Summary
Background
Gallbladder cancer (GBC), the most common malignancy of the biliary tract, shows late diagnosis and low survival rate and requires continued search for new diagnostic biomarkers and therapeutic targets. Human endogenous retroviruses (HERVs) are specifically prone to be reactivated in diverse cancers and are implicated in cancer progression and immunotherapy.
Methods
Single-cell RNA sequencing was performed on tumor tissues and paired adjacent tissues from 4 GBC patients. Dual-luciferase reporter assay was applied to measure enhancer activity of HERV sequences.
Findings
We dissected the cellular diversity and described the HERV transcriptomic landscape for GBC. We found that HERVs were transcribed in a cell type-specific manner and different HERV families were associated with diverse biological effects. HERVs could function as enhancers, presumably causing altered expression of neighboring genes. The transcription level of HERVH was gradually elevated with the malignant transformation of epithelial cells, suggesting HERVH may be a potential early diagnostic biomarker of GBC. HHLA2, a newly emerging immune checkpoint, was derived by HERVH, exhibited an expressional correlation with HERVH, and was identified as a promising target for immunotherapy.
Interpretation
Exploring the transcriptional landscape and potential functional impact of HERVs highlights the important role of HERVs in GBC and provides a fresh perspective on managing GBC.
Funding
This study was supported by the National Natural Science Foundation of China (31970176, 81972256) and the research grants from the Innovation Capacity Building Project of Jiangsu province (BM2020019).
Keywords: Gallbladder cancer, Single-cell RNA sequencing, Human endogenous retrovirus, Enhancer, Immune checkpoint, HERVH
Abbreviations: GBC, gallbladder cancer; HERV, human endogenous retrovirus; scRNA-seq, single-cell RNA sequencing; TME, tumor microenvironment; WTA, whole transcriptome analysis; DEG, differentially expressed gene; CNV, copy number variation; GO, gene ontology; NK cell, natural killer cell; NKT cell, natural killer T cell; DC, dendritic cell; ICS, intermediate cell state; HHLA2, human endogenous retrovirus-H long terminal repeat-associating 2; CD4+ Th cell, CD4+ T helper cell; IgG, immunoglobulin G; cDC, conventional DC; mo-DC, monocyte-derived DC; CAF, cancer-associated fibroblast; ECM, extracellular matrix; iCAF, inflammatory CAF; myoCAF, myo-cancer-associated fibroblast; TE, transposable element
Research in context.
Evidence before this study
Gallbladder cancer (GBC) is characterized by late-stage diagnosis and poor prognosis, so research efforts should continue to find biomarkers for early detection and innovate therapeutic approaches for improving clinical outcomes. Human endogenous retroviruses (HERVs) are remnants of ancient exogenous retroviruses and make up ∼8% of the human genome. HERVs are specifically reactivated in various cancers and are implicated in cancer development, which has spurred many studies exploring HERVs as promising diagnostic and therapeutic targets. The involvement of HERVs in GBC, which is essential for improved management of this malignancy, has not yet been systematically elucidated.
Added value of this study
We revealed the cell-type diversity of GBC by single-cell RNA sequencing and delineated the transcriptional landscape of HERVs at the single-cell level. There are substantial differences in HERV activation among different cell types, which contributes to the intratumoral heterogeneity of GBC. HERVs were demonstrated to serve as enhancers, potentially regulating the expression of neighboring genes in cancer cells. Biological functions that may be affected by those aberrantly activated HERVs were also identified for each cell population. HERV members (HERVH and HHLA2) were recognized as promising targets to help achieve early diagnosis and advance GBC immunotherapy.
Implications of all the available evidence
Exploring HERV transcription patterns and their potential impacts on cellular functions highlights the essential role of HERVs in shaping the tumor microenvironment, provides novel insights into GBC development, and offers a valuable resource for better management of GBC.
Introduction
Gallbladder cancer (GBC) is a rare malignancy and the most common type of GBC is adenocarcinoma.1,2 GBC is often diagnosed at a late stage because it usually doesn't cause noticeable signs or symptoms until it's advanced. The progression of GBC is frequently fast and the prognosis is very poor. Current therapeutic approaches, such as surgical resection, targeted therapy and immunotherapy, benefit a subset of patients with GBC.3,4 Innovative strategies to achieve early diagnosis and effective treatment are urgently needed.
Human endogenous retroviruses (HERVs), which are estimated to account for 8% of the human genome, are relics of ancestral germ-line infections by exogenous retroviruses and became stable during the course of evolution.5,6 The intact structure of HERV is the same as exogenous retroviruses: 5′LTR-gag-pro-pol-env-3′LTR, but they usually accumulated so many mutations and deletions that their viral replication is restricted in the human genome.7 Although most HERVs are silenced in normal tissues, cumulative evidence indicates that HERVs could be reactivated in diverse cancers due to dysregulated epigenetic modifications and play an important role in modulating carcinogenesis.8,9 The flanking LTRs could lead to expressional alteration of adjacent genes by acting as promoters or enhancers. For example, the expression of Krüppel-associated box (KRAB) domain–containing zinc-finger protein (KZFP) genes has been demonstrated to be induced by adjacent HERVs in tumors.10 HERV expression may trigger a 'viral mimicry' pathway, which can increase antigen presentation, expression of interferon-related genes, and expression of immune checkpoint genes, potentially sensitizing tumor cells to recognition.11,12 Furthermore, some HERVs may be translated to tumor-specific antigens that can serve as targets of T cells.13, 14, 15, 16, 17 These characteristics make them especially attractive as new therapeutic possibilities for diverse malignancies. Actually, HERVs have recently received much attention in immunotherapy and the combined use of DNA demethylation agents (resulting in HERV derepression) and immune checkpoint therapy is being tested in clinical trials in a variety of cancers.18, 19, 20
Single-cell RNA sequencing (scRNA-seq) has been widely applied to dissect the cellular composition of tissues and explore functional heterogeneity.21, 22, 23 It provides a convenient way to investigate how HERV expression varies among different cell types. Given that HERVs are dispersed in multiple copies throughout human genome, achieving accurate estimation of locus-specific expression of HERVs with scRNA-seq data is challenging.24 Tools tailored for scRNA-seq data to achieve locus quantification taking multi-mapped reads into consideration are lacking. Keeping only uniquely mapped reads is a commonly used strategy in RNA-seq analysis, although it would lead to expressional underestimation of some HERVs.10,25,26 Therefore, the findings related to HERVs should be interpreted in the context of the techniques and methods used.
Given the essential role of HERVs in other cancers and the fact that the contribution of HERVs to GBC is currently unknown, we would like to investigate whether HERVs are also reactivated and implicated in various cellular processes in GBC, aiming to find new insights into GBC management. In this study, we applied scRNA-seq to reveal the cellular composition of the tumor microenvironment (TME) and report the comprehensive single-cell transcriptome profile of HERVs in GBC tumor tissues and adjacent normal tissues. In addition, we investigated the enhancer activity of HERV-derived sequences by dual-luciferase reporter assay and explored their potential functional impacts in each cell type. These results would help us further the understating of the functional role of HERVs and provide clues to the discovery of better treatment strategies for GBC.
Methods
Ethics approval and consent to participate
4 patients with gallbladder adenocarcinoma were recruited and they all signed the consent forms. Tumor tissue and adjacent normal tissue were collected from each GBC patient. The matched adjacent normal tissue was taken from the mucosal tissue of the gallbladder at least 2 cm from the edge of the tumor. This present study was approved by the Ethics Committee of Eastern Hepatobiliary Surgery Hospital (EHBHKY2021-K-006).
Single-cell dissociation
Fresh tumor tissues and adjacent normal tissue collected were stored in MACS Tissue Storage Solution (Miltenyi Biotec) before processing. The single-cell suspension was generated as described below. First, the tissue sample was minced into small pieces that were ∼1 mm3 in size on ice after washing with phosphate-buffered saline (PBS, Gibco). Collagenase IV (Worthington) and DNase I (Worthington) were subsequently used to enzymatically dissociated these pieces and this step lasted for 30 min at 37 °C. After dissociation, the sample was passed through a 70 μm cell strainer (Falcon) and the mixture was then centrifuged for 5 min at 300×g to remove the supernatant. Next, the red blood cells were lysed with red blood cell lysis buffer (Miltenyi Biotec) and the sample was washed with PBS containing 0.04% BSA (Thermo Fisher Scientific). A 35 μm cell strainer (Falcon) was added to re-filtered cell pellets after re-suspending in PBS containing 0.04% BSA. Cell viability was assessed by staining dissociated cells through Calcein-AM (Thermo Fisher Scientific) and Draq7 (BD Biosciences). Finally, the dead cells in the single cell suspension were removed using a MACS dead cell removal kit from Miltenyi Biotec.
Single-cell library preparation and sequencing
Whole transcriptomic information of each sample was captured with the BD Rhapsody single-cell system. To realize single-cell capture, the suspension was randomly distributed across more than 200,000 microwells by limited dilution method. Beads containing oligonucleotide barcodes were required to be added excessively to make sure that almost each microwell has one bead. Trapped cells were lysed, allowing the released mRNA molecules to hybridize with barcoded capture oligos on the beads in the microwell. Beads were transferred to a single tube and then reverse transcription and ExoI (Thermo Fisher Scientific) digestion were carried out. Next, the unique molecular identifier (UMI) and cell barcode were used to tag the synthetic cDNA at the 5′ end, that is equivalent to the 3′ end of the mRNA molecule. The final single cell library was generated through several procedures including random priming and extension (RPE), RPE amplification PCR and WTA index PCR. Library quantification was performed by Agilent Bioanalyzer 2200 High Sensitivity DNA Chip and Qubit High Sensitivity DNA assay (Thermo Fisher Scientific). All generated libraries were sequenced by an illumina sequencer (Illumina, San Diego, CA) with PE150 strategy (paired-end 150bp).
Cell culture
The GBC-SD (RRID: CVCL_6903), human gallbladder carcinoma cell line was purchased from Zrbiorise (Shanghai, China). GBC-SD and HEK293T (ATCC, RRID: CVCL_0063) cells were maintained in high-glucose DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco), penicillin (100 IU/ml, Gibco) and streptomycin (100 μg/ml, Gibco) in a humidified atmosphere containing 5% CO2 at 37 °C. The GBC-SD and HEK293T cell lines were characterized by Azenta Life Sciences (Jiangsu, China) using short tandem repeat (STR) markers (Supplementary File 1).
Plasmids
PGL3-promoter, PGL3-control and pRL-TK were purchased from HedgehogBio Science and Technology Ltd (Shanghai, China). All potential enhancer sequences of HERV (Supplementary Table S4) were amplified from the HEK293T genome using nested PCR with 2 × Phanta Max Master Mix (Vazyme, P525). The PGL3-promoter was enzymatically cut using KpnI-HF (NEB, R3142S). All potential enhancer sequences of HERV were inserted into the PGL3-promoter separately using NEBuilder HiFi DNA Assembly Master Mix (NEB, E2621). The sequences of all plasmids were confirmed by Sanger sequencing.
Dual-luciferase reporter assay
A total of 5 × 104 GBC-SD cells were plated in 24-well cell culture plates (corning, 3524) and transfected with 500 ng plasmids per well by using Lipofectamine 2000 Transfection Reagent (Invitrogen, 11668019). The ratio of the experimental vector to the co-reporter vector pRL-TK was 24:1. After 48 h, the cells were collected for luciferase activity evaluation by using a dual-luciferase reporter system (HANBIO, China) according to the manufacturer's protocol.
Quantification of gene and HERV expressions
Raw sequencing data was analyzed using whole transcriptome analysis (WTA) pipeline of BD Rhapsody™ on a local installation (BD® Single-Cell Multiomics Analysis Setup User Guide, Doc ID: 47383). The steps in the WTA analysis mainly include removing reads with low quality, annotating R1 and R2 reads, collapsing reads into raw molecules, determining putative cells and generating expression matrices. For details, please refer to BD® Single-Cell Multiomics Bioinformatics Handbook (Doc ID: 54169). During the annotation process, STAR (v2.5.2b)27 was used to align reads to the human reference genome (GRCh38.p12, UCSC). Gene annotation file was downloaded from GENCODE (https://www.gencodegenes.org/, Release 31) and HERV annotation file which was compiled by RepeatMasker was obtained from UCSC Table Browser for GRCh38 (https://genome.ucsc.edu/cgi-bin/hgTables). Only the uniquely mapped reads were calculated to estimate gene and HERV expression levels. Besides, many HERV loci were found to overlap with exons of host genes, so the overlapping regions were removed for HERV loci to avoid quantification bias.
Quality control and cell type determination
The gene-cell count matrix was imported into R package Seurat (version 4.0.6) for subsequent analyses.28 Low quality cells and cell doublets or multiplets were removed based on the gene count and mitochondrial contamination (Supplementary Table S1) and genes expressed in less than 3 cells were filtered out for each sample. After applying these filtering criteria, the filtered gene-cell count matrix was normalized by the function NormalizeData() and 2000 most variable genes of each sample were selected to correct the batch effect derived from individual samples. Significant principal components identified by function ElbowPlot() were used for graph-based clustering and t-distributed stochastic neighbor embedding (tSNE) visualization. Subclustering of cell types of interest was done with the same method. The identity of each cluster was characterized by the expression of the canonical marker genes.
For HERV(locus)-cell count matrix, only cells that passed through the previous filter and HERV loci that were expressed in at least 3 cells were retained and HERV(family)-cell count matrix was calculated by aggregating counts of each HERV locus belonging to the same family. The filtered HERV-cell count matrices were also normalized by the function NormalizeData().
Comparison of HERV expression
To describe the difference of HERV expression between GBC and normal samples, the number of active HERV loci and total expression level of HERV loci were calculated and displayed. We defined a HERV locus active if it was expressed in at least 3 cells for each sample. The statistical method used for each comparison was two-sided Wilcoxon rank sum test and significance levels were indicated by these symbols: ns, not significant; ∗, p ≤ 0.05; ∗∗, p ≤ 0.01; ∗∗∗, p ≤ 0.001; ∗∗∗∗, p ≤ 0.0001.
Differential expression analysis
The specific HERV families (adjusted p-value < 0.05 and log2FC > 0.5) of each major cell type from GBC tissues were selected by the function FindAllMarkers(). Differentially expressed HERV families and HERV loci between tumor- and normal-derived cells were identified using the function FindMarkers(). HERV families or HERV loci were considered statistically significant if their adjusted p-values by Bonferroni were less than 0.05. The contribution of significantly upregulated HERV loci to the total increment in HERV expression in each cell type was calculated as the ratio of the sum of expression increments from significantly upregulated HERV loci over the sum of expression increments from all upregulated HERV loci (log2FC > 0). The expression increment of a HERV locus was defined as the average change in expression between cells from the tumor and adjacent normal tissue.
HERV-derived enhancer prediction in epithelial cells
To search for HERV loci that may have enhancer activity to improve the expression of adjacent differentially expressed genes (DEGs) (genes with adjusted p-value < 0.05), pairs between HERV loci and DEGs within 500 kb in the genome were selected as the base set.29, 30, 31 The initial screening was based on whether HERVs and DEGs were both upregulated (gene.log2FC > 0 & HERV.log2FC > 0) in GBC-derived epithelial cells. The further filtering was carried out according to these 3 aspects: co-expression between HERV locus and DEG, enhancer-gene link predicted by GeneHancer, and DNase and histone modification signal from ENCODE project. The co-expression was defined if there was a positive correlation (Spearman correlation coefficient > 0.3) between HERV and DEG expression. GeneHancer interactions were downloaded from the GeneLoc database (https://genecards.weizmann.ac.il/geneloc/index.shtml) and ENCODE Candidate Cis-Regulatory Elements (cCREs) data was obtained from the UCSC Table Browser (https://genome.ucsc.edu/cgi-bin/hgTables).
Identification of malignant cells
There are two key considerations when separating malignant cells from non-malignant epithelial cells. On the one hand, copy number variation (CNV) is thought to be a characteristic of malignant cells and inferCNV (https://github.com/broadinstitute/inferCNV) was used to detect somatic chromosomal copy number alterations. This algorithm was implemented for each patient and fibroblasts and endothelial were considered as the reference. The CNV signal of each epithelial cell was summarized as CNV score, which was the mean square of the CNV estimates across all genomic locations. On the other hand, phenotypic similarities of malignant cells will drive them to cluster together, so we performed sub-clustering for epithelial cells with a high resolution. A cell cluster composed mainly of cells with high CNV scores was designated as malignant. Here, subcluster 11 and 12 showing highest CNV scores were classified as malignant cells (Supplementary Figure S3c).
Single-cell trajectory construction
The epithelial cell trajectory was generated by Monocle (Version 2.20.0) algorithm.32 Differentially expressed genes selected by Seurat were used to define each cell's progress and “DDRTree” method was applied to reduce data dimensionality. HERV families that changed along with the developmental trajectory were calculated by function differentialGeneTest().
Identification of genes and putative pathways associated with HERV families
To investigate how dramatically upregulated HERV families (adjusted p-value < 0.05, log2FC > 0.5) potentially influence the cellular function, we performed correlation analysis to find the genes whose expression pattern was similar to that of HERV family (Spearman correlation coefficient > 0.3) and gene ontology (GO) enrichment analysis was used to identify pathways enriched by top 100 correlated genes.
HERV-gene interaction prediction
To predict the interactions between upregulated HERV loci and neighboring DEGs (within 500 kb) in GBC-derived cell types, we calculated the expression correlations between them. HERV locus and DEG were thought to be associated if HERV expression was positively correlated with DEG expression (Spearman correlation coefficient > 0.3).
Role of funders
The funders played no role in study design, data collection, data analyses, interpretation, or writing of report.
Results
Aberrant activation of HERVs in GBC
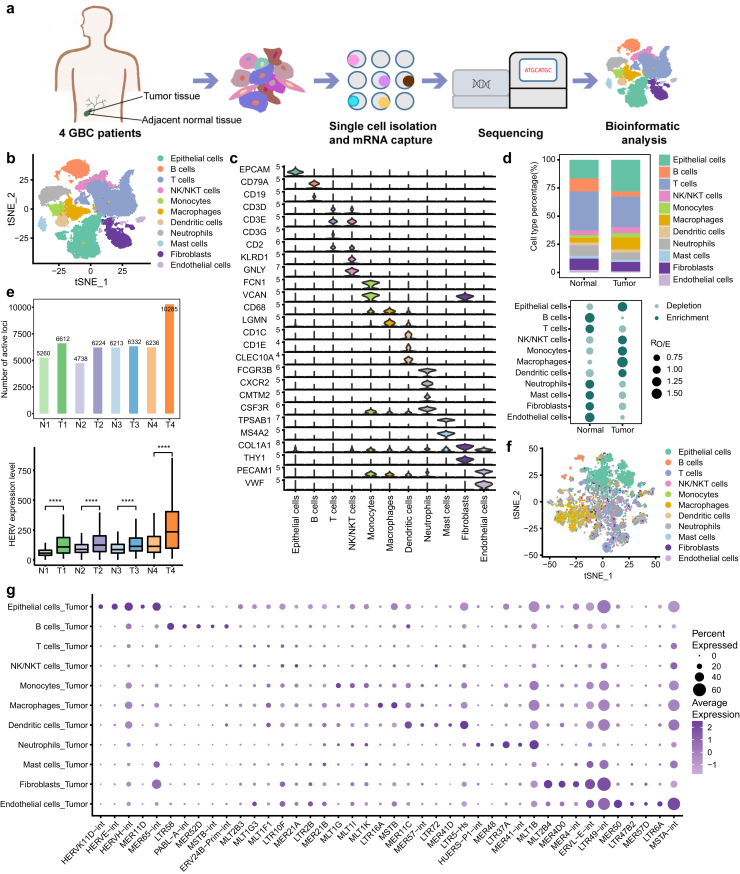
To investigate the cellular heterogeneity and characterize the molecular signature in GBC, tumor tissues and matched normal tissues were collected from 4 patients with gallbladder adenocarcinoma to perform scRNA-seq (Fig. 1a, Supplementary Table S2). After quality filtering, a total of 28,301 cells were obtained and catalogued into 11 main cell types annotated with canonical marker genes, including epithelial cells, B cells, T cells, natural killer (NK) or natural killer T (NKT) cells, monocytes, macrophages, dendritic cells (DCs), neutrophils, mast cells, fibroblasts and endothelial cells (Fig. 1b and c, Supplementary Figure S1a and b). T cells and epithelial cells were the most abundant cells in both tumor and adjacent normal tissues (Fig. 1d, Supplementary Figure S1c). Moreover, epithelial cells, monocytes and macrophages were predominantly enriched in tumor tissues.
Fig. 1.
Single-cell landscape and HERV expression patterns in GBC and adjacent normal tissues. a Overview of the study design. b tSNE projection of 28,301 single cells from tumor and adjacent normal tissues, color-coded by cell type. c Violin plot displaying expression of canonical marker genes for major cell types. d Relative proportion of major cell types in different tissue types (top). Dot plot showing the distribution of major cell types in different tissue types estimated by Ro/e,33 the ratio of observed to expected cell numbers in each cell type (bottom). e Comparison of the number of active HERV loci (top) and the overall expression of HERVs (bottom) in the matched samples. Statistical significance was evaluated by two-sided Wilcoxon rank sum test. f tSNE plot of all single cells from tumor tissues only based on the HERV expression. The top 1000 variable HERV loci were used. g Dot plot displaying expression of cell type-specific HERV families for major cell types.
We characterized the HERV expression patterns in patients with GBC and found that samples from GBC tissues had more active HERV loci and higher HERV expression levels than samples from adjacent normal tissues (Fig. 1e). The derepression of HERVs in tumor tissues implies that HERVs may play a role in the initiation and progression of GBC. Furthermore, dimension reduction analyses based on HERV expression alone showed that the cells with the same cell labels tended to cluster together, indicating that HERVs were actively transcribed in tumors in a cell type-specific manner (Fig. 1f). To identify which HERVs contribute to tumor heterogeneity in terms of cellular composition, the differential expression of all HERV families and HERV loci was calculated in each cell type (Fig. 1g, Supplementary File 3). Interestingly, HERVE, HERVK and HERVH, the most reported families in cancer research, were found to be mainly expressed in epithelial cells (Supplementary Figure S1d).
HERVs as enhancers potentially regulate adjacent DEGs
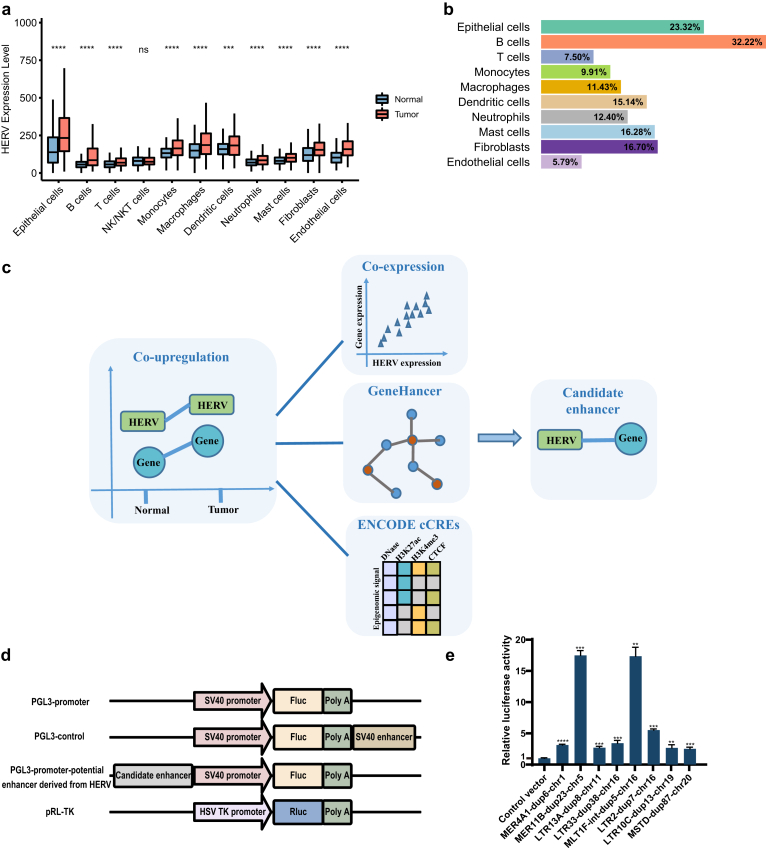
For all cell types except NK/NKT cells, HERVs were extensively activated in tumors compared to adjacent normal tissues and HERVs were expressed at the highest level in epithelial cells (Fig. 2a, Supplementary Figure S2a and b). Based on the criteria of log2FC > 0.5 and adjusted p-value < 0.05, only 4 significantly increased HERV loci were detected in T cells derived from tumors compared to T cells derived from adjacent normal tissues and only 5 HERV loci were significantly increased in GBC-derived DCs (Supplementary Figure S2c). Actually, for each cell type, the increment caused by significantly elevated HERVs loci accounted for only a small part of the total increment caused by all HERVs (Fig. 2b), suggesting that the HERV loci that have not passed the cut-off should also be considered by researchers. This also reminds us to explore the abnormal expression of HERVs in GBC at both the locus level and family level.
Fig. 2.
HERV-derived enhancers prediction. a Comparison of overall expression of HERVs between GBC and normal tissue cells for each major cell type. Statistical significance was evaluated by two-sided Wilcoxon rank sum test. b Contribution of significantly upregulated HERV loci to the increase in HERV expression. c Workflow of HERV-derived enhancers prediction. d Schematic of dual-luciferase reporter assay. The pGL3-Promoter Vector contained an SV40 promoter upstream of the luciferase gene. HERV containing putative enhancer elements can be inserted upstream of the promoter-luc+ transcriptional unit. PGL3-control containing SV40 enhancer sequences was used as a positive control. e Dual-luciferase reporter assay to test enhancer activity of the genomic fragments derived from HERV in GBC-SD. Data are shown as the means ± SD. ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001. Significance values for each comparison were calculated by Student's t-test. Use Welch's t-test when two groups don't have the same variance. At least three biological repeats were carried out.
The exaptation of HERVs as regulatory elements influencing the transcription of host genes has been established by many studies.25,34 To find HERVs which act as enhancers to drive the expression of the neighboring DEGs, scRNA-seq data for epithelial cells combined with other informative data were used to predict the possible links (Fig. 2c). First, we picked out the pairs of HERVs and neighboring DEGs in which both HERV loci and neighboring DEGs were upregulated in epithelial cells from GBC. Then, further filtering was performed based on the following criteria: 1) co-expression between HERV and neighboring DEG in GBC-derived epithelial cells; 2) predefined enhancer-gene link by GeneHancer database which predicted the associations between regulatory elements (enhancers and promoters) and target genes based on multiple information sources35; 3) HERV region showing enhancer-like signature supported by high DNase and H3K27ac with low H3K4me3 signal from ENCODE cCREs.36 As long as pairs of HERVs and neighboring DEGs met any two of the above criteria, HERVs were considered to be candidate enhancers modeling the expression of the neighboring DEGs. The predicted HERV-DEG pairs were provided in Supplementary File 4.
We selected some HERVs predicted by our pipeline to validate that the genomic fragments derived from HERVs had enhancer activity using dual-luciferase reporter assay (Fig. 2d and e, Supplementary Table S3). Interestingly, non-LTR sequence derived from HERV, such as MLT1F-int-dup5-chr16 (chr16:29915600-29916253), also showed significant enhancer activity. The neighboring genes, such as UCA1, SEPHS2 and DCTPP1, that are potentially regulated by HERV-derived enhancers, have been reported to play oncogenic roles in tumor proliferation and metastasis (Supplementary Table S4).37, 38, 39, 40 However, some neighboring genes, TMEM219 and YPEL3, mediate anti-tumor activities.41,42
HERVH expression is increased with the malignant transformation of epithelial cells
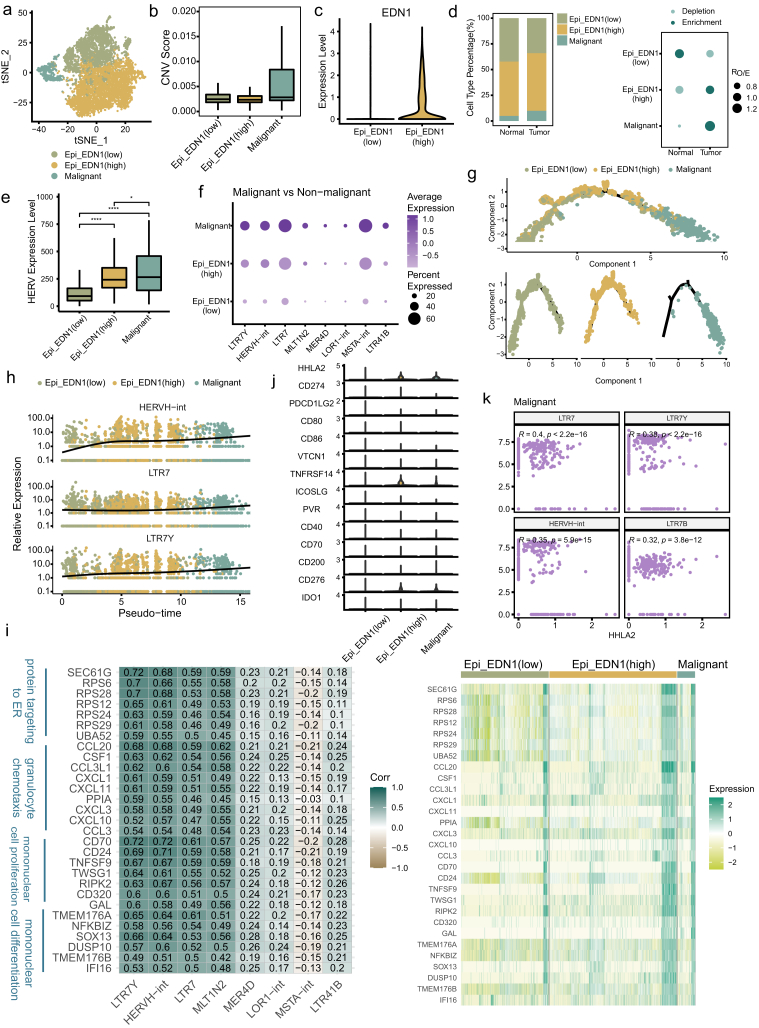
GBC originates from epithelial cells, which has been demonstrated by previous studies.43,44 To distinguish malignant and normal epithelial cells resident in GBC tissues, large-scale CNVs were inferred with stromal cells as references. The epithelial cells were divided into subclusters and the subclusters with markedly higher CNV scores were identified as malignant cells (Fig. 3a and b, Supplementary Figure S3a–c). The remaining non-malignant subclusters were further annotated as Epi_EDN1(low) and Epi_EDN1(high) according to the expression level of EDN1 (Fig. 3a and c). The Epi_EDN1(high) subtype, which simultaneously exhibited high expression of a mesenchymal marker (MMP7) and a cancer stem cell marker (CD44), was thought to represent an intermediate cell state (ICS) between normal and malignant cells (Supplementary Figure S3d) and the Epi_EDN1 (low) subtype was expected to represent normal epithelial cells. The Epi_EDN1 (high) cells at the middle stage and Epi_EDN1 (low) cells at the early stage in the transcriptional trajectory of malignant transformation confirmed these assumptions (Fig. 3g, Supplementary Figure S3h). Tumor and adjacent normal tissues contained all subtypes at different fractions (Fig. 3d, Supplementary Figure S3e). As expected, malignant cells were dominantly identified in tumors, whereas Epi_EDN1(low) cells were more enriched in adjacent normal tissues.
Fig. 3.
Identification of malignant cells and HERVH derepression in the process of malignant transformation. a tSNE projection of epithelial cells, color-coded by cell type. b Boxplot showing the CNV score for each epithelial cell subtype. c Violin plot displaying expression of EDN1 in two non-malignant subtypes. d Relative proportion of epithelial cell subtypes in different tissue types (left). Dot plot showing the distribution of epithelial cell subtypes in different tissue types estimated by Ro/e (right). e Comparison of overall expression of HERVs among epithelial cell subtypes. Statistical significance was evaluated by two-sided Wilcoxon rank sum test. f HERV families upregulated in malignant cells compared to non-malignant cells (adjusted p-value < 0.05 and log2FC > 0.5). g Differentiation trajectory of malignant and normal epithelial cells inferred by Monocle2, color-coded by cell type. h Dynamic expression changes of HERVH elements along the differentiation trajectory. i Heatmaps depicting expression correlation between gene and HERV family in malignant cells (left, Spearman correlation coefficient was also shown) and relative expression of these correlating genes across epithelial cell subtypes (right). j Violin plot displaying expression of the collected immune checkpoints in epithelial cell subtypes. k Scatter plot showing expression correlation between HHLA2 and HERVH element. Spearman correlation coefficient and two-tailed p-value were shown.
We compared the total expression level of HERVs among these subtypes and found that malignant cells exhibited the highest level (Fig. 3e). HERVs showed a higher signal of transcription in Epi_EDN1(high) than in Epi_EDN1(low), indicating that the activation of HERVs has already occurred in the epithelial cells at intermediate state before transforming into malignant cells. Next, we identified HERV families that showed differential expression in these subtypes. Strikingly, HERVH-associated elements (HERVH-int, LTR7Y) upregulated in Epi_EDN1(high) (vs. Epi_EDN1(low)) were further elevated in malignant cells (Fig. 3f, Supplementary Figure S3f, Supplementary File 5). At the locus level, 37/55 of the upregulated HERV loci in malignant cells (vs. non-malignant cells) belong to the HERVH family (Supplementary Figure S3g, Supplementary File 5). These results imply the functional importance of HERVH in GBC formation. To further explore the aberrant activation of HERVH, we constructed a transcriptional trajectory with the definitive malignant and non-malignant epithelial cells (Fig. 3g, Supplementary Figure S3h). Among the HERVs displaying transcriptional alterations with the tumor progression, HERVH-int, LTR7Y and LTR7 of the HERVH family showed the highest significance (Fig. 3h, Supplementary File 6), suggesting that HERVH may be a promising biomarker for early GBC diagnosis. To elucidate the impacts of HERVH derepression on this biological process, we identified the genes whose expression was correlated with the expression of these HERVH elements in malignant cells. These genes were highly enriched for GO terms related to protein targeting to ER, granulocyte chemotaxis, cell proliferation and differentiation (Fig. 3i, Supplementary Figure S3i). It is possible that the alteration of these genes’ expression was caused by the derepression of HERVH. HERVH has been reported to play a crucial role in pluripotency maintenance in human pluripotent stem cells25,45 and functions of HERVH in malignant cells revealed by us are consistent with that. In addition, we noticed that the gene UCA1 showed a strong association with HERVH-int and was specifically increased in malignant cells (Supplementary Figure S3j), which fit well with the fact that UCA1, which has been found to enhance the proliferation, migration and invasion of bladder cancer cell, is a lncRNA generated by HERVH elements.46
HERVs are highly valued in immunotherapy, such as immune checkpoint inhibitors, for their ability to sensitize tumor cells to immunological recognition.47 Human endogenous retrovirus-H long terminal repeat-associating 2 (HHLA2), a recently emerging immune checkpoint, is considered to be derived by HERVH.48 HHLA2 was thought to play a dual role in carcinogenesis: one for immunostimulation and another for immunosuppression.49 We examined the transcription of HHLA2 in epithelial cell subtypes, as well as some immune checkpoints potentially expressed in tumor cells (Fig. 3j). We found that HHLA2 expression was detectable in Epi_EDN1(high) (represent ICS) and malignant cells and was higher than that of PD-L1 (CD274), which is the most studied immune checkpoint. The weak signal for PD-L1 transcription may explain, at least in part, the limited efficacy of immunotherapy targeting PD-L1 in some patients with GBC. The elevated expression level of HHLA2 in intermediate and malignant cells and its dual role (one for immunostimulation and another for immunosuppression) in carcinogenesis suggest that HHLA2 may serve as a favorable candidate for GBC treatment. Further, we found an expressional correlation of HHLA2 and HERVH elements (Fig. 3k), implying that combining DNA demethylating agents that aim to induce HERVH activation with targeting HHLA2 therapy may be a promising therapeutic strategy.
Proliferative T cells show abnormally high expression of HERVs
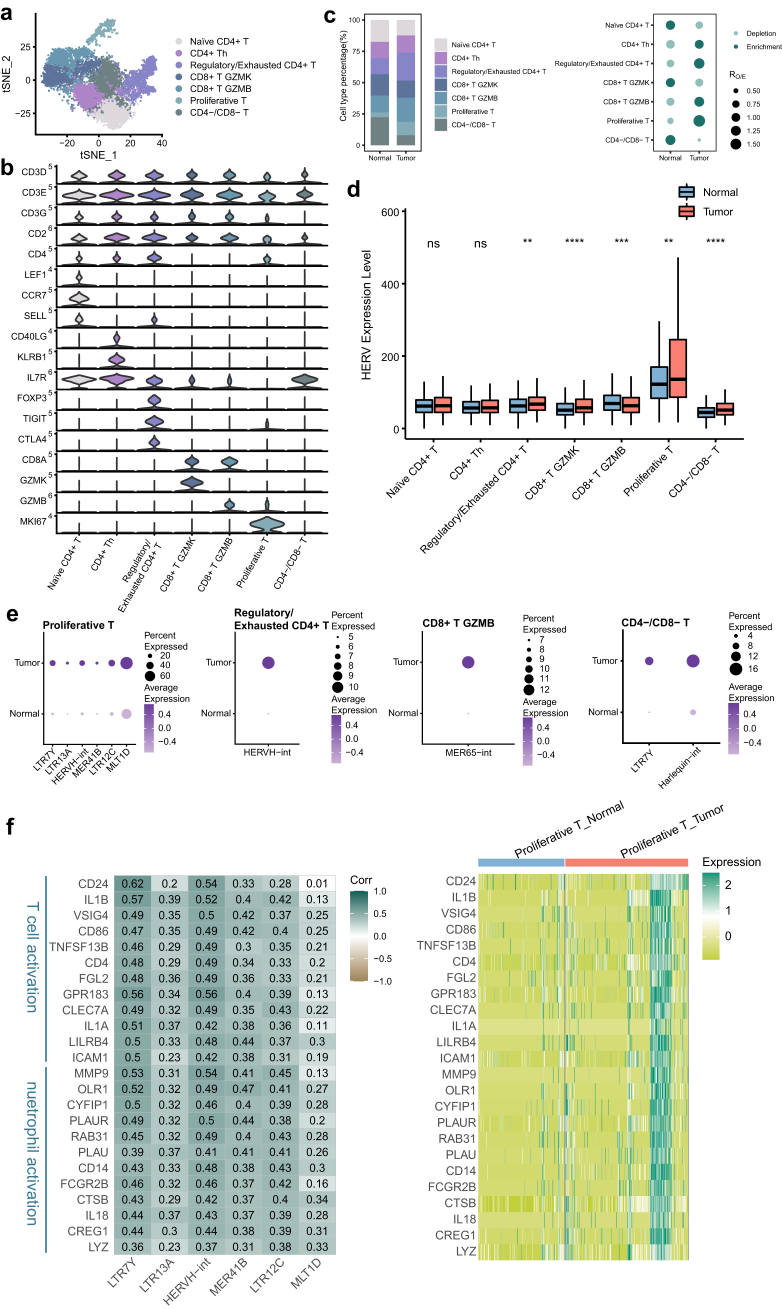
T cells display heterogeneity in cellular composition and functional states in the TME. Here, T cells were further partitioned into 7 distinct subpopulations annotated by marker genes, including naïve CD4+ T, CD4+ T helper (CD4+ Th), regulatory/exhausted CD4+ T, CD8+ T GZMK, CD8+ T GZMB, proliferative T and CD4−/CD8− T cells (Fig. 4a and b, Supplementary Figure S4a and b). Compared with the adjacent normal tissues, tumor tissues showed reduced proportions of naïve CD4+ T and CD4−/CD8− T cells and increased proportions of proliferative T and regulatory/exhausted CD4+ T cells (Fig. 4c, Supplementary Figure S4c). More regulatory/exhausted CD4+ T cells accumulated in tumor tissues, indicating a change from immune activation to immune suppression during tumor progression.
Fig. 4.
Proliferative T cells showing prominent expression levels of HERVs. a tSNE projection of T cells, color-coded by cell type. b Violin plot displaying expression of canonical marker genes for T cell subtypes. c Relative proportion of T cell subtypes in different tissue types (left). Dot plot showing the distribution of T cell subtypes in different tissue types estimated by Ro/e (right). d Comparison of overall expression of HERVs between GBC and normal tissue cells for each T cell subtype. Statistical significance was evaluated by two-sided Wilcoxon rank sum test. e HERV families upregulated in GBC-derived T cell subtypes (adjusted p-value < 0.05 and log2FC > 0.5). f Heatmaps depicting expression correlation between gene and HERV family in tumor-derived proliferative T cells (left, Spearman correlation coefficient was also shown) and relative expression of these correlating genes in all proliferative T cells (right).
HERVs were expressed at different levels in different subtypes of T cells. When we compared cells from tumor tissues with those from adjacent normal tissues, significant HERV transcription changes were observed in all T cell subtypes except naïve CD4+ T and CD4+ Th cells (Fig. 4d). Proliferative T cells were characterized by the prominent expression levels of HERVs in our data. HERVH-associated elements (LTR7Y and HERVH-int), LTR13A, MER41B, LTR12C and MLT1D were upregulated in proliferative T cells from tumor tissues (Fig. 4e). The top correlated genes with upregulated HERVH were associated with T cell activation (Fig. 4f, Supplementary Figure S4d). Strikingly, we observed that many genes involved in neutrophil behaviors, such as neutrophil activation and degranulation, were transcriptionally increased in tumor-derived proliferative T cells and displayed association with HERVH, MER41B and LTR12C. The association between neutrophil activity and one HERVK locus has been reported in peripheral blood mononuclear cells (PBMCs) from elderly people.50 Here, we hypothesized that the neutrophil state is regulated by proliferative T cells in GBC and that HERV may play an important role in this process.
Besides, high expression of HERVH-int in regulatory/exhausted CD4+ T cells, MER65-int in CD8+ T GZMB and LTR7Y and Harlequin-int in CD4−/CD8− T cells was also detected in tumor tissues (Fig. 4e).
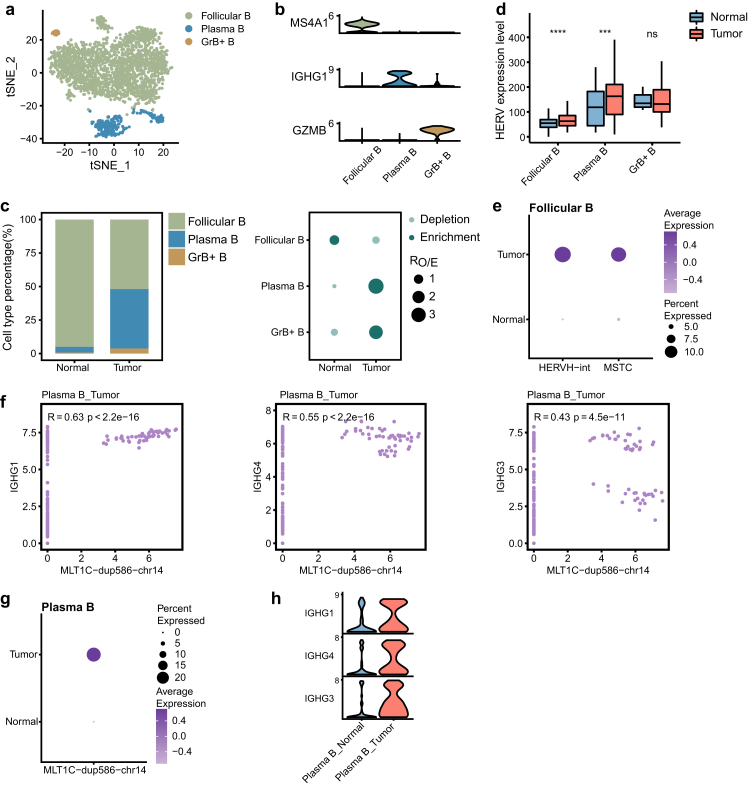
The association of IgG genes with the MLT1C locus in plasma B cells
B cells that infiltrate the TME play a multifaceted role in modulating the tumor immunity.51 Three major B cell subtypes, including follicular B cells (MS4A1), plasma B cells (IGHG1) and granzyme B-secreting B cells (GrB+ B cells, GZMB), were identified from our data based on the signature genes (Fig. 5a and b, Supplementary Figure S5a and b). The composition of these B cell subtypes was distinct between GBC and normal samples (Fig. 5c, Supplementary Figure S5c). The relative proportion of plasma B cells was observed to be increased in GBC, whereas follicular B cells were enriched in normal samples and comprised the majority of B cells in the adjacent normal tissues.
Fig. 5.
IgG genes are associated with the neighboring MLT1C locus in plasma B cells. a tSNE projection of B cells, color-coded by cell type. b Violin plot displaying expression of canonical marker genes for B cell subtypes. c Relative proportion of B cell subtypes in different tissue types (left). Dot plot showing the distribution of B cell subtypes in different tissue types estimated by Ro/e (right). d Comparison of overall expression of HERVs between GBC and normal tissue cells for each B cell subtype. Statistical significance was evaluated by two-sided Wilcoxon rank sum test. e HERV families upregulated in GBC-derived Follicular B cells (adjusted p-value < 0.05 and log2FC > 0.5). f Scatter plot showing expression correlation between the indicated gene and HERV locus. Spearman correlation coefficient and two-tailed p-value were shown. g Dot plot displaying expression level of the MLT1C locus in plasma B cells. h Violin plot displaying expression of IgG genes in plasma B cells.
Direct comparison of the expression level of HERVs in GBC and normal tissues for each B cell subtype revealed that tumor-derived follicular B and plasma B cells exhibited the increased abundance of HERV transcription (Fig. 5d). Compared with non-tumor-derived follicular B cell, cells in tumors showed higher expression levels of HERVH-int and MSTC (Fig. 5e). Plasma B cells, which are terminally differentiated B cells, can secrete antibodies that are an essential component of humoral immunity.52 Immunoglobulin G (IgG), a type of antibody, is produced and released by plasma B cells. Through our analysis, we discovered a transcriptional association between one MLT1C locus (MLT1C-dup586-chr14, chr14:105750174-105750614) and neighboring IgG genes (IGHG1, IGHG4 and IGHG3) (Fig. 5f). The expression association and upregulation of both them led us to speculate that MLT1C-dup586-chr14 may be a regulator that can promote the expression of these neighboring IgG genes (Fig. 5g and h).
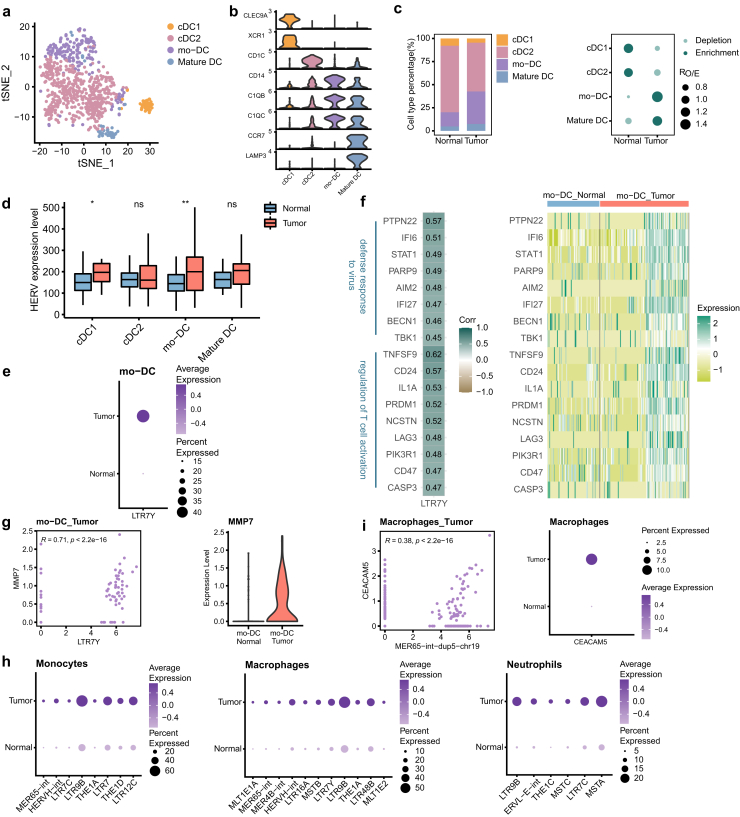
Overexpression of HERVH in all myeloid cell types
Emerging evidence emphasizes the key role of myeloid cells in modulating cancer progression.53 In our study, the myeloid cells were separated into monocytes, macrophages, DCs and neutrophils (Fig. 1b). DCs were further categorized into conventional DC (cDC1 and cDC2), monocyte-derived DC (mo–DC) and mature DC based on the prominent expression markers (Fig. 6a and b, Supplementary Figure S6a and b). For both tumor and adjacent tissues, cDC2 and mo-DC accounted for the majority of DCs. Notably, mo-DCs and mature DCs were enriched in GBC tissues, while cDC1s and cDC2s were depleted (Fig. 6c, Supplementary Figure S6c).
Fig. 6.
The activation of HERVH in myeloid cells. a tSNE projection of DCs, color-coded by cell type. b Violin plot displaying expression of canonical marker genes for DC subtypes. c Relative proportion of DC subtypes in different tissue types (left). Dot plot showing the distribution of DC subtypes in different tissue types estimated by Ro/e (right). d Comparison of overall expression of HERVs between GBC and normal tissue cells for each DC subtype. Statistical significance was evaluated by two-sided Wilcoxon rank sum test. e HERV families upregulated in GBC-derived mo-DCs (adjusted p-value < 0.05 and log2FC > 0.5). f Heatmaps depicting expression correlation between gene and HERV family in tumor-derived mo-DCs (left, Spearman correlation coefficient was also shown) and relative expression of these correlating genes in all mo-DCs (right). g Scatter plot showing expression correlation between the indicated gene and HERV family (left). Spearman correlation coefficient and two-tailed p-value were shown. Violin plot displaying expression of gene MMP7 in mo-DCs (right). h HERV families upregulated in GBC-derived myeloid cell subtypes (adjusted p-value < 0.05 and log2FC > 0.5). i Scatter plot showing expression correlation between the indicated gene and HERV locus (left). Spearman correlation coefficient and two-tailed p-value were shown. Dot plot displaying expression of gene CEACAM5 in macrophages (right).
HERV expression patterns were different among DC subtypes (Fig. 6d). cDC1s and mo-DCs in GBC showed significant elevations in HERV expression, but no significant differences were detected in cDC2s and mature DCs. Mo-DCs are believed to arise from monocytes in the context of inflammation or infection and this subpopulation can induce T cell activation in various tumor models.54,55 We found that LTR7Y were overexpressed in mo-DCs derived from GBC tissues (Fig. 6e). Genes presenting similar expression patterns with LTR7Y in mo-DCs were mainly implicated in antiviral response and T cell activation (Fig. 6f, Supplementary Figure S6d), suggesting that the enhancement of mo-DCs’ antiviral and inflammatory abilities may be regulated by LTR7Y. What's more, LTR7Y exhibited strong association with gene MMP7 (Fig. 6g), which functions as an oncogenic factor to mediate occurrence and progression of several types of cancers.56 This association was also seen in malignant cells, proliferative T cells and macrophages, which further supports the hypothesis that elevated MMP7 level in these cells may be associated with the activation of LTR7Y (Supplementary Figure S6e).
The significant increases in expression of HERVH elements were also observed in other myeloid cells, monocytes (HERVH-int, LTR7C and LTR7), macrophages (HERVH-int and LTR7Y) and neutrophils (LTR7C) (Fig. 6h). Genes associated with HERVH elements in tumor-derived monocytes and macrophages were mostly involved in immune cell activation and differentiation (Supplementary Figure S6d). These biological processes play key roles in TME. Here, we speculated that these profound perturbations occurred in myeloid cells might be related to the abnormal transcription of HERVH elements. Taken together, we discovered that HERVH elements were upregulated in almost all myeloid cell types and they may have similar impacts on myeloid cell behavior and function.
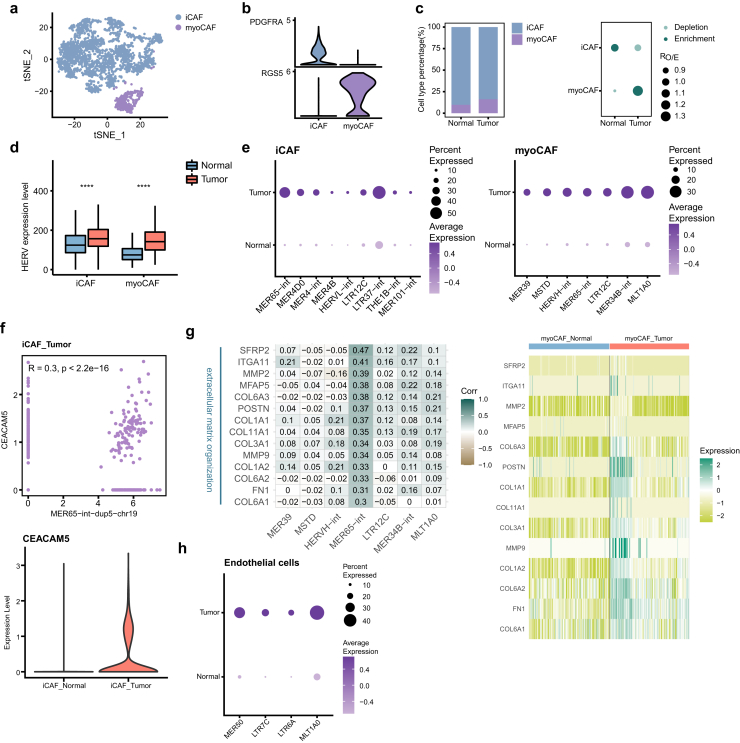
MER65-int is activated in fibroblasts and has an expressional correlation with ECM genes
Cancer-associated fibroblasts (CAFs) are one of the most dominant components of the tumor stroma and have heterogeneous phenotypes and functions. A large number of studies support that CAFs can promote tumor migration and invasion through various mechanisms, such as remodelling the extracellular matrix (ECM) and modulating the tumor immune system.57 Sub-clustering of fibroblasts revealed 2 main distinct subtypes, including inflammatory CAFs (iCAFs; FDGFRA) and myo-cancer-associated fibroblasts (myoCAFs; RGS5) (Fig. 7a and b, Supplementary Figure S7a and b). In both GBC tissues and adjacent normal tissues, iCAFs were the predominant cell type. Compared to that in normal tissues, the proportion of myoCAFs in GBC tissues was increased, whereas the level of iCAFs was slightly decreased (Fig. 7c, Supplementary Figure S7c).
Fig. 7.
The activation of MER65-int in fibroblasts. a tSNE projection of fibroblasts, color-coded by cell type. b Violin plot displaying expression of canonical marker genes for fibroblast subtypes. c Relative proportion of fibroblast subtypes in different tissue types (left). Dot plot showing the distribution of fibroblast subtypes in different tissue types estimated by Ro/e (right). d Comparison of overall expression of HERVs between GBC and normal tissue cells for each fibroblast subtype. Statistical significance was evaluated by two-sided Wilcoxon rank sum test. e HERV families upregulated in GBC-derived fibroblast subtypes (adjusted p-value < 0.05 and log2FC > 0.5). f Scatter plot showing expression correlation between the indicated gene and HERV locus (top). Spearman correlation coefficient and two-tailed p-value were shown. Violin plot displaying expression of the indicated gene in iCAFs (bottom). g Heatmaps depicting expression correlation between gene and HERV family in tumor-derived myoCAFs (left, Spearman correlation coefficient was also shown) and relative expression of these correlating genes in all myoCAFs (right). h HERV families upregulated in GBC-derived endothelial cells (adjusted p-value < 0.05 and log2FC > 0.5).
Regarding the expression level of HERVs, both iCAFs and myoCAFs in tumors presented significant elevation (Fig. 7d). HERVs that were abnormally activated in GBC were screened (Fig. 7e, Supplementary File 5). For iCAFs, MER65-int, MER4D0 and MER4-int are the top upregulated HERVs. At the locus level, MER65-int-dup5-chr19 (chr19:41729515-41730138) was associated with its neighboring gene CEACAM5 in iCAFs (Fig. 7f). CEACAM5, encoding carcinoembryonic antigen (CEA), has been used as a tumor biomarker in clinical detection.58 After checking their locations, we found that part of the MER65-int-dup5-chr19 sequence serves as the exon of CEACAM5. Besides, this association was also observed in GBC-derived macrophages (Fig. 6i). Whether the transcription of this MER65-int element induces the high expression of CEACAM5 in iCAFs and macrophages from GBC tissues deserves further investigation. For myoCAFs, MER65-int specifically activated in GBC-derived cells exhibited an association with genes related to ECM organization (Fig. 7g, Supplementary Figure S7d). This suggests the possibility that active MER65-int may be involved in dysregulation of ECM genes.
In endothelial cells from GBC tissues, MER50, LTR7C, LTR6A and MLT1A0 were upregulated (Fig. 7h). However, how these HERV families affect cellular functions is not clear.
Discussion
GBC might not be detected until it's advanced and catching the cancer early will benefit patient survival. The prognosis of GBC is poor and only some patients exhibited desired outcome. Therefore, the pursuit for early diagnostic biomarkers and more effective treatment strategies for GBC is ongoing. HERVs have received a lot of attention because of their strong association with cancer development and immunotherapy. In this study, we determined the cellular composition and presented a comprehensive single-cell transcriptional profile of HERVs for GBC tissues, highlighting that HERVs were transcribed in a cell type-specific manner. We found that HERVs, as enhancers, have the potential to alter host gene expression and further change the characteristics of tumors. We also suggested that HERVH may be a candidate for early GBC diagnosis and targeting HHLA2 might be an appealing strategy for GBC treatment. Furthermore, for each cell type, we explored which biological functions are potentially affected by various HERV families. Our study could provide a framework for future discoveries of functional HERVs as molecular and cellular therapeutic targets for GBC.
Here, dual-luciferase reporter assay was applied to demonstrate that some HERVs can act as enhancers. HERVs also work as alternative promoters, resulting in transcriptional initiation of host genes, including cancer-related genes.59 In addition to acting as transcriptional regulators, HERV generate noncoding RNA (ncRNA) or protein product to modulate the gene regulatory network, as a perpetrator or protector in carcinogenesis.5,60 These functions were not explained in our study, but deserve to be further explored. Within each cell population, we identified biological functions that may be biased by the aberrantly expressed HERVs, but whether and by which mechanisms distinct HERVs affect cellular behavior remain to be investigated.
The widespread reactivation of HERVs has been linked to some malignancies, suggesting their potential to help cancer screen.61 In our study, the expression of HERVH was progressively elevated with malignant transformation of epithelial cells and displayed the highest significance compared to other HERV families, leading us to speculate that HERVH may be an early indicator of GBC. The role of HERVH as biomarker has been discussed in several cancers, including colorectal carcinoma, prostate cancer and lung cancer.62, 63, 64 Further experiments, at the nucleic acid or protein level, are needed to verify whether HERVH is an effective diagnostic biomarker. HHLA2, a newly discovered immune checkpoint belonging to B7 family, is thought to be derived by HERVH.48 The expression of HHLA2 was detected in a subset of intermediate and malignant cells, which is higher than that of some hot immune checkpoints such as PD-L1. Considering the fact that there are substantial differences in expression of immune checkpoints among diverse cancers and even in the same type of cancer, the expression profile of immune checkpoints varies from patient to patient, although the expression of HHLA2 was modest in our data, it might be an ideal immunotherapeutic target for a specific group of GBC patients. The family diversity of HERVs contributes to their functional complexity in cells. The discovery of the association of HHLA2 with HERVH indicates that epigenetic agents specially tailored to target HERVH should be a research priority in combined epigenetic and immune therapy.
The fact that HERVs are present in multiple copies within the human genome poses a challenge for quantification.65 A common strategy used in routine analysis is to keep only reads that uniquely map to the HERV loci. The advantage of this approach is that the expressional signal for each locus can be obtained, but the disadvantage is that it tends to underestimate the transcript level of HERVs, especially evolutionarily young HERVs. Another approach, using the “multi-mapper” strategy, largely preserves the HERV-derived reads, but the transcriptional information of each HERV locus is lost.66,67 A number of computational tools, such as RepEnrich, TEtranscripts, REdiscoverTE and so on, are produced to quantify transposable element (TE, HERV belongs to TE) expression for bulk RNA-seq data and the best choice of the strategy should be guided by the specific biological question.65,68, 69, 70 Nevertheless, current resources available for TE quantification at single-cell resolution are relatively limited, although scRNA-seq technology opens the possibility to investigate TE transcription variability among different cell populations, factors driving such diversity and cellular phenotypes influenced by TE derepression.24 Pioneering pipelines, including scTE71 and a framework with assembled transcripts,72 have been generated to report the TE expression at single-cell resolution, but they both have certain limitations to application, namely counting only at the family level and counting only assembled transcripts respectively. In this study, we considered using uniquely mapping reads to achieve TE quantification to ensure mapping accuracy and obtain locus information. As the biological significance of HERVs is becoming understood, we believe that there will be a rapid advance in scRNA-seq computational pipelines tailored for HERV to be compatible with a wide variety of scRNA-seq protocols.
In summary, our work highlights the functional role of HERVs in GBC and provides a new resource for cancer diagnosis and management.
Contributors
J.W., X.J. and J.C. conceived the research. J.C. directed the analysis. J.W. and M.R. performed the analysis. J.W. and M.R. contributed equally. J.Y., M.H., X.W. and W.M. conducted the experiments. All the authors discussed the results and wrote the paper. J.C., J.W. and M.R. have verified the underlying data. All authors read and approved the final version of the manuscript.
Data sharing statement
Raw single-cell RNA sequencing data reported in this study are available in Genome Sequence Archive for Human (GSA-Human) under accession number HRA001917.
Declaration of interests
The authors declare that they have no competing interests.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (31970176, 81972256) and the research grants from the Innovation Capacity Building Project of Jiangsu province (BM2020019).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2022.104319.
Contributor Information
Xiaoqing Jiang, Email: jxq1225@sina.com.
Jie Cui, Email: jcui@ips.ac.cn.
Appendix A. Supplementary data
References
- 1.Canale M., Monti M., Rapposelli I.G., et al. Molecular targets and emerging therapies for advanced gallbladder cancer. Cancers (Basel) 2021;13(22):5671. doi: 10.3390/cancers13225671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baiu I., Visser B. Gallbladder cancer. JAMA. 2018;320(12):1294. doi: 10.1001/jama.2018.11815. [DOI] [PubMed] [Google Scholar]
- 3.De Lorenzo S., Garajova I., Stefanini B., Tovoli F. Targeted therapies for gallbladder cancer: an overview of agents in preclinical and clinical development. Expert Opin Investig Drugs. 2021;30(7):759–772. doi: 10.1080/13543784.2021.1928636. [DOI] [PubMed] [Google Scholar]
- 4.Song X., Hu Y., Li Y., Shao R., Liu F., Liu Y. Overview of current targeted therapy in gallbladder cancer. Signal Transduct Target Ther. 2020;5(1):230. doi: 10.1038/s41392-020-00324-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang M., Liang J.Q., Zheng S. Expressional activation and functional roles of human endogenous retroviruses in cancers. Rev Med Virol. 2019;29(2) doi: 10.1002/rmv.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lander E.S., Linton L.M., Birren B., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 7.Alcazer V., Bonaventura P., Depil S. Human endogenous retroviruses (HERVs): shaping the innate immune response in cancers. Cancers. 2020;12(3):610. doi: 10.3390/cancers12030610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petrizzo A., Ragone C., Cavalluzzo B., et al. Human endogenous retrovirus reactivation: implications for cancer immunotherapy. Cancers. 2021;13(9):1999. doi: 10.3390/cancers13091999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steiner M.C., Marston J.L., Iniguez L.P., et al. Locus-specific characterization of human endogenous retrovirus expression in prostate, breast, and colon cancers. Cancer Res. 2021;81(13):3449–3460. doi: 10.1158/0008-5472.CAN-20-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito J., Kimura I., Soper A., et al. Endogenous retroviruses drive KRAB zinc-finger protein family expression for tumor suppression. Sci Adv. 2020;6(43):eabc3020. doi: 10.1126/sciadv.abc3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brocks D., Schmidt C.R., Daskalakis M., et al. Erratum: DNMT and HDAC inhibitors induce cryptic transcription start sites encoded in long terminal repeats. Nat Genet. 2017;49(11):1661. doi: 10.1038/ng1117-1661c. [DOI] [PubMed] [Google Scholar]
- 12.Wolff F., Leisch M., Greil R., Risch A., Pleyer L. The double-edged sword of (re)expression of genes by hypomethylating agents: from viral mimicry to exploitation as priming agents for targeted immune checkpoint modulation. Cell Commun Signal. 2017;15(1):13. doi: 10.1186/s12964-017-0168-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang-Johanning F., Radvanyi L., Rycaj K., et al. Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients. Cancer Res. 2008;68(14):5869–5877. doi: 10.1158/0008-5472.CAN-07-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schiavetti F., Thonnard J., Colau D., Boon T., Coulie P.G. A human endogenous retroviral sequence encoding an antigen recognized on melanoma by cytolytic T lymphocytes. Cancer Res. 2002;62(19):5510–5516. [PubMed] [Google Scholar]
- 15.Mullins C.S., Linnebacher M. Endogenous retrovirus sequences as a novel class of tumor-specific antigens: an example of HERV-H env encoding strong CTL epitopes. Cancer Immunol Immunother. 2012;61(7):1093–1100. doi: 10.1007/s00262-011-1183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith C.C., Beckermann K.E., Bortone D.S., et al. Endogenous retroviral signatures predict immunotherapy response in clear cell renal cell carcinoma. J Clin Invest. 2018;128(11):4804–4820. doi: 10.1172/JCI121476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saini S.K., Orskov A.D., Bjerregaard A.M., et al. Human endogenous retroviruses form a reservoir of T cell targets in hematological cancers. Nat Commun. 2020;11(1):5660. doi: 10.1038/s41467-020-19464-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Attermann A.S., Bjerregaard A.M., Saini S.K., Gronbaek K., Hadrup S.R. Human endogenous retroviruses and their implication for immunotherapeutics of cancer. Ann Oncol. 2018;29(11):2183–2191. doi: 10.1093/annonc/mdy413. [DOI] [PubMed] [Google Scholar]
- 19.Chiappinelli K.B., Strissel P.L., Desrichard A., et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162(5):974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goel S., DeCristo M.J., Watt A.C., et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature. 2017;548(7668):471–475. doi: 10.1038/nature23465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez R.K., Gordon M.G., Subramaniam M., et al. Single-cell RNA-seq reveals cell type-specific molecular and genetic associations to lupus. Science. 2022;376(6589) doi: 10.1126/science.abf1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X., Miao J., Wang S., et al. Single-cell RNA-seq reveals the genesis and heterogeneity of tumor microenvironment in pancreatic undifferentiated carcinoma with osteoclast-like giant-cells. Mol Cancer. 2022;21(1):133. doi: 10.1186/s12943-022-01596-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hornburg M., Desbois M., Lu S., et al. Single-cell dissection of cellular components and interactions shaping the tumor immune phenotypes in ovarian cancer. Cancer Cell. 2021;39(7):928–944.e6. doi: 10.1016/j.ccell.2021.04.004. [DOI] [PubMed] [Google Scholar]
- 24.O'Neill K., Brocks D., Hammell M.G. Mobile genomics: tools and techniques for tackling transposons. Philos Trans R Soc Lond B Biol Sci. 2020;375(1795) doi: 10.1098/rstb.2019.0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J., Xie G., Singh M., et al. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature. 2014;516(7531):405–409. doi: 10.1038/nature13804. [DOI] [PubMed] [Google Scholar]
- 26.Goke J., Lu X., Chan Y.S., et al. Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell. 2015;16(2):135–141. doi: 10.1016/j.stem.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 27.Dobin A., Davis C.A., Schlesinger F., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Arensbergen J., van Steensel B., Bussemaker H.J. In search of the determinants of enhancer-promoter interaction specificity. Trends Cell Biol. 2014;24(11):695–702. doi: 10.1016/j.tcb.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marbach D., Lamparter D., Quon G., Kellis M., Kutalik Z., Bergmann S. Tissue-specific regulatory circuits reveal variable modular perturbations across complex diseases. Nat Methods. 2016;13(4):366–370. doi: 10.1038/nmeth.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corces M.R., Granja J.M., Shams S., et al. The chromatin accessibility landscape of primary human cancers. Science. 2018;362(6413):eaav1898. doi: 10.1126/science.aav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu X., Mao Q., Tang Y., et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017;14(10):979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo X., Zhang Y., Zheng L., et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med. 2018;24(7):978–985. doi: 10.1038/s41591-018-0045-3. [DOI] [PubMed] [Google Scholar]
- 34.Chuong E.B., Elde N.C., Feschotte C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science. 2016;351(6277):1083–1087. doi: 10.1126/science.aad5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fishilevich S., Nudel R., Rappaport N., et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford) 2017;2017:bax028. doi: 10.1093/database/bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Consortium E.P., Moore J.E., Purcaro M.J., et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 2020;583(7818):699–710. doi: 10.1038/s41586-020-2493-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xuan W., Yu H., Zhang X., Song D. Crosstalk between the lncRNA UCA1 and microRNAs in cancer. FEBS Lett. 2019;593(15):1901–1914. doi: 10.1002/1873-3468.13470. [DOI] [PubMed] [Google Scholar]
- 38.Carlisle A.E., Lee N., Matthew-Onabanjo A.N., et al. Selenium detoxification is required for cancer-cell survival. Nat Metab. 2020;2(7):603–611. doi: 10.1038/s42255-020-0224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nunziata C., Polo A., Sorice A., et al. Structural analysis of human SEPHS2 protein, a selenocysteine machinery component, over-expressed in triple negative breast cancer. Sci Rep. 2019;9(1) doi: 10.1038/s41598-019-52718-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu J., Dong W., He H., et al. Autophagy induced by overexpression of DCTPP1 promotes tumor progression and predicts poor clinical outcome in prostate cancer. Int J Biol Macromol. 2018;118(Pt A):599–609. doi: 10.1016/j.ijbiomac.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 41.Cai Q., Dozmorov M., Oh Y. IGFBP-3/IGFBP-3 receptor system as an anti-tumor and anti-metastatic signaling in cancer. Cells. 2020;9(5):1261. doi: 10.3390/cells9051261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J., Wen X., Ren X.Y., et al. Correction to: YPEL3 suppresses epithelial-mesenchymal transition and metastasis of nasopharyngeal carcinoma cells through the Wnt/beta-catenin signaling pathway. J Exp Clin Cancer Res. 2021;40(1):400. doi: 10.1186/s13046-021-02189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hundal R., Shaffer E.A. Gallbladder cancer: epidemiology and outcome. Clin Epidemiol. 2014;6:99–109. doi: 10.2147/CLEP.S37357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Misra S., Chaturvedi A., Misra N.C., Sharma I.D. Carcinoma of the gallbladder. Lancet Oncol. 2003;4(3):167–176. doi: 10.1016/s1470-2045(03)01021-0. [DOI] [PubMed] [Google Scholar]
- 45.Fort A., Hashimoto K., Yamada D., et al. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat Genet. 2014;46(6):558–566. doi: 10.1038/ng.2965. [DOI] [PubMed] [Google Scholar]
- 46.Wang F., Li X., Xie X., Zhao L., Chen W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett. 2008;582(13):1919–1927. doi: 10.1016/j.febslet.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 47.Wang L., Amoozgar Z., Huang J., et al. Decitabine enhances lymphocyte migration and function and synergizes with CTLA-4 blockade in a murine ovarian cancer model. Cancer Immunol Res. 2015;3(9):1030–1041. doi: 10.1158/2326-6066.CIR-15-0073. [DOI] [PubMed] [Google Scholar]
- 48.Mager D.L., Hunter D.G., Schertzer M., Freeman J.D. Endogenous retroviruses provide the primary polyadenylation signal for two new human genes (HHLA2 and HHLA3) Genomics. 1999;59(3):255–263. doi: 10.1006/geno.1999.5877. [DOI] [PubMed] [Google Scholar]
- 49.Ying H., Xu J., Zhang X., Liang T., Bai X. Human endogenous retrovirus-H long terminal repeat-associating 2: the next immune checkpoint for antitumour therapy. EBioMedicine. 2022;79 doi: 10.1016/j.ebiom.2022.103987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Autio A., Nevalainen T., Mishra B.H., Jylha M., Flinck H., Hurme M. Effect of aging on the transcriptomic changes associated with the expression of the HERV-K (HML-2) provirus at 1q22. Immun Ageing. 2020;17:11. doi: 10.1186/s12979-020-00182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sarvaria A., Madrigal J.A., Saudemont A. B cell regulation in cancer and anti-tumor immunity. Cell Mol Immunol. 2017;14(8):662–674. doi: 10.1038/cmi.2017.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nutt S.L., Hodgkin P.D., Tarlinton D.M., Corcoran L.M. The generation of antibody-secreting plasma cells. Nat Rev Immunol. 2015;15(3):160–171. doi: 10.1038/nri3795. [DOI] [PubMed] [Google Scholar]
- 53.Engblom C., Pfirschke C., Pittet M.J. The role of myeloid cells in cancer therapies. Nat Rev Cancer. 2016;16(7):447–462. doi: 10.1038/nrc.2016.54. [DOI] [PubMed] [Google Scholar]
- 54.Kuhn S., Yang J., Ronchese F. Monocyte-derived dendritic cells are essential for CD8(+) T cell activation and antitumor responses after local immunotherapy. Front Immunol. 2015;6:584. doi: 10.3389/fimmu.2015.00584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chow K.V., Lew A.M., Sutherland R.M., Zhan Y. Monocyte-derived dendritic cells promote Th polarization, whereas conventional dendritic cells promote Th proliferation. J Immunol. 2016;196(2):624–636. doi: 10.4049/jimmunol.1501202. [DOI] [PubMed] [Google Scholar]
- 56.Liao H.Y., Da C.M., Liao B., Zhang H.H. Roles of matrix metalloproteinase-7 (MMP-7) in cancer. Clin Biochem. 2021;92:9–18. doi: 10.1016/j.clinbiochem.2021.03.003. [DOI] [PubMed] [Google Scholar]
- 57.Sahai E., Astsaturov I., Cukierman E., et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer. 2020;20(3):174–186. doi: 10.1038/s41568-019-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berg K.C.G., Eide P.W., Eilertsen I.A., et al. Multi-omics of 34 colorectal cancer cell lines - a resource for biomedical studies. Mol Cancer. 2017;16(1):116. doi: 10.1186/s12943-017-0691-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lamprecht B., Walter K., Kreher S., et al. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat Med. 2010;16(5):571–579. doi: 10.1038/nm.2129. 1p following 9. [DOI] [PubMed] [Google Scholar]
- 60.Bannert N., Hofmann H., Block A., Hohn O. HERVs new role in cancer: from accused perpetrators to cheerful protectors. Front Microbiol. 2018;9:178. doi: 10.3389/fmicb.2018.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao Y., Yu X.F., Chen T. Human endogenous retroviruses in cancer: expression, regulation and function. Oncol Lett. 2021;21(2):121. doi: 10.3892/ol.2020.12382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perot P., Mullins C.S., Naville M., et al. Expression of young HERV-H loci in the course of colorectal carcinoma and correlation with molecular subtypes. Oncotarget. 2015;6(37):40095–40111. doi: 10.18632/oncotarget.5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Manca M.A., Solinas T., Simula E.R., et al. HERV-K and HERV-H env proteins induce a humoral response in prostate cancer patients. Pathogens. 2022;11(1):95. doi: 10.3390/pathogens11010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zare M., Mostafaei S., Ahmadi A., et al. Human endogenous retrovirus env genes: potential blood biomarkers in lung cancer. Microb Pathog. 2018;115:189–193. doi: 10.1016/j.micpath.2017.12.040. [DOI] [PubMed] [Google Scholar]
- 65.Lanciano S., Cristofari G. Measuring and interpreting transposable element expression. Nat Rev Genet. 2020;21(12):721–736. doi: 10.1038/s41576-020-0251-y. [DOI] [PubMed] [Google Scholar]
- 66.Treangen T.J., Salzberg S.L. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet. 2011;13(1):36–46. doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goerner-Potvin P., Bourque G. Computational tools to unmask transposable elements. Nat Rev Genet. 2018;19(11):688–704. doi: 10.1038/s41576-018-0050-x. [DOI] [PubMed] [Google Scholar]
- 68.Criscione S.W., Zhang Y., Thompson W., Sedivy J.M., Neretti N. Transcriptional landscape of repetitive elements in normal and cancer human cells. BMC Genomics. 2014;15:583. doi: 10.1186/1471-2164-15-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jin Y., Tam O.H., Paniagua E., Hammell M. TEtranscripts: a package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics. 2015;31(22):3593–3599. doi: 10.1093/bioinformatics/btv422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kong Y., Rose C.M., Cass A.A., et al. Transposable element expression in tumors is associated with immune infiltration and increased antigenicity. Nat Commun. 2019;10(1):5228. doi: 10.1038/s41467-019-13035-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.He J., Babarinde I.A., Sun L., et al. Identifying transposable element expression dynamics and heterogeneity during development at the single-cell level with a processing pipeline scTE. Nat Commun. 2021;12(1):1456. doi: 10.1038/s41467-021-21808-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shao W., Wang T. Transcript assembly improves expression quantification of transposable elements in single-cell RNA-seq data. Genome Res. 2021;31(1):88–100. doi: 10.1101/gr.265173.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.