Abstract
Apical organs are relatively simple larval nervous systems. The extent to which apical organs are evolutionarily related to the more complex nervous systems of other animals remains unclear. To identify common developmental mechanisms, we analyzed the gene regulatory network (GRN) controlling the development of the apical organ in sea urchins. We characterized the developmental expression of 30 transcription factors and identified key regulatory functions for FoxQ2, Hbn, Delta/Notch signaling, and SoxC in the patterning of the apical organ and the specification of neurons. Almost the entire set of apical transcription factors is expressed in the nervous system of worms, flies, zebrafish, frogs, and mice. Furthermore, a regulatory module controlling the axial patterning of the vertebrate brain is expressed in the ectoderm of sea urchin embryos. We conclude that GRNs controlling the formation of bilaterian nervous systems share a common origin and that the apical GRN likely resembles an ancestral regulatory program.
Transcription factors controlling formation of apical organs are expressed in nervous systems throughout Bilateria.
INTRODUCTION
Many theories have been developed over the years to reconstruct the evolutionary relationship between nervous systems of vastly different morphology and function in attempts to explain the evolution of the nervous system. One of them, proposed over a century ago, postulates that the last common ancestor of echinoderms, hemichordates, and chordates might have resembled in appearance the marine larvae of extant echinoderms (1–3). According to this theory, the central nervous system of chordates would have evolved from a common ancestor with an apical organ, which gave rise to the anterior nervous system and brain, and a ciliated band nervous system, which fused to form the neural tube. More recent comparisons based on molecular gene expression data, however, seem to offer little support for this theory (1, 4–6). Thus, there is a remarkable similarity of regulatory modules controlling the regionalization of the central nervous system along the anterior-posterior and mediolateral axes in vertebrates, arthropods, and annelids, raising the possibility of a bilaterian ancestor that already had a relatively complex regionalized nervous system (4–6). However, the absence of complexity and regionalization in the nervous system of many bilaterian clades challenges this possibility and would have to be the result of secondary losses if their ancestors had a relatively complex and partitioned nervous system (7, 8). An alternative explanation is that the conservation of axial patterning mechanisms might have occurred for reasons other than the regionalization of the brain and that these mechanisms might therefore be less suitable for understanding the evolution of the nervous system. Further insights into developmental mechanisms that control the formation of the nervous system in different clades are therefore necessary for reconstructing the evolutionary history of the nervous system.
Here, we analyzed the gene regulatory network (GRN) controlling the development of the apical organ in sea urchin embryos to identify developmental mechanisms that are used in echinoderms and that serve as a basis for evolutionary comparisons. GRNs orchestrate developmental processes including axial patterning, cell fate specification, morphogenesis, cell proliferation, and many other aspects of development (9). The comparison of developmental GRNs operating in different animals provides a way to assess the evolutionary relationship of developmental programs across considerable evolutionary distances because it is unlikely that extensive similarity in transcription factor components or regulatory circuits has evolved entirely independently. It should be noted however that even homologous GRNs may have further evolved by the co-option of additional regulatory factors and by the rewiring of regulatory circuits and thus have acquired the capacity to regulate the formation of novel morphological structures (10, 11). The promise of GRN comparisons is to reveal where transcription factor co-options and novel regulatory circuits have contributed to the evolution of novel features in the animal body plan.
Echinoderm larvae have a centralized nervous system, with neurons located either in the apical organ, which corresponds to the anterior neurogenic ectoderm, or in the ciliary band ectoderm (12, 13). Apical organs are present in several animal phyla including Cnidaria, ctenophores, annelids, and echinoderms (13, 14). The similarities between apical organs in various cnidarian and bilaterian species indicate that apical organs are homologous (14). As sister groups of chordates, echinoderms and hemichordates are in a phylogenetically important position to reveal insights into the evolution of the nervous system in early deuterostomes.
In the purple sea urchin Strongylocentrotus purpuratus, the apical organ of pluteus larvae includes four to six serotonergic neurons in addition to other neurons and other non-neural cell types (15). Most neurons in the apical organ and ciliary band are specified during the development of the larva after the onset of gastrulation. However, the precursors of serotonergic neurons form exclusively within the apical organ and are distinctly specified before the onset of gastrulation, indicating that the patterning of the apical organ occurs early during sea urchin development (16). Several developmental mechanisms contribute to the initial specification of the apical neurogenic ectoderm. Thus, SoxB transcription factors are responsible for initially activating the expression of foxq2 in the neurogenic ectoderm (17, 18). The anterior localization of the apical organ is defined by antagonism to Wnt signaling from the vegetal pole, which restricts the expression of the forkhead transcription factor (FoxQ2) and the specification of the apical neurogenic ectoderm to the animal pole (19–21). The boundaries of the apical neurogenic ectoderm are further defined by repressive cross-interactions between FoxQ2 and Emx and by expression of Wnt signaling antagonists downstream of FoxQ2 (20, 22). Additional regulatory mechanisms were identified, which control neurogenesis not only in the apical organ but also throughout the sea urchin larva, including Delta/Notch signaling, SoxC, and Brn1/2/4 (16, 18, 23, 24). However, developmental mechanisms that contribute to the development of the apical organ after definition of its progenitor field and that lead to the specification of serotonergic neurons and other apical cell fates are so far not well resolved.
In this study, we analyzed the GRN that controls the formation of the apical organ and the specification of serotonergic neurons before gastrulation. We determined the spatial and temporal expression of 31 transcription factors and signaling ligands during this process and performed perturbation experiments to test the function of key nodes. The results show that FoxQ2 and Hbn function upstream in the specification of apical cell fates and in the patterning of the apical organ. Both transcription factors together activate the expression of SoxC in the apical neurogenic ectoderm, which is subsequently restricted to neuronal precursors by Delta/Notch signaling. SoxC in turn activates the expression of several proneural transcription factors to specify neural precursors. A comparative analysis indicates that the orthologs of most transcription factors constituting the sea urchin apical GRN are expressed in the nervous system throughout Bilateria. In addition, transcription factors patterning the anterior-posterior axis of the nervous system in flies and mice are expressed in the neural and non-neural ectoderm of sea urchin embryos, indicating that this regulatory module is conserved in echinoderms even in the absence of a complex partitioned nervous system. These results suggest a common origin of developmental programs that control the formation of apical organs and more complex brains despite their differences in morphology and function. Given the absence of co-options in the apical GRN, the ancestral form of anterior nervous system might resemble the apical organs of extant echinoderm larvae, as proposed many years ago.
RESULTS
Identification of transcription factors that constitute the apical GRN
Previous systematic gene expression analyses identified 48 regulatory genes that were annotated as being expressed in the apical organ at some point during the first 3 days of sea urchin development (table S1) (15, 16, 22, 25–30). To identify transcription factors that potentially contribute to the pregastrular development of the apical organ, we analyzed the time-course expression levels of these candidate genes up to the onset of gastrulation at 30-hour postfertilization (hpf) by NanoString nCounter technology (31). Because only two to four serotonergic neurons are specified at this early stage in the apical organ, even expression levels as low as 60 mRNA molecules per embryo may result in functional levels of transcription factors. Using this threshold, we identified 43 regulatory genes that are expressed during pregastrular development, whereas five genes were expressed at levels too low to be functionally relevant and were excluded from further analyses (fig. S1A). The spatial expression of the 43 regulatory genes was further analyzed by whole-mount in situ hybridization (WMISH), showing that 31 regulatory genes are expressed in the apical neurogenic ectoderm just before gastrulation, while 12 regulatory genes are expressed elsewhere in the embryo at this stage (fig. S1B). These results show that the GRN controlling the pregastrular development of the apical organ includes at least 30 transcription factors in addition to the signaling ligand Delta.
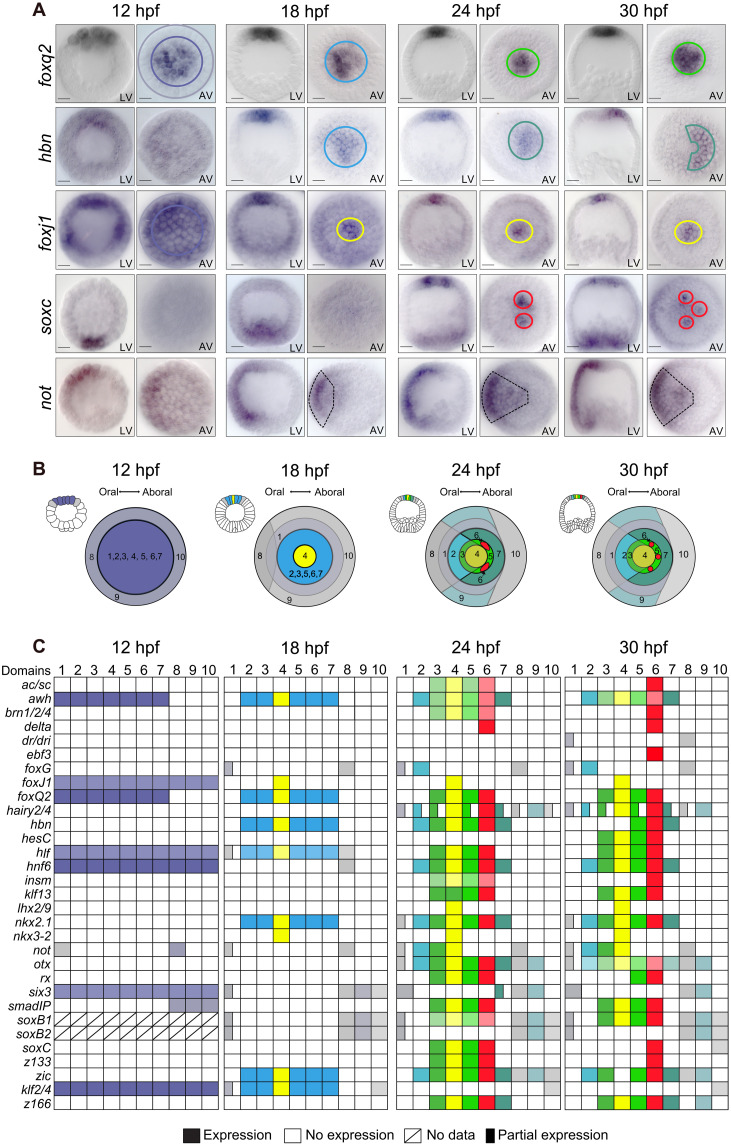
On the basis of the spatial expression patterns of these transcription factors, the apical organ includes several cell fates by 30 hpf that are organized into concentric domains, as summarized in Fig. 1. In the center of the apical organ, a central apical tuft region is marked by expression of foxj1 and nkx3-2 [Fig. 1, A (yellow circle) to C; and fig. S2]. A larger domain of the apical plate that overlaps with the tuft domain expresses 12 regulatory genes including foxq2 and nkx2.1 (Fig. 1A, green circle). Several regulatory genes are expressed in the apical organ and in surrounding ectodermal cells, including otx, zic, and soxb1 (Fig. 1B, gray domains). The apical organ shows an oral/aboral asymmetry, as indicated by expression of hbn on the aboral side (Fig. 1, A and B, blue-green half circle), and of not, six3, and foxg on the oral side [Fig. 1, A (dashed line) and B (light blue and gray oral quadrant)]. Furthermore, a few individual cells within the aboral apical organ specifically express several proneural regulatory genes including soxc, achaete-scute (ac/sc), delta, and brn1/2/4 (Fig. 1, A and B, red circles). These cells correspond to the precursors of serotonergic neurons that, in addition, express 15 transcription factors that are present more broadly in the apical organ (Fig. 1C). These results confirm that extensive patterning of the apical organ and the specification of the serotonergic neural precursors occurs before gastrulation.
Fig. 1. Developmental expression of regulatory genes in the apical neural ectoderm.
(A) Spatial gene expression of selected regulatory genes between 12 and 30 hpf indicating the specification of distinct cell fate domains in the apical neural ectoderm. Distinct cell fate domains are indicated by colored circles. Complete dataset available in fig. S2. (B) Diagram of embryos summarizing the apical cell fate domains expressing distinct regulatory states, with small lateral schemes indicating the position of the apical plate within embryos and large schemes indicating apical views. Color code as in (A). (C) Boolean expression matrix summarizing the spatial expression of all analyzed regulatory genes at indicated time points. Numbered columns indicate the regulatory state for each cell fate domain shown in matched color in (B). LV, lateral view; AV, apical view. Scale bars, 20 μm.
Dynamic specification of cell fates during development of the apical organ
Neurogenesis typically starts with the specification of a broad neurogenic ectoderm domain in which, as development proceeds, a subset of cells acquire neural cell fate identity and differentiate into neural cell types (32). To determine the progression of patterning and cell fate specification in the developing apical organ and to identify transcription factors that contribute to this process, we analyzed the expression of the apical transcription factors during earlier development of the apical organ. For the 31 regulatory genes, spatial expression was analyzed from early specification of apical neurogenic ectoderm up to gastrulation (12 to 30 hpf) at 3-hour time intervals by WMISH (Fig. 1A and fig. S2, A to E). For simplification, the resulting spatial gene expression data are summarized in a Boolean expression matrix (Fig. 1, B and C). These data indicate that several important regulatory transitions occur during the specification of the apical organ.
The initial specification of apical neurogenic ectoderm as distinct from other ectodermal cell fates occurs at 12 hpf, when foxq2 expression becomes restricted to the animal pole domain (Fig. 1A) (21, 22). In addition to foxq2, six other regulatory genes are expressed at this early stage throughout the apical neurogenic ectoderm, including foxj1, arrowhead homolog (awh), and six3, in addition to soxb1, which is broadly expressed throughout the ectoderm (Fig. 1C and fig. S2). A few hours later, expression of foxj1 becomes restricted and, by 18 hpf, specifies a central tuft domain, while expression of six3 is excluded from the apical domain (Fig. 1A). Cells of the apical neurogenic ectoderm are otherwise uniformly specified by expression of seven regulatory genes, including foxq2, nkx2.1, and hbn.
Extensive patterning of the apical organ occurs between 18 and 24 hpf, essentially establishing most apical cell fates specified before gastrulation. This process coincides with the activation of 15 additional regulatory genes in the apical neurogenic ectoderm (Fig. 1C and fig. S2). Thus by 24 hpf, the three concentric domains of the apical organ are specified: the central apical domain marked by foxj1 and nkx3-2; a broader apical domain expressing foxq2, nkx2.1, rx, and awh; and cells surrounding these two domains that express otx, six3, and zic (Fig. 1C and fig. S2). The origin of the oral-aboral asymmetry in the apical organ is provided by expression of not, hnf6, and foxg in the oral quadrant, which, at this stage, intersects all three concentric apical domains (Fig. 1, A and B, and fig. S2).
The specification of serotonergic neurons during development of the apical organ is indicated by the expression of SoxC, a transcription factor that is expressed in neurons throughout the sea urchin larva and required for neurogenesis (16, 18). Apical neural progenitors are first detected at 21 hpf when soxc expression occurs in clusters of cells located on the aboral side of the apical domain (Fig. 1A and fig. S2F). Within a few hours, expression of soxc becomes restricted to two to three individual cells, the precursors of serotonergic neurons (Fig. 1A and fig. S2). This transition in the spatial expression of soxc occurs at a time when several proneural transcription factors start being expressed in apical neural precursors, including insulinoma-associated protein (Insm), early B-cell factor 3 (EBF3), Brain 1/2/4 (Brn1/2/4), and Achaete-Scute (Ac/sc) (Fig. 1C and fig. S2). Soxc is therefore one of the earliest proneural regulatory genes expressed during development of the apical organ.
These results show that the sea urchin apical organ develops from the early specification of neurogenic ectoderm to the expression of proneural transcription factors in serotonergic neural precursors within just about 18 hours before the onset of gastrulation. Several transcription factors are expressed in key positions within this process and are likely to control the specification of neurons and other cell fates in the apical organ.
FoxQ2 controls specification of various cell fates in the apical organ
FoxQ2 is one of the earliest transcription factors expressed in the apical neurogenic ectoderm, together with Six3 (Fig. 2A and fig. S2) (22, 28, 29). Previous studies showed that the perturbation of Six3 affects the expression of many transcription factors within the apical organ (29). However, expression of six3 clears from the apical neurogenic ectoderm by 15 hpf, and Six3 is therefore unlikely to function as a direct regulator of gene expression in the apical organ beyond very early stages of development. Expression of foxq2 on the other hand is exclusively detected in the apical neurogenic ectoderm throughout embryogenesis, beyond the onset of gastrulation (28). Although several mechanisms have been identified that control the earliest expression of FoxQ2 in the apical neurogenic ectoderm (21, 22), its function during apical development is not clear.
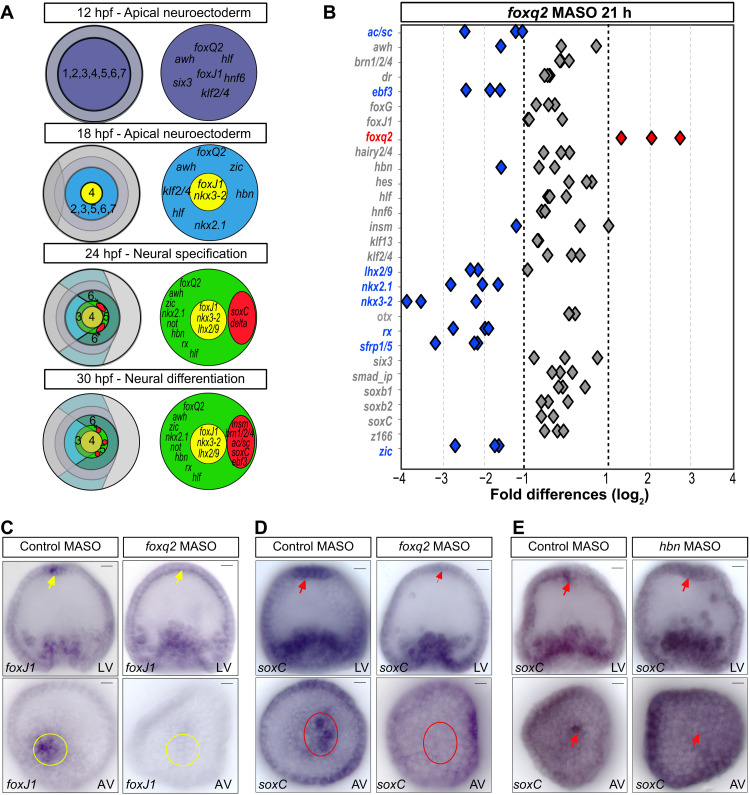
Fig. 2. Specification of neural progenitors downstream of FoxQ2 and Hbn.
(A) Summary of developmental expression of regulatory genes in apical cell fate domains, suggesting a regulatory hierarchy of neuronal specification. (B) NanoString analysis of regulatory gene expression in embryos injected with foxq2 morpholinos or control morpholinos, shown as fold differences in expression levels at 21 hpf. Gene expression of apical genes is indicated as unaffected (gray), up-regulated (red), or down-regulated (blue) by injection of foxq2 morpholinos compared to control. (C and D) Spatial expression of foxJ1 (C) and soxC (D) in embryos injected with control morpholinos and foxQ2 morpholinos and analyzed by WMISH at 30 hpf. (E) Spatial expression of soxc in embryos injected with control morpholinos and hbn morpholinos and analyzed by WMISH at 30 hpf. MASO, morpholino antisense oligonucleotides. Scale bars, 20 μm.
To determine the function of FoxQ2 in the development of the apical organ and in the specification of neural precursors, we perturbed the expression of FoxQ2 using morpholinos. Sea urchin embryos were injected at fertilization with foxq2 morpholinos or random control morpholinos, and the expression of apical regulatory genes was analyzed at 21 and 30 hpf using NanoString nCounter analysis. A probe set was used including 29 of the 31 apical regulatory genes, all except z133/fez and delta. The results show that FoxQ2 affects the expression of apical regulatory genes both at 21 and 30 hpf (Fig. 2B and fig. S3). Consistent with previous results, expression of the Wnt signaling antagonist sfrp1/5 was down-regulated and expression of foxq2 was up-regulated in embryos injected with foxq2 morpholinos (20, 22). The perturbation of FoxQ2 also affected the expression of several additional regulatory genes, including genes broadly expressed in the apical neural ectoderm (nkx2.1, zic, and rx), genes expressed in the central apical domain (nkx3-2 and lhx2-9), and genes specifically expressed in apical neural precursors (ac/sc and ebf3) (Fig. 2B). FoxQ2 is therefore required for the expression of transcription factors that are associated with several distinct cell fates within the apical organ.
If FoxQ2 is responsible for controlling the specification of different apical cell fates, then it should, particularly, regulate the expression of transcription factors that first define apical cell fate domains during development. However, the perturbation of FoxQ2 showed no effect on the expression levels of two transcription factors that mark the initial specification of their respective domains, foxj1 in the central apical domain and soxc in neural precursors. We thus examined the expression of these two genes by WMISH in embryos with perturbed FoxQ2 expression. While foxj1 was expressed in the central apical domain in embryos injected with control morpholinos, expression was not detectable in embryos injected with foxq2 morpholinos (Fig. 2C). Similarly, expression of soxc was specifically abolished in the apical neural precursors upon perturbation of FoxQ2, while soxc expression was not affected in embryos injected with control morpholinos (Fig. 2D). These results show that FoxQ2 functions upstream in the apical GRN, controlling the expression of transcription factors that pattern the apical organ and that drive the specification of apical cell fates including neural precursors.
Hbn activates soxc expression in aboral apical neurogenic ectoderm
In sea urchins, serotonergic neurons form exclusively on the aboral side of the apical organ. Because FoxQ2 is expressed evenly throughout the apical organ, other transcription factors must be responsible for the localized activation of soxc expression in the aboral apical ectoderm. The expression of FoxQ2 also precedes expression of soxc by several hours, further indicating that FoxQ2 alone is not sufficient to activate soxc expression. Three regulatory genes start being expressed between 12 and 18 hpf in the apical ectoderm just before the onset of soxc expression. Nkx2.1 and zic are broadly expressed in the apical organ and in surrounding ectoderm. Hbn on the other hand is only initially broadly expressed in the apical domain and becomes restricted to the aboral apical ectoderm after 21 hpf, when neural precursors are specified (fig. S2B). To evaluate the function of Hbn in the initial specification of apical neural precursors, embryos were injected with hbn morpholinos, and expression of soxc was analyzed by WMISH at 30 hpf. While soxc expression was detectable in neural precursors in embryos injected with control morpholinos, perturbation of Hbn abolished the expression of soxc in apical neural precursors (Fig. 2E). FoxQ2 and Hbn are therefore both necessary for the expression of soxc at the aboral boundary of the apical domain. This is consistent with a previous study showing that Hbn and FoxQ2 are required for the specification of serotonergic neurons in a related sea urchin species, Hemicentrotus pulcherrimus (33). The results here indicate that FoxQ2 and Hbn constitute an AND logic gate that regulates the expression of SoxC in apical neural precursors and thereby, although possibly indirectly and in combination with other regulators, controls the specification of serotonergic neurons.
Delta/Notch signaling restricts the specification of apical neurons by regulating soxc
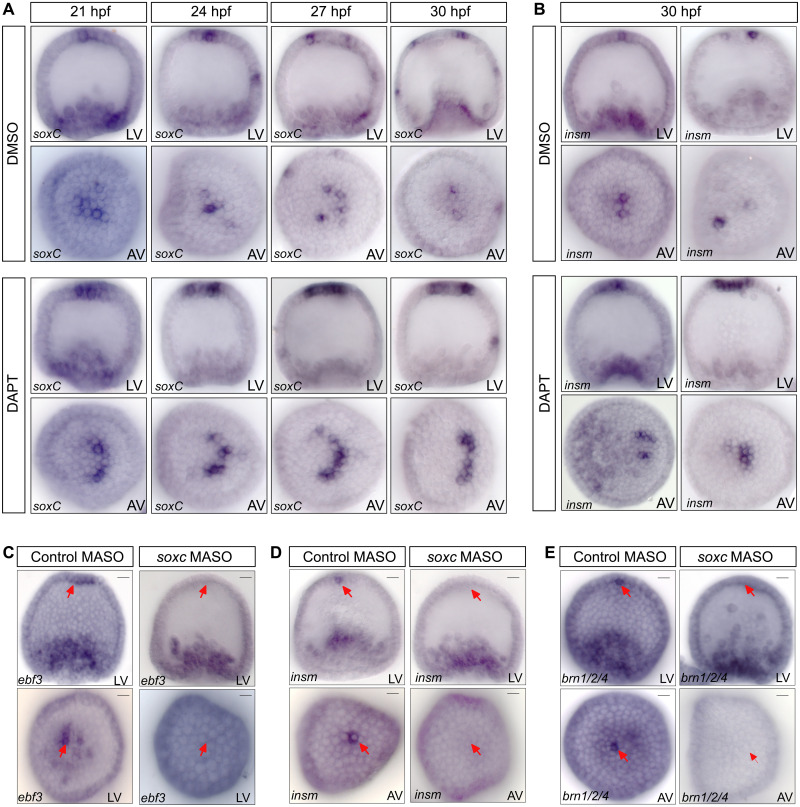
By the onset of gastrulation, the precursors of serotonergic neurons are dispersed within the aboral apical ectoderm, as shown by expression of soxc (Fig. 1A). Initially however, the expression of soxc is activated more broadly in the apical neurogenic ectoderm at 21 to 24 hpf (Fig. 1A and fig. S2D). The restriction of soxc expression thus occurs after the initial activation to specify a few individual cells that give rise to the serotonergic neurons. Although FoxQ2 and Hbn are responsible for the activation of soxc expression in the apical domain, other mechanisms must be involved in this dynamic change of the soxc expression pattern. Previous studies show that the perturbation of Delta/Notch signaling leads to an increased number of neurons throughout the sea urchin larva (23, 30). To test whether Delta/Notch signaling is responsible for controlling the developmental restriction of soxc expression to neural precursors, Delta/Notch signaling was blocked using the γ-secretase inhibitor DAPT (N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester) (34, 35). To monitor the formation of apical neural precursors, the expression of soxc and insm was analyzed by WMISH in embryos treated with DAPT or with dimethyl sulfoxide (DMSO) for control.
A developmental time-course analysis of soxc expression between 21 and 30 hpf shows that soxc expression changes from cell clusters to individual neural precursors by 27 hpf in DMSO-treated control embryos. However, in embryos with perturbed Delta/Notch signaling, soxc expression remains unchanged during the same time period and is detected in clusters of cells in the aboral apical neural ectoderm (Fig. 3A). Thus, at 30 hpf, the expression of soxc and insm occurs exclusively in single cells in the apical domain of DMSO-treated control embryos, while expression is more broadly distributed in the absence of Delta/Notch signaling (Fig. 3, A and B). The role of Delta/Notch signaling in the restriction of soxc expression is consistent with the onset of delta expression in the apical domain at 24 hpf (fig. S2A). Thus, Delta/Notch signaling among apical neural progenitors inhibits the expression of soxc in non-neural cells and restricts the continued specification of neural cell fates to individual precursors of serotonergic neurons.
Fig. 3. Function of Delta/Notch signaling and SoxC in apical neurogenesis.
(A) Soxc expression was analyzed by WMISH in embryos treated with the Delta/Notch signaling inhibitor DAPT or with DMSO for control. Delta/Notch signaling is required for restriction of soxc expression to individual neural precursors. (B) WMISH detecting expression of insm in embryos treated with DAPT or DMSO for control at 30 hpf. (C to E) WMISH showing expression of ebf3 (C), insm (D), and brn1/2/4 (E) in embryos injected with soxc morpholinos or control morpholinos and analyzed at 30hpf, showing that SoxC is required for the expression of all three genes in neural precursors. MASO, morpholino antisense oligonucleotides. Scale bars, 20 μm.
SoxC activates expression of proneural transcription factors in neural precursors
The tight combinatorial regulation of soxc expression downstream of FoxQ2, Hbn, and Delta/Notch signaling and its early activation in neural progenitors indicate that SoxC functions as a key node in the specification of serotonergic neurons. SoxC is broadly required for the formation of neurons in sea urchin embryos, including the apical serotonergic neurons (16, 24). On the basis of the expression data in fig. S2, SoxC is one of the first transcription factors specifically expressed in the apical neural progenitors (Fig. 1 and fig. S2D). The expression of several transcription factors is activated in apical neural precursors shortly after the restriction of soxc expression to neural precursors. The spatial expression data show that ac/sc, brn1/2/4, ebf3, insm, and z133 are expressed in a few single cells in the aboral apical organ at 30 hpf (Fig. 1C and fig. S2). To determine whether SoxC is required for the activation of proneural regulatory genes in apical neural precursors, expression of SoxC was perturbed using morpholinos and expression of brn1/2/4, ebf3, and insm was analyzed at 30 hpf by WMISH. In embryos injected with control morpholinos, the expression of these proneural regulatory genes was observed in apical neural precursors, while expression was not detectable in embryos injected with soxc morpholinos (Fig. 3, C to E). These results indicate that SoxC controls the specification of serotonergic neurons by activating the expression of several proneural transcription factors, possibly together with FoxQ2. The regulation of soxc expression by the upstream apical GRN therefore provides a link between early specification of neurogenic ectoderm and the specification of neural precursors in the apical organ.
Orthologs of apical transcription factors are commonly expressed in bilaterian nervous systems
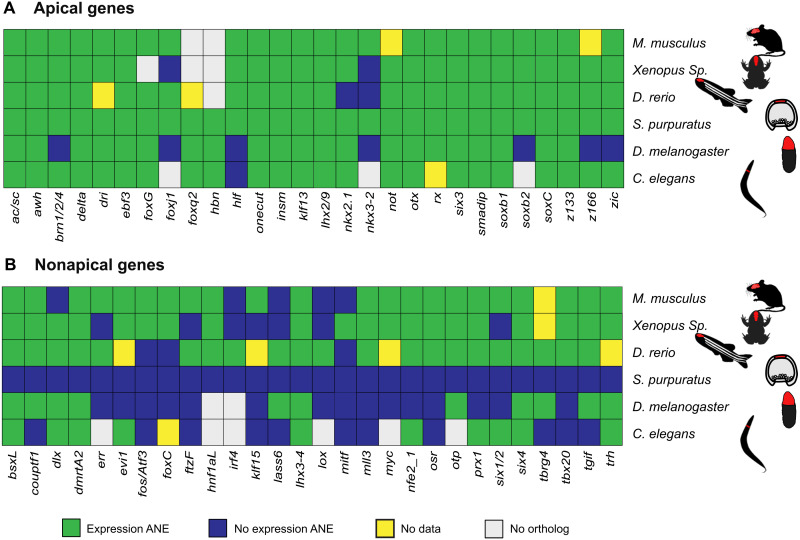
The systematic analysis of regulatory gene expression in this study identified most of transcription factors that constitute the GRN of the apical organ, offering an opportunity for a comparison between the apical GRN and GRNs associated with the nervous system of other animals. If the apical organ is evolutionarily related to more complex nervous systems, then some that overlap in transcription factor expression would be expected. Orthologs were identified for 28 of the 31 sea urchin apical transcription factors in mice, xenopus, zebrafish, Drosophila, and Caenorhabditis elegans. Expression of orthologous regulatory genes in the central nervous system of these species was analyzed using publicly available databases (see Materials and Methods and table S2). The results show that all transcription factors expressed in the apical organ have orthologs that are expressed in the nervous system of other animals (Fig. 4A). Expression in the nervous system was observed for all 24 orthologs in Mus musculus (100%), 23 of 25 orthologs in Xenopus tropicalis and Danio rerio (88%), 21 of 28 orthologs in Drosophila melanogaster (75%), and 23 of 24 orthologs in C. elegans (96%) (Fig. 4A). The orthologs of 20 sea urchin apical transcription factors (>70%) are expressed in the nervous system in every species for which orthologs were identified.
Fig. 4. Comparative analysis of transcription factor expression in the anterior nervous system.
Transcription factor expression in the anterior nervous system of five bilaterian species was analyzed using publicly available in situ data. (A) Analysis for orthologs of transcription factors expressed in the sea urchin apical domain. (B) Analysis for orthologs of transcription factors expressed in the sea urchin embryo but not in the apical domain. Of the 28 transcription factors expressed in the apical organ, 20 are expressed in the anterior nervous system of all species for which expression data are available compared to 7 transcription factors of the nonapical gene set. ANE, anterior nervous system.
Because random co-options of transcription factors might contribute to this result, we conducted the same analysis using 28 regulatory genes that were randomly selected from genes not expressed in the apical neurogenic ectoderm during sea urchin embryogenesis (table S3). This set includes transcription factors that are expressed in the endoderm, mesoderm, or ectoderm of sea urchin embryos and even includes a few known neural transcription factors such as nfe2 and six4. The results show that for nonapical regulatory genes, expression in the brain or anterior neurons was found for 22 of 27 orthologs in M. musculus, 20 of 27 orthologs in X. tropicalis, 21 of 24 orthologs in D. rerio, 11 of 26 orthologs in D. melanogaster, and 10 of 21 orthologs in C. elegans (Fig. 4B). Thus, particularly, in vertebrates, the overall likelihood of expression in the nervous system is relatively high even for randomly selected transcription factors, suggesting that frequent co-options have occurred during the evolution of the nervous system. However, only few transcription factors in this group are expressed in the nervous system of all five species. Only 7 of the 28 (25%) nonapical transcription factors are commonly expressed in the anterior nervous system of all species analyzed here, as compared to 70% for apical transcription factors. Expression of transcription factors in the nervous systems of every species examined is therefore far less likely to occur as a result of independent evolutionary events. The large fraction of transcription factors of the sea urchin apical GRN showing expression in the nervous system of several bilaterian species suggests that these transcription factors already operated as part of a GRN that was assembled before the separation of echinoderms from other bilaterian clades, indicating a deep evolutionary relationship of GRNs of the central nervous system in Bilateria.
Previous studies suggest that the apical organ might be related particularly to the vertebrate forebrain on the basis of expression of transcription factors such as Six3 (29). To test whether the expression of apical transcription factors is restricted to particular regions of the vertebrate brain, we separately identified expression in forebrain, midbrain, and hindbrain using the mouse Gene Expression Database (GXD) (36). The results show that about 60% of the transcription factors (17 of 28) are expressed in all three subdomains of the brain, while only 7 transcription factors are expressed exclusively in the mouse forebrain (see fig. S4). Thus, the expression of orthologs of apical transcription factors is generally not restricted to individual subdomains of the brain, although only the forebrain shows expression of the complete set of apical transcription factors. The similarity between apical organs and vertebrate brains in terms of transcription factor expression is therefore strongest in the forebrain but encompasses the entire brain.
Expression of nervous system axial patterning genes throughout the sea urchin ectoderm
Given the remarkable fraction of transcription factors of the apical GRN that also contribute to the development of other nervous systems of various form and function, it is conceivable that regulatory modules that are commonly expressed in animals with more complex nervous systems are also deployed in sea urchin embryos. We therefore determined the state of expression of a regulatory module that shows deep conservation throughout Bilateria and that is associated with the patterning of the anterior-posterior axis of the nervous system and its tripartite organization, particularly, in vertebrates and flies (6). How to interpret the conservation of regulatory modules over these large phylogenetic distances, however, poses certain challenges (7, 8). Because echinoderms are an early branching deuterostome clade with a relatively simple centralized larval nervous system, assessing the expression of the axial regulatory module might provide further insights into the ancestral function of this regulatory module in early deuterostomes.
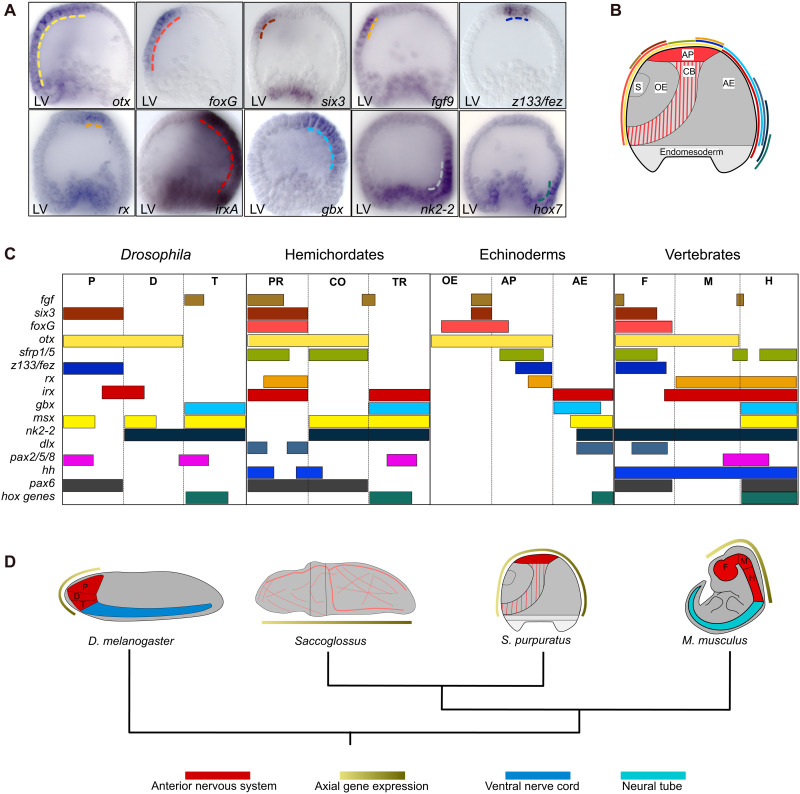
Several regulatory genes associated with the partitioning of the vertebrate brain also contribute to the patterning of the sea urchin apical organ, including six3, foxg, rx, z133/fez, and otx (Fig. 1, A to C, and fig. S2). Previous studies also showed that sfrp1/5 and msx are expressed in the apical organ and in the aboral ectoderm, respectively (19, 20, 37). In addition, we analyzed the spatial expression of fgf9, hh, irxA, gbx, nk2-2, dlx, pax2/5/8, pax6, and hox7 just before gastrulation by WMISH (Fig. 5A and fig. S5). The results show that 13 of 16 regulatory genes are expressed in specific patterns in the sea urchin ectoderm, while hh, pax2/5/8, and pax6 are not expressed in the ectoderm at this stage (fig. S5). The relative order in which the axial transcription factors are expressed within the sea urchin ectoderm is similar to the order of their expression along the anterior-posterior axis of the vertebrate brain (Fig. 5, B to D). However, in sea urchins, the axial transcription factors are expressed throughout the entire ectoderm, including the oral and aboral ectoderm, which do not give rise to neurons. Thus, six3, otx, and foxg mark the oral ectoderm and anterior/oral apical neural ectoderm; otx, sfrp1/5, fez, and rx mark the posterior/aboral apical neural ectoderm; and irx, gbx, msx, nk2-2, and hox7 mark the aboral ectoderm. Furthermore, the gene expression boundary between otx and gbx, which in vertebrates delineates the border between midbrain and hindbrain (38), separates apical neurogenic ectoderm and aboral ectoderm in sea urchins (Fig. 5C). Similarly, the boundary of expression between z133/fez and irxA, which in vertebrates separates the thalamus and prethalamus (39), is present also in sea urchins and separates the apical neurogenic ectoderm from the aboral ectoderm.
Fig. 5. Spatial expression of brain axial patterning genes in the ectoderm of sea urchin embryos.
(A) Spatial expression of sea urchin regulatory genes that are orthologous to genes associated with the anterior-posterior (A/P) axis of the anterior nervous system in vertebrates, analyzed in sea urchin embryos at 30 hpf. (B) Diagrammatic representation of a 30-hpf embryo summarizing the spatial expression data shown in (A) and fig. S5. Color code as in (A) and (C). (C) Comparison of axial gene expression in Drosophila, hemichordates, sea urchins, and mice indicating a similar order in regulatory gene expression. Expression data for msx and sfrp 1/5 in sea urchins were obtained from (19, 76). Expression data for Drosophila, hemichordates, and mice were obtained from (1, 6, 38, 39, 63, 64, 77). Color code as in (B). (D) Schemes of four bilaterian animal body plans indicating expression of axial patterning genes in respect to the localization of the nervous system. Axial gene expression occurs along the A/P axis of the brain in Drosophila and mice and throughout the ectoderm in hemichordates and sea urchins. P, D, and T, protocerebrum, deutocerebrum, and tritocerebrum; PR, CO, and TR, proboscis, collar, and trunk; OE, AP, and AE, oral ectoderm, apical organ, and aboral ectoderm; F, M, and H, forebrain, midbrain, and hindbrain. Scale bars, 20 μm.
These results indicate that the axial patterning module is conserved and functional also in echinoderms. However, its expression throughout the ectoderm, including non-neural oral and aboral ectoderm, suggests that, in sea urchin embryos, this module functions more broadly in ectodermal patterning (Fig. 5D).
DISCUSSION
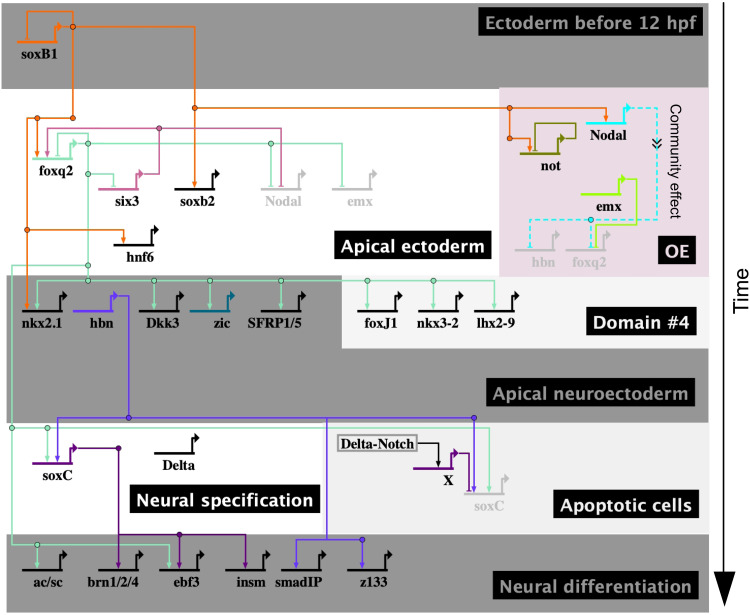
In this study, we identified most of transcription factors and several regulatory linkages that control the development of the apical organ and the specification of serotonergic neurons in sea urchin embryos. We show that in this system, the apical organ forms within just about 18 hours from the initial specification of apical neurogenic ectoderm to the formation of neural precursors, in a process that involves relatively few regulatory steps. A model of the GRN controlling the development of the apical organ, including most of its transcription factors and the regulatory linkages identified here and elsewhere, is shown in Fig. 6. Although this GRN model is far from complete in terms of regulatory interactions, several crucial regulatory transactions were identified that show how FoxQ2, Hbn, and Delta/Notch signaling control the specification of serotonergic neurons by regulating expression of a key transcription factor in this process, SoxC, which, in turn, controls the expression of several proneural transcription factors (Fig. 6).
Fig. 6. BioTapestry model of the GRN for apical neurogenesis.
A model of the apical GRN has been generated in BioTapestry (78). The specification of apical serotonergic neurons in sea urchins is controlled by a GRN that causally connects the early specification of the apical neural ectoderm to the specification of neural precursors and to neural differentiation. Different boxes indicate the various phases of neurogenesis and/or different embryonic domains. Evidence for regulatory linkages is summarized in fig. S6 and table S4 (79–82).
This study shows that the orthologs of almost all transcription factors of the apical GRN are also expressed in the central nervous system of several other bilaterian animals. These findings support a common evolutionary origin of apical organs and more complex brains, because it is very unlikely that the deployment of the entire suite of apical transcription factors in the development of more complex nervous systems is the result of random evolutionary co-options. It remains to be seen to what extent these apical transcription factors execute conserved versus newly acquired functions during neural development in different animals. For several apical transcription factors, current evidence indicates that not only expression but also regulatory functions during development of the nervous system have been conserved over remarkable evolutionary distances.
A first example for the conservation of regulatory mechanisms is provided by the upstream section of the apical GRN that controls the initial specification of the apical neurogenic ectoderm in early sea urchin embryos. The earliest specification of the apical organ depends on SoxB1 activating the expression of foxq2 (17) and on Wnt signaling from the vegetal pole, which leads to repression of neurogenic cell fates and restriction of foxq2 expression to the apical domain (19, 20). Further activation of foxq2 expression occurs downstream of Six3 (29), while FoxQ2 represses six3 and activates the expression of Wnt signaling antagonists (Fig. 6) (20, 22). Similar mechanisms lead to the specification of neurogenic ectoderm throughout Bilateria and in Cnidaria. Thus, SoxB class transcription factors are required for neural commitment in the embryonic nervous system in vertebrates (40, 41), Drosophila (42), and Nematostella (43). Similarly, antagonism to Wnt signaling contributes broadly to the specification of the anterior nervous system, in Cnidaria and Bilateria as well as in apical organs and complex brains (14, 44–46). Furthermore, six3 genes are expressed in the apical organ or anterior nervous system of cnideria, arthropods, annelids, and vertebrates, representing a common feature of Bilateria (45–47). Even the activation of foxq2 expression in neurogenic ectoderm downstream of Six3 orthologs occurs in Cnidaria and in arthropods, as well as in zebrafish photoreceptors (48–50).
In sea urchins, the initial specification of the apical organ is followed by a relatively short phase of patterning and by the specification of neural precursors. Our results show that FoxQ2 functions upstream in the apical GRN and is required for the expression of several transcription factors that pattern the apical organ. FoxQ2, together with Hbn, controls the specification of apical neural precursors by activating the expression of soxc. Similarly, orthologs of FoxQ2 are expressed in the apical organ of Cnidaria and brachiopods and in the central nervous system of centipedes and insects (48, 49, 51). FoxQ2 is also expressed in Tribolium castaneum, where it controls the specification and patterning of central brain structures, comparable to its role in the sea urchin apical organ (49, 52). However, despite the important function of FoxQ2 and Hbn in the early development of the nervous system in invertebrates, this function has been lost in vertebrates and foxq2 and hbn are not present in the genomes of tetrapods and vertebrates, respectively (53, 54).
Some of the most broadly shared regulatory features of the apical GRN are those involved in neurogenesis, which is consistent with previous comparisons showing that the neuronal expression of proneural transcription factors is widely conserved (32). Of the 20 transcription factors that are commonly expressed in the nervous system of all six species examined here, 15 are expressed in apical serotonergic neurons in sea urchins, although most of them also in other cell fates of the apical organ. Overall, about 70% of the regulatory state of apical neurons is conserved. Furthermore, Delta/Notch signaling, which is involved in the restriction of the neural cell fate to neural precursors in sea urchins, is required for the restriction of neural cell fates in many other developmental contexts. Thus, Delta/Notch-mediated lateral inhibition is commonly deployed during neurogenesis in many animals, including other echinoderms, Drosophila, and vertebrates (55–57). In sea urchins, Delta/Notch signaling controls the expression of SoxC, which is required for the specification of apical neural precursors. SoxC activates the expression of neural regulatory genes including ac/sc, ebf3, insm, and brn1/2/4. In vertebrates, orthologs of all five transcription factors are involved in neurogenesis and neuronal differentiation (58–60). In mice for instance, the SoxC class transcription factors Sox4, Sox11, and Sox12 are expressed at high levels in neural progenitors in the brain and function cooperatively with Brn2 in the activation of neural genes (40, 61, 62). Acsc1 functions as a pioneer transcription factor, recruiting other transcription factors such as Brn2 to neural genes during neuronal differentiation. The co-expression of Soxc, Brn1/2/4, and Ac/sc in apical serotonergic neurons suggests that these transcription factors constitute a conserved regulatory module with a conserved function in neural differentiation.
One of the few regulatory modules that are known to be deeply conserved among bilaterian animals is the regulatory module that patterns the anterior-posterior axis of the brain in arthropods and vertebrates and that contributes to its tripartite organization (4, 6, 38, 63, 64). However, it remains a matter of debate whether the conservation of these axial patterning modules also indicates that a complex regionalized nervous system was present in bilaterian ancestors (1, 4, 6, 8). Evidence indicating that this regulatory module might be conserved for reasons other than the tripartite organization of the nervous system stem from observations in animals lacking a tripartite brain. Thus, in hemichordates, orthologs of axial transcription factors including Otx, Pax2/5/8, Gbx2, and Hox are expressed in a similar order; however, instead of marking different parts of the nervous system, these regulators separate the ectoderm into proboscis ectoderm, collar ectoderm, anterior trunk ectoderm, and posterior trunk ectoderm (1, 8). However, because hemichordates have a diffuse nerve net that extends along almost the entire body axis, ectodermal patterning might still contribute to the specification of different neuronal cell types (8). The expression of the axial patterning module throughout the ectoderm of sea urchin embryos on the other hand shows that, in nonchordate deuterostomes, this regulatory module controls ectodermal patterning beyond a central nervous system, indicating that this function might be ancestral within deuterostomes. Accordingly, the function of this regulatory module in the tripartite organization of the vertebrate brain might be the result of the co-option of novel developmental processes downstream of this module that occurred within chordates after the separation from echinoderms and hemichordates (Fig. 5D). Similar evolutionary co-options that enabled this regulatory module to control the partitioning of the nervous system would have occurred independently within protostomes.
Together, the apical GRN displays an extensive degree of conservation of its regulatory components. Not only are a majority of its transcription factors expressed in the anterior nervous system throughout Bilateria, but also many of them appear to control similar aspects of neural development. This overlap in regulatory mechanisms in the sea urchin apical GRN and the GRNs controlling the formation of the nervous system in other animals supports the conclusion that bilaterian nervous systems share a common evolutionary origin and are therefore homologous. This homology includes not only developmental mechanisms controlling the anterior localization of the nervous system but also processes involved in the patterning of the nervous system and in the specification and differentiation of neural cell types. Because the apical GRN includes few, if any, transcription factors that are not shared with other neural GRNs, few gene co-options have occurred that are specific to apical organs, indicating that the apical GRN has undergone limited evolutionary modifications. Apical organs such as present in extant echinoderms might therefore closely resemble an ancestral form of anterior nervous system. Accordingly, the far more complex nervous systems of vertebrates have evolved by extensive co-option of dozens, if not hundreds, of additional transcription factors and signaling molecules that supported the expansion of neuronal cell types and the establishment of novel functional regions of the brain. Because the development of the vertebrate brain involves far more than the specification of a few cell types, even transcription factors of an ancestral nervous system GRN would be likely to have acquired novel regulatory functions, and as a result, ancestral GRN circuitry might not be conserved apart from individual regulatory interactions and smaller regulatory modules, as discussed above. As this study indicates, the sea urchin apical GRN might provide an important point of reference to further reconstruct the regulatory changes that led to the evolution of the vertebrate nervous system.
MATERIALS AND METHODS
NanoString nCounter assay
Gene expression levels in wild-type and perturbed embryos were analyzed by NanoString nCounter essay according to previous protocols (65). Briefly, ~200 embryos were collected for each sample, and RNA was extracted using the QIAGEN RNeasy Micro Kit (QIAGEN, Hilden, Germany). Samples were ethanol-precipitated and resuspended in 7.5 μl of nuclease-free water, and 5 μl was used for hybridization. Hybridization reactions were performed according to the manufacturer’s instructions at 65°C for at least 18 hours. Hybridized probes were recovered with the NanoString Prep Station and immediately evaluated with the NanoString nCounter. Counts were normalized by the mean of total counts, and mRNA copies per embryo for each gene was determined using linear regression of counts for the positive spike-in probes divided by the number of embryos per sample.
Whole-mount in situ hybridization
WMISH analyses were performed according to previous protocols (66). Briefly, sea urchin embryos were fixed in paraformaldehyde. Fixed embryos were incubated at 60°C overnight in hybridization buffer [50% (v/v) formamide, 5× SSC, 1× Denhardt’s, yeast transfer RNA (1 mg/ml), heparin (50 ng/ml), and 0.1% Tween-20] with digoxigenin-labeled RNA probes (0.5 to 1 ng/μl) (table S1). Embryos were washed with hybridization buffer [2× SSCT (2× SSC and 0.1% Tween-20), 0.2× SSCT, and 0.1× SSCT; each for 20 min at 60°C]. Subsequently, the antibody incubations were performed at room temperature with 1:1000 diluted anti-Digoxigenin-AP (anti-DIG) Fab fragments (Fab) (Roche). Embryos were washed 6× with maleic acid buffer containing Tween 20 (MABT) buffer (0.1 M maleic acid, 0.15 M NaCl, and 0.1% Tween-20) and 2× with alkaline phosphatase (AP) buffer [100 mM tris-Cl (pH 9.5), 100 mM NaCl, 50 mM MgCl2, and 1 mM levamisole]. 5-Bromo-4-chloro-3-indolyl-phosphate and nitro blue tetrazolium were used for staining. Embryos stained by WMISH were analyzed by microscopy.
Microinjection and perturbation analysis
Morpholino sequences targeting eve, foxq2, and soxc were previously described: foxq2, GAATTAGAGGCGGTAGATGTGTCGT; eve, CAGAAACCACTCGATCAATGTTTGC; and soxc, GAACCATCTTGAAGTCAGCATTCAC (22, 67). The sequence of the hbn morpholino was 5′-AGGAACAACGACCGCCGTCATTC-3′. Morpholinos were injected at 300 μM in 0.12 M KCl. For control, embryos were injected with random control morpholinos or left uninjected. A total of 200 to 300 morpholino-injected embryos were collected at different time points. Experiments were performed with two to three independent batches of embryos. For experiments involving WMISH, effects of perturbations were analyzed in 20 to 60 embryos, with results shown representing at least 90 to 95% of scored embryos. To block Delta/Notch signaling, the γ-secretase inhibitor DAPT (34) was dissolved in DMSO. Embryos were treated with DAPT at a final concentration of 8 μM or with DMSO as control starting at 3 hpf, as previously described (35).
Identification of orthologs
The orthologs of sea urchin transcription factors were determined using Panther least divergent ortholog (LDO) (68, 69). Orthologs were further validated using EggNOG 5.0 (70), and for D. melanogaster and C. elegans, we used the pipeline implemented in ENSEMBL metazoan (71). Genes were excluded from the analysis where the orthologous relationship was not clearly resolved (hesC and hairy2/4) and where apical expression occurs only transiently (klf2/4). In the vast majority of cases, the same set of orthologs were identified with all methods used, except when no LDO was identified or when no orthologs or >3 orthologs were identified in Panther.
Comparison of gene expression in nervous system
For each gene identified to be orthologous to one of the sea urchin regulatory genes in the apical or nonapical gene set, we evaluated expression in developing brains and/or neurons using publicly available data. For M. musculus, we used the mouse GXD to determine genes expressed in the forebrain, midbrain, or hindbrain (36). For Xenopus and zebrafish, we used Xenbase (72) and ZFIN (73), respectively, to determine whether gene orthologs are expressed in the forebrain, midbrain, or hindbrain. For D. melanogaster, we used the Berkley Drosophila Genome Project in situ database (74) and evaluated the expression of orthologs in the brain during embryogenesis. Last, for C. elegans, we used WormBase to evaluate the gene expression in anterior neurons (75).
Acknowledgments
In memory of E. H. Davidson, whose ambition to obtain a complete understanding of the GRNs for the early sea urchin embryo inspired this project.
Funding: This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development Grants HD037105 and HD095982 awarded to I.S.P.
Author contributions: Conceptualization: R.F. and I.S.P. Investigation: R.F. Formal analysis: R.F. and I.S.P. Visualization and figure preparation: R.F. Methodology: R.F. and I.S.P. Writing—original draft: R.F. and I.S.P. Writing—review and editing R.F. and I.S.P. Supervision: I.S.P. Funding acquisition: I.S.P.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Tables S1 to S4
REFERENCES AND NOTES
- 1.Lowe C. J., Wu M., Salic A., Evans L., Lander E., Stange-Thomann N., Gruber C. E., Gerhart J., Kirschner M., Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell 113, 853–865 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Garstang W., Preliminary note on a new theory of the phylogeny of the chordata. Zool. Anzeiger 22, 122–125 (1894). [Google Scholar]
- 3.Gerhart J., Lowe C., Kirschner M., Hemichordates and the origin of chordates. Curr. Opin. Genet. Dev. 15, 461–467 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Arendt D., Denes A. S., Jékely G., Tessmar-Raible K., The evolution of nervous system centralization. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 1523–1528 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denes A. S., Jékely G., Steinmetz P. R. H., Raible F., Snyman H., Prud’homme B., Ferrier D. E. K., Balavoine G., Arendt D., Molecular architecture of annelid nerve cord supports common origin of nervous system centralization in Bilateria. Cell 129, 277–288 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Hirth F., Kammermeier L., Frei E., Walldorf U., Noll M., Reichert H., An urbilaterian origin of the tripartite brain: Developmental genetic insights from Drosophila. Dev. 130, 2365–2373 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Martín-Durán J. M., Pang K., Børve A., Lê H. S., Furu A., Cannon J. T., Jondelius U., Hejnol A., Convergent evolution of bilaterian nerve cords. Nature 553, 45–50 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hejnol A., Lowe C. J., Embracing the comparative approach: How robust phylogenies and broader developmental sampling impacts the understanding of nervous system evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20150045 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.I. S. Peter, E. H. Davidson, Genomic Control Process, Development and Evolution (Academic Press/Elsevier, 2015). [Google Scholar]
- 10.Peter I. S., Davidson E. H., Evolution of gene regulatory networks controlling body plan development. Cell 144, 970–985 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davidson E. H., Erwin D. H., Gene regulatory networks and the evolution of animal body plans. Science 311, 796–800 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Hinman V. F., Burke R. D., Embryonic neurogenesis in echinoderms. Wiley Interdiscip. Rev. Dev. Biol. 7, e316 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Angerer L. M., Yaguchi S., Angerer R. C., Burke R. D., The evolution of nervous system patterning: Insights from sea urchin development. Dev. 138, 3613–3623 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marlow H., Tosches M. A., Tomer R., Steinmetz P. R., Lauri A., Larsson T., Arendt D., Larval body patterning and apical organs are conserved in animal evolution. BMC Biol. 12, 7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burke R. D., Angerer L. M., Elphick M. R., Humphrey G. W., Yaguchi S., Kiyama T., Liang S., Mu X., Agca C., Klein W. H., Brandhorst B. P., Rowe M., Wilson K., Churcher A. M., Taylor J. S., Chen N., Murray G., Wang D., Mellott D., Olinski R., Hallböök F., Thorndyke M. C., A genomic view of the sea urchin nervous system. Dev. Biol. 300, 434–460 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garner S., Zysk I., Byrne G., Kramer M., Moller D., Taylor V., Burke R. D., Neurogenesis in sea urchin embryos and the diversity of deuterostome neurogenic mechanisms. Dev. 143, 286–297 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Barsi J. C., Li E., Davidson E. H., Geometric control of ciliated band regulatory states in the sea urchin embryo. Dev. 142, 953–961 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McClay D. R., Miranda E., Feinberg S. L., Neurogenesis in the sea urchin embryo is initiated uniquely in three domains. Dev. 145, dev167742 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui M., Siriwon N., Li E., Davidson E. H., Peter I. S., Specific functions of the Wnt signaling system in gene regulatory networks throughout the early sea urchin embryo. Proc. Natl. Acad. Sci. U.S.A. 111, E5029–E5038 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Range R. C., Wei Z., An anterior signaling center patterns and sizes the anterior neuroectoderm of the sea urchin embryo. Dev. 143, 1523–1533 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yaguchi S., Yaguchi J., Angerer R. C., Angerer L. M., A Wnt-FoxQ2-nodal pathway links primary and secondary axis specification in sea urchin embryos. Dev. Cell 14, 97–107 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Li E., Cui M., Peter I. S., Davidson E. H., Encoding regulatory state boundaries in the pregastrular oral ectoderm of the sea urchin embryo. Proc. Natl. Acad. Sci. U.S.A. 111, E906–E913 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mellott D. O., Thisdelle J., Burke R. D., Notch signaling patterns neurogenic ectoderm and regulates the asymmetric division of neural progenitors in sea urchin embryos. Dev. 144, 3602–3611 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Wei Z., Angerer L. M., Angerer R. C., Neurogenic gene regulatory pathways in the sea urchin embryo. Dev. 143, 298–305 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howard-Ashby M., Materna S. C., Brown C. T., Chen L., Cameron R. A., Davidson E. H., Identification and characterization of homeobox transcription factor genes in Strongylocentrotus purpuratus, and their expression in embryonic development. Dev. Biol. 300, 74–89 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Howard-Ashby M., Materna S. C., Brown C. T., Chen L., Cameron R. A., Davidson E. H., Gene families encoding transcription factors expressed in early development of Strongylocentrotus purpuratus. Dev. Biol. 300, 90–107 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Materna S. C., Howard-Ashby M., Gray R. F., Davidson E. H., The C2H2 zinc finger genes of Strongylocentrotus purpuratus and their expression in embryonic development. Dev. Biol. 300, 108–120 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Tu Q., Brown C. T., Davidson E. H., Oliveri P., Sea urchin Forkhead gene family: Phylogeny and embryonic expression. Dev. Biol. 300, 49–62 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Wei Z., Yaguchi J., Yaguchi S., Angerer R. C., Angerer L. M., The sea urchin animal pole domain is a Six3-dependent neurogenic patterning center. Dev. 136, 1179–1189 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slota L. A., McClay D. R., Identification of neural transcription factors required for the differentiation of three neuronal subtypes in the sea urchin embryo. Dev. Biol. 435, 138–149 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geiss G. K., Bumgarner R. E., Birditt B., Dahl T., Dowidar N., Dunaway D. L., Fell H. P., Ferree S., George R. D., Grogan T., James J. J., Maysuria M., Mitton J. D., Oliveri P., Osborn J. L., Peng T., Ratcliffe A. L., Webster P. J., Davidson E. H., Hood L., Dimitrov K., Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 26, 317–325 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Hartenstein V., Stollewerk A., The evolution of early neurogenesis. Dev. Cell 32, 390–407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yaguchi J., Takeda N., Inaba K., Yaguchi S., Cooperative Wnt-nodal signals regulate the patterning of anterior neuroectoderm. PLOS Genet. 12, e1006001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hughes J. N., Dodge N., Rathjen P. D., Rathjen J., A novel role for γ-secretase in the formation of primitive streak-like intermediates from ES cells in culture. Stem Cells 27, 2941–2951 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Materna S. C., Davidson E. H., A comprehensive analysis of δ signaling in pre-gastrular sea urchin embryos. Dev. Biol. 364, 77–87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith C. M., Hayamizu T. F., Finger J. H., Bello S. M., McCright I. J., Xu J., Baldarelli R. M., Beal J. S., Campbell J., Corbani L. E., Frost P. J., Lewis J. R., Giannatto S. C., Miers D., Shaw D. R., Kadin J. A., Richardson J. E., Smith C. L., Ringwald M., The mouse Gene Expression Database (GXD): 2019 update. Nucleic Acids Res. 47, D774–D779 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su Y.-H., Li E., Geiss G. K., Longabaugh W. J. R., Krämer A., Davidson E. H., A perturbation model of the gene regulatory network for oral and aboral ectoderm specification in the sea urchin embryo. Dev. Biol. 329, 410–421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urbach R., A procephalic territory in Drosophila exhibiting similarities and dissimilarities compared to the vertebrate midbrain/hindbrain boundary region. Neural Dev. 2, 23 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irimia M., Piñeiro C., Maeso I., Gómez-Skarmeta J., Casares F., Garcia-Fernàndez J., Conserved developmental expression of Fezf in chordates and Drosophila and the origin of the zona limitans intrathalamica (ZLI) brain organizer. Evodevo 1, 7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bergsland M., Ramsköld D., Zaouter C., Klum S., Sandberg R., Muhr J., Sequentially acting Sox transcription factors in neural lineage development. Genes Dev. 25, 2453–2464 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graham V., Khudyakov J., Ellis P., Pevny L., SOX2 functions to maintain neural progenitor identity. Neuron 39, 749–765 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Ferrero E., Fischer B., Russell S., SoxNeuro orchestrates central nervous system specification and differentiation in Drosophila and is only partially redundant with dichaete. Genome Biol. 15, R74 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richards G. S., Rentzsch F., Transgenic analysis of a SoxB gene reveals neural progenitor cells in the cnidarian Nematostella vectensis. Development 141, 4681–4689 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Petersen C. P., Reddien P. W., Wnt signaling and the polarity of the primary body axis. Cell 139, 1056–1068 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Marlow H., Matus D. Q., Martindale M. Q., Ectopic activation of the canonical wnt signaling pathway affects ectodermal patterning along the primary axis during larval development in the anthozoan Nematostella vectensis. Dev. Biol. 380, 324–334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lavado A., Lagutin O. V., Oliver G., Six3 inactivation causes progressive caudalization and aberrant patterning of the mammalian diencephalon. Development 135, 441–450 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Steinmetz P. R., Urbach R., Posnien N., Eriksson J., Kostyuchenko R. P., Brena C., Guy K., Akam M., Bucher G., Arendt D., Six3 demarcates the anterior-most developing brain region in bilaterian animals. Evodevo 1, 14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sinigaglia C., Busengdal H., Leclère L., Technau U., Rentzsch F., The bilaterian head patterning gene six3/6 controls aboral domain development in a cnidarian. PLoS Biol. 11, e1001488 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitzmann P., Weißkopf M., Schacht M. I., Bucher G., A key role for foxQ2 in anterior head and central brain patterning in insects. Development 144, 2969–2981 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogawa Y., Shiraki T., Fukada Y., Kojima D., Foxq2 determines blue cone identity in zebrafish. Sci. advances 7, eabi9784 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Santagata S., Resh C., Hejnol A., Martindale M. Q., Passamaneck Y. J., Development of the larval anterior neurogenic domains of Terebratalia transversa (Brachiopoda) provides insights into the diversification of larval apical organs and the spiralian nervous system. Evodevo 3, 3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He B., Buescher M., Farnworth M. S., Strobl F., Stelzer E. H. K., Koniszewski N. D. B., Muehlen D., Bucher G., An ancestral apical brain region contributes to the central complex under the control of foxQ2 in the beetle Tribolium. eLife 8, e49065 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mazet F., Yu J. K., Liberles D. A., Holland L. Z., Shimeld S. M., Phylogenetic relationships of the Fox (Forkhead) gene family in the Bilateria. Gene 316, 79–89 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Mazza M. E., Pang K., Reitzel A. M., Martindale M. Q., Finnerty J. R., A conserved cluster of three PRD-class homeobox genes (homeobrain, rx and orthopedia) in the Cnidaria and Protostomia. Evodevo 1, 3 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haddon C., Smithers L., Schneider-Maunoury S., Coche T., Henrique D., Lewis J., Multiple δ genes and lateral inhibition in zebrafish primary neurogenesis. Development 125, 359–370 (1998). [DOI] [PubMed] [Google Scholar]
- 56.Chitnis A., Henrique D., Lewis J., Ish-Horowicz D., Kintner C., Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene δ. Nature 375, 761–766 (1995). [DOI] [PubMed] [Google Scholar]
- 57.Yankura K. A., Koechlein C. S., Cryan A. F., Cheatle A., Hinman V. F., Gene regulatory network for neurogenesis in a sea star embryo connects broad neural specification and localized patterning. Proc. Natl. Acad. Sci. U.S.A. 110, 8591–8596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wapinski O. L., Vierbuchen T., Qu K., Lee Q. Y., Chanda S., Fuentes D. R., Giresi P. G., Ng Y. H., Marro S., Neff N. F., Drechsel D., Martynoga B., Castro D. S., Webb A. E., Südhof T. C., Brunet A., Guillemot F., Chang H. Y., Wernig M., Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell 155, 621–635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pozzoli O., Bosetti A., Croci L., Consalez G. G., Vetter M. L., Xebf3 is a regulator of neuronal differentiation during primary neurogenesis in Xenopus. Dev. Biol. 233, 495–512 (2001). [DOI] [PubMed] [Google Scholar]
- 60.Farkas L. M., Haffner C., Giger T., Khaitovich P., Nowick K., Birchmeier C., Pääbo S., Huttner W. B., Insulinoma-associated 1 has a panneurogenic role and promotes the generation and expansion of basal progenitors in the developing mouse neocortex. Neuron 60, 40–55 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Dy P., Penzo-Méndez A., Wang H., Pedraza C. E., Macklin W. B., Lefebvre V., The three SoxC proteins—Sox4, Sox11 and Sox12—exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 36, 3101–3117 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen C., Lee G. A., Pourmorady A., Sock E., Donoghue M. J., Orchestration of neuronal differentiation and progenitor pool expansion in the developing cortex by soxc genes. J. Neurosci. 35, 10629–10642 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Holland L. Z., Carvalho J. E., Escriva H., Laudet V., Schubert M., Shimeld S. M., Yu J. K., Evolution of bilaterian central nervous systems: A single origin? Evodevo 4, 27 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pani A. M., Mullarkey E. E., Aronowicz J., Assimacopoulos S., Grove E. A., Lowe C. J., Ancient deuterostome origins of vertebrate brain signalling centres. Nature 483, 289–294 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Materna S. C., Nam J., Davidson E. H., High accuracy, high-resolution prevalence measurement for the majority of locally expressed regulatory genes in early sea urchin development. Gene Expr. Patterns 10, 177–184 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ransick A., Ernst S., Britten R. J., Davidson E. H., Whole mount in situ hybridization shows Endo 16 to be a marker for the vegetal plate territory in sea urchin embryos. Mech. Dev. 42, 117–124 (1993). [DOI] [PubMed] [Google Scholar]
- 67.Peter I. S., Davidson E. H., A gene regulatory network controlling the embryonic specification of endoderm. Nature 474, 635–639 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mi H., Muruganujan A., Ebert D., Huang X., Thomas P. D., PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–D426 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Altenhoff A. M., Boeckmann B., Capella-Gutierrez S., Dalquen D. A., DeLuca T., Forslund K., Huerta-Cepas J., Linard B., Pereira C., Pryszcz L. P., Schreiber F., da Silva A. S., Szklarczyk D., Train C.-M., Bork P., Lecompte O., von Mering C., Xenarios I., Sjölander K., Jensen L. J., Martin M. J., Muffato M.; Quest for Orthologs consortium, Gabaldón T., Lewis S. E., Thomas P. D., Sonnhammer E., Dessimoz C., Standardized benchmarking in the quest for orthologs. Nat. Methods 13, 425–430 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huerta-Cepas J., Szklarczyk D., Heller D., Hernández-Plaza A., Forslund S. K., Cook H., Mende D. R., Letunic I., Rattei T., Jensen L. J., von Mering C., Bork P., eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res 47, D309–D314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yates A. D., Achuthan P., Akanni W., Allen J., Allen J., Alvarez-Jarreta J., Amode M. R., Armean I. M., Azov A. G., Bennett R., Bhai J., Billis K., Boddu S., Marugán J. C., Cummins C., Davidson C., Dodiya K., Fatima R., Gall A., Giron C. G., Gil L., Grego T., Haggerty L., Haskell E., Hourlier T., Izuogu O. G., Janacek S. H., Juettemann T., Kay M., Lavidas I., le T., Lemos D., Martinez J. G., Maurel T., McDowall M., McMahon A., Mohanan S., Moore B., Nuhn M., Oheh D. N., Parker A., Parton A., Patricio M., Sakthivel M. P., Abdul Salam A. I., Schmitt B. M., Schuilenburg H., Sheppard D., Sycheva M., Szuba M., Taylor K., Thormann A., Threadgold G., Vullo A., Walts B., Winterbottom A., Zadissa A., Chakiachvili M., Flint B., Frankish A., Hunt S. E., IIsley G., Kostadima M., Langridge N., Loveland J. E., Martin F. J., Morales J., Mudge J. M., Muffato M., Perry E., Ruffier M., Trevanion S. J., Cunningham F., Howe K. L., Zerbino D. R., Flicek P., Ensembl 2020. Nucleic Acids Res. 48, D682–D688 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bowes J. B., Snyder K. A., Segerdell E., Jarabek C. J., Azam K., Zorn A. M., Vize P. D., Xenbase: Gene expression and improved integration. Nucleic Acids Res. 38, D607–D612 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruzicka L., Howe D. G., Ramachandran S., Toro S., van Slyke C. E., Bradford Y. M., Eagle A., Fashena D., Frazer K., Kalita P., Mani P., Martin R., Moxon S. T., Paddock H., Pich C., Schaper K., Shao X., Singer A., Westerfield M., The Zebrafish Information Network: New support for non-coding genes, richer Gene Ontology annotations and the Alliance of Genome Resources. Nucleic Acids Res. 47, D867–D873 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hammonds A. S., Bristow C. A., Fisher W. W., Weiszmann R., Wu S., Hartenstein V., Kellis M., Yu B., Frise E., Celniker S. E., Spatial expression of transcription factors in Drosophila embryonic organ development. Genome Biol. 14, R140 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Harris T. W., Arnaboldi V., Cain S., Chan J., Chen W. J., Cho J., Davis P., Gao S., Grove C. A., Kishore R., Lee R. Y. N., Muller H.-M., Nakamura C., Nuin P., Paulini M., Raciti D., Rodgers F. H., Russell M., Schindelman G., Auken K. V., Wang Q., Williams G., Wright A. J., Yook K., Howe K. L., Schedl T., Stein L., Sternberg P. W., WormBase: A modern model organism information resource. Nucleic Acids Res. 48, D762–D767 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ben-Taboude-Leon S., Su Y. H., Lin K. T., Li E., Davidson E. H., Gene regulatory control in the sea urchin aboral ectoderm: Spatial initiation, signaling inputs, and cell fate lockdown. Dev. Biol. 374, 245–254 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hirth F., On the origin and evolution of the tripartite brain. Brain Behav. Evol. 76, 3–10 (2010). [DOI] [PubMed] [Google Scholar]
- 78.Longabaugh W. J., BioTapestry: A tool to visualize the dynamic properties of gene regulatory networks. Methods Mol. Biol. 786, 359–394 (2012). [DOI] [PubMed] [Google Scholar]
- 79.Range R., Lapraz F., Quirin M., Marro S., Besnardeau L., Lepage T., Cis-regulatory analysis of nodal and maternal control of dorsal-ventral axis formation by Univin, a TGF-β related to Vg1. Development 134, 3649–3664 (2007). [DOI] [PubMed] [Google Scholar]
- 80.Saudemont A., Haillot E., Mekpoh F., Bessodes N., Quirin M., Lapraz F., Duboc V., Röttinger E., Range R., Oisel A., Besnardeau L., Wincker P., Lepage T., Ancestral regulatory circuits governing ectoderm patterning downstream of Nodal and BMP2/4 revealed by gene regulatory network analysis in an echinoderm. PLOS Genet. 6, e1001259 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li E., Materna S. C., Davidson E. H., Direct and indirect control of oral ectoderm regulatory gene expression by Nodal signaling in the sea urchin embryo. Dev. Biol. 369, 377–385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yaguchi J., Angerer L. M., Inaba K., Yaguchi S., Zinc finger homeobox is required for the differentiation of serotonergic neurons in the sea urchin embryo. Dev. Biol. 363, 74–83 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 to S4