Single-cell progenitor hierarchy at CMML identifies a monocytic-biased trajectory associated with disease progression, expanded by inflammation, and diminished by hypomethylation therapy.
Abstract
Myeloblast expansion is a hallmark of disease progression and comprises CD34+ hematopoietic stem and progenitor cells (HSPC). How this compartment evolves during disease progression in chronic myeloid neoplasms is unknown. Using single-cell RNA sequencing and high-parameter flow cytometry, we show that chronic myelomonocytic leukemia (CMML) CD34+ HSPC can be classified into three differentiation trajectories: monocytic, megakaryocyte-erythroid progenitor (MEP), and normal-like. Hallmarks of monocytic-biased trajectory were enrichment of CD120b+ inflammatory granulocyte–macrophage progenitor (GMP)-like cells, activated cytokine receptor signaling, phenotypic hematopoietic stem cell (HSC) depletion, and adverse outcomes. Cytokine receptor diversity was generally an adverse feature and elevated in CD120b+ GMPs. Hypomethylating agents decreased monocytic-biased cells in CMML patients. Given the enrichment of RAS pathway mutations in monocytic-biased cells, NRAS-competitive transplants and LPS-treated xenograft models recapitulated monocytic-biased CMML, suggesting that hematopoietic stress precipitates the monocytic-biased state. Deconvolution of HSPC compartments in other myeloid neoplasms and identifying therapeutic strategies to mitigate the monocytic-biased differentiation trajectory should be explored.
Significance:
Our findings establish that multiple differentiation states underlie CMML disease progression. These states are negatively augmented by inflammation and positively affected by hypomethylating agents. Furthermore, we identify HSC depletion and expansion of GMP-like cells with increased cytokine receptor diversity as a feature of myeloblast expansion in inflammatory chronic myeloid neoplasms.
This article is highlighted in the In This Issue feature, p. 476
INTRODUCTION
Myeloblasts, initially described in 1900 by Otto Naegeli, have long been regarded as the neoplastic cell and hallmark feature of acute myeloid leukemia (AML). These cells are pathologically defined by a large round nucleus composed of fine nonaggregated chromatin with a small basophilic cytoplasm. Myeloblasts are immunophenotypically characterized by the surface expression of CD34, although some CD34-negative, CD117-positive myeloblast populations have been reported (1, 2). The expansion of myeloblasts within the bone marrow and peripheral blood is uniformly associated with disease progression across all chronic myeloid neoplasms and defines transformation to AML when myeloblasts meet or exceed 20% (3). Myeloblasts are thought to represent hematopoietic stem and progenitor cells (HSPC) but are not generally resolved into stem and progenitor subpopulations during clinical evaluation. Therefore, how the malignant expansion of myeloblasts reshapes the HSPC compartment and its impact on clinical outcomes remains undefined. Understanding the composition of HSPC compartments could establish the differentiation trajectories most relevant to disease progression and provide insights into the mechanisms that promote these adverse hematopoietic states.
To address this question, we transcriptionally and immunophenotypically mapped the CD34+ hematopoietic compartment of 55 unique chronic myelomonocytic leukemia (CMML) patients with comparable clinical baseline characteristics at single-cell resolution (Supplementary Table S1; Supplementary Fig. S1). CMML is a lethal myeloid neoplasm hallmarked by the expansion of classic monocytes in the peripheral blood with no therapies that improve overall survival. Given its uniform hematopoietic output and that increases in myeloblast percentage are associated with prognostically relevant subtypes (CMML-0/1/2) and inferior survival (4), CMML is an excellent model to evaluate the composition of HSPC and its clinical consequence in myeloid neoplasms.
In this study, we investigated the transcriptome, mutational landscape, and spectrum of cytokine receptors in CD34+ CMML patient samples. Single-cell RNA sequencing (scRNA-seq) was used to evaluate HSPC transitional cellular states, and high-parameter flow cytometry was utilized to evaluate previously defined immunophenotypic HSPC states ranging from hematopoietic stem cells (HSC) to myeloid progenitors. This combined approach provided a comprehensive map of HSPC subpopulations at various stages of CMML disease progression. We identify aberrations in HSC fate and lineage determination and an inflammatory granulocyte–macrophage progenitor (GMP)–like state with increased cytokine receptor diversity. Moreover, these aberrations were associated with adverse clinical outcomes that may present putative targets of prognostic and therapeutic relevance.
RESULTS
CMML HSPC Are Characterized by Three Hematopoietic Trajectories
Until recently, hematopoiesis has been modeled as a hierarchical branching process with distinct stages defined by the expression of surface proteins (5). However, murine and human scRNA-seq data sets of bone marrow cells suggest that hematopoiesis occurs in a probabilistic manner with gradients of differentiation defined by transcriptional expression (6). To evaluate this modern framework of hematopoiesis in CMML, we sequenced 137,578 high-quality CD34+ single cells from 39 patient samples [median age: 72 years, WHO classification: CMML-0 (51.72%), CMML-1 (20.68%), CMML-2 (20.68%), AML (3.4%); more details described in Supplementary Table S1] using the 10X Genomics platform (Fig. 1A). Additionally, we integrated eight publicly available samples (6–8) containing 63,672 additional high-quality cells from normal CD34+ enriched HSPC (Fig. 1B). To avoid effects due to nonbiological differences such as lab protocols and sequencing targets, batch correction was performed with the Harmony algorithm (9).
Figure 1.
CMML HSPC are characterized by three hematopoietic trajectories. A, Excess bone marrow and aspirate specimens not utilized for pathologic evaluation were collected from CMML patients at clinically scheduled time points and enriched for CD34+ cells. Single cells [CD34 cells enriched from patient bone marrrow mononuclear cells (BMMNC)] were encapsulated for scRNA-seq via DropSeq with the 10X Chromium controller and sequenced on an Illumina NovaSeq. Data from CD34+ cells enriched from BMNNCs of 8 healthy individuals available from publicly available data sets were integrated with the 39 CMML patient samples for downstream analysis. B, UMAP visualization of the 201,250 cells with normal CD34+ cells represented in red and CMML CD34+ cells represented in gray. C, CMML single cells were projected onto a reference Normal CD34+ (6), and branch probabilities and differentiation potentials were inferred using a nearest-neighbor approach to reveal (D) three distinct hematopoietic trajectories in CMML: monocytic-biased, MEP-biased, and normal-like. Monocytic-biased samples had overdensities (as indicated in red) in the monocytic branch, MEP-biased samples had overdensities in the erythroid/megakaryocytic (Ery/Mega) branch, and normal-like samples showed neither overdensity in monocytic and erythroid branches and cell densities of similar proportion to that of the normal samples. These trajectories were classified based on the overdensities compared with the reference hematopoietic, gene signature analysis, and hierarchical clustering (in Supplementary Fig. S2). E, These trajectory biases were confirmed using previously published gene-expression profiles of HSCs, MEPs, and GMPs. Monocytic-biased patients were highly enriched for genes upregulated in GMPs, and MEP-biased patients were highly enriched for genes upregulated in MEPs. F, Hemoglobin was significantly elevated in the MEP-biased group (P = 0.037; Mann–Whitney test). G, Kaplan–Meier (KM) survival analysis of CMML patients stratified on progenitor trajectory in treatment-naïve samples treatment-naïve (n = 25; log-rank P = 0.0349). H, LDH levels obtained from clinical evaluation at the time of tissue sampling (n = 38; Fisher exact test P: 0.0378). I and J, There was a significant depletion in the fraction of cells identified as CLP across treatment-naïve samples (I; P = 0.0004; Mann–Whitney test) and in all three differentiation trajectories identified in CMML (J; Mann–Whitney comparisons; between normal and monocytic-biased, P = 0.0207; normal and MEP-bias, P = 0.0030; normal and normal-like, P = 0.0031). CLP cell-type assignment was calculated from the SingleR consensus (see Supplementary Methods).
Following batch correction, we estimated the differentiation trajectories of CD34+ cells using Palantir (6), leveraging previously published normal CD34+ samples (6) by projecting each CMML patient's CD34+ single-cell data onto a reference differentiation trajectory, as visualized in Fig. 1C. This mapping provided a probability that any given CD34+ cell would differentiate into one of six lineages: lymphoid (CLP), monocytic (Mono), erythroid (Ery), classic dendritic cells (cDC), plasmacytoid dendritic cells (pDC), or megakaryocyte (Mk). Based on the differentiation probabilities, Palantir also computes a differentiation potential as a measure of pluripotency. We visualized differences in differentiation trajectories by plotting cellular density relative to the normal reference used for projection (6), revealing areas with higher or lower than expected densities across trajectories in a pseudotime map, as shown in Fig. 1D for all treatment-naïve samples.
CMML patients could be classified into three distinct trajectory biases using this visualization. Eight of 25 (32%) patient samples were “monocytic-biased,” showing a disproportionate number of cells mapping to a monocytic differentiation probability as expected in CMML. However, 8 of the 25 (32%) patient samples had a disproportionate number of cells mapping to megakaryocytic and erythroid differentiation probabilities, termed “MEP-biased,” and 9 of 25 (36%) patients mapped to a third group with cell densities of similar proportions to that of the normal samples, termed “normal-like.”
The robustness of our classification was tested in two ways. First, we analyzed the CMML scRNA-seq data as a pseudo-bulk data set. This was accomplished by using the arithmetic mean for the scaled expression values of the top 2,000 genes (ranked by expression variability) and computing an averaged matrix consisting of unique samples and these representative genes. The resultant matrix was used to perform Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP; ref. 10) to visualize the distances between patient samples and Ward hierarchical clustering to group samples. The pseudo-bulk UMAP and Ward hierarchical clustering structure preserved the initial classification, suggesting that these three differentiation trajectories were robust, as highlighted in Supplementary Fig. S2A–S2E.
Further, we validated the bias assignment by comparing averaged RNA expression in granulocyte-GMP, megakaryocytic erythroid progenitors, and HSC gene signatures (11). As expected, monocytic-biased CMML was highly enriched for genes upregulated in GMPs, MEP-biased CMML was enriched for genes upregulated in MEPs, and normal-like showed no distinct bias (Fig. 1E). These lineage skewing trajectories were also observed when CMML samples were mapped onto a single-cell proteo-genomic reference map of normal hematopoiesis (11), whose cells had pseudotime analysis previously characterized by Slingshot (ref. 12; Supplementary Fig. S3).
Last, we evaluated whether there were clinicopathologic differences between the groups that could substantiate the clinical relevance of our classification and provide insight into the basis for these disparate differentiation trajectories. We abstracted a total of 33 clinicopathologic features from each patient and observed that MEP-biased CMML was associated with higher hemoglobin levels (median 12.20 g/dL in MEP-biased compared with 9.7 g/dL, P: 0.0037, Mann–Whitney test; Fig. 1F) consistent with the assigned bias. Although there was no association between monocytic-biased CMML and leukocytosis or monocytosis (Supplementary Fig. S4), monocytic-biased CMML was associated with an inferior survival (median survival of monocytic-biased patients was 39 months compared with 51 months; P: 0.0349, log rank; Fig. 1G) and elevated lactate dehydrogenase level, which is a common clinical indicator of inflammation (P = 0.0378, Fisher exact test; Fig. 1H). Further, the majority of differentiation projections from patient samples displayed depletion of the lymphoid (CLP) lineage independent of trajectory bias as expected in CMML (P: 0.0004, Mann–Whitney test; Fig. 1I and J).
HSC Depletion Is Associated with Myeloblast Expansion in Monocytic-Biased CMML
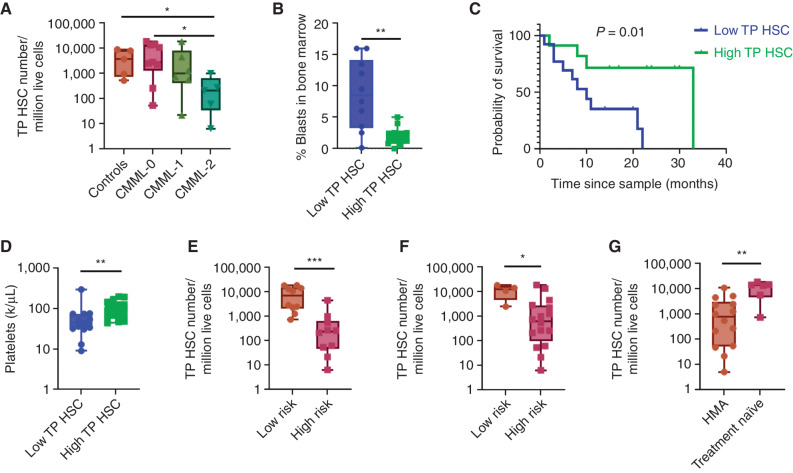
We also observed a decrease in the HSC compartment, defined as those cells assigned as transcriptionally pluripotent by Palantir, that was most pronounced in patient samples defined as “monocytic-biased” CMML (P: 0.0004, Mann–Whitney test; Supplementary Fig. S5; Fig. 1D and E). This was validated by quantifying the relative expression of genes across a variety of published HSC gene signatures (13–17) at single-cell resolution for each sample (Supplementary Fig. S6). We further validated HSC depletion by profiling well-defined, flow cytometry–based, HSC immunophenotypes that included Lin−CD34+CD38−CD45RA−CD90+CD49f+ (triple-positive HSCs); Lin−CD34+CD38−CD45RA−CD90+ (double-positive HSCs); and Lin−CD34+CD38− (single-positive HSCs; Supplementary Fig. S7) in a separate cohort of CD34+ enriched CMML bone marrow mononuclear cell samples from 20 individuals across all WHO-defined subtypes (3).
When analyzing the triple-positive HSC immunophenotype (TP HSC), defined as Lin−CD34+CD38−CD45RA−CD90+ CD49f+, a significant and progressive reduction in the frequency of HSCs was observed as the WHO subtype increased, suggesting that HSC depletion is associated with CMML disease progression. For example, CMML-2 cases had significantly reduced frequency of TP HSC cells compared with CMML-0 and controls (CMML-2 with 212 per million vs. CMML-0 with 2,827 per million, P = 0.02; vs. controls with 3,689 per million, P = 0.03, Mann–Whitney test; Fig. 2A). We observed a similar reduction in HSC frequency across WHO subtypes using the double-positive HSC immunophenotype (DP HSC), defined as Lin−CD34+CD38−CD45RA−CD90+ (CMML-2 with 1,116 per million vs. controls with 42,248 per million, P = 0.007; vs. CMML-0 with 26,450 per million, P = 0.01, Mann–Whitney test; Supplementary Fig. S8A). Lastly, we observed a reduction in the frequency of HSCs in CMML-2 cases as compared with controls in single-positive HSCs (SP HSC), defined as Lin−CD34+CD38− (CMML-2 with 17,506 per million vs. controls with 105,135 per million, P = 0.007, Mann–Whitney test; Supplementary Fig. S8B), but did not find a reduction in HSC content between CMML-0/1/2 cases, likely because this immunophenotype also captures non-HSC myeloid progenitors and hence can also be considered as HSPC.
Figure 2.
Monocytic-biased HSPC are characterized by HSC depletion. A, Comparison of HSC frequency between controls and WHO-classified CMML stages using flow cytometry showed HSC depletion with disease progression in triple-positive HSCs, n = 20 patient cases and five control cases. B, Evaluation of bone marrow blast content between low HSC and high HSC groups of patients showed that blast content was inversely correlated with HSC numbers in triple-positive HSCs, n = 20 patient cases. Data were analyzed using nonparametric Mann–Whitney test (C) KM survival analysis showed inferior survival in patients with low HSC content compared with patients with high HSC content using triple-positive HSC immunophenotype, n = 26 patient cases (log-rank P = 0.01). D, Patients with low numbers of triple-positive HSCs showed thrombocytopenia (P = 0.005), (E) were at higher risk per MDACC (P = 0.0003), and (F) Mayo (P = 0.02) prognostic scoring systems. G, Patients who underwent HMA therapy had significantly lower triple-positive HSC content than treatment-naïve patients, n = 26 patient cases. Data were analyzed using nonparametric Mann–Whitney test; P value significance represented by *, < 0.05; **, < 0.01, ***, < 0.001.
Since the WHO classification of CMML (CMML-0/1/2) is exclusively based on myeloblast expansion within the peripheral blood and bone marrow, we compared HSC numbers to pathologically observed myeloblast content. Patients were classified as high or low HSC content based on HSC immunophenotype using the median number of HSCs across all patients as a cutoff point. In the triple- and double-positive HSC immunophenotypes, a significant inverse association between myeloblast and HSC content was observed (TP HSC: 8.5% blasts-low HSC group vs. 1.5% blasts-high HSC group; P = 0.004; DP HSC: 8.5% blasts vs. 1.85% blasts; P = 0.01, Mann–Whitney test; Fig. 2B; Supplementary Fig. S8C). We observed a similar trend between high and low HSC groups using the HSPC immunophenotype, but the difference was not statistically significant (Supplementary Fig. S8D).
Consistent with the notion that HSC depletion is an adverse clinical feature, we observed that patients with low HSC content were associated with significantly inferior survival (low HSC: 10 months vs. high HSC: 33 months; P = 0.01, log-rank; Fig. 2C), thrombocytopenia (low HSC: 44 k/μL vs. high HSC: 104 K/μL; P = 0.005, Mann–Whitney test; Fig. 2D), and higher risk disease based on the MD Anderson Cancer Center (MDACC; P = 0.0003, Mann–Whitney test; Fig. 2E) and Mayo Clinic CMML prognostic scoring systems (P = 0.02, Mann–Whitney test; Fig. 2F). We also observed that patients treated with hypomethylating agents (HMA) at the time of sampling had significantly lower HSC content as compared with treatment-naïve patients (HMA: 777.8/1 × 106 vs. treatment-naïve: 13,9201 × 106; P = 0.003, Mann–Whitney test; Fig. 2G), suggesting that HMA therapy may influence HSPC states (18).
Monocytic-Biased CMML Is Associated with Expansion of Inflammatory GMP-like Cells
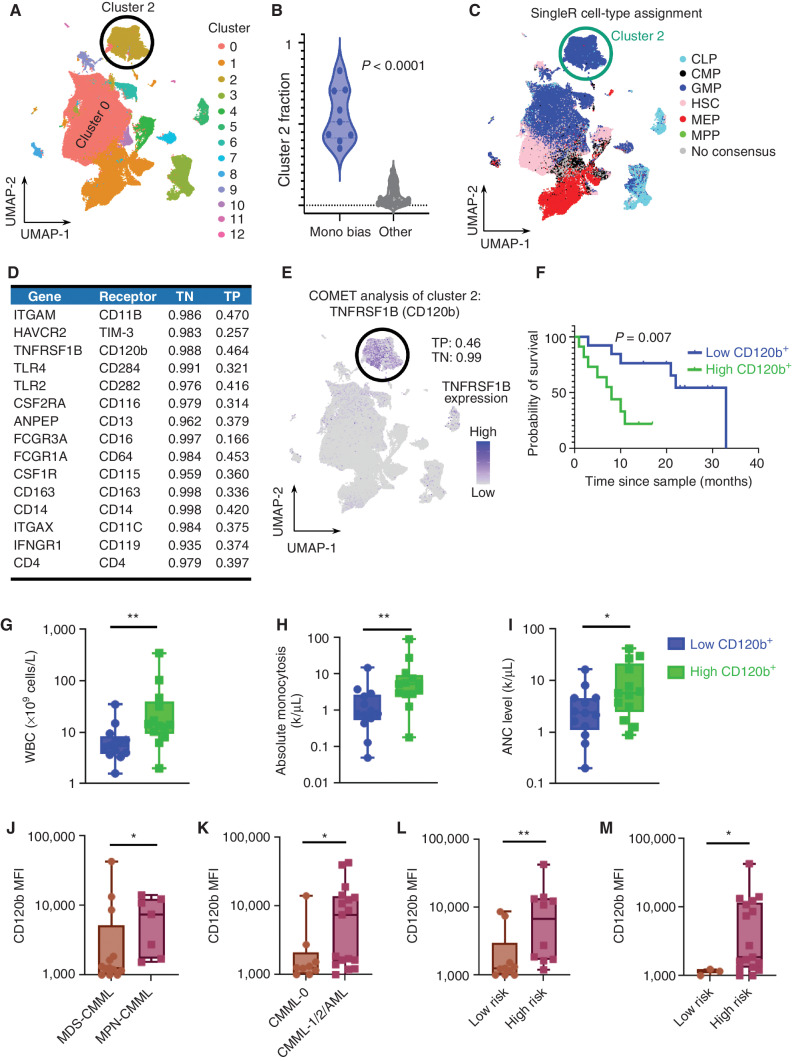
Given the adverse clinical features and HSC depletion seen in monocytic-biased CMML, we sought to define the cellular origin and mechanisms that could potentiate this abnormal HSPC state. We implemented a pipeline for scRNA-seq analysis in Seurat (19) that performed dimension reduction [principal component analysis (PCA) and UMAP (10)] and clustering (Louvain algorithm) detailed in Methods (Fig. 3A). We observed that monocytic-biased samples were significantly enriched for cluster 2 (Clus2) cells, suggesting that the fraction of cells in Clus2 is the major driver for monocytic-biased assignment (monocytic-biased: median 51.3% cells in Clus2; other: 3.9%, P < 0.0001; Fig. 3B). Further, normal samples had very few cells assigned to Clus2 (median 0.4%), suggesting that expansion of this population is a feature unique to monocytic-biased CMML (Supplementary Fig. S9).
Figure 3.
Monocytic-biased CMML is associated with the expansion of an inflammatory GMP-like HSPC population. A, Graph-based clustering of the CD34+ cohort identified 13 distinct clusters across the 201,250 single cells. B, Samples in the monocytic-biased group were enriched in Clus2. C, SingleR was used to determine cell-type assignment using previously published paired references of bulk RNA-seq of flow cytometry–sorted cells. Clus2 was enriched for GMP cell-type assignment. D, COMET was used to identify differential gene-expression markers well-suited for validation with flow cytometry, and the top-ranked markers had an average log base 2-fold change of 4.50 in the single genes from Clus2 compared with the rest of the cells, average true positive rate of 36.7%, and average true negative rate of 98.0%. E, COMET identified TNFRSF1B (encoded cell-surface marker CD120b) as the best single marker for identifying Clus2 cells with a true positive performance of 46% and true negative performance of 99%. F, The predictive power of CD120b was validated in the complementary flow cytometry data set, where individuals with high CD120+ expression had inferior survival (n = 26; log-rank P = 0.007). Clinicopathologic variables were compared in CD120+ high vs. low patients and individuals with CD120+ high expression had (G) increased white blood cell (WBC) counts (P = 0.007), (H) increased absolute monocytosis (P = 0.004), and (I) increased absolute neutrophil count (ANC; P: 0.04). Patients with high CD120b GMPs were (J) myeloproliferative (P = 0.04), (K) associated with disease progression (P = 0.02), and (L) were categorized as high risk per MDACC (P = 0.005) and (M) Mayo prognostic scoring systems (P = 0.03), n = 26 patient cases. Nonparametric Mann–Whitney tests were used to compare two group data. P value significance represented by *, < 0.05; **, < 0.01; ***, < 0.001.
Next, we implemented singleR (20), an R package that assigns cell type using RNA-seq data from flow cytometry–sorted references. Figure 3C shows the singleR cell-type assignment for all cells, using flow-sorted reference data from four distinct sources (21). Virtually all (98.8%) of the cells within Clus2 were identified as GMP, which was additionally corroborated by scoring cells based on other published GMP signatures (ref. 13; Supplementary Fig. S10). Although most Clus2 cells were GMP-like, GMP-like cells (56.6%) were also assigned to cluster 0. These non-Clus2 GMPs were clearly distinct in UMAP (Supplementary Tables S2 and S3), motivating us to explore whether Clus2 may represent a recently described GMP subtype (22). Indeed, Clus2 cells were associated with upregulation of CTNNB1 (Supplementary Fig. S11A) and low IRF8 expression (Supplementary Fig. S11B), as well as upregulation of WNT signaling transcription (Supplementary Fig. S11C) and Fc gamma receptors (FCGR) upregulation (Supplementary Fig. S12A–S12F) consistent with self-renewing so-called cluster GMPs observed to be expanded in murine models of leukemia and emergency granulopoiesis (22, 23).
To validate the cellular identity of Clus2 cells by flow cytometry, we implemented COMET (24), an established tool that approximates flow cytometry markers that can be used to identify clusters of cells from scRNA-seq data. This analysis identified ten putative markers (Fig. 3D) with a true negative rate above 98% (average, 98.9%), an average log base 2 fold change of 5.08 in the single genes from Clus2 compared with the rest of the cells, average true positive rate of 37.5%, including TNFRSF1B, which encodes the surface receptor CD120b, as the most promising receptor for approximating Clus2 cells in flow cytometry data sets (Fig. 3E; true negative 98.8%, true positive 46.4%). We then profiled CD120b expression by flow cytometry across progenitors and observed that CD120b expression was significantly higher in GMPs of patients as compared with stem and other progenitor populations [HSCs, common myeloid progenitors (CMP), and MEPs], consistent with the proposed Clus2 cell identity from our scRNA-seq analysis (Supplementary Fig. S13). Next, CD120b+ median fluorescent intensity (MFI) was calculated for each patient sample, and samples were classified as high CD120b+ or low CD120b+ based on whether their MFI was above or below the median. Using this cutoff point, patients with high CD120b+ expression in GMPs had significantly inferior survival as compared with patients with low CD120b+ (high CD120b+: 8 months vs. low CD120b+: 33 months, P = 0.007, log-rank; Fig. 3F and was also merged with the scRNA-seq cohort to increase power the log-rank P = 0.001 and hazard ratio of 2.76; Supplementary Fig. S14). Patients with high CD120b+ expression also had significantly higher WBCs (high CD120b+: 13.90 × 106 cells/L vs. low CD120b+: 5.49 × 106 cells/L; P = 0.007, Mann–Whitney test), absolute monocytosis (high CD120b+: 5.34 vs. low CD120b+: 0.81; P = 0.004, Mann–Whitney test), and neutrophils (high CD120b+: 5.7 vs. low CD120b+: 2.36; P = 0.04, Mann–Whitney test; Fig. 3G–I). CD120b+ high GMP samples were also associated with patients who were myeloproliferative (P = 0.04, Mann–Whitney test), had undergone disease progression by WHO classification (P = 0.02, Mann–Whitney test), and were at significantly higher risk by both the MDACC (P = 0.005, Mann–Whitney test) and Mayo clinic prognostic scoring systems (P = 0.03, Mann–Whitney test) consistent with monocytic-biased CMML (Fig. 3J–M). Other putative markers by COMET analysis that could be analyzed with our flow cytometry panel included TLR4, which encodes the surface receptor CD284 (Supplementary Fig. S15A). Flow cytometry–based investigation revealed that CD284 expression was significantly higher in GMPs of patients as compared with stem and other progenitor populations (Supplementary Fig. S15B). Patients with high CD284 expression also had significantly inferior survival as compared with patients with low CD284 expression (high CD284+: 10 months vs. low CD284+: undefined, P = 0.007, log-rank; Supplementary Fig. S15C). Collectively, these data suggest that the monocytic-biased CMML HSPC state is characterized by enrichment of GMP-like cells that may have self-renewal capacity and can be approximated by flow cytometry markers such as CD120b and CD284.
Clus2 Cells Are Not Clonally Distinct and Have Clinically Relevant Temporal Dynamics
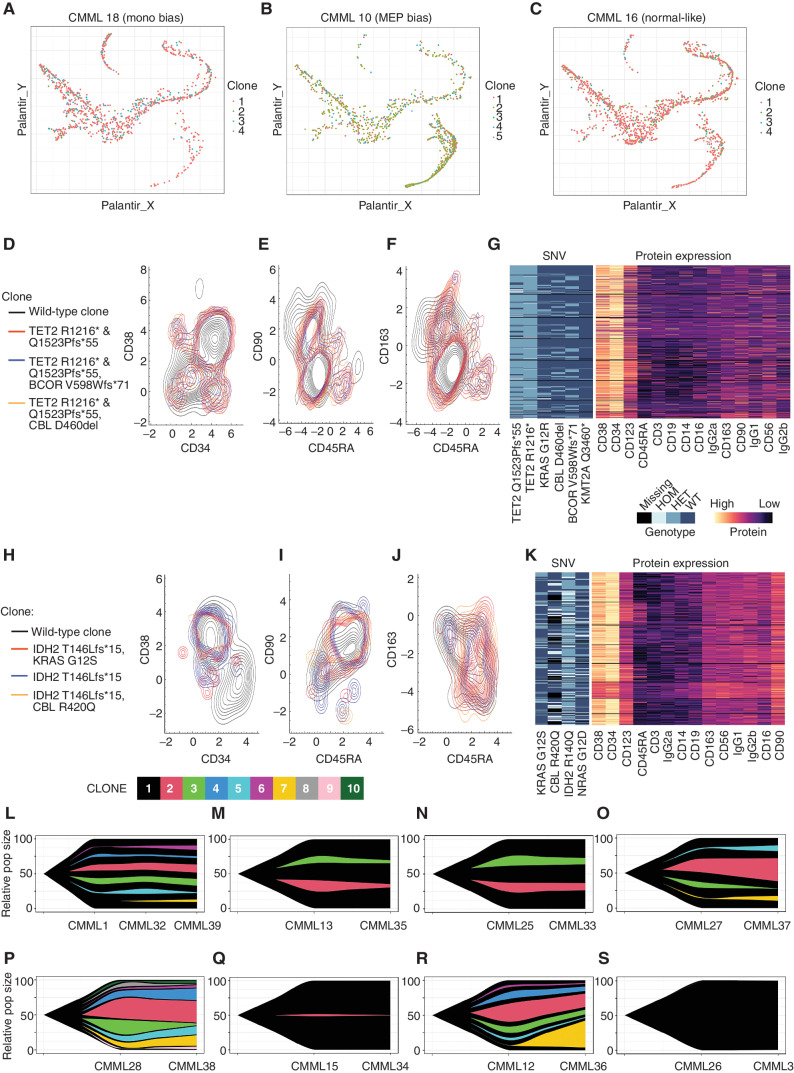
We next sought to explore the clonal origin and temporal dynamics of Clus2 cells. Given the excellent coverage of mitochondrial variants in scRNA-seq data derived from the 10X platform, we first used mitoClone (25), a tool that enables the detection of clonal populations based on mitochondrial variants, to determine whether the clonal composition of Clus2 cells was distinct from other clusters. We identified 1 to 25 clones (mean clone number 9.31) per sample and mapped these clusters onto both UMAP and pseudotime differentiation trajectories in a patient-specific manner. This analysis identified no statistical enrichment of mitochondrial variant-defined clones in Clus2 cells across all HSPC-biased subtypes (representative clonal mappings shown in Fig. 4A–C, all mappings shown in Supplementary Fig. S16). We next performed multiomic scDNA-seq coupled with immunophenotyping using a pool of 45 oligo-conjugated antibody panels in a representative nonmonocytic-biased (Fig. 4D–G) and monocytic-biased (Fig. 4H–K) CD34+ enriched BMMNC patient specimen.
Figure 4.
The monocytic-bias CMML phenotype is not driven by a clonally distinct population. Cells from monocytic-biased (A), MEP-biased (B), and normal-like (C) CMML samples plotted in a pseudotime map show differentiation trajectories. Cells are colored by their mitoClone assigned clone and do not show any association between trajectory and clone. D–G, A MEP-bias and low CD120b+ patient from the flow cytometry cohort (4-I-001) and (H–K) a monocytic-bias and high CD120b+ patient from the flow cytometry cohort (5-M-001) were selected for clonal characterization using single-cell DNA sequencing coupled with protein expression for hematologic markers with the Mission Bio Tapestri Platform. Contour plots of the distribution of distinct genotypic clones were visualized with their corresponding protein expression data. D and H, CD34 vs. CD38 (E, I), CD45RA vs. CD90, and (F, J) CD45RA vs. CD163 highlight the normal vs. malignant GMP population, where CD163 serves as a marker of monocytic-biased cells. G and K, Multiassay heatmaps highlight the variants of interest used to define clones and their corresponding protein expression at single-cell resolution. In a complementary analysis in the scRNA-seq cohort, clonal populations derived from mitochondrial RNA mutations from the scRNA-seq were also performed on sequential samples using mitoClone. Across time points, samples show very similar clonal distributions regardless of treatment with (L–O) HMA, (P–Q) ruxolitinib, (R) chemotherapy, and (S) no treatment.
We visualized the multiomic data by plotting the protein expression of markers relevant to HSC (Fig. 4D–E and Fig. 4H–I) and putative Clus2 cell (Fig. 4F and J) identity for each mutationally defined clone. Because CD120b and CD284 were not available in this multiomic platform, putative Clus2 cells were defined by CD163 expression, originally noted in COMET analysis (Fig. 3D). This enabled us to observe that HSC and putative Clus2 cell identity was not enriched in a specific clone, consistent with our mitoClone analysis. Indeed, CMML clones appeared to have protein expression phenotypes that were far more similar between each other than compared with patient-specific wild-type cells. Moreover, clustering based on a lager panel of protein expression identified no clonal subtypes (Fig. 4G and K) that could be resolved, suggesting that the hematopoietic architecture of these representative cases is not driven by a clonally distinct population.
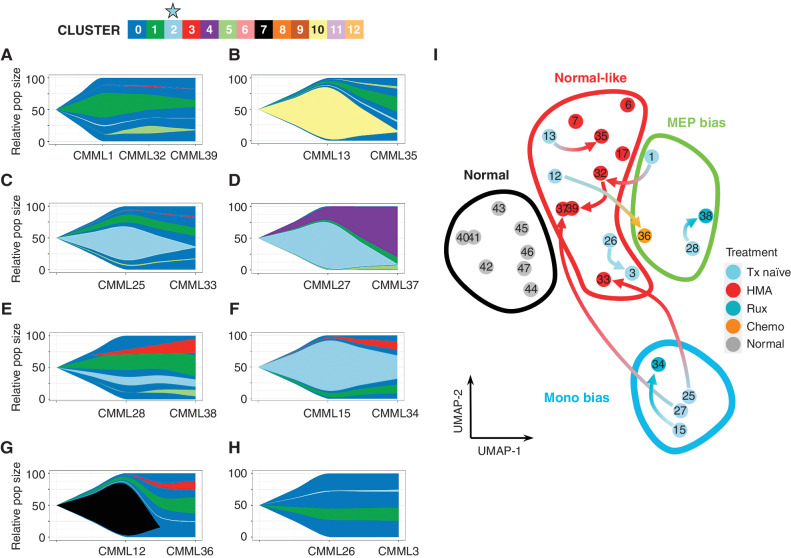
To explore the temporal dynamics of Clus2 cells, we performed scRNA-seq on eight sequential samples before and after treatment. Although treatment with ruxolitinib did not affect Clus2 cells, we observed a marked decrease in the fraction of cells in Clus2 in all HMA-treated cases (n = 4), which was most pronounced in monocytic-biased patients (n = 2). Further, clonal tracing using mitochondrial variants (25) in these cases demonstrated that although the fraction of Clus2 cells was decreased, the clonal composition was unaffected, consistent with previous work (ref. 26; clonal dynamics in Fig. 4L–S, cluster dynamics in Fig. 5A–H) and suggesting that HMA therapy can redirect the monocytic-biased differentiation trajectories (27). This was also evident in UMAP space where all unique patient samples, including sequential samples, were plotted and grouped by their hematopoietic trajectory. The arrows in this analysis denote the change in hematopoietic trajectory observed after treatment, most strikingly observed in HMA-treated cases (Fig. 5I).
Figure 5.
Hypomethylating agent treatment is associated with the depletion of Clus2 cells. Although mitoClone revealed stable clonality distribution after treatment, as demonstrated in Fig. 4L–S, the cluster dynamics changed dramatically after treatment with (A–D) HMA, (E–F) ruxolitinib, (G) chemotherapy, and (H) no treatment. Interestingly, two HMA-treated patients (C–D) were monocytic-biased in their pretreatment sample and showed a marked decrease in cluster 2 fraction (light blue) following HMA treatment. I, Sample movement in pseudo-bulk representation on UMAPs following treatment (complementary analysis in Supplementary Fig. S2; sample numbering matches the sample numbering in Fig. 4L–S and Fig. 5A–H). Three of the four pre-HMA treatment samples move (red arrows) from monocytic-biased or MEP-biased to normal-like phenotype after treatment with HMA. Additionally, three samples were collected from patients after HMA treatment (without a matched treatment-naïve sample) that were also grouped in this normal-like phenotype. Ruxolitinib treatment shows some change in transcriptomics but is not sufficient to be classified with a new differentiation trajectory following treatment.
Clus2 Cells Are Enriched for Inflammatory Transcriptional and Proteomic Programs
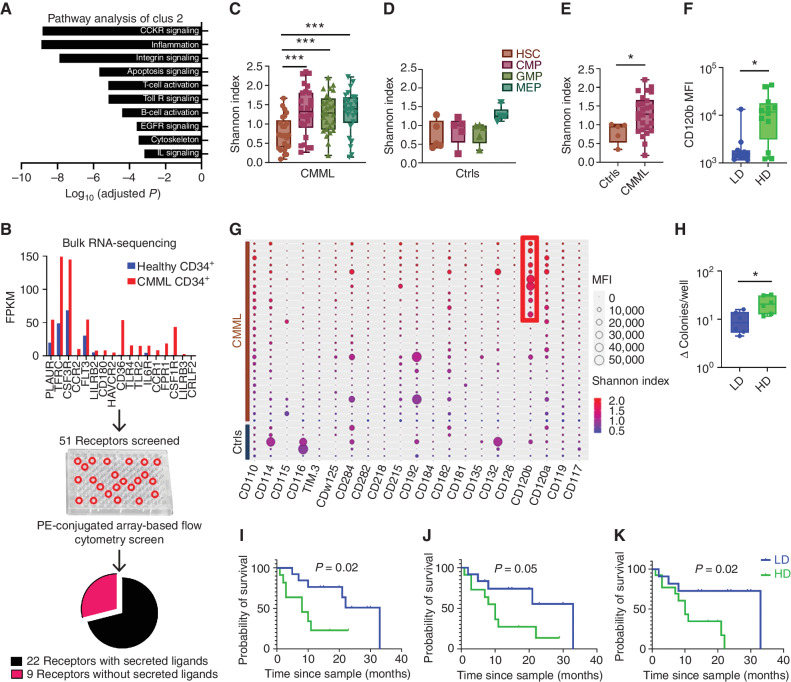
To explore the unique molecular features of Clus2 cells, we performed pathway analysis [Enricher (28) and PANTHER (29) pathways] on differentially expressed genes elevated in Clus2 and identified enrichment in inflammatory pathways to include cytokine receptor signaling as shown in Fig. 6A. We validated this inflammatory transcriptional profile by broadly profiling cytokine receptor protein expression in CMML HSPC using high-parameter flow cytometry in immunophenotypically well-defined hematopoietic populations at single-cell resolution. To build this high-parameter flow cytometry panel, we used bulk RNA-seq data from publicly available CD34+ CMML (30–34) and healthy data sets and prioritized 51 receptors as differentially expressed in CMML (Fig. 6B; Supplementary Table S4).
Figure 6.
Clus2 cells are enriched for inflammatory transcriptional and proteomic programs. A, Panther pathway analysis of genes differentially expressed in Clus2. Clus2 cells showed the most significant association with cytokine receptor signaling and inflammation. B, Analysis of publicly available bulk RNA-seq data sets showed differential regulation of receptors between healthy and CMML CD34+ cells. The shortlisted 51 receptors were evaluated for expression and frequency data using PE-conjugated flow cytometry screen (n = 4 patients, 15 healthy subjects). Based on the data from the screen, 22 cytokine receptors were shortlisted to be included in the 28-color flow cytometry panel. C and D, Comparison of CRD using the Shannon index showed an increase in CRD in myeloid progenitors of patients (CMML) compared with HSCs but not in controls (Ctrls). E, Comparison of CRD between patients and controls showed a higher Shannon index in patient GMPs (P = 0.04), n = 26 patient cases, and five control cases. F, High diversity (HD) patient GMPs showed significantly higher CD120b expression as compared with low diversity (LD) GMPs (P = 0.03), n = 26 patient cases. Data were analyzed using a nonparametric Mann–Whitney test. G, Balloon plot shows MFIs of cytokine receptors in 26 patient cases and five control cases, along with the Shannon index for each patient and control. As highlighted in the red box, the high diversity patient GMPs showed higher CD120b expression. H, Colony-forming assays with BMMNCs derived from high diversity (n = 2) and low diversity cases (n = 2) showed a significantly higher ratio of the total number of colonies per well in the methylcellulose-based medium with recombinant human cytokines/total number of colonies per well in the methylcellulose-based medium without human cytokines in high diversity as compared with low diversity cases (P = 0.02). Data were analyzed using a nonparametric Mann–Whitney test. I, KM survival analysis showed inferior survival in patients with high-diversity GMPs (log-rank P = 0.02), (J) high-diversity triple-positive HSCs (log-rank P = 0.05), and (K) high-diversity double-positive HSCs (log-rank P = 0.02) as compared with patients with the low-diversity counterparts, n = 26 patient cases. Data were analyzed using log-rank (Mantel–Cox) test. P value significance represented by *, < 0.05; **, < 0.01; ***, < 0.001.
We mapped the previously unknown relative expression intensities and frequency of our prioritized receptors in HSPC, using phycoerythrin (PE)-conjugated antibodies for all 51 receptors in an array-based format enabling relative comparison of expression and frequency in CMML and normal pooled CD34+ samples (n = 4 patients, 15 healthy subjects) as described in Methods (Fig. 6B). We further prioritized receptors by comparing their expression based on ≥10 positive cells for each respective receptor; ≥10% of Lin−CD34+CD38− population; and ≥5 MFI ratio of CMML to normal. Based on these criteria, we identified 22 receptors with cognate ligands differentially expressed in CMML versus normal (Fig. 6B). We optimized a final 30-parameter flow panel considering each receptor's expression, frequency, and spillover spreading error (35) that includes all 22 cytokine receptors and six markers to define HSPC (Supplementary Fig. S17A–S17F). We acquired data from bone marrow–derived CD34+ cells of 26 patients and five controls, performing dimensionality reduction (UMAP; ref. 10) and clustering (Phenograph; ref. 36) in stem and myeloid progenitor populations (Supplementary Fig. S17G). Lastly, to generate a summary metric that considered the entire 22 cytokine receptor panel, we calculated the Shannon index to compare cytokine receptor diversity (CRD) across all the patient samples and normal controls gated for respective stem and myeloid progenitor cell types (HSCs, CMPs, GMPs, and MEPs).
This analysis demonstrated a significant increase in CRD in myeloid progenitors compared with HSCs in patients but not in healthy controls Fig. 6C and D. Furthermore, a comparison of diversity between patients and controls identified a significant increase in CRD of GMPs (P = 0.04, Mann–Whitney test; Fig. 6E). CD120b+ expression was also significantly higher in high CRD CMML GMPs compared with those with low CRD (high diversity-CD120b MFI: 8,072 vs. low diversity CD120b MFI: 2,444; P = 0.03, Mann–Whitney test; Fig. 6F and G). Clonogenicity assays using bone marrow mononuclear cells (BMMNC) derived from high diversity and low diversity cases demonstrated that high CRD patient samples had a statistically significant increased response to a cocktail of recombinant human cytokines (37) and elevated in CMML (38–40) compared with low diversity cases (P = 0.02, Mann–Whitney test), suggesting that CRD is functionally relevant in vitro (Fig. 6H). Higher CRD was also significantly associated with inferior survival (high diversity: 8 months vs. low diversity: 33 months; P = 0.02. log rank; Fig. 6I), consistent with our scRNA-seq Clus2 analysis. A significantly inferior survival was also noted in triple- and double-positive HSCs with high CRD (triple-positive HSCs—high diversity: 10 months vs. low diversity: 33 months; P = 0.05; double-positive HSCs: P = 0.02, log-rank; Fig. 6J and K).
Competitive Bone Marrow Transplantation and Inflammatory Stress-Induced Hematopoiesis Can Induce a GMP-like Expansion in CMML Models
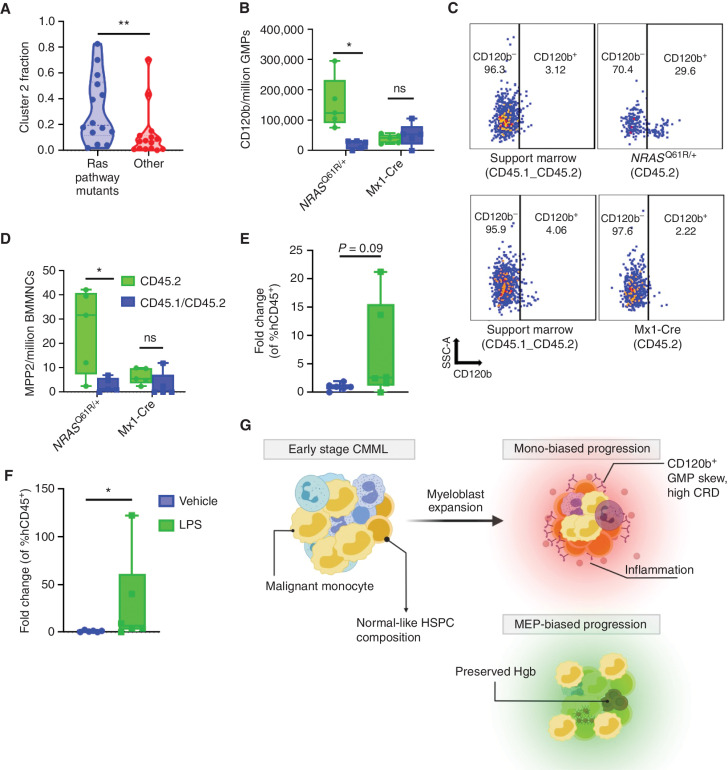
Given the clinical and molecular evidence supporting monocytic-biased CMML as an inflammatory GMP-like expansion of Clus2 cells (Figs. 3B and 6A), we hypothesized that stress/inflammatory events could contribute to the expansion of inflammatory GMP-like HSPC and other early myeloid-biased progenitor cell populations, during disease progression. We modeled proliferative stress-induced hematopoiesis by performing bone marrow transplant experiments with NRASQ61R/WT bone marrow cells and controls. An NRAS-competitive transplant model was chosen because it recapitulates CMML-like features (41), and RAS pathway mutations were associated with increased Clus2 fraction in our human scRNA-seq data set (median NRAS, KRAS, and/or CBL mutants: 19.6%, median none: 7.1%, P = 0.009; Fig. 7A). The results from the long-term competitive transplant experiment demonstrated an expansion of inflammatory GMPs (P = 0.02, multiple paired t test; Fig. 7B and C), as measured by CD120b expression, and myeloid-biased multipotent progenitor 2 (MPP2) cells (P = 0.03, multiple paired t test) compared with controls recapitulating the HSPC compartment in human monocytic-biased CMML, 41 weeks after transplantation (Fig. 7D). We did not observe significant differences in other HSPC populations (GMPs, MEPs, CMPs, MPP1-4, LSKs, and HSCs) between NRAS and controls (Supplementary Fig. S18A–S18H). To validate this observation in human CMML cells, we modeled inflammatory stress-induced hematopoiesis in patient-derived xenografts (PDX). We selected CMML samples with RAS pathway mutations and generated PDX models as previously described (42). We confirmed engraftment by performing bone marrow aspirates and measuring hCD45 content by flow cytometry and treated mice with 10 μg i.p. LPS or vehicle. We also confirmed that this dose of LPS could induce human cytokine production in our CMML PDX 6 hours after injection (Supplementary Fig. S19). After 24 hours, mice were sacrificed and evaluated for human hematopoietic reconstitution. As seen in murine models, expansion of inflammatory GMPs was noted in LPS compared with vehicle controls suggesting that human CMML cells can augment HSPC composition in response to inflammation (Fig. 7E and F).
Figure 7.
Stress-induced hematopoiesis can induce a GMP-like expansion in CMML models. A, Samples from patients with RAS pathway mutations (KRAS, NRAS, and CBL) show elevated Clus2 fraction (median RAS mutated, n = 14: 0.196; median WT, n = 15: 0.071; P: 0.009 from nonparametric Mann–Whitney test). B, Competitive BMT studies showed an expansion of CD120b GMPs in NRASQ61R/+ compared with Mx1-Cre controls. C, Representative flow plot of B. D, Competitive BMT studies showed an expansion of myeloid-biased MPP2 in NrasQ61R/+ compared with Mx1-Cre controls. Support marrow (CD45.1/CD45.2) vs. CD45.2 NrasQ61R/+;Mx1-Cre, n = 5 NrasQ61R/+; mice and 5 Mx-1Cre mice. Data were analyzed using multiple paired t test. E and F, PDX models of RAS-mutant patients (n = 2) showed an expansion of GMPs and CD120b GMPs in LPS-treated mice compared with the corresponding vehicle; n = 6 LPS mice, n = 6 vehicle mice. Data are represented as fold change over vehicle by dividing individual data points by the average of the corresponding vehicle group giving a fold change. Data were analyzed using a nonparametric Mann–Whitney test. P value significance represented by *, < 0.05; **, < 0.01; ***, < 0.001. G, Schematic showing the three distinct HSPC biases identified in CMML. CMML HSPC are characterized by HSC depletion and myeloblast expansion. Monocytic CMMLs are characterized by high expression of inflammatory cytokine receptors such as CD120b, increased CRD and an inflammatory profile.
DISCUSSION
Chronic myeloid neoplasms have been historically defined by their effect on terminal hematopoiesis. Myeloblasts, however, include the self-renewing leukemic population and therefore drive clonal evolution and disease progression. Our study sought to map this critical cellular compartment by profiling CD34+ BMMNCs at single-cell resolution in a clinically and genetically annotated CMML cohort. Although CMML is characterized by peripheral monocytosis, our study identified three distinct HSPC differentiation trajectories associated with clinically relevant features and prognosis. Although the CMML patient samples and healthy controls were not age-matched, healthy controls were used solely as a reference map and therefore our observations were unlikely driven be age-dependent hematopoietic effects.
Our study transcriptionally and immunophenotypically demonstrated that HSC depletion is a feature of disease progression and myeloblast expansion. This observation has previously been made in murine models of leukemia (43, 44). However, our study recapitulates this in human leukemia and associates HSC depletion with disease progression, suggesting that HSC content may be a prognostically relevant feature in CMML.
Although several reports have suggested that inflammation and stress-induced hematopoiesis are features that contribute to the development of myeloid neoplasms (45), our data indicate that this may be the major driver of one, but not all, differentiation trajectories in CMML. The inflammatory disease trajectory, termed monocytic-biased CMML in our study, was characterized by adverse outcomes, HSC depletion, and the expansion of a GMP-like inflammatory population (Fig. 7G), transcriptionally enriched for cytokine receptor signaling and consistent with self-renewing GMPs previously described in murine models (22, 44, 46–48). Analysis of clinical outcomes in two distinct patient cohorts profiled by scRNA-seq and flow cytometry, respectively, affirmed that inflammatory GMPs are associated with an aggressive disease phenotype, poor prognosis, and inferior survival, thereby emphasizing the relevance of inflammatory cytokine signaling in chronic myeloid neoplasms.
To demonstrate that this inflammatory phenotype extended beyond transcriptional programs, we developed a 30-parameter flow cytometry panel designed to annotate cytokine receptors in CMML HSPC. To the best of our knowledge, we observe for the first time, at single-cell resolution, that cytokine receptor upregulation is present in chronic myeloid neoplasms and that increases in CRD constitute a molecular feature of disease progression. Given the known inflammatory milieu in the CMML bone marrow microenvironment (38), CRD may represent a context-specific fitness advantage leading to adverse outcomes. Further, our observation that CMML GMPs have the highest CRD relative to other CMML populations may also suggest that differentiation from HSC to GMP during disease progression represents an evolutionary strategy to increase CRD (Fig. 7G). Clonogenicity assays also suggested that high CRD progenitors may have a more robust response to inflammatory cytokines compared with low CRD cells consistent with a potential fitness advantage in this context.
Last, we explored whether the observed cellular differentiation trajectories were malleable after the diagnosis of CMML is fully manifested. Indeed, using sequential samples, we observed that HMA-treated patients had a profound depletion of the GMP-like population that defined monocytic-biased CMML. Consistent with previous studies suggesting that clonal architecture is preserved after HMA therapy (26), our clonal tracing analysis demonstrated that clonal composition remained unchanged despite profound depletion of Clus2 cells after HMA therapy, suggesting that these nonclonal dynamics may be clinically informative in the context of treatment. Furthermore, our murine and human NRAS-based model of stress-induced hematopoiesis also showed expansion of inflammatory GMPs consistent with previous studies that have suggested that human leukemia may usurp emergency myelopoiesis programs for disease initiation (23, 45, 49).
We show that stress-induced hematopoiesis can alter HSPC composition akin to hematopoiesis seen in monocytic-biased CMML, suggesting that inflammatory events, even after the disease is fully manifested, could adversely affect disease progression. Although enriching for CD34+ cells enabled us to finely map the HSPC compartment, our study could not explore the impact of CD34− progenitors or terminally differentiating cells on the self-renewing population.
Given the discordance between enrichment in RAS pathway mutation in monocytic-biased CMML and lack of RAS mutation enrichment in Clus2 cells, it is plausible that more terminally differentiated RAS-mutated populations contribute to HSPC composition in a non–cell-autonomous manner. Further, the continued deconvolution of HSPC at the single-cell resolution of other myeloid neoplasms and strategies to mitigate triggers of stress-induced hematopoiesis to prevent the monocytic-biased state should be explored (11, 50).
METHODS
Patient Samples and Characteristics
Bone marrow samples were obtained from a total of 55 patients. Our study was skewed toward Caucasian patients consistent with the known epidemiologic demographics of this disease (51, 52). Further, there were no gender-specific differences. Patient samples were collected after obtaining written informed consent. The use of human materials was approved by the Institutional Review Board of the Moffitt Cancer Center Scientific Review Committee and the University of South Florida Institutional Review Board in accordance with the Declaration of Helsinki. For the flow cytometry cohort, control CD34+ cells were sourced from Lonza, enriched from apheresis product, and BMMNCs from subjects without hematologic malignancies. Baseline patient characteristics for patient samples used in scRNA-seq and flow cytometry studies are shown in Supplementary Tables S5 and S6, respectively.
CD34 Enrichment of Patient Samples
Sixty million cryopreserved BMMNCs per sample were obtained from the Moffitt Tissue Core. Samples were thawed, washed with 20% FBS in PBS, and treated with DNase I for 15 minutes to reduce cell clumping. First, dead cell removal was performed according to the manufacturer's instructions (Miltenyi Biotech; catalog number: 130-090-101) followed by CD34 enrichment, also according to the manufacturer's protocol (Miltenyi Biotec; catalog number: 130-046-703). For the flow cytometry cohort, CD34 cells were enriched from BMMNCs based on the manufacturer's instructions (Miltenyi Biotec: 130-046-703).
scRNA-seq and Analysis
Chromium Single Cell 3′ Library, Gel Bead and Multiplex Kit and Chip Kit (10X Genomics v3) was used to encapsulate and barcode for cDNA preparation of enriched CD34+ BMMNCs in 39 samples across 29 unique patients. The targeted cell population sizes were 5,000 cells for each sample (Supplementary Table S7). The libraries were constructed according to the manufacturer's protocol, sequenced on an Illumina NovaSeq, and mapped to the human genome (GRCh38) using CellRanger (10X Genomics). Healthy control CD34+ enriched BMMNCs from 8 unique patients were integrated from publicly available data (6–8). Raw gene-expression matrices were generated per sample using CellRanger (v.3.0.1) were combined in R(v.4.0.2) and converted to a Seurat object (19). Quality control metrics removed cells with over two standard deviations of the mean of unique molecular identifier (UMI) count, less than 200 genes per cell, and those with over 25% of the UMIs derived from the mitochondrial genome. From the remaining 137,578 cells, the gene-expression matrices were log-normalized and scaled to remove variation due to total cellular read counts. To reduce the dimensionality of the data set, the first 200 principal components were calculated based on the top 2,000 variable genes, and Harmony (9) was applied to remove batch differences. All cells were clustered using the Louvain algorithm implemented by Seurat (v.3) with a resolution of 0.05; the algorithm identified 13 communities. The data were then visualized by running the UMAP algorithm (10). CMML patient samples and normal references (Supplementary Fig. S20) were individually projected onto an established hematopoietic reference [Setty (6) rep1, denoted Normal1 in this article] in t-SNE space and assigned different hematopoietic lineages from the references using the consensus assignment of the 30 nearest neighbors in the reference. Differential gene analysis was performed on clusters of interest (e.g., Clus2) using FindMarkers in Seurat and annotated using Enrichr (28, 53), pathway analysis (54), and COMET (24) was used to identify robust markers for flow cytometry expression. SingleR (20) was implemented for cell-type annotation for each sample compared against three hematopoietic references (55–58). mitoClone (25) was used to reconstruct clones using mitochondrial reads from scRNA-seq data, and evoFreq was used to visualize these clonal assignments as fish plots (59). Additional detail on the implementation of these methods is described in the Supplementary Methods. All computational pipelines for these analyses are publicly available on GitHub: https://github.com/mcfefa/CMML-diversity.
Single-Cell DNA and Protein Sequencing Preparation, Sequencing, and Analysis
CD34+ cells from a representative non–monocytic-biased and monocytic-biased patient were thawed, washed with PBS + 10% FCS, cell staining buffer (BioLegend; catalog number: 420201), and quantified using a Countess cell counter (Invitrogen). 1.0 × 106 cells were then resuspended in cell staining buffer and incubated with TruStain FcX (BioLegend; catalog number: 422301), blocking buffer (Tapestri Protein Staining Kit; catalog number: MB51-0017) for 15 minutes on ice. Post blocking, the cells were incubated with a pool of 45 oligo-conjugated antibodies (BioLegend; catalog number: 399906; Supplementary Table S8) for 30 minutes on ice. Cells were then washed multiple times with cell staining buffer followed by resuspension of the cells in Tapestri cell buffer (Tapestri Single-Cell DNA Core Ambient Kit v2; catalog number: MB51-007) and requantification. 10,000 single cells (4,000 cells/μL; >90% viability) were encapsulated, lysed, and barcoded using the Tapestri Single-Cell DNA + Protein Sequencing platform (MissionBio, Inc.). Using the MissionBio Tapestri Single-Cell DNA Myeloid Oligo Pool (catalog number: MB03-0036), barcoded samples were then subjected to targeted PCR amplification of 312 amplicons covering 45 genes known to be involved in myeloid malignancies (Supplementary Table S9). DNA PCR products were then isolated from individual droplets and purified with AMPure XP beads (BeckmanCoulter Life Sciences; catalog number: A63881). The DNA PCR products were then used as a PCR template for library generation and repurified using MPpure XP beads. Protein PCR products (supernatant from AMPure XP bead incubation) were incubated with Tapestri pullout oligo (5 μmol/L) at 96°C for 5 minutes followed by incubation on ice for 5 minutes. Protein PCR products were then purified using the included streptavidin beads (Invitrogen) and were used as a PCR template for the incorporation of i5/i7 Illumina indices followed by purification using AMPure XP beads. All libraries, both DNA and Protein, were quantified using the Qubit dsDNA BR Assay kit (Thermo Fisher Scientific; catalog number: Q32851) and the Agilent Bioanalyzer and were pooled for sequencing on an Illumina NovaSeq 6000 S4-300 lane by the MCC Molecular Genomics Core. The FASTQ files for scDNA libraries were analyzed through the Tapestri pipeline (v2). For the protein analysis, custom scripts in python (v3.7.12) were used leveraging Mission Bio's mosaic package (v2.0) to analyze both single-cell variant and antibody expression per cell. Variants were filtered through an empirically curated list of panel-specific mutations that were guided by cooccurrence with bulk sequencing. We then sought to define genotype clones, which we identified as cells that possessed identical genotype calls for the variants of interest. Dominant clones (highlighted in Fig. 4) were defined as the top three largest mutant clones in the sample, and the ild-type clone was defined as the genotype clone that was wild-type for all variants of interest in the sample.
PE-Conjugated Flow Cytometry Screen
CD34+ HSPC from CMML patients and healthy subjects (n = 4 patients and 15 healthy subjects) were used for the flow cytometry screen. Cells were incubated in a 96-well plate with Zombie Green (BioLegend) for 20 minutes in the dark at room temperature. Cells were washed twice with flow buffer. Cells were resuspended in BriIliant stain buffer (BD Biosciences; catalog number: 566349), and Human FcR Blocking Reagent (Miltenyi Biotec; catalog number: 130-059-901) was added to each sample and incubated for 10 minutes at room temperature. Without washing, titrated volumes of antibodies were added to each sample and incubated for 20 minutes at room temperature in the dark. Samples were washed and stained with streptavidin-PE (SA-PE) for 20 minutes at room temperature in dark. The cells were washed and resuspended in a flow buffer. Data were acquired using Symphony A-5 flow cytometer (BD). Single-stained cells, Anti-Mouse Comp Beads (BD Biosciences; catalog number: 552843) were used for compensation controls (BD). Fluorescence minus one (FMO) controls were used to set population gates. Data were analyzed using FlowJo 10 and GraphPad Prism 7 software. The analysis was performed using the single-positive HSC immunophenotype. Reagent details are provided in Supplementary Table S10.
Development of a 30-Parameter Flow Cytometry Panel
The high-parameter CR flow panel was built based on receptor expression and receptor frequency derived from the PE-conjugated flow cytometry screen. Based on receptor expression (MFI) data, the receptors with high expression were paired with fluorophores with low stain index, and receptors with low expression were paired with fluorophores with high stain index (Supplementary Fig. S17A and S17B). We factored in the data regarding receptor frequency because some receptors (for instance, TLR4) were found to have high expression but only in a small percentage of cells. In such cases, it was not prudent to conjugate such receptors with a fluorophore of low stain index. Based on these data, such receptors were conjugated to fluorophores with a medium stain index (Supplementary Fig. S17C). The next step in panel development was based on spillover spreading error (SSE; ref. 35). We utilized the SSE data from erythrocyte-lysed whole blood (LWB) stained with CD4 antibody conjugates on Symphony A5 (Supplementary Fig. S17D). These data enabled us to choose fluorophore pairs with low cross-talk.
We generated spillover spreading matrix (SSM) and FMO controls specific to our panel by staining compensation particles and cells with a titred volume of respective 28 antibodies/dyes (single-cell stain controls). A few fluorophore pairs had SSE >5 and FMO/compensation controls data indicated cell spreading that could interfere with signal resolution (Supplementary Fig. S17E). We identified the sources of spillover and took measures of reducing the antibody volumes of concerned markers in consideration with the antibody titration data and also used appropriate stain blockers. Based on this approach we generated a revised SSM (Supplementary Fig. S17F). Even though we found a few fluorophore pairs with SSE >5, the analysis of FMO data and compensation controls showed that the revised approach would not interfere in resolving distinct populations of receptors (Supplementary Fig. S17G).
Staining for CR in Human HSPC
CD34+ HSPC from patients and healthy donors were incubated in 5-mL flow tubes with Zombie Green for 20 minutes in the dark at room temperature. Cells were washed twice with flow buffer. Cells were resuspended in Brilliant Stain Buffer Plus (BD Horizon; catalog number: 566385) and Human FcR blocking reagent (Miltenyi Biotec; catalog number: 130-059-901). True-stain monocyte blocker (BioLegend; catalog number: 426102) was added to each sample and incubated for 10 minutes at room temperature. Without washing, titrated volumes of antibodies were added to each sample and incubated for 20 minutes at room temperature in the dark. Samples were washed and stained with SA-PE-Cy5.5/CF-350/BV570 in a sequential manner for 20 minutes each at room temperature in dark. The cells were washed and resuspended in a flow buffer. Data were acquired using a Symphony A-5 flow cytometer (BD). Sphero rainbow calibration particles (8 peaks), 3.0–3.4 μm (BD Biosciences; catalog number: 559123) were used to calibrate laser voltages for each fluorochrome. Single-stained cells, Anti-Mouse/Anti-Comp Beads (BD Biosciences; catalog number: 552843), Anti-Rat/Anti-Hamster Comp Beads (BD Biosciences; catalog number: 552845), UltraComp eBeads (Thermo Fisher; catalog number: 01-2222-42), and Anti-REA Comp Beads (Miltenyi Biotec; catalog number: 130-104-693) were used for compensation controls. Data were analyzed as explained above. Reagent details are provided in Supplementary Table S11.
Colony-Forming Assays
Colony-forming assays were performed using BMMNCs from 2 high diversity and 2 low diversity patients per the manufacturer's instructions. 2 × 104 BMMNCs from each patient were seeded in a methylcellulose-based medium with recombinant human cytokines (STEMCELL Technologies; catalog number: H4034) and in the methylcellulose-based medium without human cytokines (STEMCELL Technologies; catalog number: H4230) in triplicate using the SmartDish (STEMCELL Technologies; catalog number: 27370) and incubated under 5% CO2 at 37°C for 14 days. At the end of 14 days, the colonies were assessed using STEMvision (STEMCELL Technologies), and the data were represented as the ratio of the total number of colonies per well in the methylcellulose-based medium with recombinant human cytokines/total number of colonies per well in the methylcellulose-based medium without human cytokines.
Animal Models
All animals were housed in accordance with institutional standards set by Moffitt Cancer Center and the University of South Florida (USF), and all procedures were approved by the USF Institutional Animal Care and Use Committee (IACUC). B6 CD45.2 (C57BL/6J, stock #000664), B6 CD45.1 (B6.SJL-Ptprca Pepcb/BoyJ, stock #002014), Mx1-Cre (B6.Cg-Tg(Mx1-cre)1Cgn/J, stock #003556), and NSG-S (NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(CMV-IL3,CSF2,KITLG)1Eav/MloySzJ, stock #013062) mice were purchased from The Jackson Laboratory. C57BL/6J NrasLSL Q61R mice were a gift from Jing Zhang. NrasLSL Q61R mice were crossed to Mx1-Cre mice to generate NrasQ61R/+ experimental mice, which were hemizygous for the Mx1-Cre transgene. B6 CD45.2 mice were crossed to B6 CD45.1 to generate B6 CD45.1/CD45.2 mice. NSG-S mice were bred in a pathogen-free environment. All xenografts used 6- to 12-week-old NSG-S mice. Genotyping was performed by Transnetyx.
Competitive Bone Marrow Transplants
Femora and tibiae were dissected from freshly killed 5- to 10-week-old experimental mice (Mx1-Cre and NrasQ61R/+; Mx1-Cre, all of which express CD45.2 alone), and bone marrow was flushed into sterile saline. Red blood cells were lysed with sterile ACK lysis buffer (8.29 g/L ammonium chloride, 1 g/L potassium bicarbonate, 37 mg/L EDTA). Pelleted cells were resuspended in sterile saline and counted. Recipient 7- to 15-week-old mice were lethally irradiated with two doses of 500 cGy, 4 hours apart. 0.5 million bone marrow cells from Mx1-Cre, NrasQ61R/+;Mx1-Cre were mixed with an equal number of support marrow cells and injected via tail vein into recipient mice. B6 CD45.1 mice were used as transplant recipients and B6 CD45.1/CD45.2 mice were used as the source of support marrow. Chimerism was assessed in peripheral blood by staining for mCD45.1 and mCD45.2 and analysis by flow cytometry starting at four weeks after transplant and was repeated every four weeks. Five weeks after transplant, mice receiving cells from Mx1-Cre strains received two i.p. injections of 250 μg pI-pC (Millipore Sigma) 48 hours apart. The mice were euthanized 41 weeks after transplantation, myeloid tissues were collected, and flow cytometry was performed using an LSR II flow cytometer (BD) and analyzed with FlowJo v10.
Murine Stem and Progenitor Panel Staining
The murine stem and progenitor pool investigations were performed on Mx1-Cre and NrasQ61R/+;Mx1-Cre mice. Bone marrow cells were isolated from mice femora and tibiae. Flow cytometry was performed using BD LSRII (BD), and data were analyzed as explained above. The following gating strategy was used to study CD120b expression in HSCs and myeloid progenitors. LSK: lineage−c-KIT+Sca-1+; HSC: lineage−c-KIT+Sca-1+CD48−CD150+CD135−CD34−; MPP1: lineage−c-KIT+Sca-1+CD48−CD150+CD135−CD34+; MPP2: lineage−c-KIT+Sca-1+ CD48+CD150+CD135−CD34+; MPP3: lineage−c-KIT+Sca-1+CD48+CD150−CD135−CD34+; MPP4: lineage−c-KIT+Sca-1+CD48+CD150− CD135+CD34+; CMP: lineage−c-KIT+Sca-1−CD34+CD16/32−; MEP: Lineage−c-KIT+Sca-1−CD34−CD16/32−; GMP: lineage−c-KIT+Sca-1−CD34+CD16/32+. Reagent details are provided in Supplementary Table S12. The comparisons were made between the support marrow (CD45.1/CD45.2) and NrasQ61R/+; Mx1-Cre (CD45.2).
PDX Generation
Two to five million, T-cell depleted, BMMNCs from 2 unique CMML patient samples were transplanted via tail vein into 6 sublethally irradiated, 6- to 10-week-old female NSG-S mice. The number of mice per patient was determined by the number of BMMNCs available.
Two weeks after transplant, bone marrow biopsies were done on the left tibia of each mouse to monitor engraftment. The procedure was done under anesthesia and administration of analgesics both pre- and post-op. Once engraftment was detected, mice were randomized to receive 10 μg LPS (E.coli, O111:B4, Sigma-Aldrich) or saline vehicle via intraperitoneal injection. Mice were euthanized 24 hours later. Myeloid tissues and peripheral blood were collected post-mortem for further analysis. FACS flow cytometry using the LSRII was used to determine leukemic engraftment. Reagent details are provided in Supplementary Table S13.
PDX Plasma Cytokine Profiling
Peripheral blood was collected by submandibular bleed into a lithium heparin microtainer 6 hours after i.p. injection of LPS or vehicle (BD; catalog number: 365965). Plasma was isolated by spinning down the peripheral blood at 7,000 × g and removing the resulting supernatant. Cytokine levels were measured using the Human Proinflammatory 7-plex Tissue Culture Kit (Supplementary Fig. S19; Meso Scale Discovery; catalog number: K15008B-1). Before running the assay, plasma was diluted 1:2 using the provided diluent buffer. The Meso QuickPlex SQ 120 was used for imaging (Meso Scale Discovery; catalog number: AI0AA-0).
Statistical Analysis
Data were analyzed using nonparametric Mann–Whitney tests, Kruskal–Wallis test, multiple paired t test, Fisher exact test, and Chi-square analysis. Survival data were analyzed using log-rank (Mantel–Cox) test.
Data Availability
The bulk RNA-seq data used in flow cytometry panel development were from healthy and CMML CD34+ cells extracted from the following publicly available data sets: Healthy CD34+ cells: the Gene-Expression Omnibus (GEO): GSE69239, GSE55689, GSE63569, accession ID: PRJEB6573; CMML CD34+: GEO: GSE76203.
The scRNA-seq data from healthy CD34+ cells were extracted from three publicly available data sets: one patient sample was from 10X Genomics at https://www.10xgenomics.com/resources/datasets/cd-34-plus-cells-1-standard-1-1-0, three patient samples available from the Human Cell Atlas data portal at https://data.humancellatlas.org/explore/projects/091cf39b-01bc-42e5-9437-f419a66c8a45, and four patient samples available from GEO under GSE133181. Normal sample characteristics are shown in Supplementary Table S14.
The scRNA-seq data for the CMML CD34+ cells analyzed here have been made publicly available in the GEO: accession number GSE211033.
Code Availability
The code and documentation describing how to run our computational analysis pipeline and reproduce our results are open-source and publicly available GitHub repository: https://github.com/mcfefa/CMML-diversity. A virtual machine producing the environment is available on Code Ocean (doi: 10.24433/CO.1315403.v1): https://codeocean.com/capsule/9823313/tree/v1.
Supplementary Material
Acknowledgments
We thank Drs. Peter Van Galen and Lars Velten for technical assistance and discussion of mutations and clonal analysis in single-cell RNA-sequencing studies and application of mitoClone in our data set. This work has been supported in part by the Molecular Genomics Facility, Flow Cytometry Core Facility, and Biostatistics and Bioinformatics Shared Resource at the H. Lee Moffitt Cancer Center and Research Institute, an NCI-designated Comprehensive Cancer Center (P30-CA076292). We acknowledge the assistance and reagent support from BD Biosciences. This work was supported by the American Cancer Society ACS-IRG award IRG-17-173-22 and ACS-RSG award RSG-19-2015-01-LIB, NCI R37 CA234021-03, and the Myelodysplastic Syndromes (MDS) Foundation and the International Working Group for the Prognosis of MDS (IWG-PM). P.M. Altrock also acknowledges funding from Richard O. Jacobson Foundation, Moffitt Cancer Center Evolutionary Therapy Center of Excellence, and the William G. ‘Bill’ Bankhead Jr and David Coley Cancer Research Program (20B06).
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors’ Disclosures
M.C. Ferrall-Fairbanks reports other support from UF Foundation and grants from the American Cancer Society during the conduct of the study. R.S. Komrokji reports grants and personal fees from BMS, personal fees from Novartis, AbbVie, Jazz, Pharma Essentia, Servio, CTI, Geron, and Taiho outside the submitted work. D.A. Sallman reports consultancies: AbbVie, Inc., Molecular Partners AG, Takeda; advisory board: Syndax, Syros, BMS, Gilead, Kite, Janssen, Novartis, Servier, and Shattuck Labs; research funding: Shattuck Labs and Jazz. R. Bejar reports grants from MDS Foundation during the conduct of the study; other support from Aptose Biosciences and personal fees from Gilead and Epizyme outside the submitted work. P.M. Altrock reports grants from the American Cancer Society and NCI/NIH during the conduct of the study; grants from Richard O. Jacobson Foundation, Moffitt Cancer Center Evolutionary Therapy Center of Excellence, William G. “Bill” Bankhead Jr and David Coley Cancer Research Program, Kite Pharma/Gilead, and personal fees from CRISPR Therapeutics outside the submitted work. E. Padron reports grants from BD Biosciences, Incyte and Taiho during the conduct of the study; personal fees from Blueprint, Novartis, grants from Kura, and Stemline outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
M.C. Ferrall-Fairbanks: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. A. Dhawan: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. B. Johnson: Data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. H. Newman: Data curation, formal analysis, investigation, methodology, writing–original draft. V. Volpe: Investigation. C. Letson: Data curation, formal analysis, investigation, methodology, writing–original draft. M. Ball: Data curation, investigation. A.M. Hunter: Investigation, methodology. M.E. Balasis: Investigation, methodology, project administration. T. Kruer: Investigation. N. Ben-Crentsil: Investigation. J.L. Kroeger: Investigation, methodology. R. Balderas: Resources, investigation, methodology. R.S. Komrokji: Resources, data curation, investigation. D.A. Sallman: Resources, data curation, investigation. J. Zhang: Resources, writing–review and editing. R. Bejar: Conceptualization, resources, funding acquisition, investigation. P.M. Altrock: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. E. Padron: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Tsao AS, Kantarjian H, Thomas D, Giles F, Cortes J, Garcia-Manero G, et al. C-kit receptor expression in acute leukemias-association with patient and disease characteristics and with outcome. Leuk Res 2004;28:373–8. [DOI] [PubMed] [Google Scholar]
- 2. Wells SJ, Bray RA, Stempora LL, Farhi DC. CD117/CD34 expression in leukemic blasts. Am J Clin Pathol 1996;106:192–5. [DOI] [PubMed] [Google Scholar]
- 3. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016;127:2391–405. [DOI] [PubMed] [Google Scholar]
- 4. Padron E, Garcia-Manero G, Patnaik MM, Itzykson R, Lasho T, Nazha A, et al. An international data set for CMML validates prognostic scoring systems and demonstrates a need for novel prognostication strategies. Blood Cancer J 2015;5:e333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 2000;404:193–7. [DOI] [PubMed] [Google Scholar]
- 6. Setty M, Kiseliovas V, Levine J, Gayoso A, Mazutis L, Pe'er D. Characterization of cell fate probabilities in single-cell data with Palantir. Nat Biotechnol 2019;37:451–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun 2017;8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hua P, Roy N, de la Fuente J, Wang G, Thongjuea S, Clark K, et al. Single-cell analysis of bone marrow-derived CD34+ cells from children with sickle cell disease and thalassemia. Blood 2019;134:2111–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 2019;16:1289–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 2019;37:38–44. [DOI] [PubMed] [Google Scholar]
- 11. Triana S, Vonficht D, Jopp-Saile L, Raffel S, Lutz R, Leonce D, et al. Single-cell proteo-genomic reference maps of the hematopoietic system enable the purification and massive profiling of precisely defined cell states. Nat Immunol 2021;22:1577–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 2018;19:477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu Z, Gao S, Diamond C, Kajigaya S, Chen J, Shi R, et al. Sequencing of RNA in single cells reveals a distinct transcriptome signature of hematopoiesis in GATA2 deficiency. Blood Adv 2020;4:2656–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Galen P, Mbong N, Kreso A, Schoof EM, Wagenblast E, Ng SWK, et al. Integrated stress response activity marks stem cells in normal hematopoiesis and leukemia. Cell Rep 2018;25:1109–17. [DOI] [PubMed] [Google Scholar]
- 15. Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med 2011;17:1086–93. [DOI] [PubMed] [Google Scholar]
- 16. Wiseman DH, Baker SM, Dongre AV, Gurashi K, Storer JA, Somervaille TC, et al. Chronic myelomonocytic leukaemia stem cell transcriptomes anticipate disease morphology and outcome. EBioMedicine 2020;58:102904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Galen P, Hovestadt V, Wadsworth Ii MH, Hughes TK, Griffin GK, Battaglia S, et al. Single-cell RNA-seq reveals AML hierarchies relevant to disease progression and immunity. Cell 2019;176:1265–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alfonso A, Montalban-Bravo G, Takahashi K, Jabbour EJ, Kadia T, Ravandi F, et al. Natural history of chronic myelomonocytic leukemia treated with hypomethylating agents. Am J Hematol 2017;92:599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 2019;20:163–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rapin N, Bagger FO, Jendholm J, Mora-Jensen H, Krogh A, Kohlmann A, et al. Comparing cancer vs normal gene expression profiles identifies new disease entities and common transcriptional programs in AML patients. Blood 2014;123:894–904. [DOI] [PubMed] [Google Scholar]
- 22. Herault A, Binnewies M, Leong S, Calero-Nieto FJ, Zhang SY, Kang YA, et al. Myeloid progenitor cluster formation drives emergency and leukaemic myelopoiesis. Nature 2017;544:53–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kang Y-A, Paik H, Zhang SY, Chen J, Warr MR, Fan R, et al. Secretory MPP3 reinforce myeloid differentiation trajectory and amplify myeloid cell production. bioRxiv 2021: 2021.09.01.458573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Delaney C, Schnell A, Cammarata LV, Yao-Smith A, Regev A, Kuchroo VK, et al. Combinatorial prediction of marker panels from single-cell transcriptomic data. Mol Syst Biol 2019;15:e9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Velten L, Story BA, Hernandez-Malmierca P, Raffel S, Leonce DR, Milbank J, et al. Identification of leukemic and pre-leukemic stem cells by clonal tracking from single-cell transcriptomics. Nat Commun 2021;12:1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Itzykson R, Kosmider O, Renneville A, Morabito M, Preudhomme C, Berthon C, et al. Clonal architecture of chronic myelomonocytic leukemias. Blood 2013;121:2186–98. [DOI] [PubMed] [Google Scholar]
- 27. Duchmann M, Yalniz FF, Sanna A, Sallman D, Coombs CC, Renneville A, et al. Prognostic role of gene mutations in chronic myelomonocytic leukemia patients treated with hypomethylating agents. EBioMedicine 2018;31:174–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, Jenkins SL, et al. Gene set knowledge discovery with enrichr. Curr Protoc 2021;1:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mi H, Ebert D, Muruganujan A, Mills C, Albou LP, Mushayamaha T, et al. PANTHER version 16: a revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res 2021;49:D394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Casero D, Sandoval S, Seet CS, Scholes J, Zhu Y, Ha VL, et al. Long non-coding RNA profiling of human lymphoid progenitor cells reveals transcriptional divergence of B cell and T cell lineages. Nat Immunol 2015;16:1282–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Woll PS, Kjallquist U, Chowdhury O, Doolittle H, Wedge DC, Thongjuea S, et al. Myelodysplastic syndromes are propagated by rare and distinct human cancer stem cells in vivo. Cancer Cell 2014;25:794–808. [DOI] [PubMed] [Google Scholar]
- 32. Dolatshad H, Pellagatti A, Fernandez-Mercado M, Yip BH, Malcovati L, Attwood M, et al. Disruption of SF3B1 results in deregulated expression and splicing of key genes and pathways in myelodysplastic syndrome hematopoietic stem and progenitor cells. Leukemia 2015;29:1092–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quek L, Otto G, Garnett C, Lhermitte L, Lau IJ, Karamitros D, et al. Functional and genetic heterogeneity of distinct leukemic stem cell populations in CD34-human acute myeloid leukemia. Blood 2014;124:15. [Google Scholar]
- 34. Unnikrishnan A, Papaemmanuil E, Beck D, Deshpande NP, Verma A, Kumari A, et al. Integrative genomics identifies the molecular basis of resistance to azacitidine therapy in myelodysplastic syndromes. Cell Rep 2017;20:572–85. [DOI] [PubMed] [Google Scholar]
- 35. Nguyen R, Perfetto S, Mahnke YD, Chattopadhyay P, Roederer M. Quantifying spillover spreading for comparing instrument performance and aiding in multicolor panel design. Cytometry A 2013;83:306–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD, et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 2015;162:184–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fujisaki T, Berger MG, Rose-John S, Eaves CJ. Rapid differentiation of a rare subset of adult human lin(-)CD34(-)CD38(-) cells stimulated by multiple growth factors in vitro. Blood 1999;94:1926–32. [PubMed] [Google Scholar]
- 38. Niyongere S, Lucas N, Zhou JM, Sansil S, Pomicter AD, Balasis ME, et al. Heterogeneous expression of cytokines accounts for clinical diversity and refines prognostication in CMML. Leukemia 2019;33:205–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang Y, He L, Selimoglu-Buet D, Jego C, Morabito M, Willekens C, et al. Engraftment of chronic myelomonocytic leukemia cells in immunocompromised mice supports disease dependency on cytokines. Blood Adv 2017;1:972–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Padron E, Painter JS, Kunigal S, Mailloux AW, McGraw K, McDaniel JM, et al. GM-CSF-dependent pSTAT5 sensitivity is a feature with therapeutic potential in chronic myelomonocytic leukemia. Blood 2013;121:5068–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang J, Liu Y, Li Z, Du J, Ryu MJ, Taylor PR, et al. Endogenous oncogenic Nras mutation promotes aberrant GM-CSF signaling in granulocytic/monocytic precursors in a murine model of chronic myelomonocytic leukemia. Blood 2010;116:5991–6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yoshimi A, Balasis ME, Vedder A, Feldman K, Ma Y, Zhang H, et al. Robust patient-derived xenografts of MDS/MPN overlap syndromes capture the unique characteristics of CMML and JMML. Blood 2017;130:397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yamashita M, Dellorusso PV, Olson OC, Passegue E. Dysregulated haematopoietic stem cell behaviour in myeloid leukaemogenesis. Nat Rev Cancer 2020;20:365–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med 2004;351:657–67. [DOI] [PubMed] [Google Scholar]
- 45. Zhao JL, Baltimore D. Regulation of stress-induced hematopoiesis. Curr Opin Hematol 2015;22:286–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bruserud O, Reikvam H, Brenner AK. Toll-like receptor 4, osteoblasts and leukemogenesis; the lesson from acute myeloid leukemia. Molecules 2022;27:735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sedger LM, McDermott MF. TNF and TNF-receptors: from mediators of cell death and inflammation to therapeutic giants—past, present and future. Cytokine Growth Factor Rev 2014;25:453–72. [DOI] [PubMed] [Google Scholar]
- 48. Heaton WL, Senina AV, Pomicter AD, Salama ME, Clair PM, Yan D, et al. Autocrine Tnf signaling favors malignant cells in myelofibrosis in a Tnfr2-dependent fashion. Leukemia 2018;32:2399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pietras EM, Reynaud D, Kang YA, Carlin D, Calero-Nieto FJ, Leavitt AD, et al. Functionally distinct subsets of lineage-biased multipotent progenitors control blood production in normal and regenerative conditions. Cell Stem Cell 2015;17:35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zeng AGX, Bansal S, Jin L, Mitchell A, Chen WC, Abbas HA, et al. A cellular hierarchy framework for understanding heterogeneity and predicting drug response in acute myeloid leukemia. Nat Med 2022;28:1212–23. [DOI] [PubMed] [Google Scholar]
- 51. Rollison DE, Howlader N, Smith MT, Strom SS, Merritt WD, Ries LA, et al. Epidemiology of myelodysplastic syndromes and chronic myeloproliferative disorders in the United States, 2001–2004, using data from the NAACCR and SEER programs. Blood 2008;112:45–52. [DOI] [PubMed] [Google Scholar]
- 52. Guru Murthy GS, Dhakal I, Mehta P. Incidence and survival outcomes of chronic myelomonocytic leukemia in the United States. Leuk Lymphoma 2017;58:1648–54. [DOI] [PubMed] [Google Scholar]
- 53. Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf 2013;14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu DC, DiBiase A, et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell 2011;147:577–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell 2011;144:296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mabbott NA, Baillie JK, Brown H, Freeman TC, Hume DA. An expression atlas of human primary cells: inference of gene function from coexpression networks. BMC Genomics 2013;14:632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Martens JH, Stunnenberg HG. BLUEPRINT: mapping human blood cell epigenomes. Haematologica 2013;98:1487–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gatenbee CD, Schenck RO, Bravo RR, Anderson ARA. EvoFreq: visualization of the evolutionary frequencies of sequence and model data. BMC Bioinf 2019;20:710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The bulk RNA-seq data used in flow cytometry panel development were from healthy and CMML CD34+ cells extracted from the following publicly available data sets: Healthy CD34+ cells: the Gene-Expression Omnibus (GEO): GSE69239, GSE55689, GSE63569, accession ID: PRJEB6573; CMML CD34+: GEO: GSE76203.
The scRNA-seq data from healthy CD34+ cells were extracted from three publicly available data sets: one patient sample was from 10X Genomics at https://www.10xgenomics.com/resources/datasets/cd-34-plus-cells-1-standard-1-1-0, three patient samples available from the Human Cell Atlas data portal at https://data.humancellatlas.org/explore/projects/091cf39b-01bc-42e5-9437-f419a66c8a45, and four patient samples available from GEO under GSE133181. Normal sample characteristics are shown in Supplementary Table S14.
The scRNA-seq data for the CMML CD34+ cells analyzed here have been made publicly available in the GEO: accession number GSE211033.



![Figure 1. CMML HSPC are characterized by three hematopoietic trajectories. A, Excess bone marrow and aspirate specimens not utilized for pathologic evaluation were collected from CMML patients at clinically scheduled time points and enriched for CD34+ cells. Single cells [CD34 cells enriched from patient bone marrrow mononuclear cells (BMMNC)] were encapsulated for scRNA-seq via DropSeq with the 10X Chromium controller and sequenced on an Illumina NovaSeq. Data from CD34+ cells enriched from BMNNCs of 8 healthy individuals available from publicly available data sets were integrated with the 39 CMML patient samples for downstream analysis. B, UMAP visualization of the 201,250 cells with normal CD34+ cells represented in red and CMML CD34+ cells represented in gray. C, CMML single cells were projected onto a reference Normal CD34+ (6), and branch probabilities and differentiation potentials were inferred using a nearest-neighbor approach to reveal (D) three distinct hematopoietic trajectories in CMML: monocytic-biased, MEP-biased, and normal-like. Monocytic-biased samples had overdensities (as indicated in red) in the monocytic branch, MEP-biased samples had overdensities in the erythroid/megakaryocytic (Ery/Mega) branch, and normal-like samples showed neither overdensity in monocytic and erythroid branches and cell densities of similar proportion to that of the normal samples. These trajectories were classified based on the overdensities compared with the reference hematopoietic, gene signature analysis, and hierarchical clustering (in Supplementary Fig. S2). E, These trajectory biases were confirmed using previously published gene-expression profiles of HSCs, MEPs, and GMPs. Monocytic-biased patients were highly enriched for genes upregulated in GMPs, and MEP-biased patients were highly enriched for genes upregulated in MEPs. F, Hemoglobin was significantly elevated in the MEP-biased group (P = 0.037; Mann–Whitney test). G, Kaplan–Meier (KM) survival analysis of CMML patients stratified on progenitor trajectory in treatment-naïve samples treatment-naïve (n = 25; log-rank P = 0.0349). H, LDH levels obtained from clinical evaluation at the time of tissue sampling (n = 38; Fisher exact test P: 0.0378). I and J, There was a significant depletion in the fraction of cells identified as CLP across treatment-naïve samples (I; P = 0.0004; Mann–Whitney test) and in all three differentiation trajectories identified in CMML (J; Mann–Whitney comparisons; between normal and monocytic-biased, P = 0.0207; normal and MEP-bias, P = 0.0030; normal and normal-like, P = 0.0031). CLP cell-type assignment was calculated from the SingleR consensus (see Supplementary Methods).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/e952/9627238/3d7b12ac1de3/536fig1.jpg)