Abstract
Abstract
Microbe (including bacteria, fungi, and virus) infection in brains is associated with amyloid fibril deposit and neurodegeneration. Increasing findings suggest that amyloid proteins, like Abeta (Aβ), are important innate immune effectors in preventing infections. In some previous studies, amyloid peptides have been linked to antimicrobial peptides due to their common mechanisms in membrane-disruption ability, while the other mechanisms of bactericidal protein aggregation and protein function knockdown are less discussed. Besides, another important function of amyloid peptides in pathogen agglutination is rarely illustrated. In this review, we summarized and divided the different roles and mechanisms of amyloid peptides against microbes in antimicrobial activity and microbe agglutination activity. Besides, the range of amyloids’ antimicrobial spectrum, the effectiveness of amyloid peptide states (monomers, oligomers, and fibrils), and cytotoxicity are discussed. The good properties of amyloid peptides against microbes might provide implications for the development of novel antimicrobial drug.
Key points
• Antimicrobial and/or microbial agglutination is a characteristic of amyloid peptides.
• Various mechanisms of amyloid peptides against microbes are discovered recently.
• Amyloid peptides might be developed into novel antimicrobial drugs.
Supplementary information
The online version contains supplementary material available at 10.1007/s00253-022-12246-w.
Keywords: Amyloid peptides, Antimicrobial activity, Microbe agglutination
Introduction
Amyloid proteins were originally thought to be misfolded proteins which accumulate and deposit, and thus cause neurodegeneration and disorders. Over 35 amyloid proteins or peptide deposition are reported to be strongly linked to human diseases, including Aβ, α-synuclein, islet amyloid polypeptide (IAPP), prion protein (PrP), and so on (Ross & Poirier 2004; Vaquer-Alicea & Diamond 2019). Amyloid proteins/peptides have the propensity to aggregate and self-assembly into amyloids, characterized by highly ordered, cross-β-sheet rich structure, where the β-strands align perpendicularly to the fibril axis. The amyloid fibrils exhibit characteristic x-ray crystallographic diffraction patterns, ultra-structural, the dye binding of Thioflavin T and Congo red, and so on (Benson et al. 2020). Except for the traditionally known pathogenic amyloid fibrils, the concept of “functional amyloid fibrils” is gradually induced these years. Functional amyloid fibrils are reported to be present throughout all kingdoms of life and exert important biological roles, as reservoirs for certain polypeptide hormones storage, structures for biofilm formation, information carriers for long-term memory, and so on (Chen et al. 2019; Otzen & Riek 2019). Besides, large numbers of non-pathogenic nature proteins or synthetic peptides have been proved to form amyloid fibrils. The effective aggregation tendency of proteins/peptides depends on many factors, such as folding efficiency, thermodynamic stability, and most important the intrinsic aggregation propensity. The intrinsic aggregation propensity is driven by the β-sheet aggregation of short specific segments composed of 5–15 residues, termed aggregation prone regions (ARPs) (Khodaparast et al. 2018). APRs are mainly hydrophobic and buried inside the hydrophobic core of the folded protein to prevent aggregation. However, when the protein is partially or completely unfolded, the APRs are exposed and triggered the aggregation (De Baets et al. 2014). APRs is a crucial concept in amyloid field, and many computational methods have been explored to identify the APRs in proteins or peptides sequences and to predict segments aggregation propensity, e.g., TANGO, ZipperDB, Waltz, Zyggregator, and PASTA2. The majority of synthetic amyloid peptides are designed based on APRs, which have been widely exploited for therapeutic medical applications, such as antitumor agent, antiviral agent, and antibacterial agent investigation (Bourgade et al. 2016; Gallardo et al. 2016; Schnaider et al. 2017; Abdelrahman et al. 2020). Moreover, the concept of APRs has also been developed in the use of biomaterials or food characteristics improvement (Housmans et al. 2021).
Recently, increasing evidences show that native amyloid peptides/proteins like Aβ possess antimicrobial (including antibacterial, antifungal, and antiviral) and pathogen agglutination properties (including bacteria, fungus, and virus agglutination), and they might be an ancient, highly conserved innate immune effectors that play important roles in anti-infections (Kagan et al. 2012; Gosztyla et al. 2018; Makin 2018). Besides, more engineered amyloid peptides have been found with antimicrobial or microbial agglutination properties (Salinas et al. 2018; Chen et al. 2021). Amyloid peptides are no longer the previously regarded useless and harmful substances, which might be developed into novel antimicrobial drugs. Though the antimicrobial effect of amyloid peptides has been reviewed by some previous studies, the antimicrobial mechanisms are not fully summarized; besides, the microbial agglutination effect is overlooked and rarely discussed (Kagan et al. 2012; Last & Miranker 2013; Zhang et al. 2014; Moir et al. 2018). This review will focus on the effects of amyloid peptides against microbes in antimicrobial activity and microbe agglutination activity, and elaborate their mechanisms as comprehensive as possible. Moreover, the range of antimicrobial spectrum, cytotoxicity, and effectiveness of amyloid peptide states (fibrils, monomers, and oligomers) will also be discussed. The good properties of amyloid peptides against microbes might provide implications for the development of novel antimicrobial drugs and shed light on new ideas against the increasingly serious microbial infections. In the end, we will take severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as an example to discuss the possible treatment implications of amyloid peptides toward SARS-CoV-2.
Effects of amyloid peptides against microbes and the possible mechanisms
Both native and engineered amyloid proteins/peptides have been found to display functional roles against microbes, including antimicrobial function and microbe agglutination function. The summarization of current reported amyloid peptides against bacteria and fungi, and against virus are listed in Table S1 and Table S2, respectively.
Antimicrobial activity and its possible mechanism
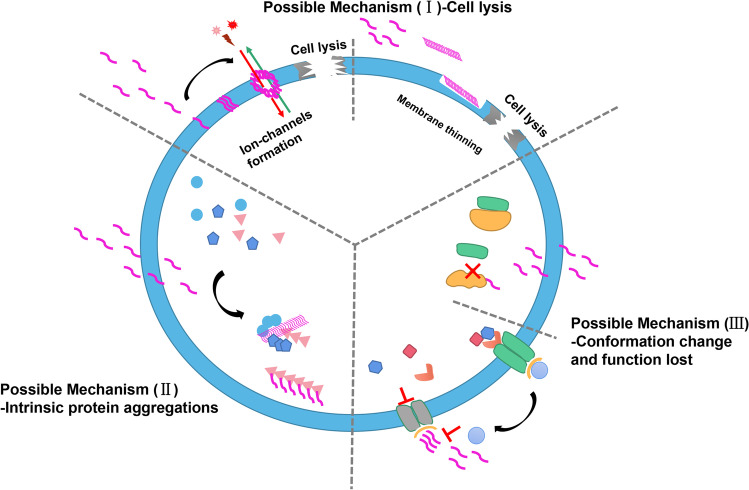
According to the different mechanisms of amyloid peptides exerting antimicrobial activities, these proteins/peptides can be further divided into three: (1) amyloid proteins/peptides interacting with membranes and inducing membrane disruption; (2) amyloid proteins/peptides internalized by microbes and causing protein aggregation; (3) amyloid proteins/peptides altering protein conformation and thus disrupting protein’s function. The classical antimicrobial effects and their possible mechanisms are illustrated in Fig. 1.
Fig. 1.
The illustrated three mechanisms of amyloid peptides in antimicrobes activities, which are membrane disruption and cell lysis, protein aggregations, protein conformation change and function lost
First antimicrobial mechanism: membrane disruption
In 2002, Hirakura et al. (2002) initially reported that serum amyloid A (SAA) exerted antimicrobial activities through forming ion channels and thus inducing lysis of bacterial cells. They found the ion channels formed by SAA in planar lipid bilayer membranes are (1) relatively non-selective, allowing the influx of toxic ions and efflux of vital cellular constituents; (2) irreversibly associated with membrane, remaining open for long periods of time, and voltage-independent, thus insuring a leak in the membrane; and (3) relatively large. In 2009, the well-known prion protein was also reported to show broad antimicrobial function (Pasupuleti et al. 2009). Again, in 2010, Soscia et al. (2010) found Aβ in vitro exerted antimicrobial activity against eight microbes, with a potency equivalent to, and in some strains even greater than the 37-residue peptide whose N-terminal sequence is LL (LL-37). The above findings gradually uncover the function of amyloid peptides in antimicrobial activities in the following years. Various researches have been done to investigate the interaction between amyloids and membrane of microbes. Using 3:1 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine:1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylglycerol to simulate the bacterial membrane, Walsh et al. (2014) found that oligomers of PrP(106–126) cause disruption of anionic membranes which proceeds through a carpet or detergent model. Caillon et al. (2013) found that it is possible that the dermaseptin S9 (Drs S9) binds to the membrane with a transmembrane insertion and induces a transient pore causing leakage. Auvynet et al. (2008) found that anionic phospholipid vesicles can promote the structural interconversion of Drs S9 from β-hairpin into β-sheet. Interaction of the peptide with alkyl chains of dimyristoylphosphatidylglycerol phospholipids consequently resulted in disturbance of the alkyl chain order of the fluid bilayer. In this way, it is likely that oligomers in the β-sheet formation exerted microbicidal activity by disturbing both the membrane interface and the hydrophobic core of the bacterial membrane. Accumulating evidences suggest that a number of antimicrobial peptides (AMPs) or host defense peptides exert antimicrobial activities also through formation of amyloid associated ion channels/pore structures, and some in vitro studies have proved that they are capable of forming amyloid-like fibrils with similar morphologies to Aβ1-42 fibrils in solution (Harris et al. 2012). These AMPs include LL-37 (Sood et al. 2008), lactoferrin (Nilsson & Dobson 2003), temporin B and temporin L (Mahalka & Kinnunen 2009), protegrin-1 (Jang et al. 2011), and so on. The shortest amyloid peptide with antimicrobial activity through membrane disruption mechanism reported thus far is diphenylalanine (Harris et al. 2012). Aside from membrane disruption by ion channels/pore structure formation, amyloid aggregates or fibrils can exert stress on membrane, leading to membrane thinning and thus ion leakage (Valincius et al. 2008; Pasupuleti et al. 2009; Zhang et al. 2014).
Second antimicrobial mechanism: protein aggregations
The second mechanism also has many supporting evidences. Bednarska et al. (2016) speculated that the amyloid peptides derived from APR of bacterial proteins might induce toxic protein aggregation more efficiently in their originated bacteria than in other organisms. By using TANGO, they screened several amyloid peptides from Staphylococcus aureus (S. aureus), among which C30 indeed formed amyloid fibrils, and C30 was internalized by S. aureus, causing many bacterial proteins aggregation, which was positively associated with bacterial death. Khodaparast et al. (2018) also identified some redundant APRs (that were not sequence unique in the genome) from Escherichia coli (E. coli) proteome, and they found these peptides induced widespread protein aggregation involving hundreds of proteins, resulting in bactericidal aggregation cascades and inclusion body formation. This reaction is bactericidal not only to E. coli, but also to Acinetobacter baumannii, including antibiotic-resistant clinical strains. Kurpe et al. (2020a, b) screened APR peptides from ribosomal S1 protein of Thermus thermophilus, which was vital and specific only for bacteria. They found R23I (modified with bacterial cell penetration segment) could coaggregate with S1 protein in vitro. Under the effective antimicrobial concentration of R23I, the S1 ribosomal protein was absent and the intensity of protein biosynthesis was suppressed, leading to the decrease of proteins number. The same strategy was also performed by Kravchenko et al. (2022) targeting S1 protein of S. aureus, and hybrid peptides R23F, R23DI, and R23EI also exhibit a broad antimicrobial activity against both Gram-positive and Gram-negative bacteria. This strategy could also be traced in the mammalian cell researches. In the study of antitumor work, Gallardo et al. (2016) predicted and designed an APR peptide sequence derived from the translocation signaling sequence of vascular endothelial growth factor receptor 2 (VEGFR2), termed vascin. They found prefibrillar-forming vascin oligomers could penetrate mammalian cells and interacted with the nascent VEGFR2 protein, resulting in its aggregation and function knockdown, and thus vascin exert antitumor functions.
By summarizing the second mechanism, a theory is apparent that the amyloidogenic peptides derived from APRs can coaggregate and interfere the functions of their “parent” or relatival proteins sharing APR sequence, like a protein knockdown mechanism.
Third antimicrobial mechanism: conformation change and function lost
The third mechanism is rarely reported. Zabrodskaya et al. (2018) reported that the amyloid peptide PB1(6–13) (6–13 residues of basic polymerase 1) monomer, derived from the influenza A virus polymerase complex PB1, was found to interact with its matrix protein PB1 subunit N-terminal region, causing a conformation of PB1 shift toward beta structures, and making PB1 lost its ability to interact with the acidic polymerase subunit; as a result, the influenza virus’ polymerase complex functionality was broken (Egorov et al. 2013; Zabrodskaya et al. 2018).
Microbe agglutination activity and its possible mechanism
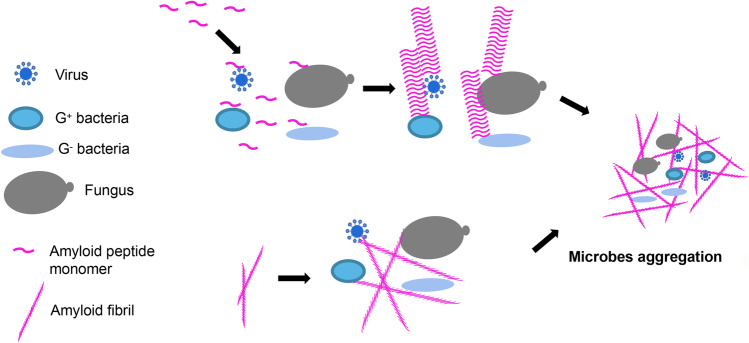
Microbe agglutination is another reaction of amyloid peptides when encountering microbes. The previous evidences of brain plaques containing microbial DNA in Alzheimer’s disease (AD) definitely suggest that Aβ formed amyloid fibrils agglutinating microbes, and it is further proved by in vivo studies (Wozniak et al. 2009; Eimer et al. 2018). Voth et al. (2020) found the amyloids, induced by pulmonary endothelial when infected by type III secretion system (T3SS) incompetent Pseudomonas aeruginosa (P. aeruginosa), can aggregate bacteria both in suspension and on solid substrate and corroborate their proclivity to form annular inclusions. Chu et al. (2012) demonstrated that human α-defensin 6 (HD6), whose effects are quite different from other host defensins, undergoes ordered self-assembly to form amyloid fibrils and nanonets that surround and entangle bacteria. Notably, HD6 only entangles bacteria and exerts adhesion inhibition effect, without bactericidal effect. Chu et al. (2012) exclude the possibility of HD6 targeting the bacterial surface carbohydrates because it lacks lectin-like activity. Since Salmonella mutant lacking both fimbriae and flagella causes neither nanofibrils nor aggregation, thus they regard that the appendages are HD6 anchoring targets that trigger HD6 nanofibril formation. Aside from the appendages binding mechanism, Kumar et al. (2016) found Aβ protected against microbial (e.g., Candida albicans and Salmonella typhimurium) infection in mouse and worm models of AD and cell lines through aggregating into amyloid fibrils agglutinating microbes and thus inhibiting microbe adhesion to cells. Heparin-binding AMPs show high affinities for some unique microbial carbohydrates, which play a key role in promoting peptides, and recognize and bind to microbial surface (Tsai et al. 2011; Kumar et al. 2016). Heparin-binding AMPs have a heparin-binding motif (XBBXBX, X represents a hydrophobic or uncharged residue, and B represents a basic residue) (Ridge et al. 2000). Kumar et al. (2016) found Aβ also contained a XBBXBX sequence between residues 12–17 (VHHQKL), and they proposed the VHHQKL mediated Aβ soluble oligomeric species targeting and binding to the bacterial wall carbohydrates and provide a nidus and anchor for Aβ fibril propagation. And then the protofibrils disturb the microbial adhesion to host cells; in the meantime, they also link and agglutinate and consequently entrap the unattached microbial cells in a protease-resistant network of β-amyloid. Previously, our group showed that some APR hexapeptides, regardless of its derived resource, possessed microbial agglutination and inhibited microbial adhesion to form biofilm, but without interfering microbial proliferation (Chen et al. 2021). We proved that the amyloid-forming hexapeptides might bind to the microbial wall carbohydrates as stated by Kumar et al., but these amyloid-forming hexapeptides did not satisfy the XBBXBX regular, indicating that heparin-binding motif was not necessarily for amyloid peptides. In the studies of virus agglutination by amyloid peptides, Eimer et al. (2018) also prove that Aβ oligomers bind herpesvirus surface glycoproteins, while Ezzat et al. (2019) demonstrate that viral protein corona directs amyloid peptides binding. Though the binding mechanisms are inconsistent among different peptides in different studies, a consistent result is that microbes can facilitate amyloid peptide aggregation in a very short time.
The outcome of microbial agglutination by amyloid fibrils should be concerned. Most evidences support the view that microbial agglutination inhibits microbial entry cells and facilitates microbial clearance. For bacteria and fungus, HD6 (Chu et al. 2012) and Aβ (Kumar et al. 2016) block bacterial and fungal biofilm formation and adhesion to host cells. For virus, Aβ is reported to inhibit herpes simplex virus type 1 (HSV1) infection of host cells in vitro and protects against HSV1 encephalitis through viral agglutination in vivo study (Eimer et al. 2018). In addition to reducing viral uptake by epithelial cells, viral agglutination by amyloid fibrils also has immune modulating effects, which increases uptake of virus by neutrophils, reducing viral protein synthesis in monocytes, and reducing interleukin-6 production (White et al. 2014). However, the fragments of the semen marker prostatic acidic phosphatase fibrils were reported to drastically enhance viral attachment and fusion to target cells, including T cells, macrophages, and epithelial cells, promoting the transmission of human immunodeficiency virus (Munch et al. 2007).
The classical agglutination activities without antibacterial effects and their possible mechanisms are illustrated in Fig. 2.
Fig. 2.
The illustrated mechanisms of amyloid peptides in microbes agglutination activities. Two microbes agglutination approaches, amyloid monomers and amyloid fibrils, are shown
As discussed above, the microbe agglutination activities are independently divided from antimicrobial activities, while some amyloid proteins/peptides exert both activities. Eosinophil cationic protein (ECP), a small cationic protein secreted during inflammation processes, is found to exert both pathogens agglutination and antimicrobial activities (Pulido et al. 2012). Torrent et al. (2012) observed ECP firstly formed amyloid-like aggregates at the bacteria surface agglutinating G− bacteria, and then induced bacterial death. They demonstrated that the formation of ECP aggregates on bacterial surface disrupted the lipopolysaccharide bilayer of G− bacteria, leading to the exposure of the internal cytoplasmatic membrane to the protein action, therefore promoting the membrane disruption and bacteria death. Spitzer et al. (2016) also reported Aβ42 and its variants Aβx-42 both exerted antimicrobial activity and microbe agglutination activities against G+ bacteria, G− bacteria, and fungi.
Range of antimicrobial spectrum
For the microbial agglutination effect, all the reported amyloid proteins/peptides show broad agglutination effect. For antimicrobial effect, most of the reported amyloid peptides/proteins show broad or narrow broad effects, e.g., Aβ and ECP. ECP displays specific activities toward G− bacteria, which is a classical narrow broad example (Tu et al. 2021). Since predicted APR can nucleate lots of protein aggregation containing the same APR, redundant APRs (that are not sequence unique in one unique genome) will also exhibit broad antimicrobial effect, like the redundant APRs derived from E. coli proteome, which are bactericidal not only to E. coli, but also to Acinetobacter baumannii. However, amyloid peptides that target specific specie are also reported. The amyloid peptide PB1(6–13), derived from the influenza A virus polymerase complex PB1, shows a specific antivirus effect toward its derived influenza virus. Besides, Michiels et al. (2020) synthesized two APR peptides derived from influenza A and Zika virus, respectively. The two peptides could inhibit virus replication with high specificity and without cross reactivity. However, the amyloid segments screened from the abovementioned phenol-soluble modulins α3-LFKFFK (PSMα3-LFKFFK) showed dose-dependent antibacterial activity against the G+ bacteria Micrococcus luteus and Staphylococcus hominis, but without toxic to its derived bacteria S. aureus (Salinas et al. 2018).
Cytotoxicity
Since amyloid fibrils are historically associated with some pathological diseases, the cytotoxicity of synthetic amyloid peptides/proteins should be clearly declared before use. According to the update and recommendations by the International Society of Amyloidosis nomenclature committee, amyloid fibrils can be categorized into 5 classes: (1) in vivo and ex vivo disease-related fibrils; (2) in vivo and ex vivo functional fibrils; (3) recombinant fibrils of disease-related proteins and of functional amyloid proteins; (4) fibrils of synthetic or non-disease related peptides; and (5) fibrils from condensates and hydrogels that give the cross-β diffraction pattern (Benson et al. 2020). Besides, it is shown that amyloid fibrils formed in vitro from recombinant protein (usually in short period) differ profoundly from those formed in vivo by the same precursor (over a long period) (Benson et al. 2020). All the synthetic peptides used in monomers or fibrils, reviewed in this study, showed no obvious or very little cell cytotoxicity. Besides, engineered amyloid fibrils are widely studied as drug carrier, gels, and biomaterials for cell adhesion and tissue engineering applications, indicating a safe use (Babych et al. 2018; Das et al. 2018; Wang et al. 2019). It seems a safe use of synthetic engineered amyloid peptides as therapeutic agents. Here, we still emphasize that the cytotoxicity of amyloid peptide monomers, fibrils, and especially their intermediate oligomers, should be well examined before in vivo use, and we recommend to design engineered amyloid peptides derived from the non-cytotoxic proteins’ APR sequences.
Which state is effective: monomers, oligomers, or fibrils?
Most of the studies focus on the effect of amyloid monomers, and prove the effectiveness of monomers in antimicrobial activities or microbe agglutination. There are also a substantial of evidence that suggest that the antimicrobial activity mainly depends on the formation of unstable oligomers which acts as an intermediate state of amyloid fibrils, while the protofibrils and fibrils of amyloid are per se noneffective (Bucciantini et al. 2002; Butterfield & Lashuel 2010; Walsh et al. 2014). Therefore, the main dispute locates on whether amyloid fibrils are effective. As for the peptides with antimicrobial effects, Wang et al. (2012) found both the two states of hIAPP monomer and protofibrils/fibrils are effective in antimicrobial, but the monomer’s antimicrobial effect was greater than those of annular protofibrils and fibrils. Auvynet et al. (2008) found that the bactericidal activity of Drs S9 is most effective in the form of oligomers, peptides (dipolymer and tetramer) attenuated, while protofibrils have no antibacterial activity. Holch et al. (2020) also found that the antimicrobial activity of β-2-microglobulin (B2M) was present in the supernatant but not in the insoluble fibrils, suggesting that the soluble B2M containing monomers and oligomers, and perhaps also the fragments, may contribute to bacterial inhibition. In addition, Juhl et al. (2020) found the antimicrobial activities of peptidyl-glycine-leucine-carboxyamide (PGLa) decreased with fibril formation. Besides, Zabrodskaya et al. (2018) found that after the amyloid peptide PB1(6–13) monomer formation into fibrils, it was no longer able to interact with PB1 N-terminal region, nor was it able to exert antiviral activity. Different antimicrobial effectiveness are reported even for the same amyloid proteins. Lysozyme is an antibacterial enzyme, known as muramidase, and it can form amyloid fibrils. Bouaziz et al. (2017) demonstrated that both lysozyme monomers and fibrils had antimicrobial activities, but the effect of fibrils was weaker than that of monomers, while Wei et al. (2021) reported that lysozyme fibrils showed significantly enhanced antibacterial activity compared to their monomers and oligomers. Therefore, it can be concluded that the monomers and oligomers have definitely antimicrobial effect, while the effect of amyloid fibrils depends.
As for the peptides with microbial agglutination effect, Kumar et al. (2016) compared the antimicrobial activities of Aβ42 monomer, soluble oligomeric ADDLs (amyloid-β-derived diffusible oligomeric ligands), and high ordered protofibril, and they found that compared to monomer, ADDLs exhibited potentiated, and protofibrils attenuated, adhesion inhibition, and agglutination activities. We previously found not only the monomers of amyloid hexapeptides derived from C123 but also their amyloid fibrils possessed pathogen agglutination abilities (Chen et al. 2021). However, the bovine serum albumin amyloid fibrils coating substrate was reported to be resistant to the nonspecific adsorption of microbes (including bacteria and fungus) onto the substrate, indicating that amyloid fibrils coating substrate show a repulsive force against microbes, not a microbe adsorption.
Implications for treating SARS-CoV-2
A link between SARS-CoV-2 infection and amyloidosis has been raised recently (Tavassoly et al. 2020a; Jana et al. 2021). Tavassoly et al. (2020a) found that a cleavage from S protein (~ 150 aa) possessed higher aggregation propensity than others (Aβ40, M45, and α-synuclein). This cleavage peptide can form toxic fibrils in the brain, or exert as a seed to facilitate the heparin and heparin binding proteins, like Aβ, aggregation into amyloid fibrils. In the hypothesis of SARS-CoV-2 invasion brain leading to neurodegeneration, Idrees et al. (Idrees & Kumar 2021) use the protein–protein docking server which proved that SARS-CoV-2 spike S1 protein receptor binding domain interacts with different amyloid proteins, and they think the interaction assists amyloid proteins binding to the viral surface and initiate amyloid proteins aggregation, which finally leads to neurodegeneration. Semerdzhiev et al. (2022) found the nucleocapsid protein (N-protein) of SARS-CoV-2, not the spike protein, considerably speeded up α-synuclein aggregation process, which might be the molecular basis for correlation between SARS-CoV-2 infection and Parkinsonism. It is likely that the interaction between N-protein and α-synuclein is mediated via the C-terminal region or the aggregation-prone central non-amyloidal component (NAC) region of α-synuclein: negatively charged α-synuclein binding to positively charged N-protein and thus exposes its aggregation-prone central NAC region and eliminates electrostatic repulsion, consequently causing formation of amyloids. According to the above findings, SARS-CoV-2 or its proteins indeed bind and facilitate amyloid protein aggregation, but whether the fibrils aggregating exert a protection against viral entry cells and facilitate viral clearance by immune cells or a harmful effect leading to amyloidosis should be investigated further.
Inhibitory virus entry into cells and stopping the transmission pathways are crucial therapeutic methods, and many researches also focus on the study of preventing virus entry into cells (Jackson et al. 2019; Tavassoly et al. 2020b). The spike S protein is the principal protein mediates SARS-CoV-2 attachment and entry into human target cells, which is an important and promising target, and most of the vaccine candidates against SARS-CoV-2 are based on spike S protein (Xia 2021). Previously, peptides derived from the membrane-proximal and membrane-distal heptad repeat region (HR) of the SARS-CoV spike protein assembled into a rod-like complex, and they markedly inhibited SARS-CoV infection cells (Bosch et al. 2004). HRs are more conserved region in SARS-CoV2 spike protein, which are promising target regions to design antiviral inhibitors dealing with virus mutation (Kurpe et al. 2020a, b). Kurpe et al. (2020a, b) use amyloid prediction software to predict the amyloidogenic region in SARS-CoV-2 and reported a noteworthy region of about 30 residues at the C-terminal of S-protein. It is hypothesized that anti-Cov peptides can bind to receptor-binding domain of S1 protein and reduce the affinity of the S1 and human angiotensin-converting enzyme 2 (hACE2) receptor of the target cell. In this way, the viral particles stick together and thus prevent the spread of virus and reduce the viral load. N-protein of SARS-CoV-2 plays an important role in controlling the replication and assembling of viruses, which is also an important target to fight against the virus. Mukherjee et al. (2022) found the amyloid proteins hIAPP and α-Synuclein can stabilize the partial folded conformation and completely folded conformation of RG-1 G4Q (RNA-G-quadruplex formed by putative G-quadruplex forming sequence RG-1 which locates in the coding sequence region of SARS-CoV-2 nucleocapsid phosphoprotein), respectively. RG1 G4Q can inhibit the translation of mRNA of the SARS-CoV-2 N-protein (Zhao et al. 2021). Therefore, hIAPP and α-Syn might influence the replication of SARS-CoV-2 by stabilizing the folded conformation of RG1 G4Q and consequently influence the expression level of N-protein. Of course, the other proteins, including membrane protein, envelope protein, or even the S1/S2 cleavage proteases, can be used as the target to screen APRs.
Aside from designing unique amyloid peptides, amyloid fibrils coated with iron oxyhydroxide nanoparticles (Fe NPs) are found to have outstanding efficacy against a broad range of viruses, including SARS-CoV-2. The β-lactoglobulin (BLG) amyloid fibrils was obtained by culturing BLG monomers under the condition of pH = 2.0 at 90 °C. Afterwards, the Fe NPs are precipitated directly on the BLG amyloid fibrils (BLG AF) by raising the pH in presence of FeCl3·6H2O, which formed the BLG AF-Fe membranes. The BLG AF-Fe membranes could eliminate viruses effectively from water by retaining and inactivating the enveloped, non-enveloped, airborne, and waterborne viruses, including SARS-CoV-2. The filtration efficiency was from ~ 106PFUml−1 of SARS-CoV-2 to below the detection limit after filtration. However, no remarkable effects were found for BLG AF or Fe alone (Palika et al. 2021). Since amyloid fibrils or modified fibrils are effective in filtrating SARS-CoV-2, the other previously found engineered amyloid peptides with nonselective and broad microbe agglutination activities deserve to be tested.
Once upon the infection occurrence, the viruses enter cell cytoplasm where the common amyloid peptides cannot reach to agglutinate them, then the amyloid peptides with antivirus properties can be considered. Modified amyloid peptides with cell-penetrating abilities have also been reported. Kokotidou et al. (2019) succeeded in developing the cell internalization amyloid peptides carrying DNA through incorporation of positively charged residues and aromatic residue at carefully selected positions. Importantly, the cell-penetrating amyloid peptides/DNA complex exhibited strong antimicrobial activity against E. coli, without significant cytotoxicity. In this scenario, engineered amyloid peptides with antiviral activities might be modified to possess the cell-penetrating abilities to treat the SARS-CoV-2-infected cells. Aside from the direct antivirus effect, amyloid peptides can also be used as carries for other antivirus agents; in this case, amyloid peptide aggregation into fibrils intend to absorb more virus and increase the local antivirus agent’s concentration, which might exert stronger antivirus effects. Amyloid peptides have recently been the hot studied drug carries candidates because of their good physical and chemical properties. Fibrils formed by amyloid peptides are stable, and can be decorated with organic or inorganic elements; besides, the self-assembly process can be controlled by modulating assembly conditions (Adler-Abramovich & Gazit 2014; Wei et al. 2017).
Conclusion and outlook
Amyloid fibrils are no longer the previously known toxic and useless substances, and more functional roles have been discovered in recent years. Amyloid peptides have been widely applied in multiple biomaterial and biomedical fields, including as therapeutic agents, carries of drugs, and tissue engineering scaffolds. The recent findings reveal that amyloid fibrils also exert antimicrobial activity, microbial agglutination activities, or with both activities. In the shortage of antibiotics today, the discovery of amyloid peptides against microbes is conducive to develop novel antimicrobial agents. Herein, we highlight the development of amyloid peptides as novel antimicrobial drugs, including anti-SARS-CoV-2.
Supplementary information
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Data availability
All data generated or analyzed during this study are included in this published article.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Dongru Chen and Xiangqi Liu contributed equally to this work
References
- Abdelrahman S, Alghrably M, Lachowicz JI, Emwas A, Hauser CAE, Jaremko M. “What doesn’t kill you makes you stronger”: future applications of amyloid aggregates in biomedicine. Molecules. 2020;25:5245. doi: 10.3390/molecules25225245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler-Abramovich L, Gazit E. The physical properties of supramolecular peptide assemblies: from building block association to technological applications. CHEM SOC REV. 2014;43:6881–6893. doi: 10.1039/C4CS00164H. [DOI] [PubMed] [Google Scholar]
- Auvynet C, El AC, Lacombe C, Bruston F, Bourdais J, Nicolas P, Rosenstein Y. Structural requirements for antimicrobial versus chemoattractant activities for dermaseptin S9. FEBS J. 2008;275:4134–4151. doi: 10.1111/j.1742-4658.2008.06554.x. [DOI] [PubMed] [Google Scholar]
- Babych M, Bertheau-Mailhot G, Zottig X, Dion J, Gauthier L, Archambault D, Bourgault S. Engineering and evaluation of amyloid assemblies as a nanovaccine against the Chikungunya virus. Nanoscale. 2018;10:19547–19556. doi: 10.1039/C8NR05948A. [DOI] [PubMed] [Google Scholar]
- Bednarska NG, van Eldere J, Gallardo R, Ganesan A, Ramakers M, Vogel I, Baatsen P, Staes A, Goethals M, Hammarstrom P, Nilsson KP, Gevaert K, Schymkowitz J, Rousseau F. Protein aggregation as an antibiotic design strategy. MOL MICROBIOL. 2016;99:849–865. doi: 10.1111/mmi.13269. [DOI] [PubMed] [Google Scholar]
- Benson MD, Buxbaum JN, Eisenberg DS, Merlini G, Saraiva M, Sekijima Y, Sipe JD, Westermark P. Amyloid nomenclature 2020: update and recommendations by the International Society of Amyloidosis (ISA) nomenclature committee. Amyloid. 2020;27:217–222. doi: 10.1080/13506129.2020.1835263. [DOI] [PubMed] [Google Scholar]
- Bosch BJ, Martina BE, Van Der Zee R, Lepault J, Haijema BJ, Versluis C, Heck AJ, De Groot R, Osterhaus AD, Rottier PJ. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc Natl Acad Sci U S A. 2004;101:8455–8460. doi: 10.1073/pnas.0400576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouaziz Z, Soussan L, Janot JM, Lepoitevin M, Bechelany M, Djebbi MA, Amara A, Balme S. Structure and antibacterial activity relationships of native and amyloid fibril lysozyme loaded on layered double hydroxide. Colloids Surf B Biointerfaces. 2017;157:10–17. doi: 10.1016/j.colsurfb.2017.05.050. [DOI] [PubMed] [Google Scholar]
- Bourgade K, Dupuis G, Frost EH, Fulop T. Anti-viral properties of amyloid-beta peptides. J ALZHEIMERS DIS. 2016;54:859–878. doi: 10.3233/JAD-160517. [DOI] [PubMed] [Google Scholar]
- Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- Butterfield SM, Lashuel HA. Amyloidogenic protein-membrane interactions: mechanistic insight from model systems. Angew Chem Int Ed Engl. 2010;49:5628–5654. doi: 10.1002/anie.200906670. [DOI] [PubMed] [Google Scholar]
- Caillon L, Killian JA, Lequin O, Khemtemourian L. Biophysical investigation of the membrane-disrupting mechanism of the antimicrobial and amyloid-like peptide dermaseptin S9. PLoS ONE. 2013;8:e75528. doi: 10.1371/journal.pone.0075528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Cao Y, Yu L, Tao Y, Zhou Y, Zhi Q, Lin H. Characteristics and influencing factors of amyloid fibers in S. mutans biofilm. AMB EXPRESS. 2019;9:31. doi: 10.1186/s13568-019-0753-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Li J, Pan T, Wu R, Tao Y, Lin H. The broad-spectrum antibiofilm activity of amyloid-forming hexapeptides. MICROB BIOTECHNOL. 2021;14:656–667. doi: 10.1111/1751-7915.13721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H, Pazgier M, Jung G, Nuccio SP, Castillo PA, de Jong MF, Winter MG, Winter SE, Wehkamp J, Shen B, Salzman NH, Underwood MA, Tsolis RM, Young GM, Lu W, Lehrer RI, Baumler AJ, Bevins CL. Human alpha-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science. 2012;337:477–481. doi: 10.1126/science.1218831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Jacob RS, Patel K, Singh N, Maji SK. Amyloid fibrils: versatile biomaterials for cell adhesion and tissue engineering applications. Biomacromol. 2018;19:1826–1839. doi: 10.1021/acs.biomac.8b00279. [DOI] [PubMed] [Google Scholar]
- De Baets G, Schymkowitz J, Rousseau F. Predicting aggregation-prone sequences in proteins. ESSAYS BIOCHEM. 2014;56:41–52. doi: 10.1042/bse0560041. [DOI] [PubMed] [Google Scholar]
- Egorov VV, Matusevich OV, Shaldzhyan AA, Skvortsov AN, Zabrodskaya YA, Garmay YP, Landa SB, Lebedev DV, Zarubayev VV, Sirotkin AK, Vasin AV, Kiselev OI. Structural features of the peptide homologous to 6–25 fragment of influenza A PB1 protein. Int J Pept. 2013;2013:370832. doi: 10.1155/2013/370832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eimer WA, Vijaya KD, Navalpur SN, Rodriguez AS, Mitchell T, Washicosky KJ, Gyorgy B, Breakefield XO, Tanzi RE, Moir RD. Alzheimer's disease-associated beta-amyloid is rapidly seeded by herpesviridae to protect against brain infection. Neuron. 2018;99:56–63. doi: 10.1016/j.neuron.2018.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzat K, Pernemalm M, Palsson S, Roberts TC, Jarver P, Dondalska A, Bestas B, Sobkowiak MJ, Levanen B, Skold M, Thompson EA, Saher O, Kari OK, Lajunen T, Sverremark EE, Nilsson C, Ishchenko Y, Malm T, Wood M, Power UF, Masich S, Linden A, Sandberg JK, Lehtio J, Spetz AL, El AS. The viral protein corona directs viral pathogenesis and amyloid aggregation. NAT COMMUN. 2019;10:2331. doi: 10.1038/s41467-019-10192-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo R, Ramakers M, De Smet F, Claes F, Khodaparast L, Khodaparast L, Couceiro JR, Langenberg T, Siemons M, Nystrom S, Young LJ, Laine RF, Young L, Radaelli E, Benilova I, Kumar M, Staes A, Desager M, Beerens M, Vandervoort P, Luttun A, Gevaert K, Bormans G, Dewerchin M, Van Eldere J, Carmeliet P, Vande VG, Verfaillie C, Kaminski CF, De Strooper B, Hammarstrom P, Nilsson KP, Serpell L, Schymkowitz J, Rousseau F. De novo design of a biologically active amyloid. SCIENCE. 2016;354(6313):aah4949. doi: 10.1126/science.aah4949. [DOI] [PubMed] [Google Scholar]
- Gosztyla ML, Brothers HM, Robinson SR. Alzheimer's amyloid-beta is an antimicrobial peptide: a review of the evidence. J ALZHEIMERS DIS. 2018;62:1495–1506. doi: 10.3233/JAD-171133. [DOI] [PubMed] [Google Scholar]
- Harris F, Dennison SR, Phoenix DA. Aberrant action of amyloidogenic host defense peptides: a new paradigm to investigate neurodegenerative disorders? FASEB J. 2012;26:1776–1781. doi: 10.1096/fj.11-199208. [DOI] [PubMed] [Google Scholar]
- Hirakura Y, Carreras I, Sipe JD, Kagan BL. Channel formation by serum amyloid A: a potential mechanism for amyloid pathogenesis and host defense. Amyloid. 2002;9:13–23. doi: 10.3109/13506120209072440. [DOI] [PubMed] [Google Scholar]
- Holch A, Bauer R, Olari LR, Rodriguez AA, Standker L, Preising N, Karacan M, Wiese S, Walther P, Ruiz-Blanco YB, Sanchez-Garcia E, Schumann C, Munch J, Spellerberg B. Respiratory ss-2-Microglobulin exerts pH dependent antimicrobial activity. VIRULENCE. 2020;11:1402–1414. doi: 10.1080/21505594.2020.1831367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housmans JAJ, Wu G, Schymkowitz J, Rousseau F (2021) A guide to studying protein aggregation. The FEBS Journal. 10.1111/febs.16312 [DOI] [PubMed]
- Idrees D, Kumar V. SARS-CoV-2 spike protein interactions with amyloidogenic proteins: potential clues to neurodegeneration. Biochem Biophys Res Commun. 2021;554:94–98. doi: 10.1016/j.bbrc.2021.03.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JW, Hancock TJ, Dogra P, Patel R, Arav-Boger R, Williams AD, Kennel SJ, Wall JS, Sparer TE (2019) Anticytomegalovirus peptides point to new insights for CMV entry mechanisms and the limitations of in vitro screenings. msphere 4(1):e00586–18 [DOI] [PMC free article] [PubMed]
- Jana AK, Greenwood AB, Hansmann UH (2021) Presence of a SARS-COV-2 protein enhances Amyloid Formation of Serum Amyloid A. J Phys Chem B 125(32):9155–9167 [DOI] [PMC free article] [PubMed]
- Jang H, Arce FT, Mustata M, Ramachandran S, Capone R, Nussinov R, Lal R. Antimicrobial protegrin-1 forms amyloid-like fibrils with rapid kinetics suggesting a functional link. BIOPHYS J. 2011;100:1775–1783. doi: 10.1016/j.bpj.2011.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhl DW, Glattard E, Lointier M, Bampilis P, Bechinger B. The reversible non-covalent aggregation into fibers of PGLa and Magainin 2 preserves their antimicrobial activity and synergism. Front Cell Infect Microbiol. 2020;10:526459. doi: 10.3389/fcimb.2020.526459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan BL, Jang H, Capone R, Teran AF, Ramachandran S, Lal R, Nussinov R. Antimicrobial properties of amyloid peptides. Mol Pharm. 2012;9:708–717. doi: 10.1021/mp200419b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodaparast L, Khodaparast L, Gallardo R, Louros NN, Michiels E, Ramakrishnan R, Ramakers M, Claes F, Young L, Shahrooei M, Wilkinson H, Desager M, Mengistu TW, Nilsson K, Hammarstrom P, Aertsen A, Carpentier S, Van Eldere J, Rousseau F, Schymkowitz J. Aggregating sequences that occur in many proteins constitute weak spots of bacterial proteostasis. NAT COMMUN. 2018;9:866. doi: 10.1038/s41467-018-03131-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokotidou C, Jonnalagadda S, Orr AA, Vrentzos G, Kretsovali A, Tamamis P, Mitraki AA. Designer amyloid cell-penetrating peptides for potential use as gene transfer vehicles. Biomolecules. 2019;10(1):7. doi: 10.3390/biom10010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravchenko SV, Domnin PA, Grishin SY, Panfilov AV, Azev VN, Mustaeva LG, Gorbunova EY, Kobyakova MI, Surin AK, Glyakina AV, Fadeev RS, Ermolaeva SA, Galzitskaya OV. Multiple antimicrobial effects of hybrid peptides synthesized based on the sequence of ribosomal S1 protein from Staphylococcus aureus. INT J MOL SCI. 2022;23(1):524. doi: 10.3390/ijms23010524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar DK, Choi SH, Washicosky KJ, Eimer WA, Tucker S, Ghofrani J, Lefkowitz A, McColl G, Goldstein LE, Tanzi RE, Moir RD. Amyloid-beta peptide protects against microbial infection in mouse and worm models of Alzheimer's disease. SCI TRANSL MED. 2016;8:340r–372r. doi: 10.1126/scitranslmed.aaf1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurpe S, Grishin S, Surin A, Selivanova O, Fadeev R, Dzhus U, Gorbunova E, Mustaeva L, Azev V, Galzitskaya O. Antimicrobial and amyloidogenic activity of peptides synthesized on the basis of the ribosomal S1 Protein from Thermus thermophilus. INT J MOL SCI. 2020;21:6382. doi: 10.3390/ijms21176382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurpe SR, Grishin SY, Surin AK, Panfilov AV, Slizen MV, Chowdhury SD, Galzitskaya OV. Antimicrobial and amyloidogenic activity of peptides. Can Antimicrobial Peptides Be Used against SARS-CoV-2? INT J MOL SCI. 2020;21:9552. doi: 10.3390/ijms21249552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last NB, Miranker AD. Common mechanism unites membrane poration by amyloid and antimicrobial peptides. Proc Natl Acad Sci U S A. 2013;110:6382–6387. doi: 10.1073/pnas.1219059110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalka AK, Kinnunen PK. Binding of amphipathic alpha-helical antimicrobial peptides to lipid membranes: lessons from temporins B and L. Biochim Biophys Acta. 2009;1788:1600–1609. doi: 10.1016/j.bbamem.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Makin S. The Amyloid Hypothesis on Trial NATURE. 2018;559:S4–S7. doi: 10.1038/d41586-018-05719-4. [DOI] [PubMed] [Google Scholar]
- Michiels E, Roose K, Gallardo R, Khodaparast L, Khodaparast L, van der Kant R, Siemons M, Houben B, Ramakers M, Wilkinson H, Guerreiro P, Louros N, Kaptein S, Ibanez LI, Smet A, Baatsen P, Liu S, Vorberg I, Bormans G, Neyts J, Saelens X, Rousseau F, Schymkowitz J. Reverse engineering synthetic antiviral amyloids. NAT COMMUN. 2020;11:2832. doi: 10.1038/s41467-020-16721-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir RD, Lathe R, Tanzi RE. The antimicrobial protection hypothesis of Alzheimer's disease. ALZHEIMERS DEMENT. 2018;14:1602–1614. doi: 10.1016/j.jalz.2018.06.3040. [DOI] [PubMed] [Google Scholar]
- Mukherjee SK, Knop JM, Winter R. Modulation of the conformational space of SARS-CoV-2 RNA quadruplex RG-1 by cellular components and the amyloidogenic peptides alpha-synuclein and hIAPP. Chemistry. 2022;28:e202104182. doi: 10.1002/chem.202104182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch J, Rucker E, Standker L, Adermann K, Goffinet C, Schindler M, Wildum S, Chinnadurai R, Rajan D, Specht A, Gimenez-Gallego G, Sanchez PC, Fowler DM, Koulov A, Kelly JW, Mothes W, Grivel JC, Margolis L, Keppler OT, Forssmann WG, Kirchhoff F. Semen-derived amyloid fibrils drastically enhance HIV infection. Cell. 2007;131:1059–1071. doi: 10.1016/j.cell.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Nilsson MR, Dobson CM. In vitro characterization of lactoferrin aggregation and amyloid formation. BIOCHEMISTRY-US. 2003;42:375–382. doi: 10.1021/bi0204746. [DOI] [PubMed] [Google Scholar]
- Otzen D, Riek R. Functional amyloids. Cold Spring Harb Perspect Biol. 2019;11(2):a033860. doi: 10.1101/cshperspect.a033860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palika A, Armanious A, Rahimi A, Medaglia C, Gasbarri M, Handschin S, Rossi A, Pohl MO, Busnadiego I, Gubeli C, Anjanappa RB, Bolisetty S, Peydayesh M, Stertz S, Hale BG, Tapparel C, Stellacci F, Mezzenga R. An antiviral trap made of protein nanofibrils and iron oxyhydroxide nanoparticles. NAT NANOTECHNOL. 2021;16:918–925. doi: 10.1038/s41565-021-00920-5. [DOI] [PubMed] [Google Scholar]
- Pasupuleti M, Roupe M, Rydengard V, Surewicz K, Surewicz WK, Chalupka A, Malmsten M, Sorensen OE, Schmidtchen A. Antimicrobial activity of human prion protein is mediated by its N-terminal region. PLoS ONE. 2009;4:e7358. doi: 10.1371/journal.pone.0007358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido D, Moussaoui M, Andreu D, Nogues MV, Torrent M, Boix E. Antimicrobial action and cell agglutination by the eosinophil cationic protein are modulated by the cell wall lipopolysaccharide structure. Antimicrob Agents Chemother. 2012;56:2378–2385. doi: 10.1128/AAC.06107-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridge RJ, Paus EJ, Novitsky TJ, Ketchum PA. Reversible binding of heparin to the loop peptide of endotoxin neutralizing protein. J Endotoxin Res. 2000;6:17–23. doi: 10.1177/09680519000060010301. [DOI] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. NAT MED. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Salinas N, Colletier JP, Moshe A, Landau M. Extreme amyloid polymorphism in Staphylococcus aureus virulent PSMalpha peptides. NAT COMMUN. 2018;9:3512. doi: 10.1038/s41467-018-05490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaider L, Brahmachari S, Schmidt NW, Mensa B, Shaham-Niv S, Bychenko D, Adler-Abramovich L, Shimon L, Kolusheva S, DeGrado WF, Gazit E. Self-assembling dipeptide antibacterial nanostructures with membrane disrupting activity. NAT COMMUN. 2017;8:1365. doi: 10.1038/s41467-017-01447-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semerdzhiev SA, Fakhree M, Segers-Nolten I, Blum C, Claessens M. Interactions between SARS-CoV-2 N-protein and alpha-synuclein accelerate amyloid formation. ACS CHEM NEUROSCI. 2022;13:143–150. doi: 10.1021/acschemneuro.1c00666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood R, Domanov Y, Pietiainen M, Kontinen VP, Kinnunen PK. Binding of LL-37 to model biomembranes: insight into target vs host cell recognition. Biochim Biophys Acta. 2008;1778:983–996. doi: 10.1016/j.bbamem.2007.11.016. [DOI] [PubMed] [Google Scholar]
- Soscia SJ, Kirby JE, Washicosky KJ, Tucker SM, Ingelsson M, Hyman B, Burton MA, Goldstein LE, Duong S, Tanzi RE, Moir RD. The Alzheimer's disease-associated amyloid beta-protein is an antimicrobial peptide. PLoS ONE. 2010;5:e9505. doi: 10.1371/journal.pone.0009505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer P, Condic M, Herrmann M, Oberstein TJ, Scharin-Mehlmann M, Gilbert DF, Friedrich O, Gromer T, Kornhuber J, Lang R, Maler JM. Amyloidogenic amyloid-beta-peptide variants induce microbial agglutination and exert antimicrobial activity. Sci Rep. 2016;6:32228. doi: 10.1038/srep32228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavassoly O, Safavi F, Tavassoly I. Seeding brain protein aggregation by SARS-CoV-2 as a possible long-term complication of COVID-19 infection. ACS CHEM NEUROSCI. 2020;11:3704–3706. doi: 10.1021/acschemneuro.0c00676. [DOI] [PubMed] [Google Scholar]
- Tavassoly O, Safavi F, Tavassoly I. Heparin-binding peptides as novel therapies to stop SARS-CoV-2 cellular entry and infection. MOL PHARMACOL. 2020;98:612–619. doi: 10.1124/molpharm.120.000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrent M, Pulido D, Nogues MV, Boix E. Exploring new biological functions of amyloids: bacteria cell agglutination mediated by host protein aggregation. PLOS PATHOG. 2012;8:e1003005. doi: 10.1371/journal.ppat.1003005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai PW, Yang CY, Chang HT, Lan CY. Human antimicrobial peptide LL-37 inhibits adhesion of Candida albicans by interacting with yeast cell-wall carbohydrates. PLoS ONE. 2011;6:e17755. doi: 10.1371/journal.pone.0017755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu W, Xue K, Lou S, Zhu C, Yu Z. Self-assembly of virulent amyloid-derived peptides into nanoantibacterials. Nanoscale. 2021;13:9864–9872. doi: 10.1039/D1NR01622A. [DOI] [PubMed] [Google Scholar]
- Valincius G, Heinrich F, Budvytyte R, Vanderah DJ, McGillivray DJ, Sokolov Y, Hall JE, Losche M. Soluble amyloid beta-oligomers affect dielectric membrane properties by bilayer insertion and domain formation: implications for cell toxicity. BIOPHYS J. 2008;95:4845–4861. doi: 10.1529/biophysj.108.130997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquer-Alicea J, Diamond MI. Propagation of protein aggregation in neurodegenerative diseases. ANNU REV BIOCHEM. 2019;88:785–810. doi: 10.1146/annurev-biochem-061516-045049. [DOI] [PubMed] [Google Scholar]
- Voth S, Gwin M, Francis CM, Balczon R, Frank DW, Pittet JF, Wagener BM, Moser SA, Alexeyev M, Housley N, Audia JP, Piechocki S, Madera K, Simmons A, Crawford M, Stevens T. Virulent Pseudomonas aeruginosa infection converts antimicrobial amyloids into cytotoxic prions. FASEB J. 2020;34:9156–9179. doi: 10.1096/fj.202000051RRR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh P, Vanderlee G, Yau J, Campeau J, Sim VL, Yip CM, Sharpe S. The mechanism of membrane disruption by cytotoxic amyloid oligomers formed by prion protein(106–126) is dependent on bilayer composition. J BIOL CHEM. 2014;289:10419–10430. doi: 10.1074/jbc.M113.515866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Liu Q, Chen JC, Cui YX, Zhou B, Chen YX, Zhao YF, Li YM. Antimicrobial activity of human islet amyloid polypeptides: an insight into amyloid peptides' connection with antimicrobial peptides. BIOL CHEM. 2012;393:641–646. doi: 10.1515/hsz-2012-0107. [DOI] [PubMed] [Google Scholar]
- Wang R, Yang X, Cui L, Yin H, Xu S. Gels of Amyloid Fibers Biomolecules. 2019;9:210. doi: 10.3390/biom9060210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Su Z, Reynolds NP, Arosio P, Hamley IW, Gazit E, Mezzenga R. Self-assembling peptide and protein amyloids: from structure to tailored function in nanotechnology. CHEM SOC REV. 2017;46:4661–4708. doi: 10.1039/C6CS00542J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Wu S, Xia J, Shao P, Sun P, Xiang N. Enhanced antibacterial activity of hen egg-white lysozyme against Staphylococcus aureus and Escherichia coli due to Protein Fibrillation. Biomacromol. 2021;22:890–897. doi: 10.1021/acs.biomac.0c01599. [DOI] [PubMed] [Google Scholar]
- White MR, Kandel R, Tripathi S, Condon D, Qi L, Taubenberger J, Hartshorn KL. Alzheimer's associated beta-amyloid protein inhibits influenza A virus and modulates viral interactions with phagocytes. PLoS ONE. 2014;9:e101364. doi: 10.1371/journal.pone.0101364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak MA, Mee AP, Itzhaki RF. Herpes simplex virus type 1 DNA is located within Alzheimer's disease amyloid plaques. J PATHOL. 2009;217:131–138. doi: 10.1002/path.2449. [DOI] [PubMed] [Google Scholar]
- Xia X. Domains and functions of spike protein in SARS-Cov-2 in the context of vaccine design. Viruses. 2021;13:109. doi: 10.3390/v13010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabrodskaya YA, Lebedev DV, Egorova MA, Shaldzhyan AA, Shvetsov AV, Kuklin AI, Vinogradova DS, Klopov NV, Matusevich OV, Cheremnykh TA, Dattani R, Egorov VV. The amyloidogenicity of the influenza virus PB1-derived peptide sheds light on its antiviral activity. BIOPHYS CHEM. 2018;234:16–23. doi: 10.1016/j.bpc.2018.01.001. [DOI] [PubMed] [Google Scholar]
- Zhang M, Zhao J, Zheng J. Molecular understanding of a potential functional link between antimicrobial and amyloid peptides. Soft Matter. 2014;10:7425–7451. doi: 10.1039/C4SM00907J. [DOI] [PubMed] [Google Scholar]
- Zhao C, Qin G, Niu J, Wang Z, Wang C, Ren J, Qu X. Targeting RNA G-Quadruplex in SARS-CoV-2: a promising therapeutic target for COVID-19? Angew Chem Int Ed Engl. 2021;60:432–438. doi: 10.1002/anie.202011419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.