Abstract
Splicing of plant mitochondrial introns is facilitated by numerous nucleus-encoded protein factors. Although some splicing factors have been identified in plants, the mechanism underlying mitochondrial intron splicing remains largely unclear. In this study, we identified a small P-type pentatricopeptide repeat (PPR) protein containing merely four PPR repeats, small PPR protein 2 (SPR2), which is required for the splicing of more than half of the introns in maize (Zea mays) mitochondria. Null mutations of Spr2 severely impair the splicing of 15 out of the 22 mitochondrial Group II introns, resulting in substantially decreased mature transcripts, which abolished the assembly and activity of mitochondrial complex I. Consequently, embryogenesis and endosperm development were arrested in the spr2 mutants. Yeast two-hybrid, luciferase complementation imaging, bimolecular fluorescence complementation, and semi-in vivo pull-down analyses indicated that SPR2 interacts with small MutS-related domain protein PPR-SMR1, both of which are required for the splicing of 13 introns. In addition, SPR2 and/or PPR-SMR1 interact with other splicing factors, including PPR proteins EMPTY PERICARP16, PPR14, and chloroplast RNA splicing and ribosome maturation (CRM) protein Zm-mCSF1, which participate in the splicing of specific intron(s) of the 13 introns. These results prompt us to propose that SPR2/PPR-SMR1 serves as the core component of a splicing complex and possibly exerts the splicing function through a dynamic interaction with specific substrate recognizing PPR proteins in mitochondria.
The interaction between two unique pentatricopeptide repeat proteins mediates the splicing of over half of the Group II introns in maize mitochondria.
Introduction
Mitochondria are eukaryotic organelles that are derived from α-proteobacteria through endosymbiosis around 1.7 billion years ago. During evolution, most genes from the α-proteobacteria progenitor were lost or migrated to the host nuclear genome, leaving roughly 5% of genes in the mitochondrial genomes of land plants (Gabaldon and Huynen, 2004). As a result of co-evolution, the expression of mitochondrial genes is highly regulated by the nuclear genome, particularly at the posttranscriptional level, including RNA editing, cleavage, maturation, stabilization, and intron splicing (Hammani and Giege, 2014).
Intron splicing is essential to the expression of the mitochondrial genes. Introns are classified into Groups I and II based on the tertiary structures (Bonen and Vogel, 2001). Group II introns exist in bacteria and organelle genomes of lower eukaryotes and plants. They are large catalytic RNAs that also act as mobile genetic elements (Lambowitz and Zimmerly, 2004, 2011). Almost all of the mitochondrial introns in plants are Group II. In bacteria, Group II introns can self-splice, which is facilitated by the intron-encoded cognate maturase (Mat) (Singh et al., 2002). However, mitochondrial Group II introns in land plants have lost the self-splicing activity caused by the loss of maturase genes in the introns and compounded by accumulated mutations in the intron sequences (de Longevialle et al., 2010; Lambowitz and Zimmerly, 2004). Only one maturase (matR) gene is found in the NADH dehydrogenase1 (nad1) intron 4 in plant mitochondrial genomes (Wahleithner et al., 1990). Instead, splicing of these Group II introns invokes a recruitment of many nucleus-encoded proteins from over seven families, which include nuclear-encoded maturases (nMATs) (Nakagawa and Sakurai, 2006; Keren et al., 2009, 2012; Cohen et al., 2014; Shevtsov-Tal et al., 2021), mitochondrial transcription termination factor (mTERF) proteins (Hsu et al., 2014; Yang et al., 2020b), DEAD-box proteins (Köhler et al., 2010), chloroplast RNA splicing and ribosome maturation (CRM) proteins (Zmudjak et al., 2013; Chen et al., 2019), plant organelle RNA recognition (PORR) proteins (Colas des Francs-Small et al., 2012), regulator of chromosome condensation (RCC1) family proteins (Kuhn et al., 2011; Cao et al., 2021), and pentatricopeptide repeat (PPR) proteins (Barkan and Small, 2014; Brown et al., 2014).
PPR proteins are characterized by having a tandem array of degenerated ∼35 amino acid PPR motifs (Lurin et al., 2004). Each PPR motif forms a hairpin structure with two anti-paralleled α-helixes, and each motif recognizes one nucleotide in RNA via interaction with the residues at 6 and 1′ in the motif (Barkan and Small, 2014). PPRs belong to a large protein family with over 600 members in maize (Zea mays) (Lurin et al., 2004; Chen et al., 2018). Based on the domain structure, PPR proteins are divided into P-subclass and PLS-subclass. Most PLS-subclass PPR proteins function in RNA editing, whereas P-subclass PPR proteins are found to function in translation initiation, RNA stabilization, RNA maturation, RNA cleavage, and intron splicing (Barkan and Small, 2014). P-subclass PPR proteins are involved in the splicing of one to a few introns in mitochondria except PPR-SMR1, a SMR domain-containing PPR that is required for the splicing of 16 Group II introns in maize mitochondria (Chen et al., 2019). For example, PPR proteins ORGANELLE TRANSCRIPT PROCESSING 43 (OTP43) and ABA overly sensitive 5 (ABO5) are required for the splicing of nad1 intron 1 and nad2 intron 3 in Arabidopsis thaliana, respectively (de Longevialle et al., 2007; Liu et al., 2010). EMPTY PERICARP16 (EMP16) functions in the splicing of nad2 intron 4 in maize (Xiu et al., 2016). Defective kernel 35 (DEK35) and PPR18 are required for the cis-splicing of nad4 intron 1 (Chen et al., 2016; Liu et al., 2020). Because of the importance of these genes in mitochondrial oxidative respiration, loss of intron splicing often cause severe defects in plant growth and development and even embryo lethality.
Despite the knowledge that some P-type PPR proteins function in mitochondrial intron splicing, the molecular mechanism remains unclear. Given the size and complexity of the PPR protein family, functions of special PPR proteins remain to be deciphered, particularly the PPRs with few PPR repeats, as these proteins cannot confer target site-binding specificity. In this study, we characterized a P-type PPR protein with merely four PPR repeats, small PPR protein 2 (SPR2). Unlike the reported PPR proteins, SPR2 is required for the splicing of 15 out of the 22 Group II introns in maize mitochondria. Surprisingly, 13 of these introns overlap with those that require small MutS-related domain protein PPR-SMR1 for splicing. We demonstrate that SPR2 interacts with PPR-SMR1, and SPR2/PPR-SMR1 interacts with other known splicing factors that are required for the splicing of these introns. These results show that SPR2 and PPR-SMR1 form a core complex, which interacts with other splicing factors to carry out intron splicing. This mechanism may account for the splicing of 13 out of the 22 Group II introns in maize mitochondria. This finding extends the understanding of the intron splicing mechanism in mitochondria.
Results
Phenotypic and genetic characterization of spr2
PPR proteins recognize RNA substrates via a one-repeat one-nucleotide mechanism (Barkan et al., 2012; Yin et al., 2013). This raises the question of how the specificity is determined for PPRs with few PPR repeats. Theoretically, a PPR protein with four PPR repeats could recognize four sites in any 1-kb mRNA, evidently too many to confer a tight specificity. To answer this question, we attempted to identify the function of PPR proteins with less than four PPR repeats. This led us to study the function of GRMZM2G122344 designated as SPR2, which contains four PPR repeats, and the mutants are available from the Maize Genetics Stock Center.
We first examined whether a loss of SPR2 would affect plant growth and development in maize. Two insertional mutants were obtained with Mu insertions at +21 and +529 bp from the translation start codon ATG of Spr2, named spr2-1 and spr2-2, respectively (Figure 1A). The selfed progeny of spr2-1/+ and spr2-2/+ heterozygous plants segregated wild-type (WT) and empty pericarp (emp) kernels at a 3:1 ratio (spr2-1, 311:102, P > 0.9; spr2-2, 330:108, P > 0.9) (Figure 1, B and C; Supplemental Figure S1A), suggesting that the emp phenotype is caused by a monogenic and recessive mutation. Linkage analysis by polymerase chain reaction (PCR) genotyping using Spr2-specific primers and the Mu-specific primer Mu-TIR8 showed that the Mu insertion is tightly linked to the emp phenotype in a population of 32 plants (Supplemental Figure S2). Reciprocal crosses between spr2-1/+ and spr2-2/+ heterozygotes produced approximately a quarter emp mutant kernels (Figure 1D), indicating that Spr2 is the causative gene for the emp phenotype. Reverse transcription-PCR (RT–PCR) analyses indicated that the WT Spr2 transcripts could not be detected in the spr2 alleles (Figure 2B), suggesting that both alleles are probably null.
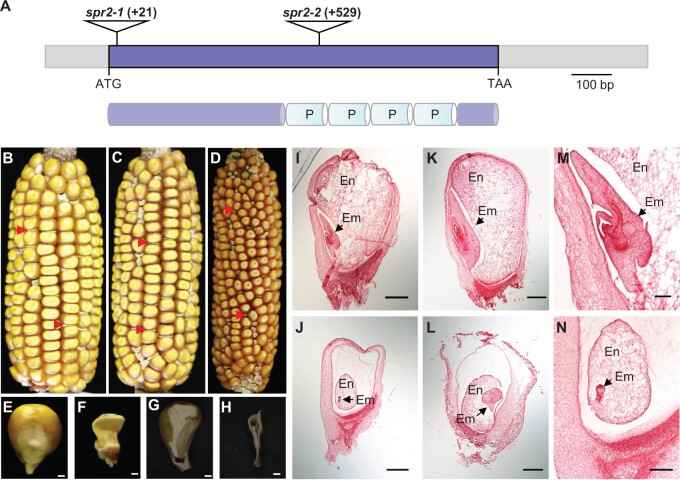
Figure 1.
Mutation of Spr2 blocks embryogenesis and endosperm development. A, The top part represents the gene structure of Spr2, and the lower represents the protein structure of SPR2. The predicted SPR2 protein contains four P-type PPR motifs (P). Mu insertions in two alleles were indicated by triangles. B, A self-pollinated spr2-1/+ ear. Arrows point to spr2-1 mutant kernels. C, A self-pollinated spr2-2/+ ear. Arrows point to spr2-2 mutant kernels. D, Allelic test of spr2-1 and spr2-2 alleles. Arrows point to spr2 mutant kernels. E and F, Mature WT (E) and spr2-1 (F) kernels. G and H, Dissection of mature WT (G) and spr2-1 (H) kernels. I–N, Comparison of WT and spr2-1 kernel development at 10 DAP and 15 DAP. WT kernels at 10 DAP (I and M) and 15 DAP (K); spr2-1 kernels at 10 DAP (J and N) and 15 DAP (L). En, endosperm; Em, embryo. Scale bars, 1 mm in (E–H); 0.5 mm in (I–N).
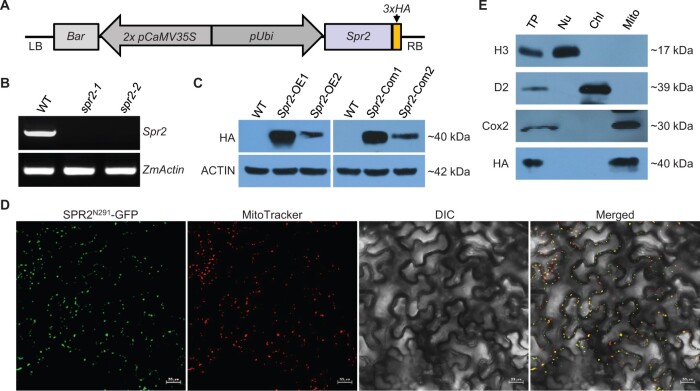
Figure 2.
Spr2 encodes a mitochondrion-localized PPR protein. A, Transgenic construct for overexpressing Spr2. LB: left border; RB: right border; 3 × HA: triple HA tag. B, Analysis of transcript levels of Spr2 in spr2 mutants by RT–PCR analysis using primer Spr2-F3 and Spr2-R3. RNAs were normalized against maize Actin gene (GRMZM2G126010). C, Analysis of protein abundance of SPR2 in overexpression lines (Spr2-OE) and complemented lines (Spr2-Com) by western blotting analysis. Maize ACTIN protein was used as a loading control. D, Subcellular localization of SPR2. Red fluorescence signals are from mitochondrial marker MitoTracker. Scale bar, 20 μm. E, Immunoblotting localization of SPR2 in total protein (TP), nuclear protein (Nu), mitochondrial protein (Mito), and chloroplast protein (Chl) extracts obtained from overexpression lines Spr2-OE1 probed with antibodies against H3 (Histone H3, nuclei marker), D2 (chloroplastic marker), Cox2 (mitochondrial marker), and HA.
To further confirm that the emp phenotype is caused by disruption of Spr2, we generated transgenic plants over-expressing Spr2 and crossed onto the mutants. The full-length coding sequence of Spr2 fused with HA tag under the control of maize ubiquitin1 promoter was transformed into the maize inbred line KN5585 (Figure 2A; Supplemental Figure S1B). Eleven independent transgenic lines (Spr2-OE1 to OE11) were obtained. RT–qPCR results showed that the expression levels of Spr2 are approximately 100–3,000 times higher in the Spr2-OE lines than that in the WT (Supplemental Figure S3A). The spr2-2 heterozygous plants were crossed with Spr2-OE1 and Spr2-OE2 to generate the F1 plants. The ones harboring both the Mu insertion in Spr2 and the transgene were selfed to obtain the F2 progeny. From the seedlings of the F2, viable homozygous spr2-2 mutants were identified by PCR using primer Ubi-F and Spr2-R1 (Supplemental Figure S4, A and B). All the spr2-2 mutants expressed Spr2, which was detected by western blotting using the HA antibody (Figure 2C). These seedlings and ears grew as normally as the WT (Supplemental Figures S1, C and S4, C). These data indicate that over-expression of Spr2 rescues the embryo-lethality of spr2, confirming that the emp phenotype is indeed caused by the loss of SPR2.
Compared with the WT, the kernel development is severely impaired in the spr2 mutants (Figure 1, B–H; Supplemental Figure S1A). No embryo or endosperm structures could be identified in spr2-1 mature kernels (Figure 1H), indicating that the embryo and endosperm development was arrested. Paraffin sectioning showed that while the WT embryo reached the coleoptilar stage (Figure 1, I and M) at 10-day after pollination (DAP), the spr2-1 embryo remained at the preembryo stage (Figure 1, J and N). At 15 DAP, the WT embryo entered the late embryogenesis stage (Figure 1K), whereas the spr2-1 embryo remained at the early transition stage (Figure 1L). The endosperm in spr2-1 was clearly underdeveloped compared with that of the WT (Figure 1, I–N). These results indicate that the Spr2 mutation arrests the embryo and endosperm development in maize.
SPR2 is targeted to mitochondria
Spr2 is an intronless gene encoding a 35-kDa protein with 320 amino acid residues. SPR2 harbors only four P-type PPR motifs as predicted by the TPRpred algorithm (http://tprpred.tebingen.mpg.de/tprpred) (Figure 1A). RT–qPCR analysis showed that Spr2 is ubiquitously expressed with a high expression level in shoots (Supplemental Figure S3B), indicating that Spr2 is a constitutive gene that may play an important role in maize growth and development.
To determine the subcellular localization of SPR2, we fused the N-terminal 291 amino acids of SPR2 with GFP at the C-terminus and then transfected Nicotiana benthamiana leaves via Agrobacterium-mediated infiltration. Green fluorescence signals were detected in dots that were merged with the red signals of MitoTracker (Figure 2D), indicating that SPR2 is targeted to mitochondria. To confirm the localization, we performed the western blotting analysis on fractions of different cellular compartments of the transgenic Spr2-OE1 plants by using the HA antibody. Histone H3, D2, and Cox2 were used as markers of nucleus, chloroplast, and mitochondrion, respectively. The results showed that SPR2 was only detected in the mitochondrial fraction (Figure 2E), demonstrating that SPR2 is localized in mitochondria.
Spr2 mutation inhibits the respiration while enhancing the AOX pathway
To assess whether the mutation of SPR2 affects mitochondrial respiration, we measured the mitochondrial respiratory rates in the WT and spr2-1. Total respiratory (Vt), cytochrome respiratory capacity (Vcyt), and alternative respiratory capacity (Valt) were quantified. As shown in Supplemental Table S1, the percentage of Valt/Vt was substantially increased in spr2-1 compared with WT, whereas the proportion of Vcyt/Vt was sharply decreased. These data indicate that the cytochrome pathway respiration is impaired, and the alternative oxidase pathway is remarkably enhanced in spr2. Western blotting analysis showed that AOX abundance was increased substantially in spr2-1 compared with WT (Figure 3E), suggesting that the AOX pathway is enhanced in spr2. The maize genome contains three AOX genes, AOX1, AOX2, and AOX3. RT–PCR and RT–qPCR results showed that the transcripts of AOX2 and AOX3 were dramatically increased in the spr2 alleles compared with WT (Supplemental Figure S5, A and B). These data indicate Spr2 loss of function impairs the cytochrome pathway and enhances the alternative oxidase pathway.
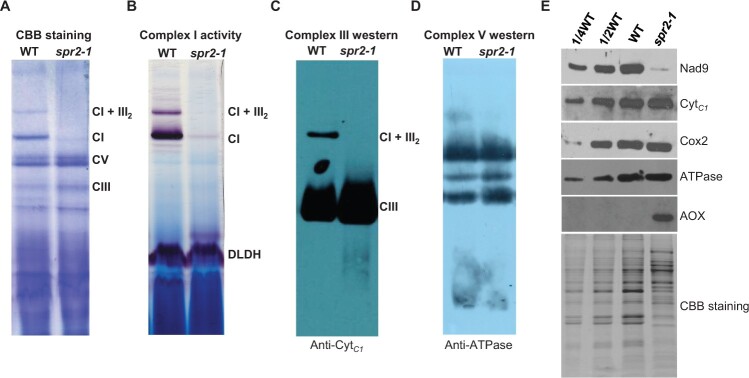
Figure 3.
The mutation of Spr2 abolished complex I assembly and activity. A, The BN-PAGE gels were stained with CBB. Crude mitochondrial extracts from endosperm and embryo of spr2-1 and WT immature kernels at 11 DAP. The positions of respiratory complexes I, III, V, and supercomplex I + III2 are indicated. CI: Complex I, CIII: Complex III, CV: Complex V. B, Detection of NADH dehydrogenase activity of complex I. Dihydrolipoamide dehydrogenase was used as a loading control. C and D, Accumulation of mitochondrial complex III and V in Spr2-1 mutant and WT by western blotting using specific antibodies against CytC1 (C) and ATPase (D). E, Immunodetection analysis with antibodies against Nad9, CytC1, Cox2, ATPase, and AOX.
Loss of Spr2 affects the assembly and activity of complex I
To determine whether the mitochondrial complexes are affected in spr2, Blue native (BN)-polyacrylamide gel electropheresis (PAGE) and western blotting were performed using crude mitochondrial proteins isolated from spr2-1 and WT kernels at 11 DAP. Coomassie Brilliant Blue (CBB) staining and In-gel NBT-NADH activity staining showed that the complex I and super-complex CI + CIII2 were completely deficient in spr2-1 (Figure 3, A and B). Complex III was slightly accumulated in spr2-1 (Figure 3, A and C), whereas complex V remained unchanged (Figure 3, A and D). In addition, we determined the protein abundance of the representative subunit of the mitochondrial complexes by Western blotting analysis using antibodies against Nad9 (NADH dehydrogenase subunit 9, complex I), Cytc1 (cytochrome c1, complex III), Cox2 (a subunit of cytochrome oxidase, complex IV), and ATPase (α subunit of ATPase, complex V). Compared with that in the WT, the abundance of Nad9 was strikingly reduced, Cox2 and ATPase unchanged, and Cytc1 slightly increased (Figure 3E). These results are consistent with the BN-PAGE analysis. Collectively, these results indicate SPR2 is vital for the assembly and activity of complex I in mitochondria.
SPR2 is required for the splicing of 15 Group II introns in mitochondria
To investigate the function of SPR2, we first compared the transcript levels of the 35 mitochondrion-encoded genes between the WT and spr2 mutant kernels at 11 DAP. RT–PCR results indicated that 30 of the mitochondrial genes showed no differences in transcript levels between the WT and spr2. However, the transcripts of nad1, nad2, nad4, nad5, and nad7 were remarkably reduced in the spr2 alleles (Figure 4). Analysis of the WT and spr2-1 kernels at 13 and 19 DAP, and spr2-2 at 17 and 19 DAP resulted in the same result, confirming that Spr2 loss of function results in a deficiency of the mature transcripts of nad1, nad2, nad4, nad5, and nad7 (supplemental Figure S6).
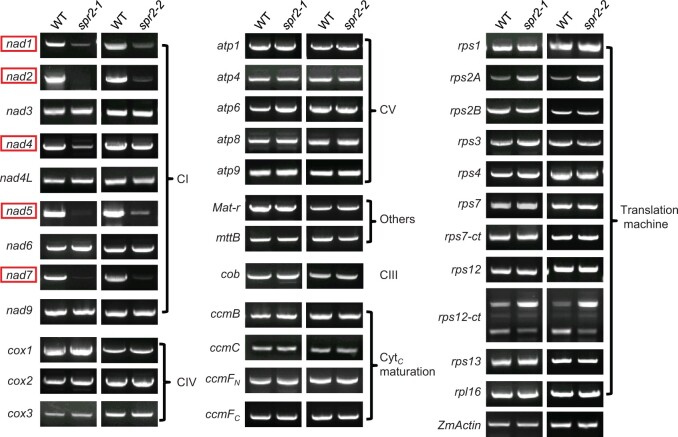
Figure 4.
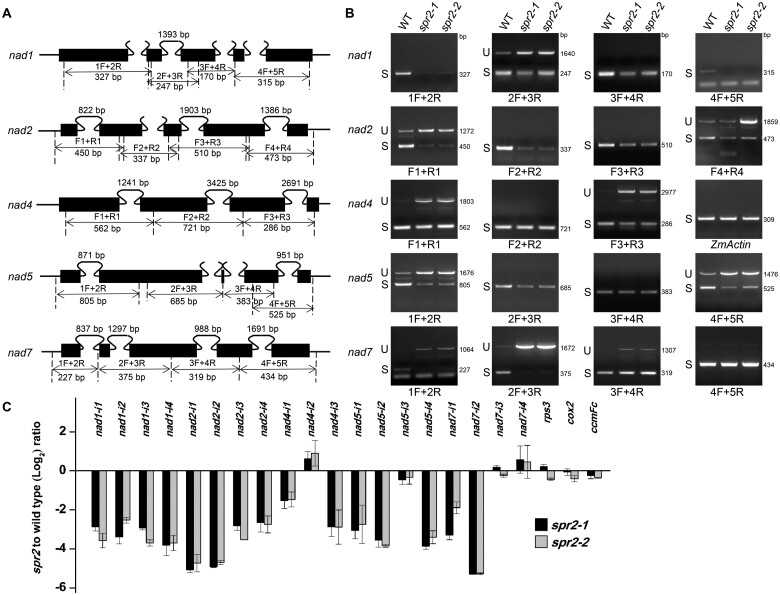
Spr2 mutants affect expression of nad1, nad2, nad4, nad5, and nad7 in the mitochondria. RT–PCR analysis of transcript levels of 35 mitochondrion-encoded genes in WT and spr2 mutants. The substantial decrease of nad1, nad2, nad4, nad5, and nad7 transcripts occurs in spr2 alleles. Normalization was performed against ZmActin (GRMZM2G126010). The RNA was isolated from the same ear segregating for WT and spr2 mutants. CI: Complex I, CIII: Complex III, CIV: Complex IV, CV: Complex V.
Maize mitochondrial nad1, nad2, nad5, and nad7 gene each contains four introns, and nad4 contains three (Figure 5A). To test whether the deficiency of the mature transcripts is caused by defects in intron splicing, we amplified fragments containing each of the introns in these nad transcripts by RT–PCR (Figure 5B;Supplemental Figure S7). Primers were designed to anchor on exons across each intron (Figure 5A). The results showed that splicing of the 15 Group II introns (nad1-int1 to 4; nad2-int1 to 4; nad4-int1, 3; nad5-int1, 2, 4 and nad7-int1, 2) was impaired in both spr2 alleles (Figure 5B;Supplemental Figure S7). In addition, we examined the intron splicing efficiency in spr2 by RT–qPCR analysis. Results indicated that the splicing of the 15 Group II introns was dramatically decreased in spr2 (Figure 5C), which is consistent with the RT-PCR results. These data demonstrate that SPR2 is required for the splicing of these 15 introns in maize mitochondria.
Figure 5.
Spr2 is involved in the splicing of multiple mitochondrial introns. A, Gene structure diagram of the maize nad1, nad2, nad4, nad5, and nad7 gene. Exons are shown as filled black boxes. The primers and expected amplification products are indicated. F: Forward primer, R: Reverse primer. B, RT–PCR analysis of inefficient splicing of mitochondrial introns of nad1, nad2, nad4, nad5, and nad7 in WT and spr2 mutants using primers as indicated in (A). Normalization was performed against ZmActin. S, intron spliced; U, intron unspliced; bp: base pair. C, RT-qPCR analysis of splicing efficiency of all 22 mitochondrial introns in spr2 alleles. The ratio of spliced to unspliced fragments was used to measure splicing efficiency. Data are means (±se) of three biological replicates.
The intron splicing efficiency in Spr2 transgenic lines (Spr2-OE1 and OE2) and complemented lines (Spr2-Com1 and Com2) were examined as well. The results showed that the splicing efficiency of these introns in Spr2 transgenic lines and complemented lines was comparable to that in the WT (Supplemental Figure S8, A and B). These results indicate that (1) over-expression of SPR2-HA restores the splicing defects in the mutants and (2) over-expression of SPR2-HA does not increase the splicing efficiency.
SPR2 interacts with PPR-SMR1 in vitro and in vivo
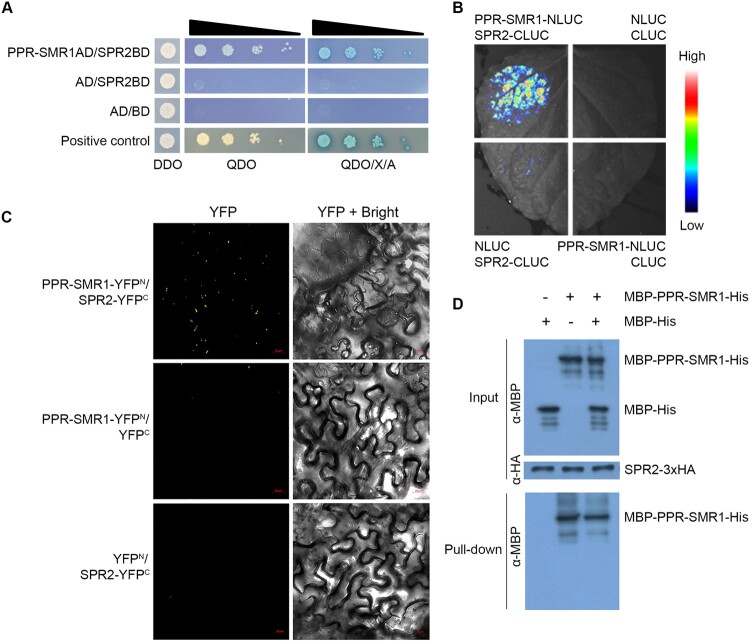
Several splicing factors have been identified in mitochondria in recent years (Dai et al., 2021), but most of these factors are involved in the splicing of only one or a few introns except PPR-SMR1. PPR-SMR1 is required for the splicing of 16 introns in maize mitochondria (Chen et al., 2019). SPR2 is probably the second PPR protein that is required for the splicing of more than half of the introns in mitochondria. Interestingly, a comparison of the introns shows that both SPR2 and PPR-SMR1 are required for the splicing of 13 mitochondrial introns (Supplemental Table S2), raising the possibility that SPR2 may interact with PPR-SMR1 to mediate the splicing. To investigate this possibility, we first tested the interaction by yeast two-hybrid (Y2H) analysis. Results showed that co-transformant expressing PPR-SMR1AD/SPR2BD could grow in QDO and QDO/X/A medium (Figure 6A), suggesting that SPR2 interacts with PPR-SMR1 in Y2H analysis. Next, we tested the SPR2/PPR-SMR1 interaction using the luciferase complementation imaging (LCI) analysis. High levels of luciferase activity were detected in the combination of co-expressing PPR-SMR1 fused to N-terminal luciferase (PPR-SMR1–NLUC) and SPR2 fused to C-terminal luciferase (SPR2-CLUC) (Figure 6B), indicating the interaction of SPR2 and PPR-SMR1. As negative controls, combinations of CLUC/PPR-SMR1–NLUC or SPR2–CLUC/NLUC showed no luciferase activity (Figure 6B). In addition, the bimolecular fluorescence complementation (BiFC) assay was used to test the SPR2/PPR-SMR1 interaction. The results showed strong fluorescence signals in N. benthamiana leaves co-infiltrated with PPR-SMR1 fused to the N-terminal yellow fluorescent protein (YFP) (PPR-SMR1–YFPN) and SPR2 fused to the C-terminal YFP (SPR2–YFPC) (Figure 6C). No YFP fluorescence signal was detected in the combinations of PPR-SMR1–YFPN/YFPC or YFPN/SPR2–YFPC (Figure 6C), indicating that SPR2 physically interacts with PPR-SMR1 in a complex in vivo. We performed semi-in vivo pull-down assays to explore SPR2/PPR-SMR1 interaction in maize (Figure 6D). Mitochondrial proteins containing the target protein SPR2-HA were extracted from the Spr2-OE1 seedlings. The recombinant proteins MBP-PPR-SMR1-His (6 × Histidine) and MBP-His (6 × Histidine) were purified from Escherichia coli. The MBP-His protein was added to the binding reaction as a negative control. MBP-PPR-SMR1-His, MBP-His, and MBP-PPR-SMR1-His/MBP-His were incubated with SPR2-HA protein and Anti-HA Magnetic Beads for 2 h, respectively. After incubation and washing 7 times, the HA beads bound with SPR2-HA protein pulled down MBP-PPR-SMR1-His, but not MBP-His (Figure 6D). The specific immunoprecipitation of PPR-SMR1 by SPR2 in the semi-in vivo pull-down assays was further confirmed in the third lane of a negative control (Figure 6D). These results suggest that SPR2 interacts with PPR-SMR1 in vitro and in vivo.
Figure 6.
SPR2 interacts with PPR-SMR1. A, Interaction analysis of SPR2 and PPR-SMR1 in Y2H assays. The combination of PPR-SMR1AD/SPR2BD was co-transfected into yeast strain Y2H Gold and spotted onto SD/–Leu/–Trp (DDO) medium and growth of diploid yeast colonies on SD/–Ade/–His/–Leu/–Trp (QDO) medium and QDO with added the X-α-gal and AbA (QDO/X/A) medium to reveal protein–protein interactions. Positive interaction was verified by growth on QDO and QDO/X/A plates. AD: GAL4 activation domain; BD: GAL4 DNA binding domain. B, LCI assays to determine interactions between SPR2 and PPR-SMR1. The combinations of PPR-SMR1–NLUC/SPR2–CLUC were co-infiltrated into 4-week-old N. benthamiana leaves. The luciferase signals were visualized using the Lumazone FA Pylon2048B system. The intensity of the fluorescent signals represents their interaction activities. C, BiFC analysis for interactions between SPR2 and PPR-SMR1. YFP is split into N-terminus (YFPN) and C-terminus (YFPC). The combination of PPR-SMR1–YFPN/SPR2–YFPC was co-infiltrated into 4-week-old N. benthamiana leaves using Agrobacterium. YFP signals were detected by a confocal laser microscope. Scale bars, 20 μm. D, Semi-in vivo pull-down assays for interactions between SPR2 and PPR-SMR1. Equal amounts of MBP-PPR-SMR1-His and MBP-His were combined with anti-HA magnetic beads preincubated with SPR2-HA. Pulled-down samples were analyzed by immunoblot with anti-MBP and anti-HA antibody. “+” and “−” indicate the presence and absence of corresponding proteins in the reactions, respectively. MBP: maltose binding protein; HA: human influenza hemagglutinin.
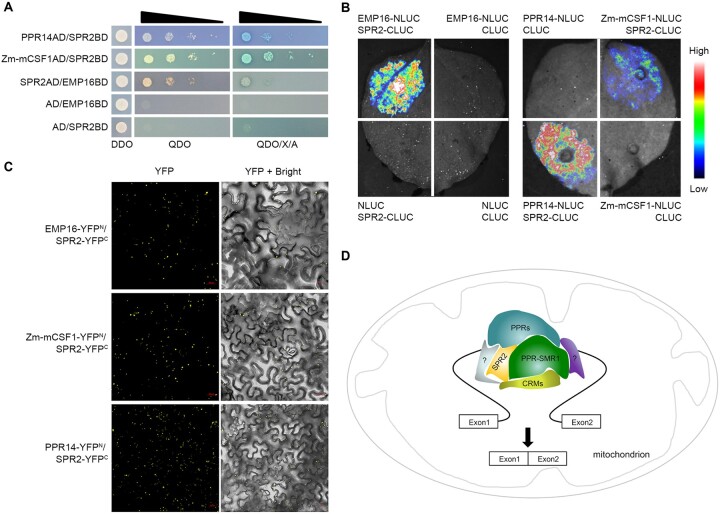
Within the 13 introns that require the function of both SPR2 and PPR-SMR1, several splicing factors are required for the splicing of one or a few of these introns. These include EMP16, PPR14, and Zm-mCSF1, with functions detailed in Supplemental Table S2 (Xiu et al., 2016; Chen et al., 2019; Wang et al., 2020). To explore the relationship between these proteins and SPR2/PPR-SMR1, protein interaction analyses were carried out. The Y2H results showed that SPR2 interacted with EMP16, PPR14, and Zm-mCSF1 (Figure 7A). LCI assays and BiFC analysis also showed that SPR2 interacted with EMP16, PPR14, and Zm-mCSF1 (Figure 7, B and C; Supplemental Figure S9). PPR-SMR1 was reported to interact with PPR14 and Zm-mCSF1 (Chen et al., 2019; Wang et al., 2020), but not EMP16 in Y2H and LCI assays (Supplemental Figure S10, A and B). Interestingly, the interaction between PPR-SMR1 and EMP16 was generated when co-expressed with SPR2 (Supplemental Figure S10B). These data indicate that SPR2 may physically interact with PPR14, EMP16, and Zm-mCSF1, whereas PPR-SMR1 may interact with PPR14 and Zm-mCSF1, but not EMP16. We further tested whether SPR2 can form dimer or multimer. The results of Y2H, LCI, and BiFC analyses showed that SPR2 could form dimer or multimer (Supplemental Figure S11).
Figure 7.
SPR2 interacts with multiple splicing factors. A, Interaction analysis of between SPR2 and EMP16, PPR14, and Zm-mCSF1 in Y2H assays. DDO medium: SD/–Leu/–Trp medium; QDO medium: SD/–Ade/–His/–Leu/–Trp medium; QDO/X/A: QDO medium with added the X-α-gal and AbA. AD: GAL4 activation domain; BD: GAL4 DNA binding domain. B, LCI assays to determine interactions between SPR2 and EMP16, PPR14, and Zm-mCSF1. The intensity of the fluorescent signals represents their interaction activities. C, BiFC analysis for interactions between SPR2 and EMP16, PPR14, and Zm-mCSF1 in N. benthamiana leaves. YFP signals were detected by a confocal laser microscope. Scale bars, 20 μm. D, A proposed working model of SPR2. Different colors represent various splicing factor. PPRs indicates other P-type PPR proteins, CRMs indicates CRM family proteins, and question mark indicates the other kind of mitochondrial splicing factors.
Discussion
SPR2 is required for the splicing of 15 introns and essential for complex I biogenesis and seed development in maize
We characterized a special P-type PPR protein SPR2 that harbors merely four PPR motifs. SPR2 is localized in mitochondria (Figure 2, D and E). Spr2 loss of function severely impaired the splicing of 15 Group II introns, which account for over half of the introns in maize mitochondria. These include introns of nad1-int1 to 4; nad2-int1 to 4; nad4-int1, 3; nad5-int1, 2, 4 and nad7-int1, 2 (Figure 5, B and C). The defects in the splicing were confirmed in two spr2 alleles, and over-expression of SPR2 in spr2 restored the splicing of these introns to the WT level (Supplemental Figure S8B). These data confirm that SPR2 is required for the splicing of 15 introns in maize mitochondria.
These 15 introns reside in the genes coding for components of mitochondrial complex I, an entry point of the mitochondrial OXPHOS electron transport chain. As a result, the loss of splicing eliminates or reduces the expression of these genes, thus impairing the assembly and activity of complex I in spr2 (Figure 3, A and B). Each of these Nad proteins has been shown to be essential to the assembly of mitochondrial complex I and is vital for the dehydrogenase activity (Klodmann et al., 2010; Ligas et al., 2019). For instance, an absence of Nad1 and Nad2 blocks complex I assembly and activity (Cai et al., 2017; Ren et al., 2017). A loss of Nad7 inhibits complex I assembly in maize (Wang et al., 2020; Xu et al., 2020). With nearly a complete deficiency in five Nad proteins (Nad1, Nad2, Nad4, Nad5, and Nad7), the loss of function of SPR2 completely blocks the assembly and activity of mitochondrial complex I. Given the essentiality of the OXPHOS chain to mitochondrial function, it explains the severely arrested embryogenesis and endosperm development of the spr2 alleles as embryogenesis and endosperm development depend on mitochondrial function. The blocked cytochrome pathway induces the operation of the AOX pathway (Figure 3E;Supplemental Figure S5).
SPR2 and PPR-SMR1 form a core complex to facilitate Group II intron splicing
Recent studies have uncovered proteins from several families that are required for the splicing of mitochondrial introns. These include nMAT, mTERF, DEAD-box proteins, CRM, PORR, RCC1, and PPR proteins. All of these are RNA-binding proteins, and the family of PPR proteins is the largest. It is found that most of these proteins are required for the splicing of one or a few introns (Kuhn et al., 2011; Colas des Francs-Small et al., 2012; Keren et al., 2012; Xiu et al., 2016). Moreover, the splicing of a specific intron may require the participation of multiple splicing factors. One example is the splicing of nad2 intron 3, which requires PPR14, PPR20, PPR231, PPR-SMR1, Zm-mCSF1, mTERF15, and DEK47 (Chen et al., 2019; Wang et al., 2020; Yang et al., 2020a, 2020b; Cao et al., 2021). How these splicing factors function in the splicing event is unclear.
As mitochondrial introns are derived from the bona fide Group II introns in bacteria (Lambowitz and Zimmerly, 2004), their splicing is believed to be catalyzed by themselves, whereby the intron folds into a catalytically active conformation, then the intron cleaves itself and ligates the exons. Compared with the nuclear DNAs, mitochondrial DNAs encounter a much higher frequency of mutations and rearrangements (Palmer et al., 2000). As such, the mitochondrial introns in land plants have lost the self-splicing capability because of their inability to form a catalytic conformation. To adapt to these changes, plants recruited various nucleus-encoded splicing factors to assist the intron folding to a catalytically active conformation. If this idea is correct, the splicing factors from multiple protein families, as mentioned above, may function to facilitate or maintain a proper intron conformation.
How these factors facilitate intron splicing is unclear. They may bind to the intron independently or form a complex and then bind to the intron. Because the functions of many splicing factors are still unknown, a clear picture invites future studies. Many known splicing factors function on a single intron or a few introns (Kuhn et al., 2011; Colas des Francs-Small et al., 2012; Keren et al., 2012; Xiu et al., 2016); however, SPR2 is involved in the splicing of 15 introns, over half of the introns in mitochondria. This finding suggests a major role of SPR2 in intron splicing in maize. Besides SPR2, a single factor required for the splicing of a large proportion of introns in mitochondria is reported with PPR-SMR1 in maize (Chen et al., 2019). PPR-SMR1, a unique PPR protein with a small MutS-related (SMR) domain at the C-terminus, is involved in the splicing of 16 introns in maize mitochondria (Chen et al., 2019). We found that 13 of the introns requiring SPR2 also require PPR-SMR1, implying that these two proteins have a highly coordinated relationship (Supplemental Table S2). Independent approaches indicate SPR2 physically interacts with PPR-SMR1 in a complex (Figure 6). These data, combined with the impaired splicing of an identical set of 13 introns in the mutants, provide compelling evidence that SPR2 and PPR-SMR1 physically interact to form a core complex to facilitate the splicing of these 13 introns in maize mitochondria.
We tried identifying sequence motifs common to SPR2 and PPR-SMR1 mediated versus nonmediated introns; however, we did not find any reasonable target sequences. One possibility is that SPR2 and PPR-SMR1 may not directly bind to RNA. SPR2 contains only four PPR repeats, which will have a loose specificity even if it binds based on the 6, 1′ code (Barkan et al., 2012; Yin et al., 2013). These two proteins may interact with other splicing factors that bind to the introns to restore or maintain the intron in a catalytic conformation. In that case, SPR2 and PPR-SMR1 may bind loosely or do not bind to the RNA. The structural defect in these introns that has to be remedied by SPR2 and PPR-SMR1 is unknown, which might be challenging to identify in sequences.
Other splicing factors interact with SPR2 and PPR-SMR1 in intron splicing
Splicing of a single intron requires the participation of many splicing factors. For instance, DEK35, DEK43, EMP8, EMP602, PPR18, and PPR-SMR1 are required for the splicing of nad4 intron 1 (Chen et al., 2016; Sun et al., 2018; Chen et al., 2019; Liu et al., 2020; Ren et al., 2020; Ren et al., 2019;). How these factors function in the splicing of these 13 introns is also unknown. One notion is that they may function independently on the splicing. Another is that they cooperate in the SPR2/PPR-SMR1 core complex. Our data support the latter model. PPR-SMR1 has been shown to interact with Zm-mCSF1 and PPR14 in maize mitochondria (Chen et al., 2019; Wang et al., 2020). EMP603, a PPR protein, was reported to interact with DEAD-box RNA helicase PMH2-5140, RAD52-like proteins ODB1-0814, and ODB1-5061, and Zm-mCSF1 (Fan et al., 2021). We have shown that SPR2 interacts with Zm-mCSF1, PPR14, and EMP16 (Figure 7, A–C), and PPR-SMR1 can interact with EMP16 through the bridge of SPR2 in LCI assay (Supplemental Figure S10). SPR2, PPR-SMR1, and Zm-mCSF1 are required for the splicing of nad2-int2, int3, and nad5-int1 (Supplemental Table S2), implying that they potentially form a protein complex in intron splicing. In addition, SPR2, PPR-SMR1, and PPR14 function in the splicing of nad2-int3 and nad7-int2 (Supplemental Table S2), and also interact with each other (Figure 7, A–C) (Wang et al., 2020), supporting a potential complex mediating the splicing of the two introns. Other splicing factors required for individual intron splicing may interact with SPR2 and/or PPR-SMR1. With the number of introns involved and many splicing factors, the splicing complex may be highly dynamic and transient. Thus, isolation of a stable complex may be challenging.
Based on the results, we propose a model for the SPR2 and PPR-SMR1 function in the Group II intron splicing in maize mitochondria (Figure 7D). SPR2 interacts with PPR-SMR1 to form a core component, which allows other intron-specific splicing factors to bind on. SPR2 and PPR-SMR1 can interact with various splicing factors to facilitate the splicing of an individual intron, such as CRM proteins and P-type PPR proteins. The PPR proteins may function predominantly to recognize specific sequences in the intron via a one-repeat one-nucleotide mechanism (Barkan et al., 2012; Yin et al., 2013). The binding of these proteins may restore or maintain the intron in a catalytically active configuration, thus initiating intron splicing. The SMR domain was reported to act as an RNA endonuclease (Liu et al., 2013a; Zhang and Lu, 2019). However, how this function is executed in intron splicing is unclear. The P-class PPR proteins and other splicing factors may play a role in recognizing the specific RNA sites and maintaining the structure of the splicing complex. Although the splicing mechanism of mitochondrial introns is still unclear, uncovering a core role of SPR2/PPR-SMR1 will facilitate the complete elucidation of the mitochondrial intron-splicing mechanism in plants.
Materials and methods
Plant materials
The Mu insertion lines spr2-1 (UFMu-03323) and spr2-2 (UFMu-05570) in the W22 genetic background were obtained from the Maize Genetic Stock Center (McCarty et al., 2005). All maize (Z. mays) plants were cultivated either in the greenhouses of Shandong University in Qingdao, or the Experimental Stations in Jinan and Sanya, China. Nicotiana benthamiana plants used for testing the GFP/YFP fluorescence intensity and luciferase signal were grown in growth chambers at 25/22°C (day/night) temperature with a 12-h light/12-h dark cycle.
To overexpress the Spr2 gene, a 960-bp Spr2 cDNA fragment fused with 3xHA tag was amplified and cloned into the pUNTF vector under the control of the maize ubiquitin1 promoter. The maize inbred line of KN5585 is used as transforming receptor and transgenic maize plants were generated by Agrobacterium tumefaciens-mediated transformation. The transformation experiments were conducted by the Wimi Biotechnology. Eleven independent transgenic plants were obtained. Spr2-OE lines crossed with spr2-2/+ heterozygotes and self-crossed for one generation, yielding the complementary lines Spr2-Com. Maize genomic DNA was extracted using the urea extraction method (Tan et al., 2011).
Light microscopy of cytological sections
The immature WT and spr2 mutant kernels at 10 and 15 DAP were collected from the same self-pollinated heterozygous F2 segregating ears. For paraffin section preparation, the kernel samples were fixed, dehydrated, infiltrated, embedded, and stained with Johansen’s Safranin O, as described previously (Liu et al., 2013b). The sections were observed and imaged with a stereo microscope (Carl-Zeiss, Germany).
Phylogenetic analysis
Construction of the phylogenetic tree was performed using MEGA version 7 software by the maximum likelihood method (Kumar et al., 2016). The sequences of SPR2 and its homologous proteins were obtained from the NCBI database.
Subcellular localization
Subcellular localization was performed as described by Liu et al. (,2013b). To express SPR2N291-GFP fusion proteins, the amplified SPR2 N-terminal first 873-bp fragment from maize inbred line W22 was cloned and recombined into the binary vector pGWB5 (a gift from Tsuyoshi Nakagawa, Shimane University) by gateway site-specific recombination (Invitrogen, Carlsbad, CA, USA). The resulting construct pGWB5-SPR2N291 was transformed into A. tumefaciens strain EHA105 and infiltrated into N. benthamiana epidermal leaves. After incubation at 25°C for 28–32 h, the fluorescence signals were detected using ZEISS LSM 900 confocal microscope (Carl-Zeiss, Oberkochen, Germany). MitoTracker Red (Thermo Fisher Scientific, Waltham, MA, USA) was used to label the mitochondria with a working concentration of 100 nM. Images were obtained using LSM 900 with GFP (excitation wavelength = 488 nm and detection wavelength = 490–540 nm) and MitoTracker Red (excitation wavelength = 555 nm and detection wavelength = 566–625 nm).
RNA extraction, RT–PCR, and RT–qPCR
Total RNA was extracted from the immature embryo and endosperm in the WT and the spr2 alleles by carefully removing the pericarp with TRIzol reagent (Invitrogen) and treated with RNase-free DNase I (New England Biolabs, Hitchin, UK) at 37°C for 2 h to remove DNA contamination. The complete removal of DNA was checked by PCR on genomic DNA. Complementary DNA (cDNA) was obtained RNA by reverse-transcribed PCR using the TranScript First-Strand cDNA Synthesis SuperMix kit (TransGen, China). RNAs were normalized by ZmActin gene (GRMZM2G126010) amplification. RT–qPCR was performed with the LightCycler 96 System (Roche, Basel, Switzerland) and the SYBR Green I Master PCR kit (Roche, Switzerland) with three biological plants. All primers and sequences used in this study are listed in Supplemental Table S3 or as described previously (Chen et al., 2016, 2019; Liu et al., 2020; Wang et al., 2020).
BN-PAGE and complex I activity assay
Crude mitochondria were extracted from the immature embryo and endosperm without pericarp of WT and spr2 kernels at 11 DAP as previously described with some modifications (Meyer et al., 2009). BN-PAGE and in-gel complex I activity assay were performed as described (Meyer et al., 2009). Totally 130 µg maize mitochondrial protein was loaded onto BN-PAGE using a Native PAGE Sample Prep kit (Thermo Fisher Scientific). The gels were stained by Coomassie Blue R-250 (Sangon, Shanghai, China) and assayed for complex I activity in assay buffer according to (Sun et al., 2015).
Immunoblot analysis
To isolate subcellular fractions, nuclear protein and chloroplastic protein were extracted from two-week-old leaves of Spr2-OE1 transgenic maize using Cytoplasmic and Nuclear Extraction kit for Cells and Chloroplast Isolation kit (Invent Biotechnologies, Plymouth, MN, USA), respectively. The mitochondrial protein was extracted from transgenic plants Spr2-OE1 as described above. The protein samples were separated on a 12.5% (w/v) sodium dodecyl sulfate polyacrylamide gel electropheresis (SDS–PAGE) gel and transferred to polyvinylidene fluoride blotting membrane (0.45 μm; Millipore, Billerica, MA, USA). After transferring membrane and blocking, antiserum against HA (1:2,000, Invitrogen), Arabidopsis Cox2 (1:5,000, Agrisera, Sweden, http://www.agrisera.com), D2 (1:5,000, a gift from Dr. Lixin Zhang, Henan University, China), H3 (Histone H3, 1:10,000), and Actin (1:5,000, Abclonal, China) were used as primary antibodies. Horseradish peroxidase-conjugated goat anti-rabbit antibody was used as secondary antibody with a 1:10,000 dilution (Sangon, China). Signals were detected using the Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific) visualized on X-ray films (Kodak, Tokyo, Japan). Other Western blotting was performed using primary antibodies against wheat Nad9 (1:3,000), cytochrome c1 (CytC1, 1:5,000), ATPase (ATPase α-subunit, 1:10,000), and AOX (1:10,000) according to Xiu et al. (2016).
Determination of respiration rate
Respiration rates were measured as previously described (Wang et al., 2015). The total respiration rate (Vt), the alternative pathway capacity (Valt), and the cytochrome pathway capacity (Vcyt) were indicated by the consumption of nmol O2 min−1 g−1 fresh weight of the maize kernels. The O2 consumption assay was performed using specific inhibitors 2-mM potassium cyanide (Sigma-Aldrich, St Louis, MO, USA) and 2-mM salicylhydroxamic acid (Sigma-Aldrich).
Y2H analysis
The Y2H analysis was performed using the Yeastmaker Yeast Transformation System 2 (Clontech, Mountain View, CA, USA) according to the manufacturer’s instructions. The cDNA corresponding to SPR2, EMP16, PPR14, PPR-SMR1, and Zm-mCSF1 without stop codon were amplified and cloned into the pGADT7-AD and pGBKT7-BD vector used as prey and bait protein, respectively. These different combinations were co-transfected into yeast (Saccharomyces cerevisiae) strain Y2H Gold (Clontech) and spotted onto SD/-Leu/-Trp (DDO) medium and growth of diploid yeast colonies on SD/-Ade/-His/-Leu/-Trp (QDO) medium with added the X-α-gal (Clontech) and AbA (Clontech) to reveal protein-protein interactions. The combinations of pGBKT7-53 (human P53)/pGADT7-T (T-antigen) was used as a positive control. The combinations of pGADT7-AD/pGBKT7-BD were used as a negative control.
LCI assay
Full length Spr2, Emp16, PPR14, PPR-SMR1, and Zm-mCSF1 without stop codon were amplified and inserted into pCAMBIA1300-nLUC (nLUC) and pCAMBIA1300-cLUC (cLUC) for the LCI assay (Chen et al., 2008; Gou et al., 2011). The resulting constructs were transformed into A. tumefaciens strain EHA105 and combinations were co-infiltrated into 4-week-old N. benthamiana leaves. The luciferase signals were visualized using the Lumazone FA Pylon2048B system after injecting with 1-mM luciferin (BIOSYNTH, Switzerland). Three biological samples were used for the assay.
BiFC analysis
The coding sequences of Spr2, Emp16, PPR14, PPR-SMR1, and Zm-mCSF1 were cloned into pSPYNE-35S or pSPYCE-35S for BiFC assays (Walter et al., 2004). The resulting constructs were co-infiltrated into N. benthamiana leaves using Agrobacterium as reported (Chen et al., 2008). The fluorescence signals were detected using ZEISS LSM 900 confocal microscope (Carl-Zeiss, Germany) using settings (excitation wavelength: 488 nm; detection wavelength: 500–580 nm). Laser intensity and detection range remained unchanged in all the BiFC assays and were similar for all pictures. This analysis was carried out with three biological replicates in different N.benthamiana plants.
Semi-in vivo pull-down assays
The semi-in vivo pull-down assays were performed as previously described with some modifications (Li et al., 2021). For SPR2/PPR-SMR1 interaction assays, the preys recombinant MBP-PPR-SMR1-His (6 × Histidine) and MBP-His (6 × Histidine) were purified from Rosetta strains of E. coli as previously described (Chen et al., 2019). The bait SPR2-HA crude mitochondrial protein was extracted from Spr2-OE1 seedlings (Meyer et al., 2009) and solubilized with IP buffer (20-mM HEPES-KOH, 40-mM KCl, 5-mM EDTA, 0.5% (v/v) Triton X-100, 1-mM PMSF, 1 × Protease Inhibitor Cocktail, 0.2% (v/v) β-mercaptoethanol) and incubated for 30 min at 4°C. The samples were centrifuged for 10 min at 20,000g. After centrifugation, the supernatant, the prey recombinant protein, and 25 μL Pierce Anti-HA Magnetic Beads (Thermo Fisher Scientific) for 2 h at 4°C and washed 7 times with wash buffer (20-mM HEPES-KOH, 40-mM KCl, 300-mM NaCl, 5-mM EDTA, 1% (v/v) Triton X-100). The MBP-His protein was added to the binding reaction as a negative control. The precipitates were eluted into 60 μL of SDS 2 × loading buffer and detected using anti-HA and anti-MBP antibody (1:5,000, New England Biolabs).
Accession numbers
Sequence data for Spr2 can be found in GenBank (http://www.ncbi.nlm.nih.gov) under accession number GRMZM2G122344.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotypes of spr2-1/+ ear, overexpressed ear Spr2-OE1, and complemented ear Spr2-Com1.
Supplemental Figure S2. Linkage analysis of spr2-1 mutant.
Supplemental Figure S3. Expression analysis of Spr2.
Supplemental Figure S4. Genetic complementation of maize spr2-2 with Spr2.
Supplemental Figure S5. Expression of AOX genes in the spr2 mutants.
Supplemental Figure S6. The spr2 mutant is deficient for mitochondrial nad1, nad2, nad4, nad5, and nad7 mature transcript.
Supplemental Figure S7. RT–PCR analysis of the splicing of the nad1, nad2, nad4, nad5, and nad7 introns in the spr2 mutants.
Supplemental Figure S8. Splicing efficiency analysis of all 22 mitochondrial introns in overexpression lines, complemented lines, and WT.
Supplemental Figure S9. Negative controls in BiFC analysis.
Supplemental Figure S10. PPR-SMR1 has no direct physical interaction with EMP16 in Y2H and LCI assays.
Supplemental Figure S11. SPR2 can form multimer.
Supplemental Table S1. Alteration of the respiration rate of WT and spr2-1.
Supplemental Table S2. List of some splicing factors which shared target intron(s) with SPR2.
Supplemental Table S3. List of the primers used in this study.
Supplementary Material
Acknowledgments
We thank the Maize Genetic Stock Center for providing the maize stocks (UFMU-03323 and UFMu-05570), Prof. Congming Lu (Shandong Agricultural University) for BiFC vectors, and Prof. Tsuyoshi Nakagawa (Shimane University, Japan) for the pGWB vector. We thank the SDU Core Facilities for Life and Environmental Sciences for the assistance in microimaging of laser scanning confocal microscopy analysis.
Funding
This research was supported by grants from the National Natural Science Foundation of China (31630053 to B.C.T. and 31900264 to R.L.).
Conflicts of interest statement. The authors declare no conflict of interest.
Contributor Information
Shi-Kai Cao, Key Laboratory of Plant Development and Environment Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, China.
Rui Liu, Key Laboratory of Plant Development and Environment Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, China; College of Life Science, Yangtze University, Jingzhou 434025, China.
Miaodi Wang, Key Laboratory of Plant Development and Environment Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, China.
Feng Sun, Key Laboratory of Plant Development and Environment Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, China.
Aqib Sayyed, Key Laboratory of Plant Development and Environment Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, China.
Hong Shi, Key Laboratory of Plant Development and Environment Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, China.
Xiaomin Wang, Key Laboratory of Cell Activities and Stress Adaptations, Ministry of Education, School of Life Sciences, Lanzhou University, Lanzhou 730000, China.
Bao-Cai Tan, Key Laboratory of Plant Development and Environment Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, China.
S.K.C., R.L., and B.C.T. designed the experiments. S.K.C. and R.L. performed most of the experiments. M.W. participated in vector construction and protein purification. F.S. contributed to the BN gel analysis. A.S. helped with the phylogenetic analysis. H.S. provided help in BiFC analysis. X.W. measured respiration rate. S.K.C., R.L., and B.C.T. analyzed data and wrote the manuscript. All authors read and approved this article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is Bao-Cai Tan (bctan@sdu.edu.cn).
References
- Barkan A, Small I (2014) Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65: 415–442 [DOI] [PubMed] [Google Scholar]
- Barkan A, Rojas M, Fujii S, Yap A, Chong YS, Bond CS, Small I (2012) A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet 8: e1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonen L, Vogel J (2001) The ins and outs of group II introns. Trends Genet 17: 322–331 [DOI] [PubMed] [Google Scholar]
- Brown GG, Colas des Francs-Small C, Ostersetzer-Biran O (2014) Group II intron splicing factors in plant mitochondria. Front Plant Sci 5: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M, Li S, Sun F, Sun Q, Zhao H, Ren X, Zhao Y, Tan BC, Zhang Z, Qiu F (2017) Emp10 encodes a mitochondrial PPR protein that affects the cis-splicing of nad2 intron 1 and seed development in maize. Plant J 91: 132–144 [DOI] [PubMed] [Google Scholar]
- Cao SK, Liu R, Sayyed A, Sun F, Song R, Wang X, Xiu Z, Li X, Tan BC (2021) Regulator of chromosome condensation 1-domain protein DEK47 functions on the intron splicing of mitochondrial nad2 and seed development in maize. Front Plant Sci 12: 695249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Li YX, Li C, Shi Y, Song Y, Zhang D, Li Y, Wang T (2018) Genome-wide analysis of the pentatricopeptide repeat gene family in different maize genomes and its important role in kernel development. BMC Plant Biol 18: 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Feng F, Qi W, Xu L, Yao D, Wang Q, Song R (2016) Dek35 encodes a PPR protein that affects cis-splicing of mitochondrial nad4 intron 1 and seed development in maize. Mol Plant 10: 427–441 [DOI] [PubMed] [Google Scholar]
- Chen Z, Wang HC, Shen J, Sun F, Wang M, Xu C, Tan BC (2019) PPR-SMR1 is required for the splicing of multiple mitochondrial introns, interacts with Zm-mCSF1, and is essential for seed development in maize. J Exp Bot 70: 5245–5258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Zmudjak M, Colas des Francs-Small C, Malik S, Shaya F, Keren I, Belausov E, Many Y, Brown GG, Small I, et al. (2014) nMAT4, a maturase factor required for nad1 pre-mRNA processing and maturation, is essential for holocomplex I biogenesis in Arabidopsis mitochondria. Plant J 78: 253–268 [DOI] [PubMed] [Google Scholar]
- Colas des Francs-Small C, Kroeger T, Zmudjak M, Ostersetzer-Biran O, Rahimi N, Small I, Barkan A (2012) A PORR domain protein required for rpl2 and ccmFC intron splicing and for the biogenesis of c-type cytochromes in Arabidopsis mitochondria. Plant J 69: 996–1005 [DOI] [PubMed] [Google Scholar]
- Dai D, Ma Z, Song R (2021) Maize kernel development. Mol Breed 41: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Longevialle AF, Small ID, Lurin C (2010) Nuclearly encoded splicing factors implicated in RNA splicing in higher plant organelles. Mol Plant 3: 691–705 [DOI] [PubMed] [Google Scholar]
- de Longevialle AF, Meyer EH, Andres C, Taylor NL, Lurin C, Millar AH, Small ID (2007) The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 intron 1 in Arabidopsis thaliana. Plant Cell 19: 3256–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan K, Ren Z, Zhang X, Liu Y, Fu J, Qi C, Tatar W, Rasmusson AG, Wang G, Liu Y (2021) The pentatricopeptide repeat protein EMP603 is required for the splicing of mitochondrial nad1 intron 2 and seed development in maize. J Exp Bot 72: 6933–6948 [DOI] [PubMed] [Google Scholar]
- Gabaldon T, Huynen MA (2004) Shaping the mitochondrial proteome. Biochim Biophys Acta 1659: 212–220 [DOI] [PubMed] [Google Scholar]
- Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW (2011) Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23: 1512–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K, Giege P (2014) RNA metabolism in plant mitochondria. Trends Plant Sci 19: 380–389 [DOI] [PubMed] [Google Scholar]
- Hsu YW, Wang HJ, Hsieh MH, Hsieh HL, Jauh GY (2014) Arabidopsis mTERF15 is required for mitochondrial nad2 intron 3 splicing and functional complex I activity. PLoS One 9: e112360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I, Bezawork-Geleta A, Kolton M, Maayan I, Belausov E, Levy M, Mett A, Gidoni D, Shaya F, Ostersetzer-Biran O (2009) AtnMat2, a nuclear-encoded maturase required for splicing of group-II introns in Arabidopsis mitochondria. RNA 15: 2299–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I, Tal L, des Francs-Small CC, Araujo WL, Shevtsov S, Shaya F, Fernie AR, Small I, Ostersetzer-Biran O (2012) nMAT1, a nuclear-encoded maturase involved in the trans-splicing of nad1 intron 1, is essential for mitochondrial complex I assembly and function. Plant J 71: 413–426 [DOI] [PubMed] [Google Scholar]
- Klodmann J, Sunderhaus S, Nimtz M, Jansch L, Braun HP (2010) Internal architecture of mitochondrial complex I from Arabidopsis thaliana. Plant Cell 22: 797–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler D, Schmidt-Gattung S, Binder S (2010) The DEAD-box protein PMH2 is required for efficient group II intron splicing in mitochondria of Arabidopsis thaliana. Plant Mol Biol 72: 459–467 [DOI] [PubMed] [Google Scholar]
- Kuhn K, Carrie C, Giraud E, Wang Y, Meyer EH, Narsai R, des Francs-Small CC, Zhang B, Murcha MW, Whelan J (2011) The RCC1 family protein RUG3 is required for splicing of nad2 and complex I biogenesis in mitochondria of Arabidopsis thaliana. Plant J 67: 1067–1080 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S (2004) Mobile group II introns. Annu Rev Genet 38: 1–35 [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S (2011) Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol 3: a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu Y, Liu H (2021) Semi-in-vivo pull-down assay for blue light-dependent protein interactions. Methods Mol Biol (Clifton, NJ) 2297: 161–166 [DOI] [PubMed] [Google Scholar]
- Ligas J, Pineau E, Bock R, Huynen MA, Meyer EH (2019) The assembly pathway of complex I in Arabidopsis thaliana. Plant J 97: 447–459 [DOI] [PubMed] [Google Scholar]
- Liu R, Cao SK, Sayyed A, Xu C, Sun F, Wang X, Tan BC (2020) The mitochondrial pentatricopeptide repeat protein PPR18 is required for the cis-splicing of nad4 intron 1 and essential to seed development in maize. Int J Mol Sci 21: 4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Melonek J, Boykin LM, Small I, Howell KA (2013a) PPR-SMRs: ancient proteins with enigmatic functions. RNA Biol 10: 1501–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, He J, Chen Z, Ren X, Hong X, Gong Z (2010) ABA overly-sensitive 5 (ABO5), encoding a pentatricopeptide repeat protein required for cis-splicing of mitochondrial nad2 intron 3, is involved in the abscisic acid response in Arabidopsis. Plant J 63: 749–765 [DOI] [PubMed] [Google Scholar]
- Liu YJ, Xiu ZH, Meeley R, Tan BC (2013b) Empty Pericarp5 encodes a pentatricopeptide repeat protein that is required for mitochondrial RNA editing and seed development in maize. Plant Cell 25: 868–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DR, Settles AM, Suzuki M, Tan BC, Latshaw S, Porch T, Robin K, Baier J, Avigne W, Lai J, et al. (2005) Steady-state transposon mutagenesis in inbred maize. Plant J 44: 52–61 [DOI] [PubMed] [Google Scholar]
- Meyer EH, Tomaz T, Carroll AJ, Estavillo G, Delannoy E, Tanz SK, Small ID, Pogson BJ, Millar AH (2009) Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiol 151: 603–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa N, Sakurai N (2006) A mutation in At-nMat1a, which encodes a nuclear gene having high similarity to group II intron maturase, causes impaired splicing of mitochondrial nad4 transcript and altered carbon metabolism in Arabidopsis thaliana. Plant Cell Physiol 47: 772–783 [DOI] [PubMed] [Google Scholar]
- Palmer JD, Adams KL, Cho Y, Parkinson CL, Qiu YL, Song K (2000) Dynamic evolution of plant mitochondrial genomes: mobile genes and introns and highly variable mutation rates. Proc Natl Acad Sci USA 97: 6960–6966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren RC, Wang LL, Zhang L, Zhao YJ, Wu JW, Wei YM, Zhang XS, Zhao XY (2020) DEK43 is a P-type PPR protein responsible for the cis-splicing of nad4 in maize mitochondria. mJ Integr Plant Biol 62: 299–313 [DOI] [PubMed] [Google Scholar]
- Ren X, Pan Z, Zhao H, Zhao J, Cai M, Li J, Zhang Z, Qiu F (2017) EMPTY PERICARP11 serves as a factor for splicing of mitochondrial nad1 intron and is required to ensure proper seed development in maize. J Exp Bot 68: 4571–4581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Fan K, Fang T, Zhang J, Yang L, Wang J, Wang G, Liu Y (2019) Maize empty pericarp602 encodes a P-type PPR protein that is essential for seed development. Plant Cell Physiol 60: 1734–1746 [DOI] [PubMed] [Google Scholar]
- Shevtsov-Tal S, Best C, Matan R, Aldrin Chandran S, Brown GG, Ostersetzer-Biran O (2021) nMAT3 is an essential maturase splicing factor required for holo-complex I biogenesis and embryo-development in Arabidopsis thaliana plants. Plant J 106: 1128–1147 [DOI] [PubMed] [Google Scholar]
- Singh RN, Saldanha RJ, D'Souza LM, Lambowitz AM (2002) Binding of a group II intron-encoded reverse transcriptase/maturase to its high affinity intron RNA binding site involves sequence-specific recognition and autoregulates translation. J Mol Biol 318: 287–303 [DOI] [PubMed] [Google Scholar]
- Sun F, Wang X, Bonnard G, Shen Y, Xiu Z, Li X, Gao D, Zhang Z, Tan BC (2015) Empty pericarp7 encodes a mitochondrial E-subgroup pentatricopeptide repeat protein that is required for ccmFN editing, mitochondrial function and seed development in maize. Plant J 84: 283–295 [DOI] [PubMed] [Google Scholar]
- Sun F, Zhang X, Shen Y, Wang H, Liu R, Wang X, Gao D, Yang YZ, Liu Y, Tan BC (2018) The pentatricopeptide repeat protein EMPTY PERICARP8 is required for the splicing of three mitochondrial introns and seed development in maize. Plant J 95: 919–932 [DOI] [PubMed] [Google Scholar]
- Tan BC, Chen Z, Shen Y, Zhang Y, Lai J, Sun SS (2011) Identification of an active new mutator transposable element in maize. G3 (Bethesda) 1: 293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahleithner JA, MacFarlane JL, Wolstenholme DR (1990) A sequence encoding a maturase-related protein in a group II intron of a plant mitochondrial nad1 gene. Proc Natl Acad Sci USA 87: 548–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang HC, Chen Z, Yang YZ, Sun F, Ding S, Li XL, Xu C, Tan BC (2020) PPR14 interacts with PPR-SMR1 and CRM protein Zm-mCSF1 to facilitate mitochondrial intron splicing in maize. Front Plant Sci 11: 814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XM, Chang N, Bi YR, Tan BC (2015) Measurement of mitochondrial respiration rate in maize (Zea mays) leaves. Bio-Protocol 5: e1483 [Google Scholar]
- Xiu Z, Sun F, Shen Y, Zhang X, Jiang R, Bonnard G, Zhang J, Tan BC (2016) EMPTY PERICARP16 is required for mitochondrial nad2 intron 4 cis-splicing, complex I assembly and seed development in maize. Plant J 85: 507–519 [DOI] [PubMed] [Google Scholar]
- Xu C, Song S, Yang YZ, Lu F, Zhang MD, Sun F, Jia R, Song R, Tan BC (2020) DEK46 performs C-to-U editing of a specific site in mitochondrial nad7 introns that is critical for intron splicing and seed development in maize. Plant J 103: 1767–1782 [DOI] [PubMed] [Google Scholar]
- Yang H, Xiu Z, Wang L, Cao SK, Li X, Sun F, Tan BC (2020a) Two pentatricopeptide repeat proteins are required for the splicing of nad5 introns in maize. Front Plant Sci 11: 732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YZ, Ding S, Wang Y, Wang HC, Liu XY, Sun F, Xu C, Liu B, Tan BC (2020b) PPR20 is required for the cis-splicing of mitochondrial nad2 intron 3 and seed development in maize. Plant Cell Physiol 61: 370–380 [DOI] [PubMed] [Google Scholar]
- Yin P, Li Q, Yan C, Liu Y, Liu J, Yu F, Wang Z, Long J, He J, Wang HW, et al. (2013) Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature 504: 168–171 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lu C (2019) The enigmatic roles of PPR-SMR proteins in plants. Adv Sci 6: 1900361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmudjak M, Colas des Francs-Small C, Keren I, Shaya F, Belausov E, Small I, Ostersetzer-Biran O (2013) mCSF1, a nucleus-encoded CRM protein required for the processing of many mitochondrial introns, is involved in the biogenesis of respiratory complexes I and IV in Arabidopsis. New Phytol 199: 379–394 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.