Abstract
Brain tumors remain one of the most difficult tumors to treat and, depending on the histology, have a poor prognosis. Of brain tumors, glioblastoma (GBM) is the most common malignant glioma and has a dismal prognosis, with only about 5% of patients alive five years after diagnosis. While advances in targeted therapies and immunotherapies are rapidly improving outcomes in a variety of other cancers, the standard of care for GBM has largely remained unaltered since 2005. There are many well-studied challenges that are either unique to brain tumors (i.e., blood-brain barrier and immunosuppressive environment) or amplified within GBM (i.e., tumor heterogeneity at the cellular and molecular levels, plasticity, and cancer stem cells) that make this disease particularly difficult to treat. While we touch on all these concepts, the focus of this review is to discuss the immense inter- and intra-tumoral heterogeneity and advances in our understanding of tumor cell plasticity and epigenetics in GBM. With each improvement in technology, our understanding of the complexity of tumoral heterogeneity and plasticity improves and we gain more clarity on the causes underlying previous therapeutic failures. However, these advances are unlocking new therapeutic opportunities that scientists and physicians are currently exploiting and have the potential for new breakthroughs.
Keywords: Glioblastoma, Heterogeneity, Plasticity, Epigenetics, Tumor Microenvironment
Introduction
A study published in 2017 by the National Cancer Institute found across-the-board improvements in 5-year survival for patients with cancer from the 1970s to 2013 (except for cervical and uterine cancer) [1]. However, even with this improvement in survival, patients with brain tumors still have only a 30% survival at 5 years after diagnosis, and this number is far worse for some tumors types, such as glioblastoma (GBM) in adults and diffuse intrinsic pontine glioma (DIPG) in children.
The brain and spinal cord are the most well-protected organs in the body and are evolutionarily designed to be protected from damage as much as possible throughout one’s lifetime. In addition, each region of the brain has a specific function and can cause serious physical or mental disability when perturbed. These truths lead to various challenges that are not seen in other regions of the body when attempting to treat tumors within the central nervous system (CNS). An obvious example of this is that neurosurgeons must first maneuver through the skull and, when in the parenchyma, must resect the tumor with as narrow margins as possible to limit debilitating comorbidities.
This review will discuss the many ways by which brain tumors have evolved and prevented scientists and clinician from developing treatments that improve long-term survival. We will focus on the high levels of heterogeneity and plasticity in GBM and other brain tumors as well as discuss new therapeutic avenues that have been exposed due to the exponential growth in our understanding of these tumors.
Overview of brain tumors
Adult brain tumors can be generally classified into three categories: peripheral tumors that metastasize to the CNS, primary benign or low-grade neoplasms, and high-grade gliomas.
It is estimated that between 10-20% of patients with cancer will be diagnosed with a brain metastasis, and autopsy studies have reported a true incidence of 30-40% in all patients with cancer[2-5]. A total of 67-80% of brain metastases come from either lung, breast, or melanoma primary tumors. In all three of these tumors, the presence of brain metastases portends a poor survival. The median survival of patients with brain metastasis has been observed to be 13 months[6], and even in patients with the best prognostic factors, the most recent data demonstrate a median survival of only 33 months after the diagnosis of a brain metastasis[6]. While more recent studies have identified genetic differences between the primary tumor and brain metastases[7], the intratumoral heterogeneity of brain metastases has not been investigated in great detail.
Primary brain tumors are classified according to the World Health Organization (WHO) classification system. Meningioma is the most common primary brain tumor, with 90% classified as grade 1 and benign. For most patients, complete surgical resection is curative, but a subset will have recurrent disease[8]. A recent study interrogated the genomic enhancer landscape of meningioma and identified novel disease subgroups[9]. Studies such as these are rapidly improving our understanding of meningiomas and our ability to determine which patients will have recurrent disease.
Gliomas are a group of brain tumors that were once thought to derive from glial cells. However, more recent evidence points to multiple cells of origin, including neural and glial progenitor cells within the brain [10-12]. Gliomas consist of astrocytomas, oligodendrogliomas and ependymomas, and until 2016, the WHO classification primarily relied on histologic features. However, the latest WHO classification system from 2016 integrates molecular data into the diagnostic strategy[13]. Grade I (rarely seen in adults) and II tumors are classified as low-grade gliomas and have a relatively favorable prognosis of around 10 years depending on the histological subtype. Most of these patients have a mutation to the isocitrate dehydrogenase (IDH1) gene, which leads to the generation of the oncometabolite 2-hydroxyglutarate. The natural course of some low-grade gliomas is to progress to high-grade gliomas, and therefore, maximal safe surgical resection of these tumors is the primary treatment. In some instances, these patients will also receive radiation or the chemotherapy temozolomide (TMZ). However, a recent retrospective study identified TMZ as a modifiable risk factor that contributes to malignant transformation along with wild-type IDH or mutated IDH with intact 1p/19q[14].
Pediatric brain tumors are now the number one cause of cancer-related death in children. These pediatric tumors include a wide range of benign and malignant tumors with various prognostic indications. These tumors include medulloblastoma, a primitive neuroectodermal tumor that accounts for approximately 20% of childhood brain tumors[15]. Medulloblastoma comprises a biologically heterogeneous group of embryonal tumors that have been subgrouped into the following four types: WNT, Sonic Hedgehog, Group 3 and Group 4[16-19]. As a result of its prevalence, the genetic profile of medulloblastoma has been well characterized, and the cell heterogeneity and plasticity of these tumors have been investigated. However, for the purposes of this review, we will focus on adult GBM as a paradigm for cellular heterogeneity and plasticity.
Finally, high-grade gliomas are made up of WHO grade III and IV tumors. Here, we will focus on WHO grade IV glioma, or GBM. Treatment for GBM includes maximal safe surgical resection, radiation, TMZ, and the recently approved tumor-treating fields (TTF), an antimitotic treatment that delivers alternating electric fields to the scalp[20,21]. Despite these therapies, the latest clinical trials have demonstrated a median survival of only 20.9 months, with a median survival outside clinical trials thought to be around 15 months[20]. There are a multitude of hypotheses for the poor prognosis of patients with GBM, including a failure of drugs to penetrate the blood-brain barrier[22], tumor cell invasion into the brain parenchyma, a hypoxic microenvironment[23], cancer stem cells (CSCs)[24] and tumor cell heterogeneity and plasticity[25].
Here, we will focus primarily on GBM and the heterogeneity observed within individual tumors and between tumors. We will investigate how advances in single-cell sequencing and epigenetic profiling are providing new insights into the complexity of the disease and discuss how CSCs can interact with other cells in the tumor microenvironment and drive treatment resistance, tumor recurrence and tumor cell plasticity. Finally, we discuss the ways that immune cells, endothelial cells, astrocytes, and neurons support the tumor microenvironment and may provide novel therapeutic targets.
Molecular heterogeneity of glioblastoma
It has long been observed that GBM possesses extensive inter- and intra-tumoral heterogeneity. Early histological studies of GBM focused on the extent of necrosis, nuclear size, astrocytic differentiation, cell size, number of mitotic cells, distribution of cell density and vascularization[26]. Through this approach and the sampling of various locations within the tumor, scientists and pathologists were able to observe significant histological variations even within the same tumor. This phenomenon can be demonstrated ex vivo, as well. In one example of this, five subclones were isolated from a single tumor, and after orthotopic xenotransplantation, each subclone had a distinct histology and tumorgenicity[27].
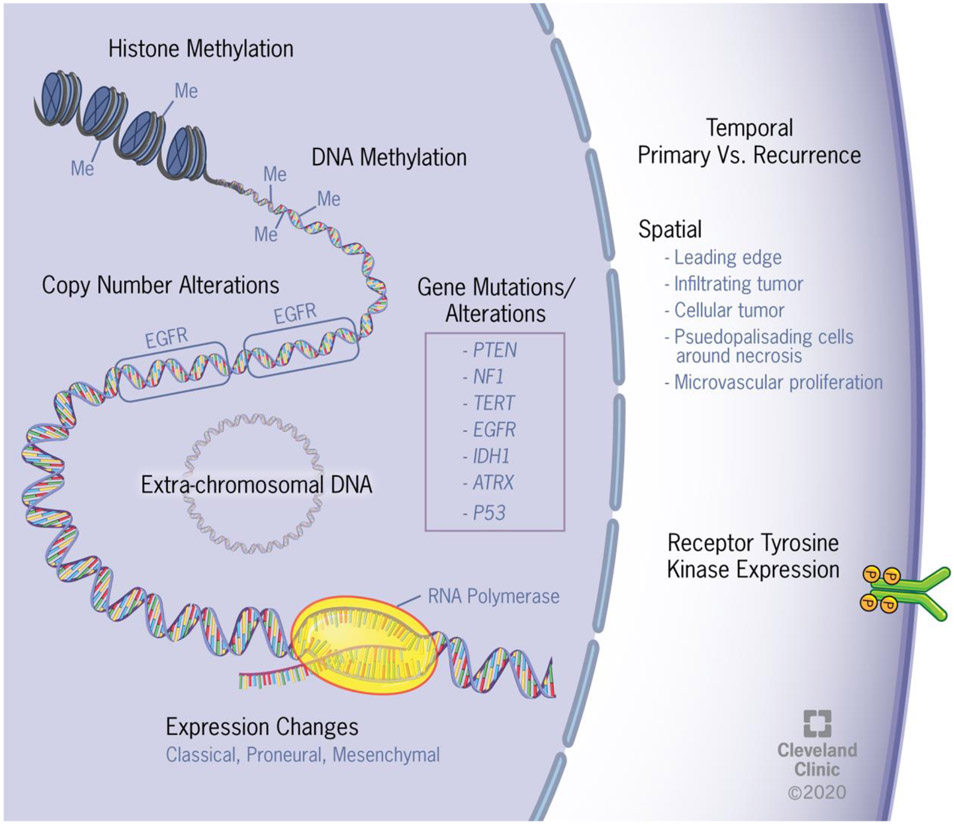
One of the first technological advancements that furthered our understanding of GBM heterogeneity was the development of whole genome amplification (WGA) methods, which enable the identification of chromosomal imbalances. Nobusawa et al. utilized WGA on 14 different primary GBM samples from two to five locations within each tumor and identified some alterations that were common among all locations and others that were region specific[28]. In another study, single molecule molecular inversion probes targeting 33 cancer genes were used to assay point mutations and gene amplifications. The authors found evidence of regional mutational heterogeneity even within the same tumor[29]. Some commonly altered genes among GBM patients include epidermal growth factor receptor (EGFR), telomerase reverse transcriptase (TERT), platelet-derived growth factor receptor alpha (PDGFRA), cyclin-dependent kinase 4 (CDK4), murine double minute-2 (MDM2), MDM4, and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA)[30]. Other common mutations or deletions of tumor suppressors include phosphatase and tensin homolog (PTEN), ATRX chromatin remodeler (ATRX), IDH1, and TP53[31,32]. Amplification of the genes encoding receptor tyrosine kinases (RTKs), especially EGFR and PDGFRA, has been well documented in GBM. Snurderl et al. demonstrated that amplification of different RTKs were rarely found in the same region of the tumor, however, different RTKs were amplified in distinct subpopulations of cells[33]. Spatially distinct RTK amplifications were then verified by two additional studies, further emphasizing the intra-tumoral heterogeneity[34,35]. Studies such as these may help to explain the lack of efficacy thus far of EGFR targeted therapies in GBM[36-39]. This lack of efficacy is in contrast to the use of EGFR inhibitors in EGFR-mutant lung cancer with CNS metastases, where one study found a 91% CNS objective response rate in patients treated with the EGFR inhibitor osmertinib[40,41]. As a result of these studies demonstrating heterogeneity within individual tumors and the lack of efficacy of RTK inhibitor monotherapy, the field has largely moved away from the use of these drugs. However, there is ongoing research on the use of RTK inhibitors as a combination therapy as well as on developing immunotherapies that target cells with high RTK expression.
Advances in sequencing technology and the initiation of The Cancer Genome Atlas (TCGA) led to a major shift in our classification of GBM. Bioinformatics analysis of gene expression profiles of tumors identified four distinct molecular subtypes of GBM: proneural, mesenchymal, classical and neural (the neural subtype is excluded from most in follow-up classifications as the neural subtype was thought to be a contamination from normal neural tissue)[42,43]. While this characterization of bulk tumor RNA revolutionized the classification of GBM, heterogeneity and plasticity within these groups were quickly identified. In the earliest challenge to this classification scheme, Sottoriva et al. sampled spatially distinct tumor fragments from 11 patients with GBM and found that within the same patient, multiple molecular subtypes could be identified[44]. The authors also found heterogeneous driver aberrations and copy number alterations (CNA) within the same tumor. In addition, by comparing CNA data and measuring mitotic distances among cells of a tissue fragment, the investigators reconstructed the phylogeny and relationships among subclones. Through this method, the authors found that loss of cyclin-dependent kinase inhibitor 2A (CDKN2A/B) and amplification of EGFR, CDK6 and MET occur early in tumor development, while alterations in PDGFRA, PTEN and TP53 are later malignant events[44]. A later study utilizing 127 multi-sector or longitudinal specimens from 52 patients confirmed these findings[45]. These authors also conducted a chemical screen of patient-derived cells and found that targeting truncal events is more efficacious in reducing tumor burden[45]. Finally, viewing GBM from a temporal standpoint, Wang et al. found that 66% of patients undergo a subtype switch at recurrence, highlighting the plasticity of these expression states[46].
Gill et al. performed RNA-seq on biopsies taken from within the contrast-enhancing core and the non-enhancing margins of tumors. The contrast-enhancing regions resembled proneural, classical, or mesenchymal subtypes, whereas the non-enhancing margin resembled a neural subtype[47]. The authors also found that the expression pattern of the non-enhancing region was influenced by non-neoplastic brain cells, which will be discussed in greater detail in later sections. Finally, patterns of cell type alterations varied among the GBM subtypes, as the non-enhancing regions of proneural tumors were enriched with oligodendrocyte progenitor genes, whereas mesenchymal tumors were enriched for astrocytic and microglial genes[47]. The importance of molecular subtyping for laboratory studies cannot be overstated; however, almost ten years after its initial development, molecular subtyping has had almost no translational impact on the clinical setting.
Another attempt to further our understanding of the spatial heterogeneity of GBM was the development of the Ivy Glioblastoma Atlas Project (Ivy GAP)[48]. The Ivy GAP database includes a comprehensive pathology-molecular map of GBM that enables the unbiased study of molecular alterations of known anatomical features. The authors utilized laser microdissection and RNA-seq in 41 patients, dissecting out the leading edge, infiltrating tumor, cellular tumor, pseudopalisading cells around necrotic regions, and microvascular proliferation regions for each tumor. In general, samples from the same anatomical feature were more like each other than like other samples from the same tumor. However, even with this complex analysis, there was no mutation associated with a particular anatomic feature that predicted overall survival better than O6-methylguanine–DNA methyltransferase (MGMT) promotor methylation status of the bulk tumor, suggesting that this known adversary remains a significant and predictive hurdle[48]. The creators of the Ivy GAP platform have deposited their data online (http://glioblastoma.alleninstitute.org/), and this data have already led to numerous additional publications[49-52].
The next technological leap that added to our understanding of intratumor heterogeneity was single-cell sequencing. In the first of these studies, Patel et al. performed single-cell RNA-seq on five different tumors. The authors confirmed previous finding by showing that GBM subtype classifiers are variably expressed across individual cells within a single tumor[53]. A more recent study combined single-cell RNA-seq, analysis of the TCGA, and experimental models to argue that malignant cells in GBM exist in four main cellular states that are reminiscent of canonical neurodevelopmental cell types. This includes astrocyte (AC)-like, oligodendrocyte progenitor cell (OPC)-like, neural progenitor cell (NPC)-like and mesenchymal (MES)-like states. Single-cell lineage tracing experiments found plasticity between these four states. The NPC-like, OPC-like, and AC-like transcriptional states correlated with copy number aberrations in specific loci: PDGFRA, CDK4 and EGFR, respectively[54].
Another question that remains to be completely resolved is treatment-induced plasticity and temporal heterogeneity. In 2014, Johnson et al. sequenced the exomes of 23 initial low-grade IDH mutant gliomas and recurrent tumors from the same patient[55]. In 43% of cases, at least half of the mutations in the initial tumor were undetected at recurrence. Additionally, 6 of 10 patients treated with TMZ at recurrence were hypermutated and harbored driver mutations in retinoblastoma and Akt-mTOR pathway genes[55]. However, more recently, the Glass consortium assembled a database of initial and recurrence samples from 222 patients. In this study, 35 patients exhibited treatment-related hypermutation at recurrence, and 70% of the cohort had an increased mutational burden after recurrence. However, the authors found that driver genes detected at the initial disease were retained at recurrence and there was little evidence of recurrence-specific gene alterations. Only 16% of IDH wild-type patients exhibited treatment-related hypermutation, similar to the 17% estimate in Wang et al.[46]. Therefore, the authors concluded that the strongest selective pressures occur early in glioma development and that current therapies shape this evolution in a stochastic manner[56]. An important difference between the two studies is that in Johnson et al. all 23 patients were IDH mutant, while only 88/222 patients in Barthel et al. were IDH mutant. Additionally, within IDH-mutant tumors without 1p19q deletion, 47% of patients had treatment-associated hypermutation, suggesting that treatment-related mutations may be dependent on IDH mutation status[56].
Study after study in GBM consistently observe a high level of intratumoral heterogeneity. Early pathologists observed this heterogeneity through histology, and with each advance in genomic technology, the heterogeneity of GBM has been further clarified on multiple molecular levels (Figure 1). In the next section, we will discuss more recent studies on the epigenetics of GBM that are providing additional insights into the intra-tumoral heterogeneity and plasticity of these cells.
Figure 1. Glioblastoma exhibits inter- and intratumoral heterogeneity at multiple levels.
Schematic depicting inter- and intratumoral heterogeneity ranging from epigenetic changes to the enhancer landscape to amplification of receptor tyrosine kinases.
Epigenetic heterogeneity in glioblastoma
While advances in our understanding of the genetics and expression patterns of GBM have enabled vertical advancements in the field, these advances have yet to result in changes in the clinical management of GBM, and the five-year survival remains around 5%[57]. Additionally, the GLASS consortium finding that mutations at recurrence are largely random suggests that mutational evolution is not driving therapeutic resistance and plasticity in GBM. This has led many in the field to investigate potential epigenetic pathways that may be contributing to therapeutic resistance and recurrence.
The most established of these epigenetic changes is MGMT promotor methylation[58]. The MGMT gene encodes a DNA repair protein that removes alkyl groups from the O6 position of guanine, an important site of DNA alkylation. Therefore, patients with a hypomethylated MGMT promotor have elevated levels of the MGMT protein and are more resistant to alkylating agents such as TMZ. MGMT methylation status remains one of the best predictors of survival in GBM.
In the first attempt at measuring global methylation patterns in GBM, researchers utilized TCGA data and observed a subset of patients with hypermethylation at a large number of loci. This study then classified a subset of patients as having a glioma-CpG island methylator phenotype (G-CIMP). These patients are generally younger and experience significantly improved outcomes[59]. To investigate intratumoral heterogeneity in methylation, Wenger et al. took three to four spatially separated biopsies from 12 GBM patients and performed genome-wide DNA methylation analysis[60]. As Sottoriva et al. found using transcriptional data, when GBM subtype was determined by methylation status, 5 of 12 patients had cells from multiple subtypes[44,60]. Ultimately, the authors concluded that GBM contains a significant variety of intratumoral DNA methylation patterns. In another study, methylation changes over time were investigated using formalin-fixed paraffin-embedded samples and bisulfate sequencing of 112 patient samples. DNA methylation was predictive of immune cell infiltration, extent of necrosis, and shape of tumor cell nuclei. Interestingly, DNA methylation erosion and subclonal heterogeneity were variable across patients but similar between primary and recurring tumors, which argues against a therapy-induced increase in epigenetic heterogeneity, as well as against a decrease in epigenetic heterogeneity due to strong selective sweeps driven by therapeutically resistant subclones[61].
Another epigenetic modification that has been identified as markedly upregulated in GBM is N6-methyladenine (N6-ma). N6-mA levels are dynamically regulated by ALKBH1, and depletion of ALKBH1 led to the transcriptional silencing of numerous oncogenic pathways by decreasing chromatin accessibility. In another study from this group, N6-methyladenosine (m6A) mRNA modifications were found to be upregulated in CSCs. The m6A reader YTHDF2 then stabilizes this modified mRNA, including the mRNA encoding MYC and VEGFA[62]. Another study investigated the super-enhancer landscape of 44 patient-derived GBM stem cells, 50 primary tumors and 10 neural stem cells. The authors found two highly distinct super-enhancer states that showed opposing patterns of H3K27ac. Group 2 was highly associated with mesenchymal features, while group 1 exhibited proneural, classical and proliferative features[63]. Finally, extrachromosomal DNA can lead to oncogene amplification that drives cancer growth[64]. In GBM, this extrachromosomal DNA is unevenly inherited by offspring cells and impacts the oncogenic potential of the daughter cells[65]. Furthermore, EGFR amplification occurs in circular extrachromosomal DNA, and these circular pieces of DNA tend to harbor epigenetic enhancer regions that topologically interact with the EGFR locus to increase EGFR gene expression[66]. Together, these epigenetic observations bolster our understanding of intratumoral heterogeneity and provide a novel avenue for therapeutics targeting the epigenetic landscape[64].
As a whole, the field of GBM epigenetics is relatively young, and there is still much to learn about how epigenetic changes at all levels impact tumor cell behavior. The use of drugs targeting epigenetic modifications either by histone deacetylase (HDAC) inhibitors, bromodomain & extra-terminal domain (BET) inhibitors, or other mechanisms was recently reviewed[25]. However, to date, only slightly more than 20 clinical trials have been initiated in the epigenetics field, and this number pales in comparison to the number of trials using chemotherapies, anti-angiogenic agents and immunotherapies. Therefore, while there is still much to learn, targeting epigenetic changes in GBM is an exciting path moving forward.
Cancer stem cells
Any discussion of GBM heterogeneity and plasticity is not complete without a discussion of the CSC population. CSCs are a population of tumor cells that are defined by their functional ability to self-renew, initiate tumors, and undergo persistent proliferation[31,67]. There is no single marker of CSCs in GBM, however the glycoprotein CD133 was initially found to mark a CSC population and is widely used for sorting tumor cells today[68,69]. Furthermore, it was discovered that those with a higher proportion of CD133+ cells present correlated with shorter survival[70,71]. CD133 has been shown to signal through AKT and Wnt to drive the CSC state[72]. However, it has been well documented that subgroups within the CD133− population can also exhibit CSC functional characteristics[73,74]. To date, various intracellular proteins (SOX2, MYC, and NESTIN) and cell surface markers (CD133, CD15, CD49f, L1CAM and CD44) have been identified as markers of CSCs in GBM[75-82]. The multiple, not necessarily overlapping, CSC markers begins to describe the vast amount of heterogeneity among the CSC population itself[74].
Drivers of the cancer stem cell state
While the CSC state has been widely studied and numerous factors have been shown to drive cells into a stem cell-like phenotype or to be necessary for CSC maintenance including microRNAs[83,84] and metabolism[85] here we will highlight two of these factors, hypoxia and treatment resistance. Hypoxia is a hallmark of the GBM microenvironment and leads to phenotypic changes[86]. Hypoxia promotes growth of CSCs and increases the expression of hypoxia-inducible factors (HIFs) and vascular endothelial growth factor (VEGF)[86-89]. In chronic hypoxic conditions, HIFs can upregulate stem cell transcription factors including KLF4, SOX2, and OCT4[88,90] that in turn affect downstream pathways. Furthermore, HIF1α can directly activate Notch and WNT signaling pathways[91]. Thus, the hypoxic conditions present in GBM can induce a more stem cell-like phenotype that leads to further propagation of tumor growth.
Current standard of care for GBM follows what is known as the Stupp protocol. This includes a gross total surgical resection followed by radiotherapy and chemotherapy using the alkylating agent TMZ and, more recently, TTF[20,21,92,93]. However, both chemotherapy and radiotherapy can cause changes to CSC phenotypes, and CSCs have elevated levels of resistance to these therapies[94,95]. CSCs contribute to the radiation resistance through increased use of DNA repair mechanisms[94]. A recent study found that exposure to ionizing radiation leads to CSCs that were initially enriched for a CD133+ proneural signature to transition into a CD109+ mesenchymal subtype. This shift to CD109 positivity often leads to a mesenchymal recurrence[96].
As previously discussed, TMZ functions by adding alkyl groups to thymine and guanine, which causes DNA damage and initiates apoptosis. However, TMZ does not work on all CSC populations, leading to divergent CSC phenotypes[95]. Further studies have also shown that after exposure to TMZ, the CSC pool increases both in vitro and in vivo. Lineage-tracing analysis demonstrated that the CSC pool had shifted toward increased expression of stem markers such as CD133, SOX2, and OCT4[30,97]. These studies highlight how ionizing radiation and TMZ treatment can induce plasticity within the CSC population and drive post-treatment changes.
Models of glioblastoma heterogeneity
Three primary models have been formulated to explain the heterogeneity within GBM: the clonal evolution model, the CSC model, and the plasticity model. The clonal evolution model suggests that certain CSCs are selected over time based on factors such as their genetic, epigenetic, and tumor microenvironment (TME) interactions. The selection of these cellular traits drives progression and increases heterogeneity, as the selective pressures are temporally and spacially distinct[98-101]. Therefore, certain CSCs evolve to be more fit to survive in hypoxic environments, while others may grow in more nutrient-dense regions[99]. The second model is the CSC model, which suggests that a small subpopulation of cells is predisposed to drive tumor progression, invasiveness, and therapeutic resistance. Thus, the observed heterogeneity is a result of the differentiation of the CSC population, which gives rise to intermediate progenitor and terminally differentiated progeny[31,100,101]. Therefore, in this model, CSCs are the source of tumor initiation and heterogeneity. Lastly, the plasticity model builds upon the CSC model and states that CSCs can interconvert between stem cell and differentiated states. Unlike the CSC model, the plasticity model suggests that even upon differentiation, the differentiated cells can convert back into CSCs[49,54,86]. Two recently published studies in GBM provide strong evidence that the behavior of CSCs is best described by the plasticity model. Dirkse et al. demonstrated that the phenotypic heterogeneity observed in GBM arises from non-hierarchical, reversible state transitions that are driven by the microenvironment[86]. In another study, Neftel et al. utilized single-cell RNA-sequencing, bulk genetics and the TCGA and found that malignant cells exist in four main cellular states that each exhibit a high level of plasticity[54]. Together, these studies and others are slowly shifting the focus of the field from identifying therapies that target CSCs to identifying therapies that target the plasticity of GBM.
Glioblastoma interaction with non-tumor cells
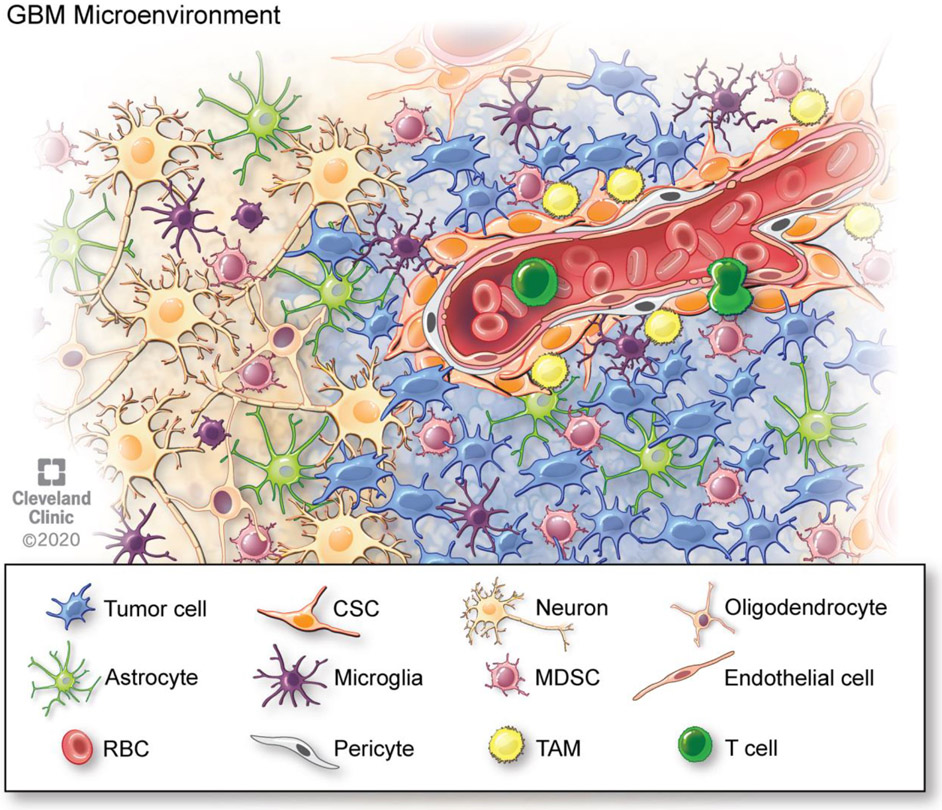
One fact that has been understood for many years is that the GBM microenvironment consists of more than merely tumor cells. Numerous other cell populations within the brain and the immune system contribute to the conditions that lead to tumor growth. These populations include endothelial cells, microglia, astrocytes, neurons, and immune cells. The failure of many tumor cell targeting therapies has led scientists and physicians to investigate the potential of targeting other cells within the tumor microenvironment (Figure 2).
Figure 2. The tumor microenvironment of glioblastoma includes various other cells types in addition to tumor cells.
Schematic depicting the crosstalk between cell types in the tumor microenvironment that drive glioblastoma progression and recurrence.
Endothelial cells
It has long been understood that malignant cells require oxygen, nutrients, and the removal of waste to proliferate and survive. Brain tumors expand their vasculature through a process known as tumor angiogenesis, where endothelial cells proliferate or are recruited from the bone marrow. In addition, endothelial cells also play an essential role in what is known as the blood-brain barrier, which functions to closely monitor and control which chemicals and cells come into and out of the brain[102]. Tumor cells and endothelial cells can interact through three primary mechanisms. First, tumor cells secrete numerous growth factors including stromal-derived factor-1 (SDF-1), platelet-derived growth factor (PDGF), transforming growth factor (TGF-β), fibroblastic growth factor 2 (FGF2) and the most well studied, VEGF[103]. Secondly, the two cell types communicate via direct contact through gap junctions, in particular through connexin 43[104-106]. Finally, indirect interactions via intermediate cells such as pericytes, astrocytes and neurons also occur. The complex relationship between endothelial cells and tumor cells was recently reviewed in further detail[103].
Brain tumors are some of the most vascularized solid tumors found in humans, and the targeting of endothelial cells in GBM was a leading effort in clinical trials in the late 2000s[107]. In 2009, after a phase II trial that demonstrated a dramatic overall radiographic response rate, the FDA granted accelerated approval to bevacizumab, a monoclonal antibody against VEGF-A[108,109]. However, two phase III trials in patients with newly diagnosed GBM failed to demonstrate any improvement in overall survival[110,111]. Numerous follow-up studies that combined bevacizumab with various chemotherapies and targeted therapies have also failed to demonstrate any overall survival benefit[112-123]. Only one phase II study of a combination of bevacizumab and lomustine in patients with recurrent GBM demonstrated a survival benefit at 9 months; however, the subsequent phase III trial did not show any improvement in survival[124]. Additionally, a multitude of other studies has investigated additional anti-angiogenic agents, but none of these treatments have led to improved overall survival, and as a result, the field has slowly shifted away from anti-angiogenic treatments[125-135]. While the endothelial-tumor cell interaction remains an active area of research, clinical efforts to target angiogenesis in GBM have for the most part come to a halt. However, the use of bevacizumab as an alternative to steroids in the setting of immunotherapy has gained traction (NCT03452579).
Neurons
The first known observation of an interaction between GBM cells and neurons came in 1938 by the Belgian pathologist Dr. H. J. Scherer. He observed that glioma cells had a tendency to grow along and around normal neurons, and he described this observation as “precocious perineural growth”[136]. Since that time, determining whether invasive tumor cells were merely traveling along a path of least resistance or whether the two cell types are actively engaging in bidirectional signaling has been the goal of numerous neuroscientists and cancer biologists[137]. When considering the latest evidence for neuron-tumor cell relationships, it is worth remembering that there is ample evidence for a close, bidirectional relationship between glial cells and neurons in normal physiology.
The first molecule that was uncovered as a pro-tumorigenic neuronal molecule is the synaptic cell adhesion protein neuroligin-3 (NLGN3). NLGN3 is proteolytically cleaved at the cell surface, releasing the N-terminal ectodomain, which in turn leads to tumor cell proliferation[138]. In mice with NLGN3 knocked out, intracranial injection of xenografts leads to failure of the tumor to progress. While this was true across numerous molecularly distinct gliomas, xenografts of breast cancer brain metastases were not impacted by NLGN3 knockout[139].
The role of neurotransmitters in glioma progression has been of interest to clinicians and scientists since the observation that the use of tricyclic antidepressants reduces the odds of developing glioma in a dose- and time-dependent manner[140]. Because tricyclic antidepressants are classically thought to act via the inhibition of serotonin and norepinephrine re-uptake, the role of neurotransmitters in glioma development is of great interest. Glioma cells have been shown express dopamine, glutamate, GABA and serotonin receptors[141-145].
In 2019, three ground-breaking studies were published that further characterized the interaction between neurons and tumor cells within the brain and the importance of glutamate. Venkataramani et al. were able to demonstrate functional bona fide chemical synapses between presynaptic neurons and postsynaptic glioma tumor cells[146]. These synapses formed with typical synaptic ultrastructure and produced postsynaptic currents via glutamate receptors of the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) subtype. Furthermore, tumor growth and invasion were reduced by anesthesia or the AMPA receptor antagonist perampanel. Venkatesh et al. observed a similar finding mediated through AMPA receptor-dependent neuronglioma synapses. They also found that neuronal activity evokes non-synaptic activity-dependent potassium currents that are amplified by gap junction-mediated tumor interconnections[147]. Finally, Zeng et al. investigated breast-to-brain metastasis and identified activation of N-methyl-D-aspartate receptors (NMDARs) by glutamate ligands as essential. Pseudo-tripartite synapses between cancer cells and glutamatergic neurons were responsible for stimulating the NMDARs[148].
Uncovering the interactions between neurons and tumor cells has unlocked an entirely new avenue of therapeutic options. One method for targeting this interaction is the utilization of neurotransmitter-blocking drugs that may mimic the phenotype observed with tricyclic antidepressants[140]. Another innovative approach that has been discussed is the implantation of electrodes into the resection cavity during craniotomy to disrupt neuron-tumor cell interactions. This would be an additional electromagnetic treatment option for GBM (in addition to TTF[20]).
Astrocytes
Tumor-associated astrocytes directly interact with GBM cells, leading to the formation of reactive astrocytes. These astrocytes facilitate tumor progression, proliferation and migration through multiple mechanisms[149]. The signaling between astrocytes and GBM tumor cells is bidirectional. Tumor cells have been shown to activate astrocytes through receptor activator of nuclear factor kappa-B ligand (RANKL), extracellular vesicles, and connexin 43 transmission of cGAMP, whereas astrocytes promote tumor growth through the secretion of IL-6, VEGF, TGF-β, growth/differentiation factor 15 (GDF-15), glutamine, tumor necrosis factor (TNF) and numerous other cytokines[149]. As evidenced by many of the anti-inflammatory molecules just mentioned, astrocytes play a significant role in driving an immunosuppressive microenvironment in GBM[150]. Astrocytes have also been shown to play a role in assisting tumor cell migration and infiltration. Connective tissue growth factor (CTGF) secreted by astrocytes mediates GBM cell infiltration and is a potential therapeutic target in GBM[151]. While less is known about astrocytes compared to other cell populations within the microenvironment, targeting astrocytes is an underexplored area and remains a viable option for GBM therapies due to their central role in regulating other cell populations.
Immune Cells
Two lines of thinking have long driven the dogma that the brain is an “immunologically privileged” organ: the existence of the blood-brain barrier and the lack of a conventional lymphatic system[152]. However, evidence is rapidly accumulating that emphasizes the importance of immune cells within the tumor microenvironment for tumor progression and the potential for immune-stimulating therapies to be the treatments of the future. However, it is worth remembering that the immune checkpoint (PD-1 and CTLA-4) inhibitors that have shown field-altering efficacy in tumors including melanoma[153] and lung cancer[154] have thus far failed to demonstrate any efficacy in GBM[155].
The different GBM subtypes outlined above have been shown to have heterogeneity within their immune populations. Utilizing the CIBERSORT method, which characterizes the cellular composition of a complex tissue from gene expression profiles[156], Wang et al. found that tumor-associated macrophages (TAMs), neutrophils and CD4+ T cells are increased in tumors enriched in the mesenchymal signature, whereas an activated dendritic cell signature was found more in the “classical” tumors [51]. Another group utilized a tissue microarray to investigate the immune cell composition of 98 tumors. Microglia and TAMs were the most prevalent cells in all four subtypes, and CD3+ cells made up only 1% of cells within the tumor, independent of the subtype[157]. Additionally, tumor-infiltrating lymphocytes (TILs) generally displayed an exhausted phenotype, and NK cells expressed reduced levels of activating receptors, highlighting the challenges of T cell- and NK cell-targeted therapies in GBM[158,159]. Studies such as these highlight the low frequency of TILs, which may account for the lack of efficacy of immune checkpoint inhibitors thus far.
The exhausted phenotype of TILs in GBM is in part due to the highly immunosuppressive tumor microenvironment of GBM. The secretion of cytokines including TGF-β, IL-6, IL-10, macrophage migration inhibitory factor (MIF) and numerous others has been demonstrated to cause a local immunosuppressive phenotype[160-163]. In addition to soluble factors, direct cell-cell communication through PD-L1, tolerogenic HLA, the apoptosis-inducing receptor FAS, and changes in glycosylation can all cause immune suppression[164-167].
Another type of immune cell found in GBM is myeloid-derived suppressor cells (MDSCs). MDSCs are a population of immune cells that have a remarkable ability to suppress T cells through a variety of mechanisms and are subdivided into monocytic (mMDSC) and granulocytic (gMDSC) subsets. MIF secreted by CSCs within the tumor leads to increased MDSCs, and targeting this cell population with low-dose capecitabine has been demonstrated to be a potential therapeutic option in clinical trials[168,169]. Additionally, it was recently observed that there is a sex difference in MDSC populations in GBM, with gMDSCs elevated in the blood of females and mMDSCs enriched in the tumors of males[170].
Finally, microglia are the resident immune cells within the brain. Numerous studies have demonstrated that microglia within GBM predominately exhibit anti-inflammatory M2 polarization[171,172]. Microglia and tumor cells cross-talk though the secretion of EGF, colony-stimulating factor-1R (CSF-1R), TGF-β and IL-10[173]. Microglia can facilitate the invasion of GBM cells through the upregulation of matrix metallopeptidase 14[174]. GBM cells also upregulate CD47 to prevent phagocytosis by microglia, and work is being done to block CD47[175]. Finally, a recent study demonstrated that microglia function in a sex-specific manner, as female microglia that were deficient in junctional adhesion molecule-A (JAM-A) succumbed to GBM more rapidly than WT females, whereas the same phenotype was not observed in males[176]. This study combined with the MDSC data above highlights the heterogeneity between males and females and opens up the prospect of sex-specific treatments[177].
While the clinical data for therapies targeting the immune system thus far have shown little efficacy, our understanding of the immune compartment of the GBM tumor microenvironment has grown since the initiation of these trials. In addition to the studies mentioned above, the role of dendritic cells[178] and B cells is actively being researched[179]. Currently, there are at least 60 active immunotherapy clinical trials for GBM that are outlined in a recent review[180]. These trials rage from novel checkpoint inhibitors, vaccines targeting specific antigens or patient tumor lysates, chimeric antigen receptor (CAR)-T and CAR-NK cell therapies, oncolytic viruses, and macrophage-based immunotherapies. Studying the many ways immune cells interact with tumor cells is an active area of research and may be the foundation of future therapies.
Conclusion
Brain tumors are among the most heterogeneous tumors to have been characterized. From early histological studies that documented regions of necrosis to more recent single-cell RNA sequencing and methylation profiling, GBM is a tumor with resounding intra- and inter-tumoral heterogeneity. In addition, heterogeneity at the level of cell type, the sex and age of the patient, and plasticity over time all add to the complexity of the disease. Unfortunately, this lack of homogeneity or single driver mutations has left GBM patients without the targeted therapies that have revolutionized treatments for tumors such as breast cancer, melanoma, lung cancer and chronic myeloid leukemia.
With each new technological advancement, new research emerges, at a significant financial price, that verifies what pathologists have known for well over 50 years. This leaves physicians and scientists with the challenge of translating laboratory advancements into changes in clinical care. Our understanding of MGMT promotor methylation and IDH mutation status has revolutionized prognostic outlook for patients, and these characteristics are utilized in the design of clinical trials. In contrast, laboratory advancements such as tumor subtyping (classical, proneural and mesenchymal) and methylation profiles have for the most part remained an academic exercise. The goal moving forward must be the integration of these advancements into clinical trial designs that will match treatment approaches with the patients most likely to benefit.
With recent findings, three topics emerge as areas that require further research and are most likely to lead to future therapies. First, a move toward growing our understanding of the plasticity of CSCs and the epigenetics of GBM as a whole is shifting our understanding of the disease. To date, CSC targeted therapies have not panned out as viable treatment opportunities; however, focusing on the plasticity of GBM and the epigenetic changes that drive this plasticity may unlock unexpected therapeutic avenues. The second area is furthering our understanding of the communication between GBM cells and other cells within the tumor microenvironment. In particular, the interaction between neurons and tumor cells provides an opportunity for pharmacologic inhibitors, as well as surgical and electromagnetic techniques, that could revolutionize the treatment of brain tumors. Finally, it is impossible to ignore the success of immunotherapy in other cancers, and therefore, furthering our understanding of the immunosuppressive microenvironment in GBM has the potential to usher in the next generation of immune-stimulating therapies.
The efficacy of drugs such as osimertinib[40] (an EGFR inhibitor) and nivolumab/ipilimumab[181] in the setting of brain metastases has demonstrated that targeted therapies and immunotherapies have the potential to reach tumors within the brain. For the reasons outlined above, GBM heterogeneity adds additional layers of complexity that must be overcome for these and other drugs to improve survival in GBM patients. However, due to advances in our understanding of the genetics and epigenetics of tumor cells as well as of the cell populations that tumor cells rely on, the prospect of making the first new meaningful developments in clinical care for only the second time since 2005 remains high[20,21].
Acknowledgements
We regret that we were unable to include all work on glioblastoma, cancer stem cells, cellular heterogeneity, and plasticity in this review due to spatial limitations. We would like to thank the members of the Lathia laboratory for insightful discussions. We thank Ms. Amanda Mendelsohn for illustration assistance and Dr. Erin Mulkearns-Hubert for editorial assistance. Work in the Lathia laboratory is supported by the Cleveland Clinic, Case Comprehensive Cancer Center, the American Brain Tumor Association, National Brain Tumor Society, and NIH R01 NS109742, R01 NS117104, and P01 CA245705. Alice Lo work was supported by a summer research scholarship provided by CWRU SOURCE with additional funding provided by the Women in Science and Engineering Roundtable.
Abbreviations.
- CNS
Central nervous system
- GBM
glioblastoma
- DIPG
diffuse intrinsic pontine glioma
- WHO
World Health Organization
- TMZ
temozolomide
- TTF
tumor-treating fields
- CSCs
cancer stem cells
- WGA
whole genome amplification
- EGFR
epidermal growth factor receptor
- IDH1
isocitrate dehydrogenase
- MDM
murine double minute
- TERT
telomerase reverse transcriptase
- PDGFRA
platelet-derived growth factor receptor alpha
- PTEN
phosphatase and tensin homolog
- ATRX
ATRX chromatin remodeler
- PIK3CA
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha
- CDK4
cyclin-dependent kinase 4
- TCGA
The Cancer Genome Atlas
- CNA
copy number alteration
- Ivy GAP
IVY Glioblastoma Atlas Project
- NPC
neural-progenitor cell
- OPC
oligodendrocyte progenitor cell
- AC
astrocyte
- G-CIMP
glioma-CpG island methylator phenotype
- N6-ma
N6-methyladenine
- m6A
N6-methyladenosine
- ALKBH1
AlkB homolog 1, histone H2A dioxygenase
- HIFs
hypoxia-inducible factors
- VEGF
vascular endothelial growth factor
- TME
tumor microenvironment
- SDF-1
stromal derived factor-1
- TGF-β
transforming growth factor-β
- FGF2
fibroblastic growth factor 2
- NLGN3
neuroligin-3
- NMDARs
N-methyl-D-aspartate receptors
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- RANKL
receptor activator of nuclear factor kappa-B ligand
- GDF-15
growth/differentiation factor 15
- CTGF
connective tissue growth factor
- TAMs
tumor-associated macrophages
- TILs
tumor-infiltrating lymphocytes
- MIF
macrophage migration inhibitory factor
- MDSCs
myeloid-derived suppressor cells
- mMDSCs
monocytic MDSCs
- mMDSCs
granulocytic MDSCs
- CSF-1R
colony-stimulating factor-1R
- JAM-A
junctional adhesion molecule-A
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
Manmeet Ahluwalia declares the following: grants/research support-Astrazeneca, Abbvie, BMS, Bayer, Incyte, Pharmacyclics, Novocure, Mimivax, Merck. Receipt of honoraria or consulting fees: Elsevier, Wiley, VBI, Vaccines, Bayer, Tocagen, Novocure. Shareholder: Doctible, Mimivax, Cytodyn.
References
- [1].Jemal A, Ward EM, Johnson CJ, Cronin KA, Ma J, Ryerson AB, Mariotto A, Lake AJ, Wilson R, Sherman RL, Anderson RN, Henley SJ, Kohler BA, Penberthy L, Feuer EJ, Weir HK, Annual Report to the Nation on the Status of Cancer, 1975–2014, Featuring Survival, J Natl Cancer Inst. 109 (2017). 10.1093/jnci/djx030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Tabouret E, Chinot O, Metellus P, Tallet A, Viens P, Gonçalves A, Recent Trends in Epidemiology of Brain Metastases: An Overview, Anticancer Res. 32 (2012) 4655–4662. [PubMed] [Google Scholar]
- [3].Hall WA, Djalilian HR, Nussbaum ES, Cho KH, Long-term survival with metastatic cancer to the brain, Med. Oncol 17 (2000) 279–286. [DOI] [PubMed] [Google Scholar]
- [4].Percy AK, Elveback LR, Okazaki H, Kurland LT, Neoplasms of the central nervous system. Epidemiologic considerations, Neurology. 22 (1972) 40–48. 10.1212/wnl.22.1.40. [DOI] [PubMed] [Google Scholar]
- [5].Tsukada Y, Fouad A, Pickren JW, Lane WW, Central nervous system metastasis from breast carcinoma. Autopsy study, Cancer. 52 (1983) 2349–2354. [DOI] [PubMed] [Google Scholar]
- [6].Sperduto PW, Mesko S, Li J, Cagney D, Aizer A, Lin NU, Nesbit E, Kruser TJ, Chan J, Braunstein S, Lee J, Kirkpatrick JP, Breen W, Brown PD, Shi D, Shih HA, Soliman H, Sahgal A, Shanley R, Sperduto WA, Lou E, Everett A, Boggs DH, Masucci L, Roberge D, Remick J, Plichta K, Buatti JM, Jain S, Gaspar LE, Wu C-C, Wang TJC, Bryant J, Chuong M, An Y, Chiang V, Nakano T, Aoyama H, Mehta MP, Survival in Patients With Brain Metastases: Summary Report on the Updated Diagnosis-Specific Graded Prognostic Assessment and Definition of the Eligibility Quotient, J. Clin. Oncol (2020) JCO2001255. 10.1200/JCO.20.01255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Brastianos PK, Carter SL, Santagata S, Cahill DP, Taylor-Weiner A, Jones RT, Van Allen EM, Lawrence MS, Horowitz PM, Cibulskis K, Ligon KL, Tabernero J, Seoane J, Martinez-Saez E, Curry WT, Dunn IF, Paek SH, Park S-H, McKenna A, Chevalier A, Rosenberg M, Barker FG, Gill CM, Van Hummelen P, Thorner AR, Johnson BE, Hoang MP, Choueiri TK, Signoretti S, Sougnez C, Rabin MS, Lin NU, Winer EP, Stemmer-Rachamimov A, Meyerson M, Garraway L, Gabriel S, Lander ES, Beroukhim R, Batchelor TT, Baselga J, Louis DN, Getz G, Hahn WC, Genomic Characterization of Brain Metastases Reveals Branched Evolution and Potential Therapeutic Targets, Cancer Discov. 5 (2015) 1164–1177. 10.1158/2159-8290.CD-15-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhao L, Zhao W, Hou Y, Wen C, Wang J, Wu P, Guo Z, An Overview of Managements in Meningiomas, Front Oncol. 10 (2020) 1523. 10.3389/fonc.2020.01523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Prager BC, Vasudevan HN, Dixit D, Bernatchez JA, Wu Q, Wallace LC, Bhargava S, Lee D, King BH, Morton AR, Gimple RC, Pekmezci M, Zhu Z, Siqueira-Neto JL, Wang X, Xie Q, Chen C, Barnett GH, Vogelbaum MA, Mack SC, Chavez L, Perry A, Raleigh DR, Rich JN, The Meningioma Enhancer Landscape Delineates Novel Subgroups and Drives Druggable Dependencies, Cancer Discov. (2020). 10.1158/2159-8290.CD-20-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhu Y, Guignard F, Zhao D, Liu L, Burns DK, Mason RP, Messing A, Parada LF, Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma, Cancer Cell. 8 (2005) 119–130. 10.1016/j.ccr.2005.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chow LML, Endersby R, Zhu X, Rankin S, Qu C, Zhang J, Broniscer A, Ellison DW, Baker SJ, Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain, Cancer Cell. 19 (2011) 305–316. 10.1016/j.ccr.2011.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Liu C, Sage JC, Miller MR, Verhaak RGW, Hippenmeyer S, Vogel H, Foreman O, Bronson RT, Nishiyama A, Luo L, Zong H, Mosaic analysis with double markers reveals tumor cell of origin in glioma, Cell. 146 (2011) 209–221. 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW, The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary, Acta Neuropathol. 131 (2016) 803–820. 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- [14].Tom MC, Park DYJ, Yang K, Leyrer CM, Wei W, Jia X, Varra V, Yu JS, Chao ST, Balagamwala EH, Suh JH, Vogelbaum MA, Barnett GH, Prayson RA, Stevens GHJ, Peereboom DM, Ahluwalia MS, Murphy ES, Malignant Transformation of Molecularly Classified Adult Low-Grade Glioma, Int J Radiat Oncol Biol Phys. 105 (2019) 1106–1112. 10.1016/j.ijrobp.2019.08.025. [DOI] [PubMed] [Google Scholar]
- [15].Wang J, Garancher A, Ramaswamy V, Wechsler-Reya RJ, Medulloblastoma: From Molecular Subgroups to Molecular Targeted Therapies, Annu Rev Neurosci. 41 (2018) 207–232. 10.1146/annurev-neuro-070815-013838. [DOI] [PubMed] [Google Scholar]
- [16].Northcott PA, Buchhalter I, Morrissy AS, Hovestadt V, Weischenfeldt J, Ehrenberger T, Gröbner S, Segura-Wang M, Zichner T, Rudneva VA, Warnatz H-J, Sidiropoulos N, Phillips AH, Schumacher S, Kleinheinz K, Waszak SM, Erkek S, Jones DTW, Worst BC, Kool M, Zapatka M, Jäger N, Chavez L, Hutter B, Bieg M, Paramasivam N, Heinold M, Gu Z, Ishaque N, Jäger-Schmidt C, Imbusch CD, Jugold A, Hübschmann D, Risch T, Amstislavskiy V, Gonzalez FGR, Weber UD, Wolf S, Robinson GW, Zhou X, Wu G, Finkelstein D, Liu Y, Cavalli FMG, Luu B, Ramaswamy V, Wu X, Koster J, Ryzhova M, Cho Y-J, Pomeroy SL, Herold-Mende C, Schuhmann M, Ebinger M, Liau LM, Mora J, McLendon RE, Jabado N, Kumabe T, Chuah E, Ma Y, Moore RA, Mungall AJ, Mungall KL, Thiessen N, Tse K, Wong T, Jones SJM, Witt O, Milde T, Von Deimling A, Capper D, Korshunov A, Yaspo M-L, Kriwacki R, Gajjar A, Zhang J, Beroukhim R, Fraenkel E, Korbel JO, Brors B, Schlesner M, Eils R, Marra MA, Pfister SM, Taylor MD, Lichter P, The whole-genome landscape of medulloblastoma subtypes, Nature. 547 (2017) 311–317. 10.1038/nature22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Northcott PA, Robinson GW, Kratz CP, Mabbott DJ, Pomeroy SL, Clifford SC, Rutkowski S, Ellison DW, Malkin D, Taylor MD, Gajjar A, Pfister SM, Medulloblastoma, Nature Reviews Disease Primers. 5 (2019) 1–20. 10.1038/s41572-019-0063-6. [DOI] [PubMed] [Google Scholar]
- [18].Taylor MD, Northcott PA, Korshunov A, Remke M, Cho Y-J, Clifford SC, Eberhart CG, Parsons DW, Rutkowski S, Gajjar A, Ellison DW, Lichter P, Gilbertson RJ, Pomeroy SL, Kool M, Pfister SM, Molecular subgroups of medulloblastoma: the current consensus, Acta Neuropathologica. 123 (2012) 465–472. 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Manoranjan B, Venugopal C, Bakhshinyan D, Adile AA, Richards L, Kameda-Smith MM, Whitley O, Dvorkin-Gheva A, Subapanditha M, Savage N, Tatari N, McKenna D, Bassey-Archibong B, Winegarden N, Hallett R, Provias JP, Yarascavitch B, Ajani O, Fleming A, Bader GD, Pugh TJ, Doble BW, Singh SK, Wnt activation as a therapeutic strategy in medulloblastoma, Nat Commun. 11 (2020) 4323. 10.1038/s41467-020-17953-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Stupp R, Taillibert S, Kanner A, Read W, Steinberg DM, Lhermitte B, Toms S, Idbaih A, Ahluwalia MS, Fink K, Meco FD, Lieberman F, Zhu J-J, Stragliotto G, Tran DD, Brem S, Hottinger AF, Kirson ED, Lavy-Shahaf G, Weinberg U, Kim C-Y, Paek S-H, Nicholas G, Bruna J, Hirte H, Weller M, Palti Y, Hegi ME, Ram Z, Effect of Tumor-Treating Fields Plus Maintenance Temozolomide vs Maintenance Temozolomide Alone on Survival in Patients With Glioblastoma: A Randomized Clinical Trial, JAMA. 318 (2017) 2306–2316. 10.1001/jama.2017.18718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJB, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO, European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups, National Cancer Institute of Canada Clinical Trials Group, Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma, N. Engl. J. Med 352 (2005) 987–996. 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- [22].Harder BG, Blomquist MR, Wang J, Kim AJ, Woodworth GF, Winkles JA, Loftus JC, Tran NL, Developments in Blood-Brain Barrier Penetrance and Drug Repurposing for Improved Treatment of Glioblastoma, Front Oncol. 8 (2018) 462. 10.3389/fonc.2018.00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Colwell N, Larion M, Giles AJ, Seldomridge AN, Sizdahkhani S, Gilbert MR, Park DM, Hypoxia in the glioblastoma microenvironment: shaping the phenotype of cancer stem-like cells, Neuro Oncol. 19 (2017) 887–896. 10.1093/neuonc/now258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gimple RC, Bhargava S, Dixit D, Rich JN, Glioblastoma stem cells: lessons from the tumor hierarchy in a lethal cancer, Genes Dev. 33 (2019) 591–609. 10.1101/gad.324301.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Suter R, Rodriguez-Blanco J, Ayad NG, Epigenetic pathways and plasticity in brain tumors, Neurobiol. Dis (2020) 105060. 10.1016/j.nbd.2020.105060. [DOI] [PubMed] [Google Scholar]
- [26].Burger PC, Green SB, Patient age, histologic features, and length of survival in patients with glioblastoma multiforme, Cancer. 59 (1987) 1617–1625. . [DOI] [PubMed] [Google Scholar]
- [27].Reinartz R, Wang S, Kebir S, Silver DJ, Wieland A, Zheng T, Küpper M, Rauschenbach L, Fimmers R, Shepherd TM, Trageser D, Till A, Schäfer N, Glas M, Hillmer AM, Cichon S, Smith AA, Pietsch T, Liu Y, Reynolds BA, Yachnis A, Pincus DW, Simon M, Brüstle O, Steindler DA, Scheffler B, Functional Subclone Profiling for Prediction of Treatment-Induced Intratumor Population Shifts and Discovery of Rational Drug Combinations in Human Glioblastoma, Clin Cancer Res. 23 (2017) 562–574. 10.1158/1078-0432.CCR-15-2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Nobusawa S, Lachuer J, Wierinckx A, Kim YH, Huang J, Legras C, Kleihues P, Ohgaki H, Intratumoral patterns of genomic imbalance in glioblastomas, Brain Pathol. 20 (2010) 936–944. 10.1111/j.1750-3639.2010.00395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kumar A, Boyle EA, Tokita M, Mikheev AM, Sanger MC, Girard E, Silber JR, Gonzalez-Cuyar LF, Hiatt JB, Adey A, Lee C, Kitzman JO, Born DE, Silbergeld DL, Olson JM, Rostomily RC, Shendure J, Deep sequencing of multiple regions of glial tumors reveals spatial heterogeneity for mutations in clinically relevant genes, Genome Biol. 15 (2014) 530. 10.1186/s13059-014-0530-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Safa AR, Saadatzadeh MR, Cohen-Gadol AA, Pollok KE, Bijangi-Vishehsaraei K, Glioblastoma stem cells (GSCs) epigenetic plasticity and interconversion between differentiated non-GSCs and GSCs, Genes & Diseases. 2 (2015) 152–163. 10.1016/j.gendis.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CLL, Rich JN, Cancer stem cells in glioblastoma, Genes Dev. 29 (2015) 1203–1217. 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ludwig K, Kornblum HI, Molecular markers in glioma, J Neurooncol. 134 (2017) 505–512. 10.1007/s11060-017-2379-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, Akhavanfard S, Cahill DP, Aldape KD, Betensky RA, Louis DN, Iafrate AJ, Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma, Cancer Cell. 20 (2011) 810–817. 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- [34].Little SE, Popov S, Jury A, Bax DA, Doey L, Al-Sarraj S, Jurgensmeier JM, Jones C, Receptor tyrosine kinase genes amplified in glioblastoma exhibit a mutual exclusivity in variable proportions reflective of individual tumor heterogeneity, Cancer Res. 72 (2012) 1614–1620. 10.1158/0008-5472.CAN-11-4069. [DOI] [PubMed] [Google Scholar]
- [35].Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y, Ozawa T, Holland EC, Huse JT, Jhanwar S, Leversha MA, Mikkelsen T, Brennan CW, Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response, Proc. Natl. Acad. Sci. U.S.A 109 (2012) 3041–3046. 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Weller M, Butowski N, Tran DD, Recht LD, Lim M, Hirte H, Ashby L, Mechtler L, Goldlust SA, Iwamoto F, Drappatz J, O’Rourke DM, Wong M, Hamilton MG, Finocchiaro G, Perry J, Wick W, Green J, He Y, Turner CD, Yellin MJ, Keler T, Davis TA, Stupp R, Sampson JH, ACT IV trial investigators, Rindopepimut with temozolomide for patients with newly diagnosed, EGFRvIII-expressing glioblastoma (ACT IV): a randomised, double-blind, international phase 3 trial, Lancet Oncol. 18 (2017) 1373–1385. 10.1016/S1470-2045(17)30517-X. [DOI] [PubMed] [Google Scholar]
- [37].Hegi ME, Diserens A-C, Bady P, Kamoshima Y, Kouwenhoven MCM, Delorenzi M, Lambiv WL, Hamou M-F, Matter MS, Koch A, Heppner FL, Yonekawa Y, Merlo A, Frei K, Mariani L, Hofer S, Pathway analysis of glioblastoma tissue after preoperative treatment with the EGFR tyrosine kinase inhibitor gefitinib--a phase II trial, Mol Cancer Ther. 10 (2011) 1102–1112. 10.1158/1535-7163.MCT-11-0048. [DOI] [PubMed] [Google Scholar]
- [38].Chakravarti A, Wang M, Robins HI, Lautenschlaeger T, Curran WJ, Brachman DG, Schultz CJ, Choucair A, Dolled-Filhart M, Christiansen J, Gustavson M, Molinaro A, Mischel P, Dicker AP, Bredel M, Mehta M, RTOG 0211: a phase 1/2 study of radiation therapy with concurrent gefitinib for newly diagnosed glioblastoma patients, Int J Radiat Oncol Biol Phys. 85 (2013) 1206–1211. 10.1016/j.ijrobp.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Haas-Kogan DA, Prados MD, Tihan T, Eberhard DA, Jelluma N, Arvold ND, Baumber R, Lamborn KR, Kapadia A, Malec M, Berger MS, Stokoe D, Epidermal growth factor receptor, protein kinase B/Akt, and glioma response to erlotinib, J Natl Cancer Inst. 97 (2005) 880–887. 10.1093/jnci/dji161. [DOI] [PubMed] [Google Scholar]
- [40].Reungwetwattana T, Nakagawa K, Cho BC, Cobo M, Cho EK, Bertolini A, Bohnet S, Zhou C, Lee KH, Nogami N, Okamoto I, Leighl N, Hodge R, McKeown A, Brown AP, Rukazenkov Y, Ramalingam SS, Vansteenkiste J, CNS Response to Osimertinib Versus Standard Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Patients With Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer, J Clin Oncol. (2018) JCO2018783118. 10.1200/JCO.2018.78.3118. [DOI] [PubMed] [Google Scholar]
- [41].Soria J-C, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, Dechaphunkul A, Imamura F, Nogami N, Kurata T, Okamoto I, Zhou C, Cho BC, Cheng Y, Cho EK, Voon PJ, Planchard D, Su W-C, Gray JE, Lee S-M, Hodge R, Marotti M, Rukazenkov Y, Ramalingam SS, Osimertinib in Untreated EGFR-Mutated Advanced Non–Small-Cell Lung Cancer, New England Journal of Medicine. (2017). 10.1056/NEJMoa1713137. [DOI] [PubMed] [Google Scholar]
- [42].Shen R, Mo Q, Schultz N, Seshan VE, Olshen AB, Huse J, Ladanyi M, Sander C, Integrative subtype discovery in glioblastoma using iCluster, PLoS ONE. 7 (2012) e35236. 10.1371/journal.pone.0035236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN, Cancer Genome Atlas Research Network, Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1, Cancer Cell. 17 (2010) 98–110. 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sottoriva A, Spiteri I, Piccirillo SGM, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavaré S, Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics, Proc Natl Acad Sci U S A. 110 (2013) 4009–4014. 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lee J-K, Wang J, Sa JK, Ladewig E, Lee H-O, Lee I-H, Kang HJ, Rosenbloom DS, Camara PG, Liu Z, van Nieuwenhuizen P, Jung SW, Choi SW, Kim J, Chen A, Kim K-T, Shin S, Seo YJ, Oh J-M, Shin YJ, Park C-K, Kong D-S, Seol HJ, Blumberg A, Lee J-I, Iavarone A, Park W-Y, Rabadan R, Nam D-H, Spatiotemporal genomic architecture informs precision oncology in glioblastoma, Nature Genetics. 49 (2017) 594–599. 10.1038/ng.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wang J, Cazzato E, Ladewig E, Frattini V, Rosenbloom DIS, Zairis S, Abate F, Liu Z, Elliott O, Shin Y-J, Lee J-K, Lee I-H, Park W-Y, Eoli M, Blumberg AJ, Lasorella A, Nam D-H, Finocchiaro G, Iavarone A, Rabadan R, Clonal evolution of glioblastoma under therapy, Nature Genetics. 48 (2016) 768–776. 10.1038/ng.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Gill BJ, Pisapia DJ, Malone HR, Goldstein H, Lei L, Sonabend A, Yun J, Samanamud J, Sims JS, Banu M, Dovas A, Teich AF, Sheth SA, McKhann GM, Sisti MB, Bruce JN, Sims PA, Canoll P, MRI-localized biopsies reveal subtype-specific differences in molecular and cellular composition at the margins of glioblastoma, Proceedings of the National Academy of Sciences of the United States of America. 111 (2014) 12550–12555. 10.1073/pnas.1405839111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Puchalski RB, Shah N, Miller J, Dalley R, Nomura SR, Yoon J-G, Smith KA, Lankerovich M, Bertagnolli D, Bickley K, Boe AF, Brouner K, Butler S, Caldejon S, Chapin M, Datta S, Dee N, Desta T, Dolbeare T, Dotson N, Ebbert A, Feng D, Feng X, Fisher M, Gee G, Goldy J, Gourley L, Gregor BW, Gu G, Hejazinia N, Hohmann J, Hothi P, Howard R, Joines K, Kriedberg A, Kuan L, Lau C, Lee F, Lee H, Lemon T, Long F, Mastan N, Mott E, Murthy C, Ngo K, Olson E, Reding M, Riley Z, Rosen D, Sandman D, Shapovalova N, Slaughterbeck CR, Sodt A, Stockdale G, Szafer A, Wakeman W, Wohnoutka PE, White SJ, Marsh D, Rostomily RC, Ng L, Dang C, Jones A, Keogh B, Gittleman HR, Barnholtz-Sloan JS, Cimino PJ, Uppin MS, Keene CD, Farrokhi FR, Lathia JD, Berens ME, Iavarone A, Bernard A, Lein E, Phillips JW, Rostad SW, Cobbs C, Hawrylycz MJ, Foltz GD, An anatomic transcriptional atlas of human glioblastoma, Science (New York, N.Y.). 360 (2018) 660–663. 10.1126/science.aaf2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Miller TE, Liau BB, Wallace LC, Morton AR, Xie Q, Dixit D, Factor DC, Kim LJY, Morrow JJ, Wu Q, Mack SC, Hubert CG, Gillespie SM, Flavahan WA, Hoffmann T, Thummalapalli R, Hemann MT, Paddison PJ, Horbinski CM, Zuber J, Scacheri PC, Bernstein BE, Tesar PJ, Rich JN, Transcription elongation factors represent in vivo cancer dependencies in glioblastoma, Nature. 547 (2017) 355–359. 10.1038/nature23000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Yu D, Khan OF, Suvà ML, Dong B, Panek WK, Xiao T, Wu M, Han Y, Ahmed AU, Balyasnikova IV, Zhang HF, Sun C, Langer R, Anderson DG, Lesniak MS, Multiplexed RNAi therapy against brain tumor-initiating cells via lipopolymeric nanoparticle infusion delays glioblastoma progression, Proc Natl Acad Sci U S A. 114 (2017) E6147–E6156. 10.1073/pnas.1701911114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wang Q, Hu B, Hu X, Kim H, Squatrito M, Scarpace L, deCarvalho AC, Lyu S, Li P, Li Y, Barthel F, Cho HJ, Lin Y-H, Satani N, Martinez-Ledesma E, Zheng S, Chang E, Sauvé C-EG, Olar A, Lan ZD, Finocchiaro G, Phillips JJ, Berger MS, Gabrusiewicz KR, Wang G, Eskilsson E, Hu J, Mikkelsen T, DePinho RA, Muller F, Heimberger AB, Sulman EP, Nam D-H, Verhaak RGW, Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment, Cancer Cell. 32 (2017) 42–56.e6. 10.1016/j.ccell.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Celiku O, Gilbert MR, Lavi O, Computational modeling demonstrates that glioblastoma cells can survive spatial environmental challenges through exploratory adaptation, Nature Communications. 10 (2019) 5704. 10.1038/s41467-019-13726-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, Louis DN, Rozenblatt-Rosen O, Suvà ML, Regev A, Bernstein BE, Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma, Science. 344 (2014) 1396–1401. 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, Richman AR, Silverbush D, Shaw ML, Hebert CM, Dewitt J, Gritsch S, Perez EM, Gonzalez Castro LN, Lan X, Druck N, Rodman C, Dionne D, Kaplan A, Bertalan MS, Small J, Pelton K, Becker S, Bonal D, Nguyen Q-D, Servis RL, Fung JM, Mylvaganam R, Mayr L, Gojo J, Haberler C, Geyeregger R, Czech T, Slavc I, Nahed BV, Curry WT, Carter BS, Wakimoto H, Brastianos PK, Batchelor TT, Stemmer-Rachamimov A, Martinez-Lage M, Frosch MP, Stamenkovic I, Riggi N, Rheinbay E, Monje M, Rozenblatt-Rosen O, Cahill DP, Patel AP, Hunter T, Verma IM, Ligon KL, Louis DN, Regev A, Bernstein BE, Tirosh I, Suvà ML, An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma, Cell. 178 (2019) 835–849.e21. 10.1016/j.cell.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, Fouse SD, Yamamoto S, Ueda H, Tatsuno K, Asthana S, Jalbert LE, Nelson SJ, Bollen AW, Gustafson WC, Charron E, Weiss WA, Smirnov IV, Song JS, Olshen AB, Cha S, Zhao Y, Moore RA, Mungall AJ, Jones SJM, Hirst M, Marra MA, Saito N, Aburatani H, Mukasa A, Berger MS, Chang SM, Taylor BS, Costello JF, Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma, Science. 343 (2014) 189–193. 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Barthel FP, Johnson KC, Varn FS, Moskalik AD, Tanner G, Kocakavuk E, Anderson KJ, Abiola O, Aldape K, Alfaro KD, Alpar D, Amin SB, Ashley DM, Bandopadhayay P, Barnholtz-Sloan JS, Beroukhim R, Bock C, Brastianos PK, Brat DJ, Brodbelt AR, Bruns AF, Bulsara KR, Chakrabarty A, Chakravarti A, Chuang JH, Claus EB, Cochran EJ, Connelly J, Costello JF, Finocchiaro G, Fletcher MN, French PJ, Gan HK, Gilbert MR, Gould PV, Grimmer MR, Iavarone A, Ismail A, Jenkinson MD, Khasraw M, Kim H, Kouwenhoven MCM, LaViolette PS, Li M, Lichter P, Ligon KL, Lowman AK, Malta TM, Mazor T, McDonald KL, Molinaro AM, Nam D-H, Nayyar N, Ng HK, Ngan CY, Niclou SP, Niers JM, Noushmehr H, Noorbakhsh J, Ormond DR, Park C-K, Poisson LM, Rabadan R, Radlwimmer B, Rao G, Reifenberger G, Sa JK, Schuster M, Shaw BL, Short SC, Smitt PAS, Sloan AE, Smits M, Suzuki H, Tabatabai G, Van Meir EG, Watts C, Weller M, Wesseling P, Westerman BA, Widhalm G, Woehrer A, Yung WKA, Zadeh G, Huse JT, De Groot JF, Stead LF, Verhaak RGW, GLASS Consortium, Longitudinal molecular trajectories of diffuse glioma in adults, Nature. 576 (2019) 112–120. 10.1038/s41586-019-1775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Delgado-López PD, Corrales-García EM, Survival in glioblastoma: a review on the impact of treatment modalities, Clin Transl Oncol. 18 (2016) 1062–1071. 10.1007/s12094-016-1497-x. [DOI] [PubMed] [Google Scholar]
- [58].Hegi ME, Diserens A-C, Gorlia T, Hamou M-F, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JEC, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R, MGMT gene silencing and benefit from temozolomide in glioblastoma, N Engl J Med. 352 (2005) 997–1003. 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- [59].Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RGW, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Van Den Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SB, Laird PW, Aldape K, Identification of a CpG Island Methylator Phenotype that Defines a Distinct Subgroup of Glioma, Cancer Cell. 17 (2010) 510–522. 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Wenger A, Ferreyra Vega S, Kling T, Bontell TO, Jakola AS, Carén H, Intratumor DNA methylation heterogeneity in glioblastoma: implications for DNA methylation-based classification, Neuro-Oncology. 21 (2019) 616–627. 10.1093/neuonc/noz011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Klughammer J, Kiesel B, Roetzer T, Fortelny N, Nemc A, Nenning K-H, Furtner J, Sheffield NC, Datlinger P, Peter N, Nowosielski M, Augustin M, Mischkulnig M, Ströbel T, Alpar D, Ergüner B, Senekowitsch M, Moser P, Freyschlag CF, Kerschbaumer J, Thomé C, Grams AE, Stockhammer G, Kitzwoegerer M, Oberndorfer S, Marhold F, Weis S, Trenkler J, Buchroithner J, Pichler J, Haybaeck J, Krassnig S, Mahdy Ali K., von Campe G, Payer F, Sherif C, Preiser J, Hauser T, Winkler PA, Kleindienst W, Würtz F, Brandner-Kokalj T, Stultschnig M, Schweiger S, Dieckmann K, Preusser M, Langs G, Baumann B, Knosp E, Widhalm G, Marosi C, Hainfellner JA, Woehrer A, Bock C, The DNA methylation landscape of glioblastoma disease progression shows extensive heterogeneity in time and space, Nat. Med 24 (2018) 1611–1624. 10.1038/s41591-018-0156-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Dixit D, Prager BC, Gimple RC, Poh HX, Wang Y, Wu Q, Qiu Z, Kidwell RL, Kim LJY, Xie Q, Vitting-Seerup K, Bhargava S, Dong Z, Jiang L, Zhu Z, Hamerlik P, Jaffrey SR, Zhao JC, Wang X, Rich JN, The RNA m6A reader YTHDF2 maintains oncogene expression and is a targetable dependency in glioblastoma stem cells, Cancer Discov. (2020). 10.1158/2159-8290.CD-20-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Mack SC, Pajtler KW, Chavez L, Okonechnikov K, Bertrand KC, Wang X, Erkek S, Federation A, Song A, Lee C, Wang X, McDonald L, Morrow JJ, Saiakhova A, Sin-Chan P, Wu Q, Michaelraj KA, Miller TE, Hubert CG, Ryzhova M, Garzia L, Donovan L, Dombrowski S, Factor DC, Luu B, Valentim CLL, Gimple RC, Morton A, Kim L, Prager BC, Lee JJY, Wu X, Zuccaro J, Thompson Y, Holgado BL, Reimand J, Ke SQ, Tropper A, Lai S, Vijayarajah S, Doan S, Mahadev V, Miñan AF, Gröbner SN, Lienhard M, Zapatka M, Huang Z, Aldape KD, Carcaboso AM, Houghton PJ, Keir ST, Milde T, Witt H, Li Y, Li C-J, Bian X-W, Jones DTW, Scott I, Singh SK, Huang A, Dirks PB, Bouffet E, Bradner JE, Ramaswamy V, Jabado N, Rutka JT, Northcott PA, Lupien M, Lichter P, Korshunov A, Scacheri PC, Pfister SM, Kool M, Taylor MD, Rich JN, Therapeutic targeting of ependymoma as informed by oncogenic enhancer profiling, Nature. 553 (2018) 101–105. 10.1038/nature25169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kim H, Nguyen N-P, Turner K, Wu S, Gujar AD, Luebeck J, Liu J, Deshpande V, Rajkumar U, Namburi S, Amin SB, Yi E, Menghi F, Schulte JH, Henssen AG, Chang HY, Beck CR, Mischel PS, Bafna V, Verhaak RGW, Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers, Nat Genet. 52 (2020) 891–897. 10.1038/s41588-020-0678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].deCarvalho AC, Kim H, Poisson LM, Winn ME, Mueller C, Cherba D, Koeman J, Seth S, Protopopov A, Felicella M, Zheng S, Multani A, Jiang Y, Zhang J, Nam D-H, Petricoin EF, Chin L, Mikkelsen T, Verhaak RGW, Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma, Nat Genet. 50 (2018) 708–717. 10.1038/s41588-018-0105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Morton AR, Dogan-Artun N, Faber ZJ, MacLeod G, Bartels CF, Piazza MS, Allan KC, Mack SC, Wang X, Gimple RC, Wu Q, Rubin BP, Shetty S, Angers S, Dirks PB, Sallari RC, Lupien M, Rich JN, Scacheri PC, Functional Enhancers Shape Extrachromosomal Oncogene Amplifications, Cell. 179 (2019) 1330–1341.e13. 10.1016/j.cell.2019.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Papaccio F, Paino F, Regad T, Papaccio G, Desiderio V, Tirino V, Concise Review: Cancer Cells, Cancer Stem Cells, and Mesenchymal Stem Cells: Influence in Cancer Development, STEM CELLS Translational Medicine. 6 (2017) 2115–2125. 10.1002/sctm.17-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A, Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma, Cancer Res. 64 (2004) 7011–7021. 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- [69].Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB, Identification of human brain tumour initiating cells, Nature. 432 (2004) 396–401. 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- [70].Pallini R, Ricci-Vitiani L, Banna GL, Signore M, Lombardi D, Todaro M, Stassi G, Martini M, Maira G, Larocca LM, Maria RD, Cancer Stem Cell Analysis and Clinical Outcome in Patients with Glioblastoma Multiforme, Clin Cancer Res. 14 (2008) 8205–8212. 10.1158/1078-0432.CCR-08-0644. [DOI] [PubMed] [Google Scholar]
- [71].Zeppernick F, Ahmadi R, Campos B, Dictus C, Helmke BM, Becker N, Lichter P, Unterberg A, Radlwimmer B, Herold-Mende CC, Stem Cell Marker CD133 Affects Clinical Outcome in Glioma Patients, Clin Cancer Res. 14 (2008) 123–129. 10.1158/1078-0432.CCR-07-0932. [DOI] [PubMed] [Google Scholar]
- [72].Manoranjan B, Chokshi C, Venugopal C, Subapanditha M, Savage N, Tatari N, Provias JP, Murty NK, Moffat J, Doble BW, Singh SK, A CD133-AKT-Wnt signaling axis drives glioblastoma brain tumor-initiating cells, Oncogene. 39 (2020) 1590–1599. 10.1038/s41388-019-1086-x. [DOI] [PubMed] [Google Scholar]
- [73].Beier D, Hau P, Proescholdt M, Lohmeier A, Wischhusen J, Oefner PJ, Aigner L, Brawanski A, Bogdahn U, Beier CP, CD133+ and CD133− Glioblastoma-Derived Cancer Stem Cells Show Differential Growth Characteristics and Molecular Profiles, Cancer Res. 67 (2007) 4010–4015. 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- [74].Chen R, Nishimura MC, Bumbaca SM, Kharbanda S, Forrest WF, Kasman IM, Greve JM, Soriano RH, Gilmour LL, Rivers CS, Modrusan Z, Nacu S, Guerrero S, Edgar KA, Wallin JJ, Lamszus K, Westphal M, Heim S, James CD, VandenBerg SR, Costello JF, Moorefield S, Cowdrey CJ, Prados M, Phillips HS, A hierarchy of self-renewing tumor-initiating cell types in glioblastoma, Cancer Cell. 17 (2010) 362–375. 10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- [75].Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, Kornblum HI, Cancerous stem cells can arise from pediatric brain tumors, Proc. Natl. Acad. Sci. U.S.A 100 (2003) 15178–15183. 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA, An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors, Nat. Genet 40 (2008) 499–507. 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Suvà ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD, Riggi N, Chi AS, Cahill DP, Nahed BV, Curry WT, Martuza RL, Rivera MN, Rossetti N, Kasif S, Beik S, Kadri S, Tirosh I, Wortman I, Shalek AK, Rozenblatt-Rosen O, Regev A, Louis DN, Bernstein BE, Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells, Cell. 157 (2014) 580–594. 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Ligon KL, Huillard E, Mehta S, Kesari S, Liu H, Alberta JA, Bachoo RM, Kane M, Louis DN, Depinho RA, Anderson DJ, Stiles CD, Rowitch DH, Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma, Neuron. 53 (2007) 503–517. 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH, A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs, Cell. 143 (2010) 313–324. 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Lathia JD, Gallagher J, Heddleston JM, Wang J, Eyler CE, MacSwords J, Wu Q, Vasanji A, McLendon RE, Hjelmeland AB, Rich JN, Integrin alpha 6 regulates glioblastoma stem cells, Cell Stem Cell. 6 (2010) 421–432. 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Bao S, Wu Q, Li Z, Sathornsumetee S, Wang H, McLendon RE, Hjelmeland AB, Rich JN, Targeting cancer stem cells through L1CAM suppresses glioma growth, Cancer Res. 68 (2008) 6043–6048. 10.1158/0008-5472.CAN-08-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Anido J, Sáez-Borderías A, Gonzàlez-Juncà A, Rodón L, Folch G, Carmona MA, Prieto-Sánchez RM, Barba I, Martínez-Sáez E, Prudkin L, Cuartas I, Raventós C, Martínez-Ricarte F, Poca MA, García-Dorado D, Lahn MM, Yingling JM, Rodón J, Sahuquillo J, Baselga J, Seoane J, TGF-β Receptor Inhibitors Target the CD44(high)/Id1(high) Glioma-Initiating Cell Population in Human Glioblastoma, Cancer Cell. 18 (2010) 655–668. 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- [83].Alrfaei BM, Clark P, Vemuganti R, Kuo JS, MicroRNA miR-100 Decreases Glioblastoma Growth by Targeting SMARCA5 and ErbB3 in Tumor-Initiating Cells, Technol Cancer Res Treat. 19 (2020) 1533033820960748. 10.1177/1533033820960748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Alvarado AG, Turaga SM, Sathyan P, Mulkearns-Hubert EE, Otvos B, Silver DJ, Hale JS, Flavahan WA, Zinn PO, Sinyuk M, Li M, Guda MR, Velpula KK, Tsung AJ, Nakano I, Vogelbaum MA, Majumder S, Rich JN, Lathia JD, Coordination of self-renewal in glioblastoma by integration of adhesion and microRNA signaling, Neuro-Oncology. 18 (2016) 656–666. 10.1093/neuonc/nov196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Zhou W, Yao Y, Scott AJ, Wilder-Romans K, Dresser JJ, Werner CK, Sun H, Pratt D, Sajjakulnukit P, Zhao SG, Davis M, Nelson BS, Halbrook CJ, Zhang L, Gatto F, Umemura Y, Walker AK, Kachman M, Sarkaria JN, Xiong J, Morgan MA, Rehemtualla A, Castro MG, Lowenstein P, Chandrasekaran S, Lawrence TS, Lyssiotis CA, Wahl DR, Purine metabolism regulates DNA repair and therapy resistance in glioblastoma, Nature Communications. 11 (2020). 10.1038/s41467-020-17512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Dirkse A, Golebiewska A, Buder T, Nazarov PV, Muller A, Poovathingal S, Brons NHC, Leite S, Sauvageot N, Sarkisjan D, Seyfrid M, Fritah S, Stieber D, Michelucci A, Hertel F, Herold-Mende C, Azuaje F, Skupin A, Bjerkvig R, Deutsch A, Voss-Böhme A, Niclou SP, Stem cell-associated heterogeneity in Glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment, Nature Communications. 10 (2019) 1787. 10.1038/s41467-019-09853-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Pistollato F, Chen H-L, Rood BR, Zhang H-Z, D’Avella D, Denaro L, Gardiman M, te Kronnie G, Schwartz PH, Favaro E, Indraccolo S, Basso G, Panchision DM, Hypoxia and HIF1α Repress the Differentiative Effects of BMPs in High-Grade Glioma, STEM CELLS. 27 (2009) 7–17. 10.1634/stemcells.2008-0402. [DOI] [PubMed] [Google Scholar]
- [88].Prager BC, Xie Q, Bao S, Rich JN, Cancer Stem Cells: The Architects of the Tumor Ecosystem, Cell Stem Cell. 24 (2019) 41–53. 10.1016/j.stem.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Soeda A, Park M, Lee D, Mintz A, Androutsellis-Theotokis A, McKay RD, Engh J, Iwama T, Kunisada T, Kassam AB, Pollack IF, Park DM, Hypoxia promotes expansion of the CD133-positive glioma stem cells through activation of HIF-1α, Oncogene. 28 (2009) 3949–3959. 10.1038/onc.2009.252. [DOI] [PubMed] [Google Scholar]
- [90].Mathieu J, Zhang Z, Zhou W, Wang AJ, Heddleston JM, Pinna CMA, Hubaud A, Stadler B, Choi M, Bar M, Tewari M, Liu A, Vessella R, Rostomily R, Born D, Horwitz M, Ware C, Blau CA, Cleary MA, Rich JN, Ruohola-Baker H, HIF Induces Human Embryonic Stem Cell Markers in Cancer Cells, Cancer Res. 71 (2011) 4640–4652. 10.1158/0008-5472.CAN-10-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Man J, Yu X, Huang H, Zhou W, Xiang C, Huang H, Miele L, Liu Z, Bebek G, Bao S, Yu JS, Hypoxic Induction of Vasorin Regulates Notch1 Turnover to Maintain Glioma Stem-like Cells, Cell Stem Cell. 22 (2018) 104–118.e6. 10.1016/j.stem.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJB, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, Gijtenbeek J, Marosi C, Vecht CJ, Mokhtari K, Wesseling P, Villa S, Eisenhauer E, Gorlia T, Weller M, Lacombe D, Cairncross JG, Mirimanoff R-O, European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups, National Cancer Institute of Canada Clinical Trials Group, Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial, Lancet Oncol. 10 (2009) 459–466. 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- [93].Stupp R, Hegi ME, Gilbert MR, Chakravarti A, Chemoradiotherapy in Malignant Glioma: Standard of Care and Future Directions, JCO. 25 (2007) 4127–4136. 10.1200/JCO.2007.11.8554. [DOI] [PubMed] [Google Scholar]
- [94].Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN, Glioma stem cells promote radioresistance by preferential activation of the DNA damage response, Nature. 444 (2006) 756–760. 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- [95].Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, Lu L, Irvin D, Black KL, Yu JS, Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma, Mol. Cancer 5 (2006) 67. 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Minata M, Audia A, Shi J, Lu S, Bernstock J, Pavlyukov MS, Das A, Kim S-H, Shin YJ, Lee Y, Koo H, Snigdha K, Waghmare I, Guo X, Mohyeldin A, Gallego-Perez D, Wang J, Chen D, Cheng P, Mukheef F, Contreras M, Reyes JF, Vaillant B, Sulman EP, Cheng S-Y, Markert JM, Tannous BA, Lu X, Kango-Singh M, Lee LJ, Nam D-H, Nakano I, Bhat KP, Phenotypic Plasticity of Invasive Edge Glioma Stem-like Cells in Response to Ionizing Radiation, Cell Reports. 26 (2019) 1893–1905.e7. 10.1016/j.celrep.2019.01.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Auffinger B, Tobias AL, Han Y, Lee G, Guo D, Dey M, Lesniak MS, Ahmed AU, Conversion of differentiated cancer cells into cancer stem-like cells in a glioblastoma model after primary chemotherapy, Cell Death & Differentiation. 21 (2014) 1119–1131. 10.1038/cdd.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].McGranahan N, Swanton C, Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future, Cell. 168 (2017) 613–628. 10.1016/j.cell.2017.01.018. [DOI] [PubMed] [Google Scholar]
- [99].Prasetyanti PR, Medema JP, Intra-tumor heterogeneity from a cancer stem cell perspective, Mol Cancer. 16 (2017) 41. 10.1186/s12943-017-0600-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Rich JN, Cancer stem cells: understanding tumor hierarchy and heterogeneity, Medicine. 95 (2016) S2. 10.1097/MD.0000000000004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Thankamony AP, Saxena K, Murali R, Jolly MK, Nair R, Cancer Stem Cell Plasticity – A Deadly Deal, Front Mol Biosci. 7 (2020). 10.3389/fmolb.2020.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Daneman R, Prat A, The Blood–Brain Barrier, Cold Spring Harb Perspect Biol. 7 (2015). 10.1101/cshperspect.a020412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Peleli M, Moustakas A, Papapetropoulos A, Endothelial-Tumor Cell Interaction in Brain and CNS Malignancies, Int J Mol Sci. 21 (2020). 10.3390/ijms21197371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Lin JH-C, Takano T, Cotrina ML, Arcuino G, Kang J, Liu S, Gao Q, Jiang L, Li F, Lichtenberg-Frate H, Haubrich S, Willecke K, Goldman SA, Nedergaard M, Connexin 43 Enhances the Adhesivity and Mediates the Invasion of Malignant Glioma Cells, J. Neurosci 22 (2002) 4302–4311. 10.1523/JNEUROSCI.22-11-04302.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Thuringer D, Boucher J, Jego G, Pernet N, Cronier L, Hammann A, Solary E, Garrido C, Transfer of functional microRNAs between glioblastoma and microvascular endothelial cells through gap junctions, Oncotarget. 7 (2016) 73925–73934. 10.18632/oncotarget.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Zhang W, DeMattia JA, Song H, Couldwell WT, Communication between malignant glioma cells and vascular endothelial cells through gap junctions, Journal of Neurosurgery. 98 (2003) 846–853. 10.3171/jns.2003.98.4.0846. [DOI] [PubMed] [Google Scholar]
- [107].Batchelor TT, Reardon DA, de Groot JF, Wick W, Weller M, Antiangiogenic Therapy for Glioblastoma: Current Status and Future Prospects, Clin Cancer Res. 20 (2014) 5612–5619. 10.1158/1078-0432.CCR-14-0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Kreisl TN, Kim L, Moore K, Duic P, Royce C, Stroud I, Garren N, Mackey M, Butman JA, Camphausen K, Park J, Albert PS, Fine HA, Phase II Trial of Single-Agent Bevacizumab Followed by Bevacizumab Plus Irinotecan at Tumor Progression in Recurrent Glioblastoma, J Clin Oncol. 27 (2009) 740–745. 10.1200/JCO.2008.16.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Friedman HS, Prados MD, Wen PY, Mikkelsen T, Schiff D, Abrey LE, Yung WKA, Paleologos N, Nicholas MK, Jensen R, Vredenburgh J, Huang J, Zheng M, Cloughesy T, Bevacizumab alone and in combination with irinotecan in recurrent glioblastoma, J Clin Oncol. 27 (2009) 4733–4740. 10.1200/JCO.2008.19.8721. [DOI] [PubMed] [Google Scholar]
- [110].Gilbert MR, Dignam JJ, Armstrong TS, Wefel JS, Blumenthal DT, Vogelbaum MA, Colman H, Chakravarti A, Pugh S, Won M, Jeraj R, Brown PD, Jaeckle KA, Schiff D, Stieber VW, Brachman DG, Werner-Wasik M, Tremont-Lukats IW, Sulman EP, Aldape KD, Curran WJ, Mehta MP, A randomized trial of bevacizumab for newly diagnosed glioblastoma, N Engl J Med. 370 (2014) 699–708. 10.1056/NEJMoa1308573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Chinot OL, Wick W, Mason W, Henriksson R, Saran F, Nishikawa R, Carpentier AF, Hoang-Xuan K, Kavan P, Cernea D, Brandes AA, Hilton M, Abrey L, Cloughesy T, Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma, N Engl J Med. 370 (2014) 709–722. 10.1056/NEJMoa1308345. [DOI] [PubMed] [Google Scholar]
- [112].Francesconi AB, Dupre S, Matos M, Martin D, Hughes BG, Wyld DK, Lickliter JD, Carboplatin and etoposide combined with bevacizumab for the treatment of recurrent glioblastoma multiforme, J Clin Neurosci. 17 (2010) 970–974. 10.1016/j.jocn.2009.12.009. [DOI] [PubMed] [Google Scholar]
- [113].Reardon DA, Desjardins A, Peters KB, Gururangan S, Sampson JH, McLendon RE, Herndon JE, Bulusu A, Threatt S, Friedman AH, Vredenburgh JJ, Friedman HS, Phase II study of carboplatin, irinotecan, and bevacizumab for bevacizumab naïve, recurrent glioblastoma, J Neurooncol. 107 (2012) 155–164. 10.1007/s11060-011-0722-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Reardon DA, Desjardins A, Vredenburgh JJ, Gururangan S, Sampson JH, Sathornsumetee S, McLendon RE, Herndon JE, Marcello JE, Norfleet J, Friedman AH, Bigner DD, Friedman HS, Metronomic chemotherapy with daily, oral etoposide plus bevacizumab for recurrent malignant glioma: a phase II study, Br J Cancer. 101 (2009) 1986–1994. 10.1038/sj.bjc.6605412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Ali SA, McHayleh WM, Ahmad A, Sehgal R, Braffet M, Rahman M, Bejjani G, Friedland DM, Bevacizumab and irinotecan therapy in glioblastoma multiforme: a series of 13 cases, J Neurosurg. 109 (2008) 268–272. 10.3171/JNS/2008/109/8/0268. [DOI] [PubMed] [Google Scholar]
- [116].Bokstein F, Shpigel S, Blumenthal DT, Treatment with bevacizumab and irinotecan for recurrent high-grade glial tumors, Cancer. 112 (2008) 2267–2273. 10.1002/cncr.23401. [DOI] [PubMed] [Google Scholar]
- [117].Kang TY, Jin T, Elinzano H, Peereboom D, Irinotecan and bevacizumab in progressive primary brain tumors, an evaluation of efficacy and safety, J Neurooncol. 89 (2008) 113–118. 10.1007/s11060-008-9599-0. [DOI] [PubMed] [Google Scholar]
- [118].Zuniga RM, Torcuator R, Jain R, Anderson J, Doyle T, Ellika S, Schultz L, Mikkelsen T, Efficacy, safety and patterns of response and recurrence in patients with recurrent high-grade gliomas treated with bevacizumab plus irinotecan, J Neurooncol. 91 (2009) 329–336. 10.1007/s11060-008-9718-y. [DOI] [PubMed] [Google Scholar]
- [119].Hasselbalch B, Eriksen JG, Broholm H, Christensen IJ, Grunnet K, Horsman MR, Poulsen HS, Stockhausen M-T, Lassen U, Prospective evaluation of angiogenic, hypoxic and EGFR-related biomarkers in recurrent glioblastoma multiforme treated with cetuximab, bevacizumab and irinotecan, APMIS. 118 (2010) 585–594. 10.1111/j.1600-0463.2010.02631.x. [DOI] [PubMed] [Google Scholar]
- [120].Desjardins A, Reardon DA, Coan A, Marcello J, Herndon JE, Bailey L, Peters KB, Friedman HS, Vredenburgh JJ, Bevacizumab and daily temozolomide for recurrent glioblastoma, Cancer. 118 (2012) 1302–1312. 10.1002/cncr.26381. [DOI] [PubMed] [Google Scholar]
- [121].Drappatz J, Lee EQ, Hammond S, Grimm SA, Norden AD, Beroukhim R, Gerard M, Schiff D, Chi AS, Batchelor TT, Doherty LM, Ciampa AS, Lafrankie DC, Ruland S, Snodgrass SM, Raizer JJ, Wen PY, Phase I study of panobinostat in combination with bevacizumab for recurrent high-grade glioma, J Neurooncol. 107 (2012) 133–138. 10.1007/s11060-011-0717-z. [DOI] [PubMed] [Google Scholar]
- [122].Cuneo KC, Vredenburgh JJ, Sampson JH, Reardon DA, Desjardins A, Peters KB, Friedman HS, Willett CG, Kirkpatrick JP, Safety and efficacy of stereotactic radiosurgery and adjuvant bevacizumab in patients with recurrent malignant gliomas, Int J Radiat Oncol Biol Phys. 82 (2012) 2018–2024. 10.1016/j.ijrobp.2010.12.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Taal W, Oosterkamp HM, Walenkamp AME, Dubbink HJ, Beerepoot LV, Hanse MCJ, Buter J, Honkoop AH, Boerman D, de Vos FYF, Dinjens WNM, Enting RH, Taphoorn MJB, van den Berkmortel FWPJ, Jansen RLH, Brandsma D, Bromberg JEC, van Heuvel I, Vernhout RM, van der Holt B, van den Bent MJ, Single-agent bevacizumab or lomustine versus a combination of bevacizumab plus lomustine in patients with recurrent glioblastoma (BELOB trial): a randomised controlled phase 2 trial, The Lancet Oncology. 15 (2014) 943–953. 10.1016/S1470-2045(14)70314-6. [DOI] [PubMed] [Google Scholar]
- [124].Wick W, Gorlia T, Bendszus M, Taphoorn M, Sahm F, Harting I, Brandes AA, Taal W, Domont J, Idbaih A, Campone M, Clement PM, Stupp R, Fabbro M, Le Rhun E, Dubois F, Weller M, von Deimling A, Golfinopoulos V, Bromberg JC, Platten M, Klein M, van den Bent MJ, Lomustine and Bevacizumab in Progressive Glioblastoma, N. Engl. J. Med 377 (2017) 1954–1963. 10.1056/NEJMoa1707358. [DOI] [PubMed] [Google Scholar]
- [125].Reardon DA, Vredenburgh JJ, Desjardins A, Peters K, Gururangan S, Sampson JH, Marcello J, Herndon JE, McLendon RE, Janney D, Friedman AH, Bigner DD, Friedman HS, Effect of CYP3A-inducing anti-epileptics on sorafenib exposure: results of a phase II study of sorafenib plus daily temozolomide in adults with recurrent glioblastoma, J Neurooncol. 101 (2011) 57–66. 10.1007/s11060-010-0217-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Pan E, Yu D, Yue B, Potthast L, Chowdhary S, Smith P, Chamberlain M, A prospective phase II single-institution trial of sunitinib for recurrent malignant glioma, J Neurooncol. 110 (2012) 111–118. 10.1007/s11060-012-0943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Gerstner ER, Eichler AF, Plotkin SR, Drappatz J, Doyle CL, Xu L, Duda DG, Wen PY, Jain RK, Batchelor TT, Phase I trial with biomarker studies of vatalanib (PTK787) in patients with newly diagnosed glioblastoma treated with enzyme inducing anti-epileptic drugs and standard radiation and temozolomide, J Neurooncol. 103 (2011) 325–332. 10.1007/s11060-010-0390-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Brandes AA, Stupp R, Hau P, Lacombe D, Gorlia T, Tosoni A, Mirimanoff RO, Kros JM, van den Bent MJ, EORTC study 26041-22041: phase I/II study on concomitant and adjuvant temozolomide (TMZ) and radiotherapy (RT) with PTK787/ZK222584 (PTK/ZK) in newly diagnosed glioblastoma, Eur J Cancer. 46 (2010) 348–354. 10.1016/j.ejca.2009.10.029. [DOI] [PubMed] [Google Scholar]
- [129].Hainsworth JD, Ervin T, Friedman E, Priego V, Murphy PB, Clark BL, Lamar RE, Concurrent radiotherapy and temozolomide followed by temozolomide and sorafenib in the first-line treatment of patients with glioblastoma multiforme, Cancer. 116 (2010) 3663–3669. 10.1002/cncr.25275. [DOI] [PubMed] [Google Scholar]
- [130].Kreisl TN, Smith P, Sul J, Salgado C, Iwamoto FM, Shih JH, Fine HA, Continuous daily sunitinib for recurrent glioblastoma, J Neurooncol. 111 (2013) 41–48. 10.1007/s11060-012-0988-z. [DOI] [PubMed] [Google Scholar]
- [131].Drappatz J, Norden AD, Wong ET, Doherty LM, Lafrankie DC, Ciampa A, Kesari S, Sceppa C, Gerard M, Phan P, Schiff D, Batchelor TT, Ligon KL, Young G, Muzikansky A, Weiss SE, Wen PY, Phase I study of vandetanib with radiotherapy and temozolomide for newly diagnosed glioblastoma, Int J Radiat Oncol Biol Phys. 78 (2010) 85–90. 10.1016/j.ijrobp.2009.07.1741. [DOI] [PubMed] [Google Scholar]
- [132].Reardon DA, Egorin MJ, Desjardins A, Vredenburgh JJ, Beumer JH, Lagattuta TF, Gururangan S, Herndon JE, Salvado AJ, Friedman HS, Phase I pharmacokinetic study of the vascular endothelial growth factor receptor tyrosine kinase inhibitor vatalanib (PTK787) plus imatinib and hydroxyurea for malignant glioma, Cancer. 115 (2009) 2188–2198. 10.1002/cncr.24213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Reardon DA, Vredenburgh JJ, Coan A, Desjardins A, Peters KB, Gururangan S, Sathornsumetee S, Rich JN, Herndon JE, Friedman HS, Phase I study of sunitinib and irinotecan for patients with recurrent malignant glioma, J Neurooncol. 105 (2011) 621–627. 10.1007/s11060-011-0631-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Reardon DA, Groves MD, Wen PY, Nabors L, Mikkelsen T, Rosenfeld S, Raizer J, Barriuso J, McLendon RE, Suttle AB, Ma B, Curtis CM, Dar MM, de Bono J, A Phase I/II Trial of Pazopanib in Combination with Lapatinib in Adult Patients with Relapsed Malignant Glioma, Clin Cancer Res. 19 (2013) 900–908. 10.1158/1078-0432.CCR-12-1707. [DOI] [PubMed] [Google Scholar]
- [135].Muhic A, Poulsen HS, Sorensen M, Grunnet K, Lassen U, Phase II open-label study of nintedanib in patients with recurrent glioblastoma multiforme, J Neurooncol. 111 (2013) 205–212. 10.1007/s11060-012-1009-y. [DOI] [PubMed] [Google Scholar]
- [136].Scherer H, Structural development in gliomas, Am J Cancer. 34 (1938) 333–351. [Google Scholar]
- [137].Gillespie S, Monje M, An active role for neurons in glioma progression: making sense of Scherer’s structures, Neuro Oncol. 20 (2018) 1292–1299. 10.1093/neuonc/noy083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Venkatesh HS, Johung TB, Caretti V, Noll A, Tang Y, Nagaraja S, Gibson EM, Mount CW, Polepalli J, Mitra SS, Woo PJ, Malenka RC, Vogel H, Bredel M, Mallick P, Monje M, Neuronal Activity Promotes Glioma Growth through Neuroligin-3 Secretion, Cell. 161 (2015) 803–816. 10.1016/j.cell.2015.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Venkatesh HS, Tam LT, Woo PJ, Lennon J, Nagaraja S, Gillespie SM, Ni J, Duveau DY, Morris PJ, Zhao JJ, Thomas CJ, Monje M, Targeting neuronal activity-regulated neuroligin-3 dependency in high-grade glioma, Nature. 549 (2017) 533–537. 10.1038/nature24014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Walker AJ, Card T, Bates TE, Muir K, Tricyclic antidepressants and the incidence of certain cancers: a study using the GPRD, Br J Cancer. 104 (2011) 193–197. 10.1038/sj.bjc.6605996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Dolma S, Selvadurai HJ, Lan X, Lee L, Kushida M, Voisin V, Whetstone H, So M, Aviv T, Park N, Zhu X, Xu C, Head R, Rowland KJ, Bernstein M, Clarke ID, Bader G, Harrington L, Brumell JH, Tyers M, Dirks PB, Inhibition of Dopamine Receptor D4 Impedes Autophagic Flux, Proliferation, and Survival of Glioblastoma Stem Cells, Cancer Cell. 29 (2016) 859–873. 10.1016/j.ccell.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Li J, Zhu S, Kozono D, Ng K, Futalan D, Shen Y, Akers JC, Steed T, Kushwaha D, Schlabach M, Carter BS, Kwon C-H, Furnari F, Cavenee W, Elledge S, Chen CC, Genome-wide shRNA screen revealed integrated mitogenic signaling between dopamine receptor D2 (DRD2) and epidermal growth factor receptor (EGFR) in glioblastoma, Oncotarget. 5 (2014) 882–893. 10.18632/oncotarget.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Mahé C, Bernhard M, Bobirnac I, Keser C, Loetscher E, Feuerbach D, Dev KK, Schoeffter P, Functional expression of the serotonin 5-HT7 receptor in human glioblastoma cell lines, Br J Pharmacol. 143 (2004) 404–410. 10.1038/sj.bjp.0705936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Labrakakis C, Patt S, Hartmann J, Kettenmann H, Functional GABA(A) receptors on human glioma cells, Eur J Neurosci. 10 (1998) 231–238. 10.1046/j.1460-9568.1998.00036.x. [DOI] [PubMed] [Google Scholar]
- [145].Gallo V, Patneau DK, Mayer ML, Vaccarino FM, Excitatory amino acid receptors in glial progenitor cells: molecular and functional properties, Glia. 11 (1994) 94–101. 10.1002/glia.440110204. [DOI] [PubMed] [Google Scholar]
- [146].Venkataramani V, Tanev DI, Strahle C, Studier-Fischer A, Fankhauser L, Kessler T, Körber C, Kardorff M, Ratliff M, Xie R, Horstmann H, Messer M, Paik SP, Knabbe J, Sahm F, Kurz FT, Acikgöz AA, Herrmannsdörfer F, Agarwal A, Bergles DE, Chalmers A, Miletic H, Turcan S, Mawrin C, Hänggi D, Liu H-K, Wick W, Winkler F, Kuner T, Glutamatergic synaptic input to glioma cells drives brain tumour progression, Nature. 573 (2019) 532–538. 10.1038/s41586-019-1564-x. [DOI] [PubMed] [Google Scholar]
- [147].Venkatesh HS, Morishita W, Geraghty AC, Silverbush D, Gillespie SM, Arzt M, Tam LT, Espenel C, Ponnuswami A, Ni L, Woo PJ, Taylor KR, Agarwal A, Regev A, Brang D, Vogel H, Hervey-Jumper S, Bergles DE, Suvà ML, Malenka RC, Monje M, Electrical and synaptic integration of glioma into neural circuits, Nature. 573 (2019) 539–545. 10.1038/s41586-019-1563-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Zeng Q, Michael IP, Zhang P, Saghafinia S, Knott G, Jiao W, McCabe BD, Galván JA, Robinson HPC, Zlobec I, Ciriello G, Hanahan D, Synaptic proximity enables NMDAR signalling to promote brain metastasis, Nature. 573 (2019) 526–531. 10.1038/s41586-019-1576-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Zhang H, Zhou Y, Cui B, Liu Z, Shen H, Novel insights into astrocyte-mediated signaling of proliferation, invasion and tumor immune microenvironment in glioblastoma, Biomed Pharmacother. 126 (2020) 110086. 10.1016/j.biopha.2020.110086. [DOI] [PubMed] [Google Scholar]
- [150].Henrik Heiland D, Ravi VM, Behringer SP, Frenking JH, Wurm J, Joseph K, Garrelfs NWC, Strähle J, Heynckes S, Grauvogel J, Franco P, Mader I, Schneider M, Potthoff A-L, Delev D, Hofmann UG, Fung C, Beck J, Sankowski R, Prinz M, Schnell O, Tumor-associated reactive astrocytes aid the evolution of immunosuppressive environment in glioblastoma, Nature Communications. 10 (2019) 2541. 10.1038/s41467-019-10493-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Edwards LA, Woolard K, Son MJ, Li A, Lee J, Ene C, Mantey SA, Maric D, Song H, Belova G, Jensen RT, Zhang W, Fine HA, Effect of Brain- and Tumor-Derived Connective Tissue Growth Factor on Glioma Invasion, J Natl Cancer Inst. 103 (2011) 1162–1178. 10.1093/jnci/djr224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Louveau A, Smirnov I, Keyes TJ, Eccles JD, Rouhani SJ, Peske JD, Derecki NC, Castle D, Mandell JW, Lee KS, Harris TH, Kipnis J, Structural and functional features of central nervous system lymphatic vessels, Nature. 523 (2015) 337–341. 10.1038/nature14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, Akerley W, van den Eertwegh AJM, Lutzky J, Lorigan P, Vaubel JM, Linette GP, Hogg D, Ottensmeier CH, Lebbé C, Peschel C, Quirt I, Clark JI, Wolchok JD, Weber JS, Tian J, Yellin MJ, Nichol GM, Hoos A, Urba WJ, Improved survival with ipilimumab in patients with metastatic melanoma, N. Engl. J. Med 363 (2010) 711–723. 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe S, O’Brien M, Rao S, Hotta K, Leiby MA, Lubiniecki GM, Shentu Y, Rangwala R, Brahmer JR, KEYNOTE-024 Investigators, Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer, N. Engl. J. Med 375 (2016) 1823–1833. 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- [155].Reardon DA, Brandes AA, Omuro A, Mulholland P, Lim M, Wick A, Baehring J, Ahluwalia MS, Roth P, Bähr O, Phuphanich S, Sepulveda JM, De Souza P, Sahebjam S, Carleton M, Tatsuoka K, Taitt C, Zwirtes R, Sampson J, Weller M, Effect of Nivolumab vs Bevacizumab in Patients With Recurrent Glioblastoma: The CheckMate 143 Phase 3 Randomized Clinical Trial, JAMA Oncol. 6 (2020) 1003–1010. 10.1001/jamaoncol.2020.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA, Robust enumeration of cell subsets from tissue expression profiles, Nat Methods. 12 (2015) 453–457. 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Martinez-Lage M, Lynch TM, Bi Y, Cocito C, Way GP, Pal S, Haller J, Yan RE, Ziober A, Nguyen A, Kandpal M, O’Rourke DM, Greenfield JP, Greene CS, Davuluri RV, Dahmane N, Immune landscapes associated with different glioblastoma molecular subtypes, Acta Neuropathol Commun. 7 (2019). 10.1186/s40478-019-0803-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Close HJ, Stead LF, Nsengimana J, Reilly KA, Droop A, Wurdak H, Mathew RK, Corns R, Newton-Bishop J, Melcher AA, Short SC, Cook GP, Wilson EB, Expression profiling of single cells and patient cohorts identifies multiple immunosuppressive pathways and an altered NK cell phenotype in glioblastoma, Clin Exp Immunol. 200 (2020) 33–44. 10.1111/cei.13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Mohme M, Schliffke S, Maire CL, Rünger A, Glau L, Mende KC, Matschke J, Gehbauer C, Akyüz N, Zapf S, Holz M, Schaper M, Martens T, Schmidt NO, Peine S, Westphal M, Binder M, Tolosa E, Lamszus K, Immunophenotyping of Newly Diagnosed and Recurrent Glioblastoma Defines Distinct Immune Exhaustion Profiles in Peripheral and Tumor-infiltrating Lymphocytes, Clin Cancer Res. 24 (2018) 4187–4200. 10.1158/1078-0432.CCR-17-2617. [DOI] [PubMed] [Google Scholar]
- [160].Otvos B, Silver DJ, Mulkearns-Hubert EE, Alvarado AG, Turaga SM, Sorensen MD, Rayman P, Flavahan WA, Hale JS, Stoltz K, Sinyuk M, Wu Q, Jarrar A, Kim S-H, Fox PL, Nakano I, Rich JN, Ransohoff RM, Finke J, Kristensen BW, Vogelbaum MA, Lathia JD, Cancer Stem Cell-Secreted Macrophage Migration Inhibitory Factor Stimulates Myeloid Derived Suppressor Cell Function and Facilitates Glioblastoma Immune Evasion, Stem Cells. 34 (2016) 2026–2039. 10.1002/stem.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Couto M, Coelho-Santos V, Santos L, Fontes-Ribeiro C, Silva AP, Gomes CMF, The interplay between glioblastoma and microglia cells leads to endothelial cell monolayer dysfunction via the interleukin-6-induced JAK2/STAT3 pathway, J Cell Physiol. 234 (2019) 19750–19760. 10.1002/jcp.28575. [DOI] [PubMed] [Google Scholar]
- [162].Kuppner MC, Hamou MF, Sawamura Y, Bodmer S, de Tribolet N, Inhibition of lymphocyte function by glioblastoma-derived transforming growth factor beta 2, J Neurosurg. 71 (1989) 211–217. 10.3171/jns.1989.71.2.0211. [DOI] [PubMed] [Google Scholar]
- [163].Bloch O, Crane CA, Kaur R, Safaee M, Rutkowski MJ, Parsa AT, Gliomas promote immunosuppression through induction of B7-H1 expression in tumor-associated macrophages, Clin Cancer Res. 19 (2013) 3165–3175. 10.1158/1078-0432.CCR-12-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Parsa AT, Waldron JS, Panner A, Crane CA, Parney IF, Barry JJ, Cachola KE, Murray JC, Tihan T, Jensen MC, Mischel PS, Stokoe D, Pieper RO, Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma, Nat Med. 13 (2007) 84–88. 10.1038/nm1517. [DOI] [PubMed] [Google Scholar]
- [165].Wischhusen J, Friese MA, Mittelbronn M, Meyermann R, Weller M, HLA-E protects glioma cells from NKG2D-mediated immune responses in vitro: implications for immune escape in vivo, J Neuropathol Exp Neurol. 64 (2005) 523–528. 10.1093/jnen/64.6.523. [DOI] [PubMed] [Google Scholar]
- [166].Didenko VV, Ngo HN, Minchew C, Baskin DS, Apoptosis of T lymphocytes invading glioblastomas multiforme: a possible tumor defense mechanism, J Neurosurg. 96 (2002) 580–584. 10.3171/jns.2002.96.3.0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Dusoswa SA, Verhoeff J, Abels E, Méndez-Huergo SP, Croci DO, Kuijper LH, de Miguel E, Wouters VMCJ, Best MG, Rodriguez E, Cornelissen LAM, van Vliet SJ, Wesseling P, Breakefield XO, Noske DP, Würdinger T, Broekman MLD, Rabinovich GA, van Kooyk Y, Garcia-Vallejo JJ, Glioblastomas exploit truncated O-linked glycans for local and distant immune modulation via the macrophage galactose-type lectin, Proc Natl Acad Sci U S A. 117 (2020) 3693–3703. 10.1073/pnas.1907921117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Alban TJ, Bayik D, Otvos B, Rabljenovic A, Leng L, Jia-Shiun L, Roversi G, Lauko A, Momin AA, Mohammadi AM, Peereboom DM, Ahluwalia MS, Matsuda K, Yun K, Bucala R, Vogelbaum MA, Lathia JD, Glioblastoma Myeloid-Derived Suppressor Cell Subsets Express Differential Macrophage Migration Inhibitory Factor Receptor Profiles That Can Be Targeted to Reduce Immune Suppression, Front. Immunol 11 (2020). 10.3389/fimmu.2020.01191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Peereboom DM, Alban TJ, Grabowski MM, Alvarado AG, Otvos B, Bayik D, Roversi G, McGraw M, Huang P, Mohammadi AM, Kornblum HI, Radivoyevitch T, Ahluwalia MS, Vogelbaum MA, Lathia JD, Metronomic capecitabine as an immune modulator in glioblastoma patients reduces myeloid-derived suppressor cells, JCI Insight. 4 (2019). 10.1172/jci.insight.130748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Bayik D, Zhou Y, Park C, Hong C, Vail D, Silver DJ, Lauko A, Roversi G, Watson DC, Lo A, Alban TJ, McGraw M, Sorensen M, Grabowski MM, Otvos B, Vogelbaum MA, Horbinski C, Kristensen BW, Khalil AM, Hwang TH, Ahluwalia MS, Cheng F, Lathia JD, Myeloid-derived suppressor cell subsets drive glioblastoma growth in a sex-specific manner, Cancer Discov. (2020). 10.1158/2159-8290.CD-19-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Kees T, Lohr J, Noack J, Mora R, Gdynia G, Tödt G, Ernst A, Radlwimmer B, Falk CS, Herold-Mende C, Régnier-Vigouroux A, Microglia isolated from patients with glioma gain antitumor activities on poly (I:C) stimulation, Neuro Oncol. 14 (2012) 64–78. 10.1093/neuonc/nor182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Li W, Graeber MB, The molecular profile of microglia under the influence of glioma, Neuro Oncol. 14 (2012) 958–978. 10.1093/neuonc/nos116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Coniglio SJ, Eugenin E, Dobrenis K, Stanley ER, West BL, Symons MH, Segall JE, Microglial stimulation of glioblastoma invasion involves epidermal growth factor receptor (EGFR) and colony stimulating factor 1 receptor (CSF-1R) signaling, Mol Med. 18 (2012) 519–527. 10.2119/molmed.2011.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Markovic DS, Vinnakota K, Chirasani S, Synowitz M, Raguet H, Stock K, Sliwa M, Lehmann S, Kälin R, van Rooijen N, Holmbeck K, Heppner FL, Kiwit J, Matyash V, Lehnardt S, Kaminska B, Glass R, Kettenmann H, Gliomas induce and exploit microglial MT1-MMP expression for tumor expansion, Proc Natl Acad Sci U S A. 106 (2009) 12530–12535. 10.1073/pnas.0804273106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Willingham SB, Volkmer J-P, Gentles AJ, Sahoo D, Dalerba P, Mitra SS, Wang J, Contreras-Trujillo H, Martin R, Cohen JD, Lovelace P, Scheeren FA, Chao MP, Weiskopf K, Tang C, Volkmer AK, Naik TJ, Storm TA, Mosley AR, Edris B, Schmid SM, Sun CK, Chua M-S, Murillo O, Rajendran P, Cha AC, Chin RK, Kim D, Adorno M, Raveh T, Tseng D, Jaiswal S, Enger PØ, Steinberg GK, Li G, So SK, Majeti R, Harsh GR, van de Rijn M, Teng NNH, Sunwoo JB, Alizadeh AA, Clarke MF, Weissman IL, The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors, Proc Natl Acad Sci U S A. 109 (2012) 6662–6667. 10.1073/pnas.1121623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Turaga SM, Silver DJ, Bayik D, Paouri E, Peng S, Lauko A, Alban TJ, Borjini N, Stanko S, Naik U, Keri RA, Connor JR, Barnholtz-Sloan JS, Rubin JB, Berens M, Davalos D, Lathia JD, JAM-A functions as a female microglial tumor suppressor in glioblastoma, Neuro Oncol. (n.d.). 10.1093/neuonc/noaa148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Yang W, Warrington NM, Taylor SJ, Whitmire P, Carrasco E, Singleton KW, Wu N, Lathia JD, Berens ME, Kim AH, Barnholtz-Sloan JS, Swanson KR, Luo J, Rubin JB, Sex differences in GBM revealed by analysis of patient imaging, transcriptome, and survival data, Science Translational Medicine. 11 (2019). 10.1126/scitranslmed.aao5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Song E, Mao T, Dong H, Boisserand LSB, Antila S, Bosenberg M, Alitalo K, Thomas J-L, Iwasaki A, VEGF-C-driven lymphatic drainage enables immunosurveillance of brain tumours, Nature. 577 (2020) 689–694. 10.1038/s41586-019-1912-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Lee-Chang C, Miska J, Hou D, Rashidi A, Zhang P, Burga RA, Jusué-Torres I, Xiao T, Arrieta VA, Zhang DY, Lopez-Rosas A, Han Y, Sonabend AM, Horbinski CM, Stupp R, Balyasnikova IV, Lesniak MS, Activation of 4-1BBL+ B cells with CD40 agonism and IFNγ elicits potent immunity against glioblastoma, J Exp Med. 218 (2021). 10.1084/jem.20200913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Daubon T, Hemadou A, Romero Garmendia I, Saleh M, Glioblastoma Immune Landscape and the Potential of New Immunotherapies, Front Immunol. 11 (2020) 585616. 10.3389/fimmu.2020.585616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Long GV, Atkinson V, Menzies AM, Lo S, Guminski AD, Brown MP, Gonzalez MM, Diamante K, Sandhu SK, Scolyer RA, Emmett L, McArthur GA, A randomized phase II study of nivolumab or nivolumab combined with ipilimumab in patients (pts) with melanoma brain metastases (mets): The Anti-PD1 Brain Collaboration (ABC)., JCO. 35 (2017) 9508–9508. 10.1200/JCO.2017.35.15_suppl.9508. [DOI] [Google Scholar]