Abstract
The heart is composed of multiple cell types, each with a specific function. Cell-type-specific approaches are necessary for defining the intricate molecular mechanisms underlying cardiac development, homeostasis, and pathology. While single-cell RNA-seq studies are beginning to define the chamber-specific cellular composition of the heart, our views of the proteome are more limited because most proteomics studies have utilized homogenized human cardiac tissue. To promote future cell-type specific analyses of the human heart, we describe the first method for cardiomyocyte isolation from cryopreserved human cardiac tissue followed by flow cytometry for purity assessment. We also describe a facile method for preparing isolated cardiomyocytes and whole cardiac tissue homogenate for bottom-up proteomic analyses. Prior experience in dissociating cardiac tissue or proteomics is not required to execute these methods. We compare different sample preparation workflows and analysis methods to demonstrate how these can impact the depth of proteome coverage achieved. We expect this how-to guide will serve as a starting point for investigators interested in general and cell-type-specific views of the cardiac proteome.
Keywords: Cardiomyocyte isolation, Proteomics, S-trap, Data independent acquisition
1. Introduction
The heart is composed of multiple cell types including cardiomyocytes, cardiac fibroblasts, endothelial cells, smooth muscle cells, and immune cells. The cell types work together to maintain cardiac homeostasis, each cell type with a specific function. Cardiomyocytes are responsible for muscular contractions and cardiac fibroblasts maintain the structural integrity of the heart [1]. Intercellular communications among these cell types (e.g. cell-to-cell and cell-to-extracellular matrix (ECM) interactions) are crucial for proper cardiac function [2]. Consequently, cell-type-specific approaches are necessary to define the intricate molecular mechanisms underlying cardiac development, homeostasis, and pathology.
Experiments using single-cell RNA-seq have recently described the gene expression profiles among major cell types in the adult and embryonic human heart [3–5]. These studies provide new insights into chamber-specific cellular composition of the heart and identify genes with previously unknown cardiac expression. Because transcriptomic strategies cannot predict the functional diversity of the proteome (e.g. post-translational modifications, protein-protein interactions, and protein degradation), mass spectrometry (MS) has been increasingly applied to study the cardiac proteome in health and disease. Top-down (MS analysis of intact proteins) and bottom-up (MS analysis of peptides) proteomics studies have identified putative biomarkers for cardiomyopathies [6,7] and sarcomeric proteoforms altered in hypertrophic cardiomyopathy [8]. In-depth analysis of the cardiac proteome revealed pathophysiological mechanisms for ischemic [9,10] and dilated cardiomyopathies [11,12], identified signaling proteins [13] from left ventricular cardiac tissue and metabolic pathways in healthy and disease mouse models [14], and elucidated the protein composition of different regions of the heart [15].
How gene expression findings from single-cell RNAseq studies relate to protein sequence and abundance in human cardiac cells is currently poorly defined, mainly because most proteomics studies have been performed using animal models or homogenized human cardiac tissue. The proteomic study by Doll et al., presented a general proteome of two explanted and cultured cell types (CD31-positive endothelial cells and cardiac fibroblasts) from atrial cardiac tissue and explanted and cultured smooth muscle cells from arteria mammaria interna. The study also used homogenized myocardial tissue for the ventricular, atrial, and septal samples, but no isolated primary cardiomyocytes were included in the analysis [15]. The proteomic study of patients with mitral valve prolapse by Linscheid et al. similarly used homogenized tissues, but isolated cardiomyocytes were not analyzed [16]. We speculate that the reason previous studies of the human cardiac proteome have not included isolated cardiomyocytes is that these cells are challenging to obtain, and previous methods for isolation have required fresh tissue [17–21].
To address these limitations, this how-to article describes methods to 1) isolate adult human cardiomyocytes from cryopreserved cardiac tissue suitable for proteomic analysis and 2) prepare cardiac tissue or isolated cardiomyocyte samples for bottom-up proteomic analysis. We introduce the principles and features of the methodologies and provide example data from non-failing adult human cardiac tissue. We describe the first method for cardiomyocyte isolation from cryopreserved human cardiac tissue followed by flow cytometry for purity assessment. We also describe facile sample preparation methods for bottom-up proteomic analysis of isolated cardiomyocytes and whole cardiac tissue homogenate. Prior experience in dissociating cardiac tissue or proteomics is not required. The samples generated from these workflows are suitable for analysis using MS instrumentation common to biomedical research laboratories and academic core facilities. We compare different proteomic sample preparation workflows, MS acquisition modes, and analysis methods to demonstrate their impact on the depth of proteome coverage achieved. This how-to guide is expected to serve as a starting point for investigators interested in general and cell-type-specific views of the cardiac proteome. The described methods will promote future studies that provide a more comprehensive view of cell-type and chamber-specific processes in healthy and diseased hearts, including post-translational modifications, protein interactions, and the impact of alternative splicing.
2. Materials and methods
To facilitate implementation of these approaches, detailed methods and reagent information are in the supplementary information. Protein digestion and peptide fractionation methods are based on detailed protocols we recently described [22].
2.1. Study design
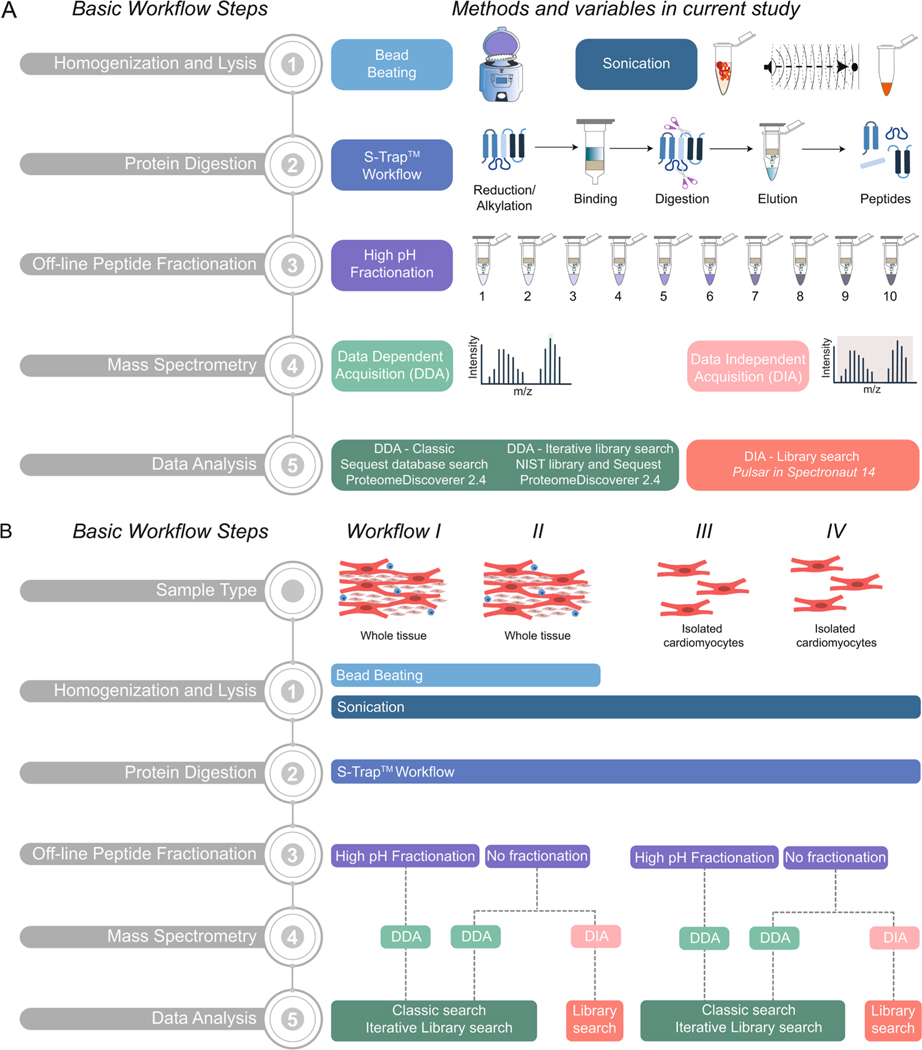
Bottom-up proteomic workflows include steps to homogenize samples, digest proteins into peptides by enzymatic or chemical means, and remove contaminants before MS analysis. Optionally, off-line peptide fractionation after digestion of complex samples generates multiple lower-complexity samples, which typically provide a greater depth of protein sequence and proteome coverage. Here, we used facile methods for homogenization and lysis, protein digestion, peptide fractionation, and different MS acquisition modes to generate high-quality MS data (Fig. 1A). We applied four different experimental workflows (Fig. 1B) to demonstrate the effects that different sample preparation and data analysis methods can have on proteome coverage when analyzing samples with varying complexity. Specimens from non-failing hearts from donors for which multiple anatomical regions were available (Table 1) were used. We compared isolated cardiomyocytes and whole cardiac tissue from different myocardial regions to show differences in the proteome among anatomic regions of the heart.
Fig. 1.
Overview of bottom-up proteomics methodologies for processing human cardiac tissue and cardiomyocytes. A) Schematic representation of five basic steps in a bottom-up proteomics workflow: (1) Homogenization and Lysis, (2) Protein Digestion, (3) Off-line Peptide Fractionation, (4) Mass Spectrometry, and (5) Data Analysis. B) Schematic representation of four workflows applied for the proteomic analysis of human cardiac tissue or isolated cardiomyocytes in the current study.
Table 1.
Patient characteristics and analysis workflows used for each specimen.
| Patient | Age | Sex | Cause of death | Heart region | Proteomic workflows |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| I |
II |
II |
III |
IV |
IV |
|||||
| DDA | DDA | DIA | DDA | DDA | DIA | |||||
| 1 | 32 | M | Suicide | Apex | ● | ● | ● | ● | ||
| LV | ● | ● | ● | ● | ||||||
| RV | ● | ● | ● | ● | ||||||
| 2 | 75 | F | Hemorrhagic stroke | Apex | ● | ● | ● | ● | ||
| LV | ● | ● | ● | ● | ||||||
| RV | ● | ● | ● | ● | ||||||
| 3 | 56 | M | Alcohol ingestion, coronary artery disease | LV | ● | ● | ● | ● | ||
| RV | ● | ● | ● | ● | ||||||
| 4 | 39 | M | Cerebrovascular accident | Apex | ● | ● | ● | ● | ||
F = female; M = male; LV = left ventricle; RV = right ventricle; proteomic workflows refer to those described in Fig. 1B; DDA = data dependent acquisition; DIA = data independent acquisition.
2.2. Cardiomyocyte isolation from frozen human tissue
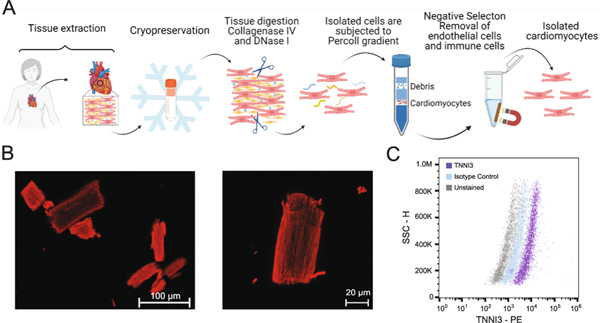
Human cardiac tissue (Table 1) was obtained under institutional review board approvals (PRO00022202; PRO643–17-EP) and cryopreserved in 10% dimethyl sulfoxide (DMSO), 10% fetal bovine serum (FBS), 80% Dulbecco’s Modified Eagle Medium (DMEM) until use. An overview of the isolation method is shown in Fig. 2A. Tissue was thawed and cut into 1 mm3 pieces while submerged in ice-cold wash solution (115 mM potassium gluconate, 2.5 mM potassium chloride (KCl), 5 mM potassium phosphate monobasic (KH2PO4), 2 mM magnesium sulfate (MgSO4), 2 mM calcium chloride (CaCl2), 30 mM sucrose, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.4). Tissue pieces were collected with a 70 μm cell strainer and washed three times with ice-cold wash solution. Tissue pieces were placed in digestion solution (300 Units/mL collagenase type 4, 5 Units/mL DNase I, and 1 mg/mL trypsin inhibitor in wash solution) in a 50 mL spinner flask at 37 °C. A total of three digestions were performed, each yielding a fraction of crude cardiomyocytes that was filtered through a 200 μm cell strainer. Crude cardiomyocytes from three digestions were combined, collected by centrifugation at 80 ×g for 5 min at 4 °C, and washed three times in ice-cold wash solution. Cellular debris was separated from the crude cardiomyocytes by Percoll gradient as described in supplementary information. Crude cardiomyocytes were further purified by removing immune cells and endothelial cells by negative selection. Dynabeads™ CD45 and Dynabeads™CD31 Endothelial Cell were washed three times in wash solution containing 0.1% FBS using an Invitrogen DynaMag™−2 magnet to collect beads. 25 μL of each bead type was added to 1 mL cell suspension, agitated with end-over-end rotation for 30 min at 4 °C, and beads were collected for 1 min against the magnet. The supernatant containing the isolated cardiomyocytes was used for further studies.
Fig. 2.
Cardiomyocyte isolation and purity assessment. A) Schematic overview of cardiomyocyte isolation workflow using cryopreserved human adult cardiac tissue. B) Representative confocal immunofluorescence images of isolated cardiomyocytes stained for cardiac actin (red). Magnification 20× (left) and 63× (right); Scale bar, 100 μm (left), 20 μm (right) C) Flow cytometry assessment of isolated cells for TNNI3 positivity. Cells gated on fluorescence intensity versus side scatter (SSC-H) result in a >80% anti-TNNI3 PE positive population as compared to isotype control. Panel A was generated with Biorender. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.3. Assessment of isolated cardiomyocytes by flow cytometry and imaging
Isolated cardiomyocytes were assessed for percent troponin I (TNNI3) positivity by flow cytometry. Cardiomyocytes were fixed, permeabilized, blocked, stained for TNNI3, and analyzed using an Attune NxT acoustic focusing flow cytometer (Thermo Fisher Scientific). Data for unstained, isotype controls, and stained cells were analyzed using FlowJo v.10.7.1 (FlowJo LLC). Visual assessment of cardiomyocytes using immunofluorescent labeled cardiac actin was performed on a Zeiss 710 confocal laser scanning microscope.
2.4. Isolated cardiomyocyte cell lysis
Isolated cardiomyocytes were lysed in 0.1–0.5 mL of cell lysis buffer (5% sodium dodecyl sulfate (SDS), 50 mM triethylammonium bicarbonate (TEAB) at pH 7.55, 2 mM magnesium chloride). 250 units of Benzonase® was added, and samples were sonicated (UP200St, Hielscher, Germany) on ice for three 10-s intervals at 30% amplitude with 1-min rest periods between intervals.
2.5. Whole tissue homogenization and cell lysis
Approximately 50 mg of heart tissue was minced and placed in homogenization buffer (5% SDS, 50 mM TEAB, pH 7.55) in 2 mL tubes pre-filled with ceramic beads. Tissue pieces were homogenized in a Precellys 24 homogenizer at 6500 rpm, 2 times at 15 s each, with 10 s of break. Samples were then sonicated as described above and clarified by centrifugation for 8 min at 13,000 ×g.
2.6. Suspension-trapping (S-trap™) processing
Total protein concentrations for each sample were measured using the Pierce 660 nm protein assay kit using a Varioskan LUX Multimode Microplate Reader with SkanIt 5.0 software (Thermo Fisher Scientific) per manufacturer instructions. 50 μg of protein from each sample was reduced with 10 mM tris(2-carboxyethyl)phosphine (TCEP) in a thermomixer for 30 min at 37 °C at 1200 rpm, followed by alkylation with 40 mM acrylamide in a thermomixer for 30 min at 37 °C at 1200 rpm. Samples were acidified with 12% phosphoric acid then diluted with 165 μL S-Trap™ binding buffer (100 mM TEAB, 90% methanol with pH 7.1). Samples were loaded onto S-Trap™ micro spin columns and washed three times with S-Trap™ binding buffer. Trypsin-LysC in 50 mM TEAB solution was added to the S-Trap™ at a ratio of 1:12 protease: protein. The S-Trap™ was incubated at 37 °C for 2 h. Peptides were eluted sequentially with 50 mM TEAB, 0.2% aqueous formic acid and 50% acetonitrile (MeCN) containing 0.2% formic acid. Combined peptide elutions were vacuum centrifuged to dryness.
2.7. High-pH reversed-phase fractionation
Peptides collected from three separate S-Trap™ digestions of 50 μg protein were combined, dried, and resuspended in 300 μL of 0.1% tri-fluoroacetic acid (TFA). Peptides were fractionated using Pierce high pH fractionation kit as described [23]. Columns were conditioned with 300 μL of MeCN twice and equilibrated with 300 μL of 0.1% TFA twice. Samples were applied to the columns and washed with purified water (18 Ω). Peptides were eluted with 5%, 7.5%, 10%, 12.5% 15%, 17.5%, 20% and 50% MeCN in 0.1% TEA, then with 15% MeCN in 5% formic acid, and finally with 50% MeCN in 5% formic acid for a total of 10 fractions. All fractions were vacuum dried and resuspended in 25 μL of 2% MeCN in 0.1% formic acid in water.
2.8. Mass spectrometry
Peptide samples were measured by Pierce Quantitative Fluorometric Peptide Assay using a Varioskan LUX multimode microplate reader and diluted to 0.2 μg/μL. Samples (5 μL; 1 μg per injection) were analyzed using a Dionex UltiMate™ 3000 RSLCnano liquid chromatography (LC) system coupled to an Orbitrap Exploris™ 480 mass spectrometer (Thermo Fisher Scientific). Data acquisition and analysis settings are in supplementary information (Supplemental Tables S4 and S5). Data-dependent acquisition (DDA) sample sets spiked with Pierce™ Retention Time Calibration mixture (Thermo Fisher Scientific), and data-independent acquisition (DIA) sample sets spiked with iRT synthetic peptide mixture (Biognosys) were blocked and acquired in randomized order with digested E. coli spiked with iRT synthetic peptide mixture injected immediately before and after every sixth sample to evaluate system performance. Consistency and reproducibility of system performance were evaluated using Skyline Daily (20.2.1.315), QuiC (3.1.200602.47994), and Proteome Discoverer 2.4 (Thermo Fisher Scientific). Data files for all experiments in Table 1 are available at MassIVE (massive.ucsd.edu; doi:10.25345/C55515) and experimental identifiers for each file name are provided in Supplemental Table S6.
2.9. MS data analysis
DDA data were analyzed in Proteome Discoverer 2.4 using either a classic search or an iterative library search (Fig. 1A, B). Classic search used Sequest [24] search engine against a human SwissProt reference proteome (42,304 sequence entries, December 2019). The iterative library search used MSPepSearch [25] for spectral matching against three consensus peptide libraries from NIST (total 1,127,970 spectra entries, May 2020). Spectra that did not match to peptides with a high probability were searched using Sequest. Target decoy analysis was performed by searching a reverse database with an overall false discovery rate (FDR) of 1% at the protein and peptide levels.
DIA data were analyzed in Spectronaut™ 14 (Biognosys, Switzerland) using directDIA™ to search against a fasta file (human SwissProt reference proteome (42,304 sequence entries, December 2019)). Additionally, fractionated peptide data from left and right ventricles - workflows I and III - were searched separately using the Pulsar™ search engine to create a spectral library. DirectDIA™ search data were added to this library. For both directDIA™ and Pulsar™, default settings were used, with the exception that propionamide (C) was used as a fixed modification. Only precursors (i.e. MS1 of peptides) and proteins with a Qvalue of 0.01 were considered identified. As part of quality control assessment, box plots for data normalization and target and decoy distribution were assessed (Fig. S1). Data were globally normalized on the median peptide signal (Supplemental Fig. S1A, B). A minimum of 3 fragments was used for peptide quantification. Proteins with a Qvalue ≤0.05 and an absolute average log2 ratio ≥ 0.58 were considered differentially abundant between chambers.
3. Results
3.1. Cardiomyocyte isolation and purity assessment
Methods for cardiomyocyte isolation from fresh adult human tissue have been described [17–21]. However, access to fresh human tissue may not be possible or practical for all studies depending on infrastructure for specimen collection and distribution. The aforementioned methods use buffer solutions that mimic the ionic composition of the extracellular environment (e.g. 1× PBS, 140 mM NaCl). When we tested these methods for isolating cardiomyocytes from cryopreserved tissue, we found that cell membrane integrity was significantly compromised. We observed increased cell swelling and damage, which we attribute to the fact that cardiomyocytes isolated from frozen tissue fail to regulate ion gradients as efficiently as freshly isolated cardiomyocytes. Also, highly tryptic digestion enzymes (e.g. collagenase type II, protease XXIV) used in the prior methods led to premature digestion of cell surface proteins into peptides, which are then removed during subsequent wash steps and thus unavailable for MS analysis.
Here, we developed a new cardiomyocyte isolation method compatible with frozen tissue that preserves cells for proteomic analysis, including plasma membrane proteins (Fig. 2A). Major differences between this method and those described for fresh tissue include 1) wash and digestion buffers are similar in molecular composition to the ionic composition of intracellular environment of cardiomyocytes (i.e. 150 mM potassium ions, 15 mM sodium ions), 2) digestion buffer contains collagenase type IV (low tryptic activity) and trypsin inhibitor to prevent premature cleavage of proteins, 3) Percoll gradient is used for gentle removal of debris, and 4) CD45 and CD31 negative selection of crude cardiomyocytes is used to remove immune and endothelial cells.
Previous methods for human adult cardiomyocyte isolation did not describe quantitative assessments of cardiomyocyte purity. Imaging can be used to rapidly assess cellular integrity of isolated cardiomyocytes (Fig. 2B), but quantification of cellular heterogeneity is challenging. Flow cytometry can provide quantitative, single-cell measurements to accurately assess population heterogeneity with high sensitivity. However, published protocols describing flow cytometry-based assessment of intact, isolated adult human cardiomyocytes are not yet available. This is likely because intact human adult cardiomyocytes are relatively large, rectangular cells (10–25 μm × 100 μm), which pose technical challenges for standard flow cytometer configurations designed to analyze smaller, spherical cells like blood cells or cultured cell lines. Here, we adopted our previous method for flow cytometry analysis of human stem cell-derived cardiomyocytes [26,27] to work with isolated human adult cardiomyocytes using an acoustic focusing flow cytometer.
On average, starting with 250–500 mg wet weight tissue, we obtain 800 μg of protein from cells that are ≥80% TNNI3-positive (Fig. 2C) based on flow cytometry assessment. During method development, we found that excluding the negative selection step resulted in samples that contained detectable levels of CD45 and CD31 by MS. CD45 and CD31 have been exclusively attributed to non-cardiomyocytes [28,29] and therefore, the negative selection is important for removing these contaminating cells.
3.2. S-trap™ for facile sample preparation
In bottom-up proteomics workflows, complete sample lysis and protein solubilization are key for experimental success. Cardiac tissue and isolated cardiomyocytes can be difficult to disrupt due to varying levels of ECM content and high abundance of contractile proteins. Traditional laboratory detergents that are effective at disrupting fibrous tissue (e.g. SDS) are incompatible with MS, requiring removal prior to injection onto the instrument. Detergent removal is commonly achieved using trichloroacetic acid/acetone precipitation or in-gel digestion, but these methods are laborious and prone to sample loss. Here, we used suspension trapping sample preparation because of the streamlined processing capabilities and ease of use [30]. Samples are solubilized in 5% SDS and loaded onto the S-Trap™ spin column. Protein digestion occurs on-column, and MS-compatible peptides are then eluted from the column. This fast and easy approach to processing bottom-up proteomic samples for MS analysis does not require specialized expertise. A single experimenter can typically generate MS-ready peptide samples in 8 h using this method, including sample homogenization, lysis, and digestion [22]. As part of reproducibility assessment, dot plots of the number of peptide groups found in individual samples of each unfractionated workflows (II and IV) were assessed (Fig. S2).
3.3. Increasing depth of proteome coverage with peptide fractionation and library searching
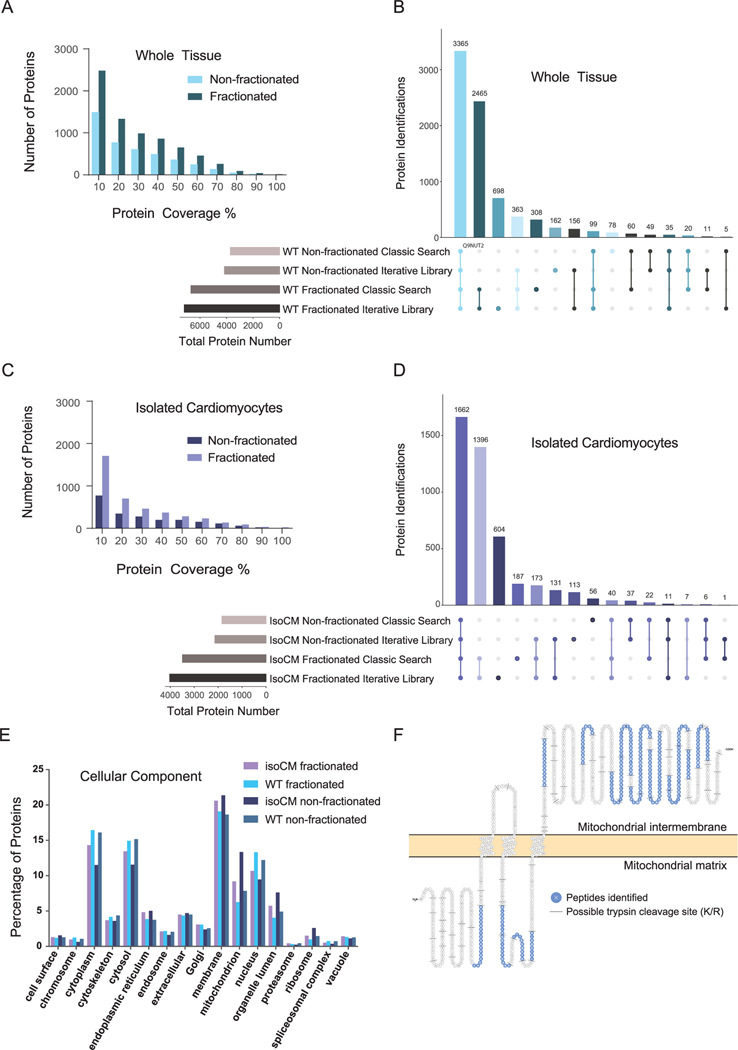
Cardiac tissue and isolated cardiomyocytes are complex samples. As such, limited solubility of membrane and ECM proteins, high molecular heterogeneity, and a large dynamic range of protein concentrations can present limits to the number of proteins identified and the observed sequence coverage of each protein. Often, discovery proteomics experiments can be augmented with off-line peptide fractionation to enhance depth of proteome coverage. This method generates multiple lower complexity samples from a single high complexity sample. Peptide fractionation requires more sample preparation and MS acquisition time but benefits to proteome coverage observed here include 42–47% more proteins identified and 10–12% increase in sequence coverage for common proteins in isolated cardiomyocytes and tissue homogenate (Fig. 3A–D). Moreover, off-line peptide fractionation datasets can be utilized to build libraries for the analysis of quantitative data to increase peptide identifications and confidence in quantitative measurements.
Fig. 3.
Overview of whole tissue and isolated cardiomyocyte proteome. A) Bar chart displaying number of proteins identified at different percentages of coverage in workflows I and II. B) UpSet plot visualizing the intersections of identified proteins from workflows I and II in DDA mode, processed with a classic search and an iterative spectral library search. C) Bar chart displaying number of proteins identified at different percentages of coverage in both workflow III and IV in DDA mode. D) UpSet plot visualizing the intersections of identified proteins from workflow III and IV in DDA mode, processed with a classic search and an iterative spectral library search. E) Percentage of proteins annotated among various cellular components based on Gene Ontology for each workflow. F) Protein sequence showing 34% coverage of mitochondrial potassium channel ATP-binding subunit (ABCB8). For each sample type, three biological replicates (donors) were each analyzed by three technical replicates by MS. UpSet plots were generated using UpSetR [34]. Note: Upset plots graph the intersections of various elements, similar to a Venn diagram. However, unlike a Venn diagram, an upset plot is not limited by a certain number of elements and provides an easier way to interpret the common and uncommon identifications in proteomic datasets.
In addition to strategic sample preparation, the workflow used to process qualitative data can also impact proteome coverage (Fig. 1). In what we define here as a classic search strategy, experimental peptides are matched to theoretical spectra predicted to arise from peptides generated by the digestion of proteins contained in a reference proteome database. In contrast, a spectral library approach matches observed spectra to experimental data previously generated and contained within a spectral library. Spectral matching can increase proteome coverage [31,32] because a spectral pattern is more specific than a database search - like a fingerprint - capable of identifying the peptide despite noise and contaminants [33].
Multiple tools and spectral libraries are available [35]. Here, we use the free software (MSPepSearch) and consensus library made from >10,000 raw files provided by The National Institute of Standards (peptide.NIST.gov) in Proteome Discoverer 2.4. The iterative library search used here begins with a spectral library search followed by a classic search for any spectra that were not matched with high confidence in the library search. We observe that for all sample types, the iterative library search provides more protein identifications than the classic search (Fig. 3B, D). Specifically, identifications increased by 495 protein groups for fractionated whole tissue and increased by 537 protein groups for fractionated isolated cardiomyocytes. The distribution of proteins identified from different subcellular locations were similar between datasets (Fig. 3E). Cell surface proteins were identified in all datasets, confirming that the current isolation protocol preserves plasma membrane proteins. Membrane proteins were the highest represented cellular component, providing evidence that the sample preparation and protein solubilization strategy is effective for this protein class. An example of sequence coverage is shown for mitochondrial potassium channel ATP-binding subunit (ABCB8), a transmembrane protein with known mitochondrial localization (Fig. 3F). Proteins identified in whole tissue with fractionation from a classic search are provided in Table S7 and from the library search in Table S8.
3.4. Analyses of isolated cardiomyocytes provides a deeper view than tissue homogenate
Cardiac tissue is dense and populated by multiple different cell types. The contribution of each cell type varies depending on anatomical region, model system (i.e. human or animal), and disease state [5,29,36,37]. The injured heart undergoes significant pathophysiological changes [38] to adapt and repair the area of insult, also referred to as cardiac remodeling. Cardiac remodeling is characterized by hypertrophy and/or loss of cardiomyocytes, cardiac fibrosis, and proliferation of non-cardiomyocytes (e.g. cardiac fibroblast and immune cells) [38,39]. The fibrotic response is attenuated by the pathological activation of cardiac fibroblasts and immune cells, leading to excessive deposition of ECM proteins and contributes to adverse ECM remodeling [40].
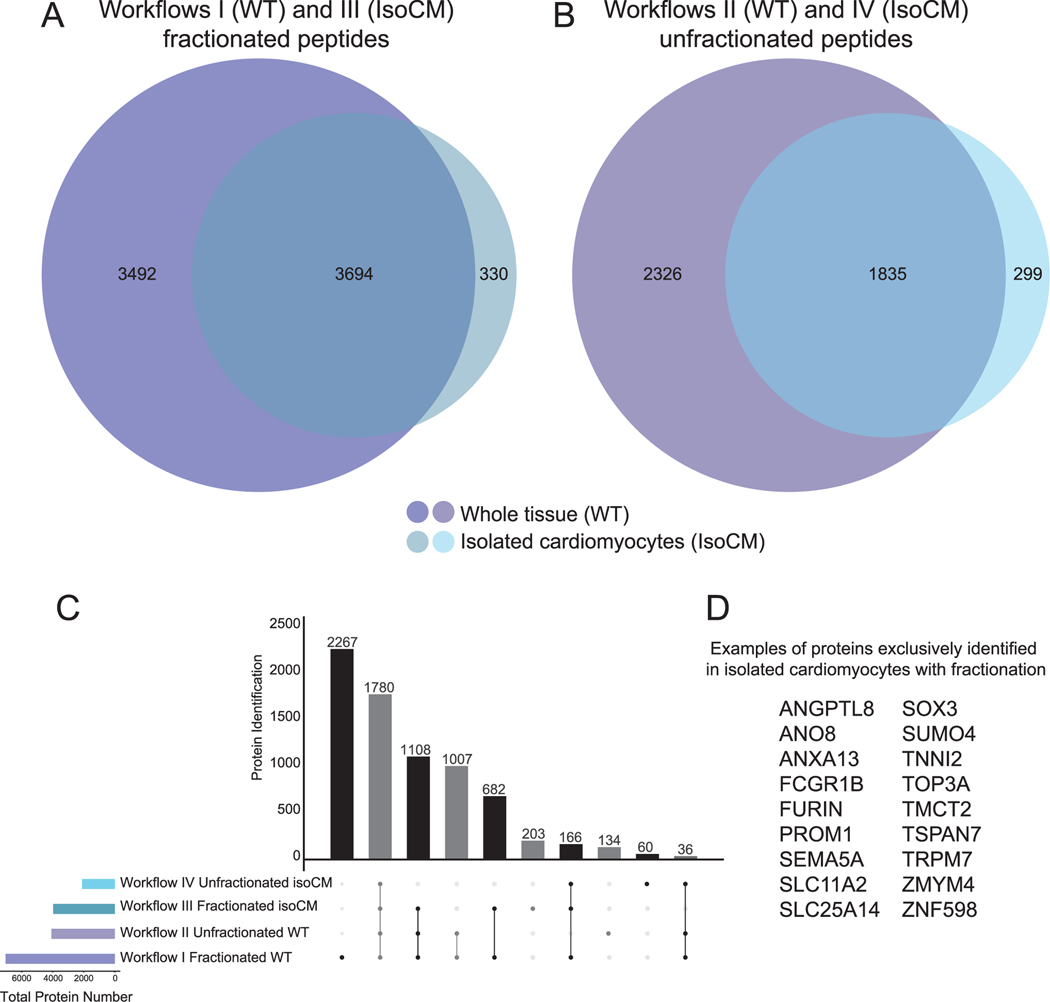
This dynamic heterogeneity can pose challenges to interpreting data from proteomic analyses of tissue homogenate as the cell-type of origin cannot be inferred. Also, as tissue homogenate provides an average view of the proteins across all cell types within the specimen, identifying lower abundance proteins present in a single cell type is challenging. The analysis of isolated cardiomyocytes provides evidence of cell-type localization and a deeper view of the cardiomyocyte proteome. We observed this improvement for fractionated and unfractionated peptide workflows when comparing whole tissue and isolated cardiomyocytes from the apex (Fig. 4A, B). 203 proteins were identified exclusively in fractionated isolated apical cardiomyocytes (Fig. 4C), providing evidence of the benefit of performing cell isolation and peptide fractionation. Of the 203 proteins, examples include proteins with known cell surface localization (e.g. FURIN, FCGR1B, PROM1, SEMA5A, SLC11A2, and TSPAN7) and mitochondrial localization (e.g. SLC25A14, TOP3A), highlighting the advantage of using isolated cells for proteomic analyses (Fig. 4D). Although peptide fractionation can increase proteome coverage (Fig. 4), this process adds additional steps of sample handling including solvent evaporation. It is possible that some peptides are lost in this process, and this could explain the 60 proteins identified uniquely in the isolated cardiomyocytes without fractionation.
Fig. 4.
Deeper view of the proteome is achieved when peptide fractionation is combined with cardiomyocyte isolation. A and B) Venn diagrams displaying total number of proteins identified in apex for each workflow by DDA searched with the iterative spectral library strategy. C) UpSet Plot (generated by UpSetR [34]) showing overlap of proteins identified in unfractionated and fractionated apex whole tissue and isolated cardiomyocytes from apex. D) Examples from among the 203 proteins identified exclusively in isolated cardiomyocytes with fractionation.
3.5. Quantitative comparisons reveal differences among chambers
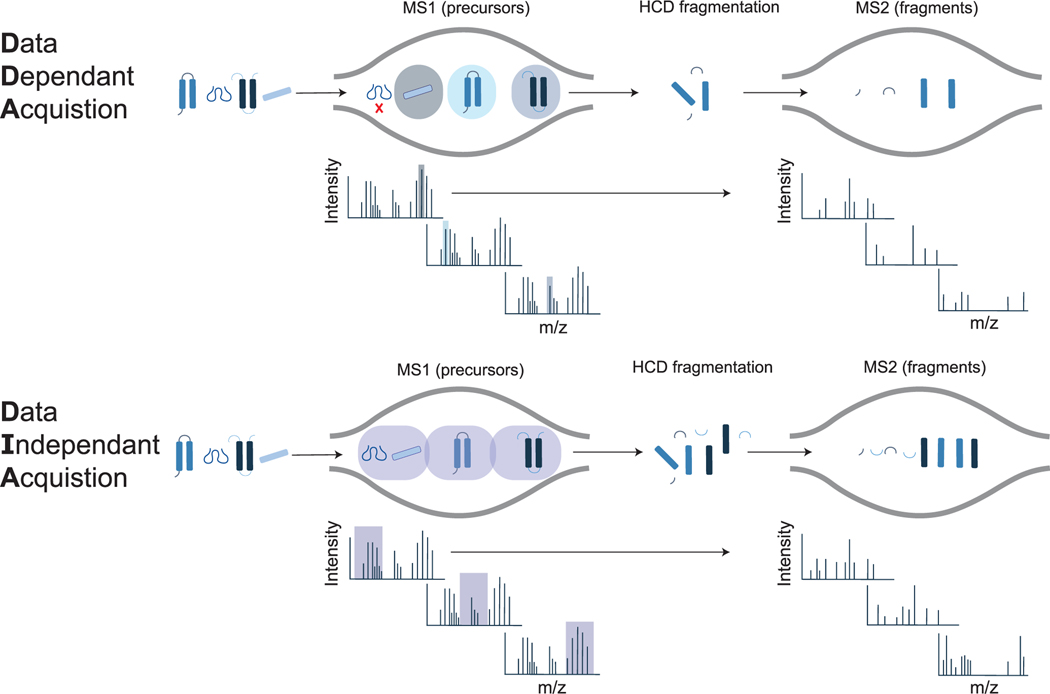
Numerous methods for relative, untargeted quantification of peptides are available, including label-based and label-free methods [41,42]. When working with human tissue, metabolic labels are typically unfeasible, especially for healthy donor hearts. Chemical labels (e. g. TMT, iTRAQ, DiLeu) are possible but are limited by the number of samples that can be compared [43–46]. In contrast, label-free methods use simplified sample preparation workflows, and allow comparison of an unlimited number of samples. However, label-free methods require more data acquisition time because they cannot be multiplexed like label-based methods. Label-free methods include data-dependent acquisition (DDA) and data-independent acquisition (DIA) strategies [47]. In DDA, a single precursor at a time is selected for gas-phase dissociation, whereas in DIA, all precursors within an m/z window are selected for simultaneous dissociation (Fig. 5). In both cases, the precursor and resulting fragment masses are used during data analysis to match the experimentally observed fragmentation pattern to a proposed peptide sequence. DIA data are more complex as the resulting fragmentation spectra are chimeras of multiple precursors, requiring different data analysis workflows than DDA. However, there are advantages to using DIA for quantification. DIA suffers from fewer missing values compared to DDA [48] and the data can be continually re-processed as spectral libraries advance to enhance proteome coverage.
Fig. 5.
Overview of DDA and DIA acquisition methods. In DDA mode, the instrument selects precursors for fragmentation one at a time, which can lead to missing features. In DIA mode, all precursors within a designated m/z window are fragmented sequentially and repeatedly throughout the acquisition, leading to fewer missing features, but complex MS2 spectra.
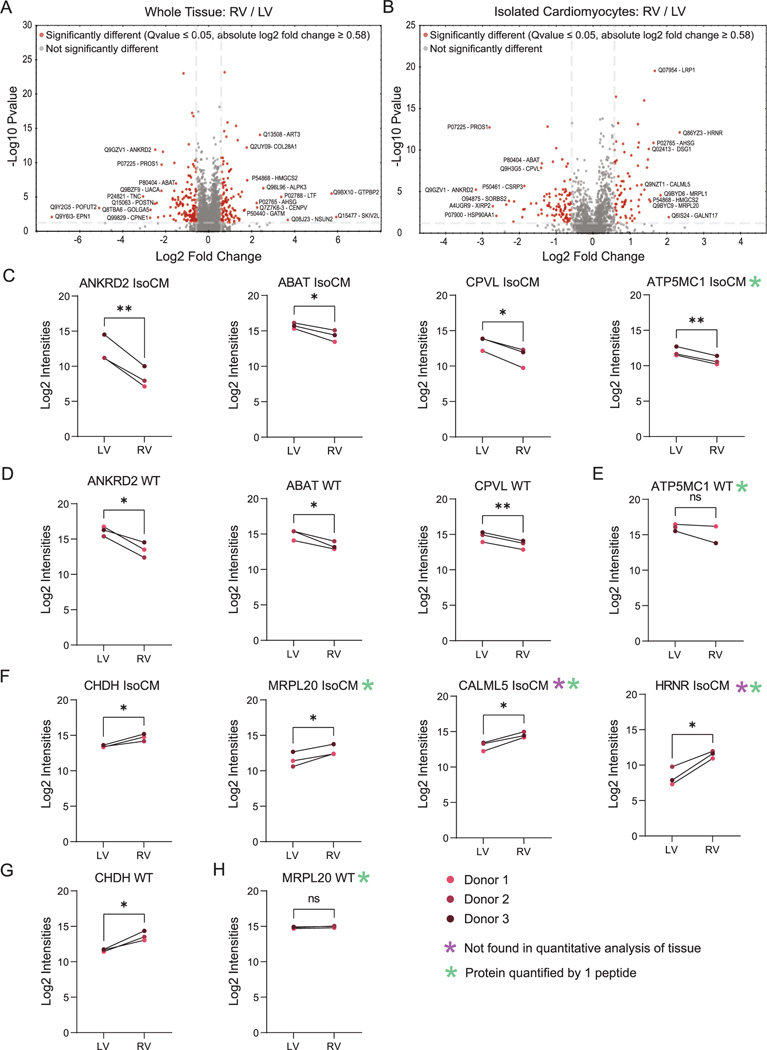
Here, we applied DIA to quantify proteins between left and right ventricle, starting with either whole tissue or isolated cardiomyocytes. Differences in the proteome between chambers can be expected because the cells from each chamber evolve developmentally from different sources (e.g. primary heart field gives rise to the left and right atria and left ventricle; secondary heart field gives rise to right ventricle and outflow tracts). The different chambers also have different functions (i.e. blood is propelled from the right atria to the right ventricle, where it is pumped to the lungs via the pulmonary arteries; from the lungs, blood enters the left atria then ventricle, which pumps oxygen-rich blood to the rest of the body). Of the 3035 protein groups quantified in whole tissue homogenate, 2784 proteins were found at relatively similar abundance between left and right ventricle (Fig. 6A), whereas 141 and 110 were more abundant in left and right, respectively. Of the 1452 protein groups quantified in isolated cardiomyocytes, 1277 proteins were found at relatively similar abundance between right and left ventricles (Fig. 6B), whereas 101 and 74 were more abundant in left and right ventricles, respectively. Of the 175 differentially abundant proteins found in isolated cardiomyocytes, 134 were only found in isolated cardiomyocytes and not in the whole tissue analyses (Supplemental Table S9). Of those proteins quantified in isolated cardiomyocytes, ANKRD2, ABAT, CPVL, and ATP5MC1 were higher in abundance in left compared to right ventricular cardiomyocytes (Fig. 6B). Although similar trends were observed for four of the proteins in whole tissue, ATP5MC1 showed no significant difference between left and right ventricular whole tissue (Fig. 6C). Examples of proteins with higher abundance in right compared with left ventricular cardiomyocytes were CHDH, MRPL20, CALML5, and HRNR (Fig. 6E). Whereas abundance of CHDH showed a similar trend in whole tissue (Fig. 6F), MRPL20 was not significantly different (Fig. 6G), and CALML5 and HRNR were not detected in the quantitative analysis of left and right whole tissue (Fig. 6H). These data demonstrate the advantages of using isolated cells in addition to whole tissue analyses for expanding the view of proteomic differences among chambers of the human heart.
Fig. 6.
Overview of differentially abundant proteins in homogenized cardiac tissue and isolated cardiomyocytes using workflow II and IV in DIA mode. Volcano plot displaying differentially abundant proteins between A) left ventricular and right ventricular tissue and B) isolated cardiomyocytes from left and right ventricles. Horizontal bar represents a significance level of Qvalue ≤0.05. Vertical bars represent a significance level of log2 fold change ≥0.58 (right) and ≤ − 0.58 (left). Log2 intensities measurements reveal proteins that were higher in abundance in C) left ventricular cardiomyocytes (ANKRD2, ABAT, CPLV, ATP5MC1) and D) tissue. E) ATP5MC1 was not significantly more abundant when comparing left and right ventricular tissue. Log2 intensities measurements reveal proteins that were higher in abundance in F) right ventricular cardiomyocytes (CHDH, MRPL20, HRNR, CALML5) and G) tissue. H) MRPL20 was not significantly more abundant when comparing left and right ventricular tissue. Purple asterisk indicates proteins not detected in the quantitative analysis of whole tissue. Green asterisk indicates proteins that were quantified by 1 peptide. All other proteins were quantified by 2 or more peptides. For each sample type, three biological replicates (donors) were analyzed by three technical replicates by MS. (pairwise t-test; *p < 0.05, **p < 0.01). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
4.1. Cardiomyocyte isolation and purity
Human adult cardiomyocytes are more challenging to isolate compared to those from animal models because the tissue is commonly derived from surgical discards and it is typically not possible to perfuse an intact human heart with the solutions and buffers used to enhance tissue dissociation in animal models. While methods for isolating human adult cardiomyocytes from fresh tissue have been described [17–21], we find these methods do not preserve plasma membrane proteins when applied to cryopreserved tissue. Here, we describe a new method to isolate human adult cardiomyocytes from 250 to 500 mg cryopreserved tissue and assess purity by flow cytometry. This method allows tissues to be collected as they become available and stored until enough samples for statistical power have been acquired. Subsequently, isolations can proceed on all samples selected for the study, which should help to minimize technical variation during the isolation.
We have not tested this protocol on freshly isolated tissue, but we expect that the protocol for isolating cardiomyocytes from fresh tissue will be similar, with the exception of the buffer solutions. Specifically, we observed that buffers with high sodium concentration (e.g. 140 mM sodium salts) led to increased swelling of cardiomyocytes and subsequently damaged cardiomyocytes. We therefore developed buffer solutions that mimic the intracellular environment (i.e. 150 mM potassium salts) to prevent swelling and damage to the cardiomyocytes. This would not be necessary when working with fresh tissue as shown previously [20,21].
Collagenase type II is often recommended for digestion of cardiac tissue because of its efficiency in digesting tough tissue. Due to its high tryptic activity, it prematurely digests cell surface proteins prior to proteomics experiments. Collagenase type IV is best suited to preserve cell surface proteins, which is consistent with recommendations from Worthington Biochemical (product datasheet), which state that collagenase type IV limits the damage to membrane proteins and receptors. Following enzymatic and mechanical digestion of cardiac tissue, subjecting crude cardiomyocytes to the Percoll gradient as well as performing CD31/CD45 negative selection on cardiomyocytes are associated with some loss of cardiomyocytes. However, in our experience it did not affect our ability to perform follow up experiments. For most preparations, there is enough material for flow cytometric analysis as well as proteomics experiments or other follow up experiments (e.g. isolation of RNA, staining of cardiomyocytes). The Percoll gradient also removes smaller cell types, but not to purity. This is evidenced by our quality control experiments where we noticed that CD31 and CD45 were identified by MS, which prompted us to include a negative selection step.
We have tested a penultimate version of this protocol on snap frozen tissue and noticed that the morphology of the cardiomyocytes was greatly compromised during the freezing process, which prevented the isolation of structurally sound cardiomyocytes. Therefore, we did not pursue this approach further. We tested this method on tissue preserved in AllProtect®, which did not produce high yield of cardiomyocytes. It was challenging to isolate cardiomyocytes from tissue preserved in AllProtect® as the tissue becomes hard and is surrounded by a rubbery layer, which makes it difficult for digestion enzymes to penetrate the tissue.
Cell purity is important when interpreting data from ‘omics approaches. The flow cytometry protocol described here is applicable for determining the percentage of cells positive for the cardiomyocyte-specific marker TNNI3 by comparing the fluorescence intensity between the antibody-stained cells and the isotype control. The method provides excellent separation between the antibody-stained cells and unstained control. However, the isotype control is shifted in fluorescence intensity from the unstained control. This suggests that further efforts to improve the method may be necessary to increase the separation between the antibody-stained cells and the isotype control.
Common non-cardiomyocyte proteins were not detected by MS in the isolated cardiomyocytes, so we do not have evidence that the isolated cardiomyocytes contain significant numbers of contaminating cell types. Common endothelial cell markers CD31 and CD105 were detected in the quantitative analysis of whole cardiac tissue of left and right ventricles but not in isolated cardiomyocytes. Common fibroblast markers vimentin and CD90 were detected in the quantitative analysis of whole cardiac tissue of left and right ventricles but not in isolated CM. Vimentin is also considered a smooth muscle marker. We did not identify the common immune cell markers CD45 and CD11b in whole cardiac tissue and isolated CM, but this could be due to the low abundance of immune cells in healthy cardiac tissue.
Thus, although imperfect at this time, this is the first method for flow cytometry analysis of intact human adult cardiomyocytes and should aid other investigators in performing this type of assessment. We expect further refinement of the method (e.g. gentler fixation and per-meabilization conditions) will improve separation of antibody-stained and isotype control and thus increase the percentage of cells considered to be TNNI3-positive. The cells isolated by this method are suitable for a wide range of proteomic experiments beyond the methodologies applied here.
4.2. Selecting a proteomics method that is right for your study
There is no one-size-fits-all proteomics method. All bottom-up proteomics methods are biased in some way - typically towards identifying peptides from proteins that are most soluble or abundant under the experimental conditions used. Results obtained here show advantages and disadvantages among the approaches – including expanded coverage obtained when combining cell isolation with fractionation at the cost of more labor and time. We expect these results will aid other investigators to select a workflow that is most apt for their study. Additionally, cardiomyocytes isolated from cryopreserved tissue may experience more stress than those isolated from fresh tissue. Although we have not detected stress pathway proteins (e.g. calpain, caspase) in the quantitative analysis of isolated cardiomyocytes, additional experiments are required to determine their activation in cardiomyocytes isolated from cardiac tissue preserved in freezing media, which may impact the utility of isolated cardiomyocytes for certain study designs.
We describe a facile method for preparation of cardiac samples for bottom-up proteomic analyses. Bottom-up sample preparation for cardiac tissue has traditionally required cumbersome methods to solubilize the fibrous tissue and remove contaminating detergents before MS analysis. The S-trap™ method used here, in conjunction with rigorous mechanical disruption, allows for solubilization of cardiac tissue proteins, enzymatic digestion, and cleanup within one workday [49]. The LC column and MS settings used here are common among laboratories, but different LC columns (e.g. different length, particle pore size) could be used and may improve sequence coverage. The DDA data generated here are available and can be used by other investigators interested in mining this discovery dataset and to build spectral libraries for future DDA or DIA analyses. We used DIA for relative peptide quantitation, but other methods are possible, and the choice will depend on study goals, access to MS instrumentation, costs, and time. The proteomics results will depend on the configuration and performance of the MS instrumentation and software used to analyze the data. During experimental planning, users are advised to consult with the MS instrumentation operator to ensure the design is appropriate for the instrumentation and software used for analysis.
4.3. Expectations for the level of expertise required to perform analyses described here
Here, we summarize the level of expertise required to perform the experiments described in this “how to” article.
Cardiomyocyte isolation – basic experience in primary cell isolation required. With more experience, the user will be able to adjust the protocol to work with lower integrity tissues.
Flow cytometry – basic experience in flow cytometry and access to a flow cytometer with capabilities of analyzing large cells is required.
Sample homogenization and digestion – no specialized experience required.
Mass Spectrometry – expertise in MS required. Non-experts should consult with MS operators before designing experiments to receive guidance specific to the instrumentation and data analysis software available.
ProteomeDiscoverer – requires MS expertise to setup the data processing, but search results can be provided to non-experts for review.
Spectronaut – requires MS expertise to setup the data processing but does not require manual peak picking, so data analysis proceeds rapidly. Validated statistical methods are included to help new users comply with standards in the field.
4.4. Future studies
Future studies can apply the described cardiomyocyte isolation and proteomic sample preparation methods to specimens from additional anatomical regions, focusing on sub-anatomical regions (e.g. epicardial versus endocardial) to generate a more precise spatial mapping of the human heart proteome. Peptide samples from these methods can be combined with strategies that enrich for specific classes of post-translational modifications (e.g. phosphorylation, ubiquitination, glycosylation) to provide enhanced detection of modifications compared to unenriched samples. It is also possible to simultaneously extract proteins and metabolites using the recently described SiTrap™ [30] to develop complementary views of the molecular landscape in the heart. Expanding the data analysis approach to include proteogenomic approaches that use RNA-seq data to define sample-specific protein sequence databases should enable precise definitions of the cardiac proteome, including the impact of alternative splicing on truncations and altered sites of post-translational modifications [50]. Finally, while the example data provided here were generated from non-failing donor hearts, the methodologies are expected to be broadly applicable to exploring protein dynamics in cardiac disease [51–53].
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health [R01-HL134010, R01-HL126785; R35-HL155460 to R.L.G.], American Heart Association [20PRE35200049 to L.B.L.]. LBL is a member of the MCW-MSTP which is partially supported by a T32 grant from NIGMS, GM080202. Funding sources were not involved in study design, data collection, interpretation, analysis or publication. Flow cytometry was performed using instrumentation in the UNMC Flow Cytometry Research Facility, administrated through the Office of the Vice Chancellor for Research, and supported by state funds from the Nebraska Research Initiative (NRI) and The Fred and Pamela Buffett Cancer Center’s National Cancer Institute Cancer Support Grant. Major instrumentation has been provided by the Office of the Vice Chancellor for Research, The University of Nebraska Foundation, the Nebraska Banker’s Fund, and by the NIH-NCRR Shared Instrument Program.
Footnotes
Declaration of competing interest
R.L.G. is on the advisory board of ProtiFi, LLC and receives no compensation of any kind for this role.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.yjmcc.2021.08.008.
References
- [1].Fountoulaki K, Cardiothoracic Intensive Care Unit, Onassis Cardiac Surgery Centre, Athens, Greece, N. Dagres, Second University Department of Cardiology, Attikon General Hospial, University of Athens, Athens, Greece, Cellular communications in the heart, Card. Fail. Rev 1 (2015) 64, 10.15420/cfr.2015.1.2.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Martins-Marques T, Hausenloy DJ, Sluijter JPG, Leybaert L, Girao H, Intercellular communication in the heart: therapeutic opportunities for cardiac ischemia, Trends Mol. Med 27 (2021) 248–262, 10.1016/j.molmed.2020.10.002. [DOI] [PubMed] [Google Scholar]
- [3].Asp M, Giacomello S, Larsson L, Wu C, Fürth D, Qian X, Wärdell E, Custodio J, Reimegård J, Salmén F, Österholm C, Ståhl PL, Sundström E, Åkesson E, Bergmann O, Bienko M, Månsson-Broberg A, Nilsson M, Sylvén C, Lundeberg J, A spatiotemporal organ-wide gene expression and cell atlas of the developing human heart, Cell 179 (2019) 1647–1660.e19, 10.1016/j.cell.2019.11.025. [DOI] [PubMed] [Google Scholar]
- [4].Tucker Nathan R., Mark Chaffin, Stephen J. Fleming, Amelia W. Hall, Victoria A. Parsons, Kenneth C. Bedi, Amer-Denis Akkad, Caroline N. Herndon, Alessandro Arduini, Irinna Papangeli, Carolina Roselli, François Aguet, Hoan Choi Seung, Kristin G. Ardlie, Mehrtash Babadi, Kenneth B. Margulies, Christian M. Stegmann, Patrick T. Ellinor, Transcriptional and cellular diversity of the human heart, Circulation 142 (2020) 466–482, 10.1161/CIRCULATIONAHA.119.045401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Litviñuková M, Talavera-López C, Maatz H, Reichart D, Worth CL, Lindberg EL, Kanda M, Polanski K, Heinig M, Lee M, Nadelmann ER, Roberts K, Tuck L, Fasouli ES, DeLaughter DM, McDonough B, Wakimoto H, Gorham JL, Samari S, Mahbubani KT, Saeb-Parsy K, Patone G, Boyle JJ, Zhang H, Zhang H, Viveiros A, Oudit GY, Bayraktar OA, Seidman JG, Seidman CE, Noseda M, Hubner N, Teichmann SA, Cells of the adult human heart, Nature 588 (2020) 466–472, 10.1038/s41586-020-2797-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kline KG, Frewen B, Bristow MR, MacCoss MJ, Wu CC, High quality catalog of proteotypic peptides from human heart, J. Proteome Res 7 (2008) 5055–5061, 10.1021/pr800239e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Westbrook JA, Wheeler JX, Wait R, Welson SY, Dunn MJ, The human heart proteome: two-dimensional maps using narrow-range immobilised pH gradients, Electrophoresis 27 (2006) 1547–1555, 10.1002/elps.200500777. [DOI] [PubMed] [Google Scholar]
- [8].Tucholski T, Cai W, Gregorich ZR, Bayne EF, Mitchell SD, McIlwain SJ, de Lange WJ, Wrobbel M, Karp H, Hite Z, Vikhorev PG, Marston SB, Lal S, Li A, dos Remedios C, Kohmoto T, Hermsen J, Ralphe JC, Kamp TJ, Moss RL, Ge Y, Distinct hypertrophic cardiomyopathy genotypes result in convergent sarcomeric proteoform profiles revealed by top-down proteomics, Proc. Natl. Acad. Sci 117 (2020) 24691–24700, 10.1073/pnas.2006764117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Xiang F, Shi Z, Guo X, Qiu Z, Chen X, Huang F, Sha J, Chen X, Proteomic analysis of myocardial tissue from the border zone during early stage post-infarct remodelling in rats, Eur. J. Heart Fail 13 (2011) 254–263, 10.1093/eurjhf/hfq196. [DOI] [PubMed] [Google Scholar]
- [10].Liu T, Chen L, Kim E, Tran D, Phinney BS, Knowlton AA, Mitochondrial proteome remodeling in ischemic heart failure, Life Sci. 101 (2014) 27–36, 10.1016/j.lfs.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nishtala K, Phong TQ, Steil L, Sauter M, Salazar MG, Kandolf R, Kroemer HK, Felix SB, Volker U, Klingel K, Hammer E, Virus-Induced Dilated Cardiomyopathy is Characterized By Increased Levels of Fibrotic Extracellular Matrix Proteins and Reduced Amounts of Energy-Producing Enzymes, 2011, p. 12. [DOI] [PubMed] [Google Scholar]
- [12].Lu D, Xia Y, Chen Z, Chen A, Wu Y, Jia J, Sun A, Zou Y, Qian J, Ge J, Cardiac proteome profiling in ischemic and dilated cardiomyopathy mouse models, Front. Physiol 10 (2019) 750, 10.3389/fphys.2019.00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Aye TT, Scholten A, Taouatas N, Varro A, Van Veen TAB, Vos MA, Heck AJR, Proteome-wide protein concentrations in the human heart, Mol. BioSyst 6 (2010) 1917, 10.1039/c004495d. [DOI] [PubMed] [Google Scholar]
- [14].Lau E, Cao Q, Ng DCM, Bleakley BJ, Dincer TU, Bot BM, Wang D, Liem DA, Lam MPY, Ge J, Ping P, A large dataset of protein dynamics in the mammalian heart proteome, Sci. Data 3 (2016) 160015, 10.1038/sdata.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Doll S, Dreßen M, Geyer PE, Itzhak DN, Braun C, Doppler SA, Meier F, Deutsch M-A, Lahm H, Lange R, Krane M, Mann M, Region and cell-type resolved quantitative proteomic map of the human heart, Nat. Commun 8 (2017) 1469, 10.1038/s41467-017-01747-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Linscheid N, Poulsen PC, Pedersen ID, Gregers E, Svendsen JH, Olesen MS, Olsen JV, Delmar M, Lundby A, Quantitative proteomics of human heart samples collected in vivo reveal the remodeled protein landscape of dilated left atrium without atrial fibrillation, Mol. Cell. Proteomics 19 (2020) 1132–1144, 10.1074/mcp.RA119.001878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kaestner L, Schwarz S, Lipp P, Isolation of Human Atrial Myocytes: Characteristics and Improvements, 2012, p. 13. [Google Scholar]
- [18].Voigt N, Zhou X-B, Dobrev D, Isolation of human atrial myocytes for simultaneous measurements of Ca2+ transients and membrane currents, J. Vis. Exp 50235 (2013), 10.3791/50235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Coppini R, Ferrantini C, Aiazzi A, Mazzoni L, Sartiani L, Mugelli A, Poggesi C, Cerbai E, Isolation and functional characterization of human ventricular cardiomyocytes from fresh surgical samples, J. Vis. Exp 51116 (2014), 10.3791/51116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Guo G, Chen L, Rao M, Chen K, Song J, Hu S, A modified method for isolation of human cardiomyocytes to model cardiac diseases, J. Transl. Med 16 (2018) 288, 10.1186/s12967-018-1649-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Fiegle DJ, Volk T, Seidel T, Isolation of human ventricular cardiomyocytes from vibratome-cut myocardial slices, J. Vis. Exp 61167 (2020), 10.3791/61167. [DOI] [PubMed] [Google Scholar]
- [22].Wojtkiewicz M, Berg Luecke L, Kelly MI, Gundry RL, Facile preparation of peptides for mass spectrometry analysis in bottom-up proteomics workflows, Curr. Protoc 1 (2021), 10.1002/cpz1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yang F, Shen Y, Camp DG 2nd, Smith RD, High-pH reversed-phase chromatography with fraction concatenation for 2D proteomic analysis, Expert Rev. Proteomics 9 (2012) 129–134, 10.1586/epr.12.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Eng JK, McCormack AL, Yates JR, An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database, J. Am. Soc. Mass Spectrom 5 (1994) 976–989, 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- [25].Zhang Z, Burke M, Mirokhin YA, Tchekhovskoi DV, Markey SP, Yu W, Chaerkady R, Hess S, Stein SE, Reverse and random decoy methods for false discovery rate estimation in high mass accuracy peptide spectral library searches, J. Proteome Res 17 (2018) 846–857, 10.1021/acs.jproteome.7b00614. [DOI] [PubMed] [Google Scholar]
- [26].Waas M, Weerasekera R, Kropp EM, Romero-Tejeda M, Poon EN, Boheler KR, Burridge PW, Gundry RL, Are these cardiomyocytes? Protocol development reveals impact of sample preparation on the accuracy of identifying cardiomyocytes by flow cytometry, Stem Cell Rep. 12 (2019) 395–410, 10.1016/j.stemcr.2018.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Berg Luecke L, Waas M, Gundry RL, Reliable protocols for flow cytometry analysis of intracellular proteins in pluripotent stem cell derivatives: a fit-for-purpose approach, Curr. Protoc. Stem Cell Biol 50 (2019), e94, 10.1002/cpsc.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Donovan A, Koretzky A, CD45 and the immune response, J. Am. Soc. Nephrol 10 (1993). [DOI] [PubMed] [Google Scholar]
- [29].Pinto AR, Ilinykh A, Ivey MJ, Kuwabara JT, D’antoni ML, Debuque R, Chandran A, Wang L, Arora K, Rosenthal NA, Tallquist MD, Revisiting cardiac cellular composition, Circ. Res 118 (2016) 400–409, 10.1161/CIRCRESAHA.115.307778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zougman A, Selby PJ, Banks RE, Suspension trapping (STrap) sample preparation method for bottom-up proteomics analysis, Proteomics 14 (2014), 10.1002/pmic.201300553, 1006–1000. [DOI] [PubMed] [Google Scholar]
- [31].Cho J-Y, Lee H-J, Jeong S-K, Kim K-Y, Kwon K-H, Yoo JS, Omenn GS, Baker MS, Hancock WS, Paik Y-K, Combination of multiple spectral libraries improves the current search methods used to identify missing proteins in the chromosome-centric human proteome project, J. Proteome Res 14 (2015) 4959–4966, 10.1021/acs.jproteome.5b00578. [DOI] [PubMed] [Google Scholar]
- [32].Fernández-Costa C, Martínez-Bartolomé S, McClatchy DB, Saviola AJ, Yu N-K, Yates JR, Impact of the identification strategy on the reproducibility of the DDA and DIA results, J. Proteome Res 19 (2020) 3153–3161, 10.1021/acs.jproteome.0c00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lam H, Building and searching tandem mass spectral libraries for peptide identification, Mol. Cell. Proteomics MCP 10 (2011), 10.1074/mcp.R111.008565.R111.008565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H, UpSet: visualization of intersecting sets, IEEE Trans. Vis. Comput. Graph 20 (2014) 1983–1992, 10.1109/TVCG.2014.2346248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Griss J, Spectral library searching in proteomics, Proteomics 16 (2016) 729–740, 10.1002/pmic.201500296. [DOI] [PubMed] [Google Scholar]
- [36].Bergmann O, Zdunek S, Felker A, Jovinge S, Zdunek S, Felker A, Dynamics of cell generation and turnover in the human heart, Cell 161 (2015) 1566–1575, 10.1016/j.cell.2015.05.026. [DOI] [PubMed] [Google Scholar]
- [37].Souders CA, Borg TK, Banerjee I, Baudino TA, Pressure overload induces early morphological changes in the heart, Am. J. Pathol 181 (2012) 1226–1235, 10.1016/j.ajpath.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Goldsmith EC, Borg TK, The dynamic interaction of the extracellular matrix in cardiac remodeling, J. Card. Fail 8 (2002) S314–S318, 10.1054/jcaf.2002.129258. [DOI] [PubMed] [Google Scholar]
- [39].Cohn JN, Ferrari R, Sharpe N, Cardiac remodeling—concepts and clinical implications: a consensus paper from an international forum on cardiac remodeling, J. Am. Coll. Cardiol 35 (2000) 569–582, 10.1016/S0735-1097(99)00630-0. [DOI] [PubMed] [Google Scholar]
- [40].Weber KT, Sun Y, Bhattacharya SK, Ahokas RA, Gerling IC, Myofibroblast-mediated mechanisms of pathological remodelling of the heart, Nat. Rev. Cardiol 10 (2013) 15–26, 10.1038/nrcardio.2012.158. [DOI] [PubMed] [Google Scholar]
- [41].Meyer JG, Schilling B, Clinical applications of quantitative proteomics using targeted and untargeted data-independent acquisition techniques, Expert Rev. Proteomics 14 (2017) 419–429, 10.1080/14789450.2017.1322904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Vidova V, Spacil Z, A review on mass spectrometry-based quantitative proteomics: targeted and data independent acquisition, Anal. Chim. Acta 964 (2017) 7–23, 10.1016/j.aca.2017.01.059. [DOI] [PubMed] [Google Scholar]
- [43].Wang H, Alvarez S, Hicks LM, Comprehensive comparison of iTRAQ and label-free LC-based quantitative proteomics approaches using two Chlamydomonas reinhardtii strains of interest for biofuels engineering, J. Proteome Res 11 (2012) 487–501, 10.1021/pr2008225. [DOI] [PubMed] [Google Scholar]
- [44].Xiang F, Ye H, Chen R, Fu Q, Li L, N,N-dimethyl leucines as novel isobaric tandem mass tags for quantitative proteomics and peptidomics, Anal. Chem 82 (2010) 2817–2825, 10.1021/ac902778d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, Burkhard PR, Sanchez J-C, Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags, Anal. Chem 80 (2008) 2921–2931, 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- [46].Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ, Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents, Mol. Cell. Proteomics MCP 3 (2004) 1154–1169, 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- [47].Liu H, Sadygov RG, Yates JR, A model for random sampling and estimation of relative protein abundance in shotgun proteomics, Anal. Chem 76 (2004) 4193–4201, 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- [48].Barkovits K, Pacharra S, Pfeiffer K, Steinbach S, Eisenacher M, Marcus K, Uszkoreit J, Reproducibility, specificity and accuracy of relative quantification using spectral library-based data-independent acquisition, Mol. Cell. Proteomics MCP 19 (2020) 181–197, 10.1074/mcp.RA119.001714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ludwig KR, Schroll MM, Hummon AB, Comparison of in-solution, FASP, and S-trap based digestion methods for bottom-up proteomic studies, J. Proteome Res 17 (2018) 2480–2490, 10.1021/acs.jproteome.8b00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lau E, Han Y, Williams DR, Thomas CT, Shrestha R, Wu JC, Lam MPY, Splice-junction-based mapping of alternative isoforms in the human proteome, Cell Rep. 29 (2019), 10.1016/j.celrep.2019.11.026, 3751–3765.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Mouton AJ, DeLeon-Pennell KY, Rivera Gonzalez OJ, Flynn ER, Freeman TC, Saucerman JJ, Garrett MR, Ma Y, Harmancey R, Lindsey ML, Mapping macrophage polarization over the myocardial infarction time continuum, Basic Res. Cardiol 113 (2018) 26, 10.1007/s00395-018-0686-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Rookyard AW, Paulech J, Thyssen S, Liddy KA, Puckeridge M, Li DK, White MY, Cordwell SJ, A global profile of reversible and irreversible cysteine redox post-translational modifications during myocardial ischemia/reperfusion injury and antioxidant intervention, Antioxid. Redox Signal 34 (2021) 11–31, 10.1089/ars.2019.7765. [DOI] [PubMed] [Google Scholar]
- [53].Pitoulis FG, Hasan W, Papadaki M, Clavere NG, Perbellini F, Harding SE, Kirk JA, Boateng SY, de Tombe PP, Terracciano CM, Intact myocardial preparations reveal intrinsic transmural heterogeneity in cardiac mechanics, J. Mol. Cell. Cardiol 141 (2020) 11–16, 10.1016/j.yjmcc.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.