Abstract
Background
High expression of the metabolic enzyme arginase-2 (ARG2) by cancer cells, regulatory immune cells, or cells of the tumor stroma can reduce the availability of arginine (L-Arg) in the tumor microenvironment (TME). Depletion of L-Arg has detrimental consequences for T cells and leads to T-cell dysfunction and suppression of anticancer immune responses. Previous work from our group has demonstrated the presence of proinflammatory ARG2-specific CD4 T cells that inhibited tumor growth in murine models on activation with ARG2-derived peptides. In this study, we investigated the natural occurrence of ARG2-specific CD8 T cells in both healthy donors (HDs) and patients with cancer, along with their immunomodulatory capabilities in the context of the TME.
Materials and methods
A library of 15 major histocompatibility complex (MHC) class I-restricted ARG2-derived peptides were screened in HD peripheral blood mononuclear cells using interferon gamma (IFN-γ) ELISPOT. ARG2-specific CD8 T-cell responses were identified using intracellular cytokine staining and ARG2-specific CD8 T-cell cultures were established by enrichment and rapid expansion following in vitro peptide stimulation. The reactivity of the cultures toward ARG2-expressing cells, including cancer cell lines and activated regulatory T cells (Tregs), was assessed using IFN-γ ELISPOT and a chromium release assay. The Treg signature was validated based on proliferation suppression assays, flow cytometry and quantitative reverse transcription PCR (RT-qPCR). In addition, vaccinations with ARG2-derived epitopes were performed in the murine Pan02 tumor model, and induction of ARG2-specific T-cell responses was evaluated with IFN-γ ELISPOT. RNAseq and subsequent GO-term and ImmuCC analysis was performed on the tumor tissue.
Results
We describe the existence of ARG2-specific CD8+ T cells and demonstrate these CD8+ T-cell responses in both HDs and patients with cancer. ARG2-specific T cells recognize and react to an ARG2-derived peptide presented in the context of HLA-B8 and exert their cytotoxic function against cancer cells with endogenous ARG2 expression. We demonstrate that ARG2-specific T cells can specifically recognize and react to activated Tregs with high ARG2 expression. Finally, we observe tumor growth suppression and antitumorigenic immunomodulation following ARG2 vaccination in an in vivo setting.
Conclusion
These findings highlight the ability of ARG2-specific T cells to modulate the immunosuppressive TME and suggest that ARG2-based immunomodulatory vaccines may be an interesting option for cancer immunotherapy.
Keywords: Immunomodulation, Arginase, Vaccination, Tumor Microenvironment
WHAT IS ALREADY KNOWN ON THIS TOPIC
Overexpression of arginase enzymes in cancer cells and immunosuppressive cells leads to the depletion of the important amino acid arginine from the tumor microenvironment, which negatively affects T cell-mediated antitumor immunity. Naturally occurring effector CD4+ T cells specific to arginase-2 (ARG2) were previously shown to be present at a high frequency in both healthy donors and patients with cancer and induced tumor growth delay on activation in a murine tumor model.
WHAT THIS STUDY ADDS
In this study, we identify CD8+ T cells specific to an ARG2-derived epitope. Moreover, we confirm that ARG2 is preferentially expressed by functional immunosuppressive Tregs, and we show that ARG2-expressing activated Tregs, along with ARG2-expressing cancer cell lines, can be specifically recognized by cytotoxic ARG2-specific CD8+ T cells. Moreover, we demonstrate tumor growth suppression and antitumorigenic immunomodulation in an in vivo model of cancer.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE, OR POLICY
This study highlights the antiregulatory function of ARG2-specific CD8+ effector T cells, which puts ARG2-based immunomodulatory vaccines forward as a novel therapeutic modality against cancer.
Introduction
The immune system plays an important role in suppression of malignancy by inhibiting tumor progression and aiding tumor elimination. However, tumors can evade immune surveillance using a number of mechanisms, including production of anti-inflammatory cytokines, downregulation of important surface receptors, recruitment of immunosuppressive cells to the tumor bed, expression of inhibitory molecules that negatively affect T-cell function and activation of metabolic pathways that deplete the tumor microenvironment (TME) of amino acids needed for T-cell function. Amino acid depletion can be caused by the upregulation of metabolic enzymes, such as tryptophan 2,3-dioxygenase, indolamine 2,3-dioxygenase (IDO), or arginases (ARG) in tumor cells, stromal cells or regulatory immune cells with immunosuppressive functions.1 2 Sufficient arginine (L-Arg) availability is essential for optimal T-cell function, and L-Arg deprivation causes CD3ζ chain downregulation and inhibits T-cell proliferation, differentiation and cytokine production, thereby dampening anticancer immune responses.3 4
ARG enzymes exist in two isoforms, arginase-1 (ARG1) and arginase-2 (ARG2), that catalyze the same biochemical reaction but differ in their subcellular localization and expression patterns.5 6 ARG1 is a cytosolic enzyme that is expressed by several immunosuppressive cell types—for example, myeloid-derived suppressor cells (MDSCs), tumor-associated macrophages (TAMs), and regulatory dendritic cells (DCs)—and increased ARG1 expression has been reported in a number of cancers.7–10 ARG2 is localized to the mitochondria and is not as well described as ARG1. Increased ARG2 expression, leading to immunosuppression, has been reported in the cells of some solid tumors,11–18 in acute myeloid leukemia (AML) blasts,19 and in neuroblastoma.20 High ARG2 expression has also been described in immunosuppressive cells of the TME, including DCs,21 regulatory T cells (Tregs),22 TAMs,12 and cancer-associated fibroblasts.12 23
We have previously described the existence of proinflammatory T cells that specifically recognize HLA-restricted epitopes from degraded intracellular self-antigens derived from immune inhibitory proteins, including IDO, programmed cell death ligand 1 (PD-L1), and ARG1.24–30 These cells have been termed ‘anti-Tregs’ due to their ability to directly target regulatory immune cells.31 Recently, we showed that vaccination with IDO and PD-L1 epitopes, in combination with anti-PD1 antibodies, has shown remarkably promising clinical efficacy as first-line treatment in a clinical phase II trial for patients with metastatic melanoma.32 Additionally, our group recently demonstrated that therapeutic vaccination against ARG1-derived epitopes, in combination with anti-PD1 blockade, facilitated antitumor immune responses and reduced tumor growth in several murine cancer models.33 Hence, immunomodulatory vaccination targeting such epitopes expressed by immune regulatory cells in the TME is a novel and very promising therapeutic modality against cancer. We also previously described the existence of ARG2-specific T cells that recognized multiple ARG2-derived epitopes.34 These T cells were present at a high frequency in both healthy donors (HDs) and patients with cancer, suggesting their natural role in immune homeostasis. Further characterization of ARG2-specific T cells has revealed that they primarily exert CD4+ responses to ARG2 and that they recognize and react to immune cells and cancer cells with intracellular ARG2 expression in an antigen-dependent manner. Simultaneous activation of CD4+ and CD8+ epitopes can result in strong synergistic protection against tumors.35 In the present study, we therefore aimed to further characterize ARG2-specific T cells to investigate whether short ARG2-derived peptides could elicit CD8+ T-cell responses. We describe the existence of cytotoxic ARG2-specific CD8+ T cells, thereby showing that the immune response against ARG2 peptides also include cytotoxic effector CD8+ T cells. Moreover, we demonstrate that ARG2-specific T cells react to Tregs with high ARG2 expression, which highlights the antiregulatory role of ARG2-specific effector T cells. Finally, we show the antitumorigenic and immunomodulatory effect of ARG2-specific T cells in an in vivo setting after vaccination with an ARG2-derived epitope.
Materials and methods
Donor material
HDs’ peripheral blood mononuclear cells (PBMCs) were isolated using density gradient separation over Lymphoprep (Alere) and cryopreserved at −150°C in fetal bovine serum (FBS, Life Technologies) supplemented with 10% dimethyl sulfide (DMSO). PBMCs from patients with cancer with solid tumors were derived from blood samples drawn within a minimum 4 weeks after administration of any therapy. PBMCs were maintained in X-vivo (BioNordika) supplemented with 5% human serum (Sigma Aldrich).
Peptides (human ARG2)
Peptide sequences were predicted using algorithms (available at www.syfpeithi.de36 and cbs.dtu.dk37 38). The list of short peptides was selected to only include peptide with a score above 24 (SYFPEITHI) or with a rank below 1.5 (NetMHC). All peptides were synthesized by Schäfer and had a purity above 90%. Peptides were dissolved in 100% DMSO to a stock concentration of 10 mM or in sterile water to a stock concentration of 2 mM. Peptides dissolved in water were sterile filtered before use. All peptides used in this study are listed in online supplemental table S1.
jitc-2022-005326supp010.pdf (119.1KB, pdf)
In vitro stimulation and interferon gamma (IFN-γ) ELISPOT
PBMCs from HDs or patients with cancer were subjected to in vitro stimulation with ARG2-derived peptides by plating the cells with 10 µM peptide. The following day, the cells received low-dose interleukin (IL)-2 treatment (120 U/mL). At 10–14 days after peptide stimulation, the cells were used for IFN-γ ELISPOT. ELISPOT plates were coated with 7.5 µg/µL IFN-γ capture antibody (MabTech) overnight. The next day, the plates were washed and blocked in X-vivo media followed by the plating of 2.5–3.5×105 PBMCs per well for ELISPOT, with and without restimulation with 5 µM peptide. A minimum of three technical replicates were set up per condition. The cells were incubated 14–16 hours before they were washed away and a biotin-conjugated secondary antibody (Mabtech) was added, followed by a 2-hour incubation. Next, the wells were washed again, followed by incubation with streptavidin-conjugated alkaline phosphatase (Mabtech) for 1 hour. Finally, spots were developed by addition of 5-Bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (BCIP/NBT) substrate (Mabtech). Tap water was used to stop the reaction. Spots corresponding to IFN-γ secretion were quantified by visualization on a CTL ImmunoSpot S6 Ultimate-V analyzer with ImmunoSpot software V.5.1. Peptide-specific IFN-γ secreting cells were calculated by subtracting the average spot count in control wells from the average spot count in peptide-stimulated wells. ELISPOT assays with ARG2-specific T cells (effector cells) and various immune cells or cancer cells as target cells were set up by adding 3–5×104 effector cells to ELISPOT wells followed by 5×103–104 target cells. Peptide pulsing was performed by incubating the target cells with 20 µM peptide for 1 hour, followed by two washes to remove unbound peptide. Effector cells with added peptide served as the positive control and target cells plated without effector cells served as the negative control. In ELISPOT assays with immune cells as target cells, wells containing only effector cells were also included as controls. The average spot count from replicate wells was subtracted from the spot count of effector cells plated with the respective target cells.
Ex vivo ELISPOT
PBMCs were thawed and rested overnight. The next day, 9×105 cells were plated per well. Control and peptide stimulations were performed in at least triplicates. The rest of the protocol was performed as described previously.
Cell lines
K562, K562-A1, FM6, FM28, and FM82 cell lines were maintained in RMPI-1640 (Gibco) supplemented with 10% FBS. OCI-M2 was maintained in Iscove’s MDM (Gibco) with 20% FBS. Cells were passaged every 2–3 days. Adherent cells (FM6, FM28, and FM82) were passaged following detachment from the flask with 0.25% trypsin (Gibco). All cells lines were confirmed to be mycoplasma negative.
Intracellular cytokine staining (ICS)
PBMCs were in vitro stimulated with peptide as described previously. At 10–14 days post simulation, the cells were used for ICS. Cells were incubated with peptide or a no-peptide control and a CD107a-PE antibody (BD) for 1 hour, followed by addition of GolgiPlug (BD). After a 4-hour incubation period, the cells were washed and stained with antibodies for extracellular markers: CD3–PC/H7, CD4–FITC, and CD8–PerCP. Dead cells were stained with FVS-510. Next, the cells were permeabilized using fixation/permeabilization buffer (Invitrogen), followed by staining with intracellular antibodies: IFN-γ–APC and tumor necrosis factor alpha (TNF-α)–BV421. Data were acquired using a BD Canto II flow cytometer and analyzed using FlowJo. The gating strategy is presented in online supplemental figure S6, and the antibodies used in this study are listed in online supplemental table S2.
jitc-2022-005326supp006.pdf (1.5MB, pdf)
jitc-2022-005326supp011.pdf (73.7KB, pdf)
Flow cytometry-based analysis of HLA expression
HLA-ABC and HLA-B8 expression levels in cell lines were analyzed by staining with HLA-ABC-FITC (BD) and HLA-B8-PE (Miltenyi Biotec) antibodies, respectively. The HLA-negative cell line K562 was used to set the gates for HLA-ABC+ and HLA-B8+ cells. Data were acquired using a BD Canto II flow cytometer and analyzed using FlowJo. The antibodies used in this study are listed in online supplemental table S2.
Generation of ARG2-specific T-cell cultures
ARG2-specific T-cell cultures were obtained by stimulation of PBMCs with A2S05 peptide, followed by low-dose IL-2 (120 U/mL) the next day. At 12 days after peptide stimulation, the cells were restimulated with peptide, and IFN-γ -secreting cells were isolated using magnetic beads (Miltenyi Biotec). Next, these cells were expanded using a rapid expansion protocol, including feeder cells, CD3 antibody, and high-dose IL-2 (3000–6000 U/mL). On days 16 and 17 after initiation of expansion, culture specificity was determined by stimulation with A2S05 peptide and determination of the cytokine release by ICS.
Activation, expansion, and isolation of Tregs and Trest
PBMCs were thawed and stimulated with CD3/CD28 Dynabeads (Gibco), following the manufacturer’s instructions, in X-vivo supplemented with 10% FBS, 1% sodium pyruvate (Gibco), and 1% non-essential amino acids (Gibco). On days 2, 5, and 7 after bead activation, IL-2 (300 U/mL) was added. On day 8, the beads were removed, and the cells were plated in medium without IL-2. On day 9, the cells were stained with CD3–APC/H7, CD4–PerCP, CD25–Pe-Cy/7, and the dead-cell stain FVS-510. An aliquot of cells was used as a fluorescence minus one control for CD127, while the remaining cells were stained with CD127–FITC. An aliquot of CD127–FITC stained cells was fixed, permeabilized and stained with an FOXP3–PE antibody. From the live population, Tregs were sorted as CD3+CD4+CD25highCD127− and Trest were sorted as CD3+CD4+CD25lowCD127+. Sorting was performed using a BD FACSMelody flow cytometer, and data were visualized using BD FACSChorus software. The gating strategy and specific sorting gates are presented in online supplemental figures S4 and S5, and the antibodies used are listed in online supplemental table S2.
jitc-2022-005326supp004.pdf (2.9MB, pdf)
jitc-2022-005326supp005.pdf (2.7MB, pdf)
Total RNA extraction
Cells were harvested, washed in PBS, and pelleted for RNA extraction. Cell pellets were stored at −80°C until RNA isolation. Total RNA isolation was performed using the RNEasy Plus Mini Kit (Qiagen), following the manufacturer’s instructions, with elution in 30 µL RNAse-free water. The RNA concentration was determined using a NanoDrop2000 Spectrophotometer (Thermo Scientific). RNA was stored at −80°C.
cDNA synthesis and RT-qPCR
cDNA was synthesized using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) with random primers and 400 ng or 1000 ng RNA (T cells and cancer cell lines, respectively). Before RT-qPCR analysis, cDNA was diluted 1:2–1:4. RT-qPCR analysis was performed using the TaqMan Gene Expression Assay on a Roche LightCycler 480 instrument. The assay was performed in technical triplicates for all primers, and the results were analyzed as previously described.39 For low-expression samples with no amplification during the assay, the Ct value was set to 40. Controls lacking reverse transcriptase were included in the primer validation analysis. The primers used in this study are listed in online supplemental table S3.
jitc-2022-005326supp012.pdf (123.2KB, pdf)
Preparation of cell lysates for western blotting
Set2, UKE-1, and sorted Treg and Trest cells were washed twice with sterile PBS before being pelleted and stored at −80°C. Cell pellets were resuspended in ice-cold RIPA Lysis Buffer (Thermo Scientific) supplemented with Halt Protease Inhibitor Cocktail (Thermo Scientific) at a 1:100 dilution. Cell lysates were placed under constant agitation for 15 min at 4°C before being centrifuged at 16,800×g at 4°C for 15 min. Supernatants were then transferred to new Eppendorf tubes and used for protein concentration measurements performed with the BCA Protein Assay Kit (Thermo Scientific) according to the manufacturer’s protocol.
Western blot analysis of ARG2 expression in sorted Treg and Trest
Volumes of cell lysate corresponding to 20 µg total protein were mixed with distilled water, Bolt Sample Reducing Agent (1:10 dilution), and Bolt LDS Sample Buffer (1:4 dilution, Invitrogen) for a total sample volume of 50 µL. The samples were incubated at 99°C for 10 min to aid denaturation and subsequently separated on Bolt 4%–12% Bis–Tris Plus gels (Invitrogen) for 20 min at 200 V using a PowerPac HV (BioRad) and Bolt MES SDS Running Buffer (Invitrogen). To allow for protein size quantification, the BioRad Precision Plus Protein Dual Color ladder was used. The gel was transferred to an iBlot 2 PVDF Ministack (Invitrogen) and electroblotted with an iBlot Gel 2 Transfer device (Invitrogen) according to the manufacturer’s guidelines. The membranes were cut in two pieces to allow for separate stainings of the ARG2 and vinculin proteins. Both parts of the membrane were blocked for 1 hour in Tris-buffered saline (TBS) buffer (Thermo Scientific) supplemented with 0.1% Tween-20 (Sigma Aldrich), known as TBST) with 5% added skimmed milk powder. Next, one part of the membrane was incubated overnight at 4°C with the primary ARG2-specific antibody diluted 1:1000 in blocking buffer, while the other half of the membrane was kept in TBST overnight. On the following day, the unstained part of the membrane was incubated for 1 hour with the vinculin-specific antibody diluted 1:100 000 in blocking buffer. Then, the membranes were washed with TBST three times for 5 min. The ARG2-stained and vinculin-stained membranes were then incubated with either anti-rabbit or anti-mouse secondary antibodies, respectively, at a 1:2000 dilution in blocking buffer for 1 hour. After three washes, the membranes were developed for 5 min with SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific) and visualized on a Gel Doc XR System (BioRad) using ImageLab software V.5.2.1. The used antibodies are listed in online supplemental table S2.
51Cr-release cytotoxicity assay
Conventional 51Cr-realease cytotoxicity assays were performed to evaluate the cytotoxicity of ARG2-specific CD8+ T cells, as previously described.40 Briefly, target cells were labeled with 100 µCi radioactive 51Cr for 1 hour, followed by two washes to remove any excess 51Cr outside the cells. Effector cells and target cells were plated at different effector-to-target (E:T) ratios and incubated for 4 hours. Next, 100 µL supernatant was recovered, and 51Cr release was determined using a 2470 Automatic γ-counter (Perkin Elmer). Maximum 51Cr release was determined in separate well by addition of 100 µL of 10% Triton-X to target cells. Spontaneous target cell lysis was determined in other wells by incubating the target cells with medium alone. Assays were set up using technical duplicates for all E:T ratios; maximum and minimum release wells were set up with six technical replicates.
In vitro Treg suppression assay
The CD8-positive fraction of the activated PBMCs used for Treg/Trest sorting was isolated on day 8 after stimulation initiation using magnetic bead separation (Miltenyi Biotec). The purity of the sorted cells was assessed by flow cytometry after staining the cells with CD3–APC/H7, CD8–FITC and FVS-510. On the following day, the CD8 T cells were stained with carboxyfluorescein succinimidyl ester (CFSE) dye (Sigma Aldrich) at a concentration of 5 µM. Next, 1.5×105 CFSE-labeled CD8 T cells were cocultured in a round-bottom 96-well plate together with 0.3×105 sorted-purified Treg or Trest cells. The cells were stimulated with human T activator anti-CD3/CD28 Dynabeads (Gibco) at a ratio of 1:25 and supplemented with IL-2 (300 U/mL) 1 day after coculture initiation. The proliferation status of the CD8 T cells was analyzed using flow cytometry on days 0 and 5 after initiating the coculture. Staining the cells with CD4–PerCP enabled segregation of the Treg/Trest cells from the CFSE-labeled CD8 T cells. For each analyzed time point, three technical replicates were set up. Data were acquired using a BD Canto II flow cytometer and analyzed using FCS Express V.7 and FlowJo V.10. The antibodies used can be found in online supplemental table S2.
Treg/Trest population analysis in cocultures of activated PBMCs with effector T cells
On day 9 following stimulation initiation, 7.5×105 activated PBMCs were incubated alone or together with either 5×105 ARG2-specific CD8 T cells or 5×105 autologous control CD8 T cells. The control CD8 T cells were isolated from activated PBMCs as described previously. The cells were cultured for 6 hours in a 48-well plate after which they were stained using the same procedure as for the sorting of Tregs and Trest as described earlier. In addition, similar gating strategies were used (see online supplemental figure S5) based on samples that only contained activated PBMCs. The Treg and Trest population sizes of the samples containing ARG2-specific CD8 T cells or control CD8 T cells were compared with the samples containing only activated PBMCs to determine the percentual decrease in the two populations. Three technical replicates were included for both conditions with added T cells, while six technical replicates were included for the condition with activated PBMCs alone.
Animal experiments
Animal experiments were conducted at the animal facility at Copenhagen University Hospital, Herlev, Department of Oncology. C57BL/6 female mice were bred in-house from a background of C57BL/6JBomTac. Daily care was performed by animal caretakers in the animal facility.
Peptide vaccinations
The murine ARG1 peptide (mARG1_169–177, ISAKDIVYI) was synthesized by Schäfer and was dissolved in DMSO to a stock concentration of 10 mM. The murine ARG2 peptide (mARG2_188–196, LSPPNIVYI) was synthesized by PepScan or Schäfer and dissolved in sterile water to a stock concentration of 2 mM. The ARG2 peptide was filtered before use by passing it through a sterile 0.22 µm filter. The purity of all synthesized peptides was >90%. All peptides used in this study are listed in online supplemental table S1. ARG2 peptide (100 µg) was suspended in a volume of 50 µL sterile water and emulsified 1:1 with Montanide ISA 51 (Seppic). Control vaccines included 50 µL sterile water, emulsified 1:1 with Montanide. Mice were vaccinated subcutaneously at the base of the tail with 100 µL of control or peptide emulsions. For tumor studies, mice were vaccinated on days 11 and 17 (Pan02), on days 0 and 7 (MC38), or on days 0 and 5 (B16–F10 and LL2) post tumor inoculation. For the remaining studies, mice were vaccinated one to three times with a minimum 1-week interval between vaccinations.
Tumor studies
Pan02, MC38, B16–F10 and LL2 cell lines were thawed 1 week prior to inoculation and cultured in DMEM (Gibco) with 10% FBS (Invitrogen) and 1% penicillin and streptavidin (P/S) (Life Technologies).
Female C57BL/6 mice received subcutaneous inoculations of 5×105 Pan02, MC38, B16-F10 or LL2 cells on the right flank. The tumor volume was measured with a digital caliper and calculated as (length×width2)/2. Experimental endpoint was defined as tumor volume above 1000 mm3 or the presence of tumorous ulceration. Investigators that performed tumor measurements were blinded to the treatment groups.
Murine IFN-γ ELISPOT
Mice were sacrificed at endpoint for tumor studies and 1 week after the last vaccination for the remaining studies. Spleens were harvested, smashed through a 70 µm filter, and red blood cells were lysed with Red Blood Cell Lysis Solution (Qiagen). 8×105 splenocytes were plated per well in IFN-γ ELISPOT plates. IFN-γ ELISPOT was performed as previously described.34 Tumors were harvested, cut into smaller pieces and enzymatically digested in RPMI-1640 medium (Gibco) containing 2.1 mg/mL collagenase type 1 (Worthington), 75 µg/mL DNase I (Worthington), 5 mM CaCl2, and 1% P/S. Cells were filtered through a 70 µm cell strainer and red blood cells were lysed as described previously. CD45+ tumor-infiltrating lymphocytes (TILs) were isolated from tumor single-cell suspensions with CD45 microbeads (for MC38 and LL2 tumors) (Miltenyi Biotec) or with CD45 (TIL) microbeads (for B16–F10 tumors) (Miltenyi Biotec). The isolated CD45+ cells were rested overnight. 5×105 CD45+ cells were plated per well in IFN-γ ELISPOT plates. For phenotyping ARG2 responses, CD8+ and CD4+ T cells were isolated from the spleen of vaccinated mice with CD8a (Ly-2) and CD4 (L3T4) microbeads (Miltenyi Biotec), respectively, following manufacturer’s instruction. Isolated T cells were rested overnight. 2.8×105 CD8+ or CD4+ T cells were plated in an IFN-γ ELISPOT plate with 6×105 splenocytes from a naïve mouse (used as antigen-presenting cells). Peptide-specific responses were reported as the difference in the average number of spots between peptide-stimulated and unstimulated wells.
Tumor RNA extraction and RNA sequencing
Tumor fragments (≤30 mg) were stored in RNAlater (Invitrogen) at −80°C. Tumors were homogenized on a TissueLyser (Qiagen), and RNA was extracted with the RNEasy Plus Mini Kit (Qiagen), according to the manufacturer’s instructions. RNA concentration was measured on a Nanodrop 2000 Spectrophotometer. Isolated RNA was stored at −80°C. RNAseq was performed on tumors from four untreated and six vaccinated mice as previously described.41 In short, 500 ng purified RNA (RIN score >7) was enriched for polyadenylated mRNA using oligo dT magnetic beads (Illumina) followed by fragmentation and cDNA synthesis using random priming (NEBNext). The cDNA was prepared for Illumina sequencing by adaptor ligation followed by indexing using PCR and size selection. Concentration of the cDNA libraries was determined by the KAPA Library Quantification Kits (Roche) and sequenced using Novaseq 6000 Illumina sequencing platform. RNAseq data is available on GEO repository (GSE212500). Alignment and quantification of reads was performed as previously described.41 Shortly, sequenced DNA was aligned to the GRCm39 reference genome assembly using STAR V.2.7.8, and reads were quantified using featureCounts,42 and the ENSEMBL Genes and Transcripts V.104. Differential gene expression was analyzed by DESeq2.43 Volcano plots were generated with EnhancedVolcano package V.1.8.0 based on the differential gene expression analysis. Biological processes associated with differentially upregulated genes were assessed with The Gene Ontology Resource (http://geneontology.org/) followed by classification of the most specific subprocesses.
ImmuCC
The computational framework of the CIBERSORT analytical tool,44 along with the developed ImmuCC signature matrix (non-tissue specific),45 suitable for the deconvolution of mouse bulk RNA-Seq data, were used to characterize and to quantify 25 immune cell subtypes. The ImmuCC signature matrix used consists of 511 genes of which 510 genes from our bulk RNA-Seq data were mapped (one missing). For the deconvolution of the bulk RNA-Seq samples with CIBERSORT, DEseq2’s median of ratios normalized data were used to produce the input mixture matrix. Additionally, the analysis included both CIBERSORT-relative and CIBERSORT-absolute modes. While CIBERSORT-relative represents immune cell fractions relative to the total immune content, thus suitable for intrasample comparisons, CIBERSORT-absolute produces a score that quantifies the abundance of each cell type, making it appropriate for intrasample comparisons between cell types as well as intersample comparisons of the same cell type. The CIBERSORT outputs were generated by performing 1000 permutations and by disabling the quantile normalization parameter. For this study, two population schemes were defined (compact and extended, online supplemental tables S6 and S7), resulting in the aggregation of some of the 25 immune subpopulations. Total absolute scores for subpopulations merged were calculated as the sum of the subpopulations. The relative fractions were recalculated based on each scheme’s new total immune content.
jitc-2022-005326supp015.pdf (37.1KB, pdf)
jitc-2022-005326supp016.pdf (108.4KB, pdf)
Statistical analysis
Statistical analysis of ELISPOT responses was performed in R studio using distribution free resampling rule as described by Moodie et al.46 The descriptive statistics from the proliferation assay and the change in Treg population size from the Treg/Trest population analysis were analyzed statistically using unpaired t-tests. Two-way analysis of variance with comparisons between multiple time points was performed to determine statistical significance for tumor growth curves. Statistical significance for the difference in peptide-specific IFN-γ-secreting cells and ImuneCC-related analysis between treatment groups were assessed with a Mann-Whitney U test and an unpaired, two-tailed t test, respectively. Statistical analyses were performed in GraphPad Prism V.9.
Results
Spontaneous CD8+ T-cell responses to ARG2
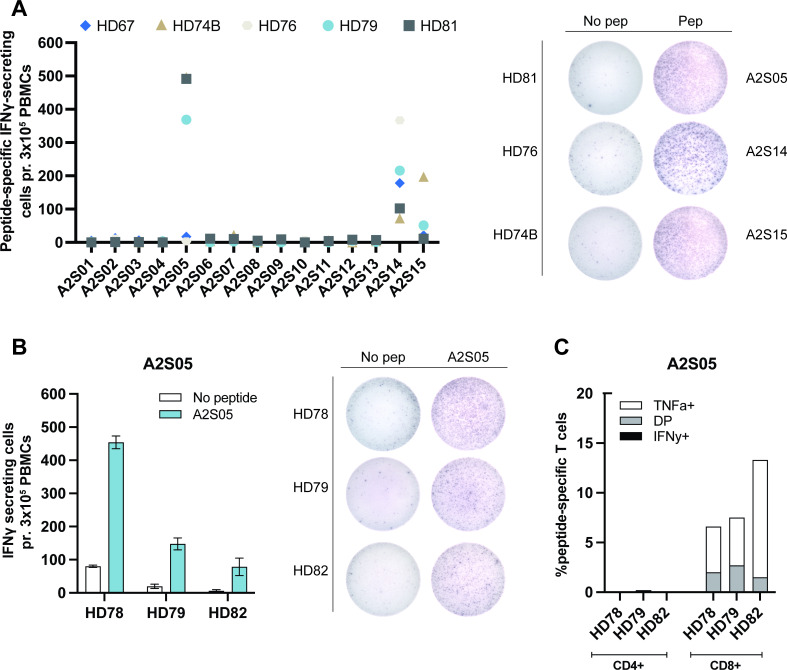
In our previous studies of ARG2 as a T-cell target, we demonstrated CD4+ T-cell responses to ARG2-derived peptides.34 Here, we set out to investigate ARG2-specific CD8+ T cells using short peptides that could be directly presented on MHC class I molecules. Using in silico HLA-prediction algorithms (available at www.syfpeithi.de36 and cbs.dtu.dk37 38), we obtained a library of 15 short ARG2-derived peptides (9mers and 10mers) predicted to strongly bind to HLA-A2 (online supplemental table S1). We screened these 15 peptides in PBMCs from five HDs who were confirmed to be HLA-A2 positive (HLA-A2+). PBMCs were stimulated with each peptide in vitro and restimulated in IFN-γ ELISPOT after 12–14 days of culture. The peptides A2S05, A2S14 and A2S15 were identified as strong candidates by eliciting significant responses in PBMCs from three or more donors (figure 1A). However, we could not validate the responses seen with A2S15, as repetition of the IFN-γ ELISPOT in HDs did not show significant responses (online supplemental figure S1A). On the other hand, A2S14 elicited strong IFN-γ ELISPOT responses in the validation set-up (online supplemental figure S1B), but further characterization of the responses by ICS for TNF-α and IFN-γ revealed that the reaction was from CD4+ T cells (online supplemental figure S1C). A2S05 was also found to elicit strong IFN-γ ELISPOT responses in a validation experiment (figure 1B) and ICS for TNF-α and IFN-γ demonstrated that the responses were from CD8+ T cells (figure 1C).
Figure 1.
Identification of an ARG2 peptide that elicits CD8+ T-cell responses. (A, left) IFN-γ ELISPOT screening of responses to 15 different HLA-A2 predicted ARG2-derived peptides in five healthy HLA-A2+ donors. 3×105 cells were plated per well. Control and peptide stimulations were done in triplicates. Peptide-specific IFN-γ secreting cells are given as the difference of spots between cells in the peptide-stimulated wells and control wells. (A, right) Representative examples of the ELISPOT responses (shown in A). (B, left) IFN-γ ELISPOT responses of three HDs to A2S05. 3×105 cells were plated per well. Control and peptide stimulations were done in triplicates. Bars represent the mean IFN-γ spot count±SD. (B, right) Representative examples of the ELISPOT responses (shown in B). (C) Representative intracellular cytokine staining for IFN-γ and TNF-α secreting in samples from three HDs stimulated with control or A2S05 gating strategy is available in online supplemental figure S6. DP: TNF-α+ IFN-γ+. DP, double positive; HD, healthy donor; IFN-γ, interferon gamma; PBMC, peripheral blood mononuclear cell; TNF-α, tumor necrosis factor alpha.
jitc-2022-005326supp001.pdf (912.3KB, pdf)
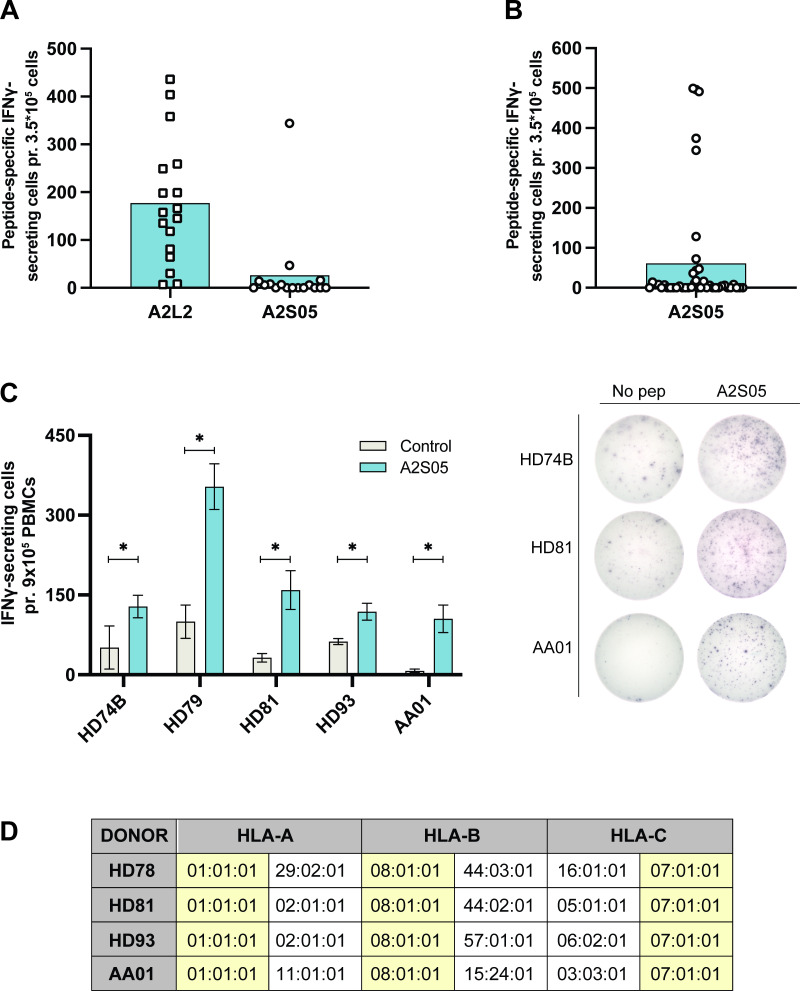
Having identified A2S05 as an ARG2-derived peptide that elicited CD8+ T-cell responses, we screened 17 HDs for response to this peptide, along with a previously described highly immunogenic ARG2-derived peptide, A2L2.34 We observed strong and frequent responses to A2L2, but only few responses to A2S05 (figure 2A). Since the HLA type of the HDs used for this assay was undetermined, we speculated that their HLA types could be incompatible with our HLA-A2 predicted peptide, A2S05. Accordingly, we screened 38 donors (13 patients with cancer with solid tumors and 25 HDs) with known HLA-A2 status (positive or negative) for response to A2S05. We primarily selected donors who were HLA-A2+ (n=29) but included some HLA-A2− donors (n=9) as controls. Again, responses to A2S05 were not frequently observed, with statistically significant responses occurring in only 10 out of the 38 donors (figure 2B). Importantly, in donors with strong in vitro responses to A2S05, we were also able to detect strong and significant responses in PBMCs ex vivo (figure 2C). Interestingly, responses to A2S05 were observed in both HLA-A2+ and HLA-A2− donors (online supplemental figure S1D), suggesting that A2S05 was not HLA-A2 restricted as predicted by the in silico algorithm. Indeed, we performed full HLA typing of four donors with strong A2S05 responses and found that only two donors were HLA-A2+, whereas all four donors shared HLA-A1, HLA-B8 and HLA-C7 haplotypes (figure 2D).
Figure 2.
A2S05 elicits responses in both HLA-A2+ and HLA-A2− donors and responses are also detectable ex vivo. (A) IFN-γ ELISPOT responses to A2L2 and A2S05 in 17 HDs. 3.5×105 cells were plated per well. Control and peptide stimulations were done in triplicates. Peptide-specific IFN-γ secreting cells are given as the difference of spots between cells in the peptide-stimulated wells and control wells. Each dot represents one donor and bars represent the mean. (B) IFN-γ ELISPOT responses to A2S05 in 30 HDs and 13 patients with cancer (2 patients with prostate cancer and 11 patients with melanoma) with known HLA types. Each dot represents one donor and bars represent the mean. (C, left) Ex vivo ELISPOT responses to A2S05. 9×105 cells were plated per well and control and peptide stimulations were done in three to six replicates. Bars represent the mean IFN-γ spot count±SD. *P≤0.05 (according to the DFR rule) (C, right) Representative examples of the ex vivo ELISPOT responses (shown in C). (D) HLA-typing data from the three HDs and one patient with cancer (AA01, melanoma) with strong responses to A2S05. HLA types shared between donors are highlighted in yellow. DFR, distribution free resampling rule; HD, healthy donor; IFN-γ, interferon gamma; PBMC, peripheral blood mononuclear cell.
Characterization of ARG2-specific CD8+ T cells
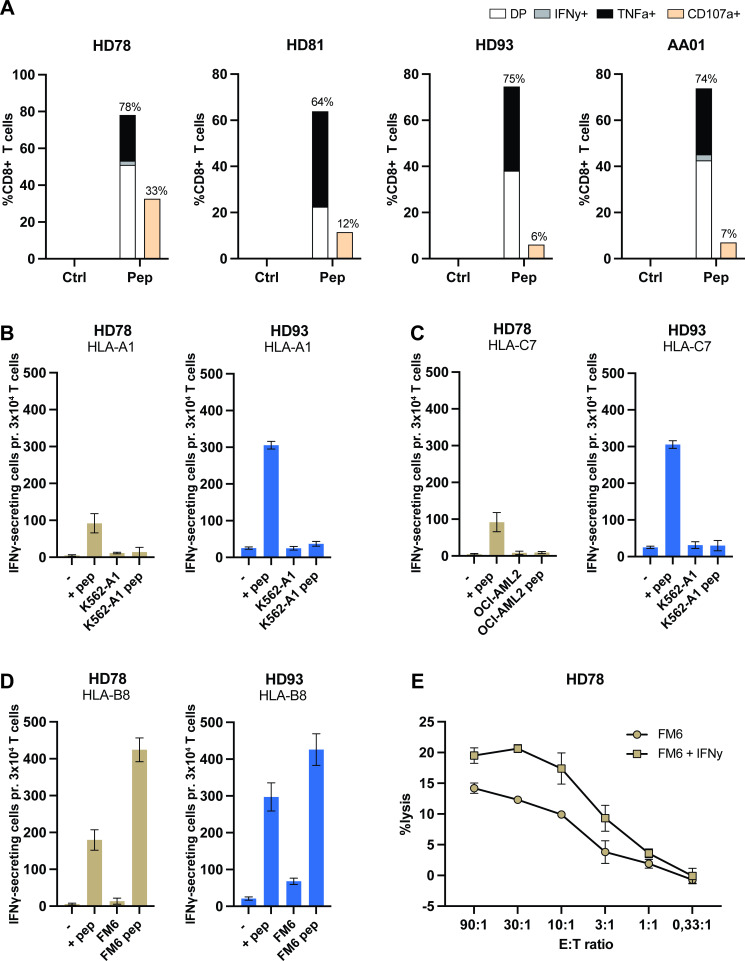
To further characterize the ARG2-specific CD8+ T cells, we established A2S05-specific T-cell cultures from four donors. Briefly, donor PBMCs were stimulated with A2S05 in vitro. Twelve days later, peptide-specific T cells were restimulated and isolated using an IFN-γ capture kit. IFN-γ-producing cells were expanded using a rapid expansion protocol and the specificity was examined at 16–17 days after expansion by ICS for IFN-γ and TNF-α. We were able to establish highly specific CD8+ T-cell cultures from all four donors (figure 3A).
Figure 3.
ARG2-specific CD8+ T cells recognize A2S05 in the context of HLA-B8. (A) Generation of A2S05-specific T-cell cultures. ARG2-specific CD8+ T cells were expanded from three HDs and one patient with cancer (AA01, melanoma). The specificity of the T-cell cultures against A2S05 was assessed by intracellular cytokine staining for TNF-α and IFN-γ. CD107a was included as a marker of cytotoxicity. Bars show the percentage of CD8+ T cells expressing CD107a and secreting IFN-γ, TNF-α or both (DP) in response to Ctrl stimulation or A2S05 stimulation (pep). (B–D) ARG2-specific T cells from two donors (HD78 and HD93) were evaluated in IFN-γ ELISPOT with cancer cells lines prepulsed with A2S05 peptide. The same cell lines without peptide stimulation were included as Ctrls. 3×104 ARG2-specific T cells were plated with 1×104 cancer cells (E:T of 3:1). The cell lines were either HLA-A1+ (B), HLA-C7+, (C) or HLA-B8+ (D). T cells plated alone (−) or T cells plated with A2S05 (+pep) served as negative and positive Ctrls, respectively. Bars represent the mean IFN-γ spot count±SD of technical triplicates. (E) 51Cr-release assay of FM6 and FM6 stimulated with IFN-γ (100 U/mL) for 24 hours prior to the assay with A2S05-specific T cells from HD78. Error bars represent mean±SD of technical duplicates. Ctrl, control; DP, double positive; E:T, effector-totarget ratio; -HD, healthy donor; IFN-γ, interferon gamma; TNF-α, tumor necrosis factor alpha.
These cultures were used to determine the HLA restriction of A2S05. IFN-γ ELISPOTs were performed by coculturing A2S05-specific T cells from HD78 and HD93 with different haplotype-relevant target cells lines prepulsed with A2S05 peptide. These experiments revealed no reactivity toward HLA-A1+ and HLA-C7+ cell lines (figure 3B, C) but showed recognition and reactivity activity toward the HLA-B8+ metastatic malignant melanoma cell line FM6 (figure 3D). FM6 was confirmed to be HLA-B8+ by flow cytometry and confirmed to express ARG2 by RT-qPCR (online supplemental figure S2). All four ARG2-specific CD8+ T-cell cultures were confirmed to show reactivity against FM6 and three additional HLA-B8+ cell lines (online supplemental figure S3). The specificity of the ARG2-specific CD8+ T-cell cultures was confirmed by the lack of reactivity against a peptide with the corresponding ARG1 sequence (ARG1_65–73) in IFN-γ ELISPOT (online supplemental figure S3). Next, we performed standard 51Cr-release cytotoxicity assays to establish whether the ARG2-specific CD8+ T cells could lyse FM6 cells. Indeed, the specific T cells lysed FM6 cells in a concentration-dependent manner (figure 3E), demonstrating the cytotoxic capacity of ARG2-specific CD8+ T cells. Stimulation with IFN-γ increased the mean fluorescence intensity of HLA-B8 expression (online supplemental figure S2) and resulted in a small increase in the lysis of FM6 cells (figure 3E).
jitc-2022-005326supp002.pdf (796.2KB, pdf)
jitc-2022-005326supp003.pdf (1.1MB, pdf)
ARG2-specific T cells specifically recognize activated Tregs
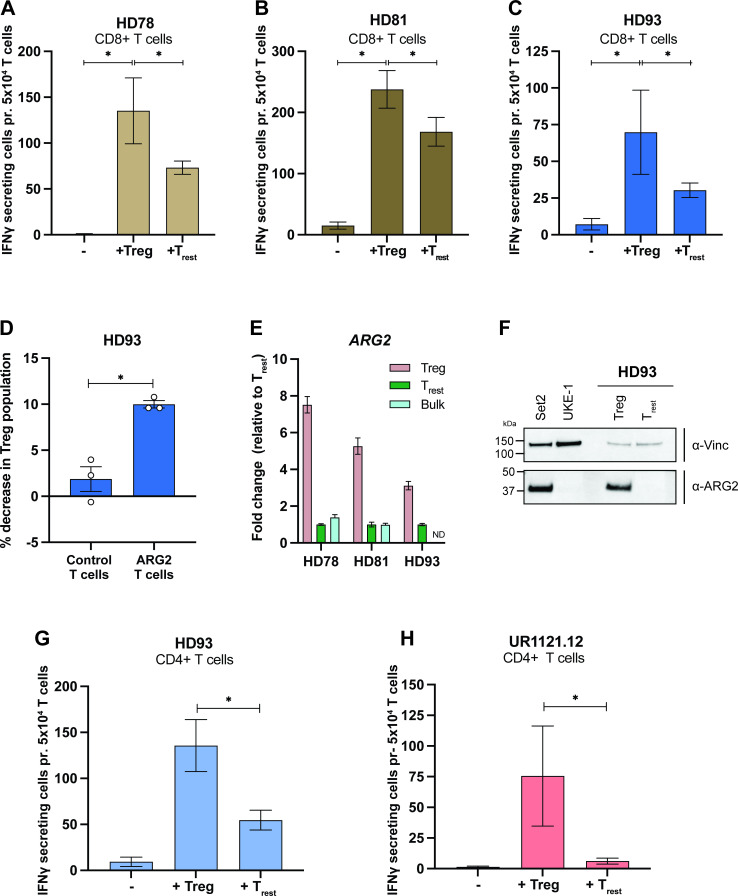
In a recent study, Lowe and colleagues found that activated Tregs from peripheral blood exhibited high ARG2 expression when compared with activated effector CD4+ T cells. Furthermore, they demonstrated an ARG2-dependent suppression of T-cell proliferation by Tregs.22 We hypothesized that if activated Tregs express higher levels of ARG2 than other T cells, they could be preferentially recognized by ARG2-specific T cells. To test this hypothesis, we activated PBMCs using anti-CD3/CD28 beads and added IL-2 on days 2, 5, and 7 based on the experiments described by Lowe et al.22 On day 9 after activation, we purified Tregs and resting T cells (Trest) using FACS (online supplemental figures S4A and S5) and plated Tregs or Trest as target cells for autologous ARG2-specific CD8+ T cells in an IFN-γ ELISPOT assay. We observed significantly higher responses to Tregs than to Trest in all three donors (figure 4A–C). Furthermore, coculture of activated PBMCs with ARG2-specific T cells led to a significant decrease in the Treg population when compared with activated PBMCs cocultured with autologous CD8+ T cells (figure 4D), suggesting cytotoxic activity of ARG2-specific T cells toward Tregs. Importantly, RT-qPCR analysis showed threefold to sevenfold higher ARG2 expression in Tregs than in Trest (figure 4E), along with minimal levels of ARG1 expression (online supplemental figure S4B), indicating ARG2-dependent recognition of Tregs. Preferable ARG2 expression in Tregs on protein level was confirmed by western blot analysis (figure 4F and online supplemental figure S4). We also compared ARG2 expression levels in Tregs and Trest to expression levels in the bulk culture of activated PBMCs from which the two T-cell subsets were isolated and found the bulk culture to express ARG2 at levels comparable to Trest (figure 4E). Moreover, ARG2 expression in the ARG2-specific T cells was comparable to the ARG2 expression in Trest (online supplemental figure S4D). Finally, we plated Tregs or Trest as target cells for autologous ARG2-specific CD4+ T cells in an IFN-γ ELISPOT assay. Interestingly, ARG2-specific CD4+ T cells also showed significantly higher responses toward Tregs than Trest (figure 4F–G).
Figure 4.
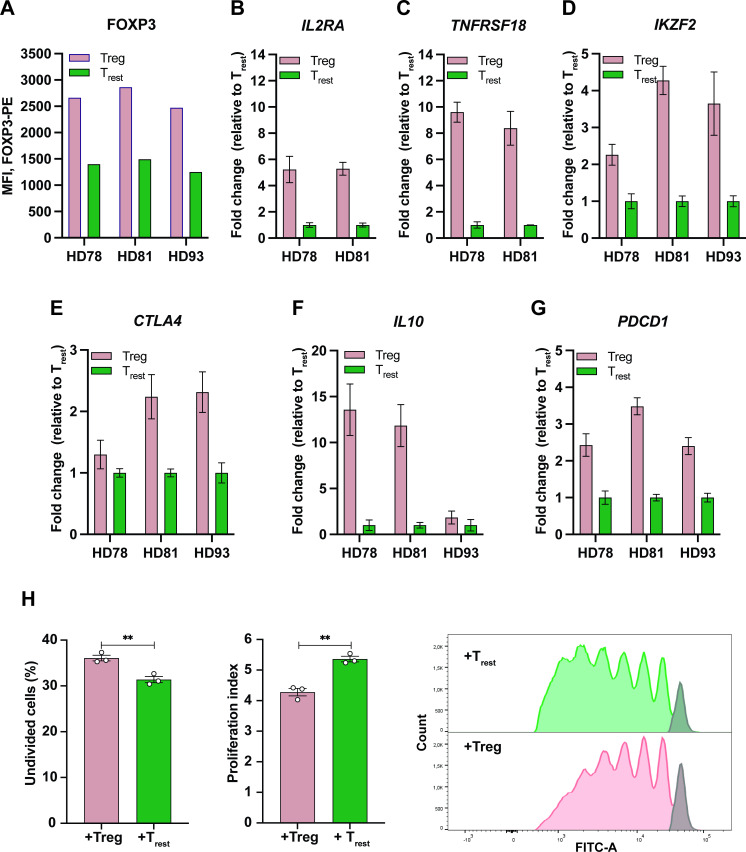
ARG2-specific T cells recognize and react to stimulated Tregs (Tregs). (A–C) IFN-γ ELISPOT responses of ARG2-specific CD8+ T cells to in vitro activated and expanded Tregs (Tregs) or resting CD4+ T cells (Trest). 5×104 ARG2-specific CD8+ T cells were plated per well with 5×103 target cells (E:T of 10:1). Bars show the mean IFN-γ spot count±SD. Background from Tregs and Trest have been corrected in the bars. All conditions were plated in six replicates. *P≤0.05 (according to the DFR). (D) Flow cytometry assessment of the Treg population after coculture of activated PBMCs with autologous CD8+ T cells (Ctrl T cells) or ARG2-specific T cells for 6 hours. Cocultures were set up with a 3:2 ratio of activated PBMCs to T cells. Activated PBMCs alone were used to set the gates for Tregs, which are similar to the gates used for sorting of Tregs for the ELISPOT experiment in (C). Bars show the mean decrease in Treg population when cocultured with Ctrl T cells or ARG2-specific T cells compared with PBMCs plated alone±SD of three technical replicates (depicted as dots). *P=0.0194 (unpaired t test). (E) RT-qPCR analysis of ARG2 expression in the Tregs and Trest used for the ELISPOT (in A–C). The bulk culture of activated PBMCs from which the Treg and Trest were isolated was also included. ARG2 expression was normalized to the housekeeping gene POL2RA and presented as fold change versus Trest. Bars show the mean±SD of technical triplicates. (F) Western Blot analysis assessing ARG2 expression in sorted Tregs and Trest from HD93. Cell lines Set2 and UKE-1 are included as ARG2 positive and negative Ctrls, respectively. (G, H) IFN-γ ELISPOT responses of ARG2-specific CD4+ T cells to in vitro activated and expanded Tregs or Trest. 5×104 ARG2-specific CD4+ T cells were plated per well with 5×103 target cells (E:T of 10:1). Bars show the mean IFN-γ spot count±SD of six technical replicates. Background from Tregs and Trest have been corrected in the bars. All conditions were plated in six replicates. *P≤0.05 (according to the DFR). Ctrl, control; DFR, distribution free resampling rule; HD, healthy donor; IFN-γ, interferon gamma; PBMC, peripheral blood mononuclear cell; Treg, regulatory T cell; Trest, resting T cell.
To confirm the immunosuppressive phenotype of the isolated, ARG2-expressing Tregs, we took an aliquot of activated PBMCs and stained intracellularly for FOXP3. Assessment of FOXP3 expression between cells sorted as Tregs and Trest showed higher FOXP3 expression in Tregs compared with Trest (figure 5A). This was also confirmed by RT-qPCR analysis of FOXP3 expression in Tregs and Trest (online supplemental figure S4C). To further validate our sort-purification strategy, we assessed the expression of several other Treg signature genes by RT-qPCR and found preferential expression of IL2RA (CD25), TNFRSF18 (GITR), IKZF2 (HELIOS), CTLA4, and IL10 in Tregs (figure 5B–F). Interestingly, Tregs also preferably expressed PDCD1 (PD-1) over Trest (figure 5G). Finally, the functional phenotype of the sort-purified Tregs was confirmed by a proliferation assay. To this end, we isolated highly pure populations of autologous CD8+ T cells (online supplemental figure S4E). We assessed proliferation of anti-CD3/CD28 and IL-2 stimulated CD8+ T cells cocultured with Trest or Tregs over the course of 5 days and found that Tregs significantly suppressed CD8+ T-cell proliferation compared with Trest (figure 5H).
Figure 5.
Characterization of sort-purified Tregs. (A) MFI of FOXP3-PE in sorted Tregs and Trest from the three donors following intracellular staining. (B–F) RT-qPCR analysis of the expression of IL2RA (B), TNFRSF18 (C), IKZF2 (D), CTLA4 (E), or IL10 (F) in the Tregs and Trest. The expression of each Treg signature gene was normalized to the housekeeping gene POL2RA and presented as fold change versus Trest with bars showing the mean±SD of technical triplicates. (G) RT-qPCR analysis of PDCD1 expression in Tregs and Trest. Data is represented as relative expression to the housekeeping gene POL2RA (arbitrary units) with bars showing the mean±SD of technical triplicates. (H) In vitro Treg suppression assay. (Left) Percentage of undivided CD8+ T cells after 5 days of co-culture with sort-purified Tregs (+Treg) or Trest (+Trest). Bars indicate the mean of three technical triplicates (depicted as dots). **P=0.0069 (unpaired t-test). (Middle) Proliferation index of CD8 T cells after 5 days of coculture with Tregs or Trest. Bars indicate the mean of three technical triplicates (depicted as dots). **P=0.0022 (unpaired t-test). (Right) CFSE staining of CD8+ T cells cocultured with Tregs or Trest for 5 days. The gray bar depicts the undivided population. CFSE, carboxyfluorescein succinimidyl ester; IL, interleukin; MFI, mean fluorescence intensity; Treg, regulatory T cell; Trest, resting T cell.
ARG2-specific T cells induced by peptide vaccination are immunomodulatory and inhibit tumor growth in a murine model of pancreatic cancer
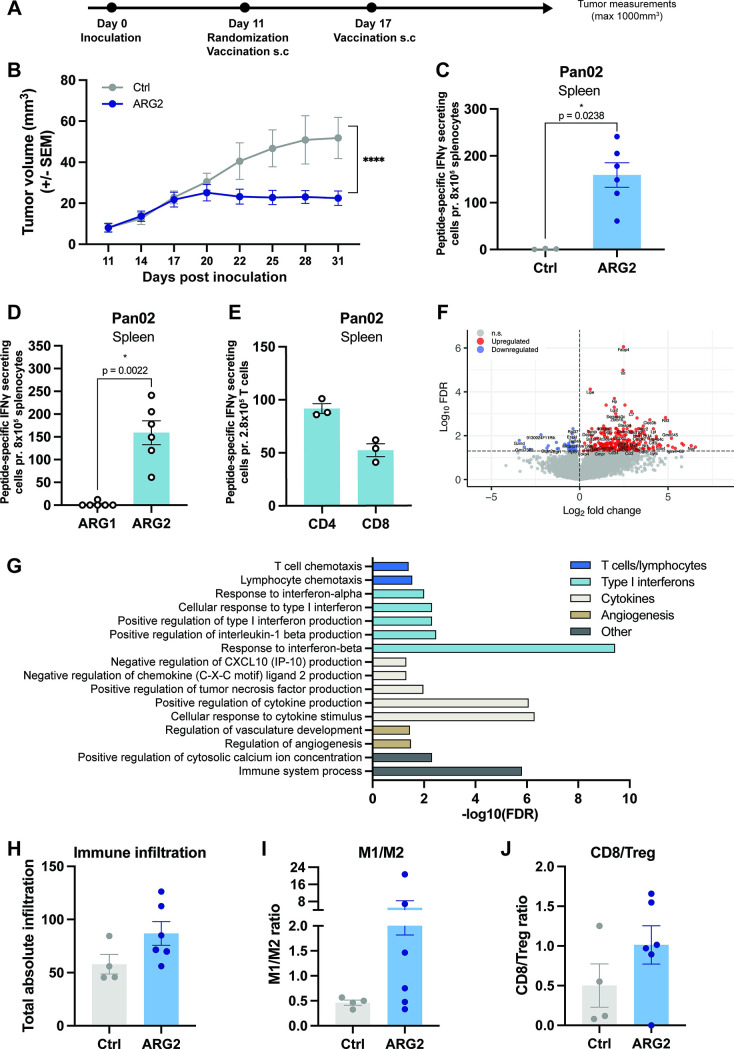
We have previously identified a highly immunogenic murine ARG2-derived epitope that elicited strong and frequent T-cell responses on a single vaccination.34 Moreover, we have shown that vaccination with the immunogenic ARG2-derived peptide induced tumor growth delay in a murine model of lung cancer.34 To confirm the antitumor effect of ARG2-derived peptide vaccination in another tumor model and to investigate the immunomodulatory function of ARG2-specific T cells, we employed the syngeneic mouse model Pan02. Pan02 is a model of pancreatic ductal adenocarcinoma (PDAC) and thereby relevant for studying ARG2-based vaccines given the previously demonstrated correlation between ARG2 expression and poor prognosis in patients with PDAC together with the demonstration of high Arg2 expression in Pan02 tumors at tumor endpoint.12 34
We challenged C57BL/6 mice with Pan02 cells, and on day 11, when tumors became palpable, we allocated mice with similar average tumor volumes into different treatment groups. The mice received either the previously described ARG2-based vaccine34 or a control vaccine on days 11 and 17 post tumor inoculation (figure 6A). Intriguingly, we observed marked inhibition of tumor growth in the ARG2-vaccinated mice; indeed, tumor growth ceased after day 20 (figure 6B). Moreover, one mouse in the ARG2-vaccinated group showed complete tumor regression (online supplemental figure S7A). These results were confirmed in an independent study (online supplemental figure S7B). The safety of the vaccine was also demonstrated with no decrease in bodyweight in the ARG2-vaccinated group (online supplemental figure S7C).
Figure 6.
An ARG2-based immunomodulatory vaccine reduces tumor growth, induces activation of ARG2-specific CD4+ and CD8+ T cells and promotes a proinflammatory tumor microenvironment. (A) Overview of the experimental timeline. (B) Average Pan02 tumor growth for mice receiving a Ctrl or ARG2-peptide immunomodulatory vaccine. Mice (n=10 per group) were inoculated and vaccinated according to the experimental overview (in A). Data are represented as mean±SEM. ****P<0.0001 (two-way analysis of variance test). (C) IFN-γ ELISPOT on splenocytes isolated from mice receiving the Ctrl (n=3) or ARG2-based (n=6) vaccine from the study in shown in B. Briefly, 8×105 splenocytes were plated per well. Ctrl and peptide stimulations were performed in triplicates. Peptide-specific IFN-γ secreting cells were quantified as the difference in the number of spots counted between the peptide-stimulated and Ctrl wells. Each dot represents one mouse and bars represent the mean±SEM. *P=0.0238 (Mann-Whitney test). (D) ARG1 and ARG2-specific IFN-γ-secreting cells among splenocytes from Pan02 tumor-bearing mice receiving the ARG2-based peptide vaccine were assayed with an IFN-γ ELISPOT. In this assay, 8×105 splenocytes were plated per well with either ARG1 or ARG2 peptide. Each dot represents one mouse, and bars represent the mean±SEM. *P=0.0022 (Mann-Whitney test). (E) ARG2-specific IFN-γ-secreting cells present in CD4+ and CD8+-sorted T cells isolated from splenocytes of Pan02 tumor-bearing mice treated with the ARG2-based peptide vaccine were assayed with an IFN-γ ELISPOT assay. In this assay, 2.8×105 T cells were plated together with 6×105 antigen-presenting cells (splenocytes from a naïve mouse) with or without ARG2 peptide. Each dot represents one sample (pooled from two mice), and bars represent the mean±SEM. (F) Randomly selected tumors from Pan02 tumor-bearing mice from the experiment (in B) were harvested (n=4–6 per group) on day 31. Tumor RNA was extracted, and bulk RNAseq was performed. Differentially expressed genes (false discovery rate (FDR) <0.05 and absolute log2 fold change >0) in Pan02 tumors from ARG2-vaccinated mice compared with Ctrl vaccinated mice were identified and presented in a volcano plot, n=282 upregulated and n=33 downregulated genes. (G) Immune-related biological processes (Gene Ontology analysis) associated with significantly upregulated genes in RNAseq data (described in F). (H–J) Bar plots showing the total absolute immune infiltration score (arbitrary unit) of all immune populations (H) or the immune score ratio of M1:M2 macrophages (I) and CD8 T cells to Tregs (J). Immune population scores were generated with ImmuCC algorithm, using bulk tumor RNAseq (as described in F). Each dot represents one mouse, and bars represent the mean±SEM. Ctrl, control; IFN-γ, interferon gamma; Treg, regulatory T cell.
jitc-2022-005326supp007.pdf (1.3MB, pdf)
Six ARG2-vaccinated mice and three control-vaccinated mice were randomly selected for ELISPOT analysis on day 31 post tumor inoculation. Here, we observed strong responses in all ARG2-vaccinated mice, and no responses in control-vaccinated mice (figure 6C). The observed immune responses were specific to ARG2 as no cross-reactivity to a peptide derived from the corresponding region of ARG1 was observed, despite partly similar sequences (figure 6D and online supplemental figure S7D). Next, we randomly selected pairs of ARG2-vaccinated mice and prepared pooled splenocyte samples. From these samples, we isolated CD4+ and CD8+ T cells. In ELISPOT assays, we observed responses in both the CD4+ and the CD8+ fractions in all three groups of ARG2-vaccinated mice (figure 6E).
Due to the small tumor sizes, limited amounts of tumor tissue were available. However, homing of ARG2-specific T cells to the tumor of mice vaccinated with the ARG2-based vaccine was shown in three other murine tumor models (online supplemental figure S7E–G), suggesting that the vaccine induces tumor infiltration of ARG2-specific T cells on vaccination. To assess the consequences of inducing ARG2-specific T cells on the transcriptional landscape in the tumor, we isolated whole tumor RNA from four mice treated with the control vaccine and six mice treated with the ARG2-based vaccine and performed RNAseq. Differential gene expression analysis identified a total of 282 upregulated and 33 downregulated genes after vaccination with ARG2 peptide (figure 6F and online supplemental table S4). Gene Ontology analysis based on biological processes showed that within the genes upregulated in mice treated with the ARG2-based vaccine, 44% of the enriched processes were associated with tumor immunology (online supplemental figure S7H and online supplemental table S5). Of these, 76% were associated with positive antitumor immune responses; 10% were associated with negative antitumor immune response and the remaining 14% were associated with immune responses to bacteria (online supplemental figure S7I and online supplemental table S5). Among the significantly enriched biological processes in mice treated with the ARG2-based vaccine, we found processes associated with both innate and adaptive immunity. We observed an enrichment of processes associated with T cells and lymphocytes, type I interferons, cytokine regulation, angiogenesis and immune system processes and calcium ion concentrations, altogether indicating the development of an immune responses in the ARG2-vaccinated mice (figure 6G and online supplemental table S5).
jitc-2022-005326supp013.xlsx (31.4KB, xlsx)
jitc-2022-005326supp014.pdf (77.5KB, pdf)
Intrigued by these observations, we used the ImmuCC algorithm to assess the relative composition of infiltrating immune cell types in the bulk tumor samples (online supplemental tables 6 and 7).45 Overall, we observed a clear trend toward a higher average infiltration of immune cells into the TME on vaccination with the ARG2-based immunomodulatory vaccine (figure 6H). Specifically, we found an increase in the absolute proportion of several immune cell types in mice receiving the ARG2-vaccination, including NK cells, and a tendency toward higher macrophage and monocyte numbers (online supplemental figure S8). Higher average M1:M2 macrophage and CD8:Treg ratios (figure 6I, J) in mice receiving the ARG2-derived peptide vaccine indicated a more proinflammatory TME and suggested an immunomodulatory capacity of ARG2-specific T cells induced on vaccination.
jitc-2022-005326supp008.pdf (1.2MB, pdf)
Discussion
In the current study, we describe the first observation of CD8+ T-cell responses specific to ARG2. We screened a library of 15 peptides that were predicted to bind strongly to HLA-A2. We identified one peptide that elicited CD8+ responses, although this response turned out to be HLA-B8 restricted. ARG2-specific T-cell responses could be detected ex vivo, suggesting that ARG2-specific T cells constitute a natural part of the immune system. We generated highly specific ARG2-targeting T-cell cultures and demonstrated that ARG2-specific T cells were able to recognize and react to relevant target cells, including a cancer cell line derived from metastatic malignant melanoma expressing ARG2. More importantly, we demonstrated that ARG2-specific T cells can recognize and react to normal immune cells with regulatory functions, as we observed preferential targeting of activated Tregs with high ARG2 expression. In addition, we showed that ARG2-based immunomodulatory vaccines induced significant delays in tumor growth in the murine Pan02 tumor model. This effect was associated with strong systemic ARG2-specific CD4+ and CD8+ T-cell responses. Interestingly, RNAseq analysis of Pan02 tumor tissue revealed upregulated immune infiltration along with higher M1:M2 and CD8:Treg ratios in ARG2-vaccinated mice, further underlining the immunodulatory capabilities of ARG2-specific T cells.
The ability of ARG2-specific T cells to recognize and react to Tregs highlights their immunomodulatory function. Moreover, their ability to target regulatory cells supports that ARG2-specific T cells are anti-Tregs, characterized by their ability to ‘regulate the regulators’.47 We previously demonstrated that IDO-specific anti-Tregs specifically lysed IDO-expressing DCs, thereby relieving IDO-mediated immune suppression.25 Moreover, we have shown that targeting of ARG1-expressing myeloid cells by ARG1-specific T cells promotes a shift from a Th2 environment to a Th1 environment due to ARG1-specific T-cell cytokine production.48 It has previously been demonstrated that ARG2 expression is preferentially induced on activation in human Tregs from peripheral blood with an effector memory phenotype, characterized by CD45RO+ expression and that these Tregs can suppress T-cell proliferation in an ARG2-dependent manner.22 The observed preferential expression of ARG2 in activated Tregs compared with the activated bulk culture or activated Trest suggests that ARG2 induction serves as a general active mechanism for increased immunosuppressive capacities of Tregs. This mechanism is thus comparable to immunosuppression exerted by MDSCs due to ARG1 expression.49 Therefore, the targeting of activated Tregs by ARG2-specific T cells, as demonstrated here, could have important immunomodulatory potential by removing the immune suppression exerted by activated Tregs with high ARG2 expression. Many tumors are characterized by high numbers of Tregs, and the targeting of activated immunosuppressive Tregs by ARG2-specific anti-Tregs could potentially relieve Treg-mediated immune suppression on TILs, thereby promoting anticancer immunity. The presence of ARG2-specific T cells in HDs suggests not only that this mechanism is relevant for treatment of malignancy, but also that targeting of Tregs by ARG2-specific T cells could occur as a mechanism for the maintenance of immune homeostasis.
We speculate that the targeting of ARG2-expressing regulatory cells is not limited to Tregs. We have previously reported the ARG2-dependent recognition of DCs,34 and ARG2 is expressed in both human and murine DCs.21 In murine DCs, ARG2 expression is negatively regulated by miRNA-155 expression, and ARG2 overexpression reportedly inhibits T-cell proliferation in vitro and in vivo.50 Therefore, the preferential targeting of ARG2-overexpressing DCs could be another immunomodulatory function of ARG2-specific T cells. Moreover, other immunosuppressive cells of the tumor stroma could be targets for ARG2-specific anti-Treg recognition, thus extending the immunomodulatory functions of ARG2-specific T cells. Indeed, the immunomodulatory function of ARG2-specific T cells was demonstrated in the murine Pan02 tumor model following ARG2-vaccination. Here, gene expression changes in Pan02 tumors from ARG2-vaccinated mice indicated the induction of an antitumor immune response in the form of increased immune cell infiltration and the establishment of a more proinflammatory microenvironment with higher M1:M2 and CD8:Treg ratios. The induction of a more immunopermissive TME on ARG2 vaccination could explain the significant inhibition of Pan02 tumor growth that was observed. Additionally, three other murine tumor models, namely, MC38, B16–F10 and LL2, were used to validate the ability of ARG2-specific T cells to infiltrate the tumor bed. Overall, these results highlight the capability of ARG2-specific T cells to alter the immune landscape in favor of an antitumorigenic immune response. Of note, the immunomodulatory effect of the ARG2-derived peptide vaccine may be mediated by other mechanisms than direct targeting of Tregs in a murine setting, as ARG2 may not be expressed in murine Tregs.22 Recently, our group demonstrated immunomodulatory effects in several murine tumor models with an ARG1-based vaccine. In particular, ARG1 vaccination in combination with a PD-1 blocking antibody also increased the M1:M2 macrophage ratio in the MC38 model.33 Although ARG1 and ARG2 catalyze the same reaction, they are differentially expressed and regulated. As such, vaccinations with both ARG1-derived and ARG2-derived epitopes could potentially provide synergistic effects by targeting different arginase-expressing cells within the tumor.
Combinations of immunomodulatory vaccines with immune checkpoint inhibitors have shown promising results in preclinical murine cancer models where synergy between a PD-1 antibody and IDO or ARG1 vaccines has been demonstrated,33 51 likely by PD-1 blockade promoting anti-Treg function. A similar synergy might exist for the combination of ARG2 vaccine with PD-1 antibody treatment. Moreover, ARG2 expression has been demonstrated in activated Tregs,22 and the RT-qPCR analysis presented in this study showed preferable expression of PD-1 in activated functional Tregs compared with Trest. A recent study suggested the balance of PD-1-expressing CD8+ T cells and PD-1-expressing Tregs to be an important indicator of responsiveness to PD-1 blockade.52 It was demonstrated that in TMEs with Tregs expressing high levels of PD-1, PD-1 blocking antibody therapy led to tumor progression by activation of PD-1+ Tregs. Conversely, if PD-1 was predominantly expressed by CD8+ effector T cells, administration of PD-1 blocking antibodies caused effector cell activation and expansion, leading to tumor regression. Since ARG2-expressing activated Tregs indeed were PD-1+ positive, targeting of Tregs by ARG2-specific T cells not only would remove ARG2+ Tregs but also could lead to a rebalance of the ratio of PD-1+CD8+ T cells to PD-1+Tregs in favor of the former and increase the likelihood of a positive therapeutic response to PD-1 blocking antibodies.
Importantly, the highly specific ARG2-targeting CD8+ T-cell culture from HD93 used in this study showed ARG2 expression that was on par with the levels observed in the sorted Trest cells and substantially lower than in the sorted Tregs (online supplemental figure S4D). In addition, we have been able to successfully sustain long-term cultures of cytotoxic ARG2-specific CD8+ T cells from multiple donors in vitro. The well-documented expression of ARG2 in T cells on activation seems to be restricted to Tregs, which explains the apparent lack of fratricide effects of activated cytotoxic ARG2-specific T cells.53
In IFN-γ ELISPOT, we used cancer cell lines as target cells to identify the HLA-B8 restriction of the A2S05 peptide by specific recognition and cytotoxic activity against the metastatic malignant melanoma cell line FM6 and three other HLA-B8+ cancer cell lines. This was surprising, given that the peptide had been predicted to bind to HLA-A2. Interestingly, it has been proposed that the HLA-B8 haplotype plays a protective role against melanoma, based on the observation of significantly decreased frequency of HLA-B8 in patients with advanced melanoma compared with the frequency in HDs.54 Moreover, in chronic myeloid leukemia (CML), HLA-B8 expression is associated with decreased incidence of CML.55 Given the ARG2 upregulation observed in Tregs and the malignant melanoma cells in metastatic melanoma,22 56 and the immunosuppressive role described for ARG2 in acute myeloid leukemia,19 the HLA-B8 restriction of the described ARG2-peptide could suggest that ARG2-specific T cells play a role in immune surveillance in melanoma and CML.
In conclusion, we have described the existence of ARG2-specific cytotoxic CD8+ anti-Tregs. These ARG2-specific T cells specifically recognized activated Tregs with high ARG2 expression. ARG2 is preferentially expressed by functional Tregs to support their immunosuppressive function. Therefore, ARG2-specific anti-Tregs possess an immunomodulatory capacity through their ability to target regulatory immune cells. In fact, activation of ARG2-specific anti-Tregs through ARG2 vaccination in a murine setting facilitated an immune-mediated antitumor response and had an inhibitory effect on tumor growth in the Pan02 model. Conclusively, the activation of ARG2-specific T cells by, for example, immunomodulatory vaccination constitutes a novel interesting therapeutic possibility for targeting immunosuppressive cells to boost anticancer immune responses in different cancer settings.
jitc-2022-005326supp009.pdf (2.1MB, pdf)
Acknowledgments
We thank Evelina Martinenaite for fruitful discussions and Merete Jonassen for expert technical support.
Footnotes
Twitter: @marper2323, @Majkensiers
SEW-B and TLL contributed equally.
Contributors: Conceptualization: MHA. Methodology: SEWB, TLL and MHA. Investigation: SEWB, TLL, MPP, AS, MLH, MS, MOH, MAJ, and LG. Visualization: SEWB, TLL, and MPP. Provision of reagents: IMS, ÖM, DHM, NØ, MD, and LG. Writing—original draft: SEWB. Writing—review and editing: SEWB, TLL, MPP, and MHA. Final review: SEWB, TLL, MPP, AS, MLH, MS, MOH, MAJ, IMS, ÖM, NØ, DHM, MD, LG, and MHA. Funding acquisition: MHA and TLL. Supervision: MHA. Guarantor: MHA.
Funding: This work was supported by the University of Copenhagen, Herlev Hospital, Tømrermester Jørgen Holm og Hustru Elisa F Hansens Mindelegat (21034), and the Danish Cancer Research Fund (FID2157728).
Competing interests: MHA is named as an inventor on various patent applications relating to the therapeutic use of arginase peptides, including ARG2 peptides, for vaccination. These patent applications have been transferred to the company IO Biotech ApS whose purpose is to develop immunomodulatory vaccines for cancer treatment. MHA is cofounder, shareholder and scientific advisor of IO Biotech ApS. IMS is cofounder, shareholder and clinical advisor of IO Biotech ApS. The remaining authors declare no conflicts of interest.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human subjects, and all patient protocols were approved by the scientific ethics committee for the Capital Region of Denmark and conducted in accordance with the provisions of the Declaration of Helsinki. The subjects gave written informed consent to participate in the study before taking part. Experiments were performed with approval from the Danish Ethics Committee on Experimental Animal Welfare (Dyreforsøgstilsynet).
References
- 1.Zhai L, Spranger S, Binder DC, et al. Molecular pathways: targeting IDO1 and other tryptophan dioxygenases for cancer immunotherapy. Clin Cancer Res 2015;21:5427–33. 10.1158/1078-0432.CCR-15-0420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munder M. Arginase: an emerging key player in the mammalian immune system. Br J Pharmacol 2009;158:638–51. 10.1111/j.1476-5381.2009.00291.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zea AH, Rodriguez PC, Culotta KS, et al. L-Arginine modulates CD3zeta expression and T cell function in activated human T lymphocytes. Cell Immunol 2004;232:21–31. 10.1016/j.cellimm.2005.01.004 [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez PC, Zea AH, DeSalvo J, et al. L-Arginine consumption by macrophages modulates the expression of CD3 zeta chain in T lymphocytes. J Immunol 2003;171:1232–9. 10.4049/jimmunol.171.3.1232 [DOI] [PubMed] [Google Scholar]
- 5.Jenkinson CP, Grody WW, Cederbaum SD. Comparative properties of arginases. Comp Biochem Physiol B Biochem Mol Biol 1996;114:107–32. 10.1016/0305-0491(95)02138-8 [DOI] [PubMed] [Google Scholar]
- 6.Caldwell RB, Toque HA, Narayanan SP, et al. Arginase: an old enzyme with new tricks. Trends Pharmacol Sci 2015;36:395–405. 10.1016/j.tips.2015.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Boniface J, Mao Y, Schmidt-Mende J, et al. Expression patterns of the immunomodulatory enzyme arginase 1 in blood, lymph nodes and tumor tissue of early-stage breast cancer patients. Oncoimmunology 2012;1:1305–12. 10.4161/onci.21678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lang S, Bruderek K, Kaspar C, et al. Clinical relevance and suppressive capacity of human myeloid-derived suppressor cell subsets. Clin Cancer Res 2018;24:4834–44. 10.1158/1078-0432.CCR-17-3726 [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez PC, Ernstoff MS, Hernandez C, et al. Arginase I-producing myeloid-derived suppressor cells in renal cell carcinoma are a subpopulation of activated granulocytes. Cancer Res 2009;69:1553–60. 10.1158/0008-5472.CAN-08-1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rotondo R, Barisione G, Mastracci L, et al. IL-8 induces exocytosis of arginase 1 by neutrophil polymorphonuclears in nonsmall cell lung cancer. Int J Cancer 2009;125:887–93. 10.1002/ijc.24448 [DOI] [PubMed] [Google Scholar]
- 11.Bronte V, Kasic T, Gri G, et al. Boosting antitumor responses of T lymphocytes infiltrating human prostate cancers. J Exp Med 2005;201:1257–68. 10.1084/jem.20042028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ino Y, Yamazaki-Itoh R, Oguro S, et al. Arginase II expressed in cancer-associated fibroblasts indicates tissue hypoxia and predicts poor outcome in patients with pancreatic cancer. PLoS One 2013;8:e55146. 10.1371/journal.pone.0055146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gannon PO, Godin-Ethier J, Hassler M, et al. Androgen-regulated expression of arginase 1, arginase 2 and interleukin-8 in human prostate cancer. PLoS One 2010;5:e12107. 10.1371/journal.pone.0012107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerutti JM, Delcelo R, Amadei MJ, et al. A preoperative diagnostic test that distinguishes benign from malignant thyroid carcinoma based on gene expression. J Clin Invest 2004;113:1234–42. 10.1172/JCI19617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bron L, Jandus C, Andrejevic-Blant S, et al. Prognostic value of arginase-II expression and regulatory T-cell infiltration in head and neck squamous cell carcinoma. Int J Cancer 2013;132:E85–93. 10.1002/ijc.27728 [DOI] [PubMed] [Google Scholar]
- 16.Singh R, Pervin S, Karimi A, et al. Arginase activity in human breast cancer cell lines: N(omega)-hydroxy-L-arginine selectively inhibits cell proliferation and induces apoptosis in MDA-MB-468 cells. Cancer Res 2000;60:3305–12. [PubMed] [Google Scholar]
- 17.Porembska Z, Luboiński G, Chrzanowska A, et al. Arginase in patients with breast cancer. Clin Chim Acta 2003;328:105–11. 10.1016/s0009-8981(02)00391-1 [DOI] [PubMed] [Google Scholar]
- 18.del Ara RM, González-Polo RA, Caro A, et al. Diagnostic performance of arginase activity in colorectal cancer. Clin Exp Med 2002;2:53–7. 10.1007/s102380200007 [DOI] [PubMed] [Google Scholar]
- 19.Mussai F, De Santo C, Abu-Dayyeh I, et al. Acute myeloid leukemia creates an arginase-dependent immunosuppressive microenvironment. Blood 2013;122:749–58. 10.1182/blood-2013-01-480129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mussai F, Egan S, Hunter S, et al. Neuroblastoma arginase activity creates an immunosuppressive microenvironment that impairs autologous and engineered immunity. Cancer Res 2015;75:3043–53. 10.1158/0008-5472.CAN-14-3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grzywa TM, Sosnowska A, Matryba P, et al. Myeloid cell-derived arginase in cancer immune response. Front Immunol 2020;11:1–24. 10.3389/fimmu.2020.00938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lowe MM, Boothby I, Clancy S, et al. Regulatory T cells use arginase 2 to enhance their metabolic fitness in tissues. JCI Insight 2019;4:1–18. 10.1172/jci.insight.129756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giatromanolaki A, Harris AL, Koukourakis MI. The prognostic and therapeutic implications of distinct patterns of argininosuccinate synthase 1 (ASS1) and arginase-2 (Arg2) expression by cancer cells and tumor stroma in non-small-cell lung cancer. Cancer Metab 2021;9:1–10. 10.1186/s40170-021-00264-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsen SK, Munir S, Woetmann A, et al. Functional characterization of Foxp3-specific spontaneous immune responses. Leukemia 2013;27:2332–40. 10.1038/leu.2013.196 [DOI] [PubMed] [Google Scholar]
- 25.Sørensen RB, Berge-Hansen L, Junker N, et al. The immune system strikes back: cellular immune responses against indoleamine 2,3-dioxygenase. PLoS One 2009;4:e6910. 10.1371/journal.pone.0006910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hjortsø MD, Larsen SK, Kongsted P, et al. Tryptophan 2,3-dioxygenase (TDO)-reactive T cells differ in their functional characteristics in health and cancer. Oncoimmunology 2015;4:968480. 10.4161/21624011.2014.968480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munir S, Andersen GH, Svane IM, et al. The immune checkpoint regulator PD-L1 is a specific target for naturally occurring CD4+ T cells. Oncoimmunology 2013;2:e23991. 10.4161/onci.23991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinenaite E, Munir Ahmad S, Hansen M, et al. CCL22-specific T cells: modulating the immunosuppressive tumor microenvironment. Oncoimmunology 2016;5:e1238541. 10.1080/2162402X.2016.1238541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinenaite E, Mortensen REJ, Hansen M, et al. Frequent adaptive immune responses against arginase-1. Oncoimmunology 2018;7:e1404215. 10.1080/2162402X.2017.1404215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmström MO, Mortensen REJ, Pavlidis AM, et al. Cytotoxic T cells isolated from healthy donors and cancer patients kill TGFβ-expressing cancer cells in a TGFβ-dependent manner. Cell Mol Immunol 2021;18:415–26. 10.1038/s41423-020-00593-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andersen MH. Immune regulation by self-recognition: novel possibilities for anticancer immunotherapy. J Natl Cancer Inst 2015;107:djv154–8. 10.1093/jnci/djv154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kjeldsen JW, Lorentzen CL, Martinenaite E, et al. A phase 1/2 trial of an immune-modulatory vaccine against IDO/PD-L1 in combination with nivolumab in metastatic melanoma. Nat Med 2021;27:2212–23. 10.1038/s41591-021-01544-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aaboe Jørgensen M, Ugel S, Linder Hübbe M, et al. Arginase 1-based immune modulatory vaccines induce anticancer immunity and synergize with anti-PD-1 checkpoint blockade. Cancer Immunol Res 2021;9:1316–26. 10.1158/2326-6066.CIR-21-0280 [DOI] [PubMed] [Google Scholar]
- 34.Weis-Banke SE, Hübbe ML, Holmström MO, et al. The metabolic enzyme arginase-2 is a potential target for novel immune modulatory vaccines. Oncoimmunology 2020;9:1771142. 10.1080/2162402X.2020.1771142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ossendorp F, Mengedé E, Camps M, et al. Specific T helper cell requirement for optimal induction of cytotoxic T lymphocytes against major histocompatibility complex class II negative tumors. J Exp Med 1998;187:693–702. 10.1084/jem.187.5.693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rammensee H, Bachmann J, Emmerich NP, et al. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 1999;50:213–9. 10.1007/s002510050595 [DOI] [PubMed] [Google Scholar]
- 37.Andreatta M, Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics 2016;32:511–7. 10.1093/bioinformatics/btv639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nielsen M, Lundegaard C, Worning P, et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci 2003;12:1007–17. 10.1110/ps.0239403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bookout AL, Cummins CL, Mangelsdorf DJ. High-throughput real-time quantitative reverse transcription PCR. Curr Protoc Mol Biol 2006;Chapter 15:15.8.1–15.8.28. 10.1002/0471142727.mb1508s73 [DOI] [PubMed] [Google Scholar]
- 40.Andersen MH, Bonfill JE, Neisig A, et al. Phosphorylated peptides can be transported by TAP molecules, presented by class I MHC molecules, and recognized by phosphopeptide-specific CTL. J Immunol 1999;163:3812 LP–8. [PubMed] [Google Scholar]
- 41.Fjæstad KY, Mette A, Rømer A, et al. Blockade of beta-adrenergic receptors reduces cancer growth and enhances the response to anti-CTLA4 therapy by modulating the tumor microenvironment. Springer US, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liao Y, Smyth GK, Shi W. featureCounts: an efficient General purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–30. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 43.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol 2014;15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Z, Huang A, Sun J, et al. Inference of immune cell composition on the expression profiles of mouse tissue. Sci Rep 2017;7:1–11. 10.1038/srep40508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moodie Z, Price L, Janetzki S, et al. Response determination criteria for ELISPOT: toward a standard that can be applied across laboratories. Methods Mol Biol 2012;792:185–96. 10.1007/978-1-61779-325-7_15 [DOI] [PubMed] [Google Scholar]
- 47.Andersen MH. Anti-regulatory T cells. Semin Immunopathol 2017;39:317–26. 10.1007/s00281-016-0593-x [DOI] [PubMed] [Google Scholar]
- 48.Martinenaite E, Ahmad SM, Bendtsen SK, et al. Arginase-1-based vaccination against the tumor microenvironment: the identification of an optimal T-cell epitope. Cancer Immunol Immunother 2019;68:1901–7. 10.1007/s00262-019-02425-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Umansky V, Blattner C, Fleming V, et al. Myeloid-derived suppressor cells and tumor escape from immune surveillance. Semin Immunopathol 2017;39:295–305. 10.1007/s00281-016-0597-6 [DOI] [PubMed] [Google Scholar]
- 50.Dunand-Sauthier I, Irla M, Carnesecchi S, et al. Repression of arginase-2 expression in dendritic cells by microRNA-155 is critical for promoting T cell proliferation. J Immunol 2014;193:1690–700. 10.4049/jimmunol.1301913 [DOI] [PubMed] [Google Scholar]
- 51.Dey S, Sutanto-Ward E, Kopp KL, et al. Peptide vaccination directed against IDO1-expressing immune cells elicits CD8+ and CD4+ T-cell-mediated antitumor immunity and enhanced anti-PD1 responses. J Immunother Cancer 2020;8:1–14. 10.1136/jitc-2020-000605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumagai S, Togashi Y, Kamada T, et al. The PD-1 expression balance between effector and regulatory T cells predicts the clinical efficacy of PD-1 blockade therapies. 21. Springer US: Nat Immunol, 2020: 1346–58. [DOI] [PubMed] [Google Scholar]
- 53.Geiger R, Rieckmann JC, Wolf T, et al. L-Arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell 2016;167:829–42. 10.1016/j.cell.2016.09.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fensterle J, Trefzer U, Berger T, et al. HLA-B8 association with late-stage melanoma – an immunological lesson? BMC Med 2006;4:1–6. 10.1186/1741-7015-4-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Posthuma EF, Falkenburg JH, Apperley JF, et al. HLA-B8 and HLA-A3 coexpressed with HLA-B8 are associated with a reduced risk of the development of chronic myeloid leukemia. the chronic leukemia Working Party of the EBMT. Blood 1999;93:3863–5. [PubMed] [Google Scholar]
- 56.Yu Y, Ladeiras D, Xiong Y, et al. Arginase-II promotes melanoma migration and adhesion through enhancing hydrogen peroxide production and STAT3 signaling. J Cell Physiol 2020;235:9997–10011. 10.1002/jcp.29814 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-005326supp010.pdf (119.1KB, pdf)
jitc-2022-005326supp006.pdf (1.5MB, pdf)
jitc-2022-005326supp011.pdf (73.7KB, pdf)
jitc-2022-005326supp004.pdf (2.9MB, pdf)
jitc-2022-005326supp005.pdf (2.7MB, pdf)
jitc-2022-005326supp012.pdf (123.2KB, pdf)
jitc-2022-005326supp015.pdf (37.1KB, pdf)
jitc-2022-005326supp016.pdf (108.4KB, pdf)
jitc-2022-005326supp001.pdf (912.3KB, pdf)
jitc-2022-005326supp002.pdf (796.2KB, pdf)
jitc-2022-005326supp003.pdf (1.1MB, pdf)
jitc-2022-005326supp007.pdf (1.3MB, pdf)
jitc-2022-005326supp013.xlsx (31.4KB, xlsx)
jitc-2022-005326supp014.pdf (77.5KB, pdf)
jitc-2022-005326supp008.pdf (1.2MB, pdf)
jitc-2022-005326supp009.pdf (2.1MB, pdf)
Data Availability Statement
Data are available upon reasonable request.