Abstract
In this article we investigate group differences in phthalate exposure profiles using NHANES data. Phthalates are a family of industrial chemicals used in plastics and as solvents. There is increasing evidence of adverse health effects of exposure to phthalates on reproduction and neurodevelopment and concern about racial disparities in exposure. We would like to identify a single set of low-dimensional factors summarizing exposure to different chemicals, while allowing differences across groups. Improving on current multigroup additive factor models, we propose a class of Perturbed Factor Analysis (PFA) models that assume a common factor structure after perturbing the data via multiplication by a group-specific matrix. Bayesian inference algorithms are defined using a matrix normal hierarchical model for the perturbation matrices. The resulting model is just as flexible as current approaches in allowing arbitrarily large differences across groups but has substantial advantages that we illustrate in simulation studies. Applying PFA to NHANES data, we learn common factors summarizing exposures to phthalates, while showing clear differences across groups.
Keywords: Bayesian, chemical mixtures, factor analysis, hierarchical model, metaanalysis, perturbation matrix, phthalate exposures, racial disparities
1. Introduction.
Exposures to phthalates are ubiquitous. They are present in soft plastics, including vinyl floors, toys and food packaging. Medical supplies such as blood bags and tubes contain phthalates. They are also found in fragrant products such as soap, shampoo, lotion, perfume and scented cosmetics. There is substantial interest in studying levels of exposure of people in different groups to phthalates and in relating these exposures to health effects. This has motivated the collection of phthalate concentration data in urine in the National Health and Nutrition Examination Survey (NHANES) (https://wwwn.cdc.gov/nchs/nhanes). Excess levels of phthalates in blood/urine have been linked to a variety of health outcomes, including obesity (Kim and Park (2014), Zhang et al. (2014), Benjamin et al. (2017)) and birth outcomes (Bloom et al. (2019)). When they enter the body, phthalate parent compounds are broken down into different metabolites; assays measuring phthalate exposures target these different breakdown products. As these chemicals are often moderately to highly correlated, it is common to identify a small set of underlying factors (e.g., Weissenburger-Moser et al. (2017)). Epidemiologists are interested in interpreting these factors and in using them in analyses relating exposures to health outcomes.
However, many research groups have noticed a tendency to estimate very different factors in applying factor analysis to similar datasets or groups of individuals. For example, Maresca et al. (2016) examined data from three children’s cohorts and noted marked differences in factor structure in one of them. James-Todd et al. (2017) found variation in phthalate exposure patterns of pregnant women by race. Bloom et al. (2019) noted differences in both phthalate exposures and in associations with the birth outcomes by race. Certainly, we would like to allow for possible differences in exposures across groups; indeed, studying such differences is one of our primary interests. Such differences may relate to questions of environmental justice and may partly explain differences with ethnicity in certain health outcomes. However, even though the levels and specific sources of phthalate exposures may vary across groups, there is no reason to suspect that the fundamental relationship between levels of metabolites and latent factors would differ. Such differences in the factor structure are more likely to arise due to statistical uncertainty and sensitivity to slight differences in the data and can greatly complicate inferences on similarities and differences across groups.
To set the stage for discussing factor modeling of multigroup data, consider a typical factor model for p-variate data Yi = (Yi1, …, Yip)T from a single group,
| (1.1) |
where it is assumed that data are centered prior to analysis, ηi = (ηi1, …, ηik)T are latent factors, Λ is a p × k factor loadings matrix and is a diagonal matrix of residual variances. Under this model, marginalizing out the latent factors ηi induces the covariance H = cov(Yi) ΛΛT + Σ.
To extend (1.1) to data from multiple groups, most of the focus has been on decomposing the covariance into shared and group-specific components. A key contribution was Joint and Individual Variation Explained (JIVE) which identifies a low-rank covariance structure that is shared among multiple data types collected for the same subject (Feng, Hannig and Marron (2015), Lock et al. (2013), Feng et al. (2018)). In a Bayesian framework, Roy, Schaich-Borg and Dunson (2019) proposed Time Aligned Common and Individual Factor Analysis (TACIFA) for a related problem. More directly relevant to our phthalate application are the multistudy factor analysis (MSFA) methods of De Vito et al. (2018, 2019). These approaches replace Ληi in (1.1) with an additive expansion containing shared and group-specific components. De Vito et al. (2018) implement a Bayesian version of MSFA (BMSFA), while De Vito et al. (2019) develop a frequentist implementation. Kim et al. (2018) propose a related approach using PCA.
The above methods are very useful in many applications but do not address our goal of improving inferences in our phthalate application by obtaining a common factor representation that holds across groups. In addition, we find that additive expansions can face issues with weak identifiability; the model can fit the data well by decreasing the contribution of the shared component and increasing that of the group-specific components. This issue can lead to slower convergence and mixing rates for sampling algorithms for implementing BMSFA and potentially higher errors in estimating the component factor loadings matrices.
We aim to identify a single set of phthalate exposure factors under the assumption that the data in different groups can be aligned to a common latent space via multiplication by a perturbation matrix. We represent the perturbed covariance in group j as , where Σ is the common covariance and Qj is the group-specific perturbation matrix. As in the common factor model in (1.1), the overall covariance Σ can be decomposed into a component characterizing shared structure and a residual variance. The utility of the perturbation model also extends beyond multigroup settings. In the common factor model the error terms only account for additive measurement error. We can obtain robust estimates of factor loadings from data in a single group by allowing for observation-specific perturbations. This accounts for both multiplicative and additive measurement error. In this case we define separate Qi’s for each data vector Yi. Here, Qi ’s are multiplicative random effects with mean Ip. Thus, E(QiYi) = E(Yi) and the covariance structure on Qi would determine the variability of QiYi.
We take a Bayesian approach to inference using Markov chain Monte Carlo (MCMC) for posterior sampling, related to De Vito et al. (2018) but using our Perturbed Factor Analysis (PFA) approach instead of their additive BMSFA model. Model (1.1) faces well-known issues with nonidentifiability of the loadings matrix Λ (Früehwirth-Schnatter and Lopes (2018), Lopes and West (2004), Ročková and George (2016), Seber (2009)); this nonidentifiability problem is inherited by multiple group extensions such as BMSFA. It is very common in the literature to run MCMC ignoring the identifiability problem and then postprocess the samples. Aßmann, Boysen-Hogrefe and Pape (2016), McParland et al. (2014) obtain a postprocessed estimate by solving an orthogonal Procrustes problem but without uncertainty quantification (UQ). Roy, Schaich-Borg and Dunson (2019) postprocess the entire MCMC chain iteratively to draw inference with UQ. Lee, Lin and McLachlan (2018) address nonidentifiability by giving the latent factors nonsymmetric distributions, such as half-t or generalized inverse Gaussian. We instead choose heteroscedastic latent factors in the loadings matrix and postprocess based on Roy, Schaich-Borg and Dunson (2019). We find this approach can also improve accuracy in estimating the covariance, perhaps due to the flexible shrinkage prior that is induced.
The next section describes the data and the model in detail. In Section 3 prior specifications are discussed. Our computational scheme is outlined in Section 4. We study the performance of PFA in different simulation setups in Section 5. Section 6 applies PFA to NHANES data to infer a common set of factors summarizing phthalate exposures, while assessing differences in exposure profiles between ethic groups. We discuss extensions of our NHANES analysis in Section 7.
2. Data description and modeling.
NHANES is a population representative study collecting detailed individual-level data on chemical exposures, demographic factors and health outcomes. Our interest is in using NHANES to study disparities across ethnic groups in exposures. Data are available on chemical levels in urine recorded from 2009 to 2013 for 2749 individuals. We consider eight phthalate metabolites—Mono-(2-ethyl)-hexyl (MEHP), Mono-(2-ethyl-5-oxohexyl) (MEOHP), Mono-(2-ethyl-5-hydroxyhexyl) (MEHHP), Mono-2-ethyl-5-carboxypentyl (MECPP), Mono-benzyl (MBeP), Mono-ethyl (MEP),Mono-isobutyl (MiBP) and Mono-n-butyl (MnBP)—and measured in participants identifying with racial and ethnic groups: Mexican American (Mex), Other Hispanic (OH), Non-Hispanic White (N-H White), Non-Hispanic Black (N-H Black) and Other Race (Other) which includes multiracial. Previous work has shown differences across ethnic groups in patterns of use of products that contain phthalates (Taylor et al. (2018)) and in measured phthalate concentrations (James-Todd et al. (2017)). Recent work has also indicated exposure effects themselves may vary across ethnic groups (Bloom et al. (2019)), but this may be difficult to disentangle if summaries of exposure lack sufficient robustness and generalizability.
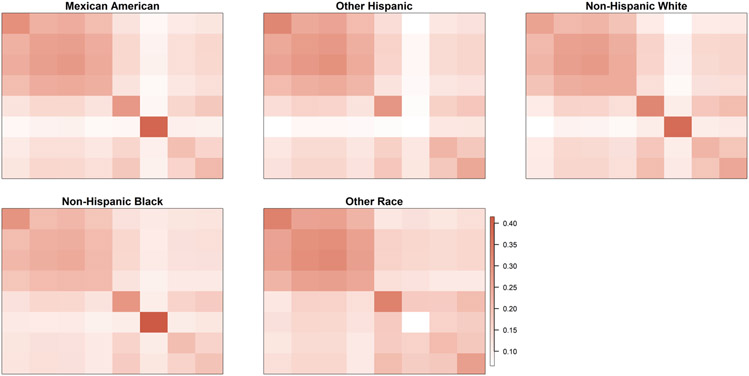
We summarize the average level of each chemical in each group in Table 1. In Table 2 we compute the Hotelling T2 statistic between each pair of groups. As noted in Bloom et al. (2019), exposure levels were generally higher among non-whites. For each phthalate variable we also fit a one-way ANOVA model to assess differences across the groups taking Mexican–Americans as the baseline. The results are included in Tables 2-6 in the Supplementary Materials (Roy et al. (2021)). For the majority of phthalates, concentrations are lowest among non-Hispanic whites. We plot group-specific covariances in Figure 1. It is evident that there is shared structure with some differences across the groups.
Table 1.
NHANES data comparison across ethnic groups in terms of number of participants and average levels of different phthalate chemicals, with standard deviations in brackets
| Mex | OH | N-H White | N-H Black | Other/multi | |
|---|---|---|---|---|---|
| Number of participants | 566 | 293 | 1206 | 516 | 168 |
| MnBP | 3.85 (1.96) | 4.31 (4.96) | 3.56 (2.17) | 4.21 (1.81) | 3.76 (2.11) |
| MiBP | 2.88 (1.36) | 3.21 (1.70) | 2.52 (1.20) | 3.47 (1.67) | 2.79 (1.21) |
| MEP | 8.32 (6.64) | 9.51 (6.99) | 6.91 (5.43) | 11.16 (9.38) | 7.33 (7.57) |
| MBeP | 2.79 (1.58) | 2.78 (1.61) | 2.70 (1.67) | 3.06 (1.77) | 2.56 (1.68) |
| MECPP | 4.80 (3.68) | 4.55 (2.68) | 4.16 (2.40) | 4.36 (2.38) | 4.34 (2.64) |
| MEHHP | 3.83 (3.01) | 3.73 (2.59) | 3.45 (2.25) | 3.79 (2.26) | 3.57 (2.45) |
| MEOHP | 3.11 (2.41) | 3.00 (1.92) | 2.79 (1.69) | 3.05 (1.71) | 2.86 (1.86) |
| MEHP | 1.58 (1.23) | 1.59 (1.29) | 1.33 (0.89) | 1.61 (0.99) | 1.58 (1.31) |
Table 2.
Hotelling T2 statistic between phthalate levels for each pair of groups in NHANES
| Mex | OH | N-H White | N-H Black | Other/multi | |
|---|---|---|---|---|---|
| Mex | 0.00 | 31.84 | 108.59 | 191.93 | 28.04 |
| OH | 31.84 | 0.00 | 156.45 | 63.48 | 28.69 |
| N-H White | 108.59 | 156.45 | 0.00 | 409.64 | 55.51 |
| N-H Black | 191.93 | 63.48 | 409.64 | 0.00 | 101.90 |
| Other/multi | 28.04 | 28.69 | 55.51 | 101.90 | 0.00 |
Fig. 1.
Covariance matrices of the phthalate chemicals across different ethnic groups for NHANES data.
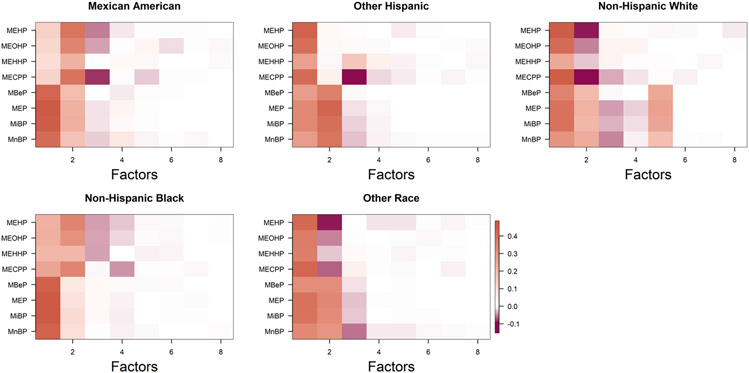
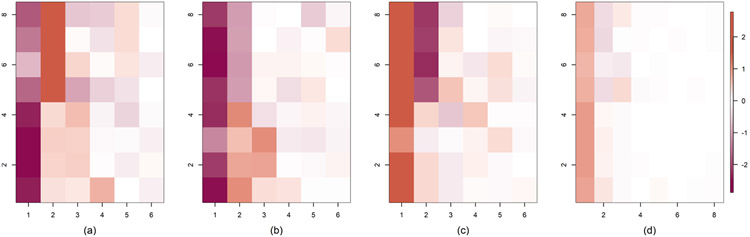
Fitting the common factor model (1.1) separately for each group, we obtain noticeably different loadings structures, as depicted in Figure 2. Throughout the article, including for Figures 1 and 2, we plot matrices using the mat2cols() function for R package IMIFA (Murphy, Viroli and Gormley (2020a, 2020b)). This function standardizes the color scale across the different matrices. We choose color palettes coral3 and deeppink4 from R package RColorBrewer (Neuwirth (2014)) for positive and negative entries, respectively. Zero entries are white, and the color gradually changes as the value moves away from zero. In the next subsection we propose a novel approach for multigroup factor analysis to identify common phthalate factors.
Fig. 2.
Estimated loadings matrices for different ethnic groups in NHANES based on applying separate Bayesian factor analyses using the approach of Bhattacharya and Dunson (2011).
2.1. Multigroup model.
We have data from multiple groups, with the groups corresponding to individuals in different racial and ethnic categories in NHANES. Each p-dimensional mean-centered response Yij, belonging to group Gj, for j ∈ 1 : J and i ∈ 1 : nj, is modeled as
| (2.1) |
The perturbation matrices Qj are of dimension p × p, and follow a matrix normal distribution, with isotropic covariance U = V αIp. The latent factors ηij are heteroscedastic, so that E is diagonal with nonidentical entries such that E = diag(e1, …, ek) with k factors in the model and Σ = diag(σ1, …, σp). We discuss advantages of choosing heteroscedastic latent factors in more detail in Section 2.3. After integrating out the latent factors, observations are marginally distributed as . If we write , then Λ is the shared loadings matrix, and ΨjΛ is a group-specific loadings matrix. We can quantify the magnitude of perturbation as ∥Ψj∥F, where ∥ · ∥F stands for the Frobenius norm. For identifiability of Qj ’s, we consider Q1 = Ip and n1 ≥ 2. We call our model Perturbed Factor Analysis (PFA).
2.1.1. Model properties.
Let Ω1 and Ω2 be two positive definite (p.d.) matrices. Then, there exist nonsingular matrices A and B, where Ω1 = AAT and Ω2 = BBT. By choosing E = AB−1, we have Ω1 = EΩ2ET. If the two matrices Ω1 and Ω2 are close, then E will be close to the identity. However, E is not required to be symmetric. In our multigroup model in (2.1), the Qjs allow for small perturbations around the shared covariance matrix H = ΛEΛT + Σ. We define the following class for the Qj ’s:
The index ϵ controls the amount of deviation across groups. In (2.1) we define U = V = αIp, for some small α, which we call the perturbation parameter. By choosing U = V to be isotropic covariances, we impose uniform perturbation across all the rows and columns around Ip. However, the perturbation matrices themselves are not required to be symmetric.
Lemma 1. P(Qi ∉ Cϵ) ≤ exp(−ϵ2/2α2).
The proof follows from Chebychev’s inequality. This result allows us to summarize the level of induced perturbation for any given α. Using this lemma, we can show the following result:
Therefore, the Kullback–Leibler divergence between the marginal distributions of any two groups j and l can be bounded by up to some constant. We define a divergence statistic between groups j and l as . This generates a divergence matrix D = ({djl}), where larger djl implies a greater difference between the two groups.
As in other factor modeling settings, it is important to carefully choose the number of factors. This is simpler for PFA than for other multigroup factor analysis models, because we only need to select a single number of factors instead of separate values for shared and individual-specific components. Indeed, we can directly apply methods developed in the single group setting (Bhattacharya and Dunson (2011)). Due to heteroscedastic latent factors, we can directly use the posterior samples of the loading matrices without applying factor rotation. Computationally, PFA tends to be faster than multigroup factor models such as BMSFA due to a smaller number of parameters and better identifiability, leading to better mixing of Markov chain Monte Carlo (MCMC).
2.1.2. Choosing the parameter α for multigroup data.
The parameter α controls the level of perturbation across groups. We use a cross-validation technique to choose α based on 10 randomly chosen 50–50 splits. We randomly divide each group 50/50 into training and test sets 10 times. Then, for a range of α values we fit the model on the training data and calculate the average predictive log-likelihood of the test set for all the 10 random splits. After integrating out the latent factors, the predictive distribution is QjYij ∼ N(0, ΛEΛT Σ). If there are multiple values of α with similar predictive log-likelihoods, then the smallest α is chosen.
Alternatively, we can take a fully Bayesian approach and put a prior on α. We call this method Fully Bayesian Perturbed Factor Analysis (FBPFA). We see that FBPFA performs similarly to PFA in practice but involves a slightly more complex MCMC implementation. PFA avoids sensitivity to the prior for α but requires the computational overhead of cross-validation, ignores uncertainty in estimating α and can, potentially, be less efficient in requiring a holdout sample.
2.2. Measurement-error model.
We can modify the multigroup model to obtain improved factor estimates in single group analyses by considering observation-level perturbations. Here, we observe Yij ’s for j = 1, …, mi, which are mi, many proxies of “true” observation Wi with multiplicative measurement errors such that Wi = QijYij. The modified model is
| (2.2) |
In this case the Qij ’s apply a multiplicative perturbation to each data vector. We have where is a matrix. Thus, here the measurement errors are and E(Uij∣Wi) = 0. This model is different from the multiplicative measurement error model of Sarkar et al. (2018). In their paper, observations Yij ’s are modeled as Yij = Wi ◦ Uij, where ◦ denotes the elementwise dot product and Uij ’s are independent of Wi with E(Uij) = 1. Thus, the measurement error in the lth component (i.e., Uijl) is dependent on Wi primarily through Wil. However, in our construction the measurement errors are a linear function of the entire Wi.
This is a much more general setup than Sarkar et al. (2018). With this generality comes issues in identifying parameters in the distributions of Qij and Yij. For simplicity, we again assume U = V = αIp. In this case we have
Thus, only the diagonal elements of H are not identifiable, and the perturbation parameter α does not influence the dependence structure among the variables. Hence, with our heteroscedastic latent factors we can still recover the loading structure. To tune α, we can use the marginal distributions QijYij ∼ N(0, α2sIp + H) to develop a cross validation technique when mi > 1 for all i = 1, …, n, as in Section 2.1.2. We split the data into training and testing sets. Then, fit the model in the training set, and find the α minimizing the predictive log-likelihood in the test set. We can alternatively estimate α as in FBPFA by using a weakly informative prior concentrated on small values, while assessing sensitivity to hyperparameter choice.
2.3. The special case Qj = Ip for all j.
For data within a single group without measurement errors (in the sense considered in the previous subsection), we can modify PFA by taking Qj = Ip for all j. Then, the model reduces to a traditional factor model with heteroscedastic latent factors,
| (2.3) |
where E is assumed to be diagonal with nonidentical entries. Integrating out the latent factors, the marginal likelihood is Yi ∼ N(0, ΛEΛT + Σ). Except for the diagonal matrix E, the marginal likelihood is similar to the marginal likelihood for a traditional factor model. As E has nonidentical diagonal entries, the likelihood is no longer invariant under arbitrary rotations. For the factor model in (1.1), (Λ, η) and (ΛR, RT η) have equivalent likelihoods for any orthonormal matrix R. This is not the case in our model unless R is a permutation matrix. Thus, this simple modification over the traditional factor model helps to efficiently recover the loading structure. This is demonstrated in Case 1 of Section 5. We also show that the posterior is weakly consistent in the Supplementary Materials (Roy et al. (2021)).
3. Prior specification.
As in Bhattacharya and Dunson (2011), we put the following prior on Λ to allow for automatic selection of rank and easy posterior computation:
The parameters ϕlk control local shrinkage of the elements in Λ, whereas τk controls column shrinkage of the kth column. We follow the guidelines of Durante (2017) to choose hyperparameters that ensure greater shrinkage for higher indexed columns. In particular, we let κ1 = 2.1 and κ2 = 3.1 which works well in all of our simulation experiments.
Suppose we initially choose the number of factors to be too large. The above prior will then tend to induce posteriors for in the later columns that are concentrated near zero, leading to λlk ≈ 0 in those columns. The corresponding factors are then effectively deleted. In practice, one can either leave the extra factors in the model, as they will have essentially no impact, or conduct a factor selection procedure by removing factors having all their loadings within °ζ of zero. Both approaches tend to have similar performance in terms of posterior summaries of interest. We follow the second strategy, motivated by our goal of obtaining a small number of interpretable factors summarizing phthalate exposures. In particular, we apply the adaptive MCMC procedure of Bhattacharya and Dunson (2011) with ζ = 1 × 10−3.
For the heteroscedastic latent factors, each diagonal element of E has an independent prior,
for some constant u. In our simulations we see that u has minimal influence on the predictive performance of PFA. However, as u increases, more shrinkage is placed on the latent factors. We choose u = 10 for most of our simulations. For the residual error variance Σ, we place a weakly informative prior on the diagonal elements,
In our simulations a weakly informative IG(0.1,0.1) prior on α works well in terms of both predictive performance and estimation of the loading structure, including in single group analyses.
4. Computation.
Because all of the parameters have conjugate full conditional distributions, posterior inference is straightforward with a Gibbs sampler. For the model in (2.1), the full conditional of the perturbation matrix Qj is
where Гj = [(V ⊗ U)−1 + Sj ⊗ H−1]−1 and . The notation ⊗ stands for Kronecker’s product. The full conditionals for all other parameters are described in Bhattacharya and Dunson (2011), replacing Yij by QjYij. For the model in (2.2), the full conditional of Qij is
where Гij = (V ⊗ U)−1 + Sij ⊗ H−1]−1 and . Other parameters can again be updated using the results in Bhattacharya and Dunson (2011), replacing Yij by QijYij. To sample the entire vec(Qij) or vec(Qj) together, we need to invert a p2 × p2 matrix at each step. Instead, we iteratively update the columns of Qij or Qj which does not require matrix inversion when U = V = αIp. For simplicity in notation, we only show the update for the columns in Qj of the model in (2.1). The full conditional of the lth column in Qj is
where and Ml = −∑k∈Gj (Qj,−lY−l,k − Ληk)Yl,k/Σ + gl/α with Σ the diagonal error covariance matrix. Here, Qj,−l denotes the p × (p − 1) dimensional matrix removing the lth column from Qj. Similarly, Y−l,k denotes the response of the kth individual from group Gj, removing the lth variable and gl the p-dimensional vector with one in the lth entry. If we put a prior on α, the posterior distribution of α is given by for the model in (2.1) and for the model in (2.2); the posterior distribution of α is . The posterior distribution of is , where Qj(l,) denotes the lth row of Qj. The posterior distribution of is .
5. Simulation study.
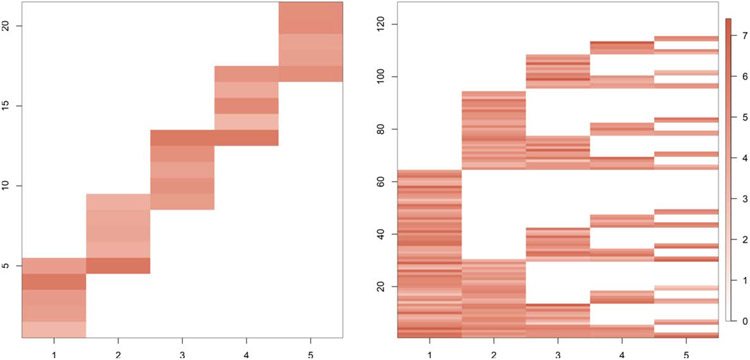
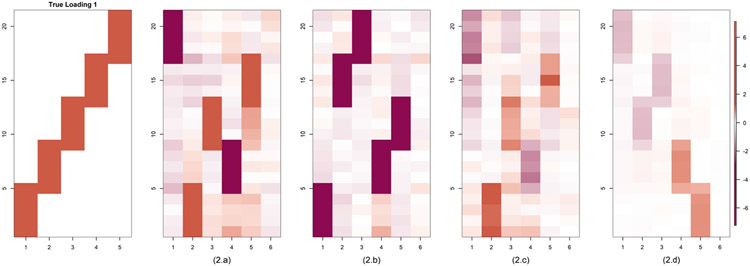
In this section we study the performance of PFA in various simulation settings. As ground truth we consider the two loading matrices given in Figure 3. These are similar to ones considered in Ročková and George (2016). R code to generate the data is provided in the github page https://github.com/royarkaprava/Perturbed-factor-model. The two matrices both have five columns, the first has 21 rows and the second has 128 rows. We compare the estimated loading matrices for different choices of the perturbation parameter α and the shape parameter u, controlling the level of shrinkage on the diagonal entries of E.
Fig. 3.
Simulation study true loading matrices of dimension 25 (Loading 1) and 128 × 5 (Loading 2), respectively.
For the single group case we compare with the method of Bhattacharya and Dunson (2011) (B&D) which corresponds to the special case of our approach that fixes the perturbation matrices and latent factor covariances equal to the identity. A point estimate of the loading matrix for B&D is calculated by postprocessing the posterior samples. We use the algorithm of Aßmann, Boysen-Hogrefe and Pape (2016) to rotationally align the samples of Λ, as in De Vito et al. (2018). In contrast, our method uses the postprocessing algorithm of Roy, Schaich-Borg and Dunson (2019). For the multigroup case we compare our estimates with Bayesian Multi-Study Factor Analysis (BMSFA). We use the MSFA package at https://github.com/rdevito/MSFA.
We compare the different methods using predictive log-likelihood. Simulated data are randomly divided 50/50 into training and test sets. After fitting each model on the training data, we calculate the predictive log-likelihood of the test set. All methods are run for 7000 iterations of the Gibbs sampler, with 2000 burn-in samples. Each simulation is replicated 30 times.
5.1. Case 1: Single group, Qj = Ip.
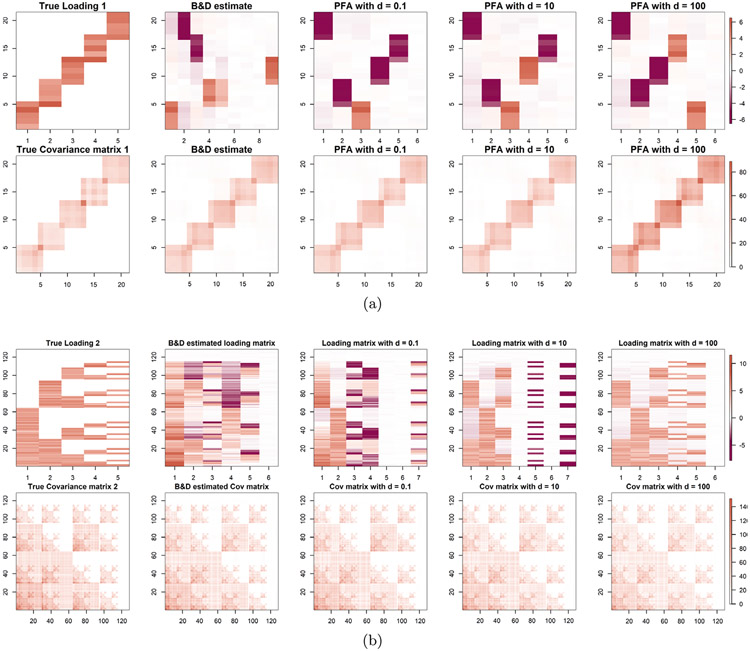
We first consider single group factor analysis. Starting with the two loading matrices in Figure 3, we simulate latent factors from N(0, I5) and generate datasets of 500 observations, with residual variance Σ = I5. We compare B&D with our method when Qj = I5, so the only adjustment is to use heteroscedastic latent factors. Note that the simulated latent factors actually have identical variances.
In Tables 3 and 4, we compare methods by MSE of the estimated vs. true covariance matrix. For the loading matrix 1, B&D had an MSE of 2.59 which is dominated by our method across a range of values of u.
Table 3.
MSE of estimated covariance matrix for loading matrix 1, across different values of u
| u | MSE |
|---|---|
| 0.1 | 1.04 |
| 10 | 1.32 |
| 100 | 1.38 |
Table 4.
MSE of estimated covariance matrix for loading matrix 2, across different values of u
| u | MSE |
|---|---|
| 0.1 | 5.21 |
| 10 | 9.45 |
| 100 | 10.32 |
For the loading matrix 2, B&D had an MSE of 10.81. Again, our method beats this across a range of values of u. The B&D model is a special case of PFA with E = Ip in (2.3). We conjecture that the gains seen for PFA are due to the more flexible induced shrinkage structure on the covariance matrix.
We compare the estimated and true loading matrices in Figure 4. Throughout the paper, PFA estimated loadings are based on , where and are the estimated loading and covariance matrix of the factors, respectively. This makes the loadings comparable to methods that use identity covariance for the latent factors. PFA performs overwhelmingly better at estimating the true loading structure compared with B&D FA. The first five columns of the estimated loadings based on PFA are very close to the true loading structure under some permutation. This gain over B&D may be due to a combination of the more flexible shrinkage structure and the different postprocessing scheme. On comparing for different choices of u, we see that the estimates are not sensitive to the hyperparameter u. Estimation MSE is better for smaller u, but the general structure of the loadings matrix is recovered accurately for all u.
Fig. 4.
Comparison of true and estimated loading matrices and covariance matrices for Case 1. Comparison of B&D FA with our method for different choices of u. Results for loading matrix 1 are in (a) and two in (b). For all the cases, we fix Qj = I .The color gradient added in the last image of each row holds for all the images in that row.
5.2. Case 2: Multigroup, multiplicative perturbation.
In this case we simulate data from the multigroup model in (2.1) for i = 1, …, 500. Observations Yi are first generated using the same method as in Case 1. The data are then split into 10 groups of 50 observations such that Gj = {Yk : 50(j – 1) ≤ k ≤ 50j} for j = 1, …, 10. The groups are perturbed using matrices Qj0 ~ MN(Ip, α0Ip, α0Ip) for different choices of α0, setting Q1 = Ip.
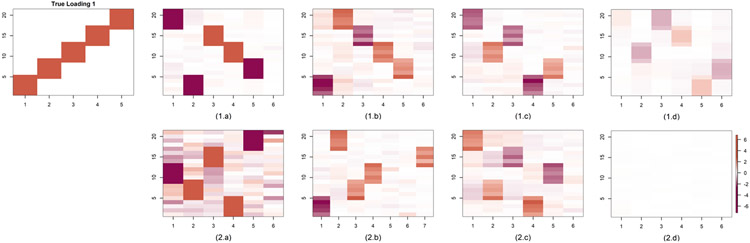
Figure 5 shows the estimated loading matrices. We obtain accurate estimates, even when assuming a higher level of perturbation than the truth, α0 ≤ α. However, the estimates are best when α = α0 = 10−4. Performance degrades more sharply when we underestimate the level of perturbation, as in the case where α = 10−4, α0 = 10−2. The BMSFA code requires an upper bound on the number of shared and group-specific factors; we choose both to be 6. The estimated loadings from PFA and FBPFA are better than BMSFA. Except for cases with a higher level of perturbation using BMSFA, the estimated loadings matrix is close to the truth under some permutation of the columns. BMSFA estimated loadings usually are of lower magnitude which may be due to the additive structure of the model. Thus they look faded in almost all the figures. The cumulative shrinkage prior used in PFA and BMSFA induces continuous shrinkage instead of exact sparsity but, nonetheless, does a good job overall of capturing the true sparsity pattern. BMSFA estimated loadings for α0 = 10−2 look blank as the estimated loading has all entries near zero. Results for the Case 2 loading structure are shown in Figure 5 of the Supplementary Material (Roy et al. (2021)).
Fig. 5.
Comparison of estimated loading matrix 1 in simulation Case 2 with different choices of α0 and α where Qj0 ∼ MN(Ip, α0Ip, α0Ip) and U = αIp = V. (a) α = 10−4, α0 = 10−4, (b) α = 10−2, α0 = 10−4, (c) FBPFA with α0 = 10−4, (d) BMSFA with α0 = 10−4, (e) α = 10−4, α0 = 10−2, (f) α = 10−2, α0 = 10−2, (g) FBPFA with α0 = 10−2, (h) for BMSFA with α0 = 10−2. True loading matrices are plotted twice in columns 1 for easier comparison with other images.
We also apply the same simulation setting with higher error variances. Figure 6 compares the estimated loadings. All the methods identify the true structure when the residual standard deviation is 5, but only PFA with correctly specified α produces good estimates when we increase it to 10.
Fig. 6.
Comparison of estimated loading matrices in simulation Case 2 with increasing error variances and α0 = 10−4: (a) α = 10−4 with error variance 25, (b) α = 10−4 with error variance 100, (c) BMSFA with error variance 25, (d) BMSFA with error variance 100, (e) FBPFA with error variance 25, (f) FBPFA with error variance 100.
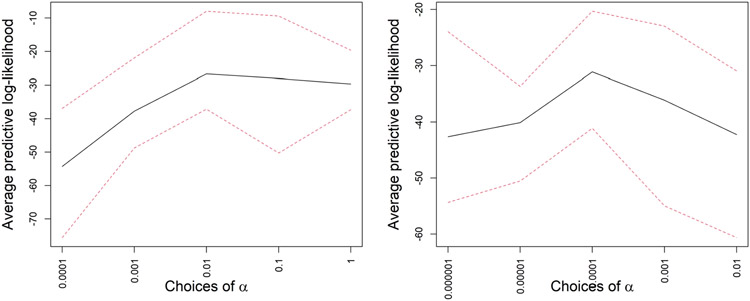
In Figure 7 we show the predictive log-likelihood averaged over all the splits under a range of α values. For each choice of α, we fit the model to a training set and calculate the predictive log-likelihood on a test set for 10 randomly chosen training-test splits. This demonstrates the utility of our cross-validation technique in finding the optimal α. In Table 5 we compare the performance of PFA with optimal α, FBPFA and BMSFA in terms of predictive likelihood. PFA and FBPFA have overwhelmingly better performance. We also show the range of predictive log-likelihoods for each choice of α in Figure 7 using dotted red lines. FBPFA is better at recovering the true loading structure for p = 21, but for the higher dimensional case with p = 128 PFA tends to be better.
Fig. 7.
Average predictive log-likelihoods in black along with two dotted red lines, denoting the range of predictive log-likelihoods across all the splits for different choices of α in simulation Case 2 for two cases—in the first case the perturbation matrices are generated using true α0 = 10−2, and in the second case α0 = 10−4. This is based on the simulation experiment of Figure 5.
Table 5.
Average predictive log-likelihood for PFA with optimal α, FBPFA and BMSFA in simulation Case 2
| True loading | α 0 | PFA for optimal α | FBPFA | BMSFA |
|---|---|---|---|---|
| Loading 1 | 10−4 | −31.65 | −25.92 | −679.36 |
| 10−2 | −30.32 | −26.01 | −921.76 | |
| Loading 2 | 10−4 | −210.43 | −251.13 | −6871.38 |
| 10−2 | −351.42 | −304.10 | −25,018.50 |
5.2.1. Case 2.1: Partially shared factors.
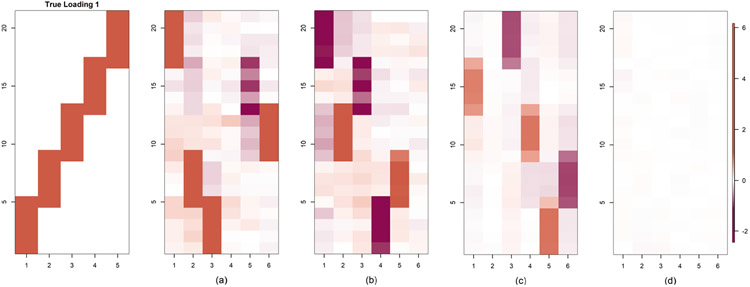
We also repeat the Case 2 simulation but modified to accommodate a partially shared structure. In particular, we generated the data as in Case 2, but for the last two groups we let for i ∈ Gj and j = 9, 10. Here, Λ01 = Λ0[, 1 : 3 is a p × 3 dimensional matrix having the first three columns from Λ0. The choices for Λ0 are given in Figure 3.
The estimated loading matrices are compared in Figure 8 for two levels of perturbations, α0 = 10−4, 10−2. FBPFA works well for any level of perturbation, while BMSFA works well for only the lower perturbation level. The estimated loading in Figure 8(d) looks blank due to near zero entries. Estimated loadings for the case 2 loading structure are in Figure 6 of the Supplementary Material (Roy et al. (2021)).
Fig. 8.
Comparison of estimated loading in the partially shared modification of simulation Case 2. Row 1 corresponds to true loading structure 1 and row 2 to true loading structure 2. (a) FBPFA with α0 = 10−4, (b) FBPFA with α0 = 10−2, (c) BMSFA with α0 = 10−4, (d) BMSFA with α0 = 10−2.
5.3. Case 3: Multigroup factor model.
In this case we generate data from the Bayesian multistudy factor analysis (BMSFA) model, as in De Vito et al. (2018),
| (5.1) |
where the group-specific loadings (Ψj ’s) are lower in magnitude in comparison to the shared loading matrix Λ. We generate the Ψj s from N(−0.5, 0.8). These Ψj matrices are the same dimension as the shared loading matrix Λ.
Figure 9 compares the estimated loadings with the true shared loadings across different methods—PFA for two different perturbation parameters, FBPFA and BMSFA. Although we generate the data from BMFSA, all the estimates are very much comparable in Figure 9. However, PFA and FBPFA again outperform BMSFA in terms of predictive log-likelihoods, as shown in Table 6. In the Supplementary Material (Roy et al. (2021)) we present another simulation setting akin to BMSFA. There, the data generating process is similar to BMSFA with minor modifications in the shared and group-specific loading structures. Estimated loadings for the Case 2 loading structure are in Figure 7 of the Supplementary Material (Roy et al. (2021)).
Fig. 9.
Comparison of estimated loading matrices in simulation Case 3 when the true data generating process follows the model in (5.1). (a) PFA with α = 10−4, (b) PFA with α = 10−2, (c) FBPFA, (d) BMSFA.
Table 6.
Average predictive log-likelihood for PFA for different choices of α, FBPFA and BMSFA in Simulation Case 3
| Generative distribution of Ψj ’s |
True loading |
PFA for α = 10−2 |
PFA for α = 10−4 |
FBPFA | BMSFA |
|---|---|---|---|---|---|
| N(−0.2, 0.2) | Loading 1 | −33.41 | −44.10 | −26.16 | −500.66 |
| Loading 2 | −336.58 | −259.79 | −265.77 | −9547.77 | |
| N(−0.5, 0.8) | Loading 1 | −49.25 | −53.32 | −37.89 | −720.35 |
| Loading 2 | −584.82 | −308.72 | −1003.02 | −14,187.36 |
5.4. Case 4: Mimic NHANES data.
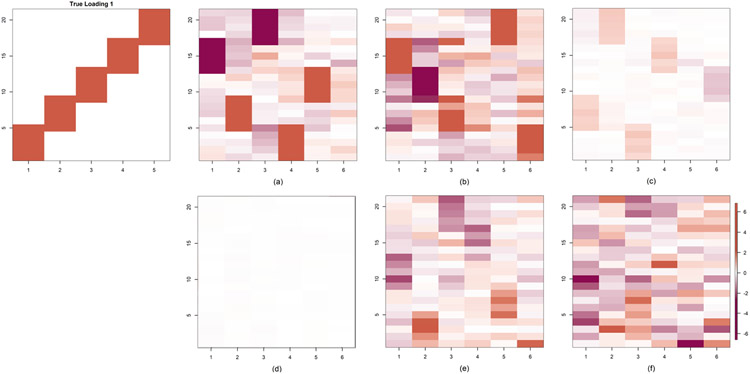
In this section we generate data from the model in (2.1). However, the model parameters Qj ’s and Λ are first estimated on the NHANES data with α = 10−2. Then, based on these estimated parameters, we generate the data following the same model. Figure 10 compares the estimated loadings across all the methods—PFA for different choices of α, FBPFA and BMSFA. We find that all the methods recover loading structures that match with our estimated loadings from Section 6. For PFA and FBPFA, we selected the number of factors, as described in Section 3, and hence these approaches produced fewer columns than BMSFA.
Fig. 10.
Comparison of estimated loading matrices in simulation Case 4 where the true loading matrix is PFA estimated loading with α = 10−2 for mean-centered NHANES data and the data are simulated from the model (2.1). (a) PFA with α = 10−2, (b) PFA with α = 10−4, (c) FBPFA, (d) BMSFA. The color gradient added in the last image holds for all the images.
6. Application to NHANES data.
We fit our model in (2.1) to the NHANES dataset, as described in Section 2, to obtain phthalate exposure factors for individuals in different ethnic groups. Before analysis the mean chemical levels across all groups are subtracted to mean-center the data. The data are then randomly split, with 2/3 in each group as a training set and 1/3 as a test set. We collect 5000 MCMC samples after a burn-in of 5000 samples. Convergence is monitored based on the predictive log-likelihood of the test set at each MCMC iteration. The hyperparameters are the same as those in Section 5. The predictive log-likelihood of the test data are used to tune α, as described in Section 2.1.2. Based on our cross-validation technique, α = 10−2 is chosen. Note that our final estimates are based on the complete data. Our BMSFA implementation sets the upper bounds at 8 for both shared and group-specific factors. Figure 4 of the Supplementary Material (Roy et al. (2021)) depicts trace plots of the loading and perturbation matrices for FBPFA.
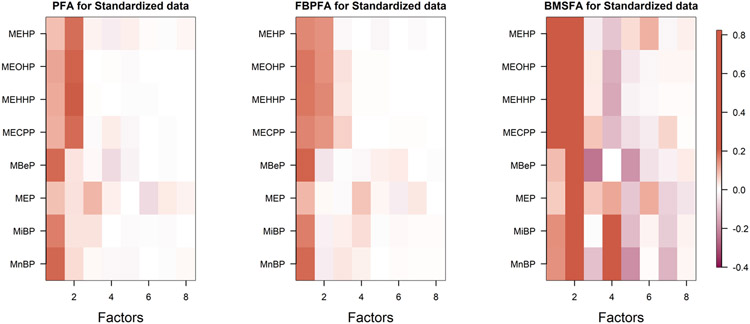
Figure 11 shows the estimated loadings using PFA, FBPFA and BMSFA. All plots show two significant factors, with some suggestions of a third factor. For PFA and FBPFA, all the chemicals load on the first factor, and the second factor is loaded on by the first four chemicals, namely MEHP, MEOHP, MEHHP and MECPP. Figure 1 also suggests that these four chemicals are related to each other more than the others. This is not surprising, as MECPP, MEHHP and MEOHP are oxidative metabolities of MEHP.
Fig. 11.
Comparison of estimated loading matrices with different methods. PFA estimates are based on α = 10−2.
Comparing the predictive log-likelihoods for PFA, FBPFA and BMSFA, we obtain values of 4.12, 5.02 and −7.99, respectively, suggesting much better performance for the PFA-based approaches. As an alternative measure of predictive performance, we also consider test sample MSE using 2/3 of the data as training and 1/3 as test. The predictive value of the test data is estimated by averaging the predictive mean conditionally on the parameters and training data over samples generated from the training data posterior. We obtain predictive MSE values of 0.05 and 0.09 for PFA with α = 10−4 and α = 10−2, respectively. FBPFA yields a value of 0.14, while BMSFA has a much larger error of 0.70. Considering that the data are normalized before analysis, the naive prediction that sets all the values to zero would yield an MSE of 1.0.
To explore similarities across groups, we calculate divergence scores and create the divergence matrix D. This matrix is provided in Table 7. To summarize D, we also provide a network plot between different groups in Figure 12. The plot uses greater edge widths between ethnic groups when the divergence statistic between the groups is small. In particular, the edge width between two nodes (j, l) is calculated as 60/djl. Thicker edges imply greater similarity. This figure implies that non-Hispanic whites differ the most in phthalate exposure profiles from the other groups, supporting the findings of our exploratory ANOVA analysis in the Section 3 of the Supplementary Material (Roy et al. (2021)) and findings in the prior literature (Bloom et al. (2019)). We find evidence of one extra factor among non-Hispanic whites relative to the other groups in Figure 2. The two Hispanic ethnicity groups are very similar in loadings as shown in Figure 2 and also their Hotelling T2 distance is relatively small in Table 2. These results support our preliminary analysis, described in Section 2.
Table 7.
Estimated divergence scores for different pairs of ethnic groups based on NHANES data
| Mex | OH | N-H White | N-H Black | Other/multi | |
|---|---|---|---|---|---|
| Mex | 0.00 | 5.86 | 41.35 | 10.22 | 3.82 |
| OH | 5.86 | 0.00 | 40.93 | 8.38 | 4.45 |
| N-H White | 41.35 | 40.93 | 0.00 | 40.06 | 41.17 |
| N-H Black | 10.22 | 8.38 | 40.06 | 0.00 | 9.48 |
| Other/multi | 3.82 | 4.45 | 41.17 | 9.48 | 0.00 |
Fig. 12.
Network plot summarizing similarity between ethnic groups based on the divergence metric with thicker edges implying smaller divergence scores. Nodes correspond to different ethnic groups.
In Table 8 we show square norm differences between the group-specific loadings, estimated using BMSFA. The (i, j)-th entry of the table is calculated as , where the matrices Ai and Aj are the estimated group-specific loadings for the groups i and j, respectively, using BMSFA. Based on these results, BMSFA-based inferences on differences in exposure profiles across ethnic groups are noticeably different from our PFA-based results reported above. In particular, all the groups seem to be approximately the same distance apart. The results using BMSFA from Table 8 are not consistent with Hotelling T2-based our exploratory data analysis results from Table 2 and PFA-based results from Table 7.
Table 8.
Square norm difference of the group-specific loadings, estimated using BMSFA
| Mex | OH | N-H White | N-H Black | Other/multi | |
|---|---|---|---|---|---|
| Mex | 0.00 | 0.75 | 0.70 | 0.59 | 0.78 |
| OH | 0.75 | 0.00 | 0.63 | 0.75 | 0.73 |
| N-H White | 0.70 | 0.63 | 0.00 | 0.68 | 0.84 |
| N-H Black | 0.59 | 0.75 | 0.68 | 0.00 | 0.91 |
| Other/multi | 0.78 | 0.73 | 0.84 | 0.91 | 0.00 |
7. Discussion.
In this paper our focus has been on identifying a common set of phthalate exposure factors that hold across different ehtnic groups, while also inferring differences in exposure profiles across groups. To accomplish this goal, we focused on data from NHANES which contain rich information on ethnicity and exposures. We found that our proposed perturbed factor analysis (PFA) approach had significant advantages over existing approaches in addressing our goals.
There are multiple important next steps to consider to expand on our analysis. A first is to include covariates. In addition to phthalates and ethnic group data, NHANES collects information that may be relevant to understanding ethnic differences in exposure profiles, including BMI, age, socioeconomic status, gender and other factors. Li and Jung (2017) consider a related problem of incorporating covariates into factor models for multiview data. Perhaps the simplest and most interpretable modification of our PFA model of the NHANES data is to allow the factor scores ηij to depend on covariates Xij = (1, Xij2, …, Xijq)T through a latent factor regression. For example, we could let ηij = βXij + ξij, with β a k × q matrix of coefficients and ξij ∼ N(0, E).
Another important direction is to extend the analysis to allow inferences on relationships between exposure profiles and health outcomes. NHANES contains rich data on a variety of outcomes that may be adversely affected by phthalate exposures. To include these outcomes in our analysis and effectively extend PFA to a supervised context, we can define separate PFA models for the exposures and outcomes, with these models having shared factors ηij but different perturbation matrices. It is straightforward to modify the Gibbs sampler used in our analyses to this case and/or the extension described above to accommodate covariates Xij; further modifications to mixed continuous and categorical variables can proceed as in Carvalho et al. (2008), Zhou et al. (2015) with the perturbations conducted on underlying variables. Relying on such a broad modeling and computational framework, it would be interesting to attempt to infer causal relationships between ethnicity and phthalates and adverse health outcomes. Mediation analysis may provide a useful framework in this respect.
Beyond this application the proposed PFA approach may prove useful in other contexts. The important features of PFA include both the incorporation of a multiplicative perturbation of the data and the use of heteroscedastic latent factors. These innovations are potentially useful beyond the multiple group setting to meta analysis, measurement error models, non-linear factor models (including Gaussian process latent variable models (Lawrence (2004), Lawrence and Candela (2006))) and variational autoencoders (Kingma and Welling (2014), Pu et al. (2016)) and even to obtaining improved performance in “vanilla” factor modeling lacking hierarchical structure.
Code is available for implementing the proposed approach and replicating our results from Section 5.2 at https://github.com/royarkaprava/Perturbed-factor-model and the Supplementary Material (Roy et al. (2021)).
Supplementary Material
Acknowledgments.
We would like to thank Roberta De Vito for sharing her source code of the Bayesian MSFA model. We would also like to thank Noirrit Kiran Chandra for his feedback on the code which heavily improved its usage both in low- and high-dimensional settings.
Funding.
This research was partially supported by grant R01-ES027498 and R01-ES028804 from the National Institute of Environmental Health Sciences (NIEHS) of the National Institutes of Health (NIH).
Footnotes
SUPPLEMENTARY MATERIAL
Contents (DOI: 10.1214/20-AOAS1435SUPPA; .pdf). There are three sections in the Supplementary Materials. In Section S-1 we establish weak consistency of the posterior distribution. Then second Section S-2 contains results from some extra simulation experiments and some exploratory analysis of the NHANES data. Section S-3 contains more supporting plots referred in Section 5.
Source code (DOI: 10.1214/20-AOAS1435SUPPB; .zip). R Codes for implementing the proposed approach and replicating our results from Section 5.2.
REFERENCES
- Assmann C, Boysen-Hogrefe J and Pape M (2016). Bayesian analysis of static and dynamic factor models: An ex-post approach towards the rotation problem. J. Econometrics 192 190–206. MR3463672 10.1016/j.jeconom.2015.10.010 [DOI] [Google Scholar]
- Benjamin S, Masai E, Kamimura N, Takahashi K, Anderson RC and Faisal PA (2017). Phthalates impact human health: Epidemiological evidences and plausible mechanism of action. J. Hazard. Mater 340 360–383. 10.1016/j.jhazmat.2017.06.036 [DOI] [PubMed] [Google Scholar]
- Bhattacharya A and Dunson DB (2011). Sparse Bayesian infinite factor models. Biometrika 98 291–306. MR2806429 10.1093/biomet/asr013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom MS, Wenzel AG, Brock JW, Kucklick JR, Wineland RJ, Cruze L, Unal ER, Yucel RM, Jiyessova A et al. (2019). Racial disparity in maternal phthalates exposure; association with racial disparity in fetal growth and birth outcomes. Environ. Int 127 473–486. [DOI] [PubMed] [Google Scholar]
- Carvalho CM, Chang J, Lucas JE, Nevins JR, Wang Q and West M (2008). High-dimensional sparse factor modeling: Applications in gene expression genomics. J. Amer. Statist. Assoc 103 1438–1456. MR2655722 10.1198/016214508000000869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vito R, Bellio R, Trippa L and Parmigiani G (2018). Bayesian multi-study factor analysis for high-throughput biological data. ArXiv preprint. Available at arXiv:1806.09896. [Google Scholar]
- De Vito R, Bellio R, Trippa L and Parmigiani G (2019). Multi-study factor analysis. Biometrics 75 337–346. MR3953734 10.1111/biom.12974 [DOI] [PubMed] [Google Scholar]
- Durante D (2017). A note on the multiplicative gamma process. Statist. Probab. Lett 122 198–204. MR3584158 10.1016/j.spl.2016.11.014 [DOI] [Google Scholar]
- Feng Q, Hannig J and Marron JS (2015). Non-iterative joint and individual variation explained. ArXiv preprint. Available at arXiv:1512.04060. [Google Scholar]
- Feng Q, Jiang M, Hannig J and Marron JS (2018). Angle-based joint and individual variation explained. J. Multivariate Anal 166 241–265. MR3799646 10.1016/j.jmva.2018.03.008 [DOI] [Google Scholar]
- FrÜehwirth-Schnatter S and Lopes HF (2018). Sparse Bayesian factor analysis when the number of factors is unknown. ArXiv preprint. Available at arXiv:1804.04231. [Google Scholar]
- James-Todd TM, Meeker JD, Huang T, Hauser R, Seely EW, Ferguson KK, Rich-Edwards JW and McElrath TF (2017). Racial and ethnic variations in phthalate metabolite concentration changes across full-term pregnancies. J. Expo. Sci. Environ. Epidemiol 27 160–166. 10.1038/jes.2016.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH and Park MJ (2014). Phthalate exposure and childhood obesity. Ann. Pediatr. Endocrinol. Metab 19 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kang D, Huo Z, Park Y and Tseng GC (2018). Meta-analytic principal component analysis in integrative omics application. Bioinformatics 34 1321–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingma DP and Welling M (2014). Auto-encoding variational Bayes. In International Conference on Learning Representations. [Google Scholar]
- Lawrence ND (2004). Gaussian process latent variable models for visualisation of high dimensional data. In Advances in Neural Information Processing Systems 16 (Thrun S, Saul LK and Schölkopf B, eds.) 329–336. MIT Press, Cambridge. [Google Scholar]
- Lawrence N and Candela JQ (2006). Local distance preservation in the GP-LVM through back constraints. In International Conference on Machine Learning’06. [Google Scholar]
- Lee SX, Lin T-I and McLachlan GJ (2018). Mixtures of factor analyzers with fundamental skew symmetric distributions. ArXiv preprint. Available at arXiv:1802.02467. [Google Scholar]
- Li G and Jung S (2017). Incorporating covariates into integrated factor analysis of multi-view data. Biometrics 73 1433–1442. MR3744555 10.1111/biom.12698 [DOI] [PubMed] [Google Scholar]
- Lock EF, Hoadley KA, Marron JS and Nobel AB (2013). Joint and individual variation explained (JIVE) for integrated analysis of multiple data types. Ann. Appl. Stat 7 523–542. MR3086429 10.1214/12-AOAS597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes HF and West M (2004). Bayesian model assessment in factor analysis. Statist. Sinica 14 41–67. MR2036762 [Google Scholar]
- Maresca MM, Hoepner LA, Hassoun A, Oberfield SE, Mooney SJ, Calafat AM, Ramirez J, Freyer G, Perera FP et al. (2016). Prenatal exposure to phthalates and childhood body size in an urban cohort. Environ. Health Perspect 124 514–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McParland D, Gormley IC, McCormick TH, Clark SJ, Kabudula CW and Collinson MA (2014). Clustering South African households based on their asset status using latent variable models. Ann. Appl. Stat 8 747–776. MR3262533 10.1214/14-AOAS726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Viroli C and Gormley IC (2020a). Infinite mixtures of infinite factor analysers. Bayesian Anal. 15 937–963. MR4132655 10.1214/19-BA1179 [DOI] [Google Scholar]
- Murphy K, Viroli C and Gormley IC (2020b). IMIFA: Infinite mixtures of infinite factor analysers and related models. R package version 2.1.3 [Google Scholar]
- Neuwirth E (2014). RColorBrewer: ColorBrewer palettes. R package version 1.1-2 [Google Scholar]
- Pu Y, Gan Z, Henao R, Yuan X, Li C, Stevens A and Carin L (2016). Variational autoencoder for deep learning of images, labels and captions. In Advances in Neural Information Processing Systems 2352–2360. [Google Scholar]
- RoČkovÁ V and George EI (2016). Fast Bayesian factor analysis via automatic rotations to sparsity. J. Amer. Statist. Assoc 111 1608–1622. MR3601721 10.1080/01621459.2015.1100620 [DOI] [Google Scholar]
- Roy A, Schaich-Borg J and Dunson DB (2019). Bayesian time-aligned factor analysis of paired multivariate time series. ArXiv preprint. Available at arXiv:1904.12103. [PMC free article] [PubMed] [Google Scholar]
- Roy A, Lavine I, Herring AH and Dunson DB (2021). Supplement to “Perturbed factor analysis: Accounting for group differences in exposure profiles.” 10.1214/20-AOAS1435SUPPA, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A, Pati D, Chakraborty A, Mallick BK and Carroll RJ (2018). Bayesian semiparametric multivariate density deconvolution. J. Amer. Statist. Assoc 113 401–416. MR3803474 10.1080/01621459.2016.1260467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seber GA (2009). Multivariate Observations 252. Wiley, New York. [Google Scholar]
- Taylor KW, Troester MA, Herring AH, Engel LS, Nichols HB, Sandler DP and Baird DD (2018). Associations between personal care product use patterns and breast cancer risk among white and black women in the sister study. Environ. Health Perspect 126 027011. 10.1289/EHP1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenburger-Moser L, Meza J, Yu F, Shiyanbola O, Romberger DJ and LeVan TD (2017). A principal factor analysis to characterize agricultural exposures among Nebraska veterans. J. Expo. Sci. Environ. Epidemiol 27 214–220. 10.1038/jes.2016.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Meng X, Chen L, Li D, Zhao L, Zhao Y, Li L and Shi H (2014). Age and sex-specific relationships between phthalate exposures and obesity in Chinese children at puberty. PLoS ONE 9 e104852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Bhattacharya A, Herring AH and Dunson DB (2015). Bayesian factorizations of big sparse tensors. J. Amer. Statist. Assoc 110 1562–1576. MR3449055 10.1080/01621459.2014.983233 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.