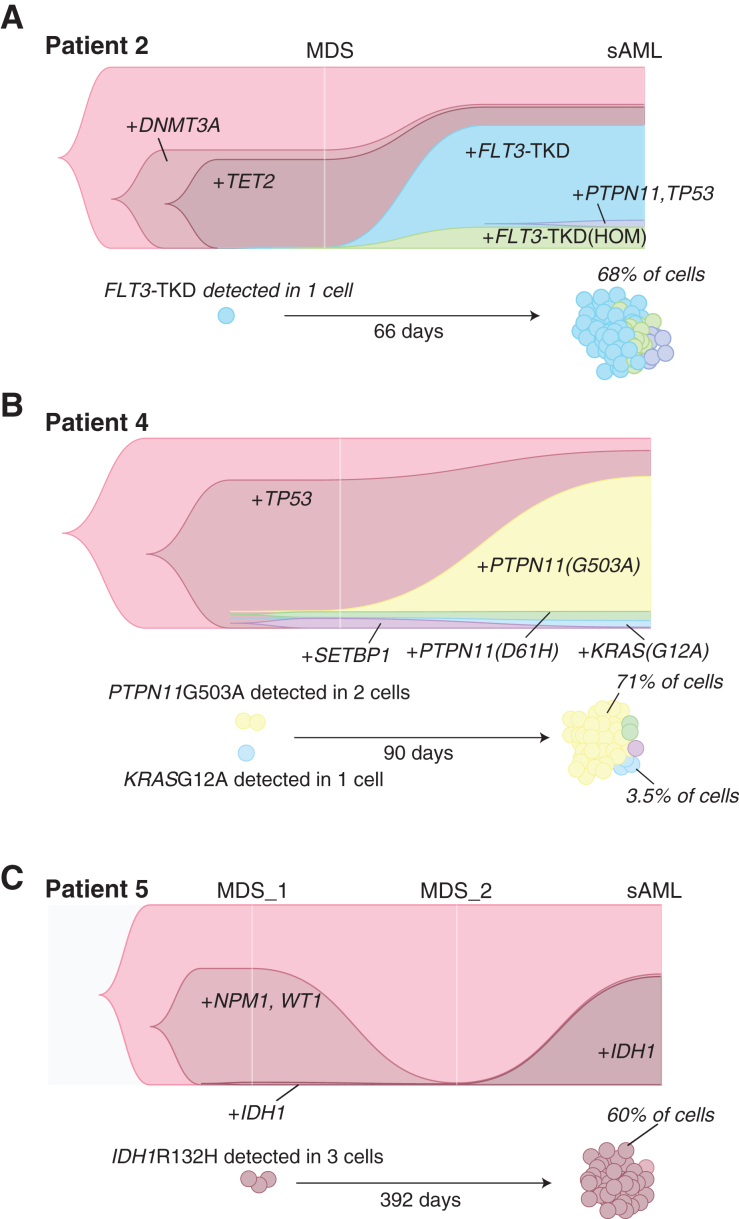
Within some cases, the variant that ultimately defined the dominant subclone at disease progression was often detected at MDS in fewer than 10 cells. However, the presence of these exact mutations in larger numbers at a second, or third, time point, allowed us to confirm these rare cells as subclones in the MDS sample. In patient 2, the MDS-to-sAML transition was characterized by the acquisition of an FLT3 mutation to the DNMT3A + TET2 clone that dominated the MDS mutational landscape (Fig. 4A). The growth of the FLT3 clone in this patient progressed rapidly, with the diagnosis of sAML made just two months later. One cell in the MDS sample was found to have an FLT3 mutation, yet this mutation was present in 68% of cells sequenced at sAML (Fig. 4A). In patient 4, the PTPN11G503A clone and the KRASG12A clone were found in two cells and one cell, respectively, at MDS, but these mutations were in 71% and 3.5% of total cells sequenced at sAML (Fig. 4B). Analysis of three samples (two MDS and one sAML) from patient 5 found only three cells at the first MDS timepoint with an IDH1R132H mutation. All mutations were undetectable at a second, posttreatment, time point, and then the IDH1 clone became the dominant clone (60% of cells) in driving recurrent disease in sAML (Fig. 4C). Despite its ability to capture some mutations in a small number of cells, scDNA-seq did not detect an FLT3-internal tandem duplication (ITD) for one patient in the Dynamic-S group (patient 17), which was present on bulk-sequencing at a VAF of 0.31 in the MDS sample. Interestingly, this patient was treated with sorafenib with azacitidine after this MDS sample was collected and subsequently gained an FLT3-TKD mutation, while losing the FLT3-ITD in sAML, a known resistance mechanism to sorafenib and other FLT3 inhibitors (37).
Figure 4.
Subclonal expansion of rare cells in dynamic architectural change. A, Patient 2 clonal prevalence and depiction of the expansion of rare clones with FLT3-TKDmut from MDS to sAML. B, Patient 4 rare cell expansion of the PTPN11mut subclone. C, Patient 5, rare cell expansion of the IDH1mut subclone.
Clonal Mutations Are Enriched in Primitive and Mature Myeloid Cells and Rare in Lymphoid Cells
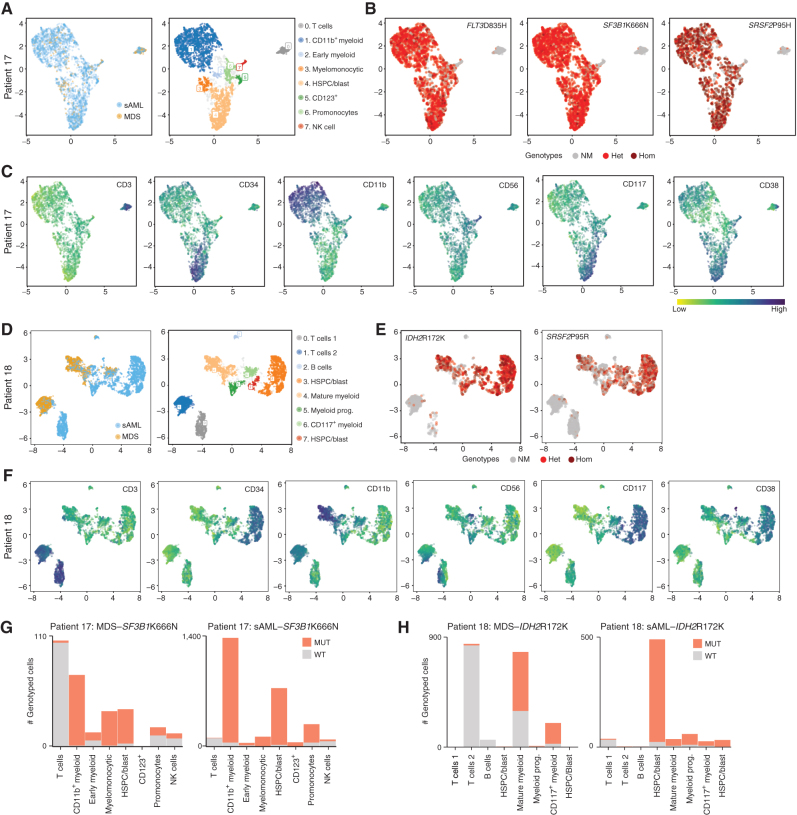
To dissect genotype/phenotype relationships, we used scDNA-seq combined with antibody–oligonucleotide conjugates (AOC) for key cell-surface markers on hematopoietic cells (Supplementary Table S8; refs. 15, 16, 38). Three MDS/sAML sample pairs (six samples) were stained, sequenced, and analyzed. Two of these sample sets were from the Dynamic-S cohort (patients 14 and 17), whereas the third was from the Static cohort (patient 18). To visualize cellular organization based on immunophenotype, we embedded AOC data in a two-dimensional map using uniform manifold approximation and projection (UMAP) combined with hierarchical density-based clustering (HDBSCAN) to visualize the data (39). Each cluster was assigned a cell type name based on the expert assessment of immunophenotype, though given the limited panel size, assignment of cell identity overlapped across clusters. For patient 17, the visualization of single-cell genotypes on the surface-marker–based UMAP revealed a strong bias of all three mutations for myeloid cells, though rare T cells and NK cells did harbor mutations (Fig. 5A and B). Single-cell visualization of surface-marker abundance identified primitive and mature myeloid cells as well as lymphocyte populations (Fig. 5C). In patient 18, primitive cells show a large expansion at sAML, including hematopoietic stem and progenitor-like cell clusters, from MDS to sAML (Fig. 5D–F). Similarly, the visualization of genotypes within individual cells revealed rare mutant T and NK cells harboring mutant genotypes (Fig. 5E and F). Analysis of mutated versus nonmutated cells for founding mutations in patients 17 and 18 (SF3B1 K666N and IDH2 R172K, respectively) in all cell clusters demonstrated rare mutations in all lymphoid cell subsets identified as well as MDS to sAML increase in more primitive cell types (Fig. 5G and H). Analysis of patient 14 data revealed a similar enrichment of mutations in the primitive (CD117+ cell cluster) and rare mutations in T cells (Supplementary Fig. S6A–S6F).
Figure 5.
Combined protein and DNA analysis reveals mutational identities of both myeloid and lymphoid lineages. A, UMAP embeddings (patient 17) mapped on cell-surface marker levels colored by samples or by HDBSCAN clusters. Clusters were identified by immunophenotype and named for cell type by immunophenotype. B, Genotypes are shown for each mapped cell (only cells for which each genotype was known). Code for each variant is no mutation (NM), heterozygous (Het), or homozygous (Hom). C, Surface-marker expression of major markers used to define cell type. D–F, Similar to A–C, but with patient 18. G and H, Cell cluster–based analysis of the proportion of mutated cells MDS to sAML for SF3B1 (patient 17) or IDH2 (patient 18).
Single-Cell RNA Sequencing Identifies Transcriptional Changes Associated with MDS Progression
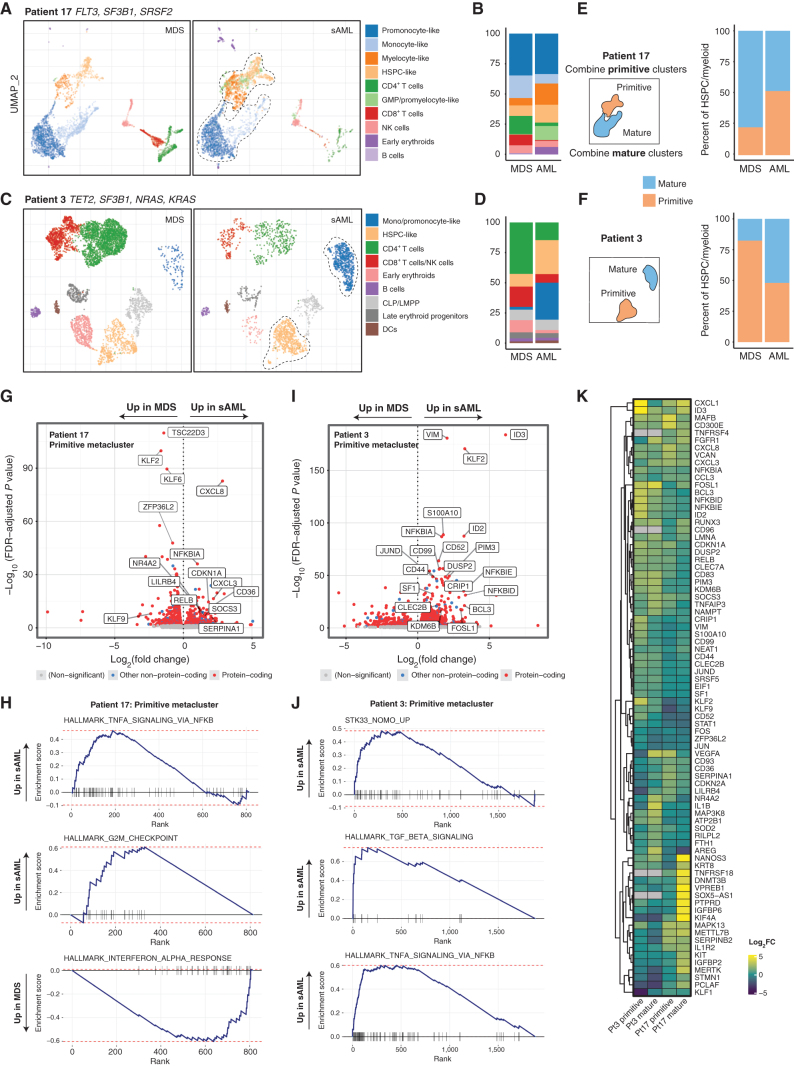
We performed single-cell RNA sequencing (scRNA-seq) on both MDS and sAML samples of patients 3 and 17 to examine transcriptional changes that define MDS therapy resistance and progression. These cases both feature dominant clone signaling mutations, but different clonal architecture changes (dynamic vs. static). Analysis of paired samples allowed each patient's MDS sample to serve as a baseline to dissect transcriptional changes in the sAML sample. We visualized samples from each patient's scRNA-seq data with UMAP, followed by clustering and expert naming of cell subsets (Fig. 6A and B; refs. 39, 40).
Figure 6.
scRNA-seq of longitudinal samples identify transcriptional gene sets that accompany disease progression. A, Gene expression-derived UMAP embedding of all cells from scRNA-seq of two samples from patient 17, clustered with HDBSCAN and then labeled by cell type according to transcriptional signature. The dotted line encapsulates primitive and mature cells (for E, F). B, Number of cells per cluster shown and change from MDS to sAML. C and D, UMAP and cluster distribution for patient 3 as in A–B. E and F, Cells mapped from each time point and patient separately with a depiction of the creation of metaclusters that are either labeled primitive or mature. G, Differentially expressed genes for primitive metacluster for patient 17, with selected top significant genes labeled. H, GSEA plots for patient 17 primitive metacluster. I, Differentially expressed genes for patient 3 primitive metacluster. J, Patient 3 GSEA plots for primitive metacluster. K, Heatmap depicting differentially expressed genes across all metaclusters with heat based on log2 fold change increase.
In patient 17, we confirmed the presence of both early and more mature myeloid leukemia cells seen in the sample with the clinical pathology data, which identified approximately 45% blast and blast equivalents in the sAML (which included myeloblasts, monoblasts, and promonocyte-like cells in this acute myelomonocytic leukemia). Clinical flow cytometry identified similar numbers of primitive myeloblasts [15% of whole bone marrow (WBM) cells, positive for CD34] and more promonocyte-like blast equivalents (19% of WBM, negative for CD34), confirming the heterogeneity seen with both of our single-cell modalities (Fig. 6A). Overall, clusters within the primitive clusters increased from MDS to sAML, whereas the mature cells decreased slightly (Fig. 6B). To facilitate a comparison of transcriptional profiles based on disease time point and cell type, we computationally merged the primitive (HSPC-, GMP/promyelocyte-, and myelocyte-like) and mature (promonocyte- and monocyte-like) myeloid clusters into two metaclusters, primitive and mature (Fig. 6C). We performed a similar analysis with patient 3, which had a static architecture with stable dominant KRAS and minor NRAS subclones, which differed from patient 17 in that both early and mature metaclusters were made up of only one subcluster and both population had large increases at sAML relative to other cell types (Fig. 6D–F). We next performed differential expression (DE) and single-cell gene set enrichment analysis (scGSEA) on pseudo-bulk transcriptional data from the primitive and mature metaclusters from MDS or sAML for each patient (Fig. 6G–K; Supplementary Fig. S7A–S7D). The expanding primitive metacluster from patient 17 demonstrated an increase in signaling molecules, including CXCL8 (IL8), CXCL3, RELB, and NR4A2. Additionally, there were increases in suppressors of signaling pathways, NFKBIA, SOCS3, and the surface receptor LILRB4, which has been implicated in leukemia immune escape (Fig. 6G; ref. 41). Downregulated in this primitive cluster were genes involved in the differentiation of myeloid cells, including KLF2 and KLF6 (42). Top gene sets for primitive cells from patient 17 sAML included those related to cell-cycle progression and TNFα signaling via NF-κB, whereas interferon-α and interferon-γ signaling were enriched in the MDS sample (Fig. 6H). Within the primitive cell cluster for patient 3, some of the top transcripts with increased expression in sAML included signaling genes (JUND, FOSL1), intermediate filament gene vimentin (VIM), and surface-marker genes associated with leukemia stem cells (CD52, CD99, and CD44; Fig. 6I; refs. 43–46). Interestingly, patient 3 demonstrated enrichment in gene sets for TGFβ and TNFα signaling in sAML, as well as those related to intermediate filament signaling (serine/threonine kinase STK33) in KRAS-mutant leukemia cells (Fig. 6J; ref. 47). Top genes in the mature metacluster for patient 17 included downregulation of several major histocompatibility complex genes (e.g., CD74, HLA-DQA1, and HLA-DPA1) with an increase in inflammatory cytokines (CXCL8, CXCL2), whereas gene set enrichment included proliferation-associated cell cycle (G2M_CHECKPOINT) and DNA replication gene sets (E2F_TARGETS), whereas interferon gene sets were enriched in MDS (Fig. 6K; Supplementary Fig. S7A and S7B). The mature metacluster for patient 3 showed an increase in inflammatory genes at sAML, including IL1B, and enrichment of interferon gene sets in the sAML sample, along with inflammatory and KRAS-associated signaling gene sets (Supplementary Fig. S7C and S7D). These data outline a framework for a personalized analysis of transcriptional enrichment to discover potentially critical pathways for disease progression.
DISCUSSION
Progression of MDS to sAML is a clinically devastating event. Since the first fundamental studies demonstrated the clonal origins of this transformation, the field has focused on defining clonal populations with ever-increasing granularity (10, 12, 13). Recent studies using scDNA-seq have focused primarily on AML, demonstrating clonal architectures that become progressively more complex as myeloid neoplasia advances (15, 16). Here we dissect patterns of change in clonal architecture and clonal evolution upon progression to sAML. Our data document differing complexity in MDS clonal transformation. Whereas some patients have relatively stable patterns of clonal complexity, others show striking changes. The former, which we refer to as the Static group, displays minimal changes in architecture while still transforming to sAML, suggesting that genomic evolution might not explain disease progression entirely. Given that all of the samples in the Static group had founder mutations in DNA methylation genes (DNMT3A/TET2/IDH1/2), it is possible that changes in the epigenome drive cell proliferation and accelerate blast growth through progressively disrupted differentiation (48–50). In contrast, the Dynamic groups possessed large clonal architectural changes, with respect to either chromosomally-defined (Dynamic-C group) or smaller variant-defined (Dynamic-S group) clonal changes. With the Dynamic-S cases, the emergent mutations tended to be those classically associated with sAML and enriched for signaling effector mutations (12, 14). These signaling effectors present potential therapeutic strategies for this group. By contrast, the Dynamic-C group was enriched for TP53 mutations and characterized by karyotypes of increasing complexity during disease progression. These changes do not present clear therapeutic targets given the lack of specific mutations beyond TP53. Although our analysis identified that clonal architecture changes do not correlate with survival, these genomic associations, namely, signaling or TP53 mutations within the dominant clone at sAML, did associate with poor survival at sAML. Though our multivariate model revealed significant effects, any survival effects are limited here and would need to be confirmed in a larger cohort. Ultimately, this study is limited by a small cohort size, heterogeneous treatment, and the possibility that an amplicon panel would miss some driver mutations, due to either technical limitations or simply not having a gene represented on the panel. With respect to subclonal structure, we demonstrated that RAS family mutations often occur in multiple competing subclones, demonstrating convergent evolution. This also raises the question about whether these subclones enable the emergence or selection of similar clones.
We also examined the relationships between genotype and phenotype in our cohort. In a subset of our cases, we show that mutations are ubiquitous in primitive and mature myeloid cells at sAML. We further show that lymphocytes may possess mutations observed in myeloid cells, even those mutations that are more prominent in sAML. The mutational involvement of lymphocytes indicates contribution by mutant stem cell clones still capable of multilineage output in clonal hematopoiesis. Although this has been observed by others in myeloid malignancies and aplastic anemia, the implications for immune function have not been fully elucidated (9, 51–53). Confirming frequency and understanding whether these mutated lymphocytes have some pathogenic or disease-promoting effects are areas for future interest. Further, our scRNA-seq analysis revealed gene expression associated with disease progression. We identified downregulation of HLA genes, upregulation of intermediate filaments, upregulation of LILRB4, and inflammatory signaling as potential mechanisms of transformation, which have been previously associated with AML (41, 54, 55). Future analysis of larger scRNA-seq data sets of sAML may further identify disease targets.
Despite the fact that approximately one third of patients diagnosed with MDS will progress to sAML, there are few therapies that alter this risk, and only a transplant offers a chance at a cure (56). Deepening our understanding of this disease progression by combining mutational identity, cellular phenotypes, and transcriptional signatures holds tremendous promise to realize new therapeutic avenues, the hope to identify preemptive measures that can prevent leukemic transformation before it occurs.
METHODS
Reagents
Tapestri-related reagents, including AOCs, were purchased from Mission Bio, Inc. Dextran sulfate was purchased from Research Products International. Custom oligonucleotides for i5/i7 indexing and 5′ Biotin were purchased from Integrated DNA Technologies (IDT). Human TruStain FcX and Ampure XP beads were purchased from BioLegend and Beckman Coulter, respectively. Streptavidin beads and Dulbecco's PBS were purchased from Thermo Fisher. Single-cell RNA reagents were purchased from 10× Genomics. Custom myeloid bulk DNA sequencing kits were purchased from Archer Dx.
Patient Samples
Patients included were diagnosed with MDS and progressed to sAML between 2015 and 2019. Pathologic diagnosis of both MDS and sAML was assigned according to the World Health Organization criteria (57). Patients were enrolled via written informed consent for sample collection, and bone marrow aspirates were collected at both stages of disease and processed by the Hematologic Tissue Repository at Vanderbilt University Medical Center (VUMC). Bone marrow mononuclear cells were cryopreserved and stored in liquid nitrogen until use. All patient tissue and data were obtained with written informed consent and used according to the protocol approved by the local Institutional Review Board and conducted in accordance with the ethical standards of the institution and with the Declaration of Helsinki.
Sample Preparation, Library Generation, and Sequencing
scDNA-seq.
Single-cell library DNA prep was performed using the Tapestri platform and reagents (MissionBio) according to the manufacturer's instructions. Briefly, cryopreserved cells were thawed, washed with PBS, and manually counted using a hemocytometer. Cells were normalized to 5,000 cells/μL using a Cell Buffer (MissionBio). Next, samples were loaded into a microfluidics cartridge where individual cells in conjunction with lysis buffer were encapsulated into water-in-oil droplets using the Tapestri instrument. Encapsulated cells were tagged with a unique barcode, and the DNA from barcoded cells was amplified via multiplex PCR using a targeted myeloid panel that included 312 amplicons across 47 genes known to be associated with myeloid malignancies (Supplementary Tables S9 and S10). Next, amplified cellular DNA was released from individual oil droplets and purified using Ampure XP beads (Beckman Coulter). Libraries were generated by incorporating dual i5/i7 indices and library template (MissionBio) with the purified PCR products during a second PCR and purified again with Ampure XP beads. The final product was quantified via Qubit fluorometer (Thermo Fisher) and assessed for quality using an Agilent Bioanalyzer. Samples were pooled prior to sequencing with a 25% spike-in of PhiX and run on a NovaSeq 6000 (Illumina) S4 flow cell to generate 150 bp paired-end reads. Sequencing was performed at the Vanderbilt Technologies for Advanced Genomics (VANTAGE) sequencing core.
scDNA-seq with Antibody–Oligonucleotide Conjugates.
Samples for combined AOC-based protein detection were prepared in the same manner as described above with the following modifications. Cells were normalized to 10,000 cells/μL in 100 μL and incubated with 10 mg/mL dextran sulfate (Research Products International), Human TruStain FcX (BioLegend), and 1× staining buffer (MissionBio) for 3 minutes at ambient temperature. Next, cells were stained with a combination of 13 AOCs (Supplementary Table S8; MissionBio) for 30 minutes at ambient temperature. Following staining, cells were washed three times in Dulbecco's PBS (Gibco), recounted, and processed as above with the addition of adding 2 μmol/L antibody tag forward primer (Tapestri) prior to the barcoding step. DNA libraries were prepared and purified as above. Protein PCR products, which exist in the supernatant from the Ampure XP bead purification step, were isolated by a 5-minute incubation with 2 μL 5′ Biotin Oligo (IDT) for 5 minutes at 96°C, followed by a 5-minute incubation on ice. Isolated proteins were washed using 2× binding and washing buffer (10 mmol/L Tris-HCL, 1 mmol/L EDTA, 2 M NaCl) and streptavidin beads (Thermo Fisher). Protein libraries were generated using the washed proteins, library template (MissionBio), and i5/i7 indices (IDT) via PCR. The protein library PCR product was cleaned again using Ampure XP beads. Both DNA and protein libraries were quantified, quality checked, and sequenced as above.
scRNA-seq.
scRNA-seq libraries were created from viable patient-derived samples using the 10X Chromium 5′ library (patient 17) or 10X Chromium 3′ v1 (patient 3) preparation kits (10X Genomics) using the manufacturer's recommendations and targeting 10,000 cells per sample. Next-generation 150 nt paired-end sequencing was performed on an Illumina Novaseq6000. Low quality reads were filtered out and CellRanger Count v3.1 (patient 17) or v6.1 (patient 3) (10X Genomics) was used to align reads onto the GRCh38 reference genome. Downstream analysis was performed as below.
Bulk DNA-seq.
Bulk sequencing from clinical sample sequencing was used when available to corroborate variants. When available, clinical bulk-sequencing results were used. In instances where this testing was not performed and remaining patient cells existed, Archer Dx kits were used to perform bulk-sequencing sample prep and library generation as described previously (Supplementary Table S11; ref. 58).
Data Analysis
Pipeline Processing and Variant Filtering.
FASTQ files from single-cell DNA samples were processed via the Tapestri Pipeline v1.8.4. Adapters were trimmed using Cutadapt (59, 60), and reads longer than 30 nt were aligned to the hg19 reference genome with BWA-MEM (61). Cells were then called based on amplicon-read completeness in each barcode. Variants were called using GATK 3.7 (62) with a joint-calling approach that follows GATK best practices (63, 64). Then, the variant lists were decomposed, filtered, and the genotype/cell matrix loaded into the Tapestri Insights software package (v.2.2) where low-quality cells and variants were removed based on genotype quality score <30, variants genotyped in <50% of cells, read depth <10 reads, cells with <50% of genotypes present, cells with genotypes of <20% alternate allele frequencies, and variants mutated in <0.5% of cells. For some pathogenic mutations that were poorly genotyped, e.g., IDH2R172 and SRSF2P95, we had to lower the “exclude variants genotyped in <50% of cells” parameter to exclude variants genotyped in <20% of cells. Using five apparent heterozygous germline variants in each sample, the allele dropout (ADO) was calculated using the following formula for each of the five variants: [(# of wild-type cells + # of homozygous cells)/total number of cells genotyped] × 100 (Supplementary Table S3). The data were further analyzed in R, including the Tapestri package.
Mutational and Clonal Analysis.
Variants filtered as above were then assessed for known pathogenicity or likely pathogenicity via ClinVar and Varsome databases (65, 66). Nonintronic, previously identified somatic variants were included in clonal analyses. The clonal architecture of each sample was determined using genotype clustering and zygosity information in Tapestri Insights (Mission Bio). Serial samples from an individual patient (MDS and sAML samples) were analyzed concurrently and compared to determine clonal progression. Clones with <10 cells were excluded unless the same clone was also observed at another time point in the same patient. Oncoprints were constructed from sample-level and cell-level variant information using the R ComplexHeatmap package (67). Set interaction analysis was visualized with the UpSetR package (68). Clonal prevalence plots of the temporal clonal evolution data were generated from clonal phylogeny and clonal prevalence at MDS and sAML time points using the Timescape package (69).
Single-Cell Protein Analysis.
FASTQ files from single-cell protein samples were processed by Mission Bio. The input FASTQ files were first validated to identify the run chemistry and check the sequence quality using the fastp tool (70). Adapters were trimmed and short reads were removed. Next, PCR handle and capture sequences were trimmed, and the antibody barcodes were extracted from R2 reads and corrected using the error correction map. Output count files were then generated. For the three patients for whom we also had antibody barcoding data (patients 14, 17, and 18), we explored the intersect of genotype call and protein marker identity, following centered log-ratio transformation of the multiomics data derived from the same barcodes.
scRNA-seq Analysis.
scRNA-seq data were obtained from a 10X Genomics Chromium-based CITE-Seq experiment. After processing via Cell Ranger, two samples from patient 17 yielded a combined 9,606 cells and 33,538 genes. The data were filtered in Seurat to include at least 500 RNA molecules per cell, at least 250 genes per cell, at least 0.8 of the log10 value of genes per UMI (log10GenesPerUMI), and a proportion of transcripts mapping to mitochondrial genes less than 15%. Only genes that had a nonzero expression value in at least 10 cells were preserved, resulting in a final data set of 6,863 cells and 15,359 genes. Samples from patient 3 were demultiplexed and filtered using the same parameters as the patient 17 analysis, resulting in a final data set of 7,100 cells and 18,099 genes.
The sample-level data were normalized using the SCTransform method (71), which utilizes a regularized negative binomial regression for normalization and variance stabilization. Furthermore, it estimates the variance of the raw filtered data, identifies the most variable genes, and regresses out mitochondrial mapping contribution as a confounding source of variation. After review, the UMAP embeddings of the normalized data revealed a misalignment of the two samples. We, therefore, integrated the data sets using Seurat's canonical correlation analysis method (72), using the top 5,000 features as the integration selection. Standard exploratory data analysis methods were used to identify cell populations and quantify gene-expression differences between these populations.
Pseudo-Bulk Differential Gene-Expression Analysis.
To mitigate the effects of false discoveries and bias toward highly expressed genes in single-cell DE analysis methods, we opted for a pseudo-bulk method (73). For patients 3 and 17 single-cell data, cells associated with primitive and mature myeloid clusters were split by condition (MDS and sAML) resulting in patient-specific single-cell data sets: primitive-MDS, primitive-sAML, mature-MDS, and mature-sAML. For each data set, we generated pseudo-samples by applying a binomial distribution to randomly distribute single cells into sample groups. Then, for each sample group, the raw counts per gene were summed to generate an ensemble of pseudo-bulk input files. Per patient, we applied DESeq2, to independently study the differential gene expression of sAML versus MDS, for primitive and mature cell populations (74).
GSEA.
GSEA was performed using Fast GSEA using preranked lists and Molecular Signatures Database v7.1 Hallmark gene sets, which represent well-defined biological states or processes (75, 76). The preranked lists were generated from the fold change and adjusted P-value results of the DESeq2-derived differentially expressed genes between sAML and MDS, on a per patient and metacluster basis.
Statistical Analysis
Statistical significance was set at P < 0.05. For pairwise comparisons, we used a two-sided Student t test or Wilcoxon rank-sum test as indicated. For comparison of multiple groups, we used a one-way analysis of variance test in R. To evaluate nonrandom associations of mutational identities in the three pattern groups, we used a Fisher exact test. Kaplan–Meier plots and Cox proportional hazard modeling were done in R using packages survival (https://github.com/therneau/survival) and survminer (https://github.com/kassambara/survminer). The Shannon diversity index was calculated as described previously using clone identity and number for each sample in place of species (77). Plots were created using the packages dplyr, tidyr, ggpubr, and ggplot2 in R (version 4.1.0) within RStudio (version 1.4.1717; ref. 78).
Data Availability Statement
Deidentified single-cell and bulk DNA sequencing data have been deposited at NCBI Sequence Read Archive in BioProject PRJNA831862. Single-cell RNA-seq data are available in the Gene Expression Omnibus (GEO) repository under accession number GSE205490.
Supplementary Material
Acknowledgments
Patient samples were supplied by the Vanderbilt Hematologic Malignancy Tissue Repository, a Vanderbilt-Ingram Cancer Center (VICC)- and VUMC-supported resource. We acknowledge Vanderbilt Technologies for Advanced Genomics (VANTAGE) for help with sequencing. We thank Angela Jones at VANTAGE for her sequencing expertise and advice throughout the project. We gratefully acknowledge Robert Durruthy-Durruthy, Jose Jacob, Yue Wang, Kelly Kaihara, and all other members of MissionBio for the technical support, analytical help, and genomics expertise. We also thank Crystal Amsberry from ArcherDx for her assistance with sequencing and analysis. This work was supported by a VICC Ambassadors Award (P.B. Ferrell), a Vanderbilt Institute for Clinical and Translation Research Clinical Translational Science Award Program 5UL1TR002243 (P.B. Ferrell and T. Guess), NIH K23HL138291 (P.B. Ferrell). M.R. Savona is supported by NIH 1R01CA262287 and 1U01OH012271, an LLS Clinical Scholar Award, the Biff Ruttenburg Foundation, the Adventure Alle Fund, the Beverly and George Rawlings Research Directorship, the E.P. Evans MDS Foundation.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors’ Disclosures
M.R. Savona reports personal fees from AbbVie, BMS, CTI, Sierra Oncology, Novartis, grants from Astex and Incyte, personal fees and other support from Karyopharm, Ryvu, personal fees from Sierra Oncology, grants and personal fees from Takeda, and TG Therapeutics outside the submitted work. P.B. Ferrell reports grants from Incyte, Astex Pharmaceuticals, and Forma Therapeutics outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
T. Guess: Conceptualization, data curation, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. C.R. Potts: Formal analysis, validation, investigation, methodology, writing–review and editing. P. Bhat: Data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. J.A. Cartailler: Data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. A. Brooks: Formal analysis, investigation. C. Holt: Software, formal analysis, investigation, visualization, writing–review and editing. A. Yenamandra: Data curation, writing–review and editing. F.C. Wheeler: Data curation, writing–review and editing. M.R. Savona: Resources, project administration, writing–review and editing. J.-P. Cartailler: Conceptualization, data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. P.B. Ferrell: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Kuykendall A, Duployez N, Boissel N, Lancet JE, Welch JS. Acute myeloid leukemia: the good, the bad, and the ugly. Am Soc Clin Oncol Educ Book 2018;38:555–73. [DOI] [PubMed] [Google Scholar]
- 2. Bennett JM. Secondary acute myeloid leukemia. Leuk Res 1995;19:231–2. [DOI] [PubMed] [Google Scholar]
- 3. Weinberg OK, Seetharam M, Ren L, Seo K, Ma L, Merker JD, et al. Clinical characterization of acute myeloid leukemia with myelodysplasia-related changes as defined by the 2008 WHO classification system. Blood 2009;113:1906–8. [DOI] [PubMed] [Google Scholar]
- 4. Nowell PC. The clonal evolution of tumor cell populations. Science 1976;194:23–8. [DOI] [PubMed] [Google Scholar]
- 5. Greaves M, Maley CC. Clonal evolution in cancer. Nature 2012;481:306–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shlush LI, Mitchell A, Heisler L, Abelson S, Ng SWK, Trotman-Grant A, et al. Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature 2017;547:104–8. [DOI] [PubMed] [Google Scholar]
- 7. Kroeger H, Jelinek J, Estécio MRH, He R, Kondo K, Chung W, et al. Aberrant CpG island methylation in acute myeloid leukemia is accentuated at relapse. Blood 2008;112:1366–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 2012;481:506–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Klco JM, Spencer DH, Miller CA, Griffith M, Lamprecht TL, O'Laughlin M, et al. Functional heterogeneity of genetically defined subclones in acute myeloid leukemia. Cancer Cell 2014;25:379–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Walter MJ, Shen D, Ding L, Shao J, Koboldt DC, Chen K, et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med 2012;366:1090–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Walter MJ, Shen D, Shao J, Ding L, White BS, Kandoth C, et al. Clonal diversity of recurrently mutated genes in myelodysplastic syndromes. Leukemia 2013;27:1275–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Makishima H, Yoshizato T, Yoshida K, Sekeres MA, Radivoyevitch T, Suzuki H, et al. Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet 2017;49:204–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen J, Kao YR, Sun D, Todorova TI, Reynolds D, Narayanagari SR, et al. Myelodysplastic syndrome progression to acute myeloid leukemia at the stem cell level. Nat Med 2019;25:103–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Menssen AJ, Walter MJ. Genetics of progression from MDS to secondary leukemia. Blood 2020;136:50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miles LA, Bowman RL, Merlinsky TR, Csete IS, Ooi AT, Durruthy-Durruthy R, et al. Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature 2020;587:477–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morita K, Wang F, Jahn K, Hu T, Tanaka T, Sasaki Y, et al. Clonal evolution of acute myeloid leukemia revealed by high-throughput single-cell genomics. Nat Commun 2020;11:5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pellegrino M, Sciambi A, Treusch S, Durruthy-Durruthy R, Gokhale K, Jacob J, et al. High-throughput single-cell DNA sequencing of acute myeloid leukemia tumors with droplet microfluidics. Genome Res 2018;28:1345–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ediriwickrema A, Aleshin A, Reiter JG, Corces MR, Kohnke T, Stafford M, et al. Single-cell mutational profiling enhances the clinical evaluation of AML MRD. Blood Adv 2020;4:943–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Taylor J, Mi X, North K, Binder M, Penson A, Lasho T, et al. Single-cell genomics reveals the genetic and molecular bases for escape from mutational epistasis in myeloid neoplasms. Blood 2020;136:1477–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kennedy AL, Myers KC, Bowman J, Gibson CJ, Camarda ND, Furutani E, et al. Distinct genetic pathways define pre-leukemic and compensatory clonal hematopoiesis in Shwachman-Diamond syndrome. bioRxiv 2020: 2020.06.04.134692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alberti-Servera L, Demeyer S, Govaerts I, Swings T, De Bie J, Gielen O, et al. Single-cell DNA amplicon sequencing reveals clonal heterogeneity and evolution in T-cell acute lymphoblastic leukemia. Blood 2021;137:801–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dillon LW, Ghannam J, Nosiri C, Gui G, Goswami M, Calvo KR, et al. Personalized single-cell proteogenomics to distinguish acute myeloid leukemia from non-malignant clonal hematopoiesis. Blood Cancer Discov 2021;2:319–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Benard BA, Leak LB, Azizi A, Thomas D, Gentles AJ, Majeti R. Clonal architecture predicts clinical outcomes and drug sensitivity in acute myeloid leukemia. Nat Commun 2021;12:7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eastburn DJ, Huang Y, Pellegrino M, Sciambi A, Ptacek LJ, Abate AR. Microfluidic droplet enrichment for targeted sequencing. Nucleic Acids Res 2015;43:e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu L, Durruthy-Durruthy R, Eastburn DJ, Pellegrino M, Shah O, Meyer E, et al. Clonal evolution and changes in two AML patients detected with a novel single-cell DNA sequencing platform. Sci Rep 2019;9:11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lindsley RC, Mar BG, Mazzola E, Grauman PV, Shareef S, Allen SL, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood 2015;125:1367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ogawa S. Genetics of MDS. Blood 2019;133:1049–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med 2016;374:2209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grove CS, Vassiliou GS. Acute myeloid leukaemia: a paradigm for the clonal evolution of cancer? Dis Model Mech 2014;7:941–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McMahon CM, Ferng T, Canaani J, Wang ES, Morrissette JJD, Eastburn DJ, et al. Clonal selection with RAS pathway activation mediates secondary clinical resistance to selective FLT3 inhibition in acute myeloid leukemia. Cancer Discov 2019;9:1050–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Menssen AJ, Khanna A, Miller CA, Srivatsan SN, Chang GS, Shao J, et al. Convergent clonal evolution of signaling gene mutations is a hallmark of myelodysplastic syndrome progression. Blood Cancer Discov 2022;3:330–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haase D, Stevenson KE, Neuberg D, Maciejewski JP, Nazha A, Sekeres MA, et al. TP53 mutation status divides myelodysplastic syndromes with complex karyotypes into distinct prognostic subgroups. Leukemia 2019;33:1747–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bernard E, Nannya Y, Hasserjian RP, Devlin SM, Tuechler H, Medina-Martinez JS, et al. Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nat Med 2020;26:1549–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. DiNardo CD, Cortes JE. Mutations in AML: prognostic and therapeutic implications. Hematology Am Soc Hematol Educ Program 2016;2016:348–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Man CH, Fung TK, Ho C, Han HH, Chow HC, Ma AC, et al. Sorafenib treatment of FLT3-ITD(+) acute myeloid leukemia: favorable initial outcome and mechanisms of subsequent nonresponsiveness associated with the emergence of a D835 mutation. Blood 2012;119:5133–43. [DOI] [PubMed] [Google Scholar]
- 38. Demaree B, Delley CL, Vasudevan HN, Peretz CAC, Ruff D, Smith CC, et al. Joint profiling of DNA and proteins in single cells to dissect genotype-phenotype associations in leukemia. Nat Commun 2021;12:1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Campello RJGB, Moulavi D, Sander J. Density-based clustering based on hierarchical density estimates. Advances in knowledge discovery and data mining. Berlin, Heidelberg: Springer Berlin Heidelberg; 2013. p. 160–72. [Google Scholar]
- 40. McInnes L, Healy J. Accelerated hierarchical density based clustering. 2017 IEEE International Conference on Data Mining Workshops (ICDMW)2017:33–42.
- 41. Deng M, Gui X, Kim J, Xie L, Chen W, Li Z, et al. LILRB4 signalling in leukaemia cells mediates T cell suppression and tumour infiltration. Nature 2018;562:605–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Humbert M, Halter V, Shan D, Laedrach J, Leibundgut EO, Baerlocher GM, et al. Deregulated expression of Kruppel-like factors in acute myeloid leukemia. Leuk Res 2011;35:909–13. [DOI] [PubMed] [Google Scholar]
- 43. Chung SS, Eng WS, Hu W, Khalaj M, Garrett-Bakelman FE, Tavakkoli M, et al. CD99 is a therapeutic target on disease stem cells in myeloid malignancies. Sci Transl Med 2017;9:eaaj2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Blatt K, Herrmann H, Hoermann G, Willmann M, Cerny-Reiterer S, Sadovnik I, et al. Identification of campath-1 (CD52) as novel drug target in neoplastic stem cells in 5q-patients with MDS and AML. Clin Cancer Res 2014;20:3589–602. [DOI] [PubMed] [Google Scholar]
- 45. Eisenwort G, Sadovnik I, Keller A, Ivanov D, Peter B, Berger D, et al. Phenotypic characterization of leukemia-initiating stem cells in chronic myelomonocytic leukemia. Leukemia 2021;35:3176–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med 2006;12:1167–74. [DOI] [PubMed] [Google Scholar]
- 47. Scholl C, Frohling S, Dunn IF, Schinzel AC, Barbie DA, Kim SY, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell 2009;137:821–34. [DOI] [PubMed] [Google Scholar]
- 48. Izzo F, Lee SC, Poran A, Chaligne R, Gaiti F, Gross B, et al. DNA methylation disruption reshapes the hematopoietic differentiation landscape. Nat Genet 2020;52:378–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang X, Su J, Jeong M, Ko M, Huang Y, Park HJ, et al. DNMT3A and TET2 compete and cooperate to repress lineage-specific transcription factors in hematopoietic stem cells. Nat Genet 2016;48:1014–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet 2011;44:23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yoshizato T, Dumitriu B, Hosokawa K, Makishima H, Yoshida K, Townsley D, et al. Somatic mutations and clonal hematopoiesis in aplastic anemia. N Engl J Med 2015;373:35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lundgren S, Keranen MAI, Kankainen M, Huuhtanen J, Walldin G, Kerr CM, et al. Somatic mutations in lymphocytes in patients with immune-mediated aplastic anemia. Leukemia 2021;35:1365–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vercauteren SM, Starczynowski DT, Sung S, McNeil K, Salski C, Jensen CL, et al. T cells of patients with myelodysplastic syndrome are frequently derived from the malignant clone. Br J Haematol 2012;156:409–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Petti AA, Williams SR, Miller CA, Fiddes IT, Srivatsan SN, Chen DY, et al. A general approach for detecting expressed mutations in AML cells using single cell RNA-sequencing. Nat Commun 2019;10:3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Christopher MJ, Petti AA, Rettig MP, Miller CA, Chendamarai E, Duncavage EJ, et al. Immune escape of relapsed AML cells after allogeneic transplantation. N Engl J Med 2018;379:2330–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Corey SJ, Minden MD, Barber DL, Kantarjian H, Wang JC, Schimmer AD. Myelodysplastic syndromes: the complexity of stem-cell diseases. Nat Rev Cancer 2007;7:118–29. [DOI] [PubMed] [Google Scholar]
- 57. Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood 2009;114:937–51. [DOI] [PubMed] [Google Scholar]
- 58. Crowgey EL, Mahajan N, Wong WH, Gopalakrishnapillai A, Barwe SP, Kolb EA, et al. Error-corrected sequencing strategies enable comprehensive detection of leukemic mutations relevant for diagnosis and minimal residual disease monitoring. BMC Med Genomics 2020;13:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 2011;17:10–2. [Google Scholar]
- 60. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 2013;43:11 0 1–0 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kopanos C, Tsiolkas V, Kouris A, Chapple CE, Albarca Aguilera M, Meyer R, et al. VarSome: the human genomic variant search engine. Bioinformatics 2018;35:1978–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 2018;46:D1062–D7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016;32:2847–9. [DOI] [PubMed] [Google Scholar]
- 68. Conway JR, Lex A, Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 2017;33:2938–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Smith MA, Nielsen CB, Chan FC, McPherson A, Roth A, Farahani H, et al. E-scape: interactive visualization of single-cell phylogenetics and cancer evolution. Nat Methods 2017;14:549–50. [DOI] [PubMed] [Google Scholar]
- 70. Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018;34:i884–i90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 2019;20:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive integration of single-cell data. Cell 2019;177:1888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Squair JW, Gautier M, Kathe C, Anderson MA, James ND, Hutson TH, et al. Confronting false discoveries in single-cell differential expression. Nat Commun 2021;12:5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, Sergushichev A. Fast gene set enrichment analysis. bioRxiv 2021:060012. [Google Scholar]
- 77. Spellerberg IF, Fedor PJ. A tribute to Claude Shannon (1916–2001) and a plea for more rigorous use of species richness, species diversity and the ‘Shannon–Wiener’ Index. Global Ecol Biogeogr 2003;12:177–9. [Google Scholar]
- 78. Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag; 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deidentified single-cell and bulk DNA sequencing data have been deposited at NCBI Sequence Read Archive in BioProject PRJNA831862. Single-cell RNA-seq data are available in the Gene Expression Omnibus (GEO) repository under accession number GSE205490.