SUMMARY
Stem cell research endeavors to generate specific subtypes of classically-defined “cell-types”. Here we generate >90% pure human artery or vein endothelial cells from pluripotent stem cells within 3–4 days. We specified artery cells by inhibiting vein-specifying signals and vice versa. These cells modeled viral infection of human vasculature by Nipah and Hendra viruses, which are extraordinarily deadly (~57–59% fatality rate) and require biosafety-level-4 containment. Generating pure populations of artery and vein cells highlighted that Nipah and Hendra viruses preferentially infected arteries; arteries expressed higher levels of their viral-entry receptor. Virally-infected artery cells fused into syncytia containing up to 23 nuclei, which rapidly died. Despite infecting arteries and occupying ~6–17% of their transcriptome, Nipah and Hendra largely eluded innate-immune detection, minimally eliciting interferon signaling. We thus efficiently generate artery and vein cells, introduce stem-cell-based toolkits for biosafety-level-4 virology, and explore the arterial tropism and cellular effects of Nipah and Hendra viruses.
In Brief:
Pure populations of human artery and vein cells are rapidly generated from pluripotent stem cells. This provides a BSL4-compatible in vitro model to study how the deadly Nipah and Hendra viruses infect blood vessels.
Graphical Abstract
INTRODUCTION
Endothelial cells (ECs)—the innermost constituents of blood vessels—pervade all tissues and have manifold roles in health and disease. In health, they control entry and exit of bloodborne nutrients, wastes and immune cells to and from their client tissues (Augustin and Koh, 2017; Coultas et al., 2005; Potente and Mäkinen, 2017; Rafii et al., 2016). In disease, ECs contribute to atherosclerosis and other afflictions that account for over half of human deaths (Augustin and Koh, 2017; Carmeliet, 2005), including viral diseases of the vasculature.
Many Risk Group-4 viruses cause deadly vascular diseases, are among the most lethal human pathogens known, and require biosafety-level-4 (BSL4) containment (CDC and NIH, 2020). Here we focus on Nipah and Hendra viruses, which respectively cause outbreaks throughout Asia (~59% fatality rate) and Australia (~57% fatality rate), and are Risk Group-4 paramyxoviruses in the genus Henipavirus (Ang et al., 2018; Chua et al., 2000; Mahalingam et al., 2012; Murray et al., 1995). Individual Nipah virus outbreaks epidemiologically vary due to viral variants and other factors: the 1999 Malaysian and 2018 Indian outbreaks respectively exhibited fatality rates of ~40% and ~91% (Ang et al., 2018; Arunkumar et al., 2019). Of exigent concern, no treatments or vaccines for either Nipah or Hendra are presently approved for human use, and these diseases are thus classified Priority Diseases by the World Health Organization (Gomez Roman et al., 2021; Mehand et al., 2018). Nipah and Hendra viruses engender severe vascular disease (multiorgan vasculitis), which can culminate in respiratory and neurological failure (Baseler et al., 2015; Geisbert et al., 2010; Murray et al., 1995; Wong et al., 2002). To understand the enigmatic vascular diseases caused by Risk Group-4 viruses, we must precisely map their target cells and effects on such cells. This pursuit has been complicated by unique constraints of BSL4 experimentation and inadequate experimental systems.
A challenge in studying ECs is that they encompass multiple subtypes—including artery, vein, capillary and lymphatic ECs—which are molecularly distinct and subserve different physiological functions (De Val and Black, 2009; Fish and Wythe, 2015; Potente and Mäkinen, 2017). ECs are well known to be targets of Nipah and Hendra in vitro (Aljofan et al., 2009; Erbar et al., 2008; Lo et al., 2010; Mathieu et al., 2012; Monaghan et al., 2014; Schountz et al., 2019) and in vivo (Baseler et al., 2015; Geisbert et al., 2010; Murray et al., 1995; Wong et al., 2002). However, whether different EC subtypes harbor distinct functions in viral diseases remains enigmatic. Artery ECs express higher levels of EFNB2 (Chong et al., 2011; Wang et al., 1998)—an entry receptor for Nipah and Hendra viruses (Bonaparte et al., 2005; Negrete et al., 2005)—leading to the hypothesis that these viruses might preferentially infect artery ECs. Nevertheless, both artery and vein ECs can reportedly be infected by Nipah and Hendra in vitro (Aljofan et al., 2009; Lo et al., 2010; Mathieu et al., 2012; Monaghan et al., 2014). Yet, an important proviso is that current systems to culture ECs do not preserve arteriovenous identity: primary ECs originally isolated from arteries or veins rapidly relinquish their arteriovenous subtype identity in vitro (Aranguren et al., 2013). This technical limitation has obscured viral tropism for, and their effects on, specific EC subtypes.
Generating well-defined populations of human artery vs. vein ECs en masse would empower us to explore viral infection of the vasculature, and in particular which EC subtype(s) are infected. Differentiation of human pluripotent stem cells (hPSCs, including embryonic and induced pluripotent stem cells) into specific cell-types (e.g., ECs) is rapidly advancing, yet generating pure populations of desired cellular subtypes (e.g., artery and vein ECs) remains quite challenging (Fowler et al., 2019). Starting from pluripotent cells, we must map the branching lineage choices through which they differentiate into cellular subtypes and the extracellular signals that control each step in their journey. Extant protocols often differentiate hPSCs towards mesoderm using BMP, FGF, TGFβ and/or WNT, and then bias differentiation towards ECs using VEGF and other signals. However, these signals are also permissive for the formation of other mesodermal lineages; consequently, a heterogeneous population containing a subset of ECs usually arises (e.g., Halaidych et al., 2018; Kurian et al., 2012; Lian et al., 2014; McCracken et al., 2019; Nguyen et al., 2016; Olmer et al., 2018; Paik et al., 2018; Patsch et al., 2015; Zhang et al., 2017). hPSC-derived ECs sometimes lack (reviewed by Lin et al., 2017) or coexpress (Halaidych et al., 2018; Olmer et al., 2018) arterial and venous markers. On the other hand, overexpression of pan-endothelial transcription factor (TF) ETV2 efficiently differentiates hPSCs into ECs, but they are also apparently bereft of arterial or venous identity (Ng et al., 2020; Wang et al., 2020).
Inspired by our developmental understanding of how VEGF and NOTCH specify artery vs. vein subtypes of ECs in vivo (Coultas et al., 2005; De Val and Black, 2009; Fang and Hirschi, 2019; Fish and Wythe, 2015; Lin et al., 2007; Potente and Mäkinen, 2017), recent protocols manipulated these two signals to bias hPSC-derived ECs towards artery or vein identity with partial efficiency in vitro (Ditadi et al., 2015; Gage et al., 2020; Park et al., 2018; Rosa et al., 2019; Sriram et al., 2015; Zhang et al., 2017), which constituted an important step forward. However, the challenges in generating artery and vein ECs in vitro indicate gaps in our understanding of how these subtypes developmentally arise and intimate that additional signals for arteriovenous specification may exist.
Here we chart a developmental roadmap for the stepwise diversification of hPSCs into primitive streak, lateral mesoderm, and subsequently into artery and vein ECs. At each step of differentiation, we reveal the extracellular signals that need to be activated, and those that must be simultaneously inhibited to prevent emergence of undesired cell-types. We created >90% pure populations of either human artery or vein ECs within 3–4 days of hPSC differentiation. We then applied these well-defined artery and vein EC populations to BSL4 virology: Nipah and Hendra viruses preferentially targeted hPSC-derived artery ECs. More broadly, hPSC-derived cell-types will provide a powerful platform to explore the biology of, and countermeasures against, Risk Group-4 viruses under BSL4 containment.
RESULTS
Efficient differentiation of hPSCs into primitive streak and lateral mesoderm by repressing paraxial mesoderm and endoderm
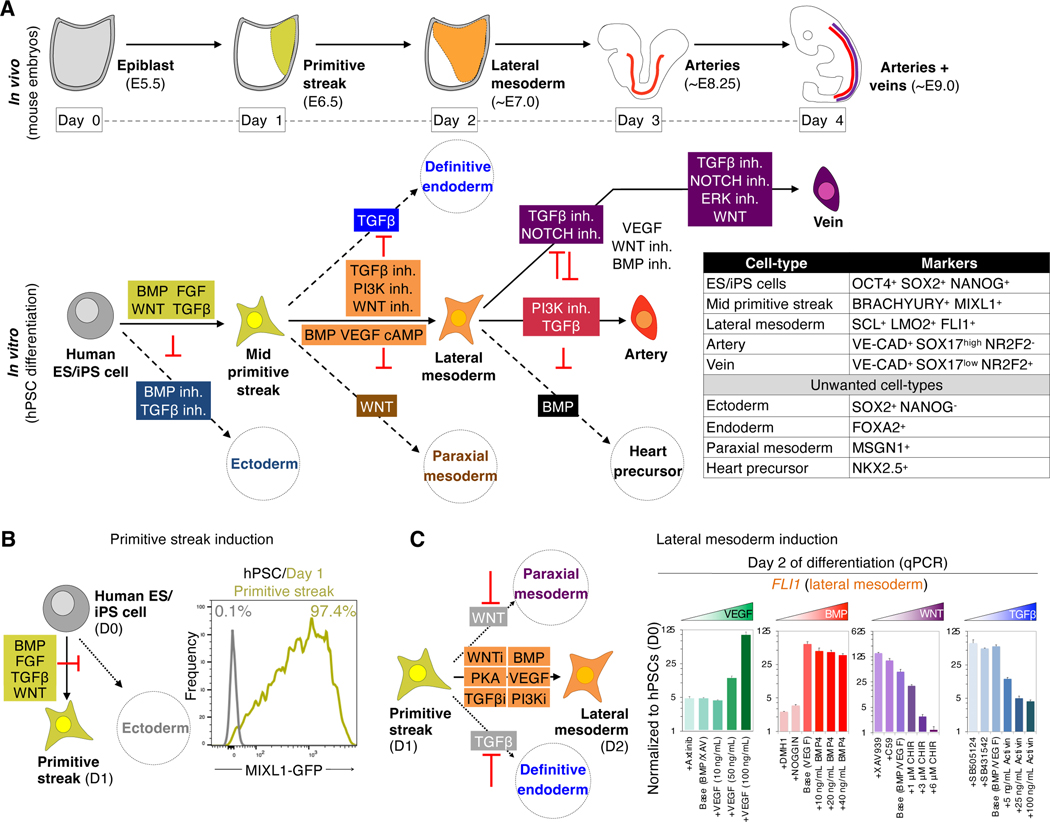
Starting from hPSCs in vitro, we sought to emulate the stepwise in vivo development of pluripotent cells (corresponding to day 5.5 of mouse embryonic development [~E5.5]) into primitive streak (~E6.5), lateral mesoderm (~E7–7.5), and finally artery (~E8.25) and vein ECs (~E9.0) (Fig. 1a) (Chong et al., 2011). First, activating BMP, FGF, TGFβ and WNT for 24 hours differentiated hPSCs into 97.3±0.3% pure MIXL1-GFP+ mid primitive streak while suppressing ectoderm formation (Fig. 1b) (Loh et al., 2016).
Figure 1: Efficient generation of human primitive streak and lateral mesoderm from hPSCs within 1 and 2 days, respectively.
A) Artery and vein development
B) Generating MIXL1+ mid primitive streak within 24 hours of hPSC differentiation (Loh et al., 2016), assayed by flow cytometry of MIXL1-GFP hPSCs (Davis et al., 2008)
C) VEGF and BMP specify, whereas TGFβ and WNT repress, day 2 lateral mesoderm. Day 1 hPSC-derived primitive streak was treated with the indicated signals (concentrations indicated, and 10 nM Axitinib; 200 ng/mL NOGGIN; 1 μM DMH1, XAV939 and C59; 2 μM SB505124 and SB431542; base conditions contained 10 ng/mL BMP4 and/or 100 ng/mL VEGF) for 24 hours; qPCR was performed on day 2.
Second, we further differentiated hPSC-derived day 1 mid primitive streak into day 2 lateral mesoderm, which contains endothelial progenitors in vivo (Ciau-Uitz et al., 2013). We induced lateral mesoderm markers SCL, LMO2 and FLI1 by 1) activating BMP, VEGF and PKA, while 2) simultaneously inhibiting WNT, TGFβ and PI3K to repress formation of unwanted paraxial mesoderm and endoderm (Loh et al., 2014; Loh et al., 2016) (Fig. 1c, Fig. S1a–e). We thus generate lateral mesoderm prior to generating ECs.
Efficient differentiation of hPSC-derived lateral mesoderm into artery ECs by repressing vein and heart fates
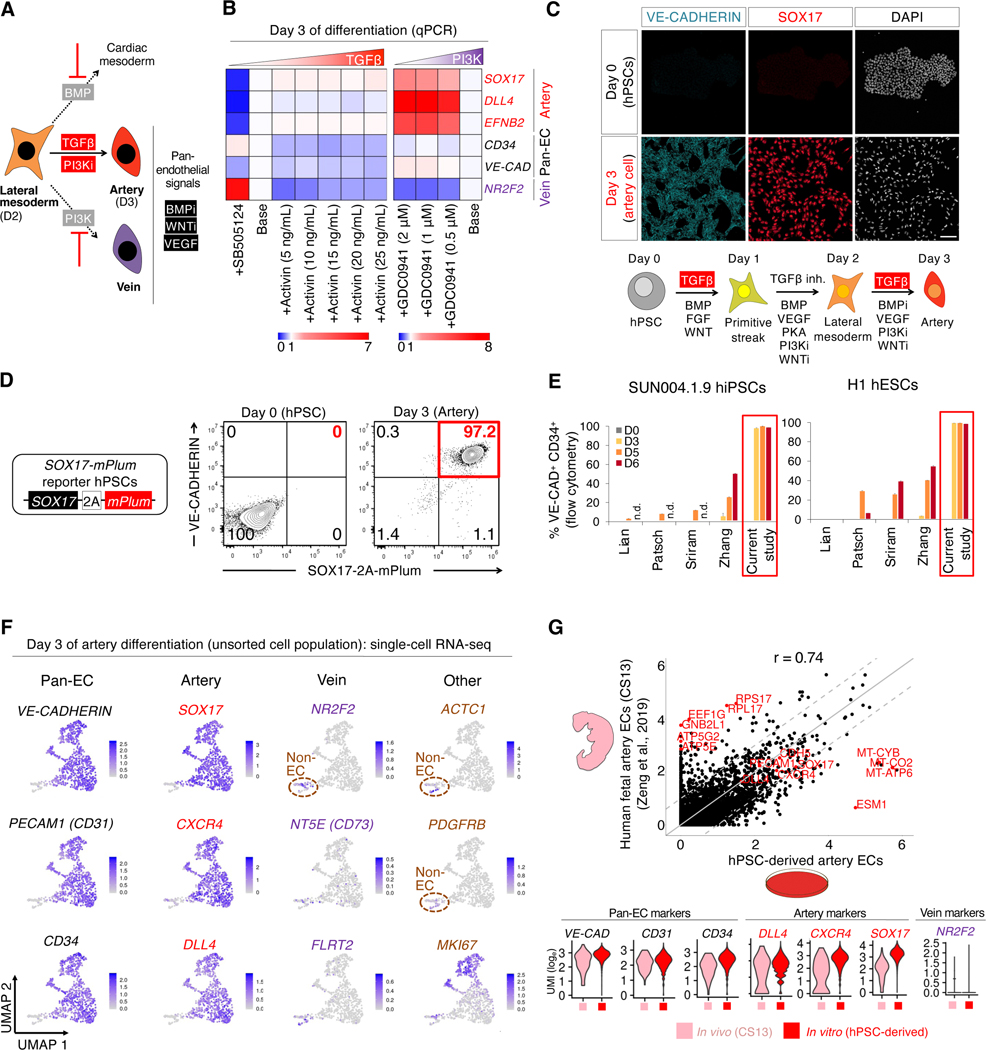
Subsequently, we identified the signals that controlled the further segregation of hPSC-derived day 2 lateral mesoderm into artery, vein or heart lineages (Fig. 2a). To differentiate lateral mesoderm towards ECs, we activated VEGF, while simultaneously inhibiting BMP and WNT, which respectively induced NKX2.5+ heart precursors (Loh et al., 2016) and CDX2+ non-ECs (Fig. S2a–c). Having inhibited formation of non-ECs, next we screened for signals that induce arterial marker SOX17 (Corada et al., 2013; Sakamoto et al., 2007) using SOX17-mCherry knock-in reporter hPSCs (Loh et al., 2014).
Figure 2: Efficient generation of human artery endothelial cells from hPSCs within 3 days.
A) Summary of present work
B) TGFβ promotes, while PI3K inhibits, day 3 artery formation. Day 2 hPSC-derived lateral mesoderm was treated with the indicated signals, including TGFβ agonist (Activin, 5–25 ng/mL), TGFβ inhibitor (SB505124, 2 μM) or PI3K inhibitor (GDC0941, 0.5–2 μM), for 24 hours; qPCR was performed on day 2 (base media: GDC0941+VEGF+XAV939+AA2P+TTNPB [left] and VEGF+XAV939+AA2P+TTNPB+Activin [right]). Gene expression was normalized to the base media lacking TGFβ or PI3K modulators.
C) Immunostaining of H1 hPSCs either before or after differentiation into day 3 artery ECs; scale = 100 μm
D) Flow cytometry of SOX17–2A-mPlum hPSCs before or after differentiation into day 3 artery ECs
E) H1 and SUN004.1.9 hPSC lines differentiated using the present artery EC induction method or 4 prevailing EC differentiation protocols (Lian et al., 2014; Patsch et al., 2015; Sriram et al., 2015; Zhang et al., 2017); flow cytometry was performed on days 0, 3, 5 and 6 of differentiation; n.d. = not determined
F) scRNAseq of H1 hPSCs differentiated towards artery ECs; the entire day 3 cell population was harvested without pre-selecting ECs; gene expression in loge UMI units
G) scRNAseq comparison of hPSC-derived day 3 artery ECs vs. Carnegie Stage 13 (CS13) human fetal artery ECs (Zeng et al., 2019); 6000 variable genes (top, each dot = 1 gene) or selected genes (bottom) shown
Additional signals (TGFβ and PI3K) controlled the bifurcation of day-2 lateral mesoderm into artery vs. vein fates: TGFβ specified arteries, whereas PI3K conversely induced veins (Fig. 2b, Fig. S2bii–iv). Consequently, we efficiently generated artery ECs by 1) inducing arterial fate through TGFβ activation, while 2) repressing vein formation by simultaneously inhibiting PI3K (Ditadi et al., 2015; Hong et al., 2006; Park et al., 2018). While previous protocols to putatively generate hPSC-derived artery ECs inhibited TGFβ (Zhang et al., 2017), we found that TGFβ inhibition in fact blocked artery specification and instead induced venous marker NR2F2 (Fig. 2b, Fig. S2bii). This thus disclosed a temporally-dynamic role for TGFβ signaling in specifying arterial fate, changing every 24 hours across 3 days of hPSC differentiation. TGFβ had to be initially activated (inducing primitive streak, day 1), then inhibited (specifying lateral mesoderm, day 2) and later re-activated (to induce arterial fate, day 3) (Fig. 2c).
Taken together, activation of TGFβ (to specify arteries) and VEGF (to specify ECs) together with simultaneous blockade of BMP, WNT and PI3K for 24 hours differentiated day-2 lateral mesoderm into day-3 artery ECs, while inhibiting unwanted differentiation into either heart or vein lineages (Fig. S2d). This delineates a complex signaling code for arterial specification beyond activation of VEGF and NOTCH (reviewed by Coultas et al., 2005; Fang and Hirschi, 2019; Fish and Wythe, 2015). We generated SOX17+ CD144+ artery ECs within 3 days of hPSC differentiation (Fig. 2c), which were >90% pure, as assayed using newly-engineered SOX17–2A-mPlum reporter hPSCs (Fig. 2d) and SOX17-mCherry hPSCs (Fig. S2e). Artery differentiation was reproducible across 4 hESC and hiPSC lines, generating 97.9±2.4% pure CD144+ DLL4+ arterial ECs within 3 days, and was more efficient and rapid than 4 published differentiation protocols (Lian et al., 2014; Patsch et al., 2015; Sriram et al., 2015; Zhang et al., 2017) (Fig. 2e, Fig. S2f,g).
Identity and uniformity of hPSC-derived artery cells, and similarities with in vivo artery cells
scRNAseq reaffirmed the efficiency of artery differentiation. After 3 days of differentiation, 91.9% of the population formed a predominant artery EC cluster that expressed artery-specific (SOX17, CXCR4 and DLL4) and pan-EC (VE-CADHERIN, CD31 and CD34) markers (Fig. 2f), which was confirmed by qPCR (Fig. S2h,i). Nevertheless, individual artery ECs varied in their cell-cycle status, as shown by MKI67 expression (Fig. 2f). ECs were artery-specific: they did not express venous marker NR2F2 (Fig. 2f), consistent with our inhibition of vein-specifying signals (e.g., PI3K). This thus contrasts with past hPSC-derived ECs that co-expressed both arterial and venous markers (Olmer et al., 2018; Paik et al., 2018). However, 8.1% of cells were non-ECs that expressed mesenchymal markers (Fig. 2f). Taken together, activation of artery-inducing signals with simultaneous inhibition of vein-specifying signals efficiently generated hPSC-derived artery ECs, with scarce mesenchyme-like cells.
Attesting to their lineage fidelity, hPSC-derived artery ECs showed overall transcriptional similarity (R=0.74) with in vivo artery ECs within 4-week-old human fetuses (Zeng et al., 2019), and expressed comparable levels of pan-EC and arterial markers (Fig. 2g). This was promising, given that in vivo vs. in vitro cells were not precisely matched in their genetic background or developmental stage. Genes that were differentially expressed by artery ECs in vivo and in vitro largely related to metabolism and protein translation (Fig. S2j, Table S1), and as such, may reflect metabolic adaptation to cell culture. Additionally, ESM1—a VEGF-induced gene (Shin et al., 2008)—was higher in vitro (Fig. S2j), perhaps reflecting VEGF levels in the media. Overall, hPSC-derived artery ECs are transcriptionally similar, but are not identical, to fetal artery ECs.
Efficient differentiation of hPSC-derived lateral mesoderm into veins by repressing artery and heart fates
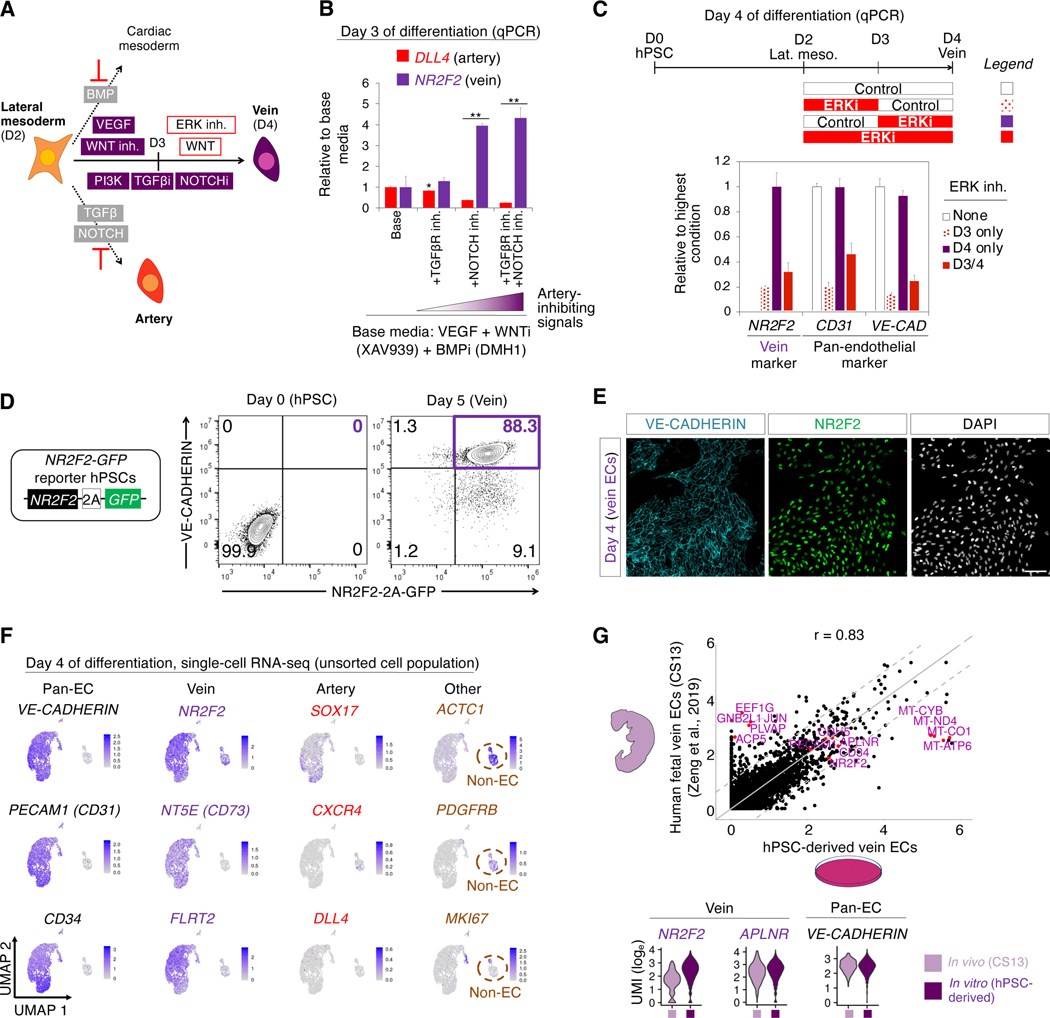
Having defined artery-specifying signals, next we asked what signals confer vein identity (Fig. 3a). Starting from day 2 hPSC-derived lateral mesoderm, we dually inhibited the artery-specifying signals TGFβ and NOTCH to block arterial differentiation (Fig. 3b). While NOTCH inhibition promotes venous fate (Ditadi et al., 2015; Duarte et al., 2004; Gage et al., 2020; Gale et al., 2004; Krebs et al., 2004; Lawson et al., 2001; Park et al., 2018), we found that NOTCH inhibition was not sufficient to efficiently induce vein ECs in vitro. We thus explored additional signals for vein specification.
Figure 3: Efficient generation of human vein endothelial cells from hPSCs within 4 days.
A) Summary of present work
B) Dually inhibiting TGFβ and NOTCH promotes day 3 vein gene expression. Day 2 hPSC-derived lateral mesoderm was further differentiated for 24 hours with the indicated signals, including TGFβ inhibitor (SB505124, 2 μM) and/or NOTCH inhibitor (RO4929097, 1 μM); qPCR performed on day 3; *P<0.05; **P<0.01
C) VEGF/ERK activation followed by inhibition is critical for vein formation. Day 2 hPSC-derived lateral mesoderm was further differentiated for 48 hours in the presence or absence of ERK inhibitor (PD0325901, 100 nM) for the indicated durations; qPCR performed on day 4 (base media: VEGF+SB505124+RO4929097+XAV939+DMH1+AA2P for days 3–4)
D) Flow cytometry of H1 NR2F2–2A-GFP hPSCs before or after differentiation into day 5 vein ECs
E) NR2F2 and VE-CADHERIN immunostaining of H1 hPSC-derived day 4 vein ECs; scale = 100 μm
F) scRNAseq of H1 hPSCs differentiated towards vein ECs; the entire day 4 cell population was harvested without pre-selecting ECs; gene expression in loge UMI units
G) scRNAseq comparison of hPSC-derived day 4 vein ECs vs. Carnegie Stage 13 (CS13) human fetal vein ECs (Zeng et al., 2019); 6000 variable genes (top, each dot = 1 gene) or selected genes (bottom) shown
We discovered that VEGF/ERK signaling had a temporally-dynamic role in vein EC specification in vitro: on day 3, it had to be activated to generate ECs, whereas on day 4, it had to be inhibited to generate vein ECs (Fig. 3c). ERK blockade on day 3 prevented EC specification (Fig. 3c). Yet, after cells acquired EC identity by day 3, subsequently inhibiting ERK on day 4 did not impair EC identity but rather was critical to instruct vein fate (Fig. 3c). Prolonged activation or inhibition of VEGF for 48 hours did not yield vein ECs, illustrating the importance of temporal VEGF/ERK modulation in vitro (Fig. 3c). Modulating VEGF/ERK signaling with such dynamism is also crucial for vein specification in vivo (Casie Chetty et al., 2017; Lawson et al., 2002; Shalaby et al., 1995), but was not fully appreciated by earlier vein differentiation protocols (Ditadi et al., 2015; Gage et al., 2020; Park et al., 2018; Rosa et al., 2019; Sriram et al., 2015; Zhang et al., 2017). Taken together, manipulating VEGF/ERK and WNT in a temporally-dynamic way, along with inhibiting artery-specifying signals, differentiated hPSCs into vein ECs (Fig. 3a, Fig. S3a,b).
Through this approach, we could efficiently generate >88% pure NR2F2+ CD144+ vein ECs, as shown using newly-engineered NR2F2–2A-GFP knock-in reporter hPSCs (Martin et al., 2019) (Fig. 3d, Fig. S3c,d) and wild-type hPSCs (Fig. 3e, Fig. S3e). scRNAseq reaffirmed the efficiency of vein differentiation: 89.3% of cells formed a vein EC cluster that expressed vein (NR2F2, FLRT2 and APLNR) and pan-EC (VE-CADHERIN, CD31 and CD34) markers (Fig. 3f). ECs were vein-specific: they minimally expressed arterial markers (Fig. 3f), consistent with our inhibition of artery-specifying signals (e.g., NOTCH, TGFβ and VEGF/ERK). There was a small fraction (10.7%) of contaminating, mesenchyme-like non-ECs (Fig. 3f). Taken together, we efficiently generated hPSC-derived vein ECs, with a minority of mesenchyme-like cells. hPSC-derived vein ECs showed high overall transcriptional similarity with in vivo artery ECs within the 4-week-old CS13 human fetus (Zeng et al., 2019) (R = 0.83); both expressed venous markers (Fig. 3g, Fig. S3f). Genes that were differentially expressed by vein ECs in vivo and in vitro largely related to metabolism and protein translation and trafficking (Fig. S3g,h, Table S2). Taken together, hPSC-derived vein ECs are transcriptionally similar to fetal vein ECs, although they are not identical.
In summary, we generate enriched populations of either artery (Fig. 2) or vein ECs (Fig. 3) within 3 or 4 days of hPSC differentiation, respectively. We find that artery ECs arise earlier than vein ECs from hPSCs, mirroring how artery ECs (~E8.25) precede vein ECs (~E9.0) in mouse embryos (Chong et al., 2011).
Transcriptional and functional differences between hPSC-derived artery and vein cells
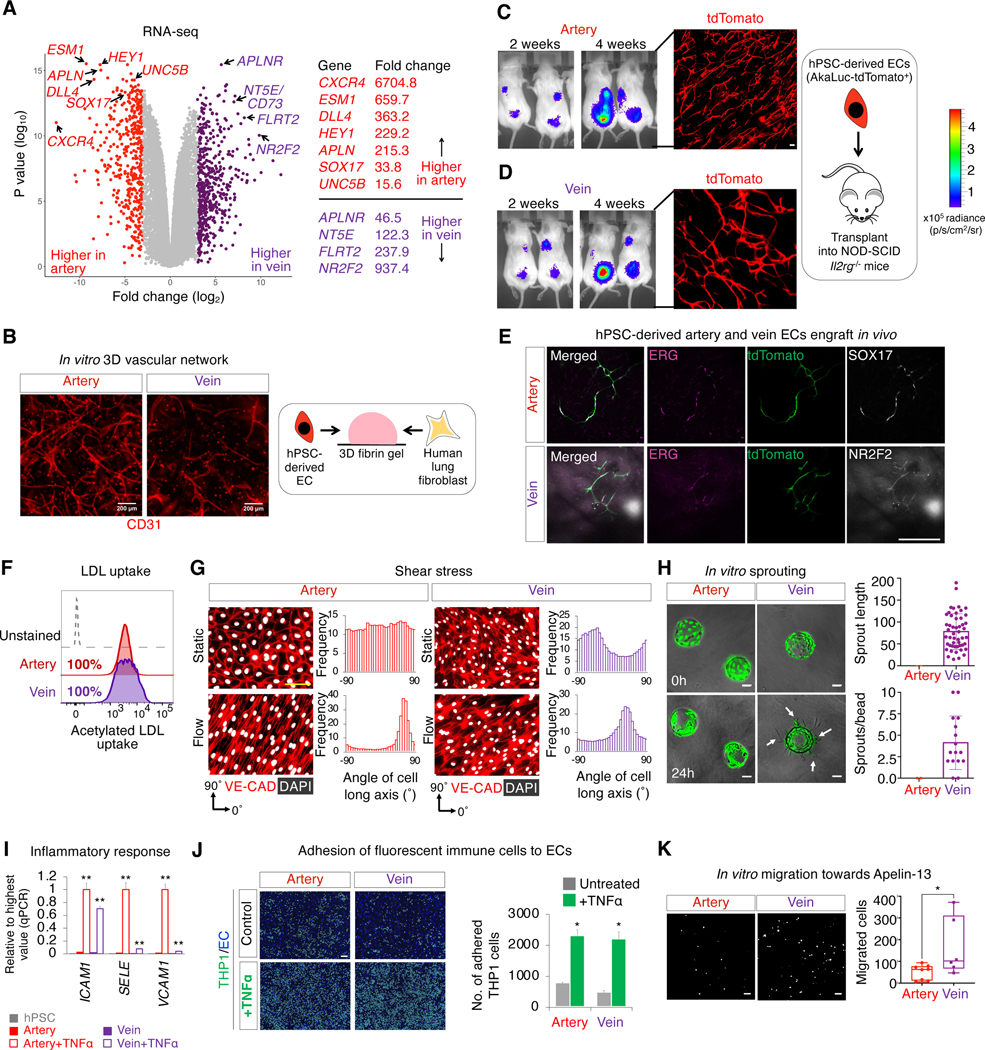
There were stark transcriptome-wide differences between hPSC-derived artery and vein ECs, including disparate expression of known arteriovenous subtype markers (Fig. 4a, Fig. S4a, Table S3). Certain EC attraction and repulsion signals showed arterial- or venous-biased expression, suggesting potential bidirectional signaling between these two EC subtypes to coordinate their spatial positions. First, hPSC-derived artery ECs expressed the chemokine APELIN, whereas vein ECs expressed its cognate receptor (APLNR; Fig. 4a, Fig. S4a), which is investigated below. In vivo, artery-secreted APELIN chemoattracts APLNR+ veins, thus positioning arteries and veins in parallel (Kidoya et al., 2015). Second, hPSC-derived vein ECs expressed FLRT2, a repulsive ligand for the arterially-expressed netrin receptor UNC5B (Fig. 4a, Fig. S4a) (Lu et al., 2004; Tai-Nagara et al., 2017), potentially contributing to why arteries and veins do not intermix. However, hPSC-derived artery and vein ECs did not express markers of organ-specific ECs (Kalucka et al., 2020) (Fig. S4b): hence, they may correspond to the earliest ECs arising during development prior to their entry into various organs (Chong et al., 2011). An interactive browser to explore scRNAseq profiles of Hpsc-derived artery and vein ECs is available at https://anglab.shinyapps.io/arteryveinscrna/.
Figure 4: Transcriptional and functional differences between hPSC-derived artery and vein endothelial cells.
A) Bulk-population RNA-seq of H1 hPSC-derived day 3 artery vs. day 4 vein ECs (CD144+ FACS sorted), each dot = 1 gene
B) 3D vascular networks formed by H1 hPSC-derived artery and vein ECs cocultured with fibroblasts in fibrin gel for 1 week (Kurokawa et al., 2017); scale = 200 μm
C-E) Subcutaneous transplantation of CAG-AkaLuciferase-tdTomato+ SUN003.1 hPSC-derived artery and vein ECs, monitored by intravital imaging (to assess cell proliferation) and microscopy (4 weeks post-transplant); scale = 150 μm
F) Flow cytometry of fluorescently-labelled, acetylated LDL uptake by H1 hPSC-derived artery and vein ECs within 4 hours
G) SUN004.1.9 hPSC-derived artery and vein ECs exposed to shear stress or static conditions for 24 hours align to the direction of fluid flow, quantified by the angle of the long axis of CD144+ cell membranes; scale = 100 μm
H) Beads coated with CAG-GFP+ SUN004.1.9 hPSC-derived vein ECs preferentially form vascular sprouts after embedding in 3D fibrin gel for 1 day, as opposed to artery EC-coated beads; scale = 100 μm
I) H1 hPSC-derived artery and vein ECs were expanded for 5 days, and then treated with TNFα (10 ng/mL) or left untreated for 4 hours, followed by qPCR to assess TNFα-induced upregulation of immune cell adhesion molecules; **P<0.01
J) H1 hPSC-derived artery and vein ECs were expanded for 6 days, and then were treated with TNFα (10 ng/mL) or left untreated for 4 hours, prior to addition of fluorescent THP1 cells for 30 minutes; scale = 100 μm; *P<0.05
K) In Transwell assay, CAG-GFP+ SUN004.1.9 hPSC-derived vein ECs preferentially migrate towards Apelin-13; images of migrated cells situated on the bottom membrane surface; scale = 100 μm; *P<0.05
Importantly, both hPSC-derived artery and vein ECs executed hallmark EC functions. First, both EC subtypes formed 3-dimensional vascular networks in vitro when cocultured with mesenchymal cells within fibrin hydrogels for 1 week (Fig. 4b, Fig. S4c,d) (Kurokawa et al., 2017). Second, artery and vein ECs likewise formed vascular networks in vivo upon subcutaneous transplantation into adult immunodeficient NOD-SCID Il2rg−/− mice for 1 month (Fig. 4c–e). Third, artery and vein ECs also incorporated LDL and acetylated LDL (Fig. 4f, Fig. S4e) and sealed a deliberate gap in monolayer cultures (Fig. S4f).
Nevertheless hPSC-derived artery and vein ECs functionally differed in various properties: 1) responses to shear stress, 2) sprouting ability, 3) responses to inflammation, and 4) migration towards chemokines, revealing differences between these two EC subtypes. First, upon 24 hours of exposure to fluid flow, both artery and vein ECs aligned themselves to the flow axis and upregulated flow-induced TF KLF2 (Fig. 4g, Fig. S4g). However, shear stress preferentially upregulated AQP1 and CX37 in artery ECs (Fig. 4g, Fig. S4g), evincing different transcriptional responses to shear stress by these two EC subtypes. Second, vein (not artery) ECs coated on beads formed vascular sprouts that protruded into fibrin gels within 24 hours (Fig. 4h). This is consistent with the notion that vein ECs preferentially sprout in vivo (Red-Horse and Siekmann, 2019; Xu et al., 2014). Third, ECs respond to inflammation by upregulating adhesion molecules to recruit immune cells (e.g., Halaidych et al., 2018; Patsch et al., 2015; Zhang et al., 2017). Upon treatment with inflammatory cytokine TNFα for 4 hours, both artery and vein ECs upregulated the immune-cell adhesion molecule ICAM1 and physically adhered to monocyte-like cells (Fig. 4i,j). Nevertheless, artery and vein ECs partially differed in their responses to inflammation: immune-cell adhesion molecules VCAM1 and ESELECTIN were specifically upregulated in artery ECs (Fig. 4i, Fig. S4h). Fourth, we found that artery and vein ECs migrated towards different chemokines. Vein ECs preferentially migrated across a porous membrane towards the endothelial chemokine APELIN (Fig. 4k). This is consistent with how vein ECs express higher levels of APLNR (Fig. 4a, Fig. S4a), which is critical for vein migration in vivo (Kidoya et al., 2015). Collectively, hPSC-derived artery and vein ECs exhibited important transcriptional and functional differences, reifying that they have acquired distinct subtype identities.
Nipah and Hendra viruses preferentially induce multinucleated syncytia from artery cells
Next we applied hPSC-derived artery and vein ECs to study the tropism of Nipah and Hendra viruses, which must be handled under BSL4 containment (CDC and NIH, 2020). Thus hPSC-derived artery and vein ECs were cryopreserved, and then thawed and briefly cultured and expanded prior to viral infection under BSL4 containment. To expand vein ECs, it was critical to continuously inhibit the artery-specifying signals TGFβ and NOTCH (Fig. S5a,b). In these conditions, artery and vein ECs could be cultured for at least 6 days, while retaining their respective subtype identities and the ability to activate interferon signaling (Fig. S5a–d).
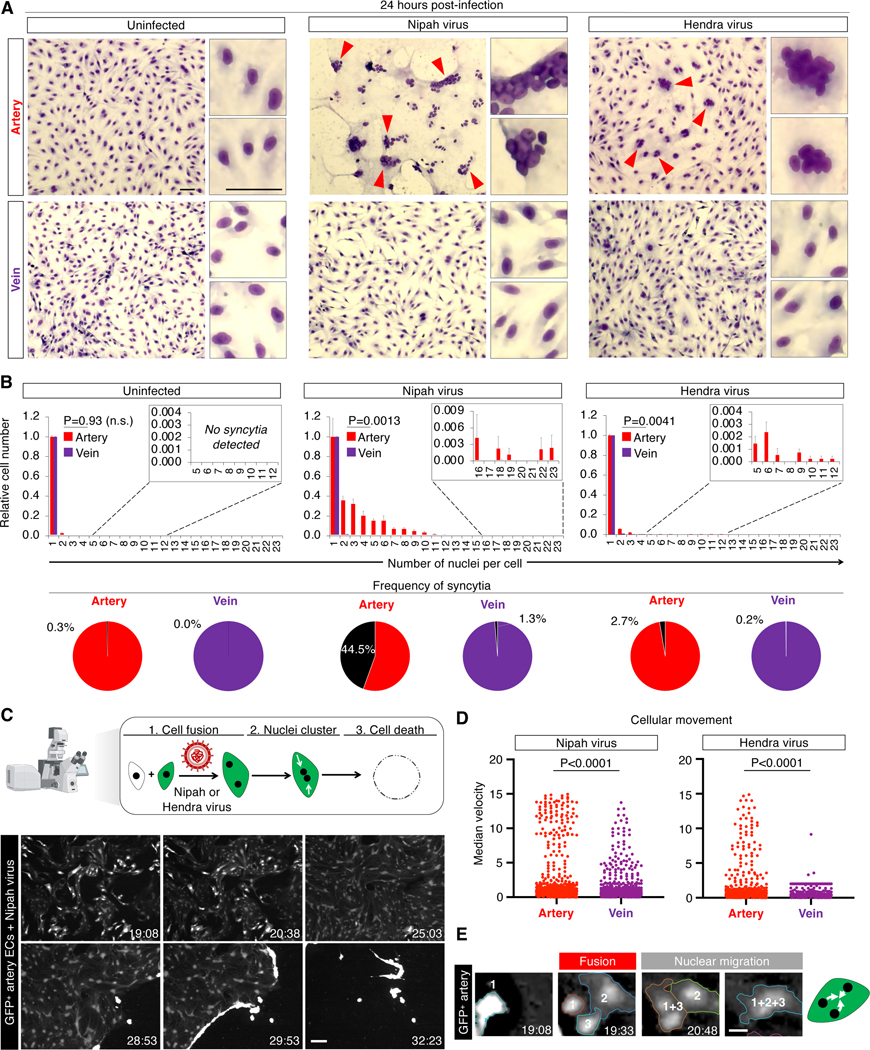
Upon infecting artery ECs, Nipah and Hendra viruses induced them to fuse into multinucleated syncytia, including prodigious syncytia harboring up to 23 nuclei per cell (Fig. 5a). Within 24 hours, Nipah virus extensively induced arterial syncytia (accounting for 44.5% of ECs), whereas vein ECs were minimally fused (1.3% of ECs; P=0.0013, Fig. 5b). Likewise, Hendra virus preferentially induced syncytia among artery ECs (P=0.0041, Fig. 5b). Nipah virus reduced artery EC numbers by 71.0% within 24 hours of infection; vein ECs were significantly less affected (P=0.0002, Fig. S5e).
Figure 5: Nipah and Hendra viruses target artery cells.
A) Syncytia (arrows) in H1 hPSC-derived artery and vein ECs inoculated with Nipah or Hendra viruses for 24 hours; scale = 25 μm
B) Numbers of nuclei per cell after inoculation with Nipah or Hendra viruses for 24 hours (top); syncytia frequency (≥3 nuclei/cell; bottom)
C) Live imaging of cocultured SUN004.1.9 GFP+ and H1 wild-type artery ECs infected with Nipah virus, tracking cell fusion into GFP+ syncytia, followed by death (scale = 200 μm, timestamp: hours:minutes)
D) Live imaging showed Nipah- and Hendra-infected artery ECs displayed significantly greater median velocity (a measure of cell fusion) than vein ECs
E) Fusion and GFP diffusion between 3 Nipah-infected artery ECs, followed by aggregation of their nuclei (scale bar = 20 μm, timestamp: hours:minutes)
By implementing live imaging in BSL4 containment, we confirmed that Nipah and Hendra viruses preferentially induced syncytia formation in artery ECs. Cocultures of GFP+ and wild-type hPSC-derived ECs were infected and cell fusion was continuously quantified by live imaging (Fig. S5f,g; Movie S1). Nipah-infected artery ECs were progressively consumed into several massive syncytia over tens of hours and eventually eliminated (Fig. 5c). Again, Nipah and Hendra viruses preferentially induced cell fusion in artery ECs, as opposed to vein ECs (Fig. 5d). Cell fusion was rapid, occurring in minutes (Fig. 5d). Strikingly, after virally-induced cell fusion, nuclei subsequently aggregated together within syncytia, forming large clusters of nuclei (Fig. 5e, Movie S1) akin to a “bunch of grapes” (Fig. 5a). This contrasts with healthy syncytia (e.g., skeletal muscle), wherein nuclei are randomly distributed across the cell (Metzger et al., 2012). Taken together, Nipah and Hendra viruses preferentially induce artery ECs to fuse together, as opposed to vein ECs.
Nipah and Hendra viruses preferentially target artery cells in an EFNB2-dependent fashion
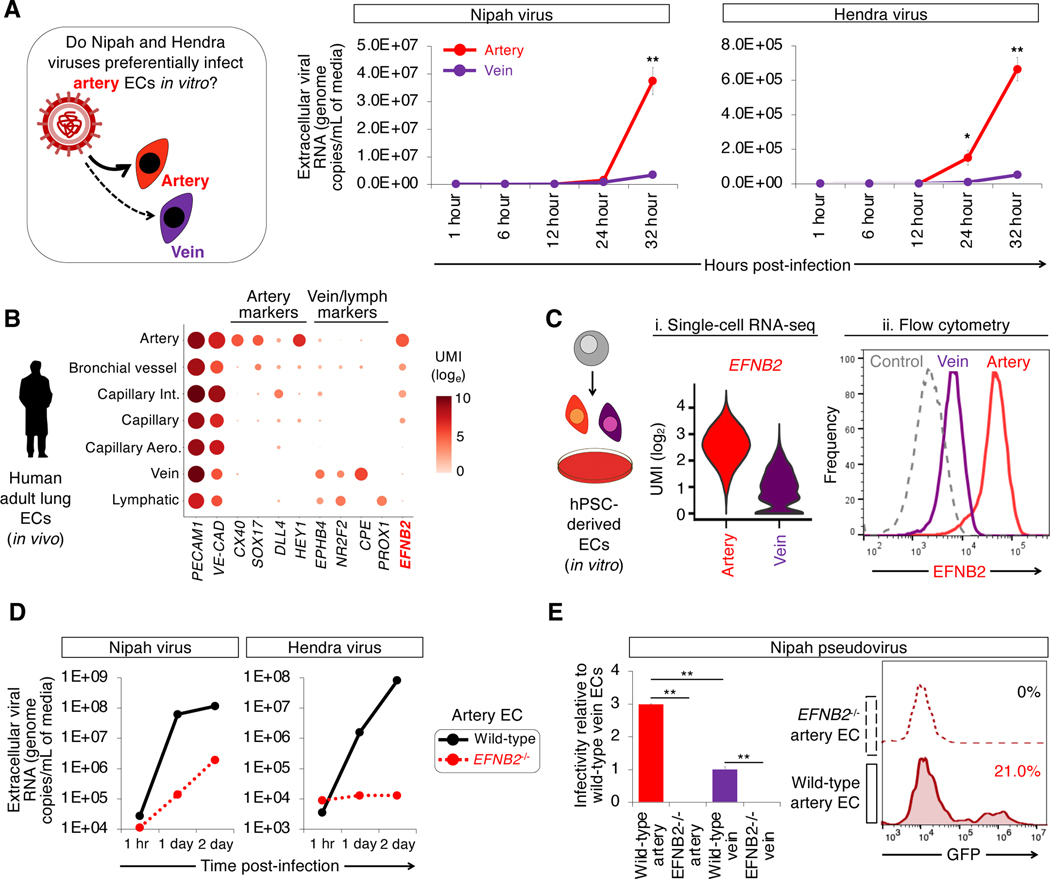
Consistent with how Nipah virus preferentially induced syncytia among artery ECs, Nipah-infected artery ECs produced 11.2-fold higher levels of Nipah viral RNA relative to vein ECs, as assayed by viral RNA levels in the culture media (P=0.01, Fig. 6a). Likewise, artery ECs produced 17.4-fold higher levels of Hendra viral RNA compared to vein ECs, (P=0.01, Fig. 6a). Intracellular viral RNA levels similarly disclosed an arterial tropism of these viruses, with the proviso that at later timepoints, Nipah RNA levels in artery ECs declined due to destruction of the cells (Fig. S6a).
Figure 6: Artery cells express higher levels of EFNB2, the Nipah and Hendra virus receptor.
A) Nipah and Hendra viral RNA replication in H1 hPSC-derived artery and vein ECs, as quantified by qPCR of culture media; *P<0.05; **P<0.01
B) scRNAseq of human adult lung ECs (Travaglini et al., 2020)
C) Higher EFNB2 levels in hPSC-derived artery ECs relative to vein ECs, as shown by i) scRNAseq of day 3 artery vs. day 4 vein ECs and ii) flow cytometry for surface EFNB2 protein expression on artery vs. vein ECs, 4 days post-expansion
D) Nipah or Hendra viral RNA replication in H1 and EFNB2−/− hPSC-derived artery and vein ECs, as assayed by qPCR of culture media
E) H1 and EFNB2−/− hPSC-derived artery and vein ECs inoculated with GFP-encoding Nipah pseudovirus (Palomares et al., 2013); **P<0.01
Why do Nipah and Hendra viruses preferentially infect artery ECs, as opposed to vein ECs? EFNB2—a known receptor for these viruses (Bonaparte et al., 2005; Negrete et al., 2005)—is expressed more highly by artery ECs in vivo, in both mice (Chong et al., 2011; Wang et al., 1998) and humans (Fig. 6b) (Travaglini et al., 2020). hPSC-derived ECs faithfully reproduced this arteriovenous difference in EFNB2 expression. hPSC-derived artery ECs expressed 8.8- and 10.4-fold higher levels of EFNB2 surface protein and mRNA, respectively, compared to vein ECs (Fig. 6c, Fig. S6b,c). By contrast, primary human artery and vein ECs expanded in conventional media relinquished arteriovenous differences (Aranguren et al., 2013): they expressed similar EFNB2 levels (Fig. S6d). In sum, hPSC-derived cell-types afford advantages (e.g., preserved subtype identity) relative to previous cellular models used in BSL4 virology.
Preferential Nipah virus entry into artery ECs was principally driven by EFNB2. Genetically deleting EFNB2 using Cas9 (Fig. S6e) (Martin et al., 2019) led to a substantial 430.3- to 6289.2-fold reduction of Nipah and Hendra virus replication in EFNB2−/− artery ECs, respectively (Fig. 6d). To precisely assay viral entry, we employed Nipah pseudoviruses, which enter cells but do not replicate or inflict virus-specific cellular derangements (Palomares et al., 2013). Indeed, Nipah pseudoviruses entered artery ECs 3.2±0.2 times more efficiently than vein ECs (Fig. 6e, Fig. S6f). Ablating EFNB2 completely abrogated Nipah pseudovirus entry into artery ECs, but did not impair entry of an unrelated vesicular stomatitis virus (VSV)-based pseudovirus (Fig. 6e, Fig. S6g). Therefore EFNB2 is necessary for Nipah and Hendra virus entry, complementing the longstanding findings that EFNB2 is sufficient for entry of these viruses (Bonaparte et al., 2005; Negrete et al., 2005).
In sum, the developmental bifurcation between artery vs. vein ECs is important for viral infection, as it culminates in differential expression of viral-entry receptors (i.e., EFNB2) between these two EC subtypes and thus differing susceptibility to Nipah and Hendra infection. Beyond EFNB2, artery and vein ECs also differ in the expression of other genes, which may contribute to preferential viral entry and replication efficiencies in artery ECs.
Nipah and Hendra viruses minimally alter gene expression in artery cells, largely eluding the antiviral program
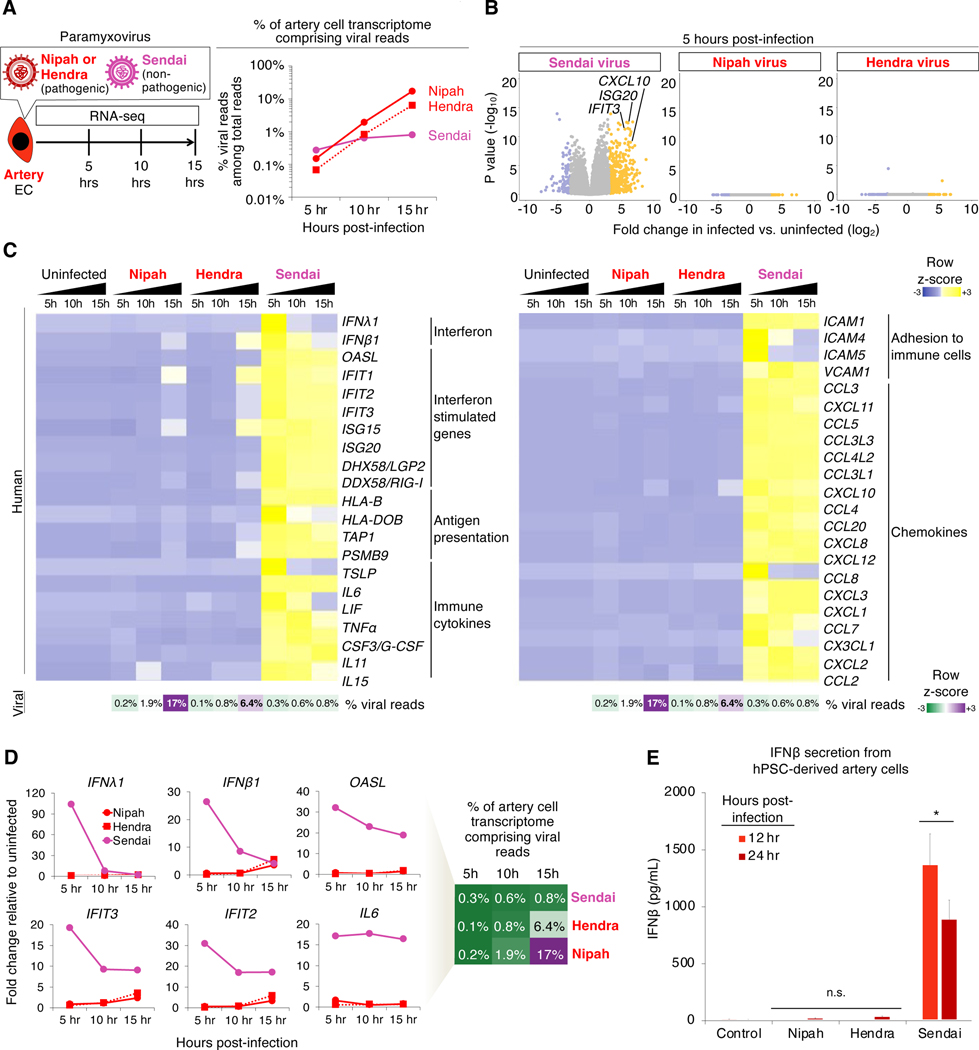
Finally, we used RNA-seq to discover how artery ECs transcriptionally react to Nipah virus, Hendra virus or a related non-pathogenic paramyxovirus, Sendai virus (Cantell strain) (López et al., 2006). Cells were infected with a high viral titer (multiplicity of infection [MOI] ≈ 5), and RNA-seq was performed at 5, 10 or 15 hours post-infection (Fig. 7a, Table S4). All three paramyxoviruses exhibited the expected 3’-to-5’ gene transcriptional gradient along their genomes (Fig. S7a,b), authenticating our RNA-seq analysis.
Figure 7: Nipah and Hendra viruses elicit minimal transcriptional changes in artery endothelial cells, despite extensive viral replication and cell fusion.
A) Percentage of hPSC-derived artery EC transcriptome comprising viral reads, as quantified by RNA-seq; averaged from H1 and SUN004.1.9 hPSCs
B) Transcriptome-wide differences between infected vs. uninfected artery ECs by RNA-seq; averaged from H1 and SUN004.1.9 hPSCs
C,D) RNA-seq of infected hPSC-derived artery ECs, including viral read percentage in transcriptome; averaged from H1 and SUN004.1.9 hPSCs
E) IFNβ secretion by hPSC-derived artery ECs; averaged from H1, SUN004.1.9 and SUN004.2 hPSCs; *P<0.05; n.s. = not significant
Between 5 and 15 hours post-infection, Nipah and Hendra virus RNA replicated extensively in artery ECs (114.5-fold and 93.2-fold increase, respectively), relative to Sendai virus, which achieved limited (2.9-fold) RNA replication (Fig. 7a). After 15 hours, Nipah occupied 17.2% of the transcriptome (~1 in 6 mRNAs encoding Nipah genes) and Hendra occupied 6.4% of the transcriptome (Fig. 7a). Therefore, Nipah and Hendra viruses replicate significantly more efficiently than the non-pathogenic Sendai virus in artery ECs.
Despite immense viral replication and cell fusion, Nipah and Hendra viruses led to surprisingly minimal host gene expression changes: they were remarkably effective at suppressing interferon production. Nipah virus massively replicated in artery ECs, yet it did not upregulate or downregulate a single gene across the entire transcriptome to a statistically significant extent, at 5–10 hours post-infection (Fig. 7b, Fig. S7c). Similar results were observed for Hendra virus (Fig. 7b, Fig. S7c). Even when Nipah and Hendra virus RNA achieved extremely high levels at later timepoints (6.4–17.2% of the transcriptome, at 15 hours post-infection), artery ECs minimally upregulated IFNβ1, IFNλ1 and interferon target genes IFIT2 and IFIT3 (all less than 6-fold upregulation; Fig. 7c,d). Despite high viral loads, Nipah- or Hendra-infected artery ECs did not secrete the antiviral cytokine IFNβ to a statistically-significant degree at 12 or 24 hours post-infection (Fig. 7e). This starkly differed from Sendai virus, which induced a 100.8-fold elevation in IFNβ secretion within 24 hours (Fig. 7e). Despite low viral loads (<0.8% of the transcriptome), Sendai potently induced antiviral genes, including type I/III interferons (López et al., 2006) and interferon response genes that restrict viral replication (Shaw et al., 2017) (Fig. 7c,d). Additionally, Sendai-infected artery ECs elaborated immune cytokines, chemokines and immune-cell adhesion molecules known to recruit immune cells; these antiviral and inflammatory genes were minimally induced by Nipah or Hendra infection (Fig. 7c,d). Normalizing to viral loads, Nipah and Hendra viruses induced IFNλ1 to a much lower extent (69.5-fold and 45.1-fold, respectively) than Sendai virus (Fig. 7d). Likewise, Nipah and Hendra viruses upregulated interferon target gene IFIT2 much less effectively (109.2-fold and 23.2-fold, respectively) than Sendai virus (Fig. 7d).
DISCUSSION
To precisely induce desired cellular subtypes, here we chart a stepwise roadmap for hPSC differentiation into artery vs. vein subtypes of ECs. We mapped the temporally-dynamic extracellular signals that drive or inhibit each step of differentiation. At the bifurcation between artery vs. vein subtypes, we precisely induced artery ECs by repressing vein-specifying signals and vice versa. This approach generated >90% pure populations of artery- vs. vein-specific ECs within 3–4 days of hPSC differentiation. The ability to efficiently and rapidly generate artery and vein ECs “on demand” should avail regenerative medicine, tissue engineering and modelling of human vascular diseases. Importantly, these hPSC-derived artery and vein ECs were distinguished by clear transcriptional and functional differences. Here we emphasize a pivotal distinction between artery and vein ECs: Nipah and Hendra viruses preferentially target artery ECs in vitro. In turn, this subtype-specific viral tropism emphasizes the importance of producing specific cellular subtypes in vitro and provides a vista on the target cells of deadly viruses.
A roadmap for human artery vs. vein development
First, we differentiated hPSCs into MIXL1+ primitive streak and then SCL+ HAND1+ lateral mesoderm within 1 and 2 days, respectively. Subsequently, hPSC-derived lateral mesoderm bifurcated into >90% pure SOX17+ artery ECs (day 3) or, alternatively, >90% pure NR2F2+ vein ECs (day 4). Thus, artery ECs arise prior to vein ECs in vitro, consistent with in vivo findings (Chong et al., 2011)—these two cellular subtypes do not arise synchronously.
Inducing cellular subtypes: blocking the formation of alternative subtypes
As opposed to generating “generic” cell-types of ambiguous subtype identity, we hypothesized it should be possible to produce desired cellular subtypes by explicitly inhibiting the signals that induce the opposing cellular subtype. At the bifurcation between artery and vein subtypes, TGFβ and NOTCH specify artery fate, whereas PI3K instructs vein identity. We induced artery ECs through a two-pronged approach: activating artery-inducing signal TGFβ, while repressing vein-specifying signal PI3K. The converse signals—TGFβ and NOTCH inhibition (together with PI3K activation)—instead induced vein ECs. This signaling logic enabled the exclusive generation of either artery or vein ECs in the absence of the opposing lineage, as quantified by scRNAseq and other means. Our findings thus reveal a more complex signaling code for artery vs. vein specification, extending beyond the current manipulation of VEGF and/or NOTCH (reviewed by Coultas et al., 2005; Fang and Hirschi, 2019; Fish and Wythe, 2015) to bias hPSCs towards arterial or venous fates (Ditadi et al., 2015; Gage et al., 2020; Park et al., 2018; Rosa et al., 2019; Sriram et al., 2015; Zhang et al., 2017). Explicitly inhibiting the signals that specify alternate subtype identities may be a general approach to precisely engineer desired cellular subtypes from stem cells.
Inducing cellular subtypes: temporally-dynamic signaling
hPSCs swiftly traverse alternating lineage paths every 24 hours and consequently, extracellular signals must be rapidly activated or inhibited to precisely guide differentiation down desired paths in preference to unwanted ones. Indeed, in the mouse embryo, cells rapidly segue between different states every 12–24 hours: E5.5 pluripotent cells → E6.5 primitive streak → ~E7-E7.5 lateral mesoderm → ~E8.25 artery ECs and ~E9.0 vein ECs (Chong et al., 2011). We found the same extracellular signal specified 3 different cell-types over the course of 3 days. For instance, TGFβ initially induced mid primitive streak on day 1; then repressed lateral mesoderm on day 2; and finally induced arterial ECs on day 3. By contrast, VEGF induced lateral mesoderm on day 2, and then induced and eventually inhibited vein ECs on days 3 and 4, respectively. Temporally-dynamic activation and repression of extracellular signals was required to efficiently transition cells from one state to the next, with two consecutive cell-types sometimes sharing diametrically opposed requirements for the same signal (e.g., VEGF) in vitro, consistent with VEGF’s temporally-dynamic role in vivo (Casie Chetty et al., 2017; Lawson et al., 2002; Shalaby et al., 1995). Consequently, prolonged activation or inhibition of a given signal generates a heterogeneous cell population. The previously-overlooked requirement for temporally-dynamic TGFβ and VEGF signals likely complicated past efforts to generate artery and vein ECs from hPSCs. It is therefore critical to map the temporal dynamics with which these signals operate, and to manipulate them with equal dynamism to precisely effect differentiation.
Artery ECs are target cells of Nipah and Hendra viruses
By virtue of their high fatality rates and a general dearth of approved treatments, Risk Group-4 viruses imperil global health: mapping their human target cells and their effects on such cells is crucial. Arteries and veins have largely been regarded synonymously in viral diseases. However, we found Nipah and Hendra viruses preferentially targeted artery ECs and fused them into multinucleated arterial ECs in vitro. This is consonant with a past study of Syrian hamsters, which suggested that Nipah primarily infected arteries in vivo, as morphologically inferred by elastic membrane staining (Baseler et al., 2015). Hence, as opposed to promiscuously targeting all ECs, we demonstrate that certain viruses may preferentially target specific endothelial subtypes. Mechanistically, this arterial tropism was driven at least in part by higher expression of viral-entry receptor EFNB2 (Bonaparte et al., 2005; Negrete et al., 2005) in artery ECs, which we demonstrated was required for viral entry, as shown by genetic deletion. The developmental diversification of artery vs. vein ECs may thus hold hitherto-unappreciated significance in viral diseases.
Past studies infected various ECs sourced from a range of species and organs with Nipah and Hendra viruses in vitro (Aljofan et al., 2009; Erbar et al., 2008; Lo et al., 2010; Mathieu et al., 2012; Monaghan et al., 2014; Schountz et al., 2019). However, these ECs were cultured in conditions wherein arteriovenous subtype identity was lost (Aranguren et al., 2013), often unbeknownst to researchers. This thus accentuates the importance of being able to generate and maintain well-defined populations of hPSC-derived artery and vein ECs in vitro to study whether viruses preferentially target specific EC subtypes and exert subtype-specific effects.
Having identified artery ECs as preferential target cells of Nipah and Hendra viruses, we systematically detailed the effects of these viruses on artery ECs using live-imaging and RNA-seq. One of the greatest surprises was that Nipah and Hendra viruses inflicted surprisingly minor transcriptional effects on artery ECs, despite extensive viral replication and prodigious cell fusion. Indeed, both viruses were “stealthy” to the innate immune system of artery ECs. Despite occupying an impressive fraction (6.4–17.2%) of an artery cell’s transcriptome, both Nipah and Hendra viruses minimally induced interferon signaling or antiviral genes. This minimal interferon response correlated with the ability of these lethal viruses to massively replicate and achieve high viral loads, in stark contrast with the non-pathogenic Sendai virus. Indeed, Nipah virus P, V and W proteins are known to silence interferon production and signaling (Childs et al., 2007; Ciancanelli et al., 2009; Rodriguez et al., 2002). Past studies reached diverging conclusions regarding whether Nipah virus activates (Elvert et al., 2020; Lo et al., 2010; Mathieu et al., 2012), or fails to elicit (Schountz et al., 2019; Virtue et al., 2011), interferon signaling in vitro. Here, we re-investigate this question using well-defined populations of artery and vein ECs. Additionally, we quantify the extent of interferon suppression relative to respective viral RNA loads, using RNA-seq. Our work also illuminates the cellular effects of Hendra virus, which hitherto were generally understudied compared to, and assumed to be similar to, Nipah virus.
A platform to study Risk Group 4 viruses using hPSC-derived cell-types
More broadly, we introduce hPSC-derived cell-types as enhanced model systems to study the target cells and cellular effects of Risk Group-4 viruses. Due to BSL4 experimental constraints, in vitro studies of Risk Group-4 viruses have hitherto exploited cancer cells (which poorly reflect native physiology) or primary cells (which are difficult to expand long-term and to genetically edit, and often relinquish features of their in vivo counterparts), neither of which may harbor clear subtype identities (e.g., arteriovenous identity). hPSC-derived cell-types can be rapidly generated in large numbers, are genetically manipulatable, and retain certain properties of their in vivo counterparts (e.g., subtype identity) (Drubin and Hyman, 2017). These decisive advantages of hPSC-derived cell-types have already been exploited to provide new insights into Risk Group 2 and 3 viruses (Harschnitz and Studer, 2021; Simoneau and Ott, 2020). Indeed, hPSC-derived ECs exemplify many of these advantages: they exhibit arteriovenous differences in EFNB2 expression (which are otherwise obscured in primary ECs cultured in conventional conditions) and are genetically tractable (as evinced by our genetic deletion of EFNB2 to confirm its role in viral entry).
Beyond Nipah and Hendra viruses, hPSC-derived artery and vein ECs could provide a powerful platform to delineate the target cells and cellular effects of additional Risk Group-4 viruses—including arenaviruses, filoviruses, flaviviruses and nairoviruses—that cause lethal and enigmatic vascular diseases. Eventually hPSC-derived cell-types could enable therapeutic and genetic screens to discover countermeasures against currently-incurable Risk Group-4 viruses, using cell-types that are more physiologically relevant than the cancer cells often used in BSL4 experiments.
Limitations of the study
The endothelial roadmap described here remains incomplete: it focuses on artery and vein ECs, but does not encompass generation of capillaries, lymphatics or organ-specific ECs (Augustin and Koh, 2017; Nguyen et al., 2021; Potente and Mäkinen, 2017). Creating an increasingly diverse array of ECs in vitro will, in turn, avail virology. Nipah and Hendra viruses infect ECs across multiple organs in vivo (Baseler et al., 2015; Geisbert et al., 2010; Murray et al., 1995; Wong et al., 2002), but how organ-specific ECs respond to these viruses remains a mystery. Finally, while we find Nipah and Hendra viruses preferentially target human artery ECs in vitro, in vivo verification is of paramount import. In this regard, it is critical to appreciate that cells in vivo and in vitro are not equivalent (Fowler et al., 2019): hPSC-derived artery and vein ECs are transcriptionally similar, but are not identical to, their respective in vivo counterparts. Going forward, we envision that hPSC-derived cell-types will provide a powerful in vitro platform to explore the tropism and biology of Risk Group-4 viruses, generating hypotheses that can then be tested in de rigueur animal models in vivo.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Requests for further information should be directed to and will be fulfilled by the Lead Contact, Kyle M. Loh (kyleloh@stanford.edu).
Materials Availability
Multiple human pluripotent stem cell lines were developed as part of this study: H1 NR2F2–2A-GFP hESCs, H1 SOX17–2A-mPlum hESCs, H1 EFNB2−/− hESCs, SUN004.1.9 CAG-eGFP hiPSCs, SUN004.2 CAG-mScarlet hiPSCs, and SUN003.1 CAG-AkaLuciferase-tdTomato hiPSCs. All cell lines will be made freely available upon request and the completion of applicable Material Transfer Agreements.
Data and Code Availability
Bulk-population RNA-seq and single-cell RNA-seq datasets generated as part of this study are available at the NCBI Sequence Read Archive, accession number PRJNA837932. Single-cell RNAseq datasets of hPSC-derived artery and vein ECs that were generated as part of this study can be interactively browsed at a custom web portal: https://anglab.shinyapps.io/arteryveinscrna/. Computational scripts used for genomics analyses conducted as part of this study are available at Github: https://github.com/LayTengAngLab/ArteryVeinEndothelial.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture
All cells in this study were cultured in standard incubator conditions (20% O2, 5% CO2 and 37 °C).
Human pluripotent stem cell lines
The following human pluripotent stem cell (hPSC) lines were used in this study: wild-type H1, H7 and H9 hESCs (WiCell); H9 SOX17-mCherry knock-in reporter hESCs (Loh et al., 2014); H1 NR2F2–2A-GFP knock-in reporter hESCs (generated in this study); H1 SOX17–2A-mPlum knock-in reporter hESCs (generated in this study); SUN004.1.9 hiPSCs carrying a constitutively-expressed CAG-GFP PiggyBac transgene (generated in this study); SUN004.2 hiPSCs carrying a constitutively-expressed CAG-mScarlet lentiviral transgene (generated in this study); and SUN003.1 hiPSCs carrying a constitutively-expressed CAG-AkaLuciferase-tdTomato lentiviral transgene (generated in this study). SUN004.1.9, SUN004.2 and SUN003.1 hiPSCs represent three clonal hiPSC lines originally obtained from two different individuals (SUN003 and SUN004). H1, SUN004.1.9, SUN004.2 and SUN003.1 hPSCs are of a XY genotype, whereas H7 and H9 hPSCs are of a XX genotype.
Undifferentiated hPSCs were propagated in mTeSR1 medium (StemCell Technologies) + 1% penicillin/streptomycin (Thermo Fisher) or alternatively, mTeSR Plus medium (StemCell Technologies) + 1% penicillin/streptomycin in monolayer cultures, on Geltrex basement membrane matrix-coated plates (described below). For mTeSR1, media was changed daily and for mTeSR Plus, media was changed every other day as per the manufacturer’s recommendations. For the sake of brevity, we refer to mTeSR1 and mTeSR Plus interchangeably as “mTeSR” for the remainder of these Methods. In order to maintain cultures of undifferentiated hPSCs, when they became partially confluent, undifferentiated hPSCs were passaged by treating them for 7 minutes with EDTA (Versene, Thermo Fisher) at room temperature. Subsequently, EDTA was removed, mTeSR was added, and then hPSCs were manually scraped off the plate to generate clumps. hPSC clumps were then seeded onto new plates that had been precoated with Geltrex basement membrane matrix (described below) in mTeSR medium + 1% penicillin/streptomycin. To reiterate, during EDTA-based maintenance passaging of undifferentiated hPSCs as clumps, ROCK inhibitor was not added.
Human pluripotent stem cell-derived endothelial cells
After differentiation of hPSCs into artery cells (within 3 days, as described below) or vein cells (within 4 days, as described below), the resultant hPSC-derived artery and vein cells could be expanded for up to 6 additional days. Day 3 hPSC-derived artery cells were expanded in EGM2 (Endothelial Cell Growth Medium 2, Lonza CC-3162), which was refreshed every 24 hours. Day 4 hPSC-derived vein cells were expanded in EGM2 + SB505124 (2 μM) + RO4929097 (2 μM), which was refreshed every 24 hours. Expansion of hPSC-derived artery and vein cells was performed in monolayer cultures on Geltrex-coated cell culture plates. Expanded hPSC-derived artery and vein cells were used for certain functional assays as well as viral infection experiments. The biological sex of hPSC-derived artery and vein cells corresponds to the biological sex of the parental hPSC line used to generate them.
Human umbilical artery and vein endothelial cells
Human umbilical artery endothelial cells (HUAEC, Sigma-Aldrich, 202–05N) and human umbilical vein endothelial cells (HUVEC, Sigma-Aldrich, 200–05N) were both propagated on Geltrex-coated plates in EGM2 (Lonza CC-3162) media, which was refreshed every 1–2 days. The biological sex of these primary human endothelial cells was not determined.
Human lung fibroblast cells
Primary human adult lung fibroblasts (normal human lung fibroblasts [NHLF], Lonza, CC-2512) were propagated in Fibroblast Growth Medium-2 (Lonza, CC-3132), with media changes every 2–3 days. Once confluent, fibroblasts were washed twice with DPBS without ions (Thermo Fisher, 14190144) and exposed to 0.05% Trypsin-EDTA (Thermo Fisher, 25300062) for 2 min. Trypsin was neutralized using fresh FGM2, and the cell solution was centrifuged at 300g in order to pellet cells for additional culture or experiments. Human fibroblasts were cocultured with endothelial cells to generate 3-dimensional vascular networks (described below). For all relevant experiments, fibroblasts between passages 3–7 were used. Where indicated, NHLFs obtained from different donors were used in experiments. The biological sex of these primary human lung fibroblasts was not determined.
THP1 cells
THP1 cells (ATCC, TIB-202) were propagated in RPMI-1640 + 10% fetal bovine serum (FBS) in suspension cultures, and were passaged every 2–3 days by adding fresh media and diluting their concentration to 3×105 cells/mL. THP1 cells were used to assess the ability of immune cells to adhere to endothelial cells (described below). THP1 cells represent a transformed human monocyte-like cell line of a XY genotype.
HEK293T/17 cells
HEK293T/17 cells (ATCC, CRL-11268) were propagated in DMEM + 10% fetal bovine serum (FBS) and were passaged every 3–4 days at a 1:10 ratio through trypsinization. HEK293 cells were used to package pseudotyped lentivirus particles (described below). HEK293T/17 cells represent a transformed human cell line that is an easily-transfected derivative of the parental HEK293T line, and are of a XX genotype.
Vero E6 cells
Vero C1008 (Vero 76, clone E6, Vero E6) cells (European Collection of Authenticated Cell Cultures (ECACC), 85020206) were propagated in DMEM containing 2% FBS, 2 mM glutamine, and 1% penicillin/streptomycin and passaged at a 1:10 ratio through trypsinization. Vero E6 cells were used to propagate Nipah and Hendra viruses in order to generate viral stocks and to quantitatively titrate infectious virus from the supernatant of virally-infected cell cultures (described below). Vero E6 cells represent a transformed African green monkey (Chlorocebus sabaeus) cell line of a XX genotype.
Mouse models
Immunodeficient NOD-SCID Il2rg−/− (NSG) mice, obtained from The Jackson Laboratory, were housed in the Lokey Stem Cell Research Building’s Barrier Mouse Facility. Adult NSG mice were transplanted with hPSC-derived artery and vein ECs (described below). NSG mice were 2–4 months old and both male and female mice were used.
Nipah virus, Malaysia variant
Nipah virus, Malaysia variant (Nipah virus/human/MY/1999/CDC; genomic sequence reported in NCBI accession number NC_002728) was originally isolated by the CDC from the cerebrum of an infected human in Malaysia in 1999, and was passaged on Vero E6 cells (Harcourt et al., 2000). We previously demonstrated the ability of Nipah virus, Malaysia variant to cause fatal disease in African green monkeys (Prescott et al., 2015) and Syrian hamsters (DeBuysscher et al., 2013), thus verifying its pathogenicity. Nipah virus is a member of the species Nipah henipavirus; genus Henipavirus; family Paramyxoviridae; order Mononegavirales. Nipah virus, Malaysia variant was obtained from CDC Viral Special Pathogens Branch reference stocks.
Hendra virus, Brisbane variant
Hendra virus, Brisbane variant (Hendra virus/horse/AU/1994/Hendra; genomic sequence reported in NCBI accession number NC_001906) was originally isolated from an infected horse in Hendra, a suburb of Brisbane, Australia, in 1994 (Murray et al., 1995). Hendra virus is a member of the species Hendra henipavirus; genus Henipavirus; family Paramyxoviridae; order Mononegavirales. Hendra virus, Brisbane variant was obtained from CDC Viral Special Pathogens Branch reference stocks.
Sendai virus, Cantell strain
The original provenance of Sendai virus, Cantell strain is unknown, but it was passaged over 100 times in chicken embryonated eggs at the Central Public Health Laboratory in Helsinki, Finland (Cantell and Hirvonen, 1981). Sendai virus, Cantell strain is known to induce interferon-β signaling in vivo and in vitro: it does not cause severe disease in mice, nor is it presently known to cause human disease (López et al., 2006; Yoshida et al., 2018). Sendai virus is a member of the species Murine respirovirus; genus Respirovirus; family Paramyxoviridae; order Mononegavirales. Sendai virus, Cantell strain was obtained from ATCC (VR-907).
METHOD DETAILS
Data visualization
Standard plots were prepared with Microsoft Excel, Microsoft PowerPoint or GraphPad Prism. Heatmaps were constructed in Heatmapper (Babicki et al., 2016) or, alternatively, the HeatMapViewer function of the online GenePattern platform (Reich et al., 2006). Flow cytometry data were visualized with FlowJo. Other types of data visualization were prepared as described in the respective sections below.
Basement membrane matrices
hPSCs were maintained and differentiated on cell culture plates that been pre-coated with either Geltrex or vitronectin basement membrane matrices. Geltrex (Thermo Fisher) was diluted 1:100–1:200 in DMEM/F12 (Thermo Fisher) and was used to coat tissue culture plastics for at least 1 hour at 37 °C. Recombinant human truncated vitronectin (Gibco, A14700; “VTN-N”) was diluted to a 10 μg/mL stock in PBS (lacking Ca2+ or Mg2+) and was used to coat tissue culture plastics for at least 1 hour at 37 °C. After coating with either Geltrex or vitronectin, the basement membrane solution was aspirated, leaving behind a thin film; subsequently, cells were plated on the Geltrex- or vitronectin-coated cell culture plastics.
Preparing CDM2 basal media
hPSCs were differentiated into artery and vein cells as described below in Chemically Defined Medium 2 (CDM2). The composition of CDM2 has been described previously (Loh et al., 2014; Loh et al., 2016): 50% IMDM + GlutaMAX (Thermo Fisher, 31980–097) + 50% F12 + GlutaMAX (Thermo Fisher, 31765–092) + 1 mg/mL polyvinyl alcohol (Sigma, P8136–250G) + 1% v/v chemically defined lipid concentrate (Thermo Fisher, 11905–031) + 450 μM 1-thioglycerol (Sigma, M6145–100ML) + 0.7 μg/mL recombinant human insulin (Sigma, 11376497001) + 15 μg/mL human transferrin (Sigma, 10652202001) + 1% v/v penicillin/streptomycin (Thermo Fisher, 15070–063). Polyvinyl alcohol was brought into suspension by gentle warming and magnetic stirring, and the media was sterilely filtered (through a 0.22 μm filter) prior to use.
hPSC differentiation into artery and vein cells
Seeding hPSCs for differentiation (Step 0). In contrast to passaging hPSCs for maintenance, a different passaging procedure was used to plate hPSCs for differentiation; notably, sparse seeding of hPSCs as single cells is paramount for efficient differentiation. To seed hPSCs for differentiation, undifferentiated hPSCs were dissociated into single cells (Accutase, Thermo Fisher) and plated into recipient wells in mTeSR supplemented with thiazovivin (1 μM, Tocris; a ROCK inhibitor, to enhance hPSC survival after passaging) onto plates precoated with either Geltrex basement membrane matrix or recombinant Vitronectin (described above), thus plating 25,000–50,000 hPSCs/cm2 (i.e., ~95,000–190,000 hPSCs/well of a 12-well plate). Freshly-seeded hPSCs were allowed to adhere and recover for 24 hours in mTeSR + 1 μM thiazovivin prior to initiating differentiation, during which the hPSCs re-formed small clumps. All subsequent differentiation steps were performed in defined, serum-free CDM2 basal media (Loh et al., 2014; Loh et al., 2016). To reiterate, hPSCs are maintained by passaging as clumps (to maintain normal karyotype) but are seeded for differentiation as single cells (to enable efficient differentiation).
Day 1 (mid primitive streak induction, 24 hours [Step 1]). Day 0 hPSCs were briefly washed (DMEM/F12, Thermo Fisher) to remove all traces of mTeSR + thiazovivin. Then, they were differentiated towards mid primitive streak in CDM2 media supplemented with Activin A (30 ng/mL, R&D Systems), BMP4 (40 ng/mL, R&D Systems), CHIR99021 (6 μM, Tocris), FGF2 (20 ng/mL, Thermo Fisher) for 24 hours, as previously described (Loh et al., 2016) (with the optional addition of PIK90 (100 nM, Calbiochem)).
Day 2 (lateral mesoderm induction, 24 hours [Step 2]). Day 1 mid primitive streak cells were briefly washed (DMEM/F12) and then differentiated towards lateral mesoderm in CDM2 media supplemented with BMP4 (40 ng/mL), GDC-0941 (2.5 μM, Cellagen Technology), Forskolin (10 μM, Tocris), SB-505124 (2 μM, Tocris), VEGF (100 ng/mL, R&D Systems), XAV939 (1 μM, Tocris) and ascorbic acid-2-phosphate (AA2P; 200 μg/mL, Sigma) for 24 hours. Subsequently, lateral mesoderm was used for artery induction (Step 3a; below) or vein induction (Step 3b; below).
Day 3 (artery induction, 24 hours [Step 3a]). Day 2 lateral mesoderm cells were briefly washed (DMEM/F12) and then differentiated towards artery ECs in CDM2 media supplemented with Activin A (15 ng/mL), DMH1 (250 nM, Tocris), GDC-0941 (2.5 μM), VEGF (100 ng/mL), XAV939 (1 μM) and AA2P (200 μg/mL) for 24 hours, thereby yielding day 3 artery ECs.
Days 3–4 (vein induction, 24 hours [Step 3b]). Day 2 lateral mesoderm cells were briefly washed (DMEM/F12) and then differentiated in CDM2 media supplemented with SB505124 (2 μM), DMH1 (250 nM), RO4929097 (2 μM, Cellagen Technology), VEGF (100 ng/mL), XAV939 (1 μM) and AA2P (200 μg/mL) for 24 hours. Subsequently, day 3 cells were briefly washed (DMEM/F12) and then differentiated in CDM2 media supplemented with SB505124 (2 μM), RO4929097 (2 μM), PD0325901 (500 nM, Tocris), CHIR99021 (1 μM) and AA2P (200 μg/mL) for 24 hours, thereby yielding day 4 vein ECs.
In most experiments, hPSCs were plated and then differentiated into artery and vein ECs using the aforementioned media with respective media changes every 24 hours (without dissociating and replating cells at intermediate timepoints). However, we subsequently found that the efficiency of vein EC differentiation was enhanced when, at the end of day 2 of differentiation, lateral mesoderm cells were dissociated and re-plated at high density (~79,000 cells/cm2, i.e., 300,000–400,000 cells/well of a 12-well plate) in day-3 vein induction media (Fig. S3b). No ROCK inhibitor was used when replating lateral mesoderm cells. After 24 hours, day-3 cells were then differentiated into vein ECs using the aforementioned day-4 vein induction media without any additional resplitting.
Maintenance of hPSC-derived artery and vein cells
After hPSC differentiation into artery cells (within 3 days) or vein cells (within 4 days) as described above, they could be maintained for at least 6 additional days in vitro on Geltrex-coated cell culture plates. Day 3 hPSC-derived artery cells were expanded in EGM2 (Endothelial Cell Growth Medium 2, Lonza CC-3162), which was refreshed every 24 hours. Day 4 hPSC-derived vein cells were expanded in EGM2 + SB505124 (2 μM) + RO4929097 (2 μM), which was refreshed every 24 hours.
Cryopreservation and thawing of hPSC-derived artery and vein cells
After hPSC differentiation into artery cells (within 3 days) or vein cells (within 4 days) as described above, they were dissociated using either Accutase (Thermo Fisher) or TrypLE Express (Thermo Fisher). hPSC-derived artery cells were of sufficient purity to be frozen directly, whereas in certain experiments, hPSC-derived vein cells were first magnetically enriched for CD34+ cells using CD34 microbead-based Magnetically Activated Cell Sorting (MACS, Miltenyi Biotec 130–046-703) prior to freezing. MACS was performed generally as per the manufacturer’s recommendations, with slight modifications: after dissociation, hPSC-derived vein cells were counted using a hemocytometer, and then resuspended in FACS buffer (PBS + BSA + EDTA) with a 6- to 7-fold excess of CD34 microbeads and Fc blocking reagent (Miltenyi Biotec 130–046-703) compared to the manufacturer’s recommendation. Vein cells were stained on ice for 30 minutes with CD34 microbeads and Fc blocking reagent, washed, and then magnetically bound to a MACS LS column (Miltenyi Biotech 130–042-401). Unbound cells were then magnetically bound to a second MACS LS column to recover any residual CD34+ cells that were not captured in the first enrichment. Subsequently, CD34+ vein cells were collected, pelleted, and frozen. Flow cytometry was performed prior to and after magnetic enrichment (both bound and unbound cell fractions) to assess the precision of cell enrichment, which yielded >95% pure CD144+ cells throughout all MACS experiments. Both hPSC-derived artery and vein cells were frozen in freezing media (90% FBS + 10% DMSO, which was sterilely filtered before use) in cryopreservation vials, and were transferred to liquid N2 for long-term storage.
When desired, hPSC-derived artery and vein cells were rapidly thawed in a 37 °C water bath, washed to remove freezing media, and then thawed onto Geltrex-coated plates. As described above, hPSC-derived artery cells were maintained in EGM2, whereas hPSC-derived vein cells were maintained in EGM2 + SB505124 (2 μM) + RO4929097 (2 μM). For the first 24 hours post-thawing, ROCK inhibitor (thiazovivin, 1 μM) was added to both artery and vein cells to enhance cell survival, and was subsequently removed.
Alternate endothelial differentiation protocols for hPSCs
hPSCs were differentiated towards ECs using the artery EC differentiation protocol developed in this study, or alternatively, 4 published methods for EC differentiation (Lian et al., 2014; Patsch et al., 2015; Sriram et al., 2015; Zhang et al., 2017) in a side-by-side comparison. As per above, hPSCs were dissociated into single cells (using Accutase) and plated at a density of 25,000–50,000 hPSCs/cm2 onto Geltrex-coated plates, in mTeSR media supplemented with thiazovivin (1 μM, Tocris). Freshly-seeded hPSCs were allowed to adhere and recover for 24 hours in mTeSR + 1 μM thiazovivin prior to initiating differentiation. Immediately before differentiation, cells were briefly washed (DMEM/F12) to remove all traces of mTeSR media, and then the following differentiation media was added:
Lian et al., 2014. As described previously (Lian et al., 2014), hPSCs were treated with CHIR99021 6 μM, Tocris) for 2 days in LaSR basal medium (Advanced DMEM/F12 [Thermo Fisher] + GlutaMAX [2.5 mM, Gibco] + AA2P [60 μg/ml, Sigma]) for 2 days. Then, cells were briefly washed (DMEM/F12) to remove all traces of CHIR99021-containing medium and cells were maintained in LaSR basal medium for 3–4 additional days.
Patsch et al., 2015. As described previously (Patsch et al., 2015), hPSCs were treated with BMP4 (25 ng/ml, R&D Systems) and CP21R7 (1 μM, Selleckchem) in Priming Medium (50% DMEM/F12 with Glutamax media + 50% Neurobasal media + N2 + B27 [all from Thermo Fisher]). After 3 days, the cells were briefly washed (DMEM/F12) to remove all traces of Priming Medium and were cultured in EC Induction Medium (StemPro-34 SFM medium [Life Technologies] + VEGF [200 ng/ml, R&D Systems] + Forskolin [2 μM, Tocris]). EC Induction Medium was renewed after one day and was maintained for an additional 2–3 days.
Sriram et al., 2015. As described previously (Sriram et al., 2015), hPSCs were treated with CHIR99021 (5 μM, Tocris) in STEMdiff APEL basal medium (StemCell Technologies) for 1 day. After 24 hours, cells were briefly washed (DMEM/F12) to remove all traces of CHIR99021-containing medium and were treated with FGF2 (50 ng/mL, R&D Systems) in STEMdiff APEL basal medium for 1 day. Then, cells were briefly washed (DMEM/F12, Thermo Fisher) to remove all traces of FGF2-containing medium and were cultured in STEMdiff APEL basal medium + BMP4 (25 ng/mL, R&D Systems) + VEGF (50 ng/mL, R&D Systems) for 3 additional days.
Zhang et al., 2017. As described previously (Zhang et al., 2017), hPSCs were cultured in E8BAC media (E8 media [Thermo Fisher] + BMP4 [5 ng/mL, R&D Systems] + Activin A [25 ng/mL, R&D Systems] + CHIR99021 [1 μM, Tocris]) for 2 days. Subsequently, cells were briefly washed (DMEM/12), and then from day 2 to day 6, cells were differentiated in E5 media + FGF2 (100 ng/ml, R&D Systems) + VEGF (50 ng/ml, R&D Systems) + SB431542 (10 μM, Tocris) + Resveratrol (10 μM, R&D Systems) + L690 (10 μM, R&D Systems).
Flow cytometry to quantify the efficiency of EC differentiation was performed on days 0, 3, 5 and 6 of differentiation.
Generation of hiPSC lines
Human peripheral blood mononuclear cells (PBMCs) obtained from two different healthy adult human volunteers (SUN003 and SUN004) were reprogrammed using non-integrating, replication-deficient Sendai viruses encoding the iPSC reprogramming factors (Nishimura et al., 2017). This yielded SUN004.1.9 hiPSCs (referring to donor 4, line 1, subclone 9), SUN004.2 hiPSCs (referring to donor 4, line 2) and SUN003.1 hiPSCs (referring to donor 3, line 1) used in this study.
Ficoll was used to isolate PBMCs from the whole blood of healthy human adult volunteers (SUN003 and SUN004), whose identities were kept anonymous. 2 million PBMCs from each donor were split between two wells of a 24-well plate, with each well containing 1 million cells in 1 mL of PBMC media (RPMI + 10% FBS + GM-CSF (10 ng/mL) + IL-3 (10 ng/mL) + IL-6 (10 ng/mL) + FLT3L (10 ng/mL) + TPO (10 ng/mL) + SCF (10 ng/mL)) for PBMC expansion; cultures were kept in standard incubator conditions (37 °C and 5% CO2). After 6 days of PBMC expansion, cells were dissociated and combined into a single well of a 24-well plate in fresh PBMC media and expansion cytokines.
Subsequently, PBMCs were inoculated with replication-deficient, auto-erasing Sendai virus encoding KLF4, OCT4 (POU5F1), SOX2 and C-MYC (a gift from Dr. Mahito Nakanishi, at the National Institute of Advanced Industrial Science and Technology, Tsukuba, Japan (Nishimura et al., 2017)) at multiplicity of infection (MOI) = 2 to initiate iPSC reprogramming. This recombinant Sendai virus has been described previously: it encodes 1) modified versions of the NP and P/C/V/W genes of Sendai virus (Nagoya strain), followed by 2) KLF4, OCT4, SOX2 and C-MYC genes, and finally 3) a modified version of the L gene of Sendai virus (C1.151 strain) whose 3’ UTR contains 4x repeats of the miR-302 target sequence (Nishimura et al., 2017; Nishimura et al., 2011). This recombinant Sendai virus does not encode the structural genes M, F or HN and thus is replication-deficient: after infecting a cell, it does not produce progeny virions (Nishimura et al., 2011). Moreover, after infected cells have become reprogrammed into iPSCs and upregulate miR-302, this recombinant Sendai virus becomes auto-erased, as it contains 4x repeats of the miR-302 target sequence (Nishimura et al., 2017).
2 days after inoculation with Sendai virus, cells were again dissociated, pelleted and resuspended in 12 mL of 50% PBMC media and 50% hiPSC media (DMEM/F12, 20% KnockOut Serum Replacement [KOSR], 1% non-essential amino acids [NEAA], 1% penicillin/streptomycin [P/S], 100 μM 2-mercaptoethanol, and 5 ng/ml FGF2) supplemented with GM-CSF (10 ng/mL), IL-3 (10 ng/mL), IL-6 (10 ng/mL), FLT3L (10 ng/mL), TPO (10 ng/mL) and SCF (10 ng/mL). The cell suspension was split among three wells of a 6-well plate that contained mouse embryonic fibroblasts (MEFs). MEFs had been seeded on the previous day by first coating wells with 0.1% porcine gelatin (which had been pre-coated for 1 hour at 37 °C), after which mitomycin-C-treated, mitotically-inactivated C57BL/6 MEFs were plated at 15,000 cells/cm2 in MEF media (DMEM with 10% FBS). After plating hiPSCs on MEFs in 50% PBMC media + 50% hiPSC media, half-media changes were performed for the next 7–10 days with hiPSC media, thereby gradually shifting the overall media composition towards hiPSC media.
Morphologically-conspicuous hiPSC colonies were then manually picked, mechanically separated by pipette trituration and placed in hiPSC media on a new MEF-coated plate with 10 μM Y-27632 (ROCK inhibitor, to enhance cell survival) for first 24 hours. After 5 days of daily media changes (using hiPSC media), hiPSCs were dissociated into single cells with TrypLE and transitioned onto a vitronectin-coated plate in E8 media supplemented with 10 μM Y-27632 for 24 hours. Undifferentiated hiPSCs were routinely passaged every 4–6 days by re-seeding them at a density of 5,000–10,000 cells/cm2. Subsequently, hiPSCs were adapted to culture on Geltrex-coated plates in mTeSR media. This procedure was used to generate three distinct, clonal hiPSC lines referred to as SUN003.1, SUN004.1 and SUN004.2 (see next section).
Fluorescent labelling of hiPSC lines
SUN003.1, SUN004.1 and SUN004.2 hiPSC lines were engineered to carry constitutively-expressed reporter transgenes to enable the tracking of differentiated progeny cells.
SUN003.1 hiPSCs carry a constitutively-expressed CAG-AkaLuciferase-tdTomato lentiviral transgene. The pCS-CAG-AkaLuciferase-tdTomato lentiviral plasmid was generated by cloning AkaLuciferase (Iwano et al., 2018) into the pCS-CAG-tdTomato plasmid (Masaki et al., 2016), such that AkaLuciferase and tdTomato are expressed together as an in-frame fusion protein. Lentivirus was then generated using the LV-MAX Lentiviral Production System (Gibco, A35684) as per the manufacturer’s protocol. SUN003.1 hiPSCs were transduced with the CAG-AkaLuciferase-tdTomato lentivirus, cultured for 7–10 days, and then tdTomato+ hiPSCs were isolated by FACS, thus yielding CAG-AkaLuciferase-tdTomato + SUN003.1 hiPSCs (referring to referring to donor 3, line 1).
SUN004.1.9 hiPSCs carry a constitutively-expressed CAG-eGFP PiggyBac transgene. To integrate the PiggyBac transposon, SUN004.1 hiPSCs were seeded into 1 well of a 6-well plate at a density of 20,000 cells/cm2, and the following day, they were then transfected with 250 μL of transfection mixture, which contained 1.25 μg of donor plasmid (CAG-GFP PiggyBac donor plasmid), 1.25 μg of helper plasmid (CMV-PBase, encoding the PiggyBac transposase enzyme) and 7.5 μL of Lipofectamine 3000 (L3000001, Thermo Fisher). 5 to 10 days post-transposition, GFP+ hiPSCs that had been successfully transposed were isolated by fluorescence-activated cell sorting (FACS). The CAG-GFP+ SUN004.1.9 hiPSC subclonal line (referring to donor 4, clone 1, subclone 9) was isolated by manually picking and expanding a single GFP+ colony.
SUN004.2 hiPSCs carry a constitutively-expressed CAG-mScarlet lentiviral transgene. The pCS-CAG-mScarlet lentiviral plasmid was generated by cloning mScarlet into the pCS-CAG-eGFP plasmid (Masaki et al., 2016). Lentivirus production and transduction of SUN004.2 hiPSCs was performed as described above, and then 7–10 days post-transduction, mScarlet+ hiPSCs were isolated by FACS, thus yielding CAG-mScarlet+ SUN004.2 hiPSCs (referring to referring to donor 4, line 2).
Flow cytometry
Undifferentiated and differentiated hPSCs were dissociated by incubation in TrypLE Express (Gibco) for 5 minutes at 37°C. Subsequently, dissociated cells in TrypLE Express were diluted 1:5–1:10 in DMEM/F12 and centrifuged (pelleted) at 500g for 5 minutes. Each cell pellet was resuspended in FACS buffer (PBS + 1 mM EDTA [Invitrogen] + 2% v/v FBS [Atlanta Bio] + 1% Penicillin/Streptomycin [Gibco]) supplemented with fluorescently-conjugated primary antibodies, and antibody staining occurred for 30 minutes on ice protected from light. After staining, cells were washed twice with FACS buffer and resuspended in 200 μL FACS buffer with DAPI (1:10,000, Biolegend) for live/dead discrimination. Flow cytometry was performed on a BD Biosciences FACSAria II flow cytometer or a Beckman Coulter CytoFlex analyzer (Stanford Stem Cell Institute FACS Core). For data analysis, cells were gated based on forward and side scatter with height and width used for doublet discrimination. Subsequently, live cells that were negative for DAPI were gated for all marker analyses and calculations of population frequency.
Quantitative PCR
Undifferentiated or differentiated hPSCs were lysed in 350 μL of RLT Plus Buffer and RNA was extracted using the RNeasy Plus Mini Kit (Qiagen) according to the manufacturer’s protocol. 300 ng of total RNA was reverse transcribed into cDNA for qPCR using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer’s protocol. qPCR was performed in 384-well format as previously described (Loh et al., 2016), using gene-specific forward and reverse primers on a QuantStudio 5 qPCR machine (Thermo Fisher). qPCR primer sequences are provided in Table S5. Expression of all genes was normalized to the levels of the reference gene YWHAZ. This qPCR procedure was only used for biosafety level 2 (BSL2) cell cultures. A separate qPCR procedure was used for biosafety level 4 (BSL4) materials, and is detailed further below.
Immunostaining
Cultured monolayer cells were fixed, permeabilized and immunostained as previously described (Loh et al., 2016). Imaging was conducted using an FV3000 confocal microscope (Olympus).
Genome editing to create knock-in fluorescent reporter hPSCs
H1 NR2F2–2A-GFP knock-in hPSCs and H1 SOX17–2A-mPlum knock-in hPSCs were genetically engineered using the Cas9/AAV6 system (Martin et al., 2019). This genomic knock-in strategy entails the electroporation of hPSCs with a Cas9-sgRNA ribonucleoprotein (RNP) complex (to create a double-stranded break at a desired genomic locus), and delivery of a donor template for homologous recombination, supplied by AAV6 (Martin et al., 2019).
In brief, hPSCs were treated with 10 μM ROCK inhibitor (Y-27632) 24 hours prior to genome editing. hPSCs at 70–80% confluence were then dissociated using Accutase (Life Technologies) followed by neutralization with ROCK inhibitor-supplemented mTeSR1 media. Prior to electroporation, RNP complex was formed by combining 5 μg of HiFi SpCas9 (Integrated DNA Technologies) and 1.75 μg of sgRNA for 10 minutes at room temperature, which was then diluted with 20 μL of P3 Primary Cell Solution buffer (Lonza). For each electroporation reaction, 500,000 hPSCs were mixed with the Cas9/sgRNA RNP-containing nucleofection solution. Nucleofection was performed using a 16-well Nucleocuvette Strip with the 4D Nucleofector system (Lonza) using the CA137 electroporation code. Following electroporation, cells were transferred into one well of a Matrigel-coated 24-well plate containing 500 μL of mTeSR1 media supplemented with 10 μM Y-27632. AAV6 donor vector (encoding NR2F2–2A-GFP and SOX17–2A-mPlum donor templates for homologous recombination) was added at 100,000 multiplicity of infection (MOI) directly to hPSCs immediately after plating. Cells were then incubated at 37°C for 24 hours. Media was changed 24 hours post-editing and 10 μM Y-27632 was removed 48 hours after.
The NR2F2 and SOX17 synthetic sgRNAs were purchased from Synthego with chemically-modified nucleotides at the three terminal positions at both the 5′ and 3′ ends. Modified nucleotides contained 2′-O-methyl 3′-phosphorothioate:
NR2F2 sgRNA: AUUUAUUGAAUUGCCAUAUA
SOX17 sgRNA: CUGCAGGCUGGGGCGGAUCA
Genome editing to create EFNB2-knockout hPSCs
The EFNB2 gene was deleted in H1 hPSCs using the aforementioned Cas9 protocol (section “Genome editing to create knock-in fluorescent reporter hPSCs” (Martin et al., 2019)), with the exception that Cas9 RNPs were used to generate indels in the EFNB2 locus, but no templates for homologous recombination were provided. This procedure was repeated twice: the initial round of gene targeting produced EFNB2+/− hPSCs, which were then re-electroporated with Cas9 RNP targeting EFNB2, thereby generating EFNB2−/− hPSCs.
sgRNAs targeting the EFNB2 locus were designed using CHOPCHOP (Labun et al., 2019) and verified using the COSMID program (Cradick et al., 2014). The EFNB2 synthetic sgRNAs was purchased from Synthego with chemically-modified nucleotides at the three terminal positions at both the 5' and 3' ends. Modified nucleotides contained 2'-O-methyl 3'-phosphorothioate. The genomic sgRNA target sequence was:
EFNB2 sgRNA: AGGCUCUAAAACUAUCGAUU
3D network formation from hPSC-derived endothelial cells
Three different types of endothelial cells were used in the formation of in vitro vascular networks in fibrin hydrogels. Where indicated, 1) hPSC-derived artery endothelial cells, 2) hPSC-derived vein endothelial cells, or 3) umbilical cord blood-derived endothelial colony forming cells (CB-ECFC, a positive control (Kurokawa et al., 2017)) were utilized.
hPSC-derived artery and vein cells were frozen and thawed (as described above), and after thawing, were cultured in EGM2 medium for 3 days (supplemented with 2 μM Thiazovivin for the first 24 hours). EGM2 media was refreshed every 24 hours for the 3-day period. After 3 days of expansion, hPSC-derived artery and vein endothelial cells and normal human lung fibroblasts (NHLFs) were used to produce 3D endothelial networks as described previously (Kurokawa et al., 2017). In brief, hPSC-derived endothelial cells and normal human lung fibroblasts (NHLFs) were respectively dissociated using 0.05% Trypsin-EDTA (Thermo Fisher, 25300062). The cells were then mixed at a ratio of 1 endothelial cell:2 fibroblasts. The resultant cell mixture was centrifuged at 300g for 5 min, and resuspended in 10 mg/mL of bovine fibrinogen (Sigma-Aldrich, F8630). 50 μL of the cell-fibrinogen solution was then placed inside of a small chamber consisting of cured polydimethylsiloxane (Sylgard 184) on glass, and clotted using thrombin from bovine plasma (Sigma-Aldrich T4648). The final thrombin concentration in the 10 mg/mL fibrin hydrogel was 2 unit/mL (which allowed for clotting of the fibrinogen into fibrin). After complete clotting of the hydrogels, the resultant fibrin hydrogels were submerged in 500 μL EGM2 and cultured in a 37 °C, 5% CO2 humidified incubator. EGM2 media changes occurred every 1–2 days with fresh, warm EGM2. After 7 days of culture to allow for network formation in EGM2, hydrogels were fixed using 10% formalin (Sigma F5554) in order to undergo immunofluorescent analysis. Briefly, hydrogels were blocked using 2% BSA (Sigma A2153) in PBS+0.1% Tween-20 (Sigma P9416), after which antibodies were used to stain the tissues overnight at 4 °C. Images were then taken on a fluorescent inverted microscope (Olympus IX83). Resultant image analysis and image preparation was performed using Fiji.
hPSC-derived endothelial cell migration in vitro in scratch assay
hPSCs were differentiated into artery and vein endothelial cells as described above. Then, hPSC-derived artery cells were cultured in EGM2 for 1 day while vein endothelial cells were cultured in EGM2 + SB505124 (2 μM) + RO4929097 (2 μM) for 1 day, until they formed a confluent monolayer. Then, media was removed and cells were washed with DMEM/F12 before being scratched with a P200 pipette tip affixed to the end of a P200 pipette. After cells were scratched, floating cells were washed away with DMEM/F12. Then, artery expansion media (EGM2) was added to artery endothelial cells and vein expansion media (EGM2 + SB505124 (2 μM) + RO4929097 (2 μM)) was added to vein endothelial cells. Media was replaced every 24 hours. Cells were imaged 0, 6, 12, 24 and 48 hours after scratching, using the Leica Paula Microscope. For image acquisition, tape was used on the microscope stage to mark the perimeter of the cell culture plate after scratching such that for images taken at different timepoints, the image was acquired at the same location every time. To quantify the closure of the gap, each image was set to scale using ImageJ. Then, using the ImageJ measurement tool, the length of the gap between both borders of the closing cell monolayer were measured at 6 different locations.
Shear stress-induced polarization of hPSC-derived endothelial cells
hPSC-derived day-3 artery endothelial cells were cultured in a 6-well plate with EGM2 medium for 24 hours, either in static culture (standard conditions) or a rotator (shear stress conditions). hPSC-derived day-4 vein endothelial cells were magnetically enriched for CD34+ cells (Miltenyi Biotec) and cultured for 1 week in EGM2 media prior to exposure to shear stress for 24 hours as per above. Shear stress conditions were described previously and entailed culturing cells on a 6-well plate on an orbital shaker (Poduri et al., 2017). After 24 hours of static or shear stress conditions, both immunostaining and qPCR was performed.
After exposure to static media or shear stress for 24 hours, immunostaining was performed to quantify the degree of cell alignment. To this end, cells were fixed in 4% paraformaldehyde (PFA) for 15 minutes at 4 °C. Then, the cells were washed with PBS for three times, waiting 5 minutes between each wash. After washing, cells were incubated in 4 °C overnight with primary antibody in 0.5% Triton X-100/PBS solution. Next, cells were washed with PBS for three times, waiting 15 minutes between each wash. Afterwards, the cells were incubated with secondary antibody in 0.5% Triton X-100/PBS solution. Secondary antibody was removed and DAPI in PBS was added and incubated for 5 minutes. Then, the cells were washed with 0.5% Triton X-100/PBS three times, waiting 15 minutes between each wash. Finally, PBS was added to the cells, and imaging was conducted using a Keyence BZ-9000 microscope.
After imaging, quantification of cell alignment was performed using ImageJ. Cell directionality analysis was done using the “Directionality” plugin, using Fourier components analysis with Nbins=90, producing a directionality histogram which starts at −90 degrees and ends at +90 degrees.
LDL and acetylated LDL uptake assay
hPSC-derived day 3 artery and day 4 vein endothelial cells were washed with DMEM/F12 (Thermo Fisher) and incubated at 37°C for 4 hours with either a 1:25 concentration of low-density lipoprotein (LDL) conjugated with DyLight550 (Cayman) or a 5 μg/mL concentration of acetylated LDL conjugated to Alexa Fluor 488 (Fisher Scientific) in their respective differentiation media. As a negative control, hPSC-derived day 3 artery and day 4 vein endothelial cells were incubated in their respective differentiation media without LDL DyLight 550 or acetylated LDL Alexa 488. After incubation, cells were dissociated by incubating them in TrypLE Express (Gibco) for 5 minutes at 37 °C. Subsequently, the dissociated cells were diluted 1:10 in DMEM/F12 and centrifuged at 500g for 5 minutes. Each cell pellet was then resuspended in FACS buffer (DPBS + 5 mM EDTA [Invitrogen] + 0.5% BSA) supplemented with anti-CD144 antibody conjugated to Alexa Fluor 647 (BD Biosciences) and incubated on ice for 30 minutes, while being protected from light. After antibody staining, cells were washed twice with FACS buffer and resuspended in 300 μL FACS buffer containing DAPI (1:10,000, Biolegend). Then the resuspended cells were passed through a 100 μm strainer (BD Biosciences) and analyzed using a FACSAria II flow cytometer (Stanford Stem Cell Institute FACS Core). During flow cytometry data analysis, we gated on CD144+ endothelial cells before quantifying fluorescent LDL or acetylated LDL uptake by endothelial cells.
Immune cell adhesion to TNFα-treated endothelial cells
The THP1 human monocyte-like cell line was fluorescently labelled with CFSE and then cocultured with hPSC-derived endothelial cells to assess immune cell adhesion to endothelial cells. Methods to assess adhesion of immune cells to endothelial cells were modified from previously-published protocols (Rosa et al., 2019; Zhang et al., 2017).
First, THP1 cells were fluorescently labelled with CFSE, largely as per the manufacturer’s recommendations. THP1 cells were resuspended in DMEM/F12 at a concentration of 1 million cells/mL and then incubated with 5 μM of CellTrace CFSE (Thermo Fisher Scientific, C34554) for 20 minutes at room temperature, protected from light, and with occasional pipetting to keep cells resuspended during the staining process. After 20 minutes of incubation, the cell suspension was diluted 1:5 in RPMI1640 + 10% FBS for 5 minutes. Then, cells were centrifuged at 300g for 5 minutes, the supernatant was aspirated, and then fresh RPMI1640 + 10% FBS was added.
Separately, hPSC-derived artery and vein endothelial cells cultured in 12-well plates were stimulated with the inflammatory cytokine TNFα (10 ng/mL) for 4 hours in their respective culture media (EGM2 for artery cells and EGM2 + RO4929097 (2μM) + SB505124 (2μM) for vein cells) at 37 °C. As a negative control, artery and vein cells were cultured in their respective culture media in the absence of TNFα.
Subsequently, Hoechst dye (1 μg/mL) was added in the dark for 5 minutes to fluorescently label the nuclei of live artery and vein cells. Then, cells were washed with DMEM/F12. After washing, CFSE-labelled THP-1 immune cells (described above) were added to each well in a 12-well plate at 1 million cells/well in EGM2 media for 30 minutes at 37 °C. After this 30-minute coculture of THP1 and endothelial cells, the wells were washed 3 times with DMEM/F12 to remove any non-adhered THP1 cells. Images were taken using a Keyence BZ-9000 fluorescence microscope and quantification of the number of adhered THP1 cells was performed using ImageJ/Fiji.
Endothelial sprouting assay
The sprouting assay was performed as previously described (Nakatsu et al., 2007), with modifications. First, CAG-GFP+ SUN004.1.9 hPSC-derived artery or vein ECs were thawed and respectively maintained in artery or vein expansion media (EGM2 for artery ECs and EGM2 + RO4929097 (2 μM) + SB505124 (2 μM) for vein ECs) for 24 hours until they were fully confluent. Second, ECs were cultured with EGM2 + 10% heat-inactivated FBS for 24 hours, prior to dissociating them for the sprouting assay. Third, hPSC-derived artery and vein ECs were dissociated using trypsin and counted. In parallel, Cytodex-3 beads (Sigma Aldrich, C3275) were allowed to settle for 5 minutes, and were washed briefly in 1 mL of warm EGM2 media before removing the media. Fourth, Cytodex-3 beads were coated with hPSC-derived ECs by incubating them at a concentration of ~400 cells/bead in 1.5 mL of warm EGM2 media. The bead/cell suspension was incubated for 4 hours at 37°C, with gentle pipetting every 20 minutes. After 4 hours, the EC-coated beads were transferred to a well of a non-tissue-culture-treated 24-well plate and left overnight in EGM2 + 10% heat-inactivated FBS. The EC-coated beads were then transferred to a 15 mL conical tube and left to settle for 5 minutes, followed by three washes with warm EGM2, and then the beads were counted on a coverslip.
To embed EC-coated beads in a 3D fibrin gel, beads were resuspended in sterilely-filtered fibrinogen solution (2.0 mg/mL, Sigma Aldrich, F8630) at a concentration of ~500 beads/mL, and 0.5 mL of the fibrinogen/bead suspension was added to each well of a 24-well plate. Then, 0.5 mL of thrombin (0.625 units/mL, Sigma Aldrich, T3399) was added and the bead-fibrinogen-thrombin mixture was triturated ~4–5 times, with care to avoid the formation of bubbles. The plate was left in the hood for 5 minutes, and then incubated at 37° C for 10 minutes to generate fibrin gels. After the gels formed, 1 mL of EGM2 media + 10% FBS was added dropwise. Gels were incubated at 37°C for 24 hours in order for GFP+ endothelial sprouts to emerge. At the end of the assay, EGM2 was removed and the gels were washed with DPBS prior to overnight fixation in 4% paraformaldyde. Gels were imaged using a fluorescence microscope. The number of sprouts per bead and the length of each sprout was quantitated on at least five beads using MetaMorph software.
Endothelial chemotaxis towards Apelin-13
The chemotaxis assay was performed as previously reported (Kälin et al., 2007), with modifications. The polycarbonate membrane of a Transwell filter (with 8 μm pores; Corning, 3422) was coated with 0.1% gelatin overnight. Separately, CAG-GFP+ SUN004.1.9 hPSC-derived artery or vein ECs were respectively maintained in artery or vein expansion media (EGM2 for artery ECs and EGM2 + RO4929097 (2 μM) + SB505124 (2 μM) for vein ECs) until they were fully confluent. hPSC-derived artery or vein ECs were then dissociated using trypsin and resuspended in Endothelial Basal Media 2 media (EBM2, Lonza) at concentration of 1.5 × 105 cells/mL. Then, 100 μL of the cell suspension was added to the upper chamber of the gelatin-coated Transwell filter, and 600 μL of EBM2 media + Apelin-13 (100 ng/mL, Tocris, 2420) was added to the lower chamber. Cells were incubated at 37°C for 4 hours to allow for cell migration towards the lower chamber. Then, non-migratory cells were removed with a cotton swab. Migratory cells on the bottom of the Transwell membrane were fixed and stained with DAPI. Cells were imaged under a fluorescence microscope and counted using ImageJ software.
Subcutaneous transplantation of hPSC-derived endothelial cells
SUN003.1 CAG-AkaLuciferase-tdTomato hPSC-derived ECs were subcutaneously transplanted into adult, immunodeficient NOD-SCID Il2rg−/− (NSG) mice. In brief, hPSC-derived endothelial cells were suspended in a 1:1 mixture of Matrigel (phenol-free, growth factor reduced) + F12 medium (supplemented with 100 ng/mL FGF2 and 50 ng/mL VEGF). NSG mice were then anesthetized with 2% isoflurane and 100% oxygen flowing at 2 liters/minute until they were unconscious and unresponsive to stimuli (i.e., a toe pinch). A fold of skin directly lateral to the spine and near the neck was swabbed with ethanol in order to disinfect it, and then was gently pinched. A volume of 50–200 μL containing ~106 cells suspended in the Matrigel + F12 + FGF2 + VEGF solution was then injected under the skin flap using a 27-gauge syringe. Anesthesia was then discontinued, and mice were allowed to recover in a warm cage until they became alert and awake.
1 month post-transplantation, NSG mice were sacrificed, and subcutaneous gels containing hPSC-derived ECs were recovered and then fixed in 4% formaldehyde overnight at 4 °C. Fixed gels were then placed in PBT buffer (PBS with 0.5% Triton-X) with primary antibodies and incubated overnight at 4 °C on a shaker. The next day, samples were washed every 1.5 hours in PBT buffer for approximately 8 hours at room temperature, and with gentle agitation. Subsequently, Alexa Fluor 488-conjugated secondary antibodies (Life Technologies) were then added in PBT buffer at a 1:250 concentration and incubated overnight at 4 °C on a shaker. Finally, the stained samples were washed every 1.5 hours with PBT buffer for approximately 8 hours and cleared using Vectashield (Vector Labs, H-1000). Samples were then placed in a microscope chamber and imaged using a Zeiss LSM-700 confocal microscope. Image processing was done in ImageJ.
Single-cell RNA-sequencing
We executed single-cell RNA-seq on H1 hPSC-derived day 3 artery ECs and day 4 vein ECs using the 10x Genomics Chromium v3 platform (Zheng et al., 2017). Libraries were prepared using the 10x Genomics Chromium 3’ Kit v3 following the manufacturer’s guidelines. Libraries were prepared with i7 indices (compatible with Illumina sequencers) and then multiplexed. Multiplexed libraries were sequenced across multiple lanes of an Illumina HiSeq 4000 sequencer by Novogene. Unique sequences in each i7 index were used for demultiplexing. Cell Ranger (10x Genomics) was used to perform read alignment to the hg38 reference genome, filtering, barcode counting and UMI counting.
Cell matrix files generated from Cell Ranger were imported into RStudio using the function “Read10x_h5” in Seurat v3 (Stuart et al., 2019). We then implemented well-established quality control procedures to exclude dying/dead cells that 1) expressed low numbers of genes, 2) displayed too low or too high mitochondrial counts, or 3) did not express reference/housekeeping genes ACTB or YWHAZ; additionally, we also 4) computationally excluded doublets by removing cells that had expressed markedly higher numbers of genes. Only high-quality single-cell transcriptomes that passed these quality control metrics were used for subsequent analyses. scRNAseq data were normalized using the “LogNormalized” function and scaled using a linear transformation in Seurat v3 (Stuart et al., 2019). Expression of marker genes was denoted by different color shades and superimposed over the entire cell population, as visualized on a Uniform Manifold Approximation and Projection (UMAP) projection (Becht et al., 2018). Importantly, all cells within the entire day 3 and day 4 differentiated populations were shown on the UMAP projection, without any pre-selection of ECs.
An interactive web portal (Ouyang et al., 2021) to explore scRNAseq profiles of hPSC-derived artery and vein ECs is available at https://anglab.shinyapps.io/arteryveinscrna/.
Comparing in vivo vs. in vivo artery and vein cells using single-cell RNA-seq
We used scRNAseq to quantify the degree of transcriptional similarity between hPSC-derived artery and vein ECs (in vitro; described above) as well as human artery and vein ECs obtained from a Carnegie Stage 13 (CS13) human fetus (in vivo). scRNAseq data of CS13 human fetal artery and vein ECs were generated previously (Zeng et al., 2019) and were downloaded from the Gene Expression Omnibus (GSE135202). Importantly, both in vivo and in vitro EC datasets were generated using the 10x Genomics Chromium platform (Zheng et al., 2017), thus mitigating concerns that would otherwise arise from a cross-platform comparison.
We performed quality control and normalization for CS13 human fetal scRNAseq data using the methods described above for hPSC-derived artery and vein ECs (section “Single-cell RNA-sequencing”). In brief, dead/dying cells as well as doublets were computationally excluded and then CS13 human fetal scRNAseq data were normalized using the “LogNormalized” function and scaled using a linear transformation in Seurat v3 (Stuart et al., 2019).
To decompose the CS13 human fetal scRNAseq dataset into its constituent cell-types, we used Seurat v3 (Stuart et al., 2019) to perform Louvain clustering (resolution 0.05), which separated the fetal cell population into 4 subsets: artery ECs (CDH5+, PECAM1+, DLL4+, CXCR4+, HEY1+); vein ECs (CDH5+, PECAM1+, NT5E+, APLNR+, NR2F2+, NRP2+); mesenchymal (TWIST2+ CXCL12+ DLK1+ HAND1+ FBLN5+ WT1+), and epithelial cells (EPCAM+, CDX2+). The existence of these 4 clusters—and the marker genes used to discriminate them—are consistent with what was reported in the original report that described these scRNAseq data (Zeng et al., 2019). Next, we focused our analysis only on the fetal ECs: we selected cells that expressed pan-endothelial genes (PECAM1 and CDH5) at a level of UMI > 1.221 (i.e., UMI e0.2) in both the CS13 artery and CS13 vein subpopulations for downstream analyses.
In parallel, we computationally isolated ECs from our scRNAseq datasets of hPSC-derived day 3 artery and day 4 vein ECs in vitro. Louvain clustering (resolution = 0.1) was applied using Seurat v3 (Stuart et al., 2019), which decomposed the day 3 population into artery ECs (CDH5+, PECAM1+, CD34+, SOX17+, CXCR4+, DLL4+) vs. non-ECs; similarly, this procedure decomposed the day 4 population into vein ECs (CDH5+, PECAM1+, CD34+, NR2F2+, NT5E+, FLRT2+) vs. non-ECs. The hPSC-derived artery ECs and vein ECs were computationally isolated and then used for subsequent analyses.
We then merged our in vitro scRNAseq dataset—composed of hPSC-derived day 3 artery ECs and day 4 vein ECs, in addition to other in vitro cell-types—with the CS13 human fetal artery and vein EC dataset. The combined scRNAseq dataset was normalized and scaled using SCTransform (Hafemeister and Satija, 2019) to remove technical variation while retaining biological variation. Next, we calculated the average expression of each gene in the SCTransform-normalized dataset (containing both in vivo and in vitro ECs) using the “AverageExpression” function in Seurat v3 (Stuart et al., 2019). We then identified the top 6000 variable genes in this merged dataset using the “CellScatter” function.
On the basis of the expression of these 6000 variable genes, we calculated the Pearson correlation coefficient 1) between our hPSC-derived artery ECs vs. CS13 human fetal artery ECs and 2) between our hPSC-derived vein ECs vs. CS13 human fetal vein ECs.
To find differentially-expressed genes between hPSC-derived vs. CS13 fetal artery and vein ECs, we used the function “FindMarkers” in Seurat v3 (Stuart et al., 2019), with parameters set to only identify genes that were expressed in at least 25% of cells within that subcluster. This identified the most differentially-expressed genes between 1) hPSC-derived vs. human fetal artery ECs and 2) hPSC-derived vs. human fetal vein ECs. Of these genes that strongly differed in expression levels between in vitro-derived vs. in vivo ECs, we manually annotated those that had been previously linked to ribosome translation (GNB2L1, EEF1G, RPL17, RPS17) and mitochondrial respiration (ATP5G2, ATP5E, ATP5L, MT-ND2, MT-ND1, MT-ND4, MT-CTB, MT-CO1, MT-ATP6, MT-CO2, MT-CO3, MT-CTB). Finally, we performed gene ontology analysis on the lists of genes differentially expressed between hPSC-derived vs. CS13 fetal ECs using the online Enrichr platform (Chen et al., 2013; Kuleshov et al., 2016).
Bulk-population RNA-seq of uninfected cells
We performed bulk-population RNA-seq on H1 hPSC-derived day 3 artery ECs and day 4 vein ECs, both of which were FACS purified for CD144+ ECs to exclude any spurious gene expression from contaminating non-ECs. Cells were lysed using Zymo RNA lysis buffer and RNA was extracted using the Zymo Quick RNA kit according to the manufacturer’s instructions. We determined the RNA integrity number (RIN) of each sample using Agilent Bioanalyzer on-chip electrophoresis, and high-quality RNA samples with RIN ≥ 8 were enriched for poly(A) sequences and used to construct RNA-seq libraries, which were then ligated to adaptors for multiplexing. RNA-seq libraries were then sequenced on the DNBSEQ-G400 sequencer using DNA nanoball and combinational probe anchor synthesis technology by BGI Global Genomic Services to generate 150-bp paired-end reads. To minimize batch effects, all libraries were pooled together into a single sample, which was then sequenced on multiple lanes.
After obtaining RNA-seq reads, we performed quality control using FastQC (Andrews, 2010). Next, we used Trim Galore (Krueger, 2012) to remove index adaptors, low-quality base-calls or reads less than 20bp, using a read quality cutoff Phred score of 33. Trimmed and filtered reads from each single library (which was originally sequenced on multiple lanes) were then merged into a pooled file, each representing a single library. Subsequently reads were aligned with the GRCh38 (hg38) Homo sapiens reference genome and gene-level RNA-seq counts were quantified using Kallisto (Bray et al., 2016). We then filtered and log2 normalized counts per million (Tidyverse, BaseR, and EdgeR) (Robinson et al., 2010; Wickham et al., 2019). Then, we performed dimensional reduction and differential gene expression analyses (Limma and EdgeR) (Law et al., 2014; Ritchie et al., 2015; Robinson et al., 2010) to identify genes differentially expressed between hPSC-derived artery vs. vein ECs. These data were visualized on volcano plots: genes that showed a log2 fold differences less than 3 (i.e., less than 8-fold differential expression) were represented in gray, while differentially expressed genes (i.e., those with greater than 8-fold expression differences) were highlighted in distinct colors.
Generation of Nipah-pseudotyped lentivirus
Non-replicating, GFP-expressing lentiviruses pseudotyped to solely display Nipah or vesicular stomatitis virus (VSV) envelope proteins were generated from HEK293T/17 packaging cells using a 3rd generation lentiviral packaging system. In brief, cell-culture plates were coated with poly-L-lysine (R&D Systems, 3438–200-01) for 5 minutes at room temperature, which was then aspirated. Then 1 million HEK293T/17 cells (ATCC, CRL-11268) were seeded onto each poly-L-lysine-coated well of a 6-well plate. 24 hours after seeding, HEK293T/17 cells were ~80–90% confluent and they were transfected using FuGENE HD (Promega, E2311).
The following lentiviral packaging plasmids encoding Nipah or VSV envelope proteins were used:
pCAGGS-coNiV-G-HA: Encodes a codon-optimized NiV-G gene whose amino acid product is identical to the protein encoded by the NiV-G reference gene (NC_002728.1: 8710–11255 nucleotides, from Nipah virus/human/MY/1999/CDC), and additionally carries a hemagglutinin (HA) tag (Palomares et al., 2013). Provided by Dr. Benhur Lee’s laboratory (Icahn School of Medicine).
pcDNA3.1-coNiV-F T5F-Au1: Encodes a codon-optimized NiV-F gene whose amino acid product is a truncated version of the protein encoded by the NiV-F reference gene (NC_002728.1: 6370–8706 nucleotides, from Nipah virus/human/MY/1999/CDC). However 1) the cytoplasmic tail of NiV-F was truncated to only carry 5 amino acid residues (a change that improves the production of pseudotyped lentiviruses) and 2) additionally it carries an AU1 tag (Palomares et al., 2013). Provided by Dr. Benhur Lee’s laboratory (Icahn School of Medicine).
VSV-G: Encodes vesicular stomatitis virus envelope protein (VSV-G). Provided by Dr. Owen Witte’s laboratory (University of California Los Angeles) (Goldstein et al., 2011).
HEK293T/17 cells were transfected using FuGENE HD with the following amounts of lentiviral packaging plasmids per well of a 6-well plate: 1.38 μg pMDL plasmid + 0.53 μg pREV plasmid + 0.76 μg envelope protein-encoding plasmid(s) + 2.13 μg pRS348 EF1A-GFP plasmid. For the envelope protein-encoding plasmid(s), the following amounts were used for: 1) VSV pseudovirus (0.76 μg VSV-G plasmid) or 2) Nipah pseudovirus (0.37 μg NiV-G plasmid + 0.37 μg NiV-F plasmid). For transfection, room temperature DMEM medium was first mixed with FuGENE, and then mixed with plasmid DNA; after 15 minute incubation at room temperature, the transfection solution was added dropwise to HEK293T/17 cells.
16–20 hours post-transfection, the culture media was replaced with 1mL of pre-warmed DMEM + 10% FBS media. 24 hours media later, the media (containing pseudotyped lentivirus particles encoding a constitutively-expressed EF1A-GFP reporter) was harvested and either immediately used for transduction experiments or frozen at −80 °C.
Transduction of endothelial cells with Nipah-pseudotyped lentivirus
After Nipah- and VSV-pseudotyped lentiviruses were generated as described above, they were used to transduce hPSC-derived artery and vein endothelial cells. First, wells in a 96-well, flat-bottom plate were coated with Geltrex (50–100 μL/well), which was then aspirated. Subsequently, hPSC-derived artery or vein cells (generated from either wild-type or EFNB2−/− hPSCs) were thawed and 16,800–67,200 cells were seeded per Geltrex-coated well of a 96-well plate in 100 μL of respective artery or vein expansion media supplemented with the ROCK inhibitor thiazovivin for 24 hours. Artery or vein expansion media was changed every 24 hours, and thiazovivin was withheld after the initial 24 hour seeding period.
72 hours post-seeding, media was removed from artery and vein cells, and then 100 μL of fresh artery or vein expansion media was added, in addition to 0.3–100 μL of viral supernatant containing pseudotyped lentiviruses (or control media lacking pseudotyped viruses). Pseudotyped lentiviruses were added for 24 hours prior to removal of culture media, and subsequently, media was replaced with fresh artery and vein media every 24 hours.
6 days post-seeding (i.e., 3 days post-transduction), each well of artery or vein cells was dissociated through the addition of 50 μL TrypLE Express (incubated at 37 °C, 3–6 minutes) and trituration. Then, 150 μL of FACS buffer was added per well, and cells were transferred into a 96-well V-bottom plate before centrifugation (at 4 °C, 5 minutes). Subsequently, supernatant was aspirated to remove the dissociation buffer, and cells were resuspended in 150 μL of FACS buffer + DAPI per well. Flow cytometry was performed using a CytoFLEX flow cytometer (Beckman Coulter Life Sciences) to enumerate the percentage of GFP+ (i.e., virally-transduced) viable cells in each well.
The percentage of GFP+ cells was used to quantify the relative efficiency with which a given pseudotyped lentivirus transduces artery vs. vein endothelial cells (i.e., the arteriovenous tropism of a given pseudotyped lentivirus). In brief, artery and vein cells were inoculated with an equal volume of lentiviral supernatant containing Nipah- or VSV-pseudotyped lentivirus and the percentage of GFP+ cells were quantified by flow cytometry, as described above. In parallel, wells of artery and vein cells were dissociated and counted to determine cell numbers, in order to correct for any differences in transduction efficiency attributable to the use of different numbers of artery vs. vein cells.
Next, the multiplicity of infection (i.e., after inoculation with a given volume of pseudotyped lentiviruses, the number of pseudotyped lentiviruses per each single cell) was calculated using Poisson statistics as follows. Assuming that, after viral inoculation, the number of viruses that transduces a single cell follows a Poisson distribution (Ellis and Delbrück, 1939), then: P(n) = (mn * e−m)/n!, where P(n) is the probability that a cell will be transduced by n number of viruses and m is the multiplicity of infection.
The frequency of non-transduced cells in a given cell population is described by P(n = 0) = e−m. The frequency of transduced cells is described by P(n > 0) = 1 – P(0). Consequently, P(n > 0) = 1 – e−m. In this case, after transduction with GFP-encoding lentiviruses, P(n > 0) is equivalent to the percentage of GFP+ cells in a given cell population (henceforth “GFP+”). In summary, the multiplicity of infection (m) can be described by: m = –ln(1 – GFP+). In this study, the percentage of GFP+ cells was always than 60% (i.e., the multiplicity of infection was always less than 0.916), thereby enabling the multiplicity of infection to be accurately determined within the linear range of this quantitative assay. In this manner, the percentage of GFP+ artery or vein cells (enumerated by flow cytometry) was used to calculate the multiplicity of infection, which was then multiplied by the number of artery or vein cells to yield the inferred number of pseudotyped lentivirus particles that were originally used to inoculate the artery or vein cells. Given that the same absolute volume of pseudotyped lentivirus was used to inoculate artery or vein cells, any differences in the inferred number of pseudotyped lentivirus particles for artery vs. vein cells must be attributable to differences in the tropism of the pseudotyped lentivirus for artery vs. vein cells. Finally, the inferred number of pseudotyped lentivirus particles used to inoculate artery cells was divided by the inferred number of pseudotyped lentivirus particles used to inoculate vein cells, thus calculating the arteriovenous tropism of Nipah- or VSV-pseudotyped lentiviruses.
Generating stocks of Nipah and Hendra viruses and viral quantification
Authentic Nipah and Hendra viruses are classified in Risk Group 4 (CDC and NIH, 2020). All experiments with Risk Group 4 viruses were conducted under maximum containment conditions in the biosafety level 4 (BSL4) laboratory of the Robert Koch Institute, in accordance with standard operating procedures institutionally approved by the Robert Koch Institute.
For this study, Nipah virus, Malaysia variant (Nipah virus/human/MY/1999/CDC) (Harcourt et al., 2000) and Hendra virus, Brisbane variant (Hendra virus/horse/AU/1994/Hendra) (Murray et al., 1995) were obtained from CDC Viral Special Pathogens Branch reference stocks. These viruses were then subsequently propagated on Vero E6 cells (Vero C1008 (Vero 76, clone E6, Vero E6), from the European Collection of Authenticated Cell Cultures (ECACC)) in DMEM + 2% FBS to generate archive stocks, which were frozen at −80°C. As needed, archive stocks were thawed and passaged once more on Vero E6 to generate working stocks of these viruses, which were frozen at −80°C. Working stocks of these viruses were used for all subsequent experiments.
These viral stocks were quantified using the tissue culture infectious dose 50% (TCID50) assay. For viral quantitation, Vero E6 cells were seeded into 96-well tissue culture plates in DMEM containing 10% FBS, 2 mM glutamine, and 1% penicillin/streptomycin, at such a density that after overnight plating the cells became 95–100% confluent the following day. Separately, serial 10-fold dilutions of virus stocks were then diluted in DMEM. Then, the Vero E6 culture media was aspirated and 100 μL of viral supernatant was added to triplicate wells of Vero E6 cells. This was incubated for 1 hour at 37 °C, 5% CO2. Subsequently, 100 μL of DMEM containing 4% FBS, 4 mM glutamine, and 2% penicillin/streptomycin was added to the wells (such that the final FBS concentration was 2%) and this was incubated for 4–5 days at 37 °C, 5% CO2. The cells were then scored for the presence of cytopathic effects (CPE). The endpoint dilution of the viral stock in which 50% of the wells exhibited CPE was calculated using the Spearman-Karber method, therefore quantifying the viral dose present in the original viral stocks (expressed as TCID50 units/mL). These titrated viral stocks were then used to infect hPSC-derived artery and vein ECs, as described below.
In certain experiments, Sendai virus, Cantell strain (Cantell and Hirvonen, 1981)—a paramyxovirus not known to cause disease (López et al., 2006; Yoshida et al., 2018)—was used as a control for Nipah and Hendra viruses, which are highly-lethal paramyxoviruses. Sendai virus is classified in Risk Group 2, but was handled in the BSL4 laboratory for consistency. Sendai virus, Cantell strain was obtained from ATCC (VR-907).
Infection of hPSC-derived artery and vein cells with Nipah or Hendra viruses
Prior to viral infection, frozen stocks of hPSC-derived artery and vein ECs were thawed as described above and propagated on Geltrex-coated 24-well plates, 48-well plates or 8-well chamber slides (Ibidi, 80826) in their respective expansion media (EGM2 for artery ECs and EGM2 + RO4929097 + SB505124 for vein ECs) until they became fully confluent, at 4–5 days post-seeding. At that point, they were then transferred into BSL4 containment.
In each experiment, hPSC-derived artery or vein ECs were then inoculated with the same infectious quantity of a given virus. This enabled a side-by-side comparison of the responses of artery vs. vein ECs. Artery and vein ECs were inoculated with viruses for 1 hour (37°C, 5% CO2) in EGM2 medium. Different amounts of virus (titrated as described above) were used in various experiments depending on the objective of the experiment:
Visualization of syncytia formation at a fixed timepoint (Fig. 5a,b, Fig. S5e): 104 TCID50 units of Nipah or Hendra virus were added per well of a 24-well plate
Visualization of syncytia formation through live-imaging (Fig. 5c–e, Fig. S5h,i): 104 TCID50 units of Nipah or Hendra virus were added per well of an 8-well chamber slide
Quantification of viral replication kinetics (Fig. 6a, Fig. S6a): 5×103 TCID50 units of Nipah or Hendra virus were added per well of a 48-well plate
RNA-seq analysis of infected cells (Fig. 7a–d, Fig. S7a–c): 105 TCID50 units of Nipah or Hendra virus, or 30 HA units of Sendai virus, were added per well of a 24-well plate
IFNβ secretion by infected cells (Fig. 7e, Fig. S7d): 5×104 TCID50 units of Nipah or Hendra virus, or 15 HA units of Sendai virus, were added per well of a 48-well plate
After 1 hour of incubation with virus, cells were washed twice in EGM2 to remove residual virus in the media. Infected artery and vein ECs were then cultured in their respective arterial or venous expansion media for various lengths of time, as described for each individual experiment. Media was changed every 24 hours.
Assessing cytopathic effects in virally-infected hPSC-derived artery and vein cells
To visualize cytopathic effects, Diff-Quik staining (Labor+Technik, LT 001) was performed on hPSC-derived ECs that had been infected with Nipah virus, Hendra virus or left uninfected for 24 hours. For each condition, 5 fields of view were randomly selected and imaged. In each image, the number of total nuclei as well as syncytia were quantified using ImageJ/Fiji. To count multinucleated cells, ImageJ/Fiji’s cell counter was used to label and tally the total number of cells with 1 nucleus and ≥2 nuclei. To count the total number of cells, multiple ImageJ/Fiji plugins were used to process the images automatically. Images were converted to 8-bit format, and then thresholded, smoothened and filtered to remove noise (despeckled). In each image, overlapping objects were separated using the function “watershed” and finally, cells/particles were counted using the function “analyze particles”. In summary, both 1) the number of nuclei per cell and 2) the frequency of multinucleated cells relative to total cell numbers were quantified.
Live imaging of Nipah and Hendra infection in BSL4 conditions
To visualize the kinetics and extent of Nipah and Hendra virus-induced cell fusion and cell death in real time, live imaging was performed under maximum containment in the BSL4 laboratory. To quantify cell fusion, GFP+ ECs (obtained from the SUN004.1.9 hiPSC line) and wild-type ECs (obtained from the H1 hESC line) were cocultured at a 4:1 ratio, such that fusion between cells of these two genotypes would manifest as diffusion of GFP from one cell to the other.
To this end, frozen stocks of wild-type H1-derived and GFP+ SUN004.1.9-derived artery and vein ECs were thawed and cocultured as described above, on Geltrex-coated 8-well chamber slides compatible with microscopy (Ibidi, 80826) in 200 μL of their respective arterial or venous expansion media. After infection with Nipah virus (104 TCID50 units/well), Hendra virus (104 TCID50 units/well) or a negative control (uninfected), live imaging of artery and vein ECs was performed on a spinning disk confocal microscope (CorrSight, FEI) for 24 hours, with a frame rate of 5 minutes, at 37°C, 5% CO2, using a 5x objective lens. For these live imaging experiments, a high virus titer was used in order to rapidly induce cytopathic effects to visualize virally-induced cell fusion and death.
Computational analysis of virally-infected live imaging data
Time-lapse videos were imported into Fiji and then analyzed using Mastodon or Trackmate v6.0.2 (Tinevez et al., 2017). As described above, cocultures of wild-type and GFP+ cells were used for live-imaging experiments. GFP+ cells were identified as spots or blobs with an estimated blob/spot size of 20 pixels (using the Laplacian of Gaussian [LoG] filter). Then, the 20-pixel spots were filtered based on a quality score above 0.26–0.28. Next, the spots were then linked from frame to frame to build track segments and linking costs of the tracks were calculated (using LAP tracker, based on the Linear Assignment Problem mathematical framework). The parameters for frame-to-frame linking, track segment gap closing, and track segment merging were all set to a maximum distance of 15 pixels. The outputs of this analysis include the median velocity of spots and cell tracks. Cell tracks were exported as .xml files and imported into Icy (de Chaumont et al., 2012) to obtain images whereby cell tracks were overlaid over the microscope field. To compare the median velocities of infected artery vs. vein cells, the Mann Whitney non-parametric t-test was performed and two-tailed p-values calculated to determine statistical significance.
Quantification of viral replication
hPSC-derived artery and vein ECs were inoculated with Nipah and Hendra viruses for 1 hour and then washed twice in EGM2 to remove excess virus (as described above). After viral inoculation, both the extracellular levels of viral RNA (encompassing viral particles budding into the culture media) and the intracellular levels of viral RNA (encompassing intracellular viral replication) were quantified across different timepoints.
To quantify viral genomes present in the media, 140 μL of the culture media (i.e., supernatant) was placed in 560 μL of lysis buffer AVL (Qiagen QIAamp Viral RNA extraction kit, Qiagen, 52904), in addition to 560 μL of 100% ethanol, in order to inactivate it for transfer out of the BSL4 laboratory. Then, RNA was extracted following the manufacturer’s instructions. To quantify intracellular viral genomes, RNA was first extracted from cellular monolayers as described below (in the section “Bulk-population RNA-seq of virally-infected cells”). Subsequently, qRT-PCR was performed using primers directed against the viral genome and the AgPath-ID™ One-Step RT-PCR Kit (Thermo Fisher, 4387391).
Bulk-population RNA-seq of virally-infected cells
H1 hPSC-derived and SUN004.1.9 hPSC-derived artery and vein ECs in 24-well plates were inoculated with Nipah virus (105 TCID50 units/well), Hendra virus (105 TCID50 units/well) or Sendai virus, Cantell strain (30 HA units/well) or left uninfected (negative control). For RNA-seq experiments, a high viral dose (multiplicity of infection [MOI] ≈ 5) was used in order to synchronize infection in a large proportion of cells. In the same experiment, hPSC-derived artery and vein ECs from two separate genetic backgrounds (H1 hESC-derived and SUN004.1.9 hiPSC-derived) were infected to ensure the consistency of any transcriptional changes that were identified. The timepoints of 5, 10 and 15 hours post-infection were chosen because they encompassed viral entry, viral replication and virus-induced cell fusion prior to extensive cellular destruction at 24 hours post-infection induced by Nipah virus (at the viral dose used in this experiment). For each timepoint, Nipah-, Hendra- or Sendai-infected cells were transcriptionally compared against uninfected cells (negative control) harvested at the same timepoint.
Cells were harvested at 5 hours, 10 hours, and 15 hours post-infection; at each timepoint, cells were lysed in 350 μL of buffer RLT from the RNeasy Micro Kit (Qiagen, 74004) and then triturated with 350 μL of 70% ethanol to inactivate infectious material for transfer out of the BSL4 laboratory. Then, RNA was extracted as per the manufacturer’s instructions. After quality control procedures, polyadenylated mRNA was enriched using oligo(dT) beads. The mRNA was then fragmented randomly in fragmentation buffer, followed by cDNA synthesis using random hexamers and reverse transcriptase. After first-strand synthesis, a second-strand synthesis buffer (Illumina) was added with dNTPs, RNase H and E. coli polymerase I to generate the second strand by nick-translation. The final cDNA library was then subjected to a round of purification, terminal repair, A-tailing, ligation of sequencing adapters, size selection and PCR enrichment. RNA-seq libraries were sequenced on an Illumina NovaSeq 6000 sequencer by Novogene.
After obtaining RNA-seq reads, quality control was performed using FastQC (Andrews, 2010) and Trim Galore (Krueger, 2012) as described above (section “Bulk-population RNA-seq of uninfected cells”). Trimmed and filtered reads from each individual library (which was originally sequenced on multiple lanes) were then merged into a single pooled file for each library.
RNA-seq profiles of virus-infected cells contained both human and viral reads. To analyze human (host cell) transcripts, reads were aligned to the GRCh38 (hg38) Homo sapiens reference genome and gene-level RNA-seq counts were quantified using Kallisto (Bray et al., 2016). These counts were imported into RStudio and differential gene expression analyses of these counts were performed using DESeq2 (Love et al., 2014). For all samples, regularized log transformation was performed on the counts to remove the dependence of variance on the mean.
To determine the overall percentage of viral reads within a given RNA-seq sample, Kallisto (Bray et al., 2016) was used to align reads to the reference genomes of Nipah virus (NCBI accession number NC_002728), Hendra virus (NCBI accession number NC_001906) or Sendai virus (NCBI accession number NC_001552). This enabled the detection of all viral reads within a given sample. Then the number of total viral counts was divided over the number of total human + viral counts to estimate what proportion of the cellular transcriptome comprised viral transcripts.
To quantify the expression of specific viral genes, Kallisto was used to align reads to the following viral genes:
For Nipah virus, reads were aligned to Nipah F (NC_002728.1:6370–8706 nucleotides), Nipah G (NC_002728.1:8710–11255 nucleotides), Nipah L (NC_002728.1:11259–18213 nucleotides), Nipah M (NC_002728.1:5008–6366 nucleotides), Nipah N (NC_002728.1:56–2297 nucleotides) and Nipah P/V/W/C (otherwise known as Nvgp2; NC_002728.1:2301–5004 nucleotides).
For Hendra virus, reads were aligned to Hendra F (NC_001906.3:6346–8676 nucleotides), Hendra G (NC_001906.3:8680–11243 nucleotides), Hendra P/V/W/C (otherwise known as Hvgp2; NC_001906.3:2283–4980 nucleotides), Hendra L (NC_001906.3:11247–18201 nucleotides), Hendra M (NC_001906.3:4984–6342 nucleotides) and Hendra N (NC_001906.3:56–2279 nucleotides).
For Sendai virus, reads were aligned to Sendai F (NC_001552.1:4866–6563 nucleotides), Sendai HN (NC_001552.1:6693–8420 nucleotides), Sendai L (NC_001552.1:8556–15242 nucleotides), Sendai M (NC_001552.1:3669–4715 nucleotides), Sendai NP (NC_001552.1:120–1694 nucleotides) and Sendai P/V/W/C (NC_001552.1:1844–3550 nucleotides).
Quantification of IFNβ secretion by virally-infected cells
H1 hPSC-derived, SUN004.1.9 hPSC-derived and SUN004.2 hPSC-derived artery and vein ECs in 48-well plates were inoculated with Nipah virus (5×104 TCID50 units/well), Hendra virus (5×104 TCID50 units/well), Sendai virus (15 HA units/well), or left uninfected (negative control), for 1 hour. For this experiment, a high viral dose (multiplicity of infection [MOI] ≈ 5) was used in order to synchronize infection in a large proportion of cells.
After 1 hour of viral infection, cells were washed twice in EGM2 medium, and then artery and vein cells were cultured in their respective growth media in standard incubator conditions (37°C, 5% CO2). 12 and 24 hours later, supernatants (culture media) were harvested, and stored at −80 °C. Media was then thawed, and the concentration of secreted interferon-β (IFNβ) protein in the undiluted media was quantified using ELISA (enzyme-linked immunosorbent assay), specifically using the Human IFNβ Quantikine QuicKit ELISA Kit (R&D Systems, QK410) following the manufacturer’s instructions. Absorbance was quantified using a TECAN Sunrise plate reader (Tecan Trading AG, Switzerland) with a spectral filter. The concentration of IFNβ in the samples was calculated by comparing the absorbance to that of a standard curve generated with human IFNβ, after subtracting background absorbance, using TECAN Magellan software. Data shown represent the average of results from three separate cell lines (H1, SUN004.1.9 and SUN004.2 hPSCs).
Calculation of case fatality rates
The case fatality rates of both Nipah and Hendra viruses are currently estimated at ~57–59%, and are referred to as such in this manuscript.
Nipah virus: In a recent review (Ang et al., 2018), Nipah virus was recognized to have caused 326 fatalities among 573 human cases in the following regions: Malaysia and Singapore (1998–1999), India and Bangladesh (2001–2012) and the Philippines (2014). Additionally, the World Health Organization has recognized 48 fatalities among 65 new human cases in Bangladesh (2013–2015) and India (2018) (https://web.archive.org/web/20180613144643/ http://www.searo.who.int/entity/emerging_diseases/links/morbidity-and-mortality-nipah-sear-2001–2018.pdf?ua=1 ). Taken together, Nipah virus is currently recognized to have caused 374 fatalities among 638 human cases (~58.6% case fatality rate).
Hendra virus: Hendra virus is currently recognized to have caused 4 fatalities among 7 human cases in Australia (~57.1% case fatality rate) (Mahalingam et al., 2012).
QUANTIFICATION AND STATISTICAL ANALYSIS
Throughout this study, all error bars represent the standard error of the mean (s.e.m.) and a two-tailed t test was employed to determine statistical significance. Statistical significance values (P values) were reported in the figures and accompanying main text. Throughout all figures in this manuscript, wherever asterisks are used to denote statistical significance, *P<0.05 and **P<0.01, as defined in the figure legends. Statistical tests were implemented in Microsoft Excel and GraphPad Prism.
For qPCR analyses of differentiated hPSCs, for each experimental condition, two wells of cells in the same cell culture plate were analyzed (representing biological replicates) and for each of these wells, qPCR was performed twice in two separate wells of a 384-well qPCR plate (representing technical replicates). The average (mean) of these four datapoints is reported in histograms, and error bars represent the standard error of the mean.
For Diff-Quik staining of virally-infected hPSC-derived artery and vein cells, for each experimental condition, microscope images of five randomly-selected fields-of-view were generated. The number of nuclei per cell was quantified to determine syncytia formation: the average (mean) of data obtained from these five fields-of-view is reported in histograms, and error bars represent the standard error of the mean.
For qPCR analyses of viral replication in wild-type hPSC-derived artery and vein cells, for each experimental condition, three independent wells of cells in the same cell culture plate were analyzed (representing biological replicates). The average (mean) of these three datapoints is reported in histograms, and error bars represent the standard error of the mean.
For RNA-seq analyses of gene expression in virally-infected hPSC-derived artery and vein cells, for each experimental condition, two independent wells of cells in the same cell culture plate were analyzed (representing biological replicates). The average (mean) of these two datapoints is reported in histograms and heatmaps, and error bars represent the standard error of the mean.
For interferon-β secretion from virally-infected hPSC-derived artery and vein cells, for each experimental condition, three independent wells of cells in the same cell culture plate were analyzed (representing biological replicates). The average (mean) of these three datapoints is reported in histograms and heatmaps, and error bars represent the standard error of the mean.
Supplementary Material
A) BMP specifies, whereas TGFβ and WNT repress, day 2 lateral mesoderm formation; i) day 1 hPSC-derived primitive streak was treated with 100 ng/mL VEGF for 24 hours in the presence or absence of BMP4 (10–40 ng/mL) or BMP inhibitors (1 μM DMH1 or 200 ng/mL NOGGIN); ii) day 1 hPSC-derived primitive streak was treated with 10 ng/mL BMP4 + 100 ng/mL VEGF for 24 hours in the presence or absence of a WNT agonist (CHIR99021, 1–6 μM) or WNT inhibitors (1 μM C59 or 1 μM XAV939); iii) day 1 hPSC-derived primitive streak was treated with 10 ng/mL BMP4 + 100 ng/mL VEGF for 24 hours in the presence or absence of a TGFβ agonist (Activin, 5–100 ng/mL) or TGFβ inhibitors (2 μM SB505124 or SB431542); qPCR was performed on day 2 cell populations
B) VEGF treatment induces day 2 lateral mesoderm; day 1 hPSC-derived primitive streak was treated with 10 ng/mL BMP4 + 1 μM XAV939 for 24 hours, in the presence or absence of VEGF (10–100 ng/mL) or a VEGFR inhibitor (Axitinib, 10 nM); qPCR was performed on day 2 cell populations (data for FLI1 is also shown in Fig. 1c)
C) PI3K inhibitors specify day 2 lateral mesoderm; day 1 hPSC-derived primitive streak was treated with BMP + VEGF + XAV939 in the presence or absence of PI3K inhibitors (PIK90 or GDC0941) for 24 hours to generate lateral mesoderm, which was then further differentiated into artery ECs for 24 additional hours (using VEGF alone); flow cytometry was performed on day 3 artery EC populations
D) PKA agonists specify day 2 lateral mesoderm; day 1 hPSC-derived primitive streak was treated with BMP + GDC0941 + VEGF + XAV939 in the presence or absence of PKA agonists (8-bromocAMP [1 mM] or Forskolin [10 μM]) or PKA inhibitor (KT5720 [1 μM]) for 24 hours to generate day 2 candidate lateral mesoderm, which was then further differentiated into artery ECs for 24 additional hours (using GDC0941 + VEGF + XAV939); flow cytometry was performed on day 3 artery EC populations
E) Immunostaining of H1 hPSC-derived day 2 lateral mesoderm, with DAPI nuclear counterstain (bottom); scale = 100 μm
A) BMP represses day 3 artery formation; day 2 hPSC-derived lateral mesoderm was treated with Activin + GDC0941 + VEGF + XAV939 + Vitamin C for 24 hours, in the presence or absence of BMP4 (5–40 ng/mL) or BMP inhibitor (DMH1); qPCR was performed on day 3 cell populations
B) TGFβ promotes, whereas WNT and PI3K repress, day 3 artery formation from SOX17-mCherry hPSCs; i,ii) day 2 hPSC-derived lateral mesoderm was further differentiated for 24 hours with VEGF, in the presence or absence of TGFβ agonist (Activin, 10 ng/mL), TGFβ inhibitor (SB505124, 2 μM), WNT agonist (CHIR99021), or WNT inhibitor (XAV939); iii,iv) day 2 hPSC-derived lateral mesoderm was further differentiated for 24 hours with VEGF+XAV939, in the presence or absence of TGFβ agonist (Activin, 10 ng/mL) or PI3K inhibitor (GDC0941); flow cytometry was performed on day 3 cell populations
C) WNT inhibits day 3 artery formation; day 2 hPSC-derived lateral mesoderm was further differentiated with VEGF for 24 hours, in the presence or absence of WNT agonist (CHIR99021) or WNT inhibitor (XAV939); qPCR (right) was performed on day 3 cell populations
D) Definition of the set of extracellular signals necessary for efficient artery differentiation; day 2 hPSC-derived lateral mesoderm was further differentiated for 24 hours into artery cells using the complete artery specification media (red; Activin + GDC0941 + VEGF + XAV939 + AA2P + DMH1) or in conditions in which each factor was individually withheld; flow cytometry was performed on day 3 cell populations
E) Flow cytometry of SOX17-mCherry hPSCs differentiated towards artery ECs for 3 days (top); subgating on SOX17+ CD34+ ECs shows that they uniformly express VE-CADHERIN and CD31 (bottom)
F) Flow cytometry of H1, H7, H9 and SUN004.1.9 hPSCs differentiated into artery ECs for 3 days reveals that >94% of cells are CD144+ CD34+ ECs
G) Side-by-side comparison of the artery EC differentiation protocol (described in this study) against 4 prevailing methods for endothelial differentiation (Lian et al., 2014; Patsch et al., 2015; Sriram et al., 2015; Zhang et al., 2017) in the H1 and SUN004.1.9 hPSC lines; flow cytometry to assess the percentage of CD34+ CD144+ endothelial cells was performed on days 3, 5 and 6 of differentiation (n.d. = not determined)
H) qPCR timecourse of hPSCs differentiated towards artery ECs reveals sequential expression of pluripotency, primitive streak, lateral mesoderm and finally, pan-endothelial and artery-specific markers; qPCR was performed on day 0 (undifferentiated) and differentiated day 1, 1.5, 2, 2.5 and 3 cell populations, with qPCR data shown normalized to the timepoint with highest expression of each given marker gene
I) qPCR of FACS-purified day 3 SOX17+ CD34+ artery ECs generated from the SOX17-mCherry hPSC line (as well as undifferentiated hPSC controls) reveals expression of artery-specific and pan-endothelial markers, while markers of alternate cell fates are minimally expressed; qPCR data are shown relative to the reference gene YWHAZ (100% = same expression as YWHAZ)
J) Transcriptional comparison of hPSC-derived day 3 artery ECs vs. Carnegie Stage 13 (CS13) human fetal artery ECs (Zeng et al., 2019) by scRNAseq to determine the top 10 genes that were differentially expressed by in vivo vs. in vitro artery ECs (left) and gene ontology (GO) analysis was performed on all differentially expressed genes (right)
A) WNT activation on day 4 of differentiation specifies vein ECs; day 3 H1 NR2F2–2A-GFP hPSC-derived ECs were further differentiated into day 4 vein ECs (base media: SB505124 + RO4929097 + PD0325901 + AA2P), in the presence or absence of WNT inhibitor (XAV939, 1 μM) or WNT agonist (CHIR99021, 1–7 μM) for 24 hours; flow cytometry was performed on day 4 vein populations to quantify the purity of NR2F2–2A-GFP+ CD144+ vein ECs
B) Re-splitting lateral mesoderm improves vein EC differentiation; day 2 hPSC-derived lateral mesoderm was differentiated into day 4 vein ECs without resplitting (“unsplit”) or, alternatively, was dissociated and then re-seeded at high density (~79,000 cells/cm2) in day 3 differentiation media, followed by treatment with day 4 vein EC media. Flow cytometry was performed at day 4 of hPSC differentiation.
C) Strategy to generate NR2F2–2A-GFP knock-in reporter hPSCs using Cas9/AAV6 genome editing (Martin et al., 2019)
D) After genome editing (Martin et al., 2019), PCR genotyping of individual hPSC lines (derived from single colonies) reveals successful integration of the NR2F2–2A-GFP knock-in reporter allele
E) Immunostaining of undifferentiated H1 hPSCs, with DAPI nuclear counterstain (negative control for Fig. 3e); scale = 100 μm
F) Transcriptional comparison of hPSC-derived day 4 vein ECs vs. Carnegie Stage 13 (CS13) human fetal vein ECs (Zeng et al., 2019) by scRNAseq; gene expression is quantified in UMI expression units (loge scale)
G) Transcriptional comparison of hPSC-derived day 4 artery ECs vs. Carnegie Stage 13 (CS13) human fetal vein ECs (Zeng et al., 2019) by scRNAseq to determine the top 10 genes that were differentially expressed by in vivo vs. in vitro vein ECs
H) Transcriptional comparison of hPSC-derived day 4 artery ECs vs. Carnegie Stage 13 (CS13) human fetal vein ECs (Zeng et al., 2019) by scRNAseq to determine gene ontology (GO) terms associated with genes differentially expressed in vivo and in vitro
A) scRNAseq of H1 hPSC-derived day 3 artery vs. day 4 vein ECs (CD144+ FACS sorted) reveals mutually-exclusive expression of hallmark arteriovenous marker genes
B) hPSC-derived day 3 artery ECs and day 4 vein ECs do not express known markers of organ-specific EC identities (Kalucka et al., 2020), as shown by scRNAseq (as positive controls, expression of reference genes YWHAZ, ACTB and GAPDH is shown); dot size indicates percentage of cells expressing a given marker and dot color reflects the level of gene expression
C) H1 hPSC-derived artery and vein ECs were either mixed with adult human lung fibroblasts (ratio of 1 EC:2 fibroblasts) or not mixed with fibroblasts, and then subsequently cultured in a 3D fibrin gel with EGM2 media for 1 week in vitro (Kurokawa et al., 2017). Only hPSC-derived ECs mixed with fibroblasts formed 3D networks, as assessed by CD31 immunostaining, demonstrating the importance of fibroblast support for vascular network formation in this experimental system; scale = 500 μm
D) SUN004.1.9 hPSC-derived artery ECs were mixed with adult human lung fibroblasts from two different individuals (#1 and #2), and then subsequently cultured in a 3D fibrin gel with EGM2 media for 1 week in vitro (Kurokawa et al., 2017). hPSC-derived ECs mixed with fibroblasts from both individuals formed 3D networks, as assessed by CD31 immunostaining; scale = 200 μm
E) H1 hPSC-derived day 3 artery ECs and day 4 vein ECs were stained for LDLR expression (left), or cultured with LDL DyLight 550 for 4 hours in their respective arterial or venous expansion media (right), prior to flow cytometry.
F) H1 hPSC-derived day 3 artery ECs and day 4 vein ECs were each expanded for 24 hours in their respective arterial or venous expansion media until they formed confluent monolayers, and then were scratched with a pipette tip. Serial imaging was performed over the next 48 hours to quantify closure of the cell monolayer; scale = 100 μm
G) SUN004.1.9 hPSC-derived artery and vein ECs exposed to shear stress or static conditions for 24 hours turn on different genes, as shown by qPCR; gene expression normalized such that for each gene, the sample expressing the highest level of the gene has an expression level = 1.0
H) H1 hPSC-derived artery and vein ECs were thawed and expanded for 6 days in their respective arterial or venous expansion media, and then were subsequently treated with TNFα (10 ng/mL) or left untreated for 4 hours. Flow cytometry was performed to assess TNFα-induced changes in immune-cell adhesion molecules CD54/ICAM1, CD62E/E-SELECTIN and CD106/VCAM1.
A) hPSC-derived artery and vein ECs can be expanded for 6 days in vitro while retaining mutually-exclusive expression of arteriovenous subtype markers. SUN004.1.9 hPSCs were differentiated into day 3 artery and day 4 vein ECs (“day 0” of expansion), frozen, and which were then respectively thawed in artery expansion media (EGM2) or vein expansion media (EGM2 + RO4929097 (2 μM; NOTCH inhibitor) + SB505124 (2 μM; TGFβ inhibitor)); qPCR of expanded artery and vein ECs was performed at multiple timepoints post-thawing, and mRNA expression is shown relative to the expression of reference gene YWHAZ (i.e., YWHAZ = 1.0)
B) Continued inhibition of artery-specifying signals TGFβ and NOTCH signals is required to expand hPSC-derived vein ECs in vitro. SUN004.1.9 hPSCs were differentiated into artery or vein ECs (“day 0” of expansion), and then frozen; vein ECs were then thawed in EGM2, or in the absence of hydrocortisone (HC) and/or presence of RO4929097 (2 μM; NOTCH inhibitor) and/or SB505124 (2 μM; TGFβ inhibitor) for 4 additional days; qPCR was then performed to determine mRNA expression, which is shown relative to the expression of reference gene YWHAZ (i.e., YWHAZ = 1.0)
C) hPSC-derived artery ECs can be expanded for 8 days in vitro. hPSCs were differentiated into artery ECs in 3 days (“day 0” of expansion), and then subsequently expanded in EGM2 for up to 8 days. Flow cytometry confirmed continued expression of pan-EC markers CD34 and CD144 and arterial marker DLL4.
D) hPSC-derived artery and vein ECs activate interferon signaling in response to a double-stranded RNA mimic, poly(I:C). SUN004.1.9 hPSCs were differentiated into artery or vein ECs, frozen, and then thawed and cultured in their respective expansion media (EGM2 for artery ECs and EGM2 + RO4929097 + SB505124 for vein ECs) for 5 days. Then, poly(I:C) (50 μg/mL) was added to the respective culture media for 24 hours and qPCR was performed, with mRNA expression shown relative to the expression of reference gene YWHAZ (i.e., YWHAZ = 1.0); **P<0.01
E) H1 hPSC-derived artery and vein ECs were inoculated with Nipah or Hendra viruses for 24 hours, and then fixed and stained with Diff-Quik to assess cytopathic effects; total cell number (including mononucleated and multinucleated cells) was then quantified and normalized to uninfected controls; n.s. = not significant
H) Cocultures of GFP+ and wild-type hPSC-derived artery and vein ECs were inoculated with either Nipah or Hendra virus; live imaging was conducted over an 8-hour interval. The trajectory of single GFP+ cells was computationally tracked by live imaging, with each single-cell trajectory colored differently; scale bar = 200 μm
I) Cocultures of GFP+ and wild-type hPSC-derived artery and vein ECs were inoculated with either Nipah or Hendra virus; live imaging was conducted over an 8-hour interval. The trajectory of single GFP+ cells was computationally tracked by live imaging, revealing two features: 1) increasing numbers of GFP+ cells (reflecting fusion of GFP+ cells with formerly wild-type cells, and leading to an apparent increase in GFP+ cells) and 2) fusion (where multiple cell tracks merged)
A) Intracellular levels of Nipah and Hendra viral RNA after inoculating H1 hPSC-derived artery and vein ECs with Nipah and Hendra viruses, as quantified by qPCR of the cell monolayer (after removing the culture media); *P<0.05; **P<0.01; †extensive cell loss
B) Significantly higher expression of EFNB2 in hPSC-derived artery ECs relative to vein ECs. hPSC-derived artery and vein ECs generated from H1, SUN004.1.9 and SUN004.2 hPSC lines were frozen, and then thawed in their respective arterial or venous expansion media for 3–6 days prior to qPCR. EFNB2 expression is shown relative to the expression of reference gene YWHAZ (i.e., YWHAZ = 1.0); **P<0.05.
C) scRNAseq of hPSC-derived artery and vein ECs revealed that among all EFNA and EFNB family members, EFNB2 was most highly expressed in artery ECs, whereas EFNB3 (another candidate entry-receptor for Nipah and Hendra viruses (Negrete et al., 2007)) was minimally expressed.
D) Primary human artery and vein ECs cultured in standard conditions relinquish arteriovenous identity in vitro. Human umbilical artery and vein ECs were expanded in EGM2, and compared against hPSC-derived day 3 artery ECs and day 4 vein ECs by qPCR.
E) Cas9-mediated deletion of EFNB2 in hPSCs, as shown by i) sequencing of the EFNB2 gene in H1 wild-type and EFNB2−/− hPSCs, revealing a 1-base deletion in both EFNB2 alleles in the latter hPSC line and ii) flow cytometry for EFNB2 expression in H1 wild-type and EFNB2−/− hPSCs prior to and after differentiation into artery ECs
F) Nipah pseudovirus preferentially enters hPSC-derived artery ECs, by comparison to vein ECs. hPSC-derived artery and vein ECs were thawed, expanded for 3 days in respective arterial or venous expansion media, and then transduced with GFP-encoding lentiviruses pseudotyped with Nipah or VSV envelope proteins for 24 hours. 48 hours later, flow cytometry was performed to enumerate the percentage of GFP+ ECs, revealing arterially-biased tropism of Nipah-pseudotyped lentivirus. As a negative control, VSV-G-pseudotyped lentivirus did not exhibit arterially-biased tropism; **P<0.01
G) EFNB2 is dispensable for VSV pseudovirus entry. H1 wild-type and EFNB2−/− hPSC-derived artery and vein ECs were thawed, expanded for 3 days, and then infected with equal infectious units of GFP-encoding, non-replicating lentiviruses pseudotyped with VSV envelope proteins. Flow cytometry was performed 3 days later, showing that EFNB2 is dispensable for infection by VSV pseudovirus; **P<0.01; n.s. = not significant
A) RNA-seq analysis of hPSC-derived artery ECs infected with either Nipah, Hendra or Sendai virus for 5, 10 or 15 hours, followed by alignment of viral reads to viral transcripts, confirms the known 3’to-5’ transcriptional gradient along the paramyxovirus genome, with 3’ most gene (N) expressed at the highest level, followed by decreasing expression of progressively more 5’ genes. The y-axis represents viral mRNA expression relative to the entire transcriptome of infected artery ECs (100% = all human + viral reads). Average data from H1 and SUN004.1.9 hPSCs shown.
B) Expression of each individual viral gene in hPSC-derived artery ECs infected with either Nipah, Hendra or Sendai virus. Percentage indicates the levels at which each individual viral gene (e.g., N) was expressed relative to the entirety of all viral gene expression (100% = all viral reads). Data was averaged from H1 and SUN004.1.9 hPSCs infected for 5, 10 or 15 hours.
C) Differentially expressed genes between infected vs. uninfected artery ECs by RNA-seq, 10 and 15 hours post-infection; averages of H1 and SUN004.1.9 hPSC-derived cell-types shown.
D) IFNβ secretion by hPSC-derived artery ECs as quantified by ELISA; averaged from H1, SUN004.1.9 and SUN004.2 hPSCs; *P<0.01; n.s. = not significant
Movie S1. Live-imaging of cocultured wild-type and GFP+ artery or vein ECs infected by Nipah or Hendra viruses, related to Figure 5.
Table S1. Gene expression differences between hPSC-derived vs. human fetal artery ECs, as assayed by single-cell RNA-seq, related to Figure 2. Single-cell RNA-seq of H1 hPSC-derived artery ECs in vitro vs. Carnegie Stage 13 human fetal artery ECs in vivo. Genes that are differentially expressed between in vitro vs. in vivo artery ECs are shown, and loge fold change is shown (i.e., positive fold change indicates higher expression in hPSC-derived cells, relative to in vivo cells). For each gene, “pct.1” and “pct.2” shows the percentage of cells that detectably express that gene, among H1 hPSC-derived artery ECs and human fetal artery ECs, respectively.
Table S2. Gene expression differences between hPSC-derived vs. human fetal vein ECs, as assayed by single-cell RNA-seq, related to Figure 3. Single-cell RNA-seq of H1 hPSC-derived vein ECs in vitro vs. Carnegie Stage 13 human fetal vein ECs in vivo. Genes that are differentially expressed between in vitro vs. in vivo vein ECs are shown, and loge fold change is shown (i.e., positive fold change indicates higher expression in hPSC-derived cells, relative to in vivo cells.) For each gene, “pct.1” and “pct.2” shows the percentage of cells that detectably express that gene, among H1 hPSC-derived vein ECs and human vein artery ECs, respectively.
Table S3. Differentially-expressed genes between hPSC-derived artery vs. vein ECs, as assayed by bulk-population RNA-seq, related to Figure 4. Bulk-population RNA-seq of H1 hPSC-derived day 3 artery ECs vs. day 4 vein ECs (both FACS sorted for CD144+ cells). All counts > 1 were normalized to the total read count and log2 transformed using Limma-Voom.
Table S4. Bulk-population RNA-seq of hPSC-derived artery and vein cells infected by Nipah, Hendra or Sendai viruses, related to Figure 7. H1 and SUN004.1.9 hPSC-derived artery and vein ECs were inoculated with Nipah, Hendra or Sendai viruses for 5, 10 or 15 hours, and bulk-population RNA-seq was performed. Replicate 1 = H1 and replicate 2 = SUN004.1.9. Counts were regularized, log2 transformed and normalized by size factors using DESeq2.
KEY RESOURCES TABLE
| Reagent or Resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| anti-CD144 Alexa Fluor 647 antibody | BD | 561567 |
| anti-human CD73 (Ecto-5'-nucleotidase) PE | Biolegend | 344004 |
| anti-human Delta-like protein 4 (DLL4) APC | Biolegend | 346508 |
| Anti-NR2F2 antibody | Abcam | ab211777 (1:100) |
| Anti-SOX17 antibody | R&D Systems | AF-1924 (1:1000) |
| Anti-CD31 | BD Biosciences | 555444 |
| Anti-ERG (ETS related gene) | Abcam | ab92513 (1:500) |
| Mouse Anti-Human CD144 (VE-Cad) | BD Bioscience | 555661 (1:500) |
| Anti-EFNB2 | R&D Systems | AF496 (1:5) |
| APC Donkey Anti-Goat | R&D Systems | F0108 (1:10) |
| Alexa Fluor 568 Donkey Anti-rabbit | Thermo Fisher Scientific | 1:250 |
| Alexa Fluor 647 Donkey Anti-mouse | Thermo Fisher Scientific | 1:250 |
| Bacterial and viral strains | ||
| Nipah virus (Malaysia variant): Nipah virus/human/MY/1999/ CDC | Centers for Disease Control and Prevention, Viral Special Pathogens Branch | (Harcourt et al., 2000) |
| Hendra virus (Brisbane variant): Hendra virus/horse/AU/1994/Hendra | Centers for Disease Control and Prevention, Viral Special Pathogens Branch | (Murray et al., 1995) |
| Sendai virus (Cantell strain) | ATCC | VR-907 |
| Sendai virus (recombinant, replication-defective, encoding KLF4/OCT4/SOX2/MYC and miR-302 target site) | Dr. Mahito Nakanishi, National Institute of Advanced Industrial Science and Technology | (Nishimura et al., 2017) |
| NEB® Stable Competent E. coli | New England BioLabs | C3040H |
| Chemicals, peptides and recombinant proteins | ||
| mTesR Plus medium | STEMCELL Technologies | #100–0276 |
| Penicillin/streptomycin | Thermo Fisher Scientific | 15–140-122 |
| KnockOut Serum Replacement | Thermo Fisher Scientific | 10828028 |
| MEM Non Essential Amino Acids Solution (100X) | Thermo Fisher Scientific | 11–140-050 |
| Geltrex LDEV-Free, hESC-Qualified, Reduced Growth Factor Basement Membrane Matrix | Thermo Fisher Scientific | A1413302 |
| Recombinant Human Vitronectin Protein, Truncated (VTN-N) | Gibco | A14700 |
| Matrigel Matrix GFR Phenol RF | BD Biosciences | 356231 |
| Gelatin Solution | Sigma | ES-006-B |
| Versene solution | Thermo Fisher Scientific | 15040066 |
| Trypsin-EDTA, 0.05% | Thermo Fisher Scientific | 25300054 or 25300062 |
| Accutase - Enzyme Cell Detachment Medium | Thermo Fisher Scientific | 00–4555-56 |
| TrypLE Express Enzyme (1X) | Thermo Fisher Scientific | 12604013 |
| Fetal Bovine Serum Premium Select | Atlanta Biologicals | S11550H |
| 2-mercaptoethanol | Gibco | 21985023 |
| RPMI 1640 Medium, GlutaMAX | Thermo Fisher Scientific | 61870036 |
| DMEM | Sigma | D6429 |
| DMEM/F12 + GlutaMAX | Gibco | 10565042 |
| IMDM + GlutaMAX | Thermo Fisher | 31980–097 |
| F12 + GlutaMAX | Thermo Fisher | 31765–092 |
| Polyvinyl alcohol | Sigma | P8136–250G |
| Chemically defined lipid concentrate | Thermo Fisher | 11905–031 |
| 1-thioglycerol | Sigma | M6145–100ML |
| Recombinant human insulin | Sigma | 11376497001 |
| Recombinant human VEGF | R&D Systems | 293-VE-0500 |
| Recombinant human FGF2 | R&D Systems | 233-FB-01M |
| Human transferrin | Sigma | 10652202001 |
| Recombinant human BMP4 | R&D Systems | 314-BP-050 |
| Penicillin/streptomycin | Thermo Fisher | 15070–063 |
| Recombinant Activin | R&D Systems | 338-AC-500/CF |
| GDC-0941 | Cellagen Technology | C4321–25 |
| Forskolin | Tocris | 1099 |
| Thiazovivin | Tocris | 3845 |
| XAV939 | Tocris | 3748 |
| Ascorbic acid-2-phosphate | Sigma | 49752–10G |
| DMH1 | Tocris | 4126 |
| SB505124 | Tocris | 3263 |
| RO4929097 | Cellagen Technology | C7649–10 |
| CHIR99201 | Tocris | 4423 |
| PD0325901 | Tocris | 4192 |
| PIK90 | Calbiochem | 528117 |
| 8-bromo-cAMP | Tocris | 1140 |
| KT5720 | Calbiochem | 420320 |
| GM-CSF | Peprotech | 100–11 |
| IL-3 | Peprotech | 200–03 |
| IL-6 | Peprotech | 200–06 |
| FLT3L | Peprotech | 300–19 |
| TPO | R&D Systems | 288-TP-005/CF |
| SCF | Peprotech | 300–07 |
| Endothelial Cell Growth Medium 2 (EGM2) | Lonza | CC-3162 |
| Endothelial Cell Growth Basal Medium 2 (EBM2) | Lonza | CC-3156 |
| Fibroblast Growth Medium 2 (FGM2) | Lonza | CC-3132 |
| Fibrinogen solution | Sigma Aldrich | F8630 |
| Thrombin | Sigma Aldrich | T3399 |
| Cytodex-3 beads | Sigma Aldrich | C3275 |
| Apelin-13 | Tocris | 2420 |
| Transwell polycarbonate membrane cell culture inserts, 6.5 mm Transwell with 8.0 μm pore polycarbonate membrane insert, 24-well | Corning | 3422 |
| Lipofectamine 3000 | Thermo Fisher Scientific | L3000001 |
| Low-density lipoprotein Dylight 550 | Cayman Chemical | 10011125 |
| Acetylated Alexa Fluor 488 conjugate low-density lipoprotein | Thermo Fisher Scientific | L-23380 |
| Recombinant Human TNFα | R&D Systems | 210-TA-020 |
| FuGENE® HD Transfection Reagent | Promega | E2311 |
| Poly-L-lysine | R&D Systems | 3438–200-01 |
| LV-MAX Lentiviral Production System | GibcoGibco | A35684 |
| Bovine serum albumin (BSA) | Sigma | A2153 |
| Tween-20 | Sigma | P9416 |
| 10% Formalin | Sigma | F5554 |
| Fibrinogen | Sigma-Aldrich | F8630 |
| Thrombin | Sigma-Aldrich | T4648 |
| Vectashield | Vector Labs | H-100 |
| Glutamax | Gibco | 35050061 |
| STEMdiff™ APEL™ | StemCell Technologies | 05270 |
| CP21R7 | Selleckchem | S7954 |
| Neurobasal medium | Thermo Fisher Scientific | 21103049 |
| B27 | Thermo Fisher Scientific | 17504044 |
| N2 | Thermo Fisher Scientific | 17502001 |
| StemPro-34 SFM medium | Thermo Fisher Scientific | 10639011 |
| Essential 8 media | Thermo Fisher Scientific | A1517001 |
| SB431542 | Tocris | 1614 |
| Resveratrol | R&D Systems | 1418/100 |
| L690 | R&D Systems | 0682/5 |
| Poly(I:C) sodium salt | Sigma | P1530 |
| Critical commercial assays | ||
| Human IFNβ Quantikine QuicKit ELISA | R&D Systems | QK410 |
| RNeasy Plus Mini Kit | Qiagen | 10010049 |
| QIAamp Viral RNA kit | Qiagen | 52904 |
| RNeasy Micro kit | Qiagen | 74004 |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems | 4368814 |
| AgPath-ID™ One-Step RT-PCR Kit | Thermo Fisher | 4387391 |
| SYBRGreen | Thomas Scientific | BIO-94050 |
| 10x Genomics Chromium single cell 3’ GEM reagent kit (v3) | 10x Genomics | 1000075 |
| Chromium Chip B Single Cell Kit 48rxns | 10x Genomics | 1000073 |
| LEGENDScreen PE-Conjugated Human Antibody Plates | Biolegend | 700007 |
| Magnetically Activated Cell Sorting (MACS) | Miltenyi Biotec | 130–046-703 |
| MACS LS column | Miltenyi Biotech | 130–042-401 |
| Fc blocking reagent | Miltenyi Biotec | 130–046-703 |
| Triton X-100 | Sigma | X100–500ML |
| PBS, pH 7.4 | Thermo Fisher | 10010049 |
| DPBS without ions | Thermo Fisher | 14190144 |
| Diff-Quik stain | Labor+Technik | LT 001 |
| 8-well chambered coverslip for live imaging | Ibidi | 80826 |
| CellTrace CFSE Cell proliferation kit | Thermo Fisher Scientific | C34554 |
| Molecular Probes DAPI (4',6 Diamidino 2 Phenylindole, Dihydrochloride) | Thermo Fisher Scientific | D1306 |
| Experimental models: cell lines | ||
| H1 hESCs | WiCell | WiCell, WA01 |
| H7 hESCs | WiCell | WiCell, WA07 |
| H9 hESCs | WiCell | WiCell, WA09 |
| H1 NR2F2–2A-GFP knock-in reporter hESCs | This study | N/A |
| H1 SOX17–2A-mPlum knock-in reporter hESCs | This study | N/A |
| H9 SOX17-mCherry knock-in reporter hESCs | This study | (Loh et al., 2014) |
| SUN004.1.9 CAG-GFP hiPSCs | This study | N/A |
| SUN004.2 CAG-tdTomato hiPSCs | This study | N/A |
| SUN003.1 CAG-AkaLuciferase-tdTomato hiPSCs | This study | N/A |
| Human umbilical artery endothelial cells (HUAEC) | Sigma-Aldrich | 202–05N |
| Human umbilical vein endothelial cells (HUVEC) | Sigma-Aldrich | 200–05N |
| Normal human lung fibroblast cells (NHLF) | Lonza | CC-2512 |
| THP-1 cells | ATCC | TIB-202 |
| HEK 293T/17 cells | ATCC | CRL-11268 |
| Vero C1008 (Vero 76, clone E6, Vero E6) | ECACC | 85020206 |
| Experimental models: organisms/strains | ||
| Mus musculus (mouse): NOD-SCID Il2rg−/− strain | The Jackson Laboratory | 005557 |
| Oligonucleotides | ||
| Primers | See Table S5 | |
| Recombinant DNA | ||
| pCAGGS-coNiV-G-HA | Shreyas Kowdle and Benhur Lee, Icahn School of Medicine at Mount Sinai | (Palomares et al., 2013) |
| pcDNA3.1+coNiV-F T5F-AU1 | Shreyas Kowdle and Benhur Lee, Icahn School of Medicine at Mount Sinai | (Palomares et al., 2013) |
| pCAGGS-coHeV-G-HA | Shreyas Kowdle and Benhur Lee, Icahn School of Medicine at Mount Sinai | (Negrete et al., 2007) |
| pCAGGS-coHeV-F-AU1 | Shreyas Kowdle and Benhur Lee, Icahn School of Medicine at Mount Sinai | (Negrete et al., 2007) |
| pVSV-G | Owen Witte, University of California Los Angeles | (Goldstein et al., 2011) |
| pMDL | Owen Witte, University of California Los Angeles | (Goldstein et al., 2011) |
| pREV | Owen Witte, University of California Los Angeles | (Goldstein et al., 2011) |
| pRS348 EF1A-GFP; mU6-CD34 sgRNA | Reuben Saunders and Jonathan Weissman, Whitehead Institute/MIT | N/A |
| PiggyBac CAG-GFP donor plasmid | Austin Smith, University of Cambridge | (Takashima et al., 2014) |
| CMV-PBase plasmid | Austin Smith, University of Cambridge | (Takashima et al., 2014) |
| pCS-CAG-mScarlet lentiviral plasmid | This study | N/A |
| pCS-CAG-AkaLuciferase-tdTomato lentiviral plasmid | This study | N/A |
| Software and algorithms | ||
| ImageJ/Fiji | (Schindelin et al., 2012) | https://imagej.net/software/fiji/ |
| Seurat v3 | (Stuart et al., 2019) | https://satijalab.org/seurat/ |
| Ggplot2 | (Wickham, 2016) | https://ggplot2.tidyverse.org/ |
| Tidyverse | (Wickham et al., 2019) | https://www.tidyverse.org/ |
| Dplyr | (Wickham et al., 2022) | https://dplyr.tidyverse.org/ |
| Limma | (Ritchie et al., 2015) | https://bioconductor.org/packages/release/bioc/html/limma.ht |
| Voom | (Law et al., 2014) | https://rdrr.io/bioc/limma/man/voom.html |
| EdgeR | (Robinson et al., 2010) | http://bioconductor.org/packages/release/bioc/html/edgeR.html |
| RStudio | RStudio Team | https://www.rstudio.com/ |
| Icy | (de Chaumont et al., 2012) | http://icy.bioimageanalysis.org/ |
| Kallisto | (Bray et al., 2016) | https://pachterlab.github.io/kallisto/ |
| Cell Ranger | 10x Genomics | https://support.10xgenomics.com/single-cell-geneexpression/software/pipelines/latest/what-is-cell-ranger |
| DESeq2 | (Love et al., 2014) | https://github.com/mikelove/DESeq2 |
| SCTransform | (Hafemeister and Satija, 2019) | https://github.com/satijalab/sctransform |
| Enrichr | (Chen et al., 2013; Kuleshov et al., 2016) | https://maayanlab.cloud/Enrichr/ |
| FastQC | (Andrews, 2010) | http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Trim Galore | (Krueger, 2012) | http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
| Trackmate v6.0.2 | (Tinevez et al., 2017) | https://imagej.net/plugins/trackmate/ |
| ShinyCell | (Ouyang et al., 2021) | https://github.com/SGDDNB/ShinyCell |
HIGHLIGHTS.
Efficient, rapid generation of >90% pure human artery and vein cells in 3–4 days
Precisely inducing artery cells by blocking vein-specifying cues and vice versa
Nipah and Hendra viruses preferentially infect artery (rather than vein) cells
Human pluripotent stem cells provide a new platform for biosafety-level-4 virology
ACKNOWLEDGMENTS
We thank A. Elefanty, E. Stanley and E. Ng for MIXL1-GFP and SOX17-mCherry hPSCs; the CDC Viral Special Pathogens Branch and M. Nakanishi for viruses; B. Lee, J. Weissman, A. Smith and O. Witte for plasmids; P. Rios, T. Nishimura, N. Pek, S. Miao, M. Gu and M. Rabinovitch for contributing to preliminary experiments; and M. Nichane, M.R. Corces, A. Kershner, A. Chang, I. Williams, P. Beachy, R. Nusse, K. Davis, B. Azizoglu, R. Saunders, W. Allen, S. Zheng, M. Blewett, I. Weissman, A. McCarty and J. Choi for reagents and advice. Infrastructure support was provided by the Stanford Stem Cell FACS Core and Animal Facility, Stanford Diabetes Genomics Analysis Core Facility (NIH P30DK116074), C. Carswell-Crumpton, M. Alvarez and L. Dunkin-Hubby. This work was supported by NIH (DP5OD024558 and R01HL128503), JDRF Northern California Center of Excellence, Additional Ventures Innovation Fund, Stanford Ludwig and Beckman Centers, Stanford Maternal and Child Health Research Institute, A*STAR Industrial Alignment Fund (H18/01/a0/017), Robert Koch Institute intramural funds, and the Anonymous, Fickel and Weintz families. This work was also supported by CIRM Bridges Program TB1-01195 (A.C. and A.T.N.); NIH T32GM119995 (R.M.M., J.L.F.) and T32GM007365 (T.C.); NDSEG and Stanford Bio-X Fellowships (J.L.F.); and the Assar Gabrielsson Foundation and Swedish Research Council (20170034, J.B.). L.T.A. is a Siebel Investigator. K.R.H. is an HHMI Investigator. K.M.L. and C.C. are Human Frontier Science Program Young Investigators (RGY0069/2019). K.M.L. is a Packard Foundation Fellow, Pew Scholar, Baxter Foundation Faculty Scholar and The Anthony DiGenova Endowed Faculty Scholar.
Footnotes
DECLARATION OF INTERESTS
K.M.L., L.T.A., A.C., A.T.N. and J.L.F. have filed patent applications related to endothelial differentiation.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aljofan M, Saubern S, Meyer AG, Marsh G, Meers J, and Mungall BA (2009). Characteristics of Nipah virus and Hendra virus replication in different cell lines and their suitability for antiviral screening. Virus research 142, 92–99. 10.1016/j.virusres.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S.(2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Ang BSP, Lim TCC, and Wang L.(2018). Nipah Virus Infection. Journal of Clinical Microbiology 56, e01875–01817-01810. 10.1128/JCM.01875-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranguren XL, Agirre X, Beerens M, Coppiello G, Uriz M, Vandersmissen I, Benkheil M, Panadero J, Aguado N, Pascual-Montano A, et al. (2013). Unraveling a novel transcription factor code determining the human arterial-specific endothelial cell signature. Blood 122, 3982–3992. 10.1182/blood-2013-02-483255. [DOI] [PubMed] [Google Scholar]
- Arunkumar G, Chandni R, Mourya DT, Singh SK, Sadanandan R, Sudan P, Bhargava B, and Group N.I.P.a.H.S. (2019). Outbreak Investigation of Nipah Virus Disease in Kerala, India, 2018. The Journal of infectious diseases 219, 1867–1878. 10.1093/infdis/jiy612. [DOI] [PubMed] [Google Scholar]
- Augustin HG, and Koh GY (2017). Organotypic vasculature: From descriptive heterogeneity to functional pathophysiology. Science 357. 10.1126/science.aal2379. [DOI] [PubMed] [Google Scholar]
- Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, and Wishart DS (2016). Heatmapper: web-enabled heat mapping for all. Nucleic Acids Research 44, W147–153. 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baseler L, de Wit E, Scott DP, Munster VJ, and Feldmann H.(2015). Syrian hamsters (Mesocricetus auratus) oronasally inoculated with a Nipah virus isolate from Bangladesh or Malaysia develop similar respiratory tract lesions. Veterinary pathology 52, 38–45. 10.1177/0300985814556189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, Ginhoux F, and Newell EW (2018). Dimensionality reduction for visualizing single-cell data using UMAP. Nature Biotechnology 37, 38–44. 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- Bonaparte MI., Dimitrov AS., Bossart KN., Crameri G., Mungall BA., Bishop KA., Choudhry V., Dimitrov DS., Wang L-F., Eaton BT., and Broder CC. (2005). Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proceedings of the National Academy of Sciences 102, 10652–10657. 10.1073/pnas.0504887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, and Pachter L.(2016). Near-optimal probabilistic RNA-seq quantification. Nature Biotechnology 34, 525–527. 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Cantell K, and Hirvonen S.(1981). Preparation and assay of Sendai virus. Methods in enzymology 78, 299–301. 10.1016/0076-6879(81)78131-x. [DOI] [PubMed] [Google Scholar]
- Carmeliet P.(2005). Angiogenesis in life, disease and medicine. Nature 438, 932–936. 10.1038/nature04478. [DOI] [PubMed] [Google Scholar]
- Casie Chetty S, Rost MS, Enriquez JR, Schumacher JA, Baltrunaite K, Rossi A, Stainier DYR, and Sumanas S.(2017). Vegf signaling promotes vascular endothelial differentiation by modulating etv2 expression. Developmental Biology 424, 147–161. 10.1016/j.ydbio.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC, and NIH. (2020). Biosafety in Microbiological and Biomedical Laboratories—6th Edition (U.S. Department of Health and Human Services; ). [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, and Ma'ayan A (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128–114. 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs K, Stock N, Ross C, Andrejeva J, Hilton L, Skinner M, Randall R, and Goodbourn S.(2007). mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology 359, 190–200. 10.1016/j.virol.2006.09.023. [DOI] [PubMed] [Google Scholar]
- Chong DC, Koo Y, Xu K, Fu S, and Cleaver O.(2011). Stepwise arteriovenous fate acquisition during mammalian vasculogenesis. Developmental Dynamics 240, 2153–2165. 10.1002/dvdy.22706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua KB, Bellini WJ, Rota PA, Harcourt BH, Tamin A, Lam SK, Ksiazek TG, Rollin PE, Zaki SR, Shieh W, et al. (2000). Nipah virus: a recently emergent deadly paramyxovirus. Science 288, 1432–1435. 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- Ciancanelli MJ, Volchkova VA, Shaw ML, Volchkov VE, and Basler CF (2009). Nipah virus sequesters inactive STAT1 in the nucleus via a P gene-encoded mechanism. Journal of Virology 83, 7828–7841. 10.1128/JVI.02610-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciau-Uitz A, Pinheiro P, Kirmizitas A, Zuo J, and Patient R.(2013). VEGFA-dependent and - independent pathways synergise to drive Scl expression and initiate programming of the blood stem cell lineage in Xenopus. Development 140, 2632–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corada M, Orsenigo F, Morini MF, Pitulescu ME, Bhat G, Nyqvist D, Breviario F, Conti V, Briot A, Iruela-Arispe ML, et al. (2013). Sox17 is indispensable for acquisition and maintenance of arterial identity. Nature Communications 4, 2609. 10.1038/ncomms3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coultas L, Chawengsaksophak K, and Rossant J.(2005). Endothelial cells and VEGF in vascular development. Nature 438, 937–945. 10.1038/nature04479. [DOI] [PubMed] [Google Scholar]
- Cradick TJ, Qiu P, Lee CM, Fine EJ, and Bao G.(2014). COSMID: A Web-based Tool for Identifying and Validating CRISPR/Cas Off-target Sites. Mol Ther Nucleic Acids 3, e214. 10.1038/mtna.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RP, Ng ES, Costa M, Mossman AK, Sourris K, Elefanty AG, and Stanley EG (2008). Targeting a GFP reporter gene to the MIXL1 locus of human embryonic stem cells identifies human primitive streak-like cells and enables isolation of primitive hematopoietic precursors. Blood 111, 1876–1884. [DOI] [PubMed] [Google Scholar]
- de Chaumont F, Dallongeville S, Chenouard N, Herve N, Pop S, Provoost T, Meas-Yedid V, Pankajakshan P, Lecomte T, Le Montagner Y, et al. (2012). Icy: an open bioimage informatics platform for extended reproducible research. Nat Methods 9, 690–696. 10.1038/nmeth.2075. [DOI] [PubMed] [Google Scholar]
- De Val S, and Black BL (2009). Transcriptional control of endothelial cell development. Dev Cell 16, 180–195. 10.1016/j.devcel.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBuysscher BL, de Wit E, Munster VJ, Scott D, Feldmann H, and Prescott J.(2013). Comparison of the pathogenicity of Nipah virus isolates from Bangladesh and Malaysia in the Syrian hamster. PLoS neglected tropical diseases 7, e2024. 10.1371/journal.pntd.0002024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditadi A., Sturgeo CM., Tobe J., Awon G., Kenned M., Yzaguirr AD., Azzol L., N ES., Stanle EG., Frenc DL., et al. (2015). Human definitive haemogenic endothelium and arterial vascular endothelium represent distinct lineages. Nature Cell Biology 17, 580–591. 10.1038/ncb3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, and Hyman AA (2017). Stem cells: the new “model organism”. Molecular Biology of the Cell 28, 1409–1411. 10.1091/mbc.E17-03-0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte A, Hirashima M, Benedito R, Trindade A, Diniz P, Bekman E, Costa L, Henrique D, and Rossant J.(2004). Dosage-sensitive requirement for mouse Dll4 in artery development. Genes & Development 18, 2474–2478. 10.1101/gad.1239004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis EL, and Delbrück M.(1939). THE GROWTH OF BACTERIOPHAGE. The Journal of general physiology 22, 365–384. 10.1085/jgp.22.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvert M, Sauerhering L, and Maisner A.(2020). Cytokine Induction in Nipah Virus-Infected Primary Human and Porcine Bronchial Epithelial Cells. The Journal of infectious diseases 221, S395–S400. 10.1093/infdis/jiz455. [DOI] [PubMed] [Google Scholar]
- Erbar S, Diederich S, and Maisner A.(2008). Selective receptor expression restricts Nipah virus infection of endothelial cells. Virology journal 5, 142–146. 10.1186/1743-422X-5-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, and Hirschi K.(2019). Molecular regulation of arteriovenous endothelial cell specification. F1000Research 8, 1208. 10.12688/f1000research.16701.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish JE, and Wythe JD (2015). The molecular regulation of arteriovenous specification and maintenance. Developmental Dynamics 244, 391–409. 10.1002/dvdy.24252. [DOI] [PubMed] [Google Scholar]
- Fowler JL, Ang LT, and Loh KM (2019). A critical look: Challenges in differentiating human pluripotent stem cells into desired cell types and organoids. Wiley Interdisciplinary Reviews: Developmental Biology 113, 891–823. 10.1002/wdev.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage BK, Liu JC, Innes BT, MacParland SA, McGilvray ID, Bader GD, and Keller GM (2020). Generation of Functional Liver Sinusoidal Endothelial Cells from Human Pluripotent Stem-Cell-Derived Venous Angioblasts. Cell Stem Cell. 10.1016/j.stem.2020.06.007. [DOI] [PubMed] [Google Scholar]
- Gale NW, Dominguez MG, Noguera I, Pan L, Hughes V, Valenzuela DM, Murphy AJ, Adams NC, Lin HC, Holash J, et al. (2004). Haploinsufficiency of delta-like 4 ligand results in embryonic lethality due to major defects in arterial and vascular development. Proceedings of the National Academy of Sciences of the United States of America 101, 15949–15954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert TW, Daddario-DiCaprio KM, Hickey AC, Smith MA, Chan Y-P, Wang L-F, Mattapallil JJ, Geisbert JB, Bossart KN, and Broder CC (2010). Development of an acute and highly pathogenic nonhuman primate model of Nipah virus infection. PLoS ONE 5, e10690. 10.1371/journal.pone.0010690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AS, Drake JM, Burnes DL, Finley DS, Zhang H, Reiter RE, Huang J, and Witte ON (2011). Purification and direct transformation of epithelial progenitor cells from primary human prostate. Nature Protocols 6, 656–667. 10.1038/nprot.2011.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez Roman R, Tornieporth N, Cherian NG, Shurtleff AC, ĽAzou Jackson M, Yeskey D, Hacker A, Mungai E, and Le TT (2021). Medical countermeasures against henipaviruses: a review and public health perspective. Lancet Infect Dis. 10.1016/S1473-3099(21)00400-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister C, and Satija R.(2019). Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biology, 1–15. 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaidych OV, Freund C, van den Hil F, Salvatori DCF, Riminucci M, Mummery CL, and Orlova VV (2018). Inflammatory Responses and Barrier Function of Endothelial Cells Derived from Human Induced Pluripotent Stem Cells. Stem Cell Reports 10, 1642–1656. 10.1016/j.stemcr.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt BH, Tamin A, Ksiazek TG, Rollin PE, Anderson LJ, Bellini WJ, and Rota PA (2000). Molecular characterization of Nipah virus, a newly emergent paramyxovirus. Virology 271, 334–349. 10.1006/viro.2000.0340. [DOI] [PubMed] [Google Scholar]
- Harschnit O., and Stude L. (2021). Human stem cell models to study host-virus interactions in the central nervous system. Nature Reviews Immunology, 1–13. 10.1038/s41577-020-00474-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CC, Peterson QP, Hong J-Y, and Peterson RT (2006). Artery/vein specification is governed by opposing phosphatidylinositol-3 kinase and MAP kinase/ERK signaling. Curr Biol 16, 1366–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwano S, Sugiyama M, Hama H, Watakabe A, Hasegawa N, Kuchimaru T, Tanaka KZ, Takahashi M, Ishida Y, Hata J, et al. (2018). Single-cell bioluminescence imaging of deep tissue in freely moving animals. Science 359, 935–939. 10.1126/science.aaq1067. [DOI] [PubMed] [Google Scholar]
- Kälin RE, Kretz MP, Meyer AM, Kispert A, Heppner FL, and Brändli AW (2007). Paracrine and autocrine mechanisms of apelin signaling govern embryonic and tumor angiogenesis. Developmental Biology 305, 599–614. 10.1016/j.ydbio.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Kalucka J, de Rooij LPMH, Goveia J, Rohlenova K, Dumas SJ, Meta E, Conchinha NV, Taverna F, Teuwen L-A, Veys K, et al. (2020). Single-Cell Transcriptome Atlas of Murine Endothelial Cells. Cell, 1–37. 10.1016/j.cell.2020.01.015. [DOI] [PubMed] [Google Scholar]
- Kidoya H, Naito H, Muramatsu F, Yamakawa D, Jia W, Ikawa M, Sonobe T, Tsuchimochi H, Shirai M, Adams RH, et al. (2015). APJ Regulates Parallel Alignment of Arteries and Veins in the Skin. Developmental Cell 33, 247–259. 10.1016/j.devcel.2015.02.024. [DOI] [PubMed] [Google Scholar]
- Krebs LT, Shutter JR, Tanigaki K, Honjo T, Stark KL, and Gridley T.(2004). Haploinsufficient lethality and formation of arteriovenous malformations in Notch pathway mutants. Genes & Development 18, 2469–2473. 10.1101/gad.1239204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F.(2012). Trim Galore. https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/. [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Research 44, W90–97. 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian L, Sancho-Martinez I, Nivet E, Aguirre A, Moon K, Pendaries C, Volle-Challier C, Bono F, Herbert J-M, Pulecio J, et al. (2012). Conversion of human fibroblasts to angioblast-like progenitor cells. Nature methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa YK, Yin RT, Shang MR, Shirure VS, Moya ML, and George SC (2017). Human Induced Pluripotent Stem Cell-Derived Endothelial Cells for Three-Dimensional Microphysiological Systems. Tissue engineering. Part C, Methods 23, 474–484. 10.1089/ten.tec.2017.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labun K, Montague TG, Krause M, Torres Cleuren YN, Tjeldnes H, and Valen E.(2019). CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res 47, W171–W174. 10.1093/nar/gkz365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, and Smyth GK (2014). voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15, R29. 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson ND, Scheer N, Pham VN, Kim CH, Chitnis AB, Campos-Ortega JA, and Weinstein BM (2001). Notch signaling is required for arterial-venous differentiation during embryonic vascular development. Development 128, 3675–3683. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Vogel AM, and Weinstein BM (2002). sonic hedgehog and vascular endothelial growth factor act upstream of the Notch pathway during arterial endothelial differentiation. Developmental Cell 3, 127–136. [DOI] [PubMed] [Google Scholar]
- Lian X, Bao X, Al-Ahmad A, Liu J, Wu Y, Dong W, Dunn KK, Shusta EV, and Palecek SP (2014). Efficient Differentiation of Human Pluripotent Stem Cells to Endothelial Progenitors via Small-Molecule Activation of WNT Signaling. Stem Cell Reports 3, 804–816. 10.1016/j.stemcr.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F-J, Tsai M-J, and Tsai SY (2007). Artery and vein formation: a tug of war between different forces. EMBO Rep 8, 920–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Gil C-H, and Yoder MC (2017). Differentiation, Evaluation, and Application of Human Induced Pluripotent Stem Cell-Derived Endothelial Cells. Arteriosclerosis, Thrombosis, and Vascular Biology, ATVBAHA.117.309962. 10.1161/ATVBAHA.117.309962. [DOI] [PubMed] [Google Scholar]
- Lo MK, Miller D, Aljofan M, Mungall BA, Rollin PE, Bellini WJ, and Rota PA (2010). Characterization of the antiviral and inflammatory responses against Nipah virus in endothelial cells and neurons. Virology 404, 78–88. 10.1016/j.virol.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Loh KM, Ang LT, Zhang J, Kumar V, Ang J, Auyeong JQ, Lee KL, Choo SH, Lim CYY, Nichane M, et al. (2014). Efficient Endoderm Induction from Human Pluripotent Stem Cells by Logically Directing Signals Controlling Lineage Bifurcations. Cell Stem Cell 14, 237–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh KM., Chen A., Koh PW., Deng TZ., Sinha R., Tsai JM., Barkal AA., Shen KY., Jain R., Morganti RM., et al. (2016). Mapping the Pairwise Choices Leading from Pluripotency to Human Bone, Heart, and Other Mesoderm Cell Types. Cell 166, 451–467. 10.1016/j.cell.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López CB, Yount JS, Hermesh T, and Moran TM (2006). Sendai virus infection induces efficient adaptive immunity independently of type I interferons. Journal of Virology 80, 4538–4545. 10.1128/JVI.80.9.4538-4545.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S.(2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, le Noble F, Yuan L, Jiang Q, De Lafarge B, Sugiyama D, Bréant C, Claes F, De Smet F, Thomas J-L, et al. (2004). The netrin receptor UNC5B mediates guidance events controlling morphogenesis of the vascular system. Nature 432, 179–186. 10.1038/nature03080. [DOI] [PubMed] [Google Scholar]
- Mahalingam S, Herrero LJ, Playford EG, Spann K, Herring B, Rolph MS, Middleton D, McCall B, Field H, and Wang LF (2012). Hendra virus: an emerging paramyxovirus in Australia. Lancet Infect Dis 12, 799–807. 10.1016/S1473-3099(12)70158-5. [DOI] [PubMed] [Google Scholar]
- Martin RM, Ikeda K, Cromer MK, Uchida N, Nishimura T, Romano R, Tong AJ, Lemgart VT, Camarena J, Pavel-Dinu M, et al. (2019). Highly Efficient and Marker-free Genome Editing of Human Pluripotent Stem Cells by CRISPR-Cas9 RNP and AAV6 Donor-Mediated Homologous Recombination. Cell Stem Cell 24, 821–828.e825. 10.1016/j.stem.2019.04.001. [DOI] [PubMed] [Google Scholar]
- Masaki H, Kato-Itoh M, Takahashi Y, Umino A, Sato H, Ito K, Yanagida A, Nishimura T, Yamaguchi T, Hirabayashi M, et al. (2016). Inhibition of Apoptosis Overcomes Stage-Related Compatibility Barriers to Chimera Formation in Mouse Embryos. Cell Stem Cell 19, 587–592. 10.1016/j.stem.2016.10.013. [DOI] [PubMed] [Google Scholar]
- Mathieu C, Guillaume V, Sabine A, Ong KC, Wong KT, Legras-Lachuer C, and Horvat B.(2012). Lethal Nipah virus infection induces rapid overexpression of CXCL10. PLoS ONE 7, e32157. 10.1371/journal.pone.0032157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken IR, Taylor RS, Kok FO, de la Cuesta F, Dobie R, Henderson BEP, Mountford JC, Caudrillier A, Henderson NC, Ponting CP, and Baker AH (2019). Transcriptional dynamics of pluripotent stem cell-derived endothelial cell differentiation revealed by single-cell RNA sequencing. European Heart Journal 385, 9963117. 10.1093/eurheartj/ehz351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehand MS, Al-Shorbaji F, Millett P, and Murgue B.(2018). The WHO R&D Blueprint: 2018 review of emerging infectious diseases requiring urgent research and development efforts. Antiviral research 159, 63–67. 10.1016/j.antiviral.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger T, Gache V, Xu M, Cadot B, Folker ES, Richardson BE, Gomes ER, and Baylies MK (2012). MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature 484, 120–124. 10.1038/nature10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan P, Green D, Pallister J, Klein R, White J, Williams C, McMillan P, Tilley L, Lampe M, Hawes P, and Wang L-F (2014). Detailed morphological characterisation of Hendra virus infection of different cell types using super-resolution and conventional imaging. Virology journal 11, 1–12. 10.1186/s12985-014-0200-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray K, Selleck P, Hooper P, Hyatt A, Gould A, Gleeson L, Westbury H, Hiley L, Selvey L, and Rodwell B.(1995). A morbillivirus that caused fatal disease in horses and humans. Science 268, 94–97. 10.1126/science.7701348. [DOI] [PubMed] [Google Scholar]
- Nakatsu MN, Davis J, and Hughes CCW (2007). Optimized fibrin gel bead assay for the study of angiogenesis. Journal of visualized experiments : JoVE, 186. 10.3791/186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrete OA., Chu D., Aguilar HC., and Lee B. (2007). Single amino acid changes in the Nipah and Hendra virus attachment glycoproteins distinguish ephrinB2 from ephrinB3 usage. Journal of Virology 81, 10804–10814. 10.1128/JVI.00999-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrete OA, Levroney EL, Aguilar HC, Bertolotti-Ciarlet A, Nazarian R, Tajyar S, and Lee B.(2005). EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature 436, 401–405. 10.1038/nature03838. [DOI] [PubMed] [Google Scholar]
- Ng AHM, Khoshakhlagh P, Rojo Arias JE, Pasquini G, Wang K, Swiersy A, Shipman SL, Appleton E, Kiaee K, Kohman RE, et al. (2020). A comprehensive library of human transcription factors for cell fate engineering. Nature Biotechnology 51, 987. 10.1038/s41587-020-0742-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen J, Lin Y-Y, and Gerecht S.(2021). The next generation of endothelial differentiation: Tissue-specific ECs. Cell Stem Cell 28, 1188–1204. 10.1016/j.stem.2021.05.002. [DOI] [PubMed] [Google Scholar]
- Nguyen MTX, Okina E, Chai X, Tan KH, Hovatta O, Ghosh S, and Tryggvason K.(2016). Differentiation of Human Embryonic Stem Cells to Endothelial Progenitor Cells on Laminins in Defined and Xeno-free Systems. Stem Cell Reports 7, 802–816. 10.1016/j.stemcr.2016.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Ohtaka M, Takada H, Kurisaki A, Tran NVK, Tran YTH, Hisatake K, Sano M, and Nakanishi M.(2017). Simple and effective generation of transgene-free induced pluripotent stem cells using an auto-erasable Sendai virus vector responding to microRNA-302. Stem Cell Research 23, 13–19. 10.1016/j.scr.2017.06.011. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Sano M, Ohtaka M, Furuta B, Umemura Y, Nakajima Y, Ikehara Y, Kobayashi T, Segawa H, Takayasu S, et al. (2011). Development of defective and persistent Sendai virus vector: a unique gene delivery/expression system ideal for cell reprogramming. The Journal of biological chemistry 286, 4760–4771. 10.1074/jbc.M110.183780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmer R, Engels L, Usman A, Menke S, Malik MNH, Pessler F, Göhring G, Bornhorst D, Bolten S, Abdelilah-Seyfried S, et al. (2018). Differentiation of Human Pluripotent Stem Cells into Functional Endothelial Cells in Scalable Suspension Culture. Stem Cell Reports 10, 1657–1672. 10.1016/j.stemcr.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang JF, Kamaraj US, Cao EY, and Rackham OJL (2021). ShinyCell: Simple and sharable visualisation of single-cell gene expression data. Bioinformatics. 10.1093/bioinformatics/btab209. [DOI] [PubMed] [Google Scholar]
- Paik DT, Tian L, Lee J, Sayed N, Chen IY, Rhee S, Rhee J-W, Kim Y, Wirka RC, Buikema JW, et al. (2018). Large-Scale Single-Cell RNA-Seq Reveals Molecular Signatures of Heterogeneous Populations of Human Induced Pluripotent Stem Cell-Derived Endothelial Cells. Circulation Research. 10.1161/CIRCRESAHA.118.312913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomares K, Vigant F, Van Handel B, Pernet O, Chikere K, Hong P, Sherman SP, Patterson M, An DS, Lowry WE, et al. (2013). Nipah virus envelope-pseudotyped lentiviruses efficiently target ephrinB2-positive stem cell populations in vitro and bypass the liver sink when administered in vivo. Journal of Virology 87, 2094–2108. 10.1128/JVI.02032-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MA, Kumar A, Jung HS, Uenishi G, Moskvin OV, Thomson JA, and Slukvin II (2018). Activation of the Arterial Program Drives Development of Definitive Hemogenic Endothelium with Lymphoid Potential. Cell Reports 23, 2467–2481. 10.1016/j.celrep.2018.04.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsch C, Challet-Meylan L, Thoma EC, Urich E, Heckel T, O'Sullivan JF, Grainger SJ, Kapp FG, Sun L, Christensen K et al. (2015). Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nature Cell Biology 17, 994–1003. 10.1038/ncb3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduri A, Chang AH, Raftrey B, Rhee S, Van M, and Red-Horse K.(2017). Endothelial cells respond to the direction of mechanical stimuli through SMAD signaling to regulate coronary artery size. Development 144, 3241–3252. 10.1242/dev.150904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potente M, and Mäkinen T.(2017). Vascular heterogeneity and specialization in development and disease. Nature Reviews Molecular Cell Biology 18, 477–494. 10.1038/nrm.2017.36. [DOI] [PubMed] [Google Scholar]
- Prescott J., DeBuysscher BL., Feldmann F., Gardner DJ., Haddock E., Martellaro C., Scott D., and Feldmann H. (2015). Single-dose live-attenuated vesicular stomatitis virus-based vaccine protects African green monkeys from Nipah virus disease. Vaccine 33, 2823–2829. 10.1016/j.vaccine.2015.03.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafii S, Butler JM, and Ding B-S (2016). Angiocrine functions of organ-specific endothelial cells. Nature 529, 316–325. 10.1038/nature17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Red-Horse K, and Siekmann AF (2019). Veins and Arteries Build Hierarchical Branching Patterns Differently: Bottom-Up versus Top-Down. BioEssays 41, e1800198. 10.1002/bies.201800198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, and Mesirov JP (2006). GenePattern 2.0. Nature Genetics 38, 500–501. 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JJ, Parisien J-P, and Horvath CM (2002). Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. Journal of Virology 76, 11476–11483. 10.1128/jvi.76.22.11476-11483.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa S, Praça C, Pitrez PR, Gouveia PJ, Aranguren XL, Ricotti L, and Ferreira LS (2019). Functional characterization of iPSC-derived arterial- and venous-like endothelial cells. Scientific Reports 9, 3826–3815. 10.1038/s41598-019-40417-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto Y, Hara K, Kanai-Azuma M, Matsui T, Miura Y, Tsunekawa N, Kurohmaru M, Saijoh Y, Koopman P, and Kanai Y.(2007). Redundant roles of Sox17 and Sox18 in early cardiovascular development of mouse embryos. Biochem Biophys Res Commun 360, 539–544. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schountz T, Campbell C, Wagner K, Rovnak J, Martellaro C, DeBuysscher B, Feldmann H, and Prescott J.(2019). Differential Innate Immune Responses Elicited by Nipah Virus and Cedar Virus Correlate with Disparate In Vivo Pathogenesis in Hamsters. Viruses 11, 291–211. 10.3390/v11030291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalaby F, Rossant J, Yamaguchi TP, Gertsenstein M, Wu XF, Breitman ML, and Schuh AC (1995). Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature 376, 62–66. [DOI] [PubMed] [Google Scholar]
- Shaw AE, Hughes J, Gu Q, Behdenna A, Singer JB, Dennis T, Orton RJ, Varela M, Gifford RJ, Wilson SJ, and Palmarini M.(2017). Fundamental properties of the mammalian innate immune system revealed by multispecies comparison of type I interferon responses. PLoS Biology 15, e2004086. 10.1371/journal.pbio.2004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JW, Huggenberger R, and Detmar M.(2008). Transcriptional profiling of VEGF-A and VEGF-C target genes in lymphatic endothelium reveals endothelial-specific molecule-1 as a novel mediator of lymphangiogenesis. Blood 112, 2318–2326. 10.1182/blood-2008-05-156331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoneau CR, and Ott M.(2020). Modeling Multi-organ Infection by SARS-CoV-2 Using Stem Cell Technology. Cell Stem Cell 27, 859–868. 10.1016/j.stem.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram G, Tan JY, Islam I, Rufaihah AJ, and Cao T.(2015). Efficient differentiation of human embryonic stem cells to arterial and venous endothelial cells under feeder- and serum- free conditions. Stem Cell Research & Therapy, 1–17. 10.1186/s13287-015-0260-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, and Satija R.(2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e1821. 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai-Nagara I., Yoshikawa Y., Numata N., Ando T., Okabe K., Sugiura Y., Ieda M., Takakura N., Nakagawa O., Zhou B., et al. (2017). Placental labyrinth formation in mice requires endothelial FLRT2/UNC5B signaling. Development 144, 2392–2401. 10.1242/dev.149757. [DOI] [PubMed] [Google Scholar]
- Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, Mansfield W, et al. (2014). Resetting Transcription Factor Control Circuitry toward Ground-State Pluripotency in Human. Cell 158, 1254–1269. 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez JY, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, and Eliceiri KW (2017). TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90. 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- Travaglini KJ, Nabhan AN, Penland L, Sinha R, Gillich A, Sit RV, Chang S, Conley SD, Mori Y, Seita J, et al. (2020). A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 587, 619–625. 10.1038/s41586-020-2922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtue ER, Marsh GA, and Wang LF (2011). Interferon Signaling Remains Functional during Henipavirus Infection of Human Cell Lines. Journal of Virology 85, 4031–4034. 10.1128/JVI.02412-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HU, Chen ZF, and Anderson DJ (1998). Molecular distinction and angiogenic interaction between embryonic arteries and veins revealed by ephrin-B2 and its receptor Eph-B4. Cell 93, 741–753. [DOI] [PubMed] [Google Scholar]
- Wang K, Lin R-Z, Hong X, Ng AH, Lee CN, Neumeyer J, Wang G, Wang X, Ma M, Pu WT, et al. (2020). Robust differentiation of human pluripotent stem cells into endothelial cells via temporal modulation of ETV2 with modified mRNA. Science Advances 6, eaba7606. 10.1126/sciadv.aba7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H.(2016). ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag New York: ). [Google Scholar]
- Wickham H, Averick M, Bryan J, and Chang W.(2019). Welcome to the Tidyverse. The Journal of Open Source Software. 10.21105/joss.01686. [DOI] [Google Scholar]
- Wickham H, François R, Henry L, and Müller K.(2022). dplyr: A Grammar of Data Manipulation. https://dplyr.tidyverse.org.
- Wong KT, Shieh W-J, Kumar S, Norain K, Abdullah W, Guarner J, Goldsmith CS, Chua KB, Lam SK, Tan CT, et al. (2002). Nipah virus infection: pathology and pathogenesis of an emerging paramyxoviral zoonosis. The American Journal of Pathology 161, 2153–2167. 10.1016/S0002-9440(10)64493-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Hasan SS, Schmidt I, Rocha SF, Pitulescu ME, Bussmann J, Meyen D, Raz E, Adams RH, and Siekmann AF (2014). Arteries are formed by vein-derived endothelial tip cells. Nature Communications 5, 5758. 10.1038/ncomms6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A, Kawabata R, Honda T, Sakai K, Ami Y, Sakaguchi T, and Irie T.(2018). A Single Amino Acid Substitution within the Paramyxovirus Sendai Virus Nucleoprotein Is a Critical Determinant for Production of Interferon-Beta-Inducing Copyback-Type Defective Interfering Genomes. Journal of Virology 92. 10.1128/JVI.02094-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, He J, Bai Z, Li Z, Gong Y, Liu C, Ni Y, Du J, Ma C, Bian L, et al. (2019). Tracing the first hematopoietic stem cell generation in human embryo by single-cell RNA sequencing. Cell Research, 1–14. 10.1038/s41422-019-0228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Chu L-F, Hou Z, Schwartz MP, Hacker T, Vickerman V, Swanson S, Leng N, Nguyen BK, Elwell A, et al. (2017). Functional characterization of human pluripotent stem cell-derived arterial endothelial cells. Proceedings of the National Academy of Sciences of the United States of America, 201702295. 10.1073/pnas.1702295114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GXY., Terry JM., Belgrader P., Ryvkin P., Bent ZW., Wilson R., Ziraldo SB., Wheeler TD., McDermott GP., Zhu J., et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nature Communications 8, 14049. 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) BMP specifies, whereas TGFβ and WNT repress, day 2 lateral mesoderm formation; i) day 1 hPSC-derived primitive streak was treated with 100 ng/mL VEGF for 24 hours in the presence or absence of BMP4 (10–40 ng/mL) or BMP inhibitors (1 μM DMH1 or 200 ng/mL NOGGIN); ii) day 1 hPSC-derived primitive streak was treated with 10 ng/mL BMP4 + 100 ng/mL VEGF for 24 hours in the presence or absence of a WNT agonist (CHIR99021, 1–6 μM) or WNT inhibitors (1 μM C59 or 1 μM XAV939); iii) day 1 hPSC-derived primitive streak was treated with 10 ng/mL BMP4 + 100 ng/mL VEGF for 24 hours in the presence or absence of a TGFβ agonist (Activin, 5–100 ng/mL) or TGFβ inhibitors (2 μM SB505124 or SB431542); qPCR was performed on day 2 cell populations
B) VEGF treatment induces day 2 lateral mesoderm; day 1 hPSC-derived primitive streak was treated with 10 ng/mL BMP4 + 1 μM XAV939 for 24 hours, in the presence or absence of VEGF (10–100 ng/mL) or a VEGFR inhibitor (Axitinib, 10 nM); qPCR was performed on day 2 cell populations (data for FLI1 is also shown in Fig. 1c)
C) PI3K inhibitors specify day 2 lateral mesoderm; day 1 hPSC-derived primitive streak was treated with BMP + VEGF + XAV939 in the presence or absence of PI3K inhibitors (PIK90 or GDC0941) for 24 hours to generate lateral mesoderm, which was then further differentiated into artery ECs for 24 additional hours (using VEGF alone); flow cytometry was performed on day 3 artery EC populations
D) PKA agonists specify day 2 lateral mesoderm; day 1 hPSC-derived primitive streak was treated with BMP + GDC0941 + VEGF + XAV939 in the presence or absence of PKA agonists (8-bromocAMP [1 mM] or Forskolin [10 μM]) or PKA inhibitor (KT5720 [1 μM]) for 24 hours to generate day 2 candidate lateral mesoderm, which was then further differentiated into artery ECs for 24 additional hours (using GDC0941 + VEGF + XAV939); flow cytometry was performed on day 3 artery EC populations
E) Immunostaining of H1 hPSC-derived day 2 lateral mesoderm, with DAPI nuclear counterstain (bottom); scale = 100 μm
A) BMP represses day 3 artery formation; day 2 hPSC-derived lateral mesoderm was treated with Activin + GDC0941 + VEGF + XAV939 + Vitamin C for 24 hours, in the presence or absence of BMP4 (5–40 ng/mL) or BMP inhibitor (DMH1); qPCR was performed on day 3 cell populations
B) TGFβ promotes, whereas WNT and PI3K repress, day 3 artery formation from SOX17-mCherry hPSCs; i,ii) day 2 hPSC-derived lateral mesoderm was further differentiated for 24 hours with VEGF, in the presence or absence of TGFβ agonist (Activin, 10 ng/mL), TGFβ inhibitor (SB505124, 2 μM), WNT agonist (CHIR99021), or WNT inhibitor (XAV939); iii,iv) day 2 hPSC-derived lateral mesoderm was further differentiated for 24 hours with VEGF+XAV939, in the presence or absence of TGFβ agonist (Activin, 10 ng/mL) or PI3K inhibitor (GDC0941); flow cytometry was performed on day 3 cell populations
C) WNT inhibits day 3 artery formation; day 2 hPSC-derived lateral mesoderm was further differentiated with VEGF for 24 hours, in the presence or absence of WNT agonist (CHIR99021) or WNT inhibitor (XAV939); qPCR (right) was performed on day 3 cell populations
D) Definition of the set of extracellular signals necessary for efficient artery differentiation; day 2 hPSC-derived lateral mesoderm was further differentiated for 24 hours into artery cells using the complete artery specification media (red; Activin + GDC0941 + VEGF + XAV939 + AA2P + DMH1) or in conditions in which each factor was individually withheld; flow cytometry was performed on day 3 cell populations
E) Flow cytometry of SOX17-mCherry hPSCs differentiated towards artery ECs for 3 days (top); subgating on SOX17+ CD34+ ECs shows that they uniformly express VE-CADHERIN and CD31 (bottom)
F) Flow cytometry of H1, H7, H9 and SUN004.1.9 hPSCs differentiated into artery ECs for 3 days reveals that >94% of cells are CD144+ CD34+ ECs
G) Side-by-side comparison of the artery EC differentiation protocol (described in this study) against 4 prevailing methods for endothelial differentiation (Lian et al., 2014; Patsch et al., 2015; Sriram et al., 2015; Zhang et al., 2017) in the H1 and SUN004.1.9 hPSC lines; flow cytometry to assess the percentage of CD34+ CD144+ endothelial cells was performed on days 3, 5 and 6 of differentiation (n.d. = not determined)
H) qPCR timecourse of hPSCs differentiated towards artery ECs reveals sequential expression of pluripotency, primitive streak, lateral mesoderm and finally, pan-endothelial and artery-specific markers; qPCR was performed on day 0 (undifferentiated) and differentiated day 1, 1.5, 2, 2.5 and 3 cell populations, with qPCR data shown normalized to the timepoint with highest expression of each given marker gene
I) qPCR of FACS-purified day 3 SOX17+ CD34+ artery ECs generated from the SOX17-mCherry hPSC line (as well as undifferentiated hPSC controls) reveals expression of artery-specific and pan-endothelial markers, while markers of alternate cell fates are minimally expressed; qPCR data are shown relative to the reference gene YWHAZ (100% = same expression as YWHAZ)
J) Transcriptional comparison of hPSC-derived day 3 artery ECs vs. Carnegie Stage 13 (CS13) human fetal artery ECs (Zeng et al., 2019) by scRNAseq to determine the top 10 genes that were differentially expressed by in vivo vs. in vitro artery ECs (left) and gene ontology (GO) analysis was performed on all differentially expressed genes (right)
A) WNT activation on day 4 of differentiation specifies vein ECs; day 3 H1 NR2F2–2A-GFP hPSC-derived ECs were further differentiated into day 4 vein ECs (base media: SB505124 + RO4929097 + PD0325901 + AA2P), in the presence or absence of WNT inhibitor (XAV939, 1 μM) or WNT agonist (CHIR99021, 1–7 μM) for 24 hours; flow cytometry was performed on day 4 vein populations to quantify the purity of NR2F2–2A-GFP+ CD144+ vein ECs
B) Re-splitting lateral mesoderm improves vein EC differentiation; day 2 hPSC-derived lateral mesoderm was differentiated into day 4 vein ECs without resplitting (“unsplit”) or, alternatively, was dissociated and then re-seeded at high density (~79,000 cells/cm2) in day 3 differentiation media, followed by treatment with day 4 vein EC media. Flow cytometry was performed at day 4 of hPSC differentiation.
C) Strategy to generate NR2F2–2A-GFP knock-in reporter hPSCs using Cas9/AAV6 genome editing (Martin et al., 2019)
D) After genome editing (Martin et al., 2019), PCR genotyping of individual hPSC lines (derived from single colonies) reveals successful integration of the NR2F2–2A-GFP knock-in reporter allele
E) Immunostaining of undifferentiated H1 hPSCs, with DAPI nuclear counterstain (negative control for Fig. 3e); scale = 100 μm
F) Transcriptional comparison of hPSC-derived day 4 vein ECs vs. Carnegie Stage 13 (CS13) human fetal vein ECs (Zeng et al., 2019) by scRNAseq; gene expression is quantified in UMI expression units (loge scale)
G) Transcriptional comparison of hPSC-derived day 4 artery ECs vs. Carnegie Stage 13 (CS13) human fetal vein ECs (Zeng et al., 2019) by scRNAseq to determine the top 10 genes that were differentially expressed by in vivo vs. in vitro vein ECs
H) Transcriptional comparison of hPSC-derived day 4 artery ECs vs. Carnegie Stage 13 (CS13) human fetal vein ECs (Zeng et al., 2019) by scRNAseq to determine gene ontology (GO) terms associated with genes differentially expressed in vivo and in vitro
A) scRNAseq of H1 hPSC-derived day 3 artery vs. day 4 vein ECs (CD144+ FACS sorted) reveals mutually-exclusive expression of hallmark arteriovenous marker genes
B) hPSC-derived day 3 artery ECs and day 4 vein ECs do not express known markers of organ-specific EC identities (Kalucka et al., 2020), as shown by scRNAseq (as positive controls, expression of reference genes YWHAZ, ACTB and GAPDH is shown); dot size indicates percentage of cells expressing a given marker and dot color reflects the level of gene expression
C) H1 hPSC-derived artery and vein ECs were either mixed with adult human lung fibroblasts (ratio of 1 EC:2 fibroblasts) or not mixed with fibroblasts, and then subsequently cultured in a 3D fibrin gel with EGM2 media for 1 week in vitro (Kurokawa et al., 2017). Only hPSC-derived ECs mixed with fibroblasts formed 3D networks, as assessed by CD31 immunostaining, demonstrating the importance of fibroblast support for vascular network formation in this experimental system; scale = 500 μm
D) SUN004.1.9 hPSC-derived artery ECs were mixed with adult human lung fibroblasts from two different individuals (#1 and #2), and then subsequently cultured in a 3D fibrin gel with EGM2 media for 1 week in vitro (Kurokawa et al., 2017). hPSC-derived ECs mixed with fibroblasts from both individuals formed 3D networks, as assessed by CD31 immunostaining; scale = 200 μm
E) H1 hPSC-derived day 3 artery ECs and day 4 vein ECs were stained for LDLR expression (left), or cultured with LDL DyLight 550 for 4 hours in their respective arterial or venous expansion media (right), prior to flow cytometry.
F) H1 hPSC-derived day 3 artery ECs and day 4 vein ECs were each expanded for 24 hours in their respective arterial or venous expansion media until they formed confluent monolayers, and then were scratched with a pipette tip. Serial imaging was performed over the next 48 hours to quantify closure of the cell monolayer; scale = 100 μm
G) SUN004.1.9 hPSC-derived artery and vein ECs exposed to shear stress or static conditions for 24 hours turn on different genes, as shown by qPCR; gene expression normalized such that for each gene, the sample expressing the highest level of the gene has an expression level = 1.0
H) H1 hPSC-derived artery and vein ECs were thawed and expanded for 6 days in their respective arterial or venous expansion media, and then were subsequently treated with TNFα (10 ng/mL) or left untreated for 4 hours. Flow cytometry was performed to assess TNFα-induced changes in immune-cell adhesion molecules CD54/ICAM1, CD62E/E-SELECTIN and CD106/VCAM1.
A) hPSC-derived artery and vein ECs can be expanded for 6 days in vitro while retaining mutually-exclusive expression of arteriovenous subtype markers. SUN004.1.9 hPSCs were differentiated into day 3 artery and day 4 vein ECs (“day 0” of expansion), frozen, and which were then respectively thawed in artery expansion media (EGM2) or vein expansion media (EGM2 + RO4929097 (2 μM; NOTCH inhibitor) + SB505124 (2 μM; TGFβ inhibitor)); qPCR of expanded artery and vein ECs was performed at multiple timepoints post-thawing, and mRNA expression is shown relative to the expression of reference gene YWHAZ (i.e., YWHAZ = 1.0)
B) Continued inhibition of artery-specifying signals TGFβ and NOTCH signals is required to expand hPSC-derived vein ECs in vitro. SUN004.1.9 hPSCs were differentiated into artery or vein ECs (“day 0” of expansion), and then frozen; vein ECs were then thawed in EGM2, or in the absence of hydrocortisone (HC) and/or presence of RO4929097 (2 μM; NOTCH inhibitor) and/or SB505124 (2 μM; TGFβ inhibitor) for 4 additional days; qPCR was then performed to determine mRNA expression, which is shown relative to the expression of reference gene YWHAZ (i.e., YWHAZ = 1.0)
C) hPSC-derived artery ECs can be expanded for 8 days in vitro. hPSCs were differentiated into artery ECs in 3 days (“day 0” of expansion), and then subsequently expanded in EGM2 for up to 8 days. Flow cytometry confirmed continued expression of pan-EC markers CD34 and CD144 and arterial marker DLL4.
D) hPSC-derived artery and vein ECs activate interferon signaling in response to a double-stranded RNA mimic, poly(I:C). SUN004.1.9 hPSCs were differentiated into artery or vein ECs, frozen, and then thawed and cultured in their respective expansion media (EGM2 for artery ECs and EGM2 + RO4929097 + SB505124 for vein ECs) for 5 days. Then, poly(I:C) (50 μg/mL) was added to the respective culture media for 24 hours and qPCR was performed, with mRNA expression shown relative to the expression of reference gene YWHAZ (i.e., YWHAZ = 1.0); **P<0.01
E) H1 hPSC-derived artery and vein ECs were inoculated with Nipah or Hendra viruses for 24 hours, and then fixed and stained with Diff-Quik to assess cytopathic effects; total cell number (including mononucleated and multinucleated cells) was then quantified and normalized to uninfected controls; n.s. = not significant
H) Cocultures of GFP+ and wild-type hPSC-derived artery and vein ECs were inoculated with either Nipah or Hendra virus; live imaging was conducted over an 8-hour interval. The trajectory of single GFP+ cells was computationally tracked by live imaging, with each single-cell trajectory colored differently; scale bar = 200 μm
I) Cocultures of GFP+ and wild-type hPSC-derived artery and vein ECs were inoculated with either Nipah or Hendra virus; live imaging was conducted over an 8-hour interval. The trajectory of single GFP+ cells was computationally tracked by live imaging, revealing two features: 1) increasing numbers of GFP+ cells (reflecting fusion of GFP+ cells with formerly wild-type cells, and leading to an apparent increase in GFP+ cells) and 2) fusion (where multiple cell tracks merged)
A) Intracellular levels of Nipah and Hendra viral RNA after inoculating H1 hPSC-derived artery and vein ECs with Nipah and Hendra viruses, as quantified by qPCR of the cell monolayer (after removing the culture media); *P<0.05; **P<0.01; †extensive cell loss
B) Significantly higher expression of EFNB2 in hPSC-derived artery ECs relative to vein ECs. hPSC-derived artery and vein ECs generated from H1, SUN004.1.9 and SUN004.2 hPSC lines were frozen, and then thawed in their respective arterial or venous expansion media for 3–6 days prior to qPCR. EFNB2 expression is shown relative to the expression of reference gene YWHAZ (i.e., YWHAZ = 1.0); **P<0.05.
C) scRNAseq of hPSC-derived artery and vein ECs revealed that among all EFNA and EFNB family members, EFNB2 was most highly expressed in artery ECs, whereas EFNB3 (another candidate entry-receptor for Nipah and Hendra viruses (Negrete et al., 2007)) was minimally expressed.
D) Primary human artery and vein ECs cultured in standard conditions relinquish arteriovenous identity in vitro. Human umbilical artery and vein ECs were expanded in EGM2, and compared against hPSC-derived day 3 artery ECs and day 4 vein ECs by qPCR.
E) Cas9-mediated deletion of EFNB2 in hPSCs, as shown by i) sequencing of the EFNB2 gene in H1 wild-type and EFNB2−/− hPSCs, revealing a 1-base deletion in both EFNB2 alleles in the latter hPSC line and ii) flow cytometry for EFNB2 expression in H1 wild-type and EFNB2−/− hPSCs prior to and after differentiation into artery ECs
F) Nipah pseudovirus preferentially enters hPSC-derived artery ECs, by comparison to vein ECs. hPSC-derived artery and vein ECs were thawed, expanded for 3 days in respective arterial or venous expansion media, and then transduced with GFP-encoding lentiviruses pseudotyped with Nipah or VSV envelope proteins for 24 hours. 48 hours later, flow cytometry was performed to enumerate the percentage of GFP+ ECs, revealing arterially-biased tropism of Nipah-pseudotyped lentivirus. As a negative control, VSV-G-pseudotyped lentivirus did not exhibit arterially-biased tropism; **P<0.01
G) EFNB2 is dispensable for VSV pseudovirus entry. H1 wild-type and EFNB2−/− hPSC-derived artery and vein ECs were thawed, expanded for 3 days, and then infected with equal infectious units of GFP-encoding, non-replicating lentiviruses pseudotyped with VSV envelope proteins. Flow cytometry was performed 3 days later, showing that EFNB2 is dispensable for infection by VSV pseudovirus; **P<0.01; n.s. = not significant
A) RNA-seq analysis of hPSC-derived artery ECs infected with either Nipah, Hendra or Sendai virus for 5, 10 or 15 hours, followed by alignment of viral reads to viral transcripts, confirms the known 3’to-5’ transcriptional gradient along the paramyxovirus genome, with 3’ most gene (N) expressed at the highest level, followed by decreasing expression of progressively more 5’ genes. The y-axis represents viral mRNA expression relative to the entire transcriptome of infected artery ECs (100% = all human + viral reads). Average data from H1 and SUN004.1.9 hPSCs shown.
B) Expression of each individual viral gene in hPSC-derived artery ECs infected with either Nipah, Hendra or Sendai virus. Percentage indicates the levels at which each individual viral gene (e.g., N) was expressed relative to the entirety of all viral gene expression (100% = all viral reads). Data was averaged from H1 and SUN004.1.9 hPSCs infected for 5, 10 or 15 hours.
C) Differentially expressed genes between infected vs. uninfected artery ECs by RNA-seq, 10 and 15 hours post-infection; averages of H1 and SUN004.1.9 hPSC-derived cell-types shown.
D) IFNβ secretion by hPSC-derived artery ECs as quantified by ELISA; averaged from H1, SUN004.1.9 and SUN004.2 hPSCs; *P<0.01; n.s. = not significant
Movie S1. Live-imaging of cocultured wild-type and GFP+ artery or vein ECs infected by Nipah or Hendra viruses, related to Figure 5.
Table S1. Gene expression differences between hPSC-derived vs. human fetal artery ECs, as assayed by single-cell RNA-seq, related to Figure 2. Single-cell RNA-seq of H1 hPSC-derived artery ECs in vitro vs. Carnegie Stage 13 human fetal artery ECs in vivo. Genes that are differentially expressed between in vitro vs. in vivo artery ECs are shown, and loge fold change is shown (i.e., positive fold change indicates higher expression in hPSC-derived cells, relative to in vivo cells). For each gene, “pct.1” and “pct.2” shows the percentage of cells that detectably express that gene, among H1 hPSC-derived artery ECs and human fetal artery ECs, respectively.
Table S2. Gene expression differences between hPSC-derived vs. human fetal vein ECs, as assayed by single-cell RNA-seq, related to Figure 3. Single-cell RNA-seq of H1 hPSC-derived vein ECs in vitro vs. Carnegie Stage 13 human fetal vein ECs in vivo. Genes that are differentially expressed between in vitro vs. in vivo vein ECs are shown, and loge fold change is shown (i.e., positive fold change indicates higher expression in hPSC-derived cells, relative to in vivo cells.) For each gene, “pct.1” and “pct.2” shows the percentage of cells that detectably express that gene, among H1 hPSC-derived vein ECs and human vein artery ECs, respectively.
Table S3. Differentially-expressed genes between hPSC-derived artery vs. vein ECs, as assayed by bulk-population RNA-seq, related to Figure 4. Bulk-population RNA-seq of H1 hPSC-derived day 3 artery ECs vs. day 4 vein ECs (both FACS sorted for CD144+ cells). All counts > 1 were normalized to the total read count and log2 transformed using Limma-Voom.
Table S4. Bulk-population RNA-seq of hPSC-derived artery and vein cells infected by Nipah, Hendra or Sendai viruses, related to Figure 7. H1 and SUN004.1.9 hPSC-derived artery and vein ECs were inoculated with Nipah, Hendra or Sendai viruses for 5, 10 or 15 hours, and bulk-population RNA-seq was performed. Replicate 1 = H1 and replicate 2 = SUN004.1.9. Counts were regularized, log2 transformed and normalized by size factors using DESeq2.
Data Availability Statement
Bulk-population RNA-seq and single-cell RNA-seq datasets generated as part of this study are available at the NCBI Sequence Read Archive, accession number PRJNA837932. Single-cell RNAseq datasets of hPSC-derived artery and vein ECs that were generated as part of this study can be interactively browsed at a custom web portal: https://anglab.shinyapps.io/arteryveinscrna/. Computational scripts used for genomics analyses conducted as part of this study are available at Github: https://github.com/LayTengAngLab/ArteryVeinEndothelial.