Abstract
Eleutherococcus nodiflorus (Dunn) S. Y. Hu is a momentous medicinal plant belonging to the Araliaceae family. In the current investigation, we determined the complete chloroplast genome of E. nodiflorus and analyzed the phylogenetic relationship among Eleutherococcus plants. The chloroplast genome of E. nodiflorus exhibited a typical quadripartite structure with a full length of 156,770 bp, including 133 genes, containing 88 protein-coding genes, 8 rRNA genes, 37 tRNA genes, and 1 presumed pseudogene (ycf1). The overall GC content observed was 37.95%, with the highest GC content of 43.02% found in the IR region. Comparative genome analysis revealed five highly variable regions among Eleutherococcus species, providing potential markers for further investigations on species identification and population genetics. A total of 44 small simple repeats were identified throughout the chloroplast genome of E. nodiflorus. The phylogenetic analysis indicated a sister relationship between E. nodiflorus and E. eleutheristylus, suggesting a close genetic relationship between the two Eleutherococcus plants. These results enhance the understanding of the plant evolution within Eleutherococcus plants and provide basic genetic resources for the development of species identification and investigation of population genetic diversity of the Eleutherococcus genus and Araliaceae.
Keywords: Eleutherococcus nodiflorus, complete chloroplast genome, diversity regions, phylogenetic analysis, Eleutherococcus genus
Eleutherococcus nodiflorus (Dunn) S. Y. Hu 1980 is a medicinal plant belonging to the Eleutherococcus genus of the Araliaceae family. The Eleutherococcus genus was comprised of more than 40 shrub species, which were mainly distributed in eastern Asia. The dried roots and barks of E. nodiflorus were used as one of the sources of Chinese medicine Acanthopanacis cortex for expelling wind and dampness, nourishing the liver and kidney, etc. Acanthopanacis cortex was also reported to exhibit anti-aging activities, regulating blood pressure, and lowering blood sugar (Yang et al. 2020). The Acankoreanogenin A from the leaves of E. nodiflorus inhibited the release of pro-inflammatory cytokine HMGB1, revealing the potential of a candidate therapy for fulminant hepatitis (Zhang et al. 2011). Due to their similar morphological characteristics, the Acanthopanacis cortex is frequently mistaken with root barks from closely related Eleutherococcus species (Wang et al. 2005). The adulterants of Acanthopanacis cortex significantly affected its clinical efficiency and brought potential safety issues (Liang et al. 2014). The complete chloroplast (cp) genomes present a powerful molecular strategy for species identification and phylogenetic relationship investigation at the general and tribe levels (Dong et al. 2021; Zhou et al. 2021). Therefore, determining the complete cp genome of E. nodiflorus is fascinating and necessary to contribute to plant authentication and taxonomic classification of the genus Eleutherococcus.
Fresh leaf samples of Eleutherococcus nodiflorus were collected from the Zhejiang Chinese Medical University’s Botanical Garden of Medicinal Plants in the Fuyang area (30°05′8″ N, 119°52′53″ E). The leaf specimen was submitted at the Medicinal Herbarium Center of the Zhejiang Chinese Medical University (https://yxy.zcmu.edu.cn; Herbarium Code: MHCZCMU; Collector: Rubin Cheng, biothcheng@hotmail.com) under the voucher number XZWJ-20200713. The extracted total genomic DNA was subjected to sequencing using the Illumina Hiseq Platform following our previous reports (He et al. 2021; Wang et al. 2021). To obtain the chloroplast genome of E. nodiflorus, the raw reads were trimmed using Trimmomatic and assembled the resulting cleaned reads using metaSPAdes 3.13.0 along with the cp genome of Eleutherococcus brachypus (NC_050832) as the reference (Bolger et al. 2014; Nurk et al. 2017). The chloroplast genome of E. nodiflorus was annotated with GeSeq and further confirmation was done using BLAST (Tillich et al. 2017). The complete cp genome of E. nodiflorus was submitted to GenBank database under accession number MZ362512.
The full-length cp genome of E. nodiflorus was 156,770 bp long, with the conserved quadripartite structure consisting of a LSC region (86,710 bp), an SSC region (18,174 bp), and two IR regions of 25,943 bp each. A total of 133 genes were identified in the cp genome of E. nodiflorus, containing 88 protein-coding genes, 8 rRNAs, 37 tRNAs, and 1 pseudogene (ycf1). It also contained 17 repetitive genes in the IR region, including 7 tRNAs, 4 rRNAs, and 6 protein-coding genes. The most frequently used amino acid in E. nodiflorus was Leucine (Leu) (10.58%), followed by Isoleucine (Ile) (8.40%), Serine (Ser) (7.74%), and Glycine (Gly) (6.99%). The tRNA genes in E. nodiflorus varied in length between 48 and 93 nucleotides, with the GC content ranging from 41.09% to 62.16%. The comparative genome analysis of Eleutherococcus species revealed five diverse regions among the cp genomes with Pi values higher than 0.01, providing potential markers for species identification and phylogenetic investigations of the Eleutherococcus genus (Supplementary Material Fig. S1). The clpP1 was the highest variable gene identified, locating in the LSC region with the Pi value of 0.01867. We also determined 44 small simple repeats in the cp genome of E. nodiflorus with length ranging from 10 to 17 bp.
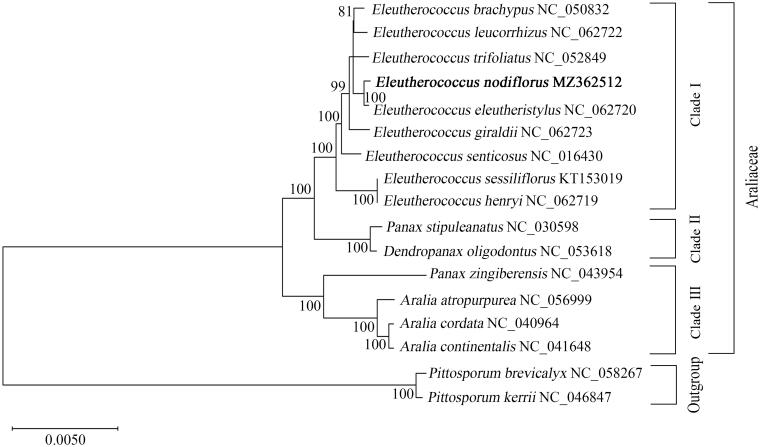
To further explore the phylogenetic relationship of Eleutherococcus, the ML tree was constructed using MEGA X. The tree results indicated that E. nodiflorus clustered with E. eleutheristylus with high support values, suggesting a relatively close genetic relationship between the two Eleutherococcus plants (Figure 1). In addition, a core group of five Eleutherococcus plants (E. brachypus, E. leucorrhzus, E. trifoliatus, E. nodiflorus, and E. eleutheristylus) was observed in the ML tree, providing insights for the taxonomic revision in Eleutherococcus genus (Figure 1). The nine Eleutherococcus species combined together to form a monophyletic group, which displayed a sister relationship with the group of the Panax and Dendropanax. However, P. stipuleanatus and P. zingiberensis were divided into two clades in the ML tree, indicating the requirement for further investigations and revisions for the Panax genus. These findings provide the basic cp genome information of E. nodiflorus, which would contribute to the development of species identification strategies in the future and promote the investigations of population genetics and evolutionary relationships within Eleutherococcus and Araliaceae plants.
Figure 1.
Phylogenetic relationship between newly sequenced Eleutherococcus nodiflorus and other representative species of Araliaceae based on complete chloroplast genome analysis. The tree was generated with 71 protein-coding genes of E. nodiflorus including the closely related plants in the Eleutherococcus and Araliaceae using the maximum likelihood (ML) method by MEGA X. The ML tree was analyzed based on the Kimura 2-parameter model. The Pittosporum brevicalyx and Pittosporum kerrii were chosen as the outgroup. The newly identified genome of Eleutherococcus nodiflorus was represented in bold. Numbers on nodes represented bootstrap values for 81, 99, and 100 replicates in the ML analysis. The GenBank accession number was listed after the species name.
Supplementary Material
Funding Statement
This work was supported by the Research Project of Zhejiang Chinese Medical University [2021JKZKTS020B] and Project of Quality Guarantee System of Chinese Herbal Medicines [Grant No. 201507002-4].
Ethics approval
Since the Project of Quality Guarantee System of Chinese Herbal Medicines, Dr. Rubin Cheng has obtained the permission by Zhejiang Chinese Medical University to collect plant samples. Because of the important medical value of E. nodiflorus, Dr. Cheng collected the specimen of E. nodiflorus for further molecular study. The plant material collection and experimental research were conducted according to the Plant Protection and Regulation of Zhejiang Chinese Medical University.
Author contributions
ML and RC conceived and designed the experiments. QG and QL contributed to the resource sampling and species identification. ML, QG and MZ performed the experiments. ML, MZ and QL analyzed the data and wrote the paper. RC revised and approved the final version of the paper. All authors have reviewed and approved the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession number MZ362512. The associated BioProject, SRA, and BioSample numbers of E. nodiflorus are PRJNA808818, SRR18086324 and SAMN26107482, respectively.
References
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, Ying Z, Yu S, Wang Q, Liao G, Ge Y, Cheng R.. 2021. Complete chloroplast genome of Stephania tetrandra (Menispermaceae) from Zhejiang Province: insights into molecular structures, comparative genome analysis, mutational hotspots and phylogenetic relationships. BMC Genomics. 22(1):880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zheng Z, Wang Q, Zhou M, Liao G, Ge Y, Cheng R.. 2021. Complete chloroplast genome sequence of the medicinal plant ramie (Boehmeria nivea L. gaud) and its phylogenetic relationship to other Urticaceae species. Mitochondrial DNA B Resour. 6(3):1136–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Deng F, Xing H, Wen H, Shi X, Martey ON, Koomson E, He X.. 2014. P-glycoprotein- and organic anion-transporting polypeptide-mediated transport of periplocin may lead to drug-herb/drug-drug interactions. Drug Des Devel Ther. 8(8):475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA.. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S.. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Huang Z, Gao C, Ge Y, Cheng R.. 2021. The complete chloroplast genome sequence of Rubus hirsutus Thunb. and a comparative analysis within Rubus species. Genetica. 149(5–6):299–311. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhang L, Sun Y.. 2005. Semipreparative separation and determination of eleutheroside E in Acanthopanax giraldii Harms by highperformance liquid chromatography. J. Chromatogr. Sci. 43(5):249–252. [DOI] [PubMed] [Google Scholar]
- Yang JB, Cai W, Li MH, Li NX, Ma SC, Cheng XL, Wei F.. 2020. Progress in chemical and pharmacological research of Acanthopanax gracilistylus. Modern Med J China. 22(04):652–662. [Google Scholar]
- Zhang BX, Li N, Zhang ZP, Liu HB, Zhou RR, Zhong BY, Zou MX, Dai XH, Xiao MF, Liu XQ, et al. 2011. Protective effect of Acanthopanax gracilistylus-extracted Acankoreanogenin A on mice with fulminant hepatitis. Int Immunopharmacol. 11(8):1018–1023. [DOI] [PubMed] [Google Scholar]
- Zhou M, Yan M, Ying Z, He X, Ge Y, Cheng R.. 2021. Characterization of the complete chloroplast genome of Oxalis corymbosa DC. (Oxalidaceae), a medicinal plant from Zhejiang Province. Mitochondrial DNA B Resour. 6(3):1138–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession number MZ362512. The associated BioProject, SRA, and BioSample numbers of E. nodiflorus are PRJNA808818, SRR18086324 and SAMN26107482, respectively.