Abstract
Objectives
A lactobacilli-dominated vaginal microbiome may protect against pelvic inflammatory disease (PID), but one dominated by Gardnerella species might increase susceptibility. Not all lactobacilli are equally protective. Recent research suggests that D(−) isomer lactic acid producing lactobacilli (Lactobacillus crispatus, Lactobacillus jensenii and Lactobacillus gasseri) may protect against infection with Chlamydia trachomatis, an important cause of PID. Lactobacillus iners , which produces L(+) isomer lactic acid, may be less protective. We investigated the microbiome in stored vaginal samples from participants who did or did not develop PID during the prevention of pelvic infection (POPI) chlamydia screening trial.
Methods
Long-read 16S rRNA gene nanopore sequencing was used on baseline vaginal samples (one per participant) from all 37 women who subsequently developed clinically diagnosed PID during 12-month follow-up, and 111 frequency matched controls who did not, matched on four possible risk factors for PID: age <20 versus ≥20, black ethnicity versus other ethnicity, chlamydia positive versus negative at baseline and ≥2 sexual partners in the previous year versus 0–1 partners.
Results
Samples from 106 women (median age 19 years, 40% black ethnicity, 22% chlamydia positive, 54% reporting multiple partners) were suitable for analysis. Three main taxonomic clusters were identified dominated by L. iners, L. crispatus and Gardnerella vaginalis. There was no association between a more diverse, G. vaginalis dominated microbiome and subsequent PID, although increased Shannon diversity was associated with black ethnicity (p=0.002) and bacterial vaginosis (diagnosed by Gram stain p<0.0001). Women who developed PID had similar relative abundance of protective D(−) isomer lactic acid producing lactobacilli to women without PID, but numbers of PID cases were small.
Conclusions
In the first-ever community-based prospective study of PID, there was no clear association between the vaginal microbiome and subsequent development of PID. Future studies using serial samples may identify vaginal microbial communities that may predispose to PID.
Keywords: pelvic inflammatory disease, microbiology, cohort studies, epidemiology, RNA
Background
Pelvic inflammatory disease (PID) is an infection-induced inflammation of the female upper reproductive tract that can lead to infertility and ectopic pregnancy. Chlamydia trachomatis and Neisseria gonorrhoeae cause up to 30% of PID, but in many cases, the aetiology is polymicrobial or unclear.1 In addition, we do not know why only some women with genital C. trachomatis infection develop upper tract infection. A healthy, low diversity, vaginal microbiome dominated by lactobacilli may protect against disease.2–5 However, a high diversity microaerophilic/anaerobic microbiome with fewer lactobacilli (as may be found in bacterial vaginosis often with Gardnerella vaginosis and/or Atopobium vaginae) may increase susceptibility to STIs by causing cervicomucosal barrier disruption and epithelial portals of entry for ascending infection.3 6
Not all lactobacilli are equal.3 A recent study suggested that lactobacilli that produce mainly D(−) isomer lactic acid (Lactobacillus crispatus, L. jensenii and L. gasseri) may be associated with long-term protection against C. trachomatis infection, possibly by reducing epithelial cell recycling.7 By contrast, a vaginal microbiome dominated by L. iners, which mainly produces L(+) isomer lactic acid, may be associated with increased susceptibility to infection.7–9
A recent systematic review called for longitudinal studies of vaginal microbiota and infections to lay the foundation for possible prevention and treatment strategies.5 We used stored baseline self-taken vaginal swabs from a cohort of 2357 ethnically diverse young female students recruited from public areas at 22 London colleges to the prevention of pelvic infection (POPI) chlamydia screening trial in 2004–2006.10Previously, 16S rRNA sequencing of 20 samples using a long-read PacBio sequencing platform had confirmed adequate DNA integrity in these samples.11
Conventional short-read 16S rRNA gene sequencing is constrained in its ability to comprehensively speciate pathogens within a sample because of the limited 16S variable regions that are usually sequenced. Long-read sequencing platforms such as PacBio and Oxford Nanopore Technologies’ MinION allow for full sequencing of the 16S rRNA gene for microbiome studies.12 13
We used long-read 16S rRNA gene nanopore sequencing to investigate bacterial communities in 37 women who developed clinical PID within the following year, and 111 frequency matched control women who did not. Since D(−) isomer lactic acid producing lactobacilli may protect against C. trachomatis,5 7 a major cause of PID, we also explored the relative abundance of D(−) isomer lactic acid producing lactobacilli in women with and without subsequent PID, and in women with and without concurrent C. trachomatis infection or bacterial vaginosis (BV).14
Methods
Study samples
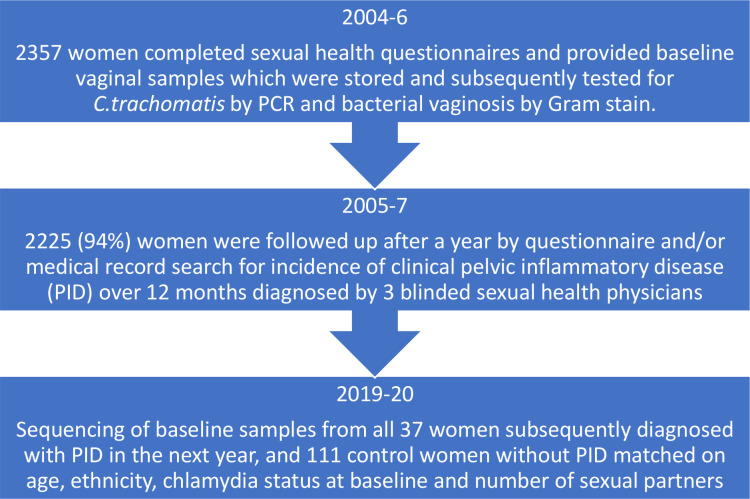
One hundred and forty-eight frozen, baseline vaginal samples (one sample per participant) were retrieved from all 37 women who subsequently developed clinically diagnosed PID in the next year and 111 controls with no PID, as part of the POPI trial10 (figure 1). At recruitment, participants had provided self-taken vaginal swabs in the college toilets, rolled the swab over a glass slide for future Gram stain analysis for BV (Nugents score ≥7) and placed the swab in a tube of Aptima transport medium (Gen Probe), which was frozen within 24 hours. Controls (three per index case of PID) were frequency matched on possible risk factors for PID15: age <20 versus ≥20, black ethnicity versus other ethnicity, chlamydia positive versus negative at baseline and ≥2 sexual partners in the previous year versus 0–1 partners. The diagnosis of PID was made by three sexual health physicians (blinded to chlamydia status) based on medical records and participant questionnaires after 12 months and using modified Hager’s criteria: pelvic pain, cervical motion tenderness and/or adnexal tenderness.10
Figure 1.
POPI vaginal microbiota study flow chart. PID, pelvic inflammatory disease; POPI, prevention of pelvic infection.
DNA preparation and nanopore sequencing
DNA was isolated from vaginal samples using QIAamp DNA Mini Kit (Qiagen), following the manufacturer’s protocol for Gram-positive and difficult-to-lyse bacteria, and quantified using Qubit 3.0 Fluorometer. 16S rRNA genes in each sample were amplified with the primers, B27F (TTTCTGTTGGTGCTGATATTGCAGRGTTYGATYMTGGCTCAG) and B1492R (ACTTGCCTGTCGCTCTATCTTCRGYTACCTTGTTACGACTT), which were modified from S-D-bact-0008-c-S20 and S-*-Univ-1492-a-A-19.16 DNA libraries, each containing 12 different clinical samples, were prepared using PCR Barcoding Kit - EXP-PBC096 and Ligation Sequencing Kit 1D - SQK-LSK108, and sequenced on MinION MK I with SpotON Flow Cell Mk I FLO-MIN106 R9.4 (Oxford Nanopore Technologies). Sequencing reads were live base called locally using the MinKNOW protocol - NC_48Hr_Sequencing_Run_FLO-MIN106_ SQK-LSK108.py. Sequence data have been submitted to the ENA database with accession number PRJEB41336.
Microbiome comparison and statistical analysis
The sequence analysis protocol was based on previously published methods.17 Briefly, reads were first filtered for length >1200 and <1800 bases, before chimeric reads were removed with yacrd.18 Remaining reads were mapped to the National Center for Biotechnology Information (NCBI) 16S Microbial database using minimap2.19 Aligned reads were merged by species using R, counted and normalised using decostand (R vegan package V.4.0.4, 2021-02-15) by dividing by the margin total. The normalised number of reads mapping to each species against that sample was shown in a heat map. Shannon alpha diversity metrics were generated with vegan, and statistical significance was determined using a Kruskal-Wallis rank sum test for six clinical characteristics: PID versus no PID, age <20 versus ≥20, black ethnicity versus other ethnicity, chlamydia positive versus negative at baseline, ≥2 sexual partners in the previous year versus 0–1 partners and bacterial vaginosis by Gram stain versus no bacterial vaginosis. P values were adjusted for multiple comparisons using the Benjamini-Hochberg method.
The relative abundance of the dominant species was explored (including a combination of D(−) isomer lactic acid producing lactobacilli L. crispatus, L. jensenii and L. gasseri), for three clinical diagnoses: pelvic inflammatory disease, bacterial vaginosis or C. trachomatis. Finally, we investigated beta diversity using non-metric multidimensional scaling (NMDS) of Bray-Curtis distances.
The study was constrained by the number of women diagnosed with PID. We estimated that a sample size of 148 women (37 who developed PID and 111 who did not develop PID) would detect a significant difference (p<0.05, power 80%) if vaginal samples from 35% of women with PID had Gardnerella vaginalis 15 versus ≤11% of women without PID. (We used G. vaginalis as a surrogate for bacterial vaginosis.)
Excluded samples
An initial heatmap representation of the relative abundances (online supplemental figure 1) showed four major clusters, one of which was dominated by the plant pathogen Burkholderia gladioli. Although a rare cause of opportunistic infection in humans,20 the presence of B. gladioli is unusual, and the Burkholderia genus has been associated with contamination in other microbiome studies.21 Further investigation showed that the majority of samples with high levels of B. gladioli were collected within a specific time period at the end of the study (online supplemental figure 2), suggesting a systematic contamination within the samples collected at the time. A similar plot for L. crispatus showed no chronological bias (data not shown). In light of this, all 42 samples containing more than 1% B. gladioli were removed from further analysis. (This included 14 samples with PID, 13 with BV and 9 with C. trachomatis.) Inclusion or exclusion of these samples had little effect on the conclusions of this study.
sextrans-2021-055260supp001.pdf (569KB, pdf)
Results
Study population
Samples from 106 participants (23 with PID) had adequate sequencing data and were included in the analysis. The median age of the women in the cohort was 19 years (range 16–27), 40% (42) described their ethnic group as black and 54% (57) reported ≥two sexual partners in the previous year. At baseline, 22% of women (23) had C. trachomatis, 21% (22) had bacterial vaginosis (diagnosed by Nugent’s score 7–10 on Gram stain, with a further three classified as intermediate), 4% (4) had Mycoplasma genitalium and 2% (2) Neisseria gonorrhoeae (diagnosed by Gen-Probe PCR). The estimated mean time from baseline sample to diagnosis of PID, where data were available (in 22 of 23 women), was 35 weeks (range 13–52 weeks).
Vaginal microbiota show three main clusters
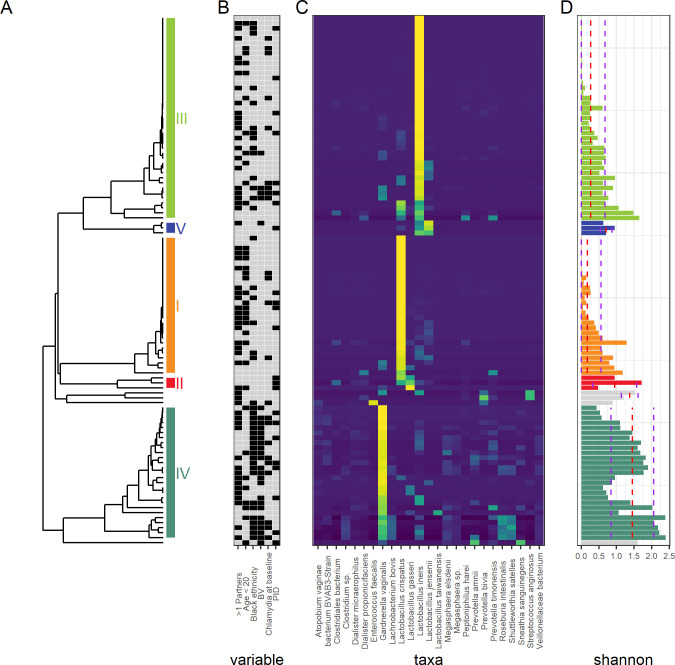
Figure 2 shows three main taxonomic clusters dominated by L. iners, L. crispatus and G. vaginalis, with a smaller number of samples dominated by L. jensenii and L. gasseri. The lactobacillus dominated clusters showed lower Shannon diversity than the G. vaginalis cluster.
Figure 2.
Heat map of relative abundances of microbial taxa in the vaginal bacterial communities of 106 women who did or did not develop pelvic inflammatory disease (PID) in the next 12 months. (A) Hierarchical clustering based on the Pearson correlation dissimilarity of the relative abundances within each sample. Clusters are labelled I-V16. (B) Clinical characteristics: black filled box indicates PID, Chlamydia positive at baseline, >1 sexual partner in the past year, age < 20 years, self-assigned black ethnic group. (C) Heatmap of relative abundances showing only the most common species. (D) Shannon diversity for each sample: red dashed line shows the median diversity for each cluster, purple dashed lines show ± the SD.
No association between increasing diversity and PID
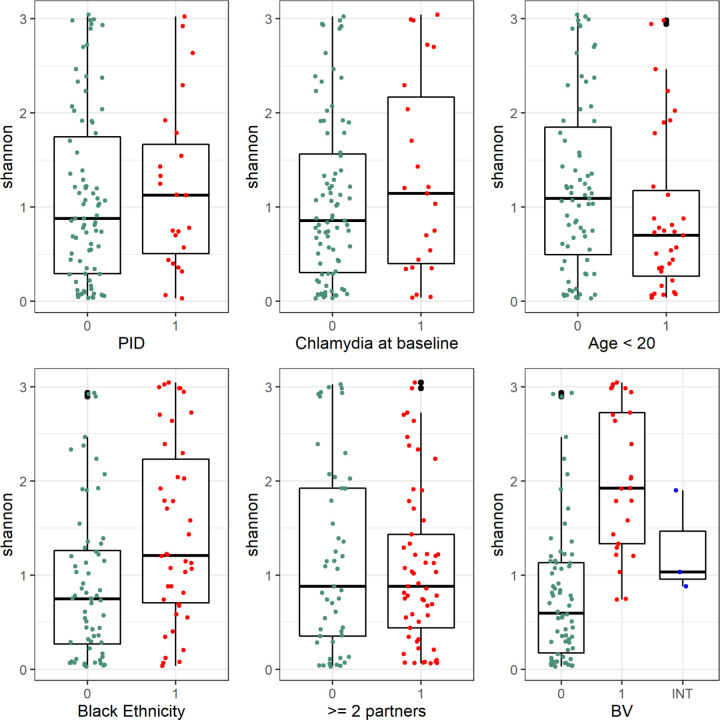
Comparing Shannon alpha diversity (figure 3) across six risk factors (PID, C. trachomatis, BV, age, ethnicity and multiple partners) showed no association between a more diverse microbiome and subsequent PID (Kruskal-Wallace rank sum test p=0.53) or concurrent C. trachomatis infection (p=0.38). However, there was a significantly higher diversity in samples from participants aged ≥20 years (p=0.048), participants of black ethnic group (p=0.002) and those with a diagnosis of bacterial vaginosis (BV, p<0.0001).
Figure 3.
Shannon diversity plotted for each clinical risk factor. Kruskal-Wallis rank sum test shows differences in Shannon diversity between groups for bacterial vaginosis (BV; n=22, p=5.4 ×x10-8), black ethnic group (n=42, p=0.002), age <20 (n=33, p=0.048 less diverse), but no differences for pelvic inflammatory disease (n=23, PID: p=0.53), Chlamydia at baseline (n=23, p=0.38), ≥2 sexual partners (n=57, p=0.88). 1 (red)=Has risk factor, 0 (green)=does not have risk factor.
PID was not associated with lower abundance of D(−) isomer lactic acid producing lactobacilli
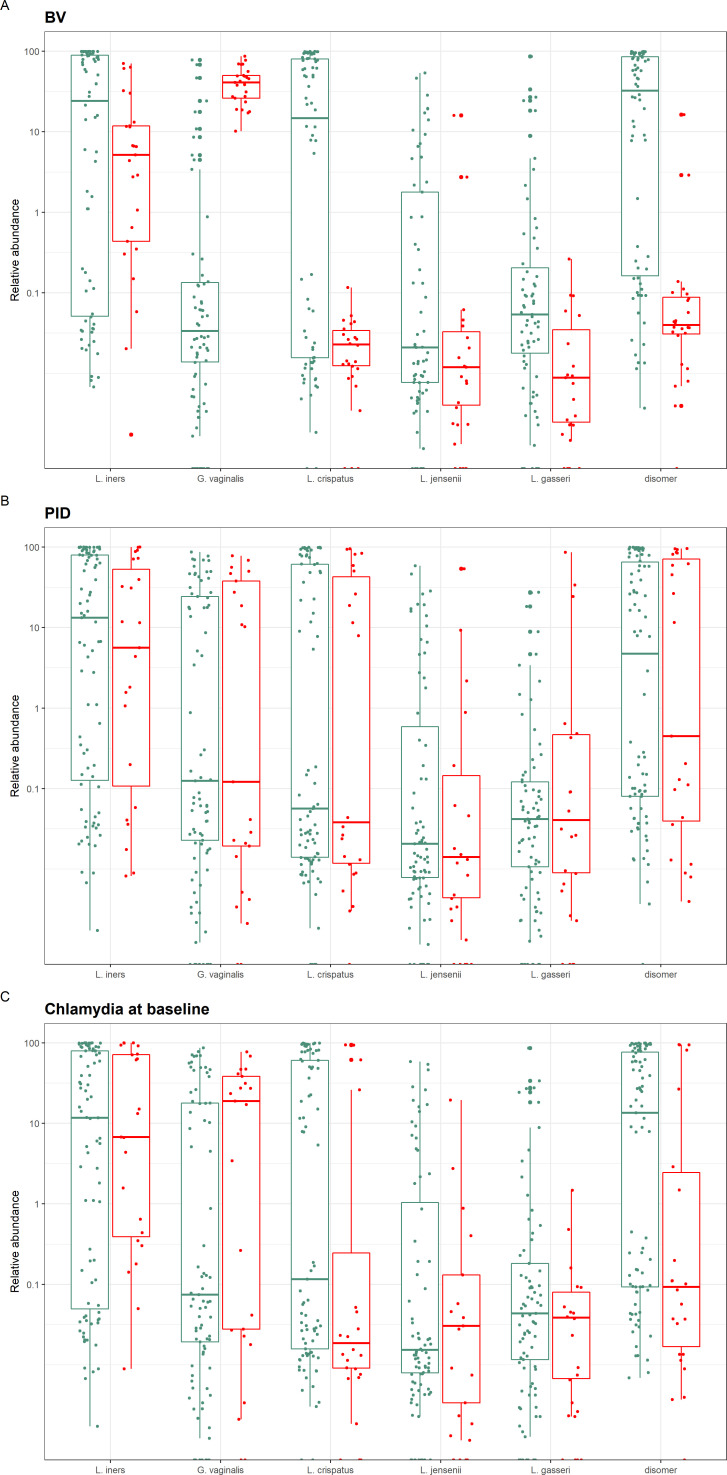
Figure 4 demonstrates the relative abundance of the five main species Lactobacillus iners, Lactobacillus crispatus, Gardnerella vaginalis, Lactobacillus jensenii and Lactobacillus gasseri (including a combination of D(−) isomer lactic acid producing lactobacilli L. crispatus, L. jensenii and L. gasseri), related to the three main clinical diagnoses:
Figure 4.
Relative abundance of the dominant species, including a combination of D(−) isomer lactic acid producing lactobacilli (Lactobacillus crispatus, L. jensenii and L. gasseri), compared for the three main clinical diagnoses: box A: bacterial vaginosis (BV); box B: pelvic inflammatory disease (PID); box C: chlamydia trachomatis at baseline. Red: has clinical diagnosis. Green: does not have clinical diagnosis. BV (A) was associated with lower abundances of D(−) isomer lactic acid producing lactobacilli species (p=4.05×10-6) and higher abundance of Gardnerella vaginosis (p = 3.06×10-9), but this did not apply to PID (B) or Chlamydia.
BV at baseline.
PID within the next 12 months.
C. trachomatis at baseline.
This shows that unlike BV, neither PID nor C. trachomatis, was associated with lower abundance of D(−) isomer lactic acid producing lactobacilli species (p=0.77 and 0.33, respectively). Online supplemental figure 3 shows similar results.
Beta diversity shows no association with PID
Visualisation of Bray-Curtis based NMDS dimensions one and two (stress=0.09 after 20 iterations (online supplemental figure 4) showed no association with subsequent PID or concurrent C. trachomatis. However for BV, three clusters correlated with the dominant species shown in figure 1 (L. iners, L. crispatus and G. vaginalis), with dimensions one and two distinguishing BV versus no BV (online supplemental figure 4A, B, respectively).
Discussion
Principal findings
In the first ever community-based, frequency-matched prospective study of vaginal microbiota in women who did or did not develop PID, we found no association between a more diverse microbiota and subsequent pelvic inflammatory disease. Women with bacterial vaginosis had lower relative abundance of protective D(−) isomer lactic acid producing lactobacilli, but this did not apply to women who developed PID.
Strengths and weaknesses
This study is unique as it is the only community-based prospective study of the vaginal microbiome and PID, and the first to explore the possible association of PID with low levels of vaginal D(−) isomer lactic acid producing lactobacilli. Previous studies were based in hospitals or clinics,5 9 22 or were cross-sectional rather than prospective.23 Forty per cent of participants were of black ethnicity, a group who may have higher rates of bacterial vaginosis8 24 and STIs10 compared with other ethnic groups. This is supported by our finding that the G. vaginalis taxonomic cluster was associated with black ethnicity, an association that lends internal validity to this study.8 The project also responds to recent calls for vaginal microbiome research to include prospective studies5 and data on sexual behaviour.2
To our knowledge, this is also the first study to use long-read sequencing on self-taken vaginal swabs. Combined with evidence that unsupervised self-collected vaginal swabs give similar microbiota results to clinician-collected swabs,25 this study supports feasibility of larger scale community-based projects. Encouragingly, the clusters we identified using long-read sequencing were similar to those found in earlier studies9 24 with three main clusters: one dominated by L. iners, one by L. crispatus and a more diverse cluster dominated by anaerobes. Further strengths include our pilot work confirming the integrity of stored samples11 and the availability of detailed information on the demographics and baseline infection status of all participants, with sequence analysis being conducted blind to these characteristics. Finally, the large number of participants (>2300) in the POPI trial ensured that controls were well matched.
The main limitation is that this was an exploratory study with a small number of PID cases. However, it is similar in size to earlier studies7 9 23 and contributes to the very limited available data on vaginal microbiota composition in relation to PID. The small sample size also meant we could not do complex epidemiological analyses. Another major weakness is the absence of negative and positive controls. The lack of laboratory negative extraction controls meant we could not be sure that B. gladioli was a contaminant. Removal of affected samples did not seem to influence the results, although it did reduce the power of the study. PID is a clinical diagnosis of low specificity, with around a third of diagnoses not confirmed on laparoscopy. This could also weaken the power of the study. However, diagnosis was confirmed by three experienced sexual health physicians.10 Although we found a significant association between BV and lower abundance of D-isomer lactic acid producing lactobacilli, we could not attribute causality or measure D(−) isomer lactic acid concentrations in these samples. We only assessed samples at one time point, and there was an estimated time gap of 3–12 months between obtaining the samples and diagnosis of PID, with only two women diagnosed with PID within 4 months of sampling. The vaginal microbiota may have changed over time in a significant proportion of women, potentially weakening the power of the study.6 However, this was unavoidable in a prospective study of PID.22
Another weakness is that the sensitivity of samples stored for 16 years may be reduced. Nonetheless, other sequencing studies using stored samples have shown reliable results.11 26 Although commonly used, the Kruskal-Wallis rank sum test is not generally considered appropriate for differential analysis of compositional data. A differential abundance analysis approach that is suited to compositional data would have been preferable. We used 16S rRNA gene sequencing, which does not cover fungi, protozoa and viruses, and may not reliably identify low loads of C. trachomatis or N. gonorrhoeae.3 However, samples had already been tested for C. trachomatis, N. gonorrhoeae and M. genitalium by PCR.10 15 We could not evaluate three recently identified novel Gardnerella species,27 as it is unclear whether full length 16S rRNA sequencing adequately differentiates the species within the Gardnerella genus, and they are not yet included in NCBI and other 16S rRNA databases. In addition, the taxon for bacterial vaginosis associated bacteria such as BVAB-1 (Candidatus Lachnocurva vaginae) may be misclassified as Shuttleworthia using the NCBI database. Finally, we analysed by the dominant species within a community state type9 rather than by actual community state type,28 but this would likely give similar results.
Comparison with other studies
There is a dearth of studies of vaginal microbiota in women with and without PID,3 23 29 and like ours, none have shown a clear relationship between the vaginal microbiome and subsequent PID. A cross-sectional study from China used high-throughput sequencing on pelvic and cervical samples from 38 women with PID and 19 controls. They found microbiota in PID could be dominated either by a single organism or by polymicrobial infection.23 Analyses of bacterial vaginosis and incident PID in the Gynecologic Infection Follow-Through study were also inconclusive, with one analysis showing no association22 and others that women with bacterial vaginosis-associated bacteria had increased risk of PID.6 14 Interestingly, microbiological analysis of POPI samples15 found bacterial vaginosis was not significantly associated with PID after adjustment for baseline C. trachomatis infection. Most recently, Trent and colleagues analysed samples from 26 women aged 13–25 years with PID who were enrolled in the Technology Enhanced Community Health Nursing study.29 Over half of participants had low abundance of Lactobacillus species indicative of bacterial vaginosis.
Other studies of the vaginal microbiome in healthy ethnically diverse women have found similar main clusters/community state types to those in our study, two dominated by lactobacilli and one with higher proportions of anaerobic organisms.9 24 A recent study of the vaginal microbiome during genital infections4 found that L. crispatus was progressively replaced by L. iners in the shift from a healthy to an infected microbiome. As in our study, this was mainly characterised by anaerobic genera such as Gardnerella, Prevotella, Megasphaera, Roseburia and Atopobium. We did not find an association between C. trachomatis and L. iners dominated vaginal microbiota,9 but numbers with C. trachomatis infection were small. Finally, our finding of lower rates of protective D(−) isomer lactic acid producing lactobacilli in women with (vs without) concurrent bacterial vaginosis confirms other studies.7 The cervicovaginal microbiota could modulate host functions to protect against infection.7
Implications
This study supports the feasibility of larger scale community-based projects, the use of self-collected vaginal swabs and the integrity of stored samples. The significantly higher abundance of D(−) isomer lactic acid producing lactobacilli in women without bacterial vaginosis is in line with other studies suggesting a protective role for these lactobacilli.3 7 30 Finally, negative studies are important. The key impact from this study is the need for more work, particularly serial sampling studies, to gain knowledge of predisposing vaginal communities that may lead to PID.
Key messages.
There are few data on vaginal microbiota and subsequent development of pelvic inflammatory disease (PID).
A lactobacilli-dominated vaginal microbiome may protect against pelvic inflammatory disease, but one dominated by Gardnerella species might increase susceptibility.
In this first ever community-based prospective study of PID, there was no clear association between vaginal microbiota and development of PID in the next 12 months.
In line with previous studies, findings highlight the need for large serial sampling studies to identify vaginal microbiota that might predispose to PID.
Acknowledgments
We would like to thank Professors David Martin and David Strachan for advice on study design.
Footnotes
Handling editor: Jo Gibbs
SK-B, LZ, AAW, STS and PO contributed equally.
Contributors: STS, PO and SKB designed the study; LZ and LP conducted the long-read 16S rRNA gene nanopore sequencing; AAW conducted the bioinformatics analysis; MF advised on statistics; PO and SKB wrote the first draft; and all authors contributed to the final version. PO, STS and AAW are responsible for the overall content as guarantor(s).
Funding: The Wellcome Trust Institution Strategic Support Fund grant number 204809/Z/16/Z funded the sequencing of samples. Additional support came from National Institute for Health Research, Invention for Innovation (i4i) grant: 'A Point of Care Antimicrobial Resistance test for Neisseria gonorrhoeae and Mycoplasma genitalium infection - Ensuring accurate therapy and antibiotic stewardship in sexual health medicine' (II-LB-0214–20005). The Prevention of Pelvic Infection trial was funded by the BUPA Foundation grant number 684/GB14B and the UK Medical Research Council grant 80 280.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as supplemental information. Not applicable.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human participants and was approved by Bromley research ethics committee reference 07/Q0705/16. Participants gave informed consent to participate in the study before taking part.
References
- 1. Haggerty CL, Totten PA, Tang G, et al. Identification of novel microbes associated with pelvic inflammatory disease and infertility. Sex Transm Infect 2016;92:441–6. 10.1136/sextrans-2015-052285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lewis FMT, Bernstein KT, Aral SO. Vaginal microbiome and its relationship to behavior, sexual health, and sexually transmitted diseases. Obstet Gynecol 2017;129:643–54. 10.1097/AOG.0000000000001932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van de Wijgert JHHM, JHHM vandeW. The vaginal microbiome and sexually transmitted infections are interlinked: consequences for treatment and prevention. PLoS Med 2017;14:e1002478. 10.1371/journal.pmed.1002478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ceccarani C, Foschi C, Parolin C, et al. Diversity of vaginal microbiome and metabolome during genital infections. Sci Rep 2019;9:14095. 10.1038/s41598-019-50410-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tamarelle J, Thiébaut ACM, de Barbeyrac B, et al. The vaginal microbiota and its association with human papillomavirus, Chlamydia trachomatis, Neisseria gonorrhoeae and Mycoplasma genitalium infections: a systematic review and meta-analysis. Clin Microbiol Infect 2019;25:35–47. 10.1016/j.cmi.2018.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ness RB, Kip KE, Hillier SL, et al. A cluster analysis of bacterial vaginosis-associated microflora and pelvic inflammatory disease. Am J Epidemiol 2005;162:585–90. 10.1093/aje/kwi243 [DOI] [PubMed] [Google Scholar]
- 7. Edwards VL, Smith SB, McComb EJ, et al. The cervicovaginal Microbiota-Host interaction modulates Chlamydia trachomatis infection. mBio 2019;10. 10.1128/mBio.01548-19. [Epub ahead of print: 13 08 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Borgdorff H, van der Veer C, van Houdt R, et al. The association between ethnicity and vaginal microbiota composition in Amsterdam, the Netherlands. PLoS One 2017;12:e0181135. 10.1371/journal.pone.0181135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van Houdt R, Ma B, Bruisten SM, et al. Lactobacillus iners-dominated vaginal microbiota is associated with increased susceptibility to Chlamydia trachomatis infection in Dutch women: a case-control study. Sex Transm Infect 2018;94:117–23. 10.1136/sextrans-2017-053133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oakeshott P, Kerry S, Aghaizu A, et al. Randomised controlled trial of screening for Chlamydia trachomatis to prevent pelvic inflammatory disease: the POPI (prevention of pelvic infection) trial. BMJ 2010;340:1642. 10.1136/bmj.c1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wagner J, Kerry-Barnard S, Sadiq ST, et al. Research benefits of storing genitourinary samples: 16S rRNA sequencing to evaluate vaginal bacterial communities. Int J STD AIDS 2017;28:315–7. 10.1177/0956462416689441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shin J, Lee S, Go M-J, et al. Analysis of the mouse gut microbiome using full-length 16S rRNA amplicon sequencing. Sci Rep 2016;6:29681. 10.1038/srep29681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Quan L, Dong R, Yang W, et al. Simultaneous detection and comprehensive analysis of HPV and microbiome status of a cervical liquid-based cytology sample using Nanopore MinION sequencing. Sci Rep 2019;9:19337. 10.1038/s41598-019-55843-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haggerty CL, Ness RB, Totten PA, et al. Presence and concentrations of select bacterial vaginosis-associated bacteria are associated with increased risk of pelvic inflammatory disease. Sex Transm Dis 2020;47:344–6. 10.1097/OLQ.0000000000001164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hay PE, Kerry SR, Normansell R, et al. Which sexually active young female students are most at risk of pelvic inflammatory disease? A prospective study. Sex Transm Infect 2016;92:63–6. 10.1136/sextrans-2015-052063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Klindworth A, Pruesse E, Schweer T, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 2013;41:e1. 10.1093/nar/gks808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cuscó A, Catozzi C, Viñes J, et al. Microbiota profiling with long amplicons using nanopore sequencing: full-length 16S rRNA gene and whole rrn operon. F1000Res 2018;7:7:1755. 10.12688/f1000research.16817.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marijon P, Chikhi R, Varré J-S. yacrd and fpa: upstream tools for long-read genome assembly. Bioinformatics 2020;36:3894–6. 10.1093/bioinformatics/btaa262 [DOI] [PubMed] [Google Scholar]
- 19. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 2018;34:3094–100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Imataki O, Kita N, Nakayama-Imaohji H, et al. Bronchiolitis and bacteraemia caused by Burkholderia gladioli in a non-lung transplantation patient. New Microbes New Infect 2014;2:175–6. 10.1002/nmi2.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Salter SJ, Cox MJ, Turek EM, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 2014;12:87. 10.1186/s12915-014-0087-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ness RB, Hillier SL, Kip KE, et al. Bacterial vaginosis and risk of pelvic inflammatory disease. Obstet Gynecol 2004;104:761–9. 10.1097/01.AOG.0000139512.37582.17 [DOI] [PubMed] [Google Scholar]
- 23. Wang Y, Zhang Y, Zhang Q, et al. Characterization of pelvic and cervical microbiotas from patients with pelvic inflammatory disease. J Med Microbiol 2018;67:1519–26. 10.1099/jmm.0.000821 [DOI] [PubMed] [Google Scholar]
- 24. Ravel J, Gajer P, Abdo Z, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 2011;108(Suppl 1):4680–7. 10.1073/pnas.1002611107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wylie KM, Blankenship SA, Tuuli MG, et al. Evaluation of patient- versus provider-collected vaginal swabs for microbiome analysis during pregnancy. BMC Res Notes 2018;11:706. 10.1186/s13104-018-3809-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bai G, Gajer P, Nandy M, et al. Comparison of storage conditions for human vaginal microbiome studies. PLoS One 2012;7:e36934. 10.1371/journal.pone.0036934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hill JE, Albert AYK, VOGUE Research Group . Resolution and Cooccurrence patterns of Gardnerella leopoldii, G. swidsinskii, G. piotii, and G. vaginalis within the vaginal microbiome. Infect Immun 2019;87. 10.1128/IAI.00532-19. [Epub ahead of print: 18 11 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. France MT, Ma B, Gajer P, et al. VALENCIA: a nearest centroid classification method for vaginal microbial communities based on composition. Microbiome 2020;8:166. 10.1186/s40168-020-00934-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Trent ME, Perin J, Matson P. Vaginal microbiota among adolescent and young adult women with pelvic inflammatory disease. Sex Transm Infect 2019:A265. [Google Scholar]
- 30. Cohen CR, Wierzbicki MR, French AL, et al. Randomized trial of Lactin-V to prevent recurrence of bacterial vaginosis. N Engl J Med 2020;382:1906–15. 10.1056/NEJMoa1915254 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
sextrans-2021-055260supp001.pdf (569KB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplemental information. Not applicable.