Abstract
Background
To gain more insight into the biological factors that mediate vulnerability to display externalizing behaviors, we leveraged genome-wide association study summary statistics on 13 externalizing phenotypes.
Methods
After data classification based on genetic resemblance, we performed multivariate genome-wide association meta-analyses and conducted extensive bioinformatic analyses, including genetic correlation assessment with other traits, Mendelian randomization, and gene set and gene expression analyses.
Results
The genetic data could be categorized into disruptive behavior (DB) and risk-taking behavior (RTB) factors, and subsequent genome-wide association meta-analyses provided association statistics for DB and RTB (Neff = 523,150 and 1,506,537, respectively), yielding 50 and 257 independent genetic signals. The statistics of DB, much more than RTB, signaled genetic predisposition to adverse cognitive, mental health, and personality outcomes. We found evidence for bidirectional causal influences between DB and substance use behaviors. Gene set analyses implicated contributions of neuronal cell development (DB/RTB) and synapse formation and transcription (RTB) mechanisms. Gene-brain mapping confirmed involvement of the amygdala and hypothalamus and highlighted other candidate regions (cerebellar dentate, cuneiform nucleus, claustrum, paracentral cortex). At the cell-type level, we noted enrichment of glutamatergic neurons for DB and RTB.
Conclusions
This bottom-up, data-driven study provides new insights into the genetic signals of externalizing behaviors and indicates that commonalities in genetic architecture contribute to the frequent co-occurrence of different DBs and different RTBs, respectively. Bioinformatic analyses supported the DB versus RTB categorization and indicated relevant biological mechanisms. Generally similar gene-brain mappings indicate that neuroanatomical differences, if any, escaped the resolution of our methods.
Keywords: Disruptive behavior, Externalizing behavior, Gene set analysis, Genetic correlation, GWAS, Mendelian randomization, N-GWAMA, Risk-taking behavior, Stratified LD-score regression
Externalizing psychopathologies are highly common in the general population (1, 2, 3, 4), and expressions of externalizing behavior including bullying, violence, delinquent activities, risk taking, and other related actions have a negative impact on individual prospects, local communities, and our society as a whole (5). It is therefore vitally important to gain more insight into the underlying (neuro)biological processes to understand why some individuals are more susceptible to displaying externalizing behaviors than others.
Externalizing symptoms generally co-occur substantially (6), which suggests commonalities in their etiologies (3,7). This is supported by indications for common genetic and environmental risk factors based on twin family studies (8, 9, 10, 11) and evidence for shared brain substrates from neurobiological studies (12, 13, 14). There is also evidence for causal relationships, particularly between substance use and (other) externalizing symptoms (15), a topic of high relevance for public health.
To broaden our understanding of common genetic and neurobiological backgrounds that underlie the co-occurrence of different externalizing behaviors, we leveraged publicly available genome-wide association study (GWAS) summary statistics on lifetime cannabis use (16), antisocial behavior (17), aggressive behavior (18), four item indicators of angriness and irritability from UK Biobank (19), and attention-deficit/hyperactivity disorder (ADHD) diagnosis (20). Furthermore, we added five items from a recent study on the genetics of risk tolerance (21). The 13 included datasets are summarized in Table 1.
Table 1.
GWAS Summary Statistics Included in the Multivariate Genome-wide Association Meta-analysis
| Phenotype | Sample Size | Source |
|---|---|---|
| Aggression | 18,988 | Pappa et al. (18) |
| Angry Outbursts | 71,196 | Sudlow et al. (19) |
| Extreme Irritable | 157,357 | Sudlow et al. (19) |
| Irritability | 501,652 | Sudlow et al. (19) |
| Irritable for 2 Days | 202,883 | Sudlow et al. (19) |
| ADHD | 53,293 | Demontis et al. (20) |
| Antisocial Behavior | 25,781 | Tielbeek et al. (17) |
| General Risk Tolerance | 466,571 | Karlsson Linnér et al. (21) |
| Drinks per Week | 414,343 | Karlsson Linnér et al. (21) |
| Ever Smoker | 518,633 | Karlsson Linnér et al. (21) |
| Number of Sexual Partners | 370,711 | Karlsson Linnér et al. (21) |
| Automobile Speeding Propensity | 404,291 | Karlsson Linnér et al. (21) |
| Lifetime Cannabis Use | 184,765 | Pasman et al. (16) |
ADHD, attention-deficit/hyperactivity disorder; GWAS, genome-wide association study.
Using these data, we first identified clusters of traits with high genetic resemblance that together comprise the higher-order externalizing behavior dimension. Subsequently, the GWAS data of the clusters were meta-analyzed using N-weighted meta-analysis (22). To characterize the genetic information in the meta-analyzed data, we evaluated the genetic relationships of the clusters with other phenotypes. In addition, for an identified cluster characterized by disruptive-type behaviors, we tested for causal relationship with smoking and alcohol consumption using bidirectional Mendelian randomization (MR). Previous studies have not been able to uncover the causal nature and direction of the relationship between disruptive behaviors (DBs) and smoking and alcohol use, two major risk factors for morbidity and mortality. Finally, biological annotation analyses were performed to identify associated brain regions and cell types.
Methods and Materials
Phenotype Selection
Based on the disinhibited externalizing spectrum described by Krueger et al. (1), we collected GWAS summary statistics for 13 externalizing phenotypes (Table 1). Only samples with a European/North-American ancestry (23) were included (see Supplement 1).
Identification of Genetic Factor Structures
To identify genetic factor structures among the 13 included phenotypes, we first applied linkage disequilibrium score regression (LDSC) (24,25) to compute pairwise genetic correlations (rg) (see Supplement 1).
Second, knowing the genetic correlations, we examined relationships using hierarchical clustering with (1 − rg) as genetic distance measures between phenotypes and with linkage based on Ward’s method (26). We followed the Calinski-Harabasz criterion to indicate the optimal number of clusters (27).
Third, in addition to hierarchical clustering, we applied factor analysis on the GWAS summary statistics using genomic structural equation modeling (28) (see Supplement 1).
Multivariate N-Weighted Genome-wide Association Meta-analysis
Univariate GWAS summary statistics were separately combined for identified factors using N-weighted genome-wide association meta-analysis (GWAMA) (22), which is robust against sample overlap (see Supplement 1).
Genetic Relationships With Other Traits
We computed pairwise genetic correlations using LDSC (24,25) for our identified factors with 61 additional phenotypes in the following categories: mental health, cognition and socioeconomic status, personality, social, substance use, cardiovascular disease risk, physical health, anthropomorphic, and reproduction (see Table S6 in Supplement 2 and Supplement 1 for details).
Mendelian Randomization
For one of the identified clusters characterized by DBs, we applied MR to test for causal relationship with substance use behaviors (see Supplement 1).
Gene Associations
We performed gene-based analyses in MAGMA version 1.08 (http://ctg.cncr.nl/software/magma) (29) with the N-weighted GWAMA summary statistics of the identified factors as input. The gene test statistics are defined as the mean single nucleotide polymorphism (SNP) association using the sum of −log (SNP p value).
Tissue-Type Associations
We used the gene associations of the two factors as input for a tissue-type analysis using MAGMA version 1.08 (29). We investigated tissue-specific gene expression values as gene properties using 53 tissues from the Genotype-Tissue Expression project v.7 (30) (see Supplement 1).
Gene Set Associations
We continued with competitive gene set analyses using MAGMA version 1.08 (29) to test whether the genes in a gene set are more strongly associated with the factor phenotypes than the other genes in the genome (see Supplement 1).
Conditional Analyses for Tissue Types and Gene Sets
For the significantly associated tissue types and gene sets, we performed conditional analyses using MAGMA version 1.08 (29) to evaluate redundancy between associations (see Supplement 1).
Stratified LDSC of Tissue Types
We applied stratified LDSC to investigate which tissues and cell types are enriched for the identified factors (see Supplement 1).
Stratified LDSC of Local Gene Expression Across the Human Brain
To identify brain regions where genes relevant for externalizing behavior factors are differently expressed, we computed stratified LD scores based on differential gene expression, using data from 3707 gene expression measurements across 211 different brain regions (31) (see Supplement 1).
Stratified LDSC of Brain Cell Types
We obtained a matrix of gene counts for single nuclei (n = 14,963) from the prefrontal cortex and hippocampus of multiple human donors studied by Habib et al. (32). We subsequently determined the differential expression of genes in seven types of brain cells: GABAergic (gamma-aminobutyric acidergic) interneurons, excitatory neurons, astrocytes, oligodendrocytes, microglia, endothelial cells, and neural stem cells (32) (see Supplement 1).
Results
Identification of Genetic Factor Structure
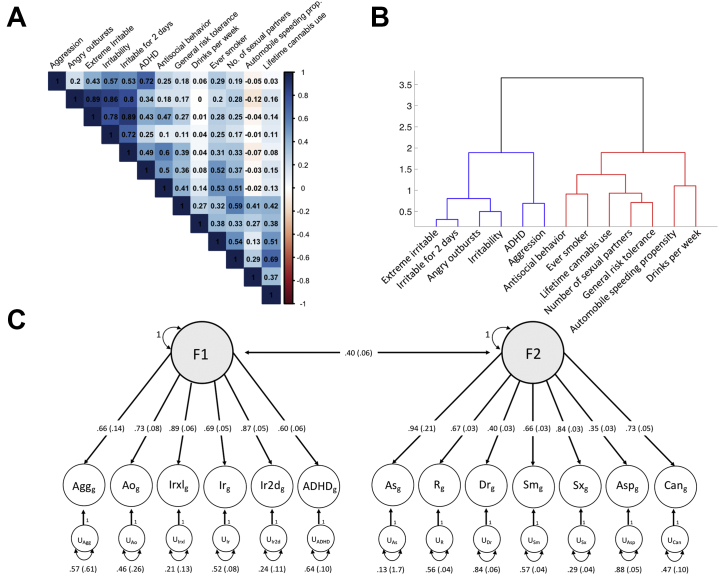
LDSC (Figure 1A) indicated substantial genetic correlations, particularly among phenotypes characterized by disruptive-type behaviors (e.g., aggression and ADHD: rg = 0.72, SE = 0.18) and phenotypes characterized by risk-taking behaviors (RTBs) (e.g., cannabis use and number of sexual partners: rg = 0.69, SE = 0.02), with less genetic overlap between pairs of phenotypes characterized by the different behavior types (e.g., aggression and cannabis use: rg = 0.03, SE = 0.13). Hierarchical clustering on genetic resemblance (Figure 1B) confirmed this categorization into DBs (aggression, angry outbursts, different measures of irritability, and ADHD) and RTBs (antisocial behavior, general risk tolerance, drinks per week, ever smoker, number of sexual partners, automobile speeding propensity, and lifetime cannabis use). Exploratory factor analysis by genomic structural equation modeling supported a division into two phenotypic clusters by demonstrating a substantial increase in explained variance from 34% for a one-factor model to 53% for a two-factor model, but a much more moderate increase to 58% for a three-factor model (Figure 1C; Tables S1–S3 in Supplement 2). The two-factor exploratory factor analysis model assigned similar phenotypes to each factor as our hierarchical clustering approach. The loadings of the two-factor model, however, did not support a clear allocation for ADHD and antisocial behavior (Table S1 in Supplement 2). For these phenotypes, we followed the hierarchical clustering results, assigning ADHD to factor 1 (DB) and antisocial behavior to factor 2 (RTB).
Figure 1.
Genetic factor structure of externalizing behaviors. (A) Genetic correlations between the externalizing phenotypes calculated by linkage disequilibrium score regression. (B) Hierarchical clustering dendrogram based on genetic resemblance of the externalizing phenotypes. Blue represents the disruptive behavior cluster, and red represents the risk-taking behavior cluster. (C) Based on the results of an exploratory factor analysis of the genetic correlations presented in panel (A), a confirmatory factor model with two correlated genetic factors was specified using genomic structural equation modeling. In this model, the common factors account for the genetic covariation among the externalizing traits, i.e., each of the two common genetic factors represents variation in genetic liability that is shared across the phenotypes that load on it. Disruptive behavior represents shared genetic liability among disorders characterized by disruptive behavior, and risk-taking behavior represents the shared liability for risk-taking behavior. One-headed arrows connecting the common genetic factors to the individual traits represent standard loadings, which can be interpreted as coefficients from a regression of the true genetic liability for the trait on the common factor. Two-headed arrows connecting the genetic components represent their correlations. Two-headed arrows connecting the genetic components of the individual traits to themselves represent residual genetic variances and correspond to the proportion of heritable variation in liability to each individual trait that is unexplained by the two factors. ADHD, attention-deficit/hyperactivity disorder; Agg, aggression; Ao, angry outbursts; As, antisocial behavior; Asp, automobile speeding propensity; Can, lifetime cannabis use; Dr, drinks per week; Irxl, extreme irritable; Ir, irritability; Ir2d, irritable for 2 days; R, general risk tolerance; Sm, ever smoker; Sx, number of sexual partners.
Multivariate GWAMA
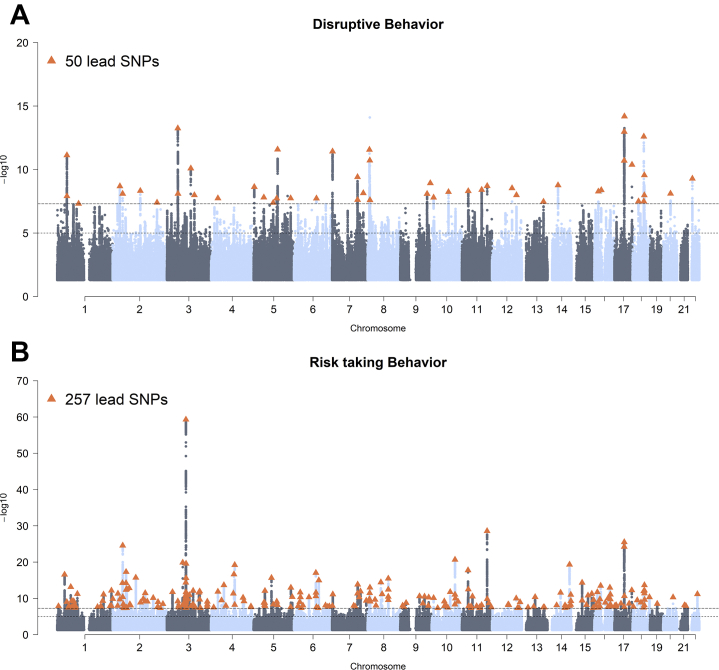
For both factors, we subsequently meta-analyzed the phenotype GWAS data using N-weighted GWAMA. For the DB factor, we identified 50 independent genome-wide significant SNPs at 42 loci (Neff = 523,150) (Figure 2A; Table S4 in Supplement 2). For RTB, we identified 257 independent genome-wide significant SNPs at 194 genomic risk loci (Neff = 1,506,537) (Figure 2B; Table S5 in Supplement 2). The LD score intercepts were close to 1 for both DB (intercept = 1.0167, SE = 0.0086; LDSC ratio = 0.0367, SE = 0.0156) and RTB (intercept = 1.0031, SE = 0.0132; LDSC ratio = 0.0242, SE = 0.0106), indicating that neither population stratification nor sample overlap, but rather an increase of polygenic signal, was driving the SNP associations. The SNP heritability as defined by LDSC was 0.0396 (SE = 0.0015) for DB and 0.022 (SE = 0.0007) for RTB. The genetic correlation between DB and RTB was 0.33 (SE = 0.02).
Figure 2.
Manhattan plots of the meta-analyzed phenotype genome-wide association study data. (A) Disruptive behavior factor. (B) Risk-taking behavior factor. The x-axis represents the chromosomal position, and the y-axis represents the significance on a −log10 scale. Each approximately independent genome-wide significant association (lead SNP) is marked by a triangle (p < 5 × 10−8). SNP, single nucleotide polymorphism.
Genetic Relationships With Other Traits
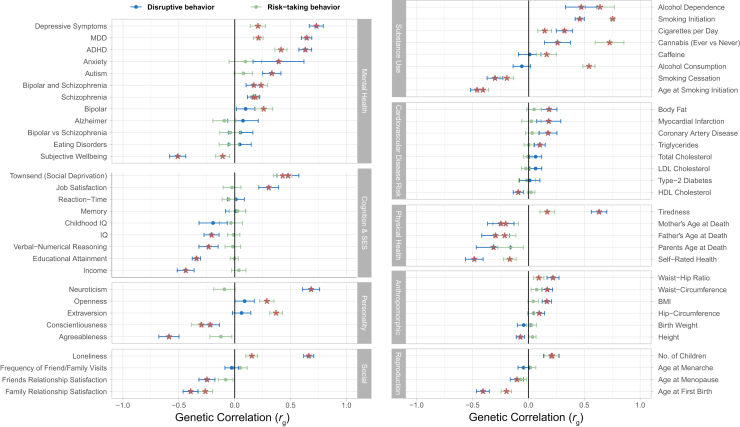
Figure 3 and Table S7 in Supplement 2 show pairwise genetic correlations for DB and RTB with 61 additional phenotypes. DB, much more than RTB, showed a pattern of genetic overlap pointing to adverse outcomes of cognition, socioeconomic status, several mental and physical health measures, and personality. For example, DB showed a negative genetic correlation with educational attainment (rg = −0.34, SE = 0.02) and income (rg = −0.44, SE = 0.04), whereas genetic correlations for RTB with these phenotypes were close to zero. In addition, DB was more positively correlated with depressive symptoms (rg = 0.73, SE = 0.03) and neuroticism (rg = 0.68, SE = 0.04) and more negatively correlated with agreeableness (rg = −0.59, SE = 0.05) compared with RTB (depressive symptoms: rg = 0.21, SE = 0.03; neuroticism: rg = −0.09, SE = 0.005; agreeableness: rg = −0.12, SE = 0.05).
Figure 3.
Genetic correlations with other phenotypes for disruptive behavior (blue) and risk-taking behavior (green) factors. Points represent the correlation estimates, and lines represent the 95% confidence intervals. Significant associations, after correction for multiple testing, are marked by orange stars. ADHD, attention-deficit/hyperactivity disorder; BMI, body mass index; HDL, high-density lipoprotein; LDL, low-density lipoprotein; MDD, major depressive disorder; SES, socioeconomic status.
Causal Relationship of DB With Smoking and Alcohol Use
We tested for bidirectional causal effects between DB (which does not include substance use phenotypes) and measures of smoking (33) and alcohol use (34) using MR. We focused on these specific relationships because DBs and substance use are particularly strongly associated, and knowledge of (the direction of) potential causal effects has major public health implications. When DB was the exposure variable, inverse variance weighted analyses provided strong evidence for causal effects such that DB increases the odds of smoking initiation (β = 0.39, 95% CI = 0.24–0.54, p = 2.0 × 10−7) and decreases the odds of being able to successfully quit smoking (β = −0.20, 95% CI = −0.33 to −0.07, p = .002) (Table 2; Figure S1 in Supplement 1). Effect sizes and statistical evidence for a causal relationship between DB and smoking initiation were broadly consistent across the different MR methods. However, for DB to smoking cessation, the weighted mode and MR-Egger did not support a causal relationship (Table 2). Some evidence for pleiotropic effects was provided by a significant Cochran’s heterogeneity test (Cochran’s Q p value = 2.4 × 10−33) (Table S8 in Supplement 2) but not supported by the MR-Egger intercept (only DB → smoking initiation) (Table S9 in Supplement 2). Steiger filtering and MR-PRESSO did not affect the results and continued to support causal effects (Tables S10–S12 in Supplement 2). No consistent results were found for a causal relationship between DB and number of cigarettes smoked per day (Table 2; Table S9 in Supplement 2).
Table 2.
Results of the Two-Sample Bidirectional Mendelian Randomization Analyses Between DB and Smoking and Alcohol Use Behavior
| Exposure | Outcome | SNPs, n | F | IVW |
Weighted Median |
Weighted Mode |
MR-Egger (SIMEX) |
GSMR |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β | 95% CI | p | β | 95% CI | p | β | 95% CI | p | β | 95% CI | p | SNPs, n | β | 95% CI | p | ||||
| DB | Smoking initiation | 40 | 37.41 | 0.39 | 0.24 to 0.54 | 2.0 × 10−7 | 0.25 | 0.13 to 0.37 | 3.2 × 10−5 | 0.15 | −0.06 to 0.36 | .180 | 0.43 | 0.09 to 0.77 | .017 | 33 | 0.342 | 0.27 to 0.41 | 1.7 × 10−20 |
| DB | Cigarettes/day | 38 | 37.61 | 0.14 | 0.01 to 0.27 | .029 | −0.01 | −0.12 to 0.10 | .895 | −0.06 | −0.16 to 0.04 | .284 | −0.31 | −0.60 to −0.02 | .041 | 34 | 0.178 | 0.09 to 0.26 | 3.3 × 10−5 |
| DB | Smoking cessation | 40 | 37.33 | −0.20 | −0.33 to −0.07 | .002 | −0.11 | −0.25 to 0.03 | .118 | 0.02 | −0.24 to 0.28 | .873 | 0.17 | −0.11 to 0.45 | .254 | 37 | −0.190 | −0.29 to −0.09 | 9.9 × 10−5 |
| DB | Alcohol/week | 40 | 37.41 | −0.01 | −0.04 to 0.02 | .472 | −0.002 | −0.03 to 0.03 | .897 | 0.01 | −0.02 to 0.04 | .545 | −0.04 | −0.11 to 0.03 | .341 | 35 | −0.020 | −0.04 to 0.00 | .043 |
| DB | Alcohol use disorder | 46 | 37.00 | 0.26 | 0.08 to 0.44 | .004 | 0.16 | −0.10 to 0.42 | .245 | 0.14 | −0.12 to 0.40 | .288 | n.a. | n.a. | n.a. | 43 | 0.310 | 0.10 to 0.52 | .003 |
| Smoking Initiation | DB | 317 | 27.53 | 0.17 | 0.14 to 0.20 | 9.9 × 10−31 | 0.15 | 0.12 to 0.18 | 8.0 × 10−21 | 0.15 | 0.05 to 0.25 | .004 | n.a. | n.a. | n.a. | 296 | 0.186 | 0.17 to 0.21 | 6.4 × 10−72 |
| Alcohol/week | DB | 80 | 28.61 | 0.04 | −0.13 to 0.21 | .632 | 0.052 | −0.12 to 0.22 | .543 | −0.17 | −2.07 to 1.73 | .859 | −0.24 | −0.54 to 0.06 | .133 | 74 | 0.0719 | −0.05 to 0.20 | .261 |
| Alcohol Dis. 5 × 10−8 | DB | 7 | 26.35 | −0.02 | −0.06 to 0.02 | .299 | −0.02 | −0.06 to 0.02 | .480 | −0.01 | −0.08 to 0.06 | .833 | n.a. | n.a. | n.a. | 7 | 0.00016 | −0.04 to 0.04 | .993 |
| Alcohol Dis. 1 × 10−5 | DB | 25 | 21.94 | −0.01 | −0.03 to 0.01 | .328 | −0.01 | −0.03 to 0.01 | .654 | −0.01 | −0.06 to 0.04 | .652 | n.a. | n.a. | n.a. | 24 | −0.0078 | −0.02 to 0.01 | .365 |
n.a. indicates that MR-Egger results were not reported because of limited reliability based on the I2 measure being <0.60. F > 10 generally indicates the instrument is sufficiently strong.
DB, disruptive behavior; Dis., disorder; F, F-statistic indicating instrument strength; GSMR, generalized summary data–based Mendelian randomization; IVW, inverse variance weighted regression; SIMEX, simulation extrapolation; SNP, single nucleotide polymorphism.
From DB to alcohol use disorder, there was evidence for a causal increasing effect (inverse variance weighted analysis: β = 0.26, 95% CI = 0.08–0.44, p = .004), which was consistent (but weaker) in direction of effect across multiple MR methods (Table 2). MR-Egger was not available owing to low reliability; however, there was no heterogeneity based on Cochran’s Q (p = .457) (Table S8 in Supplement 2). Steiger filtering and MR-PRESSO did not change the outcome (Tables S10–S12 in Supplement 2).
In the opposite direction, we found strong evidence for a causal, increasing effect of smoking initiation on DB (inverse variance weighted analysis: β = 0.17, 95% CI = 0.14–0.20, p = 9.9 × 10−31), which was consistent across weighted median, weighted mode, and generalized summary data–based MR (MR-Egger not available). Steiger filtered analyses and MR-PRESSO did not change the results (Table 2; Tables S10–S12 in Supplement 2). However, there was marked evidence for heterogeneity (Cochran’s Q p = 7.3 × 10−46) (Table S8 in Supplement 2). There was no clear evidence for causal effects of alcohol use on DB.
Biological Annotations
Gene Associations
Genome-wide gene-based association analysis identified 81 genes significantly associated with DB (Table S13 in Supplement 2) and 318 genes significantly associated with RTB (Table S14 in Supplement 2) after Bonferroni correction. CADM2 showed an exceptionally strong association (2.10 × 10−29) with RTB.
Tissue-Type Associations
We used the gene-based test statistics as input for gene set analysis in MAGMA. Starting with a tissue-type analysis, we found significant associations at the Bonferroni level for nine brain regions for DB and 13 brain regions for RTB (Tables S15 and S16 in Supplement 2). Conditional analyses indicated that only the frontal cortex showed an independent association with DB, while independent associations were limited to the frontal cortex and cerebellum for RTB.
Gene Set Associations
Subsequent gene set analyses identified three gene sets for DB involved in cell development and 13 gene sets for RTB involved in cell development, synapse formation, and transcription (Tables S17 and S18 in Supplement 2). Conditional gene set analyses on the three gene sets associated with DB showed that the three gene set associations were highly related (Table S17 in Supplement 2). In addition, the conditional analyses of the 13 gene sets identified for RTB traits revealed redundancy for multiple gene sets related to synapse and cell development and transcription, leaving seven gene sets with independent association (Table S18 in Supplement 2).
Stratified LDSC
Complementary to gene set analysis, we applied stratified LDSC to study effects on 10 general tissue-type groups. Consistent with the gene set analyses, these analyses indicated significant enrichment after Bonferroni correction of the central nervous system (CNS) for both DB (ZCNS = 6.84) (Table S19 in Supplement 2) and RTB (ZCNS = 7.97) (Table S20 in Supplement 2). It has to be noted that other tissues were associated with DB and RTB as well (e.g., liver, kidney), but these associations were considerably weaker (Tables S19 and S20 in Supplement 2). Zooming in on associations on the cell-type level for DB revealed 45 significant cell type–specific annotations after Bonferroni correction, of which the top 15 all involved the CNS (Table S21 in Supplement 2). A similar pattern was observed for RTB, where we found 96 significant cell type–specific annotations, of which 32 indicated CNS involvement (Table S22 in Supplement 2).
Differential Gene Expression
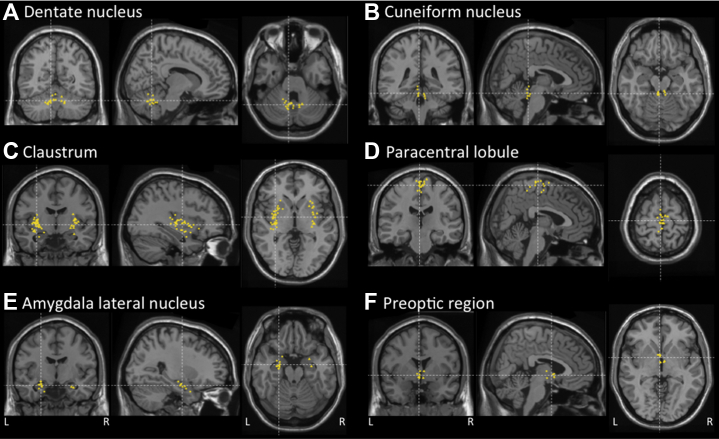
To pinpoint relevant brain areas associated with DB and RTB more accurately, we proceeded with a local differential gene expression approach using stratified LDSC (31). For DB, we identified significant enrichment after Bonferroni correction exclusively in the dentate nucleus (cerebellum, Z = 3.35, p = .0004) (Figure 4A; Table S23 in Supplement 2). For RTB, there was also enrichment in the dentate nucleus (cerebellum, Z = 3.22, p = .0006) and additional enrichment in the cuneiform nucleus (brain stem, Z = 4.74, p = 1.06 × 10−6) (Figure 4B), claustrum (cortex, Z = 4.08, p = 2.27 × 10−5) (Figure 4C), paracentral lobule (cortex, Z = 3.35, p = .0004) (Figure 4D), lateral amygdaloid nucleus (subcortex, Z = 3.12, p = .0009) (Figure 4E), and preoptic region (subcortex, Z = 3.34, p = .0004) (Figure 4F; Table S24 in Supplement 2).
Figure 4.
Brain regions with local differential gene expression enrichment for the disruptive behavior and risk-taking behavior factors in coronal, sagittal, and axial views. The locations of the samples of brain tissues that were used to measure gene expression by Hawrylycz et al. (31) are projected to a standard Montreal Neurological Institute template brain (Colin27). For every annotation, the figure is centered on the averaged Montreal Neurological Institute coordinates of the brain samples. (A) Cerebellum: dentate nucleus. (B) Brainstem: cuneiform nucleus. (C) Cortex: claustrum. (D) Cortex: paracentral lobule. (E) Subcortex: lateral amygdaloid nucleus. (F) Subcortex: preoptic region.
Single-Cell Analysis
Finally, we investigated the involvement of specific brain cell types using LDSC. For DB, this revealed enrichment in excitatory neurons of the prefrontal cortex (Z = 2.88, p = .002) and the hippocampal CA3 region (Z = 2.85, p = .002) (Table S25 in Supplement 2). For RTB, we found enrichment in excitatory prefrontal cortex neurons (Z = 3.14, p = .0008) (Table S26 in Supplement 2).
Discussion
In a bottom-up approach to evaluate overlap in genetic and neurobiological backgrounds underlying externalizing symptoms, we collected 13 publicly available GWAS summary statistics for a range of externalizing phenotypes. Assessment of genetic resemblance indicated a categorization into two externalizing-related factors, one characterized by DB and another by RTB, that together explained 53% of the total genetic variance. Meta-analyzing the factor-specific phenotypes yielded 50 loci for DB and 257 loci for RTB.
Clustering of aggression, angriness, and irritability items in the DB factor fits very well with the general finding that these are highly comorbid behaviors and also cluster together in diagnoses of conduct and oppositional defiant disorder (35). Addition of ADHD aligns with the fact that DBs coexist most commonly with ADHD (36) and with evidence of strong genetic resemblance between ADHD and conduct and oppositional defiant disorder (37). The phenotypes of the RTB factor related to pursuing risk (21). Three RTB traits (general risk tolerance, number of sexual partners, lifetime cannabis use) similarly clustered together in another recent GWAMA (38). Addition of antisocial behavior aligns with the fact that antisocial behavior frequently includes thoughtless, self-centered, and immediately rewarding acts, which are also typical for risk taking. The present RTB phenotypes related to substance use, speeding, and sexual promiscuity are closely linked to social misconduct (38,39). In genomic structural equation modeling, antisocial behavior also loaded on DB, which is concordant with observations from the Hierarchical Taxonomy of Psychopathology consortium that antisocial behavior is a broad construct and also alludes to disruptive phenotypes (1,40).
Assessment of genetic relationships with other traits indicated associations for DB with higher neuroticism and lower conscientiousness and agreeableness, corroborating previous findings (41). For RTB, we found associations with higher extraversion, higher openness to experience, and lower conscientiousness, which is also in line with earlier reports (42). Another notable result was that DB, much more than RTB, showed negative genetic associations with indices of mental and physical health. Previous studies indicated associations with mental and physical health problems for both disruptive-ADHD and risk-taking behaviors (43, 44, 45, 46). This finding of stronger genetic correlations for DB points to the possible distinction that DB phenotypes, characterized by affective instability/irritability and impaired cognitive control, are generally central to mental and physical health problems (44,47), whereas for RTB, mental health problems are more often at the core (45), resulting in risk behaviors, such as substance use, that ultimately affect physical health (46).
In addition, we applied MR to explore causal relationships between the DB factor and smoking and alcohol use. In full agreement with previous findings (15,48), we obtained evidence that DB traits promote substance use (strong evidence for an increased odds of smoking initiation and decreased odds to successfully quit smoking and weak evidence for increased odds of alcohol use disorder). This causal direction is also consistent with expectations for behavioral development, in that DB traits are more likely to occur at younger ages than risk behaviors. Smoking and alcohol use are not yet appropriate and less feasible during early childhood (15,48). However, we also replicated the observation that smoking might causally increase the risk for DB traits. This finding could have important consequences for intervention strategies but needs further inquiry. To distinguish a causal influence from horizontal pleiotropy, i.e., when a genetic variant influences the two traits through independent pathways, dose-response effects should be included in future MR analyses (taking cigarettes per day as the exposure and applying stratification on smoking status).
Biological annotation by gene set analysis and stratified LDSC converged on genetic enrichment in the brain for both DB and RTB traits. An atlas of differential brain region gene expression pointed specifically to the dentate nucleus of the cerebellum, for both DB and RTB, and the cuneiform nucleus of the brainstem, preoptic region of the hypothalamus, lateral nucleus of the amygdala, and the claustrum and paracentral cortical lobule, which were significant for RTB.
Involvement of these brain regions confirms current hypotheses on the neurobiological background of externalizing symptoms but also points to the importance of relatively underrepresented regions. Especially for the medial part of the preoptic hypothalamic region, animal studies indicate an important role in stimulating aggression to facilitate reproduction (49), as well as in modulating mesolimbic activity involved in reward processing (50). The contribution of the amygdala to emotional processing is also generally recognized (51), and there is ample evidence that amygdala–frontal cortex network abnormalities predispose to externalizing behaviors (14), including aggression and risk taking (52,53). In addition, amygdala volume reductions are commonly found in ADHD (54, 55, 56). Furthermore, a recent study showed that GABAergic neurons in the medial amygdala project to the medial preoptic area to regulate reward from social stimuli by controlling the release of dopamine in the nucleus accumbens (57).
The other brain regions have been implicated less frequently, despite previous evidence for associations. For the cerebellar dentate, a relevant role in the development of reinforcement learning relevant to addiction has been indicated (58), and there is evidence that abnormal development of corticocerebellar connections contributes to ADHD (59). The cuneiform nucleus is part of the mesencephalic locomotor control network (60) and related to autonomic fear and stress responses (61,62). The claustrum is richly interconnected with almost all regions of the cerebral cortex and hypothesized to play a role in regulating attention and resilience to distraction (63,64), which links this region to ADHD (65). The anterior section of the paracentral lobule includes motor control regions but has also been linked to executive control function and attention orienting (22,23). Single-cell analysis further elucidated the important role of the CNS for both DB and RTB and pointed to glutamatergic prefrontal neurons for DB and RTB and pyramidal hippocampal neurons for DB. Glutamatergic neurotransmission has been associated with alcohol and tobacco use (33) and RTB (21) before, but more research is needed to address the role of these specific cell types and their functions on the externalizing phenotypes.
Finally, testing for specific biological mechanisms using gene set analysis indicated associations for gene sets primarily related to neuronal development for both DB and RTB. In addition, synaptic functions and transcription regulation were identified for RTB. These three biological mechanisms have been related to a wide range of psychiatric disorders (66) and may play a broad role in behaviors and brain-related traits.
This study comes with a number of limitations. First, the two identified factors DB and RTB combine several distinct phenotypic measures, which inevitably leads to etiologic heterogeneity (reflected by the imperfect genetic correlations). In addition, phenotyping could be as poor as a single question (e.g., about regular use of a substance). Poor and heterogenous phenotyping, with the trade-off of having large sample sizes available, are a common phenomenon in GWAS meta-analyses of single traits (67) or multiple traits (22) and generally results in lower SNP heritability estimates compared with the individual traits, which was also the case here for DB (4%) and RTB (2%). Second, we incorporated three measures on irritability, which seems redundant. However, the mutual genetic correlation between these phenotypes was not perfect (rg < 1), and therefore, we note that including GWAS statistics of all three phenotypes does provide additional genetic information in GWAMAs. Third, in the MR analyses, the tested effects from smoking and alcohol use to DB did not fully comply with the expected temporal order. The aggression and ADHD variables of DB included children (which amounts to ∼5% of the total number of DB participants). This means that smoking and alcohol use were, for those variables, not valid exposures. In addition, we limited assessment of causal directions to the relationship between the DB factor and smoking and alcohol use. We also demonstrated interesting relationships of DB and RTB with several other traits, such as personality and health characteristics. Given the complexity of the included study cohorts forming the different summary statistics, it was beyond the scope of this study to also test for causal relationships with these traits. Fourth, except for ADHD, the GWAS data in our study related to behavioral extremes that did not exceed the diagnostic threshold. Given previous evidence of genetic heterogeneity between nondiagnosed and diagnosed individuals (68,69), we emphasize that the findings in this study apply in particular to general rather than excessive diagnosed externalizing behaviors. Finally, regarding the biological annotation analyses, we must consider limitations in drawing conclusions about different findings for DB and RTB. DB and RTB are moderately correlated (rg = 0.33) and therefore not completely independent. In addition, the effective sample size of RTB is ∼3 times larger than that of DB (1,506,537 vs. 523,150), so there is likely more power to detect significant associations for RTB. For example, differential gene expression indicated more brain regions with statistically significant enrichment for RTB, but we note that the same regions also showed relatively increased enrichment among the 211 brain regions tested for DB.
In summary, we extend previous findings involving externalizing behavior and provide further evidence for common genetic architectures, particularly for different DBs and RTBs. Follow-up evaluations of the data in this study indicated genetic relationships with personality traits and mental and physical health behaviors in agreement with the DB versus RTB categorization and highlighted possible bidirectional causal relationships between DB and substance use traits. Biological annotation revealed generally similar gene-brain mappings that mediate the predisposition to comorbid externalizing phenotypes. Possible subtle differences in neurobiological backgrounds between DB and RTB traits may be resolved in future studies based on higher-resolution genetic association and gene expression data that are becoming increasingly available.
Acknowledgments and Disclosures
We thank the research participants and employees of 23andMe and UK Biobank for making this work possible. In addition, we thank the Social Science Genetic Consortium, the EAGLE consortium, and the International Cannabis Consortium for making their summary statistics publicly available. Without their data sharing policy, this study was not possible.
This study used genome-wide association study summary statistics published by 23andMe and the UK Biobank. All input data used in the present study are from public sources (listed in Table 1). Output data are accessible after a data request application to ntr.datamanagement.fgb@vu.nl.
The code to perform the multivariate analyses can be found at: https://github.com/baselmans/multivariate_GWAMA
BB is supported by an NWO-ZonMW Rubicon Grant (Grant No. 45219101). All other authors report no biomedical financial interests or potential conflicts of interest.
Footnotes
Supplementary material cited in this article is available online at https://doi.org/10.1016/j.bpsgos.2021.09.007.
Supplementary Material
References
- 1.Krueger R.F., Kotov R., Watson D., Forbes M.K., Eaton N.R., Ruggero C.J., et al. Progress in achieving quantitative classification of psychopathology. World Psychiatry. 2018;17:282–293. doi: 10.1002/wps.20566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krueger R.F., South S.C. Externalizing disorders: Cluster 5 of the proposed meta-structure for DSM-V and ICD-11. Psychol Med. 2009;39:2061–2070. doi: 10.1017/S0033291709990328. [DOI] [PubMed] [Google Scholar]
- 3.Beauchaine T.P., Zisner A.R., Sauder C.L. Trait impulsivity and the externalizing spectrum. Annu Rev Clin Psychol. 2017;13:343–368. doi: 10.1146/annurev-clinpsy-021815-093253. [DOI] [PubMed] [Google Scholar]
- 4.Ahmad S.I., Hinshaw S.P. Attention-deficit/hyperactivity disorder, trait impulsivity, and externalizing behavior in a longitudinal sample. J Abnorm Child Psychol. 2017;45:1077–1089. doi: 10.1007/s10802-016-0226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.GBD 2016 Alcohol Collaborators Alcohol use and burden for 195 countries and territories, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2018;392:1015–1035. doi: 10.1016/S0140-6736(18)31310-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartels M., Hendriks A., Mauri M., Krapohl E., Whipp A., Bolhuis K., et al. Childhood aggression and the co-occurrence of behavioural and emotional problems: Results across ages 3–16 years from multiple raters in six cohorts in the EU-ACTION project. Eur Child Adolesc Psychiatry. 2018;27:1105–1121. doi: 10.1007/s00787-018-1169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mullins-Sweatt S.N., DeShong H.L., Lengel G.J., Helle A.C., Krueger R.F. Disinhibition as a unifying construct in understanding how personality dispositions undergird psychopathology. J Res Pers. 2019;80:55–61. doi: 10.1016/j.jrp.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jang K.L., Vernon P.A., Livesley W.J. Personality disorder traits, family environment, and alcohol misuse: A multivariate behavioural genetic analysis. Addiction. 2000;95:873–888. doi: 10.1046/j.1360-0443.2000.9568735.x. [DOI] [PubMed] [Google Scholar]
- 9.Krueger R.F., Hicks B.M., Patrick C.J., Carlson S.R., Iacono W.G., McGue M. Etiologic connections among substance dependence, antisocial behavior, and personality: Modeling the externalizing spectrum. J Abnorm Psychol. 2002;111:411–424. [PubMed] [Google Scholar]
- 10.Venables N.C., Hicks B.M., Yancey J.R., Kramer M.D., Nelson L.D., Strickland C.M., et al. Evidence of a prominent genetic basis for associations between psychoneurometric traits and common mental disorders. Int J Psychophysiol. 2017;115:4–12. doi: 10.1016/j.ijpsycho.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young S.E., Friedman N.P., Miyake A., Willcutt E.G., Corley R.P., Haberstick B.C., Hewitt J.K. Behavioral disinhibition: Liability for externalizing spectrum disorders and its genetic and environmental relation to response inhibition across adolescence. J Abnorm Psychol. 2009;118:117–130. doi: 10.1037/a0014657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noordermeer S.D.S., Luman M., Oosterlaan J. A systematic review and meta-analysis of neuroimaging in oppositional defiant disorder (ODD) and conduct disorder (CD) taking attention-deficit hyperactivity disorder (ADHD) into account. Neuropsychol Rev. 2016;26:44–72. doi: 10.1007/s11065-015-9315-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y., Raine A. Prefrontal structural and functional brain imaging findings in antisocial, violent, and psychopathic individuals: A meta-analysis. Psychiatry Res. 2009;174:81–88. doi: 10.1016/j.pscychresns.2009.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ameis S.H., Ducharme S., Albaugh M.D., Hudziak J.J., Botteron K.N., Lepage C., et al. Cortical thickness, cortico-amygdalar networks, and externalizing behaviors in healthy children. Biol Psychiatry. 2014;75:65–72. doi: 10.1016/j.biopsych.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Treur J.L., Demontis D., Smith G.D., Sallis H., Richardson T.G., Wiers R.W., et al. Investigating causality between liability to ADHD and substance use, and liability to substance use and ADHD risk, using Mendelian randomization. Addict Biol. 2021;26 doi: 10.1111/adb.12849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasman J.A., Verweij K.J.H., Gerring Z., Stringer S., Sanchez-Roige S., Treur J.L., et al. GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal influence of schizophrenia. Nat Neurosci. 2018;21:1161–1170. doi: 10.1038/s41593-018-0206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tielbeek J.J., Johansson A., Polderman T.J.C., Rautiainen M.R., Jansen P., Taylor M., et al. Genome-wide association studies of a broad spectrum of antisocial behavior. JAMA Psychiatry. 2017;74:1242–1250. doi: 10.1001/jamapsychiatry.2017.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pappa I., St Pourcain B., Benke K., Cavadino A., Hakulinen C., Nivard M.G., et al. A genome-wide approach to children’s aggressive behavior: The EAGLE consortium. Am J Med Genet B Neuropsychiatr Genet. 2016;171:562–572. doi: 10.1002/ajmg.b.32333. [DOI] [PubMed] [Google Scholar]
- 19.Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., et al. UK Biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12 doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demontis D., Walters R.K., Martin J., Mattheisen M., Als T.D., Agerbo E., et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019;51:63–75. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karlsson Linnér R., Biroli P., Kong E., Meddens S.F.W., Wedow R., Fontana M.A., et al. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet. 2019;51:245–257. doi: 10.1038/s41588-018-0309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baselmans B.M.L., Jansen R., Ip H.F., van Dongen J., Abdellaoui A., van de Weijer M.P., et al. Multivariate genome-wide analyses of the well-being spectrum. Nat Genet. 2019;51:445–451. doi: 10.1038/s41588-018-0320-8. [DOI] [PubMed] [Google Scholar]
- 23.Gravel S. Population genetics models of local ancestry. Genetics. 2012;191:607–619. doi: 10.1534/genetics.112.139808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bulik-Sullivan B.K., Loh P.R., Finucane H.K., Ripke S., Yang J., Schizophrenia Working Group of the Psychiatric Genomics Consortium, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bulik-Sullivan B., Finucane H.K., Anttila V., Gusev A., Day F.R., Loh P.R., et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward J.H., Jr. Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 1963;58:236–244. [Google Scholar]
- 27.Caliński T., Harabasz J. A dendrite method for cluster analysis. Commun Stat. 1974;3:1–27. [Google Scholar]
- 28.Grotzinger A.D., Rhemtulla M., de Vlaming R., Ritchie S.J., Mallard T.T., Hill W.D., et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat Hum Behav. 2019;3:513–525. doi: 10.1038/s41562-019-0566-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Leeuw C.A., Mooij J.M., Heskes T., Posthuma D. MAGMA: Generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11 doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hawrylycz M.J., Lein E.S., Guillozet-Bongaarts A.L., Shen E.H., Ng L., Miller J.A., et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Habib N., Avraham-Davidi I., Basu A., Burks T., Shekhar K., Hofree M., et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods. 2017;14:955–958. doi: 10.1038/nmeth.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu M., Jiang Y., Wedow R., Li Y., Brazel D.M., Chen F., et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51:237–244. doi: 10.1038/s41588-018-0307-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walters R.K., Polimanti R., Johnson E.C., McClintick J.N., Adams M.J., Adkins A.E., et al. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat Neurosci. 2018;21:1656–1669. doi: 10.1038/s41593-018-0275-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.American Psychiatric Association . Fifth Edition. American Psychiatric Association Publishing; Washington, DC: 2013. Diagnostic and Statistical Manual of Mental Disorders. [Google Scholar]
- 36.Kunwar A., Dewan M., Faraone S.V. Treating common psychiatric disorders associated with attention-deficit/hyperactivity disorder. Expert Opin Pharmacother. 2007;8:555–562. doi: 10.1517/14656566.8.5.555. [DOI] [PubMed] [Google Scholar]
- 37.Dick D.M., Viken R.J., Kaprio J., Pulkkinen L., Rose R.J. Understanding the covariation among childhood externalizing symptoms: Genetic and environmental influences on conduct disorder, attention deficit hyperactivity disorder, and oppositional defiant disorder symptoms. J Abnorm Child Psychol. 2005;33:219–229. doi: 10.1007/s10802-005-1829-8. [DOI] [PubMed] [Google Scholar]
- 38.Karlsson Linnér R., Mallard T.T., Barr P.B., Sanchez-Roige S., Madole J.W., Driver M.N., et al. Multivariate genomic analysis of 1.5 million people identifies genetic associations with traits related to self-regulation and addiction. Nat Neurosci. 2021;24:1367–1376. doi: 10.1038/s41593-021-00908-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mishra S., Lalumiére M.L. In: Evolutionary Forensic Psychology: Darwinian Foundations of Crime and Law. Duntley J.D., Shackelford T.K., editors. Oxford University Press; New York, NY: 2008. Risk-taking, antisocial behavior, and life histories. [Google Scholar]
- 40.Krueger R.F., Hobbs K.A., Conway C.C., Dick D.M., Dretsch M.N., Eaton N.R., et al. Validity and utility of Hierarchical Taxonomy of Psychopathology (HiTOP): II. Externalizing superspectrum. World Psychiatry. 2021;20:171–193. doi: 10.1002/wps.20844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nigg J.T., John O.P., Blaskey L.G., Huang-Pollock C.L., Willcutt E.G., Hinshaw S.P., Pennington B. Big five dimensions and ADHD symptoms: Links between personality traits and clinical symptoms. J Pers Soc Psychol. 2002;83:451–469. doi: 10.1037/0022-3514.83.2.451. [DOI] [PubMed] [Google Scholar]
- 42.McGhee R.L., Ehrler D.J., Buckhalt J.A., Phillips C. The relation between five-factor personality traits and risk-taking behavior in preadolescents. Psychology. 2012;3:558–561. [Google Scholar]
- 43.Barker E.D., Ing A., Biondo F., Jia T., Pingault J.B., Du Rietz E., et al. Do ADHD-impulsivity and BMI have shared polygenic and neural correlates? Mol Psychiatry. 2021;26:1019–1028. doi: 10.1038/s41380-019-0444-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loth A.K., Drabick D.A.G., Leibenluft E., Hulvershorn L.A. Do childhood externalizing disorders predict adult depression? A meta-analysis [published correction appears in J Abnorm Child Psychol 2014; 42:1115–1116] J Abnorm Child Psychol. 2014;42:1103–1113. doi: 10.1007/s10802-014-9867-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pailing A.N., Reniers R.L.E.P. Depressive and socially anxious symptoms, psychosocial maturity, and risk perception: Associations with risk-taking behaviour. PLoS One. 2018;13 doi: 10.1371/journal.pone.0202423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCloughen A., Foster K., Marabong N., Miu D., Fethney J. Physical health risk behaviours in young people with mental illness. Issues Ment Health Nurs. 2015;36:781–790. doi: 10.3109/01612840.2015.1036480. [DOI] [PubMed] [Google Scholar]
- 47.Derks I.P.M., Bolhuis K., Yalcin Z., Gaillard R., Hillegers M.H.J., Larsson H., et al. Testing bidirectional associations between childhood aggression and BMI: Results from three cohorts. Obesity (Silver Spring) 2019;27:822–829. doi: 10.1002/oby.22419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fluharty M.E., Sallis H., Munafò M.R. Investigating possible causal effects of externalizing behaviors on tobacco initiation: A Mendelian randomization analysis. Drug Alcohol Depend. 2018;191:338–342. doi: 10.1016/j.drugalcdep.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aleyasin H., Flanigan M.E., Russo S.J. Neurocircuitry of aggression and aggression seeking behavior: Nose poking into brain circuitry controlling aggression. Curr Opin Neurobiol. 2018;49:184–191. doi: 10.1016/j.conb.2018.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tobiansky D.J., Roma P.G., Hattori T., Will R.G., Nutsch V.L., Dominguez J.M. The medial preoptic area modulates cocaine-induced activity in female rats. Behav Neurosci. 2013;127:293–302. doi: 10.1037/a0031949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phelps E.A., LeDoux J.E. Contributions of the amygdala to emotion processing: From animal models to human behavior. Neuron. 2005;48:175–187. doi: 10.1016/j.neuron.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 52.Davidson R.J., Putnam K.M., Larson C.L. Dysfunction in the neural circuitry of emotion regulation—A possible prelude to violence. Science. 2000;289:591–594. doi: 10.1126/science.289.5479.591. [DOI] [PubMed] [Google Scholar]
- 53.O’Brien J.W., Hill S.Y. Neural predictors of substance use disorders in young adulthood. Psychiatry Res Neuroimaging. 2017;268:22–26. doi: 10.1016/j.pscychresns.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tajima-Pozo K., Ruiz-Manrique G., Yus M., Arrazola J., Montañes-Rada F. Correlation between amygdala volume and impulsivity in adults with attention-deficit hyperactivity disorder. Acta Neuropsychiatr. 2015;27:362–367. doi: 10.1017/neu.2015.34. [DOI] [PubMed] [Google Scholar]
- 55.Van Dessel J., Sonuga-Barke E., Moerkerke M., Van der Oord S., Lemiere J., Morsink S., Danckaerts M. The amygdala in adolescents with attention-deficit/hyperactivity disorder: Structural and functional correlates of delay aversion. World J Biol Psychiatry. 2020;21:673–684. doi: 10.1080/15622975.2019.1585946. [DOI] [PubMed] [Google Scholar]
- 56.Hoogman M., Bralten J., Hibar D.P., Mennes M., Zwiers M.P., Schweren L.S.J., et al. Subcortical brain volume differences in participants with attention deficit hyperactivity disorder in children and adults: A cross-sectional mega-analysis [published correction appears in Lancet Psychiatry 2017; 4:436] Lancet Psychiatry. 2017;4:310–319. doi: 10.1016/S2215-0366(17)30049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu R.K., Zuo Y., Ly T., Wang J., Meera P., Wu Y.E., Hong W. An amygdala-to-hypothalamus circuit for social reward. Nat Neurosci. 2021;24:831–842. doi: 10.1038/s41593-021-00828-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moulton E.A., Elman I., Becerra L.R., Goldstein R.Z., Borsook D. The cerebellum and addiction: Insights gained from neuroimaging research. Addict Biol. 2014;19:317–331. doi: 10.1111/adb.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shaw P., Ishii-Takahashi A., Park M.T., Devenyi G.A., Zibman C., Kasparek S., et al. A multicohort, longitudinal study of cerebellar development in attention deficit hyperactivity disorder. J Child Psychol Psychiatry. 2018;59:1114–1123. doi: 10.1111/jcpp.12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Caggiano V., Leiras R., Goñi-Erro H., Masini D., Bellardita C., Bouvier J., et al. Midbrain circuits that set locomotor speed and gait selection. Nature. 2018;553:455–460. doi: 10.1038/nature25448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Silva B.A., Gross C.T., Gräff J. The neural circuits of innate fear: Detection, integration, action, and memorization. Learn Mem. 2016;23:544–555. doi: 10.1101/lm.042812.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Korte S.M., Jaarsma D., Luiten P.G., Bohus B. Mesencephalic cuneiform nucleus and its ascending and descending projections serve stress-related cardiovascular responses in the rat. J Auton Nerv Syst. 1992;41:157–176. doi: 10.1016/0165-1838(92)90137-6. [DOI] [PubMed] [Google Scholar]
- 63.Goll Y., Atlan G., Citri A. Attention: The claustrum. Trends Neurosci. 2015;38:486–495. doi: 10.1016/j.tins.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 64.Atlan G., Terem A., Peretz-Rivlin N., Sehrawat K., Gonzales B.J., Pozner G., et al. The claustrum supports resilience to distraction. Curr Biol. 2018;28:2752–2762.e7. doi: 10.1016/j.cub.2018.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dickstein S.G., Bannon K., Castellanos F.X., Milham M.P. The neural correlates of attention deficit hyperactivity disorder: An ALE meta-analysis. J Child Psychol Psychiatry. 2006;47:1051–1062. doi: 10.1111/j.1469-7610.2006.01671.x. [DOI] [PubMed] [Google Scholar]
- 66.Cross-Disorder Group of the Psychiatric Genomics Consortium Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell. 2019;179:1469–1482.e11. doi: 10.1016/j.cell.2019.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wray N.R., Ripke S., Mattheisen M., Trzaskowski M., Byrne E.M., Abdellaoui A., et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668–681. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johnson E.C., Demontis D., Thorgeirsson T.E., Walters R.K., Polimanti R., Hatoum A.S., et al. A large-scale genome-wide association study meta-analysis of cannabis use disorder. Lancet Psychiatry. 2020;7:1032–1045. doi: 10.1016/S2215-0366(20)30339-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kranzler H.R., Zhou H., Kember R.L., Vickers Smith R., Justice A.C., Damrauer S., et al. Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations [published correction appears in Nat Commun 2019; 10:2275 and Nat Commun 2019; 10:4050] Nat Commun. 2019;10:1499. doi: 10.1038/s41467-019-09480-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.