Abstract
In addition to the significant role in physical activity, skeletal muscle also contributes to health through the storage and use of macronutrients associated with energy homeostasis. However, the mechanisms of regulating integrated metabolism in skeletal muscle are not well-defined. Here, we compared the skeletal muscle transcriptome from obese and lean control subjects in different species (human and mouse) and found that interferon regulatory factor 4 (IRF4), an inflammation-immune transcription factor, conservatively increased in obese subjects. Thus, we investigated whether IRF4 gain of function in the skeletal muscle predisposed to obesity and insulin resistance. Conversely, mice with specific IRF4 loss in skeletal muscle showed protection against the metabolic effects of high-fat diet, increased branched-chain amino acids (BCAA) level of serum and muscle, and reprogrammed metabolome in serum. Mechanistically, IRF4 could transcriptionally upregulate mitochondrial branched-chain aminotransferase (BCATm) expression; subsequently, the enhanced BCATm could counteract the effects caused by IRF4 deletion. Furthermore, we demonstrated that IRF4 ablation in skeletal muscle enhanced mitochondrial activity, BCAA, and fatty acid oxidation in a BCATm-dependent manner. Taken together, these studies, for the first time, established IRF4 as a novel metabolic driver of macronutrients via BCATm in skeletal muscle in terms of diet-induced obesity.
Introduction
Skeletal muscle tissue is capable of generating force for movement and contributes to health through the storage and use of energy homeostasis–related macronutrients (1). Disrupted skeletal muscle function has significant effects on disease risk. Emerging data have revealed that nutritional state (e.g., obesity) and certain nutrients also negatively alter the skeletal muscle metabolome of healthy groups (2). For example, compared with the healthy, individuals with obesity display lower contents of some amino acids in skeletal muscle but higher short-chain acylcarnitine species in blood (3). However, the mechanism of integrating the metabolism of different macronutrients in skeletal muscle, especially that in an obese state, is not well-known. Decoding this will be an attractive strategy for identifying novel therapeutic targets for metabolic diseases.
Obesity is closely associated with chronic inflammation. For example, inflammatory signals and free fatty acids were reported to activate Toll-like receptor 4 (TLR4) on the surface of myocytes in vivo and in vitro (4). Interferon regulatory factors (IRFs) have been shown to mediate TLR4-induced gene expression (5). Interestingly, IRF4 played key roles in the development and function of immune cells (6–8) and was also an important regulator of adipogenesis and lipid handling in adipose tissue (9,10). IRF4 served as a transcriptional partner of the cofactor PGC-1α and a critical regulator of mitochondrial biogenesis and thermogenesis in brown adipose tissue (BAT) (11,12). Moreover, the skeletal muscle IRF4 knockout (MKO) mice on a chow diet presented increased exercise capacity (13). Therefore, it is of great significance to determine the IRF4 participation in energy homeostasis of skeletal muscle.
While most amino acids are catabolized in the liver, branched-chain amino acids (BCAA) are primarily oxidized in skeletal muscle under the control of branched-chain aminotransferases (14). Mitochondrial branched-chain aminotransferase (BCATm) is highly expressed in skeletal muscle and functions to transfer NH3 from BCAA to α-ketoglutaric acid so as to form three respective branched-chain ketoacids (BCKA) and glutamate (15). Mice with whole-body BCATm deletion were lean and exhibited higher energy expenditure (EE), higher plasma BCAA concentration, lower glucose (16), and delayed growth of lymphoma tumors compared with wild-type (WT) mice (17). In addition, the BCATm inhibitor lowered weight gain but simultaneously improved glucose excursions and had a more pronounced effect on circulating BCAA levels in Zucker fatty rats (18). All of these studies suggested that BCATm greatly acted on but was not limited to BCAA catabolism. However, the regulating mechanism and targets in skeletal muscle of BCATm still remain unclear.
To address the transcriptome alterations in the skeletal muscle tissue of obese groups, we firstly analyzed the RNA sequencing (RNA-Seq) data from humans. We found that IRF4 was conservatively upregulated in obese muscle. Then, we generated skeletal muscle–specific IRF4 overexpression (MOE) and MKO mice to investigate the role of IRF4 in skeletal muscle. Finally, we demonstrated the critical role of muscle IRF4 in regulating skeletal muscle metabolism and the effects of a high-fat diet (HFD) on energy homeostasis.
Research Design and Methods
Human Subjects
A total of 10 male subjects (mean ± SEM age 31.1 ± 9.7 years) from Huashan Hospital, Fudan University, were included in this study. All subjects underwent routine physical and laboratory examinations. The subjects were divided into lean and obese groups based on BMI according to Chinese criteria. Skeletal muscle tissue near knee joint was collected with use of arthroscopy during operation. All studies were in accord with the Helsinki Declaration of 1975 and approved by the ethics committee of Huashan Hospital, Fudan University. Each subject provided written informed consent.
Animals
Mice were maintained under a 12 h light/12 h dark cycle at constant temperature (23°C) with free access to food and water. The muscle-specific Irf4 knockout mice and muscle-specific Irf4 overexpression mice were generated as previously reported (13). The 12-week-old db/db mice were used in this study. For diet-induced obesity (DIO) studies, mice were fed an HFD (D12331; Research Diets) for 12 weeks. All animal studies were approved by the Institutional Animal Care and Use Committee of UCLA.
Cell Lines, Culture Conditions, and Transfection
Human embryonic kidney (HEK)293T cells were cultured in Dulbecco’s modified Eagle medium/high glucose (DMEM/H) (Gibco) with 10% FBS (Gibco), 2 mmol/L l-glutamine (Gibco), and 1% penicillin-streptomycin (Gibco) in a 5% CO2 atmosphere at 37°C. Mouse C2C12 myoblasts were grown in DMEM/H with 10% FBS and 1% penicillin-streptomycin in a 5% CO2 atmosphere at 37°C. Lipofectamine LTX with Plus Reagent was used for transfecting according to the instructions of the manufacturer (Thermo Fisher Scientific).
Body Composition Measurement
The minispec mq10 nuclear magnetic resonance (NMR) analyzer (Bruker) was used to measure the body composition of mice according to the manufacturer’s instructions. Briefly, mice were put in an NMR tube and loaded in the NMR machine. The body composition was measured automatically by the machine.
Exercise Capacity Measurement
The exercise capacity of mice was measured with a treadmill as previously described (13). Briefly, mice adapted to the treadmill for 2 days before the exercise test. For adaptive exercise, the treadmill was set at a 20° incline and began with a 5 min bout of 0 m/min, followed by 10 m/min and 14 m/min. For the exercise test, the treadmill was set at a 10° incline, began at a speed of 10 m/min, and increased by 2 m/min every 5 min until mice were exhausted.
Food Intake
Mice were housed individually with free access to water. Before experiments, the cage was changed and the food was weighed. After 24 h, the food was reweighed, and the amount consumed was calculated by difference.
Indirect Calorimetry
The metabolic condition of mice that were HFD fed for 8 weeks was determined with the Oxymax/CLAMS Comprehensive Lab Animal Monitoring System (Columbus Instruments). Body weight–matched mice were housed individually and maintained under a 12 h light/12 h dark cycle at 23°C with free access to food and water. Oxygen consumption, carbon dioxide production, respiratory exchange ratio (RER), locomotor activity, and Energy expenditure were measured during the experiment. Body composition was measured before and after the CLAMS experiment. Data were analyzed with CalR, a Web-based analysis tool for indirect calorimetry experiments. ANCOVA was used for intergroup comparison. Total body mass was used as a covariate.
Glucose and Insulin Tolerance Tests
Glucose (GTT) and insulin (ITT) tolerance tests were performed as previously described (13). Briefly, mice were fasted overnight before GTT. Glucose (1 g/kg i.p.) was administered, and blood glucose levels were measured at 0, 15, 30, 60, and 120 min after injection. Mice were fasted for 6 h before ITT. Insulin (0.7 units/kg i.p.) was administered, and blood glucose was measured at 0, 15, 30, 60, and 120 min after injection.
BAT Temperature
The BAT temperatures of mice were measured with DAS-8007-P wireless reader system (BMDS). Mice with 7 weeks’ HFD feeding were maintained under a 12 h light/12 h dark cycle at 23°C with free access to food and water. Before temperature transponder implanting, the mice received inhaled anesthesia and the hair in the scapular area was removed with depilatory cream. The temperature transponder, IPTT-300 (BMDS), was subcutaneously implanted into the scapular area of mice according to the manufacturer’s instructions. The puncture port was closed with tissue adhesive. After adaptation for 3 days, the temperature measured in at 6:00 p.m. via the DAS-8007-P wireless reader.
BCAA Measurement
The BCAA levels of serum, skeletal muscle tissue, and C2C12 cells were measured with a BCAA kit (Sigma-Aldrich) according to the manufacturer’s instructions.
Glycogen Extraction and Measurement
Muscle tissue (10 mg) was rapidly homogenized with 200 µL double-distilled H2O for 10 min on ice. Then, the homogenates were boiled for 10 min to inactivate enzymes. After that, the homogenates were centrifuged at 18,000 rpm for 10 min at 4°C. The supernatant was collected and measured via Glycogen Assay Kit II (Abcam) according to the manufacturer’s instructions.
mtDNA Copy Number
mtDNA copy no. was determined via quantitative RT-PCR. Briefly, total DNA was isolated from the cells with use of REDExtract-N-Amp Tissue PCR Kit (Sigma-Aldrich) according to the manufacturer’s instructions. The mtDNA copy no. was calculated from the ratio of COX II (mitochondrial-encoded gene) to Actin (nuclear-encoded gene).
Mitochondria Isolation and Bioenergetics
Mitochondria were isolated from gastrocnemius muscle (GAS). The oxygen consumption of mitochondria was measured via a Seahorse analyzer (Seahorse Bioscience). In brief, the mitochondria were plated in 20 μL (pyruvate + malate and succinate + rotenone) and centrifuged onto the plate. After that, the volume was brought up to 150 μL/well with respiration buffer. The data were normalized to protein concentration.
Protein Extraction and Western Blot Analysis
Proteins were extracted with radioimmunoprecipitation assay buffer (Boston BioProducts) containing protease and phosphatase inhibitors (Thermo Fisher Scientific) and were quantified with Pierce Rapid Gold BCA Protein Assay Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Briefly, 40 μg lysate was loaded onto SDS-PAGE gels, blotted onto polyvinylidene difluoride membranes (Millipore), and incubated with antibodies. Protein amount was quantified with ImageJ Java 1.6.0_2.4 software.
Analysis of Gene Expression With Quantitative Real-time PCR
TRIzol method (Thermo Fisher Scientific) was used to extract total RNA from tissues and cells according to the manufacturer’s instructions. Briefly, 1 μg RNA was converted into cDNA using High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific), and quantitative QPCR was performed with a QuantStudio 7 Flex Real-Time PCR System (Thermo Fisher Scientific) using SYBR Green PCR Master Mix (Life Technologies) according to the manufacturer’s instructions. Tbp was used as the endogenous control.
Histology
BAT, inguinal WAT (iWAT), and epididymal WAT (eWAT) of male mice fed HFD for 12 weeks were harvested and fixed in 10% formalin (Sigma-Aldrich) and embedded in paraffin wax. Sections were cut and stained with hematoxylin-eosin according to a standard protocol. The number and size of adipocytes were measured with AdipoCount.
Plasmid Construction
pLKO.1-shIRF4, encoding shRNA targeting IRF4, was constructed through inserting shRNA sequence (CCGGGCAATGACTTTGAGG-AATTGGCTCGAGCCAATTCCTCAAAGTCATTGCTTTTTG [forward] and AATTCA-AAAAGCAATGACTTTGAGGAATTGGCTCGAGCCAATTCCTCAAAGTCATTGC [reverse]) into the AgeI/EcoRI site of pLKO.1 puro (Addgene). pCDH-BCATm was constructed through amplifying PCR products from C57BL/6J mouse cDNA and insertion into the XbaI/NotI site of pCDH (Addgene).
Dual-Luciferase Reporter Assays
The pGL4.22-BCATm-promoter (−2041/103) reporter plasmid was constructed through amplifying PCR products from C57BL/6J mouse genomic DNA and inserting it into the KpnI/NheI site of the luciferase reporter vector pGL4.22-Vector (Promega). The pGL4.22-BCATm-promoter (−807/103) reporter plasmid was constructed through amplifying PCR products from pGL4.22-BCATm-promoter (−2041/103) reporter plasmid and inserting it into the KpnI/NheI site of the luciferase reporter vector pGL4.22-Vector (Promega). The mutant reporter plasmids were constructed through amplifying the pGL4.22-BCATm-promoter (−2041/103) reporter plasmid. HEK293T cells were cultured in 24 well plates and cotransfected with pCDH-IRF4 plasmid, luciferase reporter plasmid, and pRL-TK (control reporter) through using ProFection Mammalian Transfection System (Promega) according to the manufacturer’s instructions. After 48 h, cells were harvested and measured with the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Luciferase activity was normalized to Renilla luciferase activity (control reporter).
Total RNA Extraction, Library Construction, and Sequencing
Three pairs of human skeletal muscle samples were provided for RNA-Seq. Total RNA was extracted from the skeletal muscle tissues with Trizol (Invitrogen, Carlsbad, CA) according to manual instruction. The double-stranded PCR products were heated, denatured, and circularized with the splint oligonucleotide sequence to get the final library. The final library was amplified with phi29 to make DNA nanoball (DNB), which had more than 300 copies of one molecule. The DNBs were loaded into the patterned nanoarray and pair-end 150 base reads were generated on the BGIseq500 platform (BGI, Shenzhen, China).
Sequence Alignment and Gene Expression Analysis
The sequencing data were filtered with Trim Galore! (v0.6.6) with removal of reads containing sequencing adapter and reads with low-quality base. Afterward clean reads were obtained and stored in FASTQ format. The clean reads were mapped to the reference genome (GRCh38.p13) with STAR (version 2.7.8a). Then the gene expression level was calculated with RSEM (v1.3.3). Essentially, differential expression analysis was performed with the DESeq2 (version 1.4.5) or edgeR (version 3.32.1) with P value <0.05 and |log fold change| >1.5. The heat map was drawn with pheatmap (version 1.0.12) according to the gene expression in different samples.
Serum Metabolomics Analysis
The serum samples from mice were used to measure ∼300 targeted metabolites with an ultraperformance liquid chromatography coupled to tandem mass spectrometry system (UPLS-TQMS; Metabo-Profile Biotechnology Co., Ltd, Shanghai, China). Test mixtures were used as a quality control in the metabolomics platform.
BCKDH Activity Assay
The BCKDH activity was determined with a spectrophotometer method as previously described (19). Briefly, 150 mg GAS were homogenized in extraction buffer. The supernatant was incubated with 27% (w/v) polyethylene glycol for 20 min on ice. After that, the mixture was centrifuged at 12,000g for 10 min at 4°C and the pellet was resuspended in ice-cold suspending buffer. The BCKDH activity was measured with 2× assay buffer (60 mmol/L K3PO4, 4 mmol/L MgCl2, 0.8 mmol/L thiamine pyrophosphate, 0.8 mmol/L CoA, 2 mmol/L NAD+, 0.2% (w/v) Triton X-100, 4 mmol/L dithiothreitol, and 10 units/mL dihydrolipoamide dehydrogenase, pH 7.3) and by recording of the NADH formation at 340 nm.
Adeno-Associated Virus–BCATm Production, Purification, and Injection
Adeno-associated virus (AAV)2/9-MCS-BCATm was produced and purified by Boston Children’s Hospital Viral Core. For BCATm expression experiment in vivo, a dose of 1.3 × 1011 gene copies of AAV-BCATm was injected into the GAS of MI4KO mice and the same dose of AAV-GFP was injected into the GAS of MI4KO and control mice on 4 week HFD. GTT and ITT were performed after 2 or 3 weeks of virus injection, respectively. Temperature measurement and CLAMS experiment were performed after 4 weeks of virus injection. The mice were sacrificed after CLAMS experiment.
Quantification and Statistical Analysis
Unpaired two-tailed t test or nonparametric test was performed for comparison of two groups. One-way ANOVA and post hoc multiple comparisons (least significant difference method) were performed for intergroup comparisons of more than two groups. All data except human clinical data were presented as mean ± SEM. P < 0.05 was considered statistically significant.
Data and Resource Availability
The accession no. for the RNA-Seq data reported in this article is GEO: GSE180906. Other data generated or analyzed during this study are included in the published article and Supplementary Material. The sequence of primers is provided in Supplementary Table 3. The key resource used in this study is provided in Supplementary Table 4. All bioinformatics software used in the study is publicly available. Further information and requests for resources and reagents should be directed to, and will be fulfilled by, the corresponding author.
Results
IRF4 Was Increased in Skeletal Muscle From Obese Subjects of Different Species
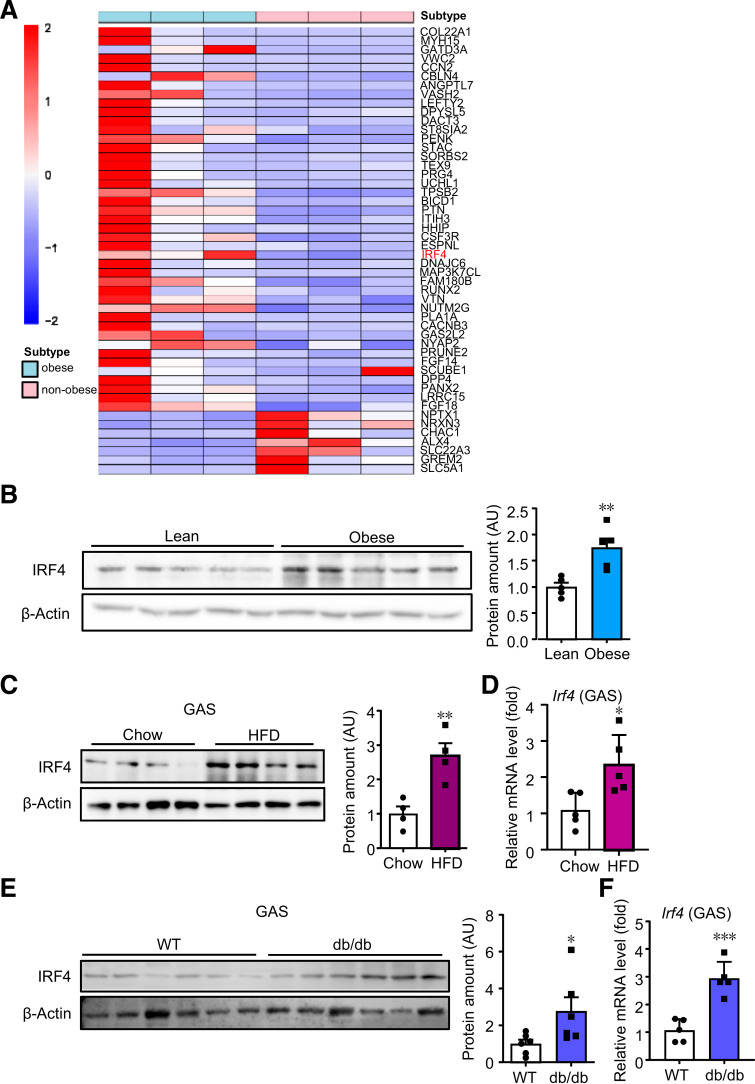
For characterization of the factor changes, transcriptome analysis was performed in the skeletal muscle of humans. Three pairs of age-matched skeletal muscle samples from males were subjected to RNA-Seq, and 133 genes were identified to be different, revealing that obesity altered transcriptomic profiles in human skeletal muscle (Fig. 1A and Supplementary Table 1). Among these genes, IRF4, a gene member of the IRF family, was one of the top changed genes. IRF4 was reported to handle lipid metabolism and thermogenesis in adipose tissue and T cells (10–12,20,21). Based on the above findings, IRF4 was chosen for further investigation, of its role in skeletal muscle metabolism. To confirm that IRF4 was the only IRF member increased in skeletal muscle of the obese, we analyzed the expression of IRF family members in skeletal muscle from microarray data of lean and obese subjects using GSE data sets. The results revealed that only Irf4 expression was significantly higher in the skeletal muscle of obese subjects than in that of lean subjects (22) (Supplementary Fig. 1A and B), consistent with our findings. Moreover, the IRF4 protein level was also higher in the human skeletal muscle of obese subjects than that of the lean (Fig. 1B and Supplementary Fig. 1C–F). To investigate whether IRF4 change was conserved in mice, we examined the mRNA and protein levels in DIO and genetic mouse models. According to the results, IRF4 mRNA and protein levels were increased in GAS of DIO mice (Fig. 1C and D) and genetically induced diabetes (Fig. 1E and F). However, IRF4 expression remained unchanged in the liver of humans and mice with obesity compared with that of lean control subjects (Supplementary Fig. 1G–I); in contrast, IRF4 expression was decreased in obese BAT of mice, as reported previously (11). These data indicated that IRF4 expression was improved in the skeletal muscle of human and mouse obese subjects.
Figure 1.
Association between skeletal muscle IRF4 and obesity. A: Heat map plot comparing top 50 different genes in human skeletal muscle between obese and nonobese groups. B: Western blot analysis of the expression of IRF4 in muscle of humans with or without obesity. Protein amount was quantified with ImageJ (n = 5) (**P < 0.01). C: Western blot analysis of the expression of IRF4 in GAS of mice on HFD or chow diet. Protein amount was quantified with ImageJ (n = 4) (**P < 0.01). D: mRNA expression of Irf4 in GAS of mice on HFD or chow (n = 5) (*P < 0.05). E: Western blot analysis of the expression of IRF4 in GAS of db/db mice and WT mice. Protein amount was quantified with ImageJ (n = 6) (*P < 0.05). F: mRNA expression of Irf4 in GAS of db/db mice and WT mice (n = 5) (***P < 0.001). All results are expressed as means ± SEM. AU, arbitrary units.
IRF4 Overexpression in Skeletal Muscle Was Susceptible to DIO
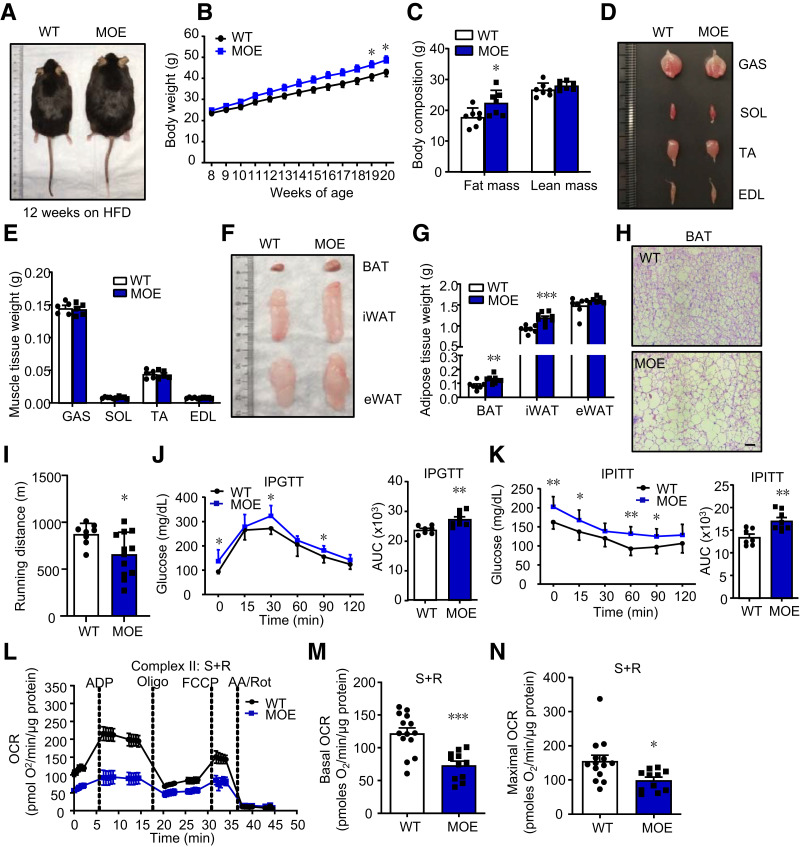
Given that obesity is associated with enhanced muscle IRF4 expression, we determined whether IRF4 was sufficient to promote obesity. Mice with skeletal muscle–specific overexpression of IRF4 were generated and showed a significant increase in body weight and fat mass on an HFD (Fig. 2A–G). Although all the fat depots were larger in MOE mice, only adipocytes from BAT were less multilocular and more hypertrophic than those from control mice (Fig. 2F–H). Afterward, the exercise capacity of MOE mice was measured, which was lower than that of control mice (Fig. 2I and Supplementary Fig. 2A). In addition, we investigated the effect of IRF4 overexpression in skeletal muscle on glucose homeostasis when the body weight of the two groups of mice was equal (Fig. 2B). We found an impaired GTT and ITT of MOE mice relative to that of control mice on an HFD (Fig. 2J and K).
Figure 2.
Skeletal muscle–specific IRF4 overexpression mice are prone to DIO. A: The morphology of male MOE and WT mice (12-week on HFD). B: Body weight of male MOE and control mice on HFD (n = 8–12) (*P < 0.05). C: Body composition of male MOE and control mice on HFD (n = 7) (*P < 0.05). D: The morphology of GAS, soleus (SOL), tibialis anterior (TA), and extensor digitorum longus (EDL) in MOE and WT mice (12 weeks on HFD). E: The weight of GAS, soleus, tibialis anterior, and extensor digitorum longus in MOE and WT mice on 12-week HFD (n = 5). F: The representative morphology of BAT, iWAT, and eWAT in MOE and WT mice (12 weeks on HFD). G: The weight of BAT, iWAT, and eWAT in MOE mice and WT mice on 12-week HFD (n = 7–9) (**P < 0.01 and ***P < 0.001). H: Hematoxylin-eosin staining and adipocyte number of BAT of MOE and WT mice on 12-week HFD (scale bars, 50 μm). I: The running distance of male MOE and WT mice (n = 7–12) (*P < 0.05). J and K: GTT and ITT in male MOE and control mice (n = 7–8) (*P < 0.05 and **P < 0.01). L: Seahorse assays of OCR in the isolated mitochondria from GAS of MOE and control mice (n = 11–14). M and N: The basal and maximal OCR of mitochondrial complex II from GAS in MOE and WT mice (n = 11–14) (**P < 0.01 and ***P < 0.01). All results are expressed as means ± SEM. AA, antimycin A; AUC, area under the curve; FCCP, carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone; IPGTT, intraperitoneal GTT; IPTT, intraperitoneal ITT; Oligo, oligonucleotide; Rot, rotenone; S, succinate.
Skeletal muscle of MOE mice showed no difference in mitochondria numbers (as measured with mtDNA) (Supplementary Fig. 2C), suggesting that IRF4 might be insufficient for mitochondrial biogenesis. However, basal and maximal ADP-stimulated mitochondrial complex II respiration rates were lessened in MOE mouse muscle (Fig. 2L–N), while no significant difference was observed in the complex I respiration rates (Supplementary Fig. 2D–F). Taken together, these results suggested that IRF4 in skeletal muscle was sufficient for DIO.
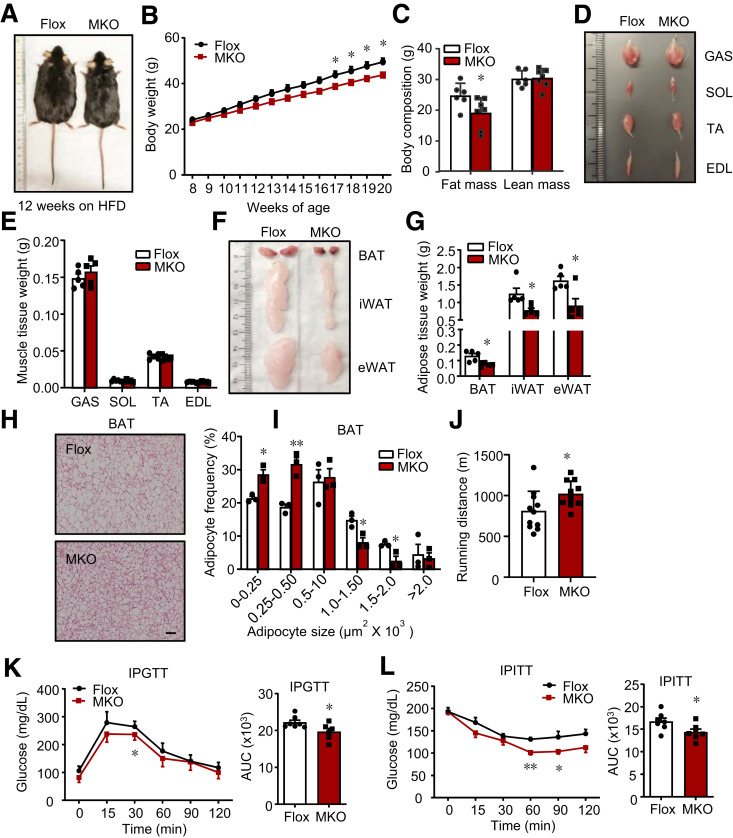
IRF4 Ablation in Skeletal Muscle Protected Mice From DIO
To evaluate the necessity of IRF4 for anti-DIO, we generated skeletal muscle–specific IRF4 knockout mice. On an HFD, the male MKO mice became less obese and presented lower body weight and less fat mass but similar lean mass compared with the controls (Fig. 3A–G). Although the skeletal muscle depots of MKO mice appeared normal and were similar to those of controls in size (Fig. 3D and E), MKO mice had significantly smaller white and brown adipose depots (Fig. 3F and G). Although no difference was found in adipocyte size in iWAT or eWAT, the brown adipocytes of MKO mice were more multilocular and smaller than those of Irf4flox/flox; Cre littermates (Fig. 3H and I). In addition, female mice showed phenotypes similar to those of the male mice (Supplementary Fig. 3A and B).
Figure 3.
Mice with IRF4 ablation in skeletal muscle are resistant to HFD-induced obesity. A: The morphology of male MKO and Flox mice on 12-week HFD. B: The body weight of male MKO and Flox mice on HFD (n = 8–10) (*P < 0.05). C: The body composition of male mice from A (n = 6–7) (*P < 0.05). D: The morphology of GAS, soleus (SOL), tibialis anterior (TA), and extensor digitorum longus (EDL) in male MKO and Flox mice on 12-week HFD. E: The weight of GAS, soleus, tibialis anterior, and extensor digitorum longus in male MKO and Flox mice on 12-week HFD (n = 5). F: The morphology of BAT, iWAT, and eWAT in male MKO and Flox mice on 12-week HFD. G: The weight of BAT, iWAT, and eWAT from mice on 12-week HFD (n = 5) (*P < 0.05). H: Hematoxylin-eosin staining and adipocyte number of BAT in male MKO and Flox mice on 12-week HFD (scale bars, 50 μm). I: The adipocyte frequency of different adipocyte size of BAT from H (n = 3) (*P < 0.05 and **P < 0.01). J: The running distance of male MKO and Flox mice (n = 10–11) (*P < 0.05). K and L: GTT and ITT in male MKO and Flox mice (n = 6–7) (*P < 0.05 and **P < 0.01). All results are expressed as means ± SEM. IPGTT, intraperitoneal GTT; IPTT, intraperitoneal ITT.
The testing results of the exercise capacity of these mice indicated that the MKO mice ran longer than the control mice (Fig. 3J and Supplementary Fig. 3C). In addition, MKO mice showed improved glucose tolerance and insulin sensitivity compared with the control mice on an HFD (Fig. 3K and L).
IRF4 Disruption in Skeletal Muscle of DIO Mice Reinforced Mitochondrial Function and Enhanced Energy Expenditure
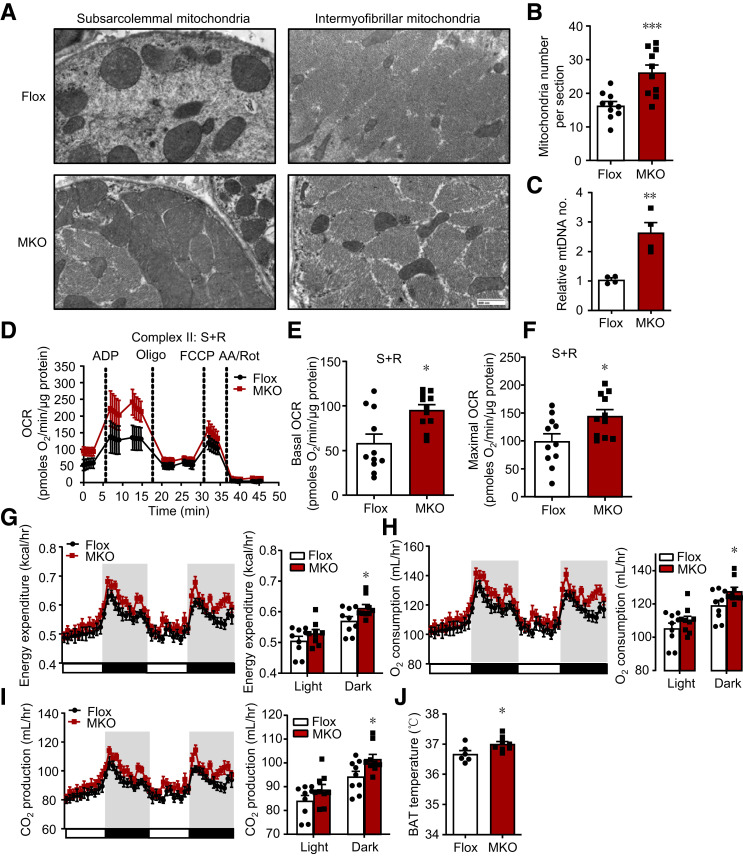
We next assessed mitochondria in skeletal muscle. First, the ultrastructure analysis by electron microscopy showed that more mitochondria were located in both subsarcolemmal and intermyofibrillar regions in MKO mice (Fig. 4A and B). This phenomenon was confirmed by increased mitochondrial DNA content (Fig. 4C). Additionally, isolated mitochondria showed a substantial enhancement in both basal and maximal oxygen consumption rate (OCR) when incubated with succinate and rotenone (Fig. 4D–F). No difference existed in mitochondrial OCR between groups of mitochondria incubated with pyruvate and malate (Supplementary Fig. 4A–C), indicating that IRF4 deletion in skeletal muscle specifically enhanced complex II (succinate dehydrogenase) activity but did not affect the activity of complex I. The above data demonstrate that IRF4 loss increased both the number and OCR of mitochondria to reinforce mitochondrial function in skeletal muscle.
Figure 4.
Ablation of IRF4 in skeletal muscle leads to increased mitochondrial function and enhanced EE. A: Mitochondrial density assessed with electron microscopy in GAS of MKO and Flox mice. Original magnification, ×7,200. B: Quantification of subsarcolemmal and intermyofibrillar mitochondria number per image area (×4,800) in the GAS (analysis of 10 images, for each group) (***P < 0.001). C: The relative mitochondria DNA copy number of GAS (n = 4) (**P < 0.01). D: Seahorse assays of mitochondrial complex II OCR in isolated mitochondria from GAS of MKO and Flox mice (n = 11). E and F: The basal and maximal OCR of mitochondrial complex II from GAS in MKO and Flox mice (n = 11) (*P < 0.05 and **P < 0.01). G: The EE of male MKO mice and Flox mice on 8-week HFD (n = 9, body weight matched) (*P < 0.05). H: Oxygen consumption of male MKO and Flox mice on 8-week HFD (n = 9) (*P < 0.05). I: CO2 production of male MKO and Flox mice on 8-week HFD (n = 9) (*P < 0.05). J: BAT temperature of male MKO and Flox mice on 8-week HFD at room temperature (n = 8) (*P < 0.05). All results are expressed as means ± SEM. AA, antimycin A; FCCP, carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone; hr, hour; Rot, rotenone; S, succinate.
Mitochondria are primary drivers of energy metabolism in skeletal muscle. Because the mitochondrial function was enhanced in IRF4-deleted muscle, we carried out the metabolic cage experiment to test EE. MKO mice on the HFD had food intake similar to that of controls (Supplementary Fig. 4D). Also, no difference was observed in the RER between MKO and Flox mice (Supplementary Fig. 4E). However, EE was significantly improved in MKO mice, with inductions in both oxygen consumption and carbon dioxide production, especially in the dark cycle (Fig. 4G–I). This difference in EE did not result from altered locomotor activity (Supplementary Fig. 4F). Interestingly, the brown fat temperature was significantly higher in the MKO mice than in the Flox mice housed at room temperature (Fig. 4J). The above EE and physiological changes were assessed after 6–8 weeks of HFD, before significant differences appeared in body weight and composition (Supplementary Fig. 4G–I), confirming that these effects were not caused by differences in body weight.
In addition, the metabolism of adipose tissue was also known to contribute to EE (23,24). On this basis, we measured key genes involved in the regulation of thermogenesis in adipose tissue, again using mice receiving 6–8 weeks of HFD before differences in body weight and composition appeared. PGC-1α, the master regulator of thermogenic genes (25), was measured. PGC-1α mRNA and its target gene Ucp1 were unaltered in BAT and iWAT of MKO mice. Other thermogenic and fatty acid oxidation (FAO) genes, (e.g., β3 adrenergic receptor and Pparα) were also unchanged (Supplementary Fig. 4J and K). Furthermore, leptin and adiponectin, two important fat-derived hormones significantly enhancing EE, were not altered in the HFD-fed MKO mice (Supplementary Fig. 4J and K). BCAA catabolism in BAT could contribute to energy homeostasis as well (26). Therefore, we tested some key genes in this pathway. Bcatm and Bckdha expression were promoted in BAT from MKO mice (Supplementary Fig. 4L), indicating that BCAA catabolism in BAT might be partially responsible for the altered EE.
IRF4 in Skeletal Muscle Could Reprogram Metabolome and Transcriptionally Regulate BCATm
To investigate the affecting mechanism of skeletal muscle IRF4 in terms of systemic metabolism, we performed targeted metabolomics analyses for amino acids, organic acids, amines, fatty acids, carbohydrates, and bile acids in the serum of MKO and control littermates (Supplementary Table 2). A total of 47 differently expressed metabolites were chosen in the MKO group (P < 0.2) for pathway analysis with MetaboAnalyst 5.0. The top nine enrichment pathways were analyzed (Fig. 5A). Among the noteworthy changes, a significant decrease was observed in serum glucose (Fig. 5B), consistent with improved insulin sensitivity of MKO mice. Though some significant fatty acid changes were not found, the pathway analysis showed changed FAO (Fig. 5A). In addition, an increase in BCAA, without changes in other essential amino acids or downstream BCAA-derived metabolites (e.g., BCKA, including 2-ketoisovaleric acid, ketoleucine, and 2-oxo-3-methylvaleric acid), was observed (Fig. 5C). These data indicated that IRF4 deficiency in skeletal muscle reprogrammed systemic metabolism, leading to anti-DIO.
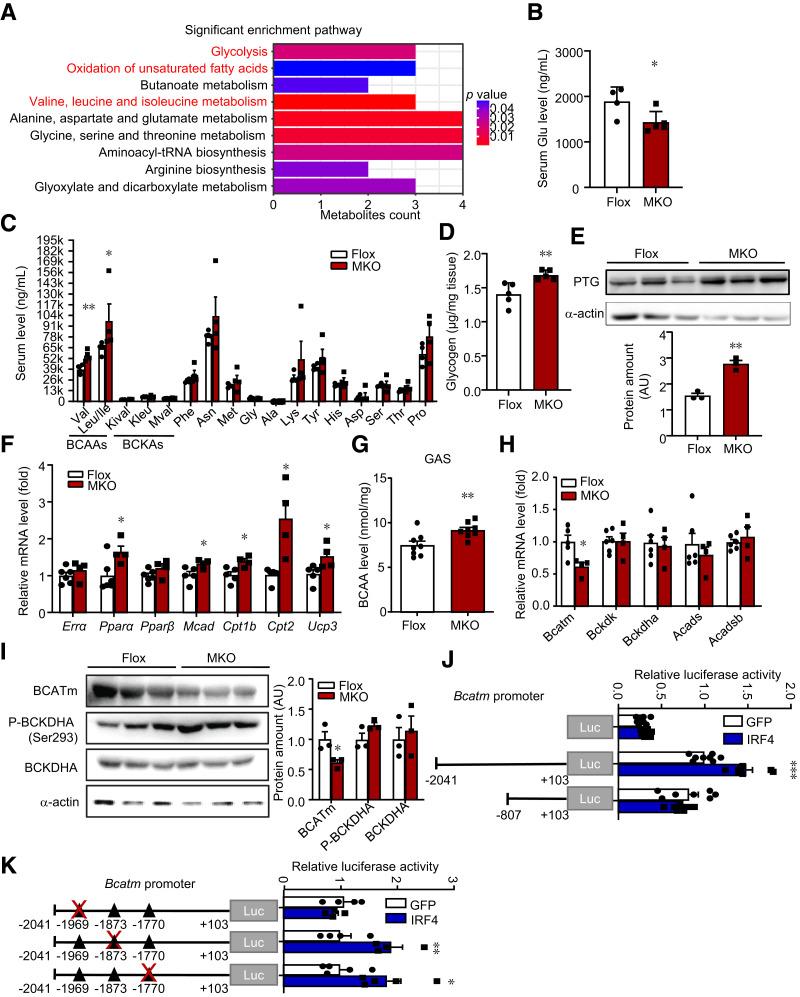
Figure 5.
IRF4 in skeletal muscle reprograms metabolome and transcriptionally regulates BCATm. A: Pathway analysis of 40 changed metabolites from targeted metabolomics data. B: Serum glucose (Glu) levels in MKO and Flox mice on HFD (n = 4–5) (*P < 0.05). C: Serum amino acids levels in MKO and Flox mice on HFD (n = 4) (*P < 0.05 and **P < 0.01). D: The glycogen level of GAS in male MKO and Flox mice (n = 5) (**P < 0.01). E: Western blot analysis of the expression of PTG in skeletal muscle of MKO and Flox mice. Protein amount was quantified with ImageJ (n = 3) (**P < 0.01). F: Quantitative PCR analysis of FAO genes in GAS from MKO and Flox mice (n = 4–6) (*P < 0.05). G: BCAA level in GAS of MKO and Flox mice (n = 6–8) (***P < 0.001). H: Quantitative PCR analysis of gene expression relative to BCAA catabolism in GAS of male MKO and Flox mice on HFD (n = 8) (*P < 0.05). I: Western blot analysis of BCAA catabolism in GAS of MKO and Flox mice. Protein amount was quantified with ImageJ (n = 3) (*P < 0.05). P-, phosphorylated. J: Dual luciferase assays of Bcatm promoter. Luciferase activity was corrected for Renilla luciferase activity and normalized to GFP group (n = 7–8) (***P < 0.001). K: The mutant Bcatm promoter fused to a luciferase reporter gene was cotransfected into HEK293T cells together with pCDH-GFP or pCDH-IRF4. Luciferase activity was corrected for Renilla luciferase activity and normalized to GFP group (n = 4–5) (*P < 0.05). All results are expressed as means ± SEM. AU, arbitrary units; Kival, 2-ketoisovaleric acid; Kleu, ketoleucine; Mval, 2-oxo-3-methylvaleric acid.
Considering the metabolites altered in serum, we further investigated related metabolites changes in skeletal muscle. Glycogen level was enhanced in skeletal muscle of MKO mice (Fig. 5D), with higher protein targeting to glycogen (PTG) expression (Fig. 5E). Next, triglycerides were also calculated in skeletal muscle. Surprisingly, muscle triglycerides showed no significant difference between MKO and Flox mice (Supplementary Fig. 5A). However, FAO genes, such as Pparα and its target genes, were upregulated in IRF4-deleted muscle (Fig. 5F). Lipolytic gene expression was thus in line with the increased FAO genes. Interestingly, Atgl and Hsl were not altered in the skeletal muscle of MKO and control mice (Supplementary Fig. 5B).
To determine the source of increased serum BCAA levels in MKO mice, we quantified BCAA levels of skeletal muscle and liver tissues, the major storage and interconnected hubs of these amino acids (18,27). According to the results, MKO mice showed a higher level of BCAA in skeletal muscle (Fig. 5G), with no difference in BCAA level in the liver compared with that of controls (Supplementary Fig. 5C). To investigate whether IRF4 was sufficient to mediate BCAA metabolism in skeletal muscle, we quantified the BCAA level of the serum and skeletal muscle of MOE mice. A decrease in BCAA level was observed in the serum and muscle of MOE mice relative to control mice (Supplementary Fig. 5D and E). Next, we examined the mRNA level of BCAA catabolic enzymes in skeletal muscle. Both BCATm mRNA expression and protein expression were significantly suppressed in MKO muscle tissue (Fig. 5H and I). After BCATm, the next step in the BCAA metabolic pathway was conducted by branched-chain α-ketoacid dehydrogenase complex (BCKDC) (Supplementary Fig. 5F). Western blot analysis demonstrated that total or phosphorylated amounts of BCKDHA were unaltered (Fig. 5I). However, BCKDC activity in IRF4-deleted muscle was lower than that of the control, as shown with ex vivo enzymatic assay (Supplementary Fig. 5G). This might explain the increased level of BCAA but not BCKA. The BCAA level was reduced, while BCATm was elevated, in the skeletal muscle of MOE mice, with no changes in the total or phosphorylated level of BCKDHA (Supplementary Fig. 5H).
BCAA could activate the mTOR pathway, which in turn regulated protein synthesis and mitochondrial function (28). On this basis we next considered whether the mTOR signaling pathway was enhanced in the skeletal muscle of MKO mice. The results showed a significantly rising phosphorylation level of the mTOR target p70S6K (T389) in the GAS of MKO mice (Supplementary Fig. 5I). Consistently, the mTOR pathway was mildly but significantly repressed in IRF4 gain-of-function muscle tissue (Supplementary Fig. 5H).
PGC-1α is a master regulator of macronutrient metabolism and mitochondrial biogenesis of skeletal muscle. For this reason, we then assessed PGC-1α expression. For PGC-1α, though the mRNA level was unchanged, protein level increased in the skeletal muscle of MKO mice (Supplementary Fig. 5J and K). In contrast, the PGC-1α protein level was downregulated in MOE skeletal muscle (Supplementary Fig. 5L).
To illustrate the molecular mechanism of how IRF4 participates in multiple-integrated metabolism, we tested the IRF4 targets. Though some key metabolic genes changed in the loss of IRF4 skeletal muscle, BCATm was the only one downregulated. Therefore, we proposed that BCATm was transcriptionally regulated by IRF4. Luciferase reporter genes containing different fragments of the BCATm promoter were constructed and cotransfected into 293T cells, together with pCDH-GFP or pCDH-IRF4. The results showed that the activity of the BCATm promoter was stimulated in the −2041/103 region but was not affected in the −807/103 region by IRF4, indicating that the element responsible for the action of IRF4 on the BCATm promoter was between the −2041 and −807 regions (Fig. 5J). We then analyzed the BCATm promoter between the −2041 and −807 regions to predict the interaction between promoters and transcription factors (https://jaspar.genereg.net). Subsequently, three potential binding sites of IRF4 (ISRE) were found within the −2041 and −807 regions of the BCATm promoter. Then, three mutant reporter genes with a disrupted ISRE motif were constructed one by one. IRF4 had no effects on the BCATm promoter with a −1969/−1955 ISRE mutation but a positive effect on the BCATm promoter with a −1873/−1859 or −1770/−1756 ISRE mutation (Fig. 5K). These data suggested that IRF4 manipulated BCATm expression via enhancing the activity of the BCATm promoter. Moreover, the −1969/−1955 ISRE region was responsible for this effect.
Overexpressed BCATm Counteracted IRF4 Ablation–Induced Metabolic Phenotype
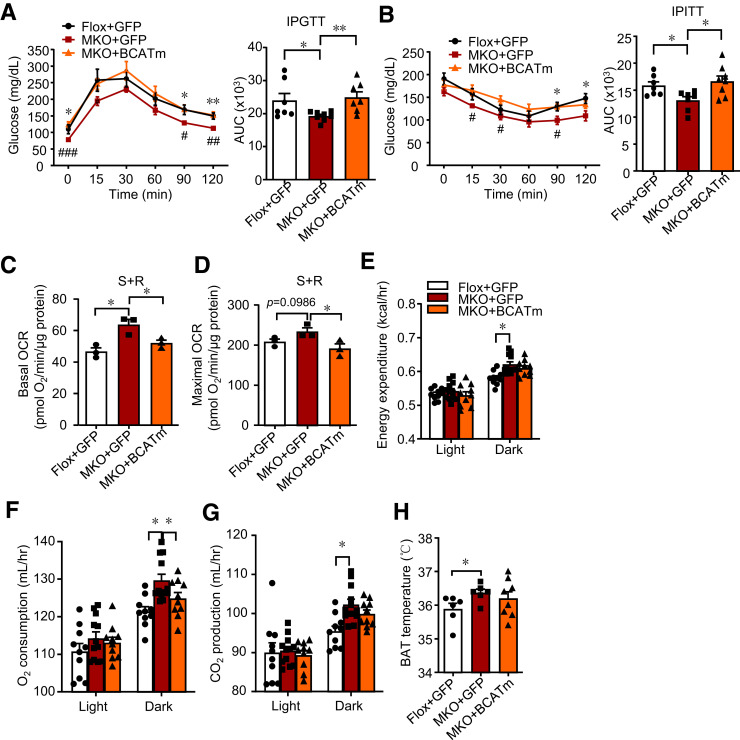
To verify the meditating role of BCATm in the effects of IRF4 in skeletal muscle in vivo, we forced overexpression of BCATm directly in the GAS of mice using AAV delivery. BCATm mRNA and protein levels significantly increased in the AAV-BCATm–treated group (Supplementary Fig. 6A and B). Four weeks after AAV injection, no difference was observed in the GAS weight of the BCATm group compared with that of the control group (Supplementary Fig. 6C). Then, the metabolic phenotype was analyzed. ITT and GTT returned to normal in MKO mice injected with AAV-BCATm (Fig. 6A and B), despite the similar body weight and composition in the three groups (Supplementary Fig. 6D and E). Furthermore, the mitochondria extracted from AAV-BCATm–injected GAS showed rescued basal and maximal OCR levels of complex II (Fig. 6C and D), while BCATm increased complex I activity (Supplementary Fig. 6F and G). Concomitant with this, BCATm injection partially reversed the rising EE and BAT temperature in MKO mice (Fig. 6E–H and Supplementary Fig. 6H–L).
Figure 6.
Overexpressed BCATm can reverse metabolic phenotype induced by IRF4 ablation. A and B: GTT and ITT in MKO+AAV-BCATm mice, MKO+AAV-GFP mice, and Flox+AAV-GFP mice on 6- or 7-week HFD (n = 7–8) (*P < 0.05 and **P < 0.01). C and D: The basal and maximal OCR of mitochondria complex II from GAS in MKO+AAV-BCATm mice, MKO+AAV-GFP mice, and Flox+AAV-GFP mice (n = 3) (*P < 0.05). E: The EE of male MKO+AAV-BCATm mice, MKO+AAV-GFP mice, and Flox+AAV-GFP mice on 8-week HFD (n = 10–12, body weight matched) (*P < 0.05). F: Oxygen consumption of male MKO+AAV-BCATm mice, MKO+AAV-GFP mice, and Flox+AAV-GFP mice on 8-week HFD (n = 10–12) (*P < 0.05). G: CO2 production of male MKO+AAV-BCATm mice, MKO+AAV-GFP mice, and Flox+AAV-GFP mice on 8-week HFD (n = 10–12) (*P < 0.05). H: BAT temperature of male MKO+AAV-BCATm mice, MKO+AAV-GFP mice, and Flox+AAV-GFP mice on 8-week HFD in the evening at room temperature (n = 6–8) (*P < 0.05). All results are expressed as means ± SEM. AUC, area under the curve; hr, hour; IPGTT, intraperitoneal GTT; IPTT, intraperitoneal ITT; R, rotenone; S, succinate.
BCATm Was Required for the Loss of IRF4-Mediated Metabolic Reprogramming in Skeletal Muscle
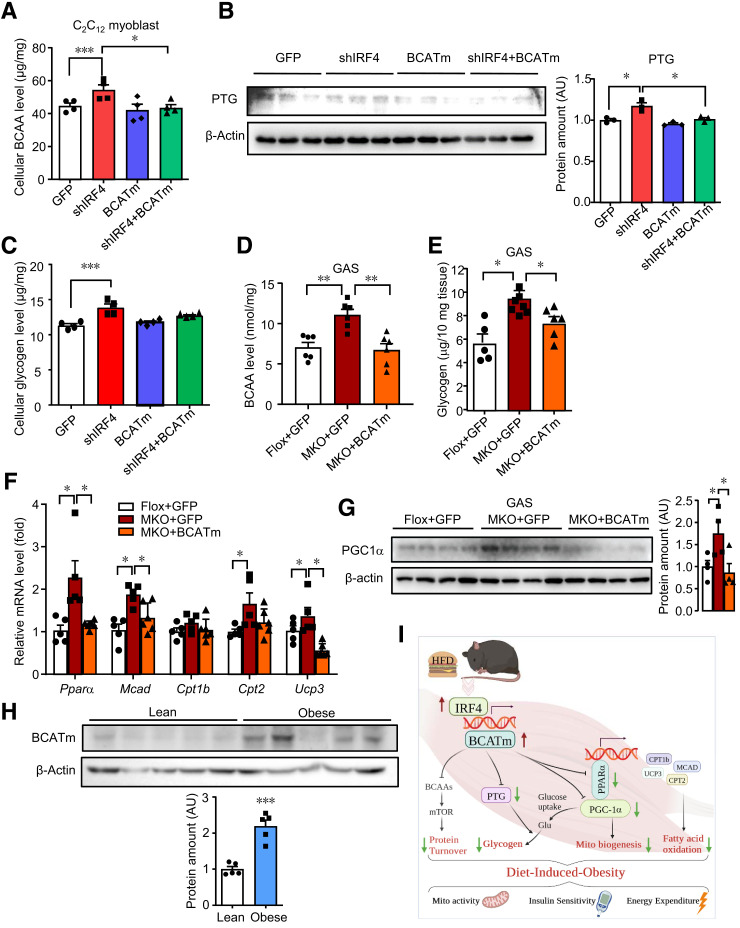
To identify the mediation of BCATm in response to the effect of disrupted IRF4 in skeletal muscle on macronutrient metabolism, we adopted a BCATm gain-of-function approach in vitro. C2C12 myoblasts were transfected with plasmids expressing at least one of shIRF4 and BCATm for 72 h (Supplementary Fig. 7A). BCATm represented a significant ascending protein expression level when transfected with BCATm plasmid (Supplementary Fig. 7B). IRF4 knockdown significantly increased the BCAA level compared with the control, whereas BCATm overexpression in the context of IRF4 knockdown showed recovered BCAA level (Fig. 7A). Next, we found that PTG expression was upregulated in IRF4 knockdown C2C12 cells, while BCATm overexpression recovered PTG expression (Fig. 7B). Consistent with this finding, the glycogen level of cells returned to normal when the cells were transfected with both vectors (Fig. 7C).
Figure 7.
BCATm is responsible for multiple-integrated metabolism in IRF4-deleted skeletal muscle. A: Cellular BCAA level of C2C12 myoblasts (n = 4) (*P < 0.05 and ***P < 0.001). B: Western blot analysis of PTG expression in C2C12 cells. Protein amount was quantified with ImageJ (n = 3) (*P < 0.05). C: Cellular glycogen level of C2C12 myoblasts (n = 4) (*P < 0.05). D and E: BCAA and glycogen levels in GAS of male MKO+AAV-BCATm mice, MKO+AAV-GFP mice, and Flox+AAV-GFP mice (n = 5–7) (*P < 0.05, **P < 0.01). F: Quantitative PCR analysis of FAO genes (n = 5) (*P < 0.05). G: Western blot analysis of the expression of PGC-1α. Protein amount was quantified with ImageJ (n = 4) (*P < 0.05). H: Western blot analysis of the expression of BCATm in muscle of humans with or without obesity. Protein amount was quantified with ImageJ (n = 5) (***P < 0.001). I: Working model. All results are expressed as means ± SEM. AU, arbitrary units; Mito, mitochondrion.
Next, we wished to confirm these findings in vivo using the AAV system. GAS BCAA and glycogen levels were normalized in AAV-BCATm–treated MKO mice compared with those of sham-injected controls (Fig. 7D and E). Surprisingly, the serum BCAA level and mTOR signaling were normalized after only GAS injection with BCATm (Supplementary Fig. 7C and D). We next measured FAO genes expression. Indeed, BCATm injection was found to offset Pparα and its target genes expression (Fig. 7F). The protein level of PGC-1α was enhanced in IRF4 disrupted muscle but lessened in BCATm-treated muscle (Fig. 7G). Taken together, these data identified that BCATm contributed to IRF4 loss–induced metabolic changes in skeletal muscle.
Finally, considering IRF4 increased in human with obesity, we tested BCATm in skeletal muscle of obese and lean subjects. The protein level of BCATm went up (Fig. 7H), whereas BCAA were not significantly increased in obese skeletal muscle (Supplementary Fig. 7E) as reported before (29,30). Therefore, our data strongly suggested a model in which IRF4 induced BCATm expression and then regulated macronutrients metabolism in skeletal muscle, eventually manipulating energy homeostasis (Fig. 7I, working model).
Discussion
Skeletal muscle can store glucose in the form of glycogen and also serves as a reservoir of amino acids stored as protein (31). Therefore, muscle is crucially important for balancing the metabolic needs of other organs. Our previous studies identified that IRF4 was required for BAT–skeletal muscle cross talk and associated with exercise exhaustion (12,13). Our discovery here shows that IRF4 in skeletal muscle regulates metabolic reprogramming via multiple-integrated metabolism in a BCATm-dependent manner. To the best of our knowledge, this is the first study to demonstrate the importance of inflammation-immune factor (i.e., IRF4) as a transcriptional regulator to coordinate the metabolism of carbohydrates, lipids, and amino acids in skeletal muscle.
Of note, IRF4 plays a key role in the development and function of immune cells (6,32). We describe that IRF4 expression in fat induces catabolic effects (improved lipolysis, antilipogenesis, increased M2 macrophage) (10,20). Generally, IRF4 expression is downregulated in adipocytes of DIO mice. However, in the current study, IRF4 expression is upregulated in the skeletal muscle of obese mice and humans. Furthermore, mice on an HFD with a specific loss of IRF4 in whole adipose tissue (FI4KO) (10) or BAT (BATI4KO) (11) show an obese phenotype, reduced EE, and impaired insulin sensitivity. In contrast, it appears to have the opposite action of IRF4 in the skeletal muscle—the MKO mice are obesity resistant with enhancement in EE, insulin sensitivity, and exercise capacity. Although skeletal muscle tissue and BAT were reported to share a common precursor and show metabolic similarities (such as high mitochondrial content and increased EE), specific deletion or overexpression of skeletal muscle IRF4 did not affect IRF expression in BAT (13). Therefore, BAT has limited benefit on metabolic outcomes, as opposed to changes in skeletal muscle metabolism. Mechanically, BAT in MKO mice may alter EE via BCAA catabolism because a BAT-specific BCAA catabolism deletion reduces BAT fuel oxidation and thermogenesis (25). These data indicate the existence of tissue-specific phenotypes associated with IRF4. This may be a precise and complex mechanism of an organism to keep a balanced and stable condition, which is intriguing, and further research on this is required for a more comprehensive understanding of life.
BCAA have been reported to have antiobesity effects. Nonetheless, circulating levels of BCAA tend to increase in individuals with obesity and are associated with unsatisfying metabolic health status, which may lead to insulin resistance or type 2 diabetes in the future. This paradox does not seem to be a problem in our studies. In the current study, the MKO mice exhibit a higher level of BCAA associated with a lean phenotype than the control mice. Numerous important factors may contribute to the contribution of excess BCAA in MKO to metabolic phenotype. First, MKO mice show an increase of BCAA content in both skeletal muscle and plasma; in contrast, other studies report that BCAA only rose in plasma, and no difference existed in muscle from DIO mice (30,33). Second, the model of dysfunctional BCAA metabolism assumed that an accumulation of BCAA metabolites resulted in dysfunction (34). In the targeted metabolomics on the MKO mouse model it is found to have an elevated level of only BCAA, with absent changes in plasma BCKA level. This may be explained by decreased BCATm expression and BCKDH activity in GAS. Third, our findings bring to mind the BCATm−/− mouse model by Hutson et al. and BCATm inhibitor in rats by Newgard et al., which revealed that deletion/inhibition of BCATm increased the circulating level of BCAA resulting in improved glycemic control, decreased adiposity, and increased thermogenesis (16,18). These findings are meaningful as we prove that BCATm is a mediator for BCAA regulation by IRF4. Finally, in addition to the role of BCATm in the futile protein turnover cycle (synthesis and degradation) (16,35), this study reveals that skeletal muscle BCATm not only exerts transamination for BCAA but also is responsible for the mitochondrial activity, glycogen metabolism, and FAO, probably independent of BCAA level.
Taken together, we firstly demonstrate that skeletal muscle IRF4 serves as a critical signaling node through which the catabolisms of lipids, glucose, and BCAA interplay. Moreover, IRF4 loss in skeletal muscle prevents DIO and insulin resistance. IRF4 could be a potential pharmacological target, with significant implications for the treatment of the obesity epidemic.
Article Information
Acknowledgments. The authors gratefully acknowledge Electron Microscopy Core of UCLA for performing electron microscopy analysis, Linsey Stiles in the Metabolism Research Theme at UCLA for performing Seahorse analysis, and KetengEdit (www.ketengedit.com) for linguistic assistance during the preparation of this manuscript.
Funding. This work was funded by the National Natural Science Foundation of China (92157203), National Institutes of Health (R00 DK106550 to X.K.), the National Key R&D Program of China (2019YFA0801900, 2018YFA0800300), the National Natural Science Foundation of China (31971074, 32150610475), Innovation Team and Talents Cultivation Program of National Administration of Traditional Chinese Medicine (ZYYCXTD-D-202001 to T.L.), Shanghai Frontiers Science Research Base of Exercise and Metabolic Health, the National Natural Science Foundation of China (82100849) and Young Foundation of Zhongshan Hospital Fudan University (2021ZSQN07 to X.Z.), American Heart Association Postdoctoral Fellowship (18POST34060000 to T.Y.), and a Natural Science Foundation of Shanghai Science and Technology Commission General project (21ZR1413200 to H.Y.).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. The experimental plan was designed by T.Y., X.Z., T.L., and X.K. T.Y., X.Z., S.G., J.Z., Z.Z., T.L., and X.K. performed experiments and analyzed data. S.G., Y.F., H.W., Y.H., J.Z., and X.K. collected the human samples and analyzed RNA-Seq data. T.Y. and X.Z. carried out the EE experiments and analyzed data. T.Y., X.Z., and X.K. wrote the manuscript incorporating edits and comments from S.D.M., P.T., T.L., and all other authors. X.K. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
See accompanying article, p. 2251.
This article contains supplementary material online at https://doi.org/10.2337/figshare.20054417.
T.Y., H.Y., X.Z., and Q.Z. contributed equally.
References
- 1. Astrup A. Macronutrient balances and obesity: the role of diet and physical activity. Public Health Nutr 1999;2:341–347 [DOI] [PubMed] [Google Scholar]
- 2. Obici S, Wang J, Chowdury R, et al. Identification of a biochemical link between energy intake and energy expenditure. J Clin Invest 2002;109:1599–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baker PR 2nd, Boyle KE, Koves TR, et al. Metabolomic analysis reveals altered skeletal muscle amino acid and fatty acid handling in obese humans. Obesity (Silver Spring) 2015;23:981–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rogero MM, Calder PC. Obesity, inflammation, Toll-like receptor 4 and fatty acids. Nutrients 2018;10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Uematsu S, Akira S. Toll-like receptors and type I interferons. J Biol Chem 2007;282:15319–15323 [DOI] [PubMed] [Google Scholar]
- 6. Nam S, Lim JS. Essential role of interferon regulatory factor 4 (IRF4) in immune cell development. Arch Pharm Res 2016;39:1548–1555 [DOI] [PubMed] [Google Scholar]
- 7. Bollig N, Brüstle A, Kellner K, et al. Transcription factor IRF4 determines germinal center formation through follicular T-helper cell differentiation. Proc Natl Acad Sci USA 2012;109:8664–8669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hagman J. Critical Functions of IRF4 in B and T Lymphocytes. J Immunol 2017;199:3715–3716 [DOI] [PubMed] [Google Scholar]
- 9. Eguchi J, Yan QW, Schones DE, et al. Interferon regulatory factors are transcriptional regulators of adipogenesis. Cell Metab 2008;7:86–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eguchi J, Wang X, Yu S, et al. Transcriptional control of adipose lipid handling by IRF4. Cell Metab 2011;13:249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kong X, Banks A, Liu T, et al. IRF4 is a key thermogenic transcriptional partner of PGC-1α. Cell 2014;158:69–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kong X, Yao T, Zhou P, et al. Brown adipose tissue controls skeletal muscle function via the secretion of myostatin. Cell Metab 2018;28:631–643.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu X, Yao T, Wang R, et al. IRF4 in skeletal muscle regulates exercise capacity via PTG/glycogen pathway. Adv Sci (Weinh) 2020;7:2001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Holeček M. Branched-chain amino acids in health and disease: metabolism, alterations in blood plasma, and as supplements. Nutr Metab (Lond) 2018;15:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hutson SM, Sweatt AJ, Lanoue KF. Branched-chain [corrected] amino acid metabolism: implications for establishing safe intakes. J Nutr 2005;135(Suppl.):1557S–1564S [DOI] [PubMed] [Google Scholar]
- 16. She P, Reid TM, Bronson SK, et al. Disruption of BCATm in mice leads to increased energy expenditure associated with the activation of a futile protein turnover cycle. Cell Metab 2007;6:181–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ananieva EA, Bostic JN, Torres AA, et al. Mice deficient in the mitochondrial branched-chain aminotransferase (BCATm) respond with delayed tumour growth to a challenge with EL-4 lymphoma. Br J Cancer 2018;119:1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. White PJ, Lapworth AL, McGarrah RW, et al. Muscle-liver trafficking of BCAA-derived nitrogen underlies obesity-related glycine depletion. Cell Rep 2020;33:108375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nakai N, Kobayashi R, Popov KM, Harris RA, Shimomura Y. Determination of branched-chain alpha-keto acid dehydrogenase activity state and branched-chain alpha-keto acid dehydrogenase kinase activity and protein in mammalian tissues. Methods Enzymol 2000;324:48–62 [DOI] [PubMed] [Google Scholar]
- 20. Eguchi J, Kong X, Tenta M, Wang X, Kang S, Rosen ED. Interferon regulatory factor 4 regulates obesity-induced inflammation through regulation of adipose tissue macrophage polarization. Diabetes 2013;62:3394–3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Man K, Miasari M, Shi W, et al. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat Immunol 2013;14:1155–1165 [DOI] [PubMed] [Google Scholar]
- 22. Park JJ, Berggren JR, Hulver MW, Houmard JA, Hoffman EP. GRB14, GPD1, and GDF8 as potential network collaborators in weight loss-induced improvements in insulin action in human skeletal muscle. Physiol Genomics 2006;27:114–121 [DOI] [PubMed] [Google Scholar]
- 23. Lu P, Zhang FC, Qian SW, et al. Artemisinin derivatives prevent obesity by inducing browning of WAT and enhancing BAT function. Cell Res 2016;26:1169–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu M, Bai J, He S, et al. Grb10 promotes lipolysis and thermogenesis by phosphorylation-dependent feedback inhibition of mTORC1. Cell Metab 2014;19:967–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab 2005;1:361–370 [DOI] [PubMed] [Google Scholar]
- 26. Yoneshiro T, Wang Q, Tajima K, et al. BCAA catabolism in brown fat controls energy homeostasis through SLC25A44. Nature 2019;572:614–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hutson SM, Harper AE. Blood and tissue branched-chain amino and alpha-keto acid concentrations: effect of diet, starvation, and disease. Am J Clin Nutr 1981;34:173–183 [DOI] [PubMed] [Google Scholar]
- 28. Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 2007;450:736–740 [DOI] [PubMed] [Google Scholar]
- 29. Lynch CJ, Adams SH. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol 2014;10:723–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou M, Shao J, Wu CY, et al. Targeting BCAA catabolism to treat obesity-associated insulin resistance. Diabetes 2019;68:1730–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Argilés JM, Campos N, Lopez-Pedrosa JM, Rueda R, Rodriguez-Mañas L. Skeletal muscle regulates metabolism via interorgan crosstalk: roles in health and disease. J Am Med Dir Assoc 2016;17:789–796 [DOI] [PubMed] [Google Scholar]
- 32. Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol 2006;6:644–658 [DOI] [PubMed] [Google Scholar]
- 33. Lee J, Vijayakumar A, White PJ, et al. BCAA supplementation in mice with diet-induced obesity alters the metabolome without impairing glucose homeostasis. Endocrinology 2021;162:bqab062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Menni C, Fauman E, Erte I, et al. Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes 2013;62:4270–4276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lynch CJ, Kimball SR, Xu Y, Salzberg AC, Kawasawa YI. Global deletion of BCATm increases expression of skeletal muscle genes associated with protein turnover. Physiol Genomics 2015;47:569–580 [DOI] [PMC free article] [PubMed] [Google Scholar]