Abstract
A highly efficient and robust multiple scales in silico protocol, consisting of atomistic Molecular Dynamics (MD), coarse-grain (CG) MD, and constant-pH CG Monte Carlo (MC), has been developed and used to study the binding affinities of selected antigen-binding fragments of the monoclonal antibody (mAbs) CR3022 and several of its here optimized versions against 11 SARS-CoV-2 variants including the wild type. Totally 235,000 mAbs structures were initially generated using the RosettaAntibodyDesign software, resulting in top 10 scored CR3022-like-RBD complexes with critical mutations and compared to the native one, all having the potential to block virus-host cell interaction. Of these 10 finalists, two candidates were further identified in the CG simulations to be the best against all SARS-CoV-2 variants. Surprisingly, all 10 candidates and the native CR3022 exhibited a higher affinity for the Omicron variant despite its highest number of mutations. The multiscale protocol gives us a powerful rational tool to design efficient mAbs. The electrostatic interactions play a crucial role and appear to be controlling the affinity and complex building. Studied mAbs carrying a more negative total net charge show a higher affinity. Structural determinants could be identified in atomistic simulations and their roles are discussed in detail to further hint at a strategy for designing the best RBD binder. Although the SARS-CoV-2 was specifically targeted in this work, our approach is generally suitable for many diseases and viral and bacterial pathogens, leukemia, cancer, multiple sclerosis, rheumatoid, arthritis, lupus, and more.
Keywords: COVID-19, Molecular dynamics, Coarse-grain simulation, Constant-pH Monte Carlo, Multiscale protocol, Electrostatic interactions
Abbreviation: Ab-Ag, antibody-antigen
Graphical Abstract
1. Introduction
The world has been in the grip of the COVID-19 pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), for roughly two years and a half with no corner on Earth being saved. To date, more than 603 million cases and 6.5 million deaths are reported with no end in sight [https://www.who.int/] at the time of writing. Vaccines have been developed in the shortest possible time and manufactured in large quantities on a global scale [1]. Yet, the number of people to be vaccinated remains quite large giving time for new mutations of the virus to develop with a relatively high potential for breakthrough cases [2].
Understanding the structural features of viruses is important in developing new therapeutic strategies. Coronaviruses have four types of glycoproteins all taking part in the pathogenesis: spike (S), membrane, nucleocapsid, and envelope. At the top of the S protein is the receptor-binding domain (RBD), which binds to the angiotensin-converting enzyme 2 (ACE2) receptor of the host cell to promote an entry to infect the human cells.
In developing the vaccines, the main choices space from mRNA vaccines releasing a synthetic mRNA sequence encoding the virus S protein from a nano-particle to viral vector vaccines continuing DNA encoding the S protein [1], [3]. Despite some promising medicines [4] including PAXLOVID, there is still no effective drug against COVID-19 and neutralizing monoclonal antibodies (mAbs) are currently the most attractive alternative. One successful approach in the past to treat various infectious diseases is convalescent plasma therapy (CPT) [5]. This treatment administered antibody-containing plasma, collected from COVID-19 recovering patients to recipient patients that still did not develop a protective immune response, reducing thus the viral load in the recipient. One important limitation of CPT in the treatment of COVID-19 is the diversity of virus variants found in the population which makes the selection of donors difficult. On the other hand, mAbs or polyclonal antibody cocktails can be specifically engineered and biotechnologically produced to fight SARS-CoV-2. Spike protein is the primary target of these mAbs with four classes of them being described to date depending on the location of their target epitope on the S protein [6]. Regardless of their source, either from convalescent blood or industrially produced, human mAbs are safe therapeutic tools and can be produced quickly. There are already more than 50 commercially available mAbs approved for the treatment of other inflammatory and immune disorders and other infectious pathogens. Several candidates against SARS-CoV-2 are by now in different trial phases [7], [8]. Patents have already been deposited (e.g., US 2021/0292393, US20210388066A1), and some mAbs are approved by regulatory agencies such as the FDA [9], [10].
Among available mAbs, CR3022 is a class IV mAb that does not bind to an RBD epitope that overlaps the ACE2 binding site, but to a conserved region in RBD when the spike homotrimer is in an “up” (or open) configuration exposing the RBD to interact with either ACE2 or binder molecules [11], [12], [13]. Exploiting such conserved regions in RBD (despite requiring the S protein to be at the upstate), class IV mAbs have broad neutralizing activity against SARS-CoV-2, its variants, and other related coronaviruses [11]. The broad sarbecovirus neutralizing activity from CR3022 has already been confirmed even for the Omicron variant [14], [15]. Due to the abrupt appearance of new variants of concerns (VOCs) [16] in a short time, improving a mAb that has its epitope on a conserved region of the RBD may constitute an advantage for developing biopharmaceuticals broadly effective against different variants of the virus. Moreover, attention has recently been turned again to CR3022 as a promising candidate for COVID-19 treatment and prevention as new experimental and computational evidence became available. Recent studies [16–19] show that CR3022 binds RBD of SARS-CoV-2 with relatively high affinity (KD ∼ 3–115 nM), although the tendency for stronger complexes formed with RBD SARS-CoV-1 is still dominant. These new data suggest that CR3022 is an appealing candidate to be subjected to affinity maturation, especially due to its cross-reactivity. Therefore, CR3022 has been chosen for this study and will be the central part of our discussions.
Modern virology, immunology, and related fields are expanding their achievements by incorporating theoretical approaches in their daily practice [17], [18], [19]. Attempts to computationally optimize existing Abs for SARS-CoV-2 were already reported in the literature [20], [21] following the work of Giron and co-authors that provided an optimized CR3022 [20]. A large number of such studies emphasize how computational tools are being routinely used for complementing experiments in the Ab design process. A large diversity of algorithms for computational Ab studies are available, from those that predict the structure of an Ab based on its primary sequence only (i.e., antibody modeling) [22], to the ones that make use of detailed structural information from X-ray diffraction data to design improved antibodies from existent ones (i.e., antibody design) [23]. Experimental structural data regarding the complex between the Ab to be improved and its antigen (Ag) greatly improves the prediction accuracy although antibody-antigen (Ab-Ag) computationally modeled structures could be used as well [24]. In short, the general workflow of computational design of novel paratopes includes: (i) generation of new complementarity-determining regions (CDRs) sequences (the part of the variable chains in immunoglobulins) from experimental antibody databases and their grafting/modeling on the antibody framework, (ii) CDRs sequence redesign introducing mutations, (iii) Ab-Ag docking according to the known epitope and relative orientation of the partners, (iv) binding energy evaluation using a scoring function. These steps should be repeated to find a better binding antibody. Common algorithms include: RosettaAntibodyDesign [25], AbDesign [26], OptCDR [27], and OptMAVEn [28]. In addition to this general protocol, other molecular modeling techniques can be applied to improve the accuracy of predictions. Molecular dynamics (MD) simulations [29], [30], represent a well-established methodology in biological sciences [31], [32], [33], [34]. Atomistic MD has been successfully used in the past in Ab design studies for affinity maturation of camelid nanobodies against alpha-synuclein, a weak immunogenic antigen [35], of bevacizumab antibody for an increased affinity to vascular endothelial growth factor A) [36], and of a toll-like receptor targeting Ab [37], to mention just a few examples. However, different steps of Ab-Ag interaction take place on different time scales, which cannot be effectively covered by conventional atomistic simulations only.
In this paper, we propose a novel multiscale approach for in silico affinity maturation of CR3022 against RBD of SARS-CoV-2 in order not only to improve binding but also to understand the molecular determinants at different times/length scales which can be exploited to further modify and modulate the binding affinity. Our approach includes an initial conventional Ab design stage, followed by complimentary evaluations of the best candidates using constant-pH (CpH) Monte Carlo (MC) calculations, constant-charge coarse-grain (CG) MD calculations, and constant-charge atomistic MD simulations. We then selected, as the result of the affinity maturation process, the candidates that showed improved affinity in all four types of evaluations. The strength of our approach is the use of complementary methods for binding affinity estimation, with different physical bases ranging from rigid-body long-range interaction assessment to local conformational rearrangements upon intimate Ab-Ag binding. Furthermore, our multiscale approach allowed us to test our candidates against 11 different strains of the SARS-CoV-2 virus. We identified key residues of the 6 CDRs in the light (L) and heavy (H) chains important for binding, located either at the interaction interface or distant to it, together with mutations that improve binding like S35K (CDR-L1), S72E (CDR-L1), Y110R (CDR-L3), and Y39W (CDR-H1). Also, the total net charge of the mAb proved to be an important parameter that should be considered when aiming to design better binders for the RBD of SARS-CoV-2.
2. Theoretical methods
2.1. Proposed framework for the current study
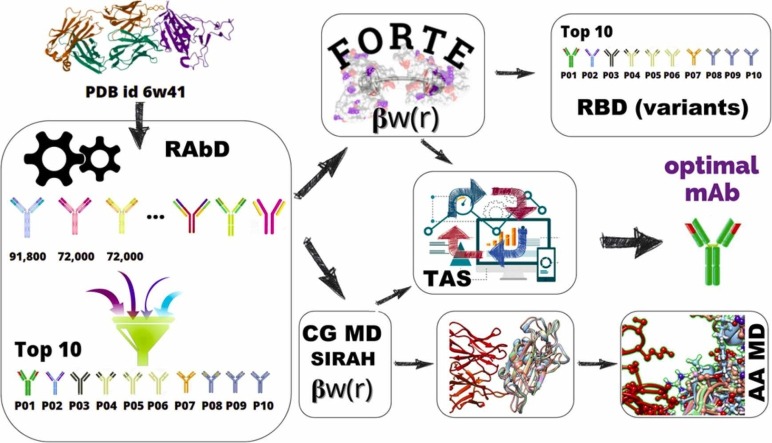
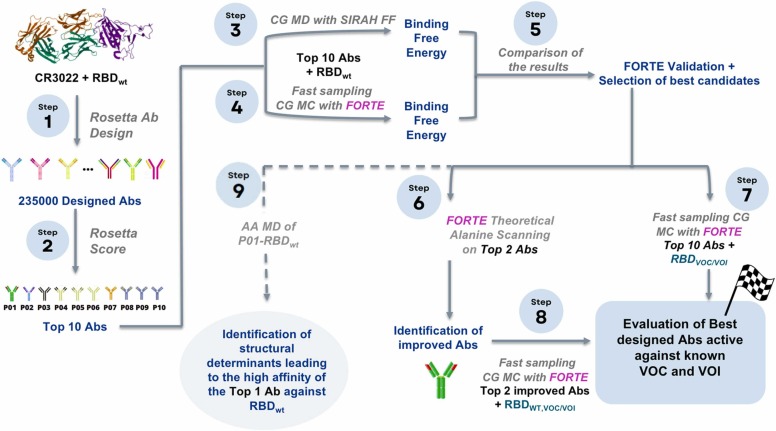
The methods combined in this work (see Fig. 1), are (steps 1–2) a structural-bioinformatics-based methodology to explore macromolecules as potential candidates with higher RBD affinity using an existing experimental RBD-CR3022 complex as a template, (step 3) classical MD simulations with an enhanced sampling to precisely quantify the free energy of interactions for the RBD-binder complexation, and (steps 4, 7–8) a quicker MC sampling of a more simplified protein-protein model to investigate a larger number of complexes and search towards an optimal binder. Additional atomistic simulations were also performed to explore more details of the Ab-Ag interface (step 9 in Fig. 1). These tools’ combination allows us to explore different aspects of the complex physical interactions involved in the Ab-Ag complexation process. The pros and cons of these approaches, together with the procedure in which they were combined here, are detailed in the Supplementary material (SM) in Section S1.
Fig. 1.
Scheme for the multiple scales in silico protocol, consisting of an initial structural-bioinformatics-based methodology to explore macromolecules as potential candidates (steps 1 and 2), constant protonation state CG MD (steps 3), constant-pH (variable protonation state) CG MC simulations (steps 4, 6–8), and an atomistic constant charge MD simulation (step 9). At the end of this cycle, an optimized mAb with a higher binding affinity is obtained. See the text and the SM for more details.
2.2. Structural-bioinformatics-based RAbD approach
The RosettaAntibodyDesign (RAbD) [25] tool was employed as the first step to suggest potential binder candidates for the next phases (see Fig. 1). Considering that any antibody has 6 CDRs (i.e., L1–3 on the light chain and H1–3 on the heavy chain) one has to decide which of these CDRs should be modified concerning the original structure (CR3022 as given by PDB id 6w41, an X-ray data with a resolution of 3.08 Å, pH 4.6 [38]). We ran 3 sets of calculations: (A) all CDR's CDRs considered for full design (using the GraftDesign and SeqDesign procedures of RAbD) [25]; (B) all CDR's CDRst L1 were considered for full design (GraftDesign and SeqDesign). (C) all CCDRs except L1 and H3 were considered for full design (GraftDesign and SeqDesign).
In total, we generated several 91,800 candidates for (A), 72,000 candidates for (B), and 72,000 candidates for (C) scenarios. From the entire pool of candidates, we have selected the best-improved mAbs using two criteria: (I) the complex must have an Ab-Ag interface score below − 150 REU (ROSETTA Energy Units) (the native interface score is −65 REU), and (II) an Ab-Ag interface surface area larger than 1900 Å2 (the native complex interface surface area is 2060 Å2). All RAbD- generated candidates were first ordered based on their ROSETTA score in ascending order. All candidates that did not fulfilled the above criteria were excluded. The remaining candidates were manually (visually) evaluated excluding the unrealistic ones. For the sake of clarity, more explanations on the choice of the above thresholds can be found in the SM files. From this final list, the first 10 best candidates (P01 to P10) from the final RAbD list plus the native complex were considered for the next analyses.
2.3. CG molecular dynamics (MD) with an enhanced sampling approach for free energy calculations
Umbrella sampling (US) constant-protonation state MD simulations were employed to evaluate the free energy of binding of CR3022 and the top ten Ab-Ag complexes selected from the above described RAbD calculations (Step 3 in Fig. 1). In these constant-charge simulations, the protonation states of titratable groups are set at the beginning of the simulation (at pH 7 here) and remain constant [31], [39]. For the efficiency of calculations, a reduced representation of the interacting partners and solvent was adopted, using the SIRAH 2.2 CG force field (FF) [40], an approach successfully used in the past for Ab-Ag complexes free energy calculations [41]. A total number of 36 windows were used for US simulations which ensured a sufficient histogram overlapping using a force constant of 1500 kJ mol−1 nm−2 applied to the center of the mass of the RBD. Ab-Ag complexes were simulated for 16 ns in each US window (the first 2 ns were discarded). US simulations were repeated 10 times. The Potentials of Mean Force (PMF) profiles were constructed using Weighted Histogram Analysis Method [42]. All simulations have been performed with the RBD with the wild-type (wt/Wuhan) sequence (NCBI sequence NC_045512) at constant temperature (300 K) and pressure (1 atm) using GROMACS 2019.3 suite [43], [44] on Beskow supercomputer at PDC Stockholm, Sweden.
2.4. A fast constant-pH coarse-grained simulation approach for free energy calculations on a large scale
The ten selected fragments of mAbs candidates obtained from the RAbD analysis were also submitted to exhaustive investigations utilizing a fast CpH CG biophysical model specially designed for protein-protein complexation [45], [46], [47], [48] – see step 4 in Fig. 1. With “constant-pH” we refer to methods where the amino acid charge is allowed to change during the simulation run as a function of the variable surrounding at a given pH (given as input parameter) [31], [49]. These CG simulations are less expensive than the US calculations with the CG SIRAH FF described above and have the benefit to allow the description of amino-acid protonation state variations occurring at constant pH. This implies that important electrostatic interactions as the charge regulation mechanism are properly included in the model [50], [51], [52], [53], [54]. This electrostatic model is called “FORTE” (Fast cOarse-grained pRotein-proTein modEl). [55], [56], [57] The core of such a model is the fast proton titration scheme [58], [59] combined with the possibility to translate and rotate the macromolecules using the Metropolis MC method [60]. The great advantage of FORTE is the proper modeling of the electrostatic and pH features with a reduced number of sites and a smoother energy landscape. This provides an optimal cost-benefit ratio for longer runs for several systems, especially for semi-quantitative comparisons. Conversely, it is less appealing to quantitatively reproduce experimental K d values. All calculations with FORTE were performed at pH 7, 150 mM of NaCl, and 298 K. After equilibration, at least 3 × 109 MC steps were run during the production phases. Three replicate runs were carried out for each simulated system.
The free energies of interactions (binding free energies), or PMF [w(r)/KBT], as a function of the macromolecule’s separation distances (r) were of primary interest. They were directly calculated from their center-center pair radial distribution functions [βw(r)= -ln g(r)] and sampled using histograms during the production phase of the MC runs. For long enough simulations, βw(r) is typically obtained with good accuracy and able to reproduce experimental trends [20].
Different sets of calculations were carried out with FORTE. Initially (see step 4 in Fig. 1), the binding properties of the 10 top candidates (protein P01 to P10) that were obtained from the structural-bioinformatics-based analysis (using RAbD) were tested to form complexes with the RBD from the SARS-CoV-2 wt: RBDwt+Px → RBDwtPx, where x ranges from 1 to 10. βw(r) was calculated for all systems. Simulations with the fragment of the original mAbs CR3022 were also performed for comparison [38]. All these simulations were repeated for RBDs built up with sequences with different mutations (see step 7 in Fig. 1) present in some relevant VOCs and variants of interest (VOI) (see Section 1 in SM for extended details).
After confirming the outcome from the US calculations with FORTE, an optimization procedure was invoked to explore the possibility of further improving the binding features of the fragment of mAbs given by RAbD (step 6 in Fig. 1). The two best binders classified as Pbest and Pbest-1 by FORTE with a higher chance to block the RBDwt were further submitted to a “theoretical alanine scanning” (TAS) [20], a technique used here to determine the contribution of a specific amino acid to the RBD-binding (see Fig. S2).
2.5. Atomistic MD simulations
To enhance our understanding of the close interactions of our best candidate with RBDwt, and to allow for local conformational re-arrangements at the interaction interface, we used high-resolution models, based on all-atom classical force fields, to simulate Ab-RBDwt complexes. Constraint-free MD simulations of the wild-type CR3022-RBD and Pbest-RBDwt (Pbest is generically used here to refer to the best mAb identified with Rosetta score and in the previous CG simulations) complexes have been performed using the Amber99SB-ILDN FF [61] for protein description and the TIP3P [62] water model (step 9 in Fig. 1). The Ab-RBDwt models have been solvated with enough solvent molecules to ensure a minimum distance between periodic images of 2.2 nm. Ions have been added to neutralize the solvated systems. The length of the simulations was 50 ns at constant pressure (1 bar) and temperature (298 K). The GROMACS 2019.3 suite [44] has been used for all atomistic MD simulations.
3. Results and discussion
3.1. ROSETTA-designed antibody structure: Identifying the top best binder candidates
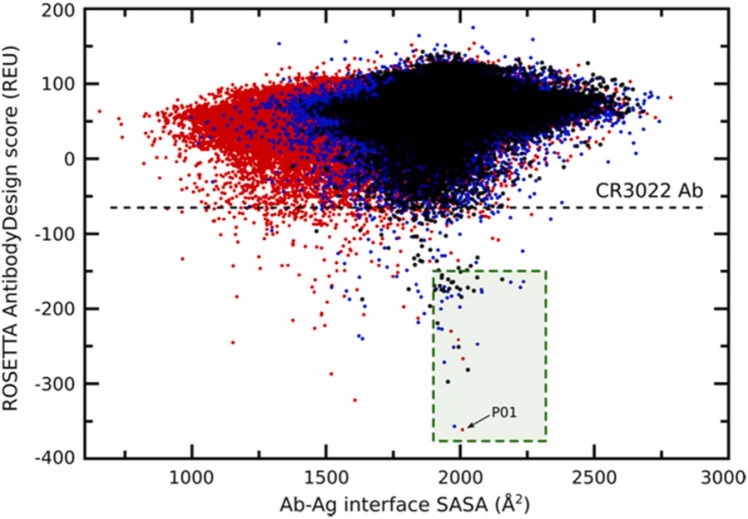
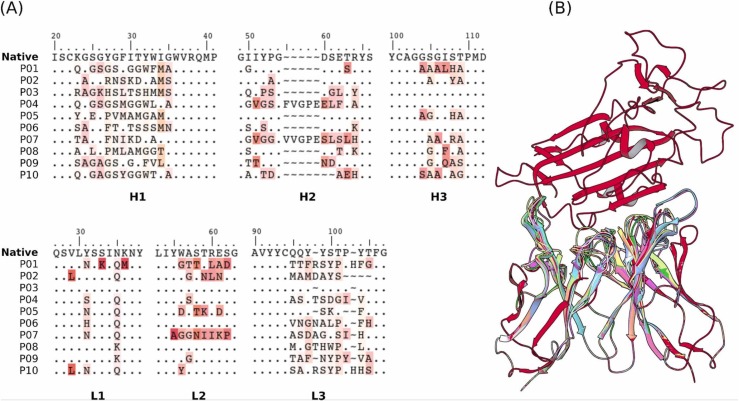
The general workflow of the antibody design procedure consisted of generating a large number of Ab-Ag structures using RAbD [25], followed by ranking of the generated models according to the ROSETTA scoring function. In short, the RAbD algorithm is meant to sample the diverse sequence, structure, and binding space of an antibody-antigen complex. We have generated 235,000 candidates who were ranked according to the ROSETTA scoring function. From the total number of designed candidates, only 383 showed better affinities compared with the native CR3022-RBDwt complex ( Fig. 2). From this list, we choose 10 complexes (according to the criteria described in the “Theoretical Methods” section) that were further evaluated by free energy (ΔG) calculations using both umbrella sampling (US) and FORTE (CDRs primary sequences are presented in Fig. 3). We then considered, as the most promising mAbs candidates, the structures that give consensus results in all three types of evaluations.
Fig. 2.
The 2D plot of RAbD score against antibody-antigen interface SASA (solvent accessible surface area). The horizontal line corresponds to the CR3022/RBDwt complex RAbD score given in Rosetta Energy Units (REU). The green square indicates the area in the two-dimensional plot that corresponds to the mAb candidates' selection criteria. The P01, i.e., the best candidate according to the Rosetta score, is indicated in this plot with an arrow.
Fig. 3.
Sequences of the CDRs (A) and the structures (B) of the 10 selected mAbs for evaluations using US/SIRAH and FORTE methods. Dots represent identities. Residue letters in (A) were colored according to similarity with their counterparts in the reference native CR3022 sequence. In (B) RBDwt (up) was colored in red and each candidate mAb (down) in a different color.
All computational methods have their limitations. Evaluating the Ab-Ag complexes with different computational approaches, and selecting the ones that are consistent in all evaluations, strongly increase the reliability of predictions. Thus, the observed improved affinity of the models could be considered to result from the real physics/chemistry of the complex interactions in the systems and not from any particular algorithm-related issues.
3.2. RBDwt-mAb free energy of interactions explored with umbrella sampling CG simulations
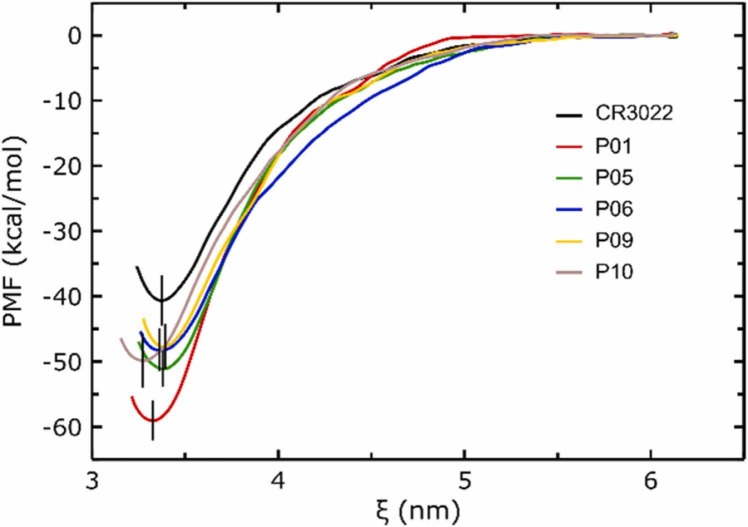
Free energy calculations using US MD calculations revealed that from the list of the top 10 best RAbD candidates only P01, P05, P06, P09, and P10 showed statistically significant better binding affinities to RBDwt compared with the native CR3022 (see Fig. 4 for their PMF profiles). However, the errors in estimating the binding free energies for P05, P06, P09, and P10 did not allow us to distinguish between them in terms of affinity. Among the candidates P01 seems the best, giving statistically significant better affinity of binding compared with both the native CR3022 and P05, P06, P09, and P10.
Fig. 4.
Averaged RBDwt-mAb PMF profiles for the candidates that give statistically significantly better binding affinity than CR3022 in the constant-protonation state MD simulations with the SIRAH CG force field using the umbrella sampling method. The vertical bars represent the estimated standard deviations of the minimum of the PMF profiles. See the text for more details.
However, caution must be taken when considering absolute values obtained by US-CG SIRAH to compare with more rigorous atomistic or experimental data. As the results of Patel et al. (2017) showed, only the ΔΔG = ΔGmutant - ΔGwild-type should be regarded quantitatively, as the granularity of the SIRAH-CG model may affect the absolute values of the calculated ΔG from the PMF profiles. On the other hand, ΔΔG computations based on SIRAH-CG did fit well with experimental data [41] and are much more efficient than atomistic US approaches, which of course give more precise absolute values for ΔG [63]. Therefore, it is meaningless to use the CG-US results for a direct comparison with experimental data like Kd.
The free energy evaluations based on CG-US showed ΔΔG values of − 18.3 kcal/mol for P01 and − 7.1 to − 10.8 kcal/mol for the P01-P05-P06-P09-P10 group relative to the CR3022 antibody. These values are comparable to experimental kinetic data found in the literature for stabilizing mutations induced in Ab-Ag complexes [64], [65], [66].
3.3. RBD-mAb free energy of interactions explored with FORTE
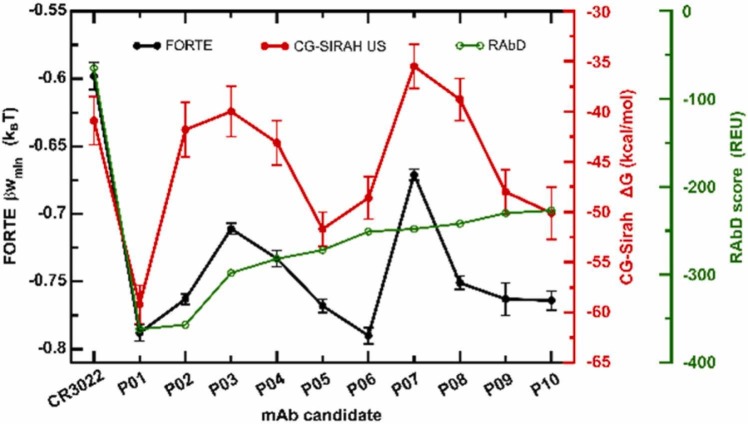
The simplified CpH CG protein-protein model can reproduce the main trends obtained using the more elaborate models (see above). Fig. 5 shows the minimum values of the free energy of interactions for the complexation between the SARS-CoV-2 RBDwt with the fragment of CR3022 and with all engineered mAbs given by RAbD from P01 to P10. These new mAbs candidates all have a higher RBDwt affinity as observed for the simulations with SIRAH FF too (see Fig. 5). They can be ranked as P01/P06 > P02/P05/P08/P09/P10 > P04 > P03 > P05 in terms of their binding affinities. It is virtually impossible to distinguish the proteins within the groups P01/P06 and P02/P05/P08/P09/P10 due to their estimated standard deviations. Conversely, there is no difficulty in determining that the two best candidates are P01 and P06. Both proteins have virtually identical free energy profiles for their complexation with the RBDwt [− 0.788(6) KBT and − 0.790(6) KBT for P01 and P06, respectively). Yet, the mAb P01 was selected as Pbest to follow the consensus with the best binder found in the previous biophysical simulations with the SIRAH FF (see Fig. 4). Since the physical description is not the same between the two models, this reinforces the conclusion that P01 has special features for a stronger binding with the RBDwt.
Fig. 5.
Minima free energy of interactions values [βwmin] measured for the SARS-CoV-2 RBDwt-mAbs complexation at pH 7 and 150 mM of NaCl by the CpH MC simulations (FORTE), CG-SIRAH Umbrella sampling (CG-SIRAH/US), and the RAbD scores, for the wildtype RBDwt and different fragments of mAbs candidates (P01 to P10). The data for the complexation with the native fragment of CR3022 is indicated as WT. The error bars were calculated using the three replicates for each simulation system. We considered as a significant difference any value larger than the estimated standard deviations (see Fig. S8 and Table S2). The minima values obtained by classical MD with SIRAH FF are included in the figure for the sake of comparison. Values from the RAbD in REU are also reported for comparison.
Although P06 was not found equivalent to P01 according to the PMF obtained with the US SIRAH MD (see Figs. 4 and 5), FORTE calculations indicate that it could still be an interesting candidate since it scores as the best together with P01. Therefore, P06 was assumed as Pbest-1, i.e., the second-best binder. The fact that FORTE provided these two binders (P01 and P06) with equivalent binding affinities instead of a single one as seen in the previous approach could be due to a couple of reasons: (a) a smoother energetic landscape in this CG model eventually is not able to describe the same roughness of the phase space, (b) the presence of the charge regulation mechanism in the FORTE approach (absent in the constant-protonation state MD simulations) that give rise to an extra attraction, c) the lack of structural rearrangements in the FORTE approach. Nevertheless, even in the US/SIRAH FF approach, P06 is a better binder than the native CR3022 (see Fig. 4).
Interestingly both CG simulations revealed some subgroups of binders with similar binding free energies among the member of each subgroup (within the error estimation). This suggests that Abs produced from previous infections or by the seroconversion of a given vaccine could have a chance to neutralize more than one strain in agreement with clinical reports [67]. It also illustrates that a weaker response can be obtained depending on the specific mutations as also expected. Equivalent reasoning can be done for therapeutic molecules designed to prevent the attachment of the spike protein via the RBD in the human cell. For instance, Wang and co-authors described a potent mAb from covalent patients that works well against 23 variants [68].
Using CG models it becomes difficult to quantitatively (not qualitatively) reproduce experimental Kd values and/or typically βw(r) values obtained by more detailed force field descriptions [20], [45], [46]. The problem described above for SIRAH FF might be more severe for FORTE due to its higher granularity. Yet, any ambiguity in the interpretation of the obtained data can be solved when results are interpreted in relative terms. For instance, comparing the measurements between a set of similar macromolecules under the same experimental conditions [20], [45]. This is exactly the case for this comparison between different pairs of RBDs and a putative binder. Both CG approaches are useful for distinguishing between stabilizing and destabilizing CDR modifications, for ranking different candidates relative to the wild-type and in-between them. Qualitative comparison can still be made as shown in Fig. 5.
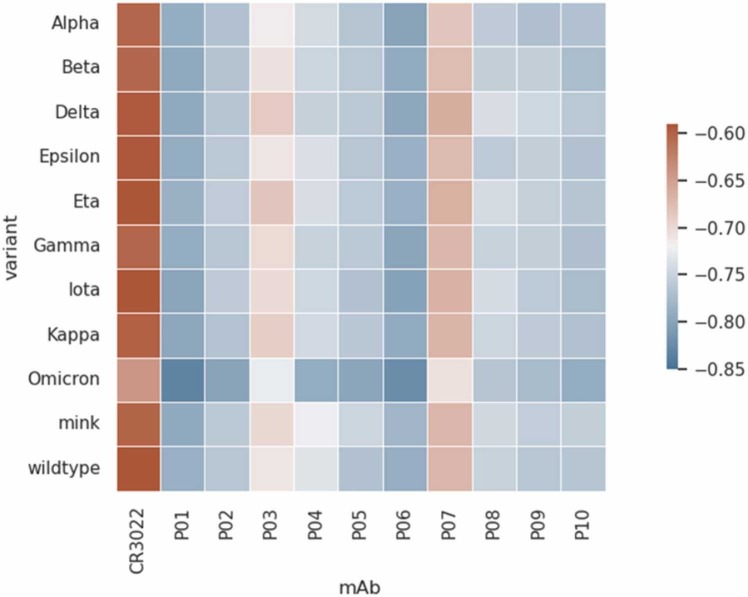
After validating the theoretical predictions obtained with FORTE by comparison with the results from the US on CG simulations with the finer-grained SIRAH FF, we expanded its analysis and tested the binding affinities of these Rosetta-designed fragments of mAbs to the selected mutated form of SARS-CoV-2 RBDs. It is particularly interesting to assess if the engineered mAbs are good binders also for the past and present VOCs (Alpha [69], Beta [70], Gamma [71], Delta [72], and Omicron/BA.1 [73]), VOIs (Epsilon, [74] Eta, [75] Iota, [76] Kappa [72]), and the so-called mink variant [77]. Such information would be vital for any practical therapeutic applications of these mAbs in the future. The free energy data at the same experimental conditions as in Fig. 5 for several pairs of RBDs and mAbs was compiled and displayed as a heatmap-style plot in Fig. 6. In this Figure, the darkest blue is the case with the highest binding affinity. Conversely, the darkest red is given to the system with the lowest binding affinity. This is observed for the native CR3022 interacting with any studied RBD. All putative Rosetta-designed mAbs improved the binding features in comparison with the native CR3022. The general behavior for all RBDs seems similar to what was observed for the interaction with the SARS-CoV-2 RBDwt (see Fig. 5). All molecules P01 to P10 have the potential to block the interaction between virus and host cell preventing the RBD to be available to anchor ACE2. The two best candidates for all these RBDs identified by FORTE were P01 and P06. This reinforces the conclusions concerning their potential ability to neutralize both the wild-type SARS-CoV-2 virus and the main present variants. Considering the estimated error bars, all Rosetta-designed proteins follow the same trend seen in Fig. 5 for RBDwt. They also tend to respond with similar affinities to all studied RBDs, i.e., a good binder for one specific RBD is a good binder for another RBD too. However, there are interesting exceptions. For example, P03 has a relatively lower affinity for RBDDelta, RBDEta, RBDKappa, and RBDmink in comparison with the RBDwt. Surprisingly, both the native CR3022 and all Rosetta-designed binders show a higher affinity for RBDOmicron. This suggests that both CR3022 and its derived-mAbs have a good chance to neutralize the Omicron variant.
Fig. 6.
Heatmap with the minima free energy of interactions values (βwmin in KBT units) for the SARS-CoV-2 RBD-mAbs complexation at pH 7 and 150 mM of NaCl by the CpH MC simulations (FORTE) for the RBD of the main critical variants and different Rosetta-designer binder candidates (P01 to P10). The reference is the native fragment of CR3022. All values of. The maximum estimate error is 0.01. See the text for a description of mutations considered in each case.
3.4. RBD-Ab local interface interactions explored with atomistic MD simulations
We have used atomistic MD simulations to comparatively evaluate P01 with the wild-type CR3022 in terms of different interaction contributions to RBDwt-Ab binding. The simulations were first analyzed for obtaining the probability distribution functions of three interface interaction descriptors (see Fig. S4), namely the number of (i) Ab-Ag salt bridges, (ii) non-polar contacts and (iii) hydrogen bonds (between polar-polar and polar-charged residues). Visual analysis of the trajectories allowed us to evaluate the impact of the introduced mutations on the local molecular environment of the RBDwt-P01 complex. The results showed that the main interaction interface contributions to the increased affinity of P01 to RBDwt, compared with the wild-type CR3022, come from an increased number of salt bridges, hydrogen bonding, and a higher hydrophobic contact area to RBDwt (see SM Section 1 for a comprehensive discussion on the contribution of individual mutated residues to the improved stability of the RBDwt-P01 complex).
3.5. Physical insights to design a more efficient monoclonal antibody for the RBD
Individual residue electrostatic contributions to the complexation RBDwt-P01 were investigated through the TAS procedure described in Section 2. All the ionizable residues of P01 were replaced by ALA one at a time, and we refer to the resulting set comprising all possible single, double, and triple mutations as {P01’}, {P01’’} and {P01’’’}, respectively. The same holds for P06. The best candidates within each set were referenced without brackets. The corresponding βw(r) functions for the complexation RBDwt-P01 ´ were estimated by the FORTE approach. The differences in the βw min values observed for P01 and P01’ [ΔΔG=βw min(P01’)-βw min(P01)] are portrayed in Fig. S5. The purple lines indicate the maximum estimated errors (0.01 KBT). Free energy shifts larger than 0.01 KBT are considered significant. Positive shifts indicate mutations that negatively impact the complexation process. These amino acids (ASP-9. L, GLU-17. L, ASP-72. L, ASP-88. L, ASP-100. L, GLU-11. H, GLU-23. H, GLU-67. H, ASP-83. H, and ASP-137. H) are critical for this molecular mechanism, i.e., replacing one of them with a neutral residue reduces the RBDwt affinity which is not good to block the virus-cell interaction. For instance, the substitution of the ASP residue in position 88 at the light chain (“ASP-88. L” at L2) by ALA decreases the binding affinity by 0.03 KBT units. Conversely, negative values of ΔΔG correspond to mutations that can improve the capability of P01 to bind the RBDwt which is potentially useful to prevent viral entry into the Note that most of the key amino acids that favor the RBDwt-P01 complexation are acid residues (ASP and GLU). There are just a couple of exceptions (e.g., ARG-77 at the light chain) but always with smaller differences than the estimated standard deviations (ΔΔG<0.01 KBT). The neutralization of a basic ionizable group (e.g., LYS-13 at the heavy chain) implies a reduction of the total net charge of the binder and an improvement in its binding features (ΔΔGLYS-13. H = −0.03 KBT). Indeed, it was noticed before that a decrease of the binder net charge gives a higher RBDwt affinity [20]. This information is a useful insight to designing a more efficient therapeutic binder that can avoid the attachment of the RBD to the human cell.
The most important cases where the single ALA mutation in P01 has a stronger influence on the SARS-CoV-2 RBDwt-P01 complexation were mapped in the molecular structure. This can be seen in Fig. S6. Interestingly not only amino acids at the antigen-antibody interface are critical for the complexation. In fact, amino acids buried inside the protein structure can also have an important influence on the complexation due to the long-range nature of the electrostatic interactions and the electrostatic coupling between the titratable groups. This is in line with a broader view of epitopes (and paratopes) defined as “electrostatic epitopes” [78], where any ionizable group can affect the interactions and drive the complexation. It can also be seen that a more negatively charged mAbs tend to have a higher RBD affinity [34]. This is further discussed in Section 3 at the SM.
3.6. Additional optimization of the best RBD binder
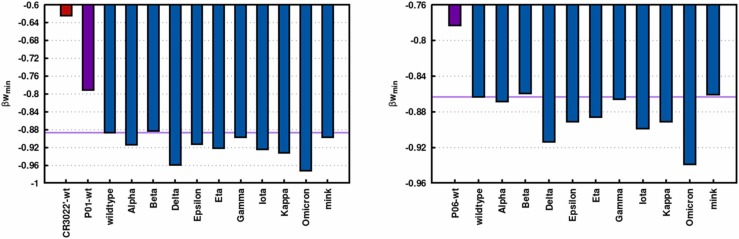
The two best candidates among the sets {P01’’’} (P01’’’: L.R18E, H.K20E, and H.K84A) and {P06’’’} (P06’’’: H.K20A, H.G61E, and H.K84A) obtained through the three cycles of electrostatic optimization pipeline (based on the TAS procedure) were tested against the most common SARS-CoV-2 VOCs and VOIs. This was done to assess if these macromolecules could have the potential to stop the virus from working efficiently targeting RBDs from common VOCs and VOIs. Fig. 7a and b show the βw min values for the complexes RBD-P01’’’ and RBD-P06’’’, respectively. The new optimized binders P01’ and P06’ can form stable molecular complexes with all studied RBDs. Due to the additional electrostatic optimization, they have improved RBD binding affinities in comparison with the initial template (P01 or P06) provided by the RAbD approach. Consequently, they have more chances for a successful neutralization in vitro/in vivo of the virus. Moreover, this result indicated that the integrated biophysical modeling multi-approach used here offers an efficient route to designing better macromolecular ligands. Intriguingly both putative mAbs P01’’’ and P06’’’ have a stronger affinity for some RBDs from variants that are threatening the immune response. For instance, P01’’’ has the highest affinity observed for both RBDDelta and RBDOmicron. This is a promising result to deal with the present crisis offering two ideal candidates to treat the pathogenic threats from different SARS-CoV-2 variants. Quite recently, a pre-print work suggested cross-reactivity specifically between antibodies for Delta and Omicron variants [79]. P01’’’ is slightly better than P06’’’ due to its higher RBD affinity for all mutants in general (e.g., βw min = −0.96(1) KBT for RBDDelta-P01’’’, and βw min = −0.92(1) KBT for RBDDelta-P06’’’). From a bioinformatics point of view, it is interesting to point out that an optimized mAb that is solely obtained by the three cycles electrostatic optimization pipeline (CR3022’) [20] is not as good as the one achieved by the present strategy.
Fig. 7.
Binding RBD affinities for optimized mAbs obtained through the three cycles electrostatic optimization pipelines. (a) Left panel: mAb P01’’’ based on the Rosetta-designed P01. (b) Right panel: mAb P06’’’ based on the Rosetta-designed P06. Data from the estimated βwmin values for the molecular complexation between RBDs from different variants with P01’’’ (P01 with three mutations L: R18E and H: K20E and K84A) and P06’’' (P06 with three mutations H: K20A, G61E, and K84A), respectively, are shown in blue bars. Data for the complexes with the original Rosetta-designed mAbs (RBDwt-P01 and RBDwt-P06) before the electrostatic optimization process and CR3022 ´ (RBDwt-CR3022 ´) are given in the purple and red bars, respectively, for comparison. The purple lines are drawn to guide the eyes for the comparison with the outcomes for the RBDwt. See the text for more details.
4. Conclusions
A highly efficient and robust multiple scales in silico protocol combing the RosettaAntibodyDesign with MD and MC molecular simulations and an electrostatic optimization pipeline was proposed and applied to CR3022. Two more efficient binder candidates, P01 and P06, were obtained and show clearly the strongest interaction.
In a closer analysis, we can find that amino acids both at the surface and also deeper inside the protein structure are critical for the complexation through electrostatic coupling between titratable groups and are characterized as “electrostatic epitopes”.
Furthermore, we can observe that the net charge of the mAbs is one important determinant in binding affinity and there is a clear nearly linear tendency for lower charged mAbs to exhibit higher affinity to RBD. The Coulombic forces are driving the antigen-antibody association, as also observed in previous studies [20], [63], while other short-range interactions are important in the close-range association as well.
Our combined multi-scale approach is found to be a fast, robust, and reliable tool to design better macromolecular ligands allowing us to identify the best candidates for the different variants of SARS-CoV-2 including Omicron. It is a pragmatic approach for a short development cycle for SARS-CoV-2 diagnosis, treatment, and prevention.
This multi-approach is a general theoretical framework towards high specific antibodies for SARS-CoV-2 and can be extended to other diseases (e.g., other viral and bacterial pathogens, leukemia, cancer, multiple sclerosis, rheumatoid arthritis, lupus) both for diagnosis and therapeutic purposes. It requires only the knowledge of the target key macromolecule and an initial putative binder, in order to improve it by theoretical design before synthesis and testing.
Credit Author Statement
Andrei Neamtu: Conceptualization, Investigation, Methodology, Software, Data curation, Writing – original draft, Writing – reviewing and editing, Francesca Mocci: Data curation, Methodology, Writing – original draft, Writing – reviewing and editing, Aatto Laaksonen: Data curation, Methodology, Writing – original draft, Writing – reviewing and editing, Fernando Barroso: Conceptualization, Investigation, Methodology, Software, Data curation, Writing – original draft, Writing – reviewing and editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work has been supported in part by the “Fundação de Amparo à Pesquisa do Estado de São Paulo” [Fapesp 2020/07158–2 (F.L.B.d.S.)] and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) [CNPq 305393/2020–0 (FLBdS)]. F.L.B.d.S. is also deeply thankful for resources provided by the Swedish National Infrastructure for Computing (SNIC) at NSC and PDC. A.L.acknowledges the Swedish Research Council for financial support, and partial support from a grant from the Ministry of Research and Innovation of Romania (CNCS - UEFISCDI, project number PN-III-P4-ID-PCCF-2016–0050, within PNCDI III). F.M. acknowledges financial support from Progetto Fondazione di Sardegna (Grant CUP: F72F20000230007).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.colsurfb.2022.112986.
Appendix A. Supplementary material
Supplementary material.
.
Data availability
Data will be made available on request.
References
- 1.Mallapaty S., Callaway E., Kozlov M., Ledford H., Pickrell J., Van Noorden R. How COVID vaccines shaped 2021 in eight powerful charts. Nature. 2021;600(7890):580–583. doi: 10.1038/d41586-021-03686-x. [DOI] [PubMed] [Google Scholar]
- 2.Gupta R.K., Topol E.J. COVID-19 vaccine breakthrough infections. Science. 2021;374(6575):1561–1562. doi: 10.1126/science.abl8487. [DOI] [PubMed] [Google Scholar]
- 3.Prates-Syed W.A., Chaves L.C.S., Crema K.P., Vuitika L., Lira A., Côrtes N., Kersten V., Guimarães F.E.G., Sadraeian M., Barroso da Silva F.L., Cabral-Marques O., Barbuto J.A.M., Russo M., Câmara N.O.S., Cabral-Miranda G. VLP-based COVID-19 vaccines: an adaptable technology against the threat of new variants. Vaccines. 2021;9(12):1409. doi: 10.3390/vaccines9121409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reis G., dos Santos Moreira-Silva E.A., Silva D.C.M., Thabane L., Milagres A.C., Ferreira T.S., dos Santos C.V.Q., de Souza Campos V.H., Nogueira A.M.R., de Almeida A.P.F.G., Callegari E.D., de Figueiredo Neto A.D., Savassi L.C.M., Simplicio M.I.C., Ribeiro L.B., Oliveira R., Harari O., Forrest J.I., Ruton H., Sprague S., McKay P., Glushchenko A.V., Rayner C.R., Lenze E.J., Reiersen A.M., Guyatt G.H., Mills E.J. Effect of early treatment with fluvoxamine on risk of emergency care and hospitalisation among patients with COVID-19: the together randomised, platform clinical trial. Lancet Glob. Health. 2022;10(1):e42–e51. doi: 10.1016/S2214-109X(21)00448-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keller M.A., Stiehm E.R. Passive immunity in prevention and treatment of infectious diseases. Clin. Microbiol. Rev. 2000;13(4):602–614. doi: 10.1128/CMR.13.4.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar S., Chandele A., Sharma A. Current status of therapeutic monoclonal antibodies against SARS-CoV-2. PLOS Pathog. 2021;17(9) doi: 10.1371/journal.ppat.1009885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., Gilchuk P., Zost S.J., Tahan S., Droit L., Turner J.S., Kim W., Schmitz A.J., Thapa M., Wang D., Boon A.C.M., Presti R.M., O’Halloran J.A., Kim A.H.J., Deepak P., Pinto D., Fremont D.H., Crowe J.E., Corti D., Virgin H.W., Ellebedy A.H., Shi P.-Y., Diamond M.S. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27(4):717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hastie K.M., Li H., Bedinger D., Schendel S.L., Dennison S.M., Li K., Rayaprolu V., Yu X., Mann C., Zandonatti M., Diaz Avalos R., Zyla D., Buck T., Hui S., Shaffer K., Hariharan C., Yin J., Olmedillas E., Enriquez A., Parekh D., Abraha M., Feeney E., Horn G.Q., CoVIC-DB Team Aldon, Ali Y., Aracic H., Cobb S., Federman R.R., Fernandez R.S., Glanville J.M., Green J., Grigoryan R., Lujan Hernandez G., Ho A.G., Huang D.D., Ingraham K.-Y.A., Jiang J., Kellam W., Kim P., Kim C., Kim M., Kong H.M., Krebs C., Lan S.J., Lang F., Lee G., Leung S., Liu C.L., Lu J., MacCamy Y., McGuire A., Palser A.T., Rabbitts A.L., Rikhtegaran Tehrani T.H., Sajadi Z., Sanders M.M., Sato R.W., Schweizer A.K., Seo L., Shen J., Snitselaar B., Stamatatos J.L., Tan L., Tomic Y., van Gils M.T., Youssef M.J., Yu S., Yuan J., Zhang T.Z., Peters Q., Tomaras B., Germann G.D., Saphire T., E. O Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: a global consortium study. Science. 2021;374(6566):472–478. doi: 10.1126/science.abh2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boggiano C., Eisinger R.W., Lerner A.M., Anderson J.M., Woodcock J., Fauci A.S., Collins F.S. Update on and future directions for use of anti–SARS-CoV-2 antibodies: national institutes of health summit on treatment and prevention of COVID-19. Ann. Intern. Med. 2021:M21–3669. doi: 10.7326/M21-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubin R. Monoclonal antibodies for COVID-19 preexposure prophylaxis can’t come fast enough for some people. JAMA. 2021;326(19):1895. doi: 10.1001/jama.2021.19534. [DOI] [PubMed] [Google Scholar]
- 11.Barnes C.O., Jette C.A., Abernathy M.E., Dam K.-M.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., Robbiani D.F., Nussenzweig M.C., West A.P., Bjorkman P.J. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588(7839):682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giron C.C., Laaksonen A., Barroso da Silva F.L. Up state of the SARS-COV-2 spike homotrimer favors an increased virulence for new variants. Front. Med. Technol. 2021;3 doi: 10.3389/fmedt.2021.694347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan R., Zhang Y., Li Y., Ye F., Guo Y., Xia L., Zhong X., Chi X., Zhou Q. Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2. Cell Res. 2021;31(6):717–719. doi: 10.1038/s41422-021-00490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao, Y.; Wang, J.; Jian, F.; Xiao, T.; Song, W.; Yisimayi, A.; Huang, W.; Li, Q.; Wang, P.; An, R.; Wang, J.; Wang, Y.; Niu, X.; Yang, S.; Liang, H.; Sun, H.; Li, T.; Yu, Y.; Cui, Q.; Liu, S.; Yang, X.; Du, S.; Zhang, Z.; Hao, X.; Shao, F.; Jin, R.; Wang, X.; Xiao, J.; Wang, Y.; Xie, X.S. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies; 2021; p 2021.12.07.470392. 10.1101/2021.12.07.470392. [DOI] [PMC free article] [PubMed]
- 15.Mader A.-L., Tydykov L., Glück V., Bertok M., Weidlich T., Gottwald C., Stefl A., Vogel M., Plentz A., Köstler J., Salzberger B., Wenzel J.J., Niller H.H., Jantsch J., Wagner R., Schmidt B., Glück T., Gessner A., Peterhoff D. Omicron’s binding to sotrovimab, casirivimab, imdevimab, CR3022, and sera from previously infected or vaccinated individuals. iScience. 2022;25(4) doi: 10.1016/j.isci.2022.104076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parums D.V. Editorial: revised world health organization (WHO) terminology for variants of concern and variants of interest of SARS-CoV-2. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2021;27:e933622–1-e933622-2. doi: 10.12659/MSM.933622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ibrahim B., McMahon D.P., Hufsky F., Beer M., Deng L., Mercier P.L., Palmarini M., Thiel V., Marz M. A new era of virus bioinformatics. Virus Res. 2018;251:86–90. doi: 10.1016/j.virusres.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Sato H., Yokoyama M., Toh H. Genomics and computational science for virus research. Front. Microbiol. 2013;4 doi: 10.3389/fmicb.2013.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma D., Priyadarshini P., Vrati S. Unraveling the web of viroinformatics: computational tools and databases in virus research. J. Virol. 2015;89(3):1489–1501. doi: 10.1128/JVI.02027-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corrêa Giron C., Laaksonen A., Barroso da Silva F.L. On the interactions of the receptor-binding domain of SARS-CoV-1 and SARS-CoV-2 spike proteins with monoclonal antibodies and the receptor ACE2. Virus Res. 2020;285 doi: 10.1016/j.virusres.2020.198021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luan B., Huynh T. In silico antibody mutagenesis for optimizing its binding to spike protein of severe acute respiratory syndrome coronavirus 2. J. Phys. Chem. Lett. 2020;11(22):9781–9787. doi: 10.1021/acs.jpclett.0c02706. [DOI] [PubMed] [Google Scholar]
- 22.Weitzner B.D., Jeliazkov J.R., Lyskov S., Marze N., Kuroda D., Frick R., Adolf-Bryfogle J., Biswas N., Dunbrack R.L., Gray J.J. Modeling and docking of antibody structures with rosetta. Nat. Protoc. 2017;12(2):401–416. doi: 10.1038/nprot.2016.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chowdhury R., Grisewood M.J., Boorla V.S., Yan Q., Pfleger B.F., Maranas C.D. IPRO+/−: computational protein design tool allowing for insertions and deletions. Structure. 2020;28(12):1344–1357. doi: 10.1016/j.str.2020.08.003. [DOI] [PubMed] [Google Scholar]
- 24.Sivasubramanian A., Sircar A., Chaudhury S., Gray J.J. Toward high-resolution homology modeling of antibody F v regions and application to antibody-antigen docking. Proteins Struct. Funct. Bioinform. 2009;74(2):497–514. doi: 10.1002/prot.22309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adolf-Bryfogle J., Kalyuzhniy O., Kubitz M., Weitzner B.D., Hu X., Adachi Y., Schief W.R., Dunbrack R.L. Rosetta antibody design (RAbD): a general framework for computational antibody design. PLOS Comput. Biol. 2018;14(4) doi: 10.1371/journal.pcbi.1006112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lapidoth G.D., Baran D., Pszolla G.M., Norn C., Alon A., Tyka M.D., Fleishman S.J. AbDesign: an algorithm for combinatorial backbone design guided by natural conformations and sequences: combinatorial backbone design in antibodies. Proteins Struct. Funct. Bioinforma. 2015;83(8):1385–1406. doi: 10.1002/prot.24779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pantazes R.J., Maranas C.D. OptCDR: a general computational method for the design of antibody complementarity determining regions for targeted epitope binding. Protein Eng. Des. Sel. 2010;23(11):849–858. doi: 10.1093/protein/gzq061. [DOI] [PubMed] [Google Scholar]
- 28.Chowdhury R., Allan M.F., Maranas C.D. OptMAVEn-2.0: De Novo design of variable antibody regions against targeted antigen epitopes. Antibodies. 2018;7(3):23. doi: 10.3390/antib7030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alder B.J., Wainwright T.E. Studies in molecular dynamics. I. General method. J. Chem. Phys. 1959;31(2):459–466. [Google Scholar]
- 30.Rahman A. Correlations in the motion of atoms in liquid argon. Phys. Rev. 1964;136(2A):A405–A411. [Google Scholar]
- 31.Barroso da Silva F.L., Carloni P., Cheung D., Cottone G., Donnini S., Foegeding E.A., Gulzar M., Jacquier J.C., Lobaskin V., MacKernan D., Mohammad Hosseini Naveh Z., Radhakrishnan R., Santiso E.E. Understanding and controlling food protein structure and function in foods: perspectives from experiments and computer simulations. Annu. Rev. Food Sci. Technol. 2020;11(1):365–387. doi: 10.1146/annurev-food-032519-051640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hollingsworth S.A., Dror R.O. Molecular dynamics simulation for all. Neuron. 2018;99(6):1129–1143. doi: 10.1016/j.neuron.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karplus M., McCammon J.A. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 2002;9(9):646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- 34.McCammon J.A., Gelin B.R., Karplus M. Dynamics of folded proteins. Nature. 1977;267(5612):585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- 35.Mahajan S.P., Meksiriporn B., Waraho-Zhmayev D., Weyant K.B., Kocer I., Butler D.C., Messer A., Escobedo F.A., DeLisa M.P. Computational affinity maturation of camelid single-domain intrabodies against the nonamyloid component of alpha-synuclein. Sci. Rep. 2018;8(1) doi: 10.1038/s41598-018-35464-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corrada D., Colombo G. Energetic and dynamic aspects of the affinity maturation process: characterizing improved variants from the bevacizumab antibody with molecular simulations. J. Chem. Inf. Model. 2013;53(11):2937–2950. doi: 10.1021/ci400416e. [DOI] [PubMed] [Google Scholar]
- 37.Ahmad B., Batool M., Kim M.-S., Choi S. Computational-driven epitope verification and affinity maturation of TLR4-targeting antibodies. Int. J. Mol. Sci. 2021;22(11):5989. doi: 10.3390/ijms22115989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan M., Wu N.C., Zhu X., Lee C.-C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020 doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Damjanović A., Wu X., García-Moreno E., Brooks B.R. Backbone relaxation coupled to the ionization of internal groups in proteins: a self-guided langevin dynamics study. Biophys. J. 2008;95(9):4091–4101. doi: 10.1529/biophysj.108.130906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Machado M.R., Barrera E.E., Klein F., Sóñora M., Silva S., Pantano S. The SIRAH 2.0 force field: altius, fortius, citius. J. Chem. Theory Comput. 2019;15(4):2719–2733. doi: 10.1021/acs.jctc.9b00006. [DOI] [PubMed] [Google Scholar]
- 41.Patel J.S., Ytreberg F.M. Fast calculation of protein–protein binding free energies using umbrella sampling with a coarse-grained model. J. Chem. Theory Comput. 2018;14(2):991–997. doi: 10.1021/acs.jctc.7b00660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hub J.S., de Groot B.L., van der Spoel D. G_wham – a free weighted histogram analysis implementation including robust error and autocorrelation estimates. J. Chem. Theory Comput. 2010;6(12):3713–3720. doi: 10.1021/ct100494z. [DOI] [Google Scholar]
- 43.Abraham, M.J.; van der Spoel, D.; Lindahl, E.; Hess, B.; and the GROMACS development team. GROMACS User Manual Version 2019, 2019.
- 44.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 45.Barroso da Silva F.L., Pasquali S., Derreumaux P., Dias L.G. Electrostatics analysis of the mutational and PH effects of the N-terminal domain self-association of the major ampullate spidroin. Soft Matter. 2016;12:5600–5612. doi: 10.1039/c6sm00860g. [DOI] [PubMed] [Google Scholar]
- 46.Delboni L., Barroso da Silva F.L. On the complexation of whey proteins. Food Hydrocoll. 2016;55:89–99. [Google Scholar]
- 47.Kurut A., Persson B.A., AAkesson T., Forsman J., Lund M. Anisotropic interactions in protein mixtures: self assembly and phase behavior in aqueous solution. J. Phys. Chem. Lett. 2012;3(6):731–734. doi: 10.1021/jz201680m. [DOI] [PubMed] [Google Scholar]
- 48.Persson B., Lund M., Forsman J., Chatterton D.E.W., Akesson Torbjörn. Molecular evidence of stereo-specific lactoferrin dimers in solution. Biophys. Chem. 2010;3(3):187–189. doi: 10.1016/j.bpc.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 49.Barroso da Silva F.L., Dias L.G. Development of constant-PH simulation methods in implicit solvent and applications in biomolecular systems. Biophys. Rev. 2017;9(5):699–728. doi: 10.1007/s12551-017-0311-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adžić N., Podgornik R. Charge regulation in ionic solutions: thermal fluctuations and Kirkwood-Schumaker interactions. Phys. Rev. E. 2015;91(2) doi: 10.1103/physreve.91.022715. [DOI] [PubMed] [Google Scholar]
- 51.Barroso da Silva F.L., Derreumaux P., Pasquali S. Protein-RNA complexation driven by the charge regulation mechanism. Biochem. Biophys. Res. Commun. 2018;498(2):264–273. doi: 10.1016/j.bbrc.2017.07.027. [DOI] [PubMed] [Google Scholar]
- 52.Barroso da Silva F.L., Lund M., Jönsson B., AAkesson T. On the interaction between protein and polyelectrolyte. J. Phys. Chem. B. 2006;110(9):4459–4464. doi: 10.1021/jp054880l. [DOI] [PubMed] [Google Scholar]
- 53.Barroso Da Silva F.L., Jönsson B. Polyelectrolyte-protein complexation driven by charge regulation. Soft Matter. 2009;5(15):2862–2868. [Google Scholar]
- 54.Lund M., Jönsson B. Charge regulation in biomolecular solution. Q. Rev. Biophys. 2013;46:265–281. doi: 10.1017/S003358351300005X. [DOI] [PubMed] [Google Scholar]
- 55.Poveda-Cuevas S.A., Etchebest C., Barroso da Silva F.L. Self-association features of NS1 proteins from different flaviviruses. Virus Res. 2022 doi: 10.1016/j.virusres.2022.198838. [DOI] [PubMed] [Google Scholar]
- 56.Giron C.C., Laaksonen A., Barroso da Silva F.L. Differences between omicron SARS-CoV-2 RBD and other variants in their ability to interact with cell receptors and monoclonal antibodies. J. Biomol. Struct. Dyn. 2022;0(0):1–21. doi: 10.1080/07391102.2022.2095305. [DOI] [PubMed] [Google Scholar]
- 57.Barroso da Silva F.L., Giron C.C., Laaksonen A. Electrostatic features for the receptor binding domain of SARS-COV-2 wildtype and its variants. Compass to the severity of the future variants with the charge-rule. J. Phys. Chem. B. 2022 doi: 10.1021/acs.jpcb.2c04225. [DOI] [PubMed] [Google Scholar]
- 58.Barroso da Silva F.L.B., da; MacKernan D. Benchmarking a fast proton titration scheme in implicit solvent for biomolecular simulations. J. Chem. Theory Comput. 2017;13(6):2915–2929. doi: 10.1021/acs.jctc.6b01114. [DOI] [PubMed] [Google Scholar]
- 59.Teixeira A.A., Lund M., Barroso da Silva F.L. Fast proton titration scheme for multiscale modeling of protein solutions. J. Chem. Theory Comput. 2010;6:3259–3266. doi: 10.1021/ct1003093. [DOI] [PubMed] [Google Scholar]
- 60.Metropolis N.A., Rosenbluth A.W., Rosenbluth M.N., Teller A., Teller E. Equation of state calculations by fast computing machines. J. Chem. Phys. 1953;21:1087–1097. [Google Scholar]
- 61.Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O., Shaw D.E. Improved side-chain torsion potentials for the amber Ff99SB protein force field. Proteins Struct. Funct. Bioinform. 2010;78(8):1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–953. [Google Scholar]
- 63.Nguyen H., Lan P.D., Nissley D.A., O’Brien E.P., Li M.S. Electrostatic interactions explain the higher binding affinity of the CR3022 antibody for SARS-CoV-2 than the 4A8 antibody. J. Phys. Chem. B. 2021;125(27):7368–7379. doi: 10.1021/acs.jpcb.1c03639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moal I.H., Fernández-Recio J. SKEMPI: a structural kinetic and energetic database of mutant protein interactions and its use in empirical models. Bioinformatics. 2012;28(20):2600–2607. doi: 10.1093/bioinformatics/bts489. [DOI] [PubMed] [Google Scholar]
- 65.Pons J., Rajpal A., Kirsch J.F. Energetic analysis of an antigen/antibody interface: alanine scanning mutagenesis and double mutant cycles on the Hyhel-10/Lysozyme interaction. Protein Sci. 1999;8(5):958–968. doi: 10.1110/ps.8.5.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pruett P.S., Air G.M. Critical interactions in binding antibody NC41 to influenza N9 neuraminidase: amino acid contacts on the antibody heavy chain. Biochemistry. 1998;37(30):10660–10670. doi: 10.1021/bi9802059. [DOI] [PubMed] [Google Scholar]
- 67.Edara V.V., Hudson W.H., Xie X., Ahmed R., Suthar M.S. Neutralizing antibodies against SARS-CoV-2 variants after infection and vaccination. JAMA. 2021;325(18):1896–1898. doi: 10.1001/jama.2021.4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang L., Zhou T., Zhang Y., Yang E.S., Schramm C.A., Shi W., Pegu A., Oloniniyi O.K., Henry A.R., Darko S., Narpala S.R., Hatcher C., Martinez D.R., Tsybovsky Y., Phung E., Abiona O.M., Antia A., Cale E.M., Chang L.A., Choe M., Corbett K.S., Davis R.L., DiPiazza A.T., Gordon I.J., Hait S.H., Hermanus T., Kgagudi P., Laboune F., Leung K., Liu T., Mason R.D., Nazzari A.F., Novik L., O’Connell S., O’Dell S., Olia A.S., Schmidt S.D., Stephens T., Stringham C.D., Talana C.A., Teng I.-T., Wagner D.A., Widge A.T., Zhang B., Roederer M., Ledgerwood J.E., Ruckwardt T.J., Gaudinski M.R., Moore P.L., Doria-Rose N.A., Baric R.S., Graham B.S., McDermott A.B., Douek D.C., Kwong P.D., Mascola J.R., Sullivan N.J., Misasi J. Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants. Science. 2021;373(6556):eabh1766. doi: 10.1126/science.abh1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang T.-J., Yu P.-Y., Chang Y.-C., Liang K.-H., Tso H.-C., Ho M.-R., Chen W.-Y., Lin H.-T., Wu H.-C., Hsu S.-T.D. Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function. Nat. Struct. Mol. Biol. 2021;28(9):731–739. doi: 10.1038/s41594-021-00652-z. [DOI] [PubMed] [Google Scholar]
- 70.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., Mlisana K., von Gottberg A., Walaza S., Allam M., Ismail A., Mohale T., Glass A.J., Engelbrecht S., Van Zyl G., Preiser W., Petruccione F., Sigal A., Hardie D., Marais G., Hsiao N., Korsman S., Davies M.-A., Tyers L., Mudau I., York D., Maslo C., Goedhals D., Abrahams S., Laguda-Akingba O., Alisoltani-Dehkordi A., Godzik A., Wibmer C.K., Sewell B.T., Lourenço J., Alcantara L.C.J., Kosakovsky Pond S.L., Weaver S., Martin D., Lessells R.J., Bhiman J.N., Williamson C., de Oliveira T. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592(7854):438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 71.Voloch C.M., da Silva Francisco R., de Almeida L.G.P., Cardoso C.C., Brustolini O.J., Gerber A.L., Guimarães A.P., de C., Mariani D., da Costa R.M., Ferreira O.C., Covid19-UFRJ Workgroup L.N.C.C., Workgroup Cavalcanti, Frauches A.C., de Mello T.S., Leitão C.M.B., de I., Galliez C., Faffe R.M., Castiñeiras D.S., Tanuri T.M.P.P., de Vasconcelos, A. T. R A. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J. Virol. 2021;95(10):e00119–e00121. doi: 10.1128/JVI.00119-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang J., Xiao T., Cai Y., Lavine C.L., Peng H., Zhu H., Anand K., Tong P., Gautam A., Mayer M.L., Walsh R.M., Rits-Volloch S., Wesemann D.R., Yang W., Seaman M.S., Lu J., Chen B. Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 delta variant. Science. 2021;374(6573):1353–1360. doi: 10.1126/science.abl9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ford C.T., Jacob Machado D., Janies D.A. Predictions of the SARS-CoV-2 omicron variant (B.1.1.529) spike protein receptor-binding domain structure and neutralizing antibody interactions. Front. Virol. 2022;2 [Google Scholar]
- 74.Zhang W., Davis B.D., Chen S.S., Sincuir Martinez J.M., Plummer J.T., Vail E. Emergence of a novel SARS-CoV-2 variant in Southern California. JAMA. 2021;325(13):1324. doi: 10.1001/jama.2021.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pereira F., Tosta S., Lima M.M., Reboredo de Oliveira da Silva L., Nardy V.B., Gómez M.K.A., Lima J.G., Fonseca V., Oliveira T., Lourenço J., Alcantara L.C., Giovanetti M., Leal A. Genomic surveillance activities unveil the introduction of the SARS‐CoV‐2 B.1.525 variant of interest in Brazil: case report. J. Med. Virol. 2021;93(9):5523–5526. doi: 10.1002/jmv.27086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.West A.P., Wertheim J.O., Wang J.C., Vasylyeva T.I., Havens J.L., Chowdhury M.A., Gonzalez E., Fang C.E., Di Lonardo S.S., Hughes S., Rakeman J.L., Lee H.H., Barnes C.O., Gnanapragasam P.N.P., Yang Z., Gaebler C., Caskey M., Nussenzweig M.C., Keeffe J.R., Bjorkman P.J. Detection and characterization of the SARS-CoV-2 lineage B.1.526 in New York. Nat. Commun. 2021;12(1):4886. doi: 10.1038/s41467-021-25168-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bayarri-Olmos R., Rosbjerg A., Johnsen L.B., Helgstrand C., Bak-Thomsen T., Garred P., Skjoedt M.-O. The SARS-CoV-2 Y453F mink variant displays a pronounced increase in ACE-2 affinity but does not challenge antibody neutralization. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Poveda-Cuevas S.A., Etchebest C., Barroso da Silva F.L. Identification of electrostatic epitopes in flavivirus by computer simulations: the PROCEED pKa method. J. Chem. Inf. Model. 2020;60(2):944–963. doi: 10.1021/acs.jcim.9b00895. [DOI] [PubMed] [Google Scholar]
- 79.Khan K., Karim F., Cele S., San J.E., Lustig G., Tegally H., Bernstein M., Ganga Y., Jule Z., Reedoy K., Ngcobo N., Mazibuko M., Mthabela N., Mhlane Z., Mbatha N., Giandhari J., Ramphal Y., Naidoo T., Manickchund N., Magula N., Abdool Karim S.S., Gray G., Hanekom W., von Gottberg A., Team C.O.M.M.I.T.-K.Z.N., Gosnell B.I., Lessells R.J., Moore P.L., de Oliveira T., Moosa M.-Y.S., Sigal A. Omicron infection enhances neutralizing immunity against the delta variant; preprint. Infect. Dis. 2021 doi: 10.1101/2021.12.27.21268439. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Data Availability Statement
Data will be made available on request.