Abstract
Introduction
Host lipids play important roles in tuberculosis (TB) pathogenesis. Whether host lipids at TB treatment initiation (baseline) affect subsequent treatment outcomes has not been well characterised. We used unbiased lipidomics to study the prospective association of host lipids with TB treatment failure.
Methods
A case–control study (n=192), nested within a prospective cohort study, was used to investigate the association of baseline plasma lipids with TB treatment failure among adults with pulmonary TB. Cases (n=46) were defined as TB treatment failure, while controls (n=146) were those without failure. Complex lipids and inflammatory lipid mediators were measured using liquid chromatography mass spectrometry techniques. Adjusted least-square regression was used to assess differences in groups. In addition, machine learning identified lipids with highest area under the curve (AUC) to classify cases and controls.
Results
Baseline levels of 32 lipids differed between controls and those with treatment failure after false discovery rate adjustment. Treatment failure was associated with lower baseline levels of cholesteryl esters and oxylipin, and higher baseline levels of ceramides and triglycerides compared to controls. Two cholesteryl ester lipids combined in a unique classifier model provided an AUC of 0.79 (95% CI 0.65–0.93) in the test dataset for prediction of TB treatment failure.
Conclusions
We identified lipids, some with known roles in TB pathogenesis, associated with TB treatment failure. In addition, a lipid signature with prognostic accuracy for TB treatment failure was identified. These lipids could be potential targets for risk-stratification, adjunct therapy and treatment monitoring.
Shareable abstract (@ERSpublications)
In this study, unbiased lipidomics were used to identify host lipids prospectively associated with TB treatment failure. These lipids, some with known roles in TB pathogenesis, could be potential targets for adjunct therapy and treatment monitoring. https://bit.ly/3vHZ0Ec
Introduction
Our understanding of host factors that contribute to tuberculosis (TB) susceptibility, pathogenesis and treatment outcomes have benefited from studies utilising various “omics” approaches [1–10]. For example, transcriptomic studies from diverse settings have implicated type I interferon-mediated immune responses as a contributor to the increased risk of active TB disease [1–3]. Similarly, multiple studies utilising untargeted metabolomics have characterised metabolites, such as various amino acids, that differ among individuals with and without TB disease [4–10]. However, the role of the host lipidome in TB disease remains poorly understood despite the important roles lipids play in cell membrane structure, signalling pathways, metabolism and susceptibility to pathogens including TB [11].
While the lipidome of the pathogen Mycobacterium tuberculosis has been characterised in detail [12–15], there are few studies focusing on the host lipidome in the context of TB [16–18]. Studies from Africa and Mexico have shown differences in circulating concentrations of various phosphatidylcholines, lysophosphatidylcholines (LPCs), cholesterol and fatty acids among individuals with and without TB disease [16–18]. Although the coverage of lipids is limited and often inadequate for some classes of lipids in untargeted metabolomics platforms [19], information is also available from metabolomics studies confirming findings from those studies (e.g. lower levels of LPCs in TB) and identify differential levels of other lipids (specific phosphatidylcholines, sphingolipids, fatty acids and oxygenated fatty acids [4–10].
Despite multiple cross-sectional studies that have focused on characterising the lipids and metabolite differences between individuals with and without TB disease, prospective studies that identify lipids prospectively associated with TB treatment failure are lacking. We do not yet know whether certain lipids, measured prior to TB treatment initiation or during early treatment, are associated with future development of treatment failure. These data could be used to identify individuals at high risk of TB treatment failure and could also inform potential therapeutics.
In order to address this gap in knowledge, we nested a longitudinal case–control study within a prospective cohort [20] of individuals with TB to study the association of baseline host plasma lipidome with subsequent adverse TB treatment failure. The objective of the study was to gain biological insights, related to host lipids, into the aetiology of differential TB treatment outcomes and to identify potential biomarkers that can predict treatment failure.
Methods
Study design and population
We nested a case–control study within the Cohort for Tuberculosis Research by the Indo-US Medical Partnership (CTRIUMPH) study [20]. The CTRIUMPH study is a cohort study of individuals (adults and children) with active pulmonary TB and extrapulmonary TB recruited from two sites in India at Byramjee Jeejeebhoy Government Medical College (BJMC; Pune) and National Institute for Research in Tuberculosis (NIRT; Chennai). As part of the same study, the study protocol, data collection instruments and operating procedures across the two sites were standardised [20]. In the parent study conducted between August 2014 and December 2017, study participants were newly diagnosed with TB based on culture, GeneXpert (Cepheid, Sunnyvale, CA, USA), microscopy or clinical diagnosis. They were enrolled within 1 week of TB treatment initiation (i.e. baseline) and were followed during treatment and through 18 months post-treatment cessation. Those with drug-resistant TB, prior TB, on anti-TB treatment longer than a week at baseline and pregnant women were excluded from the study. All participants were on standard weight-based fixed-dose first-line regimen based on Indian TB guidelines: rifampicin, isoniazid, pyrazinamide and ethambutol for 2 months followed by rifampicin, isoniazid and ethambutol for 4 months.
In this case–control study within CTRIUMPH, we used bio-banked heparin plasma samples, collected at baseline, to study the association of host plasma lipidome with development of TB treatment failure among adults (≥18 years) with pulmonary TB and standard first-line TB treatment. Cases were all adults with active pulmonary TB who developed TB treatment failure (n=46), defined either microbiologically (i. e. detection of TB by culture at end of treatment; n=42) or through microscopy/clinical (smear-positive and/or radiology/signs and symptoms highly suggestive of TB; n=4). Controls (n=146) were a random selection of adults with pulmonary TB who were “cured” of active TB, defined as the absence of microbiological (n=104) or clinical (n=42) evidence of TB disease at treatment completion, and who did not die or develop recurrent TB during the study period. In addition, we conducted a subset analysis for treatment failure by limiting the analysis only to those with culture or Xpert-confirmed pulmonary TB diagnosis at baseline as well as culture-confirmed outcomes (failure or cure). Outcomes of failure or cure were assessed between the 5th and 7th months of treatment.
The ethics statement is included in the supplementary material.
Laboratory assessment
We used two lipidomic panels to assess levels of lipids in baseline plasma samples: 1) complex lipid panel and 2) oxylipins and endocannabinoids panel. Samples from the two study sites were run in separate batches.
Complex lipids
Complex lipids, including ceramides, sphingomyelins, cholesterol esters, oxysterols, lyso- and phospholipids, mono-, di- and triacylglycerols, were semi-quantified using an untargeted approach by liquid chromatography with quadrupole time of flight mass spectrometry (LC-QTOF-MS). Lipids from plasma were extracted following protocols described previously [21] and detailed in the supplementary material.
Oxylipins
Nonesterified oxylipins, endocannabinoids, polyunsaturated fatty acids and nonsteroidal anti-inflammatory drugs were isolated by liquid extraction protocol using methanol/acetonitrile mixture (1:1 v/v) from 40 μL of plasma and quantified by ultra-performance liquid chromatography-MS/MS using internal standard methods and further detailed in the supplementary material.
Statistical analysis
Lipidomic analysis was performed on 192 individuals. 470 annotated lipids were available for analysis (as detailed in the supplementary material). We assessed lipids that were significantly different between cases (TB treatment failure) and controls. We used standard least-square regression with adjustments for baseline age, body mass index (BMI), sex, study site, diabetes, alcohol consumption, smoking and HIV to study the relationship of lipids with each outcome in Jmp v14. Lipids with a p-value <0.05 after adjusting for the Benjamini–Hochberg false discovery rate (FDR) of 0.2 were considered statistically significant. In addition to individual lipids, we assessed lipid families that were different by outcome.
We performed cross-validation with k=1 for classification by splitting the dataset into training (75% of data samples) and test (25% of data samples) sets. In log-transformed lipid data adjusted for covariates using the sva package [22], a machine-learning based random forest algorithm (randomForest package) was applied to the training set to identify the minimal variable set with high classification power to differentiate cases and controls, separately for the primary and subset analysis (further details in the supplementary material). The accuracy of the lipid models was assessed by performing the receiver operating characteristic curve and measuring the area under the curve (AUC).
Results
Study population characteristics
Our analysis (n=192) included a study population of adults with active pulmonary TB disease who went on to successfully complete treatment without subsequent TB recurrence or death (controls; n=146), or who failed treatment (cases; n=46). 44% of the study population were recruited from BJMC and 56% were recruited from NIRT (table 1). The median (interquartile range (IQR)) age of study participants was 39 (25–49) years and BMI was 17 (15.5–19) kg·m−2. 67% of study participants were male, 14% were current smokers, 49% drank alcohol, 2% had HIV (three out of four participants were on antiretroviral therapy) and 32% had diabetes (table 1). Cases and controls were different by BMI (16 versus 17 in cases compared to controls, respectively; p=0.09), but not by other study population characteristics (table 1).
TABLE 1.
Characteristics of the study population among cases and controls (n=192)
| Overall | Control | Treatment failure | p-value | |
|---|---|---|---|---|
| Subjects | 192 | 146 | 46 | |
| Age years | 39 (25–49) | 40 (25–50) | 35 (26–48) | 0.33 |
| Body mass index kg·m−2 | 17 (15.5–19) | 17 (16–20) | 16 (15–18) | 0.09 |
| Sex | ||||
| Female | 63 (33) | 48 (33) | 15 (33) | 0.99 |
| Male | 129 (67) | 98 (67) | 31 (67) | |
| Site | ||||
| BJMC | 84 (44) | 64 (44) | 20 (43) | 0.99 |
| NIRT | 108 (56) | 82 (56) | 26 (57) | |
| Smoking status | ||||
| Never | 115 (60) | 92 (63) | 23 (50) | 0.28 |
| Former | 50 (26) | 35 (24) | 15 (33) | |
| Current | 27 (14) | 19 (13) | 8 (17) | |
| Alcohol | ||||
| Yes | 94 (49) | 68 (47) | 26 (57) | 0.31 |
| No | 98 (51) | 78 (53) | 20 (43) | |
| Diabetes status | ||||
| Yes | 61 (32) | 48 (33) | 13 (28) | 0.59 |
| No | 131 (68) | 98 (67) | 33 (72) | |
| HIV | ||||
| Yes | 4 (2) | 2 (1) | 2 (4) | 0.24 |
| No | 188 (98) | 144 (99) | 44 (96) |
Data are presented as n, median (interquartile range) or n (%), unless otherwise stated. p-values were calculated using Fisher’s exact test for categorical variables and Wilcoxon rank-sum for continuous variables to determine the difference in study population characteristics by cases (treatment failure) and controls. BJMC: Byramjee Jeejeebhoy Government Medical College; NIRT: National Institute for Research in Tuberculosis.
Lipidome and TB treatment failure
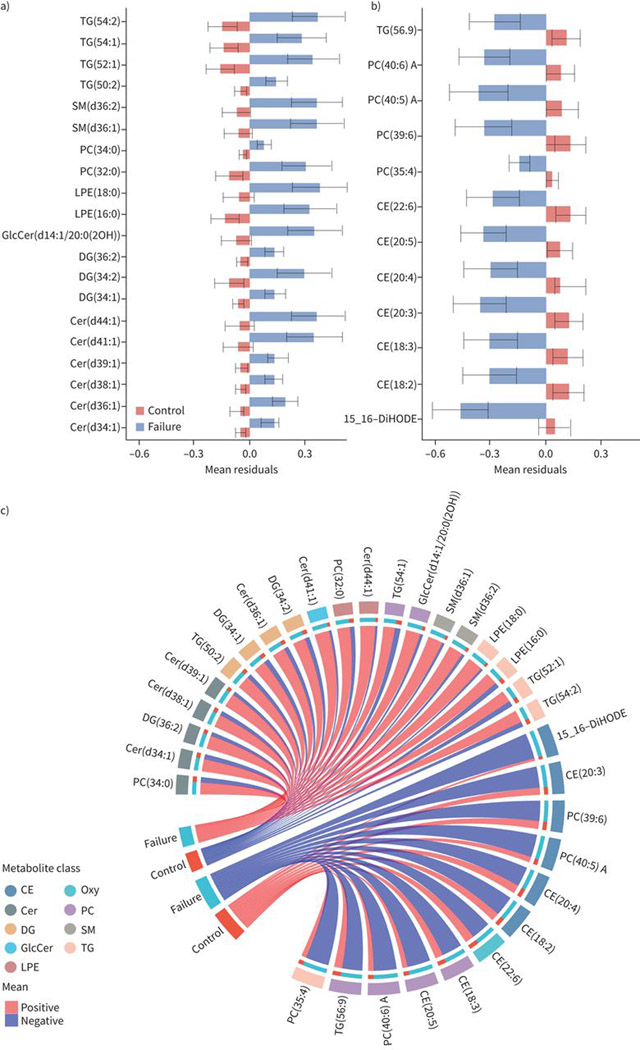
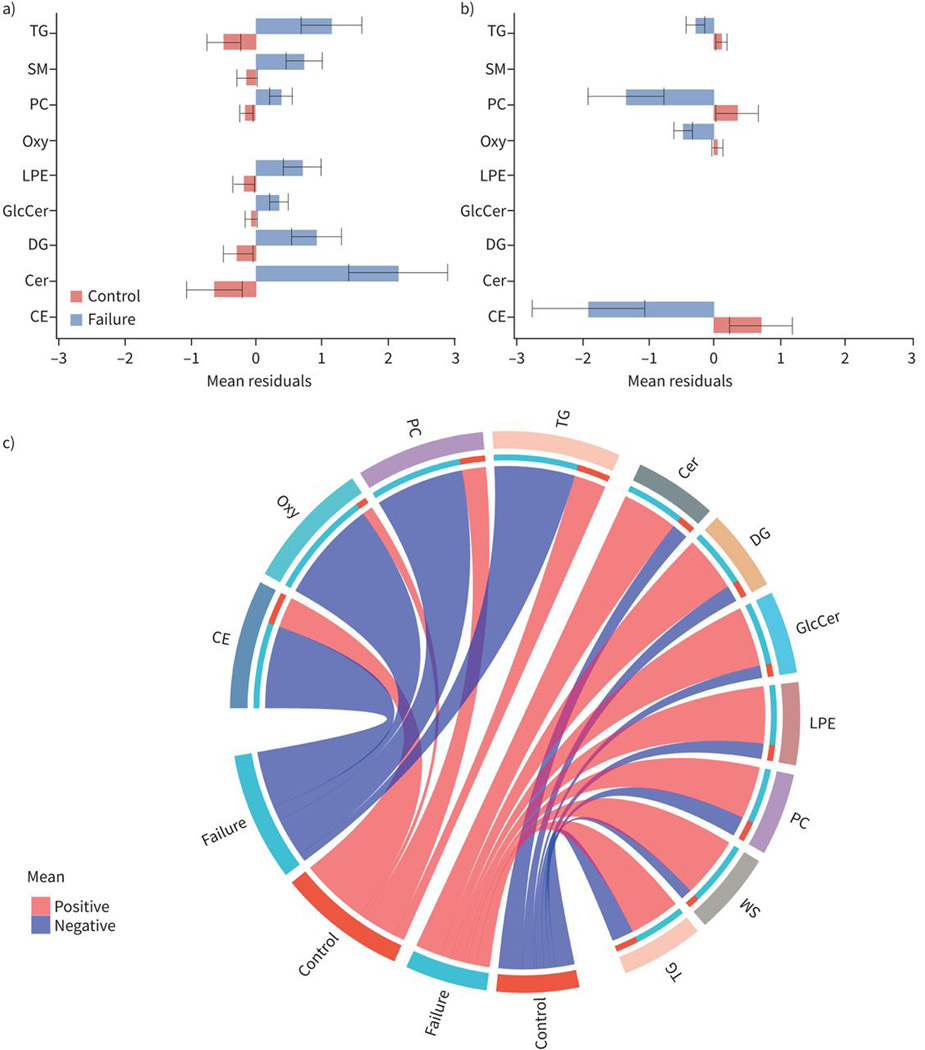
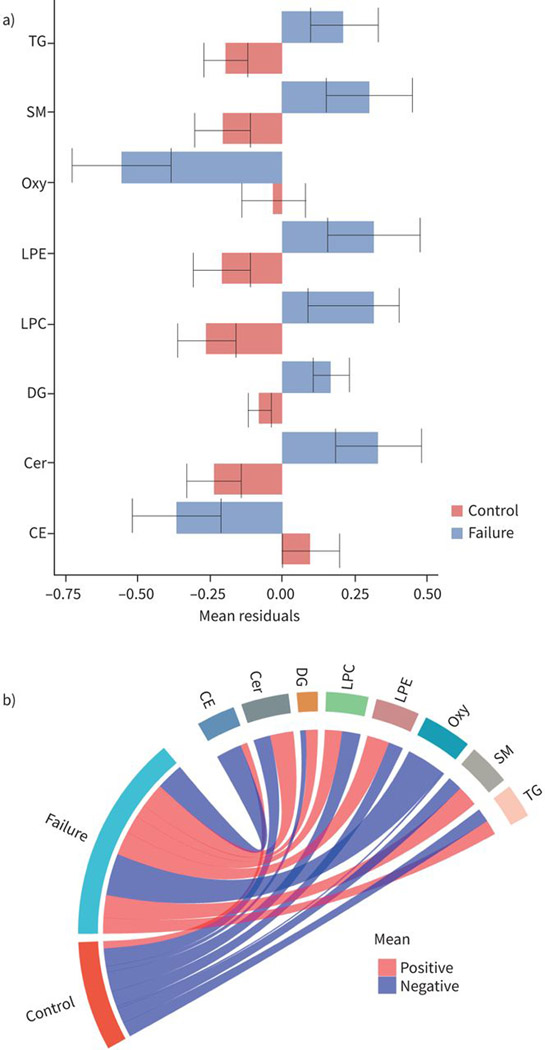
470 annotated lipids were detected with the lipidomic panels measuring complex lipids and oxylipins. In our case–control analysis of treatment failure and controls on the combined population of BJMC and NIRT, 32 lipids were significantly different (FDR threshold 0.2) between cases and controls after adjustment of covariates including age, BMI, sex, diabetes, alcohol consumption, smoking, HIV and study site (figure 1). Levels of 12 lipids were lower and 20 lipids were higher in cases compared to controls (figure 1). Those with TB treatment failure had lower levels of phosphatidylcholines (PC), cholesteryl esters (CE), oxylipins (Oxy) and triacylglycerol (TG), while they had higher levels of ceramides (Cer), diacylglycerols (DG), lyso-phosphatidyl ethanolamines (LPE), other phosphatidylcholines and triacylglycerols, and sphingomyelins (SM) compared to controls (figure 2).
FIGURE 1.
Baseline levels of individual lipids associated with tuberculosis treatment failure. Barplots showing average baseline levels of mean±SE of residuals for individual lipids that are significantly a) higher or b) lower in treatment failure compared to controls. The x-axes show the mean residuals after adjusting for body mass index, age, study site, sex, diabetes, alcohol, smoking and HIV status. Differences are considered significant if their false discovery rate-adjusted p-values <0.05. c) Circular plots showing the individual lipids and the lipid families that are increased or decreased at baseline among failures compared to controls. TG: triacylglycerol; SM: sphingomyelin; PC: phosphatidylcholine; LPE: lyso-phosphatidylethanolamine; GlcCer: glycospingolipid; DG: diacylglycerol; Cer: ceramides; CE: cholesterol esters; DiHODE: dihydroxyoctadecadienoic acid; Oxy: oxylipins.
FIGURE 2.
Baseline levels of lipid families associated with tuberculosis treatment failure. Barplots showing baseline levels of mean and standard errors of residuals for lipid families that are significantly a) higher or b) lower in treatment failure compared to controls. The x-axes show the mean residuals (sum of residuals of each individual lipid in the lipid family) after adjusting for body mass index, age, study site, sex, diabetes, alcohol, smoking and HIV status. Differences are considered significant if their false discovery rate-adjusted p-values <0.05. c) Circular plots showing the lipid families that are increased or decreased at baseline among failures compared to controls. TG: triacylglycerol; SM: sphingomyelin; PC: phosphatidylcholine; Oxy: oxylipins; LPE: lyso-phosphatidylethanolamine; GlcCer: glycospingolipid; DG: diacylglycerol; Cer: ceramides; CE: cholesterol esters.
The profile of the specific lipids within each family of lipids that were increased or decreased among those with TB treatment failure was informative. For example, six species of cholesteryl esters with an acyl chain length of 18–22 and degree of unsaturation from 2:6 were decreased in cases compared to controls (figure 1). Within the oxylipin and triacylglycerol families, only one lipid each (15,16-dihydroxyoctadecadienoic acid (DiHODE), an oxygenated derivative of α-linolenic acid (ALA); and TG (56:9), respectively) was decreased among cases compared to controls (figure 1). For phosphatidylcholines, while two species of phosphatidylcholine, with a combined chain length of 32–34 in their two acyl groups that were saturated, were increased in cases compared to controls, four longer-chain phosphatidylcholines with a combined chain length of 35–40 and higher degrees of unsaturation (four to six combined double bonds) were decreased (figure 1).
Six species of ceramides, with a combined chain length of 34–44 in their sphingoid base and acyl chain with one double bond in their acyl chain, were increased in cases compared to controls (figure 1). Monoor poly-unsaturated species of diacylglycerols (combined 34–36 chain length in their two acyl groups; three lipids) and triacylglycerols (50–54 chain length combined in their three acyl groups; four lipids) were also increased in those with treatment failure compared to controls. Other lipids that were increased among cases were saturated LPEs (16:0 and 18:0) and sphingomyelins (36:1 and 36:2) compared to controls (figure 1).
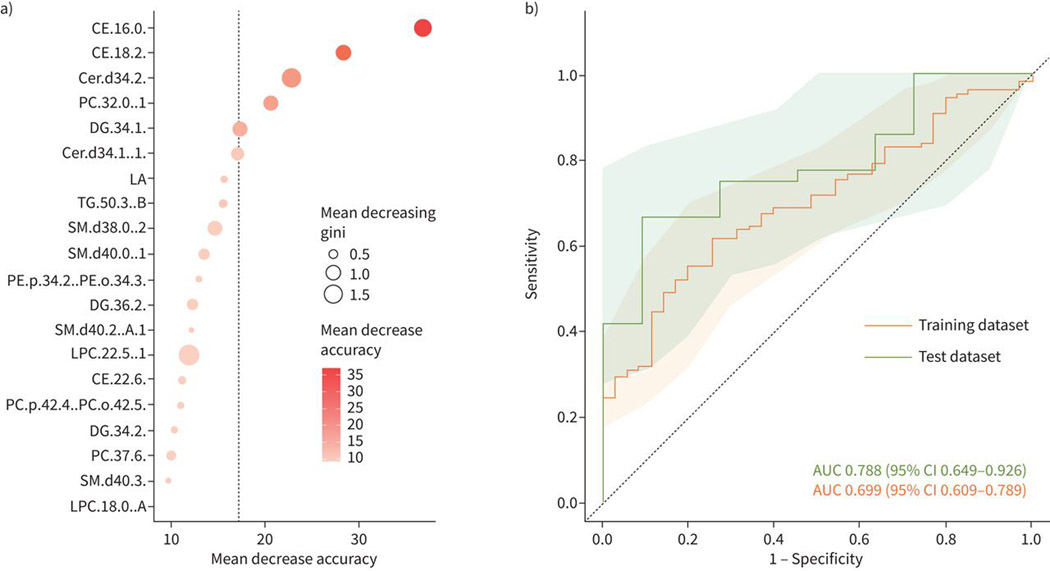
Using a machine-learning random forest algorithm, we identified CE (16:0) and CE (18:2) as the two lipids with the most classification accuracy between cases and controls (figure 3a). These lipids were combined in a unique classifier model that had an AUC (95% CI) of 0.70 (0.61–0.79) in the training dataset and 0.79 (0.65–0.93) in the test dataset (figure 3b).
FIGURE 3.
Accuracy of prediction model. a) The dot plot shows the mean decrease accuracy and mean decreasing gini values from the random forest analysis to classify treatment failure and controls (cures). b) The lipids from the random forest model are evaluated by receiver operating characteristic curve. The area under the curve (AUC) for the training and test datasets are shown and the shaded area shows the confidence intervals of the AUCs.
Subset analysis of lipidome and TB treatment failure
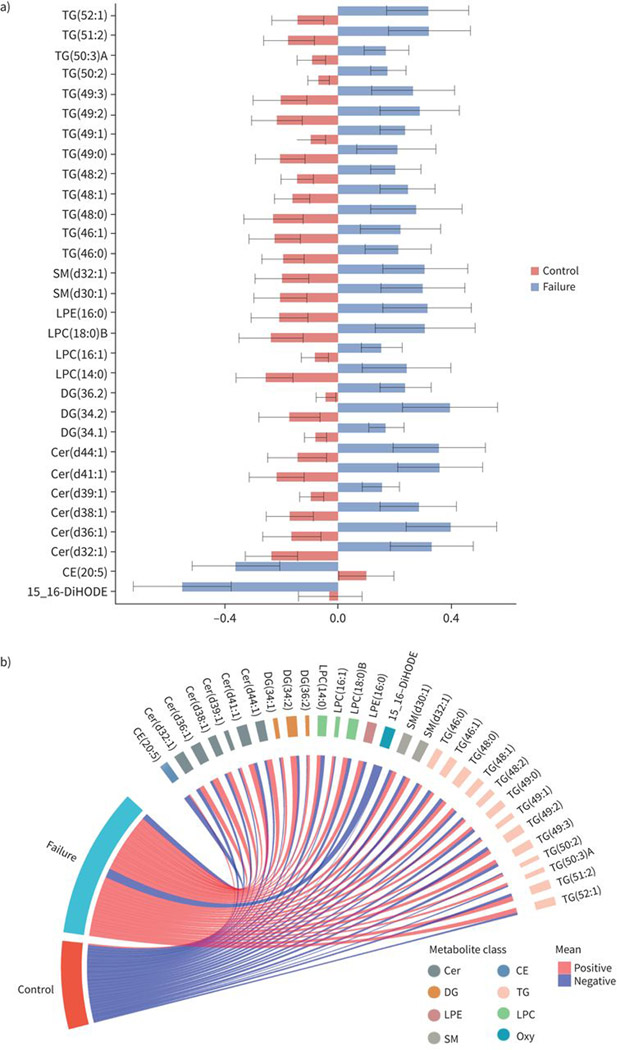
We conducted a subset analysis by limiting the analysis of TB treatment failure to adults with culture or GeneXpert-confirmed pulmonary TB at baseline and culture-confirmed treatment failure (cases; n=37) or culture-confirmed cure (controls; n=90). In this analysis, the results were similar to the primary analysis. 30 lipids were significantly different between cases and controls after adjustment for covariates (figure 4). Individuals with TB treatment failure had lower levels of oxylipins and cholesteryl esters, and higher levels of ceramides, sphingomyelins, LPC, LPE, diacylglycerol and triacylglycerol (figure 5). Of note, approximately half of the individual lipids that were significant in this analysis overlapped with the primary analysis and most of the others were lipids that were part of the same lipid family as the primary analysis.
FIGURE 4.
Baseline levels of lipids associated with tuberculosis (TB) treatment failure in subset analysis. Barplots showing average baseline levels of mean and standard error of residuals for individual lipids that are significantly a) higher or b) lower in treatment failure compared to controls in subset analysis. The subset analysis was limited to those with culture or Xpert-confirmed pulmonary TB diagnosis at baseline as well as culture-confirmed outcomes (failure or cure). The x-axis shows the mean residuals after adjusting for body mass index, age, study site, sex, diabetes, alcohol, smoking and HIV status. Differences are considered significant if their false discovery rate-adjusted p-values <0.05. c) Circular plots showing the individual lipids and the lipid families that are increased or decreased at baseline among failures compared to controls. TG: triacylglycerol; SM: sphingomyelin; LPE: lyso-phosphatidylethanolamine; LPC: lyso-phosphatidylcholine; DG: diacylglycerol; Cer: ceramide; DiHODE: dihydroxyoctadecadienoic acid; CE: cholesterol ester; Oxy: oxylipin.
FIGURE 5.
Baseline levels of lipid families associated with tuberculosis (TB) treatment failure in subset analysis. Barplots showing baseline levels of mean and standard errors of residuals for lipid families that are significantly a) higher or b) lower in treatment failure compared to controls in subset analysis. The subset analysis was limited to those with culture or Xpert-confirmed pulmonary TB diagnosis at baseline as well as culture-confirmed outcomes (failure or cure). The x-axis shows the mean residuals (sum of residuals of each individual lipid in the lipid family) after adjusting for body mass index, age, study site, sex, diabetes, alcohol, smoking and HIV status. Differences are considered significant if their false discovery rate-adjusted p-values <0.05. c) Circular plots showing the lipids families that are increased or decreased at baseline among failures compared to controls. TG: triacylglycerol; SM: sphingomyelin; Oxy: oxylipin; LPE: lyso-phosphatidylethanolamine; LPC: lyso-phosphatidylcholine; DG: diacylglycerol; Cer: ceramide; CE: cholesterol ester.
Looking at the individual lipids, only two lipids were significantly lower, and 28 lipids were significantly higher in failure compared to controls (figure 4). The lipids that were lower in failure were CE (20:5) and 15,16-DiHODE; the latter was also significant in the primary analysis. The lipids that were higher in failure in this analysis as well as the primary analysis included LPE (16:0) and multiple ceramides (d36:1, d38:1, d39:1, d41:1 and d44:1), diacylglycerols (34:1. 34:2 and 36:2) and triacylglycerols (50:2, 52:1) (figure 4). Other lipids that were significantly higher in failure in this analysis, but not in the primary analysis, included various LPCs (14:0, 16:1, 18:0 B), sphingomyelins (d30:1 and d32:1) and triacylglycerols (combined acyl chain length from 46–52 with 0–3 combined degrees of unsaturation) (figure 4). In contrast, there were various lipids that were significant in the primary analysis but not in this analysis, such as certain phosphatidylcholines.
Using machine-learning approaches, we identified four lipids (PC (37:6), PC (p-42:4)/PC (o-42:5), PC (36:1) and CE (18:2)) with the most classification accuracy between cases and controls in the subset analysis (supplementary figure S1a). When combined in a unique classifier model, these lipids that had an AUC (95% CI) of 0.74 (0.64–0.84) in the training dataset and 0.71 (0.52–0.89) in the test dataset (supplementary figure S1b).
Discussion
In our study of adults with active pulmonary TB in Pune and Chennai, India, we identified levels of various baseline lipids associated with development of TB treatment failure. Baseline levels of cholesteryl esters, oxylipin, 15,16-DiHODE (oxygenated derivative of ALA) and certain phosphatidylcholines were among the lipids that were lower in those with TB treatment failure, while lipid families with higher levels included ceramides, diacylglycerols and various triacylglycerols. Some of these lipids have recognised and important roles in susceptibility or resistance to TB pathogenesis, while others have not been studied in the context of TB. Specific cholesteryl esters were the best predictors of TB treatment failure. Our results identify specific lipids for assessment in future mechanistic studies to better understand their role in TB outcomes, along with clinical studies that seek to utilise them as predictors for TB treatment failure and potential targets for treatment monitoring and adjunct therapy.
In our primary analysis, we observed that individuals who failed treatment had lower baseline levels of various cholesteryl esters. In fact, two of the cholesteryl esters (16:0 and 18:2) combined had the best predictive accuracy for subsequent treatment failure. While higher cholesterol levels are a known risk factor for TB disease [23] and utilised by M. tuberculosis for survival [24], the roles of cholesteryl esters in TB are less clear. One potential reason for this might be inefficient conversion of free cholesterol to cholesteryl esters; we observed that sphingomyelins are increased in failures, and sphingomyelins can inhibit the esterification of cholesterol [18]. Furthermore, the esterification of cholesterol to cholesteryl ester is also reduced during the acute phase response of inflammation [18], suggesting that higher cholesteryl esters represent increased inflammation and increased risk of subsequent failure. While future studies will need to clarify the role of cholesteryl esters in TB treatment failure, our results suggest these lipids have predictive utility. Further discussion on other lipids (e.g. 15,16-DiHODE and phosphatidylcholines) different in failure are discussed in the supplementary material.
Baseline levels of various ceramides were higher in individuals who failed treatment compared to those achieving a cure. In line with our observation with Cer (d34:1), multiple studies of adults and children have observed that individuals with TB have higher levels of this ceramide compared to healthy controls or even other respiratory diseases [7,25,26]. Ceramides are sphingolipids involved in host immunity against M. tuberculosis and other pathogens by reducing bacterial burden through increased phagocytosis [27], and increased activation of macrophages [7,27] and natural killer T-cells [28]. While individuals with TB have higher levels of these ceramides, our data suggest that among individuals with TB, those with the highest levels of ceramides have an increased risk of failure. We hypothesise that baseline levels of this lipid may identify individuals with an excessive immune response, potentially due to ineffective control of M. tuberculosis. As is true for various immune mediators, while ceramides are needed to fight pathogens, unresolved and excessive levels of these mediators are linked to adverse outcomes [29,30].
Higher baseline triacylglycerol levels were observed in individuals who eventually failed treatment compared to cured controls. In addition to using triacylglycerols as an energy source, M. tuberculosis utilises host triacylglycerols to accumulate lipid droplets within macrophages, allowing them to lose acid-fast staining, become phenotypically resistant to certain TB drugs (e.g. rifampicin and isoniazid) and enter dormancy [31–33]. In fact, data suggest that triacylglycerols and triacylglycerol-rich lipid bodies can be detected in sputum samples of individuals with active TB, can be used to monitor treatment responses (e.g. quantifying “persisters”), are increased in TB strains that cause more rapid disease development and are associated with adverse treatment outcomes [34–37]. These observations provide a direct link between triacylglycerols and treatment failure, where M. tuberculosis in individuals with the highest levels of triacylglycerols is better able to resist killing by the TB drugs. Interestingly, a recent study observed that a triacylglycerol-reducing drug restricted M. tuberculosis grown in zebrafish models (further details in the supplementary material) [38]. In summary, these data identify specific triacylglycerols that are associated with increased failure and could serve as baseline indicators for increased risk of failure along with a potential utility for treatment monitoring.
While our subset analysis had some differences (e.g. phosphatidylcholines), expected based on the differences in diagnosis and outcome assessment, most of the baseline lipids (i.e. same lipid species or from same family) from the primary analysis, such as ceramides, diacylglycerols, triacylglycerols and oxygenated fatty acids remained significantly associated with treatment failure.
Our study has a few limitations. Our analysis studied the lipidome only at baseline and future studies will need to determine how the lipidome profile changes later in treatment. Additional limitations include limited sample size to conduct subset analysis by comorbidities (e.g. HIV and diabetes) and medications (e.g. metformin and statins), methodological limitations of LC-MS/MS to validate all structural details along with challenges in measuring these lipids in clinical practice (detailed in the supplementary material). Regardless, to our knowledge, this was the first study to assess the relationship of baseline lipidome with TB treatment outcomes and we identified lipids that are associated with risk of future treatment failure and should be tested in future studies for their utility in prediction and treatment monitoring. Despite these limitations, our study had strengths including nesting within a well-characterised cohort, a comprehensive and unbiased panel of lipids measured prior to development of a clinically relevant outcome and adjusting for important covariates.
In conclusion, our study identified baseline lipids associated with TB treatment failure. Some of these lipids have known roles in TB pathogenesis and our data suggest that they also probably affect treatment failure. Additional lipids that have not been studied in the context of TB should be assessed in future studies on whether and how they affect TB pathogenesis and treatment outcomes. These lipids could identify subset of individuals at high risk of treatment failure and who can be targeted for adjunct therapy. Modulating these lipids prior to or during early treatment could potentially reduce TB treatment failure.
Supplementary Material
Acknowledgements:
The authors thank the study participants for their time and contributions. The authors also acknowledge Persistent Systems for data management software support and Hewlett Packard for donation of computer equipment used in the study. The contents of this publication are solely the responsibility of the authors and do not represent the official views of the Department of Biotechnology, the Indian Council of Medical Research, the US National Institutes of Health or CRDF Global. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Support statement:
The parent study CTRIUMPH and the data in this manuscript were collected as part of the Regional Prospective Observational Research for Tuberculosis (RePORT) India Consortium. This project has been funded in whole or in part with federal funds from the Government of India’s (GOI) Department of Biotechnology (DBT), Indian Council of Medical Research (ICMR), the US National Institutes of Health (NIH), National Institute of Allergy and Infectious Diseases (NIAID), Office of AIDS Research, and distributed in part by CRDF Global (USB1-31147-XX13 CRDF/NIH). Any mention of trade names, commercial projects, or organizations does not imply endorsement by any of the sponsoring organizations. Research reported in this publication was also supported by NIAID (UM1AI069465 and R01AI097494), NICHD (R00HD089753), the Gilead Foundation, the Ujala Foundation, and the Fogarty International Center BJGMC-JHU HIV-TB Program (D43TW009574). This work was further supported by the Johns Hopkins University School of Medicine funds and Columbia University Department of Epidemiology funds. BBA was supported by Intramural research program from FIOCRUZ and from the National Institutes of Health (U01AI115940) and NIH/CRDF RePORT International Supplemental Funds. A.N. Gupte was supported by NIH Research Training Grant # D43 TW009340 funded by the NIH Fogarty International Center. B.B. Andrade is a senior investigator from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Brazil. Additional support was provided to O. Fiehn by NIH U24 DK097154 and to J.W. Newman by USDA Intramural Projects 2032-51530-022-00D and 2032-51530-025-00D. The USDA is an equal opportunity employer and provider. Funding information for this article has been deposited with the Crossref Funder Registry.
Conflict of interest:
R. Shivakoti reports grants from NIH, during the conduct of the study. J.W. Newman has nothing to disclose. L.E. Hanna has nothing to disclose. A.T.L. Queiroz has nothing to disclose. K. Borkowski has nothing to disclose. A.N. Gupte has nothing to disclose. M. Paradkar has nothing to disclose. P. Satyamurthi has nothing to disclose. V. Kulkarni has nothing to disclose. M. Selva has nothing to disclose. N. Pradhan has nothing to disclose. S.V.B.Y. Shivakumar has nothing to disclose. S. Natarajan has nothing to disclose. R. Karunaianantham has nothing to disclose. N. Gupte has nothing to disclose. K. Thiruvengadam has nothing to disclose. O. Fiehn has nothing to disclose. R. Bharadwaj has nothing to disclose. A. Kagal has nothing to disclose. S. Gaikwad has nothing to disclose. S. Sangle has nothing to disclose. J.E. Golub has nothing to disclose. B.B. Andrade has nothing to disclose. V. Mave reports grants from NIH, during the conduct of the study. A. Gupta reports grants from NIH, during the conduct of the study. C. Padmapriyadarsini has nothing to disclose.
References
- 1.Berry MP, Graham CM, McNab FW, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 2010; 466: 973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cliff JM, Kaufmann SH, McShane H, et al. The human immune response to tuberculosis and its treatment: a view from the blood. Immunol Rev 2015; 264: 88–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta RK, Turner CT, Venturini C, et al. Concise whole blood transcriptional signatures for incipient tuberculosis: a systematic review and patient-level pooled meta-analysis. Lancet Respir Med 2020; 8: 395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng S, Du YQ, Zhang L, et al. Analysis of serum metabolic profile by ultra-performance liquid chromatography-mass spectrometry for biomarkers discovery: application in a pilot study to discriminate patients with tuberculosis. Chin Med J 2015; 128: 159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiner J 3rd, Parida SK, Maertzdorf J, et al. Biomarkers of inflammation, immunosuppression and stress with active disease are revealed by metabolomic profiling of tuberculosis patients. PloS One 2012; 7: e40221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frediani JK, Jones DP, Tukvadze N, et al. Plasma metabolomics in human pulmonary tuberculosis disease: a pilot study. PLoS One 2014; 9: e108854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau SK, Lee KC, Curreem SO, et al. Metabolomic profiling of plasma from patients with tuberculosis by use of untargeted mass spectrometry reveals novel biomarkers for diagnosis. J Clin Microbiol 2015; 53: 3750–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiner J 3rd, Maertzdorf J, Sutherland JS, et al. Metabolite changes in blood predict the onset of tuberculosis. Nat Commun 2018; 9: 5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duffy FJ, Weiner J 3rd, Hansen S, et al. Immunometabolic signatures predict risk of progression to active tuberculosis and disease outcome. Front Immunol 2019; 10: 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho Y, Park Y, Sim B, et al. Identification of serum biomarkers for active pulmonary tuberculosis using a targeted metabolomics approach. Sci Rep 2020; 10: 3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayer-Barber KD, Sher A. Cytokine and lipid mediator networks in tuberculosis. Immunol Rev 2015; 264: 264–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jain M, Petzold CJ, Schelle MW, et al. Lipidomics reveals control of Mycobacterium tuberculosis virulence lipids via metabolic coupling. Proc Natl Acad Sci USA 2007; 104: 5133–5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Layre E, Al-Mubarak R, Belisle JT, et al. Mycobacterial lipidomics. Microbiol Spectr 2014; 2: 2.3.03. [DOI] [PubMed] [Google Scholar]
- 14.Crick PJ, Guan XL. Lipid metabolism in mycobacteria – insights using mass spectrometry-based lipidomics. Biochim Biophys Acta 2016; 1861: 60–67. [DOI] [PubMed] [Google Scholar]
- 15.Pal R, Hameed S, Kumar P, et al. Comparative lipidomics of drug sensitive and resistant Mycobacterium tuberculosis reveals altered lipid imprints. 3 Biotech 2017; 7: 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.López-Hernández Y, Lara-Ramírez EE, Salgado-Bustamante M, et al. Glycerophospholipid metabolism alterations in patients with type 2 diabetes mellitus and tuberculosis comorbidity. Arch Med Res 2019; 50: 71–78. [DOI] [PubMed] [Google Scholar]
- 17.Wood PL, Tippireddy S, Feriante J. Plasma lipidomics of tuberculosis patients: altered phosphatidylcholine remodeling. Future Sci OA 2017; 4: FSO255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vrieling F, Ronacher K, Kleynhans L, et al. Patients with concurrent tuberculosis and diabetes have a pro-atherogenic plasma lipid profile. EBioMedicine 2018; 32: 192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins JM, Siddiqa A, Jones DP, et al. Tryptophan catabolism reflects disease activity in human tuberculosis. JCI Insight 2020; 5: e137131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupte A, Padmapriyadarsini C, Mave V, et al. Cohort for Tuberculosis Research by the Indo-US Medical Partnership (CTRIUMPH): protocol for a multicentric prospective observational study. BMJ Open 2016; 6: e010542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matyash V, Liebisch G, Kurzchalia TV, et al. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res 2008; 49: 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet 2007; 3: 1724–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soh AZ, Chee CB, Wang YT, et al. Dietary cholesterol increases the risk whereas PUFAs reduce the risk of active tuberculosis in Singapore Chinese. J Nutr 2016; 146: 1093–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilburn KM, Fieweger RA, VanderVen BC. Cholesterol and fatty acids grease the wheels of Mycobacterium tuberculosis pathogenesis. Pathog Dis 2018; 76: fty021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andreas NJ, Basu Roy R, Gomez-Romero M, et al. Performance of metabonomic serum analysis for diagnostics in paediatric tuberculosis. Sci Rep 2020; 10: 7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam CW, Law CY. Untargeted mass spectrometry-based metabolomic profiling of pleural effusions: fatty acids as novel cancer biomarkers for malignant pleural effusions. J Proteome Res 2014; 13: 4040–4046. [DOI] [PubMed] [Google Scholar]
- 27.Anes E, Kühnel MP, Bos E, et al. Selected lipids activate phagosome actin assembly and maturation resulting in killing of pathogenic mycobacteria. Nat Cell Biol 2003; 5: 793–802. [DOI] [PubMed] [Google Scholar]
- 28.Chackerian A, Alt J, Perera V, et al. Activation of NKT cells protects mice from tuberculosis. Infect Immun 2002; 70: 6302–6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karjalainen JP, Mononen N, Hutri-Kähönen N, et al. New evidence from plasma ceramides links apoE polymorphism to greater risk of coronary artery disease in Finnish adults. J Lipid Res 2019; 60: 1622–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laaksonen R, Ekroos K, Sysi-Aho M, et al. Plasma ceramides predict cardiovascular death in patients with stable coronary artery disease and acute coronary syndromes beyond LDL-cholesterol. Eur Heart J 2016; 37: 1967–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daniel J, Maamar H, Deb C, et al. Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLoS Pathog 2011; 7: e1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daniel J, Deb C, Dubey VS, et al. Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J Bacteriol 2004; 186: 5017–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maurya RK, Bharti S, Krishnan MY. Triacylglycerols: fuelling the hibernating Mycobacterium tuberculosis. Front Cell Infect Microbiol 2018; 8: 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kayigire XA, Friedrich SO, van der Merwe L, et al. Simultaneous staining of sputum smears for acid-fast and lipid-containing Myobacterium tuberculosis can enhance the clinical evaluation of antituberculosis treatments. Tuberculosis 2015; 95: 770–779. [DOI] [PubMed] [Google Scholar]
- 35.Sloan DJ, Mwandumba HC, Garton NJ, et al. Pharmacodynamic modeling of bacillary elimination rates and detection of bacterial lipid bodies in sputum to predict and understand outcomes in treatment of pulmonary tuberculosis. Clin Infect Dis 2015; 61: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shim D, Kim H, Shin SJ. Mycobacterium tuberculosis infection-driven foamy macrophages and their implications in tuberculosis control as targets for host-directed therapy. Front Immunol 2020; 11: 910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tong J, Liu Q, Wu J, et al. Mycobacterium tuberculosis strains of the modern Beijing sublineage excessively accumulate triacylglycerols in vitro. Tuberculosis 2020; 120: 101892. [DOI] [PubMed] [Google Scholar]
- 38.McClean CM, Tobin DM. Early cell-autonomous accumulation of neutral lipids during infection promotes mycobacterial growth. PLoS One 2020; 15: e0232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.