Abstract
Chronic myeloid leukemia (CML) is a slowly progressing hematopoietic cell disorder. Sphingosine kinase 1 (SPHK1) plays established roles in tumor initiation, progression, and chemotherapy resistance in a wide range of cancers, including leukemia. However, small-molecule inhibitors targeting SPHK1 in CML still need to be developed. This study revealed the role of SPHK1 in CML and investigated the potential anti-leukemic activity of hirsuteine (HST), an indole alkaloid obtained from the oriental plant Uncaria rhynchophylla, in CML cells. These results suggest that SPHK1 is highly expressed in CML cells and that overexpression of SPHK1 represents poor clinical outcomes in CML patients. HST exposure led to G2/M phase arrest, cellular apoptosis, and downregulation of Cyclin B1 and CDC2 and cleavage of Caspase 3 and PARP in CML cells. HST shifted sphingolipid rheostat from sphingosine 1-phosphate (S1P) towards the ceramide coupled with a marked inhibition of SPHK1. Mechanistically, HST significantly blocked SPHK1/S1P/S1PR1 and BCR-ABL/PI3K/Akt pathways. In addition, HST can be docked with residues of SPHK1 and shifts the SPHK1 melting curve, indicating the potential protein-ligand interactions between SPHK1 and HST in both CML cells. SPHK1 overexpression impaired apoptosis and proliferation of CML cells induced by HST alone. These results suggest that HST, which may serve as a novel and specific SPHK1 inhibitor, exerts anti-leukemic activity by inhibiting the SPHK1/S1P/S1PR1 and BCR-ABL/PI3K/Akt pathways in CML cells, thus conferring HST as a promising anti-leukemic drug for CML therapy in the future.
Keywords: Chronic myeloid leukemia, SPHK1, Hirsuteine, Sphingolipid rheostat, SPHK1/S1P/S1PR1, BCR-ABL/PI3K/Akt
INTRODUCTION
Chronic myeloid leukemia (CML) is a slowly progressing hematological malignancy characterized by the accumulation of transformed hematopoietic progenitor cells in bone marrow and peripheral blood. The Philadelphia chromosome (Ph), which results from [t(9;22)(q34;q11.2)] reciprocal translocation, and its gene product bcr-abl are present in 95% of CML patients (Lion, 2011). Mechanistically, bcr-abl encodes BCR-ABL. BCR-ABL is an oncoprotein with consistent tyrosine kinase activity, which leads to cell transformation and uncontrolled cell growth (Osman and Deininger, 2021). As a result, Ph, bcr-abl and BCR-ABL proteins with a molecular weight of 210-kDa, serve as the basis of pathology, diagnosis, and monitoring in CML. Therefore, during the past decades, the abnormal fusion protein BCR-ABL, which has tyrosine kinase activity, represents a logical target for CML therapy (Mojtahedi et al., 2021). Tyrosine kinase inhibitors (TKIs) that selectively target BCR-ABL, such as Imatinib, nilotinib, and dasatinib, have been developed. TKIs have led to extended lifespans for many patients with CML and are considered to be the most spectacular success in the field of targeted cancer therapy (Amir and Javed, 2021). However, a considerable number of patients cannot benefit from TKIs because of their toxicity, resistance, and intolerance (Cortes and Lang, 2021). Therefore, novel treatment options are urgently required.
Sphingolipids, ubiquitous cell membrane constituents, are involved in various diseases, including cancer (Codini et al., 2021). Metabolites of sphingolipids, such as ceramide (Cer), sphingosine (Sph), and sphingosine 1-phosphate (S1P), are viewed as important signaling effectors that regulate many cellular processes, ranging from cell growth to cell apoptosis and cellular responses to stress (Velazquez et al., 2021). Cer can be deacylated to form Sph, which can be further phosphorylated to produce S1P. Evidence has identified that Cer/Sph induces cell apoptosis, whereas S1P promotes cell survival and migration. These oppositely acting molecules are interconvertible within cells, and their balance has been regarded as cellular “sphingolipid rheostat,” which determines cell fate (Green et al., 2021). Sphingosine kinases (SPHKs) catalyze the conversion of Sph to S1P and are considered to be the particular enzymes regulating the sphingolipid rheostat. SPHKs are evolutionarily conserved lipid kinases. There are two isoforms in mammalian cells, SPHK1 and SPHK2, of which SPHK1 is widely reported to have established roles in cancer initiation, progression, and drug resistance in numerous cancers, including leukemia (Green et al., 2021; Velazquez et al., 2021). Moreover, elevated SPHK1 levels have also been found in several cancers, such as breast, lung, colorectal, kidney and brain cancers (Lupino et al., 2019). Thus, SPHK1 is considered a druggable target for cancer therapy. However, targeting SPHK1 with small-molecule inhibitors in CML has rarely been reported.
Bioactive natural products are important sources of many current therapeutic agents, either in their original form or as derivatives. Due to their diverse mechanisms of action, various natural compounds derived from medicinal plants have been extensively studied to treat cancer (Wilson et al., 2020). Hirsuteine (HST) is an indole alkaloid obtained primarily from the hooks or hook-bearing branches of the oriental plant Uncaria rhynchophylla and is used in the treatment of cerebrovascular disorders and neurodegeneration (Mohd Sairazi and Sirajudeen, 2020). Qi et al. (2014) reported that HST protected normal neuronal cells from glutamate-induced cell death, thus exhibiting neuroprotective efficacy. Earlier pharmacological studies with HST have demonstrated that it possesses antihypertensive, anti-inflammatory, neuromodulatory, and protective effects, and exerts cytotoxicity and MDR-reversal activity in human hepatocellular carcinoma cells (Huang et al., 2017; Kushida et al., 2021). However, the anti-leukemic activity of HST against CML and its underlying mechanisms are not well understood.
This study aims to explore the potential anti-leukemic efficacy of HST in CML cells and the underlined mechanism.
MATERIALS AND METHODS
Reagents, antibodies and plasmids
Hirsuteine was purchased from Abphyto biotech (Chengdu, Sichuan, China). 3-(4, 5-Dimethyl-2-thiazolyl)-2, 5-diphenyl-2Htetrazolium bromide (MTT) reagent was from Amresco (Solon, OH, USA). Propidium iodide (PI) was from Sigma-Aldrich (St. Louis, MO, USA). The Annexin V-FITC/PI Apoptosis Detection Kit was purchased from Dalian Meilun Biological Product Factory (Dalian, Liaoning, China). TRIzol reagent was purchased from Absin (Shanghai, China). Human S1P ELISA kit (ELS11663), Human Cer ELISA kit (ELS11665) and Human SPHK1 ELISA Kit (ELS11669) were bought from Tianjin Dingguo Biotechnology (Tianjin, China). Fetal bovine serum (FBS) was from Biological Industries (Kibbutz Beit-Haemek, Israel). Antibodies against Cyclin B1 (1:1,000, #4135), CDC2 (1:1,000, #9116), Cyclin D1 (1:1,000, #55506), Caspase 3 (1:1,000, #9662), PARP (1:1,000, #9532), Caspase-8 (1:1,000, #9746), Caspase-9 (1:1,000, #9502), Cytochrome c (1:1,000, #4272), BCR-ABL (1:1,000, #2862), p-BCR-ABL (Y412) (1:1,000, #2865), PI3K-p110α (1:1,000, #4249), p-Akt (Ser473) (1:1,000, #3787), Akt (1:1,000, #9272), β-actin (1:1,000, #8457), anti-rabbit and anti-mouse HRP-conjugated secondary antibodies (1:2,000) were purchased from Cell Signaling Technology (Danvers, MA, USA). Antibody against Bcl-2 (1:1,000, #SC-7382) was from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Antibody against p-SPHK1 (Ser225) (1:1,000, #19561-1-AP) was from Proteintech (Wuhan, Hubei, China). Antibody against SPHK1 (1:1,000, #A0139) was from ABclonal (Wuhan, Hubei, China), Human SPHK1 overexpression plasmid was purchased from Genechem (Shanghai, China).
Cell culture and transient transfection
Human CML cell line K562 was obtained from the National Collection of Authenticated Cell Cultures (Shanghai, China), K562/G01, an Imatinib-selected multidrug resistance (MDR) cell subline, was obtained from the Institute of Hematology, Chinese Academy of Medical Sciences (Tianjin, China). Human peripheral blood mononuclear cells (PBMCs) were isolated from the blood samples of healthy volunteers with the approval of the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital using Dakewei Human Lymphocyte Separation Medium (Beijing, China). All cells were cultured in RPMI-1640 containing 10% FBS, 10 µg/mL streptomycin, and 100 U/mL penicillin, in a humidified incubator at 37°C with 5% CO2. Cells were authenticated by short tandem repeat (STR) analysis. For K562/G01 cell culture, 1 μg/mL of Imatinib was added to the culture medium to maintain the MDR characteristics. K562/G01 was further grown in a drug-free culture medium for 10 days before assay. Cells were seeded onto twelve-well plates and transfected with 1 µg of SPHK1 or empty vector control plasmid using Lipofectamine® 2000 (Invitrogen Corp., Carlsbad, CA, USA) in a condition of 37°C, 5% CO2 for 6 h. The culture medium was then replaced by fresh RPMI-1640 medium containing antibiotics and FBS before subsequent experiments.
MTT assay
MTT assay was performed as we previously reported (Yin et al., 2019). Cells were harvested and seeded at a density of 4×104 cells/well onto a 96-well plate, then treated with HST at various concentrations for 48 h or at different time for 32 µM. After further incubation with 5 mg/mL MTT at 37°C for 4 h, the medium was discarded, and the produced formazan blue was dissolved with 100 µL DMSO. The absorbance value at 490 nm was measured by using a microplate reader iMark (BIO-RAD, Hercules, CA, USA).
Flow cytometry
Cell cycle analysis and cell apoptosis were detected by flow cytometry as we reported before (Zhang et al., 2019). In brief, for cell cycle analysis, cells under detection were harvested and fixed in 75% ethanol overnight, washed with PBS, and stained with propidium iodide (PI) staining (50 μg/mL PI, 100 μg/mL RNase A and 0.5% Triton X-100) at 4°C for 30 min in the dark. For cell apoptosis, cells under detection were harvested and stained with Annexin V-FITC/PI double staining. The samples were immediately analyzed by BD Accuri C6 flow cytometer (BD Biosciences, San Jose, CA, USA).
RNA isolation and qRT-PCR
qRT-PCR was conducted as previously described (Zhou et al., 2016). Total RNA was extracted using TRIzol reagent (Life Technologies, Carlsbad, CA, USA). 500 ng of total RNA was reverse transcribed into cDNA using PrimeScript RT Master Mix Reagent Kit (TaKaRa, Tokyo, Japan). qPCR was conducted using TB Green Premix Ex Taq Reagent Kit (TaKaRa) according to the manufacturer’s instructions, and detected by a CFX96TM Real-Time PCR Detection System (BIO-RAD). The sequences of primers were listed in Supplementary Table 1. The relative gene expression was determined by the comparative Ct (ΔΔCt) method. GAPDH was used as the reference gene for normalization.
Western blotting
Western blotting was conducted as previously described (Zhou et al., 2016; Yu et al., 2018). Briefly, cells were lysed by RIPA lysis buffer, quantified, and resolved by SDS-PAGE followed by immunoblotting using specified primary antibodies and HRP-conjugated secondary antibodies, respectively. The signals were visualized using ECL reagents and quantified by Image J (Rawak Software Inc., Stuttgart, Germany). β-actin was the internal reference.
ELISA
SPHK1 activity, S1P concentration, and Cer concentration were determined by ELISA. Cells were seeded at a density of 4×105 cells/well in 6-well plates. After stepwise increasing concentrations of HST treatment for 48 h, the cells were harvested. The supernatant was taken for examination of S1P concentration using ELISA kit according to the manufacturer’s instructions. The cell precipitation was repeatedly freeze-thaw lysed and cell lysates were used to determine SPHK1 activity and Cer concentration by using Human SPHK1 ELISA Kit and Human Cer ELISA Kit, respectively, according to the manufacturer’s instructions.
Cellular Thermal Shift Assays (CETSAs)
Target engagement assay of SPHK1 was performed by CETSAs, as reported previously (Liu et al., 2020; Yin et al., 2021). Briefly, cells were treated with DMSO or 32 μM HST for 4 h. Cells were then collected, washed with PBS, and resuspended in 600 μL PBS containing protease inhibitor cocktail. Each cell suspension was divided and 100 μL aliquots were transferred into PCR tubes, heated at different temperatures for 3 min followed by cooling for 3 min at room temperature. The cells were then repeatedly freeze-thawed using liquid nitrogen. Subsequently, after centrifugation, cell lysates were separated and quantification of the remaining soluble protein was achieved by western blot analysis.
Docking simulation protocol
Docking simulation was operated using the Discovery-Studio 2017 R2 molecular modeling software (Dassault Systèmes Information Technology Co., Ltd., Shanghai, China). The three-dimensional (3D) structures of HST were generated with ChemDraw (PerkinElmer Inc., MA, USA) and were energy minimized with CHARMm force field. The initial 3D geometric coordinates of SPHK1 (PDB code: 3vzb) were obtained from the Protein Databank (PDB) (https://www.rcsb.org/structure/3VZB/). Then, the protein structure was prepared by removing water molecules and adding hydrogen. CDOCKER protocols were employed as docking approaches and calculated the predicted binding energy (kcal/mol). The complex structure with the most favorable binding-free energies was selected as the optimal docked conformation for late experimental verification.
Database of chronic myeloid leukemia patients
Clinical data can be obtained via GEO (https://www.ncbi.nlm.nih.gov/geo/) with the publically available dataset (GSE71014). The SPHK1 expression in CML patients was analyzed by the Kaplan-Meier estimate.
Statistical analysis
All experiments were repeated at least in triplicate. Results are presented as mean ± SD. Statistical analyses were determined by Student’s t-test and one-way ANOVA using GraphPad software. Relapse-free survival (RFS) rates were plotted using Kaplan-Meier analysis and compared with the long-rank test. Differences at p-value<0.05 were considered statistically significant.
RESULTS
SPHK1 is upregulated in CML cells and overexpression of SPHK1 represents poor clinical outcomes in CML patients
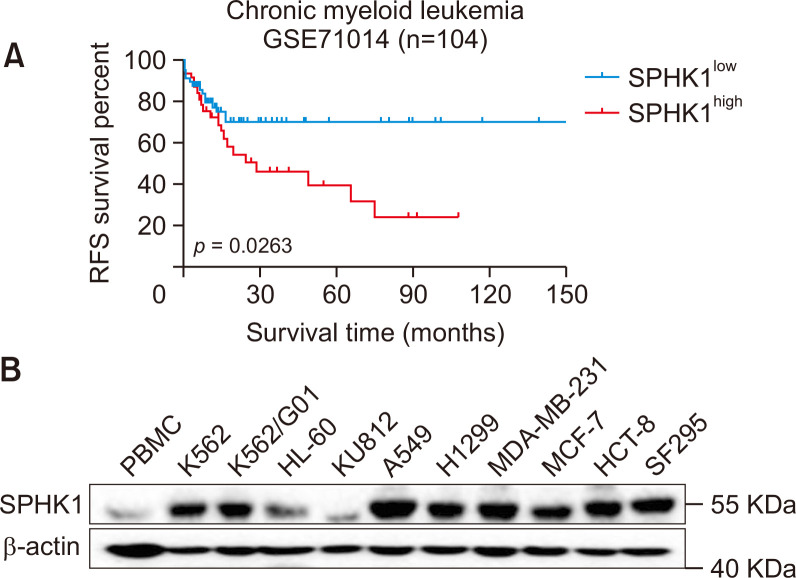
First, to evaluate the clinical correlation of SPHK1 in CML patients, a Kaplan-Meier estimate was conducted. Fig. 1A showed that CML patients with higher SPHK1 expression usually displayed poorer relapse-free survival (RFS) rates than those with lower SPHK1 expressions (GSE71014, p=0.0263). The expression of SPHK1 in human PBMCs and a panel of human cancer cell lines representing various cancers including colon, lung, breast, and brain cancers, and leukemia, was analyzed by western blotting. As shown in Fig. 1B, extremely high levels of SPHK1 protein were found in various human cancer cell lines, and SPHK1 expression in K562 and K562/G01 cells was significantly higher than that in PBMCs. These results demonstrate that SPHK1 is upregulated in CML cells and that SPHK1 overexpression correlates with poor clinical outcomes in CML patients.
Fig. 1.
SPHK1 is highly expressed in CML cells and predicts clinical outcomes. (A) Kaplan-Meier representation of the relapse-free survival (RFS) of the two groups of CML patients with different SPHK1 expression levels (GSE71014). p-value was obtained using Kaplan-Meier analysis and compared with a long-rank test. (B) The protein expression of SPHK1 in PBMCs and various cancer cell lines (n=3).
HST inhibits proliferation and induces apoptosis in CML cells
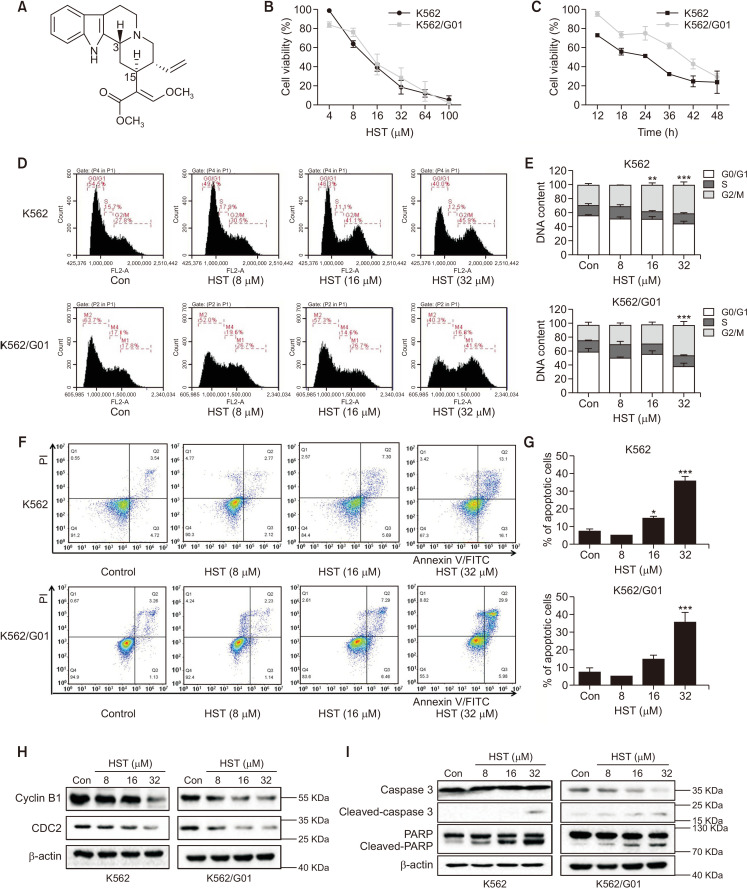
The structure of HST is shown in Fig. 2A. The MDR characteristics of K562/G01 were first determined by the MTT assay. K562/G01 cells showed obvious resistance to Imatinib (Supplementary Fig. 1). The cell growth inhibitory effect of HST on K562 and K562/G01 was assessed by the MTT assay. As shown in Fig 2B and 2C, HST potently reduced cell growth in a time-dependent (32 μM, 12-48 h) and dose-dependent (4-100 μM, 48 h) manner in both cell lines. The IC50 values of HST in K562 and K562/G01 cells were 12.33 μM and 12.77 μM, respectively. To explore the mechanism of growth inhibition of CML cells by HST, cell cycle distribution and cell apoptosis were further assessed by flow cytometry analysis after HST exposure. As shown in Fig. 2D and 2E, HST (32 μM, 48 h) increased the G2/M phase cell population to 45.9% in K562 cells and 41.6% in K562/G01 cells compared with the cells treated with vehicle, which was 27.8% in K562 cells and 17.8% in K562/G01 cells, suggesting that HST arrested the G2/M cell cycle in both CML cell lines. The results of apoptosis indicated that HST (32 μM, 48 h) exposure led to a potent increase in the apoptotic population in both K562 and K562/G01 cells (Fig. 2F, 2G, Supplementary Fig. 3), suggesting that HST promoted CML cell apoptosis. Consistently, similar results were obtained using western blotting. As shown in Fig. 2H and Supplementary Fig. 2A, HST led to the downregulation of Cyclin B1 and CDC2, and upregulation of Cyclin D1 in both cell lines. These apoptosis results were further reinforced by western blotting, in which HST induced the cleavage of PARP, Caspase 3, Caspase 8, and Caspase 9. Moreover, HST treatment led to an increase in Cytochrome c release and a decrease of Bcl-2 (Fig. 2I, Supplementary Fig. 2B). These results indicated that HST inhibits proliferation and induces apoptosis in CML cells.
Fig. 2.
HST inhibits proliferation and induces apoptosis in CML cells. (A) The chemical structure of HST. (B, C) Cells were treated with indicated doses of HST for 48 h (B) or with 32 μM HST at indicated time point (C), cell viability was determined by MTT assay. (D, E) Cell cycle distribution (D) was detected by flow cytometry after 48 h-treatment of increasing HST, percentages of cell population in each phase were shown (E). (F, G) Cell apoptosis was analyzed by flow cytometry after 48 h-treatment of increasing HST, percentages of apoptotic cells were shown (G). (H) Western blot was performed to determine cell cycle regulators, CDC 2, Cyclin B1. (I) Western blot was performed to determine cell apoptosis effectors, Caspase 3, PARP. Data represent mean value ± SD. *p<0.05, **p<0.01, ***p<0.001.
HST regulates the sphingolipid rheostat and represses SPHK1/S1P/S1PR1 and BCR-ABL/PI3K/Akt in CML cells
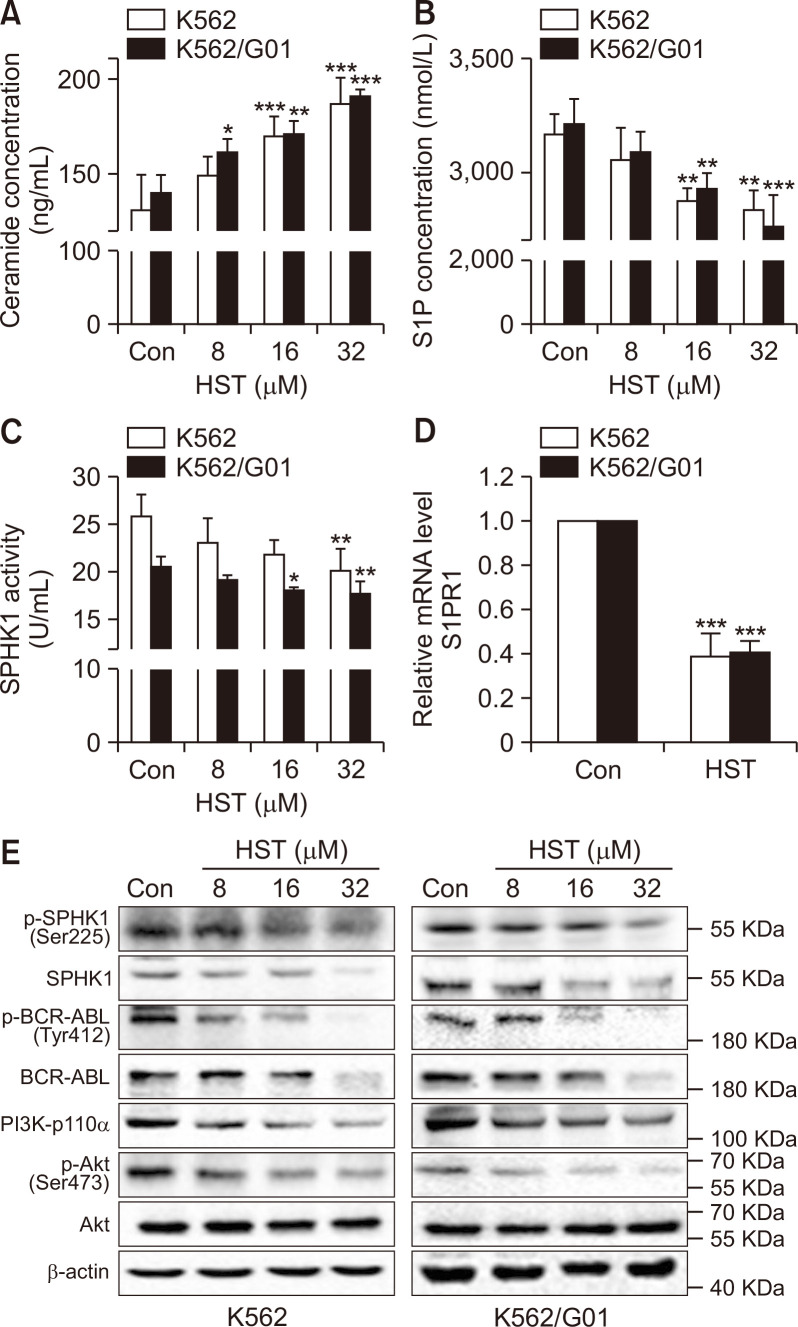
It has been known that sphingolipid rheostat play key roles in regulating many cellular processes in cancer (Zheng et al., 2019). To characterize the effects of HST on sphingolipid rheostat, K562 and K562/G01 cells were treated with HST or vehicle, and the level of S1P and Cer was determined by ELISA. As shown in Fig. 3A and 3B, in K562 and K562/G01 cells, Cer were significantly increased by HST treatment, whereas S1P were decreased, suggesting that HST shifts the sphingolipid rheostat from S1P towards Cer in CML cells. As SPHK1 is the rate-limiting enzyme governing sphingolipid rheostat activity, we hypothesized that HST might inhibit SPHK1. To test this possibility, cellular SPHK1 activity was detected by ELISA. The result in Fig. 3C showed that cellular SPHK1 activity was markedly reduced by HST in K562 and K562/G01 cells. Furthermore, it has been demonstrated that S1P regulates diverse cellular functions through extracellular ligation to S1P receptors (S1PRs). Therefore, S1PR1 expression in response to HST was examined by qRT-PCR. The results in Fig. 3D show that HST sharply repressed S1PR1 expression in both CML cell lines. As shown in Fig. 3E, HST repressed p-SPHK1 (Ser225), SPHK1, p-BCR-ABL (Tyr412), BCR-ABL, PI3K-p110α, and p-Akt (Ser473) in K562 and K562/G01 cells. In summary, the above results suggest that HST modulates sphingolipid rheostat and represses the SPHK1/S1P/S1PR1 and BCR-ABL/PI3K/Akt signaling pathways in CML cells.
Fig. 3.
HST regulates the sphingolipid rheostat, represses SPHK1/S1P/S1PR1 and BCR-ABL/PI3K/Akt in CML cells. (A, B) K562 cells and K562/G01 cells were exposed with HST for 48 h. Cer concentration (A) and extracellular S1P level (B) were detected by ELISA. (C) SPHK1 activity was detected by ELISA. (D) S1PR1 level was analyzed by qRT-PCR. (E) SPHK1 and BCR-ABL/PI3K/Akt signaling were detected by western blot. Data represent mean value ± SD. *p<0.05, **p<0.01, ***p<0.001.
HST targets at SPHK1
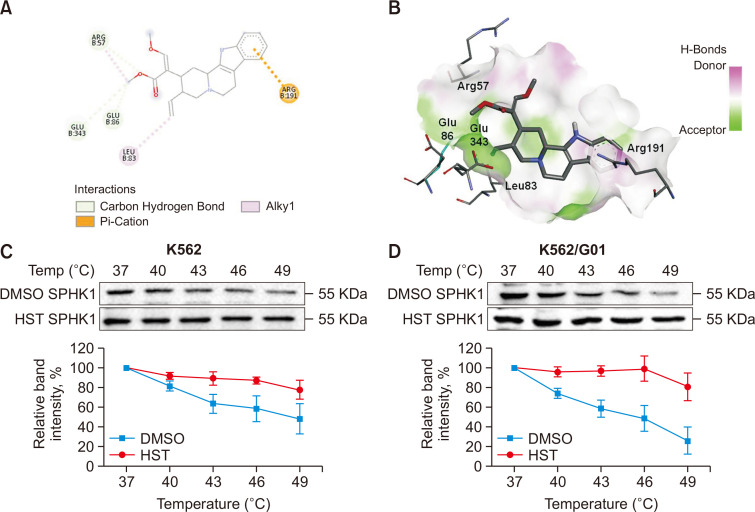
To examine whether HST potentially targets SPHK1 and to gain insights into the detailed interactions of HST with SPHK1, a molecular docking simulation was first performed. The CDOCKER docking results in Fig. 4A and 4B show that HST can be docked with the Arg57, Glu343, Glu86, Leu83 and Arg191 residues of SPHK1 (binding energy: –6.6 kcal/mol). Based on the interactions of HST with SPHK1 at the Arg191 residue, which is an ATP-binding site residue of the enzyme (Jairajpuri et al., 2020), we deduced that HST may serve as an ATP-competitive inhibitor of SPHK1. CETSAs have been widely used to investigate interactions between proteins and their ligands. To confirm these results, CETSAs were further performed. As shown in Fig. 4C and 4D, HST (32 μM) induced SPHK1 thermal stability at the indicated temperatures compared to the vehicle in K562 and K562/G01 cells, indicating potential protein-ligand interactions between SPHK1 and HST in CML cells.
Fig. 4.
HST targets at SPHK1. (A, B) Molecular docking study of HST with SPHK1. (A) Two-dimensional interaction diagram of HST and SPHK1. Only interacting residues were labeled. (B) The spatial interaction diagram of HST and SPHK1. (C, D) CETSAs were performed on K562 cells and K562/G01 cells, the SPHK1 expression was determined by western blot (C). Relative band intensity of SPHK1 (D). Experiments were repeated at least in triplicate. Data represent mean value ± SD.
HST exerts anti-leukemic effect in CML cells through SPHK1 suppression
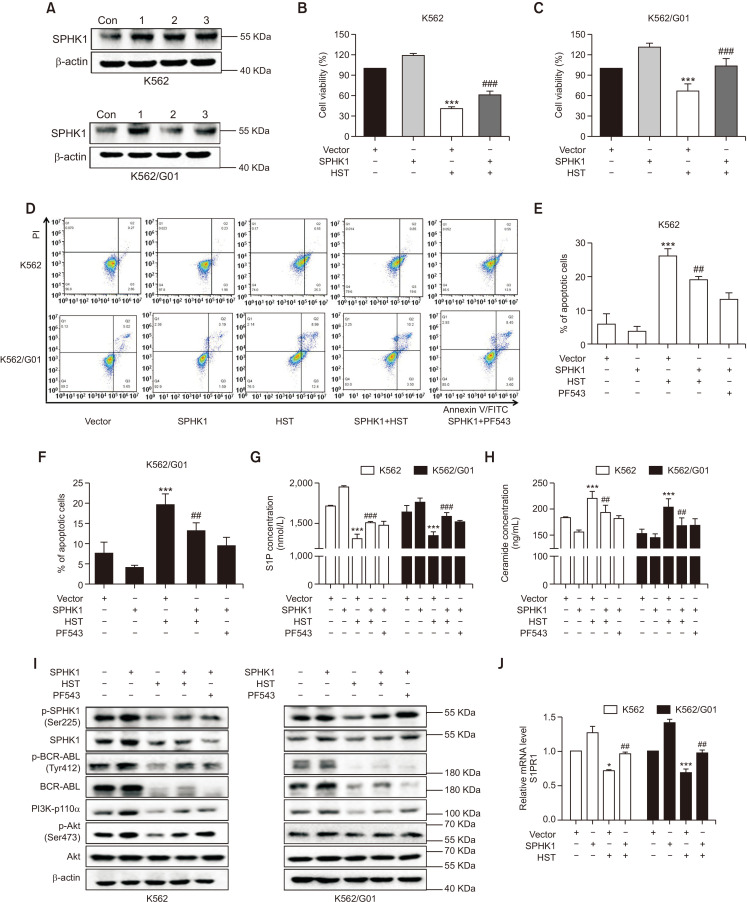
To explore the involvement of SPHK1 in the anti-leukemic properties of HST in CML cells, cell growth, apoptosis, sphingolipid rheostat, and related signaling cascades in K562 and K562/G01 cells in response to HST were examined after SPHK1 overexpression. Fig. 5A shows that SPHK1 was overexpressed in K562 and K562/G01 cells when the cells were transfected with SPHK1 vector or empty vector for 6 h. MTT assays showed that SPHK1 overexpression significantly prevented the proliferation inhibition induced by HST in K562 and K562/G01 cells (Fig. 5B, 5C). The results of apoptosis showed that HST promoted apoptosis in K562 and K562/G01 cells, whereas these effects were attenuated by SPHK1 overexpression in both cell lines (Fig. 5D-5F). Similar results were also found after treatment with PF543, a known SPHK1 inhibitor. ELISA experiments examining Cer and S1P concentrations showed that SPHK1 overexpression antagonized the effect of HST on sphingolipid rheostat in both cell lines (Fig. 5G, 5H). Protein expression indicated that p-SPHK1 (Ser225), SPHK1, p-BCR-ABL (Tyr412), BCR-ABL, PI3K-p110α, and p-Akt (Ser473) were downregulated by HST, whereas this inhibitory effect of HST was reversed by SPHK1 overexpression in both cell lines (Fig. 5I). qRT-PCR results showed that the downregulation of S1PR1 caused by HST treatment was also counteracted by SPHK1 overexpression (Fig. 5J). In summary, these data indicate that overexpression of SPHK1 attenuates the growth inhibitory and pro-apoptotic activities of HST in K562 and K562/G01 cells, suggesting that HST exerts anti-leukemic effects on CML cells probably through SPHK1 suppression.
Fig. 5.
HST exerts anti-leukemic effect in CML cells through SPHK1 suppression. K562 cells and K562/G01 cells were exposed with HST for 48 h after transfected with SPHK1 vector or empty vector for 6 h. PF543 (30 μM in K562, 40 μM in K562/G01) was used as a positive control. (A) SPHK1 expression was detected by western blot. (B, C) Cell viability was conducted using MTT. (D-F) Cell apoptosis was analyzed using flow cytometry. (G, H) Extracellular S1P level (G) and Cer concentration (H) were detected by ELISA. (I) SPHK1 and BCR-ABL/PI3K/Akt signaling were detected by western blot. (J) S1PR1 level was determined using qRT-PCR. Data represent mean value ± SD. *p<0.05, ***p<0.001 versus the empty vector-transfected cells; ##p<0.01, ###p<0.001 versus the empty vector-transfected cells treated with HST.
DISCUSSION
Although studies have already demonstrated the anticancer potential of HST, the anti-leukemic effect of HST and its underlying mechanisms remain to be explored. This study showed that HST inhibited cell proliferation, promoted cell apoptosis, and arrested the cell cycle at the G2/M phase, thus exerting potential anti-leukemic efficacy in K562 and K562/G01 cells.
Recent evidence has demonstrated the oncogenic characterization of SPHK1 in various types of cancer because of its association with many cellular activities important for cancer including growth, transformation, metastasis, and chemotherapy resistance (Pitman and Pitson, 2010; Zheng et al., 2019; Velazquez et al., 2021). Researchers have also reported overexpression of SPHK1 in a diverse array of cancers and its relationship with poor prognosis (Pitman and Pitson, 2010; Lupino et al., 2019; Velazquez et al., 2021). Therefore, SPHK1 has gained increasing attention over the past few decades as a potential drug target against cancer, including leukemia. This study showed that SPHK1 is upregulated in K562 and K562/G01 cells and overexpression of SPHK1 is closely associated with unfavorable prognosis in CML patients, consistent with the important role of SPHK1 as a novel molecular target in CML therapy.
Owing to the high level of interest in SPHK1 signaling, several SPHK1 inhibitors are currently undergoing preclinical research, and only a few agents have entered clinical trials as chemotherapeutics against cancers (Pitman and Pitson, 2010; Companioni et al., 2021). Potent and selective SPHK1 inhibitors with low toxicity obtained from natural sources are needed. Therefore, we propose that HST may be an ATP-competitive inhibitor of SPHK1. It has been reported that SPHK inhibitors mainly target three binding sites of SPHKs, namely, the Sph binding pocket, ATP binding pocket, and dimerization site (Ding et al., 2021). In our study, HST formed a close association with Arg57, Glu343, Glu86, Leu83, and Arg191 residues, in which Arg191 is an ATP-binding site residue of SPHK1 and the others are key residues for enzyme activity of SPHK1 (Jairajpuri et al., 2020; Ding et al., 2021), suggesting that HST targets the ATP-binding pocket of SPHK1 and inhibits SPHK1. Further studies may involve an experiment to show whether mutation on the residues of SPHK1 could change the current results or not, which may act as strong evidence to support the in silico study.
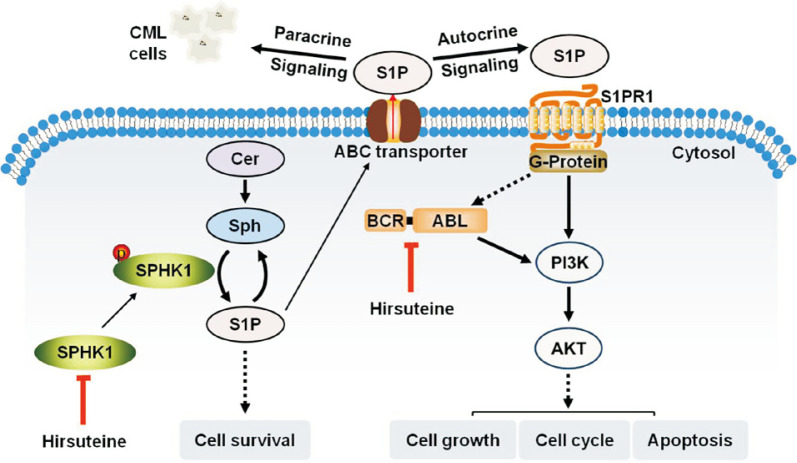
S1P and Cer are bioactive lipid mediators involved in various pathophysiological processes (Green et al., 2021). Our results showed that in both K562 and K562/G01 cells, HST shifted the rheostat from S1P towards Cer, manifesting pro-apoptotic functions, and finally exhibiting anti-leukemic efficacy in CML cells. SPHK1 is the rate-limiting enzyme in sphingolipid rheostat. Stimulation of SPHK1 in response to diverse growth factors or other stimuli mediates the intracellular conversion of Cer and Sph to S1P (Sukocheva et al., 2020; Green et al., 2021). S1P, in turn, acts either as an intracellular secondary messenger or as an extracellular ligand that activates S1PRs in an autocrine and/or paracrine manner. Extracellular S1P transported by several ATP-binding cassette (ABC) transporters accesses and activates S1PRs, and the latter, through binding with different G-proteins, such as Go, Gi, G12/13 and Rho, initiates intracellular signaling cascades such as PI3K, Akt, and MAPK, thereby participating in diverse cellular functions (Hannun and Obeid, 2018). The SPHK1/S1P/S1PR1 pathway has become a point of interest in cancer therapy, as it plays a key role in maintaining cancer survival signals and leading to cancer progression, including leukemia.
S1PRs are a family of S1P-specific G protein-coupled receptors. To date, five subtypes of S1PRs (S1PR1-5) have been reported. S1PR1-3 are ubiquitously expressed in most cells, whereas S1PR4 and S1PR5 are less abundant and are restricted to distinct cells (Blaho and Hla, 2014). In the present study, we found that, in K562 and K562/G01 cells, S1PR1-3 were commonly expressed subtypes, while S1PR4 expression was relatively low and S1PR5 was almost undetectable (Supplementary Fig. 4). Because S1PR1 was highly expressed in K562 and K562/G01 cells as well as the involvement of S1PR1 in cancer progression (Patmanathan et al., 2017), S1PR1 was selected for subsequent experiments.
Previous findings have demonstrated that SPHK1 has intrinsic catalytic activity, and phosphorylation of SPHK1 at Ser-225 increases its catalytic activity and induces its translocation to the cell membrane, which is important for the oncogenic signaling of SPHK1 (Pitson et al., 2003). In this study, HST inhibited cellular SPHK1 activity, decreased S1P and S1PR1, and downregulated p-SPHK1 (Ser225), p-BCR-ABL (Tyr412), PI3K-p110α, and p-Akt (Ser473), thereby blocking SPHK1/S1P/S1PR1 and BCR-ABL/PI3K/Akt signaling in K562 and K562/G01 cells (Fig. 6). However, SPHK1 overexpression reversed the inhibitory effect of HST on these signaling cascades in both cell lines. Notably, a previous study has reported that BCR-ABL can increase the expression and cellular activity of SPHK1 in CML cells (Li et al., 2007). There may be controversial issues regarding the relationship between SPHK1 and BCR-ABL in the cell-signaling cascade. Further studies, including whether SPHK1 is located upstream or downstream of BCR-ABL, and whether SPHK1 signals directly to BCR-ABL through S1PR1, are required to address these questions. However, our present results are consistent with the emerging oncogenic role of SPHK1 in CML and demonstrate the potential of SPHK1 inhibitors in CML therapeutics.
Fig. 6.
Scheme representing the mechanism by which HST exerts anti-leukemic activity in CML cells.
Furthermore, we also found that the protein expression of SPHK1 and BCR-ABL was decreased by HST in both cell lines. It seems difficult to explain the mechanism based on our present results. However, it is known that Cer can bind to and activate lysosomal cathepsin D, which can degrade various proteins and enzymes (Ren et al., 2010). Furthermore, it has been reported that many apoptotic stimuli can increase lysosomal permeability and induce the release of cathepsins, resulting in the degradation of various cytosolic proteins (Ren et al., 2010). As a result, it may be speculated the downregulation of SPHK1 and BCR-ABL protein expression by HST might be mediated by Cer accumulation, which could activate cathepsins and induce protein degradation. Further studies are required to address the details.
This study shows that, in K562 and K562/G01 cells, HST inhibits cell proliferation, arrests the cell cycle, and promotes cell apoptosis, possibly resulting from inhibition of SPHK1, thereby conferring HST as a potential anti-leukemic drug candidate against CML.
ACKNOWLEDGMENTS
This work was supported jointly by grants from the National Natural Science Foundation of China (No. 81973570 to YQ, Nos. 81673464 and 82073890 to DK, No. 81873089 to HY), the National Key Research and Development Program of China (No: 2021YFE0203100 to HY).
Footnotes
REFERENCES
- Amir M., Javed S. A review on the therapeutic role of TKIs in case of CML in combination with epigenetic drugs. Front. Genet. 2021;12:742802. doi: 10.3389/fgene.2021.742802.c8e9f761f7e743b59fc5801617b46c3c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaho V. A., Hla T. An update on the biology of sphingosine 1-phosphate receptors. J. Lipid Res. 2014;55:1596–1608. doi: 10.1194/jlr.R046300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codini M., Garcia-Gil M., Albi E. Cholesterol and sphingolipid enriched lipid rafts as therapeutic targets in cancer. Int. J. Mol. Sci. 2021;22:726. doi: 10.3390/ijms22020726.12c678e4ebe7412e88d0bdf48c8d5b00 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Companioni O., Mir C., Garcia-Mayea Y., LLeonart M. E. Targeting sphingolipids for cancer therapy. Front. Oncol. 2021;11:745092. doi: 10.3389/fonc.2021.745092.78a3b9d174414e78a00e849ca0aed82b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes J., Lang F. B. Third-line therapy for chronic myeloid leukemia: current status and future directions. J. Hematol. Oncol. 2021;14:44. doi: 10.1186/s13045-021-01055-9.604211664c504d9d9acff4ceeaf4a19e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding T. D., Zhi Y., Xie W. L., Yao Q. Q., Liu B. Rational design of SphK inhibitors using crystal structures aided by computer. Eur. J. Med. Chem. 2021;213:113164. doi: 10.1016/j.ejmech.2021.113164. [DOI] [PubMed] [Google Scholar]
- Green C. D., Maceyka M., Cowart L. A., Spiegel S. Sphingolipids in metabolic disease: the good, the bad, and the unknown. Cell Metab. 2021;33:1293–1306. doi: 10.1016/j.cmet.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannun Y. A., Obeid L. M. Author correction: sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018;19:673. doi: 10.1038/s41580-018-0046-6. [DOI] [PubMed] [Google Scholar]
- Huang B. Y., Zeng Y., Li Y. J., Huang X. J., Hu N., Yao N., Chen M. F., Yang Z. G., Chen Z. S., Zhang D. M., Zeng C. Q. Uncaria alkaloids reverse ABCB1-mediated cancer multidrug resistance. Int. J. Oncol. 2017;51:257–268. doi: 10.3892/ijo.2017.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jairajpuri D. S., Mohammad T., Adhikari K., Gupta P., Hasan G. M., Alajmi M. F., Rehman M. T., Hussain A., Hassan M. I. Identification of sphingosine kinase-1 inhibitors from bioactive natural products targeting cancer therapy. ACS Omega. 2020;5:14720–14729. doi: 10.1021/acsomega.0c01511.3ed453670f51465a8c689f7e5aa8ba24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushida H., Matsumoto T., Ikarashi Y. Properties, pharmacology, and pharmacokinetics of active indole and oxindole alkaloids in uncaria hook. Front. Pharmacol. 2021;12:688670. doi: 10.3389/fphar.2021.688670.5ba5a5120b1f4e39a8e715c3b38c0a50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. F., Huang W. R., Duan H. F., Wang H., Wu C. T., Wang L. S. Sphingosine kinase-1 mediates BCR/ABL-induced upregulation of Mcl-1 in chronic myeloid leukemia cells. Oncogene. 2007;26:7904–7908. doi: 10.1038/sj.onc.1210587. [DOI] [PubMed] [Google Scholar]
- Lion T. SP 108 resistance to tyrosine kinase inhibitors in chronic myeloid leukemia. Eur. J. Cancer. 2011;47:S3. doi: 10.1016/S0959-8049(11)72600-4. [DOI] [Google Scholar]
- Liu H. W., Chen Q., Lu D., Pang X., Yin S. S., Wang K. L., Wang R., Yang S. S., Zhang Y., Qiu Y. L., Wang T., Yu H. Y. HTBPI, an active phenanthroindolizidine alkaloid, inhibits liver tumorigenesis by targeting Akt. FASEB J. 2020;34:12255–12268. doi: 10.1096/fj.202000254R. [DOI] [PubMed] [Google Scholar]
- Lupino L., Perry T., Margielewska S., Hollows R., Ibrahim M., Care M., Allegood J., Tooze R., Sabbadini R., Reynolds G., Bicknell R., Rudzki Z., Lin , Hock, Zanetto U., Wei W., Simmons W., Spiegel S., Woodman C. B. J., Rowe M., Vrzalikova K., Murray P. G. Sphingosine-1-phosphate signalling drives an angiogenic transcriptional programme in diffuse large B cell lymphoma. Leukemia. 2019;33:2884–2897. doi: 10.1038/s41375-019-0478-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd Sairazi N. S., Sirajudeen K. N. S. Natural products and their bioactive compounds: neuroprotective potentials against neurodegenerative diseases. Evid. Based Complement. Alternat. Med. 2020;2020:6565396. doi: 10.1155/2020/6565396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojtahedi H., Yazdanpanah N., Rezaei N. Chronic myeloid leukemia stem cells: targeting therapeutic implications. Stem Cell Res. Ther. 2021;12:603. doi: 10.1186/s13287-021-02659-1.e6a09fb8e18347229f6e905abf131cd4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman A. E. G., Deininger M. W. Chronic myeloid leukemia: modern therapies, current challenges and future directions. Blood Rev. 2021;49:100825. doi: 10.1016/j.blre.2021.100825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patmanathan S. N., Wang W., Yap L. F., Herr D. R., Paterson I. C. Mechanisms of sphingosine 1-phosphate receptor signalling in cancer. Cell. Signal. 2017;34:66–75. doi: 10.1016/j.cellsig.2017.03.002. [DOI] [PubMed] [Google Scholar]
- Pitman M. R., Pitson S. M. Inhibitors of the sphingosine kinase pathway as potential therapeutics. Curr. Cancer Drug Targets. 2010;10:354–367. doi: 10.2174/156800910791208599. [DOI] [PubMed] [Google Scholar]
- Pitson S. M., Moretti P. A. B., Zebol J. R., Lynn H. E., Xia P., Vadas M. A., Wattenberg B. W. Activation of sphingosine kinase 1 by ERK1/2-mediated phosphorylation. EMBO J. 2003;22:5491–5500. doi: 10.1093/emboj/cdg540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W., Yue S. J., Sun J. H., Simpkins J. W., Zhang L., Yuan D. Alkaloids from the hook-bearing branch of Uncaria rhynchophylla and their neuroprotective effects against glutamate-induced HT22 cell death. J. Asian Nat. Prod. Res. 2014;16:876–883. doi: 10.1080/10286020.2014.918109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S. Y., Xin C. Y., Pfeilschifter J., Huwiler A. A novel mode of action of the putative sphingosine kinase inhibitor 2-(p-hydroxyanilino)-4-(p-chlorophenyl) thiazole (SKI II): induction of lysosomal sphingosine kinase 1 degradation. Cell. Physiol. Biochem. 2010;26:97–104. doi: 10.1159/000315110. [DOI] [PubMed] [Google Scholar]
- Sukocheva O. A., Furuya H., Ng M. L., Friedemann M., Menschikowski M., Tarasov V. V., Chubarev V. N., Klochkov S. G., Neganova M. E., Mangoni A. A., Aliev G., Bishayee A. Sphingosine kinase and sphingosine-1-phosphate receptor signaling pathway in inflammatory gastrointestinal disease and cancers: a novel therapeutic target. Pharmacol. Ther. 2020;207:107464. doi: 10.1016/j.pharmthera.2019.107464. [DOI] [PubMed] [Google Scholar]
- Velazquez F. N., Hernandez-Corbacho M., Trayssac M., Stith J. L., Bonica J., Jean B., Pulkoski-Gross M. J., Carroll B. L., Salama M. F., Hannun Y. A., Snider A. J. Bioactive sphingolipids: advancements and contributions from the laboratory of Dr. Lina M. Obeid. Cell. Signal. 2021;79:109875. doi: 10.1016/j.cellsig.2020.109875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson B. A. P., Thornburg C. C., Henrich C. J., Grkovic T., O'Keefe B. R. Creating and screening natural product libraries. Nat. Prod. Rep. 2020;37:893–918. doi: 10.1039/C9NP00068B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S. S., Qiu Y. L., Jin C. Y., Wang R., Wu S., Liu H. W., Koo S., Han L. F., Zhang Y., Gao X. M., Pang X., Wang T., Yu H. Y. 7-Deoxynarciclasine shows promising antitumor efficacy by targeting Akt against hepatocellular carcinoma. Int. J. Cancer. 2019;145:3334–3346. doi: 10.1002/ijc.32395. [DOI] [PubMed] [Google Scholar]
- Yin S. S., Yang S. S., Luo Y. M., Lu J., Hu G. Y., Wang K. L., Shao Y. Y., Zhou S. Y., Koo S., Qiu Y. L., Wang T., Yu H. Y. Cyclin-dependent kinase 1 as a potential target for lycorine against hepatocellular carcinoma. Biochem. Pharmacol. 2021;193:114806. doi: 10.1016/j.bcp.2021.114806. [DOI] [PubMed] [Google Scholar]
- Yu H. Y., Yin S. S., Zhou S. Y., Shao Y. Y., Sun J. C., Pang X., Han L. F., Zhang Y., Gao X. M., Jin C. Y., Qiu Y. L., Wang T. Magnolin promotes autophagy and cell cycle arrest via blocking LIF/Stat3/Mcl-1 axis in human colorectal cancers. Cell Death Dis. 2018;9:702. doi: 10.1038/s41419-018-0660-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chen T., Dou Y. H., Zhang S. L., Liu H. Y., Khishignyam T., Li X. F., Zuo D., Zhang Z., Jin M. H., Wang R., Qiu Y. L., Zhong Y. X., Kong D. X. Atorvastatin exerts antileukemia activity via inhibiting mevalonate-YAP axis in K562 and HL60 cells. Front. Oncol. 2019;9:1032. doi: 10.3389/fonc.2019.01032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X. J., Li W., Ren L. W., Liu J. Y., Pang X. C., Chen X. P., Kang D., Wang J. H., Du G. H. The sphingosine kinase-1/sphingosine-1-phosphate axis in cancer: potential target for anticancer therapy. Pharmacol. Ther. 2019;195:85–99. doi: 10.1016/j.pharmthera.2018.10.011. [DOI] [PubMed] [Google Scholar]
- Zhou Q. X., Chen Y. L., Chen X., Zhao W. N., Zhong Y. X., Wang R., Jin M. H., Qiu Y. L., Kong D. X. In vitro antileukemia activity of ZSTK474 on K562 and multidrug resistant K562/A02 cells. Int. J. Biol. Sci. 2016;12:631–638. doi: 10.7150/ijbs.14878. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.