Abstract
A nosocomial outbreak by cefiderocol (FDC)-resistant NDM-1-producing Klebsiella pneumoniae (NDM-Kp) occurred in a large tertiary care hospital from August 2021–June 2022 in Florence, Italy, an area where NDM-Kp strains have become endemic. Retrospective analysis of NDM-Kp from cases observed in January 2021–June 2022 revealed that 21/52 were FDC-resistant. The outbreak was mostly sustained by clonal expansion of a mutant with inactivated cirA siderophore receptor gene, which exhibited high-level resistance to FDC (MIC ≥ 32 mg/L) and spread independently of FDC exposure.
Keywords: FDC; CPE; K. pneumoniae; Enterobacterales; resistance mechanism, cirA, nosocomial infections; carbapenemase
Cefiderocol (FDC) is one of the few novel antimicrobial agents active against multidrug-resistant Gram-negative bacteria strains producing metallo-beta-lactamases (MBLs) [1-3]. Thus far, acquired FDC resistance has been occasionally reported in sporadic cases [4-6]. Here, we report a nosocomial outbreak involving 21 patients caused by NDM-1-producing Klebsiella pneumoniae (NDM-Kp) resistant to FDC, which occurred in a large tertiary care Italian hospital from August 2021 to June 2022. We describe the features of the strains involved in this outbreak.
Outbreak detection and investigation
In late 2018, an outbreak of NDM-producing Enterobacterales emerged in Tuscany in central Italy, involving several hospitals especially from the north-western area [7,8]. The outbreak was mostly sustained by strains of an ST147 NDM-Kp sublineage (named ST147-vir clone) which uniformly exhibited FDC susceptibility [9]. Since 2019, at our facility, the largest regional tertiary care hospital located in the central area of Tuscany, cases with NDM-Kp related to the regional outbreak have been recurrently observed following the transfer of infected or colonised patients from other hospitals or long-term-care facilities, with limited intra-hospital transmission.
Since its introduction in the hospital formulary (list of pharmaceutical agents and related information) in June 2021, FDC has been occasionally used for treating infections caused by multidrug-resistant Gram-negative pathogens, mostly based on presumed susceptibility. In vitro susceptibility to FDC was not routinely tested because of the accuracy issues reported with available commercial systems [10,11] and by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) [12].
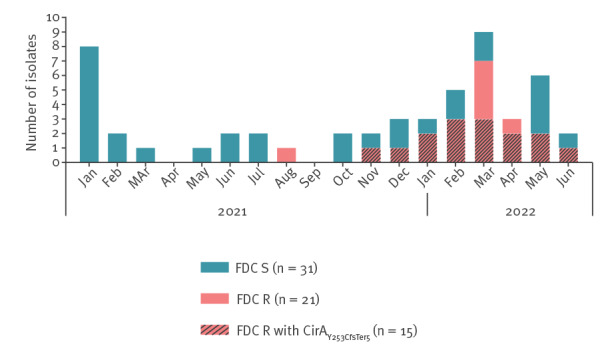
In June 2022, following the detection of an FDC-resistant NDM-Kp isolate (tested upon specific request of an infectious disease consultant), FDC susceptibility was retrospectively evaluated by reference broth microdilution (BMD) with iron-depleted medium [13] on NDM-Kp isolates from 52 cases observed since January 2021. This analysis revealed the emergence of FDC-resistant NDM-Kp in the second half of 2021, with a remarkable increase in the first months of 2022, i.e. a peak in January to March, during which 12 of 21 total FDC-resistant NDM-Kp isolates were observed (Figure 1; see Supplementary Table S1 for an overview of the clinical data of the cases). Among the resistant isolates, FDC minimum inhibitory concentrations (MICs) ranged from 4 to > 64 mg/L, with a bimodal distribution (see Supplementary Figure S1 for an overview of antimicrobial susceptibility test results).
Figure 1.
Number of retrospectively analysed non-replicate NDM-1-producing Klebsiella pneumoniae isolates by month of detection from a tertiary care hospital outbreak in Florence, Italy, January 2021–June 2022 (n = 52 isolates)
FDC R: cefiderocol-resistant Klebsiella pneumoniae isolates; FDC R CirAY253CfsTer5: cefiderocol-resistant, CirAY253CfsTer5-positive Klebsiella pneumoniae isolates; FDC S: cefiderocol-susceptible Klebsiella pneumoniae isolates.
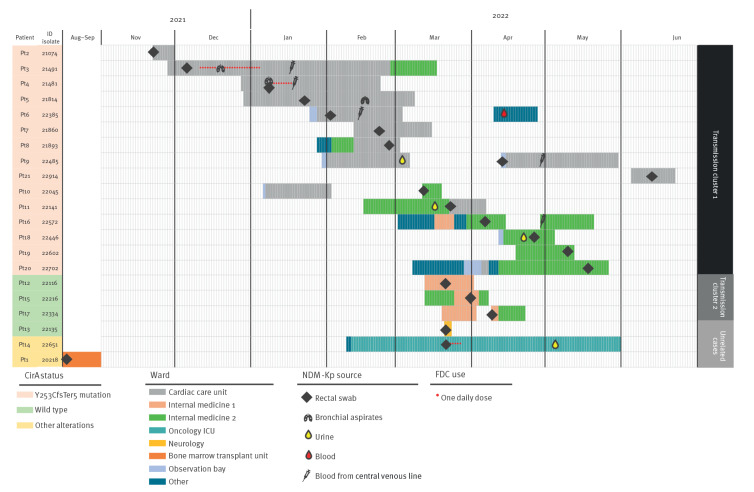
Of the 21 patients (11 men, 12 women; median age: 67 years (range: 45–91)) from which FDC-resistant NDM-Kp were isolated, all experienced colonisation and nine experienced infections including bloodstream infections (BSI; n = 5, all related to the central venous catheter) and/or urinary tract infections (UTI; n = 4) and/or lower respiratory tract infections (LRTI; n = 3) (Figure 2). Notably, none of the 21 cases had received FDC before isolation of FDC-resistant NDM-Kp.
Figure 2.
Timeline of cefiderocol-resistant NDM-1-producing Klebsiella pneumoniae-positive cultures during a tertiary care hospital outbreak in Florence, Italy, August 2021–June 2022 (n = 21 patients)
FDC: cefiderocol; ICU: intensive care unit; NDM-Kp: NDM-1-producing Klebsiella pneumoniae.
CirA alterations, hospital wards, the NDM-Kp source specimen, and the eventual cefiderocol treatment were reported for each patient. Thirteen of 15 patients, positive for a CirAY253CfsTer5 mutant, exhibited epidemiological links (overlapping period in the same ward) with the index case (Pt2) or among each other.
Genomic analysis
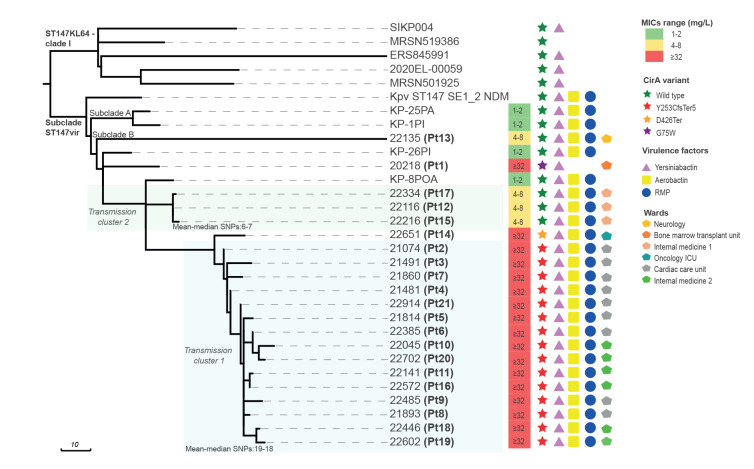
Whole genome sequencing (WGS) was carried out either on the first isolate from patients who experienced only carriage of FDC-resistant NDM-Kp, or on the first isolate from the most clinically relevant infection (BSI > LRTI > UTI), when more isolates were available from the same patient (n = 9 patients) (Figure 3). WGS was performed by Illumina Miseq (San Diego, United States) as previously described [9]. WGS analysis revealed that all the FDC-resistant NDM-Kp isolates belonged to ST147 and were related with the ST147-vir clone circulating in Tuscany [9] (Figure 3).
Figure 3.
Phylogenetic analysis of cefiderocol-resistant NDM-1-producing ST147 Klebsiella pneumoniae from tertiary care hospital outbreak in Florence, Italy, August 2021–June 2022 (n=21)
ICU: intensive care unit; MIC: minimum inhibitory concentration; RMP: regulators of mucoid phenotype.
KP-1PI, KP-25PA, KP-26PI, KP-8POA were included as comparators belonging to the ST-147-vir clone circulating in Tuscany [9]. ERS845991 (from Hungary), MRSN519386 (from Middle East region), SIKP004 (from Thailand), MRSN501925 (from Germany), Kpv_ST147_SE1_2_NDM (from United Kingdom) and 2020EL-00059 (from United States) strains were included in the analysis as outliner representants of ST147 K. pneumoniae. The short-term bacterial evolution was generated using Gubbins v. 3.2.1 and RaxML tree builder. Branch lengths were reported in substitutions (point mutations) per genome. The complete genome of K. pneumoniae KP-1PI (Accession number CP071027), that was the first NDM-Kp ST147 isolated from a Tuscan region outbreak in 2018, was used as reference FDC susceptible strains for all in silico analyses [9].
Overall, 17 isolates carried alterations of the cirA siderophore receptor gene (Figure 3). Of these, 15 carried a 7-bp duplication (c.761_767dup) causing a frameshift followed by a premature stop codon (p.Y253CfsTer5), while singletons carried either a nucleotide deletion (c.1281del) leading to a premature stop codon (p.D426Ter) or a missense mutation (c.223G > T) leading to a G75W amino acid substitution (Figure 3; see Supplementary Figure S2 for the CirA amino acid sequence comparison). Interestingly, all the NDM-Kp with cirA alterations exhibited high-level FDC resistance (MICs: ≥ 32 mg/L) (Figure 3).
The remaining four isolates showing low-level FDC resistance (MICs: 4–8 mg/L) did not carry alterations in either cirA (Figure 3) or other genes for iron uptake systems (i. e. fiu, fhuA, fepA, fbpA, efeO, exbB, exbD, fyuA, fitA, fur, iutA, iroN), but carried missense mutations in the baeS (c.539C > T, p.T180I; n = 3) or in the envZ (c.109T > A; p.F37I; n = 1) genes encoding the sensor kinases of two component systems which regulate the expression of cirA and fiu iron-uptake genes [6,14]. Interestingly, missense mutations in these genes, although of different nature, were previously associated with FDC resistance [14].
Fine-tuned phylogenomics revealed the presence of two clusters of closely related isolates, likely originating through the clonal expansion of NDM-Kp with the CirAY253CfsTer5 and with the BaeST180I alterations, respectively, while the singletons with other cirA alterations or with an altered envZ were more divergent (Figure 3).
The index CirAY253CfsTer5 NDM-Kp (#21074_Pt2) clone was isolated in November 2021 from the surveillance swab of a patient transferred from another hospital in north-western Tuscany. Of the remaining 14 patients with CirAY253CfsTer5 NDM-Kp isolates, 13 were observed in inpatients who were not colonised or infected by NDM-Kp at admission, while only one (#22045_Pt10) was found to be already positive at admission, in March 2022. Epidemiological links could be traced among the cases associated with CirAY253CfsTer5 NDM-Kp, suggesting that this clone had entered the hospital with the index patient and had thereafter spread within wards attended by patients undergoing cardiac surgery. A colonised patient (#22045_Pt10), previously admitted for cardiac surgery, was the likely source for the subsequent dissemination of the CirAY253CfsTer5 NDM-Kp clone in a medical ward, following readmission (Figure 2).
The index BaeST180I NDM-Kp strain (#22116_Pt12), isolated in March 2022 from the surveillance swab of a patient that was negative at admission, was associated with another cluster of transmission in another medical ward, while the three singletons were epidemiologically unrelated (Figure 2).
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing of isolates subjected to WGS, carried out by reference methods [15] and interpreted according to EUCAST clinical breakpoints [16], revealed that all FDC-resistant isolates were resistant to all tested beta-lactams and beta-lactam/beta-lactamase inhibitors combinations, aminoglycosides and fluoroquinolones, while 21/21 and 12/21 were susceptible to colistin and fosfomycin, respectively. The MICs of aztreonam/avibactam and tigecycline were 0.125–0.5 mg/L and ≤ 0.25–0.5 mg/L, respectively (i.e. in the susceptible range considering aztreonam clinical breakpoints and tigecycline pharmacokinetics/pharmacodynamics (PK/PD) breakpoints) (Table). Testing of FDC susceptibility by disk diffusion [17] could unambiguously categorise all FDC-resistant isolates (see Supplementary Table S1 for FDC disk diffusion test data).
Table. Minimum inhibitory concentrations (MIC) range, MIC50, MIC90 of cefiderocol-resistant NDM-producing Klebsiella pneumoniae isolates from a tertiary care hospital outbreak in Florence, Italy, August 2021–June 2022 (n = 21).
| Antibiotics | AMC | AZAa | CAZ | CAA | CEP | CRO | CTA | ERT | MEM | PTZ | AMK | GNT | CIP | LEV | CLS | FOS | T/S | TIGb |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC range (mg/L) | > 64 | 0.125 to 0.5 | > 64 | 32 to > 64 | > 16 | > 4 | > 64 | 2 to > 2 | 32 to > 64 | 64 to > 128 | 16 to > 16 | > 8 | > 1 | > 8 | ≤ 0.5 to 1 | 4 to > 128 | > 8/152 | ≤ 0.25 to 0.5 |
| MIC50 (mg/L) | > 64 | 0.25 | > 64 | > 64 | > 16 | > 4 | > 64 | > 2 | 32 | > 128 | > 16 | > 8 | > 1 | > 8 | ≤ 0.5 | 16 | > 8/152 | ≤ 0.25 |
| MIC90 (mg/L) | > 64 | 0.25 | > 64 | > 64 | > 16 | > 4 | > 64 | > 2 | > 64 | > 128 | > 16 | > 8 | > 1 | > 8 | 1 | > 128 | > 8/152 | 0.5 |
| S | 0 | 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 21 | 12 | 0 | 21 |
AMC: amoxicillin-clavulanic acid; AMK: amikacin; AZA: aztreonam-avibactam; CAA: ceftazidime/avibactam; CAZ: ceftazidime; CEP: cefepime; CIP: ciprofloxacin; CRO: ceftriaxone; CST: colistin; CTA: ceftolozane-tazobactam; ERT: ertapenem; FOS: fosfomycin; GNT: gentamicin; LEV: levofloxacin; MEM: meropenem; MIC: minimum inhibitory concentration; S: number of susceptible isolates; PTZ: piperacillin-tazobactam; T/S: trimethoprim-sulfamethoxazole; TGC: tigecycline.
a Interpreted according to aztreonam clinical breakpoints [16].
b Interpreted according to pharmacokinetics/pharmacodynamics (PK/PD) EUCAST clinical breakpoints [16].
MICs were obtained using reference broth microdilution or agar dilution for fosfomycin [15].
The resistance profile was consistent with the resistome of the isolates (see Supplementary Table S1 for the list of the acquired resistance genes revealed by the in silico analysis), which overall reflected that of the Tuscan ST147-vir clone [9], with minor inter-isolate variability.
The virulome profile of the FDC-resistant isolates (see Supplementary Table S1 for an overview of the virulence genes) was also consistent with that of the ST147-vir clone [9], except for the lack of the aerobactin siderophore genes (iucABCD-iutA) in the single isolate carrying the CirAG75W alteration (Figure 3).
Outbreak management
Following outbreak recognition in June 2022 and communication to the Regional and National Health Authorities, a bundle of interventions was implemented.
At the regional level (i) information about the outbreak was circulated among healthcare structures through the regional network for antimicrobial and diagnostic stewardship, (ii) FDC testing by disk diffusion [17] was recommended for all MBL-producing isolates of carbapenem-resistant Enterobacterales (MBL-CRE) from clinical specimens (and possibly also from surveillance specimens), followed by confirmation of results falling within the area of technical uncertainty (ATU) at reference regional laboratories and (iii) recommendations for treatment of infections by MBL-CRE were updated.
In addition, in our hospital (i) all MBL-CRE isolates categorised as FDC-resistant by disk diffusion or with a result falling within ATU were tested with reference BMD [13], (ii) enforcement of general infection prevention and control measures were emphasised and educational meetings were organised with healthcare personnel, (iii) all confirmed FDC-resistant isolates were subjected to genomic surveillance and (iv) a screening for the CirAY253CfsTer5 mutant of NDM-Kp by real-time PCR was developed to rapidly analyse the resistant isolates (primers, probes, and reaction conditions are described in Supplementary Table S3 and Figure S3).
During the subsequent period (1 July–15 October 2022), only two cases associated with an FDC-resistant NDM-Kp CirA mutants (one with Y253CfsTer5 and the other with D426Ter) were reported at our hospital, suggesting effectiveness of the intervention.
Discussion
We report a hospital outbreak caused by FDC-resistant NDM-Kp observed in an area of Tuscany, Italy, where NDM-Kp is endemic. Since FDC is among the very few treatment options available for MBL-CRE infections, this is a worrying phenomenon in terms of resistance evolution.
By genomic surveillance, the outbreak was traced mostly to NDM-Kp strains highly resistant to FDC, carrying mutated cirA siderophore receptor genes with predominance of a CirAY253CfsTer5 mutant, which was found in 15 of 21 cases, and with a minor contribution by mutants with other resistance mechanisms. All 21 FDC-resistant mutants were derivatives of the ST147 NDM-Kp sublineage involved in the large ongoing Tuscan outbreak, which emerged in late 2018 [7-9].
The FDC-resistant mutants had likely been selected outside our hospital. None of the 21 observed cases have been treated with FDC before isolation of the resistant NDM-Kp. Moreover, during this outbreak and in the preceding period (i.e. prior to introduction of FDC in the hospital formulary in June 2021), the use of FDC in our hospital remained very limited. Admission of patients already colonised by FDC-resistant NDM-Kp followed by intra-hospital transmission was the apparent source of this outbreak, underscoring the role that transfer of colonised or infected patients from other hospitals or long-term-care facilities could play in spreading of resistant pathogens, and the importance of active surveillance.
A recent study reported that a cirA mutant of NDM-Kp, selected in vitro, exhibited reduced fitness vs the parental strain in competition experiments, suggesting a low propensity to disseminate in the absence of sustained selective pressure [18]. However, our present findings indicate that at least some cirA mutants of NDM-Kp are able to spread in the hospital environment independently of a strong FDC selective pressure. Moreover, these strains can also retain a notable virulence potential, evident by the ability to cause nosocomial infections including BSI, LRTI and UTI. Further investigation regarding the fitness and virulence of the clone described in this study, as well as whether it carries compensatory mechanisms, may be necessary to explain this unexpected behaviour. One hypothesis could be that the lack of iron-catecholate outer membrane transporter CirA might be compensated by the presence of the structurally distinct siderophores yersiniabactin and aerobactin carried by the ST147-vir clone.
Conclusions
The importance of surveillance, including genomic surveillance and active surveillance, and of strict compliance with infection prevention and control measures – based on a tight collaboration between clinical and laboratory personnel – are fundamental to avoid further spread of these difficult-to-treat Gram-negative bacteria and to preserve the activity of novel antimicrobials.
Ethical statement
Ethical approval was not needed for this study, which was based on surveillance data on alert microorganisms. All data were anonymised for the outbreak investigation.
Funding statement
This study was co-supported by two research grants funded by the Italian Ministry of Education, University, and Research (2017-7J5Y3P_004) and Italian Ministry of Health (RF-2016-02364584).
Data availability
Sequence data available via BioProject PRJNA887042.
Supplementary Data
Supplementary Data
Conflict of interest: AA reports payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events from Arrow, Seegene, Qvella and Menarini, and support for attending meetings and/or travel from Symcel. TG reports grants from Merlin, Venatorx, Arrow, payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events from bioMérieux and Thermo Fisher Scientific. GMR reports grants, consulting fees, and payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events from bioMérieux, MSD, Shionogi, Zambon, Menarini, Angelini, grants from Accelerate, Cepheid, Nordic Pharma, Seegene, Arrow, Symcel, DID, Hain Lifescience GmbH, Meridian, Setlance, Qvella, Qlinea, Biomedical Service, Quidel, QuantaMatrix, SD Biosensor, consulting fees from Pfizer and Qiagen, payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events from Becton Dickinson, Cepheid and Pfizer outside the submitted work. All other authors declare no competing interests.
Authors’ contributions: LB, AF, FP and TG were involved in the initial investigation and identification of the outbreak. MG, EM, MTM, TG and GMR designed the outbreak investigation. MC, AA, CN, AB contributed to the acquisition of phenotypic and genotypic data on isolates included in the study. MC, AA and VDP performed the phylogenetic and bioinformatic analysis. LB, EM, AF, FP, MTM performed the epidemiological investigation and collected clinical data. MC, AA, CN and AB performed the analysis of microbiological and clinical data. Project administration was by MTM and GMR; MG, EM, MTM and GMR with Regional Health Authorities coordinated the infection prevention and control interventions. Funding acquisition and study supervision were by TG and GMR. GMR drafted the initial manuscript, that was critically reviewed by MC, AA, VDP and TG. All Authors reviewed and approved the final version of the manuscript.
References
- 1. Bassetti M, Di Pilato V, Giani T, Vena A, Rossolini GM, Marchese A, et al. Treatment of severe infections due to metallo-β-lactamases-producing Gram-negative bacteria. Future Microbiol. 2020;15(15):1489-505. 10.2217/fmb-2020-0210 [DOI] [PubMed] [Google Scholar]
- 2. Paul M, Carrara E, Retamar P, Tängdén T, Bitterman R, Bonomo RA, et al. European Society of Clinical Microbiology and Infectious Diseases (ESCMID) guidelines for the treatment of infections caused by multidrug-resistant Gram-negative bacilli (endorsed by European society of intensive care medicine). Clin Microbiol Infect. 2022;28(4):521-47. 10.1016/j.cmi.2021.11.025 [DOI] [PubMed] [Google Scholar]
- 3. Gaibani P, Giani T, Bovo F, Lombardo D, Amadesi S, Lazzarotto T, et al. Resistance to ceftazidime/avibactam, meropenem/vaborbactam and imipenem/relebactam in Gram-Negative MDR bacilli: molecular mechanisms and susceptibility testing. Antibiotics (Basel). 2022;11(5):628. 10.3390/antibiotics11050628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lan P, Lu Y, Chen Z, Wu X, Hua X, Jiang Y, et al. Emergence of high-level cefiderocol resistance in carbapenem-resistant Klebsiella pneumoniae from bloodstream infections in patients with hematologic malignancies in China. Microbiol Spectr. 2022;10(2):e0008422. 10.1128/spectrum.00084-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Klein S, Boutin S, Kocer K, Fiedler MO, Störzinger D, Weigand MA, et al. Rapid development of cefiderocol resistance in carbapenem-resistant Enterobacter cloacae during therapy is associated with heterogeneous mutations in the catecholate siderophore receptor cirA. Clin Infect Dis. 2022;74(5):905-8. 10.1093/cid/ciab511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Karakonstantis S, Rousaki M, Kritsotakis EI. Cefiderocol: systematic review of mechanisms of resistance, heteroresistance and in vivo emergence of resistance. Antibiotics (Basel). 2022;11(6):723. 10.3390/antibiotics11060723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Falcone M, Tiseo G, Antonelli A, Giordano C, Di Pilato V, Bertolucci P, et al. Clinical features and outcomes of bloodstream infections caused by New Delhi metallo-β-lactamase–producing Enterobacterales during a regional outbreak. Open Forum Infect Dis. 2020;7(2):ofaa011. 10.1093/ofid/ofaa011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tavoschi L, Forni S, Porretta A, Righi L, Pieralli F, Menichetti F, et al. Tuscan Clinical Microbiology Laboratory Network . Prolonged outbreak of New Delhi metallo-β-lactamase-producing carbapenem-resistant Enterobacterales (NDM-CRE), Tuscany, Italy, 2018 to 2019. Euro Surveill. 2020;25(6). 10.2807/1560-7917.ES.2020.25.6.2000085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Di Pilato V, Henrici De Angelis L, Aiezza N, Baccani I, Niccolai C, Parisio EM, et al. Tuscan Clinical Microbiology Laboratory Network . Resistome and virulome accretion in an NDM-1-producing ST147 sublineage of Klebsiella pneumoniae associated with an outbreak in Tuscany, Italy: a genotypic and phenotypic characterisation. Lancet Microbe. 2022;3(3):e224-34. 10.1016/S2666-5247(21)00268-8 [DOI] [PubMed] [Google Scholar]
- 10. Simner PJ, Patel R. Cefiderocol antimicrobial susceptibility testing considerations: the Achilles’ heel of the Trojan horse? J Clin Microbiol. 2020;59(1):e00951-20. 10.1128/JCM.00951-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morris CP, Bergman Y, Tekle T, Fissel JA, Tamma PD, Simner PJ. Cefiderocol antimicrobial susceptibility testing against multidrug-resistant Gram-Negative bacilli: a comparison of disk diffusion to broth microdilution. J Clin Microbiol. 2020;59(1):e01649-20. 10.1128/JCM.01649-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.European Committee on Antimicrobial Susceptibility Testing (EUCAST). EUCAST warnings concerning antimicrobial susceptibility testing products or procedures. Växjö: EUCAST. [Accessed: 20 Jun 2022]. Available from: https://www.eucast.org/ast_of_bacteria/warnings
- 13. Hackel MA, Tsuji M, Yamano Y, Echols R, Karlowsky JA, Sahm DF. Reproducibility of broth microdilution MICs for the novel siderophore cephalosporin, cefiderocol, determined using iron-depleted cation-adjusted Mueller-Hinton broth. Diagn Microbiol Infect Dis. 2019;94(4):321-5. 10.1016/j.diagmicrobio.2019.03.003 [DOI] [PubMed] [Google Scholar]
- 14. Simner PJ, Beisken S, Bergman Y, Ante M, Posch AE, Tamma PD. Defining baseline mechanisms of cefiderocol resistance in the Enterobacterales. Microb Drug Resist. 2022;28(2):161-70. 10.1089/mdr.2021.0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.International Organization for Standardization (ISO). Clinical laboratory testing and in vitro diagnostic test systems. Susceptibility testing of infectious agents and evaluation of performance of antimicrobial susceptibility test devices—part 1: reference method for testing the in vitro activity of antimicrobial agents against rapidly growing aerobic bacteria involved in infectious diseases. ISO20776-1:2019. Available from: https://www.iso.org/standard/70464.html
- 16.The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Clinical breakpoints - breakpoints and guidance (v.12). Växjö: EUCAST; 1 Jan 2020. Available from: https://eucast.org/clinical_breakpoints
- 17. Matuschek E, Longshaw C, Takemura M, Yamano Y, Kahlmeter G. Cefiderocol: EUCAST criteria for disc diffusion and broth microdilution for antimicrobial susceptibility testing. J Antimicrob Chemother. 2022;77(6):1662-9. 10.1093/jac/dkac080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McElheny CL, Fowler EL, Iovleva A, Shields RK, Doi Y. In vitro evolution of cefiderocol resistance in an NDM-producing Klebsiella pneumoniae due to functional loss of CirA. Microbiol Spectr. 2021;9(3):e0177921. 10.1128/Spectrum.01779-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.