Abstract
The production of the biofuel, isobutanol, in E. coli faces limitations due to alcohol toxicity, product inhibition, product recovery, and long-term industrial feasibility. Here we demonstrate an approach of combining both in vivo with in vitro metabolic engineering to produce isobutanol. The in vivo production of α-ketoisovalerate (KIV) was conducted through CRISPR mediated integration of the KIV pathway in bicistronic design (BCD) in E. coli and inhibition of competitive valine pathway using CRISPRi technology. The subsequent in vitro conversion to isobutanol was carried out with engineered enzymes for 2-ketoacid decarboxylase (KIVD) and alcohol dehydrogenase (ADH). For the in vivo production of KIV and subsequent in vitro production of isobutanol, this two-step serial approach resulted in yields of 56% and 93%, productivities of 0.62 and 0.074 g L−1 h−1, and titers of 5.6 and 1.78 g L−1, respectively. Thus, this combined biosynthetic system can be used as a modular approach for producing important metabolites, like isobutanol, without the limitations associated with in vivo production using a consolidated bioprocess.
Keywords: α-Ketoisovalerate, Isobutanol, CRISPR, CRISPRi, Bicistronic, Fed-batch
Highlights
-
•
The α-ketoisovalerate (KIV) pathway was inserted into E. coli using CRISPR.
-
•
An in vivo to in vitro system was developed to produce biofuel.
-
•
5.6 g L−1 of KIV and 1.78 g L−1 of isobutanol were produced using this coupled system.
1. Introduction
The inevitable depletion of non-renewable resources and increasing concerns about climate change create an urgent need to develop energy sources independent of fossil fuels. Biofuel production by microorganisms from renewable resources is an efficient approach to clean energy (Olcay et al., 2018). While ethanol is the most common in vivo produced biofuel, butanol is being heralded as the next generation of biofuels. Compared with ethanol, butanol has a higher energy density and is non-hygroscopic and non-corrosive, properties that make it compatible with current pumps and engines (Durre, 2007; Jawed et al., 2020).
Butanol has several isomers, with isobutanol being the most promising with respect to yield and titer (Blombach and Eikmanns, 2011). In vivo production of isobutanol is limited due to its toxicity on the strain producing it (Chen and Liao, 2016; Dong et al., 2016). Even at concentrations as low as 1% (v/v), cells have shown signs of growth inhibition (Grimaldi et al., 2016a). Attempts have been made in the past to produce isobutanol by engineering E. coli, Bacillus subtilis and Corynebacterium glutamicum using a common strategy of expressing genes encoding KIVD from Lactococcus lactis and ADH from either Saccharomyces cerevisiae, C. glutamicum, E. coli or L. lactis (encoded by adh2, adhA, yqhD and adhA, respectively), (Atsumi et al., 2008a, 2010a; Li et al., 2011; Smith et al., 2010; Blombach et al., 2011). α-Ketoisovalerate (KIV), an intermediate of the valine pathway, is a longer-chain 2-keto acid and precursor in amino acid biosynthesis; it can be converted to a variety of compounds, including isobutanol, and also has applications as a therapeutic agent (Aparicio et al., 2012).
In one example, a significant biological isobutanol production was demonstrated in E. coli using a non-fermentative pathway by diverting KIV towards higher chain alcohols (Atsumi et al., 2008a).However, this strain faced toxicity issues, hence an isobutanol-tolerant strain (SA481) was evolved that could tolerate higher concentrations of isobutanol. Nevertheless, the improved tolerance did not result in increased production rate (Atsumi et al., 2010b). Moreover, several transcriptional analyses were performed previously to study the stress caused by different alcohols including ethanol, n‐butanol, and isobutanol for E. coli (Gonzalez et al., 2003; Brynildsen and Liao, 2009; Rutherford et al., 2010). Brynildsen and Liao demonstrated that isobutanol stress disrupts quinone–membrane interactions, which lead to respiratory distress and the activation of arcA, fur, and phoB (Brynildsen and Liao, 2009). In a recent study, B. megaterium SR7 was engineered as a bio-production host under supercritical CO2 for isobutanol production using KIV as an externally fed substrate, obtaining 0.33 g L−1 titer (Boock et al., 2019).
While there has been limited success with increasing isobutanol tolerance in vivo, there are still issues with product inhibition, product removal, and long-term industrial feasibility. In vitro production has been proposed as a solution to these in vivo problems (Grimaldi et al., 2016a). By selecting pathway enzymes from cells, the toxicity effects of isobutanol can be bypassed. Product yield is increased as there is no longer competition for metabolites. Recovery is simpler as isobutanol is directly added to solution, instead of being trapped in a cellular membrane. Industrially, cellular contamination is avoided in a cell-free system (Dudley et al., 2015). A disadvantage of this approach is the increasing complexity of the cell-free system for longer pathways.
To avoid this problem and provide a coherent process from substrate to product, we describe an approach combining in vivo production of KIV, using a genetically engineered E. coli, and in vitro conversion to isobutanol using enzymes KIVD and ADH. The KIV synthesis pathway in BCD form was integrated into the E. coli genome and competitive valine pathway was downregulated using CRISPRi technology.
2. Materials and methods
2.1. Reagents
All enzymes were purchased from New England Biolabs (NEB). All oligonucleotide synthesis and DNA sequencing were performed by Genewiz, South Plainfield, NJ 07080. All chemicals and Liquid Chromatography and Mass Spectrometry (LCMS) standards for KIV were purchased from Sigma-Aldrich. All oligonucleotides were purchased from Integrated DNA Technologies, USA. β-nicotinamide adenine dinucleotide (NADH) was purchased from OYC Americas (Vista, CA). Sodium formate was purchased from VWR (Radnor, PA).
2.2. Strains and plasmids
JCL260 (BW25113/F′ traD36, proAB+, lacIq ZΔM15 ΔadhE, ΔfrdBC, Δfnr-ldhA, Δpta, ΔpflB) and vector pLS02 with the gene coding for KIVD from Lactococcus lactis were a gift from Professor James Liao (Atsumi et al., 2008b; Soh et al., 2017). JCL 260 was used as the control strain. Plasmids pX2-Cas9 (#85811), SS9gRNA (#71656), and pKDsgRNA-ack (#62654) were purchased from Addgene. Temperature-sensitive plasmid pSIM5 plasmid was a gift from the Court lab (Datta et al., 2006). Plasmid pdCas9 was a gift from Luciano Marraffini. The genes encoding for Alcohol dehydrogenase (ADH) and Formate dehydrogenase (FDH) were synthesized using gBlock gene fragments purchased from Integrated DNA Technologies (Coralville, Iowa). Plasmid pGS-21a was used as IPTG inducible expression vector (GenScript Co.) E. coli DH5α was used as host for cloning and E. coli BL21 Star (DE3) as host for plasmid expression. All the primers used in this study are listed in Supplementary Table 1.
2.3. Culture conditions
The working culture was maintained on Luria-Bertani (LB) Agar Plates supplemented with appropriate antibiotics. Antibiotic concentrations were as follows: ampicillin (Amp 100 μg mL−1) and tetracycline (Tet 15 μg mL−1) from Fisher BioReagents, spectinomycin (Spec 50 μg mL−1), chloramphenicol (Cm 25 μg mL−1 and 50 μg mL−1), kanamycin (Kan 50 μg mL−1) and streptomycin (Strep 100 μg mL−1) from Sigma Aldrich. Overnight cultures were grown in 5 ml LB Broth containing appropriate antibiotics. 1% of the overnight culture was inoculated into 10 mL of the above modified M9 medium (6 g Na2HPO4, 3 g KH2PO4, 1 g NH4Cl, 0.5 g NaCl, 1 mM MgSO4, 1 mM CaCl2, 10 mg vitamin B1 per liter of solution) with 20 g L−1 glucose, 5 g L−1 yeast extract, 15 μg ml−1 Tet and 1,000th dilution of Trace Metal Mix A5 (2.86 g H3BO3, 1.81 g MnCl2⋅4H2O, 0.222 g ZnSO4⋅7H2O, 0.39 g Na2MoO4⋅2H2O, 0.079 g CuSO4⋅5H2O, 49.4 mg Co (NO3)2⋅6H2O per liter solution) and pH was maintained at 6.8.
2.4. Genomic integration of KIV pathway using CRISPR-Cas9
All the integration and deletion experiments were performed in JCL 260. Linear template of 5.8 kb containing KIV pathway in BCD (Mutalik et al., 2013) with constitutive promoters was integrated into the E. coli genome using Lambda Red Recombineering and Selections (Bassalo et al., 2016).The homology arms corresponding to the SS9 site were added to the repair template using end primers MG1 and MG2 respectively. E. coli JCL260 was previously transformed with the pX2-Cas9 (arabinose inducible) and pSIM5 (temperature inducible) vectors. The cells were treated with 42 °C prior to co-transformation of SS9gRNA and repair template. Transformed strains were recovered in LB supplemented with 0.02 g L−1 arabinose for 3h at 37 °C and then plated in selective media for the gRNA vector (Amp100) and pX2-Cas9 (Kan50).
2.5. Curing SS9gRNA and pX2Cas9 plasmids
SS9gRNA and pX2Cas9 plasmids were cured by inserting plasmid pKDsgRNA-ack (Addgene plasmid #62654) (Reisch and Prather, 2015). Spacers were designed to cut specifically at SS9gRNA and pX2Cas9 without any off-target effect on E. coli genome (Supplementary Fig. 2a). pKDsgRNA-ack was amplified with primer set 1 (px2Cas9_sense/pKDsg_F and pX2Cas9_Antisense/pKDsg_R) for inserting pX2Cas9 spacer and primer set 2 (SS9gRNA_sense/pKDsg_F and SS9gRNA_Antisense/pKDsg_R) for SS9gRNA spacer (Supplementary Fig. 2b). The linear dsDNA products were subsequently Dpn I digested for at least 15 min and then gel-purified. The purified products carrying homology arms were assembled using NEB Gibbson master mix (Gibson et al., 2009) to create pKDsgSS9-ack and pKDsgpX2cas9-ack circular plasmids. The product was then used to directly transform chemically competent E. coli DH5α or NEB Turbo cells and recovered on LB Agar supplemented with Spec50. The insertion of the spacer was confirmed by sequencing using primer sgRNA A (Supplementary Fig. 2a). For curing SS9gRNA, pKDsgSS9-ack was transferred in JCL260 competent cells having SS9gRNA and pX2Cas9 plasmids. Cultures were induced with arabinose (0.2%) and recovered on LB agar supplemented with anhydrotetracycline (aTc 100 ng mL−1), plates with and without Amp80. The process was repeated for curing pX2Cas9 plasmid.
2.6. CRISPRi mediated repression of ilvE gene
Cognate CRISPR-deactivated Cas9 repressor plasmids (containing dCas9, tracrRNA, and a single spacer CRISPR array) for ilvE gene were constructed at two positions (i) inside the promoter region dCas9IlvE(p) and (ii) at ORF dCas9IlvE(g) using Golden Gate assembly as described before (Sanjana et al., 2012). Two complementary and slightly offset oligonucleotides (IDT) containing the spacer sequence for dCas9 targeting were phosphorylated with Polynucleotide Kinase (PNK from NEB) and annealed (37 °C for 30 min, 98 °C for 5 min, ramp down to 25 °C over 15 min) to build inserts for each repressor variant. All primers and oligonucleotides used in this work are listed in Supplementary Table 1. Phosphorylated and annealed inserts were individually cloned into recipient plasmid pdCas9 (Bikard et al., 2013) at two adjacent Bsa I sites in the minimal, single-spacer CRISPR array using a one-pot Golden Gate reaction with Bsa I (New England BioLabs) and T7 ligase (Epicentre Biotechnologies). Ligated product was transformed to DH5α cells and recover with 250 μL SOC medium for 1 h at 37 °C. Colonies were screened by PCR using primers pdCas9-M_seqF and IlvEp_dcas9_Antisense or IlvEg_dcas9_Antisense. Plasmids were verified by digestion with restriction enzymes and Sanger sequencing. Next, plasmids were transferred to chemically competent JCL260 having genome integrated KIV pathway (MG01). Growth of ilvE downregulated strains at the ORF (MG02) and at promoter region (MG03) was compared with MG01. The overnight grown cultures in LB Broth were maintained at OD 1 and further diluted 100 times with modified minimal media as described before in culture conditions and were incubated at 225 rpm and 30 °C. Optical densities (OD) were measured at 600 nm with a Synergy H1 hybrid plate reader (BioTek Instruments, Inc.).
2.7. Fed-batch process conditions
Fed-batch fermentation was performed in a 1 L stirred bioreactor (Applikon). Cells from the glycerol stock were streaked on LB plate containing chloramphenicol and tetracycline. Single colony from the fresh plate was grown overnight in 5 mL LB medium containing respective antibiotics at 37 °C and 180 rpm. Secondary culture was prepared by inoculating 1% of primary culture to 300 mL of M9 medium with 5 g L−1 yeast extract supplemented with 2% glucose and grown at 37 °C until OD600 reached to 3.0. The grown secondary culture was used as seed culture to inoculate in a bioreactor with working volume of 3 L having M9 medium along with 5 g L−1 yeast extract and 2% glucose. Dissolved oxygen (DO) was maintained at 20% saturation and pH was maintained at 6.8 via the addition of 10% (v/v) NH4OH base using a PID (proportional, integral and differential) controller. After 6 h, intermittent linear feeding of glucose solution (500 g L−1) was initiated to maintain a glucose concentration between 20 and 10 g L−1. Samples were collected at different time interval for measuring cell density and metabolite concentration.
2.8. KIV minimum inhibitory concentration
The minimum inhibitory concentration of E. coli was measured utilizing a concentration assay where the change in OD600 was measured at set KIV concentrations after a set time. The concentration for which the change in OD600 would be 0 was extrapolated from the data (Supplementary Fig. 4).
2.9. Overexpression of KIV pathway
IPTG inducible plasmid pSA69 was a gift from Professor James Liao (Atsumi et al., 2010a). The plasmid was transformed into chemically competent MG03. This newly derived strain was compared to the base strain by utilizing the prior culture conditions in 2.3.
2.10. Plasmid construction for in vitro enzyme production
For converting KIV to isobutanol, the plasmids were constructed for in vitro enzyme production as previously described (Wong et al., 2019).
2.11. Enzyme expression and purification
The enzymes ADH, KIVD, and FDH were expressed and purified as previously described (Wong et al., 2019).
2.12. Kinetic and enzyme assays
The kinetic assays for examining the activity of the enzymes were carried out as follows. Enzyme concentration was measured using Bradford assay and then added to storage buffer. 200 μL of the enzyme mix was added to each well. For KIVD, ADH, and FDH, a concentration gradient of the reagents KIV, NADH, and NAD were used respectively. KIV and NADH have an absorbance peak at 340 nm, while NAD does not. Thus, a decrease of absorbance at 340 nm over time was measured for KIVD and ADH as reagent was consumed, while an increase in absorbance at 340 nm was measured for FDH as NADH was produced. ADH and FDH kinetic assays were conducted at room temperature, while the KIVD kinetic assay was conducted at 50 °C, while shaking, to facilitate a proper comparison with previously obtained results (Soh et al., 2017).Using the initial rate of reaction for each concentration step, the kinetic parameters kcat and Km were calculated using the Michaelis-Menten equation. Enzymatic assays were conducted as previously described (Wong et al., 2019). The in vitro reaction for the conversion of KIV to isobutanol was prepared as follows. Fermentation broth was centrifuged at 14,000 rpm at 25 °C for 10 min. Supernatant was set to a pH of 7.4 by addition of NaOH. The reaction volume was 1 mL. 800 μL of supernatant was added to a 200 μL mixture of enzyme together with 1 μmol of NADH and excess formate. The reaction tube was left shaking in an end-over-end mixer for 24 h at 35 °C. The tube was then frozen at −20 until analysis.
2.13. Analytical methods
Culture samples were harvested after 72 h and pelleted by centrifugation and supernatant was removed for KIV and other metabolite analysis. KIV analysis was done using LC-MS using standard curve from 0.1 g L−1 to 1 g L−1 (Supplementary Fig. 3B). Supernatant was mixed with internal standard salicylic acid (Li et al., 2016). LC analysis was performed by injecting 5 μL of sample to Agilent 1200 HPLC system with column Agilent Zorbax Eclipse XDB-C18 4.6 × 150 mm 5-μm. Solvent A: 0.2% formic acid, Solvent B: 0.2% formic acid in acetonitrile at a flow rate of 500 μL min−1, using gradient: 0–20% B (0–1.5 min), 20–40% B (1.5–1.7 min), 40-40% B (1.7–3.5 min), 40–65% B (3.5–5 min), 65.65% B (5.0–8 min), 65-20% (8–8.1 min) and 20% B (8.1–12 min). Mass spectrometry analysis was done using LTQ Orbitrap XL from Agilent Technologies (Santa Clara, CA, USA), Electrospray ionization, positive ion mode Resolution: 30,000 Mass accuracy 3 ppm.
Other metabolites such as glucose, succinic acid, lactic acid, acetic acid, methanol and isobutanol were detected using HPLC Agilent 1200 series instrument (Agilent) with a refractive index detector. Analytes were separated using the Aminex HPX-87H anion exchange column (Bio-Rad Laboratories) with a 5 mM sulfuric acid mobile phase at 35 °C and a flow rate of 0.8 ml min−1. Commercial standards were used for quantification of experimental samples by linear interpolation of external standard curves.
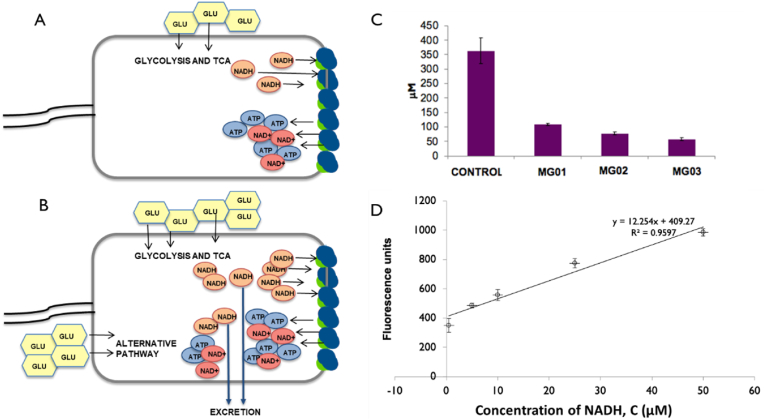
The NADH fluorescence of the extracellular culture media was measured in 1 cm quartz cuvettes (Starna, Australia) by fluorimetry equipped with a narrow band filter cut-off at 340 nm (NB340) for excitation and narrow band filter cut-off at 460 nm (NB440) for emission (Guilbault, 1973). Standards (between 5 μM and 50 μM) were prepared from analytical grade NADH (Sigma Diagnostics, USA). Arbitrary fluorescence units (AFU) were plotted as a function of NADH concentration from 0.5 to 50 μM.
3. Results
3.1. CRISPR Cas9 mediated extracellular production of KIV in shake flasks
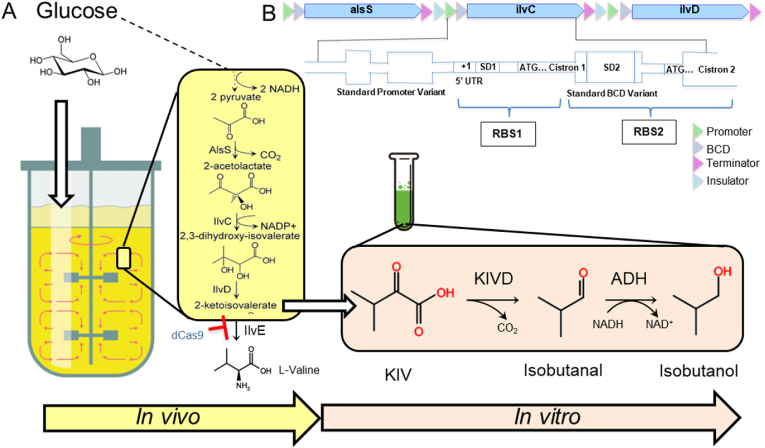
The 5.8 kb KIV pathway was integrated into the genome of E. coli JCL 260 at SS9 site using a 300 bp of homology arm to ensure library coverage (Bassalo et al., 2016). (Fig. 1). The biosynthetic pathway was constructed by cloning each gene under separate constitutive promoters and a bicistronic ribosomal binding site design (BCD). It contains a Ribosomal Binding site 1 (RBS1) motif that drives the translation of a short peptide. The short peptide's coding sequence (CDS) contained RBS2 that allows for translation initiation of the protein of interest. The stop codon of the peptide sequence overlaps with the start codon of the target CDS. This genetic architecture leads to the translational coupling of the short peptide to the protein of interest. The expression levels can be controlled with the sequence of the Shine Dalgarno and promoter sequences (Mutalik et al., 2013; Claassens et al., 2019).
Fig. 1.
Schematic illustration of isobutanol pathway (A) Pathway design for in vivo production of KIV in E. coli and in vitro production of Isobutanol in a tube. ALsS, acetolactate synthase from Bacillus subtilis; IlvC, 2-hydroxy-3-ketol-acid reductoisomerase from E. coli; IlvD, dihydroxy-acid hydratase from E. coli; KIVD, 2-keto acid decarboxylase; ADH, Alcohol dehydrogenase. (B) KIV pathway in bicistronic design (BCD) with its two Shine-Dalgarno motifs (SD1 and SD2) is shown. +1 is mRNA and 5′UTR is leader sequence. SD2 sites adhere to the BCD (Mutalik et al., 2013).
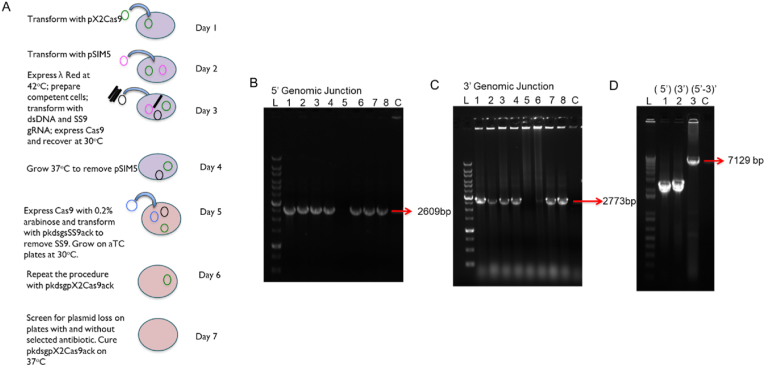
Transformed colonies were screened by colony PCR by amplification of different junctions (MG3/alsR and ilvdF/MG4) and showed 80% integration efficiency (Fig. 2BC). The complete pathway integration was further confirmed by sequencing as well as amplification of the genomic site with primers (MG3/MG4) flanking the integration region (Fig. 2D).
Fig. 2.
CRISPR Cas9 mediated integration of KIV pathway into E. coli genome (A) General outline of integration of KIV pathway and curing of plasmids (B) Screening of transformed colonies by PCR to verify upstream junction (5′) and (C) downstream junction 3’. L represents 10 kb ladder; expected sizes for correct integration are represented by arrows. PCR products are separated on 0.8% agarose gel. ‘C’ represents control (without integration) Pathway is showing 80% integration efficiency (D) PCR amplification using primers (MB67/MB68) flanking outside of the integrated region to confirm whole pathway integration 5′-3’ (lane 3).
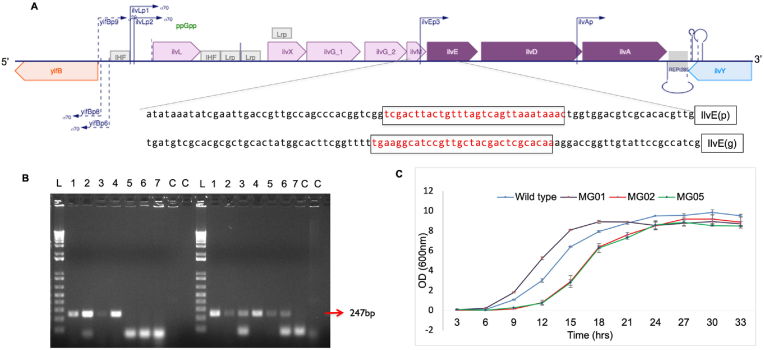
The integrated strain was successfully cured for SS9 and pX2Cas9 plasmids using pKDsgSS9ack and pKDsgpX2Cas9ack in order to overcome the extra metabolic burden (Supplementary Fig. 2AB). The resulting strain was tested for KIV production, but no significant production was observed. We hypothesized that KIV was not produced due to pathway competition for valine biosynthesis. To overcome this, the valine biosynthesis by using CRISPRi targeting the ilvE gene at two sites was inhibited: (i) in the promoter region, (ii) within the ORF of ilvE (Fig. 3AB). The downregulation was effective at both sites. Downregulation of the ilvE resulted in growth impairment when modified M9 medium was used as growth medium, a result that was expected due to the limited flux towards valine biosynthesis (Fig. 3C).
Fig. 3.
dCas9 mediated deregulation of ilvE gene (A) Schematic diagram of ilvE promoter from EcoCyc E. coli database showing spacer sequences within promoter IlvE (p) and gene region IlvE (g) designed for dCas9 targeting. (B) Colony PCR for screening of transformed colonies for successful integration of spacer regions into dCas9. L, represents 10 kb ladder; expected sizes for correct integration are represented by arrows. PCR products are separated on 1.5% agarose gel. ‘C’ represents control. (C) Effect of dCas9-mediated downregulation at the ORF using dcas9IlvE (g) primer (MG02) and promoter region using dcas9IlvE (p) primer (MG03) on the growth of E. coli having KIV pathway integrated. Data is replicate of three values. Error bars represent standard deviation.
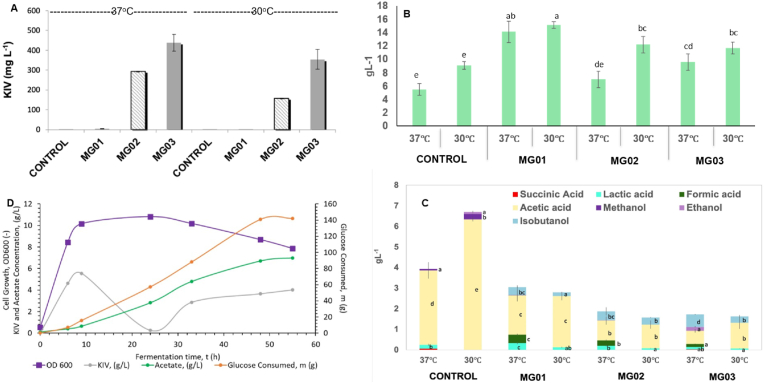
After inhibiting valine biosynthesis, the resulting strains were again tested for KIV production in shake flasks at two different temperatures (37 °C and 30 °C). The KIV peak was observed at RT 4.06 in ilvE downregulated strain during LCMS analysis (Supplementary Fig. 3A) The maximum KIV titer (400 mg L−1 KIV) in shake flasks was observed in the recombinant strain with CRISPRi targeting the promoter region of the ilvE gene (MG03) at 37 °C (Fig. 4A). In all cases, KIV production was higher at 37 °C compared to 30 °C (Fig. 4A). Glucose consumption was increased from 5.4 g L−1 to 14.1 g L−1 at 37 °C after chromosomal integration of the KIV pathway (MG01) but decreased to 6.9 g L−1 (MG02) after inhibiting valine biosynthesis (Fig. 4B). For the strain producing maximum KIV (MG03) the glucose consumption was only 9.6 g L-1 out of 20 g L−1 supplemented glucose.
Fig. 4.
2-Ketoisovalerate and other metabolites production (A) Graph showing KIV production in shake flasks at 30 °C and 37 °C before and after downregulation of ilvE gene. (B) Glucose consumption pattern of different strains at different temperatures in shake flasks. (C) Comparative metabolite profile of different metabolites in the culture supernatant of control and modified E. coli strains at two different temperatures (37 °C and 30 °C) as detected by HPLC. (D) KIV and acetate production pattern in glucose fed batch reactor at different growth stages of MG03. Values are means of three replications. Bar values (mean ± S.D) sharing same alphabet(s) do not differ significantly by LSD at P ≤ 0.05.
3.2. Analysis of other metabolites
After 72 h of growth, the supernatant was tested for the release of other metabolites e.g., succinic acid, lactic acid, formic acid, acetic acid, ethanol, methanol and isobutanol (Fig. 4C). The chromosomal integration of KIV pathway (MG01) led to decreased production of acetic acid (1.91 g L−1 at 37 °C), one of the main fermentation products from this E. coli host. It further decreased to 0.6 g L−1 and 0.9 g L−1 at 37 °C in strains MG03 and MG02 inhibited for valine biosynthesis. A small amount of methanol production (0.26 g L−1) was only observed in the control. However, a small amount of isobutanol (0.1–0.6 g L−1) and formic acid (0.1–0.4 g L−1) were detected in modified strains though these were absent in the control. No formic acid production was observed at 30 °C, whereas a small amount was detected at 37 °C.
3.3. Extracellular release of NADH
The NADH concentration was measured in the culture broth for both the control as well as the engineered strains. In the control, 0.363 mM NADH was found in the extracellular media that decreased to 0.108 mM after the integration of KIV pathway (MG01, Fig. 5C). The downregulation of valine pathway resulted in even lower concentration (0.057 mM) of NADH in the extracellular culture media (Fig. 5C).
Fig. 5.
NADH production in E. coli. (A) under slow growth rate and glucose limited condition sufficient electron transport chain exists to keep the NADH concentration low. (B) Under high growth rate electron transport chain surface limitation unable to metabolize all the NADH created, leading to NADH imbalance and excretion to outside the cell (Szenk et al., 2017) (C) NADH concentration in outer culture media in control and modified strains. (D) Standard curve showing NADH concentration with respect to fluorescence units. Values are means of three replicates. Error bars represents the standard deviation.
3.4. Fed-batch cultivation for high titer KIV production
In a 1L fed-batch reactor, cells grew until 9 h to an OD600 of 10, beyond which the culture entered stationary phase (Fig. 4D). Most of the KIV was produced in the first 6 h with KIV yield close to the theoretical maximum. Maximum titer of KIV recorded was 5.6 g L−1 after 9 h of cultivation at 0.36 g g−1 glucose, which was 56% of maximum theoretical yield (0.64 g g−1).
3.5. In vitro enzyme production and activity
For in vitro conversion of KIV to isobutanol, plasmids ADH and FDH were constructed and transformed into E. coli BL21 star (DE3) and Rosetta (DE3) respectively. The enzymes produced were purified and tested for their activity using kinetic assays to assess their efficacy in producing isobutanol, with the kinetic parameters reported in Table 1. Using the Michaelis-Menten equation, we determined that KIVD WT has a Km of 9.9 mM, a kcat of 500 s−1, and a catalytic efficiency of 50 mM−1s−1, all higher than previously reported by Soh et al., 2017). ADH WT has a Km of 30 mM, a kcat of 90 s−1, and a catalytic efficiency of 3 mM−1s−1, all higher than previously reported by Liu (Liu et al., 2012) The wildtype FDH has a Km of 0.34 mM, a kcat of 1.2 s−1, and a catalytic efficiency of 3.5 mM−1s−1.
Table 1.
Comparison of isobutanol production strategies.
| Host Cell | Substrate | Titer (g/L) | Yield (%) | Productivity (g/[Lah]) | Reference |
|---|---|---|---|---|---|
| In vivo | |||||
| C. thermocellum | Cellulose | 5.4 | 41 | 0.072 | Lin et al. (2015) |
| G. thermoglucosidqsius | Glucose | 3.3 | 22 | 0.069 | Lin et al. (2014) |
| Cellobiose | 0.6 | 8 | 0.013 | ||
| R. eutropha | Fructose | 0.2 | 5 | 0.0028 | Black et al. (2018) |
| S. cerevisiae | Glucose | 0.635 | 2 | 0.013 | Avalos et al. (2013) |
| In vitro | |||||
| Free in solution | Glucose | 0.76 | 53 | 0.032 | Guterl et al. (2012) |
| Free in solution | Glucose | 275 | 95 | 4 | Sherkhanov et al. (2020) |
| Partially immobilizeda | Keto acid | 2.59 | 54 | 0.11 | Grimaldi et al. (2016b) |
| Epoxy immobilized | Keto acid | 2 | 43 | 0.083 | Wong et al. (2019) |
| in vivo to in vitro | Keto acid | 1.78 | 93 | 0.074 | This work |
Reaction was partially immobilized: ADHWT and MBP-KIVDWT immobilized, FDHWT free in solution.
3.6. In vitro isobutanol production
In order to demonstrate that we can combine in vivo supplementation of reagent with in vitro production of biofuel product, we decided to conduct a lab-scale reaction using free in solution enzymes. The reaction step carried out by ADH is known to be reversible (Datta et al., 2006), so recycling of NADH is essential to avoid stagnation of the reaction, as excess NADH is cost prohibitive. Only a small amount of NADH (0.02 mg L−1), in combination with FDH and excess formate (0.1 g L-1), was needed for the reaction to proceed. The amount of isobutanol produced after 24h represents a titer of 1.78 g L−1 and a molar yield of 93%, compared to the starting substrate of KIV.
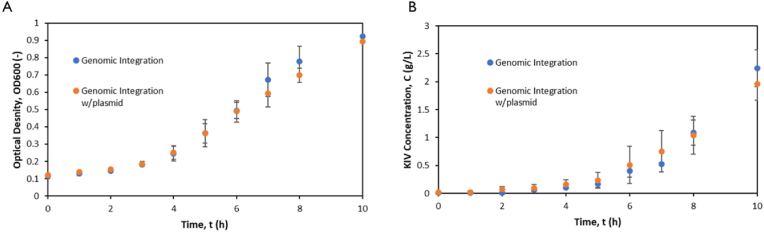
3.7. Effect of copy number on KIV production
To ascertain the effect of copy number on the production of KIV, as seen in Fig. 6, the OD600 and concentration of KIV was tracked over a period of 10 h for two cultures containing MG03 and the same strain with an inducible plasmid containing the KIV pathway. After 10 h the OD600 was 0.92 ± 0.04 and 0.89 ± 0.02 respectively; KIV concentration was 2.2 ± 0.3 and 2.0 ± 0.3 g L−1 respectively (Fig. 6). All values were within error of each other, and further measurements at 24 h mark showed no difference accounting for error.
Fig. 6.
Copy Number Effect on KIV Production: (A) OD of cultures grown at 37 °C with and without the inducible KIV pathway plasmid. (B) KIV concentration for cultures grown at 37 °C with and without the inducible KIV pathway plasmid.
4. Discussion
A multitude of issues such as increasing demand for energy, environmental concerns, political instability and depletion of non-renewable resources has ignited renewed interest in fossil fuel alternatives (Stephanopoulos, 2007). Relatively recently, microbial systems have been extensively explored for biofuel production (Ingram et al., 1999; Jarboe et al., 2007). Out of different biofuels, higher-chain alcohols such as n-butanol and isobutanol, offer several advantages such as higher energy density and lower water solubility (Atsumi et al., 2008a). Compared with n-butanol, isobutanol has higher octane number, and possibility of usage outside the fuel industry as well (Connor and Liao, 2009). Recently the isobutanol production has been investigated in engineered E. coli to reach a concentration of 22 g L−1 during aerobic cultivations (Atsumi et al., 2010a).
However, isobutanol concentrations as low as 8 g L−1 can be toxic to E. coli (Atsumi et al., 2010b). Other microorganisms, such as C. glutamicum, are more tolerant to isobutanol than E. coli but could not grow above 4 g L−1 of isobutanol in the presence of oxygen (Smith et al., 2010). KIV is thus a better target for production in E. coli, which can withstand up to 14.7 g L−1 of KIV before facing growth inhibition (Supplementary Fig. 4). By considering the bottleneck of alcohol tolerance by microorganisms, we demonstrated here a combined in vivo-in vitro approach of producing isobutanol from KIV. The KIV pathway (Fig. 1) was integrated into the genome of E. coli JCL 260 at SS9 site as SS9 site is reported as most suitable site for this pathway integration (Bassalo et al., 2016). Realizing the limitation of state-of-the-art membrane protein production, its dependency on ribosome binding site (RBS) accessibility and importance of inducer free stable protein production (Ingram et al., 1999; Salis et al., 2009), the KIV pathway was designed using BCDs (Supplementary Fig. 1). The system is based on a constitutive promoter and tuning with two Shine-Dalgarno sequences that are translationally coupled (Fig. 1B) which when combined with the gene of interest are known to reliably express within two fold of the relative target expression window (Mutalik et al., 2013). The integration of KIV pathway into the genome of JCL260 without inhibiting the competitive pathway showed no KIV production in the media. Thus, we decided to downregulate the competitive valine pathway and study the effects of targeted repression on KIV titers.
CRISPR interference (CRISPRi) technology using dCas9 was applied to repress ilvE gene encoding for branched-chain-amino-acid aminotransferase in E. coli. CRISPRi based on the catalytically dead Cas9, can efficiently repress or activate the targeted gene regulation in E. coli without altering the target sequence (Qi et al., 2013). Previous studies (Cleto et al., 2016; Lee et al., 2016) have reported the use of CRISPRi for quick and efficient metabolic pathway remodelling without any gene deletions or mutation.
The IlvE enzyme influences the flux toward L-valine formation using KIV as an intermediate (Park et al., 2007). After downregulation of ilvE gene, a KIV titer of 0.4 g L−1 with 9.6 g L−1 of glucose consumption was achieved at 37 °C at laboratory scale in MG03 that was far greater (437-fold) as compared with the control (without KIV integration). The results are supported by a study where the use of CRISPRi has improved the production of single domain antibody in E. coli (Landberg et al., 2020). The glucose consumption and KIV production was compared at 30 °C and 37 °C temperature. Lesser production of KIV and higher glucose consumption at 30 °C can be due to more overflow metabolism as compared with 37 °C. Therefore, it was necessary to analyse overflow metabolite profile of modified strains. The formation of acidic by-products is a commonly observed phenomenon in E. coli when grown under aerobic-glucose conditions (Eiteman and Altman, 2006; Xu et al., 1999). The bacterial strains were tested for the production of acetic acid, succinic acid, formic acid, lactic acid, ethanol, methanol and isobutanol. After the integration of KIV pathway, the production of acetic acid decreased from 3.61 g L−1 (in control) to 1.91 g L−1 (MG01) (Fig. 4C). This showed that integration of KIV pathway leads to reduction in unwanted bioproducts that was further reduced after inhibiting the valine pathway (0.96 g L−1 in MG02 and 0.63 g L−1 in MG03). Small amounts of formic acid (0.4 g L−1) and isobutanol (0.38 g L−1) were observed after integration of KIV pathway while these were absent in control. The reason behind this may be the presence of ALS (acetolactate synthase) gene (Fig. 1B) or some other native genes in E. coli acting like KIVD and ADH. Post hour 9, there was a decrease in KIV followed by a slow ramp increase. It's important to remember that the ilvE gene was repressed and not removed. This could lead to utilization of KIV at the height of growth for valine production followed by KIV accumulation as the cells enter the death phase. Previously, the butanol production by the native genes of E. coli has been reported (Dellomonaco et al., 2011). The NADH concentration in extracellular culture medium was measured after 72 h of growth in both control and engineered bacterial strains and was found to be higher in control cells, similar to acetate concentration (Fig. 5C). This demonstrates that the integration of KIV pathway in strain MG01 led to consumption of NADH at lower growth rate, lower glucose consumption and lesser release of acetate in the media. This overall reduction in overflow metabolism was further decreased after the downregulation of valine biosynthetic pathway in strains MG02 and MG03. The reason behind this behavior may be the interrelation between NADH production via glycolysis, the TCA cycle, acetate fermentation, and growth rate (Szenk et al., 2017). At low growth rates and less acetate production (MG02 and MG03), it may be possible that a sufficient electron transport chain exists to keep the NADH low inside the cell. At high growth rates, it might be impossible to metabolize all the NADH generated, leading to NADH imbalance and potential toxicity (Fig. 5A and B) (Vemuri et al., 2006; Imai, 2016).
The same strain was further tested for large scale production of KIV under similar temperature and minimal media conditions. In a batch reactor, we were able to produce 5.6 g L−1 KIV in 9 h, obtaining 0.36 g g−1 glucose and a productivity of 0.62 g L−1 h−1. Previously the highest Ketoisovalerate (21.8 g L-1) production was reported in C. glutamicum with a yield of 0.3 g g-1 of glucose and productivity of 0.53 g L−1 h−1 (Krause et al., 2010). The present work reported the highest Ketoisovalerate production in E. coli till date. These results are promising because apart from isobutanol production KIV also serves as a substitute for L-valine or L-leucine in chronic kidney disease patients (Aparicio et al., 2012; Chang et al., 2009; Feiten et al., 2005) and its demand is exclusively covered by chemical synthesis ((Meister and A.J.L.C.Z.G., 1983).
Integration of a genetic pathway has often been used to avoid the issues of natural plasmid loss during fermentation as well as high metabolic burden that can occur due to the number of copies of plasmid produced in a cell (Englaender et al., 2017). In this case a comparison between the integrated pathway strain MG03, which would be effectively a copy number of 1, and the same strain transformed with a low copy number plasmid containing the pathway was conducted in flasks to ascertain any gains or losses in cell growth or KIV production. As seen in Fig. 6, there was statistically no difference between strains concerning either metric. It has been shown before that a copy number as low as 1 can be the ideal for production of metabolites in E. Coli so it is not surprising that the integrated strain did not need a higher copy number to improve performance (Englaender et al., 2017).
To facilitate isobutanol production, we designed an in vivo to in vitro method using KIV as a starting material that was subjected to an enzymatic conversion to isobutanol using enzymes KIVD, ADH, and FDH. KIVD had a higher Km (9.9 mM) and kcat (500 s−1) than previously reported by Soh, (1.6 mM and 17 s−1, respectively), meaning the binding affinity of KIVD for KIV was lower but the rate at which the substrate is converted was much higher, leading to a larger catalytic efficiency (50 mM−1s−1 greater than 11 mM−1s−1) (Aparicio et al., 2012). Based on our results, we can conclude that KIVD is effective at converting KIV to aldehyde, and better than previously reported.
Next, we examined ADH. The enzyme used in our study had both a Km (30 mM) and a kcat (90 s−1) higher than previously reported by Liu (12 mM and 30 s−1, respectively), and a higher catalytic efficiency (3 mM−1s−1 greater than 2.8 mM−1s−1) (Aparicio et al., 2012). NADH is an expensive substrate, so a higher catalytic efficiency is useful since the concentration of NADH will be lower than Km. Once again, we identified an enzyme that is more efficient at converting its substrate than previously reported. As both KIVD and ADH have a high kcat, FDH is the rate limiting enzyme, with a lower kcat of 1.2 s−1. This presents a problem; despite it not being part of the main reaction of converting KIV to isobutanol, it is required for recycling NAD back to NADH. To reduce the liability of the rate limiting step on isobutanol production, we utilized an excess of relatively inexpensive formate, which allowed for high conversion of NAD back to NADH. So, while both KIVD and ADH will have lower reaction rates as their reagents begin to be consumed, NADH recycling will be maintained at a consistent rate. This led to the high conversion of KIV to isobutanol.
Table 1 showcases current isobutanol production strategies, both in vivo and in vitro. Our current isobutanol yield (93%), titer (1.78 g L−1), and production rate (0.07 g L−1 h−1) are comparable to plasmid based in vivo production (Chen and Liao, 2016) but surpassed by a purely in vitro system, such as reported in a recent publication that obtained a 275 g L−1 titer, 95% yield, and 4 g L−1 h−1 production rate of isobutanol (Sherkhanov et al., 2020). That system required challenging modifications in order to obtain this result such as scaling up the system from 300 μL to 15 mL in order to control pH which increased the total amount of purified enzyme used, continually adding additional enzyme during the reaction to maintain production due to loss of enzyme stability, and the use of 50 mL of organic solvent for a liquid-liquid extraction of the isobutanol to drive the reaction forward. Purifying and monitoring 16 enzymes is costly and a liquid-liquid separation would require non-green thermal means to extract the isobutanol from the organic solvent.
Several optimizations to our unique 2-step in vivo to in vitro system could be implemented to increase the titer. In vitro reaction time should be examined to see if it can be shortened to improve the productivity. An increase in the production of KIV will up regulate the titer, but at the potential cost of the high yield. Immobilization of the enzymes is a useful strategy to retain activity as the concentration of biofuel increases (Wong et al., 2019; Eiteman and Altman, 2006) and reduce diffusion limitations. These immobilized enzymes have higher thermal and chemical stability and can be separated from the aqueous product with ease (Sheldon, 2007). This method can thus be used in tandem with in situ removal of the biofuel product, allowing for separations that are less thermally demanding (Joseph et al., 2015). By combining these two compatible methods with a higher concentration of starting reagent, a cell-free system can be created that has both the high yield obtained so far as well as higher titer. This next step of the work is currently in progress.
5. Conclusion
We set out to show, with this two-step reaction system, the efficacy of combining in vivo supplementation of reagent with in vitro production of biofuel product in series. We have shown that this system can both work well and be optimized for future scale up, obtaining in the first step a KIV titer of 5.6 g L−1 with a yield of 56% and a productivity of 0.62 g L−1 h−1. For the second in vitro step, we obtained an isobutanol titer of 1.78 g L−1 with a yield of 93% and a productivity of 0.07 g L−1 h−1. Moreover, this approach can be expanded to a modular approach in which KIV can be used as a precursor to compounds other than isobutanol.
Additional information
Supplementary Information accompanies this paper can be found at full online version of this article.
Author contribution
MAGK and GB conceived this work; MAGK, GB, RTG, JD designed experiments. MG, MW, KJ, KG, HB, MB, CM, DE, JH, MD created the KIV producing strain using CRISPR, and carried out the in vitro production of isobutanol. MAGK, GB, and SY analyzed the data and wrote the initial draft of the manuscript. All authors read, edited, and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
MG, KJ, and SSY are grateful to SERB and Indo-US Science and Technology Forum for their financial support. MW acknowledges the Howard P. Isermann fellowship and support from the NIH training grant. We are also thankful to Dr. James Liao, University of California, Los Angeles for providing JCL260 and pLS02 strain and discussion. Authors are thankful to Dr. Dmitri V. Zagorevski for timely LCMS analysis. We are also thankful to Dr. Mirco Sorci for help with designing figures. GB acknowledges the Department of Energy (Grant No. DE/SC0006520) for funding.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2022.e00210.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Aparicio M., et al. Keto acid therapy in predialysis chronic kidney disease patients: final consensus. J. Ren. Nutr. 2012;22(2 Suppl. l):S22–S24. doi: 10.1053/j.jrn.2011.09.006. [DOI] [PubMed] [Google Scholar]
- Atsumi S., Hanai T., Liao J.C. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451(7174):86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- Atsumi S., et al. Metabolic engineering of Escherichia coli for 1-butanol production. Metab. Eng. 2008;10(6):305–311. doi: 10.1016/j.ymben.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Atsumi S., et al. Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes. Appl. Microbiol. Biotechnol. 2010;85(3):651–657. doi: 10.1007/s00253-009-2085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S., et al. Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Mol. Syst. Biol. 2010;6:449. doi: 10.1038/msb.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avalos J.L., Fink G.R., Stephanopoulos G. Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols. Nat. Biotechnol. 2013;31(4):335–341. doi: 10.1038/nbt.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassalo M.C., et al. Rapid and efficient one-step metabolic pathway integration in E. coli. ACS Synth. Biol. 2016;5(7):561–568. doi: 10.1021/acssynbio.5b00187. [DOI] [PubMed] [Google Scholar]
- Bikard D., et al. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41(15):7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black W.B., et al. Rearrangement of coenzyme A-acylated carbon chain enables synthesis of isobutanol via a novel pathway in Ralstonia eutropha. ACS Synth. Biol. 2018;7(3):794–800. doi: 10.1021/acssynbio.7b00409. [DOI] [PubMed] [Google Scholar]
- Blombach B., Eikmanns B.J. Current knowledge on isobutanol production with Escherichia coli, Bacillus subtilis and Corynebacterium glutamicum. Bioeng Bugs. 2011;2(6):346–350. doi: 10.4161/bbug.2.6.17845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blombach B., et al. Corynebacterium glutamicum tailored for efficient isobutanol production. Appl. Environ. Microbiol. 2011;77(10):3300–3310. doi: 10.1128/AEM.02972-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boock J.T., et al. Engineered microbial biofuel production and recovery under supercritical carbon dioxide. Nat. Commun. 2019;10(1):587. doi: 10.1038/s41467-019-08486-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brynildsen M.P., Liao J.C. An integrated network approach identifies the isobutanol response network of Escherichia coli. Mol. Syst. Biol. 2009;5:277. doi: 10.1038/msb.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J.H., et al. Influence of ketoanalogs supplementation on the progression in chronic kidney disease patients who had training on low-protein diet. Nephrology. 2009;14(8):750–757. doi: 10.1111/j.1440-1797.2009.01115.x. [DOI] [PubMed] [Google Scholar]
- Chen C.T., Liao J.C. Frontiers in microbial 1-butanol and isobutanol production. FEMS Microbiol. Lett. 2016;363(5):fnw020. doi: 10.1093/femsle/fnw020. [DOI] [PubMed] [Google Scholar]
- Claassens N.J., et al. Bicistronic design-based continuous and high-level membrane protein production in Escherichia coli. ACS Synth. Biol. 2019;8(7):1685–1690. doi: 10.1021/acssynbio.9b00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleto S., et al. Corynebacterium glutamicum metabolic engineering with CRISPR interference (CRISPRi) ACS Synth. Biol. 2016;5(5):375–385. doi: 10.1021/acssynbio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor M.R., Liao J.C. Microbial production of advanced transportation fuels in non-natural hosts. Curr. Opin. Biotechnol. 2009;20(3):307–315. doi: 10.1016/j.copbio.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Datta S., Costantino N., Court D.L. A set of recombineering plasmids for gram-negative bacteria. Gene. 2006;379:109–115. doi: 10.1016/j.gene.2006.04.018. [DOI] [PubMed] [Google Scholar]
- Dellomonaco C., et al. Engineered reversal of the beta-oxidation cycle for the synthesis of fuels and chemicals. Nature. 2011;476(7360):355–359. doi: 10.1038/nature10333. [DOI] [PubMed] [Google Scholar]
- Dong H., et al. Engineering Escherichia coli cell factories for n-butanol production. Adv. Biochem. Eng. Biotechnol. 2016;155:141–163. doi: 10.1007/10_2015_306. [DOI] [PubMed] [Google Scholar]
- Dudley Q.M., Karim A.S., Jewett M.C. Cell-free metabolic engineering: biomanufacturing beyond the cell. Biotechnol. J. 2015;10(1):69–82. doi: 10.1002/biot.201400330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durre P. Biobutanol: an attractive biofuel. Biotechnol. J. 2007;2(12):1525–1534. doi: 10.1002/biot.200700168. [DOI] [PubMed] [Google Scholar]
- Eiteman M.A., Altman E. Overcoming acetate in Escherichia coli recombinant protein fermentations. Trends Biotechnol. 2006;24(11):530–536. doi: 10.1016/j.tibtech.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Englaender J.A., et al. Effect of genomic integration location on heterologous protein expression and metabolic engineering in E. coli. ACS Synth. Biol. 2017;6(4):710–720. doi: 10.1021/acssynbio.6b00350. [DOI] [PubMed] [Google Scholar]
- Feiten S.F., et al. Short-term effects of a very-low-protein diet supplemented with ketoacids in nondialyzed chronic kidney disease patients. Eur. J. Clin. Nutr. 2005;59(1):129–136. doi: 10.1038/sj.ejcn.1602050. [DOI] [PubMed] [Google Scholar]
- Gibson D.G., et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6(5):343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Gonzalez R., et al. Gene array-based identification of changes that contribute to ethanol tolerance in ethanologenic Escherichia coli: comparison of KO11 (parent) to LY01 (resistant mutant) Biotechnol. Prog. 2003;19(2):612–623. doi: 10.1021/bp025658q. [DOI] [PubMed] [Google Scholar]
- Grimaldi J., Collins C.H., Belfort G. Towards cell-free isobutanol production: development of a novel immobilized enzyme system. Biotechnol. Prog. 2016;32(1):66–73. doi: 10.1002/btpr.2197. [DOI] [PubMed] [Google Scholar]
- Grimaldi J., Collins C.H., Belfort G. Towards cell‐free isobutanol production: development of a novel immobilized enzyme system. Biotechnol. Prog. 2016;32(1):66–73. doi: 10.1002/btpr.2197. [DOI] [PubMed] [Google Scholar]
- Guilbault G.G. Marcel Dekker Inc.; New York: 1973. Pratical Fluorescence: Theory, Methods and Technique. [Google Scholar]
- Guterl J.K., et al. Cell‐free metabolic engineering: production of chemicals by minimized reaction cascades. ChemSusChem. 2012;5(11):2165–2172. doi: 10.1002/cssc.201200365. [DOI] [PubMed] [Google Scholar]
- Imai S.I. The NAD World 2.0: the importance of the inter-tissue communication mediated by NAMPT/NAD(+)/SIRT1 in mammalian aging and longevity control. NPJ Syst Biol Appl. 2016;2 doi: 10.1038/npjsba.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram L.O., et al. Enteric bacterial catalysts for fuel ethanol production. Biotechnol. Prog. 1999;15(5):855–866. doi: 10.1021/bp9901062. [DOI] [PubMed] [Google Scholar]
- Jarboe L.R., et al. Development of ethanologenic bacteria. Adv. Biochem. Eng. Biotechnol. 2007;108:237–261. doi: 10.1007/10_2007_068. [DOI] [PubMed] [Google Scholar]
- Jawed K., et al. Improved butanol production using FASII pathway in e. coli. ACS Synth. Biol. 2020;9(9):2390–2398. doi: 10.1021/acssynbio.0c00154. [DOI] [PubMed] [Google Scholar]
- Joseph G., J .I., James K., Georges B. New class of synthetic membranes: organophilic pervaporation brushes for organics recovery. Chem. Mater. 2015;27:4142–4148. [Google Scholar]
- Krause F.S., Blombach B., Eikmanns B.J. Metabolic engineering of Corynebacterium glutamicum for 2-ketoisovalerate production. Appl. Environ. Microbiol. 2010;76(24):8053–8061. doi: 10.1128/AEM.01710-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landberg J., et al. CRISPR interference of nucleotide biosynthesis improves production of a single‐domain antibody in Escherichia coli. Biotechnol. Bioeng. 2020;117(12):3835–3848. doi: 10.1002/bit.27536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.J., et al. Programmable control of bacterial gene expression with the combined CRISPR and antisense RNA system. Nucleic Acids Res. 2016;44(5):2462–2473. doi: 10.1093/nar/gkw056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Wen J., Jia X. Engineering Bacillus subtilis for isobutanol production by heterologous Ehrlich pathway construction and the biosynthetic 2-ketoisovalerate precursor pathway overexpression. Appl. Microbiol. Biotechnol. 2011;91(3):577–589. doi: 10.1007/s00253-011-3280-9. [DOI] [PubMed] [Google Scholar]
- Li R., et al. A novel liquid chromatography tandem mass spectrometry method for simultaneous determination of branched-chain amino acids and branched-chain α-keto acids in human plasma. Amino Acids. 2016;48(6):1523–1532. doi: 10.1007/s00726-016-2212-5. [DOI] [PubMed] [Google Scholar]
- Lin P.P., et al. Isobutanol production at elevated temperatures in thermophilic Geobacillus thermoglucosidasius. Metab. Eng. 2014;24:1–8. doi: 10.1016/j.ymben.2014.03.006. [DOI] [PubMed] [Google Scholar]
- Lin P.P., et al. Consolidated bioprocessing of cellulose to isobutanol using Clostridium thermocellum. Metab. Eng. 2015;31:44–52. doi: 10.1016/j.ymben.2015.07.001. [DOI] [PubMed] [Google Scholar]
- Liu X., et al. Structure-guided engineering of Lactococcus lactis alcohol dehydrogenase LlAdhA for improved conversion of isobutyraldehyde to isobutanol. J. Biotechnol. 2012;164(2):188–195. doi: 10.1016/j.jbiotec.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister, A.J.L.C.Z.G. Synthesis and properties of the .alpha.-keto acids. Chem. Rev. 1983;83:321–358. [Google Scholar]
- Mutalik V.K., et al. Precise and reliable gene expression via standard transcription and translation initiation elements. Nat. Methods. 2013;10(4):354–360. doi: 10.1038/nmeth.2404. [DOI] [PubMed] [Google Scholar]
- Olcay H., et al. Techno-economic and environmental evaluation of producing chemicals and drop-in aviation biofuels via aqueous phase processing. Energy Environ. Sci. 2018;11(8):2085–2101. [Google Scholar]
- Park J.H., et al. Metabolic engineering of Escherichia coli for the production of L-valine based on transcriptome analysis and in silico gene knockout simulation. Proc. Natl. Acad. Sci. U. S. A. 2007;104(19):7797–7802. doi: 10.1073/pnas.0702609104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L.S., et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisch C.R., Prather K.L. The no-SCAR (Scarless Cas9 Assisted Recombineering) system for genome editing in Escherichia coli. Sci. Rep. 2015;5(1):1–12. doi: 10.1038/srep15096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford B.J., et al. Functional genomic study of exogenous n-butanol stress in Escherichia coli. Appl. Environ. Microbiol. 2010;76(6):1935–1945. doi: 10.1128/AEM.02323-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salis H.M., Mirsky E.A., Voigt C.A. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009;27(10):946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana N.E., et al. A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 2012;7(1):171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon R.A. Enzyme immobilization: the quest for optimum performance. Adv. Synth. Catal. 2007;349:1289–1307. [Google Scholar]
- Sherkhanov S., et al. Isobutanol production freed from biological limits using synthetic biochemistry. Nat. Commun. 2020;11(1):1–10. doi: 10.1038/s41467-020-18124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.M., Cho K.M., Liao J.C. Engineering Corynebacterium glutamicum for isobutanol production. Appl. Microbiol. Biotechnol. 2010;87(3):1045–1055. doi: 10.1007/s00253-010-2522-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh L.M.J., et al. Engineering a thermostable keto acid decarboxylase using directed evolution and computationally directed protein design. ACS Synth. Biol. 2017;6(4):610–618. doi: 10.1021/acssynbio.6b00240. [DOI] [PubMed] [Google Scholar]
- Stephanopoulos G. Challenges in engineering microbes for biofuels production. Science. 2007;315(5813):801–804. doi: 10.1126/science.1139612. [DOI] [PubMed] [Google Scholar]
- Szenk M., Dill K.A., de Graff A.M.R. Why do fast-growing bacteria enter overflow metabolism? Testing the membrane real estate hypothesis. Cell Syst. 2017;5(2):95–104. doi: 10.1016/j.cels.2017.06.005. [DOI] [PubMed] [Google Scholar]
- Vemuri G.N., et al. Overflow metabolism in Escherichia coli during steady-state growth: transcriptional regulation and effect of the redox ratio. Appl. Environ. Microbiol. 2006;72(5):3653–3661. doi: 10.1128/AEM.72.5.3653-3661.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M., et al. Cell-free production of isobutanol: a completely immobilized system. Bioresour. Technol. 2019;294 doi: 10.1016/j.biortech.2019.122104. [DOI] [PubMed] [Google Scholar]
- Xu B., Jahic M., Enfors S.O. Modeling of overflow metabolism in batch and fed-batch cultures of Escherichia coli. Biotechnol. Prog. 1999;15(1):81–90. doi: 10.1021/bp9801087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.