Abstract
SARS-CoV-2 remains an acute threat to human health, endangering hospital capacities worldwide. Previous studies have aimed at informing pathophysiologic understanding and identification of disease indicators for risk assessment, monitoring, and therapeutic guidance. While findings start to emerge in the general population, observations in high-risk patients with complex pre-existing conditions are limited.
We addressed the gap of existing knowledge with regard to a differentiated understanding of disease dynamics in SARS-CoV-2 infection while specifically considering disease stage and severity. We biomedically characterized quantitative proteomics in a hospitalized cohort of COVID-19 patients with mild to severe symptoms suffering from different (co)-morbidities in comparison to both healthy individuals and patients with non-COVID related inflammation. Deep clinical phenotyping enabled the identification of individual disease trajectories in COVID-19 patients.
By the use of the individualized disease phase assignment, proteome analysis revealed a severity dependent general type-2-centered host response side-by-side with a disease specific antiviral immune reaction in early disease. The identification of phenomena such as neutrophil extracellular trap (NET) formation and a pro-coagulatory response characterizing severe disease was successfully validated in a second cohort. Together with the regulation of proteins related to SARS-CoV-2-specific symptoms identified by proteome screening, we not only confirmed results from previous studies but provide novel information for biomarker and therapy development.
Keywords: Biomarker, COVID-19, Immune response, NETosis, Proteomics, High-risk patients
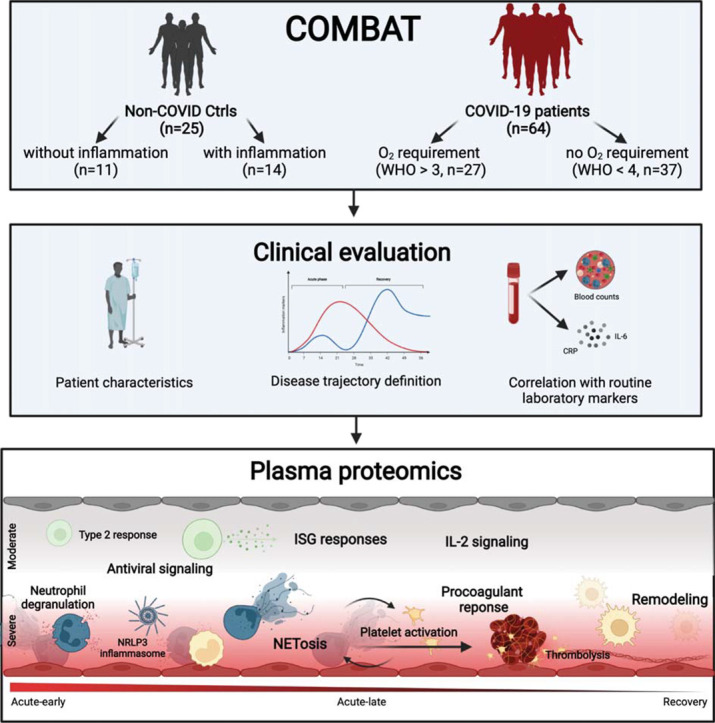
Graphical abstract
Sars-CoV-2 remains a challenging threat to our health care system with many pathophysiological mechanisms not fully understood, especially in high-risk patients. Therefore, we characterized a cohort of hospitalized COVID-19 patients with multiple comorbidities by quantitative plasma proteomics and deep clinical phenotyping. The individual patient's disease progression was determined and the subsequently assigned proteome profiles compared with a healthy and a chronically inflamed control cohort. The identified disease phase and severity specific protein profiles revealed an antiviral immune response together with coagulation activation indicating the formation of NETosis side-by-side with tissue remodeling related to the inflammatory signature.
Research in context
Evidence before this study
In the SARS-CoV-2 pandemic, studies thus far targeted both pathophysiologic understanding and biomarker identification to improve risk assessment for therapeutic guidance. While some insight was gained in the general population, high-risk COVID-19 patients with complex pre-existing conditions remain understudied. This is not only reflected by the lack of reliable, clinically applied biomarkers in this population but likely based on the use of strict time-of-infection-related disease phase assignment that cannot acknowledge the individual inflammatory response capacity in patients with significant pre-existing conditions and associated therapeutic interventions.
Added value of this study
Our study adds to the understanding of COVID-19 disease dynamics by combining advanced proteome analysis with the differentiated assignment of the patients to disease severity and stage using individual trajectories of IL-6 and CRP. Significantly supporting the identification of distinct immune phenomena, the study benefited from the comparison of expression patterns observed in COVID-19 patients to non-inflammatory and inflammatory control subjects. In the omnipresent, yet understudied patient collective with significant pre-existing conditions, we identified and validated COVID-related protein signatures that indicate antiviral response mechanisms together with NET/inflammasome activation in severely affected patients. In contrast, protein regulation in less severely diseased patients was found to be characterized by a type-2 centered immune response. In this screening approach, we not only confirmed but newly identified and validated urgently needed COVID-19 markers that reflect clinical disease characteristics while providing insight into immune pathophysiology.
Implications of all the available evidence
Addressing the important yet understudied patient collective suffering from preexisting medical conditions, our study enabled the identification of protein expression patterns that characterize disease-stage and phase in high-risk COVID-19 patients. The proteome analysis not only provides a valuable resource for further analysis in our deeply phenotyped patients but helps to identify clinically relevant pathophysiologic processes that affect clinical presentation and individual diseases susceptibility, thereby informing differentiated analysis in high-risk COVID-19.
1. Introduction
The SARS-CoV-2 pandemic continues to pose an immediate threat to global health. As of January 2022, worldwide COVID-19 cases exceed 250 million and deaths have surpassed 5,4 million [1]. Clinical manifestations vary from asymptomatic carrier to severe illness, organ dysfunction, chronic health impairment including long-COVID, and death [2].
To gain deeper insight and inform patient care, epidemiological approaches addressed clinical characteristics of different SARS-CoV-2 infection phases in the overall population and identified risk factors for adverse outcomes such as diabetes or hyperlipidemia [3], [4], [5]. Studies focused on the identification of clinical signs and early markers that reliably enable monitoring and treatment strategies [6], [7] including nationwide approaches stemming from the United Kingdom, Germany, France, Israel, and the USA [2], [8], [9]. While these attempts are already challenging in the general population [2], [10], [11], the aim has yet to be reached in cohorts of high-risk patients characterized by a complex picture of preexisting comorbidities. Care for these patients results in resource-intensive monitoring and treatment and thus remains a critical hurdle even for maximum care hospitals. Poor vaccination response rates in a large number of such complex cases and vaccine breakthroughs further complicate the picture [12], [13], [14].
Characterization of immune phenomena such as the ‘cytokine storm’ [15], [16] helped to guide treatment initiation and aided first therapeutic approaches [3], [4], [17]. Changes in human plasma protein levels have been suggested as disease indicators [18], [19], [20], in line with the implementation of protein markers for other viral diseases [21]. The analyses were furthermore used to gain pathophysiological insight in order to develop new therapeutic strategies [22], [23], [24].
To increase pathophysiologic insight and enable the identification of disease indicators in high-risk COVID-19 patients with significant preexisting conditions and to thereby inform monitoring and treatment decisions in the most important disease phases, we profiled host responses to SARS-CoV-2 infection by the use of quantitative plasma proteomics. Tracing disease trajectories by individual expression of inflammation markers enabled us to improve general time-of-infection-based approaches [25].
We herewith successfully identified and validated disease grade and disease phase-specific proteome profiles side-by-side with the regulation of characteristic routine laboratory variables in a high-risk, multimorbid patient cohort. Our study included survivors and non-survivors from COVID-19 and a range from mild to severe disease symptoms compared to patients with acute non-COVID-19 related inflammation, as well as non-inflammatory control cases. We thereby delineated both COVID-19-specific and general immune responses together with the phase-specific involvement of coagulation and remodeling processes as well as the differential regulation of proteins related to SARS-CoV-2-specific symptoms with the potential to significantly inform monitoring and treatment approaches.
2. Methods
2.1. Clinical data collection, patient grouping, and disease phase assignment
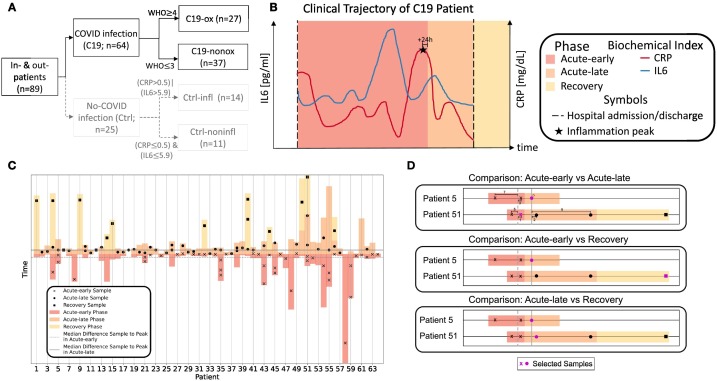
The study prospectively enrolled 64 patients with PCR confirmed SARS-CoV-2 infection during the first phase of the COVID-19 pandemic in Germany (03/2020 to 08/2020), before steroid treatment for SARS-CoV-2 was routinely prescribed. Patients were enrolled shortly before or at the onset of the acute infection phase when laboratory signs of infection and disease-specific symptoms develop. Twenty-five patients with acute (inflammatory control group; Ctrl-infl) or no/low non-COVID-19 related inflammation (healthy control group; Ctrl-noninfl) were additionally included in the study as control groups (Fig. 1A, Table 1 ). Patients and control subjects are part of the COVID-19 Registry of the LMU University Hospital Munich (CORKUM, WHO trial id DRKS00021225). Patient data were anonymized for analysis. The study was approved by the ethics committee of the Ludwig-Maximilians-Universität (LMU), Munich, Germany (Study title: “COVID-19 Register des LMU Klinikums (CORKUM)”; Project No: 20-245 (initial approval date: 03/2020; Amendment approval dates: 07/2020, 01/2021, 05/2021) as well as under the Project No: 20-259 (CPC-M bioArchive)). A second cohort of 36 patients was prospectively included for validation.
Fig. 1.
A: Patient number per study group considering C19 (black, solid) and non-C19 patients (gray, dashed); B: Exemplary COVID-19 disease trajectory based on routine biochemical indices IL-6 (blue) and CRP (red) during the hospital stay (dashed black lines) considering the respective disease phases (acute-early, acute-late, and recovery phase). C: Combined Stacked bar- and scatter plot indicating the disease phase assignment for each collected samples per patient. D: Examples of sample selection rules for group comparison in case of serial samples. When comparing acute-early and acute-late phases, for patient 5 the samples collected in the acute-early phase had a difference between sampling time point and the acute-early median of 7 or 3 day(s), whereas the sample collected during the acute-late phase showed the lowest difference of 0 days to the acute-late median. Therefore, the acute-late sample was selected for group comparison analysis. Sample selection for patient 51 from acute-early phase followed the same rules.
Table 1.
Group and phase definitions as well as study overview.
| Study groups | Description |
|---|---|
| COVID-19 (C19; n = 64) | PCR positive SARS-CoV-2 infection |
| C19 oxygen group (C19-ox; n = 27) | PCR positive SARS-CoV-2 infection; oxygen supply due to COVID-19: WHO score 4–8 |
| C19 no‑oxygen group (C19-nonox; n = 37) | PCR positive SARS-CoV-2 infection; no oxygen supply due to COVID-19: WHO score 1–3 |
| Inflammatory control group (Ctrl-infl; n = 14) | No SARS-CoV-2 infection (PCR); patient with diagnosed infection/inflammation CRP >0.5 mg/dl or IL-6 > 5.9 pg/ml |
| Healthy control group (Ctrl-noninfl; n = 11) | No SARS-CoV-2 infection (PCR); CRP ≤0.5 mg/dl and IL-6 ≤ 5.9 pg/ml |
| Inflammation peak | Time of the highest measured CRP or IL-6 value (whichever occurred later) broadened +24 h |
| Disease phase | |
| Acute-early | Interval between disease onset (symptoms and/or positive PCR test) until 24 h post inflammation peak |
| Acute-late | Interval starting 24 h post inflammation peak until hospital discharge |
| Recovery | Time after hospital discharge |
| Biospecimen | Heparin plasma |
| Sample analysis | Proteomics: Olink Explore 1536/384 |
| Routine biochemical indices | Laboratory values: Monocyte count [x109/l], Monocytes [%], Neutrophil count [x109/l], Neutrophils [%], Lymphocyte count [x109/l], Lymphocytes [%], Leukocyte count [x109/l], IL-6 [pg/ml], C-reactive protein (CRP) [mg/dl], Creatinine [mg/dl], Platelets [103/μl], Prothrombin Time, INR [sec], Fibrinogen [mg/dl], high sensitivity TroponinT (hsTroponinT) [μg/l], Partial thromboplastin time (PTT) [sec], Ferritin [μg/l], D-Dimer [μg/ml], Procalcitonin (PCT) [μg/l], glomerular filtration rate (eGFR) [ml/min/1.73m2] |
| Comorbidity (groups) | Diabetes, high cholesterol, cardiovascular disease, lung disease, kidney disease, immuno-compromised status, steroid intake during or before proteomics sampling, superinfection during proteomics sampling |
Comprehensive electronic health records of all 89 (64 COVID-19, 25 control patients) of the study cohort were provided including baseline information like age, gender, medical background about comorbidities, and medication before hospital admission. Furthermore, information about the clinical course was provided including different routine biomedical indices (e.g., blood cell measurements), different inflammation markers like C-reactive protein (CRP), IL-6, or Ferritin, coagulation markers such as platelet count or partial thromboplastin time (PTT), and other body function values (e.g., creatinine or hsTroponinT, measured at admission as well as repeatedly over the hospital stay as needed), received treatments (e.g., ventilation or medication), and adverse events (e.g., acute kidney failure, thrombosis/embolism, or death).
COVID-19 patients were classified according to ordinal scale for clinical improvement of COVID-19 infection reported by the WHO [26] and grouped into two sub-cohorts based on the need for oxygen supply, i.e., disease severity (WHO≥4 - C19-ox; WHO≤3 - C19-nonox) (Fig. 1A).
To specify the host immune response to COVID-19 infection while considering the underlying disease phase, we developed a novel approach for a high-risk, multimorbid patient cohort to improve upon general time-of-infection based approaches (e.g., [25]) or approaches using thresholds for levels of inflammation markers (e.g., [27], [28]). As IL-6 (>80 pg/ml) and CRP levels (>97 mg/l) correctly classified 80 % of patients regarding their risk of respiratory failure [27], we used these markers and extended this approach by defining disease phases based on individual trajectories while considering important clinical hallmarks. Using inflammation markers, we individually identified an inflammation peak for each patient, defined as the time point of the highest measured CRP or IL-6 value (whichever occurred later) broadened by a window of 24 h after this peak to account for individual differences in inflammation marker decline. Accordingly, a total of three disease phases were distinguished: C19-acute-early, C19-acute-late, and C19-recovery phase. The C19-acute-early phase was defined as the time between disease onset, i.e., the onset of clinical symptoms and/or first positive PCR test and the end of the inflammation peak. The C19-acute-late phase was defined as the time after the inflammation peak until hospital discharge. The C19-recovery phase was defined as the time after hospital discharge.
In cases with significant discrepancy of disease severity, i.e., maximum WHO score at admission and the individual trajectory of IL-6 and CRP, the samples were assigned to the acute-late phase, assuming a surpassed inflammation peak at admission (Figs. 1B & D, S1). Two samples (Patient 12 and 18, Fig. S1) without any IL-6 and CRP peaks were assigned based on the proteomic data, by applying a k-nearest neighbors clustering algorithm to assign the samples to their most likely disease phase while validating clinical symptoms for group assignment.
Non-C19 patients were assigned to two control groups based on the presence of inflammation: We included 14 patients with acute non-COVID related inflammation (Ctrl-infl) characterized by a maximum CRP > 0.5 mg/dl or IL-6 > 5.9 pg/ml and 11 subjects without elevated inflammation markers (Ctrl-noninfl), i.e., CRP ≤ 0.5 mg/dl and IL-6 ≤ 5.9 pg/ml (Fig. 1A). Disease phase assignment in the validation cohort followed the identical approach.
2.2. Sample collection and processing
2.2.1. Study cohort
A total of 129 samples were collected with one to five serial samples per patient across the different phases and subjected to proteomic analysis. Plasma was separated from heparinized whole blood by centrifugation at 2000g for 15 min at room temperature and immediately stored at −80 °C until preparation for proteome analysis.
2.2.2. Validation cohort
In a second cohort, 63 EDTA plasma samples were collected from 38 patients (one to two serial samples per patient) in the same time period and at the same hospital site, and subjected to proteomic analysis using the same platform.
2.3. Olink plasma proteomics
The Olink® Explore 1536 platform was used to measure protein abundance in plasma samples obtained from the study and the validation cohort. The full library consisting of four 384-plex panels (Inflammation, Oncology, Cardiometabolic, and Neurology) was used to screen 1472 proteins. Relative protein abundance was calculated from the number of matched counts on the Illumina NovaSeq 6000 run using two S1 flow cells with 2 × 50 base read lengths. The counts of protein specific-barcode sequences were transformed into Normalized Protein eXpression (NPX) units and an intensity normalization algorithm was applied to reduce the technical variation. The final data were provided in the arbitrary unit (NPX) on a log2 scale. Quality control (QC) was performed at both protein and sample levels. Three internal controls are spiked into each sample in order to monitor the quality of assay performance, as well as the quality of individual samples. Following criteria are applied to pass the sample QC: the average matched counts for each sample must exceed 500 counts; the deviation from the median value of the incubation- and amplification controls for each sample should not exceed ±0.3 NPX for either of the internal controls. We, therefore, excluded 8 samples, whose mean of the failing proteins deviated >0.5 standard deviations from the overall mean of all samples which passed QC. As a further QC instance for comparability, the three proteins TNF, CXCL8, and IL-6 were measured in each of the four Olink® Explore panels. Since all four measurements were highly correlated for each of the three proteins (TNF: r = 0.952–0.965; CXCL8: r = 0.989–0.998; IL-6: r = 0.979–0.997), we kept only one representative for each protein based on the minimal number of QC warnings, conformity in scatter plots, and population variances. Furthermore, Olink® recommends that proteins with a large proportion of samples below the limit of detection (LOD) should be excluded from the analysis. We, therefore, excluded 77 proteins that were under the LOD in >25 % of samples in all study groups. Pre-processing of the study and the validation cohort followed the same procedures.
2.4. Data analysis
We compared clinical covariates and routine biochemical indices within the first 24 h after hospital admission separately for C19-ox and C19-nonox with both control groups (Ctrl-infl and Ctrl-noninfl) in the study cohort. In addition, we analyzed routine biochemical indices and plasma proteomics by (1) comparing each phase (acute-early, acute-late, recovery) separately with both control groups (Ctrl-infl and Ctrl-noninfl) - overall (i.e., severity-independent), and within C19-ox and C19-nonox; (2) comparing the phases (acute-early, acute-late, recovery) with each other - overall (severity-independent) and within C19-ox and C19-nonox; and (3) comparing the two severity groups (C19-ox and C19-nonox) with each other in each phase (acute-early, acute-late, recovery).
Continuous and categorical variables were presented as median (interquartile range (IQR) with 25 % and 75 % percentiles) and n (%) respectively. We used the Mann-Whitney-Wilcoxon test, Welch test, Tukey's range test, X 2 test, and Dirichlet regression to compare differences between the C19 groups and the Ctrl-infl and Ctrl-noninfl control groups where appropriate. All tests were two-sided, and a P-value <0.05 was considered statistically significant. We used Python's SciPy package [29] to perform the statistical analysis. The effect sizes are described as log2 fold change (FC).
For differential proteomics analysis, one sample per patient was used to avoid autocorrelation in case serial samples were available. To select the respective sample from C19 patients, we applied the following rule: For comparisons involving either the acute-early or acute-late phase (or both), we calculated the time between each sampling time point and the inflammation peak to determine the median time between sampling and inflammation peak for both phases combined. We next determined the difference between each sampling time point and the calculated phase-medians (median difference of sampling time points and inflammation peak) and selected the sample with the smallest difference to the respective phase-median in each group or when selecting a sample from either group (see examples Fig. 1D). For calculations that included the recovery phase, we used i) the recovery phase sample in case more samples per patient were available from other phases or ii) the latest recovery phase sample in case more samples existed to achieve comparable sample numbers for each phase. The same rule was applied for the one patient with serial samples in the non-C19 group.
We used the R package limma (“Linear models for microarray data”) [30] adjusted for the following confounders: age, gender, cardiovascular diseases, diabetes, high cholesterol, lung disease, kidney disease, immuno-compromised status, superinfection during proteomics sampling, steroid treatment during hospital stay during or before proteomics sampling (Table S1) to determine differentially abundant proteins (DAP).
DAPs identified in the study cohort were subjected to statistical testing (limma) in the validation cohort while additionally considering a variable that encodes the differences in sample preparation, i.e., acquisition in EDTA (validation cohort) or heparin (study cohort), besides the aforementioned confounders.
Volcano Plots were created using the R package EnhancedVolcano [31]. We conducted overrepresentation tests (based on hypergeometric models with a minimum count of three proteins) for biological processes and pathways using ClusterProfiler [32] and ReactomePA [33], while the Enrichplot [34] package was used for visualization of the overrepresentation results. All tests for the proteomics analysis were corrected for multiple testing using Benjamini-Hochberg correction, where a P-value <0.05 was considered statistically significant.
We chose the clinical sampling closest to the proteomics sampling and accepted a range of 4 days (Fig. S2). Not all biochemical indices were available at any given time point (Fig. S3). Furthermore, no D-Dimer and IL-6 values were available in the Ctrl-noninfl group.
3. Results
3.1. Assignment to disease severity revealed differences in patient characteristics at admission
Within the enrolled 64 SARS-CoV-2 infected patients in the study cohort a maximum WHO score ≥4 [26] during the hospital stay was observed in 27 patients (C19-ox group) and ≤3 in 37 patients (C19-nonox group) (Fig. 1A). The disease severity groups showed differences in age and gender (age: C19-ox: 70 (IQR 59–79); C19-nonox: 57 (IQR 48–70); females: C19-ox: 37.0 %; C19-nonox group: 32.4 %), whereas no difference was observed for the time between symptom onset and hospitalization (C19-ox: 5 (IQR 1–8); C19-nonox: 5 (IQR 2–9) (days)) in contrast to a greater length of hospital stay in C19-ox patients (C19-ox: 12 (IQR 12–56); C19-nonox: 10 (IQR 7–17)). Most prevalent symptoms for C19-ox and C19-nonox patients were dyspnea (59.3 %; 37.8 %), fever (51.9 %; 54.1 %), fatigue (51.9 %; 29.7 %), and dry cough (48.2 %; 48.7 %). Both C19-ox and -nonox patients presented with different comorbidities including cardiovascular disease (66.7 %; 64.9 %), pre-existing lung disease (33.3 %; 13.5 %), immune compromise (37.0 %; 24.3 %), diabetes (33.3 %; 18.9 %), and hyperlipidemia (22.2 %; 18.9 %). In the course of the disease, some patients suffered from acute kidney failure (C19-ox: 11.1 %; C19-nonox: 2.7 %), whereas secondary bacterial, fungal, and/or viral infections (‘superinfection’) occurred more frequently in C19-ox patients (C19-ox: 44.4 %; C19-nonox: 27.0 %). 22.2 % and 44.4 % of patients in the C19-ox group underwent non-invasive or invasive ventilation. Therapeutic interventions including antibiotic (C19-ox: 81.5 %; C19-nonox: 64.9 %) and antithrombotic therapy (C19-ox: 59.3 %; C19-nonox: 62.2 %), parenteral nutrition (C19-ox: 37.0 %; C19-nonox: 50.0 %), non-opioid analgesics (C19-ox: 33.3 %; C19-nonox: 40.5 %) were administered in the majority of C19 patients irrespective of disease severity. Asthma therapy (C19-ox: 55.6 %; C19-nonox: 8.1 %) and antiviral treatment (C19-ox: 44.4 %; C19-nonox: 16.2 %) were used more frequently in higher disease grades.
Non-C19 patients were assigned to the two control groups based on levels of the inflammatory markers IL-6 and CRP, Ctrl-infl (n = 14) and Ctrl-noninfl (n = 11) (Fig. 1A, Table 1). Median age in years did not differ between both control groups (Ctrl-infl: 74 (IQR 61–67); Ctrl-noninfl: 69 (IQR 51–75)), whereas female patients were more frequent in the Ctrl-noninfl group (Ctrl-infl: 14.3 %; Ctrl-noninfl: 36.7 %). The median length of hospital stay (in days) was 9 (IQR 7–12) in Ctrl-infl and 1 (IQR 0–2) in Ctrl-noninfl. Ctrl-infl and Ctrl-noninfl patients were characterized by a high prevalence of comorbidities including cardiovascular disease (85.7 %; 63.6 %), pre-existing lung disease (14.3 %; 36.4 %), immune deficiency (50.5 %; 9.1 %), hyperlipidemia (28.6 %; 18.2 %), and diabetes (21.4 %; 18.2 %). Patient characteristics at hospital admission for all study groups are presented in Table S2, Table S3, details about the results of performed statistical tests are shown in Table S4.
3.2. Identification of distinct clinical trajectories for disease-severity and -phase assignment
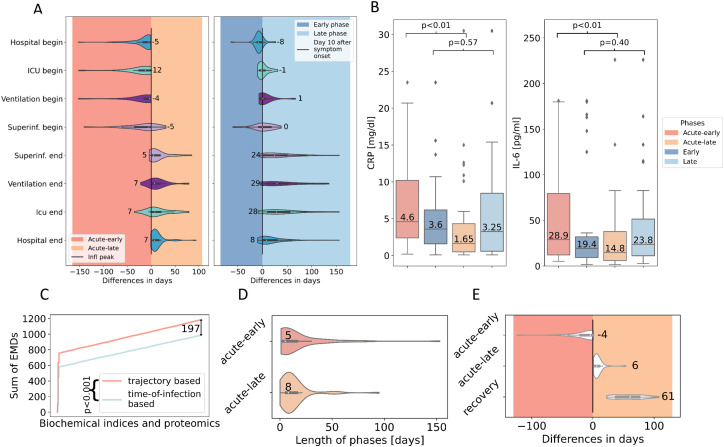
Next to disease-severity assignment according to the WHO criteria [26], we grouped samples of the 64 C19 patients of the study cohort in three distinct disease phases - acute-early, acute-late, and recovery - based on the individual trajectory of the inflammation markers IL-6 [pg/ml] and CRP [mg/dl] (Figs. 1B, S1, see Materials and Methods - Clinical Data Collection, patient grouping, and disease phase assignment). 44 samples of 35 patients were assigned to the C19 acute-early phase (interval from disease onset/or first positive PCR test until inflammation peak). 44 samples of 40 patients were assigned to the acute-late phase (interval from inflammation peak to discharge), whereas 15 samples of 13 patients were assigned to the recovery phase (after discharge). For significance tests, one sample per individual was used to avoid autocorrelation (see Material and Methods - Data analysis). This disease phase assignment was found to increase the fit of clinical symptoms and critical events, e.g., ICU admission or the initiation of oxygen treatment to the early disease phase (Fig. 2A (left)) in comparison to general time-of-infection based approaches that define disease phases by days from symptom onset (0–10 days, early phase; 10 days-discharge, late phase) [25] (Fig. 2A (right)). In addition, we showed CRP and IL-6 values to be statistically different when comparing the acute-early and acute-late disease phase (CRP: P-value<0.01; IL-6: P-value<0.01), as expected per design of phase assignment, whereas the disease phases based on the general time-of-infection approaches did not show significant differences for these routine markers (CRP: P-value = 0.57; IL-6: P-value = 0.40), indicating poor separation of subgroups (Fig. 2B).
Fig. 2.
A: Difference in days (median) between critical clinical events and disease phase assignment based on the individual trajectory-based approach (left) and the general time-of-infection-based approach (right). ICU: intensive care unit. B: Distribution of CRP and IL-6 values in different disease phases based on the individual trajectory-based (red) and general time-of-infection-based (blue) approach for disease phase assignment. C: Sum of earth mover's distance (EMD) from routine biochemical indices and proteomic levels comparing the trajectory-based (red) and time-of-infection-based (blue) approach for disease phase assignment. D: Length of disease phases (days; median) on patient level. E: Difference in days (median) between sampling time point and inflammation peak.
Quantitatively comparing the discriminatory power of the two approaches, we analyzed the overall earth mover's distance (EMD) [35] between the different phases defined by either the time-of-infection or inflammatory marker-based approach and found a significantly higher EMD (two-sided Wilcoxon Rank-sum test, P-value<0.001) for the inflammatory marker approach, indicating a clearer distinction between the different disease phases (Fig. 2C).
The median length of the acute-early phase was five (IQR 3–16) and the acute-late phase was eight (IQR 6–16) days (Fig. 2D). Sample acquisition for proteomic analysis was performed in accordance with routine procedures between four (IQR 0–19) days before to six (IQR 3–10) days after the inflammation peak as determined by Il-6 and CRP expression levels. Post-discharge (recovery) samples for proteomic analysis were obtained 61 (IQR 38–75) days after the patient surpassed the inflammation peak (Fig. 2D & E). C19 patients from all groups entered the hospital five (IQR 3–16) days before and were discharged seven (IQR 5–15) days after the inflammation peak. All patients in need of intensive care during their hospitalization were admitted to the ICU 12 (IQR 3–25) days before and discharged seven (IQR 0–20) days after the inflammation peak. In the majority of C19 patients, mechanical ventilation was initiated 4 days before the inflammation peak (IQR 2–10) and was terminated in the acute-late phase eight (IQR 4–21) days after the inflammation peak. Twenty-two C19 patients developed a secondary (super)infection (Fig. 2A (left)).
3.2.1. Disease-severity and -phase-dependent characteristics of routine laboratory values at admission and in the course of disease
At the time of admission, C19 patients of the study cohort showed significant changes in routine biochemical indices: Patients assigned to severe disease (C19-ox) showed comparable elevation of neutrophils when compared to the Ctrl-infl group, whereas patients with lower disease grades (C19-nonox) were characterized by blood cell counts within the physiologic range apart from monocytosis (Table S2, Table S3). Whereas CRP levels were found to be significantly different in all other group comparisons, CRP levels were comparable in patients from the C19-ox and Ctrl-infl group together with elevated levels of procalcitonin (PCT) and fibrinogen, thereby indicating the pathologic but non-discriminatory elevation of these parameters in C19-ox patients when compared to patients suffering from inflammation of different origin (Table S2, Table S3). Fibrinogen levels in C19-ox patients showed log2(FC) = 0.42 higher abundance when compared to the C19-nonox patients. However, ferritin levels were pathologically log2(FC) = 1.7 (C19-ox) to log2(FC) = 2.5 (C19-nonox) increased in both C19 severity groups when compared to Ctrl-infl in contrast to lower partial thromboplastin time (PTT, sec) in both C19 disease grades with most pronounced changes in C19-ox (log2(FC) = −0.27, Table S2, Table S3). Reference values for routine laboratory indices are given in Table S5.
3.2.2. Disease severity dependent differences in routine biochemical indices
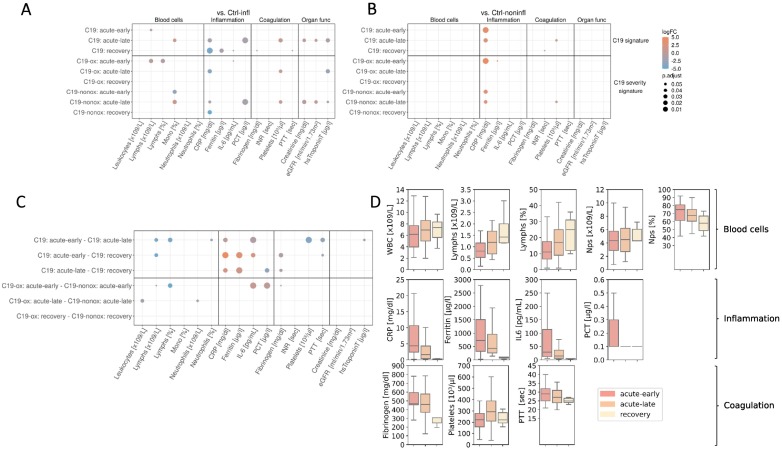
When investigating the course of the disease in the study cohort, blood cell counts in C19 patients in the acute-early phase of disease were characterized by decreased lymphocyte counts and proportion ([x109/l]: log2(FC) = −0.69, [%]: log2(FC) = −0.54) that increased in the acute-late and recovery phase, mainly driven by their downregulation in more diseased patients ([x109/l]: log2(FC) = −0.51; [%]: log2(FC) = −1.0) when compared with Ctrl-infl (Figs. 3A & C, S2, Table S6, Table S7). Whereas C19-ox patients in their acute-early and acute-late phase were comparable to the Ctrl-infl group with respect to elevated monocyte proportions, C19-nonox patients showed lower levels in the acute-early phase (log2(FC) = −3.05) that increase in the acute-late phase (log2(FC) = 0.58). Likewise, neutrophil proportions were significantly elevated in C19 patients in the acute-early phase compared to acute-late phase (log2(FC) = 0.13), with a trend to higher neutrophil levels in more diseased patients in the acute disease phase (acute-early: log2(FC) = 0.12; acute-late: log2(FC) = 0.14).
Fig. 3.
Dot plot of routine biochemical indices revealed significant (P-value ≤0.05) differences, describing a general C19 and a severity-based C19 signature when compared to the Ctrl-infl (A) and Ctrl-noninfl group (B) as well as the trajectory (C). Points represent significant level differences in the respective group comparison, where the colour of the dots describes the effect size (log2 fold change) and the size of the dots the significance niveau (adjusted P-value). D: Distribution of selected biochemical indices over the course of the disease.
In the acute-late (C19-ox: log2(FC) = −3.54, C19-nonox: log2(FC) = −1.16) and recovery phase (C19-ox: log2(FC) = −4.27, C19-nonox: log2(FC) = −4.86) CRP levels were lower in both C19 severity groups when compared to the Ctrl-infl group (Fig. 3). The overall decline in CRP values in the course of the disease in C19 patients is most pronounced in more diseased individuals to a level of the Ctr-noninfl cohort, whereas in C19-nonox patients moderately elevated CRP levels remain in the acute disease phase and normalize in the recovery phase together with the C19-ox CRP levels (Fig. 3D, Table S6, Table S7). Likewise, IL-6 values were differentially regulated through the course of the disease (Fig. 3D, Table S6, Table S7) with C19-ox patients in their acute-early phase showing significantly higher IL-6 levels (log2(FC) = 1.83, Fig. 3C) when compared to C19-nonox patients, in line with recent data [27].
Likewise, other inflammation parameters normalized in C19 patients in a disease-severity characteristic manner: While C19-ox patients in their acute-early phase showed elevated PCT levels that were indistinguishable from the Ctrl-infl group whereas C19-nonox patients did not show elevated levels in the acute-early phase (log2(FC) = 1.59; C19-ox higher). Patients from both severity groups demonstrated normalized levels as compared to Ctrl-noninfl. Additionally, we found a slow normalization of ferritin levels over the course of the disease in all C19 patients (C19: acute-early vs. recovery: log2(FC) = 3.03; C19: acute-late vs. recovery: log2(FC) = 2.30) (Fig. 3C & D), although ferritin levels still exceeded Ctrl-infl levels up until the acute-late phase (acute-early: log2(FC) = 1.42, acute-late: log2(FC) = 0.76) (Fig. 3B, Table S6, Table S7).
Accompanying the inflammatory response, platelet counts showed a physiologic niveau in more severely diseased patients in the acute-early phase to elevated levels exceeding Ctrl-infl levels in the acute-late phase (log2(FC) = 0.75). In C19-nonox patients, the analysis revealed persistently high and even further increasing platelet counts in the course of the disease when compared to the Ctrl-infl and Ctrl-noninfl groups (Ctrl-infl - acute-early: log2(FC) = 0.27, Ctrl-infl - acute-late: log2(FC) = 0.43). Elevated fibrinogen levels in the acute-early and -late phase were observed in both C19 groups compared to Ctrl-noninfl, declining in more diseased patients in the acute-late phase with C19-nonox patients reaching and C19-ox patients reaching close to physiologic levels in the recovery phase (Figs. 3C & D, S4, Table S6). Likewise, elevated PTT levels in C19-ox patients were comparable to Ctrl-infl in acute-early and normalized to physiological levels in the acute-late phase. In C19-nonox patients, moderately higher PTT levels compared to Ctrl-noninfl in acute-early normalized during the acute-late phase (Fig. 3, Table S6, Table S7).
Levels for kidney function, i.e., creatinine levels, and estimated glomerular filtration rate (eGFR) or cardiac injury, i.e., hsTroponinT did only show significant differences in the acute-late phase when compared to Ctrl-infl with creatinine and hsTroponinT reduced (creatinine: log2(FC) = −0.22, hsTroponinT: log2(FC) = −1.32) and eGFR elevated (log2(FC) = 0.29) mainly driven by changes in C19-nonox patients (Figs. 3A & C, S4, Table S6, Table S7).
3.3. Proteomic profiling tracking indicators for disease stage and severity
By comprehensively dissecting systemic immune responses at the protein level in C19 patients we significantly added to pathophysiologic insight and enabled the identification of future disease indicators using plasma proteomics obtained on the Olink® Explore platform (see Materials and Methods - Olink plasma proteomics). Disease characterization by these means found in the study cohort is outlined in the following chapters; validation of key phenomena is indicated at the end of the respective chapters.
3.3.1. Type 2 and antiviral immune response in early disease associated with subsequent activation of the coagulation cascade in C19 patients
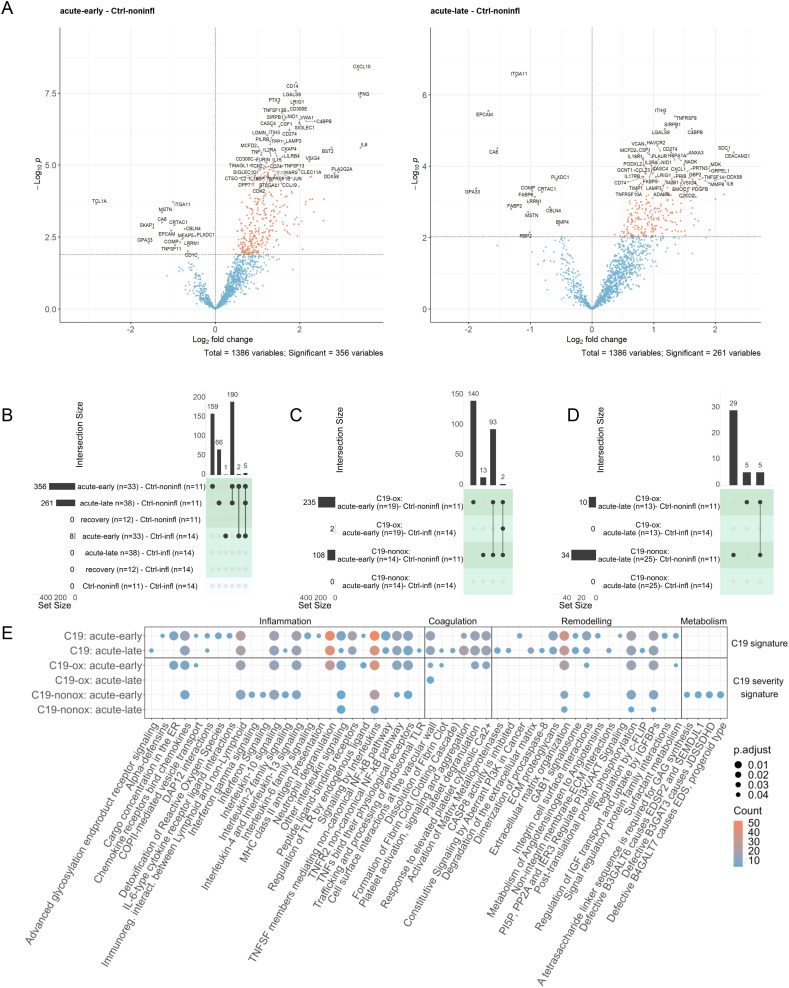
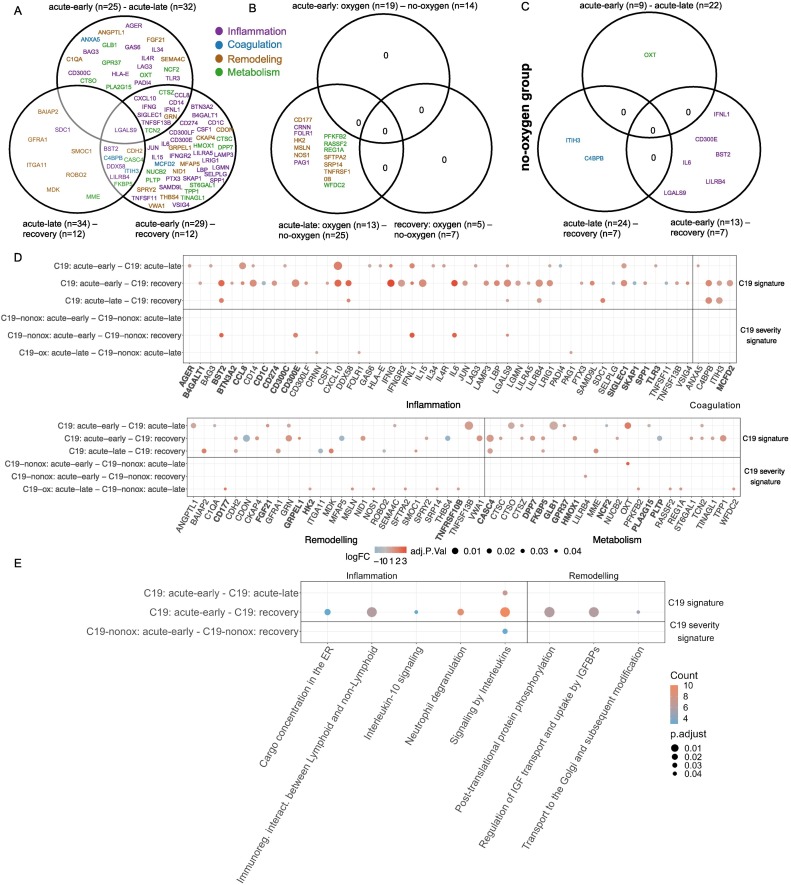
The comparison to Ctrl-noninfl patients identified the differential regulation of 356 (acute-early) and 261 (acute-late) proteins in C19 patients irrespective of disease severity. The majority of proteins were found to be upregulated (acute-early: 341; acute-late: 247) and showed a strong overlap of 195 differentially abundant proteins between the disease phases (Figs. 4A & B, 5, S5, Table S8) together with higher absolute log-fold changes in the acute-early comparison (acute-early: 1.0727 [iQR 0.833–1.362]; acute-late: 0.9683 [IQR: 0.769–2.294]). In contrast, the comparison of the recovery phase with Ctrl-noninfl did not reveal significant differences, indicating a normalization of the pathophysiologic changes after discharge in all patients.
Fig. 4.
A: Volcano plot of all phase comparisons with Ctrl-noninfl and Ctrl-infl in both severity grades C19-ox and C19-nonox. B: Overlapping significantly differentially abundant proteins in all phase comparisons with Ctrl-noninfl and Ctrl-infl overall; C: between C19-ox and C19-nonox in acute-early phase; D: between C19-ox and C19-nonox in acute-late phase. Horizontal bar graphs indicate total differentially abundant proteins (after multiple testing corrections), while vertical bar graphs indicate the number of overlapping differential proteins between different comparisons. E: Overlap of enriched pathways between all phase comparisons with Ctrl-noninfl.
Fig. 5.
Proteins involved in neutrophil degranulation and (A) NET formation, (B) IL-1 signaling, (C) TNFR cascade in all phase comparisons with Ctrl-noninfl.
Protein regulation in both acute phases was characterized by strong activation of innate and adaptive immune responses including a type-2 immune response, i.e., Interleukin-10 signaling, Interleukin-4, and Interleukin-13 signaling, and TNFR2 non-canonical NF-kB pathway signaling (i.e., TNFs bind their physiological receptors; TNF receptor superfamily (TNFSF) members mediating non-canonical NF-kB pathway), Neutrophil degranulation, DAP12 interactions, Immunoregulatory interactions between a lymphoid and a non-lymphoid cell and GPCR signaling (i.e., Chemokine receptors bind chemokines; Peptide ligand-binding receptors) when compared to Ctrl-noninfl (Fig. 4E, Table S9). The inflammation markers LGALS9 (Signaling by Interleukins, Interleukin-2 family signaling) and SIRPB1 (Neutrophil degranulation, Signal regulatory protein family interactions, DAP12 interactions) were significantly upregulated in both acute phases compared to Ctrl-noninfl, next to LRIG1, not represented in any regulated pathway. Interestingly, regulated proteins such as LGALS9 or SIRPB1 hold matrix remodeling capacities next to immune functions [36], [37].
Protein regulation in the acute-early phase was uniquely characterized by the involvement of inflammatory processes including interleukin signaling (i.e., Interleukin-6 family signaling; IL-6-type cytokine receptor-ligand interactions), innate (i.e., Alpha-defensins; Regulation of TLR by endogenous ligand), and adaptive immune responses (i.e., MHC class II antigen presentation), while Interleukin-2 family signaling and innate immune response pathways such as Advanced glycosylation endproduct receptor signaling and Trafficking and processing of endosomal TLR were only significantly regulated in the acute-late phase. The differential regulation between the acute-early and -late phase additionally comprised stress response mechanisms indicated by Detoxification of Reactive Oxygen Species, COPII-mediated vesicle transport, and Cargo concentration in the ER significantly regulated in the acute-early disease phase. The acute-early phase was furthermore characterized by regulation of the top-ranked inflammation marker CXCL10 (log2(FC) > 2, Signaling by Interleukins, Interleukin-10 signaling, Chemokine receptors bind chemokines and Peptide ligand-binding receptors), PTX3, involved in Neutrophil degranulation [38], and CD300E (Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell, DAP12 interactions), all persisting but less pronounced in the acute-late phase irrespective of disease grade. Likewise, IFNG (log2(FC) > 2, Signaling by Interleukins), TNFSF13B (TNFs bind their physiological receptors, TNFR2 non-canonical NF-kB pathway, TNF receptor superfamily (TNFSF) members mediating non-canonical NF-kB pathway), and CD14 (Regulation of TLR by endogenous ligand) were predominantly regulated in the acute-early phase in all C19 patients, whereas no differential regulation was observed in the acute-late phase when compared to Ctrl-noninfl. While TNFRSF8 (TNFs bind their physiological receptors, TNFR2 non-canonical NF-kB pathway) and HAVCR2 (Signaling by Interleukins, Interleukin-2 family signaling) were predominantly upregulated in the acute-late phase, their regulation was less pronounced in the acute-early phase when compared to Ctrl-noninfl.
Regulation of coagulation processes could be observed during both acute disease phases when compared to Ctrl-noninfl (i.e., Cell surface interactions at the vascular wall, Platelet activation, signaling and aggregation; Response to elevated platelet cytosolic Ca2+ and Platelet degranulation), accompanied by the upregulation of the coagulation marker C4BPB, not represented in any pathway, whereas Formation of Fibrin Clot (Clotting Cascade) and Dissolution of Fibrin Clot were significantly regulated in the acute-late phase, together with a downregulation of EPCAM (Cell surface interactions at the vascular wall) and upregulation of ITIH3 (Platelet degranulation, Response to elevated platelet cytosolic Ca2+, Platelet activation, signaling and aggregation), present but less pronounced in the acute-early phase, in line with observations that thrombotic events tend to appear in later disease stages of COVID-19 [39], [40].
Interestingly, a significant number of proteins associated with neutrophil activity and NET formation such as ANXA3, CCL7, HSPA1A, LCN2, LGALS9, MMP8, PPIB, PRTN3, and RETN [41], [42] were upregulated in both acute phases (acute-early: 58/78, acute-late: 54/78 NET formation proteins) compared to Ctrl-noninfl with 36 proteins similarly regulated across both acute disease phases, confirming previous reports of increased neutrophil degranulation and NETosis in C19 patients by proteome screening [43], [44], [45], [46], [47] (Fig. 5A). Similarly, the differential regulation of TNF (acute-early: 15/31, acute-late: 11/31 TNF signaling pathway proteins) and IL-1 (acute-early: 7/23, acute-late: 7/23 IL-1 signaling pathway proteins) signaling pathway associated proteins were detected in the acute disease phases when compared to Ctrl-noninfl (Fig. 5B & C). These proteomic signatures point towards the activation of inflammasome-associated processes [48], which are crucial for NET formation [49], [50].
Proteins involved in remodeling and repair processes were differentially expressed in both acute disease phases when compared to Ctrl-noninfl as indicated by the regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs), complemented by proteins involved in the Extracellular matrix organization, such as NID1 including regulation of ECM proteoglycans and Integrin cell surface interactions, eGFR signaling (i.e., GAB1 signalosome), and Post-translational protein phosphorylation. Whereas remodeling processes in the acute-early phase were characterized by tissue-cell (Metabolism of Angiotensinogen to Angiotensins; Surfactant metabolism, PI3K/AKT signaling via, i.e., PI5P, PP2A, and IER3) and cell-cell communication and remodeling (i.e., Signal regulatory protein family interactions, i.e., SIRPB1/SFTPA2/SFTPA1/SIRPA), the protein signature in the acute-late phase was dominated by processes such as the Degradation of the extracellular matrix, Activation of matrix metalloproteinases, Non-integrin membrane-ECM interactions, and regulation of the apoptosis pathways (i.e., CASP8 activity is inhibited, Regulation by c-FLIP, Dimerization of procaspase-8). These processes were accompanied by a decrease in the expression of ITGA11 (Extracellular matrix organization, Integrin cell surface interactions, top-ranked protein in the acute-late phase) and the increased expression of SDC1 (Signaling by Interleukins, Extracellular matrix organization, Other interleukin signaling, Cell surface interactions at the vascular wall). Not represented in the enriched pathways, the metabolic marker CA6 was found to be regulated in the acute-late phase. The downregulation of CA6 is strongly linked to low salivary zinc concentrations, associated with decreased taste acuity (hypogeusia) [51], and has been used in the diagnosis of Early Sjögren's Syndrome [52].
In summary, the proteomic response of C19 patients during the acute disease phase was characterized by the activation of classical inflammatory pathways, combined with markers indicating activation of the coagulation cascade and matrix remodeling when compared to Ctrl-noninfl individuals (Fig. 4, Fig. 5; S5; Table S8, Table S9). The unique combination of these processes together with the presence of specific markers, i.e., ANXA3, CCL7, HSPA1A, LCN2, LGALS9, MMP8, PPIB, PRTN3, and RETN pointed towards neutrophil degranulation and neutrophil extracellular trap (NET) formation in the acute disease phases in C19 patients, in line with previous studies describing innate immune cell activation in severe COVID-19 including neutrophil degranulation, NETosis, as well as pro-inflammatory/HLA-DRlo monocyte expansion [43], [44], [45], [46], [47] (Fig. 5). In addition, we detected a strong activation of interleukin signaling including activation of TNF signaling and a type-2 inflammation with the potential to counteract TNF-related signaling, especially in monocyte-related functions. These changes occurred together with the activation of both cytotoxic and humoral related immune defense mechanisms related to Interleukin-2 family signaling and DAP-12 in the acute-late phase, indicating the development of an adaptive immune response [53].
Whereas angiotensinogen, surfactant and SIRP metabolism, ROS regulation, and IL-6 signaling dominated protein regulation in the acute-early disease phase, the acute-later course was characterized by the differential expression of proteins indicating matrix degradation and apoptosis (Fig. 4E). Pathway enrichment analysis reflected vascular activation and organ damage that persisted into the acute-late phase together with markers of both coagulation and thrombolysis along with platelet degranulation in both acute-early and acute-late disease (Fig. 4E). However, protein regulation associated with coagulation processes was more pronounced in the acute-late phase.
The majority of the identified DAPs were confirmed in the validation cohort (acute-early: 180/356 DAP confirmed, acute-late: 100/261 DAP with 72 DAP overlapping) including key factors in inflammation (LGALS9, LRIG1, CXCL10, CD300E, IFNG, TNFRSF8, TNFS13B, CD14, HAVCR2), NETosis (SIRPB1, LRIG1, PTX3), and coagulation (LGALS9, CXCL10, C4BPB, ITIH3, NID1, SIRPB1, SFTPA1, SIRPA, ITGA11, SDC1) (Table S10).
3.3.2. Confirmation of the antiviral immune response as a C19-characteristic pattern of protein regulation
The comparison of C19 patients in the acute disease phases with Ctrl-infl patients demonstrated the differential regulation of eight proteins, all regulated in the acute-early phase irrespective of disease severity, whereas the comparison of the acute-late phase demonstrated no differentially expressed proteins (Figs. 4B, S5, Table S8). The C19-specific profile in the acute-early phase demonstrated the upregulation of antiviral signaling (DDX58, IFNL1, SAMD9L, GRN), lysosomal protein degradation (LGMN, TPP1), and Toll-like receptor signaling [54], as well as epithelial cell injury through upregulation of AGRN and downregulation of MUC16, as previously described in COVID-19 [47] of which a significant proportion was also regulated in the comparison to the Ctrl-noninfl group, i.e., DDX58, SAMD9L, LGMN, TPP1, and AGRN regulated in both acute phases; IFNL1 and GRN regulated in the acute-early phase. However, no coagulation markers or interleukin signaling-associated proteins were found differentially abundant.
The differential regulation of DDX58 (RIG-I) controls the recognition of infected cells while IFNL1 leads to the activation of the JAK/STAT signaling pathway resulting in the expression of IFN-stimulated genes (ISGs). Interestingly, these ISGs mediate the antiviral state essential for containment of SARS-CoV-2 in the upper respiratory tract [55], while loss of ISG function is associated with severe COVID-19 [56], [57]. The host response is further characterized by the regulation of apoptosis, cell cycle arrest, and DNA damage through SAMD9L, the activation of defense mechanisms involving monocyte differentiation and MHC class II presentation through LGMN, lysosomal protease functions controlled by TPP1 and activated through acidification such as GRN that holds a role in inflammation previously associated with COVID-19. Epithelial cell damage was indicated by the regulation of AGRN, as part of the lung basal membrane and MUC16, controlling mucus secretion and engaged in epithelial cell replication and apoptosis.
In summary, specific protein regulation in C19 likely related to an activation of the immune system not reflected by routine laboratory variables (i.e., CRP, IL-6, PCT, ferritin, and neutrophil proportions) that did not distinguish C19-ox patients from Ctrl-infl patients. A strong antiviral immune response side-by-side with markers indicating apoptosis and DNA damage both confirm previous findings as well as delineates the C19 immune response in the early course of the disease. The identified proteins, however, did not reach statistical significance in the validation cohort (Table S10).
3.3.3. Protein regulation in C19 patients indicates the regulation of antiviral immune response and a NETosis related activation of the coagulation cascade in more severe disease
When analyzing the general inflammatory response based on the comparison to Ctrl-noninfl cases, the acute-early phase in C19-ox patients revealed 235 significantly regulated proteins, with the majority, i.e., 227 proteins showing a strong upregulation in the acute-early phase in C19-ox patients (Figs. 4C & 5, S5, Table S8, Table S9). 140 of these 235 differentially regulated proteins characterized the general inflammatory early response in C19-ox patients and were not found to be differentially abundant within the early response in C19-nonox patients. Specific processes in higher disease severity grades of the acute-early phase involved regulation of the innate immune system via Endogenous ligand TLR signaling and Neutrophil degranulation (supported by an upregulation of the oxygen-specific markers SFTPA2, LBP, GRN, B4GALT1, and RETN in the acute-early phase) as well as stress response regulation indicated by enrichment of the pathways ER to Golgi Anterograde Transport via COPII-mediated vesicle transport and Cargo concentration in the ER. The top 10 of the 140 regulated proteins further included CCL7 (log2(FC) > 2), CCL2, JUN, AGER, NDUFS6 (log2(FC) > 2), indicating the activation of inflammatory and oxidative stress mechanisms in severe disease.
Regulated processes in severely affected C19 patients included both prothrombotic and thrombolytic processes such as Cell surface interactions at the vascular wall, Response to elevated platelet cytosolic Ca2+, Platelet degranulation, and Dissolution of Fibrin Clot. Tissue-cell interaction was represented by Surfactant metabolism, accompanied by the upregulation of the oxygen-specific marker SFTPA2 (also involved in Regulation of TLR by endogenous ligand; top 10) in the acute-early phase. All mentioned pathways apart from Dissolution of Fibrin Clot were found to be regulated in the overall comparison including both severity grades (Fig. 4E, Table S9), implying a strong contribution to the inflammatory and prothrombotic phenotype by their upregulation in patients with severe disease.
The acute-late phase in C19-ox patients revealed the differential regulation of ten proteins, eight of which were upregulated (Figs. 4D, S6, Table S8, Table S9). Inflammation markers in the acute-late phase of C19-ox patients included the upregulation of CXCL11 (log2(FC) > 2), HAVCR2, TNFRSF10B, SDC1 (log2(FC) > 2), and LGALS9, again pointing to a pronounced immune response as well as the activation of remodeling processes.
The coagulation process Cell surface interactions at the vascular wall was found to be persistently regulated in C19-ox patients in the acute-late phase, accompanied by the upregulation of the coagulation marker PLAUR, significantly involved in NET formation [58]. Likewise, involvement of remodeling as well as metabolic processes were indicated by the upregulation of CD177, MZB1, and CA6 (all three with log2(FC) > 2), as well as by the downregulation of ITGA11 in these patients. Several proteins, including the activation marker CD177, the thrombolytic PLAUR, as well as proinflammatory factors such as TNFRSF10B, HAVCR2, and MZB1 were uniquely regulated in C19-ox patients during the acute-late phase, in contrast to C19-nonox patients.
The comparison of protein regulation in C19-ox with patients from the Ctrl-infl group revealed the upregulation of DDX58 (log2(FC) > 2) and C4BPB in the acute-early phase (Figs. 4C, S5), whereas the analysis in the acute-late phase did not reveal differentially regulated proteins.
Protein regulation in patients with less severe disease (C19-nonox) revealed 108 differentially expressed proteins in the acute-early phase when compared to Ctrl-noninfl, with an upregulation in the majority of proteins, i.e., 104 (Figs. 4C, S5, Table S8, Table S9).
Inflammatory processes in the acute-early phase of C19-nonox patients comprised the enrichment of Interleukin-2 family signaling, Interferon-gamma (), and Interferon signaling (both unique in this comparison) when compared to Ctrl-noninfl. Inflammatory proteins regulated in acute-early C19-nonox, but not in C19-ox comprised the increased abundance of proteins associated with Interleukin-10 signaling (CCL3, CSF3), Chemokine receptors bind chemokines (CCL3, CCL21), and proteins not associated with enriched pathways (CCL8, CLEC6A, VSTM1). Similarly, PDGFRA was only differentially regulated in C19-nonox but not in C19-ox, whereas coagulation activation was not observed by pathway enrichment analysis.
Remodeling processes were indicated by the unique enrichment of pathways in the C19-nonox patients that related to Glycosaminoglycan metabolism (i.e., A tetrasaccharide linker sequence is required for GAG synthesis; Defective B3GAT3 causes JDSSDHD; Defective B3GALT6 causes EDSP2 and SEMDJL1; Defective B4GALT7 causes EDS, progeroid type). This was accompanied by the downregulation of the remodeling marker COMP (Extracellular matrix organization) as well as by the upregulation of TYMP, C1QTNF1, SCARB2, SIAE, and WISP2, all regulated in acute-early C19-nonox, but not in C19-ox when compared to Ctrl-noninfl. All other significantly enriched pathways in this comparison were shared with the profile observed in the acute-early phase of C19-ox patients (Fig. 4E, Table S9). When compared to Ctrl-noninfl, the acute-late phase in C19-nonox patients revealed 34 significantly regulated proteins, 25 of which were upregulated (Figs. 4D, S5, Table S8, Table S9). Specifically, 29 of those 34 proteins were regulated in the acute-late phase of C19-nonox patients only, with the top 10 ranked proteins including markers of inflammation (TNFRSF8, LRIG1), coagulation (EPCAM, C4BPB, ITIH3), remodeling (CRTAC1, NID1), and metabolic processes (FABP2, RBP2, CASC4). Pathway enrichment analysis revealed the regulation of inflammatory processes including cytokine signaling via Other interleukin signaling. Remodeling processes in C19-nonox patients during the acute-late phase included Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs), Post-translational protein phosphorylation, and Extracellular matrix organization.
All differentially expressed pathways in C19-nonox patients that were regulated in the disease independent analysis showed a shared expression with C19-ox patients indicating an equal contribution of both disease severities to the independent comparison in the acute-early disease phase. In contrast, the comparison demonstrated a dominating impact of the C19-nonox patients on the general signature in the acute late phase.
The protein profile in lower disease severity grades showed no differentially regulated proteins when comparing the acute-early and the acute-late phase to the Ctrl-infl group (Fig. 4D). Similarly, no differentially regulated proteins could be identified in the recovery phase for both oxygen-dependent and independent patients (Fig. 4B).
In summary, processes identified in C19 patients in general as outlined above were found to be more prominently or solely regulated in severely diseased C19 patients. When comparing severity groups, these changes were accompanied by a strong induction of an innate immune response in the early-acute phase indicated by Regulation of TLR through endogenous ligands, Neutrophil degranulation, and stress response mechanisms with Cargo concentration in the ER, and COPII-mediated vesicle transport. Simultaneously, procoagulant and thrombolytic phenomena, i.e., Response to elevated platelet cytosolic calcium, Platelet degranulation, Response to elevated platelet cytosolic Ca2+, Cell surface interactions at the vascular wall, and Dissolution of Fibrin Clot, could be observed in C19-ox patients (Fig. 4E).
In contrast, protein regulation in less severely diseased C19 patients was dominated by a general inflammatory response exemplified by the group-specific regulation of interleukin-2 and IFNG signaling in the acute-early phase, as well as a shared pattern, i.e., regulated in both severity groups that included remodeling processes indicated by regulation of Extracellular matrix organization, Post-translational protein phosphorylation, and Regulation of Insulin-like Growth Factor transport and uptake by Insulin-like Growth Factor Binding Proteins in the acute-early and -late phase (Fig. 4E). Interestingly, no pathways or proteins directly related to coagulation were found to be significantly regulated in less diseased patients, confirming previous clinical observations in these patients [45].
Changes observed in routine laboratory variables were reflected in protein expression patterns, i.e., elevated neutrophil numbers in more diseased patients correlated with the increased presence of markers for neutrophil degranulation and coagulation.
3.3.4. Dynamic regulation of immune and remodeling processes in the course of disease depending on disease severity
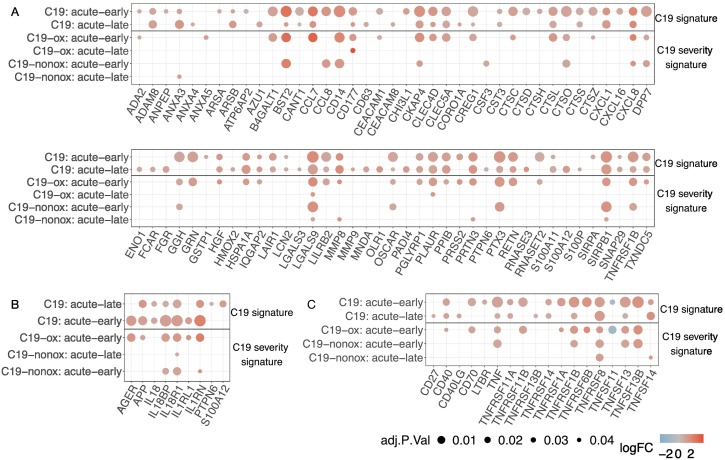
Next, we investigated the disease phase-dependent regulation of plasma proteins in C19 patients. Changes over the entire disease trajectory revealed 45 inflammation markers, four coagulation markers, 20 markers indicating remodeling processes, and 19 metabolic markers to be differentially regulated (Figs. 6A, S5, Table S10, Table S11). Specifically, 33 proteins were found to be differentially abundant in the study cohort between the acute-early and acute-late phase, yielding significant differences in interleukin signaling, but did not reach statistical significance in the validation cohort (Figs. 6A & B & E & F, S5, Table S10, Table S11). As expected, 17 of those 33 proteins showed strong differences between the acute-early and acute-late phase when compared to Ctrl-noninfl. Here, CD14, IFNG, TNFSF13B, CTSO, ANGPTL1, GRN, C1QA, AGER, IFNL1, LAG3, HLA-E, CCL8, GAS6, IL4R, CTSZ, and PLA2G15 were upregulated in the acute-early phase when compared to Ctrl-noninfl but were not significantly regulated in later phases, indicating an innate immune host response that involves vascular and matrix remodeling specific proteins and prominent interferon-related signaling in the acute-early disease phase. In contrast, PADI4 was only upregulated in the acute-late phase when compared to Ctrl-noninfl, holding a critical role in granulocyte and macrophage-dependent immune responses and a critical role in NET formation [59]. Upregulation of LGALS9, LRIG1, CXCL10, SIGLEC1, CD300C, and TCN2 in both acute phases compared to Ctrl-noninfl with significantly higher expression levels in the acute-early phase than in the acute-late phase pointed towards an innate immune response while involving factors that contribute to NET formation as well as stroke risk.
Fig. 6.
A: COVID-19 phase-dependent differential regulation of plasma proteins and their intersections. B: Differentially regulated plasma proteins and their intersections for disease severity comparisons (C19-ox/C19-nonox) considering different disease phases; purple: inflammation; blue: coagulation; brown: remodeling; green: metabolism C: Intersections of differentially abundant plasma proteins for different disease phases in C19-nonox. D: Log2 fold-change of phase-regulated proteins. Protein symbols in bold are associated with NETosis, IL-1, or TNF signaling. E: Overlapping pathways of all disease phase comparisons.
GLB1, OXT (log2(FC) > 2), FGF21, IL34, BAG3, SEMA4C, TLR3, GPR37, ANXA5 were found to be significantly upregulated in the C19 intragroup comparison of disease phases, i.e., during the acute-early disease course, whereas NCF2 was upregulated in the acute-late phase.
When comparing the acute-early to the recovery phase, we identified 59 differentially regulated proteins involved in innate and adaptive immunity, as well as remodeling processes with a predominant upregulation in the acute-early phase. The top 10 regulated proteins reflected this by the inclusion of inflammatory (CD300E, IL15, IFNG, LGALS9, CD14, CXCL10, IFNGR2, LILRB4), metabolic (CASC4), and remodeling processes (CDON9). Downregulated proteins were engaged in immune response mechanisms including cell adhesion, adaptive immune processes, cell-cell matrix interaction, and related metabolic activity (CD1C, TNFSF11, SELPLG, SKAP1 (inflammation), CDON, THBS4, MFAP5 (remodeling), PLTP (metabolism)) (Figs. 6A & B, S5).
Between the acute-late and the recovery phase, 17 proteins were found to be differentially regulated including inflammatory (BST2 (log2(FC) > 2), SDC1, DDX58, LGALS9, LILRB4), and coagulation specific (C4BPB, ITIH3) processes together with Remodeling of cardiac muscle and blood vessels (CDH2, SMOC1) and other tissues (GFRA1, MDK, BAIAP2, SMOC1), and metabolic activity (CASC4, MME, FKBP5). Whereas these proteins were upregulated in the acute-late phase, the proteins ITGA11 and ROBO2 with a role in tissue remodeling were upregulated in the recovery phase (Fig. 6A & B, bottom left circle). It has to be noted that important proteins such as BAIAP2 and GFRA1 that were identified by the phase comparison hold critical functions in the central nervous system.
While the majority of proteins, especially those associated with coagulation, showed a constant decrease in abundance over the disease trajectory, some proteins increased over time, e.g., CD1C, SELPLG (inflammation), SKAP1, TNFSF11 (inflammation), PLTP (metabolism), CDON, and MFAP5 (remodeling). Other proteins displayed more complex regulation patterns such as delayed changes, e.g., ITGA11, ROBO2, and THBS4 remained unchanged in acute disease and increased in recovery phase, while ANGPTL1 and FGF21 decreased in the acute-late and FKBP5 in the recovery phase; or alternating patterns (e.g., PADI4, SDC1, and BAIAP2) (Figs. S6 & S7).
When comparing disease phase-specific regulation for both severity grades, disease phase-dependent regulation differed for C19-ox and C19-nonox patients in the acute-late phase of the disease. Here, the comparison showed an upregulation of 14 proteins in C19-ox patients indicating the activation of innate and adaptive immune response mechanisms, energy metabolism, stress response, and remodeling including modulation of growth factor signaling (CRNN, FOLR1, PAG1 (inflammation), CD177 (log2(FC) > 2), HK2, MSLN, NOS1, SFTPA2, SRP14, TNFRSF10B (remodeling), PFKFB2, WFDC2, RASSF2, REG1A (metabolism)) (Figs. 6A & C, S5). No phase-specific regulated protein could be identified in C19-ox patients, while lower disease grades demonstrated differential regulation such as the upregulation of OXT (log2(FC) > 2, metabolism) in the acute-early phase when compared to the acute-late phase together with the inflammatory markers IFNL1, IL6, BST2 (all log2(FC) > 2), CD300E, LGALS9, and LILRB4 in comparison to the recovery phase. Additionally, coagulation and complement activation, e.g., upregulation of ITIH3 (coagulation) and C4BPB (coagulation) were observed in the acute-late phase when compared to the recovery phase. (Figs. 6A & D, S5).
Disease trajectory regulated inflammation associated pathways were primarily observed between acute-early and recovery phases, including interleukin signaling (i.e., Interleukin-10 signaling), Neutrophil degranulation, Cargo concentration in the ER, and Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell, capturing the innate and adaptive immune system, as well as ROS regulation. Remodeling processes further characterize the comparison between acute-early and recovery phase and comprised the Regulation of Insulin-like Growth Factor transport and uptake by Insulin-like Growth Factor Binding Proteins, as well as Post-translational protein phosphorylation via Transport to the Golgi and subsequent modification. Transport to the Golgi and subsequent modification was uniquely regulated in this comparison (Fig. 6F).
For proteins contributing to NET formation, a gradual decrease was detected over time for CCL8, LGALS9, ANXA5, GRN, CTSC, and MME when comparing the acute-early phase to later stages, whereas PADI4 and NCF2 showed an increase in the acute-late phase (Fig. S6), implicating neutrophil hyperactivation following the inflammatory peak.
In summary, the acute-early disease phase is specifically characterized by an innate immune, virus-related host response that involves vascular and matrix remodeling, while matrix remodeling proteins were also found to be upregulated in the recovery phase when compared to the acute-late phase (Figs. 6A & B & E & F, S5). In contrast, the critical regulator of NET formation PADI4 was differentially regulated in the acute-late phase. Protein expression pattern in both acute phases indicated regulation of innate immune defense mechanisms such as activation and recruitment of leukocytes, autophagy and indicated by the regulation of CXCL10, SIGLEC1, CD300C, NCF2, ANAX5, and BAG3 and matrix remodeling as identified through the differential expression of SEMA4, LGALS9, and GLB1, promoting mesenchymal activation and matrix formation. Interestingly, TCN2, engaged in vitamin B12 uptake, has been described to modify stroke risk [60], whereas GPR37 signaling has been shown to modulate the migration of olfactory ensheathing cells [61]. This expression pattern was most prominent in C19-ox patients who showed a strong time-dependent regulation of innate, adaptive immune, and stress response, as well as activation of the coagulation and complement system, e.g., upregulation of ITIH3 (coagulation) and C4BPB (coagulation), in the acute-late phase when compared against the recovery phase (Figs. 6A & C & D, S5).
4. Discussion
Although to date, numerous studies have described the wide range of symptoms of severe SARS-CoV-2 infection, e.g., acute respiratory distress syndrome (ARDS), lymphopenia, coagulopathy, and multi-organ damage [62], [63], [64], a detailed analysis of the underlying sequence of events is still missing. Studies that targeted protein regulation in COVID-19 patients aimed for a better understanding of disease-related processes while trying to identify potential biomarkers at the same time and have reported different immune response-related phenomena. The so-called “cytokine storm” comprised regulation of CXCL8, CXCL10, IL-6, TNFalpha, and IFNG, indicating that the synergism between TNF-α and IFNG, known to trigger inflammatory cell death and tissue damage, may account for SARS-CoV-2 mortality due to cytokine shock [15], [16], [65] and potentially addressed by existing therapies [66].
Our study addressed the gap of existing knowledge with regard to a differentiated understanding of disease dynamics while specifically considering disease stage and severity, thereby significantly adding to existing knowledge in the field (Fig. 7 ). Rooting the protein markers detected by an unbiased approach in disease pathophysiology, we achieved the identification of critical disease-stage and -phase-specific indicators in high-risk COVID-19 patients. We both confirmed as well as newly discovered urgently needed markers in a COVID-19 patient population that is omnipresent in university hospitals due to diverse preexisting conditions.
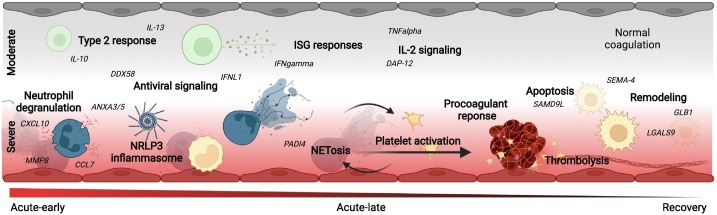
Fig. 7.
Schematic overview of the disease-phase and disease-severity-dependent pathophysiologic response in moderate (WHO≤4, upper half, C19-nonox) and severe (WHO≥4, lower half, C19-ox) C19 patients. While sharing a significant pattern of protein regulation, i.e., TNF signaling, severe disease was characterized by antiviral- and NETosis-related protein regulation including coagulation activation. Moderate disease associated patterns were dominated by a type-2 immune response and IL-2 associated signaling. Both disease severity grades progress from acute inflammation to the activation of remodeling processes.
To relate the plasma protein signatures detected by proteome screening to clinically relevant disease phases while considering internationally accepted disease severity grades, we studied COVID-19 patients of the study cohort in comparison to both non-inflammatory and inflammatory control subjects. To sensitively address the heterogeneous disease characteristics in a multimorbid patient cohort, we improved disease phase assignment by defining novel individual clinical trajectories using the inflammation markers IL-6 [pg/ml] and CRP [mg/dl]. In contrast to our approach, previous SARS-CoV-2 studies solely relied on disease phase assignment relating to the number of days after symptom onset, PCR result, or hospital admission, while other studies primarily referred to disease symptomatology [25], [67]. In general, cutoffs of inflammation markers have been used with good success in predicting COVID-19 severity at admission [27] but did not consider individual trajectories and threshold of laboratory variables that are likely of importance when studying a cohort with significant preexisting conditions and related medications.
The significant variation in the course of the disease when comparing different patient groups and treatment settings [68], including the average time for symptom resolution (2 to 71 days [69] or 10–14 (mild disease) to 21–42 (severe disease) days [70], [71]), likely renders solely ‘time-after-infection’ based disease phase assignment inaccurate, especially in patients with multiple influencing factors. In contrast to previous approaches [25], we, however, demonstrated that the trajectories based on the individual course of critical routine laboratory markers, more adequately matched clinical symptoms and critical events in the patient cohort studied, resulting in better discriminatory power for (severity-dependent) disease phase separation.
We showed that the inflammatory response in C19 patients obtained by the comparison to healthy individuals is characterized by a strong induction of innate immune response mechanisms in the early-acute phase as indicated by the regulation of TLR through endogenous ligands, Neutrophil degranulation together with cargo concentration in the ER, and COPII-mediated vesicle transport accompanied by the parallel upregulation of prothrombotic and thrombolytic phenomena, i.e., Dissolution of Fibrin Clot, Response to elevated platelet cytosolic calcium, and Platelet degranulation. The significant overlap of this protein signature with the disease-specific response revealed by the comparison to patients with signs of non-SARS-CoV-2 related inflammation the importance of interferon related antiviral signaling, i.e., DDX58 as confirmed by previous studies [72], together with the regulation of apoptosis and DNA damage, accompanied by the activation of TLR signaling, lysosomal functions and an indication of epithelial cell damage.
This response could be largely attributed to severe disease, whereas protein regulation in mild-to-moderately affected C19 patients was dominated by the disease-stage specific regulation of interleukin-2 and INFG signaling, as well as the shared regulation of remodeling processes as indicated by the regulation of Extracellular matrix organization, Post-translational protein phosphorylation, and Regulation of Insulin-like Growth Factor transport and uptake by Insulin-like Growth Factor Binding Proteins in the acute-early and late phase. However, the differential regulation indicated no activation of coagulation processes in less diseased patients in comparison to the Ctr-noninfl group although anticoagulation treatments were equally administered in both groups (C19-nonox: 62.16 %, C19-ox: 59.26 %). Interpretation of these findings, however, needs to take into account that deaths in the C19-ox group resulted in the overrepresentation of survival-related changes to protein expression in later disease phases.
The strong upregulation of proteins related to NET formation [41], [42] was observed and validated during both acute COVID phases. Activation of NET formation in context with other indicators of inflammasome activation [43], [44], [45], [46], [47], [48] specifically characterized patients with severe disease (WHO ≥4). The regulated proteins included CD177, a prominent activation marker present on the surface of circulating neutrophils [45], [73], MME (CD10) as an immaturity marker of neutrophils and previously associated with severe COVID-19 [25], [74], PDGFRA as a marker of platelet degranulation, and PADI4 as a key regulator of NETosis, whereas the classical NETosis/degranulation marker MPO was not found to be regulated in any of the comparisons. Similarly, strong activation of inflammasome related processes was indicated by the regulation of AGER as an important regulator of CASP-11 inflammasome activation [75] side-by-side with an upregulation of IL-1 and IL-18 [48], as well as IL-6 and TNF expression together with an overenrichement of TNF receptor superfamily (TNFSF) members mediating non-canonical NF-kB pathway [48]. Regulation of PLAUR points towards thromboembolic phenomena in these patients [25], [45], [76], which were controversially discussed for their dependence on disease severity [45].
When tracking the disease course, we observed the differential regulation of protein expression related to angiotensinogen, surfactant and SIRP metabolism, ROS regulation, and IL-6 signaling during early disease in the overall comparison and especially in the C19-ox patients in comparison to the non-inflammatory control group, whereas the later phase is characterized by the predominant regulation of proteins associated with matrix degradation and apoptosis.
On the one hand, we hereby show regulation of significant players in the immune host response confirming the role of inflammatory cell death and tissue damage [65]. On the other hand, we were able to add to previous studies by showing the dynamic of NETosis and inflammasome regulation [43], [44], [45], [46], [47] in severely affected patients in contrast to a type-2 centered immune response involving interleukin 4, 10, 13, and TNF signaling in both disease groups or in less severe disease only (e.g., Interleukin-2 family and IN signaling). These changes were found to be accompanied by remodeling processes.
Activation of the coagulation system was primarily detected in severely diseased patients in our cohort, although clinical reports also detected thromboembolic events in less severe disease [77], [78], potentially due to the lack of detection regarding local, organ-specific events. Activation of the coagulation system in more severely diseased patients, as well as activation of the complement system likely drives thrombo-inflammation in COVID-19 [79].
Regulation of proteins such as CA6 (associated with hypogeusia) or TCN2 (associated with stroke) identify disease characteristics, thereby supporting the significant potential of our unbiased approach to inform both pathophysiologic understanding and biomarker development.
Previous studies that employed proteome analysis mirrored our findings such as activation of the complement system, monocyte signaling (CD14, proteins of the LGAL family) and inflammation (CD48, SIRPB1) [20], as well as the regulation of different plasma protease inhibitors such as ITIH3 [19], [20], [80], [81] in COVID-19. Further in line with our findings, vascular markers such as vWF and proteins indicating coagulation activation were found to be regulated in previous studies, but in contrast to our studies described an early decrease [19], [81]. Likewise, proteins involved in metabolic processes such as lipoprotein homeostasis (PLTP) were differentially regulated in COVID-19 patients.
Enabling us to put the proteomic signatures into perspective and validate disease phase assignment, we comprehensively tracked biochemical indices. Here, comparable changes were observed in C19-ox and non-C19 related inflammation (Ctrl-infl) patients including, despite its common use in SARS-CoV2 [82], nondiscriminatory CRP levels when comparing C19-ox patients with subjects suffering from non-COVID related inflammation. Proteomic analysis, however, significantly broadened the picture by demonstrating significant differences in protein expression between these groups. In contrast, PTT levels differed between C19-ox and Ctrl-infl patients, in line with the observed proteomic pattern indicating coagulation activation in C19-ox patients. Although shortened PTT times were discussed to predict poor outcome in patients with varying diseases [83], its role in COVID-19 is still controversially discussed [84].
Changes in proteome pattern during the course of disease in each severity group were mirrored by cell numbers as well as coagulation and inflammation markers.
Regarding differential blood counts, the analysis in C19-ox patients indicated lymphopenia, low monocyte levels, and neutrophilia when compared to C19-nonox patients, in line with previous studies [2], [4], [85] but again did not differentiate well severely affected patients from subjects with inflammation of different origin (Ctrl-infl) supporting the controversial discussion of their predictive value [86]. In context with the changes in differentiated blood cell counts, the proteome changes likely reflected the activation of the immune system and again indicated the added value in delineating COVID-19 immune responses in relation to disease severity and -phase (Fig. 3, Fig. 4). Lower monocyte levels in more severely affected patients, however, could relate to the extravasation of these cells and suggest the subsequent activation of macrophages as indicated by the observed proteomic signature [87].
Association of INFG and TNF-associated monocyte polarization and the pro-fibrotic potential of monocyte-derived alveolar macrophages underline the potential of the observed signature to induce long-term remodeling [88], [89].
With regard to the impact of (co)morbidities, previous studies identified risk factors that were in part reflected in our cohort. Whereas C19-ox patients were characterized by increased age when compared to C19-nonox patients in line with previous studies [2], [4], we could not observe a higher rate of male patients in more severe disease [2], [4]. Similarly, we did not observe a significantly reduced time between symptom onset and hospitalization for more diseased patients [6], [7], but confirmed a longer hospital stay [6], [7], higher rates for ICU admission [6], [7], [90], invasive ventilation [6], [7], and adverse outcome. Further, we demonstrated a higher incidence of comorbidities, e.g., pre-existing lung disease, e.g., COPD or asthma [2], [4], hypertension [2], [4], diabetes [2], [4], kidney diseases [2], or impaired immune function [2], [4] in more diseases patients (C19-ox). In contrast, we did not find disease symptoms more prevalent in C19-ox patients compared to C19-nonox patients with the exception of fatigue, dyspnea, and an increased incidence in secondary infections, in line with previous studies [6], [7] and in part explaining the increased duration in hospital stay.
Limitations of the present study include its observational design and the retrospective analysis resulting in missing data in a small number of patients. Partially counteracting these limitations, the study benefits from homogenous and comprehensive clinical monitoring in a high-risk patient collective that continuously dominates patient admission in university hospitals during the COVID pandemic. While providing a very good basis for biomarker identification in different disease phases and severity grades, results have to be confirmed in targeted approaches in different clinical centers. These prospective studies need to include - among others - environmental or social factors not investigated in the current study while considering the impact of emerging SARS-CoV-2 variants and the effect of the potentially gender-dependent vaccination status, not present in the first pandemic wave addressed in our approach [91].
In summary, we identified a COVID-related protein signature that indicates an antiviral response together with NET/inflammasome activation predominantly driven by their regulation in severely affected patients. In contrast, regulation in less severely diseased patients was found to be characterized by a type-2 centered immune response. The findings were enabled by the newly identified disease trajectories based on the individual course of important routine laboratory variables. This approach both confirms findings from previous studies and also facilitates the identification of new proteins with significant potential to serve COVID-19 disease indicators at the same time. Translation of the results was achieved by the successful validation of the results in a second cohort, but could be further expanded through the analysis of available proteomic data sets such as Filbin et al. [92] when laboratory variables are made available. This kind of analysis can span multiple study sites and target treatment and vaccination effects. Translation into the clinical setting is supported by the ubiquitously available CRP and IL-6 that then enable to prospectively stratify COVID-19 patients according to disease severity or phase with a minimum of three serial measurements to judge the “inflammatory peak”. The identified proteomic signature will not only identify pathophysiological relevant processes and treatment targets, but improve disease stage or phase assignment.
The following are the supplementary data related to this article.
Description of confounders.
Clinical characteristics for different subgroups of the cohort at admission.
Results of subgroup comparisons for clinical characteristics and biomedical indices at admission.
Details of statistical analysis of clinical parameters.
Reference values for routine biochemical indices.
Routine biochemical indices for disease phase assignment.
Log2(fold change) of routine biochemical indices for groupwise comparisons.
Differential abundance analysis results in the study cohort: phases compared to control groups.
Pathway enrichment analysis in the study cohort: phases compared to control groups.
Differential Abundance Analysis Results in the Validation Cohort: Phases compared to control groups.
Differential abundance analysis results in the study cohort: trajectory comparisons.
Pathway enrichment analysis results in the study cohort: trajectory comparisons.
Supplementary figures
CRediT authorship contribution statement
Data Curation: Johannes C. Helmuth, Clemens Scherer, Maximilian Muenchhoff, Project administration: Benjamin Schubert and Anne Hilgendorff; Funding acquisition: Stefanie M. Hauck, Jürgen Behr, Li Deng, Narges Ahmidi, Benjamin Schubert, Anne Hilgendorff; Investigation: Johannes C. Helmuth, Clemens Scherer, Maximilian Muenchhoff, Rainer Kaiser; Methodology: Alina Bauer, Elisabeth Pachl, Benjamin Schubert; Resources: Johannes C. Hellmuth, Clemens Scherer, Maximilian Muenchhoff, Marion Frankenberger, Stefanie M. Hauck, Daniel Teupser, Nikolaus Kneidinger, Hans Stubbe, Agnese Petrera, Motaharehsadat Heydarian; Software: Alina Bauer and Elisabeth Pachl; Supervision: Narges Ahmidi, Benjamin Schubert, Anne Hilgendorff; Validation: Benjamin Schubert, Anne Higendorff, Jürgen Behr, Rainer Kaiser, Daniel Teupser; Bernhard Ryffe; Visualization: Alina Bauer and Elisabeth Pachl; Writing - original draft: Alina Bauer and Elisabeth Pachl; Writing - review and editing: Benjamin Schubert and Anne Hilgendorff; All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by COMBAT C19IR (01KI20249), funded by the Federal Ministry of Education and Research (BMBF) and the Free State of Bavaria under the Excellence Strategy of the Federal and State Government (LMUexcellent), as well as partially funded by the Bavarian Ministry for Economic Affairs, Regional Development and Energy as part of a project to support the thematic development of the Institute for Cognitive Systems (IKS). The present study was supported by the German Center for Lung Research (DZL, German Ministry of Education and Health (BMBF)) as well as the Research Training Group Targets in Toxicology (GRK2338) of the German Science and Research Organization (DFG). Additional financial support was provided by the Stiftung AtemWeg (LSS AIRR) and by the Helmholtz Association's Initiative and Networking Fund through Helmholtz AI.
Declaration of competing interest
The authors declare no conflict of interest apart from Clemens Scherer, who received speaker honoraria from AstraZeneca, outside the submitted work. All authors were involved in the decision to publish and reviewed the article before submission.
The authors declare no conflict of interest apart from Clemens Scherer, who received speaker honoraria from AstraZeneca, outside the submitted work.
Acknowledgments
We would like to thank all CORKUM investigators and staff. The authors thank the patients and their families for their participation in the CORKUM registry.
Data availability
The data that support the findings of this study are available from the corresponding authors upon request. Access to data for research purposes is possible for eligible institutes according to GDPR rules.
References
- 1.WHO Coronavirus (COVID-19) Dashboard [Internet] https://covid19.who.int/ [cited 2021 Oct 27]. Available from:
- 2.Williamson E.J., Walker A.J., Bhaskaran K., Bacon S., Bates C., Morton C.E., et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature. 2020;584(7821):430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan,China. Lancet. 2020 Feb 15;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan W.-J., Ni Z.-Y., Hu Y., Liang W.-H., Ou C.-Q., He J.-X., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J., Zhang S., Wu Z., Shang Y., Dong X., Li G., et al. Clinical outcomes of COVID-19 in Wuhan, China: a large cohort study. Ann. Intensive Care. 2020;10(1):99. doi: 10.1186/s13613-020-00706-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nachtigall I., Lenga P., Jóźwiak K., Thürmann P., Meier-Hellmann A., Kuhlen R., et al. Clinical course and factors associated with outcomes among 1904 patients hospitalized with COVID-19 in Germany: an observational study. Clin. Microbiol. Infect. 2020;26(12):1663–1669. doi: 10.1016/j.cmi.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piroth L., Cottenet J., Mariet A.-S., Bonniaud P., Blot M., Tubert-Bitter P., et al. Comparison of the characteristics, morbidity, and mortality of COVID-19 and seasonal influenza: a nationwide, population-based retrospective cohort study. Lancet Respir. Med. 2021;9(3):251–259. doi: 10.1016/S2213-2600(20)30527-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Booth A., Reed A.B., Ponzo S., Yassaee A., Aral M., Plans D., et al. Population risk factors for severe disease and mortality in COVID-19: a global systematic review and meta-analysis. PLoS One. 2021;16(3) doi: 10.1371/journal.pone.0247461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hodges G., Pallisgaard J., Schjerning Olsen A.-M., McGettigan P., Andersen M., Krogager M., et al. Association between biomarkers and COVID-19 severity and mortality: a nationwide danish cohort study. BMJ Open. 2020;10(12) doi: 10.1136/bmjopen-2020-041295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyarsky B.J., Werbel W.A., Avery R.K., Tobian A.A.R., Massie A.B., Segev D.L., et al. Antibody response to 2-dose SARS-CoV-2 mRNA vaccine series in solid organ transplant recipients. JAMA. 2021;325(21):2204–2206. doi: 10.1001/jama.2021.7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malinis M., Cohen E., Azar M.M. Effectiveness of SARS-CoV-2 vaccination in fully vaccinated solid organ transplant recipients. Am. J. Transplant. 2021;21(8):2916–2918. doi: 10.1111/ajt.16713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juthani P.V., Gupta A., Borges K.A., Price C.C., Lee A.I., Won C.H., et al. Hospitalisation among vaccine breakthrough COVID-19 infections. Lancet Infect Dis. 2021 doi: 10.1016/S1473-3099(21)00558-2. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang L., Xie X., Tu Z., Fu J., Xu D., Zhou Y. The signal pathways and treatment of cytokine storm in COVID-19. Signal. Transduct. Target Ther. 2021;6(1):255. doi: 10.1038/s41392-021-00679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buszko M., Nita-Lazar A., Park J.-H., Schwartzberg P.L., Verthelyi D., Young H.A., et al. Lessons learned: new insights on the role of cytokines in COVID-19. Nat. Immunol. 2021;22(4):404–411. doi: 10.1038/s41590-021-00901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J.-J., Dong X., Cao Y.-Y., Yuan Y.-D., Yang Y.-B., Yan Y.-Q., et al. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan,China. Allergy. 2020;75(7):1730–1741. doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- 18.Shu T., Ning W., Wu D., Xu J., Han Q., Huang M., et al. Plasma proteomics identify biomarkers and pathogenesis of COVID-19. Immunity. 2020;53(5):1108–1122. doi: 10.1016/j.immuni.2020.10.008. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Messner C.B., Demichev V., Wendisch D., Michalick L., White M., Freiwald A., et al. Ultra-high-throughput clinical proteomics reveals classifiers of COVID-19 infection. Cell Syst. 2020;11(1):11–24. doi: 10.1016/j.cels.2020.05.012. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park J., Kim H., Kim S.Y., Kim Y., Lee J.-S., Dan K., et al. In-depth blood proteome profiling analysis revealed distinct functional characteristics of plasma proteins between severe and non-severe COVID-19 patients. Sci. Rep. 2020;10(1):22418. doi: 10.1038/s41598-020-80120-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oxford K.L., Wendler J.P., McDermott J.E., White R.A., III, Powell J.D., Jacobs J.M., et al. The landscape of viral proteomics and its potential to impact human health. Expert Rev. Proteomics. 2016;13(6):579–591. doi: 10.1080/14789450.2016.1184091. [DOI] [PubMed] [Google Scholar]
- 22.Demichev V., Tober-Lau P., Lemke O., Nazarenko T., Thibeault C., Whitwell H., et al. A time-resolved proteomic and prognostic map of COVID-19. Cell Syst. 2021;12(8):780–794. doi: 10.1016/j.cels.2021.05.005. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haljasmägi L., Salumets A., Rumm A.P., Jürgenson M., Krassohhina E., Remm A., et al. Longitudinal proteomic profiling reveals increased early inflammation and sustained apoptosis proteins in severe COVID-19. Sci. Rep. 2020;10(1):20533. doi: 10.1038/s41598-020-77525-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filbin M.R., Mehta A., Schneider A.M., Kays K.R., Guess J.R., Gentili M., et al. Longitudinal proteomic analysis of severe COVID-19 reveals survival-associated signatures, tissue-specific cell death, and cell-cell interactions. Cell Rep. Med. 2021;2(5) doi: 10.1016/j.xcrm.2021.100287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182(6):1419–1440. doi: 10.1016/j.cell.2020.08.001. e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blueprint W. Novel Coronavirus. COVID-19 Therapeutic Trial Synopsis. 2020. https://www.who.int/blueprint/priority-diseases/key-action/COVID-19_Treatment_Trial_Design_Master_Protocol_synopsis_Final_18022020pdf (accessed on 5 February 2021) [Internet] Available from: https://bsitd.com.bd/wp-content/uploads/2020/06/7_an-international-randomised-trial-of-candidate-vaccines-against-covid-19.pdf.
- 27.Herold T., Jurinovic V., Arnreich C., Lipworth B.J., Hellmuth J.C., von Bergwelt-Baildon M., et al. Elevated levels of IL-6 and CRP predict the need for mechanical ventilation in COVID-19. J. Allergy Clin. Immunol. 2020;146(1):128–136. doi: 10.1016/j.jaci.2020.05.008. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LYC Chen, Hoiland R.L., Stukas S., Wellington C.L., Sekhon M.S. Confronting the controversy: interleukin-6 and the COVID-19 cytokine storm syndrome. Eur Respir J. 2020;56(4) doi: 10.1183/13993003.03006-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inglett G.E., Chen D., Liu S.X. Pasting and rheological properties of quinoa-oat composites. Int. J. Food Sci. Technol. 2015;50(4):878–884. [Google Scholar]
- 30.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7) doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blighe K., Rana S. 2019. https://github.com/kevinblighe
- 32.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu G., He Q.-Y. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol. BioSyst. 2016;12(2):477–479. doi: 10.1039/c5mb00663e. [DOI] [PubMed] [Google Scholar]
- 34.Yu G. Enrichplot: Visualization of Functional Enrichment Result. R Package Version. Vol. 1. 2018. p. 2. [Google Scholar]
- 35.Rubner Y., Tomasi C., Guibas L.J. The earth mover's distance as a metric for image retrieval. Int. J. Comput. Vis. 2000;40(2):99–121. [Google Scholar]
- 36.Hsu Y.-A., Chang C.-Y., Lan J.-L., Li J.-P., Lin H.-J., Chen C.-S., et al. Amelioration of bleomycin-induced pulmonary fibrosis via TGF-β-induced smad and non-smad signaling pathways in galectin-9-deficient mice and fibroblast cells. J. Biomed. Sci. 2020;27(1):24. doi: 10.1186/s12929-020-0616-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y.-C., Gonzalez M.E., Burman B., Zhao X., Anwar T., Tran M., et al. Mesenchymal stem/stromal cell engulfment reveals metastatic advantage in breast cancer. Cell Rep. 2019;27(13):3916–3926. doi: 10.1016/j.celrep.2019.05.084. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daigo K., Takamatsu Y., Hamakubo T. The protective effect against extracellular histones afforded by long-pentraxin PTX3 as a regulator of NETs. Front. Immunol. 2016;7(7):344. doi: 10.3389/fimmu.2016.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al-Ani F., Chehade S., Lazo-Langner A. Thrombosis risk associated with COVID-19 infection.A scoping review. Thromb. Res. 2020;192:152–160. doi: 10.1016/j.thromres.2020.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bussani R., Schneider E., Zentilin L., Collesi C., Ali H., Braga L., et al. Persistence of viral RNA, pneumocyte syncytia and thrombosis are hallmarks of advanced COVID-19 pathology. EBioMedicine. 2020;61 doi: 10.1016/j.ebiom.2020.103104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J., Li Q., Yin Y., Zhang Y., Cao Y., Lin X., et al. Excessive neutrophils and neutrophil extracellular traps in COVID-19. Front Immunol. 2020 doi: 10.3389/fimmu.2020.02063. [cited 2021 Dec 6];0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D.S., et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303(5663):1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 43.Bardoel B.W., Kenny E.F., Sollberger G., Zychlinsky A. The balancing act of neutrophils. Cell Host Microbe. 2014;15(5):526–536. doi: 10.1016/j.chom.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 44.Middleton E.A., He X.-Y., Denorme F., Campbell R.A., Ng D., Salvatore S.P., et al. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood. 2020;136(10):1169–1179. doi: 10.1182/blood.2020007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicolai L., Leunig A., Brambs S., Kaiser R., Weinberger T., Weigand M., et al. Immunothrombotic dysregulation in COVID-19 pneumonia is associated with respiratory failure and coagulopathy. Circulation. 2020;142(12):1176–1189. doi: 10.1161/CIRCULATIONAHA.120.048488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Y., Tao W., Shen F., Du W., Xu Z., Liu Z. The emerging role of neutrophil extracellular traps in arterial, venous and cancer-associated thrombosis. Front. Cardiovasc. Med. 2021;2(8) doi: 10.3389/fcvm.2021.786387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smet A., Breugelmans T., Michiels J., Lamote K., Arras W., De Man J.G., et al. A dynamic mucin mRNA signature associates with COVID-19 disease presentation and severity. JCI Insight. 2021;6(19) doi: 10.1172/jci.insight.151777. http://insight.jci.org/articles/view/151777/files/pdf Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng D., Liwinski T., Elinav E. Inflammasome activation and regulation: toward a better understanding of complex mechanisms. Cell Discov. 2020;9(6):36. doi: 10.1038/s41421-020-0167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Münzer P., Negro R., Magupalli V., Kittisopikul M., Vahabikashi A., Wong S.L., et al. Assembly of the Nlrp3 inflammasome regulates NET formation and is promoted by the vimentin intermediate filament cytoskeletal system. Arterioscler. Thromb. Vasc. Biol. 2019;39(Suppl_1):A118. [Google Scholar]
- 50.Münzer P., Negro R., Fukui S., di Meglio L., Aymonnier K., Chu L., et al. NLRP3 inflammasome assembly in neutrophils is supported by PAD4 and promotes NETosis under sterile conditions. Front. Immunol. 2021;28(12) doi: 10.3389/fimmu.2021.683803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shatzman A.R., Henkin R.I. Gustin concentration changes relative to salivary zinc and taste in humans. Proc. Natl. Acad. Sci. U. S. A. 1981;78(6):3867–3871. doi: 10.1073/pnas.78.6.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin Y., Li J., Chen J., Shao M., Zhang R., Liang Y., et al. Tissue-specific autoantibodies improve diagnosis of primary Sjögren’s syndrome in the early stage and indicate localized salivary injury. J. Immunol. Res. 2019;2019 doi: 10.1155/2019/3642937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spolski R., Li P., Leonard W.J. Biology and regulation of IL-2: from molecular mechanisms to human therapy. Nat. Rev. Immunol. 2018;18(10):648–659. doi: 10.1038/s41577-018-0046-y. [DOI] [PubMed] [Google Scholar]
- 54.Pišlar A., Mitrović A., Sabotič J., Fonović U.P., Nanut M.P., Jakoš T., et al. The role of cysteine peptidases in coronavirus cell entry and replication: the therapeutic potential of cathepsin inhibitors. PLoS Pathog. 2020;16(11) doi: 10.1371/journal.ppat.1009013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pekayvaz K., Leunig A., Kaiser R., Joppich M., Brambs S., Janjic A., Popp O., Nixdorf D., Fumagalli V., Schmidt N., Polewka V. Protective immune trajectories in early viral containment of non-pneumonic SARS-CoV-2 infection. Nat. Commun. 2022;13(1):1–21. doi: 10.1038/s41467-022-28508-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.-H., Zhang Y., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370(6515) doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370(6515) doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stavrou E.X., Fang C., Bane K.L., Long A.T., Naudin C., Kucukal E., et al. Factor XII and uPAR upregulate neutrophil functions to influence wound healing. J. Clin. Invest. 2018;128(3):944–959. doi: 10.1172/JCI92880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu X., Arfman T., Wichapong K., Reutelingsperger C.P.M., Voorberg J., Nicolaes G.A.F. PAD4 takes charge during neutrophil activation: impact of PAD4 mediated NET formation on immune-mediated disease. J. Thromb. Haemost. 2021;19(7):1607–1617. doi: 10.1111/jth.15313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hsu F.-C., Sides E.G., Mychaleckyj J.C., Worrall B.B., Elias G.A., Liu Y., et al. Transcobalamin 2 variant associated with poststroke homocysteine modifies recurrent stroke risk. Neurology. 2011;77(16):1543–1550. doi: 10.1212/WNL.0b013e318233b1f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saadi H., Shan Y., Marazziti D., Wray S. GPR37 signaling modulates migration of olfactory ensheathing cells and gonadotropin releasing hormone cells in mice. Front. Cell. Neurosci. 2019;9(13):200. doi: 10.3389/fncel.2019.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bernardes J.P., Mishra N., Tran F., Bahmer T., Best L., Blase J.I., et al. Longitudinal multi-omics analyses identify responses of megakaryocytes, erythroid cells, and plasmablasts as hallmarks of severe COVID-19. Immunity. 2020;53(6):1296–1314. doi: 10.1016/j.immuni.2020.11.017. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Faust J.S., Lin Z., Del Rio C. Comparison of estimated excess deaths in New York City during the COVID-19 and 1918 influenza pandemics. JAMA Netw. Open. 2020;3(8) doi: 10.1001/jamanetworkopen.2020.17527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324(8):782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 65.Karki R., Sharma B.R., Tuladhar S., Williams E.P., Zalduondo L., Samir P., et al. Synergism of TNF-α and IFN-γ triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell. 2021;184(1):149–168. doi: 10.1016/j.cell.2020.11.025. e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang L., Yin Z., Hu Y., Mei H. Controlling cytokine storm is vital in COVID-19. Front Immunol. 2020 doi: 10.3389/fimmu.2020.570993. [cited 2022 Feb 15] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lam S.M., Zhang C., Wang Z., Ni Z., Zhang S., Yang S., et al. A multi-omics investigation of the composition and function of extracellular vesicles along the temporal trajectory of COVID-19. Nat Metab. 2021;3(7):909–922. doi: 10.1038/s42255-021-00425-4. [DOI] [PubMed] [Google Scholar]
- 68.Tolossa T., Wakuma B., Seyoum Gebre D., Merdassa Atomssa E., Getachew M., Fetensa G., et al. Time to recovery from COVID-19 and its predictors among patients admitted to treatment center of wollega university referral hospital (WURH), Western Ethiopia: survival analysis of retrospective cohort study. PLoS One. 2021;16(6) doi: 10.1371/journal.pone.0252389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abrahim S.A., Tessema M., Defar A., Hussen A., Ejeta E., Demoz G., et al. Time to recovery and its predictors among adults hospitalized with COVID-19: a prospective cohort study in Ethiopia. PLoS One. 2020;15(12) doi: 10.1371/journal.pone.0244269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bhapkar H.R., Mahalle P.N., Dey N., Santosh K.C. Revisited COVID-19 mortality and recovery rates: are we missing recovery time period? J. Med. Syst. 2020;44(12):202. doi: 10.1007/s10916-020-01668-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jin Y.-H., Cai L., Cheng Z.-S., Cheng H., Deng T., Fan Y.-P., et al. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version) Mil. Med. Res. 2020;7(1):4. doi: 10.1186/s40779-020-0233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Winheim E., Rinke L., Lutz K., Reischer A., Leutbecher A., Wolfram L., et al. Impaired function and delayed regeneration of dendritic cells in COVID-19. PLoS Pathog. 2021;17(10) doi: 10.1371/journal.ppat.1009742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bai M., Grieshaber-Bouyer R., Wang J., Schmider A.B., Wilson Z.S., Zeng L., et al. CD177 modulates human neutrophil migration through activation-mediated integrin and chemoreceptor regulation. Blood. 2017;130(19):2092–2100. doi: 10.1182/blood-2017-03-768507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kaiser R., Leunig A., Pekayvaz K., Popp O., Joppich M., Polewka V., et al. Self-sustaining IL-8 loops drive a prothrombotic neutrophil phenotype in severe COVID-19. JCI Insight. 2021;6(18) doi: 10.1172/jci.insight.150862. https://insight.jci.org/articles/view/150862 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen R., Zhu S., Zeng L., Wang Q., Sheng Y., Zhou B., et al. AGER-mediated lipid peroxidation drives Caspase-11 inflammasome activation in sepsis. Front. Immunol. 2019;8(10):1904. doi: 10.3389/fimmu.2019.01904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26(7):1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen B., Jiang C., Han B., Guan C., Fang G., Yan S., et al. High prevalence of occult thrombosis in cases of mild/moderate COVID-19. Int. J. Infect. Dis. 2021;104:77–82. doi: 10.1016/j.ijid.2020.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Clavijo M.M., de LA Vicente Reparaz M., Ruiz J.I., Acuña M.A., Casali C.E., Aizpurua M.F., et al. Mild COVID-19 illness as a risk factor for venous thromboembolism. Cureus. 2021;13(9) doi: 10.7759/cureus.18236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Afzali B., Noris M., Lambrecht B.N., Kemper C. The state of complement in COVID-19. Nat.Rev. Immunol. 2022;22(2):77–84. doi: 10.1038/s41577-021-00665-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Geyer P.E., Arend F.M., Doll S., Louiset M.-L., Virreira Winter S., Müller-Reif J.B., et al. High-resolution serum proteome trajectories in COVID-19 reveal patient-specific seroconversion. EMBO Mol. Med. 2021;13(8) doi: 10.15252/emmm.202114167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shen B., Yi X., Sun Y., Bi X., Du J., Zhang C., et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182(1):59–72. doi: 10.1016/j.cell.2020.05.032. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu F., Li L., Xu M., Wu J., Luo D., Zhu Y., et al. Prognostic value of interleukin-6, C-reactive protein, and procalcitonin in patients with COVID-19. J. Clin. Virol. 2020;127 doi: 10.1016/j.jcv.2020.104370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reddy N.M., Hall S.W., MacKintosh F.R. Partial thromboplastin time: prediction of adverse events and poor prognosis by low abnormal values. Arch. Intern. Med. 1999;159(22):2706–2710. doi: 10.1001/archinte.159.22.2706. [DOI] [PubMed] [Google Scholar]
- 84.Devreese K.M.J. COVID-19-related laboratory coagulation findings. Int. J. Lab. Hematol. 2021;43(Suppl 1):36–42. doi: 10.1111/ijlh.13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lombardi A., Trombetta E., Cattaneo A., Castelli V., Palomba E., Tirone M., et al. Early phases of COVID-19 are characterized by a reduction in lymphocyte populations and the presence of atypical monocytes. Front. Immunol. 2020;9(11) doi: 10.3389/fimmu.2020.560330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Woodruff M.C., Ramonell R.P., Nguyen D.C., Cashman K.S., Saini A.S., Haddad N.S., et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat. Immunol. 2020;21(12):1506–1516. doi: 10.1038/s41590-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Arango Duque G., Descoteaux A. Macrophage cytokines: involvement in immunity and infectious diseases. Front. Immunol. 2014;7(5):491. doi: 10.3389/fimmu.2014.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Castro F., Cardoso A.P., Gonçalves R.M., Serre K., Oliveira M.J. Interferon-gamma at the crossroads of tumor immune surveillance or evasion. Front. Immunol. 2018;4(9):847. doi: 10.3389/fimmu.2018.00847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Misharin A.V., Morales-Nebreda L., Reyfman P.A., Cuda C.M., Walter J.M., McQuattie-Pimentel A.C., et al. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J. Exp. Med. 2017;214(8):2387–2404. doi: 10.1084/jem.20162152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Grasselli G., Greco M., Zanella A., Albano G., Antonelli M., Bellani G., et al. Risk factors associated with mortality among patients with COVID-19 in intensive care units in Lombardy,Italy. JAMA Intern. Med. 2020;180(10):1345–1355. doi: 10.1001/jamainternmed.2020.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ovies C., Semmes E.C., Coyne C.B. Pregnancy influences immune responses to SARS-CoV-2. Sci. Transl. Med. 2021;13(617) doi: 10.1126/scitranslmed.abm2070. [DOI] [PubMed] [Google Scholar]
- 92.Filbin M.R., Mehta A., Schneider A.M., Kays K.R., Guess J.R., Gentili M., Fenyves B.G., Charland N.C., Gonye A.L., Gushterova I., Khanna H.K. Longitudinal proteomic analysis of severe COVID-19 reveals survival-associated signatures, tissue-specific cell death, and cell-cell interactions. Cell Rep.Med. 2021;2(5) doi: 10.1016/j.xcrm.2021.100287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of confounders.
Clinical characteristics for different subgroups of the cohort at admission.
Results of subgroup comparisons for clinical characteristics and biomedical indices at admission.
Details of statistical analysis of clinical parameters.
Reference values for routine biochemical indices.
Routine biochemical indices for disease phase assignment.
Log2(fold change) of routine biochemical indices for groupwise comparisons.
Differential abundance analysis results in the study cohort: phases compared to control groups.
Pathway enrichment analysis in the study cohort: phases compared to control groups.
Differential Abundance Analysis Results in the Validation Cohort: Phases compared to control groups.
Differential abundance analysis results in the study cohort: trajectory comparisons.
Pathway enrichment analysis results in the study cohort: trajectory comparisons.
Supplementary figures
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon request. Access to data for research purposes is possible for eligible institutes according to GDPR rules.