ABSTRACT
Epigenetic age has emerged as an important biomarker of biological ageing. It has revealed that some tissues age faster than others, which is vital to understanding the complex phenomenon of ageing and developing effective interventions. Previous studies have demonstrated that humans exhibit heterogeneity in pace of epigenetic ageing among brain structures that are consistent with differences in structural and microanatomical deterioration. Here, we add comparative data on epigenetic brain ageing for chimpanzees, humans’ closest relatives. Such comparisons can further our understanding of which aspects of human ageing are evolutionarily conserved or specific to our species, especially given that humans are distinguished by a long lifespan, large brain, and, potentially, more severe neurodegeneration with age. Specifically, we investigated epigenetic ageing of the dorsolateral prefrontal cortex and cerebellum, of humans and chimpanzees by generating genome-wide CpG methylation data and applying established epigenetic clock algorithms to produce estimates of biological age for these tissues. We found that both species exhibit relatively slow epigenetic ageing in the brain relative to blood. Between brain structures, humans show a faster rate of epigenetic ageing in the dorsolateral prefrontal cortex compared to the cerebellum, which is consistent with previous findings. Chimpanzees, in contrast, show comparable rates of epigenetic ageing in the two brain structures. Greater epigenetic change in the human dorsolateral prefrontal cortex compared to the cerebellum may reflect both the protracted development of this structure in humans and its greater age-related vulnerability to neurodegenerative pathology.
KEYWORDS: Methylation, ageing, development, neuroscience, Pan troglodytes, Alzheimer‘s disease
Introduction
Studies over the last decade have established patterns of predictable methylation change at some CpG sites in the genome over the lifespan that is consistent across individuals [1–3]. These findings have led to the development of ‘epigenetic clocks’ that can be used to predict an individual’s chronological age from methylation levels with high accuracy [2,3]. Moreover, ‘epigenetic age,’ or an individual’s age as predicted from methylation levels using one of these epigenetic clock algorithms, is a biomarker of ageing that reflects differences in rate of development and ageing on many biological levels, including within an individual among tissues, among individuals, and among species with differing growth and senescence patterns [4–10].
In the brain, an accelerated epigenetic age has been associated with cognitive decline [11,12], age-related neuroimaging phenotypes [11], white matter tract integrity [13], decreases in neuron proportion in the prefrontal cortex [14], and Alzheimer’s disease symptoms [15,16]. Different brain regions age epigenetically at different rates [7] in ways that are consistent with observations of differences in age-related structural changes [17–19].
Humans may also differ from other species in brain region-specific differences in rate of ageing, which could provide insights into heterochrony, or divergence in pace of organismal development or ageing, across species. Heterochrony in development can result in distinct adult phenotypes and heterochrony in ageing may reflect divergence in life history strategy [20,21] and be important for understanding aetiologies of species-specific, age-related pathology [22]. Primate species show differences in tissue-specific patterns of methylation across the genome [23–25], some of which likely reflect the outcomes of species-specific developmental programmes. Brain ageing in nonhuman primates shares many features with humans [26–35] and some Alzheimer’s disease-like neuropathology has even recently been documented in some very old great apes [36–38]. However, age-related neuropathology, structural deterioration, and cognitive decline in nonhuman primates are thought to be generally milder than in humans [33,37,39–42].
To date, studies of epigenetic ageing in the brain have been limited and have not included primate species other than humans [7,16]. A cross-tissue epigenetic clock has been validated for chimpanzees [3,43], which can be applied to analyse the comparative neurobiology of ageing. To identify and investigate the human-specific aspects of epigenetic ageing, we examined comparative age-related epigenetic change in the brains of both humans and chimpanzees (Pan troglodytes).
Chimpanzees mature earlier and do not live as long as humans [44]. Our previous work [43] has found that chimpanzees have an overall faster rate of epigenetic ageing in blood than humans, which likely reflects divergence in overall organismal ageing between the two species, and a generally faster pace of biological ageing in chimpanzees, in particular. There is variation in pace of ageing and deterioration in different organs and tissues (e.g., immunosenescence, reproductive senescence, and brain ageing may be decoupled) [45]. We aimed to assess differences between humans and our closest relatives in epigenetic ageing in the brain. We specifically studied two developmentally and functionally distinct brain structures: the dorsolateral prefrontal cortex (DLPFC) and lateral cerebellum. These two structures display evolutionary changes along the human lineage and both may contribute to human cognitive capacities, making them promising candidates for comparative study [46–52]. Specifically, the DLPFC plays a role in distinctive aspects of human cognition related to executive functions, language use, and planning abilities [53,54]. The lateral cerebellum is relatively enlarged in humans and other great apes [46,47,52], and has extensive interconnections with cortical regions involved in the intricate sensorimotor control and sequencing of actions required for tool manipulation and language production [52,55].
For each of these structures, we profiled methylation in young and old adult humans and chimpanzees, and then compared age differences between structures and species. Because we were interested in differences in species patterns of epigenetic brain ageing rather than a simple comparison of rate of brain ageing between species, the chimpanzee and human subjects we selected for study for each age group were not of the same chronological age, but rather of equivalent relative ages, taking into account overall differences between the two species in life history pacing (see Figure 1(a)).
Figure 1.
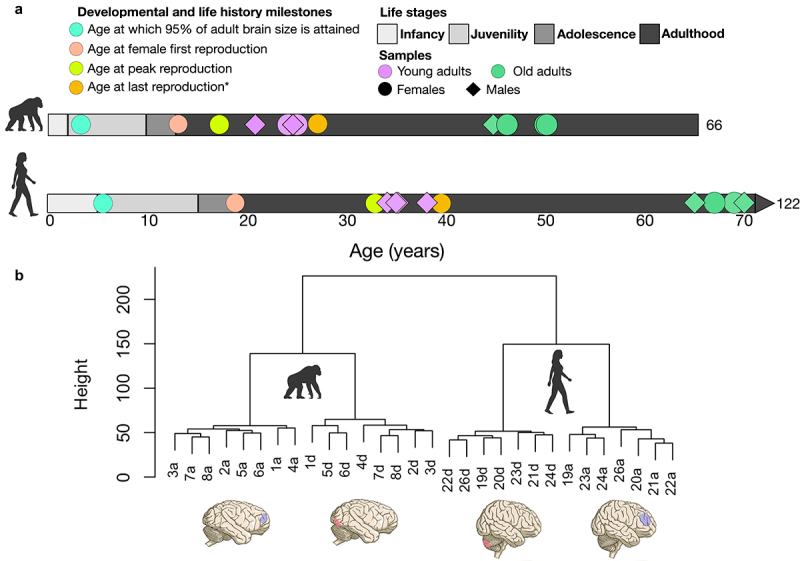
Life history stages are from [56]. Infancy is defined as birth until weaning, juvenility from weaning until menarche, adolescence from menarche until the onset of reproduction. Age at which 95% of adult brain size is attained from [57]. Age at peak reproduction is the peak proportion of females reproducing at this age and is from [58]. Chimpanzee estimate of age at first reproduction (AFR) is from wild populations and maximum lifespan (ML) estimates from captivity [44]. Human estimate of AFR based on compiled data from four contemporary hunter-gatherer groups (the Ache, Hadza, Hiwi, and!Kung) reported in [44]. Human ML (122) is also given in [44]. *Age at last reproduction is from [59]. The estimate of 39 for humans comes from the average age at last reproduction across three forager populations. Because most female chimpanzees die before reproductive senescence, these numbers are likely not equivalent for chimpanzees. [57], give an age at last birth of 42 for chimpanzees based on an average of maximum ages of last birth in four wild chimpanzee populations and 45 for humans, but which is based again on average age at last birth, not maximum. B.Brain regions analysed and dendrograms of hierarchical clustering based on full methylation data. Samples ending in ‘a’ are DLPFC samples and ending in ‘d’ are cerebellum samples. Sample numbers 1–8 are chimpanzee samples and sample numbers 19–24 and 26 are human samples. Chimpanzee and human brain figures taken from [60].
Material and Methods
Study subjects
Post-mortem brain specimens were obtained from 16 individuals (8 humans and 8 chimpanzees; see Table 1 for details). This sample size is relatively small and thus has limited power; however, it leverages precious and rare frozen tissue samples from chimpanzees. Specimens were obtained from the NIH NeuroBioBank and the National Chimpanzee Brain Resource (www.chimpanzeebrain.org). Subjects died from causes unrelated to the current research. Causes of death and clinical diagnoses are given in Table 1. Chimpanzees showed no clinical signs of neurological conditions and no neuropathology on gross inspection of the brains at autopsy. All human samples were designated as unaffected controls by the NIH NeuroBioBank staff due to the absence of neuropathology. Although some individuals had neuropsychiatric or neurological clinical diagnoses (Table 1), they were nevertheless assigned control status based on standard, extensive neuropathological examination by board certified neuropathologists that includes microanatomical inspection, microscopic neuropathological assessment, and immunohistochemical assays.
Table 1.
Study subjects.
| Species | Sex | Brain areas* | Age | Hemi-sphere | PMI (hours) | Cause of death | Clinical notes** |
|---|---|---|---|---|---|---|---|
| Pantroglodytes | F | DLPFC, cerebellum | 20.7 | L | <5 | Congestive heart failure Lana – cardiomyopathy, congestive heart failure Sherman – congestive heart failure |
|
| Pan troglodytes | M | DLPFC, cerebellum | 24 | R | <5 | Coccidioidomycosis | |
| Pan troglodytes | F | DLPFC, cerebellum | 24.5 | R | <5 | Epicardial haemorrhage | Gastroenteritis, uterine cyst |
| Pan troglodytes | M | DLPFC, cerebellum | 24.9 | L | <5 | Staph infection | |
| Pan troglodytes | M | DLPFC, cerebellum | 44.7 | L | <5 | Congestive heart failure | |
| Pan troglodytes | F | DLPFC, cerebellum | 46.1 | L | <5 | Cardiomyopathy, congestive heart failure | |
| Pan troglodytes | F | DLPFC, cerebellum | 50 | L | 2 | Euthanized for quality of life/Obesity-related respiratory compromise and severe osteoarthritis that limited mobility | Osteoarthritis, obesity, enlarged left ventricle, prediabetic, respiratory compromise |
| Pan troglodytes | F | DLPFC, cerebellum | 50 | L | <5 | Congestive heart failure | Advanced heart disease |
| Homo sapiens | M | DLPFC, cerebellum | 34 | L | 26.7 | Cardiopulmonary arrest | Acute lymphoid leukaemia, post-traumatic stress disorder |
| Homo sapiens | M | DLPFC, cerebellum | 35 | L | 7 | Cardiopulmonary arrest | Anaemia, alcohol dependence, hypertension |
| Homo sapiens | M | DLPFC, cerebellum | 35 | L | 16.2 | Malignant neoplasm of pancreas | Neoplasm of bronchus, lung, mediastinum, kidney, adrenal |
| Homo sapiens | M | DLPFC, cerebellum | 38 | L | 23.1 | Renal disease | Hyperparathyroidism, hypertension, ulcer |
| Homo sapiens | M | Cerebellum | 65 | L | 10.2 | Malignant neoplasm of bladder | Anxiety, Alzheimer’s disease |
| Homo sapiens | F | DLPFC | 69 | L | 12.6 | Cardiogenic shock | Diabetes, obesity, major depressive disorder |
| Homo sapiens | F | DLPFC, cerebellum | 67 | L | 3.75 | Cardiopulmonary arrest | Atherosclerosis, cardiac dysrhythmia |
| Homo sapiens | M | DLPFC, cerebellum | 70 | L | 10.1 | Cardiopulmonary arrest | Gout, pseudobulbar affect; multiple sclerosis, cataracts, diplopia, papilloedema, hypertension, cerebral infarction, cognitive deficits, memory loss, chronic fatigue syndrome, dysphagia, urinary frequency |
*DLPFC = Dorsolateral prefrontal cortex.
**Note on human subjects: All human subjects were designated by as free from neuropathology and characterized as unaffected controls by NIH NeuroBioBank staff following a rigorous, standardized pathology detection procedure.
We sought to match the number of males and females in each age group and species. However, we were constrained by sample availability such that males outnumbered females among the human subjects (females = 2, males = 6,) and chimpanzee females outnumbered males in the older age group (females = 5, males = 3; Table 1). For the chimpanzees, all subjects provided both DLPFC and cerebellar samples. For humans, this was the case for all but two subjects, one of which only provided a DLPFC sample and a second that only provided a cerebellar sample (Table 1). Samples representing two age groups, young adults (humans: 34–38, chimpanzees: 20–25) and old adults (humans: 65–70, chimpanzees: 44–50), were included for each species (Figure 1a; Table 1). These ages are intended to reflect broadly equivalent whole-organism life stages, as determined based on life history stages and milestones (e.g., young adult sample ages falling during the peak reproductive years; Figure 1a). Chimpanzee brain specimens were coronally sectioned following necropsy, and sections were then kept frozen at −80°C. Pre-dissected, snap-frozen, and pulverized human brain specimens were received from the NIH NeuroBioBank (Mount Sinai Brain and Tissue Repository).
Tissue dissection
Brain structures were dissected from frozen chimpanzee brain sections while kept chilled on dry ice using published species-specific brain atlases [46,61,62]. In the DLPFC, we analysed Brodmann’s area 46, which lies in the middle frontal gyrus in both humans and chimpanzees, and, in the lateral cerebellum (‘cerebellum’ hereafter), the lateral-most part of the posterior lobe (Crus I and Crus II) (Figure 1(b)). We stored dissected tissue at −20°C in microcentrifuge tubes filled with RNALater (Ambion, Austin, TX, USA) preservation buffer until we performed DNA extractions.
DNA extraction and microarray analysis
Tissues were washed with PBS buffer to remove residual RNALater and DNA was then extracted from dissected tissue using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. All DNA sample concentrations were measured on a Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and then brought to a concentration of ~70 ng/μl, either through dilution with PCR-grade water or concentration using Microcon-30kDa Centrifugal Filter Unit columns (Millipore Sigma, Burlington, MA, USA). DNA was then bisulphite-converted and assayed on the Illumina Infinium Methylation EPIC BeadChip at the Yale Center for Genome Analysis. Individuals from all species and age groups were included in each DNA extraction and on each array to avoid confounding batch effects.
Data preprocessing
We preprocessed data with the ChAMP v2.12.4 R package [63]. This included filtering the raw intensity data for probes with background-level spectral intensities that target non-CpG sites, that contain known DNA sequence variants, that target sites on sex chromosomes, and for which there were fewer than 3 reads for at least 5% of the samples. The data were normalized using Beta Mixture Quantile dilation (BMIQ), which corrects for differences in the distribution of type 1 and type 2 probes on the array by adjusting type 2 probe methylation values to fit the type 1 probe distribution [64]. We performed singular value decomposition analysis [65] to identify significant sources of covariation and batch effects in the data. The main sources of variance were species, brain structure, and age (p < 0.001), as well as individual identity (p < 0.05). Sex and batch had no significant effects. To further exclude the presence of major confounds in our data, we also performed hierarchical clustering of samples based on genome-wide methylation after filtering and normalizing using Euclidean distance between samples. Samples clustered by species and, within species, by region (Figure 1(b)).
Epigenetic clock analysis
We leveraged a dataset we generated for a prior study [66] to estimate epigenetic age of both brain structures of all human and chimpanzee individuals using an established human multi-tissue epigenetic clock, known as the ‘Horvath clock’ [3], which has been demonstrated to estimate chimpanzee age accurately from blood [3,43]. Age estimation using the Horvath clock is done via prediction using a linear equation including 353 age-predictive CpG sites (the CpGs and their coefficients can be found at the clock’s dedicated website https://horvath.genetics.ucla.edu/html/dnamage/; AdditionalFile3.csv contains the model). Age estimation with the Horvath clock also involves an age-transformation function in which age-related methylation change is logarithmic until attainment of adult age (20), after which the relationship is linear. We also estimated chimpanzee epigenetic age using the Horvath clock with adult ages of 15 and 10, which may better reflect adult age in wild and captive chimpanzees, respectively [67–69]. We additionally used a chimpanzee-specific blood epigenetic age estimator that our group recently developed [43]. The chimpanzee clock functions to predict age using a linear equation in the same way as the Horvath clock. The CpGs and coefficients comprising the chimpanzee clock can be found in the supplementary material of [43]. We used analysis of covariance to assess whether the different brain structures showed significant differences in slope, or rate of epigenetic ageing, in each species. Specifically, we used the base function aov in R v3.6.1 [70] to fit linear models with natural log-transformed epigenetic age [71,72] as the outcome variable and chronological age, region, and an interaction between chronological age and region as the predictor variables. Sex, hemisphere, post-mortem interval (PMI), and neurological diagnoses were included as covariates.
Ethics
Brain specimens were obtained from the NIH NeuroBioBank and the National Chimpanzee Brain Resource with the approval of The George Washington University Institutional Animal Care and Use Committee (Protocol #A454). No living animals were used. We followed guidelines for the ethical use of chimpanzees in research laid out by the American Psychological Association and National Institutes of Health during all aspects of this research and complied with the American Association of Biological Anthropologists Code of Ethics.
Results
In humans, the Horvath clock resulted in epigenetic age estimates that showed a moderately high correlation (rs = .74, p= 0.058) with chronological age and fairly accurate age estimates (median absolute deviation, MAD, of 5.0 years) in the DLPFC. The correlation was stronger (rs = .85, p = 0.016) but the MAD somewhat higher (6.2 years) in the cerebellum. The ages for all individuals in the older age group were underestimated by the clock in both brain structures, but much more so in the cerebellum (Figure 2). The ages of all young adults were overestimated in the cerebellum, while the ages of most young individuals (three out of four) were underestimated in the DLPFC.
Figure 2.
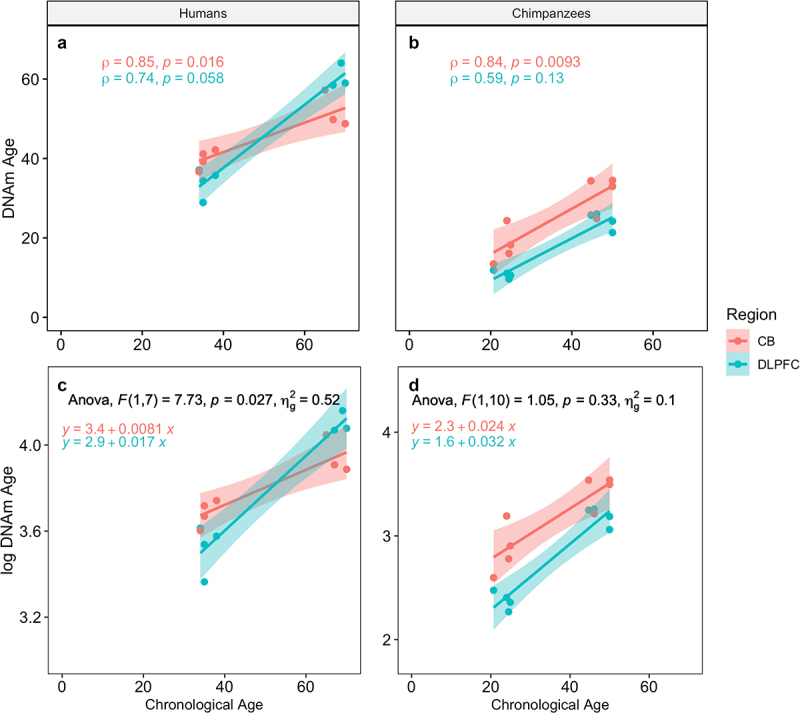
Epigenetic age estimates using the Horvath clock for humans and chimpanzees and results of the ANCOVA. A & B: Spearman’s rho is shown for each species (A = humans, B = chimpanzees) and region. C & D: Results of the ANCOVA comparing slopes of ageing for each region for (c) humans and (d) chimpanzees, along with the regression equations for each line. CB = cerebellum, DLPFC = dorsolateral prefrontal cortex.
In chimpanzees, Horvath clock epigenetic age estimates were also more highly correlated with chronological age in the cerebellum (rs = .83, p= 0.009) than in the DLPFC (rs = .59, p= 0.126). The age predictive accuracy of the Horvath clock was reduced in chimpanzees compared to humans, and was more accurate in the cerebellum (MAD = 9.37 years) than the DLPFC (MAD = 16.9 years; Figure 2). Using an age transformation with adult age set to either 15 or 10 years, to potentially reflect the earlier attainment of reproductive maturity in chimpanzees, did not improve chimpanzee age estimation in either brain structure but instead led to higher MADs (with a 15-year adult age: DLPFC = 21.3; cerebellum = 15.6; with a 10-year adult age: DLPFC = 25.8; cerebellum = 21.9). The chimpanzee-specific blood clock did not produce high correlations between predicted and chronological age in the cerebellum (rs = .56, p= 0.146) and showed a MAD of 9.56 years. Although the DLPFC showed a higher correlation between predicted and chronological age (rs = .92, p = 0.001), the MAD was 14.4 and all of the old individuals’ ages were underestimated by at least 21.5 years.
Analysis of covariance revealed a significant difference in slopes between the two brain regions in humans (F(1,7) = 7.73, p = 0.027) but not in chimpanzees (F(1,10) = 1.05, p = 0.330) using the Horvath clock (Table 2, Figure 2). A significant difference in slopes between the two brain regions was detected in chimpanzees, however, using the chimpanzee-specific blood clock (F(1,10) = 5.34, p = 0.043).
Table 2.
Results of the covariance analysis.
| Human (Homo sapiens) | |||||
|---|---|---|---|---|---|
| Effect | DFn | DFd | F | p | Partial η2 |
| Age | 1 | 7 | 30.75 | 0.000864*** | 0.815 |
| Region | 1 | 7 | 1.06 | 0.34 | 0.031 |
| Sex | 1 | 7 | 0.10 | 0.76 | 0.014 |
| Neurologic diagnosis | 1 | 7 | 0.0002 | 0.99 | 0.00002 |
| PMI | 1 | 7 | 1.24 | 0.30 | 0.150 |
| Age*Region | 1 | 7 | 7.73 | 0.027* | 0.525 |
| Chimpanzees (Pan troglodytes) | |||||
| Effect | DFn | DFd | F | p | Partial η2 |
| Age | 1 | 10 | 49.96 | 0.00003*** | 0.833 |
| Region | 1 | 10 | 19.15 | 0.001** | 0.657 |
| Sex | 1 | 10 | 2.15 | 0.174 | 0.177 |
| Hemisphere | 1 | 10 | 0.003 | 0.960 | 0.0003 |
| Age*Region | 1 | 10 | 1.050 | 0.330 | 0.095 |
Discussion
Our analysis found that human and chimpanzee brains show less age-related change in methylation compared to what has previously been observed in blood [2,43,73], which is consistent with previous studies in humans [16,74]. Within the brain, we found that in young adult humans the DLPFC tends to be epigenetically younger than the cerebellum. An epigenetically young DLPFC in early adulthood could reflect this structure’s extended development [75–77]. However, protracted development does not appear to keep the DLPFC epigenetically young after its maturation in humans. Rather, the cerebellum is epigenetically younger in old individuals (Figure 2), which is consistent with prior studies [7]. This result may be reflective of evidence in humans that the prefrontal cortex also shows earlier and greater structural, microanatomical, and neurochemical change in senescence than other neocortical areas, including white matter shrinkage, grey matter atrophy, loss of dendritic spines, reduced synaptic density, altered myelin, reduced blood flow, and reduced dopamine binding [18,29,54,78–84]. Such age-related changes of the prefrontal cortex are hypothesized to be a major driver of cognitive decline [28,34,35,54,83,85].
We found that chimpanzees also show epigenetically young DLPFCs in early adulthood, potentially reflecting that chimpanzees, like humans, also exhibit protracted aspects of neurodevelopment in the prefrontal cortex relative to macaque monkeys [86]. However, in contrast to humans, we found that in chimpanzees the DLPFC remained epigenetically younger than the cerebellum in older adulthood (Figure 2). Chimpanzees exhibit age-related grey matter atrophy, which, as in humans, is more marked in certain cortical regions, including the prefrontal cortex [62,83]. Microanatomical age-related change in the DLPFC has not been extensively studied in chimpanzees. However, some recent work has documented Alzheimer’s disease-like pathology in the prefrontal cortex in very old chimpanzees [36,37]. Chimpanzees show age-related declines in cognitive performance that have been shown to be associated with age-related grey matter structure changes in several brain regions, including the prefrontal cortex, but which are overall relatively mild [87,88]. It remains to be determined, however, whether differences in brain ageing phenotypes, for example in the prefrontal cortex, between humans and chimpanzees might contribute to potential attenuated cognitive decline in chimpanzees. Generally, more severe and prevalent pathology and cognitive decline in old humans than in nonhuman primates [33,37,39–42] has been hypothesized to be due to an extended lifespan or to the consequences of the greater burden of oxidative damage resulting from increased energy metabolism [89,90]. Improved knowledge in this area is of particular interest given our finding here of differences between humans and chimpanzees.
A number of our study’s limitations and caveats warrant discussion. Our epigenetic age estimates are only moderately correlated with chronological age in the prefrontal cortex, likely due to the small sample size; our epigenetic age estimates obtained with the Horvath clock may nevertheless be considered reasonable biomarkers of ageing given this clock was trained on brain tissue among many other tissues [3], has been validated in independent brain datasets [7], and is associated with brain-specific age-related pathology [16]. In contrast, our chimpanzee-specific epigenetic clock based on blood methylation profiles [43] does not accurately predict chimpanzee age from brain tissue, suggesting it should be considered a blood- or possibly non-neural tissue-specific ageing indicator. This is consistent with findings of poor performance in brain for human epigenetic clocks trained on blood and saliva [91]. Although we did find differences in the slope of epigenetic ageing between the prefrontal cortex and cerebellum for chimpanzees using the chimpanzee blood clock, the prefrontal cortex was nevertheless epigenetically younger than the cerebellum at both old and young ages in chimpanzees, as we found with the Horvath clock for chimpanzees and in contrast to our and others’ findings in humans [7]. Given the higher age-predictive accuracy of the Horvath clock and its validation in brain, the results using the Horvath clock are more reliable. Nevertheless, a multi-tissue or brain-specific, chimpanzee-specific clock would further clarify the degree of divergence in region-specific patterns of epigenetic ageing in humans and chimpanzees. In addition, data from more species – ideally other apes like gorillas for which CpG sites are likely to be conserved – would be necessary to confirm whether differences in patterns of ageing between brain regions, if any, are ancestral or derived.
In addition, as noted in prior studies [7], differences in cell type composition may contribute to differences among regions, as well as species, in epigenetic ageing. Humans have more glial cells per neuron in several areas of the prefrontal cortex compared to other anthropoid primates [51]. Variation in sample conditions, like storage, could also potentially produce artefacts in our data. This type of non-ideal experimental variation can unfortunately result as the consequence of the exceedingly limited availability of human and nonhuman primate tissues [92]. However, methylation fortunately has much higher ‘technical reliability,’ than, for example, RNA, meaning that it is more stable and robust to differences in PMI or storage condition [93–97]. It is thus unlikely that technical artefacts are a major issue in the current study. Finally, the differences between chimpanzees and humans may partly reflect the selected ages of individuals in each of the two age groups, which differ among species in this study (Figure 1a). We sought to select life history-equivalent ages, given differences in developmental timing and lifespan between species and previous evidence of scaling of epigenetic ageing with lifespan [43,98]. However, the contributions of absolute versus life history-scaled age to brain epigenetic ageing are unclear. Nevertheless, the older chimpanzees in our study are of chronological ages (44–50 years) at which the ageing trajectories of the two brain structures, if approximately linear, would begin to converge in humans (at the end of 5th decade; Figure 2), yet the two structures retain nearly parallel trajectories in chimpanzees (Figure 2). Sampling from additional ages would further clarify this phenomenon. A larger sample size for the two species examined here including samples representing more ages over the lifespan would also provide greater information about region-specific species differences in epigenetic ageing trajectory.
In conclusion, we found evidence for differences between humans and chimpanzees in epigenetic ageing in brain structures that may have undergone accelerated evolutionary change in the human lineage. In particular, we found that although the human DLPFC is epigenetically young early in life, its pace of age-related epigenetic change after young adulthood is fast compared to the cerebellum and relative to chimpanzees. This interspecific divergence in ageing patterns between structures may be reflective of species differences in development and senescence, and may be driven by differences in energy metabolism as well as structure-specific cell type proportions [51,89,90]. These results provide a comparative context for understanding age-related epigenetic change in the brain and indicate plasticity of ageing trajectories across tissues and species.
Acknowledgments
We thank Cheryl Stimpson for expert technical assistance; Mary Ann Cree for providing helpful information on the chimpanzees’ clinical histories; NIH NeuroBioBank (Mount Sinai Brain and Tissue Repository) for providing human tissue samples; the Dallas Zoo for chimpanzee tissue samples and for participating in the Great Ape Neuroscience Project; Mary Ann Raghanti and members of the Laboratory for Evolutionary Neuroscience for helpful discussion.
Funding Statement
This work was supported by the Center for the Advanced Study of Human Paleobiology at The George Washington University, the James S. McDonnell Foundation under grant number 220020293, the National Science Foundation under grant numbers SMA-1542848, BSC-2127961, EF-2021785, and National Institute on Aging of the National Institutes of Health under grant numbers R24 NS092988 and R01 AG067419.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
Metadata, raw spectral intensity data, and normalized beta value data are available in NCBI’s Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/sra) under accession GSE154403. R code used for all analyses is available on www.chimpanzeebrain.org.
References
- [1].Field AE, Robertson NA, Wang T, et al. DNA methylation clocks in aging: categories, causes, and consequences. Mol Cell. 2018;71:882–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Binder AM, Corvalan C, Mericq V, et al. Faster ticking rate of the epigenetic clock is associated with faster pubertal development in girls. Epigenetics. 2018;13:85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chen BH, Marioni RE, Colicino E, et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging. 8;2016:1844–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Christiansen L, Lenart A, Tan Q, et al. DNA methylation age is associated with mortality in a longitudinal Danish twin study. Aging Cell. 2016;15:149–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Horvath S, Mah V, Lu AT, et al. The cerebellum ages slowly according to the epigenetic clock. Aging (Albany NY). 2015;7:294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Marioni RE, Shah S, McRae AF, et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015a;16:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Perna L, Zhang Y, Mons U, et al. Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin Epigenet. 2016;8:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Simpkin AJ, Howe LD, Tilling K, et al. The epigenetic clock and physical development during childhood and adolescence: longitudinal analysis from a UK birth cohort. Int J Epidemiol. 2017;dyw307. DOI: 10.1093/ije/dyw307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hillary RF, Stevenson AJ, Cox SR, et al. An epigenetic predictor of death captures multi-modal measures of brain health. Mol Psychiatry. 2019;26:3806–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Marioni RE, Shah S, McRae AF, et al. The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. International Journal of Epidemiology. 2015b;44(4):1388–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hodgson K, Carless MA, Kulkarni H, et al. Epigenetic age acceleration assessed with human white-matter images. J Neurosci. 2017;37:4735–4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lu AT, Hannon E, Levine ME, et al. Genetic architecture of epigenetic and neuronal ageing rates in human brain regions. Nat Commun. 2017;8:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Grodstein F, Lemos B, Yu L, et al. The association of epigenetic clocks in brain tissue with brain pathologies and common aging phenotypes. Neurobiol Dis. 2021;105428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Levine ME, Lu AT, Bennett DA, et al. Epigenetic age of the pre-frontal cortex is associated with neuritic plaques, amyloid load, and Alzheimer’s disease related cognitive functioning. Aging (Albany NY). 2015;7:1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Braak H, Braak E. Neuropathological staging of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. https://pubmed.ncbi.nlm.nih.gov/1759558/. [DOI] [PubMed] [Google Scholar]
- [18].Jernigan TL, Archibald SL, Fennema-Notestine C, et al. Effects of age on tissues and regions of the cerebrum and cerebellum. Neurobiol Aging. 2001;22:581–594. [DOI] [PubMed] [Google Scholar]
- [19].Raz N, Lindenberger U, Rodrigue KM, et al. Regional brain changes in aging healthy adults: general trends, individual differences and modifiers. Cereb Cortex. 2005;15:1676–1689. [DOI] [PubMed] [Google Scholar]
- [20].Jones JH. Primates and the evolution of long, slow life histories. Curr Biol. 2011;21:R708–R717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sakai T, Matsui M, Mikami A, et al., 2013. Developmental patterns of chimpanzee cerebral tissues provide important clues for understanding the remarkable enlargement of the human brain. Proceedings of the Royal Society B: Biological Sciences 280, 20122398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhu Y, Sousa AMM, Gao T, et al. Spatiotemporal transcriptomic divergence across human and macaque brain development. Science. 2018;362:eaat8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hernando-Herraez I, Heyn H, Fernandez-Callejo M, et al. The interplay between DNA methylation and sequence divergence in recent human evolution. Nucleic Acids Res. 2015;43:8204–8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hernando-Herraez I, Prado-Martinez J, Garg P, et al. Dynamics of DNA methylation in recent human and great ape evolution. PLoS Genet. 2013;9. DOI: 10.1371/journal.pgen.1003763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pai AA, Bell JT, Marioni JC, et al. A Genome-Wide Study of DNA Methylation Patterns and Gene Expression Levels in Multiple Human and Chimpanzee Tissues. PLoS Genet. 2011;7:e1001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Anderson US, Stoinski TS, Bloomsmith MA, et al. Relative numerousness judgment and summation in young, middle-aged, and older adult orangutans (Pongo pygmaeus abelii and Pongo pygmaeus pygmaeus). J Comp Psychol. 2007;121:1–11. [DOI] [PubMed] [Google Scholar]
- [27].Anderson US, Stoinski TS, Bloomsmith MA, et al. Relative numerousness judgment and summation in young and old western lowland gorillas. J Comp Psychol. 2005;119:285–295. [DOI] [PubMed] [Google Scholar]
- [28].Dumitriu D, Hao J, Hara Y, et al. Selective changes in thin spine density and morphology in monkey prefrontal cortex correlate with aging-related cognitive impairment. J Neurosci. 2010;30:7507–7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Freire-Cobo C, Edler MK, Munger EL, et al., . Comparative neuropathology in aging primates: a perspective. American Journal of Primatology. 2021;83:e23299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hof PR, Morrison JH. The aging brain: morphomolecular senescence of cortical circuits. Trends Neurosci. 2004;27:607–613. [DOI] [PubMed] [Google Scholar]
- [31].Lacreuse A, Herndon JG. Nonhuman primate models of cognitive aging. Bizon JL, Woods A editors. Animal Models of Human Cognitive Aging. Humana Press: Totowa NJ. 2009. 1–30. DOI: 10.1007/978-1-59745-422-3_2 [DOI] [Google Scholar]
- [32].Lacreuse A, Russell JL, Hopkins WD, et al. Cognitive and motor aging in female chimpanzees. Neurobiol Aging. 2014;35:623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Languille S, Blanc S, Blin O, et al. The grey mouse lemur: a non-human primate model for ageing studies. Ageing Res Rev. 2012;11:150–162. [DOI] [PubMed] [Google Scholar]
- [34].Luebke J, Barbas H, Peters A. Effects of normal aging on prefrontal area 46 in the rhesus monkey. Brain Res Rev. 2010;62:212–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Peters A, Leahu D, Moss MB, et al. The effects of aging on area 46 of the frontal cortex of the rhesus monkey. Cereb Cortex. 1994;4:621–635. [DOI] [PubMed] [Google Scholar]
- [36].Edler MK, Sherwood CC, Meindl RS, et al.Aged chimpanzees exhibit pathologic hallmarks of Alzheimer’s disease. In: Neurobiol. Aging. Vol. 59; 2017. p. 107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Munger, E.L., Edler, M.K., Hopkins, W.D., Ely, J.J., Erwin, J.M., Perl, D.P., Mufson, E.J., Hof, P.R., Sherwood, C.C., Raghanti, M.A. Astrocytic changes with aging and Alzheimer’s disease-type pathology in chimpanzees. J Comp Neurol. Vol. 527. 2019. p. 1179–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Perez SE, Sherwood CC, Cranfield MR, et al. Early Alzheimer’s disease–type pathology in the frontal cortex of wild mountain gorillas (Gorilla beringei beringei). Neurobiol Aging. 2016;39:195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Autrey MM, Reamer LA, Mareno MC, et al. Age-related effects in the neocortical organization of chimpanzees: gray and white matter volume, cortical thickness, and gyrification. NeuroImage. 2014;101:59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chen X, Errangi B, Li L, et al. Brain aging in humans, chimpanzees (Pan troglodytes), and rhesus macaques (Macaca mulatta): magnetic resonance imaging studies of macro-and microstructural changes. Neurobiol Aging. 2013;34:2248–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Finch CE, Austad SN. Commentary: is Alzheimer’s disease uniquely human? Neurobiol. Aging (Albany NY). 2015;36:553–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sherwood CC, Gordon AD, Allen JS, et al. Aging of the cerebral cortex differs between humans and chimpanzees. Proc Natl Acad Sci USA. 2011;108:13029–13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Guevara E, Lawler R, Staes N, et al. Age-associated epigenetic change in chimpanzees and humans. Philos Trans Royal Soc B. 2020. DOI: 10.1098/rstb.2019.0616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gurven MD, Gomes CM. Mortality, senescence, and life span. In M. N. Muller, R. W. Wrangham, & D. R. Pilbeam (ed.,) : Chimpanzees and Human Evolution. Cambridge MA US: Belknap Press of Harvard University Press; 2017. p. 181–216. DOI: 10.4159/9780674982642-005 [DOI] [Google Scholar]
- [45].Khan SS, Singer BD, Vaughan DE. Molecular and physiological manifestations and measurement of aging in humans. Aging Cell. 2017;16:624–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Balsters JH, Cussans E, Diedrichsen J, et al. Evolution of the cerebellar cortex: the selective expansion of prefrontal-projecting cerebellar lobules. NeuroImage. 2010;49:2045–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Barton RA, Venditti C. Rapid evolution of the cerebellum in humans and other great apes. Curr Biol. 2014;24:2440–2444. [DOI] [PubMed] [Google Scholar]
- [48].Carlén M. What constitutes the prefrontal cortex? Science. 2017;358:478–482. [DOI] [PubMed] [Google Scholar]
- [49].Leiner HC, Leiner AL, Dow RS. Cognitive and language functions of the human cerebellum. Trends Neurosci. 1993;16:444–447. [DOI] [PubMed] [Google Scholar]
- [50].Raghanti MA, Stimpson CD, Marcinkiewicz JL, et al. Cholinergic innervation of the frontal cortex: differences among humans, chimpanzees, and macaque monkeys. J Comp Neurol. 2008;506:409–424. [DOI] [PubMed] [Google Scholar]
- [51].Sherwood CC, Stimpson CD, Raghanti MA, et al. Evolution of increased glia-neuron ratios in the human frontal cortex. Proc Natl Acad Sci USA. 2006;103:13606–13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Smaers JB, Turner AH, Gómez-Robles A, et al. A cerebellar substrate for cognition evolved multiple times independently in mammals. eLife. 2018;7:e35696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Miller EK. The prefontral cortex and cognitive control. Nat Rev Neurosci. 2000;1:59–65. [DOI] [PubMed] [Google Scholar]
- [54].West RL. An application of prefrontal cortex function theory to cognitive aging. Psychol Bull. 1996;120:272. [DOI] [PubMed] [Google Scholar]
- [55].Strick PL, Dum RP, Fiez JA. Cerebellum and nonmotor function. Annu Rev Neurosci. 2009;32:413–434. [DOI] [PubMed] [Google Scholar]
- [56].Gurven M. Human survival and life history in evolutionary perspective In John C. Mitani, Josep Call, Peter M. Kappeler, Ryne A. Palombit, and Joan B. Silk (eds.,): The Evolution of Primate Societies. University of Chicago Press Chicago; 2012. p. 293–314. [Google Scholar]
- [57].Robson SL, Wood B. Hominin life history: reconstruction and evolution. J Anatomy. 2008;212:394–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Caro TM, Sellen DW, Parish A, et al. Termination of reproduction in nonhuman and human female primates. Int J Primatol 16(2):205–220. [Google Scholar]
- [59].Kaplan H, Hill K, Lancaster J, et al. A theory of human life history evolution: diet, intelligence, and longevity. Evol Anthropol. 2000;9:156–185. [Google Scholar]
- [60].Sousa AM, Zhu Y, Raghanti MA, et al. Molecular and cellular reorganization of neural circuits in the human lineage. Science. 2017;358:1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Bailey P, Bonin GV, McCulloch WS. The isocortex of the chimpanzee, The isocortex of the chimpanzee. Oxford England: Illinois Press; 1950.Univ [Google Scholar]
- [62].Vickery S, Hopkins WD, Sherwood CC, et al. Chimpanzee brain morphometry utilizing standardized MRI preprocessing and macroanatomical annotations. Elife. 2020;9:e60136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Tian Y, Morris TJ, Webster AP, et al. ChAMP: updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics. 2017;33:3982–3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Teschendorff AE, Marabita F, Lechner M, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Teschendorff AE, Menon U, Gentry-Maharaj A, et al. An epigenetic signature in peripheral blood predicts active ovarian cancer. PLOS ONE. 2009;4:e8274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Guevara EE, Hopkins WD, Hof PR, et al. Comparative analysis reveals distinctive epigenetic features of the human cerebellum. PLOS Genet. 2021;17:e1009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Lammey ML, Ely JJ, Zavaskis T, et al. Effects of Aging and Blood Contamination on the Urinary Protein–Creatinine Ratio in Captive Chimpanzees (Pan troglodytes). J Am Assoc Lab Anim Sci. 2011;50:374–377. [PMC free article] [PubMed] [Google Scholar]
- [68].Videan EN, Fritz J, Murphy J. Effects of aging on hematology and serum clinical chemistry in chimpanzees (Pan troglodytes). Am J Primatol. 2008;70:327–338. [DOI] [PubMed] [Google Scholar]
- [69].Wood BM, Watts DP, Mitani JC, et al. Favorable ecological circumstances promote life expectancy in chimpanzees similar to that of human hunter-gatherers. J Hum Evol. 2017;105:41–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].R Core Team . R: a language and environment for statistical computing. Vienna Austria: R Foundation for Statistical Computing; 2019. [Google Scholar]
- [71].Box GE, Cox DR. An analysis of transformations. J R Statis Soc Ser B (Methodol). 1964;26:211–243. [Google Scholar]
- [72].Harris EK, Boyd JC. Statistical bases of reference values in laboratory medicine. Boca Raton: CRC Press; 1995. [Google Scholar]
- [73].Johansson Å, Enroth S, Gyllensten U. Continuous aging of the human DNA methylome throughout the human lifespan. PLoS ONE. 2013;8:e67378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].El Khoury LY, Gorrie-Stone T, Smart M, et al. Systematic underestimation of the epigenetic clock and age acceleration in older subjects. Genome Biol. 2019;20. DOI: 10.1186/s13059-019-1810-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Miller DJ, Duka T, Stimpson CD, et al. Prolonged myelination in human neocortical evolution. Proc Natl Acad Sci USA. 2012;109: 16480–16485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Somel M, Franz H, Yan Z, et al. Transcriptional neoteny in the human brain. Proc Natl Acad Sci USA. 2009;106:5743–5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Teffer K, Semendeferi K. Human prefrontal cortex: evolution, development, and pathology. Prog Brain Res. 2012;195:191–218. [DOI] [PubMed] [Google Scholar]
- [78].Haug H, Eggers R. Morphometry of the human cortex cerebri and corpus striatum during aging. Neurobiol Aging. 1991;12:336–338. [DOI] [PubMed] [Google Scholar]
- [79].Huttenlocher PR. Synaptic density in human frontal cortex-developmental changes and effects of aging. Brain Res. 1979;163:195–205. [DOI] [PubMed] [Google Scholar]
- [80].Scheibel ME, Lindsay RD, Tomiyasu U, et al. Progressive dendritic changes in aging human cortex. Exp Neurol. 1975;47:392–403. [DOI] [PubMed] [Google Scholar]
- [81].Seaman KL, Smith CT, Juarez EJ, et al. Differential regional decline in dopamine receptor availability across adulthood: linear and nonlinear effects of age. Hum Brain Mapp. 2019;40:3125–3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Suhara T, Fukuda H, Inoue O, et al. Age-related changes in human D1 dopamine receptors measured by positron emission tomography. Psychopharmacology (Berl). 1991;103:41–45. [DOI] [PubMed] [Google Scholar]
- [83].Tisserand DJ, Van Boxtel MP, Pruessner JC, et al. A voxel-based morphometric study to determine individual differences in gray matter density associated with age and cognitive change over time. Cereb Cortex. 2004;14:966–973. [DOI] [PubMed] [Google Scholar]
- [84].Wong DF, Wagner HN, Dannals RF, et al. Effects of age on dopamine and serotonin receptors measured by positron tomography in the living human brain. Science. 1984;226:1393–1396. [DOI] [PubMed] [Google Scholar]
- [85].Duan H, Wearne SL, Rocher AB, et al. Age-related dendritic and spine changes in corticocortically projecting neurons in macaque monkeys. Cereb Cortex. 2003;13:950–961. [DOI] [PubMed] [Google Scholar]
- [86].Bianchi S, Stimpson CD, Duka T, et al. Synaptogenesis and development of pyramidal neuron dendritic morphology in the chimpanzee neocortex resembles humans. Proc Natl Acad Sci USA. 2013;110:10395–10401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Hopkins WD, Mareno MC, Neal Webb SJ, et al. Age-related changes in chimpanzee (Pan troglodytes) cognition: cross-sectional and longitudinal analyses. Am J Primatol. 2021;83:e23214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Mulholland, M.M., Sherwood, C.C., Schapiro, S.J., Raghanti, M.A., Hopkins, W.D. Age-and cognition-related differences in the gray matter volume of the chimpanzee brain (Pan troglodytes): A voxel-based morphometry and conjunction analysis. In: Am. J. Primatol. 2021;83:e23264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Andersen JK. Oxidative stress in neurodegeneration: cause or consequence? Nat Med. 2004;10:S18–S25. [DOI] [PubMed] [Google Scholar]
- [90].Bufill E, Agustí J, Blesa R. Human neoteny revisited: the case of synaptic plasticity. Am J Hum Biol. 2011;23:729–739. [DOI] [PubMed] [Google Scholar]
- [91].Zhang Q, Vallerga CL, Walker RM, et al. Improved precision of epigenetic clock estimates across tissues and its implication for biological ageing. Genome Med. 2019;11:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Blake LE, Roux J, Hernando-Herraez I, et al. A comparison of gene expression and DNA methylation patterns across tissues and species. Genome Res. 2020;30:250–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Bock C, Halbritter F, Carmona FJ, et al. Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. In: The BLUEPRINT consortium. Nat Biotechnol. Vol. 34. 2016. p. 726–737. [DOI] [PubMed] [Google Scholar]
- [94].Bulla A, De Witt B, Ammerlaan W, et al. Blood DNA yield but not integrity or methylation is impacted after long-term storage. Biopreserv Biobank. 2016;14:29–38. [DOI] [PubMed] [Google Scholar]
- [95].Li Y, Pan X, Roberts ML, et al. Stability of global methylation profiles of whole blood and extracted DNA under different storage durations and conditions. Epigenomics. 2018;10:797–811. [DOI] [PubMed] [Google Scholar]
- [96].Teschendorff AE, Relton CL. Statistical and integrative system-level analysis of DNA methylation data. Nat Rev Genet. 2018;19:129–147. [DOI] [PubMed] [Google Scholar]
- [97].Vilahur N, Baccarelli AA, Bustamante M, et al. Storage conditions and stability of global DNA methylation in placental tissue. Epigenomics. 2013;5:341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Lowe R, Barton C, Jenkins CA, et al. Ageing-associated DNA methylation dynamics are a molecular readout of lifespan variation among mammalian species. Genome Biol. 2018;19:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Metadata, raw spectral intensity data, and normalized beta value data are available in NCBI’s Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/sra) under accession GSE154403. R code used for all analyses is available on www.chimpanzeebrain.org.


