Abstract
Amyotrophic lateral Sclerosis is an incurable, progressive neurodegenerative motor neuron disease. The disease is characterized by protein aggregates. The symptoms include weakness, denervation of muscles, atrophy and progressive paralysis of bulbar and respiratory muscles and dysphagia. Various secondary metabolites are evaluated for their ability to improve symptoms in ALS. Ginseng has been traditionally used for treating several neurodegenerative diseases. Several studies using model systems have shown a potential role of Ginseng catechins and Ginsenosides in clearing protein aggregation associated with ALS. We focus on Network pharmacology approach to understand the effect of Ginseng catechins or ginsenosides on protein aggregation associated with ALS. A catechin/ginsenoside-protein interaction network was generated and the pathways obtained were compared with those obtained from transcriptomic datasets of ALS from GEO database. Knock out of MAPK14, AKT and GSK from Catechin and BACE 1 from ginsenoside modulated pathways inhibited protein aggregation. Catechins and ginsenosides have potential as therapeutic agents in the management of ALS.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-022-03401-1.
Keywords: Amyotrophic lateral sclerosis, Ginseng, Network pharmacology, Transcriptomics, Yeast model system
Introduction
Amyotrophic lateral Sclerosis (ALS) is a progressive motor neuron disease associated with protein aggregates involving the upper motor neurons in the motor cortex and the lower motor neurons in the brain stem and spinal cord (Eisen 2009). Patients with ALS develop weakness, denervation of muscles, atrophy and progressive paralysis of bulbar and respiratory muscles and dysphagia, etc. (Brooks 1996).The incidence of ALS is reported to be between 0.6 and 3.8 per 1,00,000 individuals across the globe [3.8 in Scotland, 2.1 in Norway, 1.2 in South Korea and 0.8 in China] (Longinetti and Fang 2019) and a prevalence of 3–5 per 1,00,000 individuals (Pandey and Sarma 2017). There are very few reports on the prevalence of ALS in India. The average age of disease onset is estimated between 55 and 65 years in western literature. The same is estimated to be a decade earlier in Indian population (Vinceti 2012). Though mutations in about 853 genes are implicated, only 43 genes contribute to a vast majority of the cases (Mccann et al. 2021). Single Nucleotide Polymorphism (SNP) in the genes Super Oxide Dismutase 1 (SOD1) and Chromosome 9 Open Reading Frame 72 (C9orf72) constitute the bulk of the cases while those in Fused in Sarcoma (FUS) and Tar DNA Binding Protein(TDP-43) give rise to very aggressive forms of ALS (Guerrero et al. 2016). Various environmental and lifestyle factors are also implicated as risk factors in the disease (Malek et al. 2012). The pathogenesis involves protein aggregation, oxidative stress, mitochondrial dysfunction, neuroinflammation, excitotoxicity and deregulated metabolism (Tefera and Borges 2017).
Omic studies have been carried out to understand the global changes in transcriptome, proteome and metabolome of ALS Patients irrespective of genetic background as well as transgenic Drosophila, Zebrafish and mice model of ALS (Tokuda 2012; Blasco et al. 2013, 2016; Caballero-Hernandez et al. 2016; Kori et al. 2016; Patin et al. 2017; Morello et al. 2020). The transcriptomic analysis of ALS patients and model systems have implicated a role for transcription factors, cell signaling pathways, inflammation and metabolic pathways in the disease (Bernardini et al. 2013; Al-Chalabi et al. 2017; Ederle and Dormann 2017; Wunderlich 2019). Proteomic analysis shows changes in signaling modules while metabolomic analysis shows dysfunction in various metabolic pathways that might be critical for disease progression (Caballero-Hernandez et al. 2016). The deregulated metabolic pathways include glycolysis, citrate cycle, metabolism of branch chain amino acids, glutamine, aspartate, tryptophan, etc. (Sas et al. 2007; Ngo and Steyn 2015; Abdel-Khalik et al. 2017; Ferri and Coccurello 2017; Tefera and Borges 2017). Many transgenic model systems (mutation in SOD1, C9orf72, FUS and TDP-43) that capture the tenets of the pathophysiology of ALS have been used to understand the mechanistic aspects of the disease (Van Damme et al. 2017). The omic data as well as the data from gene knockouts, knockdowns or overexpression from these model systems show the subtle difference in disease pathology imparted by the mutant gene.
Despite these efforts, there is only one effective drug available in market while others have largely failed clinical trials (Mathis et al. 2019). Medicinal Plants which are used in traditional medicine systems for treating neurodegenerative diseases might offer potential alternatives for managing disease (Luthra and Roy 2022). These medicinal plants have a plethora of bioactive compounds like flavonoids which are polyphenolic compounds with potential antioxidant, anti-inflammatory and iron-chelating properties (Ghareeb et al. 2014; Kilani-Jaziri et al. 2016). Flavonoids are shown to mitigate symptoms and improve cognitive and motor functions in animal models of diseases like ALS, Alzheimer’s, Parkinson’s and Huntington’s Disease (Ayaz et al. 2019; Jiang et al. 2000). Resveratrol and Quercetin have by far been shown to be the most potent ones, though they have also failed in clinical trials (Bournival et al. 2009). Studies have also shown that flavonoids modulate the activity of various enzymes belonging to tryptophan pathway, glycolysis citrate cycle and signaling proteins like kinases, phosphatases, histone acetyltransferase (HAT) and Histone deacetylases (HDAC) (Spencer 2007).
The yeast model systems with knockout, over expression and inducible promoter libraries offer a high-throughput system to understand mechanistic aspects involved in protein aggregation diseases. The yeast model has been used to understand gene–protein interactions as well as toxicity to growth in various diseases like Amyotrophic Lateral Sclerosis (ALS) and Huntington’s Disease (HD) (Pradhan et al. 2022; Sai Swaroop et al. 2022). Thus, the yeast model of ALS can be used for screening the potency of flavonoids and for their role in mitigating protein aggregation.
One of the previous studies focusing on flavonoid–protein interaction has found 1400 flavonoids with about 6500 interactions, which modulate various pathways with potential implications for disease (Lacroix et al. 2018). With data on flavonoid–protein interactors, multi-Omic data from patients and model systems, a systems analysis of different flavonoids should help to identify the most effective flavonoids for managing ALS. Integrated analysis of omic data sets obtained from databases and literature as well as disease and Flavonoid–protein interaction might help not only to understand the mechanism of action of different flavonoids but also identify the potential candidate target genes for therapy to manage the disease. Further, yeast and mammalian cell culture model system could be employed to validate the results.
In the present study, we have screened Ginseng for its ability to clear protein aggregates induced by FUS and TDP-43 in a yeast model of ALS. The role of Ginseng in the treatment of ALS was further validated by using SymMap. The flavonoid content in these plants was collected from published literature (Kim 2016) and the flavonoids were used for their ability to prevent protein aggregation in the yeast system. For understanding the mechanistic aspect of flavonoids and ginsenoside mediated reduction of protein aggregation, a network pharmacology approach was employed. An interaction network was generated for flavonoids and ginsenosides that effectively cleared protein aggregates in the yeast model of ALS (Lacroix et al. 2018). The genes in the flavonoid/ginsenoside-protein interaction network was further binned into pathways which were then compared with the deregulated pathways from ALS patients and the model system. Ortholog Finder and Cytoscape were used to generate the flavonoid/ginsenoside yeast interaction network from the human network. Further, gene knockout was employed to evaluate the role of the target genes in the deregulated pathways for their ability to clear protein aggregates in the yeast model system. Our results identify the potential mechanism of action of different flavonoids and ginsenosides. Over all, the study helps to raise additional questions and help discern many core pathways which might have potential as biomarkers or therapeutic targets.
Materials and methods
Identification of medicinal plants using literature mining and SymMap database
Medicinal plants associated with ALS were identified using literature mining. Key words like Amyotrophic Lateral Sclerosis and Medicinal plants, traditional medicine and ALS were searched using Google Scholar, PubMed, Science Direct and Embase. Research articles with scientific authenticity and validity were used for the study. Some of the important results are presented in Table 1. Further, SymMap database was used to identify medicinal plants associated with ALS. SymMap creates an interaction network between symptoms, diseases, medicinal plants, molecular ingredients of medicinal plants, genes and protein targets. Enrichment analysis was performed using direct and indirect associations with the above-mentioned parameters. Bonferroni method for multiple testing correction was used to further narrow down the results. Ginseng was found to be one of the hits obtained from literature and SymMap database.
Table 1.
Table representing the anti-amyloid activity of traditional medicine plants obtained from literature mining
| Scientific name | Common name | Remarks |
|---|---|---|
| Withania somnifera | Ashwagandha |
Anti-inflammatory, anti-oxidant, anti-stress, neuro-protection, immune boosting and memorya Known to have anti-ALS activityb |
| Phyllanthus emblica | Amla | It is known to be a cognition enhancer in several neuro-degenerative diseasesc |
| Panax ginseng | Ginseng |
Immune boosting Neuro-protectiond Rich in poly-phenols and Ginsenosidese Ginsenosides Rg1 is known to reduce reactive oxygen species of dopamine, release of cytochrome c into cytosol, inhibition of caspase-3 activity and lower Nitric Oxide Synthase productionf It is known to be given for treatment of ALSg |
| Ginkgo biloba | Ginkgo |
Known to improve cognitive functionh Is known to inhibit formation of Aβ amyloidsi |
| Vitis vinifera | Grape seed extract | Grape seed extract was found to attenuate neuro-degeneration in AD micej |
| Baccopa monnieri | Brahmi |
Known to rejuvenate nerve cellsl Helps to fight oxidative stressl |
Transformation of Saccharomyces cerevisiae (yeast) with FUS and TDP-43 plasmids
BY4741 yeast strain (MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) was used for the study. BY4741 yeast cells were revived from glycerol stocks. FUS [426 Gal-FUS-YFP (Plasmid ID: 29,593), 426 Gal-FUS-RRM Mutant-YFP (Plasmid ID: 29,608)] and TDP-43 [pRS416Gal TDP-43 WT YFP (Plasmid ID: 27,447) and pRS416 Gal Q331K YFP (Plasmid ID: 27,450)] wild-type and mutant plasmids were purchased from Addgene. Plasmid preparation was performed using conventional alkaline lysis method. Transformation involves competent cell preparation, inserting the gene of interest and growing the transformed cells in selective medium. A single yeast colony of BY4741 was inoculated in 50 ml YPD broth [Yeast Extract (Himedia-RM027), Peptone (Himedia-RM001), Dextrose (Himedia- GRM016) in the ratio 1:2:2]. The cells were allowed to grow at 30 °C for 12 h. Cells were then harvested at an OD of 0.8 (600 nm). The harvested cell pellet was washed with phosphate buffer saline (PBS), resuspended in 25 ml of electroporation buffer (0.1 M Lithium acetate [Himedia, GRM1507], 10 mM Dithiothreitol (DTT) [Himedia, MB070], 10 mM Tris HCL [Himedia, MB030], 1 mM EDTA [Himedia, RM1370], ddH2O) and was incubated for an hour at 30 °C. The cell pellet was spun down and washed with 1 M Sorbitol [Himedia, GRM109]. After sorbitol wash the cell pellet was resuspended again in 100 μl of 1 M Sorbitol [Himedia, GRM109]. From the above washed pellet, 8 × 108 cells along with 2 μg/ml of plasmid DNA were transferred into the electroporation 0.2 cm cuvette (Bio-Rad, #1652086). Transformation was performed using Bio-Rad Gene Pulser Xcell using default settings (1.5 kV/25µF/200). For 3–4 days, transformed cells were incubated and allowed to grow on Himedia URA-YNB-Glucose-Agar [ (Himedia Complete supplement mixture w/o URA,G112), (Himedia Yeast nitrogen base w/o amino acids, M878) and (Himedia D-(+)-Glucose, MB037)] media at 30 °C. A single colony from each plate was seeded in Himedia URA-YNB-Glucose liquid broth. For 12 h, the yeast cells were allowed to grow. After 12 h, 200 µl of the culture was transferred into a raffinose medium containing Himedia URA-YNB-raffinose [(Himedia Complete supplement mixture w/o URA,G112), (Himedia Yeast nitrogen base w/o amino acids, M878) and (Himedia Raffinose pentahydrate, RM107)]. The cells were washed with Phosphate Buffer Saline (pH 7) and put in galactose medium for induction [(Himedia Complete supplement mixture w/o URA,G112), (Himedia Yeast nitrogen base w/o amino acids, M878) and (Himedia D-Galactose, MB177)]. Cells were grown for 7 h in galactose medium before being harvested at an OD of 0.8 (600 nm). The induced cells were used in further investigations. Induced cells were examined at 100X magnification using a Laben fluorescence microscope with an excitation and emission wavelength of 510 nm and 535 nm respectively.
Plant extraction and treatment studies
After validating transformation using fluorescence imaging techniques, we went on to study protein modulations on treatment with Ginseng extract. Metabolite and extract treatment studies for understanding protein modulation have been reported in the literature (Speldewinde and Grant 2016). Literature established method was used for extraction process (Wu et al. 2001). 200 mg of powdered Ginseng root extract powder was dissolved in 5 ml MeOH. Water bath sonication (Branson Sonicator: Model No-CPX5800H-E) was performed for 2 h. The sonication power level was set to maximum. The solvent was dried using Heidolph Rota-Evaporator. Dried extract was dissolved in DMSO and transformed yeast cells were treated with 25 µg/ml, 50 µg/ml and 100 µg/ml of the extract during the galactose induction. Appropriate controls were taken for the study. The modulation of the protein aggregates was observed using a Laben fluorescence microscope. Imaging experiments was performed with 3 biological replicates and 2 technical replicates.
Composition analysis
Ginseng root extract was found to have good anti-amyloid activity on the yeast model system for ALS. The polyphenolic composition of ginseng was obtained through literature mining (Kim 2016). Catechin derivatives were found to be in higher composition. Saccharomyces cerevisiae transformed with FUS and TDP-43 plasmids was treated with 100 µg/ml of catechin (TCI Chemicals, Cat No: G0533), epicatechin (Sigma-Aldrich, Cat No: 39263), epigallocatechin -3 gallate (TCI Chemicals, Cat No: E0694), Ginsenoside Rg1 (TCI Chemicals, Cat No: G0533), Ginsenoside Rd (TCI Chemicals, Cat No: G0550), Ginsenoside RH2 (TCI Chemicals, Cat No: G0567) and modulation of protein aggregation was studied. The polyphenols were added during the induction step and protein aggregation was studied using fluorescence microscopy and quantification techniques.
Statistical analysis
Fluorescence quantification was carried out using ImageJ software (Collins 2007). Corrected Total Cell Florescence (CTCF) was calculated using the following formula: CTCF = Integrated Density-(Area of the selected Cell * Mean Florescence of Background Readings). Further, a two-tailed t.test was performed to identify significant results (P ≤ 0.05).
Generation and analysis of protein interaction networks
Protein interaction networks for catechin, epicatechin, epigallocatechin -3 gallate were generated using literature (Lacroix et al. 2018) and Cytoscape (Shannon et al. 2003). Pathway enrichment analysis of these interacting proteins was performed using Cluego (Naik et al. 2020) and KEGG database (Kanehisa and Goto 2000). Cluego initially creates a binary gene term matrix based on the input genes and their interactions. Based on the input matrix, a term–term similarity matrix is calculated using Kappa statistics. The results from Kappa statistics help to understand the association between the genes. The created network is visualized in the form of nodes. The input genes were also grouped into pathways using KEGG database. Pathways that had a Bonferroni corrected P. Value ≤ 0.05 were used for the study. Further, genes that were directly associated with ALS were identified using DisGeNET Interaction Networks (Piñero et al. 2016) and an integrated network for catechin, epicatechin, epigallocatechin -3 gallate was generated using Cytoscape (Supplementary Fig. 3C). Biological processes, cellular components and molecular function were assessed using the GSEA toolkit (Srimadh Bhagavatham et al. 2022). GSEA toolkit uses GO Slims tool that maps the input gene annotations with a higher-level well-defined GO Slim List. Deconvolution or cell type enrichment analysis was carried out using Enrichr (Chen et al. 2013; Kuleshov et al. 2016; Xie et al. 2021) and Azimuth cell type database. Enrichr uses a combined score to identify significant pathways. Combined score is calculated by taking the Log of P-value from Fisher's exact test and multiplying it by the Z-score of the deviation from the expected rank. The pathways that have a high combined score are significant. Transcription factor and kinase enrichment analyses were carried out using X2K portal (Clarke et al. 2018). The interaction network created with polyphenols was compared with transcriptomic interaction networks of ALS patients and ALS transgenic mice obtained from the Gene Expression Omnibus database (Clough and Barrett 2016). Interaction networks of human cortex sections were created from transcriptomic datasets of post-mortem cortex sections containing 146 ALS patients and 16 controls [GSE124439 (Tam et al. 2019)]. Similar networks were constructed for FUS and TDP-43 datasets of ALS transgenic mice [GSE40652 (Lagier-Tourenne et al. 2012), GSE111775 (Neelagandan et al. 2019)]. The methodology used for transcriptomic analysis involves extraction of dataset, normalization, differential gene expression using Deseq2 and generation of interaction networks in Cytoscape (Pradhan et al. 2022; Sai Swaroop et al. 2022).The interaction networks created from ALS transcriptomic sections were overlayed with the polyphenol interaction network using Cytoscape. Common genes were identified and pathway enrichment analysis of the common genes was carried out using Enrichr. Ginsenoside interaction network was created using the STITCH database (Kuhn et al. 2007) and pathway enrichment was carried out using Enrichr and KEGG databases.
Identification of common targets between the interaction networks
Gene Set Enrichment Analysis (GSEA) and differential gene expression analysis for the transcriptomic datasets were carried out using Network analyst (Zhou et al. 2019) and GREIN (Al Mahi et al. 2019). Pathway enrichment analysis for the transcriptomic datasets of ALS human cortex, FUS transgenic mice and TDP-43 transgenic mice was also carried out using Enrichr. The common significant pathways (p ≤ 0.05) were identified using a Venn diagram plotted using Venny (Oliveros 2007) and bioinformatics.psb.ugent.be/webtools/Venn tools. Kinases that were known to be deregulated were obtained from KinMap (Eid et al. 2017). Top significant kinases obtained from kinase enrichment analysis using K2K were compared with KinMap results and common kinases were identified.
Validation of common targets
Yeast knock-outs were procured from Dharmacon (Product No. YSC1054). Yeast knock-outs of HOG1, BAR1, RIM11 and YPK1 were transformed and fluorescence imaging studies were carried out. Fluorescence quantification was carried out using Image J software.
Results
Yeast expressing wild-type and mutant FUS-EYFP or TDP-43-EYFP produce protein aggregates
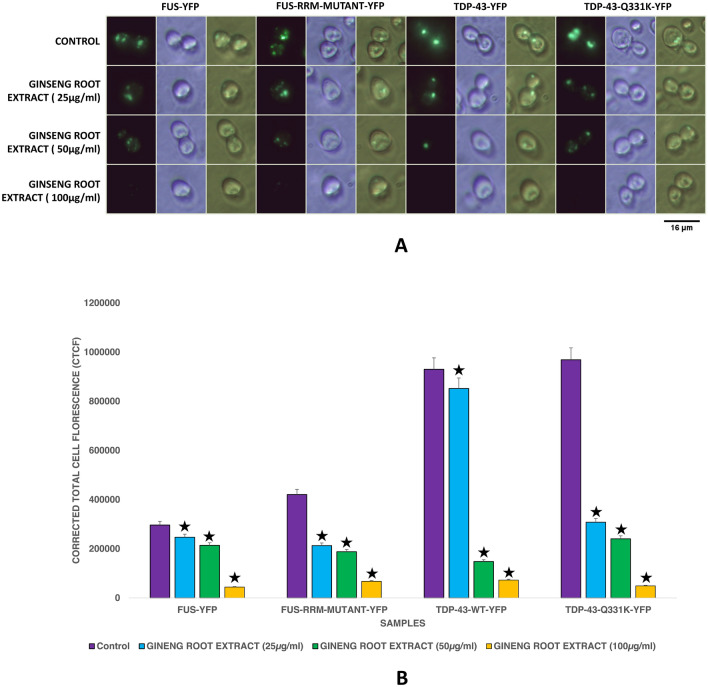
Yeast expressing wild-type FUS-YFP or mutant FUS-RRM-EYFP or wild-type TDP43-EYFP or mutant TDP-43-EYFP (Q331K) is used in the study (Fig. 1). Our results show that the FUS and TDP-43 mutants produce elevated levels of protein aggregates compared to their wild-type counterpart. The results are consistent with previous studies.
Fig. 1.
A Showing Fluorescence Microscopy results of yeast expressing FUS and TDP‒43 Wild type and Mutants exposed to different concentration of Ginseng root extract. 100 µg/ml of Ginseng root extract showed a considerable reduction in protein aggregates of FUS, TDP-43 and mutants. Imaging was carried out in the dark field (blue light) and white light. Images were arranged using PhotoScape. B: Fluorescence quantification results of yeast expressing FUS and TDP‒43 Wild type and Mutants exposed to different concentrations of Ginseng root extract. 100 µg/ml of ginseng root extract significantly reduced protein aggregates. The quantification results correlated with fluorescence imaging. Fluorescence quantification was carried out using ImageJ software (⋆ represents P ≤ 0.05)
Yeast treated with Ginseng extract shows a significant decrease in protein aggregates of FUS and TDP-43 expressing yeast model of ALS
Medicinal plants are used in traditional medical systems for the treatment of many neurodegenerative diseases (Luthra and Roy 2022). The scientific name, common name and effects of the plants used in the study are provided in Table 1. Further, the plants used to treat ALS were searched in the SymMap database which conjured Ginseng as one of the hits (Supplementary Fig. 1). We screened medicinal plants for their efficacy in clearing protein aggregates in FUS and TDP-43 expressing yeast model of ALS. 25 µg, 50 µg and 100 µg each of dried Ginseng root extract in DMSO solvent was used as given in methods and the controls were treated with DMSO vehicle alone. 100 µg/ml of ginseng was found to effectively clear protein aggregates in wild-type and mutant FUS or TDP-43 expressing yeast model of ALS (Fig. 1). The results were reconfirmed using florescence quantification technique as described in methods. Corrected Total Cell Florescence (CTCF) was calculated for the control and treatment groups. Yeast cells transformed with ALS mutant genes treated with Ginseng root extract showed significantly lesser CTCF values. Having found that ginseng cleared protein aggregates, we next looked for the composition of ginseng and the effect of these components on protein aggregate clearance in the yeast model of ALS.
Flavonoids like catechin, epicatechin, Epigallocatechin-3 gallate and Ginsenoside Rg1 cleared protein aggregates in the yeast model of ALS.
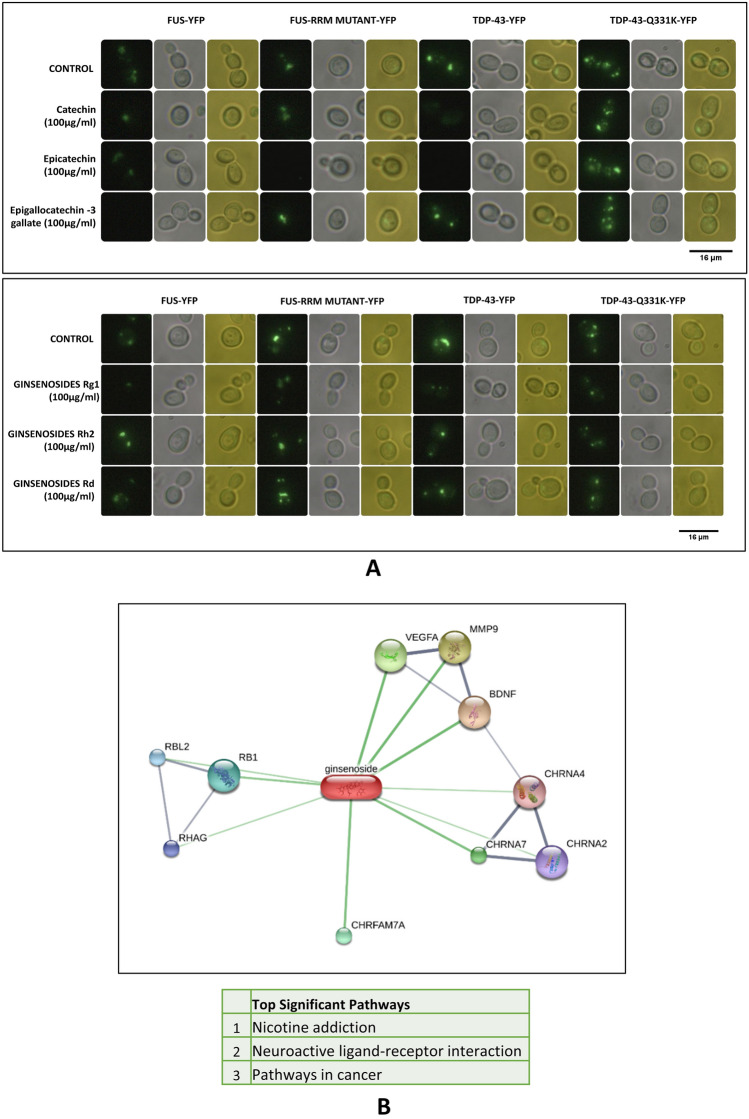
Previous studies have shown that the major components of ginseng are catechin, epicatechin, epicatechin gallate and ginsenosides (Kim 2016). Addition experiments of catechin, epicatechin or Epigallocatechin-3 gallate were carried out in a yeast model of ALS. Catechin and Epicatechin significantly decreased protein aggregates in both wild type and mutants FUS and TDP-43 yeast model of ALS (Fig. 2A). The Epigallocatechin 3 gallate though significantly reduced protein aggregates was not very effective as catechin or epicatechin (Fig. 2A and Supplementary Fig. 2A). Further, we probed the role of different Ginsenosides which is a major constituent of ginseng in clearing protein aggregates. Our analysis shows that Ginsenoside Rg1 significantly reduced protein aggregates in both wild type or mutants of FUS and TDP-43 expressing yeast model of ALS (Fig. 2A). Ginsenoside Rh2 and Rd through reduced protein aggregates, were not as significant as Rg 1 (Fig. 2A). Florescence imaging results corroborated with quantification results.
Fig. 2.
A Showing Fluorescence Microscopy results of yeast expressing FUS and TDP‒43 Wild type and Mutants exposed to different catechin derivatives and ginsenosides. Epicatechin and Rg1 showed a considerable reduction in protein aggregates of FUS and TDP-43. Imaging was carried out in the dark field (blue light) and white light. Images were arranged using PhotoScape. Fluorescence quantification was carried out using ImageJ software. Quantification results are provided in Supplementary-2. B Network file representing interacting proteins with Ginsenosides. Further, pathway enrichment analysis for the interacting proteins was carried out using Enrichr. Three pathways (nicotine addiction, neuro-active receptor–ligand interaction and pathways in cancer) were found to be significant. The network file was created using the STITCH database (stitch.embl.de)
Taken together our analysis shows catechin, epicatechin and ginsenoside Rg1 significantly reduced protein aggregation in wild type or mutant, FUS or TDP-43 expressing yeast model of ALS.
Flavonoids and Ginsenosides modulate the activity of various proteins in specific pathways with potential implications for disease.
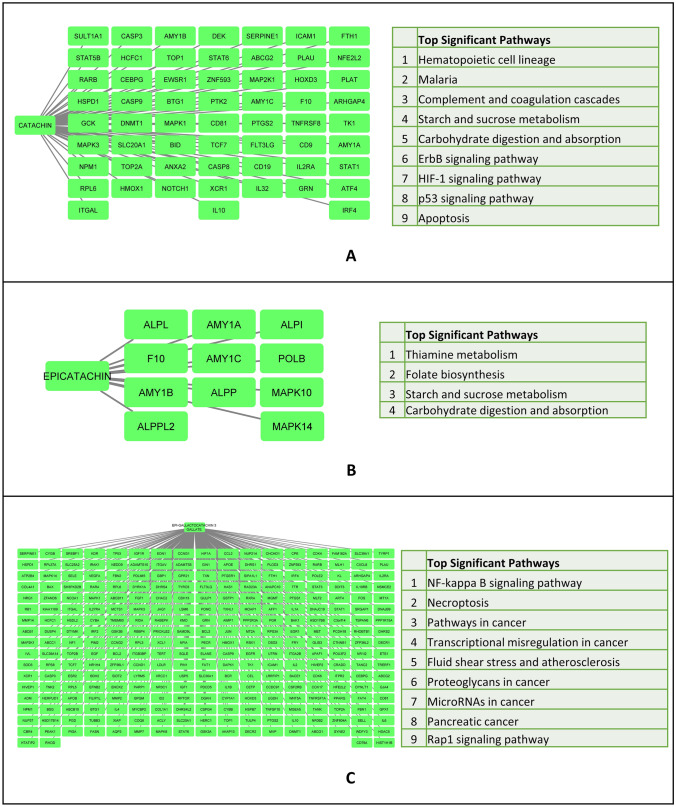
Having identified the flavonoids and ginsenoside which mediate clearance of protein aggregates in the yeast model of ALS, we looked at the pathways they modulate in humans. Interactors of Ginsenosides were obtained from the STITCH database (Fig. 2B) and an interaction network was created. The interacting genes were binned into pathways. The top significant pathways include nicotine addiction, neuro-active receptor–ligand interaction and pathways in cancer. Previous studies have catalogued flavonoids and their protein interactors (Lacroix et al. 2018). Using the flavonoids and their protein interactors an interaction network was generated using Cytoscape. Further, the interacting proteins were binned into pathways using the plug in ClueGO (Bindea et al. 2009). Our analysis shows that the interactors of catechin were binned into Hematopoietic cell lineage, malaria, complement and coagulation cascade, starch and sucrose metabolism, carbohydrate digestion and absorption, ErB, HIF1 and P53 signaling pathways and apoptosis (Fig. 3A). Similarly, the interactors of Epicatachin were binned into thiamine metabolism, folate biosynthesis, starch and sucrose metabolism as well as carbohydrate digestion and absorption (Fig. 3B). The interactors of epigallocatechin-3 gallate were binned into NFkB and Rap1 signaling pathway, necroptosis, pathways involving transcriptional mis-regulation, miRNA and proteoglycans in cancer, pancreatic cancer, fluid shear stress and arthrosclerosis (Fig. 3C).
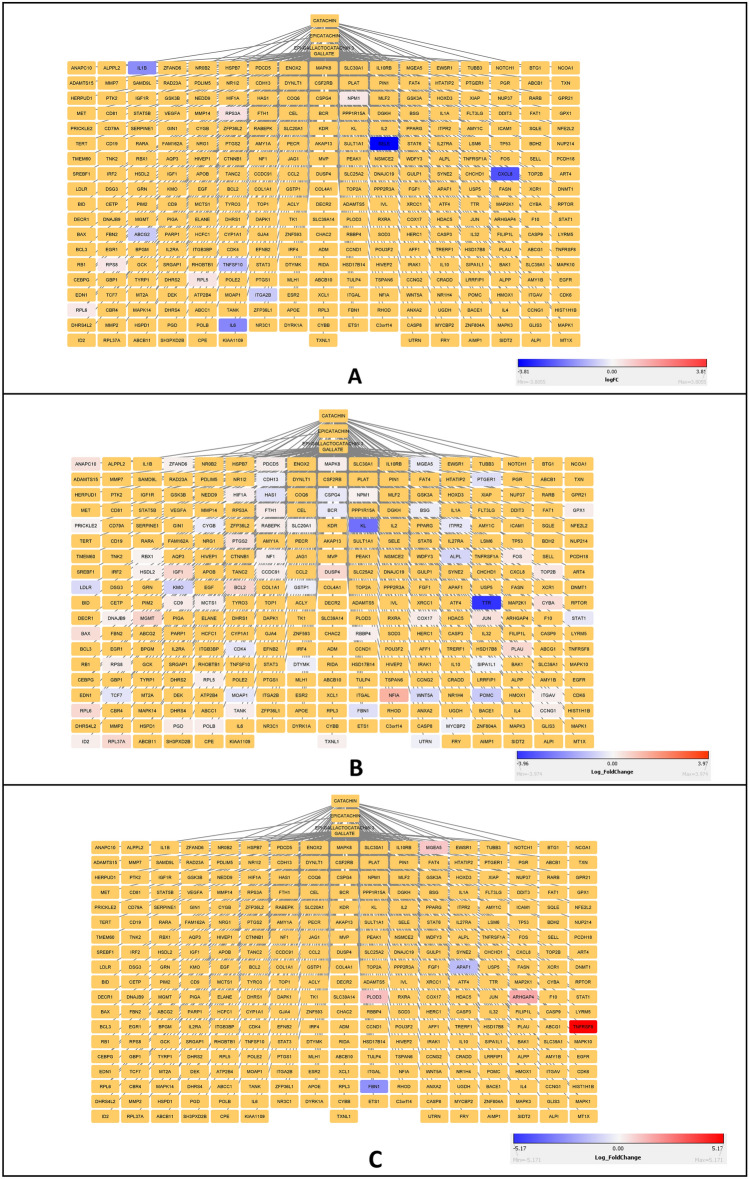
Fig. 3.
A Figure representing interacting protein networks and pathways enriched by Catechin. B Figure representing interacting protein networks and pathways enriched by Epicatechin. C Figure representing interacting protein networks and pathways enriched by Epigallactocatechin-3-gallate. Networks were made using Cytoscape and pathway enrichment was carried out using the ClueGo plugin of Cytoscape
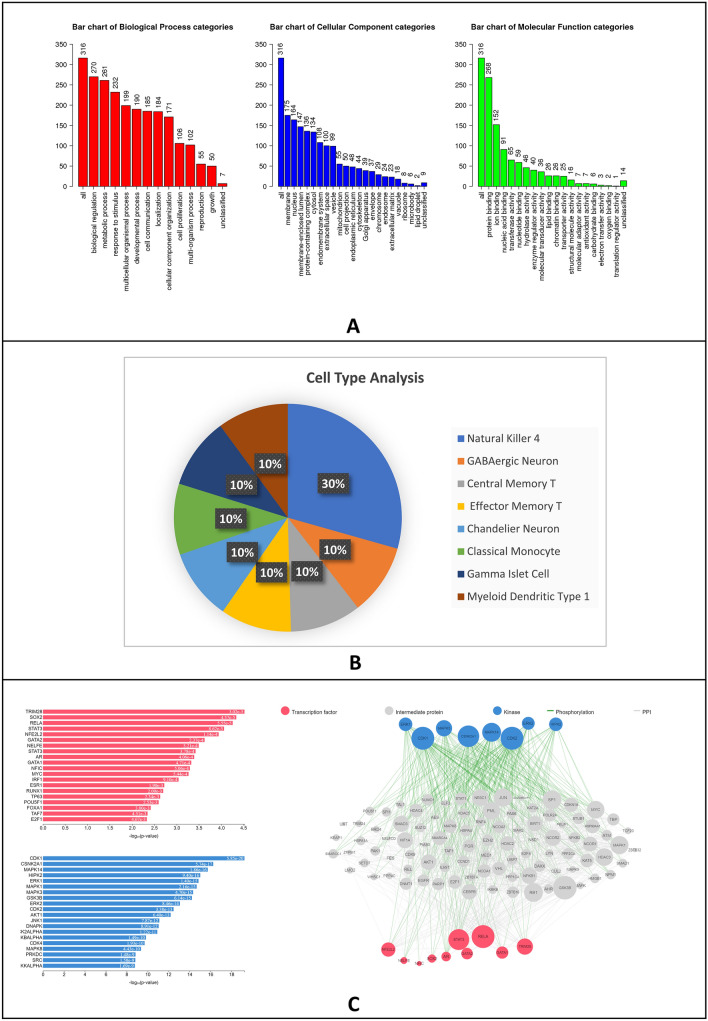
Further, the combination of catechin, epicatechin and epigallocatechin 3 gallate was subjected to combined analysis for biological process, cellular component categories and molecular function categories as well as cell type analysis. The biological process involves biological regulation, metabolic process, response to stimulus, cell communication, etc. (Fig. 4A). The cellular component included membrane nucleus and various organelles (Fig. 4A). The molecular function includes the binding of molecules like proteins, ions, nucleic acids and various classes of enzymes and regulators (Fig. 4A). The cell type analysis conjured neuronal and immune cells (Fig. 4B). The various cell types include GABAergic neurons, Chandelier neurons, Natural Killer cells, Central and effector memory T cells, monocyte and myeloid dendritic type 1 as well as gamma islet cells. On the whole, these results identify pathways, cellular processes and cell types with a potential role in the disease process. The Flavonoid interaction network generated was used for the analysis of the transcription factors and kinases that modulate their expression and function. The kinases and transcription factors were predictors from the flavonoid interaction network using X2K. Analysis of the combined interactors of ginsenoside and flavonoids shows TRIM2B, SOX2, RELA, STAT3, GATA, AR, etc. (Fig. 4C). Analysis of kinases shows CDKs, CSNK2A1, ERK, MAPK, HIPK2, JNK, etc. (Fig. 4C).
Fig. 4.
A Bar graph representing results obtained from biological processes, cellular components and molecular functions. The graphs were made using the GSEA toolkit (webgestalt.org). B Pie chart representing results obtained from Cell type analysis of interacting proteins with catechin derivatives. The pie chart was made using Enrichr, Azimuth database and Microsoft Excel 2019. C Bar graph and network file representing results obtained from transcription factor and kinase enrichment analysis of proteins that interact with catechin derivatives. Figures were made using X2K website (maayanlab.cloud/X2K)
Catechins modulate deregulated pathways in ALS which might help to achieve a favourable prognosis and better management of the disease.
To understand if catechin’s modulated genes exhibit significant changes in expression levels in ALS, the expression profile of genes from ALS patient brain cortex or Transgenic FUS or TDP-43 expressing mice model of disease were imported into the Catechin’s interaction network. The transgenic FUS mice model showed maximum changes in the expression profile of genes from the Catechin’s network (Fig. 5).
Fig. 5.
A Network file obtained from the overlay of catechin derivative interacting proteins and significant genes enriched in human post-mortem ALS cortex. Blue represents down-regulated genes while red represents up-regulated genes. B Network file obtained from the overlay of catechin derivative interacting proteins and significant genes enriched in transgenic FUS mice. Blue represents down-regulated genes while red represents up-regulated genes. C Network file obtained from the overlay of catechin derivative interacting proteins and significant genes enriched in transgenic TDP-43 mice. Blue represents down-regulated genes while red represents up-regulated genes. The network files were made using Cytoscape
Further, from the catechin’s network, the genes which showed significant changes in expression levels were binned into pathways. Our analysis shows that the genes whose expression profile changes in ALS patient's cerebral cortex were binned into pathways involved in immune cell pathways like Th1 cytokine production, regulation of astrocyte activation, glial cell proliferation, cell adhesion molecule production, gliogenesis, acute inflammatory response, leukocyte adhesion to vascular endothelial cells, etc. (Supplementary Fig. 3B). The genes whose expression profile changes in Transgenic mice expressing FUS were binned into pathways involved in negative regulation of neuroinflammatory response, regulation of fibroblast and smooth muscle proliferation, response to Iron and cadmium ion, regulation of release of cytochrome c from mitochondria, response to ROS etc. (Supplementary Fig. 3C). The genes whose expression profile changes in Transgenic mice expressing TDP-43 were binned into pathways involved in osteoclast development and differentiation, DNA damage checkpoint signaling, protein hydroxylation, eye morphogenesis, endopeptidases activity involved in the apoptotic process of cytochrome c, etc. (Supplementary Fig. 3C).
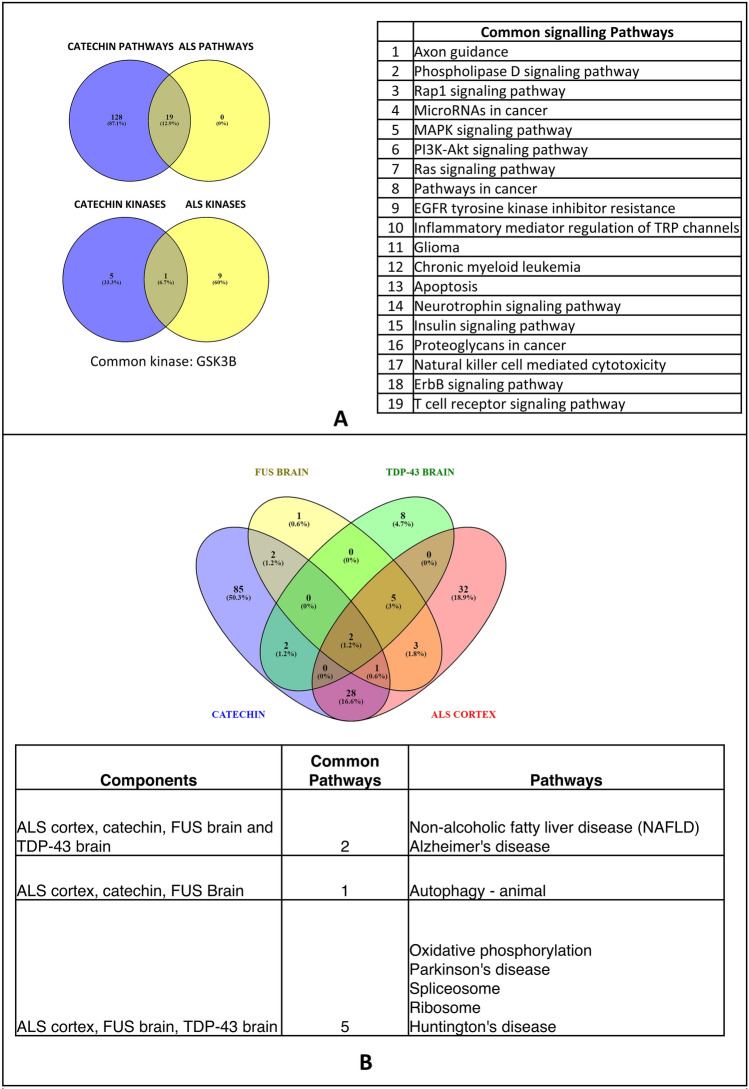
Commonality analysis between pathways obtained from the catechin–protein interaction network and those obtained from the transcriptomic analysis shows a considerable overlap of signaling pathways. The overlapping signaling pathways include Phospholipase D, RAP1, MAPK, PI3K-Akt, Ras, EGFR, Neurotrophin, insulin and ErbB signaling pathways (Fig. 6A). Common biological pathways between ALS cortex, FUS transgenic mice brain, TDP-43 transgenic mice brain and catechin-enriched pathways were obtained (Fig. 6B). Non-Alcoholic Fatty Liver Disease (NAFLD) and Alzheimer’s disease were found to be common pathways. Further, autophagy was common between catechins, ALS cortex and FUS brain. Based on the genes in the pathways and kinase, four kinases were identified as potential targets. These kinases include MAPK14, BACE2, GSK3B and AKT1 (Fig. 7).
Fig. 6.
A Venn diagram representing common signaling pathways and kinases enriched by catechin derivative interacting proteins and significant genes enriched in human post-mortem ALS cortex. B Venn diagram and table representing common biological pathways enriched by catechin derivative interacting proteins, significant genes enriched in human post-mortem ALS, significant genes enriched in transgenic FUS mice and significant genes enriched in transgenic TDP-43 mice. Venn diagrams were made using Venny (bioinfogp.cnb.csic.es/tools/venny). The individual list of enriched pathways is provided in SUPPLEMENTARY-4
Fig. 7.
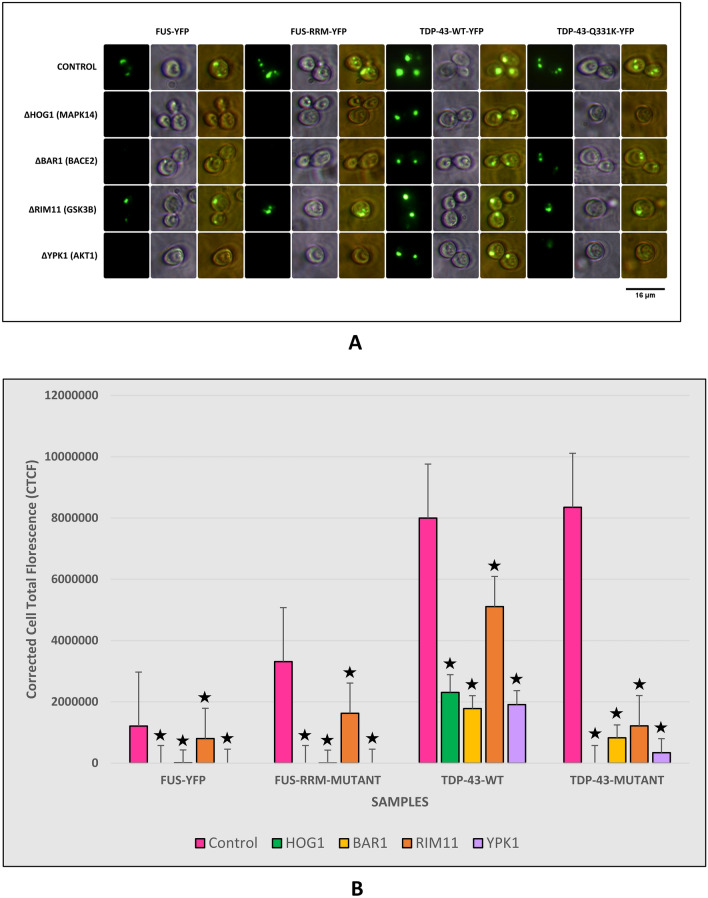
A Fluorescence Microscopy results of yeast Knock-outs expressing FUS and TDP‒43 Wild type and Mutants. All the knock-outs showed a significant reduction in protein aggregates. Imaging was carried out in the dark field (blue light) and white light. Images were arranged using PhotoScape. B Graph representing fluorescence quantification results of yeast Knock-outs expressing FUS and TDP‒43 Wild type and Mutants. The quantification results had a strong correlation with imaging results. Fluorescence quantification was carried out using ImageJ software (⋆ represents P ≤ 0.05)
Gene knock-out in pathways modulated by Catechin’s or ginsenoside mitigate protein aggregation in the yeast model of ALS.
Having identified the human kinases modulated by flavonoids in the deregulated pathways we used (flyrnai.org/cgi-bin/DRSC_orthologs.pl) an online tool to identify the corresponding yeast orthologues. The yeast orthologues of the human kinases MAPK14, BACE2, GSK3B and AKT1 were identified as HOG1, BAR1, RIM11 and YPK1.To validate the predictions yeast KO (Dharmacon) of HOG1, BAR1, RIM11 and YPK1 were employed. The kinase KO significantly reduced protein aggregation. Taken together, our results show that flavonoid-mediated clearance of protein aggregates might potentially involve kinases.
We further looked at literature to understand the potential targets of ginsenosides. Previous studies have shown that ginsenoside modulates the activity of BACE2. Further, we used the yeast BAR1 knockout which is the orthologue of BACE2. Our results show that BAR1 KO significantly reduced protein aggregates in both-wild type and mutant FUS or TDP-43 expressing yeast model of ALS (Fig. 7). Collectively, our results show that BACE2 potentially modulates aggregation of wild-type and mutant FUS and TDP-43.
Discussion
Medicinal plants from Indian, Chinese, or Tibetan traditional medicinal systems have been used to treat many neurodegenerative diseases. Similarly, Ginseng has been evaluated for its efficacy to treat many neurodegenerative diseases. Our analysis shows that Ginseng was very effective in clearing protein aggregates in both wild-type and mutant TDP-43 or FUS expressing yeast models of ALS. Further, analysis with the SymMap database showed that ginseng is used in the treatment of ALS in Chinese medicine. Previous studies have shown a beneficial effect of ginseng root in the SOD1 (G93A) transgenic mice model of ALS (Jiang et al. 2000). Having identified ginseng to mitigate protein aggregation in yeast we looked at literature to understand the composition of ginseng. Previous studies have catalogued that the major components of ginseng are Flavonoids and Ginsenosides (Kim 2016).The flavonoids include catachin, epicatechin and epigallacatachin-gallate (Kim 2016). The Ginsenosides include the ginsenoside Rg1, ginsenoside Rh2 and ginsenoside Rd (Kim 2016).
Flavonoids like catechin, epicatechin and epigallocatechin-gallate have been evaluated for their efficacy in the treatment of many Alzheimer’s and Parkinson’s diseases (Özduran et al. 2021). Studies have also shown a favourable effect of green tea catechins in the treatment of Alzheimer’s and Parkinson’s disease (Burnstock 2008). The therapeutic effects of catechins have been extensively documented in both human and animal models of diseases (Sebastiani et al. 2021). Hence, we treated the yeast model of ALS used in our study with catechin, epicatechin and epigallocatechin-gallate. Our analysis shows that catechin and epicatechin were very effective in clearing protein aggregates in the yeast model of ALS used in the study. However, epigallocatechin-gallate was not as effective as other catechins used. The catechins are shown to exert their beneficial effects by their antioxidant (metal ion chelating and ROS scavenging), anti-inflammatory and anti-apoptotic properties. Studies have also shown that catechin reduces phosphorylation of Tau and aggregation of amyloid-beta as well as reduces the levels of alpha-synuclein and increases dopamine levels (Özduran et al. 2021). They are also shown to modulate mitochondrial function and prevent the release of apoptotic proteins (Mercer et al. 2005; Schroeder et al. 2009).
Having shown that catechins could mitigate protein aggregation formation in the yeast model of ALS we looked for the proteins which are modulated by it. A previous study has catalogued the interactors of different flavonoids (Lacroix et al. 2018). Using this we generated a network of different catechin interactors and further binned them into pathways. Our analysis showed that the genes fall into many signaling and metabolic pathways. Further, we carried out cell type analysis which conjured inflammatory cells, GABAergic neurons, Chandelier neurons and gamma islet cells. The commonality analysis of pathways between catechin-modulated pathways and those obtained from the transcriptomic analysis of ALS patients showed an overlap of 19 pathways. These pathways included the MAPK, PI3K-AKT pathway, etc. The commonality analysis of kinases enriched in catechin-modulated pathways and those obtained for data sets from ALS patients showed GSK3B as the common kinase. One of the previous reviews had propounded many kinases as potential targets in ALS (Guo et al. 2020). Hence, we used the yeast model of ALS which is knocked out for the kinases of their human orthologues to evaluate their effect on protein aggregation.
Our analysis showed that knockout of HOG1 (MAPK14), RIM11 (GSK3B) and YPK1 (AKT1) significantly reduced protein aggregation. It has been demonstrated that in APP-PS1 transgenic mice brain expression of MAPK14/p38α is upregulated and genetic deficiency to MAPK14 was shown to stimulate autophagy (Alam and Scheper 2016). The increased autophagy in turn leads to degradation of BACE1 which helps to reduce protein aggregates (Alam and Scheper 2016). In ALS MAPK14/P38 is activated in motor neurons, microglia and astroglia. Similarly, in SOD1-ALS patients and SOD1 transgenic mice, MAPK14 is found to be upregulated in motor neurons and astroglia (Bendotti et al. 2004). The MAPK14 activation also leads to activation of nNOS which produces Nitric oxide which acts as a feed-forward loop through the FAS receptor to activate MAPK14 and more nNOS. Activation of MAPK14 is correlated with motor neuron cell death (Sahana and Zhang 2021). P38 activation is also linked to pathogenic aggregates in ALS involving FUS (Sama et al. 2017). Similarly, FUS and SOD1 mutant mice showed impaired axonal transport (Baldwin et al. 2016). Injection of FUS mutants into squid axoplasm led to activation of p38 and impaired axonal transport showing a toxic gain of function for FUS mutants (Sama et al. 2017). Inhibition of p38 prevents apoptosis and cyclosporine-induced cell death in the SOD1 mice model of ALS (Dewil et al. 2007). In all, these results show that inhibition of MAPK14 orthologue HOG1 could inhibit protein aggregation or amyloid clearance in the yeast model of ALS.
GSK3b emerged as one of the common kinases in the kinase enrichment analysis of both catechin interactors and ALS transcriptomic data sets. Dysregulation of GSK3b is found to be associated with many neurodegenerative diseases (Hernández et al. 2009; Duda et al. 2018). GSK3b was found to be activated in both patients and ALS model systems (Koh et al. 2011) and inhibition of GSK3b was found to be neuroprotective in ALS pathogenesis (Choi et al. 2020). Inhibition of GSK3β suppresses pathogenesis caused by SOD, TDP-43 and FUS expression in various models (Yang et al. 2013; White et al. 2021; Choi et al. 2022). Collectively our analysis shows that catechins might modulate protein aggregation through GSK3b.
Our analysis shows PI3K-AKT1 as one of the deregulated pathways in ALS. Consistent with this observation inhibition of the AKT1 homologue leads to a reduction in protein aggregates in the yeast model of ALS. Previous studies have shown that C9orf72 forms a heterodimer with SMCR8 and SMCR8 loss leading to a marked reduction of c9orf72 protein levels (Liang et al. 2019). SMCR8 loss was shown to reduce the levels of proteins in the autophagy-lysosome pathways due to activation of mTOR and AKT1 (Liang et al. 2019). Investigation of RAB11 expression and MAPK/ERK/AKT signaling in en post-mortem spinal cord specimens showed down-regulation of RAB11. RAB11 was found to be downregulated in all the ALS samples while phosphor-AKT and phosphor-S6 kinase were upregulated (Mitra et al. 2019). Loss of RAB11 is associated with ALS and overexpression of RAB11 was found to be neuroprotective (Chutna et al. 2014). However, in the mice model of ALS carboxy-terminal modulator protein which regulates AKT signaling by reducing the activity of AKT was found to be upregulated. Decreased AKT activity is attributed to muscle atrophy in ALS and is associated with increased expression of genes involved in atrophy (Wang et al. 2019). Our analysis shows a potential role for AKT in modulating protein aggregation in the yeast model of ALS.
Further, Ginsenosides are a major constituent of Ginseng. We analyzed if Ginsenoside Rg1, Ginsenoside Rh2 and Ginsenoside Rd for their ability to clear protein aggregates in FUS or TDP-43 expressing yeast model of ALS. Our analysis showed that treatment with Ginsenoside Rg1 significantly reduced protein aggregation. Previous studies have shown that Ginsenoside Re exhibited a neuroprotective effect by largely inhibiting microglial inflammation potentially involving CAMK/MAPK/NFkB signaling (Madhi et al. 2021). Ginsenoside Re also attenuated neuroinflammation in the SOD1 (G93A) symptomatic mice model of ALS (Cai and Yang 2016). Having found that Ginsenoside Rg1 clears protein aggregates in the yeast model we looked for Ginsenoside Rg1 interactors in the literature. Our literature search showed BACE2 as a potential interactor of Ginsenoside Rg1 (Mohanan et al. 2018). KO of BACE2 orthologue BAR1 in a yeast model of ALS abrogated protein aggregation. Studies have shown that BACE1 processing of amyloid precursor protein in skeletal muscle of mice model of ALS. Inhibition of BACE affects survival and motor function in mice model of ALS (Rabinovich-Toidman et al. 2012). Hence inhibition of BACE might help to achieve a favourable prognosis in ALS.
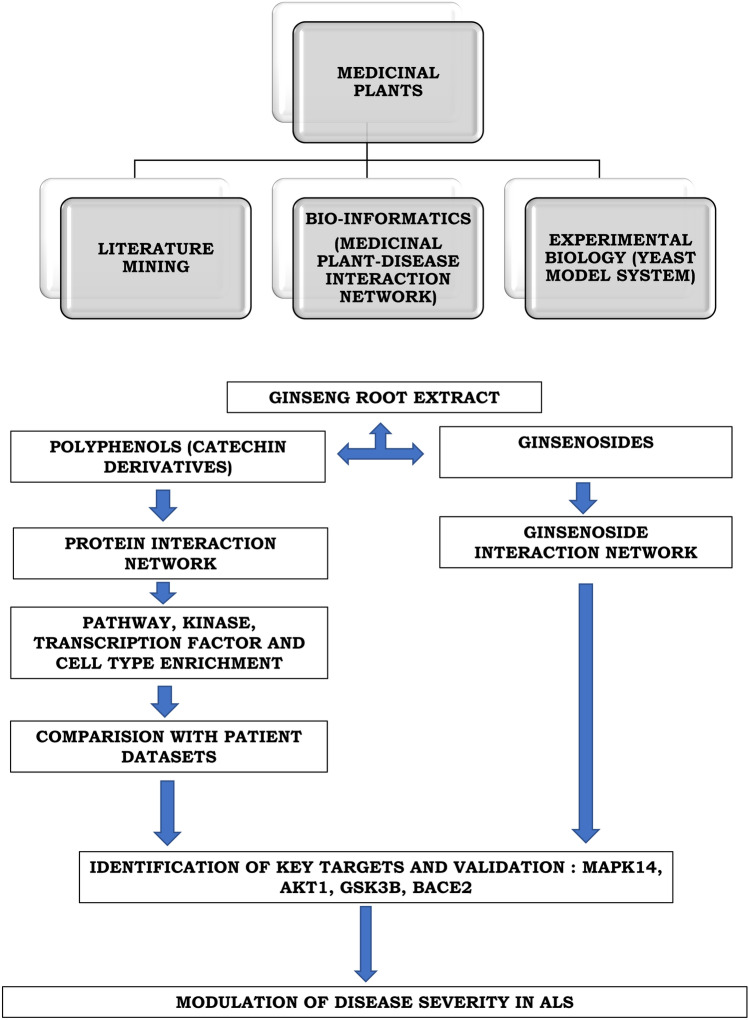
All things considered, our results show that Ginseng root extract contains secondary metabolites which could help clear protein aggregates. The catechins act by interacting and inhibiting multiple key proteins like MAPK14, GSK3b and AKT which modulate deregulated pathways in ALS thus mitigating protein aggregation in the yeast model of ALS. The Ginsenoside Rg1 interaction was also shown to clear protein aggregates in the yeast model of ALS by interacting with BAR1 an orthologue of BACE2. Overall, our results not only identified some of the active ingredients of Ginseng which helped to clear protein aggregates but also the potential targets using the network pharmacology approach. Given the identified proteins drugs targeting MAPK14, GSK3B, AKT or BACE2 could be repurposed for the treatment of ALS. The results of the study are summarized in Fig. 8.
Fig. 8.
Flow chart summarizing the methodology and the results of the study
Conclusion
Amyotrophic Lateral Sclerosis is an incurable, progressive neurodegenerative disease affecting the motor neurons. Medicinal plants from traditional medical systems used for treating neurodegenerative disease were screened for their efficacy to clear protein aggregates in yeast model of ALS. Our analysis shows that Ginseng mitigated protein aggregates in yeast model of ALS. Further, the active ingredients of ginseng which include flavonoids and ginsenosides were screened for their efficacy to clear protein aggregates. We used a network pharmacology approach to identify potential targets. Flavonoids like catechin, epicatechin and epigallocatechin-3-gallate mitigated protein aggregates in yeast model system. Ginsenosides like ginsenoside Rg1 effectively cleared protein aggregates while Rd, Rh2 had minimal effect on protein aggregates. Commonality analysis of pathways obtained from interactors of catechin, epicatechin and epigallocatechin-3-gallate as well as ginsenoside interactors overlapped with those obtained from transcriptomic analysis of ALS datasets. The common genes from the deregulated pathways were MAPK14, GSK3B, AKT and BACE1. Further Knock-outs of either of MAPK14, GSK3B, AKT and BACE1 showed clearance of protein aggregates. Our analysis shows that flavonoids like catechin, epicatechin and epigallocatechin-3-gallate and ginsenoside Rg1 potentially act through MAPK14, GSK3B, AKT and BACE1 to mitigate protein aggregation in yeast model of ALS. Flavonoids and ginsenosides could be potential therapeutic agents in ALS.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
A sincere thanks to Sri Sathya Sai Institute of Higher Learning for providing free and valuable education. We acknowledge the grant support from the Department of Biotechnology-Basic Research in Modern Biology DBT (BRB): BT/PR8226/BRB/10/1224/2013, Department of Science and Technology-The Science and Engineering Research Board–Extra Mural Research DST-SERB-EMR: EMR/2017/005381, Department of Biotechnology- Bioinformatics Infrastructure facility DBT-BIF: BT/BI/25/063/2012, Department of Science and Technology- Fund for improvement of Science and Technology Infrastructure in Higher Educational Institutions (DST-FIST): SR/FST/LSI-616/2014, University Grants Commission-Special Assistance Program (UGC-SAP III): F.3-19 /2018/DRS-III(SAP-II) for infrastructure funding.
Abbreviations
- ALS
Amyotrophic lateral sclerosis
- GEO
Gene expression omnibus
- MAPK14
Mitogen-activated protein kinase 14
- GSK
Glycogen synthase kinase
- BACE1
Beta-secretase 1
- SNP
Single nucleotide polymorphism
- SOD1
Super oxide dismutase 1
- C9orf72
Chromosome 9 open reading frame 72
- FUS
Fused in sarcoma
- TDP-43
Tar DNA binding protein 43
- HAT
Histone acetyltransferase
- HADC
Histone deacetylases
- HD
Huntington’s disease
- PD
Parkinson’s disease
- DMSO
Dimethyl sulfoxide
- GREIN
GEO RNA-seq experiments interactive navigator
- GSEA
Gene set enrichment analysis
- STITCH
Search tool for interactions of chemicals
- E-YFP
Enhanced yellow florescent protein
- URA-YNB
Uracil–yeast nitrogen base
Author contributions
The study was contributed by S.S.R. S.S.P helped in standardizing and processing several bio-informatic pipelines used for the study. D.D.V.M and K.S.P assisted in performing the yeast experiments. S.V conceptualized the entire idea, interpreted the results, and played a major role in the preparation of the manuscript.
Declarations
Conflict of interest
All authors declare no conflict of interest.
Contributor Information
R. Sai Swaroop, Email: samasthalokasukinobhavanthu@gmail.com.
Sai Sanwid Pradhan, Email: saisanwidpradhan@sssihl.edu.in.
V. M. Datta Darshan, Email: vmdattadarshan@sssihl.edu.in.
Kanikaram Sai Phalguna, Email: saiphalguna5@gmail.com.
Venketesh Sivaramakrishnan, Email: s.venketesh@gmail.com.
References
- Abdel-Khalik J, Yutuc E, Crick PJ, et al. Defective cholesterol metabolism in amyotrophic lateral sclerosis. J Lipid Res. 2017;58:267–278. doi: 10.1194/jlr.P071639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Mahi N, Najafabadi MF, Pilarczyk M, et al. GREIN: an interactive web platform for re-analyzing GEO RNA-seq data. Sci Rep. 2019;9:1–9. doi: 10.1038/s41598-019-43935-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam J, Scheper W. Targeting neuronal MAPK14/p38α activity to modulate autophagy in the Alzheimer disease brain. Autophagy. 2016;12:2516–2520. doi: 10.1080/15548627.2016.1238555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Chalabi A, Van Den Berg LH, Veldink J. Gene discovery in amyotrophic lateral sclerosis: Implications for clinical management. Nat Rev Neurol. 2017;13:96–104. doi: 10.1038/nrneurol.2016.182. [DOI] [PubMed] [Google Scholar]
- Ayaz M, Sadiq A, Junaid M, et al. Flavonoids as prospective neuroprotectants and their therapeutic propensity in aging associated neurological disorders. Front Aging Neurosci. 2019;11:155. doi: 10.3389/fnagi.2019.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin KR, Godena VK, Hewitt VL, Whitworth AJ. Axonal transport defects are a common phenotype in Drosophila models of ALS. Hum Mol Genet. 2016;25:2378–2392. doi: 10.1093/hmg/ddw105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendotti C, Atzori C, Piva R, et al. Activated p38MAPK is a novel component of the intracellular inclusions found in human amyotrophic lateral sclerosis and mutant SOD1 transgenic mice. J Neuropathol Exp Neurol. 2004;63:113–119. doi: 10.1093/jnen/63.2.113. [DOI] [PubMed] [Google Scholar]
- Bernardini C, Censi F, Lattanzi W, et al. Mitochondrial network genes in the skeletal muscle of amyotrophic lateral sclerosis patients. PLoS ONE. 2013 doi: 10.1371/journal.pone.0057739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco H, Corcia P, Pradat PF, et al. Metabolomics in cerebrospinal fluid of patients with amyotrophic lateral sclerosis: An untargeted approach via high-resolution mass spectrometry. J Proteome Res. 2013;12:3746–3754. doi: 10.1021/pr400376e. [DOI] [PubMed] [Google Scholar]
- Blasco H, Patin F, Madji Hounoum B, et al. Metabolomics in amyotrophic lateral sclerosis: How far can it take us? Eur J Neurol. 2016;23:447–454. doi: 10.1111/ene.12956. [DOI] [PubMed] [Google Scholar]
- Bournival J, Quessy P, Martinoli M-G. Protective effects of resveratrol and quercetin against MPP+-induced oxidative stress act by modulating markers of apoptotic death in dopaminergic neurons. Cell Mol Neurobiol. 2009;29:1169–1180. doi: 10.1007/s10571-009-9411-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks BR. Natural history of ALS: symptoms, strength, pulmonary function, and disability. Neurology. 1996;47:71S–82S. doi: 10.1212/WNL.47.4_Suppl_2.71S. [DOI] [PubMed] [Google Scholar]
- Burnstock G. Purinergic signalling and disorders of the central nervous system. Nat Rev Drug Discov. 2008;7:575–590. doi: 10.1038/nrd2605. [DOI] [PubMed] [Google Scholar]
- Caballero-Hernandez D, Toscano MG, Cejudo-Guillen M, et al. The “Omics” of Amyotrophic Lateral Sclerosis. Trends Mol Med. 2016;22:53–67. doi: 10.1016/j.molmed.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Cai M, Yang EJ. Ginsenoside Re attenuates neuroinflammation in a symptomatic ALS animal model. Am J Chin Med. 2016;44:401–413. doi: 10.1142/S0192415X16500233. [DOI] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14:1–14. doi: 10.1186/1471-2105-14-S18-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H-J, Cha SJ, Lee J-W, et al. Recent advances on the role of gsk3β in the pathogenesis of amyotrophic lateral sclerosis. Brain Sci. 2020;10:675. doi: 10.3390/brainsci10100675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H-J, Lee JY, Cha SJ, et al. FUS-induced neurotoxicity is prevented by inhibiting GSK-3β in a Drosophila model of amyotrophic lateral sclerosis. Hum Mol Genet. 2022;31:850–862. doi: 10.1093/hmg/ddab290. [DOI] [PubMed] [Google Scholar]
- Chutna O, Gonçalves S, Villar-Piqué A, et al. The small GTPase Rab11 co-localizes with α-synuclein in intracellular inclusions and modulates its aggregation, secretion and toxicity. Hum Mol Genet. 2014;23:6732–6745. doi: 10.1093/hmg/ddu391. [DOI] [PubMed] [Google Scholar]
- Clarke DJB, Kuleshov MV, Schilder BM, et al. eXpression2Kinases (X2K) Web: linking expression signatures to upstream cell signaling networks. Nucleic Acids Res. 2018;46:W171–W179. doi: 10.1093/nar/gky458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough E, Barrett T. Statistical genomics. Cham: Springer; 2016. The gene expression omnibus database; pp. 93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins TJ. ImageJ for microscopy. Biotechniques. 2007;43:S25–S30. doi: 10.2144/000112517. [DOI] [PubMed] [Google Scholar]
- Dewil M, dela Cruz VF, Van Den Bosch L, Robberecht W. Inhibition of p38 mitogen activated protein kinase activation and mutant SOD1G93A-induced motor neuron death. Neurobiol Dis. 2007;26:332–341. doi: 10.1016/j.nbd.2006.12.023. [DOI] [PubMed] [Google Scholar]
- Duda P, Wiśniewski J, Wójtowicz T, et al. Targeting GSK3 signaling as a potential therapy of neurodegenerative diseases and aging. Expert Opin Ther Targets. 2018;22:833–848. doi: 10.1080/14728222.2018.1526925. [DOI] [PubMed] [Google Scholar]
- Dutta K, Patel P, Rahimian R, Phaneuf D, Julien J-P. Withania somnifera reverses transactive response DNA binding protein 43 proteinopathy in a mouse model of amyotrophic lateral sclerosis/frontotemporal lobar degeneration. Neurotherapeutics. 2017;14:447–462. doi: 10.1007/s13311-016-0499-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ederle H, Dormann D. TDP-43 and FUS en route from the nucleus to the cytoplasm. FEBS Lett. 2017;591:1489–1507. doi: 10.1002/1873-3468.12646. [DOI] [PubMed] [Google Scholar]
- Eid S, Turk S, Volkamer A, et al. KinMap : a web-based tool for interactive navigation through human kinome data. BMC Bioinform. 2017 doi: 10.1186/s12859-016-1433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen A. Amyotrophic lateral sclerosis: evolutionary and other perspectives. Muscle Nerve. 2009;40:297–304. doi: 10.1002/mus.21404. [DOI] [PubMed] [Google Scholar]
- Ferri A, Coccurello R. What is “hyper” in the ALS Hypermetabolism? Mediat Inflamm. 2017 doi: 10.1155/2017/7821672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghareeb DA, ElAhwany AMD, El-Mallawany SM, Saif AA. In vitro screening for anti-acetylcholiesterase, anti-oxidant, anti-glucosidase, anti-inflammatory and anti-bacterial effect of three traditional medicinal plants. Biotechnol Biotechnol Equip. 2014;28:1155–1164. doi: 10.1080/13102818.2014.969877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero EN, Wang H, Mitra J, et al. TDP-43/FUS in motor neuron disease: complexity and challenges. Prog Neurobiol. 2016;145:78–97. doi: 10.1016/j.pneurobio.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Vandoorne T, Steyaert J, et al. The multifaceted role of kinases in amyotrophic lateral sclerosis: genetic, pathological and therapeutic implications. Brain. 2020;143:1651–1673. doi: 10.1093/brain/awaa022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández F, Nido JD, Avila J, Villanueva N. GSK3 Inhibitors and Disease. MRMC. 2009;9:1024–1029. doi: 10.2174/138955709788922647. [DOI] [PubMed] [Google Scholar]
- Jeong HY, et al. Leaf and stem of Vitis amurensis and its active components protect against amyloid β protein (25–35)-induced neurotoxicity. Arch Pharm Res. 2010;33:1655–1664. doi: 10.1007/s12272-010-1015-6. [DOI] [PubMed] [Google Scholar]
- Jiang F, DeSilva S, Turnbull J. Beneficial effect of ginseng root in SOD-1 (G93A) transgenic mice. J Neurol Sci. 2000;180:52–54. doi: 10.1016/S0022-510X(00)00421-4. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilani-Jaziri S, Mustapha N, Mokdad-Bzeouich I, et al. Flavones induce immunomodulatory and anti-inflammatory effects by activating cellular anti-oxidant activity: a structure-activity relationship study. Tumor Biol. 2016;37:6571–6579. doi: 10.1007/s13277-015-4541-5. [DOI] [PubMed] [Google Scholar]
- Kim J. Investigation of phenolic, flavonoid, and vitamin contents in different parts of Korean Ginseng (Panax ginseng C A Meyer) Prev Nutr Food Sci. 2016;21:263–270. doi: 10.3746/pnf.2016.21.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh S-H, Baek W, Kim SH. Brief review of the role of glycogen synthase kinase-3β in amyotrophic lateral sclerosis. Neurol Res Int. 2011;2011:205761. doi: 10.1155/2011/205761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konar A, Thakur MK. Science of Ashwagandha: preventive and therapeutic potentials. Berlin: Springer; 2017. Cellular and molecular targets underpinning memory enhancement by Ashwagandha; pp. 305–318. [Google Scholar]
- Kori M, Aydln B, Unal S, et al. Metabolic biomarkers and neurodegeneration: a pathway enrichment analysis of Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis. Omi A J Integr Biol. 2016;20:645–661. doi: 10.1089/omi.2016.0106. [DOI] [PubMed] [Google Scholar]
- Kuhn M, von Mering C, Campillos M, et al. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2007;36:D684–D688. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix S, Badoux JK, Parolo S, et al. OPEN A computationally driven analysis of the polyphenol-protein interactome. Sci Rep. 2018 doi: 10.1038/s41598-018-20625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Polymenidou M, Hutt KR, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15:1488–1497. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-J, et al. Bioavailability of fermented Korean red ginseng. Prev Nutr Food Sci. 2009;14:201–207. doi: 10.3746/jfn.2009.14.3.201. [DOI] [Google Scholar]
- Liang C, Shao Q, Zhang W, et al. Smcr8 deficiency disrupts axonal transport-dependent lysosomal function and promotes axonal swellings and gain of toxicity in C9ALS/FTD mouse models. Hum Mol Genet. 2019;28:3940–3953. doi: 10.1093/hmg/ddz230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Ye M, Guo H. An updated review of randomized clinical trials testing the improvement of cognitive function of Ginkgo biloba extract in healthy people and Alzheimer’s patients. Front Pharmacol. 2020;10:1688. doi: 10.3389/fphar.2019.01688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longinetti E, Fang F. Epidemiology of amyotrophic lateral sclerosis: an update of recent literature. Curr Opin Neurol. 2019 doi: 10.1097/WCO.0000000000000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López MVN, Cuadrado MPG-S, Ruiz-Poveda OMP, Del Fresno AMV, Accame MEC. Neuroprotective effect of individual ginsenosides on astrocytes primary culture. Biochim Biophys Acta (BBA) 2007;1770:1308–1316. doi: 10.1016/j.bbagen.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Luo Y, et al. Inhibition of amyloid-β aggregation and caspase-3 activation by the Ginkgo biloba extract EGb761. Proc Natl Acad Sci. 2002;99:12197–12202. doi: 10.1073/pnas.182425199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra R, Roy A. Role of medicinal plants against neurodegenerative diseases. Curr Pharm Biotechnol. 2022;23:123–139. doi: 10.2174/1389201022666210211123539. [DOI] [PubMed] [Google Scholar]
- Madhi I, Kim J-H, Shin JE, Kim Y. Ginsenoside Re exhibits neuroprotective effects by inhibiting neuroinflammation via CAMK/MAPK/NF-κB signaling in microglia. Mol Med Rep. 2021;24:1–10. doi: 10.3892/mmr.2021.12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek AM, Barchowsky A, Bowser R, et al. Pesticide exposure as a risk factor for amyotrophic lateral sclerosis: a meta-analysis of epidemiological studies: pesticide exposure as a risk factor for ALS. Environ Res. 2012;117:112–119. doi: 10.1016/j.envres.2012.06.007. [DOI] [PubMed] [Google Scholar]
- Mathis S, Goizet C, Soulages A, et al. Genetics of amyotrophic lateral sclerosis: a review. J Neurol Sci. 2019;399:217–226. doi: 10.1016/j.jns.2019.02.030. [DOI] [PubMed] [Google Scholar]
- Mccann EP, Henden L, Fifita JA, et al. Evidence for polygenic and oligogenic basis of Australian sporadic amyotrophic lateral sclerosis. J Ned Genet. 2021 doi: 10.1136/jmedgenet-2020-106866. [DOI] [PubMed] [Google Scholar]
- Mercer LD, Kelly BL, Horne MK, Beart PM. Dietary polyphenols protect dopamine neurons from oxidative insults and apoptosis: investigations in primary rat mesencephalic cultures. Biochem Pharmacol. 2005;69:339–345. doi: 10.1016/j.bcp.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Mitra J, Hegde PM, Hegde ML. Loss of endosomal recycling factor RAB11 coupled with complex regulation of MAPK/ERK/AKT signaling in postmortem spinal cord specimens of sporadic amyotrophic lateral sclerosis patients. Mol Brain. 2019;12:1–4. doi: 10.1186/s13041-019-0475-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanan P, Subramaniyam S, Mathiyalagan R, Yang D-C. Molecular signaling of ginsenosides Rb1, Rg1, and Rg3 and their mode of actions. J Ginseng Res. 2018;42:123–132. doi: 10.1016/j.jgr.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morello G, Salomone S, Agata VD, et al. From multi-omics approaches to precision medicine in amyotrophic lateral sclerosis. Front Neurosci. 2020 doi: 10.3389/fnins.2020.577755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik AA, Narayanan A, Khanchandani P, Sridharan D. Systems analysis of avascular necrosis of femoral head using integrative data analysis and literature mining delineates pathways associated with disease. Sci Rep. 2020 doi: 10.1038/s41598-020-75197-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelagandan N, Gonnella G, Dang S, et al. TDP-43 enhances translation of specific mRNAs linked to neurodegenerative disease. Nucleic Acids Res. 2019;47:341–361. doi: 10.1093/nar/gky972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo ST, Steyn FJ. The interplay between metabolic homeostasis and neurodegeneration: Insights into the neurometabolic nature of amyotrophic lateral sclerosis. Cell Regen. 2015;4(4):5. doi: 10.1186/s13619-015-0019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özduran G, Becer E, Vatansever HS. The Role and Mechanisms of Action of Catechins in Neurodegenerative Diseases. J Am Coll Nutr. 2021 doi: 10.1080/07315724.2021.1981487. [DOI] [PubMed] [Google Scholar]
- Pandey S, Sarma N. Commentary: Amyotrophic lateral sclerosis: Ongoing search for prognostic biomarkers of longevity. Neurol India. 2017;65:1155. doi: 10.4103/neuroindia.NI_7_17. [DOI] [PubMed] [Google Scholar]
- Patin F, Corcia P, Vourc’h P, et al. Omics to explore amyotrophic lateral sclerosis evolution: the central role of arginine and proline metabolism. Mol Neurobiol. 2017;54:5361–5374. doi: 10.1007/s12035-016-0078-x. [DOI] [PubMed] [Google Scholar]
- Piñero J, Bravo À, Queralt-Rosinach N, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2016;45:gkw943. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan SS, Thota SM, Saiswaroop R, et al. Integrated multi-omic analysis of Huntington disease and yeast model delineates pathways modulating protein aggregation. Dis Model Mech. 2022 doi: 10.1242/dmm.049492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich-Toidman P, Becker M, Barbiro B, Solomon B. Inhibition of amyloid precursor protein beta-secretase cleavage site affects survival and motor functions of amyotrophic lateral sclerosis transgenic mice. Neurodegener Dis. 2012;10:30–33. doi: 10.1159/000334774. [DOI] [PubMed] [Google Scholar]
- Sahana TG, Zhang K. Mitogen-activated protein kinase pathway in amyotrophic lateral sclerosis. Biomedicines. 2021;9:969. doi: 10.3390/biomedicines9080969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai Swaroop R, Akhil PS, Sai Sanwid P, et al. Integrated multi-omic data analysis and validation with yeast model show oxidative phosphorylation modulates protein aggregation in amyotrophic lateral sclerosis. J Biomol Struct Dyn. 2022 doi: 10.1080/07391102.2022.2115555. [DOI] [PubMed] [Google Scholar]
- Sama RRK, Fallini C, Gatto R, et al. ALS-linked FUS exerts a gain of toxic function involving aberrant p38 MAPK activation. Sci Rep. 2017;7:1–13. doi: 10.1038/s41598-017-00091-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarda RK, Sinha R, Sherpa M, Gupta A. Crude extracts of bacopa monnieri induces dendrite formation in rodent neural stem cell cultures: a possible use in neuronal injury. J. Neurosci Rural Pract. 2022;13:254–260. doi: 10.1055/s-0042-1743215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sas K, Robotka H, Toldi J, Vécsei L. Mitochondria, metabolic disturbances, oxidative stress and the kynurenine system, with focus on neurodegenerative disorders. J Neurol Sci. 2007;257:221–239. doi: 10.1016/j.jns.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Schroeder EK, Kelsey NA, Doyle J, et al. Green tea epigallocatechin 3-gallate accumulates in mitochondria and displays a selective antiapoptotic effect against inducers of mitochondrial oxidative stress in neurons. Antioxid Redox Signal. 2009;11:469–480. doi: 10.1089/ars.2008.2215. [DOI] [PubMed] [Google Scholar]
- Sebastiani G, Almeida-Toledano L, Serra-Delgado M, et al. Therapeutic effects of catechins in less common neurological and neurodegenerative disorders. Nutrients. 2021;13:2232. doi: 10.3390/nu13072232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speldewinde SH, Grant CM. Spermidine cures yeast of prions. Microb Cell. 2016;3:46. doi: 10.15698/mic2016.01.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer JPE. The interactions of flavonoids within neuronal signalling pathways. Genes Nutr. 2007;2:257–273. doi: 10.1007/s12263-007-0056-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srimadh Bhagavatham SK, Pulukool SK, Pradhan SS, et al. Systems biology approach delineates critical pathways associated with disease progression in rheumatoid arthritis. J Biomol Struct Dyn. 2022 doi: 10.1080/07391102.2022.2115555. [DOI] [PubMed] [Google Scholar]
- Tam OH, Rozhkov NV, Shaw R, et al. Postmortem cortex samples identify distinct molecular subtypes of ALS: retrotransposon activation, oxidative stress, and activated glia. Cell Rep. 2019;29:1164–1177. doi: 10.1016/j.celrep.2019.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefera TW, Borges K. Metabolic dysfunctions in amyotrophic lateral sclerosis pathogenesis and potential metabolic treatments. Front Neurosci. 2017 doi: 10.3389/fnins.2016.00611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda T. Biomarkers for amyotrophic lateral sclerosis. Brain Nerve. 2012;64:515–523. [PubMed] [Google Scholar]
- Uddin MS, et al. Exploring the effect of Phyllanthus emblica L on cognitive performance, brain antioxidant markers and acetylcholinesterase activity in rats: promising natural gift for the mitigation of Alzheimer’s disease. Ann Neurosci. 2016;23:218–229. doi: 10.1159/000449482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme P, Robberecht W, Van Den Bosch L. Modelling amyotrophic lateral sclerosis: Progress and possibilities. DMM Dis Model Mech. 2017;10:537–549. doi: 10.1242/dmm.029058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinceti M. The environment and amyotrophic lateral sclerosis: converging clues from epidemiologic studies worldwide. N Am J Med Sci. 2012;4:356. [PMC free article] [PubMed] [Google Scholar]
- Vishnupriya P, Padma VV. A review on the antioxidant and therapeutic potential of Bacopa monnieri. React Oxyg Spec. 2017;3:111–120. [Google Scholar]
- Wang N, et al. Ginseng polysaccharides: a potential neuroprotective agent. J Ginseng Res. 2021;45:211–217. doi: 10.1016/j.jgr.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Fry CME, Walker CL. Carboxyl-terminal modulator protein regulates Akt signaling during skeletal muscle atrophy in vitro and a mouse model of amyotrophic lateral sclerosis. Sci Rep. 2019;9:1–10. doi: 10.1038/s41598-019-40553-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MA, Massenzio F, Li X, et al. Inhibiting glycogen synthase kinase 3 suppresses TDP-43-mediated neurotoxicity in a caspase-dependant manner. Biorxiv. 2021;27:472. [Google Scholar]
- Wu J, Lin L, Chau F. Ultrasound-assisted extraction of Ginseng saponins from ginseng roots and cultured ginseng cells. Ultron Sonochem. 2001;8:347–352. doi: 10.1016/S1350-4177(01)00066-9. [DOI] [PubMed] [Google Scholar]
- Xie Z, Bailey A, Kuleshov MV, et al. Gene set knowledge discovery with Enrichr. Curr Protoc. 2021;1:e90. doi: 10.1002/cpz1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YM, Gupta SK, Kim KJ, et al. A small molecule screen in stem-cell-derived motor neurons identifies a kinase inhibitor as a candidate therapeutic for ALS. Cell Stem Cell. 2013;12:713–726. doi: 10.1016/j.stem.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Soufan O, Ewald J, et al. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47:W234–W241. doi: 10.1093/nar/gkz240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveros JC (2007) VENNY. An interactive tool for comparing lists with Venn diagrams
- Wunderlich HA (2019) ALS-associated mutations in the FUS nuclear localization signal in mice alter the cytosolic protein and RNA interactome of FUS.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.