Abstract
Background
The loss of tumor antigens and depletion of CD8 T cells caused by the PD-1/PD-L1 pathway are important factors for tumor immune escape. In recent years, there has been increasing research on traditional Chinese medicine in tumor treatment. Cycloastragenol (CAG), an effective active molecule in Astragalus membranaceus, has been found to have antiviral, anti-aging, anti-inflammatory, and other functions. However, its antitumor effect and mechanism are not clear.
Methods
The antitumor effect of CAG was investigated in MC38 and CT26 mouse transplanted tumor models. The antitumor effect of CAG was further analyzed via single-cell multiomics sequencing. Target responsive accessibility profiling technology was used to find the target protein of CAG. Subsequently, the antitumor mechanism of CAG was explored using confocal microscopy, coimmunoprecipitation and transfection of mutant plasmids. Finally, the combined antitumor effect of CAG and PD-1 antibodies in mice or organoids were investigated.
Results
We found that CAG effectively inhibited tumor growth in vivo. Our single-cell multiomics atlas demonstrated that CAG promoted the presentation of tumor cell-surface antigens and was characterized by the enhanced killing function of CD8+ T cells. Mechanistically, CAG bound to its target protein cathepsin B, which then inhibited the lysosomal degradation of major histocompatibility complex I (MHC-I) and promoted the aggregation of MHC-I to the cell membrane, boosting the presentation of the tumor antigen. Meanwhile, the combination of CAG with PD-1 antibody effectively enhanced the tumor killing ability of CD8+ T cells in xenograft mice and colorectal cancer organoids.
Conclusion
Our data reported for the first time that cathepsin B downregulation confers antitumor immunity and explicates the antitumor mechanism of natural product CAG.
Keywords: Tumor Escape, Antigen Presentation, Programmed Cell Death 1 Receptor, CD8-Positive T-Lymphocytes
WHAT IS ALREADY KNOWN ON THIS TOPIC
It has been reported that the cycloastragenol (CAG) has a variety of pharmacological effects, but its antitumor effect and mechanism are not clear.
WHAT THIS STUDY ADDS
By means of single cell multiomics sequencing, target responsive accessibility profiling, and organoid culture, we found that CAG could promote tumor antigen presentation and enhance the antitumor ability of CD8 T cells via inhibiting the degradation of major histocompatibility complex I (MHC-I) mediated by cathepsin B. Further, combined with PD-1 antibody, it has a good antitumor effect in both mouse tumor model and human colon cancer organoids.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
The antitumor mechanism of CAG reveals that the combination of blocking MHC-I degradation drug and PD-1 antibody has a better inhibitory effect on tumor.
Introduction
Colorectal cancer is one of the most common cancers worldwide. Its incidence rate and mortality rate rank among the top three, seriously threatening human life and health.1 Common treatment methods include surgery, chemotherapy and radiotherapy, targeted drugs, and immunotherapy.2–4 However, cancer cells usually exhibit loss of surface antigens and express high levels of inhibitory molecules to escape the surveillance of immune cells. Therefore, to solve the problem of tumor immune escape, researchers have developed immune checkpoint inhibitors and neoantigen therapy to block the masking of cancer cells to immune cells.5–8 Although immune checkpoint inhibitors can restore the depleted immune cells, the surface antigen of cancer cells has not been improved significantly. Therefore, it is particularly important to find drugs that can promote the presentation of tumor surface antigens.
The recognition of cancer cells by cytotoxic CD8+ T cells depends on the TCR/CD3/major histocompatibility complex I (MHC-I) pathway. TCR receives the antigen presented by the dendritic cell/cancer cell membrane protein MHC-I and transmits the signal to tightly connected CD3, which then extends deeper into the cytoplasm.9 10 At the same time, with the activation of CD28/B7 signal, CD8+ T cells can be coactivated to recognize and kill cancer cells.11 Recent studies have found that, in the process of tumor progression, lysosomes result in the reduction of MHC-I aggregation on the surface of cancer cells, which does not effectively present antigens.12 13 Liu et al reported that inhibition of PCSK9 can prevent the degradation of MHC-I in lysosomes and enable MHC-I to return to the cell membrane to present antigens.12 Therefore, restoring the antigen-presenting function of MHC-I is particularly important for blocking tumor immune escape.
An increasing number of studies has found that the application of active molecules of traditional Chinese medicine in antitumor intervention has great prospects.14 15 Cycloastragenol (CAG) is an effective active molecule in Astragalus membranaceus, which has antiviral, antibacterial, anti-inflammatory, and other pharmacological effects, but its antitumor effect is rarely reported.16–19 In this study, we found that CAG inhibited the growth of transplanted tumors of colon cancer in mice. The mechanism mainly entailed inhibiting the degradation of MHC-I, mediated by cathepsin B (CTSB), promoting the antigen presentation of cancer cells, and then enhancing the killing ability of CD8+ T cells.
Materials and methods
Antibodies and reagents
CAG was obtained from Yongjian Pharmaceutical Technology (Jiangsu, China, purity >99%). Antibodies of CTSB (Cat#12 216-1-AP, PRID:AB_2086929), HLA (Cat#15 240-1-AP, PRID:AB_1557426), Actin (Cat#66 009-1-Ig, PRID:AB_2782959), Tubulin (Cat#10 094-1-AP, PRID:AB_2210695), and Anti-mouse/rabbit Immunohistochemistry Detection Kit (Cat#PK10006) were purchased from Proteintech. Antibodies of H2-Kd (Cat#sc-53852, PRID:AB_784514), LAMP1 (Cat#sc-20011, PRID:AB_626853), and CTSB (Cat#sc-365558, PRID:AB_10842446) were purchased from Santa Cruz Biotechnology. Antibody of Ki-67 (Cat#12202, PRID:AB_2620142) was purchased from Cell Signaling Technology. Antibody of Na/K ATPase (Cat# ab3528, RRID:AB_303877) was purchased from Abcam. Flow cytometry antibodies of CD45-V500 (Cat#561487, RRID:AB_10697046), CD3-APC (Cat#345767, RRID:AB_2833003) and CD8-PerCP (Cat#345774, RRID:AB_2868802) were purchased from BD Bioscience. Flow cytometry antibody of Gzmb-FITC (Cat# 515403, RRID:AB_2114575) was purchased from BioLegend. Flow cytometry antibodies of NK1.1-PE-Cy7 (Cat#25-5941-81, RRID:AB_469664) and IFN-γ-PE (Cat#12-7311-81, RRID:AB_466192) were purchased from Thermo Fisher Scientific. DMEM medium (Cat#01-052-1ACS), Penicilin-Streptomycin (Cat#15140122) and Fetal Bovine Serum (Cat#C04001-500) were purchased from Biological Industries. Goat serum (Cat#88RNG001) and Pierce BCA protein assay Kit (Cat#23225) were purchased from Thermo Fisher Scientific. The antibodies of anti-human PD-1 (Cat#BE0188, RRID:AB_10950318) and anti-mouse PD-1 (Cat#BE0146, RRID:AB_10949053) were purchased from Bio X cell. MACS Tumor Tissue Dissociation Kit (Cat#130-095-929) was purchased from Miltenyi Biotec.
Cell culture
Mouse colon cancer cell lines MC38 and CT26 were maintained in our laboratory.20 The human colon cancer cell lines HCT-116 were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The MC38, CT26, and HCT-116 cells were cultured in DMEM medium containing 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C in a 5% CO2 incubator.
Transplantation tumor experiment
Six-eight-week-old female C57BL/6JGpt mice, BALB/c mice and BALB/c nude mice were purchased from GemPharmatech (Nanjing, China). MC38 or CT26 cancer cells (1×106) were inoculated subcutaneously into each mouse. When the tumor grows to 100 mm3, the mice were randomly divided into phosphate buffered saline (PBS) group (ig, once a day) and CAG group (ig, 50 mg/kg, once a day). Tumor volumes were determined by caliper measurement using the formula V=length×width2/2. When the tumor volume of PBS group mice reached 1000 mm3, the tumor of mice was taken out, photographed and weighed.
Single cell dissociation from mouse for single-cell RNA/ATAC-seq
Solid tumors from mice were digested using Tumor Dissociation Kit to obtain single cell suspensions. Single cell suspensions with a concentration of 1000 cells/µL were loaded on the 10×genomics chromium controller single-cell instrument following the 10×genomics manufacturer’s protocol. Reverse transcription reagents, barcoded gel beads, and partitioning oil were mixed with the cells for generating single-cell gel beads in emulsions (GEM) for reverse transcription. scRNA-seq data preprocessing and quality control
Based on the mouse reference genome GRCm38 (mm10), the CellRanger V.4.0.0 pipeline (10×Genomics) to process single-cell RNA sequence data for each experiment (GSE197229). Digital gene expression matrices were analyzed in R (V.4.0.4) using the Seurat (V.4.0.0) package.21 Prior to downstream analysis, cells were filtered by UMI number (<100,000 UMIs), gene number (<6500 genes), and mitochondrial gene percentage (‘percentage. MT’ less than 10%). Normalization was performed with the SCTransform function with regression of percentage of mitochondrial genes.22 For integration, 2000 shared highly variable genes were identified using the SelectIntegrationFeatures function.23 Integration anchors were identified based on these genes using the FindIntegrationAnchors34 function with an ‘SCT’ normalization method. The data were then integrated using the IntegrateData function. Principal component analysis (PCA) and t-distributed stochastic neighbor embedding (t-SNE) dimension reduction with the top 30 principal components were performed. A nearesteneighbor graph using the 30 dimensions of the PCA reduction was calculated using FindNeighbors, followed by clustering using FindClusters with a resolution of 0.8. Candidate Marker genes for each cell cluster were identified by the FindAllMarkers function. For each cluster of cells, group-specific differentially expressed genes were identified using the Wilcoxon Rank Sum test as implemented in FindAllMarkers.
scATAC-seq data preprocessing and quality control
The single cell ATAC-seq data (GSE197229) were preprocessed by cellranger-atac (V.2.0.0) with the count command line. For the subsequent scATAC-seq data processing and analysis, we used the ArchR (V.1.0.1) package.24 Then, we used addArchRGenome (‘mm10’) function for genome annotation and create arrow file with the createArrowFiles function with the default parameters. Next, we used the filterDoublets function to delete the potential doublets and created an ArchR project using the ArchRProject function with default parameters. We then used the Harmony package to remove the batch effect by addHarmony function.25 For dimensionality reduction, we use the addIterativeLSI function in ArchR to run with the default parameters. For single cell embedding, we selected the reducedDims object with harmony and used addTSNE function with the parameter ‘perplexity=30’ for visualization.
Integrated analysis of scRNA-seq and scATAC-seq data
In order to integrate scATAC-seq data with matched scRNA-seq data, we first used the FindTransferAnchors function from the Seurat package and aligned the data with addGeneIntegrationMatrix function in ArchR with ‘unconstrained integration’ mode. From the result, we found that most of the predicted scores >0.5. In order to improve the accuracy of the predictions so as to better integrate the two datasets, we once again integrated the scATAC-seq and scRNA-seq data using the ‘constrained integration’ mode. Briefly, we annotated the scATAC-seq data with cell types based on the gene scores of scATAC-seq. Then, we created a restricted list such that gene expression similarity was calculated only in the same cell type for both scATAC-seq and scRNA-seq. Unsupervised clustering with t-SNE revealed 13 sub cell clusters, which were annotated by known or putative markers in online supplemental table 1.
jitc-2022-004874supp002.xlsx (13KB, xlsx)
Pseudotime lineage trajectory
The cell lineage trajectory of cancer cells was inferred by using Monocle2 (V.2.18.0) R package.26 Monocytes learn the explicit master graph from single-cell genomics data by Reversed Graph Embedding to sort cells, thus solving complex biological processes robustly and accurately.27 We used the ‘‘differentialGeneTest’’ function to derive DEG from each cluster, and after constructing the cell trajectory, we detected the differentially expressed genes in the pseudotime. All pseudotime-dependent genes were visualized by the plot_pseudotime_heatmap function taking a CellDataSet object. Lineage trajectory plot and smooth expression curves based on CellDataSet were generated by plot_cell_trajectory and plot_genes_in_pseudotime, respectively.28
Single cell copy number variation calling
To identify malignant cells with clonal large-scale chromosomal copy number variations (CNV), we used the inferCNV R package to infer the genetic profiles of each cell based on the average expression of large genes sets in each chromosomal region of the tumor genome compared with normal cells.29 On a sample-by-sample basis, the immune cells were used as a reference to estimate CNVs in the cancer-related cells.
Enrichment analysis
KEGG pathway and GO annotation analyses were performed using the R package clusterProfiler (V.3.11.1) with parameters of PValueCutoff=0.05, PAdjustMethod=BH, QValueCutoff=0.05, MingsSize=10, and MaxGSSize=500.20 Gene set enrichment analysis (GSEA) was applied using 50 hallmark gene sets (h.all.V.7.4.symbols.gmt) to identify significantly enriched functional pathways via GSEA software (V.4.1.0), with screening criteria of nominal P-value<0.05 and false discovery rate q<0.250.30
Quantitative real-time PCR analysis
MC38 and HCT-116 cells were seeded in six-well plates for 6 hours and then treated with CAG for another 24 hours. The cells were washed three times with cold PBS, and centrifuged at 180 g for 5 min at 4°C. At the same time, after grinding the tumor tissue. Total RNA was extracted from MC38, HCT-116 cells and tumor tissue using the TRIzol reagent, according to the manufacturer’s instructions. The reaction volume was 20 µL containing: 1 µg RNA, 5 µL 5×Hiscript III qRT SuperMix, and RNase Free dH2O. The cDNA was subjected to quantitative PCR, with a reaction volume of 10 µL with 1 µL cDNA, 5 µL qPCR mix, 0.75 µL 5 µM primers (Forward and Reverse), and 3.25 µL RNase-free dH2O. The primers were synthesized by GenScript Biotech Corporation (Nanjing, China) according to the following sequences (online supplemental table 2).
jitc-2022-004874supp003.xlsx (10.7KB, xlsx)
Target discovery via a target-responsive accessibility profiling approach
The screening of CAG binding proteins was performed as described previously.31 32 Briefly, target-responsive accessibility profiling (TRAP) approach was employed to discover the binding proteins for CAG in cell milieu by monitoring ligand engagement-induced lysine accessibility changes on a proteome level. Briefly, two dishes of cells were treated with 10 µM CAG and dimethyl sulfoxide (DMSO), respectively. After 1-hour incubation, cells were permeabilized by M-PER buffer (Thermo Scientific) and the resultant lysates were covalently labeled by the addition of formaldehyde and borane pyridine complex that together specifically label proteinaceous lysine residues at room temperature for accessibility profiling. Then, the lysates were precipitated by organic solvent, and the collected pellets were redissolved in 8 mol/L urea, reduced by dithiothreitol (DTT) at 56℃ for 30 min, followed by alkylation using iodoacetamide (IAA) in dark for 30 min. Appropriate amount of DTT solution was added again to react with excess IAA. Subsequently, the proteome was diluted with ammonium bicarbonate solution until the final concentration of urea reaches 1 mol/L. The collected protein digests were desalted on C18 HLB columns (Waters, Milford, Massachusetts, USA), and the enriched peptides were dried and reconstituted in 0.1% formic acid (FA) aqueous solution. AnanoLC-SYNAPT G2 Si Q-TOF system (Waters) was employed to analyze the samples for quantitative profiling the lysine accessibility changes in response to CAG binding for target discovery. Data dependence acquisition in the positive mode was employed for data acquisition. Data analysis was performed using PEAKS Studio V.8.5 (BSI solutions, Waterloo, Canada). Specifically, cys alkylation was selected as fixed modification, and methionine oxidation and lysine dimethylation, achieved by TRAP labeling, were set as variable modifications. Briefly, peptides that contain TRAP-induced dimethylation and exhibited significant abundance changes with and without CAG incubation were assigned as target responsive peptides. The ratio of the abundance of each TRAP-labeled peptide indicates the extent of accessibility change, and is intimately associated with ligand-binding affinity. Student’s t-test was carried out to assess whether the detected accessibility changes of labeled peptides are statistically significant. An intergroup p value (p<0.001) and R value (TRAP ratio >2 or <0.5) was set as the cut-off to screen the target responsive peptides belonging to the CAG binding proteins from the whole quantified proteome.
Organoid culture
The human colorectal cancer organoids were constructed and cultivated by Chongqing Kingbiotech.33 Patient tissue samples acquired by surgical operation were minced into pieces as small as possible by sterile scissors. The tissue pieces were mixed thoroughly with Matrigel (Corning, Cat#356231) at the approximate ratio of 1:4 on ice. The subsequent processing referred to published protocols. Briefly, the pieces-Matrigel suspensions were seeded quickly in the multiwell plate to form hemispherical droplets and transferred to the 37°C for 15–20 min, allowing the droplets to be solidifying. Added the appropriate amount of culture medium (KingcultureTM Organoid Growth Medium, Cat#KCW-2) to each well, and changed the medium every 2–4 days.
Isolation and culture of human CD8 T cells
Add the same volume of normal saline to the peripheral blood of healthy people containing anticoagulants to dilute the whole blood. Add a certain volume of separation solution to a 15 mL centrifuge tube, slowly add the diluted whole blood along the tube wall to the top of the separation solution, and centrifuge at 750 g for 20 min. After centrifugation, use a pipette to carefully suck the monocytes in the middle white layer into a 15 mL centrifuge tube, add a certain volume of PBS to resuspend, centrifuge 250 g for 5 min, and discard the supernatant. After adding human CD8 magnetic beads to the cell precipitates, CD8 T cells were sorted by LS sorting column. CD8 T cells were added to the perforated plate pre-coated with 5 µg/mL CD3 (Thermo Fisher Scientific Cat# 16-0032-38, RRID: AB_2865578) antibody, and then 10 ng/mL IL-2 (Peprotech Cat# 212-12) and 5 µg/mL CD28 (Thermo Fisher Scientific Cat# 16-0281-81, RRID: AB_468920) functional antibodies were added cultured for 24 hours.
Coculture
After the organoids were incubated with CAG for 24 hours, the fresh culture medium was replaced, and then the organoids and activated CD8 T cells were suspended in the matrix gel, then the suspension was added to the six-well plate and placed in the 37°C incubator for 15 min, and then 2 mL serum-free complete culture medium was added for 24 hours.
Western blot
MC38 and HCT-116 cells were seeded in six-well plates for 6 hours and then treated with CAG for another 24 hours. The cells were washed three times with cold PBS, and centrifuged at 180 g for 5 min at 4°C. The total protein was lysed with WB-IP lysis containing 1% protease inhibitor for 30 min on ice. Total protein was assessed using a BCA protein quantitation kit. Protein samples were separated by 10%–12% SDS polyacrylamide gel electrophoresis and transferred to PVDF (polyvinylidene fluoride) membranes at 350 mA for 90 min. The PVDF membranes were blocked with 5% BSA for 1 hour, the strips with the indicated primary antibody overnight, and with the secondary antibody incubated for 90 min at room temperature. Finally, the strips were detected using a LumiGLO chemiluminescent substrate system (KPL, Gaithersburg, Maryland, USA).
Flow cytometry
After digestion of tumor tissue and the single-cell su spension was prepared. Surface staining was performed with surface antigen antibodies in the FCM (Flow Cytometry) buffer (PBS containing 1% FBS) and stained on ice with appropriate antibodies for 30 min. Reactive dyes (eBioscience) were used to eliminate dead cells. Intracellular cytokine staining was performed with BD cell fixative solution/extracellular membrane solution, and the cells were fixed and permeated, and then stained with antibodies against cytokines in Perm/Wash buffer (BD Biosciences).
CTSB mutant plasmids transfected
HCT-116 cells were seeded in 6-well plates for 12 hours, and then transfected with CTSB-WT-EGFP or CTSB mutant plasmids (Y75A, A77V and G198A) for 36 hours.
Transfection interference or over-expression plasmid of CTSB into MC38 cells or HCT-116 cells
MC38 cells (1×106/well) were inoculated in 6-well plates for 6 hours, then transfected with CTSB interfering RNA (sequence: Forward-GGACAUAGAUCUACCUGAATT and Reverse-UUCAGGUAGAUCUAUGUCCTT) for 48 hours, and the mRNA or protein expression levels of Ctsb and H2-k1 were detected. HCT-116 cells (1×106/well) were inoculated in 6-well plates for 6 hours, then transfected with CTSB interference (sequence: GCTGGTCAACTATGTCAACAA) or over-expression plasmid for 48 hours, and the mRNA or protein expression levels of CTSB and HLA-A were detected.
Docking technology
The 3D structure of CTSB was downloaded from the Protein Database (PDB ID: 2iPP), and the structures of CAG were downloaded from PubChem. The docking process was performed in Autodock 2 with coarse docking using a simulated annealing algorithm and a subsequent refinement using a genetic algorithm.
Cellular thermal shift assay
The MC38 cells were seeded in 10 cm plates overnight and then treated with CAG or 0.1% DMSO for another 2 hours. The cells were collected, washed with cold PBS, and centrifuged at 180 g for 5 min. The cells were then evenly divided into centrifugation tubes (70 µL each tube) and heated for 3 min at the following temperature: 46, 49, 52, 55, 58, 61, 64, and 67°C, the samples were cooled for 3 min at temperature and then kept on ice. Then, the samples were placed in a refrigerator at −80°C overnight. The samples were thawed at room temperature for 30 min and then refrigerated at −80°C for 4 hours. Finally, the samples were centrifuged at 12, 000 g for 25 min, the supernatant added to the loading buffer, and analyzed by Western blotting.
Immunohistochemical staining
Paraffin sections of tumor tissue were immersed in xylene for 20 min to dewax, and then in 100%, 75% and 50% ethanol for 10 min. After the slices were subjected to antigen repaired with sodium citrate antigen repair solution, the endogenous hydrogen peroxide was inactivated with 3% hydrogen peroxide. After blocking with 5% goat serum for 1 hour, the anti-Ki67 antibody (1:200) was added and incubated overnight at 4°C. The anti-mouse/rabbit HRP labeled polymer (100 µL) was added and samples incubated at 37°C for 30 min, followed by 100 µL of DAB working solution, and incubation at room temperature for 5 min. After 1 min of staining with hematoxylin, samples were washed in 50%, 75%, and 100% ethanol and xylene for 5 min, neutral gum was used to seal the film. The film was observed and photographed under a microscope.
Statistical analysis
Statistical analysis was performed using GraphPad Prism software (V.8.0). All results are expressed as the mean±SEM of three independent experiments. One-way analysis of variance followed by Dunnett’s post hoc test was used to evaluate the differences when there were more than two groups. The Student’s t-test was used to evaluate the significant difference between the two groups. A statistical significance was set at p<0.05.
Results
Single-cell multiomics analysis of CAG inhibiting the growth of transplanted colon cancer in mice
To investigate whether CAG has an antitumor effect, we first transplanted MC38 cancer cells into C57BL/6 mice. We noted that CAG significantly inhibited the growth of tumors (online supplemental figure S1A, B). Subsequently, we transplanted CT26 cells into BALB/c mice and found that CAG also inhibited the growth of tumors (online supplemental figure S1C, D).
jitc-2022-004874supp001.pdf (1.5MB, pdf)
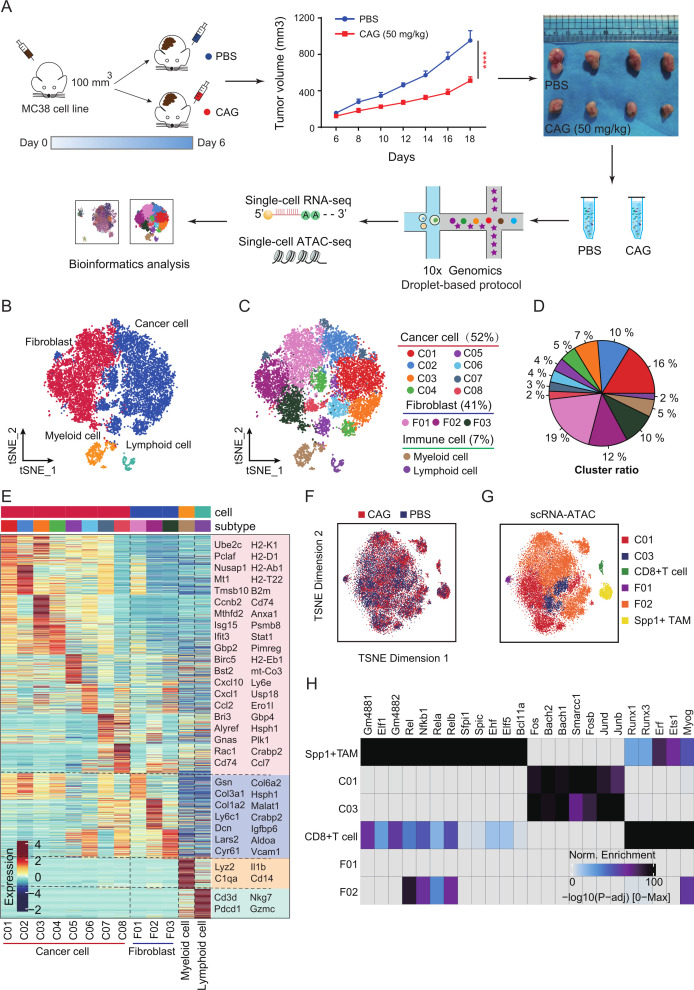
To further reveal the specific mechanism by which CAG inhibits tumor growth, we analyzed it using the scRNA-seq and scATAC-seq techniques (figure 1A). We divided the cell population into four groups: cancer cells, fibroblasts, myeloid cells, and lymphocytes (figure 1B, online supplemental figure S2A, B). We found that cancer cells (52%) and fibroblasts (41%) were mainly infiltrated in tumor tissues, while immune cells accounted for only 7%. We then divided cancer cells into eight subsets and fibroblasts into three subsets (figure 1C–E). Subsequently, the enrichment analysis of highly expressed genes in each cell population showed that MHC-I molecular pathways in cancer cells were enriched, as were the antitumor signals of T cells and NK cells (online supplemental figure S2C). Then, four groups of cells were also found using scATAC-seq and scRNA-seq integration analysis (figure 2D–G). Following comparison, we found that cancer cells (C01, C03 cells), CD8+ T cells, Spp1+ TAM cells, and fibroblasts (F01 and F02 cells) appeared in ATAC sequencing (figure 1F, G), and further, highly expressed transcription factors were enriched in these cells (figure 1H). Through these analyses, we speculate that the inhibition of tumor growth by CAG is closely related to changes in these cells.
Figure 1.
Single-cell multiomics analysis of cycloastragenol (CAG) inhibiting the growth of MC38 transplanted tumor. (A) Schematic diagram of scRNA-seq and scATAC-seq. MC38 cancer cells (1×106) were inoculated subcutaneously into each mouse (n=5). On the sixth day, the mice were randomly divided into the PBS group (ig, once a day) and the CAG group (ig, 50 mg/kg, once a day). On the 18th day, the tumor was removed, and a single-cell suspension was prepared for testing. (B) scRNA-seq analysis of the cell clusters. (C, D) Subgroup clustering and proportion. (E) Heatmap showing the classification of cell subsets and marker genes. (F, G) scATAC-seq analysis of the cell clusters. (H) Heatmap showing the transcription factors corresponding to each subgroup. Data are represented as mean±SEM. P values are determined by two-tailed Student’s t-test. ****p<0.0001. PBS, phosphate buffered saline.
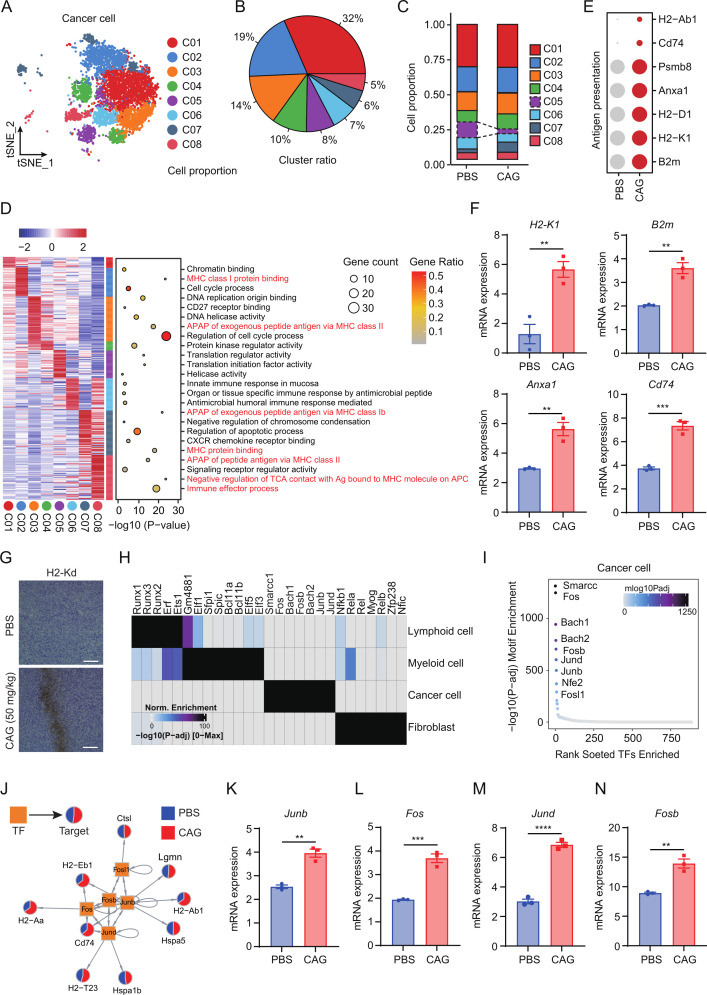
Figure 2.
CAG promotes tumor cell antigen presentation. (A–C) scRNA-seq analysis showing cancer cells clustering and proportion. (D) Left: heatmap showing expression signatures of top 50 specifically expressed genes in each cell type; the value for each gene is row-scaled Z score. Right: representative GO terms. (E) The bubble chart showing the expression of antigen presentation related genes in the two groups. (F) Expression of antigen presentation related genes in tumor tissues. (G) The expression of H2-Kd in tumor tissues detected by IHC staining. Scale bar=50 µm. (H) scATAC-seq analysis showing the transcription factors were highly expressed in each group. (I) Top 10 transcription factors in cancer cells. (J) Gene regulatory network of transcription factors and corresponding genes. (K–N) mRNA expression of transcription factors in tumor tissues. Data are represented as mean±SEM. P values are determined by two-tailed Student’s t-test. **p<0.01, ***p<0.001, ****p<0.0001. CAG, cycloastragenol.
Cag promotes tumor cell antigen presentation
To analyze the antitumor mechanism of CAG, we first analyzed the tumor cell population and divided the cancer cells into eight subgroups (figure 2A, B, online supplemental figure S3A). The results showed that, compared with the PBS group, C05 group cells decreased significantly while C07 group cells increased (figure 2C). Meanwhile, to verify the accuracy of our tumor cell clustering, we compared the CNV of lymphocytes and myeloid cells as reference cells. The results showed that the CNV of cancer cells was significantly higher than that of immune cells (online supplemental figure S3B). When analyzing the subsets of cancer cells, we found that the interferon response genes Isg15, Irf7, Ifit3, and Ifi47 were highly expressed in the C07 and C08 cell populations of the CAG group compared with the PBS group (online supplemental figure S3C). The analysis of the tumor cell population found that antigen presentation-related pathways were significantly enriched in multiple cell populations. Therefore, we speculate that CAG may promote the antigen presentation of cancer cells to play an antitumor role (figure 2D). Then, we selected the antigen presentation related genes and found that the expression level in the CAG group was significantly higher than that in the PBS group (figure 2E, online supplemental figure S3D). We also found that CAG promoted antigen presentation in tumor tissues (figure 2F, G).
Next, we used scATAC-seq to analyze the specific reasons behind CAG promoting tumor cell antigen presentation. We found that the tumor cell population highly expressed the transcription factors Fos, Junb, Jund, Fosb, and Fosl1(figure 2H, I). Subsequently, we constructed a map of the interaction between transcription factors and corresponding genes to explain that CAG promoted the expression of tumor cell antigen-presenting-related genes (figure 2J). In tumor tissues, we also found that transcription factors Junb, Fos, Jund and Fosb were significantly overexpressed in the PBS group under CAG treatment (figure 2K–N, online supplemental figure S3E-I). So far, we have found that CAG can promote the expression of transcription factors of antigen-presenting-related genes, thereby enhancing the antigen-presenting function of cancer cells. However, how these cancer cells respond to immune cell responses remains unclear.
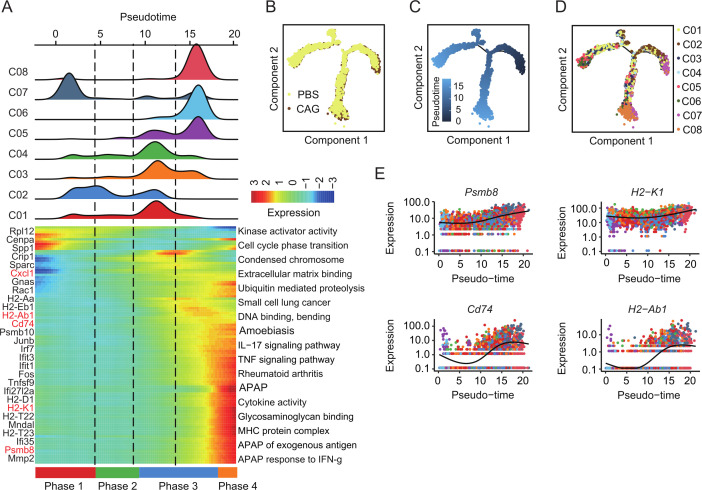
To uncover the phenomenon by which CAG promotes the antigen presentation of cancer cells, we performed trajectory analysis to investigate how cancer cells alter each other in response to immune cell responses. We found that, over time, C07 cancer cells transformed into C05, C06, and C08 cells, and gene enrichment also showed that cancer cells would transform to antigen presentation (figure 3A–E).
Figure 3.
Trajectory analysis revealed that CAG promotes the differentiation of cancer cells into antigen presentation. (A) Heatmap showing the dynamic changes in gene expression along the pseudotime (lower panel). The distribution of cancer cell subtypes during the transition (divided into four phases) along with the pseudotime. Subtypes are labeled by color (upper panel). (B–D) Pseudotime-ordered analysis of cancer cells from PBS and CAG samples. Group (B), pseudotime(C) and cancer cell subgroups (D) are labeled by colors. (E) Expression of tumor antigen presentation-related genes under pseudotemporal analysis. CAG, cycloastragenol; PBS, phosphate buffered saline.
CAG enhances the killing function of immune cells
These results show that CAG can present tumor cell antigens, so will enhancing tumor cell antigen presentation leads to the enhancement of immune cell function? To figure out this, we analyzed lymphocytes and myeloid cells, respectively. The results showed that CAG promoted the infiltration of CD8+ T cells in tumor tissues, and the infiltration of CD8+ T cells and NK cells was also increased, detected by flow cytometry (online supplemental figure S4A-F). We also observed that CAG enhanced the expression of Ifitm2, Cxcr6, and S100a6 genes in CD8+ T cells, Fcer1g, Gzmb, AW112010, and Zfp36i2 genes in NK cells, and Nfkbia and Junb genes in CD4+ T cells (online supplemental figure S4G). The expression of Ifng and Gzmb in CD8+ T cells was also significantly enhanced, as detected by flow cytometry (online supplemental figure S4H, I). Furthermore, we found that after CAG treatment, the expression of the inhibitory receptors Lag3, Tigit, and Havcr2 on the surface of CD8+ T cells decreased, and the expression of the Cd28, Cd69, Gzmk, Ccl5 and Pdcd1 genes, characterizing the activation of CD8+ T cells, increased significantly (online supplemental figure S4J). To further verify that CAG enhanced the killing function of CD8+T cells by promoting enhanced tumor antigen presentation, we transplanted CT26 cells into transplanted tumors in nude mice, and observed that CAG could not effectively inhibit the growth of tumors (online supplemental figure S4K-M).
Following this, we analyzed myeloid cells and found that Spp1+ TAM cells increased after CAG treatment, while the number of C1qc+ TAM cells decreased and the number of Il1b+ monocytes increased (online supplemental figure S5A-D). After CAG treatment, Spp1+ TAM and C1qc+ TAM cells were more susceptible to pro-inflammatory TAM transformation. Enrichment analysis found that it mainly focused on Tnf-α, Ifn-g, and the inflammatory response signaling pathway (online supplemental figure S5E-H). Based on the abovementioned results, we infer that CAG enhances the recognition and killing function of immune cells by promoting antigen presentation in cancer cells. However, how CAG is regulated is not clear.
Discovery of the CAG target protein CTSB by trap
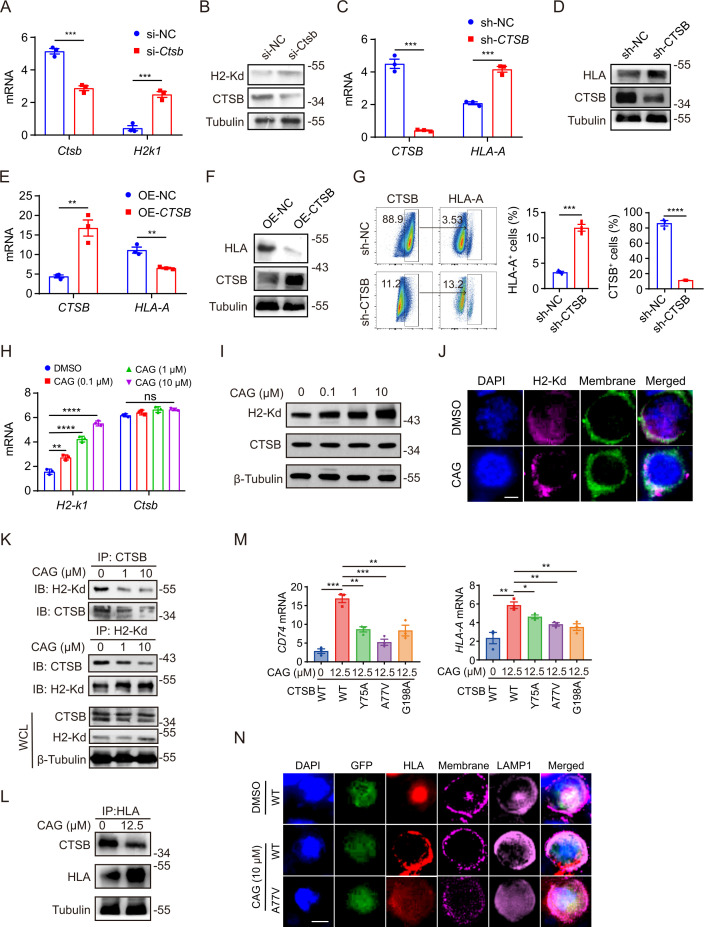
Next, we will investigate the specific CAG target protein that plays an antitumor role. We selected cancer cells as the research object, because we found that CAG can enhance the antigen presentation of cancer cells, thereby enhancing the killing function of immune cells. We first synthesized the biotin derivative of CAG and found that only 3-OH could react (online supplemental figure S6A). We then investigated whether CAG-biotin also promotes the expression of antigen presentation-related genes in the mouse MC38 tumor cell line. The results show that CAG-biotin had no effect on the expression of H2-K1, Cd74, and Anxa1 genes (online supplemental figure S6B-D).
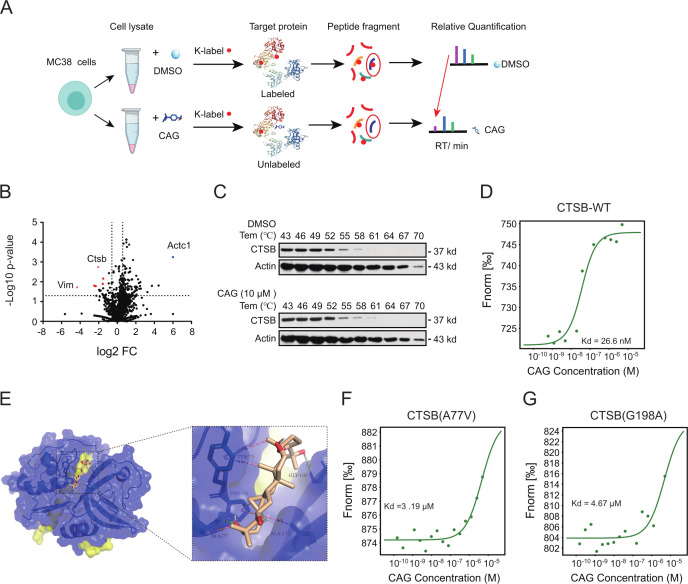
Therefore, we used the previous TRAP target search method in the laboratory.32 CAG and DMSO were both incubated with the MC38 cell line (figure 4A). We selected the protein with FC≥2 and a p≤0.05 as the candidate target protein of CAG. Following the principle that the binding of small molecules to proteins will lead to the low labeling efficiency of lysine, we selected the CTSB for the study (figure 4B). Next, we used cellular thermal shift assay (figure 4C) and microscale thermophoresis (MST) (figure 4D) to verify the binding between CAG and CTSB, and found that the affinity between CAG and CTSB was 26.6 nM (figure 4D). Subsequently, we predicted the binding sites between CAG and CTSB according to the protein crystal structure of CTSB in the PDB library. We found that the hydroxyl groups at both ends of CAG and the ALA77 and GLY198 sites of CTSB were bound by a hydrogen bond, while the TYR75, PRO76, and ALA173 sites of CAG and CTSB were bound by van der Waals force (figure 4E). In order to verify the binding site between CAG and CTSB, we transfected the mutant plasmid of CTSB in HCT-116 cells. The results show that the affinity between the mutant plasmid at A77V and G198A and CAG was hundreds of times higher than that of the WT plasmid (figure 4F, G). In the next section, we explore the specific mechanism by which CAG plays an antitumor role through CTSB.
Figure 4.
The TRAP method was used to find the target protein cathepsin B (CTSB) of CAG. (A) TRAP experiment showing the flow chart of the CAG target protein. (B) Volcano map showing differential proteins. FC=fold change (ratio of CAG to DMSO). (C) The target protein CTSB of CAG was verified by cell thermal migration. (D) HCT-116 cells were transfected with CTSB-WT-EGFP for 36 hours, and the protein lysate was taken for MST detection. (E) Docking prediction of the binding sites of CAG and CTSB (PDBID:2iPP). HCT-116 cells transfected with CTSB- A77V -EGFP (F) and CTSB-G198A-EGFP (G) for 36 hours and the protein lysate taken for MST detection. CAG, cycloastragenol; DMSO, dimethyl sulfoxide; MST, microscale thermophoresis; TRAP, target responsive accessibility profiling.
CAG inhibits CTSB-mediated MHC-I lysosomal degradation
Previous studies have demonstrated that MHC-I is mainly degraded in lysosomes during tumor progression, leading to tumor immune escape.12 13 We speculate whether CAG can also regulate the expression of MHC-I on the cell membrane through its target protein CTSB. We first used RNA interference to knock down the expression of CTSB in MC38 cell line, and the results showed that the gene and protein expression levels of H2-k1 were increased after silencing CTSB (figure 5A, B). Similarly, after knockdown and overexpression of CTSB in human HCT-116 cell line, the mRNA or protein expression levels of HLA-A increased (figure 5C, D) and decreased (figure 5E, F), respectively. At the same time, we found that tumor cells with high expression of CTSB expressed lower HLA-A levels, while in cells after CTSB knockdown increased the expression of HLA-A (figure 5G). These results indicate that CTSB exerts a negative regulatory effect on the expression of MHC-I. Then, we treated the MC38 cell line with CAG and found that CAG promoted the expression of MHC-I and had no effect on CTSB (figure 5H, I), which was consistent with the previous results of studies on tumor tissues. Moreover, we found that after CAG treatment, H2-Kd gathered from the cell to its membrane (figure 5J), which further verified our conjecture that CAG can inhibit the degradation of MHC-I molecule and make it gather at the cell membrane, which is easier to be captured by immune cells. To further validate our hypothesis, we used a Co-IP experiment to investigate whether CTSB binds to MHC-I and leads to its degradation. We found that CAG could inhibit the binding of CTSB and MHC in mouse MC38 cells (figure 5K), and the expression of MHC-I also increases significantly. We also observed the same phenomenon in human HCT116 cells (figure 5L). Next, in order to further confirm whether CAG promotes MHC-I expression and cell membrane aggregation through CTSB, we transfected CTSB mutant plasmid into human HCT-116 cells. The results showed that after transfecting Y75A, A77V, and G198V mutant plasmids, CAG’s function of enhancing CD74 and HLA-A gene expression was destroyed (figure 5M), and MHC-I molecules aggregated from cell membrane to lysosomes (figure 5N). This result showed that CAG inhibited the degradation of MHC-I in lysosomes mediated by CTSB and promoted the reaggregation of MHC-I to the cell membrane.
Figure 5.
CAG inhibits CTSB-mediated MHC-I degradation. MC38 cells (1×106/well) were inoculated in six-well plates for 6 hours, then transfected with CTSB interfering RNA for 48 hours, and the mRNA (A) and protein (B) expression levels of Ctsb and H2-k1 were detected. HCT-116 cells (1×106/well) were inoculated in six-well plates for 6 hours, then transfected with CTSB interference plasmid for 48 hours, and the mRNA (C) and protein (D) expression levels of CTSB and HLA-A were detected. HCT-116 cells (1×106/well) were inoculated in six-well plates for 6 hours, then transfected with CTSB overexpression plasmid for 48 hours, and the mRNA (E) and protein (F) expression levels of CTSB and HLA-A were detected. (G) HCT-116 cells (1×106/well) were inoculated in six-well plates for 6 hours, then transfected with CTSB interference plasmid for 48 hours, and then the expression of CTSB and HLA-A was detected by flow cytometry. (H, I) MC38 cells (1×106/well) were inoculated in six-well plates for 6 hours, then incubated with CAG (10 µM) for 24 hours, and the mRNA and protein expression levels of MHC-I and CTSB were detected. (J) The expression of MHC-I on the cell membrane of MC38 cells treated with CAG was observed via confocal microscopy. Scale bar=15 µm. (K) Coimmunoprecipitation verified that CAG inhibited the binding of CTSB and MHC-I in MC38 cells. (L) Coimmunoprecipitation verified that CAG inhibited the binding of CTSB and MHC-I in HCT-116 cells. (M) HCT-116 cells were transfected with CTSB-WT-EGFP, CTSB-Y75A-EGFP, CTSB-A77V-EGFP, and CTSB-G198A-EGFP plasmid and then incubated with CAG for 24 hours to detect the mRNA expression of CD74 and HLA-A. (N) The changes of MHC-I molecule in HCT-116 cells transfected with CTSB mutant plasmid were observed by confocal microscopy. HCT116 cells were transfected with CTSB-WT-EGFP, CTSB-A77V-EGFP, and CTSB-G198A-EGFP plasmid and then incubated with CAG for 24 hours. Scale bar=20 µm. Data are represented as mean±SEM. P values are determined by two-tailed Student’s t-test. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. CAG, cycloastragenol; CTSB, cathepsin B; MHC-I, major histocompatibility complex I; ns, not significant.
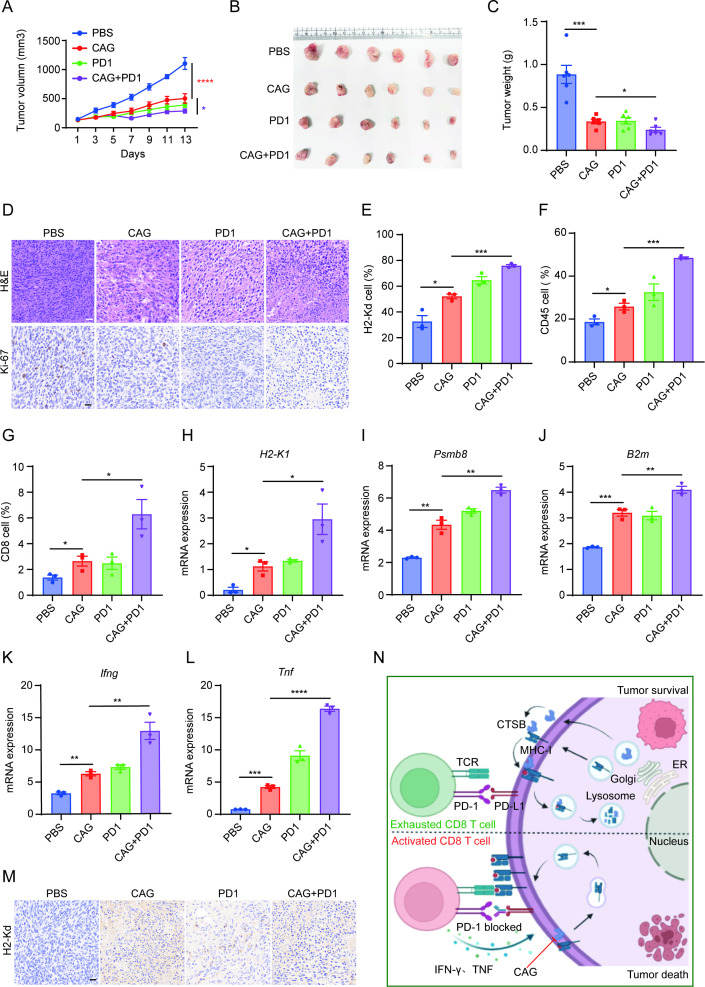
The combination of CAG and PD-1 antibody effectively enhances the tumor killing ability of CD8+ T cells
Tumor immune escape is mainly caused by the loss of the tumor cell antigen presentation function and the inhibition of immune checkpoints. According to our results, we speculated whether CAG can be used in combination with the PD-1 antibody to kill tumors, both to address the shortcomings of tumor antigen presentation and solve the depletion of CD8+ T cells caused by the PD-1/PD-L1 pathway. Thus, we transplanted MC38 cancer cells into C57BL/6 mice and formed a combination group of PD-1 antibody and CAG. The results showed that the combination group of CAG and PD-1 antibody exhibited better antitumor effect than the CAG and PD-1 groups (figure 6A–D). Moreover, we observed a significant increase in the number of infiltrated H2-Kd+ cells, CD45+ cells, and CD8+ cells in tumor tissues (figure 6E–G), and the expression of the antigen presentation-related genes H2-K1, Psmb8, and B2m was also enhanced (figure 6H–J). Meanwhile, the expression of Ifng and Tnf genes was significantly enhanced as well (figure 6K, L), and immunohistochemical staining showed that the expression of H2-Kd increased significantly (figure 6M).
Figure 6.
The combination of CAG and PD-1 antibody showed a stronger antitumor effect. (A) MC38 cancer cells (1×106) were inoculated subcutaneously into each mouse (n=6). After 5 days, the mice were randomly divided into the PBS group (ig, once a day), CAG group (ig, 5 mg/kg, once a day), PD-1 group (ip, 5 mg/kg, twice a week) and CAG+PD-1 group (CAG (50 mg/kg), ig, once a day; PD-1 (5 mg/kg), ip, twice a week). On the 18th day, the mouse tumor was removed. (B) Tumor photos. (C) Tumor weight. (D) H&E and Ki-67 staining of tumor tissue. Scale bar=50 µm. (E) H2-Kd+ cells, (F) CD45+ cells and (G) CD8+ cells infiltrated in tumor tissue. (H) H2-K1 mRNA, (I) Psmb8 mRNA, (J) B2m mRNA, (K) Ifng mRNA and (L) Tnf mRNA expression in tumor tissue. (M) IHC staining showing the expression of H2-Kd in the tumor tissue. Scale bar=15 µm. (N) Summary chart. CAG inhibits the lysosomal degradation of MHC-I mediated by CTSB, and promotes the expression of MHC-I on tumor cell membrane, which makes CD8+ T cells easier to recognize. PD-1 antibody blocks PD-1/PD-L1 pathway so that CD8+ T cells can get rid of exhaustion. Data are represented as mean±SEM. P values are determined by two-tailed Student’s t-test. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. CAG, cycloastragenol; MHC-I, major histocompatibility complex I; PBS, phosphate buffered saline.
Therefore, this study mainly clarified the antitumor mechanism of CAG, mainly by inhibiting the degradation of MHC-I mediated by CTSB, promoting the aggregation of MHC-I molecules to the cell membrane, and then enhancing the antigen presentation ability of cancer cells. Moreover, combined with PD-1 antibody, it could kill cancer cells very effectively (figure 6N).
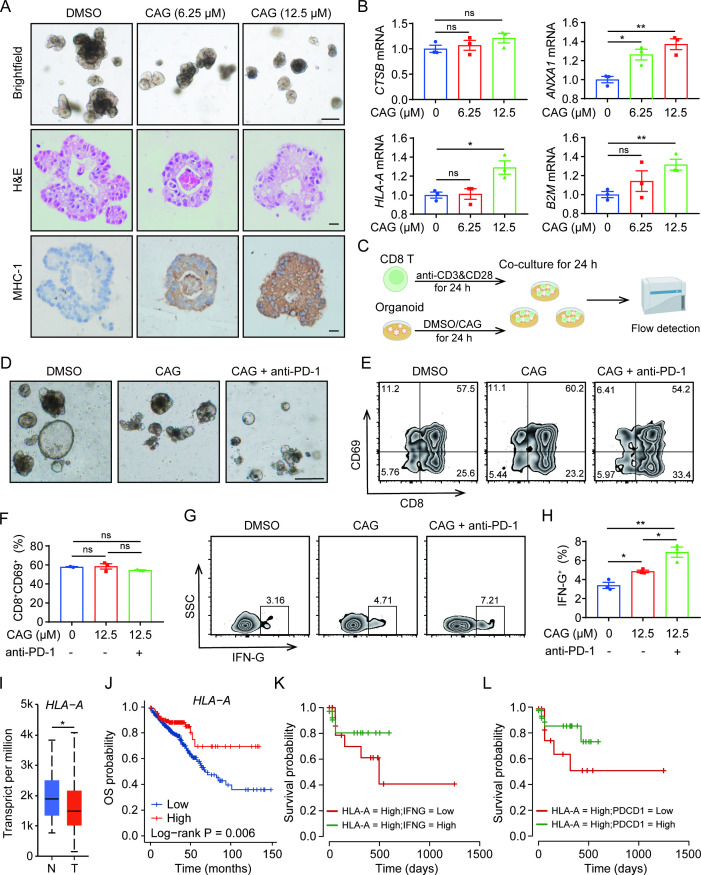
CAG promotes MHC-I expression in colorectal cancer organoids and enhances the killing ability of CD8 T cells
In order to further investigate whether the pharmacological effect of CAG can be applied clinically, human colorectal cancer organoids were used. We found that organoids incubated with CAG enhanced the expression of MHC-I (figure 7A), and the expression of antigen-presenting related genes ANXA1, B2M, and HLA-A also increased (figure 7B). In the previous experiments, we found that CAG enhanced the killing effect of CD8 T cells by promoting the antigen presentation expression of tumor cells, and the effect was superior when combined with the PD-1 antibody. To further verify these results, we collected the peripheral blood of healthy people and isolated CD8 T cells, and then incubated them with CAG-stimulated organoids for 24 hours (figure 7C). The results showed that compared with the DMSO group, CD8 T cells in tumor organoids treated with CAG no longer drifted away but were more concentrated on the surface of organoids; at the same time, there were fewer drifted away CD8 T cells in the CAG and PD-1 groups (figure 7D). We found that the IFN-G secreted by activated CD8 T cells was correspondingly enhanced (figure 7E–H).
Figure 7.
CAG promotes MHC-I expression in colon cancer organoids and enhances the killing ability of CD8 T cells. (A) After 3 days of subculture, colon cancer organoids were incubated with CAG with different concentrations for 24 hours, following which images were captured, scale bar=200 µm (up); H&E staining, scale bar=100 µm (middle) and immunohistochemistry stained with antibody of HLA-A for 1:100, scale bar=100 µm (down). (B) After 3 days of subculture, colon cancer organoids were incubated with CAG at different concentrations for 24 hours, and then the mRNA of CTSB, HLA-A, ANXA1, and B2M were detected. (C) Schematic diagram of coculture of human CD8 T cells and colon cancer tumor organoids. (D) Image acquisition after coculture of CD8 T cells with colon cancer organoids for 24 hours; the small and bright cells are CD8 T cells. The concentration of CAG was 12.5 µM and anti-PD-1 was 5 µg/mL, scale bar=100 µm. (E–H) After coculture of CD8 T cells with colon cancer organoids for 24 hours, flow cytometry was used to detect the expression of CD69 and IFN-G on CD8 T cells. (I) Analysis of HLA-A gene expression in colon cancer by TCGA database. n=Normal, T=Tumor. (J) Relationship between high and low expression of HLA-A gene and survival rate in colon cancer analyzed by TCGA database. (K) Analysis of the relationship between high expression of HLA-A gene and high expression of IFNG gene and survival rate in colon cancer by TCGA database. (L) Analysis of the relationship between high expression of HLA-A gene and high expression of PDCD1 gene and survival rate in colon cancer by TCGA database. Data are represented as mean±SEM. P values are determined by two-tailed Student’s t-test. *p<0.05, **p<0.01. CAG, cycloastragenol; MHC-I, major histocompatibility complex I; TCGA, the Cancer Genome Atlas.
So far, we found that CAG promoted the antigen-presenting expression of tumors to enhance the antitumor effect of CD8 T cells, and the combination of CAG and PD-1 antibody had a superior antitumor effect. Moreover, we analyzed the colon cancer data in the the Cancer Genome Atlas (TCGA) database and found that the antigen presentation-related genes HLA-A, HLA-B, HLA-C, CD74, and B2M showed low expression levels in tumors, and the survival rate of these genes with low expression was very poor (figure 7I, J, figure 7A–H). Therefore, we found that the up-regulated expression of HLA-A, CD74, and other genes by CAG was beneficial for tumor patients. Futhermore, we found that patients with high expression of HLA-A as well as IFNG had a higher survival rate (figure 7K); patients with high expression of HLA-A and low expression of PDCD1 also had a better survival rate (figure 7L). All of these findings suggest that CAG would be drug-candidate for the treatment of colorectal cancer.
Discussion
Immune escape is a critical reason for the failure of the immune system to control tumor growth, but how escape variants emerge during immunotherapy remains poorly understood.34 Studies have shown that the loss of tumor antigen-presenting function, epigenetic changes, the expression of antiapoptotic proteins and immunosuppressive receptors are closely related to tumor immune escape.6 7 35–37 In recent years, although immunotherapy represented by the PD-1 antibody has made great breakthroughs, the phenomena of drug resistance and unresponsive treatment of colorectal cancer has led to great dissatisfaction.38 39 Therefore, finding chemicals that can promote tumor antigen presentation and synergize with the PD-1 antibody can prevent tumor immune escape better.
Recent studies have found that active molecules from traditional Chinese medicine play an effective role in the treatment of diseases, such as celastrol in the treatment of metabolic syndrome,32 andrographolide in colitis and cancer,40 baicalin in lipid-lowering and trifolirhizin in tumor control.41 42 CAG is an active molecule in A. membranaceus and has the functions of cardiovascular diseases, liver protection, antibacterial, and treatment of abdominal aortic aneurysm.18 43–45 However, there is little research regarding it in the field of colorectal cancer. Therefore, we aimed to investigate whether and how CAG inhibits the growth of murine colon cancer.
The development of scRNA-seq and scATAC-seq technologies has greatly advanced the research of diseases.46–48 Therefore, we employed single-cell multiomics technology to analyze the specific antitumor mechanism of CAG. It was found that CAG promoted the antigen-presenting function of cancer cells, and the functions of CD8+ T cells, CD4+ T cells and NK cells in lymphocytes were enhanced to varying degrees. On the other hand, Spp1+ TAM cells in myeloid cells were more inclined to the transformation of inflammatory response and hypoxia signal was significantly inhibited after CAG treatment. These results are consistent with the results reported in previous studies, the inhibition of Spp1+ TAM hypoxia signal and enhancement of inflammatory response can inhibit the growth of tumor.49 Therefore, we speculated that CAG promotes the antigen-presenting function of cancer cells, so the corresponding CD8+ T cells and macrophages can recognize and kill cancer cells effectively. We further verified our conjecture with in vitro experiments. Flow cytometry analysis also showed that CAG increases the infiltration of CD45 immune cells, including CD8+ T cells and NK cells. In addition, the ability of IFN-γ and GZMB secreted by CD8+ T cells is also significantly enhanced by CAG. To determine whether CD8 T cells or macrophages play a major role in the antitumor experiment of CAG, we tested our conjecture with transplanted tumors in nude mice. Due to the development disorder of T cells caused by immunization of the thymus in nude mice while the function of macrophages still exists. However, the results showed that CAG could not effectively kill transplanted tumors in nude mice. These results suggest that CAG can enhance the killing effect of CD8+ T cells on tumors by promoting the antigen presentation of cancer cells.
Next, we explored how CAG promotes the antigen presentation of cancer cells by TRAP technology to find the target protein of the CAG to explain the phenomenon. CTSB is a cysteine hydrolase, which can interact with other proteins and degrade in lysosomes.50 We speculated that it might interact with antigen presentation-related proteins to cause its degradation. And it has been reported that the MHC-I molecule is degraded in lysosomes, resulting in the loss of tumor cell antigen-presenting function.13 Therefore, we suspected that CTSB might bind to the MHC-I molecule and cause its degradation in lysosomes. We verified this hypothesis that CAG could prevent the degradation of MHC-I by inhibiting the interaction of CTSB and MHC-I. Organoids is an effective model to study the progress of tumor diseases and have a great significance to promote the treatment of clinical tumor patients by investigating the effect of drugs on the growth of tumor organoids and the specific mechanism.51 52 Through our experiments, we found that combined with CAG and PD-1 antibodies have a good ability to inhibit tumor growth whether in mouse transplanted tumor model or in human colon cancer organoids. The shortage of this study is the limited number of xenograft samples and patient organoids. Although there are similar conclusions between murine models and human organoids, further investigation is needed.
Here, we describe the specific mechanism by which CAG inhibits the growth of colon cancer, mainly by inhibiting the degradation of MHC-I mediated by CTSB, to enhance the antitumor immunity of CD8+ T cells. On the one hand, it promotes the antigen presentation of cancer cells, and on the other hand, it relieves the depletion state of CD8+ T cells. The experimental results also confirmed that the combination of CAG and PD-1 antibody had a superior antitumor effect.
In conclusion, to the best of our knowledge, these findings highlight that CTSB downregulation confers antitumor immunity. Our research also explicated the specific mechanism of CAG in inhibiting the growth of colorectal cancer. It is worth mentioning that CAG has great potential as a health product in the European and American markets. Our research results provide a potential anticancer drug candidate.
Footnotes
GD, LZ and BW contributed equally.
Contributors: YS, HC and DS conceived this project and designed the study. GD, BW and XS performed the experiments and analyzed the data. LZ and HC analyzed the scRNA-seq and scATAC-seq data. DS and HC gave methodological support and conceptual advice. YS and GD wrote the manuscript. YS, as the guarantor, bears full responsibility for the work and/or conduct of the study, has the right to access the data and control the release decision.
Funding: This work was supported by National Key Research and Development Project (2017YFC1700602), National Natural Science Foundation of China (Nos. 81930117, 81872877, 82074318), the Priority Academic Programme Development of Jiangsu Higher Education Institutions and Mountain-Climbing Talents Project of Nanjing University (N/A), Natural Science Foundation Project of Shanghai, Scientific and Technological Innovation Action Plan (No. 22ZR1447400).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as online supplemental information.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
The human colon cancer organoids study was approved by the Research and Ethical Committee of Affiliated Hospital of Nanjing University of Chinese Medicine and complied with all relevant ethical regulations (2020-NL-094-02). Participants gave informed consent to participate in the study before taking part.
References
- 1.Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA A Cancer J Clinicians 2022;72:7–33. 10.3322/caac.21708 [DOI] [PubMed] [Google Scholar]
- 2.Kuipers EJ, Grady WM, Lieberman D, et al. Colorectal cancer. Nat Rev Dis Primers 2015;1:15065. 10.1038/nrdp.2015.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biller LH, Schrag D. Diagnosis and treatment of metastatic colorectal cancer. JAMA 2021;325:669–85. 10.1001/jama.2021.0106 [DOI] [PubMed] [Google Scholar]
- 4.Oliveira AF, Bretes L, Furtado I. Review of PD-1/PD-L1 inhibitors in metastatic dMMR/MSI-H colorectal cancer. Front Oncol 2019;9:396. 10.3389/fonc.2019.00396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das M, Zhu C, Kuchroo VK. Tim-3 and its role in regulating anti-tumor immunity. Immunol Rev 2017;276:97–111. 10.1111/imr.12520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang X, Wang J, Deng X, et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol Cancer 2019;18:10. 10.1186/s12943-018-0928-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhatchinamoorthy K, Colbert JD, Rock KL. Cancer immune evasion through loss of MHC class I antigen presentation. Front Immunol 2021;12:636568. 10.3389/fimmu.2021.636568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Y, Qian Y, Li Z, et al. Neoantigen‐reactive T cell: an emerging role in adoptive cellular immunotherapy. MedComm 2021;2:207–20. 10.1002/mco2.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vries JE, Yssel H, Spits H. Interplay between the TCR/CD3 complex and CD4 or CD8 in the activation of cytotoxic T lymphocytes. Immunol Rev 1989;109:119–42. 10.1111/j.1600-065X.1989.tb00022.x [DOI] [PubMed] [Google Scholar]
- 10.Huang J, Meyer C, Zhu C. T cell antigen recognition at the cell membrane. Mol Immunol 2012;52:155–64. 10.1016/j.molimm.2012.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allison JP. CD28-B7 interactions in T-cell activation. Curr Opin Immunol 1994;6:414–9. 10.1016/0952-7915(94)90120-1 [DOI] [PubMed] [Google Scholar]
- 12.Liu X, Bao X, Hu M, et al. Inhibition of PCSK9 potentiates immune checkpoint therapy for cancer. Nature 2020;588:693–8. 10.1038/s41586-020-2911-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto K, Venida A, Yano J, et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 2020;581:100–5. 10.1038/s41586-020-2229-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, Zhao Y, Luo W, et al. Celastrol induces ROS-mediated apoptosis via directly targeting peroxiredoxin-2 in gastric cancer cells. Theranostics 2020;10:10290–308. 10.7150/thno.46728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong N, Chen X, Feng J, et al. Baicalin induces ferroptosis in bladder cancer cells by downregulating FTH1. Acta Pharm Sin B 2021;11:4045–54. 10.1016/j.apsb.2021.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng G, Chen W, Wang P, et al. Inhibition of NLRP3 inflammasome-mediated pyroptosis in macrophage by cycloastragenol contributes to amelioration of imiquimod-induced psoriasis-like skin inflammation in mice. Int Immunopharmacol 2019;74:105682. 10.1016/j.intimp.2019.105682 [DOI] [PubMed] [Google Scholar]
- 17.Li M, Li S-chun, Dou B-kai, et al. Cycloastragenol upregulates SIRT1 expression, attenuates apoptosis and suppresses neuroinflammation after brain ischemia. Acta Pharmacol Sin 2020;41:1025–32. 10.1038/s41401-020-0386-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen C, Ni Y, Jiang B, et al. Anti-aging derivatives of cycloastragenol produced by biotransformation. Nat Prod Res 2021;35:2685–90. 10.1080/14786419.2019.1662011 [DOI] [PubMed] [Google Scholar]
- 19.Fauce SR, Jamieson BD, Chin AC, et al. Telomerase-based pharmacologic enhancement of antiviral function of human CD8 + T lymphocytes. J Immunol 2008;181:7400–6. 10.4049/jimmunol.181.10.7400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang B, Wang Y, Sun X, et al. CXCR6 is required for antitumor efficacy of intratumoral CD8 + T cell. J Immunother Cancer 2021;9:e003100. 10.1136/jitc-2021-003100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Butler A, Hoffman P, Smibert P, et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–20. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu G, Wang L-G, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology 2012;16:284–7. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell 2019;177:1888–902. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Granja JM, Corces MR, Pierce SE, et al. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat Genet 2021;53:403–11. 10.1038/s41588-021-00790-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korsunsky I, Millard N, Fan J, et al. Fast, sensitive and accurate integration of single-cell data with harmony. Nat Methods 2019;16:1289–96. 10.1038/s41592-019-0619-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapnell C, Cacchiarelli D, Grimsby J, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 2014;32:381–6. 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu X, Hill A, Packer J, et al. Single-cell mRNA quantification and differential analysis with census. Nat Methods 2017;14:309–15. 10.1038/nmeth.4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu X, Mao Q, Tang Y, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 2017;14:979–82. 10.1038/nmeth.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014;344:1396–401. 10.1126/science.1254257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian Y, Wan N, Ding M. Chemoproteomics maps glycolytic targetome in cancer cells. bioRxiv 2020. 10.1101/2020.11.18.387670 [DOI] [PubMed] [Google Scholar]
- 32.Zhu Y, Wan N, Shan X, et al. Celastrol targets adenylyl cyclase-associated protein 1 to reduce macrophages-mediated inflammation and ameliorates high fat diet-induced metabolic syndrome in mice. Acta Pharm Sin B 2021;11:1200–12. 10.1016/j.apsb.2020.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Driehuis E, Kretzschmar K, Clevers H. Establishment of patient-derived cancer organoids for drug-screening applications. Nat Protoc 2020;15:3380–409. 10.1038/s41596-020-0379-4 [DOI] [PubMed] [Google Scholar]
- 34.Lin K-Y, Lu D, Hung C-F, et al. Ectopic expression of vascular cell adhesion molecule-1 as a new mechanism for tumor immune evasion. Cancer Res 2007;67:1832–41. 10.1158/0008-5472.CAN-06-3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kriegsman BA, Vangala P, Chen BJ, et al. Frequent loss of IRF2 in cancers leads to immune evasion through decreased MHC class I antigen presentation and increased PD-L1 expression. J.i. 2019;203:1999–2010. 10.4049/jimmunol.1900475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomez S, Tabernacki T, Kobyra J, et al. Combining epigenetic and immune therapy to overcome cancer resistance. Semin Cancer Biol 2020;65:99–113. 10.1016/j.semcancer.2019.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mandal R, Barrón JC, Kostova I. Caspase-8: the double-edged sword. Biochim Biophys Acta Rev Cancer 1873;2020:188357. [DOI] [PubMed] [Google Scholar]
- 38.Qin S, Xu L, Yi M, et al. Novel immune checkpoint targets: moving beyond PD-1 and CTLA-4. Mol Cancer 2019;18:155. 10.1186/s12943-019-1091-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lizardo DY, Kuang C, Hao S, et al. Immunotherapy efficacy on mismatch repair-deficient colorectal cancer: from bench to bedside. Biochim Biophys Acta Rev Cancer 2020;1874:188447. 10.1016/j.bbcan.2020.188447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo W, Sun Y, Liu W, et al. Small molecule-driven mitophagy-mediated NLRP3 inflammasome inhibition is responsible for the prevention of colitis-associated cancer. Autophagy 2014;10:972–85. 10.4161/auto.28374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dai J, Liang K, Zhao S, et al. Chemoproteomics reveals baicalin activates hepatic CPT1 to ameliorate diet-induced obesity and hepatic steatosis. Proc Natl Acad Sci U S A 2018;115:E5896–905. 10.1073/pnas.1801745115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun D, Tao W, Zhang F, et al. Trifolirhizin induces autophagy-dependent apoptosis in colon cancer via AMPK/mTOR signaling. Sig Transduct Target Ther 2020;5:174. 10.1038/s41392-020-00281-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu M, Zhang S, Zhao Y, et al. Cycloastragenol improves hepatic steatosis by activating farnesoid X receptor signalling. Pharmacological Research 2017;121:22–32. 10.1016/j.phrs.2017.04.021 [DOI] [PubMed] [Google Scholar]
- 44.Wang J, Wu M-L, Cao S-P, et al. Cycloastragenol ameliorates experimental heart damage in rats by promoting myocardial autophagy via inhibition of AKT1-RPS6KB1 signaling. Biomed Pharmacother 2018;107:1074–81. 10.1016/j.biopha.2018.08.016 [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Chen C, Wang Q, et al. Inhibitory effects of cycloastragenol on abdominal aortic aneurysm and its related mechanisms. Br J Pharmacol 2019;176:282–96. 10.1111/bph.14515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JJ, Bernard V, Semaan A, et al. Elucidation of tumor-stromal heterogeneity and the ligand-receptor interactome by single-cell transcriptomics in real-world pancreatic cancer biopsies. Clin Cancer Res 2021;27:5912–21. 10.1158/1078-0432.CCR-20-3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L, Li Z, Skrzypczynska KM, et al. Single-Cell analyses inform mechanisms of Myeloid-Targeted therapies in colon cancer. Cell 2020;181:442–59. 10.1016/j.cell.2020.03.048 [DOI] [PubMed] [Google Scholar]
- 48.Gao J, Wu Z, Zhao M, et al. Allosteric inhibition reveals SHP2-mediated tumor immunosuppression in colon cancer by single-cell transcriptomics. Acta Pharm Sin B 2022;12:149–66. 10.1016/j.apsb.2021.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei J, Chen Z, Hu M, et al. Characterizing intercellular communication of pan-cancer reveals SPP1+ tumor-associated macrophage expanded in hypoxia and promoting cancer malignancy through single-cell RNA-Seq Data. Front Cell Dev Biol 2021;9:749210. 10.3389/fcell.2021.749210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen X, Yu C, Kang R, et al. Cellular degradation systems in ferroptosis. Cell Death Differ 2021;28:1135–48. 10.1038/s41418-020-00728-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Puschhof J, Pleguezuelos-Manzano C, Martinez-Silgado A, et al. Intestinal organoid cocultures with microbes. Nat Protoc 2021;16:4633–49. 10.1038/s41596-021-00589-z [DOI] [PubMed] [Google Scholar]
- 52.Lee SH, Hu W, Matulay JT, et al. Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell 2018;173:515–28. 10.1016/j.cell.2018.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-004874supp002.xlsx (13KB, xlsx)
jitc-2022-004874supp003.xlsx (10.7KB, xlsx)
jitc-2022-004874supp001.pdf (1.5MB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as online supplemental information.