Abstract
Shigella sonnei is the most common Shigella spp. in developed areas and the second most common in undeveloped regions. In this study, a multiple cross displacement amplification (MCDA) assay was used in combination with a lateral flow biosensor (LFB) assay to detect virulent S. sonnei strains containing the ipaH and wbgX genes. The multiplex MCDA-LFB assay detected wbgX at ≥1 pg/μL and ipaH at ≥10 fg/μL within 30 min in pure cultures maintained at 63°C. This assay was sensitive for ~37 CFU of virulent S. sonnei and ~3.7 CFU of Shigella spp. and enteroinvasive E. coli in stimulated fecal samples and had 100% specificity among 59 reference strains. The MCDA-LFB assay was also able to differentiate between virulent S. sonnei and other Shigella spp. and enteroinvasive E. coli among 99 clinical isolates. In summary, a multiplex MCDA-LFB assay was developed for rapid, convenient, point-of-care, and accurate identification of virulent S. sonnei within 30 min and at a constant temperature without the need for expensive lab equipment.
Keywords: virulent shigella sonnei, multiplex cross displacement amplification, lateral flow biosensor, sensitivity, specificity
Introduction
Shigella spp. is the major pathogen responsible for bacterial dysentery in both developed and developing countries, causing disease in approximately 190 million people and leading to 65,000 deaths per year (Kirk et al., 2015). Based on its biochemical and antigen characteristics, Shigella spp. is separated into four species/serogroups, S. dysenteriae, S. flexneri, S. boydii, and S. sonnei. S. flexneri and S. sonnei are the predominant Shigella spp. worldwide and S. sonnei is the main cause of shigellosis in developed areas, and the second most common in undeveloped regions (Thompson et al., 2015). Using 2004–2014 data from the National Infectious Disease Information Reporting System (NIDRS), 3,342,847 cases of bacillary dysentery were reported in China, with an average incidence rate of 22.89 cases per 100,000 person-years. S. flexneri was the most prevalent species followed by S. sonnei (2,191/6,278 isolates, 34.89%) (Chang et al., 2016).
Traditional Shigella spp. diagnosis is dependent on culture and serum agglutination, which are inefficient, expensive, and only able to detect a small proportion of actual shigellosis cases. The culture positivity rate of Shigella spp., which can cause infection with as few as 10–100 bacterial cells, depends on the number of living strains in food, fecal and environmental samples (Octavia and Lan, 2015). The conditions of sample collection and transportation, including temperature, pH, the use of antibiotics prior to specimen collection, and competition from other commensal organisms, can impact the success of a culture (Taylor and Schelhart, 1975; Vu et al., 2004). Molecular diagnostic methods including conventional, multiple, and real-time PCR have been developed to overcome the shortfalls of culture techniques (Gaudio et al., 1997; Vu et al., 2004; Lindsay et al., 2013). The gold standard method used to identify Shigella spp. is pure culture followed by serum agglutination. Slide agglutination using commercial antisera is hindered by the lack of high-quality sera, time constraints, and difficult-to-visualize results (Sun et al., 2011). Molecular methods such as multiplex PCR (Ojha et al., 2013) immunocapture PCR (Peng et al., 2002), matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) assays, (Paauw et al., 2015) and high resolution melting (HRM) assay (Pakbin et al., 2022) were developed to differentiate between Shigella spp. While these methods allow rapid and high specificity identification of Shigella, they often require expensive equipment and involve complex methodologies. Our review revealed there was no isothermal amplification technique method for detection of S. sonnei and differentiation of virulent strains at the same time.
Multiple cross displacement amplification (MCDA) technique using strand displacement nucleic acid synthesis with the Bst polymerase reacts under isothermal conditions without the requirement of the expensive apparatus (Wang et al., 2015). All ten primers are designed to recognize ten distinct regions on the purpose sequence, enhancing the specificity and sensitivity of the MCDA reaction. Amplification products are a combination of various sequences of different fragment sizes, which can be detected by turbidimeters, colorimetric agents, gel electrophoresis, and nanoparticle lateral flow biosensor (LFB) (Wang et al., 2015). Dry-proof gold nanoparticle LFB is an easy-to-use method that reliably displays the results of amplification within a short time (Li and Macdonald, 2016). While migrating over the membrane, the amplification products encounter a series of reaction zones, which creates an output in these reaction areas (Li and Macdonald, 2016). Compared with other monitoring strategies such as turbidimeters, colorimetric agents, and gel electrophoresis, the critical advantage of LFB, is possible for the multiple identifications by detecting the amplification products labeled with different biomarkers (Li and Macdonald, 2016). MCDA combined with label-based LFB allows for the identification of multiple detections of target genes in one reaction (Wang et al., 2018). Multiplex MCDA-LFB methods are now used for the detection of several infectious agents, including Salmonella spp. (Wang et al., 2017), Acinetobacter baumannii (Hu et al., 2019), and SARS−CoV−2 (Luu et al., 2021).
The invasion plasmid antigen H (ipaH) gene, multiple copies of which are located on both the chromosome and plasmid, is widely used to detect Shigella spp. and enteroinvasive Escherichia coli (EIEC) (Hartman et al., 1990). The virulent S. sonnei strains (form I O polysaccharide, phase I) comprise a large virulence plasmid, which harbors both O polysaccharide and invasion (Xu et al., 2002). The wbgX gene encodes amino sugar synthetase and is located on the S. sonnei and Plesiomonas shigelloides O17 specific virulence plasmids with more than 99% sequence similar (Shepherd et al., 2000). In this study, a simple, visual, rapid, and sensitive method was developed to detect virulent S. sonnei using multiplex MCDA-LFB that was designed to target the ipaH and wbgX genes.
Materials and methods
Strains and preparation of DNA templates
A total of 161 strains including 122 Shigella spp., 11 enteroinvasive E. coli isolates, and 28 strains of other bacterial pathogen species were used to test the analytical specificity and application of the multiplex MCDA-LFB assays ( Table 1 ). Shigella monovalent antisera (Denka Seiken, Japan) were used to serologically confirm the S. sonnei isolates. S. flexneri 2a strain 301, S. sonnei CMCC 51081 (phase I), S. sonnei CMCC M6381 (phase II), and enterohemorrhagic E. coli EDL933 (ATCC 700927) were used to develop the conditions for the MCDA-LFB assay. The bacterial strains were cultured on Brain heart infusion agar or Columbia agar plates containing 5% defibrinated sheep blood and incubated in the ambient atmosphere or 5% CO2 atmosphere at 37°C for 18–24 hours. The bacterial genomic DNA was extracted using a QIAamp DNA Mini Kit (Qiagen, Germany) following the manufacturer’s instructions, and stored at -80°C until use. Qubit 4 Fluorometer (Thermo Fisher Scientific, USA) was used to quantify the genomic DNA. The genomic DNA of S. sonnei CMCC 51081was adjusted to 10 ng/μL using AE buffer (Qiagen, Germany) and then serially diluted to seven densities from 1 ng/μL to 1 fg/μL, including 100 pg/μL, 10 pg/μL, 1 pg/μL, 100 fg/μL, 10 fg/μL and 1 fg/μL. Three dilution series were prepared and then dilutions of the same concentrations were mixed to minimize errors during dilution.
Table 1.
Strains used in this study and the results of MCDA assays.
| Bacteria | Strain no./sourcea | No. of strains | MCDA LFB resultsd | |
|---|---|---|---|---|
| ipaH | wbgX | |||
| Shigella sonnei phase I | CMCC 51081 | 1 | + | + |
| S. sonnei phase I | Isolated strain | 32 | + | + |
| S. sonnei phase II | CMCC M6381 | 1 | + | − |
| S. sonnei phase II | Isolated strain | 25 | + | − |
| S. flexneri | 301 | 1 | + | − |
| S. flexneri | Isolated strain | 32 | + | − |
| S. dysenteriae | CMCCb | 12 | + | − |
| S. boydii | CMCCc | 18 | + | − |
| enteroinvasive E. coli | CMCC 44825 | 1 | + | − |
| enteroinvasive E. coli | Isolated strain | 10 | + | − |
| enterohemorrhagic E. coli O157:H7 | EDL933 (ATCC 700927) | 1 | − | − |
| enteroaggregative E. coli | O42 | 1 | − | − |
| enteropathogenic E. coli | 2348/69 | 1 | − | − |
| enterotoxigenic E. coli | 10407 | 1 | − | − |
| uropathogenic E. coli | UPEC 536 | 1 | − | − |
| Plesiomonas shigelloides | CHPC 1.5797 | 1 | − | − |
| Listeria monocytogenes | ATCC BAA-679 | 1 | − | − |
| Salmonella enterica serovar Paratyphi A | CMCC 50001 | 1 | − | − |
| S. enterica serovar Paratyphi B | CMCC 50001 | 1 | − | − |
| S. enterica serovar Choleraesuis | ATCC BAA-664 | 1 | − | − |
| Enterobacter cloacae | ATCC 700323 | 1 | − | − |
| Vibrio parahemolyticus | ATCC 17802 | 1 | − | − |
| Yersinia enterocolitica | Isolated strain | 1 | − | − |
| Citrobacter freundii | ATCC 8090 | 1 | − | − |
| Pseudomonas aeruginosa | ATCC 27853 | 1 | − | − |
| Acinetobacter baumannii | ATCC 17978 | 1 | − | − |
| Enterococcus faecium | CGMCC1.2135 | 1 | − | − |
| E. faecalis | CGMCC1.2136 | 1 | − | − |
| E. durans | CGMCC 1.2284 | 1 | − | − |
| E. avium | CGMCC 1.2505 | 1 | − | − |
| E. hirae | CGMCC 1.2140 | 1 | − | − |
| E. mundtii | CGMCC 1.2486 | 1 | − | − |
| Staphylococcus aureus subsp. aureus | ATCC 25923 | 1 | − | − |
| Streptococcus pneumoniae | ATCC 49619 | 1 | − | − |
| S. alactolyticus | ATCC 43077 | 1 | − | − |
| S. pyogenes | ATCC 19615 | 1 | − | − |
| Stenotrophomonas maltophilia | CGMCC 1.1788 | 1 | − | − |
| Burkholderia cepacia | CGMCC 1.1813 | 1 | − | − |
aCMCC, National Center for Medical Culture Collections; ATCC, American Type Culture Collection; CGMCC, China General Microbiological Culture Collection Center; CPHC, National Pathogen Resource.
bcontaining all 12 serotypes.
ccontaining all 18 serotypes.
d+, positive; −, negative.
Design of the primers
The ipaH (GenBank accession no. M32063) and wbgX (GenBank accession no. AF285971) genes were used as targets to design the MCDA primers using Primer Primer 6.0 software and PrimerExploer V4 (Eiken Chemical Co., Ltd., Japan). Each set of primers, including two cross primers (CP1 and CP2), two displacement primers (F1 and F2), and six amplification primers (C1, C2, D1, D2, R1, and R2) was designed to recognize 10 distinct regions of each target gene. The C1 and D1 primes were labeled at 5’ end for LBF detection (Wang et al., 2017). Primer specificity was assessed using the National Center for Biotechnology Information (NCBI) BLAST. The primer sequences and the modification of optimal primers are listed in Table 2 , and the position of the primers on the target genes is shown in Supplemental Figure 1 . The oligomers were synthesized and purified using Tsingke Biological Technology (Tsingke, China).
Table 2.
Primers used in this study.
| Gene | Primers | Sequence and modification (5’-3’) | Length |
|---|---|---|---|
| ipaH | Ipa-F1 | GGCTGGAAAAACTCAGTGC | 19 |
| Ipa-F2 | TGACTTTATCCCGGGCAAT | 19 | |
| Ipa-CP1 | CATGTGAGCGCGACACGGTCAGCAGTCTTTCGCTGTTG | 39 | |
| Ipa-C1* | FITC-CATGTGAGCGCGACACGGTC | 25 | |
| Ipa-CP2 | CCTTTTCGATAATGATACCGGCGCTCCTCCAGAATTTCGAGGC | 44 | |
| Ipa-C2 | CCTTTTCGATAATGATACCGGCGC | 24 | |
| Ipa-D1* | Biotin-CTCAGTGGCATCAGCA | 23 | |
| Ipa-D2 | TCTGCTCTCCCTGGGCA | 17 | |
| Ipa-R1 | GGTTTTCCGGAGATTGTTC | 19 | |
| Ipa-R2 | GTCCATCAGGCATCAGA | 17 | |
| wbgX | Wbg-F1 | GCAATCGGTATTCATCAACTT | 21 |
| Wbg-F2 | CTTATGCAACGGTATAAAATGG | 22 | |
| Wbg-CP1 | GTGGCAATTCTTTTAACGCATCATCAGAAAGATCGATGATTTTCAGAA | 49 | |
| Wbg-C1* | FITC-GTGGCAATTCTTTTAACGCATCATC | 30 | |
| Wbg-CP2 | CTATCCGCTTAAAAACTGATTCGGC-ACAGAACAACCAATTCCAAG | 46 | |
| Wbg-C2 | CTATCCGCTTAAAAACTGATTCGGC | 25 | |
| Wbg-D1 | Dig-TTTTTGCCATTCGTTGACGT | 32 | |
| Wbg-D2 | CGCGATGATTTTATTAAGAAG | 21 | |
| Wbg-R1 | TAGGCCATTCAGGCAATTC | 19 | |
| Wbg-R2 | GATATTCATGCTTGGCATCT | 20 |
*Modification at 5′ end of the primers. a, nt, nucleotide mer; monomeric unit. FITC, fluorescein isothiocyanate; Dig, digoxigenin.
The singlex MCDA reaction
The singlex MCDA reaction for the ipaH and wbgX genes is conducted as described previously (Wang et al., 2017). WarmStart Colorimetric LAMP 2X Master Mix (New England Biolabs Inc., USA) was used to carry out the MCDA reaction. The MCDA was performed using a 25 μL mixture containing 12.5 μL of 2x reaction mixture, a 2.2 μL primer mixture including 0.4 μM each of F1 and F2, 0.8 μM each of C1 and C2, R1 and R2, D1 and D2, 1.6 μM each of CP1 and CP2, and 1 μL of the DNA template. The mixture was incubated at 63°C for 60 min and then at 80°C for 5 min to terminate the reaction. The optimal MCDA reaction temperature was determined with five temperatures ranging from 61°C to 65°C (with 1°C interval). S. flexneri 2a strain 301, S. sonnei CMCC 51081, and CMCC M6381 were used as the positive control, and enterohemorrhagic E. coli EDL933 was used as negative control for the ipaH gene. S. sonnei CMCC 51081 was used as the positive control, and S. flexneri 2a strain 301, S. sonnei CMCC M6381, and enterohemorrhagic E. coli EDL933 were used as the negative control for the wbgX gene. The mixture without the DNA template was used as the blank control. The MCDA reaction was confirmed using four monitoring techniques, including macroscopic color change observed with a color change from pink to yellow, turbidimeters (LA-320C, Eiken Chemical Co., Ltd., Japan) with the turbidity of >0.1 as positive, and 2% agarose gel electrophoresis with specific ladder DNA amplicons as positive, and LFB detection. The lateral flow dipsticks (MGHD2, Milenia HybriDetect 2T, Germany) with three lines, two test lines and one control line, were used to detect the labeled MCDA amplicons. The 5’ end of the ipaH C1 and D1 primers was labeled with fluorescein isothiocyanate (FITC) and Biotin. The FITC and Biotin amplicons were captured by the first test line using a Biotin ligand. Red lines then appeared on the first test line (T L1). The FITC and Dig (digoxigenin) amplicons used to detect the wbgX gene were caught by the second test line (T L 2) which also turned red. The control line captured the anti-FITC antibody and was used to ensure that the LFB assay was functioning. For LFB detection, 0.5 μL MCDA products were added to the sample pad of the biosensor, followed by 80 μL of HybriDetect Assay Buffer. Results were observed after incubating the sample for 2–5 min at room temperature. The MCDA operations were performed with independent laboratory equipment and experimental consumables. Blank control was added in every batch of MCDA reactions and LFB detections to avoid contamination.
MCDA assay sensitivity
The sensitivity of the MCDA-LFB assay for the ipaH and wbgX genes was respectively determined with 1 µL of serially diluted genomic DNA from S. sonnei CMCC 51081.The sensitivity of the MCDA-LFB assay in artificially contaminated human feces was determined by inoculating S. sonnei CMCC 51081 into the brain-heart infusion broth and cultivating overnight. Strain solutions (50 μL) were transferred to 5 ml brain-heart infusion broth and incubated to OD610 0.61 with agitation, which was equivalent to 3.7×108 CFU (colony-forming units)/ml using the plate count method. The cultures were then 10x serially diluted from 107–101 CFU per ml using normal saline. Seven concentrations of S. sonnei CMCC 51081 were mixed with human stool samples from healthy volunteers. The S. sonnei CMCC 51081 DNA template in spiked stool was extracted as previously described (Song et al., 2005). The experiment was independently performed in triplicate, 100 μL AE buffer (Qiagen, Germany) was used to elute the extracted DNA, and 1 μL of each supernatant was used for MCDA detection.
Optimization of multiplex MCDA primer concentration
The concentration ratio of the two sets of ipaH and wbgX primers was adjusted to ensure the simultaneous amplification of both genes according to their sensitivity. The total primer amount was fixed at 2.2 μL in a 25 μL MCDA reaction. Primer mixtures of 10 μL each of F1 and F2, 20 μL each of C1 and C2, R1 and R2, D1 and D2, and 40 μL each of CP1 and CP2 were used to make a 100μM concentration of the two genes. The five concentration ratios included 0.2–1.0 μL of the ipaH gene primer mix and 2.0–1.2 μL of the wbgX gene primer mix with a 0.2 μL interval. Reaction results were detected using the LFB assay.
MCDA assay specificity
The multiplex MCDA reactions were performed with different DNA templates from 59 reference strains including 12 S. dysenteriae, 18 S. boydii, one enteroinvasive E. coli, and 28 other bacterial pathogens ( Table 1 ) using the conditions described above to evaluate MCDA assay specificity. Each DNA template was independently analyzed in triplicate.
Use of the MCDA-LBF assay on clinical isolates
Clinical Shigella spp. isolates including 32 S. sonnei phase I, 25 S. sonnei phase II, 32 S. flexneri, and 10 enteroinvasive E. coli were used to evaluate the clinical applicability of the MCDA-LBF assay. The clinical isolates were identified using a biochemical test with API 20E (BioMérieux, France) and slide agglutination. A single colony was placed into a 1.5 ml microcentrifuge tube with 100 mL distilled water. The sample was cooked for 10 min at 100°C before cooling for 5 min on ice. The supernatant was transferred to a new 1.5 ml microcentrifuge tube, centrifuged at 12,470 g for 10 min, and used as a template to perform the multiplex MCDA-LFB assay.
Ethics statements
The study was performed following the guidelines of the Declaration of Helsinki and approved by the Ethics Committee of National Institute for Communicable Disease Control and Prevention, China CDC (No. ICDC2017-003).
Results
Functionality of the MCDA assay
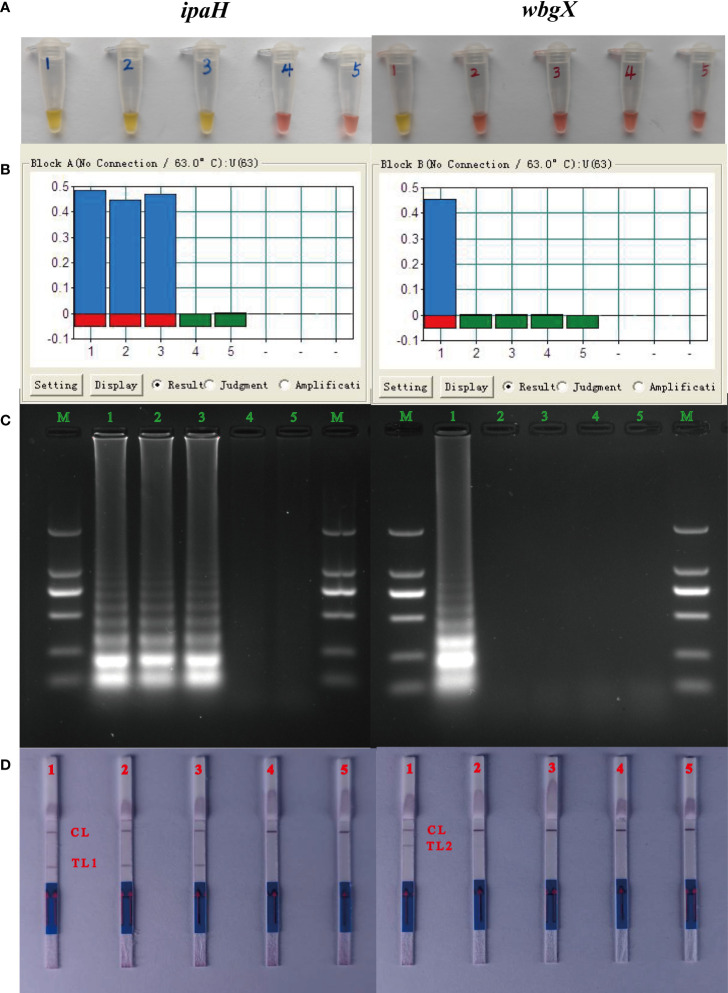
To demonstrate the functionality of the MCDA assay, genomic DNA from S. flexneri 2a strain 301, S. sonnei CMCC 51081, S. sonnei CMCC M6381, and enterohemorrhagic E. coli EDL933 was amplified for 60 min at 63°C using the ipaH and wbgX primers. Distilled water was used as a blank control. The color change from pink to yellow ( Figure 1A ), >0.1 change in turbidity ( Figure 1B ), presence of ladder DNA amplicons ( Figure 1C ), and the red test lines 1 and 2 for the ipaH and wbgX genes, respectively, were observed macroscopically. S. flexneri 2a strain 301, S. sonnei CMCC 51081, and S. sonnei CMCC M6381 were positive for the ipaH gene while enterohemorrhagic E. coli EDL933 and the blank control were negative. S. sonnei CMCC 51081 was positive for the wbgX gene while S. flexneri 2a strain 301, S. sonnei CMCC M6381, enterohemorrhagic E. coli EDL933, and the blank control were negative ( Figure 1 ). These results indicated that the ipaH and wbgX primers were appropriate candidates for developing a S. sonnei-specific multiplex MCDA-LFB assay.
Figure 1.
Verification of the MCDA products of ipaH and wbgX genes. Tubes/lanes/strips/biosensors 1-5: 1, S. sonnei CMCC 51081 (phase I); 2, S. flexneri 301; 3, S. sonnei CMCC M6381 (phase II); 4, enterohemorrhagic E. coli EDL933 (Negative control); 5, distilled water (Blank control). (A) Reaction products of the MCDA assay of ipaH and wbgX genes were visually detected by color change. (B) Turbidity in real-time using the Loopamp Real-time Turbidimeter LA-320C for the MCDA assay. (C) 2% agarose gel electrophoresis results of the MCDA assay. (D) LFB was applied for visual detection of the MCDA assay, CL (control line), T L1 (test line 1), and T L2 (test line 2). For ipaH gene, positive results were observed with Tubes 1-3 with the color changed from pink to yellow, lanes 1-3 with >0.1 turbidity changes with turbidimeters, strips 1-3 with the ladder DNA amplicons, and biosensors 1-3 with the red test line 1. Negative results were observed with Tubes/lanes/strips/biosensors 4-5 with negative control and blank control. For wbgX gene, positive results were observed with Tubes/lanes/strips 1 and biosensors 1 with the red test line 2, Negative results were observed with Tubes/lanes/strips/biosensors 4-5.
Determining the optimal temperature
Five temperatures from 61–65°C with a 1°C increment were used to determine the optimal MCDA temperature for the 60 min reaction and 10 pg S. sonnei CMCC 51081 DNA was used as a template. Positive results (>0.1 turbidities) were observed for the ipaH and wbgX genes within 20 min at all specified temperatures ( Supplemental Figure 2 ). A 20 min amplification at 63°C and 5 min termination at 80°C were used for subsequent experiments.
Sensitivity of the singlex MCDA assay for the IpaH and WbgX genes
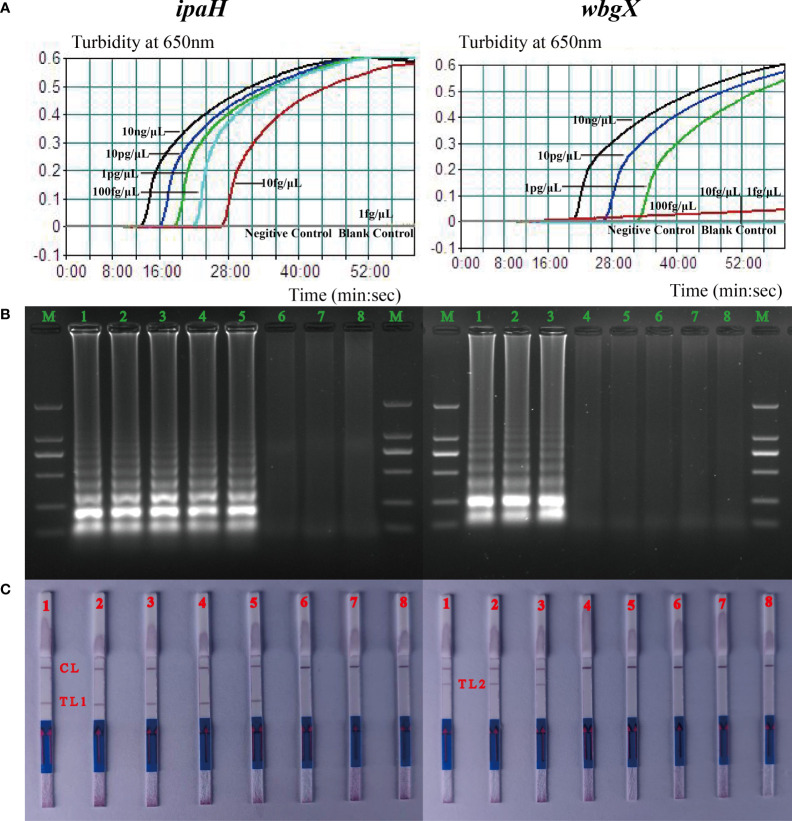
Sensitivity of the singlex MCDA assay for virulent S. sonnei was evaluated by analyzing the products yielded from the diluted S. sonnei CMCC 51081 DNA templates (10 ng/μL, 10 pg/μL, 1 pg/μL, 100 fg/μL, 10 fg/μL and 1 fg/μL) using the method previously described. Enterohemorrhagic E. coli EDL933 served as the negative control and the mixture without the DNA template was used as a blank control. The detection limits were 10 fg/μL for ipaH and 1 pg/μL for wbgX ( Figure 2A ). The MCDA products were analyzed on a 2% agarose gel by electrophoresis and LFB and the results were consistent with the turbidity curves ( Figures 2B, C ).
Figure 2.
Detection limits of the ipaH and wbgX genes of the singlex MCDA-LFB assay. The six dilution templates of S. sonnei CMCC 51081 including 10 ng, 10 pg, 1 pg, 100 fg, 10 fg, and 1 fg were subjected to MCDA reactions to detect limits of ipaH and wbgX genes. (A) Real-time sensitivity of the singlex MCDA assay of two genes monitored by measuring the turbidity (optimal density at 650 nm). The turbidity >0.1 was considered positive for the two genes. The detection limits were 10 fg/μL for ipaH and 1 pg/μL for wbgX genes. (B) Gel electrophoresis was applied for analysis of the sensitivity of ipaH and wbgX genes. Strips M, DNA marker DL2000; Strips 1–6, PCR results for six dilutions; Strips 7, negative control; Strips 8, blank control. In agarose gel analysis of the two genes, the MCDA products showed the typical ladder-like pattern amplicons, and the detection limit matched the turbidity data. (C) LFB applied for analysis of ipaH and wbgX genes MCDA products. Biosensors 1–6, MCDA results for six dilutions; Biosensor 7, negative control; Biosensor 8, blank control. CL (control line), T L1 (test line 1), T L2 (test line 2). The positive results were observed at Biosensor 5 with the red line of T L1 and Biosensor 3 with the red line of T L2 which meant the detection limits of the ipaH gene was 10 fg/μL for and wbgX gene was 1 pg/μL. The detection limits matched the turbidity data and gel electrophoresis results.
Optimization of the multiplex MCDA-LBF primer concentration
The MCDA primer concentrations were adjusted to promote simultaneous amplification of the reaction and ensure the production of reliable multiplex MCDA products given differences in the detection limit of ipaH and wbgX. Two red test lines were observed using 1.6 μL wbgX and 0.6μL ipaH primer mixtures. These concentrations were selected for the multiplex MCDA-LFB reaction conditions ( Figure 3A ).
Figure 3.

Optimization of the ratio of two primers and detection limit of multiple targets MCDA assay. (A) Biosensors 1, the primer concentration with 0.2 μL ipaH gene primers mix and 2.0 with μL wbgX gene primers mix; biosensors 2, 0.4 μL (ipaH gene) and 1.8 μL (wbgX gene); biosensors 3, 0.6 μL (ipaH gene) and 1.6 μL (wbgX gene); biosensors 4, 0.8 μL (ipaH gene) and 1.4 μL (wbgX gene); biosensors 5 1.0 μL (ipaH gene) and 1.4 μL (wbgX gene). Two red test lines appeared from the primer concentrations with 0.6μL ipaH gene 1.6 μL wbgX gene primers mixtures. (B) The sensitivity of multiple MCDA-LFB reactions with the serial dilutions genomic DNA of S. sonnei CMCC 51081. Biosensors 1–6, multiple MCDA results for six dilutions including 10 ng, 10 pg, 1 pg, 100 fg, 10 fg, and 1 fg; Biosensor 7, negative control; Biosensor 8, blank control. The sensitivity of multiple MCDA reactions was 1 pg/μL (positive results of Biosensors 3). The positive results could observe at Biosensors 5 with the concentration of 10 fg/μL with ipaH gene in the multiple MCDA reactions. No red test lines could be observed with negative control and blank control. (C) The sensitivity of the multiplex MCDA assay in artificially contaminated human feces samples. Biosensors 1–6: S. sonnei CMCC 51081 strain at 3700 CFU, 370 CFU, 37 CFU, 3.7 CFU, 0.37 CFU and 0.037 CFU per reaction; Biosensor 7, negative control; Biosensor 8, blank control. The positive results were detected for samples containing 37 CFU per reaction, and the red test lines of ipaH gene appeared in the sample with 3.7 CFU.
The sensitivity and specificity of the multiple MCDA-LBF assay
The sensitivity of the multiplex MCDA reaction was 1 pg/μL using serial dilutions of S. sonnei CMCC 51081 genomic DNA ( Figure 3B ). Positive results were observed using 10 fg/μL of the ipaH gene in a 25 min multiplex MCDA reaction. Only the control line was observed using E. coli EDL933 and the negative control. Positive results were detected for artificially contaminated human feces samples containing ≥3.7×103 CFU per ml, and the red test line representing the ipaH gene appeared in the sample containing 3.7×102 CFU ( Figure 3C ), indicating that the multiplex MCDA assay sensitivity was ~37 CFU for the wbgX gene and ~3.7 CFU for the ipaH gene.
To determine multiplex MCDA specificity, the assay was performed with DNA templates from 30 Shigella spp. reference strains including 12 S. dysenteriae, 18 S. boydii, one enteroinvasive E. coli reference strain, and 28 other bacterial pathogen species ( Table 1 ). Positive results of the ipaH gene and negative results of the wbgX gene were observed using 31 reference strains including 30 Shigella spp. strains and one enteroinvasive E. coli strain. Negative results of both the ipaH and wbgX genes were observed using 28 strains from other bacterial genera.
Application of the MCDA-LBF assay in clinical cultures
A total of 99 clinical isolates including 32 S. sonnei phase I, 25 S. sonnei phase II, 32 S. flexneri, and 10 enteroinvasive E. coli were used to evaluate the applicability of the MCDA-LBF assay. The Shigella spp. strains were identified using slide agglutination. Positive results of the ipaH gene were acquired using all isolates, while positive results of the wbgX gene were only observed using S. sonnei phase I.
Discussion
The multiplex MCDA-LFB assay has several advantages over other molecular methods including multiplex PCR, real-time PCR, and high-resolution melting (HRM) used to identify Shigella spp. First, all reactions can be performed under isothermal conditions and positive amplification can be directly observed macroscopically, thus eliminating the use of large and complex instruments required for PCR-based systems. Second, amplification efficiency is extremely high, and many products can be obtained within 25 min. Third, the reaction uses multiple primers that recognize distinct regions of the target gene, thus making it highly sensitive and specific. Finally, combined with LFB, the MCDA assay achieves multiple target applications, which increases the possible uses of this method (Wang et al., 2017). As a result, the MCDA-LFB assay is widely used for nucleic acid analysis because of its simplicity, rapidity, high efficiency, and specificity (Hu et al., 2019; Luu et al., 2021). The current study developed a multiplex MCDA-LFB assay that was able to sensitively and accurately identify virulent S. sonnei using the ipaH gene and O-antigen synthesis gene, wbgX.
A reaction temperature of 63°C was chosen to simplify the experimental conditions and allow the two target genes to react synchronously. At this temperature, two gene reactions could be completed in 25 min, allowing for the rapid detection of virulent S. sonnei. The singlex MCDA assay was able to detect DNA samples at concentrations as low as 10 fg/μL for the ipaH gene and 1 pg/μL for the wbgX gene. Because of the difference in the copy numbers, the sensitivity of the ipaH gene with MCDA assay was much high than the wbgX gene. Due to differences in the detection limits of the two genes, five primer concentrations were adjusted to achieve simultaneous amplification during the reaction and 1.6 μL wbgX gene and 0.6μL ipaH gene primer mixtures were chosen for the multiplex MCDA-LFB reaction conditions. Using this primer concentration, 1 pg/μL of the wbgX gene and 10 fg/μL of the ipaH gene from virulent S. sonnei genomic DNA was detected in the multiplex MCDA reaction after 25 min which was consistent with the singlex MCDA-LFB assay. This multiplex MCDA assay could detect as few as ~37 CFU of virulent S. sonnei and ~3.7 CFU of Shigella spp. in artificially contaminated human feces samples. The sensitivity of this MCDA-LFP assay was higher than other isothermal amplification methods such as the LAMP assay and considerably better than PCR at detecting the ipaH gene from Shigella spp. (Song et al., 2005). Positive results from spiked feces samples indicated that the multiplex MCDA-LFB could perform well using clinical samples. The analytical specificity of the multiplex MCDA-LFB assay was demonstrated for 30 Shigella spp. reference strains, one enteroinvasive E. coli strain, and 28 other bacterial pathogen species. This multiplex MCDA-LFB assay showed high specificity for Shigella spp. and enteroinvasive E. coli strains, as well as other reference strains. To determine whether the assay could be applied to clinical isolates, boiled DNA from 57 S. sonnei strains, including 32 phase I and 25 phase II strains, and 42 S. flexneri and enteroinvasive E. coli strains were analyzed. The MCDA-LFB method was able to differentiate between S. sonnei and other Shigella spp. and enteroinvasive E. coli. The detection time was only 30 min, including 20 min for amplification, 5 min for termination the reaction, and 5 min for detection.
In summary, a multiplex MCDA-LFB assay was developed for the rapid (30 min), convenient, and accurate identification of virulent S. sonnei at a constant temperature without the use of expensive equipment. This assay could also be used to detect Shigella spp. and enteroinvasive E. coli. The multiplex MCDA-LFB assay could promote the rapid and reliable clinical diagnosis and epidemiological investigation of virulent S. sonnei in the field, during point-of-care, and on-site in various laboratories including resource-challenged settings.
Data availability statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving human participants were reviewed and approved by The Ethics Committee of National Institute for Communicable Disease Control and Prevention. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author contributions
Conceptualization, DJ. Methodology, YW, ZH, and PA. Data curation, YW, SJ, and DJ. Writing—review and editing, SJ, and DJ. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by 2015011115 from Basic Research Project of Shanxi Province and 2018RU010 from Units of Discovery of Unknown Bacteria and Function.
Conflict of interest
The reviewer LG declared a shared affiliation with the authors to the handling editor at the time of review.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.1012105/full#supplementary-material
References
- Chang Z., Zhang J., Ran L., Sun J., Liu F., Luo L., et al. (2016). The changing epidemiology of bacillary dysentery and characteristics of antimicrobial resistance of shigella isolated in China from 2004-2014. BMC Infect. Dis. 16, 685. doi: 10.1186/s12879-016-1977-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudio P. A., Sethabutr O., Echeverria P., Hoge C. W. (1997). Utility of a polymerase chain reaction diagnostic system in a study of the epidemiology of shigellosis among dysentery patients, family contacts, and well controls living in a shigellosis-endemic area. J. Infect. Dis. 176, 1013–1018. doi: 10.1086/516531 [DOI] [PubMed] [Google Scholar]
- Hartman A. B., Venkatesan M., Oaks E. V., Buysse J. M. (1990). Sequence and molecular characterization of a multicopy invasion plasmid antigen gene, ipaH, of shigella flexneri. J. Bacteriol. 172, 1905–1915. doi: 10.1128/jb.172.4.1905-1915.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Niu L., Zhao F., Yan L., Nong J., Wang C., et al. (2019). Identification of acinetobacter baumannii and its carbapenem-resistant gene blaOXA-23-like by multiple cross displacement amplification combined with lateral flow biosensor. Sci. Rep. 9, 17888. doi: 10.1038/s41598-019-54465-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk M. D., Pires S. M., Black R. E., Caipo M., Crump J. A., Devleesschauwer B., et al. (2015). Correction: World health organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases 2010: A data synthesis. PLoS Med. 12, e1001940. doi: 10.1371/journal.pmed.1001940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Macdonald J. (2016). Multiplexed lateral flow biosensors: Technological advances for radically improving point-of-care diagnoses. Biosensors Bioelectronics 83, 177–192. doi: 10.1016/j.bios.2016.04.021 [DOI] [PubMed] [Google Scholar]
- Lindsay B., Ochieng J. B., Ikumapayi U. N., Toure A., Ahmed D., Li S., et al. (2013). Quantitative PCR for detection of shigella improves ascertainment of shigella burden in children with moderate-to-severe diarrhea in low-income countries. J. Clin. Microbiol. 51, 1740–1746. doi: 10.1128/JCM.02713-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu L. D. W., Payne M., Zhang X., Luo L., Lan R. (2021). Development and comparison of novel multiple cross displacement amplification (MCDA) assays with other nucleic acid amplification methods for SARS-CoV-2 detection. Sci. Rep. 11, 1873. doi: 10.1038/s41598-021-81518-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Octavia S., Lan R. (2015). “Chapter 65 - shigella and shigellosis: Genetics, epidemiology and pathogenesis,” in Molecular medical microbiology (Second edition). Eds. Tang Y.-W., Sussman M., Liu D., Poxton I., Schwartzman J. (Boston: Academic Press; ), 1147–1168. [Google Scholar]
- Ojha S. C., Yean Yean C., Ismail A., Singh K. K. (2013). A pentaplex PCR assay for the detection and differentiation of shigella species. BioMed. Res. Int. 2013, 412370. doi: 10.1155/2013/412370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paauw A., Jonker D., Roeselers G., Heng J. M., Mars-Groenendijk R. H., Trip H., et al. (2015). Rapid and reliable discrimination between shigella species and escherichia coli using MALDI-TOF mass spectrometry. Int. J. Med. Microbiol. 305, 446–452. doi: 10.1016/j.ijmm.2015.04.001 [DOI] [PubMed] [Google Scholar]
- Pakbin B., Basti A. A., Khanjari A., Brück W. M., Azimi L., Karimi A. (2022). Development of high-resolution melting (HRM) assay to differentiate the species of shigella isolates from stool and food samples. Sci. Rep. 12, 473. doi: 10.1038/s41598-021-04484-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X., Luo W., Zhang J., Wang S., Lin S. (2002). Rapid detection of shigella species in environmental sewage by an immunocapture PCR with universal primers. Appl. Environ. Microbiol. 68, 2580–2583. doi: 10.1128/AEM.68.5.2580-2583.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd J. G., Wang L., Reeves P. R. (2000). Comparison of O-antigen gene clusters of escherichia coli (Shigella) sonnei and plesiomonas shigelloides O17: sonnei gained its current plasmid-borne O-antigen genes from p. shigelloides in a recent event. Infect. Immun. 68, 6056–6061. doi: 10.1128/IAI.68.10.6056-6061.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song T., Toma C., Nakasone N., Iwanaga M. (2005). Sensitive and rapid detection of shigella and enteroinvasive escherichia coli by a loop-mediated isothermal amplification method. FEMS Microbiol. Lett. 243, 259–263. doi: 10.1016/j.femsle.2004.12.014 [DOI] [PubMed] [Google Scholar]
- Sun Q., Lan R., Wang Y., Zhao A., Zhang S., Wang J., et al. (2011). Development of a multiplex PCR assay targeting O-antigen modification genes for molecular serotyping of shigella flexneri. J. Clin. Microbiol. 49, 3766–3770. doi: 10.1128/JCM.01259-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor W. I., Schelhart D. (1975). Effect of temperature on transport and plating media for enteric pathogens. J. Clin. Microbiol. 2, 281–286. doi: 10.1128/jcm.2.4.281-286.1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. N., Duy P. T., Baker S. (2015). The rising dominance of shigella sonnei: An intercontinental shift in the etiology of bacillary dysentery. PLoS Negl. Trop. Dis. 9, e0003708. doi: 10.1371/journal.pntd.0003708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu D. T., Sethabutr O., Von Seidlein L., Tran V. T., Do G. C., Bui T. C., et al. (2004). Detection of shigella by a PCR assay targeting the ipaH gene suggests increased prevalence of shigellosis in nha trang, Vietnam. J. Clin. Microbiol. 42, 2031–2035. doi: 10.1128/JCM.42.5.2031-2035.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li H., Wang Y., Xu H., Xu J., Ye C. (2018). Antarctic Thermolabile uracil-DNA-glycosylase-supplemented multiple cross displacement amplification using a label-based nanoparticle lateral flow biosensor for the simultaneous detection of nucleic acid sequences and elimination of carryover contamination. Nano Res. 11, 2632–2647. doi: 10.1007/s12274-017-1893-z [DOI] [Google Scholar]
- Wang Y., Wang Y., Ma A.-J., Li D.-X., Luo L.-J., Liu D.-X., et al. (2015). Rapid and sensitive isothermal detection of nucleic-acid sequence by multiple cross displacement amplification. Sci. Rep. 5, 11902. doi: 10.1038/srep11902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang Y., Zhang L., Xu J., Ye C. (2017). Visual and multiplex detection of nucleic acid sequence by multiple cross displacement amplification coupled with gold nanoparticle-based lateral flow biosensor. Sensors Actuators B: Chem. 241, 1283–1293. doi: 10.1016/j.snb.2016.10.001 [DOI] [Google Scholar]
- Xu D. Q., Cisar J. O., Ambulos N., Jr., Burr D. H., Kopecko D. J. (2002). Molecular cloning and characterization of genes for shigella sonnei form I O polysaccharide: proposed biosynthetic pathway and stable expression in a live salmonella vaccine vector. Infect. Immun. 70, 4414–4423. doi: 10.1128/IAI.70.8.4414-4423.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.